Abstract
The Panel on Food Additives and Flavourings of the European Food Safety Authority was requested to evaluate the genotoxic potential of 74 flavouring substances from subgroup 1.1.1 of FGE.19 in the Flavouring Group Evaluation 200 Revision 1 (FGE.200 Rev1). In FGE.200, genotoxicity studies were provided for one representative substance, namely hex‐2(trans)‐enal [FL‐no: 05.073], and for other two substances in the same subgroup, namely 2‐dodecenal [FL‐no: 05.037] and 2‐nonenal [FL‐no: 05.171]. The Panel concluded that the concern still remains with respect to genotoxicity for the substances of this subgroup and requested an in vivo Comet assay performed in duodenum and liver for hex‐2(trans)‐enal [FL‐no: 05.073]. For the two other representative substances of subgroup 1.1.1 (nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] and oct‐2‐enal [FL‐no: 05.060]), the Panel requested a combined in vivo Comet assay and micronucleus assay. These data have been provided and are evaluated in the present opinion FGE.200 Rev1. Industry submitted genotoxicity studies on trans‐2‐octenal [FL‐no: 05.190], instead of oct‐2‐enal [FL‐no: 05.060]. Based on the available data, the Panel concluded that the concern for genotoxicity can be ruled out for hex‐2(trans)‐enal [FL‐no: 05.073], trans‐2‐octenal [FL‐no: 05.190] and nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058], therefore all the 74 substances [FL‐no: 02.020, 02.049, 02.050, 02.090, 02.112, 02.137, 02.156, 02.192, 02.210, 02.231, 05.037, 05.058, 05.060, 05.070, 05.072, 05.073, 05.076, 05.078, 05.102, 05.109, 05.111, 05.114, 05.120, 05.144, 05.150, 05.171, 05.172, 05.179, 05.184, 05.189, 05.190, 05.191, 05.195, 06.025, 06.031, 06.072, 09.054, 09.097, 09.109, 09.119, 09.146, 09.233, 09.244, 09.247, 09.276, 09.277, 09.303, 09.312, 09.385, 09.394, 09.395, 09.396, 09.397, 09.398, 09.399, 09.400, 09.410, 09.411, 09.469, 09.482, 09.489, 09.492, 09.493, 09.498, 09.678, 09.701, 09.719, 09.741, 09.790, 09.841, 09.866, 09.947, 09.948, 13.004] can be evaluated through the Procedure for flavouring substances.
Keywords: α,β‐unsaturated aldehydes; straight chain; FGE.200; flavouring substances; safety evaluation; subgroup 1.1.1; FGE.19
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
The use of flavourings is regulated under Regulation (EC) No 1334/20081 of the European Parliament and Council of 16 December 2008 on flavourings and certain food ingredients with flavouring properties for use in and on foods. On the basis of Article 9(a) of this Regulation, an evaluation and approval are required for flavouring substances.
The Union list of flavourings and source materials was established by Commission Implementing Regulation (EC) No 872/20122. The list includes a number of flavouring substances for which the safety evaluation should be completed in accordance with Commission Regulation (EC) No 1565/20003.
In February 2011, the EFSA Panel had evaluated a first dossier submitted by Industry in response to the requested data for representative substances in FGE. 200. These data were not considered adequate to alleviate the genotoxicity concern for the substance in subgroup 1.1.1 and the Panel recommended at that time ‘to perform in vivo dietary Comet assays (in drinking water or in feed, not by gavage) for the three linear representatives of subgroup 1.1.1 [FL‐no: 05.073, 05.058 and 05.060]’.
Additional data was submitted in February and June 2013 by industry related to one representative substance of subgroup 1.1.1, hex‐2(trans)‐enal [FL‐no: 05.073] and two other substances of the group.
On 21 May 2014 the EFSA CEF Panel adopted an opinion on this Flavouring Group Evaluation 200 (FGE.200). The Panel confirmed the need for an in vivo Comet assay performed in duodenum and liver for hex‐2(trans)‐enal [FL‐no: 05.073]. For the two representative substances of subgroup 1.1.1 (nona‐2(trans), 6(cis)‐dienal [FL‐no: 05.058] and oct‐2‐enal [FL‐no: 05.060]), a combined in vivo Comet assay and micronucleus assay would be required and that evidence of bone marrow exposure should be provided.
New data concerning the three representative substances of this group addressing the EFSA opinion have been submitted during 2017. The data also included updated poundage and use levels concerning these substances.
The list of the substances referred to in this letter is included in Annex II.
1.1.1. Terms of Reference
The European Commission requests the European Food Safety Authority (EFSA) to evaluate the new information submitted and, depending on the outcome, proceed to full evaluation of the substances in this group in accordance with Commission Regulation (EC) No 1565/2000. In accordance with the usual practice by the CEF panel, the first step (assessment of the genotoxicity) should be completed within nine months. An additional 9 months if necessary is also established for the second step (evaluation through the CEF Procedure).
In case the genotoxic potential cannot be ruled out or the procedure cannot be applied in the first step, EFSA is asked to quantify the exposure.
Annex II: List of flavouring substances of FGE.200 included in this evaluation
| FL‐no: | Name of the substance |
|---|---|
| 02.020 | Hex‐2‐en‐1‐ol |
| 02.049 | Nona‐2,6‐dien‐1‐ol |
| 02.050 | Pent‐2‐en‐1‐ol |
| 02.090 | Non‐2(trans)‐en‐1‐ol |
| 02.112 | Non‐2(cis)‐en‐1‐ol |
| 02.137 | Dec‐2‐en‐1‐ol |
| 02.156 | Hex‐2(cis)‐en‐1‐ol |
| 02.192 | Oct‐2‐en‐1‐ol |
| 02.210 | Undec‐2‐en‐1‐ol |
| 02.231 | trans‐2,cis‐6‐Nonadien‐1‐ol |
| 05.037 | 2‐Dodecenal |
| 05.058 | Nona‐2(trans),6(cis)‐dienal |
| 05.060 | Oct‐2‐enal |
| 05.070 | 2‐Heptenal |
| 05.072 | trans‐2‐Nonenal |
| 05.073 | Hex‐2(trans)‐enal |
| 05.076 | Dec‐2‐enal |
| 05.078 | Tridec‐2‐enal |
| 05.102 | Pent‐2‐enal |
| 05.109 | 2‐Undecenal |
| 05.111 | Octa‐2(trans),6(trans)‐dienal |
| 05.114 | 4‐Methylpent‐2‐enal |
| 05.120 | Dodeca‐2,6‐dienal |
| 05.144 | Dodec‐2(trans)‐enal |
| 05.150 | Hept‐2(trans)‐enal |
| 05.171 | Non‐2‐enal |
| 05.172 | Nona‐2(trans),6(trans)‐dienal |
| 05.179 | (E)‐Tetradec‐2‐enal |
| 05.184 | Undec‐2(trans)‐enal |
| 05.189 | 2‐Hexenal |
| 05.190 | trans‐2‐Octenal |
| 05.191 | trans‐2‐Decenal |
| 05.195 | trans‐2‐Tridecenal |
| 06.025 | 1,1‐Diethoxynona‐2,6‐diene |
| 06.031 | 1,1‐Diethoxyhex‐2‐ene |
| 06.072 | 1,1‐Dimethoxyhex‐2(trans)‐ene |
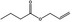
| 09.054 | Allyl butyrate |
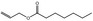
| 09.097 | Allyl heptanoate |
| 09.109 | Allyl nonanoate |
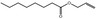
| 09.119 | Allyl octanoate |
| 09.146 | Allyl undec‐10‐enoate |
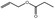
| 09.233 | Allyl propionate |
| 09.244 | Allyl hexanoate |
| 09.247 | Allyl crotonate |
| 09.276 | Oct‐2‐enyl acetate |
| 09.277 | Oct‐2(trans)‐enyl butyrate |
| 09.303 | Hept‐2‐enyl isovalerate |
| 09.312 | Allyl hexa‐2,4‐dienoate |
| 09.385 | Hept‐2‐enyl acetate |
| 09.394 | Hex‐2(E)‐enyl acetate |
| 09.395 | Hex‐2(E)‐enyl propionate |
| 09.396 | Hex‐2‐enyl butyrate |
| 09.397 | Hex‐2‐enyl formate |
| 09.398 | Hex‐2(E)‐enyl hexanoate |
| 09.399 | (2E)‐Hexenyl isovalerate |
| 09.400 | Hex‐2‐enyl phenylacetate |
| 09.410 | Allyl 2‐ethylbutyrate |
| 09.411 | Allyl cyclohexanebutyrate |
| 09.469 | Allyl cyclohexanevalerate |
| 09.482 | Allyl cyclohexaneacetate |
| 09.489 | Allyl isovalerate |
| 09.492 | Allyl cyclohexanehexanoate |
| 09.493 | Allyl 2‐methylcrotonate |
| 09.498 | Allyl cyclohexanepropionate |
| 09.678 | Pent‐2‐enyl hexanoate |
| 09.701 | Allyl phenoxyacetate |
| 09.719 | Allyl anthranilate |
| 09.741 | Allyl cinnamate |
| 09.790 | Allyl phenylacetate |
| 09.841 | 2‐Hexenyl octanoate |
| 09.866 | Allyl valerate |
| 09.947 | (E,Z)‐2,6‐Nonadienyl acetate |
| 09.948 | (2E)‐2‐Nonenyl acetate |
| 13.004 | Allyl 2‐furoate |
The following substance mentioned in the information submitted by the applicant was withdrawn from the Union List by Commission Regulation No 2017/12504 following the EFSA opinion on FGE.226 as regards its genotoxicity:
16.071 4,5‐Epoxydec‐2(trans)‐enal
This substance is therefore not included in this mandate.
2. Data and methodologies
2.1. History of the evaluation of FGE.19 substances
Flavouring Group Evaluation 19 (FGE.19) contains 360 flavouring substances from the EU Register being α,β‐unsaturated aldehydes or ketones and precursors which could give rise to such carbonyl substances via hydrolysis and/or oxidation (EFSA, 2008a).
The α,β‐unsaturated aldehyde and ketone structures are structural alerts for genotoxicity. The Panel noted that there were limited genotoxicity data on these flavouring substances but that positive genotoxicity studies were identified for some substances in the group.
The α,β‐unsaturated carbonyls were subdivided into subgroups on the basis of structural similarity (EFSA, 2008a). In an attempt to decide which of the substances could go through the Procedure, a (quantitative) structure‐activity relationship ((Q)SAR) prediction of the genotoxicity of these substances was undertaken considering a number of models that were available at that time (DEREKfW, TOPKAT, DTU‐NFI‐MultiCASE Models and ISS‐Local Models (Gry et al., 2007)).
The Panel noted that for most of these models internal and external validation has been performed, but considered that the outcome of these validations was not always extensive enough to appreciate the validity of the predictions of these models for these alpha, beta‐unsaturated carbonyls. Therefore, the Panel considered it inappropriate to totally rely on (Q)SAR predictions at this point in time and decided not to take substances through the procedure based on negative (Q)SAR predictions only.
The Panel took note of the (Q)SAR predictions by using two ISS Local Models (Benigni and Netzeva, 2007a,b) and four DTU‐NFI MultiCASE Models (Gry et al., 2007; Nikolov et al., 2007) and the fact that there are available data on genotoxicity, in vitro and in vivo, as well as data on carcinogenicity for several substances. Based on these data the Panel decided that 15 subgroups (1.1.1, 1.2.1, 1.2.2, 1.2.3, 2.1, 2.2, 2.3, 2.5, 3.2, 4.3, 4.5, 4.6, 5.1, 5.2 and 5.3) (EFSA, 2008a) could not be evaluated through the Procedure due to concern with respect to genotoxicity. Corresponding to these subgroups, 15 FGEs were established: FGE.200, 204, 205, 206, 207, 208, 209, 211, 215, 219, 221, 222, 223, 224 and 225.
For 11 subgroups, the Panel decided, based on the available genotoxicity data and (Q)SAR predictions, that a further scrutiny of the data should take place before requesting additional data from the Flavouring Industry on genotoxicity. These subgroups were evaluated in FGE.201, 202, 203, 210, 212, 213, 214, 216, 217, 218 and 220. For the substances in FGE.202, 214 and 218, it was concluded that a genotoxic potential could be ruled out and accordingly these substances were evaluated using the Procedure. For all or some of the substances in the remaining FGEs, FGE.201, 203, 210, 212, 213, 216, 217 and 220 the genotoxic potential could not be ruled out.
To ease the data retrieval of the large number of structurally related α,β‐unsaturated substances in the different subgroups for which additional data are requested, EFSA worked out a list of representative substances for each subgroup (EFSA, 2008c). In selecting the representative substances, expert judgement was applied. In each subgroup, the representative substances were selected taken into account chain length, branched chain, lipophilicity and possible additional functional groups. Likewise, an EFSA genotoxicity expert group has worked out a test strategy to be followed in the data retrieval for these substances (EFSA, 2008b).
The Flavouring Industry has been requested to submit additional genotoxicity data according to the list of representative substances and test strategy for each subgroup.
The Flavouring Industry has now submitted additional data and the present FGE concerns the evaluation of these data requested on genotoxicity.
2.2. History of the evaluation of the substances in FGE.19 subgroup 1.1.1
Subgroup 1.1.1 is one of the FGE.19 subgroups for which the Panel concluded that additional genotoxicity data are needed to perform the safety assessment of the genotoxic potential of the substances (EFSA, 2008a; EFSA CEF Panel, 2011). This conclusion was based on the in vitro and in vivo genotoxicity data available at that time (Appendix D, Tables D.1 and D.2) as well as on the outcome of the (Q)SAR predictions (Appendix C, Table C.1).
Table D.1.
Genotoxicity data (in vitro) considered by the Panel
| Register name [FL‐no] | End‐point | Test system | Concentration | Results | Reference | Remarks* |
|---|---|---|---|---|---|---|
| Nona‐2(trans),6(cis)‐dienal [05.058] | Reverse mutation | Salmonella Typhimurium TA100 | 0.01–0.1 μL/plate (8.6–86 μg/plate)(a) [4,1] | Negative | Eder et al. (1992) |
Valid Standard bacterial density was used 30‐min pre‐incubation (a)Calculated using specific gravity = 0.850–0.870 g/mL (Food and Chemical Codex, 1996) |
| Reverse mutation | S. Typhimurium TA100 |
0.005–0.15 μL/plate (4.3–129 μg/plate)(a) [4,1] 0.005–0.20 μL/plate (4.3–172 μg/plate)(a) [4,2] |
Negative | Eder et al. (1992) |
Valid Threefold bacterial cell density was used 90‐min pre‐incubation (a)Calculated using specific gravity = 0.850–0.870 g/mL (Food and Chemical Codex, 1996) |
|
| SOS chromotest | Escherichia coli PQ37 and PQ243 | 5–80 nmol (0.69–11 μg/L) | Negative | Eder et al. (1992) | Valid | |
| Sister chromatid exchange | Human lymphoblastoid Namalva cell line | 0–40 μM (0–5.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Sister chromatid exchange | Primary human blood lymphocytes | 0–50 μM (0–6.9 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Structural chromosomal aberration test | Human lymphoblastoid Namalva cell line | 0–40 μM (0–5.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Structural chromosomal aberration test | Primary human blood lymphocytes | 0–40 μM (0–5.5 μg/mL) [1] | Equivocal | Dittberner et al. (1995) | Valid | |
| Numerical chromosomal aberration test | Primary human blood lymphocytes | 0–40 μM (0–5.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Micronucleus formation | Primary human blood lymphocytes | 0–50 μM (0–6.9 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Micronucleus formation | Human lymphoblastoid Namalva cell line | 0–50 μM (0–6.9 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
|
Hex‐2(trans)‐enal [05.073] |
Reverse mutation | S. Typhimurium TA98, TA100, and TA104 | Not reported [4,5] | Positive | Kato et al. (1989) |
Validity cannot be evaluated. Abstract only According to the authors, 2‐hexenal was ‘suspected to be positive’ (Kato et al., 1989); however, no further details were provided. Liquid pre‐incubation was used |
| Reverse mutation | S. Typhimurium TA100 |
0.05–0.35 μL/plate [4,1] 0.15–0.5 μL/plate [4,2] |
Negative | Eder et al. (1992) |
Valid Standard bacterial cell density was used 30‐min pre‐incubation |
|
| Reverse mutation | S. Typhimurium TA100 |
0.01–0.15 μL/plate [4,1] 0.1–0.4 μL/plate [4,2] |
Positive | Eder et al. (1992) |
Valid Threefold bacterial cell density was used 90‐min pre‐incubation |
|
| SOS chromotest | E. coli PQ37 and PQ243 | 70–435 nmol (6.9–42.7 μg)(a) | Negative | Eder et al. (1992) |
Valid Cytotoxicity was observed at the highest dose tested (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
| Mutation |
E. coli WP2uvrA/pKM101 |
Not reported [5] | Positive | Kato et al. (1989) |
Validity cannot be evaluated. Abstract only. According to the authors, 2‐hexenal was ‘suspected to be positive’ (Kato et al., 1989); however, no further details were provided Liquid pre‐incubation was used |
|
| Micronucleus induction | Human blood lymphocytes | 5–250 μM (0.5–24.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Micronucleus induction | Lymphoblastoid Namalva cells | 5–250 μM (0.5–24.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Chromosomal aberration | Human blood lymphocytes | 5–250 μM (0.5–24.5 μg/mL) [1] | Negative | Dittberner et al. (1995) | Valid | |
| Chromosomal aberration | Lymphoblastoid Namalva cells | 5–150 μM (0.5–14.7 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Sister chromatid exchange | Human blood lymphocytes | 5–250 μM (0.5–24.5 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| Sister chromatid exchange | Lymphoblastoid Namalva cells | 5–200 μM (0.5–19.6 μg/mL) [1] | Positive | Dittberner et al. (1995) | Valid | |
| DNA repair | Rat hepatocytes | 60–600 nmol/106 cells (5.9–58.9 μmol)(a) | Positive | Griffin and Segall (1986) |
Valid Study design complies with OECD Guideline 482. UDS clearly increased at two highest concentrations with only moderate toxicity (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
|
Pent‐2‐enal [05.102] |
Reverse mutation | S. Typhimurium TA100 |
0.075–0.5 μL/plate [4,1] 0.075–0.75 μL/plate [4,2] |
Positive | Eder et al. (1992) |
Valid Standard bacterial cell density was used 30‐min pre‐incubation |
| Reverse mutation | S. Typhimurium TA100 |
0.01–0.25 μL/plate [4,1] 0.1–0.4 μL/plate [4,2] |
Positive | Eder et al. (1992) |
Valid Threefold bacterial cell density was used 90‐min pre‐incubation |
|
| SOS chromotest | E. coli PQ37 and PQ243 | 60–435 nmol (5.0–36.7 μg)(a) | Negative | Eder et al. (1992) |
Valid Cytotoxicity was observed at the highest dose tested (a)Calculated using the molecular weight of 2‐pentenal = 84.12 |
|
|
Mutation induction TG resistance |
Chinese hamster V79 cells | 0.03, 0.10 or 0.30 mM (2.5, 8.4 or 25.2 μg/mL)(a) [1] | Positive | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range. A dose‐dependent increase in the number of 6‐thioguanine mutants was observed (a)Calculated using the molecular weight of 2‐pentenal = 84.12 |
|
| Mutation induction Ouabain resistance | Chinese hamster V79 cells | 0.03, 0.10 or 0.30 mM (2.5, 8.4 or 25.2 μg/mL)(a) [1] | Negative | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range (a)Calculated using the molecular weight of 2‐pentenal = 84.12 |
|
|
Alkaline elution DNA single strand break |
Mouse leukaemia cells L1210 | 400, 600 or 800 μmol (33.648, 50.472 or 67.296 μg)(a) | Positive | Eder et al. (1993) |
Limited validity Results were not reported in detail. However, the authors stated that pentenal was slightly positive at doses at which cytotoxicity was just starting. Cytotoxicity was observed at the highest dose tested (a)Calculated using the molecular weight of 2‐pentenal = 84.12 |
|
| 2‐Heptenal [05.070] | Reverse mutation | S. Typhimurium TA104 | Up to 0.9 μmol/plate(a) (101 μg/plate)(b) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail. Liquid pre‐incubation procedure was used (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐heptenal = 112.17 |
| Reverse mutation | S. Typhimurium TA104 | Up to 4.4 μmol/plate(a) (493.5 μg/plate)(b) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail. Liquid pre‐incubation procedure was used. Addition of glutathione at 10 mM (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐heptenal = 112.17 |
|
| Reverse mutation | S. Typhimurium TA100 |
0.01–0.15 μL/plate [4,1] 0.075–0.3 μL/plate [4,2] |
Negative | Eder et al. (1992) |
Valid Standard bacterial cell density was used. 30‐min pre‐incubation. Dose‐dependent increases in mutation frequency were noted; however, these increases were never twofold higher than the spontaneous mutation frequency |
|
| Reverse mutation | S. Typhimurium TA100 |
0.005–0.1 μL/plate [4,1] 0.025–0.3 μL/plate [4,2] |
Negative | Eder et al. (1992) |
Valid Threefold bacterial cell density assay was used. 90‐min pre‐incubation Dose‐dependent increases in mutation frequency were noted; however, these increases were never two‐fold higher than the spontaneous mutation frequency |
|
| SOS chromotest | E. coli PQ37 and PQ243 | 35–270 nmol (3.9–30.3 μg)(a) | Negative | Eder et al. (1992) |
Valid Cytotoxicity was observed at the highest dose tested (a)Calculated using the molecular weight of 2‐heptenal = 112.17 |
|
|
Mutation induction TG resistance |
Chinese hamster V79 cells | 0.01, 0.03 or 0.10 mM (1.1, 3.4 or 11.2 μg/mL)(a) [1] | Positive | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range. Dose‐dependent increases in the number of 6‐thioguanine and ouabain mutants were observed (a)Calculated using the molecular weight of 2‐heptenal = 112.17 |
|
| Mutation induction Ouabain resistance | Chinese hamster V79 cells | 0.01, 0.03 or 0.10 mM (1.1, 3.4 or 11.2 μg/mL)(a) [1] | Negative | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range (a)Calculated using the molecular weight of 2‐heptenal = 112.17 |
|
| DNA single‐strand break | Mouse leukaemia L1210 cells | 200, 400 or 500 μmol (22.434, 44.868 or 56.085 μg)(a) | Positive | Eder et al. (1993) |
Limited validity Results were not reported in detail. However, the authors stated that heptenal was positive at non‐toxic doses (a)Calculated using the molecular weight of 2‐heptenal = 112.17 |
|
| trans‐2‐Nonenal [05.072] | Micronucleus formation | Rat hepatocytes | 0.1, 1, 10 or 100 μM (0.01, 0.1, 1.4 or 14.0 μg/mL) | Positive | Esterbauer et al. (1990) |
Limited validity Difficult to interpret since the result was expressed as number of cells with micronuclei per mitotic index 1.00 minus control. However, the result is considered valid. The authors of this study were the same as these from Eckl et al. (1993) |
| Micronucleus formation | Rat hepatocytes | 0.1, 10 or 100 μM (0.01, 1.4 or 14.0 μg/mL) | Equivocal | Eckl et al. (1993) |
Limited validity Cells were not under cell division at the treatment time |
|
| Chromosomal aberration | Rat hepatocytes | 0.1, 1, 10 or 100 μM (0.01, 0.1, 1.4 or 14.0 μg/mL) | Negative | Esterbauer et al. (1990) | Validity cannot fully be evaluated since types of chromosomal aberrations were not reported | |
| Chromosomal aberration | Rat hepatocytes | 0.1, 10 or 100 μM (0.01, 1.4 or 14.0 μg/mL) | Negative | Eckl et al. (1993) | Validity cannot fully be evaluated since types of chromosomal aberrations were not reported. Cells were not under cell divisions at the treatment time | |
| Sister chromatid exchange | Rat hepatocytes | 0.1, 10 or 100 μM (0.01, 1.4 or 14.0 μg/mL) | Equivocal | Eckl et al., 1993; |
Limited validity Cells were not under cell division at the treatment time |
|
| DNA repair | Rat hepatocytes | 60–600 nmol/106 cells (8.4–84.1 μg/plate) | Positive | Griffin and Segall (1986) | Valid. Study design complies with OECD Guideline 482. UDS clearly increased at four highest concentrations. Maximum at the mid‐concentration with only moderate toxicity. At increased levels of toxicity also decline in the net grain counts | |
| Non‐2‐enal [05.171] | Reverse mutation | S. Typhimurium TA104 | Up to 0.007 μmol/plate(a) (1.0 μg/plate) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail Liquid pre‐incubation procedure was used (a)Maximum non‐toxic dose |
|
Mutation induction TG resistance |
Chinese hamster V79 cells | 0.003 or 0.01 mM (0.4 or 1.4 μg/mL) [1] | Positive | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range. A dose‐dependent increase in the number of 6‐thioguanine mutants was observed |
|
|
Mutation induction Ouabain resistance |
Chinese hamster V79 cells | 0.003 or 0.01 mM (0.4 or 1.4 μg/mL) [1] | Negative | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range |
|
| 2‐Hexenal [05.189] | Reverse mutation | S. Typhimurium TA98, TA100, TA1535 and TA1537 | 3 μmol/plate (294.4 μg/plate)(a) [5] | Negative | Florin et al. (1980) |
Insufficient validity. Not in accordance with OECD Guideline 471 (inadequate study design, spot test, only one concentration tested) Isomeric composition of test substance not given (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
| Reverse mutation | S. Typhimurium TA104 | Up to 2 μmol/plate(a) (196.3 μg/plate)(b) [4,1] | Positive | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail Isomeric composition of test substance not given. Liquid pre‐incubation procedure was used (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
| Reverse mutation | S. Typhimurium TA104 | 5 μmol/plate(a) (> 490.7 μg/plate)(b) [4,1] | Positive | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail Isomeric composition of test substance not given. Liquid pre‐incubation procedure was used. Addition of 10 mM glutathione (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
| Reverse mutation | S. Typhimurium TA102 | Up to 2 μmol/plate(a) (196.3 μg/plate)(b) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail Isomeric composition of test substance not given. Liquid pre‐incubation procedure was used (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
|
Mutation induction TG resistance |
Chinese hamster V79 cells | 0.03, 0.10 or 0.30 mM (2.9, 9.8 or 29.4 μg/mL)(a) [1] | Positive | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range. A dose‐dependent increase in the number of 6‐thioguanine mutants was observed. (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
|
Mutation induction Ouabain resistance |
Chinese hamster V79 cells | 0.03, 0.10 or 0.30 mM (2.9, 9.8 or 29.4 μg/mL)(a) [1] | Negative | Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
| DNA single‐strand break | L1210 mouse leukaemia cells | 100, 250 or 500 μmol (9.814, 24.535 or 49.070 μg)(a) | Positive | Eder et al. (1993) |
Limited validity Results were not reported in detail. However, the authors stated that hexenal was positive at non‐toxic doses. Cytotoxicity was observed at the highest dose tested. Isomeric composition of test substance not given (a)Calculated using the molecular weight of 2‐hexenal = 98.14 |
|
| 2‐Octenal [05.060] | Bacterial reverse mutation | S. Typhimurium TA104 | Up to 0.8 μmol/plate(a) (101.0 μg/plate)(b) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail. Liquid pre‐incubation procedure was used (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐octenal = 126.20 |
| S. Typhimurium TA104 | Up to 4 μmol/plate(a) (504.8 μg/plate)(b) [4,1] | Negative | Marnett et al. (1985) |
Validity cannot be evaluated Results were not reported in detail. Liquid pre‐incubation procedure was used Addition of 10 mM glutathione (a)Maximum non‐toxic dose (b)Calculated using the molecular weight of 2‐octenal = 126.20 |
||
|
Mutation induction TG resistance Ouabain resistance |
Chinese hamster V79 cells | 0.01, 0.03 or 0.10 mM (1.3, 3.8 or 12.6 μg/mL) [1] |
Positive (TG resistance: HPRT mutation) Negative (Ouabain resistance) |
Canonero et al. (1990) |
Limited validity No data on cytotoxicity were provided, 2 or 3 doses were tested and unclear criteria to choose dose range The test was performed only without metabolic activation. No significant increase relative to controls was observed in the number of ouabain mutants |
|
| DNA single‐strand breaks | L1210 mouse leukaemia cells | 250, 350 μmol (44 mg/plate) | Positive | Eder et al. (1993) |
Limited validity Results were not reported in detail. However, the authors stated that hexenal was positive at 350 μmol |
OECD: Organisation for Economic Co‐operation and Development; UDS: unscheduled DNA synthesis.
[1]: Without S9 metabolic activation.
[2]: With S9 metabolic activation.
[3]: Plate incorporation method.
[4]: Pre‐incubation method.
[5]: With and without S9 metabolic activation.
*: Validity of genotoxicity studies:
Valid.
Limited validity (e.g. if certain aspects are not in accordance with OECD guidelines or current standards and/or limited documentation).
Insufficient validity (e.g. if main aspects are not in accordance with any recognised guidelines (e.g. OECD) or current standards inappropriate/not validated test system).
Validity cannot be evaluated (e.g. insufficient documentation, short abstract only, too little experimental details provided, text not in a Community language).
Table D.2.
Genotoxicity data (in vivo) considered by the Panel
| Register name [FL‐no] | End‐point | Test system | Concentration | Results | Reference | Remarks |
|---|---|---|---|---|---|---|
|
Hex‐2(trans)‐enal [05.073] |
Micronucleus induction | Human buccal mucosa cells | 10 mg/kg | Positive | Dittberner et al. (1997) |
Valid Statistically significant increases in micronuclei were observed on days 4, 5, 6 and 7 post‐administration |
Table C.1.
QSAR Predictions on Mutagenicity for 25 Aldehydes from Subgroup 1.1.1
| FL‐no JECFA‐no | EU register name | Structural formulaa | ISS local model Ames Test TA100b | MultiCASE Ames testc | MultiCASE Mouse lymphoma testd | MultiCASE Chromosomal aberration test in CHOe | MultiCASE Chromosomal aberration test in CHLf |
|---|---|---|---|---|---|---|---|
| 05.176 | Prop‐2‐enal |
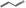
|
POS | POS | OD | NEG | OD |
|
05.102 1364 |
Pent‐2‐enal |

