Abstract
Glioblastoma is a malignant brain tumor that portends a poor prognosis. Its resilience, in part, is related to a remarkable capacity for manipulating the microenvironment to promote its growth and survival. Microglia/macrophages are prime targets, being drawn into the tumor and stimulated to produce factors that support tumor growth and evasion from the immune system. Here we show that the RNA regulator, HuR, plays a key role in the tumor-promoting response of microglia/macrophages. Knockout (KO) of HuR led to reduced tumor growth and proliferation associated with prolonged survival in a murine model of glioblastoma. Analysis of tumor composition by flow cytometry showed that tumor associated macrophages (TAMs) were decreased, more polarized toward an M1-like phenotype, and had reduced PD-L1 expression. There was an overall increase in infiltrating CD4+ cells, including Th1 and cytotoxic effector cells, and a concomitant reduction in tumor-associated polymorphonuclear myeloid-derived suppressor cells. Molecular and cellular analyses of HuR KO TAMs and cultured microglia showed changes in migration, chemoattraction, and chemokine/cytokine profiles that provide potential mechanisms for the altered tumor microenvironment and reduced tumor growth in HuR KO mice. In summary, HuR is a key modulator of pro-glioma responses by microglia/macrophages through the molecular regulation of chemokines, cytokines, and other factors. Our findings underscore the relevance of HuR as a therapeutic target in glioblastoma.
Keywords: Glioblastoma, Tumor associated macrophages and microglia, effector T cells, tumor microenvironment, cell migration, cytokines
INTRODUCTION
Glioblastoma (GB) is a highly aggressive tumor of the central nervous system, characterized by rapid growth and invasion. The tumor responds poorly to treatment and the median survival remains low at ~20 months (Desjardins et al. 2018; van den Bent et al. 2018). The malignant phenotype of GB is supported by a favorable microenvironment which it orchestrates through an active secretome. Tumor-associated macrophages and microglia (TAMs), which make up as much as 40% of the cellular content in GB, are recruited by chemoattractants that are produced by the tumor cell (Chen and Hambardzumyan 2018; Hambardzumyan et al. 2016; Sorensen et al. 2018). TAMs are then induced by the tumor to produce soluble factors such as IL-6, IL-1, TGF-β and matrix metalloproteinases (MMPs) which enhance the motility, invasion, angiogenesis and proliferation of both GB cells and glioma stem cells. TAMs can also be induced to suppress anti-tumor immune cells through production of factors such as TGF-β and PD-L1 (Hambardzumyan et al. 2016; Noguchi et al. 2017; Sun et al. 2018). The positive influence of TAMs on GB progression is underscored by clinicopathological data associating higher tumor grade and worse prognosis with increasing populations of TAMs (Sorensen et al. 2018). The molecular response of TAMs to paracrine factors released by glioma cells is clearly complex and involves numerous signaling pathways and transcriptional factors such as STATs and NF-ĸB, (Holtman et al. 2017; Li and Graeber 2012). Little attention, however, has been paid to post-transcriptional regulation of mRNAs generated by these signaling pathways. We recently demonstrated that the RNA regulator, HuR, modulates many of the downstream mRNA targets produced by these signaling pathways and has a major impact on the molecular signature of activated primary microglia (MG) (Matsye et al. 2017). HuR is a member of the Elav family of RNA binding proteins and possesses three RNA recognition motifs (Ma et al. 1996). It has a binding affinity for U- and AU-rich elements that are present in the 3’ untranslated region (UTR) of many chemokines, cytokines and growth factors (Anderson 2008; Ma et al. 1996). In general HuR positively regulates target mRNAs by stabilizing the transcript and/or augmenting translational efficiency (Anderson 2010; Brennan and Steitz 2001; Srikantan and Gorospe 2012; Wilusz and Wilusz 2004). These functions have been linked to HuR translocation to the cytoplasm and association with polysomes and/or competition with silencing miRNAs (Abdelmohsen et al. 2008; Brennan and Steitz 2001; Meisner and Filipowicz 2011). HuR can also bind to the 5’ UTR, often at or near internal ribosome entry sites, and either inhibits or augments translation (Galban et al. 2008; Kullmann et al. 2002; Meng et al. 2005). Previously, we reported on the vital role of HuR in GB where it is highly upregulated in tumor cells and promotes tumor progression in vivo (Filippova et al. 2017; Filippova et al. 2011; Nabors et al. 2001; Nabors et al. 2003). Blocking HuR either by chemical inhibition or shRNA-mediated silencing can produce a potent anti-glioma effect (Filippova et al. 2017; Filippova et al. 2011; Wang et al. 2019). In the current study, we hypothesized that HuR expression in TAMs promotes tumor progression through its role in modulating the expression of key cytokines and chemokines. Using a mouse in which HuR was deleted from TAMs, we observed a significant prolongation of survival in a syngeneic GB murine model, with a reduction of tumor size and a shift in intratumoral immune cell profiles from immunosuppressive to cytotoxic. This immune cell shift may relate to altered molecular and cellular responses of HuR-deleted TAMs to soluble factors produced by tumor cells.
MATERIALS AND METHODS
HuR Conditional Knockout Mice
All animal procedures were reviewed and approved by the UAB Institutional Animal Care and Use Committee in compliance with the National Research Council Guide for the Care and Use of Laboratory Animals. To produce a MG/macrophage HuR knockout (HuR KO), C57BL/6 HuRfl/fl mice (generously provided by Dr. Ulus Atasoy, University of Michigan, Ann Arbor, Michigan) were crossed with B6J.B6N(Cg)-Cx3cr1tm1.1(cre)Jung/J mice (Jackson Laboratory) (Figueroa et al. 2003).
Cell Culture
Murine glioma cells GL261 were a generous gift from Dr. Etty (Tika) Benveniste. Cells were grown in serum free stem cell medium [Neurobasal™-A medium supplemented with 1% B-27™ minus vitamin A, 1% CTS™ N-2, 2 mM L-glutamine, 2.5 μg/mL Amphotericin B, 50 μg / mL Gentamicin, 10 ng/ml FGF, and 10 ng/ml EGF (Gibco). To collect GL261 conditioned medium, Gl261 cells were cultured in fresh stem cell media for 24 h, and the supernatants were collected for downstream applications. For intracranial injection, GL261 cells were transduced with pGreenFire1-CMV (System Biosciences) as previously reported for green fluorescent protein (GFP) expression (Walker and Hjelmeland 2014). Primary mouse microglia used for the experiments were isolated from neonatal mouse pups (P0 – P2) using the mixed glial culture method as described previously (Matsye et al. 2017). After shaking off from the astrocyte layer, microglia were cultured in Macrophage Serum Free Medium (Macrophage-SFM) (Gibco) for downstream assays. For HuR knockdown, MG were seeded in 6-well plates at a density of 5 × 105 in Macrophage-SFM and transfected with 150 pmol SMARTpool: ON-TARGETplus Elavl1 (HuR) siRNA (Dharmacon) or siGFP control using Lipofectamine 2000 (Thermo Fisher Scientific). After 24 h, culture media were collected and used as a chemoattractant for GL261 transwell invasion assay.
Spleen Cell Isolation
After euthanasia, spleens were removed from mice and transferred to a 70-μm cell strainer placed on top of a 50 mL tube. The tissue was pushed through the strainer using a 10 mL syringe plunger and washed with 10 mL ice-cold DMEM (Corning®) to make a single cell suspension. Cells were collected by centrifuge (400 g, 5 min), and red blood cells were removed by Red Blood Cell Lysis Buffer (Biolegend). Splenic cells were then resuspended in DMEM, counted on TC20™ Automated Cell Counter (Bio-Rad Laboratories) and kept on ice for flow cytometry staining.
Brain Cell Isolation
Upon complete anesthesia by isoflurane, the mice were transcardially perfused with 15 mL ice-cold PBS at ~ 7 mL/min. The cerebrum from tumor-free naïve mice or cerebrum with tumors from the tumor-bearing mice was then extracted and cut into small (< 2 mm) pieces, and digested in 5 mL enzyme solution [PBS supplemented with 2% FBS, 1 mg/mL collagenase/dispase (Roche) and 0.5 mg/mL DNase I (Sigma-Aldrich)] for 1 hour at 37°C. After digestion, the cells were passed through a 70-μm cell strainer to make a single cell suspension, followed by centrifugation with 30% Percoll at 400 g for 30 minutes to remove myelin and cell debris. The cells were then resuspended in DMEM, counted on TC20™ Automated Cell Counter and kept on ice for flow cytometry staining.
In Vivo Tumor experiments
Eight to 12-week-old HuR KO or littermate control mice were used for the tumor intracranial injections. Upon inducing anesthesia with ketamine and xylazine cocktail, the mouse was properly positioned on the stereotaxic instrument (Stoelting Co.), and a burr hole was made 2 mm lateral (right) and 1 mm anterior to the bregma using a dental drill with a 0.45mm non-cutting bit. 104 GL261-Luc cells resuspended in DMEM were injected at a rate of 1 μL/min for 2 min using a 26G Hamilton syringe controlled by a Harvard 11 Plus Syringe Pump. For survival studies, mice were monitored twice daily until they reaching a moribund state. Survival times were recorded.
