Abstract
Following a request from EFSA, the Panel on Plant Protection Products and their Residues (PPR) developed an opinion on the state of the art of Toxicokinetic/Toxicodynamic (TKTD) models and their use in prospective environmental risk assessment (ERA) for pesticides and aquatic organisms. TKTD models are species‐ and compound‐specific and can be used to predict (sub)lethal effects of pesticides under untested (time‐variable) exposure conditions. Three different types of TKTD models are described, viz., (i) the ‘General Unified Threshold models of Survival’ (GUTS), (ii) those based on the Dynamic Energy Budget theory (DEBtox models), and (iii) models for primary producers. All these TKTD models follow the principle that the processes influencing internal exposure of an organism, (TK), are separated from the processes that lead to damage and effects/mortality (TD). GUTS models can be used to predict survival rate under untested exposure conditions. DEBtox models explore the effects on growth and reproduction of toxicants over time, even over the entire life cycle. TKTD model for primary producers and pesticides have been developed for algae, Lemna and Myriophyllum. For all TKTD model calibration, both toxicity data on standard test species and/or additional species can be used. For validation, substance and species‐specific data sets from independent refined‐exposure experiments are required. Based on the current state of the art (e.g. lack of documented and evaluated examples), the DEBtox modelling approach is currently limited to research applications. However, its great potential for future use in prospective ERA for pesticides is recognised. The GUTS model and the Lemna model are considered ready to be used in risk assessment.
Keywords: Toxicokinetic/Toxicodynamic models, aquatic organisms, prospective risk assessment, time‐variable exposure, model calibration, model validation, model evaluation
Summary
In 2008, the Panel on Plant Protection Products and their Residues (PPR Panel) was tasked by the European Food Safety Authority (EFSA) with the revision of the Guidance Document on Aquatic Ecotoxicology under Council Directive 91/414/EEC (SANCO/3268/2001 rev.4 (final), 17 October 2002).1 As a third deliverable of this mandate, the PPR Panel is asked to develop a Scientific Opinion describing the state of the art of Toxicokinetic/Toxicodynamic (TKTD) models for aquatic organisms and prospective environmental risk assessment (ERA) for pesticides with the main focus on: (i) regulatory questions that can be addressed by TKTD modelling, (ii) available TKTD models for aquatic organisms, (iii) model parameters that need to be included and checked in evaluating the acceptability of regulatory relevant TKTD models, and (iv) selection of the species to be modelled.
Chapter 2 presents the underlying concepts, terminology, application domains and complexity levels of three different classes of TKTD models intended to be used in risk assessment, viz., (i) the ‘General Unified Threshold models of Survival’ (GUTS), (ii) toxicity models derived from the Dynamic Energy Budget theory (DEBtox models), and (iii) models for primary producers. All TKTD models follow the principle that the processes influencing internal exposure of an organism, summarised under Toxicokinetics (TK), are separated from the processes that lead to damage and effects/mortality, summarised by the term Toxicodynamics (TD).
The ultimate aim of GUTS is to predict survival of individuals (as influenced by mortality and/or immobility) under untested time‐variable or constant exposure conditions. The GUTS modelling framework connects the external concentration with a so‐called damage dynamic, which is in turn connected to a hazard resulting in simulated mortality/immobility when an internal damage threshold is exceeded. Within this framework, two reduced versions of GUTS are available: GUTS‐RED‐SD based on the assumption of Stochastic Death (SD) and GUTS‐RED‐IT based on the assumption of Individual Tolerance (IT).
DEBtox modelling is the application of the Dynamic Energy Budget (DEB) theory to deal with effects of toxic chemicals on life‐history traits (sublethal endpoints). DEBtox models incorporate a dynamic energy budget part for growth and reproduction endpoints at the individual level. Therefore, DEBtox models consist of two parts, (i) the DEB or ‘physiological’ part that describes the physiological energy flows and (ii) the part that accounts for uptake and effects of chemicals, named ‘TKTD part’.
The third class of TKTD models presented are developed for primary producers. With respect to the analysis of toxic effects for primary producers, the main endpoint measured is not survival but growth. For that reason, the assessment of toxic effects on algae and vascular plants needs a submodel addressing growth as a baseline, and a connected TKTD part.
Chapter 3 deals with the problem formulation step that sets the scene for the use of the TKTD models within the risk assessment. TKTD models are species and substance specific. TKTD models may either focus on standard test species (Tier‐2C1) or also incorporate relevant additional species (Tier‐2C2). If risks are triggered in Tier‐1 (standard test species approach) and exposure is likely to be shorter than in standard tests, the development of TKTD models for standard test species is the most straightforward option. If Tier‐2A (geometric mean/weight‐of‐evidence approach) or Tier‐2B (species sensitivity distribution approach) information is also available, the development of TKTD models for a wider array of species may be the way forward to refine the risk assessment. Validated TKTD models for these species may be an option to evaluate specific risks, using available field‐exposure profiles, by calculating exposure profile‐specific LPx/EPx values (= multiplication factor to an entire specific exposure profile that causes x% Lethality or Effect), informed by an appropriate aquatic exposure assessment. Exposure profile‐specific LPx/EPx can be used in the Tier‐2C risk assessment by using the same rules and extrapolation techniques (statistical analysis and assessment factors) as used in experimental Tier‐1 (standard test species approach), Tier‐2A (geometric mean/weight‐of‐evidence approach) and Tier‐2B (species sensitivity distribution approach).
The GUTS model framework is considered to be an appropriate approach to use in the acute risk assessment scheme for aquatic invertebrates, fish and aquatic stages of amphibians. In the chronic risk assessment, it is only appropriate to use a validated GUTS model if the critical endpoint is mortality/immobility, which is not often the case. If a sublethal endpoint is the most critical in the chronic lower‐tier assessment for aquatic animals, the dynamic energy budget modelling framework combined with a TKTD part (DEBtox) is the appropriate approach to select in the refined risk assessment. TKTD models developed for primary producers may be used in the chronic risk assessment scheme with a focus on inhibition of growth rate and/or yield. Note that experimental tests and TKTD model assessments for algae and fast‐growing macrophytes like Lemna to some extent assess population‐level effects, since in the course of the test reproduction occurs.
Chapter 4 deals with the GUTS framework. This framework is considered ready for use in aquatic ERA, since a sufficient number of application examples and validation exercises for aquatic species and pesticides are published in the scientific literature, and user‐friendly modelling tools are available. Consequently, in this chapter, detailed information is provided on testing, calibration, validation and application of the GUTS modelling framework. Documentation of the formal GUTS model, and of the verification of two example implementations of the GUTS model equations in different programming languages (R and Mathematica) are presented. In addition, sensitivity analyses of both implementations are described, and an introduction is presented for GUTS parameter estimation both in the Bayesian and frequentist approach. The uncertainty, related to the stochasticity of the survival process in small groups of individuals, is discussed, and the numerical approximation of parameter confidence/credible limits is described. A checklist for the evaluation of parameter estimation in GUTS model applications is given. Descriptions of relevant GUTS modelling output are also given. Approaches to propagate the stochasticity of survival in combination with parameter uncertainty to predictions of survival over time and to LPx/EPx values are presented, allowing the calculation of corresponding confidence/credible limits. The validation of GUTS models is discussed, including requirements for the validation data sets. Qualitative and quantitative model performance criteria are suggested that appear as most suitable for GUTS, and TKTD modelling in general, including the posterior prediction check (PPC), the Normalised Root Mean Square Error (NRMSE) and the survival‐probability prediction error (SPPE). Finally, chapter 4 gives an example of the calibration, validation and application of the GUTS framework for risk assessment.
In Appendix Appendix A – Model implementation details with Mathematica and R, Appendix B – Additional result from the GUTS application example, Appendix C – The log‐logistic distribution–D, GUTS model implementations in Mathematica and R, the results of the application example with the GUT‐RED models and supporting information on the GUTS‐RED exercise are provided, respectively. Source codes of the GUTS implementations in Mathematica and R are available in Appendix E .
Chapter 5 deals with the documentation, implementation, parameter estimation and output of DEBtox modelling as illustrated with a case study on lethal and sublethal effects of time‐variable exposure to cadmium for Daphnia magna. This case study was selected since sufficiently calibrated and validated DEBtox models for pesticide and aquatic organisms were not yet available in the open literature, including raw data and programming source code to allow for re‐running all calculations. This lack of published examples of DEBtox models for pesticides and aquatic organisms, as well as the fact that no user‐friendly DEBtox modelling tools are currently available, results in the conclusion that these models are not yet ready for use in aquatic risk assessment for pesticides. Nevertheless, the DEBtox modelling approach is recognised as an important research tool with great potential for future use in prospective ERA for pesticides. The DEBtox model described in chapter 5 for cadmium and Daphnia magna illustrates the potential of the DEBtox modelling framework to deal with several kinds of data, namely survival, growth and reproduction data together with bioaccumulation data. It also proves the feasibility of estimating all DEBtox parameters from simple toxicity test data under a Bayesian framework.
In Appendix A.2.2 , a short overview of the DEBtox model implementation in R is given. The respective R code is available in an archive in Appendix E .
Chapter 6 evaluates the models currently available for primary producers, which all rely on a submodel for growth, driven by a range of external inputs such as temperature, irradiance, nutrient and carbon availabilities. The effect of the pesticide (TKTD part) on the net growth rate is described by a dose–response relationship, linking either external (the algae part) or scaled or measured internal concentrations to the inhibition of the growth rate. All experiments and tests of the models until now have been done under fixed growth conditions, as is the case for standard algae, Lemna and Myriophyllum tests. This was done because the focus has been on evaluating the model ability to predict effects under time‐variable exposure scenarios using predicted exposure profiles (e.g. FOCUS step 3 or 4) and Tier‐1 toxicity data as a starting point. The growth part of the models, however, all have the potential to incorporate changes in temperature, irradiance, nutrient and carbon availabilities in future applications.
TKTD models to describe effects of time‐variable exposures have been developed for two algal species and one PSII inhibiting herbicide. The largest drawback for implementing the algae models in pesticide risk assessment is that the flow‐through experimental setup used for model calibration/validation to simulate long‐term variable exposures of pesticides to fast growing populations of algae has not yet been standardised, nor has the robustness of the setup been ring‐tested. The current experimental setup of refined exposure tests for algae and the algae models is considered an important research tools but probably not yet mature enough to use for risk‐assessment purposes.
Lemna is the most thoroughly tested macrophyte species for which a calibrated and validated model has been documented for a sulfonyl‐urea compound. A Lemna TKTD model can be calibrated with data from the already standardised OECD Lemna test, as long as pesticide concentrations and growth are monitored several times during the exposure phase and the test is prolonged with a one week recovery period. Growth can be most easily and non‐destructively monitored by measuring surface area or frond number on a daily basis. If properly documented, the published Lemna model can be the basis for a compound‐specific Lemna model to evaluate the effects of field‐exposure profiles in Tier‐2C, particularly if in the Tier‐1 assessment Lemna is the only standard test species that triggers a potential risk. The published Myriophyllum modelling approach is not yet as well developed, calibrated, validated and documented as that for Lemna. Developing a model for Myriophyllum is complicated, as this macrophyte also has a root compartment (in the sediment) where the growth conditions (redox potential, pH, nutrient and gas availabilities, sorptive surfaces, etc.), and therefore, bioavailability of pesticides, are very different from the conditions in the shoot compartment (water column). In addition, Myriophyllum grows submerged making inorganic carbon availability in the water column a complicated affair compared to Lemna, for which access to CO2 through the atmosphere is constant and unlimited. Due to the complexity of the Myriophyllum system and the relative novelty of the published modelling approach, the available Myriophyllum model has not yet been very extensively tested and publicly assessable model codes are not yet available. OECD guidelines for conducting tests with Myriophyllum are available. In order to optimise the use of experimental data from such standardised Myriophyllum tests for model calibration, however, it is necessary that the tests are prolonged with a recovery phase in clean water and that growth is monitored over time (non‐destructively as shoot numbers and length). Although the published Myriophyllum modelling approach may be a good basis to further develop TKTD models for rooted submerged macrophytes, it currently is considered not yet fit‐for‐purpose in prospective ERA for pesticides. The currently available Myriophyllum model needs further documentation, calibration and validation.
Chapter 7 describes how TKTD models submitted in dossiers can be evaluated by regulatory authorities. Annex Annex A – Checklist for GUTS models , Annex B – Checklist for DEBtox models – C provide checklists for the evaluation of GUTS models, DEBtox models and models for primary producers. It expands on the information provided by the EFSA Opinion on Good Modelling Practice in the context of mechanistic effect models for risk assessment (EFSA PPR Panel, 2014). The chapter mainly focuses on GUTS models but also provides considerations required for DEBtox and primary producer models. The chapter covers all stages of the modelling cycle and the documentation of the model use. For GUTS models the basic model structure is always fixed and consequently several stages of the modelling cycle have been covered in this Opinion, so they do not need to be evaluated again for each use. This includes the conceptual model and the formal model. For parameter estimation of each application, all experimental data used to calibrate and validate the model should be evaluated to ensure they are of sufficient quality. The computer model can be evaluated using a combination of the ring‐test data set, a set of default scenarios and testing against an independent implementation. The regulatory model also needs to be evaluated. The environmental scenario may be covered by using standard exposure models (FOCUS), leaving the parameter estimation as the key area to evaluate. The evaluation of model analysis (sensitivity and uncertainty analysis and validation) is also described. The final stage is the evaluation of the model use that includes information about tools available to the evaluators to check the modelling.
For DEBtox models, the evaluation of the DEB (physiological) part of the model is separated from the evaluation of the TKTD part of the model. Chapter 7 focusses on the TKTD part and starts with the assumption that the DEB part has been evaluated and accepted before it is used for a regulatory risk assessment.
For primary producer models, as with DEBtox models, the evaluation of the physiological part of the model is separated from the evaluation of the TKTD part of the model. For Lemna, this has been covered in this Opinion. For other primary producers, the evaluation of the physiological part of the model needs to be completed before use in regulatory risk assessment.
Documentation of any TKTD model application should be done following Annex D .
Chapter 8 illustrates the possible use of validated TKTD models as tools in the Tier‐2C risk assessment for plant protection products. The important steps that need to be considered when conducting an ERA by means of validated TKTD models are described. The description of the approach is followed by an example data set for an organophosphorus insecticide. This case study aims to explore how GUTS modelling can be used as a Tier‐2C approach in acute ERA in combination with step 3 or step 4 FOCUSsw exposure profiles. In addition, this case study aims to compare the outcome of the experimental effect assessment tiers (standard test species approach, geometric mean approach, species sensitivity distribution approach, model ecosystem approach) with results of GUTS modelling to put the Tier‐2C approach into perspective.
Chapter 9 concludes that, based on the current state of the art (e.g. lack of documented and evaluated examples), the DEBtox modelling approach is currently limited to research applications. However, its great potential for future use in prospective ERA for pesticides is recognised. The GUTS model and the Lemna model are considered ready to be used in risk assessment.
Two examples on the evaluation of existing TKTD models (one for GUTS and one for DEBtox) used in the context of PPP authorisation are reported in Appendices F and G .
Comments received by the Pesticide Steering Network and related replies are reported in Appendix H .
Guide to the reader: the main topic concerns the implementation of modelling techniques for prospective ERA; hence its stays at the interface of different expertise areas. Taking this into account, the document was structured to allow focussing on sections linked to specific expertise.
Chapters 1, 2 and 3 provide a general context: after presenting the scope of the Scientific Opinion, general principles behind TKTD models are described and the scene for the use of the TKTD models within the risk assessment for aquatic organisms is set. Therefore, these chapters are recommended for getting a complete picture of this document.
Chapters 4, 5 and 6 focus on the description of specific TKTD models. As such the content of these chapters contain rather technical concepts and explanations, particularly addressing modellers. These chapters may be difficult for readers without modelling experience. Understanding of the technical details included in this part, however, is not critical for the reading and understanding of the following chapters.
Chapters 7 and 8 illustrate in details how TKTD models can be used in the PPP ERA context, particularly addressing risk assessors. Evaluation criteria for modelling applications are also given in chapter 7. Hence, it is recommended that this part is also carefully considered by modellers providing elaborations for the risk assessment.
Checklists for the evaluation of TKTD models are given in Annex Annex A – Checklist for GUTS models, Annex C – Checklist for primary producer models –C. Model summary for the model documentation is included in Annex D.
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
In 2008 the Panel on Plant Protection Products and their Residues (PPR) was tasked by EFSA with the revision of the Guidance Document on Aquatic Ecotoxicology under Council Directive 91/414/EEC (SANCO/3268/2001 rev.4 (final), 17 October 2002) (European Commission, 2002). As a third deliverable of this mandate, the PPR Panel is asked to develop a Scientific Opinion describing the state of the art of Toxicokinetic‐Toxicodynamic (TKTD) effect models for aquatic organisms with a focus on the following aspects:
Regulatory questions that can be addressed by TKTD modelling
Available TKTD models for aquatic organisms
Model parameters that need to be included in relevant TKTD models and that need to be checked in evaluating the acceptability of effect models
Selection of the species to be modelled.
In 2013, EFSA Panel on Plant Protection Products and their residues published the document “Guidance on tiered risk assessment for plant protection products for aquatic organisms in edge‐of‐field surface waters” as a first deliverable within the EFSA mandate of the revision of the former Guidance Document on Aquatic Ecotoxicology. This document (EFSA PPR Panel, 2013) focuses on experimental approaches within the tiered effect assessment scheme for typical (pelagic) water organisms, indicating already how mechanistic effect models could be used within the tiered approach. As a second deliverable the document “Scientific Opinion on the effect assessment for pesticides on sediment organisms in edge‐of‐field surface water” was published in 2015 (EFSA PPR Panel, 2015). This document focuses on experimental effect assessment procedures for typical sediment‐dwelling organisms and exposure to pesticides via the sediment compartment. Initially, it was emphasized that the third deliverable would focus on mechanistic effect models as tools for the prospective effect assessment procedures for aquatic organisms.
Although different types of mechanistic effect models with a focus on different levels of biological organisation are described in the scientific literature (e.g. individual‐level models, population‐level models, community‐level models, landscape/watershed‐level models), this Scientific Opinion (SO) predominantly deals with TKTD models as Tier‐2 tools in the aquatic risk assessment for pesticides. These relatively simple, mechanistic effect models are considered to be in a stage of development that might soon enable their appropriate use in the prospective environmental risk assessment for pesticides, particularly to predict potential risks of time‐variable exposures on aquatic organisms. This is of relevance since in most edge‐of‐field surface waters time‐variable exposures are more often the rule than the exception.
A consultation with Member States of the Pesticides Steering Network was held in March 2018. Comments and related replies are reported in Appendix H.
1.2. Scope of the opinion and restrictions
This SO describes the state‐of‐the‐art of TKTD models developed for aquatic organisms and exposure to pesticides in aquatic ecosystems with a focus on prospective environmental risk assessment (ERA) within the context of the regulatory framework underlying the authorisation of plant protection products in the EU. Within this context, TKTD models developed for specific pesticides and specific species of water organisms – such as fish, amphibians, invertebrates, algae and vascular plants – may be useful regulatory tools in the linking of exposure to effects in edge‐of‐field surface waters.
Where appropriate, in this SO the concepts and Tier‐1 and Tier‐2 experimental approaches already developed by EFSA PPR Panel (2013) are considered and aligned with the proposals on the regulatory use of TKTD models as tools in Tier‐2C ERA. This SO describes the potential use of TKTD models as Tier‐2C tools of the acute and chronic ERA schemes for pesticides and water organisms in edge‐of‐field surface waters. In Tier‐2, the TKTD models developed for aquatic animals focus on individual‐level responses to refine the risks of time‐variable exposure to pesticides in particular. Although TKTD models may play a role in higher‐tier ERAs as well, the coupling of TKTD models with population‐level models for aquatic invertebrates and vertebrates is not the topic of this SO. In the chronic risk assessment for algae and macrophytes like Lemna, however, a clear distinction between individual‐level and population‐level effects in Tier‐1 and Tier‐2 assessments cannot be made. Consequently, in TKTD models for these primary producers, the effects of time‐variable exposures on individuals cannot fully be separated from population‐level effects as influenced by interspecific competition between individuals in experimental test systems, the results of which are used to calibrate and validate TKTD models. TKTD models may be used to refine the risk assessment if experimental effect assessment approaches in Tier‐1 (based on standard test species) and Tier‐2 (based on standard and additional test species) in combination with an appropriate exposure assessment, trigger potential risks. The current exposure assessment for active substance approval is based on FOCUS exposure scenarios and models (2001, 2006, 2007a,b). At present, the FOCUS exposure assessment framework is under review to repair deficiencies in the current methodology. One of the main actions foreseen by the mandate,2 is that at steps 3 and 4, a series of 20 annual exposure profiles should be delivered for the edge‐of‐field surface waters of concern instead of one single year exposure profile. It is assumed that prediction of active substance concentrations in surface waters after pesticide application will be further performed using the FOCUS surface water (FOCUSsw) methodology until updated or new methods become available and will replace the existing tools. FOCUSsw is used for approval of active substances at EU level. It is also used in some Member States for product authorisation, but also different exposure assessment procedures may be used. In principle, the TKTD modelling approaches described in this SO can also be used to predict risks to aquatic organisms when linked to exposure profiles based on Member State specific exposure assessment scenarios and models.
In addition, the recommendations of the ‘Opinion on good modelling practise in the context of mechanistic effect models for risk assessment’ (EFSA PPR Panel, 2014) are, where appropriate, taken on board and operationalised for TKTD models.
Besides the regulatory use of TKTD model in the context of refined risk assessment for aquatic organisms, TKTD modelling can also enable exploration of effects of time‐variable exposure on species with trait assemblages that cannot be (so easily) tested under laboratory conditions, e.g. by extrapolating features from species with fast cycles (features of species usually tested in laboratory) to species with lower metabolic rates and longer life cycles that may become exposed more frequently. However, these more fundamental research applications of TKTD models are outside the scope of this Opinion.
2. Concepts and examples for TKTD modelling approaches
Current acute and chronic lower‐tier risk assessments for Plant Protection Products (PPPs) in edge‐of‐field surface waters (EFSA PPR Panel, 2013) rely on the quantification of treatment‐related responses from protocol tests (e.g. OECD) with standard test species or comparable toxicity tests with additional test species. According to Commission Regulation (EU) No 283/20133 and 284/20134, the L(E)C10, L(E)C20 and L(E)C50 values derived from these tests have to be reported. However, in chronic assessments for aquatic animals no observed effect concentration (NOEC) values may be used if valid EC10 values are not reported (predominantly old studies). In a tiered approach, lower‐tier assessments (i.e. Tier‐1 as described in Section 3) aim to be more conservative than higher‐tier assessments. For example, a more or less constant exposure is maintained in the standard laboratory test and pre‐defined assessment factors (AF) laid down in the uniform principles (Reg 546/20115) are applied in order to take into account a number of uncertainties, e.g. intra‐ and interspecies variability, interlaboratory variability and extrapolation from laboratory to field. Toxicity estimates (e.g. LC/EC values), however, are time‐dependent in that they are usually different for different exposure durations. In edge‐of‐field surface waters, time‐variable exposure is rather the rule than the exception (as indicated by monitoring data or modelled concentration dynamics of pesticides; see e.g. Brock et al., 2010). Consequently, if a risk is triggered in the conservative Tier‐1 approach, a refined risk assessment can be performed by considering realistic time‐variable exposure regimes. As outlined in the Aquatic Guidance Document (EFSA PPR Panel, 2013), this can be addressed experimentally or by modelling, e.g. by using TKTD modelling approaches.
TKTD modelling approaches – as outlined in more detail below – provide a modelling approach of intermediate complexity (Jager, 2017), which ranks between the simple statistical (LC/EC50) models, and fully detailed approaches focusing on the molecular level (Van Straalen, 2003). TKTD models can be used in aquatic risk assessment to link results of laboratory toxicity data to predicted (time‐variable) exposure profiles. These models may also be used to explore the changes of toxicity in time, and to explore the processes underlying the variations between species and toxicants and how they depend on environmental conditions. Some TKTD models can potentially explain links between life‐history traits, as well as explore effects of toxicants over the entire life cycle (e.g. DEBtox toxicity models derived from the Dynamic Energy Budget (DEB) theory). TKTD models for survival can, as has been recently shown (e.g. Nyman et al., 2012; Baudrot et al., 2018b; Focks et al., 2018), be parameterised based on standard, single‐species toxicity tests. These models may still provide relevant information at the individual level when extrapolating beyond the boundaries of tested conditions in terms of exposure. The parameters being used in TKTD models remain as species‐ and compound‐specific as possible and they can usually be interpreted in a physical or a biological way (see Section 2.2 for more details). In this way, parameters can be used in a process‐based context, aiding the understanding of the response of organisms to toxicants (Jager, 2017).
In the following sections, some classes of TKTD models will be explained in more detail concerning their terminology, their background and application domains, their relationships to each other, and the intrinsic or explicit complexity levels they account for (Figure 1). After a more detailed explanation of ‘Toxicokinetic modelling for uptake and internal dynamics of chemicals’ (Section 2.1), the following three sections will be dedicated to: (i) the ‘General Unified Threshold models of Survival’ (GUTS) framework for the analysis of lethal effects (Section 2.2), (ii) toxicity models derived from the Dynamic Energy Budget theory (DEBtox models) (Section 2.3), and (iii) models for primary producers (Section 2.4). An overview about strengths and weaknesses of TKTD models is given in Table 1.
Figure 1.
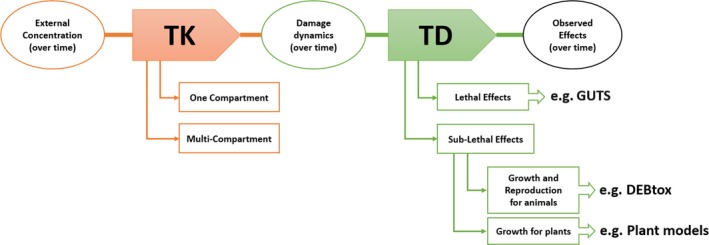
Schematic presentation of the concepts behind toxicokinetic (TK)/toxicodynamic (TD) models; GUTS stands for the General Unified Threshold model of Survival, while DEBtox stands for toxicity models derived from the Dynamic Energy Budget (DEB) theory. The damage‐dynamics concept is explained in Figure 2 and related text
Table 1.
Introduction to strengths and weaknesses of TKTD models
| Strengths | Weaknesses | |
|---|---|---|
| TKTD models (applicable to GUTS, DEBtox and models for primary producers) |
|
|
| GUTS |
|
|
| DEBtox |
|
|
| Primary producers models |
|
|
2.1. Toxicokinetic modelling for uptake and internal dynamics of chemicals
TKTD models follow one general principle: the processes that influence internal exposure of individual organisms, summarised under toxicokinetics (TK), are separated from the processes that lead to their damage and mortality, summarised by the term toxicodynamics (TD) (Figure 1). In general terms, TK processes correspond to what the organism does to the chemical substance, while the TD processes correspond to what the chemical substance does to the organism. More precisely, TK describes absorption, distribution, metabolism and elimination of hazardous substances by an organism. In aquatic systems, the main uptake routes of substances from the water phase include the organism surface and internal or external gills. Food can also be a relevant uptake route. Transport across the biological membranes can be passive (e.g. diffusion) or active (e.g. membrane transporters). Inside the organism, TK processes relate to internal partitioning of chemicals between liquid‐ and lipid‐dominated parts or between different organs of organisms. TK models result in estimates for internal exposure concentrations, which can relate to whole organism scales or to single organs. The level of detail of the TK model often depends on the purpose of the study, but also on practicalities in terms of size of the organism and single organs in question.
Specific aspects of TK, with emphasis on internal compartmentation, were discussed extensively in a recent review (Grech et al., 2017). TK models are categorised into compartment models and physiologically based TK (PBTK) models. While single organs and blood flow are explicitly considered in PBTK models, one‐ or multi‐compartment models are using a generic simplification of an organism. For aquatic invertebrates, the one‐compartment model is the most often used. One‐compartment models assume concentration‐driven transfer of the chemical from an external compartment into an internal compartment, where it is homogeneously distributed.
Transformation of molecules within the compartment can play an important role for the link from TK to the effects, as chemicals are metabolised either into inactive and excretable products (detoxification) or into active or reactive metabolites causing (toxic) effects.
TD processes are related to internal concentrations of a toxicant within individual organisms. Implicitly, biological effects are caused by the toxicant on the molecular scale, where the molecules interfere with one or more biochemical pathways. These interactions are lumped into only a few equations in TKTD models that have to handle the variability of TD processes within species. These equations especially have to provide enough degrees of freedom to capture the dynamics of responses or effects over time; the latter is the most important aspect in the current modelling context since the aim is to understand how toxic effects change over time under time‐variable exposure profiles.
2.2. Toxicodynamic models for survival
Historically, TKTD models for survival were developed and applied in a tailor‐made way to each research question. Over the years that led to a variety of different TKTD models, in parts redundant or conflicting. This conglomeration of TKTD modelling approaches led to difficulties in the communication of model applications and results and hampered the assimilation of TKTD models within ecotoxicological research and risk assessment. The publication of Jager et al. (2011), aiming at unifying the existing unrelated approaches and clarifying their underlying assumptions, tremendously facilitated the application of TKTD modelling for survival. In that paper, the General Unified Threshold models of Survival (GUTS) theory was defined and its application developed. The biggest achievement of the developed theory was probably the mathematical unification of almost all existing tailor‐made approaches under the GUTS umbrella. Recently, an update of the GUTS modelling approach was published, which works out more details of the modelling of survival and provides examples and ring‐test results; that update also suggests a slightly changed terminology, while the underlying model assumptions and equations have not been changed (Jager and Ashauer, 2018). In this scientific opinion, the updated terminology suggested by Jager and Ashauer (2018) is used, which refers to ‘scaled damage dynamics’ rather than to the previously used concept of ‘dose metrics’.
The ultimate aim of GUTS is to predict survival rate under untested exposure conditions such as time‐variable exposures, which are more likely to occur in the environment than the static exposure levels used in Tier‐1 testing. The prediction functionality of GUTS is useful for risk assessment, because in some cases, it may not be possible to test realistic time‐variable exposure profiles under laboratory conditions, e.g. for individuals characterised by a long life cycle. In addition, exposure modelling can easily create hundreds of potentially environmentally relevant exposure profiles, which would require excessive resources if tested in the laboratory (e.g. number of test animals).
All GUTS versions have in common that they connect the external concentration with a so‐called damage dynamic (see below and Jager and Ashauer (2018) for definition), which is in turn connected to a hazard resulting in simulated mortality when an internal damage threshold is exceeded (Figure 2). The unification of TKTD models for survival was made possible by creating two categories for assumptions about the death process in the TD part of GUTS: the Stochastic Death (SD) and the Individual Tolerance (IT) hypotheses that contain all the other published modelling approaches for survival.
Figure 2.
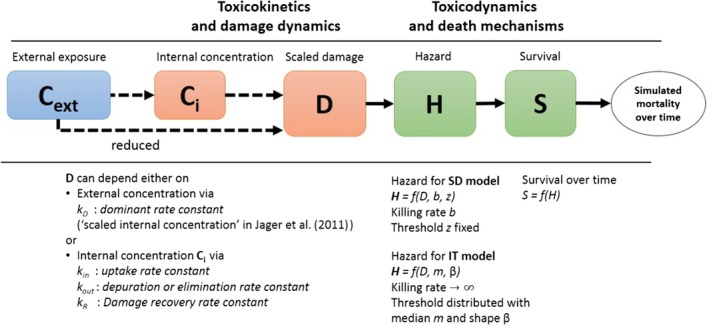
Overview of state variables in the General Unified Threshold model of Survival (GUTS) framework. The toxicokinetic part of the GUTS theory translates an external concentration into an individual damage state dynamics in a more or less (reduced model) detailed way. In the toxicodynamic part of the model, two death mechanisms are distinguished: SD for Stochastic Death and IT for Individual Tolerance; see text for more details
For models from the SD category, the threshold parameter for lethal effects is fixed and identical for all individuals of a group and a so‐called killing rate relates the probability of a mortality event in proportion to the scaled damage. Hence, death is modelled as a stochastic (random) process occurring with increased probability as the scaled damage rises above the threshold.
For models from the IT category, thresholds for effects are distributed among individuals (sensitivity varies between individuals of a population) of one group, and once an IT is exceeded, mortality of this individual follows immediately, meaning in model terms that the killing rate is set to infinity.
Both models are unified within the ‘combined GUTS’ model, in which a distributed threshold is combined with a between‐individual variable killing rate (Jager et al., 2011; Ashauer et al., 2016; Jager and Ashauer, 2018). It is assumed that all TK and TD model parameters (see Figure 2) are constant throughout the exposure profile, i.e. no effects of exposure on TK and TD dynamics are considered in addition to those already captured from the calibration experiments. Therefore, phenomena like increase in sensitivity or tolerance is not taken into account. The possibility of damages being below thresholds of lethality – inducing faster responses at a next pulse – is accounted for by the use of the scaled damage concept (see Figure 2).
Scaled damage (D in Figure 2) is the internal damage state of an organism after taking the external toxicant concentration, the uptake rate, elimination rate and potentially any damage recovery into account. The scaled damage concept translates external exposure into TD processes and finally into mortality. It links the given external concentration dynamics to the time course of the internal hazard. An ‘internal concentration’ (Ci in Figure 2) can only be considered explicitly in the model when measured internal concentrations are available or clear indications in the observed survival over time are given. The link of external concentrations to the scaled damage via a combined dominant rate constant kD leads to the ‘reduced GUTS’, which was called ‘scaled internal concentration’ in the old dose metric terminology (Jager et al., 2011). It is most often used when no measurements of internal concentrations are experimentally available as in the case of standard survival tests (Figure 2). Parameter kD can then be dominated by either elimination or by damage recovery; Chapter 4 gives the corresponding equations and details about kD estimation.
‘Damage’ is a rather abstract concept here and its level cannot be experimentally measured, but this concept still allows for further mechanistic considerations. In principle, chemicals that have entered the organism are expected to cause some damage by interference with biochemical pathways. Organisms will have capacity to repair the damage at a certain rate, but if the damage exceeds a certain threshold the organism will die. The degree to which the dynamics of internal concentrations can explain the pattern in mortality observed over time can vary. As one possibility, damage repair can be so fast that disappearance of the chemical from the organism is controlling the scaled damage and so the mortality rate. Fast damage repair in this case does not mean that there are no effects, because as long as there are high levels of scaled damage, instantaneous mortality will occur. If the damage that is caused by the chemical, however, is repaired so slowly that organisms keep dying also after elimination of the chemical, the dynamics of the internal concentrations can be accounted for in the model to capture that delay in time. For chemicals which are known to bind irreversibly to their enzymatic target site, very low or zero depuration or repair rates are supposed to be the case. However, depending on the enzyme turnover of different species, it might also happen that by synthesis of fresh, ‘clean’ enzymes, depuration or recovery could take place for such irreversibly binding compounds in case no further internal exposure takes place.
When working with small invertebrates or in some cases with fish, internal concentrations are usually measured on the whole organism level, giving an average concentration across organs and cellular compartments. Hence, measured or modelled internal concentrations might not directly reflect the concentration at a specific molecular target site, but in the GUTS modelling it is assumed that modelled internal concentrations are at least proportional to the concentration at the target sites.
Over‐parameterisation of the model could lead to multiple problems, e.g. imprecise and inaccurate parameter estimation. To avoid this, it is recommended to include the internal concentration only if measured internal concentrations are available in a data set. If internal concentrations are not explicitly modelled (‘scaled internal concentrations’ in the ‘old’ terminology), the rate constant of the scaled damage (kD) describes the rate‐limiting processes of either the chemical elimination/detoxification of the organism or the damage repair. When calibrating a GUTS model without considering the internal concentration as a state variable, it is assumed that survival data over time contain sufficient information to allow for calibration of the dominant rate constant, plus parameters for the death mechanism (SD or IT) which link the scaled damage to effects on survival. The term ‘scaled damage’ accounts for the fact that it is in general not possible to determine the absolute ‘damage’‐related values, while it is possible to assume that the scaled damage is proportional to the true (but unknown) damage and has the dimension of the external concentration. Using the scaled damage without explicit internal concentrations can still give equal or better fits to observed survival data compared with using internal concentrations, despite measured internal concentrations being available. This has been shown when testing GUTS with observed survival under time‐variable, untested exposures (Nyman et al., 2012) and holds especially according to the principle that a simpler model with equal performance is preferable to a more complex one (parsimony principle).
The choice of the specific GUTS variant is not only a conceptual decision, but has direct implications for the number of parameters and hence for the degrees of freedom for parameter estimation. Despite the existence of a theoretical framework, modelling TKTD processes with GUTS requires some experience and thorough thinking about the possible choices and degrees of freedom. The advantage of the GUTS framework is the clear definition of the concepts, of the mathematical equations and of the terminology which eases the documentation of any GUTS model application.
Model parameters for GUTS models can be interpreted in a process‐based context, despite the fact that they are not defined on a purely mechanistic basis. For example, the uptake rate constant kin has a process‐related interpretation; it quantifies the influx of chemicals into organisms, but in GUTS the uptake rate is not associated with physical processes such as membrane transport and it can account also for more than one process. This intermediate complexity of the model is chosen according to the fact that, for many compounds, purely mechanistic parameters are not usually available. Nevertheless, the chosen level of detail enables the determination of GUTS parameters from relatively simple data sets. GUTS parameters are related to dynamic processes such as uptake, elimination, death and/or physiological recovery. Hence, they can potentially be quantitatively related to biological traits such as body size, breathing mode or chemical properties such as log Kow or water solubility (Rubach et al., 2011). Understanding these relationships could make TKTD modelling a useful tool for understanding chemical toxicity across species and chemical modes of action. Based on quantitative relationships, extrapolations between species and across chemicals could in theory be possible although this is not applicable in RA based on the current state of knowledge.
More details about aspects of the GUTS models will be described and discussed in later sections of the document, including model calibration (parameter estimation), model testing and validation, deterministic and probabilistic model predictions, the domain of applicability and regulatory questions.
2.3. Toxicodynamic models for effects on growth and reproduction
The Dynamic Energy Budget (DEB) theory was originally proposed by Kooijman who started its development in 1979; see for example, Kooijman and Troost, 2007; Kooijman, 2010, where the DEB theory was used to investigate the toxicity of chemicals on daphnids and several fish species. Since then, the DEB theory became widespread and found applications in many fields of environmental sciences. The DEB theory is based on the principle that all living organisms consume resources from the environment and convert them into energy to fuel their entire life cycle (from egg to death), thus ensuring maintenance, development, growth and reproduction. In performing those activities, organisms need to follow the conservation laws for mass and energy. The DEB theory thus proposes a comprehensive set of rules that specifies how organisms acquire their energy and allocate it to the various processes. In particular, it is assumed that a fixed fraction (kappa or κ) of the mobilised energy is allocated to somatic maintenance and growth, while the rest (1 – kappa) is allocated to maturity maintenance, maturation and reproduction; this is called the kappa‐rule, a core concept within the DEB theory (Figure 3). A conceptual introduction to the DEB theory can be found in Jager (2017), while an extensive and more mathematical description is provided in Kooijman (2010).
Figure 3.
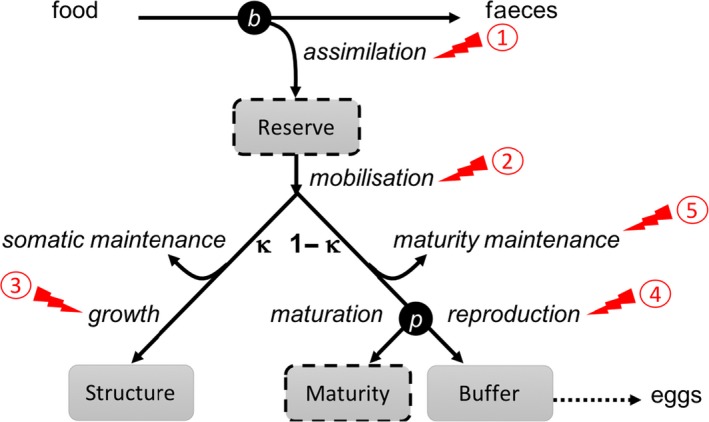
Schematic energy allocation according to the standard DEB model (adapted from Jager, 2017): ‘b’ stands for birth and ‘p’ for puberty. The reserve compartment, and consequently the maturity one (boxes with dashed borders) may be omitted in a DEBkiss model (reserve‐less DEB). Red arrows stand for the five different DEB modes of actions due to toxic stress that can be described with DEBtox models; see text and references above for more explanation on kappa (κ)
When focusing on egg‐laying ectotherm animals, a standard DEB model exists (Figure 3), which assumes that animals do not change their shape when growing (therefore, it does not cover processes such as metamorphosis), and that animals feed on one food source with a constant composition. The ‘Add‐my‐Pet’6 (AmP) database is a collection of parameters of the standard DEB model, as well as of its different variants, for more than 1,000 species, including numerous aquatic species. Conceptual overviews of the DEB theory are given in Jager et al. (2014) and Baas et al. (2018).
From the formulation of the standard DEB model, a simplification for standard laboratory toxicity tests has been derived; it is based on the following assumptions: size is a perfect proxy for maturity; size at puberty remains constant; costs per egg are constant and thus not affected by the reserve status of the mother; egg costs can only be affected by a direct chemical stress on the overhead costs; and the reserve is always in steady‐state with the food density in the environment. Under this framework, the kappa (κ) rule should not be affected by a toxicant. A reserve‐less DEB model (called ‘DEBkiss’) has recently been published (Jager et al., 2013; Jager and Ravagnan, 2016), treating biomass as a single compartment, thus leading to a simplified version of the standard DEB model making the interpretation of toxicity test data easier. However, the exclusion of the reserve compartment together with the maturation state variable (Figure 3) makes DEBkiss mainly applicable to small invertebrates that feed almost continuously.
DEBtox is the application of the DEB theory to deal with effects of toxic chemicals on life‐history traits. The original DEBtox version was published by Kooijman and Bedaux (1996), but Billoir et al. (2008) and Jager and Zimmer (2012) published updated derivations. Observing effects on life‐history traits due to chemical substances necessarily implies that one or more metabolic processes within an organism, and consequently also energy acquisition or use by it, are affected by the toxicant. In that sense, energy‐budget modelling provides a convenient way of quantitatively assessing effects on sublethal endpoints or life‐history traits without the need to go into biological details nor into details about the stressor itself. DEBtox models differ from other TKTD models in their TD part by incorporating a dynamic energy budget part for growth and reproduction endpoints at the individual level. Internal chemical exposure has an impact on the energy balance and translates into modified DEB rates. On this basis, the DEB part describing the physiological energy flows is considered as the physiological part of a DEBtox model (as used in later sections of this SO), while the part that accounts for uptake and effects of chemicals is named the TKTD part.
One of the core assumptions of DEBtox models is the existence of an internal toxicant threshold below which no measurable effect on a specified endpoint can be detected at any time, this is the so‐called no‐effect‐concentration (NEC). The value of the NEC depends on the chemical species combination; it can be modified by other stressors (e.g. other chemicals, abiotic factors, etc.) but in a DEBtox framework, the NEC is a time‐independent parameter. Although toxicity in experimental testing is related to a certain exposure duration, such time‐independent definition of the NEC is of particular interest in an ERA perspective.
DEB processes can be linked to external exposure via the concept of the scaled damage, which is often implemented as a basic, ‘linear‐with‐threshold’ relationship: i.e. the DEB model parameter value equals its control level until the internal concentration reaches the relevant NEC value, and then changes linearly dependent on the internal concentration. Simulated internal concentrations can account for constant, but also time‐variable external concentrations. DEBtox models are flexible enough to allow for the description of several modes of action via specific sets of DEBtox equations depending on the target process that is affected by the toxicant. Stated simply, in DEBtox, the toxicant can affect either the acquisition or the use of energy. In more details, typical effect targets are
Energy acquisition (assimilation, Figure 3);
Energy use from reserves (mobilisation);
Energy spent on growth;
Energy spent for reproduction; or
Allocation between somatic maintenance and reproduction.
It should be kept in mind that the definition of ‘mode of action’ in a DEB context (DEBMoA, as stated by Baas et al., 2018) is not the same as the definition of ‘mode of action’ in a general toxicity context. In a DEB context, the mode of action is mathematically formulated as a change on how the chemical affects the physiological processes accounted for in a DEB model. Hence, chemicals with different biochemical modes of action as defined by molecular target sites for, e.g. pesticides, may exhibit a similar DEB mode of action when evaluating how the pesticide affects the DEB model parameters.
Even though some tools are already available to perform parameter estimation (e.g. DEBtoxM7 and BYOM8 packages for Matlab, home‐made scripts in the R software), DEBtox models in their current form require advanced statistical skills to be fitted to experimental data. In addition, data sets that would be useful to provide best estimate of all DEB model parameters are not standardised yet. Nevertheless, in essence, DEBtox modelling approaches provide a great potential to explore toxicity far beyond classical dose‐effect approaches. A recent paper by Baas et al. (2018) fully explains all the potentiality of DEB models to assess chemical toxicity on individuals in an ERA perspective.
2.4. Models for algae and aquatic macrophytes
2.4.1. How are algae and plants different from other higher organism model?
Algae and plants are different from other higher organisms in one important aspect: they can photosynthesise and thereby create their own energy, rather than having to ingest it through food. The rate at which algae and plants photosynthesise depends on a range of environmental factors of which irradiance, inorganic carbon, nutrient availability and temperature are among the most important. This means that the quantification of energy input in a plant system is more challenging than for an organism where maximum energy uptake basically only depends on its size and food availability, and where the nutritional quality of the food is expected to balance the demands of the organism. Algae and plants need to balance their uptake of inorganic carbon, nitrogen, phosphorous and micronutrients separately to obtain the optimal tissue composition. In addition, plants may have different compartments playing different roles. In rooted aquatic macrophytes, for example, only shoots photosynthesise, while roots play an important role in the uptake of nutrients such as nitrogen and phosphorous. Other nutrients, such as potassium, are primarily taken up by the shoot (Barko and Smart, 1981).
Apart from roots and shoots being physiologically different, the environmental medium where they grow (water vs sediment) also differs immensely in terms of light, inorganic carbon, nutrient and particularly oxygen availability. This means that availabilities of different chemicals will also differ between water and sediment. Hence, if toxicity of chemicals towards plants is to be considered via uptake from both water and sediment, plant models need to account for the respective compartments (i.e. leaves/shoots and roots). For the time being, the transport of toxic chemicals between water and sediment in risk assessment is addressed with fate modelling alone. Scientifically sound and functional interfaces between fate and exposure models and effect models could help here in the future to unify such tightly connected processes. Algae and floating macrophytes get all nutrients and toxicants through the water phase; hence, for these organisms it may suffice to use one‐compartment models.
Another important difference between most heterotrophic organisms used for toxicity testing and algae and plants is that many plants, particularly aquatic macrophytes, are clonal and therefore a large part of their reproduction is vegetative. Microalgae almost exclusively reproduce by cell division, macroalgae, which are not addressed here, can have more complex life cycles. Allocation to sexual reproduction in aquatic macrophytes might take place depending on species and growth conditions, but allocation to storage organs (e.g. root stocks and vegetative propagules) that can enable the plant to survive winters or dry periods might be equally or even more important in terms of resource allocation than sexual reproduction. Hence, resource allocation patterns might be more complex in higher plants than for heterotrophic organisms. For the purpose of macrophyte TKTD modelling, macrophytes are thus considered as ‘one individual’ showing unconstrained (exponential) growth for a constrained time period or density dependent growth. Choosing density dependent growth actually means using saturation‐growth curves to describe the growth of heterotrophic organisms. Sexual reproduction or allocation to storage organs in the macrophyte growth models is not explicitly considered at this stage of plant modelling.
With respect to the analysis of toxic effects, the most obvious difference between primary producers (e.g. algae and vascular plants) and animals (e.g. invertebrates, fish and amphibians) is that for primary producers the main parameter measured is growth (OECD, 2006, 2011a, 2014a,b).
While for the analysis of mortality on invertebrates or vertebrates like fish, the survival of the individual is the normal case (or null model), this does not hold for algae and vascular plants, because they do not simply ‘die’ under exposure to a toxic chemical, but their growth dynamics can decrease or eventually completely stop. Certainly, algae and vascular plants can also die after being intoxicated, but this mortality is a process that takes much longer and is much more challenging to detect than for macroinvertebrates or fish. For microalgae, which are typically used for toxicity assessment, population density is usually measured. Hence, mortality of individual cells is difficult to distinguish from background mortality and from the reproduction of growing cells. For that reason, the assessment of toxic effects on algae and vascular plants needs growth models as a baseline or null model.
The next two sections will give an overview of how the TKTD concepts can be used on growth models for algae and the aquatic macrophyte species typically used in environmental risk assessment. An overview of the models is given in Figure 4.
Figure 4.
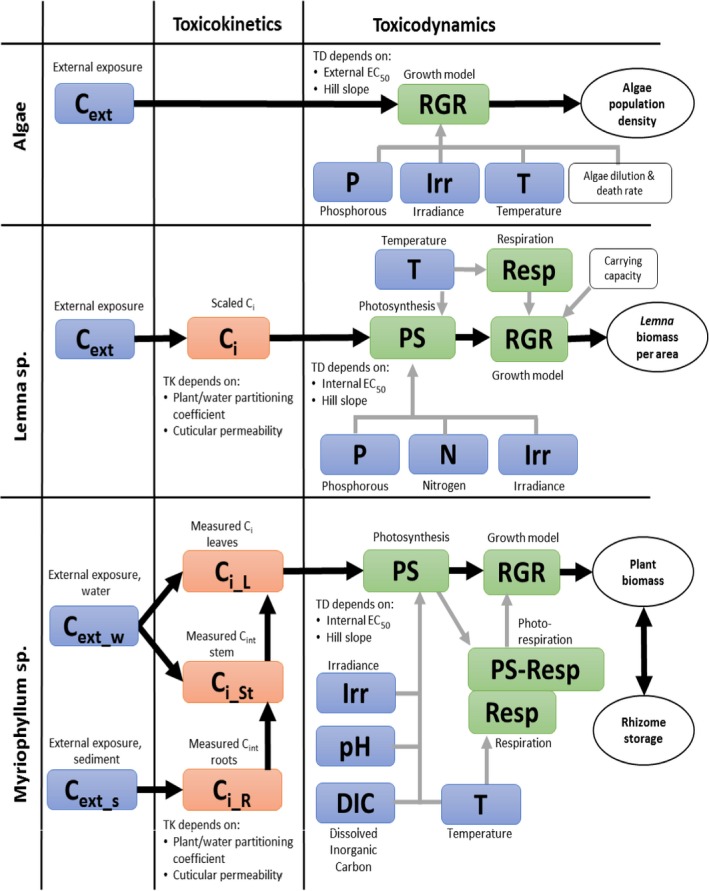
Schematic representation of the algae, Lemna spp. and Myriophyllum spp. models presented in Weber et al. (2012); Schmitt et al. (2013); Heine et al. (2014, 2015, 2016a). External factors affecting chemical uptake or growth are given in blue, internal chemical concentrations in orange and the different rate constants affecting biomass growth of the plants or algae in the respective models are given in green. White squares denote other factors included in the model such as the algae media dilution rate specific for the flow though system used in the algae test and the density‐dependent growth incorporated in the Lemna model. Model endpoints are given in white circles. The output of the growth model is relative growth rates (RGR) and the growth rates are affected by the internal concentrations via a concentration–response relationship defined by the concentration decreasing growth by 50% (EC 50) and a slope parameter of the curve (Hill slope). These two parameters are equivalent to the m and β describing the relationship between internal concentrations or damage and hazard in the IT version of GUTS (see Figure 2)
2.4.2. The TKTD concept used on pelagic microalgae
The algal species most routinely tested for risk‐assessment purposes of plant protection products is the pelagic microalga Raphidocelis subcapitata (formerly known as Pseudokirchneriella subcapitata or Selenastrum capricornutum). Models have been developed for both static (Copin and Chevre, 2015; Copin et al., 2016) and flow‐through growth systems (Weber et al., 2012), with the latter being the most elaborate model. Using flow‐through systems is the only way effects of variable exposures can be tested on microalgae growth without filtering or centrifuging the algae to separate them from the medium and keeping them in an exponential growth phase over a prolonged time span of days to weeks. As the design of the flow‐through system defines the growth conditions of the algae, it is therefore an inherent part of the model. In a flow‐through system, new growth medium is continuously being added to replace media with algae. In this way, the algae can be kept in a constant growth phase with the maximal density being determined by the growth conditions (irradiance, temperature, nutrient and carbon availability) and the flow‐through rate of the medium. Using a flow‐through system, the model can be used to simulate multiple peak/pulsed exposures such as those, e.g. from the FOCUS exposure profiles in a ‘stream scenario’. This is therefore the only algae model that will be evaluated here. Contrary to the macrophyte models, the algae model of Weber et al. (2012) does not contain a TK section, nor does it distinguish between temperature effects on photosynthesis and respiration. Instead the external chemical concentration affects the relative growth rate of the population directly, as does temperature, nutrient (only phosphorous considered) and irradiance (Weber et al., 2012). The algae model is pictured together with the two macrophyte models for comparison in Figure 4. As the TK part of the model is non‐existing, it can be argued whether the model can be categorised as a true TKTD model.
2.4.3. The TKTD concept used on aquatic macrophytes
Presently, there are two models available integrating macrophyte growth models with the TKTD principle: the Lemna model by Schmitt et al. (2013), and the Myriophyllum model by Heine et al. (2014, 2015) which is further extended in Hommen et al. (2016). The two macrophyte species are the standard test species representing an aquatic (floating) monocot (Lemna spp.) and a dicot (Myriophyllum spp.) which is rooted in the sediment. Having both a monocot and a dicot in the test battery is important, as some herbicides are selective against one of these plant groups. A rooted species will also be useful in addressing the bioavailability and toxicity of sediment‐bound pesticides. In addition, the two species represent a floating and a submerged rooted phenotype, and species with different life cycles – fast vs. more slowly growing species. In both the Lemna and Myriophyllum models, internal toxicant concentrations are modelled based on a plant/water partitioning coefficient, corresponding to a bioconcentration factor, and a measure of cuticular permeability that determine the uptake rate. Uptake by roots is not considered at present but can be implemented in the future if needed. The internal concentrations are then directly related to growth via a log‐logistic concentration‐effect model, whereby the per cent inhibition of net photosynthesis is determined. Plant growth is the sum of net photosynthesis and respiration processes, which are to different extents affected by, e.g. temperature and are, therefore, modelled as separate processes.
The main difference between the two models on the TK side is that the Lemna model works with scaled internal concentrations (described in Section 2.2), whereas the Myriophyllum model works with measured internal concentrations.
The largest difference between the two models on the TD side is the growth model. Lemna is the simplest plant: (1) it is floating, and hence the sediment compartment is not an issue, and making a distinction between roots and fronds can thus be ignored, (2) it gets its inorganic carbon mainly from CO2 in the air, which is always available compared to forms and availabilities of inorganic carbon in water and most likely not limiting for photosynthesis, and (3) it mainly reproduces vegetatively, hence allocation to reproduction or storage organs is of little importance. Assuming non‐limiting nutrient supplies, which will typically be the case in edge‐of‐field surface waters affected by agriculture, this leaves photosynthetic rates in the model being dependent only on irradiance and temperature. The influence of plant densities on irradiance is not explicitly considered in the current models, but considered indirectly through the carrying capacity. Parameters to quantify the impact of irradiance and temperature on growth can be obtained from laboratory conditions, and environmentally relevant time series of these factors can be obtained from most meteorological stations. The Lemna model does not focus on a single plant, but instead the plant‐biomass growth (photosynthesis minus respiration) per area is modelled. As the plants/fronds, when reaching a certain density, start to shade each other and block availability to inorganic carbon and nutrients, the growth will decrease, and the biomass will reach a maximum plateau. This density dependence of areal biomass growth has been built into the model in order to enable modelling of Lemna growth dynamics over a season with time‐variable exposure. The growth rate input to the model can be based both on increase in frond number or surface area over time, later calibrated to a biomass unit, as the surface area specific parameters can easily and non‐destructively be monitored over time. Converting surface area or frond number specific growth rates to biomass, however, ignores potential differences in surface to biomass ratios as a function of growth conditions.
Myriophyllum spp. are more complex plants; hence, the growth model is more complex as compared to the Lemna model. Myriophyllum is rooted, hence, the growth model needs at least root, stem and shoot compartments, which are explicitly introduced (Heine et al., 2014) and modelled in Heine et al. (2015, 2016a). In an extended Myriophyllum model (Hommen et al., 2016), rhizomes – which are energy storage organs – are also included to be able to model growth and allocation patterns over longer timescales, e.g. a whole season, which was not possible in the original model by Heine et al. (2014, 2016a). In the original growth model (Heine et al., 2014), photosynthesis takes place in the leaves and biomass is allocated to leaves, stems and roots in the fixed proportions 55:35:10%, which is an average based on literature data (Jiang et al., 2008). Uptake of chemicals can take place by leaves, stems and roots using the same cuticular permeability constant for all three tissues, but different surface to volume ratios. This in principle allows for modelling uptake from the sediment compartment in addition to uptake from the water compartment, but sediment uptake has not been implemented yet in the published Myriophyllum models. Transport between the plant parts is not well described in the literature. A rate constant value for the xylem transport of chemicals is given in the supplementary material of Heine et al. (2016a), but no references for the value is given. Phloem transport is not considered, although it could be quantified proportional to the amount of photosynthates allocated from leaves to support stem and root growth. Hence, although a growth model with different plant compartments for Myriophyllum has been built, further developments and improvements are still needed. The use of these models in risk assessment could profit from the detailed analysis of uptake and transport of organic contaminants in Myriophyllum (Diepens et al., 2014), where uptake and transport processes have been analysed experimentally and also dynamically modelled. In addition, general knowledge on water and carbon flows in aquatic plants (Pedersen and Sand‐Jensen, 1993; Best and Boyd, 1999) combined with knowledge on physicochemical properties of the pesticide could be used to enhance predictability of internal pesticide movements in Myriophyllum.
When it comes to inorganic carbon availability, Myriophyllum is also in a very different position compared to Lemna. Contrary to the aerial compartment, the availability of inorganic carbon and other gaseous compounds in the water is strongly limited by their solubility in water and by their diffusion towards the leaves. In addition, the form by which inorganic carbon exists in the water (e.g. CO2 or HCO3) is strongly pH dependent. Water pH is affected by both plant‐nutrient uptake and photosynthetic rates, which vary over the day; hence, pH can easily vary between pH 6 and 10 in a pond during a day, and to even more extreme values within dense macrophyte stands. The varying carbon availability and its effect on photosynthetic rate is incorporated in the Myriophyllum model, which is why information on both dissolved inorganic carbon (DIC) and pH is required as input parameters. As Myriophyllum has access to nutrients from the sediment, also in this model non‐limiting nutrient supply is assumed.
As for Lemna, Myriophyllum reproduces vegetatively; but in addition, it also flowers and produces seeds once or twice a year and allocates energy to rhizomes (Best and Boyd, 1999). In the extended model version (Hommen et al., 2016), the rhizomes are included but it is not explicit which principles determine allocation to rhizomes. Allocation of energy to flowers and seeds has not been incorporated (Hommen et al., 2016), as it is very minor (Best and Boyd, 1999). The typical ‘die‐off’ after sexual reproduction described in Best and Boyd (1999) is, however, incorporated in Hommen et al. (2016). Contrary to the Lemna model, density dependence of growth, when Myriophyllum reaches closed stands and thereby creates self‐shading of lower leaves, is not incorporated. This could be done, as maximum leaf area indexes for macrophyte populations exist in the literature. In the paper by Hommen et al. (2016), a ‘death rate’ has been incorporated in the Myriophyllum model, but it is not explicit how this is done mathematically and will therefore not be considered further in this opinion.
3. Problem definition/formulation
3.1. Introduction
The EFSA Opinion on good modelling practice in the context of mechanistic effect models for risk assessment (EFSA PPR Panel, 2014) says:
‘The problem formulation sets the scene for the use of the model within the risk assessment. It therefore needs to clearly explain how the modelling fits into the risk assessment and how it can be used to address protection goals’ and ‘The problem formulation needs to address the context in which the model will be used, to specify the question(s) that should be answered with the model, the outputs required to answer the question(s), the domain of applicability of the model, including the extent of acceptable extrapolations, and the availability of knowledge’.
The EFSA Aquatic Guidance Document (EFSA PPR Panel, 2013) describes that TKTD models may be used in the Tier‐2 effect assessment procedure to expand the risk assessment of PPPs based on laboratory single‐species tests with standard and additional test species (Figure 5).
Figure 5.
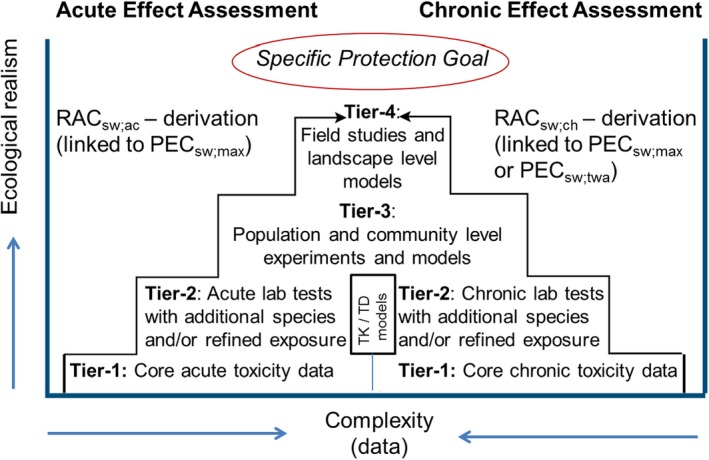
Schematic presentation of the tiered approach within the acute (left part) and chronic (right part) effect assessment for PPPs. For each PPP, both the acute and chronic effects/risks have to be assessed. The Tier‐1 and Tier‐2 effect assessments are based on single species laboratory toxicity tests, but to better address risks of time‐variable exposures the Tier‐2 assessment may be complemented with toxicokinetic/toxicodynamic (TKTD) models. Tier‐3 (population and community level experiments and models) and Tier‐4 (field studies and landscape level models) may concern a combination of experimental data and modelling to assess population and/or community level responses (e.g. recovery; indirect effects) at relevant spatiotemporal scales. All models included in such a tiered approach need to be properly tested and fulfil required quality criteria. RAC: Regulatory Acceptable Concentration; sw: surface water; ac: acute; ch: chronic; PEC: Predicted Environmental Concentration; twa: time‐weighted average
The different Tier‐2 approaches described in the Aquatic Guidance Document (EFSA PPR Panel, 2013) are schematically presented in Figure 6. For convenience, the refined exposure approach (experiments and models) for standard test species are indicated as Tier‐2C1 and when relevant additional test species are involved as Tier‐2C2.
Figure 6.
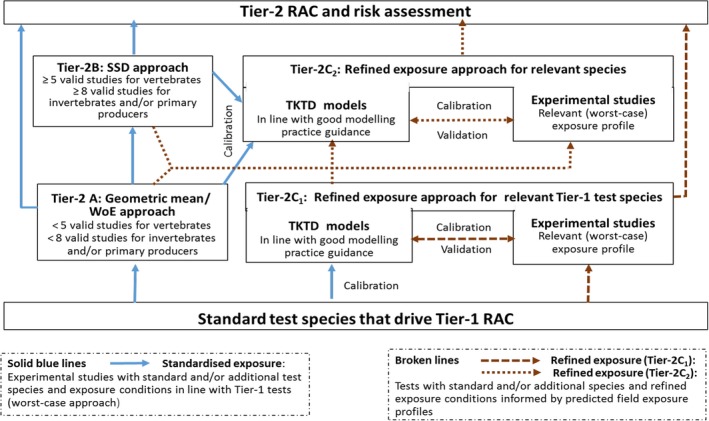
Schematic presentation of the different Tier‐2 approaches in the aquatic effect/risk assessment for PPPs. The different types of arrows indicate different approaches (solid blue lines refer to tests with standardised exposures; broken lines refer to tests with refined exposures). Further details on the geometric mean and species sensitivity distribution (SSD) approaches can be found in Chapter 8 of the Aquatic Guidance Document, while information on experimental refined exposure tests are given in Chapter 9 of the Aquatic Guidance Document (EFSA PPR Panel, 2013). Suggestions how to apply a Weight‐of‐Evidence (WoE) approach in Tier‐2 effect assessment if toxicity data for a limited number of additional species are available can be found in Section 8.2.2 of the Scientific Opinion on sediment effect assessment (EFSA PPR Panel, 2015)
Single species toxicity tests with aquatic organisms that are used in Tier‐1 (Standard test species approach), Tier‐2A (geometric mean/weight‐of‐evidence (WoE) approach) and Tier‐2B (species sensitivity distribution (SSD) approach) assessments, address concentration–response relationships based on standardised exposure conditions as laid down in test protocols. Toxicity estimates (e.g. EC50, EC10 and NOEC values) from these tests need to be expressed in terms of nominal (or initially measured) concentrations if they deviate less than 20% from the nominal (or initially measured) concentrations during the test or otherwise in mean concentrations (i.e. geometric mean or time‐weighted average concentrations) appropriately measured during the test. Consequently, these tests aim to assess the effects of a more or less constant exposure. In the field, however, time‐variable exposure regimes of PPPs in edge‐of‐field surface waters may be the rule rather than the exception. To assess toxicity estimates of more realistic time‐variable exposure profiles as predicted in the prospective exposure assessment, both refined exposure experiments and TKTD models may be used, either focussing on standard test species (Tier‐2C1) or also incorporating relevant additional species (Tier‐2C2) (see Figure 6). In the Aquatic Guidance Document (EFSA PPR Panel, 2013), it is stated that for proper use in regulatory risk assessment, experimental refined exposure studies should be based on realistic worst‐case exposure profiles informed by height, width and frequency of toxicologically dependent pulses from the relevant predicted (modelled) field exposure profiles. The realistic worst‐case exposure regimes selected for the refined exposure test thus represents those predicted exposure profiles that likely will trigger the highest risks. This also implies that the results of refined exposure tests are case‐specific. The possible implementation of another use pattern of the PPP under evaluation (e.g. due to mitigation measures) may result in a change in the relevant exposure profiles to be assessed. The main advantage of TKTD models is that they may be used as a regulatory tool to assess multiple exposure profiles.
TKTD models used as tools in Tier‐2 assessments need to be calibrated. For this, Tier‐1, Tier‐2A and/or Tier‐2B toxicity data sets as well as dedicated refined exposure tests with the selected species of concern can be used. In addition, substance‐ and species‐specific data sets derived from independent refined‐exposure experiments are required for TKTD model validation (see Figure 6). These validation experiments could be informed by the predicted field‐exposure profiles according to the worst‐case intended agricultural use of the substance. However, simulating the worst‐case exposure profile should not be considered a prerequisite for validation purpose. The validation experiment, however, should include at least two different profiles with at least two pulses each. For each pulse, at least three concentrations should be tested leading to low, medium and strong effects (see Sections 7 and 4.1.4.5 for more details). These recommendations are to address phenomena related to dynamics between internal and external exposure concentrations and possible repair of effects. To address phenomena related to toxicological dependence/independence, the individual depuration and repair time (DRT95) should be calculated and considered for the timing of the pulses; one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95 (see Section 4.1.4.5 for more details). In case DRT95 values are larger than can be realised in the duration of validation experiments, or even exceed the lifetime of the considered species, the second tested exposure profile may be defined independently from the DRT95.
