Abstract
The 3′-end processing machinery for metazoan replication-dependent histone pre-mRNAs contains the U7 snRNP and shares the key cleavage module with the canonical cleavage/polyadenylation machinery. We reconstituted an active human histone pre-mRNA processing machinery using 13 recombinant proteins and 2 RNAs, and determined its structure by cryo-electron microscopy. The overall structure is highly asymmetrical and resembles an amphora with one long handle. We captured the pre-mRNA in the CPSF73 endonuclease active site, poised for cleavage. The endonuclease and the entire cleavage module undergo extensive rearrangements for activation, triggered through the recognition of the duplex between the authentic pre-mRNA and U7 snRNA. Our studies also have significant implications for understanding canonical and snRNA 3′-end processing.
The 3′-end processing machineries for polyadenylated (1, 2) and histone pre-mRNAs (3, 4) both utilize CPSF73 to cleave pre-mRNA (5, 6), but the molecular mechanism for their functions is still poorly understood. CPSF73, CPSF100, symplekin and CstF64 comprise the histone pre-mRNA cleavage complex (HCC) (Figs. 1A,B, table S1), which is equivalent to the mammalian cleavage factor (mCF) for polyadenylated pre-mRNAs (7, 8). The cleavage site in histone pre-mRNAs is located between a conserved stem-loop (SL) that is recognized by SL binding protein (SLBP) and a histone downstream element (HDE) that base pairs with the 5′ end of U7 snRNA, forming an HDE-U7 duplex (Fig. 1A). The U7 snRNP is critical for this processing, and the Lsm11-FLASH complex recruits the HCC to the machinery (9–12) (Supplementary text).
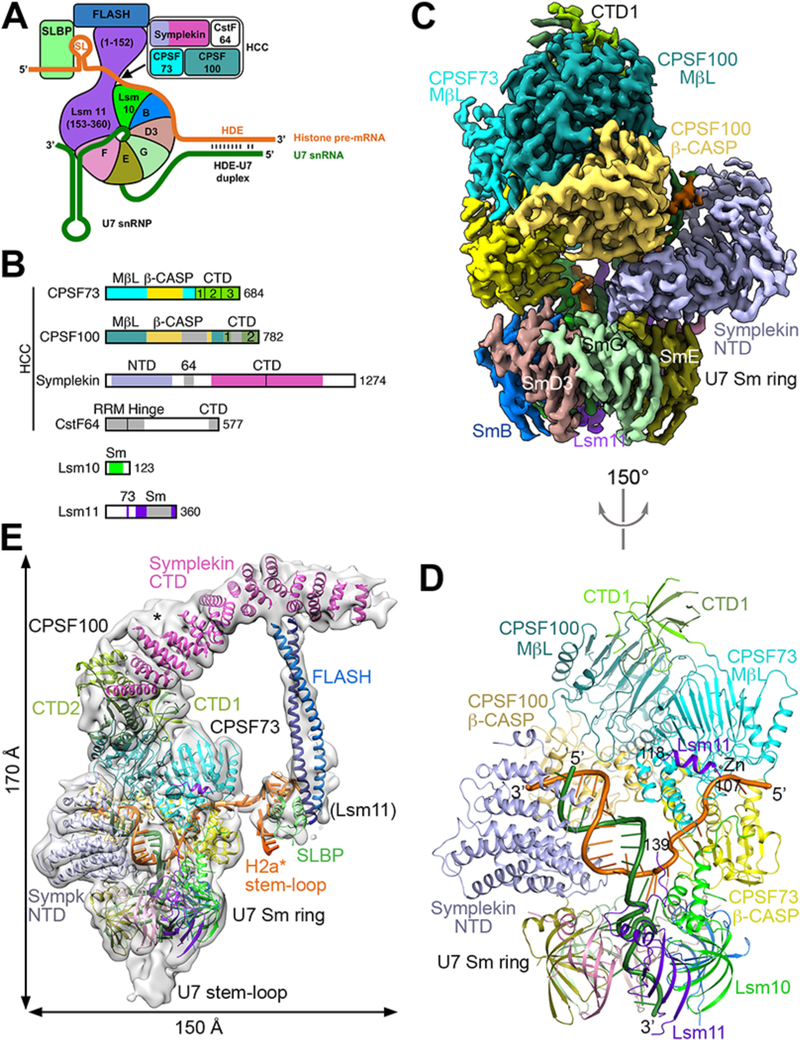
Figure 1. Overall structure of the human histone pre-mRNA 3′-end processing machinery.
