NSG1, encoding a member of the C2H2 zinc finger protein family, plays a pivotal role in maintaining organ identities in the spikelet by repressing the expression of LHS1, DL, and MFO1.
Abstract
The spikelet is an inflorescence structure unique to grasses. The molecular mechanisms underlying spikelet development and evolution are unclear. In this study, we characterized three allelic recessive mutants in rice (Oryza sativa): nonstop glumes 1-1 (nsg1-1), nsg1-2, and nsg1-3. In these mutants, organs such as the rudimentary glume, sterile lemma, palea, lodicule, and filament were elongated and/or widened, or transformed into lemma- and/or marginal region of the palea–like organs. NSG1 encoded a member of the C2H2 zinc finger protein family and was expressed mainly in the organ primordia of the spikelet. In the nsg1-1 mutant spikelet, LHS1 DL, and MFO1 were ectopically expressed in two or more organs, including the rudimentary glume, sterile lemma, palea, lodicule, and stamen, whereas G1 was downregulated in the rudimentary glume and sterile lemma. Furthermore, the NSG1 protein was able to bind to regulatory regions of LHS1 and then recruit the corepressor TOPLESS-RELATED PROTEIN to repress expression by downregulating histone acetylation levels of the chromatin. The results suggest that NSG1 plays a pivotal role in maintaining organ identities in the spikelet by repressing the expression of LHS1, DL, and MFO1.
INTRODUCTION
Inflorescence development has a major effect on crop yield and involves three types of transformation: the transition of the shoot apical meristem from vegetative to reproductive development causes a change of the branching pattern that leads to the generation of a more complex structure, the inflorescence meristem, that then produces several primary branch meristems (PBMs) until termination. Each PBM continues to generate next-order meristems laterally until they acquire floral meristem (FM) identity. Floral organs develop from the flanks of the FMs, ultimately yielding seeds (Ikeda et al., 2004). The rice (Oryza sativa) panicle includes three types of inflorescence meristems, namely, PBMs, secondary branch meristems, and spikelet meristems (SMs; Schmidt and Ambrose, 1998; McSteen et al., 2000). The grain number per panicle in rice (which is almost equal to the number of SMs) is determined mainly by the number of PBMs and secondary branch meristems. The majority of high-yield genes reported recently are involved in regulation of panicle branching, including GRAIN NUMBER1A (Gn1a), Ghd7, DENSE AND ERECT PANICLE1 (DEP1), DEP2, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE14/IDEAL PLANT ARCHITECTURE1 (OsSPL14/IPA1), and TAWAWA1 (TWA1), which have made important contributions to increases rice yield (Ashikari et al., 2005; Xue et al., 2008; Huang et al., 2009a; Jiao et al., 2010; Li et al., 2010a; Miura et al., 2010; Yoshida et al., 2013). Note that the spikelet is also an inflorescence unit and can produce one or more florets in some species of grass, which offers the possibility of increasing the number of florets per spikelet to improve yield in species that nowadays produce only a one-floret/grain spikelet, such as rice. However, to achieve this, an enhanced understanding of the genetic and molecular mechanisms of spikelet development is required.
In rice, the spikelet contains one fertile terminal floret (consisting of four whorls of floral organs: one lemma and one palea, two lodicules, six stamens, and one pistil), one pair of sterile lemmas, and one pair of rudimentary glumes. Although wild-type rice produces just a single grain per spikelet, the loss of determinacy of SMs may enable an increase in the number of florets and/or other organs per spikelet, as demonstrated from studies of mutants of genes such as SUPERNUMERARY BRACT GENE (SNB), INDETERMINATE SPIKELET1 (IDS1), and MULTI-FLORETS SPIKELET1 (MFS1; Lee and An, 2012; Ren et al., 2013). In these mutants, the transition from SM to FM is delayed, which leads to the production of an additional floret(s).
An additional possible means of increasing the number of florets per spikelet is by restoring the two lateral florets, which now are considered to be antecedents of sterile lemmas. Two prevailing hypotheses for the origin and identity of sterile lemmas (as well as rudimentary glumes) have been proposed. One hypothesis holds that sterile lemmas and rudimentary glumes are reduced glumes (Schmidt and Ambrose, 1998). The second hypothesis suggests that the spikelet of rice ancestors contained a terminal floret and two lateral florets. According to the latter proposal, the rice spikelet is a three-florets spikelet and the lateral florets degenerated into sterile lemmas during evolution (Arber, 1935; Kellogg, 2009; Yoshida et al., 2009; Kobayashi et al., 2010). Compared with the ample morphological evidence in support of the first hypothesis, abundant genetic and molecular evidence has accumulated in support of the second hypothesis from research on genes such as LONG STERILE LEMMAS (G1), EXTRA GLUME1 (EG1), PANICLE PHYTOMER2 (PAP2), ABERRANT SPIKELET AND PANICLE1 (ASP1), LEAFY HULL STERILE1 (LHS1), and LATERAL FLORET1 (LF1; Li et al., 2009a; Yoshida et al., 2009, 2012, Wang et al., 2013, 2017; Lin et al., 2014; Zhang, 2017). The loss of function of G1, EG1, PAP2, and ASP1 and overexpression of LHS1 lead to transformation of sterile lemmas into lemma-like organs, whereas the gain of function of LF1 results in development of a lateral floret (including the palea, lodicule, stamens, and pistil) in the axil of the sterile lemma. Therefore, it is possible to gain additional, complete (fertile and morphologically normal) lateral florets by combining the two types of mutants (Zhang, 2017). Nevertheless, some molecular evidence supports the first hypothesis. In the mfs1 mutant, the sterile lemma is degenerated and assumes the identity of a rudimentary glume, which implies that the sterile lemma is homologous to the rudimentary glume (Ren et al., 2013). Therefore, additional research is needed to gain an improved understanding of this issue.
In the previous study, we identified as preliminarily a spikelet mutant nonstop glumes 1-1 (nsg1-1) and mapped the NSG1 gene to a region on chromosome 4 (Wang et al., 2013). In the present study, we characterized the other two allelic recessive mutants of NSG1, namely, nsg1-2 and nsg1-3. In the three allelic mutants, the rudimentary glume, sterile lemma, palea, lodicule, and filament were elongated and/or widened and were transformed into lemma and/or marginal region of the palea (mrp)–like organs. Our data further indicate that NSG1 encodes a C2H2 zinc finger transcription repressor and plays a pivotal role in the regulation of lateral organ identities in the spikelet by repression of the expression of floral organ identity genes LHS1, DROOPING LEAF (DL), and MOSAIC FLORAL ORGANS1 (MFO1).
RESULTS
Three allelic recessive mutants of NSG1, designated nsg1-1, nsg1-2, and nsg1-3, were identified in this study. Given that nsg1-1 showed more severe defects than the other two mutants, the remainder of this study focuses on the nsg1-1 mutant.
Defective Organ Identity in the nsg1-1 Spikelet
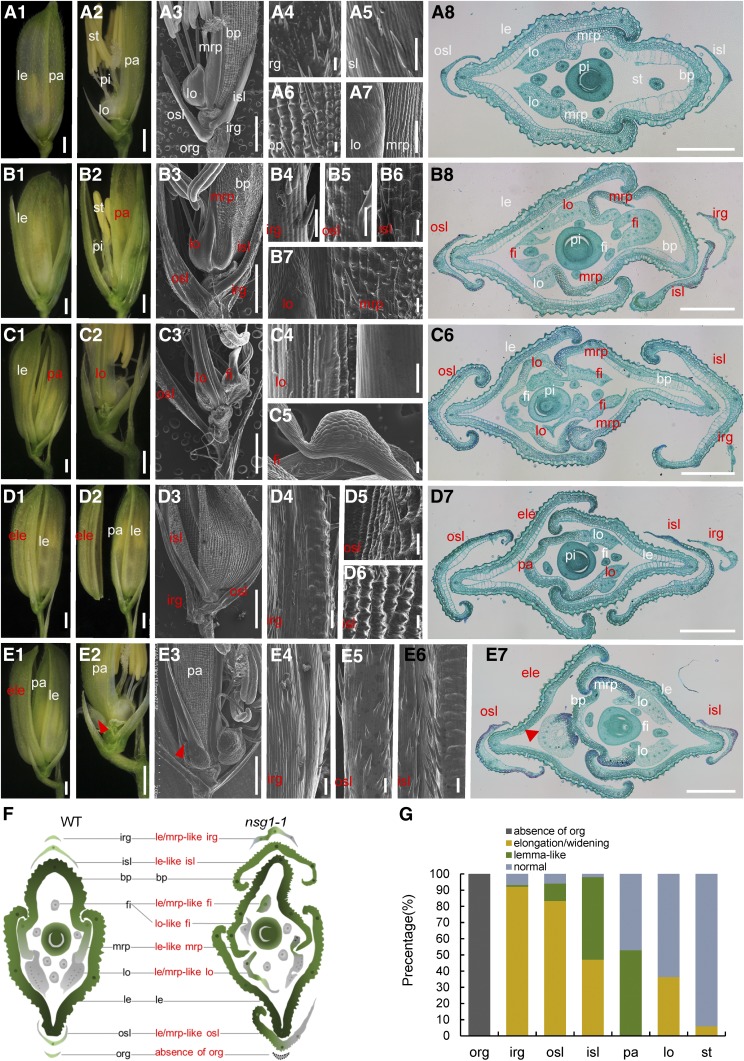
The wild-type rice spikelet is composed of one pair of rudimentary glumes, one pair of sterile lemmas (also termed empty glumes), and one terminal fertile floret. The floret consists of two additional bracts, the lemma and palea, that encase three whorls of floral organs: two lodicules, six stamens, and a central carpel (Figure 1A; Hoshikawa, 1989). The rudimentary glumes, sterile lemmas, and lemma/palea are considered to be three pairs of glume-like organs, inserted on the rachilla with distichous phyllotaxy from bottom to top. The sizes of the lemma and palea are much larger than the rudimentary glumes and sterile lemmas, and the lemma is wider than the palea (Figures 1A1 and 1A8). Compared with the lemma, the palea consists of two parts—the main body of the palea (bop) and two mrp (Figures 1A3, 1A7, and 1A8). The bop shows similar features to those of the lemma, with a silicified abaxial epidermis bearing trichomes and protrusions (Figures 1A6 and 1A8). However, the mrp structure is distinct in showing a smooth and nonsilicified abaxial epidermis due to a lack of trichomes and protrusions (Figures 1A7 and 1A8). The sterile lemma has a smooth abaxial surface with a few trichomes and is larger than the rudimentary glume (Figures 1A3 and 1A5). The two rudimentary glumes are much reduced, and abundant trichomes and protrusions are observed on the abaxial surface (Figures 1A3 and 1A4).
Figure 1.
Phenotypes of Spikelets in the Wild Type and nsg1-1 Mutant.
(A) Spikelets of the wild type. (A1) and (A2) The wild-type spikelet (A1); the lemma was removed in (A2). (A3) to (A7) The wild-type spikelet; the lemma was removed in (A3). (A4) to (A7) Surface characters of rg, sl, bop, lo and mrp, respectively. (A8) Transverse sections of the wild-type spikelet showing a similar phenotype to those in (A1) to (A7).
(B) to (E) Spikelets of the nsg1-1 mutant. (B1) and (B2) A nsg1-1 spikelet; the lemma was removed in (B2). (B3) to (B7) A nsg1-1 spikelet; the lemma was removed in (B3). (B4) to (B7) Surface characters of the elongated rg, elongated sl, lemma-like sl, and lemma-like mrp, respectively. (B8) Transverse sections of a nsg1-1 spikelet showing a similar phenotype to those in (B1) to (B7).
(C1) and (C2) A nsg1-1 spikelet (C1); the lemma and the lemma-like palea were removed in (C2).
(C3) to (C5) A nsg1-1 spikelet; the lemma and the lemma-like palea were removed in (C3). (C4) and (C5) show the surface characters of the elongated lodicule and lodicule-like stamen, respectively.
(C6) Transverse sections of a nsg1-1 spikelet showing a similar phenotype to those in (C1) to (C6).
(D1) and (D2) A nsg1-1 spikelet (D1); the additional lemma was separated in (D2).
(D3) to (D6) A nsg1-1 spikelet (D3); (D4) to (D6) show the surface characters of the elongated rg, osl, and isl, respectively.
(D7) Transverse sections of a nsg1-1 spikelet showing a similar phenotype to those in (D1) to (D6).
(E1) and (E2) A nsg1-1 spikelet (E1); the lemma and the additional lemma were removed in (E2).
(E3) to (E6) A nsg1-1 spikelet (E3); (E4) to (E6) show surface characters of the lemma-like rg, osl, and isl, respectively. (E7) Transverse sections of a nsg1-1 spikelet showing a similar phenotype to those in (E1) to (E6). Red arrowheads indicate the floral organs in the additional floret.