In regulatory decision‐making, appropriately validated TKTD models may help to reduce experimental testing when assessing different exposure profiles of the same PPP for the same species and to test more exposure profiles than can be experimentally tested in practice. In addition, calibrated TKTD models may be used as a research tool, for example to:
design refined exposure experiments to validate the model;
evaluate the possible toxicological (in)dependence of different pulses (see Section 9.3 of EFSA PPR Panel, 2013);
select the most relevant time‐frame of the annual exposure profile that should be addressed in higher‐tier effect assessments (e.g. to design the exposure regime in mesocosm tests);
explore reciprocity of effects (see Section 4.5 of EFSA PPR Panel, 2013).
In this Scientific Opinion, the focus is on the use of TKTD models as Tier 2C tools in regulatory risk assessment.
3.2. TKTD modelling and species selection
In developing TKTD models as tools to assess effects of time‐variable exposures, an important question is ‘which species to select for modelling’. The criteria for selecting species in TKTD modelling do not deviate from the criteria in selecting test species for experimental refined exposure tests in the laboratory.
In the Aquatic Guidance Document (EFSA PPR Panel, 2013), guidance is provided on the derivation of Regulatory Accepted Concentrations (RACs) for edge‐of‐field surface waters based on refined exposure laboratory toxicity tests with standard test species (experimental Tier‐2C1 in Figure 6). Indeed, current practice is that most laboratory refined exposure tests submitted for active substance approval and PPP authorisation were conducted with Tier‐1 test species. Although EFSA PPR Panel (2013) does not exclude the use of additional species in RAC derivation by means of laboratory refined exposure experiments, less guidance is provided on how to select the additional species to test. It is, however, suggested that refined exposure studies with those additional species that are identified to trigger risks in Tier‐2A and Tier‐2B might be used to evaluate the risks of the realistic worst‐case exposure profile (in Figure 6 the Tier‐2C2 RAC derivation based on experimental studies).
If risks are triggered in Tier‐1, the development of TKTD models for Tier‐1 test species is most straightforward since in every dossier for active substance approval and PPP authorisation, acute and chronic toxicity data for Tier‐1 species are usually available; these data in raw format, i.e. observations over time, can directly be used for TKTD model calibration. If only the most sensitive Tier‐1 test species (surrogate for a certain group of organisms) triggers a potential risk, it may be sufficient to calibrate and validate a substance‐specific TKTD model for the most sensitive Tier‐1 species. Validated TKTD models for these Tier‐1 species can be used to evaluate specific risks of available field‐exposure profiles by calculating exposure‐profile specific L(E)Px (= multiplication factor of an entire specific exposure profile that causes x% Mortality/Effect) values. If a potential risk is triggered for more Tier‐1 species, an option for refinement would be to calibrate and validate substance specific TKTD models for more Tier‐1 test species.
If experimental toxicity data for standard test species and additional test species are available, developing TKTD models for these standard and additional test species may be a way forward as a Tier‐2C2 approach. This may, e.g. be done when a Tier‐2A (geometric mean/WoE approach) or Tier‐2B (SSD approach) trigger potential risks. Again, validated TKTD models for these species can be used to evaluate specific risks of available field exposure profiles by calculating exposure profile‐specific EPx values. Ideally, these additional species should comprise species with lower metabolic rates and slower repair mechanisms.
If validated TKTD models for a limited number of species are made available, the exposure‐profile specific risks (Tier‐2C2 informed by Tier‐2A; see Figure 6) should be estimated with these models by using rules similar to those used when applying the geometric mean or WoE approaches based on experimental laboratory‐toxicity data. These rules concern the measurement endpoints to select, taxonomic groups that can be combined and the size of the AF to be used (see EFSA PPR Panel, 2013, 2015). If validated substance‐specific TKTD models are made available for a sufficient number of relevant species (Tier‐2C2 informed by Tier‐2B; see Figure 6), the exposure‐profile specific EPx values for the different species can be used to construct an SSD and to derive an exposure‐profile specific HP5 (= hazardous profile to 5% of the species tested). This exposure‐profile specific HP5 can be used in the risk assessment following rules similar to those used when applying the SSD approach (see EFSA PPR Panel, 2013). To ensure that this procedure is not in conflict with the requirement of the tiered approach, in the sense that lower‐tiers should be more conservative than higher‐tiers, the Tier‐2C ERAs need to be calibrated with the RACs and associated exposure profiles derived from the (surrogate) reference tier for a selected number of substances differing in exposure dynamics and toxic mode of action (see EFSA PPR Panel, 2010; see also example in Chapter 8); the (surrogate) reference tier may be an adequate Tier‐3 approach, i.e. a valid micro/mesocosm experiment or, in the future, a validated population or community‐level model.
It should always be checked whether the taxonomic groups not addressed in the refined Tier‐2C assessment remain sufficiently protected (iteration with Tier‐1 data). Also note that a Tier‐2C assessment will always concern the specific protection goal (SPG) in line with the ecological threshold option (ETO) (EFSA PPR Panel, 2013). Only if the linking of TKTD models to population‐level models can be scientifically supported in regulatory decision‐making, then also the SPG in line with the ecological recovery option (ERO) might be considered. This scientific opinion, however, focuses on the use of TKTD models to assess risks that cover lower‐tier ETO‐RAC values for water and sediment organisms in edge‐of‐field surface waters.
Note that the calibration of Tier‐2C with the (surrogate) reference tier, as mentioned above, is an important issue. Indeed previous calibration exercises performed for aquatic invertebrates exposed experimentally to insecticides have indicated that in some cases the margin of safety (difference between Tier‐1/Tier‐2 RACs and corresponding Tier‐3 RACs) can be small or insufficient, particularly in the chronic effect assessment (van Wijngaarden et al., 2015; Brock et al., 2016). Although an advantage of TKTD models is that they can predict potential effects of longer exposure periods than normally assessed in laboratory single‐species tests, the question at stake is whether shifting from standard (worst‐case) exposure (Tier‐2A/B) to refined exposure conditions (Tier‐2C), the margin of safety remains sufficient. It is thus of high relevance to check the level of protection achieved by calibrating the Tier‐2C approach with results of Tier‐3 assessments (see e.g. the example data set in chapter 8).
3.3. TKTD models and Tier‐2 risk assessment for aquatic animals
3.3.1. Regulatory context in which the models will be used
TKTD modelling may be used to address (the threshold for) individual‐level effects occurring from time‐variable exposure regimes on aquatic vertebrates and invertebrates. In principle, effects of constant exposures can also be addressed by TKTD models, e.g. to estimate a time‐independent NEC. The problem formulation for the use of TKTD modelling for predicting (the threshold of) effects should initially identify the area of the risk assessment that is being covered by the modelling and why it is needed.
The GUTS model framework is developed to address lethal effects and may be an appropriate approach to use in the acute risk assessment scheme, since lower‐tier acute toxicity tests with aquatic animals are based on the measurement endpoints mortality or immobility. Consequently, if a high risk is triggered for aquatic animals by the acute Tier‐1, Tier‐2A or Tier‐2B effect assessment, the results of suitable GUTS models (i.e. appropriately calibrated and validated) can be used. In the chronic risk assessment, however, it is only appropriate to use a validated GUTS model if the critical endpoint is mortality, which is not often the case. In chronic toxicity tests, critical measurement endpoints usually concern sublethal effects like inhibition of reproduction or growth or, in the case of insects, emergence. Consequently, only if the risk is triggered by the chronic Tier‐1, Tier‐2A or Tier‐2B effect assessment and the critical (lowest) endpoint is mortality (or immobility), the results of suitable GUTS models (i.e. appropriately calibrated and validated) for the surrogate animal species of concern can be used in the chronic risk assessment.
If a sublethal endpoint is the most critical in the chronic lower‐tier assessment, the DEB modelling framework is the appropriate TKTD approach to select in the refined risk assessment. Note, however, that DEB modelling is a generic approach that assumes isomorphic growth,9 which may be more valid for aquatic species like fish and worms, but less so for crustaceans and aquatic insects (particularly during the final moult when the aquatic stage of the insect becomes a terrestrial stage). DEBtox models focus on body mass (or energy) and length, so if the species does moult then the model may need to be adapted to account for moulting of different aquatic larval stages. For insects with both an aquatic and a terrestrial life stage (e.g. Chironomus), emergence might be a problematic sublethal endpoint to address in DEBtox modelling. Modelling of the whole life cycle may not be necessary in the chronic effect assessment, if it is demonstrated that the effect endpoint tested/modelled concerns the most sensitive life stage of the species under evaluation. A recently published DEBtox model for the whole life cycle of an endoparasitic wasp (Llandres et al., 2015) provides an example of how to approach ‘non‐continuous’ growth. Nevertheless, in order to be used as a tool in prospective ERA for PPPs, it should be demonstrated that the DEBtox model used for a specific (surrogate) species, sufficiently addresses sublethal responses such as delay in hatch or emergence if these endpoints are triggered in the chronic lower‐tier assessment.
3.3.2. Specification of the question(s) that should be answered using the model
Questions that should be answered with the application of TKTD models in risk assessment should be related to the SPG for the group being considered, covered by the EFSA Aquatic guidance document (EFSA PPR Panel, 2013) and the sediment opinion (EFSA PPR Panel, 2015). The dimensions that need to be considered in defining SPGs for PPPs and aquatic organisms are: (1) the ecological entity to protect, (2) the attribute of the selected ecological entity to consider, (3) magnitude of the tolerable effect, (4) temporal scale of the tolerable effect, (5) spatial scale of the tolerable effect, and (6) degree of certainty (EFSA PPR Panel, 2010).
According to EFSA PPR Panel (2010), the degree of certainty should always be high. In the EFSA Aquatic Guidance Document (EFSA PPR Panel, 2013), the spatial scale of the risk assessment is currently limited to local edge‐of‐field ponds, ditches and streams as defined by the FOCUS surface‐ water exposure modelling approach. Within the context of the use of TKTD models in Tier‐2, the focus is on the ETO (see Chapter 5 in EFSA PPR Panel, 2013) and not on the ERO. When deriving an ETO‐RAC, the magnitude of tolerable effects is negligible. As a consequence, the main SPG dimensions to discuss within the context of TKTD models as Tier‐2 tools in aquatic ERA for PPPs are ecological entity and attribute.
For aquatic vertebrates in edge‐of‐field surface waters (e.g. fish and aquatic stages of amphibians), the ecological entity is the individual in acute risk assessments and the population in chronic risk assessments. The attribute to protect concerns lethal effects (mortality) of individuals in the acute risk assessment and effects on population abundance/biomass in the chronic risk assessment (see EFSA PPR Panel, 2013).
For aquatic invertebrates in edge‐of‐field surface waters, the selected ecological entity is the population and the attribute abundance/biomass (see EFSA PPR Panel, 2013, 2015).
Note that TKTD models can be used to predict lethal and/or sublethal effects on individuals (including individual‐level recovery i.e. their repair of damage) of aquatic vertebrates or invertebrates following time‐varying exposure, but not to predict population‐level effects (including community interactions and population recovery). Tier‐3 and Tier‐4 assessments are needed to address population‐ and community‐level effects. Nevertheless, results of Tier‐1 and Tier‐2 approaches are assumed to provide conservative estimates of the ETO‐RACsw if applying an appropriate extrapolation technique (e.g. AFs) calibrated by comparing lower tier assessments with that of the surrogate reference tier (see EFSA PPR Panel, 2010, 2013). In the tiered effect assessment for aquatic invertebrates, this assumption can be verified by comparing lower‐tier RACsw values with ETO‐RACsw values derived from microcosm/mesocosm tests (see e.g. van Wijngaarden et al., 2015). It is usually not possible to do this for aquatic vertebrates since microcosm/mesocosm tests that study treatment‐related responses of fish and amphibians, and that can be used as surrogate reference tier to calibrate the lower‐tier effect assessment for these taxa, are rarely available. Currently, the Tier‐2 effect assessment (including the ‘geometric mean/WoE’, ‘SSD’ and ‘refined exposure’ approaches) is the highest experimental tier used for aquatic vertebrates. The use of TKTD models in the Tier 2 refined ERA for PPP may be of particular importance for vertebrates as it enables a better consideration of animal welfare issues, which is an important EU policy.
3.3.3. Specification of necessary model outputs in relation to protection goals
For TKTD modelling of lethal effects of specific exposure profiles (e.g. by means of the GUTS modelling framework), the outputs from the model are the prediction of: (i) the expected mortality/immobility (0–100%) and (ii) a multiplication factor for the exposure that is necessary to reach a certain level of effect (e.g. 10% or 50%, i.e. LPx/EPx). A more detailed description of LPX/EPX values is given in Section 4.1.4.1. This concept of the multiplication factor was originally introduced by Ashauer et al. (2013) as the ‘margin of safety’. Similarly, for predicting sublethal effects (e.g. by means of the DEBtox modelling framework), the relevant model output is the prediction of exposure‐profile‐specific sublethal effects (e.g. EP10). Within this context, the toxicological (in)dependence of different exposures (pulses) potentially occurring in edge‐of‐field surface waters within the lifespan of the individuals should be considered. The lifespan may be long for, e.g. fish, and short to long for invertebrates. Toxicological dependence of different pulses is likely to occur if (i) the internal concentration or the scaled damage is still above (or closely below) the critical threshold level when an individual experiences another pulse exposure, or (ii) when the repair of the damage caused by the earlier exposure is not yet complete (see the output of the ELINK workshop (Brock et al., 2010)). It is proposed to use the DRT95 (individual‐level depuration and repair time for 95% of the effects) as a critical threshold for the design of validation experiments. It is expected that a time‐interval < DRT95 between two exposure pulses facilitates the demonstration of toxicological dependence, while a time interval > DRT95 between pulses enables the demonstration of toxicological independence.
3.4. TKTD models and Tier‐2 risk assessment for Primary producers
3.4.1. Regulatory context in which the model will be used
TKTD models developed for primary producers could be used in the chronic risk assessment scheme with a focus on sublethal effects, since inhibition of growth is not a lethal effect. They can be used to predict (the threshold for) effects of time‐variable exposures on growth of primary producers to supplement experimental Tier‐1 and Tier‐2 assessments. It should be noted that according to EFSA PPR Panel (2013) the lower‐tier RACs are derived from estimated concentrations causing 50% inhibition of growth rate (ErC50) for primary producers (r refers to the endpoint growth rate in the EC50 estimate). In cases where proper ErC50 values are not available for primary producers, an alternative estimated concentration causing 50% inhibition of yield (EyC50) may be used (y refers to the endpoint yield in the EC50 estimate). Note that EyC50 values are usually lower than the corresponding ErC50 values. For most macrophytes, the measured endpoints (estimated ErC50 or EyC50) used in effect assessment do concern individual‐level effects; however for algae and fast‐growing floating macrophytes like Lemna, the measured endpoints do not concern individual‐level effects, since in the course of the test (72–96 h for algae; 7 days for Lemna), many new individuals have developed and effects on the algal population and Lemna as well as its recovery are ‘entangled’.
The problem formulation for the use of TKTD modelling for effects on primary producers should initially identify the species or group of algae and/or vascular plants to be considered, informed by Tier‐1 and Tier‐2 laboratory toxicity tests. Based on these tests, it may be necessary to select subgroups (e.g. the floating macrophyte Lemna spp. may be an order of magnitude more sensitive than the rooted macrophyte Myriophyllum, and the other way around; the sensitivity of the tested green algae may differ more than an order of magnitude compared with the tested diatom or blue‐green algae).
3.4.2. Specification of the question(s) that should be answered with the model
Questions that should be answered with the application of TKTD models in risk assessment should be tied to the SPG for primary producers. According to EFSA PPR Panel (2013), the ecological entity of concern is population for both algae and aquatic plants and the related attribute is abundance/biomass/growth. The recommended measurement endpoints for Tier‐1 and Tier‐2 toxicity tests, used as the input parameters for TKTD models, concern inhibition of growth rate in terms of shoot length/frond number and biomass for macrophytes and in terms of biomass (cell counts) for algae. Results of Tier‐1 and Tier‐2 tests are assumed to provide conservative estimates of the ETO‐RACsw by applying an appropriate extrapolation technique (e.g. application of AFs that are calibrated by comparing lower‐tier assessments with that based on the surrogate reference tier).
For the ETO, the magnitude of tolerable effects for algae and macrophytes is meant to be negligible in edge‐of‐field surface waters (duration therefore not relevant). As for aquatic invertebrates, the ERO will not be considered further for primary producers as it is outside the scope of this document.
3.4.3. Specification of necessary model outputs in relation to protection goals
For TKTD modelling, the outputs from the model concern exposure profile‐related magnitudes of growth inhibition, preferably of the most sensitive endpoints identified at Tier‐1 (either shoot length/frond number, biomass for macrophytes or cell counts (as surrogate for biomass) for algae). Since EFSA PPR Panel (2013) recommends using ErC50 values in the lower‐tier effect assessment, it seems logical to select also ‘growth rate’ as effect estimate in the TKTD modelling approach. From a biological and ecological point of view, the endpoint growth inhibition of biomass seems more fit‐for‐purpose for TKTD modelling approaches than growth inhibition of shoot length/frond number, since biomass is a better indicator for the energy stored in plant tissues as a net result of processes like photosynthesis and respiration. Furthermore, the relationship between shoot length/frond number and its biomass may be quite variable for the same macrophyte depending on, e.g. environmental conditions (e.g. light conditions). Nevertheless, herbicides that inhibit cell division (e.g. sulfonyl urea herbicides) or stimulate excessive cell elongation (e.g. auxin‐simulating herbicides) total shoot length of macrophytes may initially be more critical than biomass. When exposed to herbicides that inhibit cell division, the formation of new shoots may be inhibited but not necessarily the total biomass of shoots, since sugars and starch may be stored in the older tissues if photosynthesis is not directly inhibited. In the case of auxin‐simulating herbicides, the growth of the plant shoots is stimulated at the cost of its biomass so that shoots become very long but brittle. Consequently, the use of TKTD models in prospective risk assessment for time‐variable exposure of pesticides and aquatic macrophytes in particular, should be evaluated considering the critical endpoint for which the exposure profile‐specific inhibition in growth should be assessed (informed by Tier‐1). If in experimental studies the effects of pesticide exposure on growth inhibition of biomass and shoot length/frond number endpoints result in more or less similar ErC50 values with confidence intervals that overlap, then it is proposed to select biomass‐related endpoints in TKTD modelling. If not, the TKTD modelling approach should be used to predict the response for the most sensitive relevant endpoint.
3.5. Specification of the domain of applicability of the TKTD model
The domain of applicability is mentioned in the EFSA scientific opinion on Good Modelling Practise (EFSA PPR Panel, 2014) as an important aspect of model application in ERA for PPPs. In that document, it is mentioned that modelling offers the opportunity to go beyond the conditions that have been tested in experiments or observed in the field. However, care must be taken when broader conclusions are drawn from the modelling results, with respect to scales, processes and variables that are taken into account.
As mentioned already, in order to be used directly in prospective ERA for PPPs, TKTD models need to be calibrated and validated for the PPP and surrogate species of concern. Relevant aspects for the domain of applicability of the TKTD model outputs in prospective ERA, appear therefore mainly as extrapolation from surrogate to other species (same rules as for experimental data), extrapolation across different exposure profiles and extrapolation over time. No extrapolation across levels of biological organisation (e.g. from individual to population or community level) can be done by the use of TKTD models alone.
3.5.1. Intraspecies variability
Experimental data sets for calibration and validation of TKTD models may concern a specific life stage (size class) of individuals of a specific species, particularly in acute laboratory‐toxicity tests. It is assumed that if the most sensitive life stage is tested, the calibrated/validated TKTD model most likely will result in a more conservative prediction than when the experimental data set concerns a less sensitive life stage. Extrapolation to other life stages (size classes) might be possible for some cases if the TKTD processes can be corrected for changes in body size. The phenomenon of extrapolation between life stages of the same species is not TKTD specific since it also plays a role in the Tier‐1 and Tier‐2 RAC derivation based on laboratory toxicity data. This uncertainty is addressed in the AF.
3.5.2. Extrapolation between species
Essentially, the calibration and validation of a TKTD model is, at the moment, always fixed for a specific species, and currently there are no extrapolation methods available that could do species‐to‐species extrapolation of TKTD model parameters with sufficient quality to allow for the use in ERA of PPPs. Hence, there is a need for extrapolation of TKTD model predictions from a surrogate species to other species – a normal procedure in lower‐tier RAC derivation achieved e.g. by applying an appropriate AF‐ remains.
Implicitly, the question of the representativeness of one species for one or more other species is of relevance here. This, however, does not appear as being specific to TKTD modelling. In this SO, it is assumed that the AFs currently used in Tier‐1, Tier‐2A and Tier‐2B assessments, based on experimental laboratory toxicity tests, are also fit‐for‐purpose in Tier‐2C assessments using validated TKTD models, since these AF account for extrapolations to other species and from the laboratory to the field.
3.5.3. Extrapolation across exposure profiles
The lifespan of the species to be modelled can be considered for the selection of the worst‐case time window to distinguish between ecologically dependent or independent pulse exposures. The following stepwise approach might be followed:
Use the full length of the predicted exposure profile (currently, 12 months for run‐off and 16 months for drainage scenarios) in the FOCUSSW calculations as input for the TKTD model. Only if potential high risk is predicted, selecting a (realistic worst‐case) time window in accordance with the life cycle properties of the species of concern may be a refinement option as described below.
Select the worst‐case time window within the exposure profile for the species by evaluating possible effects by a ‘moving’ time‐window equal to the (realistic worst‐case) length of its life cycle (or the duration of its relevant sensitive life stage). Note that algae, multivoltine and bivoltine aquatic invertebrates and aboveground parts of most macrophytes will have a life cycle shorter than the duration of the FOCUS exposure profile. Whether this is also the case for the sensitive life stages of fish and amphibians and univoltine and semivoltine invertebrates needs to be carefully evaluated and might result in a request for a prolonged exposure profile.
If the model performs well in predicting the effects of a set of appropriately selected worst‐case time‐variable exposure profiles or worst‐case time‐frames (criteria set in the Section 7) for a specific species, it is assumed that for regulatory purposes extrapolation across different exposure profiles can be done.
3.5.4. Range of geographical areas covered by the modelling
The geographical context is predominantly implemented in the exposure assessment. The Tier‐2 effect assessment, which may be supplemented with TKTD modelling, focuses on edge‐of field scale, and it is generic for the EU. Specific regional ecological scenarios, including regional focal species, become more important in Tier‐3 considering exposure conditions at different locations and Tier‐4 assessments for risks at larger spatial and temporal scales. Extrapolation in space is implicitly done when exposure time series of different locations are evaluated by compound and species‐specific TKTD models. It should be noted that at a landscape scale addressing multiple stressors may become more relevant. TKTD models may also be used to evaluate time‐variable exposures of several pesticides to assess cumulative risks (e.g. Ashauer et al., 2017).
3.5.5. Type of substance
TKTD models can be developed for any type of PPP but for substances with specific mode of actions special consideration may be required. For example models calibrated on acute tests are a priori not suitable for insect growth regulators since the test needs to include the critical moulting phase.
In a regulatory context, TKTD models should not be used for extrapolations from one substance to another with the same MoA. Compound‐specific input parameters are needed for regulatory purposes. TKTD models can be applied independently from the MoA, but for their application it has to be checked whether unexpected mortality is observed under long‐term (chronic) exposure. In such cases, toxicity cannot be predicted based on acute testing only; calibration and validation experiments should be available on longer time scales in order to detect such potential delayed effects.
4. General Unified Threshold models of Survival (GUTS)
4.1. Definition and testing of GUTS
This section contains specific information about GUTS itself and its implementation, but not about a specific application or data set. It contains as separate subsections information about:
the formal model, that is mathematical (differential) equations and other detailed information about the model;
the model implementation, that is which programming language or environment was used, which settings of key values have been chosen, etc.;
the verification of the model implementation, that is basic tests to show that the code works as it should for some selected cases;
Sensitivity analysis of the model, that is the effects of changes in parameter values on the model output;
General information about the way model parameters have been estimated, that is how calibration routines are performed, which function is used as target for optimisation routine, etc.;
The definition of model output and how uncertainties in model predictions are handled;
Criteria for the validation of a calibrated model on an external data set.
All the above subsections are exemplified hereafter with implementations of GUTS both in the Mathematica and the R programming languages.
4.1.1. Model formalisation
4.1.1.1. Toxicokinetic model and damage dynamics
The simplest GUTS version (e.g. for an aquatic invertebrate like Daphnia magna without measurements of internal concentrations) assumes a one‐compartment model and links external concentrations directly to the scaled damage (see Section 2.3 for an introduction to the scaled damage concept). The choice of this model, called ‘reduced GUTS’ (GUTS‐RED) implies that the dominant rate constant kD (time−1) is determined directly from the raw observed survival data (without internal concentration measurements). The dynamics of the scaled damage, denoted in this case, is described by the following differential equation
| (1) |
The scaled damage is given in units of concentration, equal to the units of measurements in the external medium Cw (e.g. in mol/L). A more explicit description of the dynamics of internal concentrations, denoted (e.g. in mol/kg), accounts for the uptake of a chemical in proportion to an external concentration and simultaneous elimination of the chemical in relation to the internal concentration, as described by
| (2) |
where kin (e.g. in L/kg time) and kout (time−1) are the uptake and elimination rate constants.
Within the full GUTS framework, the simulated internal concentrations is linked to the scaled damage assuming an increasing scaled damage, denoted Di(t) (e.g. in mol/kg), according to increasing internal concentrations and a possible individual‐level repair of the scaled damage with repair rate constant kR (time−1) following
| (3) |
Possible special cases of the reduced and the full GUTS exist. In the reduced version, the external concentration can be directly used as scaled damage without accounting for repair or depuration. This might be useful in cases of very fast uptake.
As shown in Figure 7, in the full GUTS, the internal concentration can be directly used as scaled damage without accounting for damage repair, which can be useful in case of very fast damage dynamics. These cases are, however, not formulated in the standard GUTS framework and hence not further considered here. Depending on the choice of the type of the scaled damage, it is expressed in external or internal concentration units. In consequence, all related parameters, including the killing rate constant b, and the (median) threshold z and m should be given with an index for a specific application. In case the scaled damage refers to the external concentration, the index is w, in case it refers to the internal concentration, it is i. In the general case, where a concrete model has not been selected, the index can be left out. As this SO uses mainly reduced versions of GUTS, the parameters will be denoted with index w.
Figure 7.
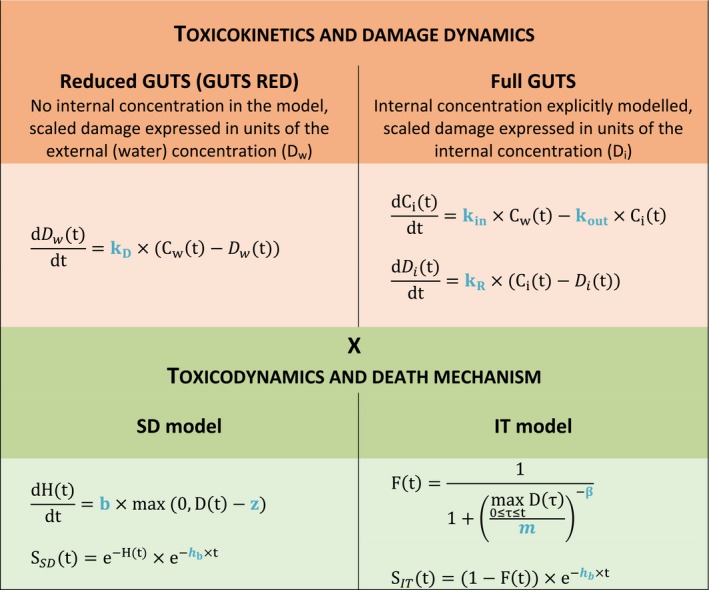
Overview about toxicokinetics, damage dynamics and survival models (stochastic death (SD) and individual tolerance (IT)) as used in the GUTS TKTD model framework. In the top panel, two formulations of the damage dynamics and the respective model equations are given. The bottom panel shows the two formulations of the death mechanism (SD and IT) together with the model equations. Parameters that must be determined from experimental data are marked in blue and explained in Chapter 2 and the following text. Combination of damage dynamics and death mechanism result in possible GUTS‐RED‐SD, GUTS‐RED‐IT, GUTS‐SD and GUTS‐IT models, which are defined by the given equations. Calibration of model parameters is explained in Section 4.1.3
4.1.1.2. Toxicodynamics and death mechanisms
The damage dynamics is connected to an individual hazard state variable, resulting in simulated mortality when an internal damage threshold is exceeded (Figure 7). Two death mechanisms are used in this scientific opinion, which are extreme cases of the GUTS theory: the SD and the IT.
For SD models, the threshold parameter for lethal effects is fixed and identical for all individuals of a group, meaning that the variance of the threshold values is zero, and the killing rate relates the probability of a mortality event in proportion to the scaled damage. In contrast, in the IT models thresholds for effects are distributed among individuals of one group, and once an individual tolerance is exceeded, mortality of this individual follows immediately, meaning in model terms that the killing rate is set to infinity. Both models are unified within the ‘combined GUTS’ model, in which a distributed threshold is combined with a between‐individual variable killing rate (Jager et al., 2011; Ashauer et al., 2016; Jager and Ashauer, 2018). For mathematical reasons, the parameter estimation of a ‘combined GUTS’ model requires more data than it can be obtained from standard survival testing. With the aim of maximising the added value of standard toxicity testing, this SO is considering the reduced GUTS versions only, with the IT and SD models as realisations of the death processes. Combinations of the choice of the scaled damage and the death mechanism give clearly defined acronyms for the different variants of GUTS, e.g. GUTS‐RED‐SD for the combination of the scaled damage without consideration of internal concentrations and the SD mechanism, or GUTS‐IT for the full GUTS model accounting for internal concentrations in combination with the IT mechanism. Please consult chapter 2 for an introduction into TKTD modelling, GUTS and the concepts of damage dynamics and death mechanisms.
In the SD model, a hazard rate is calculated following the differential equation
| (4) |
which describes hazard increasing in proportion to killing rate constant b, when the scaled damage exceeds the internal threshold concentration z. For the parameter estimation of the SD model, the killing rate constant b ([D]−1 time−1) and internal threshold concentration z ([D]) must be estimated from the survival data; [D] stands here for the unit of the scaled damage. The parameter values are kept constant during the simulated time, the processes of TK and TD are captured by the ordinary differential equations.
In the SD model, the survival probability of an individual to survive until time t is calculated as
| (5) |
where hb (time−1) is the background mortality rate constant.
In the IT model, the survival probability of an individual to survive until time t is calculated following the cumulative log‐logistic distribution of the thresholds z in a group of individuals, given by the function
| (6) |
where m is the median of the distribution of z ([D]) and β (−) is the shape parameter of the distribution.
In the IT model, the survival is related to the maximum scaled damage rather than to the actual scaled damage because death is irreversible, meaning that also under decreasing concentrations, the level of mortality in a simulated group of individuals could not become lower again. In the IT model, the survival probability of an individual to survive until time t is then calculated by
| (7) |
4.1.2. Test results for two model implementations
The implementation of a model is software‐specific. The documentation of the implementation should contain an overview of the source code files, and the version and necessary packages of the used programming environment. Testing of the implementation (‘implementation verification’) needs to be performed and documented for any model implementation, while the reasonable behaviour of the model (‘sensitivity analysis’) has been performed within this opinion for the example implementations (with Mathematica and R) and is not necessarily part of the documentation of a model implementation. The following sections contain a short overview of examples for an implementation of the GUTS TKTD model in Mathematica (Section 4.1.2.1) and in R (Section 4.1.2.2), before examples are given for the model implementation verification (Section 4.1.2.3) and the sensitivity analyses (Section 4.1.2.4).
4.1.2.1. Model implementation in Mathematica
The different GUTS‐RED versions have been implemented in Mathematica (Wolfram Research, version 11.0, http://www.wolfram.com/mathematica/). Mathematica is proprietary software for performing mathematics on a computer. It provides comprehensive methods for computation. The GUTS implementation in Mathematica uses mainly the functionality to calculate numerical solutions for ordinary differential equations (method NDSolve), to find the minimum of a given objective function (NMinimize), to read and write files of various formats (Import/Export) and to operate with lists and matrices of data. Mathematica is under continuous development; the implementation is steadily tested and verified.
The GUTS Mathematica notebooks contain applications for the single modelling steps, i.e. model calibration, model validation, predictions of surviving individuals for the exposure scenarios and probabilistic model simulations, together with all necessary data import and export functionality (see Appendix A). The source code of the Mathematica notebooks is not write‐protected, that can, however, be achieved if necessary.
Mathematica notebooks come in general in a specific format that contains both the program code and the output. Providing all notebooks and input files gives the opportunity to look into the source code and to see at the same time the output. A Mathematica installation and license will be necessary to run the program code and to redo and test calculations.
The GUTS Mathematica code has not been optimised for providing user‐friendly software. The structure of the model code enables, however, given a running Mathematica installation, to redo all steps that have been performed in the scope of the GUTS implementation, by loading and executing the GUTS‐methods notebook, followed by one of the application notebooks. Important for running the code is that input files are located in the file system as required.
4.1.2.2. Model implementation in R (package ‘morse’)
GUTS‐RED versions have also been implemented within the R‐package ‘morse’ 3.1.0 (R Core Team, 2016; Baudrot et al., 2018a). R is a free software environment for statistical computing and graphics. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS (https://www.r-project.org/). It can be downloaded from any CRAN mirror (https://cran.r-project.org/mirrors.html).
Package ‘morse’ provides tools for the analysis of survival/reproduction data collected from standard toxicity tests. It can be used to explore/visualise experimental data, and to perform estimation of LCx/ECx values by fitting concentration–response curves, or estimation of NEC values by fitting GUTS‐RED‐SD and/or ‐IT models. The LCx/ECx/NEC as well as model parameters are provided along with a quantification of their uncertainty. Package ‘morse’ can be downloaded at https://cran.r-project.org/web/packages/morse/index.html; a step‐by‐step explanation of its use is provided at https://cran.r-project.org/web/packages/morse/vignettes/tutorial.html, while a more formal description of the underlying estimation procedures is provided at https://cran.r-project.org/web/packages/morse/vignettes/modelling.pdf. The full description of all functions provided by the R‐package ‘morse’ can be found at https://cran.r-project.org/web/packages/morse/morse.pdf. The source code of these functions is accessible within R from instruction ‘morse:::function_name’. Moreover, the R‐package ‘morse’ uses the R‐package ‘rjags’ (Plummer, 2013) as an R interface to the JAGS library for Bayesian model estimation. Note that package ‘rjags’ does not include a copy of the JAGS library that needs to be installed separately (http://mcmc-jags.sourceforge.net/).
Note that basic features of the R‐package ‘morse’ are also available online within the user‐friendly web‐platform MOSAIC (Charles et al., 2017) which offers a new GUTS module specifically dedicated to the online fitting of GUTS‐RED‐SD and GUTS‐RED‐IT (http://pbil.univ-lyon1.fr/software/mosaic/guts, Baudrot et al., 2018c).
R notebooks are usually provided as readable stand‐alone text files directly usable as R scripts (.R extension). They contain all necessary code lines associated with comments, to run and get output results [See Appendix E for the code archive].
4.1.2.3. Verification of model implementation
In order to verify the model code, it would be ideal to show that the code is correctly implemented with respect to the conceptual and the formal model and that the code is error‐free. Model code verification is an important tool to build trust to model results. A rigorous verification of the complete implementation code, in the sense of a complete check of the computer code would mean an extensive effort and an enormous documentation. In order to keep the balance between effort and results, model results have been produced for a set of scenarios, representing specific classical and extreme cases, and the model outcomes have been checked for reasonable results. Results below are given side by side both for Mathematica and R implementations in order to illustrate that both independent implementations show the same results.
Classical exposure scenario
The model implementation has first been tested for a classical exposure scenario corresponding to a 4‐day period at constant concentration of a theoretical pollutant followed by a 3‐day period without exposure. For arbitrary parameter values, the survival over time (SOT) has been simulated for both the GUTS‐RED‐SD and the GUTS‐RED‐IT models, together with the scaled damage (Figure 8). As expected, this latter first increases until day 4, then decreases over the following 3‐day period.
Figure 8.
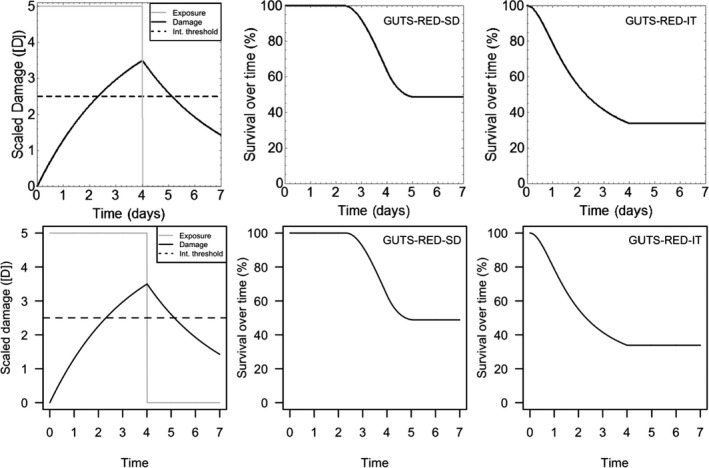
Test of the Mathematica (upper panel) and R (lower panel) implementation of GUTS‐RED‐SD and GUTS‐RED‐IT models under 4‐days constant exposure. The left panels show the scaled damage (black) and the external (gray) concentrations together with the internal threshold (dashed line). The middle panels show the survival over time for the GUTS‐RED‐SD model, parameters: hb = 0, kD = 0.3 (time−1), bw = 0.5 (L mol−1 time−1) and zw = 2.5 ([D]). The right panels show the survival over time for the GUTS‐RED ‐IT model with parameters: hb = 0, kD = 0.3 (time−1), mw = 2.5 ([D]) and β = 2 (−). Table 2 gives a full list of parameters with the explanation
Under model GUTS‐RED‐SD, the SOT first starts with a 100% value until about day 2 when the scaled damage exceeds the threshold zw value (damage curve and dashed line first intersection in Figure 8). Then, the SOT decreases until around day 5 when the scaled damage falls below the threshold (damage curve and dashed line second intersection in Figure 8). After day 5, the SOT remains constant at about 50%. Under model GUTS‐RED‐IT, survival starts to decrease from the beginning of the exposure, but the decrease stops as soon as the exposure concentration falls to zero at day 4. The final SOT under model GUTS‐RED‐IT is around 30%. Together, these results corroborate a correct implementation for both GUTS‐RED‐SD and GUTS‐RED‐IT models under constant exposure, since both model implementations show the same reasonable behaviour.
Pulsed scenarios
Another test was performed to check the Mathematica and R implementations of the GUTS‐RED‐SD and GUTS‐RED‐IT models for an arbitrary ‘pulsed’ exposure scenario. Such exposure situations, where steep increasing concentrations are followed by a period of more or less constant concentrations, which are abruptly falling down to zero concentrations, are challenging for the algorithms being used to calculate the numerical solutions of the model equations. The test for the same generic model parameterisation of GUTS‐RED‐SD and GUTS‐RED‐IT models as in Figure 8, with multiplication factors from 1 to 50, show technically correct simulation results for the scaled damage and the survival rate over time (Figure 9) (see Section 8). Most importantly, the numerical scheme and precision appears to be stable, as no indicators for numerical instability under such numerically challenging exposure schemes, i.e. abruptly changing or negative exposure concentrations, are visible. This reinforces confidence into the model implementations.
Figure 9.
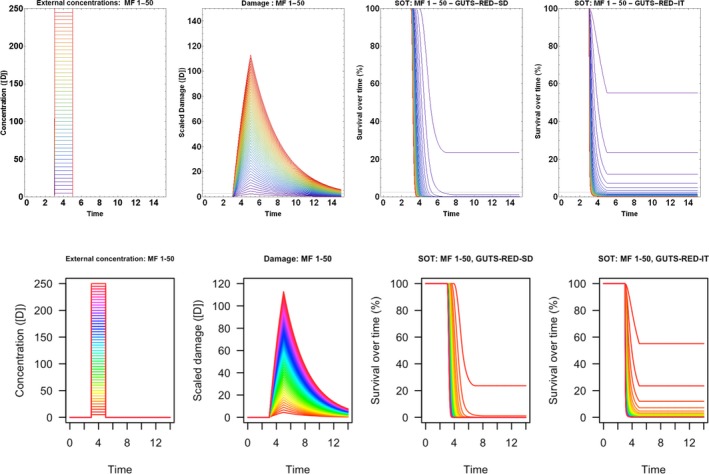
Test of the Mathematica (upper panel) and R (lower panel) implementations of GUTS‐RED‐SD and GUTS‐RED‐IT models with increasing multiplication factors for pulsed exposures, with exposure from day 3 to day 5. The external concentration of initially 5 ([D]) has been multiplied by factors from 1 to 50 (rainbow colours, first panels from the left). Second panels show the scaled damage; third panels show the survival rate over time for model GUTS‐RED‐SD and fourth panels for model GUTS‐RED‐IT. Parameters for model GUTS‐RED‐SD were: hb = 0, kD = 0.3 (time−1), bw = 0.5 (L mol−1 time−1) and zw = 2.5 ([D]); parameters for model GUTS‐RED‐IT were: hb = 0, kD = 0.3 (time−1), mw = 2.5 ([D]) and β = 2 (−). Table 2 gives a full list of parameters with the explanation
Extreme scenarios
A third and last test was performed to check the robustness of model implementations under extreme cases. Three extreme scenarios were tested as shown in Figure 10:
under a zero exposure with no background mortality, the scaled damage is equal to 0 and both the GUTS‐RED‐SD and GUTS‐RED‐IT models predict no mortality, hence survival over time equals 100%;
on the contrary, when the exposure is short but at very high concentrations (100 arbitrary units in this example), the scaled damage increases quickly during the period of exposure, then it decreases. Accordingly, the survival over time very quickly decreases from 100% to 0% in less than 1 day;
for a scenario with zero exposure but some background mortality, the damage is logically equal to 0, as in case (i), but the survival over time slightly decreases according to the value of parameter hb.
Figure 10.
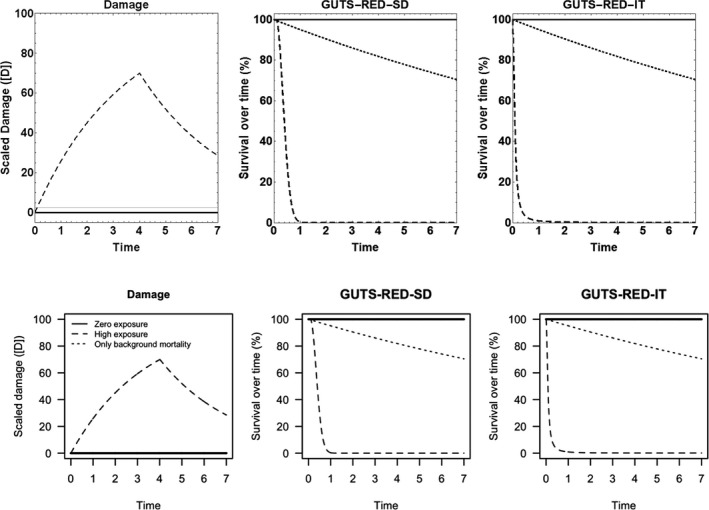
Mathematica (upper panel) and R (lower panel) implementation test under extreme cases: (i) zero exposure and no background mortality (solid lines), (ii) high exposure over days 1–4 (100 [D]) (dashed lines) and (iii) zero exposure with a slight background mortality (dotted lines). The left panels show the scaled damage; the middle panels show the survival over time under the GUTS‐RED‐SD model; the right panels show the survival over time under the GUTS‐RED‐IT model. Parameter values for the GUTS‐RED‐SD model: kD = 0.5 (time−1), bw = 0.3 (L mol−1 time−1) and zi = 2.5 ([D]); parameter values for the GUTS‐RED‐IT model: kD = 0.5 (time−1), mi = 2.5 ([D]) and β = 2 (−). Parameter hb was equal to 0 for extreme cases (i) and (ii); hb = 0.05 (time−1) for extreme case (iii). Table 2 gives with the list of parameters and their explanation
For all the tests, results of both the Mathematica as well as the R implementations of GUTS‐RED were identical and as expected, which further corroborates that the implementations of the GUTS reduced models are correct and stable.
4.1.2.4. Model sensitivity analysis
In addition to the previous numerical tests of the model implementations in Mathematica and R, a sensitivity analysis was performed. Sensitivity analyses are a tool to understand how model outputs respond to changes in parameter values. They allow the modeller to distinguish less influential parameters (that could be fixed at point values without any major change in model outputs) from the most influential parameters. The most influential parameters should receive most attention when calibrating the model.
Many methods exist to perform sensitivity analyses (Saltelli et al., 2000; Ciric et al., 2012). Here the one‐parameter‐at‐a‐time (OAT) method was used (see Figure 11), which involves varying one parameter at a time, keeping the others fixed at reference values. Excluding parameter hb from our sensitivity analysis, the toxicokinetic parameter kD was varied as well as bw and zw for the GUTS‐RED‐SD, and mw and β for the GUTS‐RED‐IT model (see Table 2 for the definition of the parameters). For more complex models with more parameter values, global sensitivity analyses are recommended in order to quantify the sensitivity of the model for changes in one parameter also in interaction with the other parameters, but for the GUTS‐RED models with three main TKTD parameters, the OAT method is still appropriate (Saltelli et al., 2000). Future sensitivity analysis could, with low effort, further investigate whether there are any interactive effects between the main model parameters.
Figure 11.
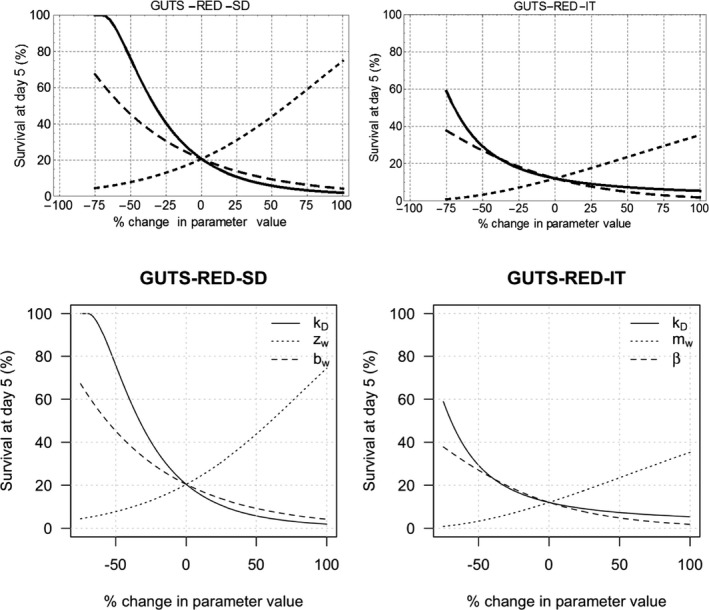
One‐at‐a‐time sensitivity analysis results for the GUTS Mathematica (upper panel) and R (lower panel) implementations of the GUTS‐RED‐SD (left) and GUTS‐RED‐IT (right) models, expressed in terms of survival rate at day 5 vs parameter values varying from −75% to +100% of their reference value; abscise 0 correspond to a parameter equal to its reference value. Reference parameter values for model GUTS‐RED‐SD: kD = 0.3 (day−1), bw = 0.5 (L mol−1 d−1) and zw = 2.5 ([D]. Reference parameter values for model GUTS‐RED‐IT: kD = 0.3 (d−1), mw = 2.5 ([D]) and β = 2 (−). Parameter hb = 0 for both models. Table 2 gives a full list of parameters with the explanation
Table 2.
Parameter symbols used for GUTS modelling in this chapter, explanation and units
| Symbol | Explanation | Example Unit |
|---|---|---|
| Dw | Scaled damage, referenced to external concentration | mol/L |
| Di | Scaled damage, referenced to internal concentration | mol/kg |
| Cw | External chemical concentration in the environment | mol/L |
| Ci | Internal chemical concentration in an organism | mol/L |
| kD | Dominant rate constant for the reduced model | day−1 (or time−1) |
| kin | Uptake rate constant for chemicals into the body | L/kg d−1 |
| kout | Elimination rate constant for chemicals from the body | day−1 |
| kR | Damage repair rate constant | day−1 |
| h | Cumulative hazard rate | day−1 |
| b | Killing rate constant | [D]/day−1 |
| bi | Killing rate constant referenced to internal concentration | kg/mol d−1 |
| bw | Killing rate constant referenced to external concentration | L/mol d−1 (or time−1) |
| z | Threshold for effects | [D] |
| zi | Threshold for effects referenced to internal concentration | mol/kg |
| zw | Threshold for effects referenced to external concentration | mol/L |
| hb | Background hazard rate | day−1 |
| mi | Median of the distribution of thresholds, ref. to internal conc. | mol/kg |
| mw | Median of the distribution of thresholds, ref. to external conc. | mol/L |
| β | Shape parameter for the distribution of thresholds | [−] |
Each parameter was varied from −75% to +100% from a reference value. This range of variation is relatively wide, but not unlikely in view of parameter confidence intervals observed in practice. In general, the range of the analysed parameter variation should be chosen according to usually observed parameter confidence/credible intervals. The influence of parameter variation on the model output was quantified considering the survival rate at day 5 (expressed in %) after a 2‐day pulse of 15 arbitrary units of external exposure from day 2 to day 4.
As shown in Figure 11, the survival rate at day 5 exponentially increases when bw (killing rate referenced to external concentration) (resp. β) decreases, while it exponentially decreases when zw (Threshold for effects referenced to external concentration) (resp. mi the median of threshold distribution referenced to internal concentration) decreases. Decreasing kD (dominant rate constant) provide an increase of the survival rate at day 5, but, in the case of model GUTS‐RED‐SD, a slight ‘saturation’ effect is noticed at very low values of kD (around −65%). In total, the influence of changes in the model parameter values was as expected. In addition, the tendency of parameter influence is very similar between both the GUTS‐RED‐SD and GUTS‐RED‐IT models, but the absolute changes in model output are weaker with the GUTS‐RED‐IT model. None of the model parameters appears more influential than the others; hence, all parameters can be considered to be important and have to be calibrated carefully. In summary, the sensitivity analyses indicate that model output changes in relation to changes in the model parameters in a continuous and reasonable way in the range between −75% and +100% of their respective reference values.
4.1.2.5. Conclusive statement on the formal and computer model
Both implementations, Mathematica and R, finally give exactly the same results when verifying the source code (Section 4.1.2.3) or analysing the sensitivity of model outputs to changes in parameter values (Section 4.1.2.4). The results corroborate the correct implementation of the formal model for both model implementations. Future implementations of GUTS could do the same checks to show correct implementation, in addition to the required ring‐test performance (see also Section 4.2, and Appendix B.6 and B.7). Moreover, OAT sensitivity analysis of the GUTS‐RED models has been performed and the results indicate that model parameters have almost equal influence on the model output, hence sensitivity analyses of the GUTS‐RED models are not required in future model applications.
4.1.3. Parameter estimation process
4.1.3.1. Introduction
Whatever the inference method (frequentist or Bayesian, see Sections 4.1.3.2 and 4.1.3.3), the goal of model parameter estimation (also named model calibration or parameter optimisation) is to get best parameter values that give an optimal fit of the modelled survival over time to the observed survival over time in the data sets that are used for calibration. According to the inference method, the steps required to get these optimal values of parameters differ, but in both cases, survival probabilities both for the SD or the IT model are calculated in the same way, with an explicit dependence on parameter vector θ, the external concentration over time (Cext(t)), and time (t). For the GUTS‐RED‐SD and the GUTS‐RED‐IT models, this is formally
| (8) |
and
| (9) |
Stochasticity of the survival process
Survival is a binary process, i.e. an individual can be either alive or dead. This fact can in small groups of individuals lead to stochastic fluctuations of predictions of survival over time, which is important to consider when laboratory test results with low numbers of tested individuals are used (e.g. typical acute test setting with 10 or 20 individuals per treatment). In such cases, survival can be modelled as a binomial process, with the number of surviving individuals being proportional to the conditional binomial distribution
| (10) |
where:
is the number of survivors in a population at time
yt is the number of individuals being alive at time t, and
is the conditional probability to survive from time t until time t + Δt, given as
| (11) |
where S(θ, Cext(t), t) is the deterministic survival rate calculated by the GUTS‐RED‐SD or the GUTS‐RED‐IT model for parameter vector θ, external concentration time course Cext(t) and time‐point t (see equations (8) and (9)).
Note that the time step is not fixed to 1 here, as the survival rates over time can be evaluated for arbitrary small or large time steps; hence, survival probabilities can be modelled as a random process using a flexible time step Δt.
For parameter vector θ, a chosen external concentration time course Cext(t) and an initial cohort size Y0, the number of surviving individuals in a group is modelled in an iterative way, starting with Y0 individuals at t = 0, by drawing for every new time step t + Δt from a conditional binomial distribution, parameterised by the number of living organisms at time t and the calculated survival rates for times t and t + Δt as given in equations (10) and (11). By performing nSmax repetitions of this procedure, nSmax realisations of the survival probabilities within a cohort of initial size Y0 are obtained, given parameter vector θ and external concentration time course Cext(t). Typically, nSmax will be chosen close to Y0. From the set of realisations of the stochastic process, statistical descriptors such as medians and percentiles are calculated. For large numbers of Y0, the stochasticity of model predictions becomes unimportant. The assessment of the stochasticity of the survival process is therefore required when for instance model predictions are compared with results from laboratory experiments with small numbers of individuals. When predictions of survival are made for environmental systems, it is supposed that the cohort size is large enough (Y0 > 100, based on expert knowledge) to ignore the stochasticity of survival.
Likelihood
Although several other methods exist either for the frequentist or the Bayesian methods, the methods here were based on the likelihood of the observed data given a parameter vector θ. The likelihood is defined as the assumed probability (density) for those observed data given parameter vector θ
| (12) |
In survival experiments, the observations at single time‐points are binomial (either alive or dead) and dependent on each other (the number of alive individuals at a single time‐point depends on the previous time‐point), so that the survival probabilities follow a multinomial distribution (or equivalently a conditional binomial distribution). Based on the multinomial distribution, Jager et al. (2011) derived the log‐likelihood function
| (13) |
where yi are experimental observations of survivors at time‐points i, and Si are simulated survival probabilities at time i, given parameter vector θ. For calculation reasons, Sn+1(θ, Cext(t), t) = 0 and .
Uncertainty
When using the model, the uncertainty in model parameter estimates comes in addition to the stochasticity of the survival process. Basically, uncertainty in parameter estimates can originate from different sources, either errors in experimental measurements or from natural biological variability.
Measurement errors and biological variability within the cohort of the individuals is accounted for within the process of the model calibration by the calculation of parameter confidence/credible intervals. While the measurement errors can be minimised by a good experimental quality, variability is intrinsic for biological systems and models are the means to explicitly address such variability. When evaluating, e.g. survival in acute toxicity tests by classical dose–response modelling, LC50 values are estimated and reported including uncertainty limits. When using TKTD modelling, uncertainty limits are approximated and reported for parameters, and the impact of these uncertainties combined with the stochasticity of the survival process are propagated to model predictions. When simulating environmental exposure scenarios, the assumption is that numbers of individuals in environmental systems are large enough to marginalise the stochasticity of survival, which results in observable differences in predicted survival only for small numbers. Still, for the prediction of environmental systems the parameter uncertainty, which captures among others part of the biological variability, has to be considered (see Section 4.1.4.2). Nevertheless, further sources of the variation of the biological response in natural systems cannot be estimated from experiments carried out under controlled conditions.
The next two sections explain the parameter optimisation process for the frequentist (Section 4.1.3.2) and for the Bayesian approach (Section 4.1.3.3), respectively. The Bayesian approach differs conceptually from the frequentist one in that it considers that the data are fixed and that the parameters are unknown random variables following a probabilistic distribution. The frequentist approach considers that the parameters are fixed and known, and that the data are considered to be one realisation of one experiment among many others that could have been performed. It should be highlighted here, that both frequentist and Bayesian approaches result in very similar parameter values, and that both approaches use approximation methods, often Monte Carlo Markov Chain (MCMC) algorithms to obtain best parameter sets and confidence/credible limits.
4.1.3.2. Frequentist approach
Parameter optimisation and likelihood
In the frequentist approach, parameters are optimised by maximising the log‐likelihood function (equation 13). In practice, often the negative of the log‐likelihood function is used because typically minimisation algorithms are used. Optimal parameter values give the best fit between the model and the observed data. Optimisation routines yield an optimal parameter vector θopt for which the log‐likelihood function shows the highest values of given the data set Y used. Optimisation results in practice depend on the starting values of the optimisation routine, so choices of starting values need to be documented, in addition to the choice and settings of the used optimisation algorithm.
For a basic understanding of this step of model calibration, that is parameter fitting via minimisation of the negative log‐likelihood, it helps to get a visual impression of what minimisation algorithms usually do (Figure 12). Parameter optimisation can be interpreted as a way to find the minimum of a ‘likelihood‐landscape’ in a n + 1‐dimensional space (n = 2 in Figure 12, but in general n is equal to the number of model parameters).
Figure 12.
Visualisation of the principle of minimising the negative log‐likelihood function under a frequentist approach. 2D (top) and 3D (bottom) view on the ‘parameter plane’ that is built by two model parameters
In the lower panel of Figure 12, the third dimension is in this case the difference between the optimal and the changed log‐likelihood (see equation 14), as introduced in Section 4.1.3.2 and also called log‐likelihood ratio and is shown as colour gradient (top) or explicit third dimension (bottom). The log‐likelihood ratio gives simply the distance of the log‐likelihood value from the optimal log‐likelihood value for an arbitrary parameter combination. Intuitively, the yellow point in the centre of the diagram represents the optimal parameter value (the minimum of the negative log‐likelihood function). During a parameter optimisation routine, red and blue points show parameter combinations that were tested with the aim to find the optimal parameter set (yellow point). All red dots represent parameter combinations which are within the confidence limits as defined by equation (14), whereas the blue dots indicate parameter combinations that were tested but which were outside of the confidence limits. Green dots represent results from the approximation scheme of the parameter confidence limits.
Optimisation routines search for a minimum in such likelihood‐landscapes (Figure 12) to find the optimal parameter combination (i.e. the parameter set with the lowest negative log‐likelihood value). Parameter confidence interval approximation (indicated by the green points in Figure 12) analyses the limits of the confidence region in a systematic way. What looks in this example smooth and straightforward can become very challenging, since not always such clearly defined minimum exists. For example, there can be multiple local minima, where the minimisation algorithm can get stuck depending on starting conditions. Hence, model calibration requires a good implementation of an appropriate optimisation routine and a comprehensive documentation of relevant aspects of the optimisation.
Examples of methods that can be used for optimisation are random walk algorithms, which search the parameter landscape with different choices of acceptance or rejection of tested parameter values. In general, stochastic optimisation algorithms (MCMCs and related methods such as Metropolis, Gibbs Sampler or Simulated Annealing), or downhill simplex methods such as Nelder–Mead are preferable over deterministic methods (e.g. Levenberg–Marquardt) because of their capability to find global minima and not to get stuck in non‐trivial optimum parameter searches.
Implementations of such algorithms are available and can be used in software such as Mathematica, where the Nminimize method gives the opportunity to choose from a variety of optimisation algorithms. Also, the OpenModel software (http://openmodel.info/) provides the opportunity of using a MCMC algorithm to find best parameters.
The convergence of optimisation algorithms is checked in the mentioned software implementations automatically, the principle is that the optimisation is starting for a number of different starting conditions (so‐called ‘chains’), and by the evaluation of differences within and between the different chains, the convergence of the algorithm is checked, e.g. by the Gelman and Rubin statistical test (Brooks and Gelman, 1998).
Numerical approximation of parameter confidence intervals
Unlike in, e.g. linear model fitting, parameter confidence limits in case of GUTS can no longer be estimated in an analytical way. They are not defined by any formula, but instead, need to be approximated by a numerical procedure. The log‐likelihood ratio method is one of the methods that can be used to approximate confidence limits for the optimal parameters θopt. Confidence limits for the single parameters for a given confidence level α were calculated as those parameter values, for which the log‐likelihood ratio fulfils the condition
| (14) |
with being the value of the chi‐square distribution for the confidence level α and the degrees of freedom of the log‐likelihood ratio df. For single parameter confidence intervals, the log‐likelihood ratio has df = 1. To find these parameter values, one value in the parameter vector, say parameter θi, is set to successively decreasing values, starting at the maximum‐likelihood estimate, i.e. the best parameter value. All parameter values with exception of parameter i are then again optimised to give a best fit to the experimental data. The value of the log‐likelihood function corresponding to parameter vector is calculated. This procedure is repeated until the constrain on is satisfied (equation 14). Likewise, this procedure is repeated for successively increasing parameter values, again starting with the optimal maximum‐likelihood estimate. First values of parameter θi leading to the violation of condition (14) are reported as confidence limits. Further reading on the numerical approximation of parameter confidence intervals are reported elsewhere e.g. Jager and Ashauer (2018).
4.1.3.3. Bayesian approach
As mentioned above, the Bayesian approach considers that data are fixed and that the parameters are unknown random variables following a probabilistic distribution. These results in the following practical implications: (i) Bayesians want to optimise the probability of parameter vector θ given the data set Y used for calibration, rather than only the likelihood (see below); (ii) the need for Bayesians to provide reasonable prior information to see the result of an experiment, then updating this information by accounting for the data. Below is a short introduction to Bayesian principles; it is recommended to readers who would like to learn more about Bayesian data analysis to consider further reading; a recommendation is, for example, Gelman (2014).
Basic principles
The keystone of the Bayesian approach is the Bayes formula
| (15) |
where Y are the observed data; P(θ|Y) is the joint posterior distribution of parameter vector θ;P(Y|θ) is the likelihood of the data given the parameters (see equation 12 and 13); P(θ) is the joint prior distribution of parameter vector θ.
Given that P(Y) is known and fixed, it is often not considered as it does not depend on θ and will not influence the posterior distribution. Hence
| (16) |
with P(θ)p(Y|θ) the unnormalised posterior density and
| (17) |
The prior distribution P(θ) expresses the available parameter information without knowing the observed data, while the posterior distribution P(θ|Y) combines this prior information (which may be more or less informative depending on what is known about the value of the parameters beforehand) with evidence from the data (expressed through the likelihood) into a posterior density probability distribution for the parameters. The overall expectation is to get a narrower posterior distribution compared to the prior (illustrated for one parameter in Figure 13, left): the difference between the two distributions reflects the information provided by the data. When the prior information is vague (translated into a flat prior distribution), and the data sufficiently informative, the results converge to those obtained by the frequentist approach (Englehardt and Swartout, 2006).
Figure 13.
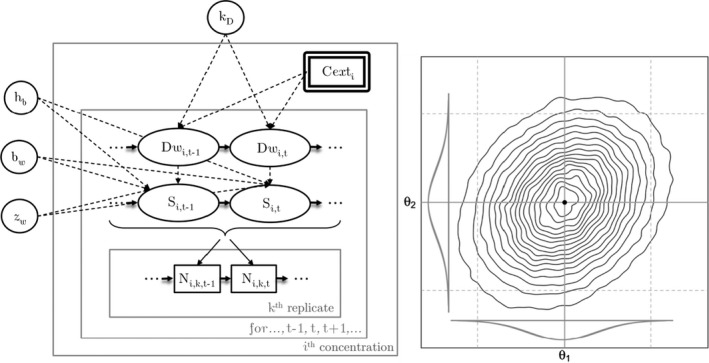
Left panel shows Directed Acyclic Graph (DAG) for the model GUTS‐RED‐SD when the exposure concentration (covariate Cext in double rectangle) is constant over time. Observed survival numbers N are represented in rectangles; ellipses correspond to latent variables (Dw for the scaled damage and S for the survival rate) and circles to parameters to estimate. Deterministic links are represented by dashed arrows, whereas stochastic links are represented by solid arrows. Grey external rectangles stand for replicate (index k), time course (index t) and exposure concentration (index i). Right panel shows theoretical binormal joint posterior distribution of parameter vector (θ1, θ2). Ellipses correspond to isoclines of the joint posterior distribution; grey distributions are marginal posterior distributions of both parameters; solid horizontal and vertical lines correspond to the medians of these marginal distributions; dashed horizontal and vertical lines correspond to the 2.5% and 97.5% quantiles of the marginal distributions. See Table 2 with the list of parameters and their explanation
Directed Acyclic Graph
Under a Bayesian framework, a model is specified by a set of prior distributions on parameters to estimate and a set of hierarchical conditional distributions linking parameters to data, which is commonly depicted in a Direct Acyclic Graph (DAG). In fact, the DAG represents a series of conditional independence assumptions, which allows the full probability model to be factorised into a product of conditional distributions, and consequently to tackle calculations. Each quantity in the model (data, covariates, parameters and latent variables10 ) corresponds to a node in the DAG, and links between nodes show direct dependences. The graph is directed, because each link is an arrow; it is acyclic because it is not possible to return to a node after leaving it by following the arrows (Lunn et al., 2000). The dependencies can be defined by deterministic (namely, mathematical functions) or stochastic (namely, probability distributions) links. For each node with no incoming arrows, prior information (Figure 13, left) should be specified.
Joint posterior distribution
The joint posterior distribution has the dimension of the number of parameters, but it can be plotted in planes of parameter pairs to visualise correlations between parameters. In an example case with two binormally distributed parameters, the joint posterior distribution can be plotted in the 2D‐parameter space as illustrated by ellipses on Figure 13 (right); in this example, parameters θ1 and θ2 appear slightly correlated. From the joint posterior distribution, the marginal posterior distributions for each parameter (as illustrated by grey normal distributions on bottom and left sides of Figure 13 (right) can be extracted. Then, from the marginal posterior distributions, some statistical summaries on parameter estimates can be extracted, usually the median (illustrated by vertical and horizontal plain grey lines on Figure 13, right) as well as 2.5% and 97.5% quantiles to serve as 95% credible intervals (illustrated by vertical and horizontal dotted grey lines on Figure 13, right). Another advantage of having the joint posterior distribution is that any posterior distribution of any function of the parameters can be obtained. In particular, when calculating an LCx,t from GUTS (for given choices of the x% and of the target‐time t), the corresponding formula depends on GUTS parameters. The uncertainty on these GUTS parameters, which is extracted from the joint posterior distribution, can be propagated to the LCx,t calculation, on which a posterior probability distribution can then also be obtained (see Section 4.1.4.4 for more details).
Parameter uncertainties
One implication of adopting a Bayesian approach is that the uncertainty on a parameter is expressed as a credible interval (also called a Bayesian confidence interval) instead of an approximate confidence interval like in the frequentist approach (see Section 4.1.4.3). The 95% credible interval delimits a range of values where the parameters should lie with a 95% probability, whereas the calculation of a confidence interval (usually a 95% confidence interval) is based on hypothetical repeated sampling from the model: if samples of the same population size are repeatedly obtained and a 95% confidence interval for each of the samples is got, it is expected that 95% of the confidence intervals will contain the true value of the parameter. Another difference is that the credibility interval is conditional to the data used to estimate the parameters, whereas the confidence interval is not.
