TRAP-seq analysis highlights the importance of translational control and mRNA decay for the successful establishment of the nitrogen-fixing symbiosis in the model legume Medicago truncatula.
Abstract
Translational control is a widespread mechanism that allows the cell to rapidly modulate gene expression in order to provide flexibility and adaptability to eukaryotic organisms. We applied translating ribosome affinity purification combined with RNA sequencing to characterize translational regulation of mRNAs at early stages of the nitrogen-fixing symbiosis established between Medicago truncatula and Sinorhizobium meliloti. Our analysis revealed a poor correlation between transcriptional and translational changes and identified hundreds of regulated protein-coding and long noncoding RNAs (lncRNAs), some of which are regulated in specific cell types. We demonstrated that a short variant of the lncRNA Trans-acting small interference RNA3 (TAS3) increased its association to the translational machinery in response to rhizobia. Functional analysis revealed that this short variant of TAS3 might act as a target mimic that captures microRNA390, contributing to reduce trans acting small interference Auxin Response Factor production and modulating nodule formation and rhizobial infection. The analysis of alternative transcript variants identified a translationally upregulated mRNA encoding subunit 3 of the SUPERKILLER complex (SKI3), which participates in mRNA decay. Knockdown of SKI3 decreased nodule initiation and development, as well as the survival of bacteria within nodules. Our results highlight the importance of translational control and mRNA decay pathways for the successful establishment of the nitrogen-fixing symbiosis.
INTRODUCTION
Translational control is a widespread mechanism that allows the cell to rapidly activate or repress translation of presynthesized mRNAs in response to endogenous or exogenous signals, without the need for de novo transcription. This mechanism thus provides flexibility and adaptability to eukaryotic organisms (Sonenberg and Hinnebusch, 2009; Merchante et al., 2017). The process of translation can be divided into four steps: initiation, elongation, termination, and ribosome recycling. Although there are some examples of regulation at the elongation and termination steps, translational regulation is exerted mainly at the initiation step (Sonenberg and Hinnebusch, 2009; Browning and Bailey-Serres, 2015).
Translational eukaryotic initiation factors recruit the mRNA to the 40S ribosomal subunit to form the initiation complex, which scans the 5′ untranslated region (UTR) until it reaches the initiation codon. At this point, the 60S ribosomal subunit joins to form the 80S ribosome, and elongation begins. Subsequently, loading of additional ribosomes forms polyribosomal complexes (polysomes, i.e., mRNAs with two or more ribosomes). Thus, quantification of the population of mRNAs associated with translating ribosomes, referred to as the translatome, provides a good estimate of the translational status of mRNAs. Translational control can affect most cellular mRNAs or act at the gene-specific level, influencing only a subset of the cellular mRNAs (Sonenberg and Hinnebusch, 2009; Roy and von Arnim, 2013). An example of general control of translation is the global repression of translational initiation observed during nutrient limitation and stress conditions that reduce ATP availability such as oxygen deprivation, heat shock, or drought (Kawaguchi et al., 2004; Branco-Price et al., 2008; Merret et al., 2017). However, specialized mechanisms have evolved that upregulate the translation of specific mRNAs (e.g., mRNAs encoding transcription factors) under these environmental conditions (Kawaguchi et al., 2004; Branco-Price et al., 2008; Mustroph et al., 2009; Juntawong et al., 2014; Bazin et al., 2017; Merret et al., 2017; Zhang et al., 2017).
Mechanisms of selective translation of certain mRNAs might involve specific RNA binding proteins (Juntawong et al., 2013; Sorenson and Bailey-Serres, 2014) and microRNA (miRNA)-containing ribonucleoprotein complexes (Li et al., 2013, 2016) but also long noncoding RNAs (lncRNAs). lncRNAs are >200 nucleotide-length transcripts with limited coding potential that participate in chromatin remodeling, splicing, protein or mRNA decoy, and translation during development, or in response to stimuli (Kung et al., 2013; Ariel et al., 2015). Translational regulation mediated by lncRNAs has been described in mammals for a nuclear-enriched antisense lncRNA (Carrieri et al., 2012) and the long intergenic noncoding RNA (lincRNA)-p21 linked to the suppression of its mRNA targets β-catenin and JunB in human cervical carcinoma cells (Yoon et al., 2012). In plants, lncRNAs have been also found associated with polysomes (Jiao and Meyerowitz, 2010; Juntawong et al., 2014; Bazin et al., 2017). In rice (Oryza sativa), a cis-natural antisense transcript enhances the translation of the cognate sense PHOSPHATE1;2 mRNA, contributing to the maintenance of phosphate homeostasis during phosphate deficiency (Jabnoune et al., 2013). Bazin et al. (2017) described three Arabidopsis (Arabidopsis thaliana) ribosome-associated cis-natural antisense transcripts whose increase in total abundance correlates with enhanced translation of their cognate sense transcripts in roots subjected to phosphate limitation. More recently, Deforges et al. (2019) identified 14 Arabidopsis lncRNAs whose expression correlated either positively or negatively with cognate sense mRNA translation under nutrient deprivation or upon exogenous treatment with different phytohormones. These examples support the notion that lncRNAs might be recruited to polysomes to regulate, either positively or negatively, the translation of their mRNA targets. However, the regulation and mechanisms of action of the majority of polysome-associated lncRNAs have remained largely unexplored.
Translational control is also connected to cytoplasmic mRNA decay (Roy and Jacobson, 2013) as translationally repressed RNAs can undergo deadenylation by different classes of deadenylases and then be degraded by the exosome complex (Chantarachot and Bailey-Serres, 2018). The exosome is a conserved multisubunit protein complex that degrades a variety of RNA substrates in the 3′-to-5′ direction (Zinder and Lima, 2017). Exosome activity requires the auxiliary cytoplasmic SUPERKILLER (SKI) complex, a tetramer composed of the SKI8 dimer, the SKI2 helicase, and the SUPERKILLER3 (SKI3) scaffold protein, which threads mRNAs into the exosome for degradation (Halbach et al., 2013). Several lines of evidence have demonstrated that cytoplasmic mRNA degradation can occur cotranslationally in the 5′-to-3′ direction by the action of the exonuclease XRN4 following removal of the 5′ mRNA m7G cap (Hou et al., 2016; Yu et al., 2016). Thus, diverse mRNA degradation mechanisms act coordinately and/or sequentially on translationally repressed mRNAs, contributing to the general control of gene expression during development, differentiation, or adaptation to changing environmental conditions.
The nitrogen-fixing symbiosis established between legumes and rhizobia is an ecologically and agronomically important interaction that results in the formation of a postembryonic root organ, the nodule, within which rhizobia reduce atmospheric nitrogen to forms that can be assimilated by the plant. Formation of functional nodules requires the activation of two independent and tightly coordinated genetic programs: rhizobial infection in the root epidermis and organogenesis of the nodule, which occurs in the inner cell layers beneath the site of infection (Oldroyd and Downie, 2008). The best-characterized mechanism of rhizobial infection involves the formation of a tubular channel, known as the infection thread, that is initiated in the root hair and ramifies and progresses toward the site where the nodule primordium will be formed. In Medicago truncatula, nodule primordium formation involves the activation of cell division in the inner root cortex and the pericycle, which gives rise to distinct nodule developmental zones: meristematic, infection, and fixation zones (Timmers et al., 1999; Xiao et al., 2014). The symbiotic genetic program requires the hierarchical activation of a set of transcription factors that orchestrate the transcriptional reprogramming in the different cell layers involved in bacterial infection and nodule organogenesis (Oldroyd, 2013). Transcriptional reprogramming has been characterized at different stages of the M. truncatula–Sinorhizobium meliloti symbiosis in whole organs (El Yahyaoui et al., 2004; Lohar et al., 2006; Benedito et al., 2008; Maunoury et al., 2010; Moreau et al., 2011), specific root cell layers (Breakspear et al., 2014; Jardinaud et al., 2016), and nodule-specific zones (Limpens et al., 2013; Roux et al., 2014). However, little is known about how translational control and mRNA stability contribute to the reprogramming of root cells for symbiosis.
In the past years, numerous studies have highlighted the importance of the translational control of gene expression during developmental processes such as seed germination (Basbouss-Serhal et al., 2015), pollen tube growth (Hofmann, 2014), and flowering (Jiao and Meyerowitz, 2010) or the adaptation to environmental challenges such as drought (Kawaguchi et al., 2004), hypoxia (Branco-Price et al., 2008; Mustroph et al., 2009; Juntawong et al., 2014), light–dark transitions (Juntawong and Bailey-Serres, 2012; Missra et al., 2015), heat shock (Merret et al., 2017; Zhang et al., 2017), low temperature (Juntawong et al., 2013), phosphate starvation (Bazin et al., 2017), and pathogens (Meteignier et al., 2017; Xu et al., 2017). In the context of root nodule symbiosis, we have previously shown that although translation is not globally affected upon rhizobial infection, specific mRNAs encoding proteins with functions in the Nod signaling pathway (i.e., Nod factor receptors and transcription factors) are selectively recruited to polysomes (Reynoso et al., 2013). To understand the molecular bases of selective translation and its relative contribution to the reprogramming of root cells for symbiosis, we compared the transcriptome and the translatome of M. truncatula roots at the genome-wide scale upon rhizobial infection using direct RNA sequencing (RNA-seq) and translating ribosome affinity purification (TRAP) combined with RNA-seq (TRAP-seq), respectively (Zanetti et al., 2005; Reynoso et al., 2015). Our analysis identified transcript variants of mRNAs and lncRNAs regulated at the translational level in roots or in root-specific cell types. We identified two alternative variants of the Trans-acting small interference RNA3 (TAS3), TAS3 and ALT TAS3, that are differentially recruited to polysomes in response to rhizobia. Enhanced polysome association of the ALT TAS3 contributes to control nodule formation and rhizobial infection. In addition, we identified two transcript variants translationally upregulated in the epidermis and cortex that encode the SKI3 subunit of the SKI complex involved in 3′-to-5′ mRNA decay. Knockdown of SKI3 revealed its positive role in nodule formation, bacterial survival, and the control of early nodulation genes.
RESULTS
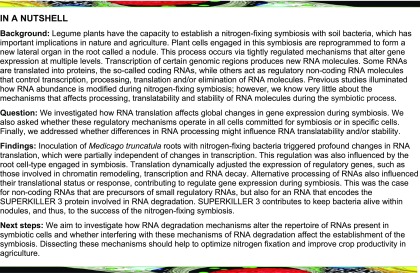
Transcriptional and Translational Changes Triggered by Rhizobia in M. truncatula Roots Are Poorly Correlated
To characterize the genome-wide effect of rhizobial infection on the expression of individual genes at both the transcriptional and translational level, we performed RNA-seq on total and TRAP RNA samples from M. truncatula roots inoculated with water (mock) or with S. meliloti. Root tissue samples from two biological replicates were collected at 48 h postinoculation (hpi). In agreement with previous studies (Lohar et al., 2006; Xiao et al., 2014; Larrainzar et al., 2015), morphological changes in response to rhizobia, such as root hair curling and cortical cell divisions, are already observed at this time point in M. truncatula roots (Figure 1A). Illumina sequencing yielded an average of 61 and 57 million paired-end 100-bp reads per library for total and TRAP samples, respectively, with nearly 90% of the reads aligned to the M. truncatula genome v4 (Table 1). Transcript abundances expressed in fragments per kilobase per million mapped reads (FPKM) were quantified for all genes (sum of all transcript variant abundances; Supplemental Data Set 1) and for all transcript variants in each biological replicate (Supplemental Data Set 2) using Cufflinks (Trapnell et al., 2012). High reproducibility (r > 0.998) was found between biological replicates in both total and TRAP samples (Supplemental Figure 1). FPKM means of both replicates were calculated for all genes (Supplemental Data Set 1); however, only genes with an FPKM mean value ≥0.5 in at least one sample were considered for further analysis. The number of genes expressed in M. truncatula roots under the conditions tested here using this criterion was 23,107 (Supplemental Data Set 1).
Figure 1.
Transcriptional and Translational Responses of Roots to S. meliloti.
(A) Schematic overview of the experimental design. Roots of M. truncatula were inoculated with water (mock) or with S. meliloti (Sm). At 48 hpi, root hair curling (RHC) and cortical cell division (CCD) were observed. Whole root extracts were prepared in a buffer that maintains polysome integrity and subjected to total RNA extraction followed by RNA-seq or polysomal RNA isolation by TRAP, also followed by RNA-seq (TRAP-seq) to characterize changes in the transcriptome or the translatome, respectively. Two completely independent biological replicates (experiments performed on different days) were obtained using root tissue from more than 100 plants per experiment and condition.
(B) Number of genes differentially regulated in Total and TRAP samples (FC ≥ 2 and ≤0.5, P < 0.05 as identified by Cuffdiff).
(C) Venn diagrams illustrating the limited overlap between significant DEGs in Total and TRAP RNA samples.
(D) Scatterplot showing the FC in total and TRAP samples in response to rhizobia. Comparison of total RNA samples from mock (water-inoculated) versus Sm-inoculated roots and of TRAP RNA samples from mock versus Sm-inoculated roots. Each dot represents the log2 of the FC in total and TRAP samples. DEGs are shown as colored dots. Selected genes are indicated: ENOD11 (Medtr3g415670), NF-YA1 (Medtr1g056530), NIN (Medtr5g099060), PUB1 (Medtr5g083030), GRAS (Medtr4g122240), AP2/ERF1 (Medtr4g086190), AP2/ERF2 (Medtr4g086165), jasmonate zim-domain protein1 (JAZ1, Medtr5g013520), JAZ2 (Medtr5g013530), Auxin Binding Protein19 (ABP19, Medtr2g019780), DXR (1-deoxy-d-xylulose 5-phosphate reductoisomerase, Medtr8g012565), Agenet domain protein (AGN, Medtr5g089140), Lipid Transfer Protein (LTP, Medtr7g094650), knotted 1 binding protein (KTN, Medtr4g118050), RmlC-like cupins superfamily protein, (Rmc-like, Medtr2g072560), and SURP and G-patch domain protein (SURP, Medtr8g036255).
