Transcriptomic analysis of Fusarium virguliforme colonization revealed extensive rewiring of infection programs, leading to asymptomatic and symptomatic phenotypes in maize and soybean, respectively.
Abstract
We exploited the broad host range of Fusarium virguliforme to identify differential fungal responses leading to either an endophytic or a pathogenic lifestyle during colonization of maize (Zea mays) and soybean (Glycine max), respectively. To provide a foundation to survey the transcriptomic landscape, we produced an improved de novo genome assembly and annotation of F. virguliforme using PacBio sequencing. Next, we conducted a high-resolution time course of F. virguliforme colonization and infection of both soybean, a symptomatic host, and maize, an asymptomatic host. Comparative transcriptomic analyses uncovered a nearly complete network rewiring, with less than 8% average gene coexpression module overlap upon colonizing the different plant hosts. Divergence of transcriptomes originating from host specific temporal induction genes is central to infection and colonization, including carbohydrate-active enzymes (CAZymes) and necrosis inducing effectors. Upregulation of Zn(II)-Cys6 transcription factors were uniquely induced in soybean at 2 d postinoculation, suggestive of enhanced pathogen virulence on soybean. In total, the data described herein suggest that F. virguliforme modulates divergent infection profiles through transcriptional plasticity.
INTRODUCTION
During host colonization, fungal plant pathogens elicit an array of symptoms in the plant, many of which stem not only from the modulation of the plant immune system but also from reprogramming of host development processes (Oliver and Ipcho, 2004; Horbach et al., 2011; Cordovez et al., 2017). Indeed, while studies surveying single trait interactions have highlighted key processes and pathways critical to pathogenesis of fungi in plants (Derntl et al., 2017; Fang et al., 2017), genomic and transcriptomic studies suggest fungi have a complex and elaborate infection program (Brown et al., 2017; Chowdhury et al., 2017). Comparative transcriptomics during fungal colonization have revealed that infection programs vary by fungal lifestyle, suggesting that induced pathways diverge within biotrophic, hemibiotrophic, and/or necrotrophic interactions. Moreover, studies investigating the interactions of fungal isolates that elicit phenotypically distinct host symptoms have led to the discovery of numerous, disparate, processes by which fungi subvert host defenses (O’Connell et al., 2012; Haueisen et al., 2018). Indeed, transcriptome-based approaches have shown that fungi regulate both host and fungal developmental programs to penetrate and colonize plants (Soanes et al., 2012; Vollmeister et al., 2012), use a constellation of secreted effector molecules (Yang et al., 2013; Haueisen et al., 2018), and express small RNAs, which modulate host defense signaling (Jiang et al., 2017; Lee Marzano et al., 2018). However, each of these studies has focused on pinpointing candidates for virulence or aggressiveness based on the interaction between the pathogen and a single host. While obviously informative, the outcomes of these approaches have left significant gaps in our understanding as to how individual pathogens infect and/or cause disease in multiple hosts.
Fungal plant pathogens often have broad host ranges, highlighting their ability to cause substantial economic losses in global agricultural systems. Although most plant pathogens colonize only a narrow range of host plants, several fungi have broad pathogenic host ranges, with additional endophytic, host ranges that involving symptomless host penetration and development (Derbyshire et al., 2017). For example, Verticillium dahliae causes diseases on more than 400 different hosts, and although it is adapted to specific hosts to cause disease, it has a much larger asymptomatic endophytic host range (Malcolm et al., 2013). This endophytic host range was more recently discovered as the causal agent of soybean (Glycine max) sudden death syndrome (SDS), caused by Fusarium virguliforme (Kolander et al., 2012). This shows that individual fungi can manipulate their genetic expression programs to enable colonization of hosts with distinct pathogenic and endophytic outcomes. Studying fungal species with both broad symptomatic and asymptomatic host phenotypes provides an opportunity to understand the transcriptional reprogramming required to promote fungal colonization of hosts and disease development.
In silico comparative studies have provided sufficient resolution to differentiate disease-eliciting plant pathogen interactions from those that are primarily endophytic (Laluk and Mengiste, 2010; Lofgren et al., 2018). However, fungal ecology-based analyses in Botrytis, Verticillium, and Fusarium species suggest that host fungal interactions exhibit a continuum of molecular crosstalk. This results in a gradation of pathogenic to mutualistic outcomes when interacting with diverse hosts, as demonstrated (Malcolm et al., 2013; Demers et al., 2015; Shaw et al., 2016). Overall, these studies demonstrate that, at least in the case of the aforementioned species, fungi can fulfill two distinct ecological niches, potentially within the same community (Selosse et al., 2018). Exploring the genomes of fungi with broad host ranges has uncovered the genomic potential that enables them to occupy diverse ecological and pathogenic niches (Ma et al., 2010; Seidl et al., 2015; Derbyshire et al., 2017). We posit that a comparison of the underlying transcriptional processes regulating a pathogenic versus endophytic lifestyle will yield novel genetic signatures promoting virulence within a susceptible host.
F. virguliforme, the causal agent of soybean SDS, is an exceptional model for analyzing fungal-plant interactions due both to its broad host range and to the severe economic loss it causes in the soybean industry. This disease is a key limitation in reaching soybean yield potential, with an estimated annual economic impact of $330 million in the United States, partly stemming from limited effective disease management practices (Koenning and Wrather, 2010; Hartman et al., 2015). F. virguliforme is an ascomycete that colonizes the roots of more than 10 plant species, stimulating leaf chlorosis and root necrosis, resulting in the eventual loss of above ground biomass (Kolander et al., 2012). However, on many monocots and weed species, F. virguliforme colonizes roots with no observable deleterious phenotype in the host (Kolander et al., 2012; Kobayashi-Leonel et al., 2017). F. virguliforme is asymptomatic in maize (Zea mays) and, in the field, may form endophytic associations between crop rotations with soybean in the same agroecosystem. Given our lack of understanding of how F. virguliforme interacts with potential host plants (e.g., soybean and maize) and subsequently occupies distinct ecological niches, we performed a systematic comparison by investigating the host-pathogen transcriptomic interface during symptomatic and asymptomatic colonization. To highlight the divergence in genetic signaling pathways underpinning fungal lifestyle, we investigated (1) the early stages of colonization between F. virguliforme and soybean and maize; (2) how early transcriptional responses of F. virguliforme colonizing maize or soybean are regulated; and (3) the potential conservation and/or distinction between asymptomatic versus symptomatic fungal transcriptomes.
RESULTS
Generation of a High-Contiguity Reference Genome for F. virguliforme
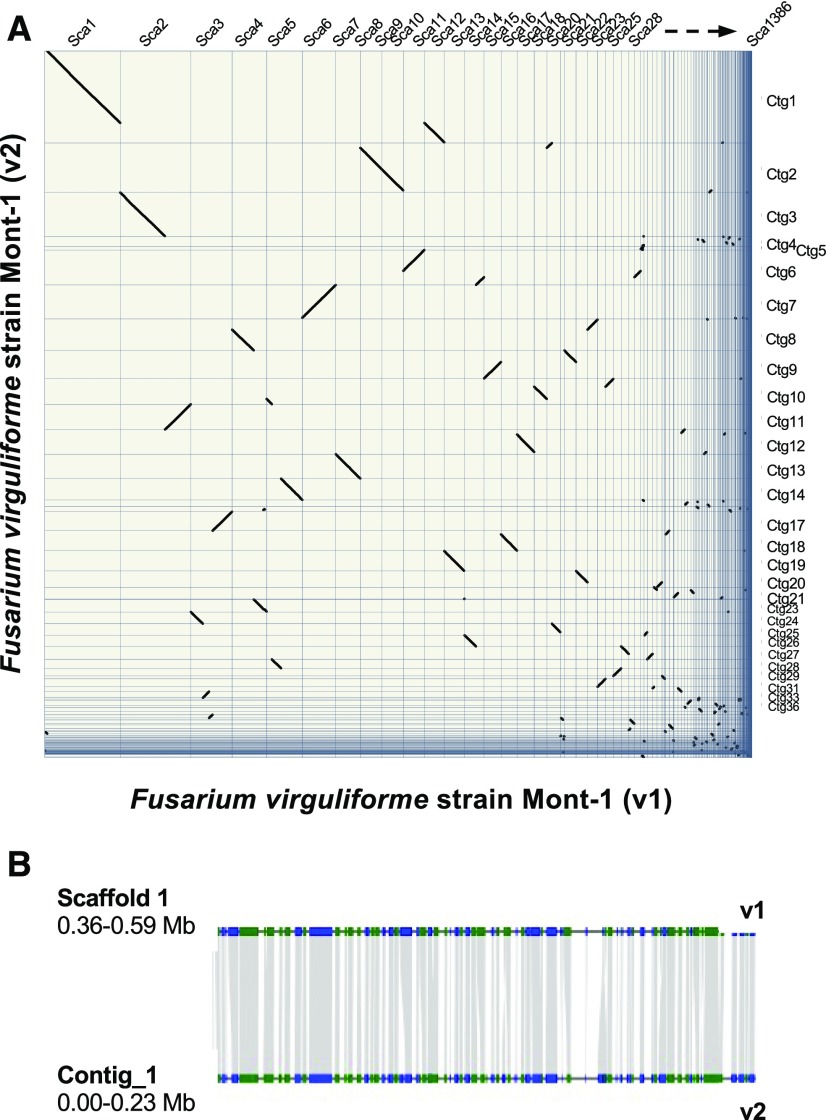
Although a draft genome of F. virguliforme is available (Srivastava et al., 2014), the current version is incomplete, thus limiting comparative and functional analyses. Therefore, we generated an improved pathogen reference genome using third-generation PacBio single-molecule sequencing technologies. We generated a high-quality F. virguliforme genome using ∼17× coverage of PacBio data (Supplemental Figure 1). Filtered reads were assembled using the long-read optimized assembler Canu (Koren et al., 2017), and resultant contigs were error-corrected using 50× Illumina data using Pilon (Walker et al., 2014). Our assembled F. virguliforme genome encompassed 52 MB with 96 contigs, with an N50 of 1.54 MB (Supplemental Table 1). The resultant genome size (Mb) was slightly larger than the version 1 (v1) draft assembly (Srivastava et al., 2014), and in this study, the contiguity and N50 were significantly improved (Supplemental Table 1). Synteny between the two genome versions was highly fragmented (Figure 1A), perhaps a result of having more than 3000 contigs in the first version of the genome. Notably, within syntenic regions, micro-collinearity between the two genomes was highly conserved (Figure 1B).
