Abstract
Cell-free DNA (cf.DNA) is a powerful noninvasive biomarker for cancer and prenatal testing, and it circulates in plasma as short fragments. To elucidate the biology of cf.DNA fragmentation, we explored the roles of deoxyribonuclease 1 (DNASE1), deoxyribonuclease 1 like 3 (DNASE1L3), and DNA fragmentation factor subunit beta (DFFB) with mice deficient in each of these nucleases. By analyzing the ends of cf.DNA fragments in each type of nuclease-deficient mice with those in wild-type mice, we show that each nuclease has a specific cutting preference that reveals the stepwise process of cf.DNA fragmentation. Essentially, we demonstrate that cf.DNA is generated first intracellularly with DFFB, intracellular DNASE1L3, and other nucleases. Then, cf.DNA fragmentation continues extracellularly with circulating DNASE1L3 and DNASE1. With the use of heparin to disrupt the nucleosomal structure, we also show that the 10 bp periodicity originates from the cutting of DNA within an intact nucleosomal structure. Altogether, this work establishes a model of cf.DNA fragmentation.
Keywords: cf.DNA, cf.DNA biology, fragmentation, fragmentomics, liquid biopsy, nuclease, DNASE1, DNASE1L3, DFFB, circulating nucleic acids
Introduction
Cell-free DNA (cf.DNA) is a rich source of information that can be applied to the diagnosis and prognostication of many physiological and pathological conditions such as pregnancy and cancer.1, 2, 3 Though circulating cf.DNA is now commonly used as a noninvasive biomarker and is known to circulate in the form of short fragments, the physiological factors governing the fragmentation and molecular profile of cf.DNA remain elusive.
Recent works have suggested that the fragmentation of cf.DNA is a non-random process associated with the positioning of nucleosomes.4, 5, 6, 7, 8 Previously, we have demonstrated that the DNASE1L3 nuclease contributes to the size profile of cf.DNA in plasma.9 In this study, we aimed to investigate the respective roles of DNASE1, DNASE1L3, and DNA fragmentation factor subunit beta (DFFB, also known as caspase-activated DNase) in cf.DNA fragmentation. We compared the cf.DNA profiles between mice deficient in each type of nuclease and their wild-type (WT) counterparts. We found that each nuclease served a different but complementary role in cf.DNA fragmentation.
Material and Methods
Murine Models
Analysis of cf.DNA from Dnase1l3−/− mice in module 2 was done using public data downloaded from the European Genome-phenome Archive (Figure 1) (EGA; EGA accession number EGAS00001003174).9 Mice carrying a targeted allele of Dnase1 (Dnase1tm1.1(KOMP)Vlcg) and mice carrying a targeted allele of Dffb (DffbC57BL/6N-Dffbem1Wtsi), both on B6 background, were obtained from the Knockout Mouse Project Repository of the University of California at Davis. The mice were maintained in the Laboratory Animal Center of The Chinese University of Hong Kong (CUHK). All experimental procedures were approved by the Animal Experimentation Ethics committee of CUHK and performed in compliance with the Guide for the Care and Use of Laboratory Animals (8th edition, 2011) established by the National Institutes of Health. Male and female mice aged 13–17 weeks were used for experiments. An analysis of the influence of sex and gender on the results was not done because their blood samples were pooled together.
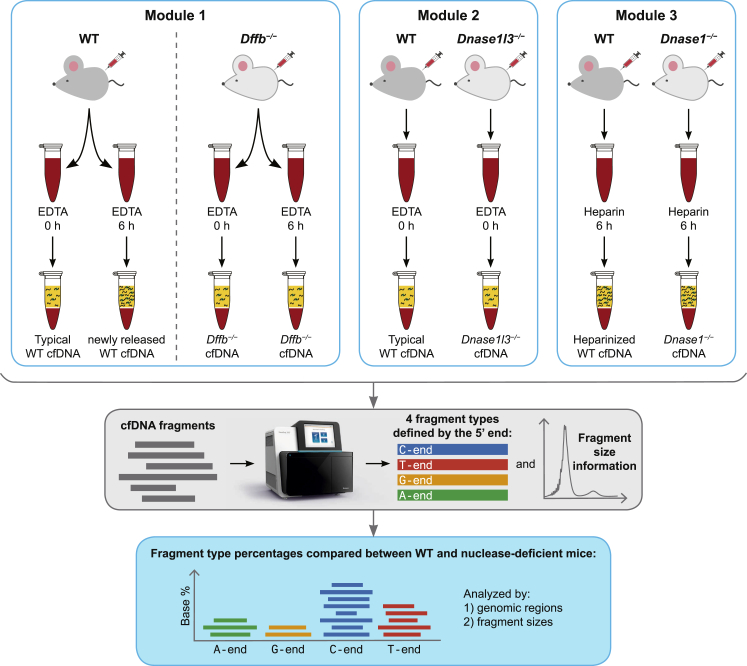
Figure 1.
Overview of the Study Methods
The data presented in this paper is divided into three modules, and each module uses cf.DNA data obtained from different nuclease-deficient mice (light gray mice) and their wild-type (WT) counterparts (dark gray mice). In module 1, blood from WT mice was subject to either 0 h or 6 h room temperature (RT) incubation in EDTA to yield typical or newly released cf.DNA, respectively. The same process was applied to blood from Dffb-deficient mice. In module 2, cf.DNA data from WT and Dnase1l3-deficient mice was analyzed. And lastly, in module 3, blood from WT and Dnase1-deficient mice underwent RT incubation in heparin for 0 h (not shown) and 6 h. The plasma cf.DNA from each condition was sequenced, and fragments were defined based on their size and the nucleotide at their 5′ end. We compared the percentages of each fragment type among the different genotypes in various genomic regions and fragment sizes in order to obtain insights into the effects of different nucleases on cf.DNA fragmentation.
Murine Sample Collection
Mice were killed and exsanguinated using cardiac puncture. Blood from each mouse was pooled and immediately distributed evenly into experimental conditions: EDTA with 0 h incubation and EDTA with 6 h incubation, or heparin with 0 h incubation and heparin with 6 h incubation (Figure 1). For the Dffb−/− experiments in module 1, a WT set and a Dffb−/− set, each with five pools of blood, were created, with each pool containing blood from 2–4 mice (Figure 1). The WT set and Dffb−/− set contained blood from 14 WT and 14 Dffb−/− mice, respectively. For the Dnase1−/− experiments in module 3, one pool was created for each genotype, from a total of 12 WT, 12 Dnase1+/−, and 11 Dnase1−/− mice in each pool (Figure 1). The EDTA tubes were commercially sourced 1.3 mL K3E micro tubes (Sarstedt). Heparin tubes were 2 mL microcentrifuge tubes with 18 IU heparin (Sigma-Aldrich) per mL blood added. Incubation was done at room temperature (RT, 15–20°C) on a rocker.
After the RT incubation time was completed, the blood samples were separated through the use of a double centrifugation protocol (1,600 × g for 10 min at 4°C, then recentrifugation of the plasma at 16,000 × g for 10 min at 4°C).10 The resulting plasma was collected, yielding 0.4–1.5 mL of plasma for each condition and time point.
Plasma DNA Extraction and Library Preparation
Plasma DNA was extracted using the QIAamp Circulating Nucleic Acid Kit (QIAGEN) according to the manufacturer’s protocol. Indexed plasma DNA libraries were constructed using a TruSeq DNA Nano Library Prep Kit according to the manufacturer’s instructions. The adaptor-ligated DNA was enriched with eight cycles of PCR and analyzed on an Agilent 4200 TapeStation (Agilent Technologies) using the High Sensitivity D1000 ScreenTape System (Agilent Technologies) for quality control and gel-based size determination. Libraries were quantified using the Qubit dsDNA high sensitivity assay kit (Thermo Fisher Scientific) before sequencing.
