Abstract
Background & Aims
Biomarkers are needed to identify patients at risk of tumor progression following chemoradiotherapy for localized esophageal cancer. These could improve identification of patients at risk for cancer progression and selection of therapy.
Methods
We performed deep sequencing (CAPP-Seq) analyses of plasma cell-free DNA collected from 45 patients before and after chemoradiotherapy for esophageal cancer, as well as DNA from leukocytes, and fixed esophageal tumor biopsies collected during esophagogastroduodenoscopy. Patients were treated from May 2010 through October 2015; 23 patients subsequently underwent esophagectomy and 22 did not undergo surgery. We also sequenced DNA from blood samples from 40 healthy individuals (controls). We analyzed 802 regions of 607 genes for single-nucleotide variants previously associated with esophageal adenocarcinoma or squamous cell carcinoma. Patients underwent imaging analyses 6–8 weeks after chemoradiotherapy and were followed for 5 years. Our primary aim was to determine whether detection of circulating tumor DNA (ctDNA) following chemoradiotherapy is associated with risk of tumor progression (growth of local, regional, or distant tumors, detected by imaging or biopsy).
Results
The median proportion of tumor-derived DNA in total cell-free DNA before treatment was 0.07%, indicating that ultrasensitive assays are needed for quantification and analysis of ctDNA from localized esophageal tumors. Detection of ctDNA following chemoradiotherapy was associated with tumor progression (hazard ratio, 18.7; P<.0001), formation of distant metastases (hazard ratio, 32.1; P<.0001), and shorter disease-specific survival times (hazard ratio, 23.1; P<.0001). A higher proportion of patients with tumor progression had new mutations detected in plasma samples collected after chemoradiotherapy than patients without progression (P=.03). Detection of ctDNA after chemoradiotherapy preceded radiographic evidence of tumor progression by an average of 2.8 months. Among patients who received chemoradiotherapy without surgery, combined ctDNA and metabolic imaging analysis predicted progression in 100% of patients with tumor progression, compared with 71% for only ctDNA detection and 57% for only metabolic imaging analysis (P<.001 for comparison of either technique to combined analysis).
Conclusions
In an analysis of cell-free DNA in blood samples from patients who underwent chemoradiotherapy for esophageal cancer, detection of ctDNA was associated with tumor progression, metastasis, and disease-specific survival. Analysis of ctDNA might be used to identify patients at highest risk for tumor progression.
Keywords: genetics, polymorphism, chemoradiotherapy, SNP
Lay Summary
Azad et al. find that detection of tumor DNA in the blood of esophageal cancer patients is associated with survival and may enable personalized treatment decisions.
Introduction
Neoadjuvant chemoradiotherapy (CRT) followed by surgery is a standard approach for treatment of localized esophageal cancers (ESCA) and has been shown to improve overall survival compared to esophagectomy alone 1. Unfortunately however, a significant fraction of patients treated with CRT and surgery will ultimately progress, with overall, distant, and locoregional progression rates of 49%, 39%, and 22%, respectively 2. Additionally, in ESCA patients who have an excellent upfront response to CRT, particularly those achieving complete responses prior to resection, surgery may not offer an additional survival benefit, potentially warranting an active surveillance approach 3–7.
Such personalization of treatment would be facilitated by the identification of a biomarker that can sensitively and noninvasively detect residual disease. Unfortunately, conventional clinical assessment and imaging, such as endoscopy, endoscopic ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) are currently not sufficiently robust methods of gauging treatment response to CRT independently 8, 9. Accordingly, there is an unmet clinical need to identify biomarkers that could sensitively detect residual disease and/or early progression in patients with ESCA.
Analysis of circulating tumor DNA (ctDNA) shortly after completion of curative intent treatment is able to detect minimal residual disease (MRD) in several carcinomas, including breast 10, colorectal 11, and lung cancers 12, 13. Preliminary studies in patients with localized squamous ESCA have suggested feasibility of ctDNA 14 or total cell-free DNA analysis 15. However, no study to date has applied ultrasensitive next generation sequencing methods to detect ctDNA MRD in ESCA patients receiving CRT and surgery. In this study we set out to test if ctDNA analysis can predict recurrence in patients with localized ESCA earlier than standard-of-care imaging. Additionally, we sought to explore whether integration of ctDNA and metabolic imaging could further improve risk stratification.
Materials and Methods
Additional methodologic details can be found in the Supplemental Methods.
Study design and patient cohort
We prospectively enrolled patients with localized ESCA at the MD Anderson Cancer Center from 5/2010 – 10/2015 with the objective of assessing the association between post-CRT ctDNA detection and freedom from progression (FFP). Inclusion criteria were: age > 18 years with previously untreated stage IA to stage IIIB EAC or ESCC being treated with definitive intent, with CRT and surgery or CRT-alone. The follow up schedule included an initial follow-up visit at 6–8 weeks from end of CRT with concurrent PET-CT or CT imaging. Patients were then monitored every three months with CT or PET-CT for the first year, every four months during the second year, and every six months in years three to five. Patient demographics and clinical characteristics are summarized in Table S1.
The study statistical plan used the following assumptions: 1) 50% of patients will develop recurrence (based on 2-year progression-free survival data from the CROSS trial neoadjuvant chemoradiotherapy arm1); 2) Sensitivity of ctDNA MRD detection will be 75% (conservative estimate based on 94% sensitivity observed in our recent study of localized lung cancer); 3) Specificity of ctDNA MRD detection will be 90% (conservative estimate based on 100% specificity observed in our recent study of localized lung cancer). Using these assumptions, the risk of progression will be 88% versus 22% for patients with positive versus negative ctDNA MRD. A total sample size of 26 patients will yield 95% power to detect this difference with a one-sided alpha of 0.05.
Eligible patients underwent a pre-treatment blood draw and genotyping with CAPP-Seq applied to micro-dissected formalin-fixed, paraffin embedded (FFPE) tumor tissue from pretreatment esophagogastroduodenoscopy (EGD) using matched germline DNA. Germline DNA was extracted from the pre-treatment blood draw. Patients then received CRT, after which a second blood draw was collected for the majority of patients (n = 32). The median time between the pre- and post-CRT draws was 86 days (IQR, 76–92). Healthy adult blood donors (n = 40) were recruited through the Stanford Blood Center (Table S5). All samples were collected with informed consent and institutional review board approval in accordance with the Declaration of Helsinki. All plasma samples were analyzed by CAPP-Seq as previously described 16, 17. A total of 218 specimens were profiled, including plasma, tumor, germline, and healthy control specimens. Healthy controls were only used to tune the detection cut-off for the ctDNA biomarker, as detailed below and were not included in survival analyses.
