Abstract
The progress of Head and Neck Squamous Cell Carcinoma (HNSCC) is dependent on both cancer stem cells (CSCs) and immune suppression. This study was designed to evaluate the distribution of CSCs and the characteristic immune suppression status in HNSCC primary tumors and lymph nodes. A total of 303 lymph nodes from 25 patients, as well as tumor and adjacent normal tissue samples, were evaluated by a quantitative PCR assay of the markers of CSCs and the characteristic immune suppression. Expressions of selected genes in The Cancer Genome Atlas (TCGA) datasets were also analyzed. In the primary tumors, we found that expressions of CSCs markers (ALDH1L1, PECAM1, PROM1) were down-regulated, while immune suppression markers FOXP3, CD47, EGFR, SOX2, and TGFB1 were up-regulated significantly when compared to that in adjacent normal tissues. In the lymph nodes, expressions of both CSCs, and immune suppression markers were upregulated significantly compared with that in primary tumors. The mRNA expression of selected CSCs and immune suppression markers exhibited the highest expression in the level II of metastasis, then declined in the level III and remained constant at a reduced value in levels IV and V of metastases. These results reveal a comprehensive understanding of the unique genetic characteristics associated with metastatic loci and potential routes of lymphatic dissemination of HNSCC, which helps to explain why the level II has a high incidence of lymph node metastasis, and why skip metastasis straight to the level IV or level V is rarely found in the clinic.
Keywords: head and neck squamous cell carcinoma (HNSCC), lymph nodes, metastasis, mRNA expression, CSCs and immune suppression markers
Introduction
Head and Neck squamous cell carcinoma (HNSCC) is ranked as the sixth most common cancer in the world, with almost 600,000 new cases occurring every year (Bray et al., 2018). More than 50% of HNSCC patients present with metastasis to local lymph nodes at the time of diagnosis. Regional lymph nodes metastasis not only indicates poor survival, but is also a major prognostic factor for the determination of the appropriate treatment (Ozdek et al., 2000; Michikawa et al., 2012). Patients with regional lymph nodes metastasis present a 30–60% 5-year survival rate compared with ~85% for patients without synchronous nodal dissemination (de Juan et al., 2013). Furthermore, regional or metastatic recurrence are more likely to arise in the patients with synchronous nodal dissemination after completing synthetic serial therapy (Wan et al., 2012). However, such regional or metastatic recurrence in HNSCC is generally considered to be incurable and resistant to conventional treatment, with almost 22 months median survival rate in patients receiving salvage surgery or irradiation, and ~12 months for those undergoing palliative chemotherapy alone (Leon et al., 2012; Ho et al., 2014).
Increasing evidence demonstrates a complex, nonlinear, branched evolution model of subclonal populations in cancers (Ginos et al., 2004). The model characterized as a dynamic process that minor subclones likely expand under the selective pressure of therapy (Ginos et al., 2004). It has been reported that hematological malignancies reveal a distinct pattern of clonal evolution in the development of therapeutic resistance and relapse (Tomasson et al., 2012). A microarray-based study has identified a number of HNSCC metastasis and recurrence associated genes in the tissue of primary or recurrent tumors, unmatched normal mucosa and lymph node metastases, but the clinical implication of these observations remains unknown (Lacko et al., 2012). Another study revealed that the mRNA expression of HNSCC in primary tumors are similar to their respective matched metastatic lymph nodes (Reis et al., 2011). These studies have provided a great insight into the genetic alterations underlying the process of metastasis in HNSCC, which will help us to identify novel therapeutic targets.
The extent of lymph node metastasis is an important prognostic factor for locally advanced HNSCC, and previous studies have reported that skip metastasis to inferior cervical lymph nodes at levels III or IV in the absence of demonstrable involvement of levels I and II is rarely found in HNSCC (Amin et al., 2017). However, the genetic alterations and underlying mechanism in the process of nodal dissemination are poorly understood and little is known about the impact of the level of lymph nodes metastasis (LNM) for patients with HNSCC. It presents a great challenge to develop more effective therapeutic strategies to prevent metastases and recurrence. For the purpose of uncovering the CSCs and immune suppression-related genetic alterations underlying metastasis in HNSCC, we chose 15 of the CSCs and immune suppression-related genes that have been reported in the primary expression of HNSCC before (Kosan and Kunz, 2002; Grosse-Gehling et al., 2013; Nor et al., 2014; Prakasam et al., 2014; Yang et al., 2014; Hartomo et al., 2015; Wu et al., 2015; Ji, 2016; Ren et al., 2016) performed RT-PCR to examine the expression levels of in cancer tissues, lymph nodes, and the matched normal tissues from the same patients with synchronous nodal metastases.
Materials and Methods
Cohorts of Enrolled HNSCC Patients
Twenty-five patients diagnosed with HNSCC and subjected to primary operation in Oral and Maxillofacial–Head and Neck Oncology Department of Shanghai Ninth People's Hospital between 2015 and 2016 were screened for the experiments. All patients recruited had not had chemotherapy or radiotherapy prior to the surgical treatment and the patients who underwent neo-adjuvant chemo or radiotherapy were excluded from our study. The mean age of 25 participants (17 men and 8 women) was 57, ranging from 52 to 74. The samples were collected during the surgery and immediately frozen, including cancer tissues, adjacent normal tissues and lymph nodes from the same patient. Adjacent normal tissues were required to be more than 2 cm from the tumor margins in the same patient. The categorization of neck dissection samples was in accordance with the topographic classification of cervical lymph node levels suggested by Gregoire et al. (2014) as IA, IB, IIA, IIB, III, IV, V. Metastatic lymph nodes were confirmed by hematoxylin-eosin (HE) staining and observed by two pathologists independently. The written informed consent of all patients was obtained, following the protocols approved by Shanghai Ninth People's Hospital Ethical Committee and the study was performed in accordance with the Declaration of Helsinki.
Exclusion Criteria
The patients in one of the following situations were excluded in this study (Zhi et al., 2015).
Patients with local recurrences or second primary tumors.
Patients who were HPV positive.
Patients who experienced chemo- or radiotherapy prior to this study.
