Current knowledge of genetic mechanisms underlying neurodegeneration is based on data “snapshots” taken at a few disease stages. Iturria-Medina et al. overcome this limitation by developing a machine-learning algorithm that can infer the intrinsic temporal information contained in large-scale neurodegenerative datasets.
Keywords: gene expression trajectories, neurodegenerative progression, unsupervised machine learning, neuropathological mechanisms, personalized treatments
Abstract
Most prevalent neurodegenerative disorders take decades to develop and their early detection is challenged by confounding non-pathological ageing processes. For all neurodegenerative conditions, we continue to lack longitudinal gene expression data covering their large temporal evolution, which hinders the understanding of the underlying dynamic molecular mechanisms. Here, we overcome this key limitation by introducing a novel gene expression contrastive trajectory inference (GE-cTI) method that reveals enriched temporal patterns in a diseased population. Evaluated on 1969 subjects in the spectrum of late-onset Alzheimer’s and Huntington’s diseases (from ROSMAP, HBTRC and ADNI datasets), this unsupervised machine learning algorithm strongly predicts neuropathological severity (e.g. Braak, amyloid and Vonsattel stages). Furthermore, when applied to in vivo blood samples at baseline (ADNI), it significantly predicts clinical deterioration and conversion to advanced disease stages, supporting the identification of a minimally invasive (blood-based) tool for early clinical screening. This technique also allows the discovery of genes and molecular pathways, in both peripheral and brain tissues, that are highly predictive of disease evolution. Eighty-five to ninety per cent of the most predictive molecular pathways identified in the brain are also top predictors in the blood. These pathways support the importance of studying the peripheral-brain axis, providing further evidence for a key role of vascular structure/functioning and immune system response. The GE-cTI is a promising tool for revealing complex neuropathological mechanisms, with direct implications for implementing personalized dynamic treatments in neurology.
Introduction
In recent decades, we have witnessed an accelerated characterization of the molecular and neuropathological mechanisms underlying neurodegenerative progression. Thanks to cutting-edge technological and methodological advances in genomic and proteomic analysis, we foresee unlimited methodological possibilities for understanding and modifying the role of genes and protein in disease (Esvelt and Wang, 2012; Tan et al., 2012; Smith et al., 2016; Mostafavi et al., 2018). Gene expression examination has been of crucial value, revealing disease-specific differentiated genes/molecular pathways and gene-gene networks with a direct effect in neuropathological and cognitive/clinical deterioration (Zhang et al., 2013; Mostafavi et al., 2018). However, neurodegenerative conditions may take decades to develop and gene expression mapping techniques are quite recent, hence the unavailability of individual gene expression datasets covering a given disease’s whole evolution. All reported studies are based on cross-sectional or short-term longitudinal data, while we continue to lack long-term datasets covering the several phases underlying neurodegeneration.
In addition, because of its highly invasive nature, brain gene expression studies in neurodegeneration are based on post-mortem tissue samples. There are major challenges associated with the translation/extrapolation of ex vivo results to in vivo conditions (Ferreira et al., 2018). This could imply that disease mechanisms (e.g. gene-gene causal networks) and potential biomarkers identified with post-mortem data may well not be entirely generalizable to live patients. In this sense, peripheral molecular measurements (e.g. plasma gene expression) may be used to cross-validate post-mortem based methodologies and findings, potentially providing minimally invasive in vivo biomarkers for accurate patient screening in the daily clinic and clinical trials implementation. Nevertheless, the lack of comprehensive longitudinal peripheral datasets, covering multiple disease stages at the individual level, makes in vivo dynamic molecular analyses unpractical. Consequently, this affects the identification of robust peripheral biomarkers across continuous disease stages and variants.
Because of the proven ability to disentangle temporal components from high-dimensional cross-sectional data, novel unsupervised machine learning techniques offer a viable opportunity for dealing with the previous limitations. The data-driven reconstruction of pseudo-temporal paths to order observations (e.g. cells, subjects) is revolutionizing ‘omics’ studies, enabling for the first time the mapping of complex dynamic processes using cross-sectional ‘snapshots’ (Magwene et al., 2003; Gupta and Bar-Joseph, 2008; Cannoodt et al., 2016; Welch et al., 2016). Based on the machine learning inference of a low dimensional space embedded in a population’s ‘omics’ data, and by creating a relative ordering of the individuals, we can accurately identify a series of molecular states that constitute a longitudinal trajectory for a process of interest (Campbell and Yau, 2018). When used in RNA-seq studies, this novel technique has provided an unprecedented insight into the evolution of multiple pathologies. It has also allowed tracking and dissecting differentiated spatiotemporal programs in single-cell analysis (Briggs et al., 2018).
Driven by the imperative of a better understanding and an earlier detection of neurodegeneration, here we extend pseudotemporal trajectory inference to the analysis of both post-mortem and in vivo (blood) gene expression neurodegenerative samples. First, to address important methodological limitations in data exploration and visualization, we introduce the contrastive trajectory inference (cTI) algorithm. This allows the unsupervised identification and ordering of enriched patterns in a diseased population (e.g. Alzheimer’s and Huntington’s diseases) relative to a comparison background population (e.g. healthy elderly). Next, we analyse gene expression samples from blood plasma of 744 subjects in the spectrum of late-onset Alzheimer’s disease (LOAD) and from 1225 autopsied brains in the spectrum of LOAD and Huntington’s disease. Our method provides molecular pathological scores that are highly predictive of neuropathological and cognitive/clinical deterioration. The results are strongly consistent for both in vivo and post-mortem data. In addition, it allows identification of genes and molecular pathways driving neurodegenerative progression, as well as analysis of (dis)similarities in molecular disease mechanisms at brain and peripheral tissue levels. The inference of contrasted genetic trajectories is a promising tool for understanding complex neuropathological mechanisms and for minimally invasive patient screening at the daily clinic, with practical implications for implementing personalized medical interventions in neurology.
Materials and methods
Study participants
This study used gene expression data (ntotal = 1969) from three large-scale databases (see Supplementary Table 1 for demographic characteristics). Each dataset was processed and analysed independently.
