Abstract
Breeding programmes to promote resistance to classical scrapie, similar to those for sheep in existing transmissible spongiform encephalopathies (TSE) regulations, have not been established in goats. The European Commission requested a scientific opinion from EFSA on the current knowledge of genetic resistance to TSE in goats. An evaluation tool, which considers both the weight of evidence and strength of resistance to classical scrapie of alleles in the goat PRNP gene, was developed and applied to nine selected alleles of interest. Using the tool, the quality and certainty of the field and experimental data are considered robust enough to conclude that the K222, D146 and S146 alleles both confer genetic resistance against classical scrapie strains known to occur naturally in the EU goat population, with which they have been challenged both experimentally and under field conditions. The weight of evidence for K222 is greater than that currently available for the D146 and S146 alleles and for the ARR allele in sheep in 2001. Breeding for resistance can be an effective tool for controlling classical scrapie in goats and it could be an option available to member states, both at herd and population levels. There is insufficient evidence to assess the impact of K222, D146 and S146 alleles on susceptibility to atypical scrapie and bovine spongiform encephalopathy (BSE), or on health and production traits. These alleles are heterogeneously distributed across the EU Member States and goat breeds, but often at low frequencies (< 10%). Given these low frequencies, high selection pressure may have an adverse effect on genetic diversity so any breeding for resistance programmes should be developed at Member States, rather than EU level and their impact monitored, with particular attention to the potential for any negative impact in rare or small population breeds.
Keywords: genetics, resistance, TSE, goats, classical, scrapie
Summary
Following a request from the European Commission, the European Food Safety Authority (EFSA) was asked to deliver a scientific opinion on genetic resistance to transmissible spongiform encephalopathies (TSEs) in goats, addressing the following terms of reference: (1) Is there sufficient scientific knowledge available to have a robust level of scientific assurance that certain polymorphisms of the prion protein gene (PRNP) present in European goat breeds confer genetic resistance to classical scrapie (i.e. to classical scrapie strains known to occur in the European Union (EU) goat population)? If this is the case, which are those polymorphisms? (2) Based on available scientific evidence, what is the frequency and distribution of PRNP polymorphisms conferring resistance to classical scrapie in European goat breeds? If possible, could EFSA produce a susceptibility ranking of goat PRNP genotypes to classical scrapie? (3) Based on available scientific evidence, what is the level of susceptibility to atypical scrapie and to BSE of the PRNP polymorphisms conferring resistance to classical scrapie? (4) What is the likely impact of measures promoting PRNP polymorphisms conferring resistance to classical scrapie in terms of susceptibility to other disease/s, of production traits and survivability, taking into account epidemiological differences between Member States (MS)? Are such polymorphisms likely to have adverse effects on genetic diversity and variability and on the maintenance of old or rare caprine breeds or those that are well‐adapted to a particular region? (5) What are EFSA's recommendations concerning strategies to apply current knowledge on genetic resistance to classical scrapie in goats in order to control and/or eradicate classical scrapie in the EU goat population?
A tool was developed to evaluate the genetic resistance to classical scrapie of alleles in the goat PRNP gene, which considers both the weight of evidence and strength of resistance for each allele of interest. The weight of evidence was based on a scale made of the combination of different types of studies, from the one that provides the least evidence, the in vitro conversion studies, to the combination of all possible studies (epidemiological studies, experimental challenge in natural host using multiple isolates, bioassays in allele specific transgenic mice using multiple isolates and in vitro conversion studies). Whether the allele of interest had been experimentally investigated against multiple scrapie isolates from geographically different origins was considered. The strength of resistance conferred by an allele was defined by scoring it, in a traffic light colour system, as demonstrating one of three levels of resistance using the wild type of the goat PRNP gene as a baseline: Red: no resistance; Amber: partial resistance; Green: resistance. To assign the strength of resistance for each allele of interest information on the presence of field cases holding the alleles of interest, the attack rates and the incubation period in the natural host or in allele‐specific transgenic mice were considered as indicators. A fourth score ‘grey’ was also given for the specific situation where there was just one study available to support the putative resistance of an allele. As a comparator, the tool was also applied to the knowledge on the role of the ovine ARR allele that was available at the time the opinion of the Scientific Steering Committee on safe sourcing of small ruminant materials was produced in 2001 (SSC, 2002).
An extensive literature review was conducted to identify relevant alleles to which the tool could be applied, to determine the frequency and distribution of such alleles within different MS and goat breeds and to assess their resistance to atypical scrapie and bovine spongiform encephalopathy (BSE). For the nine selected alleles, namely S127, M142, R143, D145, D146, S146, H154, Q211 and K222, a considerable data set has been produced to assess the levels of resistance to classical scrapie, including details of the type of evidence for or against the association with resistance to TSE. A different class of haplotype in which a nonsense polymorphism (G32stop) occurs was also reviewed, but not included in the tool since this mutation fails to translate full‐length PrP protein.
In goats, there are no published studies on any effects of PRNP polymorphisms on traits other than resistance to classical scrapie. Therefore, the scant available literature on effects of PRNP polymorphisms in sheep on other traits was used to assess, by extrapolation, the likely impact in goats.
It is concluded that the scientific knowledge related to scrapie resistance associated with goat PRNP gene polymorphisms has considerably expanded in the last 10 years. The K222, D146 and S146 alleles confer genetic resistance to classical scrapie strains known to occur in the EU goat population. The K222 polymorphism confers resistance against a variety of EU classical scrapie isolates that may reflect a variety of scrapie strains. However, there is no assurance that K222 carriers would be resistant to all TSE agent strains currently circulating in the EU goat population. The D146 and S146 alleles are associated with strong resistance against scrapie agent(s) currently circulating in Cyprus. However, data remain insufficient to assess the level of resistance that D146 and S146 might provide against other classical scrapie agents circulating in other EU goat populations. The weight of evidence for K222 is greater than that currently available for the D146 and S146 alleles and for the ARR allele in sheep at the time the 2002 SSC opinion was produced.
There are limited data available on the frequency and distribution of the PRNP alleles conferring resistance to classical scrapie as they are only known for less than 10% of the breeds listed in EU MS, and only in a restricted number of MS. Cyprus is the only MS with accurate genotype data about the whole goat population: more than 50% of the goats in Cyprus have either the D146 or the S146 allele. In other MS, these alleles are represented at low frequencies, or are absent, with some exceptions. The K222 allele is not present in Cyprus, but has been observed in most of the investigated EU breeds, although generally at low levels (< 10%). In some breeds (Cashmere, Angora, Spanish autochthonous breeds), the polymorphisms K222, D146 and S146 have not been reported.
Due to insufficient data, it is not possible to provide a ranking of susceptibility at genotype level. Based on a combination of the ‘weight of evidence’ and the ‘strength of resistance’, alleles can currently be ranked as follows, from high to low classical scrapie resistance: K222 > D146 = S146 >Q211 = H154 = M142 > S127 = H143 > wild type.
There is extremely limited data on the susceptibility of goats to either atypical scrapie or BSE for the nine alleles of interest. For atypical scrapie, it was hypothesised that the H154 allele is likely to be associated with higher risk of developing atypical scrapie. For BSE, there are currently no data available on the resistance/susceptibility to BSE infection in D146 and S146 allele carriers, but an oral challenge study indicated incomplete resistance to BSE in K222 allele carriers. There are no data available on the susceptibility of goats with the H154 allele to BSE infection.
Information on the relationship of PRNP alleles with other traits is lacking in goats and therefore the likely impact of measures promoting selection for resistance on other traits has been inferred from studies in sheep. From this literature, it is inferred that a direct effect of PRNP alleles on health and production traits is unlikely. However, in breeds with low frequencies of the favourable allele(s), and breeds with small effective population size, selection could affect other traits, with the direction of change being unpredictable. Given the low frequencies of favourable alleles in most breeds, including breeds with large populations such as Alpine and Saanen and old breeds that are well adapted to particular regions, high selection pressure is likely to have an adverse effect on genetic diversity.
The following recommendations concerning the strategies of breeding for resistance were made: (a); genetic resistance can be considered to be an effective tool to control classical scrapie in goats and may be offered as an option for MS to control classical scrapie in goats; (b) outbreak management for classical scrapie in goat herds could be based on the selection of genetically resistant animals, as defined for sheep in the Regulation 999/2001 (EC); (c) breeding for resistance programmes should be designed to take into account the starting allele frequencies with the view to preventing loss of genetic diversity and they should be developed and managed at MS level, with the impact of their implementation monitored, and with consideration being given to derogations if implementation is compulsory; (d) before developing any breeding for resistance programme, baseline surveys are needed to establish the resistant‐allele frequencies in the relevant goat populations at both MS and breed level.
Consideration was given to other aspects such as (a) informing relevant stakeholders, especially breeders, about genetic resistance to classical scrapie in goats; (b) producing guidance on how to disseminate resistant alleles in goat breeds; (c) establishing a central database to know where and how many resistant animals are available for breeding purposes at MS level; (d) encouraging the creation of a pool of resistant animals, semen, embryos and ova for dissemination of the resistant alleles in the population; (e) investigating the potential association of alleles conferring resistance to scrapie with other traits; this may be achieved through ad hoc studies or monitored during the breeding programmes.
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
As regards ovine animals, the former Scientific Steering Committee (SSC) highlighted the resistance to transmissible spongiform encephalopathies (TSE) of ARR/ARR sheep in its 2002 Scientific Opinion on safe sourcing of small ruminant materials,1 which was based on its 1999 Scientific Opinion on the policy of breeding and genotyping of sheep.2 These scientific opinions were the basis for the adoption of Regulation (EC) No 260/20033, which revised the requirements for eradication measures in case of the detection of TSE in a holding by allowing the destruction of susceptible ovine animals only and requiring the implementation of measures aimed at increasing sheep's resistance to TSEs in the infected holding. In addition, these scientific opinions were the basis for the adoption of Commission Decision 2003/100/EC4, which laid down requirements for the establishments of breeding programmes for resistance to TSE in sheep. The breeding programmes requirements were then integrated into Regulation (EC) No 999/2001 by Commission Regulation (EC) No 1923/20065 and Commission Regulation (EC) No 727/20076. In 2006, in its Opinion on the breeding programme for TSE resistance in sheep’, the Scientific Panel on Biological Hazard of the European Food Safety Authority (EFSA) confirmed the appropriateness of such a strategy (EFSA, 2006).
As regards caprine animals, until recently, there was not enough scientific knowledge on resistance to TSE in goats to adopt similar measures. However, based on the development of scientific evidence, EFSA, in its 2014 Scientific Opinion on the scrapie situation in the European Union (EU) after 10 years of monitoring and control in sheep and goats (EFSA BIOHAZ Panel, 2014), recommended that selection activities and dissemination of resistant bucks should be promoted and that formal breeding for resistance programmes, similar to those already implemented for sheep, should be initiated for goats.
In recent months, Cyprus and Italy have, respectively, submitted the attached scientific reports (see Documentation provided to EFSA), in order to substantiate the scientific basis of measures that they propose to promote resistance to TSEs in goats.
Given the long‐term effect of measures of selection and dissemination of animals with a certain genotype, it is necessary to have a solid level of scientific certainty concerning the resistance to TSE of the genotype(s) to be selected and the impact that such measures are likely to have.
EFSA is therefore requested to provide a scientific opinion on the following questions:
Is there sufficient scientific knowledge available to have a robust level of scientific assurance that certain polymorphisms of the prion protein gene (PRNP) present in European goat breeds confer genetic resistance to classical scrapie (i.e. to classical scrapie strains known to occur in the EU goat population)? If this is the case, which are those polymorphisms?
Based on available scientific evidence, what is the frequency and distribution of PRNP polymorphisms conferring resistance to classical scrapie in European goat breeds? If possible, could EFSA produce a susceptibility ranking of goat PRNP genotypes to classical scrapie?
Based on available scientific evidence, what is the level of susceptibility to atypical scrapie and to BSE of the PRNP polymorphisms conferring resistance to classical scrapie?
What is the likely impact of measures promoting PRNP polymorphisms conferring resistance to classical scrapie in terms of susceptibility to other disease/s, of production traits and survivability, taking into account epidemiological differences between Member States (MS)? Are such polymorphisms likely to have adverse effects on genetic diversity and variability and on the maintenance of old or rare caprine breeds or those that are well‐adapted to a particular region?
What are EFSA's recommendations concerning strategies to apply current knowledge on genetic resistance to classical scrapie in goats in order to control and/or eradicate classical scrapie in the EU goat population?
1.2. Interpretation of the Terms of Reference
With regard to certain points of the Terms of Reference (ToR), these are the interpretations of the Working Group (WG):
The WG will address the genetic resistance at breed level whenever possible and at population level when there is insufficient genetic uniformity in groups of goats that may constitute a breed from the legal point of view.
The definition of ‘strong genetic resistance’ proposed by the European Commission included that an animal holding a polymorphism conferring strong resistance ‘do not transmit classical scrapie during the productive life’. The WG will provide an answer on resistance to disease in individual animals, according to available knowledge considering all uncertainties and with the data available.
The impact of breeding for resistance on small populations will be assessed without referring to any particular population (ToR4).
2. Data and methodologies
2.1. Data
Scrapie data were obtained from the EU TSE database, which collects standardised surveillance data on all testing activities in all MS. The overall caseload is based on cases detected through passive and active surveillance, and during the application of eradication measures. Due to the biased and variable nature of passive surveillance, data from clinical cases were excluded from the analysis, and only the more unbiased active surveillance data, namely the animals slaughtered for human consumption (SHC) and the animals not slaughtered for human consumption (NSHC), were used for describing national trends in scrapie prevalence and for describing the occurrence by EU MS.
Information was obtained from Eurostat to describe goat demographics. In particular, the number of goats, number of breeding females, number of holdings and number of female breeding holdings were recorded for each MS in both 2005 and 2013. Eurostat was also used to obtain data on the amount of goat meat produced at slaughterhouses in the EU (2006–2015).7
The amount of goat meat produced at slaughterhouses in the EU for the period 2006–2015 was extracted from the EU statistics website, as part of the annual statistical surveys.8
Goat breed data was obtained from the EFABIS database.9
Data on the consumption of goat meat in the EU were obtained from EFSA's Comprehensive European Food Consumption database. The EFSA Comprehensive European Food Consumption Database (Comprehensive Database)10 provides a compilation of national information on food consumption at individual level. It was first built in 2010 (EFSA, 2011a; Huybrechts et al., 2011; Merten et al., 2011). Details on how the Comprehensive Database is used are published in the Guidance of EFSA (EFSA, 2011b). EFSA used its food classification system ‘FoodEx’ to categorise all foods and beverages included in the Comprehensive Database (EFSA, 2011a). Available data on goat meat consumption in the EU was extracted and summarised.
Data on the movements of live goats for breeding in 2014, 2015 and 2016 were extracted from the TRACES database.11
2.2. Methodologies
In order to address the ToR as in the mandate, and particularly provide recommendations concerning strategies for a breeding programme to control and/or eradicate classical scrapie in goats (ToR5), it is important to give an overview of the goat sector in the EU in order to put the answers into context. As a consequence of this, Section 3.1 considers information for the EU on the goat demographics in MS, goat meat production, consumption patterns, movement of live goats for breeding within the EU, their genetic diversity, and the scrapie situation in the EU.
2.2.1. Literature reviews
An evaluation of the PRNP polymorphisms associated with genetic resistance in goats (ToR 1, 2, 3) was carried out through an extensive literature review.
To retrieve data on the PRNP polymorphisms associated with genetic resistance in goats, a literature search in the PubMed database was undertaken. The publication of the first scientific paper describing polymorphisms in the PRNP gene in goats (Goldmann et al., 1996) was used as the starting date of the literature search, with a buffer of one extra year. No language restrictions were applied for the literature search (all languages included in the PubMed search engine were included: 56). The literature search was conducted on 20 July 2016. The following search string was applied: (goat* OR caprine*) AND (TSE* OR BSE OR scrapie OR PrP* OR PRNP OR prion* OR allel* OR gen*) AND (polymorph* OR breed* OR resist* OR susceptib*). These terms were searched in the titles and abstracts of books and documents, case reports, classical articles, clinical trials, comments, comparative studies, data sets, editorials, electronic supplementary materials, English abstracts, introductory journal articles, journal articles, news, newspaper articles, randomised controlled trials, reviews, scientific integrity reviews, systematic reviews, technical reports and validation studies. A total of 946 references were retrieved and a double screening (two pairs of reviewers each independently screened half of the full list) looking for potentially relevant references was conducted. Discrepancies were discussed between the two reviewers until a final shortlist of references was agreed. A subset of 50 references was selected and considered in this assessment by reviewing in full. A further 12 references were shortlisted for their relevance to other ToR during the screening process.
To retrieve data on the doppel protein (Dpl), a prion‐like protein encoded by the gene PRND, which has been found downstream of the prion gene, PRNP, in human and mice, a literature search in the PubMed database was undertaken. The time of publication was restricted to the period 1/1/1995–31/12/2016. No language restrictions were applied for the literature search (all languages included in the PubMed search engine were included: 56). The literature search was conducted on 15 December 2016. The following search string was applied: (sheep OR goat*) AND (doppel OR Doppel OR sprn OR SPRN). These terms were searched in the titles and abstracts of books and documents, case reports, classical article, clinical trial, comment, comparative study, data set, editorial, electronic supplementary materials, English abstract, introductory journal article, journal article, news, newspaper article, randomised controlled trial, review, scientific integrity review, systematic reviews, technical report and validation studies. A total of 27 references were retrieved and all of them were considered in this assessment by reviewing the full papers.
2.2.2. Analysis of surveillance data
Analysis of scrapie surveillance data was conducted separately by disease: classical scrapie (CS) vs atypical scrapie (AS). In each individual subset, descriptive frequency tables were produced showing the breakdown of animals tested and cases by country, year, and surveillance stream: SHC and NSHC. The potential for a confounding effect of stream in the case of CS became evident after comparing the stream‐specific prevalence and the different distribution of the number of tests carried out in each stream by country or by year.
A spatial description of the presence of the scrapie types was carried out by producing two sets of maps:
the occurrence of CS and AS for the period 2002–2015 by MS;
surveillance stream‐adjusted for CS and crude for AS prevalence rates for the period 2002–2015 by MS, produced through proportional symbol mapping. The adjustment on surveillance stream was carried out by means of a direct standardisation using the proportion of tests carried out in the MS in the NSHC vs SHC, in sheep and goats, respectively.
Negative binomial models were used to fit ‘count of cases detected’ and ‘year’ to estimate the country‐specific and stream‐adjusted annual prevalence ratios (PR). Significance levels of the slope of the linear function for individual MS and years were used to determine statistically significant temporal trends. Due to sparse data, BG, FI, RO and SI12 were excluded from the analyses.
The objective of this analysis was to provide a description of scrapie occurrence in goats by stream, scrapie type and MS, comparing the occurrence between years within MS.
The precision and validity of the crude PR obtained through the analysis of active surveillance data may have been affected by the targeted and sample‐based design of both the SHC and NSHC surveys. Country‐specific temporal trends are in general heterogeneous, precluding any meaningful interpretation of the overall temporal trend at the EU28 level. Therefore, the analysis and interpretation of the temporal trends has been conducted only at MS level.
3. Assessment
3.1. The goat sector and the situation of scrapie in the EU
3.1.1. Demographics, goat meat production and consumption
In 2013, there were 9,388,290 goats in the 28 EU MS distributed across 163,030 holdings (Eurostat). Figures 1 and 2 show the between‐MS variability of the number of goats (heads) and the number of holdings in each MS,12 respectively. The majority of the EU goat population is located in EL (3,213,880), ES (2,055,420) and FR (1,086,270) which accounts for around 68% of the total EU population. The EU MS with the largest number of goat holdings are: RO (40,470), EL (30,580) and ES (16,790), which represent 54% of goat holdings in the EU. Across many MS, the majority of goats are breeding females, which are reared for milk (see Table A.1 in Appendix A). According to the data, only DK and LT have < 50% of breeding females within their goat population, although the data for these two MS indicate that 90% and 73%, of holdings, respectively, are for breeding females. Holdings may have both goats and sheep present.
Figure 1.
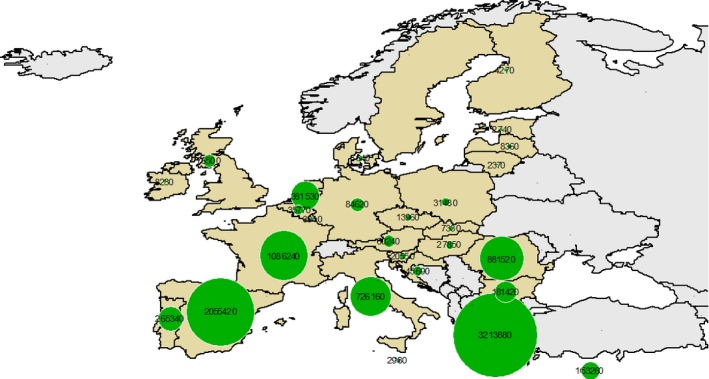
Number of goats (head) in the EU in 2013. No data from SE
Figure 2.
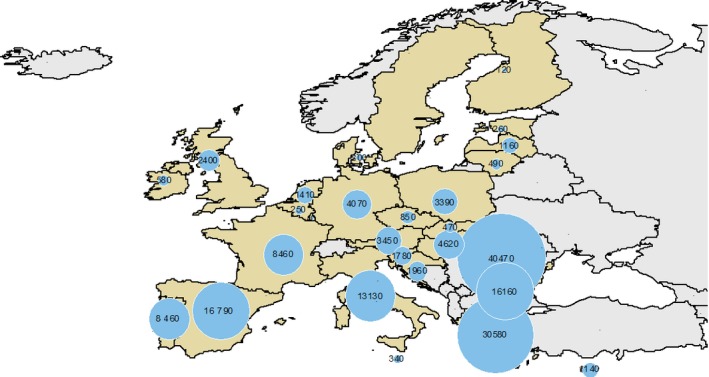
Number of goat holdings in the EU in 2013. No data from SE
Table A.1.
Comparison of goat types in 2013 (Source: Eurostat)
| Member State | Heads | Holdings | ||
|---|---|---|---|---|
| Total | Breeding females (%) | Total | Breeding females (%) | |
| AT | 60,240 | 37,480 (62.2) | 3,450 | 2,810 (81.4) |
| BE | 35,770 | 34,030 (95.1) | 250 | 220 (88.0) |
| BG | 181,420 | 166,340 (91.7) | 16,160 | 16,040 (99.3) |
| CY | 153,260 | 128,960 (84.1) | 1,140 | 1,140 (100) |
| CZ | 13,960 | 9,330 (66.8) | 850 | 700 (82.4) |
| DE | 84,620 | 55,820 (66.0) | 4,070 | 2,930 (72.0) |
| DK | 6,640 | 2,430 (36.6) | 200 | 180 (90.0) |
| EE | 2,740 | 2,280 (83.2) | 260 | 260 (100) |
| EL | 3,213,880 | 2,691,160 (83.7) | 30,580 | 30,220 (98.8) |
| ES | 2,055,420 | 1,870,610 (91.0) | 16,790 | 16,490 (98.2) |
| FI | 4,270 | 4,270 (100) | 120 | 120 (100) |
| FR | 1,086,240 | 819,570 (75.4) | 8,460 | 8,040 (95.0) |
| HR | 45,690 | 31,000 (67.8) | 1,960 | 1,950 (99.5) |
| HU | 27,850 | 14,670 (52.7) | 4,620 | 2,880 (62.3) |
| IE | 8,230 | 4,990 (60.6) | 580 | 410 (70.7) |
| IT | 726,160 | 594,230 (81.8) | 13,130 | 11,440 (87.1) |
| LT | 2,370 | 1,140 (48.1) | 490 | 360 (73.5) |
| LU | 3,940 | 2,720 (69.0) | 40 | 30 (75.0) |
| LV | 8,360 | 6,580 (78.7) | 1,160 | 1,130 (97.4) |
| MT | 2,980 | 2,660 (89.3) | 340 | 340 (100) |
| NL | 381,530 | 280,920 (73.6) | 1,410 | 1,350 (95.7) |
| PL | 31,480 | 23,690 (75.3) | 3,390 | 3,020 (89.0) |
| PT | 265,340 | 223,420 (84.2) | 8,460 | 8,400 (99.3) |
| RO | 881,520 | 769,320 (87.3) | 40,470 | 37,480 (92.6) |
| SI | 20,550 | 11,400 (55.5) | 1,780 | 1,560 (87.6) |
| SK | 7,330 | 5,280 (72.0) | 470 | 410 (87.2) |
| SE | N/A | N/A | N/A | N/A |
| UK | 76,500 | 54,930 (71.8) | 2,400 | 1,750 |
| Total | 9,388,290 | 7,849,230 | 163,030 | 151,660 |
Data also indicate that the goat population in the EU is increasing (see Table A.2 in Appendix A). Using Eurostat data from 2005 and 2013, the number of goats in the EU (for MS that reported in both years only) increased by 10.3%, with particularly high increases in RO (297.8%), SK (132.7%) and LU (123.9%). MS with the largest growth in the number of holdings were SK (135%), RO (79.7%) and MT (61.9%); however, overall number of holdings decreased by 1.86%. This is attributable to 16 MS reducing the number of holdings over this time period, with percentage decreases ranging from 4.3% (PT) to 65% (UK).
Table A.2.
Heads and holdings of goats in 2005 and 2013
| Member State | Heads | Holdings | ||
|---|---|---|---|---|
| 2005 | 2013 | 2005 | 2013 | |
| AT | 38,310 | 60,240 | 4,390 | 3,450 |
| BE | 23,500 | 35,770 | 370 | 250 |
| BG | 143,920 | 181,420 | 24,680 | 16,160 |
| CY | 283,850 | 153,260 | 1,780 | 1,140 |
| CZ | 8,600 | 13,960 | 950 | 850 |
| DE | N/A | 84,620 | N/A | 4,070 |
| DK | 3,750 | 6,640 | 150 | 200 |
| EE | 3,180 | 2,740 | 570 | 260 |
| EL | 3,151,290 | 3,213,880 | 25,680 | 30,580 |
| ES | 2,109,210 | 2,055,420 | 21,010 | 16,790 |
| FI | 5,870 | 4,270 | 220 | 120 |
| FR | 1,059,130 | 1,086,240 | 10,680 | 8,460 |
| HR | N/A | 45,690 | N/A | 1,960 |
| HU | 27,770 | 27,850 | 4,240 | 4,620 |
| IE | 8,420 | 8,230 | 650 | 580 |
| IT | 630,200 | 726,160 | 12,220 | 13,130 |
| LT | 5,220 | 2,370 | 870 | 490 |
| LU | 1,760 | 3,940 | 50 | 40 |
| LV | 5,730 | 8,360 | 750 | 1,160 |
| MT | 2,000 | 2,980 | 210 | 340 |
| NL | 257,490 | 381,530 | 1,630 | 1,410 |
| PL | 39,860 | 31,480 | 8,950 | 3,390 |
| PT | 269,120 | 265,340 | 8,840 | 8,460 |
| RO | 221,580 | 881,520 | 22,520 | 40,470 |
| SI | 18,130 | 20,550 | 1,510 | 1,780 |
| SK | 3,150 | 7,330 | 200 | 470 |
| SE | N/A | N/A | N/A | N/A |
| UK | 75,630 | 76,500 | 6,860 | 2,400 |
| Total | 8,396,670 | 9,388,290 | 159,980 | 163,030 |
The number of goats within an EU MS does not provide an overall picture of goat demographics within Europe. Many EU MS have large intensive rearing systems due to the demand for goat milk or cheese made from goat milk, for example CY, FR, EL, ES. For other EU MS, goats are reared in smaller herds on less fertile land and are often more traditional breeds, for example EL or ES. For such MS, there may not be a goat sector body working collectively to represent the industry. Such holdings may be very dependent on European agricultural subsidies (De Rancourt et al., 2006). According to the study on ‘The future of the sheep and goat sector in Europe’13 (2008) conducted by the European Commission's Directorate on Structural and Cohesion Policy, sheep and goat farming is very labour‐intensive, resilient, and lacks technical services and training, while providing a strong link with the land and the environment and a high genetic diversity.
The overall contribution of goat meat to the EU28 meat production is modest (0.1 %) and is concentrated in seven MS, representing 95.6 % of the total EU goat meat production, namely (in decreasing order of production) EL, ES, FR, RO, CY, the NL and IT. It can be described as a by‐product of dairy production.
In terms of slaughtering goats at slaughterhouses, Table B.1 of Appendix B displays the amount of goat meat produced by MS for the period 2006–2015.
