A nucleoporin interacts with an E3 ligase on the nuclear membrane, maintaining CONSTANS homeostasis and thereby controlling long-day photoperiodic flowering in Arabidopsis.
Abstract
The nuclear pore complex profoundly affects the timing of flowering; however, the underlying mechanisms are poorly understood. Here, we report that Nucleoporin96 (Nup96) acts as a negative regulator of long-day photoperiodic flowering in Arabidopsis (Arabidopsis thaliana). Through multiple approaches, we identified the E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) and demonstrated its interaction in vivo with Nup96. Nup96 and HOS1 mainly localize and interact on the nuclear membrane. Loss of function of Nup96 leads to destruction of HOS1 proteins without a change in their mRNA abundance, which results in overaccumulation of the key activator of long-day photoperiodic flowering, CONSTANS (CO) proteins, as previously reported in hos1 mutants. Unexpectedly, mutation of HOS1 strikingly diminishes Nup96 protein level, suggesting that Nup96 and HOS1 are mutually stabilized and thus form a novel repressive module that regulates CO protein turnover. Therefore, the nup96 and hos1 single and nup96 hos1 double mutants have highly similar early-flowering phenotypes and overlapping transcriptome changes. Together, this study reveals a repression mechanism in which the Nup96-HOS1 repressive module gates the level of CO proteins and thereby prevents precocious flowering in long-day conditions.
INTRODUCTION
Plants have evolved the ability to sense various environmental signals, such as seasonal daylength and temperature changes, that determine the optimal timing of flowering for successful reproduction (Michaels, 2009; de Montaigu et al., 2010; Jarillo and Piñeiro, 2011). In Arabidopsis (Arabidopsis thaliana), the prevailing molecular mechanism of daylength perception includes circadian clock-controlled transcription and proteasome-dependent protein degradation of the transcription factor CONSTANS (CO). Through these mechanisms, CO proteins are confined to accumulate at dusk in long days (Samach et al., 2000; Valverde et al., 2004; Shim et al., 2017). The accumulated CO proteins directly activate the transcription of the “florigen” gene FLOWERING LOCUS T (FT) in leaves, whose coding protein is transported to the apex and in turn induces the expression of floral meristem identity genes, such as APETALA1 and LEAFY, to initiate the formation of floral primordia (Weigel et al., 1992; Abe et al., 2005; Wigge et al., 2005). CO transcription is tightly controlled by manifold positive and negative regulators. Two different groups of transcription factors, FLOWERING BHLH (FBH) and TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR (TCP), are activators of CO, through directly binding to the CO promoter (Ito et al., 2012; Kubota et al., 2017). Both FBH1 and TCP4 physically interact with another CO gene activator, GIGANTEA (GI; Koornneef et al., 1998; Suárez-López et al., 2001; Kubota et al., 2017), but the functional dependency of FBH1 and TCP4 on GI is different. TCP4 activates CO in a GI-dependent manner, whereas FBH1 is partially independent of GI to activate CO (Kubota et al., 2017). CO expression can also be repressed by CYCLING DOF FACTOR proteins, which are able to bind DOF binding sites in the CO promoter (Imaizumi et al., 2005; Fornara et al., 2009). In addition to transcriptional regulation, posttranslational regulation of CO is also essential for measuring daylength. The best characterized posttranslational regulation of CO proteins is proteasome-dependent degradation, which utilizes different E3 ubiquitin ligases. The E3 ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) associates with SUPPRESSOR of phyA-105 to specifically degrade CO proteins in the dark (Jang et al., 2008), while another E3 ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1), mediates the degradation of CO in the morning in long days (Jung et al., 2012; Lazaro et al., 2012). Other mechanisms have also been implicated in the direct stabilization of CO proteins (Song et al., 2012, 2014).
Although numerous genes have been identified so far in flowering time regulation, the underlying molecular mechanisms for many of them are poorly understood. One example is a group of nuclear pore-associated proteins. The nuclear pore complex is embedded in the nuclear envelope, the physical barrier separating the nucleus from the cytosol, and serves as the sole channel for nucleocytoplasmic transportation of macromolecules (i.e., proteins and RNAs; Meier and Brkljacic, 2009; Wälde and Kehlenbach, 2010). The nuclear pore complex is composed of multiple copies of ∼30 different nucleoporins (Nups), which form several subcomplexes (Tran and Wente, 2006; Strambio-De-Castillia et al., 2010). In Arabidopsis, most Nups have been identified so far by genetic and biochemical approaches (Tamura et al., 2010), and some of them are reported to participate in various important developmental and resistant processes. Mutations of Nup136 (also known as Nup1), Nup160 (also known as SUPPRESSOR OF AUXIN RESISTANCE1 [SAR1]), Nup96 (SAR3 or MODIFIER OF SNC1,3 [MOS3]), and AtTPR (Arabidopsis thaliana TRANSLOCATED PROMOTER REGION, also known as NUCLEAR PORE ANCHOR) significantly promote flowering of plants (Zhang and Li, 2005; Parry et al., 2006; Jacob et al., 2007; Xu et al., 2007; Tamura et al., 2010; Parry, 2014), while plants deficient in Nup62, Nup58, and Nup54 gene functions manifest moderately early-flowering phenotypes (Zhao and Meier, 2011; Parry, 2014; Boeglin et al., 2016). Moreover, loss of function of some proteins partially distributed on the nuclear envelope also leads to an altered flowering time of plants. For example, the low expression of osmotically responsive genes4 (Gong et al., 2005), early in short days4 (Murtas et al., 2003), and hos1 (Jung et al., 2012, 2013; Lazaro et al., 2012) mutants display early-flowering phenotypes. This evidence strongly suggests that nuclear pore-associated proteins play fundamental roles in regulating flowering time in Arabidopsis. However, until now, there was no detailed evidence about the underlying molecular mechanism confirming that Nups regulate flowering.
In this study, we show that one component of the nuclear pore complex, Nup96, negatively regulates the transition to flowering in Arabidopsis. By combining a proteomic approach with a protein-protein interaction analysis, we demonstrate that flowering repressor HOS1 E3 ligase associates with Nup96 in vivo at the nuclear envelope. Loss of function of Nup96 leads to the destruction of HOS1 proteins, which results in invariable phenotypes of the nup96 and hos1 single and the nup96 hos1 double mutants in flowering time. As previously reported in hos1 mutants (Lazaro et al., 2012), the early flowering of nup96 mutants could be largely explained by an overaccumulation of CO proteins. Moreover, an unexpected reduction of the Nup96 protein level in hos1 mutants has been observed in this study. Together, these results uncover a mechanism through which the mutually stabilized Nup96-HOS1 repressor complex negatively regulates CO abundance and prevents precocious flowering of Arabidopsis in long-day conditions.
RESULTS
Nup96 Acts as a Negative Regulator of Flowering in Arabidopsis
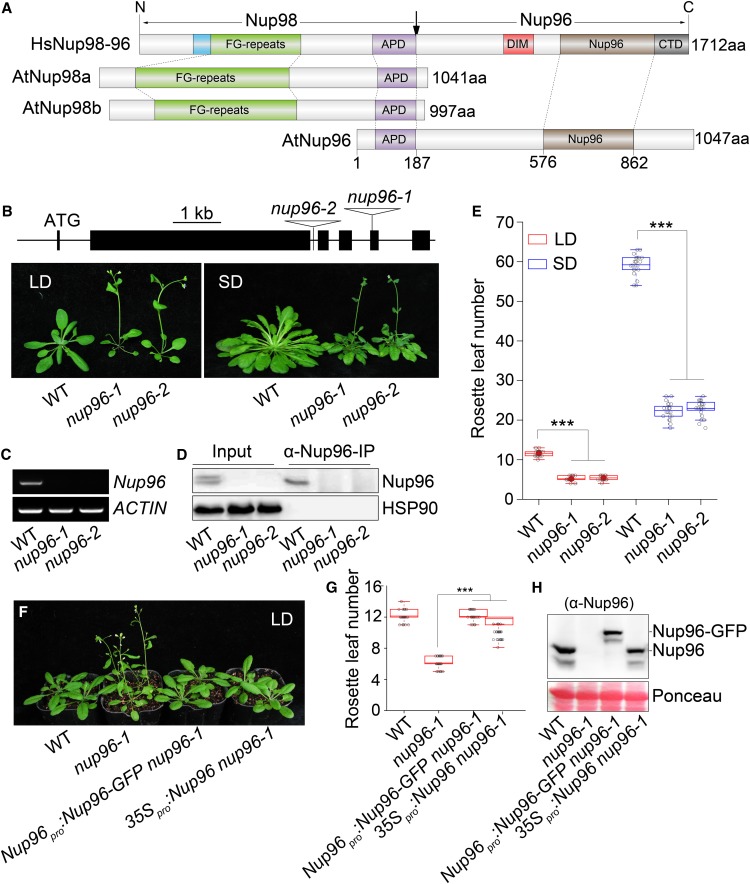
In human cells, Nup96 and Nup98 are encoded as one fusion gene and synthesized as a 186-kD precursor protein. Autoproteolytic cleavage of this precursor releases two mature nucleoporins, Nup96 and Nup98 (Fontoura et al., 1999). By contrast, Arabidopsis Nup96 (SAR3/MOS3, At1g80680) and Nup98 (Nup98a, At1g10390; Nup98b, At1g59660) genes reside in separate loci in the genome and are synthesized into proteins individually (Zhang and Li, 2005; Parry et al., 2006; Tamura et al., 2010). Noticeably, the well-conserved Autoproteolytic Processing Domain (APD) is retained in both Nup96 and Nup98 proteins in Arabidopsis (Figure 1A). Nup96 proteins from most plant species investigated present structures similar to Arabidopsis Nup96 with the conserved APD (Supplemental Figure 1; Supplemental Data Set 1), suggesting that Nup96 genes diversified independently in plant and animal lineages. Previous characterization of Nup96 in Arabidopsis has shown its crucial roles in auxin signaling and the immune response (Zhang and Li, 2005; Parry et al., 2006); although observations suggested that the nup96 mutation caused early flowering of plants (Parry et al., 2006), the underlying molecular mechanisms remained unknown.
