Abstract
OBJECTIVES
The 2019 novel coronavirus (2019-nCoV) from Wuhan, China is currently recognized as a public health emergency of global concern.
METHODS
We reviewed the currently available literature to provide up-to-date guidance on control measures to be implemented by public health authorities.
RESULTS
Some of the epidemiological characteristics of 2019-nCoV have been identified. However, there remain considerable uncertainties, which should be considered when providing guidance to public health authorities on control measures.
CONCLUSIONS
Additional studies incorporating more detailed information from confirmed cases would be valuable.
Keywords: Coronavirus, Epidemiology, Characteristics, Public health
INTRODUCTION
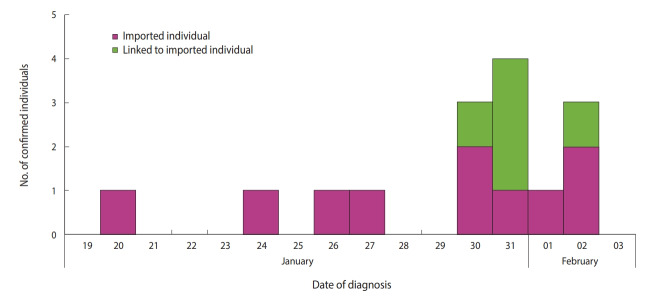
Several clusters of patients with pneumonia of unknown etiology in Wuhan, Hubei Province, China were reported to the Chinese health authorities starting on December 8, 2019, and most of these cases were epidemiologically linked to a local fish and animal market [1,2]. The pathogenic agent responsible for these clusters of pneumonia was identified as a 2019 novel coronavirus (2019-nCoV) [1]. At the very beginning of the 2019-nCoV outbreak in China, much remained unknown, except for the fact that it was transmitted by direct exposure at the market [3]. However, person-to-person transmission of 2019-nCoV has been confirmed [4], and asymptomatic individuals have been identified as potential sources of infection [5]. The number of identified cases has been steadily growing, and as of February 3, a total of 14,557 cases had been reported globally (14,411 in China and 146 in 22 other countries) [6]. Since the first laboratory-confirmed case was identified on January 20, 2020 in Korea, the number of reported cases grew to 15 as of February 3, 2020 (Figure 1 and Table 1) [7].
Figure 1.
Timeline of individuals with laboratory-confirmed 2019 novel coronavirus infections in Korea, as of February 3, 2020.
Table 1.
List of confirmed cases of 2019 novel coronavirus infection in Korea, as of February 3, 2020
| Case No. | Age (yr) | Sex | Nationality | Date of entry to Korea | Suspected infection of place or origin |
|---|---|---|---|---|---|
| #1 | 35 | Female | Chinese | Jan 19, 2020 | Wuhan, China |
| #2 | 55 | Male | Korean | Jan 22, 2020 | Wuhan, China |
| #3 | 54 | Male | Korean | Jan 20, 2020 | Wuhan, China |
| #4 | 55 | Male | Korean | Jan 20, 2020 | Wuhan, China |
| #5 | 33 | Male | Korean | Jan 24. 2020 | Wuhan, China |
| #6 | 55 | Male | Korean | - | Case #3 |
| #7 | 28 | Male | Korean | Jan 23, 2020 | Wuhan, China |
| #8 | 62 | Female | Korean | Jan 23, 2020 | Wuhan, China |
| #9 | 28 | Female | Korean | - | Case #5 |
| #10 | 54 | Female | Korean | - | Case #6 |
| #11 | 25 | Male | Korean | - | Case #6 |
| #12 | 48 | Male | Chinese | Jan 19, 2020 | Osaka, Japan |
| #13 | 28 | Male | Korean | Jan 31, 2020 | Wuhan, China |
| #14 | 40 | Female | Chinese | - | Case #12 |
| #15 | 43 | Male | Korean | Jan 20, 2020 | Wuhan, China |
There remain considerable knowledge gaps on 2019-nCoV; therefore, the public health authorities in countries with any likelihood of experiencing imported cases of 2019-nCoV should update the case definition to reflect newly updated epidemiological data. Herein, we present a review of the literature on the epidemiological characteristics of human infections with 2019-nCoV to provide an interim summary to public health authorities.
MATERIALS AND METHODS
We searched the literature for studies reporting epidemiological characteristics of 2019-nCoV, including the reproductive number, incubation period, serial interval, infectious period, generation time, latent period, and the fatality rate of hospitalized cases. The Korean Society of Epidemiology 2019-nCoV Task Force Team (KSE 2019-nCoV TFT) searched for peer-reviewed articles published from December 8, 2019 to February 1, 2020. Articles were eligible for inclusion if they reported any epidemiological characteristics of 2019-nCoV.
