Abstract
Background:
Allergic asthma is a chronic inflammatory disorder that is characterized with airway hyperreactivity (AHR) and driven by Th2 cytokine production. Group 2 innate lymphoid cells (ILC2s) secrete high amount of Th2 cytokines and contribute to the development of AHR. Autophagy is a cellular degradation pathway that recycles cytoplasmic content. However, the role of autophagy in ILC2s remains to be fully elucidated.
Objective:
We characterized the effects of autophagy deficiency on ILC2 effector functions and metabolic balance.
Methods:
ILC2s from autophagy deficient mice were isolated to evaluate proliferation, apoptosis, cytokine secretion, gene expression and cell metabolism. Also, autophagy deficient ILC2s were adoptively transferred into Rag−/−GC−/− mice, which were them challenged with IL-33 and assessed for airway hyperreactivity and lung inflammation.
Results:
We demonstrate that autophagy is extensively used by activated ILC2s to maintain their homeostasis and effector functions. Deletion of the critical autophagy gene Atg5 resulted in decreased cytokine secretion and increased apoptosis. Moreover, lack of autophagy among ILC2s impaired their ability to utilize fatty acid oxidation and strikingly promoted glycolysis as evidenced by our transcriptomic and metabolite analyses. This shift of fuel dependency led to impaired homeostasis and Th2 cytokine production, thus inhibiting the development of ILC2-mediated AHR. Notably, this metabolic reprogramming was also associated with an accumulation of dysfunctional mitochondria producing excessive reactive oxygen species.
Conclusion:
These findings provide new insights into the metabolic profile of ILC2s and suggest that modulation of fuel dependency by autophagy is a potentially new therapeutic approach to target ILC2 dependent inflammation.
Keywords: ILC2, Immune metabolism, Autophagy, Airway hyperreactivity
Graphical Abstract

CAPSULE SUMMARY
Our data illustrate that modulation of autophagy can critically affect ILC2 effector functions through metabolic reprogramming, opening avenues for novel treatment of ILC2-associated airway inflammation and asthma.
INTRODUCTION
Group 2 innate lymphoid cells (ILC2s) are a non T- and non B-lymphocyte population characterized by their lack of lineage and antigen specific markers. Upon activation by alarmins in response to tissue damage, ILC2s produce high amount of type 2 cytokines such as IL-5 and IL-13 (1–3). These cytokines promote eosinophilia and airway hyperreactivity (AHR), a cardinal feature of allergic asthma. Increased numbers of activated ILC2s have been detected in the blood and sputum of asthma patients compared to healthy controls (4–6). Moreover, increased numbers of ILC2s were also measured in the bronchoalveolar lavage (BAL) fluid of asthmatics (7). Altogether this evidence suggests that ILC2s play a critical role in the development of human allergic asthma. Exploring the mechanisms of activation and regulation of ILC2s is essential to improve the understanding of the pathogenesis of allergic asthma, which could lead to new therapeutic strategies.
Macroautophagy (hereafter referred to as autophagy) is the intracellular process by which damaged organelles are cleared and recycled, playing a critical role for cell survival (8–10). Defects in autophagy result in accumulation of damaged organelles such as mitochondria with the potential release of reactive oxygen species (ROS) that can affect apoptosis. This can in turn lead to activation of the immune system and the development of inflammatory diseases. Aside from bulk degradation, autophagy also plays a pivotal role in the regulation of metabolites levels including glucose and fatty acids (FA) and the balance of energy-generating metabolic pathways in different immune cell populations by modulating fuel dependency (11, 12). Given that immune activation is highly energy dependent and requires specific type of fuel, the mechanisms by which immune cells obtain their energy offers a new platform to modify immune functions. Unlike activated T helpers cells that favor glycolytic metabolism, memory and regulatory T cells are mainly fueled using mitochondrial oxidative phosphorylation (OXPHOS) (13–15). Similarly, innate lymphoid cells have also been shown to predominantly generate energy by fatty acid oxidation (16). Although, it has been demonstrated that autophagy plays an important role in the development of innate lymphoid cells (17), further studies are required to understand how modulation of autophagy and metabolic balance affect homeostasis and effector functions of ILC2 in the context of diseases associated with ILC2s.
In this study we showed that autophagy is extensively used by activated ILC2s to maintain their homeostasis and effector function. Deletion of the critical autophagy gene Atg5 resulted in decreased cytokine secretion, increased apoptosis, and repressed NF-κB signaling. On the other hand, induction of autophagy among WT ILC2s was associated with increased cell survival and cytokine secretion. Moreover, lack of autophagy among ILC2s impaired their ability to utilize FA oxidation and strikingly promotes glycolysis as evidenced by our transcriptomic and metabolite analyses. This shift of fuel dependency led to impaired homeostasis and Th2 cytokine production, thus inhibiting the development of ILC2-mediated AHR. Notably, this metabolic reprogramming was also associated with an accumulation of dysfunctional mitochondria producing excessive ROS. Interestingly, pyruvate treatment to promote mitochondrial respiration rescued these effector functions and restored glucose metabolism among ATG5 deficient ILC2s. Together our findings provide new insight into the metabolic profile of ILC2s and suggest that modulation of fuel dependency by autophagy is a potentially new therapeutic approach to targeting ILC2 dependent inflammation.
METHODS
Mice
Female RAG2 deficient (C.B6(Cg)-Rag2tm1.1Cgn/J) mice and RAG2 GC deficient (C; 129S4-Rag2tm1.1Flv IL2rg tm1.1Flv /J) mice (6–8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME). Rag2-deficient and Rag2 GC-deficient mice were bred in our facility at the Keck School of Medicine, University of Southern California (USC). Atg5flox/flox and LC3-GFP mice are the gift from Dr. Noboru Mizushima (Tokyo Medical and Dental University). Tfeb flox/flox mice are the gift from Dr. Babak Ranzani (Washington University School of Medicine). Atg5−/− and TfebTG mice were generated respectively by crossing Atg5flox/flox and Tfeb flox/flox mice to Rosa-Cre mice. Mice were screened by PCR and Atg5flox/flox homozygote Rosa-Cre hemizygote mice were selected for the experiments. Atg5flox/flox and Tfeb flox/flox mice were backcrossed to rosa26Cre ERT mice (Jackson Laboratories) and bred in our facility at the Keck School of Medicine, University of Southern California under protocols approved by the Institutional Animal Care and Use Committee. To induce the deletion of the Atg5 gene and the Tfeb transgene, mice received tamoxifen (800 μg/mouse/day) for five consecutive days. Animal studies were approved by the USC Institutional Animal Care and Use Committee and conducted in accordance with the USC Department of Animal Resources’ guidelines.
Lung preparation and Flow cytometry
Lungs were processed to single cell suspensions as previously described (18). Stained cells were analyzed on FACSCanto II and/or FACSARIA III systems (Becton Dickinson) and the data were analysed with FlowJo version 10 software (TreeStar, Ashland, Oregon). The following mouse antibodies were used: biotinylated anti-mouse lineage CD3e (145–2C11), CD45R (RA3–3B2), Gr-1 (RB6–8C5), CD5 (53–7.3), TCRb (H57–597), CD11c (N418), CD11b (M1/70), Ter119 (TER-119), FceRIa (MAR-1) (BioLegend) and TCR-gd (eBioGL3) (eBioscience), Streptavidin-FITC, PE-Cy7 anti-mouse CD127 (A7R34), APCCy7 anti-mouse CD45 (30-F11), APCeFluor780 anti-mouse CD11c (N418), PECy7 anti-mouse CD45 (I3/2.3) were purchased from BioLegend. PerCP-eFluor710 anti-mouse ST2 (RMST2–2) and eFluor450 anti-mouse CD11b (M1/70) were purchased from eBioscience. PE anti-mouse CCR2 (SA203G11), APC anti-mouse CCR5 (HM-CCR5), BV421 anti-mouse CCR6 (29–2L17) and PE anti-mouse CCR7 (4B12) were purchased from Biolegend. Intranuclear staining was performed using the Foxp3 Transcription Factor Staining Kit (ThermoFisher Scientific), according to the manufacturer’s instructions. PE anti-human/mouse RelA NFκB p65 (IC5078P) was purchased from R&D Systems. PE anti-mouse Phospho-IKKα/β (Ser 176/180) (16A6) was purchased from Cell Signaling Technology. eFluor-660 anti-mouse Ki-67 (SollA15) was purchased from eBioscience. Intracellular staining was performed using the BD Cytofix/ Cytoperm kit (BD Bioscience, San Jose, CA), according to the manufacturer’s instructions and as described previously (19). PE anti-mouse/human IL-5 (Biolegend) and eFluor660 anti-mouse IL-13 (eBioscience) were used. Apoptosis was assessed using an Annexin V apoptosis detection kit PE (ThermoFisher Scientific), according to the manufacturer’s instructions. For analysis of mitochondrial mass, cells were stained with 40 nM MitoTracker Green (Life Technologies) for 20 minutes or MitoSox Red (Life Technologies) for 10 minutes, respectively, at 37°C. For glucose uptake measurements, cells were incubated in media containing 50μg/ml 2-NBDG (Thermo Fisher Scientific) for 20 min at 37°C after surface antibody staining. For lipid droplet quantification, cells were incubated in media containing 1000 ng/ml Bodipy (Thermo Fisher Scientific) at 37°C for 30 min. APC/Cy7-Glut1 was purchased from N ovus Biologicals.