(A) Schematic drawing of the histone pre-mRNA 3′-end processing machinery. (B) Domain organizations of the subunits of HCC, and Lsm10 and Lsm11. The domains in CPSF100 are given slightly darker colors compared to their homologs in CPSF73. The vertical bar in the symplekin CTD marks its N-terminal half that interacts with CPSF73. Abbreviations are defined in table S1. (C) Cryo-EM density at 3.2 Å resolution for the core of the machinery. (D) Schematic drawing of the structure of the core of the machinery, viewed after a 150° rotation around the vertical axis from panel C. The proteins are colored as in Figs. 1A,1B. The U7 snRNA is in dark green, and H2a* in orange. (E) Cryo-EM density for the entire machinery (gray), low-pass filtered to 8 Å resolution to show the density of FLASH and SLBP. The possible density for CTD3 of CPSF73 is indicated with the asterisk. Structure figures are produced with PyMOL (www.pymol.org) unless noted otherwise. Panels C and E produced with Chimera (30).
To prepare a fully recombinant machinery, we reconstituted human U7 snRNP (13) and mixed it with purified human HCC, FLASH (14), and SLBP (15). Using a modified mouse histone H2a pre-mRNA (H2a*, fig. S1) as substrate, we observed robust cleavage activity generating the authentic product (Supplementary text, figs. S2,3). Strikingly, the N-terminal domain (NTD) of symplekin was essential for processing, and its binding partner Ssu72 (16) inhibited the cleavage reaction. A mutation in the active site of CPSF73 abolished the cleavage.
We purified the active machinery (fig. S3F) and obtained a cryo-EM reconstruction at 3.2 Å resolution for its core (Figs. 1C,D), and a reconstruction at 4.1 Å resolution for the entire machinery (tables S2,3, figs. S4–6). The overall structure of the machinery resembles an amphora with one long handle (Fig. 1E, fig. S6B, movie S1). The machinery core constitutes the body of the amphora, with the U7 snRNA 3′-end SL and the Sm ring at the base and the CTDs of CPSF73 and CPSF100 and the first few helical repeats of the symplekin CTD forming the mouth. CPSF73 and symplekin NTD are positioned opposite each other on the Sm ring (Fig. 1D, fig. S6A). CPSF100 interacts with both CPSF73 and symplekin, but does not directly contact the Sm ring (Fig. 1C). The symplekin CTD, FLASH dimer (14), SLBP, pre-mRNA SL, and residues 20–65 of Lsm11 form the handle of the amphora (Fig. 1E). The FLASH dimer makes an 80 Å long connection from the symplekin CTD to the SLBP-SL complex. CstF64 was not observed in the EM density, and is not required for cleavage in vitro (Supplementary text, fig. S2F).
Twelve consecutive Watson-Crick base pairs in the HDE-U7 duplex were observed in the center of the amphora (Fig. 1D, fig. S1). The metallo-β-lactamase domain of CPSF73, the β-CASP domain of CPSF100, and the concave face of the symplekin NTD (fig. S6C) surround the duplex on three sides (Figs. 1D,1E,2A). The interactions are ionic and hydrophilic in nature, but involve none of the bases in the duplex (Fig. 2B), explaining that base pairing rather than sequence is important for processing (3, 4, 13). The structure revealed an extra, U-U base pair at the bottom of the duplex (Fig. 2C, fig. S1), and analysis of histone pre-mRNA sequences suggested that U-U base pairs are common in HDE-U7 duplexes (fig. S7).
