The ubiquitin-binding protein OsDSK2a modulates the level of the gibberellin metabolism regulator EUI to coordinate plant growth and salt-stress responses in rice.
Abstract
UBL-UBA (ubiquitin-like-ubiquitin-associated) proteins are ubiquitin receptors and transporters in the ubiquitin-proteasome system that play key roles in plant growth and development. High salinity restricts plant growth by disrupting cellular metabolism, but whether UBL-UBA proteins are involved in this process is unclear. Here, we demonstrate that the UBL-UBA protein OsDSK2a (DOMINANT SUPPRESSOR of KAR2) mediates seedling growth and salt responses in rice (Oryza sativa). Through analysis of osdsk2a, a mutant with retarded seedling growth, as well as in vitro and in vivo assays, we demonstrate that OsDSK2a combines with polyubiquitin chains and interacts with the gibberellin (GA)-deactivating enzyme ELONGATED UPPERMOST INTERNODE (EUI), resulting in its degradation through the ubiquitin-proteasome system. Bioactive GA levels were reduced, and plant growth was retarded in the osdsk2a mutant. By contrast, eui mutants displayed increased seedling growth and bioactive GA levels. OsDSK2a levels decreased in plants under salt stress. Moreover, EUI accumulated under salt stress more rapidly in osdsk2a than in wild-type plants. Thus, OsDSK2a and EUI play opposite roles in regulating plant growth under salt stress by affecting GA metabolism. Under salt stress, OsDSK2a levels decrease, thereby increasing EUI accumulation, which promotes GA metabolism and reduces plant growth.
INTRODUCTION
The ubiquitin-proteasome system plays a pivotal role in regulating plant growth and abiotic stress responses (Vierstra, 2009; Wang and Deng, 2011). Protein degradation is a posttranslational process that plays key roles in various biological processes (Vierstra, 2003; Dreher and Callis, 2007). The degradation of a substrate protein by the ubiquitin-proteasome system involves four steps: the ubiquitination, recognition, delivery, and degradation of the protein by the proteasome (Tian and Xie, 2013). Emerging evidence indicates that the processes by which ubiquitinated proteins are recognized and delivered to the proteasome are finely controlled by ubiquitin-like (UBL)-ubiquitin-associated (UBA) proteins and other ubiquitin receptors in yeast (Saccharomyces cerevisiae) and humans (Kleijnen et al., 2000; Su and Lau, 2009; Rajalingam and Dikic, 2016; Ohtake et al., 2018; Romero-Barrios and Vert, 2018). UBL-UBA proteins bind to polyubiquitinated proteins and deliver them to proteasomal receptors Rpn1, Rpn10, and Rpn13 (Zhang et al., 2009; Tsuchiya et al., 2017), indicating that UBL-UBA proteins play a regulatory role in the recognition and delivery of polyubiquitinated proteins. There are three types of UBL-UBA proteins: RADIATION SENSITIVE23 (RAD23), DOMINANT SUPPRESSOR OF KAR2 (DSK2), and DNA DAMAGE-INDUCIBLE1 (DDI1). In yeast and humans, these proteins interact with the proteasome through their UBL domains and with polyubiquitinated substrate proteins through their UBA domains (Matiuhin et al., 2008). Thus, the polyubiquitination of substrate proteins is a prerequisite for their degradation.
A polyubiquitin chain can form via seven potential Lys residues within ubiquitin (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) and via the α-amino-terminus of Met 1 (Met1) of ubiquitin (Rajalingam and Dikic, 2016). The most extensively studied polyubiquitin chains are K48- and K63-linked polyubiquitin chains. K48-linked chains usually participate in 26S-proteasome–mediated degradation, and K63-linked chains are involved in DNA repair, signal transduction, endocytosis, membrane trafficking, and substrate degradation (Ohtake et al., 2018; Romero-Barrios and Vert, 2018). UBL-UBA proteins use diverse regulatory mechanisms in different species. For instance, UBL-UBA proteins in yeast are functionally redundant (Díaz-Martinez et al., 2006). In Arabidopsis (Arabidopsis thaliana), RAD23s connect polyubiquitinated proteins to the 26S proteasome (Farmer et al., 2010), whereas DSK2s mediate selective autophagy by interacting with a brassinosteroid regulator, thereby balancing plant growth and stress responses (Nolan et al., 2017). Thus, UBL-UBA proteins appear to play specific roles in growth and development.
Salinity has adverse effects on almost all aspects of a plant’s lifecycle, including seed germination, plant growth, and productivity (Deinlein et al., 2014; Yang and Guo, 2018). Plants have developed sophisticated mechanisms to withstand salt stress (Cui et al., 2015). Gibberellins (GAs) are tetracyclic diterpene phytohormones that affect plant growth and development (Schwechheimer, 2012; Davière and Achard, 2013; Hedden and Sponsel, 2015; Kwon and Paek, 2016; Wang et al., 2017) by promoting cell elongation (De-Lucas et al., 2008; Rizza et al., 2017) or cell division (Li et al., 2017) to control organ growth in the developmental context. The modulation of GA levels in response to various stresses including cold, salinity, drought stress, and submergence has been investigated over the past decade (Colebrook et al., 2014). The roles of GAs in modulating plant growth under stress conditions are complex. For example, SNORKEL and SUBMERGENCE1A (SUB1A) play opposing roles in regulating plant growth in response to flooding by stimulating or inhibiting GA responses in rice (Oryza sativa; Fukao and Bailey-Serres, 2008; Hattori et al., 2009). SEMIDWARF1 encodes a GA biosynthesis enzyme responsible for submergence-induced internode elongation (Kuroha et al., 2018). Emerging evidence indicates that the regulation in GA metabolism and signaling contributes to salt responses. For example, the accumulation of DELLA proteins under salt stress mediates growth restriction in Arabidopsis (Achard et al., 2006), and the degradation of these proteins is promoted by GA (Van De Velde et al., 2017). Furthermore, the transcriptional regulation of genes involved in GA metabolism, including those encoding Arabidopsis GA2ox7 (Magome et al., 2008) and rice GA2ox5 (Shan et al., 2014) and MYB91 (Zhu et al., 2015), mediate salt stress responses. Therefore, the regulation of GA metabolism might function in the plant response to salt stress by altering plant growth. However, how regulators of GA metabolism respond to salt stress is currently unclear. Whether UBL-UBA proteins are involved in limiting plant growth under salt stress is also unclear.
In the present study, we demonstrate that the UBL-UBA protein OsDSK2a (a homolog of DSK2) helps restrict seedling growth in rice under salt stress by modulating GA catabolism. This process is mediated by the direct interaction of OsDSK2a with polyubiquitinated ELONGATED UPPERMOST INTERNODE (EUI), a GA-deactivating enzyme (Zhu et al., 2006). This interaction results in the degradation of EUI and changes in bioactive GA levels. Salt stress restricts seedling growth by interfering with the OsDSK2a-EUI complex. Thus, the OsDSK2a-EUI module regulates GA metabolism and plant growth under salt stress.
RESULTS
The UBL-UBA Protein OsDSK2a Modulates Plant Growth
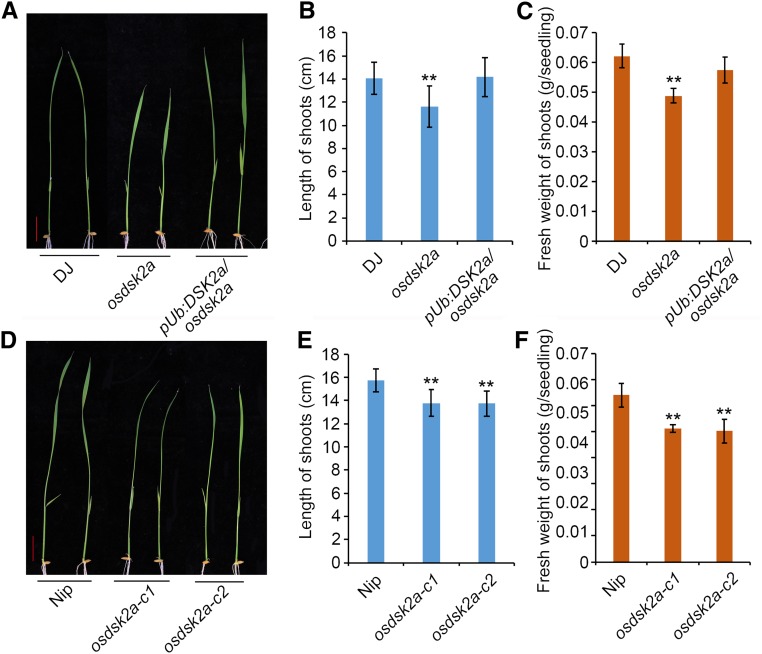
Like yeast, animals, and Arabidopsis (Farmer et al., 2010), rice contains three classes of UBL-UBA proteins, RAD23, DSK2, and DDI, each containing one N-terminal UBL and one C-terminal UBA domain (Supplemental Figure 1). To investigate the roles of rice UBL-UBA proteins in regulating plant growth and development, we screened japonica rice T-DNA insertion mutant libraries for plants with retarded growth (Jeon et al., 2000; Jeong et al., 2006). The PFG_3A-00810.L mutant, which harbors a T-DNA insertion 397 bp upstream of the ATG start codon of OsDSK2a (Supplemental Figure 2A), showed retarded growth at the seedling stage (Figure 1A). RT-PCR revealed no OsDSK2a expression in PFG_3A-00810.L, indicating that the mutant is a knockout allele of OsDSK2a, which we named osdsk2a. The expression levels of the other genes surrounding the insertion site were not altered in osdsk2a compared with wild-type Dongjin (DJ; Supplemental Figure 2B). Seedlings overexpressing OsDSK2a in the osdsk2a background (Supplemental Figure 2C) displayed recovered plant growth to wild-type levels, showing neither enhanced shoot length nor increased fresh weight (Figures 1B and 1C).
