Abstract
EFSA is requested to assess the safety of a broad range of biological agents in the context of notification for market authorisation as sources of food and feed additives, food enzymes and plant protection products. The qualified presumption of safety (QPS) assessment was developed to provide a harmonised generic pre‐assessment to support safety risk assessments performed by EFSA's scientific Panels. The safety of unambiguously defined biological agents (at the highest taxonomic unit appropriate for the purpose for which an application is intended), and the completeness of the body of knowledge are assessed. Identified safety concerns for a taxonomic unit are, where possible and reasonable in number, reflected as ‘qualifications’ in connection with a recommendation for a QPS status. The list of QPS recommended biological agents was reviewed and updated in the current opinion and therefore becomes the valid list. The 2016 update reviews previously assessed microorganisms including bacteria, yeasts and viruses used for plant protection purposes following an Extensive Literature Search strategy. The taxonomic units related to the new notifications received since the 2013 QPS opinion, were periodically evaluated for a QPS status and the results published as Statements of the BIOHAZ Panel. Carnobacterium divergens, Lactobacillus diolivorans, Microbacterium imperiale, Pasteuria nishizawae, Pediococcus parvulus, Bacillus flexus, Bacillus smithii, Xanthomonas campestris and Candida cylindracea were recommended for the QPS list. All taxonomic units previously recommended for the 2013 QPS list had their status reconfirmed as well their qualifications with the exception of Pasteuria nishizawae for which the qualification was removed. The exclusion of filamentous fungi and enterococci from the QPS evaluations was reconsidered but monitoring will be maintained and the status will be re‐evaluated in the next QPS Opinion update. Evaluation of bacteriophages should remain as a case‐by‐case procedure and should not be considered for QPS status.
Keywords: safety, QPS, food and feed, bacteria, yeast, fungi, virus
Short abstract
This publication is linked to the following EFSA Journal article: http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2017.4663/full, http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2017.e15031/full
Summary
The European Food Safety Authority (EFSA) asked the Panel on Biological Hazards (BIOHAZ) to deliver a Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food or feed. The request included three specific tasks as described in the Terms of Reference (ToR).
In 2014, the BIOHAZ Panel decided to change the evaluation procedure: instead of publishing the overall assessment of the taxonomic units previously recommended for the QPS list annually as until 2013, it is now carried out every 3 years in a Scientific Opinion of the BIOHAZ Panel (December 2016). Meanwhile, that list of microorganisms has been maintained based on the evaluation of extensive literature reviews that will be updated regularly with new publications. When an assessment for a QPS recommendation of a microbiological agent notified to EFSA is requested by the Feed Unit, the Food Ingredients and Packaging (FIP) Unit, the Nutrition Unit or the Pesticides Unit, the respective evaluations for a QPS status have been compiled and published in Panel Statements every 6 months.
The first ToR requires to keep updated the list of biological agents being notified, in the context of a technical dossier to EFSA Units (such as Feed, Food Ingredients and Packaging (FIP), Nutrition, and Pesticides), for intentional use in feed and/or food or as sources of food and feed additives, enzymes and plant protection products for safety assessment. The list has been updated with the notifications received since May of 2013 until September of 2016. The notifications received within every 6‐month period have been included in a table appended to each respective Panel Statement (five in total within this period). These new notifications were also included in Appendix E of this Opinion, compiling all microorganism notified to EFSA from the beginning of the QPS exercise in 2007. From the last notifications included in the previous QPS Opinion in 2013, 405 notifications were received between May 2013 and September 2016, of which, 137 were from Feed, 196 from FIP, 11 from Nutrition and 61 from Pesticides. For the type of microorganisms, 183 were bacteria, 177 filamentous fungi, 9 viruses and 36 yeasts TUs.
The second ToR concerns the revision of the taxonomic units previously recommended for the QPS list and their qualifications (especially the qualification regarding antimicrobial resistance (AMR)) when new information has become available and to update the information provided in the previous Opinion published in November 2013 where appropriate. The work being developed in order to meet this ToR is reflected in the current Scientific Opinion of the BIOHAZ Panel. The 2016 update reviews previously assessed microorganisms including bacteria, yeasts and viruses used for plant protection purposes following an Extensive Literature Search strategy. The list of QPS recommended biological agents was reviewed and updated in the current opinion, and therefore becomes the valid list. Information about AMR has been reviewed following recent recommendations from EFSA Opinions published in this topic.
The third ToR requires a (re)assessment of the suitability of taxonomic units notified to EFSA not present in the current QPS list for their inclusion in the updated list. The taxonomic units related to the new notifications received since the 2013 QPS opinion, were periodically evaluated for a QPS status and the results published as Statements of the BIOHAZ Panel. They have also been included in this update so that all the information about QPS microorganisms is available in a single document. Carnobacterium divergens, Lactobacillus diolivorans, Microbacterium imperiale, Pasteuria nishizawae, Pediococcus parvulus, Bacillus flexus, Bacillus smithii, Xanthomonas campestris and Candida cylindracea were recommended for the QPS list. All taxonomic units previously recommended for the 2013 QPS list had their status reconfirmed as well their qualifications with the exception of Pasteuria nishizawae for which the qualification was removed.
The QPS concept as a pre‐assessment approach for use within EFSA, that can be applied to the requests received for a safety assessment of microorganisms deliberately introduced into the food and feed chain, is discussed and refined in the light of the changes of the specific regulatory framework of the different areas covered by EFSA when dealing with those types of microorganisms. In that context, the recent experience while incorporating the QPS assessment into each specific EFSA's safety risk assessments area is described. The data and methodologies are further described, including the Extensive Literature Search approach, the verification of the identity of the main taxonomic units groups, the evaluation of the body of knowledge of the safety concerns and the possible influence of the end use of the microorganism. The workflow diagrams of the QPS process at different levels are presented.
All taxonomic units previously recommended for the 2013 QPS list had their status reconfirmed as well their qualifications with the exception of Pasteuria nishizawae for which the qualification was removed. The exclusion of filamentous fungi and enterococci from the QPS evaluations was reconsidered but monitoring will be maintained and the status will be re‐evaluated in the next QPS Opinion update. Evaluation of bacteriophages should remain as a case‐by‐case procedure and should not be considered for QPS status.
1. Introduction
1.1. Background and Terms of Reference as provided by EFSA
A wide variety of microorganisms are intentionally added at different stages into the food chain, either directly or as a source of additives or food enzymes. The European Food Safety Authority (EFSA) is requested to assess the safety of these biological agents in the context of applications for market authorisation as sources of food and feed additives, food enzymes and plant protection products.
The Scientific Committee reviewed the range and numbers of microorganisms likely to be the subject of an EFSA Opinion and in 2007 published a list of microorganisms recommended for Qualified Presumption of Safety (QPS list),1 , 2 status, consisting of 48 species of Gram‐positive non‐sporulating bacteria, 13 Bacillus species and 11 yeast species. Filamentous fungi were also assessed but these were not recommended for QPS status. The Scientific Committee recommended that a QPS approach should be implemented across EFSA and applied equally to all safety considerations of microorganisms that EFSA is required to assess. The Scientific Committee recognised that there would have to be continuing provision for reviewing and updating the QPS list. The EFSA Panel on Biological Hazards (BIOHAZ) took the prime responsibility for this and annually reviewed the existing QPS list, as recommended by the Scientific Committee.
In the first annual QPS review and update,3 the existing QPS list was reviewed and EFSA's initial experience in applying the QPS approach was described. The potential application of the QPS approach to microbial plant protection products was discussed in the 2009 review.4 In 2009, viruses and bacteriophages were assessed for the first time, leading to the addition of two virus families used for plant protection purposes to the QPS list. Bacteriophages were not considered appropriate for the QPS list. After consecutive years of updating the existing scientific knowledge, the filamentous fungi (2008–2013 updates) and enterococci (2010–2013 updates) were not recommended for the QPS list.
The 2013 update of the QPS list includes 53 species of Gram‐positive non‐sporulating bacteria, 13 Gram‐positive spore‐forming bacteria (Bacillus species), 1 Gram‐negative bacteria (Gluconobacter oxydans), 13 yeast species and 3 virus families. No QPS recommended species has been removed from the list following six (2008–2013 updates) annual reviews.
Based on the above‐mentioned information, the BIOHAZ Panel at their plenary meeting in January 2014, made a proposal for future QPS activities that was discussed at the Scientific Committee meeting in February 2014. The Scientific Committee agreed to exclude some biological groups (filamentous fungi, bacteriophages and enterococci) in future QPS activities, while the Extensive Literature Review of the QPS recommended list could be done less frequently. The deadline for the assessment of the suitability of new taxonomic units (TUs) notified to EFSA for inclusion in the QPS list would be tailored to the needs of the requesting EFSA Units and/or Scientific Panels.
ToR 1: Keep updated the list of biological agents being notified in the context of a technical dossier to EFSA Units (such as Feed, Pesticides, Food Ingredients and Packing, and Nutrition) for intentional use in feed and/or food or as sources of food and feed additives, enzymes and plant protection products for safety assessment.
ToR 2: Review taxonomic units previously recommended for the QPS list and their qualifications (especially the qualification regarding antimicrobial resistance) when new information has become available. Update the information provided in the previous opinion where appropriate.
ToR 3: (Re)assess the suitability of taxonomic units notified to EFSA not present in the current QPS list for their inclusion in that list.
1.2. Interpretation of the Terms of Reference
1.2.1. Background to the QPS assessment approach
The QPS approach was developed by the Scientific Committee of EFSA to provide a generic concept to prioritise and to harmonise risk assessment of microorganisms intentionally introduced into the food chain, in support of the respective Scientific Panels and Units in the frame of market authorisations (EFSA, 2007; Leuschner et al., 2010). The list of QPS recommended biological agents, first established in 2007, has been updated annually until 2013. Taxonomic units (TUs) (usually species for bacteria and yeasts, families for viruses) were included in the QPS list either following notifications to EFSA or proposals made by stakeholders during a public consultation in 2005, even if they were not yet notified to EFSA (EFSA, 2005). Since then and currently, the QPS assessment is only triggered when a microorganism is notified to EFSA through an application for market authorisation of regulated products (such as feed additives, food ingredients as food enzymes, novel foods and plant protection products).5
The QPS concept was first formulated by a joint working group of a number of Scientific Advisory Committees of the European Commission and placed on the website of the European Commission – Health and Food Safety Directorate General (EC – DG SANCO) in 2003.6 This concept was then developed to serve within EFSA as a tool for assessing the safety of microorganisms introduced deliberately in the food chain, obviating the need of unnecessary testing. The approaches for assessing the safety of microorganisms entering the human food chain differ considerably depending on the legislation, if any, applicable. In the view of EFSA, QPS represents a route to harmonisation of risk assessment approaches within EFSA which allows additional safety concerns (e.g. transmissible antimicrobial resistance (AMR)) to be addressed. Moreover, QPS is a pragmatic approach to risk assessment that focuses on the hazards associated with a specific microbial species and could avoid redoing unnecessary testing for already demonstrated evidence data knowledge, and therefore could allow a better use of resources without compromising safety.
A wide variety of microorganisms are intentionally used at different stages in the food chain, either directly or as a source of food and feed additives, food enzymes or used as plant protection products. In the context of applications for market authorisation of these biological agents, EFSA is requested to assess their safety. In scientific publications, the QPS system has often been misinterpreted (Songisepp et al., 2012) as the European counterpart to the Generally Recognised As Safe (GRAS) system, established by the Food and Drug Administration (FDA) in the United States. There are certain important differences between the two systems. The GRAS guidelines apply to food additives in general, whereas QPS is dedicated to microorganisms only. GRAS also concerns a specific substance or organism, i.e. it is not applicable for a whole microbial TU like the QPS system. From the opposite perspective, QPS is not applicable to single products containing a specific microbial strain, but for a TU, usually species level for bacteria and yeasts, families for viruses. As an example, Bifidobacterium longum evaluation: in the GRAS system, approval would be granted for a specific strain like B. longum strain XYZ while in the QPS system it would be applicable to the whole B. longum species. If a TU does not get QPS status, it still can get the approval after full assessment at the strain level within the respective EFSA Panel/Unit. Further details can be found in Table 1.
Table 1.
Differences between the GRAS guidelines (FDA, USA) and the QPS system (EFSA, EU) (amended from Wassenaar and Klein, 2008)
| GRAS | QPS |
|---|---|
| Applies to food additives including microorganisms | Applies to microorganisms only |
| Performed after a specific GRAS notification to the FDA | Performed for microorganisms used as a source of/contained in products assessed for the EU market authorisation |
| Determination of a GRAS status by the FDA and/or external experts | Determination of a QPS status by EFSA |
| Open to all types of food additives | Restricted only to the microorganisms related to regulated food and feed products |
| Applicants request a GRAS status | EFSA requests evaluation of new taxonomic units within the scope of an internal mandate |
| Based on history of use, body of knowledge and the absence of adverse effects at the strain level | Based on history of use, body of knowledge and the absence of adverse effects at the TU level |
| Describes specific substance or microorganism at the strain level | Describes taxonomic unit (usually species level for bacteria and yeasts, families for viruses, not at strain level) |
| Case‐by‐case safety assessment at the strain level | General safety assessment at the TU level |
| Based on specific Guidancea | Support to the safety assessment required in the Founding EU Regulationb |
| Open tool to all applicants | Internal tool only under the frame of dossiers for authorisation of regulated products by EFSA |
FDA: US Food and drug Administration; GRAS: Generally Recognised as Safe; QPS: Qualified Presumption of Safety; TU: taxonomic unit.
Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. OJ L 31, 1.2.2002, p. 1–24.
QPS entered into the European Union (EU) law with the publication of a new Commission Implementing Regulation (EU) No 562/20127 amending Commission Regulation (EU) No 234/20118 with regard to specific data required for risk assessment of food enzymes. If the microorganism used in the production of a food enzyme has a QPS status, according to the most recent list of QPS recommended biological agents adopted by the Authority (meaning EFSA), the food enzyme application could not need to provide specific toxicological test data. It should be noted that if residues, impurities, degradation products linked to the production and downstream process to obtain the food enzyme as defined in the legislation could give rise to concern, the Authority, pursuant to Article 6(1) of Regulation (EC) No 1331/20089 may request additional data for risk assessment, including toxicological test data. Specifications and scientific data needed according to a case‐by‐case basis assessment are detailed in the ‘Explanatory Note for the Guidance of the Scientific Panel of Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF) on the Submission of a Dossier on Food Enzymes'.10
1.2.2. New approach related to the present QPS mandate
In 2014, the BIOHAZ Panel decided to change the evaluation procedure: the publication of the overall assessment of the TUs previously recommended for the QPS list (EFSA BIOHAZ Panel, 2013) would be carried out after 3 years in a Scientific Opinion of the BIOHAZ Panel (the current opinion). In the meantime, that list of microorganisms would be maintained and frequently checked based on the evaluation of Extensive Literature Searches that would be regularly updated. Intermediate deliverables in the form of a Panel Statement would be produced and published, should an assessment for a QPS status of a microbiological agent notified to EFSA be requested by any other EFSA Unit, e.g. the Feed Unit, the Food Ingredients and Packaging (FIP) Unit, the Nutrition Unit or by the Pesticides Unit. Evaluations of these notifications are compiled in Panel Statements every 6 months. The conclusions of these Statements are included in this Scientific Opinion.
The Scientific Committee agreed to exclude some biological groups (filamentous fungi, bacteriophages and enterococci) notified to EFSA from the regular QPS assessment (66th plenary, 18–19 February 2014). The reason for this exclusion was that it was considered unlikely that any taxonomical units within these groups would be granted a QPS status in the foreseeable future. Thus, the assessment should be done at the strain level and therefore on a case‐by‐case basis, and should be done by the relevant EFSA Units.
EFSA asked the BIOHAZ Panel to deliver a Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food or feed (2013 update). The question included three specific tasks in the ToRs.
The first ToR required to keep updated the list of biological agents being notified, in the context of a technical dossier to EFSA Units (such as Feed, FIP, Nutrition and Pesticides), for intentional use in feed and/or food or as sources of food and feed additives, food enzymes and plant protection products for safety assessment. The notifications considered for each Panel Statement (from December 2014 until December 2016) have been published in each respective appendix. The previous list (published with the QPS 2103 update Opinion) has been updated with the corresponding notifications received between May 2013 and September 2016 (see Appendix E).
The second ToR concerns the revision of the TUs previously recommended for the QPS list and their qualifications (especially the qualification regarding AMR) when new information has become available and to update the information provided in the previous opinion (EFSA BIOHAZ Panel, 2013) where appropriate. For TUs on the QPS list, this update of the literature aims at verifying if any new safety concern has arisen that could require the removal of the TU from the list, and to verify if the qualifications still efficiently exclude safety concerns. If such a situation would have happened before the publication of the current Opinion, a Panel Statement would have been published with the explanation of the reason that lead to the exclusion of a TU or the change in a qualification. At the same time, the QPS Opinion from 2013 would have been properly changed and an erratum included. The work being developed in order to reply to this ToR is reflected in the current Opinion.
The third ToR required a (re)assessment of the suitability of TUs notified to EFSA not present in the current QPS list for their inclusion in the updated list. The current Opinion takes into consideration the outcome from the several Panel Statements published from December 2014 where the evaluation of those TUs was included. The notifications received within that period and respective evaluation for a QPS status of the TU associated have been included in Appendix E together with the previous notifications and respective evaluations. The new recommendations for a QPS status have been included in the current QPS list (Appendix A).
1.3. Additional information
1.3.1. QPS: an assessment approach for use within EFSA
QPS as a concept provides a generic safety pre‐assessment approach for use within EFSA that could be applied to all requests received by EFSA for the safety assessments of microorganisms deliberately introduced into the food and feed chain. The assessment covers risks for humans, animals and the environment. Its introduction harmonises and makes the risk assessment approach more transparent across the EFSA Scientific Panels and Units. It improves the consistency of assessments and makes better use of resources by focussing on those organisms that present the greatest risks or uncertainties (EFSA, 2005, 2009).
In the QPS concept, a safety assessment of a defined TU is considered independently of any particular specific notification in the course of an authorisation process, whenever possible. If the TU does not raise any safety concerns, or if existing safety concerns related to this TU can be clearly identified and excluded at a strain level (qualifications), a particular TU could be recommended for the QPS list. Subsequently, any specific representative of a QPS proposed TU, would not need to undergo a further safety assessment other than to satisfy any of the qualifications specified if applicable and if not required by a specific EU regulation framework. Representatives of TUs that fail to satisfy a qualification would be considered unfit for the QPS list and would remain subject to a full safety assessment, in the frame of a notification submitted to the responsible EFSA Scientific Panel/Unit (EFSA, 2007).
The QPS concept does not address hazards linked to the formulation or other processing of the products containing the microbial agents and added into the food or feed chain. Although general human safety is part of the evaluation, specific issues connected to type and level of exposure of users handling the product (e.g. dermal, inhalation, ingestion) are not addressed. Assessment of potential allergenicity to microbial residual components is beyond the QPS remit; nevertheless, in cases where there is science‐based evidence for allergenicity, it will be reported. These aspects are assessed, where applicable, separately by the EFSA Panel responsible for assessing the notification.
QPS is independent of the level of exposure. The latter is strictly related to the amount of microorganisms intentionally used in the food chain. Sometimes, a qualification ‘for production purpose only’ may apply to TUs used for the biosynthesis of specific products used in the food chain and under specific regulation. In this case, the QPS recommendation may only apply to this specific end use not including living organisms, e.g. certain enzymes, vitamins or amino acid production. This specific consideration of end use does not conflict with the generic applicability of QPS, because in this case the end use corresponds to different hazards (living organisms versus dead cells or their metabolites).
Concerning microorganisms evaluated for QPS status in previous Opinions, the continuously evolving body of knowledge possibly reveals new information that could lead to a modification of the list of QPS recommended TUs, for example to an ex‐ or inclusion of TUs on the list. Assessments of new TUs, not previously considered for the QPS list, and for which representatives are notified to EFSA are included. Microorganisms intended for usages outside the remit of EFSA, and those that have not been notified to EFSA, are not considered in this Opinion.
Acquired AMR was introduced as a possible safety concern for the assessment of the inclusion of bacterial species in the QPS list published in 2008 (EFSA, 2008). In the 2009 QPS Opinion (EFSA BIOHAZ Panel, 2009) a qualification regarding absence of antimycotic resistance for yeasts was introduced. These and other qualifications are reviewed and discussed in the present Opinion.
1.3.2. The QPS approach applied to each EFSA food and feed safety risk assessment area
The QPS approach has proved to be a useful tool to harmonise and prioritise safety assessment within EFSA. The QPS recommended list is used by EFSA's Panel on Additives and Products of Substances used in Animal Feed (FEEDAP), on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF) and on Dietetic Products, Nutrition and Allergies (NDA) and their respective current Units (Feed, FIP and Nutrition, respectively) as well as by the Pesticides Unit.
Feed additives safety assessment area
The EFSA Unit responsible for this area (Feed Unit) applies the QPS evaluation on the assessment of biological agents intended for use as feed additives or as a source of a feed additive, as defined in Regulation (EC) 1831/200311. When a biological agent is assessed for inclusion in the QPS list, the evaluation should cover the safety for the target animal species, the consumers of products derived from animals treated with the additives, and the environment. In the respective FEEDAP Opinions dealing with QPS recommended microorganisms, a standard sentence is included mentioning that the active agent in question is considered by EFSA to have a QPS status. Therefore, in such case, the FEEDAP Panel considers that no assessment of safety for the target species, consumer and the environment is required.
Following requests from applicants, the European Commission requested EFSA to provide an opinion on the implications of the deletion of the maximum dose applied to those authorised microbial products for which safety was assessed using the QPS approach and, more generally, to all microorganisms for which this approach is used. Since the QPS assessment has to take account of any reasonable use of the organism under consideration (sometimes restricted to certain types of application, e.g. enzyme production), and since QPS assessments are made independently of the dose, the FEEDAP Panel concluded that unless a specific provision relating to dose is included in the ‘qualification’ for a given TU, safety is presumed at any reasonable dose (EFSA FEEDAP Panel, 2012b).
Pesticides safety assessment area
The EFSA Unit responsible for this area (Pesticide Unit) organises the peer‐review of microbial plant protection products that are submitted for approval under Regulation (EC) No 1107/200912. The data requirements for the microorganisms (and the final product to be placed on the market) are described in the Regulations (EU) No 283/201313 and 284/201314. They request a clear identification at the strain level, information on their biological properties, on the production and uses, description of the analytical methods, investigations of effects on human health, data on residues in or on treated products, information on the fate and behaviour in the environment (persistence, multiplication and mobility in soil, water and air), and investigations of effects on non‐target organisms (birds, aquatic organisms, bees, other arthropods, earthworms, soil microorganisms). Additionally, scientific peer‐reviewed open literature published within the last 10 years before the date of submission of the dossier has to be provided, in accordance with the Guidance of EFSA.15
For the microbial pesticides approved under Regulation (EC) No 1107/2009, a period not exceeding 15 years is foreseen for the revision of the dossier including new information according to the regulatory framework. If new scientific or technical knowledge shows that the microbe no longer satisfies the approval criteria, a review of its approval can be triggered. This shows the usefulness of the QPS approach as a means of regularly updating the body of knowledge on taxonomic units of importance for EFSA Panels and Units.
In February 2016, it was agreed to improve the assessment of the QPS status and its applicability for the Pesticide Unit by taking into account the data provided to EFSA within the applicant's dossier (that is required to include an assessment of the scientific peer‐reviewed open literature). This should bring additional experts’ views on specific issues related to microorganisms being evaluated.
It is noted that, in the case of plant protection products, the QPS evaluation should be considered as addressing the safety evaluation of the risks for human consumers exposed to microorganism residues via diet. Non‐dietary human exposure during or after the application of the plant protection product represents a set of situations not normally covered by the QPS assessment. In addition, environmental risk assessment as defined by the regulation cannot be considered to be always completely covered by the QPS assessment alone, since the deliberate release of organisms into agricultural or horticultural fields or protected cropping systems before harvest (s), triggers an assessment of risk for a variety of non‐target organisms covering a wide range of taxonomic and functional groups. This assessment, contrary to that needed for food and feed additives, has to cover environmental distribution without prior digestion by farm animals or humans. There are specific regulatory data requirements that applicants must address and criteria for approval prescribed in Regulation (EC) No 1107/2009. These are not always covered by the QPS assessment process. Therefore, these non‐dietary human risk and environmental risk assessments have to be systematically considered under the process prescribed in this Regulation.
Historically microbiological agents recommended for the QPS list and proposed as plant protection products under the Council Directive 91/414/EC (Official Journal, 1991)16 were often exempted from certain data requirements, such as oral toxicity data. As an example, the QPS recommendation of the Baculoviridae family was used during the peer review of several species of baculoviruses (EFSA, 2012a,b).
Food Ingredients and Packaging safety assessment area
The EFSA Unit responsible for this area (FIP Unit) applies the QPS evaluation of those specific microbial TUs used for the production of food enzymes in agreement with the QPS approach that entered EU law with the publication of a new Commission Implementing Regulation (EU) No 562/201217 amending Commission Regulation (EU) No 234/201118 with regard to specific data required for risk assessment of food enzymes. If the microorganism used in the production of a food enzyme has a QPS status according to the most recent list of QPS recommended biological agents adopted by the Authority (meaning EFSA), the food enzyme application could not need to provide specific toxicological test data. If residues, impurities or degradation products linked to the total food enzyme production process (production, recovery and purification) could give rise to concern, the Authority, pursuant to Article 6(1) of Regulation (EC) No 1331/200819 may request additional data for risk assessment, including toxicological data. In the same legislation frame, the QPS status will also have an important consideration on the risk assessment approach applied for enzyme products derived from genetically modified microorganisms developed from strain lineage species fulfilling the recommendations for QPS status.10
Nutrition safety assessment area
The tasks of the Panel on Dietetic Products, Nutrition and Allergies (NDA) include the safety assessment of novel foods (NF) that fall under Regulation (EU) 2015/228320. ‘Novel Food’ means ‘any food that was not used for human consumption to a significant degree within the Union before 15 May 1997’. ‘Food consisting of, isolated from or produced from microorganisms, fungi or algae’ is among the categories of NF as defined by the Novel Food Regulation (EU) 2015/2283. In this case, the BIOHAZ Panel assesses whether the species would qualify for a QPS status, while the NDA Panel assesses the information provided in the novel food application on the specific strain (EFSA NDA Panel, 2016).
In the framework of Regulation (EC) No 1924/2006 on health claims made on foods (including microorganisms), the NDA Panel is also responsible for verifying the scientific substantiation (efficacy assessment) of submitted health claims. Under this framework, it should be noted that a safety assessment is not foreseen. Where relevant, the NDA Panel may recommend restrictions of use based on safety considerations.
In Table 2, the areas of assessment for the QPS approach are summarised and compared to general principles for each EFSA food and feed risk assessment area when microorganisms are considered. This table provides an overview of the principles followed for the assessment of each of the main four areas (feed, food ingredients, pesticides and nutrition). For details of the specific requirements for the safety risk assessment for each of these areas, please consult the specific EU regulations and/or guidance as described in the above specific areas under Section 1.3.2. This is not a stand‐alone table and terminology used can vary between regulatory frameworks. An effort has been made to apply the same terms as much as possible in order to improve clarity in the content of the table and to identify areas that can be considered to be generally equivalent. Where specificities could not be covered in general terms, this is reflected in the text chosen. Some of the areas are not covered by the relevant regulation or guidance and are also described in the following table.
Table 2.
Table summarising the areas of assessment for the QPS approach and for each EFSA food and feed safety risk assessment area when microorganisms are considered
| QPS assessment remit | Feed area assessment remita | Food Ingredients area assessment remitb | Pesticides area assessment remitc | Nutrition area assessment remitd |
|---|---|---|---|---|
| 1. Identity: taxonomy identification parameters |
Verification of species and strain Certificate of deposit in a culture collection |
Verification of species and strain Certificate of deposit in a culture collection |
Verification of species and strain Certificate of deposit in a culture collection |
Verification of species and strain Certificate of deposit in a culture collection (for Health Claims and Novel Foods (NF)) |
| 2.1. Body of Knowledge: history of safe use in the food and feed chain |
Other authorisations and uses Description of the genetic modifications Confirm genetic stability |
Other authorisations of the same strain lineage Description of the genetic modifications Confirm genetic stability |
Proposed uses Historical background Organisms genetically modified should comply to the GMO Regulation Confirm genetic stability |
History of safe use (for NF) |
| 2.2 Body of Knowledge: general ecology/distribution in ecosystems |
Origin to be declared Production of antimicrobial compounds |
Production of antimicrobial compounds |
Origin (geographical and place in the ecosystem) and natural occurrence (if possible at strain level) Ability to colonise available niches Production of antimicrobial compounds |
Out of the scope of the specific Regulation |
| 3.1. Safety concerns: virulence/pathogenicity/toxigenicity for humans |
Pathogenicity potential and virulence factors Production of toxins and toxic secondary metabolites |
Pathogenicity potential and virulence factors Production of toxins and toxic secondary metabolites |
Pathogenicity potential and virulence factors Possible toxicity of secondary metabolites |
Pathogenicity potential and virulence factors (for NF) |
| 3.2. Safety concerns: virulence/pathogenicity/toxigenicity for animals (domestic and wild) |
Pathogenicity potential and virulence factors Production of toxins and toxic secondary metabolites |
Out of the scope of the specific Regulation | Adverse effects of organism or metabolites on non‐target animals (in the environment) | Out of the scope of the specific Regulation |
| 3.3. Safety concerns: virulence/pathogenicity for plants | Out of the scope of the specific Regulation | Out of the scope of the specific Regulation | Adverse effects of organism or metabolites on non‐target vascular plants and algae (in the environment) | Out of the scope of the specific Regulation |
| 3.4. Safety concerns: antimicrobial resistance | Verification of the absence of antimicrobial resistance associated with acquired genes | Verification the of absence of antimicrobial resistance associated with acquired genes | Verification of the absence of antimicrobial resistance and of possible transfer of genes coding for resistance |
Verification of the absence of antimicrobial resistance associated with acquired genes (for NF) |
| 3.5. Safety concerns: environmental safety |
Genetically modified microorganisms (GMMs): deliberate release GMMs: the absence of viable cells GMMs: the absence of recombinant DNA Effect on water and soil |
Not direct applicable within Food Enzymes legislation: Microorganisms used for production purposes, including GMMs: the absence of viable cells GMMs: the absence of recombinant DNA |
Risk for non‐target organisms (which are not vertebrate animals or plants), arising from exposure to the microorganism and its secondary metabolites remaining in or on plants or plant products, in soil water and air | Out of the scope of the specific Regulation |
| 4.1. End use: intended exposure to viable cells of animals and consumers |
Tolerance studies in target animals Toxicological studies in vitro and in laboratory animals |
Not direct applicable within Food Enzymes legislation: Microorganisms used for production purposes, including GMMs: the absence of viable cells GMMs: the absence of recombinant DNA |
End use is intended exposure of a target organism, i.e. organisms that are: 1) pathogenic to or damage/consume plants or plant commodities; or 2) unwanted plants (weeds). Efficacy investigations are required for these purposes |
Intended exposure of viable cells to consumers (for NF) |
| 4.2. End use: enzymes/metabolites producer: no or limited exposure to viable cells |
Microorganisms used for production purposes, including GMMs: absence of viable cells GMMs: absence of recombinant DNA Tolerance studies in target animals Toxicological studies in vitro and in laboratory animals |
Microorganisms used for production purposes, including GMMs: absence of viable cells GMMs: absence of recombinant DNA |
When there are no viable cells in a plant protection product then it is regulated as a chemical (not a microorganism) even if it was produced by microbial fermentation |
(for NF) |
GMM: genetically modified microorganisms; NF: novel food.
Based on the specific Feed Regulation (EC) No 1831/2003 and Regulation (EC) No 429/2008.
Based on the specific FIP/GMMs Regulation: absence of recombinant DNA (under Reg. 1829/2003).
Based on the specific Pesticides Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC.
Based on the specific Health Claims Regulation (EU) No 1924/2006 and Novel foods Regulation (EU) 2015/2283.
1.3.3. Summary of the BIOHAZ Panel Statements adopted between December 2013 and December 2016
In response to ToR1, the EFSA Units (Feed, FIP, Nutrition and Pesticides Units), have been asked to update the list of biological agents being notified to EFSA. From the last notifications received for the previous QPS Opinion in 2013, 405 notifications were received between May 2013 and September 2016, of which, 137 were from Feed, 196 from FIP, 11 from Nutrition and 61 from Pesticides (see Table 3).
Table 3.
Notifications received by EFSA Units (Feed, FIP, Nutrition and Pesticides) by biological group from May 2013 until September 2016 (total numbers for the five Panel Statements)
| Unit/Panel | Not QPS | Already QPS | Grand Total | |||
|---|---|---|---|---|---|---|
| Biological group | Not evaluated | Evaluated | ||||
| Excluded in QPS 2013 update | Previously evaluated | Evaluation in stand by | ||||
| Feed/FEEDAP | 34 | 15 | 0 | 21 | 67 | 137 |
| Bacteria | 5 | 15 | 0 | 21 | 46 | 87 |
| Filamentous fungi | 29 | 0 | 0 | 0 | 0 | 29 |
| Yeasts | 0 | 0 | 0 | 0 | 21 | 21 |
| FIP/CEF | 125 | 10 | 0 | 16 | 59 | 196 |
| Bacteria | 0 | 10 | 0 | 14 | 48 | 68 |
| Filamentous fungi | 125 | 0 | 0 | 0 | 1 | 119 |
| Yeasts | 0 | 0 | 0 | 2 | 10 | 9 |
| Nutrition/NDA | 0 | 0 | 0 | 1 | 10 | 11 |
| Bacteria | 0 | 0 | 0 | 1 | 8 | 9 |
| Yeasts | 0 | 0 | 0 | 0 | 2 | 2 |
| Pesticides | 29 | 3 | 9 | 3 | 17 | 61 |
| Bacteria | 0 | 3 | 6 | 3 | 7 | 19 |
| Filamentous fungi | 29 | 0 | 0 | 0 | 0 | 29 |
| Viruses | 0 | 0 | 0 | 0 | 9 | 9 |
| Yeasts | 0 | 0 | 3 | 0 | 1 | 4 |
| Grand Total | 188 | 28 | 9 | 41 | 153 | 405 |
In response to ToR3, from those 405 notifications, 153 biological agents already had a QPS status and were not further evaluated, neither were the 188 filamentous fungi and enterococci, biological groups which have been excluded from QPS consideration (following a recommendation of the QPS 2013 update (EFSA BIOHAZ Panel, 2013). Another 28 were not included because the corresponding TUs have already been evaluated in the previous Statements during this period. Furthermore, it was agreed not to include nine notifications from Pesticides Unit as the respective dossiers (including the literature review) were not yet received (evaluation in standby). For the type of microorganism, 183 were bacteria, 177 filamentous fungi, 9 viruses and 36 yeasts TUs. The remaining 41 biological agents were assessed for the suitability of the respective TUs for inclusion in the QPS list. The assessment of the respective TUs was published in five Panel Statements, adopted every 6 months, from December 2014 until December 2016 (see Table 3).
2. Data and methodologies
2.1. Data
For the TUs associated with the notifications compiled within the time period covered by the mandate (May 2013–September 2016) and assessed for a possible QPS status within the Panel Statements adopted during this period (every six months between December 2014 and December 2016), the literature review considered the identity, the body of knowledge, history of use, and the potential safety concerns found (including AMR). Relevant databases, such as PubMed, Web of Science, CasesDatabase, CAB Abstracts or Food Science Technology Abstracts (FSTA) and Scopus, were searched and details on the search strategy, search keys and approach followed are described in each Panel Statement.
For the review of the recommendations for the QPS list (as published in 2013) and specific qualifications, an Extensive Literature Search (ELS) was run as described in Section 2.2.1 and in Appendices B and C.
2.2. Methodologies
The QPS assessment is generic regarding a notified TU intended to be intentionally added into the food chain at any stage. The QPS concept applies to microorganisms either used as viable cells in the food chain, or to produce enzymes, metabolites (e.g. amino acids), dead biomass or other specific end products that are not expected to contain live microbial cells. In this last condition, the QPS recommendation may apply only to the specific end use, e.g. enzyme production. A QPS assessment is triggered by receipt of an application dossier by EFSA that requires a safety assessment. It is intended to be independent of the specific application dossier that remains the responsibility of the EFSA Scientific Unit or Panel to which the risk assessment is mandated.
In order to illustrate how the QPS list is used or its approach considered by the four EFSA Units or by the QPS Working Group (WG), three flow charts have been prepared and included below.
The first one (Figure 1), represents how in general, EFSA Units incorporate the QPS status of a certain TU, related to a microorganism notified in an application, into their own evaluation process (risk assessment). After receiving a notification of a microorganism or their products in a new application, the relevant information is included in the ‘notifications list’. EFSA screens the respective TUs and chooses which ones are to be included in the ongoing Panel Statement to be considered for the QPS list (Figure 2). The EFSA Unit initially checks if the TU is in the QPS list and if foreseen in the respective regulatory framework, applicants may be exempt from a certain part of the data requirements and the risk assessment process may be simplified. Possible qualifications of QPS microorganisms need to be evaluated by the EFSA Unit with the information provided in the respective dossier. Only when the qualification applies, the data requirements exemption can be effective. The specific risk assessment is included in the EFSA Unit's Opinion and reference to the QPS status of the TU notified and eventual qualifications are included in that Opinion. For TUs initially not included in the QPS list, but for which a new recommendation for that list is provided from the process described in Figure 2, the inclusion in the list may still be considered for the risk assessment process of that specific EFSA Unit.
Figure 1.
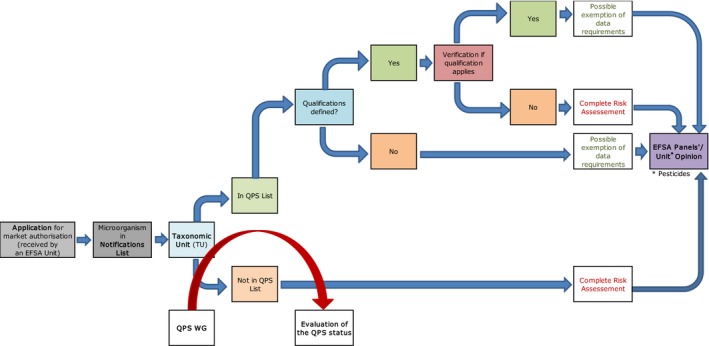
Workflow diagram describing how EFSA Units incorporate the QPS status into the safety assessment process of a microorganism notified through an application for market authorisation – overall process
- QPS: Qualified Presumption of Safety.
Figure 2.
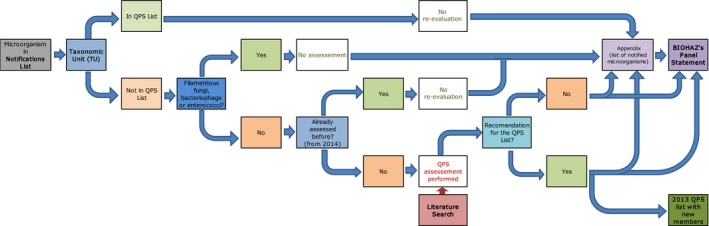
Workflow diagram describing how QPS status is assessed for the TU related to the microorganism notified to the EFSA Units under the frame of applications for market authorisation – elaboration of the BIOHAZ QPS Panel Statements
- BIOHAZ: Biological Hazards Panel; QPS: Qualified Presumption of Safety.
The second flow chart (Figure 2) represents how the evaluation of newly notified TUs, not found in the QPS list, is included in each BIOHAZ Panel Statement. EFSA screens the new TU included by the EFSA Units in the ‘notification list’ (Figure 1), checks the respective TU and chooses which are to be included in the ongoing Panel Statement to be considered for the QPS list. As explained in the background of the mandate (Section 1.1), filamentous fungi, bacteriophages or enterococci are excluded from the QPS evaluation. If the TU has already been evaluated in one of the previous Panel Statements (from December 2014), the TU is also excluded from being re‐evaluated. If a new QPS recommendation is given (and possible qualifications), the QPS list is updated. Every 6 months a new Panel Statement is published incorporating the TUs included in the ‘notification list’ from the last ones considered in the previously published Panel Statement. All respective notifications are included in appendix of each Panel Statement. If the TU is already in the QPS list, it is not evaluated at this stage but considered for the process represented in Figure 3.
Figure 3.
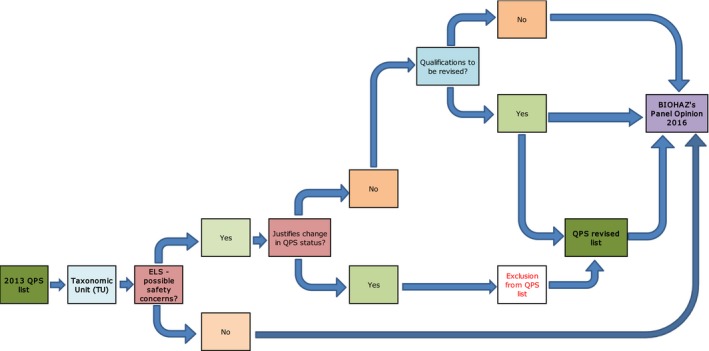
Workflow diagram describing how QPS status is reassessed for the TU included in the latest QPS list – elaboration of the BIOHAZ QPS Opinion
- BIOHAZ: Biological Hazards Panel; QPS: Qualified Presumption of Safety.
The third flow chart (Figure 3) represents how the evaluation of the TUs included in QPS list (reference to 2013 update Opinion) is performed. According to the ongoing mandate (Section 1.1), the process needs to be run and finalised every 3 years. All TUs are evaluated for possible new safety concerns that could result in excluding them from the QPS list or changing or including a possible qualification. This is done through an ELS as explained in Section 2.2.1. Any change in the QPS status or qualifications needs to be reflected in the QPS list, published in appendix to the Opinion and to each Panel Statement: if it occurs before December of 2016, the change is accomplished and the table amended within the 2013 QPS Opinion. In the current Opinion, the QPS list reflects the ELS exercise run during this period (January 2013–May 2016).
This approach to safety assessment of a defined TU (e.g. genus or, most often, species) is based on four pillars: establishing identity, body of knowledge, safety concerns (including AMR) and end use. If the TU does not raise safety concerns or, if safety concerns exist, but can be defined and excluded (the qualification), the TU can be granted QPS status. Thereafter, any strain of a microorganism, the identity of which can be unambiguously established and assigned to a QPS group, may be exempted from the need for further safety assessment, other than satisfying any qualifications specified. Microorganisms not considered suitable for QPS remain subject to a full safety assessment.
2.2.1. Review of the scientific literature
An Extensive Literature Search (ELS) with relevance screening and evaluation of studies related to safety concerns for humans, animals and/or the environment, of microorganisms recommended for the QPS 2013 list was performed.
The aim was to identify any publicly available studies reporting on safety concerns for humans, animals or the environment caused by Gram‐positive non‐sporulating bacteria, Gram‐positive sporulating bacteria, Gram‐negative bacteria, viruses used for plant protection purposes and yeasts (as identified by EFSA in the ToR and Table 1 of the 2013 Scientific Opinion) since the previous QPS review (i.e. publications from January 2013 until 6 June of 2016). The results of the ELS were part of ToR 2 of the self‐task mandate and were intended to inform this Scientific Opinion.
A short description of the methodology adopted is provided below, for a detailed protocol of the process – please refer to Annexes B and C. The process was performed according to the following main steps:
ELS for potentially relevant citations;
relevance screening to select the citations identified by the literature search, based on titles and abstract and then on full text;
evaluation of articles according to pre‐specified categories of possible safety concerns relevant to the QPS assessment.
Considering the purpose of the QPS approach, the research questions were broad in scope. The review questions were broken down into key elements using the PECO conceptual model:
Population of interest (P)
Exposure of interest (E)
Comparator (C)
Outcomes of interest (O)
The following review questions were identified:
Question 1
Is there evidence of any safety concerns, including virulence features and toxin production, for humans, animals and/or the environment associated with microbial species currently recommended for the QPS list since the previous QPS review (i.e. published since 2013 until 6 June 2016)?
The related PECO elements are specified here below:
| Population | Humans, animals and the environment |
| Exposure | Microbial species currently recommended for the QPS list since the previous QPS review |
| Comparator | Since it was expected that the prevalent study designs on this topic would lack a comparator, the latter was not included as a key element in the review question |
| Outcome | Any safety concerns, including virulence features and toxin production |
Question 2
Is there any evidence related to the presence or absence of antimicrobial resistance (AMR) or AMR genes for the same microbial species published during the same time period?
The related PECO elements are specified here below:
| Population | Humans, animals and the environment |
| Exposure | Microbial species currently recommended for the QPS list since the previous QPS review |
| Comparator | Since it was expected that the prevalent study designs on this topic would lack a comparator, the latter was not included as a key element in the review question |
| Outcome | The presence or absence of AMR or AMR genes |
The following outcomes of interest were identified:
| Question 1 |
|
| Question 2 |
|
Population, Exposure and Outcome of interest were used as eligibility criteria to select the citations identified by the literature search. In addition to them, other eligibility criteria were defined:
| Language | English |
| Period | From January 2013 until 6 June 2016 |
| Publication type | Primary research studies (i.e. studies generating new data) |
The following bibliographic sources were searched:
| Information source | Interface |
|---|---|
| Web of Science Core Collection | Web of Science, Thomson Reuters 2016 |
| CAB Abstracts | Web of Science, Thomson Reuters 2016 |
| BIOSIS Citation Index | Web of Science, Thomson Reuters 2016 |
| MEDLINE | Web of Science, Thomson Reuters 2016 |
| Food Science Technology Abstracts (FSTA) | Web of Science, Thomson Reuters 2016 |
It was decided to limit the search to the above‐mentioned bibliographic databases, without extending to the grey literature.
The search strategy used to identify relevant studies comprised two key elements, (i) a set of strings aimed at capturing articles related to the target microbiological species (Exposure); (ii) a set of strings aimed at capturing articles related to outcomes of interest (Outcome).
A total of 16,927 records were found by the search strategy that, after duplicate removal, led to the identification of 16,025 articles. Title screening led to the exclusion of 15,040 articles. Of the 985 articles eligible for Article evaluation, 765 were found not to meet the eligibility criteria and 220 were finally evaluated at full text level.
The flow of records from their identification by the different search strategies (as reported in Appendix C) until their consideration as QPS potentially relevant papers is shown in Table 4 above.
Table 4.
Flow of records by search strategy
| No | Search strategy | No records identified | No duplicate records removed | No record screened | No full text article evaluated | No of articles considered relevant for QPS |
|---|---|---|---|---|---|---|
| Gram‐positive non‐sporulating bacteria | ||||||
| 1 | Bifidobacterium | 939 | 60 | 879 | 70 | 6 |
| 2 | Corynebacterium glutamicum | 195 | 7 | 188 | 33 | 2 |
| 3 | lactobacilli | 2,432 | 171 | 2,261 | 105 | 21 |
| 4 | Lactococcus lactis | 881 | 46 | 835 | 39 | 17 |
| 5 | Leuconostoc | 160 | 21 | 139 | 26 | 9 |
| 6 | Oenococcus | 297 | 41 | 256 | 2 | 0 |
| 7 | pediococci | 815 | 125 | 690 | 11 | 2 |
| 8 | Propionibacterium | 228 | 25 | 203 | 12 | 0 |
| 9 | Streptococcus thermophilus | 352 | 17 | 335 | 12 | 0 |
| Gram‐positive sporulating bacteria | ||||||
| 10 | Bacillus | 4,176 | 85 | 4,091 | 264 | 40 |
| Gram‐negative bacteria | ||||||
| 11 | Gluconobacter oxydans | 199 | 19 | 180 | 8 | 0 |
| Yeasts | ||||||
| 12 | Debaryomyces hansenii… | 1,428 | 141 | 1,287 | 175 | 69 |
| 13 | Kluyveromyces lactis… | 1,632 | 62 | 1,570 | 89 | 14 |
| 14 | Saccharomyces cerevisiae | 2,493 | 72 | 2,421 | 117 | 27 |
| Viruses used for plant protection | ||||||
| 15 | Baculoviridae | 496 | 4 | 492 | 13 | 7 |
| 16 | Alphaflexiviridae… | 204 | 6 | 198 | 9 | 6 |
| TOTAL | 16,927 | 902 | 16,025 | 985 | 220 | |
The articles were evaluated according to the following categories of possible safety concerns:
impact on human health;
impact on animal health;
impact on the environment;
antimicrobial resistance;
other not pre‐specified concerns.
The overall results were presented in tabular format for each group/subgroup and species.
2.2.2. Identity
Information about the systematics (classification, identification and nomenclature) of the notified TU is considered in this section, including a general description of the TU. Attention is given to the inclusion of the TU name in the Official lists stemming from Taxonomy Commissions (for bacteria, yeasts and viruses) and to the use of appropriate methodologies for identification according to standardised molecular, phenotypic and chemotaxonomical methods. The occurrence of changes in the taxonomy or the use of synonyms in the taxonomical description is also highly relevant. Possible misidentifications and lack of precision within closely related taxa assignations (due to the use of phenotypic tests, etc.) are also taken into account.
In the context of a notification received by EFSA for a safety assessment, the QPS assessment is usually carried out considering taxonomic aspects, body of knowledge and safety concerns of the species (for bacteria and yeasts) or families (for virus) to which it belongs, which is referred to as the lowest taxonomic level for which QPS status can be granted (Bourdichon et al., 2012; EFSA BIOHAZ Panel, 2012).
Bacterial taxonomy
Taxonomy and nomenclature of bacteria are covered by the International Code of Nomenclature of Bacteria (1992). New TU or alteration to the taxonomy and nomenclature are published in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) (Oren and Garrity, 2016). This journal publishes a Notification List, containing all ‘validly published’ TU, i.e. the Approved List of Bacterial Names. Validly published are all taxonomic units, which are published in the IJSEM. TUs that were published outside the IJSEM are referred to as ‘effectively’ published. They appear after notification by the authors in a Validation List. Also changes in nomenclature are listed separately. These can be spelling errors in the original description or decisions of the Judicial Commission. Moreover, a comprehensive tool and up‐to‐date presentation of the current taxonomy and nomenclature of bacteria is given on the LPSN website (List of Prokaryotic names with Standing in Nomenclature, formerly List of Bacterial names with Standing in Nomenclature (LBSN)) (Euzeby, 2013).
Fungal taxonomy
The nomenclature and taxonomy of fungi, including yeasts, is covered by the International Code of Nomenclature for algae, fungi, and plants (ICN) (McNeill et al., 2012). An authoritative taxonomy of yeasts was published in 2011 (Kurtzman et al., 2011). It is still valid, although proposals for taxonomical revisions are now appearing.
The introduction of the one‐name system for pleomorphic fungi is ongoing and will undoubtedly have a strong impact on yeast nomenclature. In those cases where separate names are established and in use for both forms, the likely outcome is that one of them will eventually be given priority. The International Commission on the Taxonomy of Fungi (ICTF, 2014) has a special working group for yeasts and it is anticipated that lists of new and prioritised names will appear in the coming years. The ICN recently suggested that the perfect form (teleomorph form) is the name that should have priority.
Virus taxonomy
The taxonomy and nomenclature of viruses are the responsibility of the International Committee on Taxonomy of Viruses. Every 5–6 years a full report is made available, the most recent one, the 9th Report, is from 2012 (King et al., 2012). Annual updates are made based on proposals of study groups after adoption by the Executive Committee and are available through the ICTV website.21 The most recent update is from 2015.
Virus taxonomy is based on shared characteristics such as (i) the type of nucleic acid (RNA or DNA), (ii) the structure of the nucleic acid (single‐stranded or double‐stranded RNA or DNA), (iii) the polarity of the nucleic acid (positive stranded = translatable into proteins; negative stranded = non‐translatable into proteins) and (iv) the form of the virus (isometric, rod‐shaped, filamentous or pleiomorphic). In addition to these characteristics, the replication strategy (v) of the viruses is also taken into account and could contribute to their taxonomic position (Baltimore, 1971, 1974). Viruses are organised in orders (‐virales), families (‐viridae), genera (‐virus) and species (‐virus) by virtue of shared characteristics as described above. The species is the lowest taxon considered by the ICTV. Many viruses do not have a common ancestor; therefore phylogenetic information is only useful within taxons in directing the taxonomy of viruses. The current status of e‐viruses (computationally generated from next generation sequencing endeavours) is being discussed within the ICTV.
Plant virus taxonomy
Plant viruses cause diseases in plants and (sometimes) insects. Many of these viruses are transmitted via direct contact or by vectors (insects, nematodes, fungi). The large majority (> 90%) of plant viruses contain positive stranded (= directly translatable) RNA as genetic information. About 1,000 plant virus species have been recognised by the ICTV and they have been accommodated into two orders and 20 families (King et al., 2012).
Relevant for this report (notifications) are the Alphaflexiviridae (Order Tymovirales) accommodating seven genera encompassing 49 species in total, including the genus Potexvirus containing the species Pepino mosaic virus, and Potyviridae, encompassing eight genera, including the genus Potyvirus with 162 species including Zucchini mosaic virus.
Baculovirus taxonomy
Baculoviruses are large DNA viruses occurring in members of the insect orders Lepidoptera (moths and butterflies), Hymenoptera (sawflies) and Diptera (flies). The family Baculoviridae is subdivided into four genera, Alphabaculovirus, Betabaculovirus, Gammabaculovirus and Deltabaculovirus (Jehle et al., 2006). Fifty‐five baculovirus species have been officially recognised as species (King et al., 2012). About 700 further baculovirus isolates have been described in literature, but not yet biologically and genetically fully analysed and therefore not accepted as species by the ICTV. Baculoviruses, unlike many other virus groups, have a common ancestor assisting in the assignment of the taxonomic status of any baculovirus.
2.2.3. Body of knowledge
The body of knowledge concerning a defined TU is assessed to determine whether there is sufficient information to reach a conclusion regarding its safety. The body of knowledge includes the history of use (Constable et al. 2007; Pariza et al., 2015) and ecology of a TU in the agro‐food chain or in other sectors, the scientific literature, clinical aspects, industrial applications, and other factors as considered appropriate.
History of use in the food and feed chain
The history of use of a specific microbial species in the food chain is taken into consideration. In particular, information on the direct use of viable cells (e.g. as feed additives, food starter cultures, microorganisms as food with a health claim or plant protection products) or the use for production purposes (e.g. production of amino acids, enzymes, vitamins and polysaccharides) is examined to evaluate the history of human and animal exposure to the TU under assessment.
General ecology/distribution in ecosystems
The assessment of safety for the environment of a TU proposed for the QPS list takes into account the distribution in natural environments (e.g. in the gut of wild and farmed animals, and plants association), Information about the natural habitats of the organism and the types of samples from which it can usually be isolated is considered. Likewise, information on the geographical distribution range (e.g. does the species occur worldwide?) is valuable. It is also of interest whether the species can occur as a commensal or an endophyte. Spread and prevalence in natural environments and the survival and longevity in the food chain are considered. Properties related to colonisation ability and routes for dispersal are considered. Knowledge about its interactions with other microorganisms, especially with respect to antagonism and competitive ability, is also relevant.
2.2.4. Safety concerns
Safety concerns were investigated in the course of the first assessment of a TU proposed for the QPS list, and are regularly verified for the QPS TUs. In this Opinion, only scientific information that can be cited in a transparent manner and includes a scientifically valid description of the methodologies and the results obtained is considered (i.e. the methods used are suitable for the TU and the evaluation and can be relied on).
Virulence/pathogenicity to humans
TUs assessed for the QPS list should not represent a hazard to human health when used in the food or feed chain. Relevant information includes case reports of human diseases, particularly infections or human intoxications linked to the TU under assessment. Additional important information is whether the negative impacts affected patients with severe underlying diseases, and whether transmission occurred through food or other routes (e.g. medical devices). Studies indicating the presence of virulence factors (e.g. toxins and enzymes that may contribute to the pathogenicity of the microorganism) in the TU are also relevant for identification of potential safety concerns.
Assessment of allergenicity to microbial residual components is beyond the QPS assessment remit; nevertheless, if there is science‐based evidence for some microbial species related to well‐defined clinical cases, this may be reported.
Virulence/pathogenicity to vertebrate animals (domesticated and wild)
Reports of infection, intoxication or other diseases caused by the assessed TU on domesticated and wild animals are also a relevant set of information for identifying potential safety concerns. As with safety concerns for humans, whether diseases occur through feed or other routes (e.g. wounds, inhalation) is also relevant information. Whenever the TU has been studied as a probiotic in animals, publications reporting failure of the probiotic to, for example, promote growth of farmed animals are not considered as indicators of a safety concern.
Antimicrobial resistance
The scope of the review is to provide general background information on AMR issues concerning the TU under assessment. In particular, a generic qualification for all bacterial TUs on the QPS recommended list is that strains should not harbour any acquired gene conferring resistance to clinically relevant antimicrobials, in order to exclude the presence of potentially transferable AMR. The ability to produce antimicrobials is also relevant because these antimicrobials could select for resistance in bacterial populations. Especially important is the ability to produce antimicrobials which are used in human and veterinary practice or which are inactivated by genes conferring cross‐resistance to those. Moreover, microorganisms producing antimicrobials carry genes that confer resistance to their own compounds, which might be transferred to other bacteria and further disseminated.
In the case of yeasts, transferability of AMR determinants is not an issue, but a QPS TU should not be resistant to antimycotic compounds used in human medicine.
In this review, the bacterial TUs recommended for the 2013 QPS list were revised with regard to their potential to produce antimicrobials, and the presence of transferable or transmissible AMR genes. The fungal TUs were revised for their potential to produce antimicrobials and for resistance to antimycotic compounds.
Environmental safety
For plant protection products, as mentioned above, the QPS considerations of environmental safety do not cover all aspects of the regulatory data requirements.
The assessment of environmental safety considers information on, e.g. the capability of the species to survive, compete and proliferate in specific environments, the possibility that it may cause adverse health or environmental effects not connected to pathogenicity and infectivity to vertebrate animals and plants, and the possibility for transfer and expression of the microbial DNA in other organisms.
So far, safety of plants has not been systematically considered in the QPS assessment. In the QPS Statements and in the 2016 QPS Opinion, it was decided to consider infections and other diseases caused to plants in the QPS assessment and updates, should they appear in the literature searches.
