Abstract
Production of the Panton-Valentine leukocidin (PVL) by Staphylococcus aureus is mediated via the genes lukS-PV and lukF-PV which are carried on bacteriophage ϕSa2. PVL is associated with S. aureus strains that cause serious infections and clones of community-associated methicillin-resistant S. aureus (CA-MRSA) that have additionally disseminated widely. In Western Australia (WA) the original CA-MRSA were PVL negative however, between 2005 and 2008, following the introduction of eight international PVL-positive CA-MRSA, PVL-positive WA CA-MRSA were found. There was concern that PVL bacteriophages from the international clones were transferring into the local clones, therefore a comparative study of PVL-carrying ϕSa2 prophage genomes from historic WA PVL-positive S. aureus and representatives of all PVL-positive CA-MRSA isolated in WA between 2005 and 2008 was performed. The prophages were classified into two genera and three PVL bacteriophage groups and had undergone many recombination events during their evolution. Comparative analysis of mosaic regions of selected bacteriophages using the Alignments of bacteriophage genomes (Alpha) aligner revealed novel recombinations and modules. There was heterogeneity in the chromosomal integration sites, the lysogeny regulation regions, the defence and DNA processing modules, the structural and packaging modules and the lukSF-PV genes. One WA CA-MRSA (WA518751) and one international clone (Korean Clone) have probably acquired PVL-carrying ϕSa2 in WA, however these clones did not disseminate in the community. Genetic heterogeneity made it impossible to trace the source of the PVL prophages in the other WA clones. Against this background of PVL prophage diversity, the sequence of one group, the ϕSa2USA/ϕSa2wa-st93 group, was remarkably stable over at least 20 years and associated with the highly virulent USA300 and ST93-IVa CA-MRSA lineages that have disseminated globally.
Introduction
Staphylococcus aureus is a pandemic pathogen that is also part of the human microbiota [1]. Paramount to the success of S. aureus has been its ability to utilize mobile elements to acquire and disseminate antibiotic resistance, virulence and adaptive mechanisms amongst staphylococcal populations. In methicillin-sensitive S. aureus (MSSA) and community-associated methicillin-resistant S. aureus (CA-MRSA) the Panton-Valentine leukocidin (PVL) is a virulence factor that is carried on a bacteriophage known as ϕSa2 which is integrated into the chromosome as a prophage [2]. PVL is a bi-component, pore-forming toxin produced by co-transcribed genes, lukF-PV and lukS-PV, that targets and lyses human macrophages, polymorphonuclear leukocytes and monocytes and also incites the human inflammatory immune response [3]. Strains encoding PVL are associated with skin and soft tissue infections and dangerous invasive infections however, the role that the toxin plays in virulence is controversial and as yet, no clear-cut selective advantage has been shown for CA-MRSA that produce PVL [4–8]. Many virulent strains of MSSA and CA-MRSA do not produce PVL, however, as the pathogen evolves it is evident that those that have disseminated to cause the greatest burden of infectious disease harbor the prophage [9].
The PVL bacteriophage genome is composed of functionally colinear main modules encoding genes for lysogeny, DNA processing, head morphogenesis and packaging, tail morphogenesis, and lysis, with the lukSF-PV genes encoded between the lysis and lysogeny modules in the circularly permutated bacteriophage [10]. The lysogeny, lukSF-PV and lysis regions are well conserved with minor polymorphisms. Most diversity occurs in the DNA processing module with the head and tail morphogenesis genes showing diversity depending on the genus and PVL group. ϕSa2 can be vertically transmitted with the chromosome during replication or it can enter the lytic cycle and transmit horizontally to another cell. It has been well documented that bacteriophages undergo high rates of recombination and both these forms of transmission allow opportunity for genetic exchange, the potential mechanisms being transposition, site-specific recombination, homing endonucleases and homologous and illegitimate recombination [11]. While it is believed that horizontal gene transfer between S. aureus of different lineages is rare due to a lineage-specific type1 restriction-modification system [12], an in-vivo study revealed that bacteriophage transferred frequently during co-colonization by S. aureus of the same lineage and recombination between different bacteriophage occurred [13]. An investigation of MRSA colonisation in remote WA revealed that 8% of screening swab sets with an MRSA were colonised with multiple lineages of MRSA and 51.7% were co-colonised with an MSSA [14]. This would provide ideal opportunities for bacteriophage transmission and recombination to occur.
Five lineages of CA-MRSA, ST1-IVa (WA1), ST78-IVa (WA2), ST5-IVa (WA3), ST45-V (WA4) and ST8-IVa (WA5) emerged in remote Western Australian (WA) communities and WA1, WA2 and WA3 eventually disseminated to the capital city Perth and the eastern states of Australia [15, 16]. Unlike CA-MRSA that were being reported outside of WA, the WA strains were PVL negative [17]. There were however, two lineages of PVL-positive MSSA in remote WA communities, ST93-MSSA and ST121-MSSA [14].
In 2005, a PVL-positive strain belonging to the same lineage as WA1 was isolated, followed in 2008, by WA2-, WA3- and WA5-like PVL positive clones. In WA, all MRSA are submitted to a central facility for typing and epidemiological investigation [18] and between 2005 and 2008 eight international PVL-positive CA-MRSA were introduced into WA. The rise in the number of PVL-positive CA-MRSA in WA since the first was found in 2003 has been alarming. In 2003/2004 2.1% of CA-MRSA were PVL positive, however by 2015/2016 this had risen to 52.8%, with the predominant clones being ST93-IVa (Queensland clone, 63%), ST5-IVc (WA 121, 19.5%) and ST30-IVc (WSPP, 6.8%). WA1-, WA2- and WA3-like PVL-positive clones were still in the community in 2016 however, they had not thrived and formed lower percentages of 0.7%, 0.17% and 1.1% respectively while PVL-positive WA5 had disappeared [19].
The overall aims of this study were to investigate PVL prophages from lineages of PVL-positive MRSA isolated in WA between 2005 and 2008 firstly, to gain insights into the genetics of geographically and temporally related PVL prophages in WA and secondly, to determine if PVL bacteriophages from the international strains had horizontally transmitted into the local WA clones. A comparative analysis of the PVL prophages has been performed using conventional sequence analysis, and regions of selected prophages have been compared using the Alignments of bacteriophage genomes (Alpha) aligner, which is an application that creates a partial order of gapless alignments along the bacteriophage genomes, allowing the identification of common core sequences and modular segments [20, 21]. Heterogeneity between the bacteriophages has been investigated using Alpha aligner defined modules and coding sequence comparisons. PVL bacteriophages were induced from PVL-positive S. aureus from the WA community and attempts were made to lysogenise prototype PVL-negative WA CA-MRSA.
Materials and methods
Bacterial strains
Genotypes and year of isolation of PVL-positive clones and their PVL prophage sizes are presented in Table 1. All MRSA except WA2RNSH95 and USA300 FPR3757 were from cases of infection or colonization in the WA community [18]. WA2RNSH95 was a WA2 clone from Sydney, Australia. The USA300 clone was present in WA [22] and the prophage ϕSa2USA from FPR3757 (Genbank: NC_007793) was used for genetic comparison. MSSA isolates W17S and K25S were colonizing isolates from remote WA communities [14]. ST772-V was previously sequenced [23]. MW2 (Genbank: BA000033) was used as a lukSF-PV gene-sequencing and prophage induction control. ϕSLT (Genbank: AB045978) and ϕSa2958 (Genbank: AP009363) were lukSF-PV gene sequencing controls.