(E) FCs in the transcriptome or the translatome of symbiotic marker genes. Values represent the ratio of the FPKM mean values of rhizobium-inoculated samples/FPKM mean values of mock-inoculated samples of two biological replicates. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) according to Cuffdiff parameters. ∞ indicates that FPKM values in mock-inoculated samples were zero or near zero. Full name and references for these genes can be found in Supplemental Data Set 8.
Table 1. Description of the Libraries Subjected to RNA-Seq.
| Library Description | No. of Reads | No. of Mapped Reads (%) | Mean Read Length (bp) |
|---|---|---|---|
| Total mock Rep no. 1 | 85578290 | 80148813 (93.3) | 100 |
| Total mock Rep no. 2 | 46365630 | 37188663 (80.2) | 101 |
| Total Sm Rep no. 1 | 71569736 | 66843904 (93.4) | 100 |
| Total Sm Rep no. 2 | 42000504 | 38988734 (92.8) | 101 |
| TRAP mock Rep no. 1 | 72405126 | 66465623 (91.8) | 100 |
| TRAP mock Rep no. 2 | 42234292 | 38988734 (92.3) | 101 |
| TRAP Sm Rep no. 1 | 77669944 | 71543211 (92.1) | 100 |
| TRAP Sm Rep no. 2 | 38198292 | 32042020 (83.8) | 101 |
Total number of reads obtained in total and TRAP Illumina RNA-seq libraries per independent biological replicate (Rep). Each data set was obtained from paired-end sequencing in HiSeq 2000 and mapped against the M. truncatula genome v4 using TopHat2. Data were retrieved from align summary output files of TopHat2.
To identify genes regulated at transcriptional and translational levels in response to rhizobia, we first performed pairwise comparisons of total RNA samples from mock versus S. meliloti–inoculated roots and of TRAP RNA samples from mock versus inoculated roots, and we selected differentially expressed genes (DEGs) that met the following criteria: a fold change (FC) of at least 2 between samples and P-value ≤ 0.05. In this way, we identified 116 genes and 186 genes up- and downregulated, respectively, at the transcriptional level in response to rhizobia (Figure 1B; Supplemental Data Set 3). The number of DEGs identified here in the transcriptome is higher, but of the same order of magnitude as that previously reported for M. truncatula roots at the same time point after inoculation with S. meliloti using Affymetrix DNA microarrays (Lohar et al., 2006). In the translatome, 130 genes and 87 genes were identified as up- and downregulated, respectively (Figure 1B; Supplemental Data Set 3). The translational status of a set of genes belonging to the Nod signaling pathway correlated well with the data previously generated by RT-qPCR for these specific genes (Reynoso et al., 2013), indicating that TRAP-seq can robustly predict translational changes in response to rhizobia (Supplemental Figure 2).
The comparison of DEGs regulated in the translatome and in the transcriptome identified a set of 41 genes that were homodirectionally upregulated at both the transcriptional and translational levels in response to S. meliloti (Figure 1C). This set includes genes with key functions in rhizobial infection and/or nodule formation, for example, genes encoding the transcription factors Nodule Inception (NIN), Ethylene Response Factor required for Nodulation1 (ERN1), and Nuclear Factor YA1 (NF-YA1); the cell wall remodeling proteins Nodule Pectate Lyase (NPL) and Expansin B1 ligase (ExpB1); and the E3 ubiquitin ligases Plant U-box protein1 (PUB1) and seven in absentia (SINA; Figures 1D and 1E; Supplemental Data Set 4). Remarkably, nearly 65% (75/116) of the DEGs upregulated in the root transcriptome did not show an increase in the translatome, indicating a partial uncoupling between transcriptional and translational responses. Moreover, only 3 of the 186 genes downregulated at the transcriptional level were also significantly downregulated at the translational level (Figure 1C). Furthermore, a poor correlation was found between transcriptional and translational changes in response to S. meliloti (Pearson correlation r = 0.1014, n = 22,171), as illustrated in Figure 1D. These analyses support the notion that selective translation of mRNAs contributes to regulate gene expression at early stages of the symbiotic interaction between legumes and rhizobia.
DEGs in the transcriptome and the translatome were classified into functional categories based on Gene Ontology (GO) followed by manual inspection and classification of individual genes (Supplemental Figure 3A; Supplemental Data Set 3). Besides proteins of unknown function, the most represented categories among genes upregulated at the transcriptional level were redox processes, transport, perception, and signaling, whereas among transcriptionally downregulated genes the most represented categories were metabolism and transcriptional regulation. Genes encoding proteins involved in transcriptional regulation were also highly represented in the list of genes upregulated at the translational level, whereas genes encoding proteins involved in hormone metabolism or biosynthesis, such as five genes encoding Small Auxin Upregulated (SAUR)–like proteins, were overrepresented among translationally downregulated genes (Supplemental Figure 3B).
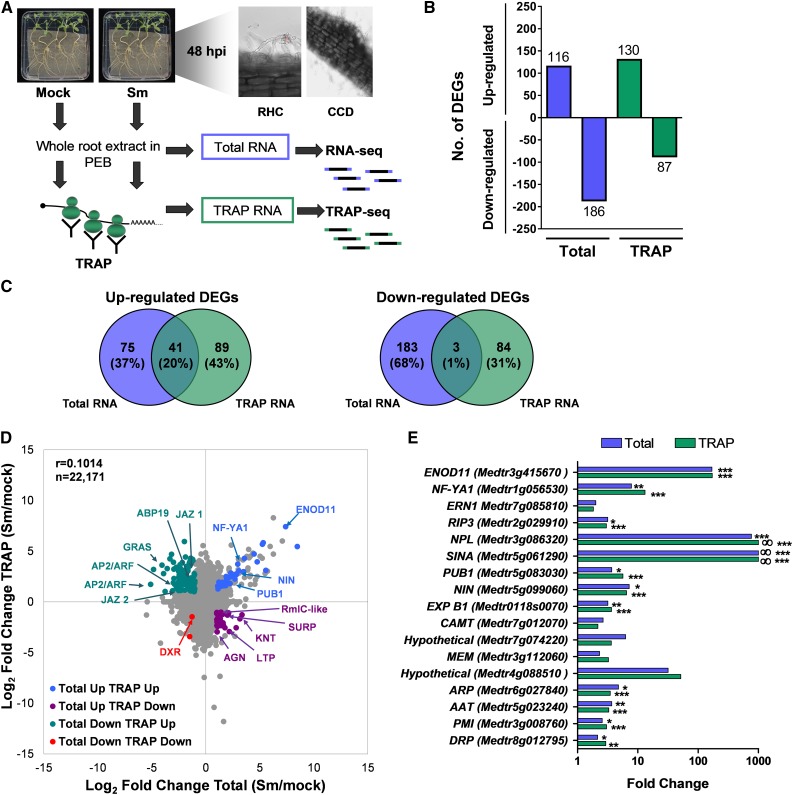
Rhizobial Infection Introduces Significant Changes in the RL of mRNAs with Functions in Transcriptional and Cotranscriptional Processes
Ribosome loading (RL, calculated as the ratio of the FPKM in TRAP RNA samples versus the FPKM in total RNA samples) is considered to be a reliable estimate of the efficiency of mRNA utilization for protein synthesis and a critical parameter to evaluate the contribution of translational regulation level. To gain insight into the dynamics of RL during rhizobial infection, RL was calculated for all expressed genes in mock- and S. meliloti–inoculated samples (Supplemental Data Set 5). RL of individual genes varied more than 250-fold in both mock- and S. meliloti–inoculated roots (Figure 2A; Supplemental Figure 4). Statistical analysis identified 532 genes (2.4% of the total) that increased (z-score > 2.0) and 503 genes (2.3%) that decreased (z-score < −2.0) their RL in response to rhizobia (Figure 2B). An important number of genes with significant changes in RL belonged to the transcriptional regulation category (Figure 2C), with 108 genes encoding transcription factors. Classification of these transcription factors revealed that APETALA2/Ethylene Response Factor (AP2/ERF) and MYB were the most represented families (Supplemental Figure 5; Supplemental Data Set 6). Examples of genes with increased RL within this functional category were JAZ proteins, a B subunit of the NF-Y complex, and MADS-box binding proteins, whereas genes with reduced RL included both general (e.g., TFIID and the cotranscriptional repressor paired amphipathic helix protein Sin3-like3) and gene-specific transcription factors of the KNT, MYB, GRAS, and bHLH families (Figures 2A and 2D), as well as genes with functions in mRNA splicing (e.g., the pre-mRNA–splicing factor ATP-dependent RNA helicases DHX16 and PRP16) or in the progression of the cell cycle (cyclin-like F box protein; Figures 2A and 2D). These results show that gene expression is strongly reprogrammed at the translational level during the progression of the symbiotic interaction.
Figure 2.
RL Changes in Response to Rhizobia.
(A) Scatterplot showing the FC in RL (calculated as FPKM in TRAP RNA samples/FPKM in total RNA) in S. meliloti (Sm)– versus mock-inoculated roots plotted against the FC in total RNA samples in Sm- and mock-inoculated roots. Genes that increase (green) or decrease (blue) in RL in response to rhizobial infection are colored.
(B) Histogram of FC in RL between Sm- and mock-inoculated samples. Genes that increase or decrease in RL, using a z-score >2 or < −2, respectively, as a threshold are indicated.
(C) Functional classification of genes with significant changes in RL, based on GO and redefined by manual inspection of individual genes.
(D) RNA-seq read coverage of selected genes that increase (top) or decrease (bottom) in RL in response to rhizobial infection. Total RNA samples are shown in blue and TRAP RNA samples in green. Numbers on the top left indicate the maximum read value of the scale, which was the same for each gene in all samples.
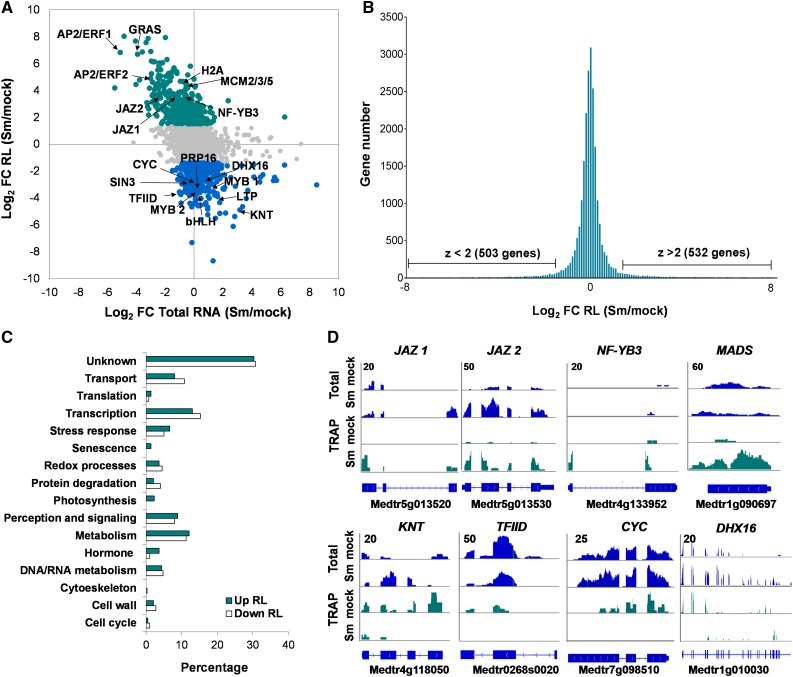
Sequence Features of mRNAs with Altered RL in Response to Rhizobia
mRNA sequence features present in the 5′ and 3′ UTRs, as well as in the coding sequences (CDSs) contribute to the modulation of the association to the translational machinery and thus affect RL of mRNAs (Kawaguchi and Bailey-Serres, 2005; Juntawong and Bailey-Serres, 2012; Juntawong et al., 2014; Lei et al., 2015; Munusamy et al., 2017; Xu et al., 2017). Sequence features of mRNAs with altered RL were compared with those of all the mRNAs expressed in M. truncatula roots (Figure 3). mRNAs that increased or decreased their RL in response to rhizobia exhibited shorter CDSs (857 or 780 nucleotides, on average, respectively) than the average of all mRNAs expressed in roots (1364 nucleotides), but also shorter 5′ UTRs and 3′ UTRs. This observation partially coincides with that reported in Arabidopsis under darkness or hypoxia conditions, where transnationally regulated genes exhibited shorter CDSs and 5′ UTRs than the average of all genes (Juntawong and Bailey-Serres, 2012). The guanine cytosine (GC) content of CDSs and 5′ UTRs from translationally regulated transcripts was lower than the average of all mRNAs, but no differences were found in the 3′ UTRs. Stable secondary structures in the 5′ UTR of cellular mRNAs affect ribosome scanning and are thought to impair mRNA translation. The potential for secondary structures measured as minimal free energy (ΔG°) of 5′ UTRs was significantly higher for those mRNAs whose RL increased or decreased in response rhizobia (−26 or −30 kcal/mol, respectively) than that estimated for all genes (−40 kcal/mol). It is noteworthy that the average ΔG° of the 3′ UTR for all genes (−89 kcal/mol) was much lower than that of the 5′ UTR, revealing that M. truncatula mRNAs tend to be more structured in their 3′ UTR than in their 5′ UTR, a feature previously observed in Arabidopsis (Kawaguchi et al., 2004). Remarkably, the ΔG° of the 3′ UTR of mRNAs that change their RL in response to rhizobia was much higher (−46 kcal/mol) than that of the average of all mRNAs. Altogether, these results indicate that mRNAs with shorter and less structured CDSs, 5′ UTR and 3′ UTR tend to be more susceptible to translational regulation during root nodule symbiosis.
