Abstract
Ineffective cellular delivery is a common problem in numerous biological applications. Developing delivery reagents that work robustly in a variety of experimental settings remains a challenge. Herein, we report how peptides derived from the prototypical cell penetrating peptide TAT can be used in combination with a small molecule, UNC7938, to deliver macromolecules into the cytosol of cells by a simple co-incubation protocol. We establish successful delivery of peptides, DNA plasmids, and a single-chain variable fragment antibody. We also demonstrate that delivery works in hard-to-transfect mammalian cells and under conditions typically inhibitory to cell-penetrating peptides. Mechanistically, UNC7938 destabilizes the membrane of endosomes. This, in turn, enhances the endosome-leakage activity of cell-penetrating peptides and facilitates the endosomal escape of macromolecules initially internalized by mammalian cells via endocytosis. This combined selective membrane-destabilization represents a new chemical space for delivery tools and provides a novel solution to the problem of endosomal entrapment that often limits the effectiveness of reagent-based delivery approaches.
Keywords: endosomal escape, endocytosis, delivery, plasma membrane translocation
Introduction.
The delivery of macromolecules into live human cells is highly desirable in several biological, biotechnological and therapeutic applications(1). These include transfection experiments for microscopy, cellular reprogramming strategies, or gene editing. To this end, strategies that use mechanical, electrical or chemical means to cross biological membranes have been developed(2). Strategies that use delivery agents are particularly appealing because they can potentially be applied both in cell cultures and in vivo. These agents typically utilize the process of endocytosis to gain initial access to the interior of cells(3). This, in principle, can circumvent the toxicity that would result from creating pores large enough to bring macromolecular cargos directly through the plasma membrane. It also enables cell-specific delivery via targeting of endocytic receptors (4, 5). However, a secondary step of endosomal escape is usually required for macromolecules to reach their intracellular targets, whether it be in the cytosolic space, nucleus, or other organelles(6, 7). For many delivery agents, including liposomes, polymer-based particles, or cell-penetrating peptides (CPPs), endosomal egress is inefficient (8–11). As a result, the majority of material administered to cells is entrapped within endosomes, with only small amounts of macromolecular cargos entering the cytosol and reaching their intended destination. Admittedly, a low cytosolic delivery efficiency may be sufficient to elicit an intracellular response when using certain cargos. However, this problem remains limiting for macromolecular cargos that require high intracellular concentrations for activity. The low endosomal escape activity of delivery agents contributes to the heterogeneous results often observed within a cell culture, as highlighted by a population of cells showing some cytosolic delivery and another population displaying none (11, 12). Overall, making the step of endosomal escape more efficient should therefore improve the outcome of delivery experiments.
TAT is a prototypical delivery agent that suffers from poor endosomal escape activity (11, 13, 14). This CPP has been used for the successful delivery of several cargos and positive results have prompted several clinical trials(15, 16). However, TAT is insufficiently active for many applications and several strategies have been used to improve its endosomal escape(15). For instance, reagents that display multiple copies of TAT leak from endosomes more than their monomeric counterpart(17–20). In particular, dimeric or trimeric analogs, including the disulfide linked dimer dfTAT (Figure 1A), are capable of mediating the release of endocytosed material from endosomes. Specifically, TAT reportedly releases less than 1% of macromolecules trapped in endosomes, while dfTAT can release more than 90%(19). Consequently, the level of macromolecules delivered into the cytosol of cells is greatly increased and their intracellular biological activities are improved. Notably, the increase in cytosolic delivery achieved with dfTAT does not yield an increase in toxicity, with cells being relatively undisturbed in their proliferation rates and gene expression profiles(19).
Figure 1.
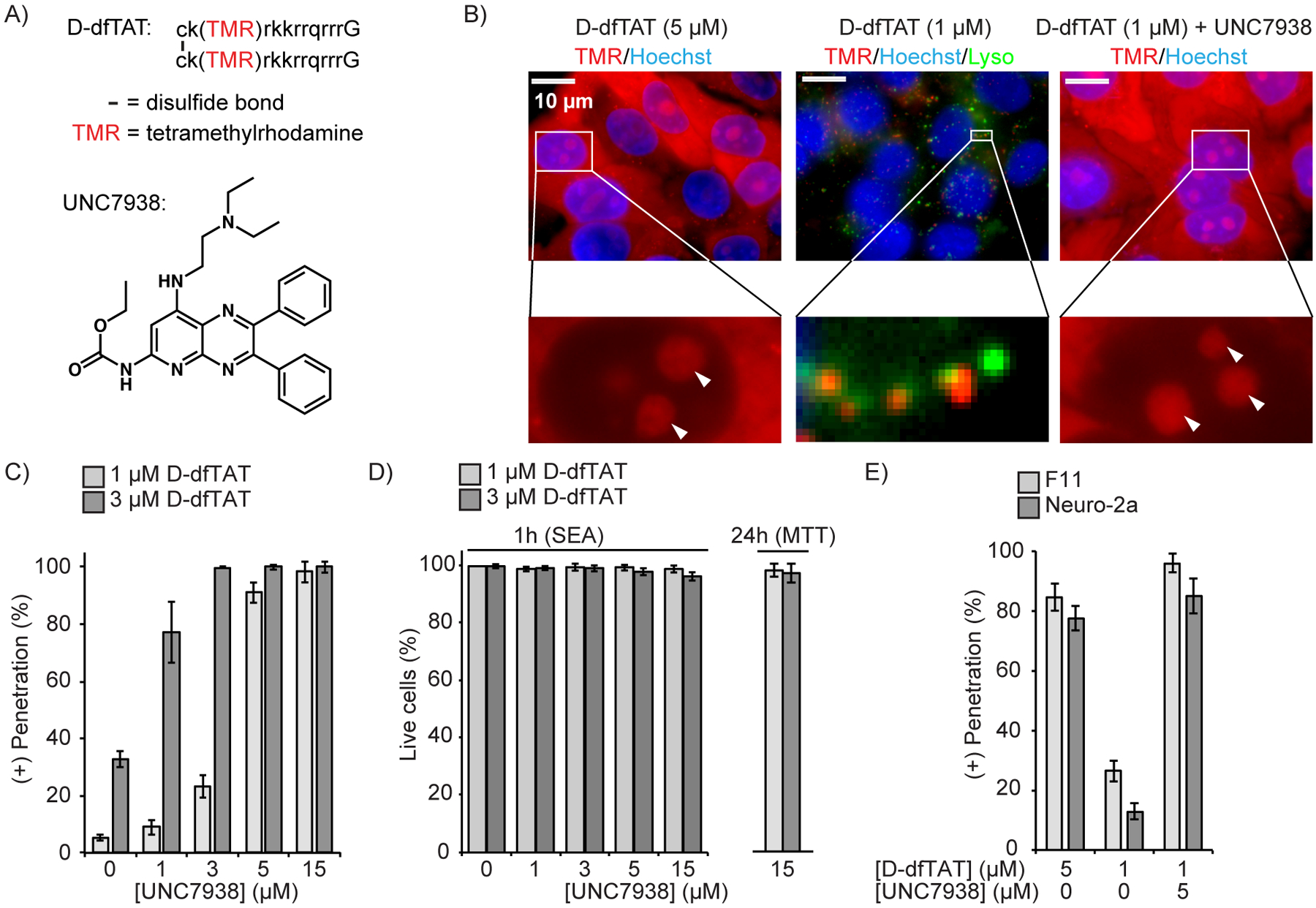
UNC7938 enhances cytosolic penetration of the CPP D-dfTAT. A) Structures of the peptide D-dfTAT and of the small molecule UNC7938. Lower case letters in the peptide sequence refer to D-amino acids. dfTAT has the identical structure but is synthesized with L-amino acids instead. B) Fluorescence microscopy images of HeLa cells treated with D-dfTAT and UNC7938 for 30 min. Images are overlays of fluorescence images pseudo-colored red for TMR and blue for Hoechst 33342. An overlay with Lysotracker Green, pseudo-colored green, is also shown for the condition showing a punctate peptide distribution. Nucleoli are highlighted with white arrows in zoomed-in images. C) HeLa cells were incubated with D-dfTAT and UNC7938 for 30 min, washed, incubated with SYTOX green and Hoechst 33342, and imaged. The cells displaying D-dfTAT positive nucleolar staining while excluding SYTOX green were counted as alive and positive for cell penetration by the peptide. The total number of cells present was established by counting all Hoechst 33342 stained nuclei. The data represented are the means of biological triplicates with corresponding standard deviations (>500 cells counted per experiment). D) Quantification of cell viability 1 and 24 h after incubation with D-dfTAT and UNC7938 (30 min incubation), as measured by a SYTOX Green exclusion assay (SEA) and MTT assay. The data represented correspond to the mean of biological triplicates (>500 cells counted per experiment). E) Effect of UNC7938 on D-dfTAT cell penetration in the F11 and neuro-2a cell lines.