Table 1. WA PVL-positive bacteriophages and lysogens.
| Bacteriophage | Size (bp) | Lysogen genotype CC, ST-SCCmec |
CloneStrain | Year of isolation | Reference |
|---|---|---|---|---|---|
| WA PVL-positive Clones | |||||
| ϕSa2wa-st1 | 45,585 | 1, ST1-IVa | WA115798 | 2005 | This study |
| ϕSa2wa-st5 | 44,823 | 5, ST5-IVa | WA318790 | 2008 | This study |
| ϕSa2wa-st8 | 45,914 | 8, ST8-IVa | WA518751 | 2008 | This study |
| ϕSa2wa-st78 | 45,878 | 88, ST78-IVa | WA2RNSH95 | 2008 | This study |
| ϕSa2wa-st93mssa | 45,913 | Singleton, ST93 | W17S | 1995 | [14] |
| ϕSa2wa-st121mssa | 45,621 | 121, ST121 | K25S | 1995 | [14] |
| International Clones | |||||
| ϕSa2wa-st22 | 38,576 | 22, ST22-IVc | 16386 | 2007 | [18] |
| ϕSa2wa-st30 | 45,780 | 30, ST30-IVc | WSPP16663 | 2002 | [25] |
| ϕSa2wa-st59 | 42,133 | 59, ST59-V | Taiwan clone16672 | 2003 | [26] |
| ϕSa2wa-st72 | 47,213 | 72, ST72-IVa | Korean clone15803 | 2006 | [18] |
| ϕSa2wa-st80 | 45,164 | 80, ST80-1Vc | European clone15395 | 2004 | [27] |
| ϕSa2wa-st93 | 45,913 | ST93-IVa | Qld clone16790 | 2003 | [28] |
| ϕSa2wa-st772 | 42,402 | 1, ST772-V | Bengal Bay clone17048 | 2007 | [23] |
| ϕSa2USA | 45,914 | 8, ST8-IVa | USA300_FPR3757 | 2003 | [29] |
| WA PVL-negative Clones | |||||
| NA | NA | 1, ST1-IVa | WA1WBG8287 | 1995 | [24] |
| NA | NA | 88, ST255-IVa | WA2WBG8366 | 1995 | [24] |
| NA | NA | 5, ST5-IVa | WA3WBG8378 | 1995 | [24] |
| NA | NA | 45, ST45-V | WA4WBG8404 | 1995 | [24] |
| NA | NA | 8, ST8-IVa | WA5WBG7583 | 1989 | [30] |
Abbreviations: bp, base pairs; NA, Not applicable; WA, Western Australian, Qld, Queensland; WSPP, Western Samoan Phage Pattern
PVL-negative WA1WBG8287, WA2WBG8366, WA3WBG8378, WA4WBG8404 and WA5WBG7583 are historic prototype clones from the WA community [24]. Bacteriophage indicator and propagating strains were RN4220, WBG248, WBG356, WBG696 and WBG286.
Sequencing of bacterial and PVL-prophage genomes and genetic analysis
Twelve bacterial genomes were sequenced using Illumina NextSeq sequence chemistry (Illumina Australia, Scoresby, Victoria 3179) and assembled with SPAdes, v3.9.0. The PVL-prophage reads were extracted and analysed using MacVector with Assembler, v15.5.3 (Accelrys, Cambridge, UK). The sequences of ϕSa2wa-st1, -st8, -st30, -st72 and -st93mssa were on single contigs, the remainder were assembled by overlapping contigs utilising the MacVector Assembler bowtie and phrap algorithms. Bacteriophage were designated as phi Sa2 Western Australia-host sequence type (ϕSa2wa-st). Except for prophages ϕSa2USA and ϕSa2wa-st772, National Centre for Biotechnology Information (NCBI) homology searches used only whole bacteriophage genome sequences for comparisons.
lukSF-PV sequencing
Isolates were cultured on brain heart infusion agar (BHIA) (Gibco Diagnostics, Gaithersburg, MD, USA), incubated at 37°C, grown in trypticase soy broth (Gibco Diagnostics, Gaithersburg, MD, USA) and incubated overnight at 37°C. DNA was extracted using the Invitrogen PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA) according to the instructions of the manufacturer with lysostaphin (Sigma-Aldrich, St. Louis, MO, USA) used to lyse the S. aureus cell wall. lukSF-PV was amplified as previously described [31]. Amplicons were purified using the Ultraclean DNA PCR Clean Up Kit (MoBio Laboratories, GeneWorks, Thebarton, SA, Australia) and sequences were compared with the lukSF-PV genes from ϕSLT (Genbank: AB045978).
Bacteriophage induction and hybridisation
Bacteriophage were induced using Mitomycin C (Sigma-Aldrich, St. Louis, MO, USA) as previously described [10]. Plaques were transferred onto nylon membranes using standard techniques [32] and DNA was cross-linked to the membrane (Amersham Biosciences, Little Chalfont, Bucks, England) using a GS Gene Linker UV Chamber (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were treated with 2 mg/mL Proteinase K (Roche Diagnostics, Mannheim, Germany). The hybridisation probe was obtained by PCR amplification of lukSF-PV using previously described primers [33]. PCR products were purified using the MoBio PCR Cleanup Kit. Probes were prepared using the DIG DNA Labelling and Detection Kit according to the manufacturer’s instructions (Boehringer Mannheim, Mannheim, Germany). Plaque hybridisation was performed as directed by the manufacturer (Boehringer Mannheim, Mannheim, Germany).
PVL-bacteriophage propagation and lysogenisation of PVL-negative WA CA-MRSA
To propagate the mitomycin C-induced PVL-positive bacteriophages the plaques were extracted and crushed with 3 drops of BHIB, and the mixture left to stand for 10 minutes. This suspension was added to 100 μL of an overnight culture of the indicator strain, 3 mL of molten 3% BHIA was added and the mixture poured onto a BHIA plus 0.004M Ca2+ base plate which was incubated overnight at 30°C. The overlay containing the bacteriophage and indicator strain was scraped off and filtered. Each of the PVL-negative strains of WA CA-MRSA were grown overnight in BHIB and lawn-inoculated onto BHIA plus 0.004M CaCl2. A drop of each PVL-bacteriophage lysate was placed on the lawn and incubated at 30°C overnight. Isolated colonies growing in the centre of plaques present on lawns of PVL-negative WA clones were picked, their total DNA was isolated and lysogeny was detected using previously described primers [33].
Results
Sequence analysis and bacteriophage classification reveal diversity amongst the prophages
Fourteen prophage genomes between flanking direct 21 base pair (bp) repeats of 5′-AGGGCAAAAAAAGGGCg/aGATT-3′ termed attL and attR were analysed (Table 1). The 12 new prophage sequences from this study have been deposited in the NCBI database under accession numbers MF580410, MK940809 and MG029509 to MG029518.
The prophages were between 38,576 and 47,213 bp in size with between 41.4 and 100% nucleotide (nt) identity, GC compositions of 31 to 33.4% and 52 to 75 protein-coding sequences of 25 or more amino acids (aa).
The prophage genomes had the organisation of Siphoviridae family Sfi21-like PVL viruses of the Caudovirales order and, according to the most recent staphylococcal bacteriophage classification criteria, were placed into two genera and three PVL bacteriophage groups (Table 2) [34–36]. ϕSa2wa-st22, -st59 and -st772 (76.1–76.7% nt identity) were placed into the 77likevirus genus of icosahedral-headed bacteriophage. ϕSa2wa-st22 and -st772 were group 1 PVL bacteriophage with 74.4% nt identity and ϕSa2wa-st59 was group 3. ϕSa2wa-st1, -st5, -st8, -st30, -st72, -st78, -st80, -st93, -st93mssa, -st121mssa and ϕSa2USA (74.2–100% nt identity) were 3alikevirus genus, prolate-headed group 2 PVL bacteriophage. ϕSa2wa-st5 was unusual in that it encoded type C DNA polymerase (Genbank: AUM57702) rather than type A (exemplified by ϕSa2wa-st93 Genbank: AUM58245) (Fig 1).
