Abstract
Rare sugar D-allulose as a substitute sweetener is produced through the isomerization of D-fructose by D-tagatose 3-epimerases (DTEases) or D-allulose 3-epimerases (DAEases). D-Allulose is a kind of low energy monosaccharide sugar naturally existing in some fruits in very small quantities. D-Allulose not only possesses high value as a food ingredient and dietary supplement, but also exhibits a variety of physiological functions serving as improving insulin resistance, antioxidant enhancement, and hypoglycemic controls, and so forth. Thus, D-allulose has an important development value as an alternative to high-energy sugars. This review provided a systematic analysis of D-allulose characters, application, enzymatic characteristics and molecular modification, engineered strain construction, and processing technologies. The existing problems and its proposed solutions for D-allulose production are also discussed. More importantly, a green and recycling process technology for D-allulose production is proposed for low waste formation, low energy consumption, and high sugar yield.
Keywords: D-allulose, D-allulose 3-epimerase, engineering strain, biological catalysis, D-tagatose 3-epimerase
D-Allulose Physic-Chemical Characters and Application
D-Allulose has a molecular formula of C6H12O6 and a molecular weight of 180.16. The structural difference between D-allulose and D-fructose is located at the paired C2–C3 atoms (Chan et al., 2012). The physical characteristics of D-allulose include an appearance as white crystalline substance, odorless, low hygroscopicity, high solubility in water, and melting point of 96°C (Zhang et al., 2016a). D-Allulose is a kind of low energy monosaccharide sugar and present in very small quantities in natural products. And it is approximately 70% relative sweetness to sucrose with 0.2 Kcal/g that was 95% calorie reduction compared to sucrose and comprises only 0.3% of energy deposit in the body of animals (Chung et al., 2012).
The United States Food and Drug Administration (FDA) has confirmed the safety of D-allulose as generally regarded as safe (GRAS) food (Zhang et al., 2017). D-Allulose contains almost the same taste, performance, and texture with other sugars but it is mostly used by health concisions people globally for its very low energy. D-Allulose can be applied in food, pharmaceutical preparations, and dietary supplements (Granström et al., 2004). D-Allulose containing formulations were less prone to retrogradation in a starch-based composite gel matrix (Ilhan et al., 2020). Manufacturers widely add D-allulose in their food products in combination with other sweeteners. D-Allulose is globally applied in beverages (soft drinks and health drinks), savory dishes (soups, sauces, toppings, salads, and pickles), pharmaceuticals as a gelling agent, bakery products, ice cream, yogurt, and other low-calorie foods. Besides, D-allulose is also used as a thickening and stabilizing agent in bread, biscuits, rye cakes, and meat dishes.
D-Allulose exhibits almost zero calories with a low degree of energy density (Matsuo et al., 2002). D-Allulose can’t raise the blood sugar levels in diabetic patients and hence has been used to be a unique metabolic regulator of fat and glucose metabolism (Matsuo and Izumori, 2006, 2009; Hossain et al., 2011; Iida et al., 2013). Besides, D-allulose has also demonstrated a variety of activities, such as inhibitory activity toward intestinal digestive enzymes (Maeng et al., 2019), antioxidant enhancement (Sun et al., 2006), competitive transport through the intestinal mucosa with glucose (Kishida et al., 2019), enhancing glucokinase translocation of from the hepatic nucleus to cytoplasm (Shintani et al., 2017), strong anti-hyperlipidemic and anti-hyperglycemic effects (Hossain et al., 2015b), anti-inflammatory actions on adipocytes (Hossain et al., 2015a), preventing obesity and type 2 diabetes mellitus (Hossain et al., 2015b), and inhibiting trichomonad development (Harada et al., 2012). D-Allulose could be used as a health food or medicine for consumers with special needs. Thus, D-allulose has a potential application value as a substitute for glucose. Many health concisions individuals are shifting their preference toward consuming healthy food products. The application range of D-allulose is much broader than that of artificial non-sugar sweeteners (Yang et al., 2019). Hence, the global D-allulose market will observe robust growth over the forecast period.
D-Allulose In Vivo Metabolism
D-Allulose belongs to a real and natural sugar rather than an artificial sweetener (Ran et al., 2019). After feeding, D-allulose hasn’t the same metabolic pathway as D-fructose in rats (Kishida et al., 2019). D-Allulose was highly stable in simulated gastric fluid, in fasted state simulated intestinal fluid and in human and rat hepatocytes, whereas D-fructose was rapidly metabolized (Maeng et al., 2019). Intestinal D-allulose transport was mediated by glucose transporter type 5 at lower affinity relative to D-fructose (Kishida et al., 2019). D-Allulose is not involved in glucose related metabolism (Iwasaki et al., 2018). Thus, D-allulose is not metabolized in an animal liver, and thus it is impossible to contribute to hepatic energy production (Maeng et al., 2019).
During the in vivo metabolism of D-allulose, a small quantity of D-allulose is decomposed into short-chain fatty acids under the action of cecal microbes (Kimoto-Nira et al., 2017). Most of D-allulose is excreted out of the body in the two forms of urine and feces after intake by oral administration or intravenous injection approaches. In the form of urine transported through the blood circulation, approximately 98% of D-allulose was excreted after intravenous injection (Whistler et al., 1974). D-Allulose is partly absorbed in the small intestine when orally administered in rats, and the remaining substance enters the blood circulation and is finally excreted out of the body via urine containing D-allulose (Iwasaki et al., 2018). Besides, in the form of feces, considerable D-allulose is excreted after the treatment of cecal microbes in the intestinum crassum of animals (Matsuo et al., 2003). In the in vivo metabolism of D-allulose, mice and dogs were previously used in animal experiments (Han et al., 2016; Nishii et al., 2016; Do et al., 2019). Human beings have a rich and developed sweat gland system that can discharge the liquid containing D-allulose in the body.
D-Allulose can’t be metabolized and in vivo converted into energy, but it plays an important role in the physiological activity of animals. D-Allulose was proved to have a significant anti-hyperglycemic effect and no abnormal clinical problems according to the result of the clinical study (Hayashi et al., 2010). Even the feeding amount reached 2 g D-allulose per Kg rat’s weight, no observed-adverse effect was shown based on D-allulose-associated reproductive toxicity (Kim et al., 2019). With dogs as the tested animals, all the dogs had a good appetite when 4 g/kg of D-allulose was administered, thus single oral dose of D-allulose didn’t show severe toxicity (Nishii et al., 2016). In addition, D-allulose could effectively decrease the activity of hepatic lipogenic enzymes and the levels of blood lipid and blood sugar (Chu et al., 2003; Kimura et al., 2005; Matsuo et al., 2015). The decreased hepatic lipogenic enzyme reduces the tissue weight of liver and abdominal adipose and inhibits fat accumulation (Chung et al., 2012). D-Allulose regulates the blood lipid by changing the activity of lipid-regulating enzymes (Han et al., 2016). Further, D-allulose could alter cholesterol metabolism by reducing serum PCSK9 levels (Kanasaki et al., 2019). All in all, the biological activities of D-allulose mainly focused on enhancing insulin tolerance (Shintani et al., 2017; Iwasaki et al., 2018), inhibiting postprandial blood glucose rise (Shintani et al., 2017), reducing abdominal fat accumulation (Do et al., 2019), and preventing diabetes (Fukada et al., 2010).