In principle, arginine-rich CPPs such as TAT or dfTAT share some of the limitations generally encountered with polycationic delivery agents. For instance, given that the delivery process first requires endocytic uptake, cells that display low endogenous levels of endocytosis are typically more refractory to reagent-based delivery methods(19). Moreover, negatively-charged species readily interact with polycationic delivery agents, thereby promoting aggregation and limiting the ability of the delivery agents to interact with cellular factors important for cell entry. For instance, albumin, a low pI protein abundantly present in fetal bovine serum (FBS) used in cell culture media, is often inhibitory to cell delivery(19). Removing FBS from cell cultures may circumvent this problem. Yet, this can be relatively deleterious to sensitive cells that require the anti-apoptotic and anti-stress stimulation provided by FBS(21). Notably, the inhibitory effect of anionic species is problematic when the macromolecule that is targeted for delivery is itself negatively charged. In particular, CPPs often fail to deliver nucleic acids(22). Overall, the combination of these various effects lead to delivery experiments that work well in some instances but poorly in others.
Recently, a small molecule, UNC7938 (Figure 1A), was shown to successfully deliver RNAs intracellularly, in vitro and in vivo (23). In particular, UNC7938 releases RNA cargos from endosomes, thereby allowing the oligonucleotides to reach the cytosol and nucleus of cells (23). UNC7938, like TAT-derived CPPs, may therefore promote endosomal membrane leakage. However, because UNC7938 and CPPs are structurally distinct, we postulated that these species may destabilize endosomal membranes in different ways. In turn, we hypothesized that these reagents, when combined, may promote endosomal leakage at levels not achievable when using these cell delivery tools individually. In particular, we envisioned that such a cocktail would provide enhanced endosomal escape activities, permit cytosolic delivery in cells refractory to penetration, and enable the delivery of macromolecular cargos undeliverable by CPPs alone.
Results.
UNC7938 enhances the cytosolic penetration of CPPs
We first examined whether UNC7938 impacts the cellular penetration of several arginine-rich CPPs: TAT, r9, dfTAT and D-dfTAT. All four peptides display a distinctive nucleolar staining of live cells when successful cytosolic entry is achieved. This staining is observed using fluorescence microscopy to assess whether the peptides have entered cells(24–26). UNC7938 induced an increase of the % of cells with nucleolar staining for all peptides (Figure S1). In contrast, UNC7938 did not promote cytosolic penetration of k9, a control peptide typically unable to escape endosomal entrapment on its own(19). UNC7938-induced cell penetration was overall more efficient for dfTAT and D-dfTAT than for TMR-TAT and r9 (27). The rest of our study therefore focuses on the combined activities between UNC7938 and these CPPs. We use D-dfTAT in experiments directly assessing the cell penetration of CPP because of the relative ease with which its nucleolar staining can be detected, and use dfTAT in cargo delivery experiments because of its degradation propensity and its subsequent innocuous effects on cells (24).
We first evaluated the effect of UNC7938 on the endosomal escape of D-dfTAT itself because efficient leakage of D-dfTAT from endosomes is typically a good predictor of the successful delivery of cargos into the cytosol of cells. In HeLa, D-dfTAT achieves approximately 80–90% cytosolic penetration at 5 μM, but less than 10 or 25% at 1 μM or 3 μM, respectively (Figure 1 B, C). The cells that do not exhibit nuclear staining display instead a punctate fluorescence indicative of the peptide being trapped inside endosomes (Figure 1B). Remarkably, the percentage of cells displaying nucleolar staining reaches 100% in the presence of increasing concentrations of UNC7938 and cells with a punctate fluorescence disappear (Figure 1C, S2). Similar results were obtained with DRG-F11 and neuro-2a cell lines (Figure 1E). The combination of D-dfTAT and UNC7938 was non-toxic to cells, as evaluated either 1 or 24 h after incubation (Figure 1D). Finally, while the prior results were obtained after co-incubation of UNC7938 and dfTAT, we found that similar effects could be observed when the two compounds were incubated in succession (Figure S2). This, in turn, indicates that dfTAT and UNC7938 can promote endosomal escape while being endocytosed independently and that interactions between UNC7938 and dfTAT outside the cell are not necessary to mediate cytosolic release.
UNC7938 enhances endosomal membrane leakage
In order to elucidate how UNC7938 enhances the cell penetration of CPPs, D-dfTAT was first preloaded in endosomes by using a 1h incubation at 1 μM (85% cells displaying endosomal entrapment as in Fig 1B). UNC7938, incubated with cells for 30 min post-CPP incubation, however, caused a redistribution of D-dfTAT with ~70% of cells displaying nucleolar staining of the peptide (Figure 2A). Moreover, cytosolic penetration was almost entirely abolished in the presence of bafilomycin, an inhibitor of endosomal acidification and endocytic trafficking(28). Together, these data indicate that UNC7938 promotes the endocytic escape of D-dfTAT. Notably, UNC7938 does not increase the total amount of D-dfTAT that enters cells (Figure 2B). This, in turn, suggests that UNC7938 does not enhance endocytic uptake of the peptide and does not favor peptide-mediated endosomal leakage by increasing how much peptide is endocytosed.
Figure 2.
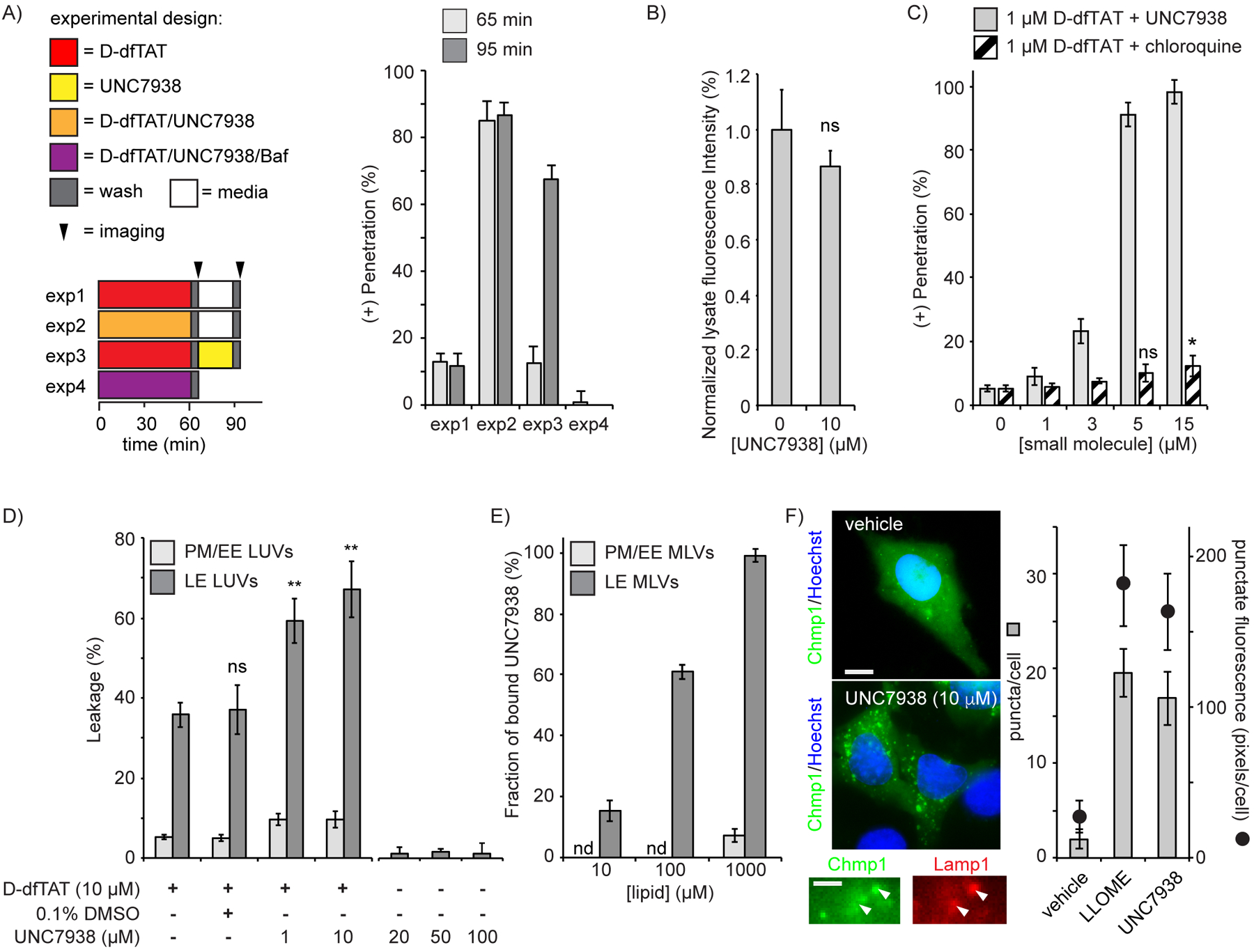
UNC7938 enhances the cytosolic entry of D-dfTAT by facilitating endosomal leakage. A) Hela cells were incubated with combinations of D-dfTAT, UNC7938, and bafilomycin for 1h. Cells were then washed and incubated for an additional 30 min. Cells were imaged after the first and second incubation period and cytosolic penetration of D-dfTAT was scored. B) Total peptide uptake of D-dfTAT (1 μM) with or without UNC7938 after 30 in incubation. C) Comparison of the effect of UNC7938 and chloroquine on D-dfTAT cytosolic penetration. D) Calcein-encapsulated LUVs were treated with D-dfTAT with or without UNC7938, or vehicle (0.1% DMSO). E) Binding of UNC7938 to multi-lamellar vesicles. The label nd (not detected) denotes conditions where no binding was detected. F) HeLa cells transfected with EGFP-Chmp1 and CFP-Lamp1 were treated with vehicle (DMSO), LLOME, or UNC7938. The number of EGFP-Chmp1 puncta present in imaged cells was counted and the total number of pixels displaying a punctate distribution quantified. Representative 100X images are overlay of EGFP-Chmp1 and Hoechst (scale bar: 10 μm). Magnified images show partial co-localization of Chmp1 puncta with Lamp1 positive endosomal organelles (scale bar: 2 μm). In all panels, the data reported corresponds to the mean of biological triplicates (>500 cells per experiment). The label ns corresponds to p>0.05, * corresponds to p≤0.05, ** corresponds to p≤0.01 by a two-tailed t-test.