Table 2. WA PVL prophage lukSF-PV polymorphisms, prophage classifications and lysogen lineages.
| Prophage | Lysogen CC, ST |
Lysogen Genus/PVL gp. | SNPs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| lukS-PV | lukF-PV | ||||||||||
| 33 | 105 | 345 | 443 | 527 | 663 | 1186 | 1396 | 1729 | |||
| ϕSLT | 30, ST30 | 3alikevirus/2 | G | T | C | G | A | G | C | A | A |
| ϕSa2wa-st30 | 30, ST30 | 3alikevirus/2 | G | T | C | G | A | G | C | A | A |
| ϕSa2wa-st772 | 1, ST772 | 77likevirus/1 | G | T | C | G | A | G | C | A | A |
| ϕSa2958 | 5, ST5 | 3alikevirus/2 | G | T | C | G | A | G | C | G | A |
| ϕSa2wa-st1 | 1, ST1 | 3alikevirus/2 | G | T | C | G | A | G | C | G | A |
| ϕSa2wa-st22 | 22, ST22 | 77likevirus/1 | G | T | C | G | A | G | C | G | A |
| ϕSa2wa-st59 | 59, ST59 | 77likevirus/3 | G | T | C | G | A | G | C | G | A |
| ϕSa2wa-st8 | 8, ST8 | 3alikevirus/2 | G | T | C | G | G | T | C | A | G |
| ϕSa2wa-st72 | 72, ST72 | 3alikevirus/2 | G | T | C | G | G | T | C | A | G |
| ϕSa2wa-st93 | S, ST93 | 3alikevirus/2 | G | T | C | G | G | T | C | A | G |
| ϕSa2wa-st93mssa | S, ST93 | 3alikevirus/2 | G | T | C | G | G | T | C | A | G |
| ϕSa2USA | 8, ST8 | 3alikevirus/2 | G | T | C | G | G | T | C | A | G |
| ϕSa2wa-st5 | 5, ST5 | 3alikevirus/2 | G | T | C | A | A | G | C | G | A |
| ϕSa2wa-st78 | 88, ST78 | 3alikevirus/2 | G | C | C | G | A | G | C | G | A |
| ϕSa2wa-st121mssa | 121, ST121 | 3alikevirus/2 | G | T | C | G | A | G | T | A | A |
| ϕSa2wa-st80 | 80, ST80 | 3alikevirus/2 | A | T | T | G | A | G | C | A | A |
| ϕSa2mw | 1, ST1 | 3alikevirus/2 | G | T | C | G | G | T | C | A | A |
Nucleotides differing from those of ϕSLT are shaded. Abbreviations: gp., group
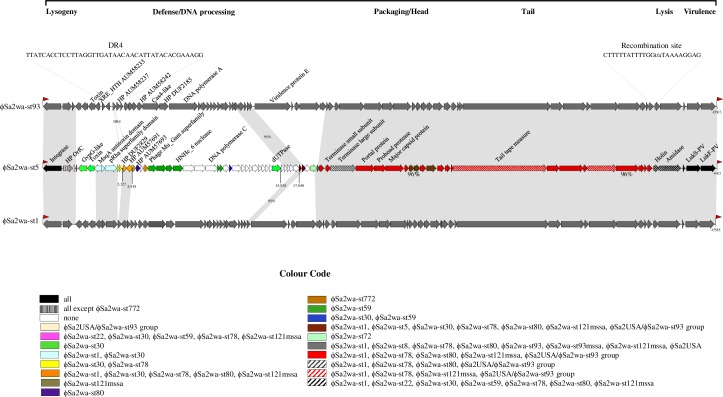
Fig 1. Diagrammatic comparison of ϕSa2wa-st5 with ϕSa2wa-st1 and ϕSa2wa-st93.
ϕSa2wa-st5 ORFs are represented as arrows indicating the direction of transcription and coloured according to the PVL prophage or groups of PVL prophages from PVL-positive S. aureus in WA that share 97 to 100% nucleotide identity. Where identity is less than 97% this is indicated under the ORFs. Regions of the genomes with 97 to 100% identity with ϕSa2wa-st5 are shaded. Where identity is less than 97% this is indicated in the shaded region. The main functional modules are indicated on a line above the genomes. Red flags indicate attL and attR sites. Genome size is indicated at the right-hand end. Proteins encoded by ORFs relevant to this study and structural proteins are indicated. Hypothetical proteins are identified by their Genbank accession number. The positions and sequences of DR4 and a widely-shared recombination site are presented. Abbreviations: DR, direct repeat; HP, hypothetical protein.
ϕSa2wa-st93, -st93mssa (100% nt identity) and -st8 (99.97% nt identity) were considered to be the same bacteriophage as the international ϕSa2USA (99.97% nt identity), with ϕSa2wa-st72 (96.6% nt identity) very closely related. These will be known as the ϕSa2USA/ϕSa2wa-st93 group in this study. The prophages found in the WA clones, WA115798 (ϕSa2wa-st1), WA2RNSH95 (ϕSa2wa-st78) and WA318790 (ϕSa2wa-st5) had identities of 80.8 to 93.8% and, although related, they were not identical to each other or any PVL bacteriophage in this study or in the NCBI database while ϕSa2wa-st8 from WA518751 had only 1 bp difference with ϕSa2USA and will be included in the ϕSa2USA/ϕSa2wa-st93 group.
The ϕSa2 chromosomal integration site was heterogenous
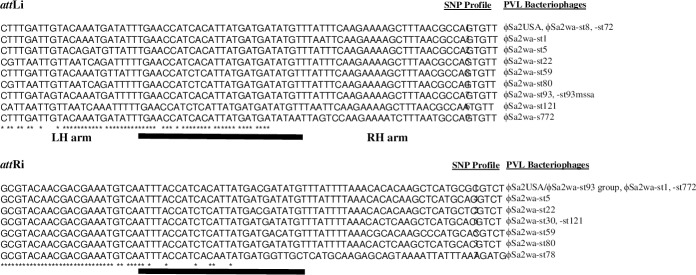
Chromosomal sequences proximal to the prophage terminals encoded the hybrid attLi and attRi sites of the attB and attP sites on the chromosome and a circularly permuted form of the bacteriophage. They consist of a 29-bp central core and 25-bp left-hand (LH) and right-hand (RH) arms (Fig 2) [37]. There were nine single nucleotide polymorphism (SNP) profiles for attLi and seven for attRi (Fig 2). Two groups of prophages shared identical attLi and attRi sites; international prophage ϕSa2USA and ϕSa2wa-st72 with ϕSa2wa-st8, and Australian international prophage ϕSa2wa-st93 with ϕSa2wa-st93mssa. Of the prophage in the WA CA-MRSA-like strains, ϕSa2wa-st8 shared attRi with the ϕSa2USA/ϕSa2wa-st93 group and ϕSa2wa-st772; ϕSa2wa-st1 had a unique attLi and ϕSa2wa-st8 shared attLi with ϕSa2wa-st72 and ϕSa2USA. ϕSa2wa-st5 and ϕSa2wa-st78 had unique integration-site sequences. attLi of ϕSa2wa-st78 could not be identified, however its attRi was reasonably similar to the ϕSa2wa prophages over the LH arm and the first 17 bp of the common core (3 bp difference) while 32 of the remaining 37 bp were different (Fig 2). attLi of ϕSa2wa-st30 was absent due to a 268 bp deletion (detected by comparison with the intact “preferred integration site” of WA2RNSH95).
Fig 2. Integration-site sequences proximal to the terminals of PVL prophages from PVL-positive S. aureus in WA (2005 to 2008).
Sequences have been aligned using ClustalW. The central core sequence is underlined with a thick black line. Left-hand and right-hand arms are indicated. SNP profiles are numbered alongside their respective bacteriophages. Identical nucleotides are indicated by an asterisk, absence of an asterisk indicates a polymorphic site. Abbreviations: LH, left hand; RH, right hand; attLi, left-hand integration site; attRi, right-hand integration site.