(F) Schematic diagrams of the wild-type (WT) and nsg1-1 spikelet structure in transverse section.
(G) Percentage of elongated or lemma-like organs in nsg1-1 spikelets.
White type indicates the normal organs, and red type indicates the abnormal organs.
Bar in *1, *2 and *3 = 1000 μm (the asterisk mean [A], [B], [C], [D], and [E]); bar in (A8), (B8), (C6), and (D7) = 500 μm; bar = 50 μm in all other images.
ele, additional lemma; fi, filament; irg, inner rudimentary glume; isl, inner sterile lemma; le, lemma; lo, lodicule; mrp, marginal region of palea; osl, outer sterile lemma; pa, palea; st, stamen.
In the nsg1-1 spikelets, the rudimentary glume and sterile lemma were elongated and/or widened to various degrees. The inner rudimentary glume was elongated to the same length as the sterile lemma in ∼93% of nsg1-1 spikelets (Figures 1B4, 1D4, and 1G), of which ∼1% showed a lemma-like epidermis with similar protrusions (Figure 1D4). The outer and inner sterile lemmas were elongated and widened in the majority of nsg1-1 spikelets, in which ∼11 and 51%, respectively, showed lemma-like features (Figures 1B to 1E and 1G). In contrast to the smooth epidermis of the wild-type sterile lemma, the nsg1-1 lemma-like sterile lemmas displayed a rough abaxial epidermis similar to that of the wild-type lemma, in which the tubercles and trichomes showed an orderly parallel arrangement (Figures 1B5, 1B6, 1D4 to 1D6, 1E4 to 1E6, and 1G).
In addition, a number of floral organs gained a lemma-like identity in the nsg1-1 spikelets. The mrps were transformed into a lemma-like structure in ∼53% of nsg1-1 spikelets. These lemma-like mrps were significantly wider than those of the wild type and showed a crooked structure resembling the wild-type lemma. Histological analysis also showed a silicified abaxial epidermis of these mrps instead of a smooth and nonsilicified one. Notably, these features of the abnormal mrps were characteristic of the wild-type lemma (Figures 1B7, 1B8, 1C6, and 1G). The lodicules were also elongated and widened, and/or gained lemma and/or mrp-like identities in more than one-third of nsg1-1 spikelets (Figures 1C4, 1B8, 1C6, and 1G). We also observed that the filaments of the stamens were broadened and were transformed into lodicule/mrp-like organs in ∼6% of nsg1-1 spikelets (Figures 1C5, 1B8, 1C6, and 1G).
Histological and qPCR Analysis of Lateral Organs in nsg1-1 Spikelets
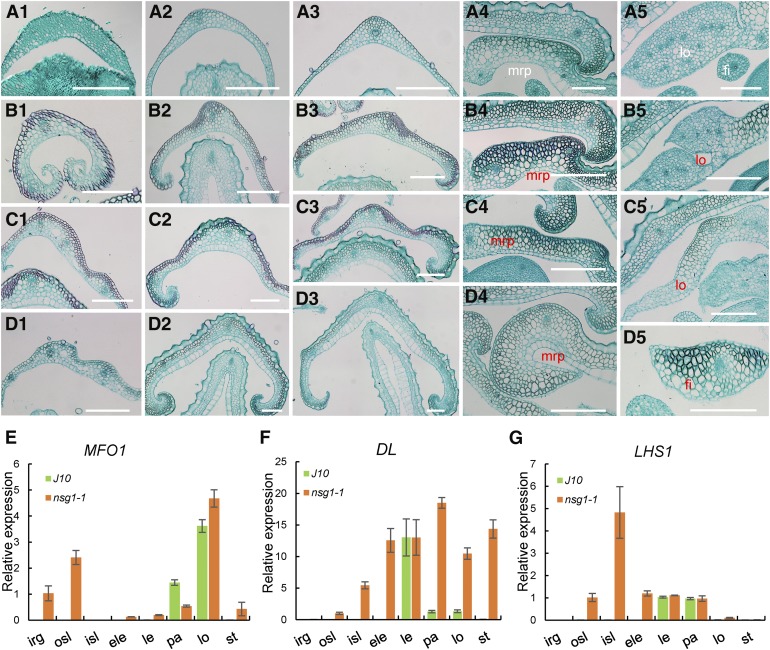
In accordance with these observations, almost all of the lateral organs (rudimentary glumes, sterile lemmas, paleas, lodicules, and stamens except for the lemmas) tended to be transformed into lemma-like organs, and the organs that were much nearer to the position of the lemmas were more similar to the lemmas (Figure 1F). We therefore further explored the histologic structure of these organs (Figure 2; Supplemental Figure 1). In a transverse section of wild-type lemma and bop, a number of vascular bundles and a four-layered structure that consisted of a silicified abaxial epidermis, several layers of fibrous sclerenchyma cells, several layers of spongy parenchyma cells, and a vacuolated abaxial epidermis were observed (Figure 1A8 and 2A4; Supplemental Figure 1B).
Figure 2.
Histological and RT-qPCR Analysis of Lateral Organs in Spikelets of the Wild Type and nsg1-1 Mutant.
(A) to (D) Transverse sections of lateral organs in a wild-type spikelet (A). Transverse sections of lateral organs in nsg1-1 spikelets (B). (A1) rg of the wild type; (B1), (C1), and (D1) show mrp-like rg in nsg1-1 spikelets. (A2) osl of the wild type; (B2), (C2), and (D2) show mrp-like and/or lemma-like osl in nsg1-1 spikelets. (A3) isl of the wild type; (B3), (C3), and (D3) show mrp-like and/or lemma-like isl in nsg1-1 spikelets. (A4) mrp of the wild type; (B4), (C4), and (D4) show lemma-like mrp in nsg1-1 spikelets. (A5) Lodicule and stamen of the wild type; (B5), (C5), and (D5) show abnormal lodicule, mrp-like lodicule, and mrp-like filament in nsg1-1 spikelets, respectively.
(E) to (G) RT-qPCR analysis of MFO1, DL, and LHS1 expression. ACTIN was used as a control. RNA was isolated from flower organs of wild-type and nsg1-1 spikelets. Error bars indicate sd. At least three replicates were performed, from which the mean value was used to represent the expression level.
White type indicates the normal organs, and red type indicates the abnormal organs. Bar = 150 μm.
Bar = 150 μm.
ele, additional lemma; fi, filament; irg, inner rudimentary glume; isl, inner sterile lemma; le, lemma; lo, lodicule; mrp, marginal region of palea; osl, outer sterile lemma; pa, palea; st, stamen.
Compared with the wild-type lemma and bop, the wild-type mrp consisted of a nonsilicified abaxial epidermis, a large number of spongy parenchyma cells, several fibrous sclerenchyma cells, and a nonvacuolated abaxial epidermis (Figure 1A8 and 2A4; Supplemental Figure 1C). With regard to the wild-type rudimentary glume, three tissues were observed, namely, an abaxial epidermis, an adaxial epidermis, and several layers of spongy parenchyma cells, and there was no obvious vascular bundle (Figure 2A1). In the wild-type sterile lemma, a single vascular bundle and three tissues similar to rudimentary glumes were observed (Figures 1A8, 2A2, and 2A3; Supplemental Figure 1A). The wild-type lodicule consisted of many parenchymatous cells and several interspersed tracheal elements (Figure 2A5). The wild-type filament showed radial organization and consisted of one central vascular bundle and several layers of parenchymatous cells (Figure 2A5).
In almost all elongated and/or widened nsg1-1 rudimentary glumes and sterile lemmas, the number of vascular bundles was increased to three to five, and several layers of ectopic fibrous sclerenchyma cells were observed between the abaxial epidermis and parenchyma cell layers (Figures 2B1 to 2B3, 2C1 to 2C3, and 2D1 to 2D3; Supplemental Figures 1D to 1F). In addition, the silicified abaxial epidermis and vacuolated adaxial epidermis specific to the wild-type lemma and bop were observed in some of the elongated and/or widened nsg1-1 sterile lemmas, but not the rudimentary glumes (Figures 2C1, 2C3, 2D2, and 2D3; Supplemental Figure 1F). Some inner sterile lemmas showed a characteristic of the wild-type lemma with same cell layers and number of vascular bundles (Figure 2D3). Therefore, the histologic characteristics of nsg1-1 rudimentary glumes were very similar to the wild-type mrp, while nsg1-1 sterile lemmas displayed the combined cell characteristics of the wild-type lemma and mrp. In some nsg1-1 mrps, lodicules, and filaments, ectopic cell layers of fibrous sclerenchyma cells were formed to various degrees (Figures 2B4, 2B5, 2C4, 2C5, 2D4, and 2D5). A silicified abaxial epidermis and vacuolated abaxial epidermis were observed in several nsg1-1 mrps (Figure 2D4). These results indicated that nsg1-1 mrps gained a partially lemma-like character and nsg1-1 lodicules and filaments gained a partially mrp-like character. Taken together, these observations demonstrated that all of the lateral organs in nsg1-1 spikelets, other than the lemma, had gained a lemma- and/or mrp-like anatomical structure by differentiation of ectopic fibrous sclerenchyma cell layers, a silicified abaxial epidermis, and/or vacuolated abaxial epidermis. In addition, we concluded that an organ gained a stronger lemma-like identity with increased proximity to the lemma.
Next, the mRNA levels of certain organ identity/marker genes were detected to further determine the identity of these defective organs, namely, DL (mainly expressed in the lemma and pistil and specifies their identity; Yamaguchi et al., 2004), LHS1 (mainly expressed in the lemma and bop and specifies their identity; Wang et al., 2010), and MFO1 (mainly expressed in the mrp and lodicule and specifies their identity; Li et al., 2011). In nsg1-1 inner rudimentary glumes, MFO1 was ectopically expressed, which suggested that these organs gained an mrp-like identity (Figure 2E). In nsg1-1 outer sterile lemmas, LHS1, DL, and MFO1 were ectopically expressed, which suggests that these organs gained a partial lemma-like and/or mrp-like identity (Figures 2E to 2G). In nsg1-1 inner sterile lemmas, MFO1 expression was not detected, whereas LHS1 and DL were ectopically expressed at an increased level, which indicated that these organs gained a stronger lemma identity (Figures 2E to 2G). In nsg1-1 paleas, upregulation of DL and downregulation of MFO1 expression were observed, which demonstrates that these organs gained a lemma-like identity (Figures 2E to 2G). In nsg1-1 lodicules, upregulation of DL expression was detected, which similarly implies that a lemma-like identity was gained (Figure 2F). In nsg1-1 stamens, DL and MFO1 expression levels were increased, suggesting that these organs gained a lemma-, mrp-, and lodicule-like identity (Figures 2F and 2G). Thus, the expression patterns of these genes were consistent with the phenotypic observations.
Similarity of Defects in the nsg1-2 and nsg1-3 Mutants to Those of the nsg1-1 Mutant
The nsg1-2 and nsg1-3 mutants showed very similar defects in morphology, histology, and expression patterns of genes associated with organ identity compared with those of the nsg1-1 mutant (Supplemental Figures 2 and 3). However, some differences were observed. Outer rudimentary glumes were absent in almost all nsg1-1 spikelets examined (Figures 1B to 1E, and 1G) and were absent in ∼78% of nsg1-3 spikelets (Supplemental Figures 2B and 3B). By contrast, this phenotype was not observed in nsg1-2 spikelets; thus, a pair of rudimentary glumes were retained in the nsg1-2 spikelets (Supplemental Figures 2A and 3A). In addition, no obvious defects were observed in the outer sterile lemma of nsg1-2 spikelets, whereas more than 90% of the outer sterile lemmas were transformed into a lemma-like organ in nsg1-1 and nsg1-3 spikelets (Figure 1G; Supplemental Figures 3A and 3B). The palea and lodicule of nsg1-3 spikelets showed defects in a few cases, whereas the mutation frequency was much higher in the nsg1-2 and nsg1-1 spikelets (Figure 1G; Supplemental Figures 3A and 3B). Taken together, these observations demonstrate that the nsg1-1 mutant showed more severe defects than the nsg1-2 and nsg1-3 mutants.
