Abstract
Severe aplastic anemia (SAA) is a rare disorder characterized by hypoplastic bone marrow and progressive pancytopenia. The etiology of acquired SAA is not understood but is likely related to abnormal immune responses and environmental exposures. We conducted a genome-wide association study of individuals with SAA genetically matched to healthy controls in discovery (359 cases, 1,396 controls) and validation sets (175 cases, 1,059 controls). Combined analyses identified linked SNPs in distinct blocks within the major histocompatibility complex on 6p21. The top SNP encodes p.Met76Val in the P4 binding pocket of the HLA class II gene HLA-DPB1 (rs1042151A>G, odds ratio [OR] 1.75, 95% confidence interval [CI] 1.50–2.03, p = 1.94 × 10−13) and was associated with HLA-DP cell surface expression in healthy individuals (p = 2.04 × 10−6). Phylogenetic analyses indicate that Val76 is not monophyletic and likely occurs in conjunction with different HLA-DP binding groove conformations. Imputation of HLA-DPB1 alleles revealed increased risk of SAA associated with Val76-encoding alleles DPB1∗03:01, (OR 1.66, p = 1.52 × 10−7), DPB1∗10:01 (OR 2.12, p = 0.0003), and DPB1∗01:01 (OR 1.60, p = 0.0008). A second SNP near HLA-B, rs28367832G>A, reached genome-wide significance (OR 1.49, 95% CI 1.22–1.78, p = 7.27 × 10−9) in combined analyses; the association remained significant after excluding cases with clonal copy-neutral loss-of-heterozygosity affecting class I HLA genes (8.6% of cases and 0% of controls). SNPs in the HLA class II gene HLA-DPB1 and possibly class I (HLA-B) are associated with SAA. The replacement of Met76 to Val76 in certain HLA-DPB1 alleles might influence risk of SAA through mechanisms involving DP peptide binding specificity, expression, and/or other factors affecting DP function.
Keywords: aplastic anemia, genome-wide association study, GWAS, bone marrow failure, etiology, HLA, HLA-DP, hematpoietic cell transplantation
Main Text
Aplastic anemia is a rare heterogeneous disorder characterized by progressive pancytopenia and bone marrow hypoplasia.1,2 An estimated 500 to 1,000 cases of severe aplastic anemia (SAA) occur in the United States each year.3 It occurs at all ages, and its etiology is not well understood. Acquired aplastic anemia is often immune-mediated and may occur after hepatitis or with certain drug or environmental exposures.1,2 A limited number of studies with small sample sizes have evaluated the potential role(s) of common single-nucleotide polymorphisms (SNPs) in aplastic anemia etiology.3, 4, 5, 6, 7, 8 For example, a study of 170 cases and 222 controls from Pakistan found two FAS and two FASL SNPs associated with SAA.9 Another study evaluated four FOXP3 SNPs in 94 aplastic anemia cases and 195 controls from Korea, and this study suggested an association with disease and response to immunosuppressive therapy (IST).10 Aplastic anemia also occurs in individuals with rare inherited bone marrow failure syndromes caused predominantly by pathogenic germline variants in DNA-repair, ribosomal, or telomere-biology genes.11 Somatic copy neutral loss of heterozygosity in chromosome 6 (chr6-CNLOH) encompassing the HLA-class I locus has been previously described in acquired SAA.12,13 Hematopoietic stem cells with chr6-CNLOH are thought to escape the cytotoxic T cell immune attack by deleting HLA alleles involved in autoantigen presentation.14
The cytopenias in aplastic anemia may progress to a life-threatening severe disease, and affected individuals are at high risk of progression to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).15 In acquired SAA, allogeneic hematopoietic cell transplantation (HCT) is the first line of therapy for young individuals with a matched sibling donor, while IST, followed by HCT in non-responders, is recommended for all others.2,16,17
We conducted a genome-wide association study (GWAS) of acquired SAA to agnostically evaluate the contribution of common germline SNPs to the etiology of this highly morbid disease. Individuals with SAA (n = 895) were identified from the Center for International Blood and Marrow Transplant Research (CIBMTR) database, and biorepository and germline DNA were collected prior to HCT between 1989 and 2015. CIBMTR is a research collaboration between the National Marrow Donor Program (NMDP) “Be The Match Registry” and the Medical College of Wisconsin; CIBMTR has more than 450 reporting HCT centers, and it represents one of the world’s largest databases and research repositories for HCT research. All individuals provided informed consent, and the use of the samples was approved by the NMDP Institutional Review Board (IRB-1991-0002). We excluded 93 individuals with inherited bone marrow failure based on clinical diagnoses reported to CIBMTR. Genotyping was conducted using the Illumina Infinium OmniExpress BeadChip array at the Cancer Genomics Research Laboratory (CGR) in the Division of Cancer Epidemiology and Genetics (DCEG) at the National Cancer Institute (NCI). The controls were derived from cancer-free subjects drawn from two cohort studies (the Prostate, Lung, Colon and Ovarian Cancer Prevention Trial [PLCO]18 and the American Cancer Society Cancer Prevention Study II [CPSII]19 previously scanned on the Illumina Omni 2.5M SNP microarray) as well as four other USA-based studies (Mayo Clinic Case-Control Study of Non-Hodgkin Lymphoma and Chronic Lymphocytic Leukemia [MAYO];20 NCI Surveillance, Epidemiology, and End Results Non-Hodgkin Lymphoma [NHL] Case-Control Study [NCI-SEER];21,22 Women’s Health Initiative [WHI];23 and the Population-based Case-Control Study in Connecticut Women [YALE]24) which were also scanned on the Infinium OmniExpress chip. In order to reduce the effect of population stratification, population substructure analyses using STRUCTURE and principal components analyses (PCA) were used to limit the study inclusion to individuals of European ancestry. The final analyses included 534 acquired SAA cases (359 in a discovery set and 175 in a validation set) and 2,455 controls (1,396 in the discovery set and 1,059 in the validation set). Individuals included in the validation set were independent from those in the discovery set, and all study participants were genetically proven to be unrelated. Additional details are available in Supplementary Methods, Table S1, and Figure S1A and S1B.
