Abstract
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a serious, debilitating disorder with a wide spectrum of symptoms, including pain, depression, and neurocognitive deterioration. Over 17 million people around the world have ME/CFS, predominantly women with peak onset at 30–50 years. Given the wide spectrum of symptoms and unclear etiology, specific biomarkers for diagnosis and stratification of ME/CFS are lacking. Here we show that actin network proteins in circulating extracellular vesicles (EVs) offer specific non-invasive biomarkers for ME/CFS. We found that circulating EVs were significantly increased in ME/CFS patients correlating to C-reactive protein, as well as biological antioxidant potential. Area under the receiver operating characteristic curve for circulating EVs was 0.80, allowing correct diagnosis in 90–94% of ME/CFS cases. From two independent proteomic analyses using circulating EVs from ME/CFS, healthy controls, idiopathic chronic fatigue, and depression, proteins identified from ME/CFS patients are involved in focal adhesion, actin skeletal regulation, PI3K-Akt signaling pathway, and Epstein-Barr virus infection. In particular, talin-1, filamin-A, and 14–3-3 family proteins were the most abundant proteins, representing highly specific ME/CFS biomarkers. Our results identified circulating EV number and EV-specific proteins as novel biomarkers for diagnosing ME/CFS, providing important information on the pathogenic mechanisms of ME/CFS.
Keywords: ME/CFS, circulating EV, non-invasive biomarkers, actin network proteins
Introduction
ME/CFS is a serious and complex debilitating disease with a wide spectrum of symptoms, including muscle pain and neurocognitive deterioration that occurs following ME/CFS development (2015; Gallagher et al., 2004; Twisk, 2014, 2018). The diagnosis of ME/CFS is based on clinical symptoms that include a broad spectrum of disease severity from mild to debilitating. Conclusive ME/CFS non-invasive diagnosis is thus difficult for clinicians to achieve using the current ME/CFS diagnostic methods and subjective symptoms, and although immunological abnormalities including impaired calcium ion channel (Brenu et al., 2013; Brenu et al., 2014; Brenu et al., 2011; Cabanas et al., 2019; Jason et al., 2009; Montoya et al., 2017), dis-regulation of the neuronal-immunological system (G and Maes, 2014; Komaroff et al., 2018), abnormalities of metabolism (Nagy-Szakal et al., 2018; Naviaux et al., 2016), and has been recognized as an important contributor to ME/CFS, the pathogenic mechanisms are not fully understood. Potential ME/CFS biomarkers that have been proposed include changes in autonomic nervous function (Van Cauwenbergh et al., 2014), circulating cytokines (Broderick et al., 2010; Moneghetti et al., 2018; Yang et al., 2019), Epstein-Barr (EB) virus (Loebel et al., 2017), energy metabolism (Castro-Marrero et al., 2013; Mikirova et al., 2012), oxidative stress (Maes et al., 2011), and sleep-wake cycle (Togo and Natelson, 2013), but additional biomarkers are needed to distinguish ME/CFS from other diseases associated with fatigue, such as idiopathic chronic fatigue (ICF) and depression. Current ME/CFS therapies, cognitive behavior therapy and graded exercise treatment, are not fully effective (Cleare et al., 2015). The discovery of objective ME/CFS biomarkers, as well as ME/CFS pathogenic mechanisms including ME/CFS etiology, represent a critical breakthrough long-awaited in the field of ME/CFS (Lloyd and Meer, 2015).
Extracellular vesicles (EVs) are released from damaged or stressed cells with cellular content, such as proteins, and circulate in the bloodstream (Yanez-Mo et al., 2015). EVs are thus recognized as non-invasive biomarkers for a variety of diseases (Yanez-Mo et al., 2015). Furthermore, EVs contribute to disease pathogenesis via their function in cell-to-cell communications and delivering EV contents from the cell or origin to target cells, resulting in modulation of cell signaling in target cells (Eguchi and Feldstein, 2018; Yanez-Mo et al., 2015). Circulating EV number and EV composition can be used for diagnosis of human diseases, including chronic liver diseases (Eguchi et al., 2019; Shah et al., 2018) as well as for metabolic status (Kobayashi et al., 2018).
MATERIALS AND METHODS
Subjects and study design
The study was approved by the ethics committees of Kansai University Welfare of Science (Approval No. 09–06) and Osaka City University Graduate School of Medicine (Approval No. 2151), and was conducted in accordance with the Declaration of Helsinki. All subjects, ME/CFS patients (n=99), ICF patients (n=6), depression patients (n=8), and healthy individuals (n=56) provided written informed consent for participation in the study before enrolment. Healthy individuals who were confirmed not to have abnormal results on any major clinical laboratory tests (hemoglobin, CRP, albumin, triglycerides, glucose, AST, ALT, or cholesterol, etc.), BMI ≥30 and <17, subjective sleep problems, problems in daily life by fatigue, and shift worker. Out of 56 recruited healthy individuals three were excluded from the analysis, as they did not meet the inclusion criteria. ME/CFS, ICF, and depression patients who visited the outpatient clinic of Osaka City University hospitals and B clinic were randomly enrolled into the study. ME/CFS and ICF were diagnosed based on the 1994 Center for Disease Control clinical criteria (Fukuda criteria)(Fukuda et al., 1994) and Canadian of Consensus Case Definition (Carruthers et al., 2011) by specialists at the Osaka City University hospitals and B clinic. Depression was assessed using a structured clinical interview for DSM-IV. Exclusion criteria were as follows: 1) neuro-inflammatory or immune disorders diagnosed by clinical laboratory tests and magnetic resonance imaging; 2) any active medical condition that could explain the presence of chronic fatigue; 3) presence of any diagnosable illness that relapsed or was not completely resolved, such as some types of malignancy or chronic cases of hepatitis B or C virus infection; 4) alcohol or other substance abuse; 5) severe obesity as defined by a body mass index ≥ 30 kg/m2; 6) pregnancy; or 7) lactation. The presence of major depressive disorder, fibromyalgia, or somatoform disorder was not a criterion for exclusion. Abdominal discomfort syndrome was not assessed. Psychiatric disorders associated with CFS symptoms were diagnosed by psychiatrists at Osaka City University Hospitals and B clinic. Blood was collected with or without anticoagulant for plasma or serum, respectively. CRP was measured using serum by LSI Medience (Tokyo, Japan).
