Abstract
Blastocystis sp. is a common eukaryotic parasite, which infects humans as well as various other animals. To date, epidemiological data regarding the detection rate and distribution of Blastocystis sp. subtypes in pet rodents are lacking in China; the present study aims to fill this gap. A total of 503 fecal samples collected from pets in different locations in southwestern China were screened for the presence of Blastocystis sp. using a nested PCR amplification of SSU rRNA method. Forty-two samples (8.35%) tested positive for Blastocystis sp. colonization. Two subtypes of Blastocystis sp. were identified based on nucleotide sequence homology and phylogenetic analysis: Blastocystis ST4 was present in 41 samples, and Blastocystis ST17 was found in 1 sample. Our results revealed robust host preference of Blastocystis ST4 and confirmed that Blastocystis ST17 can also parasitize rodents.
Keywords: Blastocystis sp., Pet rodent, Subtype, China
Highlights
-
•
This is the first survey of Blastocystis sp. from pet rodent in southwestern of China and that 503 rodents were surveyed.
-
•
Forty-two samples (8.35%) tested positive for Blastocystis sp. colonization.
-
•
Forty-one isolates were identified as subtype ST4 and one as ST17.
-
•
Subtype ST4 shows a robust host preference for rodents.
1. Introduction
Blastocystis is a genus containing common single-celled intestinal parasitic protists (Andersen and Stensvold, 2015). Blastocystis sp. commonly colonizes the gastrointestinal tracts of humans and a range of other animals (Greige et al., 2018). It is transmitted among hosts through the fecal-oral route (Asghari et al., 2019). Previous studies have shown that Blastocystis sp. is associated with irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD) (Kumarasamy et al., 2018). However, no study has been able to confirm that Blastocystis sp. is the sole etiological agent of either IBS or IBD (Stensvold and Clark, 2016; Shirvani et al., 2019). Therefore, multicenter studies are also required to further investigate the clinical implications of Blastocystis sp. with respect to IBS and IBD (Boorom et al., 2008; Wawrzyniak et al., 2013; Rojaleen et al., 2016; Lepczyńska et al., 2017; Aynur et al., 2019). Moreover, studies have indicated that Blastocystis sp. is a commensal organism that inhabits the healthy gut, rather than an organism that is only present during gut dysbiosis that is a characteristic feature of metabolic or infectious inflammatory diseases of the lower gastrointestinal tract (Scanlan et al., 2014; Yoshikawa et al., 2016). Several studies have demonstrated the presence of Blastocystis sp. in healthy, aymptomatic individuals from Europe, Asia, Africa and South America (Guimaraes and Sogayar, 1993; Moosavi et al., 2012; Pandey et al., 2015; Ben Abda et al., 2017; Nieves-Ramirez et al., 2018). Currently, the pathogenicity of Blastocystis sp. remains controversial and unclear (Li et al., 2018; Asghari et al., 2019), which makes it difficult to implement the systematic research approaches commonly used to study other infectious species (Boorom et al., 2008).
Two date, 17 different subtypes of Blastocystis sp. have been identified (Cian et al., 2017). Among them, ST1 to ST9 and ST12 have been reported in humans with varying prevalence levels (Ramírez et al., 2016). ST1 to ST8 have been identified in both humans and animals, and considered have zoonotic potential (Song et al., 2017; Xiao et al., 2019). The others strains (ST9 to ST17) have been exclusively identified in either humans or animals; for example, ST9 has only been isolated from humans (Stensvold et al., 2009; Tan, 2008). Studies conducted in different parts of the world show that ST4 is the most common subtype detected in rodents (Katsumata et al., 2018). In addition, ST1 - ST8 (with the exception of ST6), ST13, and ST17 have all been isolated from various rodents (Alfellani et al., 2013a, Alfellani et al., 2013b; Cian et al., 2017; Katsumata et al., 2018; Valença-Barbosa et al., 2019; Xiao et al., 2019) (Table 2).
Table 2.
Subtypes and positive samples of Blastocystis sp. detected from rodents in the world.
