The structure of the capsid of the gene-therapy vector AAVhu.37 was obtained at 2.56 Å resolution by single-particle cryo-electron microscopy.
Keywords: adeno-associated viruses, gene therapy, gene-delivery vectors, capsid, cryo-EM structure, AAVhu.37
Abstract
Adeno-associated viruses (AAVs) are used as in vivo gene-delivery vectors in gene-therapy products and have been heavily investigated for numerous indications. Over 100 naturally occurring AAV serotypes and variants have been isolated from primate samples. Many reports have described unique properties of these variants (for instance, differences in potency, target cell or evasion of the immune response), despite high amino-acid sequence conservation. AAVhu.37 is of interest for clinical applications owing to its proficient transduction of the liver and central nervous system. The sequence identity of the AAVhu.37 VP1 to the well characterized AAVrh.10 serotype, for which no structure is available, is greater than 98%. Here, the structure of the AAVhu.37 capsid at 2.56 Å resolution obtained via single-particle cryo-electron microscopy is presented.
1. Introduction
Adeno-associated viruses (AAVs), belonging to the Dependoparvovirus genus and the Parvoviridae family, are a group of small, non-enveloped, DNA viruses that are currently under intense study in the field of gene therapy (Mueller & Flotte, 2008 ▸; Daya & Berns, 2008 ▸). Their lack of pathogenicity and their ability to transduce both dividing and quiescent cells makes them promising candidates for the delivery of many clinically relevant cargoes; however, challenges remain regarding capsid engineering, including tissue-specific transduction, avoidance of the innate immune response and manufacturability (Mingozzi & High, 2011 ▸). The AAV capsid is composed of 60 subunits of viral protein assembled in a 25 nm icosahedral capsid that is capable of packaging a genome of approximately 4.7 kb (Berns & Parrish, 2015 ▸). Over 100 naturally occurring AAV serotypes and variants have been described and have been shown to have a variety of different properties (for example, packaging efficiency, biodistribution profile, stability etc.), which is largely attributed to the sequence diversity and conformational variability found in the variable-region (VR) loops located on the surface of the AAV capsid (Wu et al., 2006 ▸). These AAV isolates have been organized into clades (A through F) according to their sequence similarity in the hope of identifying important sequences contributing to the similar functional characteristics of a group (Gao et al., 2004 ▸).
The clade E serotype AAVhu.37 efficiently transduces the liver in mouse models and nonhuman primates (NHP) after intravenous injection (Wang, Wang et al., 2010 ▸; Greig et al., 2018 ▸). When injected directly into mouse brain, AAVhu.37 also transduces the central nervous system (CNS; Cearley et al., 2008 ▸). AAVhu.37 is 98.5% identical to AAVrh.10 and differs by only one amino acid (VP1 residue 24) from AAVrh.39. AAVrh.10 (Zhang et al., 2011 ▸; Yang et al., 2014 ▸) and AAVrh.39 (Lawlor et al., 2009 ▸) cross the blood–brain barrier (BBB) and efficiently transduce NHP CNS tissues (Rosenberg et al., 2014 ▸). Although the BBB-penetrating capacity of AAVhu.37 has not been assayed in published work, it is likely that the structural motifs in AAVhu.37 that are responsible for BBB penetration would be identical to those in AAVrh.39 and AAVrh.10. In a preclinical study for the treatment of hemophilia A, macaques administered with AAVhu.37 carrying a functional human blood coagulation factor VIII (hFVIII) gene linked to E03.TTR (liver-specific) promoter exhibited high levels of hFVIII circulating in the blood (Greig et al., 2018 ▸). While hepatic gene transfer can induce immunity to the transferred gene (Dobrzynski et al., 2006 ▸), hepatic gene transfer with AAV can induce immune tolerance (Mount et al., 2002 ▸; Mingozzi et al., 2003 ▸; Breous et al., 2009 ▸, 2011 ▸), even when the gene transfer is not systemic (Martino et al., 2009 ▸). There is an effect of the serotype on the immune tolerance level (Greig et al., 2017 ▸), and AAVhu.37 has favorable tolerance and expression effects (Greig et al., 2018 ▸). This study has been further extended into an ongoing clinical trial (ClinicalTrials.gov NCT03588299; https://clinicaltrials.gov/). Furthermore, AAVhu.37 has been reported to be favorable for high-yield, scalable manufacturing, and it binds AVB resin, providing a simple purification strategy (Wang et al., 2015 ▸; Greig et al., 2018 ▸).