2.2.5. End use
For a TU, the body of knowledge and the safety concerns may differ for the living organisms and for the dead biomass or specific compounds produced (e.g. long history of use for enzymes or amino acids production for food/feed purposes). For the majority of the TUs, the QPS approach assesses the deliberate introduction of viable microorganisms capable of multiplication in the food chains, with consequent exposure of humans and/or animals. The second circumstance does not involve a significant number of live microbial cells and only the products derived from microbial metabolism, such as cell extracts, enzyme preparations and amino acids are considered. In this latter case, the QPS recommendation may only apply to this specific end use not including living organisms, which is indicated as a qualification in the QPS list. This specific consideration of end use does not conflict with the generic applicability of QPS because in this case the end use corresponds to different hazards (living organisms versus dead cells or their metabolites). Other types of end use of these TUs might impact the dissemination of the taxonomic units and/or the exposure of humans or animals. This requires a specific case‐by‐case risk assessment.
2.2.6. Can safety concerns be excluded?
Qualifications for antimicrobial resistance
The absence of acquired genes coding for resistance to antimicrobials relevant for humans and animals in a QPS recommended bacterial TU is a generic qualification. The verification that a specific bacterial strain, notified to a certain Panel, fulfils the qualification of the absence of acquired AMR genes is conducted by the specific EFSA Unit/Panel to which the notification was assigned. Within the framework of EFSA activities, the use of interpretative criteria and methods to define and monitor AMR have been harmonised and are reflected in EFSA's guidance documents. The use of harmonised methods and epidemiological cut‐off values ensures the comparability of data over time at country level, and also facilitates the comparison of the occurrence of resistance between the Member States (EFSA FEEDAP Panel, 2012a).
In the case of yeasts, acquired AMR genes are not of relevance, but susceptibility to antimycotic compounds used in human medicine should be proved.
Qualification for the absence of toxigenic potential
Several Bacillus species are on the QPS list with the qualification ‘absence of toxigenic activity’. This is based on the observation that some strains among the Bacillus species on the QPS list have caused food‐borne intoxication in the past, which have been attributed to the production by these strains of compounds with toxic activities. Technical guidance to identify these toxic compounds among Bacillus species has been elaborated and updated by EFSA (EFSA FEEDAP Panel, 2011, 2014). The application of the qualification should permit identification of this safety concern among strains of the QPS Bacillus species. It is the purpose of the regular update of the QPS list to verify that no other relevant safety concerns have been identified for the QPS species of Bacillus.
Qualification for production purposes
The qualification ‘for production purpose only’ applies to a TU used for the biosynthesis of specific products used in the food chain and subject to a specific authorisation (e.g. feed additives – vitamins, amino acids, polysaccharides and enzymes – and food enzymes). For most of the TUs used for production, data are lacking on direct exposure to humans and animals, while there is a long history of use of their fermentation products in the food chain. Under specific regulation (e.g. Commission Regulation (EC) No 429/2008), the absence of production organisms in the additive derived from fermentation must be demonstrated.
3. Assessment
Under this section, two types of assessments have been included: the re‐evaluation of the TUs included in the QPS list published in the Opinion from 2013 (ToR2) and the assessment of the new TUs corresponding to the microorganisms notified to the EFSA Units under the frame of an application for market authorisation (ToR3). The latter have been included in the Panel Statements adopted between December 2014 and December 2016. For both types of assessments, the QPS approach has been applied in the same way and based on the four main pillars as described in Section 2.2.
For the revision of the TUs included in the QPS list and respective qualifications, the update of the body of knowledge to check for possible new safety concerns was done based on an ELS and on expert knowledge. The previous information published in 2013 (EFSA BIOHAZ Panel, 2013) was taken into consideration whenever it was recommended the monitoring of possible safety concerns.
For the evaluation of the new TUs corresponding to the microorganisms notified to the EFSA Units between May of 2013 and September of 2016, the search for information was done using the available databases and according to the expert knowledge. These TUs were not included in the ongoing ELS revision as only the ones that were already present in the 2013 Opinion were considered for this step.
3.1. Gram‐positive non‐sporulating bacteria
3.1.1. Bifidobacterium species
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Bifidobacterium species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 879; after screening at title/abstract level, 70 passed to the full text phase; of those, six were considered relevant for the QPS assessment.
Five references concerned case reports involving bifidobacteria in patients with immunosuppression and/or underlying disease. Two reports described infections with Bifidobacterium breve, i.e. a sepsis in a 2‐year‐old immunocompromised child (acute leukaemia with chemotherapy) and a 45‐year‐old patient with infection and a severe underlying disease (Suwantarat et al., 2014; Avcin et al., 2015). A further three reports describe infections with Bifidobacterium longum. The first report (Bertelli et al., 2015) includes two cases with bacteraemia in preterm infants receiving antimicrobials, identifying the infectious agent more precisely as Bifidobacterium longum subsp. infantis. Three similar cases were reported by Zbinden et al. (2015). Tena et al. (2014) describe a case of peritonitis due to B. longum in a patient with underlying disease.
Revision of antimicrobial resistance aspects
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), one report described for the first time the occurrence of the acquired erm(X) gene in B. longum subsp. longum (Luo et al., 2015).
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
The cases of infection in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection. The use of microorganisms intended to be used as ‘probiotic’ for humans as a health claim does not fall under the remit of the QPS assessment. In conclusion, the QPS status of the Bifidobacterium species previously included in the list does not change and monitoring should continue.
3.1.2. Carnobacterium divergens (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014).
Identity
The Carnobacterium genus belongs to the family Carnobacteriaceae in the order of Lactobacillales (Collins et al., 1987). The most important species is Carnobacterium maltaromaticum due to its common occurrence in foods of animal origin. C. divergens (and later also C. maltaromaticum) has been reclassified and transferred from the genus Lactobacillus to the described genus nov. Carnobacterium in 1987 (Collins et al., 1987) based on phenotypic classification. The first description was given by Holzapfel and Gerber (1983). The original strains were isolated from raw vacuum‐packaged, as well as SO2‐treated, minced beef, in the course of shelf life studies on this product (Holzapfel and Gerber, 1983). The complete genome sequence is known for some strains of Carnobacterium spp., but not for C. divergens.
Body of knowledge
The species C. divergens frequently dominates the microbiota of refrigerated meat and seafood, stored under vacuum or modified atmosphere (Laursen et al., 2005; Leisner et al., 2007; Rieder et al., 2012). For its ability to produce bacteriocins, this species has been used in food with the aim to reduce spoilage and pathogenic bacteria (Richard et al., 2003; Leisner et al., 2007; Rihakova et al., 2009). C. divergens has been also studied as probiotic for fish, such as Atlantic cod (Gadus morhua L.) (Lauzon et al., 2010), Atlantic salmon (Salmo salar L.) (Ringø et al., 2007) and rainbow trout (Oncorhynchus mykiss) (Kim and Austin, 2008), and as probiotic for chicken for fattening (Jözefiak et al., 2011).
Safety concerns
In a single study two strains of C. divergens, isolated from the blood of a newborn delivered by caesarean section and from a febrile lymphoma patient, were identified by sequencing the variable regions of the 16S rRNA gene. The two strains encode a possibly acquired new class D, β‐lactamase Oxa 63 (Meziane‐Cherif et al., 2008).
However, these infections represent extremely rare individual cases, occurring on highly vulnerable individuals, and do not suggest a risk for consumers or animals via exposure through the food and feed chain.
Conclusions on a recommendation for the QPS list
The TU is well described and the body of knowledge shows it as a common species in the food chain, especially in meat. C. divergens can be recommended for the QPS list with the qualification of absence of acquired AMR determinants.
3.1.3. Corynebacterium glutamicum
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the C. glutamicum has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 188; after screening at title/abstract level, 33 passed to the full text phase; of those, two were considered relevant for the QPS assessment.
A literature review did not reveal any new information about adverse health effects or safety concerns since the last update (EFSA BIOHAZ Panel, 2013).
Revision of antimicrobial resistance aspects
The involvement of class 1 integrons in the AMR towards streptomycin/spectinomycin and tetracycline in C. glutamicum isolates has been confirmed and reviewed by Deng et al. (2015).
No additional relevant information was published in the last year on the antimicrobial susceptibility or resistance of C. glutamicum.
Update on other qualifications
This TU has the following qualification ‘QPS only applies when the species is used for aminoacid production’. Due to a lack of knowledge in relation to history of use of the viable organisms and because other members of the same genus are pathogenic, the qualification is confirmed.
Other relevant information
No new relevant information was identified.
Conclusion regarding a QPS recommendation
The QPS recommendation is confirmed for C. glutamicum as well as the qualification.
3.1.4. Lactobacillus species
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Lactobacillus species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 2,261; after screening at title/abstract level, 105 passed to the full text phase; of those, 21 were considered relevant for the QPS assessment.
Two of them (Krishnan and Abraham, 2014; Encarnacion et al., 2016) did not provide any information on how the identification of the aetiological agents of the two endocarditis cases was performed, and another (Doern et al., 2014b) duplicated the information of Doern et al. (2014a). The 18 reports that remained described association or causality of infection on heavily debilitated/immunocompromised patients (premature newborns or elderly subjects suffering from cancer, vascular disease complicated diabetes, diverticulitis, etc.).
Two articles (Martinez et al., 2014; Lee et al., 2015) compiled cases that occurred in particular hospitals over prolonged periods. In both, a variety of lactobacilli (L. fermentum, L. gasseri, L. casei, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus, L. delbrueckii, L. salivarius) were associated with disease, but in most instances to just single cases. This illustrates the ability of microorganisms, even those considered harmless, to colonise patients suffering from life‐threatening illnesses. Out of the rest, single reports associated L. paracasei (Franko et al., 2013), L. fermentum (Chery et al., 2013), L. acidophilus (Mehta et al., 2013), L. gasseri (Sun et al., 2015a) and L. casei (Aroutcheva et al., 2016) with production of endocarditis, cholecystitis, septicaemia, surgical wound/urinary tract infection and contamination of a central catheter, respectively, in patients who have suffered from prostate cancer, coronary disease, lymphoma, simultaneous infection by a coagulase‐negative Staphylococcus and chronic lung and renal insufficiency.
Two reports addressed infections where L. plantarum was isolated (Nei et al., 2013; Tena et al., 2013) as part of polymicrobial infections. The first patient suffered from an advanced laryngeal cancer while the second had uncontrolled diabetes that caused chronic arterial ischaemia.
Finally, eight reports on infection with L. rhamnosus were detected, one of which describes two cases. In all of them, the affected patients were immunocompromised and included five newborns, out of which, one suffered an intrauterine growth restriction (Sadowska‐Krawczenko et al., 2014), two were premature children, one had suffered consecutive intestinal perforations (Brecht et al., 2016), and another had experienced respiratory distress and a previous sepsis by Staphylococcus haemolyticus (Dani et al., 2016). The fifth was a girl with a trisomy 18 and triple‐X syndromes who had previously suffered from sepsis and pneumonia (Dani et al., 2016). All of these developed bacteraemia by L. rhamnosus, as did a kidney transplant recipient (Falci et al., 2015) and two patients with ulcerative colitis who were being treated with corticosteroids and mesalazine (Vahabnezhad et al., 2013; Meini et al., 2015). The last two were a man and a baby (trisomy 21) suffering from acute myeloid leukaemia and bronchiolitis, respectively, who developed a severe oral infection (Ishihara et al., 2014) and pneumonia (Doern et al., 2014a,b). It is noticeable that at least eight of the nine patients were ingesting or have recently taken L. rhamnosus‐based probiotic preparations despite their general compromising situation and that in several cases genetic identity between pathogenic and probiotic strains was confirmed.
Revision of antimicrobial resistance aspects
A report (Jaimee and Halami, 2016) described the presence of aminoglycoside resistance determinants in a few L. plantarum isolates from meat. No evidence of transmissibility was provided. In addition, lactobacilli, as any other anaerobic organisms, tend to be intrinsically resistant to aminoglycosides which weakens the claim on causality of the observed lack of susceptibility.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed. As already noted in the 2013 Opinion, L. rhamnosus produced most of the clinical cases reported, probably due to frequent inclusion of isolates of this species into human ‘probiotic’ preparations. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection. The use of microorganisms intended to be used as ‘probiotic’ for humans as a health claim does not fall under the remit of the QPS assessment.
3.1.4.1. Lactobacillus diolivorans (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a).
Identity
The species L. diolivorans, first described in Krooneman et al. (2002) (belongs to the group of obligate heterofermentative lactobacilli. The phylogenetic analysis of the 16S rRNA, indicates that this species belong to the Lactobacillus buchneri group.
Body of knowledge
A search for the body of knowledge on L. diolivorans was done considering all years available in the literature databases, using the species name as search terms. A limited number of papers (32) were retrieved. This species was originally isolated from maize silage and it has been found in several foods of plant origin, such as apple juice, sourdough, pickles and tofu. The organism has been isolated also from kefir grains.
Safety concerns
No reports were found on safety concerns related to this Lactobacillus species in the literature database searches. Since members of this species were in the past probably assigned to L. buchneri, a species granted the QPS status, additional safety concerns related to misidentification are not expected.
Antimicrobial resistance
No information related to the presence of AMR determinants has been identified in L. diolivorans.
Conclusions on a recommendation for the QPS list
The species L. diolivorans is a natural component of bacterial communities of fermented vegetables and plant derived products. It has never been implicated in human or animal diseases and therefore can be recommended for the QPS list.
3.1.5. Lactococcus lactis
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Lactococcus species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 835; after screening at title/abstract level, 39 passed to the full text phase; of those, 18 were considered relevant for the QPS assessment.
Eight of them describe non‐reliable phenotypical identification methods, do not give enough indication on which methodology was used, or/and provide data strongly suggestive of environmental contamination (Buchelli‐Ramirez et al., 2013; Feierabend et al., 2013; Hadjisymeou et al., 2013; Rostagno et al., 2013; Inoue et al., 2014; Karanth et al., 2014; Lee et al., 2014a; Taniguchi et al., 2016). All of these describe single cases of patients suffering from very debilitating illnesses, such as necrotising pneumonia, heart or renal failure, peritonitis, uncontrolled diabetes, etc.
Of the remaining articles, three papers describe L. lactis isolates from cases of bovine mastitis (Plumed‐Ferrer et al., 2013, 2015; Werner et al., 2014) while another links strains of L. lactis to aquaculture fish problems (Ucko and Colorni, 2014). However, a definite link between these animal pathologies with L. lactis as the aetiological agent is lacking. The other communications describe association or causality of infection with L. lactis in seriously debilitated/immunocompromised patients, such as premature newborns or elderly subjects suffering from cancer, uncontrolled diabetes, heart problems, etc. These include bacteraemia (Karaaslan et al., 2015, 2016) and uncomplicated urinary infection (Newby and Ramesh, 2014).
Revision of antimicrobial resistance aspects
Three papers addressed the susceptibility resistance to antimicrobials of L. lactis isolates (Plumed‐Ferrer et al., 2015; Zycka‐Krzesinska et al., 2015; Li et al., 2016). In two of these, terR and blaCMY‐2 determinants were detected; whether these genes encoded relevant phenotypes or/and if any of them were plasmid‐encoded was not determined. In the third report, general resistance to rifampin, presumably a chromosomally encoded trait, was found among L. lactis isolates from cases of bovine mastitis.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
There is no need to change the QPS recommendation of L. lactis, as the infections reported were extremely scarce, and the affected patients already suffered from highly debilitating illnesses and/or were significantly immunodepressed. The possibility that L. lactis might be involved in bovine mastitis, albeit limited for the moment, should be monitored.
3.1.6. Leuconostoc species
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Leuconostoc species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 139; after screening at title/abstract level, 26 passed to the full text phase; of those, nine were considered relevant for the QPS assessment.
Three reports describe cases of infection in humans associated with Leuconostoc spp. All of them used unreliable phenotypical identification methods or did not give enough data on the methodology.
They refer to a case of cholecystolithiasis in an 83‐year‐old Asian woman due to Lc. lactis (Yang et al., 2015), a case with pleural empyema due to Leuconostoc mesenteroides in a healthy person handling pickled vegetables (Usta‐Atmaca et al., 2015), and a case of an endophthalmitis by vancomycin‐resistant Lc. mesenteroides after intravitreal injection of ranibizumab in an 89‐year‐old patient with cancer (Damasceno et al., 2016).
These articles mostly describe cases of infection in patients with predisposing or risks factors, with unreliable identification methods, and do not suggest a risk for the consumer via exposure through the food and feed chain. The above new information does not modify the QPS recommendation of Leuconostoc species.
Revision of antimicrobial resistance aspects
Six articles have reported the AMR and minimal inhibitory concentration (MIC) values of strains belonging to Leuconostoc isolated from different food sources. The AMR of Leuconostoc pseudomesenteroides (n = 13) isolated from Alorena green table olives to 15 antimicrobials was evaluated. No genes encoding possible transferable AMR determinants for the observed phenotypic resistance were detected by polymerase chain reaction (PCR). The AMR of 14 Lc. mesenteroides isolates from Turkish dairy products to six antimicrobials was studied (Bașbülbül et al., 2015). No AMR genes were detected by PCR in the Leuconostoc isolates. Selected Leuconostoc isolates from traditional cheeses made from raw milk were tested for AMR (Alegría et al., 2013). From the 14 isolates tested, 13 were susceptible or intrinsically resistant to a set of 16 antimicrobials while one Leuconostoc citreum strain showed an atypical resistance to ciprofloxacin. Flórez et al. (2016) determined the MIC for 16 antimicrobials in 34 isolates of dairy origin. Atypical resistances were found for several antimicrobials. Evidence of the genetic basis of atypical resistances, and interspecies transfer of erythromycin resistance were shown. In another article, the genome sequence of three Lc. mesenteroides isolates from Italian soft cheese samples were published (Campedelli et al., 2015). The isolates displayed atypical resistance to several antimicrobials. Preliminary analysis of the sequences revealed the presence of erm(B) and tet(S) in two isolates. The AMR of one Lc. citreum strain, isolated from Korean kimchi, was studied (Ji et al., 2013) and found susceptible to all antimicrobials tested.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
The cases of infections in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. The above new information does not modify the QPS recommendation of Leuconostoc species. Therefore, the QPS recommendation for Lc. mesenteroides, Lc. lactis, Lc. pseudomesenteroides and Lc. citreum was confirmed.
3.1.7. Microbacterium imperiale (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014).
Identity
Microbacterium imperiale, previously known as Brevibacterium imperiale, was included in the genus based on its close relationship with Microbacterium lacticum (Collins et al., 1983). The genus is phylogenetically coherent as determined by 16S rRNA gene sequencing and chemotaxonomic data (Takeuchi and Yokota, 1994; Rivas et al., 2004; Park et al., 2006, 2008). The bacteria of the genus Microbacterium are Gram‐positive organisms that belong to the phylum Actinobacteria (G + C ≈ 66–70%), strictly aerobic, rod shaped and usually non‐motile.
Body of knowledge
Their habitat is the soil where they thrive on plant decaying material thanks to their enzymatic potential to degrade complex polysaccharides. Xylanolytic, amilolytic and β‐glucosidase activities have been detected in different isolates of the genus (Rivas et al., 2004; Park et al., 2006, 2008; Wu et al., 2014). Endophytic and gut of caterpillar associated strains have been isolated as well (Zinniel et al., 2002; Huang et al., 2012; Gan et al., 2014), with no signs of pathology perceived in the colonised plant or animal tissues.
No records of intended use of M. imperiale cells in foods manufacturing exist. However, the enzymes produced by organisms of the genus are used in food processing. Of special interest to this evaluation is the use of the 1,4‐α‐maltotriohydrolase for the production of maltotriose, an oligosaccharide used for the production of desserts and baked pastries (Anonymous, 2000, 2011; Wu et al., 2014).
Safety concerns
In literature, no association of M. imperiale to pathology has been reported. In fact, out of the 84 species of the genus Microbacterium, only four have been described as involved in human pathological processes, the cases being extremely rare, occurring in patients with predisposing conditions and, in some cases, being part of a polymicrobial infection (Alonso‐Echanove et al., 2001; Giammanco et al., 2006; Adames et al., 2010; Enoch et al., 2011; Buss et al., 2014). The frequent need of a previous life‐threatening or immunodeficiency condition for successful Microbacterium spp. infection may indicate that no significant virulence factors are produced by the species of this genus. Finally, resistance to chemotherapy appears to be scarce, with an almost universal susceptibility to β‐lactam and glycopeptide antibiotics (Adames et al., 2010; Buss et al., 2014).
Conclusions on a recommendation for the QPS list
No record exists of intended use of any Microbacterium spp. in food processing and/or ingestion of viable cells. There is a history of use in food processing of enzymes produced by M. imperiale, therefore it can only be recommended for QPS for enzyme production.
3.1.8. Oenococcus oeni
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered O. oeni has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 265; after screening at title/abstract level, two passed to the full text phase; of those, none were considered relevant for the QPS assessment.
Revision of antimicrobial resistance aspects
No new information regarding AMR issues was retrieved during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified for O. oeni. Therefore, its QPS status does not change and monitoring should continue.
3.1.9. Pasteuria nishizawae (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015b).
Identity
The genus Pasteuria comprises endospore‐forming, Gram‐positive bacteria of the phylum Firmicutes, phylogenetically mostly related to members of the family Alicyclobacillaceae, as judged by the sequence similarity of their 16S rRNA genes (Preston et al., 2003; Noel et al., 2005). Sequencing of sporulation genes and multilocus sequence typing also place these organisms as members of the order Bacillales (Preston et al., 2003; Trotter and Bishop, 2003; Charles et al., 2005). All Pasteuria species described so far are obligate parasites of invertebrates, including plant parasitic nematodes and planktonic freshwater crustaceans of the genus Daphnia, and none have been grown in vitro. Five species have been recognised: P. ramosa (which parasitises cladoceran water‐fleas), P. hartismeri, P. penetrans, P. thornei and P. nishizawae (parasitising plant pathogenic nematodes) based on their host range, morphology and 16S rRNA signatures (Bishop et al., 2007, 2011). Since vegetative forms of P. nishizawae (and of any other nematode‐parasitic Pasteuria spp.) have so far only been found in the pseudocoelum of female worms once they have infected the roots of the susceptible plant, it has been extremely difficult to obtain pure DNA. Thus, endospores have been the only possible source for DNA to the point that this has prevented genome sequence determination.
P. nishizawae parasitises Heterodera glycines, the causal agent of soybean cyst formation, and it has been proposed as a biocontrol agent for that disease. Its endospores also attach to larvae of Heterodera schachtii, the cyst eelworm of sugarbeet, possibly indicating that its host range is wider than described (Sayre et al., 1991; Noel et al., 2005). In addition to its host specificity, P. nishizawae presents a morphologically distinctive vegetative cycle and endospores and, most important, a 16S rRNA gene sequence that is less than 96% identical to that of any other Pasteuria spp. investigated (Preston et al., 2003; Atibalentja et al., 2004; Noel et al., 2005).
Body of knowledge
The Pasteuria species show a high host specificity that is commonly restricted to one or a few related species of nematodes. Since the plant host range of their host helminths is also very narrow, each Pasteuria species can be ascribed to particular plants. The relation between the bacterium and the plant is mutualistic, because the bacterium kills an important plant parasite. This has given rise to expectations of using Pasteuria in control of pest nematodes.
In soil, endospores of Pasteuria (morphologically similar to those of bacilli) present peripheral fibres that attach to the tegument of recently hatched, free‐living juvenile worms. Upon infestation of the root by the larvae (which may be mechanically prevented in case that a high number of spores have attached to them), spore germination takes place, followed by formation of a pseudomycelium (reason why Pasteuria was proposed to belong to the phylum Actinomycetales) that becomes fragmented and expands throughout the body of the helminth, leading to formation of new microcolonies. Sporulation takes place only in the adult female worm (possibly because males abandon the plant before complete maturation) thus interfering with the externalisation of its caudal end, which, in the absence of the bacterium, will swell to form a cyst full of eggs. Up to 106 endospores can be produced per infected worm, which may amount to 107–108 shed into the soil per infected plant.
Safety concerns
A search in the Web of Science retrieved information from the Environmental Protection Agency (EPA) of the United States on ‘Exemption from the Requirement of a Tolerance’ for residues of P. nishizawae‐Pn1 in all food commodities, including drinking water (US EPA, 2012) registered as an active ingredient in products for controlling the soybean cyst nematode. The exemption was based on the absence of any signs of toxicity/pathogenicity/hypersensitivity in tests performed on laboratory animals. Other databases looked at, such as the CasesDatabase, GoogleScholar, CAB Abstracts or Food Science Technology Abstracts, provided no additional information.
QPS evaluation is not equivalent to the one performed by EPA, therefore a separate evaluation was conducted. As already stated, members of the genus Pasteuria have a narrow nematode host range and their spores only become vegetative cells after infestation of a plant root by the helminth. Thus, it seems unlikely that P. nishizawae can develop in, or harm any other organisms. Moreover, endospores resembling those of Pasteuria have been detected attached to almost all nematode species investigated and, in many cases, infection has been proven (Chen and Dickson, 1998; Tian et al., 2007). This indicates the ubiquitous nature of these bacteria and of their frequent contact with higher organisms, including humans. Nevertheless, there are no reports on pathogenicity or hypersensitivity even in farmers or crops handlers known to use Pasteuria‐based helminth control.
Antimicrobial resistance
No data on antimicrobial susceptibility/resistance are available for Pasteuria spp.
Conclusions on a recommendation for the QPS list
In December 2015, P. nishizawae was recommended for the QPS status for use as a plant protection product to combat cyst nematodiasis (EFSA BIOHAZ Panel, 2015b). This conclusion was based on the following: (i) it is an obligate parasite, unable to grow independently of its host species, H. glycines and possibly H. schachtii. In addition, available evidence indicates that this species of bacteria requires entry of the nematode into the root of a plant for vegetative growth; (ii) the ubiquity and abundance of Pasteuria spp. endospores in soils and the lack of any reports on harmful effects of these bacteria on organisms other than their hosts.
The qualification linked to this taxonomic unit was re‐evaluated and the QPS recommendation is now ascribed without the previous qualification (‘QPS only applies when used in pesticides to combat cyst nematodiasis’).
3.1.10. Pediococcus species
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Pediococcus species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 690, after screening at title/abstract level, 11 passed to the full text phase; of those, one was considered relevant for the QPS assessment, (Al‐Badah et al., 2015), describing an incidental colonisation by pediococci of endodontic roots infected with a variety of pathogens. However, unreliable phenotypic identification was used in this article.
Revision of antimicrobial resistance aspects
Yüceer and Özden Tuncer (2015) report isolation of Pediococcus acidilactici and Pediococcuspentosaceus from almost 50% of uninoculated sukuc samples (a dry, spicy sausage, also named sucuk, sujux, suxhuk, etc.), suggestive of their technological role, and determined their AMR profiles. A lack of susceptibility to vancomycin, aminoglycosides and quinolones, all of which may be considered as intrinsic/chromosomally encoded. No attempt to relate these traits to specific genes associated to mobile elements is reported.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
There is no need to change the recommendation of the QPS‐granted pediococci species because no causality of infection has been reported during the scrutinised period.
3.1.10.1. Pediococcus parvulus (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a).
Identity
Pediococcus parvulus appears to be a well‐defined species by 16S rRNA gene sequencing (Collins et al., 1990). It clusters with other species of the genus, such as P. acidilactici, P. pentosaceus and Pediococcus damnosus (Collins et al., 1990) which has been confirmed after comparison of the P. parvulus DSM20332 draft sequence (accession number: NZ_JQBE01000001) with those of representative strains of the other species (Sun et al., 2015b). Phylogenetically, the whole genus has been proposed to be allocated into the Lactobacillus genus complex together with genera Weisella, Leuconostoc, Oenococcus and Fructobacillus because in phylogenetic trees their species appear intermixed with those of Lactobacillus (Sun et al., 2015b).
Body of knowledge
P. parvulus is commonly associated to spoilage of alcoholic beverages because it commonly produces EPS and diacetaldehyde upon fermentation of the must sugars (Renouf et al., 2007; Petri et al., 2013; Delsart et al., 2016). The former confers an oily appearance to the liquid known as ropiness, while the latter generates a butter‐like flavour. In addition, some strains are histamine producers (Landete et al., 2005). The EPS synthesising strains have been proposed as suitable for production of ropy dairy products (Elizaquível et al., 2011) but they have not been included in any commercial products. Similarly, it appears that EPS‐producing strains are less susceptible to simulated and mice digestive conditions, while lowering the cholesterol levels (Immerstrand et al., 2010; Lindström et al., 2013) reason why they have been proposed as potential probiotics but never used as such or intentionally added to any food. P. parvulus is, however, frequently found in fermented foods (Mesas et al., 2011; Abriouel et al., 2012; Bağder Elmacı et al., 2015) and feed, such as silage (Tohno et al., 2012), and thus there is frequent exposure to it without any signs of pathogenicity for humans or animals.
Safety concerns
No communications on pathogenicity of P. parvulus were detected.
Antimicrobial resistance
Two reports on P. parvulus antibiotic resistance (Rojo‐Bezares et al., 2006; Danielsen et al., 2007) provide similar data. The strains examined are highly susceptible to the β‐lactams tested, but resistant to vancomycin, tetracycline, aminoglycosides, ciprofloxacin and trimethoprim. Genes conferring resistance to aminoglycosides (ant(6), aac(6’)‐aph(2’’)) and tetracyclines (tet(L)) have been detected but their expression or transmissibility was not tested.
Conclusions on a recommendation for the QPS list
P. parvulus can be granted the QPS status, being a species commonly found in fermented food and beverages and based on lack of pathogenicity as determined by the absence of any significant virulence determinants in its genome and of any reports on its role on human or animal infection.
3.1.11. Dairy propionic acid bacteria – Propionibacterium species
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered Propionibacterium species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 203, after screening at title/abstract level, 12 passed to the full text phase; of those, none was considered relevant for the QPS assessment.
Revision of antimicrobial resistance aspects
No new information regarding AMR issues was identified during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, the QPS status of the Propionibacterium species does not change.
3.1.12. Streptococcus thermophilus
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered S. thermophilus has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 335; after screening at title/abstract level, 12 passed to the full text phase; of those, none was considered relevant for the QPS assessment.
Revision of antimicrobial resistance aspects
No new information regarding AMR issues was identified during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change.
3.2. Gram‐positive spore‐forming bacteria
3.2.1. Bacillus species
Taxonomy
Bacillus paralicheniformis, a recently described species, comprises former group 2 Bacillus licheniformis strains. Among relevant features of these new species members is the absence of ability to produce lichenicidin. Nevertheless, a putative new lantipeptide and gene clusters encoding bacitracin and fengycin are common features identified in B. paralicheniformis isolates (Dunlap et al., 2015)
The often‐incorrect assignment of Bacillus pumilus isolates was recently reported (Espariz et al., 2016; Branquinho et al., 2014). Classifying strains of this species using 16S rRNA gene sequence analysis can lead to incorrect species assignment, as it demonstrates over 99.5% identity with other species in the B. pumilus group.
Update of the body of knowledge on safety concerns for QPS Bacillus species
The total number of references found through the ELS was 4,091; after screening at title/abstract level, 264 passed to the full text phase; of those, 41 were considered relevant for the QPS assessment.
The following papers were not considered for further QPS assessment because of methodological shortcomings in the method used to identify the Bacillus strains to the species level (Jaber et al., 2013; Li et al., 2013b; K?vanç et al., 2014; Garcia Hejl et al., 2015; Guo et al., 2015; Shivamurthy et al., 2015; Grass et al., 2016). Bacillus thuringiensis is not considered for QPS, and therefore papers on this species (e.g. Fagerlund et al., 2014) were not further considered. A paper considering Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus was not considered in the context of the QPS evaluation on Bacillus spp. (Wenzler et al., 2015). The paper on a transovarial transmission of a Bacillus subtilis isolate that was made from a silkworm was not considered relevant for the QPS evaluation (Rai et al., 2013). Northern Ireland disease surveillance reports (Anonymous 2013a,b,c, 2014a,b,c) mentioned the involvement of B. licheniformis in bovine and ovine abortion cases. As these cases were linked with haematogenous spread of the bacteria (Agerholm et al., 1999), they were not further considered in the QPS evaluation.
Yoo et al. (2014) investigated a set of isolates, collected over several years, for possible production of cereulides and found producing isolates of B. subtilis, B. pumilus and Bacillus megaterium. Due to the lack of information on the identification methodology used for these isolates and the unreliability of several bacterial identification methodologies, there is uncertainty associated with the identity of these isolates.
The enterotoxic potential of B. megaterium isolates has been investigated by their adherence and invading potential in enterocyte‐like Caco‐2 cells (López et al., 2013). The cytotoxic activity reported, would be detected by current methodologies for assessing toxigenic activity in Bacillus species (EFSA BIOHAZ Panel, 2014).
The B. subtilis ATCC6051 showed a weak virulence in a virulence assay on brine shrimps (Lee et al., 2014b). Because this non‐standard virulence test is not yet validated, this result was not further taken considered the QPS evaluation.
Lichenysin production by all B. licheniformis isolates tested by Madslien et al. (2013) was reported, with cytotoxic levels associated with levels above 10 μg mL−1 corroborating previous findings (Madslien et al., 2013). Moreover, the cytotoxic activity reported, would be detected by the mandatory assessment of toxin production in isolates of QPS Bacillus species intended for food use (EFSA BIOHAZ Panel, 2014).
Idelevich et al. (2013) reported a bacteraemia case of small colony variants of B. licheniformis related to a pacemaker. The infection was cured by removal of the infected pacemaker.
A B. subtilis strain producing gamma‐glutamyltranspeptidase (GGT) as a bone‐resorbing virulence factor has also been described (Kim et al., 2016).
The cases of infections in humans and animals that were reported were not taken into account because of uncertainty on the identification methodology used and/or because the cases were linked to specific predisposing factors and do not suggest a risk for the consumer or the animal via exposure through the food and feed chain.
A recently published opinion of the EFSA BIOHAZ Panel addressed the risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including B. thuringiensis in foodstuffs did not report any additional concerns about Bacillus spp. included in the QPS list (EFSA BIOHAZ Panel, 2016b).
Revision of antimicrobial resistance aspects
Several papers reporting on AMR Bacillus strains were not taken into account because of uncertainty on the identification methodology (K?vanç et al., 2014), or on the methodology used to confirm the AMR (Shweta and Joseph, 2013; Fernández‐Fuentes et al., 2014; Mohammadou et al., 2014; Sadashiv and Kaliwal, 2014)
Update on other qualifications
The above new information does not affect the Bacillus‐related QPS qualification (‘absence of toxigenic activity’).
Other relevant information
A review article on new diagnostic identification methods of Bacillus stressed the evolution from detection towards subtyping and risk‐related strain profiling (Ehling‐Schulz and Messelhäusser 2013).
The whole genome of an isolate reported as Bacillus amyloliquefaciens subsp. plantarum (UCMB5033) has been published (Niazi et al., 2014). The whole genome of an isolate from the gut of an organically reared broiler reported as B. subtilis has been published (Schyns et al., 2013). The whole genome sequences of 20 isolates reported to belong to B. anthracis, B. atrophaeus, B. cereus, B. licheniformis, B. macerans, B. megaterium, B. mycoides and B. subtilis have been published (Daligault et al., 2014).
Conclusion regarding a QPS recommendation
There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain.
3.2.1.1. Bacillus flexus (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a).
Identity
B. flexus was originally described by Batchelor in 1919, and validated as revived species name (sp. nov., nom. rev.) by Priest et al. (1988). B. flexus is most closely related to Bacillus paraflexus (16S rRNA gene sequence similarity 98.1%), although also phylogenetically closely related to B. nealsonii (95%), B. niabensis (95%) and B. azotoformans (94%) (Chandna et al., 2013). B. flexus can be identified through 16S rRNA gene sequencing and differentiated from its closest phylogenetic neighbour B. paraflexus with standard phenotypic tests.
Body of knowledge
B. flexus has been used for production of enzymes, e.g. β‐amylase in food production and of alkaline amylase, lipase and protease for detergent formulations (Niyonzima and More, 2014). It has also been used for the production of exopolysaccharides (EPS) (Singh et al., 2013), and for bioremediation (Sivaprakasam et al., 2008; Pal et al., 2014; Das et al., 2016). Zhang et al. (2014) sequenced the whole genome of B. flexus strain T6186‐2, isolated from deep‐subsurface oil reservoirs.
Safety concerns
A recent outbreak of wound infections in burned patients was reported (Uçar et al., 2016). The outbreak was associated with contaminated swabs for wound sampling. Isolate identification procedures and virulence features characterisation were not presented in the study, neither was the relation between the infection and the B. flexus strain. To our knowledge, there are no reports of any virulence feature or disease associated with B. flexus, further supporting the absence of pathogenicity potential in a non‐compromised host.
Antimicrobial resistance
Genes encoding resistance to vancomycin (vanB), fosfomycin (fosB) and tetracycline (tetA) were described in a B. flexus strain (Zhang et al., 2014). Nevertheless, it was not possible from the data presented to assess if these genes are part of the chromosomal core genome and therefore present in all members of this species or associated with mobile resistance elements. Moreover, it is not possible to infer if they confer resistance to the mentioned antimicrobials.
Conclusions on a recommendation for the QPS list
B. flexus can be recommended for the QPS list with a qualification of the absence of toxigenic activity (as applied to all strains of Bacillus species recommended to the QPS list).
3.2.1.2. Bacillus smithii (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2017).
Identity
B. smithii is a rod‐shaped, motile, spore‐forming, facultatively anaerobic and facultatively thermophilic bacterium. This species is most closely related to Bacillus coagulans, which is also a facultatively thermophilic species. The complete genome of B. smithii type strain (B. smithii DSM 4216T) is available (Bosma et al., 2016).
Body of knowledge
There is a limited body of knowledge (48 references were found). As most spore‐forming bacteria, it is ubiquitous in nature, and therefore, it is also present in many raw materials and dry ingredients of processed food such as milk products (Lücking et al., 2013). It also has potential for the production of enzymes and other compounds, e.g. nitrile hydratases (Takashima et al., 2000) and a thermophilic inulinase (Gao et al., 2009). B. smithii possesses a possible protective effect against Salmonella and Clostridium difficile (Suitso et al., 2007; Jögi et al., 2008). It has been considered a relevant microorganism for biotechnological purposes, namely for conversion of biomass to fuel or chemicals (Bosma et al., 2015).
Safety concerns
Cytotoxicity assays using Vero and HEp‐2 cells in several Bacillus spp. strains, including B. smithii, did not identify any cytotoxic components, indicating that the risk of food‐borne disease is most likely low if at all (Lücking et al., 2013). Since members of this species were in the past probably assigned to B. coagulans, a species with QPS status, additional safety concerns related to misidentification are not expected.
Antimicrobial resistance
No information related to the presence of AMR determinants in members of this taxon has been identified.
Conclusions on a recommendation for the QPS list
The species B. smithii is a natural component of bacterial communities of fermented vegetables and plant derived products. Considering the lack of evidence of pathogenicity, it can be recommended for the QPS list with a qualification of absence of toxigenic activity (as applied to all strains of Bacillus species recommended to the QPS list).
3.2.2. Geobacillus stearothermophilus
Taxonomy
The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 4,091 together with Bacillus search; after screening at title/abstract level, 264 passed to the full text phase; of those and specifically for this TU, none was considered relevant for the QPS assessment.
Revision of antimicrobial resistance aspects
No new information regarding AMR was retrieved during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change.
3.3. Gram‐negative bacteria
3.3.1. Gluconobacter oxydans
Taxonomy
Since the last update on the QPS status (EFSA BIOHAZ Panel, 2013), no new information on the taxonomy of the considered G. oxydans species has been published.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 180; after screening at title/abstract level, eight passed to the full text phase; of those, none was considered relevant for the QPS assessment.
Revision of antimicrobial resistance aspects
No new information regarding AMR issues was identified during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified that justifies changing the qualification that the QPS only applies when the species is used for vitamin production.
Other relevant information
The presence of strains of G. oxydans with putative production of antimicrobials was followed in the ATCC Online Catalogue22 as indicated in the 2013 QPS Opinion. Antimicrobial production is not mentioned in any of the isolates, apart from one described as Gluconobacter spp.
Conclusion regarding a QPS recommendation
A total of 188 references were screened for relevant information related to G. oxydans. No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, the QPS status of G. oxydans does not change.
3.3.2. Xanthomonas campestris (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015a).
Identity
X. campestris is a valid name species (Skerman et al., 1980; Vauterin et al., 1995) from the genus Xanthomonas. It is a Gram‐negative, strictly aerobic, motile rod. X. campestris is a plant pathogen originally described as causing a vascular disease of Brassica spp. The species has been subdivided into pathovars, grouping strains having the same host plants (Bradbury, 1984).
Body of knowledge
On Brassica spp., X. campestris pathovar campestris is a seed‐borne pathogen, causing a systemic vascular disease of the plant (Vicente and Holub, 2013). Different pathovars do not produce the same symptoms, some causing non systemic spots on the host's leaves (Bradbury, 1984). X. campestris have been extensively studied in relation to interactions with the plant hosts in the field, but a literature search similar to that described for the body of knowledge of B. circulans failed to find studies on its presence in the harvested plants used as food or feed.
Some strains of X. campestris produce capsular polysaccharides, giving a slimy appearance of colonies on agar media containing utilisable carbon sources. Xanthan gum is one of them and it is composed of repetitive units of d‐glucose, d‐mannose, d‐glucuronic acid with terminal groups of pyruvic and acetic acids (Bradbury, 1984). Xanthan gum is industrially produced from X. campestris grown on adequate carbon sources for a very wide range of food and non‐food applications (Palaniraj and Jayaraman, 2011). Xanthan gum is used worldwide as an additive in many processed foods (Codex Alimentarius, 2014).
The genome of a strain of X. campestris used for industrial production of xanthan gum has been sequenced (Tao et al., 2012).
Safety concerns
X. campestris is a pathogen of many plant species. A search on safety concerns similar to that done for B. circulans retrieved articles on the virulence of X. campestris on plants (search strings in Appendix A). An additional search was done on its association with animals and humans, combining search terms relating to human and some animal species with X. campestris. Only one publication was found, mentioning its isolation from a human blood sample in China (Li et al., 1990).
Antimicrobial resistance
X. campestris pv. campestris expresses a chromosomally encoded class A β‐lactamase which confers resistance to penicillin and carboxypenicillins (Yang et al., 2011, 2014). No evidence of the acquisition of an antibiotic resistance gene was retrieved in the literature search performed.
Conclusions on a recommendation for the QPS list
Xanthan gum produced by X. campestris has a long and broad history of safe use in the food industry. X. campestris is a plant pathogen. Apart from one record (Li et al., 1990), X. campestris has never been implicated in human or animal disease. However, human consumers are presumably very rarely exposed to high levels of X. campestris through food, indicating a lack of knowledge on the effect of high levels of live cells of X. campestris on animals and humans.
In all papers screened, none of them mentioned acquisition of resistance to antimicrobials. X. campestris can be recommended for the QPS list for the production of xanthan gum.
3.4. Yeasts
Fungi are unique among living organisms because they may have two valid names. The primary name is based on the sexual state or teleomorph, but a second valid name may be based on the asexual state or anamorph. This redundancy of names developed because teleomorphs have not been found for many fungi, or it has not been clear that a particular teleomorph is the same species as a particular anamorph (Kurtzman et al., 2011).
In the screened scientific reports on yeasts, alternatively the teleomorph or anamorph names (and sometimes both) are used. However, in the following evaluations of the yeast species, in general, the teleomorph name is preferentially used, but for clarity the anamorph (when known) and synonyms are also mentioned.
3.4.1. Candida cylindracea (TU included after the 2013 QPS update)
Evaluation published in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014).
Identity
C. cylindracea belongs to the Ogataea clade of the ascomycetous yeasts (Kurtzman et al., 2011; Daniel et al., 2014). The species was described by Yamada and Machida (1962), and validated by Meyer and Yarrow (1998). No synonym names have been used. Only the anamorphic form is known and described. The type strain for C. cylindracea – CBS 6330 – is also marketed under other designations, e.g. DSMZ 2031 (online) and ATCC 14930 (online). Unfortunately, in the literature on lipase‐producing yeasts, the C. cylindracea type strain has at times been referred to as Candida rugosa (e.g. Benjamin and Pandey (1998); Takaç et al. (2010)). This has caused some confusion since C. cylindracea and C. rugosa are two well‐defined species, not closely related phylogenetically (Kurtzman et al., 2011). It is also unfortunate since C. rugosa is considered an emerging, opportunistic yeast (Miceli et al., 2011). However, identification according to molecular methods can easily separate between the two species. It is therefore recommended that the species identity of lipase‐producing strains of Candida is confirmed by using such methods.
Body of knowledge
C. cylindracea has been used for a long time in industry as a lipase producer (Tomizuka et al., 1966; Brozzoli et al., 2009). The Ogataea clade to which it belongs does not include the pathogenic yeast Candida albicans (which belongs to the Lodderomyces–Spathaspora clade) or other Candida species associated with human infections, like Candida tropicalis, Candida glabrata, Candida parasilopsis or C. rugosa.
Safety concerns
A literature search for ‘Candida cylindracea’ on Thomson Reuters Web of Science (7 July 2014) gave 797 hits. The vast majority of the retrieved studies treated different aspects of enzyme production by this species. None of the studies implied a potential safety issue for C. cylindracea. No clinical reports for C. cylindracea were recovered in the search and the species is not mentioned in reviews on emerging opportunistic yeasts (e.g. Miceli et al. (2011)). C. cylindracea does not grow at 37°C (Kurtzman et al., 2011).
Conclusions on a recommendation for the QPS list
In the C. cylindracea bibliography, the species was only reported for use as an enzyme producer and no safety concerns were identified. Therefore, it was concluded that it can be recommended for the QPS status. However, since there were no reports on its use in applications involving direct consumption of C. cylindracea viable cells by humans or animals, the QPS should apply only for the production of enzymes.
3.4.2. Debaryomyces hansenii
Taxonomy
The anamorph form of D. hansenii is Candida famata. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment.
However, from these, only 37 were linked to D. hansenii and considered relevant for the QPS assessment. In the majority of the publications identified, the authors used the name of the anamorphic form, C. famata.
Only in very few of the reports, the yeast was identified by molecular techniques like PCR using specific primers (Pisa et al., 2015) or PCR‐restriction fragment length polymorphism (PCR‐RFLP) (Muadcheingka and Tantivitayakul, 2015) or other techniques like specific antibody detection (Pisa et al., 2015) or by matrix‐assisted laser desorption/ionisation time‐of‐flight mass spectrometry (MALDI‐TOF MS) (Riat et al., 2015). In the majority of the publications, physiological criteria were used or the methodologies used were not specified. Using this approach, the presence of D. hansenii has been described in patients infected by HIV (Nidhi et al., 2015; Ribeiro et al., 2015; Minea et al., 2016), with cancer (Li et al., 2013a; Nidhi et al., 2015), with post‐operational acute respiratory distress syndrome (Mun et al., 2015), with invasive candidiasis (Ghahri et al., 2013; Wang et al., 2014; Jung et al., 2015; Nieto et al., 2015), in pregnant women in Malaysia hospital (Masri et al., 2015) or vulvovaginal candidiasis (Liu et al., 2014), in patients with fungal eye infections (Ghodasra et al., 2014), with psoriasis (Sarvtin et al., 2014), with chronic diarrhoea (Banerjee et al., 2013), athlete′s foot (Chan et al., 2013) and in children with neutropenia (Haddadi et al., 2014). The value of these reports is often limited considering the limitations of the identification method.
In the big majority of cases, D. hansenii was isolated from patients with various serious underlying diseases or other immunosuppressed states, and there is no connection between the disease and consumption of the yeast. Additionally, in all the cases, the presence of D. hansenii among other species involved in the yeast infections is very low, seldom above 5%.
Revision of antimicrobial resistance aspects
Beyda et al. (2013) described a strain of D. hansenii from bloodstream infections which exhibited a reduced susceptibility to echinocandins and azoles.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
The new information regarding potential concerns for human or animal safety, or other related aspects do not imply new concerns with respect to the QPS status of D. hansenii. Therefore, its QPS status does not change.
3.4.3. Hanseniaspora uvarum
Taxonomy
The anamorph form of H. uvarum is Kloeckera apiculata. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Specifically for this TU, no further information concerning safety concerns was found.
Revision of antimicrobial resistance aspects
No new information regarding AMR issues was retrieved during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not suffer any change.
3.4.4. Kluyveromyces species
Taxonomy
Two species of the genus Kluyveromyces are included in QPS list, Kluyveromyces lactis (anamorph Candida spherica) and Kluyveromyces marxianus (anamorph Candida kefyr). The species names have not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. From these, only 43 were linked to Kluyveromyces species.
The ELS search retrieved no new studies with relevance for the QPS evaluation of K. lactis (or its anamorphic name C. spherica). For K. marxianus (including the anamorph C. kefyr), a total number of 43 references were selected after full text phase. The great majority of studies (33) deemed relevant for the QPS used the anamorph name C. kefyr. The new information on K. marxianus/C. kefyr is evaluated below.
Quite a few of the reports deemed relevant in the ELS (43) presented information demonstrating a low or very low prevalence (seldom above 5%) of K. marxianus among fungi (mainly Candida spp.) isolated from patients with various serious underlying diseases, like cancer or HIV, or other immunosuppressed states, and patients with catheters (de Freitas et al., 2013; Ghahri et al., 2013; Parmeland et al., 2013; Abrantes et al., 2014; Dufresne et al., 2014; Haddadi et al., 2014; Taj‐Aldeen et al., 2014; Youngster et al., 2014; Leuck et al., 2015; Menezes et al., 2015; Nidhi et al., 2015; Nieto et al., 2015; Sahin et al., 2015).
Sarbu et al. (2013) studied virulence factors in isolates from vulvovaginal infections. A single case included a K. marxianus isolate, but it was positive only for haemolysins and siderophore‐like compounds and negative for the other factors studied.
Muadcheingka and Tantivitayakul (2015) found K. marxianus at a low prevalence (3.6%) in Candida isolates from patients with oral candidiasis (no underlying diseases reported) in a dental clinic in Thailand.
A K. marxianus bloodstream infection in a patient with brainstem dysfunction and a case of cardiac arrest was reported by Khan et al. (2015).
Alfouzan et al. (2015) reported very low prevalence of K. marxianus among yeast isolates from patients with vaginitis (no other underlying disease reported) in Kuwait, and Madhumati et al. (2014) reported similar observations in India.
Swarajyalakshmi and Jyothilakshmi (2014) reported a case of sinusitis caused by K. marxianus (solely biochemical identification) in a woman with diabetes.
K. marxianus was moderately prevalent (5.8%) among 855 yeast isolates from various clinical specimens (mainly nail and vulva‐vagina) from different regions in Iran (Mohammadi et al., 2013). The association with disease in the subjects is not described and the significance of the study for the QPS is therefore unclear.
Khosravi et al. (2013) reported that K. marxianus constituted 6.2% of yeast species obtained from patients with nail infections.
Revision of antimicrobial resistance aspects
Some studies demonstrate antimycotic susceptibility in K. marxianus. Montagna et al. (2014) found no amphotericin resistance in the tested clinical isolates, and Yigit and Aktas (2014) similarly found no resistance against amphotericin B and azoles.
Other studies reported antimycotic resistance in K. marxianus clinical isolates. For instance, in India, Shyamala and Parandekar (2014) reported azole resistance in isolates from HIV patients, and Deorukhkar and Santosh (2013) and Sasikala et al. (2013) in isolates from patients with suspected or confirmed vulvovaginal candidiasis.
Dufresne et al. (2014) provided indications that under treatment with antimycotics, some K. marxianus isolates could develop resistance to the provided substance. Similarly, Fekkar et al. (2013) showed that a K. marxianus isolate acquired echinocandin resistance after initiation of caspofungin treatment for candidemia.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
There is no doubt that K. marxianus/C. kefyr should be considered a significant opportunistic fungus, and it has received increased attention in recent years However, reports where it has been unambiguously shown to be causative agent of infectious disease in otherwise healthy individuals are very rare. Therefore, its QPS status does not change. There is reason to be alert regarding whether there is a tendency for K. marxianus to become more common in this kind of infection.
3.4.5. Komagataella pastoris
Taxonomy
The anamorph of K. pastoris is not described. The previous name of this species is Pichia pastoris. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Specifically for this TU, no further information concerning safety concerns was found.
Revision of antimicrobial resistance aspects
No new information regarding AMR was identified during the period covered by the ELS.
Update on other qualifications
For K. pastoris, the QPS only applies when the species is used for enzyme production and no viable cells are found.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change. The qualification is unchanged.
3.4.6. Lindnera jadinii
Taxonomy
The anamorph form of L. jadinii is Candida utilis. Synonyms of this species are Pichia jadinii, Hansenula jadinii and Torulopsis utilis. The species name has not been changed since the 2013 QPS Opinion.
All studies relevant for the evaluation of this species were retrieved when using the anamorph name C. utilis, and there were no hits when using the teleomorph name L. jadinii.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. From these, only six were linked to L. jadinii.
Svobodova et al. (2016) compared biochemical identification of clinical yeast isolates to Wickerhamomyces anomalus/Candida pelliculosa and L. jadinii/C. utilis, with MALDI‐TOF MS identification of the same isolates. The MALDI‐TOF, identified the majority of the isolates as Candida fabianii, a non‐QPS species. This indicates that the prevalence of L. jadinii (and W. anomalus) in clinical samples may have been overestimated in previous studies employing biochemical identification.
Another study (Fadda et al., 2013) compared identification methods for yeast isolates from cows with mastitis. API kits for assimilation tests generally agreed well with a PCR‐RFLP method for the Candida species investigated, including L. jadinii/C. utilis.
Update of the body of knowledge on safety concerns
Minea et al. (2016) found L. jadinii in low prevalence from HIV or diabetes patients in Romania, but it the identification methods used in the study were not described. Hammad et al. (2013) reported a very low prevalence (0.6%) of L. jadinii in oral yeast flora of type II diabetics with periodontitis in Jordan. Luzzati et al. (2013) found very low prevalence (0.7%) of L. jadinii in yeast isolates from elderly, candidemia patients in Italy. Eddouzi et al. (2013) reported very low prevalence (0.25%) of L. jadinii among isolates from patients with fungal infections in Tunisian hospitals.
Scoppettuolo et al. (2014) reported a rare case of catheter‐related blood stream infection with L. jadinii in a cancer patient with a long‐term implanted venous catheter.
Revision of antimicrobial resistance aspects
Minea et al. (2016) reported antimicrobial susceptibility of clinical isolates, largely confirming previous data. A Tunisian study (Eddouzi et al., 2013) reported high antimycotic susceptibility in clinical isolates of L. jadinii.
Update on other qualifications
For L. jadinii, the QPS only applies when the species is used for enzyme production and no viable cells are found.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
Few studies reported isolation of L. jadinii/C. utilis in clinical samples. Human isolates were only recovered from people with underlying disease, and prevalence was generally low compared to other infectious yeast isolated from collections of clinical samples. No studies reported increased prevalence of antimycotic resistance. In conclusion, no information was retrieved to indicate a change in the QPS status.
3.4.7. Ogataea angusta
Taxonomy
The anamorph form of O. angusta is not described. A synonym of this species is Pichia angusta. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Specifically for this TU, no further information concerning safety concerns was found.
Revision of antimicrobial resistance aspects
No new information regarding AMR was retrieved during the period covered by the ELS.
Update on other qualifications
For Ogataea angusta, the QPS only applies when the species is used for enzyme production and no viable cells are found.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change.
3.4.8. Saccharomyces cerevisiae/species
Taxonomy
The anamorph form of S. cerevisiae is not described. A synonym of this species is Saccharomyces boulardii. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 2,421, after screening at title/abstract level, 117 passed to the full text phase; of those, 27 were considered relevant for the QPS assessment and linked to S. cerevisiae. From the searches done, for the other two searches for yeasts groups as described in Appendix C, 7 references were also found. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Therefore, a total of 34 references were identified for this TU.
During this period, new cases of fungaemia caused by S. cerevisiae were described. In the majority of these reports, the authors used molecular techniques like RFLP of the ribosomal region that included the 58S and ITS region. The reports where the authors used only physiological methods for identification or the methodology was not described, were not considered. Popiel et al. (2015) report fungaemia in a 60‐year‐old man whose orthotopic liver transplant was complicated by S. cerevisiae (Popiel et al., 2015). Pillai et al. (2014) report fungaemia caused by S. cerevisiae in a woman with chronic kidney disease and S. cerevisiae was isolated from blood, urine and stool, as well as vaginal swabs (Pillai et al., 2014).
Some other reports are associated with the synonymous S. boulardii. Santino et al. (2014) reported fungaemia caused by S. cerevisiae/boulardii in an 86‐year‐old man with gastrointestinal disorders (Clostridium difficile infection). Cohen et al. (2013) reported a case of a young drug user that injected S. cerevisiae/boulardii intravenously. She developed a transient fever but spontaneously recovered after 2 days.
S. cerevisiae also has been described in fungaemias with very low incidence levels associated with other yeast species of the genus Candida (Martos et al., 2014), including other QPS species like D. hansenii and the anamorph form C. famata, C. kefyr (anamorph form of K. marxianus), C. utilis (anamorph form of L. jadinii), also appearing at low incidence (Eddouzi et al., 2013; Fadda et al., 2013; Fekkar et al., 2013; Li et al., 2013c; Arancia et al., 2014).
An allergic reaction and positive skin test was reported in a patient who took S. cerevisiae/boulardii) as an antidiarrhoeal therapy (Kartal et al., 2014).
Llopis et al. (2014) used virulence‐associated phenotypic traits and an in vivo study in a murine model, to analyse the potential virulence of viable isolates contained in dietary supplements. One of the S. cerevisiae isolates caused death in murine models. The authors suggest a strong relationship between some of the virulence‐associated phenotypic traits (ability to grow at 39°C and pseudohyphal growth) and the in vivo virulence in a mouse model of intravenous inoculation. These data confirm previous QPS qualifications for S. cerevisiae.
