An interkingdom interaction between the endophytic red yeast Rhodotorula mucilaginosa and diazotrophic endobacteria improves N nutrition in rice.
Abstract
Nitrogen (N) limits crop yield, and improvement of N nutrition remains a key goal for crop research; one approach to improve N nutrition is identifying plant-interacting, N2-fixing microbes. Rhodotorula mucilaginosa JGTA-S1 is a basidiomycetous yeast endophyte of narrowleaf cattail (Typha angustifolia). JGTA-S1 could not convert nitrate or nitrite to ammonium but harbors diazotrophic (N2-fixing) endobacteria (Pseudomonas stutzeri) that allow JGTA-S1 to fix N2 and grow in a N-free environment; moreover, P. stutzeri dinitrogen reductase was transcribed in JGTA-S1 even under adequate N. Endobacteria-deficient JGTA-S1 had reduced fitness, which was restored by reintroducing P. stutzeri. JGTA-S1 colonizes rice (Oryza sativa), significantly improving its growth, N content, and relative N-use efficiency. Endofungal P. stutzeri plays a significant role in increasing the biomass and ammonium content of rice treated with JGTA-S1; also, JGTA-S1 has better N2-fixing ability than free-living P. stutzeri and provides fixed N to the plant. Genes involved in N metabolism, N transporters, and NODULE INCEPTION-like transcription factors were upregulated in rice roots within 24 h of JGTA-S1 treatment. In association with rice, JGTA-S1 has a filamentous phase and P. stutzeri only penetrated filamentous JGTA-S1. Together, these results demonstrate an interkingdom interaction that improves rice N nutrition.
INTRODUCTION
Nitrogen (N) is a vital macronutrient for plant growth, but the limited availability of usable N has led to the extensive use of N fertilizers, which are both energy intensive to produce and environmentally unfriendly to use. N fixation, that is the conversion of N2 to forms the plant can use, provides an attractive alternative to exogenous fertilizers. However, the ability to fix atmospheric N2 is exclusive to prokaryotes and these N2-fixing prokaryotes are termed diazotrophs. Moreover, only a monophyletic group of angiosperms of the order Eurosids I interacts with a group of specific diazotrophs to exploit the N fixed by these diazotrophs within symbiotic nodules (Soltis et al., 1995). These findings have prompted the development of synthetic biology approaches to engineer N2-fixing symbiosis in cereals (Rogers and Oldroyd, 2014).
In addition to symbiotic species, the role of free-living diazotrophs in improving N metabolism is increasingly being studied (Geurts et al., 2016), as many of these organisms colonize plants as endophytes. For example, many members of the Poaceae derive a significant fraction of their N from biological N2 fixation (Carvalho et al., 2014); however, N2 is sequestered at relatively low efficiency through these interactions (Dobbelaere et al., 2003; Carvalho et al., 2014). Other studies have reported that diazotrophs associate with fungi in nature (Gehrig et al., 1996; Wang et al., 2017). Despite this, the importance of the interactions of beneficial fungi with bacteria in plant N metabolism remains to be investigated.
Endophytes are plant-associated microbes that asymptomatically colonize the plant endosphere. The community structure of endophytes in planta is primarily dictated by the soil in which the host plant grows, with plant genotype having only a limited influence (Bulgarelli et al., 2012; Lundberg et al., 2012; Peiffer et al., 2013; Schlaeppi et al., 2014; Edwards et al., 2015). Plants growing in marginal and contaminated environments harbor specialized microbes (Barzanti et al., 2007) with properties that help their hosts cope with deficiencies of critical nutrients. The broad host ranges of endophytes make this host–microbe interaction an exciting area of study, since it opens up the possibility of transmitting the critical properties of endophytes to agriculturally valuable surrogate host plants (Khan et al., 2012; Knoth et al., 2013). Fungal and bacterial endophytes coexist within the plant endosphere, providing a niche for intimate transkingdom interactions. Such interactions are largely ignored in plant–microbe interaction studies.
Many plant-associated fungi serve as hosts to endosymbiont N2-fixing microbes. These interkingdom interactions may allow the eukaryotes (fungi and, in turn, plants) to make use of the N2 fixed by the bacterial endosymbionts. For example, Geosiphon pyriforme, an arbuscular mycorrhizal (AM) fungus, harbors N2-fixing Nostoc cyanobacteria. Intracellular bacteria have been identified in various mycorrhizal species in nature (MacDonald and Chandler, 1981; Scannerini and Bonfante, 1991) and in the mycorrhiza-like fungus Serendipita indica. Gram-negative Candidatus Glomeribacter gigasporarum and the recently sequenced Mollicutes-related coccoids (Torres-Cortés et al., 2015) are endosymbionts of AM fungi Gigaspora margarita and Dentiscutata heterogama, respectively. Ustilago maydis, the causative fungus of maize smut disease, maintains N2-fixing Bacillus pumilus as an endosymbiont (Kobayashi and Crouch, 2009; Ruiz-Herrera et al., 2015). Rhizopus sp that cause rice blight harbor the rhizoxin-producing bacterium Burkholderia rhizoxinica (Partida-Martinez and Hertweck, 2005). The interaction of the human opportunistic pathogen Candida albicans with Pseudomonas aeruginosa is another example of an interkingdom association (Hogan and Kolter, 2002).
Interkingdom interactions have primarily been studied from the standpoint of pathogenesis (Valdivia and Heitman, 2007), whereas beneficial tripartite interactions have rarely been investigated in detail. One example of such a beneficial interaction in plants is the interaction between an endophytic fungus harboring a viral endosymbiont and tropical panic grass (Dichanthelium lanuginosum), where the virus–fungus duo confers heat tolerance to the plant (Márquez et al., 2007). An example of a eukaryote that uses the N2 fixed by a diazotrophic endosymbiont within fungi for nutrition is leafcutter ants of the Amazon ecosystem. The ants grow fungal gardens for food that are enriched in N2-fixing diazotrophs dominated by Klebsiella sp (Pinto-Tomás et al., 2009). Our study shows a similar three-way interkingdom interaction that improves N nutrition of plants.
Rhodotorula mucilaginosa JGTA-S1 was isolated as a shoot endophyte of narrowleaf cattail (Typha angustifolia), the predominant flora of a tailings pond associated with a uranium mine in Jaduguda, India (22.65°N, 86.35°E). The tailings pond is a metal-contaminated, highly anaerobic geochemical environment where metal-laden sludge is disposed of following the extraction of uranium ore. JGTA-S1 was one of two endophytic yeast strains isolated from Typha, along with 10 other bacterial endophytes (Saha et al., 2016). Endophytic yeasts were previously reported in maize (Zea mays), banana (Musa acuminata), rice (Oryza sativa), tomato (Solanum lycopersicum), wheat (Triticum aestivum), poplar (Populus trichocarpa), and several other plant species (Larran et al., 2001; Cao et al., 2002; Larran et al., 2002; Tian et al., 2004; Nassar et al., 2005; Gai et al., 2009; Xin et al., 2009). None of these yeast strains were shown to have detrimental effects on their plant hosts. JGTA-S1, like Rhodotorula yeasts isolated from cottonwood (Populus trichocarpa) and hybrid poplar (P. trichocarpa × Populus deltoids), has important beneficial effects on plant growth (Xin et al., 2009; Sen et al., 2019). Rhodotorula are rare unicellular fungi from the phylum Basidiomycota. R. mucilaginosa (previously R. rubra) promotes mycorrhizal colonization in soybean (Glycine max; Sampedro et al., 2004). Rhodotorula sp also exhibit biocontrol activity in postharvest apple (Malus domestica) and strawberries (Fragaria ananassa; Li et al., 2011; Zhang et al., 2014). R. mucilaginosa showed enhanced bioprotective effects when used to inoculate rice stems (Akhtyamova and Sattarova, 2013).
Since the publication of the first plant endophytic yeast genome sequence, from Rhodotorula graminis WP1 (Firrincieli et al., 2015), the genomes of 22 Rhodotorula yeasts have been sequenced, including R. mucilaginosa JGTA-S1 (accession number PEFX00000000). The JGTA-S1 genome was recently compared with the genomes of three other Rhodotorula yeasts (R. graminis WP1, R. sp JG1b, and R. mucilaginosa C2.5t1) and Rhodosporidium toruloides NP11, revealing several unique aspects of its endophytic lifestyle and plant growth-promoting activities (Sen et al., 2019). JGTA-S1 solubilizes mineral phosphate (P), as does the endophytic yeast R. graminis (Xin et al., 2009). The genome of JGTA-S1 contains several genes potentially able to promote plant growth due to their roles in producing the plant hormone auxin and several inorganic and organic phosphatases and P-uptake transporters (Sen et al., 2019). Additionally, the JGTA-S1 genome contains the first gene of the cytokinin biosynthesis pathway and a gene encoding isopentenyl-diphosphate ∆-isomerase, an enzyme that converts cytokinin from its inactive to its active form. Biosynthetic genes for abscisic acid, gibberellin, and jasmonic acid and genes responsible for sterol biogenesis and metabolism with the potential to influence the brassinosteroid biosynthesis pathway were also identified in the JGTA-S1 genome (Sen et al., 2019).
In this study, we explored the interaction between JGTA-S1 and rice, as this yeast significantly improves rice growth and N content. Our findings reveal that JGTA-S1 has a dimorphic life cycle in association with plants and harbors a N2-fixing endobacterium, Pseudomonas stutzeri. The fungus–bacterium interaction is critical for the survival of JGTA-S1, and for its ability to fix N2. JGTA-S1 also fix N2 in planta and supply N to rice. These findings uncover a beneficial tripartite interaction involving the diazotroph P. stutzeri, the yeast JGTA-S1, and the important grain crop rice; this interaction could potentially be used to improve N nutrition in cereals.
RESULTS
Rhodotorula Colonizes Rice Endophytically and Promotes Plant Growth
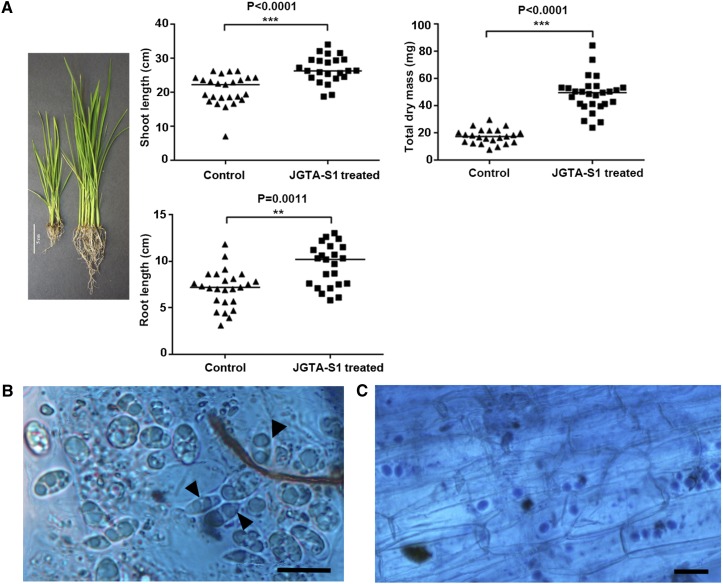
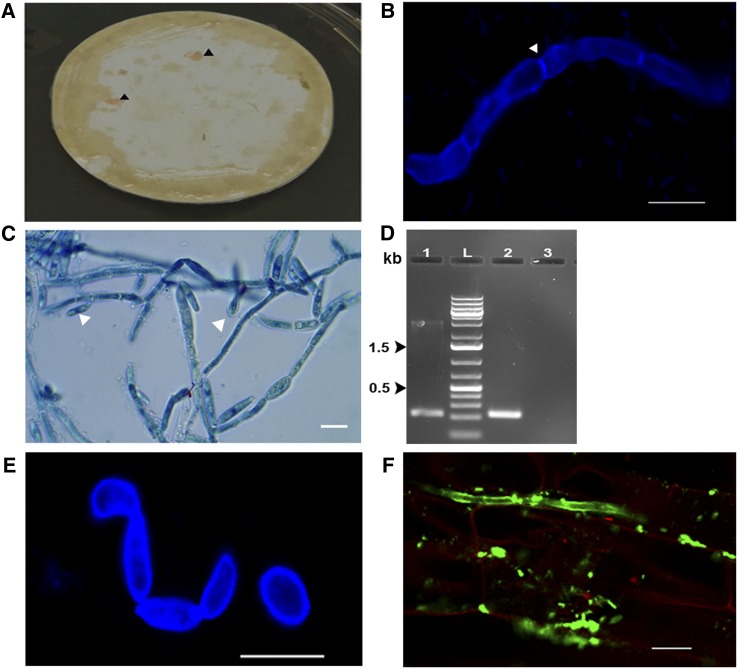
R. mucilaginosa JGTA-S1 was previously isolated as a shoot endophyte of Typha. This fungus, which forms pink colonies, was initially identified by BLAST and phylogenetic analysis of 18S rDNA (KP233783) and D1/D2 region sequences (KP233784). The JGTA-S1 genome was recently sequenced (accession number PEFX00000000). JGTA-S1 has a substantial plant growth-promoting effect on its host Typha (Sen et al., 2019), prompting us to investigate whether it would have a similar effect on rice cv Swarna MTU 7029). The plants were grown on Soilrite containing Murashige and Skoog (MS) medium (N-replete conditions) for these experiments. A Mann-Whitney test for shoot length, root length, and biomass (dry mass) in yeast-treated plants indicated that the effect on plant growth was highly significant (P-value < 0.0001 for dry mass and P-value < 0.0011 for shoot length) and substantial (fold change ∼2.8 for dry mass, ∼1.3 for shoot length, and ∼1.35 for root length) in rice plants at 3 weeks postinfection (wpi; Figure 1A). Similar results were obtained with two other varieties of rice, Bordan and IR64 (Supplemental Figures 1A and 1B).
Figure 1.
R. mucilaginosa JGTA-S1 Associates with Roots and Promotes Plant Growth in Rice.
Four- to 5-d-old rice plants of equal length were grown in Soilrite supplemented with MS medium (N-replete conditions) with or without JGTA-S1. Plant growth was measured at 3 wpi by monitoring.
(A) Shoot length, root length (control, n = 25; treated, n = 23), and dry mass (control, n = 23; treated, n = 27). (Left) Digital image of control and treated rice. Bar =5 cm. (Right) Mann-Whitney test for shoot length, root length, and total dry mass values (***P-value < 0.001, **P-value 0.001 to 0.01).