In order to further understand the structure–function relationship of AAV capsid variants across serotypes, we sought to describe the structure of the clinically relevant capsid AAVhu.37.
2. Materials and methods
2.1. Vector production
AAVhu.37 (GenBank AAS99285.1) particles containing ssDNA with a CAG promoter and encoding green fluorescent protein (GFP) were produced by triple transfection of adherent HEK293T cells. Vector particles were harvested via freeze–thawing and purified via an iodixanol gradient (Zolotukhin et al., 1999 ▸). The final product was resuspended in phosphate-buffered saline (PBS) and 0.001% Pluronic F-68. The genome titer was determined via Droplet Digital PCR (ddPCR) using primers against the rabbit β-globin polyadenylation signal of the transgene sequence as described previously (Lock et al., 2014 ▸).
2.2. Composition and purity analysis
The capsid-protein purity and the ratio of relative mass of the capsid proteins were determined by capillary gel electrophoresis. The AAV sample was denatured and reduced by heating in a solution containing sodium dodecyl sulfate and dithiothreitol. The sample was diafiltered to remove excess salt and was then electrokinetically injected into a gel-filled capillary using a PA800 Plus (SCIEX) instrument. The capsid proteins were separated based on size by applying a voltage to the capillary. Detection and quantitation of the capsid proteins were performed using detection at 214 nm.
For intact LC-MS determination, the sample was injected into a Vanquish UHPLC coupled to a Q Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The separation was performed on a UPLC BEH C4 column using reversed-phase chromatography. Proteins were deconvolved with Intact Mass v.3.6 from Protein Metrics. LC-MS/MS of digested peptides was performed as described previously (Cai et al., 2017 ▸). Peptides were searched against a database containing human UniProt sequences, common contaminants such as porcine trypsin and AAVhu.37.
2.3. CryoEM sample preparation and data collection
Pluronic F-68 was removed before vitrification by diafiltration with seven washes with PBS using an Amicon Ultra-0.5 100 kDa molecular-weight cutoff regenerated cellulose spin column (Sigma–Aldrich). Amorphous carbon film was deposited on cleaved V1 mica (Ted Pella, Redding, California, USA) by E-beam deposition using an ACE600 (Leica Microsystems GmbH) and the carbon thickness as measured by the quartz-crystal oscillation method was 2.86 nm. Carbon was floated on glow-discharged Quantifoil R2/1 grids (Quantifoil Micro Tools GmbH) within an hour of vitrification. 3.5 µl of particles diluted in PBS was applied to the grid and plunge-frozen using an EM-GP (Leica Microsystems GmbH) with Whatman No. 1 blotting paper (Sigma–Aldrich). The grid used for the final data set was blotted for 4.5 s.
Grids were imaged (Table 1 ▸) on a Talos Arctica cryoelectron microscope aligned as described previously (Herzik et al., 2017 ▸), except that AutoCTF and EPU (Thermo Fisher Scientific) were employed instead of Leginon. Images were recorded with a K2 Summit detector (Gatan) in super-resolution mode. To evaluate the presence of the genome, 15 micrographs from throughout the final data set were high-pass (1000 Å) and low-pass (28.5 Å) filtered and 1074 particles were manually scored. The mean of means for the 15 fields of view differed from the overall mean by less than 0.1%.
Table 1. Cryo-electron microscopy.
| Defocus range (90%) (nm) | 350–1720 |
| Pixel size at specimen (Å) | 1.038 |
| Super-resolution pixel size at the specimen (Å) | 0.519 |
| Total dose (e Å−2) | ∼32.8 |
| Frame rate (frames per second) | 5 |
| Micrographs collected | 736 |
| Particles extracted | 52235 |
| Particles in final reconstruction | 41849 |
| Guinier plot B factor (Å2) | 113 |
| Resolution at FSC = 0.143 | |
| Unmasked | 2.80 |
| Masked | 2.53 |
| Noise-corrected (‘true FSC’) | 2.56 |
2.4. Map reconstruction
Movies were processed with dose-weighting and rebinned to 1.038 Å per pixel using MotionCor2 (Zheng et al., 2017 ▸). Particles were boxed with EMAN2 (Tang et al., 2007 ▸). Extracted particles were imported into cryoSPARC2 and icosahedral ab initio refinement generated a data-derived initial model (Punjani et al., 2017 ▸). The structure was refined to 2.8 Å resolution with icosahedral symmetry, the handedness was flipped with reference to known parvoviral structures and a mask was generated from the structure using EMAN2. The movies were corrected again, rebinned to 0.83 Å per pixel in RELION and further refinement was pursued in both RELION and cryoSPARC: both methods produced maps that were identical by eye. The deposited map was generated by performing particle polishing in RELION (Zivanov et al., 2019 ▸) and homogeneous refinement in cryoSPARC. This map has a Fourier shell correlation (FSC) of 0.143 at 2.56 Å resolution. Representative density for residues 622–635 is shown in Fig. 3(c). We note that the resolution at FSC = 0.143 for the latter map reconstruction is greater than three times the voxel length, so it satisfies the ‘2/3 Nyquist criterion’ of van Heel (Orlova et al., 1997 ▸). In no step of map generation was a model used from any other data set.
