bHLH121 acts upstream of the iron homeostasis regulatory network and directly regulates the expression of the large majority of genes encoding proteins and peptides involved in this regulatory cascade.
Abstract
Iron (Fe) is an essential micronutrient for plant growth and development. Any defects in the maintenance of Fe homeostasis will alter plant productivity and the quality of their derived products. In Arabidopsis (Arabidopsis thaliana), the transcription factor ILR3 plays a central role in controlling Fe homeostasis. In this study, we identified bHLH121 as an ILR3-interacting transcription factor. Interaction studies showed that bHLH121 also interacts with the three closest homologs of ILR3 (i.e., basic-helix-loop-helix 34 [bHLH34], bHLH104, and bHLH115). bhlh121 loss-of-function mutants displayed severe defects in Fe homeostasis that could be reverted by exogenous Fe supply. bHLH121 acts as a direct transcriptional activator of key genes involved in the Fe regulatory network, including bHLH38, bHLH39, bHLH100, bHLH101, POPEYE, BRUTUS, and BRUTUS LIKE1, as well as IRONMAN1 and IRONMAN2. In addition, bHLH121 is necessary for activating the expression of transcription factor gene FIT in response to Fe deficiency via an indirect mechanism. bHLH121 is expressed throughout the plant body, and its expression is not affected by Fe availability. By contrast, Fe availability affects the cellular localization of bHLH121 protein in roots. Altogether, these data show that bHLH121 is a regulator of Fe homeostasis that acts upstream of FIT in concert with ILR3 and its closest homologs.
INTRODUCTION
Iron (Fe) is an essential micronutrient for plant growth and development, as it is a cofactor for several enzymes that participate in many fundamental biological processes (Hänsch and Mendel, 2009). Any defects in the maintenance of Fe homeostasis will alter plant productivity and the quality of their derived products (Briat et al., 2015). Efficient Fe uptake from soil is ensured, in dicot and nongraminaceous monocot species, by a reduction-based mechanism (Kobayashi and Nishizawa, 2012; Brumbarova et al., 2015). This process involves the reduction of Fe3+ by Fe3+-reductases such as FERRIC REDUCTION OXIDASE2 (FRO2) and the subsequent transport of the reduced Fe2+ across the rhizodermis cell plasma membrane via IRON-REGULATED TRANSPORTER1 (IRT1). Fe3+ solubilization is facilitated by two distinct mechanisms that act in concert: the acidification of the rhizosphere and the secretion of Fe3+-mobilizing coumarins (Santi and Schmidt, 2009; Fourcroy et al., 2016).
Fe homeostasis is tightly regulated at the transcriptional level by a process involving numerous basic-helix-loop-helix (bHLH) transcription factors (TFs) that form an intricate network (Gao et al., 2019). In Arabidopsis (Arabidopsis thaliana), 16 bHLH TFs (∼12% of this gene family) have thus far been shown to participate in this network. Emerging evidence derived from analysis of the molecular and genetic relationships between these TFs indicates that FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT)/bHLH29 and IAA-LEUCINE RESISTANT3 (ILR3)/bHLH105 are two important nodes in this regulatory network (Ivanov et al., 2012; Zhang et al., 2015; Tissot et al., 2019).
FIT is the Arabidopsis ortholog of tomato (Solanum lycopersicum) FER, the first cloned regulatory gene involved in Fe homeostasis (Ling et al., 2002; Colangelo and Guerinot, 2004; Yuan et al., 2005). In Arabidopsis, the fit null mutation is lethal during early seedling development without an extra supply of Fe and the induction of the Fe uptake machinery in response to Fe shortage is abolished (Colangelo and Guerinot, 2004). Interactions of FIT with bHLH38, bHLH39, bHLH100, and bHLH101 are required for inducing the expression of its target genes, including FRO2 and IRT1 (Yuan et al., 2008; Wang et al., 2013). The activity of these heterodimers is counteracted by bHLH18, bHLH19, bHLH20, and bHLH25, whose interaction with FIT promotes its degradation via the 26S proteasome pathway (Cui et al., 2018). BRUTUS LIKE1 (BTSL1) and BTSL2, two closely related RING E3 ubiquitin ligases, have also been recently proposed to negatively regulate Fe deficiency responses by directly targeting FIT, leading to its degradation via the 26S proteasome (Sivitz et al., 2011; Hindt et al., 2017; Rodríguez-Celma et al., 2019).
ILR3 and its three closest homologs (i.e., bHLH34, bHLH104, and bHLH115) can form homo- and heterodimers. These four bHLH TFs positively regulate Fe homeostasis by directly activating the expression of bHLH38, bHLH39, bHLH100, and bHLH101 (Heim et al., 2003; Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). ILR3 also acts as a transcriptional repressor when interacting with POPEYE (PYE)/bHLH47, a TF originally described as playing a negative role in the Fe deficiency responses (Long et al., 2010; Tissot et al., 2019). PYE expression is induced in response to Fe shortage by ILR3 and its closest homologs (Zhang et al., 2015; Liang et al., 2017; Kroh and Pilon, 2019; Tissot et al., 2019). It is likely that a negative feedback regulatory loop involving the ILR3-PYE complex represses PYE expression when Fe availability is not limiting (Tissot et al., 2019). ILR3 and bHLH115 interact with BRUTUS (BTS), an Fe binding E3 ubiquitin ligase closely related to BTSL1 and BTSL2, leading to their degradation via the 26S proteasome, allowing fine tuning of Fe uptake (Selote et al., 2015; Matthiadis and Long, 2016). Interestingly, a family of peptides named IRON MAN/FE-UPTAKE-INDUCING PEPTIDE (IMA/FEP) was recently shown to play a critical role in the acquisition and cellular homeostasis of Fe in plants by regulating the expression of bHLH38 and bHLH39 (Grillet et al., 2018; Hirayama et al., 2018).
In this study, we performed co-immunoprecipitation of ILR3 followed by liquid chromatography tandem–mass spectrometry (Co-IP LC-MS/MS) analyses and identified bHLH121 (a close homolog of PYE) as an ILR3-interacting TF. Interaction studies showed that bHLH121 also interacts with bHLH34, bHLH104, and bHLH115, but not with PYE, FIT, BTS, BTSL1, or BTSL2. Loss-of-function of bHLH121 (bhlh121) led to severe defects in Fe homeostasis, which were reverted by exogenous Fe supply. Expression studies and chromatin immunoprecipitation (ChIP) assays indicated that bHLH121 functions as a direct transcriptional activator of key genes involved in the Fe regulatory network, including bHLH38, bHLH39, bHLH100, bHLH101, PYE, BTS, and BTSL1 as well as IMA1 and IMA2. In addition, bHLH121 is required for the activation of FIT expression in response to Fe deficiency via an indirect mechanism. bHLH121 is expressed throughout the plant, and its expression is not affected by Fe availability. By contrast, Fe availability affects the cellular localization of bHLH121 in roots. Therefore, bHLH121 regulates Fe homeostasis in Arabidopsis by acting upstream of FIT together with ILR3 and its closest homologs.
RESULTS
bHLH121 Interacts with ILR3 in Vivo
ILR3 plays a key role in regulating Fe homeostasis in Arabidopsis (Zhang et al., 2015; Li et al., 2016; Tissot et al., 2019). We therefore conducted Co-IP LC-MS/MS to identify potential new actors involved in controlling Fe homeostasis. For this purpose, ILR3:GFP fusion protein was immunoprecipitated from ilr3-3 knockdown mutant lines complemented with the ProILR3:gILR3:GFP construct and subjected to Fe starvation (Tissot et al., 2019). Co-IP LC-MS/MS analysis led to the identification of 13 proteins (Supplemental Table). Among the potential ILR3-interacting proteins, bHLH121, a close homolog of PYE and bHLH11 (Heim et al., 2003), was identified.
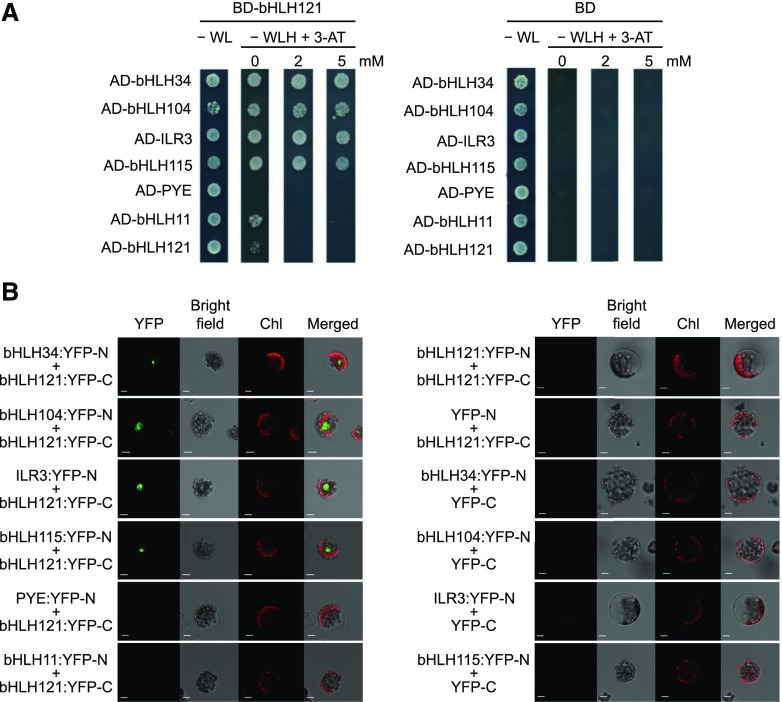
We conducted yeast two-hybrid (Y2H) assays using bHLH121 as bait (fused with the binding domain of GAL4) and ILR3 as prey (fused with the activation domain of GAL4) to confirm this interaction. We also used the closest homologs of ILR3 and bHLH121 as well as BTS, BTSL1, and BTSL2 as prey. This orientation was used since self-activation was observed for bHLH34 and bHLH104 when they were used as bait. These experiments confirmed the interaction between bHLH121 and ILR3 (Figure 1A). They also showed that bHLH121 can interact in vivo with bHLH34, bHLH104, and bHLH115, but not with PYE, BTS, BTSL1, BTSL2, or itself (Figure 1A; Supplemental Figure 1A). We also detected a weak interaction with bHLH11 (Figure 1A).