The Enzyme for D-Allulose Production
Sources of DTEase Family Enzymes
In 1994, Japanese scientist Ken Izumori discovered that D-tagatose 3-epimerase has the capability of converting D-fructose to D-allulose (Itoh et al., 1994). D-Tagatose-3-epimerases (DTEases) family is a kind of enzymes that catalyze the isomerization of C3 position of ketose monosaccharides and is also the core enzyme in the production of rare sugars (Jia et al., 2014). Normally, D-allulose is produced through the isomerization of D-fructose under the catalysis of DTEase family enzymes. DTEase family enzymes include DTEases (Yoshida et al., 2007; Zhang et al., 2009b), D-psicose 3-epimerases (DAEase) (Kim H.J. et al., 2006; Mu et al., 2013; Zhang et al., 2013a), and ketose 3-epimerase (Yoshida et al., 2019). All of these enzymes have the same characteristics that catalyze the conversion of D-fructose to D-allulose. The enzyme activity is generally calculated by measuring the amount of D-allulose with D-fructose as the catalysis substrate. One unit is defined as the amount of enzyme that catalyzes the production of 1 μmoL D-allulose per min at the optimal conditions (Zhu et al., 2019a).
DTEase from Pseudomonas cichorii ST-24 was initially identified in 1993 through hexulose catalytic epimerization (Ishida et al., 1997b). The genes coding for DTEase enzyme have been consecutively confirmed and isolated from Agrobacterium tumefaciens (Kim H.J. et al., 2006), Rhodobacter sphaeroides SK011 (Zhang et al., 2008), Clostridium cellulolyticum H10 (Mu et al., 2011), Ruminococcus sp. (Zhu et al., 2012), Thermotoga maritima (Sakuraba et al., 2009), and Clostridium scindens (Mu et al., 2013). The catalysis approaches of free enzyme and immobilization are used to convert D-fructose into D-allulose. Since DTEases were first discovered by Izumori and used to successfully catalyze the production of the rare sugars, the preparation of functional rare sugars by isomerases has become a research hotspot (Izumori and Okaya, 1993). At present, DTEases from only six strains have been identified as members of DTEases family enzymes. These strains contained Pseudomonas cichorii (Izumori and Okaya, 1993), Agrobacterium tumefaciens (Kim H.J. et al., 2006), Rhodobacter sphaeroides (Zhang et al., 2009b), Clostridium cellulolyticum (Mu et al., 2011), Ruminococcus sp. (Zhu et al., 2012), and Desmospora sp. (Zhang et al., 2013b). More DTEases sources and more efficient DTEases activity need to be further explored (Oh et al., 2007; Zhang et al., 2009a; Mu et al., 2011; Jia et al., 2014; Park et al., 2016a, b).
Enzymatic Properties
DTEase family enzymes possess a highly conserved activity center and key amino acid residues with similar features, such as optimum temperature of 40°C–70°C, optimal pH of 7.5–9.0, molecular weights of 32–34 kDa for monomer, 64–68 kDa for dimer, 128–139 kDa for tetramer (Kim H.J. et al., 2006; Zhang et al., 2008, 2009b, 2013a; Mu et al., 2013). The half-lives of DTEase family enzymes vary from 15 min (Mu et al., 2013) to 408 min (Mu et al., 2011). The optimal equilibrium ratios (D-allulose:D-fructose) from A. tumefaciens DAEase and P. cichorii DTEase are 33:67 and 20:80, respectively (Ishida et al., 1997a; Kim H.J. et al., 2006) (Table 1).
TABLE 1.
Properties of DTEase family enzymes.
| Optimum | Molecular | Optimal | Metal | Half-lifea | Equilibrium | kcat/Kmc | |
| Sources of DTEase family enzymes | temperature | weights | pH | ions | (min) | ratiob | (min–1mM–1) |
| Paenibacillus senegalensis (Yang et al., 2019) | 55°C | 33.5 | 8.0 | Mn2+ | – | 30:70 | 39 |
| Caballeronia fortuita (Li et al., 2019) | 65°C | 33 | 7.5 | Co2+ | 63 | 37.5:62.5 | – |
| Sinorhizobium sp. (Zhu et al., 2019b) | 50°C | – | 8.0 | Mn2+ | – | – | – |
| Staphylococcus aureus (Zhu et al., 2019a) | 70°C | 134.13 (tetramer) | 8.0 | Mg2+ | – | – | – |
| Grobacterium sp. ATCC 31749 (Tseng et al., 2018) | 55–60°C | 32 | 7.5–8.0 | Co2+ | 267 (55°C) | 30:70 | – |
| Arthrobacter globiformis M30 (Yoshihara et al., 2017) | 70°C | 128 (tetramer) | 7.0–8.0 | Mn2+ | – | – | – |
| Flavonifractor plautii (Park et al., 2016a) | 65°C | 33 | 7.0 | Co2+ | 130 | – | 156 |
| Bacillus subtilis (He et al., 2016) | 55°C | – | 7.5–8.0 | – | 170 | – | – |
| T. primitia ZAS-1 (Zhang et al., 2016b) | 70°C | – | 8.0 | Co2+ | ∼30 | 28:72(70°C) | 144 |
| Dorea sp. CAG317 (Zhang et al., 2015, 2018) | 70°C | 33 | 6.0 | Co2+ | ∼30 | 30:70(70°C) | 412 |
| C. bolteae (Jia et al., 2014) | 55°C | 139 (tetramer) | 7.0 | Co2+ | 156 | 32:68(60°C) | 107 |
| Clostridium sp. (Mu et al., 2013) | 65°C | 130 (tetramer) | 8.0 | Co2+ | 15 | 28:72 | 141.4 |
| C. scindens 35.704 (Zhang et al., 2013a) | 60°C | 132 (tetramer) | 7.5 | Mn2+ | 108 | 28:72(50°C) | 64.5 |
| Ruminococcus sp. (Zhu et al., 2012) | 60°C | 132 (tetramer) | 7.5–8.0 | Mn2+ | 96 | 28:72 | 51 |
| Clostridium cellulolyticum (Mu et al., 2011) | 55°C | 132 (tetramer) | 8.0 | Co2+ | 408 | 32:68 | 186.4 |
| Rhodobacter sphaeroides (Zhang et al., 2008) | 60°C | 64 (dimer) | 9.0 | Mn2+ | – | 23:77 | – |
| Agrobacterium Tumefaciens (Kim H.J. et al., 2006) | 50°C | 132 (tetramer) | 8.0 | Mn2+ | 64 | 33:67 | 205 |
| Pseudomonas cichorii (Itoh et al., 1994; Ishida et al., 1997a) | 40°C, 60°C | 68 (dimer) | 7.5 | – | – | 20:80 | – |
| Bacillus subtilis (Patel et al., 2019) | 60°C, 70°C | 6–11 | Co2+ Mn2+ | 28:72 |
a,b,cRepresent the optimum parameters of temperature, substrate, and pH, respectively. – Means not evaluated.
Given their importance for D-allulose biotransformation, the kinetic parameters reflect the substrate affinity and catalytic efficiency of the enzyme. DAEase has the highest catalytic efficiency among the DTEase family enzymes. The kinetic parameters of C. cellulolyticum and A. tumefaciens DAEases were 186.4 and 205 mM min–1, respectively, which are higher than those from Clostridium sp. DTEase (141.4 mM min–1) and Ruminococcus sp. (51 mM min–1) (Mu et al., 2011, 2013; Zhu et al., 2012).
Metal ions play a pivotal role in the conversion of D-fructose into D-allulose by anchoring the bound of D-fructose. Kim et al. (2010) analyzed DAEase from A. tumefaciens for the conversion of D-fructose to D-allulose. The distance between A. tumefaciens DAEase residue Glu150 and Mn2+ is critical to the activity of DAEase (Kim et al., 2010). In addition, Asp183 and His209 residues bounded by metal ions are involved in efficient substrate binding. C. cellulolyticum H10 DAEase Glu150 and Glu244 perform the epimerization reaction of catalysis at the C3 position of D-fructose (Chan et al., 2012). The DTEase family enzymes have distinctly different degrees of dependency on metal ions of Mn2+, Co2+, and Mg2+ (Tseng et al., 2018; Yang et al., 2019; Zhu et al., 2019a). A. tumefaciens DAEase and R. sphaeroides DTEase activity could be remarkably enhanced when a metal ion Mn2+ is involved in the catalytic system (Kim H.J. et al., 2006; Zhang et al., 2008). P. cichorii DTEase does not require metal ions as a cofactor (Itoh et al., 1994; Ishida et al., 1997a). However, C. cellulolyticum DAEase and C. scindens DTEase are strictly Co2+ and Mn2+ metal-dependent (Mu et al., 2011; Zhang et al., 2013a). These two enzymes are completely inactivated in the absence of metal ions.