Table B.1.
Goat meat produce at slaughterhouses in the EU for the period 2006–2015 (in thousands of tonnes)
| MS/year | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 |
|---|---|---|---|---|---|---|---|---|---|---|
| EU (28) | 77.26 | 80.57 | : | : | : | : | : | : | : | : |
| AT | 0 | 0 | 0.7 | 0.73 | 0.78 | 0.83 | 0.78 | 0.79 | 0.83 | 0.83 |
| BE | 0.03 | 0.02 | 0.05 | 0.03 | 0.08 | 0.05 | 0.09 | 0.11 | 0.13 | 0.19 |
| BG | 5.54 | 5.84 | 4.84 | 0.07 | : | : | : | : | : | : |
| CY | 3.92 | 4.01 | 3.87 | 2.74 | 2.32 | 2.35 | 2.68 | 2.28 | 2.08 | 2 |
| CZ | 0.11 | 0.1 | 0.12 | : | 0 | 0 | 0 | 0 | 0 | 0 |
| DE | 0.43 | 0.46 | 0.48 | 0.4 | 0.42 | 0 | 0 | 0 | 0 | 0 |
| DK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| EE | 0.02 | 0.03 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | : |
| EL | 39.03 | 37.91 | 37.55 | 36.66 | 35.59 | 33.56 | 30.14 | 24.82 | 23.89 | 21.93 |
| ES | 11.69 | 10.45 | 9.25 | 8.83 | 10.62 | 11.14 | 9.7 | 8.94 | 8.62 | 9.23 |
| FI | 0.01 | 0 | 0 | 0 | : | : | 0 | 0 | 0 | 0 |
| FR | 7.76 | 8.08 | 7.46 | 6.46 | 6.87 | 7.36 | 6.32 | 6.49 | 6.22 | 6.23 |
| HR | 0.01 | 1.29 | 0.9 | : | : | : | : | : | : | : |
| HU | 0.26 | 0.21 | 0.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IE | 0 | 0 | : | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IT | 2.62 | 2.23 | 2.29 | 1.46 | 1.2 | 1.2 | 1.22 | 1.29 | 1.32 | 1.81 |
| LT | 0.34 | 0.3 | 0.3 | 0 | 0 | : | : | : | : | : |
| LU | 0.01 | 0 | 0 | 0 | 0 | 0 | 0.01 | 0 | 0.01 | 0.01 |
| LV | 0.06 | 0.07 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MT | 0 | 0 | 0 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.02 | 0.02 |
| NL | 0.21 | 1.62 | 1.33 | 1.06 | 1.36 | 1.87 | 1.54 | 1.73 | 1.62 | 1.47 |
| PL | 0.38 | 0.44 | 0.27 | 0.09 | 0.27 | 0.05 | 0 | 0.02 | 0 | : |
| PT | 0.81 | 1.02 | 0.89 | 0.92 | 0.89 | 0.9 | 0.93 | 0.8 | 0.71 | 0.77 |
| RO | 3.6 | 6.04 | 7.1 | : | : | : | : | : | : | 0.05 |
| SI | 0 | 0 | 0 | : | 0 | 0 | 0 | 0 | 0.01 | 0.01 |
| SK | 0.32 | 0.3 | 0.3 | 0 | 0 | 0 | : | 0 | 0 | 0 |
| SE | 0.01 | 0.01 | 0.01 | 0.02 | 0 | 0.01 | 0.01 | 0.01 | 0 | 0.01 |
| UK | 0.12 | 0.14 | 0.13 | 0.13 | 0.17 | 0.19 | 0.23 | 0.22 | 0.29 | 0.27 |
: not reported.
0.00: no throughput.
Data for the consumption of goat meat in the EU is scarce. From five MS, the available data from the EFSA Comprehensive European Food Consumption Database (see Table B.2 in Appendix B) suggest that children in EL have the highest level of consumption (the mean consumption per day was less than 0.84 g with 2.74% of children included in the survey reporting consumption of goat meat) followed by adolescents and adults in ES with 0.22 g/day. No information is provided for the consumption of goat milk or other goat milk products.
Table B.2.
Consumption data of goat meat in the EU (EFSA Comprehensive European Food Consumption)
| MS/population class | Other children | Adolescents | Adults | Elderly | Lactating women |
|---|---|---|---|---|---|
| EL | 0.84 (23/838)* | – | – | – | 0.51 (1/65) |
| ES | 0.22 (2/651) | 0.22 (3/981) | – | – | |
| IT | – | – | 0.05 (3/2313) | 0.19 (1/290) | – |
| LV | – | – | 0.04 (1/1271) | – | – |
| NL | 0.03 (1/2057) | – | – |
* Mean consumption in grams/day (number of consumers/number of subjects).
According to TRACES database, in the period 2014–2016, a total of 13,607, 12,995 and 11,622 goats for breeding, respectively, were moved within the EU involving 24, 23 and 27 MS, respectively, either as importers, exporters or both. The MS with the largest number of goats exported were AT, ES, FR and the NL. Fifteen other MS also exported goats for breeding but in smaller numbers. In terms of import, BE, DE, IT and RO were the MS with the largest contingent of goats imported during that period. Seventeen other MS also imported goats for breeding but in smaller numbers. The movements of live goats for breeding within the EU according to TRACES database in 2014, 2015 and 2016 are displayed in Tables C.1, C.2 and C.3 of Appendix C.
Table C.1.
Number of goats traded between Member States in 2014 (modified from TRACES Data – Animal traded between Member States in 2014). Rows represent exports and columns imports
| Export/import | AT | BE | BU | CZ | DE | DK | EE | EL | ES | FR | HU | IE | IT | LT | LU | LV | NL | PL | PT | RO | SE | SI | SK | UK | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT | 25 | 2 | 892 | 131 | 370 | 581 | 36 | 4 | 2,041 | ||||||||||||||||
| BE | 53 | 5 | 58 | ||||||||||||||||||||||
| BU | 0 | ||||||||||||||||||||||||
| CZ | 3 | 12 | 1 | 24 | 64 | 104 | |||||||||||||||||||
| DE | 82 | 14 | 16 | 53 | 4 | 22 | 355 | 12 | 40 | 13 | 97 | 12 | 473 | 2 | 1,195 | ||||||||||
| DK | 0 | ||||||||||||||||||||||||
| EE | 0 | ||||||||||||||||||||||||
| EL | 0 | ||||||||||||||||||||||||
| ES | 559 | 1,053 | 53 | 1,414 | 560 | 3,639 | |||||||||||||||||||
| FR | 210 | 312 | 286 | 171 | 1,585 | 53 | 52 | 1,136 | 3,805 | ||||||||||||||||
| HU | 27 | 19 | 46 | ||||||||||||||||||||||
| IE | 1 | 1 | |||||||||||||||||||||||
| IT | 35 | 8 | 97 | 6 | 146 | ||||||||||||||||||||
| LT | 4 | 4 | |||||||||||||||||||||||
| LU | 2 | 2 | |||||||||||||||||||||||
| LV | 4 | 85 | 4 | 93 | |||||||||||||||||||||
| NL | 370 | 1,082 | 3 | 92 | 385 | 5 | 7 | 161 | 85 | 20 | 4 | 3 | 19 | 2,236 | |||||||||||
| PL | 12 | 12 | |||||||||||||||||||||||
| PT | 42 | 42 | |||||||||||||||||||||||
| RO | 88 | 15 | 103 | ||||||||||||||||||||||
| SE | 12 | 2 | 14 | ||||||||||||||||||||||
| SI | 1 | 1 | |||||||||||||||||||||||
| SK | 19 | 3 | 10 | 32 | |||||||||||||||||||||
| UK | 2 | 4 | 15 | 1 | 11 | 33 | |||||||||||||||||||
| Total | 487 | 1,098 | 235 | 353 | 1,283 | 385 | 9 | 571 | 266 | 165 | 244 | 15 | 3,524 | 109 | 231 | 38 | 6 | 153 | 1,482 | 2,780 | 0 | 36 | 115 | 22 | 13,607 |
Table C.2.
Number of goats traded between Member States in 2015 (modified from TRACES Data – Animal traded between Member States in 2015). Rows represent exports and columns imports
| Export/import | AT | BE | CZ | DE | EE | EL | ES | FR | HR | HU | IE | IT | LT | LU | LV | NL | PL | PT | RO | SE | SI | SK | UK | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT | 1 | 602 | 48 | 48 | 243 | 104 | 399 | 33 | 16 | 376 | 1,870 | |||||||||||||
| BE | 12 | 160 | 10 | 182 | ||||||||||||||||||||
| CZ | 22 | 24 | 30 | 76 | ||||||||||||||||||||
| DE | 22 | 1 | 113 | 1 | 1 | 138 | ||||||||||||||||||
| EE | ||||||||||||||||||||||||
| EL | ||||||||||||||||||||||||
| ES | 262 | 40 | 1,517 | 845 | 213 | 2,877 | ||||||||||||||||||
| FR | 349 | 6 | 15 | 27 | 1,674 | 3 | 72 | 597 | 2,743 | |||||||||||||||
| HR | ||||||||||||||||||||||||
| HU | 5 | 5 | 10 | |||||||||||||||||||||
| IE | 11 | 11 | ||||||||||||||||||||||
| IT | 60 | 2 | 136 | 1 | 199 | |||||||||||||||||||
| LT | 1 | 1 | ||||||||||||||||||||||
| LU | ||||||||||||||||||||||||
| LV | 3 | 18 | 21 | |||||||||||||||||||||
| NL | 2,692 | 5 | 30 | 200 | 360 | 105 | 1 | 3,393 | ||||||||||||||||
| PL | 2 | 2 | ||||||||||||||||||||||
| PT | 43 | 43 | ||||||||||||||||||||||
| RO | 227 | 700 | 400 | 1,327 | ||||||||||||||||||||
| SE | 6 | 6 | ||||||||||||||||||||||
| SI | ||||||||||||||||||||||||
| SK | 3 | 18 | 10 | 31 | ||||||||||||||||||||
| UK | 6 | 27 | 4 | 1 | 13 | 2 | 12 | 65 | ||||||||||||||||
| Total | 22 | 3,107 | 28 | 634 | 18 | 289 | 70 | 499 | 48 | 293 | 1 | 4,677 | 20 | 108 | 1 | 23 | 139 | 847 | 701 | 1 | 33 | 51 | 1,385 | 12,995 |
Table C.3.
Number of goats traded between Member States in 2016 (modified from TRACES Data – Animal traded between Member States in 2016). Rows represent exports and columns imports
| Export/import | AD | AT | BE | BG | CH | CY | CZ | DE | DK | EE | EL | ES | FI | FR | HR | HU | IE | IT | LT | LU | LV | NL | PL | PT | RO | SE | SI | SK | SM | UK | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | 44 | 44 | |||||||||||||||||||||||||||||
| AT | 42 | 128 | 6 | 215 | 82 | 451 | 6 | 257 | 55 | 2 | 1,244 | ||||||||||||||||||||
| BE | 11 | 10 | 4 | 7 | 32 | ||||||||||||||||||||||||||
| BG | 12 | 12 | |||||||||||||||||||||||||||||
| CH | 2 | 2 | |||||||||||||||||||||||||||||
| CY | |||||||||||||||||||||||||||||||
| CZ | 4 | 3 | 8 | 110 | 125 | ||||||||||||||||||||||||||
| DE | 17 | 12 | 2 | 58 | 73 | 73 | 1 | 236 | |||||||||||||||||||||||
| DK | 1 | 1 | |||||||||||||||||||||||||||||
| EE | |||||||||||||||||||||||||||||||
| EL | |||||||||||||||||||||||||||||||
| ES | 43 | 130 | 2,772 | 889 | 3,834 | ||||||||||||||||||||||||||
| FI | |||||||||||||||||||||||||||||||
| FR | 771 | 43 | 1,413 | 3 | 2,230 | ||||||||||||||||||||||||||
| HR | |||||||||||||||||||||||||||||||
| HU | |||||||||||||||||||||||||||||||
| IE | 5 | 5 | |||||||||||||||||||||||||||||
| IT | 31 | 3 | 284 | 7 | 70 | 395 | |||||||||||||||||||||||||
| LU | |||||||||||||||||||||||||||||||
| LT | 2 | 2 | |||||||||||||||||||||||||||||
| LV | 9 | 30 | 39 | ||||||||||||||||||||||||||||
| NL | 588 | 1,175 | 6 | 8 | 293 | 34 | 209 | 2,313 | |||||||||||||||||||||||
| PL | 3 | 3 | |||||||||||||||||||||||||||||
| PT | |||||||||||||||||||||||||||||||
| RO | 40 | 40 | |||||||||||||||||||||||||||||
| SE | 9 | 9 | |||||||||||||||||||||||||||||
| SI | 2 | 2 | |||||||||||||||||||||||||||||
| SK | 3 | 11 | 13 | 27 | |||||||||||||||||||||||||||
| SM | |||||||||||||||||||||||||||||||
| GB | 16 | 950 | 16 | 5 | 20 | 20 | 1,027 | ||||||||||||||||||||||||
| Total | 43 | 605 | 1,993 | 54 | 2 | 6 | 22 | 1,386 | 64 | 86 | 146 | 97 | 9 | 368 | 217 | 136 | 20 | 4,636 | 33 | 40 | 2 | 28 | 278 | 889 | 3 | 1 | 62 | 112 | 70 | 214 | 1,1622 |
AD: Andorra. GB: Great Britain. SM: San Marino. CH: Switzerland.
3.1.2. Genetic diversity
There is a large amount of genetic diversity in European goats. The EFABIS database lists 374 goat breeds in Europe, of which 273 are within EU MS. This list is, however, not completely accurate as not all breeds are listed. The list is compiled by requests to national coordinators of the European Regional Focal Point for animal genetic resources (ERFP),14 and the response differs in completeness. For example, in EL, apart from some transboundary breeds, only ‘local breeds’ are listed, without any further specification. On the other hand, some breeds are counted more than once. This occurs if transboundary breeds are reported by more than one country. For example, the Saanen breed is reported by 21 different countries. When breeds with the same or a very similar name reported by different countries are counted as one single breed, then there are 88 fewer breeds in Europe. However, some of these transboundary breeds differ considerably between countries and then may be considered as separate breeds. For example, the Dutch Toggenburger originated from bucks imported from Switzerland around 1900 that had been crossed with local landrace goats. Nowadays, they differ from the Toggenburger remaining in CH in that they have shorter hair, are less wild, and have a slightly different colour. Consequently, it can be concluded that several hundred different goat breeds are present in Europe.
As mentioned above, there are a number of transboundary breeds in Europe, the largest in number are listed in Table 1. The Saanen breed is the most common, and is the main dairy goat in Europe and in the world. The Alpine, Boer and Nubian are also high production breeds used across Europe. For these breeds there is an exchange of animals between EU countries. The pygmy goat, on the other hand, is a hobby breed often present in farms with a social function.
Table 1.
Goat breeds reported in more than 5 European countries (http://www.efabis.net)
| Breed | Number of countries reporting |
|---|---|
| Saanen | 26 |
| Alpine | 14 |
| Boer | 14 |
| Toggenburger | 14 |
| (Anglo‐) Nubian | 10 |
| Angora | 6 |
| Pygmy | 6 |
In almost every country, there are a number of local breeds. These breeds vary in population size from below 100 breeding animals to more than 50,000. The latter are nearly all Mediterranean breeds, the former including groups such as old landrace breeds.
The diversity within different goat breeds has been investigated by Canon et al. (2006). They showed that from the south‐east of Europe to the north‐west the diversity within breeds is reduced, while the breeds in the north‐west clearly diverge from the other breeds. This is mainly due to the migration from the centre of domestication close to south‐east Europe to the north‐west. Diversity in numerically smaller breeds will have been further reduced. However, these breeds generally harbour diversity not present in the mainstream breeds. There are over 40 non‐transboundary breeds listed in the EFABIS database with fewer than 100 breeding females, and more than 90 with fewer than 1,000 females. Extra care is needed to conserve the diversity in these breeds.
3.1.3. The situation of scrapie in goats in the EU
Classical scrapie in goats
Based on EU‐wide active surveillance data (2.06 million goats tested), CS in goats was detected in 10 out of 28 MS between 2002 and 2015 (Figure 3). BG, RO and the UK reported only cases of CS, whereas both CS and AS were detected in CY, EL, ES, FI, FR, IT and SI.
Figure 3.
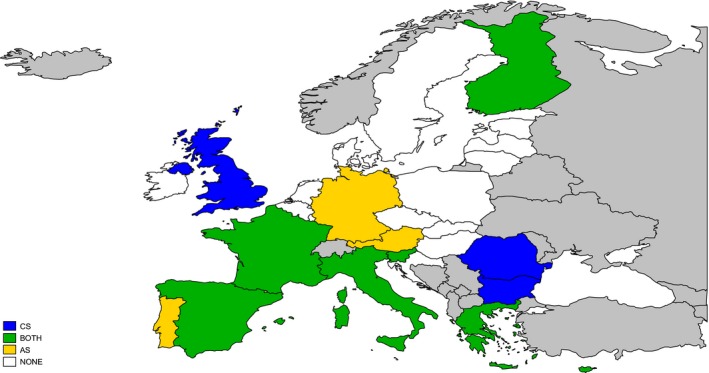
Geographical distribution of caprine classical scrapie (CS) and atypical scrapie (AS) within EU28 based on surveillance carried out between 2002 and 2015
- Green: MS that reported both CS and AS. Blue: MS that reported only CS. Yellow: MS that reported only AS. White: MS that have not reported caprine scrapie.
Since 2002, there have been 10,570 cases of CS detected (including both index cases and cases from infected herds) in these 10 MS (Figure 3). The average age at diagnosis was 50 months. Focusing on the active surveillance carried out in the MS where the disease has been detected (1.8 million goats tested), the total number of cases was 2,702, equal to an overall prevalence of 15 cases per 10,000 rapid tests. If the calculation of the prevalence is carried out after the exclusion of CY, the overall prevalence in the remaining nine countries is 2.4 cases per 10,000 rapid tests, based on 416 reported cases.
About 44% of the total number of cases (1,188) was from the SHC surveillance stream, whereas the remaining cases (1,514) were detected in NSHC animals. The overall stream‐specific prevalence in the ten MS was 13 and 17.1 cases per 10,000 rapid tests in the SHC and the NSHC, respectively. The exclusion of CY from the calculation of the stream‐specific prevalence leads to 1.2 and 3.6 cases per 10,000 rapid tests in the SHC and the NSHC, respectively.
The heterogeneous prevalence of CS in goats by MS over the period 2002–2015 is shown in Figure 4 through the stream‐adjusted prevalence by country.
Figure 4.
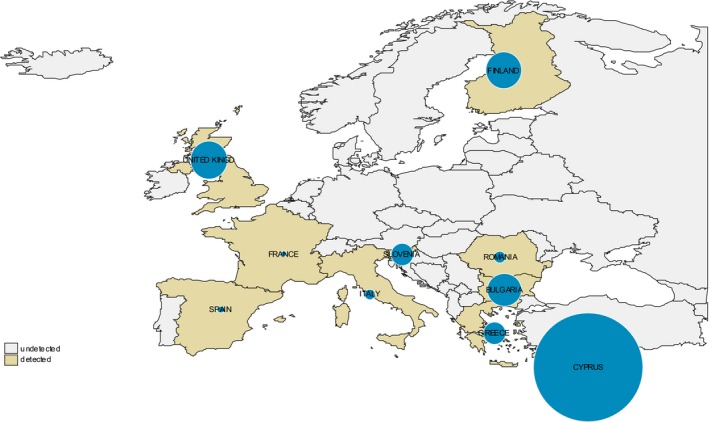
Stream‐adjusted prevalence of classical scrapie (CS) in goats within EU28 and based on active surveillance data
- Number of cases/10,000 rapid tests standardised by stream, i.e., SHC vs. NSHC during the period 2002–2015. The proportion of tests carried out in all the 28 MSs in the NSHC vs SHC in goats has been used to define the baseline population for the direct standardisation. The sizes of the blue dots are proportional to the prevalence.
Based on the result of the negative binomial models, FR (PR: 0.80; 95% CI: 0.74–0.87) and IT (PR: 0.89; 95% CI: 0.82–0.96) showed statistically decreasing trends. This was also the case for CY (PR: 0.79; 95% CI: 0.72–0.86) and the UK (PR: 0.85; 95% CI: 0.77–0.93), if the analyses were restricted to the period 2007–2015. In EL, no trend was observed, whereas in ES, although marginal, the increasing trend was statistically significant (PR: 1.09; 95% CI: 1.01–1.18) (Figure 5).
Figure 5.
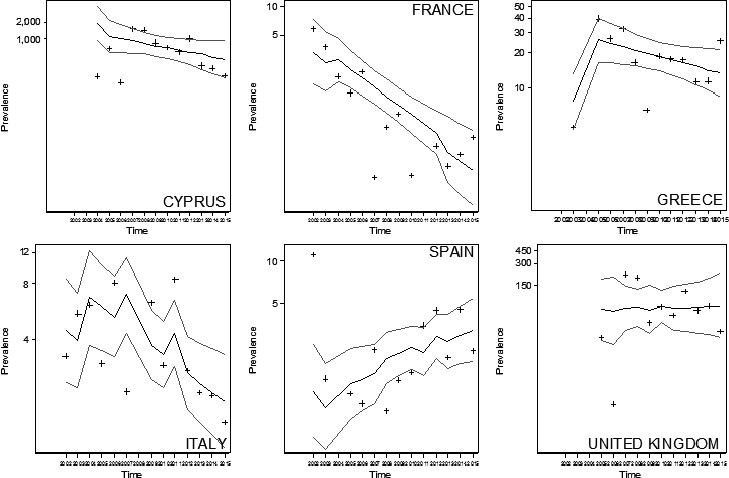
Temporal trend of CS in goats in EU MS where the disease was reported over at least 3 years during the period 2002–2015
- Crosses (+) indicate the annual stream‐adjusted prevalence (cases per 10,000 rapid tests). Lines show the linear trend (black line) and the bounds of the 95% CI (grey lines).
Atypical scrapie in goats
Over the period 2002–2015, a total of 138 cases of AS in goats were reported by 10 MS (see Figure 3). The average age at diagnosis was 82 months. In AT, DE, and PT, AS was the only caprine TSE detected whereas in CY, EL, ES, FI, FR, IT, and SI, AS appeared together with CS. With the exception of ES, FR, IT, and PT, the occurrence of cases was sporadic and, in particular, the only case reported by CY so far was detected while carrying out eradication measures in a CS outbreak.
As a consequence of the limited ability of some rapid tests to detect cases of AS in sheep in the past, this current statistical analysis was restricted to data obtained from the application of the few commercial rapid tests used mainly from 2006 onwards (the first nine cases of caprine AS were reported in 2005).
The overall prevalence over the considered period and based on active surveillance in the 10 MS (1.5 million tests) is 0.8 cases per 10,000 rapid tests. The stream‐specific PR were 0.56 and 1.1 cases per 10,000 rapid tests in the SHC and NSHC, respectively.
Figure 6 shows the temporal trends of AS in goats in the four MS for which the analysis was carried out (ES, FR, IT and PT). There was no significant trend in IT and PT, whereas both ES (PR: 1.15; 95% CI: 1.05–1.27) and FR (PR: 1.15; 95% CI: 1.04–1.26) showed a significant increase.
Figure 6.
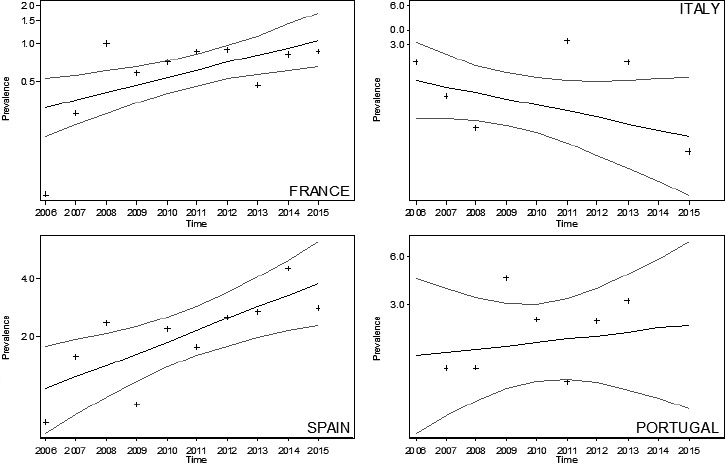
Temporal trends of AS in goats in ES, FR, IT and PT, during the period 2002–2015
- Crosses (+) indicate the annual stream‐adjusted prevalence (cases per 10,000 rapid tests). Lines show the linear trend (black line) and the bounds of the 95% CI (grey lines).
3.1.4. Epidemiology of caprine scrapie
CS in sheep and goats shows similar clinical signs, pathogenesis and pathology (Ulvund, 2006; Dustan et al., 2008; Konold et al., 2010; Acín et al., 2013). Although the involvement of both species in outbreaks is frequent (Toumazos and Alley, 1989; Agrimi et al., 1999; Billinis et al., 2002), the disease may affect goat herds where sheep are not present (Sofianidis et al., 2006; González et al., 2009).
For both sheep and goats, it is still unclear which sources of infectivity and routes of transmission are possible, and which have the greatest effect on the spread and maintenance of infection in a population. In sheep, infection with CS commonly occurs around birth mainly through oral exposure to the placenta that can accumulate large amounts of prions in ewes incubating the disease (Pattison et al., 1972; Hoinville et al., 2010). Despite the sparse accumulation of PrPSc in the placenta of goats as compared to that of sheep, placental shedding of the agent from infected animals and oral transmission have been also shown to occur goats (O'Rourke et al., 2011; Schneider et al., 2015). As in sheep, the presence of infectivity in goat milk has been recently confirmed (Konold et al., 2013). Therefore, it is likely that for both species a contaminated environment, e.g. by sharing pastures or spreading compost (Healy et al., 2004), may favour transmission (Detwiler and Baylis, 2003; Ryder et al., 2004; Seidel et al., 2007; Dexter et al., 2009), and that the disease may be introduced into populations by the movement of either ewes or does (Healy et al., 2004; McIntyre et al., 2008). In goats, in particular, it has been shown that mass restocking after the cull of a scrapie‐affected herd may play a role in re‐introducing the disease (Ortiz‐Pelaez et al., 2012). However, epidemiological studies specifically targeting the risk factors for the goat species have not been conducted to date.
3.1.5. Concluding remarks
In 2013, there were almost 10 million goats in the EU; with the majority of goats being reared for milk production. Between 2005 and 2013, the number of goats has increased by 10.3%. Some EU MS have more intensive systems, while other EU MS have smaller herds being reared using traditional methods on unfavourable land.
EU goat meat production is a by‐product of the dairy industry, accounting only for 0.1% of the total meat production in the EU28. In decreasing order, EL, ES, FR, RO, CY, the NL and IT are the biggest producers, representing 95.6% of the goat sector in the EU.
Data on the consumption of goat meat are scarce and there are no data for goat milk or other goat milk products. From the limited data provided by five MS (EL, ES, IT, LV and the NL), goat meat is not commonly eaten in those MS. Only in EL and ES the consumption of goat meat in certain age groups is greater than 0.2 g/day.
During 2014–2016, over 10,000 live goats were moved annually between EU MS for breeding. AT, ES, FR and the NL are the most active MS in terms of export, with BE, DE and IT the MS importing the largest numbers of live goats for breeding. This suggests that there is some mixing of breeding goats between MS.
There are several hundred different goat breeds in Europe. The most common breed is the Saanen, which is the main dairy goat in Europe and globally. However, in almost every MS there are local breeds, with 90 breeds having less than 1,000 breeding females. It is important that the diversity in these breeds is conserved.
Between 2002 and 2015, CS in goats was detected in 10 MS, mainly in those with the largest goat populations. In most of these MS, AS coexists with CS.
Apart from CY, where an epidemic has been ongoing over the last 10 years, the overall CS prevalence in the remaining nine MS has been low (2.4 cases per 10,000 rapid tests) with a total of a few hundred cases. A decreasing trend was identified in four MS, whereas no improvements were detectable in EL and ES.
AS cases in older goats have been detected in 10 MS; the prevalence is extremely low (0.8 cases per 10,000 rapid tests) and the trends, where they may be studied, are stable or increasing.
CS shares similar epidemiological features in sheep and goats, e.g. placental shedding of the agent from infected animals, presence of infectivity in goat milk, and susceptibility to oral transmission.
Environmental contamination, animal movements between herds and mass restocking after the cull of a CS‐infected herd are likely to play relevant roles as risk factors, but few studies are available.