Figure 1.
Loss-of-Function OsDSK2a Mutants Display Retarded Seedlings Growth.
(A) Seedling growth of T-DNA insertion osdsk2a mutant and OsDSK2a overexpression transgenic plants.
(B) and (C) Lengths and fresh weights of the shoots shown in (A).
(D) Seedling growth of osdsk2a allelic mutants generated by CRISPR/Cas9.
(E) and (F) Lengths and fresh weights of the shoots shown in (D).
Bars = 2 cm. Data are presented as mean ± sd (n = 15, **P ≤ 0.01, Student’s t test).
To evaluate the role of OsDSK2a in plant growth, we generated osdsk2a allelic mutants in the Nipponbare (Nip) background using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 (Supplemental Figure 3). Two lines with a frame-shift and premature termination of OsDSK2a, i.e., osdsk2a-c1 and osdsk2a-c2, exhibited reduced seedling growth compared with the wild type (Figure 1D). In addition, all osdsk2a alleles displayed retarded growth in various developmental processes from the seedling to heading stages (Supplemental Figure 3E and 4). During the heading stage, all internodes in the osdsk2a mutant were shortened, except for the second internode (Supplemental Figures 4C and D). Analysis of longitudinal sections of elongated regions of the uppermost internodes indicated that the cells in osdsk2a were shorter than those in DJ (Supplemental Figures 4E and F), suggesting that the shortened internodes in osdsk2a are due to longitudinally reduced cell length and not to a decrease in cell number. These results demonstrate that the knockout OsDSK2a contributes to retarded plant growth.
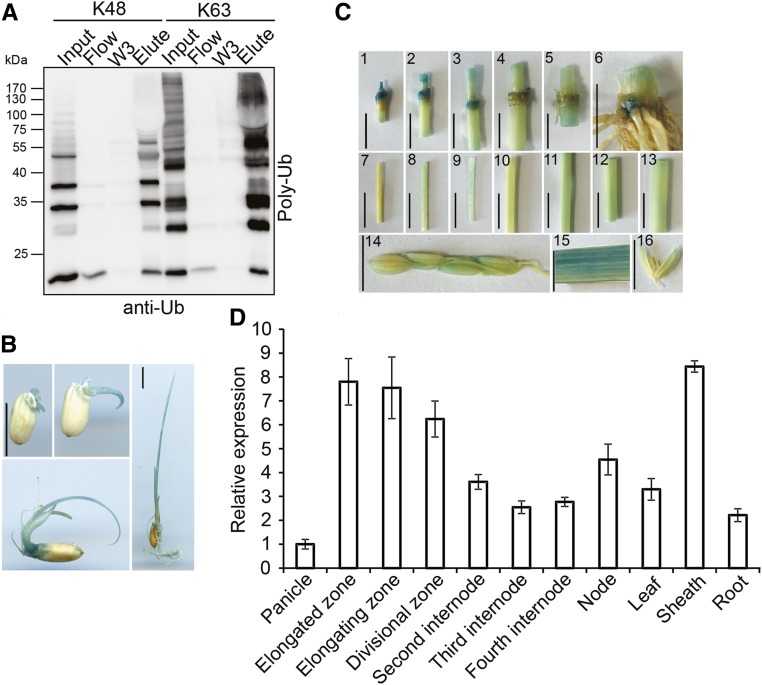
To investigate whether OsDSK2a interacts with ubiquitin or polyubiquitin chains, we expressed glutathione S-transferase (GST)-tagged OsDSK2a (GST-OsDSK2a) fusion protein in Escherichia coli (E. coli) and incubated this protein with ubiquitin mixtures containing polyubiquitin chains assembled via K48 or K63 linkages (Ub2-7). OsDSK2a bound to both K48- and K63-linked polyubiquitinated chains (Figure 2A), suggesting that OsDSK2a functions as a ubiquitin receptor and plays roles in protein degradation and signal transduction.
Figure 2.
Ubiquitin Binding Activity and Gene Expression Analysis of OsDSK2a.
(A) Binding of OsDSK2a to poly-ubiquitin (poly-Ub) chains in vitro. The OsDSK2a-GST fusion protein was precipitated by glutathione beads and incubated with a mixture of poly-Ub chains linked via K48 or K63. The flow-through was collected (Flow), and the beads were washed three times (W1–W3). The precipitated proteins were eluted (Elute) and separated by SDS-PAGE before immunoblotting with anti-Ub antibodies.
(B) pOsDSK2a: GUS expression patterns in transgenic rice plants at the seedling stage. Bars = 1 cm.
(C) pOsDSK2a: GUS expression patterns in transgenic rice plants at the heading stage. (1) to (5) Nodes from top to bottom. (6) Last internode and root. (7) Elongated zone of the uppermost internode. (8) Elongating zone of the uppermost internode. (9) Division zone of the uppermost internode. (10) to (13) Second to fifth internodes. (14) Young spikelets. (15) Flag leaf. (16) Anthers. Bars = 1 cm.
(D) qPCR analysis of OsDSK2a expression in different tissues. ACTIN was used as a control. The data represent the means ± sd (n = 3).
OsDSK2a expression was generally detected during the seedling and mature stages (Figures 2B and 2C) in transgenic pOsDSK2a:GUS plants. We performed RT-qPCR assays using total RNA from various tissues of plants at the reproductive stage, including panicles, different internodes, nodes, leaf blades, leaf sheaths, and roots. OsDSK2a expression was detected in all tissues examined but primarily in dividing tissues, including the uppermost internode, node, and leaf sheath (Figure 2D). Thus, OsDSK2a functions throughout plant growth and development.
OsDSK2a Interacts with the Gibberellin Metabolism Factor EUI and Promotes its Degradation
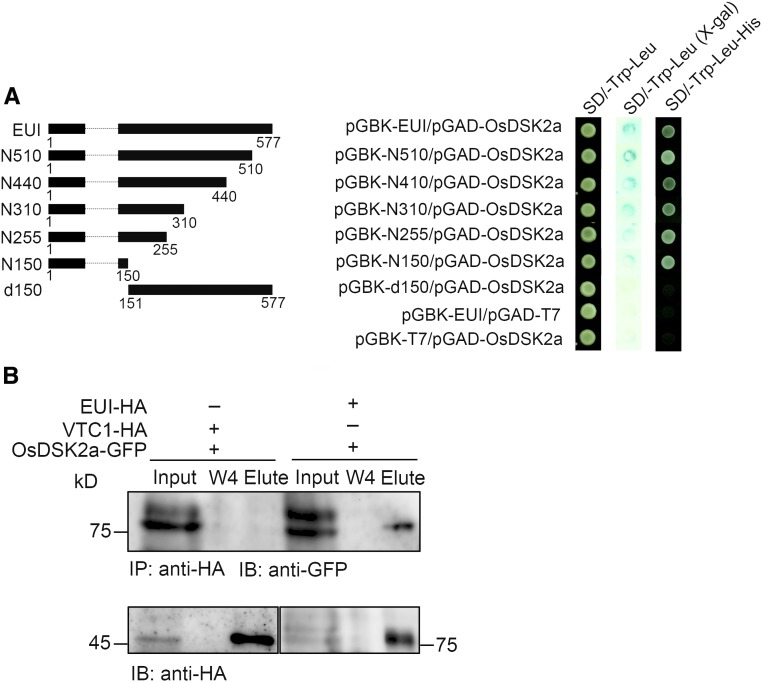
To gain insight into the mechanism by which OsDSK2a regulates plant growth, we screened for interacting factors of OsDSK2a using a yeast two-hybrid assay with full-length OsDSK2a as bait. With use of this system, ten proteins were confirmed to interact with OsDSK2a (Supplemental Table 1). Among these, we selected EUI for further study because it was previously shown to play a crucial role in GA catabolism by catalyzing the deactivation of bioactive GAs, a process thought to play a prominent role in the GA non–13-hydroxylation pathway in rice (Luo et al., 2006; Zhu et al., 2006). EUI primarily functions in more mature plants to restrict the growth of the upper internode, and it is also involved in maintaining GA homeostasis in seedlings, roots, and seeds (Zhang et al., 2008). To analyze the interaction between OsDSK2a and EUI in yeast, a series of EUI N-terminal truncated fragments were fused to GAL4-BD. The full-length fusion protein and all fragments of EUI containing the 150 N-terminal amino acids interacted with OsDSK2a, whereas an EUI truncated protein lacking the 150 N-terminal amino acids (EUI-d150) did not (Figure 3A), indicating that the 150 N-terminal amino acids of EUI are essential for its interaction with OsDSK2a.