The median age at HCT of participants with SAA was 21 years (range = 1–77), 56% were male, and the median time between SAA diagnosis and HCT was 11 months (range = 0.1–318 months). The majority of SAA-affected individuals (87%) received an unrelated donor HCT; all individuals who received a matched sibling HCT were part of the validation cohort (Table S2).
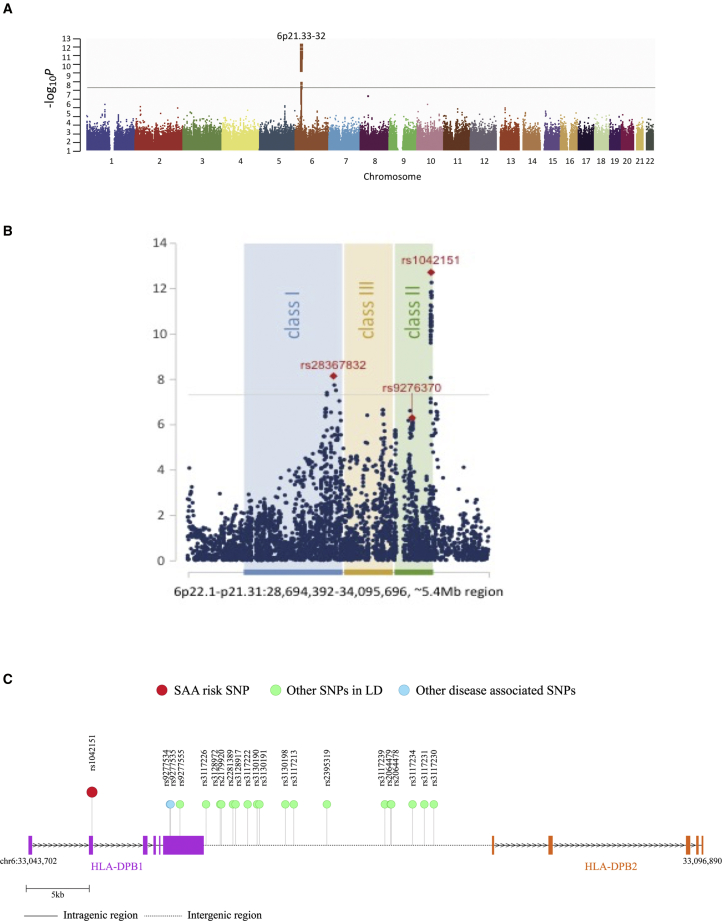
A series of strong association signals were identified across the human leukocyte antigen (HLA) genes encoded in the major histocompatibility complex (MHC) on chromosome 6p21 (Figure 1). Notably, 12 genotyped SNPs achieved genome-wide significance (pooled-p < 3.46 × 10−8; odds ratios (ORs) ranging from 1.62–1.82) (Table 1, Figure 1, and Tables S3 and S4). SNP associations with HLA class II genes were evident in two distinct regions. The top SNP, rs1042151 (pooled-p = 1.94 × 10−13), was in linkage disequilibrium with 18 other SNPs (R2 > 0.5, chr6:33,048,661-33,064,605) that were in or near HLA-DPB1 and HLA-DPB2. A second locus in HLA class II, containing 13 linked SNPs (R2 > 0.9) in or near HLA-DQA2 and HLA-DQB2 (chr6:32,707,295-32,727,905) approached genome-wide significance (top SNP, rs9276370 T>G, pooled-p = 5.01 × 10−7, OR 1.41, 95% confidence interval [CI] 1.23–1.62).
Figure 1.
Genomic Locations of Single-Nucleotide Polymorphims Associated with Severe Aplastic Anemia
(A) Manhattan plot of genome-wide association result for combined discovery and validation sets of European ancestry severe aplastic anemia cases and genomically matched controls.
(B) A portion of the Manhattan plot of the major histocompatibility complex (MHC) genomic region showing locations of SNPs in human leukocyte antigen (HLA) genes by HLA class.
