Abstract
We report an inborn error of metabolism caused by TKFC deficiency in two unrelated families. Rapid trio genome sequencing in family 1 and exome sequencing in family 2 excluded known genetic etiologies, and further variant analysis identified rare homozygous variants in TKFC. TKFC encodes a bifunctional enzyme involved in fructose metabolism through its glyceraldehyde kinase activity and in the generation of riboflavin cyclic 4′,5′-phosphate (cyclic FMN) through an FMN lyase domain. The TKFC homozygous variants reported here are located within the FMN lyase domain. Functional assays in yeast support the deleterious effect of these variants on protein function. Shared phenotypes between affected individuals with TKFC deficiency include cataracts and developmental delay, associated with cerebellar hypoplasia in one case. Further complications observed in two affected individuals included liver dysfunction and microcytic anemia, while one had fatal cardiomyopathy with lactic acidosis following a febrile illness. We postulate that deficiency of TKFC causes disruption of endogenous fructose metabolism leading to generation of by-products that can cause cataract. In line with this, an affected individual had mildly elevated urinary galactitol, which has been linked to cataract development in the galactosemias. Further, in light of a previously reported role of TKFC in regulating innate antiviral immunity through suppression of MDA5, we speculate that deficiency of TKFC leads to impaired innate immunity in response to viral illness, which may explain the fatal illness observed in the most severely affected individual.
Keywords: fructose metabolism, cyclic FMN, triokinase, TKFC, rapid genome sequencing, cataracts, cardiomyopathy, innate immunity, inborn error of metabolism, developmental delay
Main Text
Undiagnosed complex multisystem genetic disorders presenting to the pediatric intensive care unit pose a management conundrum. The need to identify (or exclude) a treatable underlying cause is of paramount importance and is a critical factor dictating management decisions. In recent years, next generation sequencing has increasingly been adopted in critically ill children,1 both to identify or exclude known treatable disorders as well as to discover novel disease-causing genes. Here we demonstrate the utility of rapid genome sequencing followed by functional studies to identify deficiency of TKFC, a bifunctional enzyme involved in fructose metabolism and the generation of riboflavin cyclic 4′,5′-phosphate (cyclic flavin mononucleotide, cFMN)2 as a cause of a fatal infantile multisystem disorder.
Affected individual 1-1 (P1-1) was the first child born to consanguineous (first cousin) parents of Gujarati ancestry at term. Concerns about eye movements led to the identification of bilateral congenital nuclear cataracts at 5 months of age. Bilateral lensectomy was performed. Subsequently she had developmental delay, with evidence of motor delay (cruising at 26 months, walking independently at 36 months but with a wide-based gait) and speech delay (babbling at 26 months, first words at 4 years, slowly improving understanding). Initial hearing assessments were normal, although there was possible low-frequency hearing impairment at 4 years of age. She had a simple febrile convulsion at 2 years of age but no other seizures and has continued to make slow developmental progress. General examination revealed microphthalmia but no specific dysmorphic features, and no organomegaly. Magnetic resonance imaging of the brain at 29 months revealed cerebellar hypoplasia, with mild delay of myelin maturation (Figure 1). Cardiac investigations including electro- and echocardiograms were normal and remained normal up to the most recent assessment at 4 years of age. Extensive metabolic investigations revealed no diagnostic abnormalities (Table S1).
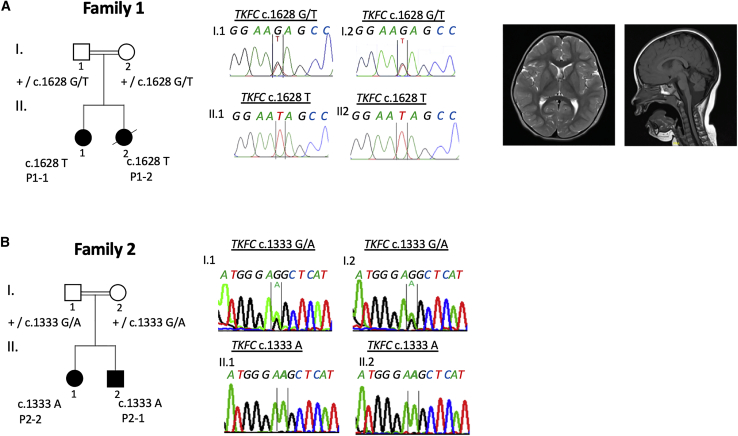
Figure 1.
Family Pedigrees, Bi-allelic TKFC Variants Observed in Affected Individuals and Brain MRI of P1-1
Representative MRI findings in affected individual P1-1 at 29 months indicating cerebellar hypoplasia. Homozygous TKFC variants segregating in each family were confirmed by Sanger Sequencing.