Numerical computation of the posterior distribution
Based on equation (16), the calculation of the posterior distribution is often tricky. Analytical solutions only exist in rare cases. Many numerical methods have been developed to approximately compute the posterior distribution in challenging cases, mainly based on simulations by MCMC sampling methods used to generate random numbers from complex joint distributions. MCMC algorithms are a general method based on drawing values of parameter vector θ from approximate distributions and then correcting those draws to better approximate the target posterior distribution, P(θ|Y). The sampling is done sequentially, with the distribution of the sampled draws depending only on the last value drawn; hence, the draws form a Markov chain. The key to the method's success, however, is not the Markov property but rather that the approximate distributions are improved at each step in the simulation, in the sense of converging to the target posterior distribution (Gelman et al., 2014). With such algorithms, the simulation process must run long enough so that the distribution of the current draws is close enough to the desired target posterior distribution; hence, the user himself must choose the number of iterations.
MCMC algorithms use random walk algorithms. Among them, the Metropolis algorithm (and its generalisation, the Metropolis–Hasting algorithm) is an adaptation of a random walk with an acceptance/rejection rule to converge to the specified target distribution (Metropolis et al., 1953; Hastings, 1970). The Gibbs sampler is a special case of the Metropolis–Hastings algorithm applicable when the joint distribution is not known explicitly, or it is difficult to sample from directly, but the conditional distribution of each parameter is known and is easy (or at least, easier) to sample from (Geman and Geman, 1984).
Several tools are available to automatically perform these computations like: JAGS for Just Another Gibbs Sampler (Plummer, 2003) as used in some parts of this opinion in combination with the R‐package rjags which provides an interface from R to the JAGS library for Bayesian data analysis; STAN, a state‐of‐the‐art platform for statistical modelling and high‐performance statistical computation (Carpenter et al., 2017); Winbugs, based on the BUGS (Bayesian inference Using Gibbs Sampling) project (Lunn et al., 2000). Today, the performance of these tools allows a wide use of the Bayesian approach, even if it is still challenging to deal with ordinary differential equations.
Convergence criteria and goodness‐of‐fit
When running an MCMC algorithm, it is suggested to run several MCMC chains in parallel (usually three). They should all converge to the same target distribution thus providing a sample of the full joint posterior distribution. These chains also allow to perform the Gelman and Rubin statistical test (Brooks and Gelman, 1998). For each parameter, the square root of the ratio between the variance of its posterior marginal distribution and the intrachain variance is calculated; it is expected to be equal to 1 if the convergence is reached. In practice, a ratio lower than 1.1 for each parameter is acceptable. Another way of checking the chain convergence is to visualise quantiles of interest of the posterior distribution (namely, 2.5%, 50% and 97.5% quantiles) over the iterations to ensure that they have stabilised at the end of the running process. Also, the autocorrelation between chain iterations should be checked (further reading in Raftery and Lewis, 1992).
If the model fits well, then the replicated data generated under the fitted model should look like the observed data. To put it in another way, the observed data should look plausible under the posterior predictive distribution, as a self‐consistency check. A basic technique for checking the fit of a model to observed data is to draw simulated values from the joint posterior predictive distribution of replicated data and compare these predicted samples to the observed data. Any systematic differences between the simulations and the observed data indicate potential failings of the model (Gelman, 2014). In practice, it is useful to examine graphical comparisons of the observed data with summaries of posterior predictive simulations, what is usually called a posterior predictive check (PPC) plot (Gelman, 2014). In PPC plots, the observed data is plotted against the corresponding estimated predictions, along with their 95% credible interval (see Figures 17 and 23 as examples).
Figure 17.
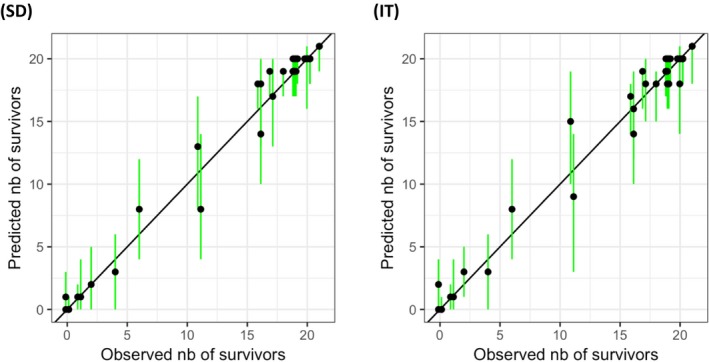
Posterior Predictive Check for the GUTS‐RED‐SD (left panel) and the GUTS‐RED‐IT (right panel) models, calibrated on a typical acute toxicity test. The x‐coordinate of black dots is the observed number of survivors while the y‐coordinate is the predicted median number of survivors. Vertical segments stand for the 95% credible intervals of the predicted values. These intervals are represented as green segments if they overlap with the line y = x
Figure 23.
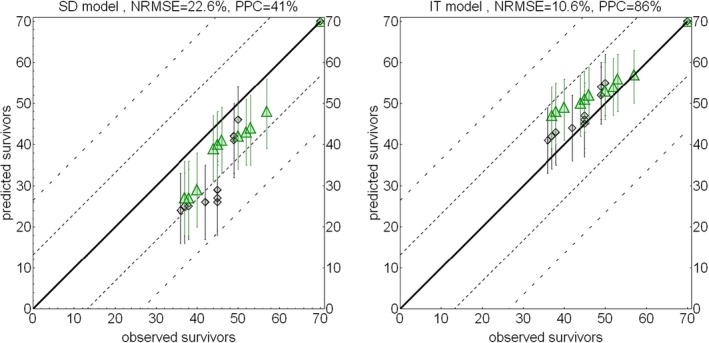
Posterior predictive plots of modelled versus observed numbers of survivors, left for the GUTS‐RED‐SD, right for the GUTS‐RED‐IT model. Triangles show results for pulse ‘A’, diamonds for pulse ‘B’. The error bars depict the 5th and 95th confidence limits for the predicted number of survivors. The solid line is the 1:1 line, the dotted line depicts the range of 25%, and the dashed line that of 50% deviation, (see Section 4.1.4.5). NRMSE and PPC values are given in the plot titles
Summary of Bayesian approach
In short, the Bayesian approach requires the following steps:
Choose the prior distributions based on previous results, literature or expert knowledge: P(θ);
Define the probabilistic model from the data, that is the random variables whose data would be one realisation assuming known values of parameters, namely the likelihood: P(Y|θ); (equation 1)
Calculate the joint posterior distribution of the parameters given the data via the Bayes formula: P(θ|Y);
Provide statistical summaries of parameter estimates (namely, appropriate quantiles);
Get any function of the parameter estimates as posterior probability distribution, like for example LCx,t calculations or predictions of new observations (see Section 4.2.2).
In essence, the Bayesian approach appears intuitive, starting from a prior distribution to derive a posterior one; this latter may then serve itself as a prior distribution to fit new data. This knowledge accumulation is an advantage of the Bayesian approach. The consideration of prior knowledge about parameters allows to use relevant information which is often available even before performing a survival experiment.
4.1.3.4. Summary on parameter estimation
The parameter optimisation process is important, because it yields the model parameters and uncertainty limits which are in consequence propagated in the model predictions. Parameter optimisation routines are built into different software and can be used without too detailed expert knowledge, but it is advantageous to have some more understanding of the details, because only this enables a safe use for regulatory risk assessment. This is also the reason why, in this section of the document, the technical details behind the parameter optimisation were outlined on a higher level of details than needed to only apply the methods. For the evaluation of parameter estimates in practice, it is convenient that algorithms are implemented in standard software, because that makes sure that the algorithms are error‐free and the convergence of the algorithm is also automatically tested.
Experience from parameter estimation gives some indication about important aspects for the calibration of GUTS models:
Calibration data should span from treatment levels with no effects up to large effects, ideally including full effects (e.g. 0% survival). This is important because parameter estimates can show reduced accuracy and precision when the experiments do not show a clear and comprehensively covered dose–response pattern.
Calibration data sets should report raw observations of mortality or immobility at least for five time‐points (initial plus four observations over time). The choice of five time‐points may be problematic in standard tests shorter than 4 days. Possible solutions are: (i) increase the number of observation in those tests; and (ii) to extend the duration of the test to for instance 96 h. If a standard 48‐h study is only available, a calibration might still be attempted but the quality of the fit (convergence, uncertainty limits and visual fit) should be carefully checked.
Attention should be paid in case the influence of time on the exertion of the effects is not fully captured in short tests (e.g. onset of effects, delayed effects, accumulated toxicity, etc.) as this is likely to result in inaccurate predictions in the validation phase.
Optimisation algorithm needs to be specified including settings (see examples in Sections 4.1.4.3 and 4.1.4.4), and also the method that was used for the approximation of the confidence/credible limits.
In general, stochastic optimisers (MCMC) are preferable over deterministic methods (e.g. Levenberg–Marquardt) because of their improved capability to find global minima.
Not only optimal parameter values, but also the log‐likelihood value, and approximated confidence or credible intervals for all parameters must be given to allow for further checks (see Section 4.2.2.1).
The ultimate test for the quality of parameter optimisation is given by the comparison between the modelled survival using the optimal parameter set and the observed survival which can be checked in plots of the survival over time or by the predictive posterior check (PPC) (see Section 4.2.2.1 for an example).
Most of these points are used also in Section 7.6.2 for the evaluation of the parameter estimation process. A checklist for assessing the quality of the parameter estimation process for a GUTS application is part of the checklist for GUTS models (Annex A).
Parameter optimisation can be done in one of the frequentist or Bayesian frameworks, which result from different ‘schools of thought’ in statistics. Both methods have advantages and disadvantages, but in practice both can be used, since they result in very similar parameter values, and both approaches depend on approximation methods such as MCMC to obtain best parameter sets and uncertainty limits.
4.1.4. Model predictions
In this section of model documentation, the specific application of the calibrated model for the calculation of predicted effects is documented as an example. It defines clearly, which and how risk assessment‐relevant endpoints are being calculated. In addition, the basic principle and the technical description of the used approach to propagate parameter uncertainty to model predictions are reported, both for the frequentist and the Bayesian approach.
4.1.4.1. Modelled endpoints
According to the definition of the regulatory questions that should be answered by the application of the model, and within the structure of the given regulatory model, the simulation model outputs have to be defined. The basis for these formulations is given in Sections 3.3.2 and 3.4.2. In case the relevant endpoints for the risk assessment are lethal effects (mortality or immobilisation), these can be calculated for time‐variable exposure profiles by using a calibrated GUTS model. Before GUTS model predictions can be considered relevant for regulatory risk assessment, the GUTS model needs to be validated by comparison of model outputs with independent observations of effects. The basic relevant model output from GUTS is the predicted mortality/immobility (immobile individuals in acute toxicity tests used for model calibration are considered ecologically dead), which is calculated per exposure pattern. If no specific information about the target time‐point of observation is given, mortality/immobility is assessed by default at the end of the analysed exposure profile. For some tested exposure profiles, no mortality/immobility will be observed in the model predictions. Here, another relevant endpoint calculated from the GUTS modelling provides useful information. Exposure profile specific multiplication factors leading to a certain effect level, e.g. 50%, at the end of the tested profile (lethal profile (LP50) for mortality, effect profile (EP50) for immobility) can be calculated for a given exposure profile P. The analogy to the LC50 or EC50 of a laboratory test on mortality under static exposure, which reports the mid‐point of the dose–response relationship and the concentration which is leading to 50% mortality or immobility is intended, but attention is needed because the LCX/ECX are concentrations, while the LPX/EPX are multiplication factors.
These LPX/EPX values are technically constructed using multiplication factors applied to the concentration time series of the exposure profile. In such a way, the whole exposure profile can be ‘shifted’ and adjusted to exactly the multiplication factor which will result in x% mortality at the end of exposure profile P. This multiplication factor is then denoted LP X. This approach is illustrated in a simple example (Figure 14).
Figure 14.
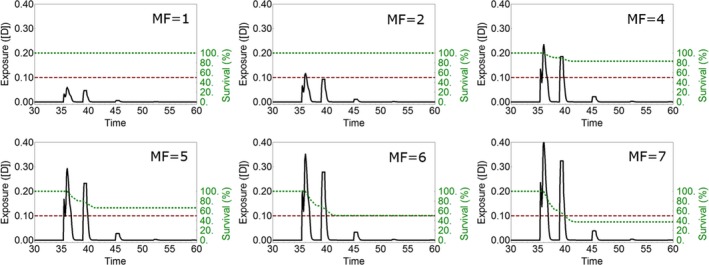
Example simulations of survival over time (dotted green lines) with the GUTS‐RED‐SD model where parameters are: kD = 0.5 ([time]−1), bw =0.05 (L mol−1 time−1) and zw = 4 ([D]), for increasing multiplication factors (MF) to an exposure profile P (solid black lines). The dashed red line indicates the 50% survival threshold that is reached for a multiplication factor of 6. Hence, in this case, LP 50 = 6. Note that the LPX is not necessarily always a whole number. Table 2 gives the list of parameters and their explanation
4.1.4.2. Consideration of uncertainty in model predictions
For any exposure profile P, mortality and LPX/EPX values can be calculated using a calibrated model. This can be done in a deterministic way, i.e. the mortality for a group (or cohort) of individuals of one species is predicted in form of the reduction of the deterministic survival rate given the optimal parameter set. In that way, average percentages of mortality/immobility in a cohort are calculated. The EFSA Scientific Opinion on Good Modelling Practice (EFSA PPR Panel, 2014), however, requires consideration of model uncertainty when performing predictions from a model (Chapter 9.2 in EFSA PPR Panel, 2014).
In general, EFSA PPR Panel (2014) lists different sources of uncertainty for model predictions such as the GUTS‐based predictions of mortalities and states that uncertainty must be differentiated from variability, the former being due to insufficient information within the model, e.g. due to inappropriate calibration data set, the latter reflecting biological variability which cannot be reduced. One source for uncertainty is the structure of the used modelling approach, which has been discussed in Chapter 2. Additional to the structural uncertainty, uncertainty comes with the exposure time series that are being used as input for the effect modelling. In the domain of fate modelling, the rationale is to construct realistic worst‐case scenarios by using simulations that show 90th percentile peak concentrations (FOCUS, 2001). Without going into detail, the basic assumption is that uncertainty in the exposure estimates is implicitly included by selecting a reasonable worst‐case for the PECmax in the time series when, e.g. using FOCUS surface water predictions as input for TKTD modelling.
As stated already in the section about parameter estimation (Section 4.1.3.1), additional sources for uncertainty in the model parameters are measurement errors and biological variability within the cohort of the individuals, which have an impact on the estimated model parameters. Whereas the measurement errors can be minimised by a good experimental quality, variability is intrinsic for biological systems and can be explicitly addressed by TKTD models. Using numerical approximations of the parameter confidence/credible intervals (see Section 4.1.3.2), the impact of remaining measurement errors and the biological variability can be propagated to model predictions.
In the following sections, the computational approach for the propagation of uncertainties in the model parameters to the model output are introduced for the frequentist and the Bayesian frameworks.
4.1.4.3. Consideration of uncertainty in model predictions in a frequentist approach
1. Step: Construction of joint parameter confidence region
In the first step, joint confidence regions for the model parameters are constructed and a corresponding set of ‘significant’ or likely parameter combinations are defined by
| (18) |
where the log‐likelihood function has been defined in equation (13), and is the value of the Chi‐square distribution for the confidence level α and the degrees of freedom of the log‐likelihood ratio df. Critical for this is the correct parameterisation of the chi‐square distribution. For the example of a three‐parameter model such as the GUTS‐RED‐SD or GUTS‐RED‐IT models ignoring the background mortality, the critical value is 7.815 (df = 3, α = 0.05). The degrees of freedom for the log‐likelihood ratio that is being used for the construction of the joint confidence region are defined by the difference in free model parameters. In this case, the degree of freedom equals 3 (df = 3), because for optimisation all three model parameters of the GUTS‐RED‐SD or GUTS‐RED‐IT models were free, whereas for the construction of the three‐dimensional joint confidence region all model parameters were fixed. In contrast, the degrees of freedom for the construction of a single parameter confidence interval is 1, because for the optimisation also three parameters were free, but only one parameter is fixed for the construction of the confidence interval (and two were free); hence for a single parameter confidence interval the degree of freedom is one (df = 3–2 = 1). The background mortality rate constant was not taken into account here, because it was not considered for the uncertainty calculations.
From such joined confidence region, sets of parameters can be constructed either by rejection sampling from previously stored model parameters that were already used for parameter estimation, or from new random values which are tested for compliance with condition equation (1).
2. Step: Simulations of survival over time in n max repetitions
In the second step, parameter uncertainty can be propagated to uncertainty in model predictions. For a given exposure profile, a number of nPmax parameter sets from the confidence region Θconf (equation 19) are drawn, and for every parameter vector, survival over time is simulated.
For small cohorts, e.g. typical numbers of fish or invertebrate individuals in a standard toxicity test, the stochasticity of survival needs to be considered in addition to the uncertainty of the survival. This would mean, that for each profile and every parameter set, survival over time is simulated as explicit conditional binomial process in nSmax iterations (see Section 4.1.3.1 ‘Stochasticity of the survival process’). The total number of simulations for small numbers of tested individuals amounts to nmax = nPmax × nSmax.
For larger cohorts of individuals, as expected for environmental systems, the stochasticity of survival does not need to be considered explicitly, and the total number of simulations is identical to the simulations using the number of parameter sets from the confidence region (nmax = nPmax).
3. Step: Simulations of survival over time in n max repetitions
These nmax realisations of the simulated numbers of surviving individuals are statistically described, e.g. the median and e.g. 5th and 95th percentiles of the distribution of the numbers of survivors are determined, in this way the uncertainty in the model predictions caused by parameter uncertainty is quantified.
This technique can also be used for the approximation of exposure profile specific multiplication factors (LPX/EPX) values. For this, for each multiplication factor, survival over time is simulated in a probabilistic way, until the selected lower percentile of the probabilistic predicted survival, e.g. the 5th percentile, is equal to the intended endpoint, e.g. 50% mortality. The multiplication factor leading to this is then the lower confidence limit for the LP50, and the procedure is repeated for the upper percentile, e.g. for the 95th.
4.1.4.4. Consideration of uncertainty in model predictions in a Bayesian approach
In addition to the stochasticity of the survival process (Section 4.1.3.1 ‘Stochasticity of the survival process’), Bayesian uncertainties on parameter estimates are contained within the joint posterior distribution. The principle to predict a variable of interest (namely, a new observation or a function of the parameters) is to consider all parameter sets from the joint posterior distribution and for each of them to calculate the variable of interest at any time‐point. This provides the posterior distribution of the variable of interest at any time‐point from which a median (50% quantile) and a 95% credible interval (the range from the 2.5% to the 97.5% quantiles) can be extracted. If the variable of interest is the survival rate over time under a given time‐variable exposure profile, then the stochasticity of the survival process will be considered through equations (10) and (11) for the calculation of the survival over time. Consequently, the final output looks like a set of curves (one for each parameter set) from which 50% quantiles are extracted leading to a median curve (orange line in Figure 15), as well as 2.5% and 97.5% quantiles to serve as a 95% credibility band (grey zone in Figure 15).
Figure 15.
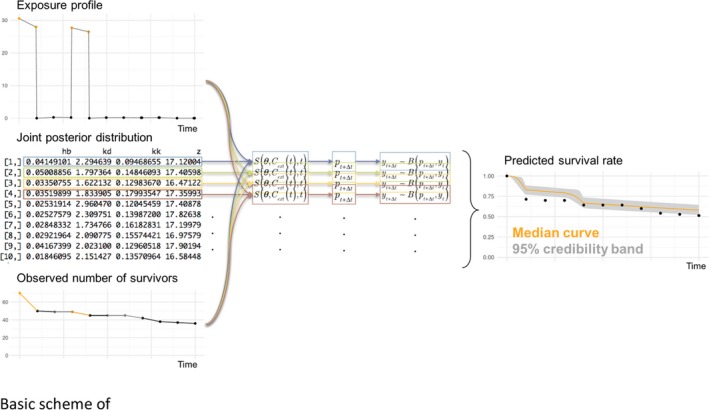
Basic scheme of the Bayesian prediction principle for a given pulsed exposure profile, a joint posterior distribution for the GUTS‐RED‐SD model and a set of observed data
4.1.4.5. Model validation
Validation data are needed to test the GUTS model performance for predictions of mortality/immobility under exposure profiles which have not been used for model calibration. The performance of the model is usually evaluated by comparing relevant model outputs with measurements (often referred to as model validation). For GUTS, relevant outputs are the simulated mortality/immobility probability over time and LPx/EPx values. For the model validation, it appears important to differentiate between invertebrates and vertebrates, because, for invertebrates, prescriptive criteria for validation data sets are requested, while for vertebrates the acceptance of already existing data for validation purpose needs to be critically checked in a case‐by‐case decision, to balance between requirements for GUTS model validation and the reduction of vertebrate testing.
Validation data check
Validation data sets should be evaluated according to the following questions
Has the effect data from experiments under time‐variable exposure, which the model predictions are compared to, been subjected to quality control? Is a description of the data available? (see also list of OECD test guidelines in chapter 7)
-
Are effect data from experiments under time‐variable exposure provided? Have the minimum requirements been matched:
-
–
At least two exposure profiles with at least two pulses each, separated by no‐exposure intervals of different duration length;
-
–
Mortality or immobility is reported at least for seven time‐points11;
-
–
The DRT95 (see below) has been calculated, and the duration of the no‐exposure intervals was defined accordingly; one of the profiles shows a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95; In case DRT95 values are larger than it can be realised in validation experiments, or even exceed the lifetime of the considered species, the second tested exposure profile may be defined independent from the DRT95.
-
–
Exposure specific dose–response curves (see Figure 22 for an example) are at least tested at three concentration levels;
-
–
Figure 22.
Multiplication factor‐response view of the GUTS‐RED‐SD (upper panel) and GUTS‐RED‐IT (lower panel) model validation results based on different pulsed or constant exposure profiles: ‘A’, ‘B’, ‘C’ and ‘Control’ (headers of single plots from left to right). Black dots depict the observed survival rate at day 10, while the solid line corresponds to the median predicted survival rate at day 10. The dashed lines delineate the 95% credibility band representing the uncertainty coming from the uncertainty on parameters estimated obtained from the calibration data set. Corresponding frequentist results are given in Figure B.6 of Appendix B
For invertebrates, this is mandatory. For vertebrates, an expert evaluation needs to identify the suitability of the validation data set on a case‐by‐case basis.
The duration of the no‐exposure time interval should be based on the DRT95, which can be calculated based on the dominant rate constant (kD) when using the GUTS‐RED models:
| (19) |
or the elimination and the repair rate constants (kout and kR, respectively) when using the full GUTS model:
| (20) |
When setting the no‐exposure interval shorter than the DRT95, some toxicological dependence can be expected; when the duration is larger than the DRT95 toxicological independence is more likely. Toxicological dependence or independence is difficult to show ultimately and is hence not recommended as mandatory criterion.
Special attention is suggested, when the compound of concern is suspected of showing increased toxicity over time (e.g. Tennekes and Sanchez‐Bayo, 2013). In such cases, specific emphasis should be put on the duration of exposure in the validation experiments, i.e. the duration of the test should be long enough to include the time‐to‐onset of maximum effects of the relevant pulsed exposure profile. For compounds that show potential latency of effects, the duration of the validation experiments should consider the question whether model calibration has been based on survival data from both acute and chronic exposure tests.
Model performance criteria
The evaluation of the quality of model predictions should be performed considering both qualitative and quantitative criteria. Qualitatively, it can be checked whether the overall response pattern in the data is matched by the model output, e.g. whether the time‐points of increasing effects in model and data correspond with each other and whether the behaviour over time is consistent. The visual match (‘visual fit’ in FOCUS Kinetics, 2006) of the model prediction quality gives a basis for the acceptability of the model predictions in comparison with the data.
Quantitative performance criteria have to be carefully defined, because many criteria are not suitable, either because their values cannot be compared with cut‐off‐criteria (e.g. log‐likelihood values) or they are not suitable for the model and question at hand (e.g. R2, chi‐squared criterion). Three different quantitative criteria are suggested for the model validation which should be considered in combination. They are applicable and can be calculated for both frequentist and Bayesian approaches.
The PPC is suggested as first criterion. The PPC concept comes from the Bayesian approach, and hence can be used naturally here. It is suggested, however, to use the confidence intervals in a similar way the credible intervals are used and to allow also frequentist results to be checked in a PPC‐like way. The PPC compares the predicted median numbers of survivors associated to their uncertainty limits with the observed numbers of survivors. This can be visualised by plotting the predicted versus the observed values and counting how frequently the confidence/credible limits intersect with the 1:1 prediction line (see Figure 23 for an example). Based on experience, PPC resulting in less than 50% of the observations within the uncertainty limits indicate poor model performance.
- The second criterion suggested is also based on the expectation that predicted and observed survival numbers matches the 1:1 line in a scatter plot. The criterion is based on the classical root‐mean‐square error (RMSE), used to aggregate the magnitudes of the errors in predictions for various time‐points into a single measure of predictive power. In order to provide a criterion expressed as a percentage, it is suggested using a normalised RMSE by the mean of the observations:
where is the mean of the n observed numbers of survivors yobs,i for i = 1, …, n. Numbers ypred,i correspond to the median of the predicted numbers of survivors at each time‐point.12 The NRMSE has the advantage to be homogeneous to a coefficient of variation (CV) where the RMSE takes the place of the standard deviation. Clear cut‐off criteria cannot be given within this SO. Nevertheless, based on experience, it is expected that the NRMSE should not exceed the upper limit of 0.5 (50%).(21) - The third criterion is based on the evaluation of the survival probabilities from the beginning to the end of the validation experiment. When the probability to survive from the beginning to the end of an experiment is given as the ratio of the number of surviving to initial individuals ytend/yinit, the difference between the observed and modelled survival probabilities, or in other words the survival probability prediction error (SPPE, Focks et al., 2018) is given as
Hence, the SPPE is suggested as indicative of model accuracy considering survival probabilities only at the end of the tested exposure profile. The SPPE indicator is negative (between 0% and −100%) for an underestimation of effects, and positive (between 0% and 100%) for an overestimation of effects. An SPPE value of 0% means an exact prediction of the observed survival probability at the end of the experiment.(22)
The suggested use of 50% as cut‐off values for the PPC and the NRMSE is based on experience, meaning the evaluation of the few existing studies available in the literature (Nyman et al., 2012; Ashauer et al., 2016; Focks et al., 2018; Jager and Ashauer, 2018). It is not possible to derive precise threshold values from mathematical or theoretical considerations, but as said above, 50% deviation in the NRMSE is according to a CV of 0.5 for the ratio between the RMSE and the mean observed survival. This CV value appears not too high, when considering that a factor of two is deemed to be acceptable for the variation of experimentally derived endpoints. Additional confidence for the suggestion of the 50% is given based on: (a) the 50% can also be on the conservative side of the risk estimate (SPPE value indicates this) and (b) it is recommended to always evaluate both the SD and IT model and to use the more conservative, if positively validated.
Finally, the suggested acceptability criteria have to be checked based on future applications of the GUTS models and possibly be adapted over time. Only when starting the use of GUTS in regulatory risk assessment, the experience basis for the formulation of validation criteria and related cut‐off values will be possible.
4.2. Documentation of GUTS model calibration, validation and application for risk assessment
This section provides an example for a possible application of the GUTS framework with respect to model calibration, validation and calculation of endpoints for risk assessment. The example is about survival of one of the most sensitive aquatic macroinvertebrate species for azole fungicides, Gammarus pulex, with special attention to the impact of time‐variable instead of constant exposure to propiconazole. In a practical application for regulatory risk assessment, the selection of the appropriate model and species would need more consideration than in the present case (see Chapter 8). As for this application the focus is more on technical aspects and documentation, the example with G. pulex is considered relevant; the endpoint of interest is survival.
The data set selected for this section is one of the three data sets, data set B1, from the GUTS ring‐test (Jager and Ashauer, 2018). Hence, the documentation of the model calibration in this section gives an example of the documentation according to the ring‐test performance that is requested for new GUTS implementations. The ring‐test documentation contains the other results for data sets A, B2 and C both for the Mathematica and the R implementations (Appendices B.6 and B.7).
4.2.1. Example data sets
4.2.1.1. Data sets used for calibration
The test data set consists of the raw data from a four‐day acute toxicity study taken from the supporting information of Nyman et al. (2012). Survival of the crustacean G. pulex under exposure to the azole fungicide propiconazole was observed. Comprehensive documentation can be found in the publication and the supporting information. The data set appears of a very good quality, with quality checks and measurement methods being accurately reported. The acute test consisted of seven pesticide concentrations between 8.2 and 37.4 nmol/mL (see Table S1–14 in Supporting Information), with two replicate beakers each, each beaker containing ten G. pulex initially. Propiconazole concentrations in water were measured and the survival of G. pulex was analysed by prodding and visual observation of daily movements for 4 days. Responses in survival for the tested concentrations ranged from 0% to 100% effects (i.e. no survival) after 4 days, so the dose–response relationship is fully covered by the set of calibration data. Raw data are provided in Appendix D Table D.1. The relatively small number of only two replicates is counterbalanced by testing seven treatment levels. The data set was used in a ring test for GUTS models and is considered realistic and sufficient for the parameterisation of GUTS modelling by more than 10 scientists from different backgrounds.
Table D.1.
Survival of G. pulex in 4‐day acute toxicity test with propiconazole and measured concentrations (from Nyman et al., 2012, supporting material)
| Day | A | B | C | D | E | F | G | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 11 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| 1 | 1 | 10 | 4 | 7 | 8 | 9 | 10 | 11 | 10 | 9 | 9 | 10 | 10 | 10 | 9 | 10 |
| 2 | 0 | 1 | 0 | 4 | 3 | 3 | 9 | 11 | 10 | 9 | 9 | 10 | 10 | 10 | 9 | 10 |
| 3 | 0 | 0 | 0 | 0 | 2 | 0 | 6 | 10 | 10 | 8 | 9 | 10 | 10 | 10 | 9 | 10 |
| 4 | 0 | 0 | 0 | 0 | 1 | 0 | 6 | 10 | 8 | 8 | 8 | 9 | 10 | 9 | 9 | 10 |
| Conc * | 35.92 | 28.93 | 24.19 | 17.87 | 13.8 | 11.91 | 8.05 | 0.00 | ||||||||
* Average measured concentrations in [μmol/L].
4.2.1.2. Data sets used for validation
The validation data taken as an example comes from the same study as the calibration data (Nyman et al., 2012), but for several variable exposure profiles in a non‐standard pulsed toxicity experiment. The pulsed toxicity experiment lasted 10 days and consisted of three exposure profiles plus a control. Each of the exposure profiles had seven replicate beakers, including one non‐solvent and one solvent control beaker. All beakers initially contained 10 G. pulex. Under exposure profiles ‘A’ and ‘B’, organisms were exposed to two 1‐day pulses (at a concentration around the LC30, 28 μmol/L). Between pulses, the organisms had a 2‐day (‘A’) or a 6‐day (‘B’) period of recovery in uncontaminated artificial pond water. In the third exposure profile (‘C’), the organisms were constantly exposed to a concentration about equal to the time‐weighted average (TWA) concentration from the pulsed exposure profiles (~ 4.6 μmol/L1). The propiconazole concentration in water was measured and the survival was observed on a daily basis. Raw data are provided in Appendix D Tables D.2 and D.3. The data set was not produced specifically for this SO, but in the scope of an earlier PhD thesis. The failure of full compliance with the minimum requirements as formulated in this SO (Section 4.1.4.5) does not prohibit its use as a demonstration data set, but the use of the data helped to derive the minimum requirements instead.
Table D.2.
Number of living G. pulex in pulsed toxicity experiments with propiconazole (from Nyman et al., 2012, supporting material)
| Day | Treatment A | Treatment B | Treatment C | Controls | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| 1 | 4 | 8 | 7 | 8 | 6 | 9 | 8 | 8 | 7 | 8 | 7 | 9 | 8 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 9 | 10 | 10 | 10 | 10 |
| 2 | 4 | 8 | 7 | 8 | 5 | 9 | 8 | 7 | 6 | 8 | 7 | 8 | 7 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 9 | 9 | 9 | 10 | 10 | 10 | 10 |
| 3 | 4 | 8 | 7 | 8 | 5 | 9 | 8 | 6 | 6 | 8 | 7 | 8 | 7 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 9 | 9 | 9 | 10 | 10 | 10 | 10 |
| 4 | 4 | 7 | 6 | 7 | 4 | 9 | 8 | 6 | 6 | 8 | 6 | 8 | 7 | 9 | 9 | 10 | 10 | 10 | 10 | 10 | 9 | 8 | 9 | 10 | 10 | 10 | 10 |
| 5 | 4 | 7 | 6 | 7 | 4 | 9 | 8 | 4 | 6 | 7 | 5 | 8 | 7 | 9 | 9 | 10 | 10 | 9 | 9 | 10 | 9 | 8 | 9 | 10 | 10 | 10 | 10 |
| 6 | 4 | 7 | 6 | 7 | 4 | 9 | 8 | 4 | 6 | 7 | 5 | 8 | 7 | 8 | 9 | 10 | 10 | 9 | 9 | 10 | 8 | 8 | 9 | 10 | 9 | 10 | 10 |
| 7 | 3 | 7 | 6 | 7 | 4 | 9 | 6 | 3 | 6 | 7 | 5 | 8 | 7 | 8 | 9 | 10 | 10 | 9 | 9 | 9 | 8 | 8 | 9 | 10 | 9 | 10 | 10 |
| 8 | 3 | 7 | 6 | 5 | 3 | 8 | 6 | 2 | 5 | 7 | 3 | 8 | 7 | 8 | 6 | 9 | 10 | 9 | 9 | 9 | 8 | 8 | 9 | 10 | 9 | 10 | 10 |
| 9 | 3 | 6 | 6 | 5 | 3 | 8 | 6 | 2 | 5 | 7 | 3 | 7 | 7 | 7 | 5 | 8 | 10 | 8 | 9 | 8 | 7 | 7 | 9 | 10 | 9 | 10 | 10 |
| 10 | 3 | 5 | 6 | 5 | 3 | 8 | 6 | 2 | 5 | 7 | 3 | 7 | 6 | 7 | 5 | 8 | 9 | 8 | 9 | 8 | 7 | 7 | 9 | 10 | 8 | 10 | 10 |
Table D.3.
Measured concentrations in the pulsed ‘validation’ experiment for G. pulex with propiconazole (from Nyman et al., 2012, supporting material)
| Time | A | B | C |
|---|---|---|---|
| [day] | [.mol L−1] | [.mol L−1] | [.mol L−1] |
| 0 | 30.56 | 28.98 | 4.93 |
| 0.96 | 27.93 | 27.66 | 4.69 |
| 1 | 0 | 0 | 4.69 |
| 1.96 | 0.26 | 0.27 | 4.58 |
| 2.96 | 0.21 | 0.26 | 4.58 |
| 3 | 27.69 | 0.26 | 4.58 |
| 3.96 | 26.49 | 0.26 | 4.54 |
| 4 | 0 | 0.26 | 4.54 |
| 4.96 | 0.18 | 0.25 | 4.58 |
| 4.97 | 0.18 | 0.25 | 4.71 |
| 5.96 | 0.18 | 0.03 | 4.71 |
| 6.96 | 0.14 | 0 | 4.6 |
| 7 | 0.14 | 26.98 | 4.6 |
| 7.96 | 0.18 | 26.28 | 4.59 |
| 8 | 0.18 | 0 | 4.59 |
| 9 | 0 | 0.12 | 4.46 |
| 9.96 | 0 | 0.12 | 4.51 |
4.2.2. Modelling results
Two model implementations were run in parallel: one under a frequentist framework with the Mathematica software (Section 4.1.2.1); another one under a Bayesian framework with the R software (Section 4.1.2.2). More implementation details are given in Appendix A Testing the implementations in Section 4.1.1.2 already indicated that both implementations give very similar outputs. The results obtained under the Bayesian framework are presented below; additional plots and results from the frequentist approach implemented in Mathematica are given in Appendix B for comparison.
4.2.2.1. Results of model calibration
The model parameters have been calibrated in a Bayesian framework as implemented either in the R‐package ‘morse’ (Baudrot et al., 2018a) or directly online within the ‘GUTS’ module of the MOSAIC web‐platform (Baudrot et al., 2018c). All details about the underlying implementation, verifications and sensitivity are given in Section 4.1.2, while details about the parameter estimation process are given in Section 4.1.3. Additional details about the parameter estimation are given in Appendix A.1 (Mathematica) and A.2 (R‐package ‘morse’).
The estimated parameter values and corresponding credible intervals are reported together with the optimal log‐likelihood values in Table 3. The background mortality rate was considered in the estimation process together with the other parameters. It was not separately calculated from the controls, because also low treatment levels may contain information about background mortality, and additionally because only in this way it is possible to check for potential correlations between all parameters of the GUTS‐RED models.
Table 3.
Parameter estimates of the GUTS‐RED‐SD and the GUTS‐RED‐IT models, expressed as medians and 95% credible intervals (range between the 2.5% and the 97.5% quantiles of the marginal posterior distributions). Respective parameter estimates in the frequentist framework are given in Table B.1 of Appendix B. Log‐likelihood values in the table are the median values of the posterior distribution of the log‐likelihood, and not the log‐likelihood calculated from the median of the parameter estimates
| GUTS‐RED‐SD parameters | Symbol | Median | 2.5% quantile | 97.5% quantile | Unit |
|---|---|---|---|---|---|
| Dominant rate constant | kD | 2.179 | 1.591 | 3.467 | day−1 |
| Background mortality | hb | 0.02683 | 0.01255 | 0.04902 | day−1 |
| Concentration threshold | zi | 16.89 | 15.41 | 18.69 | μmol/L |
| Killing rate | bi | 0.1237 | 0.0791 | 0.1889 | μmol/L per day |
| Log‐likelihood value | −125.7 | ||||
| GUTS‐RED‐IT parameters | Symbol | Median | 2.5% quantile | 97.5% quantile | |
| Dominant rate constant | kD | 0.7194 | 0.5333 | 0.9359 | day−1 |
| Background mortality | hb | 0.01579 | 0.00378 | 0.03746 | day−1 |
| Median of the threshold distribution | mi | 17.69 | 15.12 | 20.21 | μmol/L |
| Width of the threshold distribution | β | 6.700 | 4.866 | 9.065 | – |
| Log‐likelihood value | −129.6 | ||||
The dominant rate constant for both models appear different (kD = 2.207 day−1 [1.602; 3.547] for model GUTS‐RED‐SD; kD = 0.7346 day−1 [0.5385; 0.9568] for GUTS‐RED‐IT), while the internal concentration threshold in the GUTS‐RED‐SD model is very similar to the median of thresholds in the GUTS‐RED‐IT model (zw = 17.066 μmol/L [15.59; 18.87] with GUTS‐RED‐SD; mw = 17.97 μmol/L [15.34; 20.49] with GUTS‐RED‐IT). The consequence is that the time course of the internal concentration within the organisms is different between the two models, while the median thresholds at which effects occur is very close. In addition, the large value of the width of the distribution in model GUTS‐RED‐IT (β = 6.845 [5.0119; 9.295]) indicates a steep concentration–response relationship, since the concentration–response relation is directly related to the cumulated log‐logistic distribution (see Appendix C).
Figure 16 shows the survival over time (expressed as percentage of initial number of living individuals) for the calibration data set and corresponding fits as median curves and the 95% credible band. Both SD and IT models show visually a good fit to the observed survival rate, meaning that the observed survival rate as indicated by the black dots match well with the model predictions. Additional graphical results are provided in Appendix B, Figures B.1, B.2 and B.3).
Figure 16.
Survival over time (SOT) view of model calibration based on data from a typical acute toxicity study, with observations of survival under constant exposure over 4 days: (upper panel) model GUTS‐RED‐SD; (lower panel) model GUTS‐RED‐IT. The survival over time is represented as a function of time for each tested concentration (headers of single plots): black dots are the observations (observed numbers of survivors divided by the initial number of individuals), black segments show the between‐replicate variability, while the orange solid line corresponds to the median curve. The grey band is the 95% credibility band representing the uncertainty. Survival is shown as rate, meaning that stochastic influence on the number of survivors over time is not considered. Respective results including this stochasticity are shown in Appendix B.1
Figure B.2.
Comparison of priors and posteriors of the GUTS‐RED‐SD (upper panel) and GUTS‐RED‐IT (lower panel) model parameters (log10‐scale): the dotted distribution corresponds to the prior and the plain one to the posterior probability distribution of each parameter (x‐labels under single plots)
Figure B.3.
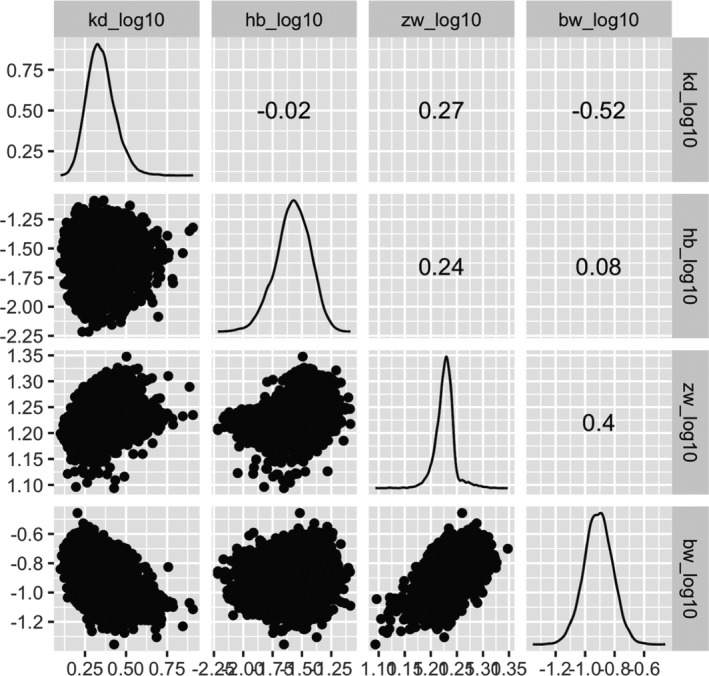
Correlation plot of the GUTS‐RED‐SD model parameters (log10‐scale)
Figure 17 illustrates the goodness‐of‐fit also by plotting the PPC, that is the predicted median numbers of survivors (y‐coordinates of black dots) as a function of the observed numbers of survivors (x‐coordinates of black dots). A good fit will result in black dots lined up along the line y = x. The estimation process also provides the uncertainty around the predicted median values as an interval (vertical segments), within which 95% of the observations are expected to lie. Visual inspection of Figure 17 does not yield any systematic bias in the deviations between modelled and observed data. The PPC results in a value 100% of the data are within the uncertainty ranges of the predictions.
Another way of looking at the calibration results is to compile a concentration–response curve at a given time‐point based on the GUTS modelling which also accounts for uncertainties (Figure 18). Under this representation, calibration results can be assessed in the same way as with a classical concentration–response relationship (e.g. a log‐logistic concentration–response model). Figure 18 shows that both GUTS‐RED‐SD and GUTS‐RED‐IT models well fit the steep slope of the observed response, meaning that the onset of effects at 17.87 μmol/L and the steep slope of the concentration–response that leads to nearly full effects already at the next tested concentration of 24.19 μmol/L are matched. See Appendix B and Figure B.5 for the same results obtained under the Mathematica implementation.
Figure 18.
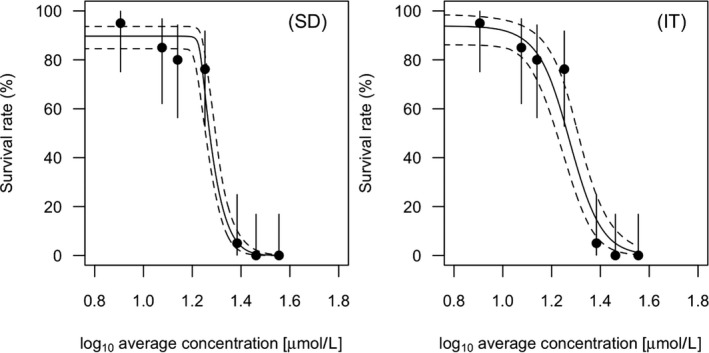
Concentration–response view of the GUTS‐RED‐SD (left panel) and GUTS‐RED‐IT (right panel) model calibration results obtained from data of a typical acute toxicity study with observations of survival under constant exposure over 4 days. Shown are the observed (black dots) and the modelled survival rates at the end of the 4‐day observation period. Please note that the model parameters have been calibrated on the survival probabilities over time, and not on the concentration–response data. The solid line corresponds to the median curve, while the dashed lines delimit the 95% credibility band representing the uncertainty. An analogue plot resulting from GUTS modelling under a frequentist framework is given in Figure B.5 of Appendix B
An additional view of the GUTS modelling results is given in Figure 19, where LC50 estimates both from classical concentration–response curve fitting at a given target time (among those of the experimental design) and from the GUTS‐RED‐SD and GUTS‐RED‐IT models are shown as function of the observation period. Such graphs illustrate the added value of GUTS models which allow estimating any LCX value at any time period (e.g. between 1 and 4 days as shown in Figure 19). There is a good accordance between LC50 estimates from a classical concentration–response curve fitting at the given target times, and those obtained from the GUTS‐RED modelling. Uncertainties appear in tendency smaller when using the GUTS instead of classical dose–response modelling, presumably because all data over time are accounted for in the GUTS estimation process.
Figure 19.
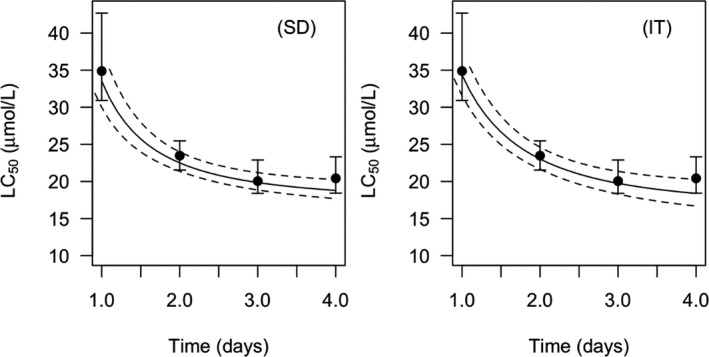
LC 50 estimates from the calibrated GUTS models (GUTS‐RED‐SD on the left panel, GUTS‐RED‐IT on the right panel) as represented by the solid lines (median curve) and the dashed lines (95% credibility band for the uncertainty), or from a classical target time analysis as represented by black dots (median estimate) and segments (95% credible intervals)
Summarising the model calibration for the example of propiconazole and G. pulex, the data appear appropriate because the concentration–response relation ranges from no up to full effects. The choice of the reduced GUTS models is appropriate because internal concentrations have not been measured so that the use of the full model cannot be recommended. The parameters in this example have been estimated under a Bayesian framework using the MCMC algorithm as implemented either in the R‐package ‘morse’ or within the web‐platform MOSAIC. Parameter values, credible limits and likelihood values have been reported (Table 3). The results of the model calibration are visualised and tested in different ways, including plotting the survival over time, the PPC and concentration–response curves.
4.2.2.2. Results of model validation
Parameter estimates from the calibrated model were used to predict expected survival of G. pulex under time‐variable exposure profiles of propiconazole (exposure profiles ‘A’, ‘B’, ‘C’ and ‘Control’ as shown in Figure 20; concentration values are available in Table D.3 of Appendix D). The validation data set has been qualitatively checked, quality control aspects are reported in the original publication (Nyman et al., 2012). The design of the experiments fulfils some basic criteria being requested for appropriate validation data sets: the observed survival was obtained from experiments under time‐variable exposure. Two time‐variable exposure profiles with two pulses each have been tested, which were separated by no‐exposure intervals of different duration lengths. The length of the no‐exposure interval was defined based on the depuration time and aimed at one exposure profile allowing for individual depuration and repair (‘B’), the other one (‘A’) not so much, despite the 95% depuration time was determined to about 10 h. Two different time‐variable exposure profiles haven been tested for one pulse height only, not at three concentration levels. Despite this, data set does not match all of the minimum requirements for validation data, these already published results, which match the calibration data in terms of substance and species but tested under time‐variable exposure, are used.
Figure 20.
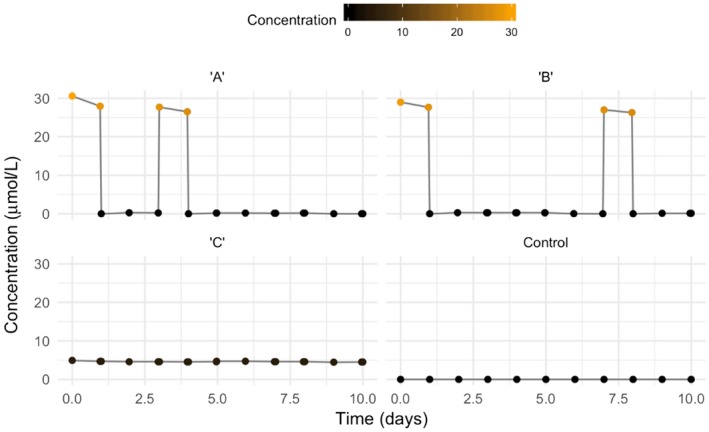
Exposure profiles of the validation data set (Nyman et al., 2012, supporting material). ‘A’: two 1‐day pulses at concentration (~ LC 30 = 28 μmol/L) and 2‐days of recovery between pulses; ‘B’: two 1‐day pulses at concentration (~ LC 30 = 28 μmol/L) and 6‐days of recovery between pulses; ‘C’: constant exposure equal to the time‐weighted average (TWA) concentration from the pulsed exposure profiles (~ 4.6 μmol/L)
Visual match
Predicted survival rates are compared with observed survival rates over time under each exposure profile (Figure 21) or at the end of the exposure period as in a classical concentration–response view (Figure 22). From Figure 21, it appears that the GUTS‐RED‐SD model is more conservative by underestimating the observed time course of the survival rate. Both models proved to be sensitive enough to detect the tendency of the data under both exposure profiles ‘A’ and ‘B’ by predicting an initial decline during the first pulse followed by a phase of reduced mortality, most probably due to background mortality, to continue with another decline after the second pulse (at day 3 in ‘A’ or day 7 in ‘B’) for the GUTS‐RED‐SD model, despite GUTS‐RED‐IT did not react on the second pulse of profile ‘B’. The experiments with exposure profile ‘C’ were performed under constant exposure. They resulted in less mortality than the pulsed profiles with a survival rate between 65% and 95% at day 10. See Figure B.6 of Appendix B for the same results obtained under the frequentist implementation. From observed survival rates, the renewal of the untreated medium in profiles ‘A’ at day 7 and ‘B’ at day 3 seemed to have affected survival. Apparently, the medium renewal caused stress to the individuals and hence led to increased mortality which could not be captured by model predictions.
Figure 21.
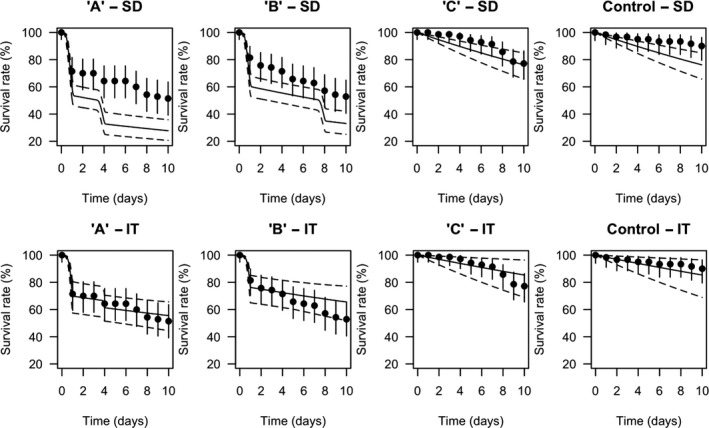
GUTS‐RED‐SD (upper panel) and GUTS‐RED‐IT (lower panel) model validation results on a typical pulsed experiment data set: the survival rate over time is represented as a function of time for each exposure profile (headers of single plots): black dots are the observed survival rates, while the solid line corresponds to the median curve. The dashed lines give the 95% credibility band representing the uncertainty coming from the uncertainty on parameters estimated obtained from the calibration data set
Under the ‘Control’ exposure profile (Figure 21), both models predict a slight decrease of the survival rate over time. The difference between both models comes from the difference in their background mortality estimates: hB = 0.02683 [0.01253; 0.04902] for GUTS‐RED‐SD, which is slightly lower than hB = 0.01570 [0.00378; 0.03749] for GUTS‐RED‐IT.
The procedure used to calculate the dose–response view as shown in Figure 22 demonstrates the potential of the GUTS modelling framework. Survival rate can be predicted for whatever multiplication factor is applied to a specific exposure profile. In this way exposure‐specific multiplication factor‐response curves can be computed, in analogy to the classical concentration–response curves used to evaluate toxicity experiments under constant conditions. For the observed survival at day 10, there is a very good correspondence between the observed and the predicted survival rate for both GUTS‐RED‐SD and GUTS‐RED‐IT models: the observed value is always within or very close to the uncertainty bands. Figure B.6 Appendix B shows the same results obtained under the frequentist implementation.
Quantitative model performance criteria
Quantitative model performance criteria were calculated, and the posterior predictive checks have been visualised (Figure 23). The PPC value for the GUTS‐RED‐SD model is 41%, the corresponding PPC value for the GUTS‐RED‐IT model is 86%, indicating that the IT model predictions match closer with the observed numbers of survivors. The PPC value for the GUTS‐RED‐IT model is in an acceptable range, while the PPC value for the GUTS‐RED‐SD model is rather low. Values for the NRMSE calculated for the GUTS‐RED‐SD (22.6%) and the GUTS‐RED‐IT (10.6%) are in very good range for a prediction error, indicating an acceptable quality of the predictions over time, both for the GUTS‐RED‐SD as well as the GUTS‐RED‐IT models.
The SPPE values measure the deviation between observed and predicted survival probabilities at the end of the tested profiles. They are calculated per exposure profile, and for the GUTS‐RED‐SD model the SPPE for scenarios ‘A’ and ‘B’ are 17.2% and 14.3%, respectively, while for the GUTS‐RED‐IT model the SPPE values for scenarios ‘A’ and ‘B’ are −7.2% and −14.3%. The positive values for the GUTS‐RED‐SD model indicate that the model predictions are overestimating mortality, while for the GUTS‐RED‐IT model the negative values indicate that mortality is underestimated in the predictions. All SPPE values are within +/− 20% of the observed survival at the end of the tested exposure profiles.
Discussion of the model validation
The three quantitative validation criteria (definition in Section 4.1.4.5) which have been calculated for the GUTS models have different emphasis. The PPC takes into account the uncertainty in the model predictions, while the NRMSE value considers the relation between median predicted and observed numbers of survivors over time, and the SPPE considers the median observed survivor number at the end of the experiment.
Overall, the quality of the model predictions appears as acceptable in this case. Quantitatively, the GUTS‐RED‐IT model gives a better match with this example validation data set, since both NRMSE as well as SPPE values are smaller. Nevertheless, qualitatively, the GUTS‐RED‐SD model would be more preferable for risk assessment, because the GUTS‐RED‐SD model predictions show values below the 1:1 line in Figure 23, hence indicating an over‐estimation of mortality and a more conservative risk assessment.
In general, acceptance of a maximum level of 50% deviation between predicted and observed numbers is suggested. This suggestion is based on the consideration that toxicological effects change on a logarithmic scale rather than on a nominal one (see also Figure 23). In addition, acceptance of 50% deviation appears protective when using GUTS predictions in combination with lower‐tier assessment factors.
The suggested use of 50% as cut‐off values for the PPC and the NRMSE is based on experience, meaning the evaluation of the few existing studies available in the literature (Nyman et al., 2012; Ashauer et al., 2016; Jager and Ashauer, 2018; Focks et al., 2018). It is not possible to derive precise threshold values from mathematical or theoretical considerations, but as said above, 50% deviation in the NRMSE is according to a CV of 0.5 for the ratio between the RMSE and the mean observed survival. This CV value appears not too high, when considering that a factor of two is deemed to be acceptable for experimentally derived endpoints. Additional confidence for the suggestion of the 50% is given by the fact that (a) the 50% can also be on the conservative side of the risk estimate (which is seen in the SPPE indicator) and (b) it is recommended to always evaluate both the SD and IT model and to use the more conservative if positively validated. Finally, suggested acceptability criteria have to be checked based on future applications of the GUTS models and possibly be adapted over time. Clearer validation criteria and related cut‐off values can be better formulated only when experience is gained with the use of GUTS in regulatory risk assessment.
Time course of scaled internal damage
One useful application of the GUTS modelling framework is to check how the simulation of the scaled damage in the GUTS‐RED models relate to the dependency of the pulses in both exposure profiles ‘A’ and ‘B’. As shown in Figure 24, the pulses in exposure profile ‘B’ appear toxicologically independent for both models, because the second peak comes only clearly later than the upper confidence limit of the DRT95 (see Section 4.1.4.5). For the exposure profile ‘A’, the GUTS‐RED‐IT model indicates toxicological dependence, since the second pulse comes later than the upper confidence limit of the DRT95, whereas the GUTS‐RED‐SD model is borderline between toxicologically dependence and independence.
Figure 24.
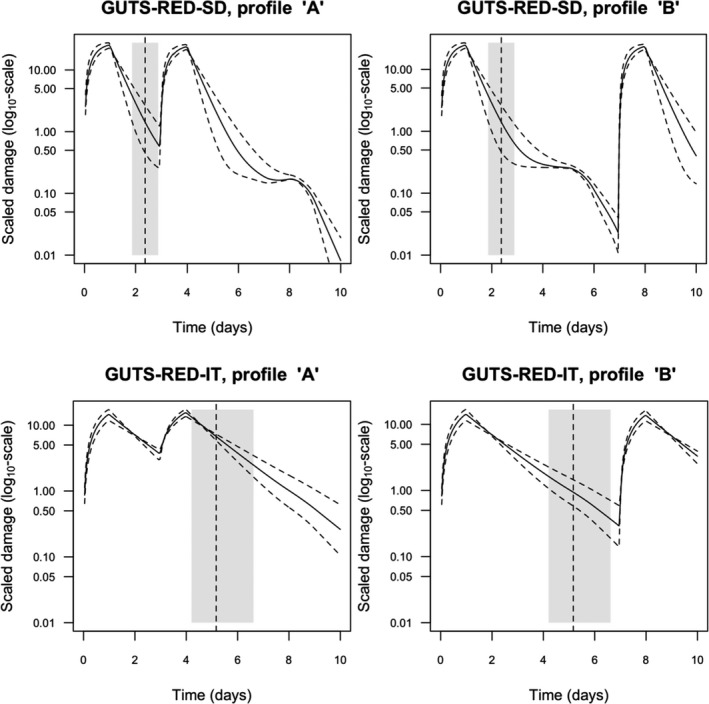
Scaled damage over time for GUTS‐RED‐SD (top panel) and GUTS‐RED‐IT (bottom panel) models under time‐variable exposure profiles ‘A’ (left panels) or ‘B’ (right panels). The vertical dashed lines correspond to the median DRT 95 (see Section 4.1.4.5), calculated based on optimal parameter values for the dominant rate constant (kD) for the GUTS‐RED‐SD and the GUTS‐RED‐IT models and their lower and upper confidence limits (grey band). The median scaled damage is simulated (solid curve) together with its uncertainty (dashed curves) with the same equation in both GUTS models. Nevertheless, the median value of the dominant rate constant differs: kD = 2.154 μmol/L per day for model GUTS‐RED‐SD; kD = 0.732 mol/L per day for model GUTS‐RED‐IT
Such simulations of the scaled damage for different exposure profiles can be of great help in designing pulsed refined experiments, in order to check if the intended pulses, in terms of height and between‐pulse period, will be toxicologically independent or not.
4.2.3. Predictions under FOCUS surface water exposure patterns
4.2.3.1. Chemical exposure data
Chemical exposure data can function as input data for GUTS model predictions. The input can consist of concentration time series as produced by the FOCUS surface water software or other exposure assessment tools.
Predicted exposure profiles in water were produced for 10 scenarios for propiconazole (Figure 25; more details on the FOCUS scenarios are given in Table 4). These exposure time series were used as input data for the GUTS‐SD and GUTS‐IT models, without assuming background mortality for any of the input scenarios.
Figure 25.
Concentration over time for 10 FOCUS exposure scenarios, ID numbers are given in the plot titles. More details on the FOCUS scenarios in Table 4
Table 4.
Analysis of risk estimation using a TER approach and based on GUTS modelling in form of LP50 values (see text for more details). TER based on an LC50 values for G. pulex of 19.2 μg/L
| Scenario | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Application | Apple | Apple | Cereals | Cereals | Cereals | Cereals | Cereals | Cereals | Cereals | Cereals |
| FOCUS SW Scenario | R1 pond | R2 stream | D1 ditch | D1 stream | D3 ditch | D4 pond | D4 stream | D5 pond | D5 stream | R4 stream |
| PECmax | 1.130 | 2.007 | 10.564 | 8.063 | 9.083 | 1.668 | 2.268 | 1.670 | 2.401 | 2.998 |
| TER | 17 | 10 | 2 | 2 | 2 | 12 | 8 | 11 | 8 | 6 |
| LP50 SD model | 17 | 44 | 3 | 20 | 12 | 12 | 205 | 12 | 195 | 39 |
| LP50 IT model | 17 | 49 | 3 | 24 | 16 | 12 | 250 | 12 | 237 | 39 |
4.2.3.2. Calculation of exposure profile specific concentration–response curves (LPX values)
The FOCUS exposure profiles were used as input data for the calibrated and validated GUTS‐RED‐SD and GUTS‐RED‐IT models. Exposure‐profile specific LP50 values were calculated by applying increasing multiplication factors to the profiles until 50% mortality at the end of the simulated exposure pattern was reached (Table 4). LP50 are compared with assessment factors as used for lower‐tier RAC values. It can nicely be seen from the table entries that, for exposure profiles where the concentration remains nearly constant (e.g. Figure 25, profiles 1, 3, 6 and 8), the LP50 is identical to the TER.
Nevertheless, in cases where the exposure profile is highly time‐variable (i.e. with some peaks and many phases with lower concentrations in between, e.g. Figure 25, profiles 2, 4, 7, 9 and 10), the LP50 values are much higher than the TER. This indicates that, for some tested exposure profiles, the risk is much lower than apparent from Tier 1 calculations. It also shows that the GUTS modelling framework can differentiate between different qualities of exposure profiles, unlike the TER calculations. The evaluation of FOCUS surface water exposure profiles by GUTS modelling confirms that the GUTS modelling of survival functions exactly as intended: Lower tier risk estimates are kept for more constant profiles, whereas the risk for time‐variable profiles is refined.
4.3. Overall summary and conclusions of the GUTS model documentation and application
The formal definition for GUTS models is standardised and documented. Examples show how the verification of any new GUTS implementation can be performed using some standard elements: default scenarios, pulsed exposure scenarios, and extreme scenarios can be simulated and checked. Sensitivity analyses of the reduced GUTS models (GUTS‐RED) have been performed and are not requested for new model implementations and applications, because the influence of the model parameters on the model outcome is known. However, results of sensitivity analyses for new applications of GUTS‐RED can be used to document a correct model implementation, and interactions between model parameters could be further investigated. For other GUTS models than the reduced, sensitivity analyses should be included and checked for future applications.
Parameter estimation is a challenging task, but standard implementations of parameter optimisation algorithms are available in software packages, both under a frequentist and a Bayesian approach. It is crucial to report not only optimal parameters, but also the optimisation method, settings of the optimisation routine and of the numerical solver, and parameter confidence limits including information on how they were derived to allow for potential expert evaluation. Results from model calibration need to be documented comprehensively. For this purpose, a corresponding checklist is provided in this Scientific Opinion. Relevant model output for risk assessment has been defined as expected mortality/immobility or exposure profile specific multiplication factors (LPx/EPx). The latter factors can be directly compared with respective assessment factors. Both mortality estimates and LPx/EPx can be calculated including confidence limits, which are calculated by propagation of model uncertainty to the model output.
For the validation of calibrated GUTS models, appropriate validation data sets are essential. Differentiating between vertebrates and invertebrates, a set of requirements is given for validation experiments, among others to test at least two different time‐variable exposure profiles. The evaluation of the quality of predictions in comparison with the observed data requires a careful combination of qualitative and quantitative criteria. Qualitatively, the visual match (‘visual fit’ in FOCUS Kinetics, 2006) between model and data is important to be checked, and quantitatively three criteria are suggested: one that takes into account the uncertainty in the model predictions (PPC), one that measures the match over time (NRMSE), and one that considers the final match between model and data (SPPE).
GUTS calibration and validation is shown for an example data set, both for a frequentist approach implemented in Mathematica and a Bayesian approach implemented in R. The results illustrate how calibration and validation could be performed and documented to allow regulators to evaluate future applications, but ideally the validation data would fulfil all minimum criteria. The application of the GUTS model to calculate refined risk estimates for concentration time series from 10 FOCUS surface water scenarios delivers very reasonable results, the GUTS modelling allows obviously to differentiate between constant exposure situations, where the refined risk is practically identical to the lower‐tier estimate, and highly variable exposure over time, where the refined risk is lower as in the lower‐tier.
One open issue for the use of GUTS in regulatory risk assessment is a question related to the constrained possibility to request new validation experiments for vertebrates. It is necessary to build up an experience base to determine which data sets are suitable for GUTS validation for vertebrates.
Another issue is related to compounds that are suspected of showing increased toxicity over time, e.g. neonicotinoids. Recent research shows that the quality of model predictions for these compounds was in some cases only acceptable when the GUTS models were calibrated based on chronic test results (Focks et al., 2018). Further classification of compounds with respect to the potential to show increased toxicity under long‐term exposure would be relevant.
Technically, one of the main questions is whether it is better to allow for user‐defined implementations of GUTS, or to aim for having one standard GUTS implementation, which only needs to be checked once. In this opinion, the decision was to consider user‐defined implementations and to give checklists and criteria at hands to allow regulators to evaluate whether a new implementation fulfils the required quality aspects.
Duality of SD and IT models and consequences for practical use in RA
A specific aspect of GUTS modelling is the duality of the SD and IT death mechanisms. As mentioned already, in theory, these two model variants are extreme cases of one overarching model and can be unified in the combined GUTS, but in practice, most often the two types of reduced GUTS models will be used. Requirements for the data for model calibration are the same, so usually both reduced models can be parameterised. In consequence, decisions are necessary about (a) which of the models is acceptable in the validation, and (b) which of them is used in the regulatory risk assessment.
