Abstract
Hsp104 is a hexameric AAA+ ring translocase, which drives protein disaggregation in nonmetazoan eukaryotes. Cryo-EM structures of Hsp104 have suggested potential mechanisms of substrate translocation, but precisely how Hsp104 hexamers disaggregate proteins remains incompletely understood. Here, we employed synchrotron X-ray footprinting to probe the solution-state structures of Hsp104 monomers in the absence of nucleotide and Hsp104 hexamers in the presence of ADP or ATPγS (adenosine 5′-O-(thiotriphosphate)). Comparing side-chain solvent accessibilities between these three states illuminated aspects of Hsp104 structure and guided design of Hsp104 variants to probe the disaggregase mechanism in vitro and in vivo. We established that Hsp104 hexamers switch from a more-solvated state in ADP to a less-solvated state in ATPγS, consistent with switching from an open spiral to a closed ring visualized by cryo-EM. We pinpointed critical N-terminal domain (NTD), NTD-nucleotide–binding domain 1 (NBD1) linker, NBD1, and middle domain (MD) residues that enable intrinsic disaggregase activity and Hsp70 collaboration. We uncovered NTD residues in the loop between helices A1 and A2 that can be substituted to enhance disaggregase activity. We elucidated a novel potentiated Hsp104 MD variant, Hsp104–RYD, which suppresses α-synuclein, fused in sarcoma (FUS), and TDP-43 toxicity. We disambiguated a secondary pore-loop in NBD1, which collaborates with the NTD and NBD1 tyrosine-bearing pore-loop to drive protein disaggregation. Finally, we defined Leu-601 in NBD2 as crucial for Hsp104 hexamerization. Collectively, our findings unveil new facets of Hsp104 structure and mechanism. They also connect regions undergoing large changes in solvation to functionality, which could have profound implications for protein engineering.
Keywords: heat-shock protein (HSP), ATPases associated with diverse cellular activities (AAA), mutagenesis, protein chemical modification, mass spectrometry (MS), α-synuclein, disaggregase, FUS, Hsp104, TDP-43
Introduction
Protein disaggregases that reverse deleterious protein misfolding and aggregation could have therapeutic utility in several fatal human neurodegenerative disorders (1–8). Yet our structural and mechanistic understanding of protein disaggregases remains incomplete (9, 10). In yeast and other nonmetazoan eukaryotes, Hsp104, a hexameric, ring-shaped AAA+ protein, couples ATP hydrolysis to the dissolution and reactivation of proteins trapped in disordered aggregates, preamyloid oligomers, phase-separated compartments, and amyloid (1, 9–13). Thus, Hsp104 confers tolerance to diverse environmental stresses, regulates the material properties of membraneless organelles, and enables tight regulation of prions for beneficial purposes (1, 12–14).
Each Hsp104 monomer is composed of an N-terminal domain (NTD),4 two AAA+ nucleotide-binding domains (NBD1 and NBD2) that bind and hydrolyze ATP, a coiled-coil middle domain (MD) inserted in NBD1, and a short C-terminal region (15). The functional Hsp104 hexamer is large and dynamic with a solvent-filled channel running the length of the assembly (16–20). In response to ATP binding and hydrolysis, the hexamer undergoes substantial changes in conformation (9, 16, 17, 19). Indeed, cryo-electron microscopy (cryo-EM) has revealed that Hsp104 hexamers populate an open “lock-washer” spiral state in the presence of ADP, but switch to closed ring structures that surround polypeptide substrate inside the channel in the presence of ATPγS (16, 17). These conformational changes are proposed to drive polypeptide translocation across the central channel, which enables the remodeling of diverse substrates, from thermally-denatured aggregates (20–22) to amyloid conformers (20, 23–27). However, to fully understand and validate the details of these conformational changes, and to understand the mechanisms that underpin substrate remodeling, Hsp104 structure and dynamics need to be probed using diverse and complementary methodologies (28–30).
Here, we employ synchrotron X-ray footprinting (XF) to probe the solution-state structure of Hsp104 (31–34). We then leverage these data to design mutations to probe novel regions that might be important for various aspects of Hsp104 function. XF uses millisecond bursts of high-flux X-rays to produce hydroxyl radicals through the radiolysis of water (31, 35, 36). Hydroxyl radicals are ideal footprinting reagents because of their similarity to water molecules, making them excellent probes for solvation (31, 35, 36). They rapidly and irreversibly react with solvent-exposed residues to provide a snapshot of the protein at the time of exposure (31, 35, 36). Here, we study the hydroxyl radical footprinting of Hsp104 in three states: monomer without nucleotide, hexamer with ADP, and hexamer with ATPγS. We used homology-modeled Hsp104 domains, based on the Thermus thermophilus ClpB (tClpB) crystal structure (37), known Hsp104 structures such as the Saccharomyces cerevisiae NTD crystal structure (38), and Hsp104 hexamer cryo-EM reconstructions (16, 17) as a foundation for our examination of the solvation state of each domain in different nucleotide-bound states. Our solvation results enabled us to explain phenomena reported in the literature, to make testable predictions about the role different regions play in Hsp104 structure and activity, and to propose how conformational changes are coupled to substrate remodeling. By identifying heavily-modified regions, as well as regions that undergo large changes in solvation between different nucleotide states, we could pinpoint specific mutations in each Hsp104 domain that modulated Hsp104 functionality.
Results
Peptide overview
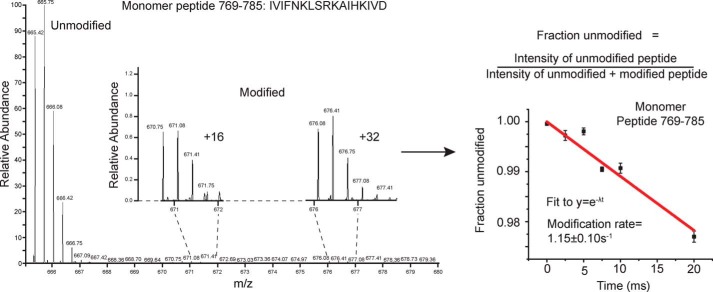
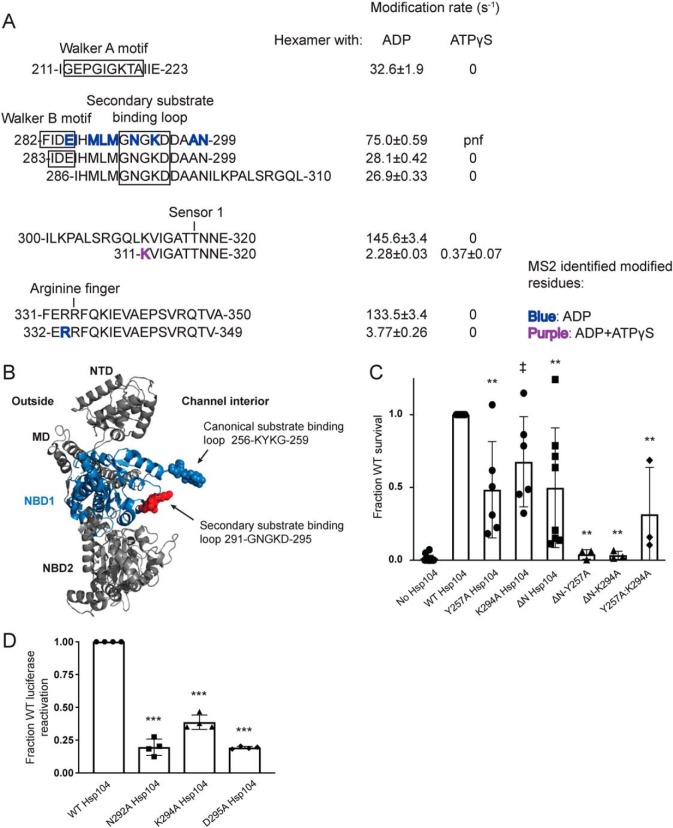
Samples of the Hsp104 monomer (hexamer dissociated by high salt) and the Hsp104 hexamer in the presence of either ADP or ATPγS were exposed to millisecond bursts of synchrotron X-rays and analyzed using MS as described under “Experimental procedures.” The monomer, hexamer with ADP, and hexamer with ATPγS have 339, 359, and 342 peptides found in all three replicates of all six time points, respectively (Table 1). Moreover, 122 (monomer), 135 (ADP), and 78 (ATPγS) of those peptides showed time-dependent modification, although some were omitted from our analysis due to poor signal to noise. After omitting these peptides, we were left with 81 (monomer), 117 (ADP), and 54 (ATPγS) peptides that showed time-dependent modification that could be fit to a first-order decay curve to yield rate information (Table 1, example of MS data and curve fitting in Fig. 1). Tables S1–S3 list all of the filtered modified and unmodified peptides for the monomer, hexamer with ADP, and hexamer with ATPγS, respectively. In instances where the modified peptide was selected for fragmentation and MS2 analysis, the identity of the specific site of modification was determined. The majority of identified peptides are unmodified. For a direct comparison of rates, we need to examine the rate information for the exact same peptide for all the states. Table 2 lists identical peptides found in two or three of the states, which cover some of the regions of interest.
Table 1.
X-ray footprinting peptide summary
Peptides reported were identified using ExMS-CL and found in all three replicates for each time point. Filtered peptides represent the peptides remaining after removal of those that had poor signal to noise.
| Sample | Peptides | Modified | Filtered modified | Final peptide count | Percent coverage |
|---|---|---|---|---|---|
| Monomer | 339 | 122 | 81 | 259 | 90% |
| Hexamer with ADP | 359 | 135 | 117 | 307 | 89% |
| Hexamer with ATPγS | 342 | 78 | 54 | 264 | 88% |
Figure 1.
X-ray exposed Hsp104 mass spectra and curve fitting. Unmodified and modified versions of the Hsp104 peptides were identified by ExMS-CL in each time point. The intensities of the singly-modified peptides were pooled, and the fraction unmodified was calculated for each time point. A pseudo–first-order decay curve extrapolated to zero was then fit to the data.
Table 2.
Selected peptide list
Peptides that are found in two or more states, which cover important regions of Hsp104, are shown. Normalized modification rates and modified residues are listed side by side for ease of comparison. ND means not determined and pnf means peptide not found.
| Start | End | Monomer |
Hexamer with ADP |
Hexamer with ATPγS |
|||
|---|---|---|---|---|---|---|---|
| Modified residues (identified by MS2) | Modification rate (s−1) | Modified residues (identified by MS2) | Modification rate (s−1) | Modified residues (identified by MS2) | Modification rate (s−1) | ||
| 8 | 38 | 0.03 ± 0.04 | 0 | 1.85 ± 0.33 | |||
| 39 | 53 | pnf | 25.02 ± 0.01 | 4.28 ± 1.50 | |||
| 55 | 64 | 0 | 3.60 ± 0.01 | 0.86 ± 0.18 | |||
| 86 | 110 | 87, 88, 90, 91, 95, 96, 100, 101, 104, 108 | 0.80 ± 0.35 | 90, 94, 96, 102, 107 | 0.59 ± 0.01 | 87, 90, 91, 94, 96, 103, 109 | 5.35 ± 0.35 |
| 121 | 134 | 0 | 6.48 ± 0.29 | 1.15 ± 0.41 | |||
| 148 | 166 | 0.22 ± 0.01 | 0.10 ± 0.002 | 0 | |||
| 211 | 223 | 0 | 32.61 ± 1.89 | 0 | |||
| 225 | 243 | 15.90 ± 11.23 | 0 | 0 | |||
| 282 | 299 | ND | 285, 286, 288, 289, 290, 292, 294, 298, 299 | 74.97 ± 0.59 | ND | ||
| 283 | 299 | ND | 28.07 ± 0.42 | 0 | |||
| 286 | 310 | ND | 26.91 ± 0.33 | 0 | |||
| 300 | 320 | ND | 145.58 ± 3.38 | 0 | |||
| 311 | 320 | ND | 311 | 2.28 ± 0.03 | 311 | 0.37 ± 0.07 | |
| 331 | 350 | 0 | 133.54 ± 3.37 | 0 | |||
| 332 | 349 | 2.37 ± 0.29 | 333 | 3.77 ± 0.26 | 0 | ||
| 405 | 414 | pnf | 21.51 ± 0.39 | 52.81 ± 9.52 | |||
| 415 | 423 | pnf | 415, 416, 417, 418, 421, 423 | 35.51 ± 0.79 | 0 | ||
| 431 | 455 | 0.22 ± 0.10 | 0.57 ± 0.01 | 0.08 ± 0.05 | |||
| 467 | 493 | 0 | 0 | 0.84 ± 0.12 | |||
| 505 | 526 | pnf | 349.14 ± 0.95 | 0 | |||
| 506 | 522 | 6.20 ± 4.30 | 20.14 ± 1.23 | 1.51 ± 1.04 | |||
| 508 | 536 | ND | 511, 517, 523 | 88.81 ± 3.41 | ND | ||
| 611 | 631 | 613, 615, 626 | 2.30 ± 0.76 | 4.94 ± 2.23 | 0 | ||
| 613 | 623 | 0 | 0 | 616, 618 | 9.54 ± 0.28 | ||
| 684 | 697 | 20.20 ± 1.33 | 684, 687 | 49.12 ± 1.27 | 0 | ||
| 733 | 744 | 1.60 ± 0.07 | 1.82 ± 0.02 | 0 | |||
| 733 | 748 | 0 | 103.06 ± 2.97 | 0 | |||
| 769 | 785 | 773, 774, 776, 778, 779, 780, 781, 782, 784 | 1.15 ± 0.10 | 772, 774, 781, 782, 783, 784 | 0.64 ± 0.00 | 772, 774, 776, 777, 780, 782 | 0.14 ± 0.02 |
| 786 | 806 | 0 | 17.55 ± 6.35 | 0 | |||
| 789 | 806 | pnf | 22.31 ± 0.31 | 0 | |||
| 789 | 809 | 2.67 ± 1.00 | 20.08 ± 0.15 | 0 | |||
| 813 | 831 | pnf | 819, 821, 822, 823 | 41.89 ± 0.18 | 0 | ||
| 843 | 857 | 0 | 843, 850, 853, 855 | 3.77 ± 0.14 | 843, 845, 846, 848, 849, 850, 852, 857 | 6.12 ± 4.49 | |
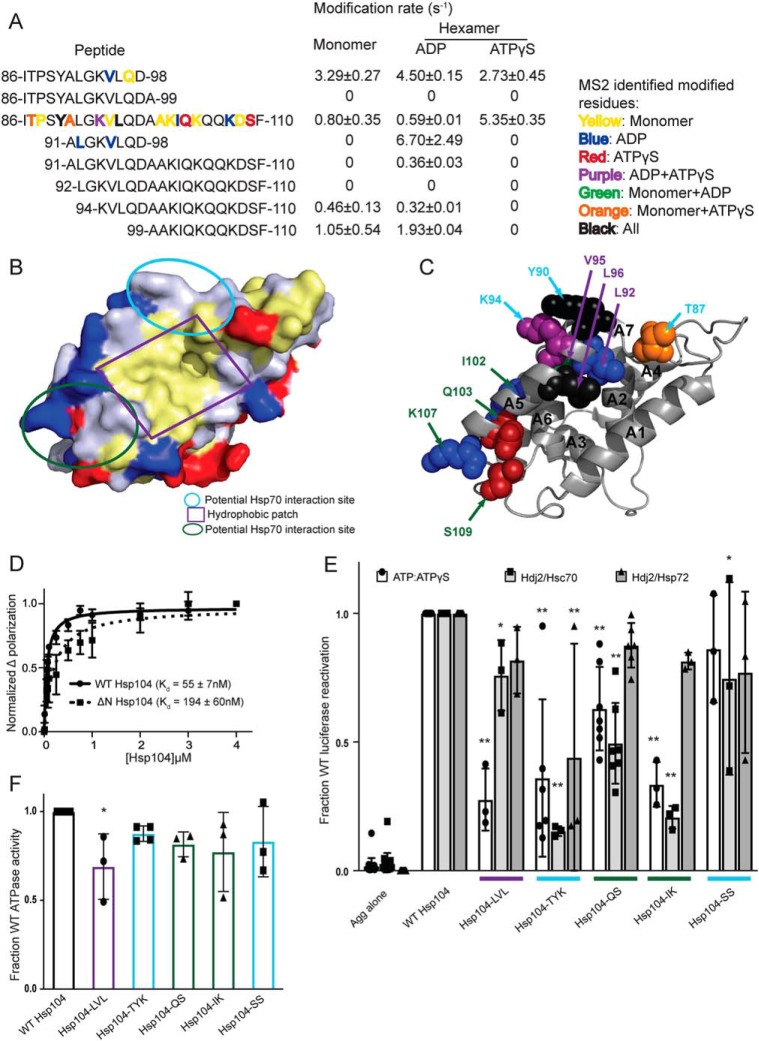
To begin our analysis of the XF solvation data of the Hsp104 hexamer and to extract relevant mechanistic implications, we assessed each Hsp104 domain in turn. We examined the following: 1) regions of known functional importance; 2) regions that are heavily modified, and therefore solvated; and 3) regions that undergo large changes in solvation between states. We then used the XF data to make mechanistic predictions about regions in each domain. These predictions were then tested via mutational analyses. An overview of the Hsp104 variants created and tested is shown in Table 3.