3.2. Strain diversity in goat TSE
3.2.1. A comparison between scrapie in sheep and goats
The occurrence of CS in goats is generally less frequent than in sheep (Chelle, 1942; MacKay and Smith, 1961; EFSA BIOHAZ Panel, 2014). The term CS is used for both species because there are similarities in neurological signs, histopathological lesions (vacuoles) and PrPSc detection by immunohistochemistry, enzyme‐linked immunosorbent assay (ELISA) and western blotting. The PrP amino acid sequence between the two species is similar, but although several polymorphisms have been described at the amino acid level in both species, the pattern and frequencies of these amino acid substitutions are different between the two, and are also breed and region dependent (Hunter and Bossers, 2006; Vaccari et al., 2009; see also Section 3.5).
Scrapie can transmit efficiently from sheep to goats following intracerebral challenge (i.c.) (Chelle, 1942; Pattison and Millson, 1961) or from goats to sheep by oral inoculation (p.o.) with infected brain material, or from the feeding of milk from scrapie‐infected goats to lambs (Konold et al., 2016). Natural infection can also spread horizontally (Stamp, 1962; Hadlow et al., 1980). In infected mixed flocks of sheep and goats, it is common that the disease occurs in both species. Indeed, goats and sheep share similar scrapie types such as CS, atypical/Nor98 scrapie, BSE and CH1641 scrapie, the latter being a rarer form of CS that shows BSE‐like PrPSc features, but can be discriminated from it (Foster and Dickinson, 1988; Eloit et al., 2005; Jeffrey et al., 2006; Colussi et al., 2008; Jacobs et al., 2011a; Spiropoulos et al., 2011). CH1641 scrapie has, until recently, not been found in goat field cases, but susceptibility was shown by i.c. infection (Foster and Dickinson, 1988).
TSE (or prion diseases) in animals and small ruminants can present different phenotypes, dependent particularly on the agent strain, and the species and genotype of the host. However, differences in phenotype may be observed even between individuals of the same species and PrP genotype. To explain this, the existence of mixtures of strains in the same individual and the emergence of new strains (mutations) have both been hypothesised and, in some cases, demonstrated (Kimberlin and Walker, 1988; Bruce and Dickinson, 1987; Bessen and Marsh, 1992; Asante et al., 2002; Beringue et al., 2008; González et al., 2010, 2012; Okada et al., 2016; Le Dur et al., 2017; Thackray et al., 2011, 2012; Simmons et al., 2015). It is therefore important to understand the range of TSE strains: the best way to accomplish this is by strain typing by bioassay in inbred animal models such as conventional mouse, bank vole lines, or transgenic mice.
3.2.2. Strain properties of TSEs in sheep and goats
Forms of TSE can be discriminated either as ‘isolate types’ when investigated in the infected case or as ‘strain types’ after inoculation and sufficient subpassaging of case material into inbred animals (usually rodents).
A few goat isolates have been fully investigated for strain characterisation, but all originate in the UK. It is unknown whether: (a) similar strains to those found in sheep are present in goats in other parts of Europe; (b) new strains are emerging; (c) or PRNP genotype of the goat plays a role in the occurrence of different strains.
3.2.2.1. Early strain typing efforts in conventional mouse lines with sheep scrapie
Historically, studies in the UK defined three classes of scrapie strains, based on their stability during subpassage within experimental rodent models using inbred mouse lines with two different PRNP polymorphisms: prn‐pa mice (encoding 108L and 189T) such as in RIII and C57Bl mice, and prn‐pb (encoding 108F and 189V) such as VM mice (Bruce and Dickinson, 1987; Westaway et al., 1987). These three classes of scrapie strain are:
Class I: 7D, ME7, 22C. 7D and ME7 probably represent independent isolations of the same strain. From scrapie‐infected goats, a strain with rapid incubation times similar to those of 7D from sheep could also be isolated (Zlotnik and Rennie, 1963).
Class II: 22A, 22F.
Class III: 87A, 31A, 51C, 125A, 138A, 153A. All of these experimentally classified ‘strains’ were isolated from different sheep, but share similar incubation times and lesion profiles and are considered to represent the same field strain.
3.2.2.2. Strain typing of European goat scrapie isolates in rodent models
The TSE regulations require that every positive TSE case in small ruminants is subject to discriminatory testing, to determine if BSE can be ruled out. This structured testing can progress to rodent bioassay, and, to date, BSE has only been confirmed in two goat field cases, one in France (Eloit et al., 2005) and one in the UK (Jeffrey et al., 2006; Spiropoulos et al., 2011).
The goat TSE strain‐typing research presented here started in 2004, and is drawn from several consecutive EU projects: a concerted action within project Neuroprion, a specific targeted research project (STREP) within the GoatBSE consortium, and an Emerging and Major Infectious Diseases of Livestock (EMIDA) project: GOAT‐TSE‐FREE. These projects enabled, in total, thirteen laboratories from seven countries (the NL, FR, the UK, IT, EL, ES, DE) to collect goat brain samples (mainly brain stem) for the investigation of TSE strain types circulating in the goat population of EU MS. At the same time, it was necessary to discriminate BSE cases accurately against a background of this variety of TSE isolates to serve as a control measure for human health protection. Over 65 goat isolates were made available from CY, ES, FR, EL, IT, the NL and the UK. A selection of 20–30 of these isolates, including one atypical/Nor98 scrapie case, was studied in depth using strain typing in rodent models. The basis for this selection was genotype variation, geographical origin, and an immunochemical assay (ELISA) (Simon et al., 2008) that can discriminate between BSE and CS. From the limited number of isolates tested, it did not appear that the different strains isolated were related to the goat PRNP genotypes (Table 2).
Table 2.
Goat TSE isolates from seven MS (see Table 4) submitted to strain typing using rodent bioassays
| Genotype | Breed and country | MS | Herd or mixed (sheep/goats) | Number of isolates |
|---|---|---|---|---|
| Wild type (Wt) | Camosciata IT, Damascus CY | IT, CY | Herd | 4 |
| Wt/PP240 | Meticcia IT, dwarfgoat NL, crossbred ES, greek indigenous EL, Damascus CY, Saanen FR, Anglo‐Nubian UK | IT, NL, ES, EL, CY, FR, UK | Mixed (1 herd in FR and 1 herd in UK) | 8 |
| Wt‐SP240 | Meticcia IT, Alpine IT, Saanen FR, Alpine FR, Alpine ES | IT, FR, ES | Herd and mixed | 9 |
| HR143, SP240 | Meticcia IT, dwarfgoat NL | IT, NL | Mixed | 2 |
| HR143, SP240 | Greek indigenous EL | EL | Mixed | 1 |
| IM142, SP240 | Alpine FR | FR | Herd | 1 |
| GS127, SP240 | UK Great Britain (milking purpose Saanen/Toggenburg/Anglo‐Nubian | UK | Herd | 3 |
| RH154,SP240 | Meticcia IT (Nor98/atypical scrapie) | IT | Herd | 1 |
| RQ211,SP240 | Saanen IT | IT | Herd | 1 |
Laboratories involved in this work: ISS‐Rome, INIA‐Madrid, WBVR‐Lelystad, FLI‐Riems, and INRA‐Tours.
For comparison, an experimental caprine BSE and a caprine CH1641 scrapie case (experimentally generated goat CH164115 were included). The murine models were: RIII mice, bank voles with wild type PrP allele, and mice transgenic for bovine, ovine and goat PRNP variants (Table 3). The goat brain samples were distributed to the testing laboratories as brain homogenate pools prepared at INRA‐Nouzilly to ensure the same starting material in each research centre. To define strains, each laboratory with its different rodent model applied phenotypic parameters according to its expertise, but they all included incubation time and Western blot pattern (the molecular mass in kDa of the non‐glycosylated PrPSc band after PK digestion in Western blotting analysis).
Table 3.
Some of the rodent models used by the GoatBSE consortium
The overall results obtained with RIII mice and bank vole bioassays in relation to the geographical strain distribution are summarised in Table 4. The results obtained for the goat field isolates showed that none of the field cases were BSE‐like (each rodent line showed susceptibility for BSE with a differential phenotype).
Table 4.
Strains per country that were found in goat TSE isolates from 7 EU MS. More than 50% of the EU's goat population is present in these countries
| Member State | RIII | RIII and bank vole | Bank vole | |||
|---|---|---|---|---|---|---|
| Classical scrapie | Atypical scrapie/Nor98 | Classical scrapie | ||||
| Cyprus (CY) | 87A | ME7 | Uk85 | |||
| France (FR) | 87A | ME7 | Uk85 | It93 | ||
| Greece (EL) | 87A | ME7 | ? | |||
| Italy (IT) | No transmission | No transmission (n = 1) | It93 | |||
| Netherlands (NL) | 87A | ME7 | Uk85 | |||
| Spain (ES) | 87A | ME7 | Uk85 | CH1641‐17K | ||
| United Kingdom (UK) | 87A | ME7 | Uk85 | CH1641‐17K | ||
Number of isolates studied per country: Cyprus (n = 2), France (n = 7), Greece (n = 1), Italy (n = 4), Netherlands (n = 3), Spain (n = 2), UK (n = 3).
? = 100% attack ratio, strain type still under investigation.
The categorisation of the strains obtained in the different models did not present a consistent overlap between the rodent models, suggesting that different scrapie ‘sub‐strains’ or ‘components’ might be present in each isolate for which each model would select its fastest strain variant, and/or that there is a host genotype effect on phenotype. Thus, every rodent line behaved differently with respect to the CS isolates. In summary:
in bank voles, strains were classified as Uk85, It93, CH1641 and 17K. In sheep, similar strains can be found (Pirisinu et al., 2013; Thackray et al., 2012);
in RIII mice, strains were characterised as 87A and its mutant ME7. These two strains have also been found regularly in sheep;
the Italian isolates failed to transmit in RIII mice;
in Tg338 mice, CS isolates were classified as CS like, except one that was classified as AS like;
in Tg110 mice, CS isolates were classified as 19K or 21K.
The geographical distribution of strains suggests that, in general, the characterised CS strains in goats are present in all the MS contributing to the study, except in IT, where only one unique strain (It93 in bank voles) was identified. It93 was also present in FR.
3.2.2.3. Biochemical typing of PrPres in goat TSE isolates
Biochemical typing of PrPres in brain tissue by triplex western blotting was used to distinguish BSE, CS and CH1641 scrapie (Jacobs et al., 2011a).
Nearly all goat isolates (3 from CY, 4 from EL, 5 from FR, 7 from IT, 2 from the NL, 2 from ES, 2 from the UK), behaved as CS, with a single PrPres population clearly different from that of experimental BSE cases (Figure 7). Within that group, there appears to be a subset of isolates representing a TSE variant of CS in IT with a somewhat higher susceptibility for proteinase K, as characterised by the removal of the PrPres N‐terminus (12B2‐ or P4‐epitope) relative to most of the other caprine and ovine CS cases. Indications for more general occurrence of a class of TSE isolates with a slightly reduced PrPres N‐terminus have also previously been reported in goat isolates from EL (Fragkiadaki et al., 2011). One out of two UK goat isolates (from the same holding) behaved as a CH1641‐like scrapie case while the other appeared as a typical CS case.
Figure 7.
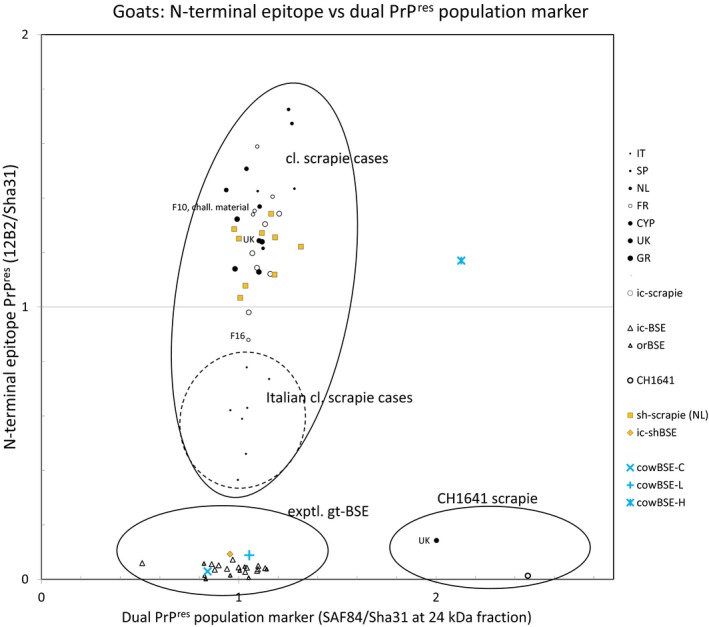
PrPres analysis of scrapie strains by triplex‐Western blotting
On the same blot, three antibodies were applied that have different epitopes on the PrP sequence: 12B2 for N‐terminus, Sha31 for the core sequence, and SAF84 somewhat more C terminal from Sha31 (Biacabe et al., 2007). By calculating ratios between epitope signals, specific properties of PrPR es appear that discriminate between the TSE types classical scrapie, BSE and CH1641. Each dot represents a separate goat TSE isolate. The IT scrapie cases exhibit a reduced N terminus epitope presence intermediate between BSE/CH1641 and the remaining classical scrapie samples (vertical axis). CH1641 can be distinguished from BSE due to the presence of a dual PrPR es triple band population (horizontal axis), only one goat isolate in the geographical study occurred clearly in the CH1641 area where also an experimental goat CH1641 isolate was located. Sheep scrapie cases from the NL (orange filled squares), experimental caprine CH1641, experimental caprine BSE, experimental caprine scrapie (Lacroux et al., 2014), and bovine classical and atypical BSE samples were used as controls.
Source: data from the GoatBSE consortium study. Graph prepared by Jan Langeveld.
3.2.3. Concluding remarks
Based on studies of a limited number of isolates from a few MS, data indicate that no goat strains that differ from the known sheep strains have been found to date. However, aberrant strains could have escaped detection, especially where negative results have been reported.
Bioassays in both wild‐type mice and bank voles, as well as in transgenic mice with bovine, ovine, or caprine PRNP, may not be able to show the full strain status of an isolate since each rodent line – even if transgenic for a certain PrP codon – will select a strain for which it is the most sensitive. That scrapie of goats (and sheep) may carry a mixture of strains cannot be excluded.
The majority of the strains that have been identified in goats were present in all the contributing MS (CY, EL, ES, FR, the NL and the UK), except in IT, where a single variant of CS was identified (It93). This variant was also present in FR.
Because of the limited number of isolates investigated, it is currently not possible to determine whether the available data on TSE agent diversity comprehensively represents the agent diversity in any of the contributing MS. No data are available on strains circulating in MS other than those contributing to the described study. Similarly, there is limited knowledge on strains circulating in sheep. Nonetheless, the set of common isolate and strain types found from analysing multiple goat isolates in different MS and their similarities to those found in sheep could be indirect evidence for the existence of a goat PRNP polymorphism(s) resistant against multiple strains.
3.3. Criteria/tools for assessing genetic resistance
3.3.1. Epidemiological studies
Following the discovery of the lower susceptibility conferred to individual sheep by certain polymorphisms of the PRNP gene (Foster and Dickinson, 1988; Hunter et al., 1993), epidemiological approaches aimed at characterising genetic resistance at population level have been widely applied. For instance, a population‐based case–control study design was used to compare the allelic variants in scrapie cases obtained from outbreaks and healthy controls from scrapie‐free flocks (Belt et al., 1995). Cross‐sectional studies were also implemented in individual sheep flocks affected with natural scrapie, exploiting the implementation of compulsory eradication measures; at one point in time culled animals were tested for TSE and submitted to genotyping. In this case, the prevalence of scrapie among animals of different genotypes and other host characteristics were compared in order to identify PRNP polymorphisms that are associated with higher/lower prevalence of disease occurrence, e.g. Vaccari et al. (2001). Finally, cohort studies were implemented by following up sheep of different genotypes in infected flocks and comparing survival times until disease, either in natural conditions (Bossers et al., 1996; Hunter et al., 1996; Baylis et al., 2002) or maintained in research facilities (Elsen et al., 1999). Reliance on the clinical status of the animals was a limitation of some of the early studies.
Because of the particularities of TSE pathogenesis (i.e. long incubation period, late neuroinvasion, need for post‐mortem disease confirmation), clinical disease occurrence underestimates the infectious status of the investigated populations. Consequently, rather than truly measuring the susceptibility/resistance to TSE infection that is associated with a particular PRNP allele or genotype, the use of clinical disease as the outcome of interest only provides a measurement of the association of genotype and incidence of clinical disease within the context of an infected flock (reduced life expectancy of individuals plus the presence of variable infectious pressure).
Cross‐sectional studies may not be the most appropriate study design since outbreaks may show very low within‐flock prevalence at the time of culling. Moreover, the low preclinical sensitivity of post‐mortem rapid testing may lead to the misclassification of infected animals. Another downside of this approach is the limited statistical power to detect significant associations of rare (low frequency) genotypes.
The inclusion of a single flock/herd or a limited number of flocks/herds from the same geographical area also has limitations. The TSE agents that are responsible for these particular outbreaks are generally not submitted to strain typing. It is therefore uncertain whether the TSE agent(s) in these flocks/herds are representative of the TSE agents in the general population.
With progress in the understanding of TSE pathogenesis and the ability to detect disease preclinically (abnormal PrP detection in the lymphoid organs and in the central nervous system (CNS)) and the acknowledgement of the presence of multiple strains of the scrapie agent in small ruminants, the design of recent epidemiological studies has evolved, providing more robustness by trying to address these caveats. For example, later studies did not rely only on clinical cases (reducing the potential for misclassification of the outcome), and analysed large data sets of sheep genotypes and all cases detected by surveillance (Baylis et al., 2004; Ortiz‐Pelaez and Bianchini, 2011) or from multiple flocks subject to stamping out measures (Vaccari et al., 2009), with the aim of estimating the relative risk of disease between genotypes, allowing for a sufficient statistical power.
Meta‐analyses that combine the quantitative outcomes of a number of studies (i.e., relative risks (RR) or odds ratios (OR 16 )) using a systematic approach, have not been performed to date, in either sheep or goats: they could offer a valuable opportunity to produce overall estimates of the effect of different genotypes on the presence of disease in populations. Differences in the study design, sample size, ascertainment bias, etc., should be considered by giving different weights to the effect estimator of each study.
In the case of goats, epidemiological studies have benefited from earlier studies of genetic resistance to scrapie in sheep, which showed significant associations between alleles and the presence of disease. Crude comparisons of genotype frequencies between a pool of positive cases and negative goats selected from surveillance or research activities and from a number of herds have been conducted in several countries (Fragkiadaki et al., 2011; Papasavva‐Stylianou et al., 2011; Acín et al., 2013). This study design had the caveats of: (a) assuming equal exposure of cases and healthy controls to scrapie; and (b) considering the true status of the (non‐tested) healthy controls as scrapie negative, with both assumptions prone to bias due to misclassification of the outcome. In some cases, it was acknowledged that the control population was only clinically negative (Goldmann et al., 2011).
One way to overcome these caveats is to cull entire infected herds and investigate genotypes and scrapie status of all animals (cross‐sectional design) or of part of them (case‐control design), either within the scope of compulsory eradication measures (Acutis et al., 2006; Vaccari et al., 2006; Barillet et al., 2009) or for research purposes (González et al., 2009; Corbière et al., 2013 Ortiz‐Pelaez et al., 2014), with a variable geographical representativeness of the scrapie strains in their source populations. In other cases, with a cohort study design, animals selected according to their genotype were left until the end of their productive life and then tested to ascertain their scrapie status (Georgiadou et al., 2017). In these studies in goats, as was the case with similar studies in sheep, where specific alleles may never be associated with disease, or cases are found only at a very low rate (e.g. ARR), it was not possible to calculate a measure of relative risk, i.e., how ‘more likely’ is the disease in an individual carrying an allele or a genotype other than the most resistant ones. However, in Georgiadou et al. (2017), a meaningful measure of absolute risk (i.e. risk difference) was provided.
3.3.2. Experimental challenge in natural host
The experimental inoculation of natural scrapie sourced from, and inoculated into, animals carrying particular PRNP genotypes is a straightforward approach to test the effect of the genetic polymorphism on the pathogenesis and/or on the susceptibility to a particular TSE (Foster et al., 1993, 1996).
The most efficient experimental transmission route is i.c. The i.c. challenge of animals harbouring different genotypes is classically used to obtain an estimate of the ability/efficiency of TSE agents to propagate in a given PrP substrate. Lack of transmission in a particular PrP variant compared to a 100% attack rate in wild‐type PrP qualifies for a low susceptibility/strong resistance to infection. Longer incubation periods of the disease in inoculated animals without a reduction of the attack rate are also compatible with an interpretation of a lower susceptibility/higher resistance to the infection. Several lines of evidence have indicated that the i.c. route allows the propagation of certain TSE agents in animals bearing genotypes that are associated with strong resistance to the disease under natural exposure conditions or following experimental challenge by the oral route. For instance in sheep, while the ARR/ARR genotype is associated with a very high resistance in animals orally challenged with CS or cattle BSE, i.c. inoculation results in clinical disease (Houston et al., 2003, 2015).
Inoculation p.o. is considered the best proxy for natural contamination with TSE agents in ruminants. It provides an estimate of the level of resistance of particular PrP alleles when challenged with a TSE agent. It encompasses not only the capacity of the agents to replicate in a particular PrP substrate, as assessed in i.c. experiments, but also the ability of the agent to infect and disseminate efficiently in hosts harbouring a particular PRNP genotype (EFSA BIOHAZ Panel, 2015). Nevertheless, oral experimental exposure cannot be considered to perfectly reproduce the natural disease (Tabouret et al., 2010).
Results obtained in sheep of a fully susceptible genotype after p.o. challenge with classical BSE (C‐type BSE) demonstrated a strong influence of the age of the recipient at inoculation on the efficacy of disease transmission (Hunter et al., 2012). A significant reduction in disease transmission was observed in animals challenged after weaning. However, successful transmission to animals several months old at challenge (Jeffrey et al., 2001; Bellworthy et al., 2005; González et al., 2014; Fast et al., in press), and in naïve adults exposed to contaminated environments (Dexter et al., 2009) shows that older animals are still susceptible to disease. These results are fully consistent with those reported for naturally scrapie affected flocks, where disease is more rarely observed in animals introduced as adults, compared to animals introduced at a younger age (Hourrigan et al., 1979).
Experimental challenge of a natural host therefore provides highly significant information concerning the relative susceptibility/resistance of a specific PRNP polymorphism to a TSE agent, but the approach remains limited by its cost and the duration of the experiment.
CS in small ruminants is caused by a variety of diverse TSE agents. Considering the time and necessary resources, the testing of several TSE agents in a natural host model is difficult to achieve. This reduces the confidence that can be put in a candidate PRNP polymorphism that would be identified in experimental challenges using a single TSE isolate.
3.3.3. Bioassays in PRNP transgenic mice
Another approach to estimate the impact of PRNP polymorphisms on the resistance/susceptibility to infection with TSE agents is the use of transgenic rodents expressing these alleles (on an endogenous PRNP knock‐out background).
The potential relevance of such a model in this application was first described in the context of variant Creutzfeldt–Jakob disease (vCJD) infection (EFSA BIOHAZ Panel, 2015). Indeed, all available vCJD epidemiological data indicate that the MM129 PrP genotype is associated with higher susceptibility to cattle BSE than the MV129 and VV129 ones. This characteristic is also observed in all the human PRNP transgenic mice, expressing these PrP variants (Bishop et al., 2006; Mead et al., 2007; Wadsworth et al., 2010; Cassard et al., 2014; Asante et al., 2015). More recently, a mouse model that expresses the human GV127 PrP variant was also shown to reproduce the protective effect associated with this allele in a Kuru‐exposed population (Asante et al., 2015).
Over the past two decades, a number of mouse lines that express different PrP variants from various animal species (cervids, sheep, etc.) have been generated (Groschup and Buschmann, 2008). Several of these mouse lines were specifically created for assessing the relative resistance/susceptibility to TSE infection associated with specific PRNP polymorphisms (Meade‐White et al., 2007; Green et al., 2008).
The use of transgenic mouse models, rather than natural host species, for bioassay offers obvious advantages in terms of cost, duration of the studies, and scope for the simultaneous testing of several TSE agents. Although in a number of instances such models produced results that were consistent with those obtained in natural hosts, in other instances they led to findings that were significantly different. For example, based on surveillance data, the ARQ allele in sheep appears to be associated with susceptibility to AS, and the VRQ allele with resistance (Moreno et al., 2007; Fediaevsky et al., 2008). However, transgenic mice expressing the sheep VRQ allele were reported to display shorter incubation periods than mice expressing the ARQ allele following challenge with AS (Le Dur et al., 2005; Arsac et al., 2009; Griffiths et al., 2010). Another example, from another species, is white‐tailed deer in which the GS96 polymorphism has been identified in epidemiological studies to be associated with potentially lower susceptibility to chronic wasting disease (CWD) (Johnson et al., 2011). Experiments in transgenic mice expressing either the glycine (G) or the serine (S) at residue 96 indicated that G96 mice were highly susceptible to infection and that S96 mice showed no evidence of disease or accumulation of PrPres during their lifespan (Meade‐White et al., 2007). However, p.o. inoculations of deer have demonstrated that S96 deer were infected, but had a longer incubation period than G96 deer before developing clinical disease (Johnson et al., 2011).
Altogether these data indicate that PRNP transgenic mouse models can provide indications concerning the impact of the expression of different PrP variants on the propagation of TSE agents. However, the data accumulated so far remain too limited to consider that such system on its own can robustly and consistently replicate the capacity of a prion to naturally propagate in a host.
3.3.4. In vitro conversion assay
According to the prion theory, the key event in the pathogenesis of TSE is the conversion of the normal cellular prion protein (PrPC, which is encoded by the PRNP gene) into an abnormal disease‐associated isoform (PrPSc). On the basis of that concept, it is generally admitted that the higher/lower resistance to prion disease associated to certain PRNP polymorphism is due (at least partly) to the differences that exist in terms of PrPC to PrPSc convertibility (thermodynamics of reactions) between the different PrP variants.
Cell‐free conversion assays are based on the in vitro conversion of recombinant PrP (substrate) following the introduction of a PrPSc seed. The cell‐free approach confirmed that different sheep PrP display a variable PrPc to PrPSc conversion efficacy. In this species, differences observed between polymorphisms, separately or in combination with cell‐free conversion assay, have a qualitative concordance with the relative level of resistance predicted by epidemiological studies (Bossers et al., 1997, 2000).
The PrPC conversion yield in cell‐free systems is usually low, making quantitative experiments difficult. Since the early 2000s, an alternative methodology that allows a highly efficient in vitro propagation of prions and does not need a partial denaturation step of the seed material has become available.
The Protein Misfolding Cyclic Amplification (PMCA) technology is thought to mimic prion replication in vitro but in an accelerated form. It is facilitated by combining a PrPC‐containing substrate with a PrPSc seed (Saborio et al., 2001; Soto et al., 2002). The newly generated PrPSc produced in homologous conversion assays (same PrPC and PrPSc amino acid sequence) shares the same biochemical, biological and structural properties as the parental isolate (Castilla et al., 2008), although exceptions have been reported (Thorne et al., 2012). As observed in vivo, the strain and source of PrPSc, in particular the PRNP genotype of seed and substrate, have a clear influence on the amplification pattern obtained by PMCA (Bucalossi et al., 2011; Taema et al., 2012; Thorne et al., 2012; Priem et al., 2014).
Results from several studies support the view that the efficiency of the in vitro PrPSc amplification (using a particular TSE agent as a seed) using PMCA in a specific substrate (PrPC harbouring particular polymorphisms) is correlated with the relative capacity of this particular TSE agent to propagate in vivo in hosts expressing the same PRNP polymorphisms. In one of these studies, an ovine CS isolate was used as a seed. Genotypes associated with susceptibility (ARQ/ARQ, ARQ/AHQ and AHQ/ARH) were able to sustain PrPSc amplification in PMCA, whereas genotypes associated with resistance to scrapie (ARQ/ARR and ARR/ARR) were unable to support the in vitro conversion (Bucalossi et al., 2011).
Altogether these results indicate that in vitro seeded PrP conversion assays (and in particular PMCA) can provide important indications of the relative sensitivity/resistance to infection by a specific TSE agent of a host‐bearing specific PRNP polymorphism. However, the data accumulated so far remain too limited to consider that such system on its own can robustly and consistently replicate the capacity of a prion to naturally propagate in a host.