In vitro stimulation of ILC2s
Murine lung ILC2s were isolated from WT control and Atg5−/− mice to >95% purity using the FACSARIA III cell sorter. Isolated murine ILC2s (5.0 × 104 per mL) were stimulated with recombinant mouse (rm)IL-33 (20 ng/mL), rmIL-2 (10 ng/mL) and rmIL-7 (10 ng/mL). In experiments involving drug treatments, etomoxir (20μg/mL Sigma) or pyruvate (2 mM Sigma) were added to the cells in complete RPMI 1640 medium (Thermo Fisher Scientific) containing 10% fetal bovine serum, L-glutamine (2mM), and a cocktail penicillin (100 μ/ml) and streptomycin (100 μg/ml) for 24 hours indicated. Controls were treated with appropriate vehicle. All cell culture experiments included at least three biological replicates per experiment and were repeated twice, unless stated otherwise in figure legends.
Confocal microscopy
Isolated lung ILC2s from LC3-GFP mice were stimulated with rmIL-33 (10 ng/mL), rmIL-2 (10 ng/mL) and rmIL-7 (10 ng/mL) for 24 hours. After incubation, cells were washed and transferred on Superfrost Plus Slides (Thermo Fisher Scientific) with a Cytospin 4 (Thermo Scientific). The cells on the slides were fixed with freshly prepared 4% paraformaldehyde in PBS (pH 8). After fixation the slides were washed and mounted Vectashield mounting medium containing DAPI (Vector Laboratories, Burlingame, California). LC3 expression was analyzed by confocal microscopy (Nikon, Instruments, Melville, NY).
Supernatant cytokine measurement
Isolated lung ILC2s were stimulated with rmIL-33 (10 ng/mL), rmIL-2 (10 ng/mL) and rmIL-7 (10 ng/mL) for 24 hours. Cytokines were measured by multiplexed fluorescent bead-based immunoassay detection (MILLIPLEX® MAP system, Millipore Corporation, Missouri U.S.A.) according to the manufacturer’s instructions, using a combination of 32-plex (MCYTMAG70KPX32) and 41-plex (HCYTMAG-60K-PX41). For each assay, the curve was performed using various concentrations of the cytokine standards assayed in the same manner and analyzed using MasterPlex2012 software (Hitachi Solutions America, Ltd.), as described by our group before (20) (21).
Metabolic flux analysis
The real-time extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using a XF 96 extracellular flux analyzer (Seahorse Bioscience). Briefly, 200,000 activated lung ILC2s were plated in Seahorse media supplemented with 1mM Pyruvate, 2mM glutamine, and 10mM Glucose. Mito stress test kit (Agilent, San Diego CA), using 1uM oligomycin, 1uM FCCP, and 0.5uM Rotenone (Rot) and Antimycin A (AA) was performed according to the manufacturer’s protocol.
Measurement of airway hyperreactivity (AHR)
Mice were administered with carrier-free recombinant mouse (rm)IL-33 intranasally (i.n.) (BioLegend, San Diego, CA, 0.5 μg per mouse in 50 μl) for 3 consecutive days. One day after the last challenge, mice were anesthetized using i.p. injection of ketamine (10 mg/mL) and xylazine (1 mg/mL). Measurements of airway resistance and dynamic compliance were conducted with the Fine Pointe RC System (Buxco Research Systems, Wilmington, NC), in which mice were mechanically ventilated using a modified version as previously described (21, 22). Mice were sequentially challenged with aerosolized PBS (baseline), followed by increasing doses of methacholine. Maximum lung resistance (RL) values were recorded during a 3-min period after each methacholine challenge.
Collection of BALF (bronchoalveolar lavage fluid) cells
After measurements of AHR, the trachea was canulated and the lungs lavaged three times with 1 ml ice cold PBS to collect BALF cells as previously described. BALF cells were stained with allophycocyanin (APC)-labeled anti-Ly-6G/Ly-6C (clone RB6–8C5, BioLegend, San Diego, CA), Alexa Fluor-labeled anti-CD19 (clone 6D5, BioLegend), phycoerythrin (PE)-labeled anti-Siglec-F (clone E50–2440, BD Pharmingen, San Diego, CA), and PE-Cy (PE-Cy7) labeled anti-CD45 (clone 30-F11, BioLegend), peridinin-chlorophyll-protein complex-Cy5.5 (PerCP-Cy5.5) labeled anti-CD3e (clone145–2C11, eBioscience, San Diego), and eFluor-450 labeled anti-CD11b (clone M1/70, eBioscience), and APC-Cy7 labeled anti-CD11c (clone N418, BioLegend).
In vivo stimulation
Rag−/− mice received two intraperitoneal (i.p.) injections of etomoxir (15 mg/kg) on days −1 and 1. Mice were also administered intranasally rm-IL-33 (BioLegend, San Diego, CA, 0.5 g per mouse in 50 μl) on days 0, 1 and 2. On day 4, AHR was assessed and BAL cells were analyzed by flow cytometry.
Adoptive Transfer
Experiments were performed as described previously (23). Briefly, activated ILC2 from IL-33 treated WT control and Atg5−/− mice were isolated as above and cultured for 48 hours in the presence of rm-IL-33 (20ng/mL) and 5×104 cells transferred i.v. in PBS 1X into Rag−/−γC−/− mice. 24 hours later, 0.5μg rmIL33 i.n. in 50μL was given once a day for three days. AHR was then measured on day 4 and BAL cells were analyzed by flow cytometry.
RNA Sequencing (RNA-seq) and data analysis
Freshly isolated lung ILC2s from WT and Atg5−/− mice after 3 intranasal injections of 0.5μg rmIL33 were stimulated (5×104/mL) with rmIL-2 (10ng/mL), rmIL-7 (10ng/mL) and rmIL-33 (20ng/mL) for 24 hours at 37°C. Total RNA was isolated using Micro RNAeasy (Qiagen, Valencia, California). 10ng of input RNA was used to produce cDNA for downstream library preparation. Samples were sequenced on a NextSeq 500 (Illumina) system. Raw reads were aligned, normalized and further analyzed using Partek® Genomics Suite® software, version 7.0 Copyright ©; Partek Inc., St Louis, MO, USA. Pathway analysis was performed using the Qiagen Ingenuity Pathway Analysis (IPA) software.
Metabolic parameter quantification
Isolated lung ILC2s from WT control and Atg5−/− mice were cultured in RPMI 1640 with 5% FBS, 2 mM L-glutamine, 100 U/ml Pen-Strep. In order to activate ILC2s, 1×106 cells/ml were seeded in 96-well plates with medium supplemented with rmIL-2 (10ng/mL) and rmIL-7 (10ng/mL) and rmIL-33 (20ng/mL) at 37°C. After 24 hours cells were homogenized in ice cold PBS. Samples were analyzed for Glucose-6-phosphate and Fructose-6-phosphate levels following the manufacturer’s instructions respectively from MAK014 and MAK020 Assay Kits (Sigma). Isolated lung ILC2s from WT control and Atg5−/− mice were cultured in RPMI 1640 with 1% FBS, 2 mM L-glutamine, 100 U/ml Pen-Strep. In order to activate ILC2s, 1×106 cells/ml were seeded in 96-well plates with medium supplemented with rmIL-2 (10ng/mL) and rmIL-7 (10ng/mL) and rmIL-33 (20ng/mL) at 37°C. After 24 hours supernatants were analyzed for L-lactate levels following the Glycolysis Cell-Based Assay Kit (#600450, Cayman Chemicals). Cells were lysed in NP40 buffer and protein was quantified using Pierce BCA Protein Assay Kit (Sigma). L-lactate levels were normalized to protein levels for each well individually. Isolated WT and Atg5−/− ILC2s were purified using MACS columns (Miltenyi Biotec). Cell count was normalized to 1×106 cells per sample and ATP levels from cell pellets were determined by using the luciferase based ATP Bioluminescence Assay Kit HS II (Roche Applied Science). Each assay is representative of at least two independent experimental repeats.
