Abstract
Hairy cell leukemia (HCL) is a purine analog-responsive B-cell malignancy containing the BRAF V600E mutation, expressing CD22, CD11c, CD103, tartrate resistant acid phosphatase (TRAP) CD25, CD123, and annexin 1A. BRAF V600E and the latter 4 markers are usually absent in the more aggressive and chemoresistant variant HCLv. To evaluate differences between HCL and HCLv, expression microarrays comparing HCL with HCLv were performed for 24694 genes using 47323 probes. Microarray data from 35 HCL and 27 HCLv purified samples showed the greatest HCL-HCLv difference in the muscle-associated gene MYF6, expressed by its 2 probes 18.5- and 10.8-fold higher in HCL than HCLv (p<0.0001). By real-time quantitative PCR (RQ-PCR), 100% of 152 classic HCL samples were MYF6-positive, vs 5 (6%) of 90 blood donors. MYF6-expression was also detected in 18 (35%) of 51 with HCLv, 11 (92%) of 12 with HCL expressing unmutated IGHV4-34, 35 (73%) of 48 with chronic lymphocytic leukemia (CLL), and 1 (8%) of 12 with mantle cell lymphoma. Hypomethylation status of MYF6 supported expression in HCL more than HCLv. Posttreatment blood samples becoming negative by flow cytometry remained MYF6+ by RQ-PCR in 42 (48%) of 87 HCL patients, and MYF6 RQ-PCR could detect 1 HCL in 105 normal cells. MYF6, universally expressed in HCL and in most CLL samples, may be a useful biomarker for these leukemias. Further studies are underway to determine the role of MYF6 in HCL.
Introduction
Classic hairy cell leukemia (HCL) is a B-cell malignancy with distinctive immunophenotype, typically having BRAF V600E mutation, and expressing CD20, CD22, CD25, CD11c, CD103, CD123, annexin A1 (Anxa1), and tartrate-resistant acid phosphatase (TRAP) [1–4]. Purine analogs achieve high rates of durable complete remissions (CR), often with minimal residual disease (MRD) [5, 6]. HCL variant (HCLv), recognized as a separate disorder [3, 7, 8], generally lacks CD25, CD123, annexin A1, TRAP, and BRAF V600E, responds more poorly to therapy, and survival from diagnosis is shorter [2, 9–12]. We reported that HCL expressing unmutated (>98% homology to germline) immunoglobulin heavy-chain variable (IGHV) rearrangement type IGHV4-34 expresses wild-type BRAF and has a poor prognosis like HCLv, whether immunophenotypically consistent with HCLv or HCL [12, 13]. Mutations within MAP2K1 encoding MEK1 have been found in HCLv and IGHV4-34+ HCL [14–16].
The human MYF6 (human Myogenic Factor 6, MRF4, or Herculin) gene is mapped to 12q21 next to another myogenesis regulated factor, MYF5 [17]. MYF6 cDNAs were isolated first from human and mouse skeletal muscle, the only tissue in which expression of the corresponding mRNA was observed [18]. The MYF6 protein is a member of a family of trans-acting transcription factors, also known as myogenic regulatory factors, including MyoD1 (Myf3) [19], myogenin (MyoG, MYF4) [20, 21] , MYF5 and MYF6 [22]. Myogenic regulatory factors are involved in the development of skeletal muscle by controlling the expression of muscle specific genes [23]. Each of these four genes encodes a highly conserved basic-helix-loop-helix (bHLH) region that is responsible for the binding of Myf proteins to E-box sites (CANNTG) located in the promoter region of muscle-specific genes. They were reported expressed in normal tissue exclusively in striated muscle [18, 20, 22].
In neoplasia, MYF6 expression was reported in 33% of rhabdomyosarcomas [23] and silent corticotroph macroadenomas [24]. MYF6 gene hypomethylation was found in non-small cell lung cancer (NSCLC), associated with stage I disease [25]. Microarray-based expression of chronic lymphocytic leukemia (CLL) samples listed MYF6 expression, associated with trisomy 12 [26], validated by real-time PCR [27]. To determine genes differentially expressed in HCL vs normal B-cells and other B-cell malignancies, samples from 10 HCL patients were compared with normal B-cells and samples from 46 patients with B-cell lymphomas or CLL, in microarray studies [28]. A total of 82 genes including MYF6 were reported upregulated in HCL, and MYF6 was one of 22 genes shown by immunohistochemistry to be expressed at the protein level [28]. Basso et al. reported MYF6 expression among 8602 other genes in 336 samples which included 16 HCL [29]. To our knowledge MYF6 expression in HCL was not further investigated, nor was it studied in HCLv. Using microarray studies, we decided to study genes upregulated in HCL as opposed to HCLv, initially without considering MYF6.
Material and methods
Patients and leukemic cells
Samples from patients with HCL, HCLv and other leukemias or controls were obtained via protocol 10-C-0066 approved by the National Cancer Institute Institutional Review Board. Patients gave written informed consent. All patients were adults and therefore did not require consent from parents or guardians. Per protocol, consent is waived in cases where patients died. Patients were consented from March 2010 to August 2019. Patients consented were either inquiring about clinical trials or had questions related to their disease and were invited to give consent to submit research samples. Demographic details of the patients are shown in Table 1. Patients included had a diagnosis of a hematologic malignancy, regardless of the status of their disease. The protocol also allowed recruitment of normal controls. Most of the samples were from patients with HCL and HCLv. The patient population was skewed toward those patients with multiply relapsed disease, although patients with newly diagnosed HCL were also consented. HCL and HCLv cells for microarray studies were obtained in sodium heparin tubes and purified by Ficoll centrifugation, followed by total B-cell isolation using the Dynabeads™ Untouched™ Human B Cells Kit (ThermoFisher Scientific). This procedure removes cells binding to CD2, CD14, CD16a, CD16b, CD36, CD43 and CD235a, including human T cells, monocytes, NK cells, macrophages, granulocytes, plasma cells, platelets, and erythrocytes (per the manual). The HCL and HCLv samples were >80% pure prior to microarray studies.
Table 1. Patients tested in microarray analysis.
| HCL | HCLv* | ||
|---|---|---|---|
| N | 35 | 27 | |
| Age range (median) | 29–75 (57) | 42–87 (70) | p < 0.0001 |
| Sex (M:F) | 31:04:00 | 22:05 | |
| Purine analog courses, range (median) | 0–8 (1) | 0–5 (1) | |
| Prior Splenectomy (% of patients) | 31% | 41% | |
| Prior Rituximab (% of patients) | 40% | 44% | |
| Unmutated IGHV4-34 (% of patients) | 0% | 41% | |
| BRAF inhibitor | 0% | 0% | |
| Leukemic cells/mm3, range (median) | 29.4–134,000 (4928) | 454–286,000 (17427) | p = 0.17 |
*For microarray analysis, the HCL group is IGHV4-34 negative, and the HCLv group includes IGHV4-34 positive (n = 11) and negative (n = 16) patients.