3.3.5. Tool for the evaluation of the genetic resistance
A tool to evaluate the genetic resistance to CS of alleles in the caprine PRNP gene has been developed, based on two main components: the weight of evidence available and the strength of resistance conferred by the alleles of interest.
3.3.5.1. Weight of evidence
The assessment of the amount and type (in terms of study design) of the evidence is based on a scale made of combination of different types of studies using Boolean terms, from the one that provides the least evidence, the in vitro conversion studies, to the combination of all possible studies as described in the above Sections 3.3.1–3.3.4. The studies should have made, at the very least, a comparison between the wild type allele and the allele of interest. In increasing order of weight of evidence, the following categories have been considered:
In vitro conversion studies
Bioassays in allele‐specific transgenic mice
Bioassays in allele‐specific transgenic mice AND in vitro conversion studies
Epidemiological studies OR experimental challenges in natural host (i.c. OR p.o.)
Epidemiological studies AND experimental challenges in natural host (i.c.)
Epidemiological studies AND experimental challenges in natural host (p.o.)
Epidemiological studies AND experimental challenges in natural host (i.c. OR p.o.) AND in vitro conversion studies
Epidemiological studies AND experimental challenges in natural host (i.c. OR p.o.) AND bioassays in allele‐specific transgenic mice AND in vitro conversion studies
Epidemiological studies AND experimental challenges in natural host (i.c. AND p.o.) AND bioassays in allele‐specific transgenic mice AND in vitro conversion studies
Epidemiological studies AND experimental challenge in natural host using multiple isolates17 (i.c. AND p.o.) AND bioassays in allele specific transgenic mice using multiple isolates AND in vitro conversion studies.
3.3.5.2. Strength of resistance
The assessment of the level of resistance conferred by an allele will be defined by scoring in a traffic light colour system one of the three levels of resistance, using the wild type of the goat PRNP gene as a baseline. The traffic light system uses the presence of field cases holding the alleles of interest, the attack rates and the incubation period in the natural host or in transgenic mice as indicators of the level of resistance. A fourth score ‘grey’ is also given for situations where sufficient studies are not available. In increasing order of resistance, the following levels have been defined as:
Grey: Evidence of resistance but only one single study available.
Red: NO RESISTANCE from studies (indicated by efficient conversion in in vitro studies, experimental transmission with a high attack rate and incubation periods the same as, or shorter than, that of challenges in wild‐type recipients), OR no significant difference in the occurrence of cases holding the allele compared with that in the wild type.
Amber: PARTIAL RESISTANCE from studies (indicated by inefficient/absent amplification in in vitro studies, experimental transmission possible, but with significantly longer incubation periods and/or a lower attack rate than that of challenges in wild‐type recipients OR inconsistent/conflicting results from different studies) AND field cases can occur.
Green: RESISTANCE from studies (indicated by lack of conversion/amplification in in vitro studies, no transmission following experimental oral challenge, and no transmission or transmission with significantly prolonged incubation periods and/or reduced attack rate than that of challenges in wild‐type recipients) AND (rare field cases OR no field cases).
Figure 8 shows a diagram of the tool showing the scale used for the assessment of the weight of evidence.
Figure 8.
Scale of the weight of evidence on genetic resistance of the caprine and ovine PRNP genes
3.3.6. Concluding remarks
The combination of epidemiological studies and experimental challenges in natural host (p.o. and/or i.c.) is pivotal for the assessment of genetic resistance.
In vitro conversion studies and bioassays in allele‐specific transgenic mice for the alleles of interest are insufficient on their own to draw robust conclusions. When combined with the other approaches (epidemiological studies and experimental challenges in natural host and/or in PRNP transgenic mice), the weight of evidence is increased.
Different epidemiological study designs (i.e. cross‐sectional studies, case‐control studies, cohort studies) have been applied to investigate the association between certain genotypes or alleles and TSE.
Epidemiological studies can suffer from various caveats such as case definitions based on clinical status, low preclinical sensitivity of diagnostic tools, coexistence of a number of different strains in the small ruminant populations, selection of unsuitable control groups (e.g. because never exposed to the agent) and small sample size.
Some of these issues were successfully addressed in more recent studies, leading to results less subject to biases and with higher precision.
At least two scrapie isolates (from geographically different origins) should be investigated experimentally to address the issue of a robust level of scientific assurance.
A tool to evaluate the genetic resistance to CS of alleles in the caprine PRNP gene has been developed, based on the weight of evidence available and the strength of resistance conferred.
3.4. Genetic resistance to TSEs in goats
3.4.1. Selected caprine PRNP gene alleles and evidence for association with classical scrapie
At least 47 amino acid polymorphisms have been reported worldwide for the goat PRNP gene so far. Only a small number of them have occurred at adequate frequencies in epidemiological studies or have been investigated by experimental studies to enable conclusions to be drawn regarding their possible association with resistance to scrapie and/or BSE.
In the scrapie‐related literature, PRNP gene haplotypes are mostly referred to as alleles, particularly in sheep where they are defined by amino acids encoded in codons 136, 154 and 171, e.g. ARR. Although this type of nomenclature is less commonly used for the caprine PRNP gene, references can be found to the wild‐type allele as IRRQS (codons 142, 154, 211, 222, 240) or similar (Corbière et al., 2013). The term wild‐type allele is used for the most common allele in a large population or in a breed or for a species; for goats, there are two equally common wild‐type PRNP alleles that differ only by polymorphism SP240 (Goldmann et al., 1996).
Most of the known amino acid polymorphisms in the caprine PRNP gene appear as single changes when compared with the wild‐type sequence, but in various combinations with amino acids S240 and P240. For example, the amino acid combinations H154‐S240 and H154‐P240 or combinations M142‐S240 and M142‐P240 have all been found in different breeds and with widely differing frequencies, while Q211‐S240 or K222‐S240 have not yet been found in the P240 combination. Reports of rare, novel allelic combinations of other polymorphisms (Srithayakumar et al., 2016) need confirmation in further studies.
The SP240 amino acid polymorphism is different from the others as its location within the protein sequence means that it will be removed after post‐translational modifications in the cell to generate the mature protein. Because it is the mature, membrane‐integrated PrP protein that undergoes the pathogenic changes during disease, the SP240 polymorphism cannot directly influence prion conversion processes. For that reason, in this report the presentation of alleles (Tables 5 and 6) does not include any codon 240 information.
Table 5.
Summary of evidence on caprine PRNP gene alleles associated with TSE susceptibility differences
| PRNP gene polymorphisms | Type of evidence for resistance association | |||||
|---|---|---|---|---|---|---|
| Codon | Amino acid polymorphisms | Amino acid associated with resistance | Epidemiological studies | Experimental inoculation in goats (i.c. or p.o.) | In vitro conversion CC* − CFC (PMCA‐QuIC)** and Mouse bioassay | Other*** |
| 127 | G‐S | S | (i.c.)
|
|||
| 142 | I‐M | M |
|
(i.c.) | CFC
|
|
| 143 | H‐R | R | CFC
|
|||
| 145 | G‐D | D |
|
(p.o.)
|
||
| 146 | N‐S | S | (i.c.)
|
CFC
|
||
| 146 | N‐D | D | (i.c.)
|
CC
CFC
|
||
| 154 | R‐H | H |
|
(i.c.) | CFC
|
|
| 211 | R‐Q | Q |
|
(i.c.) |
CFC | |
| 222 | Q‐K | K |
|
(i.c.)
|
CC |
|
| 32 | G‐stop | – |
|
|
||
* CC: cell‐based conversion assay.
** CFC: cell‐free conversion assay. Protein Misfolding Cyclic Amplification (PMCA). Quaking induced conversion (QuIC).
*** Other: Evidence from other prion studies that did not use goats.
Table 6.
Summary of the evidence for/against TSE genetic resistance in the caprine PRNP gene alleles associated with TSE susceptibility differences
| Evidence for or against resistance association of haplotypes | ||||
|---|---|---|---|---|
| Allele | Evidence | Epidemiology | Experimental inoculation in goats (i.c. or p.o.) | In vitro conversion CC* – CFC (PMCA‐ QuIC)** and Mouse bioassay |
| S127 | For |
|
|
|
| Against |
|
|
||
| M142 | For |
CFC
|
||
| Against |
|
|
||
| R143 | For |
|
|
|
| Against |
|
CFC
|
||
| D145 | For |
|
|
|
| Against |
|
|
|
|
| D146/S146 | For |
|
|
CFC
CC
|
| Against |
|
|
||
| H154 | For |
|
CC
CFC
|
|
| Against |
|
|||
| Q211 | For |
|
|
CFC
CC
|
| Against |
|
|
||
| K222 | For |
|
CFC
Bioassay Transgenic mice expressing K222 are resistant to a panel of goat scrapie isolates and to cattle BSE (Aguilar‐Calvo et al., 2014) |
|
| Against |
|
|
|
|
| 32 (G‐stop) | For |
|
|
|
| Against |
|
|
|
|
* CC: cell‐based conversion assay.
** CFC: cell‐free conversion assay. Protein Misfolding Cyclic Amplification (PMCA). Quaking induced conversion (QuIC).
Led by the results obtained in field studies, research over the past two decades has focussed on those polymorphisms that looked very promising as ‘resistant alleles’. Most alleles were not subject to these tests because they were rarely found (with allele frequencies of < 1%) in surveys and epidemiological studies, or sometimes only seen in single animals.
Consequently, for nine alleles a considerable data set has been produced covering all the criteria set out in the previous section as necessary to assess accurately the level of resistance to various TSE agents/isolates. We therefore submit amino acid substitutions in codons 127, 142, 143, 145, 146, 154, 211 and 222 as candidates that confer some degree of resistance to classical scrapie. Table 5 shows the nine caprine PRNP gene alleles associated with different susceptibility to TSE, the allele definition and studies (references) providing evidence for association with resistance. Table 6 shows a summary of the allele information in support of an association with TSE resistance, based on the studies reviewed. Table 6 also shows the information indicating allele association with TSE susceptibility similar to the wild type.
A different class of haplotype, in which a nonsense polymorphism (G32stop) results in a failure to translate full length PrP protein (PrP‐null allele), has been found in Norwegian white goats (Benestad et al., 2012). There is so far no evidence from genotyping surveys of goats in seven EU countries (section 3.6) that this nonsense mutation occurs in any other breed. Experimental data regarding scrapie challenges of goats carrying this haplotype have not yet been published. By extrapolation of the survival evidence of genetically modified mouse lines, which do not produce any PrP protein (PrP knock‐out) (Bueler et al., 1994; Manson et al., 1994), it is reasonable to assume that goats homozygous for the PrP‐null‐allele will be resistant to scrapie and BSE challenge. It is, however, not yet clear whether homozygous PrP‐null goats remain healthy throughout their lives and have no other disease phenotypes that are unrelated to TSEs. This haplotype has not been included in the tool since only animals holding haplotypes that translate functional PrP protein have been considered for this purpose. Moreover since the objective of this assessment is to inform genetic resistance to CS in goats and not genetic editing, such a haplotype should not be promoted in any breeding for resistance programme other than in the natural breed where it has been found.
Due to the frequencies of the alleles of interest in goats, field data mostly produce significant disease association only for the more common heterozygous genotypes and rarely for the scarce homozygous genotypes. Similar restrictions to the assessment of resistance association apply to low frequency genotypes representing combinations of alleles with different amino acid substitutions, e.g. genotype HR154‐QK222. According to Hardy–Weinberg equilibrium calculations, homozygotes for one rare allele (< 5%) will have a frequency of less than 0.3%, while heterozygotes for two rare alleles will have a frequency of less than 0.5%. The published surveys with full genotype information confirm the expected low frequencies: all homozygotes (excluding wild‐type alleles) were 0.48% and all double heterozygotes (no wild‐type alleles) were 0.43% (based on a total of almost 2600 genotypes) (Billinis et al., 2002; Acutis et al., 2006; Vaccari et al., 2006; Papasavva‐Stylianou et al., 2007; Bouzalas et al., 2010; Fragkiadaki et al., 2011; Goldmann et al., 2011, 2016; Papasavva‐Stylianou et al., 2011; Maestrale et al., 2015; Acín et al., 2013 Corbière et al., 2013; Barillet et al., 2009). Despite the overall problem with assigning resistance to rare homozygotes due to their observed frequencies, a protective phenotype has been described for MM142 homozygotes similar to IM142 heterozygotes (Table 5) (González et al., 2009; Goldmann et al., 2011) and also for SS146 and DD146 homozygotes compared to NS146 and ND146 heterozygotes (Georgiadou et al., 2017). There is no evidence that genotypes with combined polymorphisms significantly differ from the single polymorphism genotypes with regard to their disease resistance, but the possibility of an additive effect particularly on the incubation period lengths for some combinations cannot be excluded.
The nine alleles are partially dominant over the wild‐type allele regarding association with scrapie resistance, based on experimental challenges. They lead at least to lengthened incubation periods and in some cases to survival within the observation period. Experimental i.c. or p.o. challenges with various scrapie isolates have been performed in six heterozygous genotypes: GS127 (i.c.) (Dassanayake et al., 2015), IM142 (i.c., p.o.) (Goldmann et al., 1996; Lacroux et al., 2014), NS146 (p.o.) (White et al., 2012), NS146, ND146 (i.c., p.o.) (Cyprus, 2015),18 HR154 (i.c., p.o.) (Lacroux et al., 2014; Maestrale et al., 2015), QR211 (i.c., p.o.) (Lacroux et al., 2014; Maestrale et al., 2015); and QK222 (i.c., p.o.) (Acutis et al., 2012; White et al., 2012; Lacroux et al., 2014; Maestrale et al., 2015) and in five homozygous genotypes: DD145 (p.o.) (Maestrale et al., 2015), QQ211 (i.c.) and KK222 (i.c.) (both by Lacroux et al., 2014) and SS146 and DD146 (Cyprus, 2015) (Table 6). The i.c. inoculations were 100% successful in all challenged genotypes except in 10 QK222 and five KK222 goats, with only 20% being susceptible. I.c. challenge of NS146, ND146, SS146 and DD146 animals were successful, but with a prolonged incubation period in nearly all animals. Oral challenge was 100% successful in IM142, HH154 and QQ211 goats. Lower susceptibility was reported for RH154 and RQ211, while complete resistance within the observation periods was seen for NS146 (0/25) ND146 (0/25), SS146 (0/26), and DD146 (0/2) goats, DD145 (0/2) and QK222 (0/16) genotypes.
The dominance of the variant alleles over both wild‐type alleles would enable breeding programmes with early benefits to the reduction of disease prevalence by simply increasing the number of heterozygotes. In other words, the long‐term aim of eradicating CS cases from herds or populations by replacing all wild‐type allele carriers with variant allele carriers need not be achieved immediately.
The following section summarises and expands on the data shown in Tables 5 and 6. After describing the weight of evidence and strength of resistance for the allele D145, the other alleles have been described in order of their strength of association with CS resistance, from weakest to strongest, after the application of the two‐tier tool (Figure 9). Studies on resistance to AS and BSE are discussed later in Section 3.4.2. Note that the G‐stop has not been included in this description for the reasons explained above.
Figure 9.
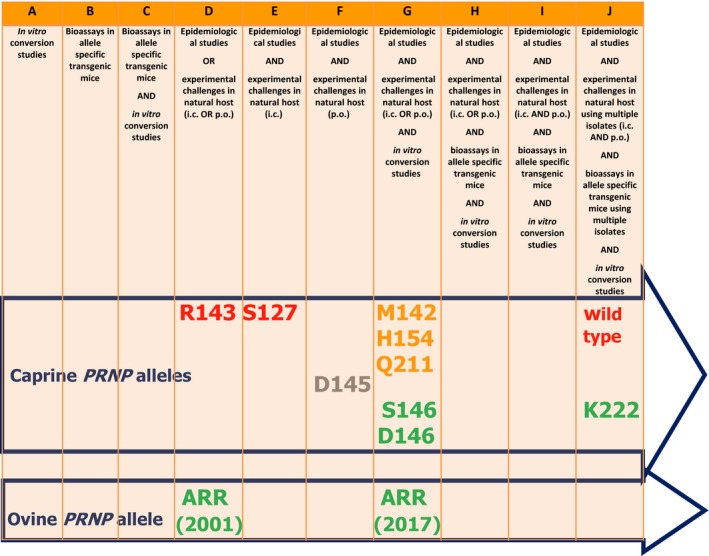
Scale of the weight of evidence on genetic resistance of the caprine PRNP gene and strength of resistance of selected alleles according to the two‐tier tool
Definitions of the two tiers are given in Sections 3.3.5.1 and 3.3.5.2. For comparison, the evidence of resistance with the ARR allele in sheep between 2001 and 2017 (see Section 3.4.3) has been added. Note: the G‐STOP has not been included in the tool.
Allele D145
This allele has recently been identified as being present in resistant goats in one Italian study involving only one goat breed combining experimental challenge and epidemiological investigation (Maestrale et al., 2015). The data, even if promising, are not supported by any other study, thus any conclusion about the level of resistance is premature. Given the current state of knowledge of disease association, this allele is of academic interest and may have only local breeding impact.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (p.o.). Column: F.
Strength of resistance: No field cases reported so far and high resistance shown in a combined field and experimental study. Nevertheless, all the data for this allele originates from a single study. Colour: GREY.
Allele R143
A moderate protective effect of this allele has been suggested from two epidemiological studies (Billinis et al., 2002; Vaccari et al., 2006), but has not been substantiated by any other study. As a consequence, the scientific knowledge is inadequate to endorse this allele as resistance associated.
Weight of evidence: Epidemiological studies OR experimental challenge in natural host (i.c. OR p.o.). Column: D.
Strength of resistance: Normal occurrence of scrapie cases holding this allele. Susceptibility is similar to the wild type. Inconsistent results in the few published studies. Field cases were reported and there is no evidence of resistance compared to wild type genotype. Colour: RED.
Allele S127
This allele appeared to give some protection against CS, both in an epidemiological study and in an experimental challenge (Goldmann et al., 2011; Dassanayake et al., 2015). The S127 delays the occurrence of clinical disease but not the development of pathological deposition of prion protein at the endpoint of disease. After experimental challenge with scrapie, S127 carriers had longer incubation periods (defined as the time between inoculation and the development of observable clinical signs). There are no in vitro conversion studies or bioassay data to refine these observations. It can be concluded that allele S127 does not promote significant resistance to CS, but that presentation of clinical disease signs is delayed in comparison to that in wild‐type animals.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (i.c.). Column: E.
Strength of resistance: Normal occurrence of scrapie cases with this allele. The susceptibility is similar to the wild type. This allele has a moderate effect but not significant increasing the incubation period and the clinical onset of the disease, but not conferring resistance compared to the wild type genotype. Colour: RED.
Allele Q211
Epidemiological studies carried out in FR, EL and Canada indicate an association with partial resistance, but positive cases have been regularly found. In contrast, Italian studies suggest no association with resistance. The divergent results suggest that the association may be scrapie strain dependent, but this has not been further investigated. In vitro studies exhibit reduced prion convertibility with two different scrapie strains. Experimental challenge with CS resulted in extended incubation periods after i.c. and p.o. inoculation.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (i.c. OR p.o.) AND in vitro conversion studies. Column: G.
Strength of resistance: It can be concluded that the Q211 allele is associated with partial resistance and extended incubation periods, but that this association is inconsistent and possibly TSE strain dependent. Field cases occur, but a protective effect compared to wild type emerged from several field studies. Also, experimental challenge and in vitro studies showed some resistance compared to wild type. Colour: AMBER.
Allele H154
This allele has been extensively investigated and appears to confer incomplete protection as implied by the occurrence of scrapie cases in both R154 and H154 goats (Billinis et al., 2002; Acutis et al., 2006; Vaccari et al., 2006; Papasavva‐Stylianou et al., 2007, 2011; Barillet et al., 2009; Fragkiadaki et al., 2011; Corbière et al., 2013). Heterozygotes show prolongation of the incubation period.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (i.c. OR p.o.) AND in vitro conversion studies. Column: G.
Strength of resistance: Field cases are present, but a protective effect compared to wild type emerged from several field studies. Experimental challenge and in vitro studies also showed some resistance compared to wild type. Colour: AMBER.
Allele M142
This allele has been intensely investigated by different means, including field data, experimental challenges, and in vitro studies for scrapie strains. Although cases of CS have been confirmed in both genotypes IM142 and MM142, the fact that some resistance is conferred by these genotypes is shown by a significant OR compared to wild type in field studies (Goldmann et al., 2011; Corbière et al., 2013) and a low in vitro conversion rate (Eiden et al., 2011). M142 carriers also have significantly extended incubation periods after intracerebral TSE challenges (Goldmann et al., 1996; Lacroux et al., 2014). It can be concluded that allele group 2 (M142) is associated with incomplete resistance to CS.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (i.c. OR p.o.) AND in vitro conversion studies. Column: G.
Strength of resistance: A protective effect, compared to wild type, was evident in the UK studies, with field cases presenting a later age of the onset of the disease, and with an increase in incubation period shown in experimental studies. Such results are not consistent when compared with field and experimental studies outside the UK. Colour: AMBER.
Alleles S146/D146
Results are available for several epidemiological studies, experimental challenges, and in vitro conversion studies for both alleles (Papasavva‐Stylianou et al., 2007, 2011; Eiden et al., 2011; White et al., 2012; Ortiz‐Pelaez et al., 2014; Niedermeyer et al., 2016; Srithayakumar et al., 2016; Georgiadou et al., 2017). In experimental challenges, strong resistance of the non‐NN146 genotypes was apparent following p.o. challenge (no positive animals after about 2,000 days post‐infection (p.i.)), while a moderate resistance was found in i.c. challenge; a significant increase of the incubation period was found in NS146, SS146 and DD146 animals, but with a 100% attack rate. No difference was identified between NN146 and ND146 goats in the i.c. challenge (Cyprus, 2015). These results indicate a strong resistance to CS, but not as strong as that of allele K222. The data have been mainly produced in Cyprus, with some data from the United States. Due to the absence or very low frequencies of alleles D146 and S146 in the rest of Europe, no data from field studies regarding resistance to other European scrapie strains are available. Moreover, no bioassays have been performed because no models transgenic for this polymorphism have been constructed. The Cyprus report (2015) supports a strong protection after p.o. scrapie challenge by both polymorphisms for DD146, SS146, ND146 and NS146 genotypes. I.c. challenge yielded no absolute resistance with any of the codon 146 allele combinations, but did result in significantly prolonged incubation periods for the DD146, SS146 and NS146 genotypes.
Weight of evidence: Epidemiological studies AND experimental challenge in natural host (i.c. OR p.o.) AND in vitro conversion studies. Column: G.
Strength of resistance: A small number of field cases were found both in NS146 and ND146 genotypes (Papasavva‐Stylianou et al., 2011) but high resistance appears from field studies. P.o. challenge showed strong resistance as well while significant prolongation of the incubation period (relative to wt controls) was observed in i.c. challenged animals of the NS146, SS146 and DD146 genotypes. Colour: GREEN.
Allele K222
There is a substantial amount of experimental and field data available for this allele supporting a strong association with resistance to CS. Most epidemiological data concur that this allele is strongly associated with high scrapie resistance in heterozygous genotypes in the five EU member states in which studies have been undertaken (Acutis et al., 2006; Vaccari et al., 2006; Barillet et al., 2009; Bouzalas et al., 2010; Fragkiadaki et al., 2011; Corbière et al., 2013; Maestrale et al., 2015). However, scrapie positive QK222 goats have been found in some of these field studies showing that this allele does not confer complete resistance (Barillet et al., 2009; Fragkiadaki et al., 2011). There are insufficient data for the homozygous KK222 genotype from these field studies to appraise the association with disease resistance, mainly due to the low frequencies of this genotype linked to the low average allele frequency.
The association with resistance is supported by experimental data. First, experimental challenges involving different scrapie isolates showed complete resistance to p.o. scrapie challenge of QK222 (White et al., 2012; Lacroux et al., 2014; Maestrale et al., 2015). A significantly reduced susceptibility of QK222 and KK222 animals to i.c. scrapie challenge was also observed; three animals died after very long incubation periods of approximately 5.5 years (Lacroux et al., 2014). Secondly, in vitro studies demonstrated a failure of prion conversion with the K222 PrP protein using one common scrapie strain (Eiden et al., 2011), although successful conversion of K222 PrP protein with another scrapie strain using a different in vitro assay may indicate strain sensitivity in this process (Kanata et al., 2016). Bioassays using transgenic mice expressing only K222 (equivalent to homozygous goat KK222 genotype) were resistant to challenge with six different scrapie isolates, and partial resistance was demonstrated by mice expressing K222 combined with wild type Q222 (approximating the caprine heterozygous genotype QK222) which succumbed to two out of four scrapie isolates. These heterozygous QK222 mice were more resistant than mice expressing only wild type Q222 (Aguilar‐Calvo et al., 2014).
Weight of evidence: Epidemiological studies AND experimental challenge in natural host using multiple isolates (i.c. AND p.o.) AND bioassays in allele specific transgenic mice using multiple isolates AND in vitro conversion studies. Column: J.
Strength of resistance: Some field cases are reported but many field studies showed a strong protective effect of the K222 allele compared to wild type. Extensive experimental challenge studies carried out by different groups in different countries showed a complete resistance to p.o. challenge and strong, although incomplete, resistance to i.c. challenge. These levels of resistance were supported by bioassays and in vitro studies. Colour: GREEN.
There are insufficient data to produce a ranking of genotypes. Therefore only a ranking of alleles has been produced. Based on both the ‘weight of evidence’ and ‘strength of resistance’, the alleles with assigned strength of resistance either red, amber or green, can be ranked as follows, from high to weak CS resistance: K222 > D146 = S146 > Q211 = H154 = M142 > S127 = H143 > wild type.19
3.4.2. Genetic resistance to atypical scrapie and BSE
Genetic association with atypical scrapie
For most of the PRNP alleles or genotypes, their associations with AS cannot be assessed due to the lack of in vivo or in vitro evidence. Nonetheless, the association with AS is strongest in HR154 and HH154 genotypes, with a high proportion of AS cases showing these genotypes in field studies from FR and IT (Arsac, 2007; Colussi et al., 2008). While results for experimental challenges in HH154 goats are not yet available, i.c. challenges of sheep with HH154 genotypes confirmed susceptibility of this genotype (Simmons et al., 2011). Data related to the K222 allele show that when mice transgenic for this allele were challenged with AS the incubation period did not alter compared to transgenic mice with caprine wild type Q222 allele (Aguilar‐Calvo et al., 2016). Moreover, one goat carrying H154 and K222 was found naturally infected by AS in Italy (Colussi et al., 2008), and another carrying the H154 and the M142 allele (Seuberlich et al., 2007).
Genetic association with BSE
The relationship between goat PRNP genetics and BSE resistance is also poorly understood, but three alleles, namely M142, R211 and K222, have been studied in goat and mouse bioassays (Goldmann et al., 1996; Konold et al., 2010; Aguilar‐Calvo et al., 2014, 2015; Fast et al., in press). The i.c. inoculation of BSE into two IM142 heterozygotes and the subcutaneous (s.c.) inoculation of one MM142 homozygote resulted in positive cases between 30 and 40 months with a 60–70% longer incubation period than the wild type. Because of the small number of challenged animals, it was not possible to calculate an average attack rate for BSE in this genotype (Goldmann et al., 1996). In a later study on codon 142 with 5 goats – two IM142 and three MM142 – all succumbed to i.c. challenge with bovine BSE between 20 and 30 months (Konold et al., 2010). A recent pathogenesis and genetic study with oral challenges in WtWt, WtM142, MM142, WtQ211 and WtK222 carriers illustrated the relatively high resistance to bovine and goat BSE infection in M142 and K222 carriers (Aguilar‐Calvo et al., 2015; Fast et al., in press).
Goat and sheep‐passaged BSE were both able to infect transgenic K222 mice (only expressing this allele), whereas these mice were resistant to cattle‐derived BSE. Transgenic mice expressing both caprine wild‐type and K222‐PrP (a condition mimicking heterozygosis) were susceptible not only to goat‐passaged BSE but also to cattle BSE, indicating that for BSE the K222 amino acid substitution does not exhibit a negative dominant effect (Aguilar‐Calvo et al., 2014).
3.4.3. Comparison of the level of knowledge for ARR in 2001 and 2017
Both the Regulation (EC) No 260/2003, which revised the requirements for eradication measures in the case of the detection of TSEs, and the Commission Decision 2003/100/EC, which laid down minimum requirements for the establishment of breeding programmes for resistance to TSE in sheep, were based on an Opinion by the former SSC on the safe sourcing of small ruminant materials (SSC, 2002). The Opinion highlighted ARR/ARR as the most resistant sheep PrP genotype to natural and experimental scrapie, which had already been concluded in a previous opinion published in 1999 on the policy of breeding and genotyping of sheep (SSC, 1999).