Bioluminescent Imaging
After injection of GL261 cells, tumor growth was measured using the IVIS® Lumina Series III In Vivo Imaging System (PerkinElmer Inc.). For imaging, mice were injected with 2.5 mg of d-luciferin substrate intraperitoneally and imaged after 10 min. Light emission from the Regions of Interested (ROI) was measured using the Living Image® Software (PerkinElmer Inc.). Photons-per-second was used for comparison between groups.
Flow Cytometry
Single cells were isolated from spleen, bone marrow, naïve brain or tumor-bearing brain as previously described. For flow cytometry, 2 × 106 cells were seeded in 96-well plate, and incubated for 20 min at 4 °C with Zombie Aqua™ Fixable Viability Kit (Biolegend). Cells were washed with staining buffer (PBS with 2% FBS) and incubated for 30 min at 4 °C with fluorescent conjugated cell surface markers (Biolegend, eBioscience), followed by one wash with staining buffer. For intracellular marker staining, cells were first fixed with the Fixation/Permeabilization Solution Kit (BD Biosciences) for 20 min, washed once with perm/wash buffer and permeabilized in perm/wash buffer overnight at 4 °C. On the following day, cells were stained with fluorescent conjugated intracellular markers (Biolegend, eBioscience) for 30 min at 4 °C. After one final wash with staining buffer, the cells were resuspended in 200 μL of staining buffer and analyzed on a BD™ LSR II Cell Analyzer (BD Biosciences). Data were analyzed using FlowJo software.
Fluorescence Activated Cell Sorting
Single cells were isolated from tumor-bearing brains as described above. All cells from one sample were collected in 5 mL Falcon® Round-Bottom Polystyrene Tubes. After a 20 min incubation with Zombie Aqua™ Fixable Viability Kit (Biolegend) at 4 °C, cells were washed with staining buffer (PBS supplemented with 2% FBS) and stained for 30 min at 4 °C with fluorescent conjugated cell surface markers (Biolegend, eBioscience), followed by one wash with staining buffer. Cells were resuspended in staining buffer and the tumor associated macrophages (CD45hi CD11b+ F4/80+) were collected on a BD™ FACS Aria II Cell Sorter (BD Biosciences).
Tissue Processing and Immunohistochemistry Staining
Upon complete anesthesia by isoflurane, mice were transcardially perfused with 15 mL ice-cold PBS at ~ 7 mL/min, followed by 20 mL ice-cold 4% paraformaldehyde (PFA) at ~ 4 mL/min. Brains were extracted and post-fixed in 4% PFA overnight at 4 °C. Fixed brains were coronally sliced by 2 mm on a Precisian Brain Slicer (Braintree Scientific), followed by dehydration in 70% EtOH for 24 h. Specimens were processed on a Leica Tissue Processor before embedding in paraffin blocks. For immunohistochemistry, 8 μm sections were stained with the following antibodies: HuR (3A2, Santa Cruz Biotechnology) at 1:200, Iba-1 (Wako) at 1:50, ki67 at 1:1000, MPO (Abcam) at 1:50 and CD4 (Abcam, 1:1,000).
RNA Isolation and qRT-PCR
Total RNA was extracted from cells using the illustra RNAspin Kit (GE Healthcare), and quantified by Nanodrop (Thermo Fisher Scientific). One microgram of RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Expression levels of target mRNAs were measured by real time qPCR using predesigned TaqMan Gene Expression Assays (Thermo Fisher Scientific), and calculated using the delta delta CT method with GAPDH as the reference gene (Livak and Schmittgen 2001).
Protein Extraction and Western Blot
Whole cell lysates were prepared using M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) and quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Fifty micrograms of protein extract were denatured with Laemmli Sample Buffer (Bio-Rad Laboratories) at 95 °C for 10 min and then resolved by electrophoresis using a 4–20% Mini-PROTEAN® TGX™ Precast Protein Gel (Bio-Rad Laboratories). Proteins were transferred to a nitrocellulose membrane and probed with an anti-HuR (3A2, Santa Cruz Biotechnology) and GAPDH (Cell Signaling Technology) antibodies.
ELISA
MG were seeded in 6-well plates at a density of 5 × 105 cells per well, and treated with GL261 conditioned media or plain stem cell media as a control for 24 h. The cultural supernatants were collected and concentrations of the following chemokines were quantified by ELISA: CXCL1, CXCL2, CCL5 (R & D Systems), CXCL10 (Boster).
Zymography
PMG were seeded in 6-well plates at a density of 5 × 105 in Macrophage-SFM. After 24 h, cells were treated with GL261 conditioned media for 24 h. Media were collected for zymography. All media samples were quantified using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Two hundred micrograms of media were mixed with Zymogram Sample buffer (Bio-Rad Laboratories) 1:2 by volume and resolved by electrophoresis using 10% Criterion™ Zymogram Gel with Gelatin (Bio-Rad Laboratories). The gels were renatured for 30 min at room temperature, and then developed overnight at 37 °C. On the following day, gels were first stained for 30 min with Coomassie brilliant blue R-250 staining solution (Bio-Rad Laboratories) and then destained for 2 h. Gels were scanned and quantified using ImageJ.
Transwell invasion and scratch wound Assays
The transwell invasion assay was performed using Corning® BioCoat™ Matrigel® Invasion Chambers with 8.0 μm membrane pores (Corning). For the MG invasion assay, a single cell suspension of 2.5 × 104 HuR KO or control MG in macrophage-SFM were added to the top chamber, and GL261 conditioned stem cell media was added to the bottom chamber as a chemoattractant. MG were allowed to migrate for 24 h in the cell culture incubator, and then fixed and stained using the Hema 3™ Manual Staining System and Stat Pack (Thermo Fisher Scientific). The inserts were removed and mounted on microscope slides and stained cells. For quantitation, 30 images were captured (throughout the filter) for each well and stained cells were counted. For GL261 invasion assay, 2.5 × 104 GL261 cells in stem cell media was added to the top chamber. MG conditioned media was collected as described above and added to the bottom chamber as the chemoattractant. GL261 cells were allowed to migrate for 24 h, then stained and quantified as described above. For the scratch wound assay, MG were seeded in 12-well plates at a density of 2 × 105 in Macrophage-SFM. After 24 h, the MG monolayer was gently scored with a 1 mL pipette tip across the center of the well. The media was replaced with GL261 conditioned media. Cells were allowed to migrate for 48 h in a cell culture chamber. Serial images were captured with the Olympus IX73 inverted microscope and cells migrating into the gaps were counted.
Statistics
Statistical analyses were performed using Graphpad Prism 7 (Graphpad Software, Inc). A two-sided student’s t-test was used for comparisons between HuR KO and control groups in all experiments. Kaplan-Meier survival curves were generated with Graphpad Prism 7. Differences between groups were considered significant when the p-value was < 0.05.