(C) Schematic of HLA-DPB1 with SNPs of interest noted.
Table 1.
Summary of combined association results for severe aplastic anemia cases and controls
| dbSNP | Gene | Locus | Position | Variant | Stage | Cases | Controls | Ip | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| rs1042151 | |||||||||
| rs1042151 | HLA-DPB1 | 6p21 | chr6: 33048661 | A > G | Discovery | 356 | 1396 | 1.68E-10 | 1.82 (1.51-2.20) |
| rs1042151 | HLA-DPB1 | 6p21 | chr6: 33048661 | A > G | Validation | 175 | 1059 | 0.0015 | 1.52 (1.17-1.97) |
| rs1042151 | HLA-DPB1 | 6p21 | chr6: 33048661 | A > G | Combined | 531 | 2453 | 1.94E-13 | 1.75 (1.50-2.03) |
| rs28367832 | |||||||||
| rs28367832 | HLA-B | 6p21 | chr6: 31305731 | G > A | Discovery | 355 | 1388 | 3.05E-06 | 1.49 (1.27-1.79) |
| rs28367832 | HLA-B | 6p21 | chr6: 31305731 | rs28367832 | Validation | 173 | 1048 | 0.001 | 1.46 (1.16-1.84) |
| rs28367832 | HLA-B | 6p21 | chr6: 31305731 | rs28367832 | Combined | 528 | 2434 | 7.27E-09 | 1.49 (1.22-1.78) |
SNPs shown are those with combined validation and discovery p < 10−8
The top SNP associated with SAA, rs1042151 A>G (combined OR 1.75, 95% CI 1.50–2.03, p = 1.94x10−13), encodes a nonsynonymous change of p.Met76Val (RefSeq accession number NP_002112.3) in the β-1 domain of HLA-DPB1. While dbSNP annotates rs1042151 as p.Met105Val because it includes the first 30 residues of the leader peptide, p.Met76Val is the appropriate numbering according to the WHO Nomenclature Committee for Factors of the HLA System (IPD-IMGT/HLA database, see Web Resources).25 rs1042151 A>G has a minor allele frequency (MAF) of 17.5% in the non-Finnish European, 8.9% in the East Asian, and 37.6% in the African/African American gnomAD populations.26 In our study participants, the MAF of rs1042151 A>G (p.Met76Val) was 18.8% in controls and 29.4% in the individuals with SAA.
DPB1 comprises the beta strand of the HLA-DP protein heterodimer located on the cell surface of antigen-presenting cells.27 Genetic variation in HLA-DPB1 has been associated with several autoimmune diseases, including rheumatoid arthritis, Grave’s disease, and multiple sclerosis.28, 29, 30 Specifically, the p.Met76Val amino acid substitution caused by the rs1042151 G variant has previously been associated with risk of aspirin-exacerbated respiratory disease.31 Residue 76 is involved in peptide binding, forming part of the P4 pocket, and the p.Met76Val substitution is likely to influence the binding repertoire.32 Two other SNPs (rs9277534 and rs9277535), located in the 3′ UTR of HLA-DPB1, approximately 6 kilobases (kb) away from rs1042151 but not genotyped in our study (Figure 1C), have been previously associated with expression of HLA-DPB1, risk and persistence of hepatitis B virus (HBV) infection, and risk of graft-versus-host disease in HCT recipients.33, 34, 35, 36
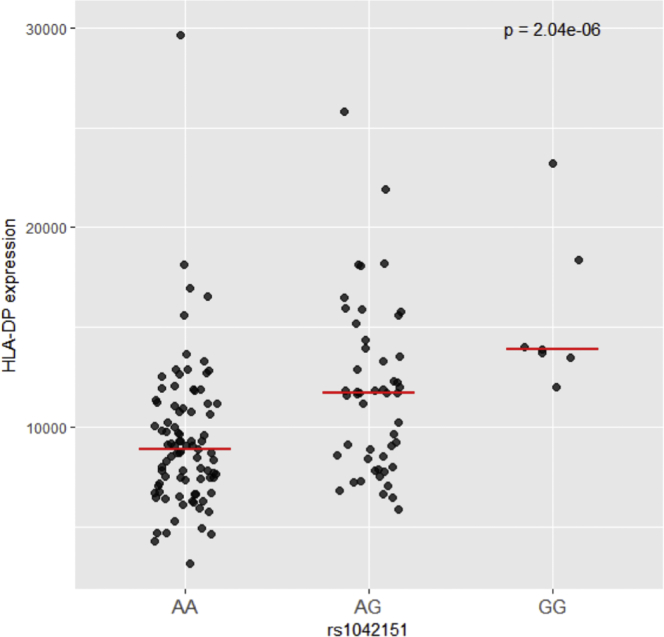
To evaluate the possible contribution of HLA-DP cell surface expression related to the association between the rs1042151 A>G genotypes and risk of SAA, we stained CD19+ cells isolated from peripheral blood drawn from 175 healthy donors with the HLA-DP-specific monoclonal antibodies B7/21 and BrafB637,38 as described previously33 (see Supplementary Methods in Supplemental Data), and we compared the median fluorescence intensity across genotypes. The risk variant, G, was associated with a significant dose-response increase in HLA-DP cell surface expression on the CD19+ cell surface (p = 2.04 × 10−6) (Figure 2); this association suggests that higher cell surface expression of HLA-DP may contribute to SAA risk. However, because of the strong linkage disequilibrium across this region, it is likely that the identified risk SNP is not solely related to cell surface expression of DP. A variant previously associated with HLA-DP cell surface expression, rs9277534 A>G,33 is also associated with SAA in our study (OR 1.64, p = 8.2 × 10−8). Additionally, rs1042151 is not the strongest HLA-DP eQTL, but based on GTExPortal (data accessed November 2019, see Web Resources),39 it is associated with the transcript level of HLA-DPB1 in transformed fibroblasts with a p value of 1 × 10−9.