The nsg1-1 Mutant Exhibited Abnormal Early Spikelet Development
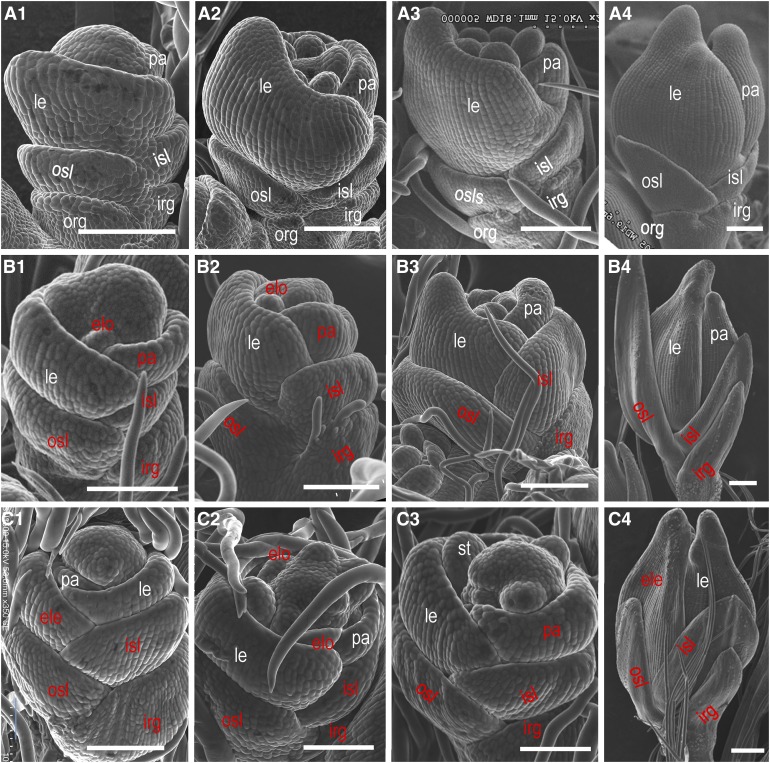
Young spikelets of both the wild-type plant and the nsg1-1 mutant were examined at different developmental stages using scanning electron microscopy (Figure 3). The wild-type sterile lemma primordia formed with alternate phyllotaxis during the spikelet 4 stage (Sp4) (Figure 3A1) and developed quickly until Sp8 (Figures 3A2 to 3A4), but developed slowly after that. The nsg1-1 sterile lemma primordia were first visible at Sp4 and thereafter differentiated and proliferated more rapidly than those of the wild type during all stages examined in this study (Figures 3B and 3C). The differentiation and proliferation of the wild-type rudimentary glume primordia were severely restricted, and no obvious change in shape and size was detected from Sp4 to Sp8, compared with other organs in the spikelet, such as the sterile lemma, lemma, and palea (Figure 3A). In nsg1-1 spikelets, however, the inner rudimentary glume primordia may overcome the developmental restriction and develop similarly to the sterile lemma and the lemma (Figures 3B and 3C). These observations indicate that the transformation of a sterile lemma and rudimentary glume into a lemma-like or lemma/sterile lemma-like organ might occur at an early developmental stage in nsg1-1 spikelets. The absence of outer rudimentary glumes in nsg1-1 spikelets was confirmed by detailed observation of spikelets at early developmental stages (Figures 3B and 3C).
Figure 3.
Scanning Electron Micrographs of Spikelets of the nsg1-1 Mutant at Early Developmental Stages.
(A) Spikelets of the wild type.
(B) nsg1-1 spikelets with relatively normal palea primordia.
(C) nsg1-1 spikelets with additional lemma or lemma-like palea primordia.
(A1),(B1), and (C1); (A2), (B2), and (C2); (A3), (B3), and (C3); and (A4), (B4), and (C4) show spikelets at the Sp4, Sp5, Sp6, and Sp8 developmental stages, respectively.
White type indicates the normal organs, and red type indicates the abnormal organs.
Bar = 100 μm.
ele, additional lemma; elo:elongated lodicule; irg, inner rudimentary glume; isl, inner sterile lemma; le, lemma; osl, outer sterile lemma; org, outer rudimentary glume; pa, palea.
The wild-type primordia of lemma and palea formed successively during Sp4. From Sp4 to Sp8, the primordia developed rapidly and finally hooked together. It was clear that the size of palea primordia was always smaller than that of lemma primordia (Figure 3A). In some nsg1-1 spikelets, however, the palea primordia was larger than that of the wild type, with the same size of lemma primordia. The growth process and appearance of these palea primordia were similar to lemma primordia as well, suggesting that the palea primordia had been transformed into lemma-like primordia (Figures 3B1, 3B2, 3C3, and 3C4). Because it was difficult to distinguish the additional lemma from lemma-like organs during the stages of primordia formation, only several additional lemmas were observed with certainty when the normal lemma and palea primordia were visible in nsg1-1 spikelets (Figure 3C1). Some defects in floral organ primordia were observed in the nsg1-1 mutant. During Sp5 to Sp7, several lodicule primordia were elongated and widened, and visible in nsg1-1 spikelets (Figures 3B2 and 3C2), but none were visible in the wild-type spikelets (Figure 3A2). Thus, the abnormal lodicule primordia might have gained a lemma-like identity. During Sp5 to Sp7, the stamen primordia were spherical in the wild type (Figures 3A2 and 3A3), whereas the shape of some stamen primordia was compressed in the nsg1-1 mutant (Figure 3C3). The abnormal stamen primordia might have gained a lodicule-like or lemma-like identity. Taken together, all the defects observed in mature spikelets were observed during stages of early spikelet development in nsg1-1 spikelets.
Identification of NSG1
In a previous study, we fine-mapped NSG1 to a region on chromosome 4, and a 13-bp insertion was identified located after +444 bp/147 amino acids of the Os04g36650 coding sequence (CDS) in the nsg1-1 mutant (Wang et al., 2013). On the basis of sequence data previously published by Wang et al. (2013), however, the chromosomal structure in the mapping region differs between two cultivars, japonica rice ‘Nipponbare’ and indica rice ‘9311’, for which whole-genome sequences are available. Between the two closest markers, the interval is ∼90 kb in 9311, but only 15 kb in Nipponbare (Bacterial Artificial Chromosome [BAC] OSJNBa0058G03), because of a large insertion located upstream of the Os04g36650 gene promoter (Wang et al., 2013). Transformation of the Os04g36650 wild-type genomic fragment from Nipponbare BAC OSJNBa0058G03 into the nsg1-1 mutant failed to complement the nsg1-1 defects, although the transformation was repeated three times during 2011 to 2014.
In 2014, we identified a T-DNA insertion line at the Os04g36650 locus, RMD_ITL-04Z11DF46, from the Rice Mutant Database (Zhang et al., 2007), which has a background of the japonica ‘Zhonghua11’ (ZH11; Figure 4A). In this insertion line, the T-DNA insertion occurred in the −1202 bp of Os04g36650 promoter; therefore, expression of the gene is almost undetectable (Supplemental Figure 4). Next, allelic verification between nsg1-1 and the insertion line was undertaken. The nsg1-1 mutant, the insertion line, and the F1 progeny displayed very similar phenotypes, which suggested that all were allelic mutants of Os04g36650 (Figure 4C). Thus, the insertion line was designated as nsg1-2.
Figure 4.
Map-Based Cloning of NSG1.
(A) Schematic illustration of the genomic structure of NSG1. The sites of the mutation in the nsg1-1, nsg1-2, and nsg1-3 mutants are shown.
(B) Structure of the NSG:GFP fusion complementary vector and the phenotypes of three independent transgenic lines.
(C) Allelism test between the nsg1-1 and nsg1-2 mutants. Three primer pairs were used to detect the distinct sites of the nsg1-1 and/or nsg1-2 mutation. Bar = 1000 μm.
We observed that the promoter sequence of nsg1-2 was located on Nipponbare BAC OSJNBb0056O04 (but not on BAC OSJNBa0058G03). The BAC OSJNBb0056O04 had been mapped to chromosome 4 in 2015 according to the genome information on the website (http://signal.salk.edu/cgi-bin/RiceGE/), and its sequence was identical to that of 9311. The complementary vector was reconstructed using DNA of the wild-type ‘Jinhui 10’ (J10) as the PCR template and primers were designed in accordance with the sequence of BAC OSJNBb0056O04. The wild-type genomic fragment of Os04g36650, which contained the 2925-bp upstream sequence from the start codon and 525-bp CDS, was cloned into the vector pCAMBIA1301 fused with the green fluorescent protein (GFP) gene and then transformed into the nsg1-1 mutant. The mutant phenotypes were rescued to various degrees in different transgenic lines, some of which were completely restored to the wild-type phenotype, such as No. 7 line. Other lines continued to develop elongated rudimentary glumes, such as No. 1 and No. 3 lines (Figure 4B). We also identified an additional allelic mutant, nsg1-3, in which a single-nucleotide substitution from A to T in the +124 bp of Os04g36650 CDS led an amino acid change from Arg to Trp and displayed extremely similar phenotypes to those of the nsg1-1 and nsg1-2 mutants (Figure 4A; Supplemental Figure 2A). Taken together, these results demonstrated that Os04g36650 was identical to NSG1.
We considered whether more than one copy of Os04g36650 was present in the genome of Nipponbare, or the copy number differed between indica and japonica rice, on account of the two BACs OSJNBa0058G03 and OSJNBb0056O04 of Nipponbare. DNA gel blot analysis was used to analyze the genomic constitution of four rice cultivars, namely, J10 (indica rice cultivar, the wild type of nsg1-1), XD1B (indica rice cultivar, the wild type of nsg1-3), ZH11 (japonica rice cultivar, the wild type of nsg1-2), and Nipponbare (japonica rice cultivar). Using the promoter sequence of NSG1, which was unique to BAC OSJNBb0056O04, as the probe, only one copy was detected among the four cultivars (Supplemental Figures 5A and 5B). Similarly, probing with the coding sequence of NSG1, which was present on the two BACs, also only one copy was detected (Supplemental Figures 5A and 5C). These results indicate that Os04g36650 was a single-copy gene in both indica and japonica rice and was located in the region corresponding to BAC OSJNBb0056O04 on chromosome 4.
NSG1 Encodes a C2H2 Zinc Finger Protein
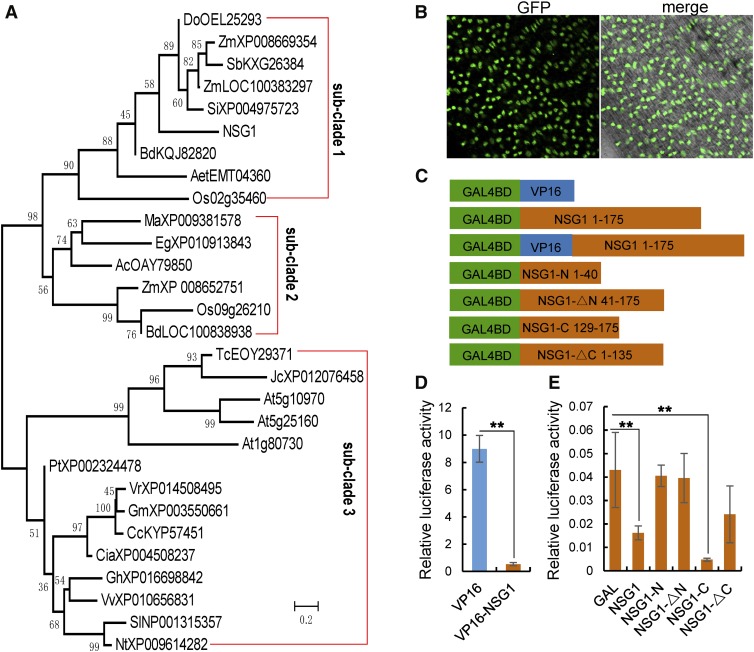
NSG1 encodes a protein of 175 amino acids and shows high similarity to transcription factors that contain a QALGGH-type single C2H2 zinc finger DNA binding domain (Figure 5A; Supplemental Figures 6 and 7B). This type of zinc finger domain has been identified in several proteins involved in flower development, such as SUPERMAN (SUP), JAGGED (JAG), and NUBBIN (NUB) in Arabidopsis (Arabidopsis thaliana) and STAMENLESS1 (SL1) in rice (Dinneny et al., 2004, 2006; Ohno et al., 2004; Xiao et al., 2009).
Figure 5.
NSG1 Encodes a Zinc Finger Protein with a Single C2H2 Motif.
(A) Phylogenetic tree for NSG1-like proteins. The phylogenetic tree was constructed using the neighbor-joining method based on the Jones–Taylor–Thornton matrix-based model. Bootstrap support values calculated from 1000 replicates are given at the branch nodes. Ac, Ananas comosus; Aet, Aegilops tauschii; Bd, Brachypodium distachyon; Ca, Capsicum annuum; Cc, Cajanus cajan; Cia, Cicer arietinum; Do, Dichanthelium oligosanthes; Eg, Elaeis guineensis; Gh, Gossypium hirsutum; Gm, Glycine max; Jc, Jatropha curcas; Ma, Musa acuminata; Nt, Nicotiana tabacum; Os, Oryza sativa; Pt, Populus trichocarpa; Sb, Sorghum bicolor; Si, Setaria italica; Sl, Solanum lycopersicum; Tc, Theobroma cacao; Vr, Vigna radiata; Vv, Vitis vinifera; Zm, Zea mays.
