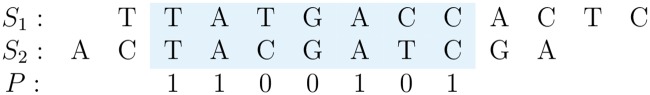Abstract
We study the number Nk of length-k word matches between pairs of evolutionarily related DNA sequences, as a function of k. We show that the Jukes-Cantor distance between two genome sequences—i.e. the number of substitutions per site that occurred since they evolved from their last common ancestor—can be estimated from the slope of a function F that depends on Nk and that is affine-linear within a certain range of k. Integers kmin and kmax can be calculated depending on the length of the input sequences, such that the slope of F in the relevant range can be estimated from the values F(kmin) and F(kmax). This approach can be generalized to so-called Spaced-word Matches (SpaM), where mismatches are allowed at positions specified by a user-defined binary pattern. Based on these theoretical results, we implemented a prototype software program for alignment-free sequence comparison called Slope-SpaM. Test runs on real and simulated sequence data show that Slope-SpaM can accurately estimate phylogenetic distances for distances up to around 0.5 substitutions per position. The statistical stability of our results is improved if spaced words are used instead of contiguous words. Unlike previous alignment-free methods that are based on the number of (spaced) word matches, Slope-SpaM produces accurate results, even if sequences share only local homologies.
Introduction
Phylogeny reconstruction is a fundamental task in computational biology [1]. Here, a basic step is to estimate pairwise evolutionary distances between protein or nucleic-acid sequences. Under the Jukes-Cantor model of evolution [2], the distance between two evolutionarily related DNA sequences can be defined as the number of nucleotide substitutions per site that have occurred since the two sequences have evolved from their last common ancestor. Traditionally, phylogenetic distances are inferred from pairwise or multiple sequence alignments. For the huge amounts of sequence data that are now available, however, sequence alignment has become too slow. Therefore, considerable efforts have been made in recent years, to develop fast alignment-free approaches that can estimate phylogenetic distances without the need to calculate full alignments of the input sequences, see [3–7] for recent review articles. Alignment-free approaches are not only used in phylogeny reconstruction, but are also important in metagenomics [8–10], to find genome rearrangements [11] and in epidemiology [12] and other medical applications, for example to identify drug-resistant bacteria [13] or to classify viruses [14, 15]. In all these applications, it is crucial to rapidly estimate pairwise similarity or dissimilarity values in large sets of sequence data.
Some alignment-free approaches are based on word frequencies [16, 17] or on the length of common substrings [18–20]. Other methods use variants of the D2 distance which is defined as the number of word matches of a pre-defined length between two sequences [15, 21–23]; a review focusing on these methods is given in [24]. kWIP [25] is a further development of this concept that uses information-theoretical weighting. Most of these approaches calculate heuristic measures of sequence (dis-)similarity that are difficult to interpret. At the same time, alignment-free methods have been proposed that can accurately estimate phylogenetic distances between sequences based on stochastic models of DNA or protein evolution, using the length of common substrings [26, 27] or so-called micro alignments [28–33].
Several authors have proposed to estimate phylogenetic distances from the number of k-mer matches between two sequences. The tools Cnidaria [34] and AAF [35] use the Jaccard index between sets of k-mers from two—assembled or unassembled—genomes to estimate the distance between them. In Mash [9], the MinHash [36] approach is used to reduce the input sequences to small ‘sketches’ which can be used to rapidly approximate the Jaccard index. Skmer [37] is a further improvement of this approach. In a previous paper, we proposed another way to infer evolutionary distances between DNA sequences based on the number of word matches between them, and we generalized this to so-called spaced-word matches [38]. Here, a spaced-word match is a pair of words from two sequences that are identical at certain positions, specified by a pre-defined binary pattern of match and don’t-care positions, see [39] for a short review of alignment-free approaches based on spaced-word matches.
The distance function proposed in [38] is now used by default in the program Spaced [40]. Theoretically, this distance measure is based on a simple model of molecular evolution without insertions or deletions. In particular, we assumed in this previous paper that the compared sequences are homologous to each other over their entire length. In practice, the derived distance values are still reasonably accurate if a limited number of insertions and deletions is allowed, and phylogenetic trees could be obtained from these distance values that are similar to trees obtained with more traditional approaches. Like other methods that are based on the number of common k-mers, however, our previous approach can no longer produce accurate results if sequences share only local regions of homology. This is a serious limitation of these methods, since many distantly related genomes share only locally limited homologies, see [41].
Recently, Bromberg et al. published an interesting new approach to alignment-free protein sequence comparison that they called Slope Tree [42]. They defined a distance measure using the decay of the number of k-mer matches between two sequences, as a function of k. Trees reconstructed with this distance measure were in accordance to known phylogenetic trees for various sets of prokaryotes. Slope Tree can also correct for horizontal gene transfer and can deal with composition variation and low complexity sequences. From a theoretical point-of-view, however, it is not clear whether the distance measure used in Slope Tree is an accurate estimator of evolutionary distances.
In the present paper, we study the number Nk of word or spaced-word matches between two DNA sequences, where k is the word length or the number of match positions of the underlying pattern for spaced words, respectively. Inspired by Bromberg’s Slope Tree approach, we study the decay of Nk as a function of k. More precisely, we define a function F(k) that depends on Nk and that can be approximated—under a simple probabilistic model of DNA evolution, and for a certain range of k—by an affine-linear function of k. The distance between two sequences—defined as the number of substitutions per site—can be estimated from the slope of F in this range, we therefore call our approach Slope-SpaM, where SpaM stands for Spaced-Word Matches. In an earlier implementation, we calculated Nk and F(k) for many values of k, in order to find the relevant affine-linear range of F and its slope therein [43]. In the present paper, we show how two values kmin and kmax can be calculated within this range such that the evolutionary distance between the compared sequences can be estimated from the values F(kmin) and F(kmax). Since it suffices to calculate the number Nk of (spaced) word matches for only two values k = kmin and k = kmax, this new implementation is much faster than the previous version of Slope-SpaM.
Using simulated DNA sequences, we show that Slope-SpaM can accurately estimate phylogenetic distances. In contrast to other methods that are based on the number of (spaced) word matches, Slope-SpaM produces still accurate results if the compared sequences contain only local homologies. In addition, we applied Slope-SpaM to infer phylogenetic trees based on genome sequences from the benchmarking project AFproject [44].