Structural Features
The thermostability of recombinant enzymes has been ascribed to the high number of hydrophobic interactions and a more rigid configuration of their amino acid sequences (Polizzi et al., 2007). The DAEases from A. tumefaciens and C. cellulolyticum H10 exhibit the tightest tetramer formation among the DTEase family enzymes. In the tetramer of DAEase, the amino acid residues form 34 hydrogen bonds between the two subunits. The high catalytic activity of DAEase is attributed to the wide interface solvent-accessible areas, which are caused by the extensive interactions between the two dimers among the DTEase family enzymes (Yoshida et al., 2007; Sakuraba et al., 2009; Li et al., 2019). In addition, DAEase expressed by the recombinant strains still exhibited excellent enzymatic properties. The heterologous expression of A. tumefaciens and C. cellulolyticum H10 DAEase exhibits long half-life, high kinetic parameters, and high thermal stability (Kim H.J. et al., 2006; Mu et al., 2011).
The A. tumefaciens DAEase is a typical representative among the DTEase family enzymes. The tetrameric arrangement of A. tumefaciens DAEase is an asymmetric unit consisting of four identical subunits of A, B, C, and D (Yoshida et al., 2007; Chan et al., 2012). The active site contains a metal ion with four residues and octahedral coordination to two water molecules. These four residues are conserved among the DTEase family enzymes (Kim K.S. et al., 2006). These four subunits are the crystallographic symmetry-related dimers, in which subunit A and D interact and both are in close contact with subunit B and C. The active site of A. tumefaciens DAEase is exposed on the same front side of the dimers. These stable dimers provide a great accessible surface to bind the substrate on the front side of the dimers.
The hydrophobic groove of the active sites and the accessible surface is located in the middle of A and B subunits. The subunit sides of the DAEase are closed and exposed at the two ends of the barrel. In the tetramer of DAEase, two dimers are enclosed together at the closed sides of the barrel. The monomer (A, B, C, and D subunits) consists of eight repeat units of (β/α)8 structure. Each monomer is composed of 13 α-helices and 8 β-strands as the main structural motif. In addition, a (β/α)8 TIM barrel also exists in the active site of the monomer (Kim K.S. et al., 2006).
Catalytic Mechanism
The catalysis of DTEase family enzymes depends on the molecular arrangement of each subunit. These subunits expose their active sites for the substrate to achieve efficient enzymatic reactions. Seven amino acid residues form hydrogen bonds with water molecules. The seven residues of A. tumefaciens DAEase/P. cichorii DTEase (Glu150/Glu152, Asp183/Asp185, His209/His211, Glu244/Glu246, Glu156/Glu158, His185/His188, and Arg215/Arg217) play an important role in substrate binding and thermal stability confirmed by site-directed mutagenesis (Yoshida et al., 2007). In addition, Trp112, Glu156, and Arg215 greatly influence the activity of the enzyme and the stabilization of cis-endiolate intermediate (Kim et al., 2010).
The epimerization in the active sites proceeds after the catalytic substrate displaces the water molecules. A proton from D-fructose C3 is removed to generate a cis-enediolate intermediate of D-allulose under the cooperation of two residues (Glu150 and Glu244) and Mn2+. In the residue sites of Glu150, Glu156, His209, and Glu244, the hydrogen bonds form between the cis-enediolate intermediate and D-allulose (Kim et al., 2010). D-Allulose is released from the position between the hydrogen bonds and water molecules in the active sites of DAEase.
Currently, the molecular modification has been performed to improve the catalytic activity and thermal stability of DTEase family enzymes. The site-directed mutagenesis was to improve the thermal stability and catalytic behavior of L-rhamnose isomerase from Caldicellulosiruptor obsidiansis for the production of D-allulose (Chen et al., 2018). The hydrophobic residues within β1-α1-loop were collectively replaced with polar amino acids. The V48N/G59N/I63N and V48N/G59N/I63N/F335S mutants respectively resulted in the increase of relative activities by 105.6 and 134.1% compared with that of the wild-type enzyme (Chen et al., 2018). Site-directed mutagenesis also improved the thermal stability of D-allulose 3-epimerase from Dorea sp. (Zhang et al., 2018). The t1/2 value of the mutant protein increased by 5.4-fold at 50°C and the Tm value increased by 17.54-fold compared with the wild-type enzyme (Zhang et al., 2018).
The recent study of the catalytic mechanism is still in the preliminary stage for DTEase family enzymes, and the relationship between enzyme structure and catalytic function needs to be deepened. Besides, some deficiencies still exist in thermal stability and substrate specificity for DTEase. The substrate specificity of DTEase family enzymes should be further utilized in the production of rare sugars to realize the efficient and green production of functional rare sugars.
Construction of Engineered Strains For D-Allulose Bio-Production
Most of DTEase family enzymes have been identified and isolated from bacteria (Table 1). The amount of enzyme expressed in the native strains is far from the requirement of the application. Thus, constructing expression vector and expressing it in heterologous organisms are of great significance in characteristics investigation and enzymic application.
E. coli Expression System
Escherichia coli expression system has the advantages of low cost and high expression efficiency. The soluble over-expression of DTEase family enzymes in E. coli was achieved and the recombined DTEase enzymes are separated and purified using affinity chromatography. The purified recombinant DTEase enzymes can be used to catalyze the production of D-allulose in a bioreactor via immobilization approach (Takeshita et al., 2000). The titer of D-allulose varies from 120 g L–1 to 218 g L–1 with a conversion yield of 24–33% (w/w) (Mu et al., 2011, 2013). With a D-fructose solution of 60% adjusted to pH = 7, approximately 25% of the substrate was converted into D-allulose (Takeshita et al., 2000). A concentration of 441 g L–1 D-allulose from 700 g L–1 D-fructose was produced with borate by immobilizing the recombinant DAEase onto duolite resins A568 and A7, whereas without borate, only 193 g L–1 D-allulose was produced (Lim et al., 2009). In addition, the production yield and conversion efficiency of recombinant E. coli cells expressing Agrobacterium tumefaciens DAEase are respectively 230 g L–1 and 33% (w/w) by using whole-cell reaction approach (Park et al., 2016b).
Bacillus sp. Expression System
The recombinant Bacillus subtilis carrying DAEase from Ruminococcus sp. could overexpress DAEase with high efficiency and low-cost. The recombinant DAEase immobilized onto the anion exchange resin matrix could facilitate stable and effective D-allulose production. The activity of recombinant DAEase expressed by B. subtilis was 58.6 U/mg, which was higher than that in E. coli (8.95 U/mg) (Table 2). Besides, the expression regulatory element of DAEase also affects the amount and activity of the enzyme. Vector pMA5-Pxy/A-RDPE could constitutively express 95 U/mL DAEase in B. subtilis. This value was higher than that of pBluescript II-SK-DTE expressed in E. coli (Ishida et al., 1997a; Chen et al., 2016). The activity of DAEase could be remarkably enhanced under the control of xylose-inducible promoter Pxy/A. The enzymatic activity reached 95 U/mL in a 7.5 L fermentor through fed-batch fermentation (Chen et al., 2016).
TABLE 2.