(B) Analysis of the subcellular localization of the NSG1 protein. Bar = 50 µm.
(C) to (E) Analysis of the transcriptional activation of NSG1 using the DLR assay system. Transactivation activity in rice protoplasts transfected with a pUAS-fLUC reporter construct, effector constructs fused with GAL4BD, and a p35S-rLUC normalization construct. (C) Schematic representation of effectors of GAL4BD plus the full-length or various truncated visions of NSG1. (D) and (E) Measurement of relative LUC activity in rice transient assay. VP16, a transcriptional activator, was used as a positive control (D), and GAL4-BD was regarded as a negative control (E). Bars and asterisks (**) represent sd and significant difference at P < 0.01, Student’s t test.
Phylogenetic analysis revealed that QALGGH-type single C2H2 zinc finger genes from angiosperms formed a NSG1-like clade that was further resolved into three subclades. Subclade 3 comprised all proteins from dicots, whereas the proteins from monocots were placed in subclades 1 and 2. NSG1, its paralog Os02g35460, and several orthologs from grasses were placed in subclade 1, whereas the paralog Os09g26210 and several orthologs from grasses and other monocots were in subclade 2 (Figure 5A; Supplemental Data Sets 1 to 3).
The protein motif prediction program MEME was used to analyze the full-length amino acid sequences of NSG1, partial NSG-like proteins, SUP, JAG, and SL1. NSG1 shared very similar motif structures (motifs 1, 2, 3, and 6) with other proteins in subclade 1, although it lacked motifs 4 and 5. Almost all NSG1-like genes shared three extremely similar motifs (motifs 1, 2, and 3), except At1g80730 with only motif 1 and motif 3. In addition, SUP and JAG shared motifs 1 and 2 with NSG1-like proteins, whereas SL1 shared only motif 1 with NSG1-like proteins (Supplemental Figure 7A). Motif 1 was characterized as a classic QALGGH-type C2H2 zinc finger DNA binding domain (Li et al., 2013), while both motifs 2 and 3, located at N and C termini, respectively, were considered as the LxLxL-type ethylene-responsive element binding factor–associated amphiphilic repression (EAR) motifs, which contain two distinct types (LxLxL and DLNxxP) and are believed to specifically interact with corepressor TOPLESS-RELATED PROTEIN (TPR) to restrict transcription of target genes (Ke et al., 2015). In the nsg1-1 mutant, the 13-bp insertion would lead to a frame shift after the first 147 amino acids, destroying the C-terminal EAR motif, while in the nsg1-3 mutant the amino acid change occurred in the conserved C2H2 zinc finger DNA binding domain (Figure 4A; Supplemental Figure 6).
To determine the subcellular localization of NSG1, the fluorescence signal of the NSG1P:NSG1:GFP fusion protein was observed directly in the complementary transgenic lines, which was confirmed by immunoblotting with anti-GFP (Supplemental Figure 7C). The GFP signals were localized in the nucleus, which suggested that NSG1 is a nucleus-targeted protein (Figure 5B). Next, a dual luciferase reporter (DLR) was used to investigate further the transcriptional regulation activity of NSG1 (Figures 5C to 5E). The positive control VP16 showed relatively high luciferase (LUC) activity, whereas VP16-NSG1 fusion protein displayed much lower activity than the positive control (Figure 5D). Meanwhile, the full-length NSG1 protein and the NSG1-C (129 to 175) truncated protein (containing motif 2) showed significantly lower LUC activity than the negative control, whereas the LUC activity in NSG1-N (1 to 40; containing motif 3), NSG1-∆N (41 to 175; lacking motif 3), and NSG1-∆C (1 to 135; lacking of motif 2) showed no significant difference compared with the negative control (Figure 5E). These findings demonstrated that NSG1 acted as a transcriptional repressor and that the C-terminal EAR motif, but not the N-terminal motif, was necessary for activity of transcriptional repression.
Expression Patterns of NSG1
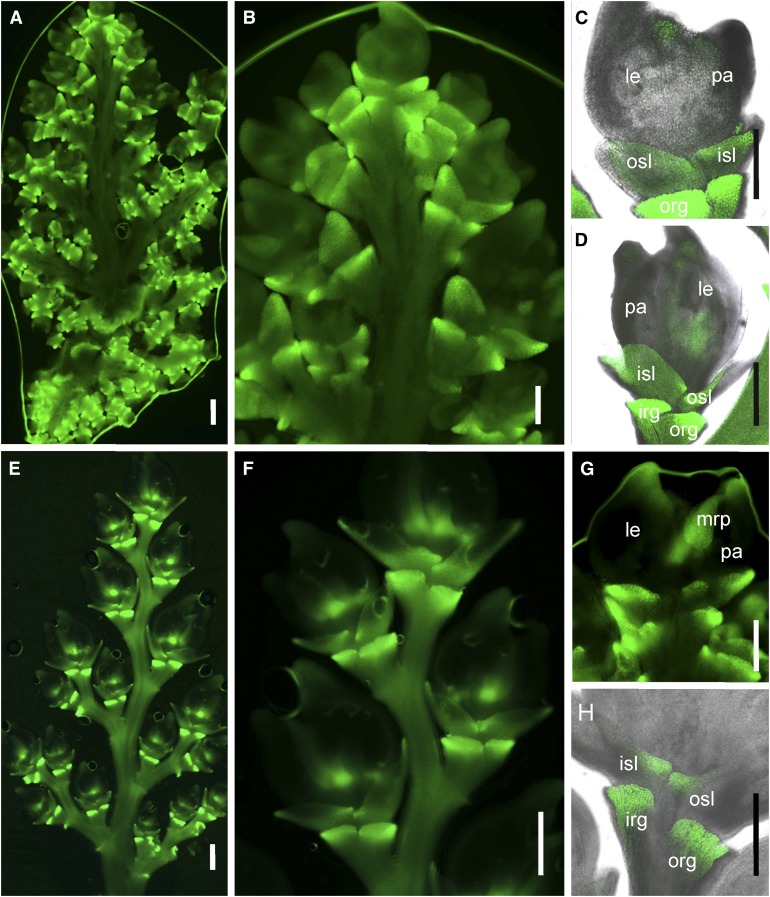
To determine the spatiotemporal expression patterns of NSG1, we used the GFP reporter gene to detect NSG1 expression in panicles at different developmental stages. In the complementation transgenic lines, the GFP expression pattern in the young panicle was examined in detail. Strong GFP signals were observed in the primordia of the rudimentary glume and sterile lemma (Figures 6A to 6D), while very weak GFP signal was detected in lemma and palea (Figures 6C and 6D). With spikelet development, the signal in both the sterile lemmas and rudimentary glumes was still high, although it was stronger in the latter (Figures 6E to 6H). At a stage after approximately Sp7, asymmetrical signal distribution was observed in sterile lemmas, in which GFP expression was stronger in the basal margin region but weaker expression was observed in other regions (Figure 6G). In addition, after about Sp7, the GFP signal was obvious at the apex of the lemma and palea primordia and in the mrp primordia (Figures 6G).
Figure 6.
Expression Pattern of NSG1.
GFP signal indicating NSG1 expression in transgenic complementation plants harboring the construct NSG1P:NSG1:GFP.
(A) A whole young panicle (∼1 cm in length).
(B) Several spikelets at developmental stages Sp4 to Sp7.
(C) A spikelet at Sp6.
(D) A spikelet at Sp7.
(E) A primary branch of a panicle (∼3.0 cm in length).
(F) Several spikelets at developmental stages Sp8 and post-Sp8.
(G) A spikelet at Sp8.
(H) A spikelet at post-Sp8.
Bar = 50 μm.
These results indicate that during spikelet development, NSG1 was always strongly expressed in the rudimentary glumes and sterile lemma, while the level of its expression in mrp and other organs/regions rose gradually. Therefore, the NSG1 expression pattern corresponded well with the function of the protein in organ identity of the spikelet.
Effects of NSG1 Mutation on Expression Patterns of Organ Identity Genes during Early Stages of Spikelet Development
To further examine the regulatory mechanism of NSG1, the expression patterns of four genes responsible for the specification of organ identity in spikelet were investigated during the early stages of spikelet development (from Sp4 to Sp8; Figure 7; Supplemental Figure 9). Quantitative real-time PCR (qPCR) analysis showed that transcription of DL was upregulated in the nsg1-1 mutant during the three defined stages of spikelet development (panicle length, < 0.5 cm, 0.5 to 1 cm, and 1 to 2 cm), LHS1 was upregulated in the nsg1-1 mutant in >0.5-cm panicles, and MFO1 transcripts were more abundant in 0.5- to 1-cm panicles of the nsg1-1 mutant, compared with those of the wild type (Supplemental Figures 8A to 8C). Transcripts of G1 exhibited a significant decrease in abundance in the nsg1-1 mutant at all defined stages of spikelet development compared with those in the wild type (Supplemental Figures 8D).
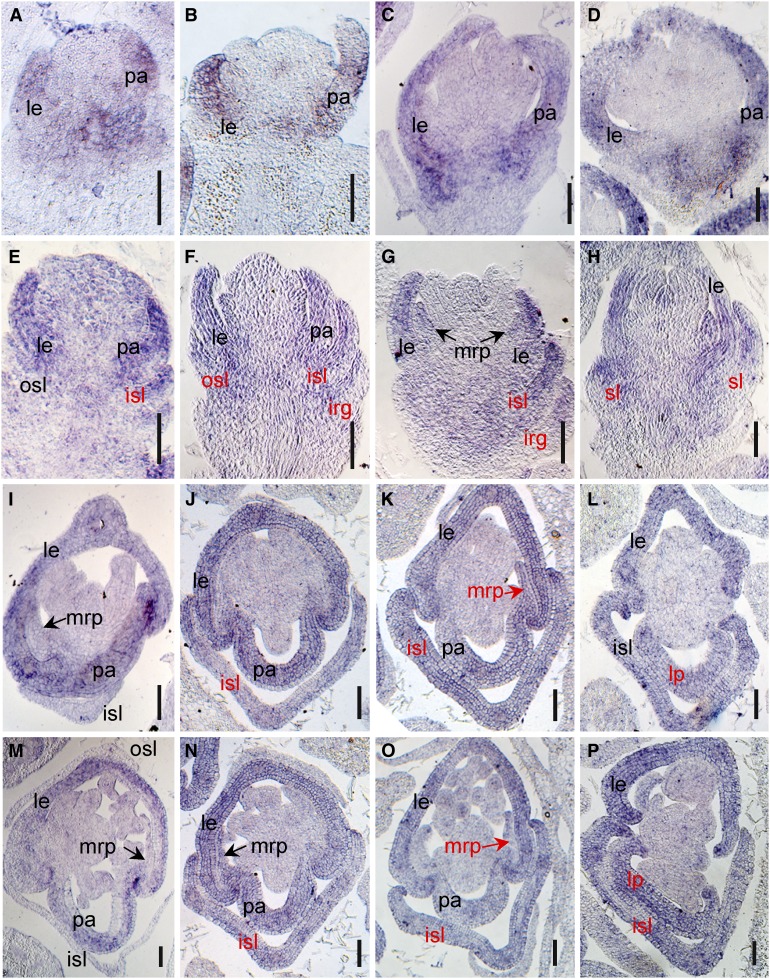
Figure 7.
Expression Pattern of LHS1 in Spikelets of the Wild Type and the nsg1-1 Mutant.
(A) to (D), (I), and (M) Expression of LHS1 in wild type spikelets, using in situ hybridization.
(E) to (H), (J) to (L), and (N) to (P) Expression of LHS1 in nsg1-1 spikelets, using in situ hybridization.
(A) and (E), (B) and (F), (C) and (G), (D) and (H) show longitudinal sections of spikelets at stages Sp5 to Sp8, respectively. (I) to (L) and (M) to (P) show transverse sections of basal region and middle region of spikelets at stages of Sp8, respectively.
Black type indicates the normal expression of genes, and red type indicates the ectopic or abnormal expression signal of genes.
Bar = 50 μm.
irg, inner rudimentary glume; isl, inner sterile lemma; le, lemma; mrp, marginal region of palea; osl, outer sterile lemma; pa, palea; sl, sterile lemma.