(B) and (C) Yeast attached to rice roots or root sections stained with lactophenol cotton blue. Binucleated telial cells attached to rice root cells (see [B], arrowheads). Bar = 5 µm. Yeast cells within the rice root section (see [C]). Bar = 10 µm.
The fungus attached to the rice root and underwent morphological changes within 24 h, demonstrating that it was completing its life cycle using rice as a host. Binucleated telial cells and diploid thick-walled teliospores were observed both outside and inside root cells within 7 d, which is typical of a Pucciniomycotina (Figure 1B). More than one yeast cell colonized a single root cell (Figure 1C), demonstrating that JGTA-S1 has an endophytic lifestyle.
Microarray Analysis of Early Changes in the Root Transcriptome upon R. mucilaginosa Treatment
We performed global transcriptomic analysis to explore the nature of the interaction between rice and JGTA-S1. Since Rhodotorula underwent morphological changes within 24 h postinfection (hpi), we analyzed changes in the rice root transcriptome at 24 hpi compared with the 0-h control to gain insight into the early genes regulated in response to JGTA-S1. A cutoff of log2 (fold change) ≥2 or −2 (P-value cutoff, 0.05) was used to identify differentially expressed genes (DEGs) between treatments (Supplemental Data Sets 2 and 3, Gene Expression Omnibus [GEO] accession number GSE99075). We performed gene clustering of all DEGs using the Multiple Experiment Viewer via Pearson correlation analysis. DEGs in plants under the same treatment showed uniform clustering (Supplemental Figure 2). The root transcriptome showed extensive changes compared with that in shoots (GEO accession number GSE64321; Supplemental Data Sets 2 and 3; Saha and Seal, 2015), with 2797 genes differentially regulated within 24 hpi in roots. The microarray data were validated by RT-qPCR of selected genes (Supplemental Figure 3).
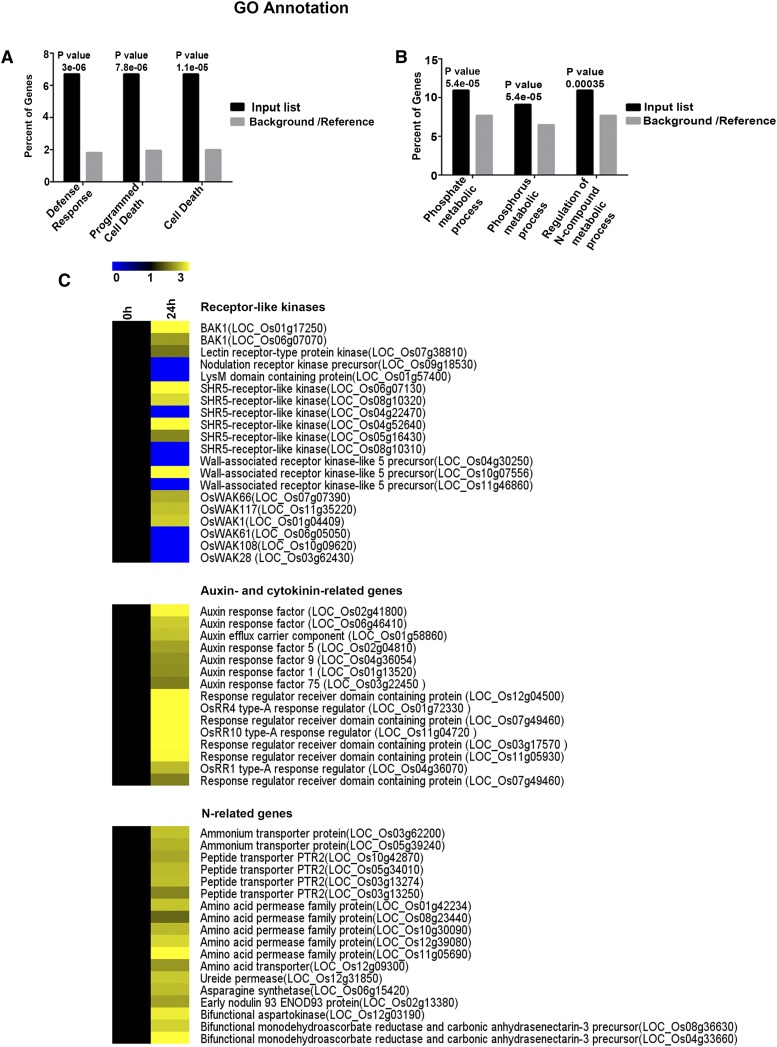
We subjected the upregulated and downregulated DEGs to Gene Ontology enrichment analysis using AgriGO (Du et al., 2010). There was clear evidence of the suppression of plant defense responses and apoptosis in the roots at 24 hpi, underscoring the notion that these processes were repressed to promote invasion and colonization (Figure 2A). This observation corroborates the finding that the JGTA-S1 genome contains multiple genes useful for escaping plant defense (Sen et al., 2019). P and N metabolic processes were significantly enriched among the DEGs, indicating that Rhodotorula promotes P and N metabolism in rice (Figure 2B). Evidence that JGTA-S1 improves P nutrition was substantiated from the genome data as well as experimentally (Sen et al., 2019), although JGTA-S1 infection did not alter total P content per plant (Supplemental Figure 4).
Figure 2.
DEGs in Rice in Response to R. mucilaginosa JGTA-S1.
(A) and (B) Downregulated (A) and upregulated (B) DEGs in rice roots at 24 hpi were subjected to AgriGO singular enrichment analysis, and enriched categories for biological function were plotted. The P-values indicating significance are listed above the bars; “e” denotes the scientific notation for power of 10. GO, Gene Ontology.
(C) Heatmap of the expression patterns of various genes.
DEGs Regulated by R. mucilaginosa in Rice Roots
Receptor-Like Protein Kinases and Defense-Related Genes
Receptor-like kinases (RLKs) represent major DEGs regulated by JGTA-S1 in rice roots (Supplemental Data Set 4). RLKs are key factors in the recognition of microbe-associated molecular patterns and regulate various processes ranging from plant development to plant defense in response to microbes. Several RLK genes, including genes for wall-associated kinases and members from SHR5-like receptor kinases were differentially regulated in rice roots and shoots (Figure 2C). Three homologs of BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 were also differentially regulated in a tissue-specific manner. A Cys-rich RLK gene was upregulated in roots, and an S-domain RLK gene was downregulated in roots. A nodulation-dependent receptor kinase precursor was downregulated in roots, and a CLAVATA-like receptor kinase precursor was downregulated in shoots upon JGTA-S1 infection (GSE64321; Saha and Seal, 2015). Genes belonging to the STRUBBELIG-RECEPTOR FAMILY of LRR kinases, which are usually involved in organ development (Chevalier et al., 2005), were upregulated in roots. We identified two LysM domain-containing protein-coding genes, one downregulated in roots and one upregulated in shoots. Putative Subtilisin protease homologs and NBS-LRR and NB-ARC genes implicated in pattern recognition and plant immunity (Pearce et al., 2010; Figueiredo et al., 2014) were also among the DEGs (Supplemental Data Set 4).
Hormone-Related Genes
Auxin- and cytokinin-regulated genes were highly upregulated in rice roots upon JGTA-S1 infection (Supplemental Data Set 4). Auxin response factor and putative cytokinin-regulated type-A response regulator genes were upregulated in roots (Figure 2C). The transcriptome data corroborate the finding that the JGTA-S1 genome sequence contains many genes potentially able to influence hormone signaling in plants (Sen et al., 2019), indicating that this yeast is able to manipulate multiple plant hormonal responses.
Genes Related to N Uptake and Metabolism
JGTA-S1 induced the expression of several genes essential for inorganic and organic N uptake and assimilation in roots. The 0- and 24-h time points represent the same phase in the circadian clock, eliminating the possibility that the up- or downregulation of the genes, especially those related to N metabolism, is diurnally controlled (Gutiérrez et al., 2008) rather than an effect of JGTA-S1 infection. Two ammonium transporter genes were upregulated in rice upon JGTA-S1 infection (Supplemental Data Set 4). Organic N transport appeared to be enhanced as well, as many genes encoding PTR2-like peptide transporters, amino acid permeases, and transporters were upregulated upon JGTA-S1 infection. An ANT1-like gene encoding aromatic and neutral amino acid transporter and a ureide permease gene were upregulated upon JGTA-S1 treatment. The upregulation of a ureide permease gene is an interesting finding, as Rhodotorula produces urease and can use urea, a commonly used N fertilizer, as the sole N source (Zimmer and Roberts, 1979). The N-assimilation–related DEGs included an Asn synthetase gene, which was upregulated in roots. An ENOD93 gene was also induced in roots. Among other genes related to N metabolism was a bifunctional aspartokinase gene (Figure 2C).
We searched for N-regulated rice genes described in two recent reports (Obertello et al., 2015 [GSE38102]; Coneva et al., 2014 [GSE61370]) among the JGTA-S1–regulated rice genes. In the GSE38102 experiment, rice plants were supplemented with KNO3 and NH4NO3 and compared with control plants supplied with KCl, while in the GSE61370 experiment, plants were grown under different nitrate regimes. We searched for DEGs regulated ≥1.5- or −1.5-fold in these studies in our data. Despite significant differences in experimental conditions, many of the N-regulated genes described in these reports, especially those differentially regulated upon N treatments (normal versus induced, GSE61370 or −N versus +N, GSE38102), were regulated in a comparable manner in our data set (Supplemental Data Sets 5 and 6). This similarity indirectly supports the positive influence of JGTA-S1 on N metabolism in rice.
N-Uptake Genes Exhibit Sustained Upregulation in Roots
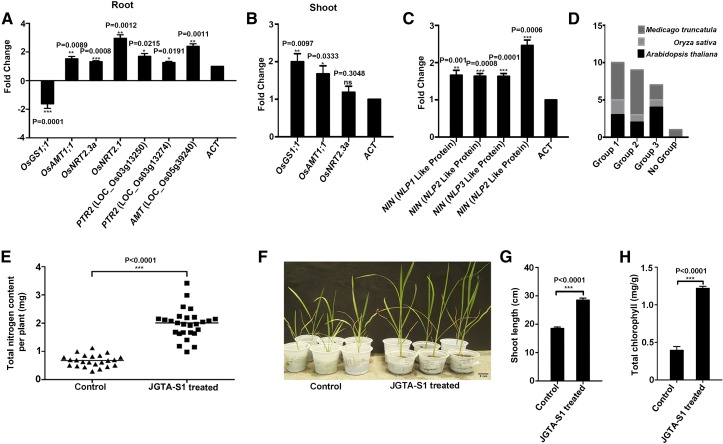
The growth-promoting effect of Rhodotorula on rice plants became exceedingly prominent at 3 to 4 wpi. We performed RT-qPCR of selected N-uptake and -assimilation genes to measure their expression levels at 3 wpi. The N-transporter genes showed sustained upregulation in JGTA-S1–treated plant roots compared with control roots. The ammonium transporter gene OsAMT1.1 (LOC_Os04g43070) was upregulated in both shoots and roots. OsNRT2.3a (LOC_Os01g50820), a nitrate transporter gene, was also induced in rice shoots compared with the untreated control. A Gln synthetase gene (OsGS1-1, LOC_Os02g50240) also maintained increased transcript levels in shoots even after 3 wpi. In roots, OsNRT2.1 and two PTR2-like genes showed sustained upregulation, pointing to a possible increase in N uptake and a persistent N-deficiency signal in the plants triggered by JGTA-S1 treatment (Figures 3A and 3B).
Figure 3.
R. mucilaginosa JGTA-S1 Improves N Metabolism in Rice.
(A) and (B) Rice plants were grown under N-replete conditions and treated with or without JGTA-S1. RNA was isolated from root and shoot tissue at 3 wpi and used for RT-qPCR. Expression levels of N-uptake and -assimilation genes at 3 wpi in (A) roots and (B) shoots.
(C) Expression levels of rice NIN-like transcription factor genes at 3 wpi.
(D) OrthoMCL clustering of rice NLP genes.
(E) Total N content per plant grown under N-replete conditions, as measured by the Kjeldahl method.
(F) to (H) Rice plants grown on artificial soil without N supplementation (low-N conditions). Effect of JGTA-S1 supplementation on rice was monitored compared with the untreated control after 10 DAI. (F) Digital image of rice plants. Shoot length (G) and chlorophyll content (H). Statistical analysis was performed using Mann-Whitney unpaired two-tailed t test. Data are given as mean ± se.
JGTA-S1 Induces the Expression of Four NIN-Like Genes
Four homologs of NODULE INCEPTION (NIN)–like genes, LOC_Os11g16290, LOC_Os09g37710, LOC_Os01g13540, and LOC_Os04g41850, were upregulated upon JGTA-S1 treatment. Only LOC_Os11g16290 crossed the threshold of twofold induction at 24 hpi. The other genes showed greater than twofold induction at 6 hpi but less than twofold induction at 24 hpi. Although first identified based on their regulatory role in nodulation, transcription factors related to NIN-like proteins (NLPs) appear to play central roles in N-(nitrate) metabolism (Ferris and Goodenough, 1997; Konishi and Yanagisawa, 2013; Yan et al., 2016) in nonnodulating plants such as Arabidopsis (Arabidopsis thaliana). Excluding LOC_Os04g41850, which was upregulated 1.3-fold at 24 hpi, all other NLPs were upregulated >1.8-fold in JGTA-S1–treated roots at 24 hpi. This upregulation was sustained even after 3weeks (Figure 3C).
Even though the roles of these rice NLP genes in nitrate responses have yet to be determined experimentally, we performed OrthoMCL analysis for comparison of orthologous groups across multiple taxa to gain insight into their likely functions. All four genes clustered into three orthologous groups compared with Medicago truncatula and Arabidopsis NLPs/NINs (Figure 3D). LOC_Os04g41850.1 and LOC_Os11g16290.1 were within the same orthologous group as AtNLP8, Medtr2g099350, and Medtr4g068000. LOC_Os01g13540.1 appears to be orthologous to AtNLP7, and Medtr0022s0430 and Medtr1g100970. LOC_Os09g37710.2 belonged to a group consisting of AtNLP1, AtNLP2, AtNLP4, and AtNLP5 and Medtr3g115400.