2.5. Model refinement
AAV8 (PDB entry 2qa0; Nam et al., 2007 ▸) was used as an initial template for modeling and had a high fit to density even before refinement. Rounds of automatic refinement in Phenix and manual refinement in Coot (Liebschner et al., 2019 ▸; Emsley et al., 2010 ▸) were conducted and the final coordinates (Table 2 ▸) were evaluated by eye and using MolProbity (Chen et al., 2010 ▸). Structural figures were generated using PyMOL (v.2.1.0; Schrödinger) or UCSF Chimera (Pettersen et al., 2004 ▸).
Table 2. Model refinement.
| No. of non-H protein atoms | 4125 |
| No. of ligand atoms | 0 |
| Average B factor (Å2) | 34 |
| R.m.s. deviations | |
| Bonds (A°) | 0.0078 |
| Angles (°) | 1.07 |
| Validation | |
| MolProbity score | 1.42 |
| Clashscore | 7.11 |
| Rotamer outliers | 1 (0.2%) |
| Ramachandran plot | |
| Favored regions | 506 (98%) |
| Additionally allowed | 10 (2%) |
| Outliers | 1 (0.2%) |
3. Results and discussion
3.1. AAVhu.37-GFP particles
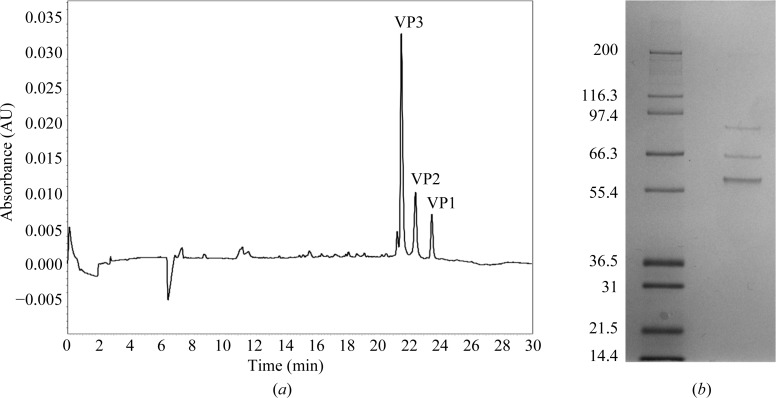
Purified AAVhu.37-GFP was titered via ddPCR against the transgene to a concentration of 7.5 × 1012 genome copies per millilitre. Capillary gel electrophoresis yielded three major peaks (Fig. 1 ▸). This reflects the generally known capsid composition of AAVs, where the capsids comprise a mixture of VP1, VP2 and VP3, with VP3 predominating (Rose et al., 1971 ▸). VP2 and VP3 are N-terminal truncations of VP1; the VP3 region is common to all three forms. The absence of other significant peaks indicates a high purity.
Figure 1.
(a) Capillary gel electrophoretic separation of an AAVhu.37 sample illustrating the presence of VP1, VP2 and VP3 proteins in the sample. (b) SDS–PAGE gel of AAVhu.37 from which three bands were cut for LC-MS/MS of the digested peptides.
We confirmed that VP1, VP2 and VP3 are the significant components of this preparation by mass spectrometry. Reversed-phase chromatography of intact capsids detected unfragmented masses matching the predicted masses of VP3 (measured 59 666.4 Da, predicted 59 667.6 Da), VP2 (measured 66 284.9 Da, predicted 66 286.0 Da) and VP1 (measured 81 491.8 Da, predicted 81 492.6 Da). SDS–PAGE analysis of the sample revealed exactly three bands (Fig. 1 ▸ b). An in-gel digest of these bands (Fig. 1 ▸ b) followed by LC-MS/MS identified all three bands as AAV capsid protein based on the presence of peptides from the shared (VP3) region. Additionally, VP1-unique peptides were detected from the highest molecular-weight band. In each of the three bands, AAV capsid protein predominated. The low-abundance contaminants mainly consisted of porcine trypsin, human heat-shock proteins and keratin.