DNA Sequencing and Alignment
Multiplexed DNA libraries were sequenced for 2 × 75 bp paired-end reads on the NextSeq 500 platform (Illumina). Sequences were assigned to their corresponding samples based on their six-base index sequence. Using the Short Oligonucleotide Alignment Program 2 (SOAP2), the paired-end reads from mouse plasma were aligned to the reference mouse genome (NCBI build 37/UCSC mm9; non-repeat-masked).11 Up to two nucleotide mismatches were allowed. Only paired-end reads aligned to the same chromosome in the correct orientation and spanning an insert size of <600 bp were retained for downstream analysis. The <600 bp cutoff was used because it is the practical readout limit of the Illumina sequencing platform. Furthermore, previous work using plasma DNA sequenced using the Pacific Bioscience Single Molecule, Real-Time (SMRT) system indicated that fragments in the 600 to 2,000 bp range accounted for less than 4.8% of the cf.DNA population.9 Paired-end reads sharing the same start and end genomic coordinates were deemed PCR duplicates and were discarded from downstream analysis. Table S1 summarizes the number of non-duplicate fragments obtained for each condition. The genome coordinates of the aligned ends were used to deduce the size of the whole fragment of the sequenced cf.DNA. The deletions of the Dnase1 and Dffb genes were observed after alignment in the Dnase1−/− and Dffb−/− mice data, respectively (Figure S1).
Base-end Analysis and Fragment Type Analysis
CCCTC-binding factor (CTCF) sites, RNA polymerase II (Pol II) binding sites, transcriptional start sites (TSS), and random genomic regions were analyzed. CTCF sites are known to be flanked by nucleosomes that have largely invariant positions in the eukaryotic genome.12 These well-positioned nucleosomes flanking the CTCF binding site allow the mapping of sequenced cf.DNA fragments in relation to nucleosomal organization. On the other hand, Pol II and TSS regions are markers of open chromatin regions.
CTCF and Pol II regions were downloaded from the mouse ENCODE project.13 The TSS of all genes in the reference mouse genome UCSC mm9 were downloaded from UCSC. 10,000 random non-overlapping regions with 10,000 bp length were randomly selected across the whole genome by BEDTools (v2.27.1).14 We used a window size of ± 500 bp. For the end density analysis, the end density of a ± 1,500 bp window of CTCF regions was normalized according to the median end counts in ± 3,000 bp CTCF regions.
For the random, CTCF, and Pol II regions, only cf.DNA fragments oriented in the direction of the Watson strand were used for analysis. For the TSS region, only cf.DNA fragments oriented in the same direction as the TSS region were used. At each position in these regions, the first nucleotide on the 5′ end was identified for each fragment, and the base-end percentage was calculated (e.g., A-end fragments/all fragments, with “all fragments” including A-end, G-end, C-end, and T-end fragments). To analyze the base-end percentages according to fragment size, both 5′ ends (on the respective Watson or Crick strands) of a cf.DNA fragment were counted per fragment, and the base-end percentages at each size were calculated.
For fragment type analysis, each fragment was assigned to a fragment type based on their two ending nucleotides. These fragments where both ends were identified were denoted with their end nucleotides and the symbol < > in between, such that a fragment with both ends as A would be designated as A < >A. The group called “all fragments” includes A< >A, A< >G, A< >C, A< >T, C< >C, C< >G, C< >T, G< >G, G< >T, T< >T fragments. Each fragment type’s percentages were calculated (e.g., A< >A fragment percent = A< >A fragments/all fragments).
cf.DNA Quantification
Heparin was found to have significant positive interference with the Qubit dsDNA high sensitivity assay (ThermoFisher Scientific) (data not shown). Instead, the Bio-Rad QX200 Droplet Digital PCR (ddPCR) platform was used for all cf.DNA quantification because the heparin interference of DNA target molecules can be ameliorated by the reaction partitioning of ddPCR.15 Heparin samples were diluted 5-fold and at least four wells per sample were used. Mouse cf.DNA was quantified through the use of the mouse TaqMan Copy number reference assay (ThermoFisher Scientific) targeting the transferrin receptor gene (Tfrc).
Statistical Analysis
Analysis was performed using custom-built programs written in Python and R languages. Statistical differences were calculated using Mann-Whitney U tests unless otherwise specified. A p value of less than 0.05 was considered statistically significant, and all probabilities were two tailed.
Results
C-End Preference in Typical Circulating cf.DNA
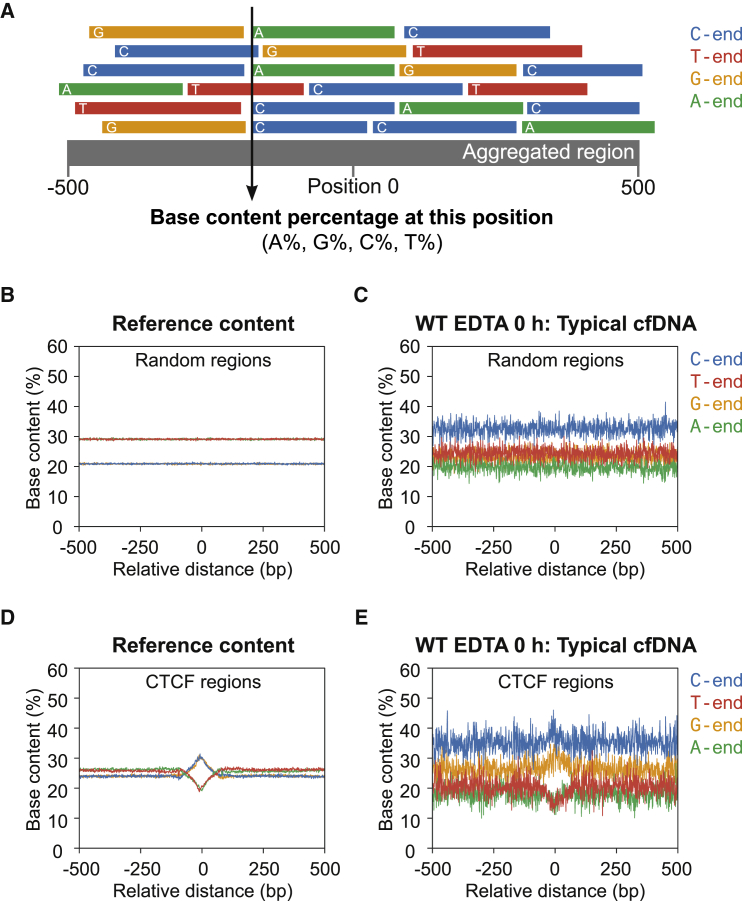
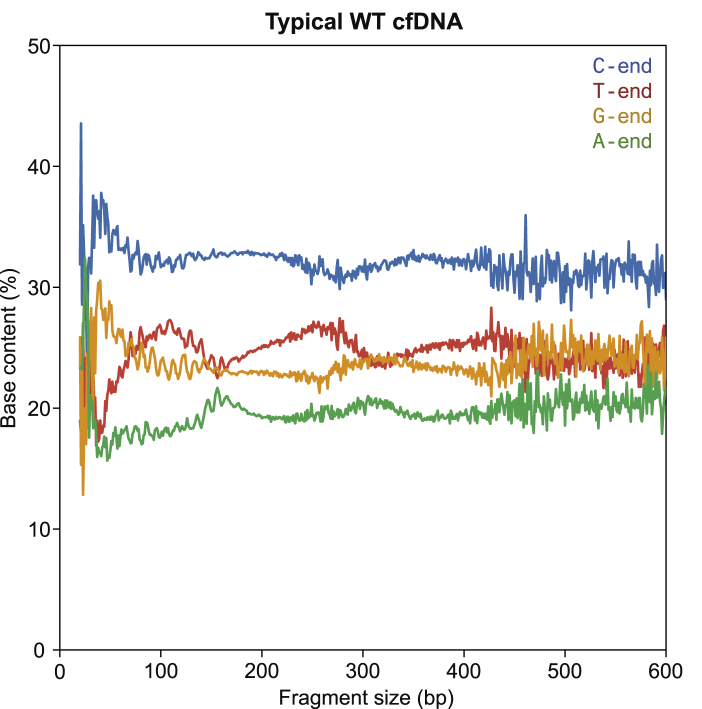
Our main study question concerns the biological basis of cf.DNA fragmentation and whether specific nucleases have any effect on the ends of cf.DNA. To this end, we compared the cf.DNA of different nuclease-deficient mice with its WT counterpart in different in vitro conditions (Figure 1). We sequenced the cf.DNA from these conditions and identified fragments according to the nucleotide base at their 5′ end and their size (Figure 1). We first analyzed the base content proportions at the 5′ end of cf.DNA fragments in different genomic regions in WT mice in order to test the hypothesis that cf.DNA fragmentation is not random (Figure 2A). If fragmentation were completely random, the end nucleotide proportions should reflect the composition of the mouse genome, which is 28.8% A, 28.8% T, 21.2% C, and 21.2% G (Figure 2B). However, the 5′ end of cf.DNA fragments in randomly selected genomic regions showed a substantial overrepresentation of C (32.6%), a slight overrepresentation of G (24.4%), and an underrepresentation of A (19.8%) and T (23.2%) (Figure 2C). This pattern of asymmetric representation was also seen in cf.DNA aligning to CTCF, TSS, and Pol II regions (Figure 2D and 2E and Figure S2A, S2B, S2D, and S2E). Because CTCF regions contain distinct nucleosomes with largely predictable positions flanking the CTCF binding site, and TSS and Pol II regions are known open-chromatin regions, we have concluded that both nucleosomal and open regions of the genome display the same C-end overrepresentation. Furthermore, when the 5′ ends are plotted across the 0–600 bp range of cf.DNA fragment sizes, the over-representation of C-ends and underrepresentation of A-ends remained evident and relatively uniform across all fragment sizes in WT cf.DNA (Figure 3). Thus, C-end predominant cf.DNA appears to be the typical cf.DNA profile in WT mice across all fragment sizes.
