MED25 regulates the alternative splicing of JAZ genes through recruiting the splicing factors PRP39a and PRP40a, thereby preventing JAZ splice variant-mediated excessive desensitization of jasmonate signaling.
Abstract
JASMONATE ZIM-DOMAIN (JAZ) transcriptional repressors are key regulators of jasmonate (JA) signaling in plants. At the resting stage, the C-terminal Jas motifs of JAZ proteins bind the transcription factor MYC2 to repress JA signaling. Upon hormone elicitation, the Jas motif binds the hormone receptor CORONATINE INSENSITIVE1, which mediates proteasomal degradation of JAZs and thereby allowing the Mediator subunit MED25 to activate MYC2. Subsequently, plants desensitize JA signaling by feedback generation of dominant JAZ splice variants that repress MYC2. Here we report the mechanistic function of Arabidopsis (Arabidopsis thaliana) MED25 in regulating the alternative splicing of JAZ genes through recruiting the splicing factors PRE-mRNA-PROCESSING PROTEIN 39a (PRP39a) and PRP40a. We demonstrate that JA-induced generation of JAZ splice variants depends on MED25 and that MED25 recruits PRP39a and PRP40a to promote the full splicing of JAZ genes. Therefore, MED25 forms a module with PRP39a and PRP40a to prevent excessive desensitization of JA signaling mediated by JAZ splice variants.
INTRODUCTION
Jasmonate (JA) is a lipid-derived hormone that regulates diverse aspects of plant immunity and development (Browse, 2009; Wasternack and Hause, 2013; Chini et al., 2016; Goossens et al., 2016; Zhai et al., 2017). In Arabidopsis (Arabidopsis thaliana), JA triggers a genome-wide transcriptional program that is largely regulated by MYC2 (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011; Kazan and Manners, 2013; Zhai et al., 2013; Chini et al., 2016; Zhai et al., 2017). The activity of this master transcription factor depends on its physical and functional interactions with MED25, a subunit of the plant Mediator transcriptional coactivator complex (Çevik et al., 2012; Chen et al., 2012; An et al., 2017; Zhai et al., 2018; Wang et al., 2019). At the resting stage, a group of JASMONATE ZIM-DOMAIN (JAZ) proteins physically interact with MYC2, thereby repressing the expression of JA-responsive genes (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Pauwels et al., 2010).
In the presence of the bioactive JA ligand, jasmonoyl-isoleucine (JA-Ile), JAZ proteins function as hormone coreceptors by forming a JA-Ile–dependent coreceptor complex with CORONATINE-INSENSITIVE1 (COI1), the F-box subunit of the ubiquitin ligase SCFCOI1 (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002; Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009; Sheard et al., 2010). SCFCOI1-dependent degradation of JAZ repressors leads to the liberation of MYC2 from transcriptional repression (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Fonseca et al., 2009; Sheard et al., 2010). Therefore, JAZ proteins can act as both transcriptional repressors (in the absence of JA-Ile) and hormone coreceptors (in the presence of JA-Ile).
In-depth investigations have provided insights into the mechanism by which JAZ proteins switch between their repressor and coreceptor functions. JAZ proteins contain a featured C-terminal JA-associated (Jas) motif, which is necessary for interactions with COI1 and the active hormone, JA-Ile (Chini et al., 2007; Thines et al., 2007; Melotto et al., 2008; Sheard et al., 2010; Pauwels and Goossens, 2011). The JAZ degron, which is located within the Jas domain, is the minimal amino acid sequence sufficient for COI1 and JA-Ile binding (Sheard et al., 2010; Pauwels and Goossens, 2011). Notably, the Jas motif is also necessary for the interactions of JAZ proteins with MYC2 (Chini et al., 2007; Melotto et al., 2008; Pauwels and Goossens, 2011). Structural studies have pointed to a critical role for the JAZ degron in switching JAZ proteins between their repressor and coreceptor functions (Zhang et al., 2015).
In addition to their repressor and coreceptor functions, emerging evidence suggests that JAZ proteins play a pivotal role in regulating the desensitization of JA signaling, which is necessary to prevent uncontrolled JA responses. For example, most Arabidopsis JAZ genes contain a highly conserved Jas intron, and alternative splicing (AS) involving the Jas intron generates a repertoire of JAZ splice variants lacking the intact C-terminal Jas motif (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013). Transgenic Arabidopsis plants expressing these JAZ splice variants show attenuated JA responses, as these JAZ splice variants still retain the ability to repress MYC2 but are more resistant to hormone-induced degradation than the wild type (Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013). A recent study has provided structural insight into the mechanism by which JAZ splice variants desensitize JA signaling: Some JAZ splice variants contain an N-terminal cryptic MYC-interaction domain (CMID) that inhibits the access of MED25 to the transcriptional activation domain of MYC transcription factors (Zhang et al., 2017).
Although Jas intron-dependent AS of JAZ genes is thought to provide a general mechanism to desensitize or deactivate JA responses, how this process is properly regulated remains elusive. For example, it is unclear how plants control the generation of dominant JAZ splice variants to proper levels to prevent excessive and/or uncontrolled desensitization of JA signaling. The splicing factors involved in Jas intron-dependent AS of JAZ genes, and how these splicing factors are recruited, remains enigmatic.
“Mediator” is an evolutionarily conserved multisubunit coactivator complex whose activity is essential for RNA polymerase II (Pol II)-dependent gene transcription (Björklund and Gustafsson, 2005; Kornberg, 2005; Malik and Roeder, 2005, 2010; Poss et al., 2013; Allen and Taatjes, 2015). Since its discovery in yeast and animals (Fondell et al., 1996), the most extensively investigated function of Mediator has been its ability to orchestrate transcription factor-dependent assembly of the Pol II preinitiation complex (PIC) via discrete interactions with signal-dependent transcription factors and Pol II (Kornberg, 2005; Malik and Roeder, 2005, 2010; Soutourina et al., 2011). In addition to its role in transcriptional initiation, novel functions are continuously being ascribed to yeast and animal Mediator in controlling almost every stage of Pol II-dependent gene transcription, including epigenetic regulation, transcriptional elongation and termination, noncoding RNA activation, chromatin loop formation, and perhaps mRNA processing (Malik and Roeder, 2010; Huang et al., 2012; Carlsten et al., 2013; Conaway and Conaway, 2013; Poss et al., 2013; Yin and Wang, 2014; Allen and Taatjes, 2015; Malik, 2016). Indeed, it has been shown that the human MED23 subunit regulates AS through interacting with the splicing factor hnRNP L (Huang et al., 2012). Recent transcriptome analysis revealed that the function of MED23 in regulating AS is conserved in Arabidopsis (Dolan and Chapple et al., 2018).
Isolation of the Arabidopsis Mediator complex revealed 21 conserved and six plant-specific subunits (Bäckström et al., 2007). Despite the identification of several plant Mediator subunits that are implicated in the regulation of plant development and adaptive responses (Kidd et al., 2011; Samanta and Thakur, 2015; Yang et al., 2016), our mechanistic understanding of the roles of plant Mediator is still in its infancy.
We have shown that the plant Mediator subunit MED25 physically and functionally interacts with MYC2, thereby playing a pivotal role in PIC formation during the activation of MYC2-mediated transcription of JA-responsive genes (Chen et al., 2012). Furthermore, MED25 is also involved in the assembly of a MYC2–MED25 functional transcription complex, which acts as an integrative hub to coordinate the actions of multiple regulators during hormone-triggered activation of MYC2 (An et al., 2017; Du et al., 2017; Liu et al., 2019; Wang et al., 2019; You et al., 2019).
Here, we report the mechanistic function of MED25 in regulating Jas intron-dependent AS of JAZ genes. We show that JA-induced production of dominant JAZ splice variants depends on the functions of MYC2 and MED25. Using affinity purification and mass spectrometry, we found that PRE-mRNA-PROCESSING PROTEIN 39a (PRP39a; Lockhart and Rymond, 1994; Wang et al., 2007; Kanno et al., 2017) and PRP40a (Kao and Siliciano, 1996; Kang et al., 2009), two subunits of the evolutionarily conserved spliceosomal U1 small nuclear ribonucleoprotein particle (snRNP) involved in cotranscriptional AS, are components of MYC2–MED25 functional transcription complex. We demonstrate that MED25 controls JA-induced recruitment of PRP39a and PRP40a to JAZ loci to facilitate the full splicing of Jas intron. Therefore, the MED25-PRP39a/PRP40a module plays a critical role in preventing JAZ splice variant-mediated excessive desensitization of JA signaling. This study exemplifies how a Mediator subunit integrates the effects of general splicing factors into a specific signaling pathway to accurately control gene expression.
RESULTS
Generation of Dominant JAZ Splice Variants Is an Integral Part of MYC2- and MED25-Dependent JA Signaling
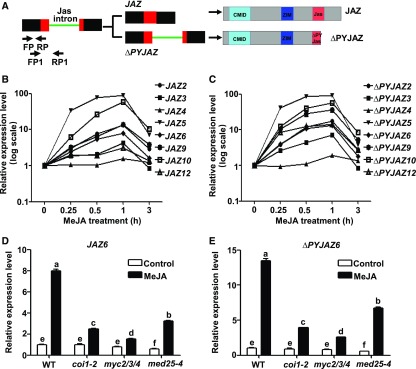
Among the Arabidopsis JAZ genes, JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ9, JAZ10, and JAZ12 contain a conserved Jas intron that splits the Jas motif into a 20 amino acid N-terminal JAZ degron and a seven amino acid C-terminal (X5PY) submotif (Chung et al., 2010; Figure 1A). Jas intron-dependent AS generates truncated JAZ splice variants (ΔPYJAZs hereafter) that play an important role in the desensitization of JA signaling (Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013; Figure 1A). To investigate how JA regulates the AS of JAZ genes, we performed RT-qPCR with RNA from methyl jasmonate (MeJA)-treated wild-type seedlings to measure fully spliced JAZ transcripts and ΔPYJAZ transcripts. We found that both fully spliced JAZ transcripts (Figure 1B) and ΔPYJAZ transcripts (Figure 1C) were quickly induced at 0.25 h after MeJA treatment and reached to a peak at 1 h after MeJA treatment, indicating that the MeJA-mediated induction kinetics of ΔPYJAZs is similar to that of the corresponding fully spliced JAZs.
Figure 1.
MeJA-Induced Generation of ∆PYJAZs Depends on MYC2 and MED25.
(A) Schematic diagram of Jas intron-dependent AS of JAZ and its corresponding translational products. In the gene structure, boxes represent exons, lines represent introns, red boxes represent Jas motif-encoding regions, and the green line represents the Jas intron. Arrows indicate forward primers (FP or FP1) and reverse primers (RP or RP1) used for RT-qPCR to amplify transcripts in which the Jas intron is spliced (FP/RP) or retained (FP1/RP1). In the protein structures of JAZ and ∆PYJAZ, cyan boxes represent CMID, blue boxes represent the ZIM domain, and red boxes represent the Jas motif.
(B) and (C) RT-qPCR showing fully spliced transcripts (B) or Jas intron-retained transcripts (C) of Jas intron-contained JAZ genes in MeJA-treated wild-type seedlings. Ten-d–old seedlings were treated without or with 100 μM of MeJA for the indicated time before RNA extraction.
(D) and (E) RT-qPCR showing JAZ6 (D) and ΔPYJAZ6 (E) expression in response to MeJA in wild type, coi1-2, myc2/3/4, and med25-4 seedlings. Ten-d–old seedlings were treated with 100 μM of MeJA for 60 min before RNA extraction.
(B) to (E) Expression levels of target genes were normalized to ACTIN7, and the expression levels in wild type without MeJA treatment were arbitrarily set to 1. Data shown are mean values of three biological repeats with sd.
(D) and (E) Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01).
We then selected JAZ6, which contains a representative Jas intron, as a model to investigate whether the AS of JAZ genes depends on the core JA signaling components including COI1, MYC2, and MED25. RT-qPCR assays indicated that both fully spliced JAZ6 (JAZ6 hereafter) and ΔPYJAZ6 were induced by MeJA treatment in the wild type (Figures 1D and 1E). Importantly, MeJA-mediated induction of both JAZ6 and ΔPYJAZ6 was largely abolished in the coi1-2 mutant, a weak mutant allele of COI1 (Figures 1D and 1E; Xu et al., 2002; Moreno et al., 2013). Parallel experiments produced similar results for other Jas-intron–containing JAZ genes (Supplemental Figure 1), indicating that MeJA-mediated induction of ΔPYJAZs strictly depends on the hormone receptor protein COI1.
Similarly, MeJA-induced expression of ΔPYJAZ6 (Figure 1E) and other ΔPYJAZs (Supplemental Figure 1) was also significantly reduced in the myc2/3/4 triple mutant (Song et al., 2014) and the med25-4 mutant (Chen et al., 2012), indicating that MeJA-mediated induction of ΔPYJAZs also depends on the MYC transcription factors and the Mediator subunit MED25.