Bio-production of D-allulose by engineered strains.
| Enzyme and | Base | Host | Expression cassette | Activities, yields, and | |
| Origin of gene | molecular weight | numbers | strains | and expression | publishing years |
| P. cichorii ST-24 | DTEase, 32.5 kDa | 873 bp | E. coli XLl-Blue | pBluescript II-SK-DTE soluble expression | 0.35 U/mL (Ishida et al., 1997a), 1997 |
| C. scindens ATCC 35704 | DTEase, 31 kDa | 864 bp | E. coli BL21 | pET-22b (+)-DTE soluble expression | 27. 9% (Xing et al., 2011), 2011 |
| C. cellulolyticum H10 ATCC35319 | DAEase, 32 kDa | 882 bp | E. coli BL21 | pET-22b(+)-Cc-dpe, soluble expression | 218 g/L (Mu et al., 2011), 2011 |
| Ruminococcus sp. 5_1_39BFAA | DAEase, 33 kDa | 876 bp | E. coli BL21 | pET-21a-dpe, soluble expression | 8.95 U/mg, 125 g/L (Zhu et al., 2012), 2012 |
| Clostridium sp. BNL1100 | DAEase, 32 kDa | 879 bp | E. coli BL21 | pET-22b(+)-Clsp-dpe, soluble expression | 120 g/L (Mu et al., 2013), 2013 |
| A. tumefaciens str. C58 | DAEase, 33 kDa | 870 bp | E. coli BL21 | pET22b (+)/pelb-dpe, secretion | 10.9 U/mL, 179 g/L (Gu et al., 2013), 2013 |
| E. coli JM109 | DTEase, 29.8 kDa | 789 bp | E. coli BL21 | pET-15b-DTE, soluble expression | No data (He et al., 2015), 2015 |
| A. tumefaciens | DAEase, 33 kDa | 870 bp | S. cerevisiae AN120 | pRS424-TEFpr-ss-xy/A soluble expression | 12.0% (Li et al., 2015), 2015 |
| A. tumefaciens ATCC 33970 | DAEase, 33 kDa | 870 bp | E. coli ER2566 | pET-24a(+)-dpe, soluble expression | 230 g/L (Park et al., 2016b), 2016 |
| Treponema primitia ZAS-1 | DAEase, 33.3 kDa | 888 bp | E. coli BL21 | pET-22b(+)-dpe, secretion | 137.5 g/L (Zhang et al., 2016b), 2016 |
| Ruminococcus sp. 5_1_39BFAA | DAEase, 33 kDa | 876 bp | B. subtilis 1A751 | pMA5-Pxy/A-RDPE, secretion | 95 U/mL, 145 g/L (Chen et al., 2016), 2016 |
| Ruminococcus sp. 5_1_39BFAA | DAEase, 33 kDa | 876 bp | B. subtilis | pNCMO2-P2-dpe, soluble expression | 58.6 U/mg (Li et al., 2018a), 2018 |
| A. tumefaciens | DAEase, 33 kDa | 870 bp | K. marxianus | pRS42H-dpe, soluble expression | 190 g L–1 (Yang et al., 2018), 2018 |
Yeast Expression System
The exogenous DAEase gene could be effectively expressed in the recombinant S. cerevisiae (Li et al., 2015) and Kluyveromyces marxianus (Yang et al., 2018). The expression vector of pRS424-TEFpr-ss-xy/A carrying A. tumefaciens DAEase could generate a 33 kDa protein in S. cerevisiae AN120 (Li et al., 2015). Both the xylose isomerase gene of T. thermophilus and A. tumefaciens DAEase gene co-expressed in yeast spores to strengthen the synergistic catalysis effect. These two recombinant enzymes were immobilized for D-allulose production with D-glucose as the substrate (Li et al., 2015). D-Fructose from D-glucose catalyzed by recombinant xylose isomerase is then converted to D-allulose by recombinant DAEase. Yang et al. (2018) have provided a valuable pathway to regenerate engineered K. marxianus cells, and thus achieve cyclic catalysis for D-allulose production. The recombinant K. marxianus produced 190 g L–1 D-allulose with 750 g L–1 D-fructose within 12 h. Approximately 100 g residual D-fructose was converted into 34 g ethanol by the engineering strains. Besides, the idea of cyclic catalysis for D-allulose production was also provided through a whole-cell reaction (Yang et al., 2018).
Bacillus sp., E. coli, and yeast are generally used to construct the recombinant system for DAEase expression. Different from E. coli, B. subtilis has no outer membrane, and the secreted protein can be released directly to the culture medium. The safety of B. subtilis is food grade. No heat source lipopolysaccharide (endotoxin) is mixed in the secreted protein products. Compared with E. coli, B. subtilis expression system is not perfect. The engineering E. coli has advantageous characteristics of clear genetic background, complete carrier receptor system, rapid growth, simple culture, and stable recombinants. Besides, the yeast expression system has its own characteristics of simple culture conditions, fast growth speed, high expression level, and simple operation. After translation, the protein can be processed and correctly modified. The deficiency of yeast expression system lies in low expression of cloned genes, long fermentation time, incorrect glycosylation of proteins and anti-cell division. Further, the high concentration of polysaccharide in supernatant is not conducive to the purification of recombinant protein.
Early researchers tend to use E. coli as host bacteria to study the expression and properties of DTEase family enzymes (Ishida et al., 1997a; Mu et al., 2011, 2013). Recently, many researchers have applied B. subtilis and yeast as host bacteria to express DTEase family enzymes for D-allulose production (Chen et al., 2016; Yang et al., 2018). The contents of D-allulose respectively reached 230 g/L (Park et al., 2016b) and 190 g L–1 (Yang et al., 2018) by using recombinant A. tumefaciens DAEase expressed in E. coli and K. marxianus. By the comparison, E. coli expression system of A. tumefaciens DAEase is the best among all the reported references shown in Table 2.
Isolation and Purification of D-Allulose
Mass production of D-allulose is systematic engineering of gene acquisition, target gene expression, microbial fermentation, catalysis, D-allulose separation and purification. The researchers pay highly attention to the development of D-allulose with high efficiency and low cost. The following two methods are mainly used for the isolation and purification of D-allulose. The first method is ion exchange resin. Both anion exchange resin matrix and synchronous moving bed chromatography are used to immobilize DAEase for D-allulose production from D-fructose. The cells of R. sphaeroides SK011 produces 6.5 g L–1 D-allulose with a productivity of 0.82 g–1h–1 from initial 50 g L–1 D-fructose by using the dialysis method based on ion exchange resin (Zhang et al., 2009a). The toluene-treated Sinorhizobium sp. produces 37 g L–1 D-allulose from 700 g L–1 D-fructose; after the treatment of anion exchange resin matrix, the purity of D-allulose is up to 99.1%. Further, the purity of D-allulose reaches 98.3% via an approach of DTF-Ca2+ cation exchange chromatography (Xing et al., 2011). For the mixed system of D-allulose and D-fructose, D-fructose is first transformed into gluconic acid, and then D-allulose of 91.2% purity is obtained by anion exchange resin (Li et al., 2018b).
The second technique for D-allulose purification is the biological method. D-Allulose is obtained after the yeast consumed residual D-fructose to produce ethanol in the mixed system of D-allulose and D-fructose. The final purity of crystal D-allulose is 85% (Takeshita et al., 2000). Besides, the purity reaches approximately 86.2% by combining pervaporation technology, cation exchange chromatography, and biological method (Song et al., 2017). The biological method is more environment-friendly than ion exchange resin.