Figure 3.
OsDSK2a Interacts with the Gibberellin Metabolism Regulator EUI.
(A) Interactions of full-length and truncated EUI proteins with OsDSK2a in yeast. At left, a schematic diagram of full-length and truncated EUI fusion proteins with GAL4-BD is shown; at right panel, the growth of transformed yeast cells on SD/-Trp-Leu and SD/-Trp-Leu-His medium, respectively. SD/-Trp-Leu (X-gal) shows a filter assay for β-galactosidase activity of the transformants. pGBK-EUI/pGAD-T7 and pGBK-T7/pGAD-OsDSK2a combinations were used as negative controls.
(B) Coimmunoprecipitation assays to detect the interaction of OsDSK2a with EUI in vivo. Total proteins extracted from callus cotransformed with EUI-HA/ OsDSK2a-GFP or VTC1-HA /OsDSK2a-GFP were immunoprecipitated with anti-HA antibodies, washed four times (W1–W4), and the eluted fractions (Elute) separated by SDS-PAGE before immunoblotting with anti-GFP antibodies.
To further confirm the interaction between OsDSK2a and EUI, we performed a coimmunoprecipitation assay using rice callus cotransformed with EUI-HA and OsDSK2a-GFP; rice callus harboring a construct containing the Arabidopsis VITAMIN C DEFECTIVE1 (VTC1) gene fused to HA was used as a negative control. Immunoblot analysis showed that OsDSK2a interacted with EUI, but not with VTC1 (Figure 3B). These results help confirm the interaction between EUI and OsDSK2a in vivo. Due to the high sequence similarity between OsDSK2a and OsDSK2b, EUI also interacted with OsDSK2b in yeast cells (Supplemental Figure 5A). However, loss-of-function OsDSK2b mutants generated by CRISPR/Cas9 displayed normal seedling growth (Supplemental Figures 5B to D), and the plant height of the osdsk2a/b double mutants was similar to that of osdsk2a-c1 (Supplemental Figure 5E). Thus, OsDSK2a and OsDSK2b play unequal roles in regulating plant growth.
EUI is generally expressed at low levels in most plant tissues (Luo et al., 2006), suggesting that its expression is inhibited in plants at the seedling stage to ensure proper plant growth. To explore this issue, we measured EUI protein levels in the leaves and sheaths of seedlings. We performed cell-free assays using protein extracts from the leaf or sheath tissues of transgenic plants overexpressing HA-tagged EUI (EUI-HA). EUI-HA was degraded more rapidly in sheaths than in leaves (Supplemental Figures 6A and B). Therefore, we used sheath tissue to examine EUI protein stability in a subsequent assay. EUI-HA was degraded more rapidly in sheaths than in leaves, which is consistent with the higher OsDSK2a transcript levels detected in sheath versus leaf tissue (Figure 2D).
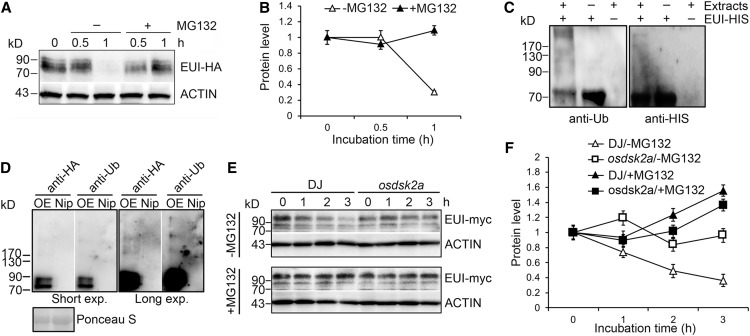
In the cell-free assay, EUI-HA produced three bands with higher molecular masses than EUI-HA. The theoretical molecular mass of EUI is ∼63 kD, and the expected molecular mass of EUI-HA is 69 kD. The molecular mass of the upper band of EUI-HA was greater than 90 kD, suggesting it might have been modified with polyubiquitin chains (Ub4). Thus, the bands shown in Supplemental Figure 6 were likely modified, possibly by ubiquitination. We therefore pretreated EUI-HA transgenic plants with MG132, an inhibitor of the ubiquitin proteasome system. In the absence of MG132, EUI-HA protein degraded after 1 h of treatment with the protein synthesis inhibitor cycloheximide (CHX). By contrast, in the presence of MG132, no obvious degradation of EUI-HA protein was observed under the same conditions (Figures 4A and 4B).
Figure 4.
OsDSK2a Promotes the Degradation of EUI through the Ubiquitin-Proteasome Pathway.
(A) Effects of MG132 on EUI degradation. Four-week-old EUI-HA transgenic plants were treated with (+) or without (−) 50 μM MG132 for the indicated time.
(B) Relative quantification of EUI-HA levels in (A). The data represent mean ± sd (n = 3).
(C) Ubiquitination of EUI in vitro. Purified EUI-HIS was incubated with (+) or without (−) wild-type DJ total protein extracts. The amount of EUI-HIS pulled down using a HIS-Select Nickel Affinity Gel was determined by immunoblotting with anti-Ub antibody (left) and anti-HIS antibody (right).
(D) Ubiquitination of EUI in vivo. Total proteins extracted from EUI-HA overexpression transgenic plants (OE) were immunoprecipitated with anti-HA antibodies; the wild type Nip was used as a negative control. The eluted fractions were separated by SDS-PAGE for immunoblotting with anti-HA antibody and anti-Ub antibody. Long exposure (Long exp.) times were used to detect the higher molecular weight ubiquitinated EUI bands. Ponceau S staining of input was used as a loading control.
(E) Promotion of EUI degradation by OsDSK2a in vitro. Protein extracts from EUI-myc overexpression transgenic plants were incubated with extracts from DJ or osdsk2a plants in the absence or presence of MG132 in cell-free assays. Protein levels were detected at different time points by immunoblotting with anti-myc antibody.
(F) Relative quantification of EUI-myc levels in (E). The data represent mean ± sd (n = 3).
One representative experiment of 3 replicates is shown in the figure.
We expressed EUI fused to HIS tag in E. coli and incubated the tagged protein with wild-type protein extracts (DJ). A poly-ubiquitin signal was observed by immunoblot analysis with anti-ubiquitin antibody, together with higher molecular mass forms of EUI-HIS (Figure 4C). The ubiquitination of EUI was further confirmed by coimmunoprecipitation analysis using EUI-HA fusion protein extracted from EUI-HA transgenic plants (Figure 4D), supporting the notion that the three bands of EUI-HA described above were modified by ubiquitin. Therefore, EUI protein can be degraded through the ubiquitin proteasome system.
Since OsDSK2a interacts with EUI and functions as a ubiquitin binding receptor, we reasoned that OsDSK2a might mediate the degradation of EUI. To test this hypothesis, we performed cell-free assays by incubating protein extracts from EUI-myc transgenic plants with extracts from wild-type DJ or osdsk2a plants. Measurement of EUI-myc protein levels showed that EUI-myc was more stable in osdsk2a than in DJ and that MG132 inhibited the decrease in EUI-myc protein levels in DJ plants (Figures 4E and 4F). Meanwhile, EUI protein was more stable in osdsk2a plants and degraded slightly after 3 h of CHX treatment in the absence of MG132 (Supplemental Figures 6C and D). Thus, OsDSK2a promotes the degradation of EUI.
The Retarded Growth of osdsk2a Plants Is Associated with Reduced Levels of Bioactive GA
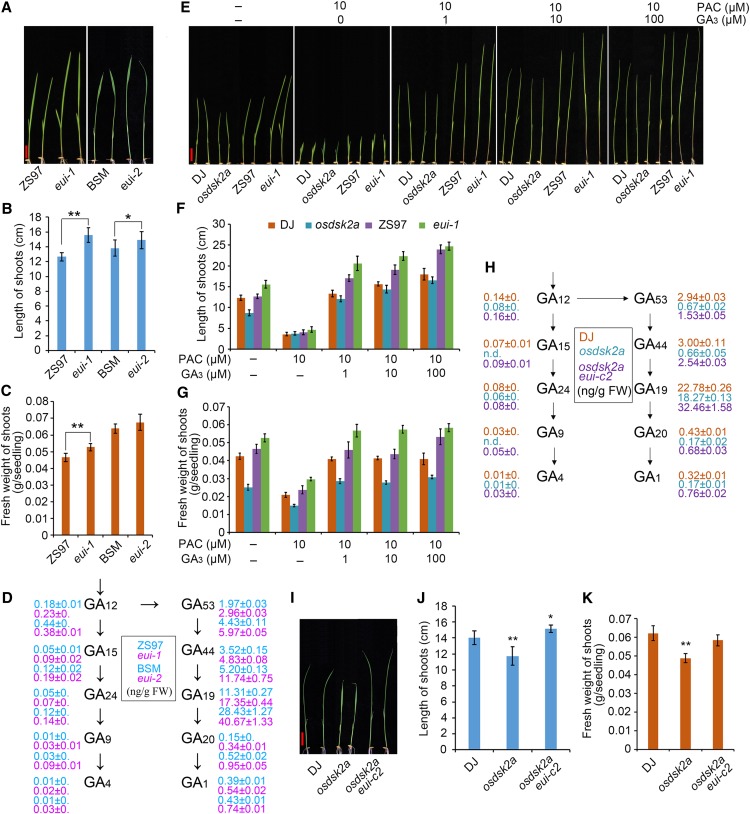
EUI contributes to GA metabolism in plants at the mature stage (Zhu et al., 2006). To explore the role of EUI in regulating plant growth at the seedling stage, we examined two different 2-week-old eui mutants, including eui-1, harboring a retrotransposon insertion in the Zhenshan 97 (ZS97) background (Zhu et al., 2006) and eui-2, with a mutation of Gly to Asp at the 500th amino acid residue of EUI in the Baishoumao (BSM) background, as identified by map-based cloning based on the elongated first internode phenotype. Both eui alleles displayed enhanced growth compared with wild-type plants at the seedling stage (Figures 5A to 5C), indicating that EUI plays a role in the growth of seedlings as well as mature plants. To investigate the role of EUI in GA metabolism, we measured the levels of bioactive GAs (GA1 and GA4) and their precursors, including GA12, GA15, GA24, GA9, GA4, GA53, GA44, GA19, GA20, and GA1, in the eui mutants and wild-type plants. Both eui mutants contained higher levels of bioactive GAs and their precursors than wild-type plants at the seedling stage (Figure 5D), which helps explain the enhanced growth of the eui mutants.