With the exception of ϕSa2wa-st78, the bacteriophages had inserted into a gene within a cluster of three or four open reading frames (ORFs) encoding a putative domain of unknown function (DUF)1672 lipoprotein [38], one downstream of the integration site and two or three upstream. The four DUF1672 domain-containing proteins of ϕSa2wa-st72 had amino acid similarity scores of 61.1–79.7% indicating they were paralogues. There was variability in the truncated ORF. ϕSa2wa-st1, -st5, -st59, -st772 and the ϕSa2USA/ϕSa2wa-st93 group had truncated the 3' end of an ORF encoding a lipoprotein_7 superfamily domain-containing protein (54.5–100% nt identity and 55.6–100% amino acid similarity) which variably also encoded a structural maintenance of the chromosome SMC_N domain (ϕSa2wa-st1, -st5, -st59 and -st72). ϕSa2wa-st22, -st30, -st80 and -st121mssa, had truncated an ORF encoding a hypothetical protein (HP) which was in the same position as the lipoprotein_7 domain ORF but lacked the lipoprotein_7 domain. ORFs truncated by ϕSa2wa-st22, -st30 and -st80 had 82.2–94% nt identity however, the ORF truncated by ϕSa2wa-st121mssa had only 26.4–36.2% nt identity. Immediately upstream of all prophages except ϕSa2wa-st78 was an ORF encoding a 62-aa HP whose sequence indicated that it was the 3' terminal of the truncated lipoprotein_7 protein ORF, when compared with the intact lipoprotein_7 domain-encoding ORF of WA2RNSH95 (not shown).
ϕSa2wa-st78 had truncated the 3' end of a 6-phospho-beta-galactosidase gene and inserted upstream of a galactose-6-phosphate degradation enzyme. The ϕSa2wa-st78 host genome, WA2RNSH95, encoded an intact lipoprotein_7 domain-encoding ORF that contained an attLi site which was homologous over the LH arm and central core (1 bp difference) with the consensus attLi but had 14 bp differences in the RH arm. This may have prevented insertion of ϕSa2wa-st78 into what appears to be a preferred site for ϕSa2.
Lysogeny regulation and modular recombination sequences
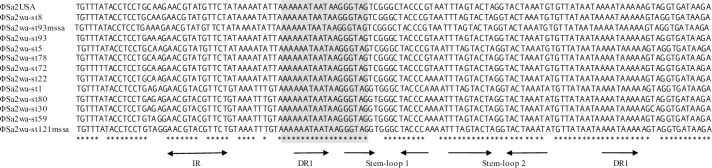
The intergenic region between the divergently transcribed integrase gene int and its associated HP ORF, originally called orfC [39] (Fig 1), contained structures indicative of involvement in regulation and lysogeny in all prophages (Fig 3). There were SNPs between the prophages, however all except ϕSa2wa-st772 had the same secondary structure which consisted of a consensus sigma factor H (SigH) binding-site [40] and a downstream inverted repeat (IR) of 5'-GAACGTAc/tGTTC-3'. Overlapping the SigH binding-site was an inverted repeat that could form a possible stem-loop structure of 5'-GGGTAGgtgggCTACCC-3' (stem-loop 1) (Fig 3). The first two nucleotides of the loop could be GT, TC or GC (ϕSa2wa-st772). There was then a previously identified and highly conserved stem-loop putative regulatory site, stem-loop 2 [41]. Both stem-loops were flanked by heptanucleotide direct repeats (DR) of 5'-AAAATAA-3 (DR1) the first of which comprised 7 bp of the SigH binding site.
Fig 3. Regulation regions of PVL prophages from PVL-positive S. aureus in WA (2005 to 2008).
Sequences have been aligned using ClustalW. The SigH binding site is shaded. Identical nucleotides in the alignment are indicated by an asterisk. Repeats are indicated by arrows. Abbreviations: IR = inverted repeat; DR = direct repeat.
ϕSa2wa-st772, which has previously been predicted to be a recombinant bacteriophage [23] had a regulation region that was somewhat different. The intergenic region was between int and a different HP ORF (exemplified by YP_00910342) transcribed on the same strand. The regulatory features however, included the SigH binding-site with its downstream IR and stem-loop 1; stem-loop 2 was absent and there was only one copy of DR1, which occurs from 24 to 33 times in the prophage genomes.
Of the prophages in the WA CA-MRSA the ϕSa2wa-st1 regulation region was identical with that of ϕSa2wa-st80 while ϕSa2wa-st5, -st8 and -st78 were identical with ϕSa2USA.
A previously described 23-bp recombination site that has been found in unrelated staphylococcal bacteriophage [42] was found downstream of the holin gene in all prophages (Fig 1). ϕSa2wa-st772 encoded the enterotoxin A gene flanked by direct repeats (DRs) of 5'-CTTTTTATTTTG-3' immediately downstream of this site thus implicating the site in the acquisition of an extra virulence factor, probably from an unrelated family ϕ3 beta haemolysin-converting bacteriophage.
ϕSa2wa-st5 has a module of 592 bp (bp 5,327–5,918) which encodes a DUF2829 protein and a HP ORF (Fig 1, Genbank: AUM57690 and AUM57691) flanked by 42-bp direct repeats (Fig 1, DR4). This module is present in ϕSa2wa-st1, -st30, -st78, -st80 and -st121mssa however, ϕSa2wa-st1, -st80 and -st121mssa lack a LH copy of DR4. Furthermore, this repeat is present as a similarly positioned single copy in all other study prophage except ϕSa2wa-st22 and -st772, indicating that the module has disseminated horizontally between bacteriophage of the same genera and the repeat is a conserved sequence that could mediate recombination, integration and excision.
lukS/F-PV sequence SNPs were not specific for PVL bacteriophage genera or lysogen genotype
A single copy of a DR that has been implicated in the deletion of the lukSF-PV and integrase module [43] was found downstream of the lukSF-PV genes in all prophages however, a second copy was not be found in any of the genomes. Nine lukSF-PV SNPs were found, six in lukS-PV and three in lukF-PV and there were eight SNP profiles (Table 2). All except the A→G (histidine→arginine) substitution at position 527 in the ϕSa2USA/ϕSa2wa-st93 group and ϕSa2MW, were synonymous. The ϕSa2USA/ϕSa2wa-st93 group SNPs were identical. ϕSa2wa-st30 and -st772 were identical to ϕSLT. ϕSa2wa-st22, -st59 and -st1 were identical to the CC5 control ϕ2958. ϕSa2wa-st78, -st80 and -st121mssa had individual lukSF-PV SNPs reported previously for their respective genetic lineages [44, 45] and ϕSa2wa-st5 had a unique lukSF-PV SNP profile.
The distribution of the SNP profiles was heterogenous. (i) Highly similar PVL prophage with the same lukSF-PV SNPs lysogenised S. aureus of three different lineages, indicating horizontal dissemination of a successful PVL bacteriophage between S. aureus of three lineages; the ϕSa2USA/ϕSa2wa-st93 group lysogenised ST8-IVa, ST72-IVa, ST93, and ST93-IVa. (ii) Different PVL prophages with different lukSF-PV SNP profiles lysogenised S. aureus of the same lineage, indicating horizontal transmission of different PVL bacteriophage into S. aureus of the same lineage; 77likevirus, PVL group 1 (ϕSa2wa-st772) and 3alikevirus, PVL group 2 (ϕSa2wa-st1 and ϕSa2mw) prophages lysogenised CC1 strains, ST772-V and ST1-IVa respectively. (iii) Different genera of PVL prophage with the same lukSF-PV SNP profile lysogenised different S. aureus lineages, indicating that different PVL bacteriophage can carry the same lukSF-PV module and that either the lukSF-PV genes disseminate horizontally between different genera of PVL bacteriophage or random substitutions occur during replication and the fittest permutations prevail regardless of the genus of PVL prophage; 77likevirus, PVL groups 1 (ϕSa2wa-st22) and 3 (ϕSa2wa-st59) lysogenised ST22-IVc and ST59-V respectively and 3alikevirus, PVL group 2 prophage (ϕ2958 and ϕSa2wa-st1) lysogenised ST5-II and ST1-IVa respectively.