Revision of antimicrobial resistance aspects
Santino et al. (2014) described an isolate of S. boulardii (synonym of S. cerevisiae) from an 86‐year‐old man that presented susceptibility to caspofungin and voriconazole but resistance to fluconazole, intraconazole and amphotericin B. According to these data, the recommendation (qualification) is to maintain the absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain.
Update on other qualifications
In the case of S. cerevisiae the qualification that applies for yeast strains able to grow above 37°C is maintained.
Other relevant information
Other papers are related to the virulence and pathogenicity characterisation of S. cerevisiae. Anoop et al. (2015) and Pérez‐Torrado and Querol (2015) overview the infection mechanisms and virulence factors in opportunistic isolates of S. cerevisiae. Pérez‐Torrado et al. (2015) performed a genomic characterisation of S. cerevisiae isolates including clinical and food isolates. The results showed increased copy numbers of immune deficiency‐like genes in opportunistic isolates, which are implicated in the de novo biosynthesis of the purine nucleotides pathway. These results demonstrated a specific mechanism present in virulent isolates of S. cerevisiae that opportunistic yeasts may use (enhanced de novo biosynthesis of the purine nucleotides pathway) to increase survival and favour infections in the host. Roig et al. (2013) reports a genetic determinant that seems to contribute to virulence in clinical isolates of S. cerevisiae, the FLO11 gene. Strope et al. (2015) performed whole genome sequencing of 93 isolates of S. cerevisiae including clinical isolates. The authors found changes in the genome sequence of the gene PDR5 (a multidrug transporter), present exclusively in clinical isolates.
Conclusion regarding the maintenance of the QPS recommendation
These new reports of S. cerevisiae appearing as an opportunistic pathogen add no further concern regarding its QPS status. Consumption of S. boulardii (synonym of S. cerevisiae) by patients with fragile health may be considered as the origin of the infection, although the use of microorganisms intended to be used as ‘probiotic’ for humans as a health claim does not fall under the remit of the QPS assessment. These new reports also confirm the previous QPS qualifications, the absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain and inability to grow above 37°C. Therefore, its QPS status does not change.
3.4.9. Schizosaccharomyces pombe
Taxonomy
There are no synonym names in common use for this species and the name has not been changed since 2013.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Specifically for this TU, one reference was identified but no further information concerning safety concerns was found.
Revision of antimicrobial resistance aspects
No new information regarding AMR was retrieved during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change.
3.4.10. Wickerhamomyces anomalus (Pichia anomala)
Taxonomy
The anamorph form of W. anomalus is Candida pelliculosa. Synonyms of this species are Hansenula anomala, Pichia anomala and Saccharomyces anomalus. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. From these, 16 references were related to W. anomalus.
Some of the reports deemed relevant in the ELS (16) reported W. anomalus/C. pelliculosa among fungi (mainly Candida spp.) isolated from patients with various serious underlying diseases and suffering infections by opportunistic microbes (Kuiper et al., 2013; Oliveira et al., 2014; Taj‐Aldeen et al., 2014; Lin et al., 2015; Nidhi et al., 2015; Tzar et al., 2015). Additionally, W. anomalus was always a minor fraction (less than 10%) of the isolates.
Svobodova et al. (2016) compared biochemical identification of clinical yeast isolates to W. anomalus and C. utilis, with MALDI‐TOF MS identification of the same isolates. MALDI‐TOF, identified the majority of the isolates as Candida fabianii, a non‐QPS species. This indicates that the prevalence of W. anomalus in clinical samples may have been overestimated in previous studies employing biochemical identification.
Deepak et al. (2015) reported that yeasts (e.g. W. anomalus) can have detoxifying activity on aflatoxin.
One study presented the MALDI‐TOF MS method that conveniently and reliably identified many yeasts that may cause opportunistic infections, including W. anomalus (Ghosh et al., 2015).
Kamoshita et al. (2015) reported a case where a 91‐year‐old woman suffered from fungal keratitis caused by W. anomalus after corneal transplantation. Similarly, Esgin et al. (2014) reported post‐operational infection by W. anomalus in the eye of an adult man in Turkey.
One study reported an outbreak of W. anomalus fungaemia, where one strain was retrieved from the bloodstream of six infants, in a neonatal intensive care unit in Taiwan (Lin et al., 2013).
Revision of antimicrobial resistance aspects
Some studies demonstrate antimycotic susceptibility in W. anomalus. Montagna et al. (2014) found no amphotericin resistance in the sole, tested clinical isolate. Shyamala and Parandekar (2014) reported that the sole tested clinical isolate of W. anomalus was susceptible to three of the tested azoles and non‐susceptible to one (itraconazole).
Update on other qualifications
For W. anomalus, the QPS only applies when the species is used for enzyme production and no viable cells are found. The absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
Comparatively few studies reported isolation of W. anomalus/C. pelliculosa in clinical samples. Human clinical isolates were mainly recovered from people with underlying disease and in these cases W. anomalus was always a minor fraction of the isolates, and there were no indications it may be food‐borne. No studies reported infection in healthy, non‐hospitalised subjects or signs of increased prevalence of antimycotic resistance. Therefore, its QPS status does not change.
3.4.11. Xanthophyllomyces dendrorhous
Taxonomy
The anamorph form of X. dendrorhous is Phaffia rhodozyma. The species name has not been changed since the 2013 QPS Opinion.
Update of the body of knowledge on safety concerns
This TU was included in a search for several TUs as described in Appendix C. The total number of references found through the ELS for these groups of TUs was 2,857; after screening at title/abstract level, 264 passed to the full text phase; of those, 143 were considered relevant for the QPS assessment. Specifically for this TU, no further information concerning safety concerns was found.
Revision of antimicrobial resistance aspects
No new information regarding AMR was retrieved during the period covered by the ELS.
Update on other qualifications
No new relevant information was identified.
Other relevant information
No new relevant information was identified.
Conclusion regarding the maintenance of the QPS recommendation
No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change.
3.5. Viruses used for plant protection
A number of viruses has been recommended for use to control plant pests. The first category encompasses ‘mild strains’ of plant viruses to mitigate the effects of ‘severe strains’ of the same virus species, the latter causing severe disease for example in tomato and squash. The viruses notified to EFSA are members of two well‐characterised plant virus families, the Alphaflexiviridae (Order Tymovirales) and Potyviridae. The second category consists of baculoviruses (family Baculoviridae) that kill specific species of pest insects.
Taxonomy
In order for viruses to be considered for QPS, the taxonomy of these pathogens should be unequivocal. The taxonomy and nomenclature of viruses are the responsibility of the International Committee on Taxonomy of Viruses. The most recent report (9th) is from November 2011 (King et al., 2012), but regular updates are published on the ICTV website, the most recent one being from 2015. The status of e‐viruses (computationally identified viruses) is being discussed. The taxonomy of the viruses relevant for QPS assessment is outlined in a previous QPS report (2013) and has not changed since.
3.5.1. Plant viruses
Viruses belonging to certain plant virus families (Alphaflexiviridae and Potyviridae) are sometimes used for cross protection purposes, i.e. the application of mild strains of a plant virus giving mild symptoms is used to protect the food or feed crop against strains of the virus giving severe symptoms and yield losses. This strategy was also previously known as premunition. The mechanistic explanation for the protective effect has not been unequivocally determined. One theory holds that the innate immunity (RNAi) response is triggered throughout the plant by mild strains and quickly controls virus attack by virus strains that would otherwise provoke a severe response. Sequence similarity between two such viruses is a prerequisite for this. Another theory postulates that the mild strains produce considerable amounts of virion coat protein. Upon uncoating of virions of a severe strain of the virus, there is a surplus of coat protein present from virions of the mild strain, hence promoting the repackaging of the uncoated virus RNA of the severe strain and thus blocking it from expression of its virulent genes.
Plant viruses do not replicate in organisms other than plants. The parts exposed to animal and/or humans are the coat protein(s) and the nucleic acid, which are, in all but a few cases, RNA. The potential effects of such viruses on animals and/or humans, when applied to food or feed, were reviewed and assessed, and the results were published in the EFSA Opinions on QPS in 2009 (EFSA, 2009, 2012b), 2010 (EFSA BIOHAZ Panel, 2010), 2011 (EFSA BIOHAZ Panel, 2011), 2012 (EFSA BIOHAZ Panel, 2012) and 2013 (EFSA BIOHAZ Panel, 2013).
3.5.1.1. Alphaflexiviridae
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 198; after screening at title/abstract level, nine passed to the full text phase; of those, six were considered relevant for the QPS assessment.
From these, four ‘alphaflexiviridae’ papers were either related to a human vaccine using an alphaflexvirus as a carrier (not related to agronomic application) or reported some hypothetical effect on the environment (EFSA BIOHAZ Panel, 2013; Yusibov et al., 2013; Duff‐Farrier et al., 2015; Minicka et al., 2015).
There was no scientific evidence that alphaflexviruses or members of the genus Potexvirus have any negative effects on animals and humans. Viruses of this family have been reported from a wide range of herbaceous and woody plants, both monocotyledons and dicotyledons. Species of this virus family are mostly plant‐specific and are transmitted from plant to plant either mechanically or through insect vectors. So, they are widely present in the environment and deliberate release will not cause additional environmental effects. In terms of safety, the viruses neither carry nor encode toxic compounds to vertebrates including humans (EFSA BIOHAZ Panel, 2013) and the familiarity principle was taken into account as well, in that, these viruses have been part of the food and feed of animals and humans via plant material.
A pathogenicity determinant of the alphaflexvirus, Pepino mosaic virus (mild strain vs severe strain) in plants was mapped to the N‐terminus of viral coat protein (Duff‐Farrier et al., 2015), but there is no evidence it can affect vertebrates. The major component of an alphaflexvirus (e.g. Pepino mosaic virus), the coat protein, was tested computationally in 2013 against a plant database (UniRef100 plant database (UniProt NREF, 2013) and did not show any homology to known toxins. None of the hits were related to the search terms ‘disease’ or ‘toxins’. No other negative impacts of alphaflexiviruses, more specifically potexviruses such as Pepino mosaic virus (genus Potexvirus) on humans or animals have been reported to date (EFSA BIOHAZ Panel, 2013).
Update on other qualifications
Not applicable.
Other relevant information
No safety concerns, including environmental effects, have been reported for members of the Alphaflexiviridae.
Conclusion regarding the maintenance of the QPS recommendation
On the basis of the scientific information identified through the ELS, the QPS recommendation on members of the Alphaflexiviridae family can be maintained.
3.5.1.2. Potyviridae
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 198; after screening at title/abstract level, nine passed to the full text phase; of those, six were considered relevant for the QPS assessment.
From the relevant reports, it was concluded that the four papers were either related to a functional, fundamental aspect of the pertinent virus or reported some ecological effect (EFSA BIOHAZ Panel, 2013; Yusibov et al., 2013; Hillung et al., 2014; Hasiow‐Jaroszewska et al., 2015).
For the potyviruses, no scientific evidence indicated that these viruses have any negative effects on animals and humans to date. In addition, the familiarity principle was taken into consideration, since these viruses have been part of the food and feed of animals and humans since they are widely present in edible plants.
Computational analysis demonstrated that the major component of a Potyvirus (Zucchini yellow mosaic virus), the coat protein, did not show any homology to known toxins (Health Canada, 1999; Kuiper et al., 2001). Such an analysis was repeated in 2012 against a plant database (UniRef100 plant database (UniProt NREF, 2013) and a general database (GenBank nt database, 2013) and none of the hits were related to ‘disease’ or ‘toxins’. Since the last major review by Kuiper et al. (2001), no new information has appeared that would compromise the conclusion drawn in 2013 (EFSA BIOHAZ Panel, 2013).
Update on other qualifications
Not applicable.
Other relevant information
No safety concerns, including environmental effects, have been reported for members of the Potyviridae.
Conclusion regarding the maintenance of the QPS recommendation
On the basis of the scientific information found, including the one retrieved from the ELS, the QPS recommendation on members of the Potyviridae family can be maintained.
3.5.2. Insect viruses
Most, but not all, insect viruses are pathogenic to insects, but only baculoviruses (family Baculoviridae) have been fully developed as bioinsecticides. The latter viruses act by killing larvae of target insects in a few days and disseminating new viruses that can affect new target insect larvae. These baculoviruses are usually species‐specific and are natural biocontrol agents, as through their lethal action on insect larvae they regulate the size of insect populations in agroecosystems. As they are sprayed on crops, the plants or fruits for consumption can be contaminated with the relevant baculovirus.
3.5.2.1. Baculoviridae
Update of the body of knowledge on safety concerns
The total number of references found through the ELS was 492; after screening at title/abstract level, 13 passed to the full text phase; of those, seven were considered relevant for the QPS assessment.
From the relevant reports, it was concluded that they either concerned fundamental and mechanistic aspects of baculoviruses or medical and/or pharmaceutical applications of baculovirus vector‐derived products, stating their safety to humans (OECD, 2002; Lapointe et al., 2012; Airenne et al., 2013; Chen et al., 2013; O'Flynn et al., 2013; Swift et al., 2013; Fujihira et al., 2014; Felberbaum, 2015; Fujita et al., 2015).
No scientific evidence indicated that these viruses have any negative effects on animals and humans to date. In addition, the familiarity principle was taken into consideration. Baculoviruses have been extensively used for over seven decades as biocontrol agents of insect pests without any report of a negative effect on humans or animals. The Organisation for Economic Cooperation and Development (OECD) already concluded in 2002 that baculoviruses were safe to use for products meant for human consumption (OECD, 2002). This opinion was supported in a more recent review by Lapointe et al. (2012).
A matter of concern has been the observation that the budded virus phenotype of baculoviruses is able to enter vertebrate cells, including mammalian cells and tissues (Hofmann et al., 1995). This baculovirus phenotype is responsible for the systemic infection of insect larvae and has been developed into an effective gene delivery vehicle (vector) for gene therapy. So far, there is no evidence that such a baculovirus‐derived vectors can transform vertebrate cells or induce malignancies in test animals or humans (Fleming and Hunt, 2000; Kost and Condreay, 2001; Chen et al., 2011; EFSA BIOHAZ Panel, 2013). In addition, this phenotype is not present in the occlusion bodies that are sprayed to control insects.
Update on other qualifications
Not applicable.
Other relevant information
The impact of using baculoviruses as insect biocontrol agents on the environment has been assessed in an OECD report in 2002 and again in a FAO report in 2007 (McWilliam, 2007). From both assessments was apparent clear that there is no major impact on the environment, beyond what is expected from an intervention, other than the elimination of pest insects. There is, for example, no effect on non‐target insects and no long‐term effects on the ecosystem. Short‐term effects may be the depletion of food (insects) for birds and reptiles, but the ecological balance is expected to be restored in the long‐term.
Conclusion regarding the maintenance of the QPS recommendation
On the basis of the scientific information found including the one retrieved through the ELS, the QPS recommendation on members of the Baculoviridae family does not change and that the family Baculoviridae is the lowest TU with QPS.
3.6. Taxonomic groups excluded from the QPS exercise
3.6.1. Bacteriophages
In the 2009 at Scientific Opinion on the maintenance of the list of QPS microorganisms intentionally added to food or feed, “phages were not considered appropriate for consideration as QPS organisms because: (i) no rational classification could be established to the species level and (ii) discarding temperate phages, which promote bacterial survival through superinfection immunity and confer new, potentially harming, properties to their hosts (lysogenic conversion) and elimination of bacterial DNA transfecting phages, had to be done on a case‐by‐case basis.
After a 5‐year period without considering phages for QPS, the working group again raised the question, based on two premises: (i) the TU to be taken into consideration may not be the species (for instance, families are being considered for insect and plant specific viruses) and (ii) the impediments associated with phage biology might be converted into qualifications.
The outcome of the discussion was, against changing the bacteriophage consideration with respect to QPS. The reasons were: (i) the lowest level phylogenetic TU should be the order Caudovirales (which includes 95% of all known phages) and which was considered to be too wide; (ii) qualifications are perceived as traits whose presence/absence in an otherwise safe organism, can be tested with procedures that provide unequivocal answers, are limited in number, and are easy to apply. Testing for virulence vs temperate, for the absence of genes that promote lysogenic conversion and deciphering the DNA packaging mechanisms as a means to distinguish between transducing and non‐transducing phages would involve thorough analysis of the genomes.
These reservations indicated that phage application on foods should remain as a case‐by‐case procedure and, consequently, that these biological entities should not be considered for the QPS status.
3.6.2. Clades of Enterococcus faecium
The QPS approach relies on the basis of the evaluation of TU, where the species/subspecies level is the lowest level of evaluation. Therefore, clades within the E. faecium species cannot be considered as a TU and cannot be evaluated separately. The specific guidance document on E. faecium produced by the FEEDAP Panel in 2012 is a valuable tool that enables a simplified evaluation without unnecessary animal experiments outside the QPS approach (EFSA FEEDAP Panel, 2012c). This approach considers that strain of E. faecium that do not contain marker genes typical of hospital‐associated isolates responsible for clinical infections or show resistance to clinically relevant antibiotics are considered safe for use as feed additives.
Thus, the conclusions of the last update on the QPS (EFSA BIOHAZ Panel, 2013) are still valid and E. faecium should be monitored and re‐evaluated in the next QPS Opinion update.
3.6.3. Filamentous fungi
Although fungal taxonomy is in a rapid development, still these studies seldom provide information about the ecological properties and the function of the TUs. The discontinuation of dual nomenclature for pleomorphic fungi has resulted in nomenclatural changes to well‐established fungal species. The increasing availability of fungal genome sequences is facilitating the discovery and characterisation of numerous novel secondary metabolites by genome mining. While knowledge of fungal secondary metabolites has grown to a big extent, information on their toxic effects in humans and animals is still evolving at a much slower rate.
Therefore, it was decided that until further notice, filamentous fungi are excluded from the QPS evaluations. Their status should be monitored and re‐evaluated in the next QPS Opinion update.
4. Conclusions
Answer to the terms of reference (ToR):
ToR 1: “Keep updated the list of biological agents being notified in the context of a technical dossier to EFSA Units (such as Feed, Pesticides, FIP and Nutrition) for intentional use in feed and/or food or as sources of food and feed additives, enzymes and plant protection products for safety assessment.”
The list of biological agents notified in the context of technical dossiers was updated. From the last notifications received for the previous QPS Opinion in 2013, 405 notifications were received between May 2013 and September 2016; of which, 137 were from Feed, 196 from FIP, 11 from Nutrition and 61 from Pesticides. For the type of microorganisms, 183 were bacteria, 177 filamentous fungi, 9 viruses and 36 yeasts TUs.
ToR 2: “Review taxonomic units previously recommended for the QPS list and their qualifications (especially the qualification regarding antimicrobial resistance) when new information has become available. Update the information provided in the previous opinion where appropriate.”
All TUs that had been previously recommended for the QPS list in the 2013 Opinion were reviewed and confirmed. The information of the previous opinion was updated and the qualifications were also confirmed. An ELS was included, which has allowed a better harmonisation and transparency of the assessment process, identifying the criteria to exclude possible safety concerns described in the articles identified within the search and screened as relevant. A more structured description of the evaluation process for the revision of TUs included in the QPS list, as well as for those evaluated for a possible QPS recommendation from new notifications (ToR3) has been applied. Information about antimicrobial resistance has been reviewed following recent recommendations from EFSA Opinions published in this topic.
ToR 3: “(Re)assess the suitability of taxonomic units notified to EFSA not present in the current QPS list for their inclusion in that list.”
Five Panel Statements have been published periodically (approximately every 6 months) in order to assess the suitability of new TU notified to EFSA and to update the list with those biological agents that were recommended for the QPS list. From a total of 405 notifications, 153 biological agents already had a QPS status and were not further evaluated, neither were the 188 filamentous fungi and enterococci, biological groups which have been excluded from QPS consideration (following a recommendation of the QPS 2013 update, 28 were not included because the corresponding TUs have already been evaluated in the previous Statements during this period. Furthermore, it was agreed not to include nine notifications from Pesticides Unit as the respective dossiers (including the literature review) were not yet received. The remaining 41 biological agents were assessed for the suitability of the respective TUs for inclusion in the QPS list.
As a result of the evaluation, C. divergens, L. diolivorans, M. imperiale, P. nishizawae, P. parvulus, B. flexus, B. smithii, X. campestris and C. cylindracea are included in this Opinion as new members since the 2013 update. This increased periodicity (instead of once per year before 2014) has allowed faster support to the EFSA Risk Assessment of regulated products, before the finalisation of the evaluation of the application for market authorisation of those products. Some changes in the evaluation done in the previous Panel Statements have been made in relation to P. nishizawae and C. divergens.
Enterococcus faecium is not recommended for the QPS list in spite of advances in recent scientific knowledge allowing a differentiation of pathogenic from non‐pathogenic strains at the clade level. The QPS approach relies on the basis of the evaluation of TU, where the species/subspecies level is the lowest level of evaluation. Therefore, clades within the E. faecium species cannot be considered as a TU and cannot be evaluated separately.
The exclusion of filamentous fungi from the QPS evaluations was reconsidered but it was recommended to keep the monitoring and to re‐evaluate it in the next QPS Opinion update.
Evaluation of bacteriophages should remain as a case‐by‐case procedure and should not be considered for QPS status.
5. Recommendations
An ELS was incorporated in the process (related to ToR2) improving the level of harmonisation and transparency of the assessment. The lessons learned during this exercise will allow identifying ways to improve and refine the searches and include further exclusion criteria during future exercises. The criteria to judge the information provided by the selected studies, as the possible safety concerns related to a certain TU, such as the identification methodologies used or the presence of microorganisms in immune‐compromised/suppressed patients, should be carefully monitored and updated in future evaluations.
While recent findings screened since 2013 do not warrant any reconsideration of the QPS status of lactic acid bacteria (LAB) and Bacillus species, further studies on both human and veterinary clinical isolates particularly from cases where there have been no predisposing factors, should be considered to find out any specific factors that might contribute to the pathogenicity.
Regarding LAB, in particular for Lactococcus lactis, further studies on both human and veterinary clinical isolates could be considered to find out any possible strain specific factors that might contribute to the pathogenicity.
Consumption of microorganisms such as Bifidobacterium species, Lactobacillus and S. boulardi (cerevisiae) by patients with immunosuppression and/or underlying disease can be the origin of the infection described in some articles. Although the use of microorganisms intended to be used as ‘probiotic’ for humans as a health claim does not fall under the remit of the QPS assessment, attention should be paid to this aspect.
The possible inclusion of E. faecium and filamentous fungi TUs in the QPS list should be monitored and re‐evaluated in the next QPS Opinion update. Bacteriophages are not foreseen to be eligible for QPS status.
Advances in AMR of microorganisms are rapidly evolving and should keep being closely monitored and taken into consideration for revision of QPS microorganisms and for the evaluation of new TUs.
More information on the absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain is needed.
Abbreviations
- AMR
antimicrobial resistance
- BIOHAZ Panel
EFSA Panel on Biological Hazards
- CEF Panel
EFSA Panel of Food Contact Materials, Enzymes, Flavourings and Processing Aids
- DG SANCO
Health and Food Safety Directorate General
- ELS
Extensive Literature Search
- EPS
exopolysaccharides
- FDA
Food and Drug Administration
- FEEDAP Panel
EFSA Panel on Additives and Products of Substances used in Animal Feed
- FIP
Food Ingredients and Packaging
- FSTA
Food Science Technology Abstracts
- GGT
gamma‐glutamyltranspeptidase
- GMM
Genetically modified microorganisms
- GRAS
Generally Recognised As Safe
- ICN
International Code of Nomenclature
- ICTF
International Commission on the Taxonomy of Fungi
- ICTV
International Commission on the Taxonomy of Viruses
- IJSEM
International Journal of Systematic and Evolutionary Microbiology
- LAB
lactic acid bacteria
- LBSN
List of Bacterial names with Standing in Nomenclature
- LPSN
List of Prokaryotic names with Standing in Nomenclature
- MALDI‐TOF MS
Matrix‐Assisted Laser Desorption/ionisation Time‐of‐Flight Mass Spectrometry
- MIC
minimal inhibitory concentration values
- NDA Panel
EFSA Panel on Dietetic Products, Nutrition and Allergies
- NF
Novel Food
- OECD
Organisation for Economic Cooperation and Development
- QPS
Qualified Presumption of Safety
- PCR
polymerase chain reaction
- PCR‐RFLP
polymerase chain reaction‐restriction fragment length polymorphism
- PECO
Population Exposure Comparator Outcome
- ToR
Term of reference
- TU
taxonomic unit
- WG
Working Group
Appendix A – The 2016 updated list of QPS Status recommended biological agents in support of EFSA risk assessments
The list of QPS status recommended biological agents is being maintained in accordance with the self‐task mandate of the BIOHAZ Panel (2017–2019). Possible additions to this list are included around every 6 months, with the first Panel Statement adopted in June 2017 and the last Panel Statement planned for adoption in December 2019. These additions are published as updates to this Scientific Opinion available at https://doi.org//10.2903/j.efsa.2017.4664 and, as of January 2018, also as supporting information linked to every Panel Statement available on the Knowledge Junction at https://doi.org//10.5281/zenodo.1146566.
6.
6.1.
6.1.1.
References
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2014. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 1: Suitability of taxonomic units notified to EFSA until October 2014. EFSA Journal 2014;12(12):3938, 41 pp. https://doi.org/10.2903/j.efsa.2014.3938
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015a. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. 2: Suitability of taxonomic units notified to EFSA until March 2015. EFSA Journal 2015;13(6):4138, 29 pp. https://doi.org/10.2903/j.efsa.2015.4138
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015b. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. 3: Suitability of taxonomic units notified to EFSA until September 2015. EFSA Journal 2015;13(12):4331, 25 pp. https://doi.org/10.2903/j.efsa.2015.4331
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2016. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 4: suitability of taxonomic units notified to EFSA until March 2016. EFSA Journal 2016;14(7):4522, 37 pp. https://doi.org/10.2903/j.efsa.2016.4522
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Herman L, Koutsoumanis K, Roland L, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G, Prieto Maradona M, Querol A, Peixe L, Suarez JE, Sundh I, Vlak JM, Aguilera‐Gómez M, Barizzone F, Brozzi R, Correia S, Heng L, Istace F, Lythgo C and Fernández Escámez PS, 2017a. Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA Journal 2017;15(3):4664, 177 pp. https://doi.org/10.2903/j.efsa.2017.4664
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Fernandez Escamez PS, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Correia S and Herman L, 2017b. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 6: suitability of taxonomic units notified to EFSA until March 2017. EFSA Journal 2017;15(7):4884, 32 pp. https://doi.org/10.2903/j.efsa.2017.4884
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Girones R, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2018. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 7: Suitability of taxonomic units notified to EFSA until September 2017. EFSA Journal 2018;16(1):5131, 42 pp. https://doi.org/10.2903/j.efsa.2018.5131
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Girones R, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Peixe L, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2018b. Statement on the update of the list of QPS recommended biological agents intentionally added to food or feed as notified to EFSA 8: Suitability of taxonomic units notified to EFSA until March 2018. EFSA Journal 2018;16(7):5315, https://doi.org/10.2903/j.efsa.2018.5315
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Álvarez Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernandez Escamez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak J, Barizzone F, Correia S and Herman L, 2019a. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 9: suitability of taxonomic units notified to EFSA until September 2019. EFSA Journal 2019;17(1):5555, 46 pp. https://doi.org/10.2903/j.efsa.2019.5555
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bolton D, Bover‐Cid S, Chemaly M, Davies R, Hilbert F, Lindqvist R, Nauta M, Peixe L, Ru G, Simmons M, Skandamis P, Suffredini E, Cocconcelli PS, Fernández Escámez PS, Maradona MP, Querol A, Suarez JE, Sundh I, Vlak JM, Barizzone F, Correia S and Herman L, 2019b. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 10: Suitability of taxonomic units notified to EFSA until March 2019. EFSA Journal 2019;17(7):5753, 79 pp. https://doi.org/10.2903/j.efsa.2019.5753
Appendix B – Extensive Literature Search, relevance screening, and article evaluation for the maintenance and update of list of QPS‐recommended biological agents intentionally added to the food or feed as notified to EFSA
This Extensive Literature Search (ELS) protocol will be used in the context of the EFSA self‐task mandate on the update of the list of QPS‐recommended biological agents intentionally added to the food or feed (EFSA‐Q‐2014‐00189).
B.1. Objective of the Extensive Literature Search
An ELS of studies related to safety concerns for humans, animals, plants and/or the environment of microorganisms recommended for the Qualified Presumption of Safety (QPS) 2016 list was performed. The aim was to identify any publicly available studies reporting on safety concerns for humans, animals or the environment caused by Gram‐positive non‐sporulating bacteria, Gram‐positive sporulating bacteria, Gram‐negative bacteria, viruses (used for plant protection purposes) and yeasts on the QPS recommended list (as identified by EFSA in the terms of reference and on Table 1 from the 2013 update QPS Scientific Opinion) since the previous QPS review (i.e. publications from 2013 until the date of the search). The results of the ELS are part of ToR 2 of the self‐task mandate and are intended to inform the BIOHAZ Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to the food or feed as notified to EFSA.
The task included the following steps:
ELS for potentially relevant citations;
Relevance screening to be used to select the citations identified by the literature search, based on titles and full‐text;
Evaluation of articles according to pre‐specified categories of possible safety concerns.
The review questions are broken down into key elements using the PECO conceptual model:
Population of interest (P)
Exposure of interest (E)
Comparator (C)
Outcomes of interest (O)
B.1.1. Target population
The populations of interest are humans, animals, plants and the environment.
B.1.2. Exposure
Citations must report on at least one species included in one of the five groups of named species specified in the EFSA QPS recommended list (Table 1) of the QPS 2013 update:
Gram‐positive non‐sporulating bacteria;
Gram‐positive sporulating bacteria;
Gram‐negative bacteria;
Viruses used for plant protection;
Yeasts.
Namely:
-
Gram‐positive non‐sporulating bacteria:
Bifidobacterium adolescentis, Bifidobacterium animalis, Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium longum, Corynebacterium glutamicum, Lactobacillus acidophilus, Lactobacillus amylolyticus, Lactobacillus amylovorus, Lactobacillus alimentarius, Lactobacillus aviaries, Lactobacillus brevis, Lactobacillus buchneri, Lactobacillus casei, Lactobacillus cellobiosus, Lactobacillus collinoides, Lactobacillus coryniformis, Lactobacillus crispatus, Lactobacillus curvatus, Lactobacillus delbrueckii, Lactobacillus farciminis, Lactobacillus fermentum, Lactobacillus gallinarum, Lactobacillus gasseri, Lactobacillus helveticus, Lactobacillus hilgardii, Lactobacillus johnsonii, Lactobacillus kefiranofaciens, Lactobacillus kefiri, Lactobacillus mucosae, Lactobacillus panis, Lactobacillus paracasei, Lactobacillus paraplantarum, Lactobacillus pentosus, Lactobacillus plantarum, Lactobacillus pontis, Lactobacillus reuteri, Lactobacillus rhamnosus, Lactobacillus sakei, Lactobacillus salivarius, Lactobacillus sanfranciscensis, Lactococcus lactis, Leuconostoc citreum, Leuconostoc lactis, Leuconostoc mesenteroides, Leuconostoc pseudomesenteroides, Oenococcus oeni, Pediococcus acidilactici, Pediococcus dextrinicus, Pediococcus pentosaceus, Propionibacterium freudenreichii, Propionibacterium acidopropionici, Streptococcus thermophilus;
-
Gram‐positive sporulating bacteria:
Bacillus amyloliquefaciens, Bacillus atrophaeus, Bacillus clausii, Bacillus coagulans, Bacillus fusiformis, Bacillus lentus, Bacillus licheniformis, Bacillus megaterium, Bacillus mojavensis, Bacillus pumilus, Bacillus subtilis, Bacillus vallismortis, Geobacillus stearothermophilus;
-
Gram‐negative bacteria:
Gluconobacter oxydans;
-
Viruses used for plant protection:
Plant viruses (Family): Alphaflexiviridae, Potyviridae;
Insect viruses (Family): Baculoviridae;
-
Yeasts:
Debaryomyces hansenii, Hanseniaspora uvarum, Kluyveromyces lactis, Kluyveromyces marxianus, Komagataella pastoris, Lindnera jadinii, Ogataea angusta, Saccharomyces bayanus, Saccharomyces cerevisiae, Saccharomyces pastorianus, Schizosaccharomyces pombe, Wickerhamomyces anomalus, Xanthophyllomyces dendrorhous.
For the yeast species, as previously, the name of the teleomorphic form is used in the list of QPS species. Important synonyms and older names were also included in the searches. For instance, names of the anamorphic growth forms were included, when such a form is known.
B.1.3. Comparator
It is expected that the prevalent study designs will be case reports or case series and studies based on surveys or isolate collections. The remaining study designs may include: studies using laboratory isolates; randomised controlled trials, field trials, or experimental designs in the laboratory; experimental designs in live animals with a deliberate disease challenge; observational study designs; animal or insect models; investigations to identify or to understand the causes of safety concerns (e.g. identification, characterisation of toxic factors, virulence mechanisms); studies to demonstrate beneficial effects but with reporting of unwanted side‐effects.
The comparator was not included as a key element in the search strategy; it would be difficult to express using free text or index terms and may not be explicitly described in the title or abstract.
B.1.4. Outcomes of interest
Outcomes of interest to this ELS are:
-
Question 1:
potential harms
safety issues
virulence or infectious characteristics
intoxication
-
Question 2:
(acquired/intrinsic) antimicrobial resistance (AMR) covering phenotypic and genotypic aspects
The QPS concept does not address hazards linked to the formulation or processing of the products based on biological agents added into the food or feed chain. Neither safety of users handling the product nor genetic modifications are taken into account.
B.1.5. Identification of the review questions
As this task involves an ELS, followed by screening for relevance and article evaluation, the research questions were broad in scope rather than focussed as appropriate to a systematic review:
Is there evidence of any safety concerns, including virulence features and toxin production, for humans, animals, plants and/or the environment associated with microbial species currently recommended for the QPS list since the previous QPS review (2013 QPS update)?
Is there evidence related to the presence or absence of antimicrobial resistance or antimicrobial resistance genes for the same microbial species published during the same time period?
B.2. Eligibility criteria for study selection
The selection of studies relevant to question 1 and 2 will be performed applying the eligibility criteria described in Table B.1.
Table B.1.
Eligibility criteria for questions 1 and 2
| Criteria | |
|---|---|
| Study design | No specific type of study design will be used to include/exclude relevant studies, although it is expected that the prevalent study designs will be case reports or case series and studies based on surveys or isolate collections |
| Study characteristics: | No exclusion will be based on study characteristics but 3 criteria for reliability of studies are proposed at the stage of article evaluation (see Section B.4 ‘Study selection and article evaluation’ below) (i) taxonomical identification carried out and well described (including method applied); (ii) clear statement on the relationship between the microorganism and the host in case of infection/disease; (iii) rationale about exposure to the microorganism is well reported and is consistent |
| Population | Humans, animals, plants and the environment |
| Exposure | Studies must report on at least one TU as identified in Section B.1.2 |
| Outcome of interest | Outcomes as listed in Section B.1.4 |
| Language | English |
| Time | 2013 until 6 June 2016 |
| Publication type | Primary research studies (i.e. studies generating new data) |
B.3. Literature searches
Searches were conducted in a range of relevant information sources to identify any evidence of safety concerns and AMR regarding the target microbial species.
Scoping searches were run to test the key terms and strategies identified, as well as the information sources selected.
Considering the results of the scoping search to handle the high number of studies identified in each group, 16 search strategies were prepared: three for yeasts, one for insect viruses, one for plant viruses, ten for Gram‐positive bacteria and one for Gram‐negative bacteria according to named species specified by EFSA in the QPS recommended list (Table A.1) of the QPS 2013 update (see Appendix A).
Table A.1.
The 2016 updated list of QPS status recommended biological agents for safety risk assessments carried out by EFSA Scientific Panels and Units
| Bacteria | |||
| Gram‐positive non‐sporulating bacteria | |||
| Species | Qualifications a | ||
|
Bifidobacterium adolescentis Bifidobacterium animalis |
Bifidobacterium bifidum Bifidobacterium breve |
Bifidobacterium longum | |
| Carnobacterium divergens f | |||
| Corynebacterium ammoniagenes r | Corynebacterium glutamicum b | QPS applies for production purposes only.n , o | |
|
Lactobacillus acidophilus Lactobacillus amylolyticus Lactobacillus amylovorus Lactobacillus animalis k Lactobacillus alimentarius Lactobacillus aviaries Lactobacillus brevis Lactobacillus buchneri Lactobacillus casei c Lactobacillus cellobiosus Lactobacillus collinoides Lactobacillus coryniformis Lactobacillus crispatus Lactobacillus curvatus |
Lactobacillus delbrueckii Lactobacillus dextrinicus s Lactobacillus diolivorans i Lactobacillus farciminis Lactobacillus fermentum Lactobacillus gallinarum Lactobacillus gasseri Lactobacillus helveticus Lactobacillus hilgardii Lactobacillus johnsonii Lactobacillus kefiranofaciens Lactobacillus kefiri Lactobacillus mucosae Lactobacillus panis |
Lactobacillus paracasei Lactobacillus paraplantarum Lactobacillus pentosus Lactobacillus plantarum Lactobacillus pontis Lactobacillus reuteri Lactobacillus rhamnosus Lactobacillus sakei Lactobacillus salivarius Lactobacillus sanfranciscensis |
|
| Lactococcus lactis | |||
|
Leuconostoc citreum Leuconostoc lactis |
Leuconostoc mesenteroides | Leuconostoc pseudomesenteroides | |
| Microbacterium imperiale f | QPS only applies when the species is used for enzyme production. | ||
| Oenococcus oeni | |||
| Pasteuria nishizawae h | |||
|
Pediococcus acidilactici |
Pediococcus parvulus i | Pediococcus pentosaceus | |
| Propionibacterium acidipropionici | Propionibacterium freudenreichii | ||
| Streptococcus thermophilus | |||
| Gram‐positive spore‐forming bacteria | |||
| Bacillus | |||
| Species | Qualifications a | ||
|
Bacillus amyloliquefaciens Bacillus atrophaeus Bacillus clausii Bacillus coagulans Bacillus flexus i |
Bacillus fusiformis Bacillus lentus Bacillus licheniformis Bacillus megaterium |
Bacillus mojavensis Bacillus pumilus Bacillus smithii j Bacillus subtilis Bacillus vallismortis |
Absence of toxigenic activity. |
| Geobacillus stearothermophilus | Absence of toxigenic activity. | ||
| Gram‐negative bacteria | |||
| Species | Qualifications a | ||
| Gluconobacter oxydans | QPS only applies when the species is used for vitamin production. | ||
| Komagataeibacter sucrofermentans p , q | QPS applies for production purposes only.n | ||
| Xanthomonas campestris g | QPS only applies when the species is used for the production of xanthan gum. | ||
| Yeasts e | |||
| Species | Qualifications | ||
| Candida cylindracea f | QPS only applies when the species is used for enzyme production. | ||
| Debaryomyces hansenii | |||
| Hanseniaspora uvarum | |||
| Kluyveromyces lactis | Kluyveromyces marxianus | ||
| Komagataella pastoris | Komagataella phaffi l | QPS only applies when the species is used for enzyme production. | |
| Lindnera jadinii | QPS only applies when the species is used for enzyme production. | ||
| Ogataea angusta | QPS only applies when the species is used for enzyme production. | ||
| Saccharomyces bayanus | Saccharomyces cerevisiae d | Saccharomyces pastorianus |
Absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain. In the case of Saccharomyces cerevisiae this qualification applies for yeast strains able to grow above 37°C. |
| Schizosaccharomyces pombe | |||
| Wickerhamomyces anomalus |
QPS only applies when the species is used for enzyme production. Absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain. |
||
| Xanthophyllomyces dendrorhous | |||
| Yarrowia lipolytica m | QPS applies for production purposes only.n | ||
| Viruses | |||
| Plant viruses | |||
| Family | |||
| Alphaflexiviridae | Potyviridae | ||
| Insect viruses | |||
| Family | |||
| Baculoviridae | |||
| Algae | |||
| Euglena gracilis r | QPS applies for production purposes only.n | ||
QPS: Qualified Presumption of Safety.
A specific representative of a QPS proposed taxonomic unit, does not need to undergo a further safety assessment other than to satisfy any of the qualifications specified if applicable. On the other hand, representatives of taxonomic units that fail to satisfy a qualification would be considered unfit for the QPS list and would remain subject to a full safety assessment, in the frame of a notification by the responsible EFSA Scientific Panel.
Generic qualification for all QPS bacterial taxonomic units: the strains should not harbour any acquired antimicrobial resistance genes to clinically relevant antimicrobials.
Brevibacterium lactofermentum is a synonym of Corynebacterium glutamicum.
The previously described species ‘Lactobacillus zeae’ has been included in the species Lactobacillus casei.
Saccharomyces cerevisiae, subtype boulardii is contraindicated for persons with fragile health, as well as for patients with a central venous catheter in place.
Yeast synonyms commonly used in the feed/food industry:
• Debaryomyces hansenii – anamorph Candida famata;
• Hanseniaspora uvarum – anamorph Kloeckera apiculata;
• Kluyveromyces lactis – anamorph Candida spherica;
• Kluyveromyces marxianus – anamorph Candida kefyr;
• Komagataella pastoris – synonym Pichia pastoris;
• Lindnera jadinii – synonyms Pichia jadinii, Hansenula jadinii, Torulopsis utilis, anamorph Candida utilis;
• Ogataea angusta – synonym Pichia angusta;
• Saccharomyces cerevisiae – synonym Saccharomyces boulardii;
• Saccharomyces pastorianus – synonym Saccharomyces carlsbergensis;
• Wickerhamomyces anomalus – synonyms Hansenula anomala, Pichia anomala, Saccharomyces anomalus, anamorph Candida pelliculosa;
• Xanthophyllomyces dendrorhous – anamorph Phaffia rhodozyma.
Microorganisms recommended in the Panel Statement published in December 2014 (EFSA BIOHAZ Panel, 2014).
Microorganisms recommended in the Panel Statement published in June 2015 (EFSA BIOHAZ Panel, 2015a).
Microorganisms recommended in the Panel Statement published in December 2015 (EFSA BIOHAZ Panel, 2015b).
Microorganisms recommended in this Panel Statement published in July 2016 (EFSA BIOHAZ Panel, 2016).
Microorganisms recommended in the Panel Statement published in March 2017 (EFSA BIOHAZ Panel, 2017a).
Microorganisms recommended in the Panel Statement published in July 2017 (EFSA BIOHAZ Panel, 2017b).
Microorganisms recommended in the Panel Statement published in January 2018 (EFSA BIOHAZ Panel, 2018).
Microorganisms recommended in the Panel Statement published in July 2018 (EFSA BIOHAZ Panel, 2018b).
The qualification ‘for production purpose only’ implies the absence of viable cells of the production organism in the final product and can also be applied for food and feed products based on microbial biomass.
Qualification that QPS only applies when the species is used for amino acid production was extended for Corynebacterium glutamicum to other production purposes in the Panel Statement published in January 2019 and July 2019 (EFSA BIOHAZ Panel, 2019a,b).
Basonym Acetobacter xylinus subsp. sucrofermentans.
Microorganisms recommended in the Panel Statement published in January 2019 (EFSA BIOHAZ Panel, 2019a).
Microorganism recommended in the Panel Statement published in July 2019 (EFSA BIOHAZ Panel, 2019b).
Lactobacillus dextrinicus (Coster and White, 1964) Haakensen et al., 2009, comb. nov., previously Pediococcus dextrinicus (Coster and White, 1964; Back, 1978). Name change indicated in the Panel Statement published in July 2019 (EFSA BIOHAZ Panel, 2019b).
The 16 subgroups of target microbial species will be searched separately.
Each search strategy will comprise two elements: the search terms (Section B.3.1) and the information sources (Section B.3.2) to be searched.
B.3.1. Search terms
The search strategies used to identify studies are given in Appendix C.
Each strategy will comprise two key elements:
Target microbial species as described in Section B.1.2 (‘Exposure’)
Safety issues as described in Section B.1.4 (‘Outcomes’)
Scoping exercises were carried out to check whether the search strategy can be run without including outcome related terms; this has the purpose of maximising the search sensitivity and should be feasible for those selected species for which the number of overall publications in the relevant time period is expected to be low.
The population of interest (humans, animals, plants or the environment) was not included as a key element in the search strategies, as it is often not explicitly described within a title or abstract. It would also have been difficult to describe adequately such a broad population using title/abstract words and/or subject headings. Population information was captured at the time of evaluating the articles (see Section 1 above).
Search terms for safety issues were identified in close collaboration with the information specialist; example of such terms, are the following: ‘toxin*’, ‘disease*’, ‘infection*’, ‘clinical*’, ‘virulen*’, ‘antimicrobial resistan*’, ‘endocarditis’.
The 16 subgroups of target microbial species were entered on separate search lines. The search line for each group will be combined with the safety terms individually. This allowed the number of results returned for each of the subgroups to be identified.
The searches were not limited by language or study design. The review period was 2013 until 6 June 2016.
B.3.2. Information sources searched
A range of information sources indexing published research were searched for studies reporting safety concerns regarding the target microbial species.
Searches to identify studies for the 2013 Qualified Presumption of Safety (QPS) list were run in the following information sources: PubMed, Web of Knowledge, CasesDatabase, Google Scholar, CAB Abstracts and Food Technology Science Abstracts. The evaluation of the results obtained revealed that CAB Abstracts and BIOSIS Citation Index appeared to be the information sources that provided the greatest added value in this topic area.
Information sources with coverage of grey literature such as Science.gov, ScienceResearch.com, OpenGREY and conference websites were not included.
Table B.2.
Information sources that were searched to identify relevant studies
| Information source | Interface |
|---|---|
| Web of Science Core Collection | Web of Science, Thomson Reuters 2016 |
| CAB Abstracts | Web of Science, Thomson Reuters 2016 |
| BIOSIS Citation Index | Web of Science, Thomson Reuters 2016 |
| MEDLINE | Web of Science, Thomson Reuters 2016 |
| Food Science Technology Abstracts (FSTA) | Web of Science, Thomson Reuters 2016 |
Search results were downloaded from the information sources and imported into EndNote® X7 bibliographic management software. For each of the 16 species groups, within‐group removal of duplicate entries was done in EndNote® X7. Following uploading of the species groups into the DistillerSR online software, removal of duplicates was again undertaken, using the Duplicate Detection feature.
B.4. Study selection and article evaluation
Studies to be included in the review were selected by a two‐step selection procedure.
B.4.1. Screening for potential relevance
To identify potentially relevant studies that will be included for article evaluation.
It will be carried out at title level.
If the information contained in the title is not relevant for the research objectives, the article is not selected for ‘Article evaluation’. Articles that will be excluded during screening this step will be stored in DistillerSR.
This step will be conducted in duplicate by experts and, if needed, EFSA staff. In case of doubts or divergences between the reviewers, the paper will proceed to step 2.
B.4.2. Article evaluation
To confirm that the article is relevant for the QPS project and, in case it is, to evaluate it.
It will be carried out at full text level.
If the information contained in the article is not relevant for the research objectives, the article is not evaluated. Articles that will not be considered relevant will be stored in DistillerSR.
This step will be conducted in duplicate by the experts. In case of divergences between the reviewers an agreement will be reached between the two experts reviewing the study.
Screeners will be trained using written documentation on study eligibility. Eligibility criteria will be pilot tested on a subset of records, and refined if prone to misinterpretation. The results of the different phases of the study selection process will be reported in a flowchart as recommended in the PRISMA statement on preferred reporting items for systematic reviews and meta‐analyses (Moher et al., 2009).
The studies and related form will be inserted in the DistillerSR software.
B.4.2.1. Questions for study selection and article evaluation
STEP 1 (Screening for potential relevance):
-
Question 1: Can this article be relevant for QPS project?
Yes or Unclear: Include and continue to Article evaluation form
No: Exclude
STEP 2 (Article evaluation):
-
Question 2: Is the full text available?
Yes: Include and continue to the Article evaluation form
No: Exclude
-
Question 3: Is the full text in English?
Yes: Include and continue to the Article evaluation form
No: Exclude
-
Question 4: Is the article relevant for QPS project?
Yes: Include and continue to the Article evaluation form
No: Exclude
-
Question 5: Identification of the microorganisms
The article will be characterised in terms of the microorganisms involved (multiple choice question: the expert can identify the microorganism/s described in the article)
-
Question 6: Safety concerns with an impact on human health
Free text
-
Question 7: Safety concerns with an impact on animal health
Free text
-
Question 8: Safety concerns with an impact on the environment
Free text
-
Question 9: Safety concerns related to AMR (antimicrobial resistance factors)
Free text
-
Question 10: Other concerns (please specify)
Free text
B.5. Presentation of the results
The overall results of the searches and evaluations of individual articles will be presented in tabular format for each group/subgroup and species.
B.6. Evidence becoming available after the first deadline for retrieving evidence
The literature search (Annexes B and C) will be repeated every 6 months.
B.7. Human resources, software and timelines for performing the ELS
Tasks for performing ELS were allocated among EFSA staff members and WG experts as shown in the table below. Provisional deadlines were agreed in advance and were subject to changes depending on the volume of data retrieved.
Table B.3.
Task allocation for performing the ELS
| WHAT | WHO | SOFTWARE | WHEN (PROVISIONAL) |
|---|---|---|---|
| Search process | EFSA staff | Endnote | Nov 2015 |
| Screening title | WG experts (with EFSA staff) | DistillerSR | Dec 2015–Jan 2016 |
| Evaluation of articles | WG experts | DistillerSR | Dec 2015–Feb 2016 |
| Presentation of the results | EFSA staff | DistillerSR | March 2016 |
References
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2013. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal 2013;11(11):3449, 108 pp. doi:10.2903/j.efsa.2013.3449
Moher D, Liberati A, Tetzlaff J and Altman DG for the PRIMS Group, 2009. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. British Medical Journal, 338, b2535. doi:10.1136/bmj.b2535
Appendix C – Search strategies
Gram‐Positive Non‐Sporulating Bacteria
Bifidobacterium
| String for species | |
| ‘Bifidobacterium adolescentis’ OR ‘Bifidobacterium animalis’ OR ‘Bifidobacterium bifidum’ OR ‘Bifidobacterium breve’ OR ‘Bifidobacterium longum’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | endocarditis OR abscess OR meningitis |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Corynebacterium glutamicum
| String for species | |
| ‘Corynebacterium glutamicum’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* OR ‘pathogen*’ |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Lactobacilli
| String for species | |
| ‘Lactobacillus acidophilus’ OR ‘Lactobacillus amylolyticus’ OR ‘Lactobacillus amylovorus’ OR ‘Lactobacillus alimentarius’ OR ‘Lactobacillus aviaries’ OR ‘Lactobacillus brevis’ OR ‘Lactobacillus buchneri’ OR ‘Lactobacillus casei’ OR ‘Lactobacillus zeae’ OR ‘Lactobacillus cellobiosus’ OR ‘Lactobacillus coryniformis’ OR ‘Lactobacillus crispatus’ OR ‘Lactobacillus curvatus’ OR ‘Lactobacillus delbrueckii’ OR ‘Lactobacillus farciminis’ OR ‘Lactobacillus fermentum’ OR ‘Lactobacillus gallinarum’ OR ‘Lactobacillus gasseri’ OR ‘Lactobacillus helveticus’ OR ‘Lactobacillus hilgardii’ OR ‘Lactobacillus johnsonii’ OR ‘Lactobacillus kefiranofaciens’ OR ‘Lactobacillus kefiri’ OR ‘Lactobacillus mucosae’ OR ‘Lactobacillus panis’ OR ‘Lactobacillus collinoides’ OR ‘Lactobacillus paracasei’ OR ‘Lactobacillus paraplantarum’ OR ‘Lactobacillus pentosus’ OR ‘Lactobacillus plantarum’ OR ‘Lactobacillus pontis’ OR ‘Lactobacillus reuteri’ OR ‘Lactobacillus rhamnosus’ OR ‘Lactobacillus sakei’ OR ‘Lactobacillus salivarius’ OR ‘Lactobacillus sanfranciscensis’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | endocarditis OR abscess OR meningitis |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | opportunistic OR virulen* |
Lactococcus lactis
| String for species | |
| ‘Lactococcus lactis’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | endocarditis OR abscess OR meningitis |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Oenococcus
| String for species | |
| ‘Oenococcus oeni’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | Not applied |
Leuconostoc
| String for species | |
| ‘Leuconostoc mesenteroides’ OR ‘Leuconostoc lactis’ OR ‘Leuconostoc pseudomesenteroides’ OR ‘Leuconostoc citreum’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Pediococci
| String for species | |
| ‘Pediococcus pentosaceus’ OR ‘Pediococcus dextrinicus’ OR ‘Pediococcus acidilactici’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | Not applied |
Proprionibacterium
| String for species | |
| ‘Propionibacterium acidipropionici’ OR ‘Propionibacterium freudenreichii’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | Not applied |
Streptococcus thermophilus
| String for species | |
| ‘Streptococcus thermophilus’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Gram‐Positive Sporulating Bacteria
Bacillus
| String for species | |
| ‘Bacillus amyloliquefaciens’ OR ‘Bacillus coagulans’ OR ‘Bacillus clausii’ OR ‘Bacillus atrophaeus’ OR ‘Bacillus fusiformis’ OR ‘Lysinibacillus fusiformis’ OR ‘Bacillus licheniformis’ OR ‘Bacillus lentus’ OR ‘Bacillus mojavensis’ OR ‘Bacillus megaterium’ OR ‘Bacillus vallismortis’ OR ‘Bacillus subtilis’ OR ‘Bacillus pumilus’ OR ‘Geobacillus stearothermophilus’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antibiotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR bacteremia OR bacteraemia OR toxin* |
| 3. Type of disease | endocarditis OR abscess OR meningitis |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | opportunistic OR virulen* |
Gram‐negative bacteria
Gluconobacter oxydans
| String for species | |
| ‘Gluconobacter oxydans’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | Not applied |
Yeasts
Debaryomyces hansenii …
| String for species | |
| ‘Debaryomyces hansenii’ OR ‘Candida famata’ OR ‘Hanseniaspora uvarum’ OR ‘Kloeckera apiculata’ OR ‘Candida spherica’ OR ‘Candida kefyr’ OR ‘Lindnera jadinii’ OR ‘Pichia jadinii’ OR ‘Hansenula jadinii’ OR ‘Torulopsis utilis’ OR ‘Ogataea angusta’ OR ‘Pichia angusta’ OR ‘Saccharomyces bayanus’ OR ‘Saccharomyces pastorianus’ OR ‘Saccharomyces carlsbergensis’ OR ‘Wickerhamomyces anomalus’ OR ‘Hansenula anomala’ OR ‘Pichia anomala’ OR ‘Candida pelliculosa’ OR ‘Xanthophyllomyces dendrorhous’ OR ‘Phaffia rhodozyma’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | Not applied |
Kluyveromyces lactis …
| String for species | |
| ‘Kluyveromyces lactis’ OR ‘Kluyveromyces marxianus’ OR ‘Komagataella pastoris’ OR ‘Pichia pastoris’ OR ‘Candida utilis’ OR ‘Saccharomyces boulardii’ OR ‘Schizosaccharomyces pombe’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antimycotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR fungemia OR fungaemia OR mycos* |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | clinical* OR death* OR morbidit* OR mortalit* OR disease* OR illness* |
| 5. Disease Risk | opportunistic OR virulen* |
Saccharomyces cerevisiae
| String for species | |
| ‘Saccharomyces cerevisiae’ | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | ‘antimicrobial resistan*’ OR ‘antimycotic resistan*’ OR ‘antimicrobial susceptibil*’ |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | infection* OR abscess* OR sepsis* or septic* OR fungemia OR fungaemia OR mycos* |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | Not applied |
| 5. Disease Risk | opportunistic or virulen* |
Viruses used for plant protection
Baculoviridae
| String for species | |
| ‘Nuclear polyhedrosis virus’ OR granulovirus OR baculoviridae | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | Not applied |
| 3. Type of disease | ‘nuclear polyhedrosis’ OR granulosis |
| 4. Mortality/Morbidity | mortalit* OR ‘safety concern*’ OR ‘health hazard’ |
| 5. Disease Risk | Not applied |
Alphaflexiviridae
| String for species | |
| Alphaflexiviridae OR Potyviridae | |
| OUTCOME | String |
| 1. Antimicrobial/Antibiotic/Antimycotic | Not applied |
| 2. Infection/Bacteraemia/Fungaemia/Sepsis | necros* |
| 3. Type of disease | Not applied |
| 4. Mortality/Morbidity | mortalit* OR ‘safety concern*’ OR ‘health hazard’ |
| 5. Disease Risk | virulen* |
Appendix D – References selected from the ELS exercice as relevant for the QPS
Gram‐Positive Non‐Sporulating Bacteria
Bifidobacterium
Avcin SL, Pokorn M, Kitanovski L, Premru MM and Jazbec J, 2015. Bifidobacterium breve Sepsis in Child with High‐Risk Acute Lymphoblastic Leukemia. Emerging Infectious Diseases, 21, 1674‐1675.
Bertelli C, Pillonel T, Torregrossa A, Prod'hom G, Fischer CJ, Greub G and Giannoni E, 2015. Bifidobacterium longum Bacteremia in Preterm Infants Receiving Probiotics. Clinical Infectious Diseases, 60, 924‐927.
Luo C, Hang X, Liu X, Zhang M, Yang X and Yang H, 2015. Detection of erm(X)‐mediated antibiotic resistance in Bifidobacterium longum subsp longum. Annals of Microbiology, 65, 1985‐1991.
Suwantarat N, Romagnoli M, Wakefield T and Carroll K, 2014. Ventriculoperitoneal shunt infection caused by Bifidobacterium breve. Anaerobe, 28, 1‐3.
Tena D, Losa C, Medina MJ and Saez‐Nieto JA, 2014. Peritonitis caused by Bifidobacterium longum: Case report and literature review. Anaerobe, 27, 27‐30.
Zbinden A, Zbinden R, Berger C and Arlettaz R. Case Series of Bifidobacterium longum Bacteremia in Three Preterm Infants on Probiotic Therapy. Neonatology, 107, 56‐59.