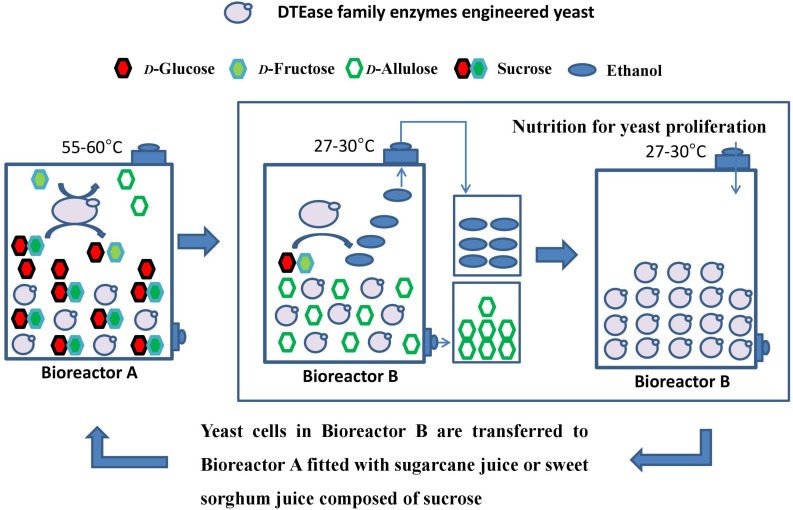
A green and recycling process technology for D-allulose production is proposed in Figure 1. The whole reaction system is carried out between Bioreactor A (for sucrose hydrolysis and D-allulose conversion) and Bioreactor B (for ethanol production, D-allulose isolation, and yeast proliferation). Crude sugarcane juice or crude sweet sorghum juice are used as material to provide sucrose. Engineered yeast contains the native invertase and integrated exogenous DTEase family enzyme genes, thus engineered yeast is capable of sucrose hydrolysis to D-glucose and D-fructose by invertase, D-fructose conversion into D-allulose at 55–60°C, and ethanol production by fermenting D-glucose and D-fructose at 27–30°C. The advantages of this method include the low cost of raw materials, utilization of intermediate products produced in the process as much as possible, reduction of waste formation, reduction of energy consumption, and improvement of sugar yield.
FIGURE 1.
Recycling process for D-allulose conversion and ethanol production by using sugarcane juice or sweet sorghum juice as materials. During catalysis at a high temperature of 55–60°C for 1–2 h in Reaction A, most of the yeast can’t stand such high temperature and die. A small number of yeast spores still survive. These surviving yeast spores proliferate and consume D-fructose and D-glucose to produce ethanol at a later lower temperature of 27–30°C in Reaction B. Besides, a small part of D-fructose was metabolized by the remaining living yeast at such high temperature, but most of D-fructose was still converted into D-allulose in Reaction A.
Prospects
The current industrial production of D-allulose has been achieved in China, Japan, South Korea, and United States. The production cost is still high due to the unsatisfactory activity of enzymes and their low reutilization frequencies. Therefore, increased enzymatic activities, stability, and catalytic frequency should be the key purposes in future research and development of DTEase family enzymes. Various advanced techniques and methods, such as directed evolution (Hong et al., 2016; Denisenko et al., 2017), screening of thermally stable enzymes (Baltaci et al., 2016), and material development of immobilized enzymes (Sahoo et al., 2016; Aldhrub et al., 2017), should be applied to modify DTEase family enzymes and thus overcome their unsatisfactory activity and stability.
The improvements of decolorization, desalination, crystallization, and drying reduce the production cost of D-allulose. The rational design or non-rational process with efficient high-throughput screening technology was the direct approach to modify the structure of DTEase family enzymes. The change of amino acid residues affects the advanced structure of DTEase, thus leading to the change of enzymatic properties. Site-directed mutagenesis was used to enhance the thermostability of DAEase from R. baltica (Mao et al., 2020) and improve the catalytic behavior of C. obsidiansis L-rhamnose isomerase (Chen et al., 2018). The improvement of DTEase family enzymes and production processing will decrease the production cost and D-allulose price, thus ensuring that D-allulose will be conveniently available to consumers.
Author Contributions
SuJ performed the data analysis and revised the manuscript. WX wrote the manuscript. PY provided ideas. SL drew the pictures. ShJ drafted the work and provided the financial support. ZZ designed the manuscript. XZ, GZ, and JL searched the literature. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported by Natural Science Foundation of Anhui Province (CN) (1908085MC80) and National Key Research and Development Program of China (2016YFD0401401-2). The funder had no role in the study design, data collection and analysis, or writing of the manuscript.
References
- Aldhrub A. H. A., Sahin S., Ozmen I., Tunca E., Bulbul M. (2017). Immobilization and characterization of human carbonic anhydrase I on amine functionalized magnetic nanoparticles. Process Biochem. 57 95–104. 10.1016/j.procbio.2017.03.025 [DOI] [Google Scholar]
- Baltaci M. O., Genc B., Arslan S., Adiguzel G., Adiguzel A. (2016). Isolation and characterization of thermophilic bacteria from geothermal areas in Turkey and preliminary research on biotechnologically important enzyme production. Geomicrobiol. J. 34 53–62. 10.1080/01490451.2015.1137662 [DOI] [Google Scholar]
- Chan H. C., Zhu Y. M., Hu Y. M., Ko T., Huang C. H., Ren F. F., et al. (2012). Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars. Protein Cell 3 123–131. 10.1007/s13238-012-2026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhu Y., Fu G., Song Y., Jin Z., Sun Y., et al. (2016). High-level intra- and extra-cellular production of D-psicose 3-epimerase via a modified xylose-inducible expression system in Bacillus subtilis. J. Indust. Microbiol. Biotechnol. 43 1–15. [DOI] [PubMed] [Google Scholar]
- Chen Z. W., Chen J. J., Zhang W. L., Zhang T., Guang C., Mu W. M. (2018). Improving thermostability and catalytic behavior of L-rhamnose isomerase from Caldicellulosiruptor obsidiansis OB47 toward D-allulose by site-directed mutagenesis. J. Agric. Food Chem. 66 12017–12024. 10.1021/acs.jafc.8b05107 [DOI] [PubMed] [Google Scholar]
- Chu C. A., Fujimoto Y., Igawa K., Grimsby J., Grippo J. F., Magnuson M. A., et al. (2003). Rapid translocation of hepatic glucokinase in response to intraduodenal glucose infusion and changes in plasma glucose and insulin in conscious rats. Am. J. Physiol. Gastrointest. Liver Physiol. 286 627–634. [DOI] [PubMed] [Google Scholar]
- Chung Y. M., Lee J. H., Kim D. Y., Hwang S. H., Hong Y. H., Kim S. B. (2012). Dietary D-psicose reduced visceral fat mass in high-fat diet-induced obese rats. J. Food Sci. 77 53–58. 10.1111/j.1750-3841.2011.02571.x [DOI] [PubMed] [Google Scholar]
- Denisenko Y. A., Gusakov A. V., Rozhkova A. M., Osipov D. O., Zorov I. N., Matys V. Y., et al. (2017). Site-directed mutagenesis of GH10 xylanase A from Penicillium canescens for determining factors affecting the enzyme thermostability. Int. J. Biol. Macromol. 104 665–671. 10.1016/j.ijbiomac.2017.06.079 [DOI] [PubMed] [Google Scholar]
- Do G. Y., Kwon E. Y., Kim Y. J., Han Y., Kim S. B., Kim Y. H., et al. (2019). Supplementation of non-dairy creamer-enriched high-fat diet with D-allulose ameliorated blood glucose and body fat accumulation in C57BL/6J mice. Appl. Sci. 9 17–20. [Google Scholar]
- Fukada K., Ishii T., Tanaka K., Yamaji M., Yamaoka Y., Kobashi K., et al. (2010). Crystal structure, solubility, and mutarotation of the rare monosaccharide D-psicose. Bull. Chem. Soc. Jpn. 83 1193–1197. 10.1246/bcsj.20100148 [DOI] [Google Scholar]
