Abstract
Objective
Regulation of food intake and energy balance depends on a group of hypothalamic neurons that release anorexigenic melanocortins encoded by the Pomc gene. Although the physiological importance of central melanocortins is well appreciated, the genetic program that defines the functional identity of melanocortin neurons and assures high levels of hypothalamic Pomc expression is only beginning to be understood. This study assessed whether the transcriptional regulator PRDM12, identified as a highly expressed gene in adult mouse POMC neurons, plays an important role in the identity and function of melanocortin neurons.
Methods
We first determined the cellular distribution of PRDM12 in the developing hypothalamus. Then we studied mutant mice with constitutively inactivated Prdm12 to evaluate possible changes in hypothalamic Pomc expression. In addition, we characterized conditional mutant mice specifically lacking Prdm12 in ISL1-positive or POMC neurons during development. Finally, we measured food intake, body weight progression up to 16 weeks of age, adiposity, and glucose tolerance in adult mice lacking Prdm12 selectively from POMC neurons.
Results
PRDM12 co-expressed with POMC in mouse hypothalamic neurons from early development to adulthood. Mice lacking Prdm12 displayed greatly reduced Pomc expression in the developing hypothalamus. Selective ablation of Prdm12 from ISL1 neurons prevented hypothalamic Pomc expression. The conditional ablation of Prdm12 limited to POMC neurons greatly reduced Pomc expression in the developing hypothalamus and in adult mice led to increased food intake, adiposity, and obesity.
Conclusions
Altogether, our results demonstrate that PRDM12 plays an essential role in the early establishment of hypothalamic melanocortin neuron identity and the maintenance of high expression levels of Pomc. Its absence in adult mice greatly impairs Pomc expression and leads to increased food intake, adiposity, and obesity.
Keywords: Proopiomelanocortin, Transcriptional regulation, Neuronal identity, Conditional mutant, Obesity, Hyperphagia
Highlights
-
•
PRDM12 co-expresses with POMC in the developing hypothalamus and determines the establishment of melanocortin neuron identity.
-
•
The onset of Pomc expression in the presumptive arcuate nucleus is impaired in mutant mice lacking Prdm12.
-
•
PRDM12 promotes the transcriptional activation of Pomc in a cell-autonomous manner.
-
•
The lack of PRDM12 from POMC neurons leads to increased food intake, adiposity and obesity in adult mice.
1. Introduction
Body weight and energy balance are regulated by brain circuits that promote diverse food intake patterns by integrating a variety of central and peripheral signals. A group of neurons located in the arcuate nucleus of the hypothalamus release the potent anorexigenic melanocortins α-, β-, and γ-melanocyte-stimulating hormones encoded by the proopiomelanocortin gene (POMC) [1]. Humans deficient in POMC [2] or melanocortin 4 receptors [3] are hyperphagic and severely obese, similar to conditions observed in mutant mice carrying null-allele mutations of these genes [4,5]. The functional relevance of central melanocortins was further appreciated after the demonstration that the genetic rescue of Pomc expression in hypothalamic Pomc-deficient mice normalized their food intake and adiposity [6], even in previously hyperphagic and obese mice [7].
Functional studies of mice carrying deleterious mutations in either coding [8] or non-coding [6,9] Pomc sequences revealed that proper food intake regulation on a standard low fat diet is intact until the hypothalamic Pomc mRNA levels fall below ∼30% of normal values. Conditional mutant mice deficient in transcription factors (TF) necessary for hypothalamic Pomc expression have shown similar results [[10], [11], [12]]. Regardless of the genetic deficit, mice expressing hypothalamic Pomc below this threshold level exhibit hyperphagia and consequently develop excessive adiposity and overweight, even when consuming regular chow. Physiological levels of hypothalamic Pomc expression are thought to be maintained by a particular set of transcription factors that selectively interact with two upstream distal enhancers known as nPE1 and nPE2 [13]. These two highly conserved enhancers evolved independently from ancient insertions of different types of retroposons, which after acquiring adaptive mutations were fixed upstream of the Pomc transcriptional unit of a common ancestor to all extant placental mammals, sometime between 300 and 90 million years ago [14,15]. Genetic and functional studies demonstrated that nPE1 and nPE2 play cooperative and partially redundant functions and only the concurrent removal of the two neuronal Pomc enhancers leads to negligible levels of hypothalamic Pomc expression, hyperphagia, and early onset extreme obesity [9].
We discovered that the concurrent presence of the homeodomain transcription factors ISL1 [10] and NKX2.1 [11] is critical to specify the identity of melanocortin neurons. The use of temporal and spatial conditional mutant mice revealed that either the absence of ISL1 [10] or NKX2.1 [11] impairs the developmental onset of hypothalamic Pomc expression. Similar deficits in adult mice greatly reduce the expression of neuronal Pomc and lead to increased adiposity and body weight. However, because the number of NKX2.1+/ISL1+ neurons greatly exceeds that of POMC neurons in the arcuate nucleus, it is evident that the co-expression of these two TFs is necessary but not sufficient to sustain neuron-specific expression of Pomc. Therefore, at least one other TF is required to specify the identity of POMC neurons and regulate Pomc expression in the arcuate nucleus. Interestingly, a suitable candidate to fulfill this role has emerged from two recent independent reports. First, an RNA-seq study designed to compare the differential expression profiles of fluorescence-assisted manually isolated AGRP or POMC arcuate neurons identified the transcriptional regulator Prdm12 within the 14 foremost differentially expressed genes in adult POMC neurons, a list that included only 2 other TFs: Nkx2.1 and Nhlh2 [16]. A more recent study designed to identify the transcriptome of individual hypothalamic POMC neurons by single-cell RNA-seq also found Prdm12 and Nhlh2 as the only two TFs among the 25 mostly enriched transcripts present in a cluster of neurons expressing Pomc at the highest levels [17].
PRDM12 is a member of the PRDM subfamily of zinc finger TFs that has attracted considerable attention after the identification of 10 homozygous nonsense point mutations in patients from 11 unrelated families suffering from congenital insensitivity to pain [18]. It was recently demonstrated that PRDM12 plays a critical role in the early differentiation of the nociceptive sensory neuronal lineage that originates in the dorsal root ganglia and is critical for pain perception [19,20].
Given the coincidental expression of Prdm12 and Pomc in the neurons of the adult arcuate nucleus and the role of PRDM12 in the early differentiation of sensory neurons in the dorsal root ganglia, we tested whether PRDM12 plays a role in specifying the identity of hypothalamic melanocortin neurons and the maintenance of Pomc expression in this brain region. By combining genetic, cellular, and functional approaches, we demonstrate that PRDM12 is critical for the activation of arcuate Pomc expression during development. In addition, PRDM12 is necessary to maintain normal levels of hypothalamic Pomc mRNA and, consequently, normal food intake, adiposity, and body weight in adult mice.