Metabolomic analysis by LC/MS/MS and GC/GC/MS
Metabolites were extracted from 5×106 isolated ILC2s from WT control and Atg5−/− mice by addition of 500 μL of ice cold 80% aqueous methanol. The supernatants were combined and filtered using a 3KD ultrafilter (Millipore), dried in a SpeedVac and then stored at −80°C. On the day of analysis, the dried extracts were re-constituted in ice cold 80% aqueous methanol. A quality control (QC) sample was made by combining 5 μL of each sample. This was injected at the start of the sequence and subsequently every 10 samples throughout the LC/MS/MS analysis. Metabolites were analyzed using a Waters Xevo TQ-XSTriple quadrupole MS coupled to a UPLC system (Waters, Elstree). The IC flow rate was 0.250 mL/min. The total run time was 37 min and the hydroxide ion gradient comprised as follows: 0mins, 0mM; 1min, 0mM; 15mins, 60mM; 25mins, 100mM; 30mins, 100mM; 30.1mins, 0mM; 37mins, 0mM. Analysis was performed in negative ion mode using a scan-range from 80–900 and resolution set to 70,000. The tune file source parameters were set as follows: Sheath gas flow 60 mL/min; Aux gas flow 20 mL/min; Spray voltage 3.6v; Capillary temperature 320°C; S-lens RF value 70; Heater temperature 450°C. AGC t arget was set to 1e6v ions and the Max IT value was 250ms. The column temperature was kept at 30°C throughout the experiment. Full scan data were acquired in continuum mode.
Raw data files were processed using ProgenesisQI (Waters). This process included alignment of retention times, identified the presence of natural abundance isotope peaks, identified adducts forms and was then used to identify compounds present. The retention times and other measurable characteristics for each metabolite were confirmed by comparing values from the experimental data with the same values from the analysis of authentic standards for each metabolite. Principal Component Analysis was also performed using ProgenesisQI from peas representing all 4215 compounds measured with a %CV < 30 in the quality control samples (normalized to all compounds).
For lipidomics, 5×106 cells were homogenized in 400 μl of 50% MeOH/H2O in a beads beater tube. Myristic-acid-14,14,14-d3 (1mg/ml) standard was spiked at ratio 1 g/5×106 cells and samples were vortexed for 5 min after adding 1 mL of tert-butyl methyl ether (MTBE). After centrifugation at 13,000 RPM supernatants were collected into a 5:1 MeOH/H2O solution. The aqueous phase was dried in a speedvac and combined with the dried MeOH pellet. For rederivatization, dried metabolite extraction was re-suspended in 20 μg/ul MOX (volume 50 μl) and reacted at 30°C for 90 min on vortex. 30 l of pyridine and 70 l of MSTFA were added to react at 60°C for 60 min before direct injection into GCxGC-MS system comprising of a Waters Synapt G2-SiQTOF MS w/ ion mobility coupled to a 2D UPLC system (Waters). The samples were injected at 280°C and the oven temperature was prog ramed from 60°C to 320°C at 10°C/min and held at 320°C for 8 min. The interface temperature to the mass spectrometer was set at 330°C and ion source was he ated at 230°C. The MS was operated at scan speeds between 5000 and 20,000 amu covering a range of m/z 45–600. Electron Ionization spectra were recorded at 70 eV.
Statistical analysis
Experiments were repeated at least 3 times (n=5–8 per group) and data are shown as the representative of 3–4 independent experiments. A two-tailed student t-test for unpaired data was used for comparisons between each group using Prism Software (GraphPad Software Inc.). The degrees of significance were indicated as: *p<0.05, **p<0.01, ***p<0.001.
Data availability
Sequence data that support the findings of this study have been deposited in Genbank with the primary accession code GSE139573. All remaining data will be made available by the corresponding author upon reasonable request.
RESULTS
Lack of autophagy in ILC2s inhibits ILC2 effector functions and ILC2-mediated AHR
To explore the role of autophagy on activated ILC2s, we first assessed the presence of active autophagy in ILC2s upon activation by confocal microscopy and flow cytometry using the LC3-GFP mouse model, which is used to measure autophagy. Naïve lung ILC2s from LC3-GFP were isolated by fluorescence-activated cell sorter (FACS) and gated as lineage− CD45+ IL-7R+ and ST2+ (Fig 1, A). Isolated ILC2s were activated with recombinant mouse (rm)IL-33 for 12 hours or PBS as control. We observed that activated ILC2s robustly upregulate LC3 expression compared to naïve ILC2s by confocal microscopy (Fig 1, B). Similarly, the quantification of LC3-GFP by flow cytometry revealed an upregulation of the LC3 protein in activated ILC2s compared to PBS treated ILC2s (Fig 1, C). These data demonstrated that the mechanism of autophagy is highly induced in ILC2s upon activation. Following the observation that autophagy is induced in activated ILC2s, we used autophagy related gene 5 (Atg5) deficient mice to study the role of autophagy pathway in ILC2s (24–26). As constitutive Atg5 deletion is embryonically lethal (27), we performed our following experiments using an inducible conditional Atg5 knockout mouse model which we refer to as Atg5−/−. To uncover the functional significance of the autophagy induction in activated ILC2s, we explored the role of autophagy on ILC2 homeostasis and effector function and in particular its production of Th2 cytokines. First, we quantified the number of activated ILC2s in the lungs of WT and Atg5−/− mice using flow cytometry. Strikingly, after 3 days of rmIL-33 intranasal (i.n.) treatment (Fig 1, D), Atg5−/− mice had less ILC2s in the lungs (Fig 1, E). Furthermore, the number of ILC2s from WT and Atg5−/− mice intranasally treated with PBS or IL-33 were quantified in bone marrow cells to assess if egress could also explain the reduced number of activated lung ILC2s. Interestingly, we observed decreased BM ILC2 numbers in Atg5−/− mice treated with either PBS or IL-33 compared to WT controls (see Fig E1). We also assessed CC chemokine receptors (CCR) expression which are crucial to ILC2 trafficking on lung activated ILC2s in WT and Atg5−/− mice. We observed no difference of CCR expression between the two groups suggesting that they do not play a role in the observed difference in ILC2 numbers (see Fig E2). These results suggest that autophagy plays a critical role in pulmonary ILC2 homeostasis during their activation. Next, to assess the effect of autophagy on ILC2 effector function, we measured the levels of cytokine secretion in isolated lung WT and Atg5−/− ILC2s. ILC2s were treated with rmIL-33 for 48 hours to induce activation. Following incubation, cytokine secretion was then measured on the cell culture supernatant by Luminex. When activated with rmIL-33, both IL-5 and IL-13 secretion were significantly decreased in Atg5−/− ILC2s (Fig 1, F) demonstrating that lack of autophagy reduces the induction of Th2 cytokine secretion. Moreover IL-9, IL-6 and GM-CSF secretion was also impaired in Atg5−/− activated ILC2 (see Fig E3, A). In contrast, we also assessed the effect of Atg5 deletion on ILC2s at steady state and we observed no effect of Atg5 deficiency on homeostasis and cytokine secretion in naïve ILC2s (see Figure E3, B and C).
Figure 1: Autophagy is required for ILC2s homeostasis and effector functions.
(A) Gating strategy of Lin−CD45+IL-7R+ST2+ ILC2 cells. LC3-GFP mice were intranasally challenged with recombinant mouse (rm)IL-33 (0.5μg) or PBS on days 1–3. On day 4 lung ILC2s were isolated and analyzed by microscopy and flow cytometry. (B) Confocal microscopy of isolated ILC2s from LC3-GFP mice (x600). (C) Mean fluorescence intensity of LC3. (D) Lung activated ILC2s were isolated from WT control and Atg5−/− mice challenged intranasally for three consecutive days with rmIL-33 according to D. (E) Total number of activated ILC2s in the lungs. (F) Lung ILC2s were isolated from WT control and Atg5−/− mice and activated with rmIL-33 for 24h. IL-5 and IL-13 were measured in the supernatants. (G) Activated ILC2s were isolated according to D. Representative flow cytometry plots of intracellular IL-5 (top panel) and IL-13 (bottom panel) in ILC2s and corresponding quantification. (H) Rag−/−GC−/− mice were adoptively transferred with ILC2s from WT control and Atg5−/− mice. After the adoptive transfer, mice were intranasally treated with rmIL-33 or PBS on days 1–3 according to H. (I) Lung resistance. (J) Total number of lung ILC2s. (K) Total number of eosinophils in BALF. Data are representative of three independent experiments, n=6. Error bars are the mean ± SEM. Student’s t-test, *p < 0.05, ***p < 0.001.