Her younger sister, affected individual 1-2 (P1-2), was diagnosed on day 1 of life with congenital cataracts, which were subsequently excised at 1 month of age. Poor feeding and difficult weight gain were noted from the neonatal period. After her first primary immunizations at 8 weeks, she developed vomiting and loose stool, and was subsequently admitted to hospital, where she was found to be hypotensive and tachypnoeic with hypoglycemia (1.6 mmol/L) and severe lactic acidosis (pH 7.15, lactate 16 mmol/L). Cardiomegaly was evident on a chest radiograph, and echocardiography demonstrated dilated cardiomyopathy with poor systolic function. Liver function was abnormal, with hypoalbuminemia (23 g/L, reference 32–52) and elevated plasma alanine transaminase (900 U/L, reference 10–25). Supportive treatment with intubation, ventilation, inotropic support, and fluid and base therapy led to improvement in the lactic acidosis. Cranial ultrasound demonstrated lenticostriatal vasculopathy, a nonspecific finding which can be seen with viral infections, lactic acidosis, and mitochondrial disease. Magnetic resonance imaging and angiography of the brain was normal at 2 months of age. Attempts at extubation led to deterioration in cardiac function and recurrence of lactic acidosis. Care was redirected and she died aged 11 weeks. Extensive metabolic investigations were unremarkable (Table S1) except for a markedly elevated fibroblast growth factor 21 (FGF21) level of 4,350 pg/mL (reference 44–1,515).
An open muscle biopsy was performed to investigate the possibility of a mitochondrial disorder, since lactic acidaemia, cataracts, cardiomyopathy, and multi-system critical illness are well-recognized features of pediatric mitochondrial diseases. The elevated level of FGF21, which has been postulated as a biomarker of mitochondrial disease,3 also indicated the possibility of an underlying mitochondrial disorder. Muscle histology did not demonstrate any characteristic features of mitochondrial disease, such as ragged-red or cytochrome oxidase-negative fibers. On electron microscopy, mitochondria appeared slightly enlarged but with a normal morphology. There was a moderate excess of lipid and glycogen content. Blood vessel endothelial and smooth muscle cells contained no atypical inclusions. Respiratory chain enzyme activities in muscle, expressed as a ratio to citrate synthase as a mitochondrial marker enzyme, were essentially within their reference ranges although there was a mild reduction of complex IV activity: complex I 0.251 (reference 0.104–0.268), complex II+III 0.111 (0.04–0.204), and complex IV 0.012 (0.014–0.034).
In view of the critical illness of P1-2 and urgent need to identify any treatable cause of her life-threatening condition, rapid trio genome sequencing of the affected individual and her parents was performed in the RaPS project1 in tandem with muscle biopsy, array CGH, and mitochondrial DNA (mtDNA) analysis. Array CGH was normal (other than extended regions of homozygosity, as expected from the pedigree) and mtDNA analysis showed no large-scale rearrangements in blood and normal mtDNA sequence in muscle. Written consent was obtained from the parents of P1-1 and P1-2 to perform trio genome sequencing (of P1-2 and parents) on a research basis under ethics number 08/H0713/82 approved by the NHS Health Research Authority NRES Committee of London Bloomsbury. For research-based whole-genome sequencing (see Supplemental Information for detailed methods), genomic DNA (gDNA) libraries were prepared using Illumina TruSeq DNA PCR-Free Library Prep (Illumina) following the manufacturer’s advice starting with 1 μg of sheared gDNA. Parental samples were pooled at equimolar concentrations and sequenced on an Illumina NextSeq 550 in High-Output Mode (29 h). The sample from individual 1-2 was sequenced on an Illumina HighSeq 2500 Dual Flow Cell, Rapid Run Mode (27 h). Mapping and variant calling were performed using a Genalice appliance running Genalice Map 2.5.5 including Mapping, Variant Calling, and the Population Calling module for trio analysis (Genalice Core BV). Genalice default configuration files were used for WGS mapping and trio variant detection. Ingenuity Variant Analysis software (QIAGEN) was used for variant filtering. No potentially pathogenic variants were detected in any known disease genes associated with primary mitochondrial disorders or other inborn errors of metabolism, cardiomyopathy, or intellectual disability. The filtering pipeline prioritized a homozygous variant c.1628G>T (p.Arg543Ile) in TKFC (DAK, ENSG00000149476, GenBank: NM_015533.3, MIM: 615844) encoding triokinase/FMN cyclase.
Using Genematcher4 we were able to identify an unrelated family with two affected individuals who also had biallelic variants in TKFC, identified by exome sequencing. Affected individual 2-2 (P2-2), a boy, is the second child of consanguineous parents of Turkish ancestry. Pregnancy and birth history, anthropometric data at birth, and the neonatal course were all unremarkable. Poor weight gain and diarrhea were noted from early infancy. Dietary exclusion of cow’s milk protein was not associated with any clinical improvement. The affected individual had oral hypersensitivity and did not tolerate introduction of solid foods. At 22 months he was noted to have global developmental delay and ophthalmological evaluation revealed bilateral cataracts. Lens extraction/replacement was performed. Based upon the exome sequencing results, a fructose- and sucrose-free diet was introduced (via gastrostomy in view of oral hypersensitivity) but did not lead to any improvement in weight gain or the diarrhea. Subsequently, the diarrhea worsened and he developed progressive non-cholestatic liver failure with fatty degeneration of the liver. He has been dependent on parenteral feeding since the age of 34 months. His clinical course has been complicated by an episode of pancreatitis. Several attempts to reintroduce significant amounts of enteral feeding failed, and currently he tolerates only 5 × 40 mL of an amino acid-based formula. At 3 years 10 months, weight (10.1 kg) and height (87 cm) are both below the first percentile. He has hepatomegaly but no splenomegaly. He cannot walk independently and has no words. An older sister, affected individual P2-1, has delayed speech development and learning difficulties. She is otherwise well, and ophthalmological examination did not show cataracts. See Table S1 for results of biochemical and hematological investigations. As the affected individual P2-1 is relatively well, she has not had detailed metabolic investigations.