Corynebacterium glutamicum
Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, Chen D, Bian H, Li Y and Yu G, 2015. Resistance integrons: class 1, 2 and 3 integrons. Annals of Clinical Microbiology and Antimicrobials, 14, 11 pp. doi: 10.1186/s12941‐015‐0100‐6
Yang L, Lu S, Belardinelli J, Huc‐Claustre E, Jones V, Jackson M and Zgurskaya HI, 2014. RND transporters protect Corynebacterium glutamicum from antibiotics by assembling the outer membrane. MicrobiologyOpen, 3, 484–496.
Lactobacilli
Aroutcheva A, Auclair J, Frappier M, Millette M, Lolans K, Montigny Dd, Carriere S, Sokalski S, Trick WE and Weinstein RA, 2016. Importance of molecular methods to determine whether a probiotic is the source of Lactobacillus bacteremia. Probiotics and Antimicrobial Proteins, 8, 31–40.
Brecht M, Garg A, Longstaff K, Cooper C and Andersen C, 2016. Lactobacillus Sepsis following a Laparotomy in a Preterm Infant: A Note of Caution. Neonatology, 109, 186–189.
Chery J, Dvoskin D, Morato FP and Fahoum B, 2013. Lactobacillus fermentum, a pathogen in documented cholecystitis. International journal of surgery case reports, 4, 662–664.
Dani C, Coviello CC, Corsini II, Arena F, Antonelli A and Rossolini GM, 2016. Lactobacillus Sepsis and Probiotic Therapy in Newborns: Two New Cases and Literature Review. AJP Reports, 6, E25–E29.
Doern CD, Nguyen ST, Afolabi F and Burnham CA, 2014. Probiotic‐Associated Aspiration Pneumonia Due to Lactobacillus rhamnosus. Journal of Clinical Microbiology, 52, 3124–3126.
Doern C, Wallace M and Burnham CA, 2014. Case Report: Lactobacillus rhamnosus Probiotic as a Cause of Pediatric Pneumonia. Abstracts of the General Meeting of the American Society for Microbiology, 114, 151.
Encarnacion CO, Loranger AM, Bharatkumar AG and Almassi GH, 2016. Bacterial Endocarditis Caused by Lactobacillus acidophilus Leading to Rupture of Sinus of Valsalva Aneurysm. Texas Heart Institute Journal, 43, 161–164.
Falci DR, Rigatto MH, Cantarelli VV and Zavascki AP, 2015. Lactobacillus rhamnosus bacteremia in a kidney transplant recipient. Transplant Infectious Disease, 17, 610–612.
Franko B, Valliant M, Recule C, Vautrin E, Brion JP and Pavese P, 2013. Lactobacillus paracasei endocarditis in a consumer of probiotics. Médecine et Maladies Infectieuses, 43, 171–173.
Ishihara Y, Kanda J, Tanaka K, Nakano H, Ugai T, Wada H, Yamasaki R, Kawamura K, Sakamoto K, Ashizawa M, Sato M, Terasako‐Saito K, Kimura S, Kikuchi M, Nakasone H, Yamazaki R, Kako S, Nishida J, Watanabe K and Kanda Y, 2014. Severe oral infection due to Lactobacillus rhamnosus during induction chemotherapy for acute myeloid leukemia. International Journal of Hematology, 100, 607–610.
Jaimee G and Halami PM, 2016. High level aminoglycoside resistance in Enterococcus, Pediococcus and Lactobacillus species from farm animals and commercial meat products. Annals of Microbiology, 66, 101–110.
Khrishnan A and Abraham A, 2014. A case of mitral valve endocarditis caused by Lactobacillus rhamnosus in an immunocompetent patient. Internal Medicine Journal, 44, doi:10.1111/imj.12435
Lee MR, Tsai CJ, Liang SK, Lin CK, Huang YT and Hsueh PR, 2015. Clinical characteristics of bacteraemia caused by Lactobacillus spp. and antimicrobial susceptibilities of the isolates at a medical centre in Taiwan, 2000‐2014. International Journal of Antimicrobial Agents, 46, 439–445.
Martinez RM, Hulten KG, Bui U and Clarridge JE, 3rd, 2014. Molecular Analysis and Clinical Significance of Lactobacillus spp. recovered from Clinical Specimens Presumptively Associated with Disease. Journal of Clinical Microbiology, 52, 30–36.
Mehta A, Rangarajan S and Borate U, 2013. A cautionary tale for probiotic use in hematopoietic SCT patients‐Lactobacillus acidophilus sepsis in a patient with mantle cell lymphoma undergoing hematopoietic SCT. Bone Marrow Transplantation, 48, 461–462.
Meini S, Laureano R, Fani L, Tascini C, Galano A, Antonelli A and Rossolini GM, 2015. Breakthrough Lactobacillus rhamnosus GG bacteremia associated with probiotic use in an adult patient with severe active ulcerative colitis: case report and review of the literature. Infection, 43, 777–781.
Nei T, Inai S, Mikami I, Sato A, Okamoto J, Yokoshima K, Nakamizo M, Haraguchi S, Sonobe K and Saito R, 2013. Descending necrotizing mediastinitis associated with Lactobacillus plantarum. BMC Infectious Diseases, 13, 398.
Sadowska‐Krawczenko I, Paprzycka M, Korbal P, Wiatrzyk A, Krysztopa‐Grzybowska K, Polak M, Czajka U and Lutynska A, 2014. Lactobacillus rhamnosus GG suspected infection in a newborn with intrauterine growth restriction. Beneficial Microbes, 5, 397–402.
Sun Y, Gautam A and Miller N, 2015. Lactobacillus gasseri Associated with Urinary and Surgical Wound Infection in a Renal Transplant Patient. American Journal of Clinical Pathology, 144, A206.
Tena D, Martinez NM, Losa C, Fernandez C, Medina MJ and Saez‐Nieto JA, 2013. Acute acalculous cholecystitis complicated with peritonitis caused by Lactobacillus plantarum. Diagnostic Microbiology and Infectious Disease, 76, 510‐512.
Vahabnezhad E, Mochon AB, Wozniak LJ and Ziring DA, 2013. Lactobacillus Bacteremia Associated With Probiotic Use in a Pediatric Patient With Ulcerative Colitis. Journal of Clinical Gastroenterology, 47, 437–439.
Lactococcus lactis
Buchelli‐Ramirez HL, Alvarez‐Alvarez C, Rojo‐Alba S, García‐Clemente M, Cimadevilla‐Suárez R, Pando‐Sandoval A and Casan‐Clará P, 2013. Necrotising pneumonia caused by Lactococcus lactis cremoris. International Journal of Tuberculosis and Lung Disease, 17, 565–567.
Feierabend D, Reichart R, Romeike B, Kalff R and Walter J, 2013. Cerebral abscess due to Lactococcus lactis cremoris in a child after sinusitis. Clinical Neurology and Neurosurgery, 115, 614–616.
Hadjisymeou S, Loizou P and Kothari P, 2013. Lactococcus lactis cremoris infection: not rare anymore? BMJ Case Reports, 2013, doi:10.1136/bcr‐2012‐008479
Inoue M, Saito A, Kon H, Uchida H, Koyama S, Haryu S, Sasaki T and Nishijima M, 2014. Subdural empyema due to Lactococcus lactis cremoris: case report. Neurologia Medico‐Chirurgica, 54, 341–347.
Karaaslan A, Soysal A, Kepenekli E and Bakir M, 2016. Lactococcus lactis spp lactis infection in infants with chronic diarrhea: two cases report and literature review in children. The Journal of Infection in Developing Countries, 10, 304–307.
Karaaslan A, Soysal A, Sarmis A, Kadayifci EK, Cerit K, Atici S, Soyletir G and Bakir M, 2015. Lactococcus lactis Catheter‐Related Bloodstream Infection in an Infant: Case Report. Japanese Journal of Infectious Diseases, 68, 341–342.
Karanth SS, Ke V, Hasan F and Acharya V, 2014. A Pregnant Woman with Lactococcus lactis Meningitis: To Treat or Not to Treat? Journal of Obstetrics and Gynaecology of India, 64, 63–64.
Lee JY, Seo MY, Yang J, Kim K, Chang H, Kim SC, Kim M‐G, Jo S‐K, Cho W and Kim HK, 2014. Polymicrobial Peritonitis with Lactococcus lactis in a Peritoneal Dialysis Patient. Chonnam Medical Journal, 50, 67–69.
Li L, Heidemann Olsen R, Ye L, Yan H, Nie Q, Meng H and Shi L, 2016. Antimicrobial Resistance and Resistance Genes in Aerobic Bacteria Isolated from Pork at Slaughter. Journal of Food Protection, 79, 589–597.
Newby B and Ramesh KK, 2014. Urinary Tract Infection in a Preterm Neonate Caused by Lactococcus lactis. The Canadian Journal of Hospital Pharmacy, 67, 453–454.
Plumed‐Ferrer C, Barberio A, Franklin‐Guild R, Werner B, McDonough P, Bennett J, Gioia G, Rota N, Welcome F, Nydam DV and Moroni P, 2015. Antimicrobial susceptibilities and random amplified polymorphic DNA‐PCR fingerprint characterization of Lactococcus lactis ssp. lactis and Lactococcus garvieae isolated from bovine intramammary infections. Journal of Dairy Science, 98, 6216–6225.
Plumed‐Ferrer C, Uusikylä K, Korhonen J and von Wright A, 2013. Characterization of Lactococcus lactis isolates from bovine mastitis. Veterinary Microbiology, 167, 592–599.
Rostagno C, Pecile P and Stefano PL, 2013. Early Lactococcus lactis endocarditis after mitral valve repair: a case report and literature review. Infection, 41, 897–899.
Taniguchi K, Nakayama M, Nakahira K, Nakura Y, Kanagawa N, Yanagihara I and Miyaishi S, 2016. Sudden infant death due to Lactococcal infective endocarditis. Legal Medicine (Tokyo, Japan), 19, 107–111.
Ucko M and Colorni A, 2014. Infections by lactic acid bacteria in marine fish from southern Israel (Red Sea): new records. Israeli Journal of Aquaculture ‐ Bamidgeh, 66, 11 pp. Available online: https://evols.library.manoa.hawaii.edu/bitstream/10524/49112/1/IJA_66.2014.939.Colorni.pdf
Werner B, Moroni P, Gioia G, Lavin‐Alconero L, Yousaf A, Charter ME, Carter BM, Bennett J, Nydam DV, Welcome F and Schukken YH, 2014. Short communication: Genotypic and phenotypic identification of environmental streptococci and association of Lactococcus lactis ssp. lactis with intramammary infections among different dairy farms. Journal of Dairy Science, 97, 6964–6969.
Zycka‐Krzesinska J, Boguslawska J, Aleksandrzak‐Piekarczyk T, Jopek J and Bardowski JK, 2015. Identification and characterization of tetracycline resistance in Lactococcus lactis isolated from Polish raw milk and fermented artisanal products. International Journal of Food Microbiology, 211, 134–141.
Leuconostoc
Alegría Á, Delgado S, Flórez AB and Mayo B, 2013. Identification, typing, and functional characterization of Leuconostoc spp. strains from traditional, starter‐free cheeses. Dairy Science & Technology, 93, 657–673.
Bașbülbül G, Özteber M and Bİyİk HH, 2015. Antibiotic resistance in lactic acid bacteria isolated from fermented dairy products and boza. Journal of Microbiology, Biotechnology and Food Sciences, 4, 513–517.
Campedelli I, Flórez AB, Salvetti E, Delgado S, Orrù L, Cattivelli L, Alegría Á, Felis GE, Torriani S and Mayo B, 2015. Draft Genome Sequence of Three Antibiotic‐Resistant Leuconostoc mesenteroides Strains of Dairy Origin. Genome Announcements, 3, e01018‐01015.
Casado Muñoz Mdel C, Benomar N, Lerma LL, Gálvez A and Abriouel H, 2014. Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally‐fermented Aloreña table olives throughout fermentation process. International Journal of Food Microbiology, 172, 110–118.
Damasceno NP, Horowitz SA and Damasceno EF, 2016. Leuconostoc as a Cause of Endophthalmitis Post‐intravitreal Injection of Ranibizumab. Ocular Immunology and Inflammation, 24, 118–119.
Flórez AB, Campedelli I, Delgado S, Alegría Á, Salvetti E, Felis GE, Mayo B and Torriani S, 2016. Antibiotic Susceptibility Profiles of Dairy Leuconostoc, Analysis of the Genetic Basis of Atypical Resistances and Transfer of Genes In Vitro and in a Food Matrix. PloS One, 11, e0145203.
Ji Y, Kim H, Park H, Lee J, Lee H, Shin H, Kim B, Franz CMAP and Holzapfel WH, 2013. Functionality and safety of lactic bacterial strains from Korean kimchi. Food Control, 31, 467–473.
Usta‐Atmaca H, Akbas F, Karagoz Y and Piskinpasa ME, 2015. A rarely seen cause for empyema: Leuconostoc mesenteroides. The Journal of Infection in Developing Countries, 9, 425–427.
Yang C, Wang D, Zhou Q and Xu J, 2015. Bacteremia Due to Vancomycin‐Resistant Leuconostoc lactis in a Patient With Pneumonia and Abdominal Infection. The American Journal of the Medical Sciences, 349, 282–283.
Oenococcus
No relevant papers selected from the retrieved references.
Pediococci
Al‐Badah AS, Ibrahim ASS, Al‐Salamah AA and Ibrahim SSS, 2015. Clonal diversity and antimicrobial resistance of Enterococcus faecalis isolated from endodontic infections. Electronic Journal of Biotechnology, 18, 175–180.
Yüceer Ö and Özden Tuncer B, 2015. Determination of antibiotic resistance and biogenic amine production la lactic acid bacteria isolated from fermented Turkish sausage (sucuk). Journal of Food Safety, 35, 276–285.
Proprionibacterium
No relevant papers selected from the retrieved references.
Streptococcus thermophilus
No relevant papers selected from the retrieved references.
Gram‐Positive Sporulating Bacteria
Bacillus
Anonymous, 2013. Northern Ireland disease surveillance, January to March 2013. Veterinary Record, 172, 657–658.
Anonymous, 2013. Regional veterinary laboratories report ‐ March 2013. Veterinary Ireland Journal, 3, 321–326.
Anonymous, 2013. Regional veterinary Laboratories Report ‐ January 2013. Veterinary Ireland Journal, 3, 189–194.
Anonymous, 2014. Regional Veterinary Laboratories report: April 2014. Veterinary Ireland Journal, 4, 371–376.
Anonymous, 2014. Regional veterinary laboratories report: March 2014. Veterinary Ireland Journal, 4, 315–320.
Anonymous, 2014. Northern Ireland disease surveillance report, October to December 2013. Veterinary Record, 174, 139–141.
Barbosa TM, Phelan RW, Leong D, Morrissey JP, Adams C, Dobson ADW and O'Gara F, 2014. A Novel Erythromycin Resistance Plasmid from Bacillus Sp. Strain HS24, Isolated from the Marine Sponge Haliclona Simulans. PloS One, 9, 11 pp. doi:10.1371/journal.pone.0115583.
Branquinho R, Meirinhos‐Soares L, Carriço JA, Pintado M and Peixe LV, 2014. Phylogenetic and clonality analysis of Bacillus pumilus isolates uncovered a highly heterogeneous population of different closely related species and clones. FEMS Microbiology Ecology, 90, 689–698.
Caetano T, Süssmuth RD and Mendo S, 2015. Impact of domestication in the production of the class II lanthipeptide lichenicidin by Bacillus licheniformis I89. Current Microbiology, 70, 364–368.
Daligault HE, Davenport KW, Minogue TD, Bishop‐Lilly KA, Broomall SM, Bruce DC, Chain PS, Coyne SR, Frey KG, Gibbons HS, Jaissle J, Koroleva GI, Ladner JT, Lo CC, Munk C, Palacios GF, Redden CL, Rosenzweig CN, Scholz MB and Johnson SL, 2014. Twenty Whole‐Genome Bacillus sp. Assemblies. Genome Announcements, 2, e00958–00914.
Ehling‐Schulz M and Messelhäusser U, 2013. Bacillus “next generation” diagnostics: moving from detection toward subtyping and risk‐related strain profiling. Frontiers in Microbiology, 4, 32.
Fagerlund A, Dubois T, Økstad OA, Verplaetse E, Gilois N, Bennaceur I, Perchat S, Gominet M, Aymerich S, Kolstø AB, Lereclus D and Gohar M, 2014. SinR controls enterotoxin expression in Bacillus thuringiensis biofilms. PloS One, 9, e87532, doi:10.1371/journal.pone.0087532.
Fernández‐Fuentes MA, Abriouel H, Ortega Morente E, Pérez Pulido R and Gálvez A, 2014. Genetic determinants of antimicrobial resistance in Gram positive bacteria from organic foods. International Journal of Food Microbiology, 172, 49–56.
Garcia Hejl C, Sanmartin N, Samson T, Soler C and Koeck JL, 2015. [Maxillary sinus infection by Bacillus licheniformis: a case report from Djibouti]. Médecine et Santé Tropicales, 25, 220–221.
Ghelardi E, Celandroni F, Salvetti S, Gueye SA, Lupetti A and Senesi S, 2015. Survival and persistence of Bacillus clausii in the human gastrointestinal tract following oral administration as spore‐based probiotic formulation. Journal of Applied Microbiology, 119, 552–559.
Grass G, Bierbaum G, Molitor E, Götte N and Antwerpen M, 2016. Genome Sequence of Bacillus pumilus Strain Bonn, Isolated from an Anthrax‐Like Necrotic Skin Infection Site of a Child. Genome Announcements, 4, 2 pp. e01741‐15, doi:10.1128/genomeA.01741‐15.
Guo FP, Fan HW, Liu ZY, Yang QW, Li YJ and Li TS, 2015. Brain Abscess Caused by Bacillus megaterium in an Adult Patient. Chinese Medical Journal (Engl.), 128, 1552–1554.
Idelevich EA, Pogoda CA, Ballhausen B, Wüllenweber J, Eckardt L, Baumgartner H, Waltenberger J, Peters G and Becker K, 2013. Pacemaker lead infection and related bacteraemia caused by normal and small colony variant phenotypes of Bacillus licheniformis. Journal of Medical Microbiology, 62, 940–944.
Jaber JJ, Kircher ML, Thorpe E, Porter RG, Sr., Leonetti JP and Marzo SJ, 2013. Recurrent post‐tympanostomy tube otorrhea secondary to aerobic endospore‐forming bacilli: a case report and brief literature review. Ear, Nose, and Throat Journal, 92, 66–72.
Kim J, Jang S, Kim A, Su H, Gunawardhana N, Jeon Y‐E, Bak EJ, Kim J‐H and Cha J‐H, 2016. Role of bacterial γ‐glutamyltranspeptidase as a novel virulence factor in bone‐resorbing pathogenesis. Journal of Microbiology, 54, 396–402.
Kıvanç SA, Kıvanç M and Güllülü G, 2014. Automated ribotyping and antibiotic resistance determining of Bacillus spp from conjunctiva of diabetic patients. Iran Journal of Basic Medical Sciences, 17, 138–144.
Lee MN, Kim SK, Li XH and Lee JH, 2014. Bacterial virulence analysis using brine shrimp as an infection model in relation to the importance of quorum sensing and proteases. Journal of General and Applied Microbiology, 60, 169–174.
Li Q, Wang C, Tang C, He Q, Li N and Li J, 2013. Bacteremia in patients with acute pancreatitis as revealed by 16S ribosomal RNA gene‐based techniques. Critical Care Medicine, 41, 1938–1950.
López AC, Minnaard J, Pérez PF and Alippi AM, 2013. In vitro interaction between Bacillus megaterium strains and Caco‐2 cells. International Microbiology, 16, 27–33.
Madslien EH, Rønning HT, Lindbäck T, Hassel B, Andersson MA and Granum PE, 2013. Lichenysin is produced by most Bacillus licheniformis strains. Journal of Applied Microbiology, 115, 1068–1080.
Malanicheva IA, Kozlov DG, Efimenko TA, Zenkova VA, Kastrukha GS, Reznikova MI, Korolev AM, Borshchevskaia LN, Tarasova OD, Sineokiĭ SP and Efremenkova OV, 2014. [New antibiotics produced by Bacillus subtilis strains]. Mikrobiologiia, 83, 445–450.
Mohammadou BA, Blay Gl, Mbofung CM and Barbier G, 2014. Antimicrobial activities, toxinogenic potential and sensitivity to antibiotics of Bacillus strains isolated from Mbuja, an Hibiscus sabdariffa fermented seeds from Cameroon. African Journal of Biotechnology, 13, 3617–3627.
Niazi A, Manzoor S, Bejai S, Meijer J and Bongcam‐Rudloff E, 2014. Complete genome sequence of a plant associated bacterium Bacillus amyloliquefaciens subsp. plantarum UCMB5033. Standards in Genomic Sciences, 9, 718–725.
Peng Q, Yuan YH and Gao M, 2013. Bacillus pumilus, a Novel Ginger Rhizome Rot Pathogen in China. Plant Disease, 97, 1308–1315.
Rai MM, Gore DG, Rathod MK and Khurad AM, 2013. Evidence of transovarial transmission of Bacillus subtilis in the silkworm, Bombyx mori L. Journal of Pharmacy Research, 7, 318–323.
Rasimus‐Sahari S, Teplova VV, Andersson MA, Mikkola R, Kankkunen P, Matikainen S, Gahmberg CG, Andersson LC and Salkinoja‐Salonen M, 2015. The Peptide Toxin Amylosin of Bacillus amyloliquefaciens from Moisture‐Damaged Buildings Is Immunotoxic, Induces Potassium Efflux from Mammalian Cells, and Has Antimicrobial Activity. Applied and Environmental Microbiology, 81, 2939–2949.
Sadashiv SO and Kaliwal BB, 2014. Isolation, characterization and antibiotic resistance of Bacillus sps. from bovine mastitis in the region of north Karnataka, India. International Journal of Current Microbiology and Applied Sciences, 3, 360–373.
Salvetti E, Orrù L, Capozzi V, Martina A, Lamontanara A, Keller D, Cash H, Felis GE, Cattivelli L, Torriani S and Spano G, 2016. Integrate genome‐based assessment of safety for probiotic strains: Bacillus coagulans GBI‐30, 6086 as a case study. Applied Microbiology and Biotechnology, 100, 4595–4605.
Schyns G, Serra CR, Lapointe T, Pereira‐Leal JB, Potot S, Fickers P, Perkins JB, Wyss M and Henriques AO, 2013. Genome of a gut strain of Bacillus subtilis. Genome Announcements, 1, 2 pp. e00184‐12, doi: 10.1128/genomeA.00184‐12.
Shivamurthy VM, Gantt S, Reilly C, Tilley P, Guzman J and Tucker L, 2016. Bacillus pumilus Septic Arthritis in a Healthy Child. Canadian Journal of Infectious Diseases and Medical Microbiology, Article ID 3265037, 3 pages, 2016. doi:10.1155/2016/3265037.
Shivamurthy V, Reilly C, Gantt S, Guzman J and Tucker L, 2015. Bacillus Pumilus; A Rare Cause of Paediatric Septic Arthritis. A Case Report. Journal of Rheumatology, 42, 1341–1341.
Shweta J and Joseph E, 2013. Identification of mastitis pathogens and their antibiogram. Veterinary Practitioner, 14, 318‐319.
Toth M, Antunes NT, Stewart NK, Frase H, Bhattacharya M, Smith CA and Vakulenko SB, 2016. Class D beta‐lactamases do exist in Gram‐positive bacteria. Nature Chemical Biology, 12, 9–14.
Wannarat W, Motoyama S, Masuda K, Kawamura F and Inaoka T, 2014. Tetracycline tolerance mediated by gene amplification in Bacillus subtilis. Microbiology, 160, 2474–2480.
Yoo JG, Chang J‐H, Kim S‐y, Ji J‐Y, Hong S‐W, Park B‐Y and Oh M‐H, 2014. Analysis of emetic toxin production by Bacillus species using cellular cytotoxicity, molecular, and chromatographic assays. Biotechnology and Bioprocess Engineering, 19, 978–983.
Gram‐negative bacteria
Gluconobacter oxydans
No relevant papers selected from the retrieved references.
Yeasts
Debaryomyces hansenii…
Abrantes P, McArthur CP and Africa CWJ, 2014. Multi‐drug resistant oral Candida species isolated from HIV‐positive patients in South Africa and Cameroon. Diagnostic Microbiology and Infectious Disease, 79, 222–227.
Alfouzan W, Dhar R, Ashkanani H, Gupta M, Rachel C and Khan ZU, 2015. Species spectrum and antifungal susceptibility profile of vaginal isolates of Candida in Kuwait. Journal de Mycologie Médicale, 25, 23–28.
Ashour SM, Kheiralla ZMH, Badawy FMI and Zaki SS, 2015. Killer Toxins of the Yeasts; Candida utilis 22 and Kluyveromyces marxianus and their Potential Applications as Biocontrol Agents. Egyptian Journal of Biological Pest Control, 25, 317–325.
Banerjee P, Kaur R and Uppal B, 2013. Study of fungal isolates in patients with chronic diarrhea at a tertiary care hospital in north India. Journal de Mycologie Médicale, 23, 21–26.
Banjara N, Nickerson KW, Suhr MJ and Hallen‐Adams HE, 2016. Killer toxin from several food‐derived Debaryomyces hansenii strains effective against pathogenic Candida yeasts. International Journal of Food Microbiology, 222, 23–29.
Beyda ND, Chuang SH, Alam MJ, Shah DN, Ng TM, McCaskey L and Garey KW, 2013. Treatment of Candida famata bloodstream infections: case series and review of the literature. Journal of Antimicrobial Chemotherapy, 68, 438–443.
Brilhante RSN, Bittencourt PV, Castelo‐Branco D, de Oliveira JS, de Alencar LP, Cordeiro RD, Pinheiro M, Nogueira EF, Pereira‐Neto WD, Sidrim JJC and Rocha MFG, 2016. Trends in antifungal susceptibility and virulence of Candida spp. from the nasolacrimal duct of horses. Medical Mycology, 54, 147–154.
Brilhante RSN, Rodrigues PHD, de Alencar LP, Riello GB, Ribeiro JF, de Oliveira JS, Castelo‐Branco D, Bandeira T, Monteiro AJ, Rocha MFG, Cordeiro RD, Moreira JLB and Sidrim JJC, 2015. Evidence of Fluconazole‐Resistant Candida Species in Tortoises and Sea Turtles. Mycopathologia, 180, 421–426.
Castanheira M, Woosley LN, Diekema DJ, Jones RN and Pfaller MA, 2013. Candida guilliermondii and Other Species of Candida Misidentified as Candida famata: Assessment by Vitek 2, DNA Sequencing Analysis, and Matrix‐Assisted Laser Desorption Ionization‐Time of Flight Mass Spectrometry in Two Global Antifungal Surveillance Programs. Journal of Clinical Microbiology, 51, 117–124.
Chan AW, Cartwright EJ, Reddy SC, Kraft CS and Wang YF, 2013. Pichia anomala (Candida pelliculosa) fungemia in a patient with sickle cell disease. Mycopathologia, 176, 273–277.
Chan GF, Sinniah S, Idris TINT, Puad MSA and Abd Rahman AZ, 2013. Multiple rare opportunistic and pathogenic fungi in persistent foot skin infection. Pakistan Journal of Biological Sciences: PJBS, 16, 208–218.
Cordeiro RDA, Bittencourt PV, Brilhante RSN, Teixeira CEC, Castelo‐Branco DDSCM, Silva STDC, De Alencar LP, Souza ERY, Bandeira TdJPG, Monteiro AJ, Sidrim JJC and Rocha MFG, 2013. Species of Candida as a component of the nasal microbiota of healthy horses. Medical Mycology, 51, 731–736.
da Silva CM, Parahym A, Leao MPC, de Oliveira NT, Amorim RDM and Neves RP, 2013. Fungemia by Candida pelliculosa (Pichia anomala) in a Neonatal Intensive Care Unit: A Possible Clonal Origin. Mycopathologia, 175, 175–179.
de Araujo PSR, Medeiros Z, de Melo FL, Maciel MA and de Melo HRL, 2013. Candida famata‐induced fulminating cholecystitis. Revista da Sociedade Brasileira de Medicina Tropical, 46, 795–796.
de Freitas EM, Nobre SAM, Pires MBD, Faria RVJ, Batista AUD and Bonan PRF, 2013. Oral Candida species in head and neck cancer patients treated by radiotherapy. Auris, Nasus, Larynx, 40, 400–404.
Deepak MB, Jhanvi SP and AnuAppaiah KA, 2015. Aflatoxin binding and detoxification by non‐saccharomyces yeast ‐ a new vista for decontamination. International Journal of Current Microbiology and Applied Sciences, 4, 310–317.
Deorukhkar SC and Saini S, 2013. Vulvovaginal candidiasis due to non albicans Candida: its species distribution and antifungal susceptibility profile. International Journal of Current Microbiology and Applied Sciences, 2, 323–328.
Dufresne SF, Marr KA, Sydnor E, Staab JF, Karp JE, Lu K, Zhang SAX, Lavallee C, Perl TM and Neofytos D, 2014. Epidemiology of Candida kefyr in Patients with Hematologic Malignancies. Journal of Clinical Microbiology, 52, 1830–1837.
Esgin H, Bulut E and Orum C, 2014. Candida pelliculosa endophthalmitis after cataract surgery: a case report. BMC Research Notes, 7, 169.
Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D and Datry A, 2013. Rapid Emergence of Echinocandin Resistance during Candida kefyr Fungemia Treatment with Caspofungin. Antimicrobial Agents and Chemotherapy, 57, 2380–2382.
Fernandez‐Ruiz M, Aguado JM, Camarena JJ, Loza A, Gadea I, Guinea J, Sierra‐Soler M, Rivas‐Gomez RA, Cuenca‐Estrella M and Candipop P, 2014. Fungemia Due to Emerging non‐Candida, non‐Cryptococcus Species: Data from a Population‐Based Surveillance in Spain. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 54, M–1105.
Ghahri M, Mirhendi H, Zomorodian K and Kondori N, 2013. Identification and antifungal susceptibility patterns of Candida strains isolated from blood specimens in Iran. Archives of Clinical Infectious Diseases, 8, e14529. doi: 10.5812/archcid.14529.
Ghodasra DH, Eftekhari K, Shah AR and VanderBeek BL, 2014. Outcomes, Impact on Management, and Costs of Fungal Eye Disease Consults in a Tertiary Care Setting. Ophthalmology, 121, 2334–2339.
Haddadi P, Zareifar S, Badiee P, Alborzi A, Mokhtari M, Zomorodian K, Pakshir K and Jafarian H, 2014. Yeast Colonization and Drug Susceptibility Pattern in the Pediatric Patients With Neutropenia. Jundishapur Journal of Microbiology, 7, 6 pp. doi:10.5812/jjm.11858.
Helinck S, Perello MC, Deetae P, de Revel G and Spinnler HE, 2013. Debaryomyces hansenii, Proteus vulgaris, Psychrobacter sp and Microbacterium foliorum are able to produce biogenic amines. Dairy Science & Technology, 93, 191–200.
Jung DS, Farmakiotis D, Jiang Y, Tarrand JJ and Kontoyiannis DP, 2015. Uncommon Candida Species Fungemia among Cancer Patients, Houston, Texas, USA. Emerging Infectious Diseases, 21, 1942–1950.
Kamoshita M, Matsumoto Y, Nishimura K, Katono Y, Murata M, Ozawa Y, Shimmura S and Tsubota K, 2015. Wickerhamomyces anomalus fungal keratitis responds to topical treatment with antifungal micafungin. Journal of Infection and Chemotherapy, 21, 141–143.
Khan Z, Ahmad S, Al‐Obaid K, Joseph L and Chandy R, 2015. Candida kefyr as a cause of bloodstream infection and adjunctive role of biomarkers in its diagnosis. Journal de Mycologie Médicale, 25, 71–75.
Khosravi AR, Shokri H, Nikaein D, Mansouri P, Erfanmanesh A, Chalangari R and Katalin M, 2013. Yeasts as important agents of onychomycosis: In vitro activity of propolis against yeasts isolated from patients with nail infection. Journal of Alternative and Complementary Medicine, 19, 57–62.
Kim SH, Shin JH, Mok JH, Kim SY, Song SA, Kim HR, Kook JK, Chang YH, Bae IK and Lee K, 2014. Misidentification of Candida guilliermondii as C. famata among Strains Isolated from Blood Cultures by the VITEK 2 System. BioMed Research International, 6 pp. doi:10.1155/2014/250408.
Kuiper JWP, van den Bekerom MPJ, van der Stappen J, Nolte PA and Colen S, 2013. 2‐stage revision recommended for treatment of fungal hip and knee prosthetic joint infections. Acta Orthopaedica, 84, 517–523.
Kumari KS, Pendru R, Harshavardhan B and Abhijit C, 2014. Distribution of Candida albicans and the non‐albicans Candida species in different clinical specimens from South India. International Journal of Microbiological Research (IJMR), 5, 1–5.
Leuck AM, Johnson JR, Hunt MA, Dhody K, Kazempour K, Ferrieri P and Kline S, 2015. Safety and efficacy of a novel silver‐impregnated urinary catheter system for preventing catheter‐associated bacteriuria: a pilot randomized clinical trial. American Journal of Infection Control, 43, 260–265.
Li D, Zhang WF, Zheng S, Ma ZY, Zhang P and Liu Z, 2013. Surveillance study of candidemia in cancer patients in North China. Medical Mycology, 51, 378–384.
Lin HC, Lin HY, Su BH, Ho MW, Ho CM, Lee CY, Lin MH, Hsieh HY, Lin HC, Li TC, Hwang KP and Lu JJ, 2013. Reporting an outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. Journal of Microbiology Immunology and Infection, 46, 456–462.
Liu XP, Fan SR, Peng YT and Zhang HP, 2014. Species distribution and susceptibility of Candida isolates from patient with Vulvovaginal candidiasis in Southern China from 2003 to 2012. Journal de Mycologie Médicale, 24, 106–111.
Madhumati B, Rani KL, Murthy DS, Reddy BKN and Ramana BV, 2014. Species distribution and antifungal susceptibility profile in vaginal candidiasis. International Journal of Pharmaceutical Research and Bio‐Science, 3, 497–502.
Masri SN, Noor SM, Nor LAM, Osman M and Rahman MM, 2015. Candida isolates from pregnant women and their antifungal susceptibility in a Malaysian tertiary‐care hospital. Pakistan Journal of Medical Sciences, 31, 658–661.
Menezes RD, Ferreira JC, de Sa WM, Moreira TD, Malvino LDS, de Araujo LB, Roeder DVD, Penatti MPA, Candido RC and Pedroso RD, 2015. Frequency of Candida species in a tertiary care hospital in Triangulo Mineiro, Minas Gerais State, Brazil. Revista do Instituto de Medicina Tropical de São Paulo, 57, 185–191.
Milanov D, Prunic B, Velhner M and Bojkovski J, 2014. Diagnosis of yeast mastitis in dairy cows. Lucrari Stiintifice ‐ Universitatea de Stiinte Agricole a Banatului Timisoara, Medicina Veterinara, 47, 56–64.
Minea B, Nastasa V, Kolecka A, Mares M, Marangoci N, Rosca I, Pinteala M, Hancianu M and Mares M, 2016. Etiologic Agents and Antifungal Susceptibility of Oral Candidosis from Romanian patients with HIV‐infection or type 1 diabetes mellitus. Polish Journal of Microbiology, 65, 123–129.
Mohammadi R, Mirhendi H, Rezaei‐Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N and Makimura K, 2013. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Medical Mycology, 51, 657–663.
Montagna MT, Lovero G, Coretti C, De Giglio O, Martinelli D, Bedini A, Delia M, Rosato A, Codeluppi M and Caggiano G, 2014. In vitro activities of amphotericin B deoxycholate and liposomal amphotericin B against 604 clinical yeast isolates. Journal of Medical Microbiology, 63, 1638–1643.
Muadcheingka T and Tantivitayakul P, 2015. Distribution of Candida albicans and non‐albicans Candida species in oral candidiasis patients: Correlation between cell surface hydrophobicity and biofilm forming activities. Archives of Oral Biology, 60, 894–901.
Mun YS, Lee MS, Park JS, Lee JW, Jung SY, Yoon HJ and Han HY, 2015. An unusual case of candidemia presenting as acute respiratory distress syndrome after a small bowel bezoar removal operation. Annals of Surgical Treatment and Research, 88, 48–51.
Neufeld PM, Melhem MDC, Szeszs MW, Ribeiro MD, Amorim EDT, da Silva M and Lazera MD, 2015. Nosocomial candidiasis in Rio de Janeiro State: Distribution and fluconazole susceptibility profile. Brazilian Journal of Microbiology, 46, 477–484.
Nidhi W, Singh SM, Nawange SR and Shruti S, 2015. Spectrum of opportunistic fungal infections in cancer/HIV patients: emerging fungal pathogens from Jabalpur Madhya Pradesh Central India. Scholars Journal of Applied Medical Sciences, 3, 1385–1390.
Nieto MC, Telleria O and Cisterna R, 2015. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagnostic Microbiology and Infectious Disease, 81, 34–40.
Oliveira VKP, Ruiz LD, Oliveira NAJ, Moreira D, Hahn RC, Melo ASD, Nishikaku AS and Paula CR, 2014. Fungemia caused by Candida species in a children's public hospital in the city of São Paulo, Brazil: Study in the period 2007‐2010. Revista do Instituto de Medicina Tropical de São Paulo, 56, 301–305.
Parmeland L, Gazon M, Guerin C, Argaud L, Lehot JJ, Bastien O, Allaouchiche B, Michallet M, Picot S, Bienvenu AL and Study G, 2013. Candida albicans and non‐Candida albicans fungemia in an institutional hospital during a decade. Medical Mycology, 51, 33–37.
Pisa D, Alonso R, Rábano A, Rodal I and Carrasco L, 2015. Different Brain Regions are Infected with Fungi in Alzheimer's Disease. Scientific Reports, 5, 15015; doi: 10.1038/srep15015 (2015).
Riat A, Rentenaar RJ, van Drongelen AM, Barras V, Bertens LC, Vlek AL, Doppenberg E, Weersink AJ, Reinders E, Vlaminckx BJ, Overbeeke N, van Burgel ND, Peterse N, Bosboom R, Boekhout T, Schrenzel J and Kusters JG, 2015. Ground steel target plates in combination with direct transfer of clinical Candida isolates improves frequencies of species‐level identification by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry in comparison with polished steel target plates. Journal of Clinical Microbiology, 53, 1993–1995.
Ribeiro Ribeiro AL, de Alencar Menezes TO, de Melo Alves‐Junior S, de Menezes SA, Marques‐da‐Silva SH and Rosário Vallinoto AC, 2015. Oral carriage of Candida species in HIV‐infected patients during highly active antiretroviral therapy (HAART) in Belém, Brazil. Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology, 120, 29–33.
Sahin SZ, Akalin H, Ersoy A, Yildiz A, Ocakoglu G, Cetinoglu ED, Dizdar OS, Kazak E and Ener B, 2015. Invasive Fungal Infections in Renal Transplant Recipients: Epidemiology and Risk Factors. Mycopathologia, 180, 43–50.
Sanchis M, Martin‐Vicente A, Capilla J and Guarro J, 2016. Antifungal therapies in murine infections by Candida kefyr Mycoses, 59, 253–258.
Sarbu I, Pelinescu D, Stoica I, Marutescu L and Vassu T, 2013. Phenotypic profiles of virulence in different Candida species isolated from vulvovaginal infections. Romanian Archives of Microbiology and Immunology, 72, 225–233.
Sarvtin MT, Hedayati MT, Abastabar M and Shokohi T, 2014. Debaryomyces hansenii colonization and its protein profile in psoriasis. Iranian Journal of Dermatology, 17, 134–137.
Sasikala G, Agatha D, Janagond AB and Thenmozhivalli PR, 2013. Characterization of Candida and its antifungal susceptibility pattern from patients with vaginal candidiasis in a tertiary care hospital in South India. Journal of Pharmaceutical and Biomedical Sciences, S1–S6.
Shyamala R and Parandekar PK, 2014. Identification and in vitro azole resistance of Candida species isolated from oropharyngeal candidiasis in human immunodeficiency virus infected patients. International Journal of Current Microbiology and Applied Sciences, 3, 816–822.
Svobodova L, Bednarova D, Ruzicka F, Chrenkova V, Dobias R, Mallatova N, Buchta V, Kocmanova I, Olisarova P, Stromerova N, Thongsri Y and Hamal P, 2016. High frequency of Candida fabianii among clinical isolates biochemically identified as Candida pelliculosa and Candida utilis. Mycoses, 59, 241–246.
Swarajyalakshmi M and Jyothilakshmi G, 2014. Candida kefyr in Invasive Paranasal Sinusitis. Indian journal of otolaryngology and head and neck surgery: official publication of the Association of Otolaryngologists of India, 66, 371–374.
Taj‐Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF and Boekhout T, 2014. Uncommon opportunistic yeast bloodstream infections from Qatar. Medical Mycology, 52, 549–553.
Tzar MN, Norazlah B and Shamsul AS, 2015. Risk Factors for Candidaemia in a Malaysian Tertiary Hospital. Sains Malaysiana, 44, 735–740.
Wang H, Wu DW, Han H, Yue JF, Zhang F, Shan TC, Guo HP and Yin M, 2014. Antibiotics exposure, risk factors, and outcomes with Candida albicans and non‐Candida albicans candidemia Results from a multi‐center study. Saudi Medical Journal, 35, 153–158.
Yigit N and Aktas E, 2014. Activities of amphotericin B, fluconazole and voriconazole against Candida bloodstream isolates determined by broth microdilution and disk diffusion methods. Turk Hijyen ve Deneysel Biyoloji Dergisi, 71, 131–140.
Youngster I, Sharma TS, Duncan CN and McAdam AJ, 2014. Yield of Fungal Surveillance Cultures in Pediatric Hematopoietic Stem Cell Transplant Patients: A Retrospective Analysis and Survey of Current Practice. Clinical Infectious Diseases, 58, 365–371.
Zarrinfar H, Kaboli S, Dolatabadi S and Mohammadi R, 2016. Rapid detection of Candida species in bronchoalveolar lavage fluid from patients with pulmonary symptoms. Brazilian Journal of Microbiology, 47, 172–176.
Zaveri J, Santamaria JA, Cohen JA and Singh K, 2015. Evaluation of Endophthalmitis in Patients with Candidemia. Investigative Ophthalmology & Visual Science, 56, 2 pp.
Kluyveromyces lactis…
Dufresne SF, Marr KA, Sydnor E, Staab JF, Karp JE, Lu K, Zhang SAX, Lavallee C, Perl TM and Neofytos D, 2014. Epidemiology of Candida kefyr in Patients with Hematologic Malignancies. Journal of Clinical Microbiology, 52, 1830–1837.
Ellouze O, Berthoud V, Mervant M, Parthiot JP and Girard C, 2016. Septic shock due to Saccharomyces boulardii. Médecine et Maladies Infectieuses, 46, 104–105.
Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D and Datry A, 2013. Rapid Emergence of Echinocandin Resistance during Candida kefyr Fungemia Treatment with Caspofungin. Antimicrobial Agents and Chemotherapy, 57, 2380–2382.
Hammad MM, Darwazeh AMG and Idrees MM, 2013. The effect of glycemic control on Candida colonization of the tongue and the subgingival plaque in patients with type II diabetes and periodontitis. Oral Surgery Oral Medicine Oral Pathology Oral Radiology, 116, 321–326.
Kartal O, Demirel F, Baysan A, Gulec M, Yesillik S, Uyanýk M, Musabak U and Sener O, 2014. An unexpected allergic reaction with Saccharomyces boulardii: a case report. Clinical and Translational Allergy, 4(Suppl 3):P100, doi:10.1186/2045‐7022‐4‐S3‐P100.
Ksouri S, Djebir S, Hadef Y and Benakhla A, 2015. Survey of Bovine Mycotic Mastitis in Different Mammary Gland Statuses in Two North‐Eastern Regions of Algeria. Mycopathologia, 179, 327–331.
Lin CC, Liu CP, Hsieh FC, Lee CM and Wang WS, 2015. Antimicrobial susceptibility and clinical outcomes of Candida parapsilosis bloodstream infections in a tertiary teaching hospital in Northern Taiwan. Journal of Microbiology Immunology and Infection, 48, 552–558.
Luzzati R, Cavinato S, Giangreco M, Grana G, Centonze S, Deiana ML, Biolo G and Barbone F, 2013. Peripheral and total parenteral nutrition as the strongest risk factors for nosocomial candidemia in elderly patients: a matched case‐control study. Mycoses, 56, 664–671.
Merseguel KB, Nishikaku AS, Rodrigues AM, Padovan AC, e Ferreira RC, de Azevedo Melo AS, Briones MR and Colombo AL, 2015. Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infectious Diseases, 15, 57, doi: 10.1186/s12879‐015‐0793‐3.
Nidhi W, Singh SM, Nawange SR and Shruti S, 2015. Spectrum of opportunistic fungal infections in cancer/HIV patients: emerging fungal pathogens from Jabalpur Madhya Pradesh Central India. Scholars Journal of Applied Medical Sciences, 3, 1385–1390.
Santino I, Alari A, Bono S, Teti E, Marangi M, Bernardini A, Magrini L, Somma SD and Teggi A, 2014. Saccharomyces cerevisiae fungemia, a possible consequence of the treatment of Clostridium difficile colitis with a probioticum. International Journal of Immunopathology and Pharmacology, 27, 143–146.
Scoppettuolo G, Donato C, De Carolis E, Vella A, Vaccaro L, La Greca A and Fantoni M, 2014. Candida utilis catheter‐related bloodstream infection. Medical Mycology Case Reports, 6, 70–72.
Taj‐Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF and Boekhout T, 2014. Uncommon opportunistic yeast bloodstream infections from Qatar. Medical Mycology, 52, 549–553.
Tyler AR, Okoh AO, Lawrence CL, Jones VC, Moffatt C and Smith RB, 2013. N‐Alkylated 2,3,3‐trimethylindolenines and 2‐methylbenzothiazoles. Potential lead compounds in the fight against Saccharomyces cerevisiae infections. European Journal of Medicinal Chemistry, 64, 222–227.
Saccharomyces cerevisiae
Al‐Ameed AI, 2013. Isolation and identification of fungi from infected milk samples obtained from cattles with mastitis and studying the antifungal activity of rosemary ethanolic extract against the main strains. Diyala Agricultural Sciences Journal, 5, En1–En13.
Anoop V, Rotaru S, Shwed PS, Tayabali AF and Arvanitakis G, 2015. Review of current methods for characterizing virulence and pathogenicity potential of industrial Saccharomyces cerevisiae strains towards humans. FEMS Yeast Research, 15, doi:10.1093/femsyr/fov057.
Arancia S, Sandini S, Graziani S, Norelli S and De Bernardis F, 2014. Use of Multiplex PCR and High‐Resolution Melting Analysis with Primers of a Gene Coding for 65 KDa Mannoprotein to Rapid Detect Candida Species in Biological Samples. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 54, M–424.
Chowdhary A, Agarwal K, Kathuria S, Gaur SN, Randhawa HS and Meis JF, 2014. Allergic bronchopulmonary mycosis due to fungi other than Aspergillus: a global overview. Critical Reviews in Microbiology, 40, 30–48.
Cohen L, Ranque S and Raoult D, 2013. Saccharomyces cerevisiae boulardii transient fungemia after intravenous self‐inoculation. Medical Mycology Case Reports, 2, 63–64.
Eddouzi J, Lohberger A, Vogne C, Manai M and Sanglard D, 2013. Identification and antifungal susceptibility of a large collection of yeast strains isolated in Tunisian hospitals. Medical Mycology, 51, 737–746.
Fadda ME, Pisano MB, Scaccabarozzi L, Mossa V, Deplano M, Moroni P, Liciardi M and Cosentino S, 2013. Use of PCR‐restriction fragment length polymorphism analysis for identification of yeast species isolated from bovine intramammary infection. Journal of Dairy Science, 96, 7692–7697.
Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D and Datry A, 2013. Rapid Emergence of Echinocandin Resistance during Candida kefyr Fungemia Treatment with Caspofungin. Antimicrobial Agents and Chemotherapy, 57, 2380–2382.
Hsu PH, Chiang PC, Liu CH and Chang YW 2015. Characterization of Cell Wall Proteins in Saccharomyces cerevisiae Clinical Isolates Elucidates Hsp150p in Virulence. Plos One, 10, doi:10.1371/journal.pone.0135174.
Lee K, 2015. A case of Saccharomyces cerevisiae fungemia associated with probiotic intake. Tropical Medicine & International Health, 20, 295–295.
Li Y, Chen W, Li X, Li H, Li H, Wang L, He L, Yang X, Wang X, Huang Y and Yao Y, 2013. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV‐infected patients in Kunming, Yunnan province of China: Candida carriage in Chinese HIV patients. BMC Infectious Diseases, 13, accessed 28 January 2013.
Llopis S, Hernandez‐Haro C, Monteoliva L, Querol A, Molina M and Fernandez‐Espinar MT, 2014. Pathogenic Potential of Saccharomyces Strains Isolated from Dietary Supplements. Plos One, 9, doi:10.1371/journal.pone.0098094.
Martos C, Muñoz P, Guinea J, Pelaez T, Marcos‐Zambrano LJ, Escribano P, Bouza E and COMIC Study Group, 2014. Fungemia Caused by Rare Yeast Species in a Spanish General Hospital. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 54, M–1092.
Mohammadi R, Mirhendi H, Rezaei‐Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N and Makimura K, 2013. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Medical Mycology, 51, 657–663.
Pana ZD, Dotis I and Roilides E, 2013. Fungal Endocarditis in Neonates: Systematic Review of 70 Cases. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 53, G–1244.
Pérez‐Torrado R, Llopis S, Perrone B, Gomez‐Pastor R, Hube B and Querol A, 2015. Comparative Genomic Analysis Reveals a Critical Role of De Novo Nucleotide Biosynthesis for Saccharomyces cerevisiae Virulence. Plos One, 10, doi:10.1371/journal.pone.0122382.
Pérez‐Torrado R and Querol A, 2015. Opportunistic Strains of Saccharomyces cerevisiae: A Potential Risk Sold in Food Products. Frontiers in Microbiology, 6, 5 pp. doi:10.3389/fmicb.2015.01522
Petrova A, Kiktev D, Askinazi O, Chabelskaya S, Moskalenko S, Zemlyanko O and Zhouravleva G, 2015. The translation termination factor eRF1 (Sup45p) of Saccharomyces cerevisiae is required for pseudohyphal growth and invasion. FEMS Yeast Research, 15, doi:10.1093/femsyr/fov033.
Pillai U, Devasahayam J, Kurup AN and Lacasse A, 2014. Invasive Saccharomyces cerevisiae infection: a friend turning foe? Saudi Journal of Kidney Diseases and Transplantation, 25, 1266–1269.
Popiel KY, Wong P, Lee MJ, Langelier M, Sheppard DC and Vinh DC, 2015. Invasive Saccharomyces cerevisiae in a liver transplant patient: case report and review of infection in transplant recipients. Transplant Infectious Disease, 17, 435–441.
Roig P, de Llanos R, Gil JV and Fernandez‐Espinar MT, 2013. FLO11 expression in clinical and non‐clinical Saccharomyces cerevisiae strains and its association with virulence. Annals of Microbiology, 63, 1423–1431.
Santino I, Alari A, Bono S, Teti E, Marangi M, Bernardini A, Magrini L, Di Somma S and Teggi A, 2014. Saccharomyces cerevisiae fungemia, a possible consequence of the treatment of Clostridium difficile colitis with a probioticum. International Journal of Immunopathology and Pharmacology, 27, 143–146.
Shively CA, Eckwahl MJ, Dobry CJ, Mellacheruvu D, Nesvizhskii A and Kumar A, 2013. Genetic Networks Inducing Invasive Growth in Saccharomyces cerevisiae Identified Through Systematic Genome‐Wide Overexpression. Genetics, 193, 1297–1310.
Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS and McCusker JH, 2015. The 100‐genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Research, 25, 762–774.
Tyler AR, Okoh AO, Lawrence CL, Jones VC, Moffatt C and Smith RB, 2013. N‐Alkylated 2,3,3‐trimethylindolenines and 2‐methylbenzothiazoles. Potential lead compounds in the fight against Saccharomyces cerevisiae infections. European Journal of Medicinal Chemistry, 64, 222–227.
Zupan J, Matos T, Cencic A and Raspor PI, 2013. Methodological approaches for unraveling ill‐natured moments of generally good‐natured Saccharomyces cerevisiae. Matica Srpska Journal for Natural Sciences, 379–396.
Viruses used for plant protection
Baculoviridae
Airenne KJ, Hu YC, Kost TA, Smith RH, Kotin RM, Ono C, Matsuura Y, Wang S and Yla‐Herttuala S, 2013. Baculovirus: an Insect‐derived Vector for Diverse Gene Transfer Applications. Molecular Therapy, 21, 739–749.
Chen CY, Lin SY, Cheng MC, Tsai CP, Hung CL, Lo KW, Hwang Y and Hu YC, 2013. Baculovirus vector as an avian influenza vaccine: Hemagglutinin expression and presentation augment the vaccine immunogenicity. Journal of Biotechnology, 164, 143–150.
Felberbaum RS, 2015. The baculovirus expression vector system: A commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnology Journal, 10, 702‐U785, doi:10.1002/biot.201400438.
Fujihira A, Suzuki T, Chang MO, Moriyama T, Kitajima M and Takaku H, 2014. Antitumor effects of baculovirus‐infected dendritic cells against human pancreatic carcinoma. Gene Therapy, 21, 849–854.
Fujita R, Ono C, Ono I, Asano S and Bando H, 2015. Analysis of the Bombyx mori nucleopolyhedrovirus ie‐1 promoter in insect, mammalian, plant, and bacterial cells. Biochemical and Biophysical Research Communications, 464, 1297–1301.
O'Flynn NMJ, Patel A, Kadlec J and Jones IM, 2013. Improving promiscuous mammalian cell entry by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. Bioscience Reports, 33, 23‐U221, doi:10.1042/bsr20120093.
Swift SL, Rivera GC, Dussupt V, Leadley RM, Hudson LC, de Ridder CMA, Kraaij R, Burns JE, Maitland NJ and Georgopoulos LJ, 2013. Evaluating Baculovirus as a Vector for Human Prostate Cancer Gene Therapy. Plos One, 8, doi:10.1371/journal.pone.0065557.
Alphaflexiviridae
Duff‐Farrier CRA, Bailey AM, Boonham N and Foster GD, 2015. A pathogenicity determinant maps to the N‐terminal coat protein region of the Pepino mosaic virus genome. Molecular Plant Pathology, 16, 308–315.
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2013. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal 2013;11(11):3449, 108 pp. doi: 10.2903/j.efsa.2013.3449.
Hasiow‐Jaroszewska B, Minicka J, Borodynko N and Pospieszny H, 2015. The genetic determinants of symptoms induction by Pepino mosaic virus in tomato. Proceedings of the IV International Symposium on Tomato Diseases, Acta Horticulturae, Orlando, Florida, USA, 39–43.
Hillung J, Cuevas JM, Valverde S and Elena SF, 2014. Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution, 68, 2467–2480.
Minicka J, Rymelska N, Elena SF, Czerwoniec A and Hasiow‐Jaroszewska B, 2015. Molecular evolution of Pepino mosaic virus during long‐term passaging in different hosts and its impact on virus virulence. Annals of Applied Biology, 166, 389–401.
Yusibov V, Streatfield SJ, Kushnir N, Roy G and Padmanaban A, 2013. Hybrid Viral Vectors for Vaccine and Antibody Production in Plants. Current Pharmaceutical Design, 19, 5574–5586.