The application for propiconazole and G. pulex shows a nice example how to handle this duality. Both the GUTS‐RED‐SD and the GUTS‐RED‐IT models fulfil validation criteria. Quantitatively, the GUTS‐RED‐IT model gives a better match with this example validation data set, but qualitatively, the GUTS‐RED‐SD model would be more preferable for risk assessment, because the GUTS‐RED‐SD model predictions lead in tendency to an over‐estimation of mortality and so to a more conservative risk assessment. It is certainly not advisable to always take the more conservative of the IT and SD models, i.e. when one of the two is clearly wrong, either by under‐ or over‐predicting mortality in the validation experiments, and fulfilment of qualitative and quantitative validation criteria is clearly failed, it should not be used for regulatory risk assessment. When, however, both models appear acceptable in the validation, the more conservative is suggested for regulatory use. The same rationale can be applied for the choice of the death mechanism for coupling of GUTS models with individual‐based models in RA.
5. DEBtox models
5.1. Documentation and implementation of the formal model
As already stated in Chapter 2, DEBtox models are rather complex mainly because they offer several ways of accounting for toxicant effect simultaneously on both growth and reproduction processes. Unlike for the GUTS models, this section cannot be comprehensive, because DEBtox model applications are still developed on a more case‐by‐case basis. However, to exemplify some aspects of the formulation and the implementation of a DEBtox model, published results by Billoir et al. (2011) are used to illustrate a case study about lethal and sublethal effects on daphnids (D. magna) exposed to time‐varying cadmium exposure concentrations within a laboratory aquatic microcosm. It should be highlighted that the example included in this chapter illustrates the model formulation, implementation, and the results of the calibration phase. A proper validation with an additional data set was not performed, and therefore could not be included here.
5.1.1. Model formulation
The dynamics of the scaled damage (called ‘scaled internal concentration’ in Billoir et al., 2011) within daphnids is derived from the total cadmium concentrations in water through a toxicokinetic model. Effects of cadmium on survival, growth and reproduction are then assumed to depend on this scaled damage. In the following, letter t refers to a general expression of time (in days), while t i refers to the experimental time‐point number i which can differ per endpoint (see Figure 27).
Figure 27.
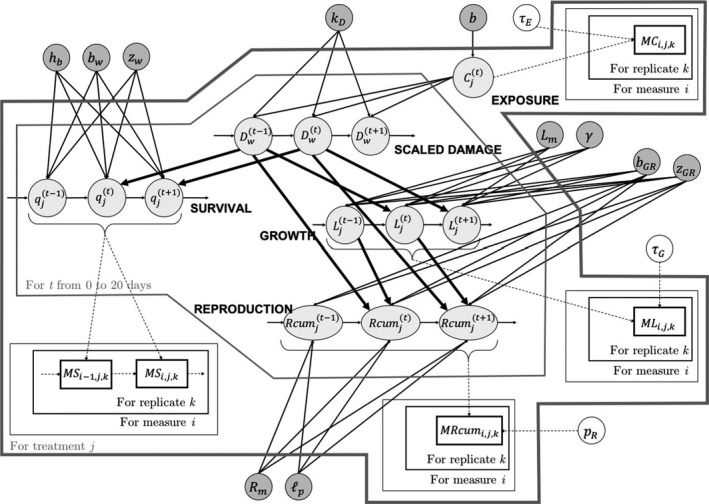
Directed acyclic graph of the DEBtox model used to describe time‐variable cadmium effect on survival, growth and reproduction of D. magna. This graph is adapted from by Billoir et al. (2011). Variable q stands for the quantity dH/dt as in equation (4)
5.1.1.1. TK models
To model the exposure concentration time profile throughout the experiment, an empirical exponential decay model with a rate b (1/d) is used
| (23) |
where Cj(t) is the time‐variable exposure cadmium concentration (μg/L) at time t with j = 0,…,4 for the control and the four treatments. Parameters (j = 0,…,4) are fixed at nominal concentrations, 0, 10, 20, 40 and 80 μg/L, that is at concentrations initially introduced 7 days before the introduction of the organisms. Consequently, C0(t) = 0, whatever t.
A normal distribution links the exposure model to the measured cadmium concentrations
| (24) |
where MCi,j,k are the measured cadmium concentrations (μg/L) at time‐point ti, treatment j and replicate k. Parameter τE is the precision (1/variance) of the measurements ((μg/L)2).
Similarly to GUTS (equation 1), for each treatment, the scaled damage at time t, Dw(t), is linked to Cj(t) through a one‐compartment model with a dominant rate constant kD (1/d)
| (25) |
with the initial condition Dw(0) = 0.
5.1.1.2. TD models
According to the DEBtox modelling approach (Jager, 2017), survival, growth and reproduction are linked to the scaled damage through ‘linear‐with‐threshold’ relationships depending on respective so‐called no‐effect‐concentrations, i.e. internal concentration thresholds below which no measurable effect can be detected (see Chapter 2). For the sake of simplicity, in this example, the NEC is assumed to be the same for growth and reproduction.
Survival is modelled with a GUTS‐RED‐SD model according to (equation 5) with parameter set θ = (hb, bw, zw). As shown on Figure 27, the observed number of survivors at time‐point ti, treatment j and replicate k is modelled by variable MSi,j,k through a conditional binomial distribution according to (equation 10) and (equation 11).
D. magna body growth is modelled using the von Bertalanffy growth model (Von Bertalanffy, 1938). Both the growth rate, γ (1/d), and the maximum body length, Lm (mm), are assumed to be affected by the scaled damage
| (26) |
| (27) |
and the initial condition Lj(0) = 1,
where Lj(t) is the body length (mm) at treatment j and time t, whereas σGR(t) is the ‘linear‐with‐threshold’ relationship applying for both growth and reproduction with ZGR the NEC (μg/L) and bGR the effect rate (μg/L per day) for both growth and reproduction. This strong assumption was made by Billoir et al. (2011), however it has to be reconsidered for other applications.
A normal stochastic link is used to relate the observed body length data to the growth model
| (28) |
where MLi,j,k are the observed body lengths (mm) at time‐point ti, treatment j and replicate k. Parameter τG is the precision (1/variance) of the observations (mm−2).
D. magna reproduction is modelled in accordance with DEB assumptions (Kooijman, 2000; see also Chapter 2). Indeed, the reproduction process depends on the growth process because organisms reproduce upon reaching their puberty length. Hence, the reproduction process is delayed when the growth process is affected by the cadmium concentration.
Among the five assumptions of the DEBtox modelling for the way the toxicant affects the daphnid energetic budget (see Chapter 2), the effect model assuming an increase in maintenance energetic costs is used to describe the reproduction process as a function of both time and cadmium concentration. The following equation, derived by Billoir et al. (2011), is given for ad libitum food conditions
| (29) |
else Rj(t) = 0.
| (30) |
with the initial condition Rcum j(0) = 0,
where Rj(t) is the daily reproduction rate per mother (#) at time t and treatment j, Rcumj(t) the time‐cumulated number of offspring per mother (#) at time t and treatment j, lp the normalised length at puberty (Lp/Lm, dimensionless, with Lp the length at puberty), Rm the maximum reproduction rate (1/d) and σGR(t) the ‘linear‐with‐threshold’ relationship defined by (equation 27).
Accounting for the count status of reproduction data, and because an increased variability of the reproduction is observed with increasing mean values (namely an over‐dispersion of the reproduction data), a negative binomial stochastic link is used to relate the observed time‐cumulated number of offspring per mother to the reproduction model. It is parameterised so that its mean equalled the reproduction model output
where MRcumi,j,k are the time and cadmium concentration mother (#) at time‐point ti, treatment j and replicate k. Parameter pR (−) accounts for the overdispersion of the reproduction data.
Figure 27 gives the directed acyclic graph of the whole model.
5.1.2. Model implementation in the R software
All endpoints (exposure, survival, growth and reproduction) were fitted simultaneously to estimate all 14 model parameters, since all corresponding observed data were linked together as shown in Figure 28. Parameters were estimated within a Bayesian framework via an R code based on JAGS and the R‐package rjags (see Appendix A, Section A.2.2 for additional information). Prior probability distributions are those reported in Table 5 according to Billoir et al. (2011). Because it is not possible to numerically integrate differential equations in JAGS, models were implemented in discrete time with one day as time step.
Figure 28.
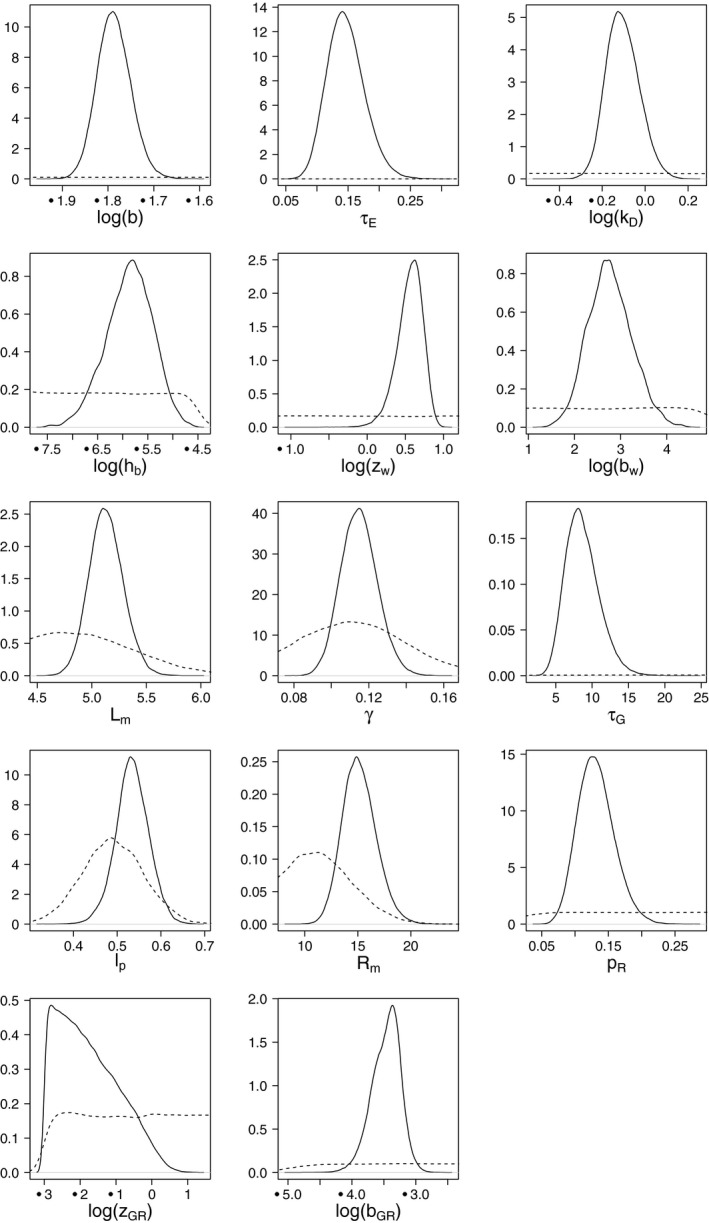
Posterior distributions of parameters obtained from simultaneous fitting of exposure, survival, growth and reproduction data (solid lines), with the corresponding prior distributions (dashed lines). Adapted from Billoir et al. (2011)
Table 5.
Parameter estimates (expressed as medians with 2.5 and 97.5% quantiles). (−) means dimensionless; # means number of offspring per mother
| Symbol | Unit | Meaning | Prior distribution | 2.5% quantile | Median | 97.5% quantile | |
|---|---|---|---|---|---|---|---|
| b | day−1 | Exponential decay rate of cadmium concentration | lognit(−5, 5) | 0.156 | 0.167 | 0.181 | |
| τE | (μg/L)−2 | Precision of exposure observations | amma(10−3, 10−3) | 0.094 | 0.145 | 0.21 | |
| k D | day−1 | Dominant rate constant | logUnit(−5, 1) | 0.785 | 0.897 | 1.06 | |
| b BG | day−1 | Background morality rate | lognit(−10, −4.5) | 0.00107 | 0.00289 | 0.00663 | |
| z w | μg/L | No‐effect‐concentration for survival | lognit(−3, 3) | 1.19 | 1.78 | 2.31 | |
| b w | μg/L per day | Survival killing rate | lognit(−5, 5) | 6.62 | 15.7 | 43.2 | |
| L m | mm | Maximum body length | (4.77, 0.59) | 4.84 | 5.13 | 5.45 | |
| γ | day−1 | Von Bertalanffy growth rate | (0.11, 0.03) | 0.0967 | 0.115 | 0.135 | |
|
|
mm−2 | Precision of body length observations | amma(10‐3, 10‐3) | 4.84 | 8.44 | 13.8 | |
| ℓp | − | Scaled body length at puberty | (0.49, 0.07) | 0.457 | 0.534 | 0.609 | |
| R m | # day−1 | Maximum reproduction rate | (10.7, 3.62) | 12.3 | 15.1 | 18.5 | |
| p R | – | Dispersion of reproduction observations | nif(0, 1) | 0.0835 | 0.13 | 0.19 | |
| z GR | μg/L | No‐effect‐concentration for growth and reproduction | lognif(−3, 3) | 0.0524 | 0.15 | 1.02 | |
| b GR | μg/L per day | Growth and reproduction killing rate | lognif(−5, 5) | 0.0192 | 0.032 | 0.0463 |
Three independent MCMC chains were run in parallel. After an initial burn‐in period of 5,000 iterations, the Bayesian algorithm was run 50,000 iterations and the corresponding sample of the joint parameter posterior distribution was recorded. The convergence of the estimation process was checked with the Gelman and Rubin statistics (Gelman and Rubin, 1992) (see Section 4.1.3.3).
The goodness‐of‐fit was assessed first by comparing prior and posterior distributions models, then with a graphical comparison of observed data and predictions in a way that account for parameter uncertainties and stochasticity of the model. Indeed, predictions are simulated at each MCMC iteration, along which the parameters vary according to their uncertainty, and with the same models than the ones considered to fit the observed data.
5.1.3. Input data
As detailed in Billoir et al. (2011), experiments were conducted in laboratory microcosms (2‐L beakers) filled with artificial sediment and gently aerated synthetic water in which cadmium (Cd) was introduced at nominal concentrations 10, 20, 40 and 80 μg/L, hereafter referred to as treatments 1 to 4. Four replicates were set up for each treatment. Microcosms were conditioned 7 days in the dark before introduction of organisms. Experiments were conducted under constant temperature, pH and light conditions.
At day 0 (7 days after the introduction of Cd), daphnids were introduced in all microcosms, then followed during 21 days. Numbers of survivors, body length and numbers of offspring were recorded at different time‐points (Figure 26). The cadmium water concentration was also regularly measured.
Figure 26.
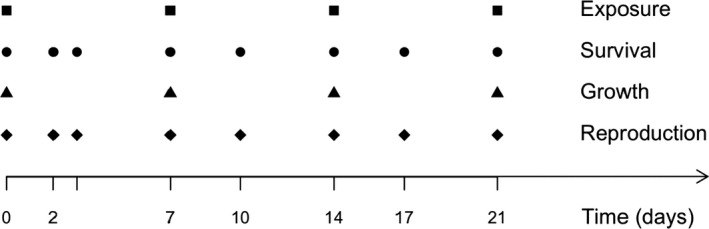
Observation time‐points (in days) of cadmium concentrations (exposure), survival, growth and reproduction of D. magna over the course of the experiment (adapted from Billoir et al., 2011)
5.2. Results of the DEBtox model
5.2.1. Parameter estimates
According to the Gelman and Rubin convergence diagnostics (Gelman and Rubin, 1992), the convergence was successfully checked. Posterior distributions of the 14 parameters are shown in Figure 28 with their corresponding priors. Posterior distributions are also summarised by statistics directly extracted from the MCMC samples: 2.5th, 50th (median) and 97.5th percentiles (Table 5). Based on Figure 28 and Table 5, the 14 model parameters were accurately estimated with posterior distributions narrower than prior ones meaning that the data allowed to reduce the uncertainty. Posterior distributions for parameter Lm (maximum body length) and Rm (maximum reproduction rate) were shifted to higher values than within the priors. This may signify that growth and reproduction of daphnids were higher in the experiments conducted by Billoir et al. (2011) than in those on which priors were based. Concerning parameters for which priors were large (e.g. b, kD, hb), narrow posteriors means that data were informative enough to provide more reliable parameter estimates. NEC threshold estimates point out that sublethal effects of cadmium were predicted to appear before lethal effects since zw = 1.78 μg/L [1.19; 2.31] is higher than zGR = 0.15 μg/L [0.0524; 1.02]. Parameter estimate kD = 0.897 day−1 [0.785; 1.06] is indicative of a rapid kinetics, inferred from the other related endpoints (survival, growth and reproduction) as no internal concentration data were available in the data set.
5.2.2. Goodness‐of‐fit
Figure 29 shows the comparison between observations and predictions for the three endpoints. Because the exposure concentration varies with time, these graphs should be interpreted in the light of the bioaccumulation kinetics (Figure 30). Almost all observed survival data were within the corresponding 95% credible interval (CI) of predicted data: 103 out of 106 data points (97.2%). For growth, 244 out of 283 data points (86.2%) were within the 95% CI, while for reproduction, 100 out of 101 data points (99%) were within the 95% CI. These results validate the choice of the stochastic links chosen according to the different biological endpoints. For example, the overdispersion of the reproduction data when time increased was taken into account by the negative binomial distribution.
Figure 29.
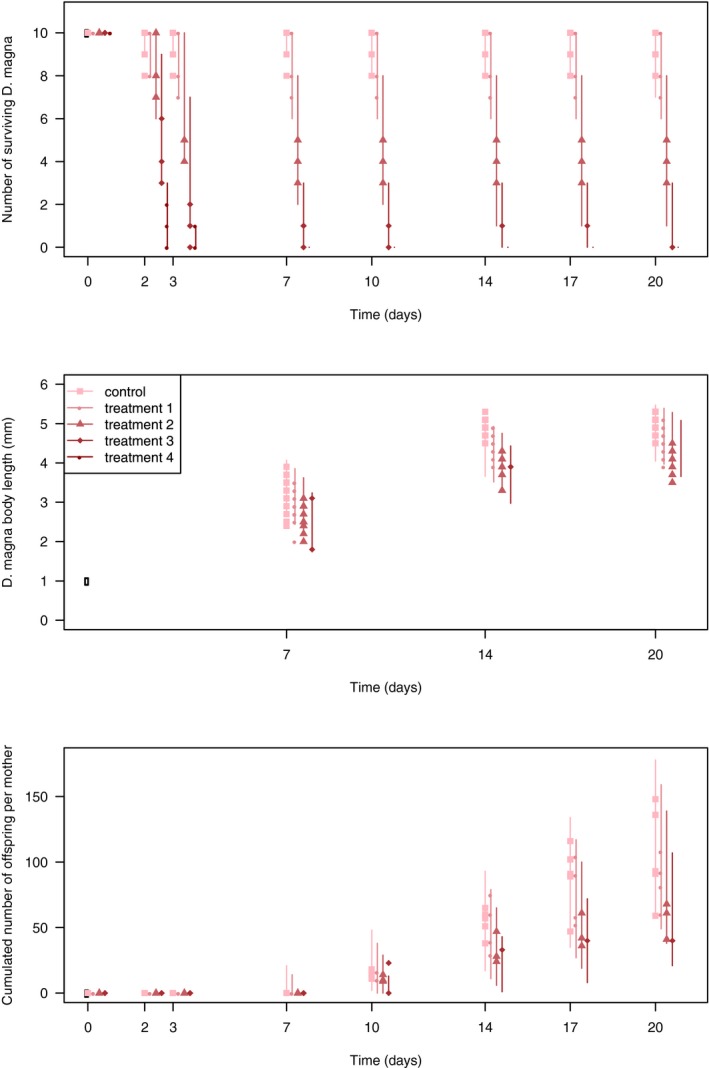
Comparison of observations and predictions for survival (upper panel), growth (middle panel) and reproduction (lower panel) D. magna data. Observed data (symbols) and corresponding 95% credible intervals of simulated data (segments) are superimposed to graphically assess the goodness‐of‐fit (Adapted from Billoir et al. (2011))
Figure 30.
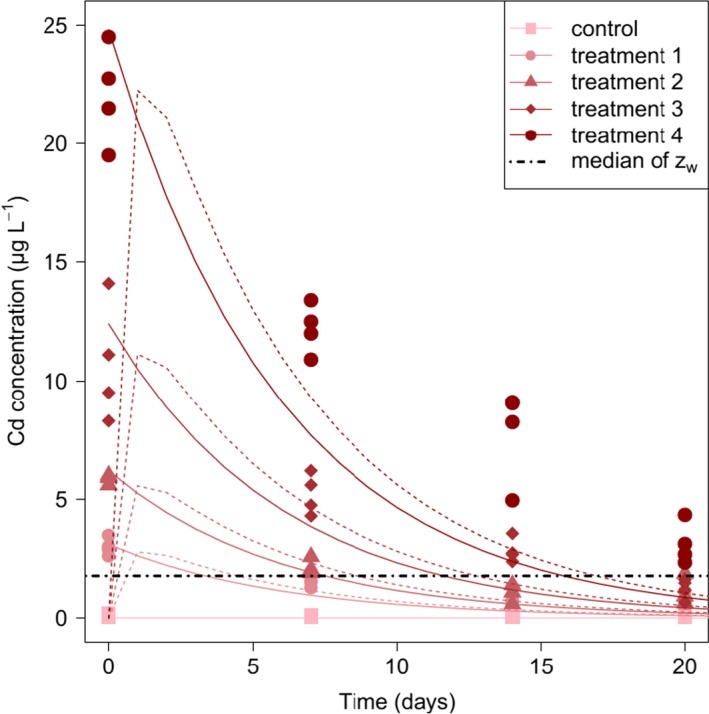
Bioaccumulation kinetics: exposure data (symbols) are superimposed to fitted median curves of both the external concentration (solid lines) and the scaled damage (dashed lines). The dotted‐dashed line stands for parameter zw (the no‐effect‐concentration threshold for survival); adapted from Billoir et al. (2011)
5.3. Concluding remarks about the DEBtox application
This case study about lethal and sublethal effects on D. magna exposed to time‐varying cadmium exposure concentrations within a laboratory aquatic microcosm illustrates the potential of a DEBtox model (with reserve in steady‐state and compound parameters, based on revised equations from Billoir et al. (2008)) to deal with several kinds of data, namely survival, growth and reproduction data together with bioaccumulation data. It also proves the feasibility of estimating all the parameters from simple toxicity test data under a Bayesian framework. Nevertheless, this case study only involved one of the five DEB modes of action (DEBMoA) that can be tested according to the chemical substance. In addition, a strong hypothesis was made about a common threshold concentration (parameter zGR) for both growth and reproduction, such simplifying a little the estimation process. A key point is also the fact that the underlying implementation remains home‐made as no user‐friendly software today exist to perform DEBtox model calibration in a simply way. Under a Bayesian framework, one of the main reason for this, is that further research is still needed to automatise the choice of the priors for the whole set of parameters, as well as to deal with ordinary differential equation in combination with MCMC calculations.
Nevertheless, the use of DEBtox model in the perspective of ERA is valuable. Particular attention should be paid to the choice of the DEBtox set of equations that needs to be argued based on knowledge on the mode of action of the chemical substance (see Table 1 and chapter 9).
6. Models for primary producers
The algae and macrophyte models have not yet been as extensively implemented and validated as the GUTS model and the standard DEB model. The three models presented in Figure 4 have each been described, calibrated and validated in one to four peer‐reviewed papers. In the following, a short overview of the pelagic microalgae model and the Lemna model described in Weber et al. (2012) and Schmitt et al. (2013) is given. These two models are the best described representatives of an algae and a macrophyte model for which also open software codes exist. The Myriophyllum model described in Heine et al. (2014, 2015, 2016a) and in Hommen et al. (2016) is less explicitly described and open software codes for the model are not yet available.
6.1. The pelagic microalgae model
6.1.1. Documentation, testing and implementation of the formal model
Here the description of the flow‐through‐system model of Weber et al. (2012) is the only given, as this is the only published algae model where variable exposures are possible to implement. In addition, this system mimics the ecological situation of algae being removed from the system by either grazing or sedimentation, or by dilution with incoming water from rain, drain or ground water sources. As already mentioned in chapter 2, the algae model by Weber et al. (2012), does not contain a TK compartment, but uses the external concentration as a direct proxy for internal concentrations assuming instantaneous equilibrium between external and internal concentrations. Hence, external pesticide concentrations, phosphorous (P), irradiance (Irr) and temperature (T) affect relative growth rates (RGR) directly through different functions. In addition, also the dilution rate of the algae and the death rate of the algae will affect the population density over time. A diagram of the model is given in Figure 31.
Figure 31.
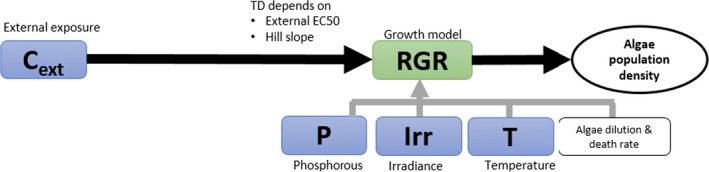
Schematic representation of the algae model presented in Weber et al. (2012). External factors affecting chemical uptake or growth are given in blue and the different rate constants affecting biomass growth rates of the algae (green box) are given with grey arrows and described in Section 6.1.1.1. The flow and effect of the toxic substance is given by black arrows and the TD‐model is described in Section 6.1.1.2
Contrary to the GUTS model, where only effects of survival are assessed, in plant models, the toxicant affects plant growth. All models of plants and algae therefore include a growth model giving the factors affecting growth.
In the model presented in Weber et al. (2012), the parameters describing nutrient availability (Q and P) have different units depending on the equation they are used in. For a better understanding of the model, the units of the model parameters and the environmental variables (Q; P and R0) were harmonised in order to keep the same unit for one parameter throughout the different equations of the model.
6.1.1.1. The growth model for algae
The growth model described in Weber et al. (2012) depends on environmental parameters such as the temperature T (°C), irradiance I (μE/m2 per s), nutrient availability Q (mg P/L), and the dilution rate of the media D (day−1) as represented in Figure 31. In addition, the concentration of the chemical stressor C (μg/L) is also affecting growth directly, describing the toxicodynamics, as toxicokinetics are ignored in this model. The parameters are implemented in the following differential equation, where A (mg fresh wt/L) represent the algae population biomass, μmax (day−1) is the maximum relative growth rate and mmax (day−1) the maximum mortality rate.
| (31) |
To describe the temperature effect on algae growth rate a skewed normal distribution is used, where Tmin, Tmax, and Topt are the minimum, maximum and optimal temperature for algal growth, respectively (°C).
| (32) |
With
The effect of irradiance on algal growth rate is given by the following equation describing a saturation curve, where Iopt (μE/m2 per s is the optimum irradiance for the algal growth rate:
| (33) |
In estimating the effect of nutrient availability to algal growth rate only phosphorous is considered by Weber et al. (2012) as a limiting nutrient, even though nitrogen and carbon availability can also limit growth. In this equation qmin (mg P/mg fresh/wt) is the minimum concentration of P in the cells to allow any cell division, while Q (mg P/L) is the internal concentration of P.
| (34) |
The internal concentration of P, Q (mg P/L), is given by the following differential equation, where vmax (mg P/mg fresh wt per day) describes the maximum uptake rate, A, mmax and D is the algal concentration (mg fresh wt/L), maximal death (day−1) rate and dilution rate (day−1), respectively.
| (35) |
The uptake of P by the algae is limited by the internal concentration of P in algae represented by f(Q, P) below. In this function qmax and qmin are the maximum and minimum internal concentration for P, respectively, (mg P/L) and ks (mg P/L) is the half‐saturated constant for extracellular P.
| (36) |
The external concentration of P is as follows with R0 (mg P/L) being the concentration of P entering the system.
| (37) |
6.1.1.2. The TD model for algae
The algal growth rate is affected by the toxicant as a function of the concentration of the chemical in the system C (μg/L) and the concentration of the toxicant causing 50% effect EC50 (μg/L), where b is the slope of the log‐logistic function.
| (38) |
The actual concentration of the chemical in the system is equivalent to the concentration of the chemical entering the system Cin (μg/L) while the degradation rate of toxicant in aquatic environments k (1 day) and the toxicant eliminated by dilution are taken into account. This relationship is given by the next differential equation:
| (39) |
6.1.1.3. Model application
The following section describes the flow‐through‐system model of Weber et al. (2012), as this is the only algae model where variable exposures are possible to implement.
Input data
The model was calibrated for two microalgae species: Desmodesmus subspicatus and Raphidocelis subcapitata (formerly known as Pseudokirscheriella subcapitata). The 10 parameters used to describe algal growth capacity and growth dependence on temperature, irradiance and P‐availability were archived through the authors own experiments combined with literature reviews on standard toxicity tests and are presented in Table C in the Supplementary Information of Weber et al. (2012) for both species. Background and further details on model development and the raw data for the calibration is given in the PhD thesis of Weber (2009) (https://publications.rwth-aachen.de/record/211799/). Calibrated parameters for the susceptibility of the algae to the herbicide isoproturon (PSII inhibitor) were obtained through standard chronic tests (OECD, 2011a), performed by the authors, with isoproturon concentration effects on growth rates being described by the Hill equation (equation 38) (Weber et al., 2012).
To validate the calibrated model, an exposure concentration profile was tested in the flow‐through‐system. The profile was based on a FOCUS D2 drainage scenario with a ditch as water body and autumn application of isoproturon of 1.5 kg a.i./ha. A time‐window of 40 days of the most critical exposure pattern was selected and the exposure profile was modified to make it applicable in the flow‐through system (Weber et al., 2012; Supplementary Information, Figure 1), keeping a focus on maintaining maximal concentration and duration of the peaks. As relatively little effects were seen for the first pulse in the experiment, pulse exposure concentrations were first doubled for the second peak and finally raised 10‐fold for the last peak, compared to the FOCUS predictions, to study recovery from a very high pulse (850 μg/L).
6.1.1.4. Model implementation
The model was implemented in Matlab version 2007b. No optimisation of parameters was performed, but model outputs were compared to the results from the flow‐through‐system employed. Model codes are not published.
6.1.1.5. Modelling results
The results of the model validation are shown as model predictions together with experimental data in Figures 32 and 33 directly copied from Weber et al. (2012).
Figure 32.
The results of the model validation for the algae Desmodesmus subspicatus showing the exposure concentrations of isoproturon in black squares (right y‐axis), and the corresponding algal biomass in black triangles (replicate A) and black circles (replicate B) (left y‐axis) together with the model prediction and its 95% confidence limits (Weber, 2009)
Figure 33.
The results of the model validation for the algae Raphidocelis subcapitata (formerly known as Pseudokirscheriella subcapitata) showing the exposure concentrations of isoproturon in black squares (right y‐axis), and the corresponding algal biomass in black circles (replicate A, Panel A) and black triangles (replicate B, Panel B) (left y‐axis) together with the model prediction and its 95% confidence limits (Weber, 2009)
6.1.1.6. Summary and discussion of the application
From a visual assessment of the model together with data, the model was able to predict both the level of growth inhibition and the rate of recovery of the population well, confirming that toxicity data from standard toxicity tests can be used to parameterise a dynamic model. The focus of the study was to predict high and damaging exposures to the algal population; hence, P‐availability, temperature and irradiance were kept constant and at close to optimal levels. It is therefore not known how well the model can also predict herbicide effects under variable growth conditions, possibly reaching extreme values in terms of P‐availability, temperature and irradiance and whether it can be equally well parameterised for other species.
The uncertainties in terms of effects under more extreme growth conditions, species differences and species interactions are, however, the same as for the static tests presently used in risk assessment.
Contrary to the static tests, the dynamic model harbours the potential of implementing effects of dynamic growth conditions on algal populations. This potential is, however, not yet extensively tested. The largest drawback for implementing the models in pesticide risk assessment is that the flow through setup used in the existing example and needed to simulate long term variable exposures of pesticides to fast growing populations of algae has not yet been standardised, nor has the robustness of the setup been ring tested. Hence, presently the setup and the models are considered as important research tools but probably not yet mature enough to use for risk assessment purposes.
In principle, dynamic conditions are also recorded in normal static/semi‐static tests whenever the test substance has a short half‐life in water. Such feature, which is normally perceived as a problem by risk assessors, since standard tests aim at maintaining constant conditions, may become an asset if the data could be used to calibrate and/or validate a TKTD model. Nevertheless, the present model has never been tested under such conditions (i.e. static tests with fast‐dissipating substances) and therefore, also in this case, its use for the risk assessment cannot be recommended at the present stage.
6.2. The Lemna model
6.2.1. Documentation, testing and implementation of the formal model
The Lemna model is described in Schmitt et al. (2013) and is summarised in Figure 34 below.
Figure 34.
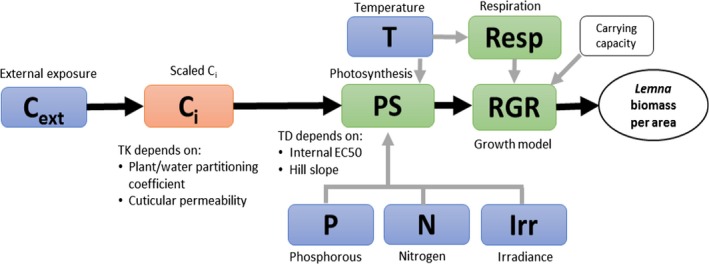
Schematic representation of the Lemna model presented in Schmitt et al. (2013). External factors affecting chemical uptake or growth are given in blue and the different rate constants affecting biomass growth of the plants (green) are given with grey arrows. The growth model is described in Section 6.2.1.1. The flow and effect of the toxic substance is given by black arrows and the TK‐compartment by an orange box. The TK model is described in Section 6.2.1.2 and the TD model in Section 6.2.1.3, respectively
The model uses scaled internal concentrations and a one‐compartment model, where the flux of pesticide in and out of the plant is governed by cuticular permeability and a plant/water partitioning coefficient, corresponding to the bioconcentration factor. A metabolic rate constant is also applied. Compared to the algae model, the growth model is extended with dependence on nitrogen availability. Temperature affects growth rates by both affecting photosynthetic rates and respirations rates, but doing so differently. Finally, growth is not exponential, but reaches a carrying capacity, when fronds start to self‐shade.
6.2.1.1. The growth model
Schmitt et al. (2013) used a differential equation to describe the growth of a Lemna population in terms of dry biomass BM (g dw/m) where (day) represent the maximum photosynthesis rate, (day) the respiration rate at reference temperature, fphoto the factor by which the maximum photosynthesis rate is reduced due to suboptimal conditions and fresp the factor by which the maximum respiration rate is reduced.
| (40) |
A set of environmental factors, θ = {T, I, P, N, D}, affects the photosynthesis and the respiration rate. The environmental factors such as temperature T (°C), light irradiation I (kJ/m2 per day), phosphorous concentration P (mg/L), nitrogen concentration N (mg/L) and population density D (g dw m−2) influence the photosynthesis and respiration factor.
The photosynthetic rate was affected by temperature using an asymmetric bell shape function including an optimum temperature for photosynthesis rate Topt (°C); a minimum temperature for photosynthesis rate Tmin (°C) and a maximum temperature for photosynthesis rate Tmax (°C), analogous to the equation used in the algae model (equation 32).
| (41) |
With
The respiration, however, increases exponentially with temperature as shown in the following equation; with Q10 (relative change caused by 10°C temperature change which is fixed to the value of 2) being the temperature dependent factor for respiration rate and Tref (°C) the reference temperature for respiration rate:
| (42) |
Relative growth rates, normalised to the maximum rate measured at different temperature conditions has been observed to increase linearly with irradiance (given as daily energy input) up to a certain level, after which it is constant. The slope of the linear phase is called α, and is given as 1/kJ m−2 day−1, where kJ m−2 day−1 is the amount of energy received per square meter per day. The intercept of the linear part of the curve is called β, and has a positive value. This is because photosynthesis is calculated as cross carbon fixation over 24 h as a function of area specific energy input over 24 h, rather than as photosynthetic rate given in moles of CO2 m‐2s−1, as a function of irradiance given as μmol m−2s−1 which are the units most often used for physiological experiments. Using daily averages means that daily energy input will never approach zero. The lowest value on the x‐axis will therefore be the shortest day with the lowest energy input allowing Lemna growth. The linear growth increase with increasing irradiation continues until the growth is light saturated at the radiation intensity Isat (kJ m−2 per day−1) from where the biomass growth stays constant with increasing irradiation.
| (43) |
The nutrient dependence is given in the following equation with nitrogen as an example. The same equation can be used for phosphorus limitation of growth. The equation includes the concentration of nitrogen [N] (mg/L) and the nitrogen concentration at which half the maximum relative growth rate is reached [N]50 (mg/L).
| (44) |
Lemna growth is also affected by the density. In the model of Schmitt et al. (2013) the relationship between growth rate and density is described by (g dw m−2), which is the limit density at which photosynthesis and respiration has the same size and net growth therefore is zero.
| (45) |
6.2.1.2. The TK model
As all parts of Lemna are in contact with water and due to the simple structure of the plant, Schmitt et al. (2013) used a one‐compartment TK model. The following differential equation gives the scaled internal concentration of the substance in the plant Ci (μg/L), in relation to the external concentration of the substance Cext (μg/L); where P (dm/day) and A (dm−2) are the plant membrane permeability and available surface area, respectively, while V (L) is the plant compartment volume. The two rates constant used in the equation represent the bioconcentration factor Kp:w and the metabolic degradation rate kmet (day−1).
| (46) |
6.2.1.3. The TD model
Schmitt et al. (2013) choose to relate the internal unbound chemical concentration to the effect using the Hill equation, rather than the calculated internal concentration (equation 46). In the equation, Emax is the maximum effect of the chemical, (μg/L) is the chemical concentration in the water phase of the plant, which is equivalent to and EC50int (μg/L) is the internal concentration causing 50% effect and b is the shape parameter. Schmitt et al. (2013) use a different parameterisation of the Hill equation than presented in for example equation 36, but it can be re‐parameterised as shown in below:
| (47) |
6.2.1.4. Model application
The Lemna model is well described in Schmitt et al. (2013) and is summarised in Figure 34.
Input data
Five types of data were used to calibrate and validate the model: (1) For calibration of the growth model, literature data were used; (2) to validate the growth model, literature data on Lemna biomass in Dutch ditches as a function of irradiance and temperature data were used; (3) to calibrate the toxicokinetic and dynamic models, standard toxicity test data (OECD, 2006) were used for the herbicide metsulfuron‐methyl; (4) to validate the TKTD model, data from a pulse exposure experiment were used; (5) finally, two FOCUS exposure scenarios for which risk assessment had failed to demonstrate safe use, using the Toxicity Exposure Ratio (TER) approach and a trigger of 10, were used to predict effects on field populations of Lemna spp. Schmitt et al. (2013).
The literature data used to calibrate the growth model were fitted by the authors to obtain the parameter values using the Optimizer‐function in MS EXCEL. The raw data, the criteria for selecting the data and the fits are shown in the supplementary material of Schmitt et al. (2013). The parameter values are given in Table 1 of Schmitt et al. (2013) together with the literature references.
The validation data set for the growth model under environmentally realistic, variable growth conditions monitored Lemna biomass in three Dutch ditches over 2 months. The data are published in Driever et al. (2005) and were combined with air temperature and radiation data taken from the European meteorological data base MARS. Mean N and P concentrations from the ditches were used within the two‐month timeframe of the experiment. Simulations were run for one year.
The plant/water partitioning coefficient for any pesticide was suggested to be calculated based on a relationship with Kow derived from Crum et al. (1999), and confirmed by other studies for a large range of chemicals with different Kow values. The cuticular permeability and the Hill parameters for the tested herbicide, metsulfuron‐methyl, were obtained using data from a standard 7‐day Lemna test (OECD, 2006), followed by a 7 day recovery period, counting frond number every 2–3 days. To validate the TKTD model, data from a pulse‐exposure experiment published by DEFRA (2005) and Boxall et al. (2013) was used.
Two FOCUS scenarios were used simulating metsulfuron‐exposure resulting from either drainage: The FOCUS scenario D2 ‘Brimstone’ or run‐off: FOCUS scenario R3 ‘Bologna’. The scenarios are graphically presented in Schmitt et al. (2013). In addition, Hommen et al. (2016) have used the model to predict effects on three FOCUS scenarios from ditches and a stream (Hommen et al., 2016).
Data used to parameterise the model on alachlor, a herbicide interacting with the gibberellin pathways in plants, and formasulfuron, another sulfonylurea herbicide inhibiting the synthesis of branched chain amino acids have been presented by Heine et al. (2016b) and Heine et al. (2017).
6.2.1.5. Model implementation
The model was implemented in R (Version 2.15.1), using the general solver for ordinary differential equations ‘ode’ of the R‐package ‘deSolve’ described in Soetaert et al. (2010) for solving the differential equations. The complete code is provided in the supplementary information of Schmitt et al. (2013). To validate the growth model, a stochastic simulation was performed varying the growth parameters in a Monte Carlo (MC) approach with 100 runs. Based on the comparison of literature data, showing very small variability in parameter values obtained in different studies, a standard deviation of 10% was used.
6.2.1.6. Modelling results
The calibration of the growth model is explicitly described in Schmitt et al. (2013) and the authors obtain values similar to those found by other authors and for other Lemna species than Lemna minor or Lemna gibba used in this study. Also, the validation of the growth model under environmentally realistic variable growth conditions predicts the growth increase in the spring well (Figure 3 in Schmitt et al., 2013; Figure 35, left panel). The authors express the wish to validate the model on field data spanning the entire year, rather than two months, to validate the model properly under field conditions. This type of data is, however, not very frequently found in the open literature.
Figure 35.
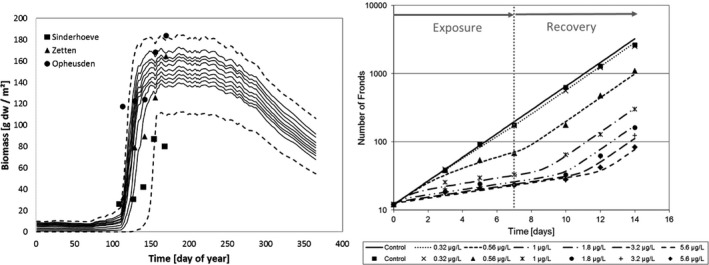
Left panel shows predicted Lemna growth (lines) over one year in comparison to observed field data from three Dutch ditches (symbols). The different lines show minimum and maximum (dashed) as well as the deciles of the biomass resulting from the Monte‐Carlo simulation with variation of model parameters. Right panel shows growth of L. gibba exposed to different concentrations of metsulfuron‐methyl. Exposure persisted until day seven with subsequent recovery in uncontaminated nutrient solution. Symbols show observed data and lines as calculated with the fitted model. Note that in the experiment the number of fronds was reduced to 15 at day seven, but the data were recalculated from day seven on by multiplication with the respective reduction factor (from Schmitt et al., 2013)
To calibrate the TK model, the plant/water partitioning coefficient was determined based on the log Kow of metsulfuron‐methyl of −0.48, while the cuticular permeability governing the uptake rate was determined from the model fits. Metabolisation of the herbicide was set to zero. The TD parameters: the internal concentration inducing 50% growth inhibition (EC50int), the slope of the Hill curve and a maximal growth inhibition was derived from fit to the standard OECD data set. All parameters were obtained with small CV's (< 1%, apart from the slope parameter, where CV was 48%). The good parameter estimates were partly due to the relatively slow initiation of effect and slow recovery, making fit of kinetic parameters possible even with a relatively low time resolution of the relative growth rates of several days (Figure 2 in Schmitt et al., 2013; Figure 35, right panel).
It is a bit surprising that a maximal inhibition of growth is introduced (being 78%), as one would expect growth inhibition to approach 100% with increasing herbicide concentration. This is most likely done, because the mode of action of metsulfuron‐methyl, inhibition of branched chain amino acid synthesis, acts really slowly, and in crops can take 4–6 weeks to fully develop. Hence, as growth will always continue for a time after uptake of the herbicide, growth inhibition measured relative to start biomass (or frond number) will never be 100% in short experiments.
The TKTD model was validated on constant and pulse exposure scenarios of Lemna grown in the lab over 42 days. The EC50int was recalibrated on the constant exposure data to account for a slightly lower sensitivity of this Lemna clone (parameterised to 0.39 rather than 0.30 μg/L), but all other parameters were kept from the first calibration data set. The model predictions described data really well for the first 3 weeks of the experiment, after which is seemed Lemna became insensitive to the herbicide growing faster than predicted by the model. This tendency was more pronounced at the constant and long pulse exposure (4‐day pulse, 3 days of clean water) compared to the short pulse exposure (2‐day pulse, 5 days of clean water), which indicated that continuous exposure leads to selectivity of resistant fronds possibly through upgrading of detoxification capacity (Figure 4 in Schmitt et al., 2013; Figure 36).
Figure 36.
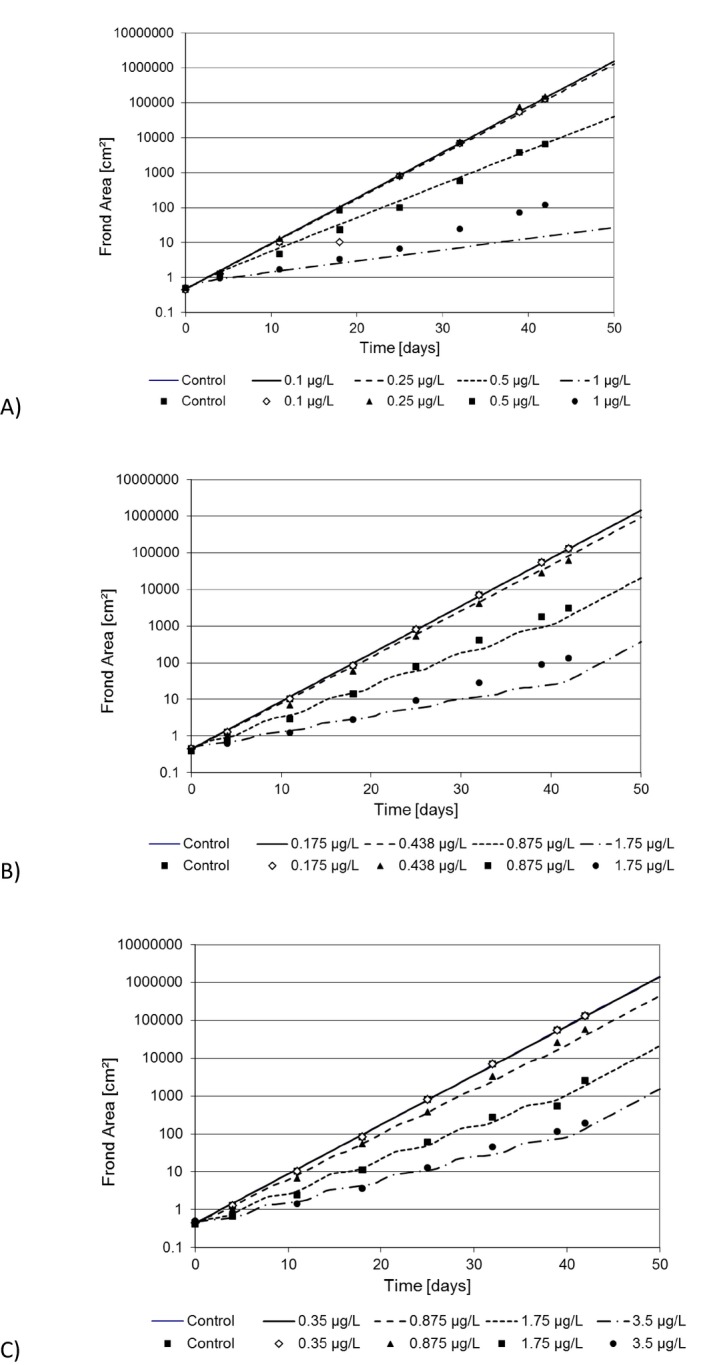
Predicted Lemna growth (lines) in comparison to observed data (symbols) from studies with three different exposure patterns: (A) continuous; (B) 4 of 7 days; and (C) 2 of 7 days exposure. Note: In all cases, control and lowest exposure level led to almost identical results in experiment and simulation and are therefore not distinguishable in the plots (from Schmitt et al., 2013)
For the prediction of effects of the different exposure profiles (called predicted environmental concentrations (PEC) in Figure 37), the effect on growth rate and biomass accumulation over time was tested both when growth rates were at their highest (considered the most sensitive growth stage) and when the plant population had achieved carrying capacity. The effects were predicted for exposure profiles (PEC's) multiplied with different factors. The maximal effect on growth of 100x PEC was 34% reduction in growth rate when application was done early in the season and 6.4% when applied after plant biomass had reached carrying capacity. The authors conclude that for macrophytes, it may not be the decrease in growth for a short period of time, but the delay in reaching maximal biomass that will have the largest ecological consequences. They therefore suggest using an effect measure based on time to reach 90% of the control population biomass. The results of two of the exposure profiles are given in Figures 6 and 7 of Schmitt et al. (2013) (Figure 37) shown below.
Figure 37.
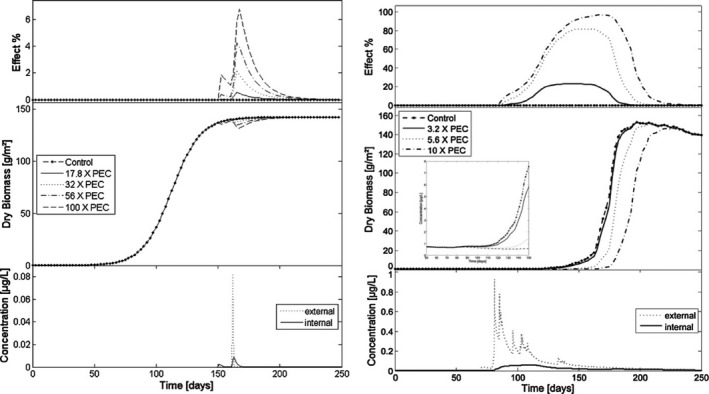
Left panel shows predicted growth of Lemna (middle) for different levels of exposure to metsulfuron‐methyl caused by run‐off occurring when the population had reached carrying capacity. Lower panel: concentration time course. Upper panel: percentage effects calculated from the difference between control and affected biomass curves. For exposure levels below 17.8x, no deviation from the control was found and the respective results were not included in the plot. Right panel shows predicted growth of Lemna (middle) for different levels of exposure to metsulfuron‐methyl caused by drainage and considering the climatic conditions also underlying the exposure simulation. The inset shows a blow up of the period 50–150 days. Lower panel: concentration time course. Upper panel: percentage effects calculated from the difference between control and affected biomass curves. For exposure levels below 3.2x, no deviation from the control was found and the respective results were not included in the plot (from Schmitt et al., 2013)
6.2.1.7. Scientific discussion of the model application
In the model presented by Schmitt et al. (2013), the use of realistic growth conditions for risk assessment purposes is emphasised, as recovery may be delayed at suboptimal conditions, and suggest a model‐based endpoint ‘time to population recovery’ as a supplement to decrease in growth rate. The concept of population recovery is outside the scope of this scientific opinion, hence not further evaluated here, but the concept of model‐based recovery times might be useful in future risk assessment. Also, the importance of increasing the time resolution of growth development for calibration and validation data is highlighted, as it makes parameter estimations more accurate. For Lemna, measuring surface growth over time, this can easily be done in a non‐destructive manner by counting fronds or measuring surface area of fronds from photos or other images. This will be of particular importance when looking at herbicides with faster modes of action than the tested sulfonylurea, as for example PSII inhibitors. The authors also suggest measuring uptake and elimination kinetics directly for more modes of action, but also to investigate if the leaf permeability decreases with decreasing temperature as has been observed for terrestrial plants. Permeability studies investigating temperature effects have, to our knowledge, not been performed. But this could potentially have a large effect on pulse exposures, as short‐term pulses may not be taken up very rapidly at colder temperatures compared to warmer temperatures if permeability is decreased at low temperatures. In addition to the issues suggested by the authors, the effect of the pKa of the herbicides combined with the pH of the water may play a role for uptake, as is seen for plant and algae uptake in general (Fahl et al., 1995; Trapp, 2004; Rendal et al., 2011). It is also known that frond size and surface/weight ratios change in Lemna when exposed to herbicides with different mode of action (Cedergreen et al., 2004; Cedergreen and Streibig, 2005). In the model presented by Schmitt et al. (2013), these ratios are assumed to be constant. It would, however, be possible to do a sensitivity analyses on the effect of variation in the frond/surface/weight ratio based on published data to investigate, if these variations are of significance for the outcome of the model.
6.2.1.8. Evaluation of the application in risk assessment
The Lemna model, its implementation, parameter estimation and validation together with sensitivity analyses of its robustness to variation in parameters and a discussion of strengths, weaknesses and wishes for future work, is documented in a scientific publication (Schmitt et al., 2013).
Physiological model parameters have been collected from the scientific literature. TKTD model parameters have been calibrated on data from a modified 14‐day OECD toxicity test by non‐linear optimisation and are reported including the parameter variability. Optimisation routine and the method used for the estimation of the parameter uncertainty limits are not clearly given in the paper or the supporting information. Variability and sensitivity analyses were made on photosynthetic rate, respiration rate, maximal density and initial biomass. Variability analysis means in this context that the authors tried to analyse how variable the most relevant model parameters are in literature. In conclusion of this analysis, the above parameters were varied within 10% CV. In the publication, it was concluded that the results of both exposure profiles are relatively robust to variation in the parameters (MC simulation of the parameters given a CV of 10% each). The uncertainty intervals of the predictions increased 1.5–2‐fold adding the variation in the parameters. Given that there is no real empirical basis for the variation of above mentioned parameters given in the paper, the chosen variation range of 10% appears rather small, especially because the parameters were partially drawn from a normal instead of a uniform distribution. Under these circumstances, a 1.5‐ to 2‐fold increase in the uncertainty intervals appears very high, since this translates to 50%–100% increase in the variation of the modelled endpoint. For future applications of the Lemna model in risk assessment, optimisation methods need to be described in more detail, and sensitivity analyses should be redone to allow for a comprehensive understanding of the variation in the model parameters.
Nevertheless, in line with the algae model, the Lemna model could potentially be used to predict effects under variable environmental conditions, as was done for the ditch validation data (Figure 35). The conceptual and the formal model appear suitable for the application of the Lemna model in risk assessment. The model was implemented in R, and the code is available, but not all input files are given. A specific implementation of verification was not included in the publication and should be included for future applications. The dynamic Lemna model harbours the possibility to predict effects also under variable growth conditions, but more validation data is needed to test the robustness of such predictions. None of the long‐term predictions of population effects of Lemna exposed to variable FOCUS exposure profiles have been validated by data from long term laboratory or field experiments. If long‐term data on Lemna growth exist from for example mesocosm studies, it could be interesting to validate the model against such data sets.
Since the publication of the model in 2013, parameterisation and validation of the model on two other herbicides with different modes of action, one affecting the gibberellin pathways in plants, and the other inhibiting branched chain amino acids, have been presented at European SETAC conferences, but since the results are not published they cannot be further checked. In addition, an application of the Lemna model for an inhibitor of carotenoid synthesis is available.
The model seems to work well for the sulfonyl‐urea compound and the validation data as presented in Schmitt et al., 2013, but the model should be validated on herbicides with other modes of action.
Summarising, the Lemna model appears suitable for use in risk assessment to evaluate effects of time‐variable exposure on Lemna growth. However, the above‐mentioned aspects of the model have to be picked up and improved to make applications acceptable for use in regulatory risk assessment (see Chapter 7 and Annex C).
6.3. The Myriophyllum model
As discussed in Section 2.4.3 the Myriophyllum model is not as completely described as the Lemna model, and source code is, to our knowledge, not publicly available. The model is described in three papers: The first one builds and parameterises the growth model, and describes how growth varies as a function of irradiance and temperature (Heine et al., 2014). The second paper focusses on describing and parameterising the TK‐processes for several herbicides, including both sulfonylurea and photo‐system II inhibitors. Also, they determine cuticular permeability for chemicals based on their log Kow (Heine et al., 2015). The final paper describes and parameterises the toxicodynamic model using the sulfonylurea iofensulfuron (Heine et al., 2016a). The fact that the model is described in three separate papers makes it difficult to get an overview of its entity, as is also discussed in Section 2.4.3 Some of the main issues not explicit in the published material on Myriophyllum are:
How the model deals with density dependent growth.
How toxicants move between compartments, both toxicants taken up by the leaves and toxicants taken up by the roots.
The Myriophyllum model has only been calibrated and validated on a single substance, iofensulfuron under constant growth conditions. Hence, its more global application to chemicals with other modes of action and to other growth conditions has yet to be seen. More detailed description of the model can be found in the PhD thesis of Heine, 2014: https://publications.rwth-aachen.de/record/459446.
A publicly available model implementation in line with that provided for the Lemna model would be a great advantage.
6.4. Summary, conclusions and recommendations for the algae and plant models
The models for the primary producers all rely on a growth model driven by a range of external inputs such as temperature, irradiance, nutrient and carbon availabilities. The effect of the pesticide on the net growth rate is described by a log‐logistic relationship, linking either external (the algae model) or scaled or measured internal concentrations to growth rate. All experiments and tests of the models up till now have been done under fixed growth conditions, as is the case for the standard algae, Lemna and Myriophyllum tests. This is because the focus has been on evaluating the model availabilities to predict effects under time‐variable exposure scenarios. The growth models do, however, all have the potential to incorporate changes in temperature, irradiance, nutrient and carbon availabilities in future applications.
For the algae, time‐variable exposures were excellently described by the model for two species of algae and one PSII inhibiting herbicide. The largest drawback for implementing the models in pesticide risk assessment is that the flow‐through setup used and needed to simulate long‐term variable exposures of pesticides to fast growing populations of algae has not yet been standardised, nor has the robustness of the setup been ring tested. Hence, for the present, the setup and the models are important research tools but probably not yet mature enough to use for risk assessment purposes.
The Lemna model is the most thoroughly tested, calibrated and validated having been verified for four different herbicides with three modes of action. In addition, it can be calibrated on data from the already standardised OECD Lemna test, as long as pesticide concentrations and growth is monitored several times during the exposure phase and the test is prolonged with a 1‐week recovery period. Growth may be monitored most easily and non‐destructively by measuring surface area or frond number, either on a daily basis or every 2 days. Calibration to biomass should be done at least at the end of the exposure phase, as some herbicides change surface to weight ratios. Validation data sets should have different exposure patterns, containing at least one but ideally two pulse exposures and be tested on a range of concentrations. If the model is properly documented and analysed according to the suggestions in Section 6.2.1.8 (See also Annex C), the Lemna model can be an important tool to evaluate the effects of time‐variable exposures of pesticides under different growth scenarios.
The Myriophyllum model is not yet as well developed, calibrated, validated and documented as the Lemna model. It is more complicated, as Myriophyllum also has a root compartment (in the sediment) where the growth conditions (redox potential, pH, nutrient and gas availabilities, sorptive surfaces etc.) and therefore also bioavailability of pesticides are very different from the conditions in the shoot compartment (water column). In addition, Myriophyllum grows submerged making carbon availability in the water column a complicated affair compared to Lemna, where access to carbon through the atmosphere is constant and unlimited. Due to the increased complexity of the system and the relative novelty of the published model, the Myriophyllum model has not yet been very extensively tested and publicly assessable model codes are not yet available. Standard guidelines for conducting tests of Myriophyllum spicatum are available (OECD 2014a,b) As for the Lemna test, the aim is to be able to use data from such standardised tests for calibration with growth being monitored over time (non‐destructively as shoot numbers and length), and including a recovery period and length/biomass calibrations. Validation data set should include at least two pulse exposures. At the present state, however, the model needs further calibration, validation and documentation to be ready to use for risk assessment purposes.
7. Evaluation of models
This section follows Chapter 10 of the EFSA Opinion on good modelling practice in the context of mechanistic effect models for risk assessment (EFSA PPR Panel, 2014) and expands on the information provided that is specific to TKTD models. The chapter focuses on GUTS modelling with additional paragraphs covering differences for DEBtox and plant models, where appropriate.
The key points that the risk assessor should pay attention to remain the same as in EFSA PPR Panel (2014):
Is the model based on commonly agreed scientific principles and/or are these principles published in the scientific literature?
Is the general behaviour of the model plausible?
Are all steps of the modelling cycle sufficiently well documented? (see Chapters 4, 5 and 6)
Is the correspondence with available independent observations acceptable?
Is the model fit for regulatory purpose? In other words, can it be used to provide an answer to the question posed in the problem definition?
Parts of these key questions have been analysed, documented and answered for some of the models already, particularly for the GUTS models. More details will be given in the following sections.
In the case of DEBtox models, there are two aspects that need to be considered: (1) how well the DEB model describes the energy budget of the species under consideration (physiological part of the model) and (2) how the effects of the chemical can be modelled through stress functions applied on DEB model parameters (TKTD part of the model).
The DEB part of the model (physiological part of the model) is usually built based on a certain amount of underlying physiological data. There are two options for the evaluation of the data that is underlying the physiological model:
If the species‐specific DEB part of the model has been previously documented and validated, then there is no need to check this for each application with a specific toxicant. The species‐specific part of the model should be able to describe the relevant endpoints observed in the controls of the validation data (e.g. weight/length or reproduction under control conditions). However, in some cases, adaptation of relevant parameters is needed to properly describe control data under specific experimental conditions. This is considered necessary for the model to be fit for purpose (see criteria for evaluating fit of the model, Section 7.7.2).
If the species‐specific DEB part of the model has not been documented and validated previously, then this needs to be properly undertaken outside of the evaluation of the TKTD part (see recommendation in Chapter 9.2).
The first of these two options is likely to be the only option applicable in risk assessment practice, so this is the focus of this SO. For an evaluation of the species‐specific DEB part of the model, the EFSA Opinion on good modelling practice in the context of mechanistic effect models for risk assessment (EFSA PPR Panel, 2014) should be used.
Models for primary producers are also divided into the physiological and the TKTD parts, which can be evaluated separately. For Lemna and some algal species, the physiological part of the model is already developed and has been evaluated in the SO, but for other aquatic plants and algal species further work is required before the physiological model and the underlying data are considered to be suitable for use in risk assessment. Similarly to DEB, the species‐specific part of the primary producer model should be able to describe the observed control data of the validation data set. Once again, when necessary, adaptation of the most relevant parameters to specific experimental conditions may be needed in order to consider the model fit for purpose (see criteria for evaluating the fit of the model, Section 7.7.2).
Every model application should be accompanied by a summary according to Annex D.
The summary checklist provided in Appendix B of EFSA PPR Panel (2014) has been adapted for TKTD models and three adapted versions (GUTS, DEBtox and plant models) are provided in Annex A, Annex B and Annex C.
When the evaluator fills in the checklist, justification should be included for all points that are not straightforward.
In Appendices F and G, the evaluation of two available model examples, one for GUTS and one for DEBtox, is reported for illustration
7.1. Evaluation of the problem definition
The chapter about the problem definition/formulation in this SO (Chapter 3) contains comprehensive information about the regulatory context in which the model is run, the regulatory questions that can be addressed with the model and the required model output to answer these questions. However, these aspects have to be defined clearly for any model application.
One important aspect that needs to be addressed as part of the problem definition is the selection of species for the modelling that has been performed (see Section 3.2 for more information about selecting species for different tiers of the risk assessment). In line with other higher‐tier approaches, the choice of the test species needs to be clearly described and justified, also considering all the available valid information. In addition, the problem definition should make clear whether the model is being used with a Tier‐1 test species, i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2. If the modelling is part of a Tier‐2C2 risk assessment, then it is very likely that TKTD models have been applied for different species, except in the rare situation where a large number of species have been tested in the laboratory at Tier‐2A/2B, and these experiments have shown that only one or a very limited number of species are sensitive to the pesticide (see Chapters 3 and 8).
7.2. Evaluation of the quality of the supporting experimental data
This part of the evaluation checks whether the experimental data which are used for the modelling (both calibration and validation data sets) have been subjected to an acceptable quality control. The focus is on data quality, i.e. the laboratory conditions, set‐up, chemical analytics and similar. All laboratory studies used to calibrate or validate the model should be summarised and evaluated considering the specific guideline that was used. Sometimes the laboratory studies providing the data cannot follow standard guidelines, if for example the design, the experimental conditions and/or the validity criteria of the study need to be modified in order to provide more appropriate data for the modelling. In such case, the study should be tailored to the modelling needs but the principles of the most closely related guideline should be followed as far as is possible. In such cases, attention should be paid to provide validity criteria, which measure whether the performance of the study is sufficient to provide reasonably reliable data. It is important that laboratory studies included test concentrations that were high enough to clearly demonstrate the effects of the toxicant. Additional specific criteria for the suitability of the data sets for model calibration and validation have been developed earlier (Sections 4.1.3.4 and 4.1.4.5) and are explained later in more detail (Sections 7.6.2 and 7.7.2, respectively).
Validity criteria and other requirements of OECD guidelines that might be considered when carrying out studies to support TKTD modelling are summarised in Table 6 (given at the end of Chapter 7), together with comments about the relevance of the criteria for data used for this purpose. Where the study design is modified in such a way that the validity criteria prescribed in the guideline are no longer applicable, the performance of the study should be carefully considered (e.g. Did the controls behave as expected? Are tested individuals numerous enough to detect effects?).
Table 6.