We further measured the cytokine secretion intracellularly by flow cytometry in vivo. After 3 days of rmIL-33 i.n. treatment, the frequencies of ILC2s that secrete IL-5 (Fig 1, G top panel) or IL-13 (Fig 1, G bottom panel) in the lungs were also decreased in Atg5 deficient mice as compared to WT control group (Fig 1, G and see Fig E3, D). Finally, to confirm the effect of autophagy in ILC2-mediated AHR, sorted lung ILC2s from WT and Atg5−/− mice were adoptively transferred to Rag2 GC double knockout mice, which lack T, B, NK cells and ILC2s. These mice were given i.n. rmIL-33 or PBS for three consecutive days, and then AHR was evaluated (Fig 1, H). As expected, i.n. administration of rmIL-33 significantly increased lung resistance in WT and Atg5−/− mice (Fig 1, I), however, lung resistance in IL-33-treated Atg5−/− mice was significantly lower than IL-33-treated WT mice, indicating that autophagy is required for IL-33-induced AHR. IL-33 treatment significantly increased the total number of ILC2s in WT and in Atg5−/− mice but the number of ILC2s was lower in Atg5−/− mice compared to WT controls in IL-33 treated groups (Fig 1, J). Similarly, IL-33 treatment significantly increased the number of eosinophils in the BAL of WT and Atg5−/− mice, although the number of eosinophils in the BAL was reduced in IL-33 treated Atg5−/− compared to WT mice, indicating that IL-33 induced inflammation is impaired in the absence of autophagy (Fig 1, K). These results suggest that autophagy is critical for ILC2 homeostasis and effector function.
Lack of autophagy inhibits the NFκ-B pathway and induces apoptosis in activated ILC2s
To investigate the molecular mechanisms of Atg5 deficiency on ILC2 survival, we next compared transcriptome analysis of ILC2 populations from WT or Atg5−/− mice, by performing RNA-sequencing (RNA-seq) analysis ex vivo. The effect of autophagy deletion on whole ILC2 transcriptome is shown as a volcanic plot based on the p-value and expression fold-change (FC) of each analyzed gene (Fig 2, A). In red and green are respectively the statistically significant genes either upregulated or downregulated by 1.5 FC in Atg5 deficient ILC2s. Notably, Ingenuity pathway analysis revealed a network of genes that altogether statistically (p-value = 5.55×10−7) induced (z-score = 1.293) the apoptosis of lymphocytes in response to Atg5 deletion (Fig 2, B). As we also previously found, the number of ILC2s were lower in Atg5−/− mice which led us to investigate whether lack of Atg5 could affect the survival or proliferation capacities of ILC2s. Atg5−/− and WT mice were i.n. challenged with rmIL-33 for three consecutive days and after 24 hours ILC2s were stained with a dead cell discrimination dye and Annexin V for analyzing cell death and apoptosis respectively. Both the number of early and late apoptotic ILC2s were significantly increased in Atg5−/− mice compared to WT mice (Fig 2, C). To evaluate whether lack of Atg5 could also affect the proliferation rate of ILC2s, we examined expression of the proliferation marker Ki-67 in WT and Atg5−/− ILC2s. The level of Ki-67 in Atg5−/− ILC2s was significantly decreased compared WT ILC2s (Fig 2, D). These data suggest that autophagy is essential for the survival and proliferation of ILC2s.
Figure 2: Lack of autophagy induces apoptosis in activated ILC2s.
Lung ILC2s were isolated from WT control and Atg5−/− mice challenged intranasally for three consecutive days with rmIL-33 (0.5μg). (A) Volcano plot comparison of whole transcriptome gene expression of WT control and Atg5−/− ILC2s, n=3. Differentially expressed genes in Atg5−/− mice (p-value<0.05) with changes of at least 1.5 fold-change (FC) are shown in red (upregulated) or green (downregulated). Notable differentially expressed genes are labeled. (B) Network analysis of upregulated (red) and downregulated (green) genes overall significantly inhibiting the apoptosis of leukocytes. (C) Representative flow cytometry plots of WT control or Atg5−/− Annexin V+7-AAD+ ILC2s and corresponding quantitation. E.A.: early apoptosis L.A.: late apoptosis (D) Representative flow cytometry plots of WT control and ATG5−/− Ki67+ ILC2s and corresponding quantitation. (E) Representative histogram of the expression of NF-κB p65 in activated WT control and Atg5−/− ILC2s. (F) Corresponding quantification presented as Mean Fluorescence Intensity (MFI) of NF-κB p65. (G) Representative histogram of the expression of P-IKKαβ in activated WT control and Atg5−/− ILC2s. (H) Corresponding quantification presented as MFI of P-IKKαβ. The level of isotype-matched stain control is shown as a gray-filled histogram. Data are representative of at least three independent experiments, n=5. Error bars are the mean ± SEM. Student’s t-test, ***p < 0.001.
Modulation of NF-κB signaling in immune cells can increase the susceptibility to apoptosis leading to decreased cell survival (28) (29) (30). Moreover, we have previously shown that loss of effector functions in ILC2s was associated with a decrease in the NF-κB pathway (20). We observed by flow cytometry that Atg5−/− deficiency reduced NF-κB p65 expression in ILC2s (Fig 2, E and F). To better establish the underlying mechanisms, we explored the signaling pathway upstream of NF-κB by measuring the activated form of IKKα/β, which is phosphorylated on serine residues 176 and 180. Consistent with our previous results, lack of autophagy reduced phosphorylated IKKα/β expression in ILC2s (Fig 2, G and H). Collectively, our data demonstrate that autophagy plays a critical role in ILC2 survival and that increased apoptosis was associated with reduced NF-κB pathway activation.
Overexpression of autophagy induces ILC2 effector functions
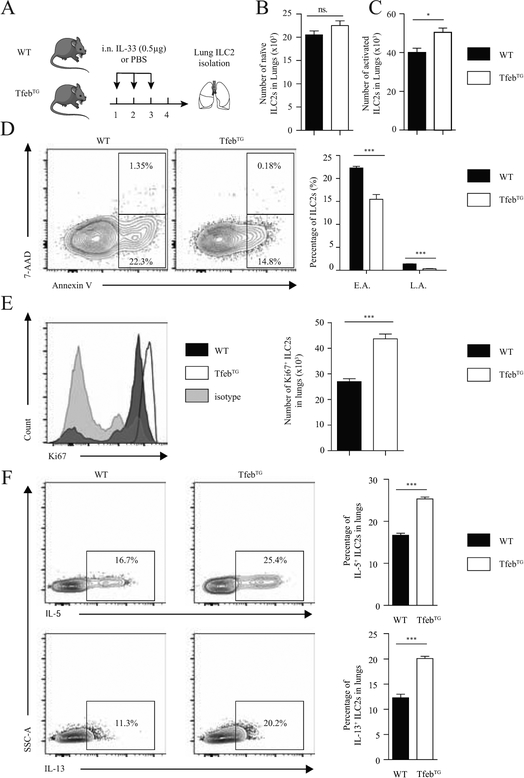
Following the characterization of the Atg5−/− mouse model, we used TfebTG mice with an overexpression of the transcription factor “EB” (Tfeb), a master regulator of autophagy-lysosomal biogenesis and critical to drive autophagy induction as previously described (31, 32). TfebTG mice have constitutively enhanced autophagy. Akin to the above experiments conducted on the Atg5−/− mice, we assessed the effect of overexpressed autophagy on ILC2 homeostasis and effector functions. We quantified the number of naïve and activated ILC2s in the lungs of WT and TfebTG mice using flow cytometry. After three days of rmIL-33 i.n. treatment (Fig 3, A), TfebTG mice had more ILC2s in the lungs whereas the number of ILC2s at steady state was not affected (Fig 3, B and C). These results which mirror those observed in Atg5−/− mice confirm the critical role of autophagy on activated ILC2s.
Figure 3: Overexpression of autophagy induces ILC2 homeostasis and effector functions.