Figure 3.
mRNA Features of Genes That Change in RL in Response to Rhizobia.
Sequence length, GC content, and ΔG° of CDS, 5′ UTR, and 3′ UTR of all mRNAs expressed in M. truncatula roots and of those mRNAs that increased or decreased in RL in response to rhizobia. a, b, and c indicate statistically significant differences in an unpaired two-tailed Student’s t test with P ≤ 0.05. nt, nucleotides.
Regulation of Selected mRNAs in Cell-Specific Translatomes in Response to Rhizobia
The TRAP methodology allows analysis of tissue-specific translatomes. This is achieved by using promoters that drive the expression of the FLAG-RPL18 protein in specific tissue types (Mustroph et al., 2009). The FLAG-RPL18 protein is incorporated into the ribosomes and polysomes of specific tissues where the promoter is active, thus facilitating affinity purification of translating ribosomes and associated mRNAs from these tissues. To characterize translational changes of individual mRNAs in specific cell types, we generated transgenic roots that express the FLAG-RPL18 under the control of the M. truncatula EXPANSIN7 (pEXP7), the Arabidopsis CORTEX SPECIFIC TRANSCRIPT (pCO2), or the SUCROSE TRANSPORT2 (pSUC2) promoters for expression in epidermis, cortex, or phloem companion cells, respectively. Epidermal root hairs are the entry points of rhizobia, whereas activation of cell division in the root inner cortex gives rise to nodule primordia (Timmers et al., 1999; Fournier et al., 2008; Xiao et al., 2014). pEXP7 and pCO2 were previously used to drive gene expression in root hairs and in differentiating cortical cells, respectively, in the context of the root nodule symbiosis (Rival et al., 2012; Vernié et al., 2015). The use of pSUC2 as a phloem-specific promoter is based on a previous report that indicated that nodule organogenesis involves symplastic continuity between the phloem and nodule initials (Complainville et al., 2003). Expression of the FLAG-RPL18 transcript was verified in these roots by RT-PCR (Supplemental Figure 6A). Cell-specific promoter activity and the expression and stability of the FLAG-RPL18 protein were verified by expressing a GFP-tagged version of FLAG-RPL18 (FLAG-GFP-RPL18) under the control of each promoter followed by fluorescence confocal microscopy observation. The FLAG-GFP-RPL18 protein accumulated in the nucleolus and cytosol of the targeted cells in both the meristematic and elongation zones of the root (Supplemental Figure 6B). Plants transformed with the various promoter:FLAG-RPL18 constructs did not show any noticeable phenotype in terms of leaf number, shoot or root length, or nodule number (Supplemental Figure 6C).
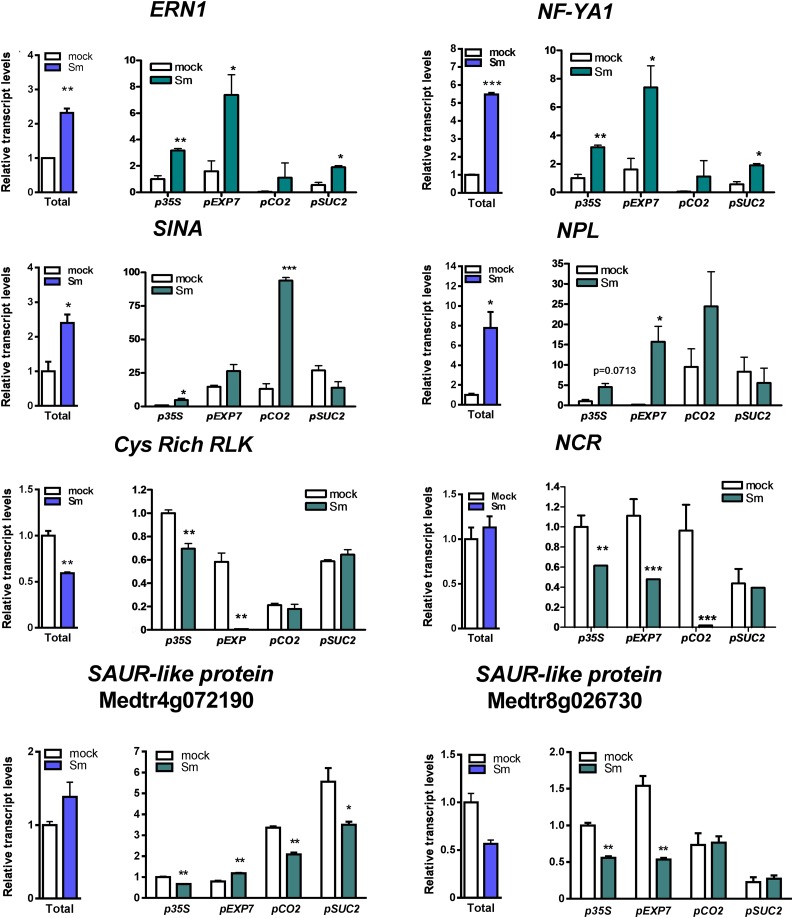
TRAP experiments were performed using transgenic roots that express FLAG-RPL18 under the control of p35S (nearly constitutive) or the three cell-specific promoters. The quality of TRAP RNA samples was evaluated by capillary electrophoresis (Supplemental Figure 7A), and the presence of mRNAs in these samples was verified by RT-PCR reactions that amplify a fragment of the HISTONE-3-LIKE (HIS3L) transcript (Supplemental Figure 7B). Transcript levels of selected DEGs were examined by RT-qPCR in the transcriptome and the translatome of the whole root (p35S), as well as in the translatomes of specific cell types. Selected DEGs included four well-known rhizobium-induced genes, ERN1, NF-YA1, NPL, and SINA, and four genes identified here as translationally downregulated, encoding a cysteine-rich receptor–like kinase (Cys-rich RLK), a Nodule cysteine-rich peptide (NCR122), and two SAUR-like proteins (Figure 4). ERN1 and NF-Y1A were enriched in the translatomes of epidermal and phloem companion cells under the symbiotic condition. NPL showed significant induction in epidermis, whereas SINA mRNAs accumulated to higher levels in the cortical translatome upon inoculation with rhizobia. The Cys-rich RLK and one of the SAUR genes (Medtr8g026730) were downregulated specifically in the epidermal translatome. NCR122 was translationally downregulated in epidermis and cortex, whereas another SAUR gene (Medtr4g072190) was downregulated only in cortex and phloem. These results indicate that TRAP-seq experiments can robustly reveal cell-type-specific gene expression, which could reflect either homodirectional regulation at transcriptional and translational levels or regulation just at the level of translation in the different cell types involved in the root nodule symbiosis.
Figure 4.
RT-qPCR Validation and Tissue-Specific Analysis of Selected Genes.
Abundance of selected gene transcripts in total and TRAP S. meliloti (Sm)– or water (mock)-inoculated samples obtained from roots expressing the FLAG-RPL18 protein in nearly all root (p35S), epidermal (pEXP7), cortical (pCO2), or phloem companion (pSUC2) cells. Total RNA samples were obtained from p35S:FLAG-RPL18 roots. Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the mock total sample or the mock p35S sample. Each bar represents the mean ± se of two biological replicates, with three technical replicates each. Asterisks indicate statistically significant differences (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11).
Changes in the Association with Polysomes of lncRNAs in Response to Rhizobia Are Influenced by the Cell-Type-Specific Context
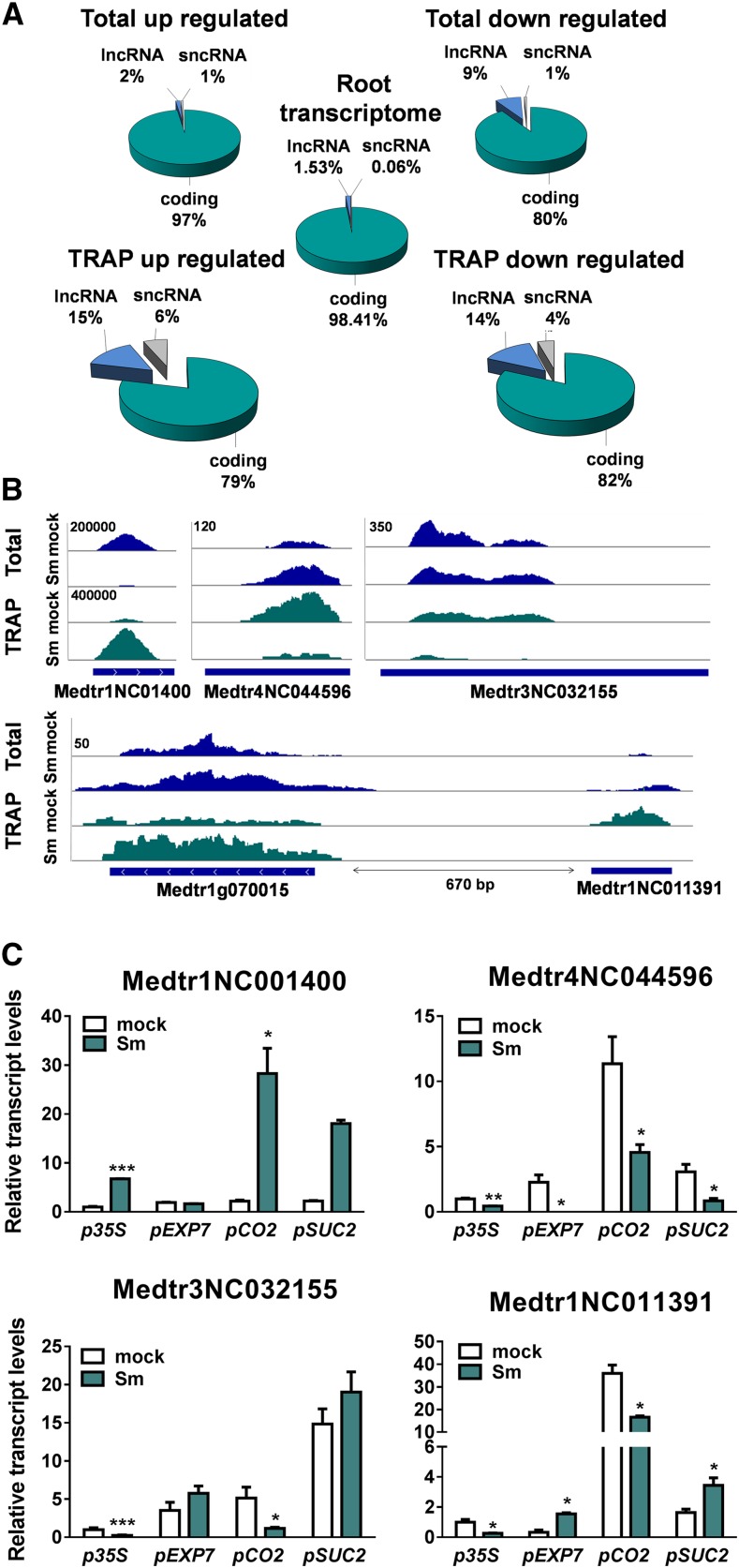
Our analysis also identified transcripts expressed in roots that had not been annotated in the M. truncatula genome or had been annotated as hypothetical proteins, but did not contain open reading frames (ORFs) longer than 100 amino acids, which could represent noncoding RNA (ncRNA) genes. We used the Coding Potential Calculator (Kong et al., 2007) and Coding Potential Assessment Tool (Wang et al., 2013) algorithms to classify nonannotated genes as protein-coding or ncRNAs. Only transcripts encoding ORFs of ≤100 amino acids were considered ncRNAs (Supplemental Data Set 7). Comparison of the differential ncRNAs identified here with the list of predicted secreted peptides described by de Bang et al. (2017) revealed no overlap between the data sets. The ncRNAs were classified as lncRNAs (≥200 nucleotides) or short ncRNAs (<200 nucleotides) based on the criteria applied to Arabidopsis ncRNAs (Liu et al., 2012). We found lncRNAs up- and downregulated in the translatome that were not differentially regulated in the whole root transcriptome (Figure 5A). lncRNAs were further classified according to their genomic location relative to nearby protein-coding genes as long intergenic ncRNAs (lincRNAs) or promoter-associated lncRNAs following the criteria recommended by Ariel et al. (2015; Supplemental Data Set 7). The lincRNAs Medtr3NC032155 and Medtr4NC044596 were downregulated, whereas Medtr1gNC01400 was upregulated in response to S. meliloti (Figures 5B and 5C). lincRNA regulation was also observed in the tissue-specific translatomes. Association of Medtr3NC032155 to the translational machinery decreased specifically in the root cortex, whereas the association of Medtr1gNC01400 was higher in cortical and phloem companion cells, but not in the epidermis, upon inoculation with S. meliloti (Figure 5C). Medtr4NC044596 was enriched in the cortical translatome of mock-inoculated roots, but downregulated in response to rhizobia in all tissues tested (Figure 5C).
Figure 5.
Differential Association of lncRNAs to Polysomes in Response to Rhizobia.
(A) Pie chart showing DEGs from total and TRAP samples and the whole root transcriptome classified as protein-coding, short (sncRNAs, ≤200 nucleotides [nt]), or long (lncRNAs, ≥200 nt) ncRNAs.
(B) RNA-seq read coverage of selected intergenic lncRNAs (top) or a promoter-associated lncRNA (bottom) that change in association to polysomes in response to rhizobial infection. Total RNA samples are shown in blue and TRAP RNA samples in green. Numbers on the top left indicate the maximum read value of the scale, which was the same for all samples.