Exome sequencing and variant prioritizing was performed in the index case subject P2-2, his sister P2-1, and parents as reported previously.5 Studies in family 2 were approved by the Ethics Committee of the Land Salzburg (number 415-E/2552/10-2019). A known homozygous variant c.941C>A (p.Pro314His) in PAH (GenBank: NM_000277.1; MIM: 612349) was identified in both children, who were both known to have mild hyperphenylalaninemia identified by newborn screening. This variant has been reported as a cause of benign hyperphenylalaninemia (MIM: 261600)6 but does not explain the additional phenotypes observed in family 2. We therefore searched for other potentially disease-causing gene variants and a homozygous variant c.1333G>A (p.Gly445Ser) was identified in TKFC in P2-1 and P2-2. Biallelic variants in TKFC were confirmed by Sanger sequencing and segregated with disease in both families (Figure 1). Genetic evidence for pathogenicity of these variants included the segregation data and the overlapping phenotypes in two unrelated families from distinct geographical regions. In silico predictions for both TKFC variants are supportive of a deleterious effect (Table 1). The c.1628G>T variant in family 1 is present in the gnomAD database at a very low frequency (2.40E−05) with no homozygous variant reported. The c.1333G>A variant is not present in gnomAD.
Table 1.
Details of TKFC Homozygous Variants Identified in Affected Members of Families 1 and 2
| Variant Details | Family 1 | Family 2 |
|---|---|---|
| Position (GRCh37) | 11:61113875 | 11:61112824 |
| Canonical transcript | NM_015533.3 | NM_015533.3 |
| cDNA change | c.1628G>T | c.1333G>A |
| Protein change | p.Arg543Ile | p.Gly445Ser |
| In Silico Predictions | ||
| SIFT prediction/score | deleterious/0 | deleterious/0.01 |
| PolyPhen prediction/score | probably damaging/1 | probably damaging/0.99 |
| Mutation taster prediction/score | disease causing/1 | disease causing/1 |
| PROVEN prediction/score | damaging/−7.11 | damaging/−5.59 |
| Condel prediction/score | deleterious/0.945 | deleterious/0.858 |
| CADD PHRED score | 32 | 32 |
| Minor Allele Frequency | ||
| gnomAD | 2.40E−05 | not in gnomAD |
| homozygous allele | none | none |
| SNP/variant accession number | dbSNP: rs547013163 | – |
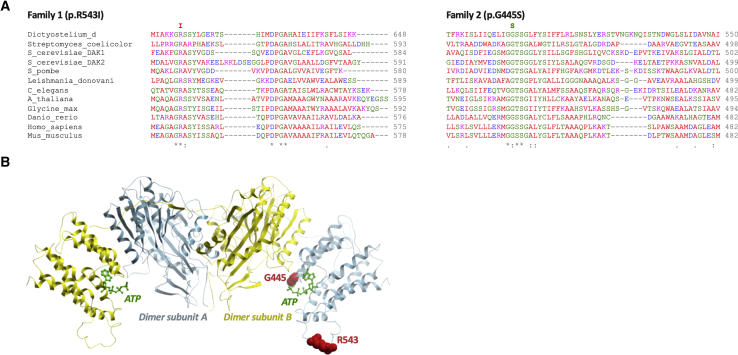
TKFC encodes a bifunctional protein that has been annotated as a homodimeric triokinase and FMN cyclase.2 Triokinase (EC 2.7.1.28) is a component of the fructose metabolism pathway first described by Hers and is responsible for the ATP-dependent phosphorylation of D-glyceraldehyde and exogenous dihydroxyacetone (DHA).7,8 Both identified variants affect evolutionarily conserved amino acid residues of TKFC (Figure 2A). Site-directed mutagenesis experiments localized amino acid residues 1–339 to the DHA kinase (K) domain and residues 359–575 to the FMN lyase (L) domain, connected by a linker region and assembling into a functional homodimer.2 Thus, the variants in families 1 and 2 affect the L domain. The variants observed in the affected individuals were modeled on the crystal structure of the Citrobacter freundii DHA kinase, which has structural and functional similarities to human TKFC (Figure 2B).9 The arginine residue at position 543 is located at the surface of the enzyme, and this position is always occupied by a basic residue (almost invariably Arg) among orthologs. Although its precise function is not known, substitution by isoleucine is likely to have impact. The glycine at 445 is close to an ATP/flavin adenine dinucleotide (FAD) binding site and substitution to serine may affect ATP/FAD binding.