Criteria for tumor-informed ctDNA detection and post-CRT monitoring
Cell-free DNA from blood samples was analyzed for presence of mutations identified by tumor genotyping. The set of mutations identified from tumor genotyping was assessed as a group in blood samples according to a previously described Monte Carlo framework 17. Detection was tuned for a specificity of 95% in a cohort of cell-free DNA from 20 independent healthy subjects using a receiver-operator curve, resulting in a ctDNA detection index < 0.06 for detection. If ctDNA detection index was ≥ 0.06, ctDNA was classified as not detected at that time point. We report absolute ctDNA levels as haploid genome equivalents per mL of plasma, the product of total plasma cell-free DNA concentration (fluorometry by Qubit, ThermoFisher Scientific, Waltham, MA) and the relative ctDNA concentration (mean allele fraction of somatic alterations). The ctDNA mutant allele fraction was calculated by averaging the mutant allele fractions for all mutations derived from tumor genotyping for that patient. All variant calls are listed in Table S9.
Somatic copy number alteration detection
Somatic copy number alterations (SCNAs) were called using method we previously described 18. In brief, SCNAs were detected using a z-score based method which involves a set of background samples to capture the region-specific variabilities across the targeted regions. For each gene, we called focal amplifications and deletions using the targeted regions as determined by the ESCA CAPP-Seq panel.
Criteria for putative emergent mutation detection
iDES-enhanced CAPP-Seq variant-calling was performed at the first post-CRT time point with matched leukocyte DNA as previously described 16. Mutations were defined as putative emergent mutations if they met the following criteria:
Non-synonymous
No variant reads in matched germline, pre-CRT plasma, or tumor and maximum of 1 variant read in the plasma of 20 healthy controls
≥1500X deduped depth in pre- and post-CRT plasma, ≥ 500X depth in matched germline, ≥ 100X depth in tumor
Recurrently mutated ESCA gene (> 5% of ESCA cBioportal samples) or ≥ 2 duplex reads
Not in TP53 (known clonal hematopoiesis-associated gene)
Statistics and analytic plan
Our primary aim was to test the hypothesis that detection of ctDNA post-CRT is associated with high risk of recurrence. The primary outcome was freedom from progression (FFP; event defined as radiographic or clinical progression). To test the main hypothesis, we employed the Kaplan–Meier method with log-rank test to estimate P values and the Cox exp(beta) method to estimate hazard ratios. The relationship of ctDNA concentration as a continuous variable with outcome was also assessed using Cox proportional hazards regression.
In exploratory analyses we considered three additional survival endpoints, distant metastasis-free survival (DMFS; event defined as distant recurrence or death), local progression-free survival (LPFS; event defined as local progression or death), and disease-specific survival (DSS; event defined as death from ESCA). For these analyses, we performed Kaplan-Meier and Cox proportional hazards analyses as above using ctDNA as a binary endpoint. To guard against guarantee-time bias,19 we used landmark analysis with the time of the post-CRT blood draw being considered the landmark.
We further constructed univariate Cox proportional hazards regressions models for baseline patient and tumor features to test if these variables were associated with the primary outcome (FFP). The Wald test was used to assess the significance of covariates, and hazard ratios were calculated by exponentiation of the model coefficients. Multiple testing adjustment was not performed for secondary analyses since they were exploratory.
Results
Profiling of ctDNA in localized ESCA before chemoradiotherapy
We retrospectively analyzed plasma samples from patients with localized esophageal adenocarcinoma (EAC, n = 35) and squamous cell carcinoma (ESCC, n = 10) who received either CRT followed by esophagectomy (n = 23) or definitive CRT (n = 22). In total, we profiled 213 blood and tissue samples from these patients and a control cohort of 40 healthy adults. Clinical characteristics of the study cohort are depicted in Figure 1B and further detailed in Table S1. Blood draws were performed before and after CRT, near the time of computed tomography (CT) and positron emission tomography–CT (PET-CT) imaging (Fig. 1A). The median time between the pre- and post-CRT draws was 86 days (IQR, 76–92).
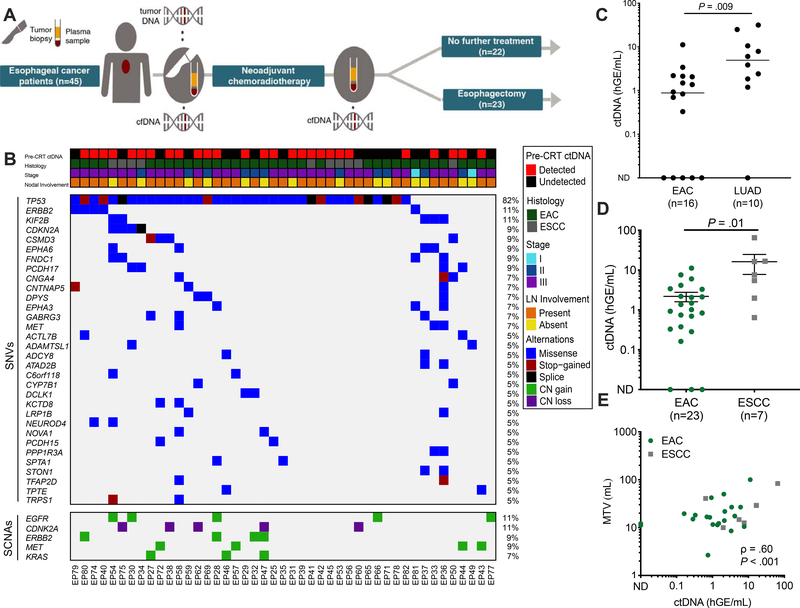
Figure 1. Pre-CRT assessment of ctDNA in patients with locally advanced ESCA.