- Granström T. B., Takata G., Tokuda M., Izumori K. (2004). Izumoring: a novel and complete strategy for bioproduction of rare sugars. J. Biosci. Bioeng. 97 89–94. [DOI] [PubMed] [Google Scholar]
- Gu L., Zhang J., Liu B. H., Wu C. D., Du G. C., Chen J. (2013). High-level extracellular production of D-psicose-3-epimerase with recombinant Escherichia coli by a two-stage glycerol feeding approach. Bioprocess Biosyst. Eng. 36 1767–1777. 10.1007/s00449-013-0952-0 [DOI] [PubMed] [Google Scholar]
- Han Y., Han H. J., Kim A. H., Choi J. Y., Cho S. J., Park Y. B., et al. (2016). D-Allulose supplementation normalized the body weight and fat-pad mass in diet-induced obese mice via the regulation of lipid metabolism under isocaloric fed condition. Mol. Nutr. Food Res. 60 1695–1706. 10.1002/mnfr.201500771 [DOI] [PubMed] [Google Scholar]
- Harada M., Kondo E., Hayashi H., Suezawa C., Suguri S., Arai M. (2012). D-Allose and D-psicose reinforce the action of metronidazole on trichomonad. Parasitol. Res. 110 1565–1567. 10.1007/s00436-011-2660-5 [DOI] [PubMed] [Google Scholar]
- Hayashi N., Iida T., Yamada T., Okuma K., Takehara I., Yamamoto T., et al. (2010). Study on the postprandial blood glucose suppression effect of D-psicose in borderline diabetes and the safety of long-term ingestion by normal human subjects. Biosci. Biotechnol. Biochem. 74 510–519. 10.1271/bbb.90707 [DOI] [PubMed] [Google Scholar]
- He W. W., Jiang B., Mu W. M., Zhang T. (2016). Production of D-allulose with D-psicose 3-epimerase expressed and displayed on the surface of Bacillus subtilis spores. J. Agric. Food Chem. 64 7201–7207. 10.1021/acs.jafc.6b03347 [DOI] [PubMed] [Google Scholar]
- He X. L., Zhou X. H., Yang Z., Xu L., Yu Y. X., Jia L. L., et al. (2015). Cloning, expression and purification of D-tagatose 3-epimerase gene from Escherichia coli JM109. Protein Exp. Purific. 114 77–79. 10.1016/j.pep.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Hong S. H., Lee S. S., Chung J. M., Jung H. S., Singh S., Mondal S., et al. (2016). Site-specific mutagenesis of yeast 2-Cys peroxiredoxin improves heat or oxidative stress tolerance by enhancing its chaperone or peroxidase function. Protoplasma 254 1–8. [DOI] [PubMed] [Google Scholar]
- Hossain A., Yamaguchi F., Hirose K., Matsunaga T., Sui L., Hirata Y., et al. (2015a). Rare sugar D-psicose prevents progression and development of diabetes in T2DM model Otsuka long-evans tokushima fatty rats. Drug Des. Dev. Ther. 9 525–535. 10.2147/DDDT.S71289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain A., Yamaguchi F., Matsuo T., Tsukamoto I., Toyoda Y., Ogawa M., et al. (2015b). Rare sugar D-allulose: potential role and therapeutic monitoring in maintaining obesity and type 2 diabetes mellitus. Pharmacol. Ther. 155 49–59. 10.1016/j.pharmthera.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Hossain M. A., Kitagaki S., Nakano D., Nishiyama A., Funamoto Y., Matsunaga T., et al. (2011). Rare sugar D-psicose improves insulin sensitivity and glucose tolerance in type 2 diabetes Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biochem. Biophys. Res. Commun. 405 7–12. 10.1016/j.bbrc.2010.12.091 [DOI] [PubMed] [Google Scholar]
- Iida T., Yamada T., Hayashi N., Okuma K., Izumori K., Ishii R., et al. (2013). Reduction of abdominal fat accumulation in rats by 8-week ingestion of a newly developed sweetener made from high fructose corn syrup. Food Chem. 138 781–785. 10.1016/j.foodchem.2012.11.017 [DOI] [PubMed] [Google Scholar]
- Ilhan E., Pocan P., Ogawa M., Oztop M. H. (2020). Role of ’D-allulose’ in a starch based composite gel matrix. Carbohydr. Polym. 228 10–16. 10.1016/j.carbpol.2019.115373 [DOI] [PubMed] [Google Scholar]
- Ishida Y., Kamiya T., Itoh H., Kimura Y., Izumori K. (1997a). Cloning and characterization of the D-tagatose 3-epimerase gene from Pseudomonas cichorii ST-24. J. Ferment. Bioeng. 83 529–534. 10.1016/s0922-338x(97)81132-4 [DOI] [Google Scholar]
- Ishida Y., Kamiya T., Izumori K. (1997b). Production of D-tagatose 3-epimerase of Pseudomonas cichorii ST-24 using recombinant Escherichia coli. J. Ferment. Bioeng. 84 348–350. 10.1016/s0922-338x(97)89257-4 [DOI] [Google Scholar]
- Itoh H., Okaya H., Khan A. R., Tajima S., Izumori S. H. K. (1994). Purification and characterization of D-tagatose 3-epimerase from Pseudomonas sp. ST-24. J. Biosci. Biotechnol. Biochem. 58 2168–2171. [Google Scholar]
- Iwasaki Y., Sendo M., Dezaki K., Hira T., Sato T., Nakata M., et al. (2018). GLP-1 release and vagal afferent activation mediate the beneficial metabolic and chronotherapeutic effects of D-allulose. Nat. Commun. 9 17–25. 10.1038/s41467-017-02488-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumori K. A., Okaya H. (1993). A new enzyme, D-ketohexose 3-epimerase from Pseudomonas sp. ST-24. Biosci. Biotechnol. Biochem. 57 1037–1039. 10.1271/bbb.57.1037 [DOI] [Google Scholar]
- Jia M., Mu W. M., Chu F. F., Zhang X. M., Jiang B., Zhou L. M., et al. (2014). A D-psicose 3-epimerase with neutral pH optimum from Clostridium bolteae for D-psicose production: cloning, expression, purification, and characterization. Appl. Microbiol. Biotechnol. 98 717–725. 10.1007/s00253-013-4924-8 [DOI] [PubMed] [Google Scholar]
- Kanasaki A., Jiang Z., Mizokami T., Shirouchi B., Iida T., Nagata Y., et al. (2019). Dietary D-allulose alters cholesterol metabolism in Golden Syrian hamsters partly by reducing serum PCSK9 levels. J. Funct. Food 60 6–9. [Google Scholar]
- Kim H. J., Hyun E. K., Kim Y. S., Lee Y. J., Oh D. K. (2006). Characterization of an Agrobacterium tumefaciens D-psicose 3-epimerase that converts D-fructose to D-psicose. Appl. Environ. Microbiol. 72 981–985. 10.1128/aem.72.2.981-985.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Yeom S. J., Kim K., Rhee S., Kim D., Oh D. K. (2010). Mutational analysis of the active site residues of a D-psicose 3-epimerase from Agrobacterium tumefaciens. Biotechnol. Lett. 32 261–268. 10.1007/s10529-009-0148-5 [DOI] [PubMed] [Google Scholar]
- Kim K. S., Kim H. J., Oh D. K., Cha S. S., Rhee S. K. (2006). Crystal structure of D-psicose 3-epimerase from Agrobacterium tumefaciens and its complex with true substrate D-fructose: a pivotal role of metal in catalysis, an active site for the non-phosphorylated substrate, and its conformational changes. J. Mol. Biol. 361 920–931. 10.1016/j.jmb.2006.06.069 [DOI] [PubMed] [Google Scholar]
- Kim Y., Park C., Sa S., Case I., Li C. M., Gao Y. L., et al. (2019). A study of D-allulose-associated reproductive toxicity in rats. Food Chem. Toxicol. 131 6–9. 10.1016/j.fct.2019.05.056 [DOI] [PubMed] [Google Scholar]
- Kimoto-Nira H., Moriya N., Hayakawa S., Kuramasu K., Ohmori H., Yamasaki S., et al. (2017). Effects of rare sugar D-allulose on acid production and probiotic activities of dairy lactic acid bacteria. J. Dairy Sci. 100 5936–5944. 10.3168/jds.2016-12214 [DOI] [PubMed] [Google Scholar]