Figure 2.
C-End Preference in Typical Circulating cf.DNA in Different Regions
(A) Schematic showing the calculation of the base content proportions at the 5′ end of cf.DNA fragments in an aggregated region.
(B and D) The reference murine genomic content of random (B) and CCCTC-binding factor (CTCF) (D) regions. A-end and T-end fragment proportions overlap, and C-end and G-end fragment proportions overlap.
(C and E) The base content proportions at the 5′ end of cf.DNA fragments of wild-type EDTA 0 h samples in random (C) and CTCF (E) regions. C and G are overrepresented and A and T are underrepresented at the 5′ ends of these typical cf.DNA fragments compared to the reference genomic content.
Figure 3.
C-End Preference in Typical Circulating cf.DNA in All Fragment Sizes
Base content proportions at the 5′ end of typical cf.DNA from wild-type EDTA 0 h samples across the range of fragment sizes.
Fragmentation Pattern in Newly Released cf.DNA in WT and Dffb-deficient Mice
A-End and G-End Preference in Newly Released cf.DNA
We explored whether this typical cf.DNA profile was created “as is” from cellular sources or produced after further digestion within the plasma. Thus, we sought to capture and analyze cf.DNA that was newly released from dying cells and to compare its profile with the typical C-end-predominant cf.DNA profile. The model system we used to explore the characteristics of newly released cf.DNA into plasma was to incubate EDTA whole blood at RT for 6 h (Module 1 of Figure 1). Exposure to RT has previously been described as a method to induce apoptotic cell death in several in vitro studies.16, 17, 18, 19 EDTA is one of the more commonly used anticoagulants for cf.DNA analysis. The anticoagulant EDTA remains largely extracellular, inhibiting both plasma DNASE1 and DNASE1L3 activities by chelating divalent cations.20, 21, 22, 23 Thus, the anticoagulant EDTA inhibits the major plasma nucleases and therefore allows newly released cf.DNA to be preserved for analysis. While such RT incubation may not necessarily reflect the in vivo reality, we believe that such an in vitro model system is nonetheless valuable in yielding insights into the biology of cf.DNA.
After 6 h whole blood RT incubation, we expected that cell death would be increased and samples would be enriched with newly released cf.DNA from white blood cells. Indeed, we found a 1.1- to 5.9-fold increase in plasma cf.DNA concentration, and an increase in long cf.DNA fragments after 6 h whole blood RT incubation compared with its paired baseline (Figure S3A and S3B).
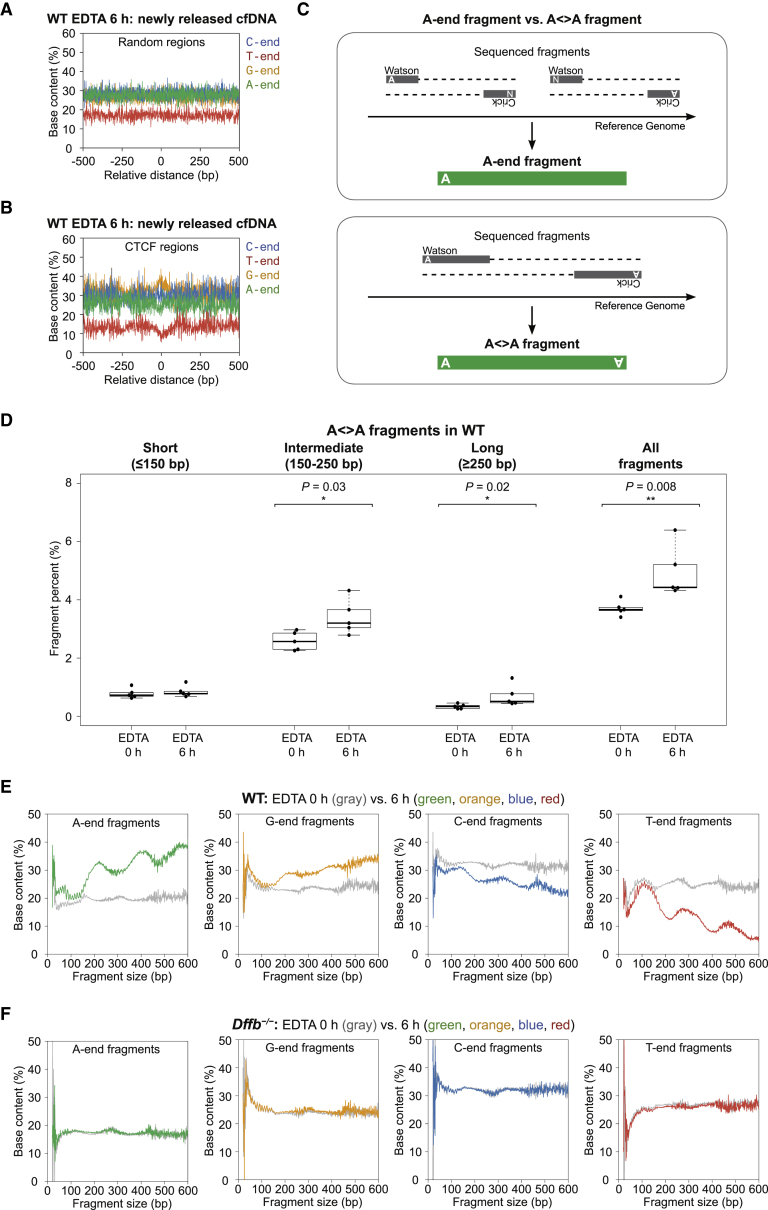
When we analyzed the 5′ ends of cf.DNA in the 6 h EDTA sample, the C-end predominance seen in typical cf.DNA was greatly diminished in the presence of newly released cf.DNA compared with its baseline 0 h incubation. C-end and T-end fragments decreased to 28.3% and 17.0%, respectively, in randomly selected genomic regions (Figure 4A). A-end and G-end fragments increased substantially to 27.7% and 27.0%, respectively, in randomly selected genomic regions (Figure 4A). These changes were also consistently visualized in the CTCF regions with nucleosomal arrays, and in TSS and Pol II regions with open chromatin regions (Figure 4B and Figure S2C and S2F). Therefore, newly released cf.DNA after whole blood incubation were enriched for A-end and G-end fragments when compared to typical cf.DNA. Because the newly released cf.DNA profile from dying cells does not appear to be similar to the typical C-end-predominant cf.DNA found in baseline samples, we inferred that the typical C-end-predominant cf.DNA would be created in a subsequent step. Because the fragment end preference is different (A-end versus C-end), we also reasoned that the generation of newly released cf.DNA likely originated from a different mechanism than that which created the typical cf.DNA.
Figure 4.