| Country | Host (scientific name) | Technique | Number of samples | Number of Positive | Prevalence (%) | Subtypes(n) | References |
|---|---|---|---|---|---|---|---|
| Brazil | Cursorial akodont (Akodon cursor) | PCR | 1 | 1 | 100 | Valença-Barbosa et al. (2019) | |
| House Rat (Rattus rattus) | PCR | 1 | 1 | 100 | ST3 (1) | ||
| Montane Grass Mouse (Akodon montensis) | PCR | 2 | 2 | 100 | |||
| Brazilian forest rodent (Atlantic Forest Nectomys) | PCR | 1 | 1 | 100 | |||
| Brazilian forest rodent (Atlantic Forest Nectomys) | PCR | 2 | 2 | 100 | ST8 (1) | ||
| Japan | Brown rat (Rattus norvegicus) | PCR | ST4 (11) | Katsumata et al. (2018) | |||
| Rat (Vole), Guinea pig (Cavia porcellus) Unclear specific host |
PCR | ST7 (2) | |||||
| Indonesia | Polynesian rat (Rattus exulans) | PCR | 12 | ? | ST4 (12) | ||
| Polynesian rat (Rattus exulans) | PCR | 77 | 10 | 13 | ST4 (9) | Yoshikawa et al. (2016) | |
| France | Norway rat (Rattus norvegicus) | qPCR | 2 | 1 | 50 | ST4 | Cian et al. (2017) |
| Capybara (Hydrochoerus hydrochaeris) | qPCR | 5 | 3 | 60 | ST2 (1),ST5 (1) | ||
| USA | Rat (Rattus sp.) | qPCR | 5 | 5 | 100 | ST4 (5) |
Noël et al. (2003) Noël et al. (2005) Yoshikawa et al. (1998) Leipe et al. (1996) |
| Guinea pig (Cavia porcellus) | qPCR | 2 | 2 | 100 | ST4 (2) | ||
| UK | Bank vole (Clethrionomys glareolus) | Sequencing | 32 | 1 | 3.13 | ST5 | Alfellani et al., 2013a, Alfellani et al., 2013b |
| Wood mouse (Apodemus sylvaticus) | Sequencing | 13 | 1 | 7.69 | ST3 | ||
| Belgium Croatia | Chinchilla (Chinchilla lanigera) | Sequencing | 5 | 2 | 40 | ST3 (2) | |
| Poland | Yellow necked mouse (Apodemus flavicollis) | Sequencing | 1 | 1 | 100 | ST3 | |
| Libya | Gundi (Ctenodactylus gundi) | Sequencing | 4 | 1 | 25 | ST17 | |
| Colombia | House Rat (Rattus rattus) | STs | 3 | 无 | ST2 (3) | Ramírez et al. (2014) | |
| China | Brown rat (Rattus norvegicus) | PCR | 108 | 4 | 3.7 | ST4 (4) | Deng et al. (2019) |
| China | Trogopterus xanthipes (Rodentia) | PCR | 69 | 21 | 30.4 | ST1 (8),ST3 (4),ST13 (9) | Xiao et al. (2019) |
| China | Eurasian Red Squirrel (Sciurus vulgaris) | PCR | 72 | 7 | 9.72 | ST4 (7) | This study |
| Eastern Chipmunk (Tamias striatus) | PCR | 171 | 8 | 4.68 | ST4 (8) | ||
| Chinchilla (Chinchilla lanigera) | PCR | 72 | 3 | 4.17 | ST4 (2),ST17 (1) | ||
| Guinea Pig (Cavia porcellus) | PCR | 90 | 12 | 13.33 | ST4 (12) | ||
| Chinese Striped Hamster (Cricetulus barabensis) | PCR | 98 | 12 | 12.24 | ST4 (12) | ||
| Eurasian Red Squirrel (Sciurus vulgaris) | PCR | 72 | 7 | 9.72 | ST4 (7) | ||
| Eastern Chipmunk (Tamias striatus) | PCR | 171 | 8 | 4.68 | ST4 (8) |
In China, over 12 provinces/municipalities have Blastocystis sp. infection reported (Wang et al., 2018; Deng et al., 2019). Blastocystis sp. has been reported in many animals, such as pigs, cattle, sheep, goats, and cats (Zhu et al., 2017; Wang et al., 2018). However, to date no genetic studies have been conducted on Blastocystis sp. isolated from pet rodents in China, and its role as reservoirs of infection for humans and other animals is unknown. Blastocystis sp. identified from rodents has been reported in literature from the USA, France, Singapore, and Japan (Katsumata et al., 2018). Pet rodents are common companion animals that live in close association with the owners, and pet rodents can harbor human pathogens (Jacob et al., 2014). The current study aimed to determine the existence and diversity of Blastocystis sp. in rodents being kept as pets in different cities of southwestern China.
2. Materials and methods
2.1. Ethical statement
This study was performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China. Before the initiation of experiments, the protocol of the current study was reviewed and approved by the Institutional Animal Care and Use Committee of the Sichuan Agricultural University under permit. No animals were harmed during the sampling process. Permission was obtained from pet owners or shop managers prior to collection of fecal specimens.