Flow cytometry analysis of circulating EVs
EVs were counted using calcein-AM via flow cytometry, as described before(Kobayashi et al., 2018). Briefly, plasma was incubated for 30 min at room temperature with final 4 μg/ml of Calcein-AM (Life Technologies, Carlsbad, CA). EV acquisition was performed using the BD LSRII Flow Cytometer System (BD Biosciences, San Jose, CA) or BD Cant II (BD Biosciences, Tokyo, Japan) for validation and data were analyzed using FlowJo software (TreeStar, Ashland, OR). Gating parameters were defined using ultraviolet 2.5 μm Alignflow alignment beads (Life Technologies) and negative controls. Forward and side scatter parameters were plotted on logarithmic scales to best cover a wide size range. Single staining controls were used to check fluorescence compensation settings and to set up positive regions. EVs were identified using a forward-scatter analysis. EV number was counted using 2.5 μm Alignflow alignment beads (Life Technologies) as the size standards.
EV size determination
Circulating EVs were ultracentrifuged at 100,000×g for 60 min at 10°C and suspended in PBS. For dynamic light-scattering analysis, entire size was measured by Zetasizer Nano ZS90 (Malvern, Worcestershire, UK) and averages were taken from 3 healthy individual or 5 ME/CFS patients. For transmission electron microscopy, EVs were adhered to 100-mesh Formvar and carbon-coated grids for 5 min at room temperature. Grids were washed once with water, stained with 1% uranyl acetate (Ladd Research Industries, Williston, VT) for 1 min, dried and viewed using a JEOL JEM-1400Plus transmission electron microscope (JEOL, Peabody, MA). Images were captured using a Gatan OneView digital camera (Gatan, Pleasanton, CA).
Oxidative stress
Both oxidation and anti-oxidation activities were measured simultaneously in serum(Fukuda et al., 2016). Briefly, oxidative activity was assessed by measuring d-ROMs (Diacron International, Grosseto, Italy) and anti-oxidative activity by measuring the Biological Antioxidant Potential (BAP) (Diacron International) using an AU480 automated analyzer (Beckman Coulter, Tokyo, Japan).
Analysis of protein composition in circulating EVs via nanoLC-MS/MS
Circulating EVs were isolated from equal amount of plasma via qEV (Izon Science, Cambridge, MA) according to the instructions from the manufacturer. Briefly, the same amount of plasma from each patient/individual was applied on the qEV column and corrected EVs as fractions. For the first group (CFS and HC; n=3 each), EVs were concentrated via Amicon ultra 3 K centrifugal filter (Millipore, Burlington, MA), followed by alkylation with iodoacetamide and trypsinization using the same amount of proteins among samples. Samples were separated using a nanoLC-MS/MS, Dina System (KYA Technologies, Tokyo, Japan) for nanoLC and AB SCIEX Triple TOF 5600 system (AB SCIEX, Tokyo, Japan) for MS/MS at Oncomics Co. (Nagoya, Japan). For the second group (CFS, ICF, and depression who were between the ages of 20 and 40: n=4 each), EVs were precipitated with trichloroacetic acid, followed by reduction, alkylation with iodoacetamide and trypsinization using the same amount of proteins among samples. Samples were separated using nanoLC-MS/MS, EASY-nLC 1200 (Thermo Fisher Scientific, Tokyo, Japan) and Q Exactive Plus (Thermo Fisher Scientific) at APRO Science Institute (Tokushima, Japan). Data from the first and second groups were analyzed using Scaffold4 (Proteome Software, Portland, OR) against the SwissProt database at APRO Science Institute. Quantitative value (normalized total spectra) on Scaffold4 was converted to log scale, and then was used for Heat map using Heatmapper software (Babicki et al., 2016). Protein interactions were generated using STRING v10.5 software (Szklarczyk et al., 2015) selecting MCL clustering and confidence levels for network edges based on experiments and databases. KEGG pathway was generated on STRING software.
Statistics
All data are expressed as mean ± standard error of the mean (SEM) unless otherwise indicated. Differences between groups were compared using the Mann-Whitney test or Kruskal-Wallis test. Correlations of EV number with CRP, BAP, and d-Roms were determined using the Spearman rank-sum test. For statistical analysis on proteomics, we converted quantitative values (normalized total spectra), which are automatically generated on Scafolld4 proteome software, to log scale, then, calculated significance by Mann-Whitney test in an initial cohort (two groups: 3 healthy individuals and 3 ME/CFS patients) or by Kruskal-Wallis test in a second subsequent independent cohort (three groups: 4 ME/CFS patients, 4 ICF patients, and 4 depression patients). Statistical analyses were performed using SPSS version 22.0J software (SPSS Japan, Tokyo, Japan). The statistical analyses for the ROC curve were performed using Prism Software (Graph Pad, La Jolla, CA).