Figure 1.
Nup96 Acts as a Negative Regulator of Flowering in Arabidopsis.
(A) Domain structures of human Nup98-96 (HsNup98-96), Arabidopsis Nup98a and Npu98b (AtNup98a and AtNup98b), and Arabidopsis Nup96 (AtNup96). Domains are indicated as follows: the GLEBS motif (blue), the Phe-Gly repeat region (FG-repeats; green), the APD (purple), the domain invasion motif (DIM; red), the Nup96 domain (Nup96; brown), and the C-terminal domain (CTD; gray). The vertical arrow indicates the sites of autoproteolytic cleavage. aa, Amino acids.
(B) Scheme of Nup96 gene structure showing the positions of two T-DNA insertions (nup96-1 and nup96-2) and flowering phenotypes of the wild type (WT) and nup96-1 and nup96-2 mutants under long-day (LD) and short-day (SD) conditions. Black lines indicate introns. Black bars represent exons. Star codon ATG is indicated. Triangles refer to T-DNAs.
(C) RT-PCR analysis of Nup96 full-length transcripts in different genotypes. ACTIN was used as the control. WT, wild type.
(D) IP assay of Nup96 proteins in different genotypes. WT, wild type.
(E) Measurement of rosette leaf numbers of different genotypes (n > 20). Asterisks indicate significant differences according to Student’s t test (***, P < 0.001). WT, wild type.
(F) Flowering phenotypes of the wild type, nup96-1, and two complementation lines. WT, wild type.
(G) Measurement of rosette leaf numbers of genotypes shown in (F); n > 20. Asterisks indicate a significant difference according to Student’s t test (***, P < 0.001). WT, wild type.
(H) Immunoblot showing protein levels of endogenous Nup96 and Nup96-GFP in genotypes shown in (F) using anti-Nup96 antibodies. WT, wild type.
To address this question, we first recharacterized the flowering phenotypes of two mutant alleles, nup96-1 (SALK_109959, also known as sar3-3 or mos3-2; Zhang and Li, 2005; Parry et al., 2006) and nup96-2 (SALK_117966, also known as mos3-3; Zhang and Li, 2005; Figure 1B) under various experimental conditions. Using RT-PCR and immunoprecipitation (IP) assays, we verified that both alleles were null, as no full-length Nup96 transcripts and proteins could be detected (Figures 1C and 1D). When grown under long-day or short-day conditions, both nup96-1 and nup96-2 mutants flowered much earlier than wild-type plants (Figures 1B and 1E), suggesting that Nup96 negatively regulates the transition to flowering in Arabidopsis. We also noticed that nup96 mutants flowered much earlier in long days than in short days, indicating a limited effect of Nup96 mutation on the plant’s sensitivity to daylength change. Next, we observed complementation of the early-flowering phenotype (Figures 1F and 1G) when nup96-1 plants were transformed with either Nup96 cDNA driven by a cauliflower mosaic virus 35S promoter or a Nup96-GFP fusion gene driven by a 2.5-kb promoter upstream of the Nup96 coding sequence, which produced normal Nup96 proteins (Figure 1H). This observation corroborated the notion that early flowering of nup96 mutants was indeed due to the loss of Nup96 function.
Evolutionarily Conserved Autoproteolytic Processing Is Not Required for the Flowering Activities of Nup96
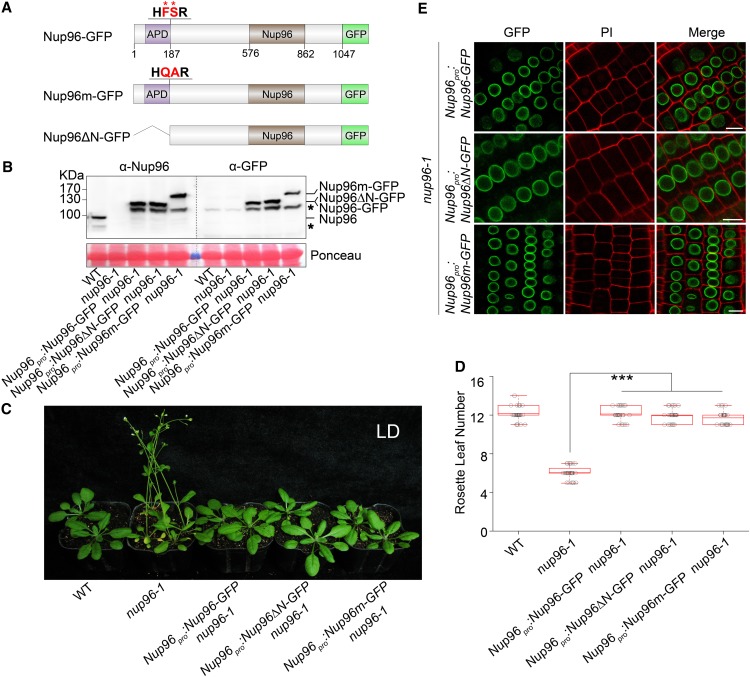
Considering that a conserved proteolytic cleavage site is located at the N terminus of Nup96 (Fontoura et al., 1999; Iwamoto et al., 2010), we wondered whether this site was active and necessary for the flowering functions of Nup96. To this end, we created mutated Nup96 proteins in which either the Phe (F187) and Ser (S188) residues in the cleavage domain (HFS, the nucleoporin2 domain in animals and yeast with pfam accession number PF04096 [Fontoura et al., 1999; Iwamoto et al., 2010]) were mutated to Gln (F187Q) and Ala (S188A), respectively, or APD (the first 187 residues) in the N terminus was deleted (Nup96Δ187; Figure 2A). Then, the wild-type Nup96, Nup96F187Q, S188A (Nup96m), and Nup96Δ187 (Nup96ΔN) genes were fused with the GFP coding sequence and expressed under the control of the 2.5-kb Nup96 promoter region (Nup96pro) in the nup96-1 background. These plants are hereafter referred to as Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP, respectively (Figure 2A). We reasoned that if the cleavage site was active in plants, the first 187 residues from the N-terminal end of Nup96 proteins would be removed. Therefore, the Nup96-GFP and Nup96ΔN-GFP transgenes should be able to produce identical proteins that are ∼20 kD smaller (∼125 kD) than the predicted size of Nup96-GFP (∼145 kD). Indeed, using an immunoblot assay, we detected recombinant proteins that were slightly smaller than 130 kD in both Nup96-GFP and Nup96ΔN-GFP plants. By contrast, a protein product larger than 130 kD was detected in Nup96m-GFP plants (Figure 2B). A similar observation was made in wild-type plants. Using an anti-Nup96-specific antibody raised in this study, which recognizes the C terminus of Nup96 (amino acids 550–900), we found that in wild-type plants, Nup96 proteins existed as an ∼100-kD form rather than the predicted 119-kD proteins (Figure 2B). These results suggested that Nup96 proteins undergo proteolytic processing in plants by removing a short fragment from its N-terminal end.
Figure 2.
Autoproteolytic Processing Is Not Required for the Physiological Activities of Nup96.
(A) Domain structures of Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP fusion proteins. Domains of Nup96 are indicated as follows: the APD (purple box), the Nup96 domain (Nup96; brown box), and the rest of the regions (white box). The GFP fused to different versions of Nup96 are shown as a green box. The autoproteolytic cleavage site is shown as “HFSR,” and the key residues (F and S) used for mutations are labeled by asterisks and highlighted as red color. The numbers below the Nup96-GFP structure denote the positions in the amino acid sequence.
(B) Immunoblots showing the levels of endogenous Nup96, Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP proteins in plants shown in (C) detected by anti-Nup96 and anti-GFP antibodies. Asterisks denote unknown bands. WT, wild type.
(C) Flowering phenotypes of the wild type, nup96-1, and transgenic lines expressing the proteins indicated in (A). WT, wild type.
(D) Rosette leaf numbers of genotypes shown in (C) at the time of flowering (n > 20). Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum. Asterisks indicate a significant difference according to Student’s t test (***, P < 0.001). WT, wild type.
(E) Subcellular distributions of Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP fusion proteins indicated in (A) in plant root cells. Bars = 10 µm.
We next investigated whether the proteolytic processing was necessary for the flowering-repression activity of Nup96 using flowering time as the readout. We observed that the early-flowering phenotype of nup96-1 mutants could be fully rescued by expressing Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP transgenes (Figures 2C and 2D), and limited phenotypic variability was found among those plants. Furthermore, we examined the subcellular distributions of Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP proteins in plant roots. All three versions of the proteins concentrated on the nuclear envelope, and no distinguishable patterns were observed (Figure 2E). Taken together, these data suggested that the proteolytic removal of the N-terminal residues was not required for the nuclear envelope-targeting and floral regulatory activities of Nup96 proteins in Arabidopsis.