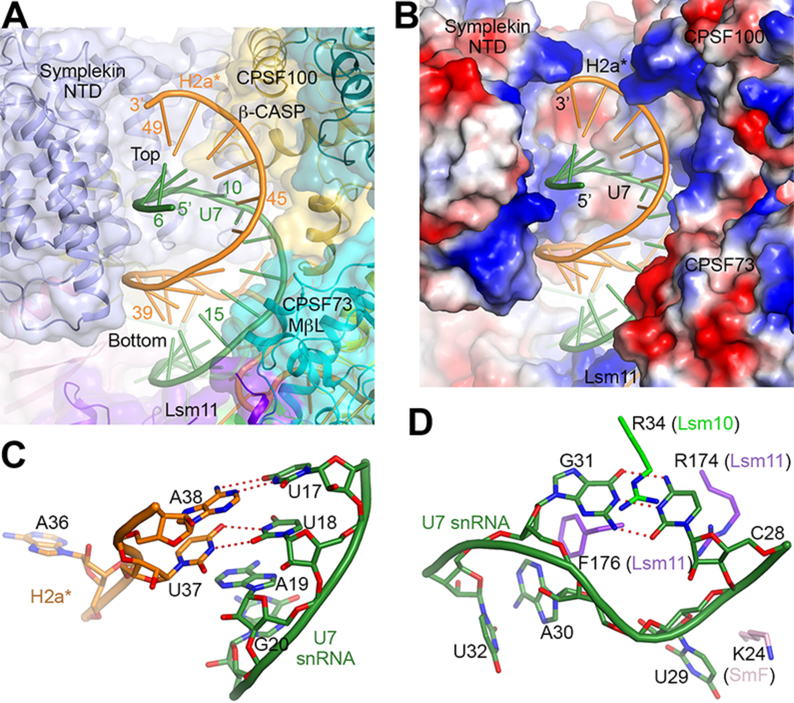
Figure 2. Recognition of the HDE-U7 duplex and the U7 Sm site.
(A) The HDE-U7 duplex is surrounded by CPSF73, CPSF100 and symplekin NTD, shown as a transparent surface. Lsm11 has interactions with the bottom of the duplex. (B) Electrostatic surface of the proteins in the duplex binding site, showing charged interactions with the backbone of the duplex. (C) A U-U base pair at the bottom of the duplex, flanked on the other face by A19 of U7 snRNA. (D) A C-G base pair in the 3′ CUAG sequence of the U7 Sm site. The base pair is flanked on one side by Arg34 of Lsm10, and on the other by Arg174 of Lsm11.
The structure also revealed a Watson-Crick base pair between C28 and G31 of the CUAG sequence at the 3′ end of the U7 Sm site (Fig. 2D, fig. S1). It is flanked by residues from Lsm10 and Lsm11 and assumes a different backbone conformation compared to other Sm sites (Fig. 2D, fig. S8A). In addition, G26 is hydrogen-bonded with C33 of H2a*, providing a direct connection between the Sm site and the pre-mRNA (figs. S1,S8B). The recognition of the first five Sm site nucleotides (21-AAUUU-25) and U27 is similar to that in spliceosomal Sm rings (figs. S1,S8B) (17, 18), although there are substantial differences in the extensions of the Sm proteins and the positions of the RNA outside the Sm ring (figs. S8C–E).
The pre-mRNA substrate (Fig. 3A) is bound in the active site of CPSF73. The correct scissile phosphate, after A26 (fig. S1), is coordinated to the two zinc ions in the active site (Fig. 3B). The A26 base has hydrogen-bonding interactions to its N1 and N6 atoms, consistent with the preference for an adenine at the cleavage site (3, 4) (figs. S9A–C). C25 has weak density (Fig. 3A) and is not recognized by CPSF73. This binding mode of the pre-mRNA clearly illuminated the molecular mechanism for the cleavage reaction. The hydroxide ion that is a bridging ligand between the two zinc ions (6) is the nucleophile that initiates the cleavage reaction (Fig. 3B), and the 3′ oxyanion of A26, the leaving group, is protonated by His396, which is activated by Glu204. Glu204, His396 and the ligands to the zinc ions are conserved among CPSF73 homologs (19, 20) including IntS11, the endonuclease for snRNA cleavage (21). Therefore, the conformation of the machinery observed here is likely poised for the cleavage reaction. Except for the brief moment during EM grid preparation, the sample was kept at 4 °C or on ice, which slowed the reaction (12) and allowed us to observe the pre-mRNA in the CPSF73 active site. There are substantial differences in the orientation of the β-CASP domain compared to that in RNase J (22, 23) (fig. S9D), and especially in the binding modes of the RNA substrate (fig. S9E).