Dataset 1
RNA expression data from the prefrontal cortex of a subset of 489 autopsied subjects were downloaded from the Religious Orders Study (ROS) (Bennett et al., 2012a) and the Memory and Aging Project Study (MAP) (Bennett et al., 2012b). These data (Bennett et al., 2018) are available at the Accelerating Medicines Partnership Alzheimer’s Disease (AMP-AD) knowledge portal (https://www.synapse.org/#, Synapse ID 3800853). ROS (Bennett et al., 2012a) and MAP (Bennett et al., 2012b) are longitudinal clinical-pathological cohort studies of ageing, Alzheimer’s disease and related disorders. Enrolment required no known sign of dementia. Upon death, a post-mortem neuropathological evaluation is performed that includes a uniform structured assessment of Alzheimer’s disease pathology, cerebral infarcts, Lewy body disease, and other pathologies common in ageing and dementia. The pathological diagnosis of Alzheimer’s disease uses NIA-Reagan and modified CERAD criteria, and the staging of neurofibrillary pathology uses Braak staging (Braak, 1991). An RNA integrity number (RIN) score >5 and a quantity threshold (5 mg) for each sample were required (Bennett et al., 2014). cRNA was hybridized to Illumina HT-12 Expression Bead Chip (48 803 transcripts) via standard protocols using an Illumina Bead Station 500GX (Webster et al., 2009; Zhang et al., 2013).
Dataset 2
Seven hundred and thirty-six individual post-mortem tissue samples from the dorsolateral prefrontal cortex Brodmann area (BA)9 of LOAD patients (n = 376), Huntington’s disease patients (n = 184) and non-demented subjects (n = 173) were collected and analysed (Zhang et al., 2013). All autopsied brains were collected by the Harvard Brain Tissue Resource Center (HBTRC; GEO accession number GSE44772), and included subjects for whom both the donor and the next-of-kin had completed the HBTRC informed consent (http://www.brainbank.mclean.org/). Correspondingly, tissue collection and the research were conducted according to the HBTRC guidelines (http://www.brainbank.mclean.org/). Post-mortem interval (PMI) was 17.8 ± 8.3 h, sample pH was 6.4 ± 0.3 and RIN was 6.8 ± 0.8 for the average sample in the overall cohort.
As previously described (Zhang et al., 2013), RNA preparation and array hybridizations applied custom microarrays manufactured by Agilent Technologies consisting of 4720 control probes and 39 579 probes targeting transcripts representing 25 242 known and 14 337 predicted genes. Arrays were quantified based on spot intensity relative to background, adjusted for experimental variation between arrays using average intensity over multiple channels, and fitted to an error model to determine significance (Emilsson et al., 2008). Braak stage, general and regional atrophy, grey and white matter atrophy and ventricular enlargement were assessed and catalogued by pathologists at McLean Hospital (Belmont, MA, USA). In addition, the severity of pathology in the Huntington’s disease brains was determined using the Vonsattel grading system (Vonsattel et al., 1985).
Dataset 3
This study used a total of 744 individuals’ data with blood gene expression information from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (adni.loni.usc.edu). The participants underwent multimodal brain imaging evaluations, including amyloid PET, tau PET and/or structural MRI. The ADNI was launched in 2003 as a public-private partnership, led by principal investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease.
The Affymetrix Human Genome U219 Array (www.affymetrix.com) was used for gene expression profiling from blood samples. Peripheral blood samples were collected using PAXgene™ tubes for RNA analysis (Saykin et al., 2015). The quality-controlled gene expression data include activity levels for 49 293 transcripts. All the participants were characterized cognitively using the Mini-Mental State Examination (MMSE), a composite score of executive function, a composite score of memory integrity (MEM) (Gibbons et al., 2012), and Alzheimer's Disease Assessment Scale-Cognitive Subscales 11 and 13 (ADAS-11 and ADAS-13, respectively). Also, they were clinically diagnosed at baseline as healthy control, early MCI, late MCI or probable Alzheimer’s disease patient (LOAD).
18F-AV-45 (amyloid-specific) and 18F-AV-1451 (tau-specific) PET images were acquired for a subset of 660 and 166 subjects, respectively. Both amyloid and tau images were preprocessed by the Jagust Laboratory (UC Berkeley, USA; Jagust et al., 2010). Using the amyloid images, subjects were categorized as amyloid positive (Aβ+) or negative (Aβ−) by applying a cut-off of 1.11 to a florbetapir composite standardized uptake value ratio (SUVR) normalized by the whole cerebellum reference (described in Landau and Jagust, 2015). Also, individual Freesurfer-defined cortical and subcortical brain regions were used to calculate weighted Flortaucipir averages for each region, which were normalized by the weighted flortaucipir at the cerebellum (described in Landau and Jagust, 2018). Based on the lobar classification topographic staging scheme for tau PET and the corresponding cut-off values proposed by Schwarz et al. (2018), the subjects were staged in Braak 0 (no tau), Braak I/II, Braak III/IV or Braak V/VI. Subsequently, they were categorized as tau negative (tau−) or positive (tau+) if they were in the stages 0 or I–VI, respectively. Structural MRI images for 741 subjects were analysed by a physician specially trained in the detection of MRI infarcts. The presence of MRI infarction was determined from the size, location and imaging characteristics of the lesion, with only lesions 3 mm or larger qualifying for consideration as cerebral infarcts (described in DeCarli et al., 2013). Finally, a subset of subjects (n = 30) was evaluated for pathological brain lesions after death. Pathological lesions were assessed using established neuropathological diagnostic criteria (described in Cairns, 2018). The analysis included histopathological assessments of amyloid-β deposits, staging of neurofibrillary tangles, scoring of neuritic plaques and assessments of co-morbid conditions such as Lewy body disease, vascular brain injury, hippocampal sclerosis, and TAR DNA-binding protein (TDP) immunoreactive inclusions (Montine et al., 2012).