Figure 1.
Syntenic Regions between Genome Versions of Fusarium virguliforme.
(A) Plot of syntenic regions retained between genome version 1 (v1) and genome version 2 (v2). Diagonal lines, including differences in lengths, illustrate distances of overlap between F. virguliforme v1 and v2 genome versions, and the syntenic regions between the scaffolds (Sca) and contigs (Ctg) in each.
(B) Micro-collinearity between scaffold 1 of genome v1 and contig 1 of genome v2 connected by shaded gray areas. Regions containing genes are highlighted in green or blue, for forward or reverse orientation, respectively.
The F. virguliforme genome generated in this study was annotated using FunGAP (Min et al., 2017), incorporating AUGUSTUS, MAKER, and BRAKER gene model prediction algorithms (Stanke et al., 2006; Cantarel et al., 2008; Hoff et al., 2016). A total of 16,050 genes from the F. virguliforme version 2.0 (v2) genome were discovered, representing an increase of 1205 genes compared with version v1. Comparisons of the coding sequences between two genome versions revealed 12,306 conserved genes with a minimum 70% gene alignment rate and 95% identity. 1422 genes were considered as misassembled or incomplete in one of the genomes. Overall, 2889 new genes that were missing in v1 were annotated in v2 and were considered novel (Supplemental Data Set 1). This gene set was enriched with genes involved in protein ubiquitination, organic compound breakdown, and porphyrin compound biosynthesis (Supplemental Table 2). Next, the completeness of genome annotation was evaluated using Benchmarking Universal Single-Copy Orthologs (Simão et al., 2015; Waterhouse et al., 2018), and we observed an ∼98% completion, with 10,162 of the 16,050 genes being supported with protein evidence. Further exploration of the genome using SignalP (v4.1; Petersen et al., 2011) and EffectorP (v2.0; Sperschneider et al., 2016) discovered 232 genes that were candidate effectors (Supplemental Data Set 2). Because F. virguliforme is a hemibiotrophic pathogen, we also searched the F. virguliforme genome for genes encoding carbohydrate active enzymes (CAZymes) and discovered 365 genes with potential functions in carbohydrate metabolism (Supplemental Data Set 3). In total, these data sets provided a resource to explore the transcriptomic variability of F. virguliforme across hosts.
Fungal Infection by F. virguliforme Produced Different Root Phenotypes on Maize versus Soybean
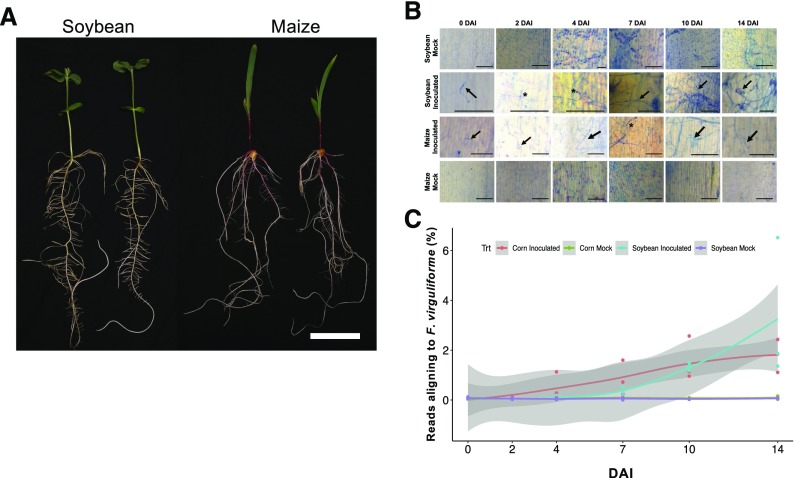
To understand transcriptome dynamics of F. virguliforme interactions with maize (asymptomatic) versus soybean (symptomatic), we profiled F. virguliforme–infected roots over a 2-week time course and collected samples for RNA-sequencing (RNA-seq) analysis. A survey of early postinoculation time points allowed us to characterize the continuum of fungal attachment, growth, penetration, differentiation, and symptom development. By the end of the 2-week time course, soybean roots showed signs of necrotrophy in both the tap and hypocotyl regions (Figure 2A). Additionally, fungal-induced root necrosis had spread to developing lateral roots adjoining the tap root. This type of symptom development is consistent with root disease progression of SDS, which begins as an asymptomatic biotrophic interaction, with the fungus depending on living plant tissue, but then turns necrotrophic, with the fungus eventually killing host tissue. In the asymptomatic host, maize, we did not observe any striking evidence of root chlorosis or necrosis over the 14-d time course of the experiment (Figure 2A).
Figure 2.
Fusarium virguliforme Pathogen Assays on Soybean and Maize.
(A) Plant growth and development of soybean and maize 14 DAI with F. virguliforme. Representative images show uninoculated (Mock) and F. virguliforme-inoculated (Inoc) plants. Bar = 4 cm.
(B) Trypan blue staining of inoculation sites on soybean cv Sloan and maize cv E13022S roots either inoculated with F. virguliforme or mock inoculated. Bars = 16 mm. Arrows point to areas of fungal growth and development; asterisks highlight appressoria-like structures.
(C) The percentage of unique RNA-seq reads aligned to F. virguliforme genome v2. Reads were trimmed by Trimmomatic v0.33 and aligned to Fusarium virguliforme genome v2 with HISAT2 v2.1.0. Each sample is indicated by a colored dot, and lines represent the mean of three biological replicates. Gray shade indicates SEM.
To monitor in planta fungal growth, we used trypan blue staining to visualize fungal hyphae on both soybean and maize roots throughout the time course. Although fungal growth and colonization were apparent in both hosts, the developmental stage varied depending on the host plant (Figure 2B). For example, following inoculation, fungal spore germination was apparent on both hosts, and by 2 d after inoculation (DAI), fungal mycelia had expanded across the root surface. Interestingly, mycelia on maize roots grew parallel to root epidermis cells, whereas mycelia growth on soybean roots did not have any apparent directional pattern of colonization. Also, by 2 DAI, round and swollen mycelial structures were observed on soybean roots, and these structures resemble penetration structures (e.g., appressoria). Support for this classification comes from documented observations of infection pegs and appressoria development during in vitro F. virguliforme infection of soybean radicals (Navi and Yang, 2008). Interestingly, these infection-like structures were also observed on maize, but not until ∼7 DAI, indicating a slower infection process. From 7 to 14 DAI, we continued to record fungal growth and development at the site of inoculation, and we observed an increase in colonization by mycelia on both hosts. By 14 DAI, however, masses of developing macroconidia were apparent on soybean roots, but not on maize roots, indicating that asexual reproduction had initiated in the symptomatic host. The transition to necrotrophy was indicated by the induction of discoloration of soybean roots at 7 DAI, followed by necrosis at 10 DAI (Figure 2B). In maize, no visible symptoms were observed throughout the time course of the experiment.
After we confirmed in planta growth of F. virguliforme on both soybean and maize, we conducted RNA-seq at six selected time intervals over the course of the infection. Additionally, we also collected samples of F. virguliforme macroconidia spores that were generated via in vitro germination. After lower quality reads and adaptors were trimmed, reads were mapped to the F. virguliforme genome v2. In our initial analysis, we identified low levels of fungal mRNA reads, representing only 0.04% to 0.13% of the total reads at 0 to 2 DAI (Figure 2C). While this was not unexpected, we generated a minimum of 200 million reads per sample (at 0 to 4 DAI), yielding read counts greater than 80,000 per biological replicate (Supplemental Figure 2; Supplemental Table 3). As expected, the percent of mRNA reads aligning varied by host over the time course. Fungal reads from maize (the asymptomatic host) increased in a linear fashion over the time course, ranging from 1.11% to 2.53% of the total reads at 7 to 14 DAI. However, fungal reads from soybean (the symptomatic host) did not increase substantially until 7 DAI (Figure 2C). At this point, the percent of fungal reads approached those observed from F. virguliforme–inoculated maize samples. In soybean, this increase coincided with both symptom development and a concomitant shift from biotrophy to necrotrophy.
Host-Induced Gene Expression Profiles in F. virguliforme
To determine whether the colonization profile of F. virguliforme differed in a manner consistent with the differing host phenotype, we used a comparative transcriptomic-based approach. We hypothesized that this approach would better position us to define the transcriptional reprogramming specific to each host. Additionally, as a function of a single, common pathogen interaction, we expected that this would also reveal the influence of the host response on fungal gene expression. First, to determine whether pathogen treatments were globally distinct from one another, we performed a principle coordinate analysis of all 39 samples from maize and soybean colonization assays, as well as samples from in vitro assays of germinating macroconidia. Using this approach, we observed that fungal responses were primarily correlated with treatment (Supplemental Figure 3) and that the germinating macroconidia formed a distinct group from samples colonizing hosts. While F. virguliforme response on hosts did form a single group, all samples were distinctly separate within this group as a function of host. Intriguingly, gene expression from both hosts were separated by time as well, with the greatest separation identified at time points between 4 and 14 DAI. Additionally, a separation from the plant samples was apparent in reads derived from 7 to 14 DAI samples of F. virguliforme–infected soybean. The grouping of samples by hosts suggests plant-fungal interactions greatly shaped F. virguliforme gene expression.