Table 3.
Overview of Hsp104 variants
Based on the X-ray footprinting solvation data, Hsp104 variants were designed and purified. Their biochemical properties were then assessed by measuring ATPase activity, intrinsic disaggregase activity (ATP/ATPγS), and disaggregase activity in collaboration with Hsp40/Hsp70 pairs (Hdj2/Hsc70 or Hdj2/Hsp72). In this table ± indicates no significant difference from WT Hsp104; - denotes a statistically significant decrease in activity down to more than 60% WT Hsp104 activity; – denotes a statistically significant decrease down to 30–60% WT Hsp104 activity, and — denotes a statistically significant decrease to less than 30% of WT Hsp104 activity. Similarly, + signifies a statistically significant increase up to less than 150% WT Hsp104 activity; and ++ signifies a statistically significant increase greater than 200% WT Hsp104 activity. ND indicates not determined.
| Variant | Residues | Location | ATPase | Disaggregation |
|||
|---|---|---|---|---|---|---|---|
| ATP | ATP/ATPγS | Hdj2/Hsc70 | Hdj2/Hsp72 | ||||
| Hsp104-LVL | L92A/V95A/L96A | NTD | - | ± | — | - | ± |
| Hsp104-TYK | T87A/Y90A/K94A | NTD | ± | ± | – | — | – |
| Hsp104-QS | Q103A/S109A | NTD | ± | ± | - | – | ± |
| Hsp104-IK | I102A/K107A | NTD | ± | ± | – | — | ± |
| Hsp104-SS | S124A/S125A | NTD | ± | ± | ± | - | ± |
| Hsp104-TR | T8A/R10A | NTD | ± | ± | — | — | - |
| Hsp104-ED | E44R/D45R | NTD | ± | ± | + | + | ± |
| Hsp104-TM | T160M | NTD-NBD1 linker | – | ± | – | — | – |
| Hsp104-RRD | R148D/R152D/R156D | NTD-NBD1 linker | ± | ± | – | — | – |
| Hsp104-DDD | D231A/D232A/D233A | NBD1 | ± | ± | – | — | — |
| Hsp104-RYD | R496A/Y497A/D498A | NBD1 | + | ++ | — | – | – |
| Hsp104-N292A | N292A | NBD1 | ND | ND | — | ND | ND |
| Hsp104-K294A | K294A | NBD1 | ND | ND | – | ND | ND |
| Hsp104-D295A | D295A | NBD1 | ND | ND | — | ND | ND |
| Hsp104-P461A | P461A | MD | - | ± | – | — | - |
| Hsp104-P557L | P557L | MD | ± | ± | — | – | - |
NTD is involved in substrate binding and productive interaction with Hsp70
We have established that the NTD of Hsp104 is crucial for hexamer cooperativity (19). The presence of the NTD enables Hsp104 to dissolve amyloid and is critical for potentiated Hsp104 variants to rescue TDP-43, FUS, and α-synuclein toxicity in yeast (19). Nonetheless, how the NTD confers these functions remains unclear. The crystal structures of the NTDs of various Hsp100 proteins, including Hsp104 from S. cerevisiae (38) and Calcarisporiella thermophila (18) and the bacterial Hsp100 homologues ClpA (39), ClpB (40), and ClpC (41), have been solved. The NTDs are highly structurally conserved despite low sequence identity (e.g. 28% sequence identity between the ClpA and ClpB NTDs). The NTDs are stable globular domains composed of two imperfect repeats of four helical bundles (R1 and R2 each contain four helices, A1–A4 and A5–A8) and are connected to NBD1 by a highly-mobile linker (18, 39).
A conserved feature of the Hsp100 NTDs is a hydrophobic patch between helices A1 and A5 (39). Interestingly, this hydrophobic patch region is solvated in all three states (peptide 86–110, Fig. 2, A and B, Table 2, and Tables S1–S3). In fact, four hydrophobic residues in this hydrophobic patch region were identified by MS2 analysis as being specific sites of hydroxyl radical modification (residues Ala-91, Leu-92, Val-95, and Leu-96, Table 2, and Table S3, Fig. 2C shown as spheres and labeled in purple). This finding indicates that this conserved region is available to interact with substrate in both the ADP and ATPγS states.
Figure 2.
Hsp104 NTD is involved in substrate and Hsp70 interactions. A, peptide sequences and modification rates for a region of the Hsp104 NTD that is highly modified. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: yellow, monomer; blue, hexamer with ADP; red, hexamer with ATPγS; purple, both hexameric states; green, monomer and hexamer with ADP; orange, monomer and hexamer with ATPγS; black, modified in all states. B, surface rendering of Hsp104 NTD from the crystal structure of the S. cerevisiae Hsp104 NTD (PDB code 5u2u). Acidic residues are shown in red, basic residues in blue, and hydrophobic residues in yellow. Circled regions are important in the structurally-conserved ClpA (PDB code 1r6c) NTD for ClpS binding as follows: site A in green, site C in light blue, and the conserved hydrophobic patch for substrate binding in purple. C, Hsp104 NTD with residues identified by MS2 as modified in a hexameric state are shown as spheres and labeled when visible. Spheres were colored by the states in which they were found modified, as in A, and labeled with colors corresponding to the regions outlined in B. D, FITC-casein, 2 mm ATPγS, and either Hsp104 or Hsp104ΔN were incubated for 20 min at 25 °C. Fluorescence polarization was measured at increasing concentrations of Hsp104, and a single site Kd was calculated using a least-squares fit to the data. Each data point represents mean ± S.E. (n = 4–5). E, urea-denatured firefly luciferase aggregates were incubated with Hsp104 or an Hsp104 variant, Hsp104-LVL (L92A/V95A/L96A), Hsp104-TYK (T87A/Y90A/K94A), Hsp104-QS (Q103A/S109A), Hsp104-IK (I102A/K107A), or Hsp104-SS (S124A/S125A) for 90 min at 25 °C in the presence of either 5.1 mm ATP or ATP/ATPγS; 2.6 mm ATP, and 2.5 mm ATPγS, or Hdj2/Hsc70; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsc70, or Hdj2/Hsp72; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsp72. Reactivation of luciferase was then determined by measuring luminescence and converted to fraction WT activity for each condition. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3–7). One-way ANOVA with Dunnett's test was performed against Hsp104-WT with * denoting p < 0.05 and ** p < 0.01. Hsp104 variants are colored based on their location as shown in B. F, ATPase activity for Hsp104 and variants. Hsp104 variants are colored based on their location in the regions highlighted in B. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3–4). One-way ANOVA with Dunnett's test comparing the variants to Hsp104-WT were performed with * denoting p < 0.05.
To test whether the NTD of Hsp104 is involved in substrate binding, we measured the affinity for a fluorescent substrate, FITC-casein, using a fluorescence polarization assay. We used the purified wildtype protein (WT) and a truncation mutant (Hsp104ΔN) missing the NTD as well as the NTD–NBD1 linker (19). Under these conditions, in the presence of ATPγS, Hsp104 binds FITC-casein robustly with a Kd of ∼55 ± 7 nm (Fig. 2D) (16, 19). In the same conditions, Hsp104ΔN binds FITC-casein with a Kd of ∼194 ± 60 nm, showing an ∼3–4-fold decrease in affinity (Fig. 2D) (19). This result supports a role for the Hsp104 NTD in binding certain substrates.
Additional residues that were identified as oxidatively modified by MS2 analysis fall into two clusters: (a) in the beginning of the A5 helix and in the loop between helices A6 and A7, (residues Thr-87, Tyr-90, Lys-94, Ser-124, and Ser-125 highlighted in light blue in Fig. 2, B and C), or (b) at the end of helix A5 and in the helix A5–A6 loop (residues Ile-102, Gln-103, Lys-107, and Ser-109 highlighted in green in Fig. 2, B and C). These two clusters of residues are in the same regions that were identified in ClpA (which lacks a MD) to interact with an adapter protein ClpS (39), which targets ClpA to aggregated substrates (42). In another Hsp100 protein, ClpC, which has a truncated version of the MD (motif 1 only), the crystal structure of the hexamer revealed that the adapter protein MecA made extensive contacts with both the MD as well as the NTD (41). Thus, we wondered whether Hsp104 might also interact with Hsp70 via the NTD in addition to proposed contacts with the MD (43–46). Indeed, one region of the Hsp104 MD found to interact with Hsp70, namely residues within motif 2 near the C-terminal end of helix L2 (47), is adjacent to the NTD in hexameric models of Hsp104 (16–18, 34, 48). Additionally, residues 100–102 (which contain the oxidatively modified Ile-102, Fig. 2, A and C) were identified as a potential Hsp70-binding site using peptide array technology (47), although this region was not investigated beyond an initial identification. NTD/Hsp70 interactions may facilitate substrate release from Hsp70 for hand-off to Hsp104. Indeed, Hsp104ΔN has reduced ability to disaggregate and reactivate chemically-denatured luciferase in the presence of Hsc70 and Hdj2 (∼40% of WT) or Hsp72 and Hdj2 (∼86% of WT) (19). Thus, the NTD enables productive interactions with Hsp70. The difference in activity between Hsp104ΔN in the presence of Hsc70 versus Hsp72 indicates that the Hsp104 hexamer may interact differently with different Hsp70 proteins. This distinction is important because Hsc70 is constitutively expressed and involved in maintaining protein homeostasis, whereas Hsp72 is induced by heat shock and facilitates response to acute cellular stress.
Next, we used the residue-specific information from the XF solvation to design several Hsp104 NTD mutants to assess contributions of specific NTD residues to interactions with substrate or Hsp70. We built five Hsp104 variants with clusters of mutations in NTD regions suggested to be important by the XF data and measured their ability to reactivate luciferase trapped in denatured aggregates. We assessed Hsp104 disaggregase activity under three conditions: (a) in the absence of Hsp70 and Hsp40, but in the presence of 1:1 ATP/ATPγS; (b) in the presence of ATP and Hsp40/Hsc70; and (c) in the presence of ATP and Hsp40/Hsp72 (19). In the presence of ATP, Hsp104 is inactive in disaggregating luciferase in the absence of Hsp70 and Hsp40 in vitro (20, 22). However, Hsp104 can be activated in the absence of Hsp70 and Hsp40 by the addition of permissive ratios of ATP/ATPγS (19, 20, 25, 49). Hsp104 has optimal activity at an ∼1:1 ratio of ATP/ATPγS (20, 49). Thus, a 1:1 ratio of ATP/ATPγS allows assessment of intrinsic Hsp104 disaggregase activity in the absence of Hsp70 and Hsp40. Hence, we could distinguish whether particular NTD mutations disrupted intrinsic Hsp104 disaggregase activity, collaboration with Hsp70, or both.
Hsp104-LVL contained mutations to hydrophobic residues found to be modified within the conserved hydrophobic patch (L92A/V95A/L96A; Fig. 2, A and C). In the presence of ATP and ATPγS, Hsp104-LVL displayed a significant defect in rescuing aggregated luciferase as expected if the region is involved in substrate binding (Fig. 2E). Hsp104-LVL ATPase activity was only mildly decreased from WT Hsp104 (∼70% WT activity), which might also contribute to reduced disaggregase activity (Fig. 2F). However, this defect in luciferase reactivation was mitigated by the presence of either Hsp70 variant (Fig. 2E). Thus, substrate-binding defects, mild ATPase defects, or both in Hsp104 can be buffered by the presence of Hsp70 and Hsp40.
Hsp104-TYK (T87A/Y90A/K94A), Hsp104-QS (Q103A/S109A), Hsp104-IK (I102A/K107A), and Hsp104-SS (S124A/S125A) harbor mutations to residues found to be modified and within regions that might be involved in interactions with Hsp70 (Fig. 2, B and C). Generally, Hsp104-SS exhibited similar disaggregase activity to WT Hsp104 although in the presence of Hdj2/Hsc70 the modest decrease was statistically significant (Fig. 2E). Hsp104-SS exhibited similar ATPase activity to WT Hsp104 (Fig. 2F). Thus, Ser-124 and Ser-125 are unlikely to play a major role in interactions with substrate or Hsp70. Thr-87, identified as oxidatively modified in the hexamer with ATPγS (Fig. 2, A and C), was an interesting target. This residue is just N-terminal to helix A5 (Fig. 2C). In ClpA, an equivalent residue, Thr-81, resides within the main ClpS-binding site (39), and in ClpC, the residue Thr-31 is essential for ClpC activity by mediating the interaction with MecA (41). Hsp104-TYK, Hsp104-QS, and Hsp104-IK all display disaggregation defects in the presence of ATP/ATPγS and Hsp40/Hsc70 (Fig. 2E). Although the defect in the presence of ATP/ATPγS was unexpected, it is more severe in the presence of Hsp40/Hsc70 (Fig. 2E). Additionally, Hsp104-TYK displays a defect in activity in the presence of Hsp40/Hsp72 (Fig. 2E). Hsp104 ATPase activity was not grossly affected by these mutations (Fig. 2F). Thus, these regions of the Hsp104 NTD play important roles in the substrate-binding and Hsp70-collaboration steps necessary to disaggregate substrate.