Contrastive trajectories inference
Given a multi-dimensional population dataset, the inference of contrasted pseudotemporal trajectories (and an individual pseudotime value) consists of three main steps:
- (i) For high-dimensional datasets (e.g. ∼40 000 transcripts), initial selection of features most likely to be involved in a trajectory across the entire population. We apply the unsupervised method proposed by Welch et al. (2016), which does not require prior knowledge of features involved in the process or differential expression analysis. Features are scored by comparing sample variance and ‘neighbourhood variance’. Specifically, for a gene transcript g, its sample variance across all samples is calculated. Then, the ‘neighbourhood variance’ is computed as:
(1)
where is the total number of gene transcripts, eij is the expression level of the jth transcript in the ith sample, N(i, j) is the jth nearest neighbour of sample i, and kc is the minimum number of neighbours needed to yield a connected graph. is similar to the sample variance computed with respect to neighbouring points rather than the mean, measuring how much g varies across neighbouring samples. Intuitively, gene transcripts most likely to be involved in a trajectory should present a more gradual variation across neighbouring points than at global scale, which would correspond to a high ratio . Thus, a threshold is applied to select those features with higher score, e.g. we kept the features with at least a 0.95 probability of being involved in a trajectory (i.e. ∼3000 gene transcripts).
(ii) Data exploration and visualization via contrastive principal component analysis (cPCA) (Abid et al., 2018). This novel technique identifies low-dimensional patterns that are enriched in a target dataset (e.g. a diseased population) relative to a comparison background dataset (e.g. demographically matched healthy subjects). By controlling the effects of characteristic patterns in the background (e.g. pathology-free and spurious associations, noise), cPCA (Abid et al., 2018) allows visualizing specific data structures missed by standard data exploration and visualization methods (e.g. PCA, Kernel PCA). Specifically, if Ctarget and Cbackground are the covariance matrices of the target and background data, the directions returned by cPCA are the singular vectors of the weighted difference of the co-variance matrices: Ctarget − α·Cbackground. The contrast parameter α represents the trade-off between having the high target variance and the low background variance. Multiple values of α are used (i.e. 100 logarithmically equally spaced points between 10−2 and 102). Instead of choosing a single α, the resulting subspaces for all the α-values are clustered (based in their proximity in terms of the principal angle and spectral clustering) (Ng et al., 2002) in a few subspaces. The data are then projected onto each of these few subspaces, revealing different trends within the target data. While the original cPCA algorithm proposes to select the final subspace via visual examination, we chose automatically the subspace that maximizes the clustering tendency in the projected target data. For this, the ‘gap’ cluster evaluation criterion, implemented in the MATLAB function evalclusters, was used.
When cPCA was applied to the selected gene expression transcripts [from step (i)], for each population, we obtained about six to eight contrasted principal components capturing the most enriched pathological properties relative to the background (i.e. subjects without cognitive deterioration and neuropathological signs). For ROSMAP, HBTRC and ADNI, sample sizes of the background populations were 177 (36%), 173 (23%) and 113 (15%), respectively. Selected α-values for these three studied datasets were: 11.76 (ROSMAP), 17.07 (HBTRC) and 11.76 (ADNI).
(iii) Subject ordering and gene expression-pseudotime calculation according to their proximity to the background population in the contrasted principal components space. For this, we first calculated the Euclidean distance matrix among all the subjects and the associated minimum spanning tree (MST). The MST was then used to calculate the shortest trajectory/path from any subject to the background subjects. Each specific trajectory consists of the concatenation of relatively similar subjects, with a given behaviour in the data’s dimensionally reduced space. The position of each subject in his/her corresponding shortest trajectory reflects the individual proximity to the pathology-free state (the background) and, if analysed in the inverse direction, to advanced disease state. Thus, to quantify the distance to these two extremes (background or disease), an individual gene expression-pseudotime score is calculated as the shortest distance value to the background’s centroid, relative to the maximum population value (i.e. values are standardized between 0 and 1). Finally, the subjects are ordered according to their gene expression-pseudotime values, from low (close to the background group) to high values (close to the most diseased subjects).
Additionally, to evaluate cPCA’s performance versus other popular dimensionality reduction techniques, we repeated step (ii) using the traditional PCA (Abdi and Williams, 2010) and the recently proposed non-linear Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) approach (McInnes et al., 2018). Subsequently, we reapplied step (iii), obtaining alternative subject orderings (and gene expression-pseudotimes) according to their proximity to the background population in the resulting PCA and UMAP components space.
Statistics
Data preprocessing
Before applying the contrastive trajectory inference (cTI) approach, each gene transcript’s activity was adjusted for relevant covariates using robust additive linear models (Street et al., 1988). Specifically, Dataset 1 gene expression was adjusted for post-mortem interval (PMI) in hours, age, gender and educational level. Dataset 2 gene expression was adjusted for PMI, sample pH, RIN, age and gender. Dataset 3 gene expression was controlled for RIN, plate number, age, gender and educational level. Also, each adjusted gene transcript activity was approximately transformed into a normal distribution via the Box-Cox transformation (Box and Cox, 1964), implemented in the MATLAB function boxcox.
Post hoc analyses
All predictive associations between grouping variables (e.g. Braak, CERAD and Vonsattel stages, clinical diagnosis) and the individual gene expression-pseudotimes were tested with ANOVA tests, familywise error (FWE)-controlled by permutations (Legendre and Legendre, 1998). For each dataset, the total contribution of each gene transcript i to the obtained reduced representation space (and the genetic trajectories) was quantified as in Abdi and Williams (2010):
| (2) |
where is the normalized eigenvalue of the contrasted principal component j, is the minimum obtained eigenvalue, is the original number of contrasted principal components, is the number of contrasted principal components with over a predefined cut-off value (i.e. 0.025), is the loading/weight of the gene transcript i on the component j, and is the total number of gene transcripts considered in the dimensionality reduction analysis. Similarly, the expected contribution value (cut-off) was calculated as in Abdi and Williams (2010):
| (3) |
The gene transcripts with total contribution over the expected contribution value were considered as highly influential to obtain the reduced representation space.
Data and code availability
The three datasets used in this study are available at the AMP-Alzheimer’s disease knowledge portal (https://www.synapse.org/#, Synapse ID 3800853), the Gene Expression Omnibus (GEO accession number GSE44772) and the ADNI database (www.adni.loni.usc.edu), respectively. We anticipate that the cTI method will be released soon as part of an open access user-friendly software. In the meantime, the MATLAB codes can be downloaded from http://www.neuropm-lab.com.