JGTA-S1 Infection Increases NUE in Rice
We measured total N levels in three different rice varieties to confirm the effect of JGTA-S1 treatment on N metabolism in rice. The total N content per plant increased significantly (fold change ∼2.95) in O. sativa cv Swarna MTU7029 (Figure 3E) upon JGTA-S1 treatment. Similar increases were observed for rice varieties IR64 (fold change ∼1.77) and Bordan (fold change ∼1.97; Supplemental Figures 5A and 5B). Since the amount of N application was identical for all plants, we calculated the relative N-use efficiency (NUE) based on the biomass of JGTA-S1–treated versus control plants (Saha et al., 2016). The relative NUE increased ∼2.62-fold upon JGTA-S1 treatment in O. sativa cv Swarna MTU7029. These results confirm the notion that JGTA-S1 improves N nutrition in rice, as predicted by the transcriptome data. This increase was independent of the rice variety examined.
JGTA-S1 Improves Rice Growth in Low-N Medium
Nutrient-regulated developmental programs in plants often influence the expression of genes involved in symbiosis. The N-regulated genes control nodulation in legumes (Benedito et al., 2008). The effect of JGTA-S1 on N nutrition was confirmed when plants were grown on low-N medium (Soilrite without supplementation of exogenous N, with or without JGTA-S1). Plants not supplemented with JGTA-S1 were supplied with N-free medium. The untreated plants turned chlorotic within 10 d counting from the day the treated plants were supplemented with JGTA-S1. We evaluated the effects of JGTA-S1 by measuring the shoot length and chlorophyll contents of the plant. Supplementing the rice plants with JGTA-S1 increased the shoot length ∼1.5-fold (P-value < 0.0001). Chlorophyll content also significantly increased (fold change ∼3.09, P-value < 0.0001; Figures 3F to 3H). The positive effect of JGTA-S1 on N metabolism is a key observation, as analysis of the JGTA-S1 genome indicated that this organism contains an incomplete nitrate assimilation pathway, suggesting that it is unable to take up nitrate as a N source or convert nitrate and nitrite to ammonium (Sen et al., 2019). However, JGTA-S1 appeared to be unique in its ability to grow in the absence of any nitrogen source, exactly like a diazotroph (Sen et al., 2019), making its effect on N nutrition in rice particularly important to investigate.
JGTA-S1 Exhibits a Dimorphic Life Cycle and Associates with Microbes in Planta
To confirm the endophytic lifestyle of JGTA-S1 in rice, we passed extracts from JGTA-S1–infected surface-sterilized plants through 0.45-µm filters. We incubated the filters as described in Methods until pink foci typical of JGTA-S1 were visible (Figure 4A). The time required for pink foci to appear varied from experiment to experiment and took 14 d or more in cold conditions (5°C). Growing the cold-tolerant JGTA-S1 (Sen et al., 2019) at 5°C provided it with a significant growth advantage over the bacteria associated with JGTA-S1, which were coisolated and difficult to eliminate. The pink JGTA-S1 material formed adherent foci on the filters. Once the foci were visible, we stained part of the material with calcofluor (a fluorochrome that binds to cellulose and chitin in plant and fungal cell walls) or lactophenol cotton blue and observed it under a confocal or bright-field microscope. Extensive filamentous structures with true septate hyphae were observed (Figures 4B and 4C). Directional budding and branching were also observed (Figure 4C, arrowhead). Such filaments were never observed when JGTA-S1 was grown in tryptic soya broth (TSB).
Figure 4.
R. mucilaginosa JGTA-S1 Shows a Transition from Yeast to Filamentous Form.
Rice plants grown under sterile conditions were supplemented with JGTA-S1. Extracts from surface-sterilized plants were passed through a 0.45-µm filter at 3 wpi. The filters were incubated on PDA plates for 3 d at 30°C, followed by 2 weeks or more at 5°C on PDA plates containing antibiotics.
(A) Pink material on filter paper (arrowhead).
(B) Calcofluor-stained JGTA-S1 showing septate hypha (arrowhead).
(C) Lactophenol cotton blue–stained JGTA-S1 filaments with budding yeast-like structures (arrowhead).
(D) Partial D1/D2 sequence amplified from genomic DNA isolated from filamentous JGTA-S1 and compared with the yeast form. Lane 1, amplicon from JGTA-S1 in filamentous form; lane L, DNA markers; lane 2, amplicon from JGTA-S1 in yeast form; lane 3, no DNA control.
(E) Calcofluor staining of JGTA-S1 filament induced by FK506.
(F) Rice roots infected with JGTA-S1 at 9 DAI stained with Alexa Fluor 488–labeled WGA and counterstained with propidium iodide. The roots were imaged under a confocal microscope. Bar = 10 µm
We amplified, cloned, and sequenced a fragment of the 26S rDNA (D1/D2) gene specific to JGTA-S1 (213 bp; Figure 4D) from the filamentous material to confirm that the hyphae belonged to JGTA-S1. Hyphal growth in the dimorphic fungus U. maydis is induced by mating pheromones, low pH, or N starvation (Martínez-Espinoza et al., 1997; Horst et al., 2012). In our case, JGTA-S1 appeared to be haploid, with no evidence of an opposite mating type (Sen et al., 2019). No hyphae formed when the yeast was grown on potato dextrose agar (PDA) plates at low pH or low temperature. Growing the JGTA-S1 cells in N-free medium resulted in the occasional development of chains of fungal cells (Supplemental Figure 6), a prerequisite for pseudohyphae formation (Gancedo, 2001), but this was distinct from the filaments observed when JGTA-S1 was extracted from plants and grown on filter paper.
JGTA-S1 grows quite rapidly in its yeast form. However, the time required to form visible pink foci containing the filamentous form of JGTA-S1 on filter paper following its extraction from the plant indicates that JGTA-S1 likely forms spores inside the plant, which germinate into slow-growing filaments following isolation from the rice endosphere. This finding demonstrates that the JGTA-S1 life cycle includes a dimorphic phase, which was not previously observed in Rhodotorula yeasts. Although filament formation was not induced by low pH, N deficiency, or low temperature, treatment with FK506, a specific inhibitor of calcineurin, induced the formation of filament-like structures in JGTA-S1 at a concentration of 1 µg/mL (Figure 4E). This observation confirms the phenomenon of dimorphism in JGTA-S1 and is consistent with the presence of an intact Mating locus in the JGTA-S1 genome, as the Mating locus is often related to the filamentous life cycle of yeasts (Sen et al., 2019).
Byr4 (g3863.t1) and Cdc16 (g5858.t1; Sen et al., 2019) homologs, which control septation in Schizosaccharomyces pombe, were also identified in the JGTA-S1 genome, supporting the notion that JGTA-S1 can form true hyphae. We stained rice seedlings infected with JGTA-S1 with Alexa Fluor 488–labeled wheat germ agglutinin (WGA), which specifically stains the fungal cell wall. JGTA-S1 filaments were observed inside the plant within 9 d after inoculation (DAI), indicating that in nature, filament formation obligatorily requires plant association (Figure 4F).
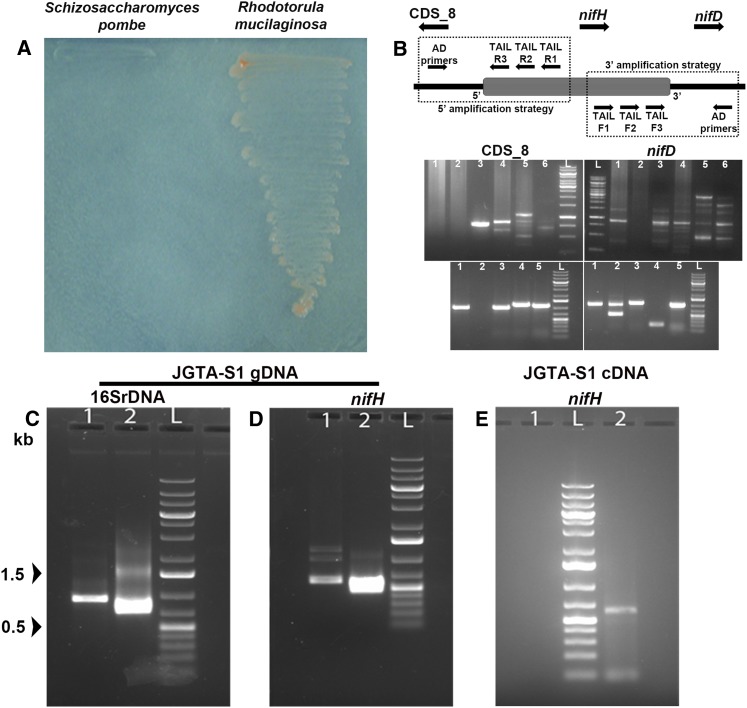
JGTA-S1 Harbors Multiple Diazotrophic Endobacteria
JGTA-S1 contains an incomplete N-metabolic pathway, yet it grew in N-free medium, whereas a laboratory strain of S. pombe did not (Figure 5A). U. maydis, the causative agent of maize smut disease, can grow in N-free medium due to the presence of the N2-fixing B. pumilus as an endosymbiont inside the fungus (Ruiz-Herrera et al., 2015). To investigate the presence of a potential N2-fixing endosymbiont in JGTA-S1, we attempted to amplify nifH genes from JGTA-S1 genomic DNA using degenerate primers (Supplemental Figure 7A). Four partial nifH sequences (MG566093 to MG566096) were cloned from JGTA-S1. BLASTX analysis of two of these nifH sequences (MG566093 and MG566095) showed that they share sequence similarity with genes from the N2-fixing Pseudomonas stutzeri (Supplemental Figure 7B), and with Bradyrhizobium japonicum (MG566094) and Bradyrhizobium sp (MG566096; Supplemental Figure 7C). Two partial nifH sequences amplified from an independent R. mucilaginosa strain (JGTA-R1) isolated from T. angustifolia roots also share similarity with genes from Bradyrhizobium sp (MG566097) and P. stutzeri (MG566098; Supplemental Figure 7B), indicating that these results were not artifacts. P. stutzeri and Bradyrhizobium sp–specific sequences were also found within the unmapped reads of the JGTA-S1 genome (Sen et al., 2019), suggesting that Bradyrhizobium sp are another potential endosymbiont of JGTA-S1. Any possibility of contamination was eliminated by subculturing the yeast several times in the presence of antibiotics, followed by microscopy.
Figure 5.
R. mucilaginosa JGTA-S1 Contains Bacteria Related to P. stutzeri as an Endosymbiont.
(A) JGTA-S1 was streaked onto N-free medium along with a laboratory strain of yeast S. pombe D18 h− to examine its diazotrophic nature.
(B) Flanking genes of P. stutzeri nifH from JGTA-S1 genomic DNA amplified using TAIL-PCR. The directions of the genes are represented as in P. stutzeri DSM4166. (Top, left) Amplification of the CDS_8-homolog. (Top, right) Amplification of the nifD-homolog using TAIL R3 or TAIL F3 gene-specific primers together with different AD primers. Lane 1, AD1; lane 2, AD2; lane 3, AD3; lane 4, AD4; lane 5, AD5; lane 6, AD6; lane L, DNA markers. (Bottom) Representative amplicons after colony PCR from putative CDS_8 and nifD clones.
(C) and (D) P. stutzeri 16SrDNA and nifH gene–specific primers were used to amplify P. stutzeri 16SrDNA (C) and nifH (D) from JGTA-S1 genomic DNA. (C) Lane 1, primary PCR product; lane 2, nested PCR with internal primers. (D) Lane 1, primary PCR product; lane 2, nested PCR with internal primer.
(E) RNA was isolated from JGTA-S1 and the cDNA used to amplify P. stutzeri–specific nifH. Lane 1, no DNA control; lane 2, nifH gene amplicon using nifH internal primers; lane L, DNA markers.
Incidentally, we previously isolated a P. stutzeri strain (P. stutzeri-JGTA-R3) along with the yeast from the Typha endosphere (Saha et al., 2016). P. stutzeri-JGTA-R3 efficiently colonized rice as an endophyte (Saha et al., 2016). Since no Bradyrhizobium strain was isolated, we studied the interaction of JGTA-S1 with P. stutzeri-JGTA-R3 in detail. We amplified and cloned full-length P. stutzeri–specific nifH and nifD sequences from JGTA-S1 genomic DNA. Thermal asymmetric interlaced (TAIL)-PCR was used for genome walking to clone the genes flanking the partial P. stutzeri nifH sequence using JGTA-S1 genomic DNA as a template. A CDS_8 gene homolog (as per P. stutzeri DSM4166 genome annotation) and nifD gene homolog flanking the nifH gene were amplified, cloned, and sequenced (Figure 5B). We further confirmed the presence of P. stutzeri in the yeast by reamplifying, cloning, and sequencing 16S rDNA (primary product 900 bp and nested product of 693 bp; Figure 5C) and nifH genes (primary product 603 bp and nested product of 454 bp; Figure 5D) from JGTA-S1 DNA using specific primers for P. stutzeri. Reads corresponding to P. stutzeri genomic sequences were also found within the raw JGTA-S1 Illumina reads not mapping to the genome (Sen et al., 2019), thereby confirming the notion that P. stutzeri is an endosymbiont of JGTA-S1. P. stutzeri–specific nifH and 16S rDNA sequences were successfully amplified from JGTA-S1 genomic DNA even after a gap of 7 years and several rounds of subculturing, confirming the stability of the association between the fungus and the endosymbiont. P. stutzeri nifH was transcribed in JGTA-S1 even when the yeast was grown in complete medium (TSB) containing sufficient organic and inorganic N sources (Figure 5E). Consistent with the presence of diazotrophic endosymbionts, R. mucilaginosa JGTA-S1 grew under anaerobic conditions (Supplemental Figure 7D) and fixed atmospheric N2. JGTA-S1 produced 0.2945 ± 0.0363 nmol of ethylene/h in an acetylene reduction assay, demonstrating why the yeast was able to survive in N-free medium. Transmission electron microscopy revealed bacteria-like structures within the yeast cells (Supplemental Figure 7E).
JGTA-S1 Contains Living Bacteria Inside Its Filaments
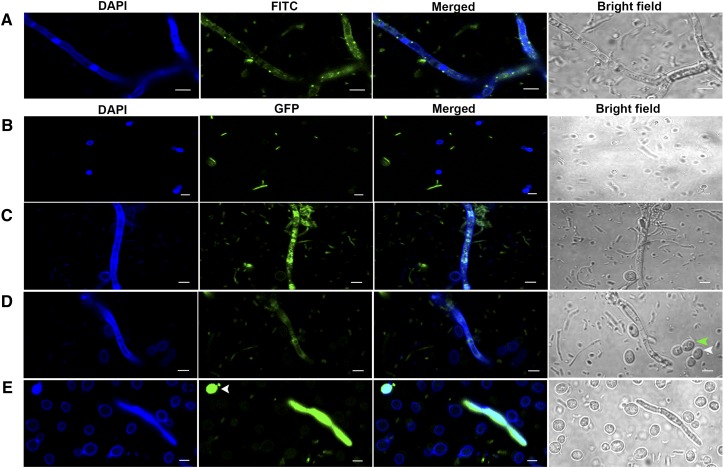
We stained the filamentous form of the fungus on filter paper using a LIVE/DEAD BacLight bacterial viability kit. Confocal microscopy revealed live bacteria in the cytoplasm of the yeast filaments (Figure 6A). When isolated from the plant, JGTA-S1 filaments were associated with many bacteria. These bacteria were likely seed-borne, as they were coisolated repeatedly even from untreated plants grown under sterile conditions (as a negative control).