The Cancer Genome Atlas (TCGA) Datasets Analysis
The selected gene expression was downloaded from mRNA expression detection platform RNA-seq version 2 (level 3) in TCGA data portal (http://cancergenome.nih.gov/). The final number of HNSCC patients included was 509. The statistical programing software R (version 2.14.1) was used to analyze the datasets with the statistical significance at P < 0.05. The normalized counts (cancer and adjacent normal) were used to compare the gene expression (RNA-Seq version 2).
mRNA Expression Profiling
Trizol reagent (Life Technologies, USA) was used to extract the total RNAs of all the acquired samples, and iScriptTMcDNA synthesis kit (Bio-Rad, CA) was used to reverse transcription (RT). The FastSYBR Green master mix with Rox (Life Technologies, USA) was used to perform the quantitative PCR. Fifteen genes were evaluated by quantitative RT-PCR. The primers for IRF1, IFNAR2, FOXP3, TMEM173, CD47, PECAM1, BMI-1, TWISTNB, ALDH1L1, PROM1, EGFR, SOX2, TGFB1, SMAD3, and STAT3 were purchased from SABiosciences. The primer sequences of the genes are listed in Table S1. The gene β-actin was chosen to be the control gene for normalization.
Statistical Analysis
Data from more than three independent experiments are represented as the mean ± standard deviation (SD). Pared t test was used to do statistical analyses comparing the genes expression between primary and adjacent normal tissues of HNSCC patients, Mann Whitney test was performed to analyze the genes expression in metastatic nodes and non-metastatic nodes and data from TCGA, Kruskal–Wallis test was used to access the expression of metastatic nodes in different levels. Mean-normalized mRNA expression value in non-metastasis lymph nodes were chosen to be control for the entire cohort of lymph nodes. The relative value of each gene was calculated as follow: The ΔΔCt = tumor ΔCt-control ΔCt, fold change of mRNA was obtained as 2(−ΔΔCt). We considered the data was significant at P < 0.05.
Results
Clinicopathologic Characteristics of HNSCC Patient Samples
Clinicopathologic characteristics for the study groups (n = 25) including age, gender, tobacco and alcohol history, tumor site, and AJCC stage [which was according to AJCC 8th Edition (Tao et al., 2006)] are summarized in Table 1. Patients' ages ranged from 52 to 74 with a mean age of 57 ± 7.8 years. The gender distribution was 68% male (17/25) and 32% female (8/25). Forty eight percent (12/25) had a history of substantial tobacco exposure (generally >20 pack years), and 44% (11/25) had documented alcohol use. The lesion sites involved were oropharynx (24%) and oral cavity (76%). 4, 36, 24, and 36% of the patients had T1, T2, T3, and T4 tumors, respectively, while 4, 20, 28, and 48% were staged to I, II, III, and IV, accordingly. The majority of the patients had subsequent therapy with radiation (92%) after the surgery. Of all the 25 patients, 13 patients were diagnosed with synchronous nodal metastasis. Metastatic carcinomatous cells were observed in 27 (2.5%) of the 303 lymph nodes. However, there were only 2 cases of skip metastases to level IV and 1 case of skip metastasis for level V. The details of these results are presented in Table 2.
Table 1.
Clinicopathological parameters of enrolled HNSCC patients.
| No. | Tobacco | Alcohol | Site of tumor origin | Pathological stage | AJCC stage |
|---|---|---|---|---|---|
| 1 | YES | YES | Tongue (oro-pharyngeal) | pT4aN0M0 | IVa |
| 2 | YES | YES | Mouth floor | pT2N0M0 | II |
| 3 | NO | NO | Buccal | pT4aN0M0 | IVa |
| 4 | NO | NO | Tongue | pT2N1M0 | III |
| 5 | YES | YES | Mouth floor | pT2N0M0 | II |
| 6 | YES | YES | Gingiva | pT2N2aMO | IVa |
| 7 | NO | NO | Gingiva | pT2N2bM0 | IVa |
| 8 | YES | YES | Mouth floor | pT3N1MO | III |
| 9 | YES | NO | Tongue | pT3N0M0 | III |
| 10 | NO | NO | Palate | pT4N0M0 | IVa |
| 11 | YES | YES | Mouth floor | pT3N1MO | III |
| 12 | YES | YES | Mouth floor | pT4aN2cM0 | IVa |
| 13 | NO | NO | Tongue | pT3N1M0 | III |
| 14 | YES | YES | Tongue (oro-pharyngeal) | pT4N0M0 | IVa |
| 15 | NO | NO | Tongue | pT3N1M0 | III |
| 16 | NO | NO | Tongue (oro-pharyngeal) | pT4N1M0 | IVa |
| 17 | YES | YES | Mouth floor | pT4N2bM0 | IVa |
| 18 | NO | YES | Gingiva | pT2N2bM0 | IVa |
| 19 | NO | NO | Tongue | pT2N0M0 | II |
| 20 | YES | YES | Tongue | pT1N0M0 | I |
| 21 | NO | NO | Tongue (oro-pharyngeal) | pT3N1M0 | III |
| 22 | YES | NO | Tongue (oro-pharyngeal) | pT4N0M0 | IVa |
| 23 | NO | NO | Tongue (oro-pharyngeal) | pT4N1M0 | IVa |
| 24 | NO | NO | Buccal | pT2N0M0 | II |
| 25 | NO | NO | Buccal | pT2N0M0 | II |
Table 2.