In addition to ΔPYJAZs, a previous study identified the JAZ10.4 splice variant from wounded leaves; JAZ10.4 encodes a dominant spliced variant lacking the intact Jas motif (ΔJasJAZ10; Chung and Howe, 2009). Our parallel experiments showed that MeJA induces the expression of JAZ10.4, and this induction is greatly impaired in coi1-2, myc2/3/4, and med25-4 (Supplemental Figure 1), indicating that MeJA-mediated induction of JAZ10.4 also depends on COI1, the MYC transcription factors, and MED25.
Overexpression of ∆PYJAZ6 Leads To Attenuated JA Responses
To evaluate the functional significance of Jas intron-dependent AS on JA signaling, we generated transgenic Arabidopsis plants (∆PYJAZ6-GFP) overexpressing ∆PYJAZ6 cDNA fused with GFP under the control of the 35S promoter (Supplemental Figure 2A). As a control, we also generated JAZ6-GFP plants overexpressing fully spliced JAZ6 cDNA fused with GFP (Supplemental Figure 2A). Transgenic lines in which the expression levels of the corresponding transgenes were largely comparable were selected for further analysis (Supplemental Figure 2B).
The selected transgenic lines were then subject to a standard JA response assay. In Arabidopsis, VEGETATIVE STORAGE PROTEIN1 (VSP1) and VSP2 are widely used marker genes for JA-regulated wounding responses, whereas PLANT DEFENSIN1.2 (PDF1.2) and THIONIN2.1 (THI2.1) are widely used marker genes for JA-regulated pathogen responses (Berger et al., 1995; Penninckx et al., 1996). JAZ genes are early JA-responsive genes that are directly targeted by the master transcription factor MYC2 (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). MeJA-induced expression of all these JA-responsive genes was not obviously altered in JAZ6-GFP plants compared with wild type (Supplemental Figures 3A to 3E), indicating that overexpressing fully spliced JAZ6 does not alter JA responses. Indeed, overexpressing several fully spliced JAZ proteins did not obviously alter JA responses in a previous study due to their rapid degradation upon JA stimulation (Chini et al., 2007). However, MeJA-induced expression of VSP1, VSP2, PDF1.2, THI2.1, and JAZ2 was significantly decreased in ∆PYJAZ6-GFP plants compared with the wild type (Supplemental Figures 3A to 3E), indicating that ∆PYJAZ6-GFP plants show reduced JA responses in terms of MeJA-induced defense gene expression.
In JA-induced root growth inhibition assays, ∆PYJAZ6-GFP plants, but not JAZ6-GFP, showed an attenuated JA response compared with their wild-type counterparts (Supplemental Figure 3F). Together, these results are consistent with previous observations that overexpression of JAZ splice variants led to attenuated JA responses (Chung and Howe, 2009; Chung et al., 2010; Moreno et al., 2013).
∆PYJAZ6 Competes with MED25 for Binding to MYC2
We then explored the mechanism by which ∆PYJAZ6 desensitizes JA signaling. First, we performed in vitro pull-down assays to compare hormone-dependent interactions of JAZ6 or ∆PYJAZ6 with COI1. In the presence of coronatine, a potent agonist of the JA-Ile receptor, purified recombinant histidine-tagged JAZ6 (His-JAZ6), but not His-∆PYJAZ6, recovered myc-tagged COI1 from leaf extracts of COI1-myc plants (Xu et al., 2002; Supplemental Figure 4A), indicating that ∆PYJAZ6 cannot interact with COI1 in the presence of coronatine. Consistently, whereas JAZ6-GFP extracted from JAZ6-GFP plants was completely degraded within 10 min of MeJA treatment, ∆PYJAZ6-GFP from ∆PYJAZ6-GFP plants remained stable even after 60 min of MeJA treatment (Supplemental Figure 4B), indicating that ∆PYJAZ6-GFP is more resistant to MeJA-induced degradation than JAZ6-GFP.
Although ∆PYJAZ6 lacks the intact C-terminal Jas motif, which is important for interactions not only with COI1 but also with MYC2, this JAZ splice variant still contains an N-terminal CMID (Figure 1A), which is important for binding to MYC transcription factors (Zhang et al., 2017). Like His-JAZ6, His-∆PYJAZ6 pulled down MYC2-myc (Chen et al., 2011) in our in vitro pull-down assays (Supplemental Figure 4C), suggesting that ∆PYJAZ6 can still interact with MYC2.
Because MYC2-regulated transcription of JA-responsive genes largely depends on its interaction with the Mediator subunit MED25 (Chen et al., 2012), and JAZ proteins exert their repressor function through competing with MED25 for interaction with MYC transcription factors (Zhang et al., 2015), we asked whether the hormone-resistant ∆PYJAZ6 could compete with MED25 for interaction with MYC2. Our firefly luciferase (LUC) complementation imaging (LCI) assays (Chen et al., 2008) showed that, in Nicotiana benthamiana leaves, the interaction between MED25 and MYC2 was dramatically reduced when ∆PYJAZ6-GFP was coexpressed with MED25-nLUC and cLUC-MYC2 (Supplemental Figures 4D and 4E), suggesting that ∆PYJAZ6 inhibits the MED25–MYC2 interaction.
To substantiate these observations, we performed in vitro pull-down experiments using purified maltose binding protein (MBP)-tagged MYC2, a glutathione s-transferase (GST)-tagged MED25 fragment containing its Middle and Activator-Interacting Domains (GST-MED25MA), and His-JAZ6 or His-∆PYJAZ6. In the absence of His-JAZ6, GST-MED25 was pulled down by MBP-MYC2 (Supplemental Figure 4F, lane 6), indicating that GST-MED25 interacts with MBP-MYC2. The interaction between GST-MED25 and MBP-MYC2 was dramatically reduced by the presence of increasing amounts of His-JAZ6 (Supplemental Figure 4F, lanes 6 to 8), suggesting that JAZ6 has an inhibitory effect on the GST-MED25–MBP-MYC2 interaction. In parallel experiments, His-∆PYJAZ6 exhibited a similar inhibitory effect on the GST-MED25–MBP-MYC2 interaction (Supplemental Figure 4F, lanes 6, 9, and 10). These results support the notion that the effects of ∆PYJAZ6 on desensitizing JA signaling are likely achieved through its interference with the MED25–MYC2 interaction.
Splicing Factors PRP39a and PRP40a Associate with MED25
Considering that MED25 plays a fundamental role in the activation of MYC2-dependent JA responses (Çevik et al., 2012; Chen et al., 2012; An et al., 2017), our findings that JA-induced generation of ∆PYJAZs depends on MED25 and that ∆PYJAZs in turn desensitize JA responses by inhibiting the MED25–MYC2 interaction revealed seemingly contradictory functions of MED25 in this process. We reasoned that, as an effective coactivator of MYC2 and other JA-inducible transcription factors (Çevik et al., 2012; Chen et al., 2012; An et al., 2017), MED25 must have evolved sophisticated mechanisms to reconcile and coordinate these opposing functions. To explore these mechanisms, we performed an affinity purification-coupled mass spectrometry assay with MED25-myc plants (Chen et al., 2012) to identify MED25-associated proteins (You et al., 2019). In this assay, PRP39a and PRP40a, together with two other RNA processing proteins, were identified as MED25-associated proteins (Supplemental Table). In Arabidopsis, PRP39 is encoded by the homologous genes PRP39a and PRP39b (Wang et al., 2007; Kanno et al., 2017; Supplemental Figure 5A), whereas PRP40 is encoded by three homologs, PRP40a, PRP40b, and PRP40c (Kang et al., 2009; Supplemental Figure 5B). It is noteworthy that PRP40a has been previously identified as the MED35 subunit of the plant Mediator complex (Bäckström et al., 2007).
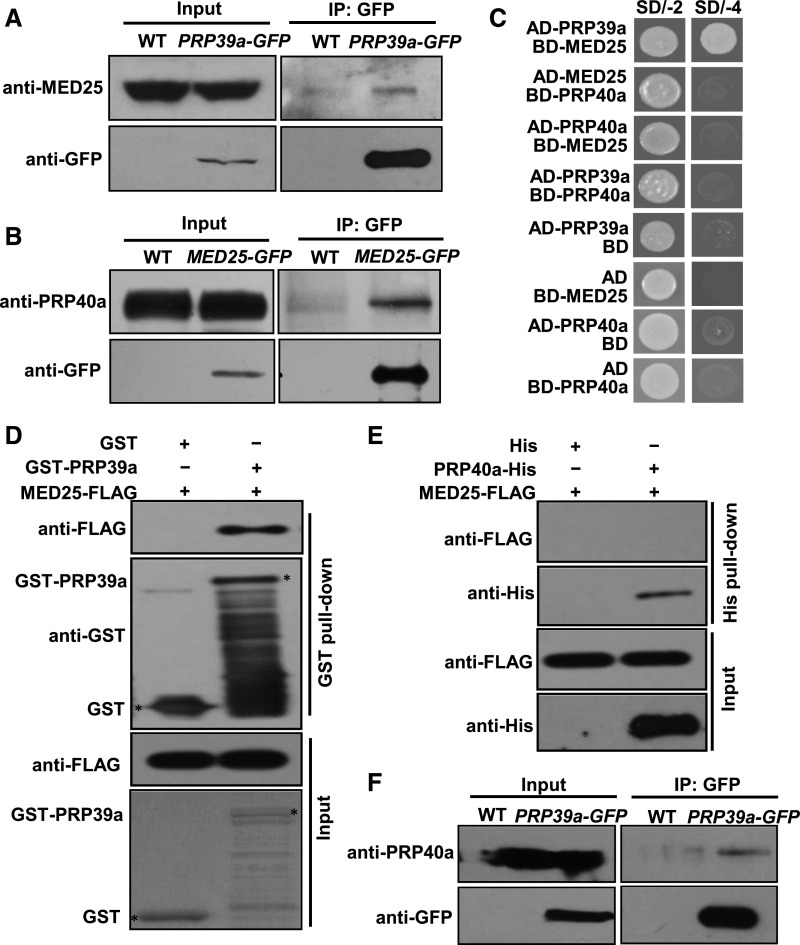
PRP39a and PRP40a associated with MED25 in the assay, suggesting that these splicing factors interact with MED25 in planta. Indeed, in co-immunoprecipitation (co-IP) assays with PRP39a-GFP plants (Supplemental Figures 6A and 6C), PRP39a-GFP pulled down native MED25 (Figure 2A). In co-IP assays with MED25-GFP plants (Chen et al., 2012), MED25-GFP pulled down native PRP40a (Figure 2B). To determine whether PRP39a or PRP40a directly interacts with MED25, we performed yeast two-hybrid (Y2H) assays, finding that PRP39a, but not PRP40a, interacted with MED25 (Figure 2C). Immunoblot analysis of the yeast strains confirmed that all the fusion proteins were expressed in yeast cells (Supplemental Figure 7), indicating that the absence of interaction cannot be attributed to a lack of protein expression. Consistently, in in vitro pull-down assays, in vitro-translated FLAG-tagged MED25 (MED25-FLAG) was pulled down by recombinant GST-PRP39a (Figure 2D), whereas in vitro-translated MED25-FLAG was not pulled down by PRP40a-His (Figure 2E). These results suggest that MED25 directly interacts with PRP39a and associates with PRP40a.
Figure 2.
PRP39a and PRP40a Associate with MED25.
(A) Co-IP assays to verify the interaction of PRP39a and MED25 in vivo. Protein extracts from 10-d–old wild type and PRP39a-GFP seedlings were immunoprecipitated with GFP antibody-bound agarose beads. Total and immunoprecipitated proteins were analyzed by immunoblotting using the indicated antibodies.
(B) Co-IP assays to verify the interaction of PRP40a and MED25 in vivo. Protein extracts from 10-d–old wild type and MED25-GFP seedlings were immunoprecipitated with GFP antibody-bound agarose beads. Total and immunoprecipitated proteins were analyzed by immunoblotting with the indicated antibodies.
(C) Y2H assays to verify the interaction of PRP39a, PRP40a, and MED25 in yeast. Transformed yeast strains with the indicated combinations of constructs were plated on SD medium lacking Leu and Trp (SD/-2) or lacking His, Ade, Leu, and Trp (SD/-4).
(D) In vitro pull-down assay to test the interaction of PRP39a and MED25. A certain amount of MED25-FLAG protein was incubated with recombinant GST-PRP39a fusion protein. Proteins were pulled down by GST Bind Resin, and the eluates were analyzed by immunoblotting with the indicated antibodies.
(E) In vitro pull-down assay to test the interaction of PRP40a and MED25. A certain amount of MED25-FLAG protein was incubated with in vitro-translated PRP40a-His fusion protein. Proteins were pulled down by Ni-NTA His Bind Resin, and the eluates were analyzed by immunoblotting with the indicated antibodies.