Appendix E – Microbial species as notified to EFSA until September 2016
| Unit EFSA/Panel | Microorganism species/strain | Intended use | EFSA Question numbera and EFSA webpage linkb | Additional information and QPS evaluation/comments |
|---|---|---|---|---|
| Bacteria | ||||
| Feed/FEEDAP |
Actinomadura roseorufa ATCC 53664 |
Production of semduramicin (coccidiostat) | EFSA‐Q‐2014‐00219 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion ‘A. roseorufa produces semduramicin, an approved coccidiostat, with antimicrobial activity and therefore cannot be considered for the QPS list. Moreover its identity is not well established’. Please refer to the complete assessment. Also notified for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). |
| Feed/FEEDAP |
Actinomadura roseorufa ATCC 53664 |
Production of semduramicin sodium | EFSA‐Q‐2015‐00714 | |
| Feed/FEEDAP | Actinomadura yumaensis | Production of maduramicin ammonium |
EFSA‐Q‐2008‐757 http://www.efsa.europa.eu/en/efsajournal/pub/1954 EFSA‐Q‐2011‐00059 |
Actinomadura yumaensis produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). |
| Feed/FEEDAP | Actinoplanes utahensis | Production of acarbose |
EFSA‐Q‐2007‐172 |
No body of knowledge, therefore it is not appropriate for QPS (EFSA, 2008). |
| Feed/FEEDAP |
Alcaligenes acidovorans = Ralstonia sp. |
Biomass for animal feed |
EFSA‐Q‐2004‐171 |
No body of knowledge, therefore it is not appropriate for QPS (EFSA, 2008). |
| FIP/CEF | Arthrobacter ramosus | Production of food enzyme 4‐α‐d‐{(1→4)a‐d‐glucano} trehalose trehalohydrolase | EFSA‐Q‐2016‐00135 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel et al., 2017) with the conclusion ‘Due to a very limited body of knowledge and the association of some Arthrobacter spp. to human disease (although not food‐borne), QPS status cannot be granted to A. ramosus’. Please refer to the complete assessment. |
| FIP/CEF | Arthrobacter ramosus | Production of food enzyme (1→4)‐α‐d‐glucan 1‐α‐d‐glucosylmutase | EFSA‐Q‐2016‐00136 | |
| Feed/FEEDAP | Bacillus amyloliquefaciens | Feed additive |
EFSA‐Q‐2007‐190 http://www.efsa.europa.eu/en/efsajournal/pub/773 EFSA‐Q‐2009‐00825 http://www.efsa.europa.eu/en/efsajournal/pub/1918 EFSA‐Q‐2011‐00389 http://www.efsa.europa.eu/en/efsajournal/pub/3042 EFSA‐Q‐2011‐00965 |
Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bacillus amyloliquefaciens, GMM strain | Production of enzyme |
EFSA‐Q‐2007‐0020 http://www.efsa.europa.eu/en/efsajournal/pub/1156 and related opinions: EFSA‐Q‐2007‐112 http://www.efsa.europa.eu/en/efsajournal/pub/1154 EFSA‐Q‐2009‐00470 http://www.efsa.europa.eu/en/efsajournal/pub/1949 Other applications: EFSA‐Q‐2010‐01295 EFSA‐Q‐2010‐01297 EFSA‐Q‐2012‐00411 |
|
| Feed/FEEDAP | Bacillus amyloliquefaciens | Technological additive | EFSA‐Q‐2016‐00646 | |
| FIP/CEF | Bacillus amyloliquefaciens | Production of food enzyme α‐amylase | EFSA‐Q‐2015‐00846 | |
| FIP/CEF | Bacillus amyloliquefaciens/AE‐GT | Production of food enzyme | EFSA‐Q‐2015‐00289 | |
| Pesticides | Bacillus amyloliquefaciens AH2 | Plant protection product |
EFSA‐Q‐2015‐00614 Application for approval |
|
| Feed/FEEDAP | Bacillus amyloliquefaciens BS 15A‐P4, LSSA01, BS2084 | Zootechnical additive |
EFSA‐Q‐2015‐00179 |
|
| Pesticides | Bacillus amyloliquefaciens strain FZB240 | Plant protection product |
EFSA‐Q‐2014‐00322 http://www.efsa.europa.eu/en/efsajournal/pub/4494 Application for approval |
|
| Pesticides | Bacillus amyloliquefaciens strain MBI600 | Plant protection product |
EFSA‐Q‐2014‐00323 http://www.efsa.europa.eu/en/efsajournal/pub/4359 Application for approval |
|
| Pesticides | Bacillus amyloliquefaciens subsp. plantarum strain D747 | Plant protection product |
EFSA‐Q‐2013‐00038 http://www.efsa.europa.eu/en/efsajournal/pub/3624 Application for approval EFSA‐Q‐2015‐00081 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
|
| FIP/CEF | Bacillus amyloliquefaciens (strain BANSC) | Production of enzyme amylase | EFSA‐Q‐2014‐00730 | |
| Feed/FEEDAP |
Bacillus brevis = Aneurinibacillus and Brevibacillus species Strains from B. brevis are now mostly Brevibacillus species and some are Aneurinibacillus species |
Biomass for animal feed |
EFSA‐Q‐2004‐171 |
No sufficient body of knowledge and safety concern because of antibiotic production. Therefore not appropriate for QPS (EFSA, 2008). It will no longer be assessed for the QPS list unless new notification to EFSA (EFSA BIOHAZ Panel, 2010). |
| Feed/FEEDAP | Bacillus cereus var. toyoi = B. cereus* | Feed additive |
EFSA‐Q‐2003‐086 http://www.efsa.europa.eu/en/efsajournal/pub/62 EFSA‐Q‐2005‐021 http://www.efsa.europa.eu/en/efsajournal/pub/288 EFSA‐Q‐2006‐037 http://www.efsa.europa.eu/en/efsajournal/pub/458 EFSA‐Q‐2007‐090 http://www.efsa.europa.eu/en/efsajournal/pub/549 EFSA‐Q‐2008‐287 http://www.efsa.europa.eu/en/efsajournal/pub/913 EFSA‐Q‐2010‐01095 and EFSA‐Q‐2011‐00832 |
QPS status inapplicable for the group of B. cereus strains (see EFSA, 2007, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion ‘Bacillus toyonensis cannot be proposed for the QPS list because it is a member of the B. cereus group, and because of the absence of evidences at the species level that it does not present safety concerns’. Please refer to the complete assessment. *The species Bacillus toyonensis (previously B. cereus var. toyoi) was recently published in the validation list no 155 (Oren and Garrity, 2014). |
| FIP/CEF | Bacillus circulans (AE‐LT) | Production of food enzyme β‐galactosidase | EFSA‐Q‐2014‐00670 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015a) with the conclusion ‘Bacillus circulans is not recommended for the QPS list due to the lack of sufficient body of knowledge on a safe history of use or presence in foods and feeds’. Please refer to the complete assessment. |
| FIP/CEF | Bacillus circulans (strain M3‐1) | Production of food enzyme β‐galactosidase | EFSA‐Q‐2016‐00210 | |
| Feed/FEEDAP | Bacillus coagulans | Feed additive | Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Bacillus coagulans | Production of lactic acid | EFSA‐Q‐2016‐00645 | |
| Feed/FEEDAP | Bacillus coagulans | Zootechnical additive | EFSA‐Q‐2014‐00832 | |
| Feed/FEEDAP | Bacillus firmus = Brevibacillus agri | Biomass for animal feed |
EFSA‐Q‐2004‐171 http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178620784006.htm |
No body of knowledge, therefore not appropriate for QPS (EFSA, 2008). It will no longer be assessed for the QPS list unless new notification to EFSA (EFSA BIOHAZ Panel, 2010). |
| Pesticides | Bacillus firmus I‐1582 | Plant protection product |
EFSA‐Q‐2011‐00999 http://www.efsa.europa.eu/en/efsajournal/pub/2868 Application for approval EFSA‐Q‐2013‐00346 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residues Limits) |
A reassessment of this species was carried out in the QPS 2012 review and it was not recommended for the QPS list Included in the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) but it was agreed not to evaluate it until the respective dossier (including the literature review) is received. |
| FIP/CEF | Bacillus flexus | Production of food enzyme β‐amylase | EFSA‐Q‐2015‐00691 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Bacillus flexus can be recommended for the QPS list with a qualification of absence of toxigenic activity (as applied to all strains of Bacillus species recommended to the QPS list)’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bacillus lentus | Feed additive |
Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. |
|
| Feed/FEEDAP | Bacillus lentus | Production of enzyme |
EFSA‐Q‐2006‐004 http://www.efsa.europa.eu/en/efsajournal/pub/412 and related question: EFSA‐Q‐2012‐00244 |
|
| European Commission SCF Opinion 2000 | Bacillus licheniformis | Production of β‐cyclodextrin (food additive carrier and stabiliser of food flavours, food colours and some vitamins) |
Scientific Committee on Food SCF/CS/ADD/AMI 52 Final (13 July) Opinion of the Scientific Committee on Food on β‐cyclodextrin produced using cycloglycosyl‐transferase from a recombinant Bacillus licheniformis (adopted on 22 June 2000) https://ec.europa.eu/food/sites/food/files/safety/docs/sci-com_scf_out58_en.pdf |
Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bacillus licheniformis | Production of enzyme |
EFSA‐Q‐2005‐090 http://www.efsa.europa.eu/en/efsajournal/pub/351 EFSA‐Q‐2006‐181 http://www.efsa.europa.eu/en/efsajournal/pub/451 EFSA‐Q‐2010‐00139 http://www.efsa.europa.eu/en/efsajournal/pub/2777 EFSA‐Q‐2008‐431 |
|
| Feed/FEEDAP | Bacillus licheniformis | Feed additive |
EFSA‐Q‐2006‐136 http://www.efsa.europa.eu/en/efsajournal/pub/2356 EFSA‐Q‐2007‐166 (withdrawn) EFSA‐Q‐2009‐00970 (withdrawn) EFSA‐Q‐2009‐00680 |
|
| Feed/FEEDAP | Bacillus licheniformis | Production of enzyme | EFSA‐Q‐2014‐00575 | |
| FIP/CEF | Bacillus licheniformis | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00911 | |
| FIP/CEF | Bacillus licheniformis, GMM strain | Production of food enzyme β‐galactosidase | EFSA‐Q‐2015‐00093 | |
| FIP/CEF | Bacillus licheniformis | Production of food enzyme subtilisin | EFSA‐Q‐2015‐00232 | |
| FIP/CEF | Bacillus licheniformis, GMM strain | Production of food enzyme glucan 1,4‐α‐maltotetraohydrolase | EFSA‐Q‐2015‐00448 | |
| FIP/CEF | Bacillus licheniformis, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2015‐00666 | |
| FIP/CEF | Bacillus licheniformis, GMM strain | Production of food enzyme pullulanase | EFSA‐Q‐2015‐00667 | |
| FIP/CEF | Bacillus licheniformis, GMM strain | Production of food enzyme acetolactate decarboxylase | EFSA‐Q‐2016‐00031 | |
| Feed/FEEDAP | Bacillus licheniformis (ATCC 53757), GMM strain | Zootechnical additive | EFSA‐Q‐2013‐00630 | |
| FIP/CEF | Bacillus licheniformis (DP‐Dzb44), GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2015‐00836 | |
| FIP/CEF | Bacillus licheniformis/DP‐Dzr46, GMM strain | Production of food enzyme glucan 1,4‐α‐maltohydrolase | EFSA‐Q‐2016‐00095 | |
| FIP/CEF | Bacillus licheniformis/DP‐Dzr50, GMM strain | Production of food enzyme glucan 1,4‐α‐maltohydrolase | EFSA‐Q‐2016‐00096 | |
| FIP/CEF | Bacillus licheniformis/DP‐Dzr52, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2016‐00093 | |
| Feed/FEEDAP | Bacillus licheniformis DSM 28710 | Zootechnical additive |
EFSA‐Q‐2015‐00346 |
|
| FIP/CEF | Bacillus licheniformis NZYM‐AC, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2013‐00586 | |
| FIP/CEF | Bacillus licheniformis (NZYM‐AN), GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2015‐00084 | |
| FIP/CEF | Bacillus licheniformis (NZYM‐AV), GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00794 | |
| FIP/CEF | Bacillus licheniformis NZYM‐BC, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2013‐00685 | |
| FIP/CEF | Bacillus licheniformis (NZYM‐BT), GMM strain | Production of food enzyme β‐galactosidase | EFSA‐Q‐2014‐00093 (withdrawn) | |
| FIP/CEF | Bacillus licheniformis (NZYM‐CE), GMM strain | Production of food enzyme xylanase | EFSA‐Q‐2015‐00064 | |
| FIP/CEF | Bacillus licheniformis/NZYM‐JA | Production of food enzyme | EFSA‐Q‐2015‐00275 | |
| FIP/CEF | Bacillus licheniformis NZYM‐KE, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2012‐00898 | |
| FIP/CEF | Bacillus licheniformis NZYM‐RH, GMM strain | Production of food enzyme serine protease | EFSA‐Q‐2014‐00292 | |
| Feed/FEEDAP | Bacillus megaterium | Production of vitamin C |
EFSA‐Q‐2010‐01290 amended to EFSA‐Q‐2011‐00250 |
Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bacillus pumilus | Feed additive | Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. | |
| FIP/CEF | Bacillus pumilus (BLXSC) | Production of food enzyme xylanase | EFSA‐Q‐2014‐00844 | |
| Pesticides | Bacillus pumilus strain QST 2808 | Plant protection product |
EFSA‐Q‐2012‐00776 http://www.efsa.europa.eu/en/efsajournal/pub/3346 Application for approval EFSA‐Q‐2014‐00359 Review of MRLs (Maximum Residue Limits) |
|
| Feed/FEEDAP | ‘Bacillus smithii’ | Production of lactic acid | EFSA‐Q‐2016‐00645 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2017) with the conclusion ‘The species Bacillus smithii is a natural component of bacterial communities of fermented vegetables and plant derived products. Considering the lack of evidence of pathogenicity, it can be recommended for the QPS list with a qualification of absence of toxigenic activity (as applied to all strains of Bacillus species recommended to the QPS list)’. Please refer to the complete assessment. |
| Pesticides | Bacillus thuringiensis subsp. aizawai (strains ABTS 1857 and GC‐91) = Bacillus thuringiensis serovar aizawai | Plant protection product |
EFSA‐Q‐2009‐00121 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00247 http://www.efsa.europa.eu/en/efsajournal/pub/3063 Application for approval |
Already considered as not appropriate for QPS (see EFSA, 2007. Considered for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). In agreement with the Pesticides Unit, this notification was kept in standby until the respective dossiers (including the literature review) are received. Mentioned in the current Scientific Opinion (refer to Section 3.2.1) ‘B. thuringiensis is not considered for QPS, and therefore papers on this species (e.g. Fagerlund et al., 2014) were not further considered’.; ‘A recently published Opinion of the EFSA BIOHAZ Panel addressed the risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs did not report any additional concerns about Bacillus spp. included in the QPS list (EFSA BIOHAZ Panel, 2016b)’. Please refer to the complete assessment. |
| Pesticides | Bacillus thuringiensis subsp. israelensis (serotype H‐14), strain AM 6552 = Bacillus thuringiensis serovar israelensis | Plant protection product |
EFSA‐Q‐2009‐00122 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00248 http://www.efsa.europa.eu/it/efsajournal/pub/3054 Application for approval |
|
| Pesticides | Bacillus thuringiensis subsp. kurstaki (strains ABTS 351, PB 54, SA11, SA 12, EG 2348) = Bacillus thuringiensis serovar kurstaki | Plant protection product |
EFSA‐Q‐2009‐00123 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00249 http://www.efsa.europa.eu/en/efsajournal/pub/2540 Application for approval |
|
| Pesticides | Bacillus thuringiensis subsp. tenebrionis (strain NB176 (TM 141)) = Bacillus thuringiensis serovar tenebrionis | Plant protection product |
EFSA‐Q‐2009‐00124 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00250 http://www.efsa.europa.eu/en/efsajournal/pub/3024 Application for approval |
|
| Feed/FEEDAP | Bacillus subtilis | Feed additive |
EFSA‐Q‐2003‐008 http://www.efsa.europa.eu/en/efsajournal/pub/6 EFSA‐Q‐2004‐174 http://www.efsa.europa.eu/en/efsajournal/pub/272 EFSA‐Q‐2005‐150 http://www.efsa.europa.eu/en/efsajournal/pub/336 EFSA‐Q‐2005‐237 http://www.efsa.europa.eu/en/efsajournal/pub/406 EFSA‐Q‐2006‐136 http://www.efsa.europa.eu/en/efsajournal/pub/2356 EFSA‐Q‐2007‐166 (withdrawn) EFSA‐Q‐2007‐040 http://www.efsa.europa.eu/en/efsajournal/pub/543 EFSA‐Q‐2008‐473 http://www.efsa.europa.eu/en/scdocs/scdoc/1314.htm EFSA‐Q‐2008‐771 http://www.efsa.europa.eu/en/efsajournal/pub/2375 EFSA‐Q‐2009‐00533 |
Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Evaluated in the current Scientific Opinion (refer to Section 3.2.1) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Bacillus species, as the few infections associated with members of the genus were linked to specific predisposing factors and do not suggest a risk for consumers or animals via exposure through the food and feed chain’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bacillus subtilis | Production of enzyme |
EFSA‐Q‐2007‐0020 http://www.efsa.europa.eu/en/efsajournal/pub/1156 EFSA‐Q‐2012‐00411 and related opinions: EFSA‐Q‐2007‐112 http://www.efsa.europa.eu/en/efsajournal/pub/1154 EFSA‐Q‐2009‐00470 http://www.efsa.europa.eu/en/efsajournal/pub/1949 Other applications: EFSA‐Q‐2010‐01298 |
|
| Feed/FEEDAP | Bacillus subtilis | Feed additive |
EFSA‐Q‐2009‐00680 http://www.efsa.europa.eu/en/efsajournal/pub/4558 EFSA‐Q‐2009‐00525 http://www.efsa.europa.eu/en/efsajournal/pub/4230 EFSA‐Q‐2010‐00814 http://www.efsa.europa.eu/en/efsajournal/pub/1867 EFSA‐Q‐2010‐01150 http://www.efsa.europa.eu/en/efsajournal/pub/2114 EFSA‐Q‐2010‐01151 http://www.efsa.europa.eu/en/efsajournal/pub/2112 EFSA‐Q‐2011‐01151 http://www.efsa.europa.eu/en/efsajournal/pub/3176 EFSA‐Q‐2012‐00246 |
|
| Feed/FEEDAP | Bacillus subtilis | Production of vitamin B2 |
EFSA‐Q‐2010‐00991 http://www.efsa.europa.eu/en/efsajournal/pub/3531 EFSA‐Q‐2010‐01319 http://www.efsa.europa.eu/en/efsajournal/pub/4349 EFSA‐Q‐2012‐00954 (withdrawn) |
|
| Feed/FEEDAP | Bacillus subtilis | Zootechnical additive | EFSA‐Q‐2014‐00587 | |
| Feed/FEEDAP | Bacillus subtilis | Zootechnical additive | EFSA‐Q‐2014‐00832 | |
| Feed/FEEDAP | Bacillus subtilis | Technological additive | EFSA‐Q‐2016‐00220 | |
| Feed/FEEDAP | Bacillus subtilis | Production of lactic acid | EFSA‐Q‐2016‐00645 | |
| FIP/CEF | Bacillus subtilis | Production of food enzyme α‐amylase | EFSA‐Q‐2016‐00133 | |
| FIP/CEF | Bacillus subtilis (strain 11096) | Production of food enzyme pectate lyase | EFSA‐Q‐2016‐00207 | |
| Feed/FEEDAP | Bacillus subtilis C‐3102 (DSM 15544) | Zootechnical additive | EFSA‐Q‐2015‐00004 | |
| Feed/FEEDAP | Bacillus subtilis C‐3102 (DSM 15544) | Zootechnical additive |
EFSA‐Q‐2015‐00005 |
|
| Feed/FEEDAP | Bacillus subtilis C‐3102 DSM 15544 | Zootechnical additive |
EFSA‐Q‐2015‐00239 |
|
| Feed/FEEDAP | Bacillus subtilis C‐3102 DSM 15544 | Zootechnical additive | EFSA‐Q‐2015‐00296 | |
| Feed/FEEDAP | Bacillus subtilis CJKB0001 | Production of vitamin B2 | EFSA‐Q‐2016‐00505 | |
| FIP/CEF | Bacillus subtilis (DP‐Ezd31), GMM strain | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2015‐00839 | |
| FIP/CEF | Bacillus subtilis (DP‐Ezg29), GMM strain | Production of food enzyme β‐galactosidase | EFSA‐Q‐2015‐00838 | |
| FIP/CEF | Bacillus subtilis (DP‐Ezm28), GMM strain | Production of food enzyme endo‐1,3(4)‐β‐glucanase | EFSA‐Q‐2015‐00828 | |
| Feed/FEEDAP | Bacillus subtilis DSM 15544 | Zootechnical additives | EFSA‐Q‐2016‐00197 | |
| Feed/FEEDAP | Bacillus subtilis (DSM 27273) | Zootechnical additive |
EFSA‐Q‐2014‐00729 |
|
| Feed/FEEDAP | Bacillus subtilis DSM 28343 | Zootechnical additive | EFSA‐Q‐2015‐00164 | |
| Feed/FEEDAP | Bacillus subtilis DSM 29784 | Zootechnical additive | EFSA‐Q‐2016‐00448 | |
| Pesticides | Bacillus subtilis strain IAB/BS03 | Plant protection product |
EFSA‐Q‐2015‐00389 Application for approval |
|
| FIP/CEF | Bacillus subtilis (LMG S‐24584) | Production of food enzyme xylanase | EFSA‐Q‐2015‐00065 | |
| FIP/CEF | Bacillus subtilis (LMGS 25520), GMM strain | Production of food enzyme aqualysin 1 | EFSA‐Q‐2014‐00920 | |
| FIP/CEF | Bacillus subtilis/LMG‐S‐27588 | Production of food enzyme | EFSA‐Q‐2015‐00408 | |
| Feed/FEEDAP | Bacillus subtilis LMG S 27588 | Production of endo‐1,4‐β‐xylanase | EFSA‐Q‐2016‐00179 | |
| FIP/CEF | Bacillus subtilis strain MAM | Production of enzyme glucan 1,4‐α‐maltohydrolase | EFSA‐Q‐2013‐00790 | |
| FIP/CEF | Bacillus subtilis MAM, GMM strain | Production of food enzyme glucans 1,4‐alpha glucosidase | EFSA‐Q‐2013‐00790 | |
| FIP/CEF | Bacillus subtilis (NBA), GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00912 | |
| FIP/CEF | Bacillus subtilis (NZYM‐CK), GMM strain | Production of food enzyme asparaginase | EFSA‐Q‐2014‐00845 | |
| FIP/CEF | Bacillus subtilis/NZYM‐DB | Production of food enzyme | EFSA‐Q‐2015‐00127 | |
| FIP/CEF | Bacillus subtilis (NZYM‐OC), GMM strain | Production of food enzyme maltogenic amylase | EFSA‐Q‐2014‐00922 | |
| FIP/CEF | Bacillus subtilis (NZYM‐SM), GMM strain | Production of food enzyme maltogenic Amylase (glucan 1,4‐α‐maltohydrolase) | EFSA‐Q‐2015‐00096 | |
| FIP/CEF | Bacillus subtilis (NZYM‐SO) | Production of food enzyme maltogenic amylase (glucan 1,4‐α‐maltohydrolase) | EFSA‐Q‐2015‐00046 | |
| Pesticides | Bacillus subtilis strain QST 713 | Plant protection product |
EFSA‐Q‐2008‐492 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) Review report for the active substance Bacillus subtilis QST 713, SANCO/10184/2003‐rev. final, July 2006 |
|
| Pesticides | Bacillus subtilis strain QST 713 | Plant protection product |
EFSA‐Q‐2016‐00172 Application for renewal of approval (AIR III) |
|
| FIP/CEF | Bacillus subtilis TD160(229), GMM strain | Production of food enzyme xylanase | EFSA‐Q‐2014‐00733 | |
| FIP/CEF |
Bacillus subtilis XAS, GMM strain |
Production of food enzyme endo 1,4‐beta xylanase |
EFSA‐Q‐2014‐00293 |
|
| Feed/FEEDAP |
Bacillus toyonensis (previously B. cereus var. toyoi) |
Zootechnical additive |
EFSA‐Q‐2014‐00043 |
Already assessed in several occasions as Bacillus cereus var. toyoi but in 2014 has been reassigned to this novel taxonomical unit. Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion ‘Bacillus toyonensis cannot be proposed for the QPS list because it is a member of the B. cereus group, and because of the absence of evidences at the species level that it does not present safety concerns’. Please refer to the complete assessment. |
| Nutrition/NDA | Pasteurised milk products fermented with Bacteroides xylanisolvens | As a novel food ingredient |
EFSA‐Q‐2014‐00301 http://www.efsa.europa.eu/en/efsajournal/pub/3955 As a Novel food ingredient in the context of Regulation (EC) No 258/97 |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Bacteroides xylanisolvens is not recommended for the QPS list, because the body of knowledge is insufficient and safety concerns cannot be totally excluded’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bifidobacterium animalis | Feed additive |
EFSA‐Q‐2006‐169 (withdrawn) EFSA‐Q‐2009‐00823 http://www.efsa.europa.eu/en/efsajournal/pub/2965 EFSA‐Q‐2009‐00817 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Evaluated in the current Scientific Opinion (refer to Section 3.1) with the conclusion: ‘The cases of infection in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection. The use of microorganisms intended to be used as “probiotic” for humans as a health claim does not fall under the remit of the QPS assessment. In conclusion, the QPS status of the Bifidobacterium species previously included in the list does not change and monitoring should continue’. Please refer to the complete assessment. |
| Feed/FEEDAP | Bifidobacterium animalis AHC7 (NCIMB 41617) | Zootechnical additive | EFSA‐Q‐2014‐00573 (withdrawn) | |
| Feed/FEEDAP | Bifidobacterium animalis subsp. animalis DSM 16284 | Zootechnical additive |
EFSA‐Q‐2014‐00224 |
|
| Nutrition/NDA | Bifidobacterium bifidum CNCM I‐3426 | Food targeted for health claims: ‘increases the proportion of healthy days by maintaining normal immune function in healthy adults during everyday life events such a moderate stress’ |
EFSA‐Q‐2014‐00673 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Bifidobacterium longum | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Evaluated in the current Scientific Opinion (refer to Section 3.1) conclusion: ‘The cases of infection in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection. The use of microorganisms intended to be used as “probiotic” for humans as a health claim does not fall under the remit of the QPS assessment. In conclusion, the QPS status of the Bifidobacterium species previously included in the list does not change and monitoring should continue’. Please refer to the complete assessment. | |
| Nutrition/NDA | A combination of four bacterial strains: Bifidobacterium longum LA 101, Lactobacillus helveticus LA 102, Lactococcus lactis LA 103 and Streptococcus thermophilus LA 104 | Food targeted for health claims: ‘improvement of bowel function by increasing stool frequency’ |
EFSA‐Q‐2013‐00893 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | BIOMIN® BBSH 797 – DSM 11798 Genus nov. (formerly Eubacterium) species nov. | Feed additive |
EFSA‐Q‐2012‐00719 |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015b): First studies revealed that the strain belongs to an undescribed new genus within the family Coriobacteriaceae. The conclusion says that ‘As the taxonomic unit of strain BBSH 797 is not identified so far, it does not qualify for QPS inclusion. A strain‐specific evaluation is therefore necessary’. Please refer to the complete assessment. |
| Feed/FEEDAP | BIOMIN® BBSH 797 – gen. nov., sp. nov. DSM 11798 | Technological additive | EFSA‐Q‐2015‐00145 | |
| GMO |
Brevibacterium lactofermentum = Corynebacterium glutamicum |
Dried killed biomass for feed | EFSA‐Q‐2007‐157 (withdrawn) |
The recipient species is QPS for production purposes only, but not for this application, therefore not appropriate for QPS (EFSA, 2008) Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.3): ‘This TU has the following qualification ‘QPS only applies when the species is used for aminoacid production’. Due to a lack of knowledge in relation to history of use of the viable organisms and because other members of the same genus are pathogenic, the qualification is confirmed’, with the conclusion: ‘The QPS recommendation is confirmed for Corynebacterium glutamicum as well as the qualification’.Please refer to the complete assessment. |
| Feed/FEEDAP | Carnobacterium divergens S1 | Zootechnical additive |
EFSA‐Q‐2013‐00996 |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion ‘The TU is well described and the body of knowledge shows it as a common species in the food chain, especially in meat. Carnobacterium divergens can be recommended for the QPS list with the qualification of absence of acquired AMR determinants’. Please refer to the complete assessment. |
| FIP/CEF | Carnobacterium maltaromaticum CNCM I‐3298 | Microbiological time temperature integrators used as ‘active and intelligent’ food contact materials |
EFSA‐Q‐2011‐00120 |
No QPS recommendation given because the species represents fish pathogens (EFSA BIOHAZ Panel, 2012). |
| FIP/CEF | Cellulosimicrobium cellulans | Production of food enzyme β‐glucanase | EFSA‐Q‐2015‐00693 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Safety concerns regarding Cellulosimicrobium cellulans have not excluded the possibility of ill effects developing during its manipulation for enzyme production. Therefore, the organism cannot be awarded QPS status’. Please refer to the complete assessment. |
| Pesticides | Chromobacterium subtsugae strain PRAA4‐1T | Plant protection product |
EFSA‐Q‐2015‐00478 Application for approval |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Due to an insufficient body of knowledge, Chromobacterium subtsugae cannot be proposed for the QPS list’. Please refer to the complete assessment. |
| FIP/CEF | Chryseobacterium proteolyticum | Production of food enzyme protein glutaminase | EFSA‐Q‐2015‐00695 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015b) with the conclusion ‘“Chryseobacterium proteolyticum” it is not a valid species name and all the studies refer to a single strain. Consequently, this taxonomical unit cannot be considered for the QPS list’. Please refer to the complete assessment. |
| Feed/FEEDAP | Clostridium butyricum | Feed additive |
EFSA‐Q‐2008‐303 http://www.efsa.europa.eu/en/efsajournal/pub/1039 EFSA‐Q‐2010‐00140 |
No history of use, possible production of botulinum toxins, therefore not appropriate for QPS (EFSA, 2008; EFSA BIOHAZ Panel, 2011). Re‐evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘The information collected supports the view that the safety of C. butyricum is only known for a few strains, therefore Clostridium butyricum is not recommended for the QPS list. Thus, no additional information supports a revision of the previous conclusion attained in 2011’. Please refer to the complete assessment. |
| Feed/FEEDAP | Clostriduim butyricum CBM 588 | Zootechnical additive |
EFSA‐Q‐2013‐00594 |
|
| Nutrition/NDA | Combination of four bacterial strains: Bifidobacterium longum LA 101, Lactobacillus helveticus LA 102, Lactococcus lactis LA 103 and Streptococcus thermophilus LA 104 | Food targeted for health claims: ‘reducing intestinal discomfort’ |
EFSA‐Q‐2013‐00892 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Corynebacterium glutamicum | Production of l‐arginine |
EFSA‐Q‐2006‐031 |
This TU has the following qualification ‘QPS only applies when the species is used for aminoacid production’. Evaluated in the current Scientific Opinion (refer to Section 3.1.3) with the conclusion: ‘The QPS recommendation is confirmed for Corynebacterium glutamicum as well as the qualification’. Please refer to the complete assessment. *Corynebacterium pekinese is not a valid species name however used in the literature(c). |
| Feed/FEEDAP | Corynebacterium glutamicum | Production of l‐tryptophan |
EFSA‐Q‐2011‐00946 |
|
| Feed/FEEDAP |
Corynebacterium glutamicum (Brevibacterium flavum) |
Production of l‐lysine HCl or sulfate |
EFSA‐Q‐2011‐00991 |
|
| Feed/FEEDAP | Corynebacterium glutamicum | Production of l‐lysine sulfate |
EFSA‐Q‐2011‐00996 |
|
| Feed/FEEDAP | Corynebacterium glutamicum | Production of l‐valine |
EFSA‐Q‐2012‐00377 |
|
| Feed/FEEDAP | Corynebacterium glutamicum | Nutritional additives (amino acid) | EFSA‐Q‐2014‐00635 (withdrawn) | |
| Feed/FEEDAP | Corynebacterium glutamicum | Production of lysine | EFSA‐Q‐2016‐00574 | |
| Feed/FEEDAP | Corynebacterium glutamicum KCCM80099 | Production l‐arginine | EFSA‐Q‐2016‐00405 | |
| Feed/FEEDAP | Corynebacterium glutamicum KCTC 10423BP | Nutritional additives (amino acid) |
EFSA‐Q‐2014‐00296 |
|
| Feed/FEEDAP | Corynebacterium pekinese* = Corynebacterium glutamicum | Production of l‐lysine sulfate |
EFSA‐Q‐2011‐00995 |
|
| Feed/FEEDAP | Ensifer adhaerens | Production of vitamin B12 |
EFSA‐Q‐2012‐00455 http://www.efsa.europa.eu/en/efsajournal/pub/4112 EFSA‐Q‐2012‐00456 |
Not recommended for the QPS list, QPS 2011 update due to insufficient body of knowledge. |
| Feed/FEEDAP | Ensifer fredii | Production of vitamin B12 | EFSA‐Q‐2012‐00456 | Not recommended for the QPS list, QPS 2011 update due to insufficient body of knowledge. |
| Feed/FEEDAP | Enterococcus faecium | Feed additive |
EFSA‐Q‐2003‐087 http://www.efsa.europa.eu/en/efsajournal/pub/207 EFSA‐Q‐2004‐001 http://www.efsa.europa.eu/en/efsajournal/pub/51 EFSA‐Q‐2004‐006 http://www.efsa.europa.eu/en/efsajournal/pub/138 EFSA‐Q‐2004‐027 http://www.efsa.europa.eu/en/efsajournal/pub/120 EFSA‐Q‐2004‐096 http://www.efsa.europa.eu/en/efsajournal/pub/206 EFSA‐Q‐2005‐020 http://www.efsa.europa.eu/en/efsajournal/pub/335 EFSA‐Q‐2006‐061 http://www.efsa.europa.eu/en/efsajournal/pub/440 EFSA‐Q‐2006‐318 http://www.efsa.europa.eu/en/efsajournal/pub/1379 EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2006‐169 (withdrawn) EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2007‐033 http://www.efsa.europa.eu/en/efsajournal/pub/521 EFSA‐Q‐2008‐289 http://www.efsa.europa.eu/en/efsajournal/pub/990 EFSA‐Q‐2008‐471 (withdrawn) EFSA‐Q‐2008‐422 http://www.efsa.europa.eu/en/efsajournal/pub/1661 EFSA‐Q‐2009‐00679 http://www.efsa.europa.eu/en/efsajournal/pub/2574 EFSA‐Q‐2009‐00969 http://www.efsa.europa.eu/en/efsajournal/pub/2118 EFSA‐Q‐2009‐00823 http://www.efsa.europa.eu/en/efsajournal/pub/2965 EFSA‐Q‐2009‐00202 (withdrawn) EFSA‐Q‐2010‐00070 http://www.efsa.europa.eu/en/efsajournal/pub/1636 EFSA‐Q‐2012‐00093 http://www.efsa.europa.eu/en/efsajournal/pub/3044 EFSA‐Q‐2010‐00009 http://www.efsa.europa.eu/en/efsajournal/pub/3097 EFSA‐Q‐2010‐00071 http://www.efsa.europa.eu/en/efsajournal/pub/3170 EFSA‐Q‐2011‐00203 http://www.efsa.europa.eu/en/efsajournal/pub/3167 EFSA‐Q‐2012‐00093 http://www.efsa.europa.eu/en/efsajournal/pub/3044 EFSA‐Q‐2012‐00421 http://www.efsa.europa.eu/en/efsajournal/pub/3175 EFSA‐Q‐2012‐00420 http://www.efsa.europa.eu/en/efsajournal/pub/3098 EFSA‐Q‐2012‐00080 http://www.efsa.europa.eu/en/efsajournal/pub/3363 EFSA‐Q‐2011‐00965 (withdrawn) EFSA‐Q‐2012‐00245 http://www.efsa.europa.eu/en/efsajournal/pub/3602 EFSA‐Q‐2012‐00454 http://www.efsa.europa.eu/en/efsajournal/pub/3727 EFSA‐Q‐2012‐00419 http://www.efsa.europa.eu/en/efsajournal/pub/4158 EFSA‐Q‐2012‐00422 |
No recommendation for QPS status (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013) Considered in the current Scientific Opinion (refer to Section 3.6.2) where it is mentioned that the conclusions of ‘the last update on QPS (EFSA BIOHAZ Panel, 2013) are still valid and E. faecium should be monitored and re‐evaluated in the next QPS Opinion update’ and in the Conclusions chapter that ‘Enterococcus faecium is not recommended for the QPS list in spite of advances in recent scientific knowledge allowing a differentiation of pathogenic from non‐pathogenic strains at the clade level. The QPS approach relies on the basis of the evaluation of TU, where the species/subspecies level is the lowest level of evaluation. Therefore, clades within the E. faecium species cannot be considered as a TU and cannot be evaluated separately.’ Please refer to the complete assessment. |
| Feed/FEEDAP | Enterococcus faecium (CECT 4515) | Zootechnical additive |
EFSA‐Q‐2014‐00827 |
|
| Feed/FEEDAP | Enterococcus faecium CECT 4515 | Zootechnical additive |
EFSA‐Q‐2015‐00055 |
|
| Feed/FEEDAP | Enterococcus faecium DSM 7134 | Zootechnical additive | EFSA‐Q‐2016‐00450 | |
| Feed/FEEDAP | Enterococcus faecium DSM 7134 | Zootechnical additive | EFSA‐Q‐2016‐00452 | |
| Feed/FEEDAP | Enterococcus faecium DSM 21913 | Zootechnical feed additive |
EFSA‐Q‐2014‐00224 |
|
| Feed/FEEDAP | Enterococcus mundtii | Feed additive | No taxonomical unit within Enterococcus can be considered as free of infectious strains. Therefore no recommendation for QPS status (EFSA, 2007). | |
| Feed/FEEDAP | Escherichia coli | Feed additive (horses) |
EFSA‐Q‐2005‐167 |
Is not recommended for the QPS list in the past. There is increasing evidence of pathogenicity (QPS 2009, 2010). Re‐evaluated for the BIOHAZ Panel statement (published December 2014) with the conclusion ‘Escherichia coli cannot be proposed for the QPS list as the safety evaluation has to be done on strain level. No further knowledge supports a revision of the previous conclusion attained in 2009’. |
| Feed/FEEDAP | Escherichia coli | Feed additive l‐lysine production |
EFSA‐Q‐2011‐00992 http://www.efsa.europa.eu/en/efsajournal/pub/3365 EFSA‐Q‐2011‐00993 http://www.efsa.europa.eu/en/efsajournal/pub/3365 EFSA‐Q‐2011‐00994 http://www.efsa.europa.eu/en/efsajournal/pub/3365 EFSA‐Q‐2011‐00995 http://www.efsa.europa.eu/en/efsajournal/pub/4052 EFSA‐Q‐2011‐00996 |
|
| GMO | Escherichia coli | Dried killed biomasses for feed |
EFSA‐Q‐2008‐412a EFSA‐Q‐2008‐669a |
|
| Feed/FEEDAP | Escherichia coli | Dried killed biomasses for feed |
EFSA‐Q‐2008‐412b EFSA‐Q‐2008‐669b |
|
| FIP/CEF | Escherichia coli | Production of food enzyme maltogenic amylase | EFSA‐Q‐2015‐00446 | |
| FIP/CEF | Escherichia coli | Production of polyhydroxyalkanoate (PHA) = from the reaction of dextrose and 1,4 butanediol | EFSA‐Q‐2011‐01080 (withdrawn) | |
| Feed/FEEDAP | Escherichia coli | Feed additive l‐threonine production |
EFSA‐Q‐2012‐00113 http://www.efsa.europa.eu/en/efsajournal/pub/4236 EFSA‐Q‐2012‐00114 http://www.efsa.europa.eu/en/efsajournal/pub/3319 EFSA‐Q‐2012‐00115 http://www.efsa.europa.eu/en/efsajournal/pub/3726 EFSA‐Q‐2012‐00116 http://www.efsa.europa.eu/en/efsajournal/pub/3674 EFSA‐Q‐2012‐00117 http://www.efsa.europa.eu/en/efsajournal/pub/4051 EFSA‐Q‐2012‐00118 |
|
| Feed/FEEDAP | Escherichia coli | Production of l‐threonine |
EFSA‐Q‐2015‐00555 EFSA Journal 2016;14(5):4470 [11 pp.] |
|
| Feed/FEEDAP | Escherichia coli | Feed additive l‐tryptophan production |
EFSA‐Q‐2011‐00946 http://www.efsa.europa.eu/en/efsajournal/pub/4238 EFSA‐Q‐2011‐00947 http://www.efsa.europa.eu/en/efsajournal/pub/3368 EFSA‐Q‐2011‐00948 http://www.efsa.europa.eu/en/efsajournal/pub/4015 EFSA‐Q‐2011‐00949 |
|
| Feed/FEEDAP | Escherichia coli (ATCC 9637) | Production of histidine | EFSA‐Q‐2016‐00304 | |
| Feed/FEEDAP | Escherichia coli (ATCC 9637) | Production of histidine | EFSA‐Q‐2016‐00305 | |
| FIP/CEF | Escherichia coli (BglA MCB3), GMM strain | Production of food enzyme β‐galactosidase | EFSA‐Q‐2015‐00622 | |
| Feed/FEEDAP | Escherichia coli CGMCC 3667 | Technological additive (production of l‐tryptophan) |
EFSA‐Q‐2015‐00251 |
|
| Feed/FEEDAP | Escherichia coli CGMCC 3667 | Production of tryptophan | EFSA‐Q‐2016‐00551 | |
| Feed/FEEDAP | Escherichia coli CGMCC 7.57 | Production of l‐lysine monohydrochloride |
EFSA‐Q‐2015‐00556 |
|
| Feed/FEEDAP | Escherichia coli CGMCC 7.59 | Production of l‐tryptophan |
EFSA‐Q‐2015‐00557 |
|
| Feed/FEEDAP | Escherichia coli/ DC231 | Nutritional/ Production of l‐lysine sulfate |
EFSA‐Q‐2014‐00003 |
|
| Feed/FEEDAP | Escherichia coli FERM BP‐10941, GMM strain | Nutritional/ Production of copper chelate of l‐lysinate‐HCl |
EFSA‐Q‐2013‐00407 http://www.efsa.europa.eu/en/efsajournal/pub/3796 EFSA‐Q‐2014‐00496 |
|
| Feed/FEEDAP | Escherichia coli K‐12 | Technological additive (production of l‐threonine) | EFSA‐Q‐2015‐00252 | |
| Feed/FEEDAP | Escherichia coli K‐12/ AG7056X | Nutritional/Production of threonine |
EFSA‐Q‐2013‐00676 |
|
| Feed/FEEDAP | Escherichia coli K‐12/ AG8012X | Nutritional/Production of tryptophan |
EFSA‐Q‐2013‐00677 |
|
| Feed/FEEDAP | Escherichia coli K‐12/INTK‐01X | Nutritional/Production of lysine |
EFSA‐Q‐2013‐00823 |
|
| Feed/FEEDAP | Escherichia coli VA‐05, GMM strain | Nutritional additives (amino acid) |
EFSA‐Q‐2014‐00299 |
|
| Feed/FEEDAP | Eubacterium sp. | Reduce toxicity of mycotoxins |
EFSA‐Q‐2003‐052 http://www.efsa.europa.eu/en/efsajournal/pub/169 EFSA‐Q‐2012‐00719 |
No body of knowledge. Not appropriate for QPS (EFSA, 2008). |
| FIP/CEF | Geobacillus caldoproteolyticus | Production of food enzyme thermolysin | EFSA‐Q‐2015‐00682 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Due to the lack of sufficient body of knowledge on a safe history of use or presence in foods and feeds, Geobacillus caldoproteolyticus (Anoxybacillus caldiproteolyticus sp. nov.) is not recommended for the QPS list’. Please refer to the complete assessment. |
| FIP/CEF | Geobacillus pallidus | Production of food enzyme 4‐α‐glucanotransferase | EFSA‐Q‐2016‐00033 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Due to the lack of sufficient body of knowledge on a safe history of use or presence in foods and feeds, Aeribacillus pallidus (ex‐Geobacillus pallidus) is not recommended for the QPS list’. Please refer to the complete assessment. |
| FIP/CEF | Geobacillus stearothermophilus | Production of food enzyme cyclomaltodextrin glucanotransferase | EFSA‐Q‐2015‐00230 | Already QPS (EFSA, 2007). Qualification: Absence of toxigenic potential (see EFSA, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore its QPS status does not change’. Please refer to the complete assessment. |
| FIP/CEF | Geobacillus stearothermophilus | Production of food enzyme cyclomaltodextrin glucanotransferase | EFSA‐Q‐2016‐00081 | |
| FIP/CEF | Geobacillus stearothermophilus | Production of food enzyme thermolysin | EFSA‐Q‐2016‐00083 | |
| FIP/CEF | Geobacillus stearothermophilus | Production of food enzyme 1,4‐α‐glucan branching | EFSA‐Q‐2016‐00100 | |
| Feed/FEEDAP | Gluconobacter oxydans | Production of vitamin C |
EFSA‐Q‐2011‐00250 |
Gluconobacter oxydans was assessed for the first time in 2013 (EFSA BIOHAZ Panel, 2013) and was recommended for the QPS list with a qualification ‘QPS only apply when the species is used for vitamin production’ which is relevant for the intended use for which the species was notified’. Re‐evaluated in the current Scientific Opinion (refer to Section 3.3) and ‘no references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore the QPS status of Gluconobacter oxydans does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Ketogulonicigenium vulgare | Production of vitamin C |
EFSA‐Q‐2011‐00250 |
Not recommended for the QPS list (EFSA BIOHAZ Panel, 2011) update due to insufficient body of knowledge. |
| FIP/CEF | Klebsiella pneumoniae | Production of food enzyme pullulanase | EFSA‐Q‐2015‐00450 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘Klebsiella pneumoniae has been implicated in human infections and can be considered a source of antibiotic resistance determinants. Therefore, this species cannot be recommended for the QPS list’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus buchneri | Feed additive |
EFSA‐Q‐2010‐01276 http://www.efsa.europa.eu/en/efsajournal/pub/213 EFSA‐Q‐2011‐00375 http://www.efsa.europa.eu/en/efsajournal/pub/2359 EFSA‐Q‐2011‐00376 http://www.efsa.europa.eu/en/efsajournal/pub/2361 EFSA‐Q‐2011‐00382 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus acidophilus | Feed additive |
EFSA‐Q‐2003‐115 http://www.efsa.europa.eu/en/efsajournal/pub/119 EFSA‐Q‐2003‐055 http://www.efsa.europa.eu/en/efsajournal/pub/52 EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2008‐377 (withdrawn) EFSA‐Q‐2010‐00071 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus acidophilus | Zootechnical additive | EFSA‐Q‐2015‐00429 | |
| Feed/FEEDAP | Lactobacillus amylolyticus | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Lactobacillus amylovorans | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Lactobacillus brevis | Feed additive |
EFSA‐Q‐2010‐01304 http://www.efsa.europa.eu/en/efsajournal/pub/4156 EFSA‐Q‐2011‐00382 http://www.efsa.europa.eu/en/efsajournal/pub/3168 EFSA‐Q‐2011‐00385 http://www.efsa.europa.eu/en/efsajournal/pub/2368 EFSA‐Q‐2012‐00086 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus brevis TAK 124‐1 NCIMB 42149 | Technological additive |
EFSA‐Q‐2015‐00280 |
|
| Feed/FEEDAP |
Lactobacillus bulgaricus = L. delbrueckii subsp. bulgaricus |
Feed additive |
EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2010‐00071 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus casei | Feed additive |
EFSA‐Q‐2011‐00381 http://www.efsa.europa.eu/en/efsajournal/pub/2884 EFSA‐Q‐2011‐00390 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus casei DSM 28872 | Technological additive | EFSA‐Q‐2016‐00237 | |
| Feed/FEEDAP | Lactobacillus casei LOCK 0915 | Zootechnical additive | EFSA‐Q‐2013‐00996 | |
| Feed/FEEDAP |
Lactobacillus casei rhamnosus = Lactobacillus rhamnosus |
Feed additive |
EFSA‐Q‐2011‐00380 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’; ‘As already noted in the 2013 Opinion, L. rhamnosus produced most of the clinical cases reported, probably due to frequent inclusion of isolates of this species into human probiotic preparations. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus cellobiosus | Feed additive |
EFSA‐Q‐2012‐00085 |
Not initially considered for QPS (see EFSA 2007, 2008). QPS recommended (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus collinoides | Feed additive |
EFSA‐Q‐2012‐00086 |
Not initially considered for QPS status (see EFSA 2007, 2008). QPS recommended (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus delbrueckii subsp. lactis | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Lactobacillus diolivorans DSM 32074 | Technological additive | EFSA‐Q‐2015‐00616 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion ‘The species Lactobacillus diolivorans is a natural component of bacterial communities of fermented vegetables and plant derived products. It has never been implicated in human or animal diseases and therefore can be recommended for the QPS list’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus farciminis | Feed additive |
EFSA‐Q‐2006‐062 http://www.efsa.europa.eu/en/efsajournal/pub/771 EFSA‐Q‐2004‐177 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus farciminis CNMA67/4R | Zootechnical additive | EFSA‐Q‐2016‐00712 | |
| Feed/FEEDAP | Lactobacillus fermentum | Feed additive |
EFSA‐Q‐2012‐00085 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| FIP/CEF | Lactobacillus fermentum | Production of food enzyme urease | EFSA‐Q‐2016‐00102 | |
| Feed/FEEDAP | Lactobacillus fermentum (NCIMB 41636) | Technological additive | EFSA‐Q‐2014‐00588 | |
| Nutrition/NDA | Lactobacillus fermentum CECT5716 | Food targeted for health claims | EFSA‐Q‐2016‐00318 | In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Lactobacillus helveticus | Feed additive |
EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2010‐00071 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus hilgardii CNMC I‐4785 | Technological additive | EFSA‐Q‐2016‐00580 | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus lactis* IBB50 | Zootechnical additive |
EFSA‐Q‐2013‐00996 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. *Should be moved to QPS Lactobacillus delbrueckii |
| Feed/FEEDAP | Lactobacillus mucosae | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Lactobacillus paracasei | Feed additive |
EFSA‐Q‐2011‐00378 http://www.efsa.europa.eu/en/efsajournal/pub/2363 EFSA‐Q‐2011‐00387 http://www.efsa.europa.eu/en/efsajournal/pub/2370 EFSA‐Q‐2012‐00082 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Nutrition/NDA | ‘Nutrimune (a heat‐treated fermented milk, fermented with Lactobacillus paracasei CBA L74)’ | Food targeted for health claims |
EFSA‐Q‐2015‐00755 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Lactobacillus pentosus | Feed additive |
EFSA‐Q‐2011‐00388 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus plantarum | Feed additive |
EFSA‐Q‐2010‐01164 http://www.efsa.europa.eu/en/efsajournal/pub/2113 EFSA‐Q‐2011‐00062 http://www.efsa.europa.eu/en/efsajournal/pub/2275 EFSA‐Q‐2011‐00186 http://www.efsa.europa.eu/en/efsajournal/pub/2408 EFSA‐Q‐2011‐00377 http://www.efsa.europa.eu/en/efsajournal/pub/2362 EFSA‐Q‐2011‐00384 http://www.efsa.europa.eu/en/efsajournal/pub/2367 EFSA‐Q‐2011‐00943 http://www.efsa.europa.eu/en/efsajournal/pub/2529 EFSA‐Q‐2011‐00374 http://www.efsa.europa.eu/en/efsajournal/pub/2732 EFSA‐Q‐2012‐00089 http://www.efsa.europa.eu/en/efsajournal/pub/2780 EFSA‐Q‐2011‐00390 http://www.efsa.europa.eu/en/efsajournal/pub/3362 EFSA‐Q‐2011‐00944 http://www.efsa.europa.eu/en/efsajournal/pub/3205 EFSA‐Q‐2011‐00125 http://www.efsa.europa.eu/en/efsajournal/pub/4397 EFSA‐Q‐2012‐00083 http://www.efsa.europa.eu/en/efsajournal/pub/3612 EFSA‐Q‐2012‐00090 http://www.efsa.europa.eu/en/efsajournal/pub/3041 EFSA‐Q‐2012‐00092 http://www.efsa.europa.eu/en/efsajournal/pub/3535 EFSA‐Q‐2012‐00094 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Nutrition/NDA | Lactobacillus plantarum 299v | Food targeted for health claims: ‘Lactobacillus plantarum 299v and increase of non‐haem iron absorption’ |
EFSA‐Q‐2015‐00696 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Nutrition/NDA | Lactobacillus plantarum TENSIA® (DSM 21380) in the semihard Edam‐type ‘heart cheese’ of Harmony™ | Food targeted for health claims: ‘maintenance of cardiovascular health through reduction of blood pressure’ |
EFSA‐Q‐2014‐00097 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Lactobacillus plantarum DSM 29024 | Technological additive | EFSA‐Q‐2015‐00627 | |
| Feed/FEEDAP | Lactobacillus plantarum DSM 29025 | Technological additive |
EFSA‐Q‐2015‐00652 |
|
| Feed/FEEDAP | Lactobacillus plantarum LOCK 0862 | Zootechnical additive |
EFSA‐Q‐2013‐00996 |
|
| Feed/FEEDAP | Lactobacillus plantarum (NCIMB 41638) | Technological additive |
EFSA‐Q‐2014‐00588 |
|
| Feed/FEEDAP | Lactobacillus plantarum TAK 59 NCIMB 42150 | Technological additive |
EFSA‐Q‐2015‐00278 |
|
| Feed/FEEDAP | Lactobacillus reuteri | Feed additive |
EFSA‐Q‐2003‐010 http://www.efsa.europa.eu/en/efsajournal/pub/229 EFSA‐Q‐2006‐169 (withdrawn) |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus rhamnosus | Feed additive |
EFSA‐Q‐2006‐062 http://www.efsa.europa.eu/en/efsajournal/pub/771 EFSA‐Q‐2011‐00380 http://www.efsa.europa.eu/en/efsajournal/pub/2365 EFSA‐Q‐2011‐00125 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’; ‘As already noted in the 2013 Opinion, L. rhamnosus produced most of the clinical cases reported, probably due to frequent inclusion of isolates of this species into human probiotic preparations. Consumption of microorganisms by patients with immunosuppression and/or underlying disease may be considered as the origin of the infection’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus rhamnosus DSM 29226 | Technological additive | EFSA‐Q‐2015‐00626 | |
| Nutrition/ NDA | Lactobacillus rhamnosus GG (ATCC 53103) and fructooligosaccharides (FOS) | Food targeted for health claims: ‘helps to reduce recurrence of lip cold sores caused by Herpes simplex virus infection in healthy susceptible individuals’ |
EFSA‐Q‐2015‐00488 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Nutrition/NDA | Synbio, a combination of Lactobacillus rhamnosus IMC 501® and Lactobacillus paracasei IMC 502® | Food targeted for health claims |
EFSA‐Q‐2014‐00567 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Lactobacillus rhamnosus (NCIMB 41640) | Technological additive |
EFSA‐Q‐2014‐00588 |
|
| Feed/FEEDAP | Lactobacillus sakei | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Lactobacillus salivarius | Feed additive |
EFSA‐Q‐2006‐169 (withdrawn) EFSA‐Q‐2009‐00823 http://www.efsa.europa.eu/en/efsajournal/pub/2965 EFSA‐Q‐2011‐00381 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.4) with the conclusion: ‘There is no requirement to change the QPS recommendation of the previously recommended Lactobacillus species, as the infections reported to be due to members of the genus were extremely scarce and affected patients that already suffered from highly debilitating illnesses and/or were significantly immunodepressed’. Please refer to the complete assessment. |
| Feed/FEEDAP | Lactobacillus salivarius spp. salivarius DSM 16351 | Zootechnical additive |
EFSA‐Q‐2014‐00224 |
|
| Feed/FEEDAP | Lactococcus lactis | Feed additive |
EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2010‐00901 http://www.efsa.europa.eu/en/efsajournal/pub/2374 EFSA‐Q‐2011‐00373 http://www.efsa.europa.eu/en/efsajournal/pub/2448 EFSA‐Q‐2011‐00383 http://www.efsa.europa.eu/en/efsajournal/pub/2366 EFSA‐Q‐2010‐00071 http://www.efsa.europa.eu/en/efsajournal/pub/3170 EFSA‐Q‐2012‐00087 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.5) with the conclusion: ‘There is no need to change the QPS recommendation of Lactococcus lactis, as the infections reported were extremely scarce, and the affected patients already suffered from highly debilitating illnesses and/or were significantly immunodepressed. The possibility that L. lactis might be involved in bovine mastitis, albeit limited for the moment, should be monitored’. Please refer to the complete assessment. |
| FIP/CEF | Lactococcus lactis (strain DGCC5920) | Production of food enzyme membrane alanyl aminopeptidase | EFSA‐Q‐2016‐00208 | |
| Feed/FEEDAP | Lactococcus lactis NCIMB 30160 | Technological additive | EFSA‐Q‐2016‐00568 | |
| FIP/CEF | Leuconostoc citreum (strain NRRL B‐30894) | Production of food enzyme alternansucrase | EFSA‐Q‐2016‐00209 | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.6) with the conclusion: ‘The cases of infections in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. The above new information does not modify the QPS recommendation of Leuconostoc species. Therefore, the QPS recommendation for Lc. mesenteroides, Lc. lactis, Lc. pseudomesenteroides and Lc. citreum was confirmed’. Please refer to the complete assessment. |
| 2001/122/EC | Leuconostoc mesenteroides | Production of dextran as NF ingredient for bakery industrial and food fermentations | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.6) with the conclusion: ‘The cases of infections in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. The above new information does not modify the QPS recommendation of Leuconostoc species. Therefore, the QPS recommendation for Lc. mesenteroides, Lc. lactis, Lc. pseudomesenteroides and Lc. citreum was confirmed’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Leuconostoc oeno = Oenococcus oeni | Feed additive | Not initially considered for QPS (see EFSA, 2007, 2008) and recommended for the QPS list (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.8) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified for Oenococcus oeni. Therefore its QPS status does not change and monitoring should continue’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Leuconostoc pseudomesenteroides | Feed additive | Not initially considered for QPS (see EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011) and recommended for the QPS list in 2012 (EFSA BIOHAZ Panel, 2012) and confirmed in 2013 and 2016. (EFSA BIOHAZ Panel, 2013, 2016a) Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.8) with the conclusion: ‘The cases of infections in humans are mostly linked to specific predisposing factors and do not suggest a risk for the consumer via exposure through the food and feed chain. The above new information does not modify the QPS recommendation of Leuconostoc species. Therefore, the QPS recommendation for Lc. mesenteroides, Lc. lactis, Lc. pseudomesenteroides and Lc. citreum was confirmed. Please refer to the complete assessment. | |
| Feed/FEEDAP | Methylococcus capsulatus | Biomass for animal feed |
EFSA‐Q‐2004‐171 |
No body of knowledge, therefore not appropriate for QPS (EFSA, 2008). |
| FIP/CEF | Microbacterium imperiale AE‐AMT | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00544 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion ‘No record exists of intended use of any Microbacterium in food processing and/or ingestion of viable cells. However, there is a history of use in food processing of enzymes produced by Microbacterium imperiale, therefore it can only be recommended for QPS for enzyme production’. Please refer to the complete assessment. |
| Feed/FEEDAP | Paenibacillus lentus DSM 28088 | Zootechnical additive (production of enzyme) | EFSA‐Q‐2014‐00115 |
Due to the absence of a body of knowledge apart from the description of the species, Paenibacillus lentus cannot be proposed for the QPS list (EFSA BIOHAZ Panel, 2014). |
| Feed/FEEDAP | Paenibacillus lentus DSM 28088 | Production of endo‐1,4‐β‐mannanase | EFSA‐Q‐2016‐00181 | |
| Opinion SCF adopted on 22/06/2000 | Paenibacillus macerans | β‐cyclodextrin production (food additive) | QPS 2009 update not recommended for QPS because of insufficient body of knowledge. Re‐evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion: ’Due to the lack of sufficient body of knowledge on a safe history of use or presence in foods and feeds, Paenibacillus macerans is not recommended for the QPS list’. Please refer to the complete assessment. | |