Figure 1.
bHLH121 Interacts in Vivo with ILR3 and with Its Closest Homologs.
(A) Y2H assays. bHLH34, bHLH104, ILR3, bHLH115, PYE, bHLH11, and bHLH121 were fused with the GAL4 activation domain (AD) and bHLH121 with the GAL4 DNA binding domain (BD) into appropriate expression vectors prior to transfer into yeast (AH109 strain). The different yeast strains were plated on nonselective medium (−WL) or on selective medium lacking histidine (−WLH) and containing various concentration of 3-amino-1,2,4-triazole (3-AT). BD alone was used as a negative control. Growing colonies representative of positive Y2H interactions were identified after 6 d of growth. H, histidine; L, leucine; W, tryptophan.
(B) BiFC assays. bHLH34, bHLH104, ILR3, bHLH115, PYE, bHLH11, and bHLH121 were fused with the N-terminal part of YFP (YFP-N) and bHLH121 with the C-terminal part of YFP (YFP-C) into appropriate expression vectors prior to transfer into Arabidopsis protoplasts and analysis by confocal microscopy. YFP-C and YFP-N alone were used as negative controls. Chl, chlorophyll fluorescence. Bar = 10 μm.
We further analyzed these interactions using bimolecular fluorescence complementation (BiFC) assays in which bHLH121 was fused to the C-terminal part of the yellow fluorescent protein (YFP) reporter gene (Figure 1B). The other TFs were fused to the YFP N-terminal part. Strong signal was observed in the nucleus when bHLH121 was assayed with bHLH34, bHLH104, bHLH115, and ILR3, whereas no signal was observed with PYE, bHLH11, or itself. This approach confirmed the interactions observed by Co-IP LC-MS/MS and Y2H experiments.
To complete this study, we performed Y2H assays using bHLH11 and FIT as bait and prey, respectively. These experiments revealed that, like bHLH121, bHLH11 interacts in vivo with ILR3 and its three closest homologs (Supplemental Figure 1B). However, it also revealed that unlike bHLH121, bHLH11 can form homodimers. By contrast, no interaction between FIT and bHLH121 was observed (Supplemental Figure 1C).
Altogether, these data indicate that bHLH121 can form heterodimers with bHLH34, bHLH104, bHLH115, and ILR3, but not with PYE, bHLH11, or FIT. In addition, these data indicate that bHLH121 cannot form homodimers and suggest that BTS, BTSL1, and BTSL2 are likely not involved in the regulation of bHLH121 stability.
bhlh121 Knockout Mutants Have Altered Fe Homeostasis
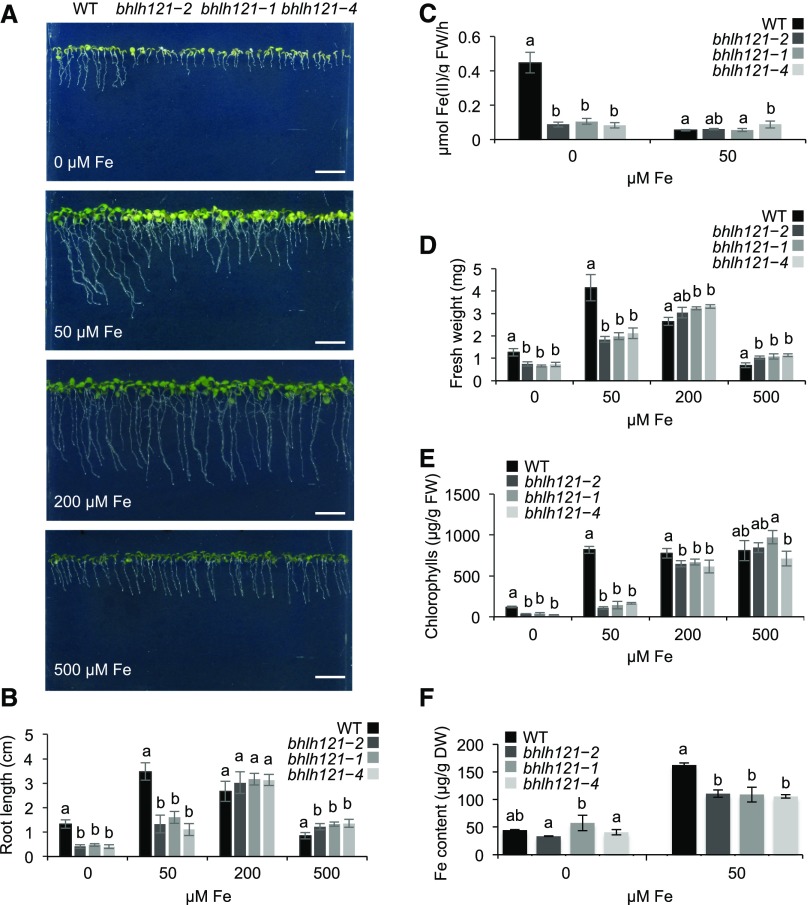
To determine whether bHLH121 plays a role in controlling Fe homeostasis, we generated three independent loss-of-function mutant alleles using the clustered regular interspaced short palindromic repeats/associated protein 9 (CRISPR-Cas9) gene editing system. One allele displayed a 656-bp deletion (bhlh121-2) leading to a truncated protein, and the two other alleles contained a single nucleotide insertion (bhlh121-1 and bhlh121-4) leading to premature stop codons (Supplemental Figure 2). All three mutations are located in the second exon, affecting the integrity of the bHLH DNA binding domain. In the absence of Fe (0 μM Fe), the bhlh121 mutants displayed a strong inhibition of primary root growth and ferric-chelate reductase (FCR) activity compared with the wild type (Figures 2A to 2C). Fresh weight and chlorophyll contents were also lower in the mutants than in the wild type (Figures 2D and 2E). When the mutants were grown in the presence of 50 μM Fe (control conditions), the root growth and fresh weight defects were partially rescued and the chlorosis symptoms were still visible. By contrast, when grown in the presence of 200 μM Fe (mild Fe excess conditions), the mutants had no discernible differences from the wild type. However, at this concentration of Fe, the primary root length and fresh weight of the wild type were slightly reduced compared with control conditions. Interestingly, the mutants appeared to be a bit less affected than the wild type by the presence of 500 μM Fe in the medium (Fe excess conditions). Fe accumulation was also compromised in mutant plants grown under control conditions, suggesting limitations in Fe uptake (Figure 2F).
Figure 2.
bhlh121 Loss-of-Function Mutants Have Decreased Tolerance to Fe Deficiency.
(A) Phenotypes of the Arabidopsis wild type (WT) and the three bhlh121 mutant alleles grown for 1 week on Fe-sufficient (50 μM Fe), Fe-deficient (0 μM Fe), or Fe-excess (200 and 500 μM Fe) medium. Bar = 1 cm.
(B) Root length of the wild type (WT) and the bhlh121 mutants grown on Fe-sufficient, Fe-deficient, and Fe-excess medium for 7 d.
(C) Ferric-chelate reductase activity of the wild type (WT) and the bhlh121 mutants grown for 1 week under control conditions and transferred to Fe-sufficient or Fe-deficient medium for 3 d.
(D) and (E) Fresh weight (FW; see [D]) and chlorophyll content (E) of the wild type (WT) and the bhlh121 mutants grown on Fe-sufficient, Fe-deficient, and Fe-excess medium for 7 d.
(F) Fe contents of the wild type (WT) and the bhlh121 mutants grown on Fe-sufficient or Fe-deficient medium for 7 d.
(B) to (F) Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 3 biological repeats from one representative experiment). Error bars show ±sd. A biological repeat comprised a pool of 12 seedlings in (B), 40 seedlings in (C), 5 seedlings in (D) and (E), and ∼80 to 100 seedlings in (F). Each experiment was repeated three times.
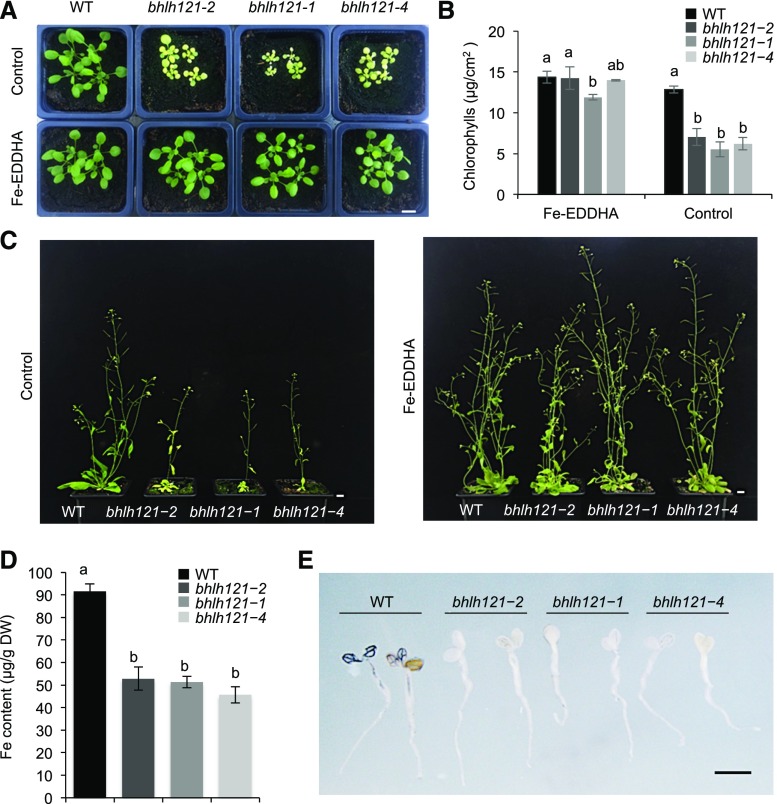
When grown in soil, the bhlh121 mutants displayed severe growth defects (e.g., reduced rosette and stem size and flower number) and chlorosis symptoms that were rescued by exogenous supply of Fe (Figures 3A and 3C). We also observed a decrease in Fe accumulation in seeds (Figure 3D). This reduced accumulation of Fe in bhlh121 seeds might explain the growth defects observed upon germination under Fe deficiency (Figure 2A). In situ analysis of Fe accumulation in the embryo confirmed that Fe content (as revealed by a darker Perls and Perls/3′-Diaminobenzidine tetrahydrochloride [DAB] staining), and not Fe localization (near the vasculature), was altered in the mutants compared with the wild type (Supplemental Figure 3). Following germination, Perls/DAB staining confirmed the latter observation (Figure 3E).