(F) Co-IP assays to verify the interaction of PRP39a and PRP40a in vivo. Protein extracts from 10-d–old wild-type and PRP39a-GFP seedlings were immunoprecipitated with GFP antibody-bound agarose beads. Total and immunoprecipitated proteins were analyzed by immunoblotting with the indicated antibodies.
Although PRP39a-GFP pulled down native PRP40a in co-IP assays (Figure 2F), we failed to detect the interaction between PRP39a and PRP40a in Y2H assays (Figure 2C; Supplemental Figure 7), suggesting that even though PRP39a and PRP40a exist in the same protein complex in vivo, they do not directly interact. This finding is consistent with the previous observation that the Prp39 and Prp40 subunits of yeast U1 snRNP do not directly interact with each other (Görnemann et al., 2011).
MYC2 and MED25 Control the JA-Induced Expression of PRP40a and its Homologous Genes
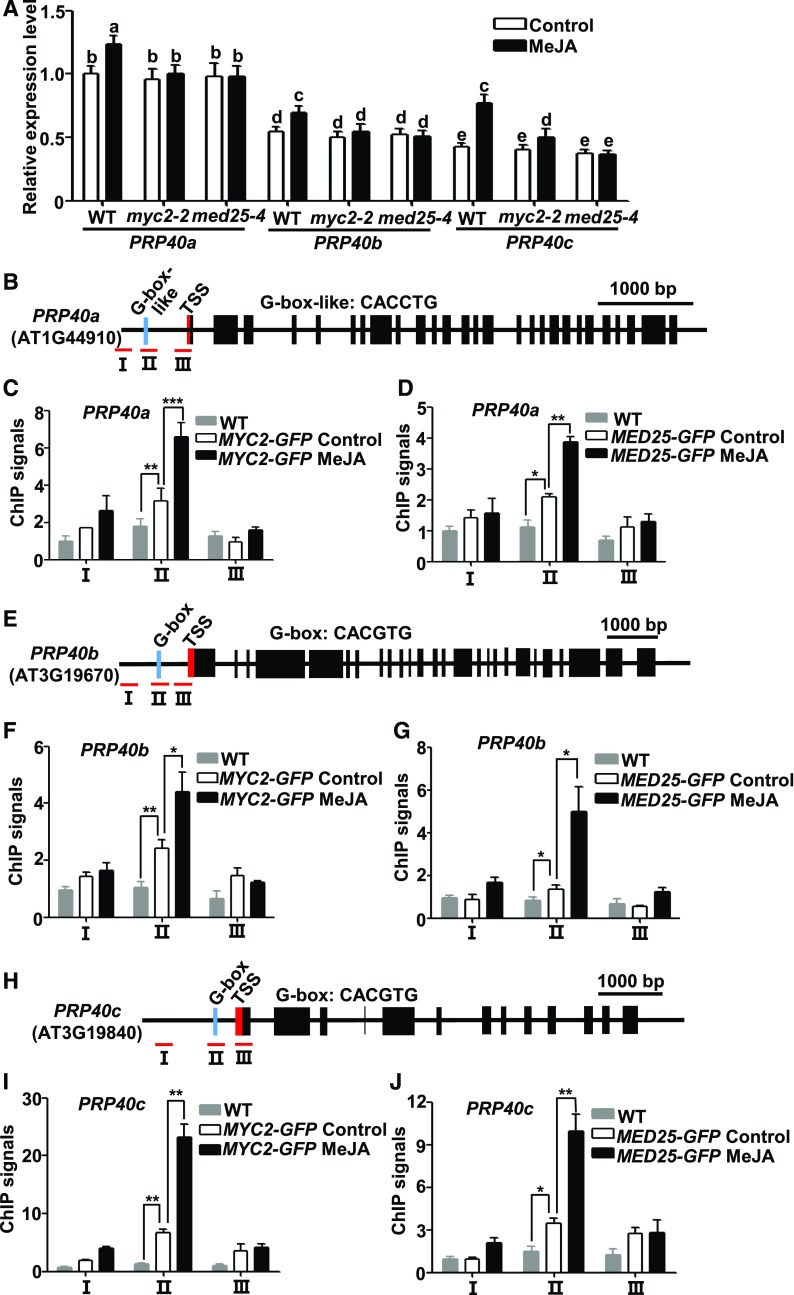
We then investigated whether JA regulates the expression of PRP39a, PRP40a, and their homologous genes. Our repeated RT-qPCR assays revealed that, whereas MeJA treatment showed negligible effects on PRP39a and PRP39b expression (Supplemental Figure 8), it had slight yet significant induction on PRP40a expression (Figure 3A). To validate this observation, we performed immunoblot analysis with anti-PRP40a antibody and found that MeJA treatment led to increased accumulation of the PRP40a protein (Supplemental Figure 9). In parallel experiments, we found that the expression levels of PRP40b and PRP40c were also significantly increased upon MeJA treatment (Figure 3A). Notably, MeJA-mediated induction of PRP40a, PRP40b, and PRP40c was significantly reduced in the myc2-2 and med25-4 mutants (Figure 3A), indicating that MeJA-mediated induction of these splicing factor genes depends on MYC2 and MED25.
Figure 3.
MeJA Induces PRP40a, PRP40b, and PRP40c through MYC2 and MED25.
(A) RT-qPCR showing the expression of PRP40a, PRP40b, and PRP40c in response to MeJA in wild type, med25-4, and myc2-2 seedlings. Ten-d–old seedlings were treated without or with 100 μM of MeJA for 30 min before RNA extraction. Expression levels of target genes were normalized to ACTIN7, and the expression levels in wild type without MeJA treatment were arbitrarily set to 1. Results shown are mean values of three biological repeats with sd. Three independent experiments with different seed batches show similar results. Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01). WT, wild type.
(B), (E), and (H) Schematic diagrams of PRP40a (B), PRP40b (E), and PRP40c (H) gene structures and PCR amplicons (indicated as I to III) used for ChIP-qPCR. Cyan box represents putative MYC2 binding G-box or G-box-like motifs; red box represents transcription start site (TSS). Scale bar = 1,000 bp.
(C), (D), (F), (G), (I), and (J) ChIP-qPCR showing the enrichment of MYC2 (C), (F), (I) and MED25 (D), (G), (J) on PRP40a (C) and (D), PRP40b (F) and (G) and PRP40c (I) and (J) chromatin upon MeJA elicitation. Ten-d–old MYC2-GFP and MED25-GFP seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking; wild-type plants without MeJA treatment were used as a negative control. Chromatin from each sample was immunoprecipitated with anti-GFP antibodies. ChIP signals were displayed as the percentage of precipitated DNA relative to input DNA. The value of amplicon I of wild type without MeJA treatment was arbitrarily set to 1. Results shown are mean values of three biological repeats with sd. Asterisks denote significance according to Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. WT, wild type.
Sequence analysis identified G-box or G-box–like motifs in the promoter regions of PRP40a, PRP40b, and PRP40c (Figures 3B, 3E, and 3H). Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assays with MYC2-GFP plants (Zhai et al., 2013) revealed enriched binding of MYC2 to the G-box or G-box–like regions of the PRP40a, PRP40b, and PRP40c promoters, especially under MeJA treatment (Figures 3C, 3F, and 3I). ChIP-qPCR assays with MED25-GFP plants (Chen et al., 2012) indicated that MeJA also enhanced the enrichment of MED25 at the same promoter regions (Figures 3D, 3G, and 3J). Together, these results indicate that MYC2 and MED25 control the JA-induced activation of PRP40a and its homologous genes.
PRP39a and PRP40a Facilitate the Splicing of Jas Intron
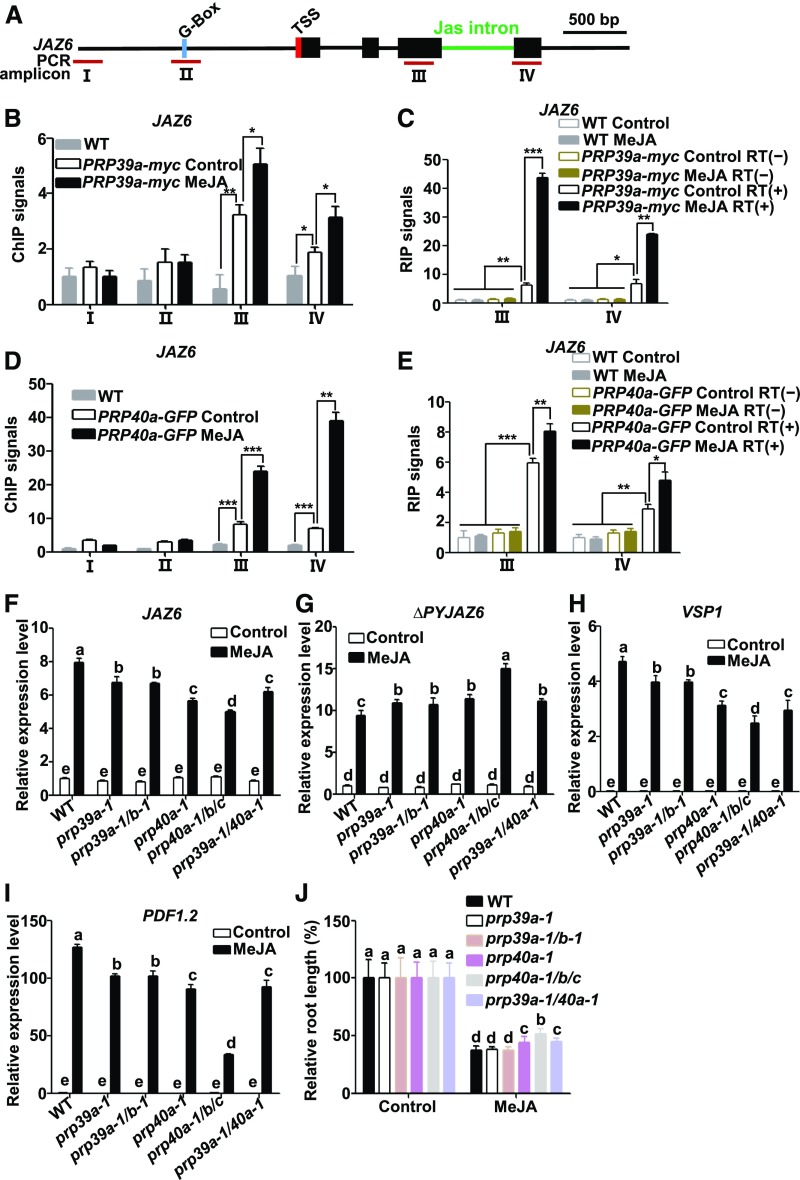
We reasoned that PRP39a and PRP40a are recruited to regulate Jas intron-dependent AS of JAZ genes. To test this hypothesis, we performed ChIP-qPCR assays with PRP39a-myc plants (Supplemental Figure 6A) and found that the enriched binding of PRP39a-myc to the gene body of JAZ6 was significantly higher than that to its promoter region (Figures 4A and 4B) and was markedly enhanced by MeJA treatment (Figure 4B). These results indicate that hormone elicitation induces the recruitment of PRP39a to the JAZ6 locus.
Figure 4.
PRP39a and PRP40a Facilitate the Full Splicing of JAZ6.
(A) Schematic diagrams of JAZ6 and PCR amplicons (indicated as I to IV) used for ChIP-qPCR and RIP-RT-qPCR. Cyan box represents the putative MYC2 binding G-box motif; green line represents the Jas intron; red box represents transcription start site (TSS). Dark-red lines labeled “III” and “IV” represent PCR amplicons used for ChIP-qPCR and RIP-RT-qPCR. Scale bar = 500 bp.
(B) and (D) ChIP-qPCR showing the enrichment of PRP39a (B) and PRP40a (D) on JAZ6 chromatin in response to MeJA. Ten-d–old PRP39a-myc or PRP40a-GFP seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking; wild-type plants without MeJA treatment were used as a negative control. Chromatin from each sample was immunoprecipitated with anti-myc or anti-GFP antibodies. ChIP signals were displayed as the percentage of precipitated DNA relative to input DNA. The value of amplicon I of wild type without MeJA treatment was arbitrarily set to 1. Data shown are mean values of three biological repeats with sd. Asterisks denote significance according to Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. WT, wild type.
(C) and (E) RIP–RT-qPCR showing the enrichment of PRP39a (C) and PRP40a (E) on JAZ6 pre-mRNA in response to MeJA. Ten-d–old PRP39a-myc or PRP40a-GFP seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking; wild-type and precipitated pre-mRNAs without reverse transcription (RT[−]) were used as negative controls. The pre-mRNA from each sample was immunoprecipitated with anti-myc or anti-GFP antibodies. Precipitated pre-mRNA and input pre-mRNA was quantified by RT-qPCR for the indicated amplicons. RIP signals were displayed as the percentage of precipitated pre-mRNA relative to input pre-mRNA. The value of amplicon III of wild type without MeJA treatment was arbitrarily set to 1. Results shown are mean values of three biological repeats with sd. Asterisks denote significance according to Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. WT, wild type.