Expression of these organ identity genes was further detected using in situ hybridization. We first detected the expression pattern of LHS1, the ectopic expression of which resulted in lemma-like sterile lemma and elongated rudimentary glumes in rice in previous study (Wang et al., 2017). In the wild-type spikelets, the strong LHS1 signal was always detected in lemma and palea primordia after approximately Sp4 (Figures 7A to 7D). It is noteworthy that LHS1 was mainly expressed in the bop, but not in the mrp (Figures 7I and 7M). In nsg1-1 spikelet, the expression of LHS1 in lemma primordia was not changed. However, the ectopic expression of LHS1 could be detected in the sterile lemma primordia frequently and in the mrp primordia occasionally (Figures 7E to 7H, 7J to 7L, and 7N to 7P). To further clarify the ectopic expression domain, we conducted in situ hybridization of LHS1 using transverse sections of wild-type and nsg1-1 spikelets at Sp8. It was very clear that LHS1 was expressed strongly in the inner lemma-like sterile lemma of nsg1-1 spikelets, and obvious signals were also detected in mrp of nsg1-1 palea (Figures 7J to 7L and 7N to 7P), yet there were no signal detected in these regions of the wild-type spikelets (Figures 7I and 7M). We also found that LHS1 was expressed in whole domain of some lemma-like palea of nsg1-1 spikelets, in which it was difficult to distinguish the mrp and bop (Figures 7L and 7P). Taken together, these results indicated that LHS1 was ectopically expressed in sterile lemma and mrp of nsg1-1 spikelets frequently and in rudimentary glumes occasionally.
The expression patterns of G1, DL, and MFO1 were also detected by in situ hybridization. Strong signals for G1 were detected in the wild-type sterile lemmas during primordium initiation and formation at approximately Sp4 and then slowly decreased but were still detectable in sterile lemmas after initiation of elongation during Sp5 to Sp8 (Supplemental Figure 9A). A signal for G1 expression was also observed in the primordia of the rudimentary glumes and palea (Supplemental Figures 9A2 to 9A4). In spikelets of the nsg1-1 mutant, however, the G1 expression signal was observed only in a minority of palea primordia and was almost undetectable in the primordia of the sterile lemmas and rudimentary glumes at all stages examined (Supplemental Figure 9B). During Sp4 and Sp5, DL expression in the wild type was detected only in lemma primordia and not in pistil primordia (Supplemental Figures 9C1 and 9C2). After Sp6, DL was also expressed in pistil primordia in the wild type (Supplemental Figures 9C3 to 9C5). In nsg1-1 spikelets, DL expression in primordia of the lemma and pistil was unchanged, and ectopic expression signals of DL were detected in primordia of the sterile lemmas and palea (Supplemental Figure 9D, indicated by red type). During Sp4 to Sp8, MFO1 expression was detected in the primordia of the mrp and lodicule in both the wild type and nsg1-1 mutant (Supplemental Figures 9E and 9F). However, ectopic expression of MFO1 was detected in the partial primordia of the sterile lemmas and rudimentary glumes (Supplemental Figure 9F, indicated by red type). In addition, the region of MFO1 expression in the FM was changed in nsg1-1 spikelets. During Sp4, MFO1 expression was restricted to the FM of the wild type (Supplemental Figure 9E1), whereas MFO1 expression was expanded to the receptacle and rachilla in spikelets of the nsg1-1 mutant (Supplemental Figures 9F1 to 9F5, indicated by red triangles).
In summary, ectopic expression of LHS1, DL, and MFO1, and absence of G1 expression in primordia of the rudimentary glumes, sterile lemmas, and palea of nsg1-1 spikelets implied that the expression of these genes was regulated negatively or positively by NSG1.
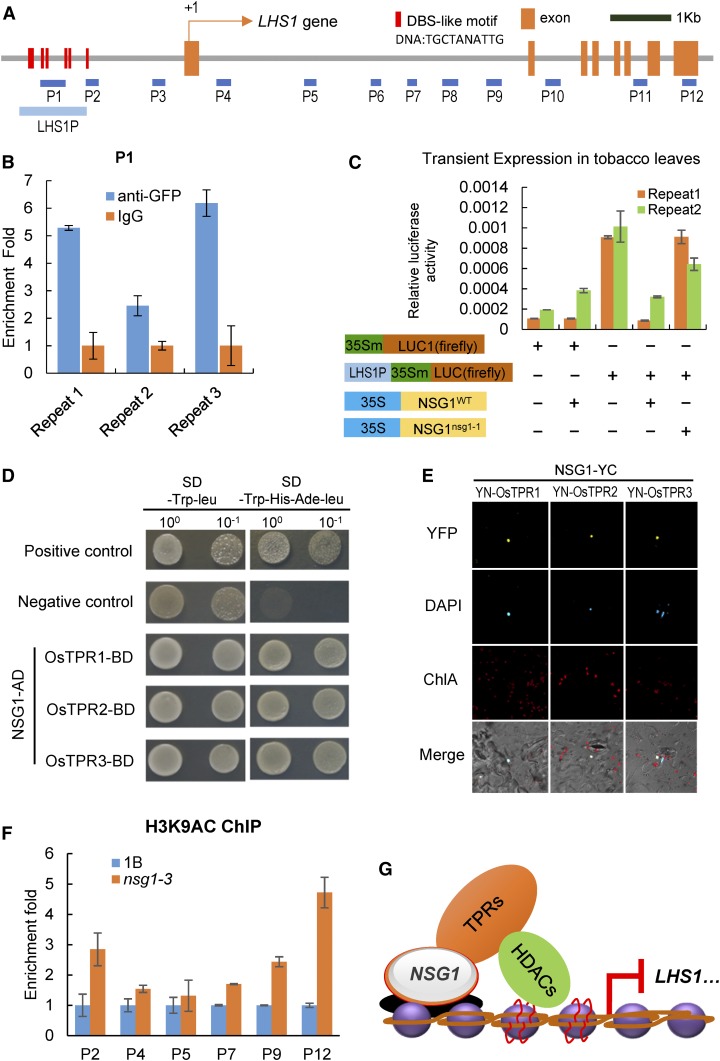
NSG1 Represses LHS1 Directly by Interacting with Corepressors OsTPRs
The above-mentioned analysis of EAR motifs, transcriptional activity, and expression patterns indicates that NSG1 may act as a repressor to regulate the transcription of DL, LHS1, and/or MFO1. It was found first that there was a cluster of DST-binding sequence (DBS)-like motifs located in the LHS1 promoter (Figure 8A; Supplemental Figure 10), which were characterized as a conserved binding site of C2H2 zinc finger domain (Huang et al., 2009b). Then, chromatin immunoprecipitation (ChIP) assays were used to examine whether NSG1 protein is able to bind to this region of the LHS1 promoter. Chromatin isolated from young panicles of NSG1P:NSG1:GFP transgenic lines was immunoprecipitated with the GFP antibody and then subjected to qPCR analysis using primer P1. The repeatable results showed that NSG1P:NSG1:GFP protein was able to bind stably to this P1 site (Figure 8B). These results suggest that NSG1 protein might be responsible for direct regulation of LHS1. Next, we detected the effect of NSG1 protein on the expression of the firefly LUC gene reporter using the DBS-like motif region of LHS1 as the promoter for transient expression assays in Nicotiana benthamiana leaves. Compared with the negative control p35Sm:LUC reporter, the LUC activity was significantly increased with the pLHS1P-35Sm:LUC reporter (Figure 8C). Coexpression of the pLHS1P-35Sm:LUC reporter with 35S:NSG1WT led to a significant decrease of the LUC activity, whereas 35S:NSG1nsg1-1 failed to downregulate the expression of the pLHS1P-35Sm:LUC reporter (Figure 8C). Therefore, these results indicated that NSG1WT protein had the ability to repress the expression of LHS1 by binding the DBS-like motif region in LHS1 promoter.
Figure 8.
Direct Regulation of LHS1 and DL Expression by NSG1.
(A) Distribution of potential binding sites in the promoter and open reading frame regions of LHS1. Blue bars indicate the DNA fragments amplified in the ChIP assays. Red bars indicate the DBS-like motifs in the promoter of LHS1.
(B) ChIP-qPCR for P1 site of LHS1 with anti-GFP antibody. ChIP enrichment compared with the input sample was tested by qPCR. Error bars indicate sd of three repeats.
(C) NSG1WT represses LHS1 expression in vivo. N. benthamiana leaves were transformed with p35Sm:LUC, p35Sm:LUC plus 35S:NSGWT, pLHS1P-35Sm:LUC, pLHS1P-35Sm:LUC plus 35S:NSGWT, or pLHS1P-35Sm:LUC plus 35S:NSGnsg1-1. Error bars indicate sd of three repeats.
(D) NSG1 interacts with OsTPR1, OsTPR2, and OsTPR3 in yeast cells by a Y2H assay.
(E) NSG1 interacts with OsTPR1, OsTPR2, and OsTPR3 in the nucleus of N. benthamiana leaf cells by a BiFC assay.
(F) ChIP-qPCR for several sites of LHS1 with anti-H3K9ac antibody between wild-type panicles and nsg1-1 panicles. Error bars indicate sd of three repeats.
(G) Model of NSG1 repressing the expression of LHS1 by recruiting TPRs-HDACs.
The proteins with EAR motifs are generally believed to interact with corepressors TPR. In rice, there are three TPR proteins (Yoshida et al., 2012): OsTPR1 (LOC_Os01g15020); OsTPR2 (LOC_Os08g06480), also named as ABERRANT SPIKELET AND PANICLE1 (ASP1); and OsTPR3 (LOC_Os03g14980). To further clarify the molecular mechanism of NSG1 regulating LHS1 expression, the interaction between NSG1 and OsTPRs was examined. First, yeast-two hybrid (Y2H) assays were performed to determine the physical interaction between NSG1 and OsTPRs. NSG1 interacted with all three OsTPR proteins in yeast cells (Figure 8D). Next, a bimolecular fluorescence complementation (BiFC) assay was conducted to detect whether the NSG1 could interact with OsTPRs in N. benthamiana leaves. The yellow fluorescent protein signal was found in the nucleus of epidermal cells of N. benthamiana leaves coexpressing the NSG1-YC fusion protein with YN-OsTPR1, YN-OsTPR2, or YN-OsTPR3 (Figure 8E). Thus, NSG1 can interact with OsTPRs in plant cells. It is well known that TPR can interact with histone deacetylases (HDACs) to mediate the acetylation level of histone protein in the targeted region (Ke et al., 2015). Therefore, we further detected the histone acetylation level in the LHS1 promoter and CDS region by ChIP-qPCR using the chromatin immunoprecipitated by an anti-H3K9ac antibody. The acetylation levels of H3K9 at P2, P4, P5, P7, P9, and P12 sites, most of them in the first intron of LHS1, were significantly increased in the nsg1-1 mutant compared with those in the wild type (Figure 8F).
Therefore, the above-described results support that NSG1 can directly bind to the promoter or other regulating region of LHS1 genes and then recruit the corepressor TPRs to repress LHS1 expression by downregulating the acetylation levels of histone on chromosomes at LHS1 (Figure 8G).
Genetic Interaction between NSG1 and LHS1
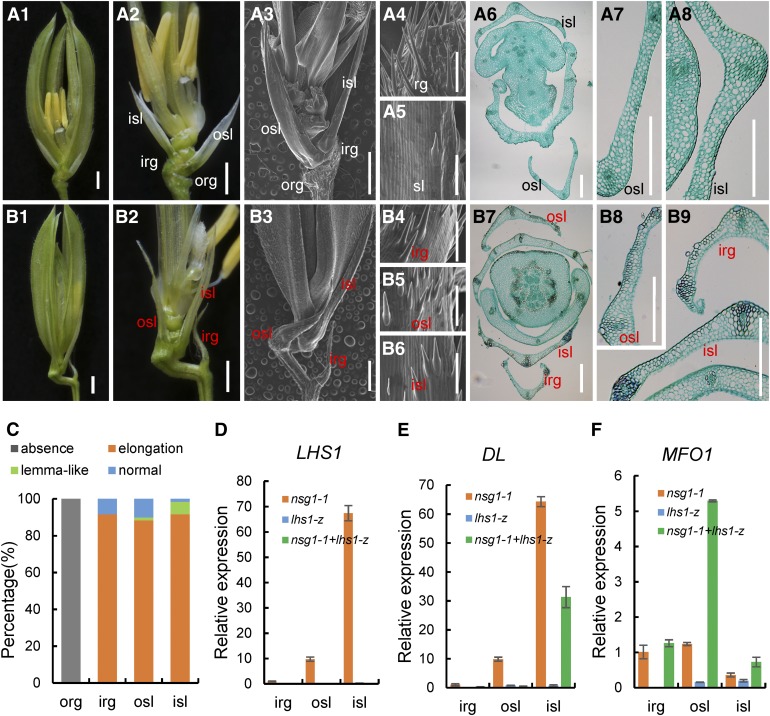
We further examined genetic interactions between NSG1 and LHS1 using nsg1-1 lhs1-z double mutants. The lhs1-z mutant is an allelic loss-of-function mutant of LHS1, as characterized by our group (Li, 2008). The mutation of LHS1 in lhs1-z and other allelic mutants led to transformation of the lemma into leaf-like organs that were longer and thinner than the wild-type lemma, owing to the absence or decreased abundance of fibrous sclerenchyma and the presence of a weaker silicified abaxial epidermis. However, the identities of the sterile lemmas and rudimentary glumes were not affected (Figure 9A; Jeon et al., 2000; Prasad et al., 2005). In the double mutant, the sterile lemmas and rudimentary glumes remained longer and wider than those of the wild type or lhs1-z, but they were distinctly shorter and thinner than those of the nsg1-1 mutant (Figures 1A1 to 1A3, 9A1 to 9A3, and B to B9B1 to 9B3). It was also observed that the abaxial surface of the inner rudimentary glumes and the outer and inner sterile lemmas were smooth, with several trichomes in the double mutant (Figures 9B4 to 9B6), which were identical to the sterile lemmas of the wild type and lhs1-z mutant (Figures 1A4, 1A5, 9A4, and 9A5).