Microarray RNA expression assay to compare HCL with HCLv
Total RNA was purified from cell pellets by the Qiagen AllPrep kit (Qiagen, Valencia, CA) following the manufacturer’s recommendations. Using the Ambion Illumina TotalPrep-96 RNA Amplification Kit (ThermoFisher Scientific, Waltham, MA), 100 ng total RNA was labeled and amplified. Use of oligo d(T) primer reverse-transcribed the RNA into cDNA from the 3-prime end. Subsequently, the cDNA underwent second strand synthesis and in vitro transcription to generate biotinylated cRNA. The labeled cRNA was hybridized to Illumina Human Ref-8 v3 Expression Bead Chips (Illumina, Inc., San Diego, CA). After washing, the Bead Chips were scanned using the Illumina Hi-Scan and images were processed and analyzed using Illumina Genome Studio v2011.1 software. All raw data were normalized with the R package Lumi using the function LumiN. Links between MYF6 and other genes including BRAF were examined using Ingenuity® Variant Analysis™ software https://www.qiagenbioinformatics.com/products/ingenuity-variant-analysis from Qiagen, Inc.
RQ-PCR for MYF6
Peripheral blood was collected using the PAXgene Blood RNA tubes (PreAnalytiX, Feldbachstrasse, Switzerland) and total RNA was extracted by the PAXgene Blood miRNA Kit (Qiagen), per manufacturer’s instruction. The 25 ul reaction mixture, containing 1–3 ug total RNA, 2 ul 10 mM dNTP mix (ThermoFisher Scientific) and 2 ul 0.5 ug/ml Oligo(dT)20 primer (ThermoFisher Scientific) was denatured at 65°C for 5 minutes and immediately chilled on ice. First strand cDNA synthesis was performed in a 20 ul reaction mixture also containing 8 ul 5x First Strand Buffer (ThermoFisher Scientific), 4 ul 0.1M DTT, 2 ul of 40 units/ul RnaseOUT and 0.5 ul of 200 U/ml SuperScriptTM III RnaseH- Reverse Transcriptase (ThermoFisher Scientific). The reaction was incubated at 50°C for 50 minutes, followed by 5 minutes at 80°C to inactivate Reverse Transcriptase, and then stored at –20°C. MYF6 RQ-PCR was performed with a QuantStudio5 thermal cycler (Applied Biosystems, Beverly, MA). Briefly, cDNA was amplified in a 25 ul total volume per reaction using the MYF6 TaqMan® Assay Hs01547104_g1 and the TaqMan GeneExpression Master Mix (ThermoScientific, Waltham, MA) per manufacture instructions. The reaction conditions were as follows: 50°C 2 min, 95°C 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The MYF6 expression level was determined relative to PGK1 gene expression level amplified using human PGK1 HEX gene specific TaqMan assay (IDT, Coralville IA) using QuantStudio Software (Applied Biosystems, Beverly, MA). The sample was considered as negative when after 40 amplification cycles the amplification signal was not detected by the thermal cycle. By limiting dilution, MYF6 RQ-PCR was able to detect 10 HCL and 20 CLL cells in 106 normal cells.
DNA methylation in HCL vs HCLv
Genome-wide DNA methylation profiling was performed on the HCL and HCLv samples using Illumina HumanMethylation450 BeadChips (Illumina) according to the manufacturer’s recommendations. Liquid handling occurred through use of a Tecan robot (Tecan Group LTD., Männedorf, Switzerland). The raw data file generated from the Illumina GenomeStudio was normalized with SWAN normalization implemented in the “lumi” R package. Two files were produced, one with the beta value for individual targets and another one with corresponding M values for the beta values. Final targets with significant cutoffs were filtered by first selecting for M values with FDR<0.05 (adjusted based on Benjamini-Hochberg procedure) unless otherwise indicated. Then absolute beta values with differences greater than 0.2 were chosen. Partek Genomics Suite, R packages of lumi, methlumi and other related R packages were used for data processing, analysis and data presentation. Expression and methylation data were uploaded to the Gene Expression Omnibus (GEO) database, NCBI tracking system #20434981
Western blot for MYF6 protein
293 cells were transfected with pCMV6-MYF6 (OriGene, Rockville, MD) using Attractene Transfection Reagent (Qiagen, Germantown, MD) and the total protein extracted by RIPA buffer (50 mM TrisHCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, ThermoScientific, Waltham, MA). For Nuclear and cytosolic protein extraction, aliquots of 8.8x106 patient cells were pelleted, then lysed with RIPA buffer at 4°C for 30 min with constant rocking. The nuclear and cytosolic proteins from the cell lines were then separated using a nuclear extraction kit (Active Motif, Carlsbad, CA) per manufacturer’s instructions. Protein content was then quantified using a DS-11 FX+ spectrophotometer (DeNovix, New Castle County, DE). For MYF6 detection, equal amounts of protein (30ug/sample) were loaded onto NuPAGE 14% Tris-Glycine gels (ThermoFisher, Waltham, MA). Nylon+ or PVDF membranes were stained with murine monoclonal antibody (Mab) SC-514379 followed by 115-035-072-anti-mouse-HRP (Jackson ImmunoResearch). Protein was detected using the K-12043-D10 Chemiluminescent Substrate kit (Advansta, San Jose, CA). Blots were then imaged using the Syngene Pxi gel documentation system (Syngene, Frederick, MD). GAPDH was detected on Nitrocellulose membranes using polyclonal antibody #9485 (Abcam, Cambridge, MA) followed by anti-mouse-HRP #115-035-166 (Jackson ImmunoResearch).
Statistics
Statistical comparison of dichotomous variables was by Fisher’s exact. Comparison of 2 groups of continuous variables was by non-parametric Wilcoxon. Comparison of groups of patients for gene expression was performed by t-test, with corrected (stepped-up) p-values for multiple comparisons. P-values were 2-sided.