(F) and (G) RT-qPCR showing JAZ6 (F) and ΔPYJAZ6 (G) expression in response to 100 μM of MeJA in wild type, prp39a-1, prp39a-1/b-1 (prp39a-1/prp39b-1), prp40a-1, prp40a-1/b/c (prp40a-1/prp40b/prp40c), and prp39a-1/40a-1 (prp39a-1/prp40a-1) seedlings. Ten-d–old seedlings were treated without or with MeJA for 30 min before RNA extraction. Expression levels of target genes were normalized to ACTIN7, and the expression levels in wild type without MeJA treatment were arbitrarily set to 1. Data shown are mean values of three biological repeats with sd.
(H) and (I) RT-qPCR showing the MeJA-induced expression of VSP1 (6-h treatment; [H]), PDF1.2 (48-h treatment; [I]) in wild type, prp39a-1, prp39a-1/b-1, prp40a-1, prp40a-1/b/c, and prp39a-1/40a-1 seedlings. Ten-d–old seedlings were treated without or with 100 μM of MeJA for the indicated time points before RNA extraction. Expression levels of target genes were normalized to ACTIN7. Results shown are mean values of three biological repeats with sd. WT, wild type.
(J) Root growth inhibition assay of 8-d–old wild type, prp39a-1, prp39a-1/b-1, prp40a-1, prp40a-1/b/c, and prp39a-1/40a-1 seedlings. Plants were grown on 1/2 MS medium containing 20 μM of JA. The root length of each genotype grown on 1/2 MS medium were arbitrarily set to 1. Results shown are the mean ± sd of measurements from 30 seedlings. WT, wild type.
(F) to (J) Three independent experiments with different seed batches show similar results. Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01).
We then performed RNA-immunoprecipitation (RIP) assays to investigate whether PRP39a associates with JAZ6 pre-mRNA. We used the myc antibody to immunoprecipitate PRP39a-myc from extracts of the PRP39a-myc transgenic line (Supplemental Figure 6A) treated without or with MeJA. The resulting immunoprecipitates of PRP39a-myc were reverse-transcribed into cDNA and measured by RT-qPCR using specific primers for JAZ6. Considering that MeJA treatment could induce the expression of JAZ6 (Chini et al., 2007; Thines et al., 2007), we present the RIP signal as the percentage of immunoprecipitated JAZ6 pre-mRNA relative to input JAZ6 pre-mRNA. As shown in Figure 4C, JAZ6 pre-mRNA was abundantly detected in immunoprecipitates from the PRP39a-myc transgenic line but not from the wild type, indicating that PRP39a indeed associates with JAZ6 pre-mRNA. Importantly, RIP signals were substantially elevated by MeJA treatment (Figure 4C), suggesting that the association of PRP39a with the JAZ6 pre-mRNA could be enhanced by MeJA treatment.
Similarly, ChIP (Figure 4D) and RIP (Figure 4E) assays with a PRP40a-GFP transgenic line (Supplemental Figures 6B and 6C) revealed that PRP40a is also recruited to the JAZ6 gene body and that this splicing factor associates with the JAZ6 pre-mRNA during JA signaling.
To examine the specificity of the recruitment of PRP39a and PRP40a, we selected three other JAZ genes for ChIP and RIP analysis. JAZ1 was selected because it does not contain a Jas intron (Supplemental Figure 10A). ChIP assays failed to detect significant enrichment of PRP39a or PRP40a to the JAZ1 locus with or without MeJA treatment (Supplemental Figures 10A, 10B, and 10D). In addition, RIP assays failed to detect the association of PRP39a or PRP40a with the pre-mRNA of JAZ1 with or without MeJA treatment (Supplemental Figures 10A, 10C, and 10E).
We then selected JAZ5 (Supplemental Figure 10F) and JAZ10 (Supplemental Figure 10K), which contain a Jas intron-like JAZ6. ChIP assays showed that the enriched binding of PRP39a-myc and PRP40a-GFP to the gene body of JAZ5 (Supplemental Figures 10F, 10G, and 10I) and JAZ10 (Supplemental Figures 10K, 10L, and 10N) were significantly higher than that to their promoter region in response to MeJA treatment. RIP assays indicated that PRP39a and PRP40a associate with the JAZ5 (Supplemental Figures 10F, 10H, and 10J) and JAZ10 (Supplemental Figures 10K, 10M, and 10O) pre-mRNAs during JA signaling. These results suggest that PRP39a and PRP40a show functional specificity in regulating the AS of JAZ genes.
Furthermore, our RT-PCR assays suggested that intron retention of PSEUDO-RESPONSE REGULATOR7 (PRR7) and ACTIN7 was not regulated by PRP39 and PRP40 (Supplemental Figure 11); therefore, we used these two genes as negative controls to show the binding specificity of PRP39a and PRP40a. We performed ChIP and RIP assays to detect whether PRPs bound to genomic DNAs and pre-mRNAs of PRR7 and ACTIN7. As shown in Supplemental Figure 10, our ChIP and RIP assays failed to detect significant enrichment of PRP39a or PRP40a to PRR7 (Supplemental Figures 10P to 10T) and ACTIN7 (Supplemental Figures 10U to 10Y), indicating that the recruitment of PRP39a and PRP40a to their targets is specific and these proteins do not randomly bind to DNA across the genome. Again, these results support that PRP39a and PRP40a exhibit specificity in regulating AS.
Next, we examined whether depletion of PRP39a or PRP40a affects JAZ6 and ∆PYJAZ6 transcript levels in response to MeJA. We obtained two T-DNA insertion mutant lines, prp39a-1 (Wang et al., 2007) and prp40a-1, from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/; Supplemental Figures 12A and 12G). These lines showed reductions in the levels of PRP39a and PRP40a (Supplemental Figures 12B and 12C, 12H to 12J). Consistent with previous observations (Wang et al., 2007), the prp39a-1 mutant showed a late-flowering phenotype under long-day conditions (Supplemental Figure 12O). However, the prp40a-1 mutant did not show obvious developmental defects (Supplemental Figures 12O and 12P). Our RT-qPCR assays revealed that the MeJA-induced expression levels of JAZ6 were significantly reduced in prp39a-1 (Supplemental Figures 12A to 12C) and prp40a-1 (Supplemental Figures 12G to 12J) compared with wild type, whereas the MeJA-induced expression levels of ∆PYJAZ6 were significantly elevated in these mutants (Figures 4F and 4G), indicating that the splicing of Jas intron of JAZ6 is impaired in these mutants. In parallel experiments, we found that while MeJA-induced expression levels of JAZ5, JAZ9, and JAZ10 were significantly lower in the prp39a-1 and prp40a-1 mutants compared with the wild type (Supplemental Figure 13), MeJA-induced expression levels of ∆PYJAZ5, ∆PYJAZ9, and ∆PYJAZ10 were significantly higher in the prp39a-1 and prp40a-1 mutants compared with the wild type (Supplemental Figure 13), indicating that the prp39a-1 and prp40a-1 mutations also impair the splicing of Jas introns of JAZ5, JAZ9, and JAZ10. Consistent with the RT-qPCR results, our PCR assays showed that the splicing forms of ΔPYJAZ5, ΔPYJAZ6, and ΔPYJAZ10 were substantially elevated in prp39a-1 and prp40a-1 mutants when compared with wild-type plants (Supplemental Figure 14B). However, our PCR failed to detect visible levels of ΔPYJAZ9 in both mutants and wild type, despite abundant levels of fully spliced JAZ9 being detected (Supplemental Figure 14B). Together, our results that depletion of PRP39a or PRP40a led to increased transcript levels of multiple JAZ splice variants suggest that these splicing factors promote the correct splicing of Jas intron during JA signaling.
Considering that our co-IP data suggest that PRP39a and PRP40a could exist in the same complex (Figure 2F), we asked whether PRP39a and PRP40a act redundantly in regulating Jas intron splicing. Interestingly, MeJA-induced expression levels of JAZ5, JAZ6, JAZ9, and JAZ10 and the ∆PY version of these JAZ genes in the prp39a-1/prp40a-1 double mutant were comparable to those in the single mutants (Figures 4F and 4G; Supplemental Figure 13), providing genetic evidence that PRP39a and PRP40a are not functionally additive in regulating Jas intron splicing.
We then asked whether PRP39a or PRP40a act redundantly with their respective paralogous copies in regulating Jas intron splicing. We generated a prp39a-1/prp39b-1 double mutant line by crossing of prp39a-1 and prp39b-1 (Supplemental Figures 12D to 12F). We found that the prp39a-1 prp39b-1 double mutant showed similar late-flowering phenotype with the prp39a-1 single mutant under long-day conditions (Supplemental Figure 12O). RT-qPCR assays revealed that the MeJA-induced expression levels of JAZ5 (Supplemental Figure 13), JAZ6 (Figure 4F), JAZ9 (Supplemental Figure 13), and JAZ10 (Supplemental Figure 13), as well as the ∆PY version of these JAZ genes in this double mutant, were comparable to those in the prp39a-1 single mutant (Figures 4F and 4G and Supplemental Figure 13)—suggesting that PRP39b does not show functional redundancy with PRP39a in regulating Jas intron splicing.
Because the PRP40b and PRP40c loci are tightly linked together, we used CRISPR/Cas9 gene editing technique (Yan et al., 2015) to engineer loss-of-function mutations of PRP40b and PRP40c in the genetic background of the prp40a-1 mutant (Supplemental Figures 12K to 12N). Sequence analyses indicated that, in the resulting prp40a-1 prp40b prp40c triple mutant, the PRP40b genomic DNA contains a T insertion at nucleotide +427, which leads to frame shift and the generation of a premature stop codon TGA (Supplemental Figures 12K and 12L); and the PRP40c genomic DNA contains a G insertion at nucleotide +643, which also leads to frame shift and the generation of a premature stop codon TAA (Supplemental Figures 12M and 12N). Although we did not find obvious developmental defects in prp40a-1 single mutants, we found that the prp40a-1 prp40b prp40c triple mutant showed late-flowering under long-day conditions (Supplemental Figure 12O), consistent with a recent observation that the prp40c mutant showed late-flowering (Hernando et al., 2019). In addition, we found that the leaf initiation rate of the prp40a-1 prp40b prp40c triple mutant was severely arrested compared with the wild type (Supplemental Figure 12P). RT-qPCR assays revealed that the MeJA-induced expression levels of JAZ6 were significantly lower in the prp40a-1 prp40b prp40c triple mutant compared with those in the prp40a-1 single mutant (Figure 4F). By contrast, the MeJA-induced expression levels of ∆PYJAZ6 were significantly higher in the triple mutant compared with those in the single mutant (Figure 4G). These results suggest that PRP40b and PRP40c act redundantly with PRP40a in regulating the splicing of the Jas intron of JAZ6. Similarly, our results support that PRP40b and PRP40c act redundantly with PRP40a to regulate the splicing of the Jas intron of JAZ5, JAZ9, and JAZ10 (Supplemental Figure 13). These observations were in line with the above results that all of the three PRP40 genes were induced by MeJA in a MYC2- and MED25-dependent manner (Figure 3).
In contrast with the effect of PRP39 and PRP40 on the splicing of the Jas intron of JAZ5, JAZ6, JAZ9, and JAZ10, we found that MeJA-induced expression of JAZ10.4 did not show significant alteration in the prp39 and prp40 mutants compared with wild-type plants (Supplemental Figure 13), suggesting that PRP39 and PRP40 might not be involved in the generation of JAZ10.4 during JA signaling, thus validating the specificity of PRP39 and PRP40 on the AS of JAZ genes.
A recent transcriptome analysis revealed that PRP40c regulates AS of 553 transcripts with various splicing events (Hernando et al., 2019). To examine the specificity of PRP39 and PRP40 in regulating AS, we performed PCR analysis to compare the AS of several randomly selected genes. Results showed that mutations of these splicing factors genes impaired the AS of RESISTANCE TO LEPTOSPHAERIA MACULANS3, NUDIX HYDROLASE HOMOLOGY7, NON-PHOTOTROPHIC HYPOCOTYL4, and RUBISCO METHYLTRANSFERASE, but showed negligible effect on the AS of ALWAYS EARLY3, PRR7, and ACTIN7 (Supplemental Figure 11). These, together with the above results that PRP39a and PRP40a specifically regulates the AS of a subset of JAZ genes (Figure 4; Supplemental Figure 13), support a scenario that the splicing factors PRP39 and PRP40 show functional specificity in regulating AS.