Figure 2.
Cell Surface Expression of HLA-DP is Associated with rs1042151 Genotype
The cell surface expression levels of specific HLA alleles were determined in healthy individuals and plotted based on their rs1042151 genotype (see Online Methods).
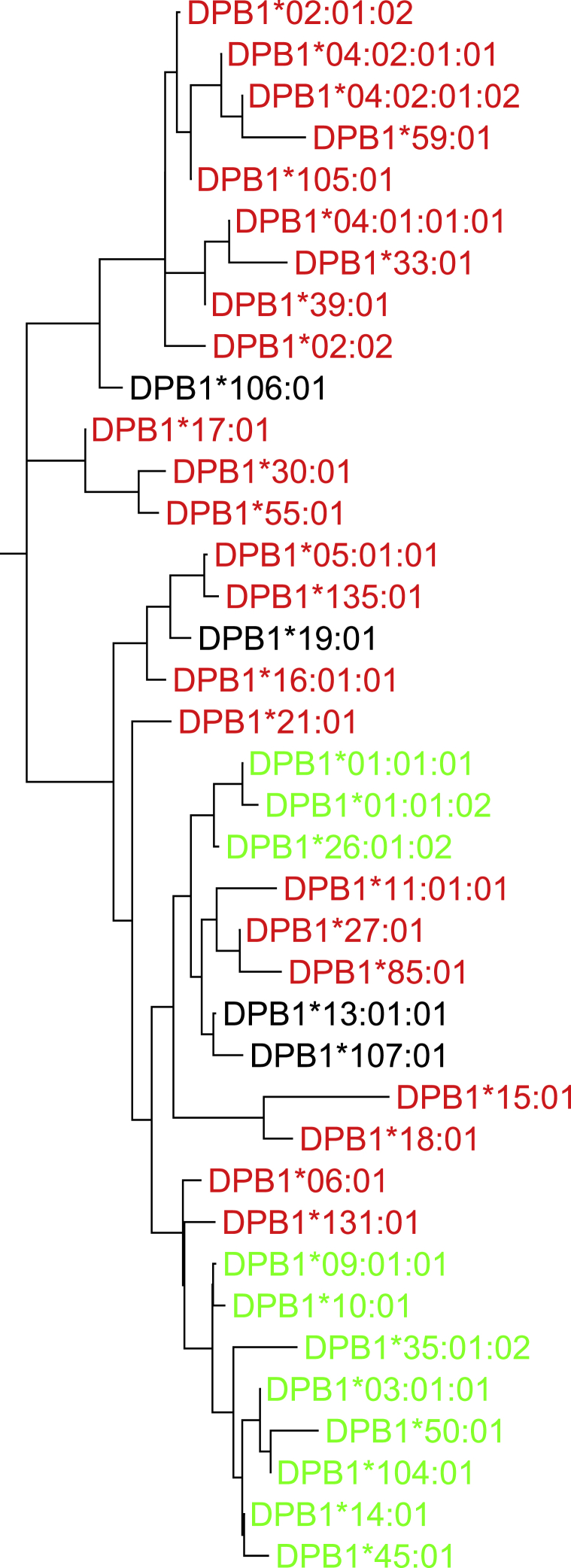
To further understand the role of HLA-DPB1 in SAA etiology, we constructed a phylogenic tree of HLA-DPB1 through the use of the neighbor-joining method based on Kimura 2-parameter distances as implemented in MEGA7 26 to visualize the distribution pattern of HLA-DP sequences as marked by Val or Met at position 76. Our results, based on both coding regions (Figure 3) and full-length gene sequences (not shown), indicate that 76Val is not monophyletic and is found in multiple distinct clades of alleles which are likely to recognize different peptide binding motifs. We then used the HIBAG software40 to impute HLA-DPB1 alleles for SAA cases and controls; comparisons with high-resolution clinical typing were available for 401 individuals with SAA and showed 92.7% concordance. SAA excess risk was associated with the HLA-DPB1 alleles containing the rs1042151 Val76 variant: DPB1∗03:01 (OR 1.66, p = 1.52 × 10−7), DPB1∗10:01 (OR 2.12, p = 0.0003), and DPB1∗01:01 (OR 1.60, p = 0.0008). On the other hand, an inverse association was present between the Met76-encoding alleles DPB1∗04:01 (OR = 0.78, p = 0.01) and DPB1∗04:02 (OR = 0.75, p = 0.02) (Table S5). Logistic regression analysis based upon the number of Val76 residues shows an additive effect; compared to zero copies of Val76-encdig alleles, two copies are associated with higher SAA risk (OR 2.32, p = 3.2 × 10−6) than one copy is (OR = 1.98, p = 1.2 × 10−11). Peptide repertoire specificity is not determined by the P4 pocket alone, and the phylogenetic distribution of Val76 (i.e., interspersed with Met76-encoding alleles) indicates that Val76 occurs on wide variety of antigen-binding groove configurations. Taken together, the data suggest that the p.Met76Val change might influence risk of SAA through a mechanism involving DP peptide binding specificity, HLA-DPB1 cell surface expression levels, and/or other factors affecting DP function. For example, the P4 pocket of DRB1 has been shown to facilitate immunogenic metabolite binding,41 a scenario that may hold true for DP, as well, resulting in risk of immune-mediated diseases such as SAA.