Results
Patient characteristics
As shown in Table 1, microarray comparison was performed for 35 HCL and 26 HCLv patients. Ten (39%) of the 26 HCLv patients had unmutated IGHV4-34+ HCLv. As expected, Table 1 shows that HCLv patients were older and had higher leukemic cells/mm3, although only the former comparison showed a significant difference. Table 2 shows clinical characteristics of the patients tested by RQ-PCR, including 154 patients with HCL, 51 with HCLv, and 12 with IGHV4-34+ HCLv. As expected, due to the higher tumor burden and poor response to treatment of HCLv, a higher percentage of patients with HCLv had prior splenectomy than patients with HCL (26 vs 6.5%, p = 0.0006). Prior splenectomy was also more common in IGHV4-34+ HCL than HCL (33% vs 6.5%, p = 0.011), consistent with the reported higher burden and chemoresistance in that variant [13].
Table 2. HCL/HCLv patients tested for Myf6 by RQ-PCR1.
| N | 154 | 51 | 12 |
|---|---|---|---|
| Age range (median) | 29–91 (53) | 40–92 (69) | 54–77 (65) |
| Sex (M:F) | 125:29:00 | 40:11:00 | 11:01 |
| Purine analog courses, range (median) | 0–6 (1) | 0–7 (1) | 0–6 (2) |
| Prior Splenectomy (% of patients) | 6.50% | 26% | 33% |
| Prior Rituximab (% of patients) | 26.00% | 29% | 33% |
| BRAF inhibitor | 0% | 2% | 0% |
| Leukemic cells/mm3, range | 0.026–38849 | 30.6–286,000 | 13.2–110,000 |
| Leukemic cells/mm3, median | 38.6 | 3384 | 1217 |
Expression of MYF6 in HCL and HCLv by gene expression profile
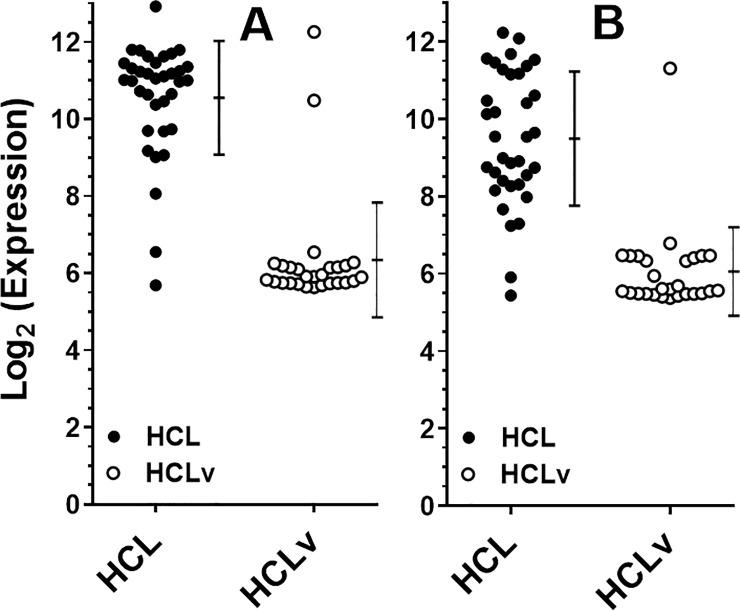
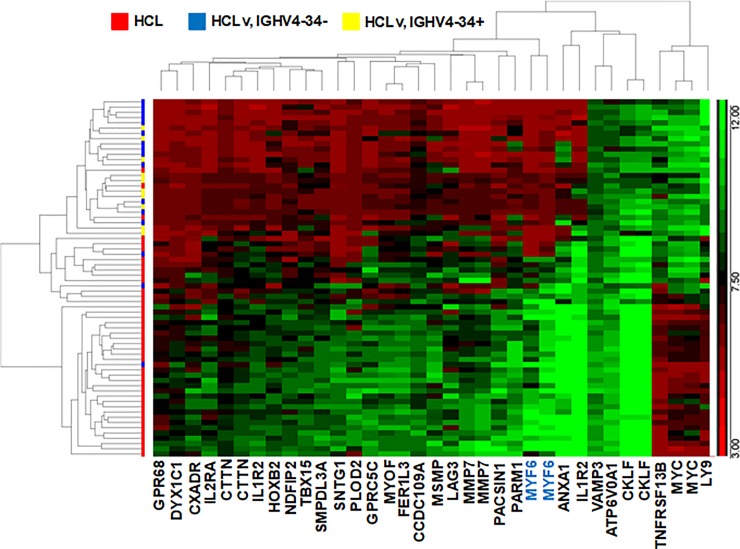
For microarray comparison of 35 patients with HCL and 27 with HCLv, leukemic cells were partially purified by CD11c sorting of the total B cell population obtained through negative B-cell selection of patient peripheral blood mononuclear cells (PBMCs). Microarray data from the 35 HCL and 27 HCLv patients showed that of 47323 probes for 24694 genes, the probe with the most difference, shown in Fig 1A and Table 3, was ILMN_1805802 for the muscle-related gene MYF6, with t-value 11.07 and stepped-up p-value 1.85 x 10−11. There was one other probe for MYF6, ILMN_2157717, which had the 13th largest difference between HCL and HCLv, with a t-value of 8.89 and stepped-up p-value 5.11 x 10−9 (Table 3, Fig 1B). By these 2 probes, MYF6 was expressed 18.5- and 10.8-fold higher in HCL than in HCLv (p<0.0001) in these 62 samples. A heat map showing the 33 probes for 30 genes with results most different between HCL and HCLv, including the 2 MYF6 probes, is shown in Fig 2. LY9, Myc, and TNFRSF13B were expressed significantly more in HCLv than classic HCL, in contrast to MYF6 and the other genes shown.
Fig 1.
Microarray expression of Myf6 in HCL (●) and HCLv (○) assessed by probe 1805802 (A) and probe 2157717 (B). Error bars indicates standard deviations around the mean.
Table 3. Microarray data for probes with absolute T-score > 7.5 with mean log2 values.