Ethics statement
The ethical approval or individual consent was not applicable.
RESULTS
Six articles were identified and included in this review (Table 2) [2,8-12] . Four relevant studies estimated the reproductive number [8,10-12]. A study of confirmed cases from Wuhan, China estimated the reproductive number to be 1.9 (95% confidence interval [CI], 1.3 to 3.2) [10]. Three other studies estimated the mean reproductive number as 0.3, 2.2, and 2.68, respectively [8,11,12]. A study reported the mean incubation period to be 6.1 days (95% CI, 3.8 to 9.7), and the mean serial interval to be 7.7 days (95% CI, 4.9 to 13.0) [10]. Two studies predicted the mean doubling time to be between 6.4 days and 8.7 days [10,11]. Three studies estimated the fatality rate of hospitalized cases as 14-15% [2,8,9]. We could not identify any studies that reported the infectious and latent periods.
Table 2.
Summary of reviews included in the study
| Study | Study setting | Reproductive (n) | Incubation period (d) | Serial interval (d) | Infectious period | Doubling time (d) | Latent period | Fatality rate among hospitalized cases (%) |
|---|---|---|---|---|---|---|---|---|
| Wu et al. [8] | Publicly-available data in China as at Jan 22, 2020 | 0.30 (95% CI: 0.17, 0.44) | NA | NA | NA | NA | NA | 14.0 (95% CI: 3.9, 32.0) |
| Huang et al. [9] | 41 confirmed cases admitted to a designated hospital in Wuhan by Jan 2, 2020 | NA | NA | NA | NA | NA | NA | 15.0 |
| Chen et al. [2] | 99 confirmed cases admitted in Wuhan Jinyintan Hosptial between Jan 1 and Jan 20, 2020 | NA | NA | NA | NA | NA | NA | 14.6 |
| Li et al. [10] | 425 confirmed cases in Wuhan as at Jan 22, 2020 | 1.9 (95% CI: 1.3, 3.2) | 6.1 (95% CI: 3.8, 9.7) | 7.7 (95% CI: 4.9, 13.0) | NA | 8.7 (95% CI: 4.8, 17.0) | NA | NA |
| Riou et al. [12] | Modelling study | 2.2 (90% CI: 1.4, 3.8) | NA | NA | NA | NA | NA | NA |
| Wu et al. [11] | Modeling study | 2.68 (95% CrI: 2.47, 2.86) | NA | NA | NA | 6.4 (95% CrI: 5.8, 7.1) | NA | NA |
CI, confidence interval; Crl, credible interval; NA, not available.
DISCUSSION
We reviewed the epidemiological characteristics of 2019-nCoV. The estimated reproductive number of 0.3 was obtained from a small number of infected persons with imperfect information in the very early stages of the outbreak [8]; therefore the reproductive number of 2019-nCoV is likely to be similar to that of the 2002/2003 severe acute respiratory syndrome (SARS) coronavirus during the pre-intervention period (range, 2 to 3) and that of the 2009 pandemic A/H1N1 influenza virus in the United States (range, 1.3 to 1.7) [13-15]. The incubation period is likely similar to that of the SARS coronavirus, but with a wider confidence interval (mean, 4.8 days; 95% CI, 4.2 to 5.5) [16]. Furthermore, it is longer than that of the 2009 pandemic A/H1N1 influenza virus (median incubation period, 1.4 days; 95% CI, 1.0 to 1.8) [17]. Therefore, the evidence reviewed above shows that the current control measures for 2019-nCoV, including a quarantine and observation period of 14 days for suspected cases, can be considered appropriate [10]. The generation time and serial interval of 2019-nCoV are longer than those of the 2009 pandemic A/H1N1 influenza virus (median generation time, 2.7 days; 95% CI, 2.0 to 3.5; and mean serial interval: range, 2.6 to 3.2) [14,18]. However, the mean serial interval of 2019-nCoV is similar to that of the SARS coronavirus (mean, 8.4 days; standard deviation, 3.8) [19]. The overall case fatality rate of 2019-nCoV was estimated by international experts to range from 3% to 14% [15,20], and it is more likely to cause infection in older age groups with commodities [2].
Epidemiological parameters are usually obtained from a consecutive timeline of the number of reported cases and contact-tracing data. However, most of the studies included in our review made estimates using data obtained from the early stages of the outbreak in Wuhan, in which the reporting of confirmed cases may have been incomplete. Furthermore, differences in the simulation methodology used in various scenarios may result in a wide spectrum of estimated values [21]. Therefore, caution is needed when interpreting these reported results.