Figure 3.
Phylogenic Tree of Coding Regions of HLA-DPB1
The HLA-DBP1 alleles are color coded according to the amino acid at residue 76 as follows: red, methionine, M; black, isoleucine, I; green, valine, V. The neighbor-joining tree is based on Kimura’s 2-parameter distances for the entire 777 base pair coding region.
One SNP in HLA class I, rs28367832 G>A, reached genome-wide statistical significance only in the combined analysis (pooled-p = 7.27 × 10−9, OR 1.49, 95% CI 1.22–1.78) (Table 1, Table S4). rs28367832 G>A is 15.9 kb telomeric of HLA-B and a common noncoding SNP with a MAF of 57.7% in the SAA cases compared with 47.5% of control in the combined analyses. This SNP was not in linkage equilibrium with our study top SNP rs1042151 (R2 = 0.0099). The association remained statistically significant in conditional analysis (p for rs28367832 = 1.82 × 10−8). Because rs28367832 G>A is located in the region possibly affected by chr6-CNLOH, we calculated the Log R ratio (LRR) and B allele frequency (BAF) in order to assess somatic chr6-CNLOH in our samples (see Figure S2 and Supplementary Methods in Supplemental Data). There were 46 individuals with SAA (8.6%) with somatic chr6-CNLOH affecting class I HLA genes (Figure S2). Next-generation sequencing of the HLA genes performed with the MIA FORA NGS kit (Immucor) confirmed these SNP array results (see Figure S3 and Supplementary Methods in Supplemental Data). Somatic chr6-CNLOH appears to be specific to SAA; none of the 2,453 controls in this study nor of 73 the individuals with Fanconi anemia in our prior work had this alteration.42 Persons with SAA and somatic chr6-CNLOH were more likely to carry the rs28367832 AA genotype, and this result suggests loss of the common rs28367832 G variant (56.1% versus 32.9%, p = 0.003). However, exclusion of SAA individuals with somatic chr6-CNLOH from the analyses did not change the germline SNP association results (data not shown).
Several additional non-HLA loci did not reach the accepted statistical genome-wide association threshold of p < 5x10−8 (Table S6). An inverse association was present between SAA and the A allele of rs7845664 on chromosome 8p11.21, a multi-allelic SNP within the ZMAT4 gene (OR 0.70, p = 3.07 × 10−7). There are little data on ZMAT4 function, but one study previously identified copy number variants (CNVs) and gene expression changes in hematologic malignancies.43 We did not identify CNVs in this region in our cohort of individuals with SAA (data not shown). Four additional non-HLA loci contained SNPs with borderline p values that may warrant follow-up in a larger study: rs12753487 (OR 1.75, pooled-p = 2.85 × 10−6), rs1731229 (OR 0.72, pooled-p = 5.26 × 10−6), rs4345355 (OR 1.54, pooled-p = 5.29 × 10−6), and rs9533317 (OR 1.4, pooled-p = 7.87 × 10−6). Of these, the intragenic rs9533317 SNP is located within EPSTI1, a gene with functions in epithelial stromal interactions.44 These results may warrant follow-up because they may identify a set of individuals in whom SAA is not immune mediated.