Design and implementation
The number of k-mer matches as a function of k
We are using standard notation from stringology as used, for example, in [45]. For a string or sequence S over an alphabet , |S| denotes the length of S, and S(i) is the i-th character of S, 1 ≤ i ≤ |S|. S [i‥j] is the (contiguous) substring of S from i to j. We consider a pair of DNA sequences S1 and S2 that have evolved under the Jukes-Cantor substitution model [2] from some unknown ancestral sequence. That is, we assume that substitution rates are equal for all nucleotides and sequence positions, and that substitution events at different positions are independent of each other. For simplicity, we first assume that there are no insertions and deletions (indels). We call a pair of positions or k-mers from S1 and S2, respectively, homologous if they go back to the same position or k-mer in the ancestral sequence. In our model, we have a nucleotide match probability p for homologous positions and a background match probability q for non-homologous nucleotides; the probability of two homologous k-mers to match exactly is therefore pk.
In our indel-free model, S1 and S2 must have the same length L = |S1| = |S2|, and positions i1 and i2 in S1 and S2, respectively, are homologous if and only if i1 = i2. Note that, under this model, a pair of k-mers from S1 and S2 is either homologous—if the k-mers start at the same respective positions in the sequences—or completely non-homologous, in the sense that none of the corresponding pairs of positions is homologous. For a pair of non-homologous k-mers from S1 and S2, the probability of an exact match is qk.
Let the random variable Xk be defined as the number of k-mer matches between S1 and S2. More precisely, Xk is defined as the number of pairs (i1, i2) for which
holds. There are (L − k + 1) possible homologous and (L − k + 1) ⋅ (L − k) possible background k-mer matches. The expected total number of k-mer matches is therefore
| (1) |
In [38], we used this equation to estimate the match probability p for two observed sequences using a moment-based approach, by replacing the expected number E(Xk) by the empirical number Nk of word matches or spaced-word matches, respectively. Although in Eq (1), an indel-free model is assumed, we could show in our previous paper, that this approach gives still reasonable estimates of p for sequences with insertions and deletions, as long as the sequences are globally related, i.e. as long as the insertions and deletions are small compared to the length of the sequences. It is clear, however, that this estimate will become inaccurate in the presence of large insertions and deletions, i.e. if sequences are only locally related.
Herein, we propose a different approach to estimate evolutionary distances from the number Nk of k-mer matches, by considering the decay of Nk if k increases. For simplicity, we first consider an indel-free model as above. From Eq (1), we obtain
| (2) |
which—for a suitable range of k—is an approximately affine-linear function of k with slope ln p. Substituting the expected value E(Xk) in the right-hand side of (2) with the corresponding empirical number Nk of k-mer matches for two observed sequences, we define
| (3) |
In principle, we can now estimate p as the exponential of the slope of F, as long as we restrict ourselves to a certain range of k. If k is too small, however, Nk will be dominated by background word matches, if k is too large, no word matches will be found at all. The range where we can use the slope of the function F to estimate the match probability p is discussed below.
The above considerations can be generalized to a model of DNA evolution with insertions and deletions and to sequences that share only local homologies. Let us consider two DNA sequences S1 and S2 of different lengths L1 and L2, respectively, that have evolved from a common ancestor under the Jukes-Cantor model, this time with insertions and deletions, and possibly including additional non-homologous parts of the sequences. Note that, unlike with the indel-free model with global homology, it is now possible that a k-mer match involves homologous as well as background nucleotide matches. Instead of deriving exact equations like (1) and (2), we therefore make some simplifications and approximations.
We can decompose S1 and S2 into indel-free pairs of ‘homologous’ substrings that are separated by non-homologous segments of the sequences. Let LH be the—unknown—total length of the homologous substring pairs in each sequence. As above, we define the random variable Xk as the number of k-mer matches between the two sequences, and p and q are, again, the homologous and background nucleotide match probabilities. For realistic data, we also have to take into account homologies between one sequence and the reverse complement of the other sequence. We therefore have twice as many background k-mer matches than in our above simplified model. All in all, the expected number of k-mer matches can be roughly estimated as
| (4) |
and we obtain
| (5) |
Similar as in the indel-free case, we define
| (6) |
where Nk is, again, the empirical number of k-mer matches. As above, we can estimate p as the exponential of the slope of F.
An important point is that these considerations remain valid if LH is small compared to L1 and L2, since LH appears only in an additive constant on the left-hand side of (5). Thus, while the absolute values F(k) do depend on the extent LH of the homology between S1 and S2, the slope of F only depends on p, q, L1 and L2, but not on LH.
The number of spaced-word matches
In many fields of biological sequence analysis, k-mers and k-mer matches have been replaced by spaced words or spaced-word matches, respectively. Let us consider a fixed word P over {0, 1} representing match positions (‘1’) and don’t-care positions (‘0’). We call such a word a pattern; the number of match positions in P is called its weight. In most applications, the first and the last symbol of a pattern P are assumed to be match positions. A spaced word with respect to P is defined as a string W over the alphabet {A, C, G, T, *} of the same length as P with W(i) = * if and only if P(i) = 0, i.e. if and only if i is a don’t-care position of P. Here, ‘*’ is interpreted as a wildcard symbol. We say that a spaced word W w.r.t P occurs in a sequence S at some position i if W(m) = S(i + m − 1) for all match positions m of P.
We say that there is a spaced-word match between sequences S1 and S2 at (i, j) if the same spaced word occurs at position i in S1 and at position j in S2, see Fig 1 for an example. A spaced-word match can therefore be seen as a gap-free alignment of length |P| with matching characters at the match positions and possible mismatches a the don’t-care positions. Spaced-word matches or spaced seeds have been introduced in database searching as an alternative to exact k-mer matches [46–48]. The main advantage of spaced words compared to contiguous k-mers is the fact that results based on spaced words are statistically more stable than results based on k-mers [8, 38, 49–52]. In short, neighbouring spaced-word matches are statistically less dependent from each other than neighbouring word matches; this is also the reason why spaced seeds lead to more sensitive results in database searching [53], compared with seeds that are defined as exact word matches as used, for example, in the original version of BLAST [54].
Fig 1. Spaced-word match between two DNA sequences S1 and S2 at (2,3) with respect to a pattern P = 1100101 representing match positions (‘1’) and don’t-care positions (‘0’).
The same spaced word TA**A*C occurs at position 2 in S1 and at position 3 in S2.