| FIP/CEF | Paenibacillus macerans | Production of food enzyme cyclomaltodextrin glucanotransferase | EFSA‐Q‐2016‐00082 | |
| Feed/FEEDAP | Paracoccus carotinifaciens Astaxanthin‐rich | Production of red carotenoids |
EFSA‐Q‐2006‐173 http://www.efsa.europa.eu/en/efsajournal/pub/546 EFSA‐Q‐2009‐00629 http://www.efsa.europa.eu/en/efsajournal/pub/1428 EFSA‐Q‐2012‐00064 |
No body of knowledge, therefore not considered for QPS (EFSA, 2008). |
| Pesticides | Pasteuria nishizawae strain Pn1 | Plant protection product |
EFSA‐Q‐2015‐00405 Application for approval |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015b) and re‐evaluated in the current Scientific Opinion (refer to Section 3.1.3) with the conclusion: ‘Pasteuria nishizawae was recommended for the QPS status for use as a plant protection product to combat cyst nematodiasis (EFSA EFSA BIOHAZ Panel, 2015b). This conclusion was based on the following: (i) it is an obligate parasite, unable to grow independently of its host species, H. glycines and possibly H. schachtii. In addition, available evidence indicates that this species of bacteria requires entry of the nematode into the root of a plant for vegetative growth; (ii) the ubiquity and abundance of Pasteuria spp. endospores in soils and the lack of any reports on harmful effects of these bacteria on organisms other than their hosts. The qualification linked to this taxonomic unit was re‐evaluated and the QPS recommendation is now ascribed without the previous qualification (“QPS only applies when used in pesticides to combat cyst nematodiasis”)’. Please refer to the complete assessment. |
| Feed/FEEDAP | Pediococcus acidilactici | Feed additive |
EFSA‐Q‐2006‐169 (withdrawn) EFSA‐Q‐2007‐205 http://www.efsa.europa.eu/en/efsajournal/pub/1037 EFSA‐Q‐2008‐421 http://www.efsa.europa.eu/en/efsajournal/pub/1038 EFSA‐2009‐00719 http://www.efsa.europa.eu/en/efsajournal/pub/1660 EFSA‐2009‐00716 http://www.efsa.europa.eu/en/efsajournal/pub/1865 EFSA‐2009‐00719 http://www.efsa.europa.eu/en/efsajournal/pub/1660 EFSA‐2009‐00716 http://www.efsa.europa.eu/en/efsajournal/pub/1865 EFSA‐Q‐2011‐00379 http://www.efsa.europa.eu/en/efsajournal/pub/2364 EFSA‐Q‐2011‐00940 http://www.efsa.europa.eu/en/efsajournal/pub/2733 EFSA‐Q‐2011‐00941 (withdrawn) EFSA‐Q‐2012‐00084 http://www.efsa.europa.eu/en/efsajournal/pub/3613 EFSA‐Q‐2012‐00253 |
Already QPS (EFSA, 2007, 2008, 2009; EFSA BIOHAZ Panel, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.10) with the conclusion: ’There is no need to change the recommendation of the QPS‐granted pediococci species because no causality of infection has been reported during the scrutinized period’. Please refer to the complete assessment. |
| Feed/FEEDAP | Pediococcus acidilactici (CNCM) MA 18/5M | Zootechnical additive |
EFSA‐Q‐2013‐00704 |
|
| Feed/FEEDAP | Pediococcus acidilactici (CNCM) MA 18/5M | Zootechnical additive |
EFSA‐Q‐2014‐00091 |
|
| Feed/FEEDAP | Pediococcus parvulus DSM 28875 | Technological additive | EFSA‐Q‐2016‐00236 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion; ‘Pediococcus parvulus can be granted the QPS status, being a species commonly found in fermented food and beverages and based on lack of pathogenicity as determined by the absence of any significant virulence determinants in its genome and of any reports on its role on human or animal infection’. Please refer to the complete assessment. |
| Feed/FEEDAP | Pediococcus pentosaceus | Feed additive |
EFSA‐Q‐2009‐00717 http://www.efsa.europa.eu/en/efsajournal/pub/1502 EFSA‐Q‐2011‐00386 http://www.efsa.europa.eu/en/efsajournal/pub/2369 EFSA‐Q‐2011‐00940 http://www.efsa.europa.eu/en/efsajournal/pub/2733 EFSA‐Q‐2012‐00091 http://www.efsa.europa.eu/en/efsajournal/pub/3284 EFSA‐Q‐2012‐00081 http://www.efsa.europa.eu/en/efsajournal/pub/3609 EFSA‐Q‐2012‐00087 http://www.efsa.europa.eu/en/efsajournal/pub/3610 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.10) with the conclusion: ‘There is no need to change the recommendation of the QPS‐granted pediococci species because no causality of infection has been reported during the scrutinized period’. Please refer to the complete assessment. |
| Feed/FEEDAP | Propionibacterium acidipropionici | Feed additive |
EFSA‐Q‐2011‐00953 |
Not proposed for QPS status (see EFSA, 2007). In 2009 recommended for the QPS list (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.11) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, its QPS status does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Propionibacterium freudenreichii shermanii | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.11) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, the QPS status of the Propionibacterium species does not change’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Propionibacterium freudenreichii shermanii | Production of vitamin B12 |
EFSA‐Q‐2012‐00456 EFSA‐Q‐2012‐00457 (withdrawn) |
|
| Feed/FEEDAP |
Propionibacterium globosum [=subspecies of Propionibacterium freudenreichii] |
Feed additive | Initially not recommended for QPS (see EFSA, 2007, Appendix A). Identical with P. freudenreichii therefore included on QPS (EFSA BIOHAZ Panel, 2010). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.11) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore, the QPS status of the Propionibacterium species does not change’. Please refer to the complete assessment | |
| FIP/CEF | Protaminobacter rubrum | Production of food enzyme isomaltulose synthase | EFSA‐Q‐2015‐00620 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion: ‘“Protaminobacter rubrum” it is not a valid species name. Consequently, this taxonomical unit cannot be considered for the QPS list.’ Please refer to the complete assessment |
| Pesticides | Pseudomonas chlororaphis strain MA342 | Plant protection product |
EFSA‐Q‐2008‐618 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) Review report for the active substance Pseudomonas chlororaphis, DG SANCO/4024/VI/98‐Final, March 2004 |
Not recommended for QPS in 2009 update (EFSA BIOHAZ Panel, 2009) because of insufficient body of knowledge and a potential risk linked to production of secondary metabolites. Re‐evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015b) with the conclusion: ‘The safe use of some strains of P. chlororaphis in plant protection products relies on the absence of colonisation of the edible part of plants. No information exists to enable assessment of the risk whether or not P. chlororaphis was used in a situation where it could produce secondary metabolites in food or feed. Because of the insufficient body of knowledge and a potential risk linked to production of secondary metabolites (EFSA BIOHAZ Panel, 2009) and to antimicrobial resistance determinants, Pseudomonas chlororaphis should not be recommended for QPS’. Please refer to the complete assessment. |
| Pesticides | Pseudomonas chlororaphis strain MA342 | Plant protection product |
EFSA‐Q‐2015‐00814 Application for renewal of approval |
|
| FIP/CEF | Pseudomonas fluorescens (BD15754), GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2016‐00200 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2017) with the conclusion: ‘The pathogenic potential of P. fluorescens demonstrated by its implication in human infections and virulence features is an important safety concern, preventing its recommendation for the QPS list. Moreover, the possibility of mupirocin‐resistant Staphylococcus aureus strains selection, as a result of P. fluorescens ability to produce mupirocin, further supports the rejection of the QPS status’. Please refer to the complete assessment. |
| Pesticides | Pseudomonas sp. strain DSMZ 13134 | Plant protection product |
EFSA‐Q‐2011‐01198 http://www.efsa.europa.eu/en/efsajournal/pub/2954 Application for approval |
Not assessed because species to be clarified (EFSA BIOHAZ Panel, 2009). |
| Pesticides | Pseudomonas sp. strain DSMZ 13134 | Plant protection product |
EFSA‐Q‐2014‐00370 Review of MRLs (Maximum Residue Limits) |
Considered for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). In agreement with the Pesticides Unit, this notification was kept in standby until the respective dossiers (including the literature review) are received. |
| FIP/CEF | Pullulanibacillus naganoensis | Production of food enzyme pullulanase | EFSA‐Q‐2015‐00451 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion: ‘The body of knowledge on Pullulanibacillus naganoensis is too limited to exclude the possibility of ill effects developing during its manipulation for enzyme production. Therefore, it cannot be awarded QPS status’. Please refer to the complete assessment. |
| Feed/FEEDAP | Rhodopseudomonas palustris | Feed additive | Insufficient body of knowledge (EFSA BIOHAZ Panel, 2009). It will no longer be assessed for the QPS list unless new notification to EFSA. | |
| Feed/FEEDAP | Serratia rubidaea | Feed additive | Insufficient body of knowledge (EFSA BIOHAZ Panel, 2009). It will no longer be assessed for the QPS list unless new notification to EFSA. | |
| Feed/FEEDAP | Streptococcus cremoris = L. lactis subsp. cremoris | Feed additive | Already QPS (EFSA, 2007). | |
| Feed/FEEDAP |
Streptococcus faecium = Enterococcus faecium |
Feed additive | No recommendation for QPS status (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013). Considered in the current Scientific Opinion (refer to Section 3.6.2) where it is mentioned that the conclusions of ‘the last update on QPS (EFSA BIOHAZ Panel, 2013) are still valid and E. faecium should be monitored and re‐evaluated in the next QPS Opinion update’ and in the Conclusions chapter that ‘Enterococcus faecium is not recommended for the QPS list in spite of advances in recent scientific knowledge allowing a differentiation of pathogenic from non‐pathogenic strains at the clade level. The QPS approach relies on the basis of the evaluation of TU, where the species/subspecies level is the lowest level of evaluation. Therefore, clades within the E. faecium species cannot be considered as a TU and cannot be evaluated separately’. Please refer to the complete assessment. | |
| Feed/FEEDAP | Streptococcus thermophilus | Feed additive |
EFSA‐Q‐2006‐135 http://www.efsa.europa.eu/en/efsajournal/pub/912 EFSA‐Q‐2010‐00071 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.1.11) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore its QPS status does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Streptomyces albus | Production of salinomycin sodium |
EFSA‐Q‐2003‐009 http://www.efsa.europa.eu/en/efsajournal/pub/75 EFSA‐Q‐2012‐00994 |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomyces albus is not recommended for the QPS list, because safety concerns cannot be excluded’. Please refer to the complete assessment. |
| Feed/FEEDAP | Streptomyces albus ATCC 21838 | Production of salinomycin sodium (coccidiostat) |
EFSA‐Q‐2013‐00706 EFSA‐Q‐2013‐00998 EFSA‐Q‐2014‐00350 (withdrawn) |
|
| Feed/FEEDAP |
Streptomyces albus NCIMB 30321 |
Production of salinomycin sodium (coccidiostat) | EFSA‐Q‐2014‐00350 (withdrawn) | |
| Feed/FEEDAP | Streptomyces aureofaciens | Production of polyether monocarboxylic acid |
EFSA‐Q‐2003‐046 |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomyces aureofaciens is not recommended for the QPS list, because the body of knowledge is limited and safety concerns cannot be excluded’. Please refer to the complete assessment. |
| Feed/FEEDAP | Streptomyces aureofaciens | Production of salinomycin |
EFSA‐Q‐2014‐00666 |
|
| Feed/FEEDAP | Streptomyces aureofaciens NRRL 8092 | Production of narasin (coccidiostat) | EFSA‐Q‐2013‐00767 | |
| Feed/FEEDAP | Streptomyces aureofaciens NRRL 8092 | Production of narasin (coccidiostat) | EFSA‐Q‐2015‐00032 | |
| Feed/FEEDAP | Streptomyces aureofaciens NRRL 8092 | Production of narasin (coccidiostat) | EFSA‐Q‐2015‐00033 | |
| Feed/FEEDAP | Streptomyces cinnamonensis | Production of monensin sodium |
EFSA‐Q‐2005‐024 http://www.efsa.europa.eu/en/efsajournal/pub/283 EFSA‐Q‐2012‐00906 EFSA‐Q‐2012‐00791 |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomyces cinnamonensis is not recommended for the QPS list, because the body of knowledge is limited and safety concerns cannot be excluded’. Please refer to the complete assessment. |
| Feed/FEEDAP | Streptomyces cinnamonensis ATCC 15413 | Production of monensin sodium (coccidiostat) | EFSA‐Q‐2013‐00752 | |
| Feed/FEEDAP | Streptomyces cinnamonensis NBIMCC 3419 | Production of monensin sodium | EFSA‐Q‐2015‐00167 (withdrawn) | |
| Feed/FEEDAP | Streptomyces lasaliensis | Production of lasalocid sodium |
EFSA‐Q‐2004‐076 |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomyces lasaliensis is not recommended for the QPS list, because its identity is not well established, the body of knowledge is limited and safety concerns cannot be excluded’. Please refer to the complete assessment. |
| Feed/FEEDAP | Streptomyces lasaliensis ATCC 31180 | Production of lasalocid A sodium (coccidiostat) | EFSA‐Q‐2013‐00813 | |
| Pesticides | Streptomyces lydicus strain WYEC 108 (ATCC 55445) | Plant protection product |
EFSA‐Q‐2012‐00775 http://www.efsa.europa.eu/en/efsajournal/pub/3425 Application for approval EFSA‐Q‐2014‐00595 Review of MRLs (Maximum Residue Limits) |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such as S. scabies. However, they produce antibiotics and may thus select for resistant bacteria. Other secondary metabolites have diverse biological activities that go from depressors of the immune system to herbicides (Butaye et al., 2003). Genome sequencing has revealed that streptomycetes carry several gene clusters for the production of secondary metabolites, many of which may be toxic, or select for antimicrobial resistance. Furthermore, the presence of specific clusters varies on a strain basis. All this precludes the consideration of any species of the genus as a QPS organism’. Please refer to the complete assessment. |
| FIP/CEF | Streptomyces mobaraensis S‐8112 | Production of food enzyme transglutaminase | EFSA‐Q‐2015‐00095 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such as S. scabies. However, they produce antibiotics and may thus select for resistant bacteria. Other secondary metabolites have diverse biological activities that go from depressors of the immune system to herbicides (Butaye et al., 2003). Genome sequencing has revealed that streptomycetes carry several gene clusters for the production of secondary metabolites, many of which may be toxic, or select for antimicrobial resistance. Furthermore, the presence of specific clusters varies on a strain basis. All this precludes the consideration of any species of the genus as a QPS organism’. Please refer to the complete assessment. |
| FIP/CEF | Streptomyces murinus | Production of food enzyme AMP deaminase | EFSA‐Q‐2015‐00683 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such as S. scabies. However, they produce antibiotics and may thus select for resistant bacteria. Other secondary metabolites have diverse biological activities that go from depressors of the immune system to herbicides (Butaye et al., 2003). Genome sequencing has revealed that streptomycetes carry several gene clusters for the production of secondary metabolites, many of which may be toxic, or select for antimicrobial resistance. Furthermore, the presence of specific clusters varies on a strain basis. All this precludes the consideration of any species of the genus as a QPS organism’. Please refer to the complete assessment. |
| FIP/CEF | Streptomyces murinus | Production of food enzyme glucose isomerase | EFSA‐Q‐2016‐00032 | |
| FIP/CEF | Streptomyces violaceoruber (strain AS‐10), GMM strain | Production of food enzyme phospholipase A2 | EFSA‐Q‐2016‐00132 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such as S. scabies. However, they produce antibiotics and may thus select for resistant bacteria. Other secondary metabolites have diverse biological activities that go from depressors of the immune system to herbicides (Butaye et al., 2003). Genome sequencing has revealed that streptomycetes carry several gene clusters for the production of secondary metabolites, many of which may be toxic, or select for antimicrobial resistance. Furthermore, the presence of specific clusters varies on a strain basis. All this precludes the consideration of any species of the genus as a QPS organism’. Please refer to the complete assessment. |
| FIP/CEF | Streptomyces violaceoruber (pChi), GMM strain | Production of food enzyme chitinase | EFSA‐Q‐2015‐00621 | |
| FIP/CEF | Streptomyces violaceoruber (strain pCol), GMM strain | Production of food enzyme microbial collagenase | EFSA‐Q‐2015‐00826 | |
| FIP/CEF | Streptomyces violaceoruber pGlu | Production of food enzyme glucanase | EFSA‐Q‐2015‐00097 | |
| Pesticides | Now unspecified Streptomyces species: ‘Streptomyces strain K 61’, formerly: Streptomyces griseoviridis | Plant protection product |
EFSA‐Q‐2009‐00134 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00295 http://www.efsa.europa.eu/en/efsajournal/pub/3061 Application for approval |
Streptomyces spp. produce antibiotics, are therefore inappropriate for QPS (EFSA, 2008). Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘Streptomycetes are essentially non‐virulent, with the exception of some plant pathogens such as S. scabies. However, they produce antibiotics and may thus select for resistant bacteria. Other secondary metabolites have diverse biological activities that go from depressors of the immune system to herbicides (Butaye et al., 2003). Genome sequencing has revealed that streptomycetes carry several gene clusters for the production of secondary metabolites, many of which may be toxic, or select for antimicrobial resistance. Furthermore, the presence of specific clusters varies on a strain basis. All this precludes the consideration of any species of the genus as a QPS organism’. Please refer to the complete assessment. |
| Feed/FEEDAP | Xanthomonas campestris | Technological additive (production of xanthan gum) |
EFSA‐Q‐2013‐01021 |
Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2015a) with the conclusion: ‘Xanthan gum produced by X. campestris has a long and broad history of safe use in the food industry. X. campestris is a plant pathogen. Apart from one record (Li et al., 1990), X. campestris has never been implicated in human or animal disease. However, human consumers are presumably very rarely exposed to high levels of X. campestris through food, indicating a lack of knowledge on the effect of high levels of live cells of X. campestris on animals and humans. In all papers screened, none of them mentioned acquisition of resistance to antimicrobials. Xanthomonas campestris can be recommended for the QPS list for the production of xanthan gum’. Please refer to the complete assessment. |
| Yeasts | ||||
| Feed/FEEDAP | Astaxanthin rich Phaffia rhodozyma = Xanthophyllomyces dendrorhous | Production of astaxanthin |
EFSA‐Q‐2004‐148 http://www.efsa.europa.eu/en/efsajournal/pub/320 EFSA‐Q‐2003‐112 |
Phaffia rhodozyma was assessed not appropriate for QPS (EFSA, 2008) because of insufficient body of knowledge. Later recommended for the QPS list (EFSA BIOHAZ Panel, 2011) as it is the imperfect form of Xanthophyllomyces dendrorhous according to the 2011 revision of the yeast taxonomy. |
| Pesticides | Aureobasidium pullulans strains DSM 14940 and DSM 14941 | Plant protection product |
EFSA‐Q‐2010‐01499 http://www.efsa.europa.eu/en/efsajournal/pub/2435 EFSA‐Q‐2011‐01200 http://www.efsa.europa.eu/en/efsajournal/pub/3183.htm Applications for approval EFSA‐Q‐2014‐00369 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
Body of knowledge insufficient (QPS 2009 update). Considered for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). In agreement with the Pesticides Unit, this notification was kept in standby until the respective dossiers (including the literature review) are received. |
| FIP/CEF | Candida cylindracea | Production of food enzyme | EFSA‐Q‐2015‐00339 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2014) with the conclusion: ‘In the Candida cylindracea bibliography, the species was only reported for use as an enzyme producer and no safety concerns were identified. Therefore it was concluded that it can be recommended for QPS status. However, since there were no reports on its use in applications involving direct consumption of Candida cylindracea viable cells by humans or animals, QPS should apply only for the production of enzymes’. Please refer to the complete assessment. |
| FIP/CEF | Candida cylindracea AE‐LAYH, GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2014‐00113 | |
| Feed/FEEDAP | Candida glabrata | Feed additive | Unsuitable for QPS (see EFSA, 2007). | |
| Feed/FEEDAP | Candida guilliermondi | Fermentation product |
EFSA‐Q‐2003‐082 |
Unsuitable for QPS (see EFSA, 2007). |
| Pesticides | Candida oleophila strain O | Plant protection product |
EFSA‐Q‐2009‐00338 http://www.efsa.europa.eu/en/efsajournal/pub/2944 Application for approval EFSA‐Q‐2013‐00039 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
Body of knowledge insufficient, therefore not appropriate for QPS (EFSA, 2008). Considered for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). In agreement with the Pesticides Unit, this notification was kept in standby until the respective dossiers (including the literature review) are received. |
| FIP/CEF | Candida rugosa | Production of food enzyme triacyglycerol lipase | EFSA‐Q‐2015‐00291 | Evaluated for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a) with the conclusion: ‘Although Candida rugosa has sometimes been reported to occur in food fermentations, due to a unclear taxonomy it may have been misidentified in several of those cases. Moreover, it has recently been described as an “emerging” human fungal pathogen and is well‐known for causing mastitis. For these reasons C. rugosa is not recommended for QPS status’. Please refer to the complete assessment. |
| Feed/FEEDAP | Hansenula polymorpha = Pichia angusta* | Production of enzymes |
EFSA‐Q‐2005‐030 |
Already QPS status applies only when species is used for enzyme production purposes (EFSA, 2008; EFSA BIOHAZ Panel, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.7) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore its QPS status does not change. The qualification is unchanged’. Please refer to the complete assessment. *Ogataea angusta: synonym Pichia angusta. |
| FIP/CEF | Hansenula polymorpha, GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2015‐00374 | |
| FIP/CEF | Hansenula polymorpha, GMM strain | Production of food enzyme hexose oxidase | EFSA‐Q‐2015‐00406 | |
| FIP/CEF | Kluyveromyces lactis | Production of food enzyme β‐galactosidase | EFSA‐Q‐2015‐00409 | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.4). The ELS search retrieved no new studies with relevance for the QPS evaluation of Kluyveromyces lactis (or its anamorphic name Candida spherica). Therefore its QPS status does not change. Please refer to the complete assessment. |
| FIP/CEF | Kluyveromyces lactis (AE‐KL) | Production of food enzyme β‐galactosidase | EFSA‐Q‐2014‐00669 | |
| FIP/CEF | Kluyveromyces lactis (CIN), GMM strain | Production of food enzyme chymosin | EFSA‐Q‐2015‐00085 | |
| FIP/CEF | Kluyveromyces lactis/CHY | Production of food enzyme | EFSA‐Q‐2015‐00129 | |
| 2148/2004/EC | Kluyveromyces marxianus var. lactisK1 | Feed additive | Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.4) with the conclusion: ‘There is no doubt that K. marxianus/C. kefyr should be considered a significant opportunistic fungus, and it has received increased attention in recent years However, reports where it has been unambiguously shown to be causative agent of infectious disease in otherwise healthy individuals are very rare. Therefore, its QPS status does not change. There is reason to be alert regarding whether there is a tendency for K. marxianus to become more common in this kind of infection’. Please refer to the complete assessment. | |
| Reg(EC)773/ 2006 Corrigendum CS | Kluyveromyces marxianus ‐fragilis | Feed additive | ||
| Feed/FEEDAP | Komagataella pastoris = Pichia pastoris, GMM strain | Production of enzyme |
EFSA‐Q‐2006‐025 http://www.efsa.europa.eu/en/efsajournal/pub/627 and related opinions: EFSA‐Q‐2009‐00804: http://www.efsa.europa.eu/en/efsajournal/pub/1550 EFSA‐Q‐2011‐00148 http://www.efsa.europa.eu/en/efsajournal/pub/2533 Other applications: EFSA‐Q‐2010‐00152 http://www.efsa.europa.eu/en/efsajournal/pub/2414 EFSA‐Q‐2013‐00022 |
Already QPS (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.5) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore its QPS status does not change. The qualification is unchanged’. ‘QPS only applies when the species is used for enzyme production and no viable cells are found’. Please refer to the complete assessment. *Komagataella pastoris (formerly named as Pichia pastoris). |
| Feed/FEEDAP | Komagataella pastoris* (DSMZ 25376), GMM strain | Zootechnical additive (production of enzyme) | EFSA‐Q‐2013‐00528 | |
| Feed/FEEDAP | Komagataella pastoris* (DSM 26643), GMM strain | Technological/production of fumonisin esterase |
EFSA‐Q‐2013‐00090 |
|
| Feed/FEEDAP | Komagataella pastoris* (DSMZ 26469), GMM strain | Zootechnical additive (production of enzyme) | EFSA‐Q‐2013‐00528 | |
| Feed/FEEDAP | Pichia pastoris* | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00829 |
|
| Feed/FEEDAP | Pichia pastoris* | Technological additive (production of enzyme for reduction of mycotoxin contamination of feed) |
EFSA‐Q‐2014‐00900 |
|
| Feed/FEEDAP | Pichia pastoris* ATCC 76273/CBS 7435/ CECT 11047 | Zootechnical additive (production of enzyme 3‐phytase) |
EFSA‐Q‐2015‐00482 |
|
| Feed/FEEDAP | Pichia pastoris* (DSM 23036) | Production of 6‐phytase | EFSA‐Q‐2016‐00291 | |
| FIP/CEF | Pichia pastoris* (PRF), GMM strain | Production of food enzyme phospholipase C | EFSA‐Q‐2016‐00201 | |
| Pesticides | Metschnikowia fructicola | Plant protection product |
EFSA‐Q‐2015‐00546 Application for approval |
Considered for the BIOHAZ Panel Statement (EFSA BIOHAZ Panel, 2016a). In agreement with the Pesticides Unit, this notification was kept in standby until the respective dossiers (including the literature review) are received. |
| GMO/GMO | Saccharomyces cerevisiae | Dried killed biomass for feed |
EFSA‐Q‐2007‐156b (withdrawn) EFSA‐Q‐2009‐00866 (withdrawn) |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.8) with the conclusion: ‘These new reports of Saccharomyces cerevisiae appearing as an opportunistic pathogen add no further concern regarding its QPS status. Consumption of Saccharomyces boulardii (synonym of S. cerevisiae) by patients with fragile health may be considered as the origin of the infection, although the use of microorganisms intended to be used as “probiotic” for humans as a health claim does not fall under the remit of the QPS assessment. These new reports also confirm the previous QPS qualifications, absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain and inability to grow above 37°C. Therefore its QPS status does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Saccharomyces cerevisiae | Feed additive |
EFSA‐Q‐2005‐025 http://www.efsa.europa.eu/en/efsajournal/pub/384 EFSA‐Q‐2005‐234 http://www.efsa.europa.eu/en/efsajournal/pub/385 EFSA‐Q‐2005‐149 http://www.efsa.europa.eu/en/efsajournal/pub/321 EFSA‐Q‐2005‐176 http://www.efsa.europa.eu/en/efsajournal/pub/370 EFSA‐Q‐2006‐003 http://www.efsa.europa.eu/en/efsajournal/pub/379 EFSA‐Q‐2006‐067 http://www.efsa.europa.eu/en/efsajournal/pub/459 EFSA‐Q‐2007‐104 http://www.efsa.europa.eu/en/efsajournal/pub/585 EFSA‐Q‐2007‐139 http://www.efsa.europa.eu/en/efsajournal/pub/772 EFSA‐Q‐2007‐165 http://www.efsa.europa.eu/en/efsajournal/pub/1353 EFSA‐Q‐2008‐009 http://www.efsa.europa.eu/en/efsajournal/pub/991 EFSA‐Q‐2008‐010 http://www.efsa.europa.eu/en/efsajournal/pub/837 EFSA‐Q‐2008‐302 http://www.efsa.europa.eu/en/efsajournal/pub/970 EFSA‐Q‐2008‐472 http://www.efsa.europa.eu/en/efsajournal/pub/1040 EFSA‐Q‐2009‐00720 http://www.efsa.europa.eu/en/efsajournal/pub/1864 EFSA‐Q‐2009‐00753 http://www.efsa.europa.eu/en/efsajournal/pub/1662 EFSA‐Q‐2009‐00818 http://www.efsa.europa.eu/en/efsajournal/pub/2439 EFSA‐Q‐2009‐00824 http://www.efsa.europa.eu/en/efsajournal/pub/1662 EFSA‐Q‐2010‐00936 http://www.efsa.europa.eu/en/efsajournal/pub/2531 EFSA‐Q‐2010‐00938 http://www.efsa.europa.eu/en/efsajournal/pub/3666 EFSA‐Q‐2010‐00992 http://www.efsa.europa.eu/en/efsajournal/pub/2173 EFSA‐Q‐2011‐00390 |
|
| Nutrition/NDA | Saccharomyces cerevisiae | Food targeted for health claims |
EFSA‐Q‐2012‐00271 |
In the framework of the EU Regulation 1924/2006 on health claims made on foods, EFSA is only requested to perform efficacy assessment (i.e. relationship between the food consumption and the claimed beneficial effect). Safety assessment is not foreseen. |
| Feed/FEEDAP | Saccharomyces cerevisiae | Organic selenium source |
EFSA‐Q‐2005‐071 http://www.efsa.europa.eu/en/efsajournal/pub/348 EFSA‐Q‐2005‐117 http://www.efsa.europa.eu/en/efsajournal/pub/430 EFSA‐Q‐2008‐381 http://www.efsa.europa.eu/en/efsajournal/pub/992 EFSA‐Q‐2009‐00524 http://www.efsa.europa.eu/en/efsajournal/pub/2279 EFSA‐Q‐2009‐00752 http://www.efsa.europa.eu/en/efsajournal/pub/2110 EFSA‐Q‐2010‐01029 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.8) with the conclusion: ‘These new reports of Saccharomyces cerevisiae appearing as an opportunistic pathogen add no further concern regarding its QPS status. Consumption of Saccharomyces boulardii (synonym of S. cerevisiae) by patients with fragile health may be considered as the origin of the infection, although the use of microorganisms intended to be used as “probiotic” for humans as a health claim does not fall under the remit of the QPS assessment. These new reports also confirm the previous QPS qualifications, absence of resistance to antimycotics used for medical treatment of yeast infections in cases where viable cells are added to the food or feed chain and inability to grow above 37°C. Therefore its QPS status does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Saccharomyces cerevisiae, GMM strain | Production of enzyme |
EFSA‐Q‐2005‐224 (withdraw) EFSA‐Q‐2009‐00534 http://www.efsa.europa.eu/en/efsajournal/pub/2451 and related application: EFSA‐Q‐2012‐00909 |
|
| Feed/FEEDAP | Saccharomyces cerevisiae | Zootechnical additive | EFSA‐Q‐2016‐00292 | |
| Feed/FEEDAP | Saccharomyces cerevisiae | Zootechnical additive | EFSA‐Q‐2016‐00297 | |
| Feed/FEEDAP | Saccharomyces cerevisiae | Zootechnical additive | EFSA‐Q‐2016‐00298 | |
| Feed/FEEDAP | Saccharomyces cerevisiae | Zootechnical additive | EFSA‐Q‐2016‐00090 | |
| FIP/CEF |
Saccharomyces cerevisiae CBS615‐94, GMM strain |
Production of food enzyme α‐galactosidase |
EFSA‐Q‐2013‐00119 |
|
| Feed/FEEDAP | Saccharomyces cerevisiae CNCM I‐1077 | Zootechnical additive | EFSA‐Q‐2014‐00029 | |
| Feed/FEEDAP | Saccharomyces cerevisiae CNCM I‐1077 | Zootechnical additive | EFSA‐Q‐2014‐00375 | |
| Feed/FEEDAP | Saccharomyces cerevisiae boulardii CNCM I‐1079 | Zootechnical additive | EFSA‐Q‐2015‐00287 | |
| Feed/FEEDAP | Saccharomyces cerevisiae CNCM I‐1079 | Zootechnical additive | EFSA‐Q‐2016‐00449 | |
| Feed/FEEDAP | Saccharomyces cerevisiae CNCM I‐3060 | Production of organic selenium | EFSA‐Q‐2016‐00138 | |
| Feed/FEEDAP | Saccharomyces cerevisiae CNCM I‐3399 | Production of histidine | EFSA‐Q‐2016‐00346 | |
| Pesticides | Saccharomyces cerevisiae strain LA02 | Plant protection product |
EFSA‐Q‐2014‐00333 http://www.efsa.europa.eu/en/efsajournal/pub/4322 Application for approval |
|
| Feed/FEEDAP | Saccharomyces cerevisiae LOCK 0141 | Zootechnical additive |
EFSA‐Q‐2013‐00996 |
|
| Feed/FEEDAP | Saccharomyces cerevisiae (MUCL 39885) | Zootechnical additive |
EFSA‐Q‐2014‐00792 |
|
| FIP/CEF | Saccharomyces cerevisiae/NA | Production of food enzyme | EFSA‐Q‐2015‐00323 | |
| Nutrition/NDA |
Saccharomyces cerevisiae (vitamin D‐enriched UV‐treated) |
As a novel food ingredient |
EFSA‐Q‐2013‐00335 http://www.efsa.europa.eu/en/efsajournal/pub/3520 As a Novel food ingredient in the context of Regulation (EC) No 258/97 |
|
| Feed/FEEDAP | Schizosaccharomyces pombe | Production of enzymes |
EFSA‐Q‐2005‐063 http://www.efsa.europa.eu/en/efsajournal/pub/350 and related questions: EFSA‐Q‐2005‐080 http://www.efsa.europa.eu/en/efsajournal/pub/404 EFSA‐Q‐2008‐272 http://www.efsa.europa.eu/en/efsajournal/pub/915 EFSA‐Q‐2011‐00835 |
Already QPS (EFSA, 2007, 2008; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a). Re‐evaluated in the current Scientific Opinion (refer to Section 3.4.9) with the conclusion: ‘No references related to possible concerns for human or animal safety, AMR or other related aspects were identified. Therefore its QPS status does not change’. Please refer to the complete assessment. |
| Feed/FEEDAP | Schizosaccharomyces pombe | Production of phytase | EFSA‐Q‐2016‐00559 | |
| Feed/FEEDAP | Trichosporon mycotoxinivorans | Feed additive | EFSA‐Q‐2010‐01030 (withdrawn) | Not recommended for the QPS list, assessed in the 2011 update (EFSA BIOHAZ Panel, 2011). |
| Filamentous fungi d | ||||
| Pesticides | Ampelomyces quisqualis strain Q10 | Plant protection product |
EFSA‐Q‐2008‐489 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) Review Report for the active substance Review Report for the active substance Ampelomyces quisqualis, SANCO/4205/VI/98‐ rev.final, October 2004 |
Not recommended for the QPS list (EFSA BIOHAZ Panel, 2011, 2012, 2013)d. |
| Pesticides | Ampelomyces quisqualis strain AQ10 | Plant protection product |
EFSA‐Q‐2015‐00021 Application for renewal of approval |
|
| Feed/FEEDAP | Ashbya gossypii | Production of vitamin B2 | EFSA‐Q‐2012‐00953 | Not recommended for the QPS list (EFSA BIOHAZ Panel, 2011, 2012, 2013). |
| FIP/CEF | Aspergillus acidus/ RF7398, GMM strain | Production of food enzyme is a endo 1,4‐β‐xylanase | EFSA‐Q‐2014‐00163 | Not recommended for the QPS listd. |
| Feed/FEEDAP | Aspergillus aculeatus | Production of enzyme |
EFSA‐Q‐2008‐432 http://www.efsa.europa.eu/en/efsajournal/pub/1186 EFSA‐Q‐2011‐00035 http://www.efsa.europa.eu/en/efsajournal/pub/2010 EFSA‐Q‐2010‐01297 http://www.efsa.europa.eu/en/efsajournal/pub/4234 EFSA‐Q‐2010‐01295 |
Potential for mycotoxin production, therefore not suitable for QPS status (see EFSA 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| FIP/CEF | Aspergillus aculeatus/ NZYM‐RE CBS 589.94 | Production of food enzyme polygalacturonase and β‐glucanase |
EFSA‐Q‐2014‐00200 EFSA‐Q‐2014‐00201 |
|
| FIP/CEF | Aspergillus fijiensis | Production of food enzyme β‐fructofuranosidase | EFSA‐Q‐2015‐00840 | Not recommended for the QPS listd. |
| FIP/CEF | Aspergillus melleus/AE‐DN | Production of food enzyme AMP deaminase | EFSA‐Q‐2014‐00326 | Not recommended for the QPS listd. |
| Feed/FEEDAP | Aspergillus niger, GMM strain | Production of enzyme |
EFSA‐Q‐2004‐068 http://www.efsa.europa.eu/en/efsajournal/pub/198 and related opinions: EFSA‐Q‐2006‐119 http://www.efsa.europa.eu/en/efsajournal/pub/474 EFSA‐Q‐2008‐418 http://www.efsa.europa.eu/en/efsajournal/pub/1155 EFSA‐Q‐2011‐00147 http://www.efsa.europa.eu/en/efsajournal/pub/2575 EFSA‐Q‐2005‐116 http://www.efsa.europa.eu/en/efsajournal/pub/369 and related opinions: EFSA‐Q‐2007‐049 http://www.efsa.europa.eu/en/efsajournal/pub/472 EFSA‐Q‐2007‐041 http://www.efsa.europa.eu/en/efsajournal/pub/544 EFSA‐Q‐2007‐189 http://www.efsa.europa.eu/en/efsajournal/pub/614 EFSA‐Q‐2008‐692 http://www.efsa.europa.eu/en/efsajournal/pub/1184 EFSA‐Q‐2009‐00603 http://www.efsa.europa.eu/en/efsajournal/pub/1427 EFSA‐Q‐2009‐00534 http://www.efsa.europa.eu/en/efsajournal/pub/2451 and related application: EFSA‐Q‐2012‐00909 EFSA‐Q‐2009‐00585 http://www.efsa.europa.eu/en/efsajournal/pub/3322 EFSA‐Q‐2008‐013 http://www.efsa.europa.eu/en/efsajournal/pub/914 and related Questions: EFSA‐Q‐2010‐00937 http://www.efsa.europa.eu/en/efsajournal/pub/2172 EFSA‐Q‐2011‐00061 http://www.efsa.europa.eu/en/efsajournal/pub/3285 EFSA‐Q‐2010‐01519 http://www.efsa.europa.eu/en/efsajournal/pub/3430 EFSA‐Q‐2012‐00411 |
Potential for mycotoxin production, therefore not suitable for QPS status (see EFSA 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Feed/FEEDAP | Aspergillus niger | Production of enzyme |
EFSA‐Q‐2008‐013a http://www.efsa.europa.eu/en/efsajournal/pub/914 and related Questions: EFSA‐Q‐2010‐00937 http://www.efsa.europa.eu/en/efsajournal/pub/2172 EFSA‐Q‐2011‐00061 http://www.efsa.europa.eu/en/efsajournal/pub/3285 EFSA‐Q‐2010‐01519 http://www.efsa.europa.eu/en/efsajournal/pub/3430 EFSA‐Q‐2012‐00411 |
|
| Feed/FEEDAP | Aspergillus niger | Production of enzyme | EFSA‐Q‐2014‐00503 | |
| Feed/FEEDAP | Aspergillus niger | Production of enzyme | EFSA‐Q‐2014‐0057 | |
| FIP/CEF | Aspergillus niger, GMM strain | Production of food enzyme carboxypeptidase C | EFSA‐Q‐2015‐00445 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme catalase | EFSA‐Q‐2015‐00449 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme cellulase, glucanase and hemicellulase covering xylanase and mannanase | EFSA‐Q‐2015‐00340 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme glucoamylase | EFSA‐Q‐2015‐00292 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme glucose oxidase and catalase | EFSA‐Q‐2013‐01018 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme inulinase | EFSA‐Q‐2015‐00827 | |
| FIP/CEF | Aspergillus niger, GMM strain | Production of food enzyme lipase | EFSA‐Q‐2015‐00561 | |
| FIP/CEF | Aspergillus niger, GMM strain | Production of food enzyme pectin lyase | EFSA‐Q‐2015‐00407 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme pectinase (polygalacturonase, pectinesterase, pectin lyase, arabanase) |
EFSA‐Q‐2015‐00038/ EFSA‐Q‐2015‐00039/ EFSA‐Q‐2015‐00040/ EFSA‐Q‐2015‐00041/ EFSA‐Q‐2015‐00042 |
|
| FIP/CEF | Aspergillus niger, GMM strain | Production of food enzyme peroxidase | EFSA‐Q‐2015‐00274 | |
| FIP/CEF | Aspergillus niger | Production of food enzyme tannase | EFSA‐Q‐2016‐00034 | |
| FIP/CEF | Aspergillus niger GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2016‐00099 | |
| FIP/CEF | Aspergillus niger GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2015‐00276 | |
| Feed/FEEDAP | Aspergillus niger | Feed additive | ||
| Feed/FEEDAP | Aspergillus niger | Zootechnical additive (production of enzymes) | EFSA‐Q‐2015‐00054 | |
| FIP/CEF | Aspergillus niger agg., GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2015‐00447 | |
| FIP/CEF | Aspergillus niger, (AE‐TGU) | Production of food enzyme α‐glucosidase | EFSA‐Q‐2014‐00800 | |
| FIP/CEF | Aspergillus niger/AGN, GMM strain | Production of food enzyme asparaginase | EFSA‐Q‐2014‐00401 | |
| FIP/CEF | Aspergillus niger (ARF) | Production of food enzyme α‐ l‐arabinofuranosidase | EFSA‐Q‐2014‐00671 | |
| FIP/CEF | Aspergillus niger (ASNSC) | Production of food enzyme pectinase | EFSA‐Q‐2014‐00839 | |
| FIP/CEF | Aspergillus niger (ASNSC) | Production of food enzyme polygalacturonase | EFSA‐Q‐2014‐00840 | |
| FIP/CEF | Aspergillus niger (ASNSC) | Production of food enzyme pectinesterase | EFSA‐Q‐2014‐00841 | |
| FIP/CEF | Aspergillus niger (ASNSC) | Production of food enzyme pectin lyase | EFSA‐Q‐2014‐00842 | |
| FIP/CEF | Aspergillus niger (ASNSC) | Production of food enzyme arabanase | EFSA‐Q‐2014‐00843 | |
| Feed/FEEDAP | Aspergillus niger (CBS 18404) | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2013‐00886 http://www.efsa.europa.eu/en/efsajournal/pub/3723 EFSA‐Q‐2014‐00291 |
|
| Feed/FEEDAP | Aspergillus niger CBS 101.672) | Preparation of 6‐phytase | EFSA‐Q‐2015‐00732 | |
| Feed/FEEDAP | Aspergillus niger (CBS 109.713) | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2013‐00886 http://www.efsa.europa.eu/en/efsajournal/pub/3723 EFSA‐Q‐2014‐00291 |
|
| Feed/FEEDAP | Aspergillus niger (CBS 109.713) | Preparation of endo‐1,4‐β‐xylanase and endo‐1,4‐β‐glucanase |
EFSA‐Q‐2016‐00302 |
|
| FIP/CEF | Aspergillus niger/DS53180, GMM strain | Production of food enzyme asparaginase | EFSA‐Q‐2013‐00895 | |
| FIP/CEF | Aspergillus niger/EPG, GMM strain | Production of food enzyme polygalacturonase | EFSA‐Q‐2014‐00402 | |
| FIP/CEF | Aspergillus niger/EPG | Production of food enzyme | EFSA‐Q‐2015‐00178 | |
| FIP/CEF | Aspergillus niger/FLOSC | Production of food enzyme | EFSA‐Q‐2015‐00130 | |
| FIP/CEF | Aspergillus niger (FLYSC), GMM strain | Production of food enzyme polygalactunase | EFSA‐Q‐2015‐00086 | |
| FIP/CEF | Aspergillus niger (FLZSC), GMM strain | Production of food enzyme polygalactunase | EFSA‐Q‐2015‐00087 | |
| FIP/CEF | Aspergillus niger (GEP), GMM strain | Production of food enzyme acid prolyl endopeptidase | EFSA‐Q‐2014‐00852 | |
| FIP/CEF | Aspergillus niger/ LFS, GMM strain | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2014‐00325 | |
| FIP/CEF | Aspergillus niger var. Macrosporus | Production of food enzyme aspergillolisin I and II | EFSA‐Q‐2015‐00623 | |
| Feed/FEEDAP | Aspergillus niger MUCL 39199 | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00229 |
|
| FIP/CEF | Aspergillus niger (PLA), GMM strain | Production of food enzyme phospholipase A2 | EFSA‐Q‐2015‐00043 | |
| FIP/CEF | Aspergillus niger (PME), GMM strain | Production of food enzyme pectinesterase | EFSA‐Q‐2015‐00044 | |
| Feed/FEEDAP | Aspergillus niger NRRL 25541 | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00503 EFSA‐Q‐2014‐00504 |
|
| FIP/CEF | Aspergillus niger/NZYM‐AK | Production of food enzyme | EFSA‐Q‐2015‐00128 | |
| FIP/CEF | Aspergillus niger/NZYM‐BE, GMM strain | Production of food enzyme glucoamylase | EFSA‐Q‐2013‐00896 | |
| FIP/CEF | Aspergillus niger/NZYM‐BF, GMM strain | Production of food enzyme glucoamylase | EFSA‐Q‐2014‐00307 | |
| FIP/CEF | Aspergillus niger/NZYM‐BR, GMM strain | Production of food enzyme amyloglucosidase | EFSA‐Q‐2013‐00686 | |
| FIP/CEF | Aspergillus niger/NZYM‐BX, GMM strain | Production of food enzyme glucan 1,4‐α‐glucosidase with activity also of an α‐amylase | EFSA‐Q‐2013‐00877 | |
| FIP/CEF | Aspergillus niger/NZYM‐DB | Production of food enzyme | EFSA‐Q‐2015‐00234 | |
| FIP/CEF | Aspergillus niger (strain NZYM‐KA) | Production of food enzyme glucose oxidase | EFSA‐Q‐2016‐00134 | |
| FIP/CEF | Aspergillus niger (NZYM‐LP), GMM strain | Production of food enzyme lysophospholipase | EFSA‐Q‐2014‐00919 | |
| FIP/CEF | Aspergillus niger/NZYM‐MC, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00306 | |
| FIP/CEF | Aspergillus niger/NZYM‐PN | Production of food enzyme | EFSA‐Q‐2015‐00407 | |
| FIP/CEF | Aspergillus niger/NZYM‐SB, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00413 | |
| FIP/CEF | Aspergillus niger (TOL), GMM strain | Production of food enzyme β‐galactosidase | EFSA‐Q‐2014‐00853 | |
| FIP/CEF | Aspergillus niger (XEA), GMM strain | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2015‐00045 | |
| FIP/CEF | Aspergillus niger/XYL, GMM strain | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2014‐00305 | |
| FIP/CEF | Aspergillus niger/ZGL | Production of food enzyme glucose oxidase | EFSA‐Q‐2013‐01005 | |
| Feed/FEEDAP |
Aspergillus niger Strains: ZLCA0323 Van Tieghem ZS9 TN‐A09 |
Production of citric acid | EFSA‐Q‐2013‐00612 | |
| Feed/FEEDAP | Aspergillus oryzae | Production of enzymes |
EFSA‐Q‐2003‐012 http://www.efsa.europa.eu/en/efsajournal/pub/66 and related opinions: EFSA‐Q‐2004‐070 http://www.efsa.europa.eu/en/efsajournal/pub/88 EFSA‐Q‐2004‐118 http://www.efsa.europa.eu/en/efsajournal/pub/132 EFSA‐Q‐2006‐060 http://www.efsa.europa.eu/en/efsajournal/pub/519 EFSA‐Q‐2007‐132 http://www.efsa.europa.eu/en/efsajournal/pub/1862 EFSA‐Q‐2009‐00535 http://www.efsa.europa.eu/en/efsajournal/pub/1915 EFSA‐Q‐2007‐133 http://www.efsa.europa.eu/en/efsajournal/pub/871 and related opinions: EFSA‐Q‐2008‐430 http://www.efsa.europa.eu/en/efsajournal/pub/1097.htm EFSA‐Q‐2009‐00536 http://www.efsa.europa.eu/en/efsajournal/pub/1634 EFSA‐Q‐2008‐419 http://www.efsa.europa.eu/en/efsajournal/pub/2790 EFSA‐Q‐2010‐00769 http://www.efsa.europa.eu/en/efsajournal/pub/2527 and related opinion: EFSA‐Q‐2011‐01172 http://www.efsa.europa.eu/en/efsajournal/pub/2730 EFSA‐Q‐2010‐01519 |
Potential for mycotoxin production, therefore not suitable for QPS status (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Feed/FEEDAP | Aspergillus oryzae | Feed additive |
EFSA‐Q‐2009‐00525 |
|
| FIP/CEF | Aspergillus oryzae/ AE‐MB | Production of food enzymes leucyl aminopeptidase, protease and amylase | EFSA‐Q‐2014‐00114 | Not recommended for the QPS listd. |
| Feed/FEEDAP | Aspergillus oryzae | Production of enzyme | EFSA‐Q‐2014‐00503 | |
| Feed/FEEDAP | Aspergillus oryzae | Technological additive | EFSA‐Q‐2016‐00220 | |
| FIP/CEF | Aspergillus oryzae, GMM strain | Production of food enzyme inulinase | EFSA‐Q‐2015‐00337 | |
| FIP/CEF | Aspergillus oryzae, GMM strain | Production of food enzyme lipase | EFSA‐Q‐2015‐00664 | |
| FIP/CEF | Aspergillus oryzae | Production of food enzyme β‐galactosidase | EFSA‐Q‐2015‐00684 | |
| FIP/CEF | Aspergillus oryzae | Production of food enzyme AMP deaminase | EFSA‐Q‐2015‐00847 | |
| FIP/CEF | Aspergillus oryzae (AE‐AA) | Production of food enzyme α‐amylase | EFSA‐Q‐2014‐00913 | |
| FIP/CEF | Aspergillus oryzae/ AE‐MB | Production of food enzymes leucyl aminopeptidase, protease and amylase | EFSA‐Q‐2014‐00114 | Not recommended for the QPS listd. |
| FIP/CEF | Aspergillus oryzae/ AE‐TL | Production of food enzymes triacylglycerol lipase and transesterase | EFSA‐Q‐2014‐00112 | |
| Feed/FEEDAP |
Aspergillus oryzae DSM 17594, GMM strain |
Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00450 |
|
| Feed/FEEDAP |
Aspergillus oryzae DSM 22594 |
Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00289 |
|
| Feed/FEEDAP | Aspergillus oryzae DSM 23104 | Zootechnical additives | EFSA‐Q‐2016‐00215 | |
| Feed/FEEDAP |
Aspergillus oryzae DSM 26372, GMM strain |
Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00447 |
|
| FIP/CEF | Aspergillus oryzae (strain L729‐48) | Production of food enzyme α‐amylase | EFSA‐Q‐2016‐00205 | |
| FIP/CEF | Aspergillus oryzae (strains NBRC 110971 and 11‐5) | Production of food enzyme tannase | EFSA‐Q‐2016‐00272 | |
| Feed/FEEDAP |
Aspergillus oryzae NRRL 66222 |
Zootechnical additive (production of enzyme) | EFSA‐Q‐2014‐00503 | |
| FIP/CEF | Aspergillus oryzae/ NZYM‐AL, GMM strain | Production of food enzyme lipase |
EFSA‐Q‐2013‐00198 |
|
| FIP/CEF | Aspergillus oryzae/ NZYM‐EX | Production of food enzyme | EFSA‐Q‐2015‐00373 | |
| FIP/CEF | Aspergillus oryzae/ NZYM‐FA, GMM strain | Production of food enzyme xylanase | EFSA‐Q‐2013‐00789 | |
| FIP/CEF |
Aspergillus oryzae strain NZYM‐FB |
Production of food enzyme xylanase |
EFSA‐Q‐2012‐00897 |
|
| FIP/CEF | Aspergillus oryzae/ NZYM‐FL, GMM strain | Production of food enzyme lipase |
EFSA‐Q‐2013‐00197 |
|
| FIP/CEF | Aspergillus oryzae/ NZYM‐KE, GMM strain | Production of food enzyme xylanase |
EFSA‐Q‐2012‐00897 |
|
| FIP/CEF | Aspergillus oryzae/ NZYM‐KP, GMM strain | Production of food enzyme glucose oxidase | EFSA‐Q‐2013‐00687 | |
| FIP/CEF | Aspergillus oryzae/ NZYM‐LH, GMM strain | Production of food enzyme lipase |
EFSA‐Q‐2012‐01009 |
|
| FIP/CEF | Aspergillus oryzae/ NZYM‐NA | Production of food enzyme α‐amylase | EFSA‐Q‐2012‐01010 | |
| FIP/CEF | Aspergillus oryzae (NZYM‐OA), GMM strain | Production of food enzyme phospholipase A1 | EFSA‐Q‐2015‐00063 | |
| FIP/CEF | Aspergillus oryzae (NZYM‐PP), GMM strain | Production of food enzyme phospholipase | EFSA‐Q‐2014‐00921 | |
| FIP/CEF | Aspergillus oryzae (strains NBRC 110971 and 11‐5) | Production of food enzyme tannase | EFSA‐Q‐2016‐00272 | |
| Feed/FEEDAP | Aspergillus oryzae DSM 23104 | Zootechnical additives | EFSA‐Q‐2016‐00215 | |
| FIP/CEF | Aspergillus oryzae/NZYM‐SP, GMM strain | Production of food enzyme asparaginase | EFSA‐Q‐2013‐00587 | |
| Pesticides | Beauveria bassiana strain 147 | Plant protection product |
EFSA‐Q‐2014‐00324 http://www.efsa.europa.eu/en/efsajournal/pub/4261 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Beauveria bassiana (ATCC‐74040 and GHA) | Plant protection product |
EFSA‐Q‐2009‐00125 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00251 and EFSA‐Q‐2009‐00252 http://www.efsa.europa.eu/en/efsajournal/pub/3031 Application for approval |
|
| Pesticides | Beauveria bassiana strain IMI389521 | Plant protection product |
EFSA‐Q‐2015‐00362 Application for approval |
|
| Pesticides | Beauveria bassiana strain NPP111B005 | Plant protection product |
EFSA‐Q‐2014‐00327 http://www.efsa.europa.eu/en/efsajournal/pub/4264 Application for approval |
|
| Pesticides | Beauveria bassiana strain PPRI5339 | Plant protection product |
EFSA‐Q‐2015‐00361 Application for approval |
|
| Pesticides | Beauveria brongniartii | Plant protection product |
EFSA‐Q‐2009‐00017 Review of MRLs (Maximum Residue Limits) |
Mycelial fungi: already considered as not appropriate for QPS. Insufficient body of knowledge, potential oosporein formation (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| AFC (as mentioned in EFSA register of questions) | Blakeslea trispora |
Production of lycopene (food colourant) Production of b‐carotene (food colourant) |
EFSA‐Q‐2004‐102 http://www.efsa.europa.eu/en/efsajournal/pub/275 EFSA‐Q‐2007‐001 |
Cannot be proposed for QPS status (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| NDA | Blakeslea trispora | Food ingredient |
EFSA‐Q‐2004‐169 http://www.efsa.europa.eu/en/efsajournal/pub/212 EFSA‐Q‐2008‐697 |
|
| Feed/FEEDAP | Blakeslea trispora | Production strain forβ‐carotene |
EFSA‐Q‐2009‐00884 |
|
| FIP/CEF | Chaetomium erraticum | Production of food enzyme dextranase | EFSA‐Q‐2015‐00685 | Not recommended for the QPS listd. |
| FIP/CEF | Chaetomium gracile | Production of food enzyme dextranase | EFSA‐Q‐2015‐00231 | Not recommended for the QPS listd. |
| Pesticides | Coniothyrium minitans | Plant protection product |
EFSA‐Q‐2008‐515 http://www.efsa.europa.eu/en/efsajourefs/pub/4458 Review of MRLs (Maximum Residue Limits) Coniotyrium minitans, SANCO/1400/ 2001‐final, July 2003 |
The body of knowledge is insufficient. Potential acrosphelide formation (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Coniothyrium minitans CON/M/91‐08 | Plant protection product |
EFSA‐Q‐2014‐00656 http://www.efsa.europa.eu/en/efsajournal/pub/4517 Application for renewal of the approval |
|
| FIP/CEF | Disporotrichum dimorphosporum/ DXL | Production of food enzymes endo‐1,4‐β‐xylanase and β‐glucanase |
EFSA‐Q‐2014‐00355 EFSA‐Q‐2014‐00356 |
Not recommended for the QPS listd. |
| Feed/FEEDAP |
Duddingtonia flagrans Alternative name: Trichothecium flagrans |
Feed additive |
EFSA‐Q‐2004‐115 http://www.efsa.europa.eu/en/efsajournal/pub/334 EFSA‐Q‐2005‐051 (withdrawn) |
Insufficient body of knowledge (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Fusarium spp., strain L 13 | Plant protection product |
EFSA‐Q‐2015‐00345 Application for approval |
Not recommended for the QPS listd. |
| FIP/CEF | Fusarium venenatum, GMM strain | Production of food enzyme trypsin | EFSA‐Q‐2014‐00412 | Not recommended for the QPS listd. |
| Pesticides |
Gliocladium catenulatum = Clonostachys rosea forma catenulata, strain J1446 |
Plant protection product |
EFSA‐Q‐2008‐559 Review of MRLs (Maximum Residue Limits) Gliocladium catenulatum, SANCO/10383/2004‐rev.4, October 2004 |
No recommendation for QPS in 2009 (EFSA BIOHAZ Panel, 2009). No new relevant information in the 2010 and 2013 updatesd. |
| Pesticides | Gliocladium catenulatum, strain J1446 | Plant protection product |
EFSA‐Q‐2015‐00582 Application for renewal of approval |
|
| FIP/CEF | Humicola isolens (NZYM‐ST) | Production of food enzyme β‐glucanase | EFSA‐Q‐2014‐00795 | Not recommended for the QPS listd. |
| FIP/CEF | Humicola isolens (NZYM‐ST) | Production of food enzyme xylanase | EFSA‐Q‐2014‐00796 | |
| FIP/CEF | Humicola isolens (NZYM‐ST) | Production of food enzyme cellulose | EFSA‐Q‐2014‐00797 | |
| Pesticides | Isaria fumosorosea, strain Apopka 97 | Plant protection product |
EFSA‐Q‐2013‐00833 http://www.efsa.europa.eu/en/efsajournal/pub/3679 Application for renewal of the approval. |
It has been formerly evaluated as Paecilomyces fumosoroseus (DG SANCO, 4203/VI/98‐final) and approved in 2001. Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2013)d. |
| Pesticides | Lecanicillium muscarium formely Verticillium lecanii, strain Ve6 | Plant protection product |
EFSA‐Q‐2009‐00130 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00255 http://www.efsa.europa.eu/en/efsajournal/pub/1446 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| FIP/CEF | Leptographium procerum | Production of food enzyme phosphodiesterase | EFSA‐Q‐2013‐01006 | Not recommended for the QPS listd. |
| Pesticides | Metarhizium anisopliae var. Anisopliae (BIPESCO 5/F52) formerly M. anisopliae | Plant protection product |
EFSA‐Q‐2009‐00131 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00253 http://www.efsa.europa.eu/en/efsajournal/pub/2498 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| FIP/CEF | Mucor javanicus | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2015‐00692 | Not recommended for the QPS listd. |
| Pesticides |
Paecilomyces fumosoroseus, strain Apopka 97, PFR 97 or CG170,ATCC20874 Current name: Isaria fumosorosea |
Plant protection product |
EFSA‐Q‐2008‐599 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2013‐00833 http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2014.3679/full Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2013)d. |
| Pesticides |
Paecilomyces fumosoroseus, strain FE 9901 (ARSEF 4490) Current name: Isaria fumosorosea |
Plant protection product |
EFSA‐Q‐2013‐00352 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00323 http://www.efsa.europa.eu/en/efsajournal/pub/2869 Application for approval |
|
| Pesticides | Paecilomyces lilacinus, strain 251 | Plant protection product |
EFSA‐Q‐2008‐600 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2010‐01337 http://www.efsa.europa.eu/en/efsajournal/pub/103r Application for approval |
Mycelial fungi: already considered as not appropriate for QPS. Potential for production of peptaibols (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Paecilomyces lilacinus, strain 251 | Plant protection product |