Figure 6.
R. mucilaginosa JGTA-S1 in Its Filamentous Form, but Not in Its Yeast Form, Associates with Bacteria.
JGTA-S1 filaments were stained with a LIVE/DEAD BacLight bacterial viability kit, counterstained with calcofluor, and imaged by confocal microscopy. DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate.
(A) JGTA-S1 filament stained with a LIVE/DEAD BacLight bacterial viability kit. JGTA-S1 was incubated with P. stutzeri-JGTA-R3-GFP in yeast form or filamentous form.
(B) Microscopy after incubation of P. stutzeri-JGTA-R3-GFP with JGTA-S1 in yeast form.
(C) Microscopy after incubation of P. stutzeri-JGTA-R3-GFP with JGTA-S1 in filamentous form.
(D) Yeast or spores generated from filaments after incubation with P. stutzeri-JGTA-R3-GFP. Arrowheads show JGTA-S1 in yeast (white) and spore form (green). JGTA-S1 isolated from plants maintained for 4 weeks on filter paper at 5°C followed by incubation with P. stutzeri-JGTA-R3-GFP.
(E) Arrowhead shows a yeast cell with GFP fluorescence. Bar = 10 µm.
P. stutzeri-JGTA-R3 Penetrates and Becomes Established within R. mucilaginosa Hyphae
To investigate whether the P. stutzeri-JGTA-R3 strain in our collection was able to associate with JGTA-S1, we incubated green fluorescent protein (GFP)–labeled P. stutzeri-JGTA-R3 with R. mucilaginosa in both its yeast and filamentous forms. Confocal microscopy revealed no association between the bacteria and the yeast form of JGTA-S1, even after 20 h of coculture (Figure 6B). However, for JGTA-S1 in filamentous form, GFP fluorescence was observed inside the filament (Figure 6C), indicating that P. stutzeri-JGTA-R3 was able to invade the fungal filaments. The pink filamentous material on filter paper appeared to be extremely slow growing, with almost no yeast form initially detected. However, after incubation with P. stutzeri-JGTA-R3 for 20 h, binucleate asexual spores (Figure 6D, green arrowhead) and cells in yeast form started to bud off of the filaments (Figure 6D, white arrowhead). This phenomenon was not observed in a control experiment in which the yeast was not coincubated with P. stutzeri-JGTA-R3. These results suggest that the bacteria promote the transition of the yeast from the hyphal form.
The filamentous form was short lived. After prolonged incubation (4 weeks or more) of filter paper containing filamentous JGTA-S1 on nutrient-rich PDA medium at 5°C, most of the hyphae converted to the fast-growing yeast form, even without coincubation with P. stutzeri-JGTA-R3 (Figure 6E). Incubation with P. stutzeri-JGTA-R3-GFP at the 4-week time point resulted in accumulation of GFP fluorescence, even inside the yeast form of JGTA-S1, as well as JGTA-S1 filaments (Figure 6E, arrowhead). This finding confirms that the yeast form originated from filaments. The filamentous form was almost nonexistent after 4 weeks. Despite the abundance of the yeast form, very few cells showed GFP fluorescence, perhaps due to the loss of the GFP plasmid from P. stutzeri JGTA-R3-GFP or the unequal vertical transmission of the bacteria into daughter yeast cells, where the amount of labeled bacteria per cell became increasingly diluted in each generation.
Levels of the Endosymbiont Pseudomonas in JGTA-S1 Is Crucial for Its Survival
Since P. stutzeri-JGTA-R3 isolated from the Typha endosphere is sensitive to tetracycline and Timentin, we used these antibiotics to remove this endobacterium from JGTA-S1 cells by repeated streaking. A strain with reduced P. stutzeri levels known as strain 11 was previously generated in this manner (Sen et al., 2019). Three other strains were generated from strain 11 by streaking these cells on medium containing the Bradyrhizobium-specific antibiotics gentamycin, kanamycin, and carbenicillin (Sen et al., 2019). The objective of these antibiotic passages was to obtain JGTA-S1 strains free of P. stutzeri and Bradyrhizobium sp.
The antibiotics gentamycin, kanamycin, and carbenicillin were chosen to cure JGTA-S1 of Bradyrhizobium sp, as most Bradyrhizobium strains are sensitive to these antibiotics (Abaidoo et al., 2002). We performed spotting assays using 59 independent colonies of JGTA-S1 strain 11 (postantibiotic treatment) on N-free medium to screen for yeast strains that showed reduced growth in the absence of any N source (Supplemental Figures 8A and 8B). Three such strains were chosen (Supplemental Figures 8A and 8B, colonies 6, 16, and 38) and named 11.1, 11.2, and 11.3, respectively. We monitored the growth of JGTA-S1 and its derived strains by spotting them onto TSB medium containing both organic and inorganic N sources or on N-free medium. We estimated the growth of the strains as a measure of their fitness. We also measured the relative number of Pseudomonas/JGTA-S1 cell by qPCR, where relative P. stutzeri 16S rDNA levels were quantified by normalizing against JGTA-S1 GAPDH as a reference gene in TSB medium. Cells to be spotted onto TSB plates were grown in TSB medium, and cells to be spotted onto N-free medium were grown in the presence of ammonium as the sole N source. Strains 11.1, 11.2, and 11.3 were maintained under antibiotic selection so that yeast cells with higher diazotrophic endobacterial levels (Pseudomonas or Bradyrhizobium) were not selected over those with low endobacterial levels during culture due to their superior growth rates. The plates used for the spotting assay did not contain antibiotics. Strains 11.1, 11.2, and 11.3 were grown in the presence of five antibiotics when grown in liquid TSB, including two for eliminating P. stutzeri and three for eliminating Bradyrhizobium sp. Only Bradyrhizobium sp–specific antibiotics were used when 11.1, 11.2, and 11.3 were grown in N-free plus (NH4)2SO4 medium before spotting onto N-free plates because these strains did not survive in the presence of only (NH4)2SO4 in the medium when all five antibiotics were used, underscoring the importance of the endobacteria especially in absence of an organic N source. The cells were washed with deionized water and spotted after normalizing the OD600 to 0.5. Strains 11.1, 11.2, and 11.3 showed reduced growth irrespective of the medium used compared with the control and strain 11 (Figures 7A and 7B). We previously proposed that the endofungal P. stutzeri levels (present per JGTA-S1 cell) respond to the N available to JGTA-S1 (Sen et al., 2019) and are under stringent control in a manner similar to that of the endobacteria of S. indica (2–20/fungal cell) and Candidatus Glomeribacter gigasporarum (10/spore). The endofungal P. stutzeri per JGTA-S1 cell in the 11.1, 11.2, and 11.3; increased slightly relative to strain 11 in response to Bradyrhizobium sp–specific antibiotics (Figure 7C). These results, together with our earlier findings, indicate that JGTA-S1 endobacteria, some of which are diazotrophs (P. stutzeri and Bradyrhizobium sp), are important for the growth and viability of JGTA-S1 cells.
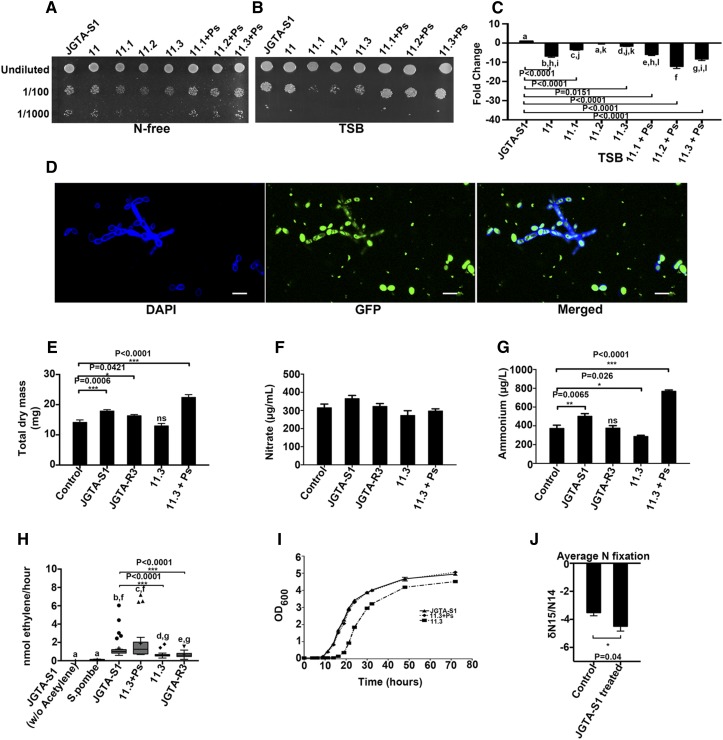
Figure 7.
JGTA-S1 Fixes N in Planta, and the Endobacterium P. stutzeri Regulates Plant Biomass and Ammonium Levels.
(A) and (B) Growth of JGTA-S1 and its derived strains 11.3 and 11.3+Ps was compared in (A) N-free medium and (B) N-replete TSB.
(C) Relative numbers of Pseudomonas/JGTA-S1 cell in these strains were measured by qPCR. Cells were grown in TSB as described in Methods, and relative levels of P. stutzeri 16S rDNA gene were quantitated against JGTA-S1 GAPDH as a reference gene.
(D) JGTA-S1 treated with FK506 incubated with GFP-labeled P. stutzeri-JGTA-R3 for 20 h. P. stutzeri outside the yeast was killed with tetracycline and Timentin, followed by confocal microscopy. The effects of JGTA-S1, P. stutzeri-JGTA-R3, 11.3, and 11.3+Ps on plant growth and inorganic N content were analyzed. Rice plants were treated with 1× MS (control), JGTA-S1, P. stutzeri-JGTA-R3, 11.3, and 11.3+Ps, and plant parameters were measured at 4 wpi. Bar = 10 µm. DAPI, 4′,6-diamidino-2-phenylindole.
(E) to (G) Total dry mass (E), nitrate content (F), and ammonium content (G). Statistical comparisons for nitrate content were performed using one-way analysis of variance with Tukey’s post hoc test (n = 3). Statistical comparisons for ammonium content (n = 6) and dry mass (control, n = 16; JGTA-S1 treated, n = 24; 11.3 treated, n = 14; 11.3+Ps treated, n = 14; JGTA-R3 treated, n = 24) were performed using Kruskal-Wallis nonparametric test followed by Dunn’s post correction test.
(H) JGTA-S1, 11.3, 11.3+Ps, and P. stutzeri-JGTA-R3 were grown in N-free medium. The OD of the cells was normalized prior to the acetylene reduction assay. S. pombe (a nondiazotrophic yeast strain) and JGTA-S1 without acetylene injection were used as negative controls. Statistical analysis was performed using the Kruskal-Wallis nonparametric test followed by Dunn’s post correction test (JGTA-S1 without acetylene, n = 10; S. pombe, n = 10; JGTA-S1, n = 32; 11.3, n = 31; 11.3+Ps, n = 27; P. stutzeri-JGTA-R3, n = 31).
(I) For the growth curve, JGTA-S1, 11.3, and, 11.3+Ps were grown in N-free medium, and OD600 was measured at different time points. Mean ± se was calculated for each time point.
(J) δ15N stable isotope analysis of rice plants treated with MS (control) and JGTA-S1. Values obtained were statistically analyzed using Student’s t test (P ≤ 0.05, n = 6). Data are given as mean ± se.
The calcium-dependent phosphatase calcineurin regulates morphogenesis in dimorphic fungi (Lee et al., 2013). We therefore treated JGTA-S1 with FK506, a calcineurin inhibitor. FK506 treatment induced the formation of filaments or pseudofilament-like structures from the yeast form, confirming its ability to generate hyphae or pseudohyphae. Therefore, dimorphism in JGTA-S1 appears to be under the control of calcineurin. P. stutzeri-JGTA-R3 invaded these artificial filaments in the same manner as natural filaments. This method allowed us to introduce P. stutzeri-JGTA-R3 into FK506-treated 11.1, 11.2, and 11.3 strains to create the +Ps strains (Figure 7D). The reduced growth of 11.1, 11.2, and 11.3 on TSB and N-free medium was restored when P. stutzeri was reintroduced into these cells (Figures 7A and 7B compare 11.1, 11.2, and 11.3 with the corresponding +Ps strains).
Endosymbiotic P. stutzeri JGTA-S1 Plays a Vital Role in Improving N Nutrition in Rice
Among the +Ps strains, we chose 11.3 +Ps (along with strain 11.3) to examine the effect of P. stutzeri-JGTA-R3 on plant growth and N nutrition compared to the control JGTA-S1. We applied JGTA-S1, P. stutzeri-JGTA-R3, 11.3, and 11.3+Ps to rice to investigate their ability to alter biomass, ammonium content, and nitrate content in plants. The growth rate of strain 11.3 was the lowest, and the growth rate of 11.3+Ps was comparable to that of JGTA-S1 (Figures 7A and 7B).
The use of these different strains of JGTA-S1 with different growth rates allowed us to compare the effects of the endobacterium Pseudomonas on N nutrition in rice through this tripartite interaction. We measured biomass, nitrate, and ammonium content in the shoots of plants treated with these Rhodotorula strains and P. stutzeri-JGTA-R3 compared to the untreated control. The increases in biomass and ammonium content of the rice plants in response to JGTA-S1 were directly related to the fitness of the yeast that, in turn, depended on the endobacterial Pseudomonas (Figures 7E and 7G). The 11.3+Ps strain was the most effective at increasing plant biomass and shoot ammonium content, whereas strain 11.3 had no effect on any of the parameters tested.