| Probe | Gene name | p-value | Stepped-Up p-value | t-score | HCL mean | HCLv mean |
|---|---|---|---|---|---|---|
| ILMN_1805802 | MYF6 | 3.90E-16 | 1.85E-11 | 11.0741 | 10.5518 | 6.34454 |
| ILMN_1812278 | LY9 | 2.39E-14 | 5.66E-10 | -9.9701 | 6.94974 | 10.4914 |
| ILMN_1772131 | IL1R2 | 3.63E-14 | 5.72E-10 | 9.86061 | 11.574 | 7.19092 |
| ILMN_1810289 | FER1L3 | 1.05E-13 | 1.24E-09 | 9.58132 | 8.87284 | 6.52241 |
| ILMN_2393712 | CTTN | 2.44E-13 | 2.11E-09 | 9.36204 | 7.56989 | 6.40702 |
| ILMN_1721580 | TBX15 | 3.51E-13 | 2.11E-09 | 9.2675 | 8.3682 | 6.65334 |
| ILMN_1810274 | HOXB2 | 3.56E-13 | 2.11E-09 | 9.26373 | 8.12313 | 6.41951 |
| ILMN_1744912 | CTTN | 5.15E-13 | 2.71E-09 | 9.16813 | 7.90934 | 6.15836 |
| ILMN_3302919 | MYOF | 8.14E-13 | 3.85E-09 | 9.04963 | 8.65623 | 6.45097 |
| ILMN_1727975 | SNTG1 | 9.68E-13 | 4.17E-09 | 9.00494 | 8.54798 | 6.01691 |
| ILMN_1685403 | MMP7 | 1.35E-12 | 5.11E-09 | 8.9186 | 8.48796 | 6.00407 |
| ILMN_2192072 | MMP7 | 1.46E-12 | 5.11E-09 | 8.89878 | 8.82602 | 6.09417 |
| ILMN_2157717 | MYF6 | 1.51E-12 | 5.11E-09 | 8.89005 | 9.48963 | 6.06077 |
| ILMN_1671142 | GPR68 | 2.17E-12 | 6.83E-09 | 8.79753 | 7.28965 | 5.87043 |
| ILMN_1677396 | NDFIP2 | 2.82E-12 | 8.35E-09 | 8.72952 | 8.20379 | 6.56636 |
| ILMN_2115135 | MSMP | 1.16E-11 | 3.23E-08 | 8.36718 | 8.83465 | 6.77264 |
| ILMN_1796349 | SMPDL3A | 2.06E-11 | 5.42E-08 | 8.22071 | 8.46603 | 6.67415 |
| ILMN_1751020 | PACSIN1 | 2.42E-11 | 6.03E-08 | 8.1798 | 9.01672 | 6.32955 |
| ILMN_1812523 | DYX1C1 | 3.35E-11 | 7.92E-08 | 8.09713 | 7.4054 | 5.99661 |
| ILMN_1813338 | LAG3 | 3.99E-11 | 8.99E-08 | 8.05234 | 8.58416 | 6.39694 |
| ILMN_1758371 | IL1R2 | 5.67E-11 | 1.22E-07 | 7.96272 | 8.1564 | 6.38335 |
| ILMN_2414027 | CKLF | 9.56E-11 | 1.97E-07 | 7.82987 | 11.8448 | 9.46051 |
| ILMN_1656560 | PARM1 | 1.11E-10 | 2.19E-07 | 7.79188 | 9.44199 | 6.99224 |
| ILMN_1680618 | MYC | 1.38E-10 | 2.60E-07 | -7.7375 | 7.22406 | 9.79519 |
| ILMN_1683774 | IL2RA | 1.62E-10 | 2.94E-07 | 7.69639 | 8.06391 | 6.03332 |
| ILMN_2184184 | ANXA1 | 2.03E-10 | 3.41E-07 | 7.63872 | 11.0697 | 7.46993 |
| ILMN_1672759 | CCDC109A | 2.07E-10 | 3.41E-07 | 7.6337 | 8.64863 | 7.24435 |
| ILMN_1759075 | TNFRSF13B | 2.09E-10 | 3.41E-07 | -7.6315 | 6.34244 | 8.68449 |
| ILMN_1712389 | CKLF | 2.46E-10 | 3.88E-07 | 7.58977 | 12.1651 | 9.88559 |
| ILMN_1752579 | ATP6V0A1 | 2.75E-10 | 4.09E-07 | 7.56097 | 10.1704 | 8.8398 |
| ILMN_2352090 | GPRC5C | 2.76E-10 | 4.09E-07 | 7.56024 | 8.71261 | 6.3762 |
| ILMN_1796925 | CXADR | 3.09E-10 | 4.38E-07 | 7.53205 | 7.70429 | 5.98888 |
| ILMN_1714527 | VAMP3 | 3.22E-10 | 4.38E-07 | 7.52127 | 9.56498 | 8.64842 |
| ILMN_1771599 | PLOD2 | 3.27E-10 | 4.38E-07 | 7.51722 | 8.45477 | 6.23114 |
| ILMN_2110908 | MYC | 3.33E-10 | 4.38E-07 | -7.5126 | 6.9804 | 9.48558 |
Fig 2. Heat map showing microarray results in HCL and HCLv.
The 22 probes for 18 genes most different between HCL and HCLv are shown. The 2 groups compared included HCL and HCLv. HCLv patients with and without the IGHV4-34 IgH rearrangement are indicated in yellow and blue, respectively.
Expression of MYF6 in classic HCL, HCLv and other B-cell malignancies by RealTime Quantitative PCR
To verify MYF6 expression by HCL, MYF6 cDNA was assessed in peripheral blood leukemic samples from 154 patients with classic HCL. As shown in Table 4, these included 147 patients with classic HCL known not to express IGHV4-34, and 7 with classic HCL not tested for IGHV4-34. In these 2 groups, 100% of the 154 patients were positive for MYF6 by RQ-PCR. Rates of MYF6 expression were lower for other hematologic malignancies, including 92% for IGHV4-34+ HCL, 32% for IGHV4-34-negative HCLv, 41% for IGHV4-34+ HCLv, 73% for CLL, 60% for marginal zone lymphoma (MZL) and 8% for mantle cell lymphoma (MCL). None of 20 patients with adult T-cell leukemia (ATL) expressed MYF6. Cells from 90 healthy donors were also tested, and 5 (6%) were positive. Thus, MYF6 was positive by RQ-PCR in all patients with HCL, in most patients with CLL, MZL, and VH4-34+ HCL, and in a minority of patients with other hematologic malignancies or normal controls, including HCLv. Patients with unmutated IGHV4-34 were more likely MYF6+ by RQ-PCR if classic HCL (92%) than HCLv (41%, p = 0.008).