Results of histopathologic examination of various cervical lymph node levels.
| Lymph node levels | ||||||||
|---|---|---|---|---|---|---|---|---|
| Numbers | Level IA | Level IB | Level IIA | Level IIB | Level III | Level IV | Level V | Total |
| Metastasis | 4 | 10 | 3 | 2 | 5 | 2 | 1 | 27 |
| Non-metastasis | 39 | 40 | 25 | 38 | 62 | 52 | 20 | 276 |
| Total | 43 | 50 | 28 | 40 | 67 | 54 | 21 | 303 |
Gene Expression Profiles in HNSCC TCGA Dataset
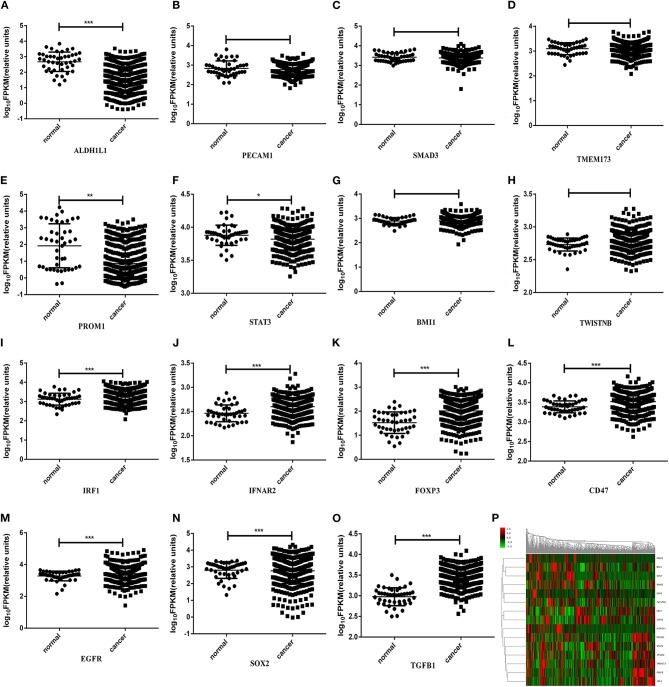
Since differentially expressed genes of the transcriptome data of HNSCC in TCGA dataset have been reported (Cancer Genome Atlas, 2015; Yan et al., 2016), we further examined the expression profiles of the selected genes (i.e., IRF1, IFNAR2, FOXP3, TMEM173, CD47, PECAM1, BMI-1, TWISTNB, ALDH1L1, PROM1, EGFR, SOX2, TGFB1, SMAD3, and STAT3) in the primary tumors of HNSCC. The Clinicopathologic characteristics of TCGA dataset was shown in Table 3. It enrolled 522 HNSCC patients and lesion sites involved 312 in oral cavity (59.8%), 115 in Larynx (22.0%), 82 in oropharynx (15.7%), 10 in hypopharynx (1.9%), and 3 in Lip (0.6%). Forty four of them were diagnosed with HPV infection. This is shown in Figure 1, in which ALDH1L1, PECAM1, SMAD3, TMEM173, PROM1, and STAT3 were expressed lower, and the rest of the genes were expressed higher in primary tumors than in adjacent normal tissues. Among them, the expression levels of IRF1, IFNAR2, FOXP3, CD47, ALDH1L1, PROM1, EGFR, SOX2, TGFB1, and STAT3 were significant (P < 0.05) between tumors and normal tissues (Figure 1).
Table 3.
Clinicopathological parameters of HNSCC patients in TCGA dataset.
| Characteristic | Number | Characteristic | Number |
|---|---|---|---|
| Gender | Age | ||
| Male | 381 | <60 | 288 |
| Female | 141 | ≥ 60 | 233 |
| Not available | 1 | ||
| Smoking status | Alcohol | ||
| Smoker | 388 | Yes | 351 |
| Nonsmoker | 121 | No | 162 |
| Not available | 13 | Not available | 9 |
| Tumor stage | Lymph node stage | ||
| I-II | 186 | N0-1 | 327 |
| III-IV | 320 | N2-3 | 173 |
| Tx | 12 | Nx | 18 |
| Not available | 4 | Not available | 4 |
| Metastasis stage | AJCC stage | ||
| M0 | 492 | I-II | 120 |
| M1 | 4 | III-IV | 388 |
| Mx | 21 | Not available | 14 |
| Not available | 5 | ||
| Primary region | Race | ||
| Gingiva | 18 | American Indian | 2 |
| Tongue | 131 | Asian | 11 |
| Hard palate | 7 | Black American | 45 |
| Floor of mouth | 61 | White American | 447 |
| Bucca | 22 | Not Available | 17 |
| Tonsil | 45 | HPV | |
| oropharynx | 37 | Positive | 44 |
| hypopharynx | 10 | Negative | 81 |
| Larynx | 115 | Not Available | 397 |
| Lip | 3 | ||
| Oral Cavity | 73 |
Figure 1.
FPKM values of ALDH1L1 (A), PECAM1 (B), SMAD3 (C), TMEM173 (D), PROM1 (E), STAT3 (F), BMI-1 (G), TWISTNB (H), IRF1 (I), IFNAR2 (J), FOXP3 (K), CD47 (L), EGFR (M), SOX2 (N), and TGFβ1 (O) expression of HNSCC patients in TCGA datasets; (P) Heatmap of gene expression profiles of HNSCC patients in TCGA datasets. Gene expression values are calculated based on the log transform of FPKM values determined from RNA sequencing analyses. High expression is depicted by red; low expression is depicted by green, normalized by row z scores (legend). *P < 0.05, **P < 0.01, ***P < 0.001.