Fragmentation Pattern in Newly Released cf.DNA in Wild-type (WT) and Dffb-deficient Mice
(A and B) Base content proportions of WT EDTA 6 h samples enriched with newly released cf.DNA in random (A) and CTCF regions (B). With newly released cf.DNA enrichment, A-end and G-end fragments increased, and C-end and T-end fragments decreased compared to the baseline base proportions in typical cf.DNA.
(C) A-end fragments versus A< > A fragments. A-end fragments are comprised of the fragments in which either the Watson or Crick strand starts with A. A< > A fragments are comprised of fragments in which both Watson and Crick strands start with A.
(D) A< > A fragment proportions compared between baseline cf.DNA (EDTA 0 h) and samples enriched with newly released cf.DNA (EDTA 6 h) in WT mice among short, intermediate, and long fragments. 6 h samples enriched with newly released cf.DNA have a significantly higher proportion of A< > A fragments in long fragment sizes >150 bp. p value calculated by Mann-Whitney U test.
(E) Percentages of cf.DNA with A-ends (green), G-ends (orange), C-ends (blue), and T-ends (red) in a WT EDTA 6 h sample enriched with newly released cf.DNA compared with the baseline representation in the EDTA 0 h sample (gray). A-end fragments and, to a lesser extent, G-end fragments have peaks ~200 bp and 400 bp.
(F) Percentages of cf.DNA with A-ends (green), G-ends (orange), C-ends (blue), and T-ends (red) in a Dffb-deficient EDTA 6 h sample compared to its baseline representation in the EDTA 0 h sample (gray). No increase in A-end fragments was observed.
A-Ends and G-Ends Among Newly Released cf.DNA of Different Sizes
We explored the A-end and G-end fragments according to their size. We identified fragments based on their two end nucleotides and analyzed the fragments in which both ends terminated with A, G, C, or T (Figure 4C). These fragments where both ends were identified were denoted with their end nucleotides and the symbol < > in between, such that a fragment with both ends as A would be designated as A< >A (Figure 4C). We compared the proportional representation of A< >A, G< >G, C< >C, and T< >T fragments among fragments of different sizes; we reasoned that any preference for cutting a particular nucleotide would be best visualized with these fragment types in which both ends encompassed the same nucleotide preference. Of these four types of fragments, 6 h samples enriched with newly released cf.DNA had a significantly higher proportion of A< >A fragments in sizes >150 bp, and the proportions increased further in long fragments ≤250 bp (Figure 4D). On the other hand, G< >G, C< >C, and T< >T fragments did not differ significantly based on size (Figure S3C). Thus, newly released cf.DNA was enriched for A-end fragments that were longer than 150 bp.
Figure 4E shows the proportion of A-end, G-end, C-end, and T-end fragments for each fragment size compared to the respective baseline unincubated EDTA levels. Surprisingly, the increase in long A-end fragments was concentrated at specific size ranges, with peaks at ∼200 bp and 400 bp that were reminiscent of nucleosomal ladder sizes. G-end fragments also had a similar but weaker periodicity at these sizes. We hypothesized that these A-end (and G-end) cf.DNA fragments were likely created by cleaving between nucleosomes, such that the full length of an intact nucleosomal DNA was retained. The peaks in periodicity would support a true preference for cutting at the inter-nucleosomal regions 5′ to an A with a slightly smaller preference for cutting 5′ to a G.
Effects of DFFB on cf.DNA with A-Ends
Because A-end long fragments were newly released from dying cells, we examined the role of apoptosis in their generation. DFFB is the major intracellular nuclease involved in DNA fragmentation during apoptosis, so we investigated samples from Dffb-deficient mice. After 6 h of EDTA incubation, cf.DNA quantity did not significantly increase and there was little or no increase in long fragments (Figure S4A and S4B). In random genomic regions and in CTCF, TSS, and Pol II regions, the A-end fragments did not increase (Figure S4C). Similarly, A< >A fragment percentages did not increase after 6 h of EDTA incubation in Dffb-deficient mice, in contrast to the results from WT mice (Figure S4D). When we compared A-end, G-end, C-end, and T-end fragment proportions at each fragment size, we found little change in Dffb-deficient mice after 6 h of EDTA incubation compared with the baseline, and there was no periodicity in the A-end and G-end fragments (Figure 4F). In comparison to the baseline EDTA 0 h blood sample between WT and Dffb-deficient mice, A-end fragments were decreased across all fragment sizes (Figure S5). Hence, these results from the WT and Dffb-deficient mice suggest that DFFB might have a major role in generating these A-end long fragments.
Effect of DNASE1L3 on Typical cf.DNA
Although we had characterized the profile of newly released cf.DNA, we had yet to clarify the process by which the typical C-end predominance was produced in plasma cf.DNA. This clear preference for C-ends in all sizes of circulating cf.DNA fragments, as seen in Figure 3, suggests the presence of a nuclease that prefers to cleave 5′ to a C. Previously, we had demonstrated that cf.DNA from WT mice had a high frequency of fragments ending in CCNN motifs (i.e., motifs consisting of two consecutive cytosines, followed by two nucleotides of any type) and that this preference for CCNN motifs in cf.DNA fragment ends was reduced in Dnase1l3-deficient mice.9 Because C-end fragments are comprised of CCNN end motifs, we hypothesized that the nuclease responsible for the C-end preference might also be DNASE1L3.
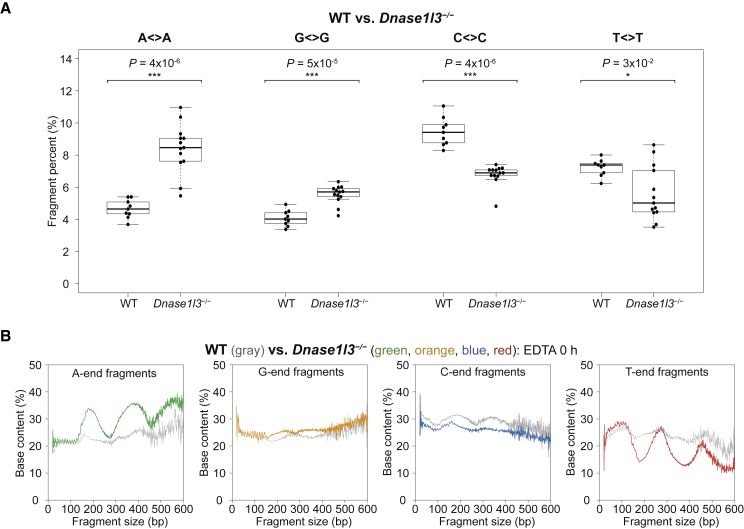
To investigate this hypothesis, we compared the specific A< >A, G< >G, C< >C, and T< >T fragment proportions between Dnase1l3-deficient mice and WT mice in the Dnase1l3-deficient data retrieved from the European Genome-phenome Archive (Module 2 of Figure 1). C< >C and T< >T fragment percentages significantly decreased in Dnase1l3-deficient mice compared to WT (Figure 5A). These results suggest that DNASE1L3 generates both C- and T-end fragments, with a greater preference for C-ends because C< >C fragment percentages are more significantly reduced.
Figure 5.
Effect of DNASE1L3 on Typical cf.DNA
(A) A< > A, G< > G, C< > C, and T< > T fragment percentages in wild-type (WT) versus Dnase1l3-deficient mice, both from EDTA 0 h samples. p value calculated by Mann-Whitney U test.
(B) Percentages of A-ends (green), G-ends (orange), C-ends (blue), and T-ends (red) in Dnase1l3-deficient EDTA 0 h cf.DNA compared with the percentages in WT EDTA 0 h cf.DNA (gray).
In the absence of DNASE1L3, the proportions of A< >A and G< >G fragments were higher than in WT (Figure 5A). When we compared the A-end, G-end, C-end, and T-end fragment proportions of each fragment size between the Dnase1l3-deficient mice and WT mice in EDTA 0 h samples, the A-end fragments demonstrated a nucleosomal periodic pattern with peaks in frequency ∼200 bp and 400 bp (Figure 5B). This nucleosomal periodic pattern of A-end fragments is similar to the one observed previously in WT EDTA 6 h samples enriched with newly released cf.DNA (Figure 3E). Hence, it appears that DNASE1L3 deficiency results in exposing the profile of newly released cf.DNA. In a substrate-enzyme-product relationship, when the enzyme is deficient, the product would decrease and the substrate would increase. Thus, DNASE1L3-deficient cf.DNA seems to have revealed its substrate cf.DNA profile, which appears to be the cf.DNA profile created by DFFB.