Table 4. Myf6 RQ-PCR results.
| Population | Total | Myf6-positive | Myf6-negative | p-value* | p-value** |
|---|---|---|---|---|---|
| HCL, non-IGHV4-34 | 147 | 147 (100%) | 0 | <0.0001 | |
| HCL, unknown VH | 7 | 7 (100%) | 0 | 1 | <0.0001 |
| HCLv, non-IGHV4-34 | 34 | 11 (32%) | 23 (68%) | <0.0001 | 0.0003 |
| HCLv, IGHV4-34 | 17 | 7 (41%) | 10 (59%) | <0.0001 | 0.0004 |
| HCL, IGHV4-34 | 12 | 11 (92%) | 1 (8%) | 0.076 | <0.0001 |
| CLL | 48 | 35 (73%) | 13 (27%) | <0.0001 | <0.0001 |
| HCL plus CLL | 3 | 3 (100%) | 0 | 1 | 0.0004 |
| Normal donors | 90 | 5 (6%) | 85 (94%) | <0.0001 | |
| ATL | 20 | 0 | 20 (100%) | <0.0001 | 0.58 |
| MCL | 12 | 1 (8%) | 11 (92%) | <0.0001 | 0.54 |
| MZL | 5 | 3 (60%) | 2 (40%) | 0.0009 | 0.0037 |
p-values by Fishers exact compared each group to HCL, non-IGHV4-34 (*) or to normal donors (**). Other comparisons included HCLv, non-IGHV4-34 vs HCLv, IGHV4-34 (p = 0.55), HCLv, IGHV4-34 vs HCL, IGHV4-34 (p = 0.0080), and HCLv, non-IGHV4-34 vs CLL (p = 0.0003).
DNA methylation in MYF6 gene region
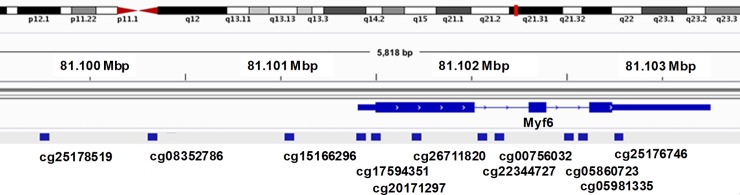
Since methylated cytosines in the context of cytosine guanine dinucleotides (CpGs) cluster at high density in regions of DNA termed CpG islands which associate with gene promoters [30], and methylation of promoter-associated CpGs is associated with transcriptional repression and gene silencing [31], we determined whether the gene for MYF6 is hypomethylated more often in HCL than in HCLv. We conducted genome-wide DNA methylation profiling using Illumina HumanMethylation450 BeadChips and 476,882 probes. We compared 34 classic HCL patients to 28 with HCLv, the HCLv group including IGHV4-34+ (n = 13) and IGHV4-34-negative (n = 15). As shown in Table 5, 8 out of 11 MYF6 cg probes had significant difference in HCL vs HCLv with respect to stepped up p-value <0.05. The 280th most different probe, cg05981335, had a t-value of -7.142, and stepped-up p-value 2.44 x 10−6. Only 1 of these 8 probes had a positive t-value indicating more hypomethylation in HCLv than HCL. This probe binds to the reverse strand and its significance is unknown. Of the remaining probes, cg08352786, cg17594351, cg15166296, and cg25178519, based on the DBTSS database (https://dbtss.hgc.jp/), bind within -2.5 kb and +1.0 kb of an annotated transcription start site (TSS) and would be considered as binding within the promotor region (Fig 3). Thus, the DNA methylation data supported more hypomethylation of MYF6 in HCL than HCLv, consistent with the higher level of expression in HCL than HCLv.
Table 5. Myf6 probes showing differences in methylation between HCL and HCLv.
| Probe | Place | P-value | Stepped-up p-value | t-value | Strand | UCSC RefGene Group | Relation to UCSC CpG Island |
|---|---|---|---|---|---|---|---|
| cg05981335 | 280 | 1.43E-09 | 2.44E-06 | -7.142 | F | Body | Island |
| cg05860723 | 295 | 1.82E-09 | 2.93E-06 | -7.082 | R | Body | Island |
| cg25176746 | 663 | 7.42E-08 | 5.34E-05 | -6.131 | F | 3'UTR | S_Shore |
| cg22344727 | 741 | 1.17E-07 | 7.52E-05 | -6.013 | R | Body | Island |
| cg08352786 | 828 | 1.75E-07 | 0.000101 | 5.908 | R | TSS1500 | N_Shore |
| cg17594351 | 2890 | 1.58E-05 | 0.002613 | -4.697 | F | TSS200 | N_Shore |
| cg00756032 | 5134 | 0.0001 | 0.009465 | -4.163 | R | Body | Island |
| cg20171297 | 6967 | 0.00025 | 0.017133 | -3.894 | R | 1stExon;5'UTR | N_Shore |
| cg15166296 | 18410 | 0.00308 | 0.079676 | -3.085 | F | TSS1500 | N_Shore |
| cg26711820 | 20008 | 0.00371 | 0.088353 | -3.02 | F | 1stExon | N_Shore |
| cg25178519 | 36755 | 0.0141 | 0.182887 | 2.529 | R | TSS1500 | N_Shore |
Fig 3. Locations for Myf6 methylation probe binding.
Blue squares showing the binding site are situated over the ‘cg’ label of each probe.
Presence of MYF6 protein in HCL cells
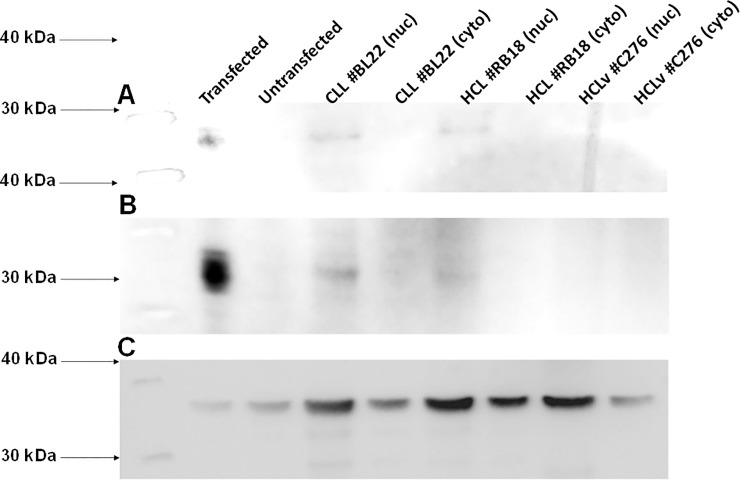
To investigate whether the high mRNA levels of MYF6 correspond to expression of MYF6 protein, we tested for the presence of MYF6 protein on HCL and HCLv cells obtained from patients, using anti-MYF6 Mab. We expected that since MYF6 is a transcription factor, its expression would be difficult to detect by western blot. In fact, other than transfected 293 cells (Fig 4, lane 1), we could not detect any MYF6 protein except for faint bands in the nuclear fraction of MYF6+ cells. The 2 patients shown with CLL and HCL were also positive by RQ-PCR and the HCL patient was also MYF6+ by microarray. As expected, we could not detect significant levels of MYF6 in the nuclear fraction of the patient with HCLv who was negative for MYF6 by RQ-PCR and microarray. The lower expression of GAPDH in lanes 1 and 2 is consistent with much higher total protein expression by 293 cells compared to primary leukemia cells, and thus a lower percentage of GAPDH in a 30 ug gel sample. Similarly, GAPDH from the cytosolic fraction of primary cells from the HCLv patient (lane 8), who had rapid disease progression, possibly constituted a lower percentage of the 30 ug of total protein loaded. We conclude that HCL cells not only express MYF6 mRNA but also its protein product.