RESULTS
Current evidence from our own and other laboratories has led us to investigate whether the number of circulating EV is increased in ME/CFS. A first group (ME/CFS 1) included 33 healthy controls (HC) and 39 ME/CFS patients diagnosed with ME/CFS based on the 1994 Center for Disease Control clinical criteria(Fukuda et al., 1994), matched with age, gender, weight, and BMI (Supplementary table 1). Circulating EVs were stained with calcein to count the intact circulating EVs (Kobayashi et al., 2018), excluding contaminated proteins in plasma and quantifying the calcein-positive circulating EVs based on the intensity of unstained circulating EVs via flow cytometry (Fig. 1A). The dot plot of gated population is shown in supplementary figure 1. The circulating EV number was significantly increased in ME/CFS patients (P < 0.001) (Fig. 1B). To confirm those results, we recruited 30 more ME/CFS patients (ME/CFS 2), excluding smokers and obese individuals (Supplementary table 1). Circulating EV number was also significantly increased in ME/CFS patients (P < 0.001) (Fig. 1B) and the mean circulating EV number was similar between ME/CFS 1 and ME/CFS 2 (Fig. 1B). An increased circulating EV number in ME/CFS was further validated in an additionally recruited 20 HC and 30 ME/CFS patients (P < 0.001) (Supplementary Fig 1B). Circulating EVs consisted of small EVs (diameter, <100 nm) and large EVs (diameter, 100–1000 nm) in both ME/CFS patients and HC as determined via dynamic light scattering analysis (Fig. 1C) and in ME/CFS patients via transmission electron microscopy (TEM) (Fig. 1D). Circulating EV number correlated significantly with serum c-reactive protein (CRP) levels (ρ = 0.442, P = 0.0007) (Fig. 1E), and this correlation persisted even in cases with CRP levels within the normal range (0 to 0.1) (ρ = 0.310, P = 0.032) (Supplementary Fig. 1C) and BAP as antioxidant potential, (ρ = −0.314, P = 0.007) (Fig. 1F), two known markers that are increased in ME/CFS(Fukuda et al., 2016). To determine specificity and sensitivity, we determined the ROC curve using circulating EV number, CRP, d-Roms, and BAP levels. Area under ROC curve (AUC) of circulating EV number in ME/CFS 1 was 0.802 (95%CI 0.70–0.90; P < 0.0001) (EVs versus CRP: P < 0.001; EVs versus BAP: P < 0.05), significantly higher than the AUC of CRP levels (0.641; 95%CI 0.49–0.79), AUC of d-Roms levels (0.540; 95%CI 0.41–0.67) and AUC of BAP levels (0.710; 95%CI 0.59–0.83; P = 0.002) (Fig. 1G). The ME/CFS 2 group was diagnosed with ME/CFS in 90–94% using several cutoff values the from AUC of EV number (Supplementary table 2).
Figure 1. Circulating EVs are diagnostic factor in ME/CFS patients correlating to CRP and BAP.
A-D) Characterization of circulating EVs from ME/CFS patients, first group (ME/CFS 1) and second group (ME/CFS 2), or healthy controls (HC) via flow cytometry. A) Histograph of unstained or stained circulating EVs from ME/CFS patient. B) Quantification of calcein-positive circulating EVs. C) Dynamic light scattering analysis of isolated EVs from ME/CFS patients (ME/CFS) or HC. D) Transmission electron microscopy of isolated EVs from ME/CFS patients. E) Dot plot of circulating EVs and CRP level. F) Dot plot of circulating EVs and BAP level. G) ROC curve with circulating EVs, CRP level, BAP level, and d-Roms level. HC: healthy individual. Values represent mean ± SEM. ***P < 0.001.
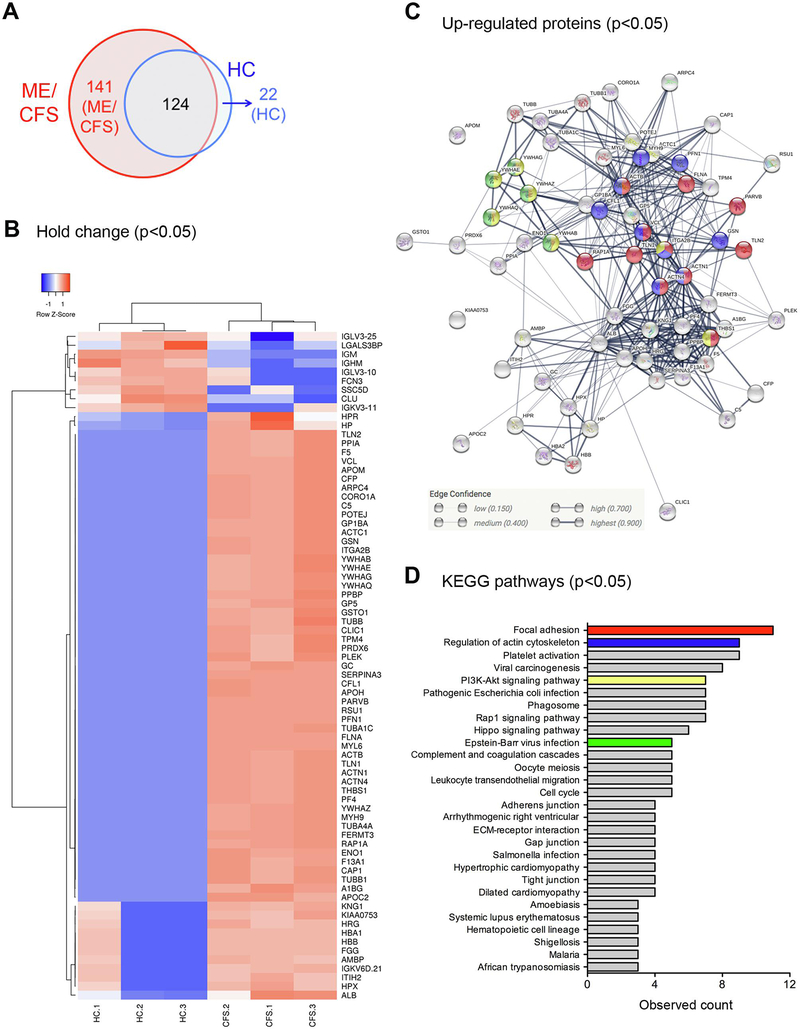
Increased levels of circulating EV number with an AUC of 0.802 for ME/CFS diagnosis led us to further investigate whether assessing EV cargo would lead to the discovery of specific EV signatures in ME/CFS patients that may represent novel biomarkers for this disease. Purified circulating EVs from three HC and three ME/CFS patients (Supplementary table 3) were ascertained in terms of the EV proteome by nano liquid chromatography tandem-mass spectrometry (nanoLC-MS/MS) analysis (Supplementary Fig 2A). A total of 124 proteins were present in EVs from both ME/CFS patients and HC, but a significant number of proteins was present only in EVs from ME/CFS patients, while 22 proteins were present only in EVs from HC (Fig. 2A, supplementary table 4). Furthermore, 75 proteins were significantly changed in EVs from ME/CFS patients compared to those from HC (P < 0.05), including 66 up-regulated proteins including actin network proteins, such as talin-1, filamin-A, actin, myosin-9, vinculin, gelsolin, and tubulin, and 9 down-regulated proteins (Fig. 2B, supplementary table 5). Notably, 63 up-regulated proteins made a cluster in protein-protein interactions (Fig. 2C) relating to several pathways, focal adhesion, regulation of the actin cytoskeletal, phosphoinositide-3-kinase (PI3K)-Akt signaling pathway, and EB virus infection (Fig. 2C and 2D, supplementary table 6).