|
POS | POS | OD | NEG | NEG |
|
05.114 1208 |
4‐Methylpent‐2‐enal |

|
POS | NEG | OD | NEG | NEG |
|
05.189 1353 |
2‐Hexenal |

|
POS | POS | OD | NEG | NEG |
| 05.073 | Hex‐2(trans)‐enal |

|
POS | POS | OD | NEG | NEG |
| Not in Register | Hex‐2(cis)‐en‐1‐al |

|
POS | POS | OD | NEG | NEG |
|
05.150 1360 |
Hept‐2(trans)‐enal |

|
POS | POS | OD | NEG | NEG |
| 05.070 | 2‐Heptenal |
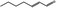
|
POS | POS | OD | NEG | NEG |
|
05.060 1363 |
Oct‐2‐enal |

|
POS | EQU | OD | NEG | NEG |
| 05.190 | trans‐2‐Octenal |

|
POS | EQU | OD | NEG | NEG |
|
05.171 1362 |
Non‐2‐enal |

|
POS | EQU | OD | NEG | NEG |
| 05.072 | trans‐2‐Nonenal |
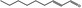
|
POS | EQU | OD | NEG | NEG |
| Not in Register | Non‐2(cis)‐en‐1‐al |

|
POS | EQU | OD | NEG | NEG |
|
05.076 1349 |
Dec‐2‐enal |

|
POS | EQU | OD | NEG | NEG |
| 05.191 | trans‐2‐Decenal |

|
POS | EQU | OD | NEG | NEG |
|
05.109 1366 |
2‐Undecenal |
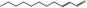
|
POS | EQU | OD | NEG | NEG |
| 05.144 | Dodec‐2(trans)‐enal |
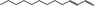
|
POS | EQU | OD | NEG | NEG |
|
05.037 1350 |
2‐Dodecenal |

|
POS | EQU | OD | NEG | NEG |
|
05.078 1359 |
Tridec‐2‐enal |

|
POS | EQU | OD | NEG | NEG |
| 05.179 | Tetradec‐2‐enal |

|
POS | EQU | OD | NEG | NEG |
|
05.111 1182 |
Octa‐2(trans),6(trans)‐dienal |
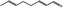
|
NEG | EQU | OD | NEG | NEG |
| Not in register | Nona‐2,6‐dien‐1‐al |

|
NEG | NEG | OD | NEG | NEG |
|
05.058 1186 |
Nona‐2(trans),6(cis)‐dienal |

|
NEG | NEG | OD | NEG | NEG |
|
05.172 1187 |
Nona‐2(trans),6(trans)‐dienal |

|
NEG | NEG | OD | NEG | NEG |
|
05.120 1197 |
Dodeca‐2,6‐dienal |

|
NEG | EQU | OD | NEG | NEG |
FEMA: Flavor and Extract Manufacturers Association; CoE: Council of Europe; CAS: Chemical Abstract Service; CHL: Chinese hamster lung; CHO: Chinese hamster ovary.
Structure subgroup.
Local model on aldehydes and ketones, Ames TA100. (NEG: Negative; POS: Positive; OD: out of domain).
MultiCase Ames test (OD: Out of domain; POS: Positive; NEG: Negative; EQU: Equivocal).
MultiCase Mouse lymphoma test (OD: Out of domain; POS: Positive; NEG: Negative; EQU: Equivocal).
MultiCase Chromosomal aberration in CHO (OD: Out of domain; POS: Positive; NEG: Negative; EQU: Equivocal).
MultiCase Chromosomal aberration in CHL (OD: Out of domain; POS: Positive; NEG: Negative; EQU: Equivocal).
Hex‐2(trans)‐enal [FL‐no: 05.073], nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058], oct‐2‐enal [FL‐no: 05.060] and 4,5‐epoxydec‐2(trans)‐enal [FL‐no: 16.071] were selected as representative substances to be tested for the subgroup 1.1.1 (EFSA, 2008c). The substance 4,5‐epoxydec‐2(trans)‐enal [FL‐no: 16.071] was subsequently considered structurally different from the other substances in subgroup 1.1.1 and was allocated to FGE.226 for evaluation on its own. The representative substances should be tested in accordance with the conditions set out in the ‘Genotoxicity Test Strategy for Substances belonging to Subgroups of FGE.19’ (EFSA, 2008b). The representative substances for subgroup 1.1.1 are shown in Table 1.
Table 1.
Representative substances for Subgroup 1.1.1 of FGE.19 (EFSA, 2008c)
| FL‐no | EU register name | Structural formula | Comments |
|---|---|---|---|
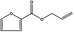
| 05.073 | Hex‐2(trans)‐enal |
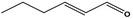
|
Data from literature and new study reports (Beevers, 2013; Bhatia et al., 2010; Dittberner et al., 1995, 1997; Durward, 2009; Eder et al., 1992; Griffin and Segall, 1986; Honarvar, 2007a; Kato et al., 1989; Sokolowski, 2007a) |
| 05.058 | Nona‐2(trans),6(cis)‐dienal |
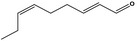
|
Data from literature (Eder et al., 1992; Dittberner et al., 1995) |
| 05.060 | Oct‐2‐enal |
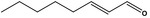
|
Data from literature (Marnett et al.,1985; Canonero et al., 1990; Eder et al., 1993) |
In October 2009, the Industry submitted the first dossier in response to the requested data (this dossier was replaced by an updated dossier in April 2010, (EFFA, 2010)).
The Panel considered these new data and its conclusion was given in an EFSA statement published in February 2011 (EFSA CEF Panel, 2011):
‘Supplementary information now provided includes both new data and arguments, which have been discussed by the Panel. Overall, the supplementary information provided by EFFA is not considered sufficient.
Although some of arguments provided by EFFA (e.g. those on metabolism and GSH‐depletion and those on the role of DNA damage) are plausible, they are not sufficient to alleviate concerns for the genotoxic and carcinogenic potential of the substances belonging to subgroup 1.1.1.
The data provided are not compliant with the ‘Genotoxicity Test Strategy for Substances in Subgroups of FGE.19’.
Therefore, the need for additional genotoxicity data has not been alleviated and genotoxicity studies should be carried out for the representative substances of subgroup 1.1.1. In line with the Genotoxicity Test Strategy (EFSA 2008b ), the Panel recommended to perform in vivo dietary Comet assays (in drinking water or in feed, not by gavage) for the three linear representatives of subgroup 1.1.1 [FL‐no: 05.073, 05.058 and 05.060]. The results may allow to identify whether there is a critical chain length for DNA damage’.
The opinion on FGE.200 (EFSA CEF Panel, 2014a) dealt with the additional genotoxicity data submitted by the International Organization of the Flavor Industry (IOFI, 2013) in response to the EFSA statement on the first dossier submitted to EFSA on FGE.200 (EFSA CEF Panel, 2011). IOFI provided additional genotoxicity studies for one representative substance in FGE.200 (hex‐2(trans)‐enal [FL‐no: 05.073]) and for other two substances in the same subgroup (2‐dodecenal [FL‐no: 05.037] and 2‐nonenal [FL‐no: 05.171] (Table 2). The CEF Panel evaluated these data and concluded that the concern still remains with respect to genotoxicity for the substances of this subgroup and their three representative substances. The Panel confirmed the need for an in vivo Comet assay performed in duodenum and liver for hex‐2(trans)‐enal [FL‐no: 05.073]. For the two other representative substances of subgroup 1.1.1 (nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] and oct‐2‐enal [FL‐no: 05.060]), a combined in vivo Comet assay and micronucleus assay was required. For the latter, evidence of bone marrow exposure was asked.
Table 2.
Overview of data submitted for subgroup 1.1.1 (IOFI, 2013)
| Test substance | Test | Test conditions | Reference |
|---|---|---|---|
Hex‐2(trans)‐enal [05.073] representative substance (purity: 98.2%)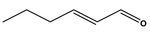
|
Bacterial reverse mutation assay | Salmonella Typhimurium strains TA98, TA100, TA102, TA1535 and TA1537 with and without metabolic activation up to 5,000 μg/plate | Sokolowski (2007a), Bhatia et al. (2010) |
| In vivo micronucleus assay |
Muta™mouse blood reticulocytes (days 1, 4 and 31) Treatment by oral gavage at doses of 120, 235 and 350 mg/kg bw per day for 28 days |
Beevers (2013) | |
| Induction of lacZ‐mutations in Muta™Mouse | Muta™Mouse treatment by oral gavage at doses of 120, 235 and 350 mg/kg bw per day for 28 days. Mutation frequencies (day 31) determined in the liver and the duodenum | Beevers (2013) | |
| In vivo micronucleus assay | Treatment by oral route at doses of 250, 500, 1,000 mg/kg bw per day. Sampling of bone marrow was done 24 and 48 h after treatment | Honarvar (2007a) | |
| In vivo rat liver unscheduled DNA synthesis (UDS) assay | Treatment by oral route at doses of 200 and 500 mg/kg bw per day. Liver was perfused at 16 and 3 h after dosing | Durward (2009) | |
2‐Dodecenal [05.037] not representative (purity: 99.4%)
|
Bacterial reverse mutation assay | S. Typhimurium strains TA98, TA100, TA102, TA1535 and TA1537 with and without metabolic activation up to 1,000 μg/plate | Sokolowski (2007b), Bhatia et al. (2010) |
| In vivo micronucleus assay | Treatment by oral route at doses of 500, 1,000 and 2,000 mg/kg bw per day. Sampling of bone marrow was done 24 and 48 h after treatment. 2,000 PCE scored at 24 h (3 doses) and 48 h (top dose) | Honarvar (2007b), Bhatia et al. (2010) | |
2‐Nonenal [05.171] not representative (purity: 96.2%)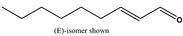
|
In vivo Micronucleus assay | Treatment by oral route at doses of 500, 1,000 and 2,000 mg/kg bw per day. Sampling of bone marrow was done 24 and 48 h after treatment. 2,000 PCE scored at 24 h (3 doses) and 48 h (top dose) | Honarvar (2008), Bhatia et al. (2010) |
bw: body weight; PCE: polychromatic erythrocytes.
Furthermore, four additional flavouring substances (trans‐2,cis‐6‐nonadien‐1‐ol [FL‐no: 02.231], undec‐2(trans)‐enal [FL‐no: 05.184], trans‐2‐octenal [FL‐no: 05.190] and trans‐2‐tridecenal [FL‐no: 05.195]) were identified which are structurally related to the substances in subgroup 1.1.1 and were evaluated within this group.
In the present revision of FGE.200 (FGE.200 Revision 1), the new required genotoxicity studies submitted by Industry (Table 3) are evaluated.
| FGE | Adopted by EFSA | Link | No. of substances |
|---|---|---|---|
| Statement on FGE.19 subgroup 1.1.1 | 21 February 2011 | http://www.efsa.europa.eu/en/efsajournal/pub/2086 | 70 |
| FGE.200 | 21 May 2014 | http://www.efsa.europa.eu/en/efsajournal/pub/3709 | 74 |
| FGE. 200Rev.1 | 13 September 2018 | http://www.efsa.europa.eu/en/efsajournal/pub/5422 | 74 |
Table 3.
List of additional genotoxicity studies evaluated in FGE.200Rev1
| Chemical name [FL‐no]: | Test system in vivo | Reference |
|---|---|---|
|
Hex‐2(trans)‐enal [FL‐no: 05.073] |
Two Comet assays in liver and duodenum | Keig‐Shevlin (2017) |
| Comet assay in liver with or without hOGG1 | ||
| Micronucleus assay in bone marrow | ||
|
Trans‐2‐Octenal [FL‐no: 05.190] |
Micronucleus assay in bone marrow | Beevers (2015a) |
| Comet assay in liver | ||
|
Nona‐2(trans), 6(cis)‐dienal [FL‐no: 05.058] |
Micronucleus assay in bone marrow | Beevers (2015b) |
| Comet assay in liver |
2.3. Presentation of the substances in flavouring group evaluation 200
FGE.200 concerns 74 straight chain, α,β‐unsaturated aldehydes, with or without additional non‐conjugated double bonds, or precursors for such structures. The 74 substances correspond to subgroup 1.1.1 of FGE.19. One former member of subgroup 1.1.1, 4,5‐epoxydec‐2(trans)‐enal [FL‐no: 16.071], was withdrawn from this subgroup and was evaluated in a new FGE (FGE.226Rev1, EFSA CEF Panel, 2017) as the Panel did not consider the substance to be sufficiently structurally related to the other 74 substances in subgroup 1.1.1. The flavouring substance [FL‐no: 16.071] was withdrawn from the Union List by Commission Regulation No 2017/12505 following the EFSA Opinion on FGE.226 (EFSA CEF Panel, 2017) as regards its genotoxicity.
The chemical structures of the substances of subgroup 1.1.1 are shown in Appendix A (Table A.1) together with their specifications.
Table A.1.
Specification Summary of the Substances in the Present Group Evaluation
| FL‐no JECFA‐no | EU register name | Structural formula | FEMA no CoE no CAS no | Phys. form Mol. formula Mol. weight | Solubilitya Solubility in ethanolb | Boiling point, °Cc Melting point, °C ID test Assay minimum | Refrac. Indexd Spec. gravitye |
|---|---|---|---|---|---|---|---|
|
02.020 1354 |
Hex‐2‐en‐1‐ol |