(C) RT-qPCR validation and tissue-specific analysis of selected lncRNAs in total and TRAP S. meliloti (Sm)– or water (mock)-inoculated samples obtained from roots expressing the FLAG-RPL18 protein in nearly all root (p35S), epidermal (pEXP7), cortical (pCO2), or phloem companion (pSUC2) cells. Total RNA samples were obtained from p35S:FLAG-RPL18 roots. Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the mock total sample or the mock p35S sample. Each bar represents the mean ± se of two biological replicates, with three technical replicates each. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11).
The promoter-associated lncRNA Medtr1NC011391 mapped ∼670 bp upstream of the transcriptional start site (TSS) of Medtr1g070015, which encodes a putative AP2/ARF transcription factor (Figure 5B). Interestingly, the association of Medtr1NC011391 transcripts to the translational machinery decreased, whereas that of Medtr1g070015 mRNAs increased upon rhizobial infection (Figures 5B and 5C). Medtr1NC011391 transcripts were enriched in the cortical translatome of mock-inoculated roots. Upon rhizobium inoculation, this lncRNA was downregulated in translatome of cortex, but upregulated in epidermal and phloem translatomes (Figure 5C). These results support the idea that the expression of lncRNAs and/or its association to the translational machinery is strongly influenced by the cell-specific context.
An Alternative Polyadenylated Short Transcript Variant of the TAS3 Gene Contributes to Modulation of Nodule Formation and Bacterial Infection
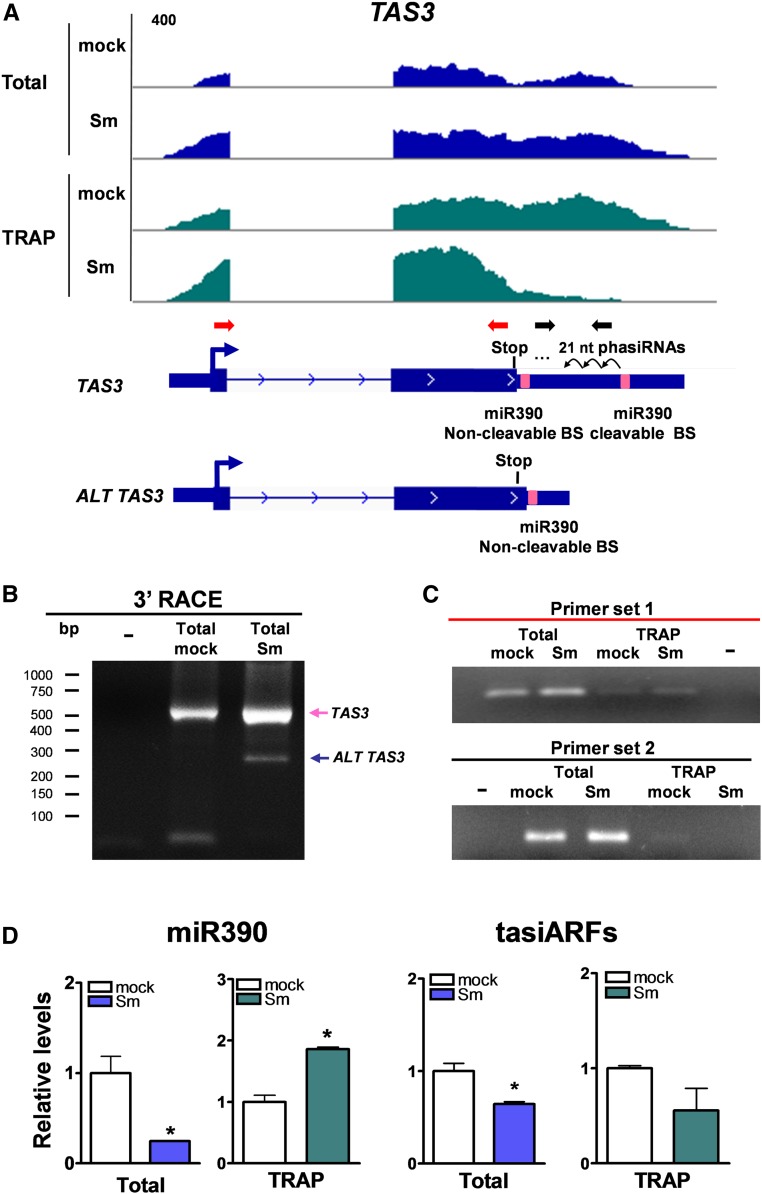
TAS3 is an evolutionarily conserved plant lncRNA that contains two target sites—one cleavable and one noncleavable—for the AGO7/miR390-containing RNA-induced silencing complex (Axtell et al., 2006; Xia et al., 2017). Cleavage of TAS3 at the target site most proximal to the 3′ end leads to the production of trans-acting small interference RNAs/phased small interference RNAs (tasiRNAs/phasiRNAs) known as tasiARFs (Montgomery et al., 2008). The M. truncatula TAS3 gene, Medtr2g033380, contains a small ORF (sORF) encoding a putative peptide of 67 amino acids, with the stop codon located eight nucleotides upstream of the noncleavable target site of miR390, which coincides with that previously reported for the Arabidopsis TAS3a, TAS3b, and TAS3c genes (Hou et al., 2016; Bazin et al., 2017). The length and position, but not the sequence of the putative peptide encoded by this sORF, are conserved among diverse plant species (Bazin et al., 2017). Moreover, it has been demonstrated that this sORF enhances the stability of the Arabidopsis TAS3a transcript (Bazin et al., 2017). Our data revealed that the M. truncatula TAS3 transcript was present in total and TRAP RNA samples (Figure 6A), supporting the notion that tasiRNAs/phasiRNAs might be produced in polysome-bound complexes (Li et al., 2016). Interestingly, RNA-seq reads were distributed along the complete TAS3 transcript; however, distribution of TRAP-seq reads of mock- and S. meliloti–inoculated roots differed, with reads predominantly on the sORF region in the rhizobium-infected roots (Figure 6A).
Figure 6.
Alternative Variants of TAS3 Are Differentially Recruited to Polysomes.
(A) Integrative Genomics Viewer images illustrating read coverage of the TAS3 gene. Schematic representation of the two transcript variants found for the TAS3 gene (Medtr2g033380) by TopHat/Cufflinks analysis is presented below read coverage. The TAS3 transcript variant contains the two miR390 binding sites (pink boxes), which leads to tasiARF production. A shorter alternative transcript variant, ALT TAS3, contains only the noncleavable miR390 binding site (BS). Both transcript variants contain an ORF that ends eight nucleotides 5′ upstream of the noncleavable miR390 binding site. Red and black arrows represent the primer pair 1 and 2, respectively, used for RT-qPCR analysis shown in (C).
(B) Nested PCR of the 3′ RACE reaction of TAS3 transcripts variants in total RNA mock- and S. meliloti–inoculated root samples. Forty cycles of PCR were performed for each PCR reaction.
(C) Expression levels of TAS3 transcripts were determined by RT-PCR using primer set 1 [red arrows in (A)] or primer set 2 [black arrows in (A)] in total and TRAP RNA samples isolated from mock- and S. meliloti (Sm)–inoculated roots. Thirty-five and 40 cycles of PCR were performed for primer set 1 and primer set 2, respectively.
(D) Relative levels of miR390 and tasiARFs in total and TRAP RNA samples isolated from mock- and S. meliloti–inoculated roots were measured by stem-loop RT-qPCR. miR390 and tasiARFs expression levels were normalized by those of U6 RNA. Each bar represents the mean ± se of two biological replicates, with three technical replicates each. Asterisks indicate statistically significant differences (*P < 0.05) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11).
TopHat/Cufflinks analyses (Trapnell et al., 2012, 2013; Kim et al., 2013) predicted two different transcript models for MtTAS3 (Figure 6A), which are consistent with alternative polyadenylation (APA). A long transcript variant containing the two miR390 target sites and a short transcript variant that excludes the cleavable miR390 target site (designated as TAS3 and ALT TAS3, respectively) were confirmed by 3′ rapid amplification of cDNA ends (RACE) and sequencing of the amplification products (Figure 6B). We used two sets of primers (indicated in Figure 6A) to verify these APA variants. Primer set 1 bound to the TAS3 ORF region and amplified a PCR product from both total and TRAP samples (Figure 6C). Primer set 2 bound specifically to the tasiARF-producing regions and amplified a PCR product from both total samples and the TRAP mock sample; however, the amplification product was undetectable in the TRAP sample from rhizobium-inoculated roots (Figure 6C). This indicates that the long TAS3 variant, which leads to the production of tasiARFs, decreased in the translatome in response to rhizobia.
We have previously shown that miR390 and tasiARF levels decrease in total RNA samples upon inoculation with S. meliloti (Reynoso et al., 2013; Hobecker et al., 2017). The ALT TAS3 variant does not lead to the production of tasiARFs but retains the noncleavable miR390 binding site. Thus, it can be speculated that the increase in the association of the ALT TAS3 variant with the translational machinery might serve to reduce tasiARF production upon rhizobial infection by functioning as an endogenous target mimic. This target mimic would associate to polysomes and sequester miR390, preventing its cleavage activity on the long TAS3 variant. Consistent with this idea, although miR390 levels decreased in total RNA samples, they increased more than twofold in TRAP samples upon rhizobial inoculation, concomitant with reduced tasiARF levels in both total and TRAP samples of S. meliloti–inoculated roots (Figure 6D). Altogether, these results suggest that differential association of the TAS3 and ALT TAS3 transcript variants in polysomal complexes may contribute to reduce tasiARFs production and inactivate the miR390/TAS3 pathway in response to rhizobial infection.
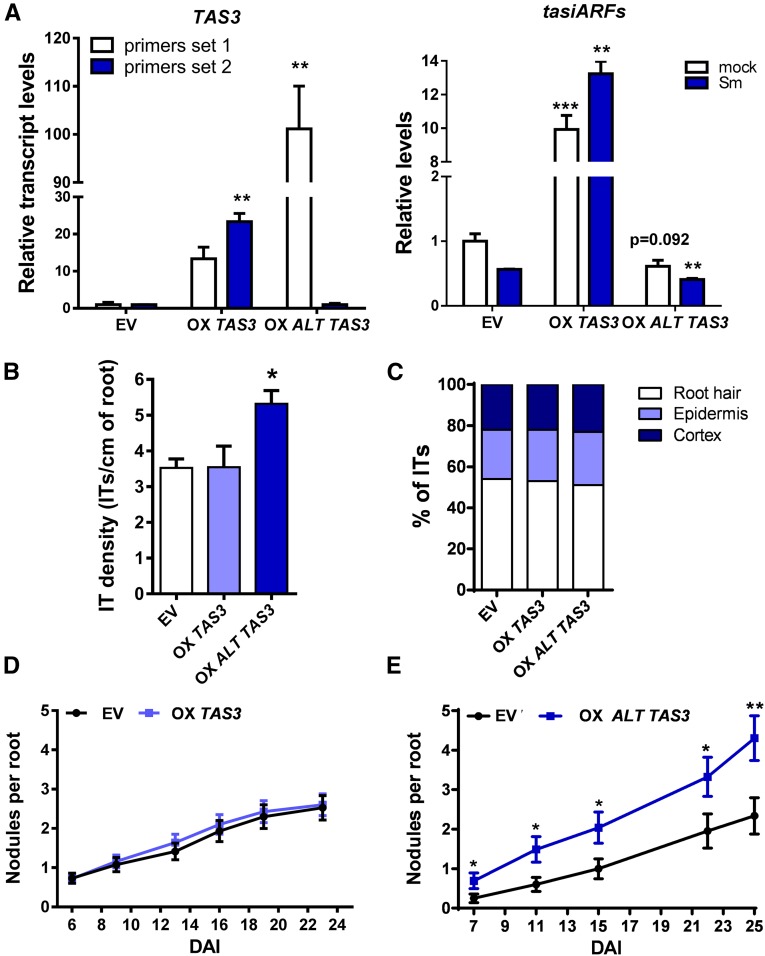
To evaluate whether the ALT TAS3 transcript variant functions as an endogenous target mimic that leads to inactivation of the miR390/TAS3 pathway, we generated transgenic roots that overexpress the ALT TAS3 variant (OX ALT TAS3) and characterized the effect on root development and nodule symbiosis. For comparison, we also overexpressed the long TAS3 variant (OX TAS3). OX TAS3 roots overproduced tasiARFs, whereas OX ALT TAS3 roots exhibited reduced production of tasiARFs compared with empty vector (EV) roots (Figure 7A). Phenotypically, OX TAS3 roots produced longer lateral roots than roots transformed with the EV similar to that previously described in Arabidopsis (Marin et al., 2010) whereas OX ALT TAS3 exhibited longer primary roots than EV roots (Supplemental Figure 8). By contrast, overexpression of ALT TAS3, but not the long TAS3, enhanced the number of nodules formed upon inoculation with S. meliloti, as well as the density of infection threads without affecting their progression (Figures 7B and 7E). The root architecture and symbiotic phenotypes observed in OX ALT TAS3 roots were reminiscent of those observed in roots that express an artificial target mimic of miR390 (MIM390) or in ago7 mutants that fail to produce tasiARFs (Hobecker et al., 2017). This reverse genetic evidence is in agreement with the hypothesis that ALT TAS3 might function as an endogenous target mimic of miR390 that associates with polysomes and sequesters miR390. Both reduction in polysome association of the long TAS3 variant and limited availability of miR390 might contribute to the inactivation of the miR390/TAS3 pathway that occurs at early stages of the symbiotic interaction.
Figure 7.
Overexpression of ALT TAS3 Enhances Nodule Formation.