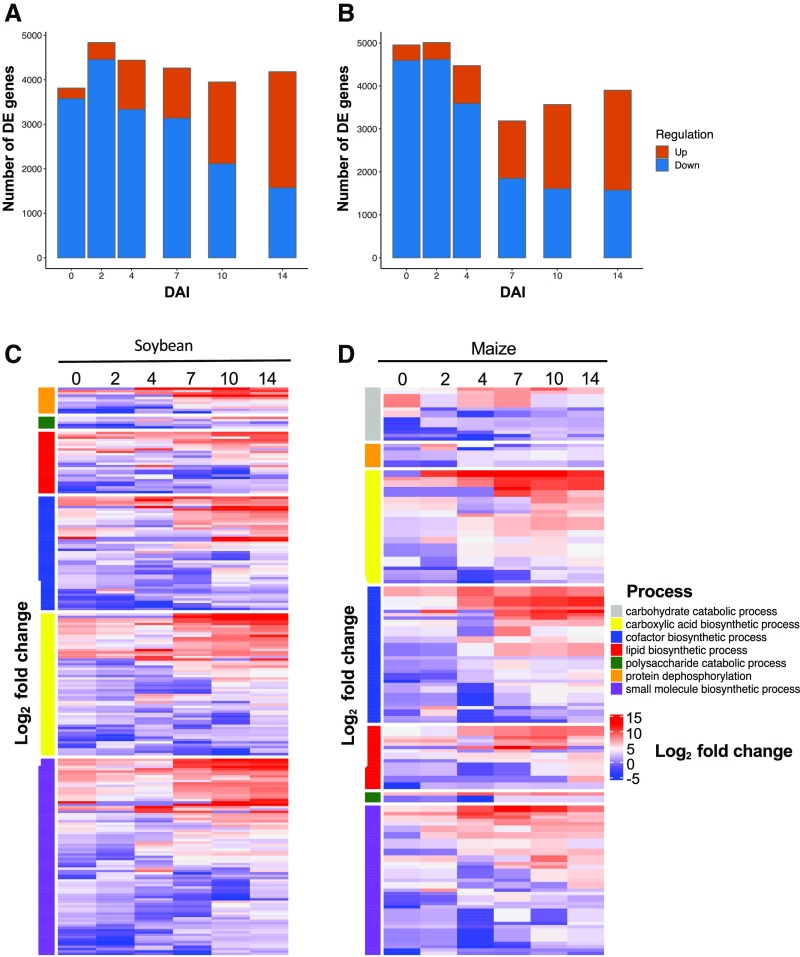
To discover F. virguliforme genes that were induced by host interaction, we next compared gene expression patterns of F. virguliforme on soybean or maize with in vitro germinated macroconidia. Using this approach, we identified 4192 and 4072 unique F. virguliforme genes that were differentially upregulated (log2 fold change > 1) in fungal samples from soybean and maize, respectively, throughout the time course. As many genes were highly induced, we filtered the differentially expressed genes (log2 fold change > 2)| to 3171 and 3010 genes to discover processes relevant to soybean or maize colonization, respectively. Of the significantly upregulated genes from F. virguliforme on soybean, the vast majority of genes were induced at 0, 7, 10, and14 DAI (Figure 3A). Similarly, the majority of induced F. virguliforme genes within maize roots were from samples derived at 7, 10, and 14 DAI (Figure 3B). Surprisingly, while the read depth was less in maize samples at 0 DAI than soybean samples, an additional 127 genes were detected as significantly upregulated at log2 fold change > 2, highlighting an elevated response in F. virguliforme to maize versus soybean. The greatest changes of unique gene upregulation occurred between 4 and 7 DAI on maize, and at 10 to 14 DAI on soybean (Supplemental Table 4). As a function of expression between both hosts and comparison of host differential gene expression between time points, we observed that only three genes were conserved. Of these three genes, one (Fvm1_12746) was functionally annotated as phosphonopyruvate decarboxylase, a component of organic acid production in fungi (Yang et al., 2015a).
Figure 3.
Temporal Expression Patterns of F. virguliforme Response Genes in Soybean and Maize Hosts versus in Germinating F. virguliforme Macroconidia.
(A) and (B) Number of differentially expressed (DE, log2(FC) > 2) F. virguliforme genes in roots of soybean (A) or maize (B) hosts versus geminating F. virguliforme macroconidia.
(C) and (D) Heat maps of significant (log2(FC) > 2) enrichment of gene ontology categories of upregulated F. virguliforme genes across pooled time points during F. virguliforme colonization of soybean (n = 233; [C]) and maize (n = 165; [D]).
We used a functional gene ontology (GO) enrichment analysis to explore the function of host induced genes in F. virguliforme. Of the more than 50 biological process categories that were enriched, several processes were consistently upregulated in both soybean and maize, including carboxylic acid, lipid, or cofactor biosynthesis, as well as polysaccharide metabolism, protein dephosphorylation, and small-molecule biosynthesis (Figures 3C and 3D; Supplemental Data Sets 4 and 5). Overall expression patterns on both hosts were similar for lipid and cofactor biosynthesis, which is not surprising, as these processes are critical for fungal growth and signaling pathways (Schrettl et al., 2007; Lysøe et al., 2008). Carboxylic acid biosynthesis was induced throughout the colonization time course, and we hypothesize that this is a critical process for secondary metabolite production in support of fungal colonization regardless of the host (Brown and Proctor, 2016). Interestingly, protein dephosphorylation and small molecule biosynthesis were enriched in fungal transcriptomic profiles and were elevated in expression when colonized soybean roots at 7 to 10 DAI.
Temporal Divergence of Gene Coexpression Upon Host Colonization by F. virguliforme
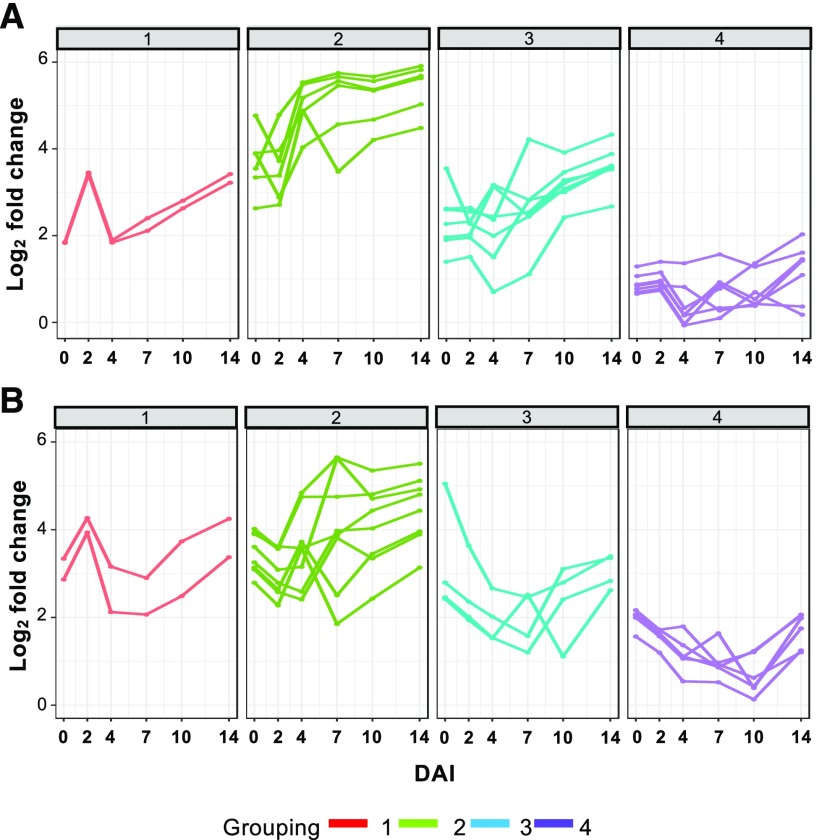
As noted above, specific genes related to processes associated with F. virguliforme colonization and development were differentially induced at distinct temporal stages during the time course. To extend our investigation, we were interested in the divergence of global gene coexpression patterns during colonization of both maize and soybean. Differential coexpression patterns could reflect F. virguliforme transcriptomic dynamics stemming from biotrophic to necrotrophic transitions during infection. To address this, we applied a weighted gene correlation network analysis on the F. virguliforme expression data collected from maize and soybean roots. Expression data sets were filtered to remove genes with low expression before construction of the coexpression network, leaving 11,112 genes after filtering. Next, we built individual networks for maize and soybean RNA-seq data, and these genes were clustered into 22 modules for F. virguliforme colonization of maize and 20 modules for F. virguliforme colonization of soybean (Supplemental Figures 4 and 5; Supplemental Data Sets 6 and 7). Then, we assigned the modules to four large groups based on their temporal patterns: (1) early induced expression at 2 DAI, but downregulated at 4 DAI; (2) elevated expression at 4–7 DAI; (3) induced expression at 7–10 DAI; and (4) downregulation of expression from 2 to 4 DAI but induced at 10 DAI (Figure 4). While these temporal patterns of coexpression modules across F. virguliforme colonization appear similar when placed within these four large groups, the gene enrichment of modules contained within these groups varied significantly by host.
Figure 4.
Distinct Gene Coexpression Groups of Host-Induced Fusarium virguliforme Response Genes.
(A) and (B) Temporal expression profile of F. virguliforme gene coexpression modules during colonization of maize (A), or soybean (B) roots across the time course. Modules were grouped into four distinct expression patterns: (1) upregulation at 2 DAI; (2) upregulation at 4–7 DAI; (3) induction at 7–10 DAI; and (4) increase in expression at 10–14 DAI. For each time point, the averaged expression of genes contained within a given module was plotted.
Gene coexpressing modules in Group 1 from the F. virguliforme–maize interaction network were enriched for putative negative regulatory elements for many functional processes, including cellular metabolism, macromolecule production, and expression of primary metabolism (Supplemental Data Set 6). This indicates that F. virguliforme was repressing secondary metabolism and using self-derived energy at early interaction times with maize roots. However, processes upregulated in F. virguliforme colonizing soybean roots at 2 DAI were enriched for reactive oxygen species (ROS) generation and oxalic acid production (Supplemental Data Set 7). Both of these processes are associated with early hemibiotrophic and necrotrophic plant fungal interactions at early time points in colonization. ROS in fungal hyphae supports the differentiation of cells for infection structures like appressoria (Heller and Tudzynski, 2011). We observed the development of appressoria like structures at 2 DAI in soybean roots (Figure 2B), suggesting that F. virguliforme is already penetrating host tissues within 48 h of contact. Oxalic acid biosynthesis genes were also enriched in the assembled modules, indicating a potential downregulation of host cell death by autophagy in order to prevent a massive necrotic response by the host, killing the fungus, similar to what was seen with Sclerotinia sclerotium (Veloso and van Kan, 2018). Taken together, these analyses suggest that F. virguliforme infects and manipulates the soybean host responses as early as 2 DAI.