This GWAS focused on individuals with SAA who required HCT. Our data suggest that idiopathic SAA may be due, in part, to germline susceptibility due to common variants in HLA class II, and possibly class I. It remains possible that some of the identified loci are associated with disease severity or lack of response to IST rather than with the etiology of SAA because all cases required HCT for their disease. However, the agreement between results from the discovery (all received unrelated donor HCT, mostly after failing IST) and validation sets (40% received HCT from matched sibling as a first line of therapy and did not receive IST) argues against results being associated primarily with IST response. The small sample size in this study is small in the scope of GWAS but very large in the context of a rare disease. We limited our analysis to individuals of European ancestry; analyses in populations of other racial backgrounds are of interest, particularly because SAA is more common in Asians.45
In summary, this GWAS of acquired SAA identified a strong association at rs1042151 A>G in the HLA-DPB1. The rs1042151 G allele encoding HLA-DPB1 Val76 occurs on three distinct alleles, DPB1∗03:01, DPB1∗10:01, and DPB1∗01:01, and this suggests that alteration of HLA-DP peptide binding specificity, as well as changes in HLA-DP cell surface expression and/or other factors affecting HLA-DP function, contribute to SAA etiology. A prospective study including the full spectrum of disease severity is warranted in order to further understand the association of HLA germline SNPs in SAA etiology.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI). The Center for International Blood and Marrow Transplant Research (CIBMTR) is supported primarily by Public Health Service Grant/Cooperative Agreement 5U24CA076518 from the NCI, the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); Grant/Cooperative Agreement 4U10HL069294 from the NHLBI and the NCI; contract HHSH250201200018C with the Health Resources and Services Administration (HRSA/DHHS); and two grants, N00014-17-1-2388 and N0014-17-1-2850, from the Office of Naval Research. The Mayo Clinic Case-Control Study of Non-Hodgkin Lymphoma and Chronic Lymphocytic Leukemia (MAYO) study is funded through grants R01 CA92153 and P50 CA97274. The NCI Surveillance, Epidemiology, and End Results Non-Hodgkin Lymphoma Case-Control Study (NCI-SEER) study is funded by the Intramural Research Program of the NCI, National Institutes of Health (NIH), and Public Health Service (N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010, and N02-PC-71105). The Population-based Case-Control Study in Connecticut Women (YALE) is funded through the NCI (CA62006 and CA165923). This project has also been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. V.R. is supported by the African Academy of Sciences (AAS) and the Royal Society, which is funded by the UK Government as part of the Global Challenge Research Fund (GCRF); and V.R. is also supported by the South African Medical Research Council (SAMRC) with funds from the Department of Science and Technology (DST). V.R. is funded in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a Developing Excellence in Leadership Training and Science (DELTAS) Africa Initiative (grant # DEL-15-006) by the AAS.
Published: January 30, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.01.004.
Accession Numbers
GWAS data will be available through the dbGAP-controlled access database accession number dbGaP: phs001710.v1.p1.
Web Resources
GTExPortal, https://www.gtexportal.org/home/gene/DPB1
Immuno Polymorphism Database—International ImMunoGeneTics project HLA (IPD-IMGT/HLA), https://www.ebi.ac.uk/ipd/imgt/hla/
National Organization for Rare Disorders (NORD) Rare Disease Information on Acquired Aplastic Anemia, https://rarediseases.org/rare-diseases/acquired-aplastic-anemia/
Supplemental Information
References
- 1.Luzzatto L., Risitano A.M. Advances in understanding the pathogenesis of acquired aplastic anaemia. Br. J. Haematol. 2018;182:758–776. doi: 10.1111/bjh.15443. [DOI] [PubMed] [Google Scholar]
- 2.Boddu P.C., Kadia T.M. Updates on the pathophysiology and treatment of aplastic anemia: a comprehensive review. Expert Rev. Hematol. 2017;10:433–448. doi: 10.1080/17474086.2017.1313700. [DOI] [PubMed] [Google Scholar]
- 3.Savage S.A., Calado R.T., Xin Z.T., Ly H., Young N.S., Chanock S.J. Genetic variation in telomeric repeat binding factors 1 and 2 in aplastic anemia. Exp. Hematol. 2006;34:664–671. doi: 10.1016/j.exphem.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Takaku T., Calado R.T., Kajigaya S., Young N.S. Interleukin-23 receptor (IL-23R) gene polymorphisms in acquired aplastic anemia. Ann. Hematol. 2009;88:653–657. doi: 10.1007/s00277-008-0666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B., Guo L., Zhang Y., Xiao Y., Wu M., Zhou L., Chen S., Yang L., Lu X., Li Y. Molecular alterations in the TCR signaling pathway in patients with aplastic anemia. J. Hematol. Oncol. 2016;9:32. doi: 10.1186/s13045-016-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., Miao M., Qiu Y., Qin Z., Wang J., Jiang Y., Ming Z., Zhang X. Association between polymorphisms in PDCD1 gene and aplastic anemia in Chinese Han population. Leuk. Lymphoma. 2013;54:2251–2254. doi: 10.3109/10428194.2013.772605. [DOI] [PubMed] [Google Scholar]