Examination of mRNA Expression in Primary Tumors by RT-PCR Detection
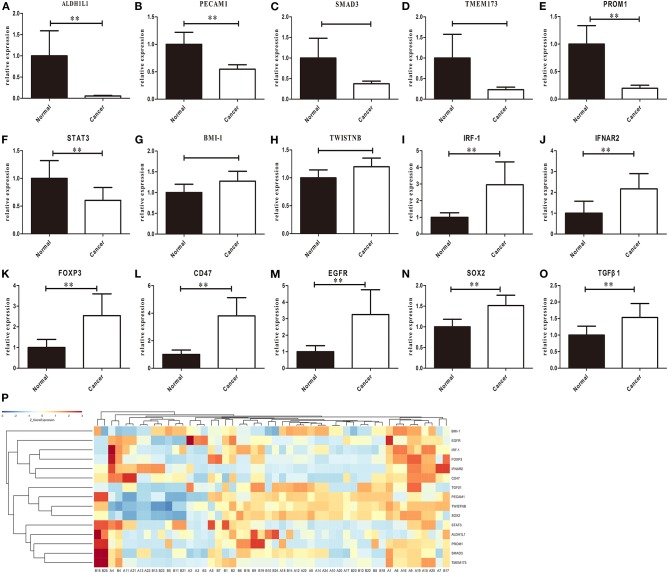
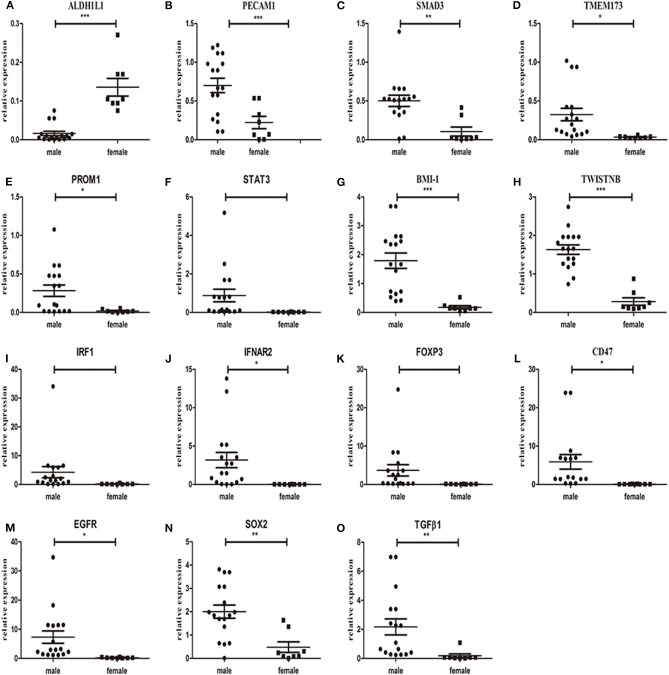
To further investigate the above-mentioned gene expression in our HNSCC samples, we validated the mRNA expression using real time RT-PCR detection. The differential levels of mRNA expression (–ΔΔCt) of the 15 genes in each of the 25 individuals HNSCC sample are shown in Figure 2. Except for PECAM1, which showed a significant decrease (P < 0.05), while the TCGA dataset revealed no significant difference, the expression levels (up or down) of the rest 14 selected genes were consistent with TCGA profiles as validated by real time RT-PCR assay for all recruited patients and a clustering analysis was performed in the primary of HNSCC (Figure 2). Furthermore, there were no significant differences between men and women in normal tissue, but it showed gender difference with male dominancy in the primary (Figure 3 and Figure S1). We also found there were no significant differences between sex and risk factors (such as smoking or drinking) (p = 0.896 and 0.694, respectively). It suggested that sex factor might play a role in the formation of CSCs and immune suppression and might be more susceptible to metastasis.
Figure 2.
RT-PCR results of ALDH1L1 (A), PECAM1 (B), SMAD3 (C), TMEM173 (D), PROM1 (E), STAT3 (F), BMI-1 (G), TWISTNB (H), IRF1 (I), IFNAR2 (J), FOXP3 (K), CD47 (L), EGFR (M), SOX2 (N), and TGFβ1 (O) expression in primary tumors. Each bar represents the average log2 gene mRNA expression level of the paired tumor and adjacent normal tissues (n = 25 per group). Eleven genes showed significant changes between paired tumor and adjacent normal tissues (P) Heatmap of gene expression profiles of HNSCC patients in the primary of HNSCC, Gene expression values are calculated based on the log transform of FPKM values determined from RNA sequencing analyses. High expression is depicted by red; low expression is depicted by green, normalized by row z scores (legend) (**p < 0.05, t-test).
Figure 3.
mRNA expression of ALDH1L1 (A), PECAM1 (B), SMAD3 (C), TMEM173 (D), PROM1 (E), STAT3 (F), BMI-1 (G), TWISTNB (H), IRF1 (I), IFNAR2 (J), FOXP3 (K), CD47 (L), EGFR (M), SOX2 (N), and TGFβ1 (O) in the primary of tumor samples between male and female patients. *P < 0.05, **P < 0.01, ***P < 0.001.
Gene Expression in Different Lymph Nodes
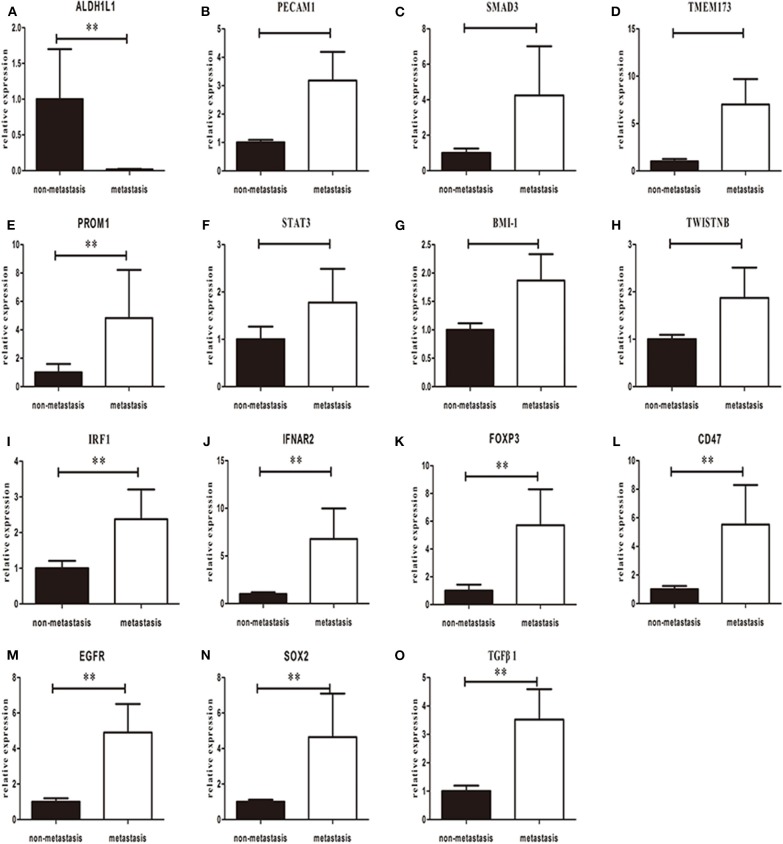
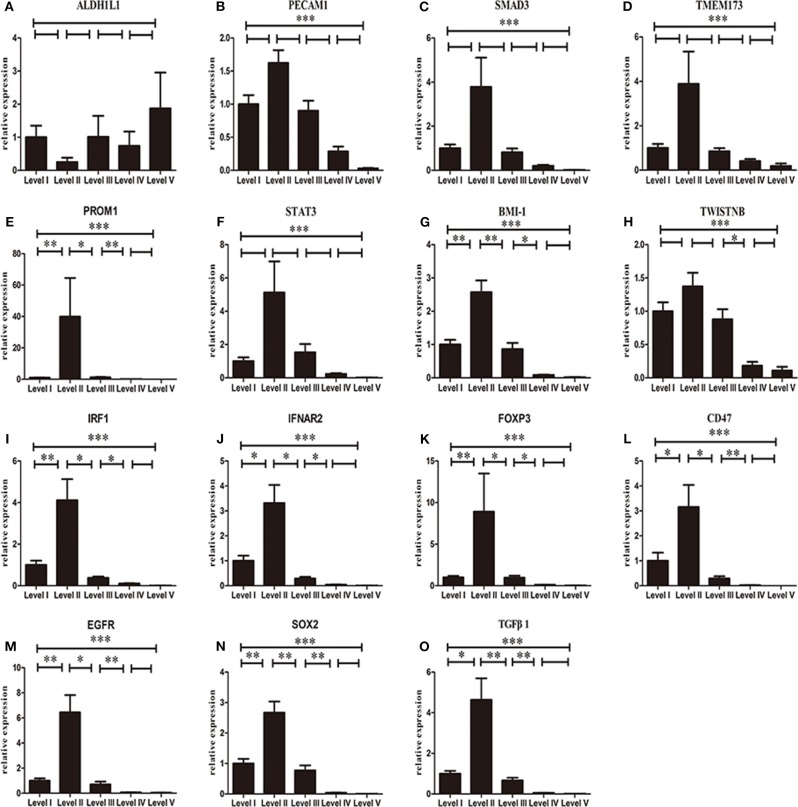
We further compared the mRNA expression of the selected genes between metastatic nodes and non-metastatic nodes in cervical lymph nodes using RT-PCR assay. Similar to the expression levels in primary tumors, the expressions of CD47, EGFR, FOXP3, IFNAR2, IRF1, SOX2, BMI-1, PROM1, and TGFB1 were upregulated significantly in metastatic nodes when compared to non-metastatic nodes (Figure 4). STAT3 and PECAM1 were decreased in primary but upregulated in lymph nodes (P > 0.05) (Figures 2, 3). One exception is ALDH1L1 that exhibited a low mRNA expression level in metastatic nodes (Figure 3). Finally, we analyzed the mRNA expression of the selected genes among different levels of metastatic lymph nodes. We found that all the selected genes except ALDH1L1 followed a similar change from level I–level V metastatic nodes, in which all the genes reached the highest expression in level II, declined in level III and remained at a constant low value in level IV and level V of metastatic nodes (Figure 5). Also, it revealed gender difference with male dominancy in the lymphatic loci of HNSCC (Figure S2).