(A) Study overview of forty-five patients with EGD-proven locally advanced ESCA enrolled in the study. Plasma samples were collected before and after chemoradiotherapy (CRT). Following CRT, patients either underwent esophagectomy (n=23) or no further treatment (n=22). (B) Clinical characteristics and tumor genotyping results. Pre-CRT ctDNA detection status and clinical characteristics, where each column is a patient and each row is a parameter (e.g. histology). Similarly, the associated co-mutation plot depicts patient-level mutational profiles of 45 tumors from patients with esophageal cancer genotyped by our ESCA-specific NGS panel. Genes mutated in at least 5% of the patients in our cohort are depicted. The fraction of tumors with mutations in each gene is denoted on the left. (C) Comparison of absolute ctDNA concentration between Stage III esophageal adenocarcinoma (EAC, n=16) and Stage III lung adenocarcinoma (LUAD, n=10). P value calculated by the Mann-Whitney test. (D) Pre-treatment ctDNA concentration in esophageal adenocarcinoma (EAC, n= 23) and esophageal squamous cell carcinoma (ESCC, n=7) patients. Patients were included if tumor-informed mutations were detected either pre- or post-CRT. Data represent mean + SEM. P value was calculated by the Mann-Whitney test. (E) Correlation of ctDNA concentration (haploid genome equivalents per mL, hGE/mL) with pre-CRT metabolic tumor volume (MTV) measured by PET-CT, stratified by histology. P value and ρ were calculated by Pearson correlation.
To interrogate the genetic alterations that are commonly found in ESCA, we developed a 185-kb cancer personalized profiling by deep sequencing (CAPP-Seq) panel targeting 802 regions from 607 genes harboring recurrent single-nucleotide variants (SNVs) in both EAC and ESCC (Table S2) by applying our previously described panel design strategy 16, 17 to two published ESCA whole-exome datasets 20, 21. We then used this panel to sequence tumor DNA from micro-dissected pre-treatment esophagogastroduodenoscopy (EGD) biopsy samples and matched leukocyte DNA (Table S3). In silico validation of the esophageal CAPP-Seq panel in independent ESCA whole exome sequencing data sets 22, 23 predicted that ≥ 1 mutation would be identified in 97.8% of EAC patients and 87.5% of ESCC patients (Fig. S1). Corresponding with these expectations, we identified ≥ 1 mutation in 44 of 45 (97.8%) tumors from our cohort, with a median of four SNVs (range, 0–23) and median of 0 copy number alterations (range, 0–3) per case (Fig. 1B, Fig. S1).Consistent with previous studies of EAC and ESCC mutation profiles 20–23, commonly mutated genes in our cohort included TP53, ERBB2, and CDKN2A.
We next applied our CAPP-Seq ESCA panel to cell-free DNA (cfDNA) extracted from pre- and post-CRT plasma samples (Table S4). Here, we used two strategies, including one informed by tumor biopsies, and a second approach naïve to these variants and directly identifying ctDNA in the blood. For the tumor-informed strategy, which is similar to that used in prior ctDNA MRD studies in other tumor types10–13, 24, 25, we identified somatic variants in tumor biopsies after consideration of germline alleles within matched leukocytes. We then applied a tumor-informed ctDNA detection approach that employs a previously described Monte Carlo-based framework to determine if a blood sample contains ctDNA 17. Using this approach, we detected ctDNA in 27 patients before CRT (Fig. 1B) for a pre-treatment sensitivity of 60%. Specificity was 95% based on analysis of cell-free DNA from 20 independent healthy subjects profiled with the same esophageal CAPP-Seq assay (Table S5). In patients with detectable ctDNA, a median of two mutations were detected at a median relative ctDNA concentration of 0.065% (range, 0–0.91%) and median absolute ctDNA concentration was 1.7 haploid genome equivalents per milliliter (hGE/mL) (range, 0–65.2). ctDNA concentrations in stage III EACs were significantly lower than those we observed in stage III lung adenocarcinomas (P = .009, Fig. 1C), even when considering only driver mutations26 (P = .004; Fig S2A), or normalizing by PET-CT-based metabolic tumor volume (P = .04; Fig. S2E). Notably, we found that patients with ESCC had on average 7-fold higher ctDNA levels than patients with EAC (P = .01; Fig. 1D and Fig. S3), consistent with observations of higher ctDNA shedding in lung squamous cell carcinomas compared to lung adenocarcinomas 13. In a multivariable regression model accounting for histology, pre-CRT metabolic tumor volume (MTV), age, sex, and nodal involvement, histology (β = 11.0, P = .03) and pre-CRT MTV (β = 0.3, P = .002) were independently associated with absolute ctDNA concentration. Absolute pre-CRT ctDNA concentration (in hGE/mL) was strongly correlated with MTV at diagnosis (ρ = 0.60, P < .001; Fig. 1E), while relative ctDNA concentration (variant allele fraction, VAF) did not (ρ = 0.29, P = .13; Fig. S4). This observation likely reflects the fact that relative ctDNA concentration can be affected by changes in the amount of cfDNA contributed by non-malignant cells. Pre-CRT MTV (OR = 1.16, P = .02) and age (OR = 0.91, P = .048) were associated with detection of ctDNA pre-CRT (Table S6).
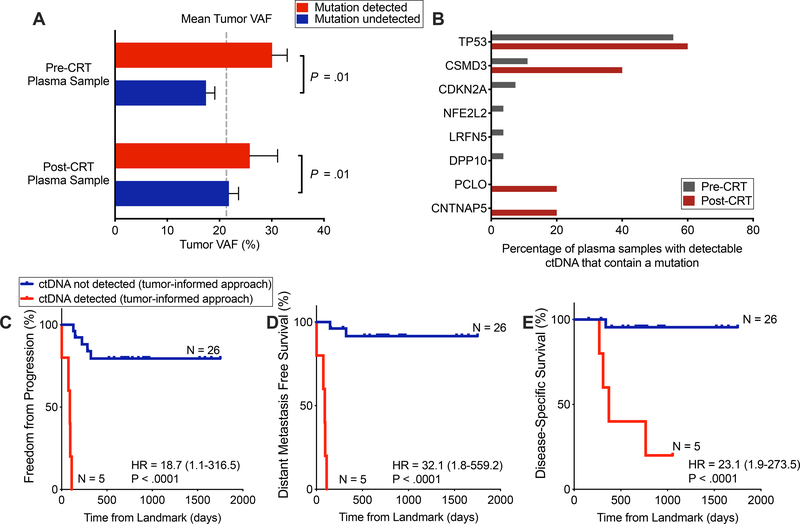
To further explore factors affecting detectability of mutations in plasma, we next asked whether lower VAF tumor mutations were less likely to be detected in plasma than higher VAF tumor mutations. Among patients with detectable ctDNA, we observed that tumor VAF was significantly higher for mutations present in cfDNA compared to mutations that were not. This was true in both pre- or post-CRT plasma (P = .01; 2-way ANOVA; Fig. 2A.
Figure 2. Detection of tumor-informed mutations following chemoradiotherapy is strongly prognostic.