Fig 4. Western blot for Myf6 protein in HCL and CLL cells.
Nylon+ (A) and PVDF (B) membranes were stained with murine Mab SC-514379 followed by anti-mouse-HRP. In C, PVDF was stained with polyclonal anti-GAPDH antibody followed by anti-mouse-HRP. Lanes include Myf6-transfected (lane 1) and untransfected (lane 2) 293 cells, nuclear and cytoplasmic fractions for CLL patient BL22 (lanes 3 & 4, respectively), HCL patient BL18 (lanes 5 & 6), and HCLv patient C276 (lanes 7 & 8). In each lane, 30 ug of total protein was added, except less in A lane 1 to obtain bands of similar intensity.
Determination of limits of detection of MRD by MYF6
To determine the sensitivity of MYF6 detection by RQ-PCR, we diluted MYF6+ HCL and CLL cells into normal peripheral blood mononuclear cells and determined the greatest dilution which could still be detected in most biologic replicates. We could reliably detect HCL at an approximate dilution of 1 in 105 normal cells, with 3 of 6 and 3 of 4 replicates positive at 3/106 and 10/106, respectively (Table 6). We could detect CLL at an approximate dilution of 2 in 105 normal cells, with 1 of 6 and 4 of 6 replicates positive at 10/106 and 20/106, respectively. Cycle threshold (CT) values are shown. Thus, in some patients with CLL and HCL, MYF6 RQ-PCR can be used to determine MRD. Of 87 HCL patients with repeat post-treatment blood samples negative for HCL by flow cytometry, 42 (48%) were still positive for MYF6. Conversely, of 76 patients with repeat post-treatment blood samples negative for MYF6 by RQ-PCR, 13 (17%) were positive by flow cytometry, at a median of 0.3 HCL cell/mm3. Thus, HCL cells from some patients may have lower expression of MYF6 and require more cells than others for positivity, while some patients with higher expression could be followed by MYF6 RQ-PCR more easily and sensitively than flow cytometry. Since Myf6 RQ-PCR could be more sensitive or less sensitive than flow cytometry in different patients, we could not report sensitivity and specificity of this assay in patients after treatment. Since 6% of normal donors were MYF6+, some patients, including those with MYF6+ monoclonal B-cell lymphocytosis (MBL), may not become MYF6 negative after resolution of circulating HCL. Additional testing will be needed to better define the sensitivity and specificity of MYF6 RQ-PCR relative to flow cytometry, or other methods including RQ-PCR using patient CDR3-specific primers [32] or deep sequencing [33].
Table 6. Sensitivity of Myf6 detection in CLL and HCL.
| Dilution HCL patient: | 1/106 | 3/106 | 10/106 | 20/106 | 50/106 | 102/106 | 103/106 |
|---|---|---|---|---|---|---|---|
| Number positive | 2 of 6 | 3 of 6 | 3 of 4 | 3 of 4 | 4 of 4 | 4 of 4 | 2 of 2 |
| PRGK1 CT (median of +’s) | 26.5 | 26.4 | 27.2 | 27 | 26.2 | 26.5 | 26.5 |
| Myf6 CT (median of +’s) | 36 | 37 | 35.8 | 37 | 36.4 | 34 | 31 |
| PRGK1-Myf6 ΔCT | 9.5 | 10.6 | 8.5 | 10.1 | 10.3 | 7.5 | 4.5 |
| Dilution CLL patient | |||||||
| Number positive | 1 of 6 | 4 of 6 | 5 of 6 | 2 of 2 | |||
| PRGK1 CT (median of +’s) | 29.4 | 26.9 | 26.1 | 25.8 | |||
| Myf6 CT (median of +’s) | 39.9 | 38.7 | 36.9 | 32.8 | |||
| PRGK1-Myf6 ΔCT | 10.5 | 11.8 | 10.8 | 6.9 |
Cycle threshold (CT) values are presented to the nearest tenth of a cycle. ΔCT values, the differences between CT values for the housekeeping gene PRGK1 and Myf6, were calculated using CT values with several decimal places.
Discussion
MYF6 is a muscle specific transcription factor normally expressed in skeletal muscle, but not other types of normal tissues. MYF6 expression has been reported for a narrow group of solid tumors including of rhabdomyosarcoma [23] and corticotroph macroadenomas [24]. Although data from microarray studies by Basso et al. in HCL was listed [28, 29], its detailed expression in HCL and HCLv was not reported. To determine genes differentially expressed in HCL as opposed to HCLv, we performed expression microarray studies for 24694 genes. We were surprised to find that a probe for MYF6 had the lowest p-value for preferential expression in HCL compared to HCLv. We found that 100% of 154 HCL samples were positive for MYF6 by RQ-PCR, with less percentages of other hematologic malignancies positive. Additionally, we discovered that most CLL samples were positive for MYF6. Increased expression of MYF6 correlated with overall hypomethylation in (CpGs)CpG probes with HCL vs HCLv. MYF6 protein could be detected in the nuclear fraction of MYF6+ HCL and CLL cells, and in these leukemias, MYF6 PQ-PCR could be used for MRD detection with sensitivities of 1/105 and 2/105, respectively.
Expression of MYF6 in HCL and other B-cell malignancies from Oncomine and Gene Expression Omnibus
The MYF6 expression data for classic HCL was available from normal and transformed array expression profiles GSE2350 contributed by Basso et al. [28, 29]. Their microarray assay studied 8603 genes in 336 normal and malignant B-cell samples, including from 10 patients with HCL. This study identified 82 genes upregulated in HCL including MYF6, and MYF6 was one of 22 proteins detectable on HCL by IHC [28]. This data set includes a median MYF6 expression level of 3.75 for 16 patients with HCL, compared to -0.6 to -1.9 for Burkitt’s Lymphoma (n = 127), centroblastic lymphoma (n = 28], CLL (n = 34], diffuse large B-cell lymphoma (n = 41], follicular lymphoma (n = 6), Hodgkin’s lymphoma (n = 4), MCL (n = 8), plasma cell leukemia (n = 3) and primary effusion lymphoma (n = 9). Median expression levels were -0.5 to -1.2 for 5 samples each of B-lymphocytes, centroblasts, memory B-cells, naïve pregerminal center B-cells, and small cleaved follicular center cells [29]. Thus, MYF6 is an important gene expressed in HCL compared to either HCLv or B-cells from malignant or benign sources. It is interesting that this reported data set shows MYF6 expression much higher in HCL (3.75, n = 16) than in CLL (-0.65, n = 34), since we found most CLL patient samples MYF6+. However, while MYF6 expression as reported by Basso et al. [28, 29] was lower in CLL than in HCL, CLL was higher than nearly all other non-HCL malignancies, and this is consistent with our data showing that 73% of CLL patients were MYF6+ by RQ-PCR. Myf6, which is expressed on chromosome 12, was reported by Porpaczy et al., to be preferentially expressed in 29 trisomy-12 CLL patients vs 32 non-trisomy-12 patients [27].