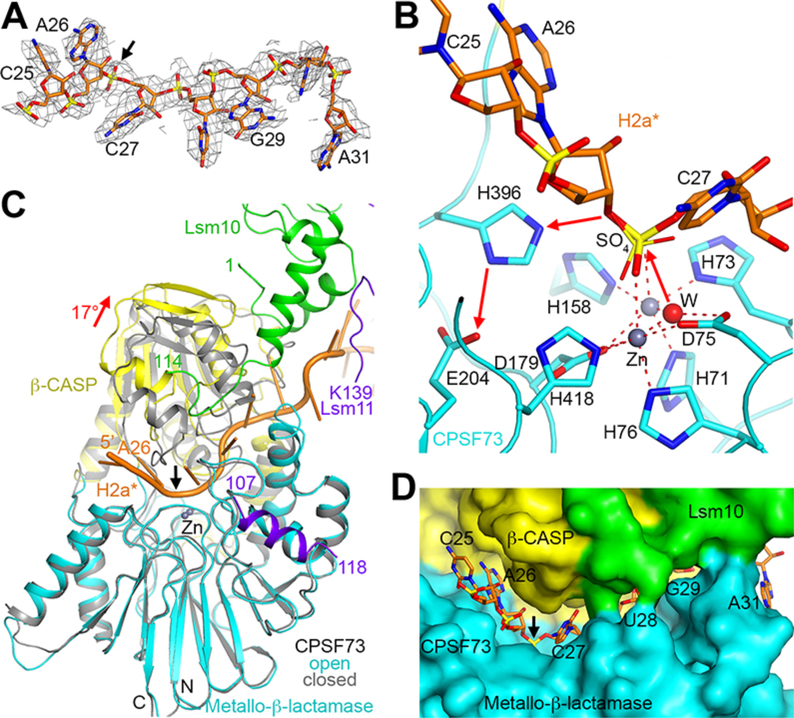
Figure 3. CPSF73 is in an active state, poised for the cleavage reaction.
(A) Cryo-EM density for H2a* nucleotides bound in the CPSF73 active site. The scissile phosphate is indicated with the black arrow. (B) The endonuclease mechanism of CPSF73. The positions of the zinc ions (gray spheres) and the bridging hydroxide (red sphere) are based on the crystal structure of CPSF73 alone (6) (PDB entry 2I7V). The position of the sulfate ion observed in the earlier structure is shown in thin sticks. (C) Overlay of the structure of CPSF73 in the active state observed here (in color) with the inactive, closed state reported earlier (gray) (6). The metallo-β-lactamase domain was used for the overlay. The rearrangement of the β-CASP domain is indicated with the red arrow, corresponding to a rotation of 17°. (D) Molecular surface of the active site region of CPSF73, colored by the domains. Lsm10 is located at the rim of the canyon, contacting nucleotides downstream of the cleavage site.
The reported structures of CPSF73 (6) and its yeast homolog Ysh1 (24) are in a closed, inactive conformation. We observed here an open, active conformation of CPSF73. A large rearrangement of its β-CASP domain relative to the metallo-β-lactamase domain, corresponding to a rotation of ~17° (Fig. 3C, fig. S9F), is necessary to create a narrow, deep canyon that is only large enough to accommodate single-stranded RNA (Fig. 3D, figs. S9A,F,G).
The N- and C-terminal extensions of Lsm10, highly conserved among vertebrate homologs (fig. S10A), have a crucial role in this conformational change for CPSF73. These extensions are placed directly against the β-CASP domain (fig. S11A) and have extensive steric clashes with its closed conformation (Fig. 3C, fig. S9F), likely helping to trigger the activation of CPSF73. In addition, a segment in the C-terminal extension of Lsm10 (residues 107–110) is positioned at the rim of the canyon (fig. S9A) and forms a part of the binding site for the 3′ portion of the substrate (Fig. 3D).
The recognition of the HDE-U7 duplex may be the critical event to initiate the conformational rearrangement in CPSF73, consistent with the requirement of symplekin NTD for cleavage. On the other hand, the NTD-Ssu72 complex is incompatible with the structure observed here, as Ssu72 would clash with the duplex as well as CPSF73 (fig. S6D), explaining the inhibitory effect of Ssu72 (Supplementary text).