Figure 4.
Overall mean-normalized mRNA expression of ALDH1L1 (A), PECAM1 (B), SMAD3 (C), TMEM173 (D), PROM1 (E), STAT3 (F), BMI-1 (G), TWISTNB (H), IRF1 (I), IFNAR2 (J), FOXP3 (K), CD47 (L), EGFR (M), SOX2 (N), and TGFβ1 (O) in lymph nodes. Each bar represents the average log2 gene mRNA expression level of the non-metastatic and metastatic lymph nodes. P-values are calculated by Wilcoxon rank-sum test. (**p < 0.05).
Figure 5.
Normalized 2 (−ΔΔCt) mRNA expression of ALDH1L1 (A), PECAM1 (B), SMAD3 (C), TMEM173 (D), PROM1 (E), STAT3 (F), BMI-1 (G), TWISTNB (H), IRF1 (I), IFNAR2 (J), FOXP3 (K), CD47 (L), EGFR (M), SOX2 (N), and TGFβ1 (O) in different levels of lymph nodes. Each bar represents the average log2 gene mRNA expression level of the metastatic lymph nodes in each individual level. P-values are calculated by Kruskal–Wallis test (*p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
The nodal dissemination of carcinomatous cell is a vital determinant of prognosis in patients with HNSCC, as the overall survival rate of patients with metastatic lymph nodes decreased by a half compared with that without nodal implicated (Yan et al., 2014). Keeping track of tumor cells through lymphatic metastasis and microenvironment in lymph nodes is particularly vital for treatment (Owens et al., 2014). As far as we know, there were no studies that investigated the mRNA expression in an individual cervical lymph node level. Hence, the TCGA database was used in our study to perform the unbiased, large-scale analysis of 15 metastasis and tumor microenvironment associated genes. The results showed a significant change of related gene expression through both the HNSCC TCGA-based and our tumor-based analyses. Furthermore, the head and neck squamous cancer (HNSCC) with HPV-negative (HPV−) always occurred in an older patient population and their clinical outcomes were unquestionably worse than HPV+ HNSCC, and few HNSCCs are associated with HPV infection. Therefore, our results provide a model to determine gene expression patterns in both the primary tumors and metastatic lymph nodes of HPV− HNSCC.
IFNAR2 encodes a protein that forms one of the two chains of a receptor for interferons alpha and beta (Zhang et al., 2015). The protein encoded by IRF1 functions as a transcription activator of interferons α-,β-, and γ-induced transcription (Choe et al., 2015) TMEM173 encodes a transmembrane protein that functions as a pattern recognition receptor that activates type I interferon responses (Schreiber and Piehler, 2015; West et al., 2015). All three genes promote the IFN-signaling in the lymph node of HNSCC (Li and Flavell, 2008). TGF-β1 encodes a secreted ligand of the transforming growth factor-beta superfamily proteins (Kudinov et al., 2016). The band of these ligands and various TGF-β receptors result in the recruitment and stimulation of SMAD3 that promotes the process of carcinogenesis (Chaturvedi et al., 2014; Wang et al., 2016). IFN-α, IFN-β and TGF-β play important roles in regulating the activity of lymph node stromal cells embracing lymphatic endothelial cells (LECs), follicular dendritic cells (DCs), and fibroblastic reticular cells (FRCs) (Yang et al., 2014; Ji, 2016). The functional stromal cells may reconstruct and remodel the lymph node, which would produce a unique microenvironment benefit for cancer metastasis (Hartomo et al., 2015). Our RT-PCR results show that are both upregulated in the lymph nodes of HNSCC patients and suggest that these hyperactive lymph node stromal cells may provide a suitable microenvironment for the metastases.