(A) Relative transcript levels of TAS3 transcripts using the set of primers 1 and 2 (red and black arrows in Figure 6A, respectively) measured in roots transformed with the EV or with constructs to overexpress TAS3 (OX TAS3) or ALT TAS3 (OX ALT TAS3). Expression levels were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the EV sample. Expression levels of tasiARFs in EV, OX TAS3, or OX ALT TAS3 roots were determined by stem-loop RT-qPCR, normalized to U6 RNA, and expressed relative to the EV sample. Each bar represents the mean ± se of three biological replicates (whole root tissue from independent experiments performed in different days), with three technical replicates each. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) in an unpaired two-tailed Student’s t test.
(B) Infection threads (ITs) per centimeter of root developed at 7 DAI in EV, OX TAS3, and OX ALT TAS3 roots. The asterisk indicates statistically significant differences compared with EV with P < 0.05 in an unpaired two-tailed Student’s t test.
(C) Progression of infection events in EV, OX TAS3, and OX ALT TAS3 roots. Infection events were classified as ITs that end in the root hair, in the epidermal cell layer, or reach the cortex at 7 DAI.
(D) Nodules per root formed in EV and OX TAS3 roots at indicated time points after inoculation with S. meliloti.
(E) Nodules per root formed in EV and OX ALT TAS3 roots at time points after inoculation with S. meliloti. Asterisks indicate statistically significant differences as compared with EV (*P < 0.05, **P < 0.01) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11). Error bars in (B) to (E) represent the mean ± se of three biological replicates (independent experiments performed in different days) each with at least 50 roots.
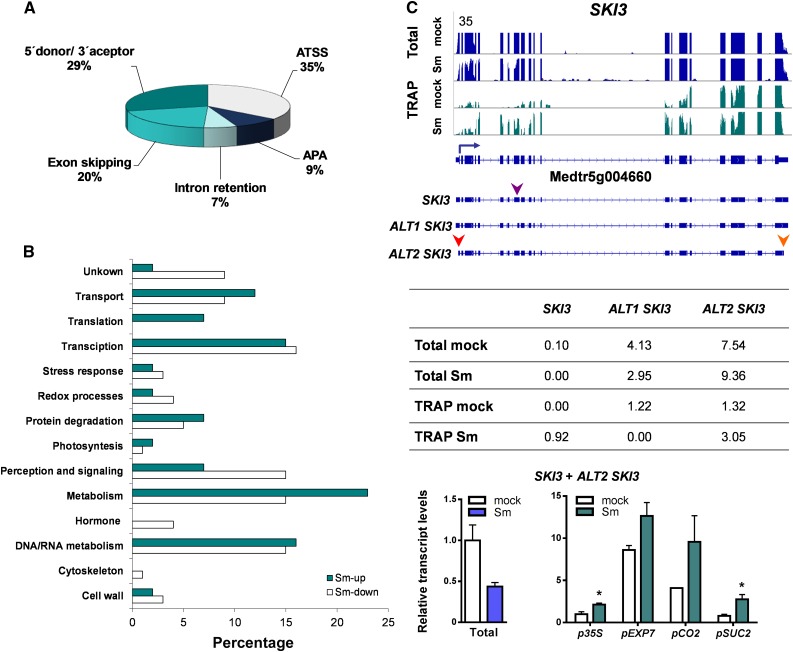
A Transcript Variant of SKI3 Functions in Modulation of mRNA Decay and Is Required for the Survival of Bacteria within Nodules
Alternative transcript variants found in eukaryotic transcriptomes are the result of alternative splicing produced either by intron retention, exon skipping/inclusion or alternative 5′ donor or 3′ acceptor splice sites (Reddy et al., 2013) as well as alternative usage of transcriptional start sites (ATSS) or APA (Reddy et al., 2013; de Lorenzo et al., 2017). Thus, we searched for transcript variants that were differentially expressed in the transcriptome and the translatome upon rhizobial inoculation, regardless of whether they were DEGs. Only transcript models supported by more than 10 reads (i.e., >10 exon junction reads or >10 reads in the alternative transcriptional start site or APA regions) were considered as alternative transcript variants. A total of 328 differentially expressed transcripts (DETs) regulated at the translational level were identified (Supplemental Data Set 8). Comparison of DEGs and DETs in the translatomes revealed that 206 DETs were also DEGs, but identified 122 genes that produced one or more DETs without changes at gene level (sum of all transcript isoforms) in response to rhizobia. Nearly one-third (42 transcripts) of these DETs were upregulated, whereas two-thirds (80 transcripts) were downregulated under symbiotic conditions (Supplemental Data Set 9). Inspection of mRNA processing events revealed that more than half of these transcripts are derived from alternative splicing, 35% display ATSS, and a minor proportion (9%) were due to APA (Figure 8A).
Figure 8.
Differential Translation of Alternative Transcript Variants.
(A) Pie chart classification of translationally regulated transcript variants produced by ATSS, APA, or alternative splicing, including intron retention, exon skipping, and alternative 5′ donor or 3′ acceptor sites.
(B) Functional classification of genes with translationally regulated alternative transcript variants based on GO and manual inspection of individual genes. The percentage of DETs are presented.
(C) Expression of alternative transcript variants of SKI3. (Top) Transcript isoforms of SKI3 and read coverage in total (blue) and TRAP (green) samples of roots inoculated with S. meliloti (Sm) or with water (mock). SKI3 gene model and its transcript variants are illustrated along the x axis. The blue arrow indicates the canonical translation initiation codon and the arrowheads indicate an alternative 3′ acceptor site (purple), an ATSS (red), and an APA (orange). (Middle) Reads corresponding to the different transcript variants of SKI3 in total and TRAP samples expressed as FPKM. (Bottom) RT-qPCR validation and tissue-specific analysis of selected transcript variants. Abundance of specific transcript variants of SKI3 in total RNA samples or in TRAP RNA samples of nearly all cells (p35S), epidermal (pEXP7), cortical (pCO2), or phloem companion (pSUC2) cells from roots inoculated with S. meliloti (Sm) or with water (mock). Total RNA samples were obtained from p35S:FLAG-RPL18 roots. Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the mock total sample or the mock p35S sample. Each bar represents the mean ± se of two independent biological replicates, with three technical replicates each. An asterisk indicates statistically significant differences in Sm compared with mock samples (*P ≤ 0.05) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11).
The analysis of GO functional categories of DETs indicated that the DNA/RNA metabolism category was overrepresented among translationally regulated DETs (Figure 8B; Supplemental Data Set 9). One of these genes (Medtr5g004660) encodes a protein highly similar to the yeast (Saccharomyces cerevisiae) SKI3 subunit, a member of SKI complex that guides RNAs to the 3′-to-5′ exoribonuclease of the exosome complex (Halbach et al., 2013; Chantarachot and Bailey-Serres, 2018). Under our experimental conditions, Medtr5g004660 produced three transcripts: SKI3 was the longest, ALT1 SKI3 was 165 nucleotides shorter (55 amino acids) due to an alternative 3′ acceptor site in exon 8, and ALT2 SKI3 was produced by ATSS and APA without affecting the coding region. Both SKI3 and ALT2 SKI3 encoded the full-length SKI3 protein, which contains multiple tetratricopeptide repeat motifs, whereas ALT1 SKI3 encoded a protein that lacks one tetratricopeptide repeat motif. The abundance of the transcript variants encoding the full-length SKI3 protein, SKI3 and ALT2 SKI3, was either reduced or not significantly affected at the steady-state mRNA level, respectively; however, both transcript variants significantly increased their association with the translational machinery upon rhizobial infection (Figure 8C). RT-qPCR analysis using primers that detected only the SKI3 and ALT2 SKI3 variants confirmed this translational upregulation in the whole root, as well as in the cell-type-specific translatomes of phloem and cortex (Figure 8C).
Inspection of previous microarray and RNA-seq data (Breakspear et al., 2014; Jardinaud et al., 2016) verified that the global abundance of SKI3 transcripts was not affected in root hairs at early time points after inoculation with S. meliloti or Nod Factor application (Supplemental Figures 9A and 9B), suggesting that SKI3 regulation is controlled at the translational level at the initial steps of the root nodule symbiosis. At later stages, that is, at 10 or 14 to 15 days after inoculation (DAI), SKI3 mRNAs accumulated to higher levels in nodules than in noninoculated roots, mainly in the fixation zone (Supplemental Figures 9C to 9E), as revealed by analysis of publicly available expression data (Benedito et al., 2008; Roux et al., 2014). This analysis indicates that translational regulation of SKI3 operates at early stages of the symbiosis and precedes changes in steady-state transcript levels.
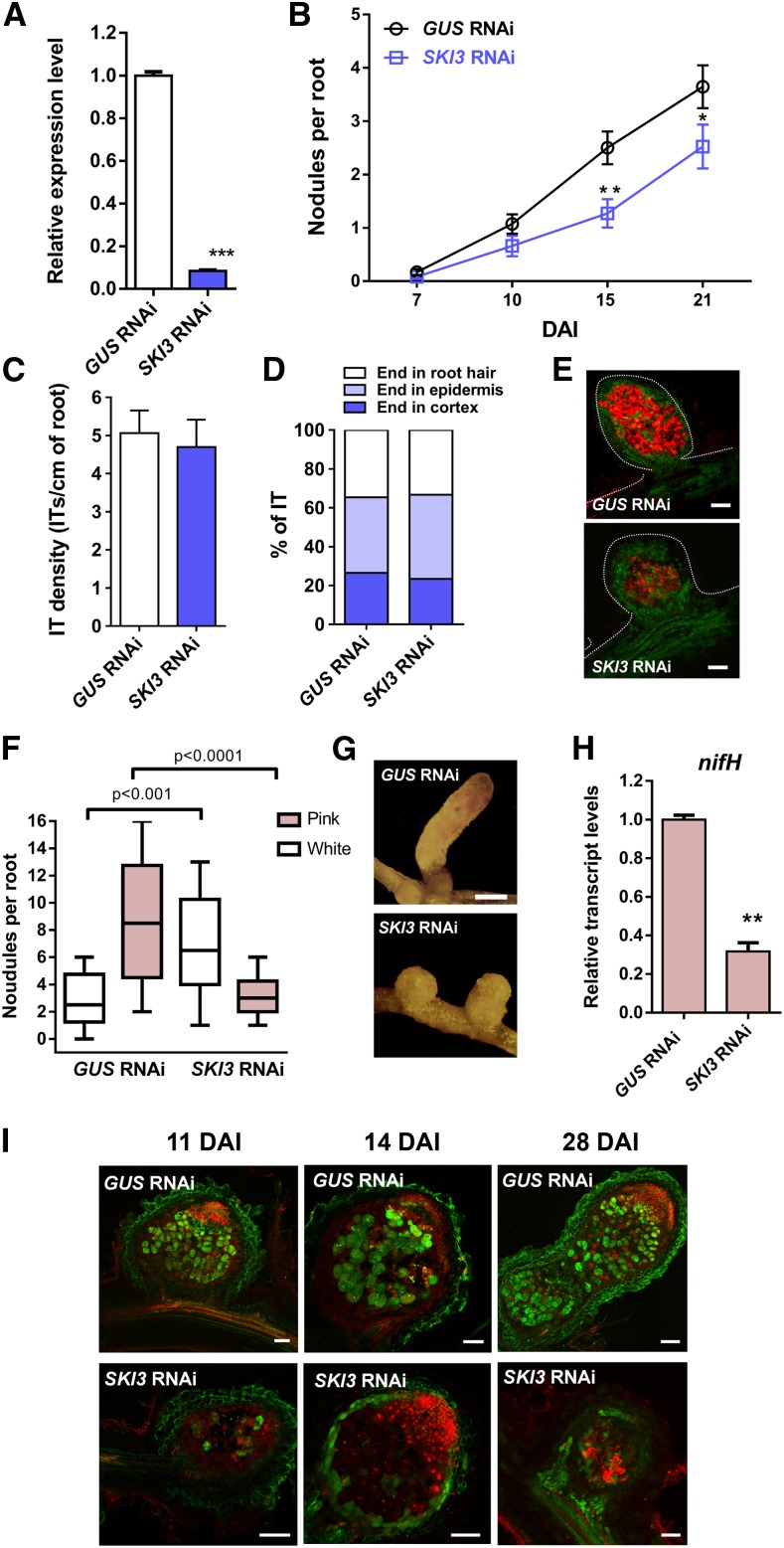
To gain insight into the function of SKI3 during root nodule symbiosis, we used an RNA interference (RNAi) approach to knock down the three SKI3 transcript variants in M. truncatula hairy roots. This strategy decreased levels of all SKI3 variants by more than 85% (Figure 9A). Knockdown of SKI3 did not affect root growth and development (Supplemental Figure 10) but caused a significant reduction in the number of nodules formed by S. meliloti at 15 and 21 DAI (50 and 40% reduction, respectively) compared with control roots transformed with a β-glucuronidase (GUS) RNAi construct (Figure 9B). The frequency of infection events or their progression to the cortical cells determined at 7 DAI was not affected by silencing of SKI3 (Figures 9C and 9D); however, nodules formed in SKI3 RNAi roots were smaller and exhibited a lower content of red fluorescent protein (RFP)–expressing bacteria than those developed in GUS RNAi (Figure 9E). An independent RNAi construct was designed to specifically knockdown the transcript variants encoding the full-length SKI3 protein (SKI3 and ALT2 SKI3), designated as SKI3 RNAi2. This construct produced a 90% reduction in transcript levels of SKI3 and ALT2 SKI3 isoforms as measured with the primer set 1, which bound specifically to these transcript isoforms; however, the primer set 2, which bound to all SKI3 isoforms, revealed only a 60% reduction, suggesting that SKI3 RNAi2 mainly affected mRNA levels of the SKI3 and ALT2 SKI3 isoforms (Supplemental Figure 11A). SKI3 RNAi2 expression produced the same phenotype as SKI3 RNAi, that is, a reduction in the number of nodules, but no effect on the number or progression of infection events (Supplemental Figure 11). Thus, we concluded that this phenotype is most likely caused by the specific silencing of SKI3 and ALT2 SKI3 isoforms.