(A) Tumor variant allele fraction (VAF), stratified by whether and at which time point a patient had detectable ctDNA (x-axis categories) and further stratified by whether a given mutation was detectable in plasma (column colors). P value calculated by a 2-way ANOVA and was significant for difference based on if a mutation was detected in plasma (P = .01), but not for if a patient was detected pre- or post-CRT. (B) Tumor mutations detected in plasma pre- and post-CRT. Denominator is the total number of patients with detectable ctDNA, pre- and post-CRT (n=27 and n=5, respectively). Genes depicted were mutated in more than 5% of tumors in cBioportal ESCA datasets. (C-E) Kaplan–Meier analyses comparing patients with detectable and undetectable ctDNA in the post-CRT sample using tumor-informed ctDNA detection for (C) freedom from progression (P<.0001, HR = 18.7 (95%CI, 1.1–316.5)), (D) distant metastasis-free survival (P<.0001, HR = 32.1 (95%CI, 1.8–559.2)) and (E) disease-specific survival (P < .0001, HR = 23.1 (95%CI, 2.0–273.5)). ctDNA negative (n =26) versus ctDNA positive (n = 5). Time (days) was measured from the landmark (post-CRT blood draw). One patient was excluded from this analysis due to an event that occurred prior to the post-CRT blood collection. P values and hazard ratios were calculated from the log-rank test.
Prognostic utility of tumor-informed post-CRT ctDNA detection in localized ESCA
Next, we evaluated the prognostic value of ctDNA detection following CRT in patients for whom a post-CRT blood draw was obtained (EAC, n = 23; ESCC, n = 8). We detected tumor-informed ctDNA at the post-CRT time point in five patients (16.1%). All five patients had EAC histology and were treated with definitive CRT alone. Consistent with pre-CRT ctDNA assessments, TP53 was the most common mutation detected in plasma (Fig. 2B). Median post-CRT relative ctDNA concentration was 0.02% (IQR, 0.008% - 0.03%) and absolute ctDNA concentration was 0.98 hGE/mL (IQR, 0.36 – 1.16). Patients with detectable ctDNA post-CRT also had significantly increased risk of disease progression (HR 18.7, P < .0001; Fig. 2C), distant metastasis (HR 32.1, P < .0001; Fig. 2D), and disease-specific death (HR 23.1, P < .0001; Fig. 2E), compared to patients with undetectable ctDNA after CRT. Detection of ctDNA post-CRT was not associated with increased risk of local progression (P = 0.17, Fig. S5), although this analysis may have been impacted by 55% of patients undergoing surgery after CRT. We further observed that consideration of ctDNA as a continuous variable (hGE/mL) was also associated with increased risk of disease progression (HR 1.62, P = .007).
Putative emergent mutations following CRT may be associated with disease progression
Among ESCA patients undergoing neoadjuvant therapy, recent evidence from a study comparing tissue samples before and after chemotherapy suggests that new mutations may emerge following treatment 27. We conducted an exploratory analysis to determine if we could observe a similar phenomenon using ctDNA. For this analysis, we performed tumor-naïve genotyping on post-CRT plasma to identify nonsynonymous mutations that were not observed in pre-CRT plasma and tumor. In order to minimize the likelihood of detecting mutations due to clonal hematopoiesis 28, 29, we excluded mutations in TP53, the only gene in our panel implicated as being recurrently involved in clonal hematopoiesis as well as any mutations that were also observed in the matched leukocyte sample (see Methods).
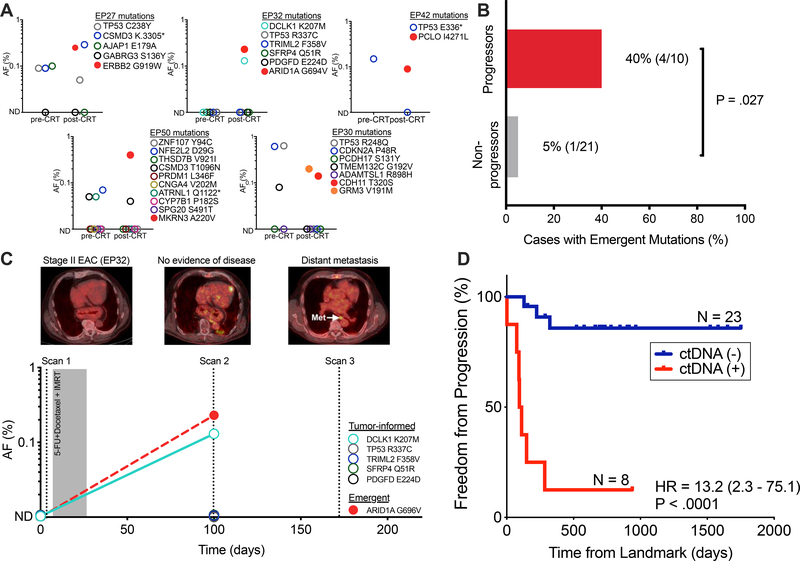
Using this approach, we observed new nonsynonymous mutations in the post-CRT plasma of 4 out of the 10 patients who progressed (Fig. 3A, B). Two new nonsynonymous mutations were detected in the post-CRT plasma of one patient (EP30) (Fig. 3A, B) who did not progress. Notably, unlike the other four patients with putative emergent mutations, EP30 underwent surgery 28 days following the post-CRT blood draw and was found to have residual tumor cells. A follow up blood draw from this patient taken after surgery did not contain these or any other new mutations. We therefore speculate that resection may have removed all residual tumor in this patient. Progressors were significantly more likely to have new mutations detected following CRT than non-progressors (P = .027, Fig. 3B). Figure 3C illustrates the clinical course of a patient with Stage II EAC who was treated with definitive CRT. Of five mutations found in the patient’s tumor, one (DCLK K207M) was detected following CRT. Emergent mutation analysis revealed a new ARID1A G696V mutation that was absent from the pre-CRT plasma, tumor, and matched leukocyte DNA. This patient developed distant metastasis 75 days following detection of this emergent variant.
Figure 3. Putative emergent mutations following CRT may be associated with disease progression.