It has been reported that ALDH1 has higher activity in the stem cell subclone of leukemia and some solid tumors (Li et al., 2016). However, ALDH1L1 showed totally different presentation in distinct types of cancers; mRNA high expressions of ALDH1L1 were reported to be correlated to higher overall survival rate for breast cancer patients but were revealed as a poor prognostic factor in gastric and prostatic cancers (Prakasam et al., 2014; Wu et al., 2015; Ren et al., 2016). Our result revealed ALDH1L1 had a lower mRNA expression level in primary and metastatic nodes and exhibited an entirely different behavior compared with other selected genes, indicated that ALDH1L1 mRNA low expressions is related with metastasis of HNSCC. Our observations showed SOX2, BMI-1, PROM1 (CD133), and TWISTNB (TWIST NEIGHBOR) upregulated in lymph nodes. SOX2, BMI-1, and PROM1 are cancer stem cells-related genes (Kosan and Kunz, 2002; Grosse-Gehling et al., 2013; Nor et al., 2014), and TWISTNB is implicated in EMT (Li and Li, 2015). Previous studies indicated that cancer stem cells (CSCs) in situ can transform into migrating cancer stem cells (MCSCs) by EMT (Schlereth et al., 2014). Subsequently, the MCSCs disseminate and form metastatic colonies (Li and Li, 2014). Furthermore, direct trans-differentiation of CSCs into LECs occurs during tumor lymphatic metastasis (Chao et al., 2012; Semenza, 2013). As previously mentioned, LECs remodel the lymph nodes that provide a comfortable microenvironment for cancer metastasis. Our observations of gene expression profiles in lymph nodes suggest that CSC may directly convert into LECs and contribute to tumor neovascularization in the metastasis process of HNSCC.
CD47 is likely involved in the process of evading immunological eradication (Chattopadhyay et al., 2005; Matlung et al., 2017), and FOXP3 is mainly considered as a biomarker of Treg cells that impede the antitumor immune responses in cancer patients (Quante et al., 2013; Triulzi et al., 2013). STAT3 overexpression in metastatic sites may restrain the immune responses to render an immunosuppressive environment (Punt et al., 2015). PECAM1 (CD31) makes up a large portion of cell intercellular junctions and loss of its function may disrupt cell adhesion (ElShamy et al., 2016). Interaction of hypoxia-surviving cells with the immunosuppressive environment influenced by newly recruited tumor-associated macrophages (TAMs), mesenchymal stromal cells (MSCs), and other types of immune cells most likely form and maintain a necrotic/hypoxic core called “aggressiveness niche” that will be the foster ground for cancer metastasis precursors (Johnson, 2001). In accordance with above reports, our study demonstrates that CD47 and FOXP3 were significantly upregulated both in primary site and lymphatic loci, but STAT3, and PECAM1 were only upregulated in lymph nodes. It indicates that there may form a more immunosuppressive environment in the lymph nodes than primary site and may exist an aggressiveness niche to foster metastasis in HNSCC patients.
A previous study revealed that male patients may have higher tendency of cancer-associated gene expression in the primary than matched adjacent normal tissue (Shores et al., 2004). Similarly, the expression of 15 selected genes showed gender differences with male dominancy not only in the primary but also in metastatic loci, with no significant differences between men and women in matched adjacent normal tissue. It indicated that males may be at a higher risk of metastasis than females in HNSCC. The mechanism underlying these expression patterns will be further studied in the future.
It have been reported that a relatively constant and sequential route might exist in the lymphatic drainage of the head and neck region (Ji, 2016). Level I and level II are the most common region of metastases in cancers of the oral cavity, while level III is most likely area of metastases in cancers of hypopharynx (Vishak and Rohan, 2014). Considering the potential mechanisms of the route of lymphatic metastasis remains unknown, our observations of the differential mRNA expressions in respective cervical levels showed that CSC and immunosuppression-related genes achieve a peak value in the level II, but maintain a constant low value in the levels IV and V, which suggests that the level II may mostly provide a necrosis-induced inflammation and hypoxia-induced immunosuppressive environment that seems to be the most fertile ground to generate the tumor cells with metastatic potentials. This supports that carcinomatous cells in the level II tend to have the highest metastatic potency, which may also offer a genetic explanation why rare skip metastases of the level IV or level V were found in HNSCC (Lydiatt et al., 2017).
The relatively small quantity of lymphatic metastasis samples, the variations of tumor sample purity, and the intratumor heterogeneity limit our current study, especially given that the expression of SMAD3, PECAM1, TMEM173, STAT3, and TWISTNB reveals no statistically significant difference among different groups. Meanwhile, our observation revealed PECAM1 was significantly decreased but it showed no significant difference in TCGA datasets. It may result from that the squamous cell cancer in oropharynx, hypopharynx, and larynx were included in TCGA dataset, and they had an entirely different biological behavior and prognosis from HNSCC. While these findings will require further validation in larger cohorts of patient samples in the future, we believe this first gene expression analysis of cervical lymph nodes in the individual level of cancer patients will provide us an important opportunity to guide the future investigations of HNSCC.