Quite obviously, approximations (4) and (5) remain valid if we define Xk to be the number of spaced-word matches for a given pattern P of weight k, and we can generalize the definition of F(k) accordingly: if we consider a maximum pattern weight K and a given set of patterns {Pk, 1 ≤ k ≤ K} where k is the weight of pattern Pk, then we can define Nk as the empirical number of spaced-word matches with respect to pattern Pk between two observed DNA sequences. F(k) can then be defined exactly as in (6), and we can estimate the nucleotide match probability p as the exponential of the slope of F.
The relevant affine-linear range of F
For simplicity, let us first assume that the two compared sequences have the same length L, and that they are homologous over their entire length. If, for a factor α, we want there to be at least α times more homologous than background word matches, we have to require L ⋅ pk ≥ α ⋅ 2 ⋅ L2 ⋅ qk. We therefore obtain a lower bound for k as
| (7) |
If, on the other hand, we want to have at least β expected word matches, we have to require L ⋅ pk ≥ β, so k would be upper-bounded by
| (8) |
Therefore, in order to ensure that the lower bound given by (7) is smaller than the upper bound in (8), we must require
| (9) |
It follows that the match probability p must be large enough for our approach to work. More specifically, a simple transformation of (9) shows that we have to require
| (10) |
As an example, with a sequence length of 1Gb, a background match probability q = 1/4 and parameters α = β = 10, we would need p > 0.57, corresponding to a Jukes-Cantor distance of around 0.64 substitutions per position in order to satisfy (9) and (10), respectively.
The inequalities derived in this subsection can be easily generalized to the situation where sequences share only a local region of homology of unknown length LH—as long as we can assume that the ratio between LH and the total sequence length is lower-bounded by some known constant.
Implementation
It is well known that the set of all k-mer matches between two sequences can be calculated efficiently by lexicographically sorting the list of all k-mers from both sequences, such that identical k-mers appear as contiguous blocks in this list. This standard procedure can be directly generalized to spaced-word matches.
To estimate the slope of F, one has to take into account that the values F(k) can be used only within a certain range of k, as explained above. If k is too small, Nk and F(k) are dominated by the number of background (spaced) word matches. If k is too large, on the other hand, the number of homologous (spaced) word matches becomes small, and for Nk < 2 ⋅ L1 ⋅ L2 ⋅ qk, the value F(k) is not even defined. For two input sequences, we therefore need to identify a range kmin, …, kmax in which F is approximately affine linear. To find such a range, we want to use inequalities (7) and (8).
There seems to be a certain difficulty in this approach, since the inequalities that we want to use to estimate the match probability p depend not only on the length of the sequences and the background match probability q, but also on the probability p that we want to estimate. It is easy to see, however, that the left-hand side of (7) is monotonically decreasing as a function of p, while the right-hand side of (8) is monotonically increasing. We can therefore calculate kmin and kmax for a fixed small probability p′ and obtain values kmin and kmax that work for any match probability p > p′, since these values are guaranteed to be within the affine-linear region that we want to use to estimate p. For a suitable small match probability p′, we therefore define
| (11) |
and
| (12) |
and estimate the slope of F as
| (13) |
The match probability p can then be estimated as the exponential of this slope, provided that p ≥ p′ holds.
In our test runs, we obtained good results with parameter values α = β = 1 and with two different values for p′, namely of p′ = 0.6 in (11) and p′ = 0.53 in (12), so are using these values by default. With a background match probability of q = 0.25, we therefore obtain default values of
| (14) |
and
| (15) |
where L is the average length of the compared sequences. With these values, we then estimate p as
| (16) |
with the function F defined as in (3). Finally, we apply the usual Jukes-Cantor correction to the estimated match probability , in order to estimate the Jukes-Cantor distance between the sequences, e.g. the number of substitutions per position that have occurred since they diverged from their last common ancestor.
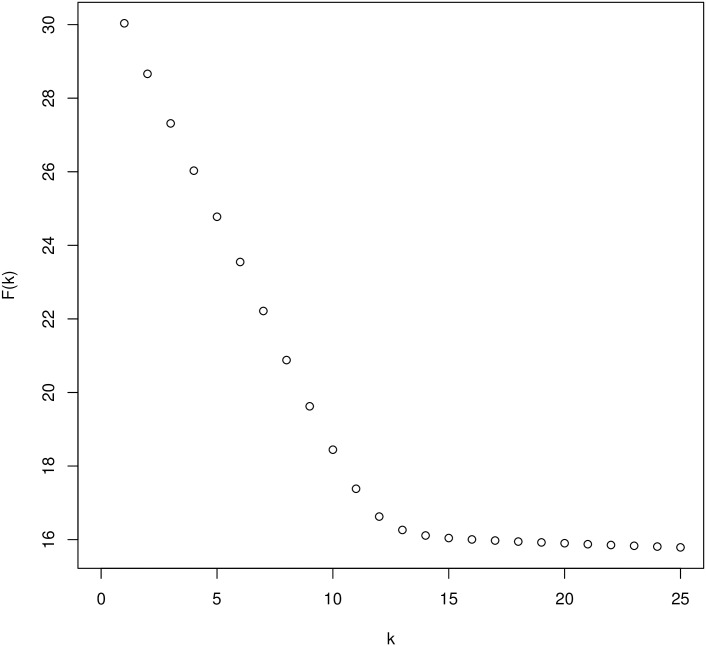
As a numerical example, to illustrate how our approach works, we applied our approach to the genomes of Shigella dysenteriae 1 197 (4.44 Mb) and E. coli strain UTI89 (5.15 Mb). In Fig 2, F(k) is plotted against the word length k for contiguous words. By inserting the average sequence length L into (14) and (15), we obtain kmin = 19 and kmax = 24. With these values, we can calculate the slope of F in the relevant range as
and we obtain an estimated match probability of . This corresponds to an average number of 0.022 substitution per position according to the Jukes-Cantor correction. With Filtered Spaced Word Matches (FSWM), as a comparison, we obtained a distance of 0.023 substitutions per position for this pair of genomes.
Fig 2. Test run on Shigella dysenteriae 1 197 (4.44 Mb) and E. coli strain UTI89 (5.15 Mb).
F(k), as defined in (3), is plotted against the word length k for contiguous words. From the length of the sequences, the values of kmin and kmax are calculated with (14) and (15) as kmin = 19 and kmax = 24.