Besides the rearrangement in CPSF73, an extensive change in the architecture of HCC is required for activation. We recently showed that mCF/HCC in an inactive state has a trilobal structure and is highly dynamic (25). In contrast, the HCC structure observed here in the active state shows dramatic differences compared to the inactive state (Fig. 4A). There are intimate contacts between CPSF73 and CPSF100 in the current structure; in fact they form a pseudo dimer (figs. S11B,C). These may be hallmarks of the active state for HCC/mCF.
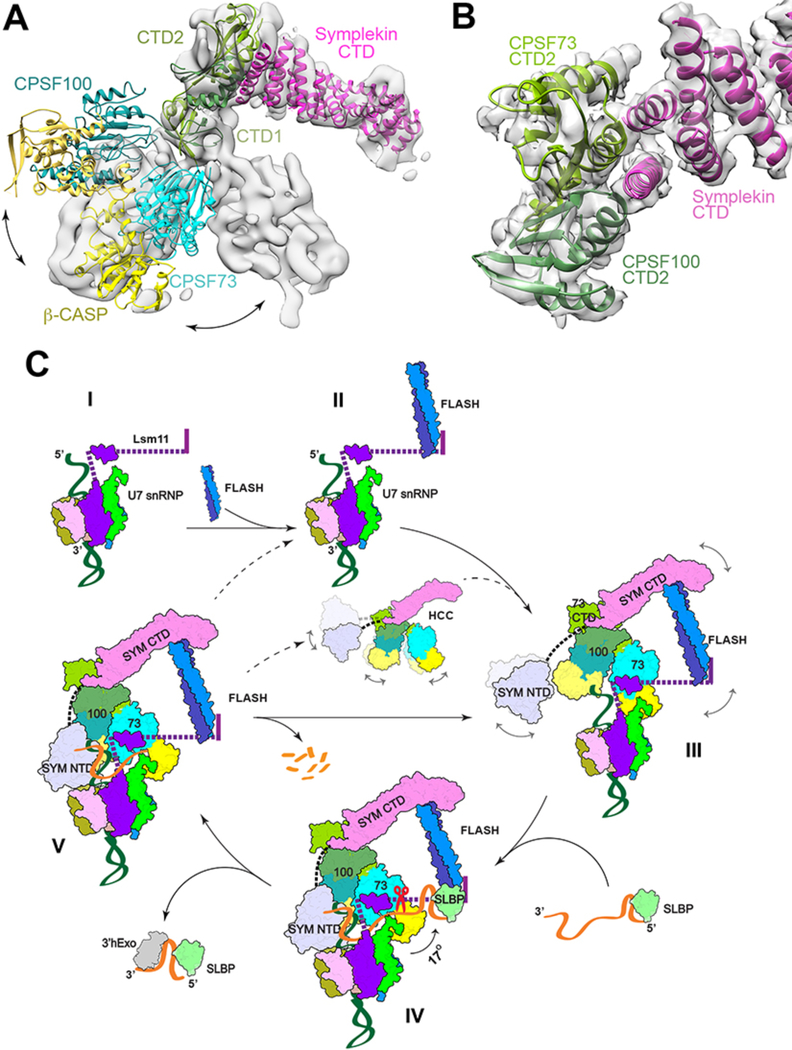
Figure 4. Schematic of histone pre-mRNA 3′-end processing cycle.
(A) Significant structural differences of HCC in an active state compared to an inactive state. The structure of HCC observed here is docked into the EM density for mCF (gray surface) (25), using the symplekin CTD as the reference. (B) Schematic drawing of the CTD2 domain complex of CPSF73 (light green) and CPSF100 (darker green) and the N-terminal segment of the symplekin CTD (magenta). The CTD complex of IntS9 and IntS11 (28) was docked into the EM density at 4.1 Å resolution (transparent surface) using Chimera. Panels A and B produced with Chimera. (C) A putative model for histone pre-mRNA 3′-end processing cycle. The machinery is assembled from the U7 snRNP (state I) with the recruitment of FLASH (II) and HCC (III), followed by the recognition of the pre-mRNA for CPSF73 and HCC activation and pre-mRNA cleavage (IV). The machinery is likely highly dynamic before the binding of the authentic pre-mRNA, and the possible flexible regions are indicated with the curved arrows and dashed lines. After the cleavage (V), the downstream product is degraded by an exonuclease activity and the machinery can be recycled directly (solid arrow), or possibly disassembled followed by re-assembly. State IV corresponds to the structure reported here, with the scissors indicating cleavage by CPSF73, and the other states are models.