2. Materials and methods
2.1. Breeding of mice
Mice of both sexes were housed in ventilated cages under controlled temperature and photoperiod (12-h light/12-h dark cycle, lights on from 7:00 AM to 7:00 PM), with tap water and laboratory chow containing 28.8% protein, 5.33% fat, and 65.87% carbohydrate available ad libitum. All of the procedures followed the Guide for the Care and Use of Laboratory Animals [21] and were approved by the INGEBI-CONICET institutional animal care and use committee. Prdm12+/lacZ(−) and Prdm12loxP mice were obtained by in vitro fertilization of C57BL/6J oocytes with thawed sperm carrying Prdm12tm2b(EUCOMM)Hmgu/Tcp or Prdm12tm2c(EUCOMM)Hmgu/Tcp alleles, respectively, generated by the European Conditional Mouse Mutagenesis Program (EUCOMM) by homologous recombination in C57BL/6N ES cells. Aliquots of cryopreserved sperm from mutant males were obtained from the Center for Phenogenomics (Toronto). Isl1-Cre [22] mice were obtained from the Jackson Laboratory (Isl1tm1(cre)Sev/J; stock no. 024242) and maintained as heterozygotes on a C57BL/6J background. Pomc-EGFP transgenic mice (C57BL/6J-Tg(Pomc-EGFP)1Low/J, JAX stock no. 009593) [23] and BAC transgenic Pomc-Cre mice [24] were also maintained as heterozygotes on a C57BL/6J background. Genotyping was performed by PCR using genomic DNA extracted from ear biopsies or embryo fragments. All of the primer sequences used are provided in Supplementary Table 1.
2.2. Tissue collection and embedding
Pregnant mice at 10.5–15.5 days post coitum (dpc, the day of detection of the vaginal plug was considered dpc 0.5) were killed by cervical dislocation and embryos were collected and washed in cold RNAse-free PBS. Fixation was performed with 4% paraformaldehyde (PFA; w/v) in phosphate buffer solution (PBS; 0.9% NaCl, 2.7 mM KCl, 10 mM K2HPO4, 2 mM KH2PO4, and pH 7.4) at 4 °C for a period of time dependent on the developmental stage. Embryos at 10.5 and 12.5 dpc were fixed for 1 h; heads from 15.5 dpc embryos were dissected and fixed for 2 h. The samples were then washed in cold PBS and cryoprotected with 10% sucrose in PBS for 16 h. The tails or posterior limbs were cut to obtain tissue for genotyping. The tissues were then stabilized in 10% sucrose/10% gelatin in PBS at 37 °C for 30 min prior to freezing. Adult mice were perfused using 4% PFA and their brains were collected and fixed for 16 h in 4% PFA at 4 °C. The whole brains were then washed in cold PBS and cryoprotected with 10% sucrose in PBS for 16 h. Hypothalamic sections were then cut grossly using a stainless steel adult brain mold prior to freezing. Gelatin blocks containing the tissues were stuck on cork sheets with Tissue-Tek OCT compound (Sakura Finetek, cat. no. 4583). The gelatin blocks were snap-frozen in 2-methylbutane (Sigma, cat. no. M32631) at −55 °C and stored at −80 °C. Sagittal (20 μm for E10.5 to E15.5) and coronal (20 μm for adult mice) sections were obtained in a cryostat (Leica 1510S) and collected on Super-Frost Plus slides (Thermo Fisher Scientific). The sections were then air dried for 16 h and stored at −20 °C until use.
2.3. X-gal staining and immunohistochemistry
Pregnant Prdm12+/lacZ mice at 13.5 dpc were cervically dislocated and the embryos were removed immediately, washed with KPBS (0.9% NaCl, 16 mM K2HPO4, 3.6 mM KH2PO4, and pH 7.4), and fixed with 4% PFA in KPBS for 20 min. Whole-mount fixed embryos were stained with 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-gal) 1 mg/ml in staining solution: PBS containing 2.12 mg/ml potassium ferrocyanide, 1.64 mg/ml potassium ferricyanide, 2 mM MgCl2, 0.01% sodium deoxycolate, and 0.02% NP-40 (pH 7.3) at 37 °C for 16 h. The embryos were then dehydrated with sucrose 10% in KPBS at 4 °C for 16 h, frozen, and sliced into 20 μm sections in a cryostat. The slides were air dried at room temperature (RT) for 16 h and then retreated with X-gal staining solution at 37 °C for 16 h. The slides were washed twice with PBS and then incubated in 1% H2O2 in KPBS for 30 min at RT, washed twice with KPBS, and incubated with rabbit polyclonal antibody for β-endorphin (1:500, H-022-33, Phoenix Pharmaceuticals) at 4 °C for 16 h. After washing with KPBS, the slides were incubated with a secondary anti-rabbit biotinylated antibody (1:200, BA-1000, Vector) for 2 h at RT, then washed with KPBS and incubated with Vectastain Elite ABC kit (Vector) 1 h at RT. Finally, the slices were rinsed in TBS (50 mM Tris–HCl and 150 mM NaCl, pH 7.5) and incubated with diaminobenzidine (25 mg/ml in TBS and 0.05% H2O2) until the reaction developed. The reaction was stopped by dipping the slides in TBS. The slides were air dried for 16 h and then coverslipped under Permount.
2.4. Immunofluorescence and cell quantification
Immunofluorescence was performed as described [10,11]. Primary antibodies used were rabbit anti-rat ACTH (1:500, National Hormone and Peptide Program); mouse anti-ISL1 (1:20, 39.4D5, Developmental Studies Hybridoma Bank, University of Iowa); rabbit anti-PRDM12 (1:50, ABE95, EMD Millipore); and chicken anti-EGFP (1:2000, GFP-1020, Aves Lab). Secondary antibodies used were goat, anti-rabbit AlexaFluor 555 (1:1000, Invitrogen); mouse, anti-rabbit AlexaFluor 488 (1:1000, Invitrogen); donkey, anti-chicken Alexa Fluor 488 (1:1000, Jackson Immuno Research, cat. no.: 703-545-155); and goat, anti-mouse AlexaFluor 647 (1:1000, Invitrogen). For PRDM12 immunofluorescence in dpc 12.5 slices and ISL1, ACTH, EGFP, and PRDM12 immunofluorescence assays in adult slices, an antigen-retrieval step prior to primary antibody incubation was performed using citrate buffer (10 mM anhydrous citric acid and 0.05% Tween-20, pH 6.0) at 70 °C for 15 min followed by two washes in KPBS. The slices were incubated for 16 h with primary antibodies at specified dilutions in 0.3% Triton-X in KPBS with 2% normal goat serum (Jackson ImmunoResearch, cat. no. 005-000-001). The chicken anti-EGFP antibody was diluted in KPBS, 0.3% Triton-X, and 2% normal donkey serum (Jackson ImmunoResearch, cat. no. 017-000-121). The next day, the slices were washed in KPBS three times and then incubated with secondary antibodies diluted in 0.3% Triton-X in KPBS for 2 h at RT. Nuclei were stained with DAPI (1 mg/l) for 10 min and then washed twice with KPBS before mounting with Vectashield (Vector Labs). Confocal images were obtained using a Leica Confocal TCS-SPE. The number of immunofluorescent ACTH+, ISL1+, PRDM12+, or EGFP+ cells were counted manually using a cell counter plug-in (ImageJ). For the expression analysis in E12.5 embryos we collected sagittal 20 μm sections spanning the whole hypothalamus and the total number of ACTH+ cells was counted in three serial sections. In E15.5 and adult mice, ACTH+, ISL1+, PRDM12+, and Pomc-EGFP+ cells were counted in two serial sections. Depending on the marker antigen, immunofluorescence experiments were repeated in 2–5 different embryos for each genotype.