The review of these two opinions provides a picture of the knowledge and uncertainties related to the selection of the ARR allele in 2003, when the decision to implement the selection of this allele in the EU sheep populations was taken.
3.4.3.1. The level of resistance provided by the ARR allele towards scrapie in sheep
The 2002 SSC opinion stated that:
‘Susceptibility to TSEs depends on the genotype of the animal and also in sheep the incidence of TSEs is linked to PrP genotype, with codons 136, 154 and 171 being of major importance. Most scrapie‐affected sheep are homozygous for glutamine (Q) at codon 171 and succumb to disease with ARQ/ARQ, VRQ/VRQ, VRQ/ARQ genotypes. However, it should be noted that, for a given genotypic configuration, different breeds may show different susceptibility levels to a TSE. An additional factor for susceptibility genotypic variation may be strain dependent’.
‘Occasionally scrapie occurs in ARQ/ARR and VRQ/ARR sheep. However, for a given level of exposure to a source of infection, the likelihood of becoming scrapie‐infected is lower than with sheep that are homozygous for glutamine (Q) at codon 171. Also, during the preclinical phase, in heterozygote ARR/VRQ or ARR/ARQ ovines, PrPSc does not seem to be easily detectable in lymph tissue or in the digestive autonomous nervous system’. These observations have been reinforced by more recent studies (Baylis et al., 2004; Corbière et al., 2007; Jacobs et al., 2011b; Langeveld et al., 2016).
‘Sheep of ARR/ARR genotype are considered to be the most resistant to scrapie. ARR/ARR animals so far have not shown to carry detectable infectivity or PrPSc, with the exception of one PrPSc positive case out of the genotyped scrapie cases. Therefore although this cannot be 100% excluded, the likelihood of this genotype to become infected with scrapie seems to be very small’. These observations have been reinforced by more recent studies (Baylis et al., 2004; Corbière et al., 2007; Groschup et al., 2007).
The SSC based its opinion on multicentric epidemiological studies that were mainly carried out in naturally affected flocks in Europe. There are a number of areas in which there were knowledge gaps at the time of inclusion of genetic resistance into the EU legislation.
Given the information on the ARR genotype at the time of the 2002 SSC opinion was produced (2001), the application of the tool to the ARR allele in sheep is as follows:
Weight of evidence: Epidemiological studies OR experimental challenge in natural host (i.c. OR p.o.). Column: D.
Strength of resistance: Data from numerous epidemiological studies showed that the ARR allele is strongly associated with resistance to scrapie in sheep (Clouscard et al., 1995; Hunter, 1997). ARQ/ARR and VRQ/ARR sheep are also resistant to scrapie but there are occasional cases. Colour: GREEN.
The current weight of evidence in 2017 for the ARR allele includes the availability of experimental challenge in natural host (i.c. OR p.o.) (O'Rourke et al., 1997; González et al., 2014; Lacroux et al., in press) showed that the ARR/ARR sheep cannot be considered to be fully resistant to CS. However, CS has very limited capacity to transmit and adapt to ARR/ARR sheep, compared to the wild type ARQ/ARQ. Thus, the weight of evidence for ARR in 2017 is column G.
For comparison purposes, the weight of evidence and strength of resistance for ARR in sheep in 2001 and 2017 have been added to the tool (Figure 9).
3.4.3.2. The level of resistance provided by the ARR allele towards the BSE agent
The 2002 SSC opinion stated that: ‘As for BSE in sheep, research data are available for only a few tens of animals, results to date indicate that the relation between sheep genotype and susceptibility to a TSE is similar for scrapie and BSE: the ARR genotypes are apparently resistant to development of clinical disease on challenge with BSE and animals carrying the glutamine (Q) allele at codon 171 are potentially susceptible to BSE and to scrapie. The influence of the genotype at codon 136 and 154 is not yet known for BSE but is being tested by direct challenge studies at IAH, UK’.
This statement was based on a limited number of experimental challenges (oral and intracerebral) (Foster et al., 2001a,b; Jeffrey et al., 2001). At the time, the SSC opinions were produced, these studies had not been published or were about to be published.
3.4.3.3. The level of resistance provided by the ARR allele towards atypical scrapie
Nor98 AS was only identified in 1998 in Norway, and not fully characterised until later, so neither the 1999 nor the 2002 SSC Opinions considered AS. The question of the resistance/susceptibility to AS associated with the ARR allele was first examined by EFSA in the opinion ‘on the Breeding programme for TSE resistance in sheep’ (EFSA, 2006).
In that opinion, the BIOHAZ Panel acknowledged that ‘There are atypical cases/infections in sheep of the ARR/ARR genotype’, but concluded that ‘given the limited data available so far, certain difference between countries and the large number of statistical tests undertaken it is too early to draw final conclusions about the effect of these alleles (ARR, ALRQ, ARH and VRQ) on atypical scrapie’.
3.4.3.4. The potential healthy carrier status of TSE agents by ARR animals
The 2002 SSC opinion stated that ‘the available information is not enough to provide an answer to the question of what is the risk that flocks of scrapie resistant sheep would carry the scrapie agent without showing clinical signs and at the same time being able to transmit the agent horizontally, vertically or via rendering. This hypothesis can therefore not be excluded. Such a situation, if shown to exist in sheep (and as yet there is no field proof of this), could lead to the maintenance of a low level of infection in a flock without any clinical signs’.
3.4.3.5. The adverse effect on health and production traits that might be associated with selection of ARR allele in sheep
Neither the 1999 nor the 2002 SSC opinions dealt with this question. The 2002 SSC opinion stated that ‘with respect to the occurrence of possible adverse effects, an effective monitoring of breed characteristics in scrapie resistant genotypes [should be carried out] to obtain reliable information on any undesirable changes (e.g. in birth weight, growth rates, strength and resistance to particular other diseases)’.
The decision to implement breeding for resistance in sheep was taken by the European Commission and did not take into consideration this issue. This topic was part of the mandate for reviewing the benefits/adverse effects of this policy that was sent to the EFSA in 2005 by the Commission. In 2006, EFSA, in its opinion ‘on the Breeding programme for TSE resistance in sheep’, concluded that ‘there was no evidence that the ARR allele had a negative impact on production traits and susceptibility to other diseases’. At that time, ‘the few reported effects of PrP seem inconsistent between studies or highly questionable due to limited sample size’ (EFSA, 2006). At the time that the EFSA opinion was produced, only one study was considered (Bossers et al., 1996).
3.4.4. Concluding remarks
At least 47 amino acid polymorphisms have been reported worldwide for the goat PRNP gene so far. Research over the past two decades has focussed on nine polymorphisms that looked promising as ‘resistant alleles’.
The K222 allele proved to be resistant to several European CS isolates, sourced from seven MS, and in several European breeds. Evidence from natural, experimental and in vitro studies supports this conclusion.
The alleles S146 and D146 are clearly associated with resistance to CS and genotypes DD146 or SS146 have so far not been reported in scrapie cases.
The alleles S146 and D146 have largely been investigated in CY, thus their resistance can be considered as proven mainly for scrapie strains circulating in CY, which include strains also found in isolates from elsewhere in Europe. Results from a small experimental study of S146 in the USA support the Cypriot data.
The presence of scrapie cases in goats holding putatively resistant alleles (K222 and D146 or S146) mirrors the situation with the ovine ARR allele, for which ARR/VRQ, ARR/ARQ and ARR/ARR CS cases have also been reported in European populations. The latter two are extremely rare, while the former carry the highly scrapie susceptible allele VRQ.
Based on the limited available data in goats and data obtained in sheep, the H154 allele is likely to be associated with higher risk of developing AS.
None of the other listed alleles (S127, M142, R143 and Q211) can be considered highly resistant to goat CS due to either some evidence of susceptibility, or the lack of sufficient scientific data (D145).
There is currently insufficient data to assess the impact of K222, D146 and S146 alleles on the susceptibility to AS and BSE in goats. Experimental oral challenge with BSE (from either cattle or goats origin) indicated a strong but incomplete resistance in K222 allele carriers.
In 2003, the EU legislation laid down a new strategy against ovine CS based on ARR as the most resistant allele available in sheep. At the time, this conclusion was based on evidence from epidemiological studies, but there was not enough evidence to exclude that ARR carriers may act as healthy carriers of TSE agents or that the ARR allele might have a negative impact on production traits and susceptibility to other diseases.
3.5. Distribution and frequency of caprine PRNP gene alleles
CY is the only country to undertake routine genotyping of the entire national goat population. Published surveys of allele frequencies have been carried out in CY, EL, ES, FR, IT, the NL and the UK, involving both transboundary and autochthonous breeds. Some additional, although not yet published, data are available from surveys carried out by the consortium of the European project GoatBSE (Appendix D). There was considerable variation in the process of selection of the tested animals as well as in the numbers of animals per breed and the number of different herds (farm holdings) sampled among these countries. Differences in the classification of animals as purebred, which was in general left to the farmer to decide, may also exist.
Below is a brief discussion of the data referring to six alleles from the nine considered in Section 3.4. Frequencies are presented as mean ± standard deviation (SD). Alleles S127 and R143 were excluded because of their low potential for providing a protective effect, based on the results from the two‐tier tool. Allele D145 is not presented in this breed analysis as it appears to be rare and has only been detected in a few Sardinian and Moroccan goats.
3.5.1. Transboundary breeds
Alpine
Animals belonging to this breed were genotyped in herds from FR, IT, ES and the UK. Allele Q211 was the most common with a mean frequency of 10 ± 2%. Alleles D146 and S146 were absent. Alleles M142 and K222 had frequencies of 6.7 ± 3.4% and 4.3 ± 3.1%, respectively. The large SD values reflect the considerable frequency differences among the Member States. While the K222 frequencies for French and Spanish herds was low (~ 7%), it was very low in British and Italian herds (1% and 2%, respectively) (Acutis et al., 2008; Barillet et al., 2009; Goldmann et al., 2011, 2016; Acín et al., 2013).
Saanen
The breed was studied in FR, IT, ES, the NL and the UK. The M142 allele was the most commonly represented allele of interest with a mean frequency of 14.7 ± 9%. As for the Alpine breed, the D146 and S146 alleles were absent, whereas the frequency of allele Q211 was similar (8.2 ± 7%). The mean frequency of allele K222 was less than half that of the Alpine (2 ± 1.9%) with a wide frequency range (0.3–4.9%) (Acutis et al., 2008; Barillet et al., 2009; Goldmann et al., 2011, 2016; Acín et al., 2013; Windig et al., 2016).
Toggenburg
Data are only available for the UK and the NL. There was a very marked difference in allele frequencies between the two MS for M142, 0% and 32%, respectively, and K222, (36% and 0.2%) for the NL and the UK, respectively. These significant differences were probably caused by founder effects borne by the selected animals, and suggest that other herds need to be genotyped for more reliable frequency estimates. Alleles D146 and S146 were absent from this breed in these studies (Goldmann et al., 2011, 2016; Windig et al., 2016).
Boer
Data are only available for the UK and the NL. Both surveys produced very high and almost identical frequencies (mean 31%) for allele D146, while allele K222 was absent. The other alleles were relatively low (0–7.6%) (Goldmann et al., 2011, 2016; Windig et al., 2016).
Nubian/Anglo‐Nubian
Data are only available for the UK and the NL. As for the Boer goats, resistance‐associated allele frequencies are generally low (range 0–7.1%) with the exception of M142 (12.9%) in the UK goats. Allele D146 had a frequency of 7.1% in Dutch Nubians but was absent in British Anglo‐Nubians. British Anglo‐Nubians showed a very low presence of allele K222 (0.4%) (Goldmann et al., 2011, 2016; Windig et al., 2016).
Cashmere and Angora
Goats from the small UK population (Goldmann et al., 2011, 2016): were genotyped. Alleles M142, H154 and Q211 were present at frequencies ranging from 0% to 5%, alleles S146, D146 and K222 were absent.
Pygmy goat
A very small number of UK goats (Goldmann et al., 2011) were genotyped. Of the nine alleles only M142 was found, with a 19.5% frequency.
3.5.2. Autochthonous breeds
CY breeds
There are no published scientific data on surveys for allele frequencies in the Cyprus goat population. However, the reply from Cyprus to the additional questions asked by the Working group indicated that the national goat population has been genotyped in full and, at present, genotypes containing alleles D146 and/or S146 constitute 53.32% of the Cypriot goat population (Appendix E).
EL breeds
In this population, all the polymorphisms were present but at low frequencies. Alleles D146, S146 and K222 had frequencies of 1.5%, 0.5% and 5.6%, respectively (Kanata et al., 2014).
ES breeds
Of the three breeds studied (Moncaina, Pirenaica and Retinta), alleles D146 and S146 were absent and allele K222 was found in Moncaina at 0.3%, but was absent from the other two. High frequencies were found for alleles M142, H154 and Q211 with means of 24.2%, 10.2% and 16%, respectively (Acín et al., 2013). In the GoatBSE consortium, the Murciano‐granadina breed was also studied and showed a high frequency of M142 (60%) and a moderate frequency of allele Q211 (18%). Alleles D146, S146 and K222 were absent in this breed.
FR breeds
In the GoatBSE project, the Corsica breed and Poitevine breed were investigated. Allele K222 had a frequency of 9.4% in the Corsica breed. The Poitevine breed has high frequencies of M142 (19.7%) and Q211 (31.3%). Allele K222 has a frequency of 3.4%.
IT breeds
The analysis for two Northern and six Southern breeds revealed different frequencies in the two parts of the country. In general, frequencies of allele K222 were high in the South (12.4 ± 5.5%) and low in the North (2.8 ± 2.1%), while the opposite is true for alleles M142 and Q211. Alleles D146 and S146 were absent in all breeds (Acutis et al., 2008; Vitale et al., 2016).
NL breeds
Two breeds were analysed, of which the Dutch Landrace had no relevant polymorphisms (alleles S127 to K222). In the Dutch Pied Original breed, alleles D146 and S146 were absent and allele K222 was present at a low frequency (5.3%) (Windig et al., 2016).
UK breeds
Small numbers of the rare Golden Guernsey breed were genotyped and they showed unusually high frequencies for alleles Q211 (54.3%) and K222 (16.7%); all other alleles were absent. The high frequency of these two alleles is most likely due to a founder effect in this small population of 3,000–4,000 animals in total (Goldmann et al., 2011, 2016).
Details of the results of various surveys across the EU showing frequencies of the alleles of interest by country and breed have been included in Appendix D. Summary figures of such surveys are shown in Table 7, Figures 10 and 11.
Table 7.
Number and frequency of goats with alleles of interest included in surveys across several EU MS
| MS | WT | Alleles of interest | Total number of isolates with a susceptible allele of interest | Total number of isolates successfully sequenced | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M142 | S146 | D146 | H154 | Q211 | K222 | ||||||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | |
| CY | 380 | 88.8 | 1 | 0.2 | 19 | 4.4 | 11 | 2.6 | 17 | 4.0 | 0 | 0.0 | 0 | 0.0 | 428 | 94.7 | 452 |
| EL | 333 | 79.9 | 7 | 1.7 | 14 | 3.4 | 2 | 0.5 | 8 | 1.9 | 26 | 6.2 | 27 | 6.5 | 417 | 88.5 | 471 |
| ES | 655 | 59.7 | 216 | 19.7 | 0 | 0.0 | 0 | 0.0 | 60 | 5.5 | 154 | 14.0 | 13 | 1.2 | 1,098 | 83.1 | 1,321 |
| FR | 414 | 69.8 | 46 | 7.8 | 0 | 0.0 | 0 | 0.0 | 13 | 2.2 | 83 | 14.0 | 37 | 6.2 | 593 | 73.2 | 810 |
| IT | 422 | 71.5 | 41 | 6.9 | 0 | 0.0 | 0 | 0.0 | 43 | 7.3 | 37 | 6.3 | 47 | 8.0 | 590 | 92.5 | 638 |
| NL | 1,115 | 77.1 | 232 | 16.0 | 1 | 0.1 | 0 | 0.0 | 6 | 0.4 | 49 | 3.4 | 43 | 3.0 | 1,446 | 94.3 | 1,534 |
| UK | 739 | 68.5 | 270 | 25.0 | 41 | 3.8 | 0 | 0.0 | 1 | 0.1 | 21 | 1.9 | 7 | 0.6 | 1,079 | 92.0 | 1,173 |
| Total | 4,058 | 71.8 | 813 | 14.4 | 75 | 1.3 | 13 | 0.2 | 148 | 2.6 | 370 | 6.5 | 174 | 3.1 | 5,651 | 88.3 | 6,399 |
Figure 10.
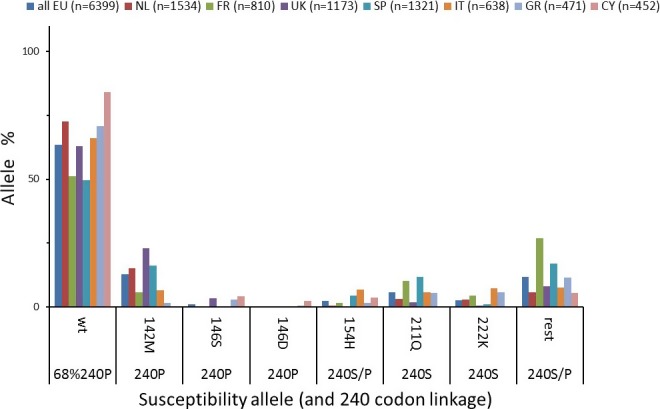
Frequency of goats with alleles of interest included in surveys across EU (GoatBSE deliverable D1.2) (http://www.goattse.eu/site/files/goatBSE_Deliverable_D1-2_21sep2011.pdf)
Figure 11.
Distribution of the codon 146 genotypes in the goat population of Cyprus for the period 2010–2016 (Veterinary Services. Ministry of Agriculture, Rural Development and Environment. Cyprus)
3.5.3. Concluding remarks
As the distribution of the PRNP alleles in goat breeds present in European MS has only been investigated in a few MS (CY, ES, IT, FR, the NL and the UK) and in a limited number of breeds, any statement about allele frequency at an EU‐wide level comes with a high degree of uncertainty. The investigations concentrated mostly on common transboundary breeds, along with a number of important autochthonous breeds. A random selection screening of frequencies at the national or regional scale would add to the knowledge of presence of alleles in goat populations in EU MS.
There is high variability in the frequency of alleles for the common transboundary breeds between MS, indicating that the number of goats studied may still have been too small to result in robust estimates of the frequencies for the EU.
The D146 and S146 alleles are present in more than 50% of the total goat population in Cyprus. In most other MS goat breeds, these alleles are absent or represented at much lower frequencies (Greek breeds, Dutch Nubian), but with some exceptions (Boer breed with 31% in the NL and 26% in the UK).
The allele K222 is widely geographically distributed in European goat populations and breeds. However, in general, its frequency is low, and it is absent in some local breeds.
3.6. The impact of promoting resistant alleles
3.6.1. The impact on traits and genetic biodiversity
In theory, there are two possible ways that promoting PRNP polymorphisms conferring resistance to CS may have an effect on other traits. The first way is a direct effect of the PRNP alleles themselves on other traits, the second way is indirectly by (chance) associations of animals carrying certain PRNP alleles also carrying polymorphisms influencing other traits (i.e. genetic drift). In goats, there are no published studies on any effects of PRNP polymorphisms on traits other than scrapie resistance. Therefore, we refer to the extensive literature on effects of PRNP polymorphisms on other traits in sheep to assess the likely impact in goats.
In sheep, the effects of PRNP polymorphisms have been studied across a range of breeds and countries. In most studies, no association between PRNP polymorphisms and other traits were found. For example, Gubbins et al. (2009) studied the association between lamb survival and PRNP genotypes in 10 sheep breeds. An association was found in only one (Charolaise) out of nine breeds. In that breed, lamb survival was higher for resistant ARR/ARR genotypes than for more susceptible ARR/VRQ genotypes. Another example is muscle depth. In many studies, no effect of PRNP alleles on muscle depth is found, as for example in Suffolk sheep (Sawalha et al., 2010). In a limited number of studies looking at a few breeds, however, an effect was found. In a Welsh mountain flock, ARR/ARR animals had greater muscle depth than ARR‐allele holding animals (Pritchard et al., 2008), while in German Black Headed Mutton sheep, ARR/ARR animals had less muscle depth than animals without ARR alleles (De Vries et al., 2004). In the Beulah breed, no effect of the number of ARR alleles was found, but animals carrying ARQ alleles had greater muscle depths (Boulton et al., 2010). Thus, few effects are found, and those that are found are not consistent with each other. A similar conclusion can be drawn for litter size. From these results, we can conclude that a direct effect of PRNP alleles on other traits is unlikely, otherwise more effects and more consistency across breeds should have been found. On the other hand, indirect trait effects of PRNP alleles do occur occasionally.
The consequence of indirect effects is that the magnitude and direction of the effect of selection for resistance alleles cannot be predicted. In general, the chance that associations occur, causing the selection for certain PRNP alleles to influence other traits, will be higher the fewer animals that carry a certain PRNP allele. In other words, genetic drift can be high when selecting for PRNP alleles present at a low frequency. The key parameter determining the magnitude of genetic drift is the effective population size (or its reciprocal, the inbreeding rate). A general guideline is that the effective population size should not be below 50 (FAO, 2012) and the inbreeding rate not above 1%.
Genetic diversity is also directly influenced by the effective population size. The lower the effective population size, the higher the loss of genetic diversity. Selection for scrapie resistance can decrease the effective population size and increase the inbreeding rate, and therefore loss of genetic diversity, especially if the starting frequencies of the resistance alleles are low. In goat populations, the frequencies of resistant alleles are below 10% in most breeds (see Section 3.5.2), thus effects on genetic diversity can be expected. There are, however, no studies quantifying the effect of selection for scrapie resistance alleles on effective population size and loss of diversity in goat populations, so extrapolation from data relating to sheep studies is necessary.
In sheep, Windig et al. (2004) investigated the effect of selection for resistance at different PRNP allele frequencies on the effective population size. The effect depended on the starting allele frequencies and the strength of selection. Selection varied from minimal (i.e. only fathers selected and both homozygous and heterozygous resistant fathers allowed), to strong (only homozygous resistant animals allowed to breed). In breeds with a low frequency of resistant alleles, strong selection resulted in a population size that was too low, with high levels of genetic diversity lost.
Alternative selection regimes can be devised to mitigate the effect of selection on loss of genetic diversity (see Figure 12). Before the selection regime can be determined, the number of breeding animals, the population structure, and the allele frequencies must all be determined. Thus, before selection can be applied without unacceptable loss of genetic diversity, a detailed inventory of the breed is needed, as well as a means to keep track of allele frequencies during the years that a breeding programme is in operation. For breeds in which production is vital for its (economic) existence, it is also important to keep track of production parameters and their possible association with resistant alleles. The only MS currently doing this is CY.
Figure 12.
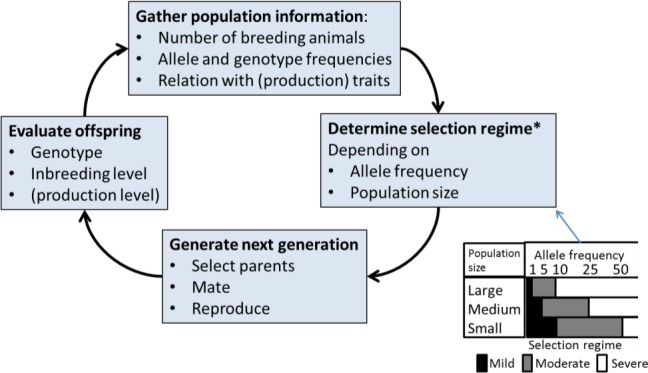
Scheme for a breeding programme for resistance to scrapie in goats, based on sheep breeds
- *The selection regime is based on allele frequency and population size (lower right corner), whereby under mild selection heterozygous and homozygous bucks are used indiscriminately, under moderate selection homozygous rams are used preferably but supplemented with heterozygous bucks, and under strong selection homozygous rams are used exclusively. Limits for classifying populations as large, medium or small have not been determined in goat breeds, but were < 750 for small breeds and > 3,750 for large breeds in sheep. Scheme designed by Jack J. Windig.
Based on information gathered about a single breed, a tailor‐made selection regime can be designed. In sheep breeds with a large population size (> 3,750 breeding females), strong selection (only homozygous resistant rams allowed to breed) could be allowed when the frequency of the resistant alleles is above 10%. Between 1% and 10% moderate selection could be applied. In this case, all homozygous resistant rams are used for breeding, but supplemented with heterozygous rams to reach the number of rams normally used for breeding. Below 1%, the advised selection regime is minimal, (i.e. both heterozygous and homozygous resistant rams can be used for breeding), with equal chances of being used. For breeds with lower population sizes, the allele frequencies at which the different selection regimes should be applied are increased. In this way, breed selection at the start is minimal, and once certain allele frequencies are reached, stronger selection methods can be applied. This regime was applied, for example, in the Fries Melkschaap in the Netherlands, and succeeded in attaining high frequencies of resistant alleles from an initial very low frequency (< 1%) without considerable loss of diversity. In goat breeds, however, the relationship between population size, allele frequency and inbreeding rates are unknown, and due to a different population structure, this relationship could be different from that in sheep breeds.
In European goats, allele frequencies differ between breeds (see Section 3.5.2), but most tested breeds have a low frequency of resistant alleles (< 10%). Moreover, frequencies are not known for many breeds and the association of resistant alleles with other traits is not known. Consequently, selection programmes promoting PRNP polymorphisms conferring resistance to CS may have considerable impact on other traits and genetic diversity. For selection programmes to succeed, i.e. raise the frequency of resistant alleles to levels high enough to combat scrapie infections, without unwanted change in other traits or large losses of diversity, careful planning is needed. In some breeds, however, resistant alleles may be completely absent, as was the case for the Dutch Landrace breed. In such a case, only introgression of resistant alleles from other breeds can introduce resistance into these breeds. This requires considerable time and careful planning to minimise the loss of genetic diversity in these breeds.
3.6.2. The doppel and shadoo proteins
The PRND gene is located 3′ of the PRNP gene at a distance of ~ 25 kilobases (kb), it encodes a protein named doppel (dpl) with sequence and structural homology to PrP (Moore et al., 1999). Although the physiological function of dpl protein is not fully understood, most studies agree on a role in sperm maturation, supported by the high level of PRND transcript observed in the testis (Behrens et al., 2002). PRND expression in the brain does not significantly alter prion disease in transgenic mice (Tuzi et al., 2002), and there is also no other experimental or epidemiological evidence to suggest an involvement of the PRND gene in prion disease. However, ectopic expression of dpl protein in mouse brain has been shown to lead to neuronal degeneration (Moore et al., 1999). Due to its close proximity to the PRNP gene, recombination between the PRNP and PRND locus will be a relatively rare event. Selection of PRNP for TSE resistance is therefore highly likely to co‐select PRND gene variants. Whether this co‐selection will have a neutral or negative effect in goats cannot be assessed yet due to the lack of data on genetic variation of the caprine PRND gene and limited knowledge of its role in male fertility.
The SPRN gene is a member of the prion protein gene family, located on chromosome 26 in goats (Lampo et al., 2007; Watts and Westaway, 2007). It encodes a protein named shadoo (Sho) with little sequence homology to the prion protein but with considerable structural similarity. Expression of SPRN transcripts is highest in brain, with overlap of PRNP expression (Gossner et al., 2009). It is unlikely that Sho can functionally replace PrP but both exhibit neurotrophic and neuroprotective activity (Watts and Westaway, 2007). Overexpression of shadoo protein in transgenic mice does not impact on the pathogenesis of scrapie (Wang et al., 2011). However, the generation of prions selectively lead to a decrease in shadoo protein in brain tissue (Watts et al., 2011). While an association with prion disease was demonstrated in human CJD (Beck et al., 2008) and in goat scrapie (Peletto et al., 2012), none was observed in sheep (Stewart et al., 2009). The SPRN effect in goats appears however to be much weaker than the association of PRNP genotype with resistance (Peletto et al., 2012).
3.6.3. Concluding remarks
While no direct effects of scrapie resistance alleles on production and other traits are to be expected, indirect effects may occur through chance association, difficult to detect, especially when frequencies of resistant alleles are low.