Depletion of PRP39a and PRP40a, and Their Homologous Genes, Leads To Attenuated JA Responses
Because prp39a-1, prp40a-1, and different combinations of these mutants accumulate elevated levels of the dominant JAZ splice variants compared with wild-type plants, we predicted that these mutants could show desensitized JA responses. Indeed, the MeJA-induced expression levels of VSP1 (Figure 4H), VSP2 (Supplemental Figure 15A), PDF1.2 (Figure 4I), and THI2.1 (Supplemental Figure 15B) were significantly reduced in prp39a-1, prp39a-1 prp39b-1, prp40a-1, prp40a-1 prp40b prp40c, and prp39a-1 prp40a-1 compared with wild type. In a JA-induced root growth inhibition assay, while the JA sensitivity of prp39a-1 and prp39a-1 prp39b-1 was comparable to that of wild type, prp40a-1, prp40a-1 prp40b prp40c, and prp39a-1 prp40a-1 were less sensitive than wild type (Figure 4J). Notably, in terms of hormone-induced defense gene expression and root growth inhibition, the prp40a-1 prp40b prp40c triple mutant showed the most severe deficiency of the JA response (Figures 4H to 4J). This observation is consistent with the finding that the three PRP40 genes show functional redundancy.
Collectively, our results support that PRP39a and PRP40a and their functional homologs promote the correct splicing of JAZ genes and thereby play an important role in preventing JAZ-splice-variants–mediated excessive desensitization of JA responses.
MeJA-Induced Recruitment of PRP39a and PRP40a Depends on MED25
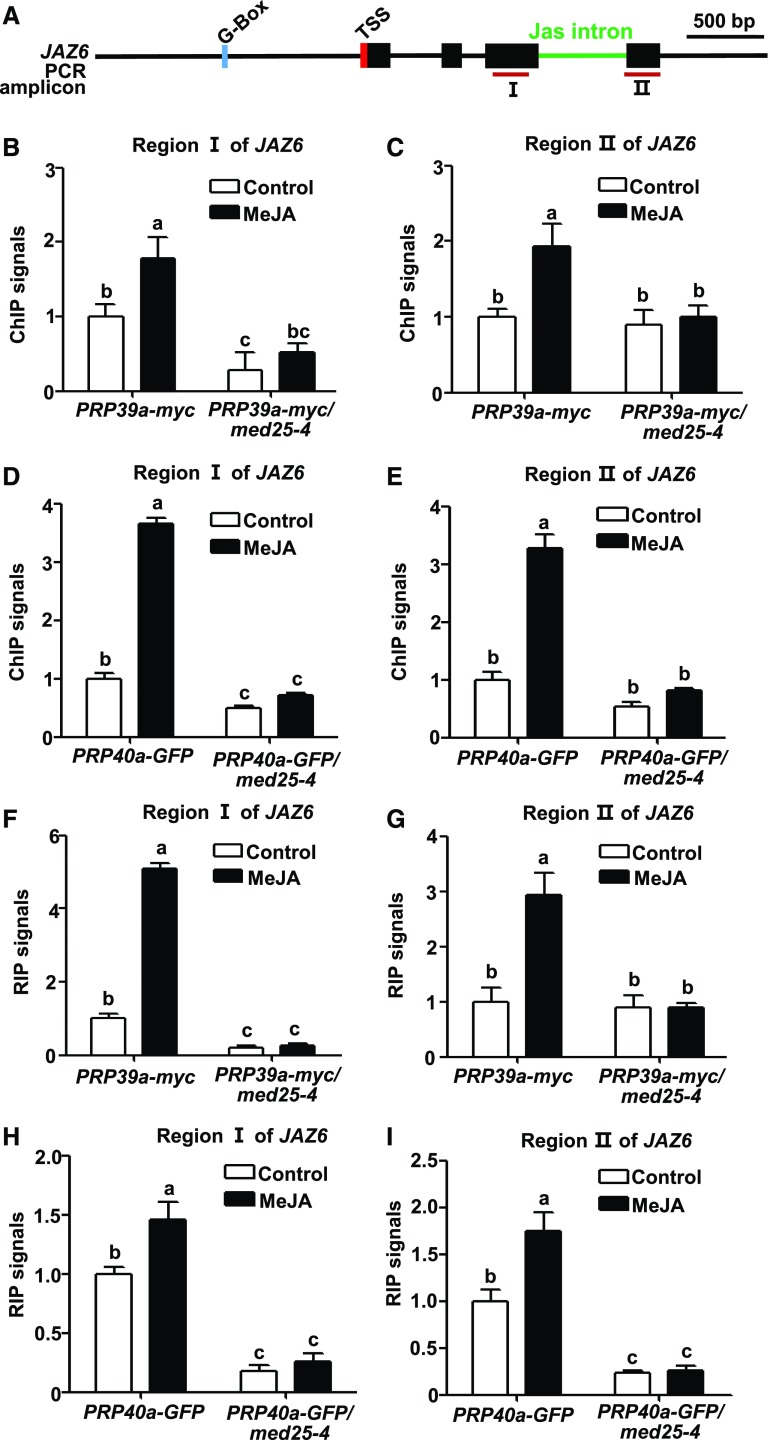
The above results support a notion that MED25 recruits the splicing factors PRP39a and PRP40a to facilitate the full splicing of Jas intron. To test this hypothesis, we examined whether the mutation of MED25 affects the recruitment of PRP39a and PRP40a to the JAZ5, JAZ6, and JAZ10 loci. For this purpose, we introduced the PRP39a-myc or PRP40a-GFP into the med25-4 mutant background through crossing. ChIP-qPCR assays showed that the MeJA-induced enhancement of PRP39a-myc (Figures 5A to 5C; Supplemental Figures 16A to 16C and 16J to 16L) and PRP40a-GFP (Figures 5A, 5D, and 5E; Supplemental Figures 16A, 16D, 16E, 16J, 16M, and 16N) enrichment to JAZ5, JAZ6, and JAZ10 was largely abolished in med25-4 compared with wild type, indicating that MeJA-induced recruitment of PRP39a and PRP40a to JAZ5, JAZ6, and JAZ10 requires the function of MED25.
Figure 5.
Depletion of MED25 Impairs MeJA-Induced Recruitment of PRP39a and PRP40a to JAZ6.
(A) Schematic diagrams of JAZ6 and PCR amplicons used for ChIP-qPCR and RIP-RT-qPCR. Cyan box represents the putative MYC2 binding G-box motif; green line represents the Jas intron; red box represents transcription start site (TSS). Dark-red lines labeled “I” and “II” represent PCR amplicons used for ChIP-qPCR and RIP-RT-qPCR. Scale bar = 500 bp.
(B) and (C) ChIP-qPCR showing that med25-4 exhibit impaired MeJA-induced enrichment of PRP39a on region I (B) and region II (C) of JAZ6. Ten-d–old PRP39a-myc and PRP39a-myc/med25-4 seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking. Chromatin from each sample was immunoprecipitated with anti-myc antibody.
(D) and (E) ChIP-qPCR showing that med25-4 exhibits impaired MeJA-induced enrichment of PRP40a on region I (D) and region II (E) of JAZ6. Ten-d–old PRP40a-GFP and PRP40a-GFP/med25-4 seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking. Chromatin from each sample was immunoprecipitated with anti-GFP antibody.
(B) to (E) ChIP signals were displayed as the percentage of precipitated DNA relative to input DNA. The value of amplicon I (B) and (D) or amplicon II (C) and (E) in PRP39a-myc (B) and (C) or PRP40a-GFP (D) and (E) without MeJA treatment was arbitrarily set to 1. Data shown are mean values of three biological repeats with sd. Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01).
(F) and (G) RIP-RT-qPCR showing med25-4 exhibit impaired MeJA-induced enrichment of PRP39a on JAZ6 pre-mRNA in region I (F) and region II (G). Ten-d–old PRP39a-myc and PRP39a-myc/med25-4 seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking. The pre-mRNA from each sample was immunoprecipitated with anti-myc antibody.
(H) and (I) RIP-RT-qPCR showing med25-4 exhibit impaired MeJA-induced enrichment of PRP40a on JAZ6 pre-mRNA in region I (H) and region II (I). Ten-d–old PRP40a-GFP and PRP40a-GFP/med25-4 seedlings were treated without or with 100 μM of MeJA for 30 min before cross linking. The pre-mRNA from each sample was immunoprecipitated with anti-GFP antibody.
(F) to (I) Precipitated pre-mRNA and input pre-mRNA was quantified by RT-qPCR for the indicated amplicons. RIP signals were displayed as the percentage of precipitated pre-mRNA relative to input pre-mRNA. The value of amplicon I (F) and (H) or amplicon II (G) and (I) in PRP39a-myc (F) and (G) or PRP40a-GFP (H) and (I) without MeJA treatment was arbitrarily set to 1. Data shown are mean values of three biological repeats with sd. Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01).
Similarly, in RIP assays with PRP39a-myc plants (Figures 5F and 5G; Supplemental Figures 16F, 16G, 16O, and 16P) or PRP40a-GFP plants (Figures 5H and 5I; Supplemental Figures 16H, 16I, 16Q, and 16R), we found that the MeJA-induced elevation of RIP signals were significantly decreased in med25-4 compared with wild type, suggesting that MED25 plays a critical role for the association of PRP39a and PRP40a with the JAZ5, JAZ6, and JAZ10 pre-mRNA during JA signaling.
DISCUSSION
The JA signaling pathway controls resource allocation between growth- and defense-related processes, and thus plays a critical role in optimizing plant acclimatization to their rapidly changing and often hostile environments. Therefore, both the activation and the deactivation/desensitization of JA responses must be under tight control. It is well recognized that JA-induced production of dominant JAZ splice variants provides a general mechanism to desensitize JA responses. However, how this process is regulated remains unclear. Here we show that the generation of JAZ splice variants depends on the Mediator subunit MED25 and that MED25 recruits the splicing factors PRP39a and PRP40a to prevent the overproduction of JAZ splice variants. Our results suggest that the MED25-PRP39a/PRP40a module acts to prevent the excessive desensitization of JA responses by promoting the full splicing of Jas intron.
PRP39a and PRP40a Promote the Full Splicing of Jas Intron
In our current understanding of the JA signaling pathway, hormone-dependent degradation of JAZ repressors leads to derepression (activation) of MYC transcription factors; subsequently, plants produce a repertoire of dominant JAZ splice variants to desensitize JA signaling. A key question is to understand how plants keep a balance between JAZ splice variant-mediated desensitization of JA signaling and JAZ degradation-mediated activation of JA signaling. We provide several lines of evidence that the splicing factors PRP39a and PRP40a are involved in the splicing of Jas intron during JA signaling. First, ChIP-qPCR assays indicated that PRP39a and PRP40a were recruited to JAZ6 in response to MeJA treatment. Second, RIP assays indicated that PRP39a and PRP40a associate with the JAZ6 pre-mRNA during JA signaling. Third, depletion of PRP39a, PRP40a, and their paralogs led to overaccumulation of JAZ splice variants and, as a consequence, attenuated JA responses. Together, these results ascribe a function of RPR39a and PRP40a in promoting the correct splicing of JAZ genes and thereby counteracting JAZ splice variant-mediated desensitization of JA signaling. Therefore, RPR39a and PRP40a are components of the JA signaling pathway that function to prevent excessive desensitization of JA responses mediated by JAZ splice variants.
Despite the fact that both PRP39a and PRP40a are involved in Jas intron-dependent AS of JAZ genes, our genetic analyses with prp39a-1 prp40a-1 double mutants indicated that the effects of PRP39a and PRP40a are not additive. In line with this observation, it is well-established that the yeast homolog of PRP39a contains a featured tetratricopeptide repeat domain and plays an important role for the stable binding of the U1 snRNP to the pre-mRNA substrate (Lockhart and Rymond, 1994). Different from PRP39a, PRP40a and its yeast homolog were considered to be potential integrators of splicing and transcription because these proteins contain featured WW domains (Bork and Sudol, 1994) and FF domains (Bedford and Leder, 1999) and show interaction with the carboxyl-terminal domain of the largest subunit of Pol II (Morris and Greenleaf, 2000; Kang et al., 2009). In addition, the yeast Prp40 protein was believed to play an essential role in early events in splice site definition (Becerra et al., 2016). These previous studies, together with our findings described herein, support a scenario that the structurally distinct splicing factors PRP39a and PRP40a play nonequivalent functions in U1 snRNP in regulating Jas intron splicing of JAZ genes.
Surprisingly, our genetic analyses failed to detect functional redundancy between PRP39a and PRP39b in regulating Jas intron splicing and JA-responses, suggesting a functional diversification of the two paralogous copies. These results are consistent with a previous observation that PRP39a and PRP39b are not functionally equivalent in regulating pre-mRNA splicing (Kanno et al., 2017).