Figure 5.
EUI Functions Downstream of OsDSK2a in GA Metabolism during the Seedling Stage.
(A) Growth of 10-d-old eui seedlings. eui-1 is a retrotransposon insertion mutant in the Zhenshan 97 (ZS97) background. eui-2 is a site mutant with a mutation at the 500th amino acid (Gly to Asp) in the BSM background.
(B) and (C) Lengths and fresh weights of the shoots shown in (A). The data represent mean ± sd (n = 15, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
(D) Contents of bioactive GAs and their precursors in 10-d-old eui seedlings. The data represent mean ± sd (n = 3).
(E) GA mediates the reduced growth of osdsk2a and enhanced growth of eui-1. Germinated DJ, osdsk2a, ZS97, and eui-1 seeds were incubated in standard liquid medium or medium supplemented with 10 μM PAC and different concentrations of GA3 (0, 1, 10, and 100 μM) for 10 d. Bar = 2 cm.
(F) and (G) Lengths and fresh weights of the shoots shown in (E). The data represent mean ± sd (n = 15).
(H) Contents of bioactive GAs and their precursors in osdsk2a and the osdsk2a eui double mutant. n.d., not detected. The data represent mean ± sd (n = 3).
(I) Seedling growth of osdsk2a eui. Bar = 2 cm.
(J) and (K) Lengths and fresh weights of the shoots shown in (I). The data represent mean ± sd (n = 15, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
The retarded growth of the osdsk2a mutants and the interaction of OsDSK2a with EUI suggested that OsDSK2a affects GA homeostasis. To further confirm the role of altered GA levels in the retarded growth of the osdsk2a mutants, we examined the growth of osdsk2a and eui-1 plants treated with paclobutrazol (PAC) and different concentrations of GA3. PAC is a GA biosynthesis inhibitor that inhibits the growth of rice seedlings. The differences in shoot length between DJ and osdsk2a seedlings and between ZS97 and eui-1 seedlings decreased under 10 μM PAC treatment. The differences in shoot length between DJ and osdsk2a seedling also decreased in response to the addition of GA3. By contrast, the shoot length of eui-1 seedlings was comparable with that of wild-type ZS97 seedlings under all treatments except 100 μM GA3 (Figures 5E to 5G). The reduced fresh weight of the seedlings was partially improved under GA treatment in osdsk2a, but not in eui-1. In addition, the reduced growth of osdsk2a plants at the tillering and heading stages was restored to wild-type levels in response to exogenous GA3 treatment (Supplemental Figure 7), indicating that the retarded growth of osdsk2a could be attributed to altered GA levels. Measurement of bioactive GAs and their precursors in seedlings showed that osdsk2a contained lower levels of all GAs except GA4 compared to wild-type plants (Figure 5H). Furthermore, the levels of several forms GAs, including GA1, GA20, and GA53, were lower in mature osdsk2a-c1 plants compared to their wild-type counterparts (Supplemental Figure 3F). These results suggest that OsDSK2a mediates the role of GA homeostasis in regulating plant growth.
To investigate the genetic relationship between OsDSK2a and EUI, we generated the osdsk2a eui double mutant in the osdsk2a background using CRISPR/Cas9. The nucleotide sequence of EUI was successfully edited in the mutants, with a 1-bp insertion and 1-bp deletion in double mutant lines osdsk2a eui-c1 and osdsk2a eui-c2, respectively (Supplemental Figures 8A to 8D). The 1-bp deletion in osdsk2a eui-c2 gave rise to a frame-shift and premature termination of EUI protein. These plants showed enhanced growth compared to the osdsk2a mutants (Figure 5I), with longer shoots but without increased fresh weight compared to wild-type seedlings (Figure 5J and 5K). In addition, the osdsk2a eui-c2 plants had longer uppermost internodes than DJ and osdsk2a plants at the heading stage, which resembles the phenotypes of the eui mutants (Supplemental Figures 8E and 8F). Measurement of the levels of bioactive GAs and their precursors in osdsk2a eui-c2 seedlings indicated that the loss-of-function of EUI in the osdsk2a mutants led to the recovery GA metabolism (Figure 5H). Finally, in agreement with the significantly elevated levels of forms of GAs including GA1, GA4, GA20, GA24, and GA53 in eui allelic mutants at the mature stage (Zhu et al., 2006), the levels of all of these GAs were significantly higher in the culms of mature osdsk2a eui-c2 versus osdsk2a plants, except for GA24 (Supplemental Figure 8G), which also suggests that EUI plays an epistatic role to OsDSK2a in modulating GA metabolism.
OsDSK2a-Promoted EUI Degradation Functions in the Salt Stress Response
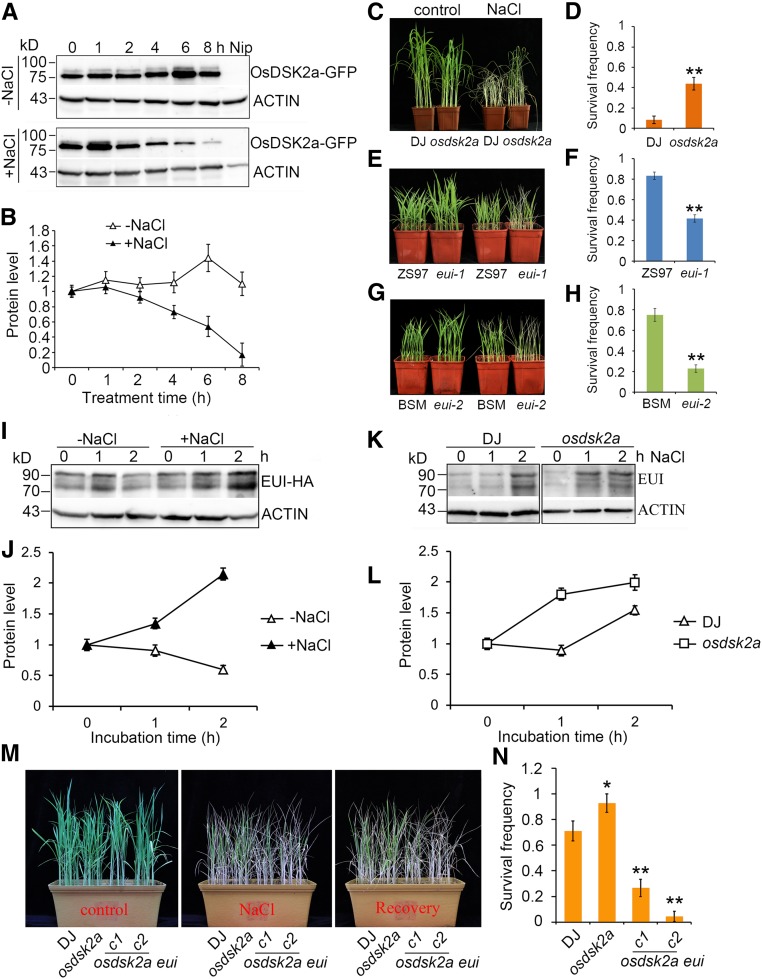
In yeast, DSK2 proteins promote the degradation of target proteins or protect these proteins from degradation, depending on DSK2 abundance (Su and Lau, 2009). Since salt stress inhibits plant growth (Achard et al., 2006; Deinlein et al., 2014; Yang and Guo, 2018) and the OsDSK2a-EUI complex contributes to seedling growth, we measured OsDSK2a protein levels in seedlings in response to salt stress to explore the regulatory role of OsDSK2a in salt-induced inhibition of seedling growth. OsDSK2a protein levels decreased in seedlings under salt stress (Figures 6A and 6B). Under high salinity stress, osdsk2a seedlings were more tolerant to salt stress than wild-type plants, with survival frequencies of ∼45% and 10% under high-salinity conditions in osdsk2a and DJ plants, respectively (Figures 6C and 6D). Correspondingly, the eui alleles were more sensitive to salt stress than wild-type plants, and the survival frequencies of eui-1 and eui-2 seedlings after salt treatment were lower than those of the corresponding wild-type seedlings (Figures 6E to 6H). These results demonstrate that OsDSK2a and EUI play opposite roles in salt tolerance. To investigate the effects of salt on seedling growth, we measured the lengths and fresh weights of the shoots of osdsk2a and eui-1 plants by treating 3-d-old seedlings with different concentrations of NaCl. The growth of osdsk2a plants was less inhibited than that of wild-type seedlings under 100 mM NaCl treatment, whereas the growth of eui-1 plants was more sensitive to salt stress compared with wild-type seedlings under 30 mM NaCl treatment (Supplemental Figure 9). Therefore, the loss-of-function of OsDSK2a helps plants adapt to salt stress.