Figure 9.
SKI3 Is Required for Nodule Formation, but Not Rhizobial Infection.
(A) SKI3 expression level in GUS and SKI3 RNAi roots. Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the GUS RNAi sample. Each bar represents the mean ± se of three biological replicates (whole root tissue from independent experiments performed in different days), with three technical replicates each. Three asterisks indicate statistically significant differences with P < 0.001 in an unpaired two-tailed Student’s t test between GUS and SKI3 RNAi roots.
(B) Nodules per root formed in GUS and SKI3 RNAi roots at 7, 10, 15, and 21 DAI with S. meliloti. Data are representative of three independent biological replicates, each with at least 50 roots. Error bars represent se of three independent biological replicates. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01) in an unpaired two-tailed Student’s t test between GUS and SKI3 RNAi roots.
(C) Infection threads (ITs) per centimeter of root developed at 7 DAI in GUS and SKI3 RNAi roots.
(D) Progression of infection events in GUS RNAi and SKI3 RNAi roots. Infection events were classified as infection threads (ITs) that end in the root hair, in the epidermal cell layer, or reach the cortex at 7 DAI. Error bars represent the mean ± se of three biological replicates (independent experiments performed in different days) each with at least 50 roots.
(E) Images of nodules developed in GUS (top) and SKI3 RNAi (bottom) roots at 28 DAI. Green and red fluorescence correspond to expression of the GFP reporter in roots and nodules and the S. meliloti expressing RFP, respectively. Bar = 0.2 mm.
(F) Number of pink and white nodules developed in GUS and SKI3 RNAi roots at 28 DAI. More than 65 nodules per construct were quantified. Error bars represent the mean ± se of three biological replicates (independent experiments performed in different days).
(G) Images illustrating fully elongated nodules developed in GUS RNAi roots compared with round and small nodules observed in SKI3 RNAi roots. Bar = 0.5 mm.
(H) Relative expression levels of S. meliloti nifH mRNA in GUS and SKI3 RNAi nodules as determined by RT-qPCR. Expression values were normalized by the sensory histidine kinase/phosphatase ntrB mRNA of S. meliloti. Each bar represents the mean ± se of three technical replicates. Two asterisks indicate statistically significant differences with P < 0.01 in an unpaired two-tailed Student’s t test between GUS and SKI3 RNAi roots (Supplemental Data Set 11).
(I) Live/dead staining of nodules developed in GUS and SKI3 RNAi roots at 11, 14, and 28 DAI with S. meliloti. Nodules were stained with SYTO9 (green) that identifies living bacteria and with propidium iodide (red), which stains nuclei of intact cells and cells with membrane damage. Bar = 50 µm.
Observation of nodules at later time points revealed that the majority of the nodules formed in SKI3 RNAi roots were white and round, in contrast to the pink and cylindrical nodules formed in GUS RNAi roots (Figures 9F and 9G), suggesting that nitrogen fixation was compromised by silencing of SKI3. This notion was further supported by the reduced expression of the S. meliloti nifH gene, which encodes a subunit of the nitrogenase complex, in SKI3 RNAi nodules (Figure 9H). The viability of S. meliloti was investigated by staining of nodule sections using the fluorescent dye SYTO9 (green), which stains living bacteria, and counterstaining with propidium iodide (red), which labels cells containing dead bacteria as well as nuclei of intact meristematic cells. In SKI3 RNAi nodules, few cells contained live bacteria and instead, many cells of the central zone were stained with propidium iodide, suggesting that silencing of SKI3 alters the viability of the bacteroids. This contrasts with that observed in control GUS RNAi nodules, where most cells of the nitrogen fixation zone were stained green by SYTO9 (Figure 9I). Transmission electron microscopy of nodules revealed that cells of the transition zone were filled with differentiated bacteroids in both GUS RNAi and SKI3 RNAi nodules; however, the symbiosomes of SKI3 RNAi nodules exhibited reduced peribacteroid space compared with those of GUS RNAi nodules. In addition, SKI3 RNAi symbiotic cells contained membrane-surrounded structures that resembled vacuoles, which were absent from GUS RNAi symbiotic cells (Supplemental Figure 12). Thus, SKI3 seems to be required for formation of functional symbiosomes and the survival of bacteria within nodules, but not for bacteroid differentiation.
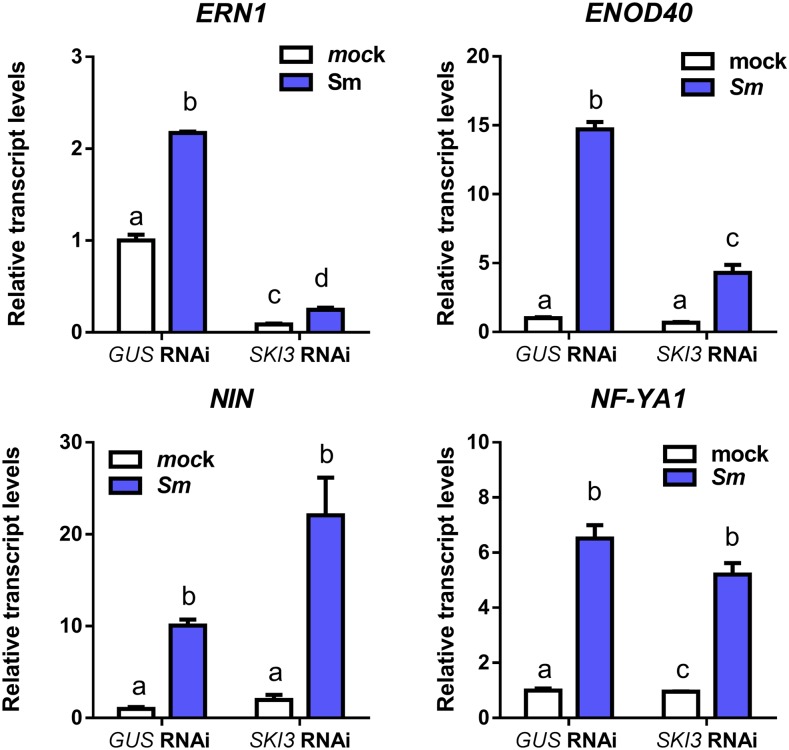
To better understand the molecular events affected by knockdown of SKI3, we tested whether the expression of different early nodulation markers was altered by silencing of SKI3. RT-qPCR experiments performed at 48 hpi with S. meliloti revealed that induction of Early Nodulin40 (ENOD40) and ERN1 in response to rhizobia was greatly reduced in SKI3 RNAi roots compared with GUS RNAi roots, whereas induction of NIN and NF-YA1 was unaffected (Figure 10). Thus, steady-state levels of two early nodulation markers related with the activation of cortical cell divisions required for nodule initiation (i.e., ENOD40 and ERN1) are significantly altered by silencing of a component of the SKI complex.
Figure 10.
Expression of Early Nodulation Markers in GUS and SKI3 RNAi Roots.
Transcripts levels of early nodulation transcripts NIN, NF-YA1, ERN1, and ENOD40 in GUS and SKI3 RNAi roots at 48 hpi with S. meliloti (Sm) or water (mock). Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the GUS RNAi mock sample. Each bar represents the mean ± se of three biological replicates with at least three technical replicates. Different letters indicate statistically significant differences in an unpaired two-tailed Student’s t test with P ≤ 0.05.
DISCUSSION
The results presented here highlight the importance of the regulation of mRNA translatability and stability for the nitrogen-fixing symbiosis between legumes and rhizobia, providing insights into the molecular mechanisms associated with rhizobial infection and nodule organogenesis. A remarkable aspect of our results was the limited correlation between transcriptome and translatome regulation observed in response to rhizobium inoculation, indicating that translational control significantly contributes to the reprogramming of gene expression in those root cells committed for symbiosis. A poor correlation between transcription and translation was also described in the immune response of Arabidopsis plants activated by either pathogen-associated molecular patterns or effectors proteins delivered by pathogenic bacteria (Meteignier et al., 2017; Xu et al., 2017), indicating that lack of correspondence between these two regulatory levels is found in different plant–microbe interactions. Meteignier et al. (2017) showed that upon activation of effector-triggered immunity, the Arabidopsis translatome exhibited a general switch from growth-related activities to defense responses. Genes upregulated at the translational level during effector-triggered immunity included those encoding proteins with functions in cell wall remodeling, production of secondary metabolites, and the hypersensitive response, whereas translationally repressed genes were those involved in primary metabolism and development. Xu et al. (2017) found that genes that change their translational status during PTI included key components of PTI signaling pathway. Our analysis of the M. truncatula root translatome at 48 hpi with S. meliloti identified an important number of upregulated symbiotic genes such as ENOD11, ERN1, NIN, and NF-YA1 among others. In addition, our analysis identified several defense-related genes downregulated upon rhizobial infection, supporting the notion that translational regulation might contribute to the suppression of defense responses at this stage of the interaction.
In addition to protein-coding genes, we identified a number of lncRNAs that changed their association with polysomes in response to rhizobia and displayed cell-specific regulation. Pervasive translation of lncRNAs has been observed in yeast (Ingolia et al., 2014), zebrafish (Danio rerio; Chew et al., 2013; Bazzini et al., 2014), mammals (Guttman et al., 2013; Bazzini et al., 2014; Wang et al., 2017), and plants (Jiao and Meyerowitz, 2010; Juntawong et al., 2014; Bazin et al., 2017), raising the question of whether widespread translation of lncRNAs is likely to be functional, producing polypeptides of biological relevance. The finding that many vertebrate and plant lncRNAs encode conserved peptides suggests that translation of certain lncRNAs might have clear evolutionary and functional implications. However, our BLAST analysis of sORFs indicated that the vast majority of the lncRNAs differentially regulated in the TRAP samples in response to rhizobia do not encode conserved peptides, suggesting that they might either encode species-specific peptides or act as regulatory RNAs during the symbiotic interaction established between legumes and rhizobia. Association with polysomes might stabilize these lncRNAs by preventing RNA decay through cellular mechanisms such as nonsense-mediated decay, decapping, and/or deadenylation.
APA of Arabidopsis TAS3 transcript has been previously suggested by Hunt (2012). Here, two TAS3 alternative transcript variants produced by APA were identified in M. truncatula under symbiotic conditions: a long TAS3 variant that results in tasiARF biogenesis and a short ALT TAS3 variant that lacks the tasiARF-producing region. Previously, we have described that levels of miR390, as well as the production of tasiARFs, decreased in roots at 48 hpi with S. meliloti (Hobecker et al., 2017). Thus, APA of TAS3 and its differential association to polysomes could function as a mechanism that contributes to reduce tasiARF production in response to S. meliloti. The results presented here indicate that the short ALT TAS3 variant might displace the long TAS3 variant from polysomes under symbiotic conditions. This short TAS3 variant contains only the noncleavable miR390 binding site; thus, it sequesters miR390 and blocks its action on the cleavage of the full TAS3 variant, reducing tasiARF production. As reduction in the production of tasiARFs is crucial for rhizobial infection and nodule development in M. truncatula (Hobecker et al., 2017), the polysome association of the ALT TAS3 transcript represents a new mechanism to regulate tasiARF production during symbiosis. Another possibility could be that the ALT TAS3 variant found in translatomes of rhizobium-inoculated roots might be the result of Dicer-like4 endonucleolytic cleavage of the double-stranded TAS3 RNA (Xie et al., 2005), which most likely occurs in the endoplasmic reticulum–bound polysomal fraction (Li et al., 2016). This seems to be unlikely since the TRAP-seq was performed on polyadenylated RNAs and sequencing of 3′ RACE products confirmed that ALT TAS3 is polyadenylated. Moreover, overexpression of the short ALT TAS3 variant, but not the long TAS3 variant, caused a phenotype similar to that observed in roots expressing an artificial target mimic of miR390 (Hobecker et al., 2017). Based on these data, we propose a model in which ALT TAS3 could act as an endogenous target mimic recruited to polysomes upon symbiotic conditions, to sequester miR390 and prevent its action on the long TAS3 variant (Supplemental Figure 13). The increase of miR390 association with polysomes under symbiotic conditions supports this hypothesis. As polysomes seem to be the site for the production of tasiRNAs (Li et al., 2016), both displacement of the long variant TAS3 from polysomes and the sequestration of miR390 by ALT TAS3 could limit the production of tasiARFs at early stages of the symbiotic interaction, releasing the repression of their targets, the auxin response factors ARF2, ARF3, and ARF4.