By taking a more detailed look at the fragment types that use both ends of a cf.DNA fragment, we found that only A< >A, A< >G, and A< >C fragments demonstrated this nucleosomal periodic pattern in both Dnase1l3-deficient samples and WT EDTA 6 h samples enriched with newly released cf.DNA (Figure S6). There were a number of notable differences between the fragments of these two sample types. In Dnase1l3-deficient mice, the periodic pattern of the A< >A, A< >G, and A< >C fragments was very prominent (Figure S6A). Because DNASE1L3 activity is absent in Dnase1l3-deficient mice, this prominence in the cf.DNA likely reflects the true preference for nucleosomal periodic cutting in the remaining active intracellular nucleases, notably DFFB. On the other hand, the periodic pattern seen in the newly released cf.DNA was attenuated, and this was especially noticeable among A< >C fragments (Figure S6B). Because DNASE1L3 activity is retained in the generation of newly released cf.DNA compared with that activity in Dnase1l3-deficient mice, this difference might suggest that DNASE1L3 would play a role in creating A< >C fragments, which might be an intermediate step to creating C< >C fragments. These results also suggested that DNASE1L3 might attenuate the preferential cutting of the DFFB nuclease by cutting after DFFB. Thus, it can be inferred that DNASE1L3 cutting might occur predominantly as a subsequent step to DFFB cutting, and that DNASE1L3 might not only have a role, but might actually be a dominant player in creating the typical profile with C-end predominance in cf.DNA (Figure 3).
Effects of DNASE1 on cf.DNA (with Heparin)
While we have demonstrated the steps involved in creating a typical cf.DNA fragment with C-end predominance, so that a full picture of the homeostasis of cf.DNA can be constructed, we wanted to explore how a cf.DNA fragment might be further digested. While C-end fragments continue to be the most prevalent even in short fragments <150 bp, we noted an enrichment of T-end fragments in sizes ∼50–150 bp and ∼250 bp in the typical cf.DNA profile (Figure 3). These peaks were not concordant with either the C-end fragments which were related to DNASE1L3 preference or the A-end fragments which were related to DFFB cutting preference. With our theory that fragment ends correlated with nuclease preference, we explored whether or not these T-ends might be related to DNASE1 preference.
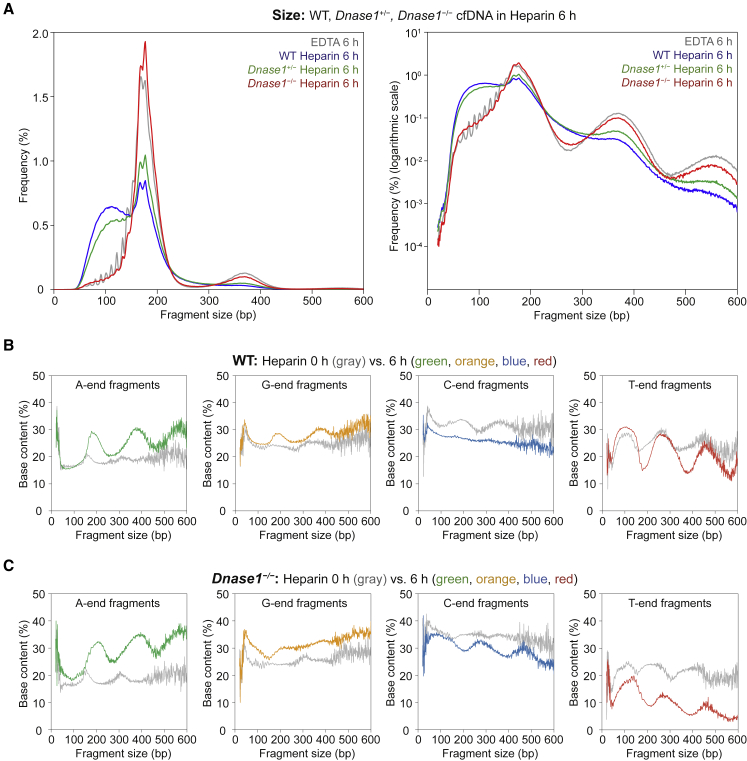
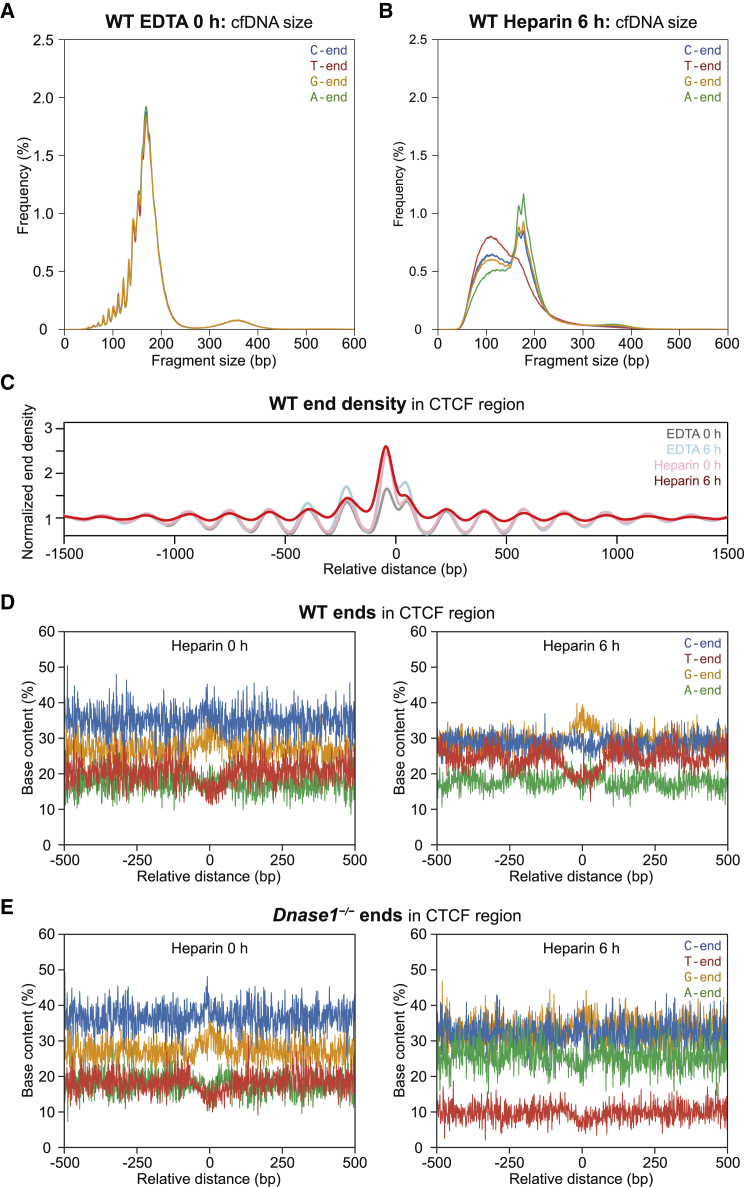
To identify DNASE1’s cutting preference, we collected whole blood from Dnase1−/−, Dnase1+/−, and WT mice, pooled the blood, and equally distributed each pool into tubes for 0 h or 6 h incubation with heparin (Module 3 of Figure 1). Heparin was used instead of EDTA because heparin is known to enhance DNASE1 activity while inhibiting DNASE1L3.23 We found that in WT and Dnase1+/− mice, 6 h of heparin incubation resulted in a striking increase in short fragments with a reduction in the 166 bp peak and a loss of nucleosomal pattern (Figure 6A). In Dnase1−/−, no size changes occurred, and the size pattern was essentially the same as in cf.DNA from EDTA blood (Figure 6A).
Figure 6.
Effect of DNASE1 on cf.DNA (with heparin)
(A) cf.DNA size profiles in wild-type (WT) (blue), Dnase1+/− (green), and Dnase1−/− (red) mice in heparin 6 h samples. Normal y axis scale (left) and logarithmic y axis scale (right).
(B) Percentages of A-ends (green), G-ends (orange), C-ends (blue), and T-ends (red) of WT heparin 6 h samples compared to its baseline representation in heparin 0 h samples (gray).