Results
Inferring enriched gene expression neurodegenerative trajectories
Gene expression, neuropathology and cognitive/clinical deterioration in 1969 demented and non-demented subjects from three large-scale studies were assessed (Fig. 1 and Datasets 1–3). Gene expression and neuropathology evaluations from both Dataset 1 (n = 489, ROSMAP Study) and Dataset 2 (n = 736, HBTRC database) were performed in autopsied brains, with genetic profiling from the prefrontal cortex. Gene expression from Dataset 3 (n = 744, ADNI database) was obtained from in vivo blood samples, with all subjects also having brain imaging evaluations including amyloid PET, tau PET and/or structural MRI.
Figure 1.
Schematic approach for gene expression trajectories analysis in neurodegeneration. (1) In vivo blood (n = 744) and post-mortem brain (n = 1225) tissues collected. (2) RNA expression for ∼40 000 transcripts (dataset-specific). (3) The high dimensional data are automatically reduced to an enriched space (about five features) via a contrastive PCA algorithm (cPCA) (Abid et al., 2018), which optimizes the exploration and visualization of the target population’s data. (4) In the contrasted principal components (cPC) space, each subject is assigned to a gene expression trajectory. The subject’s position in the corresponding gene expression trajectory reflects the individual proximity to the pathology-free state (the background) and, if analysed in the inverse direction, to the advanced disease state. An individual gene expression-pseudotime score is calculated, reflecting the distance to these two extremes (background or disease). (5) When taken as an individual molecular score of disease evolution, the gene expression-pseudotime significantly associates with neuropathological and/or cognitive measurements. (6) Both in peripheral and brain tissues, the cPCA’s loadings (or weights) allow the identification and posterior functional analysis of most informative genes in terms of pathological evolution.
Aiming to uncover the molecular reconfigurations underlying neurodegenerative evolution, we proceeded to reorder the gene expression patterns (Fig. 1). For this, we implemented a novel unsupervised algorithm for detecting enriched trajectories in a diseased population relative to a background dataset (e.g. normal controls). A distinctive feature of cTI is the use of a contrastive PCA algorithm (Abid et al., 2018), which controls by the principal components of the background data to optimize the exploration and visualization of the target. It is a generic algorithm, adaptable to different types of data (e.g. genomic, proteomic, imaging, clinical). Each gene expression dataset was first adjusted for relevant confounding covariates (e.g. RIN, age, gender and/or educational level). Next, the cTI was independently applied to the three populations, providing population-specific trajectories starting on the background data. Each trajectory was composed by the concatenation of a subset of subjects, which followed a given behaviour in the data’s dimensionally reduced space. We hypothesized that the position of each subject in these gene expression trajectories would reflect individual proximity to the pathology-free state (the background) or, if analysed in the inverse direction, proximity to advanced disease states. Correspondingly, a gene expression-pseudotime value [(0, 1) range] was calculated for each subject, with relatively low values for subjects with final positions close to the background data, and high values for subjects on the distant extremes of the population. Notice that gene expression-pseudotime could then be assumed as an individual molecular score of pathological progression, the validity of which is tested in the following sections (Fig. 1).
Post-mortem gene expression trajectories predict neurodegenerative severity
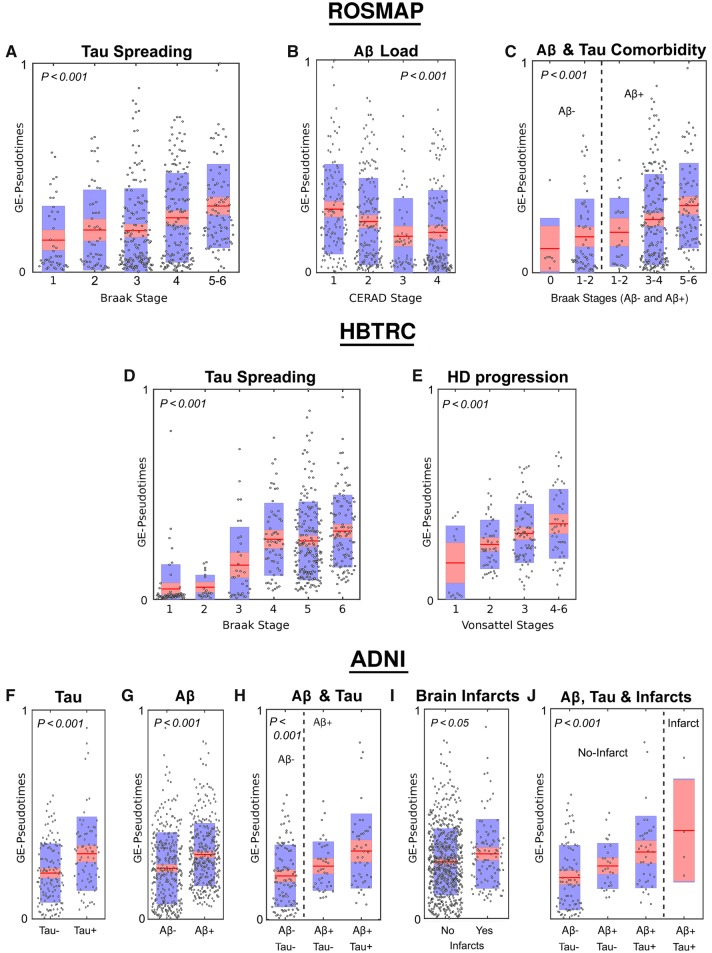
First, we analysed the gene expression trajectories obtained for the ROSMAP study (Dataset 1, n = 489). The results (Fig. 2A–C) showed a clear association between the obtained molecular disease score (gene expression-pseudotime) and the autopsied tau and amyloid assessments, with a higher gene expression-pseudotime value implying an advanced neuropathological state. Group differences in gene expression-pseudotime values were statistically tested via ANOVA tests with permutations. We found robust significant associations between the gene expression-pseudotimes and Braak stages (Fig. 2A; F = 4.09, P = 0.001, FWE-corrected), CERAD stages (Fig. 2B; F = 9.23, P < 0.001, FWE-corrected), and a composite variable (Braak+CERAD) reflecting the simultaneous presence of tau and amyloid (Fig. 2C; F = 5.97, P < 0.001, FWE-corrected).