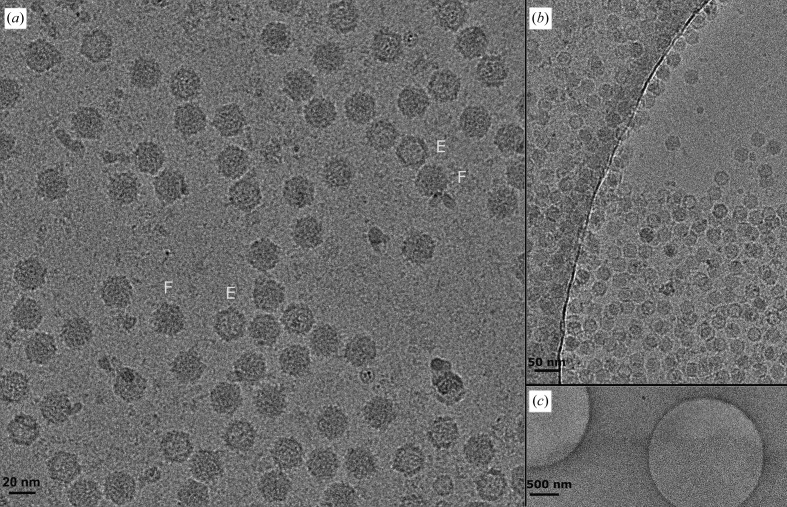
On vitrified EM grids, particles preferentially partitioned to carbon surfaces even when the solvent layer in bare holes was thick. A thin, amorphous carbon film increased the effective particle concentration (Fig. 2 ▸). 82.0% of the particles contained DNA (full or partially full), 16.3% were empty and 1.7% of the particles were insufficiently resolved to unambiguously determine the presence of the genome.
Figure 2.
(a) One randomly selected micrograph from the final data set, low-pass filtered to 8 Å resolution for display. Exemplars are labeled in cyan to represent full capsids (F) and empty capsids (E). (b) A screening micrograph in which the edge of the thin amorphous film is suspended over a hole in the carbon foil illustrates the concentrating effect of the film on the particle count. (c) The same field of view at lower magnification.
3.2. Overall AAVhu.37 structure
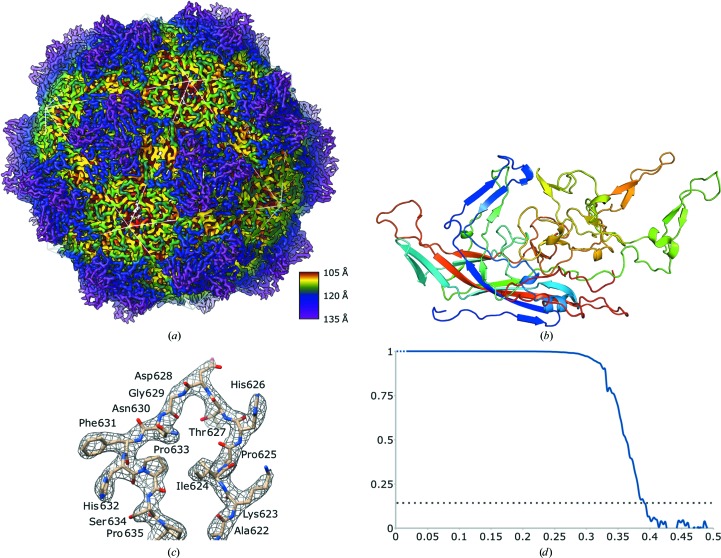
The AAVhu.37 map had an overall resolution of 2.56 Å, allowing the modeling of residues 220–738 (VP1 numbering) of the full-length capsid protein, as is typical of previously described AAV capsid structures (Fig. 3 ▸). The inability to resolve the complete N-terminus using current structure-determination methods is thought to be owing to particle-to-particle variability of the VP1 and VP2 copy number, non-uniform N-terminal orientation within the AAV capsids and areas of potential structural flexibility. A plug of density not accounted for by the model was observed in the fivefold pore beginning at a radius of 9.0 nm and terminating at a radius of 10.0 nm from the capsid center. The residues surrounding this plug were well ordered. This phenomenon is not uncommon in icosahedral maps of Parvoviridae (Subramanian et al., 2017 ▸) and may correspond to the presence of the VP1 N-terminus in an asymmetric configuration and/or ordered ions or solvent. The overall fold of the AAVhu.37 capsid is consistent with other AAV serotypes as well as with more distantly related parvovirus groups (Fig. 3 ▸ b; Mietzsch et al., 2019 ▸).