Nup96 Regulates Flowering through Modulating the Expression of Floral Integrator Genes
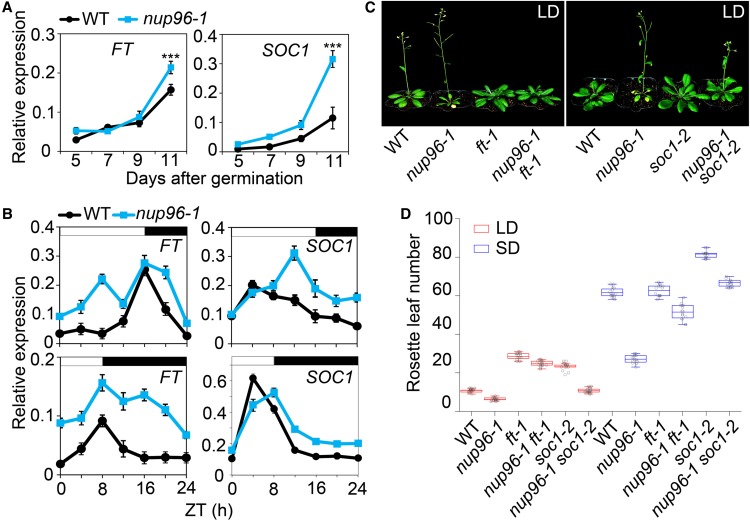
To dissect the molecular basis contributing to the early flowering of nup96-1 mutants, we first profiled the expression patterns of floral integrator genes FT and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) in both the wild type and nup96-1 mutants by qPCR. FT and SOC1 expression increased during the development of wild-type and nup96-1 seedlings grown in long days; however, both genes showed higher levels in nup96-1 on day 11 after germination (Figure 3A) when floral transition occurred (Shen et al., 2011). Then, we compared the daily expression patterns of FT and SOC1 in the wild type and nup96-1 mutants under different daylengths. The overall levels of transcripts for both genes were higher in nup96-1 than in wild-type plants, but the degrees varied among time points (Figure 3B). For example, in long days, FT appeared more abundant in the morning (Zeitgeber time 4 [ZT4]) in nup96-1 than in wild-type plants, while the level of SOC1 transcript remained higher in nup96-1 than in the wild type from noon until the end of the day (Figure 3B). These observations suggested that the early flowering of nup96 mutants was quite likely due to the upregulation of floral integrator genes. This notion was further supported by the observation that both nup96-1 ft-1 and nup96-1 soc1-2 double mutants flowered much later than nup96-1 single mutants (Figures 3C and 3D). However, it is worth noting that the ft mutation was much more effective than soc1 in terms of delaying the flowering of nup96 mutants in long days, suggesting that the elevated FT expression might be the major cause leading to the accelerated flowering of nup96 mutants in long days.
Figure 3.
Nup96 Regulates Flowering through Modulating the Expression of Floral Integrator Genes.
(A) qPCR analysis of FT and SOC1 expression in the wild type (WT) and nup96-1 mutants grown in long days for the indicated periods after germination. Values are means ± sd (n = 3 biological repeats). ***, P < 0.001.
(B) Diurnal expression of FT and SOC1 in wild-type and nup96-1 seedlings grown in long-day and short-day conditions for 9 d. Values are means ± sd (n = 3 biological repeats). White bars indicate day, and black bars represent night. WT, wild type.
(C) Flowering phenotypes of the indicated genotypes in long days. WT, wild type.
(D) Rosette leaf numbers at the time of flowering for the genotypes shown in (C); n > 20. Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum. WT, wild type.
It has been reported that the activation of FT mRNA expression mostly occurs in the leaf phloem companion cells (An et al., 2004). Given that Nup96 adversely regulated FT expression (shown above), it was likely to share a similar spatial expression pattern to FT. Therefore, we examined the spatial expression of Nup96 in transgenic plants expressing the GUS reporter driven by the Nup96 native promoter (Nup96pro). Nup96 was ubiquitously expressed throughout the plant, including the leaf vasculature, shoot apical meristem, roots, reproductive organs, and other tissues (Supplemental Figure 2A). This broad expression of Nup96 was consistent with the published transcriptomic data (Supplemental Figure 2B) and the diverse developmental abnormalities of nup96 mutants (Supplemental Figures 2C–2L).
Nup96 Interacts with HOS1 in Plants
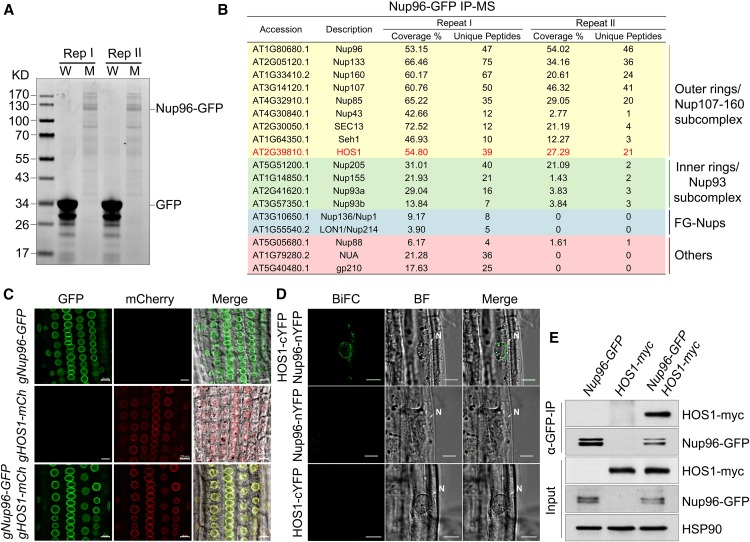
To investigate the molecular mechanisms by which Nup96 prevents plants from precocious flowering, we purified Nup96 protein complexes from Nup96-GFP/nup96-1 plants by IP using GFP affinity chromatography (Rothbauer et al., 2008), and the protein complexes were then analyzed by mass spectrometry (MS). The Nup96-GFP fusion proteins in the transgenic line we used showed comparable levels to endogenous Nup96 proteins in wild-type plants (Figures 1H and 2B). GFP-only overexpression plants were analyzed in parallel in IP-MS experiments as the negative control (Figure 4A). After excluding those identified by MS in GFP-only samples, we found that different groups of nucleoporins were specifically coprecipitated with Nup96-GFP from plant protein extracts (Figure 4B), indicating a strong association of Nup96 with other components of the nuclear pore complex. An important flowering repressor, HOS1 (Figure 4B; Jung et al., 2012; Lazaro et al., 2012), which is a RING-type E3 ubiquitin ligase and shares homology with yeast and human nucleoporin EMBRYONIC LARGE MOLECULE DERIVED FROM YOLK SAC protein (Tamura et al., 2010; Bilokapic and Schwartz, 2013), was among these proteins (Figure 4B).
Figure 4.
Nup96 Interacts with HOS1 in Plants.
(A) Coomassie Brilliant Blue staining of proteins immunoprecipitated by GFP-trap from the lysates of 35S:GFP wild-type (W) and Nup96-GFP nup96-1 (M) plants grown in long-day conditions for 9 d. Two replicates (Rep I and Rep II) were performed.
(B) Identification of the Nup96 complex by MS. HOS1 is highlighted by red color.
(C) Colocalization of Nup96-GFP and HOS1-mCherry in transgenic plant roots. Bars = 10 µm.
(D) BiFC assay showing the interaction of Nup96 and HOS1 in transgenic plants. BF, bright field. Bars = 20 µm.
(E) Co-IP assay showing the in vivo interaction of Nup96 and HOS1 in transgenic plants. GFP-trap was used to isolate Nup96-GFP from plant protein extracts, and HOS1-myc was then detected in the immunoprecipitates by anti-myc antibodies. HSP90 was used as the loading control for the input samples.
To verify the in vivo association of HOS1 and Nup96, we first examined if the two proteins had overlapping distributions in plant cells. Transgenic plants were prepared to independently express the genomic sequence of HOS1 including a 2-kb promoter region, the coding region fused with the mCherry open reading frame (gHOS1-mCh), and the entire Nup96 genomic DNA (a 2.2-kb promoter region was included) fused with the coding sequence of GFP (gNup96-GFP). Then, the dual expression lines were obtained by crossing gHOS1-mCh with gNup96-GFP plants. In agreement with the previous studies (Parry et al., 2006; Lazaro et al., 2012), we observed that both HOS1-mCherry and Nup96-GFP proteins dominantly located at the nuclear envelope either in single or dual transgenic plants (Figure 4C).
Next, we examined whether the two proteins interact with each other at the nuclear envelope using a bimolecular fluorescence complementation (BiFC) assay. We separately transformed wild-type plants with 35S:Nup96-nYFP (nYFP, N-terminal end of YFP) and 35S:HOS1-cYFP (cYFP, C-terminal end of YFP), then the two transgenes were brought together by genetic cross. We found that the BiFC signal was strongly enriched at the nuclear envelope in the double transgenic line (Figure 4D), suggesting that Nup96 and HOS1 indeed interact at the nuclear envelope. Finally, we performed a coimmunoprecipitation (co-IP) assay to detect the interaction of Nup96 and HOS1 in plants. We expressed two transgenes of HOS1-myc and Nup96-GFP simultaneously in plants resulting from the genetic cross of the transgenic line expressing 35S:HOS1-myc and the Nup96-GFP nup96-1 complementation line. Using anti-GFP antibodies, HOS1-myc proteins were successfully copurified with Nup96-GFP from the lysate of dual transgenic plants (Figure 4E), reflecting the coexistence of Nup96 and HOS1 in common protein complexes. In summary, our results demonstrate that the floral repressor Nup96 associates with HOS1 at the nuclear envelope in Arabidopsis.