To test whether UNC7938 enhances the membrane-disruption activity of D-dfTAT, in vitro leakage assays were performed. dfTAT and D-dfTAT induce the specific leakage of late endosomes by interacting with the anionic lipid BMP(24, 29). We therefore used large unilamellar vesicles (LUVs) with a lipid composition mimicking that of late endosomes (LE) as an in vitro model. LUVs with plasma/early endosome membranes (PM/EE) were used as controls. LUVs were loaded with calcein and leakage was measured by quantifying the release of this fluorescent probe in solution. As previously reported, D-dfTAT causes the leakage of LE LUVs but not of PM/EE LUVs (Figure 2D). Notably, the addition of UNC7938 causes a selective increase in LE LUV leakage in a dose-dependent manner. Leakage is however only observable in the presence of D-dfTAT, as UNC7938 alone does not cause any substantial calcein release. Furthermore, UNC7938 shows preferential binding to LE over PM/EE vesicles, suggesting that UNC7938 mediates its effect by interacting with lipid bilayers (Figure 2E).
Since the results of the in vitro assays were consistent with the hypothesis that UNC7938 enhances late endosomal membrane disruption, we next tested whether this activity could be directly observed in live cells. Charged multivesicular body protein 1 (Chmp1) participates in the repair of endosomal membranes damaged by small disruptions (i.e. those small enough to cause the cytosolic release of calcium but not of dextrans)(30, 31). We therefore reasoned that, if UNC7938 disrupts late endosomes enough to alter calcium permeation, the recruitment of Chmp1 to these organelles may be observable. Cells were transfected with Chmp1-EGFP and Lamp1-CFP, a marker of late endosomes and lysosomes. Untreated cells displayed a diffuse cytosolic fluorescence for Chmp1, with few puncta present (Figure 2F). In contrast, cells treated with UNC7838 showed a significant increase in punctate fluorescence in a pattern similar to cells incubated with LLOME, a lysosomotropic drug used as a positive control (Figure S3)(30). Notably, the fluorescent Chmp1 puncta partially co-localize with Lamp1, suggesting that UNC7938 destabilize organelles of the endocytic pathway (Figure 2F, S3).
Since UNC7938 appears to act as a facilitator of endosomal escape, we asked how its activity compares to that of chloroquine. Chloroquine, an anti-malarial drug, is used to enhance the cytosolic delivery of macromolecules externally administered to cells(11, 32, 33). Chloroquine is lysosomotropic and it disrupts endosomal membranes(34). To compare UNC7938 with chloroquine, D-dfTAT was incubated with increasing concentrations of each molecule (Figure 2C). Unlike UNC7938, chloroquine only had a modest effect on the cellular distribution of D-dfTAT. UNC7938 is therefore superior to chloroquine in promoting the cytosolic penetration of the CPP.
UNC7938 rescues the cytosolic entry of the CPP D-dfTAT when D-dfTAT alone fails
As highlighted in Figure 1, D-dfTAT alone works well in several cell types. However, we have observed that COS-7 and HEK293T, cells commonly used for protein and virus production, are resistant to D-dfTAT-mediated delivery(35). These cells exhibit low levels of endocytic uptake (Figure 3A,B), presumably reducing the concentration of CPP inside endosomes. We therefore asked whether UNC7938 would be able to rescue the activity of D-dfTAT in these cells. Hence, COS-7 and HEK293T were co-incubated with UNC7938 and D-dfTAT and the cytosolic release of D-dfTAT was assessed by fluorescence microscopy after 1 h incubation. In the absence of UNC7938, D-dfTAT entered the cytosolic space of cells in less than 20 % of COS-7, and less than 10% of HEK293T cells (Figure 3C,D). These results did not improve at higher D-dfTAT concentrations (50 μM, data not shown). In contrast, 80 to 90% cytosolic release was achieved in the presence of UNC7938 (5 μM), demonstrating that UNC7938 rescues the cytosolic penetration of D-dfTAT in refractory cells. Notably, UNC7938 did not impact the total amount of peptide taken up by the cells (Figure 3A).
Figure 3.
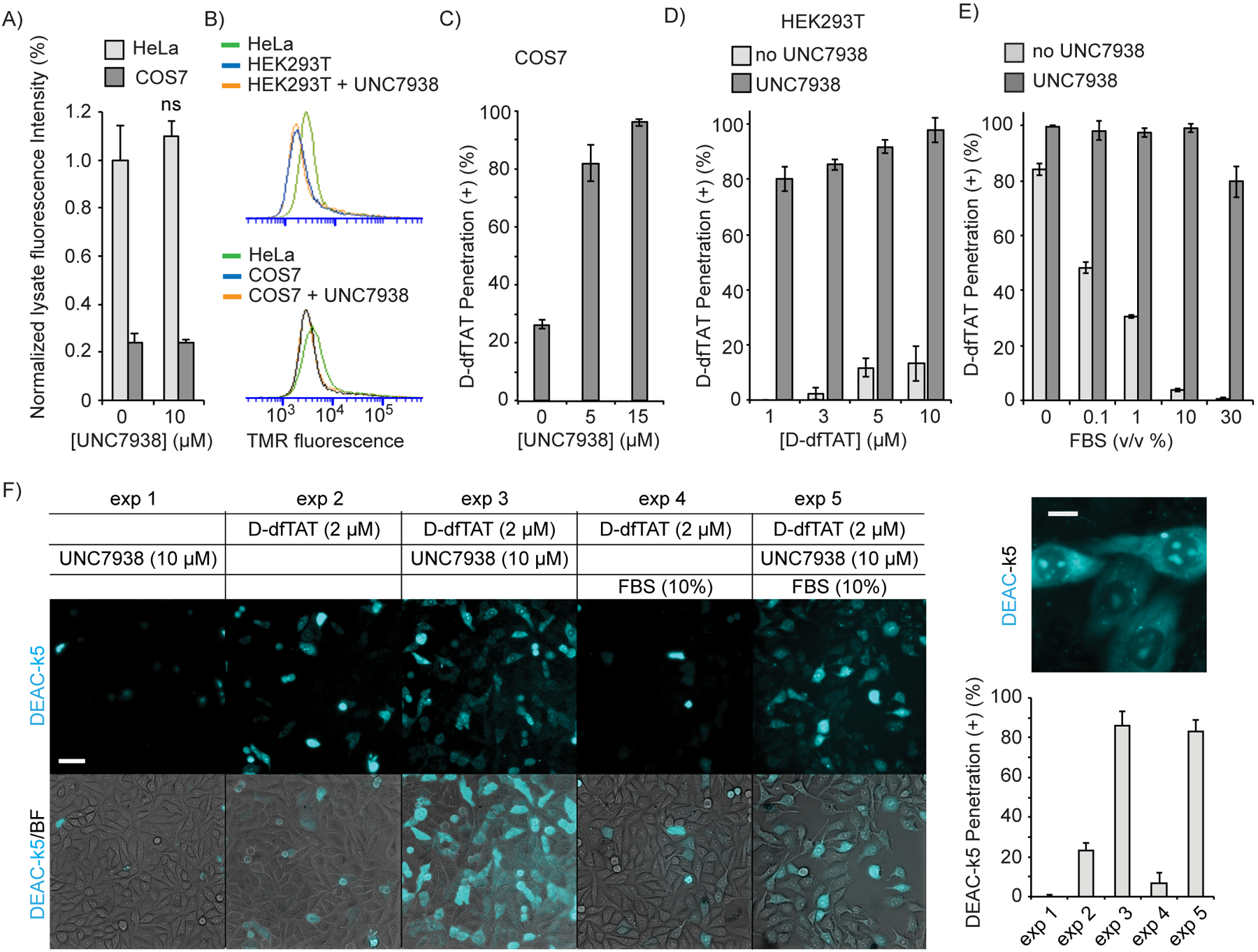
UNC7938 rescues the cytosolic penetration of D-dfTAT under conditions were CPP alone fail. A) Total uptake of D-dfTAT (5 μM) in HeLa and COS7 cells, with or without UNC7938 after 30 min incubation (ns = not significant by a two-tailed t-test). B) Uptake of TMR-r9 (5 μM) in HeLa, COS7, and HEK293T, as assessed by flow cytometry. Incubation were performed for 30 min, with or without UNC7938 (10 μM). C) Effect of UNC7938 on D-dfTAT penetration in COS7 cells. COS7 cells were treated for 30 min with D-dfTAT (5 μM) and with UNC7938. D) Effect of UNC7938 on D-dfTAT penetration in HEK293T cells. HEK293T cells were incubated for 30 min with D-dfTAT in the absence or presence of UNC7938 (5 μM). E) Effect of FBS on D-dfTAT (5 μM) penetration in HeLa cells, in the presence or absence of UNC7938 (10 μM). F) Delivery of DEAC-k5 in Hela cells with and without FBS. Cells were incubated with DEAC-k5 (10 μM), D-dfTAT (2 μM) and UNC7938 (10 μM) in L15 media with or without 10 % FBS. Representative 20X fluorescence images, pseudo-colored cyan for DEAC and overlaid with bright field images, are shown. A 100X fluorescence image of highlighting the subcellular localization of DEAC-k5 obtained after delivery with D-dfTAT and UNC7938 is provided. The cell penetration of DEAC-k5 is assessed by counting the number of cells displaying nucleolar staining of the peptide. In all panels, the data represented correspond to the mean of biological triplicates (>500 cells counted per experiment). Scale bars 20X: 50 μm, 100X: 10 μm.