Figure 9.
Spikelet Phenotypes of the lhs1 Single Mutants and the nsg1-1 lhs1-z Double Mutant.
(A) Spikelets of the lhs1-z mutant. (A1) and (A2), lhs1-z spikelet (A1); the lemma and palea were removed in (A2). (A3) to (A5) lhs1-z spikelet; the lemma and palea were removed in (A3). (A4) and (A5) show the surface characteristics of rg and sl, respectively. (A6) to (A8) Transverse sections of lhs1-z spikelet; (A7) and (A8) show the anatomical structure of osl and isl, respectively.
(B) Spikelets of the nsg1-1 lhs1-z mutant. (B1) and (B2), nsg1-1 lhs1-z spikelet; the lemma and palea were removed in (B2). (B3) to (B6) nsg1-1 lhs1-z spikelet. (B4) to (B6) Surface characters of the elongated irg, osl, and isl, respectively. (B7) to (B9) Transverse sections of nsg1-1 lhs1-z spikelet; (B8) and (B9) show the anatomical structure of irg, osl, and isl, respectively.
(C) Percentage of elongated or lemma-like organs in nsg1-1 spikelets.
(D) to (F) qPCR analysis of LHS1, DL, and MFO1 expression in spikelets of nsg1-1, lhs1-z, nsg1-1 lhs1-z double mutant. ACTIN was used as a control. RNA was isolated from rg and sl of nsg1-1, lhs1-z, nsg1-1 lhs1-z spikelets. Error bars indicate sd. At least three replicates were performed, from which the mean value was used to represent the expression level.
Bar in *1, *2, and *3 = 1000 μm (the asterisk means [A] and [B]); bar in *4, *5, and (B6) = 100 μm (the asterisk means [A] and [B]); bar in the other images = 100 μm.
irg, inner rudimentary glume; isl, inner sterile lemma; org, outer rudimentary glume; osl, outer sterile lemma; rg, rudimentary glume; sl, sterile lemma.
Histological analysis revealed that very few fibrous sclerenchyma cells and silicified abaxial epidermis cells were observed in the rudimentary glumes and sterile lemmas of the double mutant (Figures 9B7 to 9B9), whereas numerous such cells were observed in some rudimentary glumes and sterile lemmas of the nsg1-1 mutant (Figures 2B to 2D). In addition, no rudimentary glumes were transformed into lemma-like organs in the double mutant, and the number of lemma-like sterile lemmas was distinctly less than that in the nsg1-1 mutant (Figure 9C).
The expression levels of LHS1, DL, and MFO1 were detected by qPCR in rudimentary glumes and sterile lemmas of the double mutant as well as the lhs1-z and nsg1-1 single mutants (Figures 9D to 9F). Give that lhs1-z was a transposon insertion mutant of LHS1, expression of LHS1 was not detected in the lhs1-z single mutant nor the nsg1-1 lhs1-z double mutant (Figure 9D). In contrast to the ectopic expression of DL in the outer and inner sterile lemmas of the nsg1-1 spikelet, DL expression was not detected in the outer sterile lemma and was substantially decreased in the inner sterile lemma of the double mutant and was not detected in these organs in the lhs1-z mutant (Figure 9E). MFO1 was weakly expressed in rudimentary glumes and sterile lemmas of the lhs1-z mutant, similar to that of the wild type, whereas the ectopic expression level was higher in the inner rudimentary glume, and in the outer and inner sterile lemmas of the double mutant than in the nsg1-1 single mutant (Figure 9F). Collectively, these findings suggest that ectopic LHS1 activation was an important cause of ectopic formation of lemma-like tissue in the nsg1-1 sterile lemmas and rudimentary glumes.
DISCUSSION
Role of NSG1 in Specification of Sterile Lemmas and Rudimentary Glumes
In three allelic mutants of NSG1, the rudimentary glumes and the sterile lemmas were elongated and/or widened, and most of rudimentary glumes and a minority of outer sterile lemmas were transformed into the lemma and/or mrp-like organ, and most of outer sterile lemmas and inner sterile lemmas were transformed into the lemma-like organ. It was previously reported that G1 regulates sterile lemma identity. In all mutants of G1, the sterile lemma is transformed into a lemma-like organ (Yoshida et al., 2009). PAP2 regulates the specification of sterile lemmas and rudimentary glumes. In mutants of PAP2, the sterile lemmas and rudimentary glumes are elongated and show a leaf/lemma-like identity (Gao et al., 2010). These results suggest that G1, PAP2, and NSG1 are involved in maintaining the identity of rudimentary glumes and/or sterile lemmas by preventing incorrect cell differentiation.
Two prevailing hypotheses on the origin and evolution of the sterile lemmas have been proposed. One hypothesis states that a putative ancestor of Oryza had a three-florets spikelet that contained a terminal floret and two lateral florets, in which the palea and inner floral organs were absent, and the residual lemma was reduced to a sterile lemma during evolution (Arber, 1935; Kellogg, 2009; Yoshida et al., 2009; Kobayashi et al., 2010; Zhang, 2017). The second hypothesis suggests that the spikelet of Oryza species contained only one floret, and the sterile lemmas and rudimentary glumes are universally regarded as severely reduced bract structures (Schmidt and Ambrose, 1998; Hong et al., 2010). Mutation of G1, EG1, PAP2, ASP1, or NSG1, or ectopic expression of LHS1 causes homeotic transformation of the sterile lemma into a lemma to various degrees, which suggests that the sterile lemmas are homologous to lemmas, and thus partially supports the first hypothesis (Li et al., 2009a; Yoshida et al., 2009, 2012; Wang et al., 2013, 2017; Lin et al., 2014). In the lf1 mutant, lateral florets are formed in the axil of each sterile lemma, which provides strong support for the three-florets spikelet hypothesis (Zhang, 2017). However, the sterile lemma is degenerated and acquires the identity of a rudimentary glume in the mfs1 mutant. Interestingly, some elongated rudimentary glumes appeared to acquire the identity of a sterile lemma and/or lemma in the nsg1 mutants. These findings also support the opinion of sterile lemmas being homologous to rudimentary glumes in the second hypothesis.
In most grass species, the spikelet lacks sterile lemma-like organs and contains one or more florets and bract-like glume organs, which are considered to be equivalent to the rudimentary glumes of Oryza species (Yoshida et al., 2009; Hong et al., 2010). The bract-like glume organ resembles the lemma in size and structure in some grass species, such as maize and wheat (Kellogg, 2001; Yoshida et al., 2009), whereas it is severely reduced in Oryza species (Bommert et al., 2005; Li et al., 2009b). The above-mentioned results therefore imply that in rice the sterile lemma was derived from the lemma of the lateral floret, and the sterile lemmas and lemma are homologous to the rudimentary glume, which are derived from bract-like structures.
Interestingly, according to the phenotype analysis of nsg1-1 and nsg1-3 mutants, the outer rudimentary glume was absent in ∼100% of nsg1-1 spikelets and 78% of nsg1-3 spikelets (Figure 1G; Supplemental Figure 3A). In fact, we found the outer rudimentary glume was invisible at later spikelet growth stage under optical microscopy, as previously researched by Wang et al. (2013), but could not clarify whether it was degraded or absent. In this study, to further conform this, scanning electron microscopy analysis was used to carefully investigate the formation and differentiation of organ primordia in nsg1-1 spikelets at an early development stage. The results clearly indicated that the primordia of outer rudimentary glumes were not formed from the beginning (Figures 3B and 3C). There was degeneration on the glume-like organs in many rice mutants, such as lemma and palea in lhs1 and sl1 mutants, sterile lemma in mfs1 mutant (Jeon et al., 2000; Prasad et al., 2005; Xiao et al., 2009; Ren et al., 2013). However, the absence of all parts of an organ in spikelets is by no means rare. Therefore, this mutant will be useful in the future to clarify the formation mechanism of outer rudimentary glumes.
NSG1 Regulates Palea and Lodicule Development
In grass florets, the palea was considered to show an identity and origin distinct from those of the lemma. In general, the palea is considered to be homologous to the prophyll (the first leaf produced by the axillary meristem) formed on a floret axis, whereas the lemma corresponds to the bract (the leaf subtending the axillary meristem) formed on a spikelet axis (Kellogg, 2001; Ohmori et al., 2009).
Several previous studies have indicated that the specification of mrp and bop in the palea is controlled by different genes. In the depressed palea1 (dp1) mutant, the bop is lost and two mrp-like structures remain (Luo et al., 2005; Jin et al., 2011). In the retarded palea1 (rep1) mutant, development of the bop is delayed (Yuan et al., 2009). In the mfs1-1 mutant, the bop is degenerated in most florets and is absent in a minority of florets. However, in mutants of CHIMERICAL FLORAL ORGANS1 (CFO1) and MFO1, the mrp is transformed into a lemma-like structure (Ohmori et al., 2009; Li et al., 2010; Sang et al., 2012). Therefore, while MFS1, DP1, and REP1 determine bop identity, MFO1 and CFO1 are involved in regulation of mrp identity. In the nsg1-1 mutant, more than 50% of spikelets contained a lemma-like palea because the mrp gained a lemma-like identity to various degrees, and approximately one-third of spikelets showed a degenerated palea owing to degeneration or absence of the bop. These results indicate that NSG1 plays a dual role in regulation of both bop and mrp development in the palea, possibly through regulation of genes in different pathways.
The lodicule in grasses is usually considered to be the equivalent organ to the petal of dicots, which is mainly controlled by B-function genes. The rice B-class mutant superwoman1 (spw1/lhs16) and OsMADS2 OsMADS4 double RNA interference plants show transformation of the lodicules into organs that resemble the mrp (Nagasawa et al., 2003; Yadav et al., 2007; Yao et al., 2008). The mutants of CFO1 and MFO1 show lemma/pistil-like, but not mrp-like lodicules. It has been demonstrated that CFO1 regulates lodicule identity by restricting the expression of DL, which is involved in specification of the lemma and pistil. Therefore, specification of the lodicule in rice involves different regulatory pathways associated with B-function genes and CFO1–MFO1 genes (Li et al., 2010; Sang et al., 2012). In this study, the lodicules were elongated and/or widened and gained a similar identity to that of the mrp or the lemma in the nsg1 mutants. These results suggest that NSG1 is involved in the regulation of lodicule identity associated with more B-function gene pathways than the CFO1–MFO1 pathway.
NSG1 Maintains Organ Identity of the Spikelet by Regulation of LHS1, MFO1, DL, and G1 Expression
In rice spikelets, the organs can be divided into floral organs (lemma, palea, lodicule, stamens, and pistil) within the floret and nonfloral organs (rudimentary glumes and sterile lemmas) that subtend the floret. The DL gene and some MADS-box genes, including ABCE-class genes (A-class RICE APETALA 1A, B-class OsMADS2/4/16, C-class RICE AGAMOUS and OsMADS58, D-class OsMADS13, and E-class LHS1), MFO1, and CFO1, are considered to be floral organ identity genes (Li et al., 2011; Sang et al., 2012). The genes G1, EG1, PAP2, and ASP1 are involved in specification of the rudimentary glume and sterile lemma (Yoshida et al., 2009, 2012; Gao et al., 2010). The limited proper expression domain of these genes is necessary to maintain these organ identities. The interactive restriction between SPW1 and DL might be crucial for specification of stamens and the pistil (Nagasawa et al., 2003; Yamaguchi et al., 2004), and repression of CFO1 to DL is crucial for formation of the mrp, lodicule, and stamens (Sang et al., 2012). Epigenetic regulation by the EMF1-like gene, DEFORMED FLORAL ORGAN1 (DFO1)/CURVED CHIMERIC PALEA1 (CCP1), which is widely responsible for repression of OsMADS3, OsMADS58, OsMADS13, and DL, also contributes to establishment of the identity of floral organs, such as the palea, lodicule, and stamen (Yan et al., 2015; Zheng et al., 2015). However, limited information is available on gene regulation pathways for nonfloral organs of the spikelet in rice.