RESULTS
Development of a mouse model that harbors a Cx3Cr1-specific deletion of HuR
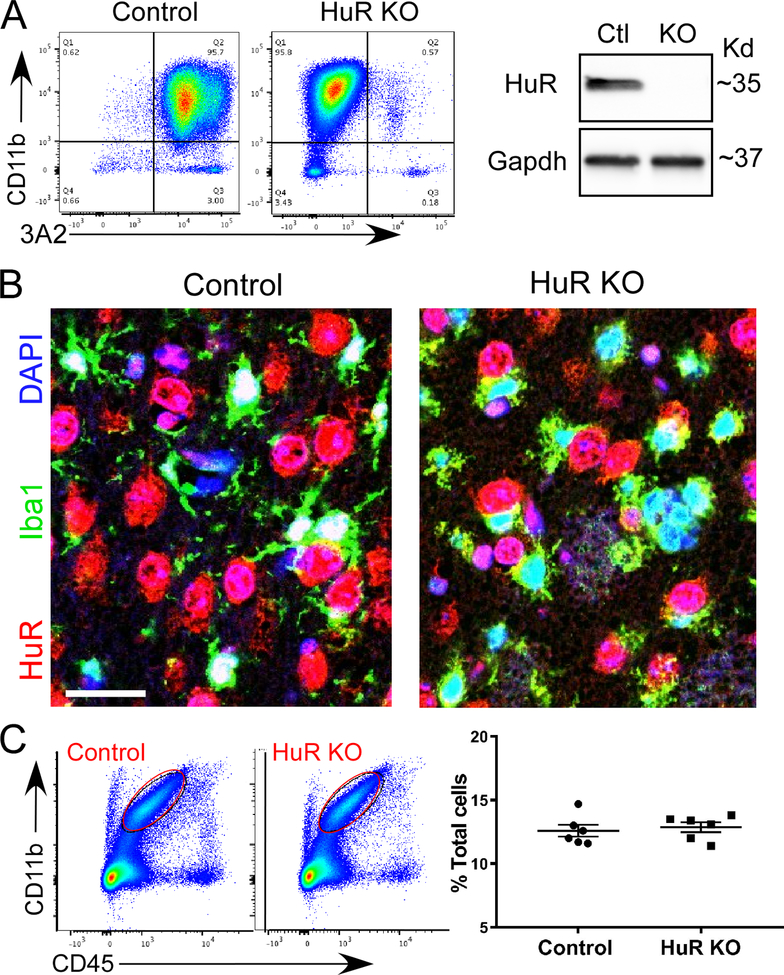
To produce a Cx3cr1 promoter-driven HuR knockout mouse, we crossed a HuRfl/fl mouse with a mouse expressing Cre recombinase under the control of the Cx3cr1 promoter (Yona et al. 2013). To verify HuR knockout, we first isolated MG from neonatal pups and assessed for HuR expression by flow cytometry and western blot (Figure 1A). No HuR was detected in CD11b+ MG from HuRfl/fl, cre+ (referred to as HuR KO) mice, whereas abundant HuR expression was detected in HuRfl/fl, cre- (referred to as control) mice. Western blot confirmed an absence of expression. We next examined mature mouse brain sections by immunohistochemistry using anti-HuR (3A2) and Iba1 antibodies (Figure 1B). In HuR KO brain, there was loss of HuR staining in cells identified by Iba1. Immunoreactivity was detected in cells adjacent to MG, which are likely neurons expressing HuR or other neuronal Hu antigens that cross-react with the 3A2 antibody. In the control brain, there was co-localized immunostaining of Iba1 and 3A2, consistent with microglial HuR expression and its typical nuclear localization (Matsye et al. 2017). To determine whether HuR KO impacted the number of MG in the brain, we quantitated them by flow cytometry in single-cell suspensions derived from whole brains (Figure 1C). The overall microglial counts (percent of total cells) were similar between control and HuR KO mice at ~12 % which is in the range of previously reported microglial counts performed by histology (Lawson et al. 1990). Since Cx3cr1 is more broadly expressed in mononuclear phagocytes, we performed flow cytometry of cells derived from spleen to determine the extent of reduced HuR expression (gating strategy shown in Figure S1). We observed a significant overall reduction of HuR mean fluorescence intensity (MFI) in CD11b+/Cx3cr1+ cells (Figure S2). Additional analysis of myeloid subsets showed a reduction of HuR MFI in macrophage (Mɸ), polymorphonuclear cell (PMN) and monocyte cell populations compared to control mice (S2) (Yona et al. 2013). In cultured MG cells there was a complete separation of peaks consistent with the absence of HuR in KO mice. Taken together, HuR expression is reduced in myeloid cell populations consistent with prior observations of Cx3cr1 activity more broadly in the mononuclear phagocyte system (Yona et al. 2013).
Figure 1. Microglial-HuR knockout mouse model.
A HuRfl/fl (control) mouse was crossed with a mouse expressing Cre recombinase under control of the Cx3cr1 promoter to produce HuRfl/fl/Cre (HuR KO) mice. (A) Cultured primary microglia were isolated from neonatal pups and assessed by flow cytometry using 3A2 (HuR) and Cd11b antibodies. Right panel shows a western blot of primary microglial cells using the same HuR antibody. (B) Immunohistochemistry of frontal cortex sections from control and HuR KO mice. Antibodies are shown to the left of the images. Scale bar, 20 μm. (C) Quantification of microglia in control and HuR KO brains by flow cytometry. To the left is a representative gating using CD11b and CD45 markers. A graph summarizing the findings for 6 control and 6 HuR KO mice is shown to the right.
HuR KO Mice show attenuated Glioma growth and prolonged survival
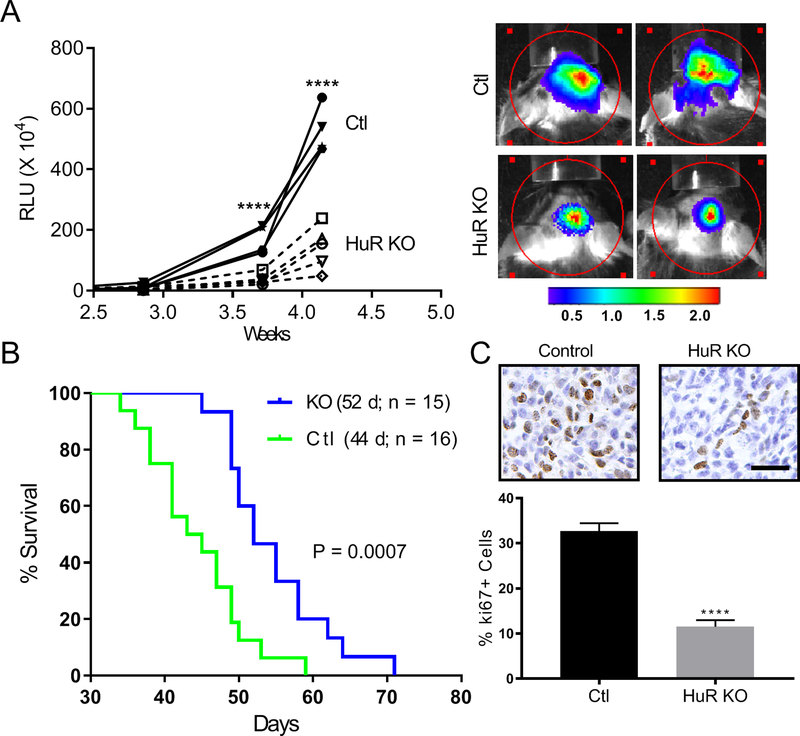
GL261 glioma cells expressing a luciferase reporter and green fluorescent protein (GFP) were injected intracranially in control and HuR KO mice to produce tumors. Serial in vivo imaging of luciferase activity showed a significant attenuation of luminescence in the HuR KO mice over time consistent with reduced tumor size (Figure 2A). This was associated with an increase in survival by 8 days (Figure 2B). Tumors were assessed for proliferation by immunohistochemical staining with an anti-ki67 antibody (Figure 2C). There was a ~three-fold decrease in positive cells from HuR KO tumors indicating a significant effect on tumor cell proliferation.
Figure 2. HuR knockout mice have attenuated GB tumor growth and prolonged survival.
(A) GL261 cells expressing a luciferase reporter were injected intracranially into HuR KO or control mice. Tumor sizes were assessed by in vivo luciferase imaging at the time points shown. To the right are representative images of each cohort. (B) Survival curves for control and HuR KO mice with median survival time shown in parentheses. (C) Serial sections from tumors were immunostained with a ki67 antibody and positive cells were counted. Data points represent the mean ± SD of random low-powered fields from 20–40 tumor sections per tumor from three independent mice per group. **** P < 0.0001; scale bar, 100 μm.
HuR KO tumor microenvironment shows altered myeloid cell populations that favor tumor inhibition
We assessed mouse brains harboring malignant gliomas by flow cytometry to quantify subpopulations of immune cells. The overall CD45hi population of immune cells was significantly lower in HuR KO mice at 60% versus 70% in control mice (Figure 3A). Within that population, there was a 50% decrease in TAMs, as defined by CD11b+ and CD45hi, in HuR KO versus control mice (Figure 3B). Immunohistochemistry of tumor sections from a control and HuR KO mouse showed an absence of HuR immunostaining in HuR KO TAMs and abundant HuR staining in the control tumor (Figure 3C). In a number of TAMs from control sections, HuR immunoreactivity merged with that of Iba1 consistent with redistribution of HuR to the cytoplasm and activation (Abdelmohsen et al. 2008; Brennan and Steitz 2001). Further analysis of TAMs with M1 and M2 markers revealed that there was a two-fold increase in the percentage of M1 TAMs in HuR KO mice (Figure 3D). On the other hand, M2-polarized TAMs were lower in HuR KO by two-fold. The MFI of MHCII was higher in HuR KO TAMs (Figure 3E). We also noted a two-fold higher MHCII expression on CD45−GFP+ tumor cells of HuR KO mice versus control mice (Figure S3). We next assessed TAMs and MG for PD-L1, a ligand that can be induced in these cells by tumors (Prima et al. 2017). This ligand was abundantly detected in TAMs, as reflected by the high MFI, but only at low levels in MG (Figure 3F). PD-L1 expression was significantly lower in TAMs from HuR KO brains compared to control.
Figure 3. Tumor-associated macrophages (TAMs) are attenuated in HuR KO mice and have altered polarization.
After tumor development, brains from control and HuR KO mice were removed and assessed for TAMs by flow cytometry. All gating was done on live cells (see Figure S1 for gating strategy). (A) Quantification of CD45hi cells by flow cytometry. (B) Quantification of TAMs (CD45hiCD11b+Gr-1loF4/80+). (C) Immunohistochemistry of representative sections from a control and HuR KO-derived tumor. Antibodies are shown to the left. Arrowheads indicate TAMS in which HuR immunoreactivity co-localizes with that of IBA-1, consistent with cytoplasmic redistribution of HuR. This is highlighted in the inset which is an enlargement of the region marked by an asterisk. Scale bar, 20 μm. (D) TAMs were further sorted into MI-polarized (MHCIIhi, CD86+) and M2-polarized (MHCIIlo, CD206+) subsets. (E) MHCII expression levels were determined based on the mean fluorescence intensity (MFI). A histogram of MFI for TAMs is shown to the right. (F) TAMs and MG (MG defined as CD45lo, CD11b+) were assessed for PD-L1 expression. A histogram of MFI for TAMs is shown to the right. For all graphs, means and SDs are shown. Each data point is representative of an individual mouse. *P < 0.05, **P < 0.01, ***, P < 0.0005, ****P < 0.0001.