NTD/NBD1/MD interactions that include the NTD–NBD1 linker contribute to Hsp104 activity
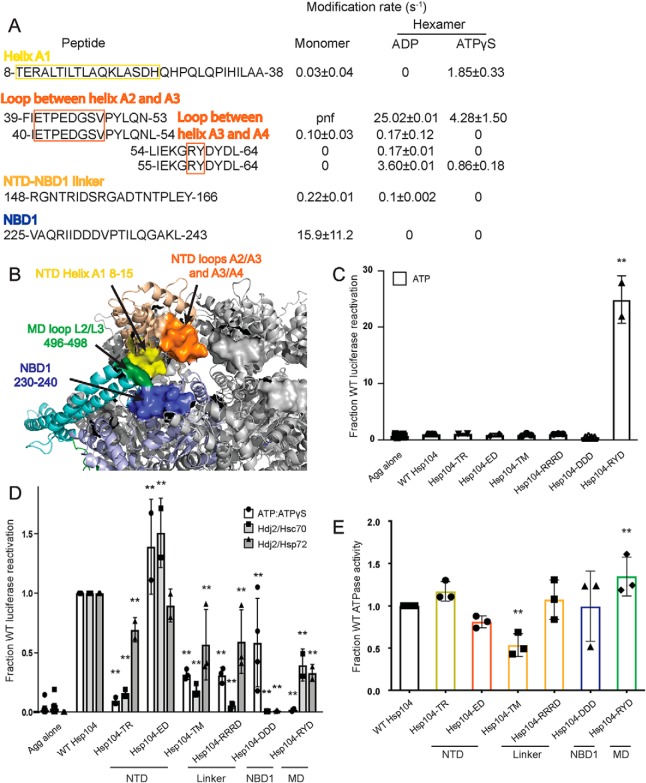
To understand how the NTD may be communicating with neighboring domains and how these interactions may change depending on the identity of the nucleotide, we first looked for regions that undergo changes in solvation between different states. These regions include the beginning of helix A1 (residues 8–15), the loop between helices A2 and A3 (residues 41–51), the loop between helices A3 and A4 (residues 58–61), and the linker between the NTD and NBD1 (residues 148–166) (Fig. 3, A and B). A peptide that covers the beginning of helix A1 starting at residue 8 appears to be unmodified in the hexamer with ADP, but it has a rate of 1.85 ± 0.33 s−1 in the hexamer with ATPγS (Table 2 and Fig. 3A). The loop between helices A2 and A3, residues 41–51, is covered by the peptide 39–53, which has rates for the ADP and ATPγS hexamers of 25 ± 0.005 and 4.3 ± 1.5 s−1, respectively (Table 2 and Fig. 3A). The loop between helices A3 and A4, residues 58–61, is covered by the peptide 55–64. This peptide is unmodified in the monomer, and it has rates for the ADP and ATPγS hexamers of 3.6 ± 0.009 and 0.85 ± 0.18 s−1, respectively (Table 2 and Fig. 3A). The linker between the NTD and NBD1 is covered by the peptide 148–166 in all three states (Table 2 and Fig. 3A). It is unmodified in the hexamer with ATPγS, has a rate of 0.101 ± 0.002 s−1 in the hexamer with ADP, and 0.215 ± 0.014 s−1 in the monomer (Table 2 and Fig. 3A).
Figure 3.
NTD–NBD1–MD interface regulates Hsp104 disaggregase activity. A and B, based on the solvation data from XF, there may be regions of the Hsp104 NTD, NBD1, and MD that make contacts that are nucleotide-dependent. In the NTD, these regions are the beginning of helix A1 in the hexamer with ADP (residues 8–15 in yellow), the A2–A3 loop (residues 39–53 in orange), and NTD–NBD1 linker (not pictured) in the hexamer with ATPγS. Based on: (1) the XF data, (2) the three monomeric tClpB crystal structures (PDB code 1qvr), and (3) the hexameric cryo-EM reconstruction (PDB code 5vy9, shown in B), we suggest that these NTD regions may be making a stable interaction with NBD1 (specifically residues 230–240 in blue) and in the hexamer with ATPγS the MD in the loop between MD helices L2 and L3 in motif 2 (residues 496–498 in green). Shifts in these contacts in response to stimuli such as Hsp70 binding, substrate binding, or ATP hydrolysis could transmit signals through the hexamer and facilitate cooperativity. In the table with rate information, pnf denotes “peptide not found.” C and D, urea-denatured firefly luciferase aggregates were incubated with either WT or an Hsp104 variant, Hsp104-TR (T8A/R10A), Hsp104-ED (E44R/D45R), Hsp104-TM (T160M), Hsp104-RRRD (R148D/R152D/R156D), Hsp104-DDD (D231A/D232A/D233A), or Hsp104–RYD (R496A/Y497A/D498A) for 90 min at 25 °C in the presence of either 5.1 mm ATP (C) or ATP/ATPγS; 2.6 mm ATP and 2.5 mm ATPγS or Hdj2/Hsc70; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsc70 or Hdj2/Hsp72; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsp72 (D). Reactivation of luciferase was then determined by measuring luminescence and converted to fraction WT activity for each condition. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 2–4). One-way ANOVA with Dunnett's test comparing the variants to Hsp104-WT were performed with ** denoting p < 0.01. E, ATPase activity for Hsp104-WT and variants. Hsp104 variants are colored based on their location as shown in A and B. Values represent mean ± S.D. (n = 3). One-way ANOVA with Dunnett's test comparing the variants to Hsp104-WT were performed with ** denoting p < 0.01.
We then compared these regions from XF to the three tClpB monomer models (37). In the crystal structure of tClpB, each monomer of the spiral trimer (designated models A–C in the PDB) has a different orientation of the NTD (37). By examining the different orientations of the NTD, we discovered regions of the NTD, NBD1, and MD that appeared capable of interacting in one or all of the three models. The homologous Hsp104 NTD residues include 8–20 found in models A and B, NBD1 residues 264–274 found in models A and C, MD residues 496–498 found in models A and C, and NTD residues 39–52, NTD/NBD1 residues 155–169, and NBD1 residues 230–240 found in all three of the models. Many of these regions display changes in solvation, as noted above. Additionally, in a hexameric model of Hsp104 generated using cryo-EM data, which models in the NTD and different MD orientations, these regions are found in close proximity (Fig. 3B) (16). These include the beginning of helix A1 residues 8–15 in particular (Fig. 3, A and B, yellow), which are protected from solvation in the hexamer with ADP, the loop between helices A2 and A3, residues 39–53 (Fig. 3, A and B, orange), which shows a decrease of solvation in the hexamer with ATPγS, and the NTD–NBD1 linker region, 148–166, which shows a protection from solvation in the hexamer with ATPγS. Consistent with this observation, formation of Hsp104 hexamers slows hydrogen–deuterium exchange (HX) of the NTD–NBD1 linker (30).
If we investigate the regions in NBD1 and the MD, we find that NBD1 residues 230–240 (Fig. 3, A and B, blue) are covered by a number of peptides, which are unmodified in both hexameric states (Tables S2 and S3). In fact, the NBD1 peptide 225–243 is unmodified in the hexamers, whereas the monomer has a modification rate of 15.9 ± 11.2 s−1 (Table 2 and Fig. 3A). NBD1 residues 262–274 display no clear trend in modification rate (Tables S1–S3), and MD residues 496–498 (Fig. 3B, green), covered by the peptide 494–504, are found unmodified in the monomer and the hexamer ATPγS, but the peptide was not found in the hexamer with ADP (Tables S1–S3). These findings indicate that the NTD, specifically the beginning of helix A1 in the hexamer with ADP, and the A2–A3 loop and NTD–NBD1 linker in the hexamer with ATPγS may be making a stable interaction with NBD1 residues 230–240 and potentially the MD, in the loop between MD helices L2 and L3 in motif 2 (Fig. 3B). Thus, different regions of the NTD can contact the same regions of NBD1 and the MD. The NTD is involved in regulating ATPase activity and is essential for global cooperativity of the hexamer (19). Thus, we suggest that these interactions may relay signals from the NTD to the rest of the hexamer that are elicited by substrate interactions, Hsp70 interactions, or both.
Further support of this model is provided by the mutant, T160M, which resides in a region of the NTD–NBD1 linker proposed to interact with NBD1, the MD, or both. T160M phenocopies the effects of NTD deletion in vivo (50). Additionally, other regions identified by peptide array as potential Hsp70-binding sites include NTD residues 34–39 (47), just N-terminal of the A2–A3 loop, and 241–249 (47), just C-terminal of the unmodified 230–240 NBD1 region, indicating that there may be a complex network of NTD–NBD1–MD/Hsp70 interactions (Fig. 3B). Mutations in this region of the MD, specifically D498V (51), T499D (52), and T499E (52), and also in the relevant region of NBD1 I230N (53), lead to potentiated Hsp104 variants, and deletion of the NTD precludes this potentiation (19). These findings suggest that this interface is important for hexamer regulation and activity. To test our proposed network of interactions, we made several Hsp104 variants with mutations in the regions suggested by the XF data, as well the T160M mutant (50).
First, we tested Hsp104-TR (T8A/R10A), which bears mutations in helix A1 (Fig. 3, A and B) and was inactive in luciferase disaggregation like WT Hsp104 in the presence of ATP alone (Fig. 3C). By contrast, Hsp104-TR displays defective luciferase disaggregase activity in the presence of ATP/ATPγS, Hdj2/Hsc70, and Hdj2/Hsp72 (Fig. 3D). Hsp104-TR displayed WT levels of ATPase activity (Fig. 3E). Thus, Thr-8 and Arg-10 in helix A1 are important for optimal intrinsic disaggregase activity and Hsp70-dependent disaggregase activity.
Next, we tested Hsp104-ED (E44R/D45R), which bears mutations in the loop between helices A2 and A3 in the NTD (Fig. 3, A and B). Unexpectedly, Hsp104-ED exhibited slightly elevated disaggregase activity in the presence of ATP/ATPγS or Hdj2/Hsc70 and WT disaggregase activity in the presence of Hdj2/Hsp72 (Fig. 3D). Hsp104-ED also displayed mildly reduced ATPase activity (Fig. 3E). Thus, the loop between helices A2 and A3 can be targeted to increase disaggregase activity under some conditions.
Hsp104-TM (T160M) and Hsp104-RRRD (R148D/R152D/R156D) are both found in the linker between the NTD and NBD1. Like WT Hsp104, Hsp104-TM and Hsp104-RRRD are inactive in luciferase disaggregation in the presence of ATP alone (Fig. 3C). They both display reduced disaggregase activity in the presence of ATP/ATPγS, Hdj2/Hsc70, and Hdj2/Hsp72 (Fig. 3D). Hsp104-TM exhibits reduced ATPase activity, whereas Hsp104-RRD does not (Fig. 3E). Thus, the NTD–NBD1 linker is crucial for optimal disaggregase activity, and the Thr-160 residue is involved in regulating the ATPase activity of the hexamer.
We next assessed Hsp104-DDD (D231A/D232A/D233A), which bears three mutations in NBD1 (Fig. 3, A and B). Like WT Hsp104, Hsp104-DDD is inactive in luciferase reactivation in the presence of ATP alone (Fig. 3C) and exhibits normal ATPase activity (Fig. 3E). Hsp104-DDD displays reduced disaggregase activity in the presence of ATP/ATPγS (Fig. 3D). Most strikingly, however, Hsp104-DDD is completely inactive in the presence of Hsp70 (Fig. 3D). By mutating these three acidic residues, we have created an Hsp104 variant that retains intrinsic disaggregase activity but is unable to collaborate with Hsp70, establishing a critical role for Asp-231, Asp-232, and Asp-233 in Hsp70 collaboration.
Hsp104–RYD (R496A/Y497A/D498A) is in the loop found between helices L2 and L3 in motif 2 of the MD. Unlike WT Hsp104, Hsp104–RYD displayed enhanced disaggregase activity in the presence of ATP alone (Fig. 3C), which is reminiscent of potentiated Hsp104 variants like Hsp104A503V (51, 52). Moreover, like potentiated Hsp104 variants, Hsp104–RYD is inactive as a disaggregase in the ATP/ATPγS condition (Fig. 3D) (49, 51). However, unlike Hsp104A503V, Hsp104–RYD had only slightly increased ATPase activity (Fig. 3E) and displayed reduced disaggregase activity compared with Hsp104 in the presence of Hsp72 or Hsc70 (Fig. 3D) (51). These data suggest that Arg-496, Tyr-497, and Asp-498 are important for optimal collaboration with Hsp70.
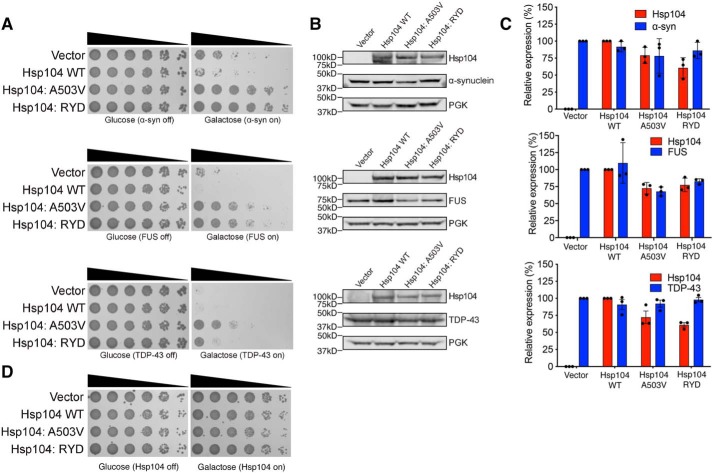
Remarkably, however, Hsp104–RYD rescued α-synuclein (which is connected to Parkinson's disease), FUS (which is connected to ALS and frontotemporal dementia (FTD)), and TDP-43 (which is also connected to ALS and FTD) toxicity in yeast, unlike WT Hsp104 (Fig. 4A). Hsp104–RYD rescued α-synuclein and FUS toxicity just as well as Hsp104A503V (51, 54), whereas rescue of TDP-43 toxicity was more robust for Hsp104A503V (Fig. 4A). Hsp104–RYD rescued the toxicity of these disease proteins, without grossly reducing their expression level (Fig. 4B). Indeed, quantification of the immunoblots revealed that Hsp104–RYD only slightly reduced α-synuclein levels to ∼86% of the vector control (Fig. 4C). Likewise, Hsp104–RYD only slightly reduced FUS levels to ∼82% of the vector control (Fig. 4C). Hsp104–RYD had no effect on TDP-43 expression (Fig. 4C). These slight reductions in α-synuclein and FUS expression are unlikely to explain the strong suppression of toxicity conferred by Hsp104–RYD. Indeed, we have previously shown that reduced FUS expression is not required for enhanced Hsp104 variants to rescue FUS toxicity (53). Moreover, Hsp104–RYD was typically expressed at a lower level of ∼61–77% of the WT Hsp104 control. Thus, even though α-synuclein and FUS expression was slightly lower, so too was expression of Hsp104–RYD. Importantly, expression of Hsp104A503V or Hsp104–RYD was not toxic to yeast in the absence of disease protein (Fig. 4D). Thus, Hsp104–RYD is a potentiated Hsp104 variant (53). This finding confirms that enhanced disaggregase activity in the presence of ATP and absence of Hsp70 is a strong indicator of enhanced activity against neurodegenerative disease proteins in yeast (51, 53, 55, 56). Collectively, these observations suggest that the proposed NTD/NBD1/MD interactions that include the NTD–NBD1 linker make important contributions to Hsp104 functionality.