(C) Percentages of A-ends (green), G-ends (orange), C-ends (blue), and T-ends (red) in heparin 6 h cf.DNA of Dnase1−/− mice compared to its baseline representation in heparin 0 h (gray).
We examined these samples for a difference in fragment end proportions. In WT and Dnase1+/− mice after 6 h heparin incubation, T-end fragment proportions increased in fragments sized ∼50-150 bp (Figure 6B, Figure S7A). In contrast, in Dnase1−/− mice, this increase was absent (Figure 6C). These observations supported our hypothesis that DNASE1 might prefer to create T-end fragments. In addition, the long A-end fragments with nucleosomal periodicity were present after 6 h heparin incubation in WT, Dnase1+/−, and Dnase1−/− mice (Figure 6B and 6C, Figure S7A). When we considered the increase in cf.DNA amounts in all three genotypes along with the literature on heparin incubation inducing apoptosis,24 we found that the presence of the A-end DFFB signature from newly released apoptotic cf.DNA was consistent (Figure S7B). Overall, heparin enhancement of DNASE1 in WT mice resulted in an increase of cf.DNA and newly released A-end fragments that were quickly digested into short T-end fragments; this result suggests that DNASE1 preferred to cut 5′ to T.
10 bp Periodicity Originates from Fragments Cut from Nucleosomes
Notably, all A-end, G-end, C-end, and T-end fragment types demonstrated a 10 bp periodicity in the short, ≥150 bp fragments among all mice genotypes (WT, Dnase1l3−/−, Dffb−/−, Dnase1+/−, and Dnase1−/−) (Figure 7A, Figure S8). Other than the C-end preference for all cf.DNA sizes, there was no particular end preference related to the 10 bp period fragments. Thus, it would be unlikely that a single particular nuclease would be responsible for the 10 bp periodicity. In fact, the prevailing theory for the 10 bp periodicity is that it is a result of nuclease digestion of DNA within an intact nucleosome. This was postulated based on the combined effect of restricted nuclease access to the DNA wrapped around histones and the periodic exposure of one strand of DNA over the other due to the 10 bp-per-turn structure of the DNA helix.25 In our heparin model, which disrupted the nucleosome structure in plasma, the 10 bp periodicity was abolished in all fragment types after 6 h heparin incubation in WT (Figure 7B).
Figure 7.
10 bp Periodicity Originates from Fragments Cut from Nucleosomes
(A) cf.DNA size profile of A-end, G-end, C-end, and T-end fragments in an EDTA 0 h wild-type (WT) sample.
(B) cf.DNA size profile of A-end, G-end, C-end, and T-end fragments in a heparin 6 h WT sample.
(C) Fragment end density in the CCCTC-binding factor (CTCF) region in the heparin 6 h sample (red) compared to the baseline samples: EDTA 0 h (gray), EDTA 6 h (light blue), and heparin 0 h (pink).
(D and E) The 5′ end base content proportions in the CTCF region of heparin 0 h (left) and 6 h (right) samples of WT (D) and Dnase1−/− (E) mice.
At the well-phased nucleosomes in the CTCF region, fragment ends within the nucleosome increase with heparin 6 h incubation in WT (Figure 7C). These results suggested that the disrupted nucleosome structure resulted in intra-nucleosomal DNA being cut as well (Figure 7C). We explored which fragment types would contribute to the intra-nucleosomal fragments in Figure 7D. In WT with heparin 6 h incubation, an increase in T-end fragments corresponding to the intranucleosomal position was apparent (Figure 7D). This effect was absent in Dnase1−/− mice (Figure 7E). These results together supported the concept that heparin would enhance DNASE1 and would disrupt the nucleosomal structure, allowing DNASE1 with T-end preference to cleave intranucleosomally.
Discussion
From this work on cf.DNA fragment ends in different mouse models, we can piece together a model that outlines the fragmentation process that generated cf.DNA (Figure 8). In our analysis of the newly released cf.DNA spontaneously created after incubating whole blood in EDTA, we have demonstrated that the newly released longer cf.DNA are enriched for A-end fragments. In particular, A< >A, A< >G, and A< >C fragments demonstrate a strong nucleosomal periodicity at ∼200 bp and 400 bp. When this same experimental model is applied to the whole blood of Dffb-deficient mice, no long A-end fragment enrichment is seen. Thus, we can conclude that DFFB is likely responsible for generating these A-end fragments.
Figure 8.
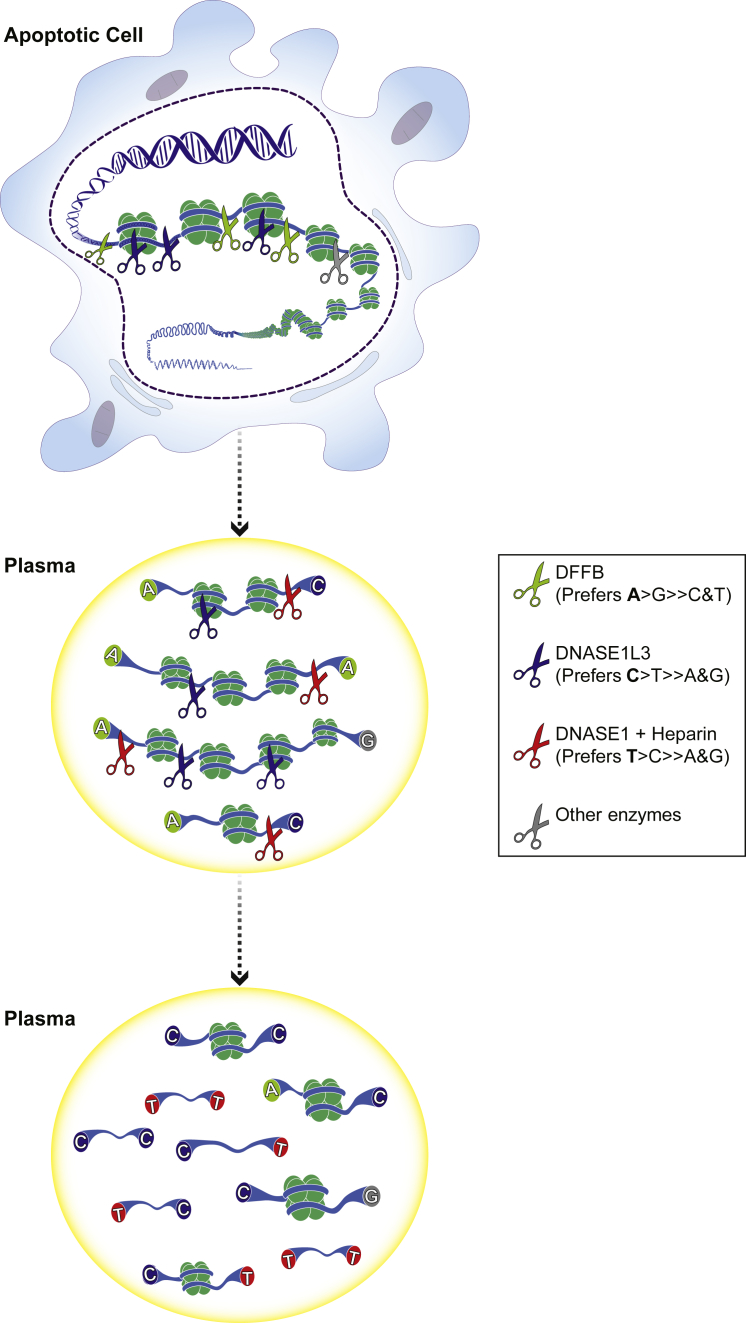
Model of cf.DNA Fragmentation
DFFB, DNASE1L3 and other intracellular enzymes form newly released cf.DNA that is A-end enriched. In plasma, DNASE1L3 generates the predominantly C-end enriched cf.DNA seen in the typical profile via its extracellular activity. DNASE1 with the help of heparin and endogenous proteases(?) can further digest cf.DNA into T-end fragments in plasma.