Figure 3.
(a) Overall capsid-surface model colored radially, with the icosahedral geometry indicated by a white-lined overlay. (b) Model of a single VP3 monomer colored from the N-terminus (blue) to the C-terminus (red). (c) Representative map section allowing unambiguous tracing of the AAVhu.37 backbone. (d) Noise-corrected Fourier shell correlation plotted against spatial frequency (Å−1) between the two half-maps of the final refinement iteration, with FSC = 0.143 indicated by a dotted line. Note that the low-resolution terms are not independent.
Intermediate-voltage microscopes are increasingly being utilized for high-resolution work. The structure described here is of higher resolution than any other peer-reviewed single-particle cryoEM structure from a 200 kV electron microscope to date, albeit by 0.03 Å (Herzik et al., 2017 ▸). Preprinted work indicates that resolutions of beyond 2 Å are attainable using commercially available 200 kV microscopes (Wu et al., 2019 ▸).
3.3. Comparison to known serotypes
Of the released structures within the Protein Data Bank (PDB), AAV8 is the most closely related capsid to AAVhu.37, with 93.5% amino-acid identity for full-length VP1. Although several AAV8 capsid structures exist in the literature, the most directly comparable entry, PDB entry 3ra4 (Nam et al., 2011 ▸), is an AAV8 capsid containing a GFP-encoding genome determined at pH 7.5 via X-ray crystallography to 2.7 Å resolution. The overall calculated r.m.s.d. of the two structures is 0.564 Å. Of the 39 amino-acid differences in VP3 between these two serotypes, 18 (46%) are in the variable-region (VR) loops that form the spikes surrounding the threefold axis of symmetry (Mietzsch et al., 2019 ▸): nine in VR-IV, four in VR-V and five in VR-VIII. The r.m.s.d.s for VR-IV (amino acids 449–477), VR-V (amino acids 493–512) and VR-VIII (amino acids 578–600) are 0.422, 0.326 and 0.302 Å, respectively. In other serotypes, these areas are known to be hotspots for binding cellular protein receptors and glycan receptors, as well as neutralizing antibodies (Zhang et al., 2019 ▸; Meyer et al., 2019 ▸; Bell et al., 2012 ▸; O’Donnell et al., 2009 ▸; Giles et al., 2018 ▸; Huang et al., 2016 ▸; Tseng & Agbandje-McKenna, 2014 ▸). Although VR-VIII has the lowest amino-acid identity of any region between AAVhu.37 and AAV8 (70%), the main chain is scarcely perturbed.
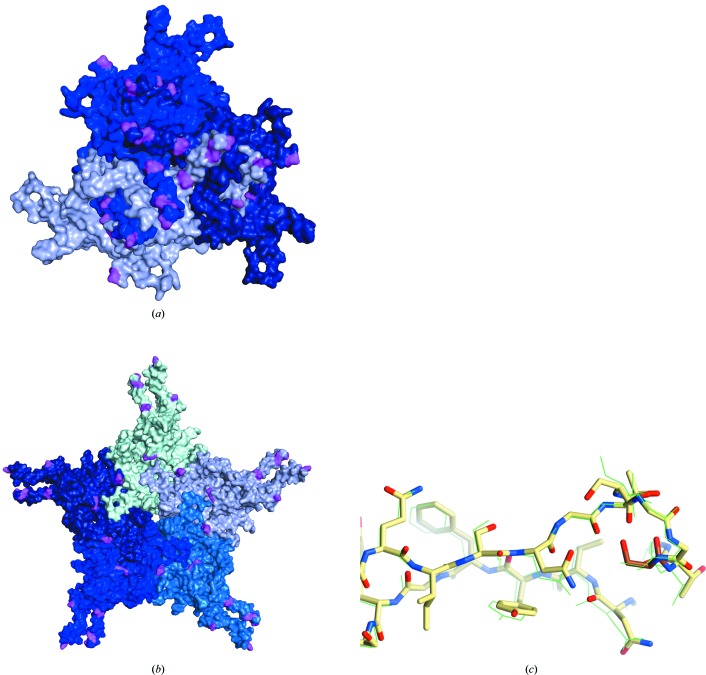
The AAVrh.10 capsid, the structure of which has not been solved, and AAVhu.37 share 98.5% amino-acid identity. Mapping AAVrh.10 residues onto the AAVhu.37 structure reveals a clustering of differential amino acids at the spikes surrounding the threefold axis of symmetry, which may be responsible for its decreased immunogenicity (Greig et al., 2018 ▸; Wang, Calcedo et al., 2010 ▸; Figs. 4 ▸ a and 4 ▸ b). Determinants of BBB penetration have been mapped to VR-I by making chimeric capsids between the BBB-penetrating AAVrh.10 and the nonpenetrating