Results
Distances between simulated DNA sequences
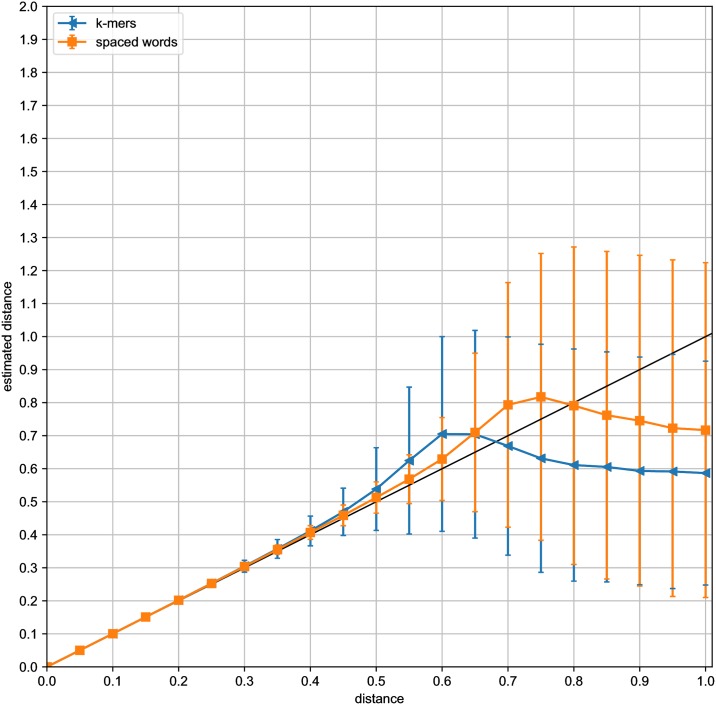
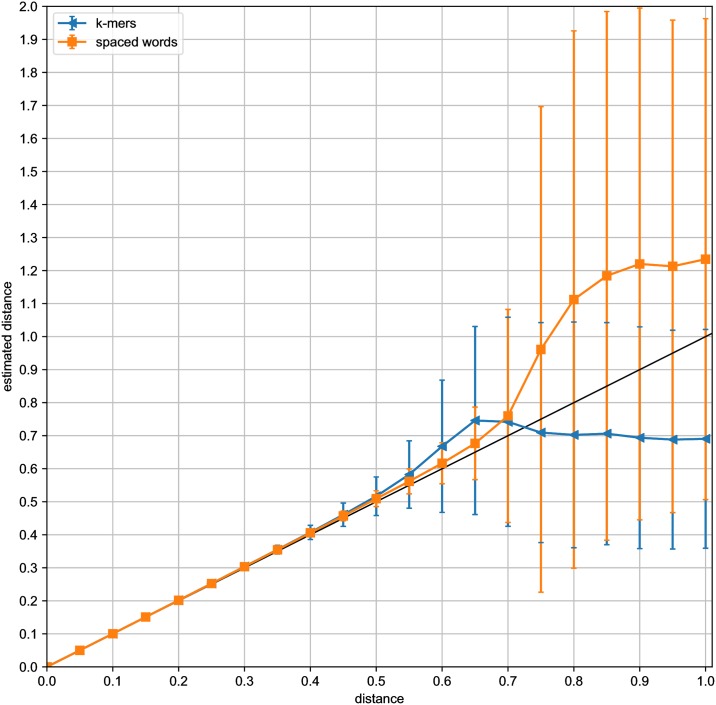
For a systematic evaluation of the distance values computed by Slope-SpaM, we generated simulated pairs of DNA sequences with Jukes-Cantor distances d between 0.05 and 1.0 substitutions per sequence position. For each value of d, we first generated 20,000 sequence pairs of length L = 100, 000 each, and we estimated their distances with Slope-SpaM. Here, we ran the program (a) with k-mers and (b) with spaced words based on randomly generated patterns with a probability of 0.5 for match positions. In Fig 3, the average estimated distances are plotted against the real distances; standard deviations are represented as error bars. Our estimates are fairly accurate for distances up to around 0.5 substitutions per site, but become statistically less stable for larger distances. We did the same test runs with simulated sequences of length L = 1, 000, 000; the results of these runs are shown in Fig 4. In both cases, the results were more accurate and statistically more stable when spaced words were used instead of contiguous k-mers.
Fig 3. For distance values d between 0.05 and 1.0, we generated pairs of simulated DNA sequences of length L = 100 kb with a Jukes-Cantor distance d, i.e. with an average of d substitutions per sequence position.
Distances between the sequences were estimated with Slope-SpaM, (a) based on k-mers and (b) based on spaced words with random patterns with a probability of 0.5 for a match position at each position. For each value of d, 20,000 sequence pairs were generated, their average estimated distances are plotted against the real distances. Standard deviations are shown as error bars.
Fig 4. Estimated vs. real distances for pairs of simulated sequences as in Fig 3, but with sequences of length L = 1Mb.
Distances between sequences with local homologies
As shown theoretically in the section Design and Implementation, our distance estimator is still applicable if the compared sequences share only local regions of homology, in contrast to other approaches that estimate phylogenetic distances from the number of word or spaced-word matches. To verify this empirically, we generated pairs of semi-artificial sequences consisting of local homologies, embedded in non-related random sequences. In each test run, the random sequences that we added to the homologous sequences had the same length in both sequences, but we carried out test runs with random sequences of varying length. We then estimated phylogenetic distances between these sequences with Slope-SpaM, as well as with Mash, Skmer and Spaced, three other alignment-free methods that are also based on the number of word or spaced-word matches. To see how these tools are affected by the presence of non-homologous sequence regions, we compared the distance values estimated from the semi-artificial sequences to the distances estimated from the original sequences by the same tool.