This hypothesis is substantiated by literature published on the DFFB enzyme, which plays a major role in DNA fragmentation during apoptosis.26,27 Enzyme characterization studies have shown that DFFB creates blunt double-strand breaks in open internucleosomal DNA regions, and that this process has a preference for A and G nucleotides (purines).26,28,29 This biology of blunt double-stranded cutting only at internucleosomal linker regions would explain the nucleosomal patterning in A< >A, A< >G, and A< >C fragments.
In this work, we have also demonstrated that typical cf.DNA in plasma obtained before incubation predominantly end in C across all fragment sizes; this C-end overrepresentation is consistent in multiple different regions across the genome. Because the typical profile of cf.DNA is so different from the profile enriched with newly released cf.DNA, we can infer that (1) one or more additional nuclease(s) create(s) this profile, (2) this nuclease or these nucleases dominate(s) the cleaving process in typical cf.DNA, and (3) this process largely occurs after the generation of newly released A-end fragments.
Because this C-end predominance is lost in Dnase1l3-deficient mice, we believe that one nuclease responsible for creating this C-end fragment overrepresentation is DNASE1L3. Although there is no existing enzymatic study that investigates the specific nucleotide cleavage preference of DNASE1L3, DNASE1L3 is known to highly efficiently cleave chromatin to almost undetectable levels without proteolytic help.30,31 The fairly uniform abundance of C-end fragments among all fragment sizes suggests that DNASE1L3 can cleave all DNA, even intranucleosomal DNA, efficiently.
DNASE1L3 has interesting properties: it is expressed in the endoplasmic reticulum to be secreted extracellularly as one of the major serum nucleases, and it translocates to the nucleus upon cleavage of its endoplasmic reticulum-targeting motif after apoptosis is induced.23,32 It has been suggested that, in its role as an apoptotic intracellular endonuclease, DNASE1L3 cooperates with DFFB in DNA fragmentation.32,33 When we compared the fragment end profiles of newly released cf.DNA with that of Dnase1l3-deficient mice, we found a noticeable attenuation of the periodicity in A-end fragments, and especially in the A< >C fragment. We suspect that this attenuation is due to the coexisting intracellular activity of DNASE1L3 during the generation of newly fragmented DNA from apoptosis in WT versus in Dnase1l3-deficient mice.
As a plasma nuclease, DNASE1L3 would help digest the DNA in circulation that had escaped phagocytosis after apoptosis. Hence, DNASE1L3 would likely exert its effect on fragmented cf.DNA after intracellular fragmentation had occurred. In a theoretical two-step process, inhibiting the second step should reveal the usually transient outcome of the first step. So, in essence, the plasma of Dnase1l3-deficient mice would have this second step of DNASE1L3 action inhibited and would expose the cf.DNA profile of the first step, the intracellular DNA fragmentation from apoptosis. This is exactly what we found: the cf.DNA fragment profile was remarkably similar to that found in newly released cf.DNA. Thus, DNASE1L3 digestion within the plasma might be a subsequent step that would result in the typical homeostatic cf.DNA.
Although we previously found that the size profile of cf.DNA from Dnase1-deficient mice did not appear to be substantially different from that of WT mice, DNASE1 is known to prefer cleaving “naked” DNA, and it can only cleave chromatin with proteolytic help in vivo.31,34 Using heparin to replace the function of in vivo proteases to enhance DNASE1 activity, we have demonstrated that DNASE1 prefers to cut DNA into T-end fragments. The increase in T-end fragments with heparin incubation is predominantly among subnucleosomally sized (50–150 bp) fragments, suggesting that DNASE1 has a role in generating short <150 bp fragments. Knowing that DNASE1 prefers to cleave naked DNA into T-end fragments, we can infer from the typical cf.DNA profile that the T-end fragment peaks in the 50–150 bp and 250–300 bp ranges may be mostly naked. This may be possible because these sizes correspond to subnucleosomal fragments or linker fragments; however, more studies should be done to further investigate this hypothesis.
The use of heparin incubation and end analysis has also provided a unique insight into the origin of the 10 bp periodicity. Because every fragment type demonstrates a 10 bp periodicity, we show that no one specific nuclease is completely responsible for the 10 bp periodicity in short fragments. Instead, we demonstrate that for all fragment types, the 10 bp periodicity is abolished when heparin is used. In addition to enhancing DNASE1 activity, heparin disrupts the nucleosomal structure.35 Although many have postulated that the 10 bp periodicity originates from the cutting of DNA within an intact nucleosomal structure, we believe that this work provides supportive evidence by showing that no 10 bp periodicity occurs in the presence of a disrupted nucleosome.
Recently, Watanabe et al. induced in vivo hepatocyte necrosis with acetaminophen overdose and apoptosis with anti-Fas antibody treatments in mice deficient in Dnase1L3 and Dffb.36 Although Watanabe et al. claim to have shown that cf.DNA is generated by DNASE1L3 and DFFB, their data only show that serum cf.DNA does not appear to increase after hepatocyte injury in Dnase1l3- and Dffb-double knockout mice. Even then, the degree of hepatocyte injury from their methods is hugely variable even in WT, and there is surprisingly low correlation with the cf.DNA amount in their apoptotic anti-Fas antibody experiments. In addition to these inconsistencies that give uncertainty to the degree of apoptosis induced in their knockout mice, they have none of the detail on fragment ends that is offered in this study. While our RT incubation system is an in vitro model, and the in vivo reality is likely to be more complicated, we believe the insights generated would nonetheless be valuable.
In this study, we have demonstrated that the typical cf.DNA fragment might be created in two major steps: (1) intracellular DNA fragmentation by DFFB, intracellular DNASE1L3, and other apoptotic nucleases, and (2) extracellular DNA fragmentation by serum DNASE1L3. Then, likely with in vivo proteolysis, DNASE1 can further degrade cf.DNA into short T-end fragments. We believe that this first model has included a number of key nucleases involved in cf.DNA generation, but the model can be further refined in the future. For example, other potential apoptotic nucleases include endonuclease G, AIF, topoisomerase II, and cyclophilins, and there are probably more to be discovered.37, 38, 39 Further studies into these nucleases with double knockout models would further refine this model and may reveal a nuclease with G-end preference. In essence, in this work, we have definitively linked the action of distinct nucleases to the cf.DNA fragment end profile, clarifying the fundamental biology and biography of cf.DNA fragments.
With this link between nuclease biology and cf.DNA physiology established, there are many important and practical implications to the field of cf.DNA. First, aberrations in nuclease biology with pathological consequences may be reflected in abnormal cf.DNA profiles.40, 41, 42 Second, plasma end motif analysis is a powerful approach for investigating cf.DNA biology and may have diagnostic applications. And lastly, the pre-analytical variables such as anticoagulant type and time delay in blood separation are vital confounders to bear in mind when mining cf.DNA for epigenetic and genetic information.
Declaration of Interests
D.S.C.H., M.N., R.W.K.C., and Y.M.D.L. have filed patent applications based on this work. Y.M.D.L. is a scientific co-founder and shareholder of Grail. Y.M.D.L. and R.W.K.C. are co-founders and shareholders of DRA Limited and Take2 Holdings Limited. Y.M.D.L. and R.W.K.C. are consultants to Grail. Y.M.D.L. is an advisor to Decheng Capital. R.W.K.C. is an advisor to Illumina. R.W.K.C. is a shareholder of Grail. Y.M.D.L. and R.W.K.C. receive royalties from Illumina, Sequenom, DRA, and Grail.
Acknowledgments
We would like to thank Ms. Lee Wing Shan and Ms. Jin Yongjie for their technical assistance.
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region (SAR) Government under the theme-based research scheme (T12-403/15-N, T12-401/16-W), by a collaborative research agreement from Grail, and by the Vice Chancellor’s One-Off Discretionary Fund of The Chinese University of Hong Kong (VCF2014021). Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation.
Published: January 30, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.01.008.
Accession Numbers
The accession number for the datasets reported in this paper is at the European Genome-Phenome Archive (EGA): EGAS00001003514.