- Kimura S., Zhang G., Nishiyama A., Nagai Y., Nakagawa T., Miyanaka H., et al. (2005). D-Allose, an all-cis aldo-hexose, suppresses development of salt-induced hypertension in Dahl rats. J. Hypertens. 23 1887–1894. 10.1097/01.hjh.0000182523.29193.e3 [DOI] [PubMed] [Google Scholar]
- Kishida K., Martinez G., Iida T., Yamada T., Ferraris R. P., Toyoda Y. (2019). D-Allulose is a substrate of glucose transporter type 5 (GLUT5) in the small intestine. Food Chem. 277 604–608. 10.1016/j.foodchem.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Li C., Lin J. Q., Guo Q. Q., Zhang C. J., Du K., Lin H. B., et al. (2018a). D-Psicose 3-epimerase secretory overexpression, immobilization, and D-psicose biotransformation, separation and crystallization. J. Chem. Technol. Biotechnol. 93 350–357. 10.1002/jctb.5360 [DOI] [Google Scholar]
- Li C., Zhang C. J., Lin J. N., Gao L., Lin H. B., Lin J. Q. (2018b). Enzymatic fructose removal from D-psicose bioproduction model solution and the system modeling and simulation. J. Chem. Technol. Biotechnol. 93 1249–1260. 10.1002/jctb.5483 [DOI] [Google Scholar]
- Li S. N., Chen Z. W., Zhang W. L., Guang C. E., Mu W. M. (2019). Characterization of a D-tagatose 3-epimerase from Caballeronia fortuita and its application in rare sugar production. Int. J. Biol. Macromol. 138 536–545. 10.1016/j.ijbiomac.2019.07.112 [DOI] [PubMed] [Google Scholar]
- Li Z., Li Y., Duan S., Liu J., Yuan P., Nakanishi H., et al. (2015). Bioconversion of D-glucose to D-psicose with immobilized D-xylose isomerase and D-psicose 3-epimerase on Saccharomyces cerevisiae spores. J. Indus. Microbiol. Biotechnol. 42 1117–1128. 10.1007/s10295-015-1631-8 [DOI] [PubMed] [Google Scholar]
- Lim B. C., Kim H. J., Oh D. K. (2009). A stable immobilized D-psicose 3-epimerase for the production of D-psicose in the presence of borate. Process Biochem. 44 822–828. 10.1016/j.procbio.2009.03.017 [DOI] [Google Scholar]
- Maeng H. J., Yoon J. H., Chun K. H., Kim S. T., Jang D. J., Park J. E., et al. (2019). Metabolic stability of D-allulose in biorelevant media and hepatocytes:comparison with fructose and erythritol. Foods 8 13–18. 10.3390/foods8100448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S., Cheng X., Zhu Z., Chen Y., Li C., Zhu M., et al. (2020). Engineering a thermostable version of D-allulose 3-epimerase from Rhodopirellula baltica via site-directed mutagenesis based on B-factors analysis. Enzyme Microb. Technol. 132 1–8. 10.1016/j.enzmictec.2019.109441 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Baba Y., Hashiguchi M., Takeshita K., Izumori K., Suzuki H. (2015). Dietary D-psicose, a C-3 epimer of D-fructose, suppresses the activity of hepatic lipogenic enzymes in rats. Asia Pacific J. Clin. Nutr. 10 233–237. 10.1046/j.1440-6047.2001.00246.x [DOI] [PubMed] [Google Scholar]
- Matsuo T., Izumori K. (2006). Effects of dietary D-psicose on diurnal variation in plasma glucose and insulin concentrations of rats. Biosci. Biotechnol. Biochem. 70 2081–2085. 10.1271/bbb.60036 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Izumori K. (2009). D-Psicose inhibits intestinal alpha-glucosidase and suppresses the glycemic response after ingestion of carbohydrates in rats. J. Clin. Biochem. Nutr. 45 202–206. 10.3164/jcbn.09-36 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Matsuo T., Suzuki H., Hashiguchi M., Izumori K. (2002). D-Psicose is a rare sugar that provides no energy to growing rats. J. Nutr. Sci. Vitaminol. 48 77–80. 10.3177/jnsv.48.77 [DOI] [PubMed] [Google Scholar]
- Matsuo T., Tanaka T., Hashiguchi M., Izumori K., Suzuki H. (2003). Metabolic effects of D-psicose in rats: studies on faecal and urinary excretion and caecal fermentation. Asia Pacific J. Clin. Nutr. 12 225–231. [PubMed] [Google Scholar]
- Mu W., Zhang W., Fang D., Zhou L., Jiang B., Zhang T. (2013). Characterization of a D-psicose-producing enzyme, D-psicose 3-epimerase, from Clostridium sp. Biotechnol. Lett. 35 1481–1486. 10.1007/s10529-013-1230-6 [DOI] [PubMed] [Google Scholar]
- Mu W. M., Xing Q. C., Yu S. H., Zhou L., Jiang B. (2011). Cloning, expression, and characterization of a D-psicose 3-epimerase from Clostridium cellulolyticum H10. J. Agric. Food Chem. 59 7785–7792. 10.1021/jf201356q [DOI] [PubMed] [Google Scholar]
- Nishii N., Nomizo T., Takashima S., Matsubara T., Tokuda M., Kitagawa H. (2016). Single oral dose safety of D-allulose in dogs. J. Vet. Med. Sci. 78 1079–1083. 10.1292/jvms.15-0676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. K., Kim N. H., Kim H. J., Park C. S., Kim S. W., Ko M., et al. (2007). D-Psicose production from D-fructose using an isolated strain, Sinorhizobium sp. World J. Microbiol. Biotechnol. 23 559–562. [Google Scholar]
- Park C. S., Kim T., Hong S. H., Shin K. C., Kim K. R., Oh D. K. (2016a). D-Allulose production from D-fructose by permeabilized recombinant cells of Corynebacterium glutamicum cells expressing D-allulose 3-epimerase Flavonifractor plautii. PLoS One 11:e0160044. 10.1371/journal.pone.0160044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. S., Park C. S., Shin K. C., Oh D. K. (2016b). Production of D-psicose from D-fructose by whole recombinant cells with high-level expression of D-psicose 3-epimerase from Agrobacterium tumefaciens. J. Biosci. Bioeng. 121 186–190. 10.1016/j.jbiosc.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Patel S. N., Kaushal G., Singh S. (2019). Characterization of a novel D-allulose 3-epimerase gene from the metagenome of a thermal aquatic habitat and D-allulose production by Bacillus subtilis whole-cell catalysis. Appl. Environ. Microbiol. 1 1–8. 10.1128/AEM.02605-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi K. M., Bommarius A. S., Broering J. M., Chaparroriggers J. F. (2007). Stability of biocatalysts. Curr. Opin. Chem. Biol. 11 220–225. [DOI] [PubMed] [Google Scholar]
- Ran G. Q., Tan D., Zhao J. P., Fan F., Zhang Q., Wu X. J., et al. (2019). Functionalized polyhydroxyalkanoate nano-beads as a stable biocatalyst for cost-effective production of the rare sugar D-allulose. Bioresour. Technol. 289 9–18. 10.1016/j.biortech.2019.121673 [DOI] [PubMed] [Google Scholar]
- Sahoo B., Dutta S., Dhara D. (2016). Amine-functionalized magnetic nanoparticles as robust support for immobilization of lipase. J. Chem. Sci. 128 1131–1140. 10.1016/j.msec.2019.01.070 [DOI] [PubMed] [Google Scholar]
- Sakuraba H., Yoneda K., Satomura T., Kawakami R., Ohshima T. (2009). Structure of a D-tagatose 3-epimerase-related protein from the hyperthermophilic bacterium Thermotoga maritima. Acta Crystallogr. Sec. F Struc. Biol. Cryst. Commun. 65 199–203. 10.1107/S1744309109002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Yamada T., Hayashi N., Iida T., Nagata Y., Ozaki N., et al. (2017). Rare sugar syrup containing D-allulose but not high-fructose corn syrup maintains glucose tolerance and insulin sensitivity partly via hepatic glucokinase translocation in wistar rats. J. Agric. Food Chem. 65 2888–2894. 10.1021/acs.jafc.6b05627 [DOI] [PubMed] [Google Scholar]