- 7.Ming Z.J., Hui H., Miao M., Qiu Y.H., Zhang X.G. Polymorphisms in PDCD1 gene are not associated with aplastic anemia in Chinese Han population. Rheumatol. Int. 2012;32:3107–3112. doi: 10.1007/s00296-011-2127-0. [DOI] [PubMed] [Google Scholar]
- 8.Chang H., Zeng F., Zhang J.Y., Mu X.Y., Meng W.T., Ma H.B., Liu T. Association of the interferon-gamma single nucleotide polymorphism +874(T/A) with response to immunosuppressive therapy in patients with severe aplastic anemia. Blood Cells Mol. Dis. 2010;45:313–316. doi: 10.1016/j.bcmd.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Rehman S., Saba N., Naz M., Ahmed P., Munir S., Sajjad S., Tabassum S., Naseem L. Single-Nucleotide Polymorphisms of FAS and FASL Genes and Risk of Idiopathic Aplastic Anemia. Immunol. Invest. 2018;47:484–491. doi: 10.1080/08820139.2018.1458106. [DOI] [PubMed] [Google Scholar]
- 10.In J.W., Lee N., Roh E.Y., Shin S., Park K.U., Song E.Y. Association of aplastic anemia and FoxP3 gene polymorphisms in Koreans. Hematology. 2017;22:149–154. doi: 10.1080/10245332.2016.1238645. [DOI] [PubMed] [Google Scholar]
- 11.West A.H., Churpek J.E. Old and new tools in the clinical diagnosis of inherited bone marrow failure syndromes. Hematology (Am. Soc. Hematol. Educ. Program) 2017;2017:79–87. doi: 10.1182/asheducation-2017.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katagiri T., Sato-Otsubo A., Kashiwase K., Morishima S., Sato Y., Mori Y., Kato M., Sanada M., Morishima Y., Hosokawa K., Japan Marrow Donor Program Frequent loss of HLA alleles associated with copy number-neutral 6pLOH in acquired aplastic anemia. Blood. 2011;118:6601–6609. doi: 10.1182/blood-2011-07-365189. [DOI] [PubMed] [Google Scholar]
- 13.Betensky M., Babushok D., Roth J.J., Mason P.J., Biegel J.A., Busse T.M., Li Y., Lind C., Papazoglou A., Monos D. Clonal evolution and clinical significance of copy number neutral loss of heterozygosity of chromosome arm 6p in acquired aplastic anemia. Cancer Genet. 2016;209:1–10. doi: 10.1016/j.cancergen.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaimoku Y., Takamatsu H., Hosomichi K., Ozawa T., Nakagawa N., Imi T., Maruyama H., Katagiri T., Kishi H., Tajima A. Identification of an HLA class I allele closely involved in the autoantigen presentation in acquired aplastic anemia. Blood. 2017;129:2908–2916. doi: 10.1182/blood-2016-11-752378. [DOI] [PubMed] [Google Scholar]
- 15.Shimamura A. Aplastic anemia and clonal evolution: germ line and somatic genetics. Hematology (Am. Soc. Hematol. Educ. Program) 2016;2016:74–82. doi: 10.1182/asheducation-2016.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacigalupo A. How I treat acquired aplastic anemia. Blood. 2017;129:1428–1436. doi: 10.1182/blood-2016-08-693481. [DOI] [PubMed] [Google Scholar]
- 17.Scheinberg P. Recent Advances and Long-Term Results of Medical Treatment of Acquired Aplastic Anemia: Are Patients Cured? Hematol. Oncol. Clin. North Am. 2018;32:609–618. doi: 10.1016/j.hoc.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 19.Amundadottir L., Kraft P., Stolzenberg-Solomon R.Z., Fuchs C.S., Petersen G.M., Arslan A.A., Bueno-de-Mesquita H.B., Gross M., Helzlsouer K., Jacobs E.J. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerhan J.R., Fredericksen Z.S., Wang A.H., Habermann T.M., Kay N.E., Macon W.R., Cunningham J.M., Shanafelt T.D., Ansell S.M., Call T.G. Design and validity of a clinic-based case-control study on the molecular epidemiology of lymphoma. Int. J. Mol. Epidemiol. Genet. 2011;2:95–113. [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee N., Hartge P., Cerhan J.R., Cozen W., Davis S., Ishibe N., Colt J., Goldin L., Severson R.K. Risk of non-Hodgkin’s lymphoma and family history of lymphatic, hematologic, and other cancers. Cancer Epidemiol. Biomarkers Prev. 2004;13:1415–1421. [PubMed] [Google Scholar]
- 22.Wang S.S., Cerhan J.R., Hartge P., Davis S., Cozen W., Severson R.K., Chatterjee N., Yeager M., Chanock S.J., Rothman N. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;66:9771–9780. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 23.Anderson G.L., Manson J., Wallace R., Lund B., Hall D., Davis S., Shumaker S., Wang C.Y., Stein E., Prentice R.L. Implementation of the Women’s Health Initiative study design. Ann. Epidemiol. 2003;13(9, Suppl):S5–S17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Hughes K.J., Zahm S.H., Zhang Y., Holford T.R., Dai L., Bai Y., Han X., Qin Q., Lan Q. Genetic variations in xenobiotic metabolic pathway genes, personal hair dye use, and risk of non-Hodgkin lymphoma. Am. J. Epidemiol. 