|
2562 69 2305‐21‐7 |
Liquid C6H12O 100.16 |
Very slightly soluble Soluble |
158–160 IR 95% |
1.437–1.442 0.836–0.841 |
|
02.049 1184 |
Nona‐2,6‐dien‐1‐ol |
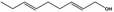
|
2780 589 7786‐44‐9 |
Liquid C9H16O 140.23 |
Insoluble Soluble |
196 IR NMR MS 95% |
1.463–1.465 0.860–0.880 |
|
02.050 1793 |
Pent‐2‐en‐1‐ol |
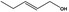
|
665 20273‐24‐9 |
Liquid C5H10O 86.13 |
Freely soluble |
141 MS 95% |
1.427–1.433 0.844–0.850 |
|
02.090 1365 |
Non‐2(trans)‐en‐1‐ol |
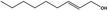
|
3379 10292 31502‐14‐4 |
Liquid C9H18O 142.23 |
Insoluble Soluble |
105 (16 hPa) IR 95% |
1.444–1.448 0.835–0.845 |
|
02.112 1369 |
Non‐2(cis)‐en‐1‐ol |
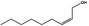
|
3720 10292 41453‐56‐9 |
Liquid C9H18O 142.23 |
Slightly soluble Soluble |
96 (13 hPa) NMR 96% |
1.447–1.453 0.841–0.847 |
|
02.137 1794 |
Dec‐2‐en‐1‐ol |

|
11750 22104‐80‐9 |
Liquid C10H20O 156.27 |
Freely soluble |
117 (19 hPa) MS 95% |
1.446–1.452 0.842–0.848 |
|
02.156 1374 |
Hex‐2(cis)‐en‐1‐ol |
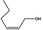
|
3924 69 928‐94‐9 |
Liquid C6H12O 100.16 |
Insoluble Soluble |
65 (0.7 hPa) NMR 92% |
1.437–1.445 0.845–0.853 |
| 02.192 | Oct‐2‐en‐1‐ol |
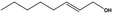
|
3887 11804 22104‐78‐5 |
Liquid C8H16O 128 |
Insoluble Soluble |
88 (hPa) MS 96% |
1.4371–1.4571 0.8384–0.8584 |
|
02.210 1384 |
Undec‐2‐en‐1‐ol |
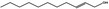
|
4068 37617‐03‐1 |
Liquid C11H22O 170.30 |
Insoluble Soluble |
100–102 (3 hPa) IR 95% |
1.447–1.453 0.838–0.848 |
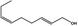
| 02.231 | trans‐2, cis‐6‐Nonadien‐1‐ol |
|
2780 589 28069‐72‐9 |
Liquid C9H16O 140.23 |
Insoluble Soluble |
196 MS 95% |
1.463–1.465 0.860–0.880 |
|
05.037 1350 |
2‐Dodecenal |

|
2402 124 4826‐62‐4 |
Liquid C12H22O 182.31 |
Practically insoluble or insoluble Freely soluble |
272 IR 93% |
1.452–1.458 0.839–0.849 |
|
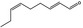
05.058 1186 |
Nona‐2(trans),6(cis)‐dienal |
|
3377 659 557‐48‐2 |
Liquid C9H14O 138.21 |
Insoluble Soluble |
94 IR 92% |
1.470–1.475 0.850–0.870 |
|
05.060 1363 |
Oct‐2‐enal |
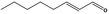
|
3215 663 2363‐89‐5 |
Liquid C8H14O 126.20 |
Slightly soluble Soluble |
84–86 (25 hPa) IR 92% |
1.449–1.455 0.835–0.845 |
|
05.070 1360 |
2‐Heptenal |
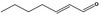
|
3165 730 2463‐63‐0 |
Liquid C7H12O 112.17 |
Practically insoluble or insoluble Freely soluble |
166 IR MS 97% |
1.428–1.434 0857–0.863 |
|
05.072 1362 |
trans‐2‐Nonenal |
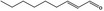
|
3213 733 18829‐56‐6 |
Liquid C9H16O 140.22 |
Practically insoluble or insoluble Freely soluble |
90 (1,2T) 1.333 IR MS 92% |
1.454–1.460 0.855–0.865 |
|
05.073 1353 |
Hex‐2(trans)‐enal |

|
2560 748 6728‐26‐3 |
Liquid C6H10O 98.14 |
Very slightly soluble Freely soluble |
47 (1.7T) 2.266 NMR MS 92% |
1.443–1.449 0.841–0.848 |
|
05.076 1349 |
Dec‐2‐enal |

|
2366 2009 3913‐71‐1 |
Liquid C10H18O 154.25 |
Insoluble Soluble |
229 IR 92% |
1.452–1.458 0.836–0.846 |
|
05.078 1359 |
Tridec‐2‐enal |

|
3082 2011 7774‐82‐5 |
Liquid C13H24O 196.33 |
Insoluble Soluble |
115–118 (13 hPa) IR 92% |
1.455–1.461 0.842–0.862 |
|
05.102 1364 |
Pent‐2‐enal |

|
3218 10375 764‐39‐6 |
Liquid C5H8O 84.11 |
Insoluble Soluble |
124 NMR 98% |
1.440–1.447 (21°) 0.850–0.856 (21°) |
|
05.109 1366 |
2‐Undecenal |
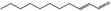
|
3423 11827 2463‐77‐6 |
Liquid C11H20O 168.27 |
Insoluble Soluble |
115 (13 hPa) NMR 98% |
1.452–1.459 0.837–0.847 |
|
05.111 1182 |
Octa‐2(trans),6(trans)‐dienal |
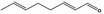
|
3466 10371 56767‐18‐1 |
Liquid C8H12O 124.19 |
Insoluble Soluble |
97–99 (5 hPa) IR NMR 96% |
1.469–1.475 0.835–0.841 |
|
05.114 1208 |
4‐Methylpent‐2‐enal |

|
3510 10364 5362‐56‐1 |
Liquid C6H10O 98.14 |
Slightly soluble Soluble |
126–130 IR NMR 97% |
1.435–1.445 0.858–0.866 |
|
05.120 1197 |
Dodeca‐2,6‐dienal |

|
3637 21662‐13‐5 |
Liquid C12H20O 180.28 |
Insoluble Soluble |
130 (7 hPa) NMR 97.5% |
1.425–1.431 0.987–0.993 |
| 05.144 | Dodec‐2(trans)‐enal |

|
2402 20407‐84‐5 |
Liquid C12H22O 182.31 |
|||
|
05.150 1360 |
Hept‐2(trans)‐enal |

|
3165 730 18829‐55‐5 |
Liquid C7H12O 112.17 |
Insoluble Soluble |
165–167 IR 97% |
1.428–1.434 0.857–0.863 |
|
05.171 1362 |
Non‐2‐enal |

|
3213 733 2463‐53‐8 |
Liquid C9H16O 140.22 |
Insoluble Soluble |
88–90 (16 hPa) IR 92% |
1.454–1.460 0.855–0.865 |
|
05.172 1187 |
Nona‐2(trans),6(trans)‐dienal |
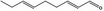
|
3766 17587‐33‐6 |
Liquid C9H14O 138.21 |
Insoluble Soluble |
88 (14 hPa) NMR 97% |
1.439–1.445 0.856–0.864 |
|
05.179 1803 |
Tetradec‐2‐enal |

|
4209 51534‐36‐2 |
Solid C14H26O 210.36 |
Freely soluble |
88 (0.3 hPa) 35 MS 95% |
1.455–1.562 n.a. |
| 05.184 | Undec‐2(trans)‐enal |
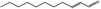
|
3423 11827 53448‐07‐0 |
Liquid C11H20O 168.27 |
Insoluble Soluble |
115 (1.3 hPa) MS 98% |
1.452–1.459 0.837–0.847 |
| 05.189 | 2‐Hexenal |

|
748 505‐57‐7 |
Liquid C6H10O 98.14 |
|||
| 05.190 | trans‐2‐Octenal |
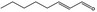
|
3215 2548‐87‐0 |
Liquid C8H14O 126.2 |
Soluble Soluble |
96 (2.5 hPa) MS 92% |
1.449–1.455 0.835–0.845 |
| 05.191 | trans‐2‐Decenal |

|
2366 3913‐81‐3 |
Liquid C10H18O 154.25 |
|||
| 05.195 | trans‐2‐Tridecenal |
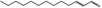
|
3082 7069‐41‐2 |
Liquid C13H24O 196.33 |
Insoluble Soluble |
117 (1.3 hPa) MS 92% |
1.455–1.462 0.842–0.862 |
|
06.025 946 |
1,1‐Diethoxynona‐2,6‐diene |
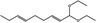
|
3378 660 67674‐36‐6 |
Liquid C13H24O2 212.33 |
Insoluble Miscible |
125 (5 hPa) IR 90% |
1.441–1.448 0.860–0.868 |
|
06.031 1383 |
1,1‐Diethoxyhex‐2‐ene |
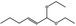
|
4047 2135 54306‐00‐2 |
Liquid C10H20O2 172.27 |
Practically insoluble or insoluble Freely soluble |
66 (8T) 10.6657 MS 95% |
1.418–1.426 0.843–0.849 |
|
06.072 1728 |
1,1‐Dimethoxyhex‐2(trans)‐ene |
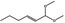
|
18318‐83‐7 |
Liquid C8H16O2 144.21 |
Freely soluble |
158 NMR 95% |
1.420–1.424 0.867–0.871 |
|
09.054 2 |
Allyl butyrate |
|
2021 280 2051‐78‐7 |
Liquid C7H12O2 128.17 |
Insoluble Soluble |
44–45 (20 hPa) IR 98% |
1.412–1.418 0.897–0.902 |
|
09.097 4 |
Allyl heptanoate |
|
2031 369 142‐19‐8 |
Liquid C10H18O2 170.25 |
Freely soluble |
210 IR 97% |
1.426–1.430 0.880–0.885 |
|
09.109 6 |
Allyl nonanoate |

|
2036 390 7493‐72‐3 |
Liquid C12H22O2 198.31 |
Insoluble Soluble |
241–242 IR 96.5% |
1.430–1.436 0.872–0.880 |
|
09.119 5 |
Allyl octanoate |
|
2037 400 4230‐97‐1 |
Liquid C11H20O2 184.28 |
Insoluble Soluble |
222 IR 97% |
1.432–1.434 0.872–0.880 |
|
09.146 9 |
Allyl undec‐10‐enoate |

|
2044 441 7493‐76‐7 |
Liquid C14H24O2 224.34 |
Insoluble Soluble |
180 (39 hPa) IR 98% |
1.448 at 30° 0.8802 at 30° |
|
09.233 1 |
Allyl propionate |
|
2040 2094 2408‐20‐0 |
Liquid C6H12O2 114.15 |
122–123 IR 99% |
1.4105 0.914 at 20° |
|
|
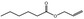
09.244 3 |
Allyl hexanoate |
|
2032 2181 123‐68‐2 |
Liquid C9H16O2 156.22 |
Insoluble 1 mL in 6 mL 70% ethanol |
185 IR 98% |
1.422–1.426 0.884–0.890 |
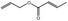
| 09.247 | Allyl crotonate |
|
4072 2222 20474‐93‐5 |
Liquid C7H10O2 126.15 |
Freely soluble |
146 MS 95% |
0.932–0.937 |
|
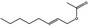
09.276 1367 |
Oct‐2‐enyl acetate |
|
3516 11906 3913‐80‐2 |
C10H18O2 170.25 |
|||
|
09.277 1368 |
Oct‐2(trans)‐enyl butyrate |

|
3517 11907 84642‐60‐4 |
Liquid C12H22O2 198.30 |
Insoluble Soluble |
112–113 (10 hPa) IR NMR MS 96% |
1.433–1.439 0.890–0.896 |
|
09.303 1799 |
Hept‐2‐enyl isovalerate |

|
4126 10664 253596‐70‐2 |
Liquid C12H22O2 198.30 |
Freely soluble |
263 NMR 95% |
0.868–0.873 |
|
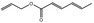
09.312 8 |
Allyl hexa‐2,4‐dienoate |
|
2041 2182 7493‐75‐6 |
Liquid C9H12O2 152.19 |
Soluble |
67 IR 99% |
1.506 0.945–0.947 |
|
09.385 1798 |
Hept‐2‐enyl acetate |

|
4125 10661 16939‐73‐4 |
Liquid C9H16O2 156.22 |
Freely soluble |
193 MS 95% |
1.428–1.434 0.889–0.895 |
|
09.394 1355 |
Hex‐2(trans)‐enyl acetate |

|
2564 643 2497‐18‐9 |
Liquid C8H14O2 142.20 |
Very slightly soluble Soluble |
165–166 IR 90% |
1.424–1.430 0.890–0.897 |
|
09.395 1378 |
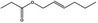
Hex‐2(trans)‐enyl propionate |
|
3932 11830 53398‐80‐4 |
Liquid C9H16O2 156.23 |
Insoluble Soluble |
91 (26 hPa) NMR 95% |
1.426–1.433 0.885–0.895 |
|
09.396 1375 |
Hex‐2‐enyl butyrate |

|
3926 53398‐83‐7 |
C10H18O2 170.25 |
|||
|
09.397 1376 |
Hex‐2‐enyl formate |
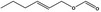
|
3927 11858 53398‐78‐0 |
C7H12O2 128.17 |
|||
|
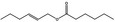
09.398 1381 |
Hex‐(2E)‐enyl hexanoate |
|
3983 53398‐86‐0 |
C12H22O2 198.31 |
|||
|
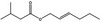
09.399 1377 |
(2E)‐Hexenyl isovalerate |
|
3930 68698‐59‐9 |
Liquid C11H20O2 184.28 |
Insoluble Soluble |
105 (26 hPa) NMR 96% |
1.425–1.435 0.875–0.885 |
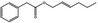
| 09.400 | Hex‐2‐enyl phenylacetate |
|
68133‐78‐8 |
Solid C14H18O2 218.29 |
Practically insoluble or insoluble Freely soluble |
336 37 NMR 95% |
n.a. n.a. |
|
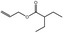
09.410 11 |
Allyl 2‐ethylbutyrate |
|
2029 281 7493‐69‐8 |
Liquid C9H16O2 156.23 |
Insoluble Soluble |
165–167 IR 99% |
1.422–1.427 0.882–0.887 |
|
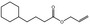
09.411 14 |
Allyl cyclohexanebutyrate |
|
2024 283 7493‐65‐4 |
Liquid C13H22O2 210.31 |
Insoluble Soluble |
104 (1 hPa) NMR 98% |
1.4608 at 20.5° 0.943–0.949 |
|
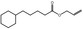
09.469 15 |
Allyl cyclohexanevalerate |
|
2027 474 7493‐68‐7 |
Liquid C14H24O2 224.34 |
Insoluble Soluble |
119 (1 hPa) IR 98% |
1.4605 at 22° 0.942–0.947 |
|
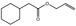
09.482 12 |
Allyl cyclohexaneacetate |
|
2023 2070 4728‐82‐9 |
Liquid C11H18O2 182.26 |
Soluble |
60 (1 hPa) NMR 96% |
1.455–1.499 0.945–0.965 |
|
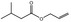
09.489 7 |
Allyl isovalerate |
|
2045 2098 2835‐39‐4 |
Liquid C8H14O2 142.20 |
Insoluble Freely soluble |
155 IR 98% |
1.413–1.418 0.879–0.884 |
|
09.492 16 |
Allyl cyclohexanehexanoate |

|
2025 2180 7493‐66‐5 |
Liquid C14H28O2 238.37 |
Insoluble Soluble |
128 (2 hPa) NMR 98% |
1.462 0.941–0.947 |
|
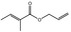
09.493 10 |
Allyl 2‐methylcrotonate |
|
2043 2183 7493‐71‐2 |
Liquid C8H12O2 140.18 |
Slightly soluble |
153 IR 98% |
1.451–1.454 0.939–0.943 |
|
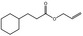
09.498 13 |
Allyl cyclohexanepropionate |
|
2026 2223 2705‐87‐5 |
Liquid C12H20O2 196.29 |
Insoluble 1 mL in 4 mL 80% ethanol |
91 (1 hPa) IR 98% |
1.457–1.462 0.945–0.950 |
|
09.678 1795 |
Pent‐2‐enyl hexanoate |
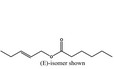
|
4191 74298‐89‐8 |
Liquid C11H20O2 184.28 |
Freely soluble |
241 MS 95% |
1.425–1.435 0.885–0.895 |
|
09.701 18 |
Allyl phenoxyacetate |
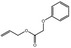
|
2038 228 7493‐74‐5 |
Liquid C11H12O3 192.22 |
100–102 (1 hPa) IR 97.5% |
1.512–1.519 1.00–1.11 |
|
|
09.719 20 |
Allyl anthranilate |
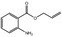
|
2020 254 7493‐63‐2 |
Liquid C10H11O2N 177.21 |
Almost insoluble |
105 (3 hPa) IR 98% |
1.569–1.577 1.12 |
|
09.741 19 |
Allyl cinnamate |
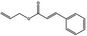
|
2022 334 1866‐31‐5 |
Liquid C12H12O2 188.22 |
Insoluble Miscible |
286 IR 97% |
1.562–1.569 1.050–1.056 |
|
09.790 17 |
Allyl phenylacetate |
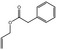
|
2039 2162 1797‐74‐6 |
Liquid C11H12O2 176.22 |
89–93 (4 hPa) IR 99% |
1.5122 at 13.5° 1.033–1.041 |
|
|
09.841 1796 |
2‐Hexenyl octanoate |
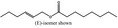
|
4135 85554‐72‐9 |
Liquid C14H26O2 226.36 |
Freely soluble |
309 MS 95% |
|
| 09.866 | Allyl valerate |
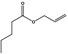
|
4074 6321‐45‐5 |
Liquid C8H14O2 142.20 |
Freely soluble |
58 (16 hPa) MS 95% |
0.999–1.005 |
|
09.947 1188 |
(E,Z)‐2,6‐Nonadienyl acetate |
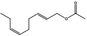
|
3952 68555‐65‐7 |
Liquid C11H18O2 182.26 |
Sparingly soluble Soluble |
231 IR NMR MS 95% |
1.448–1.458 0.905–0.907 |
| 09.948 | (2E)‐2‐Nonenyl acetate |