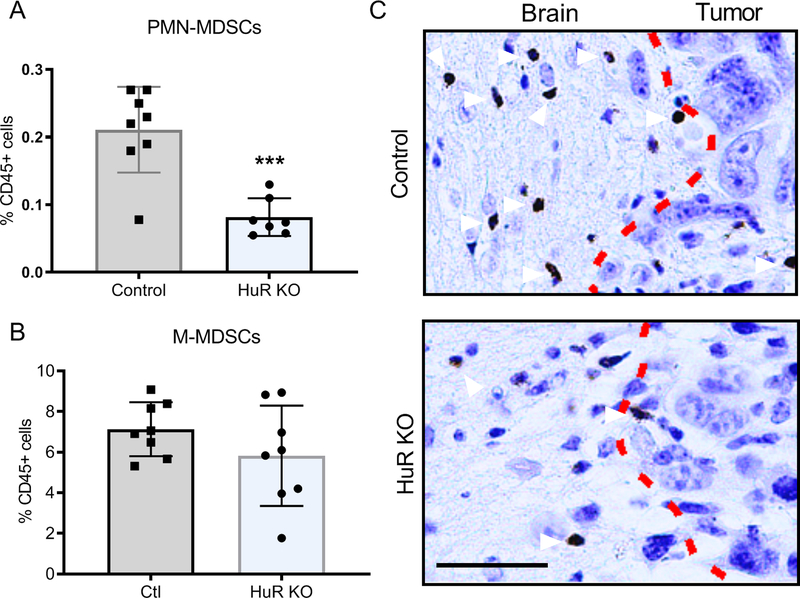
We investigated Gr1hiLy6G+ polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) and observed a 2.5-fold reduction in HuR KO brain tumors (Figure 4A). Monocytic-MDSCs, on the other hand, were not altered (Figure 4B). A representative tumor section immunostained with an anti-myeloperoxidase antibody shows a cluster of PMNs along the tumor border in a control mouse that was not seen in HuR KO mice, consistent with the flow cytometry data (Figure 4C). Taken together, HuR KO altered myeloid populations in the tumor microenvironment, increasing pro-inflammatory and suppressing anti-inflammatory profiles.
Figure 4. Polymorphonuclear myeloid-derived suppressor cells (PMN MDSCs) are attenuated in HuR KO tumors.
(A) Brain PMN MDSCs (CD45hi, CD11b+,Gr1hi,Ly6Clow, Ly6G+CD49d−) were quantified by flow cytometry. (B) Brain monocytic MDSCs (CD45hiCD11b+Gr1midLy6ChiLy6G−CD49d+) were quantified by flow cytometry. (C) Immunostaining for neutrophils (arrowheads) in brain sections using an MPO antibody. Dotted line represents the border between normal brain and tumor. Each data point is representative of an individual mouse. Means and SDs are shown. ***P = 0.0003. Scale bar, 50 μm.
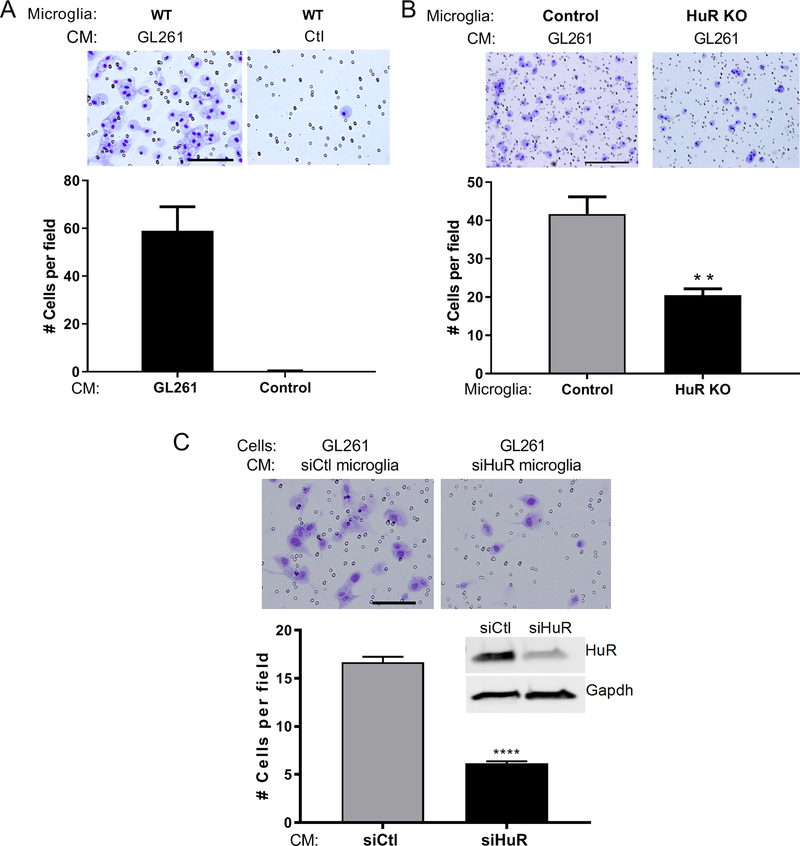
HuR knockout in MG suppresses migration and glioma cell chemotaxis
With the reduction in TAMs observed in HuR KO mice, we next determined if loss of HuR suppressed migration or invasion properties in response to chemotactic signals from glioma cells. We performed this experiment with cultured control and HuR KO MG from naïve mice. Using a matrigel-coated transwell system, we placed MG in the upper chamber and conditioned media (CM) from GL261 cells in the lower chamber. With wild-type MG, we observed an abundant migration toward GL261 CM but little to no movement with control (unconditioned) media consistent with prior observations that glioma cells secrete potent chemotactic factors for MG (Figure 5A) (Bettinger et al. 2002). With GL261 CM, migration/invasion of HuR KO MG was suppressed by more than two-fold (Figure 5B). Using a scratch assay, we observed a similar ~two-fold attenuation of HuR KO MG movement into the gap after exposure to GL261 CM (Figure S4). We next assessed the impact of HuR knockdown on the ability of MG to chemoattract glioma cells. With CM from siHuR-treated MG, we observed a nearly 3-fold suppression of migrated GL261 cells compared to control (Figure 5C). Taken together, these findings indicate that HuR plays an important role in microglial migration/chemoattraction toward glioma cells and chemoattraction of glioma cells toward MG. Dysregulated migratory profiles of these cells may contribute to the altered microenvironment and reduced tumor growth in HuR KO mice.
Figure 5. HuR knockout in microglia suppresses migration and glioma cell chemotaxis.
(A) Migration of wild-type microglia was assessed in a transwell system. Microglia were seeded in the upper chamber and conditioned media (CM) from GL261 cells or unconditioned media (control) was placed in the lower chamber for 24 h. Migrated cells were identified by hematoxylin staining and counted (see Materials and Methods). Representative micrographs are shown. Scale bar,50 μm. (B) Control and HuR KO microglia were seeded in the upper chamber with GL261 CM in the lower chamber. **P <0.005. Scale bar, 100 μm. (C) GL261 cells were seeded in the upper chamber and CM from wild-type microglia transfected with siHuR or control siRNA was placed in the lower chamber. Migrated cells were counted 24 h later. Scale bar, 50 μm. A western blot of siRNA-transfected microglia is shown along with the antibodies used. ****P < 0.0001. All experiments were performed in triplicate and the data points are the mean ± SD of counted cells in at least 10 high-powered fields.