Figure 2.
Gene expression-based predictions of neurodegenerative severity for ROSMAP, HBTRC and ADNI populations. (A–E) Gene expression-pseudotime predictive associations with Braak (A and D), CERAD (B), Braak for Aβ−/Aβ+ (C), and Vonsattel (E) stages in ROSMAP (A–E) and HBTRC (D and E). (F–J) Gene expression-pseudotime predictive associations with tau positivity (F), Aβ positivity (G), tau-Aβ co-morbidity (H), cerebral infarct occurrence (I) and tau-Aβ-infarct co-morbidity (J) in ADNI population. Points are laid over a 1.96 standard error of the mean (SEM) (95% confidence interval) in red and at 1 SD in blue. All P-values are FWE-corrected (see reported values in the ‘Results’ section). HD = Huntington’s disease.
Next, we explored the generalizability of these results in the considerably more heterogeneous database from HBTRC (Dataset 2, n = 736), including two different disorders (LOAD and Huntington’s disease) and non-demented controls. As with the previous findings, we observed a positive association between the individual molecular disease score and the levels of neuropathological affectation in both disorders (Fig. 2D and E). The gene expression-pseudotimes were significantly associated with the Braak stages (Fig. 2D; F = 11.17, P < 0.001, FWE-corrected) and the Vonsattel stages (Fig. 2E; F = 9.04, P < 0.001, FWE-corrected). The fact that this population included multiple disorders did not seem to affect the robustness of the subject ordering in relation to disease progression, which supports the identification of a promising biomarker for the analysis of co-morbid neurological conditions.
Importantly, when compared with the individual molecular disease scores obtained using the contrastive PCA algorithm (Abid et al., 2018) with those obtained using the traditional PCA and the novel non-linear UMAP approaches, we observed that cPCA-based results significantly outperformed PCA- and UMAP-based results. Essentially, the gene expression-pseudotimes obtained with PCA and UMAP did not show any significant association with neuropathological variables (all P > 0.3, FWE-corrected) (Supplementary Fig. 1). This finding strongly supports the key advantage of considering the enriched patterns in the population of interest relative to the background dataset (Abid et al., 2018).
Blood gene expression as a robust biomarker of in vivo neuropathological severity and clinical deterioration
Next, we aimed to investigate if the unsupervised ordering of gene expression patterns present in the blood can reflect neuropathological severity and, importantly, if it could be used as a marker of present and future clinical deterioration. If successful, the latter could have strong implications for the in vivo detection of future disease evolution in the clinic and to decide if a patient should be therapeutically treated or not. To test this, we identified the enriched gene expression trajectories in the plasma of 744 participants in the spectrum of LOAD from ADNI (Dataset 3), taking as background 113 subjects without cognitive/clinical alterations or any evidence of amyloid deposition or cerebral infarcts.
In line with our previous findings with the ROSMAP and HBTRC post-mortem data, the ADNI-based results (Fig. 2F–J) showed a significant predictive power of pathological severity. The individual gene expression-pseudotime values vastly reflected the differences in tau positivity (Fig. 2F; F = 17.64, P < 0.001, FWE-corrected), amyloid positivity (Fig. 2G; F = 28.22, P < 0.001, FWE-corrected), tau-amyloid co-morbidity (Fig. 2H; F = 9.58, P < 0.001, FWE-corrected), brain infarcts (Fig. 2I; F = 5.32, P < 0.05, FWE-corrected), and tau-amyloid infarcts co-morbidity (Fig. 2J; F = 7.49, P < 0.001, FWE-corrected).
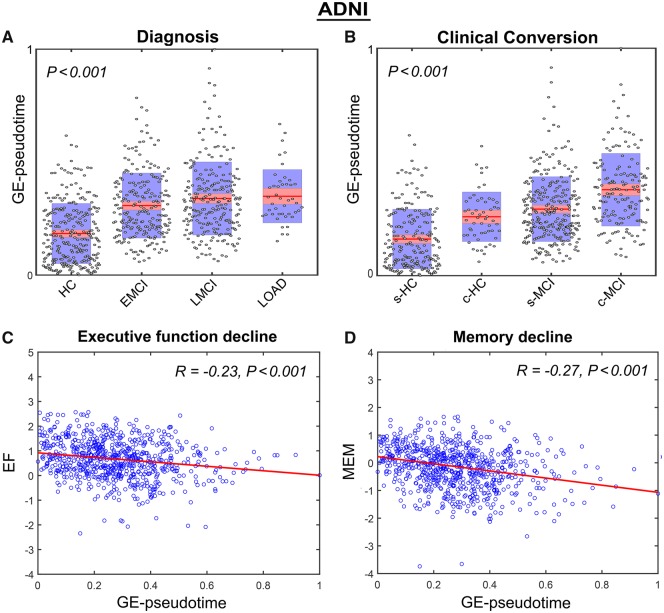
In addition, we tested if the identified subject ordering based on enriched gene expression patterns was predictive of the individual clinical and cognitive properties (Fig. 3A–D). We observed that the molecular disease score values were significantly associated with the individual clinical diagnosis (Fig. 3A; F = 56.72, P < 0.001, FWE-corrected). Importantly, they were also significantly associated with the individual clinical conversion (Fig. 3B; F = 56.61, P < 0.001, FWE-corrected). Subjects with a same clinical diagnosis at baseline, but significantly higher gene expression-pseudotimes, were consistently progressing to a more advanced disease state in an average period of 3.18 years [standard deviation (SD) 2.33]. The molecular disease score values were also significantly associated with executive function (Fig. 3C; R = 0.23, P < 0.001) and memory performance (Fig. 3D; R = 0.27, P < 0.001). However, the associations with these continuous cognitive metrics were characterized by a low predictive power, only explaining ∼5.3–7.3% of the population variance, respectively. We attribute this to both the lack of highly precise metrics for evaluating memory and executive function and the inability of the gene expression-pseudotimes to reflect specific aspects/components of each individual’s cognitive deterioration.
Figure 3.