|
4552 30418‐89‐4 |
Liquid C11H20O2 184.79 |
Sparingly soluble Very soluble |
228 IR NMR MS 98% |
1.4325–1.4425 0.874–0.894 |
|
13.004 21 |
Allyl 2‐furoate |
|
2030 360 4208‐49‐5 |
Liquid C8H8O3 152.15 |
206–209 IR 98% |
1.4945 1.181 (23°) |
FL‐no: FLAVIS number; JECFA: Joint FAO/WHO Expert Committee on Food Additives; FEMA: Flavor and Extract Manufacturers Association; CoE: Council of Europe; CAS: Chemical Abstract Service; ID: Identity; IR: infrared; NMR: nuclear magnetic resonance; MS: mass spectra.
Solubility in water, if not otherwise stated.
Solubility in 95% ethanol, if not otherwise stated.
At 1,013.25 hPa, if not otherwise stated.
At 20°C, if not otherwise stated.
At 25°C, if not otherwise stated.
Section 2.4 of the present Opinion reports the same information that was presented in the FGE. 200. Section 3 reports the evaluation of the new data.
2.4. Data evaluated by Panel in FGE.2006
In February 2011, the Panel evaluated the first dossier submitted by the Industry in response to the requested data for representative substances in FGE.200. These data were not considered adequate to alleviate the genotoxicity concern for the substance in subgroup 1.1.1 and concluded: ‘the Panel recommended to perform in vivo dietary Comet assays (in drinking water or in feed, not by gavage) for the three linear representatives of subgroup 1.1.1 [FL‐no: 05.073, 05.058 and 05.060]’.
In February and June 2013, the Industry (IOFI, 2013) submitted the second dossier which included additional data on one [FL‐no: 05.073] of the three representative substances originally selected by the Panel and supporting information to the data already submitted in the first dossier. In Table 2, the newly submitted data are listed.
2.4.1. In vitro genotoxicity tests
Bacterial reverse mutation assays
Hex‐2(trans)‐enal [FL‐no: 05.073]
Hex‐2(trans)‐enal (purity: 98.2%) was tested at concentrations up to 5,000 μg/plate (but concentrations higher than 200 μg/plate were bacteriostatic) in the Salmonella Typhimurium strains TA98, TA100, TA102, TA1535 and TA1537, in a Good Laboratory Practice (GLP) study performed according to OECD Guideline 471 (OECD, 1997a), with or without metabolic activation (Sokolowski, 2007a; Bhatia et al., 2010). A small but concentration‐dependent increase in revertant colony numbers was observed using the pre‐incubation method in strain TA100 without metabolic activation (concentrations tested 1–2,500 μg/plate). Toxic effects at higher concentrations reduced the number of revertants. Smaller increases (< 2‐fold) were also seen in the presence of S9‐mix. Therefore, a follow‐up experiment, again using the pre‐incubation method, was performed in strain TA100 over a narrow range of concentrations up to 200 μg/plate. In this follow‐up experiment, a moderate concentration‐dependent increase in revertant colony numbers was again observed without metabolic activation at 50 and 100 μg/plate. Based on the reproducibility of this effect, the author concluded a positive mutagenic outcome for this test. While the magnitude of the increase in revertant colony numbers is not substantial, these results do not exclude possible mutagenic potential in strain TA100 (Sokolowski, 2007a).
Kato et al. (1989) tested hex‐2(trans)‐enal (unknown purity) in the S. Typhimurium strains TA98, TA100 and TA104 and in Escherichia coli strain WP2uvrA/pKM101 with and without metabolic activation using the pre‐incubation method (20 min at 37°C). According to the authors, hex‐2(trans)‐enal was ‘suspected to be positive’; however, no further details were provided and the validity of this study is limited.
2‐Dodecenal [FL‐no: 05.037]
At concentrations up to 1,000 μg/plate with and without metabolic activation (but concentrations ≥ 100 μg/plate were bacteriostatic) 2‐dodecenal (purity: 99.4%) was not mutagenic in the S. Typhimurium strains TA98, TA100, TA102, TA1535 and TA1537 in a GLP study performed according to OECD Guideline 471; the limiting factor was the bacteriostatic activity (Sokolowski, 2007b). Toxic effects (reduction in revertant numbers) were seen at the higher concentrations in all parts of the study. No genotoxic effect was noted with and without metabolic activation in the five strains.
The same data for the bacterial reverse mutation assay reported by Sokolowski (2007a, b) for hex‐2(trans)‐enal [FL‐no: 05.073] and 2‐dodecenal [FL‐no: 05.037] were presented in a poster abstract (Bhatia et al., 2010).
Summary of the bacterial reverse mutation assays for both hex‐2(trans)‐enal [FL‐no: 05.073] and 2‐dodecenal [FL‐no: 05.037] are reported in Appendix D, Table D.3.
Table D.3.
Additional genotoxicity data (in vitro) considered by the Panel in FGE.200
| Register name [FL‐no] | Test system | Test object | Concentration | Result | Reference | Comments |
|---|---|---|---|---|---|---|
|
Hex‐2(trans)‐enal [05.073] |
Reverse mutation | Salmonella Typhimurium TA98, TA100, TA102, TA1535, TA1537 | 3–5,000 μg/plate [3,5] | Negative | Sokolowski (2007a) | A moderate concentration –dependent increase in revertant colony number was observed in strain TA100, in the absence of S9‐mix. Study design complies with OECD Guidelines 471 |
| S. Typhimurium TA100 | 1–2,500 μg/plate [4,5] | Positive [4,1] | ||||
| S. Typhimurium TA100 | 25–200 μg/plate [4,5] | Positive [4,1] | ||||
| Reverse mutation | S. Typhimurium TA98, TA100, TA102, TA1535, TA1537 | 3–5,000 μg/plate [3,5] | Negative | Bhatia et al. (2010) | Same test as Sokolowski (2007a) | |
| S. Typhimurium TA100 | 1–2,500 μg/plate [4,5] | Positive [4,1] | ||||
| S. Typhimurium TA100 | 25–200 μg/plate [4,1] | Positive [4,1] | ||||
| 2‐Dodecenal [05.037] | Reverse mutation | S. Typhimurium TA98, TA100, TA102, TA1535, and TA1537 | 3–5,000 μg/plate [3,5] | Negative | Sokolowski (2007b) | Concentrations up to 5,000 μg/plate were used in a pre‐experiment test. Toxic effects as a reduction in the number of revertants were observed at the higher concentrations |
|
0.1–100 μg/plate [3,1] 1–1,000 μg/plate [3,2] |
Negative | |||||
| 0.3–1,000 μg/plate [4,5] | Negative | |||||
| Reverse mutation | S. Typhimurium TA98, TA100, TA102, TA1535, and TA1537 |
1–1,000 μg/plate [3,2] 0.1–100 μg/plate [3,1] |
Negative | Bhatia et al. (2010) | Same test as Sokolowski (2007b) |
OECD: Organisation for Economic Co‐operation and Development.
[1]: Without S9 metabolic activation.
[2]: With S9 metabolic activation.
[3]: Plate incorporation method.
[4]: Pre‐incubation method.
[5]: With and without S9 metabolic activation.
2.4.2. In vivo genotoxicity tests
Hex‐2(trans)‐enal [FL‐no: 05.073]
On the basis of the in vitro bacterial reverse mutation assay results reported above for hex‐2(trans)‐enal, it was considered most appropriate to probe its genotoxic potential using a Muta™Mouse (lacZ/GalE) assay with an in vivo micronucleus component included (Beevers, 2013). The assay was carried out in transgenic mice. This combined approach minimises the number of animals used in the experiments. Micronuclei were measured in peripheral blood, and in the mutation arm of the experiment, the liver and the duodenum were chosen as the most appropriate tissues, in order to address the potential for mutation at the site of most significant metabolism and at the site of first contact, respectively. Therefore, groups of Muta™Mouse CD2‐lacZ80/HazfBR mice were administered hex‐2(trans)‐enal via gavage and the liver, duodenum and peripheral blood were analysed for the potential induction of DNA damage in a GLP study performed according to OECD Guidelines 474 (OECD, 1997b) and 488 (OECD, 2011). However, the Panel noted that there were some deviations from OECD guideline 474 (see below and Appendix D, Table D.4).
Table D.4.
Additional genotoxicity data (in vivo) considered by the Panel in FGE.200
| Register name [FL‐no] | Test system in vivo | Test object | Doses | Result | Reference | Comments |
|---|---|---|---|---|---|---|
|
trans‐2‐Hexenal [05.073] |
Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 250, 500 and 1,000 mg/kg bw | Negative | Honarvar (2007a) | Study design complies with OECD Guideline 474 |
| Unscheduled DNA synthesis | Male rats hepatocytes | 200 or 500 mg/kg bw | Negative | Durward (2009) | Purity and isomer were not specified. Study design complies with OECD Guideline 486 | |
| Micronucleus assay | Transgenic Muta™Mouse (CD2‐lacZ80/HazfBR) blood erythrocytes and reticulocytes | 120, 235 and 350 mg/kg per day | Negative | Beevers (2013) | Mice were treated by gavage for 28 days. The dose of 350 mg/kg per day was identified as MTD. Deviations from OECD Guideline 474 were identified | |
| Induction of lacZ − mutation | Transgenic Muta™Mice (CD2‐lacZ80/HazfBR) liver and duodenum | 120, 235 and 350 mg/kg per day | Negative | Beevers (2013) | Mice were treated by gavage for 28 days. The dose of 350 mg/kg per day was identified as MTD. Liver and duodenum were analysed. Study design complies with OECD Guideline 488 | |
| Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 250, 500 and 1,000 mg/kg bw | Negative | Bhatia et al. (2010) | Same test as Honarvar (2007a) | |
|
2‐Nonenal [05.171] |
Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 500, 1,000 and 2,000 mg/kg bw | Negative | Honarvar (2008) | Study design complies with OECD Guideline 474 |
| Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 500, 1,000 and, 2,000 mg/kg bw | Negative | Bhatia et al. (2010) | Same test as Honarvar (2008a) | |
| 2‐Dodecenal [05.037] | Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 500, 1,000 and 2,000 mg/kg bw | Negative | Honarvar (2007b) | Study design complies with OECD Guideline 474 |
| Micronucleus assay | Mouse bone marrow polychromatic erythrocytes | 500, 1,000 and 2,000 mg/kg bw | Negative | Bhatia et al. (2010) | Same test as Honarvar (2007b) |
OECD: Organisation for Economic Co‐operation and Development; MTD: maximum tolerated dose.
An initial range‐finding study was conducted to estimate the maximum tolerated dose (MTD) of hex‐2(trans)‐enal (purity 99.5%) after administration by oral gavage to groups of three male and three female Muta™Mouse mice. Doses of 500 mg/kg body weight (bw)/day were clearly toxic to mice, with one animal being killed in extremis on day 4 and the rest of the animals exhibiting signs of toxicity (piloerection, hunched posture) but surviving to day 7. Further groups of animals were also dosed at 250 and 350 mg/kg bw per day. No clinical signs of toxicity were observed at 250 mg/kg bw per day, but at 350 mg/kg bw per day 1 animal showed signs of clinical toxicity (hunched posture, decreased activity and dyspnoea). As a result, 350 mg/kg bw per day was identified as the MTD. As no significant gender differences in clinical signs of toxicity were observed, it was concluded that male mice alone could be used in the main experiment. Two lower doses of 120 and 235 mg/kg bw per day were also selected for testing.
Groups of six male Muta™Mouse mice were treated daily by oral gavage with hex‐2(trans)‐enal at doses of 120, 235 and 350 mg/kg bw per day, including a vehicle control (corn oil) for 28 days with a 3‐day recovery period prior to sacrifice. Concurrent positive control animals were not included in this study. Tissue‐matched positive control DNA was included in all packaging reactions in order to confirm correct assay functioning. The positive control DNA originated from animals dosed with ethylnitrosurea. All individual packaging reaction resulted in at least 30,000 plaque‐forming unit (PFU) and at least one mutant plaque. For all animals, data were generated for at least 200,000 PFU per tissue, from at least three independent packaging reactions. At least 1 million PFU were obtained per group, per tissue from a minimum of five animals. No significant increases in mutation frequency (MF) or significant dose‐related trends were observed in the liver or the duodenum. Some of the hex‐2(trans)‐enal treatment groups showed duodenum MF that exceeded laboratory historical controls but were comparable to concurrent vehicle control values. The testing laboratory had a limited number of datasets that comprise the historical control data for the duodenum in this assay and considered its historical control for the duodenum in the Muta™Mouse assay to be narrow at the time of drafting this report.
Hex‐2(trans)‐enal was evaluated in a micronucleus assay in peripheral normochromatic erythrocytes (NCE) and reticulocytes for its ability to induce chromosomal damage (micronuclei (MN)) in mice on days 4 and 31 after 28 days of dosing, using a flow cytometry method. Where possible, 20,000 reticulocytes were analysed from each blood sample. No significant differences were observed in the frequency of peripheral blood reticulocytes (% RET) in all treatment groups on day 4 or 31 after 28 days of dosing. There were no significant increases in the frequency of micronuclei compared to concurrent controls on day 4 or 31 after 28 days of dosing. On day 31, it was noted that there was a significant linear trend in micronucleated reticulocyte (% MN‐RET) frequency (p ≤ 0.05); however, as the MN‐RET frequencies for all treated animals (0.37 ± 0.04, 0.39 ± 0.05, 0.39 ± 0.06, 0.46 ± 0.09 at doses of 0, 120, 235, 350 mg/kg bw per day respectively) were highly consistent with the day 1 background levels of MN‐RET (0.38 ± 0.04, 0.39 ± 0.05, 0.41 ± 0.05, 0.42 ± 0.05, at doses of 0, 120, 235, 350 mg/kg bw per day respectively), the significant linear response was considered to be an artefact and was not indicative of any accumulation of micronuclei over time (Beevers, 2013).
The Panel noted that in the micronucleus arm of the study, the peripheral blood was sampled 72 h after the treatment while the OECD Guideline 474 recommends: ‘once between 36 and 48 h following the final treatment for the peripheral blood’. This point limited the reliability of the results obtained in the micronucleus part of the assay.
Hex‐2(trans)‐enal (purity: 98.2%) was evaluated in a micronucleus assay in bone marrow polychromatic erythrocytes (PCE) for its ability to induce chromosomal damage (MN) in mice in a GLP study performed according to OECD Guideline 474 (OECD, 1997b). Hex‐2(trans)‐enal dissolved in corn oil as a carrier was given orally to animals (5 males and 5 females) at doses of 250, 500 and 1,000 mg/kg bw. The high dose was determined in a preliminary toxicity study. Mice from all dose groups were sampled 24 h after dosing, and mice from the top‐dose and control groups were also sampled 48 h after dosing (Honarvar, 2007a).
Cyclophosphamide (40 mg/kg bw) was given as the positive control and mice were sampled at 24 h. At least 2,000 PCE were scored for each animal for MN. At the highest dose given, two males and two females died, which indicates that higher doses could not have been used. Also, in the highest dose group the numbers of PCE were clearly decreased (−35% at 24 h) as compared to the mean value of PCE of the vehicle control. This indicates that hex‐2(trans)‐enal exerts cytotoxic effects in the bone marrow at this dose level and demonstrates, in the absence of toxicokinetic measures, that the target tissue was exposed. In comparison to the corresponding vehicle controls, there was no statistically significant increase in the frequency of the detected micronuclei at any preparation interval after administration of the test item with any dose level used (Honarvar, 2007a).
2‐Hexenal (unspecified isomer and purity) was evaluated in an in vivo unscheduled DNA synthesis assay using oral administration in a GLP study performed according to OECD Guideline 486 (OECD, 1997c) (Durward, 2009). Male rats were given 200 or 500 mg/kg bw 2‐hexenal. The top dose was proposed by the sponsor, and a preliminary test by the testing facility demonstrated no deaths at this dose. As no other dose levels were used, it is not clear that this was the MTD, and perhaps a higher dose could have been used. In one experiment, livers were perfused approximately 16 h after dosing, and in a second experiment, 3 h after dosing. Following perfusion, hepatocytes were processed and areas of nucleus and cytoplasm scored for autoradiographic grains in 150 cells/animal at each sampling time using automated image analysis. A control group was given only corn oil, and the positive control groups were administered 2‐acetylaminofluorene (16 h) or N,N’‐dimethylhydrazine (3 h). Net nuclear grain counts were < 0 at the two harvest times and the percentage of cells in repair was low in all animals dosed with 2‐hexenal at the 3‐h harvest time. The percentage of cells in repair at the 16‐h harvest time was weakly increased with 1.8 ± 1.7% and 2.2 ± 0.6% cells in repair at 200 and 500 mg/kg, respectively, vs 0.4 ± 0.6% in the concurrent control; however, these values are low and within those generally observed. In the absence of an increase in the number of net grain per cell, these variations have no meaning in term of genotoxic effect. There was therefore no evidence of induction of unscheduled DNA synthesis in animals dosed with the test material at either time point.
2‐Dodecenal [FL‐no: 05.037]
2‐Dodecenal (purity: 99.4%) was evaluated in a micronucleus assay in bone marrow PCE for its ability to induce chromosomal damage in mice in a GLP study performed according to OECD Guideline 474 (OECD, 1997b). 2‐Dodecenal, dissolved in corn oil as a carrier, was given orally to animals (5 males and 5 females) at doses of 500, 1,000 and 2,000 mg/kg bw. The top dose of 2,000 mg/kg bw is a limit dose for non‐toxic substances. Mice from all dose groups were sampled 24 h after dosing, and mice from the top‐dose and control groups were sampled also at 48 h after dosing. Cyclophosphamide (40 mg/kg bw) was given as the positive control and mice were sampled at 24 h. At least 2,000 PCE were scored for each animal for MN. No cytotoxic effects were observed at any dose, based on the ratio between PCE and NCE in each treated sample versus vehicle controls.
In comparison to the corresponding vehicle controls, there was no statistically significant increase in the frequency of the detected micronuclei at any preparation interval after administration of the test item with any dose level used (Honarvar, 2007b).
2‐Nonenal [FL‐no: 05.171]
2‐Nonenal (purity: 96.2%) was evaluated in a micronucleus assay in bone marrow PCE for its ability to induce chromosomal damage in mice in a GLP study performed according to OECD Guideline 474 (OECD, 1997b). 2‐Nonenal, dissolved in corn oil as a carrier, was given orally to animals (5 males and 5 females) at doses of 500, 1,000 and 2,000 mg/kg bw. The top dose of 2,000 mg/kg was estimated as suitable by a preliminary study on acute toxicity. Mice from all dose groups were sampled 24 h after dosing, and mice from the top‐dose and control groups were sampled also at 48 h after dosing. Cyclophosphamide (40 mg/kg bw) was given as the positive control and mice were sampled at 24 h. At least 2,000 PCE were scored for each animal for MN. The numbers of PCE were slightly decreased, mainly in the top dose group at both sampling times, as compared to the mean value of PCE of the vehicle control (−13% at 24 and 48 h sampling times). However, the decrease in % PCE was small. In comparison to the corresponding vehicle controls, there was no statistically significant increase in the frequency of the detected micronuclei at any preparation interval after administration of the test item with any dose level used (Honarvar, 2008).
For both 2‐dodecenal and 2‐nonenal tested through micronucleus assays in mouse bone marrow PCE (Honarvar, 2007b, 2008), there was no direct confirmation that the bone marrow was exposed, as no toxicokinetic measures of the test substance in plasma were made.
Micronucleus data for hex‐2(trans)‐enal (Honarvar 2007a), 2‐nonenal (Honarvar, 2008) and 2‐dodecenal (Honarvar, 2007b) were reported also in a poster abstract (Bhatia et al., 2010).
The results of in vivo studies are summarised in Table D.4.
2.4.3. DNA adduct and related studies
DNA adduct studies in vitro
The ability of the α,β‐unsaturated aldehydes to bind to isolated nucleosides and nucleotides in vitro has been reported (Eder et al., 1993; Eisenbrand et al., 1995; Golzer et al., 1996; Stout et al., 2008). 2‐Hexenal and related α,β‐unsaturated aldehydes are capable of forming 1,N 2‐cyclic deoxyguanosine and 7,8‐cyclic guanosine adducts.
DNA adduct studies in vivo on hex‐2(trans)‐enal [FL‐no: 05.073]
Using a 32P‐post‐labelling method based on nuclease P1 enrichment and thin‐layer chromatography (TLC) separation of the labelled adducts,7 in vivo studies on hex‐2(trans)‐enal report adducts formation. In a first study, Schuler et al. (1999) administered hex‐2(trans)‐enal at a single dose of 500 mg/kg bw by oral route to F344 male rats. No adducts were found in the control rats. In treated rats, an adduct (1,N 2‐propanodeoxyguanosine (Hex‐PdG)) was detected in the liver. Highest Hex‐PdG adduct levels were found 2 days after gavage. Four days after gavage, the Hex‐PdG adducts level was one‐third of the maximum level but it was even higher than Hex‐PdG adducts found after 1 day. No adducts were detected 8 h after gavage. This study demonstrates that after one single high dose of hex‐2(trans)‐enal, formation of DNA adducts were induced, that there was a delay before apparition of adducts in the liver and that these adducts were repaired only slowly.
Schuler and Eder (1999) detected Hex‐PdG adducts in the forestomach, liver, oesophagus and kidneys of F344 rats at relatively high single doses, i.e. 200 and 500 mg/kg bw of hex‐2(trans)‐enal by gavage. At 50 mg/kg bw, Hex‐PdG adducts were quantified only in the oesophagus. The covalent binding index was 0.06, 0.22 and 0.62 at 50, 200 and 500 mg/kg bw, respectively (Schuler and Eder, 1999).
In the study performed by Stout et al. (2008), using a liquid chromatography with tandem mass spectrometry (LC–MS/MS)8 method, no adduct formation was reported at 50 mg/kg bw of hex‐2(trans)‐enal except in forestomach DNA of one rat exposed to a single dose and sacrificed 2 days after (Stout et al., 2008). Quantifiable levels of Hex‐PdG adducts were reported in the forestomach of animals exposed to 100 mg/kg bw per day of hex‐2(trans)‐enal for 1 or 4 weeks (once daily for 5 days per week) and at 200 mg/kg bw of hex‐2(trans)‐enal in single doses. However, Hex‐PdG was not quantifiable in forestomach DNA of rats after exposure to 0, 10 or 30 mg/kg for 1 or 4 weeks (Stout et al., 2008). These data are indicative of a dose‐ and time dependence on DNA adducts formation with hex‐2(trans)‐enal. Hex‐PdG was not quantifiable in liver DNA after exposure to 100 mg/kg for 1 or 4 weeks. These findings suggest that the genotoxicity of hex‐2(trans)‐enal was limited to the site of contact (forestomach) and DNA adduct formation occurred in the setting of severe tissue damage as demonstrated by histopathological observations. At these cytotoxic doses, cell proliferation was noted. The Panel noted that no DNA adducts were observed at 30 mg/kg per day and below.
2.4.4. Data on toxicokinetic
Analogous to other α,β‐unsaturated aldehydes, trans‐2‐hexenal is readily oxidised in vitro to trans‐2‐hexenoic acid in the cytosolic fraction of mouse liver cells (Lamé and Segall, 1986) and by isoenzymes of rat aldehyde dehydrogenase (ALDH) present in mitochondrial, cytosolic and microsomal fractions (Mitchell and Petersen, 1987). In general, the members of the ALDH superfamily demonstrate higher catalytic activity in vitro for higher molecular weight and more lipophilic aldehydes (Nakayasu et al., 1978).
Prior to absorption, 15% of a 100 mg/kg bw dose of trans‐2‐nonenal given to rats was oxidised to trans‐2‐nonenoic acid (Grootveld et al., 1998).
Linear α,β‐unsaturated aldehydes are rapidly absorbed, distributed, metabolised and excreted in the urine and, to a lesser extent, in the faeces. In in vivo experiments with trans‐2‐nonenal and trans‐2‐pentenal, male Wistar albino rats were administered a bolus dose of 100 mg/kg bw of one of the aldehydes by gavage in unheated olive oil. A control group of rats received only the unheated olive oil. Urine samples were collected prior to and after administration. Proton nuclear magnetic resonance (1H‐NMR) analysis indicated that both trans‐2‐nonenal and trans‐2‐pentenal entered systemic circulation from the gastrointestinal tract and were metabolised in the fatty acid pathway or were conjugated with glutathione (GSH) to yield the C‐3 mercapturate conjugate that is excreted mainly in the urine within 24 h. Trace amounts of trans‐2‐nonenal and trans‐2‐pentenal were detected in the faeces (Grootveld et al., 1998).
PBK/D model
A recent physiologically based kinetic/dynamic (PBK/D) study supports a dose‐dependent effect on hex‐2(trans)‐enal detoxification and development of DNA adducts (Kiwamoto et al., 2012, 2013). The detoxification of trans‐2‐hexenal proceeds via three pathways: oxidation to 2‐hexenoic acid by ALDH, reduction to 2‐hexen‐1‐ol by aldose reductase (AR), conjugation with reduced GSH either chemically or catalysed by glutathione S‐transferase (GST) (Eisenbrand et al., 1995). Kiwamoto et al. (2012) developed a PBK/D model in rats determining in vitro kinetic parameters (e.g. Km, Vmax and catalytic efficiency) for each detoxification pathway. Performance of the model was evaluated against available in vivo data from literature on rats exposed to high doses of trans‐2‐hexenal (Schuler and Eder, 1999; Stout et al., 2008). In this study, it was shown that when hex‐2(trans)‐enal is incubated with S9‐mix fractions of rat liver and rat small intestine, in the presence of NAD+, both fractions predominantly convert the substrate to 2‐hexenoic acid which does not readily form DNA conjugates and is efficiently eliminated from the urine in the form of glucuronic acid conjugates. This model predicts that the conversion of trans‐2‐hexenal at doses of 0.04 mg/kg bw (predicted human dietary exposure) and 200 mg/kg bw (dose at which DNA adduct formation in the liver was reported in rats, by Schuler and Eder, 1999) is complete within 3 h. At 0.04 mg/kg bw, GSH concentration is not affected both in liver and small intestine. At 200 mg/kg bw, GSH concentration in the small intestine (predicted as the most important detoxification pathway in this tissue) dropped rapidly and amounted to only 65% of the initial level after 24 h; also in the liver, GSH concentration is depleted, but restored within 24 h. The model suggests that at low doses of trans‐2‐hexenal, protective levels of GSH are unaffected, while at high doses significant GSH depletion occurs. The model predicts that at doses below 80 mg/kg bw all the three pathways contribute to trans‐2‐hexenal detoxification in the liver. The PBK/D model predicts that hex‐2(trans)‐enal is readily detoxified through GSH conjugation at 30 mg/kg bw and below. The same model was further developed to examine dose‐dependent detoxification and DNA adducts formation in humans upon dietary exposure (Kiwamoto et al., 2013). In this study, the kinetic parameters were derived from literature or calculated through in vitro reactions using human tissue fractions, taking into account interindividual differences. The model reveals that rapid in vivo detoxification of hex‐2(trans)‐enal at levels of average dietary exposure (0.04 mg/kg bw) makes DNA adduct formation negligible (Kiwamoto et al., 2013). Additionally, EFFA estimated a daily exposure of 0.01 mg/kg bw per day for hex‐2(trans)‐enal (EFFA, 2010) which is below the concentrations predicted to induce DNA adduct formation.