Figure 3.
bhlh121 Loss-of-Function Mutants Have Decreased Tolerance to Fe Deficiency and Accumulate Less Fe in Seeds Than the Wild Type.
(A) Phenotype of the wild-type (WT) and bhlh121 loss-of-function mutant seedlings grown in soil for 3 weeks and watered or not with ferric ethylenediamine di-(o-hydroxyphenylacetate) [Fe-EDDHA, 1‰ (w/v)], a form of Fe easily assimilated by plants.
(B) Chlorophyll content of the wild type (WT) and bhlh121 mutants grown in soil for 3 weeks and watered or not with ferric ethylenediamine di-(o-hydroxyphenylacetate) (Fe-EDDHA).
(C) Phenotypes of the wild type (WT) and bhlh121 mutants grown in soil for 6 weeks and watered or not with ferric ethylenediamine di-(o-hydroxyphenylacetate) (Fe-EDDHA).
(D) Fe content in seeds of the wild type (WT) and bhlh121 mutants grown in soil in the absence of ferric ethylenediamine di-(o-hydroxyphenylacetate) (Fe-EDDHA).
(E) Fe distribution (Perls/DAB staining) in 4-d-old wild-type (WT) and bhlh121 seedlings. Fe accumulation appears in black.
(B) and (D) Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 3 biological repeats from one representative experiment). Error bars show ±sd. A biological repeat comprised a pool of leaf disc samples from three seedlings in (B) and ∼10 mg of seeds in (D). Each experiment was repeated three times. Bar in (A) and (C) = 1 cm; bar in (E) = 1 mm.
To determine how the mutation of bHLH121 affects Fe accumulation in roots and/or shoots, we grew the mutant and wild-type plants in hydroponic solution and subjected them to Fe deficiency or control conditions. A decrease in Fe accumulation was observed in the roots of the mutants regardless of whether the plants were grown in the presence or absence of Fe (Supplemental Figure 4A). Such a decrease was also observed in the shoots (but to a lesser extent) of mutant plants grown under control conditions, whereas no difference was detected when grown under Fe deficiency (Supplemental Figure 4B).
bHLH121 Regulates the Biosynthesis of Fe-Mobilizing Coumarins
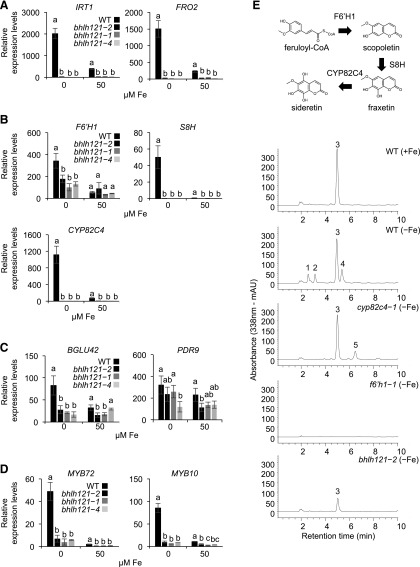
Since Fe accumulation and FCR activity were compromised in the bhlh121 mutant lines, we analyzed IRT1 and FRO2 expression. We used 1-week-old seedlings subjected or not to Fe deficiency for this experiment. As expected, RT-qPCR revealed that IRT1 and FRO2 expression was reduced in bhlh121 seedlings grown under control conditions compared with the wild type and that Fe deficiency–induced expression of both genes was also compromised (Figure 4A).
Figure 4.
Coumarin Biosynthesis Is Affected in the bhlh121 Loss-of-Function Mutants.
(A) Relative expression of IRT1 and FRO2 (high-affinity Fe uptake system).
(B) Relative expression of F6’H1, S8H, and CYP82C4 (coumarin biosynthesis).
(C) Relative expression of BGLU42 and PDR9 (coumarin secretion).
(D) Relative expression of MYB10 and MYB72 (transcriptional control of coumarin biosynthesis and secretion).
(A) to (D) Relative expression was determined by RT-qPCR in 1-week-old Arabidopsis seedlings grown on Fe-sufficient or Fe-deficient medium. Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 3 technical repeats from one representative experiment). Error bars show ±sd. Each experiment (biological repeat) comprised pooled RNA extracted from ∼30 seedlings and was independently repeated three times.
(E) (Top) Schematic representation of the coumarin biosynthetic pathway. (Bottom) Representative absorbance chromatograms obtained at 338 nm for root extracts of Arabidopsis wild type (WT), cyp8c24-1, f6’h1-1, and bhlh121-2 mutant seedlings grown for 7 d on half-strength Murashige and Skoog medium agar plates at pH 5.5. +Fe, control; −Fe, Fe deficiency. Numbered peaks correspond to sideretin-glycosides (1 and 2), scopolin: scopoletin glycoside (3), sideretin (4), and fraxin: fraxetin glycoside (5).
We then investigated the expression of genes that participate in the coumarin biosynthesis and secretion pathway (Figures 4B and 4C). This analysis revealed that the induction of F6’H1 (the first gene committed to the biosynthesis of coumarins) expression in response to Fe deficiency was reduced in all three bhlh121 mutant alleles compared with the wild type (Rodríguez-Celma et al., 2013; Schmid et al., 2014). This was also observed for S8H and CYP82C4, encoding enzymes involved in the biosynthesis of the main Fe-mobilizing coumarins (fraxetin and sideretin, respectively; Rajniak et al., 2018; Siwinska et al., 2018; Tsai et al., 2018). We therefore assayed the expression of PDR9 (ABCG transporter) and BGLU42 (β-glucosidase), two genes necessary for the secretion of coumarins in the rhizosphere (Fourcroy et al., 2014; Zamioudis et al., 2014). PDR9 expression in response to Fe deficiency was not significantly affected in the mutants compared with the wild type, whereas BGLU42 expression was reduced. We also analyzed the expression of MYB10 and MYB72, encoding two TFs required for plant growth under Fe deficiency, and the production and excretion of coumarins in the rhizosphere (by regulating for instance BGLU42 expression; Figure 4D; Palmer et al., 2013; Zamioudis et al., 2014). Both genes displayed a lack of induction in response to Fe deficiency in the mutants. These data suggest that bHLH121 is a positive regulator of fraxetin and sideretin biosynthesis and excretion in the rhizosphere.
We measured coumarin accumulation in the roots of the wild type, f6’h1-1, cyp82c4-1, and bhlh121-2 plants subjected to Fe deficiency at pH 5.5, the pH at which sideretin biosynthesis and secretion preferentially occurs (Rajniak et al., 2018). As expected, the wild-type plants grown under this condition accumulated more sideretin and sideretin-glycosides in their roots than the wild-type plants grown under control conditions (Figure 4E; Supplemental Figure 5). Similar defects that were previously described for coumarin accumulation in cyp82c4-1 (i.e., increased fraxin [fraxetin glycoside] accumulation and the absence of sideretin and sideretin-glycosides) and in f6’h1-1 (i.e., no coumarin accumulation) were also confirmed. In the roots of the bhlh121-2 mutant, sideretin and sideretin-glycosides were not detected, and the accumulation of scopolin (scopoletin glycoside) was reduced. As expected, when grown at pH 7, Fe deficiency led to an increase in esculin and fraxin levels in the wild-type roots compared with the wild-type plants grown under control conditions (Supplemental Figure 6). By contrast, very little esculin and fraxin were detected in bhlh121-2 at pH 7. Scopolin accumulation was also reduced at this pH. These coumarin measurements, which support the results of expression analysis, indicate that bHLH121 is a transcriptional activator of fraxetin and sideretin biosynthesis.
bHLH121 Is Required to Trigger the Transcriptional Regulatory Cascade That Controls Fe Deficiency Responses
We performed RT-qPCR experiments to help decipher the positioning of bHLH121 within the transcriptional regulatory network that controls Fe homeostasis. This was achieved by analyzing the mRNA abundance of the major genes involved in controlling this pathway in 1-week-old seedlings subjected or not to Fe deficiency.
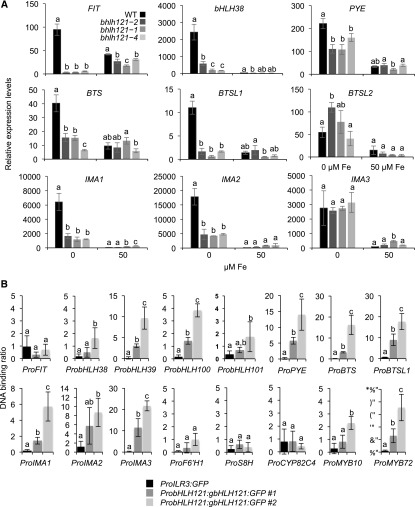
These experiments showed that the induction of bHLH38, bHLH39, bHLH100, bHLH101, and PYE expression in response to Fe deficiency was strongly reduced in all three mutants compared with the wild type, indicating that bHLH121 acts as an activator upstream of these TFs (Figure 5A; Supplemental Figure 7). By contrast, the expression levels of bHLH34, bHLH104, bHLH115, and ILR3 in the mutants were similar to those of the wild type (Supplemental Figure 7). The induction of FIT expression in response to Fe deficiency was abolished in all three bhlh121 mutant alleles, indicating that bHLH121 acts as an activator upstream of FIT (Figure 5A). The induction of BTS, BTSL1, IMA1, and IMA2 expression in response to Fe deficiency was also reduced in the mutants compared with the wild type, whereas no difference was observed for BTSL2 or IMA3 (Figure 5A).
Figure 5.
Expression of Several Genes Involved in the Fe Homeostasis Transcriptional Regulatory Network Is Compromised in the bhlh121 Loss-of-Function Mutants.
(A) Relative expression of FIT, bHLH38, PYE, BTS, BTSL1, IMA1, IMA2, and IMA3. Relative expression was determined by RT-qPCR in 1-week-old Arabidopsis seedlings grown on Fe-sufficient or Fe-deficient medium. Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 3 technical repeats from one representative experiment). Error bars show ±sd. Each experiment (biological repeat) comprised pooled RNA extracted from ∼30 seedlings and was independently repeated three times.