Numerous coexpression modules were upregulated at 4–7 DAI in both maize- and soybean-inoculated with F. virguliforme. Interestingly, most of these modules were also highly expressed for the remainder of the colonization time course (Figure 4A; Supplemental Figure 4) and were further enriched for processes associated with primary metabolism, similar to Group 1 (Table 1). However, processes associated with response to the host defenses were also enriched. For example, at 4 DAI, carboxylic acid biosynthesis-associated genes and related processes were upregulated, suggesting that toxin production was occurring (Supplemental Data Set 6). Conversely, the same processes from F. virguliforme within soybean highlighted a faster colonization program. Enrichment of processes associated with cellular catabolic processes of cellulose, pectin, and polysaccharides in the soybean infection samples were identified. Of interest and relevance to the host-association and symptomatic nature of the F. virguliforme–soybean interaction, we also observed an enrichment at 4 DAI in small molecule biosynthesis, including those potentially associated with the function of necrotrophic effectors (Supplemental Data Set 7; Chang et al., 2016a). In total, these observations highlight the initial transition from a biotrophic to necrotrophic lifestyle, coincident with the modification and breakdown of host tissue to enable further proliferation (Laluk and Mengiste, 2010).
Table 1. Selected Gene Ontology Enrichment of Distinct Gene Coexpression Groups Identifies Manipulation of the Fusarium virguliforme Transcriptome for Host Colonization.
| Grouping | Host | |
|---|---|---|
| Maize | Soybean | |
| 1 | Negative regulation of biological processes | ROS |
| Oxalic acid production | ||
| 2 | Primary metabolism |
Catabolic processes of cellulose |
| Defense to host | Small molecule biosynthesis | |
| 3 | Amino acid sugar catabolism |
Antibiotic catabolic process |
| Oxidation-reduction process | Response to ROS | |
| 4 | Nitrate transport |
Killing of cells of other organism |
| Amino acid sugar catabolism | Cell redox homeostasis | |
A full list of coexpression module gene ontology enrichment within formulated groups is provided in Supplemental Data Sets 6 and 7.
One striking observation from this analysis is that numerous diverse processes were enriched in modules upregulated at 7–10 DAI in F. virguliforme–maize samples. Of these, the upregulation of NADP stood out, as this process has been previously associated with hyphal differentiation initiation for infection structures (Heller and Tudzynski, 2011). This is consistent with our phenotypic observations shown in Figure 2B, at 7 DAI. The upregulation of gene processes in F. virguliforme associated with catabolism of amino acid sugars also suggests access to plant-derived compounds, likely via direct penetration of the host tissue by the fungus. Concomitant with this, upregulation of processes associated with chemical stimulus likely indicates F. virguliforme was sensing host defense response involving the production and secretion of antimicrobial compounds. During this same time frame, while F. virguliforme from maize was activating nutrient access-associated processes, F. virguliforme upregulated protection-associated mechanisms in soybean, including antibiotic catabolism, response to ROS, and chemical to host defense activation.
By 14 DAI, processes associated with Group 4 (Figure 4) were expressed as a function of host colonization. For example, processes involving primarily amino acid, sugar, and nitrate acquisition were induced in samples derived from maize. However, at the same time point in samples from soybean, necrotrophic processes had initiated, with an enrichment in functions associated with cell killing, organic acid transport, and self-protection from host induced ROS by cell redox homeostasis. These process enrichments were supported by the observed necrotrophic envelopment of the soybean tap root at 14 DAI (Figure 2B).
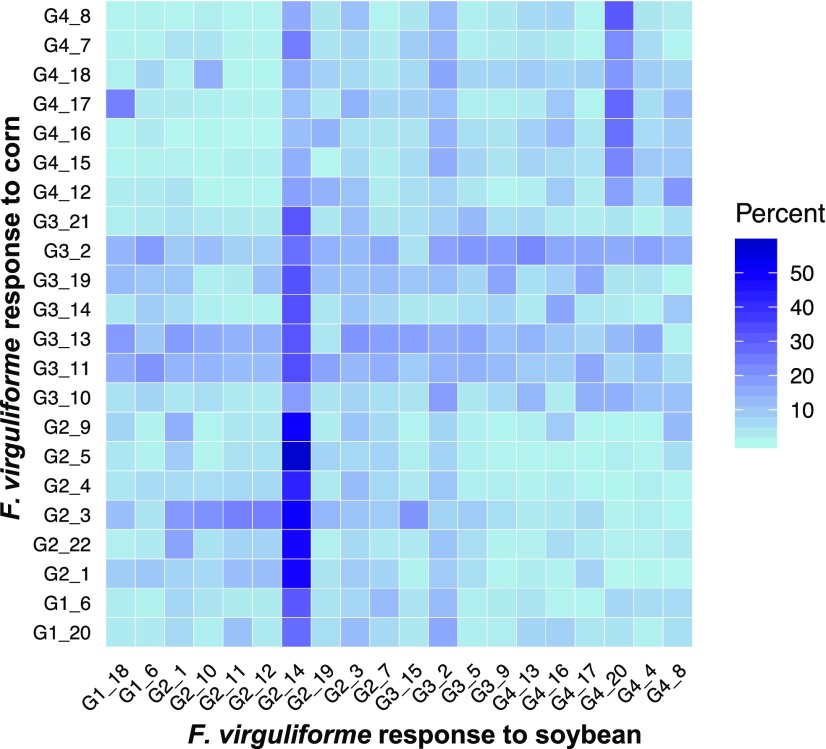
The lack of temporal conservation of enriched processes between colonization of these two hosts highlights plasticity of the F. virguliforme transcriptome. Overall, few genes were coexpressed in a similar manner within modules when compared between these two hosts (Figure 5). Moreover, only 8% of genes were conserved between coexpression networks of F. virguliforme–inoculated soybean and maize. Comparison of gene overlap highlights that processes enriched in group 3 of coexpression in maize contain more genes from the temporal of F. virguliforme across all soybean coexpression groups. Interestingly, Module 14 in Group 2 of F. virguliforme within soybean contained the greatest overlap with several early (2 to 4 DAI) induced maize modules. Module 14 was the largest coexpression module, with 3503 genes, and it may contain many genes relevant to basic cellular functions needed for viability and growth (Supplemental Figure 5; Supplemental Data Set 7).
Figure 5.
Symptomatic and Asymptomatic Hosts Reveal Transcriptomic Plasticity during F. virguliforme Colonization.
The percent of overlapping genes from the weighted gene correlation network analysis modules was calculated as the ratio of the number of shared genes between F. virguliforme expression on each host divided by the total number of genes within the module that contained the fewest number of genes. Modules are annotated with grouping assignments as shown in Figure 4. The increasing shade of blue represents increasing percent module overlap of the gene count shared between the two modules.
Host-Specific Gene Expression Patterns During Root Colonization
As noted above, we observed a temporal divergence of biological processes enriched by respective host colonization. To further explore this, we next asked if these induction responses were host specific. Previous work comparing the infection profiles of phytopathogenic Zymoseptoria tritici on wheat revealed temporal variation of isolate infection (Haueisen et al., 2018). To determine whether this is also the case in F. virguliforme, we directly compared F. virguliforme gene expression from each host at each time point (Figure 6A). The majority of genes (81%; 9002) in the F. virguliforme transcriptome were not differentially regulated in cross-species colonization. Of the proportion of genes that were differentially induced, 43% (924 genes) were uniquely upregulated during maize root colonization, 56% (1186 genes) were uniquely upregulated upon colonization of soybean, and 0.1% were consistently upregulated at multiple time points during the infection time course. Of these differentially induced genes, the vast majority were induced at 10 and 14 DAI (Supplemental Table 5; Supplemental Data Set 8). While fewer genes were differentially upregulated at early time points, these genes highlight specific processes underlying temporally distinct stages of fungal colonization. Interestingly, genes highly upregulated (log2 fold change > 20) at 0 DAI within F. virguliforme colonizing soybean roots were related to DNA methylation, suggesting that this process was induced by host signals in soybean roots. No biological processes were uniquely enriched during maize root colonization at 0 DAI. It is likely F. virguliforme began to respond to host-induced antimicrobial metabolites at 2 DAI by upregulating ABC transporters (Gupta and Chattoo, 2008) and by initializing toxin secretion using terpene synthases as the fungus grew in the maize roots. Based on the unique set of upregulated genes during soybean colonization at 2 DAI, F. virguliforme was penetrating roots via ROS production and upregulation of Zn(II)-Cys6 fungal transcription factors (Brown et al., 2007).
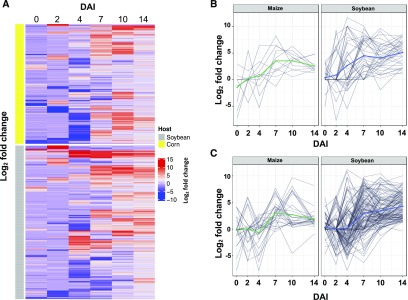
Figure 6.
Unique Host Genes Induced within Fusarium virguliforme Highlight Disease Development.
(A) Heat map of expression profiles of significantly upregulated genes (log2(FC) > 1) at a single time point in F. virguliforme colonizing soybean or maize (n = 2099). Yellow color indicates differentially upregulated genes from F. virguliforme colonization of maize, and gray color indicates differentially upregulated genes from F. virguliforme colonization of soybean.
(B) Expression patterns of significantly upregulated candidate effector genes at a single time point in maize or soybean over the infection time course. The colored lines indicate the means of all genes in the plot. Gray lines represent individual genes.
(C) Expression patterns of significantly upregulated candidate carbohydrate active enzymes at a single time point in maize or soybean over the infection time course. The colored lines indicate the means of all genes in the plot. Gray lines represent individual genes.
F. virguliforme colonization of soybean roots resulted in the activation of marked fungal defense signals at ∼4 DAI, as indicated by the rapid and strong induction (log2 fold change > 10) of various cytochrome oxidase genes. Interestingly, these genes were not upregulated at the same time in samples derived from F. virguliforme–maize colonization, suggesting that the fungus had not penetrated the root, and/or a lack of antimicrobial metabolite accumulation. However, at 7 DAI, cytochrome oxidases and NADP was upregulated in F. virguliforme maize interactions, thus indicating cellular differentiation of hyphal penetration structures (Heller and Tudzynski, 2011). At the same time, cellular degradation and nutrient access-associated processes were significantly upregulated (log2 fold change > 10) in F. virguliforme–colonized soybean samples, as indicated by the expression of glycoside hydrolases and pectinases, as well as various nutrient transporters. A larger set of genes was differentially induced in F. virguliforme between hosts at 10 and 14 DAI. At these time points, samples from maize revealed an enrichment of genes associated with secretion and catabolic processes (Supplemental Data Set 9), while those from soybean revealed a shift to processes indicative of fungal growth (e.g., glycerolipid and lipoprotein biosynthesis; Takahashi et al., 2009). This is in support of our observation of asexual production at 14 DAI upon soybean roots (i.e., Figure 2B).