2.5. mRNA quantification by qRT-PCR
Adult hypothalami or embryo heads of either sex were dissected and collected in ice-cold TriPure RNA Isolation Reagent (Sigma, cat. no. 11667165001) and stored at −80 °C until RNA extraction, which was performed following the manufacturer's instructions. RNA integrity was assessed by gel electrophoresis; clear 28S and 18S rRNA were observed in an approximate 2:1 ratio. Quantification was performed using a Nanodrop and 260/280 and 260/230 ratios were checked to assess purity. One μg of RNA was treated with DNAse I (Ambion, cat. no. AM2222) for 45 min at 37 °C and used for first-strand cDNA synthesis using a High Capacity Reverse Transcription kit with random primers (Applied Biosystems, cat. no. 4368814). Pomc and Prdm12 mRNAs were quantified using specific primers relative to the internal control β-actin (Actb). All of the primer sequences used are listed in Supplementary Table 1. The samples were run in triplicate in an Applied Biosystems 7500 Real-Time PCR System using Power Up SYBR Green Master Mix (Applied Biosystems, cat. no. A25472). Melt curves were analyzed to confirm the specificity of the PCR product. Relative quantification was done by interpolating Ct values in standard curves or using Real-Time PCR Miner [25].
2.6. Physiological measurements
Body weight was monitored regularly until 14 weeks of age. The mice were then individually housed for a week for acclimation and then their daily food intake was measured over 3 consecutive days with normal chow supplied ad libitum. The males were fasted for 18 h (5:00 P.M.-11:00 A.M.) and subjected to an intraperitoneal glucose tolerance test (GTT). Glucose was measured at time 0 using a OneTouch Ultra2 glucometer after cutting off the tip of the tail. D-glucose (1 g/kg; G5767, Sigma; i.p.) was administered and their blood glucose measured at 15, 30, 60, and 120 min. At week 16, male and female mice were euthanized via cervical dislocation. Body length was measured from the nose to the tail base. Bilateral subcutaneous (inguinal) and visceral (retroperitoneal and gonadal) white adipose tissue, interscapular brown adipose tissue, and livers were dissected and weighed.
2.7. Statistics
All of the data presented are the mean ± SEM and were analyzed by repeated measures one-way ANOVA or Student's t-test using GraphPad Prism Software (Version 5.01, 2007 GraphPad Software Inc., San Diego, CA, USA). Post hoc comparisons between groups were performed using Tukey's test. The normality of the distributions was assessed by the Shapiro–Wilk test (p > 0.05) and the equality of the variance with Bartlett's test or the F-test (p > 0.05).
3. Results
3.1. PRDM12 and POMC co-express in the developing and adult arcuate nucleus of the hypothalamus
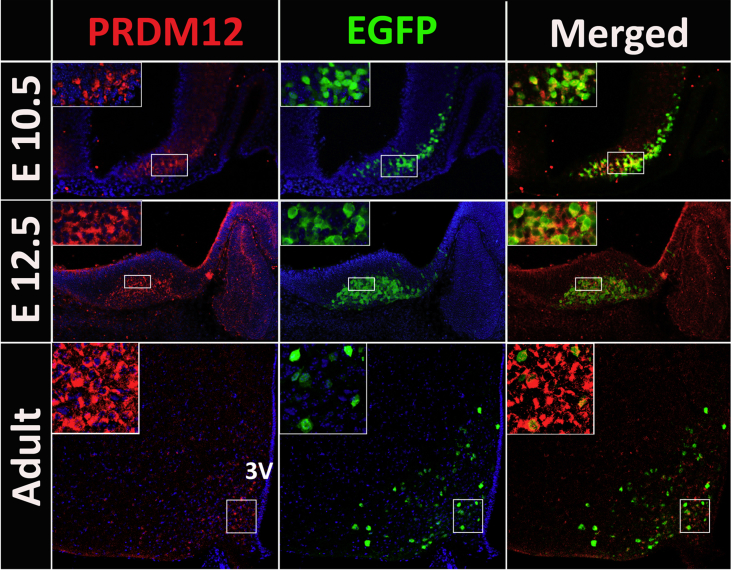
A necessary condition to support PRDM12 as a transcriptional regulator that controls the early differentiation of the melanocortin lineage is that it co-expresses with POMC in the same neurons of the developing hypothalamus. We used polyclonal anti-PRDM12 antibody to follow the expression of PRDM12 during development and adulthood in the hypothalamic sections of the validated transgenic mouse line Pomc-EGFP [10,11,23] and found that the expression domains of PRDM12 and POMC are coincidental at all time points (Figure 1). PRDM12 and POMC co-express since the initial onset of hypothalamic Pomc expression at E10.5 [10,11,26] (Figure 1, top). At this early time point, almost all of the incipient Pomc-EGFP neurons were immunopositive for PRDM12 and only a few PRDM12+ cells did not show EGFP labeling (Figure 1, top). Two days later (E12.5), at the peak of neurogenesis of the POMC neurons in the mouse embryo [10], we detected PRDM12 immunofluorescence in the entire growing population of POMC neurons that accumulate in the mantle zone of the median anterobasal nucleus [10,11] (Figure 1, middle). In the adult mouse arcuate nucleus, most POMC neurons co-express PRDM12 (Figure 1, bottom). The expression domain of PRDM12 in adults is broader than that of POMC, as previously found for ISL1 [10] and NKX2.1 [11]. Together, these results show that POMC neurons express PRDM12 in the ventromedial hypothalamus during development and adulthood.
Figure 1.
PRMD12 and POMC co-express in the developing and adult mouse hypothalamus. Expression analysis of PRDM12 in sagittal cryosections of E10.5 (top) and E12.5 (middle) Pomc-EGFP mouse embryos at the level of the hypothalamic mantle zone and coronal sections of adult Pomc-EGFP mice at the level of the arcuate nucleus (bottom). Insets are magnified views showing that EGFP+ cells co-express PRDM12. 3V, third ventricle.