2.2. Study sites
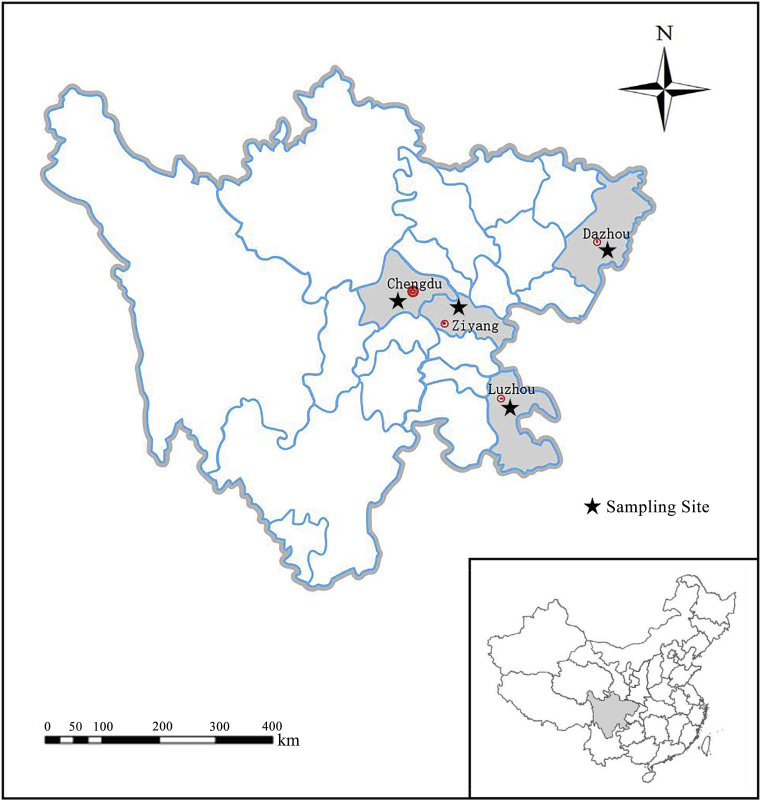
The study was carried out in Sichuan Province, China. This province covers an area of over 486,000 square kilometers, with approximately 83 million people. The sample collection was conducted in four regions of the province (Chengdu, Ziyang, Luzhou, and Dazhou) (Fig. 1).
Fig. 1.
Sampling sites in Sichuan Province of China.
2.3. Sampling
A total of 503 samples were collected from the following four regions of Sichuan province: Chengdu (n = 311), Luzhou (n = 98), Ziyang (n = 63), and Dazhou (n = 31). The animals sampled were eurasian red squirrel (Sciurus vulgaris), eastern chipmunk (Tamias striatus), chinchillas (Chinchilla lanigera), and guinea pig (Cavia porcellus), and Chinese Hamster (Cricetulus barabensis) (Table 1). The fecal samples were collected between September 2018 and May 2019 in Sichuan Province, China. Each rodent was kept in a separate cage. Approximately 200 mg of fresh fecal samples were collected using sterile gloves from the excrement disc at the bottom of the cage immediately after defecation. Samples were then transferred to sterile plastic containers marked with the species and sampling date. The fecal samples were transported to the laboratory by storing along with ice packs within 24 h of collection. All study animals were examined, and no pronounced clinical signs were apparent during sampling.
Table 1.
Detection rate and subtypes of Blastocystis sp in rodents from different sources in Southwestern China.
| Location | Host | Scientific name | No. of examined | No. of positive | Detection rate (%) | Species (n) |
|---|---|---|---|---|---|---|
| Chengdu | Eurasian Red Squirrel | Sciurus vulgaris | 72 | 7 | 9.72 | ST4 (7) |
| Eastern Chipmunk | Tamias striatus | 108 | 3 | 2.78 | ST4 (3) | |
| Chinchilla | Chinchilla lanigera | 72 | 3 | 4.17 | ST4 (2), ST17 (1) | |
| Guinea pig | Cavia porcellus | 59 | 10 | 16.95 | ST4 (10) | |
| Subtotal | 311 | 23 | 7.40 | ST4 (22), ST17 (1) | ||
| Luzhou | Chinese Striped Hamster | Cricetulus barabensis | 98 | 12 | 12.24 | ST4 (12) |
| Ziyang | Eastern Chipmunk | Tamias striatus | 63 | 5 | 7.94 | ST4 (5) |
| Dazhou | Guinea pig | Cavia porcellus | 31 | 2 | 6.45 | ST4 (2) |
| Total | 503 | 42 | 8.35 | ST4 (41), ST17 (1) |
2.4. DNA extraction
Genomic DNA was extracted directly from fecal samples (approximately 200 mg) using the QIAamp DNA Stool Mini Kit (Qiagen GmbH, Hilden, Germany), in accordance to the procedures recommended by the manufacturer. The extracted DNA was stored at −20 °C until PCR analysis.
