Figure 5.
Ca++ Signaling Pathway Components Mediate the Downstream Cellular Response to CDTa
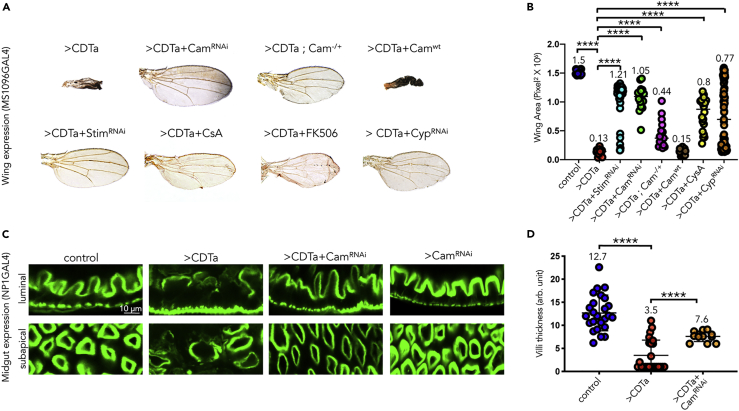
(A and B) (A) Wings from adults of indicated genotypes. Male flies expressing CDTa alone under the control of wing-specific MS1096 GAL4 (combined with tubGAL80ts) (>CDTa) or in combination with a UAS-Calmodulin RNAi transgene (>CDTa + CamRNAi), a heterozygous Cam mutant allele n339 (>CDTa; Cam+/-), a UAS-Stim RNAi transgene (>CDTa + StimRNAiE), a wild-type Cam transgene (>CDTa + CamWT), a UAS-Cyp RNAi transgene, as well as CDTa-expressing flies treated during development with either Cyclosporin A 40μM (Sigma) (>CDTa + CsA) or FK506 50μM (Selleckchem.com) (>CDTa + FK506) were grown at 25°C. (B) Surface area from wings shown in (A) were quantified with Photoshop and analyzed using Prism8 (F = 35.55, df = 7, p < 0.0001). See Figure S5 for additional controls.
(C) One-week-old males of the following genotypes: tubGAL80ts NP1/+ (control), tubGAL80ts NP1>CDTa (>CDTa), tubGAL80ts NP1>CDTa + Calmodulin RNAi transgene (>CDTa + CamRNAi), and tubGAL80ts NP1>CamRNAi (CamRNAi) were grown at RT, exposed to 31°C for 1 day, and dissected. Guts were stained with Phalloidin for F-actin and imaged by confocal microscopy. Blocking Calmodulin expression partially rescues CDTa effects on F-actin network. Scale bar represents 10 μm.
(D) Gut villi thickness was quantified using Photoshop (F = 49.05, df = 2, p < 0.0001). In (B and D) Data are represented as mean ± SD. p-values were obtained from one-way ANOVA tests.
See Figure S5.