3.2. PRMD12 is critical for the early establishment of hypothalamic melanocortin neuron identity
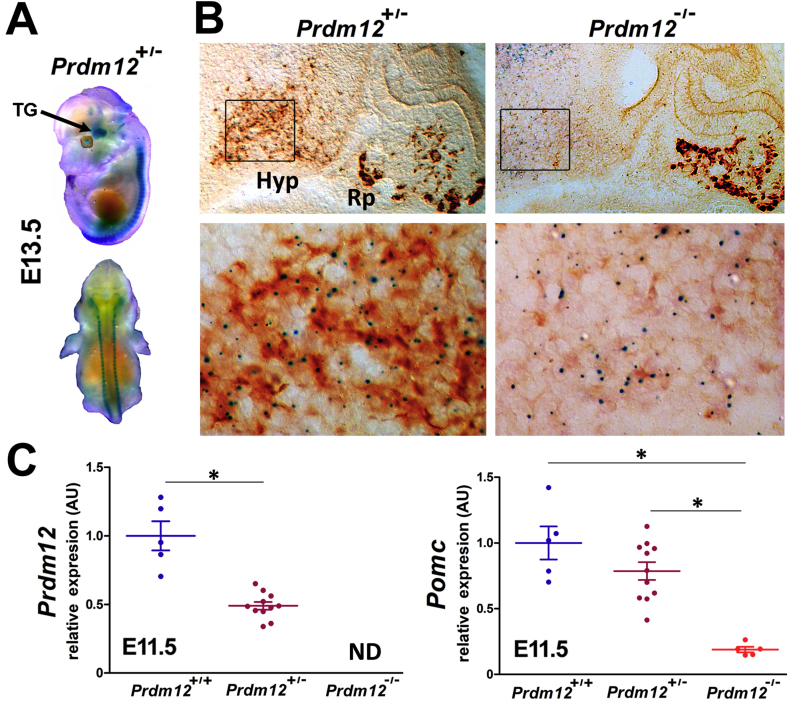
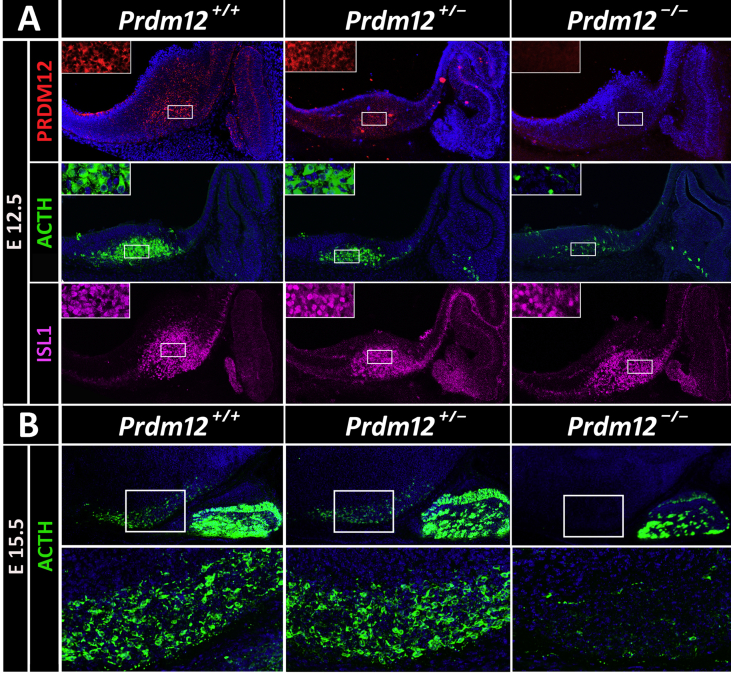
To investigate whether PRDM12 plays a role in the early establishment of the melanocortin lineage, we analyzed the developmental expression of Pomc in the hypothalamus of constitutive Prdm12−/− mice, a Prdm12 null-allele mutant strain in which the lacZ reporter gene carrying a polyadenylation site was inserted downstream of Prdm12 exon 1. After mating the Prdm12+/− mice, we collected embryos of the three genotypes at different developmental time points. Whole mount expression analysis of the reporter gene performed in E13.5 Prdm12+/− embryos demonstrated β-gal expression in the dorsal root ganglia, trigeminal ganglion, and other brain areas (Figure 2A) as previously observed in another Prdm12+/lacZ(−) knockin strain Desiderio et al. [20]. X-gal staining followed by β-endorphin immunohistochemistry performed on sagittal sections of E13.5 heterozygous Prdm12+/− embryos confirmed PRDM12 and POMC co-expression in the arcuate nucleus of the hypothalamus (Figure 2B) while in the pituitary, only POMC expression was detected (Figure 2B). Analysis of E13.5 Prdm12−/− homozygous mutant embryos showed that in the absence of PRDM12, POMC expression was greatly diminished in the hypothalamus while pituitary POMC remained intact (Figure 2B). To determine the level of hypothalamic Pomc mRNA in mice lacking Prdm12, we performed quantitative RT-PCR from heads of E11.5 embryos 1 day prior to the onset of pituitary Pomc expression [26] to avoid the presence of pituitary Pomc mRNA in the tissue samples. Homozygous Prdm12−/− embryos with undetectable levels of Prdm12 mRNA (Figure 2C, left) expressed only 18.9% of hypothalamic Pomc mRNA compared to wild-type levels (Figure 2C, right), while heterozygous Prdm12+/− E11.5 embryos expressing 49.0% of Prdm12 (Figure 2C, left) showed 78.6% of Pomc mRNA levels (Figure 2C, right) compared to wild-type Prdm12+/+ embryo heads. Immunofluorescence assays performed on sagittal sections of E12.5 Prdm12−/− embryos revealed that in the absence of PRDM12 (Figure 3A), the number of POMC+ neurons identified with an anti-ACTH antibody was markedly reduced compared with wild-type and Prdm12+/− embryos (Figure 3A; Prdm12−/−: 24.6 ± 3.1% and Prdm12+/−: 75.6 ± 6.6% compared to wild-types; one-way ANOVA: n ≥ 3 per genotype, F = 57.9; P < 0.0001 followed by Tukey's post hoc test: P < 0.05). Ablation of Prdm12 did not impair the expression of the developmental marker ISL1 (Figure 3A), suggesting that the neurons where Pomc normally expresses in the mantle zone of the hypothalamus were still present. Three days later, at E15.5, the number and brightness of the POMC immunolabeled hypothalamic neurons in embryos lacking PRDM12 were also greatly diminished (Figure 3B). Postnatal assessment of Pomc expression in Prdm12−/− mice was precluded by their perinatal lethality. Altogether, these results demonstrate that PRDM12 is necessary for the developmental expression of Pomc in the ventromedial hypothalamus.
Figure 2.
Ablation of Prdm12 impairs the developmental onset of hypothalamic Pomc expression. (A) Lateral and dorsal view of a heterozygous Prdm12+/− E13.5 embryo expressing lacZ under the transcriptional control of a null-Prdm12 allele after whole mount X-gal staining. TG, trigeminal ganglion. (B) Sagittal sections of a heterozygous Prdm12+/- and a homozygous Prdm12−/− E13.5 mutant embryo showing lacZ expression (blue dots) and ACTH immunoreactivity (brown) in the mediobasal hypothalamus and pituitary region. Note that lacZ staining is prominent in the basal hypothalamus (Hyp) but not in Rathke's pouch (Rp). Magnified views of the hypothalamic area show that in the absence of PRDM12, Pomc expression is negligible. (C) qRT-PCR of Prdm12 and Pomc mRNAs obtained from heads of wild-type, heterozygous Prdm12+/−, and homozygous Prdm12−/− E11.5 littermate embryos show that PRDM12 is necessary for the onset of hypothalamic Pomc expression. AU, arbitrary units. At least n = 5 per group. Error bars correspond to SEM. ND, non-detectable. *P < 0.0001, one-tailed Student's t-test, t = 6.314, df = 14 for Prdm12 and one-way ANOVA (F = 20) followed by Tukey's post hoc test for Pomc.
Figure 3.
Hypothalamic Pomc expression is impaired in Prdm12 knockout mice. (A) Immunofluorescence analysis using anti-PRDM12 (red) and anti-ACTH (green) antibodies in sagittal cryosections of wild-type, heterozygous Prdm12+/−, and homozygous Prdm12−/− E12.5 mouse embryos reveal that the absence of PRDM12 prevents Pomc expression in the developing hypothalamus. ISL1 immunofluorescence (magenta) labels the neurons present in the mantle zone where Pomc is normally expressed. Insets are magnified views of the indicated boxes. (B) At E15.5, hypothalamic POMC immunolabeled neurons continued to be negligible in Prdm12 knockout mice. Bottom, magnified views of the hypothalamic areas embedded in the figures above.