Central to defoliation of the host during F. virguliforme infection is the secretion of proteinaceous phytotoxins produced by the pathogen, resulting in host foliar SDS symptom development (Chang et al., 2016a). In this study, genes involved in toxin and secreted protein production were induced in a time-dependent manner, as were their levels of expression, when compared across hosts (Figures 6B and 6C). Moreover, while nearly triple the number of predicted effector-encoding genes were upregulated at 0–2 DAI in soybean, none of the candidate effector-encoding genes were differentially induced between hosts at these time points. By 4 DAI, almost four times as many candidate effectors were upregulated in soybean roots (Supplemental Figure 6), including those with predicted functional domains associated with pectin lyases, glycoside hydrolases, and necrosis-inducing proteins. However, similar to the above patterns, all but three genes were not considered differentially expressed when compared with F. virguliforme colonization of maize (Supplemental Data Set 10). Until this point in the infection process, candidate CAZymes expression profiles were induced in similar patterns in the two hosts and no differentially expressed CAZymes genes were detected at 0, 2, and 4 DAI (Supplemental Figure 7). However, at 4 DAI, pectin lyases and glycoside hydrolases were expressed at much higher levels (>10 log2) by F. virguliforme in soybean roots (Supplemental Data Set 11). This trend was exacerbated at 7 DAI, with numerous CAZymes and effectors related to pectin lyases being upregulated in soybean roots colonized by F. virguliforme. This suggests divergence in fungal colonization programs between maize and soybean at 7 DAI, potentially stemming from the shift of biotrophic to necrotrophic, as visible symptoms started to initialize at 7 DAI. A necrotrophic lifestyle was evident by 10 and 14 DAI, with 14 and 28 candidate effectors, and 37 and 114 candidate CAZymes, respectively, targeting cellular breakdown of soybean roots by F. virguliforme. Conversely, few pectin lyases were expressed during F. virguliforme colonization of maize roots. We posit that reduced expression of lyases may stem from the basic physiological differences between maize and soybean. Indeed, monocot roots contain only 10% pectin, whereas eudicots contain up to 30% (Caffall and Mohnen, 2009). The effectors that were uniquely induced between 7 and 10 DAI in maize roots are currently putative effectors with no known function.
DISCUSSION
Comparative systems-based approaches using pathogenic and nonpathogenic fungal isolates have allowed us to identify genetic signatures associated with pathogenicity and compatible host-pathogen interactions. With the use of single-pathogen/single-host approaches, a more complete understanding of the continuum of pathogenic and endophytic niches determining host range is emerging. We used a high-resolution transcriptome-based approach to define how a single fungal pathogen can rewire infection processes and host defense programs to promote either symptomatic or asymptomatic colonization, depending on the host. To accomplish this, we assembled and annotated the F. virguliforme genome de novo, generated 39 mRNA transcriptomes across in vitro and in planta time courses to identify infection program modulation across two hosts—a symptomatic soybean host and an asymptomatic maize host. We found distinct changes in root phenotypes as a function of host during F. virguliforme colonization. For example, maize roots remain asymptomatic, whereas soybean roots turn chlorotic and eventually become necrotic. Underlying these phenotypic distinctions are myriad of host-dependent transcriptional programs.
In this study, we observed that temporal changes in transcriptional dynamics during F. virguliforme colonization of maize roots were largely gradual over the infection time course. Conversely, F. virguliforme colonization of soybean caused dramatic changes in transcriptional activity at 4–10 DAI, as exhibited by coexpression module gene enrichment. This is illustrated by our observation of signaling associated with small molecule secretion and host cell death processes. These observed changes are consistent with previously described transcriptional dynamics of hemibiotrophic plant pathogens (O’Connell et al., 2012). For example, small molecules secreted by plant pathogens are hypothesized to target host defense machinery and/or processes to modulation immune responses to the fungus (Jennings et al., 2000; Chang et al., 2016a). In parallel, dephosphorylation of plant plasma membrane-localized proteins by fungal pathogens is regarded as a critical process to prevent signaling cascades that normally stimulate host defense responses (Yang et al., 2015b). In total, the upregulation of these processes in F. virguliforme during soybean colonization suggests an infection strategy to reduce host immune responses, more so, than when F. virguliforme colonizes maize roots.
Interestingly, the identified differences in transcriptomes does not appear to be a result of unique gene expression by each host, but rather is a result of the temporal induction of genes with respect to the degree of host colonization. In support of this, gene coexpression networks highlighted the temporal processes unique to each host through varying stages of fungal growth, infection, and proliferation, each of which coincided with changes in pathogen access to nutrient sources. The rapid growth and infection of F. virguliforme on soybean roots by 2 DAI indicates a rapid recognition of the host surface and initiation of the early infection program (Elliott, 2016). However, F. virguliforme gene expression patterns on maize roots through the early time course of infection were enriched for negative regulation of biological processes and primary metabolism, suggesting that this fungus was not immediately stimulated to infect the maize host. Upregulation of processes indicative of maize infection did not occur until 7 DAI, illustrating a delay in host-fungal recognition (Giovannetti et al., 1994). In soybean, upregulation of recognition-associated processes occurred at 2 DAI.
By the time F. virguliforme had penetrated maize roots (∼7 DAI), infection on soybean had already begun to transition from a biotrophic lifestyle to a necrotrophic infection. Upregulation of small protein secretion and fungal-derived toxin production demonstrates host cell modification by F. virguliforme in soybean roots, key events associated with, and required for, nutrient access (Sahu et al., 2017). Throughout the remainder of the time course of infection, we observed a general increase in the gene expression associated with cell death and pathogenicity in F. virguliforme–infected soybean. Conversely, in maize, we observed the expression of fewer catabolic process-associated genes, indicating a general reduction in processes associated with nutrient acquisition. Based on these observations, we surmise that the comparative analysis of the interaction of F. virguliforme with two hosts supports our hypothesis of a divergence in the transcriptome of this fungus. Indeed, while the vast majority of the transcriptome was expressed during fungal colonization of both maize and soybean, the induction of genes underlying distinct, often divergent, biological processes were temporally distinct. This observation agrees with previous studies which identified temporal changes of gene expression during colonization of hosts by the same fungus exhibiting different lifestyles (Lahrmann et al., 2013; Lorrain et al., 2018), and this may suggest a reduction in a shift to necrotrophy on maize.
Because transcriptome expression of F. virguliforme varied during colonization of soybean versus maize, our study offers a unique perspective to identify processes critical for necrotrophy on soybean. For example, previous analyses concluded that the upregulation of genes at 4 DAI that encode effectors and CAZymes highlights the start of the transition from biotrophy to necrotrophy (Chang et al., 2016a; Chang et al., 2016b; Ngaki et al., 2016). In the current study, we also observed an increase in the expression of CAZyme- and pectin lyase-associated transcripts in soybean compared with maize. Moreover, expression of the Necrosis Inducing Protein was highly upregulated during soybean colonization over the time course of analysis, suggesting a possible dicot-specific response, as previously hypothesized by Bae et al. (2006). Indeed, fungal effectors and CAZymes were expressed in temporally distinct waves at 2, 4, and 7 DAI in soybean-infected roots, yet not in maize. Moreover, each wave increased in intensity of gene expression. In total, these expression patterns agree with previous data, further supporting the hypothesis that the upregulation of cell-degrading and necrosis-inducing peptides is a key step in the shift from biotrophy to necrotrophy (Kleemann et al., 2012; Haueisen et al., 2018).
While a hemibiotrophic infection program ensued in soybean, infection was delayed, and catabolic activities, as inferred by transcriptional analysis, were lower when F. virguliforme colonized maize. Either a lack of host recognition by F. virguliforme (Giovannetti et al., 1994) or an upregulation of host defenses from pattern triggered immunity (Bagnaresi et al., 2012; Zhang et al., 2018) would slow fungal growth and downregulate development. Once inside the host, fewer effectors and CAZymes were uniquely expressed in maize roots and were, more often than not, downregulated after induction. In total, the lower level of expression, along with the decrease in CAZyme diversity, suggests that the cellular environment within maize roots did not stimulate prolific upregulation of necrosis-inducing peptides. This may stem from the physiological differences in cell structure between monocots and dicots (Caffall and Mohnen, 2009). Additionally, as the primary hosts for F. virguliforme are legumes, F. virguliforme may not be as adapted to colonize monocots (Zhao et al., 2013).
The full understanding of how processes that are required for, and regulate, the transition from biotrophy to necrotrophy during the hemibiotrophic stage of fungal growth are regulated has remained elusive (Rai and Agarkar, 2016; Chowdhury et al., 2017; Haueisen et al., 2018). However, it is hypothesized that transcription factors likely play a critical role in these transitions, including recent work from several groups that has shown that members of the Zn(II)-Cys6 and C2H2 zinc-finger family of transcription factors alter pathogenicity and growth (Chen et al., 2017; Sang et al., 2019). In this study, we found that several Zn(II)-Cys6 genes were uniquely upregulated during early soybean colonization, a process we hypothesize may lead to an enhancement of pathogenicity of F. virguliforme on soybean. Our current study points to several key processes (e.g., transcriptional regulation of pectin lyase biosynthesis genes and fungal toxin biosynthesis genes) that are specifically associated with the lifestyle transition from biotrophy to necrotrophy in association with soybean. Conversely, these same transcriptional transitions were significantly reduced in F. virguliforme when associated with the asymptomatic host, maize.
We propose that these gene expression profiles highlight the transcriptional plasticity of a single fungal isolate on multiple hosts. In this regard, the analysis highlights the significance of rewiring during host-pathogen interactions, including the temporal expression of distinct gene networks underpinning the development of asymptomatic and symptomatic programs. While additional work remains, this study illustrates the significance of two distinct niches within agroecosystems of this important pathogen (Rai and Agarkar, 2016): one niche that supports the maintenance and survival of a pathogen in the absence of a compatible host, and another that supports proliferation and spread of a pathogen resulting in significant yield losses of an important staple crop. The ability of F. virguliforme to function in these two distinct roles suggests the need to consider the genomic potential and ability of plant pathogens to express a gradation of transcriptional programs, which in turn, imparts lifestyle plasticity on a broad range of hosts.