Figure 4.
Hsp104–RYD is a potentiated variant. A, Hsp104–RYD antagonizes α-synuclein, FUS, and TDP-43 toxicity. Empty vector, Hsp104 (WT), Hsp104A503V, or Hsp104–RYD plasmids were transformed into W303aΔhsp104 yeast strains integrated with galactose-inducible α-synuclein-YFP, FUS, or TDP-43. Yeast were serially diluted 5-fold and spotted in duplicate onto galactose (inducing) and glucose (noninducing) media. B, samples were also processed for immunoblotting to assess Hsp104, PGK1 (loading control), and α-synuclein–YFP, FUS, or TDP-43 expression. Molecular mass markers are indicated (left). C, quantification of immunoblots from B. Hsp104 levels (red bars) were normalized to PGK (loading control), and the relative expression level compared with Hsp104 WT was determined. α-Synuclein–YFP, FUS, and TDP-43 levels (blue bars) were normalized to PGK (loading control), and the relative expression level compared with vector control was determined. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3). D, Hsp104–RYD is not toxic to yeast at 30 °C. Empty vectors, Hsp104 (WT), Hsp104A503V, or Hsp104–RYD plasmid were transformed into W303aΔhsp104 yeast strains. Yeast were serially diluted 5-fold and spotted in duplicate onto galactose (inducing) and glucose (noninducing) media.
NBD1 AAA+ motifs are more solvent-exposed in hexamers with ADP than ATPγS
NBD1 is a high-affinity, high-turnover site for ATP binding and hydrolysis (57). It is the main site of ATP hydrolysis for the Hsp104 hexamer, but it does not drive hexamerization, which is controlled by NBD2 (57, 58). ATP binding and hydrolysis are required for Hsp104 disaggregase activity (22, 23, 59, 60). Thus, we examined changes in solvation of the conserved AAA+ motifs between the hexamer with ADP and hexamer with ATPγS. Overall, NBD1 is more modified in the hexamer with ADP than the hexamer with ATPγS (Fig. 5A, Table 2, and Tables S2 and S3), particularly in the regions involved in the conserved ATP-binding pocket. This finding is consistent with a switch from an open spiral hexamer in the presence of ADP to a closed ring hexamer in the presence of ATPγS suggested by cryo-EM (16–18) and corroborated by HX studies (30).
Figure 5.
Second substrate-binding loop in NBD1 is essential for Hsp104 disaggregase activity. A, peptide sequences and modification rates for regions of interest of Hsp104 NBD1. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: blue, hexamer with ADP; purple, both hexameric states. B, NBD1 homology modeled on the tClpB crystal structure (PDB code 1qvr) shown in context of a rigid body fit Hsp104 monomer. NBD1 is shown in blue, with the canonical substrate-binding loop in blue spheres and the secondary substrate-binding loop in red spheres. C, after incubation at 37 °C for 30 min to induce Hsp104 expression, W303aΔhsp104 yeast carrying either empty vector or a plasmid encoding the indicated Hsp104 variant were heat-shocked for 20 min at 50 °C, immediately transferred to ice for 2 min, plated on SD-ura plates, and after a 2-day incubation at 30 °C, colonies were counted using an acolyte automated colony counter. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3–14). Equal expression levels were confirmed by immunoblot. A one-way ANOVA with Dunnett's test was used to compare WT Hsp104 to the variants with ‡ denoting p = 0.05 and ** denoting p < 0.01. D, urea-denatured firefly luciferase aggregates were incubated with either WT or an Hsp104 substrate-binding loop 2 (291GNGKD295) variant, for 90 min at 25 °C in the presence of 2.6 mm ATP, 2.5 mm ATPγS, and an ATP-regenerating system (1 mm creatine phosphate and 0.25 μm creatine kinase). Reactivation of luciferase was then determined by measuring luminescence and converted to fraction WT activity for each condition. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3). A one-way ANOVA with Dunnett's test comparing variants to Hsp104-WT was performed with *** denoting p < 0.0001.
The Walker A motif, residues 212–220, the Walker B motif, residues 280–285, the sensor-1 residue, Thr-317, and the arginine finger, Arg-334, are covered by peptides that suggest that in the hexamer with ATPγS the ATP-binding pocket is protected from solvation (Fig. 5A). The peptide 211–223 shows no modification in the ATPγS hexamer, but a rate of 32.6 ± 1.9 s−1 in the hexamer with ADP (Table 2 and Fig. 5A). Consistent with this observation, HX shows that ATPγS induces a tight restructuring of the Walker A motif (30). The peptides 282–299, 283–299, and 286–310 also show no modification in the hexamer with ATPγS but rates of 75 ± 0.58, 28 ± 0.42, and 27 ± 0.3 s−1, respectively, in the hexamer with ADP (Table 2 and Fig. 5A). The peptides 300–320 and 311–320, which cover the sensor-1 residue, Thr-317, show that the hexamer with ADP is much more solvated than the hexamer with ATPγS, with rates of 145.6 ± 3.4 and 2.8 ± 0.03 s−1 for ADP and 0 and 0.37 ± 0.07 s−1 for ATPγS, respectively (Table 2 and Fig. 5A). Finally, the peptides 331–350 and 332–349 that cover the arginine finger, Arg-334, are unmodified in the hexamer with ATPγS and have rates of 133.5 ± 3.4 and 3.8 ± 0.3 s−1 in the hexamer with ADP, respectively (Table 2 and Fig. 5A). Additionally, residues in these regions were identified as modified by MS2 in the hexamer with ADP, including the essential Walker B glutamate, residue Glu-285, as well as Arg-333, the residue directly preceding the arginine finger Arg-334 (Table 2 and Fig. 5A), which is proposed to act like a sensor 2 residue in NBD1 (NBD1 lacks a canonical sensor 2 residue unlike NBD2) (9, 16, 61). These differences in solvation (Fig. 5A) indicate that the six conserved ATP-binding pockets of NBD1 in the hexamer with ATPγS are either all occupied by nucleotide consistent with cryo-EM studies (16) or closed (i.e. unoccupied, but shielded from solvent). The situation is different for the hexamer with ADP. In the presence of ADP, a large degree of solvation is detected in NBD1, which is consistent with the open spiral ring of Hsp104 hexamers with ADP observed by cryo-EM and HX where NBD1 from protomer 1 is highly exposed to solvent (16, 30).
NBD1 contains a conserved loop with a role in substrate interaction
Although two substrate-binding loops have been identified in Hsp104 (16, 62), the partial defect in thermotolerance when the NBD1 loop is mutated (62) indicates that there may be additional regions of NBD1 involved in substrate binding and translocation across the channel. The canonical NBD1 substrate-binding loop, 256KYKG259 (62), is oxidatively modified in both hexameric states, indicating that the loop is solvated and capable of substrate interaction (Tables S2 and S3). HX shows that this pore-loop becomes more protected in the presence of ATPγS, indicating a gain in local structure in preparation for substrate binding (30). Although rate information for identical peptides is unavailable for both states, peptides that cover the loop have much faster rates of modification in the hexamer with ATPγS than for the hexamer with ADP: compare 10.6 ± 5.3 and 18.2 ± 0.37 s−1 for peptides 249–277 and 248–261 from the hexamer with ATPγS to 0.51 ± 0.12 and 1.15 ± 0.08 s−1 for peptides 249–278 and 249–271 from the hexamer with ADP (Tables S2 and S3). As the Hsp104 hexamer primarily binds substrate in the ATP state (16, 29, 63, 64), it is anticipated that the loop would be more exposed in the hexamer with ATPγS than in the hexamer with ADP. Indeed, this loop is not visible by cryo-EM in the presence of ADP, but becomes visible in the presence of ATPγS and engages casein (16).
Although Tyr-257 has been identified as one of two substrate-binding residues in Hsp104, thermotolerance assays show that mutation of the tyrosine to alanine results in only minor defects in Hsp104-mediated survival (62). This finding is in contrast to mutation of the NBD2 tyrosine, Tyr-662 to alanine, which results in large defects in survival after heat shock (62). This finding led us to hypothesize that regions of the NTD as well as other sites within NBD1 may contribute to substrate binding. Indeed, the NTD is involved in substrate binding (Fig. 2D). This finding is further supported by thermotolerance data showing that although either deletion of the NTD or mutation of Tyr-257 displays only mild defects in thermotolerance, the combined truncation and mutation variant, ΔN/Y257A Hsp104, is severely compromised in its ability to mediate survival after heat shock (Fig. 5C).
To determine whether there are other substrate-binding regions of NBD1, we looked for regions that change in solvation in the different hexameric states. A striking example stood out immediately. In the homology model of the Hsp104 NBD1, there are two loops that point into the interior of the hexameric Hsp104 channel (Fig. 5B). One is the canonical 256KYKG259 loop and the second is the 291GNGKD295 loop. Peptides covering the 291GNGKD295 loop are unmodified in the hexamer with ATPγS, but highly modified in the hexamer with ADP (the peptides that cover the Walker B site also cover this loop) (Fig. 5A and Table 2). In addition to the peptides (282–299, 283–299, and 286–310, Table 2 and Fig. 5A) displaying high rates of modification in the hexamer with ADP (75 ± 0.58, 28 ± 0.42, and 27 ± 0.3 s−1, respectively, Table 2 and Fig. 5A), three residues within and near the 291GNGKD295 loop were identified as modified by MS2, including Asn-292 and Lys-294 (Fig. 5A and Table 2). This loop is homologous to the ClpC 286GAGGA290 (41) loop, as well as the ClpA 292GAGAA296 (65) loop, which are secondary substrate-binding loops. Mutation or deletion of this loop results in severe defects in ClpC (41) and ClpA (65) activity. Cryo-EM has revealed that the 291GNGKD295 loop becomes ordered and in close apposition to translocating substrate in the presence of ATPγS (16). Likewise, HX has revealed that this loop becomes more tightly structured in the presence of ATPγS (30).
In vivo thermotolerance assays revealed that the K294A Hsp104 variant displayed a slight but statistically significant defect in mediating survival after heat shock compared with Hsp104 (Fig. 5C). When the K294A mutation was combined with the NTD truncation, survival after heat shock was greatly reduced and similar to the ΔN/Y257A variant (Fig. 5C). The double mutant Y257A/K294A variant was less active than the single Y257A, K294A, or ΔN mutants, but more active than the single mutants combined with the NTD truncation (Fig. 5C). These data support a role for the 291GNGKD295 loop in substrate translocation.
Next, we tested the 291GNGKD295 loop mutants in an in vitro assay with minimal components. Thus, we performed luciferase reactivation assays in the presence of a 1:1 ratio of ATP/ATPγS. Here, the loop mutants N292A, K294A, and D295A were defective in luciferase reactivation, retaining only 20, 39, and 19% of WT Hsp104, respectively (Fig. 5D). This finding confirms the importance of this loop for productive protein disaggregation.
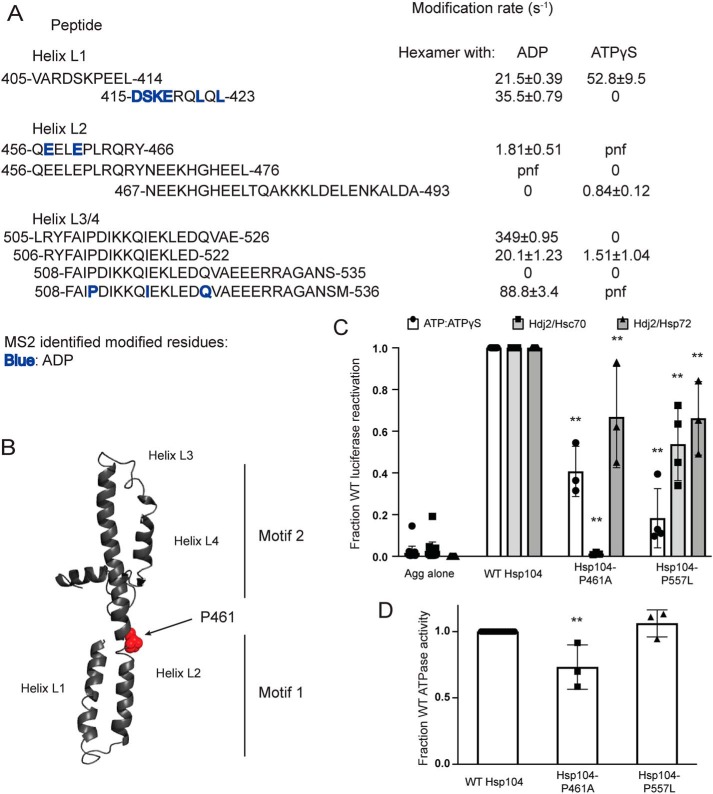
Solvation of the MD changes dramatically in the presence of different nucleotides
Next, we utilized our solvation data from the XF experiments to understand how local and global dynamics of the MD determine its role in regulating the hexamer (Fig. 6). The homology model of the Hsp104 anti-parallel coiled-coil MD, based on the tClpB structure, retains the leucine zipper-like interactions between helices L1–L4 (Fig. 6B). The MD has been implicated in regulating the ATPase and disaggregase activity of the hexamer (44–46, 51, 52, 66–74), transmitting signals between NBD1 and NBD2 (66, 67, 73), and mediating the interaction with Hsp70 (34, 43–47). Interactions with Hsp70 proteins may take place in motif 2, specifically residues 480KKK482 and residue Arg-496 (47). We have discussed in the preceding sections how these regions are poised to interact with the NTD as well as NBD1 (Fig. 3). A number of interactions between motif 2 of the MD and NBD1 have been identified (37, 45, 72, 74), and stabilization of these interactions leads to repression of the disaggregase (37, 45, 72), whereas destabilization elicits hyperactivity (37, 51, 72, 74). XF data as well as published cross-linking (34, 72) and structural data (19, 72) support a model in which the domain is highly mobile, undergoing interactions with both NBD1 (37, 45, 72, 74) and NBD2 (34). Additionally, it is likely that MDs within a given Hsp104 hexamer occupy a variety of positions (16–18, 72).
Figure 6.