Alpha alignment of colinear regions of ϕSa2wa-st1, -st5, -st59 and -st93 identified novel modules and heterogenous genes
The DNA processing main module is the most variable region in PVL bacteriophages and Fig 4 presents Alpha alignments of colinear sections of ϕSa2wa-st1, -st5, -st59 and -st93 from the 5' end of the bacteriophages. The region encodes conserved genes associated with lysogeny, and a variable region of early transcribed genes associated with lysogeny, bacteriophage defence and regulation. Variable genes and different colinear modules with similar functions can be detected in this graphical representation of heterogeneity.
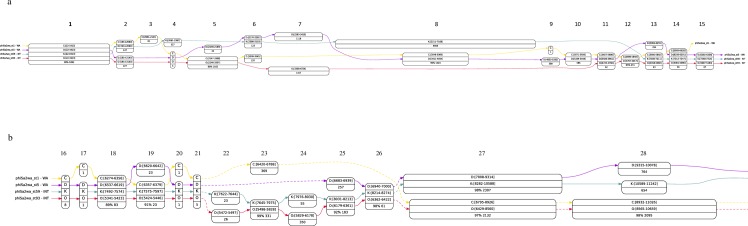
Fig 4.
A and B. Alpha alignment of colinear sections of ϕSa2wa-st1, ϕSa2wa-st5, ϕSa2wa-st59 and ϕSa2wa-st93. Each genome in the alignment is assigned an uppercase letter. Alignment positions for the corresponding genome are indicated in parentheses alongside the letter. Anchor sequences are similar segments of significant length shared by all genomes in the alignment. Nodes are gapless alignments specific for individual genomes in the alignment; they display the length and percent identity of the aligned region; unless otherwise indicated identity is 100%. Anchors and nodes are connected by color-coded arrows, one color for each genome and numbered sequentially on the figure. Dotted arrows replace nodes of less than 20 bp. Abbreviations: C, ϕSa2wa-st1; D, ϕSa2wa-st5; K, ϕSa2wa-st59; O, ϕSa2wa-st93; WA, Western Australian; INT, International.
Fig 4A has 15 alignment nodes. The anchor sequences are nodes 1 and 14 which encode int with the 5' end of orfC (exemplified by ϕSa2wa-st5, Genbank: AUM57679 and AUM57680) and the 5' terminal of a helix-turn-helix (HTH) domain-encoding ORF (Genbank: AUM57693) respectively (Fig 1). The genomes diverge at bp 1,913 (Alpha alignment node 1). Following two or three HPs of unknown function ϕSa2wa-st1 (c3081-3542), -st5 (c2946-3416) and -st93 (c3080-3550) encode a putative toxin gene, (Genbank: AUM57684, 72–89% nt identity, 82.8–94.9% aa similarity) (Fig 1). The ϕSa2wa-st1 and -st93 putative toxin sequences (77.3% nt identity) diverge at bp 3,308 and 3,307 respectively (Alpha alignment node 5). All polymorphisms in these ORFs are in the 5' ends and most (27/38) are non-synonymous. Non-random distribution of polymorphisms such as this indicates homologous recombination between divergent genes has probably occurred. ϕSa2wa-st5 has a gas vesicle protein G (GvpG) domain-encoding ORF (Genbank: AUM57683) downstream of the toxin gene (Genbank: AUM57684, Alpha alignment node 7) which has 89% nt identity with the ϕSa2wa-st1 toxin gene. The ϕSa2wa-st1 and -st5 toxin genes are followed by a xenobiotic response element (XRE)-HTH putative transcriptional regulator ORF which also encodes a MqsA antitoxin superfamily domain (Genbank: AUM57685, Alpha alignment node 8, 99.4% nt identity), while ϕSa2wa-st93 encodes a XRE-HTH transcriptional regulator (Genbank: AUM58233, Alpha alignment node 7). ϕSa2wa-st1 and -st5 then encode a XRE_HTH family regulator followed by a bacteriophage pRha superfamily domain-containing protein that interferes with infection of strains that lack integration host factor (Genbank: AUM57686 and AUM57687, Alpha alignment node 8).
When compared with all the WA-PVL prophages the node 8 module of ϕSa2wa-st1 and -st5 is shared with only ϕSa2wa-st30, and the node 7 module of ϕSa2wa-st93 is shared with only the ϕSa2USA/ϕSa2wa-st93 group. ϕSa2wa-st59 encodes a unique 4,658 bp section (bp 2511–7168), which encodes two XRE family transcriptional regulators, a bacteriophage anti-repressor and 10 putative HPs (AUM57878 to AUM57891, Alpha alignment node 8). Nodes 13, 14 and 15 encode the 5' end of a HP ORF (Genbank: AUM58237) with a common core (Alpha alignment node 14) and divergence by ϕSa2wa-st5 and ϕSa2wa-st1 indicated by the graph.
Following two to four heterogenous colinear HP ORFs the Fig 4B Alpha alignment node 27 reveals two unrelated colinear modules. ϕSa2wa-st5 and -st59 encode ORFs for a bacteriophage Mu Gam-like protein which protects double stranded DNA from exonuclease degradation, two overlapping single-stranded binding proteins and a putative HNHc_6 superfamily nuclease (Genbank: AUM57696 to AUM57698) which are not shared by any other WA PVL prophage (Fig 1). ϕSa2wa-st1 and -st93 encode three overlapping ORFs encoding a HP, a Cas4-like protein and a DUF2185 protein (Genbank: AUM58242, AUM58243 and AUM58244). This module may be a defence system or part thereof against the bacterial CRISPR-Cas system and it is shared by all study prophage (97–100% nt identity) except ϕSa2wa-st5, -st22, -st59 and -st772. ϕSa2wa-st1 and -st93 then encode DNA polymerase A (Alpha alignment node 28) while ϕSa2wa-st5 (Fig 1) and -st59 encode DNA polymerase C.
Alpha aligner-defined nodes reveal extensive mosaicism and recombination in PVL bacteriophage from WA
The singleton ST93 genome is stable and well-adapted in the geographical region. To further investigate the mosaicism in the prophages Table 3 presents the nodes of ϕSa2wa-st93 as determined by the Alpha aligner in Fig 4A and 4B, identifies the putative proteins or protein sections and shows the local prophages that encode the same sequence with 97 to 100% nt identity. ϕSa2wa-st8, -st93mssa and ϕSa2USA are almost identical to ϕSa2wa-st93 and have been excluded from the Table. There is evidence of extensive recombination. The only prophage that was identical in this region was the closely related ϕSa2wa-st72 and the only prophage not to share any module was ϕSa2wa-st772. Most of the shared sequence involved the prolate-headed 3alikevirus prophages however, there was also evidence of recombination with the 77likevirus icosahedral-headed ϕSa2wa-st59. The Alpha aligner defined modules consist of split genes, single genes, groups of genes and intergenic regions, some shared by several prophages and others by only one or two. At 96.6% nt identity ϕSa2wa-st72 is a member of the ϕSa2USA/ϕSa2wa-st93 group in this study and the Table 3 modules with homology only with ϕSa2wa-st72 represent modules encoding functions that, amongst the prophages in this study, are unique to this successful bacteriophage.