A certain loss of genetic diversity is to be expected when selecting for scrapie resistance, especially for breeds with low population numbers and/or low frequencies of resistance alleles. This loss can be restricted, but requires considerable effort to monitor allele frequencies and their association with traits.
Selection of PRNP for TSE resistance is likely to co‐select PRND gene (encoding the doppel protein) variants but the neutral or negative effect in goats cannot be assessed.
The dominance of the variant alleles over the wild‐type alleles will enable breeding regimes with early benefits to the reduction of disease prevalence by simply increasing the number of heterozygotes.
4. Answers to the Terms of Reference (ToR)
4.1. Answer to Term of Reference 1
Is there sufficient scientific knowledge available to have a robust level of scientific assurance that certain polymorphisms of the prion protein gene (PRNP) present in European goat breeds confer genetic resistance to classical scrapie (i.e. to classical scrapie strains known to occur in the EU goat population)? If this is the case, which are those polymorphisms?
Since 2009 (date of the previous goat opinion), the scientific knowledge related to scrapie resistance associated with goat PRNP gene polymorphisms has considerably expanded. This knowledge is now sufficient to consider that at least one polymorphism confers a robust genetic resistance against CS strains known to occur in the EU goat population.
The quality and certainty of the field and experimental data available for the K222, the D146 and the S146 alleles are greater than those available in the public domain for the ARR allele in sheep when the 2002 SSC opinion on safe sourcing of small ruminant materials was produced (2001).
All evidence supports the view that the K222 allele confers resistance against a variety of EU CS isolates that may reflect a variety of scrapie strains. According to the available knowledge, K222 provides a level of resistance in goats that is equivalent to that associated with the ARR allele in sheep. However, there is no assurance that K222 carriers would be resistant to all TSE agent strains currently circulating in the EU goat population.
The data also provide evidence for association of the D146 and S146 alleles with strong resistance against scrapie agent(s) currently circulating in Cyprus. Data remain insufficient to assess the level of resistance that the D146 and S146 alleles might provide against other CS agents circulating in other EU goat populations.
Some of the other goat PRNP polymorphisms, such as the D145, are potentially associated with resistance against CS. However, at this stage, there is insufficient knowledge to provide a robust level of scientific assurance.
4.2. Answer to Term of Reference 2
Based on available scientific evidence, what is the frequency and distribution of PRNP polymorphisms conferring resistance to classical scrapie in European goat breeds? If possible, could EFSA produce a susceptibility ranking of goat PRNP genotypes to classical scrapie?
Frequencies and distributions of PRNP alleles conferring resistance to CS are only known for less than 10% of the breeds listed in the EU MS, and only in a restricted number of MS.
The major studies were conducted in the MS with the largest goat populations (EL, ES, FR, IT) except RO, and in three other MS (CY, the NL and the UK). Data on the remaining MS (accounting for about 20% of the EU goat population) are absent.
CY is the only MS with accurate genotype data about the whole goat population. For the other investigated MS, data are only available for allele and genotype frequencies in some breeds.
Individual goat breeds show divergence in their genetic structure among different MS, making it difficult to develop breed‐specific PrP selection at the EU level.
In most of the investigated European breeds, the resistance‐associated alleles (D146, S146, K222), have a frequency between 0% and 10%. In some breeds (Cashmere, Angora, Spanish autochthonous breeds), none of these alleles have been reported.
More than 50% of the goats in CY hold either the D146 or the S146 alleles. In other MS, these alleles are represented at low frequencies, or are absent, with some exceptions.
The K222 allele has been observed in most of the investigated EU breeds, with notable exceptions of Cypriot and some Spanish goat breeds. In Saanen and Alpine, the D146 and S146 alleles are absent, while in a few other breeds they are present at higher frequencies (Dutch‐Nubian 7%, and Boer 30%).
Ranking of genotypes is not possible, because of insufficient data. At this stage, ranking can only be provided at allele level.
Based on both the ‘weight of evidence’ and ‘strength of resistance’, alleles can be ranked as follows, from high to weak CS resistance: K222 > D146 = S146 > Q211 = H154 = M142 > S127 = H143 > wild type.
4.3. Answer to Term of Reference 3
Based on available scientific evidence, what is the level of susceptibility to atypical scrapie and to BSE of the PRNP polymorphisms conferring resistance to classical scrapie?
Atypical scrapie:
There is currently insufficient data to assess the impact of the K222, D146 or S146 alleles on the susceptibility to AS in goats.
Based on the limited available data in goats and data obtained in sheep, the H154 allele is likely to be associated with higher susceptibility to AS compared to R154.
BSE:
The amount of data related to the level of susceptibility to BSE conferred by goats PRNP polymorphisms is limited.
Experimental oral challenge with BSE indicated an incomplete resistance in K222 allele carriers.
There are no data available about the susceptibility to BSE infection of S146 and D146 allele carriers.
There are no data available on the susceptibility of goats with the H154 allele to BSE infection.
4.4. Answer to Term of Reference 4
What is the likely impact of measures promoting PRNP polymorphisms conferring resistance to classical scrapie in terms of susceptibility to other disease/s, of production traits and survivability, taking into account epidemiological differences between Member States? Are such polymorphisms likely to have adverse effects on genetic diversity and variability and on the maintenance of old or rare caprine breeds or those that are well‐adapted to a particular region?
Information on the relationship of PRNP polymorphisms with other traits is lacking in goats. The likely impact of measures promoting selection for resistance on other traits has been inferred from studies in sheep.
Given the low frequencies of favourable alleles in most breeds, including breeds with large populations such as Alpine and Saanen, and old breeds that are well adapted to particular regions, high selection pressure is likely to have an adverse effect on genetic diversity.
A direct effect of PRNP alleles on health and production traits is unlikely. In breeds with low frequencies of the favourable allele(s), and breeds with small effective population size, selection could affect other traits, with the direction of change being unpredictable.
4.5. Answer to Term of Reference 5
What are EFSA's recommendations concerning strategies to apply current knowledge on genetic resistance to classical scrapie in goats in order to control and/or eradicate classical scrapie in the EU goat population?
Genetic resistance can be considered to be an effective tool to control CS in goats. Therefore, breeding for resistance could be offered as an option for MS to control CS in goats.
Outbreak management for CS in goat herds could be based on the selection of genetically resistant animals, as defined for sheep in the Regulation 999/2001 (EC).
If breeding for resistance programmes are implemented, they should be designed to take into account the starting allele frequencies with the view to prevention of loss of genetic diversity. If such programmes were made compulsory, derogations should be considered in the case of rare or small population breeds, or in those where no alleles of interest are present.
There are limited data available on the PRNP allele frequencies in many MS and breeds. Before developing any breeding for resistance programme, baseline surveys are needed to establish the resistant‐allele frequencies in the relevant goat populations at both MS and breed level.
Available data indicate differences in PRNP allele frequencies amongst MS for the same goat breed. Therefore, if breeding for resistance programmes are implemented, they should be developed and managed at MS level and the impact of their implementation should be monitored.
5. Other recommendations
Even if formal breeding plans are not implemented, it would be advisable to inform relevant stakeholders, especially breeders, about genetic resistance to CS in goats.
Before breeding for resistance programmes are promoted or enforced, it would be advisable to produce guidance on how to disseminate resistant alleles in goat breeds (see EFSA BIOHAZ Panel, 2014).
If breeding for resistance programmes are implemented, a central database should be established that allows MS to know where and how many resistant animals are available for breeding purposes and to evaluate breeders’ interest. Breed organisations should be encouraged to create a pool of resistant animals, semen, embryos and ova for dissemination of the resistant alleles in the population.
The association of alleles conferring resistance to scrapie with other traits should be investigated through ad hoc studies or monitored during the breeding programmes.
Documentation provided to EFSA
The K222 allele of the goat PRNP gene as candidate for selective culling in scrapie outbreaks and for future breeding programs for TSE resistance in Italian goat breeds Contributors: Pierluigi Acutis, Umberto Agrimi, Cristina Bona, Romolo Nonno, Giuseppe Ru, Gabriele Vaccari. Istituto Zooprofilattico Sperimentale de Piemonte, Liguria e Valle d'Aosta. Istituto Superiore di Sanità ‐ Dipartimento di Sanità Pubblica Veterinaria e Sicurezza Alimentare.
The effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with scrapie by different routes. Veterinary Services, Cyprus.
Reply letter to the Questions to the Cypriot challenges studies in goats with 146 codon alleles. 10 February 2017. Dr. Penelope Papasavva‐Stylianou. Senior Veterinary Officer. Veterinary Services. Nicosia 1417, Cyprus.
Glossary and Abbreviations
- Allele
One of two or more forms of the DNA sequence of a particular gene. Each gene can have different alleles and different alleles can result in different traits.
- Haplotype
A set of markers (polymorphisms) on a single chromosome that tend to be inherited together. A haplotype can refer to a combination of alleles or to a set of single nucleotide polymorphisms (SNPs).
- Hardy–Weinberg equilibrium
Allele and genotype frequencies in a population will remain constant from generation to generation in the absence of selection, mutation, genetic drift, or other forces.
- Isolate
A primary source of TSE from the natural disease. It may be passaged in the natural host or in another species. It may contain one or more ‘strain’ (EFSA, 2011)
- Polymorphism
Genetic variation within a population, upon which natural selection can operate.
- Strain
A source which has been characterised phenotypically in a host and which behaves as a single entity within that host, as far as can be demonstrated experimentally. Several serial passages in the same host species and PrP genotype are usually required to establish the phenotypic properties of strains. (EFSA, 2011a,b)
- AS
atypical scrapie
- AT
Austria
- BE
Belgium
- BG
Bulgaria
- BSE
bovine spongiform encephalopathy
- CC
cell‐based conversion assay
- CFC
cell‐free conversion assay
- CI
confidence interval
- CNS
central nervous system
- CS
classical scrapie
- CWD
chronic wasting disease
- CY
Cyprus
- CZ
Czech Republic
- DE
Germany
- DK
Denmark
- dpl
doppel protein
- EE
Estonia
- EFABIS
European Farm Animal Biodiversity Information System
- EL
Greece
- ELISA
Enzyme‐linked immunosorbent assay
- ERFP
European Regional Focal Point for Animal Genetic Resources
- ES
Spain
- FI
Finland
- FR
France
- HR
Croatia
- HU
Hungary
- i.c.
Intracerebral
- IE
Ireland
- IT
Italy
- Kb
kilobases
- KDa
kilodalton
- KO
knock‐out
- LT
Lithuania
- LU
Luxembourg
- LV
Latvia
- MS
Member State
- MT
Malta
- NL
The Netherlands
- NSHC
non‐slaughter for human consumption
- OR
odds Ratio
- p.i.
Post‐inoculation
- PK
Proteinase K
- PL
Poland
- PMCA
Protein Misfolding Cyclic Amplification
- p.o.
Per os (Oral administration)
- PR
prevalence rate
- PRND
Gene encoding the prion‐like protein doppel
- PRNP
gene encoding for the major prion protein PrP (for prion protein or protease‐resistant protein), also known as CD230 (cluster of differentiation 230)
- PrPC
Normal cellular prion protein
- PrPres
PrPSc digested by proteinase K treatment before molecular immunoanalysis by western blotting
- PrPSc
abnormal protease resistant isoform or prion protein
- PT
Portugal
- QuIC
Quaking induced conversion
- RO
Romania
- RR
relative risk
- s.c.
Subcutaneous
- SD
Standard deviation
- SE
Sweden
- Sho
shadoo protein
- SHC
Slaughter for human consumption
- SI
Slovenia
- SK
Slovakia
- SPRN
Gene encoding protein shadoo
- SSC
Scientific Steering Committee
- ToR
Term of reference
- TSE
transmissible spongiform encephalopathies
- UK
United Kingdom
- vCJD
Variant Creutzfeldt‐Jakob Disease
- WG
Working group
- WT
wild type
Appendix A – Goat population in the EU
1.
Appendix B – Goat meat production and consumption in the EU
1.
In terms of consumption, the available data of consumption of goat data in the EU as per November 2016 is displayed in Table B.2
Appendix C – Movement of live goats for breeding within the EU in the period 2014–2016
1.
Appendix D – Summary results of goat genetic surveys in the EU
1.
Table D.1.
Frequency of goats with some of the alleles of interest (M142, H154, D146, S146, Q211, K222) included in surveys across EUa and linkage to the 240 alleles. Alleles S127 and R143 were excluded because of the low interest for their protective effect, resulted from the application of the tool based on the weight of evidence and the strength of resistance
| Allele frequency (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number animals sequenced | Wt | 142 | 146 | 146 | 154 | 211 | 222 | 240P, 240S in wt | |||
| Country | Breed/production type | wt240P, 240S | M240P | S240P | D240P | H240S | Q240S | K240S | P/(S + P)×100% | Other alleles(if > 4% percentage is mentioned) | |
| NL | Dairy (Saanen/Alpine) | 1,534 | 72.7 | 15.1 | 0.06 | 0 | 0.4 | 3.2 | 2.8 | 60 | R1015.7%; S106; S1275.9%; S153; L 220 |
| FR | One H154240P; other alleles not investigated | ||||||||||
| Alpine bucks | 200 | 76.2 | 3.9 | 0 | 0 | 5.4 | 7.1 | 7.4 | 54 | ||
| Saanen bucks | 194 | 67.4 | 8.7 | 0 | 0 | 0.5 | 18.5 | 4.9 | 68 | ||
| Corsica bucks | 94 | 88.6 | 0.5 | 0 | 0 | 1 | 0.5 | 9.4 | 64 | ||
| Poitevine bucks | 104 | 45.6 | 19.7 | 0 | 0 | 0 | 31.3 | 3.4 | 42 | ||
| Total | 592 | ||||||||||
| UK | D(airy) | 932 | 61.8 | 26.6 | 0.2 | 0 | 0 | 2.3 | 0.7 | 76 | R101, S127, R143 |
| M(eat) | 157 | 56.1 | 13.2 | 24.5 | 0 | 0.3 | 0.3 | 0 | 30 | S127, R143, L218 | |
| F(fibre, wool) | 84 | 89.2 | 1.2 | 0 | 0 | 0.6 | 0 | 0 | 63 | R143, L218 | |
| Total | 1,173 | 63.0 | 23 | 3.5 | 0 | 0.1 | 1.8 | 0.6 | 69 | ||
| Alpine | 102 | 62.7 | 20.6 | 0 | 0 | 0.5 | 0.2 | 102 | 62.7 | S127 | |
| Angora | 84 | 97.6 | 0.6 | 0 | 0 | 0 | 0 | 84 | 97.6 | R143 | |
| Anglo‐Nubian | 55 | 91.8 | 5.5 | 0 | 0 | 0.9 | 0 | 55 | 91.8 | S127, R143 | |
| Anglo‐Nubian cross | 16 | 62.5 | 37.5 | 0 | 0 | 0 | 0 | 16 | 62.5 | ||
| Boer | 146 | 54.8 | 13.7 | 26 | 0 | 0 | 0 | 146 | 54.8 | R143, H154, L218 | |
| Cashmere | 30 | 75 | 1.7 | 0 | 0 | 0 | 0 | 30 | 75 | R143, H154, L218 | |
| Saanen | 272 | 58.6 | 24.1 | 0 | 0 | 2 | 0.4 | 272 | 58.6 | S127 | |
| Saanen cross | 332 | 63.6 | 30.6 | 0.5 | 0 | 2.6 | 0.9 | 332 | 63.6 | S127, R143, S146 | |
| Toggenburg | 46 | 58.7 | 31.5 | 0 | 0 | 4.3 | 0 | 46 | 58.7 | S127 | |
| Toggenburg cross | 45 | 68.9 | 23.3 | 0 | 0 | 2.2 | 0 | 45 | 68.9 | S127 | |
| Total | 1,128 | ||||||||||
| ES | R18; V37; D74; T112; S127; I137; S139, F141; H151; R215; W232 | ||||||||||
| Moncaina | 386 | 62.6 | 12.6 | 0 | 0 | 14 | 12.5 | 0.3 | 65 | ||
| Pirenaica | 44 | 44.1 | 38.1 | 0 | 0 | 13.6 | 6.8 | 0 | 94 | ||
| Retinta | 71 | 50.8 | 21.8 | 0 | 0 | 3.1 | 28.2 | 0 | 70 | ||
| Alpine | 39 | 76.9 | 3.8 | 0 | 0 | 2,6 | 10.3 | 6.4 | 58 | ||
| Saanen | 42 | 69 | 20.2 | 0 | 0 | 1.2 | 6 | 1.2 | 65 | ||
| Murciano Granadina | 50 | 23 | 60 | 0 | 0 | 0 | 18 | 0 | 83 | ||
| Alpine (scrapie herd) | 51 | 62.7 | 5.9 | 0 | 0 | 4.9 | 17.6 | 5.2 | 56 | ||
| Saanen (scrapie herd) | 52 | 67.3 | 6.7 | 0 | 0 | 0 | 25 | 0 | 66 | ||
| Cross breed (scrapie herd) | 10 | 70 | 15 | 0 | 0 | 15 | 0 | 0 | 55 | ||
| Cross breed (scrapie herd) | 563 | 58 | 16.1 | 0 | 0 | 7.7 | 17.9 | 0 | 63 | ||
| Scrapie affected | 13 | 70.8 | 15.4 | 0 | 0 | 0 | 11.5 | 0 | 84 | ||
| Total | 1,321 | ||||||||||
| IT | New alleles? | ||||||||||
| Garganica | 60 | 59.7 | 2.4 | 0 | 0 | 11.3 | 0 | 17.7 | 82.4 | V37, P110, T142, R143 | |
| Maltese | 60 | 58.9 | 0 | 0 | 0 | 9.7 | 0 | 7.3 | 87.6 | V3721.8%, R143, Q168 | |
| Red Mediterranean | 60 | 78.9 | 0 | 0 | 0 | 10.5 | 0 | 4.4 | 85.5 | V37, P110, R143, Q168 | |
| IT (North) | Camosciata delle Alpi | 84 | 62.5 | 8.9 | 0 | 0 | 11.3 | 13.7 | 2.4 | 52.3 | S127 |
| Saanen | 69 | 77.5 | 7.2 | 0 | 0 | 0 | 10.2 | 3 | 75.7 | P110, S127 | |
| Roccaverano | 70 | 70 | 5.7 | 0 | 0 | 3.6 | 13.6 | 4.3 | 63.3 | S127, Q168 | |
| Valdostana | 77 | 54.5 | 28.2 | 0 | 0 | 0 | 9.6 | 1.3 | 37.6 | S127 | |
| IT (South) | Garganica | 58 | 61.3 | 2.6 | 0 | 0 | 11.2 | 0 | 17.2 | 83.2 | V37, P110, R143 |
| Maltese | 25 | 62 | 0 | 0 | 0 | 6 | 2 | 12 | 83.7 | V3714%, R1434% | |
| Ionica | 27 | 72.2 | 0 | 0 | 0 | 7.4 | 1.9 | 7.3 | 82 | V375.6%, I137, R143 | |
| Mediterranean | 28 | 81.4 | 0 | 0 | 0 | 5.3 | 1.8 | 5.4 | 73.9 | R1435% | |
| South cross | 20 | 65 | 0 | 0 | 0 | 5 | 0 | 22.5 | 80.8 | R1435% | |
| Total | 638 | ||||||||||
GOATBSE consortium. http://www.goattse.eu/site/files/goatBSE_Deliverable_D1-2_21sep2011.pdf
Appendix E – Annex List of questions to Cypriot studies to authors
1.
The members of the ad hoc EFSA Working group on the genetic resistance to TSEs in goats, under the mandate EFSA‐Q‐2016‐00268, have studied the Cyprus report (Cyprus, 2015), and the accompanying data files and publications, included in the mandate submitted by the European Commission to EFSA. The Working group has considered it necessary to request from the authors of the report additional data and information on different aspects, in order to address the scientific robustness of the results, as follows:
1.1.
1.1.1.
Universality of the resistance associated to the D146 and the S146 alleles
1) What is the knowledge about the inoculum that was used in the study (single/several strains)?
The numbers of animals in the current experimental design required the pooling of brain material from several donor animals (12 goats from seven different flocks were sourced) to produce sufficient inoculum. To ensure that this pool was both representative of a major outbreak strain in CY, individual isolates were characterised by western blot prior to being pooled and BSE was excluded (Figure E.1). The pooled inoculum used for these challenges has also been characterised in Tg338 and tgShpXl mice (Figure E.2).
Figure E.1.
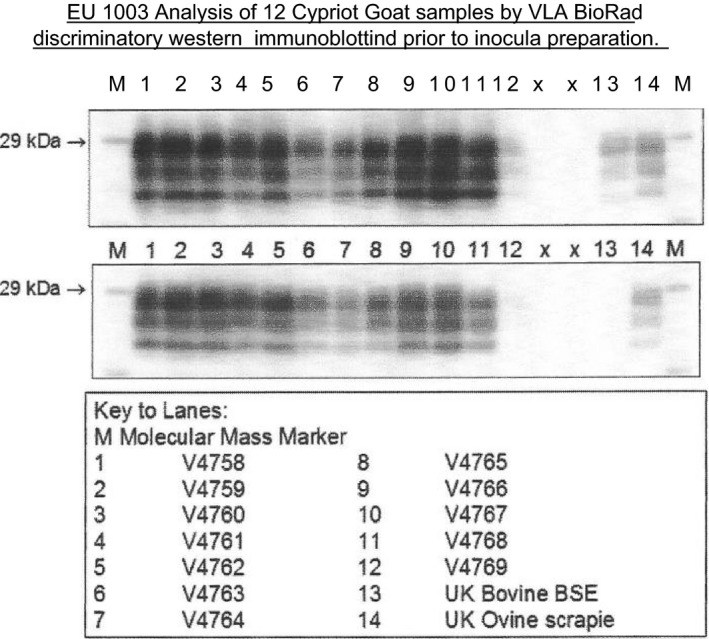
Discriminatory western blot of the 12 isolates that were used in the homogenate for the oral and intracerebral challenges
Figure E.2.
Lesion profiles from Tg338 and tgShpXl mice challenged with the homogenate that was used to challenge the goats
- Only clinically positive mice contributed to the lesion profiles. Ten mice of each mouse line were challenged. Numbers in brackets indicate the average IP of the mice which contributed to the profile. n = number of mice which contributed to the profile.
2) What is the current knowledge about the strain diversity in Cyprus and how Cypriot strains compare with mainland EU strains?
The data for this issue was presented at the 2012 EFSA Working Group (EFSA, 2012), but is not in the public domain. We can supply the information if requested provided it will not be published or distributed outside the confines of the WG.
It must be noted here that classical scrapie has an extensive variability of phenotypes in the original host and that host genetics can also influence these phenotypes. Therefore, exact strain identification of classical scrapie strains based on data directly derived from the host (WB, IHC, histopathology, PMCA, etc.) is not possible. This is why murine bioassays using either transgenic or wild‐type mice have been employed. However, these bioassays are expensive and as a result only a limited number of isolates have been described from any country.
Therefore, the current knowledge about strain diversity of classical scrapie in CY is incomplete as is the case for almost every country with extensive small ruminant population where classical scrapie is endemic (with the possible exemption of Italy where it is believed that a single strain is prevalent).
3) Can BSE be excluded in any of the identified isolates, based on the results of the IHC and discriminatory western blotting? Have these tests been performed on the identified isolates? If so, how are the results compared to the existing knowledge?
The identified isolates which were used for the preparation of the inoculum were individually characterised by discriminatory Western blot prior to being pooled and BSE was excluded (Figure E.3).
Figure E.3.
Distribution of the codon 146 genotypes in the goat population of Cyprus for the period 2010–2016
In addition to the above, CY has never reported a BSE case in either large or small ruminants despite extensive testing in accordance to EU regulations.
4) How homogenous is the Cypriot goat population?
The goat population in CY, when breeds are considered, is heterogenous. According to the Annual Review of Breeding Sheep and Goats in CY of the Department of Agriculture for the year 2015, the majority of the animals (about 64%) belong to cross bred (Damascus X Local breeds), another 25% of the animals belong to Damascus breed and the remaining 11% belongs to animals of French Alpine, Saanen and Local breeds.
However, the goat population in Cyprus is the only national goat population for which the genotype of every animal is known. Figure E.3 shows the codon 146 genotype distribution of the goat population in Cyprus for the period 2010–2016.
The intracerebral challenge
5) The final lengthening of the incubation period observed with some of the genotypes (S146 and D146 heterozygotes) following IC challenge is moderate. What is the level of statistical significance in the association between genotype and scrapie comparing each non‐NN146 genotype at the 146 codon and the NN146 genotype?
Kaplan–Meier survival curves were built for animals showing TSE clinical signs using as date of entry or exposure the date of inoculation; and as date of exit the date the animals were culled due to evidence of TSE clinical signs. Log‐rank test comparing the survival distributions of NN vs all other genotype groups individually was applied. Kaplan–Meier survival curves of the four pairs compared are shown below:
Results
The results showed that the incubation periods (between inoculation and disease confirmed until being culled due to evidence of clinical signs) in the groups of goats inoculated of DD, SS and NS genotypes are significant longer than that of the NN goats, pairwise. However, the incubation period of ND goats is not significantly longer when compared to NN goats.
Results of the individual tests:
The log‐rank test comparing the survival distribution of the NN146 group with the ND146 was not significant (χ2 = 0.06 p = 0.8) (Figure E.4).
Figure E.4.
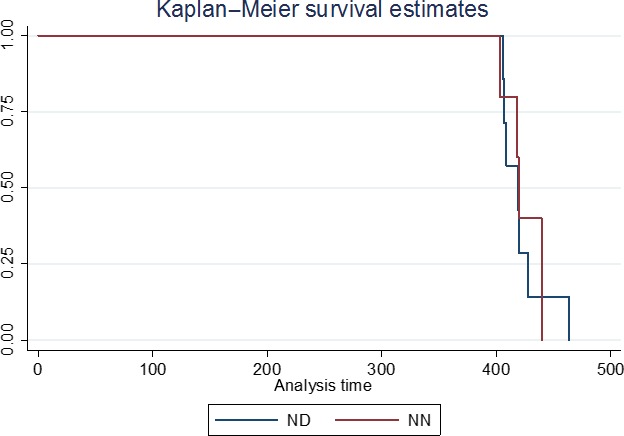
Survival curves of NN146 and ND146 groups
The log‐rank test comparing the survival distribution of the NN146 group with the NS146 was significant (χ2 = 13.83 p < 0.001) (Figure E.5).
Figure E.5.
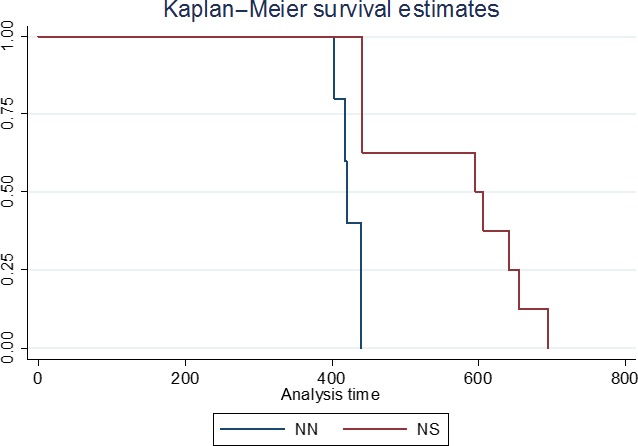
Survival curves of NN146 and NS146 groups
The log‐rank test comparing the survival distribution of the NN146 group with the DD146 was significant (χ2 = 9.86 p = 0.001) (Figure E.6).
Figure E.6.
Survival curves of NN146 and DD146 groups
The log‐rank test comparing the survival distribution of the NN146 group with the SS146 was significant (χ2 = 12.24 p < 0.001) (Figure E.7).
Figure E.7.
Survival curves of NN146 and SS146 groups
6) Has the increase in the incubation period with respect to the wild‐type genotype of the Cypriot studies been compared to the increase obtained in similar experiments for I142, Q211 and K222?
Although we have attempted to do this analysis, the data published elsewhere are not presented in a consistent way so direct comparisons are not feasible.
In addition, polymorphism at codons 142, 211 and 222 do not exist in the Cypriot goat population. On the other hand, the polymorphism at 146 is unique in the goats of this island. Therefore, combinations of polymorphisms at different codons are not possible. Under these circumstances, comparisons of incubation period variations relating to polymorphisms at specific codons are not informative – particularly when intracerebral challenges using different sources of inocula are involved as explained in our reply to question 5.
The request to compare the increase in the incubation period obtained in similar experiments looking at other polymorphisms is not straightforward. We are not aware of many studies in which i.c. challenge in the natural host have been conducted. Only direct comparisons of i.c. challenges in goats holding other genotypes of interest and the wild type at 146 codon would be appropriate.