In this study, the identities of all organs except the pistil and lemma were affected in the nsg1-1 spikelet and were all transformed into lemma/mrp-like organs to various degrees. In the nsg1-1 spikelet, LHS1, DL, and MFO1 were ectopically expressed in two or more organs, including the rudimentary glume, sterile lemma, palea, lodicule, and stamen, whereas G1 was expressed in the rudimentary glume and sterile lemma. We further verified that the NSG1 protein could bind to the sites in the promoter of LHS1, interact with rice corepressors OsTPRs, and then recruit HDACs to mediate the histone acetylation level in the targeted region. Collectively, these results suggest that NSG1 maintained organ identities in the spikelet by direct epigenetic repression of LHS1 and indirect regulation of DL, MFO1, and G1 (Figure 10). Following loss of function of NSG1 in the nsg1 mutants, these organ identity genes were ectopically expressed or down-regulated, which led to the transformation of the organs into lemma-like organs (Figure 10).
Figure 10.
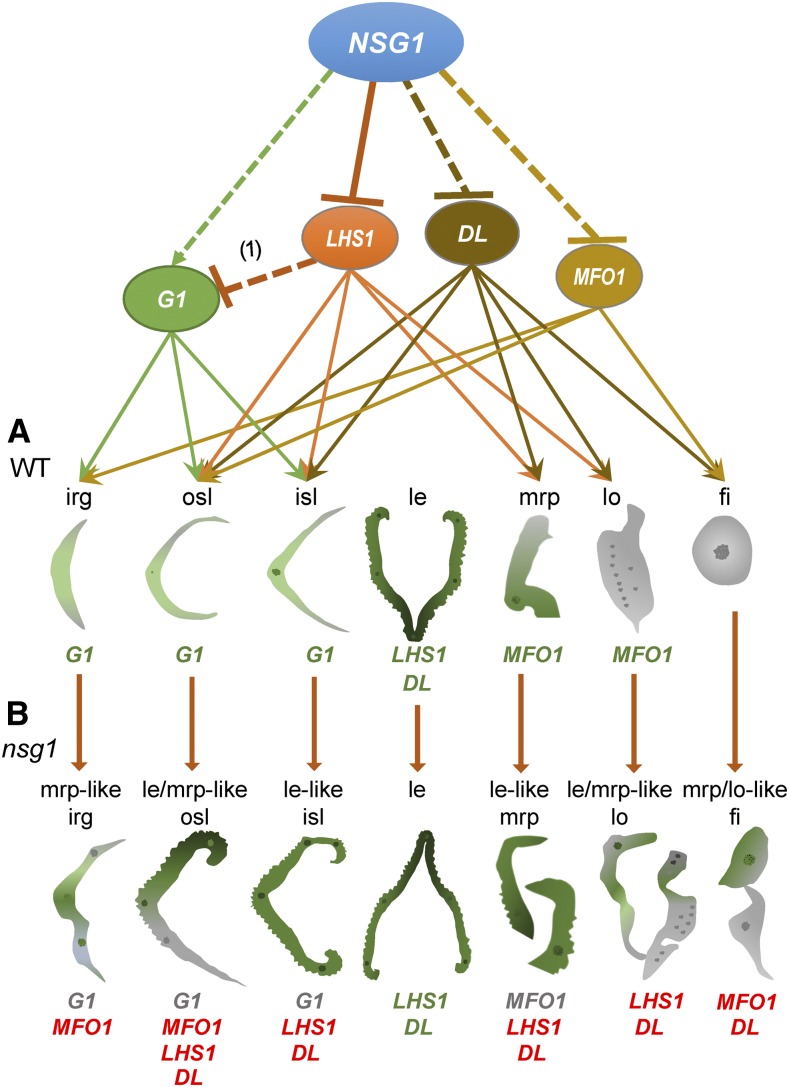
Roles of NSG1 in the Specification of Organ Identity in the Rice Spikelet.
(A) In the wild-type spikelet, G1, LHS1, DL, and MFO1 play pivotal roles in specification of lateral organ identities (G1 for rg and sl, LHS1 for le, DL for le, and MFO1 for mrp and lo). NSG1 acts as a repressor to regulate the specification of lateral organs, including rg, sl, mrp, lo, and st, by directly repressing LHS1 and DL, as well as MFO1, and indirectly activating G1 through LHS1. (1) indicates a study that suggests LHS1 represses G1 (Wang et al., 2017).
(B) In the nsg1 spikelet, with the loss of NSG1 function, LHS1, DL, and MFO1 were ectopically expressed in the rg, sl, mrp, lo, or st, and G1 expression was under-regulated in the rg and sl, which led to transformation of other lateral organs into lemma-like organs. In the nsg1-1 lhs1-z mutant, the lemma-like identity was lost completely in the nsg1-1 sterile lemmas and rudimentary glumes, suggesting that ectopic LHS1 activation was a key cause of ectopic formation of lemma-like tissue in these organs. The gene symbols in green type show a normal expression domain, those in red type show an ectopic expression domain, and those in gray type show an under-regulation expression domain.
fi, filament; irg, inner rudimentary glume; isl, inner sterile lemma; le, lemma; le-like isl, lemma like inner rudimentary glume; le-like mrp, lemma like marginal region of palea; le/mrp-like fi, lemma and/or marginal region of palea like filament; le/mrp like lo, lemma and/or marginal region of palea like lodicule; le/mrp-like osl, lemma and/or marginal region of palea like outer sterile lemma; lo, lodicule; mrp-like irg, marginal region of palea like inner rudimentary glume; mrp, marginal region of palea; osl, outer sterile lemma.
METHODS
Plant Materials
Two mutants of rice (Oryza sativa), nsg1-1 and nsg1-3, were identified from among ethyl methanesulfonate–treated plants of the wild-type xian-type (indica) restorer line J10 and the wild-type xian-type (indica) maintainer line ‘Xida1B’, respectively. An additional mutant, nsg1-2, was derived from a T-DNA insertion library (Rice Mutant Database; Zhang et al., 2007) and had the geng-type (japonica) background of ZH11. All plants were cultivated in paddies at Chongqing, China.
Microscopy
Panicles were collected at different developmental stages and fixed in 50% (v/v) ethanol, 0.9 M glacial acetic acid, and 3.7% (v/v) formaldehyde over 16 h at 4°C. The fixed samples were dehydrated with a graded ethanol series, infiltrated with xylene, and embedded in paraffin (Sigma). The 8-μm sections were transferred onto poly-l-lysine–coated glass slides, deparaffinized in xylene, and dehydrated through an ethanol series. The sections were stained sequentially with 1% (w/v) safranin O (Amresco) and 1% (w/v) Fast Green (Amresco) and then dehydrated through an ethanol series, infiltrated with xylene, and finally mounted beneath a cover slip. Light microscopy was performed using an BX53 microscope (Olympus). For scanning electron microscopy, fresh samples were examined using a Hitachi SU3500 scanning electron microscope with a −20°C cool stage. The stages of early spikelet development were identical to those defined previously (Ikeda et al., 2004).
RNA Isolation and RT-qPCR Analysis
Total RNA was isolated from young panicles and organs within the spikelet from mutants and the wild type using the RNAprep Pure Plant RNA Purification Kit (Tiangen). The first-strand cDNA was synthesized from 2 µg of total RNA using oligo(dT)18 primers in a 25-μL reaction volume using the SuperScript III Reverse Transcriptase Kit (Invitrogen). Half a microliter of the reverse-transcribed RNA was used as the PCR template with gene-specific primers (Supplemental Table 1). qPCR analysis was performed with a CFX96 Real-Time PCR Detection System (Bio-Rad) and the TB Green Premix Ex Taq II (Tli RNaseH Plus, catalog no. RR820A, Takara). PCR cycling conditions for amplification were 95°C for 30 s followed by 40 cycles of 95°C for 5 s, 60°C for 30 s. Relative expression levels were determined compared with wild-type levels using the 2(−ΔCt) analysis method. ACTIN was used as an endogenous control. At least three replicates were performed, from which the mean value was used to represent the expression level.
Complementation Test
For the complementation test, the wild-type genomic fragment of NSG1 (Os04g36650), which consisted of the 2925-bp upstream sequence from the start codon and 525-bp CDS, was amplified using the primers NSG1com-F2 and NSG1com-R1. Given that the GC content in the fragment was extremely high, the high-fidelity thermostable DNA polymerases KOD FX and KOD neo (Toyobo) were used to amplify the fragment of NSG1 (Os04g36650). The intermediate vector 35S:GFP (S65T):NOS (pCAMBIA1301) was digested using HindIII and BamHI to generate the construct pCAMBIA1301:GFP:NOS. Using the method of homologous recombination, the resulting PCR products were inserted into the vector framework pCAMBIA1301:GFP:NOS to generate the complementary vector pCAMBIA1301-NSG:GFP:NOS (NSG:GFPcom). The recombinant plasmids were introduced into the nsg1-1 mutant using the Agrobacterium-mediated transformation method as described previously (Sang et al., 2012).
For examination of GFP fluorescence in the transgenic plants, young panicles and other tissues were transferred to a glass slide and then examined under an Olympus MVX10 microscope and an Olympus FluoView 1000 confocal laser scanning microscope. The primer sequences are listed in Supplemental Table 1.
DNA Gel Blot Analysis
For DNA gel blot detection, genomic DNA (15 to 20 μg) extracted from J10, XD1B, ZH11, and Nipponbare was used. The plasmid NSG:GFPcom was linearized with EcoRI as a positive control. The probes of NSG1 promoter and its open reading frame were amplified using the primer pairs NSG1sb-PF and NSG1sb-PR and NSG1sb-OF and NSG1sb-OR, respectively, and labeled using the PCR DIG Probe Synthesis Kit (Roche), in accordance with the manufacturer’s recommendations. Transfer to the membrane was performed using the iBlot 2 Gel Transfer Device (Invitrogen). Prehybridization, hybridization, and washing were performed in accordance with the manufacturer’s recommendations and as reported by Southern (Alwine et al., 1977). Detection was performed using the DIG-High Prime DNA Labeling and Detection Starter Kit II (chemiluminescent detection), in accordance with the manufacturer’s recommendations.
Protein Sequence and Phylogenetic Analysis
Protein sequences were obtained by searching the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) using the NSG1 protein sequence as a query. Using MEGA 5.0, a phylogenetic tree was constructed using the neighbor-joining method based on the Jones–Taylor–Thornton matrix-based model. Bootstrap support values for the tree topology were calculated from 1000 replicates (Felsenstein, 1985; Saitou and Nei, 1987; Jones et al., 1992; Tamura et al., 2011). Protein motifs were predicted using MEME (http://alternate.meme-suite.org/).
Analysis of Transcriptional Activity
Using the DLR assay system, we analyzed the transcriptional activity of NSG1 in rice protoplasts. The DLR assay system was used with a GloMax 20-20 luminometer (Promega) to measure the relative LUC activity (Ren et al., 2018). The coding frame of the NSG1 cDNA was fused to the GAL4 DNA binding domain (BD), driven by the 35S promoter. VP16, a transcriptional activator, was used as a positive control, and GAL4-BD was regarded as a negative control. The VP16, VP16-NSG, BD-NSG1, and GAL4-BD effectors were transiently expressed in rice protoplasts. The primers used are listed in Supplemental Table 1.
In Situ Hybridization
The gene-specific NSG1 probe was amplified with the primers NSG1-HF and NSG1-ishSP6HR and labeled using the DIG RNA Labeling Kit (SP6/T7, Roche). Probes for known floral organ genes were prepared using the same method. Pretreatment of sections, hybridization, and immunological detection were performed as previously described by Sang et al. (2012). The primer sequences are listed in Supplemental Table 1.
Protein Interaction Analyses
Y2H assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). The full-length coding region of NSG1 was amplified and ligated into the yeast expression vector pGADT7 (Clontech) to produce pGADT7-NSG1. The full-length coding region of TPRs was introduced into pGBKT7 (BD) (Clontech) to produce pGBKT7-TPRs. The yeast strain used in Y2H assays was Y2HGOLD. The pGADT7-T+pGBKT7-lam served as negative control and the pGADT7-T+pGBKT7-53 as positive control. These plasmids were cotransformed into Y2HGOLD strain in an AD-BD–coupled manner. Detailed procedures are described in the manufacturer’s instructions (Yeast Protocols Handbook, PT3024-1; Clontech). To conduct BiFC assays, NSG and TPRs were amplified by specific primers (Supplemental Table 1) with KOD-neo polymerase and then ligated into BiFC vectors, including pSCYNE (SCFP3A N-terminus, modified) and pSCYCE (SCFP3A C-terminus, modified; Waadt et al., 2008). Next, vector pairs were cotransformed into Nicotiana benthamiana leaves. The fluorescence signals were captured under a confocal laser scanning microscope (LSM 800, Zeiss).