Figure 6.
OsDSK2a and EUI Regulate the Salt Response in an Opposite Manner, which Corresponds with the Different Responses of OsDSK2a and EUI Protein Levels to Salt Stress.
(A) Detection of OsDSK2a-GFP protein levels in plants under salt stress. Two-week-old OsDSK2a-GFP transgenic plants were treated with or without 120 mM NaCl for the indicated time. The seedling extracts were subjected to immunoblotting with anti-GFP antibodies. ACTIN was used as a loading control.
(B) Relative quantification of OsDSK2a-GFP levels in (A). The OsDSK2a-GFP level at 0 h was defined as 1. The data represent mean ± sd (n = 3).
(C) to (H) Salt tolerance of osdsk2a and eui seedlings. Two-week-old seedlings grown in soil were treated with or without 120 mM NaCl for 7 d [(C), (E), (G)], followed by analysis of survival frequencies after recovery [(D), (F), (H)]. The data represent mean ± sd (n = 3 biological replicates, 16 plants per assay, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
(I) Effects of salt stress on EUI protein levels. Four-week-old EUI-HA transgenic plants grown in liquid medium were transferred to medium with or without 120 mM NaCl for the indicated time.
(J) Relative quantification of EUI-HA levels in (I).
(K) EUI protein levels in DJ and osdsk2a seedlings under salt stress. Two-week-old seedlings grown in standard liquid medium were transferred to medium supplemented with 120 mM NaCl. Protein levels in sheaths extracted at different time points were detected by immunoblotting with anti-EUI antibodies. ACTIN was used as a loading control.
(L) Relative quantification of EUI levels in (K). The three bands in (I) and (K) were used to quantify modified EUI protein levels. The EUI level at 0 h was defined as 1. The data represent mean ± sd (n = 3).
(M) Salt tolerance of osdsk2a eui seedlings. Two-week-old seedlings grown in soil were treated with 120 mM NaCl for 7 d, followed by 1 week of recovery.
(N) Survival frequencies of the seedlings in (M) after NaCl treatment. The data represent mean ± sd (n = 3, 16 plants per assay, *P ≤ 0.05, **P ≤ 0.01, Student’s t test).
Based on the role of OsDSK2a in modulating EUI degradation and the observation that salt stress decreases OsDSK2a accumulation, we examined the effects of salt on EUI protein stability using EUI-HA transgenic plants. EUI protein strongly accumulated in plants under salinity treatment (Figures 6I and 6J) without a corresponding increase in EUI transcript levels (Supplemental Figure 10). To compare EUI protein levels between DJ and osdsk2a seedlings under salt stress, we performed immunoblot analysis of the seedlings using a newly generated anti-EUI polyclonal antibody (Supplemental Figure 11). EUI protein levels significantly increased after 2 h of salt-stress treatment in DJ seedlings but after only 1 h of salt stress treatment in osdsk2a seedlings (Figures 6K and 6L), suggesting that salinity stress induces EUI protein accumulation more rapidly in osdsk2a than in DJ. To determine whether the increased salt tolerance of osdsk2a is associated with the increased accumulation of EUI, we examined salt sensitivity in osdsk2a eui plants. The survival frequency was significantly lower in the double mutants than in wild type and osdsk2a plants (Figures 6M and 6N), indicating that the phenotypes of osdsk2a eui plants were similar to those of eui. Accordingly, increases in shoot length were more strongly inhibited under lower levels of salinity stress in osdsk2a eui versus osdsk2a plants (Supplemental Figures 9B and C). Thus, OsDSK2a and EUI protein levels represent a regulatory node in the plant response to salt stress. In addition, the suppression of OsDSK2a protein accumulation in plants under salt stress leads to the salt-induced accumulation of EUI.
The Modulation of GA Metabolism by OsDSK2a Contributes to the Inhibitory Effects of Salt on Seedling Growth
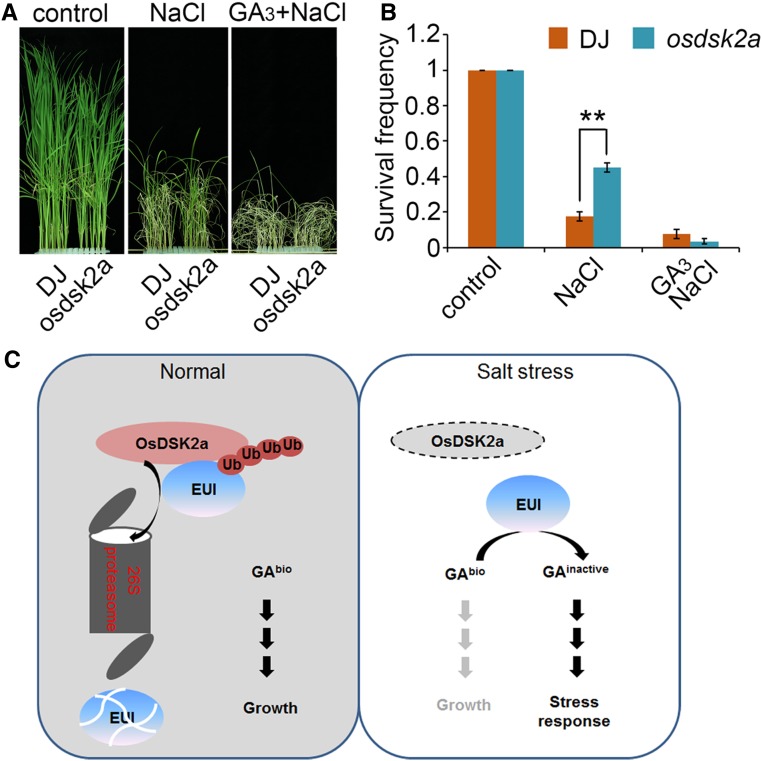
To confirm the effect of GA on the salt tolerance of the osdsk2a mutant, we pretreated 10-d-old seedlings with GA3 for 3 d, followed by treatment with 120 mM NaCl for 2 d. The survival frequencies of DJ and osdsk2a seedlings under salt stress were 20% and 45%, respectively, which decreased to no more than 10% in the seedlings of both lines subjected to GA3 pretreatment (Figures 7A and 7B). These results demonstrate that OsDSK2a responds to salt stress by interfering with GA metabolism.
Figure 7.
GA Contributes to the Salt Tolerance of osdsk2a.
(A) GA3 rescues the responses of osdsk2a to salt stress. Two-week-old DJ and osdsk2a seedlings grown in standard liquid medium were transferred to 120 mM NaCl with or without pretreatment with 100 μM GA3 for 3 d.
(B) Survival frequencies of seedlings after NaCl treatment in (A). The data represent the mean ± sd (n = 3 biological replicates, 40 plants per assay, **P ≤ 0.01, Student’s t test).
(C) Model of the role of OsDSK2a-EUI in mediating of plant growth and salt stress responses. Under normal conditions, OsDSK2a interacts with EUI and promotes its degradation to maintain GA homeostasis, thereby contributing to vegetative growth. Under salt stress, OsDSK2a protein levels decrease, thereby releasing EUI protein and allowing it to decrease bioactive GA levels, leading to inhibited plant growth and increased survival.
Since GA restricts the growth of Arabidopsis plants exposed to salinity stress, a process mediated by DELLA proteins (Achard et al., 2006), we explored whether this mechanism is conserved between monocot and dicot plants. We examined the growth of six rice cultivars, including three japonica and three indica cultivars, in response to various concentrations of NaCl (Supplemental Figure 12A). The lengths and fresh weights of shoots decreased under 50 mM NaCl treatment in all cultivars. When the NaCl concentration increased to 100 mM, the fresh weights of shoots decreased by 50% (Supplemental Figures 12B and C). To confirm the effects of the DELLA protein SLR1 on salt-inhibited growth, we explored whether GA induces the degradation of SLR1 in rice (Supplemental Figures 12D and E). Consistently, SLR1 protein levels strongly increased in plants under salt-stress treatment (Supplemental Figures 12F and G). These results indicate that the accumulation of SLR1 mediates the inhibitory effects of salt stress on seedling growth, which is consistent with previous reports about the regulatory effects of SLR1 on plant growth in rice (Ikeda et al., 2001; Asano et al., 2009). Thus, the DELLA protein SLR1 mediates the salt stress-induced inhibition of seedling growth in rice, suggesting that plant growth under salt stress is tightly regulated to coordinate growth and stress responses. Finally, we demonstrated that the absence of SLR1 accumulation under salt stress in osdsk2a plants confers OsDSK2a-mediated changes in GA metabolism to help the plants respond to salinity stress (Supplemental Figure 13). Overall, our observations indicate that OsDSK2a-mediated EUI degradation helps maintain plant growth under nonstress conditions and restricts plant growth under salt-stress conditions by modulating GA metabolism.