Validity criteria in OECD aquatic studies and their relevance for TKTD
| Guideline | Validity criteria in the unmodified test guideline (plus some additional requirements) | Relevance of these criteria for data underlying TKTD modelling and where changes are needed |
|---|---|---|
| Fish acute toxicity test (OECD, 1992) | The mortality in the control(s) should not exceed 10% (or one fish if less than 10 are used) at the end of the test |
This criterion is directly relevant as it demonstrates suitable experimental conditions for the fish This is also relevant for calibration of background mortality |
| Constant conditions should be maintained as far as possible throughout the test and, if necessary, semi‐static or flow‐through procedures should be used (see Annex 1 of the guideline for definitions) | This criterion is relevant for all conditions except where variation is intentional (i.e. to generate pulses of exposure) | |
| The dissolved oxygen concentration must have been at least 60% of the air saturation value throughout the test | This criterion is directly relevant as it relates to ensuring suitable conditions | |
| There must be evidence that the concentration of the substance being tested has been satisfactorily maintained, and preferably it should be at least 80% of the nominal concentration throughout the test. If the deviation from the nominal concentration is greater than 20%, results should be based on the measured concentration |
Concentrations over time should be measured so it is clear what the fish were exposed to. Instead of using a mean measured concentration the whole exposure profile is relevant (whether the study aims at constant or time‐variable exposure) Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Fish early life stage toxicity test (OECD, 2013) | The dissolved oxygen concentration should be > 60% of the air saturation value throughout the test | This criterion is directly relevant as it relates to ensuring suitable conditions |
| The water temperature should not differ by more than + 1.5°C between test chambers or between successive days at any time during the test, and should be within the temperature ranges specified for the test species (Annex 2 of the guideline) |
This criterion is directly relevant as it relates to ensuring suitable conditions If a physiological model that includes variable temperature is used this could also be applied to the study |
|
| The analytical measure of the test concentrations is compulsory | This is particularly important for TKTD modelling where the whole exposure profile is relevant | |
| Overall survival of fertilised eggs and post‐hatch success in the controls and, where relevant, in the solvent controls should be greater than or equal to the limits defined in Annex 2 of the guideline. | This criterion is directly relevant as it demonstrates suitable conditions for the fish | |
| Daphnia sp. acute toxicity test (OECD, (2004a) | In the control, including the control containing the solubilising agent, not more than 10% of the daphnids should have been immobilised |
This criterion is directly relevant as it demonstrates suitable conditions for the daphnids This is also relevant for calibration of background mortality |
| The dissolved oxygen concentration at the end of the test should be ≥ 3 mg/L in control and test vessels | This criterion is directly relevant as it relates to ensuring suitable conditions | |
|
ALSO (not a validity criteria): The concentration of the test substance should be measured, as a minimum, at the highest and lowest test concentration, at the beginning and end of the test |
It is important that the test concentrations are measured, but the minimum requirements are likely to be greater for TKTD modelling as information is required on the whole exposure profile Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Chironomus sp. acute immobilisation test (OECD, 2011b) | In the control, including the solvent control if appropriate, not more than 15% of the larvae should show immobilisation or other signs of disease or other stress (e.g. abnormal appearance or unusual behaviour, such as trapping at the water surface) at the end of the test | This criterion is directly relevant as it demonstrates suitable conditions for the chironomids |
| The dissolved oxygen concentration at the end of the test should be ≥ 3 mg/L in control and test vessels | This criterion is directly relevant as it relates to ensuring suitable conditions | |
|
ALSO (not a validity criteria): The concentration of the test substance should be measured, as a minimum, in the control(s), the highest and lowest test concentration, but preferably in all treatments, at the beginning and end of the test. It is recommended that results be based on measured concentrations. However, if evidence is available to demonstrate that the concentration of the test substance has been satisfactorily maintained within ± 20% of the nominal or measured initial concentration throughout the test, then the results can be based on nominal or measured initial values. |
It is important that the test concentrations are measured, but the minimum requirements are likely to be greater for TKTD modelling as information is required on the whole exposure profile Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Sediment‐water Chironomid toxicity test using spiked water (OECD, 2004b) | The emergence in the controls must be at least 70% at the end of the test | This criterion is directly relevant as it demonstrates suitable conditions for the chironomids |
| C. riparius and C. yoshimatsui emergence to adults from control vessels should occur between 12 and 23 days after their insertion into the vessels; for C. tentans, a period of 20–65 days is necessary | This criterion is directly relevant as it demonstrates suitable conditions for the chironomids | |
| At the end of the test, pH and the dissolved oxygen concentration should be measured in each vessel. The oxygen concentration should be at least 60% of the air saturation value (ASV) at the temperature used, and the pH of overlying water should be in the 6–9 range in all test vessels | This criterion is directly relevant as it relates to ensuring suitable conditions | |
| The water temperature should not differ by more than ±1.0°C. The water temperature could be controlled in an isothermal room and in that case the room temperature should be confirmed for appropriate time intervals |
This criterion is directly relevant as it relates to ensuring suitable conditions If a physiological model that includes variable temperature is used this could also be applied to the study |
|
|
ALSO (not a validity criteria): As a minimum, samples of the overlying water, the pore water and the sediment must be analysed at the start (preferably 1 h after application of test substance) and at the end of the test, at the highest concentration and a lower one |
It is important that the test concentrations are measured, but the minimum requirements in terms of frequency are likely to be greater for TKTD modelling as information is required on the whole exposure profile. Which compartments need to be measured will depend on the behaviour of the chemical and the model being used (i.e. if the model only uses water concentrations this should be the focus of the measurements | |
| Daphnia magna reproduction test (OECD, 2012) | The mortality of the parent animals (female Daphnia) does not exceed 20% at the end of the test. | This criterion is directly relevant as it demonstrates suitable conditions for the daphnids. |
| The mean number of living offspring produced per parent animal surviving at the end of the test is ≥ 60. | This criterion is directly relevant as it relates to ensuring suitable conditions. | |
|
ALSO (not a validity criteria): During the test, the concentrations of test substance are determined at regular intervals In semi‐static tests where the concentration of the test substance is expected to remain within ± 20% of the nominal, it is recommended that, as a minimum, the highest and lowest test concentrations be analysed when freshly prepared and at the time of renewal on one occasion during the first week of the test For tests where the concentration of the test substance is not expected to remain within ± 20% of the nominal, it is necessary to analyse all test concentrations, when freshly prepared and at renewal If a flow‐through test is used, a similar sampling regime to that described for semi‐static tests is appropriate (but measurement of ‘old’ solutions is not applicable in this case) |
It is important that the test concentrations are measured, but the requirements will be different for TKTD modelling as information is required on the whole exposure profile. Therefore the information in the guideline should be adapted to suit the experimental conditions and exposure profile being used Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Freshwater alga and cyanobacteria growth inhibition test (OECD, 2011a) | The biomass in the control cultures should have increased exponentially by a factor of at least 16 within the 72‐h test period. This corresponds to a specific growth rate of 0.92 day−1. For the most frequently used species the growth rate is usually substantially higher (see Annex 2 of the guideline). This criterion may not be met when species that grow slower than those listed in Annex 2 of the guideline are used. In this case, the test period should be extended to obtain at least a 16‐fold growth in control cultures, while the growth has to be exponential throughout the test period. The test period may be shortened to at least 48 hours to maintain unlimited, exponential growth during the test as long as the minimum multiplication factor of 16 is reached | This criterion is directly relevant as it demonstrates suitable conditions to allow sufficient growth are maintained. However, this criterion relates to the standard static study where low number of algae is inoculated allowing exponential growth. Since the study design will need to be different for time‐variable exposure studies, a more detailed consideration of how to determine suitable control performance will be needed for flow‐through systems |
| The mean coefficient of variation for section‐by‐section specific growth rates (days 0–1, 1–2 and 2–3, for 72‐hour tests) in the control cultures (See Annex 1 of the guideline under – coefficient of variation‖) must not exceed 35%. See paragraph 49 for the calculation of section‐by‐section specific growth rate. This criterion applies to the mean value of coefficients of variation calculated for replicate control cultures | This criterion helps to demonstrate that the controls do not experience increasing stress during the study, but that growth is stable and unlimited. A similar quality criterion should be set up for flow through systems, if they are to be used to test time‐variable exposures | |
| The coefficient of variation of average specific growth rates during the whole test period in replicate control cultures must not exceed 7% in tests with Pseudokirchneriella subcapitata and Desmodesmus subspicatus. For other less frequently tested species, the value should not exceed 10% | This criterion is important because it helps to demonstrate that the controls are sufficiently similar to allow detection of differences in the treatment. A similar quality criterion could be applied for flow‐through systems monitoring control growth over time. The number of replicates and time‐points considered will, however, need reconsideration due to the different and more elaborate setup of flow‐through systems | |
|
ALSO (not a validity criteria): Provided an analytical procedure for determination of the test substance in the concentration range used is available, the test solutions should be analysed to verify the initial concentrations and maintenance of the exposure concentrations during the test Analysis of the concentration of the test substance at the start and end of the test of a low and high test concentration and a concentration around the expected EC50 may be sufficient where it is likely that exposure concentrations will vary less than 20% from nominal values during the test. Analysis of all test concentrations at the beginning and at the end of the test is recommended where concentrations are unlikely to remain within 80–120% of nominal. For volatile, unstable or strongly adsorbing test substances, additional samplings for analysis at 24 hour intervals during the exposure period are recommended in order to better define loss of the test substance |
It is important that the test concentrations are measured, but the requirements will be different for flow‐through systems as information is required on the whole exposure profile. Therefore, the information in the guideline should be adapted to suit the experimental conditions and exposure profile being used Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Lemna sp. Growth inhibition test (OECD, 2006 ) | For the test to be valid, the doubling time of frond number in the control must be less than 2.5 days (60 h), corresponding to approximately a sevenfold increase in seven days and an average specific growth rate of 0.275 day−1. Using the media and test conditions described in this Guideline, this criterion can be attained using a static test regime. It is also anticipated that this criterion will be achievable under semi‐static and flow‐through test conditions. Calculation of the doubling time is shown in paragraph 49 | This criterion is directly relevant as it demonstrates suitable conditions to allow sufficient growth. Monitoring surface area/frond number on a daily bases allows checking if this criterion is met also during longer duration experiments where change of media and/or thinning is implemented |
|
ALSO (not a validity criteria): During the test, the concentrations of the test substance are determined at appropriate intervals. In static tests, the minimum requirement is to determine the concentrations at the beginning and at the end of the test In semi‐static tests where the concentration of the test substance is not expected to remain within ±20% of the nominal concentration, it is necessary to analyse all freshly prepared test solutions and the same solutions at each renewal If a flow‐through test is used, a similar sampling regime to that described for semi‐static tests, including analysis at the start, mid‐way through and at the end of the test, is appropriate, but measurement of ‘spent’ solutions is not appropriate in this case |
It is important that the test concentrations are measured, but the requirements will be different for TKTD modelling, as information is required on the whole exposure profile. Hence, for static systems more measuring times may have to be included to properly be able to model exposure profiles, and for semi‐static or pulse exposures this is even more important. Therefore the information in the guideline should be adapted to suit the experimental conditions and exposure profile being used Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Sediment free Myriophyllum spicatum toxicity test (OECD, 2014a) | For the test to be valid, the doubling time of main shoot length in the control must be less than 14 days. Using the media and test conditions described in this Guideline, this criterion can be attained using a static or semi‐static test regime | This criterion is directly relevant as it demonstrates suitable conditions to allow sufficient growth are maintained |
| The mean coefficient of variation for yield based on measurements of shoot fresh weight (i.e. from test initiation to test termination) and the additional measurement variables (see paragraph 37 of this guideline) in the control cultures do not exceed 35% between replicates | This criterion is important because it helps to demonstrate that the controls are sufficiently similar to allow detection of differences in the treatment | |
| More than 50% of the replicates of the control group are kept sterile over the exposure period of 14 days, which means visibly free of colonisation by other organisms such as algae, fungi and bacteria (clear solution). Note: Guidance on how to assess sterility is provided in the ring‐test report referenced in the guideline | This criterion is important because colonisation by other organisms could affect the results, increasing variability between replicates and making it harder to detect effects | |
|
ALSO (not a validity criteria): During the test, the concentrations of the test substance(s) are determined at appropriate intervals. In static tests, the minimum requirement is to determine the concentrations at the beginning and at the end of the test In semi‐static tests where the concentrations of the test substance(s) are not expected to remain within ± 20% of the nominal concentration, it is necessary to analyse all freshly prepared test solutions and the same solutions at each renewal |
It is important that the test concentrations are measured, but the requirements will be different for TKTD modelling as information is required on the whole exposure profile. Therefore the information in the guideline should be adapted to suit the experimental conditions and exposure profile being used Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
|
| Water sediment Myriophyllum spicatum toxicity test (OECD, 2014b) | For the test results to be valid, the mean total shoot length and mean total shoot fresh weight in control plants at least double during the exposure phase of the test. In addition, control plants must not show any visual symptoms of chlorosis and should be visibly free from contamination by other organisms such as algae and/or bacterial films on the plants, at the surface of the sediment and in the test medium | This criterion is directly relevant as it demonstrates suitable conditions to allow sufficient growth are maintained and because contamination could affect the results, increasing variability between replicates and making it harder to detect effects |
| The mean coefficient of variation for yield based on measurements of shoot fresh weight (i.e. from test initiation to test termination) in the control cultures does not exceed 35% between replicates | This criterion is important because it helps to demonstrate that the controls are sufficiently similar to allow detection of differences in the treatment | |
|
ALSO (not a validity criteria): The correct application of the test chemical should be supported by analytical measurements of test chemical concentrations Water samples should be collected for test chemical analysis shortly after test initiation (i.e. on the day of application for stable test chemicals or one hour after application for substances that are not stable) and at test termination for all test concentrations Concentrations in sediment and sediment pore‐water should be determined at test initiation and test termination, at least in the highest test concentration, unless the test substances are known to be stable in water (> 80% of nominal). Measurements in sediment and pore‐water might not be necessary if the partitioning of the test chemical between water and sediment has been clearly determined in a water/sediment study under comparable conditions (e.g. sediment to water ratio, application method, sediment type) See full information about analytical measurements in paragraphs 69–79 |
It is important that the test concentrations are measured, but the requirements will be different for TKTD modelling as information is required on the whole exposure profile. Therefore the information in the guideline should be adapted to suit the experimental conditions and exposure profile being used If a water sediment system is used this is likely to be extremely complicated for use in TKTD modelling because the pulses in the water and sediment will not coincide, so additional consideration of the route of exposure and internal transportation of chemicals between compartments is required Maintaining constant exposure is not required to use the study for TKTD but the exposure profile should be described. This applies even if the study aimed for constant exposure but did not achieve it |
When the toxicity studies for calibration and validation purposes are evaluated, it is in general important to check that the actual exposure profile in the study matches the intended profile in the test (+/− 20%); if it does not, then measured concentrations are used for the modelling, instead of nominal ones. In the case of fast dissipating substances, actual measured exposure profile in the test can be used.
If multiple experiments of reasonable quality are available and suitable for either calibrating or validating the TKTD model, cherry‐picking of data is not acceptable. In case one relevant data set is excluded from both calibration and validation, the choice should be appropriately justified.
In the case of GUTS, it is likely that the supporting data for both calibration and validation of the model will be laboratory toxicity studies with a range of exposure conditions (including Tier‐1 acute and chronic constant exposure and Tier‐2 time‐variable exposure) that are evaluated as part of the data package. As demonstrated in Jager (2014), the use of TKTD models for dose–time–response analysis invites a whole new and flexible view on optimal test design. In particular, classical LC50 estimates can be derived with a GUTS‐RED‐SD model even from experimental set‐ups that would have been considered as unsuitable for classical Tier‐1 regression analyses.
In the case of DEBtox models, the supporting data for the physiological part of the model were either already assessed beforehand, or need a separate evaluation than the one outlined in this SO. The only data to be evaluated for DEBtox model applications (once the physiological part of the model has been evaluated as acceptable to use in the risk assessment) are hence most likely toxicity studies focussing on sublethal effects for a range of exposure conditions, in order to calibrate and to validate the TKTD part of the model Depending on the DEB model being used, it is pivotal that experimental factors like temperature, food conditions, etc. are well documented for all valid data sets.
Models for primary producers are also divided into the physiological and the TKTD part. As with DEBtox models, once the validation of the physiological part of the model has been successfully achieved, the only additional data to be evaluated are the toxicity studies with a range of exposure conditions, in order to calibrate and validate the TKTD part of the primary producer model. It is very important that experimental conditions influencing growth (i.e. temperature, irradiance, nutrient media composition, handling and thinning, etc.) and growth calculations (frequency and type of measurements, calibration between surface and weight data, etc.) are suitably described and documented.
7.3. Evaluation of the conceptual model
For GUTS, the conceptual model is standardised in the sense that the same processes are used whatever the toxicant‐species combination. When the model is used to address lethal/immobility effects in fish or invertebrates, the conceptual model is considered suitable to address the SPGs; so, no further evaluation is required (see Chapters 3.3 and 4). If a GUTS model is being used for any other reason, then consideration as to whether it is a suitable model to address the question being asked is required and whether the question is appropriate in the context of risk assessment.
For DEBtox, the conceptual models outlined in Section 2.3 of this SO can be used as basis for the documentation of the conceptual model, but the conceptual model for the species under consideration needs to be explicitly checked (see Chapter 10.2 of EFSA PPR Panel, 2014). Even if the DEB (physiological) part of the model has been previously tested and validated for the species under consideration, there are different ways the toxicant can impact the life cycle (namely how the toxicant impacts the energy acquisition or use). The processes impacted by the toxicant should be carefully considered. Based on the available information, all possible processes where the toxicant could have an impact need to be considered (see Figure 3 in Chapter 2). In case environmental factors (e.g. temperature) are explicitly considered, their influence on the physiological part and TKTD processes needs to be documented in the conceptual model.
For primary producers, the conceptual models outlined in Section 2.4 of this SO can be used as a basis for the documentation of the conceptual model. For Lemna spp. and algae, this SO has evaluated the conceptual models and they are considered suitable to use without further evaluation, but for Myriophyllum spp. and other aquatic plants, the conceptual model still needs to be checked (see Chapter 10.2 of EFSA PPR Panel, 2014). For all primary producer species, the influence of environmental factors (e.g. light, temperature) on growth and TKTD processes in the conceptual model and the linking of those processes in the model need to be documented.
7.4. Evaluation of the formal model
The formal model documents the equations and algorithms that form the basis for the model.
For GUTS models, the equations are standardised; so, no further check is necessary. It has to be documented, however, which GUTS model version is used (full or reduced model).
For DEBtox models, a large body of equations has been defined in the scientific literature, but no standardisation was achieved up to now. In addition, at the moment, internal exposure is linked with toxicant effects in a tailor‐made modus operandi. Hence, a DEBtox model application should document all equations that are used for the modelling. Evaluation of the formal model would need to be achieved on a case‐by‐case basis by experts, until standardised DEBtox modelling including links between internal exposure and toxic effects becomes available. All variables and parameters should be listed and both the deterministic and stochastic parts of the model should be fully described (see Section 5.1).
Primary producer models are formulated in a species‐specific fashion, considering biological/life‐history traits that have an influence on the conceptual and hence also on the formal model, e.g. whether plants are rooted in sediment, or they occur/grow predominantly at the water surface or submerged in the water column. For each model application, all equations that are used for the modelling should be documented, and evaluation of the formal model would need to be achieved on a case‐by‐case basis by experts, until standardised models become available, likely specific for plant/algae species.
7.5. Evaluation of the computer model
For GUTS models, checking the implementation of the computer model should include three lines of evidence:
The proposed implementation should be tested against the ring‐test data set (Jager and Ashauer, 2018). The performance of the model with the ring‐test data sets can be used to confirm that the computer model is working as it should. It is efficient and safe to test the computer code by showing that a given model implementation produces similar outputs (model parameters, confidence limits, model predictions) as previous GUTS implementations. Results of the GUTS implementation in the R package ‘morse’ (Appendix B.6) and Mathematica (B.7) are provided as an example;
A set of scenarios (default, pulsed and ‘extreme’ cases, see Section 4.1.2.3) should be simulated and checked;
An independent implementation of GUTS (e.g. an Excel sheet or a web‐based shinyApp – http://lbbe-shiny.univ-lyon1.fr/guts-shinyapp/) should be used by the evaluator to test whether the output of the evaluated model implementation can be reproduced for some parameter sets.
In addition, the computer code of the model implementation should be made available to allow further checks by experts.
A ring‐test equivalent to the one available for GUTS models is not available for DEBtox or primary producer models yet. Therefore Chapter 10.4 of EFSA PPR Panel (2014) should be used to check the computer model code. Such an evaluation includes basically similar steps as described in Section 4.1.2 and needs expert assessment.
7.6. Evaluation of the regulatory model
The regulatory model is the combination of the computer model with the environmental scenarios and the model parameters. Figure 5 in EFSA PPR Panel (2014) has been adapted to show the relevant parts of the general regulatory model that apply to TKTD models (Figure 38).
Figure 38.
Schematic representation of a TKTD regulatory model (orange boxes). The environmental scenario feeds into the exposure model in combination with information on pesticide properties and uses. The exposure model in turn delivers the exposure profile, which is used as input by the TKTD models. Altogether, the regulatory model delivers an output, i.e. the endpoint of the assessment. For DEBtox and primary producers’ models, as indicated by the dotted arrow and the associated box, the recommendation is, for the time being, to fix the abiotic parameters to the laboratory conditions of the experiment used for calibrating the model. If in the future the relationship between toxicity and environmental conditions will be better understood and described, such recommendation can be revised. The figure represents an adaptation of Figure 5 in EFSA PPR Panel (2014)
7.6.1. Evaluation of the environmental scenarios
One of the benefits of TKTD modelling is that all the data (simulated time series instead of maximum or average concentrations) from the predicted exposure profiles (e.g. from the various FOCUS scenario) can be used as input for simulations.
For GUTS models using FOCUS scenarios as input, no further definition of the environmental conditions is needed, since the model parameters will be based on data obtained from experiments performed under standard laboratory conditions, and pesticide concentrations will have been generated using the relevant FOCUS scenarios, which consider factors such as soil type, rainfall and agronomic practice.
For DEBtox models, a basic environmental scenario has to be defined including temperature and food conditions. The physiological part of the model has the potential to extrapolate predictions of growth and reproduction to variable environmental conditions, in terms of e.g. temperature and food availability. Basically, accounting for variable energy intake is one of the main ideas behind DEB modelling. The application of the DEB theory for the prediction of toxicological effects, however, requires environmental scenarios to be fixed to the laboratory conditions of the experiments used for calibrating the TKTD part of the model. This appears necessary, because it is not possible to assess whether the extrapolation of toxicity to other environmental conditions would work. In case appropriate validation data for variable temperature or food conditions is provided, explicit extrapolation to variable environmental conditions and hence the definition of different environmental scenarios could be possible.
It is considered appropriate to fix abiotic factors of the environmental scenarios to the laboratory conditions of the experiments used for calibrating the TKTD part of the model, and also to ignore ecological factors such as species interactions. This is because the modelling will be used with the equivalent of Tier‐1 or Tier‐2 Assessment Factors, which should already cover for the extrapolation from laboratory to field conditions.
For primary producer models, a basic environmental scenario has to be defined including temperature and/or light conditions, and nutrients availability. The conditions used in the model should mirror the conditions in the experiments used for calibration, since this is in line with Tier‐2A and Tier‐2B, where additional species or refined exposure scenarios are tested in laboratory experiments without also using realistic light conditions and nutrient levels.
7.6.2. Evaluation of parameter estimation
Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine and the comprehensive documentation of methods and results. Supporting data for TKTD models have to be of sufficient quality (Section 7.2), be relevant for the risk assessment and fulfil a set of basic criteria. One important aspect about the data requirements is to have experimental observations of the relevant endpoints (e.g. mortality or reproduction) for a sufficient number of time‐points in order to have an appropriate degree of freedom to calibrate the model. The number of observations can vary per TKTD model, suggestions are given in the respective checklists (Annex A, Annex B and Annex C). Another important aspect for the calibration data set is that the response in the observed endpoint should range from no effects up to strong effects, ideally from 0% to 100%. The model can only capture the quantitative relationship between exposure and effects when a clear effect is visible in the underlying data. Parameter estimates can show reduced accuracy and precision when the experiments do not show a clear and comprehensively covered dose–response pattern. It is difficult to define a minimum required level of response in the data, but a maximum effect of less than 50% in acute assessment for effects on invertebrates and chronic effect on plants, and of less than 90% in chronic assessments for effects on aquatic invertebrates and fish, should be well justified. In addition, the time course of the experiment used for calibration should be adjusted to capture the full toxicity of the pesticide (attention should be paid to time needed for the onset of effects, occurrence of delayed effects, accumulated toxicity, etc.).
It is possible that more than one suitable data set for calibration of the model does exist, since all available reliable data sets (apart from those used for validation) should be used (even non‐standard test data when available and considered reliable). In such a case, parameter optimisation can be done separately per data set, resulting in one optimal parameter set per data set. There are at least two strategies how to continue from there. One option is to predict the validation data set based on each parameter set and to select the parameter set that shows the best performance (see Section 7.7.2). The other option is to test whether the distributions of the parameter values are significantly different, to merge the corresponding data sets and to re‐calibrate the parameters for the merged data set in case they are not.
Some TKTD model parameters may also be taken from the literature. If this is the case, it should be checked whether they are reasonable, and if uncertainty limits are also provided.
Model parameters are always estimated for a specific combination of species and compound (see Chapter 3 for background information). For the evaluation of parameter estimation, also called model calibration or fitting, the method used has to be documented in detail, including settings of optimisation routines, the numerical solver (including settings) that was used for solving the differential equations, and the method for the calculation of parameter confidence/credible limits. Knowledge about these settings is important to allow experts to further evaluate parameter estimation when there is uncertainty about the quality of the results. Detailed background information on parameter estimation is given in Section 4.1.3, and example documentations for parameter estimation process and results are given in Section 4.2.2.1 and Appendix A.1 and A.2.
The results of the parameter optimisation process are not limited to the optimal parameter set alone. The optimal parameter values must be given together with confidence or credible intervals to allow evaluation of their uncertainty. Values of the objective function for calibration (e.g. log‐likelihood function) need to be documented with sufficient numerical precision to allow for independent checks of the optimisation process. For an evaluation of the model calibration, plots of the calibrated models in comparison with the calibration data over time should be available and the visual match should be reasonable. Additional goodness‐of‐fit criteria can be reported, at least a posterior predictive check should be available and documented (example in Figure 17).
One specific of the parameter estimation for GUTS is how the background mortality rate is considered in the optimisation. The standard way is to estimate all parameters simultaneously, including the background mortality in the experiments because control data may contain information about TKTD parameters; in addition, it is also possible to check for potential correlations between model parameters. Alternatively, the background mortality rate constant can be calibrated to the observed mortality in the controls and be fixed in the calibration of the other GUTS parameters to data from the treatments.
All aspects in this section are condensed into corresponding checklists in Annex A (GUTS), Annex B (DEBtox) and Annex C (primary producers).
7.7. Evaluation of model analysis
Model analysis includes sensitivity analysis, uncertainty analysis and evaluation of the model by comparison of the model output with independent experimental data (model validation).
7.7.1. Sensitivity and uncertainty analysis
Sensitivity analysis quantifies the influence of parameters on the model outputs.
For the reduced GUTS models, the influence of the model parameters on the model results is described in this SO (see Section 4.1.2.4) and does not need to be reported on a case‐by‐case basis. Results of sensitivity analyses for GUTS can, if contained, demonstrate that the model implementation is done correctly in the process of implementation verification. For other GUTS model versions than the reduced (see Section 4.1.1), sensitivity analyses should be included for future applications.
Uncertainty analysis aims at identifying how uncertain the model output is, given that the conceptual model is accepted. Uncertainty analysis is used to relate parameter uncertainty as captured in the parameter confidence/credible intervals to uncertainty in the model output.
Model calibration needs to report estimated parameter values including their uncertainty, i.e. the confidence/credible limits. It is demonstrated in Section 4.1.4.2 how the information about parameter uncertainty can be used to estimate the uncertainty of predicted model outputs. For example, for GUTS models, calculated exposure profile specific LPx or EPx values can be generated including uncertainty limits, that explicitly quantify the range of uncertainty in the model output that is caused by experimental variation and biological variability as captured in the model parameter estimates.
For DEBtox models, the sensitivity analysis of the physiological part, i.e. the discussion of the sensitivity of the DEB model parameters, should have already been performed and evaluated beforehand. A sensitivity analysis of the parameters for the TKTD part and a related discussion, however, is mandatory in the context of every regulatory risk assessment.
For primary producer models, the sensitivity analysis of the physiological part should already have been performed and evaluated beforehand. An example for Lemna is discussed shortly in Section 6.2.1.8, concluding that for future applications of the Lemna model in risk assessment, sensitivity analyses should be redone to allow for a comprehensive understanding of the variation in the model parameters. The sensitivity analysis of the TKTD part of primary producer models, however, is mandatory in the context of every regulatory risk assessment.
For DEBtox and primary producer models, similar approaches to quantify the uncertainty in model output as outlined in Section 4.1.4.2 are possible. Depending on the number of (relevant) model parameters, the calculation of confidence/credible intervals for the modelled endpoints may need considerable calculation time and effort; therefore, in some cases, simplified ways of uncertainty assessment may be acceptable, e.g. by reducing the analyses to a subset of the most relevant parameters. This is especially the case for DEBtox models, where it is possible that only parameters directly related to the TK and TD processes have to be considered for uncertainty analysis.
7.7.2. Evaluation of the model by comparison with independent experimental measurements (model validation)
Relevant model output
The performance of a model is usually evaluated by comparing relevant model outputs with independent measurements, a process often referred to as model validation. Relevant outputs in the context of model use in risk assessment are certainly those being used for answering the regulatory question, e.g. simulated survival and related LPx/EPx values in case of GUTS survival modelling, and predictions of reproduction and growth and EPx values for DEBtox and primary producer models.
Evaluation of the experimental data sets
Considering the extrapolation of effects from constant to time‐variable exposure patterns, effect data from the experiments with time‐variable exposure appear to be the most meaningful and relevant for the evaluation of TKTD models. Ideally, laboratory studies which tested different time‐variable exposure profiles for the compound, species and endpoint to be modelled, (e.g. invertebrate survival in case of GUTS, or growth and reproduction data for DEBtox and plant models) should be available for calibration For all model types, calibrated models could be used to design validation experiments, which can in turn generate data sets that cover the most important aspects of model validation.
The quality of the experimental data, which are used for comparison of the model, needs to be properly assessed. To that purpose, the availability of comprehensive documentation is important (see Section 7.2). Particular attention should be paid on the duration of the experiment, which should be adjusted to capture the full toxicity of the pesticide (time needed for the onset of effects, occurrence of delayed effects, accumulated toxicity, etc.).
For the evaluation of experimental data set for validation, it is important that the test design should be tailored to the question at hand. Nevertheless, it appears important to differentiate between invertebrates and primary producers on one side, and aquatic vertebrates (e.g. fish) on the other.
For invertebrates and primary producers, new experiments for model validation can be performed as needed, with reasonable time, effort and costs. Ideally, as mentioned before, the validation experiments are designed on the basis of the calibrated model, in order to address the most critical aspects.
For vertebrates like fish, on the other hand, ethical concerns imply that the number of experiments should be minimised as much as possible and should only be undertaken when no other methods are available (see also Regulation 1107/200913 ). When evaluating the adequacy of validation data for fish, the rationale should be set to ensure as much use of existing data as possible while additional studies should only be generated if really essential to produce and validate a suitably reliable model for use in the risk assessment. Given the relevance of the Tier‐2 risk assessment, it is still possible to argue for, and justify, a new fish test under time‐variable exposure conditions. Nevertheless, the number of tested scenarios and replicates should be reduced to the minimum needed to gain the necessary information, so may be lower in comparison to invertebrate or primary producers’ testing. A decision whether data sets for the validation of TKTD modelling for fish are appropriate and sufficient should still be made on a case‐by‐case basis, considering the relevance of the tested exposure profiles and the tested life stages among other aspects. In general, in light of the Regulation requirement about the reduction of vertebrate testing, the use of TKTD modelling for fish with the only scope of reducing or eliminate the use of risk mitigation measures, is discouraged as this would imply further vertebrate testing for the validation.
Criteria for the validation data sets are described in Section 4.1.4.5, but it is noted that whilst the criteria set out need to be met for non‐vertebrates (invertebrates, plants and algae) this is not necessarily essential for vertebrates.
Accounting for the fact that new experiments for model validation probably have to be performed, a set of standard requirements for validation data sets for invertebrates and primary producers is developed as follows. Two exposure profiles with at least two pulses each, separated by no‐exposure intervals of different duration should be tested on their effects. From these tests, the effect observations are reported for an appropriate number of time‐points, for example mortality or immobility should be reported at least for seven time‐points in the validation data set (see Section 4.1.4.5). The duration between the peaks of the two exposure profiles should be defined in relation to the individual depuration and repair time (DRT95; see Section 4.1.4.5 and Figure 39), as far as applicable.14 For animals, DRT95 values should be calculated and the duration of the no‐exposure intervals defined accordingly: one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile larger than the DRT95. In the case that DRT95 values are larger than it can be realised in validation experiments, or even exceed the lifetime of the considered species, the second tested exposure profile may be defined independent from the DRT95. For algae, internal concentrations are considered in fast equilibrium with the external concentrations and hence depend on them. For the macrophytes, the ‘life time’ is defined as the realistic duration of an experiment following standard guidelines. For Lemna spp. the duration is approximately two weeks, with water change and thinning after one week, while for Myriophyllum spp. the maximal duration will be approximately three weeks. These two different exposure profiles should be tested for at least three concentration levels, corresponding to low, medium, and strong effects in the specific dose–response curves (see Figure 22 for an example). Suggested are peak concentrations that lead to exposure profile specific effects of 10%, 50% and 90% at the end of the respective experiment.
Figure 39.
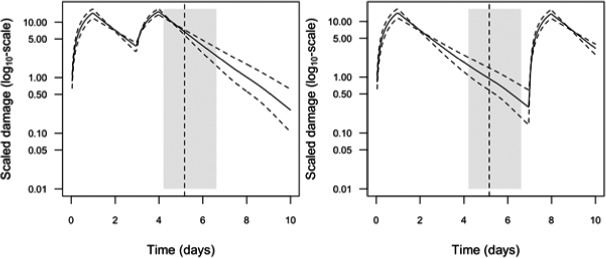
Example for exposure profiles that appear as toxicological dependent (left), or independent (right) regarding the simulated scaled damage (the solid line corresponds to the median curve, the dashed lines to the uncertainty limits). The dashed vertical line corresponds to the DTR 95, while the grey band corresponds to its 95% uncertainty range. In this example, kD = 0.732 day−1
It should be born in mind that failure to fully comply with the anticipated behaviour (e.g. no exposure intervals shorter/longer than DRT95, which is equivalent of having two toxicologically dependent/independent peaks) should not necessarily lead to rejection of the data set.
The evaluation of model validation for vertebrates needs to account for minimisation of vertebrate testing. The evaluator must also consider the following aspects when deciding if sufficient validation information has been provided:
Can existing data provide a reasonable data set (e.g. standard chronic studies might show sufficient mortality to be used as part of a GUTS validation even under standard exposure conditions)? This will also depend on the exposure profile in the standard study as the aim in the study is to maintain the concentration at 80% or above of nominal, however this may not always be achieved, particularly if a semi static design was used. In this case the standard study (with sufficient measurements of achieved concentrations) may provide a time‐variable exposure profile.
Can one study be designed to provide sufficient confidence to avoid testing two exposure profiles?
Can the number of animals used in the studies be reduced whilst still providing robust information?
When evaluating whether vertebrate model validation is sufficient, the evaluator needs to be careful to avoid the request of additional vertebrate data unless it is essential for decision‐making. For example, a greater level of uncertainty would be acceptable in the model output if the predicted LPx/EPx is high, because even if the model slightly under predicted risk, the actual risk would still not be high. As a minimum requirement, at least one validation experiment where time‐variable exposure was tested should be available.
Attention should be paid when different life stages are tested in the calibration and the validation experiments. As a general rule of thumb, early life stages are considered more sensitive. Hence, in the case of fish, a model calibrated on adult organisms could still be validated using an experiment carried out with swim‐ups. Nevertheless, very different life stages (e.g. adult fish and eggs) are likely to be subject to quite different toxicokinetic and toxicodynamic processes; therefore, a model calibrated on one life stage should not be used for predicting effects on the other, unless evidence is provided that this results in a worst‐case prediction.
Refinement options for vertebrates that do not require additional vertebrate testing are limited, so when TKTD modelling is provided, it can be used to obtain the maximum possible information from those studies that have been conducted.
Matching of the control data
For DEBtox and primary producer models, not only the toxicokinetic and toxicodynamic part, but also the physiological processes are included. Hence, modelled physiological states can be compared with independent control data, to corroborate the adequacy of the model to describe the physiological processes. Such states could be, e.g. for DEBtox the size at puberty. In some cases, relevant model parameters might need to be adapted to match the observed control dynamics.
Model performance criteria
Once the suitability and quality of the data set(s) used for model validation were evaluated, criteria for the model performance need to be defined and applied. For the optimisation of parameters, objective functions are used (e.g. the log‐likelihood function), but absolute values of these goodness‐of‐fit measures depend very often on the specifics of the experiments that were used for parameterisation (number of measurements, number of treatments) and hence cannot be used for a comparison with absolute evaluation criteria.
A combination of qualitative and quantitative criteria is suggested. Qualitatively, it can be checked whether the overall response pattern in the data is matched by the model output, e.g. whether the time‐points of increasing effects in model and data correspond with each other and whether the behaviour over time is consistent. The visual match (‘visual fit’ in FOCUS Kinetics, 2006) of the model prediction quality gives a basis for the acceptability of the model predictions in comparison with the data.
In addition to the qualitative visual match (‘visual fit’ in FOCUS Kinetics, 2006), three quantitative criteria are suggested for the evaluation of model validation results: the PPC takes into account the uncertainty in the model predictions, while the NRMSE value considers the relationship between median predicted and observed numbers of survivors over time, and the SPPE considers the absolute deviation of the median survivor number at the end of the experiment (see Section 4.1.4.5 for the definition of these criteria). These indicators need to be evaluated in concert, and an overall conclusion should consider that a validation study which shows clear agreement with all three quantitative criteria is for sure acceptable, while it might be tolerated if one indicator is close to the critical limit (e.g. PPC around 50%), when the other two indicators indicate a good validation quality. As another example, a deviation of the SPPE value could be tolerated if the model predictions overestimate the effects, since this would be conservative for the use in risk assessment, as long as the two other criteria indicate acceptable quality.
For the GUTS models, these quantitative performance criteria have to be checked with care, because GUTS models are fitted to deaths‐per‐time‐interval, hence comparing observed and predicted survival frequencies is formally not appropriate for testing the model performance (see Chapter 4). However, survival frequencies are more relevant for ERA, and if the fit is good, both comparisons will yield a good performance.
Parameter uncertainty is precisely captured in the parameter confidence/credible limits and is propagated to modelled outputs. It is also considered when evaluating model predictions using the PPC criterion (see Section 7.6.1).
While the suggested model performance criteria have been tested and documented with GUTS, their adequacy and performance for the other TKTD model types needs to be tested in the future, and possible adjustments of the model performance criteria may appear suitable. It is, for the time being, suggested to use the same performance criteria as suggested for GUTS (Section 4.1.4.5).
7.8. Evaluation of model use
According to EFSA PPR Panel Opinion on good modelling practice (EFSA PPR Panel 2014), the risk assessor should be able to re‐run the modelling using either the parameter values provided or alternative values as a way to both verify the submitted results and check whether the model behaves reasonably. A full access to the model allows the risk assessor to run alternative exposure scenarios in case they consider that the applicant has not provided the required exposure profiles. This requires that either the model source code is provided in case the model is run in a standard software (e.g. in R or Mathematica), or alternatively, an executable file or web‐based access to the model is provided.
Ideally, standard software should be available, so that applicants and Member State assessors can run the models, both for calibration and testing scenarios. This would provide a high level of confidence in the underlying model application, allowing the evaluation to focus on the specific model application (in the way FOCUS models are used for the exposure).
In the absence of standard software, it would still be of benefit to both the applicant and the assessor if an implementation of the model can be provided. One key benefit is that, should there be any concerns about the exposure profile(s) used; the assessor would have the opportunity to re‐run the modelling with alternative profiles. Although this is considered to be beneficial, it is not considered to be essential, noting the difficulty in providing the implementations that can be used by non‐TKTD modelling experts. In case executable implementation of the model is not made available to the assessor, at least the source code should be provided.
For the use of the GUTS model in regulatory risk assessment, levels of background mortality as observed in the calibration or validation data sets should be noted, but for simulations, background mortality is assumed to be 0, since the only interest is in the toxic effects. For DEBtox and primary producer models, the control data for calibration and validation may show different behaviour, which makes the adaptation of some model parameters necessary. In such a case, the influence of the adapted model parameter on the relevant model endpoints needs to be evaluated. When such influence is substantial, the parameter set producing the worst‐case prediction should be used for risk assessment.
It is acknowledged that standard software suitable for general (non‐expert) use is not yet at hand, although for GUTS there are some options available. MOSAIC (MOdeling and StAtistical tools for ecotoxicology) developed at the University of Lyon contains a GUTS tool (http://pbil.univ-lyon1.fr/software/mosaic/guts) which can be used for calibration. Moreover, GUTS‐ShinyApp (http://lbbe-shiny.univ-lyon1.fr/guts-shinyapp/) can be used to simulate predictions of survival for different exposure profiles with different TKTD parameter values for both GUTS‐RED‐SD and GUTS‐RED‐IT models.
The summary sheet (Annex D) provided by an applicant can be a helpful tool for the evaluators to highlight the critical information and to guide them through what is likely to be a complex evaluation. So it is highly recommended for applicants to provide such model summary.
For the time being, evaluators would not always be able to re‐run all modelling. Therefore, it is very important that the documentation is clear and can be checked by the evaluator. The modelled exposure scenario(s) must be fully documented. If FOCUS modelling is used for the TKTD risk assessment the following items need to be checked:
Are the input values used for FOCUS clearly stated?
Which FOCUS Step is used? (It is expected that the results will be from Step 3 or Step 4 – if Step 4, is it clear what risk mitigation has been used).
Are these the same values that have been used in the Tier‐1, Tier‐2A or Tier‐2B risk assessment? If not, has the reason for any differences been clearly justified?
Has the modelling used for all tiers been accepted in the Environmental Fate evaluation? If not, the modelling will usually have to be redone using the accepted inputs.
Overall, the evaluation needs to conclude whether the use of the model is appropriate to answer the regulatory question identified in the problem formulation section.
8. Use of TKTD models in Tier‐2C risk assessment
8.1. Introduction
The aim of this chapter is to illustrate the possible use of calibrated and validated TKTD models as tools in the Tier‐2C risk assessment for plant protection products. This will be done by describing in a concise way the important steps that need to be considered when conducting an ERA by means of calibrated and validated TKTD models. The description of the approach is followed by an example data set for an organophosphorus insecticide (Organophosphate A) that aims to compare the outcome of the different ERA tiers, in order to put the use of TKTD modelling as a Tier‐2C approach into perspective (see Figure 6 in Chapter 3). The use of TKTD models as Tier‐2C tools to refine the risks of time‐variable exposure will require exposure profiles of an active ingredient as input. These input exposure profiles should be sufficiently realistic in terms of the use of the pesticide in a certain crop and related soil, weather, field topography and types of edge‐of‐field surface waters present, as well as consider relevant risk mitigation measures when implemented. Consequently, in case the FOCUSsw methodology is used, the exposure profiles produced in steps 3 and 4 are most appropriate.
8.2. Steps that need to be considered in Tier‐2C ERA based on TKTD models
The step‐wise approach described in the decision schemes (Figures 40 and 41) and text below is developed for illustration of the use of TKTD modelling in ERA. These steps are thus restricted to TKTD modelling and do not address experimental studies to directly derive Tier‐2 RACs (see Figure 6 in Chapter 3). These Tier‐2C refined exposure tests are described in greater detail in EFSA PPR Panel (2013).
Figure 40.
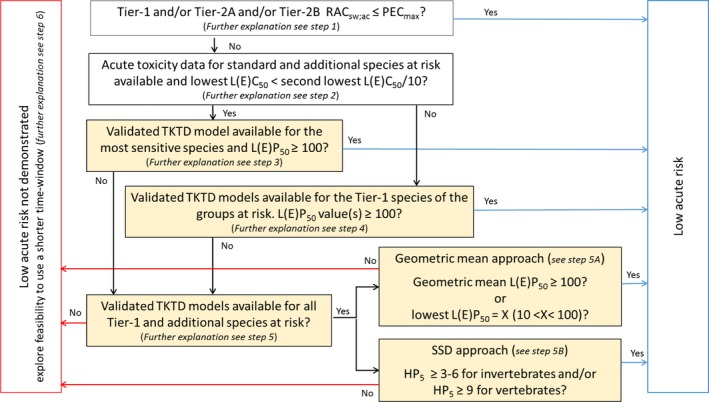
Decision scheme for the use of TKTD models and estimated L(E)P50 and HP 5 values in acute ERA. The boxes based on TKTD modelling are shaded. For further explanations, see the different steps (ad steps 1–6) described below
Figure 41.
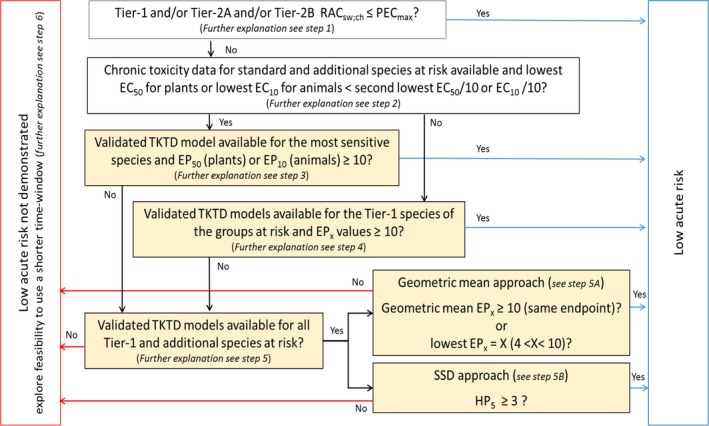
Decision scheme for the use of TKTD models and estimated EP 10/EP 50 and HP 5 values in chronic ERA. The boxes based on TKTD modelling are shaded. For further explanations, see the different steps (ad steps 1–6) described below
Step 1
Identify the taxonomic groups of aquatic organisms potentially at risk in the acute and chronic risk assessment for the substance of concern, informed by the relevant PECs (e.g. FOCUS step 3 or 4) and RACs derived by means of the Tier‐1, and if available, the Tier‐2A and/or Tier‐2B effect assessment approaches. Collect the annual exposure profiles for all relevant exposure scenarios and determine whether a Tier‐2C approach using TKTD modelling may be a suitable approach to better describe the risks of time‐variable exposures in every situation. However, TKTD modelling is more likely to be helpful to refine the risk if the exposure profiles are characterised by one or more pulse durations associated with the PECmax that are smaller than the durations of the toxicity tests that drive the risk. If the PECsw;max is larger but the corresponding PECsw;twa (time‐window smaller than the duration of the toxicity test that drives the risk) is smaller than the Tier‐1 RACsw;ch, this information can be used as an indication that TKTD modelling may be an appropriate Tier‐2C approach. If TKTD modelling is considered to be helpful to refine the risks, go to step 2.
Step 2
If, based on laboratory tests, the toxicity of the most sensitive additional test species (for the taxonomic group identified to be at risk) deviates more than an order of magnitude from any other Tier‐1 or additional test species, a risk assessment using a validated TKTD model for that particularly sensitive species (at least one order of magnitude more sensitive) is considered a cost‐effective and appropriate approach. If not, in first instance, a Tier‐2C1 assessment based on TKTD models for the Tier‐1 standard test species at risk should be explored.
Step 3 (Tier‐2C2 based on TKTD modelling for species that are at least one order of magnitude more sensitive than others)
A validated TKTD model for the particularly sensitive species and the substance of concern is used. Use in first instance the complete annual exposure profiles to calculate LPx and/or EPx values. In the acute risk assessment, the EP50 value for this species (or LP50 value for a fish/amphibian) for each relevant annual exposure profile should at least be larger than 100. In the chronic risk assessment, the EP10 (or currently EP50 for primary producers) should at least be larger than 10.
Step 4 (Tier‐2C1 based on TKTD models for standard test species)
If only toxicity estimates for Tier‐1 standard test species are available, or the lowest toxicity estimate for additional test species (Tier‐2A; Tier‐2B) of the sensitive taxonomic group does not deviate more than one order of magnitude from any other test species, a Tier‐2C1 assessment is proposed. In this approach, validated TKTD models for the standard (Tier‐1) test species of the taxonomic groups at risk and the substance of concern are used. Use the complete annual exposure profiles to calculate LPx and/or EPx values, i.e. LP50 for fish and amphibians and/or EP50 values for invertebrates (considering immobility) in the acute risk assessment, and in the chronic risk assessment EP10 values for aquatic animals and EP50 values for aquatic algae and vascular plants.
In the acute risk assessment, the exposure profile‐specific LP50/EP50 value for all relevant Tier‐1 species should at least be equal or larger than the acute Tier‐1 AF of 100 for risks to be considered low. In the chronic risk assessment, the exposure profile‐specific EP10 values (all relevant Tier‐1 animals) and the exposure‐profile‐specific EP50 values (relevant algae and/or aquatic macrophytes) should at least be equal or larger than the chronic Tier‐1 AF of 10 for risks to be considered low. Note that in a future update of the Aquatic Guidance Document, the effect assessment procedure for aquatic primary producers may be sharpened by using other criteria in lower‐tier assessments (i.e. a different ErCx and/or a different AF); the exposure‐profile‐specific EPx values would then be selected accordingly.
Step 5 (Tier‐2C2 based on TKTD models including standard and additional test species)
For the substance of concern, use validated TKTD models for the Tier‐1 and additional test species of the sensitive taxonomic group(s) at risk.
If for less than eight aquatic invertebrates and/or primary producers or for less than five fish species, appropriate TKTD models are made available, the calculated LP50/EP50/EP10 values for the different species and relevant annual exposure profiles may be used by adopting the geometric mean approach (currently predominantly in acute risk assessment only) and/or a WoE approach (go to step 5A).
If for at least eight aquatic invertebrates and/or primary producers or for at least five fish species, appropriate TKTD models are made available, the calculated LP50/EP50/EP10 values for the different species and relevant annual exposure profiles may be used by adopting the SSD approach (go to step 5B).
Step 5A (Tier‐2C2; geometric mean approach and weight‐of‐evidence approach
When using the acute Tier‐2C2 geometric mean approach, the rules as described in EFSA PPR Panel (2013) should be followed. In this approach, the geometric mean LP50/EP50 values are calculated for all relevant taxonomic groups separately (e.g. in insecticide risk assessments for crustaceans and insects separately, while this may be more taxonomic groups for fungicides). The exposure profile‐specific geometric mean LP50/EP50 values for all relevant taxonomic groups should be related to a multiplication factor (margin of safety) greater than or equal to 100 (the Tier‐1 AF) for risks to be considered acceptable.
In principle, the WoE approach might also be used in the acute risk assessment as an alternative (or validity check) for the geometric mean approach. Then, the lowest EP50 (or LP50 in case of fish) should be equal or larger than e.g. the AF used in the acute Tier‐2A WoE approach (see proposal by EFSA PPR Panel, 2015). So, depending on the number of species for which TKTD models are available for the substance of concern, the lowest LP50/EP50 for each relevant annual exposure profile should at least be larger than 10 but may be smaller than 100.
When using the chronic Tier‐2C2 WoE approach, the suggestions by EFSA PPR Panel (2015) concerning a reduced AF and the recommendations of EFSA PPR Panel (2013) concerning the endpoints to select, should be considered. In this approach, for each relevant exposure profile the lowest EP10 (or currently EP50 for primary producers) value calculated for the different species is selected. In the chronic risk assessment, the lowest EP10 (or currently EP50 for primary producers) of each relevant exposure profile should be equal to or larger than the AF that normally would be used in the chronic Tier‐2A WoE approach. So, depending on the number of species for which TKTD models are available for the substance of concern, the lowest EP10 (or currently EP50 for primary producers) should at least be larger than 4 but may be smaller than 10 (see proposal by EFSA PPR Panel, 2015).
In the near future, guidance should be developed on the rationale underlying the reduction of the AF when using the WoE approach in the acute and chronic risk assessment, based on the quantity and quality of the additional toxicity data made available.
Step 5B (Tier‐2C2; SSD approach)
When using the acute Tier‐2C2 SSD approach, the rules as described in EFSA PPR Panel (2013) should be followed. In this approach, for each relevant annual exposure profile the LP50/EP50 or EP10 calculated for the different species are used to construct exposure profile‐specific SSD graphs. The HP5 from these SSDs (exposure profile‐specific HP5) is the exposure profile derived from the modelled exposure profile that causes a specified effect on 5% of the species tested. For risk assessment, the median HP5 is selected. In the acute risk assessment for aquatic invertebrates, this median HP5 (based on acute toxicity data) should at least be equal to or larger than 3–6 (the AF used in the acute Tier‐2B effect assessment), while for aquatic vertebrates this median HP5 should at least be equal or larger than 9. In the chronic risk assessment (based on chronic toxicity data), the median HP5 should at least be greater than or equal to 3 (the AF used in the chronic Tier‐2B effect assessment), both for aquatic plants and animals.
Step 6
If risks on the aquatic taxonomic groups of concern cannot be excluded in Tier‐2C1 or Tier‐2C2 assessments when considering the whole annual exposure profile, the substance and species‐specific validated TKTD models may be used to evaluate the potential risks of a shorter time window within the annual exposure profile. Indeed, in the first step, it is considered that the same individual (for animals) or entity (cell for algae, frond for Lemna, shoot for rooted macrophytes) is present over the whole year whereas some species have a shorter life expectancy. Therefore, as a second step, it is proposed to use a reduced time‐window which should however be larger than the worst‐case estimate of the maximum life expectancy of individuals of the test species of concern that drive the risk. The full annual exposure profile should be evaluated by a moving time‐window approach and selecting the lowest LP50/EP50/EP10 value for use in the risk assessment as described in Tier‐2C1 or Tier‐2C2. It can be argued that considering yearly profiles even for short living organisms rather than selecting a reduced time‐window would be a conservative approach that enables to address potential transfer of PPP to the next generations. This however may be in conflict with other tiers within the same assessment scheme, since potential multigeneration effects are not considered in Tier‐1, Tier‐2A and Tier‐2B, nor in Tier‐3 micro/mesocosm experiments for those species that have a life cycle longer than the duration of these semi‐field tests (e.g. univoltine and semivoltine taxa).
8.3. Example data set on how to use GUTS in the ERA for Organophosphate A
8.3.1. Introduction
This example data set is based on realistic ecotoxicological data for an organophosphorus insecticide selected as benchmark compound to scientifically underpin the tiered aquatic risk assessment procedure. The presented field exposure concentrations are semi‐realistic in that they are based on realistic exposure concentrations for the benchmark compound but adapted by introducing mitigation measures in such a way that they better fit the goal of the case study. This case study aims to explore how GUTS modelling can be used as a Tier‐2C approach in the environmental risk assessment. In addition, this case study aims to compare the outcome of the different effect assessment tiers to put the use of TKTD modelling as a Tier‐2C approach into perspective.
8.3.2. Exposure concentrations in surface waters
FOCUS predictions for six relevant exposure scenarios (A–F) for different crops are selected, four drift (A, C, D and E) and two run‐off scenarios (B and F). In addition, five of these scenarios represent streams (scenarios A, B, C, D and F) and one ditches (scenario E) (see Table 7 and Figure 42).
Table 7.
Characteristics of the exposure scenarios for the example data set of Organophosphate A
| Scenario | Crop | FOCUS water body type and scenario | PECsw;max (μg/L) | PECsw;7d‐twa (μg/L) |
|---|---|---|---|---|
| A | Legumes | Stream D5 | 0.035 | 0.00019 |
| B | Vines | Stream R3 | 0.034 | 0.00033 |
| C | Pome | Stream D5 | 0.031 | 0.00014 |
| D | Maize | Stream D5 | 0.030 | 0.00035 |
| E | Maize | Ditch D6 | 0.029 | 0.00082 |
| F | Maize | Stream R3 | 0.029 | 0.00054 |
Figure 42.
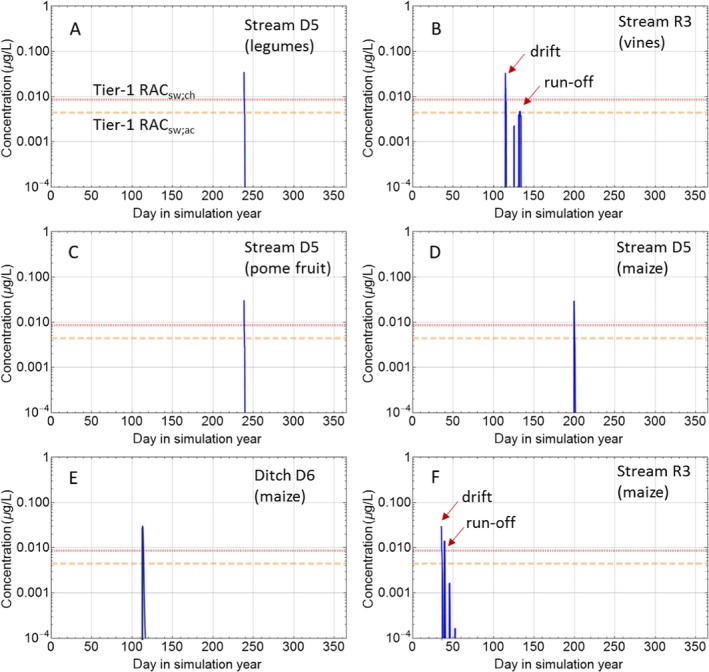
Exposure profiles for scenarios A–F (see Table 10). The Tier‐1 RACs are plotted in the profiles (see Section 8.4.1)
The insecticide is applied once a year and a spray drift causes the peak exposure, but in the run‐off scenarios the drift exposure peaks are followed by lower run‐off exposure peaks. The calculated PECsw;max values for the scenarios vary from 0.029 to 0.035 μg/L (Table 7). The highest 7‐day time‐weighted average PECs (PECsw;7d‐twa) values vary from 0.00019 to 0.00082 μg/L; a 7‐d time‐window is considered suitable in this context dealing with chronic tests on invertebrates. Although the PECsw;max value for ditch scenario E is lower than that for most stream scenarios, its PECsw;7d‐twa value is the highest. This can be explained by the much slower water flow in the ditch D6 scenario than in the stream scenarios. Exposure profiles for the six scenarios are presented in Figure 42.
8.3.3. Laboratory toxicity data for aquatic organisms
Laboratory toxicity data for standard aquatic species as requested by the data requirements (Reg. 283/2013) are presented in Table 8. In the acute data set, the EC50 values concern the effect endpoint based on mortality or immobility. As expected for an Organophosphate, the most sensitive standard test species in the acute and chronic Tier‐1 data set belong to the group of aquatic arthropods (Table 8). This case study has its focus on the application of the GUTS modelling approach since the lowest RAC is for the acute toxicity, but if the modelling is able to demonstrate a low acute risk the focus of the risk assessment would then return to the chronic risk which would also need to be addressed.
Table 8.
Toxicity data and Tier 1 RACs for standard aquatic species considered valid in the dossier of Organophosphate A and used in the Tier‐1 effect assessment
| Species | Taxonomy | Timescale and endpoint | Toxicity (μg/L) | Assessment Factor | RAC (μg/L) |
|---|---|---|---|---|---|
| Daphnia magna | Crustacea; Cladocera | 48‐h EC50 | 0.48 (0.34–0.69) | 100 | 0.0048 |
| Chronic NOEC (μg/L) | 0.085 | 10 | 0.0085 | ||
| Chironomus riparius | Insecta; Chironomidae | 48‐h EC50 | 0.44 (0.32–0.59) | 100 | 0.0044 |
| Chronic NOEC (μg/L) | 0.096 | 10 | 0.0096 | ||
| Oncorhynchus mykiss | Fish | 96‐h LC50 | 18.8 | 100 | 0.188 |
| Chronic NOEC (μg/L) | 0.57 | 10 | 0.057 | ||
| Pseudokirchneriella subcapitata | Green alga | Chronic EC50 (μg/L) | 298 (197–603) | 10 | 29.8 |
Some acute toxicity values are also available for additional aquatic arthropods. All data from this acute data set (standard and additional, 13 in total) are presented in Table 9.
Table 9.
Acute EC50 (95% confidence limit) data for standard and additional aquatic species considered valid in the dossier of Organophosphate A. NC = a reliable 95% confidence limit could not be calculated
| Species | Taxonomy | 48‐h EC50 (μg/L) | 72‐h EC50 (μg/L) | 96‐h EC50 (μg/L) | |
|---|---|---|---|---|---|
| 1 | Daphnia magna | Crustacea; Cladocera | 0.48 (0.34–0.69) | 0.25 (0.19–0.32) | 0.17 (0.12–0.23) |
| 2 | Asellus aquaticus | Crustacea; Isopoda | 6.16 (4.89–7.76) | 5.27 (4.07–6.82) | 3.43 (2.75–4.26) |
| 3 | Gammarus pulex | Crustacea; Amphipoda | 0.38 (0.20–0.70) | 0.24 (0.04–1.34) | 0.23 (0.20–0.25) |
| 4 | Neocaridina denticulata | Crustacea; Decapoda | 327 (NC) | 237 (147–381) | 171 (NC) |
| 5 | Procambarus sp. | Crustacea; Decapoda | 1.70 (1.03–2.80) | 1.29 (0.82–2.01) | 1.20 (0.75–1.93) |
| 6 | Chironomus riparius | Insecta; Chironomidae | 0.44 (0.32–0.59) | 0.32 (0.22–0.47) | 0.18 (0.07–0.43) |
| 7 | Anax imperator | Insecta; Anisoptera | 3.13 (NC) | 1.66 (1.55–1.77) | 1.63 (NC) |
| 8 | Cloeon dipterum | Insecta; Ephemeroptera | 0.76 (NC) | 0.41 (0.33–0.50) | 0.31 (0.26–0.38) |
| 9 | Notonecta maculata | Insecta; Heteroptera | 9.07 (7.18–11.5) | 6.06 (4.46–8.31) | 2.78 (NC) |
| 10 | Paraponyx stratiotata | Insecta; Lepidoptera | 2.94 (1.65–5.24) | 3.87 (2.14–6.92) | 2.86 (1.17–6.97) |
| 11 | Plea minutissima | Insecta; Heteroptera | 2.65 (2.06–3.39) | 1.55 (NC) | 1.29 (0.92–1.80) |
| 12 | Ranatra linearis | Insecta; Heteroptera | 12 (NC) | 4.40 (2.80–7.10) | 3.33 (2.95–3.76) |
| 13 | Sialis lutaria | Insecta; Megaloptera | 1.55 (0.25–9.58) | 1.07 (0.96–1.20) | 0.96 (NC) |
8.4. Tiered acute effect and risk assessment in line with the EFSA Aquatic Guidance Document
In this case study, the use of GUTS models in the acute risk assessment scheme will be illustrated. As far as the available data set allows, all possible tiers within the acute risk assessment scheme as proposed by the EFSA Aquatic Guidance Document (EFSA PPR Panel, 2013) will be given, to place the Tier‐2C ERA based on TKTD models into perspective.
8.4.1. Tier‐1 acute effect and risk assessment
Tier‐1 acute effect assessment
The Tier‐1 arthropods (D. magna and Chironomus riparius) are more sensitive than the Tier‐1 fish Oncorhynchus mykiss (Table 9). By applying an AF of 100 to the lowest toxicity value of 0.44 μg/L (48‐h EC50) for Chironomus riparius, the Tier‐1 RAC sw;ac becomes 0.0044 μg/L.
Tier‐1 acute risk assessment
The acute Tier‐1 RAC is always compared with PECsw;max values. The outcome of the Tier‐1 risk assessment procedure is presented in Table 10. If the PEC:RAC ratio is lower than 1, then the environmental risk is considered low. From the results presented in Table 10 it appears that when comparing the acute Tier‐1 RACs with the PECsw;max, potential environmental risks are triggered for all exposure scenarios.
Table 10.
Tier‐1 risk assessment procedure for Organophosphate A. If the PEC:RAC ratio is lower than 1, then the environmental risk is considered low
| Scenario | Acute risk assessment | ||
|---|---|---|---|
| PECsw;max (μg/L) | Tier‐1 RACsw;ac (μg/L) | PECsw;max:RACsw;ac ratio | |
| A | 0.035 | 0.0044 | 7.95 |
| B | 0.034 | 0.0044 | 7.73 |
| C | 0.031 | 0.0044 | 7.05 |
| D | 0.030 | 0.0044 | 6.82 |
| E | 0.029 | 0.0044 | 6.59 |
| F | 0.029 | 0.0044 | 6.59 |
8.4.2. Tier‐2A effect and risk assessment
A Tier‐2A approach may comprise the geometric mean approach based on experimental data as proposed in the EFSA PPR Panel Aquatic Guidance Document (EFSA PPR Panel, 2013) or a WoE approach as proposed in the EFSA PPR Panel sediment opinion (EFSA PPR Panel, 2015) when the Geometric mean is not appropriate (e.g. if different endpoints are measured in tests within species of the same relevant taxonomic group).
Tier‐2A acute effect assessment (Geometric Mean approach)
In total for 13 aquatic arthropods (5 crustaceans and 8 insect taxa) 96‐h EC50 values are available (Table 11). Consequently, this data set allows the Tier‐2B (SSD) approach (see Section 8.4.3). However, to illustrate the Geometric Mean approach, and to make this case more realistic, the 96‐h EC50 values for a lower number of aquatic species are selected in Table 11. The selected additional species are frequently tested in toxicity experiments, since they are abundant in freshwater ecosystems and can be easily kept under laboratory conditions.
Table 11.
Acute EC50 (95% confidence limit) data for standard and additional aquatic species considered realistic to explore the Tier‐2A effect assessment approach
| Species | Taxonomy | 96‐h EC50 (μg/L)a | |
|---|---|---|---|
| 1 | Daphnia magna | Crustacea; Cladocera | 0.17 (0.12–0.23) |
| 2 | Asellus aquaticus | Crustacea; Isopoda | 3.43 (2.75–4.26) |
| 3 | Gammarus pulex | Crustacea; Amphipoda | 0.23 (0.20–0.25) |
| 4 | Chironomus riparius | Insecta; Chironomidae | 0.18 (0.07–0.43) |
| 5 | Cloeon dipterum | Insecta; Ephemeroptera | 0.31 (0.26–0.38) |
| 6 | Plea minutissima | Insecta; Heteroptera | 1.29 (0.92–1.80) |
| Geometric mean 96‐h EC 50 for crustaceans | 0.51 | ||
| Geometric mean 96‐h EC 50 for insects | 0.42 | ||
Note that for Daphnia magna and Chironomus riparius the 48 h endpoint is normally selected for risk assessment but in this case the 96 h endpoints were used.
The geometric mean 96‐h EC50 value for the three crustaceans is 0.51 μg/L. The geometric mean 96‐h EC50 value for the three insects is 0.42 μg/L. Selecting the lowest value (0.42 μg/L for insects) and applying an AF of 100 results in a Tier‐2A RAC sw;ac of 0.0042 μg/L.
Tier‐2A acute risk assessment (geometric mean approach)
The Tier‐2A RAC based on standard and additional laboratory toxicity data is always compared with PECsw;max values. The outcome of the acute Tier‐2A risk assessment procedure is presented in Table 12. If the PEC:RAC ratio is lower than 1, then the environmental risk is considered to be low. From the results presented in Table 12, it appears that when comparing the acute Tier‐2A RACs with the PECsw;max, potential environmental risks are triggered for all exposure scenarios.
Table 12.
Tier‐2A risk assessment procedure for Organophosphate A. If the PEC:RAC ratio is lower than 1, then the environmental risk is considered low
| Scenario | Acute risk assessment | ||
|---|---|---|---|
| PECsw;max (μg/L) | Tier‐2A RACsw;ac (μg/L) | PECsw;max:RACsw;ac ratio | |
| A | 0.035 | 0.0042 | 8.33 |
| B | 0.034 | 0.0042 | 8.10 |
| C | 0.031 | 0.0042 | 7.38 |
| D | 0.030 | 0.0042 | 7.14 |
| E | 0.029 | 0.0042 | 6.90 |
| F | 0.029 | 0.0042 | 6.90 |
8.4.3. Tier‐2B effect and risk assessment
A Tier‐2B approach (SSD approach) is possible for the acute data set since for more than 8 different taxa of the sensitive taxonomic group (arthropods) laboratory toxicity data are available.
Tier‐2B acute effect assessment (SSD approach)
For the SSD approach, the 96‐h EC50 data mentioned in Table 9 are used. The SSD constructed with the 96‐h EC50 values for the 13 different aquatic arthropods is presented in Figure 43.
Figure 43.