Figure 2.
Multiple Sequence Alignment of TKFC Showing Evolutionary Conservation of Affected Amino Acids, and In Silico Modeling of TKFC Dimer
(A) Multiple sequence alignment of TKFC among different representative species and location of homozygous variants within TKFC domains. Homozygous protein variants detected in family 1 (p.Arg543Ile) and family 2 (p.Gly445Ser) are indicated and shown to affect highly conserved amino acid residues.
(B) Dimer of TKFC subunits (with K domain shown in yellow and L domain in blue), based on in silico modeling of human TKFC structure.2 The homozygous TKFC variants identified in the two families are located within the FMN lyase (L) domain (blue). For simplicity, the two mutation sites (red spheres) are shown only for the dimer subunit A. The ATP ligands from both subunits are represented in green sticks. The p.Gly445Ser variant may moderately affect ATP/FAD binding site due to its close proximity. The p.Arg453Ile variant may have a more severe effect due to predominant preference for only Arg among orthologs.
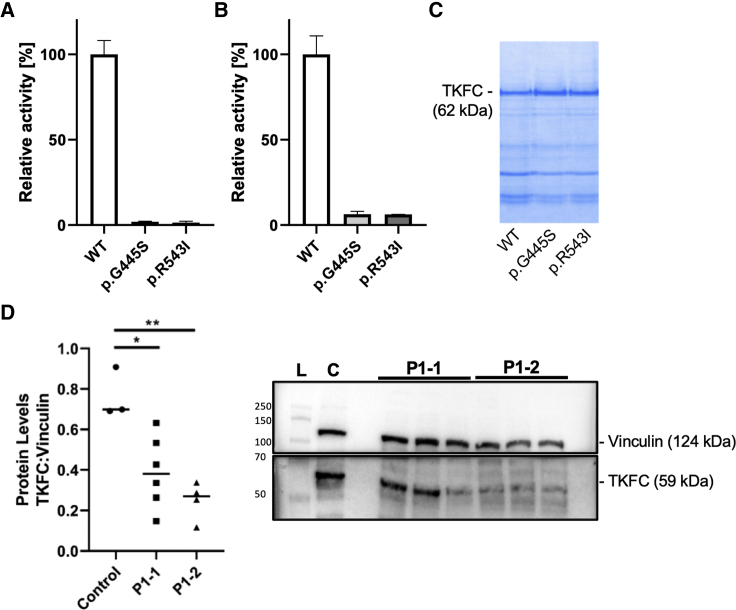
In order to quantify the enzyme activities, recombinant human TKFC proteins were expressed and purified from Escherichia coli (for details see Supplemental Information). TKFC activity was quantified using a coupled spectrophotometric assay and either D-glyceraldehyde or dihydroxyacetone as substrates.2 As shown in Figure 3A the TKFC activities of p.Gly445Ser and p.Arg543Ser were reduced to 1.92% ± 0.31% and 1.45% ± 0.75% of wild-type, respectively, when using D-glyceraldehyde as substrate. When using dihydroxyacetone as substrate, the relative activities of p.Gly445Ser and p.Arg543Ser were 6.33% ± 1.67% and 6.22% ± 0.04% of wild-type, respectively (Figure 3B). Equal amounts of protein were used for this enzyme activity assay (Figure 3C). In addition, the TKFC homozygous variant in affected individuals P1-1 and P1-2 (family 1) results in significantly reduced protein levels in both individuals as indicated in the western blot showing reduced protein content in cultured skin fibroblasts (Figure 3D). Taken together, these results confirm the deleterious effect of these variants on protein function.
Figure 3.
Enzyme Activity of Recombinant Human TKFC Protein Expressed and Purified from Escherichia coli, and Western Blot of TKFC in Subject Fibroblasts
(A–C) Enzyme activity was measured using either 10 mmol/L D-glyceraldehyde (A) or 10 mmol/L dihydroxyacetone (B) as substrate. Equal amounts of recombinant protein was used for activity assays as adjusted by polyacrylamide electrophoresis (representative gel in C). Bars show average activity, error bars standard deviation. Recombinant protein was isolated in three replicates in case of wild-type (WT) and the p.Gly445Ser variant and in two replicates for the p.Arg543Ile variant.
(D) TKFC protein studies in family 1 show significant reduction of TKFC protein levels in P1-1 and P1-2 compared to metabolic disease control subjects. Protein levels were calculated as a ratio of TKFC levels to vinculin levels. Data are expressed as median with individual data points. One-way ANOVA with Tukey’s multiple comparisons test post hoc was performed to determine significance as indicated by ∗p < 0.05 and ∗∗p < 0.01. Representative western blot shows lane 1, protein ladder (L); lane 2, metabolic control (C); lanes 4–6, affected individual P1-1; lanes 7–9: affected individual P1-2.