The conformational dynamics of HCC/mCF is due to flexibility in its core, formed by the CTDs of CPSF73, CPSF100 and symplekin (10–12, 26, 27) (Figs. 1E,4B). The CTD of CPSF73 likely has three sub-domains (CTD1–3), and that of CPSF100 has two sub-domains (Figs. 1B,E, fig. S12A). The CTD1 sub-domains of CPSF73 and CPSF100 form a six-stranded β barrel-like structure (fig. S12B). The CTD2 sub-domains form a separate complex, which makes only a small contact with the CTD1 complex, contributing to the flexibility in HCC/mCF. The overall structure of the CTD2 complex is similar to that of IntS11 and IntS9 (28). The first two helices of the symplekin CTD pack against the helices in the CTD2 complex (Fig. 4B) in the core of HCC/mCF.
The structure showed that HCC is recruited to the machinery directly by both FLASH and Lsm11 through two tethering contacts. Residues in FLASH prior to the coiled-coil domain, including the LDLY motif (10), interact with the symplekin CTD (fig. S10C), and residues 107–118 of Lsm11 interact with CPSF73 (figs. S10B,S11D) (Supplementary text). These observations explain earlier data showing the importance of the LDLY motif in FLASH and residues 65–130 in Lsm11 for HCC recruitment (10, 11, 29).
Mutagenesis and biochemical experiments supported the structural observations (Supplementary text). HCC recruitment was abolished by mutating the FLASH LDLY motif or symplekin CTD (fig. S13A). Removing the N- and C-terminal extensions of Lsm10 greatly reduced the cleavage activity without affecting U7 snRNP or machinery assembly (figs. S13B–D). Moreover, the Lsm10 mutants showed mis-processing of the pre-mRNA. Therefore, these extensions may also have a crucial role in correctly positioning CPSF73 for the cleavage reaction. Mutating as few as two symplekin NTD residues that interact with the HDE-U7 duplex (fig. S6C) greatly reduced the cleavage activity (fig. S13E). Finally, the experiments also provided evidence for an Lsm11-FLASH-SLBP-SL quaternary complex (Fig. 1E, fig. S13F).
The structure of the machinery suggests how it may be assembled for processing (Fig. 4C, movie S2, Supplementary text) and provides a molecular foundation to understand and explain the large body of biochemical and functional data on histone pre-mRNA 3′-end processing (3, 4). The structure also has significant implications for understanding canonical pre-mRNA and snRNA 3′-end processing. The binding mode of the histone pre-mRNA in CPSF73 is likely similar for canonical pre-mRNAs and snRNAs, and the active conformation of mCF for canonical pre-mRNAs is likely to be the same as that of HCC observed here. The comparison to the structure of mCF in an inactive state suggested that its correct architecture is another critical requirement for the activation of the processing machineries.
Supplementary Material
A movie showing the composite cryo-EM map and atomic model of the overall histone pre-mRNA 3′-end processing machinery.
A movie showing the putative histone pre-mRNA 3′-end processing cycle. The subunits in the machinery are represented by their structures. Conformational changes in the HCC and CPSF73 for their activation are also indicated.
Acknowledgments
We thank Laura Yen, Daija Bobe, Ed Eng, and Bob Grassucci for data collection at the New York Structural Biology Center; Mark Ebrahim and Johanna Sotiris for grids screening at the Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center at The Rockefeller University; Kehui Xiang and Dazhi Tan for initial studies on this project.
Funding. This research is supported by NIH grants R35GM118093 (to LT) and R01GM029832 (to WFM and ZD). WSA was also supported by a fellowship from the Raymond and Beverley Sackler Center for Research at Convergence of Disciplines at Columbia University Medical Center. The Simons Electron Microscopy Center at the New York Structural Biology Center is supported by grants from the Simons Foundation (349247), NYSTAR, NIH (GM103310, S10 OD019994), and Agouron Institute (F00316).