Through gene expression analyses, we found that the mRNA expression of selected CSCs and immune suppression markers exhibit the highest expression in the level II metastatic lymph nodes, then declined in the level III and remained constant at a reduced value in levels IV and V metastatic lymph nodes. These results help to explain the reason why the level II has a high incidence of lymph node metastasis, and skip metastasis to the level IV or level V is rarely found in the clinic. It will help to increase the understanding of the genetic characteristics associated with metastatic loci and potential routes of lymphatic dissemination of HNSCC, and may aid the clinical diagnosis and treatment of HNSCC.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Shanghai Ninth People's Hospital Ethical Committee. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
SQ wrote the paper. ZH, RY and LL carried out experimental studies. LM and SL were involved in statistical analysis. GR and JH modified the paper and designed this study concepts. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Support for this work was obtained from the National Natural Science Foundation of China (Grant Nos. 31140007, 81472516, 81600902).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00003/full#supplementary-material
References
- Amin M. B., Greene F. L., Edge S. B., Compton C. C., Gershenwald J. E., Brookland R. K., et al. (2017). The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 67, 93–99. 10.3322/caac.21388 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. P., Weissman I. L., Majeti R. (2012). The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Curr. Opin. Immunol. 24, 225–232. 10.1016/j.coi.2012.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S., Chakraborty N. G., Mukherji B. (2005). Regulatory T cells and tumor immunity. Cancer Immunol. Immunother. 54, 1153–1161. 10.1007/s00262-005-0699-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Gilkes D. M., Takano N., Semenza G. L. (2014). Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment. Proc. Natl. Acad. Sci. U.S.A. 111, E2120–E2129. 10.1073/pnas.1406655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C. H., Park I. S., Park J., Yu K. Y., Jang H., Kim J., et al. (2015). Transmembrane protein 173 inhibits RANKL-induced osteoclast differentiation. FEBS Lett. 589, 836–841. 10.1016/j.febslet.2015.02.018 [DOI] [PubMed] [Google Scholar]
- de Juan J., Garcia J., Lopez M., Orus C., Esteller E., Quer M., et al. (2013). Inclusion of extracapsular spread in the pTNM classification system: a proposal for patients with head and neck carcinoma. JAMA Otolaryngol. Head Neck Surg. 139, 483–488. 10.1001/jamaoto.2013.2666 [DOI] [PubMed] [Google Scholar]
- ElShamy W. M., Sinha A., Said N. (2016). Aggressiveness niche, can it be the foster ground for cancer metastasis precursors? Stem Cells Int. 2016:4829106. 10.1155/2016/4829106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginos M. A., Page G. P., Michalowicz B. S., Patel K. J., Volker S. E., Pambuccian S. E., et al. (2004). Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 64, 55–63. 10.1158/0008-5472.CAN-03-2144 [DOI] [PubMed] [Google Scholar]
- Gregoire V., Ang K., Budach W., Grau C., Hamoir M., Langendijk J. A., et al. (2014). Delineation of the neck node levels for head and neck tumors: a 2013 update. DAHANCA, EORTC, HKNPCSG, NCIC CTG, NCRI, RTOG, TROG consensus guidelines. Radiother. Oncol. 110, 172–181. 10.1016/j.radonc.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Grosse-Gehling P., Fargeas C. A., Dittfeld C., Garbe Y., Alison M. R., Corbeil D., et al. (2013). CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J. Pathol. 229, 355–378. 10.1002/path.4086 [DOI] [PubMed] [Google Scholar]
- Hartomo T. B., Van Huyen Pham T., Yamamoto N., Hirase S., Hasegawa D., Kosaka Y., et al. (2015). Involvement of aldehyde dehydrogenase 1A2 in the regulation of cancer stem cell properties in neuroblastoma. Int. J. Oncol. 46, 1089–1098. 10.3892/ijo.2014.2801 [DOI] [PubMed] [Google Scholar]
- Ho A. S., Kraus D. H., Ganly I., Lee N. Y., Shah J. P., Morris L. G. (2014). Decision making in the management of recurrent head and neck cancer. Head Neck 36, 144–151. 10.1002/hed.23227 [DOI] [PubMed] [Google Scholar]
- Ji R. C. (2016). Lymph nodes and cancer metastasis: new perspectives on the role of intranodal lymphatic sinuses. Int. J. Mol. Sci. 18:E51. 10.3390/ijms18010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. (2001). Tobacco use and oral cancer: a global perspective. J. Dent. Educ. 65, 328–339. [PubMed] [Google Scholar]
- Kosan C., Kunz J. (2002). Identification and characterisation of the gene TWIST NEIGHBOR (TWISTNB) located in the microdeletion syndrome 7p21 region. Cytogenet. Genome Res. 97, 167–170. 10.1159/000066618 [DOI] [PubMed] [Google Scholar]
- Kudinov A. E., Deneka A., Nikonova A. S., Beck T. N., Ahn Y. H., Liu X., et al. (2016). Musashi-2 (MSI2) supports TGF-beta signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. U.S.A. 113, 6955–6960. 10.1073/pnas.1513616113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacko M., De Herdt M. J., Jansen J. C., Brakenhoff R. H., Slootweg P. J., Takes R. P., et al. (2012). Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J. Clin. Oncol. 30, 4104–4110. 10.1200/JCO.2011.40.4509 [DOI] [PubMed] [Google Scholar]
- Leon X., Martinez V., Lopez M., Garcia J., Venegas Mdel P., Esteller E., et al. (2012). Second, third, and fourth head and neck tumors. A progressive decrease in survival. Head Neck 34, 1716–1719. 10.1002/hed.21977 [DOI] [PubMed] [Google Scholar]
- Li K., Guo X., Wang Z., Li X., Bu Y., Bai X., et al. (2016). The prognostic roles of ALDH1 isoenzymes in gastric cancer. Onco. Targets Ther. 9, 3405–3414. 10.2147/OTT.S102314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. O., Flavell R. A. (2008). Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity 28, 468–476. 10.1016/j.immuni.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Li S., Li Q. (2014). Cancer stem cells and tumor metastasis (Review). Int. J. Oncol. 44, 1806–1812. 10.3892/ijo.2014.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li Q. (2015). Cancer stem cells, lymphangiogenesis, and lymphatic metastasis. Cancer Lett. 357, 438–447. 10.1016/j.canlet.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Lydiatt W. M., Patel S. G., O'Sullivan B., Brandwein M. S., Ridge J. A., Migliacci J. C., et al. (2017). Head and Neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 122–137. 10.3322/caac.21389 [DOI] [PubMed] [Google Scholar]