TKFC is widely expressed with highest expression noted in the liver and small intestine. This is consistent with a role of TKFC in fructose metabolism, since liver and small intestine are the main fructose-metabolizing tissues.10 Functional relevance of observed high expression of TKFC in the adrenal glands is less straightforward to explain, but fructose metabolism has been implicated in corticosteroid hormone production.11
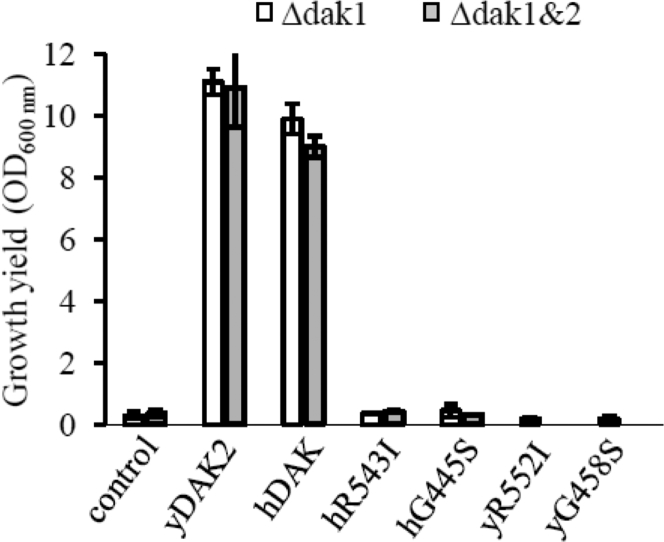
Since it was not possible to identify a robust phenotype in cells of affected individuals (cultured skin fibroblasts), functional confirmation of pathogenicity of the mutant alleles was performed using yeast models of TKFC deficiency. We utilized the sequence and predicted functional similarity of TKFC to two yeast DHA kinases Dak1 and Dak212 (Figure S1) to interrogate the functional significance of the TKFC variants identified in the affected individuals presented here. First we tested yeast wild-type and mutants containing a deletion of DAK1, DAK2, or both DAK1 and DAK2, as well as yeast cells overexpressing DAK1 or DAK2. We observed that overexpression of DAK1 or DAK2 in yeast (WT, Δdak1, Δdak2, Δdak1&2) allowed the cells to use DHA as a carbon source, as previously reported.12 We then expressed wild-type human TKFC in yeast and demonstrated that human TKFC allowed the transformed yeast cells to use DHA as carbon source (Figure 4), confirming the functional overlap of human TKFC and yeast Dak1/2. Finally, we tested the effect of the TKFC variants observed in the affected individuals, namely R543I and G445S (R552I and G458S in yeast Dak2), in the yeast model. Yeast cells overexpressing mutant human TKFC or mutant yeast DAK2 failed to grow on DHA as a carbon source (Figure 4). This result thus directly demonstrates the biochemical effect of the variants on TKFC/DAK function, from which we can infer their likely pathogenicity.
Figure 4.
Functional Studies in Yeast
Effect of mutations in wild-type and mutated human and yeast DAK on yeast growth using DHA as sole carbon source. Yeast cells freshly grown on pre-culture plates were inoculated in DHA medium at an initial OD600 nm of 0.2. The cultures were incubated for 4 days at 28°C with vigorous agitation. The OD600 nm were then recorded. The growth experiments were repeated at least twice and the data averaged. Error bars represent standard deviation. Control, cells (Δdak1 or Δdak1&Δdak2) without DAK overexpressing plasmid; yDAK2, cells overexpressing yeast DAK2; hDAK, cells overexpressing the WT human DAK/TKFC gene; hR543I and hG445S, cells overexpressing mutated human DAK/TKFC; yR552I and yG458S, cells overexpressing mutated yeast DAK.
The exact function of TKFC remains unclear to a certain extent. The two yeast orthologs Dak1 and Dak2 are expressed in stress conditions, such as heat or osmotic stress, and encode an enzyme involved in detoxification of DHA.12 A single study investigating the function of this enzyme in humans revealed that it is a bifunctional enzyme with triokinase and FMN cyclase activities,2 but it is possible that it has other roles. Three inborn errors of fructose metabolism are known: essential fructosuria (fructokinase deficiency [MIM: 229800], a rare non-disease), hereditary fructose intolerance (aldolase B deficiency [MIM: 229600]), and fructose-1,6-bisphosphatase deficiency (MIM: 229700).13 In hereditary fructose intolerance, fructose ingestion may trigger acute liver failure and proximal renal tubular dysfunction. Fructose-1,6-bisphosphatase deficiency is a disorder of gluconeogenesis and presents with hypoglycaemia and lactic acidosis. The most severely affected individual in our study, P1-2, had not been weaned or exposed to fructose, so it seems unlikely that toxicity from fructose or its metabolites played a significant role in disease pathogenesis. In P2-2 a fructose-free diet did not lead to clinical improvement.