Footnotes
Competing interests. The authors declare no competing interests.
Data availability. The atomic coordinates and the EM maps can be accessed in the Protein Data Bank (accession number 6V4X).
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Zhao J, Hyman L, Moore CL, Formation of mRNA 3’ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev 63, 405–445 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, Doublie S, Structural biology of poly(A) site definition. Wiley Interdiscip. Rev. RNA 2, 732–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominski Z, Marzluff WF, Formation of the 3’ end of histone mRNA: getting closer to the end. Gene 396, 373–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romeo V, Schumperli D, Cycling in the nucleus: regulation of RNA 3’ processing and nuclear organization of replication-dependent histone genes. Curr. Opin. Cell Biol 40, 23–31 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Dominski Z, Yang X-C, Marzluff WF, The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123, 37–48 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Mandel CR et al. , Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature 444, 953–956 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schonemann L et al. , Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Develop. 28, 2381–2393 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan SL et al. , CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3’ processing. Genes Develop. 28, 2370–2380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X-C, Burch BD, Yan Y, Marzluff WF, Dominski Z, FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3’ end processing of histone pre-mRNAs. Mol. Cell 36, 267–278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X-C et al. , A complex containing the CPSF73 endonuclease and other poyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3’-end processing. Mol. Cell. Biol 33, 28–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabath I et al. , 3’-end processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 19, 1726–1744 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skrajna A, Yang X-C, Dadlez M, Marzluff WF, Dominski Z, Protein composition of catalytically active U7-dependent processing complexes assembled on histone pre-mRNA containing biotin and a photo-cleavable linker. Nucl. Acids Res. 46, 4752–4770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucholc K et al. , Composition and processing activity of a semi-recombinant holo U7 snRNP. Nucl. Acids Res, doi: 10.1093/nar/gkz1148(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aik WS et al. , The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3’-end processing. PLoS One 12, e0186034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan D, Marzluff WF, Dominski Z, Tong L, Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3’hExo ternary complex. Science 339, 318–321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang K et al. , Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo Y, Oubridge C, van Roon AMM, Nagai K, Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5’ splice site recognition. eLife 4, e04986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Leung AKW, Kondo Y, Oubridge C, Nagai K, Re-refinement of the spliceosomal U4 snRNP core-domain structure. Acta Cryst. D72, 131–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P, Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucl. Acid Res 30, 3592–3601 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominski Z, Carpousis AJ, Clouet-d’Orval B, Emergence of the β-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 1829, 532–551 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Baillat D, Wagner EJ, Integrator: surprisingly diverse functions in gene expression. Trends Biochem. Sci. 40, 257–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorleans A et al. , Molecular basis for the recognition and cleavage of RNA by the bifunctional 5’−3’ exo/endoribonuclease RNase J. Structure 19, 1252–1261 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y et al. , Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J. Nucl. Acids Res. 43, 5550–5559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill CH et al. , Activation of the endonuclease that defines mRNA 3’ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell 73, 1217–1231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Sun Y, Shi Y, Walz T, Tong L, Structural insights into the human pre-mRNA 3’-end processing machinery. Mol. Cell, doi: 10.1016/j.molcel.2019.11.005(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullivan KD, Steiniger M, Marzluff WF, A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34, 322–332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michalski D, Steiniger M, In vivo characterization of the Drosophila mRNA 3’ end processing core cleavage complex. RNA 21, 1404–1418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Albrecht TR, Baillat D, Wagner EJ, Tong L, Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance. Proc. Natl. Acad. Sci. USA 114, 4394–4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skrajna A et al. , Mapping the interaction network of key proteins involved in histone mRNA generation: a hydrogen/deuterium exchange study. J. Mol. Biol 428, 1180–1196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF et al. , UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Sari D et al. , The MultiBac baculovirus/insect cell expression vector system for producing complex protein biologics. Adv. Exp. Med. Biol 896, 199–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung AKW et al. , Use of RNA tertiary interaction modules for the crystallisation of the spliceosomal snRNP core domain. J. Mol. Biol 402, 154–164 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Ohi M, Li Y, Cheng Y, Walz T, Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A, Grigorieff N, CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zivanov J et al. , RELION-3: new tools for automated high-resolution cryo-EM structure determination. bioRxiv, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Kucukural A, Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan KD, Coot: model-building tools for molecular graphics. Acta Cryst. D60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Adams PD et al. , PHENIX: building a new software for automated crystallographic structure determination. Acta Cryst. D58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Madeira F et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucl. Acids Res 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouet P, Courcelle E, Stuart DI, Metoz F, ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Yang X-C et al. , FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5’ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol 31, 1492–1502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strub K, Galli G, Busslinger M, Birnstiel ML, The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 3, 2801–2807 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mowry KL, Steitz JA, Identification of the human U7 snRNP as one of several factors involved in the 3’ end maturation of histone premessenger RNA’s. Science 238, 1682–1687 (1987). [DOI] [PubMed] [Google Scholar]
- 46.Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B, Purified U7 snRNAs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20, 5470–5479 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillai RS et al. , Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Develop. 17, 2321–2333 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolev NG, Steitz JA, Symplekin and multiple other polyadenylation factors participate in 3’-end maturation of histone mRNAs. Genes Develop. 19, 2583–2592 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimm C, Stefanovic B, Schumperli D, The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12, 1229–1238 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolev NG, Steitz JA, In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat. Struct. Mol. Biol 13, 347–353 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Zhang J et al. , Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc. Natl. Acad. Sci. USA 111, E2937–E2946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skrajna A et al. , U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein. RNA 23, 938–951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghazy MA, He X, Singh BN, Hampsey M, Moore C, The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensible for pre-mRNA 3’-end processing. Mol. Cell. Biol 29, 2296–2307 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streit A, Koning TW, Soldati D, Melin L, Schumperli D, Variable effects of the conserved RNA hairpin element upon 3’ end processing of histone pre-mRNA in vitro. Nucl. Acids Res. 21, 1569–1575 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominski Z, Zheng LX, Sanchez R, Marzluff WF, Stem-loop binding protein facilitates 3’-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol 19, 3561–3570 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wani S et al. , Vertebrate Ssu72 regulates and coordinates 3’-end formation of RNAs transcribed by RNA polymerase II. PLoS One 9, e106040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He X et al. , Functional interactions between the transcription and mRNA 3’ end processing machineries mediated by Ssu72 and Sub1. Genes Develop. 17, 1030–1042 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krishnamurthy S, He X, Reyes-Reyes M, Moore CL, Hampsey M, Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D, Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell 22, 91–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scharl EC, Steitz JA, The site of 3’ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13, 2432–2440 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scharl EC, Steitz JA, Length suppression in histone messenger RNA 3’-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. USA 93, 14659–14664 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolev NG, Yario TA, Benson E, Steitz JA, Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3’-end maturation. EMBO Rep. 9, 1013–1018 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatomer DC et al. , Drosophila symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus 5, 613–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gick O, Kramer A, Vasserot A, Birnstiel ML, Heat-labile regulatory factor is required for 3’ processing of histone precursor mRNAs. Proc. Natl. Acad. Sci. USA 84, 8937–8940 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X-C, Sullivan KD, Marzluff WF, Dominski Z, Studies of the 5’ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol 29, 31–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eaton JD et al. , Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Develop. 32, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatomer DC et al. , Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J. Cell. Biol 213, 557–570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goddard TD et al. , UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marz M, Mosig A, Stadler BMR, Stadler PF, U7 snRNAs: a computational survey. Geno. Prot. Bioinfo 5, 187–195 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Armon A, Graur D, Ben-Tal N, ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol 307, 447–463 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Crooks GE, Hon G, Chandonia J-M, Brenner SE, WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furger A, Schaller A, Schumperli D, Functional importance of conserved nucleotides at the histone RNA 3’ processing site. RNA 4, 246–256 (1998). [PMC free article] [PubMed] [Google Scholar]
- 73.Qu X et al. , The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3’-end processing. J. Biol. Chem 282, 2101–2115 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A movie showing the composite cryo-EM map and atomic model of the overall histone pre-mRNA 3′-end processing machinery.
A movie showing the putative histone pre-mRNA 3′-end processing cycle. The subunits in the machinery are represented by their structures. Conformational changes in the HCC and CPSF73 for their activation are also indicated.