Hsp104 MD hinge region is crucial for Hsp70 collaboration and intrinsic Hsp104 disaggregase activity. A, peptide sequences and modification rates for regions of interest of Hsp104 MD. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: blue, hexamer with ADP. B, isolated MD modeled off the tClpB crystal structure (PDB code 1qvr). Helices L1–L4 are labeled, and residue Pro-461 is shown as spheres. C, urea-denatured firefly luciferase aggregates were incubated with either WT or an Hsp104 variant, Hsp104-P461A or Hsp104-P557L for 90 min at 25 °C in the presence of either 5.1 mm ATP or ATP/ATPγS; 2.6 mm ATP and 2.5 mm ATPγS, or Hdj2/Hsc70; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsc70, or Hdj2/Hsp72; 5.1 mm ATP, 1 μm Hdj2, and 1 μm Hsp72. Reactivation of luciferase was then determined by measuring luminescence and converted to fraction WT activity for each condition. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3). One-way ANOVA with Dunnett's test comparing the variants to Hsp104-WT were performed with ** denoting p < 0.01. D, ATPase activity for Hsp104-WT and variants. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3). One-way ANOVA with Dunnett's test comparing the variants to Hsp104-WT were performed with ** denoting p < 0.01.
Interestingly, the XF data of the MD suggested large changes in solvation between the hexamer bound to ADP and the hexamer bound to ATPγS (Fig. 6A). This finding likely reflects that MD is highly dynamic, and restricting its movement through disulfide cross-linking greatly impairs Hsp104 activity (34, 37, 45). In general, the hexamer with ADP displays greater solvation than the hexamer with ATPγS (Fig. 5A, Table 2, and Tables S2 and S3). One part of the MD where this is not the case is the first three residues of the MD, 412–414, found on the peptide 405–414, which mainly covers a portion of NBD1 (Table 2 and Fig. 6A). This peptide is solvated in both hexameric states, but more so in the presence of ATPγS (compare 21.5 ± 0.4 with 52.8 ± 9.5 s−1, Table 2 and Fig. 6A). Modification rates change at the adjacent peptide. The peptide 415–423 shows that the hexamer with ATPγS is unmodified, whereas the hexamer with ADP has a modification rate of 35.5 ± 0.8 s−1 and contains several residues identified as modified by MS2: Asp-415, Ser-416, Lys-417, Glu-418, Leu-421, and Leu-423 (Table 2 and Fig. 6A).
The 415–423 peptide covers the first half of helix L1 and is adjacent to a region of helix L2, residues 456–466, which is also unmodified in the hexamer with ATPγS (Fig. 6A). This peptide, 456–466, has a modification rate of 1.8 ± 0.5 s−1 in the hexamer with ADP and contains residues identified as modified by MS2, Glu-457 and Glu-460 (Table 2 and Fig. 6A). A region more C-terminal in helix L2, residues 467–474, is unmodified in the hexamer with ADP, but is modified in the hexamer with ATPγS. A peptide that covers this region, 467–493, is unmodified in the hexamer with ADP and has a modification rate of 0.84 ± 0.1 s−1 in the hexamer with ATPγS (Table 2 and Fig. 6A). The changes in solvation indicate that transitions in this region of helix L2 may play a role in transmission of conformational changes from motif 1 to motif 2 in response to stimuli, e.g. interaction with Hsp70, binding of substrate in the NTD and NBD1, and ATP hydrolysis at NBD1 or NBD2. Indeed, the MD is crucial for communication between NBD1 and NBD2 (66, 67, 73). Additionally, a mutation in this region, L462R, disrupts intersubunit collaboration (20), suggesting that this region is critical for communication within the Hsp104 hexamer.
Several cross-links between motif 2 of the MD and NBD1 have been found in ClpB (37, 45, 72), and in Hsp104 there may be corresponding salt bridges (74). Models proposing that this motif 2/NBD1 interaction is dynamic are supported by the XF solvation data, which show large increases in solvation in the hexamer with ADP compared with the hexamer with ATPγS. These increases in solvation include the C-terminal end of helix L2 (Tables S2 and S3) as well as the C-terminal end of helix L3 through the end of the MD: peptides 505–526, 506–522, and 508–536 with rates for the hexamer with ADP of 350 ± 0.9, 20 ± 1.2, and 89 ± 3.4 s−1, respectively, versus 0 and 1.5 ± 1 s−1, and undetermined for the hexamer with ATPγS (Table 2 and Fig. 6A). Residues 494–504 are not found in the hexamer with ADP but are unmodified in the hexamer with ATPγS. This finding might indicate that helices L3 and L4 of motif 2 are partially shielded from solvent in hexamer bound to ATPγS. If this protection from solvent is representative of the repressive motif 2–NBD1 contacts, it would support a model where this repressed state of the hexamer is populated in the hexamer with ATPγS. Additionally, several potentiating mutations are found in helices L3 and L4 (37, 51, 72, 74) indicating that large changes in solvation between nucleotide-bound states may indicate mechanistically important regions of the Hsp104 hexamer.
To explore how conformational changes might be transmitted through the MD, we focused on a region of helix L2 that undergoes large changes in solvation. Within this region there is a break in the helix, and within this break there is a proline residue. We predicted that this residue, Pro-461, may serve as a “hinge” allowing large changes in the position of the MD that are important for functionality (Fig. 6B). Thus, we made Hsp104–P461A, which displayed reduced disaggregase activity in the presence of ATP/ATPγS, Hsc70 and Hdj2, and Hsp72 and Hdj2 (Fig. 6C), whereas ATPase activity was only modestly reduced (Fig. 6D). Reduced disaggregase activity was most severe in the presence of Hsc70 and Hdj2 (Fig. 6C), indicating that Pro-461 is critical for collaboration with specific Hsp70 chaperones.
We also assessed Hsp104–P557L, a mutant found just outside the MD at the end of the NBD1 small subdomain. Hsp104–P557L conveys normal thermotolerance in vivo but is unable to propagate [PSI+] prions (75, 76). Hsp104–P557L also showed decreased levels of luciferase reactivation in the presence of ATP/ATPγS, Hdj2/Hsc70, and Hdj2/Hsp72 (Fig. 6C), while retaining WT levels of ATPase activity (Fig. 6D). Here, the defect in disaggregase activity was most severe in the presence of ATP/ATPγS, indicating that intrinsic disaggregase activity was affected in a way that could be mitigated by Hsp70 and Hsp40.
NBD2: AAA+ motifs are more solvent-exposed in hexamers with ADP than ATPγS
Next, we examined modification of the conserved NBD2 ATP-binding motifs as well as the region that contains the nuclear localization signal (NLS) (77). There is no coverage of either the arginine finger (Arg-765) or the sensor 1 (Asn-728) residue. Similar to our findings in NBD1, a peptide 684–697 that covers the Walker B motif is unmodified in the hexamer with ATPγS but has a rate of 49 ± 1.3 s−1 in the hexamer with ADP (Table 2). Indeed, Leu-684 and the conserved Walker B Glu-687 are identified by MS2 as being modified in the hexamer with ADP (Table 2). Additionally, the peptide 813–831 that covers the sensor 2 residue (Arg-826) is unmodified in the hexamer with ATPγS and has a rate of 42 ± 0.2 s−1 in the hexamer with ADP (Table 2). These findings are consistent with the transition from open spiral hexamers in the presence of ADP to closed rings in the presence of ATPγS observed by cryo-EM (16) and HX (30). Indeed, the open spiral ring of Hsp104 hexamers with ADP would leave NBD2 from protomer 6 highly exposed to solvent (16).
Just C-terminal of the Walker B motif is a peptide 688–696, which in the hexamer with ATPγS has a modification rate of 57 ± 9.4 s−1, and a peptide 613–623, which covers the Walker A motif and has a rate of 9.5 ± 0.3 s−1 in the hexamer with ATPγS with residues 616 and 618 having been identified by MS2 as modified (Table 2 and Table S3). These data indicate that in contrast to NBD1, ATPγS binding in NBD2 does not fully shield the nucleotide-binding pockets from solvent. This difference may be due to the presence of empty, incompletely closed sites, and/or less stable ATPγS binding, which would result in partial solvation of some of the sites in the Hsp104 hexamer population. Differences between NBD1 and NBD2 might reflect that these NBDs originate from different AAA+ clades (78) and are responsible for divergent functions within the Hsp104 hexamer (57).
NBD2 NLS is generally solvent-exposed
The NLS in Hsp104, residues 773–789 (77), appears to be solvated in the monomer as well as both of the hexameric states, with the hexamer with ATPγS the least modified (Table 2). The NLS also includes the NBD2 hinge 773NKLS776, which connects the large and small subdomains of NBD2. Peptides that cover this regions include 769–785 and 786–806 with rates for the monomer, the hexamer with ADP, and the hexamer with ATPγS of 1.15 ± 0.1, 0.64 ± 0.003, and 0.14 ± 0.02 s−1, respectively, for 769–785 and 0, 17.6 ± 6.3, and 0 s−1, respectively, for 786–806 (Table 2). There are many MS2-identified and modified residues in this region, including lysine, which when all mutated to alanine result in a loss of nuclear Hsp104 (77), as well as residues that are in the NBD2 hinge. Lys-774 may or may not be necessary for nuclear localization; because it is in the NBD2 hinge mutating it to glutamate disrupts Hsp104 stability (79). The MS2-identified residues include (all found on the peptide 769–785): Asn-773, Lys-774, Ser-776, Lys-778, Ala-779, Ile-780, His-781, Lys-782, and Val-784 in the monomer; Phe-772 (just before NLS), Lys-774, His-781, Lys-782, Ile-783, and Val-784 found in the hexamer with ADP; and Phe-772, Lys-774, Ser-776, Arg-777, Ile-780, and Lys-782 found in the hexamer with ATPγS (Table 2). It would appear that Lys-782 is always exposed to solvent, and in the monomeric state the hinge is highly exposed (three of four NBD2 hinge residues were identified as modified, compared with the hexamer with ATPγS (two of four) and the hexamer with ADP (one of four)). The third essential lysine of the NLS, Lys-789 (77) is not on the peptide 769–785 and therefore we have not confirmed its solvation state. Nonetheless, these data suggest that the Hsp104 NLS is generally exposed to solvent and thus can likely be engaged by nuclear–import receptors for nuclear import.
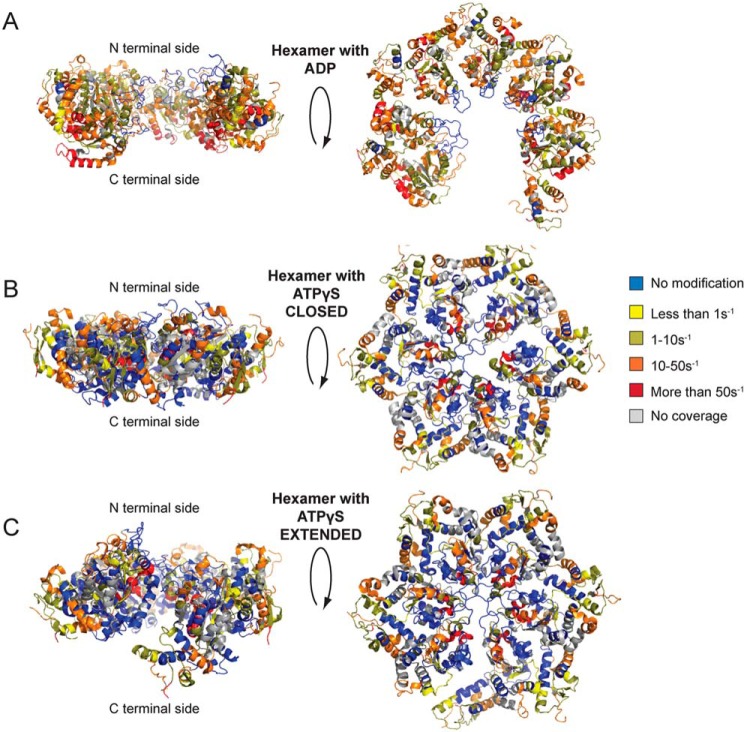
Cryo-EM hexameric models of NBD2 are consistent with the XF solvation data
NBD2 is critical for Hsp104 hexamerization (80). We have used the hexameric NBD2 model from cryo-EM studies (16) and the XF solvation data to identify a key region involved in the protomer/protomer interface (Figs. 7, A and B, and 8). We first used the XF modification rate information to assess the validity of the proposed hexameric NBD2 structures from cryo-EM studies (16). The NBD2 hexameric models have been colored based on the XF modification rates for the hexamer with ADP as well as the hexamer with ATPγS in both the closed and extended conformations (Fig. 8). Both of the states agree well overall with the models (Fig. 8); the hexamer with ADP has more regions accessible to solvent, which is consistent with an open spiral hexamer in ADP and a closed ring in the presence of ATPγS (16).
Figure 7.
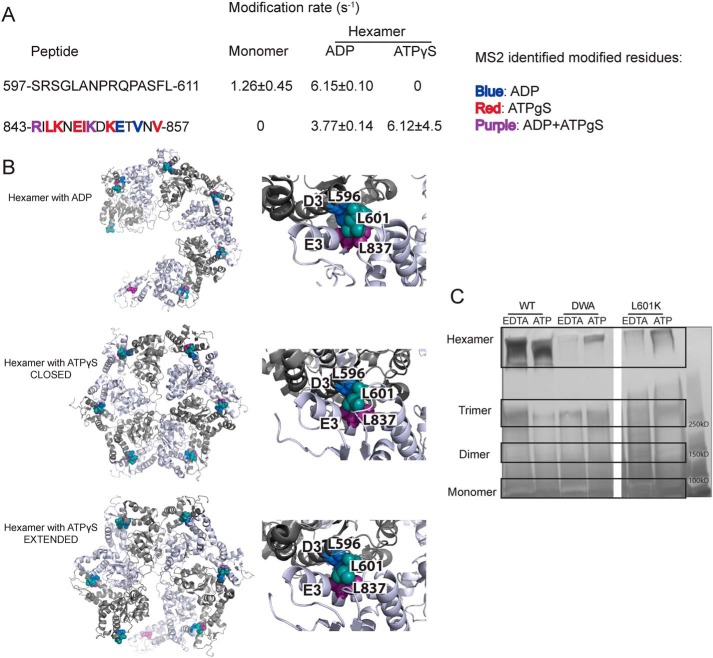
Leu-601 plays an important role in Hsp104 hexamerization. A, peptide sequences and modification rates for regions of interest of Hsp104 NBD2. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: blue, hexamer with ADP; red, hexamer with ATPγS; purple, both hexameric states. B, hexameric models (hexamer with ADP, PDB code 5vy8; hexamer with ATPγS closed, PDB code 5vjh, and extended, PDB code 5vya, conformations) of NBD2 generated from cryo-EM studies (16) showing the location of hydrophobic residues found on helices D3 and E3. On the right-hand side are higher magnification views of the interface between residues 586AIKAVSNAVRLSRSGL601 of the large subdomain of subunit 1 (helix D3) and residues 836ILNKLALRILKNEI849 of the small domain of subunit 2 (helix E3). Residues Leu-601 (large domain D3, teal), Leu-596 (large domain D3, blue), and Leu-837 (small domain E3, purple) are shown as spheres. C, glutaraldehyde cross-linking. Hsp104 shows a robust ability to form hexamers even in the presence of EDTA and absence of ATP. The double Walker A (DWA) mutant displays defects in hexamerization even in the presence of ATP. The mutant L601K, which resides in the proposed hexamer interface but outside of any secondary structure, also displays defects in the ability to form hexamers.