Cataracts were present in three of the four individuals identified to have homozygous TKFC variants. Pathogenicity of TKFC deficiency may be by glyceraldehyde formation from impaired fructose catabolism. Glyceraldehyde is a reactive molecule (for instance with hydroxyl and amino groups) and increased production may lead to dysfunction of multiple proteins. Although P1-2 was not exposed to exogenous fructose, she did receive lactose/galactose in milk, so endogenous fructose production via the sorbitol pathway14 may have contributed to her symptomatology. The observation of mildly increased urinary galactitol (Table S1) in P1-1 is interesting, given the primary role of galactitol in cataract development in the galactosaemias (MIM: 230400, 230200, 230350). The polyol pathway normally metabolizes glucose to sorbitol to fructose, the first step being catalyzed by aldose reductase using NADPH—> NADP+, and the second step by sorbitol dehydrogenase reducing NAD+ to NADH. Galactose is also a substrate for the pathway; aldose reductase can convert galactose to galactitol, but sorbitol dehydrogenase cannot metabolize the galactitol that accumulates, possibly accounting for the cataract formation in affected individuals with TKFC deficiency since aldose reductase is expressed in the lens. The pathway of endogenous fructose production from sorbitol is not well characterized and may be differentially expressed in different tissues and between different individuals, possibly accounting for the phenotypic variability observed in our affected individuals with TKFC deficiency. Another possibility is that the cataracts could be caused by accumulation of DHA due to deficiency in TKFC’s DHA kinase activity. Endogenous increase in DHA levels leads to accumulation of advanced glycation end products15 which are linked to cataract formation.16,17 Similarly, DHA can itself be a glycation end product or may be metabolized to methylglyoxal, a metabolite previously associated with cataract formation.18 Additionally, DHA accumulation was shown to induce mitochondrial stress19 and alter mitochondrial membrane potential leading to apoptotic cell death20 which might explain the phenotypes mimicking a mitochondrial disorder. Levels of fructose metabolism and endogenous fructose production are cell specific, and metabolism of DHA also appears to be tissue specific.21
The critical illness of P1-2 following a febrile illness in early infancy suggests a vital function of TKFC, at least at this developmental stage. The homozygous p.Arg543Ile mutation in this individual affects the cFMN synthase domain. The function of cFMN remains completely unknown, but it has been speculated that it may be a signaling molecule, act as a minor redox flavocoenzyme (although enzymes which might use cFMN as a flavocoenzyme have not been identified)22 or simply be an intermediate in FAD degradation.23 TKFC has been shown to be a negative regulator of melanoma differentiation-associated gene-5 (MDA5, encoded by IFIH1 [MIM: 606951]),24,25 which is involved in RNA and virus-mediated type I interferon (IFN) production and antiviral responses.26 Gain-of-function IFIH1 variants cause heterogeneous phenotypes associated with upregulated type I IFN signaling (MIM: 615846, 182250),27 while MDA5 deficiency impairs viral double-stranded RNA (dsRNA) sensing and is associated with susceptibility to severe pediatric respiratory syncytial virus and rhinovirus infections in humans.28,29 Three other genes have been associated with the gene ontology (GO) term “negative regulation of MDA-5 signalling” (GO: 0039534): C1QBP (MIM: 601269), RIOK3 (MIM: 603579), and DHX58 (MIM: 608588).30, 31, 32 Biallelic C1QBP variants in humans cause combined oxidative phosphorylation deficiency 33 (MIM: 617713), a highly variable multisystem mitochondrial disorder with phenotypes ranging from death in infancy to adult-onset progressive external ophthalmoplegia and myopathy. A common finding is cardiomyopathy and increased serum lactate,33 phenotypes observed in P1-2 in the present study. RIOK3 phosphorylates MDA5, interfering with its assembly and attenuating the innate immune response.31 DHX58 was shown to function in the RIG-I/MDA5/MAVS protective IFN response in rotavirus-infected intestinal epithelium in mice.32 The mild loss of complex IV activity observed in the muscle biopsy of P1-2 might reflect secondary damage, for example as a result of oxidative stress following immune response activation. Taking all of these findings into consideration, we postulate that TKFC, via cFMN generation, may modulate innate immune signaling, and that lack of cFMN production in response to a viral insult in a critical developmental window may have led to the fatal illness of P1-2. The TKFC-MDA5 signaling pathway may represent a potential therapeutic target for individuals with TKFC deficiency.