Table 3. Alpha aligner defined nodes of ϕSa2wa-st93 and node-associated ORFs or intergenic regions having 97–100% sequence identity with other WA PVL prophages.
| ϕSa2wa-st93 Position/Node* | Protein(s) accession no’s, HPs or regions | WA PVL bacteriophages with 97–100% sequence identity |
|---|---|---|
| 22-1913/1 | Integrase, AUM58227; Split HP, AUM58228 | ϕSa2wa-st1, -st5, -st22, -st30, -st59, -st72, -st78, -st80 -st121mssa |
| 1914-2140/2 | Split HP, AUM58228; split HP AUM58229 | ϕSa2wa-st5, -st72, -st78 |
| 2146-3307/5 | Split HP, AUM58229; HP, AUM58230; HP, AUM58231; split Toxin, AUM58232 | ϕSa2wa-st1, -st72 |
| 3308-4724/7 | Split Toxin, AUM58232; HP, AUM58233; HP, AUM58234; HP, AUM58235 | ϕSa2wa-st72 |
| 4725-4766/11 | Intergenic region | ϕSa2wa-st1, -st5, -st30, -st59, -st72, -st78, -st80, -st121mssa |
| 4767-5017/12 | HP, AUM58236 | ϕSa2wa-st72 |
| 5018-5060/13 | Split HP, AUM58237 | ϕSa2wa-st1, -st30, -st59, -st72, -st78 |
| 5061-5096/14 | Split HP, AUM58237 | ϕSa2wa-st1, -st5, -st30, -st59, -st72, -st78, -st80, ϕ -st121mssa |
| 5097-5163/15 | Split HP, AUM58237 | ϕSa2wa-st5, -st30, -st59, -st72, -st78, -st80, -st121mssa |
| 5341-5423/18 | Split DUF1270, AUM58238 | ϕSa2wa-st72, -st78 |
| 5424-5446/19 | Split DUF1270, AUM58238 | ϕSa2wa-st72, -st78 |
| 5472-5497/22 | Intergenic region | ϕSa2wa-st72 |
| 5498-5828/23 | HP, AUM58239; split DUF2482 HP, AUM58240 | ϕSa2wa-st59, -st72 |
| 5829-6178/24 | Split DUF2482 HP, AUM58240; split DUF1108 HP, AUM58241 | ϕSa2wa-st72 |
| 6179-6361/25 | Split DUF1108 HP, AUM58241 | ϕSa2wa-st72 |
| 6362-6422/26 | Split DUF1108 HP, AUM58241 | ϕSa2wa-st5, -st59, -st72 |
| 6429-8560/27 | HP, AUM58242; Cas4-like, AUM58243; DUF2815 HP, AUM58244 | ϕSa2wa-st1, ϕ -st30, -st72, -st78, -st80, -st121mssa |
| 8565-10659/28 | DNA polymerase A, AUM58245; split DUF3113 HP, AUM58246 | Sa2wa-st1, -st30, -st72, -st78, -st80, -st121mssa |
Proteins and hypothetical proteins are indicated by their Genbank protein-id number. Genbank domains of unknown function are indicated; Split proteins represent split open reading frames. Abbreviations: DUF, domain of unknown function; HP, hypothetical protein; no’s, numbers
*As presented in Fig 4
With between 77% and 82.9% nt identity ϕSa2wa-st5 from WA3 was the most distantly related of the 3alikevirus group and was successfully induced and therefore probably transmissible (Table 4). It had highest homology with ϕSa2958 (Genbank: AP009363; 99% nt identity over 72% of the genome) however, this was essentially in the structural morphology and lysis, virulence and lysogeny modules. Fig 1 presents a diagrammatic comparison of ϕSa2wa-st5, -st93 and -st1 with the ϕSa2wa-st5 ORFs coloured according to the WA PVL-prophages that shared 97–100% sequence identity. With the exception of the bp 15,838 to 17,040 module which was homologous with non-PVL bacteriophage 53 (Genbank: AY954952; 100% nt identity) the unshared regions of ϕSa2wa-st5 encoding multiple ORFs were unique. As well as random mutations that occur during chromosomal replication this heterogeneity indicates that horizontal recombination has occurred between the bacteriophage during their evolution.
Table 4. Bacteriophage induction and lysogenisation of historic PVL-negative WA CA-MRSA.
| Lysogen | Lysogenised recipients Total pfu/mL |
PVL positive plaques | Induced PVL bacteriophage | PVL-negative CA-MRSA lysogenised | |
|---|---|---|---|---|---|
| RN4220 | WBG286 | ||||
| MW2 | >1x105 | 0 | >100 | ϕSa2mw | WA5WBG7583 |
| WA115798 | 0 | 0 | 0 | 0 | NA |
| WA2RNSH95 | 0 | 1x103 | 0 | 0 | NA |
| WA318790 | 2x103 | 0 | 20 | ϕSa2wa-st5 | None |
| W17S | 0 | 1x102 | 1 | ϕSa2wa-st93mssa | Not tested |
| K25S | 3x102 | 0 | 3 | ϕSa2wa-st121mssa | Not tested |
| Qld Clone16790 | 0 | 1x102 | 1 | ϕSa2wa-st93 | WA5WBG7583 |
Abbreviations: NA, not applicable; pfu, plaque forming units.
in-vitro induction of PVL-positive ϕSa2 from Australian S. aureus and lysogenisation of historic PVL-negative WA CA-MRSA
To test the transmissibility of PVL-positive ϕSa2 lysogenising the Australian S. aureus and the lysogenic capabilities of the historic PVL-negative WA CA-MRSA (Table 1), in-vitro induction, propagation and lysogenisation experiments were performed (Table 4).
Bacteriophage were induced from all isolates tested except WA115798. Overall, only two of the five indicator strains, RN4220 and WBG286 were lysogenised and specifically, only one in each induction experiment, demonstrating some specificity of lysogenisation (Table 4). Hybridisation of the plaques revealed that ϕSa2wa-st5, -st93mssa, -st93, -st121mssa and the control, ϕSa2mw, were induced out of their lysogens. ϕSa2wa-st78 may not have been induced because it lacked an evident attLi integration site (Fig 1) however, the reason why ϕSa2wa-st1 was not induced is currently unclear.
ϕSa2wa-st121mssa could not be propagated to a sufficiently high titre in-vitro however, ϕSa2wa-st5, -st93 and the control, ϕSa2mw, were tested for their ability to lysogenise all of the historic PVL-negative WA CA-MRSA. WA5WBG7583 was lysogenised with ϕSa2wa-st93 and the control, ϕSa2mw, but not ϕSa2wa-st5. None of the other PVL-negative WA CA-MRSA were lysogenised in-vitro with any of the induced and propagated bacteriophages.
Discussion
The genomes of PVL prophages from temporally and geographically related S. aureus of local and international origin have revealed an unexpected amount of diversity that has made it difficult to trace their origins. There has been recombination between bacteriophage of the same and different genera as well as genetic diversity in the chromosomal integration sites, the regulation regions, the defence, DNA-processing, structural and packaging modules and the lukSF-PV genes. There was no evidence that the icosahedral-headed prophages from international clones of CA-MRSA had transferred to the WA clones. The prolate-headed prophage formed the largest group however, with the exception of ϕSa2wa-st8, they were so diverse it was not possible to determine if there had been horizontal transmission of whole bacteriophages. There has been recombination between the international and local prolate-headed bacteriophage and, to a lesser extent, also between prolate- and icosahedral-headed bacteriophage that are present in WA at some stage during their evolution.
With 99.97% sequence identity, it is evident that WA518751 has probably acquired ϕSa2wa-st8 in the WA community from either a ST93 S. aureus or USA300. ϕSa2wa-st93 was induced in-vitro and then it lysogenised PVL-negative WA5WBG7583 demonstrating that this clone can accept the bacteriophage. On-the-other-hand, WA518751 and USA300 had identical ϕSa2 integration-site sequences indicating that the bacteriophage could also have been horizontally transmitted from USA300.