HuR KO alters the chemokine and MMP profile of MG in response to GL261 CM
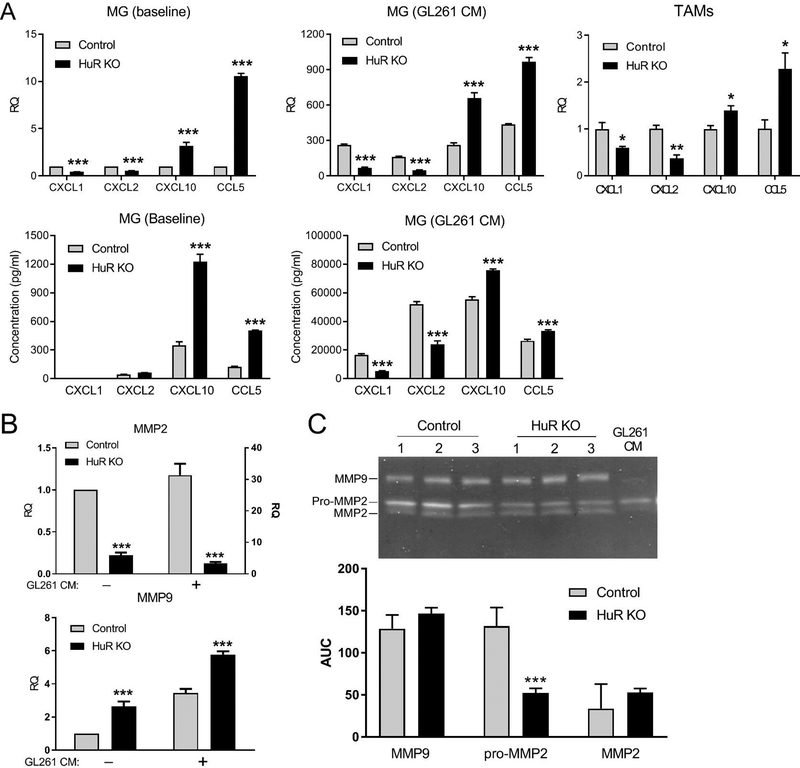
We first analyzed expression of chemokines and matrix metalloproteinases (MMPs) by MG that may influence the migration/chemoattraction profiles of both MG and glioma cells, as we observed above. We performed an initial screen for affected chemokines with multiplex analysis of HuR KO MG treated with GL261 CM for 24 h (not shown). Glioma-relevant chemokines that were significantly altered were then further analyzed by qPCR and ELISA (Figure 6A). CXCL1 and CXCL2 mRNAs were attenuated at baseline in HuR KO MG by two-fold, whereas CXCL10 and CCL5 were increased by 3- and 11-fold. With GL261 CM treatment, there was a robust induction of chemokine mRNA in control MG ranging from 160 to 430-fold over baseline. The pattern was altered in HuR KO MG similar to unstimulated cells, although more pronounced for CXCL1 and 2 (three to four-fold decrease). To determine whether the pattern was similar in vivo, we isolated TAMs from primary tumors by fluorescence-activated cell sorting and assessed changes in mRNA by qPCR. We found that the differential pattern of chemokine expression matched that of cultured MG. We next measured these chemokines by ELISA in media from cultured MG before and after GL261 CM treatment. At baseline, there was a marked ~four-fold increase in CXCL10 and CCL5 in HuR KO MG. With GL261 treatment, CXCL1 and 2 were detected at high levels in control but suppressed by two to three-fold in HuR KO MG. CXCL10 and CCL5 remained increased in HuR KO MG but by smaller margins. We next looked at MMP-2 and 9, two key metalloproteinases linked to glioma cell migration and invasion (Figure 6B) (Bjorklund and Koivunen 2005). MMP2 mRNA was suppressed by more than four-fold at baseline. With exposure to GL261 CM there was a 30-fold induction in control MG which was reduced to three-fold in HuR KO MG. MMP-9 mRNA, on the other hand, was increased at baseline by ~2.6-fold in HuR KO MG. A similar pattern was observed after GL261CM treatment although the overall induction was considerably smaller than MMP-2 (~three-fold in control MG). Zymography was performed for both MMPs using CM from MG after treatment with GL261 CM (Figure 6C). There was a 2.5-fold attenuation of pro-MMP2 in HuR KO MG and no significant change with MMP-9. Taken together, these findings indicate that loss of HuR in MG had a mixed effect on chemokine and MMP molecular responses to secreted factors from glioma cells.
Figure 6. HuR KO alters the chemokine and MMP profiles of microglia in response to GL261 conditioned media.
Cultured primary microglial cells (MG) were treated with CM from GL261 cells for 24 h and then harvested. (A) Upper row: mRNA for different chemokines was assessed by qPCR. These mRNAs were also measured in TAMs isolated by FACS from three control and three HuR KO mice with GL261-induced tumors. Lower row: chemokines in MG CM were measured by ELISA. (B) qPCR and ELISA analysis of MG as in a. The right y axis for MMP2 qPCR is for CM-stimulated cells. Data points for a and b represent the mean ± SD of three independent tests, each assayed in duplicate. (C) Zymogram of MMP2 and MMP9 from CM of MG stimulated with GL261 CM. The graph below shows the mean densitometric values (± SEM) for the bands expressed as area under the curve (AUC). *P < 0.05; **P < 0.005; ***P < 0.0001.
HuR KO alters production of cytokines linked to M1- and M2 phenotypes
In MG treated with GL261 CM, we examined cytokines associated with M1- and M2-like phenotypes to determine the impact of HuR deletion (Figure 7). At baseline, mRNAs of markers classically associated with M1-like activation were low but increased by more than a 1000-fold for IL-1β and IL-6 after GL261 CM treatment and 15-fold for TNF-α. HuR KO significantly attenuated all three inductions, most strikingly with IL-6 (three-fold). COX-2 was suppressed with GL261 CM but was not altered in HuR KO MG. ELISA of MG CM showed an overall marked increase in these proteins, with a 50% suppression of IL-1 (α and β) and a ~30% reduction in IL-6 for HuR KO MG. No changes were seen with TNF-α. For M2-associated markers, there was a mixed effect. TGF-β1 was suppressed overall after GL261 CM treatment, but by 25% more in HuR KO MG. YM1 was attenuated in HuR KO MG by 50% after CM treatment. IL-10 and arginase 1 were increased (70% and 50% respectively). ELISA of MG CM paralleled the mRNA patterns showing an attenuation of TGF-β1 (37%) and a marked increase in IL-10 (250%). In addition to these markers, we looked at PD-L1 and VEGF, two factors that promote tumor progression. We observed suppression in HuR KO MG with both mRNAs after GL261 CM treatment, more pronounced for PD-L1 (50% reduction). This reduction is consistent with that observed in TAMs in vivo (Figure 3). Secreted VEGF was decreased by 50%. Taken together, the loss of HuR in MG led to suppression of cytokines classically linked to an M1-like phenotype and several tumor-promoting factors in response to secreted factors from GL261 cells, but had a mixed effect on markers classically linked to an M2-like phenotype.
Figure 7. HuR KO alters molecular profiles associated with microglial polarization and tumor progression after exposure to GL261 CM.
Cultured primary microglial cells were treated with GL261 CM for 24 h and assessed by qPCR and ELISA. Profiles associated with classic M1- and M2-like polarization are shown as well as factors associated with tumor progression (TP). Data points represent the mean ± SD of three independent tests. **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
Cytotoxic effector T cells are increased in the HuR KO tumor microenvironment
The findings that HuR KO mice had reduced intratumoral PMN-MDSCs, an increased M1 over M2 population of TAMs, elevated MHCII expression in TAMs and tumor cells, and an increase in MG production of the T-cell associated chemokine CXCL10 prompted us to determine if there were changes in tumor-infiltrating T cells that may contribute to the attenuated tumor phenotype. We assessed T cell populations by flow cytometry in control and HuR KO brains 14 days after glioma cell injection. The gating strategy used for T cell analysis is shown in Figure S5. With CD8+ T cells, we observed nearly a three-fold increase in the Granzyme B+ (GranB) subpopulation in HuR KO mice whereas there was no quantitative change in the overall CD8 population (Figure 8). On the other hand, there was nearly a four-fold increase in the percentage of CD4+ T cells in HuR KO mice. This increase could also be seen by immunostaining with an anti-CD4 antibody. Although intratumoral FoxP3+ CD4+Treg cells were increased in HuR KO mice, there was a four-fold increase in the subpopulation of FoxP3− CD4+ effector T cells and CD4+ effector T cells expressing GranB were nearly two-fold higher in HuR KO mice. The increase in IFNγ+ cells was even greater. Accordingly, multifunctional CD4+ effector T cells expressing both GranB and IFNγ in the tumors were increased in HuR KO mice. Taken together, there were increases in tumor-infiltrating cytotoxic effector T cells for both CD8+ and CD4+ T cell compartments which may underlie the enhanced antitumor responses in HuR KO mice.
Figure 8: Effector T cells are increased in HuR KO GB tumors.
Flow cytometry was performed for CD4 and CD8 populations on cells isolated from control and HuR KO brains of GB-bearing mice. The gating strategy is shown in Figure S5. Means and SDs are shown. Each data point is representative of one independent mouse. *P < 0.05; **P <0.01. Immunohistochemistry with an anti-CD4 antibody was performed on a representative tumor section from Control and HuR KO mice. Dotted line demarcates tumor (Tu) and brain (Br) parenchyma border. Scale bar, 50 μm.
DISCUSSION
In this report we show that HuR plays an important role in GB progression through its role in modulating the molecular signature of TAMs. Deletion of HuR in these cells altered the tumor microenvironment, leading to a significant attenuation of GB tumor growth and prolonged survival. In HuR KO tumors, TAMs were shifted toward an M1-like phenotype with higher levels of MHCII expression and there was a decrease in PMN-MDSCs. Cytotoxic effector T cell populations increased within the tumor microenvironment. Loss of HuR led to changes in production of key cytokines, chemokines, and other factors that drove these cellular shifts and reduced tumor growth. Our findings underscore the molecular plasticity of TAMs and how that can impact glioma progression.