METHODS
Genome Sequencing, Assembly, and Annotation for Fusarium virguliforme
PacBio and Illumina sequencing were performed using high molecular weight DNA extracted from lyophilized (FreeZone 2.5, Labconco) Fusarium virguliforme Mont-1 mycelia grown for 4 weeks in potato dextrose broth (Millipore-Sigma; catalog no. P6685). The F. virguliforme Mont-1 isolate has been extensively used as a model for the advancement of our understanding of soybean (Glycine max) SDS, including serving as a model for genomics and transcriptomics (Srivastava et al., 2014; Ngaki et al., 2016; Sahu et al., 2017) for the analysis of pathogen effector biology (Chang et al., 2016a; Chang et al., 2016b). DNA was extracted using a modified cetyl trimethylammonium bromide procedure, with 1% polyvinylpyrrolidone (Lade et al., 2014). A PacBio library was constructed at the University of Georgia Genomics and Bioinformatics Core and size selected for 15- to 20-kb fragments using the BluePippin system (Sage Scientific). The library was sequenced on a Sequel Platform, and the single smart cell yielded 6.5 Gb of read data.
For error correction, Illumina TruSeq Nano DNA libraries were prepared and sequenced on an Illumina MiSeq v3 for a lane of 2 × 300 nucleotides and HiSeq 4000 for a lane of 2 × 150 nucleotides at Michigan State University Research Technology Support Facility. PacBio reads were assembled and error corrected using Canu (v1.8; Koren et al., 2017) using default parameters with several modification including: minReadLength = 2000, GenomeSize = 51Mb, minOverlapLength = 1000. A default max error rate of 0.24 was used for alignment during error correction and 0.045 for overlap and assembly. The max target coverage was set at 40×. A k-mer size of 16 was used for overlapping during error correction, and a k-mer size of 22 was used for overlapping during assembly.
The genome size estimate for assembly was extrapolated from the previous F. virguliforme Mont-1 draft genome of 50.9 Mb (Srivastava et al., 2014). Due to the presence of contaminating bacterial DNA in the initial assembly, draft contigs were compared with the published F. virguliforme genome assembly with LAST (v912; Kiełbasa et al., 2011), and novel contigs were validated for fungal origin by BLAST+ (v2.2.30; Camacho et al., 2009) against the nonredundant National Center for Biotechnology Information (NCBI) database. The genome graph structure was visualized in Bandage (https://academic.oup.com/bioinformatics/article/31/20/3350/196114; Wick et al., 2015) to survey contiguity and ambiguities (Supplemental Figure 1). Assembled contigs were error-corrected using Pilon (v1.22; Walker et al., 2014) and default settings, using a total of 50× coverage of Illumina paired-end 300 nt and 150 nt data for F. virguliforme. Paired end reads were adaptor and quality trimmed using Trimmomatic (v0.33; Bolger et al., 2014) and then were aligned to the draft contigs using Bowtie2 (v2.2.6; Langmead and Salzberg, 2012) with default settings. Pilon was run five times sequentially until limited (i.e., <100) corrections were found. The new genome assembly was compared to the previous genome assembly by QUAST (v3.0; Gurevich et al., 2013) and is referred to as F. virguliforme genome v2.
Transcript evidence for gene predictions was acquired from both the NCBI (Short Read Archive [SRA] SRR1382101) and germinating macroconidia from the F. virguliforme RNA-seq time course (see below). Reads were adaptor and quality trimmed using Trimmomatic (v0.33; Bolger et al., 2014) for all transcript evidence. These reads were then analyzed using Breaker, MAKER, and Augustus gene model prediction algorithms housed within FunGAP (v1.0; Min et al., 2017) as transcript evidence. The parameters for running FunGAP were set as –sister_proteome: Fusarium,–augustus_species fusarium_graminearum, with transcript reads provided as–trans_read_single. The resulting annotation from FunGAP consisted of 16,050 genes. Single-copy fungal orthologs from Benchmarking Universal Single-Copy Orthologs (v3; Simão et al., 2015) were used to assess the completeness of the genome annotation.
Functional annotation was completed using Trinotate (v3.1.1; Bryant et al., 2017). Trinotate-annotated gene models with evidence from several databases (NCBI nonredundant protein database, Swissprot-Uniprot database, GO, and InterpoScan) with BlastX finding single hit at an E-value threshold of 1E−5 (Altschul et al., 1990). We used this information to predict protein domains with HMMER (v3.1; Finn et al., 2011), transmembrane proteins with TMHMM (v2.0; Krogh et al., 2001), rRNA with RNAmmer (v1.2; Lagesen et al., 2007), secreted proteins with SignalP (v4.1; Petersen et al., 2011), and GO with GOseq (Young et al., 2010). Additionally, EffectorP (v2.0; Sperschneider et al., 2016) was used to predict fungal effectors within the secreted proteins, and dbCAN (Yin et al., 2012) was used to identify F. virguliforme CAZymes and secondary metabolism genes.
Comparative Genomics with F. virguliforme
MCSCAN toolkit (v1.1; Wang et al., 2012) was used to identify syntenic gene pairs between the second version (v2) and the first version (v1) of the F. virguliforme genome. Conserved gene blocks were discovered through LAST alignment. Plots of macro- and micro-synteny were created by the MCScan in python.
To discover novel and retained genes, the v2 F. virguliforme genome was compared to the v1 F. virguliforme genome. Coding sequences for gene models were extracted from the v1 genome by gffread (Trapnell et al., 2010). Next, gene sequences were reciprocally compared by BLASTn (v2.2.26; Altschul et al., 1990). Genes with an e-value below 1E−5, greater than 70% gene alignment, and 95% gene identity were classified as retained genes. If a gene was aligned with 95% identity with an e-value below 1E−5, but less than 70% gene alignment, it was denoted as a misassembled gene. Genes that were annotated in v2 that did not have an alignment to the v1 genome were considered novel.
Plant and F. virguliforme Assays
Soybean cv Sloan (provided by Martin Chilvers, Michigan State University), and maize (Zea mays) hybrid E13022S (Epley Brothers Hybrids Inc, provided by Martin Chilvers) were surface sterilized for 30 s in 70% (v/v) ethanol for 30 s and 10% (v/v) bleach for 20 min, and then triple rinsed in sterile distilled water for 1 min. After sterilization, soybean seeds were placed inside a Petri dish containing two sheets of sterile 100-mm Whatman filter paper soaked in 5 mL of sterile water. Soybean seeds were incubated for 5 d in total darkness at 21°C to enable germination. Maize seeds were incubated in sterile water for 24 h in darkness and placed between two sheets of sterile 100 mm Whatman filter paper with 5 mL of sterile water inside a Petri dish. Seeds were incubated for 5 d in total darkness at 21°C to ensure germination.
F. virguliforme Mont-1 was propagated on potato dextrose agar (Difco) for 7 weeks. Spores of asexual macroconidia were collected, diluted to 105 macroconidia mL−1 in sterile water, and sprayed onto germinated maize or soybean seedlings with an elongated a radicle using a 3-oz travel spray bottle. Twenty-five sprays were applied to the seedlings at angles of 0°, 90°, 180°, and 270° to that ensure seeds were thoroughly inoculated. For mock inoculated samples, sterile distilled water was sprayed onto the seedlings. After inoculated seedlings were incubated for 30 min, excess inoculum was removed, and seedlings were incubated for an additional hour. Following incubation, three maize or soybean seedlings were placed into sterilized seed germination plastic pouches (CYG gemination pouch; 16.5 cm × 18 cm; Mega International). The pouches were moistened with 25 mL of sterile distilled water. Pouches containing seedlings were placed in a BioChambers Bigfoot Series Model AC-60 growth chamber with 140 μE m−2 s−1 and 14:10 h light/dark cycle at 12°C for 7 d and then 25°C for 7 d. Plants were watered as needed with sterile distilled water. Samples from tap roots from soybean or radical roots from maize were taken at the same time of day (16:00 h) from the original 4-cm inoculation site throughout the time course. The 2-week time course was repeated three independent times in the same growth chamber, with sampling of six pooled plants for RNA isolation and three plants for DNA isolation at 0, 2, 4, 7, 10, and 14 DAI in each independent time. Time point 0 (0 DAI) was sampled immediately after completion of fungal or mock inoculation and before transferring seedlings to pouches. Germinating macroconidia were sampled from an aliquot of the fungal inoculum used above for each independent experiment that was centrifuged at 2000 × g to collect the germinated spores. Six pooled root samples were generated for each independent run for each plant species (n = 6 for soybean or maize), and this was repeated for three independent runs (n = 18 for soybean and maize). Additionally, a sample of germinating macroconidia was collected for each independent run (n = 3). In total this provided 39 RNA samples (Supplemental Figure 8). Plant growth and disease symptomology was recorded at each time point by photographing with a Nikon D50 camera.
Fungal Colonization Analyses
To visualize fungal growth on samples, microscopic analyses of maize and soybean roots were conducted at each time point for all treatments. Roots were cleared in 100% ethanol, followed by staining in a 0.05% (w/v) trypan blue (Millipore Sigma; catalog no. T6146) solution containing equal parts of water, glycerol, and lactic acid (Savory et al., 2012). Fungal structures were observed using an MZ16 dissecting scope (Leica).
RNA Extraction
Total RNA was isolated from 200 mg of ground flash-frozen germinating macroconidia and plant root samples and subsequently used for mRNA (mRNA) sequencing with an miRNeasy Mini Kit (Qiagen). Genomic DNA was removed using the TURBO DNase Free kit (Invitrogen). Extracted RNA was quantified using the Qubit RNA BR kit (Invitrogen), and RNA quality was determined by gel electrophoresis using the 2100 Bioanalyzer (Agilent) with the Agilent RNA 6000 Pico kit.
mRNA Library Preparation and Sequencing
Libraries were prepared using the Illumina TruSeq mRNA Library Preparation Kit from three biological repeats of samples collected at each time point of F. virguliforme– or mock-inoculated maize or soybean or germinating macroconidia samples by the Michigan State University Research Technology Support Facility. Pooled samples were sequenced on the Illumina HiSeq 4000 (single end 50 nucleotide mode). Base calling was performed using Illumina Real Time Analysis (RTA) v2.7.7 and the output of RTA was demultiplexed and converted to FastQ format with Illumina bcl2fastq v2.19.1.