2.5. Subtyping of Blastocystis sp.
All DNA preparations were screened for the presence of Blastocystis sp. by PCR amplification of the barcode region (a fragment of ~510 bp) of the SSU rRNA gene. The primers and cycling parameters were in accordance to those described by Scicluna et al. (2006). TaKaRa Taq DNA polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all of the PCR reactions. A negative control with no DNA added was included in all of the PCR tests. PCR products were subjected to electrophoresis in a 1.5% agarose gel and visualized by staining the gel with ethidium bromide.
2.6. Sequence analysis
All positive PCR products were directly sequenced on an ABI PRISMTM 3730 DNA Analyzer (Applied Biosystems, Foster, CA, USA), using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Nucleotide sequences obtained in the present study were subjected to BLAST (http://www.ncbi.nlm.nih.gov/blast/)., aligned with each other, and analyzed. Reference sequences were downloaded from the GenBank (http://www.ncbi.nlm.nih.gov). The sequences were aligned using Clustal X 2.0 (http://www.clustal.org/) to determine the Blastocystis sp. subtype. The nucleotide sequences generated in the present study have been deposited in GenBank (Table S1).
2.7. Phylogenetic analyses
A neighbor-joining tree was constructed to assess the genetic relationship among the Blastocystis subtypes obtained in the present study and those identified in previous studies using the software Mega 7 (http://www.megasoftware.net/). The sequences of the barcode region of Blastocystis sp. were trimmed using trimAl (Capella-Gutiérrez et al., 2009). Evolutionary distances were calculated using the Kimura two-parameter model. The reliability of the trees was assessed by bootstrap analysis with 1000 replicates.
2.8. Statistical analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc., Chicago, IL, USA). A chi-squared test was used to compare the occurrence of Blastocystis sp. in different pet markets and different species. Differences were considered statistically significant at p < 0.05.
3. Results
3.1. Detection rate of Blastocystis sp. in pet rodents
The overall minimum prevalence of Blastocystis sp. in pet rodents was 8.35% (42/503; 95% CI: 0.06–0.11). The detection rate in different rodent species ranged from 4.17 to 13.33% (Table 1), and differed significantly between the five pet species (χ2 = 9.699, df = 4, p < 0.05). There was no significant difference in the prevalence of Blastocystis sp. between the four locations tested, which ranged from 6.45 to 12.24% (χ2 = 2.473, df = 3, p > 0.05).
3.2. Distribution of Blastocystis sp. subtypes in pet rodents
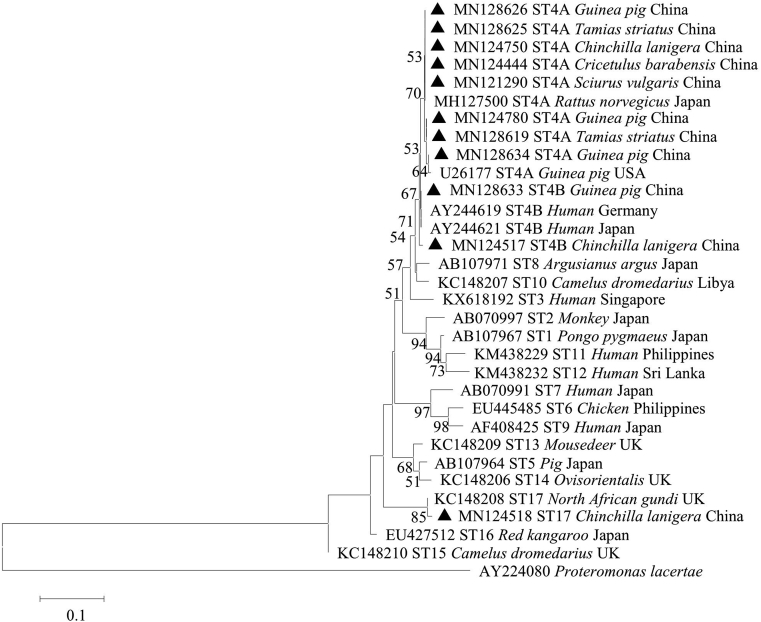
Two Blastocystis sp. subtypes were successfully sequenced, and all SSU rRNA –positive PCR samples were sequenced and phylogenetically analyzed (Fig. 2). Two subtypes (ST4 and ST17) were identified based on the phylogenetic tree (ST4, ST17), both of which are known to have zoonotic potential (Table 1, Fig. 2). ST4 was the predominant subtype, and was widely distributed among the different species of rodents and locations (Table 1). Additionally, ST17 was discovered in a Chinchilla, which is a newly identified host for this subtype.
Fig. 2.