3.3. PRDM12 expression in ISL1 neurons is critical for hypothalamic Pomc expression
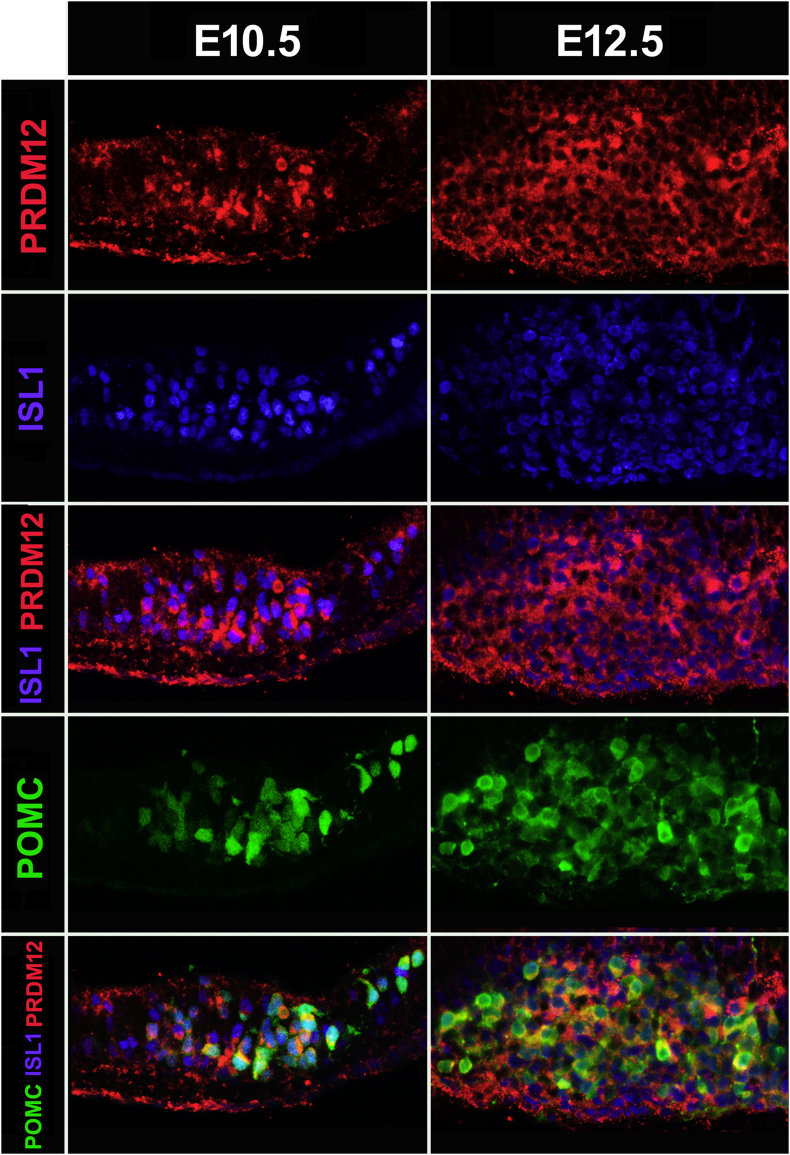
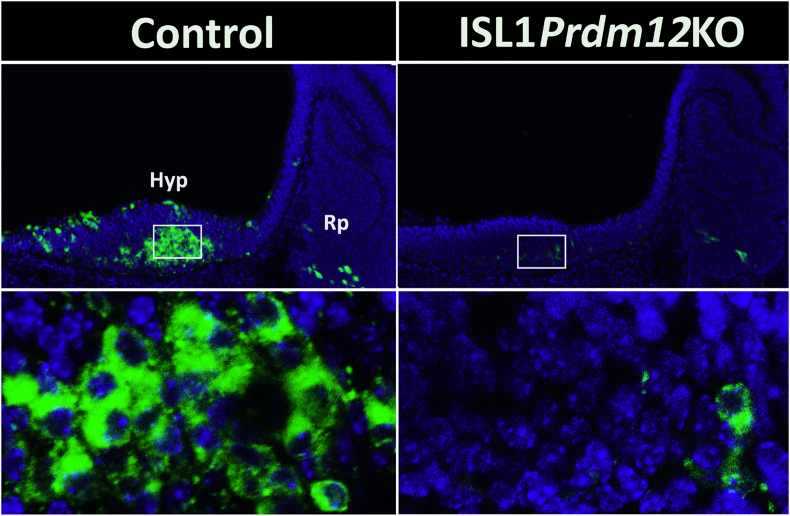
To limit Prdm12 ablation to the lineage leading to arcuate POMC neurons, we inactivated Prdm12 alleles specifically in ISL1+ neurons. PRDM12 and ISL1 co-expressed at E10.5 in the developing arcuate nucleus (Figure 4), and Pomc-EGFP-positive cells co-expressed ISL1 and PRDM12 (Figure 4). A similar pattern of triple co-expression between POMC, ISL1, and PRDM12 was observed in E12.5 embryos (Figure 4). We then mated mice carrying conditional null Prdm12 alleles (Prdm12loxP/loxP) with Isl1-Cre mice, a knockin strain that expresses Cre under the transcriptional control of Isl1 [22]. After two successive breedings, we obtained Prdm12loxP/loxP.Isl1+/Cre mice that we called ISL1Prdm12KO. ISL1Prdm12KO embryos at E11.5 showed only 14.1 ± 0.01% (n = 8) of hypothalamic Pomc mRNA compared to Prdm12loxP/loxP control littermates (n = 6) as determined by quantitative RT-PCR (one-tailed Student's t-test, t = 9.507, df = 12, P < 0.0001). Immunofluorescence analysis of ISL1Prdm12KO embryos at E12.5 showed a considerable reduction in ACTH+ neurons (Figure 5) due to the loss of PRDM12 in this region (ISL1Prdm12KO: 24.2 ± 2.1%, n = 5 relative to controls, n = 5, one-tailed Student's t-test, t = 13.34, df = 8, P < 0.0001). Thus, the lack of Prdm12 expression specifically in ISL1+ neurons impairs hypothalamic Pomc expression and confirms the critical role of PRDM12 in the early establishment of melanocortin neuron identity.
Figure 4.
Overlapping expression patterns of PRDM12, ISL1, and POMC in the developing hypothalamus. Immunofluorescence using anti-PRDM12 (red) and anti-ISL1 (blue) antibodies in sagittal cryosections of E10.5 (left) and E12.5 (right) Pomc-EGFP mouse embryos. Confocal images showing superimposed PRDM12 and ISL1 immunopositive neurons (middle lane) and triple merged with Pomc-EGFP-immunoreactive neurons (bottom).
Figure 5.
Specific deletion of Prdm12 from ISL1 neurons impairs the onset of Pomc expression. Immunofluorescence performed in sagittal cryosections of E12.5 embryos using an anti-ACTH antibody in the hypothalamic mantle zone. ISL1Prdm12KO embryos show highly reduced numbers of POMC neurons compared with Prdm12loxP/loxP controls. Bottom, magnified views of the hypothalamic areas boxed in the figures above.