TAMs represent a large component of GB and actively migrate to the tumor in response to tumor-secreted chemoattractants such as CSF-1, MCP-1, and GDNF (Broekman et al. 2018; Hambardzumyan et al. 2016). The majority of CD45+ cells in control and HuR KO mouse tumors were CD45hi consistent with being tissue Mɸ and characteristic for GB (Chen et al. 2017). In KO mice, HuR was absent from IBA1+ cells within the GB tumor (Figure 3) and diminished peripherally in Mɸ (Figure S2) indicating that both MG and Mɸ express Cre recombinase. This is consistent with prior observations that peripheral Mɸ expression of Cx3Cr1 increases after recruitment to the glioma tumor (Chen et al. 2017; Yona et al. 2013). The reduction of TAMs in HuR KO mice is likely multifactorial, reflecting a combination of intrinsic defects in migration/chemoattraction and altered cross talk with glioma cells. The potent chemoattractant effect of GB-produced soluble factors was highlighted in the transwell data which showed a ~60-fold increase in migrated MG with GL261 CM (Figure 5). The marked reduction in migration of HuR KO MG toward GL261 CM in the transwell and scratch assays (Figure S4) suggests a defect in chemoattraction and/or migration. This finding is consistent with our prior observations with siRNA-induced HuR knockdown in MG using ATP as a chemoattractant (Matsye et al. 2017). In that report, RNA sequencing analysis of lipopolysaccharide (LPS)-stimulated and HuR-silenced MG revealed significant attenuation of several mRNAs that regulate chemokine responses including CCR2, a key receptor for recruitment of MG/Mɸ to GB tumors and BAIAP2, an adapter protein important for filopodia formation and cell migration (Ahmed et al. 2010; Feng et al. 2015; Matsye et al. 2017). Thus, our finding of reduced MG migration may stem from impaired sensing of chemotactic factors or impairment of migration. A third possibility is reduced breakdown of the extracellular matrix (ECM) related to an attenuation of ECM modifiers. This could affect both MG and glioma cell migration (Figure 5). For example, MMP-2 was markedly attenuated in HuR KO MG. The 3’ UTR of MMP-2 contains AU-rich elements to which HuR can bind and regulate expression (Kong et al. 2017). The marked suppression of MMP-2 may have a larger impact on GB in vivo since MMP-2 plays a key role in breaking down the ECM to facilitate migration and invasion of glioma cells (Bjorklund and Koivunen 2005; Hambardzumyan et al. 2016). Interestingly, MMP-9, which increased at the RNA level with HuR knockout, also has AREs in the 3’ UTR and is positively regulated by HuR in other cell systems (Akool et al. 2003; Huwiler et al. 2003). These contrasting effects may reflect the pleiotropic functions of HuR or be an indirect effect of HuR KO on other gene targets (Abdelmohsen and Gorospe 2010). We previously showed that HuR positively regulates other ECM modulators in MG, including MMP12, ITGB3, Adamts1, and Thrombospondin 1 which also contribute to MG and/or glioma cell migration/invasion (Ferrer et al. 2018; Smolders et al. 2019).
The reduced TAMs observed in HuR KO mice may stem from altered cross-talk with glioma cells. TAMs produce secreted factors that modulate production of chemokines by glioma cells (Hambardzumyan et al. 2016). IL-1β, for example, which was robustly induced in MG by GL261 CM (Figure 7), stimulates glioma cells to produce CCL2 which in turn enhances recruitment of TAMs (Feng et al. 2015). The attenuation of IL-1β expression in HuR-deleted MG could disrupt this feed-forward loop and reduce TAM recruitment.
In addition to quantitative changes, there was a shift in TAM polarization toward an M1-like phenotype in HuR KO mice. GBs and other tumors have a mixture of M1 and M2-like TAMs, and often these cells have elements of both phenotypes (Broekman et al. 2018). Typically, M2-like TAMs accumulate in GB in response to secreted factors such as M-CSF, thereby promoting immunosuppression and tumor progression (Roesch et al. 2018). M1-like TAMs, however, are cytotoxic to tumors, so the shift we observed likely contributed to the attenuated tumor growth and prolonged survival in HuR KO mice (Chen and Hambardzumyan 2018; Mantovani et al. 2017). In fact, re-programming TAMs to an M1-like phenotype represents a therapeutic approach being pursued in the development of treatments for GB (Roesch et al. 2018). In addition to the increased numbers, HuR KO TAMs had higher overall MHCII expression which would facilitate their antigen presenting activity to CD4+ T cells and enhance anti-tumor responses (Accolla et al. 2014). This is particularly relevant to GB since glioma-associated TAMs generally have suppressed MHCII expression (Qian et al. 2018; Schartner et al. 2005). Interestingly, glioma cells in HuR KO tumors showed higher MHCII expression (Figure S3) which can facilitate antigen presentation and make them targets of CD4+ cytotoxic effector cells (Accolla et al. 2014; Takeuchi and Saito 2017; Thibodeau et al. 2012). Regardless of the source, MHCII molecules within tumors are key to triggering antitumor immune responses (Accolla et al. 2014). CD4+ cytotoxic T cells can develop from multiple subsets including Tregs (Takeuchi and Saito 2017). Through an MHCII-dependent manner, CD4+ cytotoxic T cells can directly reject a tumor (Quezada et al. 2010). Activated effector cells can also antagonize the immunosuppressive effects of Tregs which could mitigate the impact of an increased presence of these cells in HuR KO mice (Roychoudhuri et al. 2015). A feed forward loop is also potentially at play as an increase in IFNγ-producing effector cells would further promote M1-like polarization of TAMs. Anti-tumor T cell activation in HuR KO tumors was likely further potentiated by the downregulation of PD-L1 in TAMs (Figure 3). PD-L1 is a transmembrane protein, expressed by tumor cells and TAMs that engages with PD-1 receptors on T cells and inhibits their activation and anti-tumor immunity (Sun et al. 2018). While the relative contribution of PD-L1 (tumor cell versus TAM) as a source of immunosuppression varies among tumor types, TAMs can sustain PD-L1 expression and provide extended immunosuppression (Chen and Hambardzumyan 2018; Juneja et al. 2017; Noguchi et al. 2017). PD-L1 is detected in the majority of human GB tumors, including TAMs, and correlates with tumor grade, markers of proliferation and angiogenesis, and worse prognosis (Wöhrer et al. 2014; Xue et al. 2017a; Xue et al. 2017b). Interestingly, TAMs in HuR KO mice expressed much lower levels of PD-L1, and in vitro testing of MG indicated that PD-L1 induction by GL261 CM was markedly attenuated by HuR deletion (Figure 7). The PD-L1 transcript harbors AU-rich sequences in the 3 ‘UTR which are putative HuR binding motifs (Brennan and Steitz 2001). A recent report demonstrated that oncogenic RAS signaling upregulates PD-L1 expression by blocking the RNA destabilizer, TTP, which binds to similar motifs in the 3’ UTR (Coelho et al. 2017; Fu and Blackshear 2017). A positive regulatory role of HuR is supported by our recent finding that PD-L1 mRNA was attenuated when glioma xenolines were treated with the small molecule inhibitor of HuR, MS-444 (Wang et al. 2019).