ϕSa2wa-st8, -st72, -st93, -st93mssa and ϕSa2USA probably represent a single bacteriophage that has transmitted between lineages of S. aureus. USA300 and the Queensland clone are two of the most virulent and widely disseminated CA-MRSA and in this and a previous study [46] it is evident that there has been horizontal transmission of a ϕSa2USA/ϕSa2wa-st93-type bacteriophage between their CC8 and Singleton 93 ancestors, however there is no indication of when or where this occurred. USA300 acquired ϕSa2USA in North America following importation of its ancestor in the early 20th century [47]. ϕSa2wa-st93mssa was present in ST93-MSSA, the most prevalent colonizer in remote WA in 1995, and this clone was the ancestor of the Queensland clone that emerged in Queensland, Australia in the early 2000’s [14, 48, 49]. ϕSa2wa-st93mssa was well adapted in ST93-MSSA and Australia before the clone acquired the SCCmec and before USA300 was imported into Australia [22]. Against the background of PVL prophage diversity revealed in this study it is extraordinary that the ϕSa2USA/ϕSa2wa-st93 bacteriophage has remained stable over at least 20 years in different geographic and genetic environments. To gain insights into the success of this bacteriophage it would be informative to investigate the putative proteins of unknown function encoded by the unique modules of ϕSa2wa-st93 revealed in Table 3.
The international Korean CA-MRSA clone is characteristically PVL-negative [50] and has probably acquired ϕSa2wa-st72 in the WA community. This may represent a recent acquisition of a ϕSa2USA/ϕSa2wa-st93 bacteriophage with the prophage undergoing gradual changes as it adapts to a CC72 background and different geographical conditions. As with the WA PVL-positive CA-MRSA the PVL-positive Korean clone has not thrived and forms only 0.02% of CA-MRSA in the WA community [19].
The diversity in shared genes such as the hypothetical proteins that have been split in Table 3 according to their homologies with all the prophage in the study is interesting. In the putative toxin genes, all of the nucleotide differences were in the 5' end of the ORF and most resulted in different amino acids. This may be a defence strategy that has involved recombination within genes resulting in proteins with the same function but different antigenic profiles.
With the exception of two pairs of prophage all had distinct integration-site sequences however, the impact of this on the specificity of lysogenisation is currently unknown. The effect of lysogeny by ϕSa2 on host fitness could not be determined. The preferred insertion site was within a lipoprotein_7 domain-encoding ORF within a paralogous cluster of three or four ORFs that encoded a DUF1672 lipoprotein. As has been previously reported, the ORFs truncated by ϕSa2wa-st22, -st30 and -st80 type bacteriophage [37] and now ϕSa2wa-st121, were different, however they were similarly positioned within the same DUF1672 lipoprotein cluster. Lipoproteins serve as transporters of nutrients and contribute to virulence and fitness in S. aureus and increased complements have been associated with particularly pathogenic strains [38]. The impact of truncation of the ϕSa2 target genes on host fitness requires further investigation.
Prophage lysogenising 11 lineages of S. aureus have been investigated in this study and adaptation to different genetic backgrounds is undoubtedly one of the reasons for the diversity observed. Investigation of more genomes of PVL prophage from S. aureus belonging to the same genetic lineage is now required. The low occurrence of PVL-positive variants of established PVL-negative CA-MRSA in the WA community suggests that the clones may not have adapted well to the acquisition of PVL-positive ϕSa2.
Acknowledgments
This work was funded by grants from the Health Department of WA and Curtin University. The authors would like to acknowledge Tam Le who performed the bacteriophage PVL gene typing and induction experiments, the scientists of the Australian Collaborating Centre for Enterococcus and Staphylococcus Species (ACCESS) Typing and Research for typing the isolates and provision of epidemiological information and Warren Grubb for critical reading of the manuscript.
Data Availability
All relevant data are within the manuscript.
Funding Statement
This work was funded by Curtin University and the Health Department of Western Australia.
References
- 1.Wertheim HFL, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–62. 10.1016/S1473-3099(05)70295-4 WOS:000233691900020. [DOI] [PubMed] [Google Scholar]
- 2.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359(9320):1819–27. 10.1016/s0140-6736(02)08713-5 . [DOI] [PubMed] [Google Scholar]
- 3.Yoong P, Pier GB. Immune-activating properties of Panton-Valentine leukocidin improve the outcome in a model of methicillin-resistant Staphylococcus aureus pneumonia. Infect Immun. 2012;80(8):2894–904. Epub 2012/06/06. 10.1128/IAI.06360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremieux AC, Dumitrescu O, Lina G, Vallee C, Cote JF, Muffat-Joly M, et al. Panton-Valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. Plos One. 2009;4(9):e7204 Epub 2009/09/26. 10.1371/journal.pone.0007204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery CP, Daum RS. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for Panton-Valentine leukocidin? Infect Immun. 2009;77(5):2159–67. Epub 2009/02/25. 10.1128/IAI.00021-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boakes E, Kearns AM, Badiou C, Lina G, Hill RL, Ellington MJ. Do differences in Panton-Valentine leukocidin production among international methicillin-resistant Staphylococcus aureus clones affect disease presentation and severity? J Clin Microbiol. 2012;50(5):1773–6. Epub 2011/12/30. 10.1128/JCM.06421-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehrhahn MC, Robinson JO, Pascoe EM, Coombs GW, Pearson JC, O’Brien FG, et al. Illness severity in community-onset invasive Staphylococcus aureus infection and the presence of virulence genes. J Infect Dis. 2012;205:1840–8. 10.1093/infdis/jis279 [DOI] [PubMed] [Google Scholar]
- 8.Shallcross LJ, Fragaszy E, Johnson AM, Hayward AC. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: a systematic review and meta-analysis. Lancet Infect Dis. 2013;13(1):43–54. Epub 2012/10/30. 10.1016/S1473-3099(12)70238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlemann A, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2015;21:563–74. 10.1016/jmeegid.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma XX, Ito T, Kondo Y, Cho M, Yoshizawa Y, Kaneko J, et al. Two different Panton-Valentine leukocidin phage lineages predominate in Japan. J Clin Microbiol. 2008;46(10):3246–58. Epub 2008/08/08. 10.1128/JCM.00136-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1(4):298–303. Epub 2011/10/29. 10.1016/j.coviro.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldron DE, Lindsay JA. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188(15):5578–85. Epub 2006/07/21. 10.1128/JB.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol. 2014;6(10):2697–708. Epub 2014/09/28. 10.1093/gbe/evu214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien FG, Coombs GW, Pearman JW, Gracey M, Moss F, Christiansen KJ, et al. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Antimicrob Chemother. 2009;64(4):684–93. 10.1093/jac/dkp285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien FG, Lim TT, Chong FN, Coombs GW, Enright MC, Robinson DA, et al. Diversity among community isolates of methicillin-resistant Staphylococcus aureus isolates in Australia. J Clin Microbiol. 2004;42:3185–90. 10.1128/JCM.42.7.3185-3190.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombs GW, Nimmo GR, Bell JM, Huygens F, O’Brien FG, Malkowski MJ, et al. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J Clin Microbiol. 2004;42(10):4735–43. 10.1128/JCM.42.10.4735-4743.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40(11):4289–94. 10.1128/JCM.40.11.4289-4294.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coombs GW, Monecke S, Pearson JC, Tan HL, Chew YK, Wilson L, et al. Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region. BMC Microbiol. 2011;11:215 10.1186/1471-2180-11-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coombs GW, Pearson JC, Robinson O. Western Australian Methicillin-Resistant Staphylococcus aureus (MRSA) Epidemiology and Typing Report. Gram-positive Bacteria Typing Laboratory MD, Fiona Stanley Hospital, PathWest Laboratory Medicine—WA; 2016.