3.4. Selective ablation of Prdm12 in POMC neurons impairs Pomc expression and increases body weight
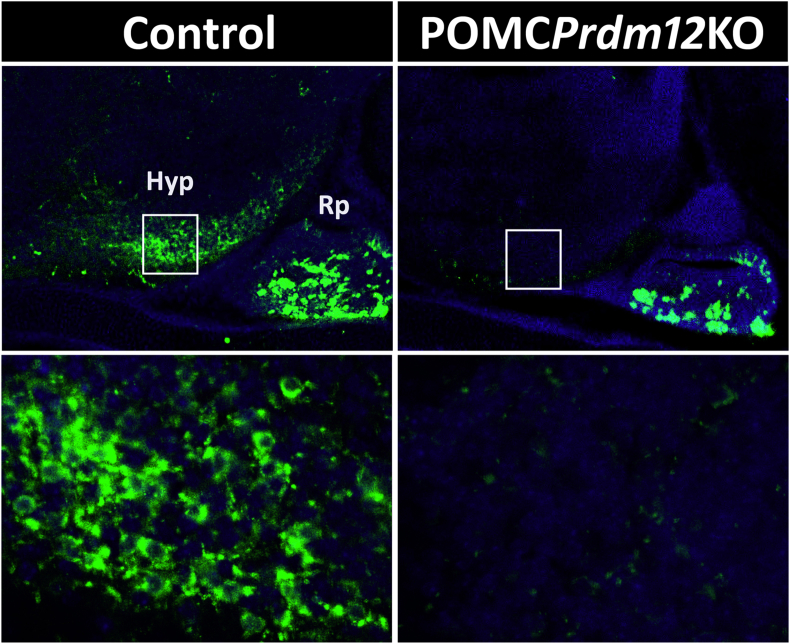
Because the ISL1Prdm12KO mice were embryonic lethal, we further restricted Prdm12 ablation selectively to POMC neurons. To this end, we crossed Prdm12loxP/loxP mice with a BAC Pomc-Cre transgenic line [24] previously used in our laboratory [10,11], and after two successive breedings obtained Prdm12loxP/loxP. Pomc-Cre mice that we called POMCPrdm12KO. To evaluate the loss of PRDM12 from POMC neurons, we performed an immunofluorescence study in E15.5 POMCPrdm12KO embryos, a developmental time point selected to allow for Cre-induced ablation of Prdm12 alleles and further clearance of PRDM12 and POMC peptides. Ablation of Prdm12 from POMC precursor neurons induced a significant reduction in the labeling of ACTH+ neurons (Figure 6), with only a few dimly fluorescent neurons remaining in the developing hypothalamus. These results further indicate that PRDM12 plays a crucial role in the developmental expression of hypothalamic Pomc and the early specification of melanocortin neuron identity in a cell-autonomous manner.
Figure 6.
Specific deletion of Prdm12 from POMC neurons impairs hypothalamic Pomc expression. Immunofluorescence performed in sagittal cryosections of E15.5 embryos using an anti-ACTH antibody in the mediobasal hypothalamic (Hyp) and Rathke's pouch (Rp) areas. POMCPrdm12KO embryos show less and very dim POMC neurons compared with Prdm12loxP/loxP controls whereas POMC cells in the future pituitary remain intact. Bottom, magnified views of the hypothalamic areas boxed in the figures above.
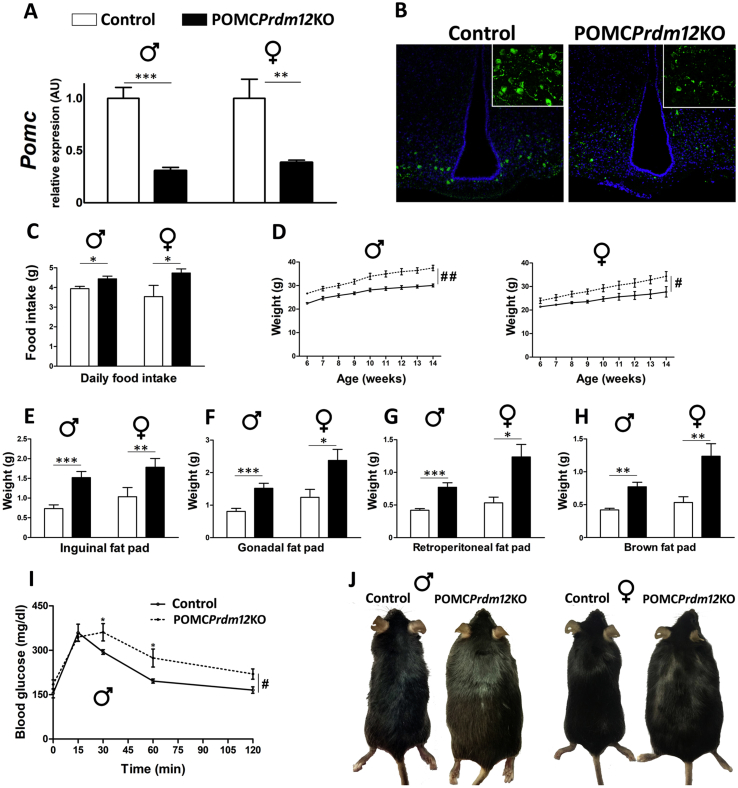
Given that PRDM12 and POMC also co-express in the arcuate nucleus in adulthood (Figure 1), we evaluated whether the selective lack of PRDM12 from POMC neurons in POMCPrdm12KO mice also affects Pomc expression and melanocortin function in adult mice. Hypothalamic Pomc mRNA levels dropped to 30.7 ± 3.2% in 16-week-old POMCPrdm12KO male mice (n = 8) compared to those found in Prdm12loxP/loxP control littermates (n = 5, one-tailed Student's t-test, P < 0.0001) (Figure 7A, left). Similar results were observed in female mice (POMCPrdm12KO 38.6 ± 2.2%, n = 5; Prdm12loxP/loxP, n = 4, one-tailed Student's t-test, P < 0.005) (Figure 7A, right). Immunofluorescence performed in coronal brain sections confirmed a marked reduction in ACTH immunoreactivity in the arcuate nucleus of POMCPrdm12KO mice compared with their control siblings (Figure 7B). In agreement with the reduction in Pomc mRNA levels, 14-week-old POMCPrdm12KO males and females were hyperphagic, consuming 12.6% and 33.9% more food daily at that age, respectively, compared to their sibling controls (Figure 7C). As a result, the POMCPrdm12KO males and females displayed higher body weight than their control littermates, with 28.3% and 32.5% overweight at 14 weeks of age (Figure 7D). Adiposity in the POMCPrdm12KO mice was highly increased as demonstrated by their enlarged inguinal, gonadal, and retroperitoneal white fat pads (Figure 7E–G) as well as interscapular brown fat (Figure 7H). The weight of the livers, in contrast, showed no statistical difference between genotypes in both sexes. A glucose tolerance test performed on 15-week-old males fasted for 18 h showed that the POMCPrdm12KO mice cleared glucose from their blood at a slower rate than their control littermates. At 16 weeks of age, the male mice showed a significantly longer nose to tail length (POMCPrdm12KO: 9.9 ± 0.1 cm, n = 15, and controls: 9.5 ± 0.1 cm, n = 9, P < 0.05, one-tailed Student's t-test: t = 2.313, df = 22, P < 0.05), whereas no genotype difference was observed in the females. Altogether, our results demonstrate that PRDM12 participates in a cell-autonomous manner to support normal Pomc expression levels in the adult arcuate nucleus and, consequently, in the regulation of food intake and normal body weight homeostasis.
Figure 7.