Nup96 and HOS1 Are Mutually Stabilized in Plants
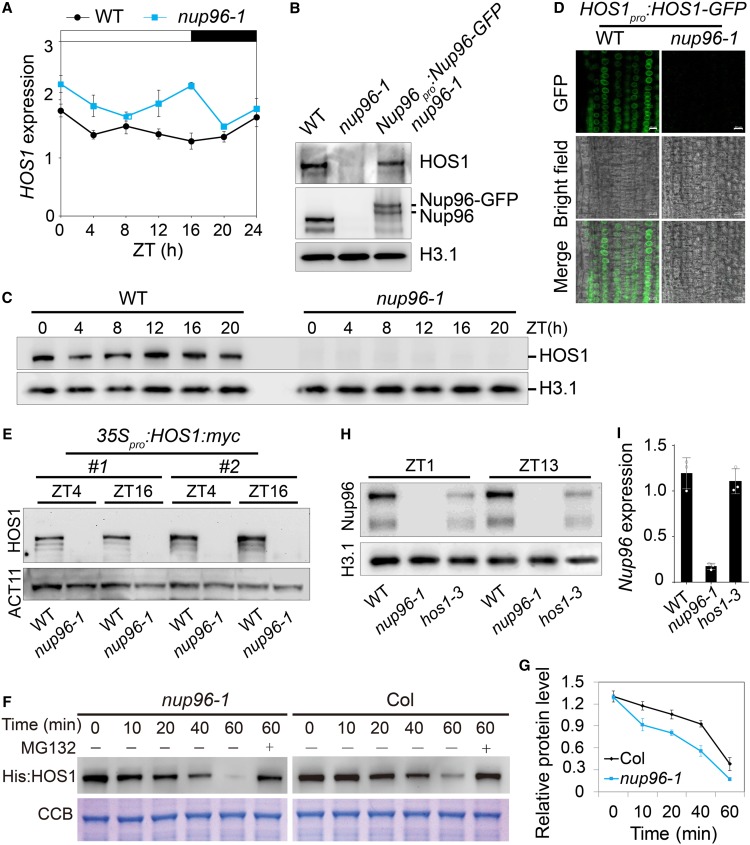
To evaluate how loss of function of Nup96 influences the activities of HOS1, we first investigated if the transcription of HOS1 was altered in nup96 mutants. qPCR results revealed that HOS1 expression was slightly elevated in nup96-1 mutants compared with that in wild-type plants (Figure 5A), suggesting that Nup96 mutation has limited effects on HOS1 expression. Next, we examined HOS1 protein levels in different genotypes using anti-HOS1 antibodies, which clearly detected endogenous HOS1 proteins in wild-type plants. However, we surprisingly found that HOS1 proteins could barely be detected in the nup96-1 background, and this loss of HOS1 proteins was restored in the Nup96-GFP nup96-1 complementation line (Figure 5B).
Figure 5.
Nup96 Stabilizes HOS1 Proteins in Plants.
(A) Diurnal expression of HOS1 in wild-type (WT) and nup96-1 seedlings grown in long-day conditions for 9 d. Values are means ± sd (n = 3 biological repeats).
(B) Immunoblot shows HOS1 levels in the nuclear extracts from the WT, nup96-1, and Nup96-GFP nup96-1 complementation plants. Histone H3.1 (H3.1) was used as the loading control.
(C) Levels of endogenous HOS1 proteins in the nuclear extracts from WT and nup96-1 seedlings grown in long-day conditions for 9 d, detected by anti-HOS1 antibodies. Histone 3.1 was used as the loading control.
(D) HOS1-GFP signals in wild-type and nup96-1 seedling root cells. Bars = 10 µm.
(E) Levels of HOS1-myc proteins in the nuclear extracts from WT and nup96-1 seedlings grown in long-day conditions for 9 d. ACTIN11 (ACT11) was used as the loading control.
(F) Assay of HOS1 protein stability in vitro. His:HOS1 proteins were purified from Escherichia coli and incubated with the total extracts of nup96-1 or Col-0 seedlings in the presence or absence of MG132. His:HOS1 proteins were probed by His antibody on an immunoblot (upper panel). Coomassie Brilliant Blue (CBB) staining is shown in the lower panel.
(G) Quantitative assay of HOS1 protein levels from three biological repeats with a representative result shown in (F).
(H) Levels of endogenous Nup96 proteins in the nuclear extracts from WT, hos1-3, and nup96-1 seedlings grown in long-day conditions for 9 d.
(I) qPCR analysis of Nup96 expression in WT, hos1-3, and nup96-1 seedlings grown in long-day conditions for 9 d. Values are means ± sd (n = 3 biological repeats).
We then performed a detailed comparison of the HOS1 protein levels between the wild type and nup96-1 mutants in a time-of-day manner. As expected, HOS1 proteins were undetectable at all time points in the nup96-1 mutant (Figure 5C), although the overall level of HOS1 mRNA was slightly higher in the nup96-1 mutant than in wild-type plants (Figure 5A). This observation was confirmed by two other experiments. First, we created transgenic plants expressing the HOS1-GFP fusion gene driven by the HOS1 native promoter (including the 5′ untranslated region of HOS1) in wild-type plants (HOS1pro:HOS1-GFP wild type) and introduced this transgene into the nup96-1 background by genetic cross. HOS1-GFP proteins could be microscopically visualized in wild-type plants but not in nup96-1 mutants (Figure 5D). Second, we expressed the HOS1-myc coding sequence under the control of the 35S promoter in the wild-type background (35S:HOS1myc wild type), and two such lines (no. 1 and no. 2) were selected to cross with nup96-1 mutants. Then, we compared HOS1-myc protein levels in the wild type with those of nup96-1 mutants at two time points (ZT4 and ZT16) by immunoblot assays. The results showed that HOS1-myc proteins were completely destroyed in nup96-1 mutants (Figure 5E).
To further investigate the relationship between Nup96 and HOS1, we performed an in vitro assay (Ruan et al., 2019) to detect the effect of Nup96 on the stability of HOS1 proteins by incubating purified recombinant His-HOS1 proteins with tissue extracts from the nup96-1 mutant and wild-type plants. Not surprisingly, the recombinant HOS1 proteins were much more sensitive to the seedling extract of the nup96-1 mutant, compared with the seedling extract of wild-type Col-0, and degraded much quickly (Figures 5F and 5G).
Thus, all the evidence above strongly suggests that Nup96 is required for stabilizing HOS1 proteins in Arabidopsis. However, we could not rule out the possibility that translation inefficiency is also a contributing factor for the deaccumulation of HOS1 proteins in nup96 mutants. This unexpected finding prompted us to examine whether HOS1 had a similar effect on Nup96 protein stability. In an immunoblot assay using anti-Nup96 antibodies, the endogenous Nup96 level was reduced in hos1-3 mutants (Figure 5H), but with unchanged Nup96 mRNA abundance (Figure 5I). Together, these results suggested a mechanism of mutual stabilization of HOS1 and Nup96 proteins in plants.
Nup96 and HOS1 Overlappingly Regulate Flowering Time in Plants
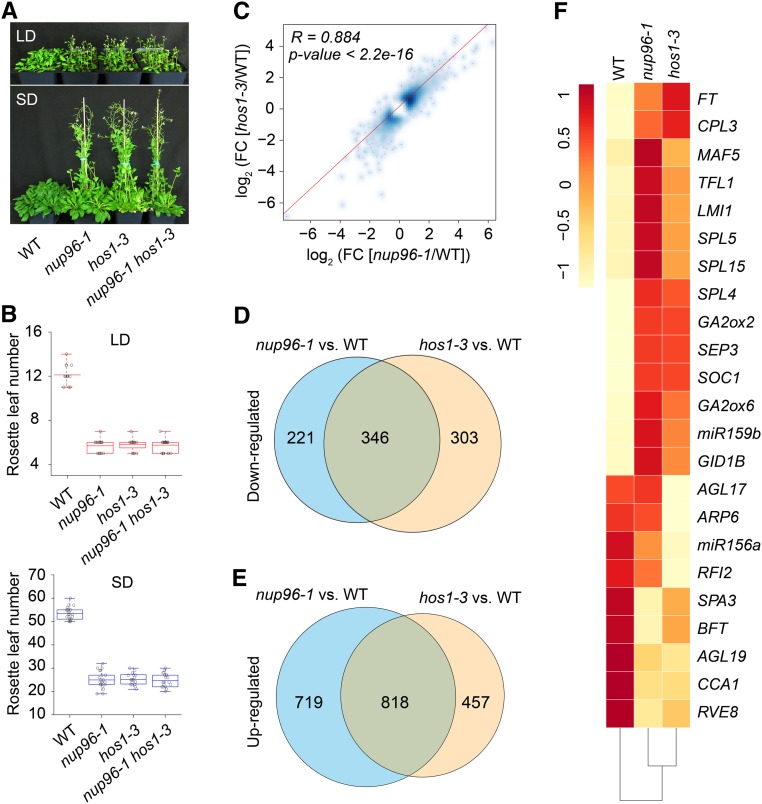
The above results showed that Nup96 and HOS1 may have overlapping functions in flowering regulation in Arabidopsis. To test this hypothesis using genetic and molecular approaches, we first constructed the nup96-1 hos1-3 double mutant by crossing. While nup96-1 mutants recapitulated the early-flowering phenotype of plants defective in HOS1 function (hos1-3), the nup96-1 hos1-3 double mutant did not display a much earlier flowering phenotype than either of its parental lines in both long days and short days (Figures 6A and 6B), suggesting that a similar molecular mechanism might be shared by both single and double mutants in photoperiodic flowering.
Figure 6.
Nup96 and HOS1 Mutations Cause Similar Transcriptomic Changes in Plants.
(A) Flowering phenotypes of the wild type (WT), nup96-1 and hos1-3 single mutants, and nup96-1 hos1-3 double mutants grown in long days (LD) and short days (SD).
(B) Rosette leaf numbers at the time of flowering for the genotypes shown in (A); n > 20. Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum. Red boxes and blue boxes present the data in long days and short days, respectively. WT, wild type.