Phylogenetic relationships among nucleotide sequences of Blastocystis partial small subunit ribosomal RNA (SSU rRNA) genes. The neighbor-joining method was used to construct the trees from the Kimura-2-parameter model. Branch numbers represent percent bootstrapping values from 1000 replicates, with values of more than 50% shown in the tree. Each sequence is identified by its accession number, subtypes, host origin, and country. Blastocystis subtypes identified in the present study are indicated in bold-type.▲ are subtypes in this study.
3.3. Phylogenetic analyses
The sequences identified in this study were aligned to known sequences downloaded from GenBank. Thirty-six sequences clustered with subtype ST4A (Genebank accession number MH127500 form Rattus norvegicus in Japan). Five sequences clustered with ST4B, which has been identified in human in Germany and Japan (AY244619, AY244621). However, only one sequence clustered with ST17, which has been identified in the North African gundi in the UK (Genebank accession number KC148208) (Fig. 2).
4. Discussion
Blastocystis sp. is the most frequent parasite colonizing in humans and a variety of animals (Meloni et al., 2011; Yoshikawa et al., 2012). Some studies have found that infection with Blastocystis sp is linked to gastrointestinal and nutritional disorders in both developing and developed countries (Seguí et al., 2018). However, other studies have shown that the presence of Blastocystis may be an indicator of good intestinal health (Andersen and Stensvold, 2015).
Our results revealed a Blastocystis sp. prevalence of 8.35% in non-diarrheal pet rodents, which is lower than that found in wild rodents in an Indonesian community (13%) (Yoshikawa et al., 2016). The difference is likely due to the studying animals were wild rodents in Indonesia and pet rodents in China. Generally, shopkeepers in this study cleaned rodent cages regularly, provided clean water, used chlorine for disinfection, and have good sanitary conditions, which may explain the low prevalence of Blastocystis sp. in this study.
Previous studies have shown the global prevalence of ST4 in rodents. This subtype predominates in rodents such as brown rats in China, Indonesia, the Philippines, and Japan, and guinea pigs in the UK (Leipe et al., 1996; Abe, 2004; Belleza et al., 2016; Katsumata et al., 2018; Wang et al., 2018). Four subtypes, ST1–ST4 have the highest prevalence (more than 90%) in humans (Cian et al., 2017). Recent studies have revealed that ST4 is the common subtype in Europe, but is rare in other countries (Forsell et al., 2016, 2017; Deng et al., 2019; Gong and Liu, 2019). A few studies have examined the prevalence of ST4 in humans in the Zhejiang and Yunan provinces of China (Deng et al., 2019). Another study in China detected only three known subtypes (ST1, ST3, and ST13) in flying squirrels (Xiao et al., 2019). Previous studies have shown that ST4 has a peculiar geographical distribution and that ST4 is a subtype of Blastocystis that is most influenced by geography and lifestyle (Beghini et al., 2017; Forsell et al., 2017). Further studies are required to investigate the mode of transmission of the Blastocystis ST4 subtype in China. Unlike other Blastocystis subtypes that are commonly found in humans, rodents appear to constitute the main animal reservoir of ST4 (Stensvold et al., 2009). In China, Blastocystis sp. may be transmitted by contaminated water to humans indicating that ST4 is more likely to spread between humans and animals (Deng et al., 2019) (Table 2).
To our knowledge, this was the first study on Blastocystis sp. subtypes in pet rodent hosts in China. ST4 was the most common subtype of Blastocystis sp. in the rodents studied. Additionally, this was the first study to subtype Blastocystis sp. from chinchillas; none of the previous studies have reported the presence of Blastocystis sp. ST17 in China. ST17 was only detected in only one chinchilla from a pet store in Chengdu; however, the existence of this subtype should be further studied by examining additional samples from this and other geographical origins (AbuOdeh et al., 2019; Martinez-Hernandez et al., 2020). The role of pet rodents in transmitting ST4 and ST17 subtypes should be further evaluated. Our findings suggest that pet rodents may act as potential reservoirs for zoonotic Blastocystis sp. Further studies are needed to determine the distribution of Blastocystis subtypes in the pet and human populations in this region.
Declaration of competing interest
There is no conflict of interests.