In an acetylene reduction assay, the N2-fixing ability of 11.3+Ps did not appear to be significantly higher than that of JGTA-S1, despite a positive shift in the median and mean values of ethylene produced per hour (Figure 7H). However, strain 11.3+Ps had a much better N2-fixing ability compared with strain 11.3 from which it was derived. The low N2-fixing ability of 11.3 mirrored its effect on biomass and ammonium content in rice. These results strongly suggest that (1) JGTA-S1 is a much better N2 fixer than free-living P. stutzeri-JGTA-R3 and (2) P. stutzeri, an endobacteria of JGTA-S1, played a significant role in increasing both the ammonium content and biomass of rice plants treated with JGTA-S1 (Figures 7E and 7G). We performed an acetylene reduction assay using S. pombe D18 h− (an unrelated fungus) and JGTA-S1 without acetylene injection as negative controls (Figure 7H). Under these conditions, no acetylene reduction took place. We constructed a growth curve of JGTA-S1, 11.3, and 11.3+Ps to quantitatively confirm their ability to grow in the absence of a N source. The JGTA-S1 and 11.3+Ps cells showed similar growth patterns, but 11.3 showed a more prolonged lag phase before reaching the exponential phase. Strain 11.3 also showed a lower growth rate and attained lower growth maxima than the other strains (Figure 7I). This growth curve analysis confirmed the ability of the yeast cells to grow without any external N supply.
Despite the upregulation of NIN-like transcription factor genes, potentially indicating a heightened nitrate response, no significant change in nitrate content was observed in JGTA-S1–treated plants versus the controls (Figure 7F). Finally, we measured the N derived from biological N2 fixation (as δ15N values) in JGTA-S1–treated plants compared to the untreated controls. This analysis indicated that the yeast-treated plants derived more N2 from the atmosphere than the controls. This atmospheric N2 was obtained via biological N2 fixation, and fixed N was supplied to the rice plants (Figure 7J).
DISCUSSION
NUE is a complex trait governed collectively by the uptake, assimilation, and remobilization of available N in the plant. NUE is an essential parameter for controlling plant yields. Finding ways to improve NUE in plants is crucial for sustainable agriculture, as it would control the indiscriminate use of N fertilizers. Given the inability of most agriculturally important plants to nodulate as legumes do, biological nitrogen fixation by diazotrophic endophytes applied to plants appears to be a useful way to improve N metabolism. The drawback, of course, lies in the fact that biological nitrogen fixation by free-living diazotrophs is much less efficient compared to the efficiency at which rhizobia can fix N2 within the sophisticated architecture of a symbiotic root nodule.
Mycorrhizae are the only fungi reported to improve N nutrition in plants, but the proposed mode of this improvement is increased N supply. AM fungi induce changes in root architecture in rice concurrent with the upregulation of several inorganic or organic N-uptake genes (Gutjahr et al., 2015). However, despite the presence of diazotrophs in the mycorrhizosphere, their participation in the improvement of N metabolism in plants has not been studied (Barbieri et al., 2005, 2007).
The property of N2 fixation is absent in eukaryotes, but they have circumvented this deficiency by associating with N2-fixing microbes (Kneip et al., 2007), allowing both partners to exploit challenging niches. Nitrogen appears to be at least one of the critical factors controlling symbiotic and pathogenic fungi–bacteria associations in plants. Examples of interkingdom interactions with the potential to improve N nutrition in the associated eukaryotes include B. pumilus–U. maydis–plant, cyanobacteria–G. pyriforme–plant (Gehrig et al., 1996), and Klebsiella sp–fungi–leafcutter ant interactions. In each of these cases, a fungus (containing diazotrophs) is one of the partners. Such interactions may serve as alternative ways of improving N transfer to the host plant. In the case of the Allomerus decemarticulatus (ant)–Trimmatostroma sp (fungus)–Hirtella physophora (plant) three-way interaction, transfer of N from the fungus to the plant has been demonstrated (Leroy et al., 2017). Various species of the Gigasporaceae AM fungi maintain bacteria from Burkholderia genera, which are rich in plant-associated N2-fixing bacteria (Estrada-De Los Santos et al., 2001). For example, Azospirillum brasilense (a member of Burkholderiaceae and an endosymbiont of G. margarita) contains a nif operon (Minerdi et al., 2001). The mycorrhiza-like basidiomycete S. indica also appears to form close associations with bacteria (Sharma et al., 2008), indicating that such associations are the rule in nature rather than the exception.
The effect of R. mucilaginosa JGTA-S1 on rice growth and N nutrition is significant and comparable to that of nodule symbiosis on N nutrition in legumes. Treatment with R. mucilaginosa JGTA-S1 resulted in a significant increase in plant biomass and the sustained upregulation of N-metabolism–related genes in rice. While this upregulation could have resulted from induced N deficiency due to an arms race between the plant and the yeast for available N (particularly ammonium), the interaction was not limited to mere upregulation of N-uptake genes. JGTA-S1 treatment did not have any deleterious effects on plant health; instead, it resulted in improved N content and NUE in rice. The yeast efficiently fixed atmospheric N2, increased free ammonium levels in planta due to the presence of diazotrophs within the yeast, one of which was confirmed to be P. stutzeri. Bradyrhizobium sp and other diazotrophs are potential JGTA-S1 endosymbionts (Sen et al., 2019) that likely play roles in the JGTA-S1–mediated improvement of N metabolism in plants as well. Since no Bradyrhizobium sp was available among the Typha endophytes, these interactions could not be confirmed in this study.
Rhodotorula has long been known for its ability to grow in the absence of N (Brown and Metcalfe, 1957). Although the authors did not report any endosymbiont in their study, Firrincieli et al., 2015 reported that R. graminis WP1 contains a dinitrogen reductase gene (nifH; Knoth et al., 2014).
Eukaryotes exploit different mechanisms to acquire and transmit their endosymbionts. In the case of Rhizopus, B. rhizoxinica can enter the fungal hyphae actively through a local disruption of the fungal cell wall (Moebius et al., 2014). Vegetative spore formation in Rhizopus is compromised in the absence of the endobacteria (Partida-Martinez et al., 2007). The endobacteria facilitate spore formation as a strategy to ensure their vertical transmission. Similarly, mycorrhizal endobacteria are transmitted through spores. The vertical transmission of bacteria to the yeast form was previously observed when JGTA-S1 filaments were mixed with P. stutzeri-JGTA-R3. The endobacteria were essential for the survival and propagation of JGTA-S1, as they helped the yeast maintain a bacterial pool despite antibiotic treatments (Sen et al., 2019).
Another important observation is the dimorphic transition from yeast to filamentous form discovered in JGTA-S1. Such transitions are often part of the sexual life cycles of human yeast pathogens Histoplasma, Cryptococcus, and Candida, resulting in the formation of adhesive and invasive filaments inside the host, which help them to penetrate host barriers (Sánchez-Martínez and Pérez-Martin, 2001). This process is triggered by a wide variety of stimuli (Gauthier, 2015). The importance of dimorphism is further highlighted by the finding that it may facilitate the association with bacteria. Candida associates with P. aeruginosa but only in filamentous rather than yeast form (Hogan and Kolter, 2002). The best-studied examples of dimorphism are found in C. albicans and the plant pathogen U. maydis, both of which share strikingly similar life strategies with R. mucilaginosa JGTA-S1. This filamentous transition is a new observation in Rhodotorula, which was previously thought to be an asexually budding yeast. Treatment with the calcineurin inhibitor FK506 induced a filament-like transition in JGTA-S1. Calcineurin was previously demonstrated to be a critical regulator of the dimorphic transition (Lee et al., 2013). Our finding, however, is in contrast to that observed in the dimorphic zygomycetes Mucor circinelloides, whose hyphal transition is inhibited by FK506 (Lee et al., 2013). Therefore, the effects of FK506 could vary depending on the fungus or dosage.
JGTA-S1 primarily associated with bacteria in the filamentous form and not in the yeast form, which is similar to reports on the Candida–Pseudomonas interaction (Hogan and Kolter, 2002). P. stutzeri-JGTA-R3, also isolated from the Typha endosphere, entered the Rhodotorula hyphae, like B. rhizoxinica, which enters Rhizopus microsporus filaments (Moebius et al., 2014). These findings indicate that JGTA-S1 likely acquired the Pseudomonas from the endosphere of Typha in free-living form, suggesting that its association with the fungus is facultative from the bacterial side. However, this is not the case for yeast, since the loss of the diazotrophic endobacteria had a fitness cost. JGTA-R3 was horizontally acquired and vertically transmitted in spore or yeast form, which budded off the filaments. The association of JGTA-S1 with its endobacteria was remarkably stable, which is in agreement with the recent finding that disrupting endosymbiotic associations with a nutrient amendment is never without fitness costs (Fisher et al., 2017).
Unlike Rhizopus, Ustilago, or Candida, JGTA-S1 appeared to associate with various bacteria within the plant. Despite the conservation of life strategies with other pathogenic fungi of subphylum Pucciniomycotina, JGTA-S1 appears to be beneficial to rice (Sen et al., 2019).
Rice genes transcriptionally responding to Rhodotorula included many genes known to be regulated in the plant in its encounter with bacteria. The downregulation of an RLK gene belonging to the SHR5 family is directly related to the plant’s interaction with diazotrophs (Vinagre et al., 2006). Several hundred RLK genes are induced in sugarcane (Saccharum officinarum) in response to the endophytes Herbaspirillum and Gluconacetobacter diazotrophicus (Nogueira et al., 2001). The flagellin receptor FLS2 is also induced in response to beneficial interactions (Trdá et al., 2014). These findings point to the existence of additional generic mechanisms by which plants respond to microbes, irrespective of the latter being pathogenic or symbiotic (Vandenkoornhuyse et al., 2015). In this study, homologs of the LRR-RLK BAK1 were upregulated in rice roots and shoots upon JGTA-S1 infection. BAK1 is a coreceptor protein for the brassinosteroid receptor BRI1 and FLS2. BAK1, which has strong effects on immunity-related genes, was upregulated in rice roots supplemented with N (Obertello et al., 2015). Nutrition, immunity, and symbiosis are closely interconnected, where nutrition is believed to regulate immunity and interkingdom symbiosis (Hiruma et al., 2016; Fisher et al., 2017).
A notable observation was the downregulation of a CLAVATA-like gene in JGTA-S1–treated rice shoots. CLAVATA1 and CLAVATA3 are implicated in shoot-derived systemic suppression of nodulation (autoregulation of nodulation; Reid et al., 2011). Although not proven conclusively, these proteins are also implicated in the autoregulation of mycorrhization (Reid et al., 2011; Staehelin et al., 2011). Whether the systemic downregulation of this CLAVATA-like gene or other CLAVATA-like kinases facilitates the colonization of yeast is an important fundamental issue to investigate. The upregulation of these microbe-related plant genes indicates that the effects of JGTA-S1 on the rice transcriptome could represent cumulative effects of the yeast and bacteria rather than the yeast alone. The observation that Rhizobium radiobacter, an endobacterium of S. indica, has a growth-promoting effect similar to that of the fungus Sharma et al. (2008) suggests that the beneficial effects of the fungi on the plant are a direct consequence of the endobacteria. The increase in biomass and free ammonium content of plants harboring various JGTA-S1 strains differing in P. stutzeri content reveals a similar correlation.
In most terrestrial systems, nitrate is the predominant form of N in the soil. JGTA-S1 contains an incomplete nitrate assimilation pathway, unlike R. graminis WP1 and related pink yeasts, making JGTA-S1 dependent on organic and ammonium N for survival even when nitrate is abundant. Additionally, JGTA-S1 was isolated from a mine wetland containing metal-laden sludge, an environment that was likely deficient in available ammonium and organic N, with levels too low for the fungus to maintain its free-living state. These genetic and environmental factors likely made the fungus entirely dependent on the host plant or its endosymbiotic diazotrophs for its N supply. The observation that P. stutzeri nifH was expressed in JGTA-S1 even under N-replete conditions indicates that JGTA-S1 uses diazotroph-derived fixed N2 even when N is abundant in its external environment. The N nutrition–related rice genes upregulated by JGTA-S1 included both N-uptake and -assimilation genes, such as an ENOD93-like gene that was upregulated in rice upon JGTA-S1 treatment. The overexpression of an ENOD93-like gene increased the NUE and amino acid transport to shoots in rice (Bi et al., 2009). The most significant transcriptomic change in rice upon JGTA-S1 treatment was the sustained upregulation of four NLPs, encoding central regulators of the nitrate response (Konishi and Yanagisawa, 2013; Yan et al., 2016). However, the roles of rice NLPs in nitrate metabolism and the contribution of the tripartite interaction to the nitrate response (if any) were not clear. Overall, the yeast appeared to have a complex effect on N metabolism in rice. The yeast acquired N2 from the atmosphere via biological N2 fixation (aided by its endosymbionts) and provided the fixed N products to the plant, potentially modulating the nitrate response (although not the nitrate concentration per se) and upregulating inorganic and organic N-uptake genes in the plant.
Collectively, the novel tripartite interaction involving R. muculaginosa-JGTA-S1, its endosymbiont diazotroph P. stutzeri, and rice uncovered in this study improves the NUE in rice, and P. stutzeri plays a vital role in this phenomenon. JGTA-S1 was more competent as an N2 fixer than free-living P. stutzeri, thereby making JGTA-S1 a better plant growth-promoting endophyte. It is tempting to speculate that the environment within JGTA-S1 facilitates the N2 fixation of P. stutzeri in a manner similar to the N2 fixation of Rhizobium within the root nodule. Rhodotorula is axenically culturable and fast growing, and its microbiome could potentially be synthetically designed. The implications of such interactions are great, paving the way for improving N metabolism in non-nodulating plants of agricultural importance without the need for genetic manipulation.
METHODS
Isolation and Identification of R. mucilaginosa JGTA-S1 from Plants
Rhodotorula mucilaginosa JGTA-S1 was isolated from surface-sterilized Typha angustifolia shoots as described by Saha et al. (2016). After sterilization, 100 μL of solution from the last wash was plated as a negative control. Debris was removed from the extract obtained from surface-sterilized plants and plated. The plates were incubated for 3 d at 30°C. Pink Rhodotorula colonies were repeatedly streaked onto TSB agar plates to obtain a pure culture. To isolate JGTA-S1 from rice, plants were grown in sterile glass jars containing Soilrite (Keltech Energies) mixed with 1× MS medium, pH 6. Extracts from surface-sterilized plants at 3 wpi were passed through a 0.45-µm filter. The filters were incubated on PDA plates for 3 d at 30°C and transferred to new PDA plates containing carbenicillin (100 µg/mL) and Timentin (250 µg/mL). The plates were incubated at 5°C for 2 weeks or until pink growth of Rhodotorula was visible. The pink material was stained with calcofluor and observed under a confocal microscope (FluoView 1200, Olympus). Genomic DNA was isolated from the filaments as described by Bolano et al. (2001). Primers were designed to amplify a JGTA-S1–specific fragment of the 26SrDNA or D1/D2 gene. Primer sequences are given in Supplemental Data Set 7. The amplified product was cloned and sequenced.