Blood gene expression-based predictions of clinical and cognitive deterioration for ADNI data. (A and B) Gene expression-pseudotime associations with clinical diagnosis (A) and future clinical conversion (B). (C and D) Scatter plots showing negative associations between molecular disease progression (reflected in gene expression-pseudotime) and measurements of cognitive integrity: memory integrity (MEM) (C) and executive function (EF) (D). In A and B, points are laid over a 1.96 SEM (95% confidence interval) in red and at 1 SD in blue, and P-values are FWE-corrected. In B, categories included are: stable healthy control (s-HC), converter healthy control (c-HC), stable MCI (s-MCI) and converter MCI (c-MCI). EMCI = early MCI; LMCI = late MCI.
Altogether, these results support that, in the context of in vivo LOAD and the ADNI population, the subject’s temporal ordering based on enriched blood gene expression patterns is strongly reflective of neuropathological and overall clinical deterioration, as well as future disease progression. It is, however, a considerably less powerful predictor of the detailed alterations observed in memory and executive function.
In vivo and post-mortem molecular pathways underlying LOAD progression
Next, we aimed to identify the genes, molecular functions and pathways responsible for the accurate prediction of neurodegenerative progression in LOAD. We also intended to clarify if similar predictive mechanisms were common to the periphery (blood) and brain tissues. In this context, the gene expression (GE)-cTI can provide a quantitative mapping of the most influential genes during the process of diseased trajectories inference. Specifically, the cPCA’s loadings (or weights) reflect how much each specific gene, in the original high dimensional space (i.e. ∼40 000 transcripts), contributed to the reduced low dimensional space from which the trajectories were obtained. Thus, we used these weights to select the genes most influential on the subject’s ordering, i.e. those genes driving the observed population differences predictive of neuropathological and cognitive/clinical alterations across the disease’s evolution. Based on the dataset-specific identified genes, we then performed large-scale gene functional analyses with the protein annotation through evolutionary relationship (PANTHER) classification system (Mi et al., 2013). In addition, using a recently reported comprehensive meta-analysis of brain cell type gene signatures (Mckenzie et al., 2018), we identified the cell types consistently associated with the most predictive genes in the brain. Of note, because these analyses were restricted to LOAD evolution, Huntington’s disease patients were excluded when using the HBTRC database.
For the ROSMAP brains, we found 845 highly influential genes with 88 functional pathways (Fig. 4A, E and Supplementary Tables 2 and 3). These Gene Ontology (GO) over-represented pathways were highly sensitive for the detection of biological processes that are commonly associated with neuropathological and cognitive deterioration mechanisms, including axon guidance, histamine H1 receptor mediation, angiogenesis, inflammation mediated by chemokine and cytokine signalling, Wnt and VEGF signalling, apoptosis, p53 pathway, and Alzheimer’s disease-amyloid secretase. For HBTRC brains, we found 416 highly influential genes with 74 functional pathways (Fig. 4B, E and Supplementary Tables 2 and 4). Eighty-nine per cent of these pathways (i.e. 66) were also among the most relevant pathways detected in ROSMAP brains. Correspondingly, the GO over-represented pathways in HBTRC brains were also highly sensitive for the detection of biological processes commonly associated with neurodegeneration.
Figure 4.
Ontology analysis of top predictor genes of LOAD development. Significant molecular functions (A–C), cell types (D) and molecular pathways (E) identified in the brain’s prefrontal cortex (PFC) (ROSMAP) and BA9 (HBTRC) areas, and the blood plasma (ADNI). (A–C) Bars show the number of genes associated to the main GO over-represented categories. In E, only common pathways identified across the three populations are shown (for complete lists of genes and pathways, see Supplementary Tables 2–5). The colour scale indicates the presence level (in %) of each functional pathway (e.g. dark blue for absent pathways, red for highly represented pathways).
The highly predictive genes in both ROSMAP and HBTRC were consistently related to five different cell types (Fig. 4D), including astrocytes, endothelial cells, microglia, neurons, and oligodendrocyte precursor cells. Interestingly, the patterns of identified cell types differed between these two brain datasets (Fig. 4D). Astrocytes (38%) and oligodendrocyte precursor cells (35%) were the most abundantly identified cell types in ROSMAP, while microglia cells were under-represented (4%). On the contrary, almost half of the identified cell types in HBTRC corresponded to microglia (48%), although astrocytes and oligodendrocyte precursor cells still represented significant proportions (22% and 17%, respectively). As discussed below, the observed inter-dataset differences in molecular pathways and brain cell types may respond to multiple causes, such as the systematic (study-specific) sampling at distinct brain locations, the use of different gene expression mapping techniques with dissimilar sensitivity/specificity capacities, and different population characteristics.
Notably, 85% and 90% of the highly predictive molecular pathways in the neurodegenerating brain (ROSMAP and HBTRC, respectively) were also among the most relevant pathways detected in the blood data (ADNI; Fig. 4C, E and Supplementary Tables 2 and 5). The common blood–brain functional pathways relevant for LOAD progression included blood coagulation, angiogenesis (linked to the formation of new blood vessels), p53 (modulating the cell cycle and playing a major role in inhibition of angiogenesis), B cell activation (involved in immune system response), and Wnt signalling (related to signal transduction), among others (Fig. 4E and Supplementary Tables 3–5). The finding of these common pathways is evidence of the direct relationship between the CNS and the body, both in health and in disease. Their unsupervised data-driven identification, therefore, supported the crucial importance of studying the periphery-brain axis (e.g. immune and vascular interactions with brain integrity) for a better understanding of systemic pathological mechanisms underlying neurodegeneration.
Interestingly, we also found another 15% and 10% of highly predictive molecular pathways in the blood that were not identified in the brain (ROSMAP and HBTRC, respectively) (Fig. 4E and Supplementary Tables 3–5). Similarly, 11% of the most predictive pathways identified in HBTRC were not common with the pathways identified in ROSMAP, and a clear difference in each pathway’s presence level across the three datasets was also noticed (Fig. 4E and Supplementary Tables 3–5). These findings may be associated with several reasons, including increased pathological co-morbidity in the periphery relative to the brain and/or crucial methodological limitations, such as the analysis of three different populations with divergent disease characteristics, and the use of different gene expression mapping techniques with dissimilar sensitivity/specificity capacities.