The Panel noted that all the metabolic parameters were obtained from in vitro studies using rat (Kiwamoto et al., 2012) or human (Kiwamoto et al., 2013) liver S9‐mix or small intestine S9‐mix and cofactors or liver mitochondrial fraction to determine the kinetic constants for ALDH‐mediated oxidation, AR‐mediated reduction and GST‐catalysed conjugation of GSH with trans‐2‐hexenal in these different tissue fractions. Due to the fact that data were obtained only in vitro, such a model is limited. The Panel noted that for these reasons, this model should be considered with cautions.
2.4.5. Discussion of mutagenicity/genotoxicity and related relevant data
In Ames assays, positive results in TA100 and TA104, were reported for several of the substances in subgroup 1.1.1, particularly when pre‐incubation conditions were used. Slight concentration‐dependent increase in revertant colony numbers was observed with hex‐2(trans)‐enal [FL‐no: 05.073] and pent‐2‐enal [FL‐no: 05.102] but not with nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] and 2‐octenal [FL‐no: 05.060]. When using a threefold bacterial cell density, pent‐2‐enal and hex‐2(trans)‐enal were clearly mutagenic with and without metabolic activation; hept‐2(trans)‐enal induced a weak and concentration‐dependent mutagenic effect with metabolic activation and it was clearly mutagenic without S9‐mix. In this assay, it was demonstrated that mutagenicity decreased and toxicity increased with increasing length of the alkyl chain in β‐position (Eder et al., 1992). The authors suggested that the dependence of cell toxicity on the increasing β‐chain length could be related to the increasing lipophilicity. A double bound in the β‐alkyl chain conjugated with that of the acrolein moiety exerted a special effect: it increases the mutagenicity significantly (Eder et al., 1992). This has been confirmed in recent GLP studies (Sokolowski, 2007a,b).
Five alk‐2‐enals, penta‐2‐enal, hex‐2‐enal, hept‐2‐enals, oct‐2‐enal and non‐2‐enal (isomers not specified), were tested for mutagenic activity in V79 Chinese hamster cells. All five alk‐2‐enals induced a concentration‐dependent increase of 6‐thioguanine (TG)‐resistant mutants with a statistically significant increase at 0.3 mM for penta‐2‐enal and hex‐2‐enal, at 0.1 mM for hept‐2‐enal and oct‐2‐enal and at 0.01 mM for non‐2‐enal. The authors reported that a significant increase in mutation frequency is caused by alkyl‐2‐enal concentrations that caused cytotoxicity. Both mutagenicity and cytotoxicity seems directly related to the chain length of the compound. Only hept‐2‐enal induced a statistically significant increase in the frequency of mutations to ouabain resistance in the same cell line (Canonero et al., 1990). This study was considered of limited validity because there is no information about the cytotoxicity levels at each concentration tested, the number of tested concentrations is limited (2 or 3) and the criteria for their choices not clearly presented.
Hex‐2(trans)‐enal [FL‐no: 05.073] and trans‐2‐nonenal [FL‐no: 05.072] were positive in an in vitro unscheduled DNA synthesis (UDS) assay performed in primary cultures of rat hepatocytes. Concentrations of both compounds from 60 to 600 nmol/106 cells (equal to 70–700 nmol/mL) showed a concentration‐dependent increase of cells positive for UDS (Griffin and Segall, 1986).
In the study by Eder et al. (1992), it is shown that in the presence of S9‐mix there is a shift in toxicity toward higher chemical concentrations, suggesting that S9‐mix could lead to partial detoxification. Also, Marnett et al. (1985) reported that toxicity is an important factor in the detection of enals as mutagens. The authors observed positive results only in the presence of GSH and attributed this effect to a partial detoxification that allows survival of bacteria and the growth of revertant colonies.
In the TA104 strain (which carry one non‐sense mutation TAA in the main DNA and not on a plasmid like TA102 strain), 2‐hexenal [FL‐no: 05.189] was mutagenic, but 2‐heptenal [FL‐no: 05.070], 2‐octenal [FL‐no: 05.060] and 2‐nonenal [FL‐no: 05.171] were not mutagenic. No mutagenic activity was observed in the TA102 strain (Marnett et al., 1985).
Positive evidence of genotoxicity was also reported in other assays (sister chromatid exchange (SCE), chromosomal aberrations (ABS), MN, hypoxanthine guanine ribosyl transferase (HPRT) mutations and UDS) in mammalian cells, but more particularly in cell lines that have low detoxification capacity, e.g. Namalva cells and V79 cells (Griffin and Segall, 1986; Canonero et al., 1990; Esterbauer et al., 1990; Eckl et al., 1993).
Hex‐2(trans)‐enal [FL‐no: 05.073] (concentrations tested from 5 to 250 μM) and nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] (concentrations tested from 5 to 40 or 50 μM) were tested in a human lymphoblastoid Namalva cell line and in human lymphocytes for SCE, ABS and MN induction without metabolic activation. Both aldehydes increased the frequency of SCE in the two cell types. The treatment with hex‐2(trans)‐enal induced a statistically significant increase in SCE from 40 μM on lymphocytes and 20 μM for Namalva cells. Nona‐2(trans),6(cis)‐dienal induced a statistically significant increase in SCE from 20 μM on lymphocytes and 10 μM for Namalva cells. Nona‐2(trans),6(cis)‐dienal was more cytotoxic than hex‐2(trans)‐enal. In human lymphocytes, neither hex‐2(trans)‐enal nor nona‐2(trans),6(cis)‐dienal induced statistically significant increase of structural chromosomal aberrations. On the contrary, in Namalva cells, both hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal induced structural chromosomal aberrations from 100 μM and 5 μM respectively. Hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal induced aneuploidies in human lymphocytes from 40μM. Hex‐2(trans)‐enal increased the frequencies of MN both in lymphocytes and in Namalva cells in a concentration‐dependent manner. The increase of MN frequency, induced by hex‐2(trans)‐enal, was statistically significant in lymphocytes from 50 μM and in Namalva cells from 150 μM, while for nona‐2(trans),6(cis)‐dienal a statistically significant increase of MN frequency was observed from 20 μM in lymphocytes and from 40 μM in Namalva cells. Using fluorescent in situ hybridisation, both lymphocytes and Namalva cells showed significantly enhanced frequencies of centromere‐positive MN for both hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal, which is coherent with the observation of aneuploidy inductions in the cytogenetic assay. This study shows that both hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal gave equivocal results in lymphocytes and positive results in Namalva cells, for structural aberrations. While for aneugenicity hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal were positive in both cell types. The Namalva cells were generally more sensitive than lymphocytes. These cells have been found poor or even totally deficient in many detoxifying enzymes and they also contain only rather low concentrations of GSH and of glutathione‐related enzymes (Dittberner et al., 1995).
Using an alkaline elution method, Eisenbrand et al. (1995) demonstrated that Namalva cells were significantly more sensitive than primary rat hepatocytes to the induction of DNA strand breaks by hexenal. In hepatocytes, about 3–5 times higher concentrations of aldehydes were necessary to induce significant effects compared to Namalva cells. The authors explained this difference by the better enzymatic activity (GSH transferase, ALDH) in primary rat hepatocytes compared to Namalva cells. In this study, the authors demonstrated that hexenal induced DNA binding in a range of doses from 1 to 5 mM (Eisenbrand et al., 1995).
Dittberner et al. (1997) performed studies on exfoliated cells of human oral mucosa. Seven healthy non‐smoking volunteers rinsed their mouth four times per day for 3 days with 100 mL of hex‐2(trans)‐enal [FL‐no: 05.073] solution at the concentration of 10 ppm, which represents a possible concentration in food. Results showed at least a doubling of MN frequency in exfoliated cells of human oral mucosa during one of the next 4 days, then the MN number dropped down to nearly the control level. In a second study, seven other volunteers were observed before and after eating 3–6 bananas that contained 35 ppm hex‐2(trans)‐enal. Six of the seven volunteers showed at least a doubling of the MN frequency during one of the next 6 days (Dittberner et al., 1997). The Panel noted that the results were statistically significant and that the protocol was consistent with standard protocols recently developed for biomonitoring studies. Therefore, the results are considered reliable. However, the Panel also noted that this kind of studies is not validated for regulatory purposes.
Primary rat hepatocytes were treated for 3 h with 0, 0.1, 1.0, 10 and 100 μM of trans‐2‐nonenal [FL‐no: 05.072] followed by a 48‐h recovery period. trans‐2‐Nonenal induced an increase (p < 0.01) in MN at 10 and 100 μM. At a concentration of 100 μM, the mean value of chromosomal aberrations was 2.7‐fold higher than in the controls, but due to the high standard deviations, these increases were not statistically significant (Esterbauer et al., 1990).
Primary rat hepatocytes were seeded and after 20 h treated with trans‐2‐nonenal [FL‐no: 05.072] at 0, 0.1, 1, 10 or 100 μM for 3 h. Then the culture medium was replaced by fresh medium added with epidermal growth factor (EGF) and bromodeoxyuridine (BrdU) (Eckl et al., 1993). Forty‐eight hours after the end of the treatment, cells were treated with colcemid, and sampled 3 h later. Slides treated with Hoechst 33258 were used for determination of SCE and the other for chromosomal aberration. trans‐2‐Nonenal induced no significant toxicity at the highest concentration tested. trans‐2‐Nonenal increased neither chromosomal aberrations nor the frequency of micronuclei. The Panel noted that in this study, EGF was added to induce cell division, but cells were not in division during the period of treatment, this deviation could result in a bias compared to recommended protocols. The Panel noted that cells used to determine the induction of chromosomal aberrations and micronuclei were pre‐treated with BrdU which weakens the chromosomes. The testing for chromosomal aberrations and micronuclei was done after a short treatment, followed by a long recovery time which does not appear to be an optimum protocol and is a deviation from the OECD Guidelines. Moreover, hepatocytes do not divide all since the mitotic index in control cultures ranged from 0.41% to 1.94%, and no method (such as the addition of cytochalasin B) was used to determine the frequency of micronuclei only in cells that divided which reduces the sensitivity of this test (Eckl et al., 1993).
Chung et al. (1999) reported that formation of cyclic propano adducts are common products from reactions of enals with DNA bases. Enals derive from lipid peroxidation of cell membrane, but the contribution from environmental sources, cannot be excluded. The mutagenicity of enals and the mutations observed in site‐specific mutagenesis studies, using a model for Hex‐PdG adducts, suggest that these adducts are potential promutagenic lesions. The authors showed that tissue GSH plays an important role in protecting DNA from cyclic adduction by enals.
Coles and Ketterer (1990) reported that 4‐hydroxynon‐2‐enal is a substrate of different classes of rat glutathione transferases that detoxified this compound. But the authors concluded that these enzymes do not provide a perfect protection and cytotoxic or genotoxic damage cannot always be avoided.
Kelson et al. (1997) isolated and characterised a human microsomal fatty ALDH, which is a distinct human ALDH isozyme that acts on a variety of medium‐ and long‐chain aliphatic substrates with a high activity towards saturated and unsaturated aliphatic aldehydes ranging from 6 to 24 carbons in length.
In cell lines poor in detoxification capacity, there is an opportunity for high concentrations (20–40 μM) of α,β‐unsaturated aldehydes to either interact directly with DNA or indirectly forming DNA adducts due to oxidative stress, leading to single DNA strand breaks but no cross‐linking of DNA. The depletion of GSH by high concentrations of α,β‐unsaturated aldehydes is known to lead to oxidative stress and to the release of nucleocytolytic enzymes, causing DNA fragmentation, cellular damage and apoptosis (see Sections 2.3 and 2.4). Hex‐2(trans)‐enal, 2‐nonenal and 2‐dodecenal did not induce MN in mice in robust GLP studies (Honarvar, 2007a, 2007b, 2008). However, only for hex‐2(trans)‐enal exposure of the target tissue was demonstrated. In addition, hex‐2(trans)‐enal did not induce unscheduled DNA synthesis in rat hepatocytes at doses up to 500 mg/kg bw in a GLP study (Durward, 2009).
However, this type of assays does not address the potential clastogenicity in the gastrointestinal tract. While such studies have not been conducted, due to structure similarity it seems to be possible that lifetime gavage administration of high concentrations of hex‐2(trans)‐enal [FL‐no: 05.073] to rats might result in carcinogenicity in the forestomach or oesophagus, similar to that observed for 2,4‐hexadienal (subgroup 1.1.4 of FGE.19, EFSA CEF Panel, 2014a,b). It is also likely that the ulcerative and necrotising lesions and consequent regenerative cell proliferation in the forestomach produced under these unique conditions would be associated with increased DNA adducts, as was observed in the hexenal DNA adduct study (Stout et al., 2008). However, production of exocyclic guanine adducts following GSH depletion may be involved, but the evidence from the 2‐hexenal and 2,4‐hexadienal studies suggests that these events are associated with significant tissue damage (ulceration, inflammation and hyperplasia) related to high bolus dosing by gavage. The inflammation and tissue damage could affect the normal biochemical processes involved in the metabolism of α,β‐unsaturated aldehydes and their detoxification. The reduced metabolic activity could increase the probability of a direct reaction between aldehydes and DNA nucleotides.
A recent PBK/D model shows that 2‐hexenal is rapidly detoxified predominantly by conjugation with GSH by GSTs, and that the rapid detoxification of 2‐hexenal reduces the risk arising from 2‐hexenal exposure through the diet. Thus, dietary exposure to doses that do not deplete GSH, and therefore do not lead either to tissue damage or DNA adducts would not be expected to pose a mutagenic or carcinogenic hazard (Kiwamoto et al., 2012, 2013). However, the Panel considered that PBK/D studies are not sufficient due to the lack of validation.
Hex‐2(trans)‐enal induced weak gene mutations in bacteria. When tested in the Muta™Mouse assay up to the MTD of 350 mg/kg bw per day, hex‐2(trans)‐enal was not mutagenic in the tissues of the duodenum, presumably the first point of contact for the test material upon transit from the glandular stomach. These results are also supported by no indication of mutagenic activity in the liver, primary point of metabolism.
Hex‐2(trans)‐enal tested for chromosomal aberrations in mammalian cell lines showed positive results, but resulted negative in the in vivo micronucleus test performed in peripheral blood reticulocytes (Beevers, 2013) and in bone marrow (Honavar, 2007a).
In summary, the available data indicate that at high concentrations of hex‐2(trans)‐enal no gene mutations were induced in the liver and duodenum of transgenic mice after a daily treatment for 28 days up to 350 mg/kg bw per day (Beevers, 2013). DNA adducts were detected in the forestomach, liver, oesophagus and kidneys of rats treated with hex‐2(trans)‐enal by gavage at relatively high single doses, i.e. 200 and 500 mg/kg bw and at 50 mg/kg in the oesophagus. DNA adducts were not quantifiable in forestomach DNA of rats after exposure to 10 or 30 mg/kg bw for 1 or 4 weeks (Schuler and Eder, 1999). However, in the same experimental condition, DNA adducts were detected locally (duodenum and oesophagus) and systemically (kidney and liver) at doses lower than the dose that proved no induction of gene mutation. The Panel noted that overall the available experimental data from animals and humans, while not showing an induction of gene mutations, do not allow to assess the potential clastogenic activity of hex‐2(trans)‐enal at the first site of contact and in the liver, where high levels of DNA adducts were observed.
2.4.6. Conclusion
The new data submitted are related only to one representative substance of subgroup 1.1.1, hex‐2(trans)‐enal [FL‐no: 05.073].
For hex‐2(trans)‐enal [FL‐no: 05.073], gene mutations were observed in vitro in S. Typhimurium TA100 and TA104, and chromosomal aberrations were observed in vitro likewise. In addition, a biomonitoring study in human buccal cells showed a statistically significant increase in the frequency of micronuclei at concentrations that might be relevant for the use of hex‐2(trans)‐enal as flavouring substances. The new submitted study performed on a Muta™Mouse model does not cover these endpoints adequately. The Panel noted that overall the available experimental data from animals and humans, while not showing an induction of gene mutations, do not allow to assess the potential clastogenic activity of hex‐2(trans)‐enal at the first site of contact and in the liver where higher levels of DNA adducts were observed than in other tissues investigated. Therefore, the new data provided by the Industry do not rule out the genotoxicity concern for the substances of subgroup 1.1.1.
For both 2‐dodecenal and 2‐nonenal tested through micronucleus assays in mouse bone marrow PCE (Honarvar, 2007b, 2008), there was no direct confirmation that the bone marrow was exposed, as no toxicokinetic measures of the test substance in plasma were made.
Under these conditions, the Panel confirms, the need for an in vivo Comet assay performed in duodenum and liver for hex‐2(trans)‐enal [FL‐no: 05.073]. For the two other representative substances of subgroup 1.1.1 (nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] and oct‐2‐enal [FL‐no: 05.060]), a combined in vivo Comet assay and micronucleus assay would be required. For the latter, evidence of target tissue exposure should be provided.
3. Assessment
3.1. New data evaluated in FGE. 200 Revision 1
The applicant has submitted data from published studies and new in vivo genotoxicity studies for: hex‐2(trans)‐enal [FL‐no: 05.073], trans‐2‐octenal [FL‐no: 05.190], nona‐2(trans)‐6(cis)‐dienal [FL‐no: 05.058]. Industry submitted genotoxicity studies on trans‐2‐octenal [FL‐no: 05.190], instead of oct‐2‐enal [FL‐no: 05.060] (configuration not specified); however, the Panel considered it acceptable because it is not expected that the different isomer will affect the outcome of the genotoxicity studies. The studies submitted are evaluated in the present revision of FGE.200 (FGE.200 Rev1). A summary of results is reported in Appendix E, Table E.1. All these studies were performed in accordance with respective OECD guidelines and in compliance with GLP.
Table E.1.
Genotoxicity data (in vivo) evaluated by the Panel in FGE.200Rev1
| Register name [FL‐no] | Test system in vivo | Test object | Route | Dose mg/kg bw per day | Result | Reference | Comments |
|---|---|---|---|---|---|---|---|
| Hex‐2(trans)‐enal 05.073 | Micronucleus bone marrow | Male Han Wistar rats | Gavage | 87.5, 175, 350 | Negative | Keig‐Shevlin (2017) | Reliable without restriction. Study performed in accordance with OECD TG 474 |
| Comet liver | Negative | Reliable without restriction. Study performed in accordance with OECD TG 489 | |||||
| Comet duodenum | Negative | Reliable without restriction. Study performed in accordance with OECD TG 489 | |||||
| trans‐2‐Octenal 05.190 | Micronucleus bone marrow | Male Han Wistar rats | Gavage | 25, 500, 1,000 | Negative | Beevers (2015a) | Reliable without restriction. Study performed in accordance with OECD TG 474 |
| Comet liver | Negative | Reliable without restriction. Study performed in accordance with OECD TG 489 | |||||
| Nona‐2(trans), 6(cis)‐dienal 05.058 | Micronucleus bone marrow | Male Han Wistar rats | Gavage | 175, 350, 700 | Negative | Beevers (2015b) | Reliable without restriction. Study performed in accordance with OECD TG 474 |
| Comet liver | Negative | Reliable without restriction. Study performed in accordance with OECD TG 489 |
OECD: Organisation for Economic Co‐operation and Development.
3.2. Hex‐2(trans)‐enal [FL‐no: 05.073] ‐ in vivo combined Comet and Micronucleus assay
Hex‐2(trans)‐enal [FL‐no: 05.073] was tested in a combined micronucleus assay in bone marrow and comet assay in duodenum and liver of Han Wistar rats by gavage (Keig‐Shevlin, 2017). Hex‐2(trans)‐enal [FL‐no: 05.073] was stored at 15–25°C, under nitrogen and protected from light. Two different batches were tested, purity was 99.5% and 99.2%, respectively. The study was performed in accordance with GLP, OECD TG 474 (OECD, 2014a) and OECD TG 489 (OECD, 2016).
In a dose range‐finding study, groups of three males and three females were given three administrations at 0 (day 1), 24 h (day 2) and 45 h (day 3) of hex‐2(trans)‐enal [FL‐no: 05.073] at 500 and 350 mg/kg bw per day, via oral gavage. Clinical signs of toxicity (e.g. piloerection, decreased activity, ptosis and hunched posture) were observed at the highest dose tested, following the second and the third administration. Following the third administration, at 4 h post‐dose, one female was found dead. In animals dosed at 350 mg/kg bw per day, signs of toxicity including piloerection and anogenital soiling were observed after the third administration.
Based on this dose range‐finding experiment, where no differences in response between female and male rats were seen, a MTD of 350 mg/kg bw per day was established; the two lower doses tested were 175 and 87.5 mg/kg bw per day.
Hex‐2(trans)‐enal [FL‐no: 05.073] was tested in three main experiments where the following assays were conducted: in the first experiment, micronucleus assay in bone marrow and comet assay in liver and duodenum; in the second experiment, comet assay in liver and duodenum; in the third experiment, comet assay in liver including a modified comet assay with human 8‐hydroxyguanine DNA‐glycosylase 1 (hOGG1) treatment.
In the main experiment, male rats (six animals per group) were dosed at 0 (day 1), 24 h (day 2) and 45 h (day 3) – by oral gavage at dose levels of 0 (corn oil), 87.5, 175 and 350 mg/kg bw per day. Animals were sampled at 48 h. Three male rats were given 150 mg/kg bw per day ethyl methanesulfonate (EMS) as a positive control. Corn oil was used as a vehicle following the same treatment schedule.
No clinical signs of toxicity where observed in the main experiments.
In experiments 1 and 2, dose‐related decreases in bodyweight were observed at 350 mg/kg bw per day. No findings in clinical chemistry and histopathology were observed.
A total of at least 500 PCE and NCE was scored to calculate the degree of bone marrow toxicity by the relative decrease in PCE. Four thousand PCE per animal were scored for the presence of MN. Although a slight decrease in PCE was observed in the bone marrow of rats treated with hex‐2(trans)‐enal [FL‐no: 05.073], this was not marked enough to indicate bone marrow toxicity. Group mean results of MN frequencies were similar to the concurrent vehicle control and no statistically significant increases in MN frequency were seen for any of the dose groups. The positive control group showed statistically significant increases in MN frequencies. Negative and positive control values were within the laboratory's historical control data.
3.2.1.
Comet assay in liver – experiment 1
In the groups of animals administered with hex‐2(trans)‐enal, the mean tail intensity was higher than the vehicle control, but the increase was not statistically significant and it was not dose‐related. All values for the mean tail intensity were within the range of the vehicle historical control (95% reference range 0.04–5.50). The authors of the study noted that in each group there were one or two animals displaying elevated %tail intensity and tail moment values which increased the group mean, therefore, the experiment was repeated.
Comet assay in liver – experiment 2
In the second experiment, it was observed that the group mean %tail intensity values for all groups treated with hex‐2(trans)‐enal exceeded the group mean vehicle control data. At 350 mg/kg bw per day, these increases were found to be statistically significant (p ≤ 0.05) and there was also a statistically significant linear trend (p ≤ 0.01). Individual animal %tail intensities in the 87.5 mg/kg bw per day dose group were in the range of 0.21–1.46, the 175 mg/kg bw per day dose group were in the range of 0.43–1.16 and the 350 mg/kg bw per day dose group were in the range of 0.25–2.68 compared to the vehicle control range of 0.08–1.03 (the 95% reference range of the historical control data was 0.04–5.50). At least three animals in each test article treated group were found to overlap with the concurrent vehicle control and all animals fell within the laboratory's historical control data.
In order to investigate the potential oxidative DNA damage of hex‐2(trans)‐enal, a modified Comet assay was conducted in a third experiment.
Comet assay in liver – experiment 3
The third comet assay was conducted with and without hOGG1. In the experiment without hOGG1, %tail intensities and tail moments were similar to the concurrent vehicle control group and fell within the laboratory's historical vehicle control data. There were no statistically significant increases in %tail intensity for any of the groups administered with hex‐2(trans)‐enal compared to the concurrent vehicle control.
In the experiment with hOGG1 modification, no statistically significant increase in mean tail intensity was observed in any groups treated with hex‐2(trans)‐enal compared to the concurrent vehicle control group confirming that hex‐2(trans)‐enal did not induce oxidative DNA damage.
The authors of the study considered that small increases in %tail intensity were observed in sporadic animals over two separate experiments in the liver, but only achieved statistical significance at one dose in one of these experiments. In the third experiment, no increases in %tail intensity were observed and hOGG1 modification did not reveal any evidence of oxidative DNA damage. The increases were considered by the authors as isolated not reproducible and therefore not biologically relevant. The Panel agrees with the conclusions by the authors.
Comet assay in duodenum – experiment 1
In the groups of animals administered with hex‐2(trans)‐enal, the mean %tail intensity values were lower than the vehicle control, but the decrease was not statistically significant and it was not clearly dose related. All individual animal data at all dose levels were generally consistent with the vehicle control animals and fell within the laboratory's historical control data. The only exceptions to this were one animal at 175 mg/kg bw per day and two animals at 350 mg/kg bw per day which fell below the laboratory's historical control 95% reference range. However, mean %tail intensity values for all test article treated groups were within the laboratory's historical vehicle ranges.