R. mucilaginosa Growth in N-Free Medium
To investigate growth on N-free medium, R. mucilaginosa JGTA-S1 and Schizosaccharomyces pombe D18 h− (negative control) were streaked onto Norris Glucose Nitrogen Free Medium (Himedia India) and incubated at 30°C for 7 d.
Plant Growth and Treatments
Rice (Oryza sativa cv Swarna MTU7029, Bordan, or IR64) seeds were surface sterilized and germinated on filter paper. Four- to 5-d-old seedlings of equal length were transferred to Soilrite containing 2× MS salt mixture (N-replete conditions, 15 mL of medium/30 g of soil) with (107 cfu JGTA-S1 suspended in 10 mL of 1× MS) or without JGTA-S1 unless otherwise mentioned. Control plants were supplied with MS only. Plants were grown in a climate-controlled room under a 24-h cycle consisting of 16 h of light at 30°C and 8 h of dark at 25°C at a humidity level of 70%. Plants were harvested at 3 wpi and used to measure root and shoot length (control, n = 25; treated, n = 23 from three independent experiments) or biomass (control, n = 23; treated, n = 27 from two independent experiments). Tissue from each plant was treated as an independent biological replicate. Plant samples were dried at 60°C for 48 h before measuring the dry weight. For microarray analysis, roots were harvested from 10 3-week-old plants at 0 or 24 h posttreatment with JGTA-S1, frozen in liquid nitrogen, and stored at −80°C. These pooled tissues were treated as independent biological replicates. To assess plant growth without exogenous N supplementation (low N), plants were grown in Soilrite supplemented with N-free medium as described by Saha et al. (2016). JGTA-S1 or N-free medium (low N) was added dropwise around the roots of 5-d-old plants. Plant parameters were measured at 10 DAI. Tissue from each plant was treated as an independent biological replicate (n = 18). To measure chlorophyll content, 400 mg of fresh leaf tissue was extracted overnight in 8 mL of 80% (v/v) acetone, and the absorbance at 645 and 663 nm was measured using a UV-2450 UV-visible spectrophotometer (Shimadzu). Total chlorophyll was calculated as total chlorophyll (mg/g) = 20.2 (A645) + 8.02 (A663) × V/1000 × 1/W (Arnon, 1949).
RNA Isolation
For microarray analysis, RNA was isolated in two replicates each from pooled root tissue obtained from 10 plants. Total RNA was isolated from the frozen tissues using a HiPurA Plant and Fungal RNA Miniprep Purification Spin Kit (Himedia India). The RNA was treated with DNase I (1 unit/2 µg of RNA, Thermo Fisher Scientific) to remove genomic DNA contamination.
Microarray Hybridization and Analysis
Total RNA isolated from rice roots (O. sativa cv Swarna MTU7029) was used for microarray analysis. The analysis was performed using an Affymetrix GeneChip Rice St 1.0, corresponding to 146,918 probes representing 41,770 known genes, expressed sequence tags, or clusters (with 15 median probes per gene) following the manufacturer’s protocol. The genes were functionally classified based on the complete genome sequence of O. sativa cv japonica in the Rice Genome Annotation Project (rice.plantbiology.msu.edu), the National Center for Biotechnology Information database, and the European Bioinformatics Institute database (O. sativa cv indica). Ensemble Plant and Phytozome 11 Biomart and RAP_DB tool (rapdb.dna.affrc.go.jp) were used to complete the annotation and to identify O. sativa cv japonica homologs of the genes.
RT-qPCR Analysis
RNA was isolated in three biological replicates. Each biological replicate contained root and shoot tissue pooled from 10 plants. Two micrograms of total RNA (as measured in a NanoDrop spectrophotometer, Thermo Fisher Scientific) was used to synthesize cDNA with Random Hexamer Primer (Fermentas) using RevertAid Reverse Transcriptase (200 units/2 µg of RNA, Fermentas) as per the manufacturer’s protocol. To validate the microarray data, 0- and 24-h–treated samples were used. To study the differential expression of genes related to N metabolism, RNA was isolated from plants at 3 wpi and compared with RNA from the uninfected controls. Each 10-μL RT-qPCR contained 5 μL of DyNAmoColorFlash SYBR Green I Master Mix (Thermo Fisher Scientific), 0.5 µM each primer, and 25 ng of cDNA. RT-qPCR was performed using a StepOnePlus real-time PCR machine (Applied Biosystems). The thermal step-up was 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Data analysis was performed using ExpressionSuite software v1.0.4 (Applied Biosystems). Fold change of expression was calculated using the 2−ΔΔCt method and plotted relative to the control using Prism 5.02 (GraphPad Software). Primer sequences are given in Supplemental Data Set 7. Primer efficiencies were checked using LinRegPCR version 2016.0 (Ramakers et al., 2003).
Predicting Rice Orthologs of NIN-Like Genes
Arabidopsis (Arabidopsis thaliana) orthologs of O. sativa NLP genes upregulated by JGTA-S1 were predicted using OrthoMCL (Li et al., 2003) and grouped into an orthologous group with Arabidopsis and Medicago sequences.
Measuring Total N Content and Relative NUE
Oven-dried (60°C overnight) plant samples were ground into a fine powder and digested with concentrated H2SO4 (Merck), and total N concentration was estimated by the Kjeldahl method in a KEL PLUSKES digestion and Distyl EMVA distillation unit (Pelican). The same amount of N was added to the soil for the control. The relative NUE of JGTA-S1 was measured as described by Saha et al. (2016) by calculating the ratio of the biomass of control (n = 23) and treated plants (n = 27); n represents individual plants.
Amplification of P. stutzeri nifH, Flanking Regions, and 16S rDNA Sequences from JGTA-S1 Genomic DNA
Partial nifH genes were amplified from R. mucilaginosa JGTA-S1 genomic DNA strictly under sterile conditions according to Steward et al. (2004). The products were cloned into the pJET1.2 vector (Thermo Fisher Scientific) according to the manufacturer’s protocol and sequenced. Full-length P. stutzeri nifH was cloned from JGTA-S1 genomic DNA as a template using modified TAIL-PCR (Liu et al., 1995). Arbitrary degenerate primers (AD1 to AD6) and nifH gene–specific primers (TAIL F1 to F3 and TAIL R1 to R3) were used in the primary and secondary PCR. The secondary PCR product was used as a template to amplify the 5′ and 3′ ends of the P. stutzeri nifH gene using specific forward and reverse primers. The flanking genes of P. stutzeri nifH were cloned by standard TAIL-PCR using JGTA-S1 genomic DNA as a template. The tertiary PCR products were cloned directly into pJET1.2 and sequenced. For specific amplification of P. stutzeri 16S rDNA and nifH sequences, primers were designed using the Primer-BLAST program (Ye et al., 2012). For nifH RT-PCR, total RNA was isolated from R. mucilaginosa cells grown in TSB medium using a Hipura Plant and Fungal RNA Isolation Kit. The RNA was treated with DNase I to remove genomic DNA prior to cDNA synthesis with Random Hexamer Primer and RevertAid RT enzyme. RT-PCR was performed using P. stutzeri–specific nifH primers. A reaction was conducted without reverse transcriptase to ensure the absence of DNA contamination. A negative control without DNA template was conducted whenever bacterial genes were amplified from JGTA-S1 DNA.
RNA Isolation and Amplification of P. stutzeri nifH from JGTA-S1
Total RNA was isolated from JGTA-S1 cells using the TRIzol method (TRIzol reagent, Invitrogen) as described previously (Sen et al., 2019). The RNA was subjected to DNase I treatment and transcribed to cDNA with Moloney Murine Leukemia Virus reverse transcriptase and random hexamers (Thermo Fisher Scientific) per the manufacturer’s protocol. cDNA (6.2 ng) was used to amplify P. stutzeri nifH. The RT-PCR cycle was 5 min at 98°C followed by 40 cycles of 30 s at 98°C, 15 s at 62°C, and 19 s at 72°C with a final extension of 7 min at 72°C. Minus RT and minus DNA controls were performed as described above.
Acetylene Reduction Assay
The acetylene reduction assay was performed using a gas chromatography with flame ionization detector system (Shimadzu GC-2010 equipped with HP-PLOT ‘S’ Al2O3 50 m, 0.53-mm column; Agilent Technologies). JGTA-S1 without the injection of acetylene and S. pombe D18 h− (an unrelated nondiazotrophic yeast) were used as negative controls. JGTA-S1 and other strains were grown at 30°C for 48 h in TSB medium, and S. pombe D18 h− cells were grown in YES medium. Approximately 107 cells were centrifuged, washed twice with sterile water, suspended in 100 μL of sterile water, and used to inoculate 5 mL of N-free medium (for JGTA-S1 cells: Glc, 5 g; yeast extract, 0.1 g; CaCl2, 0.15 g; Na2MoO4, 0.005 g; MgSO4·7H2O, 0.2 g; FeSO4·7H2O, 0.04 g; K2HPO4, 0.5 g; CaCO3, 1.0 g; and distilled water to 1 L, pH 7.0) and YES medium (for S. pombe D18 h− cells) that had been grown for 72 h at 30°C. The cells were harvested and cell number determined, and 107 cells were inoculated in N-free medium (same as above with 3 g/L Phytoagar [Duchefa]) in 16-mL airtight glass vials and sealed with Suba-Seal septa (Sigma-Aldrich). One milliliter of pure acetylene gas was injected through the Suba-Seal septa and incubated for 4 h without shaking. Ethylene production was measured by gas chromatography. JGTA-S1 cells were grown from six different cultures and maintained separately for the biological replicates, with each set containing three to six technical replicates. The total number of replicates was denoted as n. Results represent data for a total of n = 10 experiments for JGTA-S1 without acetylene, n = 10 for S. pombe D18 h−, n = 32 for JGTA-S1, n = 27 for 11.3+Ps, n = 31 for 11.3, and n = 31 for P. stutzeri-JGTA-R3.
Effect of N Deficiency and Low pH on R. mucilaginosa
Morphological variation of JGTA-S1 due to N depletion was examined by growing the cells on the same N-free medium used for the C2H2 reduction assay for 7 d. The effect of pH was checked by growing the cells on PDA at different pH levels (pH 3, 3.5, 4, and 4.5) maintained using citrate buffers of the same pH.
Staining of JGTA-S1 Filaments and Interaction with Endobacteria
Plants were grown in a sterile environment with or without JGTA-S1 treatment. The extract from surface-sterilized 3 wpi plants (Saha et al., 2016) was passed through 0.45-µm filters as described above. The pink material that formed on filter paper was suspended in 1× PBS with (when interaction with P. stutzeri-JGTA-R3-GFP was studied) or without tetracycline. The JGTA-S1 filaments were stained with calcofluor. To induce filament formation, 1 µg/mL FK506 (Tacrolimus, Panacea Biotech) was added to TSB broth while growing the fungus. The cells were imaged 3 DAI. Alexa Fluor 488–labeled WGA or lactophenol cotton blue was used when the colonization of JGTA-S1 in planta was studied. For WGA staining, seedlings infected by JGTA-S1 were infiltrated with Hank’s balanced salt solution buffer containing 5 µg/mL WGA and incubated overnight in the dark at room temperature. The plant root was counterstained with propidium iodide (excitation, 559 nm and emission, 570 to 670 nm) and imaged under a confocal microscope using an Alexa Fluor 488 filter (excitation, 473 nm and emission, 485 to 545 nm). To visualize endobacteria, the filament was stained with a LIVE/DEAD BacLight bacterial viability kit, counterstained with calcofluor, and observed under a confocal microscope with 4′,6-diamidino-2-phenylindole (excitation, 405 nm and emission, 425 to 460 nm) and fluorescein isothiocyanate filters (excitation, 473 nm and emission, 485 to 545 nm). To study P. stutzeri and JGTA-S1 interaction, P. stutzeri-JGTA-R3-GFP was grown overnight in the presence of tetracycline (10 µg/mL). P. stutzeri cells were mixed with the yeast filaments and incubated at 20°C for 20 h without shaking. The JGTA-S1 in yeast form was similarly mixed with P. stutzeri-JGTA-R3-GFP. The cells were counterstained with calcofluor and observed under a confocal microscope with 4′,6-diamidino-2-phenylindole and GFP (excitation, 473 nm and emission, 485 to 585 nm) filters.
Creating Endobacteria-Free JGTA-S1 Cells and Reintroducing Endobacteria
JGTA-S1 strains cured of endobacteria were created as described in Sen et al. (2019). In short, JGTA-S1 cells were streaked five times in N-free medium with 5 g/L (NH4)2SO4 in the presence of tetracycline (10 µg/mL) and Timentin (250 µg/mL) to obtain strain 11. Strain 11 was streaked in the presence of 30 µg/mL gentamycin, 50 µg/mL kanamycin, and 100 µg/mL carbenicillin in the same medium. Single colonies of JGTA-S1 were revived once by growing them on TSB plates without antibiotics followed by another round of streaking in the presence of gentamycin, kanamycin, and carbenicillin (Sen et al., 2019). Fifty-nine colonies obtained in this manner were screened by growing them in N-free medium plus (NH4)2SO4, followed by spotting onto N-free medium (after normalization to OD600 at 0.5) to search for strains whose growth was significantly reduced compared to the control (this study). To reintroduce P. stutzeri into strains 11.1, 11.2, and 11.3 and to obtain strains 11.1+Ps, 11.2+Ps, and 11.3+Ps, respectively, strains 11.1, 11.2, and 11.3 were grown in the presence of 1 µg/mL FK506 for 3 d, mixed with P. stutzeri-JGTA-R3 (Ps) for 20 h, and plated onto medium containing tetracycline (10 µg/mL) and Timentin (250 µg/mL) to remove any free Pseudomonas (this study).