Figure 8.
Hsp104 NBD2 cryo-EM models colored based on XF modification rates. A–C, hexameric NBD2 domains were modeled based on cryo-EM studies (16) and colored based on the XF modification rates with blue as unmodified, yellow less than 1 s−1, dark yellow 1–10 s−1, orange 10–50 s−1, red more than 50 s−1, and gray is no coverage for the hexamer with ADP (A, PDB code 5vy8) and ATPγS in the closed (B, PDB code 5vjh) and extended conformations (C, PDB code 5vya).
Using the cryo-EM models and the XF solvation data, we proposed a crucial hexameric interface. This interface includes helix D3 and part of its C-terminal loop in the large NBD2 subdomain (residues 586AIKAVSNAVRLSRSGL601, with the conserved hydrophobic residues in bold), and the last helix, E3, and part of its C-terminal loop in the neighboring NBD2 small subdomain (residues 836ILNKLALRILKNEI849, with conserved hydrophobic residues in bold) (Fig. 7B) (16). Additionally, HX reveals that there is a large increase in protection in helix D3 upon hexamerization and a smaller increase in helix E3 (30).
There is a large number of conserved, hydrophobic residues in these regions indicating that they may be mediating the subunit/subunit interaction. If the subunit/subunit interaction was driven by hydrophobic interactions, it would explain a number of previously published phenomena. The protomer interface undergoes large changes in conformation during the functional cycle of the hexamer (16–18, 30), and cross-linking the small and large subdomains results in stable hexamers with diminished function (81). If the interface was composed of rigid salt bridges, it may hinder rapid and smooth movements of the domains. If, instead, the region were a network of hydrophobic residues, then the interface could easily change register upon conformational changes of nearby regions. The proposed interface is well-suited to respond to changes in nucleotide. Indeed, the C-terminal end of the small NBD2 helix E3 contains a number of conserved hydrophobic residues that we propose to be involved in the hexamer interface, whereas the N-terminal end contains a region called the sensor and substrate discrimination motif, which contains the sensor 2 arginine (Arg-826) (37, 61). Additionally, helix D3 and loop in the NBD2 large domain that we propose to be involved in the interface is just N-terminal to the short, highly-conserved β-strand d1 that immediately precedes the conserved Walker A motif (37).
A hydrophobic interface would also explain why the Hsp104 hexamer is sensitive to salt (57, 58). As salt concentrations increase, the Hsp104 hexamer becomes destabilized (57, 58). Upon cursory inspection, this finding would make it seem unlikely that the interface was mediated by hydrophobic interactions; however, Hsp104 undergoes continual and rapid monomer exchange (20). Upon mixing Hsp104 hexamers with different (measurable) properties, monomer exchange occurs on a timescale of just a few minutes to yield an ensemble of hexamers containing subunits of different Hsp104 variants explained by the binomial distribution (20). This phenomenon of monomer exchange explains why high-salt conditions would disrupt hexamer integrity. As a monomer is released from the hexamer into high-salt conditions, there will be a propensity to make intramolecular contacts to bury the newly-exposed hydrophobic hexamer interface. We find support for this hypothesis by examining the solvation information from the XF modification data. In the monomeric sample, which was created by adding 500 mm NaCl to the buffer, there are two regions near the proposed hydrophobic interface that undergo changes in solvation. Although there is no coverage of 569–583 (just N-terminal to the D3 helix) in the hexamer with ATPγS, it is highly modified in the hexamer with ADP (Tables S2 and S3), likely due to exposure to solvent upon shifting to the “open” lock-washer conformation (Figs. 7B and 8) (16). Peptide 597–611, which covers the C-terminal end of the proposed D3-loop portion of the interface, has rates of 1.26 ± 0.45, 6.15 ± 0.10, and 0 s−1 for the monomer, hexamer with ADP, and hexamer with ATPγS, respectively (Fig. 7A and Tables S1–S3). Peptide 843–857, near the E3 portion of the proposed interface, is unmodified in the monomer and modified in both of the hexamers, with a number of residues identified as modified by MS2 (3.8 ± 0.14 s−1 with residues Arg-843, Lys-850, Glu-853, and Val-855 for the hexamer with ADP; and 6.1 ± 4.5 s−1 with residues Arg-843, Leu-845, Lys-846, Glu-848, Ile-849, Lys-850, Lys-852, and Val-857 for the hexamer with ATPγS, Table 2 and Fig. 7A). Both of these regions also contain highly-conserved hydrophobic residues (37), supporting the possibility that they may be involved in intramolecular interactions that block hexamerization in high salt. These residues in helix D3 and helix E3 are protected in the closed state but become solvated in the hexamer with ADP and in the extended conformation hexamer with ATPγS (Fig. 7B) (16).
Next, we tested whether we could disrupt the hexameric interactions by mutating a conserved hydrophobic residue in the proposed protomer interface to a charged residue. We chose Leu-601 because it is in a loop region, so it was unlikely to grossly perturb secondary structure, and it is conserved in Hsp100 proteins from plants to bacteria to yeast (37). Using a glutaraldehyde cross-linking assay (58, 80), we assessed the ability of low concentrations of Hsp104, the double Walker A mutant (DWA; bearing K218/:K620T mutations that disrupt hexamerization (58, 80)), and an L601K variant to form hexamers under two conditions, adding either EDTA or ATP. Addition of EDTA chelates the magnesium essential for ATP binding and therefore disrupts the ability of Hsp104 to form hexamers, whereas addition of ATP promotes the formation of hexamers (58). After glutaraldehyde cross-linking, samples were fractionated by SDS-PAGE to assess hexamerization (58, 80). As expected, Hsp104 exists as a mixture of monomer, dimer, trimer, and hexamer in the presence of EDTA, and upon addition of ATP shifts entirely to the hexamer (Fig. 7C) (58, 80). Hsp104–DWA, our negative control, shows very little hexamer in the presence of EDTA, and upon addition of ATP remains predominantly a mixture of monomer, dimer, and trimer (Fig. 7C) (58, 80). The L601K Hsp104 mutant appears to strongly resemble the Hsp104–DWA variant, and even upon addition of ATP, there is very little hexamer but instead mostly smaller oligomers and monomer (Fig. 7C). These data support the placement of Leu-601 near a hydrophobic hexamer interface and strongly suggest that Leu-601 enables hexamerization.
Discussion
Using XF oxidative modification data in combination with XF-guided mutational analysis, we have uncovered regions of Hsp104 crucial for its activity. Our methodology and findings are summarized in Fig. 9, and establish the following. 1) Hsp104 hexamers switch from a more solvated state in ADP to a less solvated state in ATPγS, consistent with switching from an open spiral to a closed ring visualized by cryo-EM and HX (16, 30). 2) Specific regions of the Hsp104 NTD contribute to substrate binding and Hsp70 collaboration. 3) NTD residues (Glu-44 and Asp-45) can be mutated to enhance Hsp104 disaggregase activity. 4) Novel NTD/NBD1/MD interaction sites enable optimal Hsp104 disaggregase activity. 5) The NTD–NBD1 linker enables optimal Hsp104 disaggregase activity. 6) Critical residues in NBD1 (Asp-231, Asp-232, and Asp-233) enable Hsp104 to collaborate with Hsp70. 7) Hsp104–RYD (R496A/Y497A/D498A) is a novel potentiated Hsp104 MD variant that rescues α-synuclein, TDP-43, and FUS toxicity in yeast. 8) An additional pore-loop in NBD1 (291GNGKD295) likely engages substrate during translocation and collaborates with the NTD and the primary pore-loop in NBD1 (256KYKG259) to enable optimal Hsp104 disaggregase activity. 9) An important region of the MD (the hinge region of helix L2) enables optimal Hsp104 disaggregase activity. Substitution of Pro-461 in this hinge region specifically ablates Hsp104 disaggregation activity in the presence of Hsc70 (versus Hsp72). This finding indicates a crucial role in the MD for interactions with specific Hsp70 variants that may mediate the response of Hsp104 to diverse substrate classes. 10) A hydrophobic interaction in NBD2 that involves Leu-601 mediates Hsp104 hexamer formation.
Figure 9.
Summary of methodology and major findings. Hsp104 monomer and hexamer with ADP or ATPγS were subjected to increasing exposure times of synchrotron-generated X-rays. Mass spectrometer analysis of the samples yielded oxidative modification rates for pepsin-derived peptides, and MS2 data identified specific sites of modification. Using the modification rates as a proxy for solvation, Hsp104 variants were designed with substitutions in residues found to be highly solvated or in regions that displayed large changes in solvation across the different states. Activity of these Hsp104 variants was then assessed using in vitro and in vivo assays to determine their role in Hsp104 function.
We have also shown that our XF solvation data fit well with published cryo-EM structures (16, 17), HX data (30), and can explain a number of published Hsp104 variant phenotypes. Indeed, by combining information from static cryo-EM structures, backbone and secondary structure information from HX-MS, and side-chain solvation information from XF-MS, we are able to piece together a comprehensive picture of how the Hsp104 hexamer changes and the regions involved in its auto-regulation and activity. Our findings suggest a model in which the energy of ATP binding to Hsp104 induces the formation of stable secondary structure as well as minimizing large dynamic movements, as seen by decreased solvation by XF. This decrease in conformational dynamics in response to ATP binding can be thought of as a pre-payment of the entropic cost of substrate binding and may have important implications for discrimination between soluble and aggregated substrates. When binding a large, stable aggregate, the main entropic penalty would come from decreases in mobility of the Hsp104 hexamer. Conversely, when binding soluble, potentially properly folded substrates, the main entropic penalty would come from the substrate. This mechanism of limiting the conformational dynamics of the Hsp104 hexamer in preparation for substrate binding could contribute to discriminating between aggregated substrates and soluble proteins that could, but should not, be remodeled.
The large size and dynamic behavior of Hsp104 has made it difficult to determine structural details of the hexamer and to pin down regions involved in its protein-remodeling activities. Here, we have shown that using solvation data from XF experiments to focus our mutational analyses has dramatically decreased the number of variants we need to screen to unveil new regions crucial to Hsp104 function. Our findings will help inform rational design of Hsp104 variants with specific activities. Importantly, out of the 17 Hsp104 variants we designed, built, and tested based on the XF solvation data, only one of them, Hsp104-SS (S124A/S125A), did not display an interesting phenotype. Thus, there is a strong positive correlation between the regions undergoing large changes in solvation and important functional regions of a protein, which may have profound implications for protein engineering.
Experimental procedures
Protein expression and purification
Hsp104 variants were generated using QuikChange lightning mutagenesis (Agilent Technologies). Hsp104 variants were expressed and purified as N-terminally His6-tagged constructs in a modified pPROEX HTb vector as described (82) or as untagged constructs in a pNOTAG vector (61, 73). Unless otherwise stated, Hsp104 concentrations refer to the hexamer.
Luciferase reactivation
Luciferase aggregation and reactivation were performed as described (20). Briefly, firefly luciferase (50 μm) was incubated in LRB (25 mm HEPES-KOH, pH 7.4, 150 mm KOAc, 10 mm MgOAc, 10 mm DTT) with 8 m urea at 30 °C for 30 min to form aggregates. After a rapid 100-fold dilution in LRB, the aggregates were flash-frozen and stored at −80 °C until use. Reactivation assays were carried out with Hsp104 (1 μm), Hsp70 (Hsc70 or Hsp72 at 1 μm), Hsp40 (Hdj2, 1 μm), 5.1 mm ATP, and an ATP-regenerating system (1 mm creatine phosphate, 0.25 μm creatine kinase) for 90 min at 25 °C. Alternatively, Hsp70, Hsp40, and 5.1 mm ATP were replaced with 5.1 mm nucleotide of a 1:1 ratio of ATP/ATPγS. Luciferase activity was assessed using a luciferase assay system from Promega. Luminescence was measured on a Tecan Infinite M1000 or Safire2 plate reader.
In vivo thermotolerance assay
Yeast thermotolerance assays were performed as described (51). Briefly, W303 Δhsp104 (MATa, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3-1, ade2-1, hsp104:kanMX4) yeast was transformed with a centromeric pHSE plasmid encoding an Hsp104 variant (83). The strains were grown in SD-ura media to an OD600 of 0.5 and incubated at 37 °C for 30 min to induce Hsp104 expression. Cells were then heat-shocked for 0–20 min at 50 °C, immediately transferred to ice for 2 min, and then spotted on SD-ura plates in a 5-fold dilution series. After a 2-day incubation at 30 °C, plates were imaged for analysis. Alternatively, the thermotolerance products were plated on SD-ura plates, and after a 2-day incubation at 30 °C colonies were counted using an acolyte automated colony counter (Synbiosis).
α-Synuclein, FUS, and TDP-43 toxicity in yeast
Yeast were WT W303a (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1) or the isogenic strain W303aΔhsp104 (51). Media were supplemented with 2% glucose, raffinose, or galactose as specified. The yeast strains W303aΔhsp104 303GAL-α-synuclein-YFP-304GAL-α-synuclein-YFP, W303aΔhsp104 303GAL-FUS, and W303aΔhsp104 303GAL-TDP-43 have been previously described (51, 54, 85). QuikChange site-directed mutagenesis (Agilent) was used to create mutations in the pRS416GAL-Hsp104 plasmid, and all mutations were confirmed by DNA sequencing. Yeast transformations were performed using standard PEG and lithium acetate procedures (86). For the spotting assays, yeast were grown to saturation in raffinose-supplemented dropout media overnight at 30 °C. The saturated overnight cultures were serially diluted 5-fold, and a 96-bolt replicator tool (frogger) was used to spot the strains in duplicate onto both glucose and galactose dropout plates. These plates were grown at 30 °C and imaged after 72 h to assess suppression of disease–protein toxicity. In some experiments, Hsp104 variants were transformed into W303aΔhsp104 without any disease protein and growth was assessed at 30°C on glucose or galactose dropout plates.
Immunoblotting and quantification
Yeast were grown in raffinose media to saturation, diluted to an OD600 of 0.6 (A600 nm = 0.6), and induced for 5 h (FUS and TDP-43 samples) or 8 h (α-synuclein samples). Samples were normalized to an OD600 of 0.6 (A600 nm = 0.6) post-induction, and 6 ml of cells were pelleted and resuspended with 0.1 m NaOH and then with 1× SDS sample buffer. Boiled lysates were separated by SDS-PAGE (4–20% gradient, Bio-Rad) and transferred to a polyvinylidene difluoride membrane. Primary antibodies used were anti-Hsp104 polyclonal (Enzo Life Sciences), anti-PGK monoclonal (Invitrogen), anti-GFP polyclonal (Sigma), anti-FUS polyclonal (Bethyl Laboratories), and anti-TDP-43 polyclonal (Proteintech). Secondary antibodies used were anti-rabbit (Li-Cor) and anti-mouse (Li-Cor). LI-COR Odyssey FC Imaging system was used to image blots. Western blottings were quantified using ImageJ software. Hsp104 levels were normalized to PGK (loading control) and Hsp104 WT. α-Synuclein-YFP, FUS, and TDP-43 levels were normalized to PGK (loading control) and strains not expressing Hsp104 (empty vector).