AAV1 (Albright et al., 2018 ▸). The AAVrh.10 VR-I residues that were identified as sufficient to confer BBB penetration on AAV1 are absolutely conserved in AAVhu.37. The VR-I of the nonpenetrating AAV8 differs at only one site: AAVhu.37 and AAVrh.10 contain a serine at position 269, while AAV8 contains an alanine. Superimposition of AAVhu.37 with previously published structures of AAV8 (Nam et al., 2007 ▸, 2011 ▸) reveals no evidence of differences in the VR-I loop conformation (Fig. 4 ▸ c). An important caveat is that VR-I of AAV2 rearranges during AAV receptor (AAVR) engagement (Meyer et al., 2019 ▸). Bound AAVR would sterically clash with VR-I of AAVhu.37 unless it was pushed aside during binding. Hence, we cannot exclude the possibility that the serine/alanine polymorphism could lead to a biologically relevant difference in loop flexibility or in a cofactor-engaged structure. On balance, we hypothesize that while splicing an AAVrh.10 VR-I epitope into AAV1 may be sufficient to confer BBB penetration to AAV1, one or more additional AAV8-specific factors are likely to block BBB penetration. This finding encourages further inspection of capsid elements which may confer or inhibit the ability to cross the BBB and promotes the rational design of novel capsids for more efficient BBB penetration.
Figure 4.
AAVhu.37 VP3 monomers shown in shades of blue, highlighting amino-acid differences (pink) between AAVhu.37 and AAVrh.10 relative to the (a) threefold and (b) fivefold axes of symmetry. (c) Comparison of the VR-I loop of AAV8 (shown in green as lines) and AAVhu.37 (shown as sticks with C atoms colored yellow; amino acids 262–275). Residue 269, the single amino-acid difference in VR-I between the two serotypes, is indicated by darker coloring (orange C atoms).
Supplementary Material
PDB reference: AAVhu.37 capsid, 6u95
EMDB reference: AAVhu.37 capsid, EMD-20693
Acknowledgments
We thank Haiyan Zheng and Amenah Soherwardy at the Rutgers Biological Mass Spectrometry Facility for digested LC-MS/MS. We thank the REGENXBIO analytical development department for their support on this project: Vurghun Ahmadov for capillary gel electrophoresis, Shugui Chen for undigested LC-MS, Tomoko Maekawa for ddPCR, and Zhuchun Wu and Zhenhong Li for technology development and oversight. We would also like to thank April Giles for bioinformatic analysis of the capsid sequence.
Funding Statement
This work was funded by REGENXBIO grant .
References
- Albright, B. H., Storey, C. M., Murlidharan, G., Castellanos Rivera, R. M., Berry, G. E., Madigan, V. J. & Asokan, A. (2018). Mol. Ther. 26, 510–523. [DOI] [PMC free article] [PubMed]
- Bell, C. L., Gurda, B. L., Van Vliet, K., Agbandje-McKenna, M. & Wilson, J. M. (2012). J. Virol. 86, 7326–7333. [DOI] [PMC free article] [PubMed]
- Berns, K. I. & Parrish, C. R. (2015). Fields Virology, 6th ed., edited by D. M. Knipe & P. M. Howley, pp. 1768–1791. Philadelphia: Lippincott Williams & Wilkins.