MYF6 expression by normal cells
We found that 5 of 90 normal donors were MYF6+ by RQ-PCR. Since we found that MYF6 in CLL can be detected at 2 cells per 105, it is possible that some of the normal donors may have had low levels of monoclonal B-cells. It has been reported that monoclonal B-lymphocytes can be detected in ~5% of adults over the age of 40 [34], which probably explains the Myf6 positivity of 6% of uncharacterized normal donors. Myf6 would not be an accurate target for MRD in HCL patients with MBL, but the presence of MBL is easily detected by flow cytometry prior to treatment, and the remaining ~95% of HCL patients would be evaluable. Normal blood donors were anonymous and flow cytometry was not performed on each sample. We found no MYF6 expression in malignant T-cells (Table 4). Our data showing that samples from 94% of normal donors are Myf6 negative seem inconsistent with data from Porpaczy et al. that 6 of 6 normal donors had CD19+ cells which were low+ for Myf6 [27]. The reason for the discrepancy may relate to the fact that RQ-PCR on our 90 normal donors, like our 305 leukemic patients, was performed on RNA purified from PaxGene tubes of whole blood; it is possible that RQ-PCR on CD19-selected B-cells could give different results.
Expression of MYF6 in IGHV4-34+ HCL, and investigation of the function of Myf6
We reported that patients with unmutated IGHV4-34+ HCL, indistinguishable from classic HCL immunophenotypically, have clinical features more like HCLv than HCL, including poor response to single-agent purine analog [13]. This molecularly defined variant was reported as negative for BRAF V600E [12, 35]. We found in this study that MYF6 was positive in a higher percentage of IGHV4-34+ HCL patients than patients with HCLv, 11 of 12 vs 18 of 51 (p = 0.0007). Yet it was not associated with IGHV4-34 in particular, since similar rates of Myf6 positivity were observed in IGHV4-34 positive (41% of 17) and negative (32% of 34) HCLv (Table 4, p = 0.55). We consider patients HCL rather than HCLv if their CD25 is + or bright positive, regardless of BRAF status, since the latter is not universally measured, while patients wild-type for BRAF and negative or dim+ for CD25 are considered as HCLv. To determine a possible link between MYF6 and either BRAF V600E or other MAPK pathways involved in HCL pathogenesis, we interrogated the Ingenuity software application for all proteins possibly interacting with Myf6, and did not find interactions with any known proteins of the MAPK pathway. This together with our experiments indicates that the expression of MYF6 is not dependent on the BRAF V600E mutation and may be related to other factors including CD25 expression. While the function of Myf6 in HCL is unknown, it is unlikely to have a causative role since it is observed in some other hematologic malignancies like CLL. We determined using the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) whether Myf6 expression correlates to markers which are selective for HCL compared to HCLv. Using 191 hematologic cell lines, there was no correlation between Myf6 and either CD25, CD134, TRAP, or Annexin 1a. There are no cell lines resembling classic HCL to the extent that they express BRAF V600E, although there are several resembling HCLv [36, 37]. None of the 191 hematologic cell lines examined were HCL or HCLv-like.
Additional work will be required to address the question of why MYF6, a muscle specific transcription factor, is expressed by all classic HCL. MYF6 involvement in pathways specific for HCL but not most cases of HCLv would suggest an important pathogenesis effect. Efforts to produce MYF6 knockout cells are among studies underway to investigate this hypothesis.
Supporting information
(PPTX)
Acknowledgments
We would like to recognize our data specialist Barbara Debrah, the research nurses who helped obtain samples, and the patients and clinical staff of the NIH.
Data Availability
Expression and methylation data were uploaded to the Gene Expression Omnibus (GEO) database, NCBI tracking system #20434981.
Funding Statement
This study was supported by the National Cancer Institute, Intramural program. All authors were working for and receiving salary from the National Institutes of Health (NIH).