(A) In five patients we detected six new nonsynonymous mutations in post-CRT plasma that were absent pre-CRT. Column dot plots indicates allele fractions of both tumor-informed (circles) and emergent (squares) mutations. These mutations were absent in pre-CRT plasma and matched germline, tumor, and in 20 healthy controls. (B) Presence of putative emergent mutations in patients who developed disease progression (progressors; n = 10) versus those who did not (non-progressors; n = 20). P value derived from Fisher’s exact test. (C) Patient (EP32) with non-FDG avid stage II EAC treated with CRT-alone who developed an emergent ARID1A G696V mutation following CRT. This patient went on to develop distant metastasis (para-aortic lymph node). (D) Kaplan–Meier analysis of freedom from progression (P < .0001, HR = 13.2 (95%CI, 2.3–75.1)) comparing patients with or without detectable ctDNA post-CRT (n=8 and 23, respectively). Detectable ctDNA is defined as detection of tumor-informed or putative emergent mutations following CRT. Time (days) was measured from the landmark (post-CRT blood draw). P values and hazard ratios (HR) were calculated from the log-rank test. AF, allele fraction; CRT, chemoradiotherapy; IMRT, intensity modulated radiotherapy; EAC, esophageal adenocarcinoma.
To further explore the potential utility of emergent mutation analysis we combined tumor-informed and tumor-naïve detection and considered plasma samples as positive if ctDNA was detected using either approach. As above, we found that ctDNA detection at the MRD timepoint was strongly prognostic for FFP (HR 13.2, P < .0001, Fig. 3D), DMFS (HR 28.0, P < .0001, Fig. S6A), and DSS (P < .0001, Fig. S6B). In univariable and multivariable Cox regressions, ctDNA detection at the MRD timepoint was independently associated with inferior FFP (HR 15.3, P = .0012; Table S7–8). Taken together, our findings suggest that residual ctDNA after CRT for ESCA is a strong negative prognostic marker and enables identification of patients at risk for developing recurrence.
Post-CRT ctDNA detection enables earlier identification of recurrence compared to PET-CT
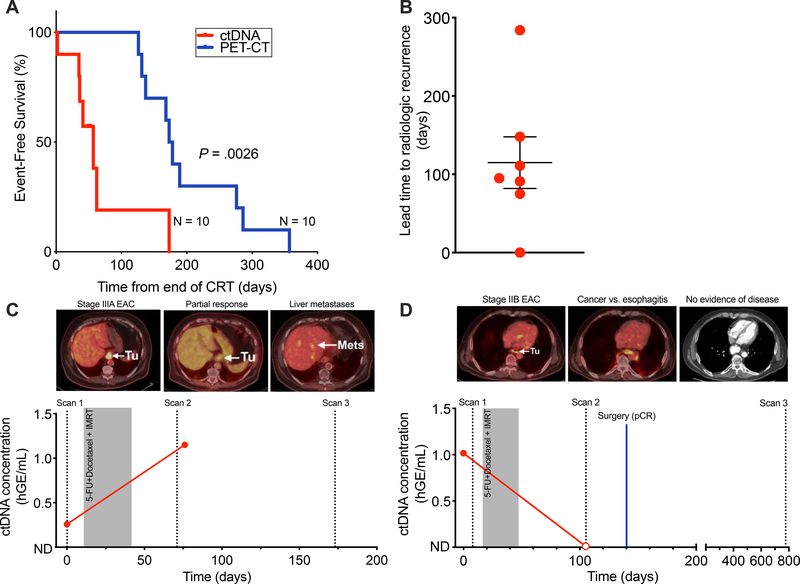
Recurrence was detected significantly earlier with ctDNA than standard-of-care PET-CT (P = .0026; Fig. 4A). Detection of ctDNA preceded recurrence detected by PET-CT in 70% of patients with an average lead time of 114.9 days (standard error of the mean, 32.9; Fig. 4B). For example, patient EP58 presented with stage IIIA EAC and had detectable pre-treatment ctDNA (Fig. 4C). The first post-CRT PET/CT at 36 days after completion of therapy revealed partial response to therapy and ctDNA was detectable in the blood draw at this time. At the subsequent follow up, 131 days after CRT, the patient was found to have distant metastasis to the liver. In contrast, patient EP29 presented with stage IIB EAC, was treated with neoadjuvant CRT, and noted to have an equivocal post-CRT PET/CT (65 days post-CRT). ctDNA from a blood draw at this same timepoint was undetectable. This patient went on to receive esophagectomy (81 days post-CRT) and histological analysis revealed a pathologic complete response (pCR) (Fig. 4D). The patient remains disease free two years post-CRT. ctDNA detection may enable earlier identification of distant metastasis compared to standard-of-care imaging.
Figure 4. ctDNA allows earlier and more robust detection of recurrence compared to PET-CT imaging.
(A) Kaplan–Meier analysis for event-free survival (P = .0026), comparing ctDNA detection at the post-CRT time point with standard-of-care PET-CT imaging (n=10). Time to event (days) was measured from end of CRT. P values and hazard ratios (HR) were calculated from the log-rank test. (B) Column dot plot of the lead time to radiographic recognition of recurrence for ctDNA detection (n=7), with the bars representing mean (114.9) and standard error of the mean (32.9). (C) Patient (EP58) with stage IIIA EAC with partial response to CRT on surveillance imaging and a decrease in post-CRT MTV but had increased levels of ctDNA post-CRT and experiences distant liver metastasis. (D) Patient (EP29) with stage IIB EAC with equivocal surveillance imaging, increased post-CRT MTV, and undetectable post-CRT ctDNA who achieves long-term survival.
Integration of ctDNA and PET-CT in patients receiving definitive CRT
Although the preferred treatment for localized ESCA is CRT followed by surgery, a significant subgroup of patients are medically unfit to undergo surgery or refuse it and for these patients, definitive CRT is an accepted treatment option 30, 31. In our cohort, we identified 12 patients receiving only CRT with both ctDNA and PET-CT evaluable following treatment (EAC, n = 10; ESCC, n = 2). Among these patients, those with detectable ctDNA at the MRD timepoint had a significantly higher risk of disease progression (HR 6.4, P = .009; Fig. 5A) and disease-specific death (HR 9.1, P = .015; Fig. 5B). Post-CRT ctDNA detection enabled robust detection of distant metastasis (4/4 patients) but was less informative for detection of isolated locoregional recurrence (1/3 patients, Fig. S7A).
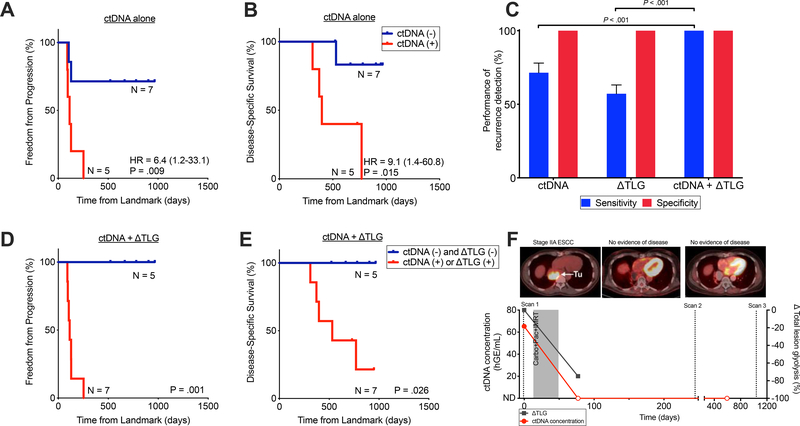
Figure 5. Integrating ctDNA and ΔTLG enables improves detection of recurrence among patients receiving CRT alone.