Web Resources
BEDTools (v2.27.1) (AR Quinlan et al., 2010), https://bedtools.readthedocs.io/en/latest/
mm9, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.18/
European Genome-Phenome Archive, https://www.ebi.ac.uk/ega/home
SOAP2 (Ruiqiang Li et al., 2009), https://github.com/gigascience/bgi-soap2
Supplemental Information
References
- 1.Lo Y.M.D., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W.G., Wainscoat J.S. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Chiu R.W.K., Chan K.C.A., Gao Y., Lau V.Y.M., Zheng W., Leung T.Y., Foo C.H.F., Xie B., Tsui N.B.Y., Lun F.M.F. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan K.C.A., Woo J.K.S., King A., Zee B.C.Y., Lam W.K.J., Chan S.L. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017;377:513–522. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 4.Sun K., Jiang P., Cheng S.H., Cheng T.H.T., Wong J., Wong V.W.S., Ng S.S.M., Ma B.B.Y., Leung T.Y., Chan S.L. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res. 2019;29:418–427. doi: 10.1101/gr.242719.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder M.W., Kircher M., Hill A.J., Daza R.M., Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov M., Baranova A., Butler T., Spellman P., Mileyko V. Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. BMC Genomics. 2015;16(Suppl 13):S1. doi: 10.1186/1471-2164-16-S13-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrananda D., Thorne N.P., Bahlo M. High-resolution characterization of sequence signatures due to non-random cleavage of cell-free DNA. BMC Med. Genomics. 2015;8:29. doi: 10.1186/s12920-015-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo Y.M.D., Chan K.C.A., Sun H., Chen E.Z., Jiang P., Lun F.M.F. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Science Translational Medicine. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. 61ra91. [DOI] [PubMed] [Google Scholar]
- 9.Serpas L., Chan R.W.Y., Jiang P., Ni M., Sun K., Rashidfarrokhi A., Soni C., Sisirak V., Lee W.-S., Cheng S.H. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. USA. 2019;116:641–649. doi: 10.1073/pnas.1815031116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu R.W.K., Poon L.L.M., Lau T.K., Leung T.N., Wong E.M.C., Lo Y.M.D. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin. Chem. 2001;47:1607–1613. [PubMed] [Google Scholar]
- 11.Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y., Sinha M., Peterson C.L., Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59:1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brauchle E., Thude S., Brucker S.Y., Schenke-Layland K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 2014;4:4698. doi: 10.1038/srep04698. 4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimura M., Okuma E., Yuo A., Sasaki T., Mukai C., Takaku F., Ishizaka Y. Room temperature-induced apoptosis of Jurkat cells sensitive to both caspase-1 and caspase-3 inhibitors. Cancer Lett. 1998;132:7–16. doi: 10.1016/s0304-3835(98)00116-5. [DOI] [PubMed] [Google Scholar]
- 18.Shimura M., Ishizaka Y., Yuo A., Hatake K., Oshima M., Sasaki T., Takaku F. Characterization of room temperature induced apoptosis in HL-60. FEBS Lett. 1997;417:379–384. doi: 10.1016/s0014-5793(97)01327-6. [DOI] [PubMed] [Google Scholar]
- 19.Bergman M., Bessler H., Salman H., Djaldetti M. Relationship between temperature and apoptosis of human peripheral blood mononuclear cells. Int. J. Hematol. 2003;77:351–353. doi: 10.1007/BF02982642. [DOI] [PubMed] [Google Scholar]
- 20.Lanigan R.S., Yamarik T.A. Final report on the safety assessment of EDTA, calcium disodium EDTA, diammonium EDTA, dipotassium EDTA, disodium EDTA, TEA-EDTA, tetrasodium EDTA, tripotassium EDTA, trisodium EDTA, HEDTA, and trisodium HEDTA. Int. J. Toxicol. 2002;21(Suppl 2):95–142. doi: 10.1080/10915810290096522. Lanigan, R.S. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurti C., Saryan L.A., Petering D.H. Effects of ethylenediaminetetraacetic acid and 1,10-phenanthroline on cell proliferation and DNA synthesis of Ehrlich ascites cells. Cancer Res. 1980;40:4092–4099. [PubMed] [Google Scholar]
- 22.Flora S.J.S., Pachauri V. Chelation in metal intoxication. Int. J. Environ. Res. Public Health. 2010;7:2745–2788. doi: 10.3390/ijerph7072745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napirei M., Wulf S., Eulitz D., Mannherz H.G., Kloeckl T. Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3) Biochem. J. 2005;389:355–364. doi: 10.1042/BJ20042124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manaster J., Chezar J., Shurtz-Swirski R., Shapiro G., Tendler Y., Kristal B., Shasha S.M., Sela S. Heparin induces apoptosis in human peripheral blood neutrophils. Br. J. Haematol. 1996;94:48–52. doi: 10.1046/j.1365-2141.1996.6202063.x. [DOI] [PubMed] [Google Scholar]
- 25.Klug A., Lutter L.C. The helical periodicity of DNA on the nucleosome. Nucleic Acids Res. 1981;9:4267–4283. doi: 10.1093/nar/9.17.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen B.D., Sørensen C.S. The caspase-activated DNase: apoptosis and beyond. FEBS J. 2017;284:1160–1170. doi: 10.1111/febs.13970. [DOI] [PubMed] [Google Scholar]
- 27.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widlak P., Garrard W.T. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J. Cell. Biochem. 2005;94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 29.Widlak P., Li P., Wang X., Garrard W.T. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- 30.Sisirak V., Sally B., D’Agati V., Martinez-Ortiz W., Özçakar Z.B., David J. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Napirei M., Ludwig S., Mezrhab J., Klöckl T., Mannherz H.G. Murine serum nucleases--contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3) FEBS J. 2009;276:1059–1073. doi: 10.1111/j.1742-4658.2008.06849.x. [DOI] [PubMed] [Google Scholar]
- 32.Errami Y., Naura A.S., Kim H., Ju J., Suzuki Y., El-Bahrawy A.H., Ghonim M.A., Hemeida R.A., Mansy M.S., Zhang J. Apoptotic DNA fragmentation may be a cooperative activity between caspase-activated deoxyribonuclease and the poly(ADP-ribose) polymerase-regulated DNAS1L3, an endoplasmic reticulum-localized endonuclease that translocates to the nucleus during apoptosis. J. Biol. Chem. 2013;288:3460–3468. doi: 10.1074/jbc.M112.423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyama R., Arai T., Kijima M., Sato S., Miura S., Yuasa M., Kitamura D., Mizuta R. DNase γ, DNase I and caspase-activated DNase cooperate to degrade dead cells. Genes Cells. 2016;21:1150–1163. doi: 10.1111/gtc.12433. [DOI] [PubMed] [Google Scholar]
- 34.Cheng T.H.T., Lui K.O., Peng X.L., Cheng S.H., Jiang P., Chan K.C.A. DNase1 does not appear to play a major role in the fragmentation of plasma DNA in a knockout mouse model. Clin. Chem. 2018;64:406–408. doi: 10.1373/clinchem.2017.280446. [DOI] [PubMed] [Google Scholar]
- 35.Villeponteau B. Heparin increases chromatin accessibility by binding the trypsin-sensitive basic residues in histones. Biochem. J. 1992;288:953–958. doi: 10.1042/bj2880953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe T., Takada S., Mizuta R. Cell-free DNA in blood circulation is generated by DNase1L3 and caspase-activated DNase. Biochem. Biophys. Res. Commun. 2019;516:790–795. doi: 10.1016/j.bbrc.2019.06.069. [DOI] [PubMed] [Google Scholar]
- 37.Samejima K., Earnshaw W.C. Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 38.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagata S. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 2018;36:489–517. doi: 10.1146/annurev-immunol-042617-053010. [DOI] [PubMed] [Google Scholar]
- 40.Jiménez-Alcázar M., Rangaswamy C., Panda R., Bitterling J., Simsek Y.J., Long A.T., Bilyy R., Krenn V., Renné C., Renné T. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017;358:1202–1206. doi: 10.1126/science.aam8897. [DOI] [PubMed] [Google Scholar]
- 41.Ozçakar Z.B., Foster J., 2nd, Diaz-Horta O., Kasapcopur O., Fan Y.S., Yalçınkaya F., Tekin M. DNASE1L3 mutations in hypocomplementemic urticarial vasculitis syndrome. Arthritis Rheum. 2013;65:2183–2189. doi: 10.1002/art.38010. [DOI] [PubMed] [Google Scholar]
- 42.Al-Mayouf S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N., Al Sonbul A., Sewairi W., Qari A., Abdallah E. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 2011;43:1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.