References
- 1.Falini B, Tiacci E, Liso A, Basso K, Sabattini E, Pacini R, et al. Simple diagnostic assay for hairy cell leukaemia by immunocytochemical detection of annexin A1 (ANXA1). Lancet. 2004;363(9424):1869–70. 10.1016/S0140-6736(04)16356-3 [DOI] [PubMed] [Google Scholar]
- 2.Matutes E. Immunophenotyping and differential diagnosis of hairy cell leukemia. Hematol Oncol Clin North Am. 2006;20(5):1051–63. 10.1016/j.hoc.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4 ed: World Health Organization; 2008. October 31, 2008. [Google Scholar]
- 4.Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–15. 10.1056/NEJMoa1014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman GR, Beutler E, Saven A. Cladribine in the treatment of hairy-cell leukaemia. Best Pract Res Clin Haematol. 2003;16(1):101–16. 10.1016/s1521-6926(02)00089-0 [DOI] [PubMed] [Google Scholar]
- 6.Hockley SL, Morgan GJ, Leone PE, Walker BA, Morilla A, Else M, et al. High-resolution genomic profiling in hairy cell leukemia-variant compared with typical hairy cell leukemia. Leukemia. 2011;25(7):1189–92. 10.1038/leu.2011.47 [DOI] [PubMed] [Google Scholar]
- 7.Cawley JC, Burns GF, Hayhoe FG. A chronic lymphoproliferative disorder with distinctive features: a distinct variant of hairy-cell leukaemia. Leuk Res. 1980;4(6):547–59. 10.1016/0145-2126(80)90066-1 [DOI] [PubMed] [Google Scholar]
- 8.Ott G, Balague-Ponz O, de Leval L, de Jong D, Hasserjian RP, Elenitoba-Johnson KS. Commentary on the WHO classification of tumors of lymphoid tissues (2008): indolent B cell lymphomas. Journal of hematopathology. 2009;2(2):77–81. 10.1007/s12308-009-0037-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreitman RJ, Wilson W, Calvo KR, Arons E, Roth L, Sapolsky J, et al. Cladribine with immediate rituximab for the treatment of patients with variant hairy cell leukemia. Clin Cancer Res. 2013;19(24):6873–81. 10.1158/1078-0432.CCR-13-1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robak T. Hairy-cell leukemia variant: recent view on diagnosis, biology and treatment. Cancer Treat Rev. 2011;37(1):3–10. 10.1016/j.ctrv.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 11.Tiacci E, Schiavoni G, Forconi F, Santi A, Trentin L, Ambrosetti A, et al. Simple genetic diagnosis of hairy cell leukemia by sensitive detection of the BRAF-V600E mutation. Blood. 2012;119(1):192–5. 10.1182/blood-2011-08-371179 [DOI] [PubMed] [Google Scholar]
- 12.Xi L, Arons E, Navarro W, Calvo KR, Stetler-Stevenson M, Raffeld M, et al. Both variant and IGHV4-34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood. 2012;119(14):3330–2. 10.1182/blood-2011-09-379339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4-34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114(21):4687–95. 10.1182/blood-2009-01-201731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waterfall JJ, Arons E, Walker RL, Pineda M, Roth L, Killian JK, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46(1):8–10. 10.1038/ng.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durham BH, Getta B, Dietrich S, Taylor J, Won H, Bogenberger JM, et al. Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations. Blood. 2017;130(14):1644–8. 10.1182/blood-2017-01-765107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason EF, Brown RD, Szeto DP, Gibson CJ, Jia Y, Garcia EP, et al. Detection of activating MAP2K1 mutations in atypical hairy cell leukemia and hairy cell leukemia variant. Leuk Lymphoma. 2017;58(1):233–6. 10.1080/10428194.2016.1185786 [DOI] [PubMed] [Google Scholar]
- 17.Cupelli L, Renault B, Leblanc-Straceski J, Banks A, Ward D, Kucherlapati RS, et al. Assignment of the human myogenic factors 5 and 6 (MYF5, MYF6) gene cluster to 12q21 by in situ hybridization and physical mapping of the locus between D12S350 and D12S106. Cytogenetics and cell genetics. 1996;72(2–3):250–1. 10.1159/000134201 [DOI] [PubMed] [Google Scholar]
- 18.Braun T, Bober E, Winter B, Rosenthal N, Arnold HH. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. The EMBO journal. 1990;9(3):821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. 10.1016/0092-8674(87)90585-x [DOI] [PubMed] [Google Scholar]
- 20.Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik KH, Arnold HH. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. The EMBO journal. 1989;8(12):3617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56(4):607–17. 10.1016/0092-8674(89)90583-7 [DOI] [PubMed] [Google Scholar]
- 22.Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. The EMBO journal. 1989;8(3):701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark J, Rocques PJ, Braun T, Bober E, Arnold HH, Fisher C, et al. Expression of members of the myf gene family in human rhabdomyosarcomas. British journal of cancer. 1991;64(6):1039–42. 10.1038/bjc.1991.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosoyama T, Nishijo K, Garcia MM, Schaffer BS, Ohshima-Hosoyama S, Prajapati SI, et al. A Postnatal Pax7 Progenitor Gives Rise to Pituitary Adenomas. Genes & cancer. 2010;1(4):388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Zhou H, Ma K, Sun J, Feng X, Geng J, et al. Abnormal methylation of seven genes and their associations with clinical characteristics in early stage non-small cell lung cancer. Oncology letters. 2013;5(4):1211–8. 10.3892/ol.2013.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haslinger C, Schweifer N, Stilgenbauer S, Dohner H, Lichter P, Kraut N, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22(19):3937–49. 10.1200/JCO.2004.12.133 [DOI] [PubMed] [Google Scholar]
- 27.Porpaczy E, Bilban M, Heinze G, Gruber M, Vanura K, Schwarzinger I, et al. Gene expression signature of chronic lymphocytic leukaemia with Trisomy 12. European journal of clinical investigation. 2009;39(7):568–75. 10.1111/j.1365-2362.2009.02146.x [DOI] [PubMed] [Google Scholar]
- 28.Basso K, Liso A, Tiacci E, Benedetti R, Pulsoni A, Foa R, et al. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors. The Journal of experimental medicine. 2004;199(1):59–68. 10.1084/jem.20031175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–90. 10.1038/ng1532 [DOI] [PubMed] [Google Scholar]
- 30.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90(24):11995–9. 10.1073/pnas.90.24.11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. [DOI] [PubMed] [Google Scholar]
- 32.Arons E, Margulies I, Sorbara L, Raffeld M, Stetler-Stevenson M, Pastan I, et al. Minimal residual disease in hairy cell leukemia patients assessed by clone-specific polymerase chain reaction. Clin Cancer Res. 2006;12(9):2804–11. 10.1158/1078-0432.CCR-05-2315 [DOI] [PubMed] [Google Scholar]
- 33.Perrot A, Lauwers-Cances V, Corre J, Robillard N, Hulin C, Chretien ML, et al. Minimal residual disease negativity using deep sequencing is a major prognostic factor in multiple myeloma. Blood. 2018;132(23):2456–64. 10.1182/blood-2018-06-858613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strati P, Shanafelt TD. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood. 2015;126(4):454–62. 10.1182/blood-2015-02-585059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain P, Ok CY, Konoplev S, Patel KP, Jorgensen J, Estrov Z, et al. Relapsed Refractory BRAF-Negative, IGHV4-34-Positive Variant of Hairy Cell Leukemia: A Distinct Entity? J Clin Oncol. 2016;34(7):e57–60. 10.1200/JCO.2013.50.9661 [DOI] [PubMed] [Google Scholar]
- 36.Sasaki M, Aritaka N, Tsukune Y, Kawahara S, Masuda A, Tsutsui M, et al. Establishment of a hairy cell leukemia variant cell line, HCLv-07. Acta Haematol. 2009;121(1):63–6. 10.1159/000210064 [DOI] [PubMed] [Google Scholar]
- 37.Weston-Bell NJ, Hendriks D, Sugiyarto G, Bos NA, Kluin-Nelemans HC, Forconi F, et al. Hairy cell leukemia cell lines expressing annexin A1 and displaying B-cell receptor signals characteristic of primary tumor cells lack the signature BRAF mutation to reveal unrepresentative origins. Leukemia. 2013;27(1):241–5. 10.1038/leu.2012.163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX)
Data Availability Statement
Expression and methylation data were uploaded to the Gene Expression Omnibus (GEO) database, NCBI tracking system #20434981.