DISCUSSION
A recent study revealed that a UBL-UBA transporter binds to a specific substrate to mediate plant growth and survival in Arabidopsis (Nolan et al., 2017). Therefore, we reasoned that additional specific substrates modulated by UBL-UBA transporters could be identified. In the present study, we demonstrated that OsDSK2a, a UBL-UBA protein in rice, contributes to plant growth by interacting with the GA metabolism factor EUI and promoting its degradation. OsDSK2a interacts with 150 N-terminal amino acids of EUI, which lack amino acids that are potentially modified by ubiquitination. Furthermore, the binding of OsDSK2a with EUI, along with the observed polyubiquitin and ubiquitination modification of EUI, imply that the OsDSK2a-promoted degradation of EUI is ubiquitination dependent. Additionally, OsDSK2a-EUI coordinately respond to salt stress, and the degradation of EUI mediated by OsDSK2a is released under salt stress. These findings are consistent with the observation that salt stress restricts plant growth. Indeed, many components of the ubiquitin-proteasome system, such as ubiquitin ligases (E3; Lyzenga and Stone, 2012), the ubiquitin-specific protease UBP16 (Zhou et al., 2012), and the ubiquitin conjugase UBC32 (Cui et al., 2012), are important regulators of the plant response to salt stress. This study provides strong evidence that OsDSK2a modulates plant growth and salt responses by regulating EUI stability and GA metabolism.
GA is an essential phytohormone that affects seed development and germination, stem and root growth, cell elongation and division, and vesicle trafficking (Schwechheimer, 2012; Davière and Achard, 2013; Kwon and Paek, 2016; Salanenka et al., 2018). Various developmental, hormonal, and environmental signals modulate both the GA biosynthesis and deactivation pathways (Yamaguchi, 2008). Bioactive and inactive forms of GAs have been identified in plants, and their homeostasis was shown to be regulated by a series of deactivation enzymes in rice (Thomas et al., 1999; Zhu et al., 2006). EUI functions as an inhibitor of plant growth whose activity is tightly regulated during the vegetative growth stage. EUI transcript levels are low in plants at the seedling stage (Zhang et al., 2008). The expression of EUI is directly regulated by the AP2 transcription factor OsAP2-39, which mediates the crosstalk between GA and abscisic acid signaling (Yaish et al., 2010). The transcription factor HOX12 also regulates the expression of EUI at the transcriptional level to modulate panicle exertion in rice (Gao et al., 2016). Thus, the transcription of EUI is tightly regulated. However, whether and how EUI is modulated at that posttranscriptional level have been unclear.
Here, we uncovered a mechanism that regulates EUI at the posttranscriptional levels and its role in GA homeostasis, increasing our understanding of the modulation of GA metabolism. We identified OsDSK2a as an interacting protein with EUI that promotes its degradation. Given that EUI is localized to the endoplasmic reticulum in rice (Zhu et al., 2006) and that DSK2 colocalizes with its target proteins in autophagic bodies under stress treatment in Arabidopsis (Nolan et al., 2017), we speculated that the interaction between OsDSK2a and EUI could occur in the cytoplasm, where proteasome and autophagic bodies are also located. However, the subcellular localization of this interaction, which might be associated with plant responses to stress conditions, remains to be determined (Marshall and Vierstra, 2019). Since OsDSK2a is a UBL-UBA protein, which E3 ligase also participates in, the ubiquitination of EUI remains to be determined. Among the proteins that interacted with OsDSK2a in a yeast two-hybrid screening was a zinc finger- and RING domain-containing protein of unknown function, which might have E3 ligase activity. The interaction between OsDSK2a and the potential E3 ligase mediating EUI degradation remains to be examined.
Plants use sophisticated mechanisms to adapt and respond to salt stress. The salt-activated repression of plant growth may represent a positive mechanism to help plants adapt to salinity stress (Achard et al., 2006; Munns and Tester, 2008). Phytohormones play important roles in adjusting plant growth rates to environmental conditions. SUB1A restricts plant growth by augmenting the accumulation of SLR1 in response to submergence (Fukao and Bailey-Serres, 2008; Fukao et al., 2011). The remorin protein OsREM4.1 coordinates the interlinking of abscisic acid and brassinosteroid signaling to balance plant growth and environmental responses in rice (Gui et al., 2016). The phosphorylation of abscisic acid receptors by TOR kinases tightly controls the switch between stress responses and the regrowth of plants under unstressed conditions (Rosenberger and Chen, 2018; Wang et al., 2018). Therefore, in addition to typical stress-responsive hormones, growth-promoting hormones play pivotal roles in plant stress responses (Verma et al., 2016).
In this study, we demonstrated that OsDSK2a helps maintain GA homeostasis and thereby sustains plant growth based on the inhibitory effect of EUI accumulation. This inhibition is released under salt stress due to reduced OsDSK2a abundance, leading to decreased levels of GA and retarded plant growth. EUI protein levels were higher in the loss-of-function osdsk2a mutant than in wild-type seedlings, and salt stress accelerated EUI accumulation in osdsk2a seedlings, giving rise to increased tolerance to high salinity stress and reduced sensitivity to salt-inhibited seedling growth in this mutant. We therefore extended our knowledge of the function of EUI, which not only negatively regulates seedling height, but also positively regulates the salt response, indicating that EUI positively regulates the salt stress response by regulating the levels of bioactive GAs. The accumulation of EUI under salt stress suggests that the stability of EUI modulates the inhibitory effect of salinity stress on plant growth. Therefore, we propose that OsDSK2a-promoted degradation of EUI serves as a control mechanism to preserve plant growth and salt stress responses.
In this study, we revealed that a UBL-UBA protein (a component of the ubiquitin-proteasome system) contributes to plant growth and salt stress responses by modulating GA metabolism. Salt stress activates the growth-repressive effects of DELLA protein in rice, suggesting that plant growth is tightly controlled under salt stress to avoid the excessive use of energy and that plant growth resumes upon the restoration of favorable conditions. Similarly, the controlled growth of osdsk2a plants under salt stress due to decreased bioactive GA levels provides the plants with increased salt tolerance, indicating that actively reduced plant growth under normal conditions confers increased tolerance to salt stress. Although DSK2 mediates the degradation of its substrates through the autophagy pathway in Arabidopsis (Nolan et al., 2017), we determined that OsDSK2a interacts directly with EUI, promoting its degradation through the ubiquitin proteasome pathway (Figure 7C). This finding improves our understanding of the degradation pathways of substrates regulated by DSK2 proteins.
Salt-induced decreases in OsDSK2a protein levels give rise to increased EUI accumulation under salinity stress (Figure 7C); similarly, the DELLA protein SLR1 improves plant survival under salt stress by reducing plant growth (Achard et al., 2006). Moreover, the loss-of-function osdsk2a mutants exhibited retarded growth, whereas plants overexpressing OsDSK2a did not exhibit enhanced growth compared to wild-type plants. Perhaps the excessive abundance of OsDSK2a does not lead to the increased degradation of its substrate, which is also dependent on the presence of an assembled proteasome complex. Taken together, our findings demonstrate that OsDSK2a-EUI coordinates plant growth and salt stress responses, thereby allowing plants to adjust to continuously changing environmental conditions.
METHODS
Plant Materials and Growth Conditions
The T-DNA insertion rice (Oryza sativa) mutant osdsk2a and wild-type rice cv DJ were obtained from RiceGE (Rice Functional Genomic Express Database, http://signal.salk.edu/cgibin/RiceGE). osdsk2a allelic mutants and osdsk2b mutants in the Nip background were generated using CRISPR/Cas9 (Xu et al., 2017), in which the target regions 5′-CTGCACATCCGGTGCACCAA-3′ of OsDSK2a and 5′-CTACAAGGGCCGGATCCTCA-3′ of OsDSK2b were introduced into the pHUN4c12 vector backbone, and the recombinant vector was transformed into Agrobacterium strain EHA105-pSOUP for rice transformation. The osdsk2a/b double mutant was generated by cross-breeding between osdsk2a-c1 and osdsk2b-c, followed by screening selfed progeny by genomic PCR. The eui-1 mutant in the ZS97 background was previously described (Zhu et al., 2006). The site mutant eui-2 was isolated by screening our ethyl methyl sulfone mutation library in the BSM background. To generate the osdsk2a eui double mutant, the target region 5′-GATCGCCGGGCTGTGCATTA-3′ of EUI genomic DNA was edited via the CRISPR/Cas9 method in the osdsk2a background. To generate the overexpression transgenic plants, the coding sequences of OsDSK2a and EUI were amplified by PCR and cloned in-frame with an HA or myc tag into pCAMBIA 1307 under the control of the CaMV 35S promoter (Wang et al., 2013) or a modified pCAMBIA 3301 vector under the control of the Ubiquitin promoter. The resulting plasmids were introduced separately into Nip or osdsk2a via Agrobacterium-mediated transformation. To generate the pOsDSK2a:GUS vector, the promoter region of OsDSK2a (2143 bp upstream of ATG) was amplified by PCR using Nipponbare genomic DNA as the template and inserted into the pCAMBIA1381Z (Cambia) binary vector. The primers used are listed in Supplemental Table 2.