Different from the PRP39a case, our data indicated that PRP40b and PRP40c act redundantly with PRP40a in regulating Jas intron splicing. First, like PRP40a, PRP40b, and PRP40c were induced by MeJA treatment. Second, overaccumulation of JAZ splice variants was enhanced in the prp40a-1 prp40b prp40c triple mutant compared with the prp40a-1 single mutant. Third, attenuation of JA responses was enhanced in the prp40a-1 prp40b prp40c triple mutant compared with the prp40a-1 single mutant. It will be interesting to elucidate the detailed action mechanisms of PRP39a and PRP40a as well as their functional paralogs in regulating Jas intron splicing in future studies.
In addition to the generation of JAZ splice variants, plants have evolved other mechanisms that ensure the proper desensitization/attenuation of already activated JA responses. For example, plants employ an evolutionarily conserved metabolic network to convert the bioactive JA-Ile into an inactive or less active form (Miersch et al., 2008; Kitaoka et al., 2011; Koo et al., 2011; VanDoorn et al., 2011; Heitz et al., 2012), thus providing an efficient route to switch off the JA signaling (Koo and Howe, 2012). Furthermore, plants activate a group of MYC-related transcription factors, termed JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3, which negatively regulate JA responses (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014). Although we currently do not exactly understand whether JAZ splice variants operate synergistically with or independent of the induction JA-Ile catabolic pathways or negative transcription factors, the existence of multiple desensitization/attenuation mechanisms could provide an explanation that depletion of PRP39s and/or PRP40s only leads to mild JA response deficiency.
MED25 Controls the JA-Induced Recruitment of PRP39a and PRP40a to JAZ Loci
We elucidated a mechanism by which PRP39a and PRP40a are recruited to JAZ loci. First, multiple protein–protein interaction assays indicated that PRP39a and PRP40a associate with MED25 in planta. Second, MeJA-mediated induction of PRP40a expression depends on MED25. Third, ChIP assays indicated that depletion of MED25 impairs MeJA-induced recruitment of PRP39a and PRP40a to JAZ6. Collectively, these results demonstrate that MED25 plays a critical role in hormone-induced recruitment of PRP39a and PRP40a to JAZ loci. Therefore, our results revealed that MED25 forms a module with PRP39a and PRP40a to facilitate the full splicing of the Jas intron to generate the full-length form of JAZs. In other words, the MED25-PRP39a/PRP40a module functions to prevent JAZ-splice-variants–mediated excessive desensitization of JA responses.
Intriguingly, our results indicated that the JA-induced generation of JAZ splice variants, whose function is to desensitize JA responses, also requires the function of MED25. In the context that MED25 plays a pivotal role in the initial activation of MYC2-dependent transcription of JA-responsive genes (i.e. PIC formation; Chen et al., 2012), our results suggest that both the JAZ splice variant-dependent desensitization of JA responses and the PRP39a/PRP40a-dependent anti-desensitization of JA responses (i.e. through reducing the generation of JAZ splice variants) are default mechanisms that are preprogrammed by MED25 during the initial induction phase of the JA signaling.
Notably, MED25 exhibited distinct recruitment modes for the heterogeneous splicing factors PRP39a and PRP40a. For example, our multiple protein interaction assays supported the notion that MED25 directly interacts with PRP39a but not PRP40a, suggesting that PRP39a, but not PRP40a, is physically recruited by MED25 to the JAZ loci. On the other hand, our gene expression assays revealed that the transcription of PRP40a and its paralogs, but not PRP39a and its paralogs, is induced by JA in a MYC2- and MED25-dependent manner.
Furthermore, we found that, while MeJA-induced expression of JAZ3 and ΔPYJAZ3 was markedly reduced in the med25-4 mutant (Supplemental Figure 1), depletion of PRP39a and/or PRP40a and their paralogs only has subtle effects on the Jas intron-dependent AS of JAZ3 (Supplemental Figure 13). It is reasonable to speculate that MED25 might also recruit as yet unidentified splicing factors for Jas intron-dependent AS of JAZ3 and other JAZ genes. Together, these observations reveal the striking functional versatility of MED25 in modulating Jas intron-dependent AS of JAZ genes—a highly important finding that should prompt further studies of the underlying mechanisms.
MED25 Acts as a Central Coactivator of MYC2-Mediated Transcription of JA-Responsive Genes
Our recent studies revealed that, in addition to bridging the communications between MYC2 and the Pol II transcriptional machinery (Chen et al., 2012), MED25 also physically and functionally interacts with different signaling components to coordinate multiple steps of MYC2-mediated transcription of JA-responsive genes. For example, at the resting stage, MED25 physically brings COI1 to MYC2 target promoters, thereby facilitating COI1-dependent degradation of JAZ repressors (An et al., 2017). Upon hormone elicitation, MED25 physically cooperates with the epigenetic regulator HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1, which selectively regulates hormone-induced histone-3 Lys-9 acetylation of MYC2 target promoters (An et al., 2017). At the same time, MED25 also physically recruits the conserved transcriptional coregulator LEUNIG_HOMOLOG, which acts as a scaffold to stabilize the MYC2-MED25-HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY1 transcription complex (You et al., 2019). Clearly, these aspects of functions of MED25 all favor the activation of MYC2-dependent JA responses.
Here we describe another aspect of mechanistically related function of MED25 in preventing JAZ splice variants-mediated excessive desensitization of JA responses. We demonstrate that this function of MED25 is achieved through cooperating with the splicing factors PRP39a and PRP40a. In line with previous observations that the yeast and mammalian homologs of PRP39a and PRP40a are involved in cotranscriptional AS (Kotovic et al., 2003; Listerman et al., 2006; Görnemann et al., 2011), our ChIP-qPCR data revealed that PRP39a and PRP40a were recruited to the gene body of JAZ6. These, together with our findings that the generation of ∆PYJAZs depends on MED25 and that the initiation of MYC2-dependent transcription of JA-responsive genes depends on MED25 (Chen et al., 2012), suggest that Jas intron-dependent AS of JAZ genes is a cotranscriptional process. Collectively, these results support a scenario that the “multitalented” MED25 acts as a central coactivator of MYC2-dependent transcriptional regulation of JA signaling.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild type. The following previously described plant materials were used in this study: coi1-2 (Xu et al., 2002), myc2-2 (Boter et al., 2004), med25-4 (Chen et al., 2012), 35Spro:MED25-myc (MED25-myc; Chen et al., 2012), 35Spro:COI1-myc (COI1-myc; Xu et al., 2002), 35Spro:MYC2-myc (MYC2-myc; Chen et al., 2011), myc2/3/4 (Song et al., 2014), 35Spro: MED25-GFP (MED25-GFP; Chen et al., 2012), and 35Spro:MYC2-GFP (MYC2-GFP; Zhai et al., 2013), prp39a-1 (Salk_133733C), prp39b-1 (Salk_142600), and prp40a-1 (Salk_021070) were ordered from the Arabidopsis Biological Resource Center. The prp40a-1 prp40b prp40c triple mutant was generated by introducing the CRISPR/Cas9 technology-mediated prp40b and prp40c in the prp40a-1 background. All Arabidopsis plants were grown on 1/2-strength Murashige and Skoog (1/2 MS) medium at 22°C under long-day conditions (16-h light/8-h dark; light intensity, 120 μmol photons m−2 s−1). For RT-qPCR, 10-d–old seedlings were treated without or with 100 μM of MeJA before RNA extraction. For ChIP and RIP assays, 10-d–old seedlings were treated without or with 100 μM of MeJA before cross linking. For MeJA treatment, 8 mL of liquid 1/2 MS medium containing MeJA at a final concentration of 100 μM was added to the plate for the indicated times. The JA-mediated root growth inhibition assays were performed as described in Chen et al. (2012), with seedlings that were grown on 1/2 MS medium containing 20 μM of JA. Nicotiana benthamiana was grown under long-day conditions (16-h light [28°C]/8-h dark [22°C]).
DNA Constructs and Plant Transformation
To generate the 35Spro:JAZ6-GFP (JAZ6-GFP) and 35Spro:∆PYJAZ6-GFP (∆PYJAZ6-GFP) constructs, the coding sequence of GFP was amplified and cloned into pCAMBIA1300 to obtain pCAMBIA1300-GFP, and the coding sequences of JAZ6 and truncated JAZ6 were amplified and cloned into pCAMBIA1300-GFP to obtain JAZ6-GFP or ∆PYJAZ6-GFP, respectively. To construct 35Spro:PRP39a-myc (PRP39a-myc), the coding sequence of PRP39a was amplified and cloned into Gateway vector pGWB17 (Nakagawa et al., 2007). To construct 35Spro:PRP39a-GFP (PRP39a-GFP) and 35Spro:PRP40a-GFP (PRP40a-GFP), the coding sequences of PRP39a and PRP40a were amplified and cloned into Gateway vector pGWB5 (Nakagawa et al., 2007). Primers used for plasmid construction are listed in the Supplemental Data Set. The constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis plants by the floral dip method. Transformants were selected based on their resistance to hygromycin. T3 homozygous lines were used for further experiments. PRP39a-myc and PRP40a-GFP were introduced into the med25-4 background by crossing. Homozygous plants were selected by genotyping.
Generation of prp40b and prp40c Using CRISPR/Cas9 Technology
Twenty-bp fragments of the PRP40b CDS (127 to 146 bp) and PRP40c CDS (280 to 299 bp) were used as the targeting sequence for genome editing of PRP40b and PRP40c, respectively. The designed targeting sequences were cloned into the BsaI site of the AtU6-26-sgRNA-SK vector (Yan et al., 2015) to generate AtU6-26-PRP40b-targetsgRNA and AtU6-26-PRP40c-targetsgRNA. The AtU6-26-PRP40b-targets-gRNA was digested by SpeI and NheI, and the cassette was cloned into the SpeI position of the pYAO:hSpCas9 vector (Yan et al., 2015) to generate pYAO:hSpCas9-PRP40b-targetsgRNA. Similarly, the AtU6-26-PRP40c-targetsgRNA was digested by SpeI and NheI, and the cassette was cloned into the SpeI position of the pYAO:hSpCas9-PRP40b-targetsgRNA to generate pYAO:hSpCas9-PRP40b-PRP40c-targetsgRNA. The construct was transformed into A. tumefaciens strain GV3101, which was used to transform Arabidopsis prp40a-1 plants by the floral dip method. Further selection was based on their resistance to hygromycin and DNA sequencing. The Cas9-free plants with mutations in the T2 progeny were identified for further experiments.
Y2H Assays
To investigate protein interactions in yeast, the coding sequences of PRP39a, PRP40a, and MED25 were fused to the AD domain in pGADT7 and the BD domain in pGBKT7, respectively. The primers used are listed in the Supplemental Data Set. The constructs were cotransformed into yeast (Saccharomyces cerevisiae) strain AH109. The presence of transgenes was confirmed by growth on SD/-Leu/-Trp plates. To assess protein–protein interactions, transformed yeast cells were suspended in liquid SD/-Leu/-Trp to OD600 = 1.0. Five-microliter samples of suspended yeast cells were spread onto plates containing SD/-Ade/-His/-Leu/-Trp medium (Clontech). Interactions were observed after 3 d of incubation at 30°C.
Purification of the MED25-Containing Protein Complex and Tandem Mass Spectrometry Analysis
Five-gram samples of 10-d–old MED25-myc seedlings were harvested, ground in liquid N2, and lysed in 10 mL of ice-cold extraction buffer (50 mM of Tris-HCl, 150 mM of NaCl, 5 mM of EDTA, 0.1% [v/v] Triton X-100, 0.2% [v/v] NP-40, 0.6 mM of PMSF, 1× Protein inhibitor, and 20 μM of MG132). After vigorous vortexing for 30 s, the samples were centrifuged at 13,000g for 10 min at 4°C. For each sample, 30 μL of the supernatant was subjected to immunoblot analysis, and the remainder was incubated with 50 μL of anti-myc agarose beads (MBL Life Science) for 4 h at 4°C with gentle shaking. The beads were collected and washed four times with extraction buffer and once with 50 mM of Tris-HCl at pH 7.5. The precipitate was eluted by adding 1× SDS loading buffer and separated by 10% SDS-PAGE. The gel was stained with GelCode Blue Safe Protein Stain (Thermo Fisher Scientific) and washed with double-distilled water.
The gel (containing the sample) was cut into small pieces and destained in buffer containing 25 mM of ammonium bicarbonate and 50% (v/v) acetonitrile. The proteins were reduced with 10 mM of DTT at 37°C for 1 h and alkylated with 25 mM of iodoacetamide at room temperature for 1 h in the dark. In-solution trypsin digestion was performed at 37°C overnight using a trypsin/substrate ratio of 1:50. Peptides were extracted from the gel with buffers containing 5% (v/v) trifluoroacetic acid and 50% (v/v) acetonitrile via two rounds of ultrasonication. The samples were freeze-dried in a SpeedVac (Thermo Fisher Scientific), and the peptides were resolubilized in 0.1% (v/v) formic acid and filtered through a 0.45-μM centrifugal filter.