(B) ChIP-qPCR analysis of the binding of bHLH121 to the promoters of FIT, bHLH38, bHLH39, bHLH100, bHLH101, PYE, BTS, BTSL1, IMA1, IMA2, IMA3, F6’H1, S8H, CYP82C4, MYB10, and MYB72. Chromatin from the two complemented bhlh121-2 lines expressing the ProbHLH121:gbHLH121:GFP construct subjected to Fe deficiency was extracted using anti-GFP antibodies. Seedlings expressing GFP under the control of the ILR3 promoter (ProILR3:GFP) were used as a negative control. qPCR was used to quantify enrichment of bHLH121 on the selected gene promoters. Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 4 to 6 technical repeats from one representative experiment). Error bars show ±sd. Each experiment (biological repeat) comprised pooled chromatin immunoprecipitated from ∼500 seedlings (2 g) and was independently repeated two times.
We generated transgenic plants overexpressing bHLH121 under the control of the Arabidopsis UBIQUITIN10 promoter (ProUBI). Based on root length measurements, we selected one representative line (ProUBI:bHLH121-Ox3) for gene expression analysis (Supplemental Figure 8). RT-qPCR highlighted the notion that IRT1 and FRO2 were expressed at higher levels in lines carrying the ProUBI:bHLH121-Ox3 transgene under Fe deficiency than in the wild type (Supplemental Figure 9). A similar pattern of expression was observed for FIT, bHLH38, bHLH39, bHLH101, and PYE. Although not significant, bHLH100 expression also appeared to follow the same trend. These results further support the notion that bHLH121 is a key transcriptional activator that acts upstream of the Fe homeostasis network.
We then performed ChIP-qPCR experiments to determine whether bHLH121 directly interacts with the promoters of its potential targets. These experiments were conducted on two bhlh121-2 mutant lines carrying the ProbHLH121:gbHLH121:GFP transgene. These two lines were selected because the growth defects observed in bhlh121-2 at the seedling and mature plant stages were rescued, indicating that the ProbHLH121:gbHLH121:GFP transgene was functional and sufficient to complement the bhlh121-2 mutation (Supplemental Figures 10A, 11 and 12). The expression of IRT1, FIT, bHLH39, and BGLU42 was also rescued in these two complemented lines (Supplemental Figure 10B). The results presented in Figure 5B and Supplemental Figure 13 support the in vivo binding of bHLH121 to the promoters of bHLH38, bHLH39, bHLH100, bHLH101, PYE, BTS, BTSL1, IMA1, IMA2, and IMA3. These results also suggest that bHLH121 does not likely interact with the promoter of FIT. Notably, these data also indicate that bHLH121 interacts with the promoters of bHLH38, bHLH39, bHLH100, bHLH101, and PYE at the same locus as bHLH104, bHLH115, and ILR3 (Zhang et al., 2015; Liang et al., 2017). In addition, these data support the notion that bHLH121 directly interacts in vivo with the promoters of MYB10 and MYB72. Indeed, these results also suggest that bHLH121 most likely does not interact with the promoters of F6’H1, S8H, or CYP82C4.
We conducted additional complementation experiments to further investigate the role of bHLH121 within the Fe homeostasis regulatory network. For this purpose, FIT and bHLH38 (coding sequences) were overexpressed in bhlh121-2. These two TFs, which are known to function as heterodimers, were selected since expression data suggested that both genes act downstream of bHLH121. None of the transgenic lines was able to overcome the bhlh121-2 growth defects (Supplemental Figure 14). This observation further confirms the notion that bHLH121 acts as a transcriptional activator of both FIT and bHLH38.
Since bHLH121 interacts in vivo with bHLH34, bHLH104, bHLH115, and ILR3 and since these four TFs target similar regulatory loci, we performed comparative primary root growth analysis using the corresponding single mutants grown under control and Fe deficiency conditions. Under control conditions, bhlh121-2 was the sole mutant that displayed growth defects (Supplemental Figure 15A). Under Fe deficiency, the growth defects observed in bhlh121-2 were more pronounced than those in any other mutant (Supplemental Figure 15B).
Altogether, these results indicate that bHLH121 is a direct activator of bHLH38, bHLH39, bHLH100, bHLH101, PYE, BTS, BTSL1, IMA1, and IMA2 expression and that bHLH121 regulates the biosynthesis and secretion of Fe-mobilizing coumarins, partly by directly regulating the expression of MYB10 and MYB72 and indirectly regulating the expression of FIT. Since bHLH121 binds to the promoter of IMA3 based on ChIP-qPCR experiments, and since IMA3 transcript levels in the bhlh121 mutants are as high as in the wild type, these data also suggest that another TF might contribute to the regulation of IMA3 expression alongside bHLH121.
Cellular Localization of bHLH121 Depends on Fe Availability
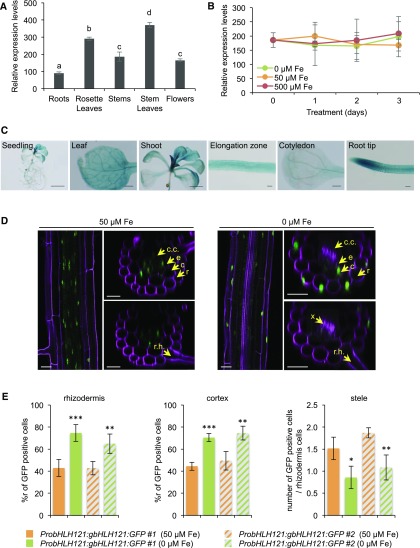
RT-qPCR analysis revealed that bHLH121 expression is not restricted to any specific organ in mature plants (Figure 6A). The highest level of mRNA accumulation was detected in leaves, that is, approximately three times higher than that in roots. RT-qPCR analysis also revealed that bHLH121 expression was not affected by any changes in Fe availability (i.e., deficiency or excess; Figure 6B).
Figure 6.
bHLH121 Expression Pattern and Protein Localization.
(A) bHLH121 expression pattern in 5-week-old Arabidopsis wild-type (WT) plants.
(B) Temporal expression pattern of bHLH121 in 1-week-old wild-type (WT) seedlings grown under Fe deficiency (0 μM) or excess (500 μM) conditions.
(A) and (B) Relative expression was determined by RT-qPCR. Means within each condition with the same letter are not significantly different according to one-way ANOVA followed by post hoc Tukey test, P < 0.05 (n = 3 technical repeats from one representative experiment). Error bars show ±sd. Each experiment (biological repeat) comprised pooled RNA extracted from tissues collected from three independent plants in (A) and from 30 seedlings in (B).
(C) bHLH121 promoter activity in seedlings grown under control conditions. Bar = 5 mm (seedling), 2 mm (shoot), 500 μm (cotyledon and leaf), and 100 μm (elongation zone and root tip).
(D) bHLH121:GFP localization in the roots (differentiation zone) of a complemented bhlh121-2 line (ProbHLH121:gbHLH121:GFP #1) subjected or not to Fe deficiency. Bar = 25 μm. c, cortex; c.c., central cylinder; e, endodermis; r, rhizodermis; r.h., root hair; x, xylem.
(E) (Left) Percentage of cells displaying GFP fluorescence in the rhizodermis cell layer (ratio between GFP-positive cells in the rhizodermis and total rhizodermis cells). (Middle) Percentage of cells displaying GFP fluorescence in the cortex cell layer (ratio between cortex GFP-positive cells and total cortex cells). (Right) Ratio between stele GFP-positive cells and total rhizodermis cells. The roots (differentiation zone) of four seedlings from two complemented bhlh121-2 lines (ProbHLH121:gbHLH121:GFP #1 and #2), subjected or not to Fe deficiency, were analyzed for each measurement. Error bars show ±sd. t test significant difference: **P < 0.01; ***P < 0.001.
To further describe the expression pattern of bHLH121, we cloned its promoter, fused it to the β-glucuronidase (GUS) reporter gene (ProbHLH121:GUS), and assessed the activity of ProbHLH121 in seedlings grown under control, Fe deficiency, and Fe excess conditions. GUS analysis confirmed the ubiquitous expression of bHLH121 (Figure 6C; Supplemental Figure 16). This analysis also suggested that GUS activity was, compared with control conditions, slightly enhanced in the cotyledons of seedlings grown under Fe deficiency and slightly repressed at the root tip when grown under Fe excess (Supplemental Figure 16).
To investigate whether bHLH121 protein accumulation and/or localization is affected by Fe availability, we subjected two independent complemented bhlh121-2 lines carrying the ProbHLH121:gbHLH121:GFP transgene to GFP fluorescence analysis by confocal microscopy. Under control conditions, GFP fluorescence in the root differentiation zone was mainly observed in the nuclei of cells localized to the central cylinder and endodermis (Figures 6D and 6E; Supplemental Figures 17 and 18). By contrast, under Fe deficiency, GFP fluorescence was primarily detected in nuclei of cells located at the rhizodermis and cortex. These experiments highlight the notion that the location of bHLH121 within the roots depends on Fe availability.
Notably, these experiments also suggested that the diffusion of propidium iodide (PI) within the root differentiation zone is affected by Fe availability, since xylem staining was observed under Fe deficiency and not under Fe replete conditions (Figures 6D and 6E; Supplemental Figures 17 and 18). Because the diffusion of PI into the stele is blocked upon the appearance of Casparian strips, Fe deficiency might alter the chemical or structural properties of this diffusion barrier, as has been shown for the suberization of the endodermis (Alassimone et al., 2010; Barberon et al., 2016). Whether there is a direct link between the diffusion properties of PI and the cellular localization of bHLH121, according to iron availability, remains to be elucidated.
Altogether, these results indicate that bHLH121 is ubiquitously expressed within the plant and that its expression is not affected by Fe availability. However, bHLH121 protein localization within the roots is modulated by the availability of Fe.
DISCUSSION
bHLH121-Dependent Complexes Are Required to Maintain Fe Homeostasis in Arabidopsis
The regulation of Fe homeostasis in plants relies on an intricate regulatory network involving several bHLH TFs (Gao et al., 2019). Several studies suggest that this network is composed of two interconnected subnetworks in Arabidopsis, with FIT (bHLH29) playing a predominant role in one network and ILR3 (bHLH105) in the other. Although overall the molecular connection between the FIT and ILR3 subnetworks is well characterized (Yuan et al., 2008; Wang et al., 2013; Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Kroh and Pilon, 2019; Tissot et al., 2019), it is still unclear how the expression of both subnetworks is synchronized in order to adapt Fe uptake to plant requirements for this micronutrient to sustain its growth and development.