In addition, our analysis of differentially translated alternative transcript variants identified SKI3, a protein component of the 3′-to-5′ mRNA decay pathway. This surveillance mechanism acts in normal turnover of cellular mRNA, but also in the RNA quality control that selectively degrades aberrant RNAs to prevent the translation of nonfunctional proteins (Schmid and Jensen, 2008). The plant SKI complex functions as a suppressor of sense posttranscriptional gene silencing by blocking the production of secondary small interference RNA from transgenes and endogenous miRNA targets, a critical mechanism that controls developmental processes in the aerial part of Arabidopsis plants (Branscheid et al., 2015; Yu et al., 2015; Zhang, 2015; Zhao and Kunst, 2016). Here, we showed that silencing of SKI3 alters nodule formation and the survival of the bacteria within the nodule cells, but not the infection process. Thus, we propose that SKI3 plays a role mainly in nodule organogenesis. Notably, translational upregulation of SKI3 was observed at 48 hpi with S. meliloti, much earlier that the nodule phenotype is revealed. However, cortical and pericycle cell divisions are already active at 48 hpi in M. truncatula roots (Xiao et al., 2014). Moreover, we found that rhizobial induction of both ENOD40 and ERN1, two genes associated to cell divisions (Charon et al., 1997; Middleton et al., 2007), is impaired at 48 hpi in SKI3 RNAi roots. The lack of induction of these two early symbiotic genes might contribute to explain the reduced number of nodules formed in SKI3 RNAi roots. Compromising SKI complex activity in plants leads to extensive silencing through siRNA amplification and/or miRNA transitivity (Martínez de Alba et al., 2011; Zhang et al., 2015). In addition, a recent study found that the yeast SKI complex might act on polysome-bound mRNAs by physically interacting with 80S ribosomes and polysomes in an RNA-dependent manner (Schmid and Jensen, 2008). Thus, we hypothesize that SKI3 might play a role in the discrimination of mRNAs that will be translated on polysomes or subjected to mRNA decay, but also to control posttranscriptional gene silencing of specific genes at early stages of nodule development, which is later reflected in the number and occupancy of the nodules once they are visible. Further identification of transcript species subjected to RNA decay via SKI/exosome complexes will help test this hypothesis.
In conclusion, our comparative analysis revealed that both protein-coding RNAs and lncRNAs are subjected to regulation at the transcriptional, transcript processing, and translational levels at early stages of the nitrogen-fixing symbiosis. Moreover, cell-type-specific translational regulation of these RNAs might influence the developmental fate of cells that are committed for symbiosis. Two key regulatory events, the alternative processing of a TAS3 transcript to produce an endogenous target mimic of miR390 and the translational regulation of a component of the SKI/exosome mRNA decay pathway, were identified to control specific steps of the root nodule symbiosis.
METHODS
Biological Material and Vectors
Medicago truncatula Jemalong A17 seeds were obtained from Institut National de la Recherche Agronomique, Montpellier, France (http://www.montpellier.inra.fr). The p35S:FLAG-MtRPL18 construct was previously generated by Reynoso et al. (2013). The pEXP7:FLAG-RPL18 construct was generated by PCR amplification of the EXPANSIN7 (pEXP7) promoter of M. truncatula using the MtEXP EcoRI F and MtEXP SacI R primers listed in Supplemental Data Set 10 and genomic DNA as template. The PCR product was introduced into p35S:FLAG-RPL18 by replacement of the p35S promoter using EcoRI and SacI digestion. The pCO2:FLAG-RPL18 and pSUC2:FLAG-RPL18 constructs were generated by replacing the ORF of Arabidopsis thaliana (Arabidopsis) RPL18 in the pCO2:FLAG-AtRPL18B and pSUC2:FLAG-AtRPL18B plasmids (Mustroph et al., 2009) by the corresponding M. truncatula RPL18 ORF using BamHI and XbaI restriction enzymes. The GUS RNAi construct was generated by amplification of a GUS fragment using the pKGWFS7.0 vector (Karimi et al., 2002) as template and the GUSRNAi F and GUSRNAi R primers listed in Supplemental Data Set 10. SKI3 RNAi constructs were generated by amplification of SKI3 fragments using M. truncatula cDNA as template and the MtSKI3RNAi F and MtSKI3RNAi R or the MtSKI3RNAi2 F and MtSKI3RNAi2 R primers listed in Supplemental Data Set 10. The GUS and SKI3 amplified fragments were cloned into the pTOPO/ENTR vector (Thermo Fisher Scientific) and recombined into the destination vector pK7GWIWG2D (II; Karimi et al., 2007). TAS3 and ALT TAS3 constructs were generated by PCR amplification of TAS3 fragments using M. truncatula cDNA as template and the MtTAS3 F and MtTAS3 R or the MtALTTAS3 F and MtALTTAS3 R primers listed in Supplemental Data Set 10. The TAS3- and ALT TAS3–amplified fragments were cloned into the pENTR/D-TOPO vector and then recombined into the destination vector pK7WG2D,1 (Karimi et al., 2002). The EV pK7WG2D,1 was used as a control. pK7GWIWG2D (II) and pK7WG2D,1 vectors carry EgfpER as a screenable marker for early visualization and selection of the transgenic roots. Binary vectors were introduced into Agrobacterium rhizogenes ARqua1 (Quandt et al., 1993) by electroporation. Sinorhizobium meliloti strain 1021 (Meade and Signer, 1977) or the same strain expressing RFP (Tian et al., 2012) was used for root inoculation as described previously by Hobecker et al. (2017).
Plant Growth Conditions, Hairy Root Transformation, and Rhizobium Inoculation
Seeds were surface sterilized and germinated on water–agar plates at 20°C in the dark for 24 h. Germinated seedlings were transferred to Petri dishes containing agar Fahraeus media (Fahraeus, 1957) covered with sterile paper. Transgenic roots were generated by Agrobacterium rhizogenes–mediated transformation as described previously (Boisson-Dernier et al., 2001). Plants that developed hairy roots were transferred to slanted boxes containing Fahraeus media free of nitrogen. Seedlings were grown at 25°C and 75% humidity with a long-day period (16-h-day/8-h-night cycle) and a photosynthetically active radiation of 200 μmol m−2 s−1 using mixed lighting containing four OSRAM cool daylight L36W/765 tubes and one OSRAM FLUORA L36W/77 tube. Roots were inoculated with 10 mL of a 1:1000 dilution of S. meliloti 1021 (Meade and Signer, 1977) culture grown in liquid TY media until OD600 reached 0.8 or with 10 mL of water as a control (mock treatment). One hour later, the excess liquid was discarded, and seedlings were incubated vertically under the growth conditions described above. Alternatively, seedlings were transferred to pots containing perlite:sand (3:1) and inoculated with 10 mL of a 1:1000 dilution of S. meliloti 1021. For RNA isolation, root tissue was harvested, frozen in liquid N2, and stored at −80°C.
Isolation of Polysomes by TRAP
Isolation of polysomes by TRAP was accomplished as described previously (Zanetti et al., 2005; Mustroph et al., 2009; Reynoso et al., 2013). Five milliliters of packed frozen root tissue was used for p35S and pSUC2 samples and 15 mL for pCO2 and pEXP7 samples. TRAP material was subjected to RNA extraction using TRIzol following the manufacturer’s recommendations (Thermo Fisher Scientific).
RNA-Seq Library Preparation and Data Analysis
RNA-seq libraries were prepared from total and TRAP RNA samples isolated from roots inoculated with water (mock) or S. meliloti using two biological replicates (i.e., the whole root systems of more than 100 composite plants were collected and pooled in independent experiments performed on different days). Total RNA was digested with RNase-free DNase (Promega). RNA quality was evaluated by capillary gel electrophoresis on an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Pico kit. Libraries were prepared using 0.4 µg of total and TRAP RNA samples and the Illumina TruSeq RNA Sample Preparation kit (v2; Illumina). SuperScript II reverse transcriptase (Thermo Fisher Scientific) was used for cDNA synthesis. PCR fragments were purified using AMPure XP beads (Beckman Coulter Genomics). The size of DNA fragments was verified on an Agilent 2100 Bioanalyzer using the DNA-1000 kit (Agilent). Libraries were sequenced using the Illumina HiSeq 1500 platform at the Craig Venter Institute (http://www.jcvi.org), which yields paired-end 100-bp reads. The number of reads obtained for each condition is listed in Table 1.
Data Analysis
Total and TRAP RNA-seq reads were aligned to the M. truncatula genome v4 using TopHat2 (Kim et al., 2013). Transcript assembly was performed using Cufflinks and DEGs and transcripts were identified using Cuffdiff with default parameters (Trapnell et al., 2012, 2013). Only genes and transcripts with at least a twofold change between samples, P < 0.05, and FPKM values greater than 0.5 in at least one sample were considered. Functional classification of genes was based on GO annotations using Blast2GO (http://www.blast2go.com/) and further manually redefined using BLAST searches and database annotations. Transcript variants along with reads aligned to the genome were visualized using the Integrative Genomic Viewer (Thorvaldsdóttir et al., 2013).
GC content and ΔG° of CDS, 5′ and 3′ UTRs were determined using The Visual Gene Developer (Jung and McDonald, 2011) available at http://www.visualgenedeveloper.net. Coding potential analysis was performed using two different alignment-free methods: the support vector machine-based classifier Coding Potential Calculator (http://cpc.cbi.pku.edu.cn; Kong et al., 2007) and the Coding Potential Assessment Tool (http://lilab.research.bcm.edu/cpat; Wang et al., 2013). A cutoff of coding potential score less than −0.5 was used to distinguish coding RNAs from ncRNAs. Sequences of lncRNAs and pre-miRNAs were analyzed using BLASTX (www.ncbi.nlm.nih.gov). ORFs of lncRNAs were predicted using the National Center for Biotechnology Information Open Reading Frame Finder (www.ncbi.nlm.nih.gov/orffinder/) using ATG as start codon and ≥30 nucleotides and the Sequence Manipulation Suite ORF Finder (http://www.bioinformatics.org; Stothrd, 2000) using ATG as start codon and ≥10 nucleotides. ORFs of lncRNAs were analyzed using BLASTP (www.ncbi.nlm.nih.gov).
RT-qPCR and 3′ RACE
Total RNA was extracted using TRIzol (Thermo Fisher Scientific), following the manufacturer’s recommendations, and digested with RNase-free DNase (Promega). Total and TRAP RNA samples were subjected to first-strand cDNA synthesis using Moloney Murine Leukemia Virus reverse transcriptase (Promega). Expression analysis by RT-qPCR was performed using the iQ SYBR Green Supermix kit (Bio-Rad) and the CFX96 qPCR system (Bio-Rad) as described previously (Blanco et al., 2009). For each pair of primers, the presence of a unique PCR product of the expected size was verified in agarose gels. The M. truncatula HIS3L was selected as reference transcript for normalization of RT-qPCR data based on previously reported geNORM analysis (Reynoso et al., 2013) as well as on the analysis of Arabidopsis cell-specific translatome data revealing that steady-state levels of HIS3L do not significantly changes across different root cell types (Mustroph et al., 2009). The S. meliloti Sensory histidine kinase/phosphatase NtrB transcript was used to normalize nifH mRNAs levels in nodule samples. Primers used are listed in Supplemental Data Set 10, and the annealing temperature for each primer set was 52°C. tasiARF quantification was performed by stem-loop RT-qPCR as described previously (Hobecker et al., 2017) using the primers listed in Supplemental Data Set 10. U6 RNA was used as reference transcript for normalization.
3′ RACE experiments were conducted using the 3′ RACE System for Rapid Amplification of cDNA Ends using the Adapter Primer for the first-strand cDNA synthesis (Supplemental Data Set 10) according to the protocol provided by the manufacturer (Thermo Fisher Scientific). PCR amplification of the target was performed using the MtTAS3 F and the universal amplification primers. A second amplification step using a nested primer and the primary PCR product was performed using the MtTAS3N F and the universal amplification primers (Supplemental Data Set 10). Amplified DNA fragments were analyzed by electrophoresis.
Phenotypic Analysis
Nodule number was recorded at different time points after inoculation with S. meliloti as described previously by Hobecker et al. (2017). Nodules were observed under a dissection microscope (MZ8, Leica Microsystems), and digital images were captured using a DFC 480 camera. Nodule size was measured from digital pictures using Photoshop CS5 at 16 DAI using more than 65 nodules per construct. Nodules and infection events were quantified in at least 50 independent roots per construct inoculated with an S. meliloti strain expressing RFP (Tian et al., 2012). Confocal microscopy was performed on nodules of 21 DAI with an S. meliloti strain expressing RFP on roots transformed using an inverted SP5 confocal microscope (Leica Microsystems) with a 20× objective. Microcolonies and infections threads were visualized, quantified, and imaged at 6 DAI in an IX51 inverted microscope (Olympus). Three biological replicates were performed for each experiment. The statistical significance of the differences for each parameter was determined by unpaired two-tailed Student’s t tests for each construct. For SYTO9/propidium iodide staining, individual nodules were excised and embedded in 6% (w/v) low melting agarose. Nodule sections of 60 mm were obtained using a VT1000 S vibratome (Leica Microsystems). Staining was performed for 30 min at room temperature on fresh nodule sections using 5 µM SYTO9 and 30 µM propidium iodide. Images of nodule sections were acquired with an inverted SP5 confocal microscope (Leica Microsystems). Images were processed with the LAS Image Analysis software (Leica Microsystems). For electronic microscopy, individual nodules were fixed in 50 mM potassium phosphate buffer, pH 7.4, containing 2% (v/v) paraformaldehyde during 2 h at 48°C. During fixation, samples were subjected to short pulses of gentle vacuum until they sank. Nodules were postfixed in 50 mM potassium phosphate buffer, pH 7.4, containing 1% (w/v) osmium tetroxide for 1 h at 48°C, rinsed three times in the same buffer, dehydrated by passing through a series of graded ethanol washes, and embedded in epoxy resin. Ultrathin sections (70 nm) were obtained with a microtome, stained with uranyl acetate and lead citrate, and observed in a JEM 1200 EX II transmission electron microscope (JEOL USA).
Accession Numbers
The data set supporting the conclusions of this article is available in the Gene Expression Omnibus under series entry number GSE133510 (RNA-seq and TRAP-seq). Raw sequence files (fastq files) and processed data files (outputs of Cufflinks) were deposited. All processed data are included in Supplemental Data Sets. TAS3 and SKI3 accession numbers are Medtr2g033380 and Medtr5g004660, respectively. All other accession numbers for major genes studied in this work are included in Supplemental Data Sets 3, Supplemental Data Sets 4 or Supplemental Data Sets 8.