The albumin present in FBS binds cationic CPPs (Figure S4). This decreases productive accumulation in endosomes and inhibits cytosolic entry(19). Based on the results obtained above, we tested whether UNC7938 would rescue D-dfTAT penetration in the presence of FBS. Cells were incubated with D-dfTAT (5 μM) in media containing increasing amounts of FBS. Without UNC7938, 1% FBS was sufficient to inhibit the cytosolic penetration of D-dfTAT by more than 50% (Figure 3E). Moreover, 10% FBS, a standard cell culture condition, inhibited the cytosolic penetration of D-dfTAT almost completely. In contrast, addition of UNC7938 (10 μM) yield CPP cytosolic penetration in more than 98% of cells with 1 or 10% FBS (Figure 3E). The UNC7938/D-dfTAT combination was also able to deliver the model peptide DEAC-k5 (901 Da, 5 positive charges) into live cells in the presence of FBS, indicating that the rescue of D-dfTAT cell penetration by UNC7938 also correlates with the enhanced cytosolic delivery of other potential cargos (Figure 3F).
CPP and UNC7938 deliver macromolecular cargos in live cells
The positive effects of UNC7938 on CPP-mediated endosomal escape should in principle be beneficial for the delivery of macromolecular cargos. Given that UNC7938 overcomes CPP inhibition by anionic albumin, we wondered whether UNC7938 would also rescue the delivery activity of CPPs when using nucleic acids as cargos. We therefore attempted to deliver plasmids encoding mTurquoise-H2A or GFP with dfTAT. dfTAT alters the electrophoretic mobility of DNA plasmids, indicating binding between peptide and nucleic acid (Figure 4A). In contrast, UNC7938 does not have a detectable effect on plasmid electrophoretic mobility, in the absence or presence of dfTAT. When co-incubated with cells, plasmids block the cytosolic penetration of dfTAT, indicating that DNA/CPP interactions inhibit endosomal escape (Figure 4B). Reciprocally, dfTAT alone is unable to transfect DNA (Figure 4C). Remarkably, addition of UNC7938 to the CPP/DNA sample restored peptide cytosolic access and yielded successful transfection. In HeLa cells, the transfection efficiency was similar to that of Lipofectamine2000 (Figure 4C). In contrast, UNC7938/dfTAT outperformed this commercial reagent in Jurkat cells, a cell line typically refractory to transfection protocols (Figure 4D)(36).
Figure 4.
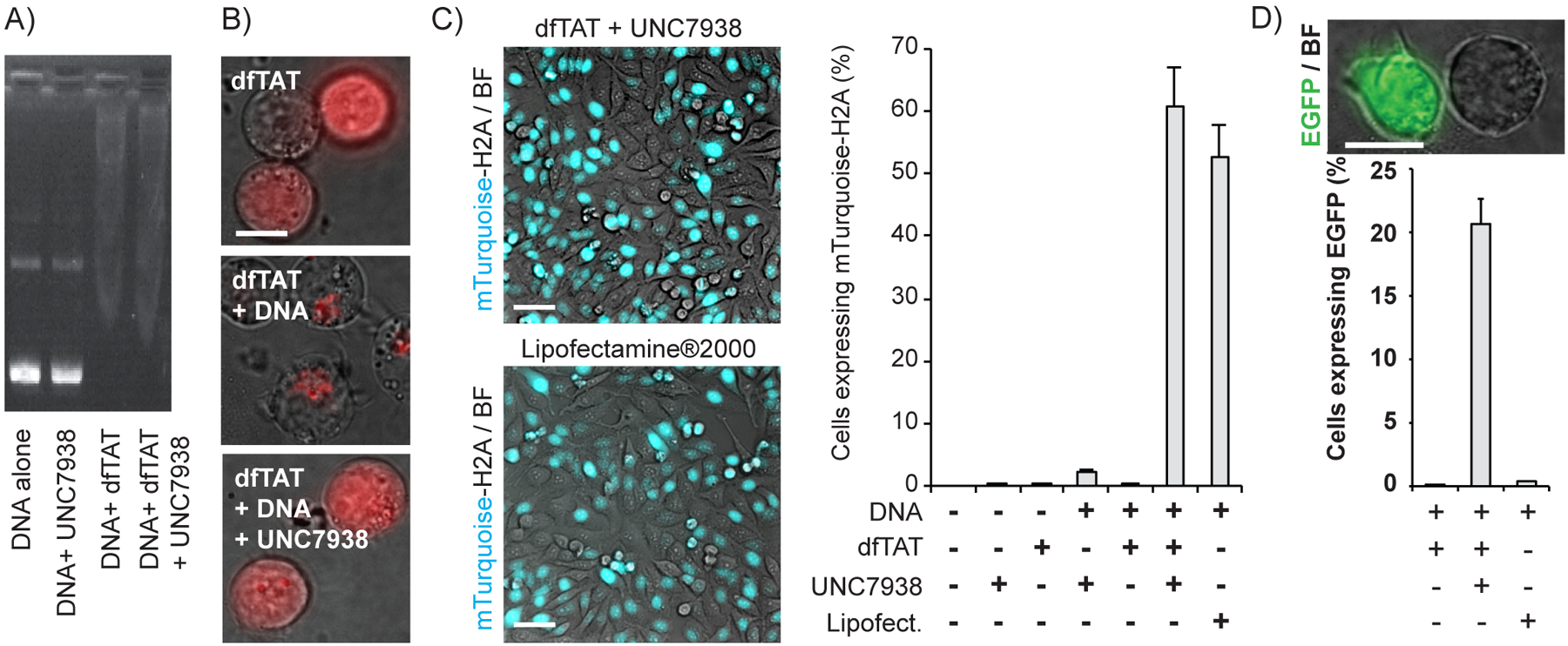
A dfTAT/UNC7938 cocktail mediates the transfection of DNA into cells. A) Gel mobility shift assay. The mTurquoise-H2A plasmid (280 ng) was mixed with dfTAT (3 μM) and UNC7938 (10 μM) and loaded in an agarose gel. Samples were then separated by electrophoresis and DNA was visualized by staining with ethidium bromide. B) Representative images of Jurkat cells incubated with dfTAT, the Gwiz-GFP DNA plasmid, and UNC7938. Images are overlay of bright field images and of dfTAT fluorescence, pseudocolored red. Scale bar 100X: 10 μm. C) Transfection experiments in Hela cells with the mTurquoise-H2A plasmid. Representative images of HeLa (20X) transfected with either lipofectamine®2000 or dfTAT/UNC7938. Images are overlays of fluorescence images from the mTurquoise signal and bright field images. Scale bars 20X: 50 μm. Histograms represent the percentage of cells transfected 24h after incubation with the reagents, as monitored by fluorescence microscopy. The data represented are the average of biological triplicates and the corresponding standard deviation. D) Transfection experiments in Jurkat T lymphocytes with Gwiz-GFP. Data were acquired as in C, with the exception that Jurkat are suspension cells. Live cell imaging was performed after letting cells settle at the bottom of a culture dish. Scale bars 100X: 10 μm.