Kaplan–Meier analysis for (A) freedom from progression (P=.0093, HR 6.4 (95% CI 1.2–33.1)) and (B) disease-specific survival (P=.015, HR 9.1 (95% CI 1.4–60.8)) stratified by post-CRT ctDNA detection among patients receiving CRT alone (n=12). Time to event (days) was measured from post-CRT PET imaging. P values and hazard ratios (HR) were calculated from the log-rank test. (C)Mean sensitivity and specificity of ctDNA, ΔTLG, and both, for any recurrence detection (n=12). ΔTLG is defined as the percent change in TLG in response to CRT. Mean and standard deviation calculated using leave-one-out approach. P values calculated by the Mann-Whitney test. Kaplan–Meier analysis for (D) freedom from progression (P=.0011) and (E) disease-specific survival (P=.026) stratified by integration of ctDNA detection and ΔTLG at the post-CRT time point (n=12). Patients in the red curve (n=7) either had detectable ctDNA following CRT or had ΔTLG ≥ −48.5% while patients in the blue curve (n=5) had undetectable ctDNA after CRT and had ΔTLG < −48.5%. Time to event (days) was measured from post-CRT PET imaging. P values were calculated from the log-rank test. Hazard ratios are not reported as they are undefined given the absence of any events in the ctDNA and ΔTLG negative group. (F) Patient (EP34) with stage IIA ESCC treated with CRT-alone who achieved long-term survival with no evidence of disease. Patient had detectable ctDNA pre-treatment and remains undetectable following CRT.
Given the observation that ctDNA analysis appeared to be more sensitive for prediction of future distant metastasis than local recurrence, we hypothesized that integration of ctDNA with an imaging biomarker capable of detecting residual local disease would further improve performance. Prior studies have suggested that changes in FDG uptake on PET-CT after treatment for localized ESCA are associated with response to therapy and survival 32–34. In an exploratory analysis, we idenitifed a cutpoint for total lesion glycolysis (TLG) that optimally stratified patients who recurred from those who did not. Specifically, we employed a leave-one-out croos-validation approach to identifiy a percent TLG change (ΔTLG) of −48.5% as best predicting ultimate recurrence (Supplementary Methods). ΔTLG displayed a sensitivity of 57.1% and specificity of 100% for recurrence prediction, compared to a sensitivity of 71.4% and specificity of 100% for ctDNA. Integration of ctDNA and PET-CT improved performance to a sensitivity of 100% and specificity of 100%, significantly higher than either ctDNA or ΔTLG alone (P < .001; Fig. 5C).
Patients with detectable ctDNA post-CRT or ΔTLG ≥ −48.5% had significantly increased risk of disease progression (P = .001; Fig. 5D), death from ESCA (P = .026; Fig. 5E), and local progression (P = .02; Fig. S7B), compared to patients with undetectable ctDNA and ΔTLG < −48.5%. Figure 5F illustrates the case of a patient with Stage IIA ESCC treated with definitive CRT who had detectable ctDNA pre-treatment and was negative by the integrated metric post-CRT. This patient remains disease free nearly three years after diagnosis. These preliminary data suggest that an integrated ctDNA-imaging biomarker may allow effective risk stratification of patients treated with CRT alone.
Discussion
Aggressive treatment with trimodality therapy consisting of CRT and esophagectomy confers a significant survival benefit but also results in considerable morbidity35, 36. For example, in the pivotal ChemoRadiotherapy for Oesophageal cancer followed by Surgery Study (CROSS), 29% of all patients (ESCC, 49%; EAC, 23%) had a pCR following CRT 1, suggesting that these patients may be candidates for treatment de-escalation. Conversely, the CROSS trial also demonstrated an overall long-term recurrence rate of ~50% 2, indicating that a significant fraction of patients could potentially benefit from further escalation of therapy. Therefore, biomarkers that can distinguish between patients at highest risk for recurrence despite trimodality therapy from those with complete response to CRT alone are a major unmet need.
Our results suggest ctDNA analysis appears to be a useful approach for risk stratification of patients with ESCA. Specifically, we found that detection of ctDNA MRD following CRT was strongly prognostic for FFP and DSS. Moreover, ctDNA detection post-CRT was more strongly predictive of distant metastasis, the most common pattern of recurrence after trimodality therapy 2, than local progression. However, it should be noted that the results for patient EP30 suggest that post-CRT ctDNA may also be able to detect local residual disease in some cases. In this case, mutations detected following CRT were absent in post-surgery plasma and this patient did not develop recurrence, suggesting resection contributed to oncologic control. We also found that ctDNA detection post-CRT was able to detect recurrence nearly three months earlier than PET-CT imaging. In an exploratory analysis of patients receiving CRT-alone, we found that integration of ctDNA analysis and PET-CT response assessment may allow identification of patients who could avoid esophagectomy.
We observed a pre-CRT median ctDNA VAF of 0.07% (IQR, 0.03 – 0.14), which after adjusting for volume is nearly 10-fold lower than in our recent study of localized lung cancer even after controlling for tumor volume 12. This suggests that ESCAs shed low amounts of ctDNA and that ultrasensitive approaches such as CAPP-Seq will be required to reliably detect ctDNA in patients with localized disease. It is important to note that our approach is unique compared to most other currently available ctDNA detection assays because it involves a tumor mutation-informed bioinformatic approach that maximizes sensitivity while maintaining high specificity. The low levels of ctDNA we observed suggest that ctDNA detection methods based only on plasma analysis (i.e. tumor-naïve) may be insufficiently sensitive to robustly detect ctDNA in localized ESCA.