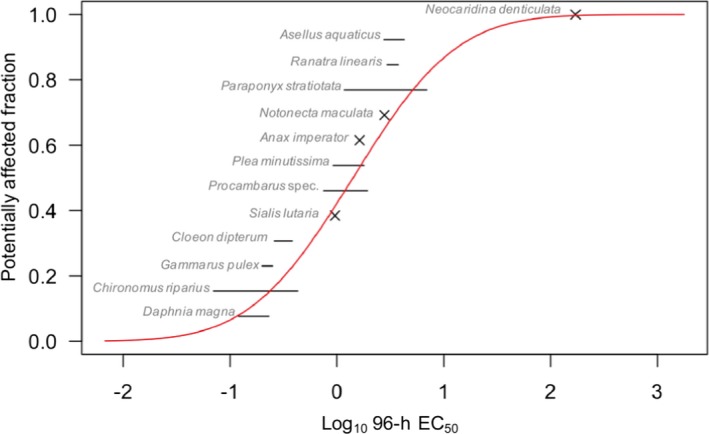
SSD graph constructed with 96‐h EC 50 values for 13 different arthropods presented in Table 9
Several valid approaches can be used to construct SSDs and to calculate HC5 values. As an example the computer program described by Charles et al. (2017) to calculate HC5 values is selected, since it allows to use censored data and 95% confidence limits of toxicity estimates as input data. Using the approach described in Charles et al. (2017) and a log‐normal distribution, the median HC5 value (and 95% confidence interval) on basis of 96‐h EC50 values for all aquatic arthropods (n = 13) is: 0.079 (0.031–0.370) μg/L. According to EFSA PPR Panel (2013) an AF of 3 to 6 should be applied to the median HC5. This results in a Tier‐2B RAC sw;ac of 0.0132–0.0263 μg/L.
Tier‐2B acute risk assessment (SSD approach)
The Tier‐2B RAC based on standard and additional laboratory toxicity data is always compared with PECsw;max values. The outcome of the acute Tier‐2B risk assessment procedure is presented in Table 13. If the PEC:RAC ratio is lower than 1, then the environmental risk is considered low. It appears that when comparing the acute Tier‐2B RACs with the PECsw;max, potential environmental risks are triggered for all exposure scenarios.
Table 13.
Tier‐2B risk assessment procedure for Organophosphate A. If the PEC:RAC ratio is < 1, then the environmental risk is considered low
| Scenario | Acute risk assessment | ||
|---|---|---|---|
| PECsw;max (μg/L) | Tier‐2B RACsw;ac (μg/L) | PECsw;max:RACsw;ac ratio | |
| A | 0.035 | 0.0132–0.0263 | 1.33–2.65 |
| B | 0.034 | 0.0132–0.0263 | 1.29–2.58 |
| C | 0.031 | 0.0132–0.0263 | 1.18–2.35 |
| D | 0.030 | 0.0132–0.0263 | 1.14–2.27 |
| E | 0.029 | 0.0132–0.0263 | 1.10–2.20 |
| F | 0.029 | 0.0132–0.0263 | 1.10–2.20 |
8.4.4. Tier‐2C acute risk assessment
The EFSA Aquatic Guidance Document (EFSA PPR Panel, 2013) allows experimental refined exposure tests to refine the RAC. The refined exposure experiments available for Organophosphate A were performed to validate the GUTS models. This was done by testing responses of test organisms subject to different pulses (around their 24‐h and 48‐h EC50s), that differed in duration (12–24 h) and intervals between repeated pulses. An example of the type of results thus obtained is presented in Figure 44 for the standard test species Chironomus riparius and D. magna. These experimental data can be used to validate the compound‐ and species‐specific GUTS models. It is noted that the validation data set does not meet all the validation criteria as set out in Section 4.2 (only 1 concentration level tested for each profile instead of 3). However, for illustrative purpose the available data set was still considered suitable as it is the most extensive data set currently available.
Figure 44.
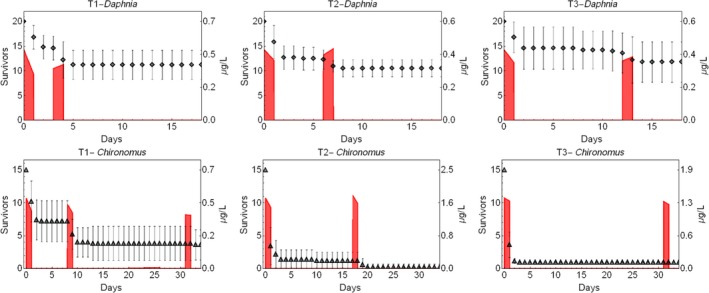
Results of experimental refined exposure tests with Organophosphate A and the aquatic standard test species Daphnia magna (top row) and Chironomus riparius (bottom row). These experiments were conducted with the aim to validate the corresponding GUTS models. The upper graphs present observed mobility over time (symbols; error bars give standard deviation based on 5 replicates) for the three treatment profiles T1, T2 and T3. Exposure profiles are plotted in red and concentration levels are indicated on the right hand axes per panel
The available experimental data allowed development of calibrated and validated GUTS models for all aquatic arthropods based on raw data delivering the relevant endpoint (calibration data as those delivering effect values reported in Table 9; validation data of the type shown in Figure 44). These GUTS models were used to assess the potential environmental risks for annual exposure profiles A–F.
Tier‐2C 1 acute risk assessment
Since at first, TKTD models for Tier‐1 species will be usually used in refined risk assessment (see the steps described in Section 8.2), the GUTS modelling approach was used to evaluate the potential risks of the time‐variable exposure profiles on the standard test species D. magna and Chironomus riparius (the acute Tier‐2C 1 approach). This is done by calculating EP50 values (based on the sum of dead and immobile individuals) for these species. In order not to be in conflict with the Tier‐1 effect assessment approach, the EP50 values should at least be equal to or larger than the multiplication factor (margin of safety) of 100 (the acute Tier‐1 AF). The EP50 values calculated by the GUTS‐RED‐SD and GUTS‐RED‐IT models for the standard test species D. magna and C. riparius are presented in Table 14.
Table 14.
Calculated EP50 values and 95% confidence limits for the Tier‐1 aquatic arthropods and exposure profiles A–F by using the GUTS‐RED‐SD and GUTS‐RED‐IT model
| Exposure scenario | ||||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| EP 50 values for the different exposure scenarios and GUTS‐RED‐SD | ||||||
| Daphnia magna | 13 (7–61) | 5 (2–33) | 18 (11–83) | 7 (4–35) | 3 (2–14) | 5 (3–25) |
| Chironomus riparius | 93 (75–122) | 38 (28–52) | 127 (103–163) | 51 (40–67) | 23 (18–30) | 37 (30–48) |
| EP 50 values for the different exposure scenarios and GUTS‐RED‐IT | ||||||
| Daphnia magna | 122 (97–145) | 39 (32–49) | 161 (127–191) | 66 (54–78) | 28 (23–32) | 40 (33–47) |
| Chironomus riparius | 93 (70–111) | 33 (26–44) | 127 (99–143) | 51 (42–60) | 22 (17–26) | 33 (27–39) |
When using the GUTS‐RED‐SD models, it appears that for all scenarios (A–F), risks are triggered (EP50 values for both standard test species should be greater than or equal to 100). The output of the GUTS‐RED‐IT models indicates that the risk is low in exposure scenario C only.
Tier‐2C 2 acute risk assessment
A more advanced approach is to calculate EP50 values for standard and additional aquatic taxa for which GUTS models are already developed (the acute Tier‐2C 2 approach). The Tier‐2C2 Geometric Mean approach normally will be used to assess the risks for the insecticide (Organophosphate A) if for less than eight aquatic arthropods calibrated/validated GUTS models are available. In Tables 15 (GUTS‐RED‐SD model calculations) and 16 (GUTS‐RED‐IT model calculations), this is explored based on the species also presented in Table 11.
Table 15.
Calculated EP50 values and 95% confidence limits for aquatic crustaceans and insects mentioned in Table 11 and exposure profiles A–F by using the GUTS‐RED‐SD models to explore the geometric mean approach
| Exposure scenarios | |||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| EP50 values GUTS‐RED‐SD | |||||||
| 1 | Daphnia magna | 13 (7–61) | 5 (2–33) | 18 (11–83) | 7 (4–35) | 3 (2–14) | 5 (3–25) |
| 2 | Asellus aquaticus | 762 (250–1,285) | 391 (92–635) | 1,035 (340–1,682) | 420 (138–682) | 181 (58–305) | 303 (85–587) |
| 3 | Gammarus pulex | 37 (6–62) | 13 (1–23) | 49 (6–85) | 20 (2–34) | 9 (0.1–15) | 13 (1–23) |
| Geometric mean Crustacea | 71.6 | 29.4 | 97.0 | 38.9 | 17.0 | 27.1 | |
| 4 | Chironomus riparius | 93 (75–122) | 38 (28–52) | 127 (103–163) | 51 (40–67) | 23 (18–30) | 37 (30–48) |
| 5 | Cloeon dipterum | 98 (75–110) | 57 (40–64) | 132 (101–153) | 54 (42–61) | 24 (18–27) | 49 (30–57) |
| 6 | Plea minutissima | 444 (392–491) | 271 (231–299) | 596 (531–661) | 247 (223–272) | 112 (91–125) | 247 (171–286) |
| Geometric mean Insecta | 159.4 | 83.7 | 215.9 | 88.0 | 39.5 | 76.5 | |
Comparing the Geometric mean EP50 values for crustaceans and insects presented in Tables 15 and 16, it appears that the GUTS‐RED‐SD models trigger risks for all scenarios (particularly the EP50 values for crustaceans are smaller than 100). For risks to be considered low, the Geometric mean EP50 values for both the crustaceans and insects need to be greater than or equal to 100. The results of the GUTS‐RED‐IT models indicate that risks are low for scenarios A, C and D only. Another observation in this example is that in GUTS‐RED‐IT modelling the EP50 values for insects are smaller than for crustaceans, while that appears to be the other way around in GUTS‐RED‐SD modelling.
Table 16.
Calculated EP50 values and 95% confidence limits for aquatic crustaceans and insects mentioned in Table 11 and exposure profiles A–F by using the GUTS‐RED‐IT models to explore the Geometric Mean approach
| Exposure scenarios | |||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| EP50 values GUTS‐RED‐IT | |||||||
| 1 | Daphnia magna | 122 (97–145) | 39 (32–49) | 161 (127–191) | 66 (54–78) | 28 (23–32) | 40 (33–47) |
| 2 | Asellus aquaticus | 1,445 (1,221–1,626) | 488 (416–671) | 1,953 (1,648–2,319) | 801 (651–901) | 342 (283–390) | 488 (412–580) |
| 3 | Gammarus pulex | 66 (49–80) | 22 (16–37) | 88 (69–110) | 35 (29–46) | 15 (11–19) | 22 (18–27) |
| Geometric mean Crustacea | 226.6 | 74.8 | 302.5 | 122.8 | 52.4 | 75.5 | |
| 4 | Chironomus riparius | 93 (70–111) | 33 (26–44) | 127 (99–143) | 51 (42–60) | 22 (17–26) | 33 (27–39) |
| 5 | Cloeon dipterum | 134 (117–155) | 49 (45–62) | 181 (158–203) | 73 (64–85) | 32 (28–36) | 48 (45–57) |
| 6 | Plea minutissima | 576 (514–641) | 208 (184–334) | 781 (691–854) | 317 (281–352) | 135 (122–153) | 205 (183–256) |
| Geometric mean Insecta | 192.9 | 69.5 | 261.8 | 105.7 | 45.6 | 68.7 | |
For more than eight different aquatic arthropods (in total 13 taxa; see Table 8) GUTS models were developed allowing a Tier‐2C2 risk assessment using the Species Sensitivity Distribution (SSD) approach. In Tables 17 (GUTS‐RED‐SD model calculations) and 18 (GUTS‐RED‐IT model calculations) this is explored.
Table 17.
Calculated EP50 (endpoint mortality plus immobility) and corresponding HP5 values (and 95% confidence limits) for all 13 aquatic arthropods (see Table 9) and exposure profiles of exposure scenarios A‐F by using the GUTS‐RED‐SD model
| Exposure scenarios | |||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| EP50 values GUTS‐RED‐SD | |||||||
| 1 | Daphnia magna | 13 (7–61) | 5 (2–33) | 18 (11–83) | 7 (4–35) | 3 (2–14) | 5 (3–25) |
| 2 | Asellus aquaticus | 762 (250–1,285) | 391 (92–635) | 1,035 (340–1,682) | 420 (138–682) | 181 (58–305) | 303 (85–587) |
| 3 | Gammarus pulex | 37 (6–62) | 13 (1–23) | 49 (6–85) | 20 (2–34) | 9 (0.1–15) | 13 (1–23) |
| 4 | Neocaridina denticulata | 42,500 (29,219–63,750) | 14,375 (9,883–26,504) | 57,500 (39,531–86,250) | 22,500 (16,172–36,563) | 9,688 (6,660–15,894) | 14,375 (9,883–24,482) |
| 5 | Procambarus spec. | 264 (206–330) | 142 (106–174) | 352 (275–467) | 146 (119–192) | 67 (54–92) | 129 (105–162) |
| 6 | Chironomus riparius | 93 (75–122) | 38 (28–52) | 127 (103–163) | 51 (40–67) | 23 (18–30) | 37 (30–48) |
| 7 | Anax imperator | 469 (432–501) | 291 (272–304) | 630 (582–674) | 261 (243–286) | 121 (114–126) | 278 (241–283) |
| 8 | Cloeon dipterum | 98 (75–110) | 57 (40–64) | 132 (101–153) | 54 (42–61) | 24 (18–27) | 49 (30–57) |
| 9 | Notonecta maculata | 1,162 (890–1,344) | 542 (333–805) | 1,572 (1,203–1,824) | 640 (492–748) | 276 (216–328) | 437 (315–628) |
| 10 | Paraponyx stratiotata | 557 (533–598) | 344 (331–363) | 747 (695–817) | 313 (300–333) | 143 (139–150) | 320 (288–360) |
| 11 | Plea minutissima | 444 (392–491) | 271 (231–299) | 596 (531–661) | 247 (223–272) | 112 (91–125) | 247 (171–286) |
| 12 | Ranatra linearis | 1,709 (1,282–2,016) | 1,025 (793–1,170) | 2,285 (2,000–2,633) | 947 (760–1,111) | 417 (313–484) | 796 (569–1,057) |
| 13 | Sialis lutaria | 109 (60–177) | 50 (27–100) | 146 (96–239) | 60 (39–98) | 26 (13–420) | 40 (26–78) |
| Median HP5 (log‐normal) (95% confidence interval) | 15 (5–80) | 7.3 (2–40) | 21 (6.6–110) | 8.5 (2.6–47) | 3.6 (0.9–20) | 6.7 (1.9–35) | |
In the Tier‐2C2 SSD approach, median HP5 values are calculated for each exposure profile. This median HP5 is the exposure profile‐specific EP50 that affects 5% of the taxa tested. In order not to be in conflict with the acute Tier‐2B effect assessment approach the median HP5 should at least be equal to or larger than a selected value in the range 3–6 (the acute Tier‐2B AF‐range).
The results of GUTS‐RED‐SD modelling (Table 17) show that risk might occur for scenarios E (median HP5 = 3.6) depending on the size of the AF (3–6) that is selected when applying the SSD approach. When using the GUTS‐RED‐IT (Table 18) models the median HP5 values are equal or larger than 8 (i.e. larger than the upper value of the AF) for all exposure scenarios, suggesting low risks of the time‐variable exposure regimes evaluated on mortality plus immobility of aquatic arthropods.
Table 18.
Calculated EP50 (endpoint mortality plus immobility) and corresponding HP5 values (and 95% confidence limits) for all 13 aquatic arthropods (see Table 8) and exposure profiles of exposure scenarios A–F by using the GUTS‐RED‐IT model
| Species | Exposure scenarios | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| EP50 values GUTS‐RED‐IT | |||||||
| 1 | Daphnia magna | 122 (97–145) | 39 (32–49) | 161 (9,127–191) | 66 (54–78) | 28 (23–32) | 40 (33–47) |
| 2 | Asellus aquaticus | 1,445 (1,221–1,626) | 488 (416–671) | 1,953 (1,648–2,319) | 801 (651–901) | 342 (283–390) | 488 (412–580) |
| 3 | Gammarus pulex | 66 (49–80) | 22 (16–37) | 88 (69–110) | 35 (29–46) | 15 (11–19) | 22 (18–27) |
| 4 | Neocaridina denticulata | 50,000 (39,063–65,625) | 16,875 (12,590–26,631) | 67,500 (51,680–90,901) | 27,500 (21,484–37,813) | 11,875 (8,906–15,586) | 16,875 (12,887–23,203) |
| 5 | Procambarus spec. | 352 (264–461) | 166 (135–228) | 479 (377–598) | 195 (153–244) | 85 (69–111) | 132 (107–181) |
| 6 | Chironomus riparius | 93 (70–111) | 33 (26–44) | 127 (99–143) | 51 (42–60) | 22 (17–26) | 33 (27–39) |
| 7 | Anax imperator | 620 (530–673) | 347 (280–390) | 830 (706–913) | 339 (297–373) | 149 (130–158) | 264 (235–298) |
| 8 | Cloeon dipterum | 134 (117–155) | 49 (45–62) | 181 (158–203) | 73 (64–85) | 32 (28–36) | 48 (45–57) |
| 9 | Notonecta maculata | 1,758 (1,636–1,978) | 605 (563–691) | 2,383 (2,215–2,606) | 967 (897–1,069) | 413 (382–455) | 605 (568–681) |
| 10 | Paraponyx stratiotata | 566 (419–743) | 352 (286–468) | 762 (583–934) | 332 (228–415) | 142 (112–181) | 293 (253–403) |
| 11 | Plea minutissima | 576 (514–641) | 208 (184–334) | 781 (691–854) | 317 (281–352) | 135 (122–153) | 205 (183–256) |
| 12 | Ranatra linearis | 2,031 (1,746–2,380) | 674 (590–1,179) | 2,754 (2,410–3,098) | 1,113 (939–1,287) | 474 (408–548) | 684 (555–801) |
| 13 | Sialis lutaria | 264 (204–338) | 88 (69–110) | 352 (275–439) | 144 (113–180) | 61 (49–76) | 88 (69–110) |
| Median HP 5 log‐normal (95% confidence interval) | 34 (15–120) | 14 (5.9–54) | 45 (20–160) | 19 (8.2–69) | 8.0 (3.4–30) | 13 (5.4–48) | |
In the example data set, the use of the GUTS‐RED‐SD model results in a more conservative risk assessment than the use of the GUTS‐RED‐IT model.
8.4.5. Tier‐3 risk assessment
In the dossier, information on a mesocosm study (experimental ponds) conducted with Organophosphate A is available. In this experimental pond study, the insecticide was applied once and four exposure concentrations were studied (measured peak concentrations 0.1, 0.9, 6 and 44 μg a.s./L) (Figure 45).
Figure 45.
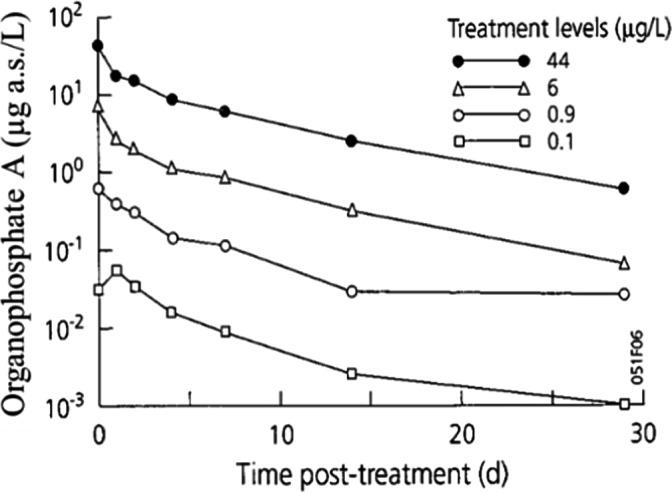
Dynamics of mean concentrations of Organophosphate A in depth‐integrated water samples in experimental ponds treated once with 0.1, 0.9, 6 and 44 μg a.s./L
The experimental pond study had a diverse community of aquatic invertebrates and a sufficient number of aquatic arthropods were present with a high enough minimum detectable difference (MDD) to consider the study valid. Effect classes (see EFSA PPR Panel, 2013) for the different treatments and most sensitive endpoints are provided in Table 19.
Table 19.
Effect classes (in μg a.s./L) of the most sensitive community‐ and population‐level endpoints in the experimental pond study that studied the impact of a single application of the insecticide Organophosphate A
| Treatment‐level (measured peak exposure of Organophosphate A in μg a.s./L) | ||||
|---|---|---|---|---|
| 0.1 | 0.9 | 6 | 44 | |
| Macroinvertebrate community | Effect class 1 | Effect class 3A | Effect class 5B | Effect class 5B |
| Most sensitive arthropod population (Gammarus pulex) | Effect class 2 | Effect class 5B | Effect class 5B | Effect class 5B |
At the community level (using Principal Response Curves) a NOEC (Effect class 1) of 0.1 μg a.s./L could be derived for both the zooplankton and macroinvertebrate community but the macroinvertebrate community showed an overall slower recovery than the zooplankton community. At the population‐level the most sensitive aquatic arthropod (G. pulex) showed a small treatment‐related effect on an isolated sampling in the 0.1 μg a.s./L treatment while all other invertebrate taxa did not show treatment‐related effects. In the 0.9 μg a.s./L treatment the microcrustacean Daphnia galeata and the insects Cloeon diperum and Caenis horaria showed short‐term responses on several consecutive samplings (Effect class 3A) while G pulex showed a long‐term response (Effect class 5B). At the two highest treatment levels, several arthropod populations showed long‐term effects.
Assuming that the exposure profile studied in the experimental ponds was realistic‐worst case and following EFSA PPR Panel (2013), a Tier‐3 ETO‐RACsw (ecological threshold option) can be derived from the experimental pond study by applying an AF of 2–3 to the Effect class 2 concentration of 0.1 μg a.s./L, resulting in a Tier‐3 ETO‐RAC sw of 0.033–0.05 μg a.s./L. A comparison of the Tier‐3 RACsw (0.033–0.05 μg/L) with the PECsw;max values as calculated for the different exposure scenarios illustrate that the environmental risks are most likely small to negligible for the exposure scenarios evaluated (see Table 20).
Table 20.
Tier‐3 risk assessment procedure for Organophosphate A. If the PEC:RAC ratio is < 1, then the environmental risk is considered acceptable
| Scenario | Acute risk assessment | ||
|---|---|---|---|
| PECsw;max (μg/L) | Tier‐3 ETO‐RACsw (μg/L) | PECsw;max:ETO‐RACsw ratio | |
| A | 0.035 | 0.033–0.05 | 1.06–0.70 |
| B | 0.034 | 0.033–0.05 | 1.03–0.68 |
| C | 0.031 | 0.033–0.05 | 0.94–0.62 |
| D | 0.030 | 0.033–0.05 | 0.91–0.66 |
| E | 0.029 | 0.033–0.05 | 0.88–0.58 |
| F | 0.029 | 0.033–0.05 | 0.88–0.58 |
The available experimental pond study can also be used to put the Tier‐2C risk assessment based on GUTS models into perspective of the tiered approach. According to the principles of the tiered approach, a lower‐tier assessment should be more conservative than a higher‐tier assessment. Consequently, the Tier‐2C risk assessment based on GUTS models should be more conservative than a Tier‐3 risk assessment (based on semi‐field (micro/mesocosm) data). This can be evaluated by using the exposure profile with a peak concentration of 0.1 μg/L simulated in the experimental pond study (see Figure 45) as input for the available GUTS models used in the Tier‐2C2 SSD approach. For a proper comparison, the exposure profile of the 0.1 μg/L treatment (the Effect class 2 concentration) needs to be divided by a factor of 2 or 3 (corresponding to the range in AFs used to derive a Tier‐3 ETO‐RAC based on an Effect class 2 concentration proposed by EFSA PPR Panel, 2013) to get EP50 values that correspond with the Tier‐3 RACsw (Table 21).
Table 21.
Calculated EP50 values (and 95% confidence limits) for aquatic arthropods and the exposure profile corresponding with the ETO‐RAC (based on Effect class 2 concentration of 0.1 μg/L) derived from the experimental pond study by using the GUTS‐RED‐SD and GUTS‐RED‐IT models as well as the corresponding HP5 values (and 95% confidence limits)
| EP50 values corresponding to exposure profile of 0.1 μg/L treatment | EP50 values corresponding to exposure profile of 0.1 μg/L treatment divided by an AF of 2 (liberal ETO‐RAC option) | EP50 values corresponding to exposure profile of 0.1 μg/L treatment divided by an AF of 3 (conservative ETO‐RAC option) | ||||
|---|---|---|---|---|---|---|
| Species | GUTS‐RED‐SD | GUTS‐RED‐IT | GUTS‐RED‐SD | GUTS‐RED‐IT | GUTS‐RED‐SD | GUTS‐RED‐IT |
| Daphnia magna | 0.5 (0.3–2.9) | 3.7 (3.0–4.6) | 1.0 (0.7–5.8) | 7.3 (6.0–9.3) | 1.5 (0.9–8.8) | 11.0 (8.9–13.7) |
| Asellus aquaticus | 36.6 (9.9–88.1) | 46.4 (40.6–63.3) | 73.2 (19.5–17.4) | 92.8 (78.3–134) | 110 (29.2–264) | 139 (120–209) |
| Gammarus pulex | 1.4 (0.2–2.5) | 2.1 (1.6–4.0) | 2.9 (0.5–5.3) | 4.1 (3.3–8.0) | 4.3 (0.5–7.7) | 6.1 (5.0–11.8) |
| Neocaridina denticulata | 1,484 (928–3,850) | 1,563 (1,257–2,832) | 2,969 (1,856–7,857) | 3,125 (2,539–5,859) | 4,375 (3,008–11,758) | 4,688 (3,516–8,345) |
| Procambarus spec. | 20.8 (11.6–24.6) | 17.1 (13.4–23.2) | 41.5 (23.3–52.3) | 34.2 (28.0–51.3) | 61.0 (33.6–75.5) | 51.3 (41.7–76.9) |
| Chironomus riparius | 5.5 (3.6–7.9) | 3.4 (2.7–4.4) | 11.0 (7.6–15.5) | 6.7 (5.5–8.4) | 16.5 (10.3–22.5) | 10.1 (8.2–13.8) |
| Anax imperator | 39.4 (36.3–42.1) | 42.4 (30.5–48.7) | 78.7 (70.1–83.7) | 84.8 (65.0–95.4) | 117 (107–127) | 127 (82.8–149) |
| Cloeon dipterum | 6.7 (3.6–8.6) | 5.0 (4.6–6.3) | 13,4 (7.2–17.4) | 10.1 (9.4–12.6) | 20.1 (10.8–26.8) | 15.3 (13.9–19.3) |
| Notonecta maculata | 53.4 (32.5–88.9) | 58.0 (54.4–70.0) | 107 (65.5–179) | 116 (109–138) | 160 (96.1–27.1) | 175 (163–224) |
| Paraponyx stratiotata | 47.3 (47.0–52.0) | 48.8 (42.7–73.2) | 94.6 (93.0–104) | 97.7 (85.6–146) | 143 (141–153) | 146 (128–215) |
| Plea minutissima | 35.4 (24.1–41.8) | 20.8 (17.9–39.9) | 70.8 (46.7–84.1) | 41.5 (35.7–77.7) | 106 (71.3–124) | 62.3 (54.5–109) |
| Ranatra linearis | 115 (86.7–151) | 63.5 (53.5–115) | 231 (163–304) | 127 (111–234) | 349 (248–447) | 190 (167–333) |
| Sialis lutaria | 4.8 (3.1–10.4) | 8.1 (6.6–10.6) | 9.7 (4.3–21.5) | 16.2 (12.1–20.2) | 14.5 (6.3–32.2) | 24.4 (18.3–33.6) |
| Median HP 5 log‐normal (95% confidence interval) | 0.88 (0.26–4.6) | 1.4 (0.26–4.6) | 1.8 (0.54–9.9) | 2.8 (1.2–11) | 2.5 (0.65–14) | 4.4 (1.8–17) |
It appears that the median HP5 values (1.8–2.8) calculated from the SSD of the EP50 values corresponding to the liberal ETO‐RAC (using and AF of 2 to extrapolate the Effect class 2 concentration) and the two types of GUTS models are smaller than 3‐6, the range in AF used in the Tier‐2C2 SSD approach (see Section 8.4.4). Also, when using the GUTS‐RED‐SD model, the median HP5 value (2.5) obtained for EP50 values corresponding to the conservative ETO‐RAC (using and AF of 3 to extrapolate the Effect class 2 concentration) is smaller than 3‐6. Only when using the GUTS‐RED‐IT model, the median HP5 value (4.4) obtained for EP50 values corresponding to the conservative ETO‐RAC (using and AF of 3 to extrapolate the Effect class 2 concentration) is higher than 3 but smaller than 6.
Overall, it can be concluded that the Tier‐2C2 risk assessment approach used in this case study seems to be worst‐case relative to the risk assessment using the Tier‐3 RACsw. This is consistent with the principles of the tiered approach, as mentioned above.
8.4.6. Concluding remarks on the use of GUTS models as part of the acute effect and risk assessment for the example substance Organophosphate A
The example data set for the substance Organophosphate A and the assessments presented above illustrate that our proposals on the use of validated GUTS models in Tier‐2C1 (based on standard test species) and Tier‐C2 assessments are not in conflict with the principle of the tiered approach that lower‐tiers should be more conservative than higher‐tiers. It is recommended that similar exercises are conducted with a representative number of substances differing in field exposure dynamics and toxic mode of action.
9. Conclusions and recommendations
In this Scientific Opinion, three different types of TKTD models are described, viz. (i) the GUTS, (ii) DEBtox models and (iii) models for primary producers. All these TKTD models follow the principle that the processes influencing internal exposure of an organism, summarised under TK, are separated from the processes that lead to damage and effects/mortality, summarised by the term TD. TKTD models are substance and species specific.
9.1. Conclusions
The GUTS modelling framework
GUTS models can be used to predict lethal effects under untested (time‐variable or constant) exposure conditions. GUTS models can be parameterised using standard single‐species toxicity test data, still providing relevant information at the individual level when extrapolating beyond the boundaries of tested conditions in terms of exposure. Simple model formulation and strictly defined terminology provides the basis for standardisation. Model simplicity allows for scanning large numbers of scenarios without in‐depth modelling experience of the user. Parameter estimation requires some statistical background and computational effort. Nevertheless, in the case of GUTS‐RED, the calibration can be achieved by using easy‐ and ready‐to‐use existing implementations of GUTS. Application examples and validation exercises (including pesticides) are available in the literature.
The DEBtox modelling framework
DEBtox models consider the links between life‐history traits such as growth and reproduction and explore the effects of toxicants over time, even over the entire life cycle under constant or time‐variable exposure profiles. DEBtox models can be used to predict both lethal and sublethal effects under untested exposure conditions. These models consist of two parts, (i) the DEB or ‘physiological’ part that describes the physiological energy flows and (ii) the part that accounts for uptake and effects of chemicals, named ‘TKTD part’. Model calibration requires combination of time series for growth and reproduction, not typically available from the standard data sets. DEBtox models require quite advanced knowledge in modelling and statistics. Although no user‐friendly generic tool is currently available to simply fit a DEBtox model, case‐specific implementations exist that can either be used to estimate parameters for a similar case study, or that can be adapted for new case studies.
TKTD modelling framework for primary producers
With respect to the analysis of toxic effects of time‐variable exposures on primary producers, the main relevant parameter is growth. A TKTD model for primary producers consists of a part addressing growth as a baseline, connected to a TKTD part addressing the toxic effects. Data for model parameter estimation can be obtained by reporting raw, non‐destructive, high‐time‐resolution data of standard tests (including an extended recovery period). For algae models, it is concluded that a drawback for implementing them in pesticide risk assessment is that the flow‐through setup used and needed to simulate long‐term variable exposures of pesticides to fast growing populations of algae has not yet been standardised, nor has the robustness of the setup been ring tested. Hence, for the present, the setup and the models are important research tools but probably not yet mature enough to use for risk assessment purposes. Use of Lemna and Myriophyllum models require quite advanced knowledge in modelling and statistics, as no generic user‐friendly software tool is currently available. Nevertheless, the already existing implementations allow the analysis of newly collected data under similar experimental conditions, or they can be adapted to be fit for other purposes.
Use of TKTD models in aquatic risk assessment for pesticides
To assess toxicity estimates for time‐variable exposure profiles as predicted by e.g. FOCUSsw steps 3 and 4, calibrated and validated TKTD models may be used, either focussing on standard test species (Tier‐2C1) or also incorporating relevant additional species (Tier‐2C2). A Tier‐2C assessment based on TKTD models will always evaluate effects relative to the SPG in accordance with the ETO.
For calibration, Tier‐1 (standard test species approach) but also Tier‐2A and/or Tier‐2B (standard and additional test species) toxicity data sets can be used. For validation, substance and species‐specific data sets derived from independent refined‐exposure experiments are required. Validated TKTD models for these species may be used to evaluate specific risks of available field exposure profiles by calculating exposure profile‐specific specific LPx/EPx values (= multiplication factor to a certain predicted exposure Profile that causes x% Lethality or Effect). These LPx/EPx values can be used in Tier‐2C risk assessment using the same rules and extrapolation techniques as used in experimental Tier‐1 (standard test species approach), Tier‐2A (geometric mean/WoE approach) and Tier‐2B (SSD approach). In practice, this means that the estimated LPx/EPx and HP5 values should be greater than or equal to the current AFs used in Tier‐1 and Tier‐2 approaches as described in the Aquatic Guidance Document.
For parameter estimation of TKTD models, it is crucial to report not only optimal parameters with their uncertainty limits, but also the optimisation method and the settings of the optimisation and of the numerical solver. In validation exercises, the effect predictions by means of a calibrated TKTD model need to be reported with their confidence/credible limits, obtained by propagation of the uncertainty associated with model parameters. The evaluation of the quality of the match of predictions with the observed data requires a careful combination of qualitative and quantitative criteria. Qualitatively, the visual match between model predictions and data is required. Quantitatively, three criteria are suggested: one that takes into account the uncertainty in the model predictions (PPC), one that measures the match over time (NRMSE), and one that considers the final match between model prediction and data (SPPE).
In a prospective ERA, for linking time‐variable exposure profiles to validated TKTD models, standard exposure models and scenarios should be selected (e.g. step 3 or 4 FOCUSsw) and their use should be evaluated as part of the standard risk assessment; if any other exposure models and scenarios are used, these would need to be evaluated separately. The calculated LPX/EPX values for the relevant exposure profiles should be reported with their confidence/credible limits.
Use of GUTS in aquatic risk assessment for pesticides
The conceptual (basic principles) and formal model (equations) of GUTS is considered to be sufficiently evaluated in this Opinion. Other aspects of GUTS model applications are recommended to be sufficiently documented in order to address the items in the checklist. The GUTS model framework is developed to address individual‐level lethal effects and may be an appropriate approach to use in the refined acute risk assessment scheme for aquatic invertebrates, fish and aquatic stages of amphibians. In the chronic risk assessment scheme, validated GUTS models can be used to predict effects of long‐term exposure on survival of the species of concern. This is only relevant if mortality is the critical endpoint in the chronic toxicity test. The formal definition for GUTS models is standardised and documented. Verification of GUTS model implementations can be performed using some standard elements: default scenarios, pulsed exposure scenarios, extreme scenarios. Sensitivity analyses of the GUTS models is not a necessity for new applications (new species‐compound combination), because the influence of the model parameters on the model outcome has already been analysed. Successful model validation, with observations of survival/mobility which have not been used for model calibrations and performed according to the recommendations provided in this SO, are a precondition for application in aquatic risk assessment.
The suggested acceptability criteria for the model validation have to be checked based on future applications of the GUTS models and possibly be adapted over time. Clearer validation criteria and related cut‐off values can be better formulated only when experience is gained with the use of GUTS in regulatory risk assessment.
Use of DEBtox in aquatic risk assessment for pesticides
The DEBtox modelling framework, based on the DEB theory, is developed to address individual‐level lethal and sublethal chronic effects. It may be an appropriate approach to use in the refined chronic risk assessment scheme for aquatic invertebrates, fish and aquatic stages of amphibians, particularly when sublethal endpoints are most critical in chronic toxicity tests. The physiological DEB part of DEBtox models needs to be evaluated separately from the TKTD part and this should be done ahead of submission of DEBtox models for regulatory use (e.g. by a group of experts at EU level). To date, in the scientific literature sufficiently documented examples of DEBtox modelling for pesticides and relevant aquatic organisms could not be found. Consequently, at this stage a proper evaluation of a species and pesticide‐specific DEBtox model could not yet be done in this scientific opinion. DEBtox model applications are recommended to be sufficiently documented to address all items in the provided checklist.
Use of primary producer models in aquatic risk assessment for pesticides
Validated TKTD models developed for primary producers can be used in the chronic risk assessment scheme with a focus on growth rate and/or yield. TKTD model assessments for algae and fast‐growing macrophytes such as Lemna assess some population‐level effects, since in the course of the test many new individuals (algal cells; Lemna fronds) have developed. The physiological part of TKTD models for primary producers needs to be evaluated separately from the TKTD part and this should be done ahead of submission of these models for regulatory use (e.g. by a group of experts at EU level). This Opinion has completed this stage of the physiological part evaluation for the available Lemna model, but not for the Myriophyllum model. The conceptual and formal model for the Lemna model can be considered to be sufficiently evaluated in this Opinion, in contrast to that of Myriophyllum and algae. Primary producers model applications are recommended to be sufficiently documented to address all items in the provided checklist.
Use of TKTD model output in aquatic risk assessment
In Tier‐2C1 acute risk assessment, the exposure profile‐specific LP50/EP50 value for all relevant standard test species should at least be greater than or equal to the acute Tier‐1 AF of 100 for risks to be considered low. In the Tier‐2C1 chronic risk assessment, the exposure profile‐specific EP10 values (all relevant standard test animals) and the exposure‐profile‐specific EP50 values (relevant standard test algae and/or aquatic macrophytes) should at least be greater than or equal to the chronic Tier‐1 AF of 10 for risks to be considered low.
If for less than eight aquatic arthropods and/or primary producers or for less than five fish species, appropriate TKTD models are made available, the calculated LP50/EP50/EP10 values for the different species and relevant annual exposure profiles may be used by adopting the geometric mean approach (currently predominantly in acute risk assessment) and/or a WoE approach. When using the acute Tier‐2C2 geometric mean approach, the rules as described in EFSA PPR Panel (2013) should be followed. The exposure profile‐specific geometric mean LP50/EP50 values for all relevant taxonomic groups should be greater than or equal to 100 (the Tier‐1 AF) for risks to be considered acceptable. When using the acute Tier‐2C2 WoE approach, the lowest LP50/EP50 of the tested species for each relevant annual exposure profile should at least be larger than 10 but may be smaller than 100, depending on the number of species for which TKTD models are available for the substance of concern. When using the chronic Tier‐2C2 WoE approach our proposal is that the lowest EP10 of the tested animal species, or currently the lowest EP50 for primary producers, should at least be larger than 4 but may be smaller than 10.
If for at least eight aquatic arthropods and/or primary producers or for at least five fish species, appropriate TKTD models are made available, the calculated LP50/EP50/EP10 values for the different species and relevant annual exposure profiles may be used by adopting the SSD approach. In the SSD approach based on TKTD models, the median HP5, derived from SSDs constructed with relevant LP50 or EPx values, is used in the risk assessment. In the acute risk assessment for aquatic invertebrates, this median HP5 should at least be greater than or equal to the AF selected in the range of 3–6 (similar to the range in AF used in SSD approach to derive a RACsw;ac) to demonstrate low risk, while for aquatic vertebrates this median HP5 should at least be greater than or equal to 9. In the chronic risk assessment, the median HP5 should at least be greater than or equal to 3, both for aquatic plants and animals.
If in Tier‐2A or Tier‐2B studies, the most sensitive additional test species is an order of magnitude more sensitive than any other test species, it may be justified to base, in first instance, the refined Tier‐2C2 assessment on a TKTD model for this species and the substance of concern. In the acute risk assessment, the EP50 value for this species (or LP50 value for fish/amphibians) for each relevant annual exposure profile should at least be larger than 10. In the chronic risk assessment, it is proposed that the EP10 (or currently EP50 for primary producers) should at least be greater than 4.
9.2. Recommendations and future perspectives
Validating TKTD models
The minimum requirements for validation experiments are the testing of two concentration profiles with at least two pulses each to address phenomena related to the modelled internal concentration or damages states (e.g. dynamics between internal and external exposure concentrations) and repair of possible effects. The DRT95 should be calculated and considered for selecting the timing of the pulses; one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile should clearly be larger than the DRT95, if feasible within time constraints of laboratory testing.
In order to minimise experimental studies with aquatic vertebrates for animal‐welfare reasons, the Panel recommends investigation as to whether the results of chronic Tier‐1 vertebrate studies can be used for validation of compound and species specific TKTD models.
GUTS models
The current state of science in the GUTS framework is sufficient to facilitate the use of these models in the aquatic risk assessment for pesticides and to initiate the development of OECD guidelines for their use in ERA.
For compounds that are suspected of showing increased lethal effects over time (incipient toxicity not reached in the acute tests), it may be necessary also to calibrate the GUTS models with mortality/survival data obtained from prolonged or chronic tests. Further classification of compounds with respect to the potential to show increased toxicity under long‐term exposure would be relevant.
Technically, one of the main questions is whether it is better to allow for user‐defined implementations of GUTS, or to request one specific GUTS implementation, which only needs to be checked once. In this opinion, the decision was to consider user‐defined implementations and to give checklists and criteria to allow regulators to evaluate whether a new implementation fulfils the required quality aspects.
DEBtox models
The lack of published examples of DEBtox models for pesticides and aquatic organisms, as well as the fact that no user‐friendly DEBtox modelling tools are currently available, result in the conclusion that these models are not yet ready for use in aquatic risk assessment for pesticides. Nevertheless, the DEBtox modelling approach is recognised as an important research tool with great potential for future use in prospective ERA for pesticides.
To facilitate wider use, DEBtox models should be made more accessible to non‐advanced model users, e.g. by creating user‐friendly tools specifically dedicated to the calibration of these models, and by promoting the use of simplified versions when applicable.
Once validated DEBtox models are available, it would be useful to develop an example of its application in risk assessment as done for GUTS.
TKTD models for primary producers
The largest drawback for implementing the published algae models in pesticide risk assessment is that the flow‐through experimental setup used for model calibration/validation to simulate long‐term variable exposures of pesticides to fast growing populations of algae, has not yet been standardised, nor has the robustness of the setup been ring‐tested. Hence, presently the experimental setup of refined exposure tests for algae and the algae models are considered as important research tools but probably not yet mature enough to use for risk assessment purposes.
The published Lemna model can be the basis for a compound–specific Lemna model. Such a model, when properly tested and documented, can be used to evaluate the effects of predicted exposure profiles in Tier‐2C, if in the Tier‐1 assessment Lemna is the only standard test species that triggers a potential risk and potential risk to rooted macrophytes can be excluded.
Although the published Myriophyllum modelling approach may be a good basis to further develop TKTD models for rooted submerged macrophytes, it is currently considered not yet fit‐for‐purpose in prospective ERA for pesticides. The currently available Myriophyllum model needs further documentation, calibration and validation.
Growth models, particularly for Myriophyllum, would benefit from including more natural growth conditions. Also, a more detailed experimental analysis of uptake, transport and elimination processes of organic contaminants in aquatic macrophytes and their incorporation in the dynamic model would strengthen the credibility of the models. A modification of the standard Lemna and Myriophyllum test by including more frequent monitoring of growth and a recovery phase would provide adequate data for initial fits of plant models.
Once validated models for primary producers are available, it would be useful to develop an example of their application in risk assessment as done for GUTS.
If the effects of pesticide exposure on growth inhibition of biomass and shoot length/frond number endpoints result in similar ErC50 values with overlapping confidence intervals, biomass‐related endpoints are always used in TKTD modelling. If the morphological endpoints prove to be significantly more sensitive, alternative approaches may need to be developed or the time span of the test may need to be extended for the full effect to take place on biomass‐related endpoints.
Use of TKTD models in aquatic ERA for pesticides
While evaluation of GUTS models and the Lemna model should be possible by non‐modelling experts using this Opinion, it would be beneficial to set up an expert group to help regulatory authorities of Member States to evaluate submissions including these models, at least until more experience has been gained.
For a selected number of substances differing in exposure dynamics and toxic mode of action, Tier‐2C risk assessments by means of TKTD models need to be compared with the risk assessments based on other tiers to check the consistency of the tiered approach (lower‐tiers should be more conservative than higher‐tiers, while respecting the same specific protection goal in all tiers).
In the near future, guidance should be developed on the rationale underlying the reduction of the AF when using the WoE approach in the acute and chronic risk assessment, based on the quantity and quality of the additional toxicity data made available.
It is important to state that the evaluation of TKTD models in the context of this SO focusses on Tier‐2 refinement, and not on the use of TKTD models in a broader environmental context. This implies that models which have been evaluated positively on their physiological or TKTD parts, would need additional testing when they are to be applied in the context of variable environmental conditions (e.g. as Tier‐3 tools in combination with population‐level modelling).
In support of daily activities of risk assessors, user‐friendly tools would need to be made available as done for GUTS, to allow regulatory authorities to re‐run Tier‐2C assessments based on DEBtox or primary producer models. In support of their future development, a privileged collaborative partnership between academia and regulatory authorities could beneficially help to fill in this gap.
Glossary
- Bayesian inference
An approach to estimate parameter values from data, based on prior knowledge on the parameter. In essence, this approach uses the prior knowledge and the comparison between measured data and model results to get a new probability distribution of the parameter value called posterior distribution. Often, the parameter value with the highest posterior probability is used for further modelling. The posterior distribution also gives a measure of the uncertainty of the parameter value; the wider it is, the less certain is the value
- Compartment models
Toxicokinetic models which use a generic idealisation of an organism as one‐ or multi‐compartment. For aquatic invertebrates, the one‐compartment model is the most often used. One‐compartment models assume concentration‐driven transfer of the chemical from an external compartment into an internal compartment, where it is homogeneously distributed. For example, the simplest GUTS (reduced GUTS – GUTS‐RED) for invertebrates (e.g. Daphnia magna) assumes one‐compartment model and links external concentrations directly to the scaled damage (see also damage and scaled damage concept)
- Conceptual model
A hypothesis regarding the structures and important factors that govern the behaviour of an object or process of interest. This can be an interpretation or working description of the characteristics and dynamics of a physical system
- Confidence interval (see also definitions of parameter estimation and frequentist inference)
The range of parameter values that would all be accepted under the assumed statistical error distribution and the chosen significance level
- Credibility interval (see also definitions of parameter estimation and Bayesian inference)
The interval that contains the given parameter value with a certain probability (e.g. 95%). It can be constructed from the posterior distribution in a Bayesian inference
- Damage
In GUTS theory, damage is a state variable that decouples toxicokinetics from the dynamics of the effect (immobility/survival). The internal concentration leads to damage, which is also repaired at a certain rate. Damage dynamics is therefore the central part of the GUTS model that translates external exposure into toxicodynamic processes and finally the death mechanism. Note that the ‘damage’ is a rather abstract concept and its level cannot be experimentally measured
- Dynamic Energy Budget theory (DEB theory)
The DEB theory is based on the fact that all living organisms consume resources from the environment and convert them into energy (following the conservation laws for mass and energy) to fuel their entire life cycle (from egg to death), thus ensuring maintenance, development, growth and reproduction
- DEBtox models
The application of the DEB theory to predict the effects of toxic chemicals on life‐history traits or, in ecotoxicological sense – lethal and sublethal endpoints. DEBtox models differ from GUTS models by incorporating a DEB part for growth and reproduction endpoints at the individual level. The DEB part describing the physiological energy flows is in this Opinion termed as the ‘physiological part’ of a DEBtox model, while the part which accounts for uptake and effects of chemicals is named the ‘TKTD part’ of the DEBtox model
- Exposure profile‐specific LPx/EPx
Multiplication factor to an entire specific exposure profile that causes X% lethality/sublethal effect (see also multiplication factor)
- Exposure profile‐specific Hazardous Profile (HP5)
Hazardous Profile to 5% of the species tested (i.e. Potentially Affected Fraction). If for a sufficient number of relevant species validated substance‐specific TKTD models are made available, the exposure profile‐specific EPx values for the different species can be used to construct an SSD. The exposure profile‐specific HP5 can be used in the risk assessment in the same way as when applying the SSD approach based on experimental data
- Evaluation (of the model)
The process used to generate information to determine whether a model and its results are of a quality sufficient to serve as the basis for a regulatory decision. It includes assessment of model parameter estimation, sensitivity analysis and validation
- Formal model (Model formalisation)
Mathematical equations and other detailed information about the model used
- Frequentist inference (also known as classical inference)
An approach to estimate parameter values from data. The goal is to find the best parameter value, defined as the one that shows the closest fit between measurements and the outcome of the simulation or statistical model. The uncertainty of this value can be described by the confidence interval (or confidence region). Most often, the error distribution (i.e. the distribution describing the random variation in the measurements) is assumed to be normal and independent for the different measurements. Prior knowledge on the parameter value (e.g. the biologically reasonable range or values from previous experiments) is not taken into account during the estimation
- General Unified Threshold models of Survival theory (GUTS)
Modelling approach used to simulate lethal effects under various exposure conditions. GUTS connects the external concentration with a so‐called damage dynamics (see also damage and scaled damage concept) which results in simulated mortality when an internal damage threshold is exceeded. Reduced GUTS (GUTS‐RED) is a modelling approach which does not account for internal concentration of the toxicant; scaled damage is based on the external (water) concentration of the toxicant. In full GUTS, internal concentration of the toxicant is explicitly modelled and scaled damage is based on the internal concentration of the toxicant
- GUTS‐IT/GUTS‐RED‐IT
GUTS models (see respective entry) based on Individual Tolerance (IT).Thresholds for effects (immobility/mortality) are distributed among individuals as sensitivity varies between individuals of a population so the death of the individual occurs once an individual tolerance is exceeded. Combinations of the choice of the scaled damage and the death mechanism give clearly defined acronyms for the different variants of GUTS, e.g. GUTS‐RED‐IT for the combination of the scaled damage without consideration of internal concentrations and the IT mechanism, or GUTS‐IT for the full GUTS model accounting for internal concentrations in combination with the IT mechanism
- GUTS‐SD/GUTS‐RED‐SD
GUTS models (see respective entry) based on Stochastic Death (SD). Death (mortality) is modelled as a stochastic (random) process occurring with increased probability as the scaled damage rises above the threshold, which is fixed and identical for all individuals in a group. Combinations of the choice of the scaled damage and the death mechanism give clearly defined acronyms for the different variants of GUTS, e.g. GUTS‐RED‐SD for the combination of the scaled damage without consideration of internal concentrations and the SD mechanism, or GUTS‐SD for the full GUTS model accounting for internal concentrations in combination with the SD mechanism
- Implementation of a model (Model implementation)
Definition of a given model code in a software (e.g. R, Mathematica). A model can have several implementations. Implementation of a model is done in a particular software or programming environment. The documentation of the implementation should contain an overview of the source code files, and the version and necessary packages of the used programming environment. Testing of the implementation (‘implementation verification’) needs to be performed and documented for any model implementation
- Kappa‐rule
A core concept of DEB theory (see respective entry). It is assumed that a fixed fraction (kappa) of the mobilised energy is allocated to somatic maintenance and growth, while the rest is allocated to maturity maintenance, maturation and reproduction. Note that in standard DEB framework, the kappa‐rule should not be affected by a toxic compound
- Multiplication factor
(Originally introduced by Ashauer et al. (2013) as the ‘margin of safety’) – Factor applied to an entire specific exposure profiles leading to a certain effect level, e.g. 50%, at the end of the tested profile (lethal profile (LP50) for mortality, effect profile (EP50) for e.g. immobility). The analogy to the LC50 or EC50 of a laboratory test on mortality/immobility under static exposure is intended, but attention is needed because the LCX/ECX are concentrations, while the LPX/EPX are multiplication factors. By using a specific multiplication factors the whole exposure profile can be ‘shifted’ and adjusted to a level that will result in x% mortality at the end of exposure profile P. This multiplication factor is then denoted LPX. Similarly, multiplication factor relevant model output is the prediction of exposure profile‐specific sublethal effects (e.g. multiplication factor to exposure profile causing 10% effect; EP10)
- Parameters
Terms in the model that are fixed when conducting a model run or simulation (but can be changed in the model development step, as a method for conducting sensitivity analysis or to achieve calibration goals)
- Parameterisation sensu stricto
Parameter definition, the process of defining parameters that are used to represent the (biological) processes in a model. They determine the interactions and controls among model mechanisms
- Parameterisation sensu lato
A word that modellers use for selecting values for a model's parameters. Other equivalent words are parameter estimation or parameter inference (see respective entries)
- Prior distribution (see also parameter estimation, Bayesian inference)
The formalised knowledge of a parameter value before parameter estimation, e.g. taken from the literature, expert judgement or previous experiments. For some parameters, there might be a lot of knowledge available, so that a narrow prior can be constructed (e.g. as a normal distribution with the mean representing the value that was measured most often and the variation representing the spread in the measurements). In other cases nearly nothing might be known (e.g. for the competitive strength of different species) and a vague, flat prior needs to be used that allows for a very wide range of potential values
- Parameter estimation (model parameter estimation, also named model calibration or parameter optimisation)
The process of adjusting model parameters within physically defensible ranges until the resulting predictions give the best possible fit to the observed data. General information about the way model parameters have been estimated, that is how calibration routines are performed, which function is used as target for optimisation routine, etc. Needs to be performed and documented for any model implementation documentation. Inference method could be frequentist or Bayesian (see respective entries)
- Physiologically based toxicokinetic (PBTK) models
Toxicokinetic models where single organs and blood flow are explicitly considered
- Regulatory model
The regulatory model is a package consisting of the following components (i) the mechanistic exposure‐effects model, (ii) programs for pre‐ and post‐processing, often made available in the form of graphical user interfaces, (iii) model parameters, and (iv) environmental scenarios. By combining the computer model with scenarios, the model will address a certain goal and can therefore be used regulatory purposes
- Scaled damage concept
A general concept that links the external concentration dynamics and the time course of the internal hazard. If measured internal concentrations are not available (the general case with all standard toxicity test), the dominant rate constant kD (previously termed ‘scaled internal concentration’) links external concentrations to the scaled damage
- Sensitivity
The degree to which the model outputs are affected by changes in selected input parameters
- Sensitivity analysis
The quantification of the effect of changes in input values or assumptions (including boundaries and model functional form) on the outputs. By investigating the relative sensitivity of model parameters, a user can become knowledgeable about the relative importance of parameters in the model
- Simulation
Development of a solution by incrementing steps through the model domain. Simulations are often used to obtain solutions for models that are too complex to be solved analytically. For most situations, where a differential equation is being approximated, the simulation model will use finite time step (or spatial step) to simulate changes in state variables over time (or space)
- Toxicokinetic (TK)
All processes that influence the dynamics in internal exposure of an individual to the toxic compound, and include absorption, distribution, metabolism and elimination. Toxicokinetic models are used to estimate internal exposure concentrations
- Toxicodynamics (TD)
All processes that lead to the damage and/or mortality of the organism exposed to toxic compounds. Biological effects are caused by the toxic compounds on the molecular level, where the molecules of the toxic compound interfere with one or more biochemical pathways. Toxicodynamic part of TKTD models integrate all those processes into only a few equations that capture the dynamics of responses or effects over time
- Toxicological (in)dependence
If the 95% depuration time of maximum of the internal concentration is larger than the time of the next exposure pulse, the two pulses are assumed to be toxicologically dependent, if the next pulse comes only after the depuration time, pulses are assumed independent
- Uncertainty
The term is used to describe the lack of knowledge about models, parameters, constants, data and beliefs. There are many sources of uncertainty, including the science underlying a model, uncertainty in model parameters and input data, observation error and code uncertainty. Additional studies and collecting more information allow error that stems from uncertainty to be minimised/reduced (or eliminated). In contrast, variability (see definition) is irreducible but can be better characterised or represented with further studies
- Uncertainty analysis
Investigation of the effects of lack of knowledge or potential errors on the model (e.g. the uncertainty associated with parameter values). When combined with sensitivity analysis (see definition), uncertainty analysis allows a model user to be more informed about the confidence that can be placed in model results
- Variable
A measured or estimated quantity that describes an object that can be observed in a system and that is subject to change. Two kinds of variables can be distinguished. The state variables (e.g. body mass) are the dependent variables calculated within a model, which are also often the performance indicators of the models that change over the simulation. The forcing variable corresponds to input data to the model (e.g. toxicity of the substance). This input data may be defined in the exposure/ecological scenario
- Variability
Observed differences attributable to true heterogeneity or diversity. Variability is the result of natural random processes and is usually not reducible by further measurement or study (although it can be better characterised)
- Validation (of the model; model validation)
The process of establishing that the model is a sufficiently accurate representation of the real world to be used as the basis for regulatory decisions. It assesses how well the model fits relevant data patterns and if the model provides predicted endpoint/output values with an acceptable error range for risk assessment. This last step is performed through the comparison of model or submodel outputs with independent empirical data or the data that were not used for parameter estimation (calibration)
- Verification (of the code/model implementation)
Examination of the algorithms and numerical technique in the computer code to ascertain that they truly represent the conceptual model and that there are no inherent numerical problems with obtaining a solution. Refers to basic tests to show that the source code works as it should for selected cases
Abbreviations
- a.i.
active ingredient
- a.s.
active substance
- AF
assessment factor
- DAG
Direct Acyclic Graph
- DEB
Dynamic Energy Budget
- DEBkiss
Reserve‐less DEB (Keep It Simple, Stupid)
- DEBtox
TKTD model based on Dynamic Energy Budget theory
- DIC
dissolved inorganic carbon
- DRT95
individual‐level depuration and repair time for 95% of the effects
- ECx
effective concentration (causing X% effect)
- EFSA PPR Panel
Plant Protection Products and their Residues
- ERA
Environmental Risk Assessment
- ERC
Ecotoxicologically Relevant exposure Concentration
- ERO
ecological recovery option
- ETO
ecological threshold option
- FOCUS
FOrum for the COordination of Pesticide fate models and its USe.
- GUTS
General Unified Threshold models of Survival theory
- GUTS‐IT
GUTS models based on Individual Tolerance
- GUTS‐RED‐IT
reduced GUTS based on Individual Tolerance
- GUTS‐RED
reduced GUTS
- GUTS‐RED‐SD
reduced GUTS based on Stochastic Death
- GUTS‐SD
GUTS models based on Stochastic Death
- HP
Hazardous Profile
- IT
Individual Tolerance
- JAGS
Just Another Gibbs Sampler
- Kow
octanol/water partition coefficient
- LCx
lethal concentration (causing X% mortality)
- LP/EPx
Exposure Profile causing X% mortality/effect
- MCMC
Monte Carlo Markov Chain
- MC
Monte Carlo
- MF
multiplication factor
- MOA
mode of action
- NEC
no‐effect‐concentration
- NOEC
no observed effect concentration
- NRMSE
Normalised Root Mean Square Error
- OAT
One‐parameter‐At‐a‐Time method
- OECD
Organisation for Economic Cooperation and Development
- PEC
predicted environmental concentration
- PPC
Posterior Predictive Check plot
- PPP
Plant Protection Product
- PPR EFSA
Panel on Plant Protection Products and their Residues
- RAC
Regulatory Acceptable Concentration
- RAC
Regulatory Acceptable Concentration
- RGR
relative growth rate
- SD
Stochastic Death
- SO
Scientific Opinion
- SOT
survival over time
- SOT
survival over time
- SPG
specific protection goal
- SPPE
survival‐probability prediction error
- SSD
species sensitivity distribution
- sw
surface water
- TER
Toxicity Exposure Ratio
- TKTD
toxicokinetics/toxicodynamics
- twa
time‐weighted average
Appendix A – Model implementation details with Mathematica and R
A.1. Information for the Mathematica implementation example
Implementation details for GUTS in Mathematica
A.1..1.
Programming
The GUTS TKTD models have been implemented in Mathematica (Wolfram Research, version 11.0). Mathematica is a proprietary software for performing mathematics on a computer. It provides comprehensive methods for computation. The GUTS implementation uses mainly the functionality to calculate numerical solutions for ordinary differential equations (method NDSolve), to find the minimum of a given objective function (NMinimize), to read and write files of various formats (Import/Export) and to operate with lists and matrices of data. Mathematica is under continuous development, the implementation is steadily tested and verified.
The source code of the GUTS implementation in Mathematica is available, containing implementations of all necessary program routines. Additional Mathematica notebooks contain example applications for the GUTS Ring test, and the single modelling steps, i.e. model calibration, model validation, predictions of survival rates for the given exposure scenarios including propagation of uncertainties, together with all necessary data import and export functionality.
Testing and verification of the implementation
The implemented code used in this study has continuously been tested for programming errors and the used implementation has participated in the GUTS ring test with very good results (Jager and Ashauer, 2018). The implemented GUTS‐RED‐SD model is defined by equations (1), (4) and (5), and the GUTS‐RED‐IT model by equations (1), (6) and (7). The Mathematica implementation was extensively tested, including sensitivity analyses and model code verification (see Section 4.1.2), the corresponding source code for the tests is available. An archive containing source code can be found in Appendix E.
Methods for model calibration and parameter estimation
The reduced GUTS models, i.e. the GUTS‐RED‐SD and GUTS‐RED‐IT models, were chosen for the example application to the G. pulex and propiconazole data and calibrated (see Section 4.2.1.1). Optimal parameter values were estimated by using the NMinimize method as available in Mathematica. Optimal parameter values giving the best fit between data and model simulations were identified and are reported in Appendix B – Table B.1. Figure B.5 shows calibrated model in comparison to the data over time and in a dose–response view.
Settings of the optimisation routine
For the model calibration, the negative of the log‐likelihood function was minimised as target for the minimisation, which is equivalent to the maximisation of the log‐likelihood function. The target function was minimised using NMinimize by choosing Simulated Annealing as optimisation algorithm as variant of the Metropolis algorithm. NMinimize was used with the following settings: {“SimulatedAnnealing”, “PerturbationScale” → 3, “SearchPoints” →50}; GeneralOptions → {PrecisionGoal → 12, MaxIterations → 10}.
Parameter confidence intervals
Characterisation of parameter confidence limits by likelihood profiling was done as described in Section 4.1.3.2. Settings for the stepwise optimisation for NMinimize were {“SimulatedAnnealing”, “PerturbationScale” ‐> 3, “SearchPoints” ‐> 20}; GeneralOptions:{PrecisionGoal ‐> 6, MaxIterations ‐> 10}. About 10.000 parameter samples were tested for the model calibration and confidence approximation.
Numerical solver
The numerical solver of the ordinary differential equations is important for stability of the numerical solutions. Exposure time series need to be interpolated and both sudden large shifts of the exposure as well as strongly fluctuating values challenge the numerical stability of the solver. For the tested exposure time series, the NDSolve method of Mathematica was used, with application of the Runge–Kutta solver with adaptive step width. NDSolve provides a collection of different methods, it typically solves differential equations by going through several different stages, depending on the type of equations. For the solutions, the AccuracyGoal option was set to 12, what gives 12 digits absolute precision, and the Option Method → {“StiffnessSwitching”} was used to account for the abrupt changes in concentration levels under pulsed exposure.
Choice of starting values for the optimisation
Since the location of global optimal model parameters in a multi‐dimensional parameter space is a challenging task, optimal parameter sets depend to a certain degree on the starting values for the optimisation procedure. For the application to the G. pulex and propiconazole data sets, random numbers between 0 and 5 were used as initially (between 0 and 0.5 for the background mortality). The optimisation was repeated three times, where for the second and third optimisation run random numbers between 0 and the ceiling of the optimal value of the optimisation run before were used as starting values. There were no constraints on the parameter intervals, only they had to be positive (> 0). No further assumptions were made to identify the optimal parameter values.
List of files for the GUTS implementation in Mathematica
The Mathematica implementation of GUTS models consists of the following files:
-
GUTS‐Methods‐v24‐07062018.nb: contains the source code which implement the GUTS models and all necessary routines, i.e. in detail:
-
–
the implementation of the GUTS‐SD and GUTS‐IT models;
-
–
the implementation of the objective function for parameter optimisation;
-
–
the implementation of the parameter optimisation;
-
–
the routines for parameter confidence limit approximation;
-
–
the routines for uncertainty analysis, i.e. for probabilistic simulations;
-
–
the calculation of other goodness‐of‐fit measures;
-
–
GUTS‐Implementation‐Check‐AF‐new.nb: several tests and verification of the model implementation functions (see 4.1.2 for the corresponding results).
-
GUTS‐EFSA‐propiconazole‐FittingAndFirstSteps.nb: application of the implemented GUTS models to the data set B of the GUTS ring test (propiconazole and G. pulex; see Jager and Ashauer, 2018). The following steps are done:
-
–
Load data
-
–
Parameter optimisation for the GUTS‐RED‐SD
-
–
Parameter optimisation for the GUTS‐RED‐IT
-
–
Confidence limit approximation for the GUTS‐RED‐SD
-
–
Confidence limit approximation for the GUTS‐RED‐IT
-
–
Compile figures of the model calibration over time including uncertainty (Figure B.5)
-
–
Calculate LC50 values for constant exposure at different time‐points
-
–
Create dose–response relationship for the calibration data (Figure B.5)
-
–
Predictions of the validation data set
-
–
Compile figures of the model validation over time including uncertainty (Figure B.6)
-
–
Create and plot dose–response relationship for the validation data (Figure B.6)
-
–
Create a PPC plot (Figure 23)
-
–
-
GUTS‐EFSA‐propiconazole‐exposureAnalysis.nb: calculates LP50 values for the application example in Section 4.2.3 (Predictions under FOCUS surface water exposure patterns)
-
–
reads in parameter information for propiconazole and G. pulex (optimal values and samples)
-
–
reads in exposure time series
-
–
predicts LP50 values (Table 4)
-
–
A.2. Information for the R implementations
A.2.1. GUTS models
A.2.1..1.
Programming
GUTS models (RED versions) are implemented within the R package ‘morse’ 3.1.0 (https://CRAN.R-project.org/package=morse), which provides easy‐to‐use R functions to explore/visualise experimental data and to fit models under a Bayesian framework for getting concentration thresholds (No Effect Concentration) estimates associated to their uncertainty (see chapter 4 for methodological aspects). The source code is available and consists of the file list provided below. Saved R objects (.RData extension) are also available to exactly reproduce all results (Tables and Figures). This avoids getting slight differences in the results due to the implemented stochastic processes when running scripts by its own. All the calculation can also be done directly on‐line through a web‐browser from the web‐platform MOSAIC and its specifically dedicated module GUTS: http://pbil.univ-lyon1.fr/software/mosaic/guts.
Testing and verification of the implementation
As detailed in chapter 4, all R functions used to get results from the calibration and the validation data sets have been extensively tested for their robustness and the relevance of their results. This implementation has also participated in the GUTS ring‐test (Jager and Ashauer, 2018). The corresponding R code is provided within the file GUTS‐implementation‐check.R (Appendix E).