- 20.Swenson KM, Guertin P, Deschenes H, Bergeron A. Reconstructing the modular recombination history of Staphylococcus aureus phages. BMC Bioinformatics. 2013;14 Suppl 15:S17 10.1186/1471-2105-14-S15-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berard S, Chateau A, Pompidor N, Guertin P, Bergeron A, Swenson KM. Aligning the unalignable: bacteriophage whole genome alignments. BMC Bioinformatics. 2016;17:30 Epub 2016/01/14. 10.1186/s12859-015-0869-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monecke S, Ehricht R, Slickers P, Tan HL, Coombs G. The molecular epidemiology and evolution of the Panton-Valentine leukocidin-positive, methicillin-resistant Staphylococcus aureus strain USA300 in Western Australia. Clin Microbiol Infec. 2009;15(8):770–6. Epub 2009/06/30. 10.1111/j.1469-0691.2009.02792.x . [DOI] [PubMed] [Google Scholar]
- 23.Monecke S, Baier V, Coombs GW, Slickers P, Ziegler A, Ehricht R. Genome sequencing and molecular characterisation of Staphylococcus aureus ST772-MRSA-V, "Bengal Bay Clone". BMC Res Notes. 2013;6 http://www.biomedcentral.com/1756-0500/6/548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien FG, Pearman JW, Gracey M, Riley TV, Grubb WB. Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. J Clin Microbiol. 1999;37(9):2858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adhikari RP, Cook GM, Lamont I, Lang S, Heffernan H, Smith JM. Phenotypic and molecular characterization of community occurring, Western Samoan phage pattern methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2002;50(6):825–31. 10.1093/jac/dkf242 . [DOI] [PubMed] [Google Scholar]
- 26.Coombs GW, Monecke S, Ehricht R, Slickers P, Pearson JC, Tan HL, et al. Differentiation of clonal complex 59 community-associated methicillin-resistant Staphylococcus aureus in Western Australia. Antimicrob Agents Ch. 2010;54(5):1914–21. Epub 2010/03/10. 10.1128/AAC.01287-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegger M, Wirth T, Andersen PS, Skov RL, De Grassi A, Simoes PM, et al. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. MBio. 2014;5(5):e01044–14. Epub 2014/08/28. 10.1128/mBio.01044-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coombs GW, Goering RV, Chua KY, Monecke S, Howden BP, Stinear TP, et al. The molecular epidemiology of the highly virulent ST93 Australian community Staphylococcus aureus strain. Plos One. 2012;7(8):e43037 Epub 2012/08/18. 10.1371/journal.pone.0043037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367(9512):731–9. Epub 2006/03/07. 10.1016/S0140-6736(06)68231-7 . [DOI] [PubMed] [Google Scholar]
- 30.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. 10.1016/0195-6701(93)90100-e [DOI] [PubMed] [Google Scholar]
- 31.Wolter DJ, Tenover FC, Goering RV. Allelic variation in genes encoding Panton-Valentine leukocidin from community-associated Staphylococcus aureus. Clin Microbiol Infec. 2007;13(8):827–30. Epub 2007/07/06. 10.1111/j.1469-0691.2007.01763.x . [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual press Csh, editor. New York: 1989. [Google Scholar]
- 33.Fey PD, Said-Salim B, Rupp ME, Hinrichs SH, Boxrud DJ, Davis CC, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob Agents Ch. 2003;47(1):196–203. 10.1128/AAC.47.1.196-203.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deghorain M, Van Melderen L. The staphylococci phages family: an overview. Viruses. 2012;4(12):3316–35. Epub 2013/01/25. 10.3390/v4123316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez D, Adriaenssens EM, Martinez B, Rodrigtuez A, Lavigne R, Kropinsky AM, et al. Three proposed new bacteriophage genera of staphylococcal phages: "3likevirus", "77likevirus" and "Phietalikevirus". Arch Virol. 2014;159:389–98. 10.1007/s00705-013-1833-1 [DOI] [PubMed] [Google Scholar]
- 36.Zhang M, Ito T, Li S, Jin J, Takeuchi F, Yang YH, et al. Identification of a third type of PVL phage in ST59 methicillin-resistant Staphylococcus aureus (MRSA) strains. FEMS Microbiol Letts. 2011;323:20–8. [DOI] [PubMed] [Google Scholar]
- 37.Boakes E, Kearns AM, Ganner M, Perry C, Hill RL, Ellington MJ. Distinct bacteriophages encoding Panton-Valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J Clin Microbiol. 2011;49(2):684–92. Epub 2010/11/26. 10.1128/JCM.01917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shahmirzadi SV, Nguyen MT, Gotz F. Evaluation of Staphylococcus aureus Lipoproteins: Role in Nutritional Acquisition and Pathogenicity. Front Microbiol. 2016;7:1404 Epub 2016/09/30. 10.3389/fmicb.2016.01404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll D, Kehoe MA, Cavanagh D, Coleman DC. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi 13 and phi 42. Mol Microbiol. 1995;16(5):877–93. Epub 1995/06/01. 10.1111/j.1365-2958.1995.tb02315.x . [DOI] [PubMed] [Google Scholar]
- 40.Tao L, Wu X, Sun B. Alternative sigma factor SigmaH modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog. 2010;6(5):e1000888 Epub 2010/05/21. 10.1371/journal.ppat.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002;289(1–2):109–18. Epub 2002/05/31. 10.1016/s0378-1119(02)00481-x . [DOI] [PubMed] [Google Scholar]
- 42.Kraushaar B, Hammerl JA, Kienol M, Heinig ML, Sperling N, Dinh Thanh M, et al. Acquisition of virulence factors in livestock-associated MRSA: Lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci Rep. 2017;7(1):2004 Epub 2017/05/19. 10.1038/s41598-017-02175-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Haddad L, Moineau S. Characterization of a Novel Panton-Valentine leukocidin (PVL)-encoding staphylococcal phage and its naturally PVL-lacking variant. App Env Microbiol. 2013;79(8):2828–32. Epub 2013/02/12. 10.1128/AEM.03852-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otter JA, Kearns AM, French GL, Ellington MJ. Panton-Valentine leukocidin-encoding bacteriophage and gene sequence variation in community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol and Infect: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010;16(1):68–73. Epub 2009/08/28. 10.1111/j.1469-0691.2009.02925.x . [DOI] [PubMed] [Google Scholar]
- 45.Dumitrescu O, Tristan A, Meugnier H, Bes M, Gouy M, Etienne J, et al. Polymorphism of the Staphylococcus aureus Panton-Valentine leukocidin genes and its possible link with the fitness of community-associated methicillin-resistant S. aureus. J Infect Dis. 2008;198:792–4. 10.1086/590914 [DOI] [PubMed] [Google Scholar]
- 46.McCarthy AJ, Witney AA, Lindsay JA. Staphylococcus aureus temperate bacteriophage: carriage and horizontal gene transfer is lineage associated. Front Cell Infect Mi. 2012;2:6 Epub 2012/08/25. 10.3389/fcimb.2012.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss L, Stegger M, Akpaka PE, Alabi A, Breurec S, Coombs G, et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. P Natl Acad Sci USA. 2017;114(49):E10596–E604. Epub 2017/11/22. 10.1073/pnas.1702472114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinear TP, Holt KE, Chua K, Stepnell J, Tuck KL, Coombs G, et al. Adaptive change inferred from genomic population analysis of the ST93 epidemic clone of community-associated methicillin-resistant Staphylococcus aureus. Genome Biol Evol. 2014;6(2):366–78. Epub 2014/02/01. 10.1093/gbe/evu022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hal SJ, Steinig EJ, Andersson P, Holden MTG, Harris SR, Nimmo GR, et al. Global scale dissemination of ST93: A divergent Staphylococcus aureus epidemic lineage that has recently emerged from remote northern Australia. Front Microbiol. 2018;9:1453 Epub 2018/07/25. 10.3389/fmicb.2018.01453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim ES, Song JS, Lee HJ, Choe PG, Park KH, Cho JH, et al. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother. 2007;60(5):1108–14. Epub 2007/09/22. 10.1093/jac/dkm309 . [DOI] [PubMed] [Google Scholar]