ATPase assay
Hsp104 variants (0.25 μm monomer) were incubated for 5 min at 25 °C with ATP (1 mm) in LRB (25 mm HEPES-KOH, pH 7.4, 150 mm KOAc, 10 mm MgOAc, 10 mm DTT). ATPase activity was assessed using a malachite green phosphate detection kit (Innova).
Fluorescence polarization
Fluorescence polarization experiments with Hsp104 variants and FITC-casein were performed as described (51). Briefly, Hsp104 variants in increasing concentrations (10 nm to 4 μm) were added to FITC-casein (6 nm) and ATPγS (2 mm) in LRB. After a 20-min incubation, fluorescence polarization of the FITC-casein was measured using a Tecan Infinite M1000 plate reader.
Glutaraldehyde cross-linking
Glutaraldehyde cross-linking experiments were carried out as described (58). Briefly, Hsp104 variants were diluted to 0.04 mg/ml in 40 mm HEPES-KOH, pH 7.4, 140 mm KCl, 10 mm MgCl2, 1 mm DTT plus either 20 mm EDTA or 5 mm ATP. The samples were incubated with glutaraldehyde (0.1%) for 12 min. The cross-linking reaction was quenched by addition of 1 m glycine, pH 6, and the proteins were precipitated using TCA. After two washes with chilled acetone, the samples were run on an SDS-polyacrylamide gel, and reaction products were visualized using a silver stain kit (Invitrogen).
Synchrotron hydroxyl radical footprinting (XF)
WT Hsp104 was purified as in Ref. 82, with a final step of gel filtration using a Superdex 200 column (GE Healthcare) equilibrated in 50 mm cacodylate, pH 7.0, 140 mm KCl, 10 mm MgCl2 (for the hexamer) or 50 mm cacodylate, pH 7.0, 500 mm KCl, and 20 mm MgCl2 (for the monomer, as confirmed by size-exclusion chromatography followed by multiangle light scattering (Wyatt)). Samples were kept on ice and transported to Brookhaven National Laboratory (Upton, NY). All samples were kept on ice and diluted at the beamline 10 min prior to use to 10 μm Hsp104 in 50 mm cacodylate, pH 7.0, 140 mm KCl, 10 mm MgCl2, and 1 mm ADP or ATPγS (for the hexamer) or 50 mm cacodylate, pH 7.0, 500 mm KCl, and 20 mm MgCl2 (for the monomer). X-ray dose-dependent oxidation of Alexa488 (Invitrogen) in the three buffer conditions was used to determine optimal exposure times and provide normalization constants. The presence of nucleotide in our buffer affects the effective hydroxyl radical population (i.e. nucleotide quenches the oxidation of the sample). Therefore, all of the rates have been normalized to the monomer sample based on the Alexa488 decay data (87, 88) with normalization factors of 2.35 and 2.04 for the hexamers with ADP and ATPγS, respectively. The samples were exposed to a mirror-focused synchrotron X-ray beam (5.5 mrad angle, focus value of 6 mm) at the X28C beamline of the National Synchrotron Light Source at Brookhaven National Laboratory for 0–20 ms. The exposure time of the samples was controlled by flow rate through the flow cell of a KinTek (Austin, TX) stopped-flow apparatus (87). Oxidation was immediately quenched by the addition of 10 mm methioninamide, and samples were frozen with dry ice, transported to the University of Pennsylvania, and stored at −80 °C.
Mass spectrometry on the XF samples
The irradiated samples were thawed on ice and diluted to 1 μm in 5 mm HEPES-NaOH, pH 7.0, 140 mm KCl, 10 mm MgCl2, 1 mm DTT, 1% TFA and immediately injected onto an in-line fragmentation–separation/MS analysis system. Samples were first passed through an immobilized pepsin column onto a C8 trap column and washed for 3 min (2% acetonitrile, 1% formic acid, pH 2, 0 °C). Samples were then flowed onto an analytical C18 column, eluted using a gradient optimized for the highly-charged Hsp104 peptides (6 μl/min linear 2–30% acetonitrile gradient over 20 min, followed by a linear 30–50% acetonitrile gradient over 10 min, 0.1% formic acid, pH 2, and 0 °C), and injected by electron spray ionization into an linear quadrupole ion trap (LTQ) Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The four most abundant peptides from each scan were selected for fragmentation by collision-induced dissociation and measured in the LTQ stage.
Identification of oxidatively modified peptides
A modified version of the ExMS (89) program, called ExMS-CL (which stands for “covalent labeling”), was used to process and analyze the X-ray footprinting MS/MS data to identify oxidative modifications on the protein samples. ExMS-CL makes a peptide list of the target protein, including unmodified and modified peptides. The unmodified peptides are obtained from searching MS/MS runs (similar to SEQUEST), with a user-set score threshold (in this case a p score cut-off of 0.01) to build a “peptide pool.” From here, ExMS-CL identified modified and unmodified peptides, and their intensities were assigned and calculated as described in Ref. 90. Importantly, the user-set shift in the RT search window for modified peptides (−3 min to +5 min) was determined by manually searching the datasets for modified peptides using SEQUEST. The ExMS-CL output contains the peptide sequence, all the modifications found with their individual position on the peptide, the peptide intensity quantification, and the number of modification possibilities.
Analysis of modification rate of peptides using Matlab
Output of ExMS-CL was read into Matlab (Mathworks) and analyzed using custom scripts. Modified and unmodified versions of peptides and their intensities were read from the ExMS-CL output files and matched up. Fraction unmodified was calculated for each MS/MS sample using the peptide intensities (the sum of unmodified versions of the peptide divided by the sum of all modified and unmodified versions of the peptide). For each time point, the MS/MS run outputs were averaged together, and standard deviation was calculated. For each peptide, the average fraction unmodified was reported for each time point. Once the time-dependent modification was calculated, each peptide was binned into one of three categories by the Matlab program: no time-dependent modification (0–1 time points after time 0 were modified with respect to time 0); sporadic modification (2 time points after time 0 were modified with respect to time 0); and time-dependent modification (greater than 2 time points after time 0 were modified with respect to time 0). For the peptides that displayed time-dependent modification, the program used the first four time points (0, 2.5, 5, and 7.5 ms) to determine the rate of modification by fitting to a first-order exponential decay. For those peptides that did display time-dependent modification but were classified as sporadically modified by the Matlab program, Origin 8.1 was used to manually fit the curve. In a few cases, there does appear to be time-dependent modification; however, a good fit to the data was impossible. In those cases, the rate of modification is listed as not determined.
Because of the presence of sporadic modifications on certain peptides in the unexposed samples, we took care to remove peptides that displayed modification in the absence of X-ray exposure. Additionally, we limited our analysis to singly-modified peptides. This eliminated the possibility that a preexisting modification (independent of the X-ray footprinting experiment) was causing local conformational changes or unfolding events that would affect the analysis. Deviations from a linear response curve that could be extrapolated to zero indicated a buildup of multiple modified species that may or may not be solvation-dependent in the later time points of some peptides. Additionally, the amount of singly-modified peptides began to decrease in the 20-ms time point, again indicating a shift to multiple modifications. Therefore, we limited our analysis to the 0–7.5-ms time points, which did not display these deviations.
Comparisons of the modification rates of peptides in different states (monomer, hexamer with ADP, and hexamer with ATPγS) were done by the following: 1) comparing the rates of identical peptides, and 2) comparing the rates of overlapping peptides. Because oxidative modification can theoretically alter the cleavage preference of pepsin, it was important to take into account the modification rate of all overlapping peptides and to use these values as a relative guide of solvation rather than an absolute value.
Homology modeling and cryo-EM model assessment
Hsp104 was homology-modeled domain by domain using SWISS-MODEL (84, 91, 92) based on the S. cerevisiae Hsp104 NTD (PDB code 5u2u) (38) and ClpB crystal structure PDB code 1qvr (NBD1, MD, and NBD2) (37). To assess large-scale changes in solvation between the hexameric states, the cryo-EM models, PDB codes 5vy8, 5vjh, and 5vya (16), were colored based on the peptide with the highest rate of modification for each residue.
Author contributions
E. A. S. and J. S. conceptualization; E. A. S., S. W. E., and J. S. resources; E. A. S. data curation; E. A. S., M. A. S., and Z.-Y. K. software; E. A. S. formal analysis; E. A. S., Z.-Y. K., L. M., S. W. E., and J. S. supervision; E. A. S. and J. S. funding acquisition; E. A. S. validation; E. A. S., A. T., E. G., M. S. G., and Z.-Y. K. investigation; E. A. S., A. T., and J. S. visualization; E. A. S., Z.-Y. K., L. M., S. W. E., and J. S. methodology; E. A. S. and J. S. writing-original draft; E. A. S. and J. S. writing-review and editing.
Supplementary Material
Acknowledgments
We thank JiaBei Lin, Xiang Ye, Korrie Mack, Zachary March, Edward Chuang, and Ryan Cupo for comments on the manuscript. We also thank Kushol Gupta for assistance in performing the XF experiments. This research used beamline X28C of the National Synchrotron Light Source, a United States Department of Energy (DOE) Office of Science User Facility operated by Brookhaven National Laboratory under Contract No. DE-AC02-98CH10886.
This work was supported by an American Heart Association predoctoral fellowship (to E. A. S.), National Institutes of Health Grants T32GM008275 (to E. A. S. and M. A. S.) and R01GM099836 and DP2OD002177 (to J. S.), a Muscular Dystrophy Association Research Award (to J. S.), an ALS Association Award (to J. S.), the Life Extension Foundation (to J. S.), a Linda Montague Pechenik Research Award (to J. S.), the Packard Center for ALS Research at The Johns Hopkins University (to J. S.), and Target ALS (to J. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S3.
- NTD
- N-terminal domain
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- MD
- middle domain
- XF
- X-ray footprinting
- PDB
- Protein Data Bank
- ANOVA
- analysis of variance
- FUS
- fused in sarcoma
- YFP
- yellow fluorescent protein
- PGK
- phosphoglycerate kinase
- FTD
- frontotemporal dementia
- ALS
- amyotrophic lateral sclerosis
- FITC
- fluorescein isothiocyanate
- HX
- hydrogen–deuterium exchange
- DWA
- double Walker A mutant
- NLS
- nuclear localization signal.
References
- 1. Shorter J. (2008) Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals 16, 63–74 10.1159/000109760 [DOI] [PubMed] [Google Scholar]
- 2. Jackrel M. E., and Shorter J. (2017) Protein-remodeling factors as potential therapeutics for neurodegenerative disease. Front. Neurosci. 11, 99 10.3389/fncom.2017.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mack K. L., and Shorter J. (2016) Engineering and evolution of molecular chaperones and protein disaggregases with enhanced activity. Front. Mol. Biosci. 3, 8 10.3389/fmolb.2016.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shorter J. (2016) Engineering therapeutic protein disaggregases. Mol. Biol. Cell 27, 1556–1560 10.1091/mbc.E15-10-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shorter J. (2017) Designer protein disaggregases to counter neurodegenerative disease. Curr. Opin. Genet. Dev. 44, 1–8 10.1016/j.gde.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. March Z. M., Mack K. L., and Shorter J. (2019) AAA+ protein-based technologies to counter neurodegenerative disease. Biophys. J. 116, 1380–1385 10.1016/j.bpj.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo L., Fare C. M., and Shorter J. (2019) Therapeutic dissolution of aberrant phases by nuclear-import receptors. Trends Cell Biol. 29, 308–322 10.1016/j.tcb.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo L., Kim H. J., Wang H., Monaghan J., Freyermuth F., Sung J. C., O'Donovan K., Fare C. M., Diaz Z., Singh N., Zhang Z. C., Coughlin M., Sweeny E. A., DeSantis M. E., Jackrel M. E., et al. (2018) Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 10.1016/j.cell.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shorter J., and Southworth D. R. (2019) Spiraling in control: structures and mechanisms of the Hsp104 disaggregase. Cold Spring Harb. Perspect. Biol. 11, a034033 10.1101/cshperspect.a034033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sweeny E. A., and Shorter J. (2016) Mechanistic and structural insights into the prion-disaggregase activity of Hsp104. J. Mol. Biol. 428, 1870–1885 10.1016/j.jmb.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wallace E. W., Kear-Scott J. L., Pilipenko E. V., Schwartz M. H., Laskowski P. R., Rojek A. E., Katanski C. D., Riback J. A., Dion M. F., Franks A. M., Airoldi E. M., Pan T., Budnik B. A., and Drummond D. A. (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S. (2015) Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kroschwald S., Munder M. C., Maharana S., Franzmann T. M., Richter D., Ruer M., Hyman A. A., and Alberti S. (2018) Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Rep. 23, 3327–3339 10.1016/j.celrep.2018.05.041 [DOI] [PubMed] [Google Scholar]
- 14. Newby G. A., and Lindquist S. (2013) Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol. 23, 251–259 10.1016/j.tcb.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 15. DeSantis M. E., and Shorter J. (2012) The elusive middle domain of Hsp104 and ClpB: location and function. Biochim. Biophys. Acta 1823, 29–39 10.1016/j.bbamcr.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gates S. N., Yokom A. L., Lin J., Jackrel M. E., Rizo A. N., Kendsersky N. M., Buell C. E., Sweeny E. A., Mack K. L., Chuang E., Torrente M. P., Su M., Shorter J., and Southworth D. R. (2017) Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 10.1126/science.aan1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yokom A. L., Gates S. N., Jackrel M. E., Mack K. L., Su M., Shorter J., and Southworth D. R. (2016) Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat. Struct. Mol. Biol. 23, 830–837 10.1038/nsmb.3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Michalska K., Zhang K., March Z. M., Hatzos-Skintges C., Pintilie G., Bigelow L., Castellano L. M., Miles L. J., Jackrel M. E., Chuang E., Jedrzejczak R., Shorter J., Chiu W., and Joachimiak A. (2019) Structure of Calcarisporiella thermophila Hsp104 disaggregase that antagonizes diverse proteotoxic misfolding events. Structure 27, 449–463.e7 10.1016/j.str.2018.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sweeny E. A., Jackrel M. E., Go M. S., Sochor M. A., Razzo B. M., DeSantis M. E., Gupta K., and Shorter J. (2015) The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57, 836–849 10.1016/j.molcel.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeSantis M. E., Leung E. H., Sweeny E. A., Jackrel M. E., Cushman-Nick M., Neuhaus-Follini A., Vashist S., Sochor M. A., Knight M. N., and Shorter J. (2012) Operational plasticity enables Hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell 151, 778–793 10.1016/j.cell.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsell D. A., Kowal A. S., Singer M. A., and Lindquist S. (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 10.1038/372475a0 [DOI] [PubMed] [Google Scholar]