In terms of analytical approaches, similar methods have been applied in the same context, targeting alleles other than D146 or S146.
Acutis et al. (2012) showed that the probability of survival of the QK222 goats vs the QQ222 goats was significantly higher (p = 0.002). In Maestrale et al. (2015), comparison of goats survivorship was applied using three susceptibility levels after o.c. challenge: resistant (DD145, QK222, and QK222‐S240?), susceptible with long post‐infection time (RH154, RH154‐SP240, PQ168‐PP240, RQ211, and RQ211‐SP240), and susceptible with short post‐infection time (wild‐type, GV37, OO168‐PP240, and SP240). The rank test comparing the survivorship between the three levels of susceptibility was highly significant (chi square: 46.9; p < 0.000). There were not pairwise comparisons conducted at individual allele level. The analysis in this study does not include testing the association of the potential resistance with a particular genotype, since they were pooled for analysis, In our study, we were able to provide pair wise comparisons.
In transgenic mice, there have been a number of studies measuring incubation periods of Tg mice expressing alleles of interest. For example, Aguilar‐Calvo et al. (2014) looked at i.c. challenged mice expressing K222. Incubation periods are not comparable between bioassay and natural host. Moreover, these authors did not provide any statistical result comparing i.p. between KK222, KQ222 and QQ222.
Lacroux et al. (2014) did a similar study using an inoculum form a single scrapie case to inoculate 1.c. and o.c. goats expressing different combination of alleles (142, 154, 211 and 222). The authors did not produce any statistical results showing difference in incubation periods between genotypes. However, they showed that it is possible the same allele to provide resistance after o.c. and susceptibility after i.c. challenge as it has been observed in the current study.
The oral challenge
7) The age at challenging was ≫ 5 months in most animals, if we understand correctly. What could be the impact of the post weaning exposure vs. natural exposure around birth?
The difficulties of sourcing sufficient animals of the appropriate genotypes to allow for five challenged animals in each genotype/time point group (a number consistent with other studies) meant that the animals on the oral challenge study could not be exposed perinatally. However, the comparison of relative susceptibility in the different groups is considered valid because all genotype groups were of a similar age range when they were dosed.
Previous studies investigating susceptibility of goats to oral challenge with scrapie have inoculated recipient animals within 48 h of birth (Lacroux et al., 2014). While this models the likely earliest field exposure (through milk from infected dams e.g Konold et al., 2016) and reflects challenge via the gut when it is still effectively monogastric, the effects of age at challenge on susceptibility are more generally linked to the age‐related involution of the GALT, which is considered to be a portal of infection.
However, several scrapie and BSE experimental challenges in sheep have shown that older animals can be successfully inoculated orally, and exposure of naïve older or adult sheep to scrapie infected environments can also result in successful infection (Dexter et al., 2009). Goats have also been successfully challenged orally after weaning at 2–3 months of age (Maestrale et al., 2015).
The susceptible NN146 positive control group in the present study all succumbed following challenge at around 6 months of age. In all NN146 goats, there was peripheral distribution of PrPSc in the LRS including GALT tissues. LRS infectivity was detected as early as 6 months pi where LRS was the only affected tissue, tissue in agreement with the pathogenesis of scrapie under natural conditions. In contrast in the GALT tissue from all other genotypes which were challenged at similar age as the NN146 animals, no PrPSc was detected 24 months pi.
This evidence further suggests that under natural conditions selection against the N146 allele could provide a means for resistance selection in the goats of Cyprus. This was the original objective of the project and has been addressed successfully.
8) Similarly, the late age at challenge of a majority of the goats might have led to a poor transmission in the non‐wild type codon 146 variants. What are the arguments in favour of reliable protection?
Please see answer in question 7
9) What is the current status of the remaining 16 live animals in the oral challenge, more than nine months since the submission of the report?
As per 20 December 2016, six out of the 16 challenged goats that were reported alive in our report, had died. There are 10 animals still alive (3 SS146, 3 DD146, 3 NS146, 1 ND146) (Table Table D.2).
Table Table D.2.
Summary of the status of the sixteen goats in the oral challenge goats still alive
| No. | Date of death | Days post inoculation (days) | Inoculation (years) | Genotype at 146 codon | BioRad TeSeE SAP ELISA | Examined tissues |
|---|---|---|---|---|---|---|
| 1 | 1/612016 | 2,275 | 6.23 | NS146 | Negative | All examined tissues were negative |
| 2 | 18/6/2016 | 2,126 | 5.82 | ND146 | Negative | All examined tissues were negative |
| 3 | 23/6/2016 | 2,131 | 5.83 | ND146 | Negative | All examined tissues were negative |
| 4 | 9/7/2016 | 2,147 | 5.88 | DD146 | Negative | All examined tissues were negative |
| 5 | 6/8/2016 | 2,175 | 5.96 | DD146 | Negative | All examined tissues were negative |
| 6 | 10/8/2016 | 2,179 | 5.97 | ND146 | Negative | All examined tissues were negative |
| 7 | Still alive | 2,477 | 6.8 | ND146 | ||
| 8 | Still alive | 2,477 | 6.8 | NS146 | ||
| 9 | Still alive | 2,477 | 6.8 | NS146 | ||
| 10 | Still alive | 2,477 | 6.8 | NS146 | ||
| 11 | Still alive | 2,311 | 6.3 | DD146 | ||
| 12 | Still alive | 2,311 | 6.3 | DD146 | ||
| 13 | Still alive | 2,311 | 6.3 | DD146 | ||
| 14 | Still alive | 2,307 | 6.3 | SS146 | ||
| 15 | Still alive | 2,307 | 6.3 | SS146 | ||
| 16 | Still alive | 2,302 | 6.3 | SS146 |
Other issues
10) Has the IDEXX test been applied to any of the tissues from challenged goats?
YES, the IDEXX test has been applied to brain tissue from i.c. challenged animals to resolve discrepancies that arose between BioRad and IHC.
Specifically, it was noticed that a number of samples that were BioRad negative were diagnosed TSE positive with IHC. The discrepancies were only observed in resistant phenotypes. All NN146 animals were successfully detected by BioRad.
Application of the IDEXX test identified as positive all the goats that had also been diagnosed positive with IHC and also identified as negative two intercurrent deaths.
11) Has the inoculum used in the experimental challenges and/or the new isolates from the challenged goats been subject to bioassay?
The answer for the first part of the question is YES. The inoculum used in the experimental challenges has been subject to mouse bioassay. The inoculum used for these challenges has been inoculated into Tg338 and tgShpXl mice. Please also see answer 1.
The answer for the second part is NO. The new isolates from the challenged goats have not been subject to bioassay. It was out of the aims and scope of the study.
However, some of these isolates based on route of inoculation, genotype and TSE phenotype will be subjected to bioassay in transgenic mice at Animal and Plant Health Agency (APHA).
12) What are the details of the breeding for resistance programme to scrapie in goats of Cyprus? Are there any data available showing the progress of the programme and the impact on the scrapie incidence? Can you provide it to the Working Group?
In the framework of the implementation of a surveillance programme and the control and eradication measures applied according to (EN) Regulation 999/2001, it has been observed a considerable reduction of the TSE index cases in herds and in mixed flocks. Figure E.8 shows the trend of the index cases per category of farm.
Figure E.8.
Annual number of TSE index cases per category of farm in CY
Suggested citation: EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) , Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Gironés R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Skandamis P, Speybroeck N, Simmons M, Kuile BT, Threlfall J, Wahlström H, Acutis P‐L, Andreoletti O, Goldmann W, Langeveld J, Windig JJ, Ortiz‐Pelaez A and Snary E, 2017. Scientific Opinion on genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA Journal 2017;15(8):4962, 79 pp. 10.2903/j.efsa.2017.4962
Requestor: European Commission
Question number: EFSA‐Q‐2016‐00268
Panel members: Antonia Ricci, Ana Allende, Declan Bolton, Marianne Chemaly, Robert Davies, Pablo Salvador Fernández Escámez, Rosina Gironés, Lieve Herman, Kostas Koutsoumanis, Roland Lindqvist, Birgit Nørrung, Lucy Robertson, Moez Sanaa, Panagiotis Skandamis, Emma Snary, Niko Speybroeck, Benno Ter Kuile, Giuseppe Ru, Marion Simmons, John Threlfall and Helene Wahlström.
Declared interests: Marion Simmons and Giuseppe Ru recused themselves from the discussion during Working Group meetings that involved two documents to which they had contributed and which were submitted by the European Commission along with the mandate. Both documents are listed in the section Documentation provided to EFSA on p. 53 (numbers 1 and 2) of this Opinion.
Acknowledgements: The Panel wishes to thank the members of the GoatBSE and GOAT‐TSE‐FREE consortia, and the Cypriot Veterinary authorities.
Reproduction of the images listed below is prohibited and permission must be sought directly from the copyright holder: Figure 11 and Figures in Appendix E: © Cypriot Veterinary Services
Adopted: 5 July 2017
Notes
Commission Regulation (EC) No 260/2003 of 12 February 2003 amending Regulation (EC) No 999/2001 of the European Parliament and of the Council as regards the eradication of transmissible spongiform encephalopathies in ovine and caprine animals and rules for the trade in live ovine and caprine animals and bovine embryos. J L 37, 13.2.2003, p. 7–11.
Commission Decision of 13 February 2003 laying down minimum requirements for the establishment of breeding programmes for resistance to transmissible spongiform encephalopathies in sheep (Text with EEA relevance) (notified under document number C(2003) 498).
Regulation (EC) No 1923/2006 of the European Parliament and of the Council of 18 December 2006 amending Regulation (EC) No 999/2001 laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies (Text with EEA relevance) OJ L 404, 30.12.2006, p. 1–8.
Commission Regulation (EC) No 727/2007 of 26 June 2007 amending Annexes I, III, VII and X to Regulation (EC) No 999/2001 of the European Parliament and of the Council laying down rules for the prevention, control and eradication of certain transmissible spongiform encephalopathies.
Two‐letter codes for EU and EFTA countries haves been used throughout the scientific opinion. See Glossary.
http://www.europarl.europa.eu/RegData/etudes/etudes/join/2008/397253/IPOL-AGRI_ET(2008)397253_EN.pdf
Material supplied by Thierry Baron (ANSES, Lyon, FR).
For distinction of the Boolean term OR, the abbreviation of Odds Ratio appears in italics (OR).
Multiple isolates implies that several – preferably more than two – scrapie isolates from geographically different origins have been investigated experimentally.
Data extracted from the document ‘The effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with scrapie by different routes’ attached to the mandate (see Documents provided to EFSA), and from the reply letter to the Questions to the Cypriot challenges studies in goats wit h146 codon alleles (Appendix E) are cited in this opinion as follows: Cyprus, 2015.
> greater than; = equal than.
References
- Acín C, Martin‐Burriel I, Monleon E, Lyahyai J, Pitarch JL, Serrano C, Monzon M, Zaragoza P and Badiola JJ, 2013. Prion protein gene variability in Spanish goats. Inference through susceptibility to classical scrapie strains and pathogenic distribution of peripheral PrP(sc.). PLoS ONE, 8, e61118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G and Caramelli M, 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. Journal of General Virology, 87, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Acutis PL, Colussi S, Santagada G, Laurenza C, Maniaci MG, Riina MV, Peletto S, Goldmann W, Bossers A, Caramelli M and Cristoferi I, 2008. Genetic variability of the PRNP gene in goat breeds from Northern and Southern Italy. Journal of Applied Microbiology, 104, 1782–1789. [DOI] [PubMed] [Google Scholar]
- Acutis PL, Martucci F, D'Angelo A, Peletto S, Colussi S, Maurella C, Porcario C, Iulini B, Mazza M, Dell'atti L, Zuccon F, Corona C, Martinelli N, Casalone C, Caramelli M and Lombardi G, 2012. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Veterinary Research, 43, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrimi U, Ru G, Cardone F, Pocchiari M and Caramelli M, 1999. Epidemic of transmissible spongiform encephalopathy in sheep and goats in Italy. Lancet, 353, 560–561. [DOI] [PubMed] [Google Scholar]
- Aguilar‐Calvo P, Espinosa JC, Pintado B, Gutierrez‐Adan A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O and Torres JM, 2014. Role of the goat K222‐PrP(C) polymorphic variant in prion infection resistance. Journal of Virology, 88, 2670–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar‐Calvo P, Fast C, Tauscher K, Espinosa JC, Groschup MH, Nadeem M, Goldmann W, Langeveld J, Bossers A, Andreoletti O and Torres JM, 2015. Effect of Q211 and K222 PRNP polymorphic variant in the susceptibility of goats to oral infection with goat bovine spongiform encephalopathy. Journal of Infectious Diseases, 212, 664–672. [DOI] [PubMed] [Google Scholar]
- Aguilar‐Calvo P, Espinosa JC, Andreoletti O, González L, Orge L, Juste R and Torres JM, 2016. Goat K222‐PrPC polymorphic variant does not provide resistance to atypical scrapie in transgenic mice. Veterinary Research, 47, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsac J, 2007. Similar biochemical signatures and prion protein genotypes in atypical scrapie and Nor98 Cases, France and Norway. Emerging Infectious Disease Journal 13, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsac JN, Betemps D, Morignat E, Feraudet C, Bencsik A, Aubert D, Grassi J and Baron T, 2009. Transmissibility of atypical scrapie in ovine transgenic mice: major effects of host prion protein expression and donor prion genotype. PLoS ONE, 4, e7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante EA, Smidak M, Grimshaw A, Houghton R, Tomlinson A, Jeelani A, Jakubcova T, Hamdan S, Richard‐Londt A, Linehan JM, Brandner S, Alpers M, Whitfield J, Mead S, Wadsworth JD and Collinge, J , 2015. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature, 522, 478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante EA, Linehan JM, Desbruslais M, Joiner S, Gowland I, Wood AL, Welch J, Hill AF, Lloyd SE, Wadsworth JDF and Collinge J, 2002. BSE prions propagate as either variant CJD‐like or sporadic CJD‐like prion strains in transgenic mice expressing human prion protein. The EMBO Journal, 21, 6358–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami‐Goudarzi K, Rupp R, Babilliot JM, Lacroux C, Lugan S, Schelcher F, Chartier C, Corbière F, Andreoletti O and Perrin‐Chauvineau C, 2009. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222 and 240 in French Alpine and Saanen breeds and their association with classical scrapie. Journal of General Virology, 90, 769–776. [DOI] [PubMed] [Google Scholar]
- Baylis M, Goldmann W, Houston F, Cairns D, Chong A, Ross A, Smith A, Hunter N and McLean AR, 2002. Scrapie epidemic in a fully PrP‐genotyped sheep flock. Journal of General Virology, 83, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, Tongue S and Gravenor MB, 2004. Risk of scrapie in British sheep of different prion protein genotype. Journal of General Virology, 85, 2735–2740. [DOI] [PubMed] [Google Scholar]
- Beck JA, Campbell TA, Adamson G, Poulter M, Uphill JB, Molou E, Collinge J and Mead S, 2008. Association of a null allele of SPRN with variant Creutzfeldt‐Jakob disease. Journal of Medical Genetics, 45, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Genoud N, Naumann H, Rülicke T, Janett F, Heppner FL, Ledermann B and Aguzzi A, 2002. Absence of the prion protein homologue Doppel causes male sterility. The EMBO Journal, 21, 3652–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellworthy SJ, Hawkins SAC, Green RB, Blamire I, Dexter G, Dexter I, Lockey R, Jeffrey M, Ryder S, Berthelin‐Baker C and Simmons MM, 2005. Tissue distribution of bovine spongiform encephalopathy infectivity in Romney sheep up to the onset of clinical disease after oral challenge. The Veterinary Record, 156, 197–202. [DOI] [PubMed] [Google Scholar]
- Belt PB, Muileman IH, Schreuder BEC, Bos‐de Ruijter J, Gielkens ALJ and Smits MA, 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. Journal of General Virology, 76, 509–517. [DOI] [PubMed] [Google Scholar]
- Benestad SL, Austbo L, Tranulis MA, Espenes A and Olsaker I, 2012. Healthy goats naturally devoid of prion protein. Veterinary Research, 43, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringue V, Vilotte JL and Laude H, 2008. Prion agent diversity and species barrier. Veterinary Research, 39, 47. [DOI] [PubMed] [Google Scholar]
- Bessen RA and Marsh RF, 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. Journal of Virology, 66, 2096–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacabe AG, Jacobs JG, Bencsik A, Langeveld JP and Baron TG, 2007. H‐type bovine spongiform encephalopathy: complex molecular features and similarities with human prion diseases. Prion, 1, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biljan I, Giachin G, Ilc G, Zhukov I, Plavec J and Legname G, 2012. Structural basis for the protective effect of the human prion protein carrying the dominant‐negative E219K polymorphism. The Biochemical Journal, 446, 243–251. [DOI] [PubMed] [Google Scholar]
- Billinis C, Panagiotidis CH, Psychas V, Argyroudis S, Nicolaou A, Leontides S, Papadopoulos O and Sklaviadis T, 2002. Prion protein gene polymorphisms in natural goat scrapie. Journal of General Virology, 83, 713–721. [DOI] [PubMed] [Google Scholar]
- Bishop MT, Hart P, Aitchison L, Baybutt HN, Plinston C, Thomson V, Tuzi NL, Head MW, Ironside JW, Will RG and Manson JC, 2006. Predicting susceptibility and incubation time of human‐to‐human transmission of vCJD. The Lancet Neurology, 5, 393–398. [DOI] [PubMed] [Google Scholar]
- Bossers A, Schreuder BE, Muileman IH, Belt PB and Smits MA, 1996. PrP genotype contributes to determining survival times of sheep with natural scrapie. Journal of General Virology, 77, 2669–2673. [DOI] [PubMed] [Google Scholar]
- Bossers A, Belt P, Raymond GJ, Caughey B, de Vries R and Smits MA, 1997. Scrapie susceptibility‐linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease‐resistant forms. Proceedings of the National Academy of Sciences of the United States, 94, 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossers A, de Vries R and Smits MA, 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. Journal of Virology, 74, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton K, Moore RC and Bishop SC, 2010. Associations of PrP genotype with lamb production traits in four commercial breeds of British upland and crossing sheep. Livestock Science, 127, 155–163. [Google Scholar]
- Bouzalas IG, Dovas CI, Banos G, Papanastasopoulou M, Kritas S, Oevermann A, Papakostaki D, Evangelia C, Papadopoulos O, Seuberlich T and Koptopoulos G, 2010. Caprine PRNP polymorphisms at codons 171, 211, 222 and 240 in a Greek herd and their association with classical scrapie. Journal of General Virology, 91, 1629–1634. [DOI] [PubMed] [Google Scholar]
- Bruce ME and Dickinson AG, 1987. Biological evidence that scrapie agent has an independent genome. The Journal of General Virology, 68(Pt. 1), 79–89. [DOI] [PubMed] [Google Scholar]
- Bucalossi C, Cosseddu G, D'Agostino C, Di Bari MA, Chiappini B, Conte M, Rosone F, De Grossi L, Scavia G, Agrimi U, Nonno R and Vaccari G, 2011. Assessment of the genetic susceptibility of sheep to scrapie by protein misfolding cyclic amplification and comparison with experimental scrapie transmission studies. Journal of Virology, 85, 8386–8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueler H, Raeber A, Sailer A, Fischer M, Aguzzi A and Weissmann C, 1994. High prion and PrPSc levels but delayed onset of disease in scrapie‐inoculated mice heterozygous for a disrupted PrP gene. Molecular Medicine, 1, 19–30. [PMC free article] [PubMed] [Google Scholar]
- Canon J, García D, García‐Atance MA, Obexer‐Ruff G, Lenstra JA, Ajmone‐Marsan P and Dunner S, 2006. Geographical partitioning of goat diversity in Europe and the Middle East. Animal Genetics 37, 327–334. [DOI] [PubMed] [Google Scholar]
- Cassard H, Torres JM, Lacroux C, Douet JY, Benestad SL, Lantier F, Lugan S, Lantier I, Costes P, Aron N, Reine F, Herzog L, Espinosa JC, Beringue V and Andreoletti O, 2014. Evidence for zoonotic potential of ovine scrapie prions. Nature Communications, 5, 5821. [DOI] [PubMed] [Google Scholar]
- Castilla J, Gutierrez Adan A, Brun A, Pintado B, Ramirez MA, Parra B, Doyle D, Rogers M, Salguero FJ, Sanchez C, Sanchez‐Vizcaino JM and Torres JM, 2003. Early detection of PrPres in BSE‐infected bovine PrP transgenic mice. Archives of Virology, 148, 677–691. [DOI] [PubMed] [Google Scholar]
- Castilla J, González‐Romero D, Saa P, Morales R, De Castro J and Soto C, 2008. Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell, 134, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelle PL, 1942. A case of scrapie in goats. Bulletin de l'Académie Vétérinaire de France, 15, 294–295. [Google Scholar]
- Clouscard C, Beaudry P, Elsen JM, Milan D, Dussaucy M, Bounneau C, Schelcher F, Chatelain J, Launay JM and Laplanche JL, 1995. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. Journal of General Virology, 76, 2097–2101. [DOI] [PubMed] [Google Scholar]
- Colussi S, Vaccari G, Maurella C, Bona C, Lorenzetti R, Troiano P, Casalinuovo F, Di Sarno A, Maniaci MG, Zuccon F, Nonno R, Casalone C, Mazza M, Ru G, Caramelli M, Agrimi U and Acutis PL, 2008. Histidine at codon 154 of the prion protein gene is a risk factor for Nor98 scrapie in goats. Journal of General Virology, 89, 3173–3176. [DOI] [PubMed] [Google Scholar]
- Corbière F, Barillet F, Andreoletti O, Fidelle F, Laphitz‐Bordet N, Schelcher F and Joly P, 2007. Advanced survival models for risk‐factor analysis in scrapie. Journal of General Virology, 88, 696–705. [DOI] [PubMed] [Google Scholar]
- Corbière F, Perrin‐Chauvineau C, Lacroux C, Costes P, Thomas M, Bremaud I, Martin S, Lugan S, Chartier C, Schelcher F, Barillet F and Andreoletti O, 2013. PrP‐associated resistance to scrapie in five highly infected goat herds. Journal of General Virology, 94, 241–245. [DOI] [PubMed] [Google Scholar]
- Dassanayake RP, White SN, Madsen‐Bouterse SA, Schneider DA and O'Rourke KI, 2015. Role of the PRNP S127 allele in experimental infection of goats with classical caprine scrapie. Animal Genetics, 46, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rancourt MFN, Lavín MP, Tchakérian E and Vallerand F, 2006. Mediterranean sheep and goats production: an uncertain future. Small Ruminant Reserach, 62, 167–179. [Google Scholar]
- De Vries F, Borchers N, Hamann H, Drögemüller C, Reinecke S, Lüpping W and Distl O, 2004. Associations between the prion protein genotype and performance traits of meat breeds of sheep. The Veterinary Record, 155, 140–143. [DOI] [PubMed] [Google Scholar]
- Detwiler LA and Baylis M, 2003. The epidemiology of scrapie. Revue scientifique et technique (International Office of Epizootics), 22, 121–143. [DOI] [PubMed] [Google Scholar]
- Dexter G, Tongue SC, Heasman L, Bellworthy SJ, Davis A, Moore SJ, Simmons MM, Sayers AR, Simmons HA and Matthews D, 2009. The evaluation of exposure risks for natural transmission of scrapie within an infected flock. BMC Veterinary Research, 5, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bari MA, Chianini F, Vaccari G, Esposito E, Conte M, Eaton SL, Hamilton S, Finlayson J, Steele PJ, Dagleish MP, Reid HW, Bruce M, Jeffrey M, Agrimi U and Nonno R, 2008. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. Journal of General Virology, 89, 2975–2985. [DOI] [PubMed] [Google Scholar]
- Dustan BH, Spencer YI, Casalone C, Brownlie J and Simmons MM, 2008. A histopathologic and immunohistochemical review of archived UK caprine scrapie cases. Veterinary Pathology Online, 45, 443–454. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2006. Opinion of the Scientific Panel on Biological Hazards on “the breeding programme for TSE resistance in sheep”. EFSA Journal 2006;4(7):382, 46 pp. 10.2903/j.efsa.2006.382 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2011a. Evaluation of the FoodEx, the food classification system applied to the development of the EFSA Comprehensive European Food Consumption Database. EFSA Journal 2011;9(3):1970, 27 pp. 10.2903/j.efsa.2011.1970 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2011b. Use of the EFSA Comprehensive European Food Consumption Database in Exposure Assessment. EFSA Journal 2011;9(3):2097, 34 pp. 10.2903/j.efsa.2011.2097 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012. Scientific and technical assistance on the provisional results of the study on genetic resistance to Classical scrapie in goats in Cyprus. EFSA Journal 2012;10(11):2972, 27 pp. 10.2903/j.efsa.2012.2972 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2014. Scientific Opinion on the scrapie situation in the EU after 10 years of monitoring and control in sheep and goats. EFSA Journal 2014;12(7):3781, 155 pp. 10.2903/j.efsa.2014.3781 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015. Scientific Opinion on a request for a review of a scientific publication concerning the zoonotic potential of ovine scrapie prions. EFSA Journal 2015;13(8):4197, 58 pp. 10.2903/j.efsa.2015.4197 [DOI] [Google Scholar]
- Eiden M, Soto EO, Mettenleiter TC and Groschup MH, 2011. Effects of polymorphisms in ovine and caprine prion protein alleles on cell‐free conversion. Veterinary Research, 42, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloit M, Adjou K, Coulpier M, Fontaine JJ, Hamel R, Lilin T, Messiaen S, Andreoletti O, Baron T, Bencsik A, Biacabe AG, Beringue V, Laude H, Le Dur A, Vilotte JL, Comoy E, Deslys JP, Grassi J, Simon S, Lantier F and Sarradin P, 2005. BSE agent signatures in a goat. The Veterinary Record, 156, 523–524. [DOI] [PubMed] [Google Scholar]
- Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, Khang JV, Poivey JP, Lantier F and Laplanche JL, 1999. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Archives of Virology, 144, 431–445. [DOI] [PubMed] [Google Scholar]
- FAO (Food and Agriculture Organization), 2012. Phenotypic characterization of animal genetic resources. FAO Animal Production and Health Guidelines No. 11. Rome.