- Song Y., Nguyen Q. A., Wi S. G., Yang J., Bae H. J. (2017). Strategy for dual production of bioethanol and D-psicose as value-added products from cruciferous vegetable residue. Bioresour. Technol. 223 34–39. 10.1016/j.biortech.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Sun Y. X., Hayakawa S., Chuamanochan M., Fujimoto M., Innun A., Izumori K. (2006). Antioxidant effects of Maillard reaction products obtained from ovalbumin and different D-aldohexoses. Biosci. Biotechnol. Biochem. 70 598–605. 10.1271/bbb.70.598 [DOI] [PubMed] [Google Scholar]
- Takeshita K., Suga A., Takada G., Izumori K. (2000). Mass production of D-psicose from D-fructose by a continuous bioreactor system using immobilized D-tagatose 3-epimerase. J. Biosci. Bioeng. 90 453–455. 10.1016/s1389-1723(01)80018-9 [DOI] [PubMed] [Google Scholar]
- Tseng W. C., Chen C. N., Hsu C. T., Lee H. C., Fang H. Y., Wang M. J., et al. (2018). Characterization of a recombinant D-allulose 3-epimerase from Agrobacterium sp ATCC 31749 and identification of an important interfacial residue. Int. J. Biol. Macromol. 112 767–774. 10.1016/j.ijbiomac.2018.02.036 [DOI] [PubMed] [Google Scholar]
- Whistler R. L., Singh P. P., Lake W. C. (1974). D-Psicose metabolism in the rat. Carbohydr. Res. 34 200–202. 10.1016/s0008-6215(00)80385-7 [DOI] [PubMed] [Google Scholar]
- Xing Q. C., Mu W. M., Jiang B., Zhou L. M., Zhang T. (2011). Cloning, expression, purification and characterization of D-tagatose-3-epimerase from Clostridium scindens ATCC 35704. Food Ferment. Indus. 37 6–10. [Google Scholar]
- Yang J. G., Tian C. Y., Zhang T., Ren C. X., Zhu Y. M., Zeng Y., et al. (2019). Development of food-grade expression system for D-allulose 3-epimerase preparation with tandem isoenzyme genes in Corynebacterium glutamicum and its application in conversion of cane molasses to D-allulose. Biotechnol. Bioeng. 116 745–756. 10.1002/bit.26909 [DOI] [PubMed] [Google Scholar]
- Yang P. Z., Zhu X. X., Zheng Z., Mu D. D., Jiang S. T., Luo S. Z., et al. (2018). Cell regeneration and cyclic catalysis of engineered Kluyveromyces marxianus of a D-psicose-3-epimerase gene from Agrobacterium tumefaciens for D-allulose production. World J. Microbiol. Biotechnol. 34 7–13. 10.1007/s11274-018-2451-6 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Yamada M., Nishitani T., Takada G., Izumori K., Kamitori S. (2007). Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose. J. Mol. Biol. 374 443–453. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Yoshihara A., Suzuki T., Morimoto K., Kamitori S. (2019). X-ray structure of a novel ketose 3-epimerase from Shinella zoogloeods which is capable of D-allulose production. FEBS Open Biol. 9 257–265. [Google Scholar]
- Yoshihara A., Kozakai T., Shintani T., Matsutani R., Ohtani K., Iida T., et al. (2017). Purification and characterization of (D)-allulose 3-epimerase derived from Arthrobacter globiformis M30, a GRAS microorganism. J. Biosci. Bioeng. 123 170–176. 10.1016/j.jbiosc.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Zhang L., Jiang B., Mu W. M., Zhang T. (2009a). Bioproduction of D-psicose using permeabilized cells of newly isolated Rhodobacter sphaeroides SK011. Front. Chem. Eng. China 3:393 10.1007/s11705-009-0252-z [DOI] [Google Scholar]
- Zhang L., Mu W., Jiang B., Zhang T. (2009b). Characterization of D-tagatose-3-epimerase from Rhodobacter sphaeroides that converts D-fructose into D-psicose. Biotechnol. Lett. 31 857–862. 10.1007/s10529-009-9942-3 [DOI] [PubMed] [Google Scholar]
- Zhang L. T., Jiang B., Mu W. M., Zhang T. (2008). Characterization of D-tagatose 3-epimerase from Rhodobacter sphaeroides SK011. J. Biotechnol. 136 726–729. [DOI] [PubMed] [Google Scholar]
- Zhang W., Fang D., Xing Q., Zhou L., Jiang B., Mu W. (2013a). Characterization of a novel metal-dependent D-psicose 3-epimerase from Clostridium scindens 35704. PLoS One 8:e0062987. 10.1371/journal.pone.0062987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Fang D., Zhang T., Zhou L., Jiang B., Mu W. M. (2013b). Characterization of a metal-dependent D-psicose 3-epimerase from a novel strain, Desmospora sp. 8437. J. Agric. Food Chem. 61 11468–11476. 10.1021/jf4035817 [DOI] [PubMed] [Google Scholar]
- Zhang W. L., Li H., Zhang T., Jiang B., Zhou L., Mu W. M. (2015). Characterization of a D-psicose 3-epimerase from Dorea sp. CAG317 with an acidic pH optimum and a high specific activity. J. Mol. Catal. B-Enzym. 120 68–74. 10.1016/j.molcatb.2015.05.018 [DOI] [Google Scholar]
- Zhang W. L., Yu S. H., Zhang T., Jiang B., Mu W. M. (2016a). Recent advances in D-allulose: physiological functionalities, applications, and biological production. Trends Food Sci. Technol. 54 127–137. 10.1016/j.tifs.2016.06.004 [DOI] [Google Scholar]
- Zhang W. L., Zhang T., Jiang B., Mu W. M. (2016b). Biochemical characterization of a D-psicose 3-epimerase from Treponema primitia ZAS-1 and its application on enzymatic production of D-psicose. J. Sci. Food Agri. 96 49–56. 10.1002/jsfa.7187 [DOI] [PubMed] [Google Scholar]
- Zhang W. L., Zhang T., Jiang B., Mu W. M. (2017). Enzymatic approaches to rare sugar production. Biotechnol. Adv. 35 267–274. 10.1016/j.biotechadv.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Zhang W. L., Zhang Y. M., Huang J. W., Chen Z. W., Zhang T., Guang C., et al. (2018). Thermostability improvement of the D-allulose 3-epimerase from Dorea sp CAG317 by site-directed mutagenesis at the interface regions. J. Agri. Food Chem. 66 5593–5601. 10.1021/acs.jafc.8b01200 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Men Y., Bai W., Li X., Zhang L., Sun Y., et al. (2012). Overexpression of D-psicose 3-epimerase from Ruminococcus sp. in Escherichia coli and its potential application in D-psicose production. Biotechnol. Lett. 34 1901–1906. 10.1007/s10529-012-0986-4 [DOI] [PubMed] [Google Scholar]
- Zhu Z. L., Gao D. K., Li C., Chen Y., Zhu M. L., Liu X., et al. (2019a). Redesign of a novel D-allulose 3-epimerase from Staphylococcus aureus for thermostability and efficient biocatalytic production of D-allulose. Microb. Cell Fact. 18 10–16. 10.1186/s12934-019-1107-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. L., Li C., Liu X., Gao D. K., Wang X. Y., Tanokura M., et al. (2019b). Biochemical characterization and biocatalytic application of a novel D-tagatose 3-epimerase from Sinorhizobium sp. RSC Adv. 9 2919–2927. 10.1039/c8ra10029b [DOI] [PMC free article] [PubMed] [Google Scholar]