- Breous, E., Somanathan, S., Bell, P. & Wilson, J. M. (2011). Gastroenterology, 141, 348–357. [DOI] [PMC free article] [PubMed]
- Breous, E., Somanathan, S., Vandenberghe, L. H. & Wilson, J. M. (2009). Hepatology, 50, 612–621. [DOI] [PMC free article] [PubMed]
- Cai, N., Bai, Z., Nanda, V. & Runnels, L. W. (2017). Sci. Rep. 7, 42739. [DOI] [PMC free article] [PubMed]
- Cearley, C. N., Vandenberghe, L. H., Parente, M. K., Carnish, E. R., Wilson, J. M. & Wolfe, J. H. (2008). Mol. Ther. 16, 1710–1718. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Daya, S. & Berns, K. I. (2008). Clin. Microbiol. Rev. 21, 583–593. [DOI] [PMC free article] [PubMed]
- Dobrzynski, E., Fitzgerald, J. C., Cao, O., Mingozzi, F., Wang, L. & Herzog, R. W. (2006). Proc. Natl Acad. Sci. USA, 103, 4592–4597. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gao, G., Vandenberghe, L. H., Alvira, M. R., Lu, Y., Calcedo, R., Zhou, X. & Wilson, J. M. (2004). J. Virol. 78, 6381–6388. [DOI] [PMC free article] [PubMed]
- Giles, A. R., Govindasamy, L., Somanathan, S. & Wilson, J. M. (2018). J. Virol. 92, e01011-18. [DOI] [PMC free article] [PubMed]
- Greig, J. A., Nordin, J. M. L., White, J. W., Wang, Q., Bote, E., Goode, T., Calcedo, R., Wadsworth, S., Wang, L. & Wilson, J. M. (2018). Hum Gene Ther. 29, 763–770. [DOI] [PMC free article] [PubMed]
- Greig, J. A., Wang, Q., Reicherter, A. L., Chen, S.-J., Hanlon, A. L., Tipper, C. H., Clark, K. R., Wadsworth, S., Wang, L. & Wilson, J. M. (2017). Hum. Gene Ther. 28, 392–402. [DOI] [PubMed]
- Herzik, M. A. Jr, Wu, M. & Lander, G. C. (2017). Nat. Methods, 14, 1075–1078. [DOI] [PMC free article] [PubMed]
- Huang, L. Y., Patel, A., Ng, R., Miller, E. B., Halder, S., McKenna, R., Asokan, A. & Agbandje-McKenna, M. (2016). J. Virol. 90, 5219–5230. [DOI] [PMC free article] [PubMed]
- Lawlor, P. A., Bland, R. J., Mouravlev, A., Young, D. & During, M. J. (2009). Mol. Ther. 17, 1692–1702. [DOI] [PMC free article] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Lock, M., Alvira, M. R., Chen, S.-J. & Wilson, J. M. (2014). Human Gene Therapy Methods, 25, 115–125. [DOI] [PMC free article] [PubMed]
- Martino, A. T., Nayak, S., Hoffman, B. E., Cooper, M., Liao, G., Markusic, D. M., Byrne, B. J., Terhorst, C. & Herzog, R. W. (2009). PLoS One, 4, e6376. [DOI] [PMC free article] [PubMed]
- Meyer, N. L., Hu, G., Davulcu, O., Xie, Q., Noble, A. J., Yoshioka, C., Gingerich, D. S., Trzynka, A., David, L., Stagg, S. M. & Chapman, M. S. (2019). eLife, 8, e44707. [DOI] [PMC free article] [PubMed]
- Mietzsch, M., Penzes, J. J. & Agbandje-McKenna, M. (2019). Viruses, 11, 362. [DOI] [PMC free article] [PubMed]
- Mingozzi, F. & High, K. A. (2011). Nat. Rev. Genet. 12, 341–355. [DOI] [PubMed]
- Mingozzi, F., Liu, Y.-L., Dobrzynski, E., Kaufhold, A., Liu, J. H., Wang, Y., Arruda, V. R., High, K. A. & Herzog, R. W. (2003). J. Clin. Invest. 111, 1347–1356. [DOI] [PMC free article] [PubMed]
- Mount, J. D., Herzog, R. W., Tillson, D. M., Goodman, S. A., Robinson, N., McCleland, M. L., Bellinger, D., Nichols, T. C., Arruda, V. R., Lothrop, C. D. Jr & High, K. A. (2002). Blood, 99, 2670–2676. [DOI] [PubMed]
- Mueller, C. & Flotte, T. R. (2008). Gene Ther. 15, 858–863. [DOI] [PubMed]
- Nam, H.-J., Gurda, B. L., McKenna, R., Potter, M., Byrne, B., Salganik, M., Muzyczka, N. & Agbandje-McKenna, M. (2011). J. Virol. 85, 11791–11799. [DOI] [PMC free article] [PubMed]