Methods for the model calibration
Even if internal concentrations were measured in the Nyman et al., study, the most widely used versions of GUTS models, namely GUTS‐RED‐SD and GUTS‐RED‐IT, were chosen to fit data from the calibration data set. Parameters were estimated within a Bayesian framework according to Section 4.1.3.3 Three independent MCMC chains were run in parallel. After an initial burn‐in period of 5,000 iterations, the Bayesian algorithm (from the JAGS software) was run 11,238 iterations and the corresponding sample of the joint parameter posterior distribution was recorded. The convergence of the estimation process was checked with the Gelman and Rubin statistics (see chapter 4, Section 4.1.3.3 for details).
List of files for the GUTS implementation in R
The R implementation of GUTS models consists of the following files:
-
GUTS‐implementation‐check.R: contains the source code to perform testing and verification of R functions implemented within package ‘morse’ (see chapter 4 for the corresponding results). This file produces the following outputs (.png for Figures):
-
–
test‐SD‐IT‐cst.png: test of the R implementation of GUTS‐RED‐SD and GUTS‐RED‐IT models under constant exposure;
-
–
test‐SD‐IT‐var.png: test of the R implementation of GUTS‐RED‐SD and GUTS‐RED‐IT models with increasing multiplication factors for pulsed exposures
-
–
sensi‐SD‐IT‐surv5.png: R one‐at‐a‐time sensitivity analysis of models GUTS‐RED‐SD and GUTS‐RED‐IT;
-
–
sensi‐SD‐IT‐extreme.png: R implementation test of GUTS‐RED‐SD and GUTS‐RED‐IT models under extreme cases.
-
–
-
GUTS‐run‐fit.R: contains the source code to fit GUTS GUTS‐RED‐SD and ‐IT models on the calibration data set, to get graphical results for checking the convergence of the Bayesian process leading to parameter estimates, and to get plots of the results under various shapes as shown below. This file produces the following outputs (.png for Figures, .txt for text files, .RData for R objets):
-
–
GUTS‐calibration‐results.txt: parameter estimates of the GUTS GUTS‐RED‐SD and ‐IT models fitted on the calibration data set;
-
–
plot‐fit.cstSD.png: GUTS GUTS‐RED‐SD model calibration on a typical acute toxicity test: the survival over time is represented as a function of time of each tested concentration;
-
–
plot‐fit.cstIT.png: GUTS GUTS‐RED‐IT model calibration on a typical acute toxicity test: the survival over time is represented as a function of time of each tested concentration;
-
–
plot‐ppc.cstSD.png: Posterior Predictive Check for the GUTS GUTS‐RED‐SD model, calibrated on a typical acute toxicity test;
-
–
plot‐ppc.cstIT.png: Posterior Predictive Check for the GUTS GUTS‐RED‐IT model, calibrated on a typical acute toxicity test;
-
–
plot‐Nsurv‐SD.png: GUTS GUTS‐RED‐SD model calibration on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each tested concentration;
-
–
plot‐Nsurv‐IT.png: GUTS GUTS‐RED‐IT model calibration on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each tested concentration;
-
–
surv‐dose‐response.png: GUTS GUTS‐RED‐SD and ‐IT model calibration on a typical acute toxicity test: the survival rate (%) is represented as a function of the concentration at the end of the experiment (day 4);
-
–
Nsurv‐dose‐response.png: GUTS GUTS‐RED‐SD and ‐IT model calibration on a typical acute toxicity test: the number of survivors is represented as a function of the concentration at the end of the experiment (day 4);
-
–
LC50‐versus‐time.png: LC50 estimates from the calibrated GUTS SIC models
-
–
quant‐Nsurv‐SD.txt: quantiles of the simulated numbers of survivors under the GUTS GUTS‐RED‐SD model;
-
–
quant‐Nsurv‐IT.txt: quantiles of the simulated numbers of survivors under the GUTS GUTS‐RED‐IT model.
-
–
fit.cstSD.RData: fit results of model GUTS‐RED‐SD on calibration data;
-
–
fit.cstIT.RData: fit results of model GUTS‐RED‐IT on calibration data;
-
–
Nsurv‐SD.Rdata: number of survivors simulated from model GUTS‐RED‐SD under the constant exposure scenario of the calibration data set;
-
–
surv‐SD.Rdata: survival rates simulated from model GUTS‐RED‐SD under the constant exposure scenario of the calibration data set;
-
–
Nsurv‐IT.Rdata: number of survivors simulated from model GUTS‐RED‐IT under the constant exposure scenario of the calibration data set;
-
–
surv‐IT.Rdata: survival rates simulated from model GUTS‐RED‐IT under the constant exposure scenario of the calibration data set.
-
–
-
GUTS‐validation.R: contains the source code to perform simulation under the validation data set, that is under time‐variable exposure concentration profiles, and to get also graphical results. This file produces the following outputs:
-
–
validation‐profiles.png: exposure concentration profiles of the validation data set;
-
–
predict‐SOT‐SD.png: GUTS GUTS‐RED‐SD model validation on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each exposure scenario;
-
–
predict‐SOT‐IT.png: GUTS GUTS‐RED‐IT model validation on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each exposure scenario;
-
–
predict‐MF‐SD‐SOT.png: GUTS GUTS‐RED‐SD model validation on a typical acute toxicity test: the number of survivors at the end of the experiment (day 10) is represented as a function of a multiplication factor applied to each exposure;
-
–
predict‐MF‐IT‐SOT.png: GUTS GUTS‐RED‐IT model validation on a typical acute toxicity test: the number of survivors at the end of the experiment (day 10) is represented as a function of a multiplication factor applied to each exposure.
-
–
A.2.2. DEBtox model
Please refer to Table A.1 below to make easier the connection between parameter symbols in this chapter and their corresponding names within R code files.
Table A.1.
Correspondence between parameter symbols in the chapter and names used in R code
| Symbol | Unit | Meaning | R name | |
|---|---|---|---|---|
| b | 1/day | Exponential decay rate of cadmium concentration | cCext | |
|
|
(g.L−1)−2 | Precision of exposure observations | tauE | |
| k D | 1/day | Dominant rate constant | ke | |
| h b | 1/day | Background morality rate | cSDm | |
| z w | .g.L−1 | No‐effect‐concentration for survival | necSDm | |
| b w | ·g−1.L.day−1 | Survival killing rate | kSDm | |
| L m | mm | Maximum body length | LmGDm | |
| γ | 1/day | von Bertalanffy growth rate | gamGDm | |
|
|
mm−2 | Precision of body length observations | tauGDm | |
| lp | – | Scaled body length at puberty | lpRDm | |
| R m | #/day | Maximum reproduction rate | RmRDm | |
| p R | – | Dispersion of reproduction observations | p | |
| z GR | ·g.L−1 | No‐effect‐concentration for growth and reproduction | necRDm | |
| k GR | ·g−1.L.day−1 | Growth and reproduction killing rate | kRDm |
The source code of the DEBtox implementation in R is available and consists of the following files:
DEBtox‐data.R: data formatted to be used by rjags;
DEBtox‐main.R: the main R code to run the Bayesian process and make predictions;
DEBtox‐model.bug: the model code in JAGS;
DEBtox‐raw‐data.txt: the observed data as they were provided in their raw format;
DEBtox‐get‐param.R: the R code to get parameter estimates within a separate.txt file;
DEBtox‐results.R: the R code to check convergence and get graphical results of the goodness‐of‐fit. This file also provides the R code for Figures 3, 4 and 5 of Billoir et al. (2011).
These files provide outputs in two folders:
-
MCMC with two MCMC objects:
-
–
DEBtox‐MCMC‐param.RData containing the joint posterior distribution of the model fit to the observed data;
-
–
DEBtox‐MCMC‐pred.RData containing the predictions.
-
–
-
FIGS with graphical results:
-
–
DEBtox‐Cext‐Cint.pdf: fitting results with exposure data (Figure 4 from Billoir et al., 2011);
-
–
DEBtox‐density.pdf: densities of marginal posterior distributions;
-
–
DEBtox‐obs‐pred‐SGRDm.pdf: comparisons between observations and predictions for survival, growth and reproduction (Figure 5 from Billoir et al., 2011);
-
–
DEBtox‐pairs.pdf: visualisation of the joint posterior distribution, notably correlations between parameters;
-
–
DEBtox‐prior‐posterior.pdf: graphical comparison of prior and posterior distributions (Figure 3 from Billoir et al., 2011);
-
–
DEBtox‐trace.pdf: trace over the iterations of the MCMC chains.
-
–
In addition to these files, a file entitled DEBtox‐param.txt is provided to format Table A.1 as below.
Appendix B – Additional result from the GUTS application example
B.1. Fit plots as numbers of survivors
Figure B.1.
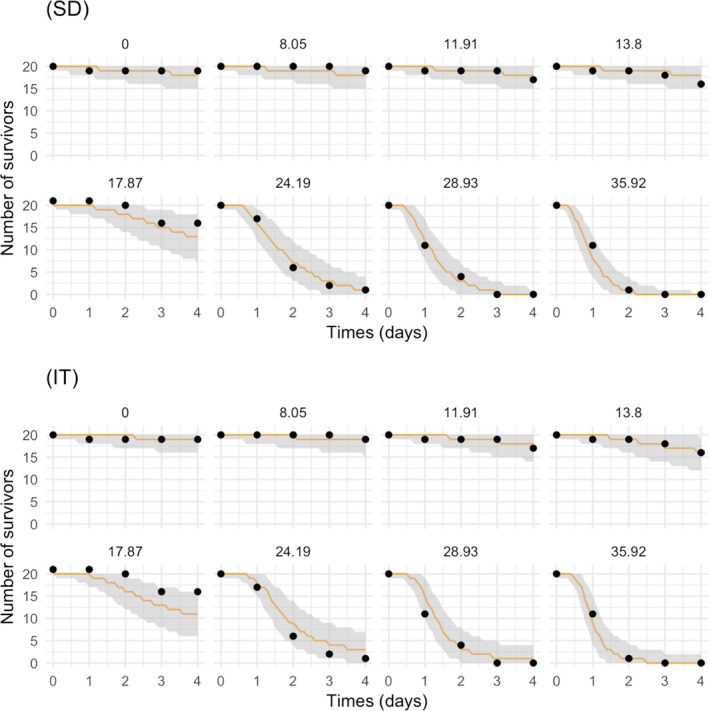
Fit plot of the GUTS‐RED‐SD (upper panel) and GUTS‐RED‐IT (lower panel) model calibration results on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each tested concentration (headers of single plots): black dots are the observed numbers of survivors, while the orange plain line corresponds to the predicted median numbers of survivors. The grey band is the 95% credibility band representing the uncertainty
B.2. Prior‐posterior comparison
B.2..1.
B.2..1.1.
Correlation plots
Figure B.4.
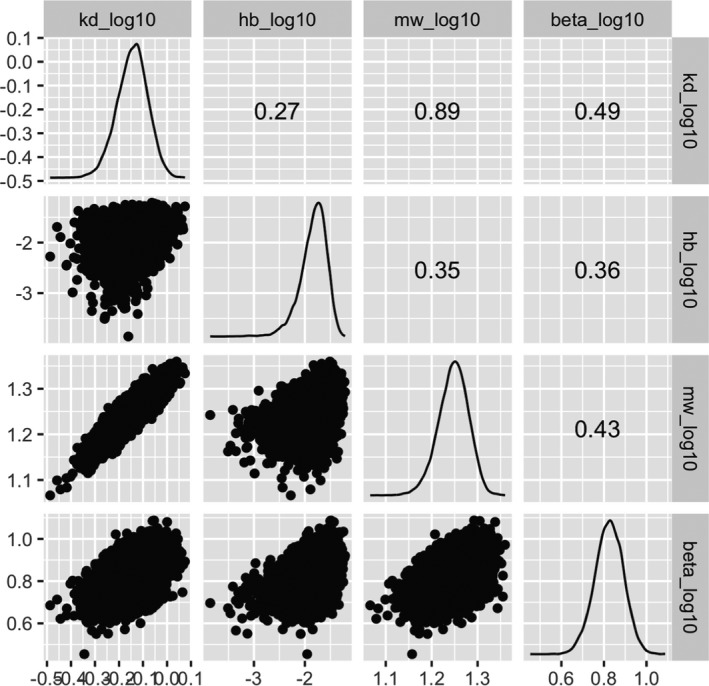
Correlation plot of the GUTS‐RED‐IT model parameters (log10‐scale)
B.3. Parameter values as optimised within the frequentist approach
Table B.1.
Optimal parameter values (column ‘Value’) as obtained in the frequentist framework (Section 4.1.3.2 and Table 3), and lower (5%) and upper (95%) confidence limits. Values obtained by application of the SimulatedAnnealing optimisation routine (see Appendix A.1 for more details). The results can be compared with results from the Bayesian framework reported in Table 3
| Symbol | Value | 5% CL | 95% CL | Units | |
|---|---|---|---|---|---|
| GUTS‐SIC‐SD | |||||
| Background hazard rate | hb | 0.028 | 0.021 | 0.031 | 1/day |
| Dominant rate constant | kD | 2.154 | 1.758 | 2.333 | 1/day |
| Killing rate | bw | 0.132 | 0.092 | 0.138 | μmol/L per day |
| Threshold | zw | 17.067 | 15.96 | 18.19 | μmol/L |
| Negative ln likelihood | ln L | 123.8307592 | |||
| GUTS‐SIC‐IT | |||||
| Background hazard rate | hb | 0.018 | 0.005 | 0.042 | 1/day |
| Dominant rate constant | kD | 0.732 | 0.576 | 0.909 | 1/day |
| Median of threshold distributions | mw | 17.83 | 16.25 | 20.34 | μmol L−1 |
| Slope of the distribution | β | 6.958 | 5.486 | 7.845 | [−] |
| Negative ln likelihood | ln L | 127.7684792 | |||
B.4. Concentration–response view in the frequentist framework
Figure B.5.
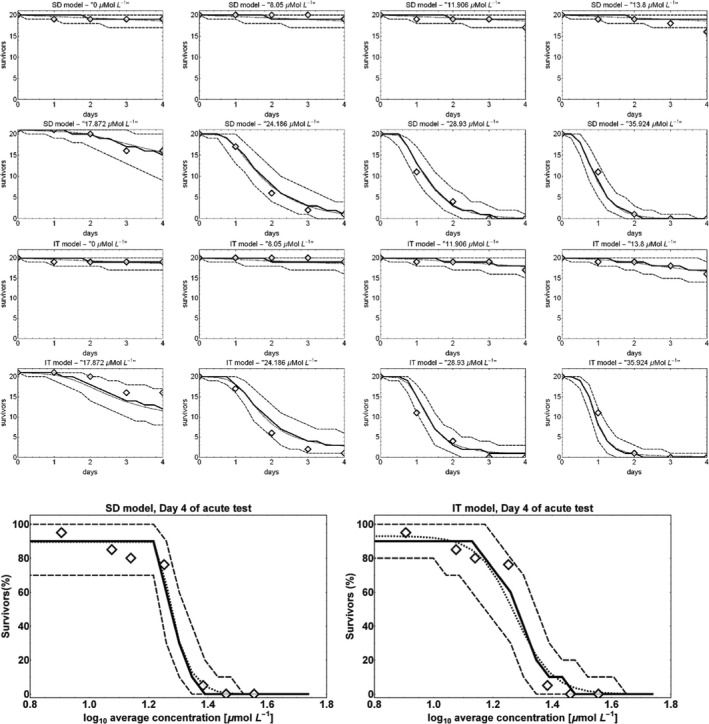
GUTS‐RED model calibration for the frequentist approach. Data from a typical acute toxicity study with propiconazole and G. pulex (see Section 4.2), with observation of survival under constant exposure over 4 days.
- (Top panels) Diamond symbols depict the survivors over time for the experimentally tested concentrations given in the plot titles. The solid lines show the modelled number of survivors, dashed lines are the 5th and 95th percentiles of model uncertainty.
- (Bottom panels) Observed (diamonds) and modelled survival at the end of the 4‐day observation period in the concentration–response view. The solid lines show the modelled survival at day 4, dashed lines are the 5th and 95th percentiles of model uncertainty. Dotted lines show the deterministic survival rate.
B.5. Validation results in the frequentist framework
Figure B.6.
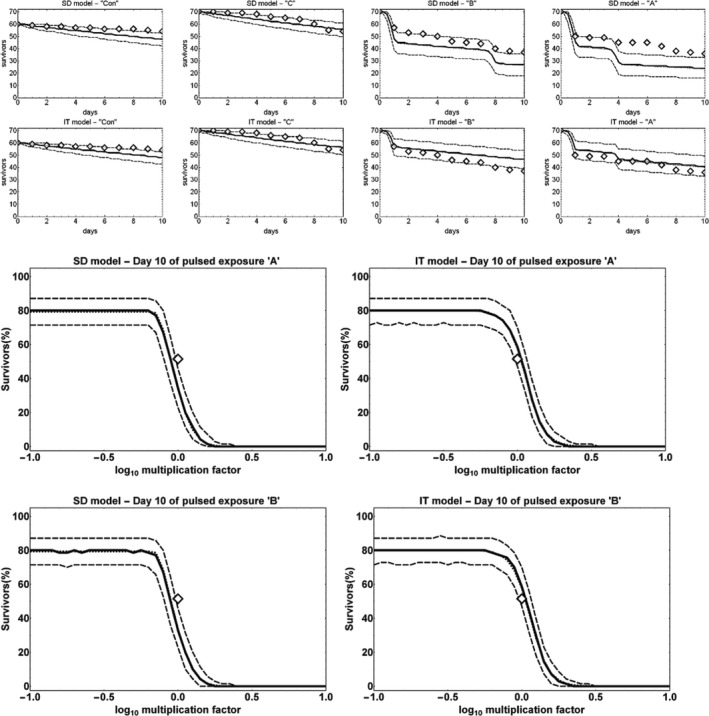
GUTS‐RED model validation for the frequentist approach (see Section 4.2.2.2)
- (Top panels) Diamond symbols depict the survivors over time for the experimentally tested scenarios given in the plot titles. The solid lines show the modelled number of survivors, dashed lines are the 5th and 95th percentiles of model uncertainty.
- (Bottom panels) Observed (diamonds) and modelled survival at the end of the 10‐day observation period in the multiplication factor ‐response view. The solid lines show the modelled survival at day 4, dashed lines are the 5th and 95th percentiles of model uncertainty. Dotted lines show the deterministic survival rate.
B.6. Documentation of the GUTS ring test results for the Bayesian framework
B.6.1. Preamble
This document quickly summarises parameter estimates and log‐likelihood value from the GUTS ring‐test. Results were obtained with the R‐package ‘morse’. R‐code lines are given with the first data set; they can be copied and pasted to repeat the analysis with the other data sets. All results can be identically reproduced through the MOSAIC web interface available at http://pbil.univ-lyon1.fr/software/mosaic/guts.
Details about the ring‐test are available in chapter 7 of the GUTS e‐book from Tjalling JAGER and Roman ASHAUER (version 1.0, 2018) downloadable at https://leanpub.com/guts_book.
The parameter names of the GUTS‐RED models are the following:
| GUTS‐RED symbol | Morse symbol | Meaning |
|---|---|---|
| kd | Dominant rate constant | |
| mw | z (model SD) | (Median) threshold |
| or α (model IT) | ||
| bw | Killing rate | |
| β | Slope factor (Fs = 2) | |
| hb | Background hazard rate |
Calibration exercises with data set A.
Data set A consists of synthetic data, generated with GUTS‐RED‐SD and GUTS‐RED‐IT. Below are the parameter values used to simulate the synthetic data sets A for SD and IT (n.a. means not applicable).
| Symbol | True value SD | True value IT |
|---|---|---|
| kd | 0.8 | 0.8 |
| mw | 3 | 5 |
| bw | 0.6 | n.a. |
| β | n.a. | 5.3 |
| hb | 0.01 | 0.02 |
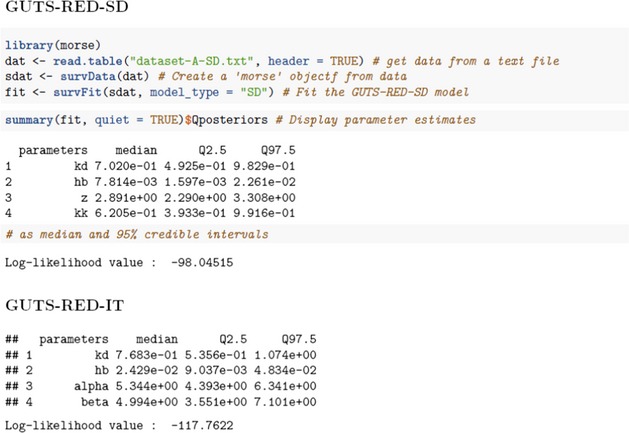
B.6.2. Calibration exercises with data set B
Data set B consists of the raw data from a typical four‐day acute toxicity study and from a non‐standard pulsed toxicity experiment taken from the supporting information of Nyman et al. (2012).
B.6.2..1.
Calibration of GUTS‐RED models using only constant exposure data

Calibration of GUTS‐RED models using only time‐variable exposure data

B.6.3. Calibration exercises with data set C
Data set C is taken from the fathead minnow toxicity database (Geiger et al., 1988). It represents a typical fish acute toxicity study, from which the raw data are used for calibration.
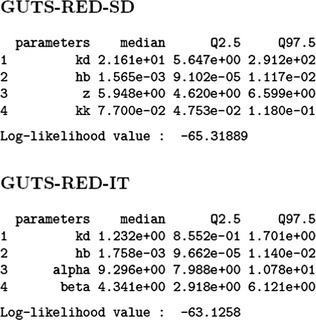
B.7. Documentation of the GUTS ring test results for the frequentist framework
B.7.1. Preamble
This section summarises parameter estimates and log‐likelihood value from the GUTS ring‐test obtained with Mathematica implementation. Example code is given below, which shows a call of the minimisation routine.
Details about the ring‐test are available in Chapter 7 of the GUTS e‐book from Tjalling Jager and Roman Ashauer (version 1.0, 2018) downloadable at https://leanpub.com/guts_book.
In the following example code lines, the optimisation routine (NMinimize) is called three times, first time with Random values between 0 and 5, and the 2nd and 3rd time with adapted starting values. The SimulatedAnnealing method was used, with 50 optimisation queues in parallel. The variable SOT$dataA contains the observed data.


B.7.2. Calibration exercises with data set A
Data set A consists of synthetic data, generated with GUTS‐RED‐SD and GUTS‐RED‐IT. Below are the parameter values used to simulate the synthetic data sets A for SD and IT (n.a. means not applicable).
| Symbol | True value SD | True value IT |
|---|---|---|
| kd | 0.8 | 0.8 |
| mw | 3 | 5 |
| bw | 0.6 | n.a. |
| β | n.a. | 5.3 |
| hb | 0.01 | 0.02 |
| GUTS‐SIC‐SD | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.008 | 0.0049 | 0.0229 | 1/day |
| Dominant rate constant | kd | 0.71 | 0.52 | 0.96 | 1/day |
| Killing rate | bw | 0.62 | 0.49 | 0.79 | L/(μmol*day) |
| Threshold | mw | 2.89 | 2.36 | 2.96 | μmol/L |
| ln likelihood | ln L | −96.4464909 |
| GUTS‐SIC‐IT | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.0262 | 0.0102 | 0.0518 | 1/day |
| Dominant rate constant | kd | 0.79 | 0.66 | 1.08 | 1/day |
| Threshold | mw | 5.42 | 4.73 | 6.41 | L/(μmol*day) |
| Slope | β | 5.19 | 4.01 | 5.26 | – |
| ln likelihood | ln L | −116.021090 |
B.7.3. Calibration exercises with data set B
Data set B consists of the raw data from a typical 4‐day acute toxicity study and from a non‐standard pulsed toxicity experiment taken from the supporting information of Nyman et al. (2012).
| GUTS‐SIC‐SD | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.0276 | 0.0211 | 0.0314 | 1/day |
| Dominant rate constant | kd | 2.15 | 1.76 | 2.33 | 1/day |
| Killing rate | bw | 0.13 | 0.09 | 0.14 | L/(μmol*d) |
| Threshold | mw | 17.06 | 15.96 | 18.19 | μmol/L |
| ln likelihood | ln L | −123.830759 |
| Parameters of GUTS‐SIC‐IT | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.0180 | 0.0048 | 0.0416 | 1/day |
| Dominant rate constant | kd | 0.73 | 0.58 | 0.91 | 1/day |
| Threshold | mw | 17.83 | 16.25 | 20.33 | L/(μmol*day) |
| Slope | β | 6.96 | 5.49 | 7.85 | – |
| ln likelihood | ln L | −127.768479 |
Calibrate GUTS‐RED model using only time‐variable exposure data
| Parameters of GUTS‐SIC‐SD | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.0231 | 0.0176 | 0.0335 | 1/day |
| Dominant rate constant | kd | 1.81 | 1.70 | 6.89 | 1/day |
| Killing rate | bw | 0.33 | 0.07 | 0.46 | L/(μmol*day) |
| Threshold | mw | 20.20 | 18.60 | 25.24 | μmol/L |
| ln likelihood | ln L | −329.031332 |
| Parameters of GUTS‐SIC‐IT | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0.0221 | 0.0159 | 0.0394 | 1/day |
| Dominant rate constant | kd | 0.20 | 0.10 | 9.47 | 1/day |
| Threshold | mw | 12.15 | 9.79 | 19.07 | L/(μmol*day) |
| Slope | β | 1.80 | 1.35 | 3.45 | – |
| ln likelihood | ln L | −333.932485 |
B.7.4. Calibration exercises with data set C
Data set C is taken from the fathead minnow toxicity database (Geiger et al., 1988). It represents a typical fish acute toxicity study, from which the raw data are used for calibration.
| GUTS‐SIC‐SD | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0 | 0 | 0 | 1/day |
| Dominant rate constant | kd | 51.344 | 7.549 | n.d. | 1/day |
| Killing rate | bw | 0.083 | 0.051 | 0.122 | L/(μmol*day) |
| Threshold | mw | 6.159 | 4.836 | 6.632 | μmol/L |
| ln likelihood | ln L | −63.2640180 |
| GUTS‐SIC‐IT | Symbol | Value | Lower CI | Upper CI | Units |
|---|---|---|---|---|---|
| Background hazard rate | hb | 0 | 0 | 0 | 1/day |
| Dominant rate constant | kd | 1.26 | 0.91 | 1.69 | 1/day |
| Threshold | mw | 9.33 | 8.62 | 10.62 | L/(μmol*day) |
| Slope | β | 4.50 | 3.12 | 6.25 | – |
| ln likelihood | ln L | −61.3141725 |
Appendix C – The log‐logistic distribution
1.
The log‐logistic distribution is a continuous probability distribution for a non‐negative random variable. It is the probability distribution of a random variable whose logarithm has a logistic distribution.
The cumulative distribution function is
where α is the median and β the shape parameter of the distribution.
The probability density function is
As a reminder, the survival probability of an individual to survive until time t under model IT is calculated by
Consequently, at a given time‐point t, the survival rate only depends on function F.
Below is the way p(x) and 1 − F(x) change as a function of x.
Hence, higher is ·, narrower is p(x) and steeper is 1 − F(x).
Appendix D – Supporting information for GUTS model implementation
Tables of raw data
1.
1.1.
List of files for model implementations
R implementation
The R implementation of GUTS models consists of the following files:
-
GUTS‐implementation‐check.R: contains the source code to perform testing and verification of R functions implemented within package ‘morse’ (see Chapter 4 for the corresponding results). This file produces the following outputs (.png for Figures):
-
–
test‐SD‐IT‐cst.png: test of the R implementation of GUTS‐RED‐SD and GUTS‐RED‐IT models under constant exposure;
-
–
test‐SD‐IT‐var.png: test of the R implementation of GUTS‐RED‐SD and GUTS‐RED‐IT models with increasing multiplication factors for pulsed exposures;
-
–
sensi‐SD‐IT‐surv5.png: R one‐at‐a‐time sensitivity analysis of models GUTS‐RED‐SD and GUTS‐RED‐IT;
-
–
sensi‐SD‐IT‐extreme.png: R implementation test of GUTS‐RED‐SD and GUTS‐RED‐IT models under extreme cases.
-
–
-
GUTS‐run‐fit.R: contains the source code to fit GUTS GUTS‐RED‐SD and ‐IT models on the calibration data set, to get graphical results for checking the convergence of the Bayesian process leading to parameter estimates, and to get plots of the results under various shapes. This file produces the following outputs (.png for Figures, .txt for text files,.RData for R objects):
-
–
plot‐fit.cstSD.png: GUTS GUTS‐RED‐SD model calibration on a typical acute toxicity test: the survival over time is represented as a function of time of each tested concentration;
-
–
plot‐fit.cstIT.png: GUTS GUTS‐RED‐IT model calibration on a typical acute toxicity test: the survival over time is represented as a function of time of each tested concentration;
-
–
plot‐ppc.cstSD.png: Posterior Predictive Check for the GUTS GUTS‐RED‐SD model, calibrated on a typical acute toxicity test;
-
–
plot‐ppc.cstIT.png: Posterior Predictive Check for the GUTS GUTS‐RED‐IT model, calibrated on a typical acute toxicity test;
-
–
plot‐Nsurv‐SD.png: GUTS GUTS‐RED‐SD model calibration on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each tested concentration;
-
–
plot‐sot‐SD.png: GUTS GUTS‐RED‐SD model calibration on a typical acute toxicity test: the survival rate over time is given as a function of time for each tested concentration;
-
–
plot‐Nsurv‐IT.png: GUTS GUTS‐RED‐IT model calibration on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each tested concentration;
-
–
plot‐sot‐IT.png: GUTS GUTS‐RED‐IT model calibration on a typical acute toxicity test: the survival rate over time is given as a function of time for each tested concentration;
-
–
surv‐dose‐response.png: GUTS GUTS‐RED‐SD and ‐IT model calibration on a typical acute toxicity test: the survival rate (%) is represented as a function of the concentration at the end of the experiment (day 4);
-
–
Nsurv‐dose‐response.png: GUTS GUTS‐RED‐SD and ‐IT model calibration on a typical acute toxicity test: the number of survivors is represented as a function of the concentration at the end of the experiment (day 4);
-
–
LC50‐versus‐time.png: LC50 estimates from the calibrated GUTS SIC models;
-
–
GUTS‐calibration‐results.txt: parameter estimates of the GUTS GUTS‐RED‐SD and ‐IT models fitted on the calibration data set;
-
–
quant‐Nsurv‐SD.txt: quantiles of the simulated numbers of survivors under the GUTS GUTS‐RED‐SD model;
-
–
quant‐surv‐SD.txt: quantiles of the simulated survival rate over time under the GUTS GUTS‐RED‐SD model;
-
–
quant‐Nsurv‐IT.txt: quantiles of the simulated numbers of survivors under the GUTS GUTS‐RED‐IT model;
-
–
quant‐surv‐IT.txt: quantiles of the simulated survival rate over time under the GUTS GUTS‐RED‐IT model;
-
–
fit.cstSD.RData: fit results of model GUTS‐RED‐SD on calibration data;
-
–
fit.cstIT.RData: fit results of model GUTS‐RED‐IT on calibration data;
-
–
Nsurv‐SD.Rdata: number of survivors simulated from model GUTS‐RED‐SD under the constant exposure scenario of the calibration data set;
-
–
surv‐SD.Rdata: survival rates simulated from model GUTS‐RED‐SD under the constant exposure scenario of the calibration data set;
-
–
Nsurv‐IT.Rdata: number of survivors simulated from model GUTS‐RED‐IT under the constant exposure scenario of the calibration data set;
-
–
surv‐IT.Rdata: survival rates simulated from model GUTS‐RED‐IT under the constant exposure scenario of the calibration data set.
-
–
-
GUTS‐validation.R: contains the source code to perform simulation under the validation data set, that is under time‐variable exposure concentration profiles, and to get also graphical results. This file produces the following outputs:
-
–
validation‐profiles.png: exposure concentration profiles of the validation data set;
-
–
predict‐SOT‐SD.png: GUTS GUTS‐RED‐SD model validation on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each exposure scenario;
-
–
predict‐MF‐SD‐SOT.png: GUTS GUTS‐RED‐SD model validation on a typical acute toxicity test: the number of survivors at the end of the experiment (day 10) is represented as a function of a multiplication factor applied to each exposure;
-
–
predict‐SOT‐IT.png: GUTS GUTS‐RED‐IT model validation on a typical acute toxicity test: the number of survivors over time is represented as a function of time for each exposure scenario;
-
–
predict‐MF‐IT‐SOT.png: GUTS GUTS‐RED‐IT model validation on a typical acute toxicity test: the number of survivors at the end of the experiment (day 10) is represented as a function of a multiplication factor applied to each exposure;
-
–
internal‐conc.png: simulation of the scaled internal concentration under exposure profiles ‘A’ and ‘B’ for both k D median estimates of model GUTS‐SD and GUTS‐IT.
-
–
-
GUTS‐fit‐addons.R: contains the source code to get additional fitting results of the GUTS‐SD and the GUTS‐IT models on the calibration data set. This file produces the following outputs:
-
–
prior‐posterior‐SD.png: comparison between priors and posteriors of the GUTS‐SD model parameters;
-
–
prior‐posterior‐IT.png: comparison between priors and posteriors of the GUTS‐IT model parameters;
-
–
correlation‐SD.png: correlation plot of GUTS‐SD mode parameters;
-
–
correlation‐IT.png: correlation plot of GUTS‐IT mode parameters.
-
–
Appendix E – Repository of codes
1.
Appendix E can be found in the online version of this output (‘supporting information’ section).
Appendix F – Example of the evaluation of an available GUTS model
1.
In the context of the approval of the active substance Benzovindiflupyr, a TKTD model was submitted to predict survival of the two most sensitive species of fish under more realistic exposure (time‐variable) to the fungicide. The GUTS model was used. The documentation of the model is available in the RAR of benzovindiflupyr (https://www.efsa.europa.eu/it/efsajournal/pub/4043). Additional information is also provided in the paper by Ashauer et al. (2013).
| ASPECT OF THE MODEL TO BE EVALUATED BY THE RISK ASSESSOR – GUTS model application for lethal effectsPlease note that when a Yes/No answer is not possible or not applicable, the box is highlighted in yellow | Yes | No | |
|
1. Evaluation of the problem definition The problem definition needs to explain how the modelling fits into the risk assessment and how it can be used to address the specific protection goals. For GUTS, questions to be answered are likely to be those that are set out in Section 3. Nevertheless, the problem definition should make clear the following points: | |||
| (a) | Is the regulatory context for the model application documented? | X | |
| (b) | Is the question that has to be answered by the model clearly formulated? | X | |
| (c) | Is the model output suitable to answer the formulated questions? | X | |
| (d) | Was the choice of the test species clearly described and justified, also considering all the available valid information (including literature)? | X | |
| (e) | Is the species to be modelled specified? – Is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species), i.e. Tier‐2C2? | X | |
|
2. Evaluation of the quality of the supporting experimental data In this part of the evaluation, it is checked whether the experimental data with which the model is compared (both calibration and validation data sets) have been subjected to quality control. The focus is on the data quality, i.e. the laboratory conditions, set‐up, chemical analytics and similar. Additional specific criteria for the suitability of the data sets for model calibration and validation are evaluated later in more detail (Sections 7 and 9 of this checklist). | |||
| (a) | Has the quality of the data used been considered and documented? (see list of OECD test guidelines in Section 7, Table 6) | X | |
| (b) | Have all available data been used (either for calibration or for validation)? If not, is there a justification why some information has not been used? | X | |
| (c) | Is it checked whether the actual exposure profile in the study matches the intended profile in the test (+/− 20%); if not, are then measured concentrations used for the modelling, instead of nominal ones? | X | |
|
3. Evaluation of the conceptual model Providing GUTS models are being used to address mortality/immobility effects in fish or invertebrates, the conceptual model will be suitable to address the specific protection goals; so, no further evaluation is required (see Sections 2.1, 2.2 and 4.1). | |||
|
4. Evaluation of the formal model The formal model contains the equations and algorithms to be used in the model. For GUTS models, the equations are standardised, so that no further check is necessary (see Section 4.1.1). It has to be documented, however, which GUTS model version is used (full or reduced model). | |||
|
5. Evaluation of the computer model The formal model is converted into a model that can run on a computer (the computer model). For GUTS models, the computer model can be tested by showing the model performance for the GUTS ring‐test data and performing some further checks (see Section 7.5). | |||
| (a) | Is the used implementation of GUTS tested against the ring‐test data set (see Section 4.2 | X | |
| (b) | Were GUTS parameters estimated for the ring‐test data and compared to the reference values, including confidence or credible intervals (Appendix B.6 and B.7)? | X | |
| (c) | Is a set of default scenarios (e.g. standard scenarios, extreme cases, see Section 4.1.2) simulated and checked? | X | |
| (d) | Are all data and parameters provided to allow an independent implementation of GUTS to be run? | X | |
|
6. Evaluation of the regulatory model – the environmental scenarios For GUTS models using FOCUS simulations (or other Member State‐specific exposure simulations) as exposure input, no further definition and check of the environmental conditions are needed, since pesticide concentrations will be generated using the relevant FOCUS simulations (or MS‐specific exposure simulations), which consider factors such as soil, rainfall and agronomic practice, and the (effect) model will have been calibrated from data collected under standard laboratory conditions. Fixing the environmental scenarios to the conditions of calibration experiments is appropriate because the modelling will be used with the equivalent of Tier‐1 or Tier‐2 assessment factors, so an extrapolation from laboratory to field conditions is already covered. | |||
|
7. Evaluation of the regulatory model – parameter estimation Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine, and the comprehensive documentation of methods and results. Model parameters are always estimated for a specific combination of species and compound (see Chapter 3 for background information).Supporting data for GUTS models are mortality or immobility data, have to be of sufficient quality (Section 2 in this checklist) and fulfil a set of basic criteria. Please check the following items to evaluate the calibration data, and the parameter optimisation process and the results (see Sections 4.1.3.1 and 7.6.2): | |||
| (a) | Is it clear which parameters have been taken from literature or other sources and which have been fitted to data?If used, are values from literature reasonable and justified? | X | |
| (b) | Are raw observations of mortality or immobility reported for at least five time‐points? | X | |
| (c) | Does calibration data span from treatment levels with no effects up to strong effects, ideally up to full effects (e.g. 0% survival)? | X | |
| (d) | Have all data available for calibration been used? If not, is there a justification why this information has not been used? | X | |
| (e) | Has attention been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide? | X | |
| (f) | Has the model parameter estimation been adequately documented, including settings of optimisation routines, and type and settings of the numerical solver that was used for solving the differential equations? | X | |
| (g) |
If Bayesian inference has been used, are priors on model parameters reported? If a frequentist approach has been used, are starting values for the optimisation reported? |
X | |
| (h) | Are the estimated parameter values reported including confidence/credible intervals? | X | |
| (i) | Is the method to get these limits reported and documented? | X | |
| (j) | Are the optimal values of the objective function for calibration (e.g. Log‐likelihood function) as the result of the parameter optimisation reported? | X | |
| (k) | Are plots of the calibrated GUTS models in comparison with the calibration data over time provided, and does the visual match appear of acceptable quality? | X | |
| (l) | Has a posterior predictive check been performed and documented? | X | |
|
8. Evaluation of the sensitivity and uncertainty analysis For the reduced GUTS models, the influence of the model parameters on the model results are known well enough. Results of sensitivity analyses can, if contained, demonstrate that the model implementation is done correctly. For other GUTS models than the reduced, sensitivity analyses should be included for future applications and be checked by the following list. | |||
| (a) | Has a sensitivity analysis been performed and adequately documented? (The range of parameter variation in the sensitivity analysis should be justified by an analysis of the expected variation of model parameters) | X | |
| (b) | Are the results of the sensitivity analysis presented so that the most sensitive parameters can be identified? | X | |
| (c) | Is the parameter uncertainty for the most important TKTD model parameters propagated to the model outputs and the results of the uncertainty propagation been documented? | X | |
| (d) | Are the model outputs reported including confidence/credible intervals? | X | |
|
9. Evaluation of the model by comparison with independent measurements (model validation) Validation data are used to test the model performance for predictions of mortality/immobility under exposure profiles which have not been used for model calibration. The performance of the model is usually evaluated by comparing relevant model outputs with measurements (often referred to as model validation). For GUTS, relevant outputs are the simulated mortality/immobility probability or LPX/EPX values. The following checklist is mandatory only for invertebrates; for vertebrates, a case‐by‐case basis check needs to be done (see also Sections 7.7.2 and 4.1.4.5) | |||
| (a) | Are effect data available from experiments under time‐variable exposure? | X | |
| (b) | Is mortality or immobility reported at least for 7 time‐points in the validation data set? | X | |
| (c) | Are two exposure profiles tested with at least two pulses each, separated by no‐exposure intervals of different duration length? | X | |
| (d) | Is the individual depuration and repair time (DRT95) calculated, and is the duration of the no‐exposure intervals defined accordingly?[Ideally one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95] | X | |
| (e) | Is each profile tested at least at 3 concentration levels, in order to obtain low, medium and high effects at the end of the respective experiment? | X | |
| (f) | Has attention been paid to the duration of the experiments considering the time course of development in toxicity of the specific pesticide? | X | |
| (g) | Does the visual match (‘visual fit’ in FOCUS Kinetics (2006)) of the model prediction quality indicate acceptability of the model predictions in comparison with the validation data? | X | |
| (h) | Do the reported quantitative model performance criteria (e.g. PPC, NRMSE, SPPE) indicate a sufficient model performance? | X | |
| (i) | Has the performance of the model been reported in an objective and reproducible way? | X | |
|
10. Evaluation of model use When using a TKTD model for regulatory purposes, the inputs of species‐ and compound specific model parameters and of exposure profile data are required to run the model under new conditions. In this stage, it is important that the model is well documented and that it is clear how the model works. Please check the following items: | |||
| (a) | Is the use of the model sufficiently documented? | X | |
| (b) | Is an executable implementation of the model made available to the reviewer, or Is at least the source code provided? | X | |
| (c) | Has a summary sheet been provided by the applicant? The summary sheet should provide quick access to the comprehensive documentation with sections corresponding to the ones of this checklist. | X | |
| (d) | Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data? For example if FOCUS Step 3 maximum values were used at Tier‐1, are the exposure profiles used Tier‐2C from the same FOCUS Step 3 modelling? If the exposure profile comes from any other source (e.g. different scenarios, different inputs, different model) has this been checked? | X | |
| (e) |
Further points to be checked by evaluators Use an independent implementation of GUTS to test whether the output of the evaluated model implementation can be reproduced for some parameter sets.The MOSAIC_GUTS web‐platform (http://pbil.univ-lyon1.fr/software/mosaic/guts) can be used to test the model calibrationThe GUTS Shiny App (http://lbbe-shiny.univ-lyon1.fr/guts-shinyapp/) to test model predictions under a specific constant or time‐variable exposure profile given the set of model parameters can be used. |
||
1.1.
1.1.1.
Explanation of the evaluation
1. Evaluation of the problem definition
It was clear from the model documentation, that the study was conducted as a higher‐tier approach for risk refinement, addressing lethal effects, as part of a weight‐of‐evidence approach, hence it is concluded that the regulatory context for the model application is not well enough documented. A clear question (objective) to be answered by the model was formulated: ‘The objective of this study was the prediction of acute toxicity towards fish under fluctuating and pulsed exposure patterns’. The model output was not recognised as suitable to answer the formulated questions, because the current definition recommends using the multiplication factor leading to 50% mortality (LP50) in the context of acute risk assessment together with a lower‐tier assessment factor. In general, there is a specific issue with the selected tolerated effect level of 10% (as used for the multiplication factor analysis), which is not in accordance with the SPG for vertebrates in the AGD. When the given effect level would have been used together with a lower‐tier safety factor it would have been tolerable, but that was not the case for the study: ‘The safety margins ranged from a factor of 26 to 500. Thus, the concentrations in the original FOCUS‐SW exposure profiles are a factor 26–500 below levels for the onset of mortality in the analysed combinations of fish species and application patterns’. The modelled species have been presented and consist of five standard fish species for acute toxicity tests. Models have been parameterised for all five fish species, but predictions of survival under time‐variable exposure profiles were carried out for the two most sensitive fish species (Cyprinus carpio & Pimephales promelas). The choice of the test species is considered to be clearly described and justified. In principle, it would have been possible to calculate LP50 values for 5 fish species and by using the SSD approach to calculate the corresponding HP5 values. These HP5 values should be larger than the AF of 9 as proposed for acute fish SSDs in the Aquatic Guidance Document. In the current example, however, validation results were presented for only one species.
The question ‘is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2’ is difficult to be answered, because this example was produced before this Opinion was written, so the concept of Tier‐2C1 and Tier‐2C2 had not been developed. The two most sensitive species were modelled, but this isn't strictly in line with the proposals for Tier‐2C1 (only standard species) or Tier‐2C2 (all available species unless there is a clear difference in sensitivity).
2. Evaluation of the quality of the supporting experimental data
Standard laboratory studies were provided on several fish and were evaluated in the DAR for benzovindiflupyr. The table below lists the studies provided and whether they were considered suitable for use in the risk assessment (which includes suitable quality to use for the TKTD modelling, but does not address whether the study type/design is useful for the TKTD mode) when evaluated by the RMS.
| Species | Study | Is it of suitable quality? |
|---|---|---|
| Rainbow trout | Acute, OECD 203 | Yes |
| Carp | Acute, OECD 203 | Yes |
| Fathead minnow | Acute, OECD 203 | Yes |
| Sheepshead minnow | Acute, OECD 203 | Yes |
| Bluegill sunfish | Acute, OECD 203 | Yes |
| Fathead minnow | ELS, OECD 210 | Yes |
The most sensitive species tested (acute tests) was the carp (Cyprinus carpio), followed by the fathead minnow (Pimephales promelas). The difference in sensitivity of all five species tested in acute tests was within an order of magnitude (LC50s 3.5–27 μg a.s./L). It is concluded that the quality of the data used has been considered and documented.
The acute toxicity studies for the two species modelled were used for the model calibration. The chronic toxicity study was used for model validation. It is concluded that all available data had been used (although it is noted that no validation data set was available for the carp).
The acute toxicity study on carp was conducted under flow through conditions and measured concentrations were between 86% and 110% of nominal. The endpoint was calculated on the basis of mean measured concentrations. The acute toxicity study on fathead minnow was conducted under flow through conditions and measured concentrations were between 86% and 94% of nominal. The endpoint was calculated on the basis of mean measured concentrations. The ELS toxicity study on fathead minnow was conducted under flow through conditions for 32 days and measured concentrations were between 88% and 120% of nominal. The endpoint was calculated on the basis of mean measured concentrations. It is concluded that the actual exposure profile in the study matches the intended profile in the test (+/−20%) (section B.9.2.1 (acute toxicity studies) and B.9.2.3 (chronic toxicity studies) of the DAR for benzovindiflupyr).
3. Evaluation of the conceptual model
The GUTS models were used to address mortality/immobility effects in fish, and the standard GUTS framework was applied, hence the conceptual model is considered to be suitable to address the specific protection goals.
4. Evaluation of the formal model
The GUTS standard model equations using the reduced versions of the SD and IT model have been used, so no further check of the formal model is necessary.
5. Evaluation of the computer model
The study was performed using a GUTS implementation in ModelMaker. It had been submitted before the ring test data were provided, hence it was impossible to test the model performance against the GUTS ring‐test data or to compare parameters to the reference values from the GUTS ring test. No additional testes, for example with standard or extreme scenarios have been provided to show the correct implementation of the model. A one‐at‐a‐time sensitivity analysis was performed and documented (P. 161 of DAR). All parameters values are given in the model documentation, but detailed information about the exposure profiles is missing in the model documentation.
6. Evaluation of the regulatory model – the environmental scenarios
Since the applied GUTS models used FOCUS surface water simulations as exposure input, no further definition and check of the environmental conditions are needed.
7. Evaluation of the regulatory model – parameter estimation
Some aspects of the parameter estimation have been evaluated positively. For model calibration, GUTS model parameters have been fitted to data and raw observations of mortality or immobility for five time‐points (including initial abundance). The calibration data shows strong effects in the highest treatment levels, and all data available for calibration can be considered to be used. Parameter values have been estimated and calibrated including confidence intervals, and optimal values of the objective function for calibration (log‐likelihood) as the result of the parameter optimisation were reported (Table 1 in Ashauer et al., 2013). Plots of the calibrated GUTS models in comparison with the calibration data over time are provided, and the visual match appear of good quality (pp. 149–151 in the DAR). The question whether attention has been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide is answered with ‘Yes’, because the ELS was performed under chronic exposure (28 days) and used for validation. Hence, delayed effects would have been detected in the validation experiments.
Some aspects of the model parameter estimation have been evaluated negatively, including the following aspects. The model parameter estimation has not been adequately documented, since settings of optimisation routines and type and settings of the numerical solver that were used for solving the differential equations are not documented. Also, starting values for the optimisation are not reported. The method to obtain parameter confidence limits is not adequately reported and documented, since it is a simple reference to a publication. A posterior predictive check has not been performed and documented.
8. Evaluation of the sensitivity and uncertainty analysis
The reduced GUTS models have been used, hence influence of the model parameters on the model results are considered to be known well enough. Despite this, a one‐at‐a‐time sensitivity analysis was performed and documented and it demonstrates that the model implementation is done correctly.
An analysis of uncertainty in the model output was not performed in a technical sense, since the uncertainty in the parameter has not been propagated to the model output.
It is acknowledged, that the study had been performed before the respective computational approach was developed, and that a comprehensive discussion of the role of uncertainty for the model output was included in the model documentation (p. 161/162 of the DAR).
9. Evaluation of the model by comparison with independent measurements (model validation)
The data used for validation is an Early Life Stage test with fathead minnow according to OECD 210 under constant exposure. More than 3 treatment levels have been tested, and the number of time‐points of reported mortality is larger than 7.
The use of early life stage test data can be considered to deliver conservative results, since in general larvae tend to be more sensitive than the adult fish, and fish early life stage tests tend to be more sensitive due to size and the prolonged exposure duration. Uncertainty about a positive answer to the abovementioned key question is added by considerations, whether fish fry is really a more sensitive indicator as compared to adult fish in this specific case. For the adult fathead minnow, an exposure to 4.4 μg/L led to about 40% mortality after 4 days. For the fry, exposure to a similar level of 4 μg/L did not indicate any effect on survival at day 4, also not on day 5, but only at day 6 roughly 40% mortality was observed.
The key question for the evaluation of the validity of the validation study is whether the combination of calibration and validation data allows for a conclusion about the confidence in the model performance for time‐variable exposure profiles. The scientific opinion suggests evaluating the validation data set for vertebrates on a case‐by‐case basis in order to make use of the available data and to limit as much as possible additional vertebrate testing. It is, however, also recommended to have at least one new fish test under time‐variable exposure conditions.
In conclusion, in this specific example many of the validation aspects cannot be evaluated positive. Most significantly, the validation data have not been obtained from time‐variable exposure. In addition, assuming that the data used for validation had been considered sufficient, this would only be applicable to fathead minnow since no further validation data for any other species were tested. Hence model predictions for carp and other species would have not been considered relevant for regulatory risk assessment.
10. Evaluation of model use
While overall the modelling is well described there are aspects that have not been well described, therefore it is concluded that the use of the model is not sufficiently well documented. The question ‘Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data?’ has been answered with ‘no’ because insufficient information has been provided to identify what exposure was used. Output from FOCUS modelling was used but it is unclear whether a) identical inputs were used by the applicant for the standard risk assessment and for the TKTD Tier‐2 risk assessment and b) if the submitting was accepted without change or whether the RMS reran the FOCUS modelling. It is necessary to include sufficient information with the TKTD report to be able to identify the source of the exposure profiles.
11. Conclusion
Overall, the documentation of the GUTS model and its application to predict the survival of fish under exposure to time‐variable profiles of benzovindiflupyr does not comply with the requirements as announced in this Scientific Opinion. Many aspects of the model documentation were evaluated positive, but there were a number of issues. Most critical issues are about the documentation of the computer model, the documentation of the parameter estimation method, the analysis of uncertainty in the model output, and the choice of the data set for model validation. It is of course acknowledged, that the model application was performed long time before the EFSA TKTD SO has been published.
Appendix G – Example of the evaluation of an available DEBtox model
1.
In the context of the approval of beta‐cyfluthrin, a DEBtox model was submitted with the scope of investigating the reasons leading to different results, in terms of survival and growth, obtained the available chronic studies (one carried out under constant exposure and another with repeated peak exposure) on rainbow trout. The documentation is available in the RAR of beta‐cyfluthrin (in publication).
| ASPECT OF THE MODEL TO BE EVALUATED BY THE RISK ASSESSOR – DEBtox model applications for sublethal effectsPlease note that when a Yes/No answer is not possible or not applicable, the box is highlighted in yellow. | Yes | No | |
|
0. Evaluation of the physiological DEB part DEBtox models consist of two parts: (iii) the physiological DEB part describing the basic metabolic processes combined with species‐specific details; this part is assumed to have been evaluated and approved beforehand; (iv) the TKTD part, describing the effects of toxicant on life‐history traits through changes in the DEB parameters. This checklist is adapted to the evaluation of the TKTD part of the DEBtox models (see Sections 2.3 and 5), but the first thing to check is whether the physiological part, meaning the DEB part, was evaluated beforehand for the chosen species. Without that check, the use of a DEBtox model cannot be suggested, even if the TKTD model part is evaluated positively. | |||
| (a) | Was the evaluation of the physiological DEB part conducted by a regulatory authority or a group delegated to this at the EU level? | X | |
| (b) | If (a) is yes, did the above evaluation of the physiological DEB part conclude that it is suitable for use in risk assessment? | ||
| (c) | If (a) and (b) are yes, can the physiological DEB part of the model describe the behaviour of the control data? | ||
|
1. Evaluation of the problem definition The problem definition needs to explain how the modelling fits into the risk assessment and how it can be used to address the specific protection goals (Chapter 3). Please check if due attention is paid to the following points: | |||
| (a) | Is the regulatory context for the model application documented? | X | |
| (b) | Is the question that has to be answered with the model clearly formulated? | X | |
| (c) | Is the model output suitable to answer the formulated questions? | X | |
| (d) | Is the choice of the test species clearly described and justified, also considering all the available valid information (including literature)? | X | |
| (e) | Is the species to be modelled specified? – Is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2? | X | |
|
2. Evaluation of the quality of the supporting experimental data In this part of the evaluation, it is checked whether the experimental data with which the model is compared (both calibration and validation data sets) have been subjected to quality control. The focus is on the data quality, i.e. the laboratory conditions, set‐up, chemical analytics and similar. Additional specific criteria for the suitability of the data sets for model calibration and validation are evaluated later in more detail (Sections 7 and 9 of this checklist). | |||
| (a) | Are all types of data fully described, meaning that factors like temperature, food conditions, measurements, handling, etc. are documented? | X | |
| (b) | Has the quality of the used data been considered and documented? (see the list of OECD test guidelines in Section 7, Table 6). | X | |
| (c) | Have all available data been used (either for calibration or for validation)? If not, is there a justification why some information has not been used? | X | |
| (d) | Is it checked whether the actual exposure profile in the study matches the intended profile in the test (+/‐ 20%); if not, are then measured concentrations used for the modelling, instead of nominal ones? | X | |
|
3. Evaluation of the conceptual model The conceptual model provides a general and qualitative description of the system to be modelled. The physiological DEB part should have been evaluated and approved beforehand (see Section 0 of this checklist). Please check the following items for the TKTD part (refer to Section 7.3): | |||
| (a) | Is the reference to the evaluation of the physiological DEB part given? | X | |
| (b) | Is the potential mode of action of the toxicant specified (effects on assimilation, growth, maintenance costs, reproductive output and/or survival)? | X | |
| (c) | Are the links between the scaled damage or the internal concentration and the DEB parameters logical? | X | |
| (d) | In case environmental factors (e.g. temperature) are explicitly considered, is their influence on the physiological and/or TKTD processes documented in the conceptual model? | X | |
| (e) | Are the modelling endpoints relevant to the specific protection goal? | X | |
|
4. Evaluation of the formal model The formal model contains equations used in the model. The physiological DEB part should have been evaluated and approved beforehand (see Section 0 of this checklist). Nevertheless, all equations (from either the physiological or the TKTD part) that include one or more parameters that are impacted by the effects of the toxicant need to be documented. Please check the following items: | |||
| (a) | Are all mathematical equations relevant for the effect of the toxicant described, including the equations of the toxicokinetic (TK) part and the equations relating the used scaled damage or internal concentrations with the DEB parameter(s)? | X | |
| (b) | Is there a list or a summary of all variables and parameters including their meaning and unit? | X | |
| (c) | Are both the deterministic part (equations describing the mean tendency of the data) and the stochastic part (the probability law describing the variability around the mean tendency) of the model fully described? See Section 5.1 for an example. | X | |
|
5. Evaluation of the computer model The computer model corresponds to the implementation of the formal model that can run on a computer. Please check the following items: | |||
| (a) | Is the computer code available, including explaining comments for the most important functions? | X | |
| (b) | Is enough information provided to allow non‐expert user to re‐run the model independently (e.g. parameter sets, input data)? | X | |
| (c) | Is it demonstrated that the mathematical model is correctly implemented (model verification), e.g. by checking and documenting the internal consistency of the model results based on a set of default or extreme cases (see e.g. Section 4.1.2.3 for an example with GUTS, ‘reality check’ in chapter 10.4 of EFSA PPR Panel, 2014)? | X | |
|
6. Evaluation of the regulatory model – the environmental scenarios The environmental scenarios determine the environmental context in which the model is run. For the application of DEBtox for the prediction of toxicological effects in a regulatory context, the environmental scenario need to be fixed to the laboratory conditions of the experiments used for calibrating the TKTD part of the model. Please check the following items: | |||
| (a) | Are all the relevant conditions, recorded in the experiments used for calibrating the TKTD part of the model, used consistently for the simulations (i.e. for generating EPx) used in the risk assessment? | ||
|
7. Evaluation of the regulatory model – parameter estimation Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine, and the comprehensive documentation of methods and results. Model parameters are always estimated for a specific combination of species and compound (see Section 3 for background information). Supporting data for DEBtox have to be of sufficient quality (Section 2 in this checklist), be relevant to the risk assessment problem and fulfil a set of basic criteria. Typical supporting data for the TKTD part of DEBtox models are experimental toxicity data for growth or growth + reproduction, to which mortality or immobility data can also be added (see Sections 2.3 and 7.2 for background information, Section 5.1.3 for an example). Specific requirements on the time resolution and temporal scale for the calibration data cannot be given, but in general, the number of time‐points has to be sufficient to provide enough degrees of freedom for the model calibration. Please check the following items to evaluate the calibration data, the parameter optimisation process and the results (see Sections 4.1.3 and 7.6.2): | |||
| (a) | Is it clear which parameters have been taken from literature or other sources and which have been fitted to data?If used, are values from literature reasonable and justified? | X | |
| (b) | Are the calibration data sufficient to provide enough degrees of freedom for the model calibration, i.e. are endpoints at least reported at several intermediate time‐points? | X | |
| (c) | Does calibration data span from treatment levels with no effects up to strong effects, ideally up to full effects (e.g. no growth, no reproduction)? | X | |
| (d) | Have all data available for calibration been used? If not, is there a justification why this information has not been used? | X | |
| (e) | Has attention been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide? | X | |
| (f) | Has the model parameter estimation been adequately documented, including settings of optimisation routines, and type and settings of the numerical solver that was used for solving the differential equations? | X | |
| (g) | If Bayesian inference has been used, are priors on model parameters reported?If a frequentist approach has been used, are starting values for the optimisation reported? | X | |
| (h) | Are the estimated parameter values reported including confidence/credible limits? | X | |
| (i) | Is the method to get these limits reported and documented? | X | |
| (j) | Are the optimal values of the objective function for calibration (e.g. log‐likelihood function) as the result of the parameter optimisation reported? | X | |
| (k) | Are plots of the calibrated DEBtox model in comparison with the calibration data over time provided, and does the visual match appear of acceptable quality? | X | |
| (l) | Has a posterior predictive check been performed and documented? | X | |
|
8. Evaluation of the sensitivity and uncertainty analysis It is assumed that sensitivity and uncertainty analyses for the physiological DEB model part have been evaluated and approved beforehand (see Section 0 of this checklist).Sensitivity analysis identifies the influence of the parameters on the model outputs and can hence identify the most influential parameters. Uncertainty analysis aims at identifying how uncertain the model output is regarding the uncertainty in parameter estimates.Please check the following items for the TKTD part: | |||
| (a) | Has a sensitivity analysis for the TKTD part been performed and been adequately documented? (The range of parameter variation in the sensitivity analysis should be justified by an analysis of the expected variation of model parameters.) | X | |
| (b) | Are the results of the sensitivity analysis presented so that the most sensitive parameters can be identified? | X | |
| (c) | Is the parameter uncertainty for the most important TKTD model parameters propagated to the model outputs and the results of the uncertainty propagation been documented? | X | |
| (d) | Are the model outputs reported including confidence/credible intervals? | X | |
|
9. Evaluation of the model by comparison with independent measurements (model validation) The model performance is usually evaluated by comparing relevant model outputs with independent experimental measurements (i.e. data from other experiments than those used for calibration, often referred to as model validation). These independent measurements (or validation data sets) are used to test the model performance in predicting the chosen endpoints under time‐variable exposure profiles. For DEBtox models, relevant outputs may be simulated growth and/or reproduction, sometimes together with simulated survival, or EPx values. The following checklist is mandatory only for invertebrates, for vertebrates a case‐by‐case basis check needs to be done (see also Sections 7.7.2 and 4.1.4.5): | |||
| (a) | Are effect data available from experiments under time‐variable exposure? | X | |
| (b) | If non‐destructive measurement is possible, are sufficient endpoint measurements provided in order to cover at least before, during and after each pulse exposure?Are these time‐points enough to allow for evaluation of changes in growth rates different from the control treatment? | X | |
| (c) | Are two exposure profiles tested with at least two pulses each, separated by no‐exposure intervals of different duration length? | X | |
| (d) | Is the individual depuration and repair time (DRT95) calculated based on toxicokinetic parameters, and is the duration of the no‐exposure intervals defined accordingly?[Ideally one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95] | X | |
| (e) | Is each profile tested at least at 3 concentration levels, in order to obtain low, medium and high effects at the end of the respective experiment? | X | |
| (f) | Has attention been paid to the duration of the experiments considering the time course of development in toxicity of the specific pesticide? | X | |
| (g) | Does the visual match (‘visual fit’ in FOCUS Kinetics, (2006)) of the model prediction quality indicate acceptability of the model predictions in comparison with the validation data? | X | |
| (h) | Do the reported quantitative model performance criteria (e.g. PPC, NRMSE, SPPE) indicate a sufficient model performance?For DEBtox models, the adequacy of the quantitative criteria listed above (set on basis of GUTS models) needs still to be fully tested and may need future adaptation. However, for the time being, this set of performance criteria is also suggested for DEBtox models (see Section 7.7.2 ‘Model performance criteria’). | X | |
| (i) | Has the performance of the model been reported in an objective and reproducible way? | X | |
|
10. Evaluation of model use When using a TKTD model for regulatory purposes, the inputs of species‐ and compound‐specific model parameters and of exposure profile data are required to run the model under new conditions. In this stage, it is important that the model is well documented and that it is clear how the model works. Please check the following items: | |||
| (a) | Is the use of the model sufficiently documented? Is a user manual available? See items provided in Section 10.7 of the EFSA PPR Panel (2014). | X | |
| (b) | Is an executable implementation of the model made available to the reviewer, or Is at least the source code provided? | X | |
| (c) | Has a summary sheet been provided by the applicant? The summary sheet should provide quick access to the comprehensive documentation with sections corresponding to the ones of this checklist. | X | |
| (d) | Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data? For example, if FOCUS Step 3 maximum values were used at Tier‐1 are the exposure profiles used Tier‐2C from the same FOCUS Step 3 modelling? If the exposure profile comes from any other source (e.g. different scenarios, different inputs, different model) has this been checked? | ||
| (e) | In case parameters of the DEB (physiological) part of the model were changed from calibration to validation in order to better describe control data:‐ Is the difference in the parameters inducing considerable difference in the model predictions?And, if yes:‐ Are the model simulations relevant for the risk assessment carried out with the parameter set producing the more conservative (worst‐case) predictions? | ||
1.1.
1.1.1.
Explanation
0. Evaluation of the physiological DEB part
The physiological DEB part of the model has never been reviewed by a regulatory authority or a group delegated to this at the EU level. Hence, no conclusion can be drawn regarding this part of the model.
1. Problem definition
The regulatory context of the model application has been defined in the original study report. Reference is made to several EFSA guidance documents, and particularly to the EFSA aquatic guidance document (EFSA PPR Panel, 2013), where mechanistic effect models in general and TKTD models in particular are described as refinement options for pesticide risk assessment.
The author proposes to use the model for investigating the reasons leading to different results (on survival and growth) obtained with two ELS experiments with rainbow trout, one carried out under constant exposure, and another with repeated peak exposure. While the question is clearly formulated, and it is of course of scientific interest, there are doubts that the question as it is can be used to directly inform the risk assessment for regulatory purposes.
Indeed, the model output consists of predictions of different endpoints (development, growth, reproductive, metabolic) which cannot be directly used in the risk assessment. Furthermore, predictions were carried out only for the exposure profile which was experimentally tested (i.e. for the exposure profile from the validation data set). Hence, the output of the model does not provide new endpoint estimations compared to the experimental ones.
The choice of the species is clear (rainbow trout), but it was not really discussed in a risk assessment context. However, considering all the available laboratory experiments for beta‐cyfluthrin, rainbow trout was consistently the most sensitive fish species in both the acute and chronic tests. Hence the choice appears reasonable but ideally should be better justified.
Rainbow trout is a Tier‐1 test species, but its use for TKTD modelling was not put into a specific assessment tier. However, it should be highlighted that this would have been extremely difficult if not impossible, as the position of TKTD models in the tiered risk assessment scheme have been specifically proposed in this SO for the first time.
2. Supporting data
The supporting data consist of two ELS experiments with rainbow trout, one carried out under constant exposure (used for calibration), and another with repeated peak exposure (used for validation). Test conditions are fully described in the specific reports hence full information is available regarding temperature, water chemistry, food conditions, biological measurements, etc.
The study used for calibration is valid according to the current OECD 210 guideline, with some minor deviations, which was assessed as not critical for the outcome of the study. The dissolved oxygen concentration stayed within the targeted limits of 6.5–11.9 ppm (three occasions above that range). The temperature during the study ranged from 8.3 to 11.9°C (recommended 8.5–11.5). The study used for validation was conducted in accordance with OECD Test Guideline No. 210 with adaptations in order to fulfil the objectives of the experiment (time‐variable exposure). Quality criteria as set out in the respective OECD test guidelines were overall considered fulfilled. Nevertheless, this information was not really accounted for in the phases of the model calibration and validation. It would be desirable that quality criteria are explicitly addressed when developing a model, in order to account for relevant deviations which may have had an impact on the biological measurements.
All available data on early life stage of rainbow trout were using for either calibrating or validating the model.