In this study, we conducted Co-IP LC-MS/MS using ILR3 as bait to identify proteins whose activity might modulate Fe homeostasis and reunite the FIT and ILR3 subnetworks. bHLH121, a close homolog of PYE, was identified as a potential ILR3 protein interaction partner. The study of three bhlh121 loss-of-function mutant alleles revealed that bHLH121 is necessary to set up appropriate responses to Fe deficiency and to maintain Fe homeostasis during the entire life span of the plant (Figures 2 and 3; Supplemental Figures 3 to 6). bHLH121 acts as a transcriptional activator of key genes involved in controlling Fe homeostasis, including FIT, bHLH38, bHLH39, bHLH100, bHLH101, PYE, MYB10, MYB72, BTS, and BTSL1 as well as IMA1 and IMA2 (Figures 4C and 5A; Supplemental Figure 7). By contrast, the effect of bHLH121 overexpression on plant growth and gene expression was weak, suggesting that bHLH121 activity requires partners whose abundance is limiting (Supplemental Figures 8 and 9).
Y2H and BiFC experiments revealed that bHLH121 can interact not only with ILR3 but also with its three closest homologs: bHLH34, bHLH104, and bHLH115 (Figure 1; Supplemental Figure 1). When we compared the phenotypes of the loss-of-function mutants of these five TFs, bhlh121-2 presented stronger defects in root development than bhlh34, bhlh104-1, ilr3-3, and bhlh115-2 (Supplemental Figure 15). Interestingly, a previous study showed that the bhlh34 bhlh104 bhlh115 triple mutant displayed stronger defects in primary root growth than the single mutants in a manner similar to that of bhlh121-2 (Liang et al., 2017). Previous studies also showed that the expression of bHLH38, bHLH39, bHLH100, bHLH101, PYE, MYB10, MYB72, and BTS was moderately reduced in the bhlh34, bhlh104, bhlh115, and ilr3 single mutants and that this decrease was stronger in bhlh34 bhlh104 double mutants (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). Similar observations were made for FIT except that FIT expression was not affected by the bhlh115 mutation. These similarities between bhlh121, the bhlh34 bhlh104 double mutant, and the bhlh34 bhlh104 bhlh115 triple mutant suggest that bHLH121 might form heterodimers with bHLH34, bHLH104, bHLH115, and ILR3 that function in the same pathway as transcriptional activators. This hypothesis was further supported by ChIP-qPCR experiments that showed that ILR3 and its closest homologs, such as bHLH121, directly interact with the promoters of bHLH38, bHLH39, bHLH100, bHLH101, and PYE at the same locus. This approach also showed that bHLH121 directly interacts with the promoters of MYB10, MYB72, BTS, and BTSL1 as well as the promoters of IMA1 and IMA2 (Figure 5B).
Fe Availability Modulates the Cellular Localization of bHLH121
While the overall expression of bHLH121 is not significantly affected by the Fe status of the plant (Figures 6A to 6C; Supplemental Figure 16), analysis of bHLH121:GFP fluorescence in roots revealed that Fe availability modifies the localization of bHLH121 to specific cell types within this tissue. When Fe was not limiting, GFP fluorescence was mainly detected in the nuclei of cells in the stele, whereas under Fe deficiency, the signal was mainly observed in the cortex and rhizodermis cells (Figures 6D to 6F; Supplemental Figures 17 and 18). Several hypotheses could explain why GFP fluorescence is preferentially observed in certain cell types depending on Fe availability. For example, some posttranscriptional mechanism could modulate the stability and/or translation of bHLH121 mRNA in a cell type– and Fe-dependent manner, or perhaps some posttranslational mechanism regulates bHLH121 localization or cell-to-cell movement. Whether one or both of these mechanisms are involved in this process is still to be determined and should be studied in the future. Nevertheless, these results suggest that the genes that are targeted by bHLH121 differ depending on Fe availability.
Interestingly, ILR3 and PYE localization in roots also depends on Fe availability, as revealed by detecting GFP fluorescence from translational fusions. When Fe is not limiting, ILR3 is detected in all cell types, but at a lower level in the epidermis and cortex than in the stele, whereas only traces of PYE can be detected in the stele (Long et al., 2010; Samira et al., 2018; Gao et al., 2019). Under Fe deficiency, both proteins accumulate in all root cell types. Unlike bHLH121 and ILR3, PYE expression and promoter activity are strongly induced in response to Fe deprivation (Long et al., 2010; Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Samira et al., 2018; Tissot et al., 2019). Notably, the overlap between ILR3, PYE, and bHLH121 accumulation patterns in root cells varies with Fe availability (Supplemental Figure 19). For instance, when Fe is not limiting, bHLH121 and ILR3 are mainly present in the stele. The precise function of this dimer in this tissue is still to be determined. However, based on the bhlh121 mutant phenotypes, it is likely that the bHLH121-dependent dimers act to maintain the basal expression levels of genes potentially involved in Fe transport, partitioning, and/or storage. Indeed, since PYE is barely present in the stele when Fe is not limiting, the repressive activity of ILR3 must be low (Rampey et al., 2006; Tissot et al., 2019). This idea is in agreement with the expression patterns of ferritin genes: the expression of these genes alongside the vasculature is induced in the presence of Fe but repressed by ILR3/PYE dimers when Fe is limiting (Reyt et al., 2015; Kroh and Pilon, 2019; Tissot et al., 2019). By contrast, when Fe is not limiting, ILR3, PYE, and bHLH121 are barely detectable in the cortex and rhizodermis cells, which is consistent with the low levels of expression of genes encoding the Fe uptake machinery (Figure 4A; Kroh and Pilon, 2019). The accumulation of ILR3 and PYE in all root cell types when Fe availability is low likely allows the storage of Fe in vacuoles and ferritins to be inhibited, while the accumulation of bHLH121 in the cortex and rhizodermis favors Fe uptake.
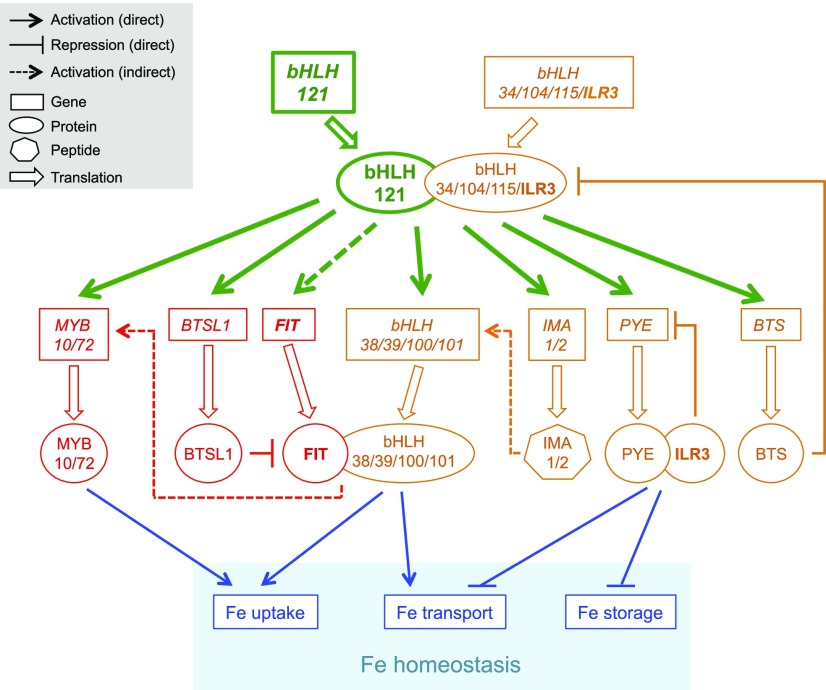
bHLH121 Acts Upstream of the Fe Homeostasis Network
The current results indicate that bHLH121 acts upstream of the Fe homeostasis network by forming heterodimers with ILR3 and its closest homologs to activate the expression of the large majority of genes encoding proteins and peptides involved in controlling Fe homeostasis (Figure 7). In the proposed model, when Fe availability is low, the bHLH121-dependent complexes activate the expression of FIT (indirectly) and the expression of bHLH38, bHLH39, bHLH100, and bHLH101 (directly). Considering that the overexpression of IMA3/FEP1 is sufficient to induce bHLH38 and bHLH39 expression, it could be hypothesized, given the structural similarity between the IMA peptides, that the concomitant induction of IMA1 and IMA2 expression (direct) by bHLH121 reinforces the induction of bHLH38 and bHLH39 expression (Grillet et al., 2018; Hirayama et al., 2018). Once induced, the encoded FIT protein heterodimerizes with bHLH38, bHLH39, bHLH100, and bHLH101 to promote the expression of structural genes involved in Fe uptake and transport. bHLH121-dependent complexes also directly activate the expression of BTSL1, whose encoded protein is predicted to target FIT to the 26S proteasome for degradation. The bHLH121-dependent complexes also directly induce MYB10 and MYB72 expression, enabling the biosynthesis and secretion of fraxetin and sideretin into the rhizosphere to improve Fe uptake.
Figure 7.
bHLH121 Acts Upstream of the Fe Homeostasis Network.