Discussion
Because of the typically long developing period of most prevalent neurodegenerative disorders, we lack exhaustive longitudinal datasets covering the continuous molecular transitions underlying disease progression. Consequently, almost all of our knowledge of the subjacent pathological mechanisms is based on data ‘snapshots’ taken and analysed at a few disease stages. Here, we aimed to overcome this crucial gap by inferring the intrinsic temporal information contained in large-scale neurodegenerative datasets. For that, we implemented a novel pattern analysis method that detects enriched gene expression trajectories in a diseased population (e.g. subjects progressing towards dementia) relative to a background population (e.g. a clinically normal control group). Our results in three different gene expression datasets (ROSMAP, HBTRC, ADNI) support the strong predictive power of this technique for identifying individual neuropathological stages and/or cognitive deterioration. This may well have broad implications for uncovering the dynamic mechanisms of molecular pathology, patient stratification in the clinic, and monitoring response to personalized treatments in neurodegeneration.
A minimally invasive molecular test for neurodegeneration could lead to better treatment and therapies (Ray et al., 2007; Park et al., 2019). An additional aim of this study was to identify an in vivo peripheral biomarker able to predict the individual’s pathophysiology and cognitive decline. When tested in 744 blood samples from ADNI, the proposed GE-cTI showed a significant association with amyloid, tau and infarcts positivity (Fig. 2F–J). Furthermore, it was significantly associated with clinical deterioration and conversion (Fig. 3A and B). The fact that the proposed machine learning model is unsupervised (i.e. the neuropathological and clinical variables are not used to train a predictive model), guarantees absence of possible circularity or data overfitting. Consequently, we can infer that the obtained genetic trajectories and the associated gene expression-pseudotime values are direct measures of molecular integrity, obtained independently of phenotypic variables, and would therefore be useful as unbiased biomarkers in clinical applications.
Our analysis of most relevant molecular pathways for predicting LOAD progression revealed a striking similarity between peripheral and intrabrain pathological mechanisms. Eighty-five to ninety per cent of the most predictive molecular pathways identified in the post-mortem brains were also identified as top predictors in the blood. These pathways support the importance of studying the peripheral-brain axis, providing further evidence for a key role of vascular structure and functioning (Bell and Zlokovic, 2009; Iturria-Medina et al., 2016, 2017), and immune system response (Gendelman, 2002; Streit et al., 2004; Labzin et al., 2018). The multi-tissue analysis based on genetic trajectories may be particularly useful for clarifying both local (tissue-specific) and systemic (inter-organs) neurodegenerative mechanisms.
Our method built on the pseudotemporal trajectory inference field (Magwene et al., 2003; Gupta and Bar-Joseph, 2008; Cannoodt et al., 2016; Welch et al., 2016). Modelling the dynamics of gene regulation, rather than focusing on static time points, is crucial for clarifying cellular transitions and what goes wrong in the case of disease (Cannoodt et al., 2016). We attempted to extend previous models by incorporating the use of a novel contrastive dimensionality reduction technique (cPCA; Abid et al., 2018), which allows detecting enriched patterns in the population of interest while adjusting by confounding components in the background population (e.g. concurrent ageing effects). We observed that this technique (cPCA) was significantly more sensitive to detecting disease progression than other popular dimensionality reduction methods (i.e. PCA and UMAP) (Supplementary Fig. 1). In a set of complementary analyses (data not shown), we observed that, in comparison with other state-of-the-art trajectory inference methods (Welch et al., 2016; Campbell and Yau, 2018), this extension provides a considerably higher sensitivity to detect diseased gene expression components (i.e. other methods could not predict neuropathology, nor clinical deterioration). In addition to uncovering disease dynamics, cTI may enable the data-driven identification of new subpopulations within a heterogeneous neurodegenerative population (Trapnell et al., 2014; Trapnell, 2015; Cannoodt et al., 2016), with strong implications for precision medicine and the selective enrolment of patients in clinical trials. Furthermore, once the data are ordered, it could also improve the inference of causative regulatory interactions underlying a disorder (Cannoodt et al., 2016).
Another advantage of cTI (and trajectory inference in general) is the ability to deal with high dimensional data. This is a key feature for the concurrent analysis of ‘multi-omics’, potentially allowing the exploration of multiple and complementary modalities, such as transcriptomics, proteomics, metabolomics and epigenomics. Contrastive trajectory inference can also be applied to the analysis of data from other fields, including multimodal brain imaging, environmental and cognitive/clinical information. Finally, although our study focused on neurodegenerative evolution, in general, cTI can be applicable to the study of multiple neurological and neuropsychiatric conditions.
Supplementary Material
Acknowledgements
We thank the two anonymous reviewers whose comments/suggestions helped improve and clarify this manuscript. Also, Dataset-1 (ROSMAP) was provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago.