For what concerns the calibration data, the actual profile in the study matched the nominal concentrations reasonably well (mean measured concentrations between 95% and 112% of the nominal). For the validation experiment, on the other hand, measured peak concentrations differed in some cases substantially from the nominal ones. From Figures 2, 3–4, it appears rather clear that measured concentrations in time were used for the estimation of the concentration profiles in the experiment. Nevertheless, the report does not explicitly address this issue, while a more straightforward consideration would have been desirable.
3. Conceptual model
As pointed out under Section 0, the physiological DEB part of the model has never been reviewed by a regulatory authority or a group delegated to this at the EU level. Hence, no reference could be given to a pre‐existing evaluation.
In this specific model application, the author specifies that the toxicant acts by impairing the feeding behaviour, and hence by reducing the food assimilation, and thus lead to growth reduction in the early life stages, and may lead to a reduction in reproduction in the adult stage. Hence, the mode of action was specified. In addition, the links between the scaled damage and the DEB parameters appear logical. Temperature and food input are reported to be two forcing variables of the model. However, the influence of these parameters in the conceptual model is not clearly described. The influence of temperature on the hatching time is reported by mean of a scatterplot, but it is unclear whether this and other environmental factors may act in other ways within the conceptual model.
The modelling endpoints are related to lethal and sublethal effects (e.g. development, growth) of fish. Hence, they are relevant for the specific protection goals. Reproduction was not considered in the model, as the focus was on early life stages.
4. Formal model
All mathematical equations relevant for the effect of the toxicant are reported. All variables together with the relevant units are described in the text, nevertheless, a list/summary is not available for the state variables, but only for the parameters (together with their mean estimate) used in the model. The stochastic part of the model was not described at all.
5. Computer model
The computer code is fully available for the MatLab software, although explaining comments are rather limited. Some output graphs are missing or cannot be identically reproduced. All parameter values are well reported (except that uncertainty limits are missing). Other input data such as biological observations and analytical measurements from the calibration and the validation experiments were not available in the modelling report, but could be found in the respective experiment report.
Model verification was performed for the DEB part by means of model simulations for different food level and temperature. The outcome of these simulations confirmed that the computer model correctly implemented the influence of the two forcing variables. Model verification for the TKTD part was not performed. The influence of different exposure scenarios (e.g. default or ‘extreme case’) was not evaluated.
6. Regulatory model: environmental scenario
As no simulation with exposure profiles other than the one tested in the experiment was run, there was no consideration of particular environmental scenario (i.e. environmental parameters having an influence on the model output).
7. Regulatory model: parameter estimation
All parameter related to the DEB part of the model were taken from the literature. On the other hand, TKTD parameters were fitted to the calibration data set.
The calibration data set consisted of five tested concentrations plus the control, two replicates each. Survival was measured weekly, and raw data are available from the original study report (although presented in a cumulative way, no distinction between replicates). Hence, for this endpoint, sufficient measurements were available. Weight was only measured at the end of the test (biomass measured per chamber, not individual fish) and was presented as mean value of the replicates. While it is acknowledged that obtaining intermediate measurements for biomass might be complex and might require destructive sampling, the available data are not considered ideal for having a fully reliable parameter estimation.
Calibration data span from no effect (no significant growth inhibition, survival comparable to control) up to full effect (100% mortality). As already specified, all data available for this life stage were used either in the calibration or the validation of the model.
The dossier of beta‐cyfluthrin is particularly data rich for what concerns aquatic organisms. The available information does not suggest issues related to delayed effects or particular temporal pattern in the manifestation of the effects. Hence, the time course of the experiment seems appropriate.
For the TKTD part of the model, parameter estimation has not been explicitly documented: settings of optimisation routines, settings of the numerical solver used for solving the differential equations, starting values used for the optimisation, and optimal values of the objective function were all included in the Matlab code, which was made available. Nevertheless such information is not easily retrievable as it was not explicitly reported in the report.
None of the estimated parameter was reported together with the respective confidence limits.
Plots comparing the results of the calibrated DEBtox model and the calibration data over time were provided for survival. For growth, comparison was only possible at test termination. Overall, the visual match appears to be acceptable.
A posterior predictive check was not included in the model documentation.
8. Sensitivity and uncertainty analysis
No sensitivity analysis was available for the TKTD part of the models. Hence, the most influential parameter could not be identified. Parameter uncertainty was not reported and it was not propagated to the model output. In fact, no estimation of the uncertainty was presented for the model outputs.
9. Model validation
The model was validated against independent experimental data obtained under time‐variable exposure.
In principle, observations on survival were available with a daily resolution. Nevertheless, significant mortality was not observed in the validation experiment at any concentration, hence the comparison with model prediction in time is not very informative. Observations on growth were only available at the end of the experiment, hence no intermediate measurements were available.
Study design requirements included in this Scientific Opinion were not fulfilled (as these have been formulated afterwards). Nevertheless, one profile with two peaks was tested on three different life stages of rainbow trout. It is unclear whether the no‐exposure interval was shorter than the DRT95. According to the model output for swim‐ups (the most sensitive life stage) the two peaks were not toxicologically independent.
Five different exposure levels had been tested in the validation experiment. Nevertheless, no significant effect on either mortality or growth was observed at any of those levels. Ideally, tested levels should induce low, medium and large effects.
As already specified for the calibration data, the available information does not suggest issues related to delayed effects or particular temporal pattern in the manifestation of the effects for beta‐cyfluthrin. Hence, it is considered that the time course of the experiment was appropriate.
The visual match of the model prediction with the experimental measurements was good, but, due to the lack of any effect, not really informative for assessing the predictive ability of the model. For growth (length and biomass), comparison was only possible for one time‐point (experiment termination). Only control data (mean and standard deviation) were superimposed to the predicted curves at the end of the pulsed exposure profile.
No quantitative model performance criteria were proposed to check the predictions of the model.
Overall, the available data set was not considered appropriate for validating model prediction, nevertheless, the performance of the model was reported in an objective and reproducible way.
10. Evaluation of model use
Although a proper user manual is not available, the use of the model in this specific case is sufficiently documented. There is no executable version of the model available for reproducing the results. The code was provided in matlab, hence in principle the analysis can be reproduced. Nevertheless, when this code was run during the review of the model, the output plots could not be exactly reproduced.
No summary sheet was provided.
It has to be noted that the calibrated model was not used to predict effects related to the estimated exposure profiles deriving from the representative uses of beta‐cyfluthrin (i.e. no simulation with FOCUS profiles as input).
The scaled functional response f was adapted to match the growth of the control of the different cohorts in the validation experiment. However no information is given on the way the match has been done. Hence, there is no way to assess whether the difference in the parameters would induce significant difference in the model predictions.
11. Conclusion
Overall, the documentation of the DEBtox model and its application does not comply with the requirements as announced in this Scientific Opinion. Many aspects of the model documentation were evaluated positive, but there were a number of issues. In particular, no conclusion can be drawn for the physiological DEB part of the model since that has never been reviewed by a regulatory authority or a group delegated to this at the EU level. The model output is considered not relevant for risk assessment and the validation data set was not considered appropriate for validating model predictions.
Appendix H – Outcome of the consultation on the Draft Opinion with the Pesticide Steering Network
1.
A consultation with the Pesticide Steering Network (mainly Member States representatives) on the Draft Scientific Opinion was held in March 2018. Overall, comments were received by six member States. Comments and related replies are available in the online version of this output (‘Supporting information’ section).
Annex A – Checklist for GUTS models
1.
| ASPECT OF THE MODEL TO BE EVALUATED BY THE RISK ASSESSOR – GUTS model application for lethal effects | Yes | No | |
|
1. Evaluation of the problem definition The problem definition needs to explain how the modelling fits into the risk assessment and how it can be used to address the specific protection goals. For GUTS, questions to be answered are likely to be those that are set out in Chapter 3. Nevertheless, the problem definition should make clear the following points: | |||
| (a) | Is the regulatory context for the model application documented? | ||
| (b) | Is the question that has to be answered by the model clearly formulated? | ||
| (c) | Is the model output suitable to answer the formulated questions? | ||
| (d) | Was the choice of the test species clearly described and justified, also considering all the available valid information (including literature)? | ||
| (e) | Is the species to be modelled specified? – Is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2? | ||
|
2. Evaluation of the quality of the supporting experimental data In this part of the evaluation, it is checked whether the experimental data with which the model is compared (both calibration and validation data sets) have been subjected to quality control. The focus is on the data quality, i.e. the laboratory conditions, set‐up, chemical analytics and similar. Additional specific criteria for the suitability of the data sets for model calibration and validation are evaluated later in more detail (Sections 7 and 9 of this checklist). | |||
| (a) | Has the quality of the data used been considered and documented? (see list of OECD test guidelines in Chapter 7, Table 6) | ||
| (b) | Have all available data been used (either for calibration or for validation)? If not, is there a justification why some information has not been used? | ||
| (c) | Is it checked whether the actual exposure profile in the study matches the intended profile in the test (+/− 20%); if not, are then measured concentrations used for the modelling, instead of nominal ones? | ||
|
3. Evaluation of the conceptual model Providing GUTS models are being used to address mortality/immobility effects in fish or invertebrates, the conceptual model will be suitable to address the specific protection goals; so, no further evaluation is required (see Chapters 2.1, 2.2 and 4.1). | |||
|
4. Evaluation of the formal model The formal model contains the equations and algorithms to be used in the model. For GUTS models, the equations are standardised, so that no further check is necessary (see Chapter 4.1.1). It has to be documented, however, which GUTS model version is used (e.g. full or reduced model). | |||
|
5. Evaluation of the computer model The formal model is converted into a model that can run on a computer (the computer model). For GUTS models, the computer model can be tested by showing the model performance for the GUTS ring‐test data and performing some further checks (see Section 7.5). | |||
| (a) | Is the used implementation of GUTS tested against the ring‐test data set (see Section 4.2)? | ||
| (b) | Were GUTS parameters estimated for the ring‐test data and compared to the reference values, including confidence or credible intervals (Appendices B.6 and B.7)? | ||
| (c) | Is a set of default scenarios (e.g. standard scenarios, extreme cases, see Section 4.1.2) simulated and checked? | ||
| (d) | Are all data and parameters provided to allow an independent implementation of GUTS to be run? | ||
|
6. Evaluation of the regulatory model – the environmental scenarios For GUTS models using FOCUS simulations (or other Member State‐specific exposure simulations) as exposure input, no further definition and check of the environmental conditions is needed, since pesticide concentrations will be generated using the relevant FOCUS simulations (or MS‐specific exposure simulations), which consider factors such as soil, rainfall and agronomic practice, and the (effect) model will have been calibrated based on data collected under standard laboratory conditions. Fixing the environmental scenario to the conditions of the calibration experiments is considered appropriate because the modelling will be used with the equivalent of Tier‐1 or Tier‐2 Assessment Factors, so an extrapolation from laboratory to field conditions is already covered. | |||
|
7. Evaluation of the regulatory model – parameter estimation Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine, and the comprehensive documentation of methods and results. Model parameters are always estimated for a specific combination of species and compound (see Chapter 3 for background information). Supporting data for GUTS models are mortality or immobility data, have to be of sufficient quality (Section 2 in this checklist) and fulfil a set of basic criteria. Please check the following items to evaluate the calibration data, and the parameter optimisation process and the results (see Sections 4.1.3.1 and 7.6.2): | |||
| (a) |
Is it clear which parameters have been taken from literature or other sources and which have been fitted to data? If used, are values from literature reasonable and justified? |
||
| (b) | Are raw observations of mortality or immobility reported for at least five time‐points15? | ||
| (c) | Does calibration data span from treatment levels with no effects up to strong effects, ideally up to full effects (e.g. 0% survival)? | ||
| (d) | Have all data available for calibration been used? If not, is there a justification? | ||
| (e) | Has attention been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide? | ||
| (f) | Has the model parameter estimation been adequately documented, including settings of optimisation routines, and type and settings of the numerical solver that was used for solving the differential equations? | ||
| (g) |
If Bayesian inference has been used, are priors on model parameters reported? If a frequentist approach has been used, are starting values for the optimisation reported? |
||
| (h) | Are the estimated parameter values reported including confidence/credible intervals? | ||
| (i) | Is the method to get these limits reported and documented? | ||
| (j) | Are the optimal values of the objective function for calibration (e.g. log‐likelihood function) as the result of the parameter optimisation reported? | ||
| (k) | Are plots of the calibrated GUTS models in comparison with the calibration data over time provided, and does the visual match appear of acceptable quality? | ||
| (l) | Has a posterior predictive check been performed and documented? | ||
|
8. Evaluation of the sensitivity and uncertainty analysis For the reduced GUTS models, the influence of the model parameters on the model results are known well enough. Results of sensitivity analyses can, if contained, demonstrate that the model implementation is done correctly. For other GUTS models than the reduced, sensitivity analyses should be included for future applications and be checked by the following list. | |||
| (a) | Has a sensitivity analysis been performed and adequately documented16? | ||
| (b) | Are the results of the sensitivity analysis presented so that the most sensitive parameters can be identified? | ||
| (c) | Is the parameter uncertainty for the most important TKTD model parameters propagated to the model outputs and the results of the uncertainty propagation been documented? | ||
| (d) | Are the model outputs reported including confidence/credible intervals? | ||
|
9. Evaluation of the model by comparison with independent measurements (model validation) Validation data are used to test the model performance for predictions of mortality/immobility under exposure profiles which have not been used for model calibration. The performance of the model is usually evaluated by comparing relevant model outputs with measurements (often referred to as model validation). For GUTS, relevant outputs are the simulated mortality/immobility probability or LPX/EPX values. The following checklist is mandatory only for invertebrates; for vertebrates, a case‐by‐case basis check needs to be done (see also Sections 7.7.2 and 4.1.4.5) | |||
| (a) | Are effect data available from experiments under time‐variable exposure? | ||
| (b) | Is mortality or immobility reported at least for 7 time‐points in the validation data set? | ||
| (c) | Are two exposure profiles tested with at least two pulses each, separated by no‐exposure intervals of different duration length? | ||
| (d) | Is the individual depuration and repair time (DRT95) calculated, and is the duration of the no‐exposure intervals defined accordingly?17 | ||
| (e) | Is each profile tested at least at 3 concentration levels, in order to obtain low, medium and high effects at the end of the respective experiment? | ||
| (f) | Has attention been paid to the duration of the experiments considering the time course of development in toxicity of the specific pesticide? | ||
| (g) | Does the visual match (‘visual fit’ in FOCUS Kinetics (2006)) of the model prediction quality indicate acceptability of the model predictions in comparison with the validation data? | ||
| (h) | Do the reported quantitative model performance criteria (e.g. PPC, NRMSE, SPPE) indicate a sufficient model performance? | ||
| (i) | Has the performance of the model been reported in an objective and reproducible way? | ||
|
10. Evaluation of model use When using a TKTD model for regulatory purposes, the inputs of species‐ and compound specific model parameters and of exposure profile data are required to run the model under new conditions. In this stage, it is important that the model is well documented and that it is clear how the model works. Please check the following items: | |||
| (a) | Is the use of the model sufficiently documented? | ||
| (b) | Is an executable implementation of the model made available to the reviewer, or Is at least the source code provided? | ||
| (c) | Has a summary sheet been provided by the applicant? The summary sheet should provide quick access to the comprehensive documentation with sections corresponding to the ones of this checklist. | ||
| (d) | Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data? For example, if FOCUS Step 3 maximum values were used at Tier‐1, are the exposure profiles used Tier‐2C from the same FOCUS Step 3 modelling? If the exposure profile comes from any other source (e.g. different scenarios, different inputs, different model) has this been checked? | ||
| (e) |
Further points to be checked by evaluators Use an independent implementation of GUTS to test whether the output of the evaluated model implementation can be reproduced for some parameter sets. The MOSAIC_GUTS web‐platform (http://pbil.univ-lyon1.fr/software/mosaic/guts) can be used to test the model calibration The GUTS Shiny App (http://lbbe-shiny.univ-lyon1.fr/guts-shinyapp/) to test model predictions under a specific constant or time‐variable exposure profile given the set of model parameters can be used. |
||
It is recommended that the evaluator justifies the replies that are not straightforward.
See also Appendix F – for an example on how to evaluate GUTS models.
Annex B – Checklist for DEBtox models
1.
| ASPECT OF THE MODEL TO BE EVALUATED BY THE RISK ASSESSOR – DEBtox model applications for sublethal effects | Yes | No | |
|
0. Evaluation of the physiological DEB part DEBtox models consist of two parts:
This checklist is adapted to the evaluation of the TKTD part of the DEBtox models (see Sections 2.3 and 5), but the first thing to check is whether the physiological part, meaning the DEB part, was evaluated beforehand for the chosen species. Without that check, the use of a DEBtox model cannot be suggested, even if the TKTD model part is evaluated positively. | |||
| (a) | Was the evaluation of the physiological DEB part conducted by a regulatory authority or a group delegated to this at the EU level? | ||
| (b) | If (a) is yes, did the above evaluation of the physiological DEB part conclude that it is suitable for use in risk assessment? | ||
| (c) | If (a) and (b) are yes, can the physiological DEB part of the model describe the behaviour of the control data? | ||
|
1. Evaluation of the problem definition The problem definition needs to explain how the modelling fits into the risk assessment and how it can be used to address the specific protection goals (Section 3). Please check if due attention is paid to the following points: | |||
| (a) | Is the regulatory context for the model application documented? | ||
| (b) | Is the question that has to be answered with the model clearly formulated? | ||
| (c) | Is the model output suitable to answer the formulated questions? | ||
| (d) | Is the choice of the test species clearly described and justified, also considering all the available valid information (including literature)? | ||
| (e) | Is the species to be modelled specified? – Is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2? | ||
|
2. Evaluation of the quality of the supporting experimental data In this part of the evaluation, it is checked whether the experimental data with which the model is compared (both calibration and validation data sets) have been subjected to quality control. The focus is on the data quality, i.e. the laboratory conditions, set‐up, chemical analytics and similar. Additional specific criteria for the suitability of the data sets for model calibration and validation are evaluated later in more detail (Sections 7 and 9 of this checklist). | |||
| (a) | Are all types of data fully described, meaning that factors like temperature, food conditions, measurements, handling, etc. are documented? | ||
| (b) | Has the quality of the used data been considered and documented? (see the list of OECD test guidelines in Section 7, Table 6). | ||
| (c) | Have all available data been used (either for calibration or for validation)? If not, is there a justification why some information has not been used? | ||
| (d) | Is it checked whether the actual exposure profile in the study matches the intended profile in the test (+/− 20%); if not, are then measured concentrations used for the modelling, instead of nominal ones? | ||
|
3. Evaluation of the conceptual model The conceptual model provides a general and qualitative description of the system to be modelled. The physiological DEB part should have been evaluated and approved beforehand (see Section 0 of this checklist). Please check the following items for the TKTD part (refer to Section 7.3): | |||
| (a) | Is the reference to the evaluation of the physiological DEB part given? | ||
| (b) | Is the potential mode of action of the toxicant specified (effects on assimilation, growth, maintenance costs, reproductive output and/or survival)? | ||
| (c) | Are the links between the scaled damage or the internal concentration and the DEB model parameters logical? | ||
| (d) | In case environmental factors (e.g. temperature) are explicitly considered, is their influence on the physiological and/or TKTD processes documented in the conceptual model? | ||
| (e) | Are the modelling endpoints relevant to the specific protection goal? | ||
|
4. Evaluation of the formal model The formal model contains equations used in the model. The physiological DEB part should have been evaluated and approved beforehand (see Section 0 of this checklist). Nevertheless, all equations (from either the physiological or the TKTD part) that include one or more parameters that are impacted by the effects of the toxicant need to be documented. Please check the following items: | |||
| (a) | Are all mathematical equations relevant for the effect of the toxicant described, including the equations of the toxicokinetic (TK) part and the equations relating the used scaled damage or internal concentrations with the DEB model parameter(s)? | ||
| (b) | Is there a list or a summary of all variables and parameters including their meaning and unit? | ||
| (c) | Are both the deterministic part (equations describing the mean tendency of the data) and the stochastic part (the probability law describing the variability around the mean tendency) of the model fully described? See Section 5.1 for an example. | ||
|
5. Evaluation of the computer model The computer model corresponds to the implementation of the formal model that can run on a computer. Please check the following items: | |||
| (a) | Is the computer code available, including explaining comments for the most important functions? | ||
| (b) | Is enough information provided to allow non‐expert user to re‐run the model independently (e.g. parameter sets, input data)? | ||
| (c) | Is it demonstrated that the mathematical model is correctly implemented (model verification), e.g. by checking and documenting the internal consistency of the model results based on a set of default or extreme cases (see e.g. Section 4.1.2.3 for an example with GUTS, ‘reality check’ in chapter 10.4 of EFSA PPR Panel, 2014)? | ||
|
6. Evaluation of the regulatory model – the environmental scenarios The environmental scenarios determine the environmental context in which the model is run. For the application of DEBtox for the prediction of toxicological effects in a regulatory context, the environmental scenario need to be fixed to the laboratory conditions of the experiments used for calibrating the TKTD part of the model. Please check the following items: | |||
| (a) | Are all the relevant conditions, recorded in the experiments used for calibrating the TKTD part of the model, used consistently for the simulations (i.e. for generating EPx) used in the risk assessment? | ||
|
7. Evaluation of the regulatory model – parameter estimation Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine, and the comprehensive documentation of methods and results. Model parameters are always estimated for a specific combination of species and compound (see Chapter 3 for background information). Supporting data for DEBtox have to be of sufficient quality (Section 2 in this checklist), be relevant to the risk assessment problem and fulfil a set of basic criteria. Typical supporting data for the TKTD part of DEBtox models are experimental toxicity data for growth or growth + reproduction, to which mortality or immobility data can also be added (see Chapters 2.3 and 7.2 for background information, Section 5.1.3 for an example). Specific requirements on the time resolution and temporal scale for the calibration data cannot be given, but in general, the number of time‐points has to be sufficient to provide enough degrees of freedom for the model calibration. Please check the following items to evaluate the calibration data, the parameter optimisation process and the results (see Sections 4.1.3 and 7.6.2): | |||
| (a) |
Is it clear which parameters have been taken from literature or other sources and which have been fitted to data? If used, are values from literature reasonable and justified? |
||
| (b) | Are the calibration data sufficient to provide enough degrees of freedom for the model calibration, i.e. are endpoints at least reported at several intermediate12 time‐points? | ||
| (c) | Does calibration data span from treatment levels with no effects up to strong effects, ideally up to full effects (e.g. no growth, no reproduction)? | ||
| (d) | Have all data available for calibration been used? If not, is there a justification why this information has not been used? | ||
| (e) | Has attention been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide? | ||
| (f) | Has the model parameter estimation been adequately documented, including settings of optimisation routines, and type and settings of the numerical solver that was used for solving the differential equations? | ||
| (g) |
If Bayesian inference has been used, are priors on model parameters reported? If a frequentist approach has been used, are starting values for the optimisation reported? |
||
| (h) | Are the estimated parameter values reported including confidence/credible limits? | ||
| (i) | Is the method to get these limits reported and documented? | ||
| (j) | Are the optimal values of the objective function for calibration (e.g. Log‐likelihood function) as the result of the parameter optimisation reported? | ||
| (k) | Are plots of the calibrated DEBtox model in comparison with the calibration data over time provided, and does the visual match appear of acceptable quality? | ||
| (l) | Has a posterior predictive check been performed and documented? | ||
|
8. Evaluation of the sensitivity and uncertainty analysis It is assumed that sensitivity and uncertainty analyses for the physiological DEB model part have been evaluated and approved beforehand (see Section 0 of this checklist). Sensitivity analysis identifies the influence of the parameters on the model outputs and can hence identify the most influential parameters. Uncertainty analysis aims at identifying how uncertain the model output is regarding the uncertainty in parameter estimates. Please check the following items for the TKTD part: | |||
| (a) | Has a sensitivity analysis for the TKTD part been performed and been adequately documented? (The range of parameter variation in the sensitivity analysis should be justified by an analysis of the expected variation of model parameters). | ||
| (b) | Are the results of the sensitivity analysis presented so that the most sensitive parameters can be identified? | ||
| (c) | Is the parameter uncertainty for the most important TKTD model parameters propagated to the model outputs and the results of the uncertainty propagation been documented? | ||
| (d) | Are the model outputs reported including confidence/credible intervals? | ||
|
9. Evaluation of the model by comparison with independent measurements (model validation) The model performance is usually evaluated by comparing relevant model outputs with independent experimental measurements (i.e. data from other experiments than those used for calibration, often referred to as model validation). These independent measurements (or validation data sets) are used to test the model performance in predicting the chosen endpoints under time‐variable exposure profiles. For DEBtox models, relevant outputs may be simulated growth and/or reproduction, sometimes together with simulated survival, or EPx values. The following checklist is mandatory only for invertebrates, for vertebrates a case‐by‐case basis check needs to be done (see also Sections 7.7.2 and 4.1.4.5): | |||
| (a) | Are effect data available from experiments under time‐variable exposure? | ||
| (b) |
If non‐destructive measurement is possible, are sufficient endpoint measurements provided in order to cover at least before, during and after each pulse exposure? Are these time‐points enough to allow for evaluation of changes in growth rates different from the control treatment? |
||
| (c) | Are two exposure profiles tested with at least two pulses each, separated by no‐exposure intervals of different duration length? | ||
| (d) | Is the individual depuration and repair time (DRT95) calculated based on toxicokinetic parameters, and is the duration of the no‐exposure intervals defined accordingly?17 | ||
| (e) | Is each profile tested at least at 3 concentration levels, in order to obtain low, medium and high effects at the end of the respective experiment? | ||
| (f) | Has attention been paid to the duration of the experiments considering the time course of development in toxicity of the specific pesticide? | ||
| (g) | Does the visual match (‘visual fit’ in FOCUS Kinetics, (2006)) of the model prediction quality indicate acceptability of the model predictions in comparison with the validation data? | ||
| (h) |
Do the reported quantitative model performance criteria (e.g. PPC, NRMSE, SPPE) indicate a sufficient model performance? For DEBtox models, the adequacy of the quantitative criteria listed above (set on basis of GUTS models) needs still to be fully tested and may need future adaptation. However, for the time being, this set of performance criteria is also suggested for DEBtox models (see Section 7.7.2 ‘Model performance criteria’). |
||
| (i) | Has the performance of the model been reported in an objective and reproducible way? | ||
|
10. Evaluation of model use When using a TKTD model for regulatory purposes, the inputs of species‐ and compound specific model parameters and of exposure profile data are required to run the model under new conditions. In this stage, it is important that the model is well documented and that it is clear how the model works. Please check the following items: | |||
| (a) |
Is the use of the model sufficiently documented? Is a user manual available? See items provided in Section 10.7 of the EFSA PPR Panel (2014). |
||
| (b) | Is an executable implementation of the model made available to the reviewer, or Is at least the source code provided? | ||
| (c) | Has a summary sheet been provided by the applicant? The summary sheet should provide quick access to the comprehensive documentation with sections corresponding to the ones of this checklist. | ||
| (d) | Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data? For example if FOCUS Step 3 maximum values were used at Tier‐1 are the exposure profiles used Tier‐2C from the same FOCUS Step 3 modelling? If the exposure profile comes from any other source (e.g. different scenarios, different inputs, different model) has this been checked? | ||
| (e) |
In case parameters of the DEB (physiological) part of the model were changed from calibration to validation in order to better describe control data: ‐ Is the difference in the parameters inducing considerable difference in the model predictions? And, if yes: ‐ Are the model simulations relevant for the risk assessment carried out with the parameter set producing the more conservative (worst‐case) predictions? |
||
It is recommended that the evaluator justifies the replies that are not straightforward.
See also Appendix G – for an example on how to evaluate DEBtox models.
Annex C – Checklist for primary producer models
1.
| ASPECT OF THE MODEL TO BE EVALUATED BY THE RISK ASSESSOR – Models for primary producers | Yes | No | |
|
0. Evaluation of the growth model All models for primary producers consist of two parts:
This checklist is adapted to the evaluation of the TKTD part of the primary producer models (See Section 2.4 and Chapter 6), but the first thing to check is whether the physiological growth part was evaluated beforehand for the chosen species. Without that check, the use of the whole model cannot be suggested, even if the TKTD model part is evaluated positively. | |||
| (a) | Was the evaluation of the (physiological) growth model part conducted by a regulatory authority or a group delegated to this at the EU level? | ||
| (b) | If (a) is yes, did the above evaluation of the physiological DEB part conclude that it is suitable for use in risk assessment? | ||
| (c) | If (a) and (b) are yes, can the physiological DEB part of the model describe the behaviour of the control data? | ||
|
1. Evaluation of the problem definition The problem definition needs to explain how the modelling fits into the risk assessment and how it can be used to address the specific protection goals (Chapter 3). Please check if due attention is paid to: | |||
| (a) | Is the regulatory context for the model application documented? | ||
| (b) | Is the question that has to be answered by the model clearly formulated? | ||
| (c) | Is the model output suitable to answer the formulated questions? | ||
| (d) | Is the choice of the test species clearly described and justified, also considering all the available valid information (including literature)? | ||
| (e) | Is the species to be modelled specified? – Is it clear whether the model is being used with a Tier‐1 test species i.e. Tier‐2C1 or with one or more relevant species (which might include the Tier‐1 species) i.e. Tier‐2C2? | ||
|
2. Evaluation of the quality of the supporting experimental data In this part of the evaluation, it is checked whether the experimental data with which the model is compared (both calibration and validation data sets) have been subjected to quality control. The focus is on the data quality, i.e. the laboratory conditions, setup, chemical analytics and similar. Additional specific criteria for the suitability of the data sets for model calibration and validation are evaluated later in more detail (Sections 7 and 9 of this checklist). | |||
| (a) | Are growth conditions (temperature, irradiance, nutrient media composition, handling and thinning, etc.) and growth calculations (frequency and type of measurements, calibration between surface and weight data, etc.) described and documented? | ||
| (b) | Has the quality of the used data been considered and documented? (see the list of OECD test guidelines Chapter 7, Table 6). | ||
| (c) | Have all available data been used (either for calibration or for validation)? If not, is there a justification why some information has not been used? | ||
| (d) | Is it checked whether the actual exposure profile in the study matches the intended profile in the test (+/− 20%); if not, are then measured concentrations used for the modelling, instead of nominal ones? | ||
|
3. Evaluation of the conceptual model The conceptual model provides a general and qualitative description of the system to be modelled. The conceptual model for physiological part is assumed to have been evaluated and approved beforehand ‘see Section 0 of this checklist); but the TKTD part, describing the toxicant effects on the physiological model needs to be documented. | |||
| (a) | Is the reference to the evaluation of the growth (physiological) model part given? | ||
| (b) | Is the mode of action of the toxicant specified (effects on e.g. assimilation or growth)? | ||
| (c) | Are the links between the external or internal concentration and the growth model logical? | ||
| (d) | In case environmental factors (e.g. nutrient levels, temperature) are explicitly considered, is their influence on the physiological and/or TKTD processes documented in the conceptual model? | ||
| (e) | Are the modelling endpoints relevant to the specific protection goal? | ||
|
4. Evaluation of the formal model The formal model contains equations used in the model. For each model application, all equations that are used for the modelling should be documented: | |||
| (a) | Are all mathematical equations described, including the equations of the toxicokinetic (TK) part, and the equations relating the used internal concentration with the growth (physiological) model parameter(s)? | ||
| (b) | Is there a list or summary of the variables and parameters including their meaning and unit? | ||
| (c) | Are both the deterministic part (equations describing the mean tendency of the data) and the stochastic part (the probability law describing the variability around the mean tendency) of the model fully described? See Section 5.1 for an example with DEBtox. | ||
| 5. Evaluation of the computer modelThe computer model corresponds to the implementation of the formal model that can run it on a computer. Please check the following items: | |||
| (a) | Is the computer code available, including explaining comments for the most important functions? | ||
| (b) | Is enough information provided to allow any user to re‐run the model independently (e.g. parameter sets, input data)? | ||
| (c) | Is it demonstrated that the mathematical model is correctly implemented (model verification), e.g. by checking and documenting the internal consistency of the model results based on a set of default or extreme cases (see e.g. Section 4.1.2.3 for an example with GUTS, ‘reality check’ in chapter 10.4 of the EFSA PPR Panel (2014))? | ||
|
6. Evaluation of the regulatory model – the environmental scenarios For the application of primary producers TKTD models for the prediction of toxicological effects in a regulatory context, the environmental scenario need to be fixed to the laboratory conditions of the experiments used for calibrating the TKTD part of the model. Please check the following items: | |||
| (a) | Are all the relevant conditions, recorded in the experiments used for calibrating the TKTD part of the model, used consistently for the simulations (i.e. for generating EPx) used in the risk assessment? | ||
|
7. Evaluation of the regulatory model – parameter estimation Parameter estimation requires a suitable data set, the correct application of a parameter optimisation routine, and the comprehensive documentation of methods and results (see Chapter 3 for background information). Model parameters are always estimated for a specific combination of species and compound. Supporting data for primary producers have to be of sufficient quality (Section 2 in this checklist), be relevant to the risk assessment problem and fulfil a set of basic criteria. Typical supporting data for the TKTD part of primary producer models are toxicity test data for growth measured either on the basis of frond number, surface area or biomass over time (see Chapters 2.4 for background information; Chapter 6 for examples). Specific requirements on the time resolution and temporal scale for the calibration data cannot be given, but in general the number of time‐points has to be sufficient to provide enough degrees of freedom for the model calibration. Please check the following items to evaluate the calibration data, the parameter optimisation and the results (see Sections 4.1.3 and 7.6.2): | |||
| (a) |
Is it clear which parameters have been taken from literature or other sources or which have been fitted to data? If used, are values from literature reasonable and justified? |
||
| (b) | Are the calibration data sufficient to provide enough degrees of freedom for the model calibration, i.e. are endpoints reported at several18 intermediate time‐points? | ||
| (c) | Does calibration data span from treatment levels with no effects up to strong effects, ideally up to full effects (e.g. no growth)? | ||
| (d) | Have all data available for calibration been used? If not, is there a justification why this information has not been used? | ||
| (e) | Has attention been paid in terms of adjusting the time course of the experiment to capture the full toxicity of the pesticide? | ||
| (f) | Has the model parameter estimation been adequately documented, including settings of optimisation routines, and type and settings of the numerical solver that was used for solving the differential equations? | ||
| (g) |
If Bayesian inference has been used, are priors on model parameters reported? If a frequentist approach has been used, are starting values for the optimisation reported? |
||
| (h) | Are the estimated parameter values reported including confidence/credible limits? | ||
| (i) | Is the method to get these limits reported and documented? | ||
| (j) | Are the optimal values of the objective function for calibration (e.g. Log‐likelihood function) as the result of the parameter optimisation reported? | ||
| (k) | Are plots of the calibrated TKTD models in comparison with the calibration data over time provided, and does the visual match appear of acceptable quality? | ||
| (l) | Has a posterior predictive check been performed and documented? | ||
|
8. Evaluation of the sensitivity and uncertainty analysis It is assumed that sensitivity and uncertainty analyses for the physiological model part have been evaluated and approved beforehand. Sensitivity analysis identifies the influence parameters have on the model outputs and can hence identify the most relevant model parameters. Uncertainty analysis aims at identifying how uncertain the model output is regarding the uncertainty in parameter estimates. Please check the following items for the TKTD part: | |||
| (a) | Has a sensitivity analysis for the TKTD part been performed and been adequately documented? (The range of parameter variation in the sensitivity analysis should be justified by an analysis of the expected variation of model parameters). | ||
| (b) | Are the results of the sensitivity analysis presented so that the most sensitive parameters can be identified? | ||
| (c) | Is the parameter uncertainty for the most important TKTD model parameters propagated to the model outputs and the results of the uncertainty propagation been documented? | ||
| (d) | Are the model outputs reported including confidence/credible intervals? | ||
|
9. Evaluation of the model by comparison with independent measurements (model validation) The model performance is usually evaluated by comparing relevant model outputs with experimental measurements (often referred to as model validation). Independent measurements (or validation data sets) are used to test the model performance in predicting the chosen endpoints under time‐variable exposure profiles. These data have not been used for the model calibration. For models for primary producers, relevant outputs are biomass or biomass‐proxies such as cell number or chlorophyll content for algae at a specific point in time. Please check the following items: | |||
| (a) | Are effect data available from experiments under time‐variable exposure? | ||
| (b) |
Are sufficient endpoint measurements provided in order to cover at least before, during and after each pulse exposure? Are these time‐points enough to allow for evaluation of changes in growth rates different from the control treatment? |
||
| (c) | Are two exposure profiles tested with at least two pulses each, separated by no‐exposure intervals of different duration length? | ||
| (d) |
‐ point not relevant for algae ‐ Is the individual depuration and repair time (DRT95; see Section 4.1.4.5) calculated based on toxicokinetic parameters, and is the duration of the no‐exposure intervals defined accordingly? [Ideally one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95] |
||
| (e) | Is each profile tested at least at 3 concentration levels, in order to obtain low, medium and high effects at the end of the respective experiment? | ||
| (f) | Has attention been paid to the duration of the experiments considering the time course of development in toxicity of the specific pesticide? | ||
| (g) | Does the visual match (‘visual fit’ in FOCUS Kinetics (2006); see Section 4.1.4.5) of the model prediction quality indicate acceptability of the model predictions in comparison with the validation data? | ||
| (h) |
Do the reported quantitative model performance criteria (e.g. PPC, NRMSE, SPPE) indicate a sufficient model performance? For primary producer models, the adequacy of the quantitative criteria listed above (set on basis of GUTS models (see Section 4.1.4.5)) needs still to be fully tested, and may need future adaptation. However, for the time being, this set of performance criteria is also suggested for primary producer models (see Section 7.7.2 ‘Model performance criteria’). |
||
| (i) | Has the performance of the model been reported in an objective and reproducible way? | ||
|
10. Evaluation of model use When using a TKTD model for regulatory purposes, the inputs of species‐ and compound specific model parameters and of exposure profile data are required to run the model under new conditions. In this stage, it is important that the model is well documented and that it is clear how the model works. Please check the following items: | |||
| (a) |
Is the use of the model sufficiently documented? Is a user manual available? See items provided in Section 10.7 of the EFSA PPR Panel (2014). |
||
| (b) | Is an executable implementation of the model made available to the reviewer, or Is at least the source code provided? | ||
| (c) | Has a summary sheet been provided by the applicant? The summary sheet should provide quick access to the comprehensive documentation with sections corresponding to the ones of this checklist. | ||
| (d) | Does the exposure profile used with the TKTD model come from the same source as the PEC used with the Tier‐1 effects data? For example if FOCUS Step 3 maximum values were used at Tier‐1, are the exposure profiles used Tier‐2C from the same FOCUS Step 3 modelling? If the exposure profile comes from any other source (e.g. different scenarios, different inputs, different model) has this been checked? | ||
| (e) |
In case parameters of the growth (physiological) model were changed from calibration to validation, in order to better describe control data:‐ Is the difference in the parameters inducing considerable difference in the model predictions? And, if yes:‐ Are model simulations relevant for the risk assessment carried out with the parameter set producing the more conservative (worst‐case) predictions? |
||
It is recommended that the evaluator justifies the replies that are not straightforward.
Annex D – Model Summary template
1.
For TKTD model applications, it is strongly suggested to document the study according to the requirements announced in the EFSA TKTD SO. The applicant needs to document in the following summary template, that the model‐specific checklists (Annex A, B and C of the EFSA TKTD SO) have been considered. Clarifications are required on how the aspects listed in the model specific checklists are reflected in the model documentation. References to the parts of the DAR/RAR or other documents where the points are fully explained should be included. This summary should provide sufficient detail to allow the reader to understand the key points about how the modelling was conducted. In addition, it should be suitable to provide the basis for the study summary that RMS will produce for the DAR/RAR. The aim is to ensure the risk assessor that all aspects of the modelling cycle are captured and also to help the model evaluators to find all the information they need in the model documentation.
| Aspect | Reference to section in the model documentation or assessment report |
|---|---|
| 1. Problem definition | |
| 2. Supporting data | |
| 3. Conceptual model[Link] | |
| 4. Formal model1 | |
| 5. Computer model2 | |
| 6. Regulatory model – the environmental scenario3 | |
| 7. Regulatory model – parameter estimation | |
| 8. Regulatory model – Sensitivity and uncertainty analysis | |
| 9. Regulatory model – Comparison with measurements | |
| 10. Reality/problem – Model use |
1 For GUTS models it is sufficient to identify which version of GUTS has been used and to refer to this Scientific Opinion. This can be done since the conceptual and formal models are standardised following Jager and Ashauer (2018).
2 If standard software implementations become available (and have been checked) it will be sufficient to identify which software and version was used.
3 For GUTS models, if output from FOCUS surface water standard software has been used, it is sufficient to document which FOCUS model versions and scenarios have been used.
Supporting information
Repository of codes
Outcome of the consultation on the Draft Opinion with the Pesticide Steering Network
Suggested citation: EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues) , Ockleford C, Adriaanse P, Berny P, Brock T, Duquesne S, Grilli S, Hernandez‐Jerez AF, Bennekou SH, Klein M, Kuhl T, Laskowski R, Machera K, Pelkonen O, Pieper S, Smith RH, Stemmer M, Sundh I, Tiktak A, Topping CJ, Wolterink G, Cedergreen N, Charles S, Focks A, Reed M, Arena M, Ippolito A, Byers H and Teodorovic I, 2018. Scientific Opinion on the state of the art of Toxicokinetic/Toxicodynamic (TKTD) effect models for regulatory risk assessment of pesticides for aquatic organisms. EFSA Journal 2018;16(8):5377, 188 pp. 10.2903/j.efsa.2018.5377
Requestor: EFSA
Question number: EFSA‐Q‐2012‐00960
Panel members: Paulien Adriaanse, Philippe Berny, Theodorus Brock, Sabine Duquesne, Sandro Grilli, Antonio F Hernandez‐Jerez, Susanne Hougaard, Michael Klein, Thomas Kuhl, Ryszard Laskowski, Kyriaki Machera, Colin Ockleford, Olavi Pelkonen, Silvia Pieper, Robert Smith, Michael Stemmer, Ingvar Sundh, Ivana Teodorovic, Aaldrik Tiktak, Chris J Topping and Gerrit Wolterink
Acknowledgements: The Panel on Plant Protection Products and their Residues wishes to thank for the support provided to this scientific output the member of the Working Group (WG): Theodorus Brock, Nina Cedergreen, Sandrine Charles, Sabine Duquesne, Andreas Focks, Michael Klein, Melissa Reed and Ivana Teodorovic and EFSA staff member: Maria Arena, Alessio Ippolito and Harry Byers. In addition the PPR Panel wishes to thank Roman Ashauer, Tjalling Jager and Thomas Preuss for their support to the WG as hearing experts.
Adopted: 27 June 2018
Notes
Council Directive 91/414/EEC of 15 July 1991 concerning the placing of plant protection products on the market.
Commission Regulation (EU) No 283/2013 of 1 March 2013 setting out the data requirements for active substances, in accordance with Regulation (EC) No 1107/2009 of the European Parliament and of the Council concerning the placing of plant protection products on the market. OJ L 93, 3.4.2013, p. 1–94.
Commission Regulation (EU) No 284/2013 of 1 March 2013 setting out the data requirements for plant protection products, in accordance with Regulation (EC) No 1107/2009 of the European Parliament and of the Council concerning the placing of plant protection products on the market. OJ L 93, 3.4.2013, p. 85–152.
Commission Regulation (EU) No 546/2011 of 10 June 2011 implementing Regulation (EC) No 1107/2009 of the European Parliament and of the Council as regards uniform principles for evaluation and authorisation of plant protection products. OJ L 155, 11.6.2011, p. 127–175.
DEBtoxM is a collection of Matlab scripts and functions to analyse toxicity data in a DEB framework that are provided by T. Jager online: http://www.debtox.info/debtoxm.html
Build Your Own Model (BYOM) is a flexible set of Matlab scripts and functions to help building, simulating and fitting its own DEB models; it is maintained by T. Jager and available online: http://www.debtox.info/byom.html
Isomorphic growth is growth that occurs at the same rate for all parts of an organism ‐ this means that the organisms shape stays the same even though the organism gets bigger.
Variables that are not observed but exist within the model, e.g. scaled damage.
The choice of seven time‐points is based on the fact that observations are usually done at the beginning and the end of a pulse, and there are two pulses per treatment and two intermediate time‐points of observations between the two pulses. In addition, a last time‐point at the end of the experiment is considered.
Theoretically it is possible to have the NRMSE as a probability distribution.
Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC.
For example, for algae, external concentrations are linked directly to effects so that the depurations time would not be relevant.
The choice of five time‐points may be problematic in standard tests shorter than 4 days. Possible solutions are: (i) increase the number of observation in those tests; (ii) to extend the duration of the test to for instance 96 hours. If a standard 48‐hour study is only available, a calibration might still be attempted but the quality of the fit (convergence, uncertainty limits and c) should be carefully checked.
Time‐points usually differ, with classically one measurement per week for growth (even sometimes only one measurement of growth at the end of the experiment) and measurements twice or three times per week for reproduction.
The choice of five time‐points may be problematic in standard tests shorter than 4 days. Possible solutions are: (i) increase the number of observation in those tests; (ii) to extend the duration of the test to for instance 96 hours. If a standard 48‐hour study is only available, a calibration might still be attempted but the quality of the fit (convergence, uncertainty limits and visual fit) should be carefully checked.
The range of parameter variation in the sensitivity analysis should be justified by an analysis of the expected variation of model parameters.
Ideally one of the profiles should show a no‐exposure interval shorter than the DRT95, the other profile clearly larger than the DRT95.
Growth should be non‐destructively monitored by measuring surface area or frond number on a daily basis or every two days. Biomass should be measured at least at the end of the exposure phase.
References
- Ashauer R, Thorbek P, Warinton JS, Wheeler JR and Maund S, 2013. A method to predict and understand fish survival under dynamic chemical stress using standard ecotoxicity data. Environmental Toxicology and Chemistry, 32, 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashauer R, Albert C, Augustine S, Cedergreen N, Charles S, Ducrot V, Focks A, Gabsi F, Gergs A, Goussen B, Jager T, Kramer NI, Nyman AM, Poulsen V, Reichenberger S, Schafer RB, Van den Brink PJ, Veltman K, Vogel S, Zimmer EI and Preuss TG, 2016. Modelling survival: exposure pattern, species sensitivity and uncertainty. Scientific Reports, 6, 29178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashauer R, O'Connor I and Esher BI, 2017. Toxic mixtures in time – the sequence makes the poison. Environmental Science and Technology, 51, 3084–3092. [DOI] [PubMed] [Google Scholar]
- Baas J, Augustine S, Marques GM and Dorne JL, 2018. Dynamic energy budget models in ecological risk assessment: from principles to applications. Science of the Total Environment, 628–629, 249–260. [DOI] [PubMed] [Google Scholar]
- Barko J and Smart R, 1981. Sediment‐based nutrition of submersed macrophyte. Aquatic Botany, 10, 339–352. [Google Scholar]
- Baudrot V, Charles S, Delignette‐Muller ML, Duchemin W, Kon‐Kam‐King G, Lopes C, Philippe Ruiz P and Veber P, 2018a. Morse: Modelling Tools for Reproduction and Survival Data in Ecotoxicology. Version 3.1.0. Available online: https://CRAN.R-project.org/package=morse.
- Baudrot V, Preux S, Ducrot V, Pave A and Charles S, 2018b. New insights to compare and choose TKTD models for survival based on an interlaboratory study for Lymnaea stagnalis exposed to Cd. Environmental Science and Technology, 52, 1582–1590. [DOI] [PubMed] [Google Scholar]
- Baudrot V, Veber P, Gence G and Charles S, 2018c. Fit GUTS reduced models online: from theory to practice. Integrated Environmental Assessment and Management, 10.1002/ieam.4061 [DOI] [PubMed] [Google Scholar]
- Best EPH and Boyd WA, 1999. A simulation model for growth of the submersed aquatic macrophyte Eurasian Watermilfoil (Myriophyllum spictum L.). Aquatic Plant Control Research Program Technical Report No. A‐99‐3.
- Billoir E, Delignette‐Muller ML, Pery AR and Charles S, 2008. A Bayesian approach to analyzing ecotoxicological data. Environmental Science and Technology, 42, 8978–8984. [DOI] [PubMed] [Google Scholar]
- Billoir E, Delhaye H, Clement B, Delignette‐Muller ML and Charles S, 2011. Bayesian modelling of daphnid responses to time‐varying cadmium exposure in laboratory aquatic microcosms. Ecotoxicology and Environmental Safety, 74, 693–702. [DOI] [PubMed] [Google Scholar]
- Boxall AB, Fogg LA, Ashauer R, Bowles T, Sinclair CJ, Colyer A and Brain RA, 2013. Effects of repeated pulsed herbicide exposures on the growth of aquatic macrophytes. Environmental Toxicology and Chemistry, 32, 193–200. [DOI] [PubMed] [Google Scholar]
- Brock TCM, Alix A, Brown CD, Capri E, Gottesbüren BFF, Heimbach F, Lythgo CM, Schulz R and Streloke M, 2010. Linking Aquatic Exposure and Effects. Risk Assessment of Pesticides (ELINK). CRC Press, Taylor and Francis Group, Boca Raton, Florida, USA. 410 pp. [Google Scholar]
- Brock TCM, Bhatta R, Van Wijngaarden RPA and Rico A, 2016. Is the chronic Tier‐1 effect assessment approach for insecticides protective for aquatic ecosystems? Integrated Environmental Assessment and management, 12, 747–758. [DOI] [PubMed] [Google Scholar]
- Brooks SP and Gelman A, 1998. General methods for monitoring convergence of iterative simulations. Journal of Computational and Graphical Statistics, 7, 434–455. [Google Scholar]
- Carpenter B, Gelman A, Hoffman M, Lee D, Goodrich B and Betancourt M, 2017. Stan: a probabilistic programming language. Journal of Statistical Software, 76, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergreen N and Streibig JC, 2005. The toxicity of herbicides to non‐target aquatic plants and algae: assessment of predictive factors and hazard. Pest Management Science, 61, 1152–1160. [DOI] [PubMed] [Google Scholar]
- Cedergreen N, Spliid NH and Streibig JC, 2004. Species‐specific sensitivity of aquatic macrophytes towards two herbicides. Ecotoxicology and Environmental Safety, 58, 314–323. [DOI] [PubMed] [Google Scholar]
- Charles S, Veber P and Delignette‐Muller ML, 2017. MOSAIC: a web‐interface for statistical analyses in ecotoxicology. Environmental Science and Pollution Research International, 12, 11295–11302. [DOI] [PubMed] [Google Scholar]
- Ciric C, Ciffroy P and Charles S, 2012. Use of sensitivity analysis to identify influential and non‐influential parameters within an aquatic ecosystem model. Ecological Modelling, 246, 119–130. [Google Scholar]
- Copin PJ and Chevre N, 2015. Modelling the effects of pulse exposure of several PSII inhibitors on two algae. Chemosphere, 137, 70–77. [DOI] [PubMed] [Google Scholar]
- Copin PJ, Perronet L and Chevre N, 2016. Modelling the effect of exposing algae to pulses of S‐metolachlor: how to include a delay to the onset of the effect and in the recovery. Science of the Total Environment, 541, 257–267. [DOI] [PubMed] [Google Scholar]
- Crum SJ, van Kammen‐Polman AM and Leistra M, 1999. Sorption of nine pesticides to three aquatic macrophytes. Archives of Environmental Contamination and Toxicology, 37, 310–316. [DOI] [PubMed] [Google Scholar]
- DEFRA , 2005. Final report of the research project “Assessing the ecotoxicological impact to aquatic organisms from pulsed exposures to pesticides – PN0946.
- Diepens NJ, Arts GHP, Focks A and Koelmans AA, 2014. Uptake, translocation, and elimination in sediment‐rooted macrophytes: a model‐supported analysis of whole sediment test data. Environmental Science and Technology, 48, 12344–12353. [DOI] [PubMed] [Google Scholar]
- Driever SM, van Nes EH and Roijackers RMM, 2005. Growth limitation of Lemna minor due to high plant density. Aquatic Botany, 81, 245–251. [Google Scholar]
- EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues), 2010. Scientific Opinion on the development of specific protection goal options for environmental risk assessment of pesticides, in particular in relation to the revision of the Guidance Documents on Aquatic and Terrestrial ecotoxicology (SANCO/3268/2001 and SANCO/10329/2002). EFSA Journal 2010;8(10):1821, 55 pp. 10.2903/j.efsa.2010.1821 [DOI] [Google Scholar]
- EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues), 2013. Guidance on tiered risk assessment for plant protection products for aquatic organisms in edge‐of‐field surface waters. EFSA Journal 2013;11(7):3290, 268 pp. 10.2903/j.efsa.2013.3290 [DOI] [Google Scholar]
- EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues), 2014. Scientific Opinion on good modelling practice in the context of mechanistic effect models for risk assessment of plant protection products. EFSA Journal 2014;12(3):3589, 92 pp. 10.2903/j.efsa.2014.3589 [DOI] [Google Scholar]
- EFSA PPR Panel (EFSA Panel on Plant Protection Products and their Residues), 2015. Scientific Opinion on the effect assessment for pesticides on sediment organisms in edge‐of‐field surface water. EFSA Journal 2015;13(7):4176, 145 pp. 10.2903/j.efsa.2015.4176 [DOI] [Google Scholar]
- Englehardt JD and Swartout J, 2006. Predictive Bayesian microbial dose‐response assessment based on suggested self‐organization in primary illness response: Cryptosporidium parvum . Risk Analysis, 26, 543–554. [DOI] [PubMed] [Google Scholar]
- European Commission , 2002. Guidance Document on Aquatic Ecotoxicology Under Council Directive 91/414/EEC. SANCO/3268/2001 rev 4 (final), 17 October 2002.
- Fahl GM, Kreft L, Altenburger R, Faust M, Boedeker W and Grimme LH, 1995. pH‐dependent sorption, bioconcentration and algal toxicity of sulfunylurea herbicides. Aquatic Ecotoxicology, 31, 175–187. [Google Scholar]
- Focks A, Belgers D, Boerwinkel M‐C, Buijse L, Roessink I and Van den Brink P, 2018. Calibration and validation of toxicokinetic‐toxicodynamic models for three neonicotinoids and some aquatic macroinvertebrates. Ecotoxicology, 1–16. 10.1007/s10646-018-1940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOCUS (Forum for the Co‐ordination of Pesticide Fate Models and their Use), 2001. FOCUS Surface Water Scenarios in the EU Evaluation Process under 91/414/EEC. Report of the FOCUS Working group on Surface Water Scenarios. EC Document Reference SANCO/4802/2001 rev. 2, 245 pp.
- FOCUS (Forum for the Co‐ordination of Pesticide Fate Models and their Use), 2006. Guidance document on estimating persistence and degradation kinetics from environmental fate studies on pesticides in EU registration. Report of the FOCUS Working Group on Degradation Kinetics. EC Document Reference SANCO/10058/2005 version 2.0, 434 pp.
- FOCUS (Forum for the Co‐ordination of Pesticide Fate Models and their Use), 2007a. Landscape and mitigation factors in aquatic risk assessment. Volume 1. Extended Summary and Recommendations’. Report of the FOCUS Working Group on Landscape and Mitigation Factors in Ecological Risk Assessment, EC Document Reference SANCO/10422/2005 v2.0. 169 pp.
- FOCUS (Forum for the Co‐ordination of Pesticide Fate Models and their Use), 2007b. Landscape and Mitigation Factors In Aquatic Risk Assessment. Volume 2. Detailed Technical Reviews. Report of the FOCUS Working Group on Landscape and Mitigation Factors in Ecological Risk Assessment, EC Document Reference, SANCO/10422/2005 v2.0. 436 pp.
- Geiger DL, Call DJ and Brooke LT, 1988. Acute Toxicities of Organic Chemicals to Fathead Minnow (Pimephales promelas), vol. IV. Center for Lake Superior Environmental Studies, University of Wisconsin‐Superior, Superior, WI, USA. [Google Scholar]
- Gelman A, 2014. How Bayesian analysis cracked the red‐state, blue‐state problem. Statistical Science, 29, 26–35. [Google Scholar]
- Gelman S and Rubin D, 1992. Inference from iterative simulation using multiple sequences. Statistical Science, 7, 457–511. [Google Scholar]
- Geman S and Geman D, 1984. Stochastic relaxation, gibbs distributions, and the bayesian restoration of images. IEEE Transactions on Pattern Analysis and Machine Intelligence, 6, 721–741. [DOI] [PubMed] [Google Scholar]
- Grech A, Brochot C, Dorne JL, Quignot N, Bois FY and Beaudouin R, 2017. Toxicokinetic models and related tools in environmental risk assessment of chemicals. The Science of the Total Environment, 578, 1–15. [DOI] [PubMed] [Google Scholar]
- Hastings W, 1970. Monte Carlo sampling methods using Markov chains and their application. Biometrika, 57, 97–109. [Google Scholar]
- Heine S, 2014. Development and specification of a toxicokinetic and toxicodynamic growth model of Myriophyllum spicatum for use in risk assessment. PhD Thesis, 132 pp. Available online: https://publications.rwth-aachen.de/record/459446
- Heine S, Schmitt W, Goerlitz G, Schaeffer A and Preuss TG, 2014. Effects of light and temperature fluctuations on the growth of Myriophyllum spicatum in toxicity tests. Environmental Science and Pollution Research, 21, 9644–9654. [DOI] [PubMed] [Google Scholar]
- Heine S, Schmitt W, Schaeffer A, Goerlitz G, Buresova H, Arts G and Preuss TG, 2015. Mechanistic modelling of toxicokinetic processes within Myriophyllum spicatum . Chemosphere, 120, 292–298. [DOI] [PubMed] [Google Scholar]
- Heine S, Schild F, Schmitt W, Krebber R, Goerlitz G and Preuss TG, 2016a. A toxicokinetic and toxicodynamic modeling approach using Myriophyllum spicatum to predict effects caused by short‐term exposure to a sulfonylurea. Environmental Toxicology and Chemistry, 35, 376–384. [DOI] [PubMed] [Google Scholar]
- Heine S, Kuhl K, Solga A, Bruns E, Preuss T and Goerlitz G, 2016b. Aquatic macrophyte modelling ‐ increasing the realism of risk assessments. Proceedings of the SETAC Europe, 26th annual meeting. Environmental contaminants from land to sea: continuities and interface in environmental toxicology and chemistry, Nantes, France, 395 pp.
- Heine S, Bolekhan A, Görlitz G, Schäfer D, Bruns E, Solga A and Preuss TG, 2017. Linking time‐variable exposure to effects‐ effect modelling as a tool at different assessment levels. Proceedings of the SETAC Europe, 27th annual meeting. Environmental quality through transdisciplinary collaboration, Brussels, Belgium, 402 pp.
- Hommen U, Schmitt W, Heine S, Brock TCM, Duquesme S, Manson P, Meregalli G, Ochoa‐Acuna H, van Vliet P and Arts G, 2016. How TK‐TD and population models for aquatic macrophytes could support the risk 1 assessment for plant protection products. Integrated Environmental Assessment and Management, 12, 82–95. [DOI] [PubMed] [Google Scholar]
- Jager T, 2014. Reconsidering sufficient and optimal test design in acute toxicity testing. Ecotoxicology, 23, 38–44. [DOI] [PubMed] [Google Scholar]
- Jager T, 2017. Making Sense to Chemical Stress. Leanpub, 117 pp. Available online: http://www.debtox.info/book.php [Google Scholar]
- Jager T and Ashauer R, 2018. Modelling survival under chemical stress. Lean, 185 pp. Available online: https://leanpub.com/guts_book [Google Scholar]
- Jager T and Ravagnan E, 2016. Modelling growth of Northern krill (Meganyctiphanes norvegica) using an energy‐budget approach. Ecological Modelling, 325, 28–34. [Google Scholar]
- Jager T, Albert C, Preuss TG and Ashauer R, 2011. General unified threshold model of survival–a toxicokinetic‐toxicodynamic framework for ecotoxicology. Environmental Science and Technology, 45, 2529–2540. [DOI] [PubMed] [Google Scholar]
- Jager T and Zimmer E, 2012. T. JSimplified dynamic energy budget model for analysing ecotoxicity data. Ecological Modelling, 225, 74–81. [Google Scholar]
- Jager T, Martin BT and Zimmer EI, 2013. DEBkiss or the quest for the simplest generic model of animal life history. Journal of Theoretical Biology, 328, 9–18. [DOI] [PubMed] [Google Scholar]
- Jager T, Barsi A, Hamda NT, Martin BT, Zimmer EI and Ducrot V, 2014. Dynamic energy budgets in population ecotoxicology: applications and outlook. Ecological Modelling, 280, 140–147. [Google Scholar]
- Jiang JH, Zhou CF, An SQ, Yang HB, Guan BH and Cai Y, 2008. Sediment type, population density and their combined effect greatly charge the short‐time growth of two common submerged macrophytes. Ecological Engineering, 34, 79–90. [Google Scholar]
- Kooijman SALM, 2000. Dynamic energy and mass budgets in biological systems, 2nd edition Cambridge University Press, Cambridge, UK: p. 426. [Google Scholar]
- Kooijman SA, 2010. Dynamic Energy Budget Theory for Metabolic Organisation. Cambridge University Press, Cambridge, United Kingdom. 490 pp. [Google Scholar]
- Kooijman SA and Bedaux JJM, 1996. Analysis of toxicity tests on fish growth. Water Research, 30, 1633–1644. [Google Scholar]
- Kooijman SA and Troost TA, 2007. Quantitative steps in the evolution of metabolic organisation as specified by the Dynamic Energy Budget theory. Biological Reviews of the Cambridge Philosophical Society, 82, 113–142. [DOI] [PubMed] [Google Scholar]
- Llandres ALAL, Marques GMGM, Maino JL, Kooijman SALM, Kearney MR and Casas J, 2015. A dynamic energy budget for the whole life‐cycle of holometabolous insects. Ecological Monographs, 85, 353–371. [Google Scholar]
- Lunn DJ, Thomas A, Best N and Spiegelhalter D, 2000. WinBUGS – a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing, 10, 325–337. [Google Scholar]
- Metropolis N, Rosenbluth A, Rosenbluth M, Teller A and Teller E, 1953. Equations of state calculations by fast computing machines. The Journal of Chemical Physics, 21, 1087–1092. [Google Scholar]
- Nyman AM, Schirmer K and Ashauer R, 2012. Toxicokinetic‐toxicodynamic modelling of survival of Gammarus pulex in multiple pulse exposures to propiconazole: model assumptions, calibration data requirements and predictive power. Ecotoxicology, 21, 1828–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 1992. Fish acute toxicity test (No 203). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2004a. Daphnia sp. acute toxicity test (No 202). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2004b. Sediment‐water Chironomid toxicity test using spiked water (No 2018). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2006. Lemna sp. Growth Inhibition Test (No 221). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2011a. Freshwater Alga and Cyanobacteria, Growth Inhibition Test (No 201). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2011b. Chironomus sp. acute immobilisation test (No 235). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2012. Daphnia magna reproduction test (No 211). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2013. Fish, Early‐life Stage Toxicity Test (No 210). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2014a. Water‐Sediment Myriophyllum spicatum toxicity test (No 239). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- OECD (Organisation for Economic Cooperation and Development), 2014b. Sediment‐free Myriophyllum spicatum toxicity test (No 238). OECD Guidelines for the Testing of Chemicals. OECD, Paris, France. [Google Scholar]
- Pedersen O and Sand‐Jensen K, 1993. Water transport in submerged macrophytes. Aquatic Botany, 44, 385–406. [Google Scholar]
- Plummer M, 2003. JAGS: A Program for Analysis of Bayesian Graphical Models Using Gibbs Sampling. Proceedings of the Proceeding of the 3rd International Workshop on distributed statistical computing Vienna.
- Plummer M, 2013. JAGS Version 3.4.0 user manual. Available online: http://sourceforge.net/projects/mcmc-jags/files/Manuals/3.x/jags_user_manual.pdf/download.
- R Core Team , 2016. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: Available online: https://www.R-project.org/. [Google Scholar]
- Raftery AE and Lewis SM, 1992. How many iterations in the Gibbs Sampler? In: Bernardo JM, Berger JO, Dawid AP, Smith AFM. (eds.). Bayesian Statistics. Oxford University Press, Oxford, United Kingdom: pp. 763–773. [Google Scholar]
- Rendal C, Kusk KO and Trapp S, 2011. The effect of pH on the uptake and toxicity of the bivalent weak base chloroquine tested on Salix viminalis and Daphnia magna . Environmental Toxicology and Chemistry, 30, 354–359. [DOI] [PubMed] [Google Scholar]
- Rubach MN, Ashauer R, Buchwalter DB, De Lange H, Hamer M, Preuss TG, Topke K and Maund SJ, 2011. Framework for traits‐based assessment in ecotoxicology. Integrated Environmental Assessment and Management, 7, 172–186. [DOI] [PubMed] [Google Scholar]
- Saltelli A, Chan K and Scott EM, 2000. Sensitivity Analysis. John Wiley & Sons, Chichester. [Google Scholar]
- Schmitt WH, Bruns E, Dollinger M and Sowig P, 2013. Mechanistic TK/TD‐model simulating the effect of growth inhibitors on Lemna populations. Ecological Modelling, 255, 1–10. [Google Scholar]
- Soetaert K, Petzoldt T and Setzer RW, 2010. Solving differential equations in R. The R Journal, 2, 5–15. [Google Scholar]
- Tennekes HA and Sanchez‐Bayo F, 2013. The molecular basis of simple relationships between exposure concentration and toxic effects with time. Toxicology, 309, 39–51. [DOI] [PubMed] [Google Scholar]
- Trapp S, 2004. Plant uptake and transport models for neutral and ionic chemicals. Environmental Science and Pollution Research International, 11, 33–39. [DOI] [PubMed] [Google Scholar]
- Van Straalen NM, 2003. Ecotoxicology becomes stress ecology. Environmental Science and Technology, 37, 324A–330A. [DOI] [PubMed] [Google Scholar]
- Von Bertalanffy L, 1938. A quantitative theory of organic growth (injuries on growth laws. II). Human Biology, 10, 181–213. [Google Scholar]
- Weber D, 2009. Measuring and predicting the effects of time‐variable exposure of pesticides on populations of green algae: combination of flow‐through studies and ecological modelling as an innovative tool for refined risk assessments. Available online: https://publications.rwth-aachen.de/record/211799/, 213 pp
- Weber D, Schäffer D, Dorgerloh M, Bruns E, Görlitz G, Hammel K, Greuss T and Ratte H, 2012. Combination of a higher‐tier flow‐through system and population modeling to assess the effects of time‐variable exposure of isoproturon on the green algae Desmodesmus subspictatus and Pseudokirchneriella subcapitata . Environmental Toxicology and Chemistry, 31, 899–908. [DOI] [PubMed] [Google Scholar]
- van Wijngaarden RP, Maltby L and Brock TC, 2015. Acute tier‐1 and tier‐2 effect assessment approaches in the EFSA Aquatic Guidance Document: are they sufficiently protective for insecticides? Pest Management Science, 71, 1059–1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Repository of codes
Outcome of the consultation on the Draft Opinion with the Pesticide Steering Network