In Arabidopsis, two interconnected regulatory modules control Fe homeostasis: the first module (red) depends on the activity of FIT and the second module (orange) on the activity of ILR3 and its closest homologs (i.e., bHLH34, bHLH104, and bHLH115). The results presented in this study indicate that bHLH121 acts upstream of the Fe homeostasis network by forming heterodimers with ILR3 and its closest homologs to activate the expression of genes encoding several regulatory proteins belonging to both modules (green). In the proposed model, the bHLH121-dependent complexes indirectly activate FIT expression in response to Fe deficiency. Once induced, the encoded FIT protein heterodimerizes with bHLH38, bHLH39, bHLH100, and bHLH101 to induce the expression of structural genes involved in Fe uptake and transport (blue). bHLH121-dependent complexes are direct activators of MYB10 and MYB72 expression (two TFs regulating coumarin biosynthesis and secretion when Fe availability is limiting and whose expression is indirectly regulated by FIT). Another direct target of these bHLH121-depdendent complexes is BTSL1. BTSL1 is a RING E3 ubiquitin ligase that negatively regulates Fe deficiency responses by targeting FIT to the 26S proteasome for degradation. On the other hand, the bHLH121-dependent complexes directly activate the expression of bHLH38, bHLH39, bHLH100, and bHLH101 in response to Fe deficiency. The direct induction of IMA1 and IMA2 expression in response to Fe deficiency by the bHLH121-dependent complexes might reinforce the induction of bHLH38 and bHLH39 expression. The potential role of bHLH121 in regulating IMA3 expression remains to be elucidated. PYE (bHLH47) and BTS (E3 ubiquitin ligase) are two additional direct targets whose expression is also induced in response to Fe deficiency. Once induced, PYE heterodimerizes with ILR3 to repress the expression of genes involved in Fe transport and storage (blue) and most probably PYE itself, in a negative feedback regulatory loop. By contrast, BTS specifically targets bHLH115 and ILR3, leading to their degradation via the 26S proteasome to fine tune Fe uptake and avoid Fe excess that could be detrimental to the plant.
On the other hand, the bHLH121-dependent complexes directly activate the expression of PYE (bHLH47) and BTS (E3 ubiquitin ligase). Once induced, PYE heterodimerizes with ILR3 to repress the expression of genes involved in Fe transport and storage, and likely PYE itself, in a negative feedback regulatory loop. By contrast, BTS specifically targets bHLH115 and ILR3, leading to their degradation via the 26S proteasome to fine tune Fe uptake and avoid Fe excess that could be detrimental to the plant.
Indeed, further experiments will be necessary to fully demonstrate this model, in particular colocalization analyses with all the major TFs, regulatory proteins (i.e., BTS, BTSL1, BTSL2) and peptides (i.e., IMA1, IMA2) involved in controlling Fe homeostasis to document the spatial distribution of the different actors depending on Fe availability. Another important question that will have to be addressed is how bHLH121 is regulated at the posttranscriptional and/or posttranslational level. Since the activity of bHLH121 depends on its interaction with ILR3 and its closet homologs, determining how the stability of bHLH34 and bHLH104 is modulated may help answer this question.
METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. bhlh121-1, bhlh121-2, and bhlh121-4 homozygous loss-of-function mutant alleles (CRISPR-Cas9 in Col-0) were generated during this study (Supplemental Figure 2). In addition, the following mutant lines were used: bhlh34 (Li et al., 2016), bhlh104-1 (Zhang et al., 2015), ilr3-3 (Li et al., 2016; Tissot et al., 2019), and bhlh115-2 (Liang et al., 2017).
Growth Conditions
In Vitro Culture
Seedlings were germinated and grown under long-day conditions (16-h-light/8-h-dark cycle; light intensity, 120 µmol/cm2/s provided by Osram 18-W 840 Lumilux neon tubes) on half-strength Murashige and Skoog medium containing 0.05% (w/v) MES, 1% (w/v) Suc, and 0.7% (w/v) agar for 7 d. The Fe concentration was 0 µM (deficiency), 50 µM (control), 200 µM (mild excess), or 500 µM (excess), and it was provided as Fe(III)-EDTA. For the ChIP experiments, 1-week-old seedlings grown under control conditions were exposed to Fe deficiency for 3 d prior to analysis. For the GUS experiments, 1-week-old seedlings grown under control conditions were exposed to Fe deficiency for 5 d or Fe excess for 1 d prior to analysis (control seedlings were maintained at 50 µM Fe).
Hydroponic Culture
Plants were grown for 3 weeks under short-day conditions (8-h-light/16-h-dark cycle; light intensity, 120 µmol/cm2/s provided by a mix of sodium-vapor and metal halide 400-W lamps) in the presence of 50 µM Fe-EDTA as described previously (Fourcroy et al., 2016). The plants were grown for 10 more days in the presence (50 µM) or absence of Fe.
Measurement of Root Length
After the growing period, roots were scanned and measured using ImageJ 1.52a software (National Institutes of Health).
Cloning
Homozygous bhlh121 mutant alleles (loss-of-function in the Col-0 genetic background) were obtained by CRISPR-Cas9 gene editing. For this purpose, two single guide RNAs (sgRNAs) were designed and cloned into the pRM-Cas9 binary vector prior to plant transformation (Wang et al., 2015). For the design of the sgRNA, the data from four different websites were merged: CRISPRSCAN (http://www.crisprscan.org/), WU-Crispr (http://crispr.wustl.edu/), CHOP CHOP (http://chopchop.cbu.uib.no/), and CRISPR RGEN tool (http://www.rgenome.net/cas-designer/). The sgRNAs located in exons that were the most highly represented among these four databases and close to the translation initiation site (ATG) were manually selected. The number of potential off-targets of these selected sgRNAs was determined using the Cas-OFFinder tool of rgenome (http://www.rgenome.net/cas-offinder/). Finally, the two sgRNAs presenting the smallest number of putative off-targets were chosen. Three different homozygous mutant alleles were obtained following plant transformation (Bechtold et al., 1993), including one displaying a 78-bp deletion (bhlh121-2) in the coding sequence (656 bp at the genomic level) and two alleles with a single nucleotide insertion (bhlh121-1 and bhlh121-4; Supplemental Figure 2A). To ensure that the generated constructs were not generating off-targets and thus affecting the integrity of key genes involved in the transcriptional regulation of Fe homeostasis, the regions between the start and stop codons of bHLH11, bHLH34, bHLH104, bHLH115, ILR3, and FIT were sequenced. No differences from the wild-type sequences were observed (Supplemental Figures 20 to 22).
The bHLH121 promoter (ProbHLH121, 2586 bp prior to the start codon) was fused to GUS in the pGWB3 binary vector (Nakagawa et al., 2007) as described in Xu et al. (2013). The wild-type plants were transformed (agroinfiltration) with ProbHLH121:GUS, and nine independent lines were assayed for GUS activity. The same procedure was used to fuse the bHLH121 promoter and genomic region (without the stop codon) to GFP (pGWB4 binary vector; ProbHLH121:gbHLH121:GFP). bhlh121-2 mutant plants were transformed with ProbHLH121:gbHLH121:GFP, and complemented lines were selected for GFP fluorescence analysis and ChIP-qPCR experiments. To overexpress FIT and bHLH38 in bhlh121-2, the corresponding coding sequences were cloned downstream of the strong, ubiquitous promoter of the Arabidopsis UBIQUITIN10 gene (pUB-GFP-Dest binary vector; Grefen et al., 2010). Lines carrying ProUBI:FIT and ProUBI:bHLH38 transgenes (10 independent lines for each construct) were analyzed for bhlh121-2 phenotypic complementation.
All PCR products were obtained using high-fidelity Phusion DNA polymerase, and each construct was sequenced to ensure its integrity. The primers used and the sequences of the guide RNAs are described in Supplemental Data Set 1.
Biochemical Analysis
Chlorophyll Content
Chlorophylls from 25 mg of leaf tissue (fresh weight) or five leaf discs (diameter, 0.35 cm) were extracted in 1 mL of 100% acetone in the dark under agitation. The absorbance (A) at 661.8 and 644.8 nm was then measured. Total chlorophyll content was assessed using the following equation: and was expressed as micrograms per gram fresh weight or micrograms per square centimeter (Lichtenthaler, 1987).
Iron Measurements
Twenty milligrams of ground material (dry weight) per sample was mixed with 750 μL of nitric oxide (65% [v/v]) and 250 μL of hydrogen peroxide (30% [v/v]) prior to homogenization. Following 10 min of incubation at room temperature, the samples were mineralized using the Microwave digestion systems (Berghof). Once mineralized, the nitric oxide proportion present in the samples was adjusted to 5 to 10% of the final volume by adding ultrapure water. Fe content present in the samples was then measured by microwave plasma atomic emission spectroscopy (Agilent Technologies).
Histochemical GUS Detection
Seedlings expressing ProbHLH121:GUS gene fusions were transferred into a 100 mM phosphate buffer, pH 7.5, solution containing 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 0.1% (v/v) Triton X-100, and 10 mM Na2-EDTA. Prior to incubating the samples at 37°C in the dark (overnight), a 1-h vacuum treatment (room temperature) was applied. Following GUS staining, chlorophylls were removed by gently shaking the samples in a clearing solution of acetic acid:ethanol (14:86). The samples were stored in 70% (v/v) ethanol prior to observation under a light microscope.
FCR Activity
Ten to 20 mg of fresh root tissue was incubated for 1 h in the dark with gentle shaking in 2 mL of FCR buffer (100 µM Fe3+-EDTA, 300 µM ferrozine, and 10 mM MES, pH 5.5). An identical assay without plant samples was used as a blank. The concentration of Fe2+-ferrozine complex (which displays a purple coloration) was determined by reading absorbance at 560 nm using a Xenius microplate reader.
Iron Staining by Perls/DAB
Iron staining was performed according to Roschzttardtz et al. (2009). The embryos were dissected from dry seeds that were previously imbibed in distilled water for 3 h. Isolated embryos or 4-d-old seedlings were vacuum infiltrated with a solution containing 2% (v/v) HCl and 2% (w/v) potassium ferrocyanide for 15 min and incubated for 30 min at room temperature. After washing with distilled water, the embryos were incubated in a methanol solution containing 10 mM NaN3 and 0.3% (v/v) H2O2 for 1 h and washed with 100 mM Na-phosphate buffer, pH 7.4. For the intensification reaction, the embryos were incubated for 10 min in 100 mM Na-phosphate buffer, pH 7.4, solution containing 0.025% (w/v) DAB hydrate, 0.005% (v/v) H2O2, and 0.005% (w/v) CoCl2·2H2O. The reaction was stopped by rinsing with distilled water. The embryos were visualized under a stereoscopic microscope (Nikon SMZ800) and imaged with a Coolpix 4500 charge-coupled device (CCD) digital camera (Nikon).