Acknowledgments
The study was financially supported by the National Science and Technology Department “13th five-year” Special Subproject of China (No.2016YFD0501009) and Chengdu Giant Panda Breeding Research Foundation (CPF2017-12, CPF2015-09). The funders contributed to the study design and data collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2020.01.012.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abe N. Molecular and phylogenetic analysis of Blastocystis isolates from various hosts. Vet. Parasitol. 2004;120:235–242. doi: 10.1016/j.vetpar.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Alfellani M.A., Stensvold C.R., Vidal-Lapiedra A., Onuoha E.S., Fagbenro-Beyioku A.F., Clark C.G. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126 doi: 10.1016/j.actatropica.2012.12.011. 11–8. [DOI] [PubMed] [Google Scholar]
- AbuOdeh R., Ezzedine S., Madkoura M., Stensvoldb R.C., Samiec A., Nasrallahd G., AlAbsid E., ElBakria A. Molecular subtyping of Blastocystis from diverse animals in the United Arab Emirates. Protist. 2019;170:125679. doi: 10.1016/j.protis.2019.125679. [DOI] [PubMed] [Google Scholar]
- Alfellani M.A., Taner-Mulla D., Jacob A.S., Imeede C.A., Yoshikawa H., Stensvold C.R., Clark C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Andersen L.O., Stensvold C.R. Blastocystis in health and disease-are we moving from a clinical to a public health perspective? J. Clin. Microbiol. 2015;54:524. doi: 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghari A., Sadraei J., Pirestanib M., Mohammadpoura I. First molecular identification and subtype distribution of Blastocystis sp. isolated from hooded crows (Corvus cornix) and pigeons (Columba livia) in Tehran Province, Iran. Comp. Immunol. Microbiol. Infect. Dis. 2019;62:25–30. doi: 10.1016/j.cimid.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Aynur Z.E., Güçlü Ö., Yıldız İ., Aynur H., Ertabaklar H., Bülent B. Molecular characterization of Blastocystis in cattle in Turkey. Parasitol. Res. 2019;118:1055–1059. doi: 10.1007/s00436-019-06243-8. [DOI] [PubMed] [Google Scholar]
- Beghini F., Pasolli E., Truong T.D., Putignani L., Segata N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11 doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleza M.L.B., Reyes J.C.B., Tongol-Rivera P.N., Rivera W.L. Subtype analysis of Blastocystis sp. isolates from human and canine hosts in an urban community in the Philippines. Parasitol. Int. 2016;65:291–294. doi: 10.1016/j.parint.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Boorom K.F., Smith H., Nimri L., Viscogliosi E., Spanakos G., Parkar U., Li L.H., Zhou X., Ülgen Z.O., Leelayoova S., Jones M.S. Oh my aching gut: irritable bowel syndrome, Blastocystis, and asymptomatic infection. Parasites Vectors. 2008;1:40. doi: 10.1186/1756-3305-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian A., Safadi D.E., Osman M., Moriniere R., Gantois N., Benamrouzvanneste S., Viscogliosi P.D., Guyot K., Li L., Monchy S., Noe C., Poirier, Nourrisson C., Wawrzyniak I., Delbac F., Bosc S., Chabé M., Petit T., Certad G., Viscogliosi E. Molecular epidemiology of Blastocystis sp. in various animal groups from two French zoos and evaluation of potential zoonotic risk. PloS One. 2017;12 doi: 10.1371/journal.pone.0169659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Chai Y., Zhou Z., Liu H., Zhong Z., Hu Y., Fu H., Yue C., Peng G. Epidemiology of Blastocystis sp. infection in China: a systematic review. Parasite. 2019;26:41. doi: 10.1051/parasite/2019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell J., Bengtsson-Palme J., Angelin M., Johansson A., Evengård B., Granlund M. The relation between Blastocystis and the intestinal microbiota in Swedish travellers. BMC Microbiol. 2017;17:231. doi: 10.1186/s12866-017-1139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell J., Granlund M., Samuelsson L., Koskiniemi S., Edebro H., Evengård B. High occurrence of Blastocystis sp. subtypes 1-3 and Giardia intestinalis assemblage B among patients in Zanzibar, Tanzania. Parasites Vectors. 2016;9:370. doi: 10.1186/s13071-016-1637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong B., Liu X. Prevalence and subtype distribution of Blastocystis in ethnic minority groups on both sides of the China–Myanmar border, and assessment of risk factors. Parasite. 2019;26:46. doi: 10.1051/parasite/2019046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greige S., Safadi D.E., Bécu N., Gantois N., Pereira B., Chabé M., Benamrouz-Vanneste S., Certad G., Hage R.E., Chemaly M., Hamze M., Viscogliosi E. Prevalence and subtype distribution of Blastocystis sp. isolates from poultry in Lebanon and evidence of zoonotic potential. Parasites Vectors. 2018;11:389. doi: 10.1186/s13071-018-2975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes S., Sogayar M.I.L. Blastocystis hominis: occurrence in children and staff members of municipal day-care centers from Botucatu, S?o Paulo State, Brazil. Mem. Inst. Oswaldo Cruz. 1993;88:427–429. doi: 10.1590/s0074-02761993000300012. Rio de Janeiro. [DOI] [PubMed] [Google Scholar]