Our observation of low ctDNA levels in localized ESCA has important implications for future studies. Notably, the median ctDNA VAF we observed in localized ESCA (0.07%), is below the reported detection limits of most commercially available ctDNA assays. Sensitivity for ctDNA detection is determined by 1) sequencing depth (i.e. amount of cfDNA); 2) number of mutations tracked; and 3) assay background.16 Potential approaches for increasing sensitivity include increasing sequencing panel size or designing personalized ctDNA assays based on exome or whole genome sequencing of tumor tissue to track more mutations as we and others have previously reported for other tumor types.13, 16
To our knowledge, this is the first study to systematically evaluate the prognostic utility of ctDNA in a cohort of patients with localized ESCA treated with CRT and esophagectomy. Few studies have focused on analysis of ctDNA in ESCA 37–40. Two of these focused solely on copy number variants and reported associations between CNVs and survival outcomes 40, 41. We instead focused on detection of SNVs since these have a lower limit of detection than CNVs 18 and are thus better suited for the low levels of ctDNA observed in our early stage cohort. Two other studies utilized NGS-based methods for ctDNA detection, though both focused on ESCC, included advanced stage patients, had smaller sample sizes, and did not explore utility of ctDNA for detection of MRD after CRT 14, 42. Our findings of the strong prognostic power of residual ctDNA after CRT is consistent with recent studies in other solid tumors and supports the broad utility of ctDNA for MRD detection 10–12, 25.
Based on our findings, response assessment via integration of ctDNA and imaging biomarkers appears a promising strategy for risk stratification in localized ESCA treated with CRT. Our data suggest that ctDNA analysis following CRT has high sensitivity for detection of occult disseminated disease but is less sensitive for detection of residual locoregional tumor deposits. Explanations for this observation include that micro metastases shed more ctDNA than residual local disease, or that the total burden of micro metastases tends to be greater than that of residual local disease. Imaging-based response assessment, such as analysis of ΔTLG, may be complementary by allowing identification of patients with incomplete local responses. Patients with favorable ctDNA and imaging responses may therefore be ideal candidates for personalized treatment approaches attempting to avoid esophagectomy. Since our analysis was exploratory, future studies will be needed to validate our findings, including the ΔTLG cut-point. Given prior challenges in finding reproducible PET-CT based predictors of local recurrence 8, 9, the combination of MRI-based response assessment with ctDNA should also be investigated. Addtionally, it would be interesting to explore combining ctDNA analysis with other potential clinical or molecular predictors of outcome in multiparmeter models that might further improve outcome prediction.
Limitations of our study include that it was retrospective and had a relatively modest cohort size. However, due to the large hazard ratio associated with ctDNA MRD in our prior study of localized lung cancer12 that served as the basis for our statistical design, the number of patients analyzed was larger than the required sample size. Furthermore, the number of patients we analyzed is consistent with those evaluated for ctDNA MRD in recent studies of other tumor types10–13, 24, 25. Additionally, we had to employ leave-one-out cross-validation in the exploratory anlaysis integrating ctDNA with PET-CT. While this is a commonly used approach for establishing sensitivity and specificity in cohorts of this size, our findings should be considered exploratory and hypothesis generating and future studies in independent cohorts will be required to validate our findings. Separately, we were unable to explore the potential utility of other collection time points such as early during CRT or after completion of surgery due to lack of sample availability. Future studies should explore these time points. We also were unable to compare the performance of ctDNA with protein-based biomarkers, such as CA19–9, and suggest this should be done in future studies. Lastly, our observation of putative emergent mutations in a subset of patients must be interpreted with caution given that we cannot be entirely certain that these mutations are originating from tumor cells. However, given the strong association with disease recurrence it seems likely that the majority of emergent mutations arise from clonal selection of residual tumor cells. Future studies investigating emergent mutations should profile metastatic/recurrent tumor deposits in addition to pre- and post-treatment plasma samples to conclusively establish the presence of these mutations in tumor cells.
In conclusion, we found that post-CRT ctDNA analysis can identify patients with localized ESCA at markedly increased risk of recurrence and death. Moreover, we explore integration of ctDNA and imaging assessment of local treatment response which may allow identification of patients treated with CRT who could be candidates for active surveillance instead of esophagectomy. Prospective studies will be required to validate our findings and to test the clinical utility of such personalized treatment approaches.
Supplementary Material
What You Need to Know
BACKGROUND AND CONTEXT
Biomarkers are needed to identify patients at risk of tumor progression following chemoradiotherapy for localized esophageal cancer. These might help identify at risk of tumor progression and in selection of therapy.
NEW FINDINGS
In an analysis of tumor DNA, germline DNA, and plasma cell-free DNA from patients who underwent chemoradiotherapy for esophageal cancer, detection of tumor DNA in blood (circulating tumor DNA, ctDNA) was associated with tumor progression, metastasis, and shorter survival times.
LIMITATIONS
This was a retrospective analysis; prospective studies and validation in independent cohorts are required.
IMPACT
Assays to measure ctDNA in blood samples of patients who underwent chemoradiotherapy for esophageal cancer might be used to identify those at highest risk for tumor progression.
Acknowledgments
Grant support
This work was supported by grants from the National Cancer Institute (M.D., A.A.A., R01CA188298), the US National Institutes of Health Director’s New Innovator Award Program (M.D., 1-DP2-CA186569), a Stanford Cancer Institute-Developmental Cancer Research Award (M.D. and A.A.A.), Doris Duke Charitable Foundation Clinical Research Mentorship Fellowship (T.D.A.), the Stanford Medical Scholars Program (T.D.A.), the Ludwig Institute for Cancer Research (M.D., A.A.A.), and the CRK Faculty Scholar Fund (M.D.), a Stanford Society for Physician Scholars Grant (T.D.A., A.A.C.), a Radiological Society of North America Resident/Fellow Grant (A.A.C.), the American Society of Clinical Oncology Young Investigator Award (A.A.C.), the Mabuchi Research Fund (S.H.L.) and the R. Lee Clark Clinical Innovator Award (S.H.L).
Funding
We acknowledge funding support from the National Cancer Institute, National Institutes of Health, Stanford Cancer Institute, Doris Duke Charitable Foundation, Ludwig Institute for Cancer Research, CRK Faculty Scholar Fund, Radiological Society of North America, American Society of Clinical Oncology, and Mabuchi Research Fund.