HPLC Analysis of Root Extracts
For HPLC analysis of coumarins, 30 mg of root material frozen in liquid nitrogen was ground in the presence of glass beads and extracted with 400 μL of methanol:water [80:20 (v/v)]. The supernatants were vacuum dried and resuspended in 100 μL of water:acetonitrile [90:10] containing 0.1% (v/v) of formic acid. HPLC-fluorescence analysis was performed using a 1220 Infinity II LC system (Agilent Technologies) coupled to a Prostar 363 fluorescence detector (Varian). Separation was done on an analytical HPLC column (Aeris 3.6 µm WIDEPORE XB-C8 200 Å, 100 × 2.1 mm; Phenomenex), with a gradient mobile phase made with 0.1% (v/v) formic acid in water (A) and 0.1% (v/v) formic acid in acetonitrile (B) and a flow rate of 0.25 mL/min. The gradient program started at 8% B for 2 min and increased linearly to 30% B in 13 min and then to 50% B in 1 min. This proportion was maintained for 4 min and returned linearly to initial conditions in 1 min. The column was allowed to stabilize for 9 min at the initial conditions. Absorbance was monitored at λ = 338 nm. Fluorescence was monitored at λexc 365 and λem 460 nm. External calibration was done using commercial coumarins: esculin (Sigma), esculetin (Sigma), fraxetin (Sigma) scopoletin (Sigma), scopolin (TargetMol), and fraxin (TargetMol). Sideretin and sideretin-glycosides identification was confirmed by LC-MS/MS.
Co-IP MS Analyses
Co-IP Analysis
Experiments were performed in triplicate for the bait ProILR3:gILR3:GFP and for the control ProILR3:GFP. For protein extraction, 0.7 g of root tissue was suspended in 1.5 mL of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% [v/v] glycerol, 5 mM MgCl2, 0.1% [w/v] IGEPAL CA-630, 2 mM DTT, 1× Complete Mini EDTA-Free Protease Inhibitor Cocktail Tablet [Roche], and 1× Phosphatase Inhibitor Cocktail 3 [Sigma]). The lysates were cleared by centrifugation at 10,000g, and IPs were performed using an µMACS GFP isolation kit (MACS purification system, Milteny Biotech) according to the manufacturer’s instructions, except that lysis buffer was used for all bead washes.
Eluted proteins were loaded for a short run (15 min at 100 V) on a 10% Mini-PROTEAN TGX pre-cast gel (Bio-Rad). The whole lane was manually excised and sequentially rinsed with water, 25 mM ammonium bicarbonate, and 50% (v/v) acetonitrile in 25 mM ammonium bicarbonate and then dehydrated with acetonitrile and dried at room temperature. Proteins were reduced with 10 mM DTT for 45 min at 56°C and then alkylated with 55 mM iodoacetamide for 30 min at room temperature in the dark. Excess iodoacetamide was removed, and gel slices were rinsed twice with 50% (v/v) acetonitrile in 25 mM ammonium bicarbonate, dehydrated with acetonitrile, and dried at room temperature. Proteins were digested by trypsin (Sequencing Grade Modified, Promega) in 25 mM ammonium bicarbonate overnight at 37°C. Peptides were eluted by step elutions with 2% (v/v) formic acid and twice with 50% (v/v) acetonitrile in 2% (v/v) formic acid. The supernatants were pooled and evaporated in a vacuum centrifuge. Peptides were resuspended in 8 μL of 2% (v/v) formic acid, and 6 μL samples were injected for LC-MS/MS analysis.
MS Analysis
The LC-MS/MS analyses were performed using an Ultimate 3000 RSLC nano system (Thermo Fisher Scientific) interfaced online with a nano easy ion source and a Q Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific). The samples were analyzed in data-dependent acquisition mode. The protein digests were loaded onto a pre-column (PepMap 100 C18, 5-µm particle size, 100-Å pore size, 300-µm i.d. × 5-mm length; Thermo Fisher Scientific) at a flow rate of 10 µL/min for 3 min. The peptides were separated in a reverse-phase column (PepMap C18, 2-µm particle size, 100-Å pore size, 75-µm i.d. × 50-cm length; Thermo Fisher Scientific) at a flow rate of 300 nL/min.
The loading buffer (solvent A) was 0.1% (v/v) formic acid in water, and the elution buffer (solvent B) was 0.1% (v/v) formic acid in 80% (v/v) acetonitrile. The linear gradient used was 2 to 25% of solvent B in 103 min, followed by 25 to 40% of solvent B from 103 to 123 min and 40 to 90% of solvent B from 123 to 125 min. The total run time was 150 min, including a high organic wash step and re-equilibration step.
The Q Exactive Plus mass analyzer was operated in positive ESI mode at 1.8 kV. In data-dependent acquisition mode, the top 10 precursors were acquired between 375 and 1500 m/z with a 2-Thomson selection window, dynamic exclusion of 40 s, normalized collision energy of 27, and resolutions of 70,000 for MS and 17,500 for MS2.
Data Analysis
Spectra were recorded with Xcalibur software (4.3.31.9; Thermo Fisher Scientific). The raw files were analyzed with MaxQuant version 1.5.5.1 (Tyanova et al., 2016) using default settings. The minimal peptide length was set to 6. The criteria “Trypsin/P” was chosen as the digestion enzyme. Carbamidomethylation of Cys was selected as a fixed modification and oxidation of Met and acetylation (protein N terminus) as variable modifications. Up to two missed cleavages were allowed. The mass tolerance for the precursor was 20 and 4.5 ppm for the first and main searches, respectively, and that for the fragment ions was 20 ppm. The files were searched against an in-house modified Arabidopsis 10 database (35,417 entries). Identified proteins were filtered according to the following criteria: at least one different trypsin peptides with at least one unique peptide and at least one razor peptide. Minimum score for modified peptides was set to 20. A peptide-spectrum match false discovery rate and a protein false discovery rate below 0.05 were required. Using the above-mentioned criteria, the rates of false peptide sequence assignment and false protein identification were lower than 5%. Proteins were selected as potential ILR3 interactors if they were identified in the three IP replicates (i.e., ILR3:GFP) and if they were not identified in any of the control IPs (i.e., GFP).
Y2H Assays
Experiments were performed as descried in Touraine et al. (2019) using the yeast AH109 (Saccharomyces cerevisiae) reporter strain and the pDEST22 and pDEST32 vectors (Invitrogen).
GFP Fluorescence Analysis by Confocal Microscopy
Roots of 7-d-old bhlh121-2 seedlings expressing ProbHLH121:gbHLH121:GFP were imaged under an LSM 880 microscope (Zeiss) with a W Plan Apochromat 20×/1.0 objective. Before imaging, roots were stained with PI for 5 min. GFP and PI were detected by excitation with an argon laser at 488 nm. The emission filter was set at 500 to 550 nm for GFP and 633 to 695 nm for PI.
BiFC Assays
Experiments were performed as described in Couturier et al. (2014). Results are representative of the observations made on at least 20 cells. All PCR products were obtained using high-fidelity Phusion DNA polymerase, and each construct was sequenced to ensure its integrity. All primers used are described in Supplemental Data Set 1.
Gene Expression Analysis
Total RNAs were extracted from the samples using the Tri-Reagent (Molecular Research Center) method. Briefly, each sample was homogenized in 1 mL of Tri-Reagent solution mixed with 160 μL of chloroform:isoamyl alcohol (24:1). Following centrifugation (10 min, 13,000 rpm, 4°C), total RNAs present in the aqueous phase were precipitated by the addition of 400 μL of isopropanol followed by another centrifugation. The pellets were washed twice with 70% ethanol and dried prior to resuspension in RNase-free water. For each sample, 1 µg of total RNA treated with DNase was reverse transcribed into cDNA using a RevertAid kit (Thermo Fisher Scientific). RT-qPCR analyses were performed using a LightCycler 480 (Roche) and LC480-SYBR-Green master I reaction mix (Roche). PROTEIN PHOSPHATASE2A SUBUNIT A3 (PP2AA3) was used as a reference gene (Czechowski et al., 2005). Expression levels were calculated using the comparative threshold cycle method. The primers used are described in Supplemental Data Set 1.
ChIP Assays
Experiments were performed as described by Gendrel et al. (2002), with modifications: (1) nuclei were isolated with the following buffer: 20 mM PIPES-KOH, pH 7.6, 1 M hexylene glycol, 10 mM MgCl2, 0.1 mM EGTA, 15 mM NaCl, 60 mM KCl, 0.5% (v/v) Triton X100, 5 mM β-mercaptoethanol, protease inhibitor cocktail (complete tablets EASYpack, Roche), and (2) after immunoprecipitation using antibodies raised against GFP (1/1000 dilution; ab290, Abcam), DNA was purified with an IPURE Kit (Diagenode). The resulting DNA was analyzed by qPCR analysis using a LightCycler 480 (Roche) and LC480-SYBR-Green master I reaction mix (Roche). Data are presented as promoter target enrichment over input, using the following formula: (Supplemental Data Sets 2 and 3). All primers used are described in Supplemental Figure 23 and Supplemental Data Set 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: bHLH11 (At4g36060); bHLH29/FIT (At2g28160); bHLH34 (At3g23210); BGLU42 (At5g36890); bHLH38 (At3g56970); bHLH39 (At3g56980); bHLH47/PYE (At3g47640); bHLH100 (At2g41240); bHLH101 (At5g04150); bHLH104 (At4g14410); bHLH105/ILR3 (At5g54680); bHLH115 (At1g51070); bHLH121 (At3g19860); BTS (At3g18290); BTSL1 (At1g74770); BTSL2 (At1g18910); CYP82C4 (At4g31940); FRO2 (At1g01580); IMA1 (At1g47400); IMA2 (At1g47395); IMA3 (At2g30766); IRT1 (At4g19690); MYB10 (At3g12820); MYB72 (At1g56160); PDR9 (At3g53480); PP2AA3 (At1g13320); S8H (At3g12900). All raw MS data and Maxquant files generated have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD014620. The ANOVA tables are displayed in Supplemental Data Set 4.
Supplemental Data
Supplemental Figure 1. bHLH11 interacts with ILR3 and with its closest homologs and forms homodimers.
Supplemental Figure 2. bhlh121 loss-of-function mutations.
Supplemental Figure 3. Fe staining of wild type and bhlh121 embryos.
Supplemental Figure 4. The bhlh121 loss-of-function mutant plants have decreased accumulation of Fe in roots and shoots.
Supplemental Figure 5. Coumarin accumulation in wild type, cyp8c24-1, f6’h1 and bhlh121-2.