(C) Scatterplot showing the expression correlation of differentially expressed genes in nup96-1 and hos1-3 mutants compared with the WT. Differentially expressed genes from RNA-seq data are defined as those with a fold change (FC; mutant/wild type) > 1.5 or < 0.67 (Fisher’s exact test, P < 0.01 and FDR < 0.01; Supplemental Data Set 2). The red line is the linear regression. R, Pearson correlation coefficient.
(D) and (E) Venn diagrams depicting the overlaps between the downregulated genes (D) and the upregulated genes (E) in nup96-1 and hos1-3 mutants. WT, wild type.
(F) Heatmap presenting the misregulated flowering time genes in nup96-1 or hos1-3 mutants compared with WT.
To further investigate the functional relationship of Nup96 and HOS1 genes in flowering control in plants, we performed RNA sequencing (RNA-seq) to interrogate the transcriptomic changes caused by Nup96 or HOS1 mutation. In comparison with wild-type plants, 2204 and 1924 genes were differentially expressed in the nup96-1 and hos1-3 mutants (Supplemental Data Set 2), respectively, whose expression levels showed a clear positive correlation (Pearson correlation coefficient = 0.884; P < 2.2 × 10−16; Figure 6C). Moreover, >50% of either downregulated or upregulated genes in nup96-1 and hos1-3 mutants were overlapping (Figures 6D and 6E). We also found that, consistent with the phenotypic observations, many flowering-related genes were misregulated in both mutants (Figure 6F). Overall, most of the differentially expressed floral promoting genes, including FT, GA2ox2, and SOC1, were upregulated, whereas the floral repressing genes, such as miR156, CCA1, and RVE, were downregulated, indicating that various flowering time genes may contribute to the early flowering of both nup96 and hos1 mutants. These results are in strong agreement with the notion that Nup96 and HOS1 partially share a common mechanism in flowering regulation in Arabidopsis.
Gene Ontology (GO) analyses revealed that the top 20 significantly enriched terms (ranked by P < 0.01) of down-regulated genes in both mutants are almost defense response-related (Supplemental Figures 3A and 3C), while the upregulated genes elicited by two gene mutations were shown to respond to various abiotic environmental stimuli, phytohormones, and chemical compounds (Supplemental Figures 3B and 3D), in agreement with previous observations (Zhang and Li, 2005; Dong et al., 2006; Parry et al., 2006; Lee et al., 2012; Parry, 2014; Lazaro et al., 2015; Lee and Seo, 2015; MacGregor and Penfield, 2015).
In summary, our data demonstrate that Nup96 and HOS1 are mutually stabilized and have overlapping functions not only in flowering regulation but also in other multiple physiological processes.
Loss of Nup96 Functions Strikingly Promotes CO Accumulation
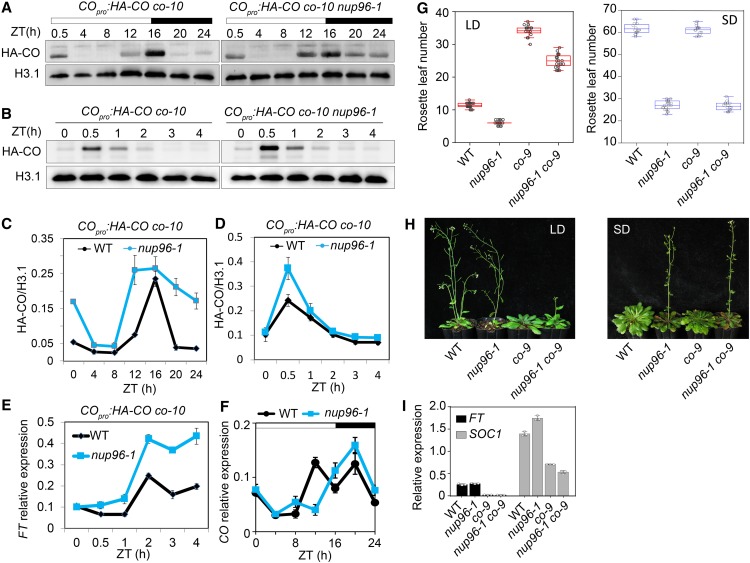
HOS1, functioning as an E3 ubiquitin ligase, is involved in the proteasome-dependent degradation of CO proteins in response to different environmental factors (Jung et al., 2012; Lazaro et al., 2012, 2015). Therefore, we reasoned that if HOS1 was inactivated in nup96 mutants, then CO proteins would overaccumulate in this background, which might lead to early flowering. To test this hypothesis, we introduced the HA-CO transgene into the nup96-1 background by crossing the nup96-1 mutant with the COpro:HA-CO co-10 line (Sarid-Krebs et al., 2015). In both the nuclear (Figures 7A–7D) and the whole (Supplemental Figure 4) protein extracts from plants grown in long days, CO proteins indeed exhibited higher levels in nup96-1 mutants than in the wild type (Columbia [Col]), specifically from dusk to the early morning. Consistent with the high abundance of CO proteins, FT was expressed at high levels in the late part of this phase (Figure 7E), suggesting that the early-flowering phenotype of the nup96-1 mutant was the result of the accumulation of CO proteins in the morning. However, the CO transcript level was not noticeably altered by nup96 mutation (Figure 7F). To determine whether the overaccumulated CO proteins were responsible for the early flowering of nup96 plants, we generated the nup96-1 co-9 double mutant and observed that the flowering time of nup96-1 mutants grown in long days was significantly delayed by the co mutation (Figures 7G and 7H). However, in short days, the nup96-1 co-9 double mutant flowered as early as the nup96-1 single mutant (i.e., not having the later-flowering character of the co-9 mutant), reminiscent of the effect of co-2 on hos1-2 in flowering regulation (Lazaro et al., 2012), where the flowering time of hos1-2 co-2 is similar to that of the hos1-2 mutant but not to that of the co-2 mutant. Such a phenotype supported the hypothesis that Nup96 and HOS1 are involved in the regulation of flowering in a similar CO-dependent pathway in long-day conditions but not in short-day conditions, because CO is confirmed to be an activator of Arabidopsis flowering only in long days but not in short days (Rédei, 1962; An et al., 2004; Datta et al., 2006). Furthermore, FT and SOC1 expression in nup96-1 co-9 double mutants was as low as that in co-9 mutants (Figure 7I). These data strongly suggested that CO acts genetically downstream of Nup96 and HOS1, and the early flowering of nup96 mutants could be largely attributed by the increased CO protein level. Therefore, the results revealed a novel repression mechanism of flowering, where Nup96 and HOS1 proteins were mutually stabilized to control CO protein homeostasis to gate flowering and loss of one of them (Nup96 or HOS1) led to degradation of the other proteins conferring early flowering in Arabidopsis (Figure 8).
Figure 7.
Loss of Nup96 Function Promotes CO Accumulation, Resulting in an Early-Flowering Phenotype in Long Days.
(A) and (B) CO protein levels in nuclear extracts from the indicated transgenic plants (COpro:HA-CO co-10 versus COpro:HA-CO co-10 nup96-1). Histone H3.1 (H3.1) was used as the loading control.
(C) and (D) Quantification of CO levels in immunoblots of (A) and (B), respectively. Values are means ± se (n = 3 biological repeats). WT, wild type.
(E) Expression levels of FT in the morning phase as in (B) and (D) in COpro:HA-CO co-10 versus COpro:HA-CO co-10 nup96-1 in long-day conditions for 9 d. Values are means ± se (n = 3 biological repeats). IPP2 (At3g02780) was used as a reference gene. WT, wild type.
(F) Diurnal expression of CO in the wild type (WT) and nup96-1 mutants grown in long-day conditions for 9 d. Values are means ± se (n = 3 biological repeats). TIP41 (At4g34270) was employed as a control gene.
(G) Rosette leaf numbers at the time of flowering for the genotypes of WT, nup96-1 and co-9 single mutants, and nup96-1 co-9 double mutants in long-day (LD) and short-day (SD) conditions (n > 20). Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum.
(H) Flowering phenotypes of WT, nup96-1 and co-9 single mutants, and nup96-1 co-9 double mutants grown in long days (left) and short days (right).
(I) Expression analysis of FT and SOC1 in different genotypes indicated in (H). Values are means ± sd (n = 3 biological repeats). WT, wild type.
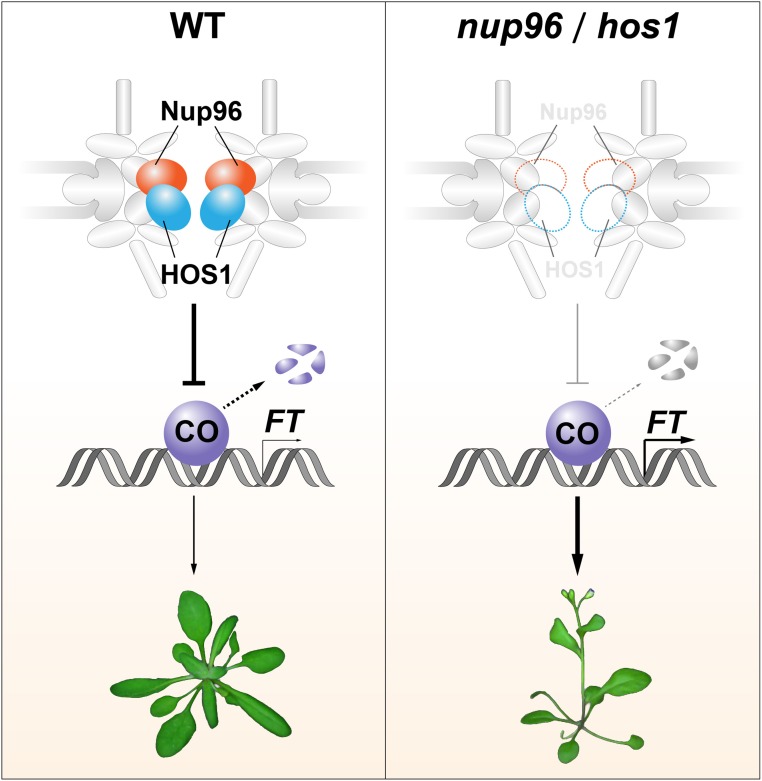
Figure 8.