Next, we tested whether UNC7938/dfTAT would deliver Phalloidin in COS-7 cells shown to be mostly resistant to dfTAT delivery alone. Phalloidin is an actin-binding bicyclic heptapeptide produced by the death cap mushroom(37). Fluorescein-phalloidin, a fluorescent analog of phalloidin, is commonly used as a cytoskeleton staining reagent. However, because fluorescein-phalloidin is not cell permeable, staining protocols require cell fixation. Cytoplasmic delivery of phalloidin with endosomal escape reagents would therefore be useful for live cell imaging applications. Incubation of COS-7 with fluorescein-phalloidin plus either UNC7938 or dfTAT led to endosomal entrapment of the cargo, as shown by the punctate distribution of green fluorescence (Figure 5A). In contrast, the combination of dfTAT and UNC7938 led to a distinct staining to cytoskeletal structures.
Figure 5.
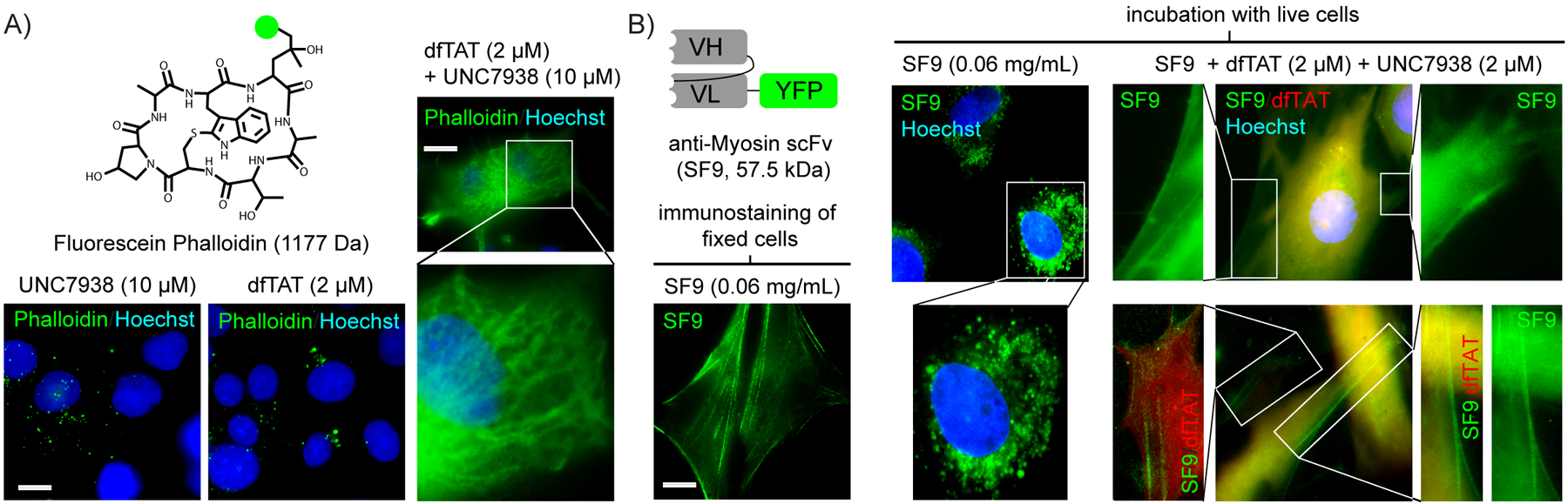
A dfTAT/UNC7938 cocktail delivers a cyclic peptide and a scFv antibody into the cytosol of live human cells. A) Delivery of fluorescein-phalloidin in COS7 cells. Cells were incubated with fluorescein-phalloidin (10 μM, fluorescein is represented by a green disc) and D-dfTAT (2 μM), UNC7938 (10 μM), or both. Representative 100X overlay fluorescence images, pseudo-colored green for the fluorescein signal and blue for Hoechst 33342, are shown. A magnified image displaying the cytoskeletal staining of phalloidin upon cell entry is provided. Scale bars 100X: 10 μm. B) Delivery of anti-NM II scFV SF9 into MCH58 human skin fibroblasts. SF9 was either incubated with fixed and permeabilized cells or with live cells, in the absence or presence of dfTAT and UNC7938. Images are overlay fluorescence images pseudocolored green for SF9, red for dfTAT and blue for Hoechst. Two different representative images of SF9 delivered into live cells are provided. Magnified images are shown and their contrast is adjusted to emphasize the staining of fiber-like structures. Scale bars 100X: 10 μm.
Antibodies and antibody fragments are useful tools for the targeted binding to specific antigens. Because antibodies are not cell-permeable, their use is generally limited to the exterior of live cells or to fixed cells. To test whether UNC7938/dfTAT could deliver antibodies into live cells, a single-chain variable fragment (scFv) that selectively binds non muscle myosin IIA (NM II) was chosen as a model(38). NM II is an actin-binding protein with central roles in cell adhesion and migration(39). A previous study showed that several commercially available delivery reagents fail to deliver this scFv (SF9) into live cells(10). To test the specificity of SF9, the protein was first incubated with fixed and permeabilized MCH58 cells. SF9, tagged with EYFP, shows staining of fiber-like structures consistent with cytoskeletal filaments, as previously reported(10, 38). When incubated with live cells, SF9 is trapped in endosomes, as shown by the punctate distribution of the EYFP signal. In contrast, co-incubation of SF9 with UNC7938 and dfTAT leads to a cytosolic distribution of the EYFP signal accompanied by the staining of fiber-like structures seen in fixed cells (Figure 5B). Overall, these results indicate that at least a substantial fraction of SF9, a 57.5 kDa fusion protein with three domains, two of which contain disulphide bonds, has successfully reached its intracellular target with EYFP and antibody domains still functional.
Discussion.
CPPs are delivery vectors used in applications ranging from basic research to clinical trials(40). While occasionally capable of direct membrane permeation, CPPs often need to mediate the membrane permeation of endosomes in order to deliver macromolecules(27). This activity is in turn dependent on the intrinsic capacity of the CPP to disrupt endosomal membrane as well as its ability to reach the endosomal lumen. Multiple variables can however prevent this from taking place. In particular, the low endocytic uptake intrinsic to certain cell types limits the accumulation of CPPs inside endosomes, keeping the luminal concentration of CPP below the threshold required for endosomal escape and cytosolic delivery. Similarly, extracellular components, including macromolecular cargo themselves, can inhibit cytosolic delivery if they interfere with the ability of the CPP to interact with the cellular factors that are responsible for endocytic uptake and endosomal escape. Overall, these problems highlight the inherent limitations shared by approaches that rely on endocytic pathway as a route of cytosolic penetration. Remarkably, the small molecule UNC7938 rescues the endosomal escape activity of the CPPs used in this study under all of these inhibitory conditions.
UNC7938 did not stimulate endocytic uptake. Therefore, UNC7938 does not appear to enhance the activity of dfTAT or D-dfTAT by increasing the concentration of the CPPs inside endosomes. Instead, UNC7938 destabilizes the membrane of endosomes. This activity is not sufficient to cause substantial membrane leakage and allows for detectable cytosolic access of the macromolecular cargos used herein. However, it dramatically facilitates the CPP-induced leakage of endosomes and improves the penetration of the peptides by lowering the threshold concentration of peptide needed for endosomal membrane permeation.
In practice, the rescue activity of UNC7938 expands the usefulness of CPP-mediated cellular delivery to new conditions and to new cargos. In particular, by enhancing cytosolic penetration efficiencies, the effect of the small molecule is to dampen the variability observed among cells types and culture conditions. This in turn leads to protocols that are robustly efficient, reproducible and reliable, and that work across a broader range of experimental settings. The small molecule also permits the development of tissue culture protocols that are efficient for cell-impermeable small molecules, peptides, proteins, and nucleic acids (Mw from 1.2 kDa to 3.5 MDa; from polycationic, neutral, and polyanionic species). This is important because current delivery techniques (i.e. commercially available transfection reagents or electroporation) still suffer from efficiency and toxicity limitations(41).