The seeds were submerged in water at 37°C for 48 h. The germinated seeds were sown in a bottomless 96-well plate in a container of Yoshida’s culture solution (Cui et al., 2015) or in growing trays filled with soil. The plants were housed in a growth chamber under a 14-h light (30°C)/10-h dark (25°C) photoperiod, with a light intensity of ∼150 µmol m−2 s−1 (white light) and 60% relative humidity. To compare the growths rates of plants of different genotypes or under different concentrations of NaCl, 3-d-old seedlings grown in liquid culture were treated with the corresponding concentrations of NaCl and the lengths and fresh weights of shoots measured after 6 d. To examine the effects of GA on seedling growth, the germinated seeds were incubated in liquid culture in medium containing 10 µM PAC and different concentrations of GA3 for 10 d. Plants at the tillering and heading stages were sprayed with 100 µM GA3. To investigate salt tolerance, 2-week-old soil-grown seedlings were watered with 120 mM NaCl for 7 d and the survival frequencies after recovery recorded. To test the effects of GA on salt tolerance in seedlings, plants were pretreated in culture solution containing 100 µM GA3 for 3 d, followed by treatment with 120 mM NaCl. Statistical significance in seedling growth or survival frequencies was analyzed by unpaired Student’s t test for two group data. The details of statistical analysis are shown in Supplemental Data Set 1.
Phylogenetic Analysis
UBL-UBA proteins were identified in rice by searching the protein NCBI database (www.ncbi.nlm.nih.gov/protein) using UBA and UBL sequences as queries. Full-length protein sequences were aligned using the ClustalX 2.0.11 multiple sequence alignment mode, followed by phylogenetic tree construction using MEGA4.0.2 (Tamura et al., 2007) with the neighbor-joining method with complete deletion of gaps, Poisson correction, and default assumptions that the patterns among lineages were homogeneous. Bootstrap replication (500 replications) was used for statistical support of the nodes in the phylogenetic tree. Alignments used to generate the phylogeny presented in Supplemental Figure 1A are listed in Supplemental Data Set 2.
Ubiquitin Chain Binding Assays
OsDSK2a-GST was purified using Glutathione Sepharose 4B (GE Healthcare). The beads were washed with buffer A (50 mM Tris- HCl, pH 7.5; 100 mM NaCl; 1 mM Na2EDTA; and 0.1% (v/v) Nonidet P-40), incubated at 4°C for 2 h with 5 mg of purified Lys-48- (K48) or Lys-63- (K63) linked chains (Ub2-7; BIOMOL) in buffer A, and extensively washed with buffer A. Bound proteins were eluted by heating the beads in SDS-PAGE sample buffer and being subjected to SDS-PAGE and immunoblot analysis with anti-Ub antibody (Merck, ST1200, USA, 1:2000 dilution; Farmer et al., 2010).
RT-qPCR Analysis and GUS Staining
Total RNA was extracted from various tissues using an Ultrapure RNA Kit (CWBIO). Total RNA (1 to 2 µg) was used as a template for cDNA synthesis with HiScript II Q RT SuperMix (Vazyme). qPCR amplification was performed with SYBR Green Real-time PCR Master Mix reagent (Toyobo) using a real-time PCR detection system according to the manufacturer’s instructions (Bio-Rad iQ5). Samples for three biological replicates were collected from three independent plants, followed by RNA extraction and independent RT-qPCR. Each qPCR amplification contained three technical replicates. The rice housekeeping gene ACTIN was used as an internal reference. The primers used for PCR are listed in Supplemental Table 2. For GUS staining, different tissues were collected from pOsDSK2a:GUS transgenic plants, followed by incubation in staining solution (50 mM sodium phosphate, pH 7.0; 10 mM EDTA; 0.5 mM K3[Fe (CN)6]; 0.5 mM K4[Fe (CN)6]; 0.1% [v/v] Triton X-100; and 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid) for 12 h at 37°C. After the samples were dehydrated in 75% (v/v) ethanol to remove the chlorophyll, photomicrographs were taken under a stereoscopic microscope.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the Clontech Yeast Two-Hybrid System (pGBKT7 and pGADT7). Full-length and truncated EUI were cloned into pGBKT7, and OsDSK2a was cloned into pGADT7. The experiment was performed according to the manufacturer’s instructions (Clontech). Leaky expression of the reporter gene in yeast Y190 cells was blocked using 10 mM 3-amino-1,2,4-triazole.
In Vivo Coimmunoprecipitation Analysis
For coimmunoprecipitation analysis, OsDSK2a-GFP was transferred into rice callus simultaneously with EUI-HA or VTC1-HA, with a construct harboring the Arabidopsis gene VTC1 fused with HA used as a negative control. Total proteins were extracted in immunoprecipitation (IP) buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 5 mM EDTA, 5% (v/v) glycerol, and 0.1% (v/v) Nonidet P-40 (IP1 buffer) and combined with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× complete protease inhibitor cocktail (Sigma-Aldrich, P9599). After centrifugation at 13,000 g for 15 min at 4°C, the supernatants were filtered through 0.2-mm filters and the protein concentrations determined using Bradford assays. For the pull-down assays, 1 mg of total proteins was incubated with 50 μL of GFP-Trap Magnetic Agarose (ChromoTek) for 3 h at 4°C on a rotary shaker. The beads were washed three times with IP1 buffer plus 1 mM PMSF and 1× complete protease inhibitor cocktail and twice with 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5% (v/v) glycerol. All washing steps were performed at 4°C for 10 min on a rotary shaker. The immunoprecipitated proteins were released by boiling in 2× SDS sample buffer. Protein blot analysis of the immunoprecipitates was performed with using anti-GFP (Abmart, M20004, 1:4000 dilution) or anti-HA (Abmart, M20003, 1:4000 dilution) antibodies.
In Vitro Pull-Down and Ubiquitination Assays
HIS tag-fused EUI was expressed in E. coli BL21. EUI-HIS was purified using a His-Select Nickel Affinity Gel (Sigma-Aldrich). Plant proteins were collected in extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% (v/v) Nonidet P-40, 4 M urea, 1 mM PMSF, and 1× complete protease inhibitor cocktail (Sigma-Aldrich, P9599). Approximately 10 µg of purified EUI-HIS was mixed with 1 mg of total proteins from wild-type DJ seedlings, followed by incubation with a His-Select Nickel Affinity Gel for 2 h at 4°C. The gel was washed three times with protein extraction buffer and the proteins released by boiling in 5× SDS sample buffer. The sample was subjected to SDS-PAGE and immunoblot analysis using anti-HIS (CWBIO, CW0286M, 1:5000 dilution) and anti-Ub (Merck, ST1200, 1:2000 dilution) antibodies separately.
Measurement of Protein Levels
For the in vivo degradation assay of EUI, EUI-HA overexpressing transgenic plants were treated with 75 µM CHX with or without 50 µM MG132 (Sigma-Aldrich, C2211). Total proteins were extracted from the plants, followed by immunoblot analysis using anti-HA antibodies. To detect SLR1 and EUI levels in plants under salt stress and GA treatment, wild-type DJ or EUI-HA-overexpressing transgenic plants were treated with 120 mM NaCl or 100 μM GA3. To assay the in vitro degradation of EUI promoted by OsDSK2a, EUI-myc protein was extracted from EUI-myc overexpression transgenic plants and combined with extracts from DJ or osdsk2a seedlings. The mixtures were incubated at 30°C with or without 50 μM MG132, followed by immunoblot analysis using anti-myc antibody (Abmart, M20002, 1:4000 dilution). To examine the effect of OsDSK2a on the stability of EUI in vivo, total proteins were extracted from osdsk2a, followed by immunoblot analysis using anti-EUI antibody. To compare salt-activated EUI accumulation between DJ and osdsk2a, both seedlings were treated with 120 mM NaCl. Total proteins were extracted from the sheath tissue of 4-week-old seedlings in degradation buffer containing 25 mM Tris-HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 1 mM PMSF, and 5 mM DTT, followed by the addition of 50 μM MG132. Detection of ACTIN levels with anti-ACTIN antibodies (Abmart, M20009, 1:4000 dilution) was used as loading controls. All mock controls consisted of an equal amount of solvent relative to each drug. Protein levels were quantified using Image Gauge V3.12 (Fujifilm). All immunoblot experiments were repeated at least three times, essentially with the same conclusions, and representative results are shown.
Quantification of Endogenous GA Levels
To measure GA levels, seedlings (including leaves and sheaths) were harvested and used to detect GA12, GA15, GA24, GA9, GA4, GA53, GA44, GA19, GA20, and GA1 levels, whereas the culms of plants at the heading stage were harvested and used to detect GA24, GA4, GA53, GA20, and GA1 levels. The detection method was previously described (Jiang et al., 2016). Each series of experiments was performed in biological triplicates.