Prdm12-specific deletion in POMC neurons increases food intake, adiposity, and leads to obesity. (A) Hypothalamic Pomc mRNA expression in POMCPrdm12KO male (n = 8) and female (n = 5) mice 16 weeks of age relative to Prdm12loxP/loxP controls (males, n = 5, and females, n = 4) assessed by qRT-PCR and expressed in arbitrary units (AU). One-tailed Student's t-test. For males, t = 7.568, df = 11, and P < 0.001. For females, t = 3.745, df = 7, and P < 0.01. (B) Immunofluorescence analysis of POMC neurons using an anti-ACTH antibody (green) in coronal hypothalamic sections of adult male Prdm12loxP/loxP (control) and POMCPrdm12KO mice. DAPI staining is shown in blue. (C) Average daily food intake measured over 3 consecutive days at 14 weeks of age in POMCPrdm12KO males (n = 7) and females (n = 4) and their control littermates (n = 5 and n = 3, respectively). One-tailed Student's t-test; males: t = 2.602, df = 10, females: t = 2.220, df = 5, P < 0.05. (D) Body weight curves of male POMCPrdm12KO (n = 15) and Prdm12loxP/loxP (n = 9) and female (KO, n = 6; control, n = 4) mice. POMCPrdm12KO mice display overweight (RMA, genotype × time effect. #P < 0.05 and ##P < 0.01 for POMCPrdm12KO vs Prdm12loxP/loxP). (E–H) Determination of inguinal, gonadal, and retroperitoneal white fat pad and interscapular brown fat pad weights in 16-week-old male POMCPrdm12KO (n = 15) and Prdm12loxP/loxP (n = 9) and female POMCPrdm12KO (n = 6) and Prdm12loxP/loxP (n = 4) mice. (I) Glucose tolerance test (GTT) in 15-week-old male POMCPrdm12KO (n = 8) and Prdm12loxP/loxP (n = 5) 18 h fasted mice receiving a 1 g/kg (i.p.) injection of D-glucose. (J) Pictures of representative obese POMCPrdm12KO male and female mice compared with Prdm12loxP/loxP siblings at 16 weeks of age. Values represent mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student's t-test).
4. Discussion
In this study, we combined developmental, cellular, and functional genetics approaches to investigate the participation of the transcriptional regulator PRDM12 in the establishment of melanocortin neuron identity and in the initiation and maintenance of Pomc expression in the arcuate nucleus. In particular: 1) PRDM12 co-expresses with POMC in neurons of the arcuate nucleus of the hypothalamus from the onset of Pomc expression at E10.5 and throughout the entire lifespan; 2) PRDM12 expression in the developing ventral hypothalamus is necessary for the onset and later maintenance of Pomc expression in this brain region; 3) the expression domain of PRDM12 in the hypothalamus is broader than that of POMC, indicating that PRDM12 is necessary but not sufficient to activate Pomc expression; 4) PRDM12 integrates a distinctive set of transcriptional regulators including NKX2.1 and ISL1 that together dictates neuronal-specific expression of Pomc in the arcuate nucleus; 5) selective ablation of Prdm12 from ISL1 neurons prevents hypothalamic Pomc expression, demonstrating that its presence is critical in the lineage leading to melanocortin neurons; 6) selective ablation of Prdm12 from POMC neurons greatly reduces the developmental expression of hypothalamic Pomc; and 7) in adult mice, the lack of PRDM12 from POMC neurons induces a considerable reduction in Pomc mRNA levels that leads to increased food intake, adiposity, and body weight.
PRDM12 is a member of the PRDM family of transcriptional regulators that participate in a variety of developmental cell fate decisions and differentiation pathways [27]. Although PRDM proteins carry a variable number of zinc finger domains, they are not considered typical transcription factors that bind to specific DNA motifs present in enhancers and/or promoters but rather function as transcriptional regulators that promote or repress gene expression by interacting with a variety of histone-modifying enzymes such as methyltransferases, deacetylases, or acetyltransferases [28]. For example, PRDM16 was identified as a transcriptional regulator that must be continuously present in brown adipose tissue cells to maintain their differentiation state while repressing skeletal myoblast and WAT lineages [29]. In the brain cortex, PRDM16 controls the proliferation of radial glia progenitors and migration of differentiated neurons to top cortical layers by modifying the epigenetic state of enhancers and repressors of different genes. In this cell type, however, Prdm16 expression is halted once neurons attain their final position in the brain cortex [30]. In turn, PRDM12 specifies the cell fate of V1 interneurons in the Xenopus spinal cord by acting as a transcriptional repressor of Dbx1 and Nkx6 [31]. It was recently demonstrated that PRDM12 plays a key role in the genetic program that determines the cell fate and functional identity of nociceptive sensory neurons that originate in the dorsal root ganglia from neural crest cells [19,20]. Mouse embryos lacking Prdm12 fail to express trkA, a high-affinity nerve growth factor receptor that acts as a critical functional marker for the terminal differentiation of nociceptive sensory neurons. As a result, Prdm12 knockout embryos poorly develop this class of sensory neurons [19,20]. In humans, PRDM12 has attracted considerable attention after the identification of patients from 11 unrelated families with congenital insensitivity to pain. The probands carry diverse homozygous or compound heterozygous mutations of PRDM12 containing different polyalanine expansions or missense point mutations [18]. The clinical and histological analysis of these patients suggested that these PRDM12 mutations prevent the normal development of sensory neurons destined to become nociceptors. Based on our observations and those of others [19,20] that Prdm12 knockout mice die perinatally, it is likely that the human coding mutations observed in patients with congenital insensitivity to pain lead to hypomorphic rather than null-allele PRDM12 variants. It would be interesting to assess if some of these patients are hyperphagic and/or obese.
Our study presents a novel and a previously unsuspected role for PRDM12, first as a key developmental regulator of the cellular differentiation of melanocortin neurons and later as a critical factor that maintains high Pomc expression levels in the hypothalamus. Furthermore, in adult mice, PRDM12 is essential to assure sufficiently elevated levels of hypothalamic Pomc expression to sustain normal satiety. Thus, PRDM12 together with ISL1 [10] and NKX2.1 [11] integrate a unique set of early transcription factors necessary to initiate neuronal specific Pomc expression in the hypothalamus, such that the absence of any of these three factors prevents the developmental onset of Pomc expression. Later in embryonic development and perinatal life, this triad of transcriptional activators that specify the early identity of melanocortin neurons is complemented by the T-box transcription factor Tbx3, which further raises hypothalamic Pomc expression to the higher physiological levels demanded in postnatal life to regulate food intake [12]. The partial redundancy and functional synergism that these four TFs exert on the overall transcriptional activity of hypothalamic Pomc can be appreciated in individual conditional mutants carrying null-alleles specifically in POMC cells, as has been shown in this report on Prdm12 and in previous studies of Isl1 [10], Nkx2.1 [11], and Tbx3 [12]. Interestingly, adult mice lacking Prdm12, Isl1, Nkx2.1, or Tbx3 from POMC neurons (PomcPrdm12KO, PomcIsl1KO, PomcNkx2.1KO, or PomcTbx3KO mice) display varying reduced levels of hypothalamic Pomc mRNA and commensurate degrees of increased adiposity and body weight that go from mild overweight in POMCNkx2.1KOs (males were 15% heavier than controls) to severe obesity, as found in the other three conditional mutant models. Although the combined presence of PRDM12, ISL1, and NKX2.1 is necessary to activate Pomc expression in the developing hypothalamus, the number of neurons co-expressing these three TFs in the arcuate nucleus considerably exceeds the number of Pomc expressing neurons. Therefore, at least one other factor yet to be discovered likely completes the distinctive full set of transcriptional regulators that determines the identity of melanocortin neurons. Although POMC neurons project to different target areas and participate in anatomically and physiologically distinct brain circuits, they all depend on the same set of transcriptional regulators to express Pomc, their common functional marker. Finally, our demonstration that the absence of Prdm12 in POMC neurons leads to hyperphagia, increased adiposity, and obesity may be of biomedical relevance. Potentially, human carriers of polymorphisms in PRDM12 leading to hypomorphic alleles may have impaired arcuate POMC expression and therefore compromised control of food intake and energy balance under permissive environmental conditions.
Author contributions
C.E.H., D.R., D.P.O, M.J.L., and M.R. designed this study; C.E.H., D.R., and M.R. conducted the research; M.J.L. and M.R. contributed new reagents and analytic tools; C.E.H., D.R., and M.R. analyzed the data and prepared the figures; M.R. wrote the paper; and C.E.H., D.R., D.P.O, M.J.L., and M.R. revised the paper.