Overall, our results suggest that the combination of two membrane-disrupting species facilitates endosomal escape and cell delivery. The use of chloroquine was an early example of such approach. However, as shown herein, chloroquine is poorly active when compared to UNC7938. Notably, chloroquine and UNC7938 share common features, including conjugated rings (quinoline vs pyrido[2,3-b]pyrazine, respectively), and a protonable amine appendage. Such features are typical of hydrophobic weak base (HWB) therapeutics(42). These compounds are membrane permeable when deprotonated but become sequestered in endosomes and lysosomes upon protonation in the acidic lumen of these organelles(42). UNC7938 may therefore follow similar principles to selectively disrupt endosomal membranes. More importantly, it also suggests that CPPs may benefit in combination with other HWBs. This chemical space remains unexplored but could yield cocktails that improve a variety of cell delivery applications.
Materials and Methods.
Peptide design, synthesis and purification
All peptides were synthesized on a rink amide MBHA resin (Novabiochem, San Diego, CA) by solid phase peptide synthesis (SPPS) using previously described protocols(19, 24). Briefly, Fmoc-Lys(Mtt)-OH, Fmoc-Lys(Boc)-OH, Fmoc-D-Lys(Boc)-OH, Fmoc-D-Lys(Dde)-OH, Fmoc-Gly-OH, Fmoc-Arg(Pbf)-OH, Fmoc-D-Arg(Pbf)-OH, Fmoc-Gln(Trt)-OH, Fmoc-D-Gln(Trt)-OH, Fmoc-Cys(Trt)-OH (Novabiochem) were used to assemble the peptides using standard Fmoc protocols. Boc-D-Cys(Trt)-OH (Novabiochem) was added to the N-terminus of D-fTAT (single chain monomeric peptide) and the Boc protecting group was removed upon TFA cleavage. The peptides fTAT (NH2-CK(ε-NH-TMR)RKKRRQRRRG-CONH2, D-fTAT (NH2-CK(ε-NH-TMR)rkkrrqrrrG-CONH2), TMR-TAT (TMR-NH-RKKRRQRRRG-CONH2), TMR-r9 (TMR-NH-rrrrrrrrr-CONH2), TMR-k9 (TMR-NH-kkkkkkkkk-CONH2), and DEAC-k5 (DEAC-NH-kkkkk-CONH2) were analyzed and purified by reverse-phase HPLC on a Hewlett-Packard 1200 series instrument and an analytical Vydac C18 column (5 μm, 4 × 150 mm). The flow rate was 1 mL/min, and detection was at 214 nm and 550 nm. Semi-preparative HPLC was performed on a Vydac C18 10 × 250 mm column. The flow rate was 4 mL/min, and detection was at 214 nm and 550 nm. All runs used linear gradients of 0.1% aqueous TFA (solvent A) and 90% acetonitrile, 9.9% water, and 0.1% TFA (solvent B). The correct identity of the peptides was confirmed by MALDI-TOF performed with a Shimadzu/Kratos instrument (AXIMA- CFR, Shimadzu, Kyoto), as reported(19, 24).
Generation of dfTAT and D-dfTAT by dimerization of L-fTAT and D-fTAT respectively
fTAT or D-fTAT (0.3 mg, 1.5 × 10−4 mmol) were dissolved in 5 mL of aerated phosphate buffer saline (PBS) (Fisher) at pH 7.4. The reaction was agitated on a nutator overnight (100% yield based on HPLC analysis). dfTAT and D-dfTAT were purified using analytical reverse-phase HPLC. Expected mass (MALDI-TOF): 4078.27, observed mass dfTAT and D-dfTAT were 4080.87 and 4078.87 respectively
Cell culture
Unless otherwise indicated, cells were obtained from ATCC. All cells were incubated in a humidified atmosphere containing 5% CO2 at 37 °C.
The DRG-F11 cell line (a hybrid of rat dorsal root ganglion and mouse neuroblastoma) were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Fisher) supplemented with 20 mM HEPES, 10% FBS and 1X penicillin/streptomycin (P/S) (Fisher).
MDA-MB-231 (breast cancer-derived epithelial), HeLa (human epithelial), neuro-2a (mouse neuroblast), MCH58 (immortalized human fibroblast), and COS-7 (African green monkey kidney fibroblast-like) cells were cultured in DMEM supplemented with 10% FBS and 1X P/S.
Jurkat (immortalized human T lymphotcyte) cells were cultured in RPMI 1640 (Fisher) supplemented with 10% FBS and 1X P/S.
HEK293F (modified human embryonic kidney) cells were cultured, shaking, between a density of 5*105 to 5*106 cells/mL in Freestyle 293 chemically defined media (Thermo Fisher).
Delivery of peptides into live cells
Mammalian cells were seeded in an 8-well dish or a 48-well dish. The cells were grown to 80–90% confluency in a 37°C humidified atmosphere containing 5% CO2. The cells were washed three times with PBS and once with Leibovit’s L15 media (L15) (Fisher). Cells were then incubated with the peptide D-dfTAT, dfTAT, D-fTAT or TMR-r9 (concentrations specified in text) in the presence and absence of UNC7938 (concentrations specified in text and vary based on the cell line tested) at 37°C for 1 h (unless otherwise specified). This was followed by washing the cells three times with L15 supplemented with heparin (Sigma) (1 mg/mL) and an extra L15 wash. Cells were treated with the cell impermeable nuclear stain SYTOX® BLUE (Thermo) to take into account cells that had a compromised plasma membrane (i.e. dead cells). Alternatively, the cell permeable dye Hoechst 33342 (Thermo) was used for nuclear staining and LysoTracker green (Thermo) (500 nM) was used to stain acidified endocytic organelles. Cells were imaged using an inverted epifluorescence microscope (Model IX81, Olympus, Center Valley, PA). The microscope is equipped with a heating stage maintained at 37 °C. All images were collected using a Rolera-MGI Plus back-illuminated EMCCD camera (Qimaging, Surrey, BC, Canada). Images were acquired using bright field imaging and four standard fluorescence filter sets: CFP (Ex = 436 ± 10 nm / Em= 480 ± 20 nm), RFP (Ex = 560 ± 20 nm / Em= 630 ± 35 nm), FITC(Ex = 488 ± 10 nm / Em= 520 ± 20 nm), DAPI (Ex = 350 ± 50 nm / Em= 460 ± 25 nm). For image viewing and processing individual the SlideBook 4.2 software was used (Olympus, Center Valley, PA) was used. As previously reported, cells that displayed a cytosolic and nucleolar peptide distribution were considered positive for cytosolic penetration (penetration(+))(24). The percentage of penetration (+) cells was calculated by dividing the number of cells that show a fluorescent nuclear staining (20X images) by the total number of cells present (acquired using bright field imaging). Cells that displayed a punctate distribution consistent with endosomal entrapment of the peptides, were considered were not counted (negative penetration). As for the cells that were stained by the SYTOX dyes, they were considered dead and were not counted for the analysis. For the quantitative experiments, an average of at least three 20X pictures were taken, representing 300–400 cells. The reproducibility of all the experiments was assessed by performing experiments with independent batches of cell cultures on three different days (i.e. biological triplicates).
Quantitative determination of peptide uptake
The mammalian cells were seeded in a 48-well dish and grown to 80% confluency. Cells were treated with D-dfTAT +/− UNC7938 (concentration indicated in texts) for 1h at various concentrations. The total peptide uptake was measured by a whole cell lysate analysis. Cells were washed with heparin (1mg/mL) and then trypsinized (50 μL) for 3 min. Cells were resuspended in L15 (350 μL) and spun down at 4°C, 2,500 rpm for 10 min. The supernatant was discarded and the cell pellet was resuspended in L15 (50 μL). An aliquot from the resuspended cells (3 μL) was withdrawn to determine the total number of cells per sample by analysis using flow cytommetry. The remaining aliquot of resuspended cells were lysed by addition of 50 μL of a lysis buffer (components: 50mM Tris, pH7.5, 2 mM EDTA, 4mM DTT, 20% Triton X-100 (Fisher) and protein inhibitor cocktail) and 3 min of vortexing. A volume of 70 μL of the lysed cells for each condition was placed in a 96 well plate. The fluorescence emission intensity was measured using a plate reader equipped with a fluorescence module (Ex=525, Em=580–640 nm) (GloMax®-Multi+ Detection System, Promega, Fitchburg, WI). To normalize, the aliquot of cells (3 μL) was resuspended in L15 (197 μL) and the total amount of cells per samples was determined using flow cytometry.