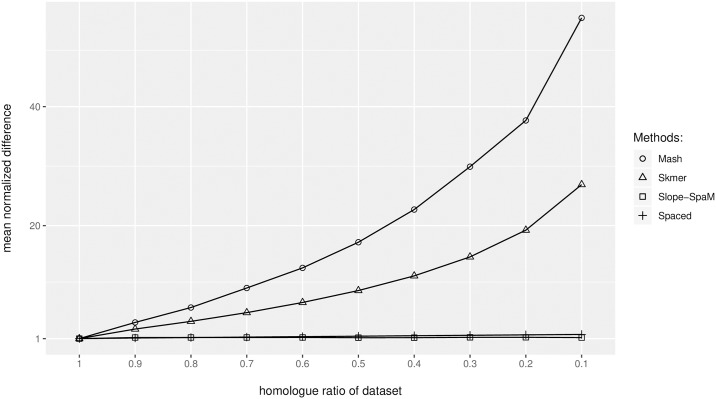
To generate these sequence sets, we started with a set of 19 homologous genomic sequences from different strains of the bacterium Wolbachia from Gerth et al. [55]. These sequences contain 252 genes; the length of the sequences varies between 165kb and 175kb in the different strains. We then generated 9 additional data sets by adding unrelated i.i.d. random sequences of varying length to the left and to the right of each of the homologous genome sequences. This way, we generated 10 sets of sequences where the proportion of homology was 1.0 in the first set—the original sequences without added random sequences—, 0.9 in the second set, 0.8 in the third set, …, and 0.1 in the 10th set. Within each of these 10 sequence sets, we estimated all pairwise distances with the four programs that we evaluated. To find out to which extent these programs are affected by adding the non-related random sequences, we calculated for each data set and for each sequence pair the ratio between the estimated distance value and the distance estimated for the respective original, fully homologous sequence pair. The average ratio of these two distance values over all 171 sequence pairs is plotted against the proportion of homology in the semi-artificial sequences in Fig 5.
Fig 5. Distances estimated by the alignment-free programs Mash [9], Skmer [37], Slope-Spam (this paper) and Spaced [40] between semi-artificial sequences consisting of 252 homologous genes from 19 strains of Wolbachia, embedded in non-related random sequences.
Ten data sets were generated by adding random sequences of different lengths. The x-axis is the proportion of the homologous sequence within the semi-artificial sequences, the y-axis is the ratio between the distance estimated from the semi-artificial sequences and the distances estimated from the original, homologous gene sequences, see the main text for more details.
As can be seen, distances calculated with Mash are heavily affected by including random sequences to the original homologous sequences. For the 10th data set, where the original homologous gene sequences account for only 10% of the generated sequences, and 90% of the sequences are unrelated random sequences, the distances calculated by Mash are, on average, more than 50 times as high as for the original gene sequences that are homologous over their entire length. Skmer and Spaced are also affected by the added random sequences, although to a lesser extent. By contrast, Slope-SpaM was hardly affected at all by the presence of the non-homologous sequences.
Phylogenetic tree reconstruction
In addition to the above artificial and semi-artificial sequence pairs, we used sets of real genome sequences with known reference phylogenies from the as benchmark data for phylogeny reconstruction AFproject [44]. AFproject is a collaborative project to systematically benchmark and evaluate software tools for alignment-free sequence comparison in different application scenarios. The web page of the project provides a variety benchmark sequence sets, and the output of arbitrary alignment-free methods on these sequences can be uploaded to the server. This way, the quality of these methods can be evaluated and compared to a large number of other alignment-free methods, among them the state-of-the-art methods.
To evaluate Slope-SpaM, we downloaded all data sets from the AFproject server that are relevant for our study, namely five sets of full-genome sequences that are available in the categories genome-based phylogeny and horizontal gene transfer: (1) a set of 29 E.coli/Shigella genomes [28], (2) a set of 14 plant genomes [56], (3) a set of 25 mitochondrial genomes from different fish species of the suborder Labroidei [57], (4) another set of 27 E.coli/Shigella [58] and (5) a set of 8 genomes of different strains of Yersinia [59]. These data sets have been used previously by developers of alignment-free tools to benchmark their methods.
We ran Slope-SpaM on the above sets of genomes and uploaded the obtained distance matrices to the AFproject web server for evaluation. For these categories of benchmark data, the AFproject server calculates phylogenetic trees from the produced distance matrices using Neighbor Joining [60]. It then compares the obtained trees to trusted reference trees of the respective data sets under the normalized Robinson-Foulds (nRF) metric. That is, the standard Robinson-Foulds (RF) distance [61] that measures the dissimilarity between two tree topologies is divided by the maximal possible RF distance of two trees with the respective number of leaves; the nRF distances can therefore take values between 0 and 1. On the AFproject server, the benchmark results are ranked in order of increasing nRF distance, i.e. in order of decreasing quality. The results of our test runs with the AFproject benchmark sequences are shown in Table 1.
Table 1. Test results on five sets of genome sequences from AFproject.
Pairwise distance values calculated with different alignment-free methods were used as input for Neighbor-Joining [60]; the table contains normalized Robinson-Foulds distances between the resulting trees and reference trees. Thus, the smaller the values are, the more similar are the produced trees to the reference trees. The table also shows the median results from 74 methods evaluated in the AFproject study. Three of the best performing programs in this study are shown (all results, except for Slope-SpaM, are taken from [44]).
| Genome-Based Phylogeny | Horizontal Gene Transfer | ||||
|---|---|---|---|---|---|
| # genomes | mtDNA | E. coli | Plants | E. coli | Yersinia |
| 25 | 29 | 14 | 27 | 8 | |
| Slope-SpaM (k-mers) | 0.32 | 0.50 | 1.00 | 0.58 | 0.20 |
| Slope-SpaM (spaced words) | 0.09 | 0.35 | 0.64 | 0.50 | 0.80 |
| median AFproject | 0.09 | 0.54 | 0.64 | 0.50 | 0.00 |
| FFP | 0.09 | 0.23 | 0.43 | 0.21 | 0.80 |
| co-phylog | 0.09 | 0.12 | 0.23 | 0.08 | 0.80 |
| Mash | 0.05 | 0.15 | 0.14 | 0.12 | 0.80 |
| Skmer | 0.09 | 0.15 | 0.25 | 0.17 | 0.80 |
It should be mentioned that the performance of the compared methods on the set of eight Yersinia genomes is in stark contrast to their performance on the other four data sets. This has also been observed for other methods evaluated in AFproject: methods that performed well on other data sets, performed badly on Yersinia and vice versa. It may therefore be advisable to look at this data set and the reference phylogeny used in AFproject in more detail.
Program run time
Table 2 shows the run time of Slope-SpaM on three of the data sets from AFproject that we used in the previous subsection. We used the set of 25 mitochondrial genomes from different fish species (total size 412 kb), the set of 29 E. coli genomes (total size 138 Mb) and the set of 14 plant genomes (total size 4.58 Gb).
Table 2. Runtime in seconds of Slope-SpaM (with spaced words) and six other alignment-free programs on three different sets of genomes that were used as benchmark data in AFproject [44].