- Nam, H.-J., Lane, M. D., Padron, E., Gurda, B., McKenna, R., Kohlbrenner, E., Aslanidi, G., Byrne, B., Muzyczka, N., Zolotukhin, S. & Agbandje-McKenna, M. (2007). J. Virol. 81, 12260–12271. [DOI] [PMC free article] [PubMed]
- O’Donnell, J., Taylor, K. A. & Chapman, M. S. (2009). Virology, 385, 434–443. [DOI] [PMC free article] [PubMed]
- Orlova, E. V., Dube, P., Harris, J. R., Beckman, E., Zemlin, F., Markl, J. & van Heel, M. (1997). J. Mol. Biol. 271, 417–437. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. (2017). Nat. Methods, 14, 290–296. [DOI] [PubMed]
- Rose, J. A., Maizel, J. V., Inman, J. K. & Shatkin, A. J. (1971). J. Virol. 8, 766–770. [DOI] [PMC free article] [PubMed]
- Rosenberg, J. B., Sondhi, D., Rubin, D. G., Monette, S., Chen, A., Cram, S., De, B. P., Kaminsky, S. M., Sevin, C., Aubourg, P. & Crystal, R. G. (2014). Hum. Gene Ther. Clin. Dev. 25, 164–177. [DOI] [PMC free article] [PubMed]
- Subramanian, S., Organtini, L. J., Grossman, A., Domeier, P. P., Cifuente, J. O., Makhov, A. M., Conway, J. F., D’Abramo, A. Jr, Cotmore, S. F., Tattersall, P. & Hafenstein, S. (2017). Virology, 510, 216–223. [DOI] [PMC free article] [PubMed]
- Tang, G., Peng, L., Baldwin, P. R., Mann, D. S., Jiang, W., Rees, I. & Ludtke, S. J. (2007). J. Struct. Biol. 157, 38–46. [DOI] [PubMed]
- Tseng, Y.-S. & Agbandje-McKenna, M. (2014). Front. Immunol. 5, 9. [DOI] [PMC free article] [PubMed]
- Wang, L., Calcedo, R., Wang, H., Bell, P., Grant, R., Vandenberghe, L. H., Sanmiguel, J., Morizono, H., Batshaw, M. L. & Wilson, J. M. (2010). Mol. Ther. 18, 126–134. [DOI] [PMC free article] [PubMed]
- Wang, L., Wang, H., Bell, P., McCarter, R. J., He, J., Calcedo, R., Vandenberghe, L. H., Morizono, H., Batshaw, M. L. & Wilson, J. M. (2010). Mol. Ther. 18, 118–125. [DOI] [PMC free article] [PubMed]
- Wang, Q., Lock, M., Prongay, A. J., Alvira, M. R., Petkov, B. & Wilson, J. M. (2015). Mol. Ther. Methods Clin. Dev. 2, 15040. [DOI] [PMC free article] [PubMed]
- Wu, M., Lander, G. C. & Herzik, M. A. (2019). bioRxiv, 855643.
- Wu, Z., Asokan, A. & Samulski, R. J. (2006). Mol. Ther. 14, 316–327. [DOI] [PubMed]
- Yang, B., Li, S., Wang, H., Guo, Y., Gessler, D. J., Cao, C., Su, Q., Kramer, J., Zhong, L., Ahmed, S. S., Zhang, H., He, R., Desrosiers, R. C., Brown, R., Xu, Z. & Gao, G. (2014). Mol. Ther. 22, 1299–1309. [DOI] [PMC free article] [PubMed]
- Zhang, H., Yang, B., Mu, X., Ahmed, S. S., Su, Q., He, R., Wang, H., Mueller, C., Sena-Esteves, M., Brown, R., Xu, Z. & Gao, G. (2011). Mol. Ther. 19, 1440–1448. [DOI] [PMC free article] [PubMed]
- Zhang, R., Cao, L., Cui, M., Sun, Z., Hu, M., Zhang, R., Stuart, W., Zhao, X., Yang, Z., Li, X., Sun, Y., Li, S., Ding, W., Lou, Z. & Rao, Z. (2019). Nat. Microbiol. 4, 675–682. [DOI] [PubMed]
- Zheng, S. Q., Palovcak, E., Armache, J.-P., Verba, K. A., Cheng, Y. & Agard, D. A. (2017). Nat. Methods, 14, 331–332. [DOI] [PMC free article] [PubMed]
- Zivanov, J., Nakane, T. & Scheres, S. H. W. (2019). IUCrJ, 6, 5–17. [DOI] [PMC free article] [PubMed]
- Zolotukhin, S., Byrne, B. J., Mason, E., Zolotukhin, I., Potter, M., Chesnut, K., Summerford, C., Samulski, R. J. & Muzyczka, N. (1999). Gene Ther. 6, 973–985. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AAVhu.37 capsid, 6u95
EMDB reference: AAVhu.37 capsid, EMD-20693