Estimating the Fitness of Endobacteria-Cured Cells
Control and 11.1, 11.2, 11.3, 11.1+Ps, 11.2+Ps, and 11.3+Ps strains were grown in N-free medium plus (NH4)2SO4 or TSB medium with or without antibiotic treatment as described below. All media were prepared in 18.3 MΩ water. The 11.1, 11.2, 11.3 cells were grown in the presence of five antibiotics, tetracycline (10 µg/mL), Timentin (250 µg/mL), gentamycin (30 µg/mL), kanamycin (50 µg/mL), and carbenicillin (100 µg/mL), when grown in TSB before they were spotted onto TSB. When the cells were spotted onto N-free medium, the cells were grown in N-free medium supplemented with (NH4)2SO4 in the presence of three antibiotics, gentamycin, kanamycin, and carbenicillin, at the same concentrations. Control, strain 11, and the +Ps strains in which P. stutzeri was reintroduced were grown without antibiotics. In all cases, the pellets were suspended and washed twice with deionized water before spotting to remove any carried-over medium. The OD600 was normalized, and the cells were spotted onto TSB or N-free medium at different dilutions (undiluted [OD600 of 0.5], 1:100, 1:1000).
Estimating the P. stutzeri Concentration per JGTA-S1 Cell
To estimate P. stutzeri levels per yeast cell, qPCR was performed. Genomic DNA was isolated from JGTA-S1 (control), 11.1, 11.2, 11.3, 11.1+Ps, 11.2+Ps, and 11.3+Ps strains grown in TSB. Cells were grown in the presence of five antibiotics as described in the previous section. A P. stutzeri–specific 16S rDNA primer was designed using primer BLAST software, with P. stutzeri DSM 4166 as the organism. An equal amount of genomic DNA was used for qPCR. Ten microliters of qPCR reaction contained 5 μL of DyNAmoColorFlash SYBR Green I Master Mix (Thermo Fisher Scientific), 0.5 µM of each primer, and 1.5 ng of genomic DNA. The nucleic acids were measured using Qubit assay kits. qPCR was performed in a 7500 real-time PCR machine (Applied Biosystems). The thermal step-up was 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C. The fold change was calculated by 2−ΔΔCt method using JGTA-S1 GAPDH gene as a reference gene. The results correspond to four independent biological replicates. Each qPCR was done in triplicate (experimental replicate). The primer efficiencies were checked using LinReg PCR 2017.1.0.0 software and found to be >90 for every primer pair. Primer sequences are given in Supplemental Data Set 7.
Measuring Dry Mass and Nitrate and Ammonium Contents in Rice Shoot Tissue
Seven-day-old rice plants were grown in N-replete medium and treated with JGTA-S1, P. stutzeri-JGTA-R3 (106 cfu/plant), 11.3, and 11.3+Ps strains as described above. Control plants were treated with 1× MS only. The treatments were repeated at 3 wpi, after which the plants were grown for an additional week. Shoot tissue from two individual plants was dried and pooled as a single biological replicate for nitrate measurements. Three such replicates (n = 3) were used. Experiments were performed in triplicate from each of these replicates. Nitrate was estimated as described by Cataldo et al., (1975), with the following modifications. Ten milligrams of dried shoot tissue was extracted in 1 mL of deionized water and incubated at 45°C for 1 h with intermittent vortexing. The sample was centrifuged, and 20 μL of the supernatant was mixed with 80 μL of 5% (w/v) salicylic acid and 1.9 mL of 2 N NaOH. The absorbance at 410 nm was measured in a spectrophotometer against a standard curve with known concentrations (0 to 300 µg/mL) of KNO3. Salicylic acid was omitted from the negative controls. Six individual plants were used as a biological replicate (n = 6) for ammonium measurements. The experiments were performed in triplicate. Free ammonium was measured from 100 mg of fresh shoot tissue that had been snap-frozen in liquid nitrogen and stored at −80°C. The tissue was homogenized in liquid nitrogen and combined with 1 mL of water. The extract was incubated at 80°C for 10 min, followed by centrifugation at 4000 rpm for 30 min at 4°C. Forty microliters of phenol-alcohol reagent (10% [v/v] phenol in 95% alcohol), 40 μL of freshly prepared sodium nitroprusside, and 100 μL of oxidizing solution (Koroleff, 1976) were added to 1 mL of supernatant and incubated for 2.5 h in the dark at room temperature. The OD630 was measured, and ammonium was quantified against known amounts of (NH4)2SO4 as described previously (Koroleff, 1976). For dry mass measurements, plant shoot tissue was dried at 60°C for 5 d and weighed. The number of plants was as follows: control, n = 16; JGTA-S1 treated, n = 24; 11.3 treated, n = 14; 11.3+Ps treated, n = 14; and JGTA-R3 treated, n = 24, where n represents individual plants.
Growth Curve Analysis of JGTA-S1 and Its Derived Strains
Three independent cultures of JGTA-S1, 11.3, and 11.3+Ps were grown in N-free medium supplemented with (NH4)2SO4 for 2 d and treated as independent biological replicates. The cells were washed with deionized water and subcultured in N-free medium for two more days. On the day of the experiment, the cultures were washed three times with deionized water and inoculated in N-free medium at an initial OD600 of 0.05. The cultures were grown at 28°C at 185 rpm, and OD600 was measured at different time points, as indicated in the graph. The experiment was conducted for 72 h. For each strain and each time point, three to six OD600 values were taken for every biological replicate. Mean ± se was calculated for individual time points for every strain.
δ15N Stable Isotope Analyses in Rice Shoots
Rice plants were treated with MS (control) or JGTA-S1. The treatments were repeated at 3 wpi, and the plants were grown for one more week. The shoot tissues were separated and dried as described above. Dried shoot tissue from 10 plants was pooled to obtain 1 g each of the biological replicate.
The stable isotope analyses were performed at the Archeology Department, University of Cape Town, with six such replicates per treatment. Values were expressed relative to the standard for atmospheric N for δ15N, as (%), according to the following equation:
where Z is the heavy isotope of N, and R is the ratio of heavier to lighter isotope for the sample and standard (15N/14N). The oven-dried plant components were milled and the combusted in a Flash 2000 organic elemental analyzer. Three different in-house standards (Merck Gel, lentil [Lens culinaris], and Acacia saligna) were used and calibrated against the standards of the International Atomic Energy Agency. N is expressed relative to atmospheric N.
Estimation of P Levels in Plant Tissues
Plant samples were oven dried (60°C for 48 h) and pulverized. Each 100-mg plant sample was digested with 2.5 mL of concentrated HNO3 (65% [v/v], Merck) and 2.5 mL of H2O2 (30% [v/v], Merck). The digested samples were filtered through Whatman 110-nm filter paper followed by a 0.22-µm filter. The total P in the digest was measured by inductively coupled plasma optical emission spectroscopy (iCAP duo 6500, Thermo Fisher Scientific; radio frequency power, 1150 W; pump rate, 50 rpm; auxiliary gas flow, 1 L/min; nebulizer gas flow, 0.6 L/min; and coolant gas flow, 12 L/min). National Institute of Standards and Technology River Sediment Sample Standard Reference Material 1645 digested in the same manner as the experimental samples was used as a standard.
Transmission Electron Microscopy of Yeast Cells
Prefixative solution (2× 0.4 M Pipes, pH 6.8, 0.4 M sorbitol, 4 mM MgCl2, 4 mM CaCl2, 8% [v/v] glutaraldehyde, and water to 20 mL) was added to a JGTA-S1 culture (OD600 = 0.6) at a 1:1 (v/v) ratio, followed by incubation for 5 min at room temperature. The cells were centrifuged at 1500g for 5 min at room temperature. The supernatant was discarded, and the cells were suspended in 1× prefixative and incubated overnight at 4°C. The cells were washed three times with 1× prefixative without glutaraldehyde at 4°C and resuspended in 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1.4 M sorbitol, and 0.5% (v/v) 2-mercaptoethanol plus 0.15 mg/mL zymolyase for 10 min at room temperature. The cells were washed with 1× prefixative without glutaraldehyde and fixed in 0.5% (w/v) osmium tetroxide and 0.8% (w/v) potassium ferrocyanide (Bauer et al., 2001) in distilled water two times for 5 min each time at 4°C. The cells were washed five to six times with 1× prefixative without glutaraldehyde, stored at 4°C, sectioned, and viewed under a microscope.
Statistical Analysis
All graphing and statistical analyses were performed using Prism 7.0 (GraphPad Software). Normality of variables was evaluated by D’Agostino-Pearson Omnibus test. The growth-promoting effects of JGTA-S1 treatment on rice (increases in total dry mass, root and shoot length, chlorophyll content, P content per plant, and total N content per plant) were analyzed using Mann-Whitney unpaired two-tailed t test. Statistical analysis of the RT-qPCR data was performed using unpaired t test. Nitrate contents in JGTA-S1–, 11.3-, 11.3+Ps-, and JGTA-R3–treated plants were statistically analyzed by one-way ANOVA followed by Tukey’s post hoc test. Ammonium content and biomass of JGTA-S1–, 11.3-, 11.3+Ps-, and JGTA-R3–treated plants were statistically analyzed by Kruskal-Wallis nonparametric test followed by Dunn’s post correction test. Acetylene reduction data were also analyzed using Kruskal-Wallis nonparametric test followed by Dunn’s post correction test and Mann-Whitney U-test. The values were plotted in box plots. Dunn’s multiple comparisons were performed to compare groups. Normally distributed two-sample groups were compared by Student’s unpaired t test. For δ15N stable isotope analysis, the values were statistically analyzed by Student’s t test. For the growth curve, mean ± se was calculated per individual time point for every strain. A value of P value < 0.05 was considered statistically significant. Bars with different letters indicate significant differences among groups. All data are given as mean ± se. A summary of the test statistics is provided in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Rhodotorula mucilaginosa JGTA-S1 promotes the growth of rice varieties IR64 and Bordan.
Supplemental Figure 2. Cluster analysis of the expression levels of DEGs in shoots and roots using Multiple Experiment Viewer with Pearson Correlation.
Supplemental Figure 3. Validation of the microarray data with selected genes.
Supplemental Figure 4. Total phosphate levels in rice do not significantly change in response to JGTA-S1 inoculation.
Supplemental Figure 5. R. mucilaginosa JGTA-S1 improves N metabolism in rice irrespective of rice variety.
Supplemental Figure 6. JGTA-S1 forms chains when grown in the absence of N.
Supplemental Figure 7. JGTA-S1 contains endobacteria, some of which are diazotrophs.
Supplemental Figure 8. Removal of bacteria reduces JGTA-S1 fitness in N-free medium.
Supplemental Data Set 1. Summary of statistical tests.
Supplemental Data Set 2. Differentially regulated rice shoot transcripts at 24 hpi with JGTA-S1.
Supplemental Data Set 3. Differentially regulated rice root transcripts at 24 hpi with JGTA-S1.
Supplemental Data Set 4. Major DEGs in rice regulated upon JGTA-S1 treatment.
Supplemental Data Set 5. Comparison of data with Coneva et al. 2014 (GSE61370): Normal versus induced N and 24 hpi JGTA-S1 treated versus control root data.
Supplemental Data Set 6. Comparison with Obertello et al. 2015 (GSE38102): +N versus –N and 24 hpi JGTA-S1 treated versus control root data.
Supplemental Data Set 7. List of primers used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Maitrayee Dasgupta, Department of Biochemistry, Calcutta University, for the LIVE DEAD bacterial stain. We thank Madhumanti Das for her help in the characterization of JGTA-S1. We also thank the Department of Biotechnology-Interdisciplinary Programme of Life Sciences for advanced Research and Education [DBT-IPLS], Calcutta University, for the use of the confocal microscopy facility and Arijit Pal and Souvik Roy for their technical assistance in microscopy. Funding was provided by the Department of Biotechnology, Ministry of Science and Technology (Project BT/PR15410/BCE/08/861/2011) and by Calcutta University (University Grants Commission-University with Potential for Excellence [UGC-UPE] to K.P.).
AUTHOR CONTRIBUTIONS
C.S. and K.P. performed most of the experiments, helped in preparing the article, and prepared the figures. C.S. prepared the graphs in GraphPad Prism. All figures were created by K.P. M.N. performed some of the JGTA-S1 and JGTA-R3 interaction analysis, M.N. and G.M. participated in nitrate measurements; H.N. participated in the spotting assay; D.M., S.S., S.M., and M.P.S. performed the acetylene reduction assays; D.S. and S.Tripathy performed JGTA-S1 genome analysis and OrthoMCL study; N.N. and S.L. measured P; S. Tripathi helped measure total N; and D.B. helped with some RT-PCR studies. M.K. and M.B. performed transmission electron microscopy of JGTA-S1. A.K. and A.J.V. performed the N2-isotope analysis. U.D.G. helped with sample collection and initial analysis of JGTA-S1. A.S. designed the experiments, created the strains, and wrote the article.