The number of confirmed cases is increasing in China and in other countries, including Korea. Furthermore, the likelihood of local transmission is increasing as a result of cases entering from China.
In light of a report describing a case of human-to-human transmission in the asymptomatic period [5], it is necessary to consider updating the case definition for surveillance; however, more detailed studies presenting evidence on the epidemiological nature, clinical presentation, and pathogenesis of 2019-nCoV are necessary to provide information for public health decision-making.
Acknowledgments
A full list of the members of KSE 2019-nCoV TFT (alphabetically ordered): Byung Chul Chun (Department of Preventive Medicine, Korea University College of Medicine, Seoul); Dong Hyun Kim (Department of Social and Preventive Medicine, Hallym University College of Medicine, Chuncheon); Hae-Kwan Cheong (Sungkyunkwan University School of Medicine, Suwon); Heeyoung Lee (Center for Preventive Medicine and Public Health, Seoul National University Bundang Hosptial, Seongnam); Hyunjin Son (Busan Center for Infectious Disease Control and Prevention, Busan National University Hospital); Ji-Hyuk Park (Department of Preventive Medicine, Dongguk University, Gyeongju), Jong-Hun Kim (Sungkyunkwan University School of Medicine, Suwon); Kwan Lee (Department of Preventive Medicine, Dongguk University, Gyeongju); Kwang-Pil Ko (Department of Preventive Medicine, Gachon University, College of Medicine, Incheon); Sukhyun Ryu (Department of Preventive Medicine, Konyang University, Daejeon); Sung-il Cho (Graduate School of Public Health, Seoul National University, Seoul); Young June Choe (Department of Social and Preventive Medicine, Hallym University College of Medicine, Chuncheon).
Footnotes
The authors have no conflicts of interest to declare for this study.
FUNDING
None.
AUTHOR CONTRIBUTIONS
Conceptualization: BCC, SR. Data curation: KSE 2019-nCoV TFT. Formal analysis: BCC, SR. Methodology: BCC, SR. Writing - original draft : BCC, SR. Writing - review & editing: BCC, KSE 2019-nCoV TFT, SR.
SUPPLEMENTARY MATERIALS
Korean version is available at http://www.e-epih.org/.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Lancet Emerging understandings of 2019-nCoV. Lancet. 2020;395:311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization Novel Coronavirus (2019-nCoV) situation reports. [cited 2020 Feb 3]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 7.Korean Ministry of Health Welfare 2019-nCoV. [cited 2020 Feb 3]. Available from : http://ncov.mohw.go.kr/tcmBoardList.do?brdId=&brdGubun=&dataGubun=&ncvContSeq=&contSeq=&board_id=(Korean)
- 8.Wu P, Hao X, Lau EH, Wong JY, Leung KS, Wu JT, et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020;25:2000044. doi: 10.2807/1560-7917.ES.2020.25.3.2000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020;25:2000058. doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Sugimoto JD, Halloran ME, Basta NE, Chao DL, Matrajt L, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326:729–733. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Centre for Disease Prevention and Control Risk assessment: outbreak of acute respiratory syndrome associated with a novel coronavirus, China: first local transmission in the EU/EEA − third update. 2020 Jan 31; [cited 2020 Feb 3]. Available from: https://www.ecdc.europa.eu/en/publications-data/risk-assessment-outbreak-acute-respiratory-syndrome-associated-novel-1.
- 16.Wong TW, Tam W. Estimating SARS incubation period. Emerg Infect Dis. 2004;10:1503–1504. doi: 10.3201/eid1008.040284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lessler J, Reich NG, Cummings DA, New York City Department of Health and Mental Hygiene Swine Influenza Investigation Team. Nair HP, Jordan HT, et al. Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med. 2009;361:2628–2636. doi: 10.1056/NEJMoa0906089. [DOI] [PubMed] [Google Scholar]
- 18.Balcan D, Hu H, Goncalves B, Bajardi P, Poletto C, Ramasco JJ, et al. Seasonal transmission potential and activity peaks of the new influenza A(H1N1): a Monte Carlo likelihood analysis based on human mobility. BMC Med. 2009;7:45. doi: 10.1186/1741-7015-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020 doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Angelis D, Presanis AM, Birrell PJ, Tomba GS, House T. Four key challenges in infectious disease modelling using data from multiple sources. Epidemics. 2015;10:83–87. doi: 10.1016/j.epidem.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Korean version is available at http://www.e-epih.org/.