Supplemental Figure 6. Coumarin accumulation in wild type and bhlh121-2.
Supplemental Figure 7. bHLH34, bHLH104, bHLH115 and ILR3 expression is not compromised in the bhlh121 loss-of-function mutants.
Supplemental Figure 8. Phenotypes of bHLH121 overexpressing lines.
Supplemental Figure 9. bHLH121 overexpression induces the expression of several Fe homeostasis-related genes.
Supplemental Figure 10. Complementation of the bhlh121-2 seedling defects with the ProbHLH121:gbHLH121:GFP transgene.
Supplemental Figure 11. Complementation of the bhlh121-2 seedling defects with the ProbHLH121:gbHLH121:GFP transgene.
Supplemental Figure 12. Complementation of the bhlh121-2 defects with the ProbHLH121:gbHLH121:GFP transgene .
Supplemental Figure 13. bHLH121 binding to the promoters of FIT, bHLH38, bHLH39, bHLH100, bHLH101, PYE, BTS, BTSL1, IMA1, IMA2, IMA3, F6’H1, S8H, CYP82C4, MYB10 and MYB72 by ChIP-qPCR.
Supplemental Figure 14. bhlh121-2 complementation assays by overexpressing FIT and bHLH38.
Supplemental Figure 15. Root growth phenotypes of bhlh121-2, bhlh34, bhlh104-1, ilr3-3 and bhlh115-2.
Supplemental Figure 16. bHLH121 promoter activity.
Supplemental Figure 17. bHLH121 protein localization in longitudinal sections of roots.
Supplemental Figure 18. bHLH121 protein localization in transverse sections of roots.
Supplemental Figure 19. Schematic representation of the localization patterns of PYE, ILR3 and bHLH121 (GFP translational fusion) in the presence or absence of Fe in the maturation zone of Arabidopsis roots.
Supplemental Figure 20. Blastn sequence comparisons between bhlh121-2 and the wild type at the bHLH11 and bHLH34 loci (cds regions).
Supplemental Figure 21. Blastn sequence comparisons between bhlh121-2 and the wild type at the bHLH104 and bHLH115 loci (cds regions).
Supplemental Figure 22. Blastn sequence comparisons between bhlh121-2 and the wild type at the ILR3 and FIT loci (cds regions).
Supplemental Figure 23. Promoter structure diagrams for the genes assayed in ChIP-qPCR experiments.
Supplemental Table. ILR3 interacting proteins identified by Co-IP LC-MS/MS.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. ChIP-qPCR experiment #1.
Supplemental Data Set 3. ChIP-qPCR experiment #2.
Supplemental Data Set 4. ANOVA tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Carine Alcon, Geneviève Conejero, and the Montpellier Rio-Imaging platform (PHIV platform, La Gaillarde, Montpellier, France) for expertise and assistance with microscopy and Sandrine Chay (Biochimie et Physiologie Moléculaire des Plantes, SAME platform, Montpellier, France) for technical support with plant Fe determination. We also thank Guillaume Cazals for LC-MS/MS analysis for the identification of coumarins (“Laboratoire de Mesures Physiques,” analytical facilities of Montpellier University, France). We thank Nathalie Berger and Alexandre Martinière for help in preparing this article. Support was provided by the Agence Nationale pour la Recherche (ANR-17-CE20-0008-01 to K.R.) and from the China Scholarship Council (to F.G.). This work was also supported by the Comisión Nacional de Investigación Científica y Tecnológica-Environmental Compliance Services (CONICYT-ECOS project C18B04).
AUTHOR CONTRIBUTIONS
F.Gao, K.R., M.B., F.V., E.I., and C.D. conceived and designed the experiments; F.Gao, K.R., M.B., N.N., H.R., V.R., F.V., and E.I. performed the experiments; F.Gao, K.R., M.B., F.Gaymard, H.R., F.V., E.I., and C.D. analyzed the data; V.S., F.Gaymard, F.V., E.I., and C.D. contributed reagents/materials/analysis tools; and F.Gao, K.R., F.Gaymard, F.V., E.I., and C.D. wrote the article.
References
- Alassimone J., Naseer S., Geldner N. (2010). A developmental framework for endodermal differentiation and polarity. Proc. Natl. Acad. Sci. USA 107: 5214–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Vermeer J.E., De Bellis D., Wang P., Naseer S., Andersen T.G., Humbel B.M., Nawrath C., Takano J., Salt D.E., Geldner N. (2016). Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164: 447–459. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316: 1194–1199. [Google Scholar]
- Briat J.F., Dubos C., Gaymard F. (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20: 33–40. [DOI] [PubMed] [Google Scholar]
- Brumbarova T., Bauer P., Ivanov R. (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 20: 124–133. [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J., Wu H.C., Dhalleine T., Pégeot H., Sudre D., Gualberto J.M., Jacquot J.P., Gaymard F., Vignols F., Rouhier N. (2014). Monothiol glutaredoxin-BolA interactions: Redox control of Arabidopsis thaliana BolA2 and SufE1. Mol. Plant 7: 187–205. [DOI] [PubMed] [Google Scholar]
- Cui Y., Chen C.L., Cui M., Zhou W.J., Wu H.L., Ling H.Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 11: 1166–1183. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcroy P., Sisó-Terraza P., Sudre D., Savirón M., Reyt G., Gaymard F., Abadía A., Abadia J., Alvarez-Fernández A., Briat J.F. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201: 155–167. [DOI] [PubMed] [Google Scholar]
- Fourcroy P., Tissot N., Gaymard F., Briat J.F., Dubos C. (2016). Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+) transport system. Mol. Plant 9: 485–488. [DOI] [PubMed] [Google Scholar]
- Gao F., Robe K., Gaymard F., Izquierdo E., Dubos C. (2019). The transcriptional control of iron homeostasis in plants: A tale of bHLH transcription factors? Front. Plant Sci. 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Grillet L., Lan P., Li W., Mokkapati G., Schmidt W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 4: 953–963. [DOI] [PubMed] [Google Scholar]
- Hänsch R., Mendel R.R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12: 259–266. [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Hindt M.N., Akmakjian G.Z., Pivarski K.L., Punshon T., Baxter I., Salt D.E., Guerinot M.L. (2017). BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9: 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Lei G.J., Yamaji N., Nakagawa N., Ma J.F. (2018). The putative peptide gene FEP1 regulates iron deficiency response in Arabidopsis. Plant Cell Physiol. 9: 1739–1752. [DOI] [PubMed] [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. (2012). Fitting into the harsh reality: Regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5: 27–42. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N.K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152. [DOI] [PubMed] [Google Scholar]
- Kroh G.E., Pilon M. (2019). Connecting the negatives and positives of plant iron homeostasis. New Phytol. 223: 1052–1055. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang H., Ai Q., Liang G., Yu D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170: 2478–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang H., Li X., Ai Q., Yu D. (2017). bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68: 1743–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler F.W. (1987). Karl Freudenberg, Burckhardt Helferich, Hermann O. L. Fischer: A centennial tribute. Carbohydr. Res. 164: 1–22. [DOI] [PubMed] [Google Scholar]
- Ling H.Q., Bauer P., Bereczky Z., Keller B., Ganal M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. USA 99: 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.A., Tsukagoshi H., Busch W., Lahner B., Salt D.E., Benfey P.N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthiadis A., Long T.A. (2016). Further insight into BRUTUS domain composition and functionality. Plant Signal. Behav. 11: e1204508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Palmer C.M., Hindt M.N., Schmidt H., Clemens S., Guerinot M.L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9: e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajniak J., Giehl R.F.H., Chang E., Murgia I., von Wirén N., Sattely E.S. (2018). Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol. 14: 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R.A., Woodward A.W., Hobbs B.N., Tierney M.P., Lahner B., Salt D.E., Bartel B. (2006). An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyt G., Boudouf S., Boucherez J., Gaymard F., Briat J.F. (2015). Iron- and ferritin-dependent reactive oxygen species distribution: Impact on Arabidopsis root system architecture. Mol. Plant 8: 439–453. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J., Connorton J.M., Kruse I., Green R.T., Franceschetti M., Chen Y.T., Cui Y., Ling H.Q., Yeh K.C., Balk J. (2019). Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. USA 116: 17584–17591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J., Lin W.D., Fu G.M., Abadía J., López-Millán A.F., Schmidt W. (2013). Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 162: 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H., Conéjéro G., Curie C., Mari S. (2009). Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol. 151: 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samira R., Li B., Kliebenstein D., Li C., Davis E., Gillikin J.W., Long T.A. (2018). The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. 97: 297–309. [DOI] [PubMed] [Google Scholar]
- Santi S., Schmidt W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schmid N.B., Giehl R.F., Döll S., Mock H.P., Strehmel N., Scheel D., Kong X., Hider R.C., von Wirén N. (2014). Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 164: 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D., Samira R., Matthiadis A., Gillikin J.W., Long T.A. (2015). Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167: 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A., Grinvalds C., Barberon M., Curie C., Vert G. (2011). Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J. 66: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Siwinska J., Siatkowska K., Olry A., Grosjean J., Hehn A., Bourgaud F., Meharg A.A., Carey M., Lojkowska E., Ihnatowicz A. (2018). Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J. Exp. Bot. 69: 1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot N., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 223: 1433–1446. [DOI] [PubMed] [Google Scholar]
- Touraine B., et al. (2019). Iron-sulfur protein NFU2 is required for branched-chain amino acid synthesis in Arabidopsis roots. J. Exp. Bot. 70: 1875–1889. [DOI] [PubMed] [Google Scholar]
- Tsai H.H., Rodríguez-Celma J., Lan P., Wu Y.C., Vélez-Bermúdez I.C., Schmidt W. (2018). Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol. 177: 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11: 2301–2319. [DOI] [PubMed] [Google Scholar]
- Wang N., Cui Y., Liu Y., Fan H., Du J., Huang Z., Yuan Y., Wu H., Ling H.Q. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6: 503–513. [DOI] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198: 59–70. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan Y.X., Zhang J., Wang D.W., Ling H.Q. (2005). AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 15: 613–621. [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Hanson J., Pieterse C.M. (2014). β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204: 368–379. [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu B., Li M., Feng D., Jin H., Wang P., Liu J., Xiong F., Wang J., Wang H.B. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27: 787–805. [DOI] [PMC free article] [PubMed] [Google Scholar]