Comet assay in duodenum – experiment 2
In the groups of animals administered with hex‐2(trans)‐enal, group mean %tail intensity values were comparable with the group mean vehicle control data. There were no statistically significant differences in %tail intensity between treated groups and the vehicle control group. All individual animal data at all dose levels were generally consistent with the vehicle control animals and fell within the laboratory's historical control data. The only exceptions to this were three animals in the dose group of 87.5 mg/kg bw per day and one animal in the dose group of 350 mg/kg bw per day which fell below the laboratory's historical control 95% reference range, but one animal in the control group was also below. However, mean %tail intensity values for all test article treated groups were within the laboratory's historical vehicle ranges.
This result that did not confirm the decrease of tail intensity in animals treated with hex‐2(trans)‐enal observed in experiment 1, allowed to conclude that this compound did not induce DNA damage in the duodenum of treated male rats.
The Panel noted that the clinical signs of toxicity observed in the dose range‐finding test (e.g. piloerection, hunched posture and decreased activity), can be considered as lines of evidence for a systemic exposure of rats after oral administration. Moreover, the Panel considered additional evidence of systemic exposure from toxicokinetic studies already evaluated in FGE.200 (see Sections 2.4.2–4.2). Therefore, the Panel concluded that hex‐2(trans)‐enal [FL‐no: 05.073] induced no chromosomal damage in the rat bone marrow as demonstrated in the in vivo micronucleus assay. In the comet assay in the liver, the first experiment was negative and the second one equivocal. The third experiment was negative and no DNA strand breaks and no primary DNA damage due to oxidative damage were induced. The Panel concluded that hex‐2(trans)‐enal [FL‐no: 05.073] induced no primary DNA damage neither in liver nor in duodenum of rats treated by oral route.
3.3. trans‐2‐Octenal [FL‐no: 05.190] ‐ in vivo Combined Comet and Micronucleus assay
trans‐2‐Octenal [FL‐no: 05.190] (purity 97.8%) was tested in a combined micronucleus assay in bone marrow and comet assay in liver of Han Wistar rats by gavage (Beevers, 2015a). The study was performed in accordance with GLP, OECD TG 474 (OECD, 2014a) and OECD TG 489 (OECD, 2014b).
In a dose range‐finding study, groups of three males and three females were given three administrations at 0 (day 1), 24 h (day 2) and 45 h (day 3) of trans‐2‐octenal, at 2,000 and 1,400 mg/kg bw per day, via oral gavage. Clinical signs of toxicity (e.g. piloerection, ptosis and hunched posture) were observed at the highest dose tested. On day 3, one female animal was found dead prior to dosing. All other animals showed piloerection and hunched posture. In animals dosed at 1,400 mg/kg bw per day, signs of toxicity including decreased activity, piloerection, hunched posture and ptosis were observed. Body weight loss was recorded in all animals.
Based on this dose range‐finding experiment, where no differences in response between female and male rats were seen, a MTD of 1,000 mg/kg bw per day was established; the two lower doses tested were 500 and 250 mg/kg bw per day.
In the main experiment, male rats (six animals per group) were dosed at 0 (day 1), 24 h (day 2) and 45 h (day 3) by oral gavage at dose levels of 0 (corn oil), 250, 500 and 1,000 mg/kg bw per day. Animals were sampled at 48 h. Three male rats were given 150 mg/kg bw per day EMS, as a positive control. Corn oil was used as a vehicle following the same treatment schedule.
Test animals were examined daily for signs of toxicity. No clinical signs of toxicity were seen at any of the test conditions applied in the main experiment. Dose‐related decreases in body weight gain were observed, resulting in body weight loss at 1,000 mg/kg bw per day. At this dose, it was observed an increase in total protein due to an increase in globulins, resulting in a small decrease in the albumin:globulin ratio. It was observed a small decrease in sodium concentration and an increase in phosphate and creatinine. An increase in urea was observed in animals administered with the two higher doses. Glucose was increased in animals given 1,000 mg/kg bw per day, and to a lesser extent in animals given 500 mg/kg bw per day.
In the liver, it was observed a decrease in glycogen vacuolation in animals dosed with 500 and 1,000 mg/kg bw per day.
A total of at least 500 PCE and NCE were scored to calculate the degree of bone marrow toxicity by the relative decrease in PCE. Four thousand PCE per animal were scored for the presence of MN.
No decrease in PCE was observed in the bone marrow of rats treated with trans‐2‐octenal [FL‐no: 05.190], indicating no evidence of bone marrow toxicity.
Group mean results of MN frequencies were similar to the concurrent vehicle control and no statistically significant increases in MN frequency were seen for any of the dose groups. The positive control group showed statistically significant increases in MN frequencies.
Negative and positive control values were within the laboratory's historical control data.
The comet assay in liver did not show statistically significant differences in tail intensity and tail moment between the treated and control groups. All individual animal data at all dose levels were consistent with the vehicle control animals and fell within the laboratory's historical control data.
The authors of the study concluded that trans‐2‐octenal [FL‐no: 05.190] did not induce micronuclei in bone marrow and it did not induce DNA damage in the liver of rats.
The Panel noted that the clinical signs of toxicity observed in the dose range‐finding test and modification of clinical biochemistry and histopathological parameters observed in the main experiment in rats, can be considered as lines of evidence of systemic bioavailability of trans‐2‐octenal [FL‐no: 05.190] after oral administration. The Panel concluded that trans‐2‐octenal [FL‐no: 05.190] induced no primary DNA damage in the liver of rats when administered up to 1,000 mg/kg bw per day, by oral route, as demonstrated in the comet assay and that it induced no chromosomal damage in the rat bone marrow as demonstrated by the in vivo micronucleus assay.
3.4. Nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] ‐ in vivo Combined Comet and Micronucleus assay
Nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] (purity 97.92%) was tested in a combined micronucleus assay in bone marrow and comet assay in liver of Han Wistar rats by gavage (Beevers, 2015b). The study was performed in accordance with GLP, OECD TG 474 (OECD, 2014a) and OECD TG 489 (OECD, 2014b).
Based on an oral gavage dose range‐finding experiment with doses up to 2,000 mg/kg bw per day, where no differences in response between female and male rats were seen, a MTD of 700 mg/kg bw per day was established; the two lower doses tested were 350 and 150 mg/kg bw per day. In this dose range‐finding experiment, clinical signs of toxicity (e.g. decreased activity, piloerection and hunched posture) were observed at the highest dose tested. Approximately 2 h after the second dose administrations all animals were found dead or humanely killed due to the severity of their observations. Also in animals, dosed at 1,000 mg/kg bw per day signs of toxicity including decreased activity, piloerection, hunched posture, arched gait and diarrhoea were observed. Although no mortality was observed, the animal conditions deteriorated after each administration. After the first administration at 700 mg/kg bw per day, male animals showed similar signs of toxicity. After the second and third administration, clinical signs of toxicity in both males and females were limited to soiling and soft faeces; no mortality occurred.
In the main experiment, male rats (six animals per group) were dosed at 0 (day 1), 24 h (day 2) and 45 h (day 3) – by oral gavage at dose levels of 0 (corn oil), 175, 350 and 700 mg/kg bw per day. Animals were sampled at 48 h. Three male rats were given 150 mg/kg bw per day of EMS, as a positive control. Corn oil was used as a vehicle following the same treatment schedule.
Test animals were examined daily for signs of toxicity. No clinical signs of toxicity were seen at any of the test conditions applied in the main experiment. At 700 mg/kg bw per day, only soft/loose brown faces were observed. Dose‐related decreases in body weight gain were seen. The authors noted small decrease in calcium and increase in phosphate in animals dosed at 700 mg/kg bw per day. Small increase in urea was observed in animals dosed at the two higher doses. There was an increase in glucose concentration in one animal given 700 mg/kg per day. Lipaemia was observed in several serum samples. In the liver, a dose‐related decrease in glycogen vacuolation was observed in all animals dosed with nona‐2(trans),6(cis)‐dienal.
A total of at least 500 PCE and NCE were scored to calculate the degree of bone marrow toxicity by the relative decrease in PCE. Four thousand PCE per animal were scored for the presence of MN.
No decrease in PCE was observed in the bone marrow of rats treated with nona‐2(trans),6(cis)‐dienal, indicating no evidence of bone marrow toxicity.
Group mean results of MN frequencies were similar to the concurrent vehicle control and no statistically significant increases in MN frequency were seen for any of the dose groups. There was no significant linear trend. The positive control group showed statistically significant increases in MN frequencies.
Negative and positive control values were within the laboratory's historical control data. Data are summarised in Appendix E, Table E.1.
The authors of the study noted that two animals treated at 350 and 700 mg/kg bw per day, respectively, had individual hedgehog values that exceeded weakly the 95% reference range of the historical control data. These were considered to be chance events that did not affect data interpretation.
The comet assay in liver did not show statistically significant differences in tail intensity between treated and control groups. All individual animal data at all dose levels were consistent with the vehicle control animals and fell within the laboratory's historical control data.
The authors of the study concluded that nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058] did not induce micronuclei in bone marrow and it did not induce DNA damage in the liver of rats.
The Panel noted that the clinical signs of toxicity observed in the dose range‐finding test and the modification of clinical biochemistry and histopathological parameters observed in the main experiment in rats, can be considered as lines of evidence of systemic bioavailability of nona‐2(trans),6(cis)‐dienal after oral administration. The Panel concluded that nona‐2(trans),6(cis)‐dienal induced no primary DNA damage in the liver of rats when administered up to 700 mg/kg per day by oral route as demonstrated in the comet assay and that it induced no chromosomal damage in the rat bone marrow as demonstrated by the in vivo micronucleus assay.
3.5. Additional studies from literature
2‐Hexenal was tested in an Ames test with pre‐incubation and in the absence of metabolic activation. To decrease the toxicity of long‐chain enals, GSH was added following the preincubation period and before plating (Grúz et al., 2017). A parallel experiment with S. Typhimurium strains TA100, TA104, TA2637, TA97, TA98 deficient of DNA polymerase RI (DNA Pol RI) was carried out in order to investigate its role in the potential mutagenic activity. 2‐Hexenal induced base substitutions in the S. Typhimurium strains TA100 and TA104 in a concentration dependent manner. This mutagenic activity was dependent by the DNA Pol RI activity, because it was not observed in strains deficient of DNA Pol RI. The authors considered that probably the DNA Pol RI activity is needed to replicate DNA despite the possible presence of DNA adducts. No mutagenic activity was demonstrated in all the other tested strains under the same testing conditions.
Induction of oxidative DNA damage was investigated in mammalian cells (V79 and Caco‐2), as a consequence of GSH depletion induced by 2‐alkenals, including E‐(2)‐hexenal (Janzowski et al., 2003). Oxidative DNA breakage was studied through in vitro Comet assay, using treatment with formamidopyrimidine DNA glycosylase (FPG). After 1 h, incubation of V79 cells with E‐(2)‐hexenal (100 μM), the total cellular glutathione (tGSH) was depleted (< 20% compared to control; viability > 85%). During 3‐h (post‐treatment) of cell incubation without E‐(2)‐hexenal, a significant increase in tGSH was observed only in the cells treated with the lowest concentration tested (30 μM). After 1h of incubation, DNA damage was observed only at the highest concentration tested (100 μM); FPG‐sensitive sites were not observed. During 3‐h (without test compound), direct DNA damage was reduced and FPG‐sensitives sites were detectable at 100 μM. Direct DNA breakage was markedly diminished, most probably by repair processes, and tGSH concentrations were observed to increase again within 3‐h post‐treatment. In Caco‐2 cells, E‐(2)‐hexenal induced less direct DNA damage compared with V79, but after 3‐h post‐treatment, direct DNA damage in Caco‐2 cells had increased compared to V79 cells. These results suggest that alkenal‐mediated oxidative stress can contribute to cytotoxic/genotoxic cell damage.
(E)‐2‐Hexenal, (E)‐2‐octenal, (E)‐2‐nonenal and (2E,6Z)‐2,6‐nonadienal were tested in mammalian cell lines (V79 and Caco‐2) and in primary human colon cells (Glaab et al., 2001). Inhibition of cell growth (IC50) and cytotoxicity (LC50) were measured in both V79 and Caco‐2 cells showing similar results. Cytotoxicity was observed after 1 h incubation in V79 cells with LC50 of 3,666, 545, 271 and 270 μM for (E)‐2‐hexenal, (E)‐2‐octenal (E)‐2‐nonenal and (2E,6Z)‐2,6‐nonadienal, respectively. If the incubation time was prolonged up to 24 h, an IC50 of 17, 6, 9 and 5 μM was obtained for (E)‐2‐hexenal, (E)‐2‐octenal, (E)‐2‐nonenal and (2E,6Z)‐2,6‐nonadienal, respectively. DNA damage (both strand breaks and oxidised purines) was investigated through the in vitro comet assay.
With (E)‐2‐hexenal and (E)‐2‐octenal, concentration‐dependent DNA damage was induced in V79 cells and in Caco‐2 cells. In both cell lines, (E)‐2‐nonenal did not induce DNA damage up to the cytotoxic concentration limit (viability 70%), while (2E,6Z)‐2,6‐nonadienal was the most genotoxic. This substance tested in primary human colon cells showed a potency at least twofold lower compared to V79 and Caco‐2 cells.
When tested in a modified comet assay, no induction of FPG sensitive sites in V79 cells were identified at concentrations that depleted up to 25% GSH. In primary human colon cells, (E)‐2‐hexenal did not induce direct DNA damage, neither FPG sensitive sites.
In V79 cells, all the substances induced depletion of total GSH down to about 20% of control after 1 h incubation at concentrations up to 50 or 100 μM. In Caco‐2 cells and in primary human colon cells, about fivefold higher concentrations were needed to observe similar GSH depletion.
(E)‐2‐Hexenal, additionally tested in primary human colon cells, depleted GSH and increased the sensitivity towards oxidative stress.
The authors of this study concluded that an elongation of alkyl chain length of 2‐alkenals results in an increase of cytotoxicity which might be related to lipophilicity. This correlation was less evident for the DNA damage. No DNA oxidative damage was noted in this study despite the decrease of cell glutathione.
3.6. Discussion
In FGE.200 (EFSA CEF Panel, 2014a), the Panel concluded that the gene mutation induced in vitro by hex‐2(trans)‐enal [FL‐no: 05.073] was not expressed in vivo. However, the available data did not allow to assess the potential clastogenic activity.
Studies from literature suggest that the toxicity and mutagenicity of 2‐alkenals are related to the chain length (Canonero et al., 1990; Eder et al., 1992; Glaab et al., 2001). The mechanism of genotoxicity seems to be related to the formation of DNA adducts (Schuler and Eder, 1999; Schuler et al., 1999; Stout et al., 2008) and probably to base substitution (Grúz et al., 2017). Genotoxic activity has been observed especially in cells that have low detoxification capacity showing also a decrease in GSH (Eisenbrand et al., 1995; Glaab et al., 2001; Janzowski et al., 2003). Some studies suggest that the genotoxic effects are due to DNA oxidative damage, but results are inconsistent (Glaab et al., 2001; Janzowski et al., 2003).
In the study by Dittberner et al. (1995), it is suggested an aneugenic mechanism for both hex‐2(trans)‐enal and nona‐2(trans),6(cis)‐dienal. However, the Panel noted that MN are observed in a range of concentrations too wide for an aneugenic substance; in addition, the lowest concentration at which a statistically significant increase in MN is observed (50 μM for hexenal and 20 μM for nonadienal) is not the same at which the increase of aneuploidy is observed (40 μM for both hexenal and nonadienal). Therefore, the Panel concluded that the mechanism of genotoxicity was not adequately investigated in this study and that the available data suggest more a clastogenic than an aneugenic activity.
Based on the new available results from a combined in vivo micronucleus assay in bone marrow and comet assay in duodenum and liver of Han Wistar rats, the Panel considered that for hex‐2(trans)‐enal [FL‐no: 05.073] no chromosomal damage was observed in the micronucleus assay in the bone marrow and no primary DNA damage in the comet assay in duodenum. In addition, no direct DNA strand breaks and no primary DNA damage due to oxidative damage were observed in the comet assay performed in liver of rats administered via oral gavage. Proof of systemic exposure was provided.
For trans‐2‐octenal [FL‐no: 05.190], no genotoxic effect was observed in the micronucleus assay in bone marrow and no DNA damage was noted in the comet assay in liver. The changes of clinical biochemistry and histopathological parameters can be considered as lines of evidence of systemic exposure of rats after oral administration.
For nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058], no genotoxic effect was observed in the micronucleus assay in bone marrow and no DNA damage was noted in the comet assay in liver. The changes of clinical biochemistry and histopathological parameters can be considered as lines of evidence of systemic exposure of rats after oral administration.
The extrapolation from 3 representative substances to 71 candidate substances is intrinsically associated with the introduction of uncertainty. In this opinion, only one toxicological endpoint is addressed (genotoxicity) related to one well defined structural alert, therefore the uncertainty is considered to be limited. Other toxicological endpoints will be addressed in separate opinions.
3.7. Conclusions
Based on these data, the Panel concluded that the concern for genotoxicity can be ruled out for the representative substances hex‐2(trans)‐enal [FL‐no: 05.073], trans‐2‐octenal [FL‐no: 05.190], nona‐2(trans),6(cis)‐dienal [FL‐no: 05.058], therefore all the 74 substances [FL‐no: 02.020, 02.049, 02.050, 02.090, 02.112, 02.137, 02.156, 02.192, 02.210, 02.231, 05.037, 05.058, 05.060, 05.070, 05.072, 05.073, 05.076, 05.078, 05.102, 05.109, 05.111, 05.114, 05.120, 05.144, 05.150, 05.171, 05.172, 05.179, 05.184, 05.189, 05.190, 05.191, 05.195, 06.025, 06.031, 06.072, 09.054, 09.097, 09.109, 09.119, 09.146, 09.233, 09.244, 09.247, 09.276, 09.277, 09.303, 09.312, 09.385, 09.394, 09.395, 09.396, 09.397, 09.398, 09.399, 09.400, 09.410, 09.411, 09.469, 09.482, 09.489, 09.492, 09.493, 09.498, 09.678, 09.701, 09.719, 09.741, 09.790, 09.841, 09.866, 09.947, 09.948, 13.004] can be evaluated through the Procedure for flavouring substances.
Documentation provided to EFSA
Beevers C, 2013. trans‐2‐Hexenal: Induction of lacZ mutations in the liver and duodenum of treated Muta™Mice. Covance Laboratories Ltd. Study no. 8262052. July 2013. Unpublished study report submitted by EFFA to FLAVIS Secretariat.
Beevers C, 2015a. Oct‐2‐enal: Rat Micronucleus and Alkaline Comet Assay. Covance Laboratories Ltd. Study no. 8312070. June 2015. Unpublished study report.
Beevers C, 2015b. Nonadienal Trans, Cis‐2,6: Rat Micronucleus and Alkaline Comet Assay. Covance Laboratories Ltd. Study no. 8312068. August 2015. Unpublished study report.
Benigni R and Netzeva T, 2007a. Report on a QSAR model for prediction of genotoxicity of α,ß‐unsaturated aldehydes in S. typhimurium TA100 and its application for predictions on α,ß‐unsaturated aldehydes in Flavouring Group Evaluation 19 (FGE.19). Unpublished report submitted by FLAVIS Secretariat to EFSA.
Benigni R and Netzeva T, 2007b. Report on a QSAR model for prediction of genotoxicity of α,ß‐unsaturated ketones in S. typhimurium TA100 and its application for predictions on α,ß‐unsaturated aldehydes in Flavouring Group Evaluation 19 (FGE.19). Unpublished report submitted by FLAVIS Secretariat to EFSA.
Durward R, 2009. In vivo liver unscheduled DNA synthesis (UDS) assay. 2‐Hexenal. Harlan Laboratories Ltd. Project no. 1834/0010. July 20, 2009. Unpublished report submitted by EFFA to FLAVIS Secretariat.
EFFA (European Flavour Association), 2010. Submission by the European Flavour Association to the European Food Safety Authority. Flavouring Group Evaluation 19 Subgroup 1.1.1 (corresponding to FGE.200): Submission of additional data related to FGE.19 subgroup 1.1.1. 25 Flavouring Substances (Flavouring Substances) of the Chemical Group 3 (Annex I of 1565/2000/EC) Structurally Related to Straight‐Chain Aliphatic Acyclic alpha,beta‐Unsaturated Aldehydes, with or without non‐Conjugated Double Bonds, Used as Flavouring Substances. 14 April 2010.
EFFA (European Flavour Association), 2018. Submission by the European Flavour Association to the European Food Safety Authority. Submission of recent poundage (volume of use) information for flavouring substances of FGE.200 (FGE.19 subgroup 1.1.1). 31 August 2018.
Gry J, Beltoft V, Benigni R, Binderup M‐L, Carere A, Engel K‐H, Gürtler R, Jensen GE, Hulzebos E, Larsen JC, Mennes W, Netzeva T, Niemelä J, Nikolov N, Nørby KK and Wedebye EB, 2007. Description and validation of QSAR genotoxicity models for use in evaluation of flavouring substances in Flavouring Group Evaluation 19 (FGE.19) on 360 α,ß‐unsaturated aldehydes and ketones and precursors for these. Unpublished report submitted by FLAVIS Secretariat to EFSA.
Honarvar N, 2007a. Micronucleus assay in bone marrow cells of the mouse with trans‐2‐Hexenal. Cyotest Cell Research GmbH, Germany. Study no. 1064907. December 07, 2007. Unpublished Report to the Research Institute for Fragrance Materials, Inc., Woodcliff Lake, NJ.
Honarvar N, 2007b. Micronucleus assay in bone marrow cells of the mouse with trans‐2‐dodecenal. Cyotest Cell Research GmbH, Germany. Study no. 1064909. September 17, 2007. Unpublished Report to the Research Institute for Fragrance Materials, Inc., Woodcliff Lake, NJ.
Honarvar N, 2008. Micronucleus assay in bone marrow cells of the mouse with 2‐nonenal. Cyotest Cell Research GmbH, Germany. Study no. 1114009. May 14, 2008. Unpublished Report to the Research Institute for Fragrance Materials, Inc., Woodcliff Lake, NJ.
IOFI (International Organization of the Flavor Industry), 2013. Flavouring Group Evaluation 19 Subgroup 1.1.1: 74 Flavouring Substances (Flavouring Substances) of the Chemical Group 3 (Annex I of 1565/2000/EC) Structurally related to straight‐chain aliphatic acyclic α,ß‐unsaturated aldehydes, with or without non‐conjugated double bonds, used as flavouring substances. FGE.200. 25/06/2013. FLAVIS/8.192.
Keig‐Shevlin Z, 2017. Trans‐2‐hexenal: rat micronucleus and alkaline Comet assay. Covance Laboratories Ltd. Study no. 8321904. August 2017. Unpublished study report.
Nikolov N, Jensen GE, Wedebye EB and Niemelä J, 2007. Report on QSAR predictions of 222 α,ß‐unsaturated aldehydes and ketones from Flavouring Group Evaluation 19 (FGE.19) on 360 α,ß‐unsaturated aldehydes and ketones and precursors for these. Unpublished report submitted by FLAVIS Secretariat to EFSA.
Sokolowski A, 2007a. Salmonella typhimurium reverse mutation with trans‐2‐hexenal. RCC Cytotest Cell Research GmbH. RIFM report no. 54280. Study no. 1064901. April 16, 2007. Unpublished Report to the Research Institute for Fragrance Materials, Inc., Woodcliff Lake, NJ.
Sokolowski A, 2007b. Salmonella typhimurium reverse mutation with trans‐2‐dodecenal. RCC Cytotest Cell Research GmbH. RIFM report no. 54281. Study no. 1064902. April 19, 2007. Unpublished Report to the Research Institute for Fragrance Materials, Inc., Woodcliff Lake, NJ.
Abbreviations
- ABS
chromosomal aberrations
- ALDH
aldehyde dehydrogenase
- AR
aldose reductase
- BrdU
bromodeoxyuridine
- bw
body weight
- CAS
Chemical Abstract Service
- CEF
Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids
- CHL
Chinese hamster lung (cells)
- CHO
Chinese hamster ovary (cells)
- CoE
Council of Europe
- DNA
Pol RI DNA polymerase RI
- EFFA
European Flavour Association
- EGF
epidermal growth factor
- EMS
ethyl methanesulfonate
- FAF
Panel on Food Additives and Flavourings
- FAO
Food and Agriculture Organization of the United Nations
- FEMA
Flavor and Extract Manufacturers Association
- FGE
Flavouring Group Evaluation
- FLAVIS
(FL) Flavour Information System (database)
- FPG
formamidopyrimidine DNA glycosylase
- GLP
Good Laboratory Practice
- GSH
glutathione
- GST
glutathione S‐transferase
- Hex‐PdG
1,N 2‐propanodeoxyguanosine
- HPLC
high‐performance liquid chromatography
- HPRT
hypoxanthine guanine ribosyl transferase
- hOGG1
human 8‐hydroxyguanine DNA‐glycosylase 1
- ID
Identity
- IOFI
International Organization of the Flavor Industry
- IR
infrared spectroscopy
- JECFA
Joint FAO/WHO Expert Committee on Food Additives
- LC–MS/MS
liquid chromatography with tandem mass spectrometry
- MF
mutation frequency
- MN
micronuclei
- MS
mass spectra
- MTD
maximum tolerated dose
- MSDI
maximised survey‐derived daily intake
- NAD+
(oxidised) β‐nicotinamide adenine dinucleotide phosphate
- NCE
normochromatic erythrocytes
- NMR
nuclear magnetic resonance
- NOAEL
no‐observed‐adverse‐effect‐level
- No
Number
- OECD
Organisation for Economic Co‐operation and Development
- PBK/D
physiologically based kinetic/dynamic
- PCE
polychromatic erythrocytes
- PFU
plaque‐forming unit
- (Q)SAR
(Quantitative) Structure Activity Relationship
- RET
reticulocytes
- SCE
sister chromatid exchange
- SOD
superoxide dismutase
- TG
6‐thioguanine
- tGSH
total cellular glutathione
- TLC
thin‐layer chromatography
- UDS
unscheduled DNA synthesis
- WHO
World Health Organization
Appendix A – Specification summary of the substances in the Flavouring Group Evaluation 200Rev1
1.
Appendix B – Summary of safety evaluation applying the Procedure
1.
Table B.1.
Summary of Safety Evaluation of the JECFA Substances in the Present Group (JECFA, 1996, 1999, 2001, 2003, 2005, 2007, 2009)
| FL‐no JECFA‐no | EU register name | Structural formula | EU MSDIa US MSDI (μg/capita per day) | Classb Evaluation procedure pathc | JECFA outcome on the named compoundd ore | EFSA conclusion on the named compound |
|---|---|---|---|---|---|---|
|
02.020 1354 |
Hex‐2‐en‐1‐ol |