(A) A cohort of WT control and TfebTG mice were intranasally treated with rmIL-33 (0.5μg) or PBS on days 1–3. On day 4 lung ILC2s were isolated and analyzed by flow cytometry according to the protocol in A. Total number of naïve (B) and activated (C) ILC2s in lungs. (D) Representative flow cytometry plots of lung Annexin V+7-AAD+ ILC2s from WT control or TfebTG mice (n=5) and corresponding quantitation. E.A.: early apoptosis L.A.: late apoptosis (E) Representative flow cytometry plots of lung Ki67+ ILC2s from WT control or TfebTG mice (n=5) and corresponding quantitation. The level of isotype-matched stain control is shown as a gray-filled histogram. (F) A cohort of WT control and TfebTG mice were intranasally treated with rmIL-33 (0.5mg) on days 1–3. On day 4 lung ILC2s were isolated and analyzed by flow cytometry. Representative flow cytometry plots of intracellular IL-5 (top panel) and IL-13 (bottom panel) in lung ILC2s and corresponding quantification. Data are representative of at least three independent experiments, n=5. Error bars are the mean ± SEM. Student’s t-test, *p < 0.05, ***p < 0.001.
Since the number of ILC2 was higher in TfebTG mice, we investigated whether overexpression of Tfeb could affect the survival or proliferation capacities of ILC2s, as we had observed with Atg5 deletion. TfebTG and WT mice were i.n. challenged with rmIL-33 for three consecutive days and after 24 hours ILC2s were stained with dead cell discrimination dye and Annexin V for analyzing cell death and apoptosis respectively. Both the number of early apoptotic and late apoptotic ILC2s were significantly decreased in TfebTG mice compare to WT mice (Fig 3, D). To evaluate whether overexpression of Tfeb could also affect the proliferation rate of ILC2s, we examined the expression of Ki-67 in WT and TfebTG ILC2s. The expression level of Ki-67 in TFebTG ILC2s was significantly increased compared to WT ILC2s (Fig 3, E). These data suggest that overexpression of Tfeb increases both the survival and proliferation of ILC2s. Additionally, we also measured cytokine secretion intracellularly by flow cytometry. After three days of rmIL-33 i.n. treatment, ILC2s that secrete IL-5 (Fig 3, F top panel) or IL-13 (Fig 3, F bottom panel) in the lungs were also increased in TfebTG mice as compared to the WT control group. Taken together, the results observed between overexpression and deletion of autophagy are consistent and highlight the essential role of autophagy in the regulation of activated ILC2 homeostasis and effector functions.
Lack of autophagy induces glycolytic metabolism in activated ILC2s
It has been previously shown in other immune cells that autophagy can affect immunometabolism and induce shifts in energy dependency (12). Furthermore, genetic ablation of autophagy can affect the regulation of metabolic pathways leading to metabolic reprogramming and dysfunction of lipid metabolism (12). To determine whether differences exist in the expression of key metabolic genes in response to Atg5 deletion, we next analyzed the gene expression profile of isolated activated WT and Atg5−/− ILC2s by performing RNA-seq analysis ex vivo. Key gene expression of fatty acid oxidation and glycolytic pathways were depicted in a heat plot (Fig 4 A). We found that although the expression of genes critical for commitment to glycolysis was higher in Atg5−/− compared to WT ILC2s, genes involved in fatty acid (FA) metabolism had a lower expression (Fig 4, A). For example, Cpt1c and Ecox3 involved in fatty acid oxidation (FAO) were downregulated, whereas glycolytic genes like G6pc3 and Eno3 were upregulated in response to autophagy deletion.
Figure 4: Lack of autophagy induces glycolytic metabolism in activated ILC2s.
Lung ILC2s were isolated from WT control and Atg5−/− mice challenged intranasally for three consecutive days with rmIL-33 (0.5μg). (A) Heat plot of selected differentially expressed genes (p-value <0.05), n=2. (B) Lung ILC2s isolated from WT or Atg5−/− mice were treated with rmIL-33 for 24 hours in vitro, n=6. Representative histogram of the expression of Glut1 (left) and corresponding quantification (right) were shown. Isotype control was shown as white-filled histogram. (C) Representative histogram of the expression of 2NBDG (left) and corresponding quantification (right) were shown. (D) Enzymatic quantification of lactate accumulation in the supernatant of cultured ILC2s. (E) Glucose-6-phosphate and (F) Fructose-6-phosphate measured in cell extracts from cultured ILC2s. (G) Oxygen consumption rate (OCR) was measured under basal conditions and in response to indicated drugs in the supernatant of cultured ILC2s, n=4. (H) Spare respiratory capacity presented as the difference in OCR after FCCP treatment and basal respiration. (I) Cell energy phenotype presented as OCR against extracellular acidification rate (ECAR). (J) ATP concentration measured in cultured ILC2s, n=6. Data are representative of at least three independent experiments. Error bars are the mean ± SEM. Student’s t-test, **p < 0.01, ***p < 0.001.
To confirm that enhanced expression of glycolytic genes had functional effects on activated ILC2s, we measured glucose uptake and utilization ex vivo. Transport of glucose through Glut1 is a key regulator of glycolytic rate and has been shown to be critical in T cell homeostasis (33). To quantify the expression of this transporter, we assessed Glut1 expression by flow cytometry. We observed that activated Atg5−/− ILC2s have increased surface levels of the Glut1 transporter compared with WT ILC2s (Fig 4, B). We next quantified glucose uptake using the fluorescent D-glucose analogue 2-NBDG. 2-NBDG was significantly more accumulated in Atg5−/− ILC2s than in WT ILC2s (Fig 4, C). In addition we also quantified L-lactate, an end-product of glycolysis in the supernatant of cultured activated WT and Atg5−/− ILC2s. Consistently, Atg5−/− ILC2s secreted more L-lactate than WT ILC2s (Fig 4, D). We also measured the concentration of G6P and F6P, products of the first two steps of glycolysis, in isolated WT and Atg5−/− ILC2s. As expected, the concentration of these two metabolites were higher in Atg5−/−compared to WT ILC2s (Fig 4, E).
Interestingly, quantification of oxygen consumption rate (OCR) reflective of active oxidative phosphorylation (OXPHOS) showed a decrease in spare respiratory capacity (difference in increased OCR after addition of FCCP compared with baseline) in Atg5−/− ILC2s (Fig 4, G and H). Notably, this was confirmed by cell energy phenotype profiling where we observed that Atg5−/− ILC2s had a higher extracellular acidification rate (ECAR) and a lower OCR compared to WT ILC2s in response to stress (FCCP and oligomycin treatment) (Fig 4, I). ILC2s mainly rely on lipid breakdown and fatty acid oxidation (FAO) to supply acetyl-CoA metabolites for OXPHOS. Atg5−/− ILC2s also showed significantly reduced concentration of ATP which can be generated by mitochondrial OXPHOS (Fig 4, J).
To evaluate the direct effect of Atg5 deletion on mitochondrial morphology, we quantified the mitochondrial content of Atg5−/− and WT ILC2s with the MitoTracker Green dye by flow cytometry. Interestingly, lack of autophagy was associated with an increase in mitochondrial size compared to WT ILC2s (see Fig E4, A). Previous reports have shown that cells defective in autophagy accumulate reactive oxygen species (ROS) (34, 35). To examine ROS levels in the mitochondria, we used the mitochondrial-specific ROS indicator MitoSOX to selectively detect superoxides in the mitochondria of live cells. MitoSOX is targeted to the mitochondria and its oxidation by superoxide leads to generation of red fluorescence. Our results indicated that MitoSOX fluorescence was enhanced in Atg5−/− ILC2s compared with WT ILC2s (see Fig E4, B). Combined with the fact that there are more mitochondria and higher ROS accumulation, these data suggest that autophagy is required for mitochondrial function. Overall, the metabolic switch that occurs in activated Atg5−/− ILC2s, engage glycolysis but rather limit mitochondrial respiration which could be explained by mitochondrial dysfunction.