EFSA‐Q‐2015‐00520 Application for renewal of approval (AIR III) |
|
| FIP/CEF | Penicillium camemberti (AE‐LG) | Production of food enzyme acylglycerol lipase | EFSA‐Q‐2014‐00668 | Not recommended for the QPS listd. |
| FIP/CEF | Penicillium citrinum/AE‐RP | Production of food enzyme ribonuclease P | EFSA‐Q‐2015‐00288 | Not recommended for the QPS listd. |
| FIP/CEF | Penicillium citrinum | Production of food enzyme aspergillus nuclease S1 | EFSA‐Q‐2015‐00845 | |
| FIP/CEF | Penicillium decumbes | Production of food enzyme α‐l‐rhamnosidase | EFSA‐Q‐2015‐00756 | Not recommended for the QPS listd. |
| Feed/FEEDAP | Penicillium funiculosum | Production of enzyme |
EFSA‐Q‐2005‐281 http://www.efsa.europa.eu/en/efsajournal/pub/471 EFSA‐Q‐2010‐01287 http://www.efsa.europa.eu/en/efsajournal/pub/3321 EFSA‐Q‐2011‐00881 |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| FIP/CEF | Penicillium funiculosum (DP‐Lzc35) | Production of food enzyme cellulase | EFSA‐Q‐2016‐00098 | |
| Feed/FEEDAP | Penicillium funiculosum (Talaromyces versatilis sp. nov. DSM 26702), GMM strain | Zootechnical additive |
EFSA‐Q‐2013‐00750 http://www.efsa.europa.eu/en/efsajournal/pub/3793 EFSA‐Q‐2014‐00463 |
|
| Feed/FEEDAP | Penicillium funiculosum (Talaromyces versatilis IMI 378536) | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2013‐00750 http://www.efsa.europa.eu/en/efsajournal/pub/3793 EFSA‐Q‐2014‐00463 |
|
| Feed/FEEDAP | Penicillium funiculosum (Talaromyces versatilis sp. nov.) | Preparation of endo‐1,4‐β‐xylanase (EC 3.2.1.8) and endo‐1,3(4)‐β‐glucanase (EC 3.2.1.6) |
EFSA‐Q‐2015‐00615 |
|
| FIP/CEF | Penicillium multicolour | Production of food enzyme β‐glucosidase | EFSA‐Q‐2015‐00273 | Not recommended for the QPS listd. |
| FIP/CEF | Penicillium roqueforti AE‐LRF | Production of food enzymes triacylglycerol lipase | EFSA‐Q‐2014‐00545 | Not recommended for the QPS listd. |
| Pesticides | Phlebiopsis gigantea (14 different strains) | Plant protection product |
EFSA‐Q‐2009‐00132 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00285 http://www.efsa.europa.eu/en/efsajournal/pub/3033 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS. Insufficient body of knowledge (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Pseudozyma flocculosa, strain ATCC 64874 | Plant protection product |
EFSA‐Q‐2009‐00315 http://www.efsa.europa.eu/en/efsajournal/pub/4250 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Purpureocillium lilacinum, strain PL 11 | Plant protection product |
EFSA‐Q‐2015‐00176 Application for approval |
Not recommended for the QPS listd. |
| FIP/CEF | Rhizomucor miehei | Production of food enzyme mucorpepsin | EFSA‐Q‐2015‐00233 | Not recommended for the QPS listd. |
| FIP/CEF | Rhizomucor miehei | Production of food enzyme mucorpepsin | EFSA‐Q‐2016‐00030 | |
| FIP/CEF | Rhizomucor miehei (strain 29547) | Production of food enzyme mucorpepsin | EFSA‐Q‐2015‐00761 | |
| FIP/CEF | Rhizomucor oryze | Production of food enzyme glucoamylase | EFSA‐Q‐2015‐00272 | Not recommended for the QPS listd. |
| FIP/CEF | Rhizopus miehei | Production of food enzyme microbial rennet | EFSA‐Q‐2014‐00851 | Not recommended for the QPS listd. |
| FIP/CEF | Rhizopus niveus (strain AE‐N) | Production of food enzyme triacylglycerol lipase | EFSA‐Q‐2014‐00732 | Not recommended for the QPS listd. |
| FIP/CEF | Rhizopus niveus/ AE‐N | Production of food enzyme rishopuspepsin | EFSA‐Q‐2015‐00452 | |
| FIP/CEF | Rhizopus oryzae/ AE‐PER | Production of food enzymes leucyl aminopeptidase | EFSA‐Q‐2014‐00354 | Not recommended for the QPS listd. |
| FIP/CEF | Talaromyces cellulolyticus | Production of food enzyme cellulase | EFSA‐Q‐2015‐00370 | Not recommended for the QPS listd. |
| FIP/CEF | Talaromyces emersonii | Production of food enzyme is endo‐1,3(4)‐β‐glucanase | EFSA‐Q‐2014‐00801 | Not recommended for the QPS listd. |
| FIP/CEF | Talaromyces emersonii | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2014‐00802 | |
| FIP/CEF | Talaromyces emersonii | Production of food enzyme cellulase | EFSA‐Q‐2014‐00803 | |
| FIP/CEF | Talaromyces versatilis | Production of food enzyme endo‐1,4‐β‐glucanase | EFSA‐Q‐2015‐00663 | Not recommended for the QPS listd. |
| FIP/CEF | Trametes hirsuta | Production of food enzyme laccase | EFSA‐Q‐2015‐00694 | Not recommended for the QPS listd. |
| Pesticides |
Trichoderma asperellum, strains ICC 012, T25 and TV1 FormerlyTrichoderma viride T25 and TV1 |
Plant protection product |
EFSA‐Q‐2009‐00136 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00300 http://www.efsa.europa.eu/en/efsajournal/pub/3036 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Trichoderma asperellum, strain T‐34 | Plant protection product |
EFSA‐Q‐2011‐00899 http://www.efsa.europa.eu/en/efsajournal/pub/2666 Application for approval EFSA‐Q‐2013‐00013 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Trichoderma atroviride I‐1237 | Plant protection product |
EFSA‐Q‐2011‐00900 http://www.efsa.europa.eu/en/efsajournal/pub/2706 Application for approval EFSA‐Q‐2013‐00039 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Trichoderma atroviride IMI 206040 and T11 | Plant protection product |
EFSA‐Q‐2009‐00137 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00297 http://www.efsa.europa.eu/en/efsajournal/pub/3056 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Pesticides | Trichoderma atroviride SC1 | Plant protection product |
EFSA‐Q‐2014‐00334 http://www.efsa.europa.eu/en/efsajournal/pub/4092 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Feed/FEEDAP | Trichoderma citrinoviride | Production of enzyme |
EFSA‐Q‐2010‐00036 |
This was submitted as Trichoderma longibrachiatum but the assessment revealed that should be classified differently. Not recommended for the QPS listd. |
| Feed/FEEDAP | Trichoderma citrinoviride | Production of enzyme | EFSA‐Q‐2014‐00575 | |
| Feed/FEEDAP | Trichoderma citrinoviride (IMI SD 135) | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2013‐00809 |
|
| Feed/FEEDAP | Trichoderma citrinoviride (IMI SD 142) | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00297 |
|
| FIP/CEF | Trichoderma citrinoviride/TCLSC | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2014‐00543 | |
| Pesticides |
Trichoderma gamsii, strain ICC 080, formerly Trichoderma viride ICC080 |
Plant protection product |
EFSA‐Q‐2009‐00138 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2012‐00424 http://www.efsa.europa.eu/en/efsajournal/pub/3062 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d |
| Pesticides | Trichoderma harzianum, Rifai strains T22 and ITEM 908 | Plant protection product |
EFSA‐Q‐2009‐00139 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00298 http://www.efsa.europa.eu/en/efsajournal/pub/3055 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Feed/FEEDAP | Trichoderma koningii | Production of enzyme |
EFSA‐Q‐2008‐288 |
This was submitted as Trichoderma longibrachiatum but the assessment revealed that should be classified as koningii. New assessment for QPS 2013 updated. |
| Feed/FEEDAP | Trichoderma longibrachiatum | Feed additive | Ineligible for QPS status (see EFSA 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. | |
| Feed/FEEDAP | Trichoderma longibrachiatum | Production of enzyme |
EFSA‐Q‐2005‐276 http://www.efsa.europa.eu/en/efsajournal/pub/405 and related opinion: EFSA‐Q‐2006‐320 http://www.efsa.europa.eu/en/efsajournal/pub/520 EFSA‐Q‐2010‐01532 http://www.efsa.europa.eu/en/efsajournal/pub/3528 EFSA‐Q‐2008‐288 http://www.efsa.europa.eu/en/efsajournal/pub/2843 EFSA‐Q‐2010‐00036 http://www.efsa.europa.eu/en/efsajournal/pub/3105 EFSA‐Q‐2010‐01025 http://www.efsa.europa.eu/en/efsajournal/pub/3207 EFSA‐Q‐2010‐01295 http://www.efsa.europa.eu/en/efsajournal/pub/4235 EFSA‐Q‐2010‐01297 http://www.efsa.europa.eu/en/efsajournal/pub/4234 EFSA‐Q‐2012‐00411 |
|
| Feed/FEEDAP | Trichoderma longibrachiatum MUCL 39203 | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2014‐00228 |
|
| Feed/FEEDAP | Trichoderma longibrachiatum SD 135 | Preparation of endo‐1,4‐beta xylanase | EFSA‐Q‐2015‐00834 | |
| Pesticides | Trichoderma polysporum, strain IMI 206039 | Plant protection product |
EFSA‐Q‐2009‐00140 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00299 http://www.efsa.europa.eu/en/efsajournal/pub/3035 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Feed/FEEDAP | Trichoderma reesei | Production of enzyme |
EFSA‐Q‐2006‐137 http://www.efsa.europa.eu/en/efsajournal/pub/548 and related opinions: EFSA‐Q‐2007‐0020 http://www.efsa.europa.eu/en/efsajournal/pub/1156 EFSA‐Q‐2007‐109 http://www.efsa.europa.eu/en/efsajournal/pub/586 EFSA‐Q‐2007‐112 http://www.efsa.europa.eu/en/efsajournal/pub/1154 EFSA‐Q‐2007‐185 http://www.efsa.europa.eu/en/efsajournal/pub/2930 EFSA‐Q‐2009‐00470 http://www.efsa.europa.eu/en/efsajournal/pub/1949 EFSA‐Q‐2010‐00141 http://www.efsa.europa.eu/en/efsajournal/pub/1916 EFSA‐Q‐2009‐00802 http://www.efsa.europa.eu/en/efsajournal/pub/2008 EFSA‐Q‐2011‐01171 http://www.efsa.europa.eu/en/efsajournal/pub/2739 EFSA‐Q‐2007‐120 http://www.efsa.europa.eu/en/efsajournal/pub/712 and related question: EFSA‐Q‐2010‐00142 http://www.efsa.europa.eu/en/efsajournal/pub/2277 EFSA‐Q‐2012‐00065 http://www.efsa.europa.eu/en/efsajournal/pub/3432 EFSA‐Q‐2012‐00693 http://www.efsa.europa.eu/en/efsajournal/pub/3364 EFSA‐Q‐2012‐00905 |
Ineligible for QPS status (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d |
| Feed/FEEDAP | Trichoderma reesei | Production of enzyme |
EFSA‐Q‐2008‐308 http://www.efsa.europa.eu/en/efsajournal/pub/1094 and related questions: EFSA‐Q‐2010‐00018 http://www.efsa.europa.eu/en/efsajournal/pub/2278 EFSA‐Q‐2008‐432 http://www.efsa.europa.eu/en/efsajournal/pub/1186 EFSA‐Q‐2008‐748 http://www.efsa.europa.eu/en/efsajournal/pub/1380 and related opinion: EFSA‐Q‐2010‐0069 http://www.efsa.europa.eu/en/efsajournal/pub/1553 EFSA‐Q‐2010‐00141 http://www.efsa.europa.eu/en/efsajournal/pub/1916 EFSA‐Q‐2011‐00112 http://www.efsa.europa.eu/en/efsajournal/pub/2111 EFSA‐Q‐2010‐00700 http://www.efsa.europa.eu/en/efsajournal/pub/1919 EFSA‐Q‐2011‐00035 http://www.efsa.europa.eu/en/efsajournal/pub/2010 EFSA‐Q‐2011‐00804 http://www.efsa.europa.eu/en/efsajournal/pub/2728 EFSA‐Q‐2012‐00668 http://www.efsa.europa.eu/en/efsajournal/pub/3172 EFSA‐Q‐2012‐00727 http://www.efsa.europa.eu/en/efsajournal/pub/3171 EFSA‐Q‐2012‐00085 |
|
| Feed/FEEDAP | Trichoderma reesei | Production of enzyme |
EFSA‐Q‐2014‐00574 |
|
| Feed/FEEDAP | Trichoderma reesei | Production of enzyme | EFSA‐Q‐2014‐00575 | |
| Feed/FEEDAP | Trichoderma reesei | Production of enzyme | EFSA‐Q‐2014‐00586 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme 4‐phytase | EFSA‐Q‐2015‐00665 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme α‐amylase | EFSA‐Q‐2015‐00681 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme aspergillopepsin I | EFSA‐Q‐2015‐00371 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme cellulase | EFSA‐Q‐2015‐00454 | |
| FIP/CEF | Trichoderma reesei | Production of food enzyme cellulose | EFSA‐Q‐2014‐00804 | |
| FIP/CEF | Trichoderma reesei | Production of food enzyme endo‐1,3(4)‐β‐glucanase | EFSA‐Q‐2014‐00805 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme endo‐1,4‐ β‐glucanase | EFSA‐Q‐2015‐00563 | |
| FIP/CEF | Trichoderma reesei | Production of food enzyme endo‐1,4‐β‐xylanase | EFSA‐Q‐2014‐00806 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme lysophospholipase C | EFSA‐Q‐2015‐00410 | |
| FIP/CEF | Trichoderma reesei, GMM strain | Production of food enzyme xylanase | EFSA‐Q‐2015‐00094 | |
| Feed/FEEDAP | Trichoderma reesei (ATCC SD‐6528), GMM strain | Zootechnical additive (production of enzyme) |
EFSA‐Q‐2013‐00997 |
|
| FIP/CEF | Trichoderma reesei/DP‐Dzh34, GMM strain | Production of food enzyme glucan 1,4‐α‐glucosidase | EFSA‐Q‐2016‐00097 | |
| FIP/CEF | Trichoderma reesei (DP‐Nzd22, GMM strain | Production of food enzyme endo‐1,4‐β‐xylanase from A. niger | EFSA‐Q‐2014‐00667 | |
| FIP/CEF | Trichoderma reesei/DP‐Nzh49, GMM strain | Production of food enzyme glucan 1,4‐α‐glucosidase | EFSA‐Q‐2016‐00094 | |
| FIP/CEF | Trichoderma reesei (RF5427), GMM strain | Production of food enzyme is a xylanase | EFSA‐Q‐2013‐00876 | |
| FIP/CEF | Trichoderma reesei (strain RF5427), GMM strain | Production of food enzyme xylanase | EFSA‐Q‐2014‐00735 | |
| FIP/CEF | Trichoderma reesei/ RF5703, GMM strain | Production of food enzyme endo 1,4‐β‐xylanase | EFSA‐Q‐2014‐00410 | |
| FIP/CEF | Trichoderma reesei (RF6197), GMM strain | Production of food enzyme polygalacturonase | EFSA‐Q‐2014‐00798 | |
| FIP/CEF | Trichoderma reesei/ RF6199 | Production of food enzyme pectine lyase | EFSA‐Q‐2014‐00164 | |
| FIP/CEF | Trichoderma reesei (RF6201), GMM strain | Production of food enzyme pectin esterase | EFSA‐Q‐2014‐00799 | |
| FIP/CEF | Trichoderma reesei (RF6232), GMM strain | Production of food enzyme Mannan endo‐1,4‐β‐mannosidase (β‐mannanase) | EFSA‐Q‐2014‐00094 (withdrawn) | |
| FIP/CEF | Trichoderma reesei/RF8793, GMM strain | Production of food enzyme phospholipase A2 | EFSA‐Q‐2014‐00411 | |
| Feed/FEEDAP | Trichoderma viride | Production of enzyme |
EFSA‐Q‐2010‐01295 http://www.efsa.europa.eu/en/efsajournal/pub/4235 EFSA‐Q‐2010‐01297 |
Mycelial fungi: already considered as not appropriate for QPS (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| FIP/CEF | Trichoderma viride (AE‐CT), GMM strain | Production of food enzyme cellulase | EFSA‐Q‐2015‐00067 | |
| Pesticides |
Verticillium albo‐atrum formerly Verticillium dahliae |
Plant protection product |
EFSA‐Q‐2009‐00141 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00303 http://www.efsa.europa.eu/en/efsajournal/pub/3059 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS. Potential production of alboatrin (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Oomycetes 38 | ||||
| Pesticides | Pythium oligandrum M1 | Plant protection product |
EFSA‐Q‐2009‐00133 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00287 http://www.efsa.europa.eu/en/efsajournal/pub/3034 Application for approval |
Mycelial fungi: already considered as not appropriate for QPS. Insufficient body of knowledge (see EFSA, 2007; EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013)d. |
| Algae | ||||
| Feed/FEEDAP | Haematococcus pluvialis | Production of astaxanthin | No body of knowledge except for this strain. Therefore not considered for QPS (EFSA, 2008). | |
| Bacteriophages | ||||
| Feed/FEEDAP | Clostridium sporogenes phage | Feed additive | QPS updates (EFSA BIOHAZ Panel, 2009, 2010), no recommendation to the QPS list because phages are subject to a case‐by‐case assessment. Considered in the current Scientific Opinion (refer to Section 3.6.1) where it is mentioned that the ‘phage application on foods should remain as a case‐by‐case procedure and, consequently, that these biological entities should not be considered for QPS status’. Please refer to all section. | |
| Feed/FEEDAP | Clostridium tyrobutyricum phage | Feed additive | ||
| BIOHAZ | Listeria monocytogenes phage | Food surface decontamination |
EFSA‐Q‐2011‐00959 |
|
| Viruses | ||||
| Pesticides | Adoxophyes orana granulovirus, strain BV‐0001 | Plant protection product |
EFSA‐Q‐2009‐00324 http://www.efsa.europa.eu/en/efsajournal/pub/2654 Application for approval EFSA‐Q‐2012‐00894 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.2) where it is mentioned that ‘On the basis of the scientific information including the ELS analysis, the QPS recommendation on members of the Baculoviridae family does not change and that the family Baculoviridae is the lowest taxonomic unit with QPS’. Please refer to the complete assessment. |
| Pesticides | Cydia pomonella granulovirus Mexican isolate | Plant protection product |
EFSA‐Q‐2009‐00126 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) EFSA‐Q‐2009‐00254 http://www.efsa.europa.eu/it/efsajournal/pub/2655 Application for approval |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.2) where it is mentioned that ‘On the basis of the scientific information including the ELS analysis, the QPS recommendation on members of the Baculoviridae family does not change and that the family Baculoviridae is the lowest taxonomic unit with QPS’. Please refer to the complete assessment. |
| Pesticides | Helicoverpa armigera nucleopolyhedrovirus | Plant protection product |
EFSA‐Q‐2009‐00341 http://www.efsa.europa.eu/en/efsajournal/pub/2865 Application for approval EFSA‐Q‐2013‐00348 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013, 2016a) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.2) where it is mentioned that ‘On the basis of the scientific information including the ELS analysis, the QPS recommendation on members of the Baculoviridae family does not change and that the family Baculoviridae is the lowest taxonomic unit with QPS’. Please refer to the complete assessment. |
| Pesticides | Pepino Mosaic Virus | Plant protection product | New assessment for QPS 2013 update (EFSA BIOHAZ Panel, 2013). Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.1) where it is mentioned that ‘On the basis of the scientific information identified through the ELS, the QPS recommendation on members of the Alphaflexiviridae family can be maintained’. Please refer to the complete assessment. | |
| Pesticides | Pepino mosaic virus, strain CH2, isolate 1906 | Plant protection product |
EFSA‐Q‐2014‐00054 http://www.efsa.europa.eu/en/efsajournal/pub/3977 Application for approval |
|
| Pesticides | Pepino mosaic virus, strain CH2, isolate 1906 | Plant protection product |
EFSA‐Q‐2015‐00483 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
|
| Pesticides | Pepino mosaic virus, isolate VC1 | Plant protection product |
EFSA‐Q‐2014‐00474 Application for approval |
|
| Pesticides | Pepino mosaic virus, strain isolate VX1 | Plant protection product |
EFSA‐Q‐2014‐00472 Application for approval |
|
| Pesticides |
Spodoptera exigua nuclear polyhedrosis virus Current name: nucleopolyhedrovirus |
Plant protection product |
EFSA‐Q‐2008‐630 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) Review Report for the active substance Spodoptera exigua nuclear polyhedrosis virus, SANCO/T14/2007‐rev.final, March 2007 |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.1) where it is mentioned that ‘On the basis of the scientific information identified through the ELS, the QPS recommendation on members of the Alphaflexiviridae family can be maintained’. Please refer to the complete assessment. |
| Pesticides | Spodoptera littoralis nucleopolyhedrovirus | Plant protection product |
EFSA‐Q‐2009‐00507 http://www.efsa.europa.eu/en/efsajournal/pub/2864 Application for approval EFSA‐Q‐2013‐00347 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.1) where it is mentioned that ‘On the basis of the scientific information identified through the ELS, the QPS recommendation on members of the Alphaflexiviridae family can be maintained’. Please refer to the complete assessment. |
| Pesticides | Zucchini yellow mosaic virus, weak strain | Plant protection product |
EFSA‐Q‐2009‐00346 http://www.efsa.europa.eu/en/efsajournal/pub/2754 Application for approval EFSA‐Q‐2013‐00012 http://www.efsa.europa.eu/en/efsajournal/pub/4458 Review of MRLs (Maximum Residue Limits) |
QPS updates (EFSA BIOHAZ Panel, 2009, 2010, 2011, 2012, 2013) recommended for the QPS list. Re‐evaluated in the current Scientific Opinion (refer to Section 3.5.1) where it is mentioned that ‘On the basis of the scientific information identified through the ELS, the QPS recommendation on members of the Potyviridae family can be maintained’. Please refer to the complete assessment. |
To find more details on specific applications please access the EFSA website – Register of Questions: http://registerofquestions.efsa.europa.eu/roqFrontend/ListOfQuestionsNoLogin?0&panel=ALL
Where no link is given this means that the risk assessment has not yet been published.
[http://www.ncbi.nlm.nih.gov/pubmed/?term=corynebacterium+pekinense e.g. Ma W, Zhao Z, Wang Y, Zhang Y, Ding J. Wei Sheng Wu Xue Bao. 2012 Nov 4;52(11):1344–51. Chinese]
Filamentous fungi were considered in the current Scientific Opinion (refer to Section 3.4.5) with the conclusion: ‘Although fungal taxonomy is in a rapid development, still these studies seldom provide information about the ecological properties and the function of the taxonomic units. The discontinuation of dual nomenclature for pleomorphic fungi has resulted in nomenclatural changes to well‐established fungal species. The increasing availability of fungal genome sequences is facilitating the discovery and characterization of numerous novel secondary metabolites by genome mining. While knowledge of fungal secondary metabolites has grown to a big extent, information on their toxic effects in humans and animals is still evolving at a much slower rate. Therefore it was decided that, until further notice, filamentous fungi are excluded from QPS evaluations. Their status should be monitored and re‐evaluated in the next QPS Opinion update’.
Supporting information
The 2016 updated list of QPS Status recommended biological agents in support of EFSA risk assessments.
Suggested citation: EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) , Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G (deceased), Prieto Maradona M, Querol A, Peixe L, Suarez JE, Sundh I, Vlak JM, Aguilera‐Gómez M, Barizzone F, Brozzi R, Correia S, Heng L, Istace F, Lythgo C and Fernández Escámez PS, 2017. Scientific Opinion on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA Journal 2017;15(3):4664, 177 pp. doi: 10.2903/j.efsa.2017.4664
The BIOHAZ Panel would like to dedicate this opinion to the memory of Günter Klein – see Obituary in the March 2017 issue of the EFSA Journal.
Requestor: EFSA
Question number: EFSA‐Q‐2014‐00189
Panel members: Ana Allende, Declan Bolton, Marianne Chemaly, Robert Davies, Pablo Salvador Fernández Escámez, Rosina Girones, Lieve Herman, Konstantinos Koutsoumanis, Roland Lindqvist, Birgit Nørrung, Antonia Ricci, Lucy Robertson, Giuseppe Ru, Moez Sanaa, Marion Simmons, Panagiotis Skandamis, Emma Snary, Niko Speybroeck, Benno Ter Kuile, John Threlfall and Helene Wahlström.
Acknowledgements: The BIOHAZ Panel wishes to thank the EFSA staff member: Mirena Ivanova for the support provided to this scientific output.
Amendment: 1. (6 June 2017) The name of one author has been corrected on page 1 and in the suggested citation paragraph; text under the additional information column has been added for E. coli rows on page 115. In Table A.1, p. 70, the text on the qualification of the taxonomic unit Pasteuria nishizawae has been deleted, following the decision taken by the BIOHAZ Panel at the meeting held in November‐December 2016. 2. (12 September 2017) in Appendix A the paragraph text has been slightly re‐edited; deletion of the text ‘and no viable cells are found’ in the Yeasts section. 3. (24 January 2018) Appendix A has been updated, following the revision of the QPS list adopted at the BIOHAZ Panel meeting held in December 2017; at the end of the Appendix a reference list has been added. 4. (25 May 2018). On page 71 in Appendix A the footnote letter attached to ‘Yeasts’ has been corrected from (j) to (e). 5. (17 July 2018) On pages 71–73 Appendix A has been updated, following the revision of the QPS list adopted at the BIOHAZ Panel meeting held in June 2018. 6. (24 January 2019) On pages 70–72 Appendix A has been updated, following the revision of the QPS list adopted at the BIOHAZ Panel meeting held in December 2018. 7. (15 July 2019) On pages 70–73 Appendix A has been updated, following the revision of the QPS list adopted at the BIOHAZ Panel meeting held in June 2019. These editorial corrections do not materially affect the contents or outcome of this scientific output. To avoid confusion, the older version has been removed from the EFSA Journal, but is available on request, as is a version showing all the changes made.
Erratum: 1. (12 September 2017) Table A.1 in Appendix A was revised to correct an erroneous footnote on “Paenibacillus lentus” and to add a new footnote ‘d’ on Lactobacillus animalis; in Appendix E the reference to Paenibacillus lentus has been deleted on p. 98; text on Paenibacillus lentus under column Additional information on page 132 for EFSA‐Q‐2016‐00181 has been revised. 2. (24 January 2018) Table A.1 in Appendix A was revised to substitute “Paenibacillus lentus” with “Bacillus lentus”. To avoid confusion, the older version has been removed from the EFSA Journal, but is available on request, as is a version showing all the changes made.
Adopted: 30 November 2016
Updated: 6 June 2017
Amended: 12 September 2017
This publication is linked to the following EFSA Journal article: http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2017.4663/full, http://onlinelibrary.wiley.com/doi/10.2903/j.efsa.2017.e15031/full
Amended: 24 January 2018
Updated: 25 May 2018
Amended: 17 July 2018
Amended: 24 January 2019
Amended: 15 July 2019
Notes
Opinion of the Scientific Committee on a request from EFSA related to a generic approach to the safety assessment by EFSA of microorganisms used in food/feed and the production of food/feed additives. The EFSA Journal 2005, 226, 1–12.
Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA – Opinion of the Scientific Committee. The EFSA Journal 2007, 293, 1–85.
Scientific Opinion of the Panel on Biological Hazards on a request from EFSA on the maintenance of the list of QPS microorganisms intentionally added to food or feed. The EFSA Journal 2008, 923, 1–48.
Scientific Opinion of the Panel on Biological Hazards (BIOHAZ) on the maintenance of the list of QPS microorganisms intentionally added to food or feed (2009 update). EFSA Journal 2009;7(12):1431, 92 pp. doi:10.2903/j.efsa.2009.1431
See more information in http://www.efsa.europa.eu/en/applications/regulatedproducts
European Commission (2003). On a generic approach to the safety assessment of microorganisms used in feed/food and feed/food production. A working paper open for comment. Available online: http://europa.eu.int/comm/food/fs/sc/scf/out178_en.pdf
Commission Implementing Regulation, 2012. Commission Implementing Regulation (EU) No 562/2012 of 27 June 2012 amending Commission Regulation (EU) No 234/2011 with regard to specific data required for risk assessment of food enzymes. OJ L 168, 28.6.2012, p. 21–23.
Commission Regulation, 2011. Commission Regulation (EU) No 234/2011 of 10 March 2011 implementing Regulation (EC) No 1331/2008 of the European Parliament and of the Council establishing a common authorisation procedure for food additives, food enzymes and food flavourings. OJ L 64, 11.3.2011, p. 15–24.
Regulation, 2008. Regulation (EC) No 1331/2008 of the European Parliament and of the Council of 16 December 2008 establishing a common authorisation procedure for food additives, food enzymes and food flavourings. OJ L 354, 31.12.2008, p. 1–6.
European Food Safety Authority, 2014; Explanatory Note for the Guidance of the Scientific Panel of Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF) on the Submission of a Dossier on Food Enzymes. EFSA supporting publication 2014:EN‐689. 22 pp.
Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003 on additives for use in animal nutrition. OJ L 268, 18.10.2003, p. 29–43.
Regulation (EC) No 1107/2009 of the European Parliament and of the Council of 21 October 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC. OJ L 309, 24.11.2009, p. 1–49.
Commission Regulation (EU) No 283/2013 of 1 March 2013 setting out the data requirements for active substances, in accordance with Regulation (EC) No 1107/2009. OJ L 93, 3.4.2013, p. 1–84.
Commission Regulation (EU) No 283/2013 of 1 March 2013 setting out the data requirements for active substances, in accordance with Regulation (EC) No 1107/2009. OJ L 93, 3.4.2013, p. 1–84. Commission Regulation (EU) No 284/2013 of 1 March 2013 setting out the data requirements for plant protection products, in accordance with Regulation (EC) No 1107/2009. OJ L 93, 3.4.2013, p. 85–152.
EFSA, 2011: Submission of scientific peer‐reviewed open literature for the approval of pesticide active substances under Regulation (EC) No 1107/2009 (EFSA Journal 2011;9(2):2092).
Council Directive of 15 July 1991 concerning the placing of plant protection products on the market (91/414/EEC). OJ L 230, 19.8.1991, p. 1–32.
Commission Implementing Regulation (EU) No 562/2012 of 27 June 2012 amending Commission Regulation (EU) No 234/2011 with regard to specific data required for risk assessment of food enzymes. OJ L 168, 28.6.2012, p. 21–23.
Commission Regulation (EU) No 234/2011 of 10 March 2011 implementing Regulation (EC) No 1331/2008 of the European Parliament and of the Council establishing a common authorisation procedure for food additives, food enzymes and food flavourings. OJ L 64, 11.3.2011, p. 15–24.
Regulation (EC) No 1331/2008 of the European Parliament and of the Council of 16 December 2008 establishing a common authorisation procedure for food additives, food enzymes and food flavourings. OJ L 354, 31.12.2008, p. 1–6.
Regulation (EU) 2015/2283 of the European Parliament and of the Council of 25 November 2015 on novel foods, amending Regulation (EU) No 1169/2011 of the European Parliament and of the Council and repealing Regulation (EC) No 258/97 of the European Parliament and of the Council and Commission Regulation (EC) No 1852/2001. OJ L 327, 11.12.2015, p. 1–22.
See ATCC website at: https://www.lgcstandards-atcc.org/Products/Cells_and_Microorganisms.aspx?geo_country=it
References
- Abrantes P, McArthur CP and Africa CWJ, 2014. Multi‐drug resistant oral Candida species isolated from HIV‐positive patients in South Africa and Cameroon. Diagnostic Microbiology and Infectious Disease, 79, 222–227. [DOI] [PubMed] [Google Scholar]
- Abriouel H, Benomar N, Cobo A, Caballero N, Fernández Fuentes MÁ, Pérez‐Pulido R and Gálvez A, 2012. Characterization of lactic acid bacteria from naturally‐fermented Manzanilla Alorena green table olives. Food Microbiology, 32, 308–316. [DOI] [PubMed] [Google Scholar]
- Adames H, Baldovi S, Martin‐Cleary C, Ortiz A and Esteban J, 2010. Peritonitis due to Microbacterium sp in a patient on cycler peritoneal dialysis. Peritoneal Dialysis International, 30, 669–670. [DOI] [PubMed] [Google Scholar]
- Agerholm JS, Jensen NE, Dantzer V, Jensen HE and Assrestrup FM, 1999. Experimental infection of pregnant cows with Bacillus licheniformis bacteria. Veterinary Pathology, 36, 191–201. [DOI] [PubMed] [Google Scholar]
- Airenne KJ, Hu YC, Kost TA, Smith RH, Kotin RM, Ono C, Matsuura Y, Wang S and Yla‐Herttuala S, 2013. Baculovirus: an insect‐derived vector for diverse gene transfer applications. Molecular Therapy, 21, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Badah AS, Ibrahim ASS, Al‐Salamah AA and Ibrahim SSS, 2015. Clonal diversity and antimicrobial resistance of Enterococcus faecalis isolated from endodontic infections. Electronic Journal of Biotechnology, 18, 175–180. [Google Scholar]
- Alegría Á, Delgado S, Flórez AB and Mayo B, 2013. Identification, typing, and functional characterization of Leuconostoc spp. strains from traditional, starter‐free cheeses. Dairy Science & Technology, 93, 657–673. [Google Scholar]
- Alfouzan W, Dhar R, Ashkanani H, Gupta M, Rachel C and Khan ZU, 2015. Species spectrum and antifungal susceptibility profile of vaginal isolates of Candida in Kuwait. Journal de Mycologie Médicale, 25, 23–28. [DOI] [PubMed] [Google Scholar]
- Alonso‐Echanove J, Shah SS, Valenti AJ, Dirrigl SN, Carson LA, Arduino MJ and Jarvis WR, 2001. Nosocomial outbreak of Microbacterium species bacteremia among cancer patients. Journal of Infectious Diseases, 184, 754–760. [DOI] [PubMed] [Google Scholar]
- Anonymous , 2000. Opinion of 13 April 1999 issued on behalf of the Supreme Council of Public Hygiene (Section for Food and Nutrition) on the draft Order of 2 October 1997 amending the additives which may be used in the manufacture of foodstuffs for human consumption. Official Bulletin of Competition, Consumption and Repression of Fraud no 8 of 25 July 2000. Available online: http://www.economie.gouv.fr/files/files/directions_services/dgccrf/boccrf/00_08/a0080024.htmv
- Anonymous , 2011. Order of 20 August 2013 amending the order of 19 October 2006 on the use of processing aids in the manufacture of certain foodstuffs. Official Journal of the French Republic, no 0211 of 11 September 2013, p. 15292, text no 45. Available online: http://www.legifrance.gouv.fr/affichTexte.do?cidTexte=eORFTEXT000027941379&categorieLien=nd
- Anonymous , 2013a. Regional veterinary laboratories report – January 2013. Veterinary Ireland Journal, 3, 189–194. [Google Scholar]
- Anonymous , 2013b. Northern Ireland disease surveillance, January to March 2013. Veterinary Record, 172, 657–658. [DOI] [PubMed] [Google Scholar]
- Anonymous , 2013c. Regional veterinary laboratories report – March 2013. Veterinary Ireland Journal, 3, 321–326. [Google Scholar]
- Anonymous , 2014a. Northern Ireland disease surveillance report, October to December 2013. Veterinary Record, 174, 139–141. [DOI] [PubMed] [Google Scholar]
- Anonymous , 2014b. Regional veterinary laboratories report: March 2014. Veterinary Ireland Journal, 4, 315–320. [Google Scholar]
- Anonymous , 2014c. Regional veterinary laboratories report: April 2014. Veterinary Ireland Journal, 4, 371–376. [Google Scholar]
- Anoop V, Rotaru S, Shwed PS, Tayabali AF and Arvanitakis G, 2015. Review of current methods for characterizing virulence and pathogenicity potential of industrial Saccharomyces cerevisiae strains towards humans. FEMS Yeast Research, 15. doi: 10.1093/femsyr/fov057 [DOI] [PubMed] [Google Scholar]
- Arancia S, Sandini S, Graziani S, Norelli S and de Bernardis F, 2014. Use of multiplex PCR and high‐resolution melting analysis with primers of a gene coding for 65 KDa mannoprotein to rapid detect Candida species in biological samples. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 54, M‐424. [Google Scholar]
- Aroutcheva A, Auclair J, Frappier M, Millette M, Lolans K, Montigny D, Carriere S, Sokalski S, Trick WE and Weinstein RA, 2016. Importance of molecular methods to determine whether a probiotic is the source of Lactobacillus bacteremia. Probiotics and Antimicrobial Proteins, 8, 31–40. [DOI] [PubMed] [Google Scholar]
- Atibalentja N, Jakstys BP and Noel GR, 2004. Life cycle, ultrastructure, and host specificity of the North American isolate of Pasteuria that parasitizes the soybean cyst nematode, heterodera glycines. Journal of Nematology, 36, 171–180. [PMC free article] [PubMed] [Google Scholar]
- Avcin SL, Pokorn M, Kitanovski L, Premru MM and Jazbec J, 2015. Bifidobacterium breve sepsis in child with high‐risk acute lymphoblastic leukemia. Emerging Infectious Diseases, 21, 1674–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back W, 1978. Elevation of Pediococcus cerevisiae subsp. dextrinicus Coster and White to Species Status [Pediococcus dextrinicus (Coster and White) comb. nov.]. International Journal of Systematic and Evolutionary Microbiology, 28, 523–527. 10.1099/00207713-28-4-523 [DOI] [Google Scholar]
- Bağder Elmacı S, Tokatlı M, Dursun D, Özçelik F and Şanlıbaba P, 2015. Phenotypic and genotypic identification of lactic acid bacteria isolated from traditional pickles of the Çubuk region in Turkey. Folia Microbiologica, 60, 241–251. [DOI] [PubMed] [Google Scholar]
- Baltimore D, 1971. Expression of animal virus genomes. Bacteriological Reviews, 35, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D, 1974. The strategy of RNA viruses. Harvey Lectures, 70 Series, 57–74. [PubMed] [Google Scholar]
- Banerjee P, Kaur R and Uppal B, 2013. Study of fungal isolates in patients with chronic diarrhea at a tertiary care hospital in north India. Journal de Mycologie Médicale, 23, 21–26. [DOI] [PubMed] [Google Scholar]
- Bașbülbül G, Özteber M and Bİyİk HH, 2015. Antibiotic resistance in lactic acid bacteria isolated from fermented dairy products and boza. Journal of Microbiology, Biotechnology and Food Sciences, 4, 513–517. [Google Scholar]
- Benjamin S and Pandey A, 1998. Candida rugosa lipases: molecular biology and versatility in biotechnology. Yeast, 14, 1069–1087. [DOI] [PubMed] [Google Scholar]
- Bertelli C, Pillonel T, Torregrossa A, Prod'hom G, Fischer CJ, Greub G and Giannoni E, 2015. Bifidobacterium longum bacteremia in preterm infants receiving probiotics. Clinical Infectious Diseases, 60, 924–927. [DOI] [PubMed] [Google Scholar]
- Beyda ND, Chuang SH, Alam MJ, Shah DN, Ng TM, McCaskey L and Garey KW, 2013. Treatment of Candida famata bloodstream infections: case series and review of the literature. Journal of Antimicrobial Chemotherapy, 68, 438–443. [DOI] [PubMed] [Google Scholar]
- Bishop AH, Gowen SR, Pembroke B and Trotter JR, 2007. Morphological and molecular characteristics of a new species of Pasteuria parasitic on Meloidogyne ardenensis . Journal of Invertebrate Pathology, 96, 28–33. [DOI] [PubMed] [Google Scholar]
- Bishop AH, 2011. Pasteuria penetrans and its parasitic interaction with plant parasitic nematodes. Endospore‐forming Soil Bacteria. N. A. Logan and P. DeVos, Springer‐Verlag, Berlin Heidelberg, Volume 27, 181–201. [Google Scholar]
- Bosma EF, van de Weijer AH, Daas MJ, van der Oost J, de Vos WM and van Kranenburg R, 2015. Isolation and screening of thermophilic bacilli from compost for electrotransformation and fermentation: characterization of Bacillus smithii ET 138 as a new biocatalyst. Applied and Environmental Microbiology, 81, 1874–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma EF, Koehorst JJ, van Hijum SA, Renckens B, Vriesendorp B, van de Weijer AH, Schaap PJ, de Vos WM, van der Oost J and van Kranenburg R, 2016. Complete genome sequence of thermophilic Bacillus smithii type strain DSM 4216(T). Standards in Genomic Sciences, 11, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdichon F, Casaregola S, Farrokh C, Frisvad JC, Gerds ML, Hammes WP, Harnett J, Huys G, Laulund S, Ouwehand A, Powell IB, Prajapati JB, Seto Y, Ter Schure E, Van Boven A, Vankerckhoven V, Zgoda A, Tuijtelaars S and Hansen EB, 2012. Food fermentations: microorganisms with technological beneficial use. International Journal of Food Microbiology, 154, 87–97. [DOI] [PubMed] [Google Scholar]
- Bradbury JF, 1984. Genus II. Xanthomonas In: Krieg NR. and Holt JG. (eds.). Bergey's Manual of Systematic Bacteriology. Vol. 1. Williams and Wilkins, Baltimore: pp. 199–210. [Google Scholar]
- Branquinho R, Meirinhos‐Soares L, Carrico JA, Pintado M and Peixe LV, 2014. Phylogenetic and clonality analysis of Bacillus pumilus isolates uncovered a highly heterogeneous population of different closely related species and clones. FEMS Microbiology Ecology, 90, 689–698. [DOI] [PubMed] [Google Scholar]
- Brecht M, Garg A, Longstaff K, Cooper C and Andersen C, 2016. Lactobacillus sepsis following a laparotomy in a preterm infant: a note of caution. Neonatology, 109, 186–189. [DOI] [PubMed] [Google Scholar]
- Brozzoli V, Crognale S, Sampedro I, Federici F, D'Annibale A and Petruccioli M, 2009. Assessment of olive‐mill wastewater as a growth medium for lipase production by Candida cylindracea in bench‐top reactor. Bioresource Technology, 100, 3395–3402. [DOI] [PubMed] [Google Scholar]
- Buchelli‐Ramirez HL, Alvarez‐Alvarez C, Rojo‐Alba S, Garcia‐Clemente M, Cimadevilla‐Suarez R, Pando‐Sandoval A and Casan‐Clara P, 2013. Necrotising pneumonia caused by Lactococcus lactis cremoris . International Journal of Tuberculosis and Lung Disease, 17, 565–567. [DOI] [PubMed] [Google Scholar]
- Buss SN, Starlin R and Iwen PC, 2014. Bacteremia caused by Microbacterium binotii in a patient with sickle cell anemia. Journal of Clinical Microbiology, 52, 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye P, Devriese LA and Haesebrouck F, 2003. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram‐positive bacteria. Clinical Microbiology Reviews, 16, 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campedelli I, Flórez AB, Salvetti E, Delgado S, Orrù L, Cattivelli L, Alegría Á, Felis GE, Torriani S and Mayo B, 2015. Draft genome sequence of three antibiotic‐resistant Leuconostoc mesenteroides strains of dairy origin. Genome Announcements, 3, e01018‐01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GF, Sinniah S, Idris TINT, Puad MSA and Abd Rahman AZ, 2013. Multiple rare opportunistic and pathogenic fungi in persistent foot skin infection. Pakistan Journal of Biological Sciences: PJBS, 16, 208–218. [DOI] [PubMed] [Google Scholar]
- Chandna P, Mayilraj S and Kuhad RC, 2013. Bacillus paraflexus sp. nov., isolated from compost. International Journal of Systematic and Evolutionary Microbiology, 63, 4735–4743. [DOI] [PubMed] [Google Scholar]
- Charles L, Carbone I, Davies KG, Bird D, Burke M, Kerry BR and Opperman CH, 2005. Phylogenetic analysis of Pasteuria penetrans by use of multiple genetic loci. Journal of Bacteriology, 187, 5700–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX and Dickson DW, 1998. Review of Pasteuria penetrans: biology, ecology, and biological control potential. Journal of Nematology, 30, 313–340. [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wu HH, Chen CP, Chern SR, Hwang SM, Huang SF, Lo WH, Chen GY and Hu YC, 2011. Biosafety assessment of human mesenchymal stem cells engineered by hybrid baculovirus vectors. Molecular Pharmaceutics, 8, 1505–1514. [DOI] [PubMed] [Google Scholar]
- Chen CY, Lin SY, Cheng MC, Tsai CP, Hung CL, Lo KW, Hwang Y and Hu YC, 2013. Baculovirus vector as an avian influenza vaccine: hemagglutinin expression and presentation augment the vaccine immunogenicity. Journal of Biotechnology, 164, 143–150. [DOI] [PubMed] [Google Scholar]
- Chery J, Dvoskin D, Morato FP and Fahoum B, 2013. Lactobacillus fermentum, a pathogen in documented cholecystitis. International Journal of Surgery Case Reports, 4, 662–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codex Alimentarius , 2014. GSFA Online. Mis à jour jusqu’à la 37ème session de la Commission du Codex Alimentarius (2014). Renseignement détaillés sur l'additif alimentaire, Gomme xanthane (415). Available online: http://www.codexalimentarius.net/gsfaonline/additives/details.html?id=48&lang=fr&print=true
- Cohen L, Ranque S and Raoult D, 2013. Saccharomyces cerevisiae boulardii transient fungemia after intravenous self‐inoculation. Medical Mycology Case Reports, 2, 63–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Jones D and Kroppenstedt RM, 1983. Reclassification of Brevibacterium imperiale (Steinhaus) and “Corynebacterium laevaniformans” (Dias and Bhat) in a Redefined Genus Microbacterium (Orla‐Jensen), as Microbacterium imperiale comb. nov. and Microbacterium laevaniformans nom. rev.; comb. nov. Systematic and Applied Microbiology, 4, 65–78. [DOI] [PubMed] [Google Scholar]
- Collins MD, Farrow JAE, Phillips BA, Ferusu S and Jones D, 1987. Classification of Lactobacillus divergens, Lactobacillus Piscicola, and some catalase‐negative, asporogenous, rod‐shaped bacteria from poultry in a new genus, Carnobacterium . International Journal of Systematic Bacteriology, 37, 310–316. [Google Scholar]
- Collins MD, Williams AM and Wallbanks S, 1990. The phylogeny of Aerococcus and Pediococcus as determined by 16S rRNA sequence analysis: description of Tetragenococcus gen. nov. FEMS Microbiology Letters, 58, 255–262. [DOI] [PubMed] [Google Scholar]
- Constable A, Jonas D, Cockburn A, Davi A, Edwards G, Hepburn P, Herouet‐Guicheney C, Knowles M, Moseley B, Oberdorfer R and Samuels F, 2007. History of safe use as applied to the safety assessment of novel foods and foods derived from genetically modified organisms. Food and Chemical Toxicology, 45, 2513–2525. [DOI] [PubMed] [Google Scholar]
- Coster E and White HR, 1964. Further Studies of the Genus Pediococcus., 37, 15–31. 10.1099/00221287-37-1-15 [DOI] [PubMed] [Google Scholar]
- Daligault HE, Davenport KW, Minogue TD, Bishop‐Lilly KA, Broomall SM, Bruce DC, Chain PS, Coyne SR, Frey KG, Gibbons HS, Jaissle J, Koroleva GI, Ladner JT, Lo CC, Munk C, Palacios GF, Redden CL, Rosenzweig CN, Scholz MB and Johnson SL, 2014. Twenty whole‐genome Bacillus sp. assemblies. Genome Announcements, 2, e00958‐00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damasceno NP, Horowitz SA and Damasceno EF, 2016. Leuconostoc as a cause of endophthalmitis post‐intravitreal injection of ranibizumab. Ocular Immunology and Inflammation, 24, 118–119. [DOI] [PubMed] [Google Scholar]
- Dani C, Coviello CC, Corsini II, Arena F, Antonelli A and Rossolini GM, 2016. Lactobacillus sepsis and probiotic therapy in newborns: two new cases and literature review. AJP Reports, 6, E25–E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel HM, Lachance MA and Kurtzman CP, 2014. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek, 106, 67–84. [DOI] [PubMed] [Google Scholar]
- Danielsen M, Simpson PJ, O'Connor EB, Ross RP and Stanton C, 2007. Susceptibility of Pediococcus spp. to antimicrobial agents. Journal of Applied Microbiology, 102, 384–389. [DOI] [PubMed] [Google Scholar]
- Das S, Jean JS, Chou ML, Rathod J and Liu CC, 2016. Arsenite‐oxidizing bacteria exhibiting plant growth promoting traits isolated from the rhizosphere of Oryza sativa L.: Implications for mitigation of arsenic contamination in paddies. Journal of Hazardous Materials, 302, 10–18. [DOI] [PubMed] [Google Scholar]
- Deepak MB, Jhanvi SP and AnuAppaiah KA, 2015. Aflatoxin binding and detoxification by non‐saccharomyces yeast – a new vista for decontamination. International Journal of Current Microbiology and Applied Sciences, 4, 310–317. [Google Scholar]
- Delsart C, Grimi N, Boussetta N, Miot Sertier C, Ghidossi R, Vorobiev E and Mietton Peuchot M, 2016. Impact of pulsed‐electric field and high‐voltage electrical discharges on red wine microbial stabilization and quality characteristics. Journal of Applied Microbiology, 120, 152–164. [DOI] [PubMed] [Google Scholar]
- Deng Y, Bao XR, Ji LL, Chen L, Liu JY, Miao J, Chen DQ, Bian HW, Li YM and Yu GC, 2015. Resistance integrons: class 1, 2 and 3 integrons. Annals of Clinical Microbiology and Antimicrobials, 14, 11 pp. doi: 10.1186/s12941-015-0100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deorukhkar SC and Santosh S, 2013. Vulvovaginal candidiasis due to non albicans Candida: its species distribution and antifungal susceptibility profile. International Journal of Current Microbiology and Applied Sciences, 2, 323–328. [Google Scholar]
- Doern C, Wallace M and Burnham CA, 2014a. Case report: Lactobacillus rhamnosus probiotic as a cause of pediatric pneumonia. Abstracts of the General Meeting of the American Society for Microbiology, 114, 151. [Google Scholar]
- Doern CD, Nguyen ST, Afolabi F and Burnham CAD, 2014b. Probiotic‐associated aspiration pneumonia due to Lactobacillus rhamnosus . Journal of Clinical Microbiology, 52, 3124–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DSMZ (German Collection of Microorganisms and Cell Cultures), online. Candida cylindracea Yamada & Machida. Available online: http://www.dsmz.de/catalogues/details/culture/DSM-2031.html
- Duff‐Farrier CRA, Bailey AM, Boonham N and Foster GD, 2015. A pathogenicity determinant maps to the N‐terminal coat protein region of the Pepino mosaic virus genome. Molecular Plant Pathology, 16, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne SF, Marr KA, Sydnor E, Staab JF, Karp JE, Lu K, Zhang SAX, Lavallee C, Perl TM and Neofytos D, 2014. Epidemiology of Candida kefyr in patients with hematologic malignancies. Journal of Clinical Microbiology, 52, 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap CA, Kwon SW, Rooney AP and Kim SJ, 2015. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. International Journal of Systematic and Evolutionary Microbiology, 65, 3487–3492. [DOI] [PubMed] [Google Scholar]
- Eddouzi J, Lohberger A, Vogne C, Manai M and Sanglard D, 2013. Identification and antifungal susceptibility of a large collection of yeast strains isolated in Tunisian hospitals. Medical Mycology, 51, 737–746. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2005. Opinion of the Scientific Committee on a request from EFSA related to a generic approach to the safety assessment by EFSA of microorganisms used in food/feed and the production of food/feed additives. EFSA Journal 2005;3(4):226, 12 pp. doi: 10.2903/j.efsa.2005.226 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2007. Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA – Opinion of the Scientific Committee. EFSA Journal 2007;5(12):587, 16 pp. doi: 10.2903/j.efsa.2007.587 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2008. Scientific Opinion of the Panel on Biological Hazards on the maintenance of the list of QPS microorganisms intentionally added to food or feed. EFSA Journal 2008;6(12):923, 48 pp. doi: 10.2903/j.efsa.2008.923 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2009. Transparency in Risk Assessment – Scientific Aspects Guidance of the Scientific Committee on Transparency in the Scientific Aspects of Risk Assessments carried out by EFSA. Part 2: General Principles. EFSA Journal 2009;7(5):1051, 22 pp. doi: 10.2903/j.efsa.2009.1051 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012a. Conclusion on the peer review of the pesticide risk assessment of the active substance Adoxophyes orana granulovirus. EFSA Journal 2012;10(4):2654, 32 pp. doi: 10.2903/j.efsa.2012.2654 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2012b. Conclusion on the peer review of the pesticide risk assessment of the active substance Cydia pomonella granulovirus (CpGV). EFSA Journal 2012;10(4):2655, 40 pp. doi: 10.2903/j.efsa.2012.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2009. Scientific Opinion on the maintenance of the list of QPS microorganisms intentionally added to food or feed (2009 update). EFSA Journal 2009;7(12):1431, 92 pp. doi: 10.2903/j.efsa.2009.1431 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2010. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2010 update). EFSA Journal 2010;8(12):1944, 56 pp. doi: 10.2903/j.efsa.2010.1944 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2011. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2011 update). EFSA Journal 2011;9(12):2497, 82 pp. doi: 10.2903/j.efsa.2011.2497 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2012. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2012 update). EFSA Journal 2012;10(12):3020, 84 pp. doi: 10.2903/j.efsa.2012.3020 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2013. Scientific Opinion on the maintenance of the list of QPS biological agents intentionally added to food and feed (2013 update). EFSA Journal 2013;11(11):3449, 108 pp. doi:110.2903/j.efsa.2013.3449 [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2014. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 1: Suitability of taxonomic units notified to EFSA until October 2014. EFSA Journal 2014;12(12):3938, 41 pp. doi: 10.2903/j.efsa.2014.3938 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015a. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. 2: Suitability of taxonomic units notified to EFSA until March 2015. EFSA Journal 2015;13(6):4138, 29 pp. doi: 10.2903/j.efsa.2015.4138 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2015b. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA. 3: Suitability of taxonomic units notified to EFSA until September 2015. EFSA Journal 2015;13(12):4331, 25 pp. doi: 10.2903/j.efsa.2015.4331 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2016a. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 4: suitability of taxonomic units notified to EFSA until March 2016. EFSA Journal 2016;14(7):4522, 37 pp. doi: 10.2903/j.efsa.2016.4522 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), 2016b. Scientific Opinion on the risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA Journal 2016;14(7):4524, 93 pp. doi: 10.2903/j.efsa.2016.4524 [DOI] [Google Scholar]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Herman L, Koutsoumanis K, Roland L, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G, Prieto Maradona M, Querol A, Peixe L, Suarez JE, Sundh I, Vlak JM, Barizzone F, Correia S and Fernández Escámez PS, 2017. Statement on the update of the list of QPS‐recommended biological agents intentionally added to food or feed as notified to EFSA 5: Suitability of taxonomic units notified to EFSA until September 2016. EFSA Journal 2017;15(3):4663, 20 pp. doi: 10.2903/j.efsa.2017.4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), 2011. Technical Guidance on the assessment of the toxigenic potential of Bacillus species used in animal nutrition. EFSA Journal 2011;9(11):2445, 13 pp. doi: 10.2903/j.efsa.2011.2445 [DOI] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), 2012a. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal 2012;10(6):2740, 10 pp. doi: 10.2903/j.efsa.2012.2740 [DOI] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), 2012b. Scientific Opinion on the removal of a maximum dose from the authorisation of microbial products assessed using the Qualified Presumption of Safety approach. EFSA Journal 2012;10(5):2680, 8 pp. doi: 10.2903/j.efsa.2012.2680 [DOI] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), 2012c. Guidance on the safety assessment on Enterococcus faecium in animal nutrition. EFSA Journal 2012;10(5):2682, 10 pp. doi: 10.2903/j.efsa.2012.2682 [DOI] [Google Scholar]
- EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed), 2014. Guidance on the assessment of the toxigenic potential of Bacillus species used in animal nutrition. EFSA Journal 2014;12(5):3665, 10 pp. doi: 10.2903/j.efsa.2014.3665 [DOI] [Google Scholar]
- EFSA NDA Panel (EFSA Panel on Dietetic Products, Nutrition and Allergies), 2016. Guidance on the preparation and presentation of an application for authorisation of a novel food in the context of Regulation (EU) 2015/2283 on Novel Foods. EFSA Journal 2016;14(10):4590, 30 pp. doi: 10.2903/j.efsa.2016.4590 [DOI] [Google Scholar]
- Ehling‐Schulz M and Messelhäusser U, 2013. Bacillus “next generation” diagnostics: moving from detection toward subtyping and risk‐related strain profiling. Frontiers in Microbiology, 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizaquível P, Sánchez G, Salvador A, Fiszman S, Dueñas MT, López P, Fernández de Palencia P and Aznar R, 2011. Evaluation of yogurt and various beverages as carriers of lactic acid bacteria producing 2‐branched (1,3)‐beta‐D‐glucan. Journal of Dairy Science, 94, 3271–3278. [DOI] [PubMed] [Google Scholar]