Funding
This research was undertaken thanks in part to funding from: the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives Initiative, the Ludmer Centre for Neuroinformatics and Mental Health, and the Brain Canada Foundation and Health Canada support to the McConnell Brain Imaging Center at the Montreal Neurological Institute. Dataset-1 collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, U01AG46152, and the Illinois Department of Public Health. ROSMAP data can be requested at www.radc.rush.edu. In addition, Dataset 3 collection and sharing for this project was funded by ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- cPCA =
contrastive principal component analysis
- GE-cTI =
gene expression contrastive trajectory inference
- HBTRC =
Harvard Brain Tissue Resource Center
- LOAD =
late-onset Alzheimer’s disease
- MCI =
mild cognitive impairment
- ROSMAP =
Religious Orders Memory and Aging Project Studies
References
- Abdi H, Williams LJ. Principal component analysis. Wires Comp Stat 2010; 2: 433–59. [Google Scholar]
- Abid A, Zhang MJ, Bagaria VK, Zou J. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat Commun 2018; 9: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Zlokovic BV. Neurovascular mechanisms and blood-brain barrier disorder in Alzheimer’s disease. Acta Neuropathol 2009; 118: 103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA. Religious orders study and rush memory and aging project. J Alzheimer's Dis 2018; 64: S161–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Schneider J, Arvanitakis Z, Wilson R. Overview and findings from the religious orders study. Car 2012a; 9: 628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D, Schneider J, Buchman A, Barnes L, Boyle P, Wilson R. Overview and findings from the rush memory and aging project. Curr Alzheimer Res 2012b; 9: 646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Yu L, De Jager PL. Building a pipeline to discover and validate novel therapeutic targets and lead compounds for Alzheimer’s disease. Biochem Pharmacol 2014; 88: 617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box GEP, Cox DR. An analysis of transformations. J R Stat Soc Ser B 1964; 26: 211–52. [Google Scholar]
- Braak HB. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–59. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 2018; 360: eaar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ. Neuropathology Data - Methods. 2018. Available from https://adni.loni.usc.edu/wp-content/themes/freshnews-dev-v2/documents/neuro/ADNI_Methods_Neuropathology_Core_03-06-2018-2.pdf (8 January 2019, date last accessed). [Google Scholar]
- Campbell KR, Yau C. Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data. Nat Commun 2018; 9: 2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannoodt R, Saelens W, Saeys Y. Computational methods for trajectory inference from single-cell transcriptomics. Eur J Immunol 2016; 46: 2496–506. [DOI] [PubMed] [Google Scholar]
- DeCarli C, Carmichael O, He J. MRI infarct assessment in ADNI. 2013. Available from https://adni.bitbucket.io/reference/docs/MRI_INFARCTS/UCD ADNI MRI Infarct Assessment Method.pdf (8 January 2019, date last accessed). [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, et al. Genetics of gene expression and its effect on disease. Nature 2008; 452: 423–8. [DOI] [PubMed] [Google Scholar]
- Esvelt K, Wang H. Genome‐scale engineering for systems and synthetic biology. Mol Syst Biol 2012; 9: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PG, Muñoz-Aguirre M, Reverter F, Sá Godinho CP, Sousa A, Amadoz A, et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat Commun 2018; 9: 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE. Neural immunity: friend or foe? J Neurovirol 2002; 8: 474–9. [DOI] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, et al. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav 2012; 6: 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bar-Joseph Z. Extracting dynamics from static cancer expression data. IEEE/ACM Trans Comput Biol and Bioinf 2008; 5: 172–82. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y, Carbonell FM, Sotero RC, Chouinard-Decorte F, Evans AC. Multifactorial causal model of brain (dis)organization and therapeutic intervention: application to Alzheimer’s disease. Neuroimage 2017; 152: 60–77. [DOI] [PubMed] [Google Scholar]
- Iturria-Medina Y, Sotero RC, Toussaint PJ, Mateos-Perez JM, Evans AC, Initiative T. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat Commun 2016; 7: 11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s Dement 2010; 6: 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin L, Heneka M, Latz E. Innate immunity and neurodegeneration. Annu Rev Med 2018; 69: 437–49. [DOI] [PubMed] [Google Scholar]
- Landau S, Jagust W. Florbetapir processing methods. 2015. Available from https://adni.bitbucket.io/reference/docs/UCBERKELEYAV45/UCBERKELEY_AV45_Methods_12.03.15.pdf (7 January 2019, date last accessed). [Google Scholar]
- Landau S, Jagust W. Flortaucipir (AV-1451) processing methods. 2018. Available from https://adni.bitbucket.io/reference/docs/UCBERKELEYAV1451/UCBERKELEY_AV1451_Methods_Aug2018.pdf (7 January 2019, date last accessed). [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. 2nd edn. Amsterdam: Elsevier Science BV; 1998. [Google Scholar]
- Magwene PM, Lizardi P, Kim J. Reconstructing the temporal ordering of biological samples using microarray data. Bioinformatics 2003; 19: 842–50. [DOI] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv.org 2018. Available from https://arxiv.org/abs/1802.03426v2.
- Mckenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep 2018; 8: 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8: 1551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, et al. National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol 2012; 123: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, et al. Decline of Alzheimer’s disease. Nat Neurosci 2018; 21: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AY, Jordan MI, Weiss Y. On spectral clustering: analysis and an algorithm. Adv Neural Inf Process Syst 2002; 14: 849–56. [Google Scholar]
- Park J, Han S, Yi D, Byun MS, Lee JH, Jang S, et al. Plasma tau/amyloid-β 1–42 ratio predicts brain tau deposition and neurodegeneration in Alzheimer’s disease. Brain 2019; 142: 771–86. [DOI] [PubMed] [Google Scholar]
- Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, Blennow K, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med 2007; 13: 1359–62. [DOI] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimer’s Dement 2015; 11: 792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz AJ, Shcherbinin S, Slieker LJ, Risacher SL, Charil A, Irizarry MC, et al. Topographic staging of tau positron emission tomography images. Alzheimer’s Dement 2018; s9-II: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Mill J, Smith RG, Lunnon K. Neuroepigenetics elucidating novel dysfunctional pathways in Alzheimer’s disease by integrating loci identified in genetic and epigenetic studies. NEPIG 2016; 6: 32–50. [Google Scholar]
- Street JO, Carroll RJ, Ruppert D. A note on computing robust regression estimates via iteratively reweighted least squares. Am Stat 1988; 42: 152–4. [Google Scholar]
- Streit WJ, Mrak RE, Griffin W. Microglia and neuroinflammation: a pathological perspective. J Neuroinflammation 2004; 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Carlson D, Walton MW, Fahrenkrug S, Hackett P. Precision editing of large animal genomes. Adv Genet 2012; 80: 37–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. Defining cell types and states with single-cell genomics. Genome Res 2015; 25: 1491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014; 32: 381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel J, Myers R, Stevens T, Ferrante R, Bird E, Richardson J. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol 1985; 44: 559–77. [DOI] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet 2009; 84: 445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Hartemink AJ, Prins JF. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol 2016; 17: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea L-G, Wang Z, McElwee J, Podtelezhnikov A, et al. Integrated systems approach identifies genetic nodes and networks in LOAD 2013; 153: 707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three datasets used in this study are available at the AMP-Alzheimer’s disease knowledge portal (https://www.synapse.org/#, Synapse ID 3800853), the Gene Expression Omnibus (GEO accession number GSE44772) and the ADNI database (www.adni.loni.usc.edu), respectively. We anticipate that the cTI method will be released soon as part of an open access user-friendly software. In the meantime, the MATLAB codes can be downloaded from http://www.neuropm-lab.com.