In conclusion, this study demonstrates the power of genome sequencing to exclude a treatable cause of a complex multisystem disorder rapidly in a critical care setting. Extensive metabolic investigation did not reveal any diagnostic clues other than an elevated FGF21 level. This, combined with the clinical features of cataract and cardiomyopathy reminiscent of Sengers syndrome,5 led to an initial suspicion of mitochondrial disease in P1-2. However, further investigation was not in keeping with a primary mitochondrial disorder. This study demonstrates the utility of genome sequencing and data sharing via Genematcher in the identification of an inborn error of metabolism. Yeast biology is extremely helpful in dissecting the contribution of variants in housekeeping genes to human disease and here supports the pathogenicity of biallelic TKFC variants as a cause of human multisystem disease, variably including cataracts, developmental delay, liver dysfunction, microcytic anemia, cerebellar hypoplasia, and fatal cardiomyopathy with lactic acidosis.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank both families for their participation in this study. Studies in family 1 were funded by National Institute of Health Research (NIHR) Great Ormond Street Hospital (GOSH) Biomedical Research Centre (BRC) NIHR GOSH/UCL BRC: ormbrc-2012-1 and in family 2 by the E-Rare project GENOMIT FWF-I2741B26 for J.A.M. S.R. also receives grant funding from Great Ormond Street Hospital Children’s Charity and the Lily Foundation. We are grateful to Drs. Kimberly Gilmour, Elizabeth Ralph, and Mesfer Al Shahrani for performing the FGF21 assays.
Published: January 30, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.01.005.
Web Resources
gnomAD Browser, https://gnomad.broadinstitute.org/
GTEx Portal, https://gtexportal.org/home/
OMIM, https://www.omim.org/
Supplemental Data
References
- 1.Mestek-Boukhibar L., Clement E., Jones W.D., Drury S., Ocaka L., Gagunashvili A., Le Quesne Stabej P., Bacchelli C., Jani N., Rahman S. Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 2018;55:721–728. doi: 10.1136/jmedgenet-2018-105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues J.R., Couto A., Cabezas A., Pinto R.M., Ribeiro J.M., Canales J., Costas M.J., Cameselle J.C. Bifunctional homodimeric triokinase/FMN cyclase: contribution of protein domains to the activities of the human enzyme and molecular dynamics simulation of domain movements. J. Biol. Chem. 2014;289:10620–10636. doi: 10.1074/jbc.M113.525626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyynismaa H., Carroll C.J., Raimundo N., Ahola-Erkkilä S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjøsund K.E., Tulkki V. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 4.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayr J.A., Haack T.B., Graf E., Zimmermann F.A., Wieland T., Haberberger B., Superti-Furga A., Kirschner J., Steinmann B., Baumgartner M.R. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am. J. Hum. Genet. 2012;90:314–320. doi: 10.1016/j.ajhg.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbade S.F., Shen N., Himmelreich N., Haas D., Trefz F.K., Hoffmann G.F., Burgard P., Blau N. Allelic phenotype values: a model for genotype-based phenotype prediction in phenylketonuria. Genet. Med. 2019;21:580–590. doi: 10.1038/s41436-018-0081-x. [DOI] [PubMed] [Google Scholar]
- 7.Hers H.G., Kusaka T. The metabolism of fructose-1-phosphate in the liver. Biochim. Biophys. Acta. 1953;11:427–437. doi: 10.1016/0006-3002(53)90062-6. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues J.R., Cameselle J.C., Cabezas A., Ribeiro J.M. Closure of the Human TKFC Active Site: Comparison of the Apoenzyme and the Complexes Formed with Either Triokinase or FMN Cyclase Substrates. Int. J. Mol. Sci. 2019;20:20. doi: 10.3390/ijms20051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siebold C., Arnold I., Garcia-Alles L.F., Baumann U., Erni B. Crystal structure of the Citrobacter freundii dihydroxyacetone kinase reveals an eight-stranded α-helical barrel ATP-binding domain. J. Biol. Chem. 2003;278:48236–48244. doi: 10.1074/jbc.M305942200. [DOI] [PubMed] [Google Scholar]
- 10.Jang C., Hui S., Lu W., Cowan A.J., Morscher R.J., Lee G., Liu W., Tesz G.J., Birnbaum M.J., Rabinowitz J.D. The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab. 2018;27:351–361.e3. doi: 10.1016/j.cmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinote A., Faria J.A., Roman E.A., Solon C., Razolli D.S., Ignacio-Souza L.M., Sollon C.S., Nascimento L.F., de Araújo T.M., Barbosa A.P. Fructose-induced hypothalamic AMPK activation stimulates hepatic PEPCK and gluconeogenesis due to increased corticosterone levels. Endocrinology. 2012;153:3633–3645. doi: 10.1210/en.2012-1341. [DOI] [PubMed] [Google Scholar]
- 12.Molin M., Norbeck J., Blomberg A. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J. Biol. Chem. 2003;278:1415–1423. doi: 10.1074/jbc.M203030200. [DOI] [PubMed] [Google Scholar]