The peptides were analyzed using a TripleTOF 5600 Mass Spectrometer (AB Sciex) coupled online to an Eksigent nanoLC Ultra HPLC system (AB Sciex) in Information Dependent Mode. The LC gradient (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile) was 5% to 90% B for 90 min at a flow rate of 300 nL/min.
The peptides were identified from the tandem mass spectrometry spectra using the software ProteinPilot v4.2 by searching against the Arabidopsis International Protein Index database (https://www.arabidopsis.org/download_files/Proteins/TAIR10_protein_lists/TAIR10_pep_20101214). The fixed modification was carbamidomethylation of Cys residues. Trypsin was specified as the proteolytic enzyme, with two missing cleavages allowed. Mass tolerance was set to 0.05 D, and the maximum false discovery rate for proteins and peptides was 1%.
Antibody Production
The coding sequence of the N-terminal region of PRP40a (amino acids 1–350, PRP40a1-350) was amplified from wild-type cDNA using gene-specific primers (Supplemental Data Set). The resulting PCR product was cloned into the BamHI and EcoRI sites of pGEX-6P-1 (GE Healthcare Life Sciences) to express GST-PRP40a1-350 truncated protein in Escherichia coli strain BL21 (DE3). The recombinant fusion protein was used to raise polyclonal antibody in mouse. Anti-PRP40a antibody were used for immunoblotting at a final concentration of 1:1,500.
Protein Expression and In Vitro Pull-Down Assays
To produce GST-PRP39a, the coding sequence of PRP39a was amplified and cloned into the pGEX-6P-1 vector. Protein expressed in E. coli BL21 (DE3) was purified using GST Bind Resin (Novagen). To produce MED25-FLAG and PRP40a-His, the coding sequences of MED25 and PRP40a were amplified and cloned into the pF3K WG (BYDV) Flexi Vector (Promega). Proteins were in vitro-translated using the TnT SP6 High-Yield Wheat Germ Protein Expression System (Promega). For the pull-down assay, 5 μL of the in vitro-translated MED25-FLAG protein was incubated with 1 μg of immobilized GST-PRP39a or GST (or 5 μL of PRP40a-His) at 4°C in binding buffer (25 mM of Tris-HCl at pH 7.5, 100 mM of NaCl, 1 mM of DTT, and Roche protease inhibitor cocktail) for 2 h. Proteins retained on the beads were analyzed by immunoblotting with anti-FLAG (Abmart,1:3,000) and anti-His (Abmart, 1:2,000) antibodies.
To produce His-JAZ6 and His-ΔPYJAZ6, the coding sequences of JAZ6 and truncated JAZ6 were amplified and cloned into the pET28a vector. Protein expressed in E. coli BL21 (DE3) was purified using Ni-NTA resin (Novagen). Pull-down assays to test the COI1–JAZ6 and COI1–ΔPYJAZ6 interactions were performed as described in Thines et al. (2007) and An et al. (2017). Ni-NTA resin (Novagen) was used to bind His-JAZ6 or His-ΔPYJAZ6. Proteins retained on the beads were detected by immunoblotting with anti-myc (Abmart, 1:3,000) and anti-His (Abmart, 1:2,000) antibodies. To test the interaction of MYC2 with JAZ6 or ΔPYJAZ6, total proteins extracted from 10-d–old MYC2-myc seedlings using extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 5 mM of EDTA, 0.1% [v/v] Triton X-100, 0.2% [v/v] Nonidet P-40, 0.6 mM of PMSF, Roche protease inhibitor cocktail, and 20 μM of MG132) were incubated with Ni-NTA–bound His-JAZ6 or His-ΔPYJAZ6 at 4°C for 1 h. Proteins retained on the beads were detected by immunoblotting with anti-myc (Abmart, 1:3,000) and anti-His antibodies (Abmart, 1:2,000).
To produce MBP-MYC2 or GST-MED25MA, the full-length coding sequences of MYC2 and truncated MED25 (MED25MA, 227 to 680 amino acids) were amplified and cloned into pMAL-c2X or pGEX-4T-1. The resulting constructs were transformed into E. coli BL21 (DE3) cells, and the recombinant proteins were purified using Amylose Resin (New England BioLabs) and GST Bind Resin (Novagen), respectively. For the pull-down assay, 1 μg of purified His-JAZ6, His-ΔPYJAZ6, or GST-MED25MA fusion protein was incubated with 1 μg of immobilized MBP-MYC2 or MBP at 4°C in binding buffer (25 mM of Tris-HCl at pH 7.5, 100 mM of NaCl, 1 mM of DTT, and Roche protease inhibitor cocktail) for 2 h. For the competitive pull-down assay, 1 μg of GST-MED25MA with 1 or 5 μg of His-JAZ6 or His-ΔPYJAZ6 was incubated with immobilized MBP-MYC2 (1 μg) at 4°C for 2 h. Proteins retained on the beads were analyzed by immunoblotting with anti-His (Abmart, 1:2,000), anti-GST (Abmart, 1:3,000), and anti-MBP (New England BioLabs, 1:10,000) antibodies.
LCI Assays
LCI assays were performed as described in Chen et al. (2008). The coding sequences of MYC2 were cloned into pCAMBIA1300-cLUC and MED25 was cloned into pCAMBIA1300-nLUC, respectively. The constructs expressing GFP, JAZ6-GFP, and ΔPYJAZ6 were described above. The primers used are summarized in the Supplemental Data Set. A. tumefaciens GV3101 carrying the indicated constructs were incubated in Luria–Bertani medium at 28°C overnight and transferred to fresh Luria–Bertani medium containing 10 mM of 2-(n-morpholino)-ethanesulfonic acid (at pH 5.6) and 40 μM of acetosyringone (1:100 ratio, v/v) for 16 h. The culture was pelleted and resuspended in 10 mM of MgCl2 containing 0.2 mM of acetosyringone to a final concentration of OD600 = 1.5. The bacteria were incubated at room temperature for at least 3 h without shaking. For coinfiltration, equal volumes of A. tumefaciens suspensions carrying the indicated constructs were infiltrated into N. benthamiana leaves. After infiltration, the plants were incubated for 72 h under a long-day condition (16-h light [28°C]/8-h dark [22°C]) before LUC activity measurements. Low-light–cooled charge-coupled device imaging apparatus (NightOWL II LB983; Berthold) was used to capture the LUC image. The leaves were sprayed with 0.5 mM of luciferin and incubated in the dark for 3 min before luminescence detection.
Co-IP Assays
Co-IP assays were performed according to a published procedure (An et al., 2017) with minor modifications. In brief, 10-d–old MED25-GFP or PRP39a-GFP seedlings were homogenized in extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 0.1% [v/v] Triton X-100, 0.2% [v/v] Nonidet P-40, 0.6 mM of PMSF, and 20 μΜ of MG132 with Roche protease inhibitor cocktail). Wild-type seedlings were used as a negative control. After protein extraction, 20 μL of protein A/G plus agarose (Santa Cruz Biotechnology) was added to the 2-mg extracts to reduce nonspecific immunoglobulin binding. After 1 h of incubation, the supernatant was transferred to a new tube. GFP antibody-bound agarose beads (MBL Life Science) were then added to the samples and incubated for 4 h at 4°C with gentle rocking. The precipitated samples were washed at least four times with protein extraction buffer, and bound proteins were eluted by heating the beads in 1× SDS protein loading buffer at 95°C for 8 min. MED25 protein was detected by immunoblotting using anti-MED25 antibody (Chen et al., 2012). PRP40a protein was detected by immunoblotting using anti-PRP40a (1:1,500) antibody.
Immunoblot Assays
To analyze PRP40a protein levels, protein extraction was performed by homogenizing 10-d–old seedlings in extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 1% [v/v] Nonidet P-40, 1 μM of DTT, 10 μM of MG132, and Roche protease inhibitor cocktail). For immunoblot analysis, SDS sample buffer was added to the protein extracts. The protein samples were boiled for 5 min, separated on SDS-PAGE gels, and transferred to polyvinylidene fluoride membranes. Immunoblots were probed with anti-PRP40a antibody. Nonspecific protein band was used as a control. To analyze the MED25, MYC2, JAZ6, and ΔPYJAZ6 protein levels in LCI assays, N. benthamiana leaves infiltrated with indicated constructs were homogenized in extraction buffer (50 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 0.1% [v/v] Nonidet P-40, 0.2% [v/v] Triton X-100, 5 μM of DTT, 10 μM of MG132, and Roche protease inhibitor cocktail). Immunoblots were probed with anti-MED25 (1:3,000), GFP (YTHX, 1:3,000), and cLUC (Sigma, 1:5,000) antibodies. To analyze the protein expression in Y2H assays, the yeast were boiled with 1× SDS loading buffer at 95°C for 8 min, then proteins were separated on SDS-PAGE gels and immunoblotted using anti-HA (AD- fusion proteins; Abmart, 1:2,000) and anti-myc (BD-fusion proteins; Abmart, 1:2,000) antibodies.
ChIP-qPCR Assays
Ten-d–old MYC2-GFP, MED25-GFP, PRP39a-myc, PRP40a-GFP, PRP39a-myc/med25-4, and PRP40a-GFP/med25-4 seedlings were treated without or with 100 μM of MeJA for the indicated times, and untreated wild-type seedlings were used as negative control. Two grams of each sample was harvested and cross linked in 1% (v/v) formaldehyde at room temperature for 10 min, followed by neutralization with 0.125 M of Gly. The chromatin complex was isolated, resuspended in lysis buffer (10 mM of Tris-HCl at pH 8.0, 20 mM of EDTA, 400 mM of NaCl, 1% [v/v] Triton X-100, and 2 mM of PMSF with 1× Roche protease inhibitor cocktail), and sheared by sonication to reduce the average DNA fragment size to ∼500 bp; 50 μL of sheared chromatin was removed for use as an input control. GFP antibody ab290 (Abcam) or myc antibody (Millipore) was incubated with Dynabeads Protein G (Invitrogen) at 4°C for at least 6 h and added to the remaining chromatin for incubation at 4°C overnight. The immunoprecipitated chromatin complex was washed with low-salt buffer (20 mM of Tris-HCl at pH 8.0, 2 mM of EDTA, 150 mM of NaCl, 0.5% [v/v] Triton X-100, and 0.2% [w/v] SDS), high-salt buffer (20 mM of Tris-HCl at pH 8.0, 2 mM of EDTA, 500 mM of NaCl, 0.5% [v/v] Triton X-100, and 0.2% [w/v] SDS), LiCl buffer (10 mM of Tris-HCl at pH 8.0, 1 mM of EDTA, 0.25 M of LiCl, 0.5% [v/v] NP-40, and 0.5% [w/v] sodium deoxycholate), and TE buffer (10 mM of Tris-HCl at pH 8.0 and 1 mM of EDTA). After washing, the immunoprecipitated chromatin was eluted with elution buffer (1% [w/v] SDS and 0.1 M of NaHCO3). Protein–DNA cross linking was reversed by incubating the immunoprecipitated complexes with 20 μL of 5 M of NaCl at 65°C overnight. DNA was recovered using a QIAquick PCR Purification Kit (Qiagen) and analyzed by qPCR. ChIP signals were displayed as the percentage of precipitated DNA relative to input DNA. The value of the indicated wild-type amplicon without MeJA treatment was arbitrarily set to 1. Three independent biological repeats were performed. Error bars represent sd. Statistical analysis was performed by Student’s t test. The primers used for qPCR are listed in the Supplemental Data Set.
RIP–RT-qPCR Assays
The RIP assay was performed as previously described by Wierzbicki et al. (2008) and Zheng et al. (2009) with minor modifications. Ten-d–old wild-type, PRP39a-myc, PRP40a-GFP, PRP39a-myc/med25-4, and PRP40a-GFP/med25-4 seedlings were treated without or with 100 μM MeJA and collected for cross linking at the indicated times to detect the association of PRP39a and PRP40a with JAZ6 pre-mRNA. After cross linking in 1% (v/v) formaldehyde, each sample was ground and resuspended in Honda buffer (0.44 M of Suc, 1.25% [w/v] Ficoll, 2.5% [w/v] dextran, 20 mM of HEPES KOH, 10 mM of MgCl2, 0.5% [v/v] Triton X-100, 5 mM of DTT, 1 mM of PMSF, protease inhibitor cocktail, and 8 U/mL of RNase inhibitor) to lyse the plant cells. The nuclear protein was sonicated in nuclear lysis buffer (50 mM of Tris-HCl, 10 mM of EDTA, 1% [w/v] SDS, 1 mM of PMSF, protease inhibitor cocktail, and 160 U/mL of RNase inhibitor). Before immunoprecipitation, the sonication products were treated with 20 U/mL DNase I and incubated with myc (Roche) or GFP (Abcam) antibodies overnight at 4°C. Immunoprecipitated complexes were collected using protein A beads (Millipore) and washed with immunoprecipitation elution buffer. The associated pre-mRNA and input pre-mRNA were subjected to DNase I treatment again using a RT-PCR kit with genomic DNA eraser (TaKaRa) and quantified by RT-qPCR with gene-specific primers for the pre-mRNAs after the reversal of cross linking. Pre-mRNA precipitated from wild type was used as a negative control, while pre-mRNA isolated before precipitation was used as an input control. RIP signals were displayed as the percentage of precipitated pre-mRNA relative to input pre-mRNA. The value of the indicated wild-type amplicon without MeJA treatment was arbitrarily set to 1. Three independent biological repeats were performed. Error bars represent sd. Statistical analysis was performed using one-way ANOVA or evaluated by Student’s t test (Supplemental File). Primers are listed in the Supplemental Data Set.