- Matlung H. L., Szilagyi K., Barclay N. A., van den Berg T. K. (2017). The CD47-SIRPalpha signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 276, 145–164. 10.1111/imr.12527 [DOI] [PubMed] [Google Scholar]
- Michikawa C., Uzawa N., Kayamori K., Sonoda I., Ohyama Y., Okada N., et al. (2012). Clinical significance of lymphatic and blood vessel invasion in oral tongue squamous cell carcinomas. Oral. Oncol. 48, 320–324. 10.1016/j.oraloncology.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Nor C., Zhang Z., Warner K. A., Bernardi L., Visioli F., Helman J. I., et al. (2014). Cisplatin induces Bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia 16, 137–146. 10.1593/neo.131744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens T., Khorooshi R., Wlodarczyk A., Asgari N. (2014). Interferons in the central nervous system: a few instruments play many tunes. Glia 62, 339–355. 10.1002/glia.22608 [DOI] [PubMed] [Google Scholar]
- Ozdek A., Sarac S., Akyol M. U., Unal O. F., Sungur A. (2000). Histopathological predictors of occult lymph node metastases in supraglottic squamous cell carcinomas. Eur. Arch. Otorhinolaryngol. 257, 389–392. 10.1007/s004050000231 [DOI] [PubMed] [Google Scholar]
- Prakasam A., Ghose S., Oleinik N. V., Bethard J. R., Peterson Y. K., Krupenko N. I., et al. (2014). JNK1/2 regulate Bid by direct phosphorylation at Thr59 in response to ALDH1L1. Cell Death Dis. 5:e1358. 10.1038/cddis.2014.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt S., Houwing-Duistermaat J. J., Schulkens I. A., Thijssen V. L., Osse E. M., de Kroon C. D., et al. (2015). Correlations between immune response and vascularization qRT-PCR gene expression clusters in squamous cervical cancer. Mol. Cancer 14:71. 10.1186/s12943-015-0350-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quante M., Varga J., Wang T. C., Greten F. R. (2013). The gastrointestinal tumor microenvironment. Gastroenterology 145, 63–78. 10.1053/j.gastro.2013.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis P. P., Waldron L., Perez-Ordonez B., Pintilie M., Galloni N. N., Xuan Y., et al. (2011). A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer 11:437. 10.1186/1471-2407-11-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z. H., Zhang C. P., Ji T. (2016). Expression of SOX2 in oral squamous cell carcinoma and the association with lymph node metastasis. Oncol. Lett. 11, 1973–1979. 10.3892/ol.2016.4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth S. L., Refaian N., Iden S., Cursiefen C., Heindl L. M. (2014). Impact of the prolymphangiogenic crosstalk in the tumor microenvironment on lymphatic cancer metastasis. Biomed. Res. Int. 2014:639058. 10.1155/2014/639058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G., Piehler J. (2015). The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 36, 139–149. 10.1016/j.it.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Semenza G. L. (2013). Cancer-stromal cell interactions mediated by hypoxia-inducible factors promote angiogenesis, lymphangiogenesis, and metastasis. Oncogene 32, 4057–4063. 10.1038/onc.2012.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shores C. G., Yin X., Funkhouser W., Yarbrough W. (2004). Clinical evaluation of a new molecular method for detection of micrometastases in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 130, 937–942. 10.1001/archotol.130.8.937 [DOI] [PubMed] [Google Scholar]
- Tao L., Lefevre M., Ricci S., Saintigny P., Callard P., Perie S., et al. (2006). Detection of occult carcinomatous diffusion in lymph nodes from head and neck squamous cell carcinoma using real-time RT-PCR detection of cytokeratin 19 mRNA. Br. J. Cancer 94, 1164–1169. 10.1038/sj.bjc.6603073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasson M. H., Shannon W. D., Payton J. E., Kulkarni S., Westervelt P., Walter M. J., et al. (2012). Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triulzi T., Tagliabue E., Balsari A., Casalini P. (2013). FOXP3 expression in tumor cells and implications for cancer progression. J. Cell Physiol. 228, 30–35. 10.1002/jcp.24125 [DOI] [PubMed] [Google Scholar]
- Vishak S., Rohan V. (2014). Cervical node metastasis in T1 squamous cell carcinoma of oral tongue- pattern and the predictive factors. Indian J. Surg. Oncol. 5, 104–108. 10.1007/s13193-014-0301-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X. C., Egloff A. M., Johnson J. (2012). Histological assessment of cervical lymph node identifies patients with head and neck squamous cell carcinoma (HNSCC): who would benefit from chemoradiation after surgery? Laryngoscope 122, 2712–2722. 10.1002/lary.23572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qiu T., Shi J., Liang J., Wang Y., Quan L., et al. (2016). Gene expression profiling analysis contributes to understanding the association between non-syndromic cleft lip and palate, and cancer. Mol. Med. Rep. 13, 2110–2116. 10.3892/mmr.2016.4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., et al. (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. 10.1038/nature14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Xue W., Huang X., Yu X., Luo M., Huang Y., et al. (2015). Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 36, 2421–2426. 10.1007/s13277-014-2852-6 [DOI] [PubMed] [Google Scholar]
- Yan J., Xue F., Chen H., Wu X., Zhang H., Chen G., et al. (2014). A multi-center study of using carbon nanoparticles to track lymph node metastasis in T1-2 colorectal cancer. Surg. Endosc. 28, 3315–3321. 10.1007/s00464-014-3608-5 [DOI] [PubMed] [Google Scholar]
- Yan L., Zhan C., Wu J., Wang S. (2016). Expression profile analysis of head and neck squamous cell carcinomas using data from the cancer genome Atlas. Mol. Med. Rep. 13, 4259–4265. 10.3892/mmr.2016.5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. Y., Vogt T. K., Favre S., Scarpellino L., Huang H. Y., Tacchini-Cottier F., et al. (2014). Trapping of naive lymphocytes triggers rapid growth and remodeling of the fibroblast network in reactive murine lymph nodes. Proc. Natl. Acad. Sci. U.S.A. 111, E109–E118. 10.1073/pnas.1312585111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. J., Jiang D. S., Li H. (2015). The interferon regulatory factors as novel potential targets in the treatment of cardiovascular diseases. Br. J. Pharmacol. 172, 5457–5476. 10.1111/bph.12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X., Lamperska K., Golusinski P., Schork N. J., Luczewski L., Kolenda T., et al. (2015). Gene expression analysis of head and neck squamous cell carcinoma survival and recurrence. Oncotarget 6, 547–555. 10.18632/oncotarget.2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.