For total fluorescence and uptake measurement using the flow cytometer, cells were trypsinized and resuspended in L15 medium. Cells were analyzed using a BD Accuri C6 flow cytometer equipped with the FL2 filter (Ex = 488 nm/Em = 533 ± 30 nm). All data was acquired at flow rate of 66 μL/min with detection of a minimum of 40,000 events. The geometric mean of the FL2 signal for each experiment was determined using the Flowjo software. The data reported represent the average and corresponding standard deviations of three independent experiments for each peptide concentration.
Delivery of cargo into live cells using D-dfTAT and UNC7938
Cells were incubated with 5 μM dfTAT, +/− 10 μM UNC7938 at various FBS concentrations (indicated in text). After the 1 h incubation, the cells were washed and imaged using 20X and 100X objectives. The percent penetration was quantified as described earlier. For delivery of DEAC-k5 cargo, HeLa cells incubated with 5 μM dfTAT, 10 μM DEAC-k5, +/− 10 μM UNC7938 and +/− 10% FBS concentrations for 1 h at 37° C. The cells were then washed and imaged using 20 X and 100X objective. The percent DEAC-k5 delivery was determined by quantifying the number of cells that display DEAC-K5 nucleolar staining divided by the total number of cells per image. For all quantitative experiments performed, three to five 20X pictures were taken per condition, representing 300–500 cells. The reproducibility of all the experiments was assessed by performing experiments with independent batches of cell cultures on three different days (i.e. biological triplicates).
DNA transfection using dfTAT and UNC7938
For cellular transfection experiments using dfTAT and UNC7938, the plasmid mTurquoise-H2A (mTurquoise-H2A-10, Addgene plasmid # 55556) was selected. HeLa cells were incubated with 280 ng mTurquoise-H2A, 10 uM UNC7938 and 3 μM dfTAT in L15 (200 μL). The cells were incubated for 2 h, and then washed. The L15 was then replaced with DMEM, since the cells were placed back into the incubator overnight. As a positive control for the transfection efficiency, cells were transfected using Lipofectamine 2000. The plasmid (280 ng) was mixed with Lipofectamine 2000 (1 μL) in opti-MEM (Fisher) medium and allowed to incubate for 10 min at room temperature. The DNA/lipofectamine complex was then incubated for 2h at 37°C with HeLa cells that were seeded the previous day in an eight-well or 48-well dish. All experiments comparing dfTAT/UNC7938 and Lipofectamine transfection were performed at the same time and imaged after 24 h.
On the day of the Jurkat cell transfections, 7×10^5 cells were harvested per condition from the main culture flask. Cells were washed twice with PBS and resuspended in L15 to be distributed evenly into individual sterile micro-centrifuge tubes for each condition. Every subsequent step consists of 200 μL volume. Cells were transfected for 2 hours with gWiz (Aldevron) by incubation at 37°C in L15 with Lipofectamine 2000 or dfTAT/UNC7938 at 5/10 μM respectively. Following the transfection, cells were washed once with heparin in L15 at 1 mg/mL followed by L15 alone, and finally resuspended in 200 μL of the RPMI 1640 growth media. The micro-centrifuge tube lids were punctured to allow for carbon dioxide exchange and all tubes were allowed to continue growth for ~20 hours. In some cases, 40 μL of the cell suspension from each condition were removed and placed in a glass-bottom 384-well plate for imaging of dfTAT penetration immediately after the two hour transfection period. After a 16–24 hour growth period, 40 μL of the cell suspension was placed in a 384-well glass-bottom plate and imaged for GFP or mTurquoise-H2A expression.
To determine the transfection efficiency, the cells were quantified using either microscopy or flow cytometry (BD Accuri C6). HeLa cells displaying a blue fluorescence in the nucleus, as a result of the expression of mTurquoise-H2A were counted positive for transfection. This number was divided by the total number of cells per image and used to determine the percent cells positive for transfection. For all quantitative experiments performed, an average of at least three 20X pictures were taken, representing 300–400 cells. The reproducibility of all the experiments was assessed by performing experiments with independent batches of cell cultures on three different days (i.e. biological triplicates).
The remainder of transfected Jurkat cells following imaging were injected into a BD Accuri C6 flow cytometer using a flow rate of 24 μL/min and a core size of 13 μm. Healthy cell events were gated based on a sample of untreated Jurkat cells. Cells with successful transfection of the gWiz plasmid were detected on the FL1 channel where the threshold for successful GFP expression was defined as any event with an FL1 signal higher than the maximum fluorescence detected from a dfTAT/UNC7938 negative control.
Liposome Preparation
The lipids used in the experiments consisted of: 1,2-dioleoyl-s-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), sn-(3-oleoyl-2-hydroxy)-glycerol-1-phospho-sn-1’-(3’-oleoyl-2’-hydroxy-glycerol) (BMP), cholesterol (chol) (Avanti Polar Lipids). Liposomes were prepared by transferring the volume of lipids dissolved in chloroform (stock solutions of known concentrations) into scintillation vials. For liposomes mimicking the intraluminal late endosomes vesicles (L.E.) the molar ratios of lipids consisted of 77:19:4 BMP:PC:PE. For liposomes mimicking the plasma membrane or early endosomes, the lipid mixture was 65:15:20 PC:PE:Chol. The lipid film was prepared by removing the chloroform from the lipid mixture using a N2 (g) and then placing the vial in a desiccator overnight. To hydrate the lipids, buffer containing 100 mM NaCl, 10 mM NaH2PO4 pH7.4, with or without 60 mM calcein was added. The lipids were then mixed vigorously and swelled for 1 h at 42°C under N2 to obtain multilamellar vesicles (MLVs). To obtain unilamellar vesicles (LUVs), the MLVS were extruded (20 passes) through a 100 nm pore size polycarbonate membrane (Whatman) using a Mini-Extruder (Avanti Polar Lipids). Dynamic light scattering was used to determine the average diameter size distribution of the liposomes using a Zeta Sizer (Malvern instrument). The liposomes encapsulated with calcein were purified by gel filtration using a Sephadex G-50 (GE Healthcare) column (2.5 × 17.5 cm) to separate the liposomes from free calcein. The eluate was collected in a 96 well plate and the plate was read using a Promega GloMax-Multi plate reader (Promega) at 450 nm and 750 nm corresponding to the wavelength of detection for calcein and liposome respectively.
Liposome leakage assays
Purified calcein-loaded LUVs were mixed with D-dfTAT in presence or absence of UNC7938 (1–100 μM) at a 1:50 peptide: lipid ratio for 1 h at room temperature in 100 mM NaCl, 10 mM NaH2PO4 pH5.5. UNC7938 stock solution Samples were centrifuged for 1 min at 4,000 rpm. To measure the amount of leaked calcein and separate soluble liposomes from released calcein, the supernatants were purified using an illustra NAP-10 Sephadex G-25 column (GE Healthcare) (the elution volumes of liposomes and free calcein were determined independently with pure samples). Fractions were collected in a 96-well plate and the fluorescence of calcein was measured using a Promega GloMax-Multi plate reader (Ex 490nm, Em 520–560nm). Note, since UNC7938 stock is dissolved in 100% DMSO. The control of D-dfTAT+ 0.1% DMSO (concentration of DMSO in the UNC7938 incubations) with liposomes was performed as a control. 100% leakage was established by treating liposomes with the detergent Triton X-100.
Liposome Binding Assays
To determine the binding affinity of UNC7938 to MLVs of different lipid compositions the MLVs were incubated for 20 min with UNC7938 (45 μM) at different small molecule:lipid ratio in the buffer composition of 100 mM NaCl, 10 mM NaH2PO4 pH 5.5. Samples were centrifuged at 87,000 rpm (rotor radius: rmin= 30.0mm rav= 34.5mm; rmax = 38.9). The supernatant was removed to measure the amount of unbound UNC7938 using the absorbance at 375 nm.
Determination of DNA/ dfTAT/ UNC7938 complex formation
To determine whether dfTAT, UNC7938 and the plasmid DNA form a complex, samples were run on an agorase gel. First, 5 μM dfTAT, 10 μM UNC7938 and 70 μg mTurquoise-H2A (in a total volume of 50 μL) were incubated for 20 min at 37°C. The sample was run on a 0.8% agarose gel (to which Ethidium Bromide (EtBr) was added) at 100 mV using TBE buffer (89mM Tris (pH 7.6), 89mM Boric Acid, 2mM EDTA) for 1 h and imaged using a gel doc imager (Amersham). As a control, mTurquoise-H2A (DNA) alone, DNA+ UNC7938 and DNA+ dfTAT were all run on the gel at the same concentrations mentioned above.
ScFv-EYFP delivery into MCH58 cells
The day before the experiment, the cells were split and seeded to reach a target confluency of ~60% the following day in a chambered glass 8-well plate (Fisher). On the day of the experiment, cells were washed three times with L15. Immediately after washing, the cells were introduced to the incubation cocktail containing various combinations of the scFv-EYFP construct, dfTAT, and UNC7938, in L15. The cells were left to incubate with the mixture for one hour at 37°C. Following the incubation, cells were washed twice with heparin at 1 mg/mL in L15, and once more with L15 alone before the addition of 5 μM Hoechst 33342. The cells were left in this solution for ten minutes to allow adequate time for the nuclear stain to permeate the cells before imaging.