Model Depicting the Molecular Mechanism of Nup96/HOS1-Mediated Flowering Regulation.
In the wild type (left), Nup96 mutually associates with and stabilizes the floral repressor HOS1 to facilitate the degradation of CO proteins, preventing precocious flowering. Loss of either Nup96 or HOS1 function (right) results in the destruction of HOS1 or Nup96 proteins, leading to overaccumulation of CO proteins in plants. The elevated CO abundance enhances FT expression, causing early flowering. WT, wild type.
DISCUSSION
As plants live in temporally fluctuating environments, the negative regulatory mechanisms that prevent precocious flowering are critical. Here, we present a set of data that elucidates the involvement of nucleoporin Nup96 in flowering regulation in Arabidopsis. Autocleavage of Nup96 proteins is conserved in all eukaryotes (Fontoura et al., 1999; Iwamoto et al., 2010), but it is not necessary for both the subcellular localization and flowering activity of Arabidopsis Nup96 proteins (Figure 2), suggesting that the significance of autocleavage of Nup96 proteins in plants remains to be uncovered. We further reveal a novel floral repression mechanism in Arabidopsis, where a nuclear pore complex component, Nup96, interacts with and stabilizes a floral repressor, HOS1, to balance the activity of flowering promotion pathways, allowing plants to eventually flower at the most advantageous time (Figure 8).
It has been reported that hos1 mutants exhibit early-flowering phenotypes under various light and temperature regimes (Ishitani et al., 1998; Jung et al., 2012; Lazaro et al., 2012; Lee et al., 2012). Very much like hos1 mutants, Nup96 mutation also causes early flowering under both long days and short days, with quite similar leaf numbers to that of hos1-3 mutants (Figures 1B and 1E). Moreover, no additive effect was observed when comparing the flowering time of the nup96-1 hos1-3 double mutant with either of its parental lines (Figures 6A and 6B). Molecular mechanisms of HOS1-dependent flowering repression have been well studied recently (Jung et al., 2012, 2013; Lazaro et al., 2012, 2015), one of which involves HOS1-mediated CO protein degradation. Under long days, the flowering time of the hos1 mutant could be strongly suppressed by CO mutation, but under short days, hos1 co double mutants flower as early as hos1 (Lazaro et al., 2012), suggesting that HOS1 regulates flowering time through a CO-dependent mechanism in long days but a CO-independent mechanism in short days. A similar observation was made in nup96 co double mutants (Figures 7G and 7H). As observed in hos1, increased CO abundance was seen in nup96 mutants, but with slightly different patterns (Figures 7A–7D; Supplemental Figure 4). In long day-grown nup96-1, CO accumulated from dusk to the early morning and resulted in the activation of FT in this phase to trigger flowering, whereas CO showed higher levels in hos1 plants in the day but not in the evening (Jung et al., 2012; Lazaro et al., 2012). One possible explanation for this discrepancy might be that Nup96 modulates CO degradation not only through HOS1 but also through another E3 ligase of CO, such as COP1, which functions late in the day (Jang et al., 2008; Liu et al., 2008; Sarid-Krebs et al., 2015). Therefore, more studies might be needed to test if COP1 activity is compromised in nup96 mutants. In summary, the evidence strongly suggests that Nup96 and HOS1 might regulate photoperiodic flowering through similar mechanisms. In addition, our data from RNA-seq (Figure 6) clearly demonstrate that many other genes, such as GA2ox2, SOC1, miR156, CCA1, and RVE, may function in the regulation of the nup96 and hos1 early-flowering phenotype. Therefore, CO may not be the only regulator of flowering in the nup96 and hos1 mutants in long days. In addition to early flowering, HOS1-deficient plants also show a lengthened period of circadian clock (MacGregor et al., 2013). It would be particularly interesting to investigate if Nup96 is also required for maintaining the normal circadian clock periodicity and if the altered circadian clock function in hos1 contributes to its early-flowering phenotype.
A previous study by Tamura et al. (2010) shows that conserved components of the nuclear pore complex, RNA EXPORT FACTOR1 (RAE1) and Nup43, are able to interact with HOS1 in plants, suggesting an association of HOS1 with the nuclear pore complex. In our study, by using Nup96 as the bait, we found that Nup96 interacted with HOS1 at the nuclear envelope in vivo (Figure 5D). Therefore, Nup96 is only one of several HOS1-interacting proteins and may serve as one of the docking sites of HOS1 proteins on the nuclear membrane. Interestingly, RAE1, Nup43, and Nup96 are all located at the outer ring of the nuclear pore. Nup43 and Nup96 are components of the Nup107-160 subcomplex (Boruc et al., 2012; González-Aguilera and Askjaer, 2012), while RAE1 has been reported to stably associate with Nup98 and the mRNA export factor TAP in Xenopus laevis egg extracts (Blevins et al., 2003). Interactions of HOS1 with diverse proteins of the nuclear pore complex might suggest that nuclear pore targeting of HOS1 is linked to its multiple molecular activities via dependent or independent mechanisms. Other studies also reveal that HOS1 interacts with ICE1, CO, FVE, and HDA6 in the nucleus (Dong et al., 2006; Jung et al., 2012, 2013; Lazaro et al., 2012), indicating that HOS1 might dynamically shuttle between the nuclear pore and nucleoplasm. It would be interesting to investigate how and when Nup96 interacts with HOS1 proteins and to determine their functions in the nucleoplasm and how HOS1 shuttles between the nuclear pore complex and nucleoplasm. However, the precise molecular functions of the Nup96-HOS1 module are not fully understood. The only evidence linking the activities of Nup96 and HOS1 at the molecular level is that both proteins play essential roles in mRNA nuclear export (Parry et al., 2006; MacGregor et al., 2013). In this regard, it is reasonable to infer that Nup96 and HOS1 work in concert to regulate mRNA nuclear export. However, the exact biological relevance of this molecular activity remains unclear. Another key aspect of HOS1 function is to modulate gene expression either through degrading transcription factors or interacting with transcription regulators. An additional study also demonstrates the association of HOS1 with chromatin in plants (Jung et al., 2013). Moreover, an interesting finding is showcased in the recent work that nonrandom regions on the chromosome are anchored at the nuclear periphery in Arabidopsis (Bi et al., 2017). Our evidence shows that HOS1 and Nup96 regulate the expression of an overlapping set of genes (Figure 6). Based on these lines of evidence, it would be interesting to establish if Nup96-HOS1 have the capability to anchor the chromosome at the nuclear periphery and thereby exert their influence on gene transcription activities.
The interaction of Nup96 and HOS1 not only links the activities of both genes but also leads to an unexpected mutual stabilization of the two proteins (Figure 5). Our results revealed that the nup96 mutation resulted in the destruction of endogenous HOS1 proteins (Figures 5C, 5E, and 5F). In hos1 plants, the Nup96 protein level was significantly reduced (Figure 5H), suggesting that HOS1 is directly or indirectly important for Nup96 protein turnover. Strikingly, we noticed that the decrease of HOS1 in nup96-1 was much more severe than that of Nup96 in hos1-3, suggesting that Nup96 is more important for HOS1 stability than vice versa and that some other factors may be involved in regulating Nup96 protein stability. However, how both proteins are degraded is still unknown. The similar mutual stabilization mechanism is also found in the ZTL-GI protein pair for maintaining the robustness of the circadian system in plants (Kim et al., 2007). The Nup96-HOS1 complex may limit the CO protein level to below a certain threshold in unfavorable conditions or at various developmental stages. For example, at early developmental stages, if CO proteins reach the threshold, plants would go into reproductive growth (early flowering) and produce a limited number of seeds due to limited vegetative growth, as was observed in mutant lines with an early-flowering phenotype, such as nup96 and hos1 mutants, so that it would be challenging for a species to survive. In this case, it would be interesting to determine how the activity of Nup96 and HOS1 is finely controlled.
Our findings define a novel regulatory mechanism that adds more complexity to the posttranscriptional processes that contribute to flowering time control in Arabidopsis. Interestingly, Arabidopsis plants deficient in another component of the Nup107-160 complex, Nup160 (also known as SAR1), have a similar early-flowering phenotype to that of nup96 and hos1 (Parry et al., 2006; Parry, 2014), suggesting the need to investigate if HOS1 proteins are diminished in the nup160 background. Given that most components of the nuclear pore complex are conserved among plants and animals, it would also be interesting to test if the mutual stabilization mechanism of Nup96-HOS1 is conserved among their counterparts in different evolutionary lineages. As several nucleoporins can interact with HOS1, future work should examine the specificity between HOS1 and different nucleoporins, crosstalk among these protein interactions, and their contributions to different cellular processes.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this work are of the Col accession. nup96-1 (SALK_109959), nup96-2 (SALK_117966), hos1-3 (SALK_069312), and co-9 (SAIL_24_H04) were obtained from the ABRC (Ohio State University). Other mutants were described previously: ft-1 (Yoo et al., 2005), soc1-2 (Lee et al., 2000), and co-10 (Laubinger et al., 2006). Seeds were placed on a moist filter paper and stratified at 4°C for 2 d, then transferred to soil and grown in either long-day (16 h of light/8 h of dark) or short-day (8 h of light/16 h of dark) conditions at 22°C. GreenPower LED toplighting (Philips Horticulture LED), with an intensity of 250 μmol m−2 s−1, was used. For measuring flowering time, the numbers of rosette and cauline leaves were counted when plants were bolting. More than 20 plants were counted and averaged for each measurement.