Lack of autophagy inhibits lipolysis in activated ILC2s
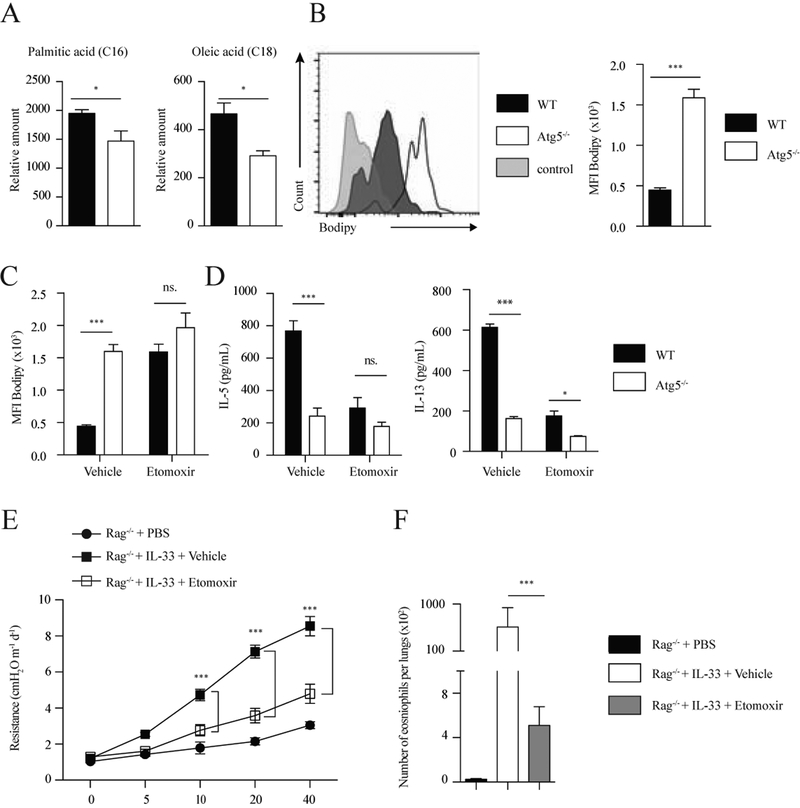
Previous studies have demonstrated that ILC2s rely predominantly on fatty acid oxidation for metabolism and lipid mobilization plays a crucial role in fueling ILC2 function (16). On the other hand, autophagy has also been described as a major pathway for lipid breakdown by targeting lipids to autophagosomes for hydrolysis via lysosomal lipases (lipophagy) (36). Lipophagy can contribute significantly to mobilize cellular lipids for provision of energy (37, 38) and is dependent on the autophagy machinery, suggesting that lack of autophagy could lead to a disruption of this mechanism. Our results have shown lack of autophagy induces a shift in the balance of energy-generating metabolic pathways from FAO toward increased glycolytic activity. As glycolysis is increased and FA metabolism is impaired in Atg5 deficient ILC2s, we hypothesized that breakdown of lipids might be affected and therefore unable to support FAO. Using lipidomics analysis, we measured levels of fatty acids in activated ILC2s of the lung. Interestingly, we observed depletion of free fatty acids, in particular palmitic acid (PA) and oleic acid (OA), in activated Atg5−/− ILC2s compared to WT ILC2s (Fig 5, A). In parallel, we used the neutral lipid-specific dye Bodipy to label lipid droplets for quantification by flow cytometry (Fig 5, B). We observed that Atg5−/− ILC2s accumulated more lipid compared to WT ILC2s (Fig 5, B). To confirm if this aberrant accumulation of lipids in Atg5−/− ILC2s could be associated with defects in FAO, we treated activated ILC2s isolated from WT and Atg5−/− mice with etomoxir, a FAO inhibitor, and assessed lipid uptake with Bodipy. In response to etomoxir, lipid uptake in WT ILC2s was significantly increased and reached the same level as in Atg5−/− ILC2s. Notably, etomoxir had no effect on the lipid uptake of Atg5−/− ILC2s. These results suggest that lack of autophagy affected metabolic pathway regulation by limiting FAO (Fig 5, C). To address this question, WT and Atg5−/− ILC2s of the lung were activated in the presence of rmIL-33, treated with etomoxir or vehicle, and the secretion of IL-5 and IL-13 measured in the supernatant. Our results show that lack autophagy reduces both IL-5 and IL-13 secretion in activated ILC2s. However in the presence of etomoxir both cytokines were no longer inducible in activated WT or Atg5−/− ILC2s (Fig 5, D). These results demonstrate the FAO pathway is critical for ILC2 cytokine production and effector function. These results also suggest that defects in supply of FFAs for FAO could play an important role in the impairment of effector functions in Atg5-deficient ILC2s.
Figure 5: Lack of autophagy inhibits lipolysis in activated ILC2s.
(A) Free fatty acid abundance in ILC2s isolated from WT control and Atg5−/− mice quantified by GCxGC/MS. Data represent 4 biological replicates per genotype. (B) Bodipy histogram (left) and quantification as MFI (right) in WT control and Atg5−/− ILC2s from lungs treated with rmIL-33 (10ng/mL) for 24 hours. The level of stain control is shown as a gray-filled histogram. (C) Bodipy quantification as MFI in WT control and Atg5−/− ILC2s from lungs were treated with rmIL-33 (10ng/mL) and stimulated with vehicle or etomoxir (20ng/μL) for 24 hours. WT control and Atg5−/− ILC2s from lungs were treated with rmIL-33 (10ng/mL) and stimulated with vehicle or etomoxir (20ng/μL) for 24 hours. (D) The levels of IL-5 (left) and IL-13 (right) were measured by ELISA on the culture supernatants, n=6. A cohort of Rag−/− mice was intranasally treated with PBS or rmIL-33 (0.5μg) on days 2–4. Mice were i.p. injected with etomoxir (15mg/kg) or vehicle on days 1 and 3. Measurement of lung function and BALF analysis followed on day 5. (E) Lung resistance. (F) Total number of eosinophils in BALF. Data are representative of at least three independent experiments, n=6. Error bars are the mean ± SEM. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
We showed that defective lipolysis was sufficient to block ILC2 activation by treating ILC2 with the FAO inhibitor etomoxir. To assess the role of FAO on ILC2s in vivo, we examined the effects of etomoxir on IL-33 induced airway hyperreactivity (AHR) and eosinophil recruitment in Rag−/− mice that lack mature B and T cells. Rag−/− mice were intranasally treated with PBS or rmIL-33 in the presence or absence of etomoxir on days 1–3 followed by measurement of lung function and sample acquisition on day 4. Lung function data show that IL-33 increases lung resistance (Fig 5, E). In contrast, IL-33 treated mice that also received etomoxir had significantly lower lung resistance than vehicle treated mice (Fig 1, G). Similarly, the number of eosinophils in bronchoalveolar lavage (BAL) was significantly lower in etomoxir treated group when compared to vehicle control group (Fig 5, F). Taken together these results indicate lipid metabolism is critical for ILC2 effector functions and that autophagy can regulate this metabolic pathway. Moreover, our results clearly establish that both active lysosomal lipolysis and subsequent oxidation in mitochondria are required for ILC2 activation and metabolic homeostasis.
Lack of autophagy inhibits TCA cycle in activated ILC2s
Our results suggest that Atg5−/− ILC2s have impaired breakdown of lipids (Fig 5, A) leading to defects in ATP production mitochondrial and respiration (Fig 4, G, and E4) and increased glycolytic pathways (Fig 4, B–F). We therefore characterized the impact of Atg5 deficiency on these metabolic pathways. During ILC2 activation, we observed profound metabolic reprogramming and a clear overall difference in the metabolome of activated Atg5−/− ILC2s compared to WT IlC2s, as illustrated in the principal component analysis (PCA) of 4,215 metabolites (Fig 6, A). By metabolomics analysis, we found Atg5−/− ILC2s had a significant reduction in TCA cycle metabolites compared with WT ILC2s (Fig 6, B and C). Indeed, among these metabolites the levels of citrate, α-ketoglutarate, fumarate and malate which are essential intermediates of the TCA cycle were significantly decreased in ATG5−/− ILC2s of the lung (Fig 6, B). We hypothesized that fatty acids are used during ILC2 activation as energy substrates for FAO and OXPHOS and tested whether pyruvate, an energy metabolite that can substitute fatty acid-derived acetyl-CoA to directly fuel OXPHOS, can also rescue the effector functions of autophagy deficient ILC2s. Pyruvate treatment alone was sufficient to restore defective glucose uptake (Fig 6, D) and cytokine secretion (Fig 6, E) of Atg5–/– ILC2s. Similarly, we observed that PA and OA treatment could also restore cytokine secretion of Atg5–/– ILC2s (see Fig E5). Altogether, these results show that administration of OXPHOS-fueling substrate pyruvate or FFA are sufficient to restore effector functions of ILC2s, supporting the notion that lipophagy provides FFAs for mitochondrial ATP production in order to drive normal ILC2 activation.
Figure 6: Lack of autophagy inhibits TCA cycle in activated ILC2s.