|
653.48 291 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.049 1184 |
Nona‐2,6‐dien‐1‐ol |

|
9.07 1 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.050 1793 |
Pent‐2‐en‐1‐ol |
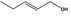
|
2.44 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.090 1365 |
Non‐2(trans)‐en‐1‐ol |
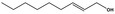
|
0.02 0.03 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.112 1369 |
Non‐2(cis)‐en‐1‐ol |
|
0.01 2 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.137 1794 |
Dec‐2‐en‐1‐ol |
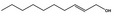
|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.156 1374 |
Hex‐2(cis)‐en‐1‐ol |
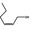
|
0.01 10 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 02.192 | Oct‐2‐en‐1‐ol |
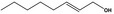
|
7.71 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
02.210 1384 |
Undec‐2‐en‐1‐ol |

|
0.01 1 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 02.231 | trans‐2, cis‐6‐Nonadien‐1‐ol |
|
8.73 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.037 1350 |
2‐Dodecenal |
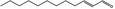
|
1.19 2 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.058 1186 |
Nona‐2(trans),6(cis)‐dienal |
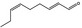
|
15.78 24 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.060 1363 |
Oct‐2‐enal |

|
0.84 0.9 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.070 1360 |
2‐Heptenal |
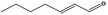
|
8.17 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.072 1362 |
trans‐2‐Nonenal |
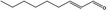
|
1.70 0.12 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.073 1353 |
Hex‐2(trans)‐enal |

|
2761 409 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.076 1349 |
Dec‐2‐enal |

|
12.94 6 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.078 1359 |
Tridec‐2‐enal |

|
0.97 0.7 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.102 1364 |
Pent‐2‐enal |

|
0.37 0.1 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.109 1366 |
2‐Undecenal |

|
0.65 0.4 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.111 1182 |
Octa‐2(trans),6(trans)‐dienal |
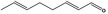
|
0.01 0.007 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.114 1208 |
4‐Methylpent‐2‐enal |

|
0.01 0.2 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.120 1197 |
Dodeca‐2,6‐dienal |

|
0.01 0.009 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 05.144 | Dodec‐2(trans)‐enal |
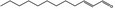
|
0.75 | No evaluation | Not evaluated by the JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.150 1360 |
Hept‐2(trans)‐enal |

|
16.27 30 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.171 1362 |
Non‐2‐enal |

|
9.89 0.4 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.172 1187 |
Nona‐2(trans),6(trans)‐dienal |
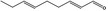
|
6.52 0.007 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
05.179 1803 |
Tetradec‐2‐enal |

|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 05.184 | Undec‐2(trans)‐enal |
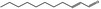
|
0.84 ND |
No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 05.189 | 2‐Hexenal |
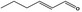
|
1.22 409 |
Class I No evaluation |
Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure. |
| 05.190 | trans‐2‐Octenal |
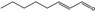
|
0.79 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 05.191 | trans‐2‐Decenal |

|
8.10 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 05.195 | trans‐2‐Tridecenal |
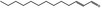
|
0.12 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
06.025 946 |
1,1‐Diethoxynona‐2,6‐diene |
|
0.01 0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
06.031 1383 |
1,1‐Diethoxyhex‐2‐ene |
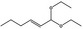
|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
06.072 1728 |
1,1‐Dimethoxyhex‐2(trans)‐ene |
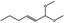
|
0.01 ND |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.054 2 |
Allyl butyrate |
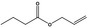
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.097 4 |
Allyl heptanoate |
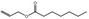
|
12.92 28 |
Class II A3: Intake above threshold, A4: Endogenous |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.109 6 |
Allyl nonanoate |
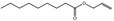
|
0.01 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.119 5 |
Allyl octanoate |
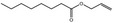
|
0.01 1.3 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.146 9 |
Allyl undec‐10‐enoate |
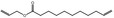
|
0.01 < 0.01 |
Class II A3: Intake above threshold, A4: Endogenous |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.233 1 |
Allyl propionate |
|
0.57 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.244 3 |
Allyl hexanoate |
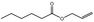
|
3583.8 820 |
Class II B3: Intake above threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
| 09.247 | Allyl crotonate |
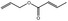
|
0.04 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.276 1367 |
Oct‐2‐enyl acetate |

|
0.03 0.7 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.277 1368 |
Oct‐2(trans)‐enyl butyrate |

|
0.15 0.7 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.303 1799 |
Hept‐2‐enyl isovalerate |

|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.312 8 |
Allyl hexa‐2,4‐dienoate |
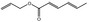
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.385 1798 |
Hept‐2‐enyl acetate |

|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.394 1355 |
Hex‐2(trans)‐enyl acetate |

|
272.73 56 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.395 1378 |
Hex‐2(trans)‐enyl propionate |
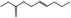
|
0.08 4 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.396 1375 |
Hex‐2‐enyl butyrate |
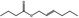
|
5.62 4 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.397 1376 |
Hex‐2‐enyl formate |

|
0.01 7 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.398 1381 |
Hex‐2‐enyl hexanoate |

|
1.36 0.09 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.399 1377 |
Hex‐2‐enyl isovalerate |
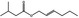
|
1.44 4 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 09.400 | Hex‐2‐enyl phenylacetate |
|
0.01 |
Class I No evaluation |
Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.410 11 |
Allyl 2‐ethylbutyrate |
|
0.01 0.02 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.411 14 |
Allyl cyclohexanebutyrate |
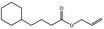
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.469 15 |
Allyl cyclohexanevalerate |
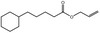
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.482 12 |
Allyl cyclohexaneacetate |
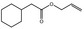
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.489 7 |
Allyl isovalerate |
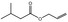
|
0.06 0.19 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.492 16 |
Allyl cyclohexanehexanoate |
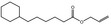
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.493 10 |
Allyl 2‐methylcrotonate |
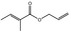
|
0.01 < 0.01 |
Class II B3: intake below threshold B4: adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.498 13 |
Allyl cyclohexanepropionate |
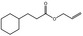
|
96.57 110 |
Class II B3: intake below threshold B4: adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.678 1795 |
Pent‐2‐enyl hexanoate |
|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.701 18 |
Allyl phenoxyacetate |
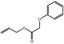
|
9.93 2.5 |
Class III B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.719 20 |
Allyl anthranilate |
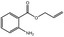
|
0.01 0.09 |
Class III B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.741 19 |
Allyl cinnamate |
|
0.01 0.28 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.790 17 |
Allyl phenylacetate |
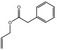
|
0.01 < 0.01 |
Class II B3: Intake below threshold, B4: Adequate NOAEL exists |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. Evaluated by JECFA before 2000 |
|
09.841 1796 |
2‐Hexenyl octanoate |
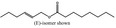
|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 09.866 | Allyl valerate |
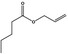
|
0.01 | No evaluation | Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
09.947 1188 |
(E,Z)‐2,6‐Nonadienyl acetate |
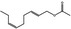
|
0.01 |
Class I A3: Intake below threshold |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
| 09.948 | (2E)‐2‐Nonenyl acetate |