Nup96 Antibody Preparation and Verification by IP
The peptide (amino acid residues from 505 to 959) of Nup96 protein was used as the antigen to develop Nup96-specific antibody by Abmart. To verify the specificity of Nup96 antibody, 12-d-old seedlings of Col-0, nup96-1, and nup96-2 were collected and ground to fine powder in liquid nitrogen. Three volumes of extraction buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM DTT, and 1× protease inhibitor mixture [Roche]) was added to the sample. After homogenization on ice for 20 min, the mixture was centrifuged at 13,000g at 4°C for 10 min. The supernatant was transferred to a new tube, 10% of which was saved as “input” sample. IP was performed as follows: 5 μL of Nup96 antibody was added to the cleared protein extracts. After incubation at 4°C for 3 h, 30 μL of Protein A/G-agarose (Abmart, A10001) was added and incubated for another 2 h at 4°C. After washing Protein A/G-agarose with extraction buffer (without protease inhibitor cocktail) five times, the proteins were eluted with 2× SDS-PAGE loading buffer by heating at 95°C for 5 min and separated on a 4 to 20% SurePAGE precast gel (Genscript, M00657) followed by immunoblot analyses.
nup96 Complementation and Autocleavage Site Analyses
The 2.5-kb region upstream of the ATG start codon was used as the promoter sequence of Nup96, which was inserted into the Fu76 entry vector (Wang et al., 2013). To identify the autocleavage of Nup96, the different versions of Nup96 (Nup96, Nup96△N [a deletion mutant of 187 amino acids of the N-terminal end], and Nup96m [two point mutations, F187Q and S188A]) were cloned into the Fu28 entry vector (Wang et al., 2013). The 2.5-kb promoters and these versions of Nup96 sequences were integrated into the Fu39-2 destination vector (Wang et al., 2013) by LR Clonase (Invitrogen, 11791020). The resulting binary vectors were transformed into the nup96-1 mutant, respectively. The 12-d-old seedlings were used for analyzing autocleavage sites by immunoblot assays. The 2-d-old seedlings were used for examining protein subcellular localizations under an LSM 710 microscope (Carl Zeiss). Propidium iodide (20 μg/mL) was used for cell wall staining.
Protein Complex Identification by LC-MS/MS
The Nup96pro:Nup96-GFP nup96-1 and 35S:GFP wild-type seedlings were grown in long-day conditions for 12 d, and two replicate samples (∼4 g for each sample) for each genotype were harvested. The seedlings were pooled and frozen in liquid N2 at ZT4 for protein extraction. GFP-trap agarose beads were used for IP of Nup96-GFP and GFP protein complexes as described previously (Wang et al., 2015). After elution from the beads, the IP samples were subjected to LC-MS/MS analysis (Shanghai Applied Protein Technology).
Gene Expression Analysis
For gene expression analyses, 12-d-old seedlings grown on MS medium supplemented with 3% sucrose and 0.6% agar were collected for RNA extraction with an EasyPure Plant RNA Kit (Transgen Biotech, ER301-01). Total RNA concentration was measured by Nanodrop 2000 (Thermo Fisher Scientific NanoDrop). Total RNA (1 μg) was used for first-strand cDNA synthesis using FastKing genomic DNA Dispelling RT SuperMix (Tiangen, KR170801). qPCR analyses were performed with SYBR Premix Ex Taq (TaKaRa) on a StepOne Plus Realtime PCR system (Applied Biosystems) as described previously (Marshall et al., 2016). To detect the full-length transcripts of Nup96 in both wild-type plants and nup96 mutants, regular RT-PCR was performed using cDNA described above as the template. ACTIN was used as the internal control in RT-PCR, while either IPP2 or PP2A was used as the internal control for qPCR. All sequences of primers are listed in the Supplemental Table.
GUS Staining
Seedlings were soaked in GUS staining solution (50 mM sodium phosphate, pH 7.2, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 20% [v/v] methyl alcohol, and 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, cyclohexylammonium salt] and subjected to a vacuum for 15 min, then incubated at 37°C for 16 to 20 h in the dark. After incubation, chlorophyll was removed from seedlings by washing in the destaining buffer (75% [v/v] ethanol and 25% [v/v] acetic acid) three to five times. The seedlings were observed using a stereomicroscope with a SZ2-ILST CCD camera (Olympus).
BiFC
HOS1 full-length coding sequence and cYFP were cloned into Fu66 (Wang et al., 2013) to generate Fu66-HOS1-cYFP, which was cloned into Fu39-2-35S by the LR reaction to generate Fu39-2-35S:HOS1-cYFP binary vector. Nup96 full-length coding sequence and nYFP were cloned into Fu66 (Wang et al., 2013) as well to generate Fu66-Nup96-nYFP, which was cloned into Fu39-2-35S by the LR reaction to generate Fu39-2-35S:Nup96-nYFP binary vector. Fu39-2-35S:Nup96-nYFP and Fu39-2-35S:HOS1-cYFP were transformed into Col-0 separately. The two resulting transgenic lines were crossed to generate plants coexpressing 35S:Nup96:nYFP and 35S:cYFP:HOS1, which were used to detect the signal of reconstituted YFP fluorescence by laser scanning confocal microscopy (LSM 710, Carl Zeiss).
Co-IP
For co-IP analyses, pGWC-HOS1 was integrated into pGWB20 (Tanaka et al., 2012) to obtain pGWB20-35S:HOS1-myc, which was used to transform wild-type Arabidopsis. Nup96pro:Nup96:GFP nup96-1 and 35S:HOS1-myc wild type were crossed to generate a transgenic line expressing both Nup96pro:Nup96:GFP and 35S:HOS1-myc. Protein extraction and IP were performed as described for LC-MS/MS analyses. For the immunoblot assay, anti-GFP antibody (MBL, 598) and anti-myc antibody (Abmart, M20002) were used to detect Nup96-GFP and HOS1-myc, respectively. Anti-HSP90 antibody (Sigma-Aldrich, SAB1305541) was used to detect endogenous HSP90 proteins as the loading control.
HOS1 Protein Analysis
To generate the HOS1pro:HOS1-GFP construct, the 2.78-kb genomic sequence upstream of the start codon was cloned into pFu76 (Wang et al., 2013) as the HOS1 promoter entry clone. Full-length HOS1 coding sequence was cloned into pFu28 (Wang et al., 2013) to produce the HOS1-GFP gene entry clone. These two plasmids were integrated into the pFu39-2 destination vector (Wang et al., 2013) to generate the HOS1pro:HOS1-GFP binary vector, which was transformed into Col-0. Then, HOS1pro:HOS1-GFP wild type was introduced into nup96-1 mutants by genetic cross. The HOS1pro:HOS1:GFP nup96-1 homozygous line was used to analyze HOS1 protein stability. To examine the colocalization of HOS1 and Nup96 proteins, we generated transgenic lines separately expressing gHOS1:mCherry and gNup96:GFP transgenes (gHOS1 and gNup96 denote genomic DNA of HOS1 and Nup96, respectively, including promoter sequences and coding sequences). Then, two single transgenic lines were crossed to generate the plants coexpressing gHOS1:mCherry and gNup96:GFP. gHOS1:mCherry and gNup96-GFP single and dual transgenic lines were used to observe the protein subcellular localizations by laser scanning confocal microscopy (LSM 710, Carl Zeiss).
In Vitro Stability Assay for HOS1 Proteins
The 12-d-old seedlings of the nup96-1 mutant and wild-type Col-0 grown in long days were harvested at ZT4 for total protein extraction with Tris-HCl extraction buffer (25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP) according to a previous report (Ruan et al., 2019). The coding sequence of HOS1 was cloned into pET28a with BamHI and SalI, and the resulting vector was transformed into Escherichia coli BL21(DE3) for recombination protein expression. The His:HOS1 proteins was purified by Ni-NTA agarose (Qiagen, 30230). The reaction for stability assay was set up by mixing His-HOS1 proteins (100 ng) and seedling extracts (400 μg of proteins) with or without MG132 (a final concentration of 50 μM). The reaction was stopped in time by adding SDS-loading buffer and subjected to immunoblot analysis, which was probed by His antibody (Abmart, M20020) for detecting His-HOS1 proteins. Coomassie Brilliant Blue staining was applied to show the amount of loading proteins. At least three independent biological repeats were performed.
Analysis of HA-CO Protein Abundance in Vivo
Seedlings grown under long-day conditions for 12 d were harvested and ground to fine powder in liquid nitrogen. Nuclear proteins were extracted as previously described (Hayama et al., 2017). Briefly, ∼500 μL of powder was mixed with 1 mL of nuclear extraction buffer (20 mM Tris-HCl, pH 6.8, 20 mM MgCl2, 5% sucrose, 40% glycerol, 0.3% Triton X-100, 0.08% β-mercaptoethanol, 0.2% plant protease inhibitor, 1 mM DTT, and 1.3 mM PMSF). The samples were then centrifuged at 3800g for 5 min at 4°C. The pellet was washed with nuclear extraction buffer three to four times until it became colorless. The pellet was heated at 95°C for 10 min in 2× SDS-PAGE loading buffer and then loaded on a precast gel (8% SurePAGE precast gel, M00663, Genscript). Anti-HA antibody (Roche, 11867423001) was used to detect HA-CO proteins. Anti‐histone H3.1 (Abmart, P30266) and anti-ACT11 (Abmart, M2009) antibodies were used as loading controls. Signals on blots were quantified using ImageJ software.