ChIP-qPCR
NSG1:GFPcom, the wild-type, and mutant plants were used in the ChIP examination. Young panicles (<2 cm) were collected for isolation of nuclear extracts. The EpiQuik Plant ChIP Kit (P-2014-48, Epigentek), anti-GFP antibody (ChIP grade; ab290, Abcam), and anti-Histone H3 (acetyl K9) antibody (ChIP grade; ab10812, Abcam) were used for ChIP assays. All PCR experiments were conducted using 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s in a reaction mixture containing 10 pmol of each primer and 1 μL of DNA from ChIP or control or 1 μL of input DNA diluted 20-fold (per biological replicate) as template. More than three biological repeats (1 g of panicle sample each) with three technical repeats each were used to produce data for statistical analysis. Experimental procedures for ChIP-qPCR were performed as previously described by Xu et al. (2010). The primer sequences are listed in Supplemental Table 1.
Transient Expression Regulation Assays in N. benthamiana Leaves
The LHS1 promoter and a mini Cauliflower mosaic virus 35S promoter were amplified with the primer pairs in Supplemental Table 1 and cloned into pGreenII0800-LUC double-reporter vector. The full-length coding region of NSG1 from J10 and nsg1-1 mutant were PCR amplified with gene-specific primers (Supplemental Table 1) and then recombined with the vector pCAMBIA1300 (Cambia) with two Cauliflower mosaic virus 35S promoters to generate the 2x35Spro:NSG1WT and 2x35Spro:NSG1nsg1-1constructs. The above-mentioned constructs were then transformed into Agrobacterium tumefaciens strain GV3101. The Agrobacterium strains containing different constructs were incubated, harvested, and resuspended in infiltration buffer (10 mM MES, 0.2 mM acetosyringone, and 10 mM MgCl2) to an ultimate concentration of OD600 = 0.6. Equal volumes of different combinations of Agrobacterium strains were mixed and coinfiltrated into N. benthamiana leaves using a needleless syringe. Plants were placed at 25°C for 48 h.
LUC and renilla LUC activities were measured using the Dual Luciferase Assay Kit (Promega). The analysis was executed using the Luminoskan Ascent microplate luminometer (Thermo Fisher Scientific) according to the instructions of the manufacturer. The results were calculated by the ratio of LUC/REN (Ba et al., 2016). At least six transient assay measurements were made for each assay.
Accession Numbers
Sequence data from this article can be accessed in the GenBank database under the following accession numbers: NSG, G1, DL, LHS1, MFO1, OsTPR1, OsTPR2, and OsTPR3 are GQ999998, AB512480, AB106553, NM_001055911, FJ666318, AP014957, AK111830 and AP014959, respectively. Locus identifications in the Rice Genome Annotation Project Database are as follows: NSG1 (LOC_Os04g36650), G1 (LOC_Os07g04670), DL (LOC_Os03g11600), LHS1 (LOC_Os03g11614), MFO1 (LOC_Os02g45770), OsTPR1 (LOC_Os01g15020), OsTPR2 (LOC_Os08g06480), and OsTPR3 (LOC_Os03g14980); BAC OSJNBa0058G03 (OSJNBa0058G03), and BAC OSJNBb0056O04 (OSJNBb0056O04).
Supplemental Data
Supplemental Figure 1. Structures of cell layers of partial organs in spikelets of the wild type and nsg1-1 mutant.
Supplemental Figure 2. Phenotypes of spikelets in the wild type and the nsg1-2 and nsg1-3 mutants.
Supplemental Figure 3. Percentage of elongated or lemma-like organs and relative expression levels of organ identity genes in spikelets of the nsg1-2 and nsg1-3 mutants.
Supplemental Figure 4. NSG1 expression in the nsg 1-2 mutant.
Supplemental Figure 5. DNA gel blot of NSG1 in the genome of four rice cultivars.
Supplemental Figure 6. Protein sequence analysis of candidate gene in wild type and nsg1 mutants.
Supplemental Figure 7. Protein motif analysis of NSG1-like protein and immunoblots in GFP-fusion transgenic plants.
Supplemental Figure 8. qPCR analysis of organ identity genes in spikelets of the wild-type and the nsg1-1 mutant.
Supplemental Figure 9. Expression pattern of organ identity genes in spikelets of the wild-type and the nsg1-1 mutant.
Supplemental Figure 10. Promoter sequence of LHS1 highlighting the DBS-like motif.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. NSG1 tree.fas alignment file.
Supplemental Data Set 2. NSG1 tree.meg alignment file.
Supplemental Data Set 3. NSG1 tree.mts alignment file.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Keming Luo (College of Life Sciences, Southwest University) for kindly providing the pGreenII0800-LUC double-reporter vector, pSCYNE (SCN, modified) and pSCYCE (SCC, modified) vectors. This work was supported by the National Natural Science Foundation of China (31730063, 31271304, and 31971919), the National Key Program for Research and Development (2017YFD0100202), the Project of CQ CSTC (CSTC2017jcyjBX0062), the Graduate Student Scientific Research Innovation Projects in Chongqing (CYS2015066), and the Fundamental Research Funds for the Central Universities (XDJK2016A013).
AUTHOR CONTRIBUTIONS
Y.-F.L. and G.-H.H. designed the research. H.Zhuang performed ChIP analysis. Y.-F.L., H.C. and H.Zhuang performed the mapping-based clone and phenotype analysis. H.-L.W., T.Z., H.Zhuang, X.-Q.Z., and Y.-F.L. performed the analysis of the expression pattern and protein activation. Z.-W.W. performed the vector construction. J.Z., H.Zheng, J.T., and Z.-L.Y contributed data analysis. Y.-F.L, H.Zhuang., and T.Z. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Alwine J.C., Kemp D.J., Stark G.R. (1977). Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. USA 74: 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber A. (1935). The Gramineae: A study of cereal, bamboo, and grasses. Empire For. J. 14: 143–146. [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Ba L.J., Kuang J.F., Chen J.Y., Lu W.J. (2016). MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. J. Agric. Food Chem. 64: 738–745. [DOI] [PubMed] [Google Scholar]
- Bommert P., Satoh-Nagasawa N., Jackson D., Hirano H.Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46: 69–78. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Weigel D., Yanofsky M.F. (2006). NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–1655. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. (2004). The role of JAGGED in shaping lateral organs. Development 131: 1101–1110. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., Guo C., Kong H., Xue H., Zhang D. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153: 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Qian Q., Zhu K., Tang D., Huang Z., Gao L., Li M., Gu M., Cheng Z. (2010). ELE restrains empty glumes from developing into lemmas. J. Genet. Genomics 37: 101–115. [DOI] [PubMed] [Google Scholar]
- Hoshikawa K. (1989). The Growing Rice Plant. (Tokyo: Nosan GyosonBunka Kyokai; ). [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., Xia G., Chu C., Li J., Fu X. (2009a). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Huang X.Y., Chao D.Y., Gao J.P., Zhu M.Z., Shi M., Lin H.X. (2009b). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Sunohara H., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54: 147–156. [Google Scholar]
- Jeon J.S., Jang S., Lee S., Nam J., Kim C., Lee S.H., Chung Y.Y., Kim S.R., Lee Y.H., Cho Y.G., An G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Jin Y., et al. (2011). An AT-hook gene is required for palea formation and floral organ number control in rice. Dev. Biol. 359: 277–288. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Ke J.Y., Ma H.L., Gu X., Thelen A., Brunzelle J.S., Li J.Y., Xu H.E., Melcher K. (2015). Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 1: e1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125: 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A. (2009). The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2: 1–14. [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.Y., An G. (2012). Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69: 445–461. [DOI] [PubMed] [Google Scholar]
- Li F., Liu W., Tang J., Chen J., Tong H., Hu B., Li C., Fang J., Chen M., Chu C. (2010a). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20: 838–849. [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Hu Y., Zhu L., Yin C., Xu J., Dreni L., Kater M.M., Zhang D. (2011). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liang W., Jia R., Yin C., Zong J., Kong H., Zhang D. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20: 299–313. [DOI] [PubMed] [Google Scholar]
- Li H., Xue D., Gao Z., Yan M., Xu W., Xing Z., Huang D., Qian Q., Xue Y. (2009a). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Xiong G., Li R., Cui J., Tang D., Zhang B., Pauly M., Cheng Z., Zhou Y. (2009b). Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 60: 1055–1069. [DOI] [PubMed] [Google Scholar]
- Li S., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 110: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.F. (2008). Map cloning and functional analysis of LEAFY HULL STERILE 1 (LHS1–3) and PISTILLOID-STAMEN (PS) PhD dissertation (Chongqing, China: Southwest University; ) . [Google Scholar]
- Lin X., Wu F., Du X., Shi X., Liu Y., Liu S., Hu Y., Theißen G., Meng Z. (2014). The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 202: 689–702. [DOI] [PubMed] [Google Scholar]
- Luo Q., Zhou K., Zhao X., Zeng Q., Xia H., Zhai W., Xu J., Wu X., Yang H., Zhu L. (2005). Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221: 222–230. [DOI] [PubMed] [Google Scholar]
- McSteen P., Laudencia-Chingcuanco D., Colasanti J. (2000). A floret by any other name: Control of meristem identity in maize. Trends Plant Sci. 5: 61–66. [DOI] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718. [DOI] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., Nagato Y., Yoshida H. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno C.K., Reddy G.V., Heisler M.G.B., Meyerowitz E.M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122. [DOI] [PubMed] [Google Scholar]
- Prasad K., Parameswaran S., Vijayraghavan U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43: 915–928. [DOI] [PubMed] [Google Scholar]
- Ren D., et al. (2018). FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69: 4853–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li Y., Zhao F., Sang X., Shi J., Wang N., Guo S., Ling Y., Zhang C., Yang Z., He G. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sang X., Li Y., Luo Z., Ren D., Fang L., Wang N., Zhao F., Ling Y., Yang Z., Liu Y., He G. (2012). CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol. 160: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.J., Ambrose B.A. (1998). The blooming of grass flower development. Curr. Opin. Plant Biol. 1: 60–67. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., Gu M., Xue Y., Cheng Z. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zeng X.-Q., Zhuang H., Shen Y.-L., Chen H., Wang Z.-W., Long J.-C., Ling Y.-H., He G.-H., Li Y.-F. (2017). Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice. Plant Growth Regul. 81: 433–442. [Google Scholar]
- Wang N., Li Y.F., Sang X.C., Ling Y.H., Zhao F.M., Yang Z.L., He G.H. (2013). nonstop glumes (nsg), a novel mutant affects spikelet development in rice. Genes Genomics 35: 149–157. [Google Scholar]
- Xiao H., Tang J., Li Y., Wang W., Li X., Jin L., Xie R., Luo H., Zhao X., Meng Z., He G., Zhu L. (2009). STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 59: 789–801. [DOI] [PubMed] [Google Scholar]
- Xu J., Yang C., Yuan Z., Zhang D., Gondwe M.Y., Ding Z., Liang W., Zhang D., Wilson Z.A. (2010). The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yadav S.R., Prasad K., Vijayraghavan U. (2007). Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Zhang X., Zhang L., Ye S., Zeng L., Liu J., Li Q., He Z. (2015). Curved chimeric palea 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J. 82: 12–24. [DOI] [PubMed] [Google Scholar]
- Yao S.G., Ohmori S., Kimizu M., Yoshida H. (2008). Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 49: 853–857. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Ohmori Y., Kitano H., Taguchi-Shiobara F., Hirano H.Y. (2012). Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 70: 327–339. [DOI] [PubMed] [Google Scholar]
- Yoshida A., et al. (2013). TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 110: 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Suzaki T., Tanaka W., Hirano H.Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. USA 106: 20103–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Gao S., Xue D.W., Luo D., Li L.T., Ding S.Y., Yao X., Wilson Z.A., Qian Q., Zhang D.B. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13 804 T-DNA flanking sequences from an enhancer-trap mutant library. Plant J. 49: 947–959. [DOI] [PubMed] [Google Scholar]
- Zhang T., et al. (2017). LATERAL FLORET 1 induced the three-florets spikelet in rice. Proc. Natl. Acad. Sci. USA 114: 9984–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., et al. (2015). DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol. 206: 1476–1490. [DOI] [PubMed] [Google Scholar]