Quantification of RNA-seq Expression and Differential Analysis
Reads were trimmed for adapter presence and quality score by Trimmomatic (v0.33; Bolger et al., 2014). The trimmed reads were uniquely mapped to the corresponding reference genome of F. virguliforme (Fv_v2) with HISAT2 (v 2.1.0; Kim et al., 2015) using the following parameters–dta–rna-strandness F. Hits from HISAT2 were converted from SAM to BAM format by Picard (v 2.18.1; http://broadinstitute.github.io/picard/). Alignments per gene model were counted by HTSeq (v0.6.1; Anders et al., 2015 with the following options: –minaqual 50 -m intersection-strict -s reverse–idattr=gene_id. Gene counts were imported into the R program DESeq2 (v1.22.2; Love et al., 2014), normalized for library size, and log2 transformed to determine correlation of biological replicates within each time point. To assess biological reproducibility, we compared gene expression across biological replicates and we found >90% reproducibility of the fungal gene expression profiles for the last time point of the infection time course, indicating the biological response of the fungus within each host was highly consistent (Supplemental Figure 2).
To determine differential gene expression patterns, DESeq2 (v1.22.2) with raw HTSeq counts was used. Genes with less than 10 total raw counts across all samples were excluded. To identify differentially expressed genes (P ≤ 0.05), DESeq2 was used. Through this approach, two types of pairwise comparison were performed: (1) maize or soybean F. virguliforme in planta samples against germinating F. virguliforme macroconidia across time point at a log2 fold change >2 to identify differentially induced genes; and (2) F. virguliforme gene expression patterns between hosts, (i.e., maize at DAI 0 versus soybean at 0 DAI, across time points) at a |log2 fold change > 1| to identify differentially induced genes.
Analysis of Gene Coexpression Networks
Genes were filtered for weighted gene correlation network analysis (Langfelder and Horvath, 2008) analysis (R Development Core Team, 2010) for 90% of genes with less than 10 reads across all samples. The resultant 11,112 genes were variance-stabilized, transformed for importation, and F. virguliforme–signed coexpression networks of maize or soybean were constructed separately. A soft threshold power of 7 and tree cut height of 0.15 were applied to both networks. Gene expression was clustered into 22 modules for F. virguliforme colonization of maize and 20 modules for F. virguliforme colonization of soybean. Modules were plotted and visualized using the R package ggplot2 (v3.1.1; Wickham, 2016).
GO Enrichment Analysis
To annotation GO terms for each protein annotated for the F. virguliforme v2 genome, unique GO terms from InterPro Scan (Jones et al., 2014) were extracted with a custom script (https://doi.org/10.5061/dryad.41ns1rn9q). Gene lists from either differential analysis or clusters from coexpression analysis were analyzed by TopGO (2.34.0) conducted (Alexa and Rahnenfuhrer, 2018) in R. Fisher’s Exact Test was conducted on each gene set with a P ≤ 0.05 to determine significance of enrichment.
Accession Numbers
The raw reads from the PacBio data, Illumina DNA-seq, and mRNA-seq reads can be downloaded at the NCBI SRA. The PacBio reads and Illumina DNA-seq and RNA-seq reads are deposited to the NCBI SRA under BioProject PRJNA551448 and PRJNA549951, respectively. The F. virguliforme genome assembly and annotation can be found at the Dryad Digital Repository (https://doi.org/10.5061/dryad.41ns1rn9q).
Supplemental Data
Supplemental Figure 1. Cartoon illustrating the genome assembly of Fusarium virguliforme.
Supplemental Figure 2. Biological consistency of samples from different time courses.
Supplemental Figure 3. Samples of fungal plant colonization group by treatment.
Supplemental Figure 4. Expression profiles of weighted gene coexpression network modules from F. virguliforme temporal colonization of maize.
Supplemental Figure 5. Expression profiles of weighted gene coexpression network modules from F. virguliforme temporal colonization of soybean.
Supplemental Figure 6. Temporal counts of Fusarium virguliforme candidate effector genes within soybean and maize hosts.
Supplemental Figure 7. Temporal counts of Fusarium virguliforme carbohydrate active enzyme related genes within soybean and maize hosts.
Supplemental Figure 8. Overview of methodology to determine transcriptomic profiles of Fusarium virguliforme during colonization of maize or soybean.
Supplemental Table 1. Genome assembly metrics of Fusarium virguliforme Versions 1 and 2.
Supplemental Table 2. GO enrichment of genes contained only in the Fv_v2 genome.
Supplemental Table 3. Number of quality trimmed reads uniquely aligned to the Fusarium virguliforme genome v2.
Supplemental Table 4. Host-specific differential gene expression across time points of Fusarium virguliforme colonization of soybean or maize against germinating macroconidia.
Supplemental Table 5. Host-specific upregulation of gene expression within time points of Fusarium virguliforme colonization of soybean or maize at log2 fold change > 1.
Supplemental Data Set 1. Gene ID conversion between v1 and v2 of the Fusarium virguliforme genome of conserved, misassembled genes, and novel or missing genes in the genome.
Supplemental Data Set 2. Gene list of candidate effectors generated from EffectorP and SingalP.
Supplemental Data Set 3. Gene list of carbohydrate active enzymes generated from CanDB.
Supplemental Data Set 4. Gene Ontology enrichment of Fusarium virguliforme > 2 log2 fold change significant genes compared to germinating macroconidia on maize host.
Supplemental Data Set 5. Gene Ontology enrichment of Fusarium virguliforme > 2 log2 fold change significant genes compared to germinating macroconidia on soybean host.
Supplemental Data Set 6. Gene Ontology enrichment of Fusarium virguliforme induced gene coexpression modules during maize root colonization.
Supplemental Data Set 7. Gene Ontology enrichment of Fusarium virguliforme induced gene coexpression modules during soybean root colonization.
Supplemental Data Set 8. Differentially expressed genes at each time point of Fusarium virguliforme colonization between soybean and maize, with maize as a comparison base.
Supplemental Data Set 9. Gene Ontology enrichment of Fusarium virguliforme temporally differentially expressed genes within either maize or soybean root colonization.
Supplemental Data Set 10. Effector list and Interpro scan annotation.
Supplemental Data Set 11. CAZyme List and Interpro scan annotation.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Kevin Childs and John Johnston for server access and computational assistance, and Marty Chilvers for providing the Fusarium virguliforme Mont-1 isolate. We would also like to recognize the support staff at the Michigan State University (MSU) Institute for Cyber enabled Research High Performance Computing Cluster for assistance in software optimization. This research was supported by the MSU project GREEEN (Generating Research and Extension to meet Economic and Environmental Needs; grant no. GR16-008), by the C.S. Mott Foundation (for fellowship support of A.B.-Y.), and by the MSU Plant Resilience Institute (grant no. GR100125-Bean2).
AUTHOR CONTRIBUTIONS
A.B.-Y. conducted experiments; A.B.-Y. and B.D. wrote the article; A.B.Y., C.M.W., R.V-B., B.D. designed the framework, analyzed data, and provided comments and editorial input during article preparation and revision.
Footnotes
Articles can be viewed without a subscription.
References
- Alexa A., Rahnenfuhrer J. (2018). topGO: Enrichment analysis for gene ontology. Bioconductor. Accessed Nov. 12, 2018.
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H., Kim M.S., Sicher R.C., Bae H.-J., Bailey B.A. (2006). Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol. 141: 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnaresi P., Biselli C., Orrù L., Urso S., Crispino L., Abbruscato P., Piffanelli P., Lupotto E., Cattivelli L., Valè G. (2012). Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS One 7: e51609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.W., Butchko R.A., Busman M., Proctor R.H. (2007). The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot. Cell 6: 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.W., Proctor R.H. (2016). Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal Genet. Biol. 89: 37–51. [DOI] [PubMed] [Google Scholar]
- Brown N.A., Evans J., Mead A., Hammond-Kosack K.E. (2017). A spatial temporal analysis of the Fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Mol. Plant Pathol. 18: 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., et al. (2017). A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Reports 18: 762–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344: 1879–1900. [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. (2008). MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.X., Domier L.L., Radwan O., Yendrek C.R., Hudson M.E., Hartman G.L. (2016a). Identification of multiple phytotoxins produced by Fusarium virguliforme including a phytotoxic effector (FvNIS1) associated with sudden death syndrome foliar symptoms. Mol. Plant Microbe Interact. 29: 96–108. [DOI] [PubMed] [Google Scholar]
- Chang H.X., Yendrek C.R., Caetano-Anolles G., Hartman G.L. (2016b). Genomic characterization of plant cell wall degrading enzymes and in silico analysis of xylanases and polygalacturonases of Fusarium virguliforme. BMC Microbiol. 16: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Le X., Sun Y., Li M., Zhang H., Tan X., Zhang D., Liu Y., Zhang Z. (2017). MoYcp4 is required for growth, conidiogenesis and pathogenicity in Magnaporthe oryzae. Mol. Plant Pathol. 18: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Basu A., Kundu S. (2017). Biotrophy-necrotrophy switch in pathogen evoke differential response in resistant and susceptible sesame involving multiple signaling pathways at different phases. Sci. Rep. 7: 17251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordovez V., Mommer L., Moisan K., Lucas-Barbosa D., Pierik R., Mumm R., Carrion V.J., Raaijmakers J.M. (2017). Plant phenotypic and transcriptional changes induced by volatiles from the fungal root pathogen Rhizoctonia solani. Front. Plant Sci. 8: 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers J.E., Gugino B.K., Jiménez-Gasco M.M. (2015). Highly diverse endophytic and soil content genus-species Fusarium oxysporum populations associated with field-grown tomato plants. Appl. Environ. Microbiol. 81: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire M., et al. (2017). The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9: 593–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derntl C., Kluger B., Bueschl C., Schuhmacher R., Mach R.L., Mach-Aigner A.R. (2017). Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc. Natl. Acad. Sci. USA 114: E560–E569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C.E. (2016). Control of gene expression in phytopathogenic Ascomycetes during early invasion of plant tissue In Biochemistry and Molecular Biology. The Mycota (A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research), D. Hoffmeister, ed, Volume III (Cham: Springer; ). [Google Scholar]