Figure 2. Detected proteins in circulating EVs from ME/CFS and HC via proteome analysis.
A) Detected protein number. B) Heat map analysis of significantly changed proteins between ME/CFS and HC (p<0.05). C) Protein interaction using significantly up-regulated proteins in ME/CFS patients (p<0.05). The colors correspond to figure D. D) KEGG pathway for significantly up-regulated proteins in ME/CFS patients. Arrhythmogenic right ventricular, Arrhythmogenic right ventricular cardiomyopathy.
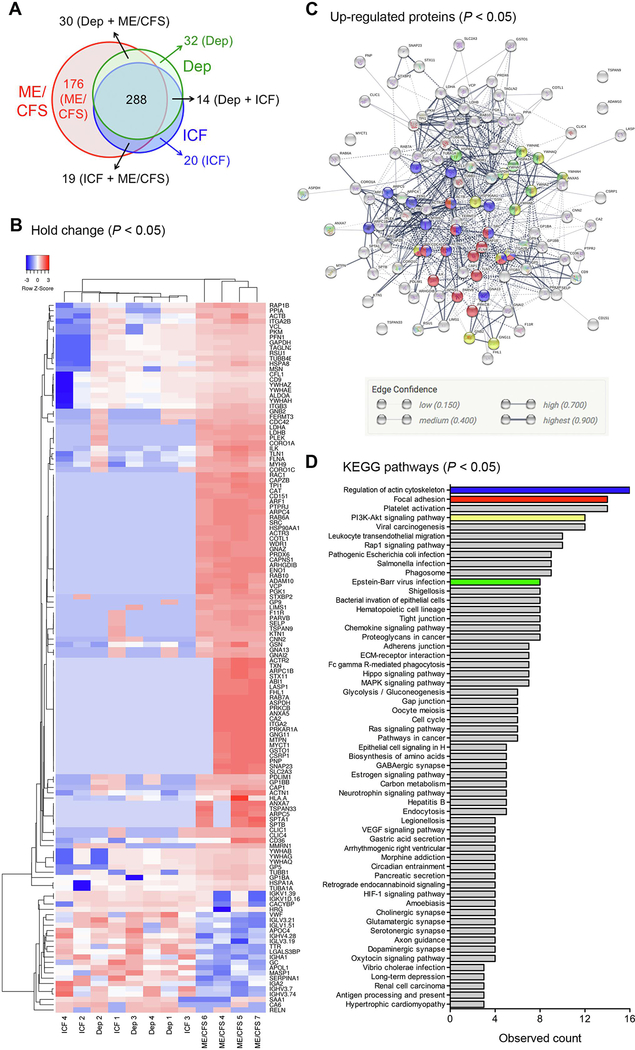
We further explored whether identified proteins in EVs from ME/CFS can distinguished from ICF and depression two condition also associated with fatigue. The number of circulating EVs was similar among the three groups (Supplementary Fig 3A). Purified circulating EVs from four ICF, four depression, and four ME/CFS patients (Supplementary table 7) were ascertained for the EV proteome by nanoLC-MS/MS analysis (Supplementary Fig 3B). A total of 579 proteins were present in EVs from ME/CFS, ICF, and depression patients. A significant number of proteins (176 of 579 proteins) were present only in EVs from ME/CFS patients, while 20 and 32 proteins were present only in EVs from ICF and depression patients, respectively (Fig. 3A, supplementary table 8). In addition, 134 proteins were significantly changed in EVs from ME/CFS patients compared to those from ICF and depression patients (P < 0.05) and there were 111 up-regulated and 23 down-regulated proteins (Fig. 3B, supplementary table 9). Notably, actin network proteins such as talin-1, filamin-A, actin, myosin-9, vinculin, gelsolin, and tubulin, were also significantly up-regulated in EVs from ME/CFS patients compared to those from ICF and depression (Fig. 3B). Of the 111 up-regulated proteins, 105 were associated with core protein-protein interactions (Fig. 3C) and involved in focal adhesion, regulation of the actin cytoskeleton, PI3K-Akt signaling pathway, and EB virus infection (Fig. 3C and 3D, supplementary table 10).
Figure 3. Proteins detected in circulating EVs from ME/CFS, ICF, and depression via proteome analysis.
A) Detected protein number. B) Heat map analysis of significantly changed proteins between ME/CFS and HC (p<0.05). C) Protein interaction using significantly up-regulated proteins in ME/CFS patients (p<0.05). The colors correspond to figure D. D) KEGG pathway in significantly up-regulated proteins in ME/CFS patients. ICF, idiopathic chronic fatigue; Arrhythmogenic right ventricular, Arrhythmogenic right ventricular cardiomyopathy; Epithelial cell signaling in H, Epithelial cell signaling in Helicobacterpylori infection.