References
- Abaidoo R.C., Keyser H.H., Singleton P.W., Borthakur D. (2002). Comparison of molecular and antibiotic resistance profile methods for the population analysis of Bradyrhizobium spp. (TGx) isolates that nodulate the new TGx soybean cultivars in Africa. J. Appl. Microbiol. 92: 109–117. [DOI] [PubMed] [Google Scholar]
- Akhtyamova N., Sattarova R.K. (2013). Endophytic yeast Rhodotorula rubra strain TG-1: Antagonistic and plant protection activities. Biochem. Physiol. 2: 104. [Google Scholar]
- Arnon D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E., Bertini L., Rossi I., Ceccaroli P., Saltarelli R., Guidi C., Zambonelli A., Stocchi V. (2005). New evidence for bacterial diversity in the ascoma of the ectomycorrhizal fungus Tuber borchii Vittad. FEMS Microbiol. Lett. 247: 23–35. [DOI] [PubMed] [Google Scholar]
- Barbieri E., Guidi C., Bertaux J., Frey-Klett P., Garbaye J., Ceccaroli P., Saltarelli R., Zambonelli A., Stocchi V. (2007). Occurrence and diversity of bacterial communities in Tuber magnatum during truffle maturation. Environ. Microbiol. 9: 2234–2246. [DOI] [PubMed] [Google Scholar]
- Barzanti R., Ozino F., Bazzicalupo M., Gabbrielli R., Galardi F., Gonnelli C., Mengoni A. (2007). Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 53: 306–316. [DOI] [PubMed] [Google Scholar]
- Bauer C., Herzog V., Bauer M.F. (2001). Improved technique for electron microscope visualization of yeast membrane structure. Microsc. Microanal. 7: 530–534. [DOI] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513. [DOI] [PubMed] [Google Scholar]
- Bi Y.-M., et al. (2009). Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling. Plant Cell Environ. 32: 1749–1760. [DOI] [PubMed] [Google Scholar]
- Bolano A., Stinchi S., Preziosi R., Bistoni F., Allegrucci M., Baldelli F., Martini A., Cardinali G. (2001). Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Res. 1: 221–224. [DOI] [PubMed] [Google Scholar]
- Brown M.E., Metcalfe G. (1957). Nitrogen fixation by a species of Pullularia. Nature 180: 282. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95. [DOI] [PubMed] [Google Scholar]
- Cao L.X., You J.L., Zhou S.N. (2002). Endophytic fungi from Musa acuminata leaves and roots in South China. World J. Microbiol. Biotechnol. 18: 169–171. [Google Scholar]
- Carvalho T.L., Balsemão-Pires E., Saraiva R.M., Ferreira P.C., Hemerly A.S. (2014). Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 65: 5631–5642. [DOI] [PubMed] [Google Scholar]
- Cataldo D.A., Maroon M., Schrader L.E., Youngs V.L. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant Anal. 6: 71–80. [Google Scholar]
- Chevalier D., Batoux M., Fulton L., Pfister K., Yadav R.K., Schellenberg M., Schneitz K. (2005). STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 9074–9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V., Simopoulos C., Casaretto J.A., El-Kereamy A., Guevara D.R., Cohn J., Zhu T., Guo L., Alexander D.C., Bi Y.M., McNicholas P.D., Rothstein S.J. (2014). Metabolic and co-expression network-based analyses associated with nitrate response in rice. BMC Genomics 15: 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere S., Vanderleyden J., Okon Y. (2003). Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci. 22: 107–149. [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N.K., Bhatnagar S., Eisen J.A., Sundaresan V. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112: E911–E920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-De Los Santos P., Bustillos-Cristales R., Caballero-Mellado J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67: 2790–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Goodenough U.W. (1997). Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo A., Monteiro F., Sebastiana M. (2014). Subtilisin-like proteases in plant-pathogen recognition and immune priming: A perspective. Front. Plant Sci. 5: 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firrincieli A., Otillar R., Salamov A., Schmutz J., Khan Z., Redman R.S., Fleck N.D., Lindquist E., Grigoriev I.V., Doty S.L. (2015). Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Front. Microbiol. 6: 978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.M., Henry L.M., Cornwallis C.K., Kiers E.T., West S.A. (2017). The evolution of host-symbiont dependence. Nat. Commun. 8: 15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai C.S., Lacava P.T., Maccheroni W. Jr., Glienke C., Araújo W.L., Miller T.A., Azevedo J.L. (2009). Diversity of endophytic yeasts from sweet orange and their localization by scanning electron microscopy. J. Basic Microbiol. 49: 441–451. [DOI] [PubMed] [Google Scholar]
- Gancedo J.M. (2001). Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 107–123. [DOI] [PubMed] [Google Scholar]
- Gauthier G.M. (2015). Dimorphism in fungal pathogens of mammals, plants, and insects. PLoS Pathog. 11: e1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig H., Schüssler A., Kluge M. (1996). Geosiphon pyriforme, a fungus forming endocytobiosis with Nostoc (cyanobacteria), is an ancestral member of the Glomales: Evidence by SSU rRNA analysis. J. Mol. Evol. 43: 71–81. [DOI] [PubMed] [Google Scholar]
- Geurts R., Xiao T.T., Reinhold-Hurek B. (2016). What does it take to evolve a nitrogen-fixing endosymbiosis? Trends Plant Sci. 21: 199–208. [DOI] [PubMed] [Google Scholar]
- Gutiérrez R.A., Stokes T.L., Thum K., Xu X., Obertello M., Katari M.S., Tanurdzic M., Dean A., Nero D.C., McClung C.R., Coruzzi G.M. (2008). Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 105: 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Sawers R.J., Marti G., Andrés-Hernández L., Yang S.Y., Casieri L., Angliker H., Oakeley E.J., Wolfender J.L., Abreu-Goodger C., Paszkowski U. (2015). Transcriptome diversity among rice root types during asymbiosis and interaction with arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA 112: 6754–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Gerlach N., Sacristán S., Nakano R.T., Hacquard S., Kracher B., Neumann U., Ramírez D., Bucher M., O’Connell R.J., Schulze-Lefert P. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D.A., Kolter R. (2002). Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 296: 2229–2232. [DOI] [PubMed] [Google Scholar]
- Horst R.J., Zeh C., Saur A., Sonnewald S., Sonnewald U., Voll L.M. (2012). The Ustilago maydis Nit2 homolog regulates nitrogen utilization and is required for efficient induction of filamentous growth. Eukaryot. Cell 11: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z., Guelich G., Phan H., Redman R., Doty S. (2012). Bacterial and yeast endophytes from poplar and willow promote growth in crop plants and grasses. ISRN Agron. 2012: 1–11. [Google Scholar]
- Kneip C., Lockhart P., Voss C., Maier U.G. (2007). Nitrogen fixation in eukaryotes--New models for symbiosis. BMC Evol. Biol. 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth J.L., Kim S.-H., Ettl G.J., Doty S.L. (2013). Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. Glob. Change Biol. Bioenergy 5: 408–418. [Google Scholar]
- Knoth J.L., Kim S.-H., Ettl G.J., Doty S.L. (2014). Biological nitrogen fixation and biomass accumulation within poplar clones as a result of inoculations with diazotrophic endophyte consortia. New Phytol. 201: 599–609. [DOI] [PubMed] [Google Scholar]
- Kobayashi D.Y., Crouch J.A. (2009). Bacterial/fungal interactions: From pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 47: 63–82. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4: 1617. [DOI] [PubMed] [Google Scholar]
- Koroleff F. (1976). Determination of ammonia In Methods of Seawater Analysis, Grasshoft K., ed (Weinheim: Verlag Chemie; ), pp. 126–133. [Google Scholar]
- Larran S., Mónaco C., Alippi H.E. (2001). Endophytic fungi in leaves of Lycopersicon esculentum Mill. World J. Microbiol. Biotechnol. 17: 181–184. [Google Scholar]
- Larran S., Perelló A., Simón M.R., Moreno V. (2002). Isolation and analysis of endophytic microorganisms in wheat (Triticum aestivum L.) leaves. World J. Microbiol. Biotechnol. 18: 683–686. [Google Scholar]
- Lee S.C., Li A., Calo S., Heitman J. (2013). Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 9: e1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C., Jauneau A., Martinez Y., Cabin-Flaman A., Gibouin D., Orivel J., Séjalon-Delmas N. (2017). Exploring fungus-plant N transfer in a tripartite ant-plant-fungus mutualism. Ann. Bot. 120: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J. Jr., Roos D.S. (2003). OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhang H., Liu W., Zheng X. (2011). Biocontrol of postharvest gray and blue mold decay of apples with Rhodotorula mucilaginosa and possible mechanisms of action. Int. J. Food Microbiol. 146: 151–156. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463. [DOI] [PubMed] [Google Scholar]
- Lundberg D.S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald R.M., Chandler M.R. (1981). Bacterium-like organelles in the vesicular-arbuscular mycorrhizal fungus Glomus caledonius. New Phytol. 89: 241–246. [Google Scholar]
- Márquez L.M., Redman R.S., Rodriguez R.J., Roossinck M.J. (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315: 513–515. [DOI] [PubMed] [Google Scholar]
- Martínez-Espinoza A.D., León C., Elizarraraz G., Ruiz-Herrera J. (1997). Monomorphic nonpathogenic mutants of Ustilago maydis. Phytopathology 87: 259–265. [DOI] [PubMed] [Google Scholar]
- Minerdi D., Fani R., Gallo R., Boarino A., Bonfante P. (2001). Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 67: 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebius N., Üzüm Z., Dijksterhuis J., Lackner G., Hertweck C. (2014). Active invasion of bacteria into living fungal cells. eLife 3: e03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar A., El-Tarabily K., Sivasithamparam K. (2005). Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol. Fertil. Soils 42: 97–108. [Google Scholar]
- Nogueira E.M., Vinagre F., Masuda H.P., Vargas C., Pádua V.L.M., Silva F.R., Santos R.V., Baldani J.I., Ferreira P.C.G., Hemerly A.S. (2001). Expression of sugarcane genes induced by inoculation with Gluconacetobacter diazotrophicus and Herbaspirillum rubrisubalbicans. Genet. Mol. Biol. 24: 199–206. [Google Scholar]
- Obertello M., Shrivastava S., Katari M.S., Coruzzi G.M. (2015). Cross-species network analysis uncovers conserved nitrogen-regulated network modules in rice. Plant Physiol. 168: 1830–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partida-Martinez L.P., Hertweck C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- Partida-Martinez L.P., Monajembashi S., Greulich K.O., Hertweck C. (2007). Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17: 773–777. [DOI] [PubMed] [Google Scholar]
- Pearce G., Yamaguchi Y., Barona G., Ryan C.A. (2010). A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 107: 14921–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L., Buckler E.S., Ley R.E. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Tomás A.A., Anderson M.A., Suen G., Stevenson D.M., Chu F.S.T., Cleland W.W., Weimer P.J., Currie C.R. (2009). Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326: 1120–1123. [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339: 62–66. [DOI] [PubMed] [Google Scholar]
- Reid D.E., Ferguson B.J., Hayashi S., Lin Y.H., Gresshoff P.M. (2011). Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 108: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C., Oldroyd G.E. (2014). Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J. Exp. Bot. 65: 1939–1946. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., León-Ramírez C., Vera-Nuñez A., Sánchez-Arreguín A., Ruiz-Medrano R., Salgado-Lugo H., Sánchez-Segura L., Peña-Cabriales J.J. (2015). A novel intracellular nitrogen-fixing symbiosis made by Ustilago maydis and Bacillus spp. New Phytol. 207: 769–777. [DOI] [PubMed] [Google Scholar]
- Saha C., Seal A. (2015). Early changes in shoot transcriptome of rice in response to Rhodotorula mucilaginosa JGTA-S1. Genom. Data 6: 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha C., Mukherjee G., Agarwal-Banka P., Seal A. (2016). A consortium of non-rhizobial endophytic microbes from Typha angustifolia functions as probiotic in rice and improves nitrogen metabolism. Plant Biol. (Stuttg.) 18: 938–946. [DOI] [PubMed] [Google Scholar]
- Sampedro I., Aranda E., Scervino J.M., Fracchia S., García-Romera I., Ocampo J.A., Godeas A. (2004). Improvement by soil yeasts of arbuscular mycorrhizal symbiosis of soybean (Glycine max) colonized by Glomus mosseae. Mycorrhiza 14: 229–234. [DOI] [PubMed] [Google Scholar]
- Sánchez-Martínez C., Pérez-Martín J. (2001). Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis--Similar inputs, different outputs. Curr. Opin. Microbiol. 4: 214–221. [DOI] [PubMed] [Google Scholar]
- Scannerini S., Bonfante P. (1991). Bacteria and bacteria like objects in endomycorrhizal fungi (Glomaceae) In Symbiosis as Source of Evolutionary Innovation: Speciation and Morphogenesis, Margulis L., and Fester R., eds (Cambridge: MIT Press; ), pp. 273–287. [Google Scholar]
- Schlaeppi K., Dombrowski N., Oter R.G., Ver Loren van Themaat E., Schulze-Lefert P. (2014). Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 111: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., Paul K., Saha C., Mukherjee G., Nag M., Ghosh S., Das A., Seal A., Tripathy S. (2019). A unique life-strategy of an endophytic yeast Rhodotorula mucilaginosa JGTA-S1-A comparative genomics viewpoint. DNA Res. 26: 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Schmid M., Rothballer M., Hause G., Zuccaro A., Imani J., Kämpfer P., Domann E., Schäfer P., Hartmann A., Kogel K.H. (2008). Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell. Microbiol. 10: 2235–2246. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Morgan D.R., Swensen S.M., Mullin B.C., Dowd J.M., Martin P.G. (1995). Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. USA 92: 2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C., Xie Z.-P., Illana A., Vierheilig H. (2011). Long-distance transport of signals during symbiosis: Are nodule formation and mycorrhization autoregulated in a similar way? Plant Signal. Behav. 6: 372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward G.F., Zehr J.P., Jellison R., Montoya J.P., Hollibaugh J.T. (2004). Vertical distribution of nitrogen-fixing phylotypes in a meromictic, hypersaline lake. Microb. Ecol. 47: 30–40. [DOI] [PubMed] [Google Scholar]
- Tian X.L., Cao L.X., Tan H.M., Zeng Q.G., Jia Y.Y., Han W.Q., Zhou S.N. (2004). Study on the communities of endophytic fungi and endophytic actinomycetes from rice and their antipathogenic activities in vitro. World J. Microbiol. Biotechnol. 20: 303–309. [Google Scholar]
- Torres-Cortés G., Ghignone S., Bonfante P., Schüßler A. (2015). Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: Transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc. Natl. Acad. Sci. USA 112: 7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trdá L., Fernandez O., Boutrot F., Héloir M.C., Kelloniemi J., Daire X., Adrian M., Clément C., Zipfel C., Dorey S., Poinssot B. (2014). The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 201: 1371–1384. [DOI] [PubMed] [Google Scholar]
- Valdivia R.H., Heitman J. (2007). Endosymbiosis: The evil within. Curr. Biol. 17: R408–R410. [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206: 1196–1206. [DOI] [PubMed] [Google Scholar]
- Vinagre F., Vargas C., Schwarcz K., Cavalcante J., Nogueira E.M., Baldani J.I., Ferreira P.C., Hemerly A.S. (2006). SHR5: A novel plant receptor kinase involved in plant-N2-fixing endophytic bacteria association. J. Exp. Bot. 57: 559–569. [DOI] [PubMed] [Google Scholar]
- Wang R., Dong L., Chen Y., Qu L., Wang Q., Zhang Y. (2017). Esteya vermicola, a nematophagous fungus attacking the pine wood nematode, harbors a bacterial endosymbiont affiliated with Gammaproteobacteria. Microbes Environ. 32: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin G., Glawe D., Doty S.L. (2009). Characterization of three endophytic, indole-3-acetic acid-producing yeasts occurring in Populus trees. Mycol. Res. 113: 973–980. [DOI] [PubMed] [Google Scholar]
- Yan D., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7: 13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ge L., Chen K., Zhao L., Zhang X. (2014). Enhanced biocontrol activity of Rhodotorula mucilaginosa cultured in media containing chitosan against postharvest diseases in strawberries: Possible mechanisms underlying the effect. J. Agric. Food Chem. 62: 4214–4224. [DOI] [PubMed] [Google Scholar]
- Zimmer B.L., Roberts G.D. (1979). Rapid selective urease test for presumptive identification of Cryptococcus neoformans. J. Clin. Microbiol. 10: 380–381. [DOI] [PMC free article] [PubMed] [Google Scholar]