ILC2s were isolated from WT control and Atg5−/− mice challenged intranasally for three consecutive days with rmIL-33 (0.5μg). (A) Principal component analysis of activated WT control or Atg5−/− ILC2s were scored based on the 4,215 detected metabolites (with Hotellings T2 ellipse at 0.95), n=4. (B) Relative levels of metabolites in the TCA cycle pathway analyzed using an LC-MS/MS system to determine the abundance of cellular metabolites, n=4. (C) Network analysis of metabolites in sorted WT control and Atg5−/−ILC2s from lungs treated with rmIL-33 (10ng/mL) for 24 hours in vitro, n=4. (D) 2-NBDG quantification as MFI in WT control and Atg5−/− ILC2s from lungs treated with rmIL-33 (10ng/mL) and stimulated with vehicle or pyruvate (2mM) for 24 hours. WT control and Atg5−/− ILC2s from lungs were treated with rmIL-33 (10ng/mL) and stimulated with vehicle pyruvate (2mM) for 24 hours. (E) The levels of IL-5 (left) and IL-13 (right) were measured by ELISA on the culture supernatants, n=4. Data are representative of at least three independent experiments. Error bars are the mean ± SEM. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
Previous studies have shown a crucial role for ILC2s in allergen-induced asthma in mouse models (39–41). Similarly, human studies have shown that the prevalence of ILC2s in the blood was increased in subjects with allergic asthma, and PBMCs from asthmatic patients produced higher amounts of IL-5 and IL-13 in response to IL-25 or IL-13 compared to healthy subjects (4, 42). Together these studies demonstrate an essential role for ILC2s in allergic asthma.
Several reports have indicated that both ILC2s and lipid mediators have increased levels in tissues from patients with asthma and contribute to the development of asthma in mouse models (43–45). Interestingly, bioactive lipid mediators are able to regulate ILC2 activity and accumulation, making them critical modulators of ILC2 function in patients with allergic diseases (46–49). These results suggest that through environmental stimuli ILC2 functions can be modulated. Overall these studies indicate that one of the mechanisms to modulate ILC2 function and to inhibit ILC2-induced AHR is to target the metabolic pathways regulating ILC2 effector function.
Accumulating evidence indicates that autophagy, a conserved lysosomal degradation pathway affects cellular immune responses in particular T lymphocyte survival and activation (50–53). Moreover, recently the role of autophagy in modulating immune responses through metabolic reprogramming has been highlighted (11, 12, 54–56). It has been established that ILC2s rely on FFA usage to fuel themselves (16), however the role of autophagy on ILC2 effector functions and its effect on the ILC2 energy-balance have not been explored.
In this study we demonstrate not only that autophagy is induced in activated ILC2s but this mechanism is also critical for ILC2 homeostasis and effector functions in vivo. Indeed our results show that the number of lung ILC2s is decreased in response to Atg5 deficiency in mice treated with IL-33 but not at steady state. However, our results suggests that Atg5−/− ILC2s in the bone marrow both in PBS and IL-33-challenged mice are significantly reduced. Previous studies show that at steady state there is a significant and continuous production of IL-33 in the bone marrow, by variety of cells including osteoclasts (57, 58). It seems that this level of activation and expansion is necessary for sufficient maintenance of ILC2s in the bone marrow. Therefore, even in naïve mice we do see a significantly lower number of ILC2s in the BM of Atg5 deficient mice, as autophagy is required for IL-33 dependent survival and expansion in the bone marrow. However, additional experiments are required to rule out the effect of lower BM output upon activation and their putative influence on the number of lung ILC2 in IL-33-challenged mice.
We also found that lack of autophagy induces lower ILC2-mediated AHR and lung inflammation in Rag−/−GC−/− mice in response to IL-33 challenge. In the absence of autophagy, ILC2 apoptosis is induced whereas ILC2 proliferation is decreased resulting in a reduced number of total ILC2s. In contrast, in the overexpressing autophagy TfebTG mouse model, we observed mirrors effects with an increased number of total ILC2s associated with increased type 2 cytokine secretion.
Interestingly, in absence of autophagy, ILC2s undergo a shift of the fuel-dependency towards the glycolytic metabolism. Our results further show that autophagy provides FFAs through lipid droplet degradation in order to maintain energy balance. While glucose uptake and glycolysis metabolite levels in ILC2s were increased in response to lack of autophagy, OXPHOS pathway and TCA cycle metabolite levels were decreased. Although the glycolysis mechanism was likely induced as a secondary compensation effect, this metabolic pathway is not enough to maintain ATP level. This depletion in ATP can explain how the lack of autophagy results in the inhibition of ILC2 effector functions.
It has been previously reported that NF-κB modulation can regulate energy homeostasis, in particular inhibition of NF-κB pathway led to an increased glycolytic metabolism and decreased cell survival (59, 60). In our study, we observed that decreased ILC2 survival was also associated with an inhibition of the NF-κB pathway. These findings consistent with previous reports demonstrate that inhibition of the NF-κB activity can translate to reduced mitochondrial respiration and enhanced glycolysis. To identify if the metabolic reprogramming is triggered by NF-κB inhibition or whether the decreased NF-κB pathway is the source of the fuel-dependency shift would require further analysis.
A causal link has been described between mitochondrial dynamic, metabolic reprogramming and effector functions in immune cells (61, 62). Mitochondria are essential organelles for metabolic activity and cell survival that constantly remodel their structure. Modulation of mitochondrial dynamic by fission-fusion can lead to increased ROS production (63), mediate mitophagy (64, 65), affect cell survival (66) and support mitochondrial OXPHOS respiration (62). In this study, we also found that lack of autophagy was associated with a metabolic shift towards glycolysis and an accumulation of defective mitochondria producing increased mitochondrial ROS.
In parallel, the inhibition of FAO in ILC2s led to accumulation of lipid droplets at similar levels observed in autophagy deficient ILC2s and also resulted in low type 2 cytokine secretion. Atg5 deficient ILC2s also had a depletion of their FFA levels compared to control cells. Altogether these results suggest that the lack of autophagy affected the FAO by limiting the access of FFA from the lipid droplet to enter in the FAO pathway. As the FAO pathway is inhibited and the acetyl-CoA produced by glycolysis is not enough, the TCA cycle cannot be fueled in order to provide ILC2s with ATP. Collectively, our data demonstrate the FAO-OXPHOS pathway under the regulation of autophagy is essential to supply sufficient ATP to fuel ILC2 effector functions. This notion is consistent with recent reports describing the complex interplay between fuel-dependency balance and immune responses (55, 67). It is noteworthy to mention that the mechanism of lipophagy, defined as the autophagic degradation of intracellular droplets through the usage of the autophagic machinery still requires further exploration (68). We further investigated whether this induced metabolic shift could be corrected by adding pyruvate in the culture media of ILC2s to fuel the physiological metabolic pathways. Strikingly, we observed that exogenous pyruvate treatment efficiently restores ILC2 glucose levels and effector functions. Similarly, addition of of FFA, such as PA and OA also rescued the impaired cytokine secretion in autophagy deficient ILC2s. These results suggest that this metabolite can correct ILC2 activation, most probably by providing substrate to fuel the TCA cycle.
In summary, our findings provide experimental support for the concept that autophagy modulation can result in metabolic reprogramming. Therefore, our results suggest a protective role of autophagy deficiency in the development of ILC2-mediated asthma and present autophagy modulators as therapeutic candidates to regulate allergic asthma through metabolic reprogramming of innate immune cells such as ILC2s.
Supplementary Material
CLINICAL IMPLICATIONS.
This study highlights the importance of autophagy on metabolic wiring of the immune cells associated with allergic lung inflammation and asthma.
ACKNOWLEDGEMENTS
We thank the Bioinformatics Service from USC in particular Dr. Yibu Chen and Meng Li for technical support.
Funding
This article was financially supported by National Institutes of Health Public Health Service grants R01 ES025786, R01 ES021801, R01 HL144790, R21 AI109059 (O.A.). B.P.H. is supported by the Swiss National Science Foundation for Early Postdoc.Mobility #181286.
Abbreviations
- AA
antimycin A
- AHR
airway hyperreactivity
- ATP
adenosine triphosphate
- BALF
bronchoalveolar lavage fluid
- BM
bone marrow
- CCR
CC chemokine receptor
- CoA
coenzyme A
- E.A.
early apoptosis
- ECAR
extracellular acidification rate
- F6P
fructose 6-phosphate
- FAO
fatty acid oxidation
- FC
fold-change
- FCCP
Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- FFA
free fatty acid
- G6P
glucose 6-phosphate
- IKKα/β
inhibitor of nuclear factor kappa-B kinase subunit alpha/beta
- ILC2s
group 2 innate lymphoid cells
- I.N.
intranasal
- I.P.
intraperitoneal
- L.A.