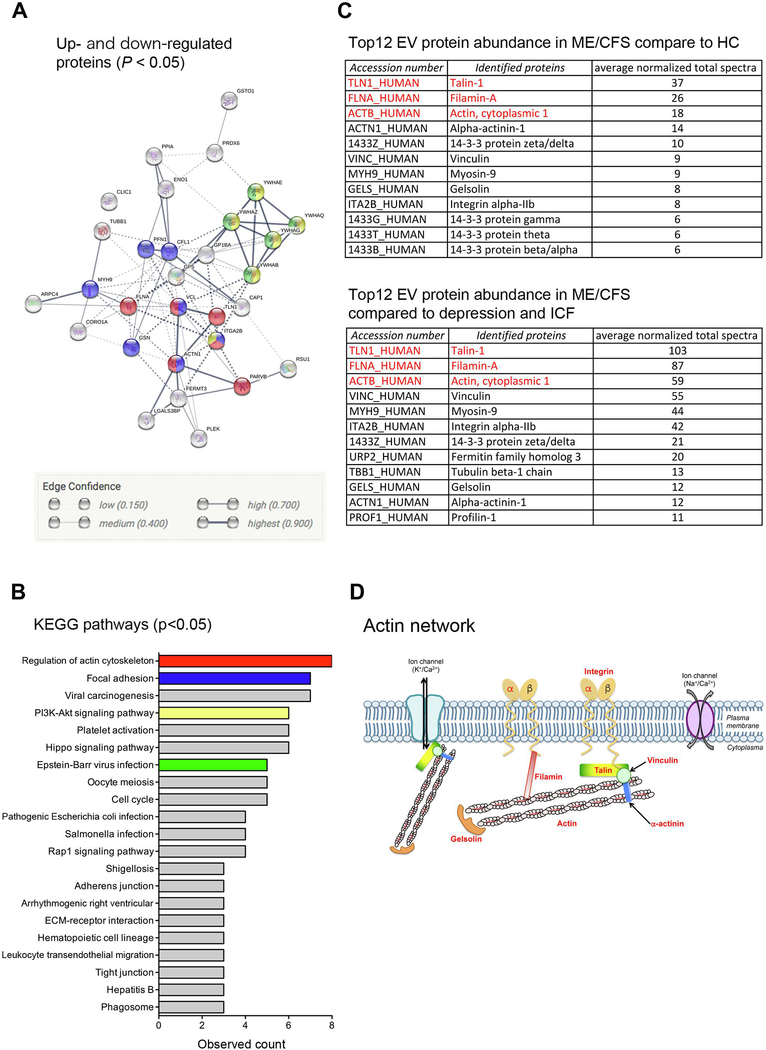
A specific circulating EV profile, from two independent experiments using ME/CFS and HC or ME/CFS, ICF, and depression allowed us to identify specific proteins in blood EVs for ME/CFS diagnosis. From the list of significantly changed proteins in EVs from ME/CFS patients, 31 proteins were identified as common proteins in EVs from ME/CFS patients, including 30 up-regulated and 1 down-regulated protein (supplementary table 11). Of the 31 identified proteins in EVs, 29 formed a cluster in protein-protein interactions (Fig. 4A), involved in regulation of the actin cytoskeleton, focal adhesion, PI3K-Akt signaling pathway, and EB virus infection (Fig. 4B, supplementary table 12). The 12 most abundant proteins in EVs were closely matched in the two independent experiments and part of the actin network protein family (talin-1, filamin-A, actin, actinin, vinculin, gelsolin, and integrin) (Fig. 4C and 4D) and 14–3-3 family proteins (Fig. 4C). In particular, the top three proteins in EVs, talin-1, filamin-A, and actin, were exactly the same in two independent experiments (Fig. 4C). These results revealed that actin network proteins in EVs are novel non-invasive biomarkers for ME/CFS diagnosis.
Figure 4. Identified proteins for specific non-invasive biomarker in ME/CFS.
A) Protein interaction using significantly changed proteins from two independent proteome analyses, ME/CFS vs HC or ME/CFS vs ICF and depression. The colors correspond to figure B. B) KEGG pathway for significantly changed proteins from two independent proteome analyses, ME/CFS vs HC or ME/CFS vs ICF and depression. C) Top 12 proteins from each proteome analysis. D) Scheme for detected actin network proteins, with red letters. ICF, idiopathic chronic fatigue; Arrhythmogenic right ventricular, Arrhythmogenic right ventricular cardiomyopathy.
DISCUSSION
This is the first report to find that actin network proteins including talin-1 and filamin-A in circulating EVs can be used for specific ME/CFS diagnosis, distinguishing from ICF and depression. We also showed that the number of circulating EVs was significantly increased in ME/CFS compared to healthy controls, confirming the findings recently reported (Castro-Marrero et al., 2018). Current potential biomarkers reported with AUC 0.7–0.8 for ME/CFS diagnosis include peripheral blood mononuclear cell gene expression (Frampton et al., 2011), plasma neuropeptide Y for symptom severity (Fletcher et al., 2010a), natural killer cell function (Fletcher et al., 2010b), cytokines in women (Fletcher et al., 2009), metagenomic or metabolomics (Nagy-Szakal et al., 2018), and blood pressure/peripheral pulse characteristics (Allen et al., 2012; Frith et al., 2012), which are also changed with orthostatic disturbance and heart diseases. Overall the current evidence based on many ME/CFS studies measuring cytokines suggest that changes in circulating cytokines do not seem to explain the core characteristic of ME/CFS (Blundell et al., 2015). We discovered new pathways regulating actin cytoskeletal and focal adhesion, and detected several other pathways (PI3K-Akt signaling pathway, and EB virus infection) that have been reported in ME/CFS studies (Navaneetharaja et al., 2016). Actin network proteins, talin-1, actin, alpha-actinin-1, vinculin, gelsolin, and integrin are essential proteins for cytoskeletal connections and ion channels connections resulting in effect on intracellular signaling (Dalghi et al., 2018; Sasaki et al., 2014), indicating that this profile may be matched to a wide variety of ME/CFS symptoms, including muscle weakness/pain, endocrine disorder, and brain inflammation. Actin network proteins also play important roles in the skeletal muscle as follows: 1) talin 1 regulates the stability of myotendinous junctions through the vinculin-talin-integrin system in skeletal muscle (Conti et al., 2008); and 2) human serum gelsolin is mainly derived from skeletal muscle (Kwiatkowski et al., 1988). Our results suggest that skeletal muscle damage may be involved in ME/CFS pathology, since ME/CFS patients lose mobility with progression of the disease and graded exercise is one of the treatments proven to have an impact on these patients (Pietrangelo et al., 2018). Actin network proteins including talin, vinculin filamin, actin, and actinin, also have an important role in cardiac myocyte and heart function (Zemljic-Harpf et al., 2009), thus the results in this study may additionally reflect a potential abnormality of heart function in ME/CFS patients. Furthermore, we also detected 14–3-3 family proteins that are associated with virus infections including EB virus. Interestingly, EB virus components were detected in a large cohort of ME/CFS patients (Pedersen et al., 2019). EB virus components trigger systemic autoimmune diseases (Draborg et al., 2016) and deficient EBV-specific B- and T-cell response were observed in ME/CFS patients (Loebel et al., 2014). This evidence led us to hypothesize that EVs may be involved in ME/CFS progression or etiology through EB virus infection and subsequent triggering of autoimmune disease in skeletal muscles, although future studies will need to investigate this hypothesis further.