Staining of fixed/permeabilized MCH58 cells with scFv-EYFP
The day before the experiment, MCH58 cells were split and seeded to reach a target confluency of ~60% the following day in a chambered glass 8 well plate (Fisher). All subsequent steps were performed at room temperature. For fixation, cells were washed three times with PBS and incubated for 15 minutes in 4% paraformaldehyde (Sigma) in PBS. Paraformaldehyde was washed away with three PBS washes and the cells were permeabilized using 0.1% Triton X-100 in PBS for 15 minutes. Following permeabilization, cells were incubated with 10% FBS in PBS for one hour before adding the scFv-EYFP construct in 10% FBS in PBS to incubate for two hours. After the incubation with scFv-EYFP, cells were washed twice with fresh PBS to remove background fluorescence before imaging.
Supplementary Material
Acknowledgments.
We are grateful to R. Chapkin for access to the Flow sight in his laboratory. We thank L. Dangott and the protein chemistry lab for aid with mass spectrometry. We thank Dr. Ting-Yi Wang for synthesis of the TMR-k9 peptide. We thank Dr. Jyoti Jaiswal for the Chmp1b-GFP plasmid, Dr. Michael Davidson for the mTurquoise-H2A plasmid, Dr. Jennifer Lippincott-Schwartz for the Cerulean-LAMP1 plasmid, Dr. Franck Perez scFv(SF9)-EYFP plasmid. We also thank Dr. J. Sacchettinni for Neuro-2a cells, Dr. Stephen Safe for Jurkat cells, Dr. S. Musser for HeLa cells, Dr. E. Shoubridge for MCH58 cells, Dr. Philippe Sarret for DRG-F11 cells, and Dr. Pingwei Li for HEK293F. This work was supported by a grant (award number R01GM110137) from the National Institutes of Health and a grant (award number RP100819) from the Cancer Prevention and Research Institute of Texas.
Footnotes
References
- 1.Stewart MP et al. , In vitro and ex vivo strategies for intracellular delivery. Nature 538, 183–192 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Stewart MP, Langer R, Jensen KF, Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chemical Reviews 118, 7409–7531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller J, Kretzschmar I, Volkmer R, Boisguerin P, Comparison of Cellular Uptake Using 22 CPPs in 4 Different Cell Lines. Bioconjugate Chemistry 19, 2363–2374 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Dissanayake S, Denny WA, Gamage S, Sarojini V, Recent developments in anticancer drug delivery using cell penetrating and tumor targeting peptides. Journal of Controlled Release 250, 62–76 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Higa M et al. , Identification of a novel cell-penetrating peptide targeting human glioblastoma cell lines as a cancer-homing transporter. Biochemical and biophysical research communications 457, 206–212 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Erazo-Oliveras A, Muthukrishnan N, Baker R, Wang TY, Pellois JP, Improving the Endosomal Escape of Cell-Penetrating Peptides and Their Cargos: Strategies and Challenges. Pharmaceuticals 5, 1177–1209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madani F et al. , Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. Journal of Biophysics 2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torchilin VP, Recent advances with liposomes as pharmaceutical carriers. Nature reviews. Drug discovery 4, 145–160 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Funhoff AM et al. , Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules 5, 32–39 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Marschall AL et al. , Delivery of antibodies to the cytosol: debunking the myths. MAbs 6, 943–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadia JS, Stan RV, Dowdy SF, Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature Medicine 10, 310–315 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Takayama K et al. , Enhanced intracellular delivery using arginine-rich peptides by the addition of penetration accelerating sequences (Pas). Journal of Controlled Release 138, 128–133 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Kaplan IM, Wadia JS, Dowdy SF, Cationic TAT peptide transduction domain enters cells by macropinocytosis. Journal of Controlled Release 102, 247–253 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Lee YJ, Datta S, Pellois JP, Real-time fluorescence detection of protein transduction into live cells. Journal of the American Chemical Society 130, 2398–2399 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Rizzuti M, Nizzardo M, Zanetta C, Ramirez A, Corti S, Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discovery Today 20, 76–85 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Munyendo WLL, Lv H, Benza-Ingoula H, Baraza LD, Zhou J, Cell Penetrating Peptides in the Delivery of Biopharmaceuticals. Biomolecules 2, 187–202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angeles-Boza AM, Erazo-Oliveras A, Lee YJ, Pellois JP, Generation of endosomolytic reagents by branching of cell-penetrating peptides: tools for the delivery of bioactive compounds to live cells in cis or trans. Bioconjugate Chemistry 21, 2164–2167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura KS, Sung M, Bolewska-Pedyczak E, Gariépy J, Probing the Impact of Valency on the Routing of Arginine-Rich Peptides into Eukaryotic Cells†. Biochemistry 45, 1116–1127 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Erazo-Oliveras A et al. , Protein delivery into live cells by incubation with an endosomolytic agent. Nature Methods 11, 861–867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brock DJ et al. , Efficient cell delivery mediated by lipid-specific endosomal escape of supercharged branched peptides. Traffic 10.1111/tra.12566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis GL, Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology 62, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alhakamy NA, Nigatu AS, Berkland CJ, Ramsey JD, Noncovalently associated cell-penetrating peptides for gene delivery applications. Therapeutic Delivery 4, 741–757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B et al. , High-throughput screening identifies small molecules that enhance the pharmacological effects of oligonucleotides. Nucleic acids research 43, 1987–1996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Najjar K, Erazo-Oliveras A, Brock DJ, Wang TY, Pellois JP, An l- to d-Amino Acid Conversion in an Endosomolytic Analog of the Cell-penetrating Peptide TAT Influences Proteolytic Stability, Endocytic Uptake, and Endosomal Escape. Journal of Biological Chemistry 292, 847–861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fretz Marjan M. et al. , Temperature-, concentration- and cholesterol-dependent translocation of L- and D-octa-arginine across the plasma and nuclear membrane of CD34+ leukaemia cells. Biochemical Journal 403, 335–342 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vivès E, Brodin P, Lebleu B, A Truncated HIV-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. Journal of Biological Chemistry 272, 16010–16017 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Wang TY et al. , Membrane Oxidation Enables the Cytosolic Entry of Polyarginine Cell-penetrating Peptides. Journal of Biological Chemistry 291, 7902–7914 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson LS, Dunn KW, Pytowski B, McGraw TE, Endosome acidification and receptor trafficking: bafilomycin A1 slows receptor externalization by a mechanism involving the receptor’s internalization motif. Molecular biology of the cell 4, 1251–1266 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erazo-Oliveras A et al. , The Late Endosome and Its Lipid BMP Act as Gateways for Efficient Cytosolic Access of the Delivery Agent dfTAT and Its Macromolecular Cargos. Cell chemical biology 23, 598–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI, Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheffer LL et al. , Mechanism of Ca(2)(+)-triggered ESCRT assembly and regulation of cell membrane repair. Nature Communication 5, 5646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caron NJ, Quenneville SP, Tremblay JP, Endosome disruption enhances the functional nuclear delivery of Tat-fusion proteins. Biochemical and biophysical research communications 319, 12–20 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Slater AF, Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacology & therapeutics 57, 203–235 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Varkouhi AK, Scholte M, Storm G, Haisma HJ, Endosomal escape pathways for delivery of biologicals. Journal of Controlled Release 151, 220–228 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Durocher Y, Perret S, Kamen A, High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic acids research 30, E9 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao N et al. , Transfecting the hard-to-transfect lymphoma/leukemia cells using a simple cationic polymer nanocomplex. Journal of Controlled Release 159, 104–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chazotte B, Labeling cytoskeletal F-actin with rhodamine phalloidin or fluorescein phalloidin for imaging. Cold Spring Harbor protocols 2010, pdb prot4947 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Nizak C et al. , Recombinant antibodies against subcellular fractions used to track endogenous Golgi protein dynamics in vivo. Traffic 4, 739–753 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR, Non-muscle myosin II takes centre stage in cell adhesion and migration. Nature Reviews Molecular Cell Biology 10, 778–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guidotti G, Brambilla L, Rossi D, Cell-Penetrating Peptides: From Basic Research to Clinics. Trends in Pharmacological Sciences 38, 406–424 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Brock DJ, Kondow-McConaghy HM, Hager EC, Pellois JP, Endosomal Escape and Cytosolic Penetration of Macromolecules Mediated by Synthetic Delivery Agents. Bioconjugate Chemistry 30, 293–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhitomirsky B, Assaraf YG, Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget 6, 1143–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