late apoptosis
- MFI
mean fluorescence intensity
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- OA
oleic acid
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- PA
palmitic acid
- PCA
principal component analysis
- QC
quality control
- RM
recombinant mouse
- RNA-seq
RNA-sequencing
- ROS
reactive oxygen species
- Rot
Rotenone
- TCA
tricarboxylic acid cycle
- WT
wild type
- NS
Not significant
Footnotes
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
REFERENCES
- 1.Hazenberg MD, Spits H. Human innate lymphoid cells. Blood. 2014;124(5):700–9. [DOI] [PubMed] [Google Scholar]
- 2.Walker JA, McKenzie AN. Development and function of group 2 innate lymphoid cells. Curr Opin Immunol. 2013;25(2):148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurrell BP, Shafiei Jahani P, Akbari O. Social Networking of Group Two Innate Lymphoid Cells in Allergy and Asthma. Front Immunol. 2018;9:2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol. 2014;134(3):671–8 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagakumar P, Denney L, Fleming L, Bush A, Lloyd CM, Saglani S. Type 2 innate lymphoid cells in induced sputum from children with severe asthma. J Allergy Clin Immunol. 2016;137(2):624–6 e6. [DOI] [PubMed] [Google Scholar]
- 6.Smith SG, Chen R, Kjarsgaard M, Huang C, Oliveria JP, O’Byrne PM, et al. Increased numbers of activated group 2 innate lymphoid cells in the airways of patients with severe asthma and persistent airway eosinophilia. J Allergy Clin Immunol. 2016;137(1):75–86 e8. [DOI] [PubMed] [Google Scholar]
- 7.Christianson CA, Goplen NP, Zafar I, Irvin C, Good JT Jr., Rollins DR, et al. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J Allergy Clin Immunol. 2015;136(1):59–68 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke AJ, Riffelmacher T, Braas D, Cornall RJ, Simon AK. B1a B cells require autophagy for metabolic homeostasis and self-renewal. J Exp Med. 2018;215(2):399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riffelmacher T, Clarke A, Richter FC, Stranks A, Pandey S, Danielli S, et al. Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity. 2017;47(3):466–80 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186(6):3299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208(7):1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20(11):1327–33. [DOI] [PubMed] [Google Scholar]
- 16.Wilhelm C, Harrison OJ, Schmitt V, Pelletier M, Spencer SP, Urban JF Jr., et al. Critical role of fatty acid metabolism in ILC2-mediated barrier protection during malnutrition and helminth infection. J Exp Med. 2016;213(8):1409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Sullivan TE, Geary CD, Weizman OE, Geiger TL, Rapp M, Dorn GW 2nd, et al. Atg5 Is Essential for the Development and Survival of Innate Lymphocytes. Cell Rep. 2016;15(9):1910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerzerho J, Maazi H, Speak AO, Szely N, Lombardi V, Khoo B, et al. Programmed cell death ligand 2 regulates TH9 differentiation and induction of chronic airway hyperreactivity. J Allergy Clin Immunol. 2013;131(4):1048–57, 57 e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galle-Treger L, Sankaranarayanan I, Hurrell BP, Howard E, Lo R, Maazi H, et al. Costimulation of type-2 innate lymphoid cells by GITR promotes effector function and ameliorates type 2 diabetes. Nat Commun. 2019;10(1):713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galle-Treger L, Suzuki Y, Patel N, Sankaranarayanan I, Aron JL, Maazi H, et al. Nicotinic acetylcholine receptor agonist attenuates ILC2-dependent airway hyperreactivity. Nat Commun. 2016;7:13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigas D, Lewis G, Aron JL, Wang B, Banie H, Sankaranarayanan I, et al. Type 2 innate lymphoid cell suppression by regulatory T cells attenuates airway hyperreactivity and requires inducible T-cell costimulator-inducible T-cell costimulator ligand interaction. J Allergy Clin Immunol. 2017;139(5):1468–77 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maazi H, Patel N, Sankaranarayanan I, Suzuki Y, Rigas D, Soroosh P, et al. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 2015;42(3):538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maazi H, Banie H, Aleman Muench GR, Patel N, Wang B, Sankaranarayanan I, et al. Activated plasmacytoid dendritic cells regulate type 2 innate lymphoid cell-mediated airway hyperreactivity. J Allergy Clin Immunol. 2018;141(3):893–905 e6. [DOI] [PubMed] [Google Scholar]
- 24.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol. 2012;74:377–401. [DOI] [PubMed] [Google Scholar]
- 26.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–6. [DOI] [PubMed] [Google Scholar]
- 28.Senftleben U, Li ZW, Baud V, Karin M. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity. 2001;14(3):217–30. [DOI] [PubMed] [Google Scholar]
- 29.Claudio E, Brown K, Siebenlist U. NF-kappaB guides the survival and differentiation of developing lymphocytes. Cell Death Differ. 2006;13(5):697–701. [DOI] [PubMed] [Google Scholar]
- 30.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, et al. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25(51):6781–99. [DOI] [PubMed] [Google Scholar]
- 31.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sergin I, Evans TD, Zhang X, Bhattacharya S, Stokes CJ, Song E, et al. Exploiting macrophage autophagy-lysosomal biogenesis as a therapy for atherosclerosis. Nat Commun. 2017;8:15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20(1):61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3(4):337–46. [DOI] [PubMed] [Google Scholar]
- 35.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106(8):2770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, et al. Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J Allergy Clin Immunol. 2012;129(1):191–8 e1–4. [DOI] [PubMed] [Google Scholar]
- 40.Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 2012;36(3):451–63. [DOI] [PubMed] [Google Scholar]
- 41.Klein Wolterink RG, Kleinjan A, van Nimwegen M, Bergen I, de Bruijn M, Levani Y, et al. Pulmonary innate lymphoid cells are major producers of IL-5 and IL-13 in murine models of allergic asthma. Eur J Immunol. 2012;42(5):1106–16. [DOI] [PubMed] [Google Scholar]
- 42.Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol. 2012;188(3):1503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavagnero K, Doherty TA. Cytokine and Lipid Mediator Regulation of Group 2 Innate Lymphoid Cells (ILC2s) in Human Allergic Airway Disease. J Cytokine Biol. 2017;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doherty TA. At the bench: understanding group 2 innate lymphoid cells in disease. J Leukoc Biol. 2015;97(3):455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schauberger E, Peinhaupt M, Cazares T, Lindsley AW. Lipid Mediators of Allergic Disease: Pathways, Treatments, and Emerging Therapeutic Targets. Curr Allergy Asthma Rep. 2016;16(7):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013;132(1):205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang JE, Doherty TA, Baum R, Broide D. Prostaglandin D2 regulates human type 2 innate lymphoid cell chemotaxis. J Allergy Clin Immunol. 2014;133(3):899–901 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doherty TA, Broide DH. Lipid regulation of group 2 innate lymphoid cell function: Moving beyond epithelial cytokines. J Allergy Clin Immunol. 2018;141(5):1587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konya V, Mjosberg J. Lipid mediators as regulators of human ILC2 function in allergic diseases. Immunol Lett. 2016;179:36–42. [DOI] [PubMed] [Google Scholar]
- 50.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186(9):5313–22. [DOI] [PubMed] [Google Scholar]
- 51.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186(3):1564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, Te Velde AA, et al. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology. 2012;142(7):1493–503 e6. [DOI] [PubMed] [Google Scholar]
- 54.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213(1):15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeVorkin L, Pavey N, Carleton G, Comber A, Ho C, Lim J, et al. Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell Rep. 2019;27(2):502–13 e5. [DOI] [PubMed] [Google Scholar]
- 57.da Luz FA, Oliveira AP, Borges D, Brigido PC, Silva MJ. The physiopathological role of IL-33: new highlights in bone biology and a proposed role in periodontal disease. Mediators Inflamm. 2014;2014:342410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang P, Li X, Meng Y, Yuan B, Liu T, Jiao M, et al. Interleukin-33 regulates hematopoietic stem cell regeneration after radiation injury. Stem Cell Res Ther. 2019;10(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tornatore L, Thotakura AK, Bennett J, Moretti M, Franzoso G. The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends Cell Biol. 2012;22(11):557–66. [DOI] [PubMed] [Google Scholar]
- 60.Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13(10):1272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein Geltink RI, O’Sullivan D, Corrado M, Bremser A, Buck MD, Buescher JM, et al. Mitochondrial Priming by CD28. Cell. 2017;171(2):385–97 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell. 2016;166(1):63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci USA. 2006;103(8):2653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823(12):2297–310. [DOI] [PubMed] [Google Scholar]
- 65.Toyama EQ, Herzig S, Courchet J, Lewis TL Jr., Loson OC, Hellberg K, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351(6270):275–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6(8):657–63. [DOI] [PubMed] [Google Scholar]
- 67.Pearce EJ, Pearce EL. Immunometabolism in 2017: Driving immunity: all roads lead to metabolism. Nat Rev Immunol. 2018;18(2):81–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulze RJ, Sathyanarayan A, Mashek DG. Breaking fat: The regulation and mechanisms of lipophagy. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862(10 Pt B):1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in Genbank with the primary accession code GSE139573. All remaining data will be made available by the corresponding author upon reasonable request.