CONCLUSION
In conclusion, we revealed that circulating EV levels are significantly increased in ME/CFS patients. These EVs contain a specific protein cargo, particularly actin network proteins and 14–3-3 family proteins, which represent novel-specific ME/CFS biomarkers and can distinguish this condition from ICF and clinical depression, which are two highly challenging differential diagnoses in the clinical arena. Future studies including larger cohorts that would allow for matching the various conditions by key variables such as age and gender as well as external validation studies are warranted.
The novel findings of this study may open new windows to reveal ME/CFS pathogenic mechanisms and may aid in the development of better ME/CFS biomarkers and effective therapies.
Supplementary Material
Supplementary figure 1. Validation of increased circulating EVs in ME/CFS patients. A) Flow cytometry analysis of dot plot in circulating calcein positive EVs from ME/CFS patient. B) Quantification of calcein-positive circulating EVs in additionally recruited ME/CFS and HC with patient cohort. HC: healthy controls. Values represent mean ± SEM. ***P < 0.001. C) Dot plot of circulating EVs and CRP level (range, 0–0.10 mg/dL).
Supplementary table 1. Cohort characterization.
Supplementary table 2. Diagnosis of ME/CFS 2 group using cutoff values from the ME/CFS 1 group.
Supplementary table 3. Cohort characterization for proteomic analysis between healthy controls and ME/CFS.
Supplementary table 4. Proteomic analysis of total proteins in circulating EVs from healthy control and ME/CFS.
Supplementary table 5. Protein list for heat map analysis in circulating EVs from healthy control and ME/CFS.
Supplementary table 6. KEGG pathway in circulating EVs from ME/CFS compared to healthy control.
Supplementary table 7. Cohort characterization for proteomic analysis in ICF, depression, and ME/CFS.
Supplementary table 8. Proteomic analysis of total proteins in circulating EVs from ICF, depression, and ME/CFS.
Supplementary table 9. Protein list for heat map analysis in circulating EVs from ICF, depression, and ME/CFS.
Supplementary table 10. KEGG pathway in circulating EVs from ME/CFS compared to ICF and depression.
Supplementary Table 11. Common proteins in circulating EVs from ME/CFS.
Supplementary table 12. Common KEGG pathway in circulating EVs from ME/CFS.
Circulating EV number was increased in ME/CFS patients correlating to CRP and BAP
AUROC for circulating EVs was 0.802 allowing correct diagnosis in 90–94% of ME/CFS
Proteins in actin skeletal regulation and EB virus infection were identified in ME/CFS patients
Talin-1, filamin-A and 14–3-3 proteins were the most abundant proteins representing highly specific ME/CFS
Acknowledgements
The authors would like to thank Dr. Marilyn Farquhar for the use of the UCSD/CMM electron microscopy facility, and Timo Meerloo and Ying Jones for electron microscopy sample preparation. UCSD/CMM electron microscopy facility was received the JEOL through NIH equipment grant, 1S10OD023527-01. The authors also thank Dr. Gerald Pao to assist heat map analysis and kunihiko Tanaka, Hitomi Hisano, and Tomoko Hakariya for technical support.
Funding: The work was partly supported by Japanese KAKEN 17K01831 to AE and SF; Japanese AMED grant Gapfree2 16822729 to AE, SF, HK, JN, YN and YW, NIH grants U01 AA022489 and DK082451 to AEF.
Abbreviations
- ME/CFS
myalgic encephalomyelitis/chronic fatigue syndrome
- EVs
extracellular vesicles
- CRP
C-reactive protein
- BAP
biological antioxidant potential
- d-Roms
diacron-reactive oxygen metabolites
- AUC
area under the receiver operating characteristic curve
- ICF
idiopathic chronic fatigue
Footnotes
Financial disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 2015. Committee on the Diagnostic Criteria for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome, Board on the Health of Select Populations, Institute of Medicine: Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Redefining an illness. The National Academies Press, Washington D.C. [PubMed] [Google Scholar]
- Allen J, Murray A, Di Maria C, Newton JL, 2012. Chronic fatigue syndrome and impaired peripheral pulse characteristics on orthostasis--a new potential diagnostic biomarker. Physiological measurement 33, 231–241. [DOI] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS, 2016. Heatmapper: web-enabled heat mapping for all. Nucleic acids research 44, W147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell S, Ray KK, Buckland M, White PD, 2015. Chronic fatigue syndrome and circulating cytokines: A systematic review. Brain, behavior, and immunity. [DOI] [PubMed] [Google Scholar]
- Brenu EW, Hardcastle SL, Atkinson GM, van Driel ML, Kreijkamp-Kaspers S, Ashton KJ, Staines DR, Marshall-Gradisnik SM, 2013. Natural killer cells in patients with severe chronic fatigue syndrome. Auto Immun Highlights 4, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenu EW, Huth TK, Hardcastle SL, Fuller K, Kaur M, Johnston S, Ramos SB, Staines DR, Marshall-Gradisnik SM, 2014. Role of adaptive and innate immune cells in chronic fatigue syndrome/myalgic encephalomyelitis. International immunology 26, 233–242. [DOI] [PubMed] [Google Scholar]
- Brenu EW, van Driel ML, Staines DR, Ashton KJ, Ramos SB, Keane J, Klimas NG, Marshall-Gradisnik SM, 2011. Immunological abnormalities as potential biomarkers in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Journal of translational medicine 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick G, Fuite J, Kreitz A, Vernon SD, Klimas N, Fletcher MA, 2010. A formal analysis of cytokine networks in chronic fatigue syndrome. Brain, behavior, and immunity 24, 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanas H, Muraki K, Balinas C, Eaton-Fitch N, Staines D, Marshall-Gradisnik S, 2019. Validation of impaired Transient Receptor Potential Melastatin 3 ion channel activity in natural killer cells from Chronic Fatigue Syndrome/ Myalgic Encephalomyelitis patients. Molecular medicine 25, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers BM, van de Sande MI, De Meirleir KL, Klimas NG, Broderick G, Mitchell T, Staines D, Powles AC, Speight N, Vallings R, Bateman L, Baumgarten-Austrheim B, Bell DS, Carlo-Stella N, Chia J, Darragh A, Jo D, Lewis D, Light AR, Marshall-Gradisnik S, Mena I, Mikovits JA, Miwa K, Murovska M, Pall ML, Stevens S, 2011. Myalgic encephalomyelitis: International Consensus Criteria. Journal of internal medicine 270, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Marrero J, Cordero M, Saez-Francas N, Jimenez-Gutierrez C, Aguliar-Montilla F, Aliste L, Alegre-Martin J, 2013. Could mitochondrial dysfunction be a differentiating marker between chronic fatigue syndrome and fibromyalgia? Antioxidants & redox signaling 19, 1855–1860. [DOI] [PubMed] [Google Scholar]