- Fast C, Goldmann W, Berthon P, Tauscher K, Niedermeyer S, Andréoletti O, Lantier I, Rossignol C, Bossers A, Jacobs JG, Hunter N, Groschup MH, Lantier F and Langeveld JPM, in press. Protecting effect of PrP codons M142 and K222 in goats orally challenged with bovine spongiform encephalopathy prions. [DOI] [PMC free article] [PubMed]
- Fediaevsky A, Tongue SC, Noremark M, Calavas D, Ru G and Hopp P, 2008. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Veterinary Research, 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JD and Dickinson AG, 1988. The unusual properties of CH1641, a sheep‐passaged isolate of scrapie. The Veterinary Record, 123, 5–8. [DOI] [PubMed] [Google Scholar]
- Foster JD, Hope J and Fraser H, 1993. Transmission of bovine spongiform encephalopathy to sheep and goats. The Veterinary Record, 133, 339–341. [DOI] [PubMed] [Google Scholar]
- Foster JD, Bruce M, McConnell I, Chree A and Fraser H, 1996. Detection of BSE infectivity in brain and spleen of experimentally infected sheep. The Veterinary Record, 138, 546–548. [DOI] [PubMed] [Google Scholar]
- Foster JD, Parnham DW, Chong A, Goldman W and Hunter N, 2001a. Clinical signs, histopathology and genetics of experimental transmission of BSE and natural scrapie to sheep and goats. The Veterinary Record, 148, 165–171. [DOI] [PubMed] [Google Scholar]
- Foster JD, Parnham DW, Hunter N and Bruce M, 2001b. Distribution of the prion protein in sheep terminally affected with BSE following experimental oral transmission. Journal of General Virology, 82, 2319–2326. [DOI] [PubMed] [Google Scholar]
- Fragkiadaki EG, Vaccari G, Ekateriniadou LV, Agrimi U, Giadinis ND, Chiappini B, Esposito E, Conte M and Nonno R, 2011. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Veterinary Research, 42, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou S, Ortiz‐Pelaez A, Simmons MM, Dawson M, Windl O, Neocleous P and Papasavva‐Stylianou P, 2017. Goats with aspartic acid or serine at codon 146 of the PRNP gene remain scrapie‐negative after lifetime exposure in affected herds in Cyprus. Epidemiology and Infection 145, 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, Dawson M and Hunter N, 1996. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. Journal of General Virology, 77(Pt. 11), 2885–2891. [DOI] [PubMed] [Google Scholar]
- Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, Fernandez N, Saunders G, Windl O, González L, Bossers A and Foster J, 2011. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Veterinary Research, 42, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann W, Marier E, Stewart P, Konold T, Street S, Langeveld J, Windl O and Ortiz‐Pelaez A, 2016. Prion protein genotype survey confirms low frequency of scrapie‐resistant K222 allele in British goat herds. The Veterinary Record, 178, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González L, Martin S, Siso S, Konold T, Ortiz‐Pelaez A, Phelan L, Goldmann W, Stewart P, Saunders G, Windl O, Jeffrey M, Hawkins SA, Dawson M and Hope J, 2009. High prevalence of scrapie in a dairy goat herd: tissue distribution of disease‐associated PrP and effect of PRNP genotype and age. Veterinary Research, 40, 6. [DOI] [PubMed] [Google Scholar]
- González L, Sisó S, Monleon E, Casalone C, van Keulen LJ, Balkema‐Buschman A, Ortiz‐Pelaez A, Iulini B, Langeveld JPM, Hoffmann C, Badiola JJ, Jeffrey M and Acín C, 2010. Variability in disease phenotypes within a single PRNP genotype suggests the existence of multiple natural sheep scrapie strains within Europe. Journal of General Virology, 91, 2630–2641. [DOI] [PubMed] [Google Scholar]
- González L, Jeffrey M, Dagleish MP, Goldmann W, Sisó S, Eaton SL, Martin S, Finlayson J, Stewart P, Steele P, Pang Y, Hamilton S, Reid HW and Chianini F, 2012. Susceptibility to scrapie and disease phenotype in sheep: cross‐PRNP genotype experimental transmissions with natural sources. Veterinary Research, 43, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González L, Pitarch JL, Martin S, Thurston L, Simmons H, Acín C and Jeffrey M, 2014. Influence of polymorphisms in the prion protein gene on the pathogenesis and neuropathological phenotype of sheep scrapie after oral infection. Journal of Comparative Pathology, 150, 57–70. [DOI] [PubMed] [Google Scholar]
- Gossner AG, Bennet N, Hunter N and Hopkins J, 2009. Differential expression of Prnp and Sprn in scrapie infected sheep also reveals Prnp genotype specific differences. Biochemical and Biophysical Research Communications, 378, 862–866. [DOI] [PubMed] [Google Scholar]
- Green KM, Browning SR, Seward TS, Jewell JE, Ross DL, Green MA, Williams ES, Hoover EA and Telling GC, 2008. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. Journal of General Virology, 89, 598–608. [DOI] [PubMed] [Google Scholar]
- Griffiths PC, Spiropoulos J, Lockey R, Tout AC, Jayasena D, Plater JM, Chave A, Green RB, Simonini S, Thorne L, Dexter I, Balkema‐Buschmann A, Groschup MH, Beringue V, Le Dur A, Laude H and Hope J, 2010. Characterization of atypical scrapie cases from Great Britain in transgenic ovine PrP mice. Journal of General Virology, 91, 2132–2138. [DOI] [PubMed] [Google Scholar]
- Groschup MH and Buschmann A, 2008. Rodent models for prion diseases. Veterinary Research, 39, 1–13. [DOI] [PubMed] [Google Scholar]
- Groschup MH, Lacroux C, Buschmann A, Lühken G, Mathey J, Eiden M, Lugan S, Hoffmann C, Espinosa JC, Baron T and Torres JM, 2007. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerging Infectious Diseases, 13, 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbins S, Cook CJ, Hyder K, Boulton K, Davis C, Thomas E, Haresign W, Bishop SC, Villanueva B and Eglin RD, 2009. Associations between lamb survival and prion protein genotype: analysis of data for ten sheep breeds in Great Britain. BMC Veterinary Research, 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlow WJ, Kennedy RC, Race RE and Eklund CM, 1980. Virologic and neurohistologic findings in dairy goats affected with natural scrapie. Veterinary Pathology, 17, 187–199. [DOI] [PubMed] [Google Scholar]
- Healy AM, Hannon D, Morgan KL, Weavers E, Collins JD and Doherty ML, 2004. A paired case–control study of risk factors for scrapie in Irish sheep flocks. Preventive Veterinary Medicine, 64, 73–83. [DOI] [PubMed] [Google Scholar]
- Hoinville LJ, Tongue SC and Wilesmith JW, 2010. Evidence for maternal transmission of scrapie in naturally affected flocks. Preventive Veterinary Medicine, 93, 121–128. [DOI] [PubMed] [Google Scholar]
- Hourrigan J, Klingsporn A, Clark WW and De Camp M, 1979. Epidemiology of scrapie in the United States. Slow Transmissible Diseases of the Nervous System, 1, 331–356. [Google Scholar]
- Houston F, Goldmann W, Chong A, Jeffrey M, González L, Foster J, Parnham D and Hunter N, 2003. Prion diseases: BSE in sheep bred for resistance to infection. Nature, 423, 498. [DOI] [PubMed] [Google Scholar]
- Houston F, Goldmann W, Foster J, González L, Jeffrey M and Hunter N, 2015. Comparative susceptibility of sheep of different origins, breeds and PRNP genotypes to challenge with bovine spongiform encephalopathy and scrapie. PLoS ONE, 10, e0143251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N and Bossers A, 2006. The PrP genotype as a marker for scrapie susceptibility in sheep In: Hörnlimann B, Riesner D. and Kretzschmar HA. (eds.). Prions in Humans and Animals. De Gruyter, Berlin: pp. 640–643. [Google Scholar]
- Hunter N, Goldmann W, Benson G, Foster JD and Hope J, 1993. Swaledale sheep affected by natural scrapie differ significantly in PrP genotype frequencies from healthy sheep and those selected for reduced incidence of scrapie. Journal of General Virology, 74, 1025–1031. [DOI] [PubMed] [Google Scholar]
- Hunter N, Foster JD, Goldmann W, Stear MJ, Hope J and Bostock C, 1996. Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Archives of Virology, 141, 809–824. [DOI] [PubMed] [Google Scholar]
- Hunter N, Goldmann W, Foster JD, Cairns D and Smith G, 1997. Natural scrapie and PrP genotype: case‐control studies in British sheep. The Veterinary Record, 141, 137–140. [DOI] [PubMed] [Google Scholar]
- Hunter N, Houston F, Foster J, Goldmann W, Drummond D, Parnham D, Kennedy I, Green A, Stewart P and Chong A, 2012. Susceptibility of young sheep to oral infection with bovine spongiform encephalopathy decreases significantly after weaning. Journal of Virology 86, 11856–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huybrechts ISI, Boon PE, Ruprich J, Lafay L, Turrini A, Amiano P, Hirvonen T, De Neve M, Arcella DMJ, Westerlund A, Ribas‐Barba L, Hilbig A, Papoutsou S, Christensen T, Oltarzewski MVS, Rehurkova I, Azpiri M, Sette S, Kersting M, Walkiewicz A, Serra‐ Majem LVJ, Trolle E, Tornaritis M, Busk L, Kafatos A, Fabiansson S, De Henauw S and Van Klaveren JD, 2011. Dietary exposure assessments for children in Europe (the EXPOCHI project): rationale, methods and design. Archives of Public Health, 69, 1169–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JG, Sauer M, van Keulen LJ, Tang Y, Bossers A and Langeveld JP, 2011a. Differentiation of ruminant transmissible spongiform encephalopathy isolate types, including bovine spongiform encephalopathy and CH1641 scrapie. The Journal of General Virology, 92, 222–232. [DOI] [PubMed] [Google Scholar]
- Jacobs JG, Bossers A, Rezaei H, van Keulen LJM, McCutcheon S, Sklaviadis T, Lantier I, Berthon P, Lantier F, van Zijderveld FG and Langeveld JPM, 2011b. Proteinase K‐resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein. Journal of Virology, 85, 12537–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey M, Ryder S, Martin S, Hawkins SAC, Terry L, Berthelin‐Baker C and Bellworthy SJ, 2001. Oral inoculation of sheep with the agent of Bovine Spongiform Encephalopathy. 1. Onset and distribution of disease‐specific prp accumulation in brain and viscera. Journal Comparative Pathology, 124, 280–289. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Martin S, González L, Foster J, Langeveld JP, van Zijderveld FG, Grassi J and Hunter N, 2006. Immunohistochemical features of PrP(d) accumulation in natural and experimental goat transmissible spongiform encephalopathies. Journal of Comparative Pathology, 134, 171–181. [DOI] [PubMed] [Google Scholar]
- Johnson CJ, Herbst A, Duque‐Velasquez C, Vanderloo JP, Bochsler P, Chappell R and McKenzie D, 2011. Prion protein polymorphisms affect chronic wasting disease progression. PLoS ONE, 6, e17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanata E, Humphreys‐Panagiotidis C, Giadinis ND, Papaioannou N, Arsenakis M and Sklaviadis T, 2014. Perspectives of a scrapie resistance breeding scheme targeting Q211, S146 and K222 caprine PRNP alleles in Greek goats. Veterinary Research, 45, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanata E, Arsenakis M and Sklaviadis T, 2016. Caprine PrP variants harboring Asp‐146, His‐154 and Gln‐211 alleles display reduced convertibility upon interaction with pathogenic murine prion protein in scrapie infected cells. Prion 10, 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin RH and Walker CA, 1988. Incubation periods in six models of intraperitoneally injected scrapie depend mainly on the dynamics of agent replication within the nervous system and not the lymphoreticular system. Journal of General Virology, 69(Pt 12), 2953–2960. [DOI] [PubMed] [Google Scholar]
- Konold T, Bone GE, Phelan LJ, Simmons MM, González L, Sisó S, Goldmann W, Cawthraw S and Hawkins SA, 2010. Monitoring of clinical signs in goats with transmissible spongiform encephalopathies. BMC Veterinary Research, 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konold T, Simmons HA, Webb PR, Bellerby PJ, Hawkins SA and González L, 2013. Transmission of classical scrapie via goat milk. Veterinary Record, 172, 455. [DOI] [PubMed] [Google Scholar]
- Konold T, Thorne L, Simmons HA, Hawkins SA, Simmons MM and González L, 2016. Evidence of scrapie transmission to sheep via goat milk. BMC Veterinary Research, 12, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C, Perrin‐Chauvineau C, Corbière F, Aron N, Aguilar‐Calvo P, Torres JM, Costes P, Bremaud I, Lugan S, Schelcher F, Barillet F and Andreoletti O, 2014. Genetic resistance to scrapie infection in experimentally challenged goats. Journal of Virology, 88, 2406–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroux C, Cassard H, Simmons H, Douet JY, Corbière F, Lugan S, Coste P, Aron N, Huor A, Tillier C, Schelcher F and Andreoletti O, in press. Classical scrapie transmission in ARR/ARR genotype sheep. [DOI] [PubMed]
- Lampo E, Van Poucke M, Hugot K, Hayes H, Van Zeveren A and Peelman LJ, 2007. Characterization of the genomic region containing the Shadow of Prion Protein (SPRN) gene in sheep. BMC Genomics, 8, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld JPM, Jacobs JG, Hunter N, van Keulen LJM, Lantier F, van Zijderveld FG and Bossers A, 2016. Prion type‐dependent deposition of PRNP allelic products in heterozygous sheep. Journal of Virology, 90, 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dur A, Beringue V, Andreoletti O, Reine F, Lai TL, Baron T, Bratberg B, Vilotte JL, Sarradin P, Benestad SL and Laude H, 2005. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proceedings of the National Academy of Sciences of the United States of America, 102, 16031–16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dur A, Lai TL, Stinnakre MG, Laisne A, Chenais N, Rakotobe S, Passet B, Reine F, Soulier S, Herzog L, Tilly G, Rezaei H, Beringue V, Vilotte JL and Laude H, 2017. Divergent prion strain evolution driven by PrPC expression level in transgenic mice. Nature Communications, 8, 14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay JMK and Smith W, 1961. (referred by Dickinson, McKay and Zlotnik J Comp Pathol 1964, 74: 250‐254). A case of scrapie in an uninoculated goat–a natural occurrence or a contact infection. The Veterinary Record, 73, 394–396. [Google Scholar]
- Maestrale C, Cancedda MG, Pintus D, Masia M, Nonno R, Ru G, Carta A, Demontis F, Santucciu C and Ligios C, 2015. Genetic and pathological follow‐up study of goats experimentally and naturally exposed to a sheep scrapie isolate. Journal of Virology, 89, 10044–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I and Hope J, 1994. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Molecular Neurobiology, 8, 121–127. [DOI] [PubMed] [Google Scholar]
- McIntyre KM, Gubbins S, Goldmann W, Hunter N and Baylis M, 2008. Epidemiological characteristics of classical scrapie outbreaks in 30 sheep flocks in the United Kingdom. PLoS ONE, 3, e3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, Joiner S, Desbruslais M, Beck JA, O'Donoghue M, Lantos P, Wadsworth JD and Collinge J, 2007. Creutzfeldt‐Jakob disease, prion protein gene codon 129VV, and a novel PrPSc type in a young British woman. Archives of Neurology, 64, 1780–1784. [DOI] [PubMed] [Google Scholar]
- Meade‐White K, Race B, Trifilo M, Bossers A, Favara C, Lacasse R, Miller M, Williams E, Oldstone M, Race R and Chesebro B, 2007. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. Journal of Virology, 81, 4533–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten C, Ferrari P, Bakker M, Boss A, Hearty A, Leclercq C, Lindtner O, Tlustos C, Verger P, Volatier JL and Arcella D, 2011. Methodological characteristics of the national dietary surveys carried out in the European Union as included in the European Food Safety Authority (EFSA) Comprehensive European Food Consumption Database. Food Additives and Contaminants Part A, 28, 975–995. [DOI] [PubMed] [Google Scholar]
- Moore RC, Lee IY, Silverman GL, Harrison PM, Strome R, Heinrich C, Karunaratne A, Pasternak SH, Chishti MA, Liang Y, Mastrangelo P, Wang K, Smit AF, Katamine S, Carlson GA, Cohen FE, Prusiner SB, Melton DW, Tremblay P, Hood LE and Westaway D, 1999. Ataxia in prion protein (PrP)‐deficient mice is associated with upregulation of the novel PrP‐like protein doppel. Journal of Molecular Biology, 292, 797–817. [DOI] [PubMed] [Google Scholar]
- Moreno CR, Moazami‐Goudarzi K, Laurent P, Cazeau G, Andreoletti O, Chadi S, Elsen JM and Calavas D, 2007. Which PrP haplotypes in a French sheep population are the most susceptible to atypical scrapie? Archives of Virology, 152, 1229–1232. [DOI] [PubMed] [Google Scholar]
- Niedermeyer S, Eiden M, Toumazos P, Papasavva‐Stylianou P, Ioannou I, Sklaviadis T, Panagiotidis C, Langeveld J, Bossers A, Kuczius T, Kaatz M, Groschup MH and Fast C, 2016. Genetic, histochemical and biochemical studies on goat TSE cases from Cyprus. Veterinary Research, 47, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Miyazawa K, Masujin K and Yokoyama T, 2016. Coexistence of two forms of disease‐associated prion protein in extracerebral tissues of cattle infected with H‐type bovine spongiform encephalopathy. Journal of Veterinary Medical Science, 78, 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke KI, Holyoak GR, Clark WW, Mickelson JR, Wang S, Melco RP, Besser TE and Foote WC, 1997. PrP genotypes and experimental scrapie in orally inoculated Suffolk sheep in the United States. Journal of General Virology, 78, 975–978. [DOI] [PubMed] [Google Scholar]
- O'Rourke KI, Zhuang D, Truscott TC, Yan H and Schneider DA, 2011. Sparse PrP Sc accumulation in the placentas of goats with naturally acquired scrapie. BMC Veterinary Research, 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Pelaez A and Bianchini J, 2011. The impact of the genotype on the prevalence of classical scrapie at population level. Veterinary Research 42, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Pelaez A, Kelly L and Adkin A, 2012. The risk of introducing scrapie from restocking goats in Great Britain. Preventive Veterinary Medicine, 107, 222–230. [DOI] [PubMed] [Google Scholar]
- Ortiz‐Pelaez A, Georgiadou S, Simmons MM, Windl O, Dawson M, Arnold ME, Neocleous P and Papasavva‐Stylianou P, 2014. Allelic variants at codon 146 in the PRNP gene show significant differences in the risk for natural scrapie in Cypriot goats. Epidemiology and Infection, 143, 1304–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasavva‐Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P and Loucaides P, 2007. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Veterinary Journal, 173, 459–462. [DOI] [PubMed] [Google Scholar]
- Papasavva‐Stylianou P, Windl O, Saunders G, Mavrikiou P, Toumazos P and Kakoyiannis C, 2011. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie. Veterinary Journal, 187, 245–250. [DOI] [PubMed] [Google Scholar]
- Pattison IH and Millson GC, 1961. Scrapie produced experimentally in goats with special reference to the clinical syndrome. Journal of Comparative Pathology, 71, 101–109. [DOI] [PubMed] [Google Scholar]
- Pattison IH, Hoare MN, Jebbett JN and Watson WA, 1972. Spread of scrapie to sheep and goats by oral dosing with foetal membranes from scrapie‐affected sheep. Veterinary Record, 90, 465–468. [DOI] [PubMed] [Google Scholar]
- Peletto S, Bertolini S, Maniaci MG, Colussi S, Modesto P, Biolatti C, Bertuzzi S, Caramelli M, Maurella C and Acutis PL, 2012. Association of an indel polymorphism in the 3′UTR of the caprine SPRN gene with scrapie positivity in the central nervous system. Journal of General Virology, 93, 1620–1623. [DOI] [PubMed] [Google Scholar]
- Pirisinu L, Marcon S, Di Bari MA, D'Agostino C, Agrimi U and Nonno R, 2013. Biochemical characterization of prion strains in bank voles. Pathogens, 2, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priem J, Langeveld JP, van Keulen LJ, van Zijderveld FG, Andreoletti O and Bossers A, 2014. Enhanced virulence of sheep‐passaged bovine spongiform encephalopathy agent is revealed by decreased polymorphism barriers in prion protein conversion studies. Journal of Virology, 88, 2903–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard TC, Cahalan CM and Ap Dewi I, 2008. Association between PrP genotypes and performance traits in a Welsh Mountain flock. Animal, 2, 1421–1426. [DOI] [PubMed] [Google Scholar]
- Ryder S, Dexter G, Bellworthy S and Tongue S, 2004. Demonstration of lateral transmission of scrapie between sheep kept under natural conditions using lymphoid tissue biopsy. Research in Veterinary Science, 76, 211–217. [DOI] [PubMed] [Google Scholar]
- Saborio GP, Permanne B and Soto C, 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature, 411, 810–813. [DOI] [PubMed] [Google Scholar]
- Sawalha RM, Villanueva B, Brotherstone S, Rogers PL and Lewis RM, 2010. Prediction of prion protein genotype and association of this genotype with lamb performance traits of Suffolk sheep. Journal of Animal Science, 88, 428–434. [DOI] [PubMed] [Google Scholar]
- Schneider DA, Madsen‐Bouterse SA, Zhuang D, Truscott TC, Dassanayake RP and O'Rourke KI, 2015. The placenta shed from goats with classical scrapie is infectious to goat kids and lambs. Journal of General Virology, 96, 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel B, Thomzig A, Buschmann A, Groschup MH, Peters R, Beekes M and Terytze K, 2007. Scrapie agent (strain 263 K) can transmit disease via the oral route after persistence in soil over years. PLoS ONE, 2, e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuberlich T, Botteron C, Benestad SL, Brunisholz H, Wyss R, Kihm U, Schwermer H, Friess M, Nicolier A, Heim D and Zurbriggen A, 2007. Atypical scrapie in a Swiss goat and implications for transmissible spongiform encephalopathy surveillance. Journal of Veterinary Diagnostic Investigation, 19, 2–8. [DOI] [PubMed] [Google Scholar]
- Simmons MM, Moore S, Konold T, Thurston L, Terry LA, Thorne L, Lockey R, Vickery C, Hawkins SAC, Chaplin MJ and Spiropoulos J, 2011. Experimental oral transmission of atypical scrapie to sheep. Emerging Infectious Diseases, 17, 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MM, Moore SJ, Lockey R, Chaplin MJ, Konold T, Vickery C and Spiropoulos J, 2015. Phenotype shift from atypical scrapie to CH1641 following experimental transmission in sheep. PLoS ONE, 10, e0117063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Nugier J, Morel N, Boutal H, Creminon C, Benestad SL, Andreoletti O, Lantier F, Bilheude JM, Feyssaguet M, Biacabe AG, Baron T and Grassi J, 2008. Rapid typing of transmissible spongiform encephalopathy strains with differential ELISA. Emerging Infectious Diseases, 14, 608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofianidis G, Psychas V, Billinis C, Spyrou V, Argyroudis S, Papaioannou N and Vlemmas I, 2006. Histopathological and immunohistochemical features of natural goat scrapie. Journal of Comparative Pathology, 135, 116–129. [DOI] [PubMed] [Google Scholar]
- Soto C, Saborio GP and Anderes L, 2002. Cyclic amplification of protein misfolding: application to prion‐ related disorders and beyond. Trends in Neurosciences, 25, 390–394. [DOI] [PubMed] [Google Scholar]
- Spiropoulos J, Lockey R, Sallis RE, Terry LA, Thorne L, Holder TM, Beck KE and Simmons MM, 2011. Isolation of prion with BSE properties from farmed goat. Emerging Infectious Diseases, 17, 2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srithayakumar V, Mitchell GB and White BN, 2016. Identification of amino acid variation in the prion protein associated with classical scrapie in Canadian dairy goats. BMC Veterinary Research, 12, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SSC , 1999. Scientific Opinion on the policy of breeding and genotyping of sheep on the policy of breeding and genotyping of sheep, i.e. the issue of whether sheep should be bred to be resistant to scrapie, adopted by the Scientific Steering Committee at its meeting of 22–23 July 1999.
- SSC , 2002. Opinion on safe sourcing of small ruminant materials (safe sourcing of small ruminant materials should BSE in small ruminants become probable: genotype, breeding, rapid TSE testing, flocks certification and specified risk materials), adopted by the Scientific Steering Committee at its meeting of 4–5 April 2002.
- Stamp JT, 1962. Scrapie: a transmissible disease of sheep. The Veterinary Record, 74, 357–362. [Google Scholar]
- Stewart P, Shen C, Zhao D and Goldmann W, 2009. Genetic analysis of the SPRN gene in ruminants reveals polymorphisms in the alanine‐rich segment of shadoo protein. Journal of General Virology, 90, 2575–2580. [DOI] [PubMed] [Google Scholar]
- Tabouret G, Lacroux C, Lugan S, Costes P, Corbière F, Weisbecker JL, Schelcher F and Andréoletti O, 2010. Relevance of oral experimental challenge with classical scrapie in sheep. Journal of General Virology 91, 2139–2144. [DOI] [PubMed] [Google Scholar]
- Taema MM, Maddison BC, Thorne L, Bishop K, Owen J, Hunter N, Baker CA, Terry LA and Gough KC, 2012. Differentiating ovine BSE from CH1641 scrapie by serial protein misfolding cyclic amplification. Molecular Biotechnology, 51, 233–239. [DOI] [PubMed] [Google Scholar]
- Thackray AM, Hopkins L, Lockey R, Spiropoulos J and Bujdoso R, 2011. Emergence of multiple prion strains from single isolates of ovine scrapie. Journal of General Virology, 92, 1482–1491. [DOI] [PubMed] [Google Scholar]
- Thackray AM, Lockey R, Beck KE, Spiropoulos J and Bujdoso R, 2012. Evidence for co‐infection of ovine prion strains in classical scrapie isolates. Journal of Comparative Pathology, 147, 316–329. [DOI] [PubMed] [Google Scholar]
- Thorne L, Holder T, Ramsay A, Edwards J, Taema MM, Windl O, Maddison BC, Gough KC and Terry LA, 2012. In vitro amplification of ovine prions from scrapie‐infected sheep from Great Britain reveals distinct patterns of propagation. BMC Veterinary Research, 8, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toumazos P and Alley MR, 1989. Scrapie in goats in Cyprus. New Zealand Veterinary Journal, 37, 160–162. [DOI] [PubMed] [Google Scholar]
- Tuzi NL, Gall E, Melton D and Manson JC, 2002. Expression of Doppel in the CNS of mice does not modulate transmissible spongiform encephalopathy disease. Journal of General Virology, 83, 705–711. [DOI] [PubMed] [Google Scholar]
- Ulvund MJ, 2006. Clinical findings in scrapie In: Hörnlimann B, Riesner D. and Kretzschmar H. (eds.). Prions in Humans and Animals. De Gruyter, Berlin: pp. 398–407. [Google Scholar]
- Vaccari G, Petraroli R, Agrimi U, Eleni C, Perfetti MG, di Bari MA, Morelli L, Ligios C, Busani L, Nonno R and Di Guardo G, 2001. PrP genotype in Sarda breed sheep and its relevance to scrapie. Archives of Virology, 146, 2029–2037. [DOI] [PubMed] [Google Scholar]
- Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palazzini N, Troiano P, Petrella A, Di Guardo G and Agrimi U, 2006. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. Journal of General Virology, 87, 1395–1402. [DOI] [PubMed] [Google Scholar]
- Vaccari G, Scavia G, Sala M, Cosseddu G, Chiappini B, Conte M, Esposito E, Lorenzetti R, Perfetti G, Marconi P and Scholl F, 2009. Protective effect of the AT137RQ and ARQK176 PrP allele against classical scrapie in Sarda breed sheep. Veterinary Research, 40, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilotte JL, Soulier S, Essalmani R, Stinnakre MG, Vaiman D, Lepourry L, Da Silva JC, Besnard N, Dawson M, Buschmann A and Groschup M, 2001. Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. Journal of Virology, 75, 5977–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale M, Migliore S, La Giglia M, Alberti P, Di Marco Lo Presti V and Langeveld JP, 2016. Scrapie incidence and PRNP polymorphisms: rare small ruminant breeds of Sicily with TSE protecting genetic reservoirs. BMC Veterinary Research, 12, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth JD, Asante EA and Collinge J, 2010. Review: contribution of transgenic models to understanding human prion disease. Neuropathology and Applied Neurobiology, 36, 576–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wan J, Wang W, Wang D, Li S, Liao P, Hao Z, Wu S, Xu J, Li N, Ouyang H and Gao H, 2011. Overexpression of Shadoo protein in transgenic mice does not impact the pathogenesis of scrapie. Neuroscience Letters, 496, 1–4. [DOI] [PubMed] [Google Scholar]
- Watts JC and Westaway D, 2007. The prion protein family: diversity, rivalry, and dysfunction. Biochimica et Biophysica Acta: Protein Structure and Molecular Enzymology, 1772, 654–672. [DOI] [PubMed] [Google Scholar]
- Watts JC, Stohr J, Bhardwaj S, Wille H, Oehler A, Dearmond SJ, Giles K and Prusiner SB, 2011. Protease‐resistant prions selectively decrease Shadoo protein. PLoS Pathogens, 7, e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway D, Goodman PA, Mirenda CA, McKinley MP, Carlson GA and Prusiner SB, 1987. Distinct prion proteins in short and long scrapie incubation period mice. Cell, 51, 651–662. [DOI] [PubMed] [Google Scholar]
- White SN, Reynolds JO, Waldron DF, Schneider DA and O'Rourke KI, 2012. Extended scrapie incubation time in goats singly heterozygous for PRNP S146 or K222. Gene, 501, 49–51. [DOI] [PubMed] [Google Scholar]
- Windig JJ, Eding H, Moll L and Kaal L, 2004. Effects on inbreeding of different strategies aimed at eliminating scrapie sensitivity alleles in rare sheep breeds in The Netherlands. Animal Science, 79, 11–20. [Google Scholar]
- Windig JJ, Hoving RA, Priem J, Bossers A, Keulen LJ and Langeveld JP, 2016. Variation in the prion protein sequence in Dutch goat breeds. Journal of Animal Breeding and Genetics, 133, 366–374. [DOI] [PubMed] [Google Scholar]
- Zlotnik I and Rennie JC, 1963. Further observations on the experimental transmission of scrapie from sheep and goats to laboratory mice. Journal of Comparative Pathology, 73, 150–162. [DOI] [PubMed] [Google Scholar]