On the largest data set, the set of plant genomes, co-phylog and kmacs were unable to produce results.
| mtDNA | E. coli | Plants | |
|---|---|---|---|
| # genomes | 25 | 29 | 14 |
| Total size | 412 kb | 138 Mb | 4.58 Gb |
| Slope-SpaM | 1 | 629 | 9,551 |
| Mash | 0.7 | 15 | 417 |
| Skmer | 6 | 938 | 3,224 |
| FSWM | 8 | 32,798 | 28,690,489 |
| andi | 3 | 325 | 13,813 |
| co-phylog | 9 | 533 | – |
| kmacs | 5 | 55,736 | – |
Availability and future directions
A number of alignment-free methods have been proposed in recent years that estimate the nucleotide match probability p in an (unknown) alignment of two DNA sequences from the number Nk of word or k-mer matches for a fixed word length k. This can be done by comparing Nk to the total number of words in the compared sequences [9] or—equivalently—to the length of the sequences [27]. A certain draw-back of these approaches is that they assume that the compared sequences are homologous to each other over their entire length. Unfortunately, these methods cannot be easily applied to sequences that share only local homologies with each other. If two sequences share only a limited region of homology and are unrelated outside this region, a given number Nk of word matches could indicate a high match probability p in an alignment of those homologous regions—while the same number of word matches would correspond to a lower p if the homology would extend over the entire length of the sequences.
In the present paper, we introduced Slope-SpaM, an alternative approach to estimate the match probability p between two DNA sequences—and thereby their Jukes-Cantor distance—from the number of word matches. The main difference between Slope-SpaM and most other word-based methods is that, instead of using only one single word length k, our program considers a function F such that the values F(k) depend on the number Nk of word matches of length k. We showed in that there is a certain range of k where F is affine-linear, and the nucleotide match probability p in an alignment of the input sequences can be estimated from the slope of F. Further, we showed that one can calculate two values kmin and kmax within the relevant affine-linear range, such that it suffices to calculated F(kmin) and F(kmax) in order to calculate the slope of F in this range. The runtime of our program is therefore essentially determined by the time required to calculate the number of k-mer matches for only two values of k.
From the definition of F, it is easy to see that the slope of F, and therefore the estimated match probability p, are not affected if the compared sequences share only local homologies. This was confirmed in our test runs with semi-artificial sequences, obtained by adding non-related random sequences to truly homologous sequences. We generalized this approach to spaced-word matches, i.e. word matches with possible mismatches at pre-defined positions, specified by a binary pattern of match and don’t-care positions.
To evaluate phylogenetic distances estimated by our approach, we used simulated DNA sequences with known Jukes-Cantor distances. For these data, we could show that distance estimates obtained with Slope-SpaM are highly accurate for distances up to around 0.5 substitutions per sequence position. By using spaced words instead of contiguous k-mers, we could improve the statistical stability of our results. To evaluate Slope-SpaM and other alignment-free methods on phylogenetic tree reconstruction, we used all relevant data sets from the benchmark project AFproject. As in the original AFproject study [44], we applied Neighbor Joining to the distance matrices produced by the approaches that we evaluated, and we compared the resulting phylogenetic tree topologies to the reference topologies that are available from the AFproject web page. This is a common way of evaluating alignment-free methods. As shown in Table 1, the performance of Slope-SpaM on these data was in the medium range, compared to the other methods that have been evaluated in AFproject, if our program was used with spaced words. We obtained better tree topologies with spaced words than with contiguous words. In terms of topological accuracy of the produced trees, however, Slope-SpaM was outperformed by some of the top methods evaluated in the AFproject study.
An advantage of Slope-SpaM, compared to most other alignment-free methods, is its high speed. It may be possible to further improve the run time of the program by using sketching techniques [62], to reduce the number of (spaced-)word matches that are to be considered. Conversely, if the number of word matches is too small for short or distantly related sequences, spaced-word matches based on multiple patterns may help to increase the range of sequences where our method can be applied.
In the last few years, a number of alignment-free approaches have been proposed that are able to use unassembled short sequencing reads as input [9, 28, 35, 37, 63, 64], as a basis for sequence clustering [34], for phylogenetic tree reconstruction [65] or for phylogenetic placement [64]. We are planning to evaluate systematically if Slope-SpaM can be used to find the position of unassembled reads in existing phylogenies, or to estimate phylogenetic distances between species based on sequencing reads instead of assembled genomes.
Since the approach implemented in Slope-SpaM is novel and rather different from existing alignment-free approaches, improvements may be possible by systematically analyzing the influence of the parameters of the program. In particular, there may be more sophisticated ways of finding suitable values of kmin and kmax to further improve the accuracy of Slope-SpaM. Also, in our study we used the Jukes-Cantor model, the simplest possible model for nucleotide substitutions. Using more sophisticated substitution models may also lead to improvements in future versions of the program.
The source code of our software is freely available through GitHub under the GNU GPLv3 license: https://github.com/burkhard-morgenstern/Slope-SpaM.
Acknowledgments
We thank Fengzhu Sun for his useful comments on an earlier version of this manuscript, Andrzej Zielezinski and Wojchiech Karlowski for making the AFproject server available, Christoph Bleidorn and Micha Gerth for discussions about the Wolbachia genomes, Chris-André Leimeister, Christoph Elfmann and Peter Meinicke for discussions on the project and four reviewers for very helpful comments and suggestions.
Data Availability
The source code of our software is freely available through GitHub under the GNU GPLv3 license: https://github.com/burkhard-morgenstern/Slope-SpaM.