- Castro-Marrero J, Serrano-Pertierra E, Oliveira-Rodriguez M, Zaragoza MC, Martinez-Martinez A, Blanco-Lopez MDC, Alegre J, 2018. Circulating extracellular vesicles as potential biomarkers in chronic fatigue syndrome/myalgic encephalomyelitis: an exploratory pilot study. Journal of extracellular vesicles 7, 1453730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleare AJ, Reid S, Chalder T, Hotopf M, Wessely S, 2015. Chronic fatigue syndrome. BMJ clinical evidence 2015. [PMC free article] [PubMed] [Google Scholar]
- Conti FJ, Felder A, Monkley S, Schwander M, Wood MR, Lieber R, Critchley D, Muller U, 2008. Progressive myopathy and defects in the maintenance of myotendinous junctions in mice that lack talin 1 in skeletal muscle. Development 135, 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalghi MG, Ferreira-Gomes M, Rossi JP, 2018. Regulation of the Plasma Membrane Calcium ATPases by the actin cytoskeleton. Biochemical and biophysical research communications 506, 347–354. [DOI] [PubMed] [Google Scholar]
- Draborg A, Izarzugaza JM, Houen G, 2016. How compelling are the data for Epstein-Barr virus being a trigger for systemic lupus and other autoimmune diseases? Current opinion in rheumatology 28, 398–404. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Feldstein AE, 2018. Extracellular vesicles in non-alcoholic and alcoholic fatty liver diseases. Liver research 2, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A, Kostallari E, Feldstein AE, Shah VH, 2019. Extracellular vesicles, the liquid biopsy of the future. Journal of hepatology 70, 1292–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher MA, Rosenthal M, Antoni M, Ironson G, Zeng XR, Barnes Z, Harvey JM, Hurwitz B, Levis S, Broderick G, Klimas NG, 2010a. Plasma neuropeptide Y: a biomarker for symptom severity in chronic fatigue syndrome. Behavioral and brain functions : BBF 6, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher MA, Zeng XR, Barnes Z, Levis S, Klimas NG, 2009. Plasma cytokines in women with chronic fatigue syndrome. Journal of translational medicine 7, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher MA, Zeng XR, Maher K, Levis S, Hurwitz B, Antoni M, Broderick G, Klimas NG, 2010b. Biomarkers in chronic fatigue syndrome: evaluation of natural killer cell function and dipeptidyl peptidase IV/CD26. PloS one 5, e10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D, Kerr J, Harrison TJ, Kellam P, 2011. Assessment of a 44 gene classifier for the evaluation of chronic fatigue syndrome from peripheral blood mononuclear cell gene expression. PloS one 6, e16872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith J, Zalewski P, Klawe JJ, Pairman J, Bitner A, Tafil-Klawe M, Newton JL, 2012. Impaired blood pressure variability in chronic fatigue syndrome--a potential biomarker. QJM : monthly journal of the Association of Physicians 105, 831–838. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A, 1994. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Annals of internal medicine 121, 953–959. [DOI] [PubMed] [Google Scholar]
- Fukuda S, Nojima J, Motoki Y, Yamaguti K, Nakatomi Y, Okawa N, Fujiwara K, Watanabe Y, Kuratsune H, 2016. A potential biomarker for fatigue: Oxidative stress and anti-oxidative activity. Biological psychology 118, 88–93. [DOI] [PubMed] [Google Scholar]
- G, M., Maes M, 2014. Oxidative and Nitrosative Stress and Immune-Inflammatory Pathways in Patients with Myalgic Encephalomyelitis (ME)/Chronic Fatigue Syndrome (CFS). Curr Neuropharmacol. 12, 168–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher AM, Thomas JM, Hamilton WT, White PD, 2004. Incidence of fatigue symptoms and diagnoses presenting in UK primary care from 1990 to 2001. Journal of the Royal Society of Medicine 97, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LA, Porter N, Herrington J, Sorenson M, Kubow S, 2009. Kindling and Oxidative Stress as Contributors to Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. J Behav Neurosci Res 7, 1–17. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Eguchi A, Tempaku M, Honda T, Togashi K, Iwasa M, Hasegawa H, Takei Y, Sumida Y, Taguchi O, 2018. Circulating extracellular vesicles are associated with lipid and insulin metabolism. American journal of physiology. Endocrinology and metabolism. [DOI] [PubMed] [Google Scholar]
- Komaroff AL, Takahashi R, Yamamura T, Sawamura M, 2018. [Neurologic Abnormalities in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: A Review]. Brain and nerve = Shinkei kenkyu no shinpo 70, 41–54. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Mehl R, Izumo S, Nadal-Ginard B, Yin HL, 1988. Muscle is the major source of plasma gelsolin. The Journal of biological chemistry 263, 8239–8243. [PubMed] [Google Scholar]
- Lloyd AR, Meer JW, 2015. The long wait for a breakthrough in chronic fatigue syndrome. Bmj 350, h2087. [DOI] [PubMed] [Google Scholar]