- Fang Y.L., Peng Y.L., Fan J. (2017). The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants. Sci. Rep. 7: 4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. (2011). HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39: W29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Sbrana C., Citernesi A.S., Avio L., Gollotte A., Gianinazzi-Pearson V., Gianinazzi S. (1994). Recognition and infection process, basis for host specificity of arbuscular mycorrhizal fungi In Impact of Arbuscular Mycorrhizas on Sustainable Agriculture and Natural Ecosystems, S. Gianinazzi, H. Schüepp, eds (Basel: ALS Advances in Life Sciences. Birkhäuser; ). [Google Scholar]
- Gupta A., Chattoo B.B. (2008). Functional analysis of a novel ABC transporter ABC4 from Magnaporthe grisea. FEMS Microbiol. Lett. 278: 22–28. [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman G.L., Chang H.X., Leandro L.F. (2015). Research advances and management of soybean sudden death syndrome. Crop Prot. 73: 60–66. [Google Scholar]
- Haueisen J., Möller M., Eschenbrenner C.J., Grandaubert J., Seybold H., Adamiak H., Stukenbrock E.H. (2018). Highly flexible infection programs in a specialized wheat pathogen. Ecol. Evol. 9: 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J., Tudzynski P. (2011). Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annu. Rev. Phytopathol. 49: 369–390. [DOI] [PubMed] [Google Scholar]
- Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M. (2016). BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbach R., Navarro-Quesada A.R., Knogge W., Deising H.B. (2011). When and how to kill a plant cell: Infection strategies of plant pathogenic fungi. J. Plant Physiol. 168: 51–62. [DOI] [PubMed] [Google Scholar]
- Jennings J.C., Apel-Birkhold P.C., Bailey B.A., Anderson J.D. (2000). Induction of ethylene biosynthesis and necrosis in weed leaves by a Fusarium oxysporum protein. Weed Sci. 48: 7–14. [Google Scholar]
- Jiang X., Qiao F., Long Y., Cong H., Sun H. (2017). MicroRNA-like RNAs in plant pathogenic fungus Fusarium oxysporum f. sp. niveum are involved in toxin gene expression fine tuning. 3 Biotech. 7: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., et al. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res. 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann J., Rincon-Rivera L.J., Takahara H., Neumann U., Ver Loren van Themaat E., van der Does H.C., Hacquard S., Stüber K., Will I., Schmalenbach W., Schmelzer E., O’Connell R.J. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum [published correction appears in PLoS Pathog. 2012 Aug;8(8)]. PLoS Pathog. 8: e1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Leonel R., Mueller D., Harbach C., Tylka G., Leandro L. (2017). Susceptibility of cover crop plants to Fusarium virguliforme, causal agent of soybean sudden death syndrome, and Heterodera glycines, the soybean cyst nematode. J. Soil Water Conserv. 72: 575–583. [Google Scholar]
- Koenning S.R., Wrather J.A. (2010). Suppression of soybean yield potential in the continental United States by plant diseases from 2006 to 2009. Plant Health Prog. 11: 10.1094/php-2010–1122–1001-rs. [Google Scholar]
- Kolander T.M., Bienapfl J.C., Kurle J.E., Malvick D.K. (2012). Symptomatic and asymptomatic host range of Fusarium virguliforme, the causal agent of soybean sudden death syndrome. Plant Dis. 96: 1148–1153. [DOI] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. (2017). Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Lade B.D., Patil A.S., Paikrao H.M. (2014). Efficient genomic DNA extraction protocol from medicinal rich Passiflora foetida containing high level of polysaccharide and polyphenol. Springerplus 3: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.-H., Rognes T., Ussery D.W. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahrmann U., Ding Y., Banhara A., Rath M., Hajirezaei M.R., Döhlemann S., von Wirén N., Parniske M., Zuccaro A. (2013). Host-related metabolic cues affect colonization strategies of a root endophyte. Proc. Natl. Acad. Sci. U.S.A. 110: 13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K., Mengiste T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? Arabidopsis Book 8: e0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Marzano S.-Y., Neupane A., Domier L. (2018). Transcriptional and small RNA responses of the white mold fungus Sclerotinia sclerotiorum to infection by a virulence-attenuating hypovirus. Viruses 10: 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren L.A., LeBlanc N.R., Certano A.K., Nachtigall J., LaBine K.M., Riddle J., Broz K., Dong Y., Bethan B., Kafer C.W., Kistler H.C. (2018). Fusarium graminearum: Pathogen or endophyte of North American grasses? New Phytol. 217: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain C., Marchal C., Hacquard S., Delaruelle C., Pétrowski J., Petre B., Hecker A., Frey P., Duplessis S. (2018). The rust fungus Melampsora larici-populina expresses a conserved genetic program and distinct sets of secreted protein genes during infection of its two host plants, larch and poplar. Mol. Plant Microbe Interact. 31: 695–706. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysøe E., Bone K.R., Klemsdal S.S. (2008). Identification of up-regulated genes during zearalenone biosynthesis in Fusarium. Eur. J. Plant Pathol. 122: 505–516. [Google Scholar]
- Ma L.J., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm G.M., Kuldau G.A., Gugino B.K., Jiménez-Gasco Mdel. M. (2013). Hidden host plant associations of soilborne fungal pathogens: an ecological perspective. Phytopathology 103: 538–544. [DOI] [PubMed] [Google Scholar]
- Min B., Grigoriev I.V., Choi I.G. (2017). FunGAP: Fungal Genome Annotation Pipeline using evidence-based gene model evaluation. Bioinformatics 33: 2936–2937. [DOI] [PubMed] [Google Scholar]
- Navi S.S., Yang X.B. (2008). Foliar symptom expression in association with early infection and xylem colonization by Fusarium virguliforme (formerly F. solani f. sp. Glycines), the causal agent of soybean sudden death syndrome. Plant Heal. Prog. 10.1094/PHP-2008–0222–01-RS. [Google Scholar]
- Ngaki M.N., Wang B., Sahu B.B., Srivastava S.K., Farooqi M.S., Kambakam S., Swaminathan S., Bhattacharyya M.K. (2016). Transcriptomic study of the soybean-Fusarium virguliforme interaction revealed a novel ankyrin-repeat containing defense gene, expression of whose during infection led to enhanced resistance to the fungal pathogen in transgenic soybean plants. PLoS One 11: e0163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.J., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R.P., Ipcho S.V. (2004). Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens. Mol. Plant Pathol. 5: 347–352. [DOI] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2010). R: A language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- Rai M., Agarkar G. (2016). Plant-fungal interactions: What triggers the fungi to switch among lifestyles? Crit. Rev. Microbiol. 42: 428–438. [DOI] [PubMed] [Google Scholar]
- Sahu B.B., Baumbach J.L., Singh P., Srivastava S.K., Yi X., Bhattacharyya M.K. (2017). Investigation of the Fusarium virguliforme transcriptomes induced during infection of soybean roots suggests that enzymes with hydrolytic activities could play a major role in root necrosis. PLoS One 12: e0169963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H., Chang H.-X., Chilvers M.I. (2019). A Sclerotinia sclerotiorum transcription factor involved in sclerotial development and virulence on pea. MSphere 4: e00615–e00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savory E.A., Adhikari B.N., Hamilton J.P., Vaillancourt B., Buell C.R., Day B. (2012). mRNA-Seq analysis of the Pseudoperonospora cubensis transcriptome during cucumber (Cucumis sativus L.) infection. PLoS One 7: e35796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., Wallner A., Arst H.N. Jr., Haynes K., Haas H. (2007). Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3: 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl M.F., Faino L., Shi-Kunne X., van den Berg G.C.M., Bolton M.D., Thomma B.P.H.J. (2015). The genome of the saprophytic fungus Verticillium tricorpus reveals a complex effector repertoire resembling that of its pathogenic relatives. Mol. Plant Microbe Interact. 28: 362–373. [DOI] [PubMed] [Google Scholar]
- Selosse M.-A., Schneider-Maunoury L., Martos F. (2018). Time to re-think fungal ecology? Fungal ecological niches are often prejudged. New Phytol. 217: 968–972. [DOI] [PubMed] [Google Scholar]
- Shaw M.W., Emmanuel C.J., Emilda D., Terhem R.B., Shafia A., Tsamaidi D., Emblow M., van Kan J.A. (2016). Analysis of cryptic, systemic Botrytis infections in symptomless hosts. Front. Plant Sci. 7: 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Soanes D.M., Chakrabarti A., Paszkiewicz K.H., Dawe A.L., Talbot N.J. (2012). Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8: e1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J., Gardiner D.M., Dodds P.N., Tini F., Covarelli L., Singh K.B., Manners J.M., Taylor J.M. (2016). EffectorP: Predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210: 743–761. [DOI] [PubMed] [Google Scholar]
- Srivastava S.K., Huang X., Brar H.K., Fakhoury A.M., Bluhm B.H., Bhattacharyya M.K. (2014). The genome sequence of the fungal pathogen Fusarium virguliforme that causes sudden death syndrome in soybean. PLoS One 9: e81832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. (2006). AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 34: W435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H.K., Toledo M.S., Suzuki E., Tagliari L., Straus A.H. (2009). Current relevance of fungal and trypanosomatid glycolipids and sphingolipids: Studies defining structures conspicuously absent in mammals. An. Acad. Bras. Cienc. 81: 477–488. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso J., van Kan J.A.L. (2018). Many shades of grey in Botrytis host plant interactions. Trends Plant Sci. 23: 613–622. [DOI] [PubMed] [Google Scholar]
- Vollmeister E., Schipper K., Baumann S., Haag C., Pohlmann T., Stock J., Feldbrügge M. (2012). Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 36: 59–77. [DOI] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., Earl A.M. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., Kissinger J.C., Paterson A.H. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R.M., Seppey M., Simão F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Schultz M.B., Zobel J., Holt K.E. (2015). Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 31: 3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant graphics for data analysis. (Springer-Verlag New York). Accessed April 11, 2019.
- Yang F., Li W., Jørgensen H.J.L. (2013). Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS One 8: e81606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lübeck M., Lübeck P.S. (2015a). Effects of heterologous expression of phosphoenolpyruvate carboxykinase and phosphoenolpyruvate carboxylase on organic acid production in Aspergillus carbonarius. J. Ind. Microbiol. Biotechnol. 42: 1533–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Xie L., Xue B., Goodwin P.H., Quan X., Zheng C., Liu T., Lei Z., Yang X., Chao Y., Wu C. (2015b). Comparative transcriptome profiling of the early infection of wheat roots by Gaeumannomyces graminis var. tritici. PLoS One 10: e0120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. (2012). dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40: W445-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tian L., Yan D.-H., He W. (2018). Genome-wide transcriptome analysis reveals the comprehensive response of two susceptible poplar sections to Marssonina brunnea infection. Genes (Basel) 9: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Liu H., Wang C., Xu J.-R. (2013). Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]