RNA-Seq and Data Analysis
For the RNA-seq experiment, seedlings grown in long-day conditions for 12 d were harvested at ZT15, pooled for each genotype, frozen in liquid N2, and stocked at −80°C. The samples were ground in liquid N2 and then divided into three parts for each genotype (considered as three repeats). The three repeats of each sample were subjected to RNA isolation and library construction independently. Total RNAs were prepared as described above, and the quality of RNA samples was analyzed with a 2100 Bioanalyzer (Agilent). Barcoded cDNA libraries were constructed using Illumina Poly-A Purification TruSeq library reagents and protocols. Samples of three biological repeats were sequenced on Illumina HiSeq 2500 V4 (paired-end 125-bp run). The paired-end reads were aligned to the Arabidopsis genome (TAIR10) using tophat-2.0.11 with an anchor length of more than eight nucleotides for spliced alignments. Only the reads that were uniquely aligned were retained for subsequent analyses. The relative abundance of mRNA was normalized and presented as fragments per kilobase of transcript per million mapped reads. The P value and false discovery rate (FDR) were calculated using the edgeR package of Bioconductor. Genes that showed larger than 1.5-fold difference (fold change > 1.5) in the relative mRNA abundance with P < 0.01 and FDR < 0.01 were considered differentially expressed. GO enrichment calculation was performed with the gProfiler web server (https://biit.cs.ut.ee/gprofiler/gost). The top 20 most significantly enriched GO terms for each gene set were retrieved, and the −log10 P values were plotted by the heatmap function in R. RNA-seq data were submitted to the National Center for Biotechnology Information with Gene Expression Omnibus accession number GSE138706.
Phylogenetic Analysis
The amino acid sequences of 40 Nup96 proteins identified from 37 plant species were derived from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html; Supplemental Data Set 1). The sequences were aligned by the ClustalW program. An unrooted phylogenetic tree was then generated by the neighbor-joining method using MEGA 5 software. Bootstrap values shown at each branch were calculated from 1000 replicates.
Statistical Analysis
All experiments in this study were performed at least three times (i.e., three biological repeats), which means three experiments with independent pools of plants prepared at different times except for LC-MS/MS and RNA-seq, which are indicated in their own sections. All statistical analyses were performed using the SPSS software package, and representative data are shown. Asterisks indicate significant difference according to Student’s t test (***, P < 0.001; **, P < 0.01; and *, P < 0.05). All results are summarized in Supplemental Data Set 3.
Accession Numbers
Accession numbers are as follows: Nup96 (AT1G80680), HOS1 (AT2G39810), CO (AT5G15840), FT (AT1G65480), SOC1 (AT2G45660), ACTIN (AT5G09810), IPP2 (AT3G02780), and PP2A (AT1G13320).
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of Nup96 proteins in green lineages.
Supplemental Figure 2. Nup96 is broadly expressed and functions in diverse developmental processes in plants.
Supplemental Figure 3. nup96 and hos1 mutations have common transcriptomic changes in multiple pathways.
Supplemental Figure 4. CO proteins over-accumulate in nup96 mutants.
Supplemental Table. List of primers used in this study.
Supplemental Data Set 1. Alignment used to generate the phylogeny presented in Supplemental Figure 1.
Supplemental Data Set 2. Differentially expressed genes identified using RNA-seq.
Supplemental Data Set 3. Statistical analysis in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ilha Lee (Seoul National University), Ji Hoon Ahn (Korea University), and the ABRC for kindly providing soc1-2, ft-1, and T-DNA insertion lines as well as George Coupland (Max Planck Institute for Plant Breeding) for kindly providing COpro:HA:CO co-10 lines and pLEELA and GW-MCS-NOS-pGREEN vectors. We also thank Lianfeng Gu (Fujian Agriculture and Forestry University) for RNA-seq analysis and Meiqin Liu for technical assistance with confocal microscopy. This work was supported by the National Natural Science Foundation of China (grants 30670189 and 31370324).
AUTHOR CONTRIBUTIONS
Y.F. and X.W. conceived the project, designed the experiments, and analyzed the results; Z.C., X.Z., P.H., L.L., and G.H. designed and performed all the experiments and analyzed the results; J.Z., F.Z., and Y.M. helped to prepare Nup96 overexpression and tissue-specific expression plants; Y.F., X.W., and Z.C. created all figures; Y.F., X.W., and L.L wrote the article.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- Bi X., Cheng Y.J., Hu B., Ma X., Wu R., Wang J.W., Liu C. (2017). Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 27: 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Schwartz T.U. (2013). Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 21: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins M.B., Smith A.M., Phillips E.M., Powers M.A. (2003). Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 278: 20979–20988. [DOI] [PubMed] [Google Scholar]
- Boeglin M., Fuglsang A.T., Luu D.T., Sentenac H., Gaillard I., Chérel I. (2016). Reduced expression of AtNUP62 nucleoporin gene affects auxin response in Arabidopsis. BMC Plant Biol. 16: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Zhou X., Meier I. (2012). Dynamics of the plant nuclear envelope and nuclear pore. Plant Physiol. 158: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H.C.M., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montaigu A., Tóth R., Coupland G. (2010). Plant development goes like clockwork. Trends Genet. 26: 296–306. [DOI] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B.M., Blobel G., Matunis M.J. (1999). A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144: 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C., Askjaer P. (2012). Dissecting the NUP107 complex: Multiple components and even more functions. Nucleus 3: 340–348. [DOI] [PubMed] [Google Scholar]
- Hayama R., Sarid-Krebs L., Richter R., Fernández V., Jang S., Coupland G. (2017). PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 36: 904–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Lee H., Stevenson B., Zhu J.K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Song Y.H., Josephson-Day A.R., Miller R.J., Breton G., Olmstead R.G., Imaizumi T. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Asakawa H., Hiraoka Y., Haraguchi T. (2010). Nucleoporin Nup98: A gatekeeper in the eukaryotic kingdoms. Genes Cells 15: 661–669. [DOI] [PubMed] [Google Scholar]
- Jacob Y., Mongkolsiriwatana C., Veley K.M., Kim S.Y., Michaels S.D. (2007). The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiol. 144: 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M. (2011). Timing is everything in plant development: The central role of floral repressors. Plant Sci. 181: 364–378. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Park J.H., Lee S., To T.K., Kim J.M., Seki M., Park C.M. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25: 4378–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo P.J., Park C.M. (2012). The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287: 43277–43287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Blankestijn-de Vries H., Hanhart C.J., Peeters A.J. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A., et al. (2017). TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 13: e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Marchal V., Le Gourrierec J., Wenkel S., Adrian J., Jang S., Kulajta C., Braun H., Coupland G., Hoecker U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222. [DOI] [PubMed] [Google Scholar]
- Lazaro A., Mouriz A., Piñeiro M., Jarillo J.A. (2015). Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27: 2437–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim J.J., Kim S.H., Cho H.J., Kim J., Ahn J.H. (2012). The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 53: 1802–1814. [DOI] [PubMed] [Google Scholar]
- Lee K., Seo P.J. (2015). The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal. Behav. 10: e1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor D.R., et al. (2013). HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell 25: 4391–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor D.R., Penfield S. (2015). Exploring the pleiotropy of hos1. J. Exp. Bot. 66: 1661–1671. [DOI] [PubMed] [Google Scholar]
- Marshall C.M., Tartaglio V., Duarte M., Harmon F.G. (2016). The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28: 2560–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2009). Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12: 752–759. [DOI] [PubMed] [Google Scholar]
- Michaels S.D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas G., Reeves P.H., Fu Y.F., Bancroft I., Dean C., Coupland G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G. (2014). Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J. Exp. Bot. 65: 6057–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei G.P. (1962). Supervital mutants of Arabidopsis. Genetics 47: 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7: 282–289. [DOI] [PubMed] [Google Scholar]
- Ruan W., Guo M., Wang X., Guo Z., Xu Z., Xu L., Zhao H., Sun H., Yan C., Yi K. (2019). Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant 12: 1060–1074. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K.C., Fornara F., Takahashi Y., Hayama R., Jang S., Tilmes V., Valverde F., Coupland G. (2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 84: 451–463. [DOI] [PubMed] [Google Scholar]
- Shen L., Kang Y.G., Liu L., Yu H. (2011). The J-domain protein J3 mediates the integration of flowering signals in Arabidopsis. Plant Cell 23: 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.S., Kubota A., Imaizumi T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Estrada D.A., Johnson R.S., Kim S.K., Lee S.Y., MacCoss M.J., Imaizumi T. (2014). Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 111: 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. (2010). The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11: 490–501. [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Kimura T., Hikino K., Goto S., Nishimura M., Mano S., Nakagawa T. (2012). Gateway vectors for plant genetic engineering: Overview of plant vectors, application for bimolecular fluorescence complementation (BiFC) and multigene construction In Genetic Engineering: Basics, New Applications and Responsibilities, Barrera-Saldaña H., ed (London: IntechOpen; ), pp. 35–58. [Google Scholar]
- Tran E.J., Wente S.R. (2006). Dynamic nuclear pore complexes: Life on the edge. Cell 125: 1041–1053. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Wälde S., Kehlenbach R.H. (2010). The part and the whole: Functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 20: 461–469. [DOI] [PubMed] [Google Scholar]
- Wang Q., Barshop W.D., Bian M., Vashisht A.A., He R., Yu X., Liu B., Nguyen P., Liu X., Zhao X., Wohlschlegel J.A., Lin C. (2015). The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol. Plant 8: 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fan C., Zhang X., Zhu J., Fu Y.F. (2013). BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol. 13: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859. [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. (2007). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X. (2005). A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Meier I. (2011). Identification and characterization of the Arabidopsis FG-repeat nucleoporin Nup62. Plant Signal. Behav. 6: 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]