- Loebel M, Eckey M, Sotzny F, Hahn E, Bauer S, Grabowski P, Zerweck J, Holenya P, Hanitsch LG, Wittke K, Borchmann P, Ruffer JU, Hiepe F, Ruprecht K, Behrends U, Meindl C, Volk HD, Reimer U, Scheibenbogen C, 2017. Serological profiling of the EBV immune response in Chronic Fatigue Syndrome using a peptide microarray. PloS one 12, e0179124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel M, Strohschein K, Giannini C, Koelsch U, Bauer S, Doebis C, Thomas S, Unterwalder N, von Baehr V, Reinke P, Knops M, Hanitsch LG, Meisel C, Volk HD, Scheibenbogen C, 2014. Deficient EBV-specific B- and T-cell response in patients with chronic fatigue syndrome. PloS one 9, e85387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Kubera M, Uytterhoeven M, Vrydags N, Bosmans E, 2011. Increased plasma peroxides as a marker of oxidative stress in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Med Sci Monit 17, SC11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikirova N, Casciari J, Hunninghake R, 2012. The assessment of the energy metabolism in patients with chronic fatigue syndrome by serum fluorescence emission. Alternative therapies in health and medicine 18, 36–40. [PubMed] [Google Scholar]
- Moneghetti KJ, Skhiri M, Contrepois K, Kobayashi Y, Maecker H, Davis M, Snyder M, Haddad F, Montoya JG, 2018. Value of Circulating Cytokine Profiling During Submaximal Exercise Testing in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Scientific reports 8, 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JG, Holmes TH, Anderson JN, Maecker HT, Rosenberg-Hasson Y, Valencia IJ, Chu L, Younger JW, Tato CM, Davis MM, 2017. Cytokine signature associated with disease severity in chronic fatigue syndrome patients. Proceedings of the National Academy of Sciences of the United States of America 114, E7150–E7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy-Szakal D, Barupal DK, Lee B, Che X, Williams BL, Kahn EJR, Ukaigwe JE, Bateman L, Klimas NG, Komaroff AL, Levine S, Montoya JG, Peterson DL, Levin B, Hornig M, Fiehn O, Lipkin WI, 2018. Insights into myalgic encephalomyelitis/chronic fatigue syndrome phenotypes through comprehensive metabolomics. Scientific reports 8, 10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaneetharaja N, Griffiths V, Wileman T, Carding SR, 2016. A Role for the Intestinal Microbiota and Virome in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)? Journal of clinical medicine 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviaux RK, Naviaux JC, Li K, Bright AT, Alaynick WA, Wang L, Baxter A, Nathan N, Anderson W, Gordon E, 2016. Metabolic features of chronic fatigue syndrome. Proceedings of the National Academy of Sciences of the United States of America 113, E5472–5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M, Asprusten TT, Godang K, Leegaard TM, Osnes LT, Skovlund E, Tjade T, Oie MG, Wyller VBB, 2019. Predictors of chronic fatigue in adolescents six months after acute Epstein-Barr virus infection: A prospective cohort study. Brain, behavior, and immunity 75, 94–100. [DOI] [PubMed] [Google Scholar]
- Pietrangelo T, Fulle S, Coscia F, Gigliotti PV, Fano-Illic G, 2018. Old muscle in young body: an aphorism describing the Chronic Fatigue Syndrome. European journal of translational myology 28, 7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Yui N, Noda Y, 2014. Actin directly interacts with different membrane channel proteins and influences channel activities: AQP2 as a model. Biochimica et biophysica acta 1838, 514–520. [DOI] [PubMed] [Google Scholar]
- Shah R, Patel T, Freedman JE, 2018. Circulating Extracellular Vesicles in Human Disease. The New England journal of medicine 379, 958–966. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C, 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo F, Natelson BH, 2013. Heart rate variability during sleep and subsequent sleepiness in patients with chronic fatigue syndrome. Autonomic neuroscience : basic & clinical 176, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twisk FN, 2014. The status of and future research into Myalgic Encephalomyelitis and Chronic Fatigue Syndrome: the need of accurate diagnosis, objective assessment, and acknowledging biological and clinical subgroups. Front Physiol. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twisk FN, 2018. Myalgic Encephalomyelitis (ME) or What? An Operational Definition. Diagnostics 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenbergh D, Nijs J, Kos D, Van Weijnen L, Struyf F, Meeus M, 2014. Malfunctioning of the autonomic nervous system in patients with chronic fatigue syndrome: a systematic literature review. European journal of clinical investigation 44, 516–526. [DOI] [PubMed] [Google Scholar]
- Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O, 2015. Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Yang Y, Wang D, Li C, Qu Y, Guo J, Shi T, Bo W, Sun Z, Asakawa T, 2019. The clinical value of cytokines in chronic fatigue syndrome. Journal of translational medicine 17, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf A, Manso AM, Ross RS, 2009. Vinculin and talin: focus on the myocardium. J Investig Med 57, 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Validation of increased circulating EVs in ME/CFS patients. A) Flow cytometry analysis of dot plot in circulating calcein positive EVs from ME/CFS patient. B) Quantification of calcein-positive circulating EVs in additionally recruited ME/CFS and HC with patient cohort. HC: healthy controls. Values represent mean ± SEM. ***P < 0.001. C) Dot plot of circulating EVs and CRP level (range, 0–0.10 mg/dL).
Supplementary table 1. Cohort characterization.
Supplementary table 2. Diagnosis of ME/CFS 2 group using cutoff values from the ME/CFS 1 group.
Supplementary table 3. Cohort characterization for proteomic analysis between healthy controls and ME/CFS.
Supplementary table 4. Proteomic analysis of total proteins in circulating EVs from healthy control and ME/CFS.
Supplementary table 5. Protein list for heat map analysis in circulating EVs from healthy control and ME/CFS.
Supplementary table 6. KEGG pathway in circulating EVs from ME/CFS compared to healthy control.
Supplementary table 7. Cohort characterization for proteomic analysis in ICF, depression, and ME/CFS.
Supplementary table 8. Proteomic analysis of total proteins in circulating EVs from ICF, depression, and ME/CFS.
Supplementary table 9. Protein list for heat map analysis in circulating EVs from ICF, depression, and ME/CFS.
Supplementary table 10. KEGG pathway in circulating EVs from ME/CFS compared to ICF and depression.
Supplementary Table 11. Common proteins in circulating EVs from ME/CFS.
Supplementary table 12. Common KEGG pathway in circulating EVs from ME/CFS.