- 13.Steinmann B.S.R. Disorders of Fructose Metabolism. In: Saudubray J.M., Walter J., editors. Inborn Metabolic Diseases, B.M. Springer; Berlin, Heidelberg: 2016. pp. 161–168. [Google Scholar]
- 14.Hannou S.A., Haslam D.E., McKeown N.M., Herman M.A. Fructose metabolism and metabolic disease. J. Clin. Invest. 2018;128:545–555. doi: 10.1172/JCI96702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molin M., Pilon M., Blomberg A. Dihydroxyacetone-induced death is accompanied by advanced glycation endproduct formation in selected proteins of Saccharomyces cerevisiae and Caenorhabditis elegans. Proteomics. 2007;7:3764–3774. doi: 10.1002/pmic.200700165. [DOI] [PubMed] [Google Scholar]
- 16.Franke S., Dawczynski J., Strobel J., Niwa T., Stahl P., Stein G. Increased levels of advanced glycation end products in human cataractous lenses. J. Cataract Refract. Surg. 2003;29:998–1004. doi: 10.1016/s0886-3350(02)01841-2. [DOI] [PubMed] [Google Scholar]
- 17.Hashim Z., Zarina S. Advanced glycation end products in diabetic and non-diabetic human subjects suffering from cataract. Age (Dordr.) 2011;33:377–384. doi: 10.1007/s11357-010-9177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shamsi F.A., Lin K., Sady C., Nagaraj R.H. Methylglyoxal-derived modifications in lens aging and cataract formation. Invest. Ophthalmol. Vis. Sci. 1998;39:2355–2364. [PubMed] [Google Scholar]
- 19.Smith K.R., Hayat F., Andrews J.F., Migaud M.E., Gassman N.R. Dihydroxyacetone Exposure Alters NAD(P)H and Induces Mitochondrial Stress and Autophagy in HEK293T Cells. Chem. Res. Toxicol. 2019;32:1722–1731. doi: 10.1021/acs.chemrestox.9b00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K.R., Granberry M., Tan M.C.B., Daniel C.L., Gassman N.R. Dihydroxyacetone induces G2/M arrest and apoptotic cell death in A375P melanoma cells. Environ. Toxicol. 2018;33:333–342. doi: 10.1002/tox.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marco-Rius I., von Morze C., Sriram R., Cao P., Chang G.Y., Milshteyn E., Bok R.A., Ohliger M.A., Pearce D., Kurhanewicz J. Monitoring acute metabolic changes in the liver and kidneys induced by fructose and glucose using hyperpolarized [2-13 C]dihydroxyacetone. Magn. Reson. Med. 2017;77:65–73. doi: 10.1002/mrm.26525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasubramaniam S., Christodoulou J., Rahman S. Disorders of riboflavin metabolism. J. Inherit. Metab. Dis. 2019;42:608–619. doi: 10.1002/jimd.12058. [DOI] [PubMed] [Google Scholar]
- 23.Cabezas A., Costas M.J., Pinto R.M., Couto A., Cameselle J.C. Identification of human and rat FAD-AMP lyase (cyclic FMN forming) as ATP-dependent dihydroxyacetone kinases. Biochem. Biophys. Res. Commun. 2005;338:1682–1689. doi: 10.1016/j.bbrc.2005.10.142. [DOI] [PubMed] [Google Scholar]
- 24.Diao F., Li S., Tian Y., Zhang M., Xu L.G., Zhang Y., Wang R.P., Chen D., Zhai Z., Zhong B. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc. Natl. Acad. Sci. USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro A., Bamming D., Horvath C.M. Negative regulation of cytoplasmic RNA-mediated antiviral signaling. Cytokine. 2008;43:350–358. doi: 10.1016/j.cyto.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing H., Su H.C. New immunodeficiency syndromes that help us understand the IFN-mediated antiviral immune response. Curr. Opin. Pediatr. 2019;31:815–820. doi: 10.1097/MOP.0000000000000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice G.I., Del Toro Duany Y., Jenkinson E.M., Forte G.M., Anderson B.H., Ariaudo G., Bader-Meunier B., Baildam E.M., Battini R., Beresford M.W. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asgari S., Schlapbach L.J., Anchisi S., Hammer C., Bartha I., Junier T., Mottet-Osman G., Posfay-Barbe K.M., Longchamp D., Stocker M. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. USA. 2017;114:8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamborn I.T., Jing H., Zhang Y., Drutman S.B., Abbott J.K., Munir S., Bade S., Murdock H.M., Santos C.P., Brock L.G. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 2017;214:1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L., Xiao N., Liu F., Ren H., Gu J. Inhibition of RIG-I and MDA5-dependent antiviral response by gC1qR at mitochondria. Proc. Natl. Acad. Sci. USA. 2009;106:1530–1535. doi: 10.1073/pnas.0811029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takashima K., Oshiumi H., Takaki H., Matsumoto M., Seya T. RIOK3-mediated phosphorylation of MDA5 interferes with its assembly and attenuates the innate immune response. Cell Rep. 2015;11:192–200. doi: 10.1016/j.celrep.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Broquet A.H., Hirata Y., McAllister C.S., Kagnoff M.F. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 33.Feichtinger R.G., Oláhová M., Kishita Y., Garone C., Kremer L.S., Yagi M., Uchiumi T., Jourdain A.A., Thompson K., D’Souza A.R. Biallelic C1QBP Mutations Cause Severe Neonatal-, Childhood-, or Later-Onset Cardiomyopathy Associated with Combined Respiratory-Chain Deficiencies. Am. J. Hum. Genet. 2017;101:525–538. doi: 10.1016/j.ajhg.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.