Funding Statement
The project was funded by VW Foundation, project VWZN3157 to BM, and by the Portuguese national funds through Foundation for Science and Technology (FCT), in the context of the project UID/CEC/00127/2019. We acknowledge support by the Open Access Publication Funds of Göttingen University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Felsenstein J. Inferring Phylogenies. Sunderland, MA, USA: Sinauer Associates; 2004. [Google Scholar]
- 2. Jukes TH, Cantor CR. Evolution of Protein Molecules. Munro HN, editor. New York: Academy Press; 1969. [Google Scholar]
- 3. Haubold B. Alignment-free phylogenetics and population genetics. Briefings in Bioinformatics. 2014;15:407–418. 10.1093/bib/bbt083 [DOI] [PubMed] [Google Scholar]
- 4. Song K, Ren J, Reinert G, Deng M, Waterman MS, Sun F. New developments of alignment-free sequence comparison: measures, statistics and next-generation sequencing. Briefings in Bioinformatics. 2014;15:343–353. 10.1093/bib/bbt067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zielezinski A, Vinga S, Almeida J, Karlowski WM. Alignment-free sequence comparison: benefits, applications, and tools. Genome Biology. 2017;18:186 10.1186/s13059-017-1319-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bernard G, Chan CX, Chan YB, Chua XY, Cong Y, Hogan JM, et al. Alignment-free inference of hierarchical and reticulate phylogenomic relationships. Briefings in Bioinformatics. 2019;22:426–435. 10.1093/bib/bbx067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kucherov G. Evolution of biosequence search algorithms: a brief survey. Bioinformatics. 2019;35:3547–3552. 10.1093/bioinformatics/btz272 [DOI] [PubMed] [Google Scholar]
- 8. Břinda K, Sykulski M, Kucherov G. Spaced seeds improve k-mer-based metagenomic classification. Bioinformatics. 2015;31:3584–3592. 10.1093/bioinformatics/btv419 [DOI] [PubMed] [Google Scholar]
- 9. Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biology. 2016;17:132 10.1186/s13059-016-0997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linard B, Swenson K, Pardi F. Rapid alignment-free phylogenetic identification of metagenomic sequences. Bioinformatics. 2019;35:3303–3312. 10.1093/bioinformatics/btz068 [DOI] [PubMed] [Google Scholar]
- 11.Hosseini M, Pratas D, Morgenstern B, Pinho AJ. Smash++: an alignment-free and memory-efficient tool to find genomic rearrangements. bioRxiv. 2019. [DOI] [PMC free article] [PubMed]
- 12. Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, Weiser JN, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Research. in press;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Břinda K, Callendrello A, Cowley L, Charalampous T, Lee RS, MacFadden DR, et al. Lineage calling can identify antibiotic resistant clones within minutes. bioRxiv. 2018.
- 14. Zhang Q, Jun SR, Leuze M, Ussery D, Nookaew I. Viral Phylogenomics Using an Alignment-Free Method: A Three-Step Approach to Determine Optimal Length of k-mer. Scientific Reports. 2017;7:40712 10.1038/srep40712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahlgren NA, Ren J, Lu YY, Fuhrman JA, Sun F. Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Research. 2017;45:39–53. 10.1093/nar/gkw1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qi J, Luo H, Hao B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Research. 2004;32(suppl 2):W45–W47. 10.1093/nar/gkh362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sims GE, Jun SR, Wu GA, Kim SH. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proceedings of the National Academy of Sciences. 2009;106:2677–2682. 10.1073/pnas.0813249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ulitsky I, Burstein D, Tuller T, Chor B. The average common substring approach to phylogenomic reconstruction. Journal of Computational Biology. 2006;13:336–350. 10.1089/cmb.2006.13.336 [DOI] [PubMed] [Google Scholar]
- 19. Leimeister CA, Morgenstern B. kmacs: the k-mismatch average common substring approach to alignment-free sequence comparison. Bioinformatics. 2014;30:2000–2008. 10.1093/bioinformatics/btu331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horwege S, Lindner S, Boden M, Hatje K, Kollmar M, Leimeister CA, et al. Spaced words and kmacs: fast alignment-free sequence comparison based on inexact word matches. Nucleic Acids Research. 2014;42:W7–W11. 10.1093/nar/gku398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reinert G, Chew D, Sun F, Waterman MS. Alignment-Free Sequence Comparison (I): Statistics and Power. Journal of Computational Biology. 2009;16:1615–1634. 10.1089/cmb.2009.0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wan L, Reinert G, Sun F, Waterman MS. Alignment-free sequence comparison (II): theoretical power of comparison statistics. Journal of Computational Biology. 2010;17:1467–1490. 10.1089/cmb.2010.0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song K, Ren J, Zhai Z, Liu X, Deng M, Sun F. Alignment-Free Sequence Comparison Based on Next-Generation Sequencing Reads. Journal of Computational Biology. 2013;20:64–79. 10.1089/cmb.2012.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ren J, Bai X, Lu YY, Tang K, Wang Y, Reinert G, et al. Alignment-Free Sequence Analysis and Applications. Annual Review of Biomedical Data Science. 2018;1:93–114. 10.1146/annurev-biodatasci-080917-013431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murray KD, Webers C, Ong CS, Borevitz J, Warthmann N. kWIP: The k-mer weighted inner product, a de novo estimator of genetic similarity. PLOS Computational Biology. 2017;13:e1005727 10.1371/journal.pcbi.1005727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haubold B, Pfaffelhuber P, Domazet-Loso M, Wiehe T. Estimating Mutation Distances from Unaligned Genomes. Journal of Computational Biology. 2009;16:1487–1500. 10.1089/cmb.2009.0106 [DOI] [PubMed] [Google Scholar]
- 27. Morgenstern B, Schöbel S, Leimeister CA. Phylogeny reconstruction based on the length distribution of k-mismatch common substrings. Algorithms for Molecular Biology. 2017;12:27 10.1186/s13015-017-0118-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yi H, Jin L. Co-phylog: an assembly-free phylogenomic approach for closely related organisms. Nucleic Acids Research. 2013;41:e75 10.1093/nar/gkt003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haubold B, Klötzl F, Pfaffelhuber P. andi: Fast and accurate estimation of evolutionary distances between closely related genomes. Bioinformatics. 2015;31:1169–1175. 10.1093/bioinformatics/btu815 [DOI] [PubMed] [Google Scholar]
- 30. Leimeister CA, Sohrabi-Jahromi S, Morgenstern B. Fast and Accurate Phylogeny Reconstruction using Filtered Spaced-Word Matches. Bioinformatics. 2017;33:971–979. 10.1093/bioinformatics/btw776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leimeister CA, Schellhorn J, Dörrer S, Gerth M, Bleidorn C, Morgenstern B. Prot-SpaM: Fast alignment-free phylogeny reconstruction based on whole-proteome sequences. GigaScience. 2019;8:giy148 10.1093/gigascience/giy148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dencker T, Leimeister CA, Gerth M, Bleidorn C, Snir S, Morgenstern B. Multi-SpaM: a Maximum-Likelihood approach to phylogeny reconstruction using multiple spaced-word matches and quartet trees. NAR Genomics and Bioinformatics. 2020;2:lqz013 10.1093/nargab/lqz013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klötzl F, Haubold B. Phylonium: Fast Estimation of Evolutionary Distances from Large Samples of Similar Genomes. Bioinformatics. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aflitos SA, Severing E, Sanchez-Perez G, Peters S, de Jong H, de Ridder D. Cnidaria: fast, reference-free clustering of raw and assembled genome and transcriptome NGS data. BMC Bioinformatics. 2015;16:352 10.1186/s12859-015-0806-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan H, Ives AR, Surget-Groba Y, Cannon CH. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics. 2015;16:522 10.1186/s12864-015-1647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broder AZ. Identifying and Filtering Near-Duplicate Documents. In: Proceedings of the 11th Annual Symposium on Combinatorial Pattern Matching. COM’00. Berlin, Heidelberg: Springer-Verlag; 2000. p. 1–10.