EUI Antibody Preparation
To prepare the anti-EUI polyclonal antibodies, a peptide comprising residues 149 to 163 of EUI was synthesized in vitro and used to immunize rabbit (prepared by CWBIO of China). The anti-EUI antibodies used in immunoblot analysis were diluted 1:1000 before analysis.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: OsRAD23a, LOC_Os09g24200; OsRAD23b, LOC_Os02g08300; OsRAD23c, LOC_Os08g33340; OsRAD23d, LOC_Os06g15360; OsDSK2a, LOC_Os03g03920; OsDSK2b, LOC_Os10g39620; OsDDI1, LOC_Os04g55150; EUI, LOC_Os05g40384; and SLR1, LOC_03g49990.
Supplemental Data
Supplemental Figure 1. Phylogenetic relationships and domain architectures of UBL-UBA proteins in rice.
Supplemental Figure 2. Identification of the osdsk2a mutant and OsDSK2a overexpression transgenic plants.
Supplemental Figure 3. Identification of osdsk2a alleles generated by CRISPR/Cas9.
Supplemental Figure 4. The osdsk2a mutants show retarded plant growth.
Supplemental Figure 5. OsDSK2a and OsDSK2b play unequal roles in seedling growth.
Supplemental Figure 6. Stability of EUI protein in different tissues and in the osdsk2a mutants.
Supplemental Figure 7. GA restores the reduced growth of osdsk2a plants at the tillering and heading stages.
Supplemental Figure 8. Identification of the osdsk2a eui double mutant generated by CRISPR/Cas9.
Supplemental Figure 9. Salt stress has different effects on the growth of osdsk2a and eui seedlings.
Supplemental Figure 10. Expression of EUI in plants under salt stress.
Supplemental Figure 11. Detection of the specificity of anti-EUI antibody.
Supplemental Figure 12. GA contributes to salt-induced decreases in plant growth.
Supplemental Figure 13. Effects of OsDSK2a on SLR1 accumulation in plants under salt stress.
Supplemental Table 1. OsDSK2a-interacting proteins identified by yeast two-hybrid screening.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. Results of statistical analyses.
Supplemental Data Set 2. Text file of the UBL-UBA protein sequence alignment used in Supplemental Figure 1A.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Zuhua He (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing the eui-1 mutant and Xing Wang Deng (School of Advanced Agricultural Sciences and School of Life Sciences, Peking University) for providing the anti-SLR1 antibodies. We thank Baodong Cai (Greensword Creation Technology Co.) for the GA measurements. This work was supported by the National key research and development program of China (grant 2016YFD0100604), the National Natural Science Foundation of China (grants 31470366 and 31771706), and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
AUTHOR CONTRIBUTIONS
J.W. and R.H. designed the research and wrote the article; J.W., H.Q., S.Z., P.W., and H.Z. performed the research; J.W., H.Q., H.Z., Y.Z., Y.M., and R.H. analyzed and discussed the data.
Footnotes
Articles can be viewed without a subscription.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- Asano K., Hirano K., Ueguchi-Tanaka M., Angeles-Shim R.B., Komura T., Satoh H., Kitano H., Matsuoka M., Ashikari M. (2009). Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genomics 281: 223–231. [DOI] [PubMed] [Google Scholar]
- Colebrook E.H., Thomas S.G., Phillips A.L., Hedden P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217: 67–75. [DOI] [PubMed] [Google Scholar]
- Cui F., Liu L., Zhao Q., Zhang Z., Li Q., Lin B., Wu Y., Tang S., Xie Q. (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L.G., Shan J.X., Shi M., Gao J.P., Lin H.X. (2015). DCA1 Acts as a transcriptional co-activator of dst and contributes to drought and salt tolerance in rice. PLoS Genet. 11: e1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Deinlein U., Stephan A.B., Horie T., Luo W., Xu G., Schroeder J.I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Martínez L.A., Kang Y., Walters K.J., Clarke D.J. (2006). Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 1: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99: 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer L.M., Book A.J., Lee K.H., Lin Y.L., Fu H., Vierstra R.D. (2010). The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Bailey-Serres J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 105: 16814–16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23: 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Fang J., Xu F., Wang W., Chu C. (2016). Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 28: 680–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J., Zheng S., Liu C., Shen J., Li J., Li L. (2016). OsREM4.1 Interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 38: 201–213. [DOI] [PubMed] [Google Scholar]
- Hattori Y., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Hedden P., Sponsel V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34: 740–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Ueguchi-Tanaka M., Sonoda Y., Kitano H., Koshioka M., Futsuhara Y., Matsuoka M., Yamaguchi J. (2001). Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22: 561–570. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45: 123–132. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Xu G., Jing Y., Tang W., Lin R. (2016). Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 7: 12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih A.H., Zhou P., Kumar S., Soccio R.E., Kedersha N.L., Gill G., Howley P.M. (2000). The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6: 409–419. [DOI] [PubMed] [Google Scholar]
- Kuroha T., et al. (2018). Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding. Science 361: 181–186. [DOI] [PubMed] [Google Scholar]
- Kwon C.T., Paek N.C. (2016). Gibberellic acid: A key phytohormone for spikelet fertility in rice grain production. Int. J. Mol. Sci. 17: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Torres-Garcia J., Latrasse D., Benhamed M., Schilderink S., Zhou W., Kulikova O., Hirt H., Bisseling T. (2017). Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number. Plant Cell 29: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47: 181–191. [DOI] [PubMed] [Google Scholar]
- Lyzenga W.J., Stone S.L. (2012). Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63: 599–616. [DOI] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 56: 613–626. [DOI] [PubMed] [Google Scholar]
- Marshall R.S., Vierstra R.D. (2019). Dynamic regulation of the 26S proteasome: From synthesis to degradation. Front. Mol. Biosci. 6: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y., Kirkpatrick D.S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S.P., Reis N., Glickman M.H. (2008). Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell 41: 33–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F., Tsuchiya H., Saeki Y., Tanaka K. (2018). K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 115: E1401–E1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam K., Dikic I. (2016). SnapShot: Expanding the ubiquitin code. Cell 164: 1074–1074.e1. [DOI] [PubMed] [Google Scholar]
- Rizza A., Walia A., Lanquar V., Frommer W.B., Jones A.M. (2017). In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants 3: 803–813. [DOI] [PubMed] [Google Scholar]
- Romero-Barrios N., Vert G. (2018). Proteasome-independent functions of lysine-63 polyubiquitination in plants. New Phytol. 217: 995–1011. [DOI] [PubMed] [Google Scholar]
- Rosenberger C.L., Chen J. (2018). To grow or not to grow: TOR and SnRK2 coordinate growth and stress response in Arabidopsis. Mol. Cell 69: 3–4. [DOI] [PubMed] [Google Scholar]
- Salanenka Y., Verstraeten I., Löfke C., Tabata K., Naramoto S., Glanc M., Friml J. (2018). Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc. Natl. Acad. Sci. USA 115: 3716–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C. (2012). Gibberellin signaling in plants - the extended version. Front. Plant Sci. 2: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C., Mei Z., Duan J., Chen H., Feng H., Cai W. (2014). OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 9: e87110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su V., Lau A.F. (2009). Ubiquitin-like and ubiquitin-associated domain proteins: Significance in proteasomal degradation. Cell. Mol. Life Sci. 66: 2819–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Phillips A.L., Hedden P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96: 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Xie Q. (2013). Non-26S proteasome proteolytic role of ubiquitin in plant endocytosis and endosomal trafficking(F). J. Integr. Plant Biol. 55: 54–63. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H., Ohtake F., Arai N., Kaiho A., Yasuda S., Tanaka K., Saeki Y. (2017). In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome In Mol Cell, Volume 66, p. 488–502.e7. [DOI] [PubMed] [Google Scholar]
- Van De Velde K., Ruelens P., Geuten K., Rohde A., Van Der Straeten D. (2017). Exploiting DELLA signaling in cereals. Trends Plant Sci. 22: 880–893. [DOI] [PubMed] [Google Scholar]
- Verma V., Ravindran P., Kumar P.P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142. [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397. [DOI] [PubMed] [Google Scholar]
- Wang F., Deng X.W. (2011). Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 21: 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yu Y., Zhang Z., Quan R., Zhang H., Ma L., Deng X.W., Huang R. (2013). Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69: 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao J., Lu W., Deng D. (2017). Gibberellin in plant height control: Old player, new story. Plant Cell Rep. 36: 391–398. [DOI] [PubMed] [Google Scholar]
- Xu R., Wei P., Yang J. (2017). Use of CRISPR/Cas genome editing technology for targeted mutagenesis in rice. Methods Mol. Biol. 1498: 33–40. [DOI] [PubMed] [Google Scholar]
- Yaish M.W., El-Kereamy A., Zhu T., Beatty P.H., Good A.G., Bi Y.M., Rothstein S.J. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6: e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217: 523–539. [DOI] [PubMed] [Google Scholar]
- Zhang D., Chen T., Ziv I., Rosenzweig R., Matiuhin Y., Bronner V., Glickman M.H., Fushman D. (2009). Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol. Cell 36: 1018–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu Y., Peng Y., Yan D., Li Q., Wang J., Wang L., He Z. (2008). Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 18: 412–421. [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Yang Y., Chen C., Liu Y., Jin X., Chen L., Li X., Deng X.W., Schumaker K.S., Guo Y. (2012). Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24: 5106–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Cheng S., Liu X., Du H., Dai M., Zhou D.X., Yang W., Zhao Y. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236: 146–156. [DOI] [PubMed] [Google Scholar]
- Zhu Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]