- Jacob J., Ulrich R.G., Freise J., Schmolz E. Monitoring von gesundheitsgefähr-denden Nagetieren. Bundesgesundheitsblatt - Gesundheitsforsch. - Gesundheitsschutz. 2014;57:511–518. doi: 10.1007/s00103-013-1924-x. [DOI] [PubMed] [Google Scholar]
- Katsumata M., Yoshikawa H., Tokoro M., Mizuno T. Molecular phylogeny of Blastocystis isolates from wild rodents captured in Indonesia and Japan. Parasitol. Res. 2018;117:2841–2846. doi: 10.1007/s00436-018-5973-9. [DOI] [PubMed] [Google Scholar]
- Kumarasamy V., Anbazhagan D., Subramaniyan V., Vellasamy S. Blastocystis sp., parasite associated with gastrointestinal disorders: an overview of its pathogenesis, immune modulation and therapeutic strategies. Curr. Pharmaceut. Des. 2018;24:3172–3175. doi: 10.2174/1381612824666180807101536. [DOI] [PubMed] [Google Scholar]
- Lepczyńska M., Białkowska J., Dzika E., Piskorz-Ogorek K., Korycins J. Blastocystis: how do specific diets and human gut microbiota affect its development and pathogenicity? Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1531–1540. doi: 10.1007/s10096-017-2965-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.C., Wang K., Gu Y. Occurrence of Blastocystis sp. and Pentatrichomonas hominis in sheep and goats in China. Parasit.Vectors. 2018;11:93. doi: 10.1186/s13071-018-2671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D.D., Tong S.M., Goggin C.L., Slemenda S.B., Pieniazek N.J., Sogin M.L. 16S-like rDNA sequences from Developayella elegans, Labyrinthuloides haliotidis,and Proteromonas lacerate confirm that the stramenopiles are a primarily heterotrophic group. Eur. J. Protistol. 1996;32:449–458. [Google Scholar]
- Martinez-Hernandez F., Martinez-Ibarra J.A., Lopez-Escamilla E., Villanueva-Garcia C., Muñoz-Garcia C.I., Rendon-Franco E., Maravilla P., Villalobos G. Molecular genotyping of Blastocystis spp. in wild mammals from Mexico. Parasitol. Res. 2020;119:97–104. doi: 10.1007/s00436-019-06530-4. [DOI] [PubMed] [Google Scholar]
- Meloni D., Sanciu G., Poirier P., Alaoui E.H., Chabé M., Delhaes L., Dei-Cas E., Delbac F., Fiori L.P., Cave D.D., Viscogliosi E. Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol. Res. 2011;109:613–619. doi: 10.1007/s00436-011-2294-7. [DOI] [PubMed] [Google Scholar]
- Moosavi A., Haghighi A., Mojarad E.N., Zayeri F., Alebouyeh M., Khazan H., Kazemi B., Zali M.R. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol. Res. 2012;111:2311–2315. doi: 10.1007/s00436-012-3085-5. [DOI] [PubMed] [Google Scholar]
- Noël C., Peyronnet C., Gerbod D., Edgcomb V.P., Delgado-Viscogliosi P., Sogin M.L., Capron M., Viscogliosi E., Zenner L. Phylogenetic analysis of Blastocystis isolates from different hosts based on the comparison of smallsubunit rRNA gene sequences. Mol. Biochem. Parasitol. 2003;126:119–123. doi: 10.1016/s0166-6851(02)00246-3. [DOI] [PubMed] [Google Scholar]
- Noël C., Dufernez F., Gerbod D., Edgcomb V.P., Delgado-Viscogliosi P., Ho L.-C., Singh M., Wintjens R., Sogin M.L., Capron M., Pierce R., Zenner L., Viscogliosi E. Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species and zoonosis. J. Clin. Microbiol. 2005;43:348–355. doi: 10.1128/JCM.43.1.348-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves-Ramirez M.E., Partida-Rodriguez O., Laforest-Lapointe I., Reynolds L.A., Brown E.M., Valdez-Salazar A., Morán-Silva P., Rojas-Velázquez L., Morien E., Parfrey L.W., Jini M., Walter J., Torres J., Arrieta M.C., Ximénez-García C., Finlay B.B. Asymptomatic intestinal colonization with protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018;3:e00007–18. doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P.K., Verma P., Marathe N., Shetty S., Bavdekar A., Patole M.S., Stensvold C.R., Shouche Y.S. Prevalence and subtype analysis of Blastocystis in healthy Indian individuals. Infect. Genet. Evol. 2015;31:296–299. doi: 10.1016/j.meegid.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez L.V., Bautista D.C., Corredor A.F., Flórez A.C., Stensvold C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014;22:223–228. doi: 10.1016/j.meegid.