- 37. Sarmashghi S, Bohmann K, P Gilbert MT, Bafna V, Mirarab S. Skmer: assembly-free and alignment-free sample identification using genome skims. Genome Biology. 2019;20:34 10.1186/s13059-019-1632-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morgenstern B, Zhu B, Horwege S, Leimeister CA. Estimating evolutionary distances between genomic sequences from spaced-word matches. Algorithms for Molecular Biology. 2015;10:5 10.1186/s13015-015-0032-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgenstern B. Sequence Comparison without Alignment: The SpaM approaches. bioRxiv. 2019. [DOI] [PubMed]
- 40. Leimeister CA, Boden M, Horwege S, Lindner S, Morgenstern B. Fast Alignment-Free sequence comparison using spaced-word frequencies. Bioinformatics. 2014;30:1991–1999. 10.1093/bioinformatics/btu177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications. 2018;9:5114 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bromberg R, Grishin NV, Otwinowski Z. Phylogeny Reconstruction with Alignment-Free Method That Corrects for Horizontal Gene Transfer. PLOS Comput Biol. 2016;12:e1004985 10.1371/journal.pcbi.1004985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Röhling S. Slope-SpaM—an alignment free sequence analysis approach [Bachelor’s Thesis]. University of Göttingen. Germany; 2019.
- 44. Zielezinski A, Girgis HZ, Bernard G, Leimeister CA, Tang K, Dencker T, et al. Benchmarking of alignment-free sequence comparison methods. Genome Biology. 2019;20:144 10.1186/s13059-019-1755-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gusfield D. Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology. Cambridge, UK: Cambridge University Press; 1997. [Google Scholar]
- 46. Brown DG. A survey of seeding for sequence alignment In: I Mǎndoiu AZ, editor. Bioinformatics Algorithms: Techniques and Applications. New York: Wiley-Interscience; 2008. p. 126–152. [Google Scholar]
- 47. Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. 10.1093/bioinformatics/18.3.440 [DOI] [PubMed] [Google Scholar]
- 48. Li M, Ma B, Kisman D, Tromp J. PatternHunter II: Highly Sensitive and Fast Homology Search. Genome Informatics. 2003;14:164–175. [PubMed] [Google Scholar]
- 49. Ilie L, Ilie S, Bigvand AM. SpEED: fast computation of sensitive spaced seeds. Bioinformatics. 2011;27:2433–2434. 10.1093/bioinformatics/btr368 [DOI] [PubMed] [Google Scholar]
- 50. Egidi L, Manzini G. Design and analysis of periodic multiple seeds. Theoretical Computer Science. 2014;522:62—76. 10.1016/j.tcs.2013.12.007 [DOI] [Google Scholar]
- 51. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 52. Noé L. Best hits of 11110110111: model-free selection and parameter-free sensitivity calculation of spaced seeds. Algorithms for Molecular Biology. 2017;12:1 10.1186/s13015-017-0092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li M, Ma B, Zhang L. Superiority and complexity of the spaced seeds. In: Proceedings of the Seventeenth Annual ACM-SIAM Symposium on Discrete Algorithm. SODA’06. Philadelphia, PA, USA: Society for Industrial and Applied Mathematics; 2006. p. 444–453.
- 54. Altschul SF, Gish W, Miller W, Myers EM, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 55. Gerth M, Gansauge MT, Weigert A, Bleidorn C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nature Communications. 2014;5:5117 10.1038/ncomms6117 [DOI] [PubMed] [Google Scholar]
- 56. Hatje K, Kollmar M. A phylogenetic analysis of the brassicales clade based on an alignment-free sequence comparison method. Frontiers in Plant Science. 2012;3:192 10.3389/fpls.2012.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fischer C, Koblmüller S, Gülly C, Schlötterer C, Sturmbauer C, Thallinger GG. Complete mitochondrial DNA sequences of the threadfin cichlid (Petrochromis trewavasae) and the blunthead cichlid (Tropheus moorii) and patterns of mitochondrial genome evolution in cichlid fishes. PLOS One. 2013;8(6):e67048 10.1371/journal.pone.0067048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Skippington E, Ragan MA. Within-species lateral genetic transfer and the evolution of transcription al regulation in Escherichia coli and Shigella. BMC Genomics. 2011;12:532 10.1186/1471-2164-12-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Darling AE, Miklós I, Ragan MA. Dynamics of Genome Rearrangement in Bacterial Populations. PLOS Genetics. 2008;4(7):e1000128 10.1371/journal.pgen.1000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- 61. Robinson DF, Foulds L. Comparison of phylogenetic trees. Mathematical Biosciences. 1981;53:131–147. 10.1016/0025-5564(81)90043-2 [DOI] [Google Scholar]
- 62. Rowe WPM. When the levee breaks: a practical guide to sketching algorithms for processing the flood of genomic data. Genome Biology. 2019;20:199 10.1186/s13059-019-1809-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lu YY, Tang K, Ren J, Fuhrman JA, Waterman MS, Sun F. CAFE: aCcelerated Alignment-FrEe sequence analysis. Nucleic Acids Research. 2017;45:W554–W559. 10.1093/nar/gkx351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balaban M, Sarmashghi S, Mirarab S. APPLES: Fast Distance-based Phylogenetic Placement. Systematic Biology. doi.org/101093/sysbio/syz063;. [DOI] [PMC free article] [PubMed]
- 65. Lau AK, Dörrer S, Leimeister CA, Bleidorn C, Morgenstern B. Read-SpaM: assembly-free and alignment-free comparison of bacterial genomes with low sequencing coverage. BMC Bioinformatics. 2019;20:638 10.1186/s12859-019-3205-7 [DOI] [PMC free article] [PubMed] [Google Scholar]