RNA Extraction, Reverse Transcription, and RT-qPCR
For RT-qPCR analysis of gene expression, total RNA was extracted from 10-d–old seedlings treated without or with 100 μM of MeJA for the indicated times using Trizol (Invitrogen) reagent. The cDNA was prepared from 2 μg of total RNA with a PrimeScript RT Reagent Kit (TaKaRa) and quantified on a LightCycler 480 (Roche Life Science) with KAPA SYBR FAST qPCR Master Mix (Sigma-Aldrich) according to the manufacturer’s instructions. The expression levels of target genes were normalized against ACTIN7. Three independent biological repeats were performed. Error bars represent sd. Statistical analysis was performed using one-way ANOVA (Supplemental File). Primers are listed in the Supplemental Data Set.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (https://www.arabidopsis.org/) under the following accession numbers: MED25, At1g25540; MYC2, At1g32640; MYC3, At5G46760; MYC4, At4G17880; VSP1, At5g24780; VSP2, At5g24770; PDF1.2, At5g44420; THI2.1, At1g72260; JAZ2, At1g74950; JAZ3, At3G17860; JAZ4, At1G48500; JAZ5, At1G17380; JAZ6, At1g72450; JAZ9, At1G70700; JAZ10, At5G13220; JAZ12, At5G20900; ACTIN7, At5g09810; PRP39a, At1G04080; PRP39b, At5G46400; PRP40a, At1G44910; PRP40b, At3G19670; PRP40c, At3G19840; RESISTANCE TO LEPTOSPHAERIA MACULANS3, At4G16990; NUDIX HYDROLASE HOMOLOGY7, At4G12720; NON-PHOTOTROPHIC HYPOCOTYL4, At5G20730; RUBISCO METHYLTRANSFERASE, At5G14260; ALWAYS EARLY3, At3G21430; PRR7, At5G02810.
Supplemental Data
Supplemental Figure 1. MeJA-induced generation of ∆PYJAZs and JAZ10.4 depends on MYC2 and MED25.
Supplemental Figure 2. Generation and characterization of JAZ6-GFP and ∆PYJAZ6-GFP transgenic plants.
Supplemental Figure 3. Overexpression of ∆PYJAZ6 attenuates JA responses.
Supplemental Figure 4. ∆PYJAZ6 competes with MED25 for interaction with MYC2.
Supplemental Figure 5. Sequence alignment of PRP39a or PRP40a with their respective paralogs in Arabidopsis.
Supplemental Figure 6. Generation of PRP40a-GFP, PRP39a-GFP, and PRP39a-myc transgenic plants.
Supplemental Figure 7. Immunoblot analysis of the PRP39a, PRP40a, and MED25 proteins in yeast strains used for Y2H assays.
Supplemental Figure 8. RT-qPCR showing the expression of PRP39a and PRP39b in response to MeJA in wild-type, myc2-2, and med25-4 seedlings.
Supplemental Figure 9. Immunoblotting analysis showing the protein accumulation of PRP40a in response to MeJA.
Supplemental Figure 10. ChIP and RIP RT-qPCR showing the enrichment of PRP39a and PRP40a on JAZ1, JAZ5, JAZ10, PRR7, and ACTIN7 chromatin and Pre-mRNA in response to MeJA.
Supplemental Figure 11. The AS deficiency phenotypes of prp39 and prp40 mutants.
Supplemental Figure 12. Identification of the prp39 and prp40 mutants.
Supplemental Figure 13. PRP39 and PRP40 promote the splicing of Jas intron.
Supplemental Figure 14. PCR-based detection of alternative Jas intron retention of JAZ5, JAZ6, JAZ9, and JAZ10.
Supplemental Figure 15. The JA responses are attenuated in prp39 and prp40 mutants.
Supplemental Figure 16. Depletion of MED25 impairs MeJA-induced recruitment of PRP39a and PRP40a to JAZ5 and JAZ10.
Supplemental Table. MED25-associating proteins identified with liquid chromatography–tandem mass spectrometry.
Supplemental Data Set. Primers Used in this Study.
Supplemental File. Statistical Analysis.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by the National Key Research and Development Program of China (grant 2016YFD0100600), the National Natural Science Foundation of China (grants 31730010, 31771448, 31770303, and 31601759), the Chinese Academy of Sciences Youth Innovation Promotion Association (grant 2014082), and the Tai-Shan Scholar Program from the Shandong Provincial Government (grant tsxk20150901).
AUTHOR CONTRIBUTIONS
C.Y.L., F.M.W., and Q.Z.Z designed the research; F.M.W., L.D., Q.C., and J.H.Z. performed the research; C.Y.L. and L.D. analyzed the data; L.D., Q.Z.Z., and C.Y.L. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Allen B.L., Taatjes D.J. (2015). The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Li L., Zhai Q., You Y., Deng L., Wu F., Chen R., Jiang H., Wang H., Chen Q., Li C. (2017). Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 114: E8930–E8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729. [DOI] [PubMed] [Google Scholar]
- Becerra S., Andrés-León E., Prieto-Sánchez S., Hernández-Munain C., Suñé C. (2016). Prp40 and early events in splice site definition. Wiley Interdiscip. Rev. RNA 7: 17–32. [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Leder P. (1999). The FF domain: A novel motif that often accompanies WW domains. Trends Biochem. Sci. 24: 264–265. [DOI] [PubMed] [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J.E. (1995). Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27: 933–942. [DOI] [PubMed] [Google Scholar]
- Björklund S., Gustafsson C.M. (2005). The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244. [DOI] [PubMed] [Google Scholar]
- Bork P., Sudol M. (1994). The WW domain: A signalling site in dystrophin? Trends Biochem. Sci. 19: 531–533. [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Carlsten J.O., Zhu X., Gustafsson C.M. (2013). The multitalented Mediator complex. Trends Biochem. Sci. 38: 531–537. [DOI] [PubMed] [Google Scholar]
- Çevik V., Kidd B.N., Zhang P., Hill C., Kiddle S., Denby K.J., Holub E.B., Cahill D.M., Manners J.M., Schenk P.M., Beynon J., Kazan K. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160: 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Chini A., Gimenez-Ibanez S., Goossens A., Solano R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33: 147–156. [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (2013). The Mediator complex and transcription elongation. Biochim. Biophys. Acta 1829: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466. [DOI] [PubMed] [Google Scholar]
- Dolan W.L., Chapple C. (2018). Transcriptome analysis of four Arabidopsis thaliana Mediator tail mutants reveals overlapping and unique functions in gene regulation. G3 (Bethesda) 8: 3093–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29: 1883–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Ge H., Roeder R.G. (1996). Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93: 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Fernández-Calvo P., Fernández G.M., Díez-Díaz M., Gimenez-Ibanez S., López-Vidriero I., Godoy M., Fernández-Barbero G., Van Leene J., De Jaeger G., Franco-Zorrilla J.M., Solano R. (2014). bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One 9: e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens J., Fernández-Calvo P., Schweizer F., Goossens A. (2016). Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91: 673–689. [DOI] [PubMed] [Google Scholar]
- Görnemann J., Barrandon C., Hujer K., Rutz B., Rigaut G., Kotovic K.M., Faux C., Neugebauer K.M., Séraphin B. (2011). Cotranscriptional spliceosome assembly and splicing are independent of the Prp40p WW domain. RNA 17: 2119–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., Pinot F. (2012). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287: 6296–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando C.E., García Hourquet M., de Leone M.J., Careno D., Iserte J., Mora Garcia S., Yanovsky M.J. (2019). A role for pre-mRNA-PROCESSING PROTEIN 40C in the control of growth, development, and stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 10: 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li W., Yao X., Lin Q.J., Yin J.W., Liang Y., Heiner M., Tian B., Hui J., Wang G. (2012). Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell 45: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.H., Feng Y., Vikram M., Jeong I.S., Lee J.R., Bahk J.D., Yun D.J., Lee S.Y., Koiwa H. (2009). Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 484: 30–38. [DOI] [PubMed] [Google Scholar]
- Kanno T., Lin W.D., Fu J.L., Chang C.L., Matzke A.J.M., Matzke M. (2017). A genetic screen for pre-mRNA SPLICING MUTANTS of Arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics 207: 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.Y., Siliciano P.G. (1996). Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol. Cell. Biol. 16: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: The master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kidd B.N., Cahill D.M., Manners J.M., Schenk P.M., Kazan K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22: 741–748. [DOI] [PubMed] [Google Scholar]
- Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011). Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52: 1757–1765. [DOI] [PubMed] [Google Scholar]
- Koo A.J., Cooke T.F., Howe G.A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc. Natl. Acad. Sci. USA 108: 9298–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J., Howe G.A. (2012). Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235–239. [DOI] [PubMed] [Google Scholar]
- Kotovic K.M., Lockshon D., Boric L., Neugebauer K.M. (2003). Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23: 5768–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I., Sapra A.K., Neugebauer K.M. (2006). Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13: 815–822. [DOI] [PubMed] [Google Scholar]
- Liu Y., Du M., Deng L., Shen J., Fang M., Chen Q., Lu Y., Wang Q., Li C., Zhai Q. (2019). MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 31: 106–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart S.R., Rymond B.C. (1994). Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol. Cell. Biol. 14: 3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S. (2016). Eukaryotic transcription regulation. Getting to the heart of the matter: Commentary on Mediator architecture and RNA polymerase II function by Plaschka et al. J. Mol. Biol. 428: 2575–2580. [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. (2005). Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30: 256–263. [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. (2010). The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O., Neumerkel J., Dippe M., Stenzel I., Wasternack C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177: 114–127. [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Shyu C., Campos M.L., Patel L.C., Chung H.S., Yao J., He S.Y., Howe G.A. (2013). Negative feedback control of jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 162: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.P., Greenleaf A.L. (2000). The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275: 39935–39943. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakata M., Mitsuda N., Herde M., Koo A.J.K., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Eggermont K., Terras F.R., Thomma B.P., De Samblanx G.W., Buchala A., Métraux J.P., Manners J.M., Broekaert W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss Z.C., Ebmeier C.C., Taatjes D.J. (2013). The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48: 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S., Thakur J.K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6: 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y., Jikumaru Y., Obayashi T., Saito H., Masuda S., Kamiya Y., Ohta H., Shirasu K. (2013). Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 163: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Fan M., Zhang X., Gao H., Huang H., Wu D., Guo H., Xie D. (2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9: e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J., Wydau S., Ambroise Y., Boschiero C., Werner M. (2011). Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331: 1451–1454. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- VanDoorn A., Bonaventure G., Schmidt D.D., Baldwin I.T. (2011). Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 190: 640–652. [DOI] [PubMed] [Google Scholar]
- Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., Olsen O.A. (2007). The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26: 1357–1366. [DOI] [PubMed] [Google Scholar]
- Wang H., Li S., Li Y., Xu Y., Wang Y., Zhang R., Sun W., Chen Q., Wang X.J., Li C., Zhao J. (2019). MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling. Nat. Plants 5: 616–625. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., Xie Q. (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8: 1820–1823. [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li L., Qu L.J. (2016). Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 58: 106–118. [DOI] [PubMed] [Google Scholar]
- Yin J.W., Wang G. (2014). The Mediator complex: A master coordinator of transcription and cell lineage development. Development 141: 977–987. [DOI] [PubMed] [Google Scholar]
- You Y., Zhai Q., An C., Li C. (2019). LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31: 2187–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Li L., An C., Li C. (2018). Conserved function of mediator in regulating nuclear hormone receptor activation between plants and animals. Plant Signal. Behav. 13: e1403709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan C., Li L., Xie D., Li C. (2017). Jasmonates In Hormone Metabolism and Signaling in Plants, Smith M.S., Li C., and Li J., eds (London: Elsevier; ), pp. 243–272. [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ke J., Zhang L., Chen R., Sugimoto K., Howe G.A., Xu H.E., Zhou M., He S.Y., Melcher K. (2017). Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. National Acad. Sci. USA 114: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., et al. (2015). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525: 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]