The altered immune cell profiles in HuR KO mice may relate to other changes in the molecular signature of TAMs. There was a robust induction of CXCL10 (Figure 6) which can boost recruitment and accumulation of CD4+ and CD8+ cells and polarize them to enhance anti-tumor activity (e.g. through GranB and IFNϒ production) (Karin and Razon 2018). CXCL10 can also directly inhibit glioma cell proliferation and promote apoptosis (Wang et al. 2018). On the other hand, CCL5 was also increased, and this cytokine has been linked to tumor progression in a number of cancers including glioma (Aldinucci and Casagrande 2018; Pan et al. 2017). The loss of HuR in MG had a mixed effect on other activation markers that do not typically align with a pro-inflammatory or anti-glioma phenotype. Markers associated with alternative activation (M2-like), for example, were differentially affected by HuR deletion. IL-10 and AG1 showed significant upregulation whereas YM1 and TGF-β1 were downregulated (Figure 7). On the other hand, classic markers associated with an M1-like phenotype, including IL-1 (α and β), IL-6 and TNF-α, were all suppressed with HuR deletion. These cytokines have been linked to glioma progression and their suppression may have contributed to the attenuation of proliferation and overall tumor growth in HuR KO mice (Albulescu et al. 2013; Han et al. 2015; Wang et al. 2012; Yeung et al. 2013). Moreover, IL-6 and TGF-β produced by MG/Mɸ promote glioma growth by supporting GSCs (Dzaye et al. 2016; Han et al. 2015; Wang et al. 2012). VEGF, although not a classic activation marker, was suppressed in HuR KO MG in response to glioma cell CM. TAMs are a source of VEGF in the tumor microenvironment in response to signaling from glioma cells and its attenuation with HuR deletion would further contribute to reduced tumor growth and proliferation in HuR KO mice (Li and Graeber 2012). Interestingly, COX-2 mRNA, which has been linked to a pro-inflammatory state in macrophages, was attenuated after treatment with glioma CM, but was not further affected by HuR deletion (Ellert-Miklaszewska et al. 2013; Parisi et al. 2018). We previously showed it to be positively regulated by HuR in colon cancer, again indicating that the impact of HuR on specific target expression is cell- and likely stimulus-dependent (Dixon et al. 2001). The upregulation of IL-10, on the other hand, was unexpected based on the presence of AREs in its 3’ UTR and the primary role of HuR as a positive regulator (Abdelmohsen and Gorospe 2010; Stoecklin et al. 2008). IL-10 has pleiotropic effects in GB and other cancers, either promoting or inhibiting tumor progression, depending upon immune cell composition within the tumor and cytokines in the milieu (Mannino et al. 2015; Perng and Lim 2015). The mixed effect of HuR on regulating markers associated with M1- and M2-like phenotypes underscores the notion that MG/Mɸ activation is multi-dimensional and cannot be categorized as a linear spectrum between these two polarization states (Ransohoff 2016). The concomitant upregulation of MHCII in HuR KO TAMs and reduced expression of “classic” M1 markers underscores this concept. In malignant glioma, many secreted cytokines associated with a classic M1-like phenotype actually promote tumor progression (e.g. IL-1, IL-6, and TNF-α) whereas increased MHCII, also associated with an M1-like phenotype, promotes anti-tumor immune cell activity (Accolla et al. 2014; Hambardzumyan et al. 2016). While we used CM from glioma cells to induce MG for these experiments, a potential caveat is that in vitro conditions do not fully recapitulate the tumor microenvironment. The concordance between molecular responses in TAMs and cultured MG (Figure 6), however, mitigates against this possibility.
Another major shift in immune cell profiles within the HuR KO tumors that could enhance anti-tumor activity was the reduction of PMN-MDSCs. Neutrophilia is a common feature of patients with GB and intra-tumoral infiltration of PMNs correlates with glioma grade (Massara et al. 2017). PMN-MDSCs promote tumor progression primarily through immunosuppression of T cell function and their immunodepletion suppresses glioma growth (Fujita et al. 2011; Gabrilovich 2017). Several possibilities emerge from our data to explain the reduction in PMN-MDSCs. First, CXCL1 and CXCL2, which are major cancer-related chemoattractants for these cells, were significantly attenuated in HuR KO TAMs in vivo and HuR KO MG in vitro (Acharyya et al. 2012; Massara et al. 2017). In our prior work, we saw a marked decline in chemotaxis of neutrophils in vitro using conditioned media from HuR-silenced MG stimulated with LPS (Matsye et al. 2017). It is also possible that reduced HuR expression in the PMN population (Figure S2) affected their mobility or chemotaxis. There was no reduction in the M-MDSC population, however, and monocytes also had a similar decrease in HuR KO mice. Finally, it is possible that the reduced PMN infiltration may be related to some other chemoattractant altered in HuR KO TAMs or by glioma cells as a result of an altered tumor microenvironment.
In conclusion, HuR knockout in TAMs had a significant influence on the tumor microenvironment in GB, leading to tumor suppression and prolonged survival. HuR deletion altered the molecular signature of TAMs which promoted an anti-tumor phenotype through changes in infiltrating immune cells and their activity, and/or loss of trophic support. We have previously shown that glioma-derived HuR is integral to tumor cell survival and progression (Filippova et al. 2017; Filippova et al. 2011; Wang et al. 2019), and the findings in this report further underscore its importance in glioma progression and potential as a therapeutic target in this devastating disease.
Supplementary Material
Main Points.
HuR deletion in tumor-associated microglia/macrophages (TAMS) reduces malignant glioma growth and prolongs survival.
HuR-deleted TAMs are reduced in glioma tumors and develop an M1-like phenotype and an alteration of cytokine and chemokine profiles.
HuR-deleted TAMs alter the glioma microenvironment with an increase in Infiltrating cytotoxic CD4+ and CD8+ T cells and a decrease in polymorphonuclear myeloid derived suppressor cells (PMN-MDSC).
Acknowledgements
This work was supported by the Department of Veterans Affairs under a Merit Review BX001706 (PHK), DOD W81XWH-18-1-0315 (JL), and NIH RO1NS104339 (ABH). We wish to thank Marion Spell and the UAB Center for AIDS Research flow cytometry core for their assistance with FACS analysis, and Terry Lewis, PhD, and the UAB Neuroscience Molecular Detection Core Facility (P30 NS47466) for assistance with immunohistochemistry. We also thank Dr. Jason Warram of the UAB Animal Imaging Core for his assistance with luminescence imaging.
Footnotes
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abdelmohsen K, Gorospe M. 2010. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 1:214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Yuki K, Kim HH, Gorospe M. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem 389:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accolla RS, Lombardo L, Abdallah R, Raval G, Forlani G, Tosi G. 2014. Boosting the MHC Class II-Restricted Tumor Antigen Presentation to CD4+ T Helper Cells: A Critical Issue for Triggering Protective Immunity and Re-Orienting the Tumor Microenvironment Toward an Anti-Tumor State. Frontiers in oncology 4:32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris Patrick G, Manova-Todorova K, Leversha M, Hogg N, Seshan Venkatraman E and others. 2012. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Cell 150:165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Goh WI, Bu W. 2010. I-BAR domains, IRSp53 and filopodium formation. Seminars in Cell & Developmental Biology 21:350–356. [DOI] [PubMed] [Google Scholar]
- Akool E-S, Kleinert H, Hamada FMA, Abdelwahab MH, Forstermann U, Pfeilschifter J, Eberhardt W. 2003. Nitric Oxide Increases the Decay of Matrix Metalloproteinase 9 mRNA by Inhibiting the Expression of mRNA-Stabilizing Factor HuR. Mol Cell Biol 23:4901–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albulescu R, Codrici E, Popescu ID, Mihai S, Necula LG, Petrescu D, Teodoru M, Tanase CP. 2013. Cytokine Patterns in Brain Tumour Progression. Mediators of Inflammation 2013:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldinucci D, Casagrande N. 2018. Inhibition of the CCL5/CCR5 Axis against the Progression of Gastric Cancer. International journal of molecular sciences 19:1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9:353–9. [DOI] [PubMed] [Google Scholar]
- Anderson P. 2010. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10:24–35. [DOI] [PubMed] [Google Scholar]
- Bettinger I, Thanos S, Paulus W. 2002. Microglia promote glioma migration. Acta Neuropathol 103:351–5. [DOI] [PubMed] [Google Scholar]
- Bjorklund M, Koivunen E. 2005. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta 1755:37–69. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell Mol Life Sci 58:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekman ML, Maas SLN, Abels ER, Mempel TR, Krichevsky AM, Breakefield XO. 2018. Multidimensional communication in the microenvirons of glioblastoma. Nat Rev Neurol 14:482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Feng X, Herting CJ, Garcia VA, Nie K, Pong WW, Rasmussen R, Dwivedi B, Seby S, Wolf SA and others 2017. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res 77:2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hambardzumyan D. 2018. Immune Microenvironment in Glioblastoma Subtypes. Front Immunol 9:1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K and others 2017. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 47:1083–1099 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A, Gromeier M, Herndon JE, Beaubier N, Bolognesi DP, Friedman AH, Friedman HS, McSherry F, Muscat AM, Nair S and others. 2018. Recurrent Glioblastoma Treated with Recombinant Poliovirus. New England Journal of Medicine 379:150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest 108:1657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzaye OD, Hu F, Derkow K, Haage V, Euskirchen P, Harms C, Lehnardt S, Synowitz M, Wolf SA, Kettenmann H. 2016. Glioma Stem Cells but Not Bulk Glioma Cells Upregulate IL-6 Secretion in Microglia/Brain Macrophages via Toll-like Receptor 4 Signaling. J Neuropathol Exp Neurol 75:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellert-Miklaszewska A, Dabrowski M, Lipko M, Sliwa M, Maleszewska M, Kaminska B. 2013. Molecular definition of the pro-tumorigenic phenotype of glioma-activated microglia. Glia 61:1178–1190. [DOI] [PubMed] [Google Scholar]