2009;170:1222–1230. doi: 10.1093/aje/kwp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P., Marsh S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020;48(D1):D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burek Kamenaric M., Maskalan M., Grubic Z., Mikulic M., Serventi Seiwerth R., Durakovic N., Vrhovac R., Stingl Jankovic K., Zunec R. HLA-DPB1 matching in unrelated hematopoietic stem cell transplantation program contributes to a higher incidence of disease relapse. Hum. Immunol. 2017;78:665–671. doi: 10.1016/j.humimm.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Orozco G., Barton A., Eyre S., Ding B., Worthington J., Ke X., Thomson W. HLA-DPB1-COL11A2 and three additional xMHC loci are independently associated with RA in a UK cohort. Genes Immun. 2011;12:169–175. doi: 10.1038/gene.2010.57. [DOI] [PubMed] [Google Scholar]
- 29.Field J., Browning S.R., Johnson L.J., Danoy P., Varney M.D., Tait B.D., Gandhi K.S., Charlesworth J.C., Heard R.N., Australia and New Zealand Multiple Sclerosis Genetics Consortium A polymorphism in the HLA-DPB1 gene is associated with susceptibility to multiple sclerosis. PLoS ONE. 2010;5:e13454. doi: 10.1371/journal.pone.0013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y., Momozawa Y., Ashikawa K., Kanai M., Matsuda K., Kamatani Y., Takahashi A., Kubo M. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet. 2015;47:798–802. doi: 10.1038/ng.3310. [DOI] [PubMed] [Google Scholar]
- 31.Park B.L., Kim T.H., Kim J.H., Bae J.S., Pasaje C.F., Cheong H.S., Kim L.H., Park J.S., Lee H.S., Kim M.S. Genome-wide association study of aspirin-exacerbated respiratory disease in a Korean population. Hum. Genet. 2013;132:313–321. doi: 10.1007/s00439-012-1247-2. [DOI] [PubMed] [Google Scholar]
- 32.Crivello P., Zito L., Sizzano F., Zino E., Maiers M., Mulder A., Toffalori C., Naldini L., Ciceri F., Vago L., Fleischhauer K. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2015;21:233–241. doi: 10.1016/j.bbmt.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Thomas R., Thio C.L., Apps R., Qi Y., Gao X., Marti D., Stein J.L., Soderberg K.A., Moody M.A., Goedert J.J. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J. Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersdorf E.W., Malkki M., O’hUigin C., Carrington M., Gooley T., Haagenson M.D., Horowitz M.M., Spellman S.R., Wang T., Stevenson P. High HLA-DP Expression and Graft-versus-Host Disease. N. Engl. J. Med. 2015;373:599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brien T.R., Kohaar I., Pfeiffer R.M., Maeder D., Yeager M., Schadt E.E., Prokunina-Olsson L. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 2011;12:428–433. doi: 10.1038/gene.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda H., Hiramatsu K., Akazawa Y., Nosaka T., Saito Y., Ozaki Y., Hayama R., Takahashi K., Naito T., Ofuji K. Genetic polymorphism and decreased expression of HLA class II DP genes are associated with HBV reactivation in patients treated with immunomodulatory agents. J. Med. Virol. 2018;90:712–720. doi: 10.1002/jmv.25011. [DOI] [PubMed] [Google Scholar]
- 37.Horejsí V., Chorváth B., Polákova K., Duraj J., Sedlák J., Karpatová M. Characterization of a new murine monoclonal antibody against human DP antigens. Tissue Antigens. 1988;32:6–11. doi: 10.1111/j.1399-0039.1988.tb01630.x. [DOI] [PubMed] [Google Scholar]
- 38.Watson A.J., DeMars R., Trowbridge I.S., Bach F.H. Detection of a novel human class II HLA antigen. Nature. 1983;304:358–361. doi: 10.1038/304358a0. [DOI] [PubMed] [Google Scholar]
- 39.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X., Shen J., Cox C., Wakefield J.C., Ehm M.G., Nelson M.R., Weir B.S. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra M.K., Damotte V., Hollenbach J.A. Structure-based selection of human metabolite binding P4 pocket of DRB1∗15:01 and DRB1∗15:03, with implications for multiple sclerosis. Genes Immun. 2019;20:46–55. doi: 10.1038/s41435-017-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Zhou W., Alter B.P., Wang T., Spellman S.R., Haagenson M., Yeager M., Lee S.J., Chanock S.J., Savage S.A., Gadalla S.M. Chromosomal Aberrations and Survival after Unrelated Donor Hematopoietic Stem Cell Transplant in Patients with Fanconi Anemia. Biol. Blood Marrow Transplant. 2018;24:2003–2008. doi: 10.1016/j.bbmt.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan J., Gao Y., Zhao X., Wu Q., Fu X., Shao Y., Yang H., Guan M., Yu B., Zhang W. The association between the copy-number variations of ZMAT4 and hematological malignancy. Hematology. 2011;16:20–23. doi: 10.1179/102453311X12902908411751. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y.H., Lee J.R., Hahn M.J. Regulation of inflammatory gene expression in macrophages by epithelial-stromal interaction 1 (Epsti1) Biochem. Biophys. Res. Commun. 2018;496:778–783. doi: 10.1016/j.bbrc.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 45.Kojima S. Why is the incidence of aplastic anemia higher in Asia? Expert Rev. Hematol. 2017;10:277–279. doi: 10.1080/17474086.2017.1302797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.