Acknowledgments
The authors thank Marta Treimun and Carolina Álvarez for excellent technical assistance and Flávio de Souza and Lucía Franchini for constructive ideas. This study was supported by National Institutes of Health Grant R01 DK068400 (to M.J.L. and M.R.) and Agencia Nacional de Promoción Científica y Tecnológica (to M.R.). C.E.H., D.R., and D.P.O received doctoral fellowships from CONICET.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.01.007.
Contributor Information
Malcolm J. Low, Email: mjlow@umich.edu.
Marcelo Rubinstein, Email: mrubins@dna.uba.ar.
Conflict of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rubinstein M., Low M.J. Molecular and functional genetics of the proopiomelanocortin gene, food intake regulation and obesity. FEBS Letters. 2017;591:2593–2606. doi: 10.1002/1873-3468.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krude H., Biebermann H., Luck W., Horn R., Brabant G., Grüters A. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nature Genetics. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 3.Farooqi I.S., Yeo G.S.H., Keogh J.M., Aminian S., Jebb S.A., Butler G. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. Journal of Clinical Investigation. 2000;106:271–279. doi: 10.1172/JCI9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaswen L., Diehl N., Brennan M.B., Hochgeschwender U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nature Medicine. 1999;5:1066–1070. doi: 10.1038/12506. [DOI] [PubMed] [Google Scholar]
- 5.Huszar D., Lynch C.A., Fairchild-Huntress V., Dunmore J.H., Fang Q., Berkemeier L.R. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 6.Bumaschny V.F., Yamashita M., Casas-Cordero R., Otero-Corchón V., de Souza F.S.J., Rubinstein M. Obesity-programmed mice are rescued by early genetic intervention. Journal of Clinical Investigation. 2012;122:4203–4212. doi: 10.1172/JCI62543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhabra K.H., Adams J.M., Jones G.L., Yamashita M., Schlapschy M., Skerra A. Reprogramming the body weight set point by a reciprocal interaction of hypothalamic leptin sensitivity and Pomc gene expression reverts extreme obesity. Molecular Metabolism. 2016;5:869–881. doi: 10.1016/j.molmet.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challis B.G., Coll A.P., Yeo G.S.H., Pinnock S.B., Dickson S.L., Thresher R.R. Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide-YY3-36. Proceedings of the National Academy of Sciences of the USA. 2004;101:4695–4700. doi: 10.1073/pnas.0306931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam D.D., de Souza F.S.J., Nasif S., Yamashita M., López-Leal R., Otero-Corchon V. Partially redundant enhancers cooperatively maintain mammalian Pomc expression above a critical functional threshold. PLoS Genetics. 2015;11:1–21. doi: 10.1371/journal.pgen.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasif S., de Souza F.S.J., González L.E., Yamashita M., Orquera D.P., Low M.J. Islet 1 specifies the identity of hypothalamic melanocortin neurons and is critical for normal food intake and adiposity in adulthood. Proceedings of the National Academy of Sciences of the USA. 2015;112:E1861–E1870. doi: 10.1073/pnas.1500672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orquera D.P., Tavella M.B., De Souza F.S.J., Nasif S., Low M.J., Rubinstein M. The homeodomain transcription factor NKX2.1 is essential for the early specification of melanocortin neuron identity and activates Pomc expression in the developing hypothalamus. Journal of Neuroscience. 2019;39:4023–4035. doi: 10.1523/JNEUROSCI.2924-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quarta C., Fisette A., Xu Y., García-Cáceres C., Legutko B., Colldén G. Functional identity of hypothalamic melanocortin neurons depends on Tbx3. Nature Metabolism. 2019;1:222–235. doi: 10.1038/s42255-018-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Souza S.J., Santangelo A.M., Bumaschny V., Avale E., Smart J.L., Low M.J. Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Molecular and Celluar Biology. 2005;25:3076–3086. doi: 10.1128/MCB.25.8.3076-3086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santangelo A.M., De Souza F.S.J., Franchini L.F., Bumaschny V.F., Low M.J., Rubinstein M. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genetics. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franchini L.F., Lopez-Leal R., Nasif S., Beati P., Gelman D.M., Low M.J. Convergent evolution of two mammalian neuronal enhancers by sequential exaptation of unrelated retroposons. Proceedings of the National Academy of Sciences of the USA. 2011;108:15270–15275. doi: 10.1073/pnas.1104997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry F.E., Sugino K., Tozer A., Branco T., Sternson S.M. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam B.Y.H., Cimino I., Polex-Wolf J., Nicole Kohnke S., Rimmington D., Iyemere V. Heterogeneity of hypothalamic pro-opiomelanocortin-expressing neurons revealed by single-cell RNA sequencing. Molecular Metabolism. 2017;6:383–392. doi: 10.1016/j.molmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y.-C., Auer-Grumbach M., Matsukawa S., Zitzelsberger M., Themistocleous A.-C., Strom T.M. Transcriptional regulator PRDM12 is essential for human pain perception. Nature Genetics. 2015;47:803–808. doi: 10.1038/ng.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartesaghi L., Wang Y., Fontanet P., Wanderoy S., Berger F., Wu H. PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Reports. 2019;26:3484–3492. doi: 10.1016/j.celrep.2019.02.098. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desiderio S., Vermeiren S., Van Campenhout C., Kricha S., Malki E., Richts S. Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Reports. 2019;26:3522–3536. doi: 10.1016/j.celrep.2019.02.097. e5. [DOI] [PubMed] [Google Scholar]
- 21.Committee on Care and Use of Laboratory Animals . Natl Inst Health; Bethesda: 1996. Guide for the care and use of laboratory animals. DHHS Publ No (NIH) 85-23. [Google Scholar]
- 22.Yang L., Cai C.-L., Lin L., Qyang Y., Chung C., Monteiro R.M. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowley M.A., Smart J.L., Rubinstein M., Cerdán M.G., Diano S., Horvath T.L. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 24.Xu A.W., Kaelin C.B., Takeda K., Akira S., Schwartz M.W., Barsh G.S. PI3K integrates the action of insulin and leptin on hypothalamic neurons. Journal of Clinical Investigation. 2005;115:951–958. doi: 10.1172/JCI24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Sheng, Fernald Russell D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of Computational Biology. 2005;12:1045–1062. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Japón M.A., Rubinstein M., Low M.J. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. Journal of Histochemistry and Cytochemistry. 1994;42:1117–1125. doi: 10.1177/42.8.8027530. [DOI] [PubMed] [Google Scholar]
- 27.Hohenauer T., Moore A.W. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- 28.Fog C.K., Galli G.G., Lund A.H. PRDM proteins: important players in differentiation and disease. Bioessays. 2012;34:50–60. doi: 10.1002/bies.201100107. [DOI] [PubMed] [Google Scholar]
- 29.Chi J., Cohen P. The multifaceted roles of PRDM16: adipose biology and beyond. Trends in Endocrinology and Metabolism. 2016;27:11–23. doi: 10.1016/j.tem.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Baizabal J.-M., Mistry M., García M.T., Gómez N., Olukoya O., Tran D. The epigenetic state of PRDM16-regulated enhancers in radial glia controls cortical neuron position. Neuron. 2018;98:945–962. doi: 10.1016/j.neuron.2018.04.033. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thélie A., Desiderio S., Hanotel J., Quigley I., Van Driessche B., Rodari A. Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Development. 2015;142:3416–3428. doi: 10.1242/dev.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.