- Encarnacion CO, Loranger AM, Bharatkumar AG and Almassi GH, 2016. Bacterial endocarditis caused by Lactobacillus acidophilus leading to rupture of sinus of Valsalva Aneurysm. Texas Heart Institute Journal, 43, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch DA, Richardson MP, Hill RL, Scorer PM and Sismey A, 2011. Central venous catheter‐related bacteraemia due to Microbacterium paraoxydans in a patient with no significant immunodeficiency. Journal of Clinical Pathology, 64, 179–180. [DOI] [PubMed] [Google Scholar]
- Esgin H, Bulut E and Orum C, 2014. Candida pelliculosa endophthalmitis after cataract surgery: a case report. BMC Research Notes, 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euzeby JP, 2013. List of prokaryotic names with standing nomenclature (LPSN). Available online: http://www.bacterio.cict.fr/ [Accessed: 28 June 2012]
- Fadda ME, Pisano MB, Scaccabarozzi L, Mossa V, Deplano M, Moroni P, Liciardi M and Cosentino S, 2013. Use of PCR‐restriction fragment length polymorphism analysis for identification of yeast species isolated from bovine intramammary infection. Journal of Dairy Science, 96, 7692–7697. [DOI] [PubMed] [Google Scholar]
- Fagerlund A, Dubois T, Økstad OA, Verplaetse E, Gilois N, Bennaceur I, Perchat S, Gominet M, Aymerich S, Kolstø AB, Lereclus D and Gohar M, 2014. SinR controls enterotoxin expression in Bacillus thuringiensis biofilms. PLoS ONE, 9, e87532, doi: 10.1371/journal.pone.0087532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falci DR, Rigatto MH, Cantarelli VV and Zavascki AP, 2015. Lactobacillus rhamnosus bacteremia in a kidney transplant recipient. Transplant Infectious Disease, 17, 610–612. [DOI] [PubMed] [Google Scholar]
- Feierabend D, Reichart R, Romeike B, Kalff R and Walter J, 2013. Cerebral abscess due to Lactococcus lactis cremoris in a child after sinusitis. Clinical Neurology and Neurosurgery, 115, 614–616. [DOI] [PubMed] [Google Scholar]
- Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D and Datry A, 2013. Rapid emergence of echinocandin resistance during Candida kefyr fungemia treatment with Caspofungin. Antimicrobial Agents and Chemotherapy, 57, 2380–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felberbaum RS, 2015. The baculovirus expression vector system: A commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnology Journal, 10, 702–U785, doi: 10.1002/biot.201400438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Fuentes MA, Abriouel H, Ortega Morente E, Pérez Pulido R and Gálvez A, 2014. Genetic determinants of antimicrobial resistance in Gram positive bacteria from organic foods. International Journal of Food Microbiology, 172, 49–56. [DOI] [PubMed] [Google Scholar]
- Fleming D and Hunt DL, 2000. Biological Safety: Principles and Practices. 3rd Edition, ASM Press, Washington: 784 pp. [Google Scholar]
- Flórez AB, Campedelli I, Delgado S, Alegría Á, Salvetti E, Felis GE, Mayo B and Torriani S, 2016. Antibiotic susceptibility profiles of dairy Leuconostoc, analysis of the genetic basis of atypical resistances and transfer of genes in vitro and in a food matrix. PLoS ONE, 11, e0145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko B, Valliant M, Recule C, Vautrin E, Brion JP and Pavese P, 2013. Lactobacillus paracasei endocarditis in a consumer of probiotics. Médecine et Maladies Infectieuses, 43, 171–173. [DOI] [PubMed] [Google Scholar]
- de Freitas EM, Nobre SAM, Pires MBD, Faria RVJ, Batista AUD and Bonan PRF, 2013. Oral Candida species in head and neck cancer patients treated by radiotherapy. Auris, Nasus, Larynx, 40, 400–404. [DOI] [PubMed] [Google Scholar]
- Fujihira A, Suzuki T, Chang MO, Moriyama T, Kitajima M and Takaku H, 2014. Antitumor effects of baculovirus‐infected dendritic cells against human pancreatic carcinoma. Gene Therapy, 21, 849–854. [DOI] [PubMed] [Google Scholar]
- Fujita R, Ono C, Ono I, Asano S and Bando H, 2015. Analysis of the Bombyx mori nucleopolyhedrovirus ie‐1 promoter in insect, mammalian, plant, and bacterial cells. Biochemical and Biophysical Research Communications, 464, 1297–1301. [DOI] [PubMed] [Google Scholar]
- Gan HY, Gan HM, Savka MA, Triassi AJ, Wheatley MS, Smart LB, Fabio ES and Hudson AO, 2014. Whole‐genome sequences of 13 endophytic bacteria isolated from shrub willow (Salix) grown in Geneva, New York. Genome Announcements, 2, doi: 10.1128/genomeA.00288-00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Bao Y, Liu Y, Zhang X, Wang J and An L, 2009. Characterization of thermo‐stable endoinulinase from a new strain Bacillus smithii T7. Applied Biochemistry and Biotechnology, 157, 498–506. [DOI] [PubMed] [Google Scholar]
- Garcia Hejl C, Sanmartin N, Samson T, Soler C and Koeck JL, 2015. Maxillary sinus infection by Bacillus licheniformis: a case report from Djibouti. Médecine et Santé Tropicales, 25, 220–221. [DOI] [PubMed] [Google Scholar]
- Ghahri M, Mirhendi H, Zomorodian K and Kondori N, 2013. Identification and antifungal susceptibility patterns of Candida strains isolated from blood specimens in Iran. Archives of Clinical Infectious Diseases, 8, e14529, doi: 10.5812/archcid.14529 [DOI] [Google Scholar]
- Ghodasra DH, Eftekhari K, Shah AR and VanderBeek BL, 2014. Outcomes, impact on management, and costs of fungal eye disease consults in a tertiary care setting. Ophthalmology, 121, 2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Paul S, Sood P, Rudramurthy SM, Rajbanshi A, Jillwin TJ and Chakrabarti A, 2015. Matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clinical Microbiology and Infection, 21, 372–378. [DOI] [PubMed] [Google Scholar]
- Giammanco GM, Pignato S, Grimont PA, Grimont F, Santangelo C, Leonardi G, Giuffrida A, Legname V and Giammanco G, 2006. Interstitial pulmonary inflammation due to Microbacterium sp. after heart transplantation. Journal of Medical Microbiology, 55, 335–339. [DOI] [PubMed] [Google Scholar]
- Grass G, Bierbaum G, Molitor E, Götte N and Antwerpen M, 2016. Genome sequence of Bacillus pumilus Strain Bonn, isolated from an anthrax‐like necrotic skin infection site of a child. Genome Announcements, 4, doi: 10.1128/genomeA.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FP, Fan HW, Liu ZY, Yang QW, Li YJ and Li TS, 2015. Brain abscess caused by Bacillus megaterium in an adult patient. Chinese Medical Journal, 128, 1552–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddadi P, Zareifar S, Badiee P, Alborzi A, Mokhtari M, Zomorodian K, Pakshir K and Jafarian H, 2014. Yeast colonization and drug susceptibility pattern in the pediatric patients with neutropenia. Jundishapur Journal of Microbiology, 7, 6 pp. doi: 10.5812/jjm.11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjisymeou S, Loizou P and Kothari P, 2013. Lactococcus lactis cremoris infection: not rare anymore? BMJ Case Reports, 2013, doi: 10.1136/bcr-2012-008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad MM, Darwazeh AMG and Idrees MM, 2013. The effect of glycemic control on Candida colonization of the tongue and the subgingival plaque in patients with type II diabetes and periodontitis. Oral Surgery Oral Medicine Oral Pathology Oral Radiology, 116, 321–326. [DOI] [PubMed] [Google Scholar]
- Hasiow‐Jaroszewska B, Minicka J, Borodynko N and Pospieszny H, 2015. The genetic determinants of symptoms induction by Pepino mosaic virus in tomato Proceedings of the IV International Symposium on Tomato Diseases, Acta Horticulturae. Orlando, Florida, USA, 39–43. [Google Scholar]
- Health Canada , 1999. Novel food information – food biotechnology. Virus resistant squash line CZW‐3. Available online: http://www.hc-sc.gc.ca/fn-an/alt_formats/hpfb-dgpsa/pdf/gmf-agm/ofb-098-106-a-eng.pdf
- Hillung J, Cuevas JM, Valverde S and Elena SF, 2014. Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution, 68, 2467–2480. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P and Strauss M, 1995. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proceedings of the National Academy of Sciences of the United States of America, 92, 10099–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel WH and Gerber ES, 1983. Lactobacillus divergens sp. nov., a new heterofermentative Lactobacillus species producing L(+)‐lactate. Systematic and Applied Microbiology, 4, 522–534. [DOI] [PubMed] [Google Scholar]
- Huang S, Sheng P and Zhang H, 2012. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). International Journal of Molecular Sciences, 13, 2563–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTF (International Commission on the Taxonomy of Fungi) , 2014. Yeasts Working group (affiliated with IUMS International Commission on Yeasts). Available online: http://www.fungaltaxonomy.org/subcommissions
- Idelevich EA, Pogoda CA, Ballhausen B, Wullenweber J, Eckardt L, Baumgartner H, Waltenberger J, Peters G and Becker K, 2013. Pacemaker lead infection and related bacteraemia caused by normal and small colony variant phenotypes of Bacillus licheniformis . Journal of Medical Microbiology, 62, 940–944. [DOI] [PubMed] [Google Scholar]
- Immerstrand T, Paul CJ, Rosenquist A, Deraz S, Martensson OB, Ljungh A, Blucher A, Oste R, Holst O and Karlsson EN, 2010. Characterization of the properties of Pediococcus parvulus for probiotic or protective culture use. Journal of Food Protection, 73, 960–966. [DOI] [PubMed] [Google Scholar]
- Inoue M, Saito A, Kon H, Uchida H, Koyama S, Haryu S, Sasaki T and Nishijima M, 2014. Subdural empyema due to Lactococcus lactis cremoris: case report. Neurologia Medico‐Chirurgica, 54, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Code of Nomenclature of Bacteria , 1992. International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR. and Clark WA. (eds.). ISBN‐10: 1‐55581‐039‐X. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- Ishihara Y, Kanda J, Tanaka K, Nakano H, Ugai T, Wada H, Yamasaki R, Kawamura K, Sakamoto K, Ashizawa M, Sato M, Terasako‐Saito K, Kimura S, Kikuchi M, Nakasone H, Yamazaki R, Kako S, Nishida J, Watanabe K and Kanda Y, 2014. Severe oral infection due to Lactobacillus rhamnosus during induction chemotherapy for acute myeloid leukemia. International Journal of Hematology, 100, 607–610. [DOI] [PubMed] [Google Scholar]
- Jaber JJ, Kircher ML, Thorpe E, Porter RG, Leonetti JP and Marzo SJ, 2013. Recurrent post‐tympanostomy tube otorrhea secondary to aerobic endospore‐forming bacilli: a case report and brief literature review. Ear Nose & Throat Journal, 92, 66–72. [PubMed] [Google Scholar]
- Jaimee G and Halami PM, 2016. High level aminoglycoside resistance in Enterococcus, Pediococcus and Lactobacillus species from farm animals and commercial meat products. Annals of Microbiology, 66, 101–110. [Google Scholar]
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM and Vlak JM, 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Archives of Virology, 151, 1257–1266. [DOI] [PubMed] [Google Scholar]
- Ji Y, Kim H, Park H, Lee J, Lee H, Shin H, Kim B, Franz C and Holzapfel WH, 2013. Functionality and safety of lactic bacterial strains from Korean kimchi. Food Control, 31, 467–473. [Google Scholar]
- Jögi E, Nurk A, Suitso I, Talpsep E, Naaber P and Lõivukene K, 2008. Bacillus smithii strain tbmi12 mscl p737 and use of endospores thereof as a probiotic or a food supplement. WO 2008067827 A1. European Patent Office. [Google Scholar]
- Jözefiak D, Sip A, Rawski M, Steiner T and Rutkowski A, 2011. The dose response effects of liquid and lyophilized Carnobacterium divergens AS7 bacteriocin on the nutrient retention and performance of broiler chickens. Journal of Animal and Feed Sciences, 20, 401–411. [Google Scholar]
- Jung DS, Farmakiotis D, Jiang Y, Tarrand JJ and Kontoyiannis DP, 2015. Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA. Emerging Infectious Diseases, 21, 1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoshita M, Matsumoto Y, Nishimura K, Katono Y, Murata M, Ozawa Y, Shimmura S and Tsubota K, 2015. Wickerhamomyces anomalus fungal keratitis responds to topical treatment with antifungal micafungin. Journal of Infection and Chemotherapy, 21, 141–143. [DOI] [PubMed] [Google Scholar]
- Karaaslan A, Soysal A, Sarmis A, Kadayifci EK, Cerit K, Atici S, Soyletir G and Bakir M, 2015. Lactococcus lactis catheter‐related bloodstream infection in an infant: case report. Japanese Journal of Infectious Diseases, 68, 341–342. [DOI] [PubMed] [Google Scholar]
- Karaaslan A, Soysal A, Kadayifci EK, Yakut N, Demir SO, Akkoc G, Atici S, Sarmis A, Toprak NU and Bakir M, 2016. Lactococcus lactis spp lactis infection in infants with chronic diarrhea: two cases report and literature review in children. Journal of Infection in Developing Countries, 10, 304–307. [DOI] [PubMed] [Google Scholar]
- Karanth SS, Ke V, Hasan F and Acharya V, 2014. A pregnant woman with Lactococcus lactis meningitis: to treat or not to treat? Journal of Obstetrics and Gynaecology of India, 64, 63–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartal O, Demirel F, Baysan A, Gulec M, Yesillik S, Uyanýk M, Musabak U and Sener O, 2014. An unexpected allergic reaction with Saccharomyces boulardii: a case report. Clinical and Translational Allergy, 4(Suppl. 3), P100. doi: 10.1186/2045-7022-4-S3-P100 [DOI] [Google Scholar]
- Khan Z, Ahmad S, Al‐Obaid K, Joseph L and Chandy R, 2015. Candida kefyr as a cause of bloodstream infection and adjunctive role of biomarkers in its diagnosis. Journal de Mycologie Médicale, 25, 71–75. [DOI] [PubMed] [Google Scholar]
- Khosravi AR, Shokri H, Nikaein D, Mansouri P, Erfanmanesh A, Chalangari R and Katalin M, 2013. Yeasts as important agents of onychomycosis: In vitro activity of propolis against yeasts isolated from patients with nail infection. Journal of Alternative and Complementary Medicine, 19, 57–62. [DOI] [PubMed] [Google Scholar]
- Kim DH and Austin B, 2008. Characterization of probiotic carnobacteria isolated from rainbow trout (Oncorhynchus mykiss) intestine. Letters in Applied Microbiology, 47, 141–147. [DOI] [PubMed] [Google Scholar]
- Kim J, Jang S, Kim A, Su H, Gunawardhana N, Jeon YE, Bak EJ, Kim JH and Cha JH, 2016. Role of bacterial gamma‐glutamyltranspeptidase as a novel virulence factor in bone‐resorbing pathogenesis. Journal of Microbiology, 54, 396–402. [DOI] [PubMed] [Google Scholar]
- King AMQ, Adams MJ, Carstens EB and Lefkowitz EJ, 2012. Virus Taxonomy: Classification And Nomenclature Of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA: Taxonomy updates are provided on this web site: http://ictvonline.org/ [Google Scholar]
- Kıvanç SA, Kıvanç M and Güllülü G, 2014. Automated ribotyping and antibiotic resistance determining of Bacillus spp from conjunctiva of diabetic patients. Iranian Journal of Basic Medical Sciences, 17, 138–144. [PMC free article] [PubMed] [Google Scholar]
- Kost TA and Condreay PJ, 2001. Innovations – biotechnology: baculovirus vectors as gene transfer vectors for mammalian cells: biosafety considerations. Science, 192, 167–169. [Google Scholar]
- Krishnan A and Abraham A, 2014. A case of mitral valve endocarditis caused by Lactobacillus rhamnosus in an immunocompetent patient. Internal Medicine Journal, 44, doi: 10.1111/imj.12435. [DOI] [Google Scholar]
- Krooneman J, Faber F, Alderkamp AC, Elferink SJ, Driehuis F, Cleenwerck I, Swings J, Gottschal JC and Vancanneyt M, 2002. Lactobacillus diolivorans sp. nov., a 1,2‐propanediol‐degrading bacterium isolated from aerobically stable maize silage. International Journal of Systematic and Evolutionary Microbiology, 52, 639–646. [DOI] [PubMed] [Google Scholar]
- Kuiper HA, Kleter GA, Noteborn HP and Kok EJ, 2001. Assessment of the food safety issues related to genetically modified foods. Plant Journal, 27, 503–528. [DOI] [PubMed] [Google Scholar]
- Kuiper JWP, van den Bekerom MPJ, van der Stappen J, Nolte PA and Colen S, 2013. 2‐stage revision recommended for treatment of fungal hip and knee prosthetic joint infections. Acta Orthopaedica, 84, 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW and Boekhout T, 2011. The Yeasts, a Taxonomic Study, 5th Edition Elsevier, London. 2354 pp. [Google Scholar]
- Landete JM, Ferrer S and Pardo I, 2005. Which lactic acid bacteria are responsible for histamine production in wine? Journal of Applied Microbiology, 99, 580–586. [DOI] [PubMed] [Google Scholar]
- Lapointe R, Thumbi D and Lucarotti CJ, 2012. Recent advances in our knowledge of baculovirus molecular biology and its relevance for the registration of baculovirus‐based products for insect control In: Soloneski S. (ed.). Integrated Pest Management and Pest Control – Current and Future Tactics. InTech Europe, Rijeka, Croatia: pp. 481–522. [Google Scholar]
- Laursen BG, Bay L, Cleenwerck I, Vancanneyt M, Swings J, Dalgaard P and Leisner JJ, 2005. Carnobacterium divergens and Carnobacterium maltaromaticum as spoilers or protective cultures in meat and seafood: phenotypic and genotypic characterization. Systematic and Applied Microbiology, 28, 151–164. [DOI] [PubMed] [Google Scholar]
- Lauzon HL, Gudmundsdottir S, Steinarsson A, Oddgeirsson M, Petursdottir SK, Reynisson E, Bjornsdottir R and Gudmundsdottir BK, 2010. Effects of bacterial treatment at early stages of Atlantic cod (Gadus morhua L.) on larval survival and development. Journal of Applied Microbiology, 108, 624–632. [DOI] [PubMed] [Google Scholar]
- Lee JY, Seo MY, Yang J, Kim K, Chang H, Kim SC, Kim M‐G, Jo S‐K, Cho W and Kim HK, 2014a. Polymicrobial peritonitis with Lactococcus lactis in a peritoneal dialysis patient. Chonnam Medical Journal, 50, 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Kim SK, Li XH and Lee JH, 2014b. Bacterial virulence analysis using brine shrimp as an infection model in relation to the importance of quorum sensing and proteases. Journal of General and Applied Microbiology, 60, 169–174. [DOI] [PubMed] [Google Scholar]
- Lee MR, Tsai CJ, Liang SK, Lin CK, Huang YT and Hsueh PR, 2015. Clinical characteristics of bacteraemia caused by Lactobacillus spp. and antimicrobial susceptibilities of the isolates at a medical centre in Taiwan, 2000–2014. International Journal of Antimicrobial Agents, 46, 439–445. [DOI] [PubMed] [Google Scholar]
- Leisner JJ, Laursen BG, Prevost H, Drider D and Dalgaard P, 2007. Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiology Reviews, 31, 592–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuck AM, Johnson JR, Hunt MA, Dhody K, Kazempour K, Ferrieri P and Kline S, 2015. Safety and efficacy of a novel silver‐impregnated urinary catheter system for preventing catheter‐associated bacteriuria: a pilot randomized clinical trial. American Journal of Infection Control, 43, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner RGK, Robinson TP, Hugas M, Cocconcelli PS, Richard‐Forget F, Klein G, Licht TR, Nguyen‐The C, Querol A, Richardson M, Suarez JE, Thrane U, Vlak JM and von Wright A, 2010. Qualified presumption of safety (QPS): a generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA). Trends in Food Science & Technology, 21, 425–435. [Google Scholar]
- Li ZX, Bian ZS, Zheng HP, Yue YS, Yao JY, Gong YP, Cai MY and Dong XZ, 1990. First isolation of Xanthomonas campestris from the blood of a Chinese woman. Chinese Medical Journal (Engl.), 103, 435–439. [PubMed] [Google Scholar]
- Li D, Zhang WF, Zheng S, Ma ZY, Zhang P and Liu Z, 2013a. Surveillance study of candidemia in cancer patients in North China. Medical Mycology, 51, 378–384. [DOI] [PubMed] [Google Scholar]
- Li QR, Wang CY, Tang C, He Q, Li N and Li JS, 2013b. Bacteremia in patients with acute pancreatitis as revealed by 16S Ribosomal RNA gene‐based techniques. Critical Care Medicine, 41, 1938–1950. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen W, Li X, Li H, Li H, Wang L, He L, Yang X, Wang X, Huang Y and Yao Y, 2013c. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV‐infected patients in Kunming, Yunnan province of China: Candida carriage in Chinese HIV patients. BMC Infectious Diseases, 13, accessed 28 January 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olsen RH, Ye L, Yan H, Nie Q, Meng HC and Shi L, 2016. Antimicrobial resistance and resistance genes in aerobic bacteria isolated from pork at Slaughter. Journal of Food Protection, 79, 589–597. [DOI] [PubMed] [Google Scholar]
- Lin HC, Lin HY, Su BH, Ho MW, Ho CM, Lee CY, Lin MH, Hsieh HY, Lin HC, Li TC, Hwang KP and Lu JJ, 2013. Reporting an outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. Journal of Microbiology Immunology and Infection, 46, 456–462. [DOI] [PubMed] [Google Scholar]
- Lin CC, Liu CP, Hsieh FC, Lee CM and Wang WS, 2015. Antimicrobial susceptibility and clinical outcomes of Candida parapsilosis bloodstream infections in a tertiary teaching hospital in Northern Taiwan. Journal of Microbiology Immunology and Infection, 48, 552–558. [DOI] [PubMed] [Google Scholar]
- Lindström C, Xu J, Oste R, Holst O and Molin G, 2013. Oral administration of live exopolysaccharide‐producing Pediococcus parvulus, but not purified exopolysaccharide, suppressed Enterobacteriaceae without affecting bacterial diversity in ceca of mice. Applied and Environmental Microbiology, 79, 5030–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XP, Fan SR, Peng YT and Zhang HP, 2014. Species distribution and susceptibility of Candida isolates from patient with Vulvovaginal candidiasis in Southern China from 2003 to 2012. Journal de Mycologie Médicale, 24, 106–111. [DOI] [PubMed] [Google Scholar]
- Llopis S, Hernandez‐Haro C, Monteoliva L, Querol A, Molina M and Fernandez‐Espinar MT, 2014. Pathogenic potential of Saccharomyces Strains isolated from dietary supplements. PLoS ONE, 9, doi: 10.1371/journal.pone.0098094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López AC, Minnaard J, Pérez PF and Alippi AM, 2013. In vitro interaction between Bacillus megaterium strains and Caco‐2 cells. International Microbiology, 16, 27–33. [DOI] [PubMed] [Google Scholar]
- Lücking G, Stoeckel M, Atamer Z, Hinrichs J and Ehling‐Schulz M, 2013. Characterization of aerobic spore‐forming bacteria associated with industrial dairy processing environments and product spoilage. International Journal of Food Microbiology, 166, 270–279. [DOI] [PubMed] [Google Scholar]
- Luo C, Hang XM, Liu XL, Zhang M, Yang X and Yang H, 2015. Detection of erm(X)‐mediated antibiotic resistance in Bifidobacterium longum subsp longum . Annals of Microbiology, 65, 1985–1991. [Google Scholar]
- Luzzati R, Cavinato S, Giangreco M, Grana G, Centonze S, Deiana ML, Biolo G and Barbone F, 2013. Peripheral and total parenteral nutrition as the strongest risk factors for nosocomial candidemia in elderly patients: a matched case‐control study. Mycoses, 56, 664–671. [DOI] [PubMed] [Google Scholar]
- Madhumati B, Rani KL, Murthy DS, Reddy BKN and Ramana BV, 2014. Species distribution and antifungal susceptibility profile in vaginal candidiasis. International Journal of Pharmaceutical Research and Bio‐Science, 3, 497–502. [Google Scholar]
- Madslien EH, Rønning HT, Lindbäck T, Hassel B, Andersson MA and Granum PE, 2013. Lichenysin is produced by most Bacillus licheniformis strains. Journal of Applied Microbiology, 115, 1068–1080. [DOI] [PubMed] [Google Scholar]
- Martinez RM, Hulten KG, Bui U and Clarridge JE, 2014. Molecular analysis and clinical significance of Lactobacillus spp. Recovered from clinical specimens presumptively associated with disease. Journal of Clinical Microbiology, 52, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martos C, Muñoz P, Guinea J, Pelaez T, Marcos‐Zambrano LJ, Escribano P and Bouza E and COMIC Study Group , 2014. Fungemia caused by rare yeast species in a Spanish General Hospital. Abstracts of the Interscience Conference on Antimicrobial Agents and Chemotherapy, 54, M‐1092. [Google Scholar]
- Masri SN, Noor SM, Nor LAM, Osman M and Rahman MM, 2015. Candida isolates from pregnant women and their antifungal susceptibility in a Malaysian tertiary‐care hospital. Pakistan Journal of Medical Sciences, 31, 658–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Burdet HM, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prd'homme van Reine WF, Smith GF, Wiersema J and Turland N, 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Koeltz Scientific Books, 140 pp. Available online: http://herbario.udistrital.edu.co/herbario/images/stories/international%20code%20of%20nomenclature.pdf [Google Scholar]
- McWilliam A, 2007. Environmental impact of baculoviruses. FAO Report 7299. 34 pp. Available online: http://www.fao.org/docs/eims/upload/agrotech/2003/R7299_FTR_anx3.pdf
- Mehta A, Rangarajan S and Borate U, 2013. A cautionary tale for probiotic use in hematopoietic SCT patients‐Lactobacillus acidophilus sepsis in a patient with mantle cell lymphoma undergoing hematopoietic SCT. Bone Marrow Transplantation, 48, 461–462. [DOI] [PubMed] [Google Scholar]
- Meini S, Laureano R, Fani L, Tascini C, Galano A, Antonelli A and Rossolini GM, 2015. Breakthrough Lactobacillus rhamnosus GG bacteremia associated with probiotic use in an adult patient with severe active ulcerative colitis: case report and review of the literature. Infection, 43, 777–781. [DOI] [PubMed] [Google Scholar]
- Menezes RD, Ferreira JC, de Sa WM, Moreira TD, Malvino LDS, de Araujo LB, Roeder DVD, Penatti MPA, Candido RC and Pedroso RD, 2015. Frequency of Candida species in a tertiary care hospital in Triangulo Mineiro, Minas Gerais State, Brazil. Revista do Instituto de Medicina Tropical de São Paulo, 57, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesas JM, Rodriguez MC and Alegre MT, 2011. Characterization of lactic acid bacteria from musts and wines of three consecutive vintages of Ribeira Sacra. Letters in Applied Microbiology, 52, 258–268. [DOI] [PubMed] [Google Scholar]
- Meyer SA and Yarrow D, 1998. Validation of the names of three Candida species. Mycotaxon, 66, 99–101. [Google Scholar]
- Meziane‐Cherif D, Decré D, Høiby EA, Courvalin P and Périchon B, 2008. Genetic and biochemical characterization of CAD‐1, a chromosomally encoded new class A penicillinase from Carnobacterium divergens . Antimicrobial Agents and Chemotherapy, 52, 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MH, Díaz JA and Lee SA, 2011. Emerging opportunistic yeast infections. Lancet Infectious Diseases, 11, 142–151. [DOI] [PubMed] [Google Scholar]
- Minea B, Nastasa V, Kolecka A, Mares M, Marangoci N, Rosca I, Pinteala M, Hancianu M and Mares M, 2016. Etiologic agents and antifungal susceptibility of oral candidosis from Romanian patients with HIV‐infection or type 1 diabetes mellitus . Polish Journal of Microbiology, 65, 123–129. [DOI] [PubMed] [Google Scholar]
- Minicka J, Rymelska N, Elena SF, Czerwoniec A and Hasiow‐Jaroszewska B, 2015. Molecular evolution of Pepino mosaic virus during long‐term passaging in different hosts and its impact on virus virulence. Annals of Applied Biology, 166, 389–401. [Google Scholar]
- Mohammadi R, Mirhendi H, Rezaei‐Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N and Makimura K, 2013. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Medical Mycology, 51, 657–663. [DOI] [PubMed] [Google Scholar]
- Mohammadou BA, Gl B, Mbofung CM and Barbier G, 2014. Antimicrobial activities, toxinogenic potential and sensitivity to antibiotics of Bacillus strains isolated from Mbuja, an Hibiscus sabdariffa fermented seeds from Cameroon. African Journal of Biotechnology, 13, 3617–3627. [Google Scholar]
- Moher D, Liberati A, Tetzlaff J and Altman DG for the PRIMS Group, 2009. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. British Medical Journal, 338, b2535. doi: 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna MT, Lovero G, Coretti C, De Giglio O, Martinelli D, Bedini A, Delia M, Rosato A, Codeluppi M and Caggiano G, 2014. In vitro activities of amphotericin B deoxycholate and liposomal amphotericin B against 604 clinical yeast isolates. Journal of Medical Microbiology, 63, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muadcheingka T and Tantivitayakul P, 2015. Distribution of Candida albicans and non‐albicans Candida species in oral candidiasis patients: correlation between cell surface hydrophobicity and biofilm forming activities. Archives of Oral Biology, 60, 894–901. [DOI] [PubMed] [Google Scholar]
- Mun YS, Lee MS, Park JS, Lee JW, Jung SY, Yoon HJ and Han HY, 2015. An unusual case of candidemia presenting as acute respiratory distress syndrome after a small bowel bezoar removal operation. Annals of Surgical Treatment and Research, 88, 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei T, Inai S, Mikami I, Sato A, Okamoto J, Yokoshima K, Nakamizo M, Haraguchi S, Sonobe K and Saito R, 2013. Descending necrotizing mediastinitis associated with Lactobacillus plantarum . BMC Infectious Diseases, 13, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby B and Ramesh KK, 2014. Urinary tract infection in a preterm neonate caused by Lactococcus lactis . The Canadian Journal of Hospital Pharmacy, 67, 453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi A, Manzoor S, Bejai S, Meijer J and Bongcam‐Rudloff E, 2014. Complete genome sequence of a plant associated bacterium Bacillus amyloliquefaciens subsp. plantarum UCMB5033. Standards in Genomic Sciences, 9, 718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidhi W, Singh SM, Nawange SR and Shruti S, 2015. Spectrum of opportunistic fungal infections in cancer/HIV patients: emerging fungal pathogens from Jabalpur Madhya Pradesh Central India. Scholars Journal of Applied Medical Sciences, 3, 1385–1390. [Google Scholar]
- Nieto MC, Telleria O and Cisterna R, 2015. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagnostic Microbiology and Infectious Disease, 81, 34–40. [DOI] [PubMed] [Google Scholar]
- Niyonzima FN and More SS, 2014. Concomitant production of detergent compatible enzymes by Bacillus flexus XJU‐1. Brazilian Journal of Microbiology, 45, 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel GR, Atibalentja N and Domier LL, 2005. Emended description of Pasteuria nishizawae . International Journal of Systematic and Evolutionary Microbiology, 55, 1681–1685. [DOI] [PubMed] [Google Scholar]
- OECD (Organisation for Economic Co‐operation and Development), 2002. Consensus document on information used in the assessment of environmental applications involving baculoviruses. Series on Harmonisation of Regulatory Oversight in Biotechnology, 20. ENV/JM/MONO(2002)1. 90 pp. Available online: http://www.oecd.org/science/biotrack/46815698.pdf
- O'Flynn NMJ, Patel A, Kadlec J and Jones IM, 2013. Improving promiscuous mammalian cell entry by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. Bioscience Reports, 33, 23–U221. doi: 10.1042/bsr20120093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira VKP, Ruiz LD, Oliveira NAJ, Moreira D, Hahn RC, Melo ASD, Nishikaku AS and Paula CR, 2014. Fungemia caused by Candida species in a children's public hospital in the city of São Paulo, Brazil: Study in the period 2007–2010. Revista do Instituto de Medicina Tropical de São Paulo, 56, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A and Garrity GM, 2014. List of new names and new combinations previously effectively, but not validly, published. International Journal of Systematic and Evolutionary Microbiology, 64, 3603–3606. [DOI] [PubMed] [Google Scholar]
- Oren A and Garrity GM, 2016. Notification of changes in taxonomic opinion previously published outside the IJSEM. International Journal of Systematic and Evolutionary Microbiology, 66, 7–8. [DOI] [PubMed] [Google Scholar]
- Pal KC, Mondal NK, Chatterjee S, Ghosh TS and Datta JK, 2014. Characterization of fluoride‐tolerant halophilic Bacillus flexus NM25 (HQ875778) isolated from fluoride‐affected soil in Birbhum District, West Bengal, India. Environmental Monitoring and Assessment, 186, 699–709. [DOI] [PubMed] [Google Scholar]
- Palaniraj A and Jayaraman V, 2011. Production, recovery and applications of xanthan gum by Xanthomonas campestris . Journal of Food Engineering, 106, 1–12. [Google Scholar]
- Park HY, Kim KK, Jin L and Lee ST, 2006. Microbacterium paludicola sp. nov., a novel xylanolytic bacterium isolated from swamp forest. International Journal of Systematic and Evolutionary Microbiology, 56, 535–539. [DOI] [PubMed] [Google Scholar]
- Park MJ, Kim MK, Kim HB, Im WT, Yi TH, Kim SY, Soung NK and Yang DC, 2008. Microbacterium ginsengisoli sp. nov., a beta‐glucosidase‐producing bacterium isolated from soil of a ginseng field. International Journal of Systematic and Evolutionary Microbiology, 58, 429–433. [DOI] [PubMed] [Google Scholar]
- Pariza MW, Gillies KO, Kraak‐Ripple SF, Leyer G and Smith AB 2015. Determining the safety of microbial cultures for consumption by humans and animals. Regulatory Toxicology and Pharmacology, 73, 164–171. [DOI] [PubMed] [Google Scholar]
- Parmeland L, Gazon M, Guerin C, Argaud L, Lehot JJ, Bastien O, Allaouchiche B, Michallet M, Picot S, Bienvenu AL and Study G , 2013. Candida albicans and non‐Candida albicans fungemia in an institutional hospital during a decade. Medical Mycology, 51, 33–37. [DOI] [PubMed] [Google Scholar]
- Pérez‐Torrado R and Querol A, 2015. Opportunistic strains of Saccharomyces cerevisiae: a potential risk sold in food products. Frontiers in Microbiology, 6, 5 pp. doi: 10.3389/fmicb.2015.01522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Torrado R, Llopis S, Perrone B, Gomez‐Pastor R, Hube B and Querol A, 2015. Comparative genomic analysis reveals a critical role of De Novo Nucleotide biosynthesis for Saccharomyces cerevisiae virulence. PLoS ONE, 10, doi: 10.1371/journal.pone.0122382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri A, Pfannebecker J, Frohlich J and Konig H, 2013. Fast identification of wine related lactic acid bacteria by multiplex PCR. Food Microbiology, 33, 48–54. [DOI] [PubMed] [Google Scholar]
- Pillai U, Devasahayam J, Kurup AN and Lacasse A, 2014. Invasive Saccharomyces cerevisiae infection: a friend turning foe? Saudi Journal of Kidney Diseases and Transplantation, 25, 1266–1269. [DOI] [PubMed] [Google Scholar]
- Pisa D, Alonso R, Rabano A, Rodal I and Carrasco L, 2015. Different brain regions are infected with fungi in Alzheimer's Disease. Scientific Reports, 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumed‐Ferrer C, Uusikyla K, Korhonen J and von Wright A, 2013. Characterization of Lactococcus lactis isolates from bovine mastitis. Veterinary Microbiology, 167, 592–599. [DOI] [PubMed] [Google Scholar]
- Plumed‐Ferrer C, Barberio A, Franklin‐Guild R, Werner B, McDonough P, Bennett J, Gioia G, Rota N, Welcome F, Nydam DV and Moroni P, 2015. Antimicrobial susceptibilities and random amplified polymorphic DNA‐PCR fingerprint characterization of Lactococcus lactis ssp lactis and Lactococcus garvieae isolated from bovine intramammary infections. Journal of Dairy Science, 98, 6216–6225. [DOI] [PubMed] [Google Scholar]
- Popiel KY, Wong P, Lee MJ, Langelier M, Sheppard DC and Vinh DC, 2015. Invasive Saccharomyces cerevisiae in a liver transplant patient: case report and review of infection in transplant recipients. Transplant Infectious Disease, 17, 435–441. [DOI] [PubMed] [Google Scholar]
- Preston JF, Dickson DW, Maruniak JE, Nong G, Brito JA, Schmidt LM and Giblin‐Davis RM, 2003. Pasteuria spp.: systematics and phylogeny of these bacterial parasites of phytopathogenic nematodes. Journal of Nematology, 35, 198–207. [PMC free article] [PubMed] [Google Scholar]
- Priest FG, Goodfellow M and Todd C, 1988. A numerical classification of the genus Bacillus . Journal of General Microbiology, 134, 1847–1882. [DOI] [PubMed] [Google Scholar]
- Rai MM, Gore DG, Rathod MK and Khurad AM, 2013. Evidence of transovarial transmission of Bacillus subtilis in the silkworm, Bombyx mori L. Journal of Pharmacy Research, 7, 318–323. [Google Scholar]
- Renouf V, Claisse O and Lonvaud‐Funel A, 2007. Inventory and monitoring of wine microbial consortia. Applied Microbiology and Biotechnology, 75, 149–164. [DOI] [PubMed] [Google Scholar]
- Riat A, Rentenaar RJ, van Drongelen AM, Barras V, Bertens LCM, Vlek ALM, Doppenberg E, Weersink AJL, Reinders E, Vlaminckx BJM, Overbeeke N, van Burgel ND, Peterse N, Bosboom R, Boekhout T, Schrenzel J and Kusters JG, 2015. Ground steel target plates in combination with direct transfer of clinical Candida isolates improves frequencies of species‐level identification by matrix‐assisted laser desorption ionization‐time of flight mass spectrometry in comparison with polished steel target plates. Journal of Clinical Microbiology, 53, 1993–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro ALR, Menezes TOD, Alves SD, de Menezes SAF, Marques‐da‐Silva SH and Vallinoto ACR, 2015. Oral carriage of Candida species in HIV‐infected patients during highly active antiretroviral therapy (HAART) in Belém, Brazil. Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology, 120, 29–33. [DOI] [PubMed] [Google Scholar]
- Richard C, Brillet A, Pilet MF, Prevost H and Drider D, 2003. Evidence on inhibition of Listeria monocytogenes by divercin V41 action. Letters in Applied Microbiology, 36, 288–292. [DOI] [PubMed] [Google Scholar]
- Rieder G, Krisch L, Fischer H, Kaufmann M, Maringer A and Wessler S, 2012. Carnobacterium divergens – a dominating bacterium of pork meat juice. FEMS Microbiology Letters, 332, 122–130. [DOI] [PubMed] [Google Scholar]
- Rihakova J, Belguesmia Y, Petit VW, Pilet MF, Prevost H, Dousset X and Drider D, 2009. Divercin V41 from gene characterization to food applications: 1998–2008, a decade of solved and unsolved questions. Letters in Applied Microbiology, 48, 1–7. [DOI] [PubMed] [Google Scholar]
- Ringø E, Salinas I, Olsen RE, Nyhaug A, Myklebust R and Mayhew TM, 2007. Histological changes in intestine of Atlantic salmon (Salmo salar L.) following in vitro exposure to pathogenic and probiotic bacterial strains. Cell & Tissue Research, 328, 109–116. [DOI] [PubMed] [Google Scholar]
- Rivas R, Trujillo ME, Sanchez M, Mateos PF, Martinez‐Molina E and Velazquez E, 2004. Microbacterium ulmi sp. nov., a xylanolytic, phosphate‐solubilizing bacterium isolated from sawdust of Ulmus nigra . International Journal of Systematic and Evolutionary Microbiology, 54, 513–517. [DOI] [PubMed] [Google Scholar]
- Roig P, de Llanos R, Gil JV and Fernandez‐Espinar MT, 2013. FLO11 expression in clinical and non‐clinical Saccharomyces cerevisiae strains and its association with virulence. Annals of Microbiology, 63, 1423–1431. [Google Scholar]
- Rojo‐Bezares B, Sáenz Y, Poeta P, Zarazaga M, Ruiz‐Larrea F and Torres C, 2006. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. International Journal of Food Microbiology, 111, 234–240. [DOI] [PubMed] [Google Scholar]
- Rostagno C, Pecile P and Stefano PL, 2013. Early Lactococcus lactis endocarditis after mitral valve repair: a case report and literature review. Infection, 41, 897–899. [DOI] [PubMed] [Google Scholar]
- Sadashiv SO and Kaliwal BB, 2014. Isolation, characterization and antibiotic resistance of Bacillus sps. from bovine mastitis in the region of north Karnataka, India. International Journal of Current Microbiology and Applied Sciences, 3, 360–373. [Google Scholar]
- Sadowska‐Krawczenko I, Paprzycka M, Korbal P, Wiatrzyk A, Krysztopa‐Grzybowska K, Polak M, Czajka U and Lutynska A, 2014. Lactobacillus rhamnosus GG suspected infection in a newborn with intrauterine growth restriction. Beneficial Microbes, 5, 397–402. [DOI] [PubMed] [Google Scholar]
- Sahin SZ, Akalin H, Ersoy A, Yildiz A, Ocakoglu G, Cetinoglu ED, Dizdar OS, Kazak E and Ener B, 2015. Invasive fungal infections in renal transplant recipients: epidemiology and risk factors. Mycopathologia, 180, 43–50. [DOI] [PubMed] [Google Scholar]
- Santino I, Alari A, Bono S, Teti E, Marangi M, Bernardini A, Magrini L, Di Somma S and Teggi A, 2014. Saccharomyces cerevisiae fungemia, a possible consequence of the treatment of Clostridium difficile colitis with a probioticum. International Journal of Immunopathology and Pharmacology, 27, 143–146. [DOI] [PubMed] [Google Scholar]
- Sarbu I, Pelinescu D, Stoica I, Marutescu L and Vassu T, 2013. Phenotypic profiles of virulence in different Candida species isolated from vulvovaginal infections. Roumanian Archives of Microbiology and Immunology, 72, 225–233. [PubMed] [Google Scholar]
- Sarvtin MT, Hedayati MT, Abastabar M and Shokohi T, 2014. Debaryomyces hansenii colonization and its protein profile in psoriasis. Iranian Journal of Dermatology, 17, 134–137. [Google Scholar]
- Sasikala G, Agatha D, Janagond AB and Thenmozhivalli PR, 2013. Characterization of Candida and its antifungal susceptibility pattern from patients with vaginal candidiasis in a tertiary care hospital in South India. Journal of Pharmaceutical and Biomedical Sciences, May (Supplement 1); 30(30), S1–S6. [Google Scholar]
- Sayre RM, Wergin WP, Schmidt JM and Starr MP, 1991. Pasteuria nishizawae sp. nov., a mycelial and endospore‐forming bacterium parasitic on cyst nematodes of genera Heterodera and Globodera . Research in Microbiology, 142, 551–564. [DOI] [PubMed] [Google Scholar]
- Schyns G, Serra CR, Lapointe T, Pereira‐Leal JB, Potot S, Fickers P, Perkins JB, Wyss M and Henriques AO, 2013. Genome of a gut strain of Bacillus subtilis . Genome Announcements, 1, e00184‐00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoppettuolo G, Donato C, De Carolis E, Vella A, Vaccaro L, La Greca A and Fantoni M, 2014. Candida utilis catheter‐related bloodstream infection. Medical Mycology Case Reports, 6, 70–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivamurthy V, Reilly C, Gantt S, Guzman J and Tucker L, 2015. Bacillus pumilus; A rare cause of paediatric septic arthritis. Case report. Journal of Rheumatology, 42, 1341–1341. [Google Scholar]
- Shweta J and Joseph E, 2013. Identification of mastitis pathogens and their antibiogram. Veterinary Practitioner, 14, 318–319. [Google Scholar]
- Shyamala R and Parandekar PK, 2014. Identification and in vitro azole resistance of Candida species isolated from oropharyngeal candidiasis in human immunodeficiency virus infected patients. International Journal of Current Microbiology and Applied Sciences, 3, 816–822. [Google Scholar]
- Singh RP, Shukla MK, Mishra A, Reddy CR and Jha B, 2013. Bacterial extracellular polymeric substances and their effect on settlement of zoospore of Ulva fasciata . Colloids and Surfaces B: Biointerfaces, 103, 223–230. [DOI] [PubMed] [Google Scholar]
- Sivaprakasam S, Mahadevan S, Sekar S and Rajakumar S, 2008. Biological treatment of tannery wastewater by using salt‐tolerant bacterial strains. Microbial Cell Factories, 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V and Sneath PHA, 1980. Approved lists of bacterial names. International Journal of Systematic Bacteriology, 30, 225–420. [Google Scholar]
- Songisepp E, Hutt P, Ratsep M, Shkut E, Koljalg S, Truusalu K, Stsepetova J, Smidt I, Kolk H, Zagura M and Mikelsaar M, 2012. Safety of a probiotic cheese containing Lactobacillus plantarum Tensia according to a variety of health indices in different age groups. Journal of Dairy Science, 95, 5495–5509. [DOI] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS and McCusker JH, 2015. The 100‐genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Research, 25, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suitso I, Jögi E, Talpsep E, Naaber P and Nurk A, 2007. P1763 Bacillus smithii TBMI12 spores as a potential competitive exclusion agent against Salmonella enteritidis . International Journal of Antimicrobial Agents, 29, S501. doi: 10.1016/S0924-8579(07)71602-3 [DOI] [Google Scholar]
- Sun Y, Gautam A and Miller N, 2015a. Lactobacillus gasseri associated with urinary and surgical wound infection in a renal transplant patient. American Journal of Clinical Pathology, 144, A206. [Google Scholar]
- Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Paul Ross R, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H and O'Toole PW, 2015b. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nature Communications, 6, 8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwantarat N, Romagnoli M, Wakefield T and Carroll KC, 2014. Ventriculoperitoneal shunt infection caused by Bifidobacterium breve . Anaerobe, 28, 1–3. [DOI] [PubMed] [Google Scholar]
- Svobodova L, Bednarova D, Ruzicka F, Chrenkova V, Dobias R, Mallatova N, Buchta V, Kocmanova I, Olisarova P, Stromerova N, Thongsri Y and Hamal P, 2016. High frequency of Candida fabianii among clinical isolates biochemically identified as Candida pelliculosa and Candida utilis . Mycoses, 59, 241–246. [DOI] [PubMed] [Google Scholar]
- Swarajyalakshmi M and Jyothilakshmi G, 2014. Candida kefyr in invasive paranasal sinusitis. Indian Journal of Otolaryngology and Head and Neck Surgery, 66, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift SL, Rivera GC, Dussupt V, Leadley RM, Hudson LC, de Ridder CMA, Kraaij R, Burns JE, Maitland NJ and Georgopoulos LJ, 2013. Evaluating baculovirus as a vector for human prostate cancer gene therapy. PLoS ONE, 8, doi: 10.1371/journal.pone.0065557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj‐Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF and Boekhout T, 2014. Uncommon opportunistic yeast bloodstream infections from Qatar. Medical Mycology, 52, 549–553. [DOI] [PubMed] [Google Scholar]
- Takaç S, Unlu AE and Erdem B, 2010. Oxygen transfer strategy modulates the productions of lipase and esterase enzymes by Candida rugosa . Journal of Molecular Catalysis B‐Enzymatic, 64, 150–154. [Google Scholar]
- Takashima Y, Kawabe T and Mitsuda S, 2000. Factors affecting the production of nitrile hydratase by thermophilic Bacillus smithii SC‐J05‐1. Journal of Bioscience and Bioengineering, 89, 282–284. [DOI] [PubMed] [Google Scholar]
- Takeuchi M and Yokota A, 1994. Phylogenetic analysis of the genus Microbacterium based on 16S rRNA gene sequences. FEMS Microbiology Letters, 124, 11–16. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Nakayama M, Nakahira K, Nakura Y, Kanagawa N, Yanagihara I and Miyaishi S, 2016. Sudden infant death due to Lactococcal infective endocarditis. Legal Medicine, 19, 107–111. [DOI] [PubMed] [Google Scholar]
- Tao F, Wang X, Ma C, Yang C, Tang H, Gai Z and Xu P, 2012. Genome sequence of Xanthomonas campestris JX, an industrially productive strain for Xanthan gum. Journal of Bacteriology, 194, 4755–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena D, Martinez NM, Losa C, Fernandez C, Medina MJ and Saez‐Nieto JA, 2013. Acute acalculous cholecystitis complicated with peritonitis caused by Lactobacillus plantarum . Diagnostic Microbiology and Infectious Disease, 76, 510–512. [DOI] [PubMed] [Google Scholar]
- Tena D, Losa C, Medina MJ and Saez‐Nieto JA, 2014. Peritonitis caused by Bifidobacterium longum: case report and literature review. Anaerobe, 27, 27–30. [DOI] [PubMed] [Google Scholar]
- Tian B, Yang J and Zhang KQ, 2007. Bacteria used in the biological control of plant‐parasitic nematodes: populations, mechanisms of action, and future prospects. FEMS Microbiology Ecology, 61, 197–213. [DOI] [PubMed] [Google Scholar]
- Tomizuka N, Ota Y and Yamada K, 1966. Studies on Lipase from Candida cylindracea Part I. Purification and properties. Agricultural and Biological Chemistry, 30, 576–584. [Google Scholar]
- Tohno M, Kobayashi H, Nomura M, Kitahara M, Ohkuma M, Uegaki R and Cai Y, 2012. Genotypic and phenotypic characterization of lactic acid bacteria isolated from Italian ryegrass silage. Animal Science Journal, 83, 111–120. [DOI] [PubMed] [Google Scholar]
- Trotter JR and Bishop AH, 2003. Phylogenetic analysis and confirmation of the endospore‐forming nature of Pasteuria penetrans based on the spo0A gene. FEMS Microbiology Letters, 225, 249–256. [DOI] [PubMed] [Google Scholar]
- Tzar MN, Norazlah B and Shamsul AS, 2015. Risk factors for Candidaemia in a Malaysian Tertiary Hospital. Sains Malaysiana, 44, 735–740. [Google Scholar]
- Uçar AD, Ergin ÖY, Avcı M, Arı A, Yıldırım M and Erkan N, 2016. Bacillus flexus outbreak in a tertiary burn center. Burns, 42, 948–949. [DOI] [PubMed] [Google Scholar]
- Ucko M and Colorni A, 2014. Infections by lactic acid bacteria in marine fish from southern Israel (Red Sea): new records. Israeli Journal of Aquaculture – Bamidgeh, 66, 11 pp. Available online: https://evols.library.manoa.hawaii.edu/bitstream/10524/49112/1/IJA_66.2014.939.Colorni.pdf [Google Scholar]
- US EPA (US Environmental Protection Agency), 2012. Pasteuria nishizawae‐Pn1; exemption from the requirement of a tolerance. Federal Register; 77: 8736–8741. Available online: http://www.gpo.gov/fdsys/pkg/FR-2012-02-15/html/2012-3586.htm [Google Scholar]
- Usta‐Atmaca H, Akbas F, Karagoz Y and Piskinpasa ME, 2015. A rarely seen cause for empyema: Leuconostoc mesenteroides . Journal of Infection in Developing Countries, 9, 425–427. [DOI] [PubMed] [Google Scholar]
- Vahabnezhad E, Mochon AB, Wozniak LJ and Ziring DA, 2013. Lactobacillus Bacteremia associated with probiotic use in a pediatric patient with Ulcerative Colitis. Journal of Clinical Gastroenterology, 47, 437–439. [DOI] [PubMed] [Google Scholar]
- Vauterin L, Hoste B, Kersters K and Swings J, 1995. Reclassification of Xanthomonas . International Journal of Systematic Bacteriology, 45, 472–489. [Google Scholar]
- Vicente JG and Holub EB, 2013. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Molecular Plant Pathology, 14, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu DW, Han H, Yue JF, Zhang F, Shan TC, Guo HP and Yin M, 2014. Antibiotics exposure, risk factors, and outcomes with Candida albicans and non‐Candida albicans candidemia results from a multi‐center study. Saudi Medical Journal, 35, 153–158. [PubMed] [Google Scholar]
- Wassenaar TM and Klein G, 2008. Safety aspects and implications of regulation of probiotic bacteria in food and food supplements. Journal of Food Protection, 71, 1734–1741. [DOI] [PubMed] [Google Scholar]
- Wenzler E, Kamboj K and Balada‐Llasat, JM , 2015. Severe sepsis secondary to persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus bacteremia. International Journal of Infectious Diseases, 35, 93–95. [DOI] [PubMed] [Google Scholar]
- Werner B, Moroni P, Gioia G, Lavin‐Alconero L, Yousaf A, Charter ME, Carter BM, Bennett J, Nydam DV, Welcome F and Schukken YH, 2014. Short communication: Genotypic and phenotypic identification of environmental streptococci and association of Lactococcus lactis ssp. lactis with intramammary infections among different dairy farms. Journal of Dairy Science, 97, 6964–6969. [DOI] [PubMed] [Google Scholar]
- Wu C, Zhou X, Xu Y, Li H, Tian Y, Xu X and Jin Z, 2014. Characterization and mechanism of action of Microbacterium imperiale glucan 1,4‐alpha‐maltotriohydrolase. Carbohydrate Research, 384, 46–50. [DOI] [PubMed] [Google Scholar]
- Yamada K and Machida H, 1962. Studies on the production of lipase by microorganisms. Part I. The selection and identification of a new strain (in Japanese). Nippon Nogeikagaku Kaishii, 36, 858–860. [Google Scholar]
- Yang TC, Tsai MJ, Tsai JJ and Hu RM, 2011. Induction of a secretable beta‐lactamase requires a long lag time in Xanthomonas campestris pv. campestris str. 17. Research in Microbiology, 162, 999–1005. [DOI] [PubMed] [Google Scholar]
- Yang TC, Chen TF, Tsai JJ and Hu RM, 2014. NagZ is required for beta‐lactamase expression and full pathogenicity in Xanthomonas campestris pv. campestris str. 17. Research in Microbiology, 165, 612–619. [DOI] [PubMed] [Google Scholar]
- Yang C, Wang D, Zhou Q and Xu JC, 2015. Bacteremia due to vancomycin‐resistant Leuconostoc lactis in a patient with pneumonia and abdominal infection. American Journal of the Medical Sciences, 349, 282–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit N and Aktas E, 2014. Activities of amphotericin B, fluconazole and voriconazole against Candida bloodstream isolates determined by broth microdilution and disk diffusion methods. Turk Hijyen ve Deneysel Biyoloji Dergisi, 71, 131–140. [Google Scholar]
- Yoo J‐G, Chang J‐H, Kim S, Ji J‐Y, Hong S‐W, Park B‐Y and Oh M‐H, 2014. Analysis of emetic toxin production by Bacillus species using cellular cytotoxicity, molecular, and chromatographic assays. Biotechnology and Bioprocess Engineering, 19, 978–983. [Google Scholar]
- Youngster I, Sharma TS, Duncan CN and McAdam AJ, 2014. Yield of fungal surveillance cultures in pediatric hematopoietic stem cell transplant patients: a retrospective analysis and survey of current practice. Clinical Infectious Diseases, 58, 365–371. [DOI] [PubMed] [Google Scholar]
- Yüceer O and Özden Tuncer B, 2015. Determination of antibiotic resistance and biogenic amine production of lactic acid bacteria isolated from fermented Turkish sausage (sucuk). Journal of Food Safety, 35, 276–285. [Google Scholar]
- Yusibov V, Streatfield SJ, Kushnir N, Roy G and Padmanaban A, 2013. Hybrid viral vectors for vaccine and antibody production in plants. Current Pharmaceutical Design, 19, 5574–5586. [DOI] [PubMed] [Google Scholar]
- Zbinden A, Zbinden R, Berger C and Arlettaz R, 2015. Case series of Bifidobacterium longum Bacteremia in three preterm infants on probiotic therapy. Neonatology, 107, 56–59. [DOI] [PubMed] [Google Scholar]
- Zhang F, Jiang X, Chai L, She Y, Yu G, Shu F, Wang Z, Su S, Wenqiong W, Tingsheng X, Zhang Z, Hou D and Zheng B, 2014. Permanent draft genome sequence of Bacillus flexus strain T6186‐2, a multidrug‐resistant bacterium isolated from a deep‐subsurface oil reservoir. Marine Genomics, 18 Pt B, 135–137. [DOI] [PubMed] [Google Scholar]
- Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG and Vidaver AK, 2002. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Applied and Environmental Microbiology, 68, 2198–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zycka‐Krzesinska J, Boguslawska J, Aleksandrzak‐Piekarczyk T, Jopek J and Bardowski JK, 2015. Identification and characterization of tetracycline resistance in Lactococcus lactis isolated from Polish raw milk and fermented artisanal products. International Journal of Food Microbiology, 211, 134–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 2016 updated list of QPS Status recommended biological agents in support of EFSA risk assessments.