Disclosure of Potential Conflicts of Interest:
A.A.C. reports honoraria and travel support from Roche Sequencing Solutions and Foundation Medicine. A.A.C. has received research funding from Roche Sequencing Solutions and served as a consultant for Oscar Health. A.M.N., A.A.A., and M.D. are co-inventors on patent applications related to cancer biomarkers. A.M.N. reports consultanct with CiberMed. A.A.A., M.D., and D.M.K. are consultants for Roche Molecular Systems and A. L. is employed by Roche Molecular Systems. J.J.C. served as a consultant with Lexent Bio. A.A.A. has served as a consultant for Genentech, Roche, Chugai, Gilead, and Celgene. A.A.A. reports ownership interest in CiberMed and FortySeven. M.D. has served as a consultant for Roche, AstraZeneca, and BioNTech. M.D. and A.A.C have received research funding from Varian Medical Systems. M.D. also reports ownership interest in CiberMed. S.H.L has received research funding from Genentech, New River Labs, BeyondSpring Pharmaceuticals, Hitachi Chemical Diagnostics, and Elekta. S.H.L. serves on an advisory board of AstraZeneca.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Author names in bold designate shared co-first authorship
- 1.van Hagen P, Hulshof MC, van Lanschot JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012;366:2074–84. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro J, van Lanschot JJB, Hulshof M, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): long-term results of a randomised controlled trial. Lancet Oncol 2015;16:1090–1098. [DOI] [PubMed] [Google Scholar]
- 3.Noordman BJ, Spaander MCW, Valkema R, et al. Detection of residual disease after neoadjuvant chemoradiotherapy for oesophageal cancer (preSANO): a prospective multicentre, diagnostic cohort study. Lancet Oncol 2018. [DOI] [PubMed] [Google Scholar]
- 4.Donahue JM, Nichols FC, Li Z, et al. Complete pathologic response after neoadjuvant chemoradiotherapy for esophageal cancer is associated with enhanced survival. Ann Thorac Surg 2009;87:392–8; discussion 398–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedenne L, Michel P, Bouche O, et al. Chemoradiation followed by surgery compared with chemoradiation alone in squamous cancer of the esophagus: FFCD 9102. J Clin Oncol 2007;25:1160–8. [DOI] [PubMed] [Google Scholar]
- 6.Stahl M, Stuschke M, Lehmann N, et al. Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. J Clin Oncol 2005;23:2310–7. [DOI] [PubMed] [Google Scholar]
- 7.Berger AC, Farma J, Scott WJ, et al. Complete response to neoadjuvant chemoradiotherapy in esophageal carcinoma is associated with significantly improved survival. J Clin Oncol 2005;23:4330–7. [DOI] [PubMed] [Google Scholar]
- 8.Schneider PM, Metzger R, Schaefer H, et al. Response evaluation by endoscopy, rebiopsy, and endoscopic ultrasound does not accurately predict histopathologic regression after neoadjuvant chemoradiation for esophageal cancer. Ann Surg 2008;248:902–8. [DOI] [PubMed] [Google Scholar]
- 9.Westerterp M, van Westreenen HL, Reitsma JB, et al. Esophageal cancer: CT, endoscopic US, and FDG PET for assessment of response to neoadjuvant therapy--systematic review. Radiology 2005;236:841–51. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- 11.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo H, Li H, Hu Z, et al. Noninvasive diagnosis and monitoring of mutations by deep sequencing of circulating tumor DNA in esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2016;471:596–602. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh CC, Hsu HS, Chang SC, et al. Circulating Cell-Free DNA Levels Could Predict Oncological Outcomes of Patients Undergoing Esophagectomy for Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giobbie-Hurder A, Gelber RD, Regan MM. Challenges of guarantee-time bias. J Clin Oncol 2013;31:2963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013;45:478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2012;2:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91–5. [DOI] [PubMed] [Google Scholar]
- 24.Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey M, Tokheim C, Porta-Pardo E, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018;173:371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Findlay JM, Castro-Giner F, Makino S, et al. Differential clonal evolution in oesophageal cancers in response to neo-adjuvant chemotherapy. Nat Commun 2016;7:11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park IH, Kim JY. Surveillance or resection after chemoradiation in esophageal cancer. Ann Transl Med 2018;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cronin-Fenton DP, Sharp L, Carsin AE, et al. Patterns of care and effects on mortality for cancers of the oesophagus and gastric cardia: a population-based study. Eur J Cancer 2007;43:565–75. [DOI] [PubMed] [Google Scholar]
- 32.Jayachandran P, Pai RK, Quon A, et al. Postchemoradiotherapy positron emission tomography predicts pathologic response and survival in patients with esophageal cancer. Int J Radiat Oncol Biol Phys 2012;84:471–7. [DOI] [PubMed] [Google Scholar]
- 33.Monjazeb AM, Riedlinger G, Aklilu M, et al. Outcomes of patients with esophageal cancer staged with [(1)(8)F]fluorodeoxyglucose positron emission tomography (FDG-PET): can postchemoradiotherapy FDG-PET predict the utility of resection? J Clin Oncol 2010;28:4714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roedl JB, Colen RR, Holalkere NS, et al. Adenocarcinomas of the esophagus: response to chemoradiotherapy is associated with decrease of metabolic tumor volume as measured on PET-CT. Comparison to histopathologic and clinical response evaluation. Radiother Oncol 2008;89:278–86. [DOI] [PubMed] [Google Scholar]
- 35.Alghamedi A, Buduhan G, Tan L, et al. Quality of life assessment in esophagectomy patients. Ann Transl Med 2018;6:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vellayappan BA, Soon YY, Ku GY, et al. Chemoradiotherapy versus chemoradiotherapy plus surgery for esophageal cancer. Cochrane Database Syst Rev 2017;8:CD010511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez A, Harada K, Mizrak Kaya D, et al. Liquid biopsies in gastrointestinal malignancies: when is the big day? Expert Rev Anticancer Ther 2018;18:19–38. [DOI] [PubMed] [Google Scholar]
- 38.Creemers A, Krausz S, Strijker M, et al. Clinical value of ctDNA in upper-GI cancers: A systematic review and meta-analysis. Biochim Biophys Acta 2017;1868:394–403. [DOI] [PubMed] [Google Scholar]
- 39.Openshaw MR, Richards CJ, Guttery DS, et al. The genetics of gastroesophageal adenocarcinoma and the use of circulating cell free DNA for disease detection and monitoring. Expert Rev Mol Diagn 2017;17:459–470. [DOI] [PubMed] [Google Scholar]
- 40.Andolfo I, Petrosino G, Vecchione L, et al. Detection of erbB2 copy number variations in plasma of patients with esophageal carcinoma. BMC Cancer 2011;11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu S, Ichikawa D, Hirajima S, et al. Clinical impact of predicting CCND1 amplification using plasma DNA in superficial esophageal squamous cell carcinoma. Dig Dis Sci 2014;59:1152–9. [DOI] [PubMed] [Google Scholar]
- 42.Ueda M, Iguchi T, Masuda T, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget 2016;7:62280–62291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.