|
0.01 |
Class I No evaluation |
Not evaluated by JECFA | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
|
13.004 21 |
Allyl 2‐furoate |
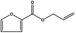
|
0.01 < 0.01 |
Class III B3: Intake below threshold, B4: No adequate NOAEL JECFA evaluated at step B5: intake below 1.5 μg/person per day |
d | Evaluated in FGE.200Rev1 as of no genotoxicity concern. The substance can be evaluated through the Procedure |
JECFA: The Joint FAO/WHO Expert Committee on Food Additives; FL‐no: FLAVIS number; MSDI: maximised survey‐derived daily intake; FGE: Flavouring Group Evaluation; NOAEL: no‐observed‐adverse‐effect‐level; ND: not determined.
EU MSDI: Amount added to food as flavour in (kg/year) × 10E9/(0.1 × population in Europe (= 375 × 10E6) × 0.6 × 365) = μg/capita per day. EU MSDI values calculated based on the most recent EFFA poundage information (kg/year) for the year 2015 (EFFA, 2018).
Thresholds of concern: Class I = 1,800 μg/person per day, Class II = 540 μg/person per day, Class III = 90 μg/person per day.
Procedure path A substances can be predicted to be metabolised to innocuous products. Procedure path B substances cannot.
No safety concern based on intake calculated by the MSDI approach of the named compound.
Data must be available on the substance or closely related substances to perform a safety evaluation.
Appendix C – (Q)SAR predictions on mutagenicity for aldehydes from subgroup 1.1.1
1.
Appendix D – Genotoxicity studies evaluated in FGE.200
1.
Appendix E – Genotoxicity data evaluated in FGE.200Rev1
1.
Suggested citation: EFSA FAF Panel (EFSA Panel on Food Additives and Flavourings) , Younes M, Aquilina G, Castle L, Engel K‐H, Fowler P, Frutos Fernandez MJ, Fürst P, Gundert‐Remy U, Husøy T, Mennes W, Moldeus P, Oskarsson A, Rainieri S, Shah R, Waalkens‐Berendsen I, Wölfle D, Binderup M‐L, Bolognesi C, Marcon F, Marzin D, Mosesso P, Carfì M, Vianello G and Gürtler R, 2018. Scientific Opinion on Flavouring Group Evaluation 200, Revision 1 (FGE.200 Rev.1): 74 α,β‐unsaturated aliphatic aldehydes and precursors from chemical subgroup 1.1.1 of FGE.19. EFSA Journal 2018;16(10):5422, 60 pp. 10.2903/j.efsa.2018.5422
Requestor: European Commission
Question numbers: EFSA‐Q‐2018‐00148, EFSA‐Q‐2018‐00149, EFSA‐Q‐2018‐00150, EFSA‐Q‐2018‐00151, EFSA‐Q‐2018‐00152, EFSA‐Q‐2018‐00153, EFSA‐Q‐2018‐00154, EFSA‐Q‐2018‐00155, EFSA‐Q‐2018‐00156, EFSA‐Q‐2018‐00157, EFSA‐Q‐2018‐00158, EFSA‐Q‐2018‐00159, EFSA‐Q‐2018‐00160, EFSA‐Q‐2018‐00161, EFSA‐Q‐2018‐00162, EFSA‐Q‐2018‐00163, EFSA‐Q‐2018‐00164, EFSA‐Q‐2018‐00165, EFSA‐Q‐2018‐00166, EFSA‐Q‐2018‐00167, EFSA‐Q‐2018‐00168, EFSA‐Q‐2018‐00169, EFSA‐Q‐2018‐00170, EFSA‐Q‐2018‐00171, EFSA‐Q‐2018‐00172, EFSA‐Q‐2018‐00173, EFSA‐Q‐2018‐00174, EFSA‐Q‐2018‐00175, EFSA‐Q‐2018‐00176, EFSA‐Q‐2018‐00177, EFSA‐Q‐2018‐00178, EFSA‐Q‐2018‐00179, EFSA‐Q‐2018‐00180, EFSA‐Q‐2018‐00181, EFSA‐Q‐2018‐00182, EFSA‐Q‐2018‐00183, EFSA‐Q‐2018‐00184, EFSA‐Q‐2018‐00185, EFSA‐Q‐2018‐00186, EFSA‐Q‐2018‐00187, EFSA‐Q‐2018‐00188, EFSA‐Q‐2018‐00189, EFSA‐Q‐2018‐00190, EFSA‐Q‐2018‐00191, EFSA‐Q‐2018‐00192, EFSA‐Q‐2018‐00193, EFSA‐Q‐2018‐00194, EFSA‐Q‐2018‐00195, EFSA‐Q‐2018‐00196, EFSA‐Q‐2018‐00197, EFSA‐Q‐2018‐00198, EFSA‐Q‐2018‐00199, EFSA‐Q‐2018‐00200, EFSA‐Q‐2018‐00201, EFSA‐Q‐2018‐00202, EFSA‐Q‐2018‐00203, EFSA‐Q‐2018‐00204, EFSA‐Q‐2018‐00205, EFSA‐Q‐2018‐00206, EFSA‐Q‐2018‐00207, EFSA‐Q‐2018‐00208, EFSA‐Q‐2018‐00209, EFSA‐Q‐2018‐00210, EFSA‐Q‐2018‐00211, EFSA‐Q‐2018‐00212, EFSA‐Q‐2018‐00213, EFSA‐Q‐2018‐00214, EFSA‐Q‐2018‐00215, EFSA‐Q‐2018‐00216, EFSA‐Q‐2018‐00217, EFSA‐Q‐2018‐00218, EFSA‐Q‐2018‐00219, EFSA‐Q‐2018‐00220 and EFSA‐Q‐2018‐00626
Panel members: Gabriele Aquilina, Laurence Castle, Karl‐Heinz Engel, Paul Fowler, Maria Jose Frutos Fernandez, Peter Fürst, Rainer Gürtler, Ursula Gundert‐Remy, Trine Husøy, Wim Mennes, Peter Moldeus, Agneta Oskarsson, Sandra Rainieri, Romina Shah, Ine Waalkens‐Berendsen, Detlef Wölfle and Maged Younes.
Adopted: 13 September 2018
Notes
Regulation (EC) No 1334/2008 of the European Parliament and of the Council of 16 December 2008 on flavourings and certain food ingredients with flavouring properties for use in and on foods and amending Council Regulation (EEC) No 1601/91, Regulations (EC) No 2232/96 and (EC) No 110/2008 and Directive 2000/13/EC. OJ L 354, 31.12.2008, p. 34–50.
Commission implementing Regulation (EU) No 872/2012 of 1 October 2012 adopting the list of flavouring substances provided for by Regulation (EC) No 2232/96 of the European Parliament and of the Council, introducing it in Annex I to Regulation (EC) No 1334/2008 of the European Parliament and of the Council and repealing Commission Regulation (EC) No 1565/2000 and Commission Decision 1999/217/EC. OJ L 267, 2.10.2012, p. 1–161.
Commission Regulation (EC) No 1565/2000 of 18 July 2000 laying down the measures necessary for the adoption of an evaluation programme in application of Regulation (EC) No 2232/96. OJ L 180, 19.7.2000, p. 8–16.
Commission Regulation (EU) 2017/1250 of 11 July 2017 amending Annex I to Regulation (EC) No 1334/2008 of the European Parliament and of the Council as regards removal from the Union list of the flavouring substance 4,5‐epoxydec2(trans)‐enal. OJ L 179, 12.7.2017, p. 3–5.
Commission Regulation (EU) 2017/1250 of 11 July 2017 amending Annex I to Regulation (EC) No 1334/2008 of the European Parliament and of the Council as regards removal from the Union list of the flavouring substance 4,5‐epoxydec‐2(trans)‐enal. OJ L 179, 12.07.2017, p. 3–5.
The data presented in Section 2.4 are cited from the Scientific Opinion FGE.200.
Detection limit 0.03 adducts per 106 nucleotides.
The limit of quantitation was 0.015 fmol Hex‐PdG/μg DNA (200 μg DNA) or 0.006 fmol Hex‐PdG/μg DNA (500 μg DNA).
References
- Bhatia SP, Politano VT and Api AM, 2010. Genotoxicity tests conducted on a group of structurally related aldehydes. Presentation at the Society of Toxicology 49th Annual Meeting, March 7‐11, 2010, Salt Lake City. [Abstract reported in: Toxicologist, 114(1), 216].
- Canonero R, Martelli A, Marinari UM and Brambilla G, 1990. Mutation induction in Chinese hamster lung V79 cells by five alk‐2‐enals produced by lipid peroxidation. Mutation Research, 224(2), 153–156. [DOI] [PubMed] [Google Scholar]
- Chung F‐L, Nath RG, Nagao M, Nishikawa A, Zhou G‐D and Randerath K, 1999. Endogenous formation and significance of 1, N2‐propanodeoxyguanosine adducts. Mutation Research, 424, 71–81. [DOI] [PubMed] [Google Scholar]
- Coles B and Ketterer B, 1990. The role of glutathione and glutathione transferases in chemical carcinogenesis. Critical Reviews in Biochemistry and Molecular Biology, 25, 47–70. [DOI] [PubMed] [Google Scholar]
- Dittberner U, Eisenbrand G and Zankl H, 1995. Genotoxic effects of the α,β‐unsaturated aldehydes 2‐trans‐butenal, 2‐trans‐hexenal and 2‐trans,6‐cis‐nonadienal. Mutation Research, 335, 259–265. [DOI] [PubMed] [Google Scholar]
- Dittberner U, Schmetzer B, Golzer P, Eisenbrand G and Zankl H, 1997. Genotoxic effects of 2‐trans‐hexenal in human buccal mucosa cells in vivo. Mutation Research, 390, 161–165. [DOI] [PubMed] [Google Scholar]
- Eckl PM, Ortner A and Esterbauer H, 1993. Genotoxic properties of 4‐hydroxyalkenals and analogous aldehydes. Mutation Research, 290(2), 183–192. [DOI] [PubMed] [Google Scholar]
- Eder E, Deininger C, Neudecker T and Deininger D, 1992. Mutagenicity of beta‐alkyl substituted acrolein congeners in the Salmonella typhimurium strain TA100 and genotoxicity testing in the SOS chromotest. Environmental and Molecular Mutagenesis, 19, 338–345. [DOI] [PubMed] [Google Scholar]
- Eder E, Scheckenbach S, Deininger C and Hoffman C, 1993. The possible role of α, ß‐unsaturated carbonyl compounds in mutagenesis and carcinogenesis. Toxicology Letters, 67, 87–103. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2008a. Minutes of the 26th Plenary meeting of the Scientific Panel on Food Additives, Flavourings, Processing Aids and Materials in Contact with Food, Held in Parma on 27 ‐ 29 November 2007. Parma, 7 January 2008. Available online: http://www.efsa.europa.eu/en/events/event/afc071127.htm.
- EFSA (European Food Safety Authority), 2008b. Statement of the Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF) on Genotoxicity Test Strategy for Substances belonging to Subgroups of FGE.19. EFSA Journal 2008;6(12):854, 1–5. Available online: 10.2903/j.efsa.2008.854 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2008c. Statement of the Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF) on List of alpha,beta‐unsaturated aldehydes and ketones representative of FGE.19 substances for genotoxicity testing. EFSA Journal 2008;6(12):910, 1–7. Available online: 10.2903/j.efsa.2008.910 [DOI] [Google Scholar]
- EFSA CEF Panel (EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids), 2011. Statement on data provided for subgroup 1.1.1 of FGE.19. EFSA Journal 2011;9(2):2086, 5 pp. 10.2903/j.efsa.2011.2086 [DOI] [Google Scholar]
- EFSA CEF Panel (EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids), 2014a. Scientific Opinion on Flavouring Group Evaluation 200 (FGE.200): 74 α,β‐unsaturated aldehydes and precursors from subgroup 1.1.1 of FGE.19. EFSA Journal 2014;12(6):3709, 57 pp. 10.2903/j.efsa.2014.3709 [DOI] [Google Scholar]
- EFSA CEF Panel (EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids), 2014b. Scientific Opinion on Flavouring Group Evaluation 203, Revision 1 (FGE.203Rev1): α,β‐Unsaturated aliphatic aldehydes and precursors from chemical subgroup 1.1.4 of FGE.19 with two or more conjugated double‐bonds and with or without additional non‐conjugated double‐bonds. EFSA Journal 2014;12(4):3626, 31 pp. 10.2903/j.efsa.2014.3626 [DOI] [Google Scholar]
- EFSA CEF Panel (EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids), 2017. Scientific Opinion on Flavouring Group Evaluation 226 Revision 1 (FGE.226Rev1): consideration of genotoxicity data on one α,β‐unsaturated aldehyde from chemical subgroup 1.1.1(b) of FGE.19. EFSA Journal 2017;15(5):4847, 24 pp. 10.2903/j.efsa.2017.4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbrand G, Schuhmacher J and Gölzer P, 1995. The influence of glutathione and detoxifying enzymes on DNA damage induced by 2‐alkenals in primary rat hepatocytes and human lymphoblastoid cells. Chemical Research in Toxicology, 8, 40–46. [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Eckl P and Ortner A, 1990. Possible mutagens derived from lipids and lipid precursors. Mutation Research, 238, 223–233. [DOI] [PubMed] [Google Scholar]
- Florin I, Rutberg L, Curvall M and Enzell CR, 1980. Screening of tobacco smoke constituents for mutagenicity using the Ames’ test. Toxicology, 18, 219–232. [DOI] [PubMed] [Google Scholar]
- Food Chemical Codex , 1996. Committee on Food Chemicals Codex. Food and Nutrition Board. Institute of Medicine, National academy of Sciences. National Academy press, Washington, DC. [Google Scholar]
- Glaab V, Collins AR, Eisenbrand G and Janzowski C, 2001. DNA‐damaging potential and glutathione depletion of 2‐cyclohexene‐1‐one in mammalian cells, compared to food relevant 2‐alkenals. Mutation Research, 497, 185–197. [DOI] [PubMed] [Google Scholar]
- Golzer P, Janzowski C, Pool‐Zobel BL and Eisenbrand G, 1996. (E)‐2‐Hexenal‐induced DNA damage and formation of cyclic 1, N2‐(1,3‐propano)‐2’‐deoxyguanosine adducts in mammalian cells. Chemical Research in Toxicology, 9, 1207–1213. [DOI] [PubMed] [Google Scholar]
- Griffin DS and Segall HJ, 1986. Genotoxicity and cytotoxicity of selected pyrrolizidine alkaloids, a possible alkenal metabolite of the alkaloids, and related alkenals. Toxicology and Applied Pharmacology, 86, 227–234. [DOI] [PubMed] [Google Scholar]
- Grootveld M, Atherton MD, Sheerin AN, Hawkes J, Blake DR, Richens TE, Silwood CJ, Lynch E and Claxson AW, 1998. In vivo absorption, metabolism, and urinary excretion of α, β‐Unsaturated aldehydes in experimental animals. Relevance to the development of cardiovascular diseases by the dietary ingestion of thermally stressed polyunsaturate‐rich culinary oils. The Journal of Clinical Investigation, 101, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grúz P, Shimizu M, Sugiyama KI and Honma M, 2017. Mutagenicity of omega‐3 fatty acid peroxidation products in the Ames test. Mutation research, 819, 14–19. [DOI] [PubMed] [Google Scholar]
- Janzowski C, Glaab V, Mueller C, Straesser U, Kamp HG and Eisenbrand G, 2003. Alpha, beta‐unsaturated carbonyl compounds: induction of oxidative DNA damage in mammalian cells. Mutagenesis, 18, 465–470. [DOI] [PubMed] [Google Scholar]
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 1996. Geneva, Switzerland. Evaluation of certain food additives and contaminants: forty‐sixth report of the Joint FAO/WHO Expert Committee on Food Additives.
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 1999. Geneva, Switzerland. Evaluation of certain food additives and contaminants: forty‐ninth report of the Joint FAO/WHO Expert Committee on Food Additives. [PubMed]
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 2001. Rome, Italy. Evaluation of certain food additives and contaminants: fifty‐seventh report of the Joint FAO/WHO Expert Committee on Food Additives.
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 2003. Rome, Italy. Evaluation of certain food additives and contaminants: sixty‐first report of the Joint FAO/WHO Expert Committee on Food Additives.
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 2005. Geneva, Switzerland. Evaluation of certain food additives: sixty‐third report of the Joint FAO/WHO Expert Committee on Food Additives.
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 2007. Geneva, Switzerland. Evaluation of certain food additives and contaminants: sixty‐eighth report of the Joint FAO/WHO Expert Committee on Food Additives.
- JECFA (Joint FAO/WHO Expert Committee on Food Additives), 2009. Rome, Italy. Evaluation of certain food additives: sixty‐ninth report of the Joint FAO/WHO Expert Committee on Food Additives.
- Kato F, Araki A, Nozaki K and Matsushima T, 1989. Mutagenicity of aldehydes and diketones. Mutation Research, 216, 366–367. [Google Scholar]
- Kelson TL, Secor McVoy JR and Rizzo WB, 1997. Human liver fatty aldehyde dehydrogenase: microsomal localization, purification, and biochemical characterization. Biochimica et Biophysica Acta, 1335, 99–110. [DOI] [PubMed] [Google Scholar]
- Kiwamoto R, Rietjens MCM and Punt A, 2012. A physiologically based in silico model for trans‐2‐hexenal detoxification and DNA adduct formation in rat. Chemical Research in Toxicology, 25(12), 2630–2641. [DOI] [PubMed] [Google Scholar]
- Kiwamoto R, Spenkelink A and Rietjens MCM, 2013. A physiologically based in silico model for trans‐2‐hexenal detoxification and DNA adduct formation in human including interindividual variation indicates efficient detoxification and a negligible genotoxicity risk. Archives of toxicology, 87, 1725–1737. [DOI] [PubMed] [Google Scholar]
- Lamé MW and Segall HJ, 1986. Metabolism of the pyrrolizidine alkaloid metabolite trans‐4‐hydroxy‐2‐hexenal by mouse liver aldehyde dehydrogenases. Toxicology and Applied Pharmacology, 82, 94–103. [DOI] [PubMed] [Google Scholar]
- Marnett LJ, Hurd HK, Hollstein MC, Levin DE, Esterbauer H and Ames BN, 1985. Naturally‐occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutation Research, 148, 25–34. [DOI] [PubMed] [Google Scholar]
- Mitchell DY and Petersen DR, 1987. The oxidation of α‐β unsaturated aldehydic products of lipid peroxidation by rat liver aldehyde dehydrogenases. Toxicology and Applied Pharmacology, 87, 403–410. [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Mihara K and Sato R, 1978. Purification and properties of a membrane‐bound aldehyde dehydrogenase from rat liver microsomes. Biochemical and Biophysical Research Communications, 83, 697–703. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co‐operation and Development), 1997a. Test No. 471: Bacterial Reverse Mutation Test. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 1997b. Test No. 474: Mammalian Erythrocyte Micronucleus Test. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 1997c. Test No. 486: Unscheduled DNA Synthesis (UDS) Test with Mammalian Liver Cells in vivo. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 2011. Test No. 488: Transgenic Rodent Somatic and Germ Cell Gene Mutation Assays. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 2014a. Test No. 474: Mammalian Erythrocyte Micronucleus Test. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 2014b. Test No. 489: in vivo mammalian alkaline comet assay. OECD Guidelines for the Testing of Chemicals, Section 4.
- OECD (Organisation for Economic Co‐operation and Development), 2016. Test No. 489: in vivo mammalian alkaline comet assay. OECD Guidelines for the Testing of Chemicals, Section 4.
- Schuler D and Eder E, 1999. Detection of 1, N2‐propanodexoyguanosine adducts of 2‐hexenal in organs of Fisher 344 rats by a 32P‐post‐labeling technique. Carcinogenesis, 20, 1345–1350. [DOI] [PubMed] [Google Scholar]
- Schuler D, Budiawan DS and Eder E, 1999. Development of a 32P‐postlabeling method for the detection of 1, N2‐propanodeoxyguanosine adducts of 2‐hexenal in vivo. Chemical Research in Toxicology, 12, 335–340. [DOI] [PubMed] [Google Scholar]
- Stout MD, Bodes E, Schoonhoven R, Upton PB, Travlos GS and Swenberg JA, 2008. Toxicity, DNA binding and cell proliferation in male F344 rats following short‐term gavage exposures to trans‐2‐hexenal. Toxicologic Pathology, 36, 232–246. [DOI] [PubMed] [Google Scholar]