Supplemental Data
Supplemental Figure 1. Reproducibility between biological replicates and between TRAP and Total samples.
Supplemental Figure 2. Correlation between RNA-seq (this study) and RT-qPCR data obtained from Reynoso et al. (2013) of Nod signaling pathway genes.
Supplemental Figure 3. Functional classification of differentially expressed genes (DEGs) in the transcriptome and the translatome.
Supplemental Figure 4. Changes in RL in response to rhizobia.
Supplemental Figure 5. Classification of transcription factors (TFs) that change their RL in response to rhizobia.
Supplemental Figure 6. Expression of FLAG-RPL18 in specific tissue types.
Supplemental Figure 7. TRAP RNA quality evaluation.
Supplemental Figure 8. Phenotypic analysis of root architecture in OXTAS3 and OX ALT TAS3 composite plants.
Supplemental Figure 9. Steady state levels of SKI3 mRNAs in M. truncatula.
Supplemental Figure 10. Phenotypic analysis of root and shoot architecture in GUS RNAi and SKI3 RNAi.
Supplemental Figure 11. Phenotypic analysis of nodule formation and infection events in SKI3 RNAi2.
Supplemental Figure 12. Transmission electron microscopy (TEM) of the transition zones of GUS and SKI3 RNAi nodules.
Supplemental Figure 13. Model explaining how the association of ALT TAS3 affects the biogenesis of tasiARFs and the stability of their target mRNAs ARF2, ARF3 and ARF4.
Supplemental Data Set 1. FPKM and Log2 FC for all genes in Total and TRAP RNA samples.
Supplemental Data Set 2. FPKM and Log2 FC for all transcripts in Total and TRAP RNA samples.
Supplemental Data Set 3. Differential expressed genes in Total and TRAP RNA samples.
Supplemental Data Set 4. Known rhizobium-induced genes.
Supplemental Data Set 5. Ribosome loading for all expressed genes in mock- and S. meliloti-inoculated samples.
Supplemental Data Set 6. Genes with changes in Ribosome loading.
Supplemental Data Set 7. Non-coding RNAs found as DEGs in Total and TRAP RNA samples.
Supplemental Data Set 8. Differentially expressed transcripts (DETs) in Total and TRAP RNA samples.
Supplemental Data Set 9. Genes with differentially expressed alternative transcripts in Total and TRAP RNA samples.
Supplemental Data Set 10. List of primers used in this study.
Supplemental Data Set 11. Summary of Statistical tests.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Martin Crespi for extremely fruitful discussion and critical comments on the manuscript. We also thank Gonzalo Torres-Tejerizo for technical assistance with histological staining and sectioning and Antonio Lagares, Jr. and María Eugenia Salas for providing S. meliloti primers. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (grants PICT2013-0384, PICT2014-0321, and PICT2016-0582) and by the National Science Foundation (RNA biology grant MCB-1716913). M.E.Z. and F.A.B. are members of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). S.T., K.H. M.A.R., and M.L. were recipients of a CONICET Fellowship. S.T. was also awarded with a short-term Fulbright fellowship to visit J.B.-S.’s laboratory at University of California, Riverside.
AUTHOR CONTRIBUTIONS
M.E.Z., F.B., C.T., and J.B.-S. conceptualized the study. S.T., M.A.R., K.H., M.H., F.B., M.L., and M.E.Z. created the methodology. S.T., M.A.R., K.H., M.L., and M.E.Z performed the research. S.T. and M.E.Z. wrote the original draft of the article. S.T., F.B., C.T., J.B.-S., and M.E.Z. reviewed and edited the article. Funding was acquired by M.E.Z., F.B., and J.B.-S, and resources were provided by B.R. and C.T.. M.E.Z., F.B., and J.B.-S. supervised the study.
References
- Ariel F., Romero-Barrios N., Jégu T., Benhamed M., Crespi M. (2015). Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 20: 362–371. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Basbouss-Serhal I., Soubigou-Taconnat L., Bailly C., Leymarie J. (2015). Germination potential of dormant and nondormant Arabidopsis seeds is driven by distinct recruitment of messenger RNAs to polysomes. Plant Physiol. 168: 1049–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J., Baerenfaller K., Gosai S.J., Gregory B.D., Crespi M., Bailey-Serres J. (2017). Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 114: E10018–E10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini A.A., Johnstone T.G., Christiano R., Mackowiak S.D., Obermayer B., Fleming E.S., Vejnar C.E., Lee M.T., Rajewsky N., Walther T.C., Giraldez A.J. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513. [DOI] [PubMed] [Google Scholar]
- Blanco F.A., Meschini E.P., Zanetti M.E., Aguilar O.M. (2009). A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21: 2797–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Branco-Price C., Kaiser K.A., Jang C.J., Larive C.K., Bailey-Serres J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56: 743–755. [DOI] [PubMed] [Google Scholar]
- Branscheid A., Marchais A., Schott G., Lange H., Gagliardi D., Andersen S.U., Voinnet O., Brodersen P. (2015). SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 43: 10975–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E., Downie J.A., Murray J.D. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K.S., Bailey-Serres J. (2015). Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13: e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Chantarachot T., Bailey-Serres J. (2018). Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. plant physiol. 176: 254–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C., Johansson C., Kondorosi E., Kondorosi A., Crespi M. (1997). enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc. Natl. Acad. Sci. USA 94: 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew G.L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E. (2013). Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140: 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Complainville A., Brocard L., Roberts I., Dax E., Sever N., Sauer N., Kondorosi A., Wolf S., Oparka K., Crespi M. (2003). Nodule initiation involves the creation of a new symplasmic field in specific root cells of medicago species. Plant Cell 15: 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bang T.C., et al. (2017). Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 175: 1669–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforges J., Reis R.S., Jacquet P., Sheppard S., Gadekar V.P., Hart-Smith G., Tanzer A., Hofacker I.L., Iseli C., Xenarios I., Poirier Y. (2019). Control of cognate sense mRNA translation by cis-natural antisense RNAs. Plant Physiol. 180: 305–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L., Sorenson R., Bailey-Serres J., Hunt A.G. (2017). Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell 29: 1262–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yahyaoui F., Küster H., Ben Amor B., Hohnjec N., Pühler A., Becker A., Gouzy J., Vernié T., Gough C., Niebel A., Godiard L., Gamas P. (2004). Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 136: 3159–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16: 374–381. [DOI] [PubMed] [Google Scholar]
- Fournier J., Timmers A.C., Sieberer B.J., Jauneau A., Chabaud M., Barker D.G. (2008). Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148: 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F., Reichelt P., Rode M., Conti E. (2013). The yeast ski complex: Crystal structure and RNA channeling to the exosome complex. Cell 154: 814–826. [DOI] [PubMed] [Google Scholar]
- Hobecker K.V., Reynoso M.A., Bustos-Sanmamed P., Wen J., Mysore K.S., Crespi M., Blanco F.A., Zanetti M.E. (2017). The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 174: 2469–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann N. (2014). No scalpel needed: Translatome of pollen tubes growing within the flower in Arabidopsis. Plant Cell 26: 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C.Y., Lee W.C., Chou H.C., Chen A.P., Chou S.J., Chen H.M. (2016). Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.G. (2012). RNA regulatory elements and polyadenylation in plants. Front. Plant Sci. 2: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S. (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8: 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q., Poirier Y. (2013). A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardinaud M.F., et al. (2016). A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by nod factors in the Medicago truncatula root epidermis. Plant Physiol. 171: 2256–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., McDonald K. (2011). Visual gene developer: a fully programmable bioinformatics software for synthetic gene optimization. BMC Bioinformatics 12: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Girke T., Bazin J., Bailey-Serres J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P., Sorenson R., Bailey-Serres J. (2013). Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 74: 1016–1028. [DOI] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Bailey-Serres J. (2005). mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 33: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38: 823–839. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G. (2007). CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35: W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung J.T., Colognori D., Lee J.T. (2013). Long noncoding RNAs: Past, present, and future. Genetics 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E., et al. (2015). Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 169: 233–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., et al. (2015). Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 84: 1206–1218. [DOI] [PubMed] [Google Scholar]
- Li S., Le B., Ma X., You C., Yu Y., Zhang B., Liu L., Gao L., Shi T., Zhao Y., Mo B., Cao X., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 5: e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Moling S., Hooiveld G., Pereira P.A., Bisseling T., Becker J.D., Küster H. (2013). Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS One 8: e64377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar D.P., Sharopova N., Endre G., Peñuela S., Samac D., Town C., Silverstein K.A., VandenBosch K.A. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, et al. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. The Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba A.E., Jauvion V., Mallory A.C., Bouteiller N., Vaucheret H. (2011). The miRNA pathway limits AGO1 availability during siRNA-mediated PTGS defense against exogenous RNA. Nucleic Acids Res. 39: 9339–9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunoury N., et al. (2010). Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 5: e9519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H.M., Signer E.R. (1977). Genetic mapping of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 74: 2076–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Stepanova A.N., Alonso J.M. (2017). Translation regulation in plants: An interesting past, an exciting present and a promising future. Plant J. 90: 628–653. [DOI] [PubMed] [Google Scholar]
- Merret R., Carpentier M.C., Favory J.J., Picart C., Descombin J., Bousquet-Antonelli C., Tillard P., Lejay L., Deragon J.M., Charng Y.Y. (2017). Heat shock protein HSP101 affects the release of ribosomal protein mRNAs for recovery after heat shock. Plant Physiol. 174: 1216–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteignier L.V., El Oirdi M., Cohen M., Barff T., Matteau D., Lucier J.F., Rodrigue S., Jacques P.E., Yoshioka K., Moffett P. (2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J. Exp. Bot. 68: 2333–2344. [DOI] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A., Ernest B., Lohoff T., Jia Q., Satterlee J., Ke K., von Arnim A.G. (2015). The circadian clock modulates global daily cycles of mRNA ribosome loading. Plant Cell 27: 2582–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141. [DOI] [PubMed] [Google Scholar]
- Moreau S., Verdenaud M., Ott T., Letort S., de Billy F., Niebel A., Gouzy J., de Carvalho-Niebel F., Gamas P. (2011). Transcription reprogramming during root nodule development in Medicago truncatula. PLoS One 6: e16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munusamy P., Zolotarov Y., Meteignier L.V., Moffett P., Strömvik M.V. (2017). De novo computational identification of stress-related sequence motifs and microRNA target sites in untranslated regions of a plant translatome. Sci. Rep. 7: 43861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Quandt H.J., Pühler A., Broer I. (1993). Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 6: 699–706. [Google Scholar]
- Reddy A.S., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso M.A., Blanco F.A., Bailey-Serres J., Crespi M., Zanetti M.E. (2013). Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J. 73: 289–301. [DOI] [PubMed] [Google Scholar]
- Reynoso M.A., Juntawong P., Lancia M., Blanco F.A., Bailey-Serres J., Zanetti M.E. (2015). Translating ribosome affinity purification (TRAP) followed by RNA sequencing technology (TRAP-SEQ) for quantitative assessment of plant translatomes. Methods Mol. Biol. 1284: 185–207. [DOI] [PubMed] [Google Scholar]
- Rival P., de Billy F., Bono J.-J., Gough C., Rosenberg C., Bensmihen S. (2012). Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development 139: 3383–3391. [DOI] [PubMed] [Google Scholar]
- Roux B., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77: 817–837. [DOI] [PubMed] [Google Scholar]
- Roy B., Jacobson A. (2013). The intimate relationships of mRNA decay and translation. Trends Genet. 29: 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B., von Arnim A.G. (2013). Translational regulation of cytoplasmic mRNAs. The Arabidopsis Book 11: e0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Jensen T.H. (2008). The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci. 33: 501–510. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson R., Bailey-Serres J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothrd P P. (2000). The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102–1104. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C.F., Garnerone A.M., Mathieu-Demazière C., Masson-Boivin C., Batut J. (2012). Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti-Medicago symbiosis. Proc. Natl. Acad. Sci. USA 109: 6751–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T., Kim J., Frances L., Ding Y., Sun J., Guan D., Niebel A., Gifford M.L., de Carvalho-Niebel F., Oldroyd G.E.D. (2015). The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang Y., Xie S., Liu Y., Xie Z. (2017). Global and cell-type specific properties of lincRNAs with ribosome occupancy. Nucleic Acids Res. 45: 2786–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Xu J., Meyers B.C. (2017). The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants. Plant Cell 29: 1232–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. (2014). Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Greene G.H., Yoo H., Liu L., Marqués J., Motley J., Dong X. (2017). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545: 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. (2012). LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Saudemont B., Bouteiller N., Elvira-Matelot E., Lepère G., Parent J.S., Morel J.B., Cao J., Elmayan T., Vaucheret H. (2015). Second-site mutagenesis of a hypomorphic argonaute1 allele identifies SUPERKILLER3 as an endogenous suppressor of transgene posttranscriptional gene silencing. Plant Physiol. 169: 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Willmann M.R., Anderson S.J., Gregory B.D. (2016). Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mRNA cap-binding complex in co-translational RNA decay in Arabidopsis. Plant Cell 28: 2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti M.E., Chang I.F., Gong F., Galbraith D.W., Bailey-Serres J. (2005). Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu X., Gaikwad K., Kou X., Wang F., Tian X., Xin M., Ni Z., Sun Q., Peng H., Vierling E. (2017). Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell 29: 1952–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., et al. (2015). Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348: 120–123. [DOI] [PubMed] [Google Scholar]
- Zhao L., Kunst L. (2016). SUPERKILLER complex components are required for the RNA exosome-mediated control of cuticular wax biosynthesis in Arabidopsis inflorescence stems. Plant Physiol. 171: 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder J.C., Lima C.D. (2017). Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 31: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]