- Feng X, Szulzewsky F, Yerevanian A, Chen Z, Heinzmann D, Rasmussen RD, Alvarez-Garcia V, Kim Y, Wang B, Tamagno I and others 2015. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget 6:15077–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer VP, Moura Neto V, Mentlein R. 2018. Glioma infiltration and extracellular matrix: key players and modulators. Glia 66:1542–1565. [DOI] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. 2003. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol 23:4991–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova N, Yang X, Ananthan S, Sorochinsky A, Hackney JR, Gentry Z, Bae S, King P, Nabors LB. 2017. Hu antigen R (HuR) multimerization contributes to glioma disease progression. J Biol Chem 292:16999–17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. 2011. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res 9:648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Blackshear PJ. 2017. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 17:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Kohanbash G, Fellows-Mayle W, Hamilton RL, Komohara Y, Decker SA, Ohlfest JR, Okada H. 2011. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res 71:2664–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI. 2017. Myeloid-Derived Suppressor Cells. Cancer Immunol Res 5:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galban S, Kuwano Y, Pullmann R, Martindale, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO and others. 2008. RNA-Binding Proteins HuR and PTB Promote the Translation of Hypoxia-Inducible Factor-1α. Mol Cell Biol 28:93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Gutmann DH, Kettenmann H. 2016. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 19:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Alvarez-Breckenridge CA, Wang QE, Yu J. 2015. TGF-beta signaling and its targeting for glioma treatment. Am J Cancer Res 5:945–55. [PMC free article] [PubMed] [Google Scholar]
- Holtman IR, Skola D, Glass CK. 2017. Transcriptional control of microglia phenotypes in health and disease. J Clin Invest 127:3220–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwiler A, Akool el S, Aschrafi A, Hamada FM, Pfeilschifter J, Eberhardt W. 2003. ATP potentiates interleukin-1 beta-induced MMP-9 expression in mesangial cells via recruitment of the ELAV protein HuR. J Biol Chem 278:51758–69. [DOI] [PubMed] [Google Scholar]
- Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH. 2017. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. The Journal of Experimental Medicine 214:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin N, Razon H. 2018. Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine 109:24–28. [DOI] [PubMed] [Google Scholar]
- Kong J, Zhang Y, Liu S, Li H, Liu S, Wang J, Qin X, Jiang X, Yang J, Zhang C and others. 2017. Melatonin attenuates angiotensin II-induced abdominal aortic aneurysm through the down-regulation of matrix metalloproteinases. Oncotarget 8:14283–14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M, Gopfert U, Siewe B, Hengst L. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5’UTR. Genes Dev 16:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, Gordon S. 1990. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39:151–70. [DOI] [PubMed] [Google Scholar]
- Li W, Graeber MB. 2012. The molecular profile of microglia under the influence of glioma. Neuro-Oncology 14:958–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–8. [DOI] [PubMed] [Google Scholar]
- Ma W-J, Cheng S, Campbell C, Wright A, Furneaux H. 1996. Cloning and Characterization of HuR, a Ubiquitously Expressed Elav-like Protein. J Biol Chem 271:8144–8151. [DOI] [PubMed] [Google Scholar]
- Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. 2015. The paradoxical role of IL-10 in immunity and cancer. Cancer Letters 367:103–107. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. 2017. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massara M, Persico P, Bonavita O, Mollica Poeta V, Locati M, Simonelli M, Bonecchi R. 2017. Neutrophils in Gliomas. Frontiers in Immunology 8:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsye P, Zheng L, Si Y, Kim S, Luo W, Crossman DK, Bratcher PE, King PH. 2017. HuR promotes the molecular signature and phenotype of activated microglia: Implications for amyotrophic lateral sclerosis and other neurodegenerative diseases. Glia 65:945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner N-C, Filipowicz W. 2011. Properties of the Regulatory RNA-Binding Protein HuR and its Role in Controlling miRNA Repression In: Großhans H, editor: Springer New York. p 106–123. [DOI] [PubMed] [Google Scholar]
- Meng Z, King PH, Nabors LB, Jackson NL, Chen C-Y, Emanuel PD, Blume SW. 2005. The ELAV RNA-stability factor HuR binds the 5’-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucl Acids Res 33:2962–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabors LB, Gillespie GY, Harkins L, King PH. 2001. HuR, an RNA stability factor, is expressed in malignant brain tumors and binds to adenine and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 61:2154–2161. [PubMed] [Google Scholar]
- Nabors LB, Suswam E, Huang Y, Yang X, Johnson MJ, King PH. 2003. Tumor necrosis factor alpha induces angiogenic factor up-regulation in malignant glioma cells: a role for RNA stabilization and HuR. Cancer Res 63:4181–7. [PubMed] [Google Scholar]
- Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, Selby MJ, Graziano RF, Mardis ER, Korman AJ and others. 2017. Temporally Distinct PD-L1 Expression by Tumor and Host Cells Contributes to Immune Escape. Cancer Immunology Research 5:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Smithson LJ, Ma Y, Hambardzumyan D, Gutmann DH. 2017. Ccl5 establishes an autocrine high-grade glioma growth regulatory circuit critical for mesenchymal glioblastoma survival. Oncotarget 8:32977–32989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi L, Gini E, Baci D, Tremolati M, Fanuli M, Bassani B, Farronato G, Bruno A, Mortara L. 2018. Macrophage Polarization in Chronic Inflammatory Diseases: Killers or Builders? Journal of Immunology Research 2018:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng P, Lim M. 2015. Immunosuppressive Mechanisms of Malignant Gliomas: Parallels at Non-CNS Sites. Frontiers in oncology 5:153–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. 2017. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proceedings of the National Academy of Sciences 114:1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Luo F, Yang J, Liu J, Liu R, Wang L, Wang C, Deng Y, Lu Z, Wang Y and others. 2018. TLR2 Promotes Glioma Immune Evasion by Downregulating MHC Class II Molecules in Microglia. Cancer Immunol Res 6:1220–1233. [DOI] [PubMed] [Google Scholar]
- Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA and others. 2010. Tumor-reactive CD4+ T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. The Journal of Experimental Medicine 207:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff RM. 2016. A polarizing question: do M1 and M2 microglia exist? Nat Neurosci 19:987–91. [DOI] [PubMed] [Google Scholar]
- Roesch S, Rapp C, Dettling S, Herold-Mende C. 2018. When Immune Cells Turn Bad-Tumor-Associated Microglia/Macrophages in Glioma. International journal of molecular sciences 19:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhuri R, Eil RL, Restifo NP. 2015. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol 33:101–11. [DOI] [PubMed] [Google Scholar]
- Schartner JM, Hagar AR, Van Handel M, Zhang L, Nadkarni N, Badie B. 2005. Impaired capacity for upregulation of MHC class II in tumor-associated microglia. Glia 51:279–85. [DOI] [PubMed] [Google Scholar]
- Smolders SM, Kessels S, Vangansewinkel T, Rigo JM, Legendre P, Brone B. 2019. Microglia: Brain cells on the move. Prog Neurobiol:101612. [DOI] [PubMed] [Google Scholar]
- Sorensen MD, Dahlrot RH, Boldt HB, Hansen S, Kristensen BW. 2018. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol 44:185–206. [DOI] [PubMed] [Google Scholar]
- Srikantan S, Gorospe M. 2012. HuR function in disease. Front Biosci (Landmark Ed) 17:189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. 2008. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem:M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Mezzadra R, Schumacher TN. 2018. Regulation and Function of the PD-L1 Checkpoint. Immunity 48:434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Saito T. 2017. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front Immunol 8:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau J, Bourgeois-Daigneault MC, Lapointe R. 2012. Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology 1:908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bent MJ, Klein M, Smits M, Reijneveld JC, French PJ, Clement P, de Vos FYF, Wick A, Mulholland PJ, Taphoorn MJB and others. 2018. Bevacizumab and temozolomide in patients with first recurrence of WHO grade II and III glioma, without 1p/19q co-deletion (TAVAREC): a randomised controlled phase 2 EORTC trial. The Lancet Oncology 19:1170–1179. [DOI] [PubMed] [Google Scholar]
- Walker K, Hjelmeland A. 2014. Method for Efficient Transduction of Cancer Stem Cells. J Cancer Stem Cell Res 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hjelmeland AB, Nabors LB, King PH. 2019. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu Z, Balivada S, Shrestha T, Bossmann S, Pyle M, Pappan L, Shi J, Troyer D. 2012. Interleukin-1beta and transforming growth factor-beta cooperate to induce neurosphere formation and increase tumorigenicity of adherent LN-229 glioma cells. Stem Cell Res Ther 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Peng X, Zhang J, Wang Z, Meng J, Cen B, Ji A, He S. 2018. LncRNA-135528 inhibits tumor progression by up-regulating CXCL10 through the JAK/STAT pathway. Apoptosis 23:651–666. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet 20:491–7. [DOI] [PubMed] [Google Scholar]
- Wöhrer A, Berghoff AS, Hainfellner JA, Kiesel B, Widhalm G, Dieckmann K, Marosi C, Filipits M, Preusser M, Wick W. 2014. Analysis of PD1 and PD-L1 Expression in Glioblastoma. Neuro-Oncology 16:ii12–ii12. [Google Scholar]
- Xue S, Hu M, Li P, Ma J, Xie L, Teng F, Zhu Y, Fan B, Mu D, Yu J. 2017a. Relationship between expression of PD-L1 and tumor angiogenesis, proliferation, and invasion in glioma. Oncotarget 8:49702–49712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Song G, Yu J. 2017b. The prognostic significance of PD-L1 expression in patients with glioma: A meta-analysis. Sci Rep 7:4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung YT, McDonald KL, Grewal T, Munoz L. 2013. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol 168:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A and others. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.