- 22. Glover J. R., and Lindquist S. (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 10.1016/S0092-8674(00)81223-4 [DOI] [PubMed] [Google Scholar]
- 23. Shorter J., and Lindquist S. (2004) Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793–1797 10.1126/science.1098007 [DOI] [PubMed] [Google Scholar]
- 24. Shorter J., and Lindquist S. (2006) Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell 23, 425–438 10.1016/j.molcel.2006.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doyle S. M., Shorter J., Zolkiewski M., Hoskins J. R., Lindquist S., and Wickner S. (2007) Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 14, 114–122 10.1038/nsmb1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shorter J., and Lindquist S. (2008) Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 27, 2712–2724 10.1038/emboj.2008.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shorter J. (2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6, e26319 10.1371/journal.pone.0026319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Durie C. L., Lin J., Scull N. W., Mack K. L., Jackrel M. E., Sweeny E. A., Castellano L. M., Shorter J., and Lucius A. L. (2019) Hsp104 and potentiated variants can operate as distinct nonprocessive translocases. Biophys. J. 116, 1856–1872 10.1016/j.bpj.2019.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weaver C. L., Duran E. C., Mack K. L., Lin J., Jackrel M. E., Sweeny E. A., Shorter J., and Lucius A. L. (2017) Avidity for polypeptide binding by nucleotide-bound Hsp104 structures. Biochemistry 56, 2071–2075 10.1021/acs.biochem.7b00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye X., Lin J., Mayne L., Shorter J., and Englander S. W. (2019) Hydrogen exchange reveals Hsp104 architecture, structural dynamics, and energetics in physiological solution. Proc. Natl. Acad. Sci. U.S.A. 116, 7333–7342 10.1073/pnas.1816184116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takamoto K., and Chance M. R. (2006) Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 10.1146/annurev.biophys.35.040405.102050 [DOI] [PubMed] [Google Scholar]
- 32. Kiselar J. G., Maleknia S. D., Sullivan M., Downard K. M., and Chance M. R. (2002) Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int. J. Radiat. Biol. 78, 101–114 10.1080/09553000110094805 [DOI] [PubMed] [Google Scholar]
- 33. Maleknia S. D., Kiselar J. G., and Downard K. M. (2002) Hydroxyl radical probe of the surface of lysozyme by synchrotron radiolysis and mass spectrometry. Rapid Commun. Mass Spectrom. 16, 53–61 10.1002/rcm.543 [DOI] [PubMed] [Google Scholar]
- 34. DeSantis M. E., Sweeny E. A., Snead D., Leung E. H., Go M. S., Gupta K., Wendler P., and Shorter J. (2014) Conserved distal loop residues in the Hsp104 and ClpB middle domain contact nucleotide-binding domain 2 and enable Hsp70-dependent protein disaggregation. J. Biol. Chem. 289, 848–867 10.1074/jbc.M113.520759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sclavi B., Sullivan M., Chance M. R., Brenowitz M., and Woodson S. A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279, 1940–1943 10.1126/science.279.5358.1940 [DOI] [PubMed] [Google Scholar]
- 36. Bohon J. (2019) Development of synchrotron footprinting at NSLS and NSLS-II. Protein Pept. Lett. 26, 55–60 10.2174/0929866526666181128125125 [DOI] [PubMed] [Google Scholar]
- 37. Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., and Tsai F. T. (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 10.1016/S0092-8674(03)00807-9 [DOI] [PubMed] [Google Scholar]
- 38. Wang P., Li J., Weaver C., Lucius A., and Sha B. (2017) Crystal structures of Hsp104 N-terminal domains from Saccharomyces cerevisiae and Candida albicans suggest the mechanism for the function of Hsp104 in dissolving prions. Acta Crystallogr. D Struct. Biol. 73, 365–372 10.1107/S2059798317002662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia D., Esser L., Singh S. K., Guo F., and Maurizi M. R. (2004) Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J. Struct. Biol. 146, 166–179 10.1016/j.jsb.2003.11.025 [DOI] [PubMed] [Google Scholar]
- 40. Li J., and Sha B. (2003) Crystal structure of the E. coli Hsp100 ClpB N-terminal domain. Structure 11, 323–328 10.1016/S0969-2126(03)00030-3 [DOI] [PubMed] [Google Scholar]
- 41. Wang F., Mei Z., Qi Y., Yan C., Hu Q., Wang J., and Shi Y. (2011) Structure and mechanism of the hexameric MecA–ClpC molecular machine. Nature 471, 331–335 10.1038/nature09780 [DOI] [PubMed] [Google Scholar]
- 42. Dougan D. A., Reid B. G., Horwich A. L., and Bukau B. (2002) ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9, 673–683 10.1016/Ser-1097-2765(02)00485-9 [DOI] [PubMed] [Google Scholar]
- 43. Miot M., Reidy M., Doyle S. M., Hoskins J. R., Johnston D. M., Genest O., Vitery M. C., Masison D. C., and Wickner S. (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. U.S.A. 108, 6915–6920 10.1073/pnas.1102828108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sielaff B., and Tsai F. T. (2010) The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 402, 30–37 10.1016/j.jmb.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haslberger T., Weibezahn J., Zahn R., Lee S., Tsai F. T., Bukau B., and Mogk A. (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol. Cell 25, 247–260 10.1016/j.molcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 46. Seyffer F., Kummer E., Oguchi Y., Winkler J., Kumar M., Zahn R., Sourjik V., Bukau B., and Mogk A. (2012) Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 19, 1347–1355 10.1038/nsmb.2442 [DOI] [PubMed] [Google Scholar]
- 47. Lee J., Kim J. H., Biter A. B., Sielaff B., Lee S., and Tsai F. T. (2013) Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. U.S.A. 110, 8513–8518 10.1073/pnas.1217988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee S., Sielaff B., Lee J., and Tsai F. T. (2010) CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 8135–8140 10.1073/pnas.1003572107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torrente M. P., Chuang E., Noll M. M., Jackrel M. E., Go M. S., and Shorter J. (2016) Mechanistic insights into Hsp104 potentiation. J. Biol. Chem. 291, 5101–5115 10.1074/jbc.M115.707976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hung G. C., and Masison D. C. (2006) N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173, 611–620 10.1534/genetics.106.056820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jackrel M. E., DeSantis M. E., Martinez B. A., Castellano L. M., Stewart R. M., Caldwell K. A., Caldwell G. A., and Shorter J. (2014) Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156, 170–182 10.1016/j.cell.2013.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tariq A., Lin J., Noll M. M., Torrente M. P., Mack K. L., Murillo O. H., Jackrel M. E., and Shorter J. (2018) Potentiating Hsp104 activity via phosphomimetic mutations in the middle domain. FEMS Yeast Res. 18, 10.1093/femsyr/foy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tariq A., Lin J., Jackrel M. E., Hesketh C. D., Carman P. J., Mack K. L., Weitzman R., Gambogi C., Hernandez Murillo O. A., Sweeny E. A., Gurpinar E., Yokom A. L., Gates S. N., Yee K., Sudesh S., et al. (2019) Mining protein-disaggregase sequence space to safely counter TDP-43, FUS, and α-synuclein proteotoxicty. Cell Rep. 28, 2080–2095.e6 10.1016/j.celrep.2019.07.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jackrel M. E., and Shorter J. (2014) Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis. Model. Mech. 7, 1175–1184 10.1242/dmm.016113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jackrel M. E., and Shorter J. (2015) Engineering enhanced protein disaggregases for neurodegenerative disease. Prion 9, 90–109 10.1080/19336896.2015.1020277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jackrel M. E., Yee K., Tariq A., Chen A. I., and Shorter J. (2015) Disparate mutations confer therapeutic gain of Hsp104 function. ACS Chem. Biol. 10, 2672–2679 10.1021/acschembio.5b00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hattendorf D. A., and Lindquist S. L. (2002) Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21, 12–21 10.1093/emboj/21.1.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Parsell D. A., Kowal A. S., and Lindquist S. (1994) Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J. Biol. Chem. 269, 4480–4487 [PubMed] [Google Scholar]
- 59. Parsell D. A., Sanchez Y., Stitzel J. D., and Lindquist S. (1991) Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353, 270–273 10.1038/353270a0 [DOI] [PubMed] [Google Scholar]
- 60. Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., and Liebman S. W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 10.1126/science.7754373 [DOI] [PubMed] [Google Scholar]
- 61. Hattendorf D. A., and Lindquist S. L. (2002) Analysis of the AAA sensor-2 motif in the C-terminal ATPase domain of Hsp104 with a site-specific fluorescent probe of nucleotide binding. Proc. Natl. Acad. Sci. U.S.A. 99, 2732–2737 10.1073/pnas.261693199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lum R., Tkach J. M., Vierling E., and Glover J. R. (2004) Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279, 29139–29146 10.1074/jbc.M403777200 [DOI] [PubMed] [Google Scholar]
- 63. Lum R., Niggemann M., and Glover J. R. (2008) Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J. Biol. Chem. 283, 30139–30150 10.1074/jbc.M804849200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bösl B., Grimminger V., and Walter S. (2005) Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 280, 38170–38176 10.1074/jbc.M506149200 [DOI] [PubMed] [Google Scholar]
- 65. Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., and Horwich A. L. (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–1041 10.1016/j.cell.2005.04.012 [DOI] [PubMed] [Google Scholar]
- 66. Watanabe Y. H., Nakazaki Y., Suno R., and Yoshida M. (2009) Stability of the two wings of the coiled-coil domain of ClpB chaperone is critical for its disaggregation activity. Biochem. J. 421, 71–77 10.1042/BJ20082238 [DOI] [PubMed] [Google Scholar]
- 67. Watanabe Y. H., Takano M., and Yoshida M. (2005) ATP binding to nucleotide binding domain (NBD)1 of the ClpB chaperone induces motion of the long coiled-coil, stabilizes the hexamer, and activates NBD2. J. Biol. Chem. 280, 24562–24567 10.1074/jbc.M414623200 [DOI] [PubMed] [Google Scholar]
- 68. Schirmer E. C., Homann O. R., Kowal A. S., and Lindquist S. (2004) Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol. Biol. Cell 15, 2061–2072 10.1091/mbc.e02-08-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wendler P., Shorter J., Plisson C., Cashikar A. G., Lindquist S., and Saibil H. R. (2007) Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131, 1366–1377 10.1016/j.cell.2007.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mogk A., Schlieker C., Strub C., Rist W., Weibezahn J., and Bukau B. (2003) Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278, 17615–17624 10.1074/jbc.M209686200 [DOI] [PubMed] [Google Scholar]
- 71. Dulle J. E., Stein K. C., and True H. L. (2014) Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS ONE 9, e87521 10.1371/journal.pone.0087521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oguchi Y., Kummer E., Seyffer F., Berynskyy M., Anstett B., Zahn R., Wade R. C., Mogk A., and Bukau B. (2012) A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 19, 1338–1346 10.1038/nsmb.2441 [DOI] [PubMed] [Google Scholar]
- 73. Cashikar A. G., Schirmer E. C., Hattendorf D. A., Glover J. R., Ramakrishnan M. S., Ware D. M., and Lindquist S. L. (2002) Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760 10.1016/Ser-1097-2765(02)00499-9 [DOI] [PubMed] [Google Scholar]
- 74. Lipińska N., Zietkięwicz S., Sobczak A., Jurczyk A., Potocki W., Morawiec E., Wawrzycka A., Gumowski K., Ślusarz M., Rodziewicz-Motowidl̸o S., Chruściel E., and Liberek K. (2013) Disruption of ionic interactions between the nucleotide binding domain 1 (NBD1) and middle (M) domain in Hsp100 disaggregase unleashes toxic hyperactivity and partial independence from Hsp70. J. Biol. Chem. 288, 2857–2869 10.1074/jbc.M112.387589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kurahashi H., and Nakamura Y. (2007) Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol. Microbiol. 63, 1669–1683 10.1111/j.1365-2958.2007.05629.x [DOI] [PubMed] [Google Scholar]
- 76. Takahashi A., Hara H., Kurahashi H., and Nakamura Y. (2007) A systematic evaluation of the function of the protein-remodeling factor Hsp104 in [PSI+] prion propagation in S. cerevisiae by comprehensive chromosomal mutations. Prion 1, 69–77 10.4161/pri.1.1.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tkach J. M., and Glover J. R. (2008) Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic 9, 39–56 10.1111/j.1600-0854.2007.00666.x [DOI] [PubMed] [Google Scholar]
- 78. Erzberger J. P., and Berger J. M. (2006) Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 10.1146/annurev.biophys.35.040405.101933 [DOI] [PubMed] [Google Scholar]
- 79. Tkach J. M., and Glover J. R. (2004) Amino acid substitutions in the C-terminal AAA+ module of Hsp104 prevent substrate recognition by disrupting oligomerization and cause high temperature inactivation. J. Biol. Chem. 279, 35692–35701 10.1074/jbc.M400782200 [DOI] [PubMed] [Google Scholar]
- 80. Schirmer E. C., Queitsch C., Kowal A. S., Parsell D. A., and Lindquist S. (1998) The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 273, 15546–15552 10.1074/jbc.273.25.15546 [DOI] [PubMed] [Google Scholar]
- 81. Biter A. B., Lee S., Sung N., and Tsai F. T. (2012) Structural basis for intersubunit signaling in a protein disaggregating machine. Proc. Natl. Acad. Sci. U.S.A. 109, 12515–12520 10.1073/pnas.1207040109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sweeny E. A., DeSantis M. E., and Shorter J. (2011) Purification of hsp104, a protein disaggregase. J. Vis. Exp. 3190 10.3791/3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sanchez Y., and Lindquist S. L. (1990) HSP104 required for induced thermotolerance. Science 248, 1112–1115 10.1126/science.2188365 [DOI] [PubMed] [Google Scholar]
- 84. Peitsch M. C. (1995) Protein modeling by E-mail. Nat. Biotech. 13, 658–660 [Google Scholar]
- 85. Jackrel M. E., Tariq A., Yee K., Weitzman R., and Shorter J. (2014) Isolating potentiated Hsp104 variants using yeast proteinopathy models. J. Vis. Exp. e52089 10.3791/52089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
- 87. Gupta S., Sullivan M., Toomey J., Kiselar J., and Chance M. R. (2007) The beamline X28C of the center for synchrotron biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J. Synchrotron Radiat. 14, 233–243 10.1107/S0909049507013118 [DOI] [PubMed] [Google Scholar]
- 88. Bohon J., Jennings L. D., Phillips C. M., Licht S., and Chance M. R. (2008) Synchrotron protein footprinting supports substrate translocation by ClpA via ATP-induced movements of the D2 loop. Structure 16, 1157–1165 10.1016/j.str.2008.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kan Z. Y., Mayne L., Chetty P. S., and Englander S. W. (2011) ExMS: data analysis for HX-MS experiments. J. Am. Soc. Mass Spectrom. 22, 1906–1915 10.1007/s13361-011-0236-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kaur P., Kiselar J. G., and Chance M. R. (2009) Integrated algorithms for high-throughput examination of covalently labeled biomolecules by structural mass spectrometry. Anal. Chem. 81, 8141–8149 10.1021/ac9013644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 92. Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37, D387–D392 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.