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Sánchez A., Hernández C., Flórez C., Bernal M.C., Giraldo J.C., Reyes P., López M.C., García L., Cooper P.J., Vicuña Y., Mongi F., Casero R.D. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- Rojaleen D., Shehla K., Mirdha B.R., Makharia G.K., Siddharta D., Rama C. Molecular characterization and subtyping of Blastocystis species in irritable bowel syndrome patients from north India. PloS One. 2016;11 doi: 10.1371/journal.pone.0147055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan P.D., Stensvold C.R., Rajilic-Stojanovic M., Heilig H.G.H.J., De Vos W.M., O'Toole P.W., Cotter P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- Scicluna S.M., Tawari B., Clark C.G. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Seguí R., Muñoz-Antoli C., Klisiowicz D.R., Oishi C.Y., Köster P.C., Lucio A.D., Hernández-de-Mingo M., Puente P., Toledo R., Esteban J.G., Carmena D. Prevalence of intestinal parasites, with emphasis on the molecular epidemiology of Giardia duodenalis and Blastocystis sp., in the Paranaguá Bay, Brazil: a community survey. Parasit.Vectors. 2018;11:490. doi: 10.1186/s13071-018-3054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirvani G., Fasihi-Harandi M., Raiesi O., Bazargan N., Zahedi M.J., Sharifi I., Kalantari-Khandani B., Nooshadokht M., Shabandoust H., Mohammadi A.M., Ebrahimipour M., Babaei Z. Prevalence and molecular subtyping of Blastocystis from patients with irritable bowel syndrome, inflammatory bowel disease and chronic urticaria in Iran. Acta Parasitol. 2019 doi: 10.2478/s11686-019-00131-y. 2019 Oct 10. [DOI] [PubMed] [Google Scholar]
- Song J., Hu R., Fan X., Wang S., Zhang H., Zhao G. Molecular characterization of Blastocystis from pigs in Shaanxi province of China. Acta Trop. 2017;173:130–135. doi: 10.1016/j.actatropica.2017.06.014. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Alfellani M.A., Nørskov-Lauritsen S., Pripa K., Victory E.L., Maddox C., Nielsen H.V., Clark C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009;39:473–479. doi: 10.1016/j.ijpara.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Stensvold C.R., Clark C.G. Current status of Blastocystis: a personal view. Parasitol. Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Tan K.S.W. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008;21:639–665. doi: 10.1128/CMR.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valença-Barbosa C., Bomfim T.C.B.D., Teixeira B.R., Gentile R., Neto S.F.D.C., Magalhães B.S.N., Balthazar D.D.A., Silva F.A.D., Biot R., Levy C.M.A., Santos H.L.C. Molecular epidemiology of Blastocystis isolated from animals in the state of Rio de Janeiro, Brazil. PloS One. 2019;14 doi: 10.1371/journal.pone.0210740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Gong B., Yang F., Zhang W., Zheng Y., Liu A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China's Heilongjiang Province. Infect. Genet. Evol. 2018;57:171–176. doi: 10.1016/j.meegid.2017.11.026. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I., Poirier P., Viscogliosi E., Meloni D. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Therap. Adv. Infect. Dis. 2013;1:167–178. doi: 10.1177/2049936113504754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zhou S., Jiang N., Tian D., Zhou Z., Zhang M., Ke H., Jiang X., Lv W., Gao Q. First record of Leptospira and Blastocystis infections in captive flying squirrels (Trogopterus xanthipes) from Enshi County, China. Acta Trop. 2019;197:105065. doi: 10.1016/j.actatropica.2019.105065. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Nagano I., Wu Z., Yap E.H., Singh M., Takahashi Y. Genomic polymorphism among Blastocystis hominis strains and development of subtypespecific diagnostic primers. Mol. Cell. Probes. 1998;12:153–159. doi: 10.1006/mcpr.1998.0161. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Dogruman-Al F., Turk S., Kustimur S., Balaban N., Sultan N. Erratum to: evaluation of DNA extraction kits for molecular diagnosis of human Blastocystis subtypes from fecal samples. Parasitol. Res. 2012;110 doi: 10.1007/s00436-011-2342-3. 1063-1063. [DOI] [PubMed] [Google Scholar]
- Yoshikawa H., Tokoro M., Nagamoto T., Arayama S. Molecular survey of Blastocystis sp. from humans and associated animals in an Indonesian community with poor hygiene. Parasitol. Int. 2016;65:780–784. doi: 10.1016/j.parint.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Zhu W., Tao W., Gong B., Yang H., Li Y., Song M., Lu Y., Li W. First report of Blastocystis infections in cattle in China. Vet. Parasitol. 2017;246:38–42. doi: 10.1016/j.vetpar.2017.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.