Abstract
Doxorubicin is a widely used cancer therapeutic, but its effectiveness is limited by cardiotoxic side effects. Evidence suggests cardiotoxicity is due not to doxorubicin, but rather its metabolite, doxorubicinol. Identification of the enzymes responsible for doxorubicinol formation is important in developing strategies to prevent cardiotoxicity. In this study, the contributions of three murine candidate enzymes to doxorubicinol formation were evaluated: carbonyl reductase (Cbr) 1, Cbr3, and thioredoxin reductase 1 (Tr1). Analyses with purified proteins revealed that all three enzymes catalyzed doxorubicin-dependent NADPH oxidation, but only Cbr1 and Cbr3 catalyzed doxorubicinol formation. Doxorubicin-dependent NADPH oxidation by Tr1 was likely due to redox cycling. Subcellular fractionation results showed that doxorubicin-dependent redox cycling activity was primarily microsomal, whereas doxorubicinol-forming activity was exclusively cytosolic, as were all three enzymes. An immunoclearing approach was used to assess the contributions of the three enzymes to doxorubicinol formation in the complex milieu of the cytosol. Immunoclearing Cbr1 eliminated 25% of the total doxorubicinol-forming activity in cytosol, but immunoclearing Cbr3 had no effect, even in Tr1 null livers that overexpressed Cbr3. The immunoclearing results constituted strong evidence that Cbr1 contributed to doxorubicinol formation in mouse liver but that enzymes other than Cbr1 also played a role, a conclusion supported by ammonium sulfate fractionation results, which showed that doxorubicinol-forming activity was found in fractions that contained little Cbr1. In conclusion, the results show that Cbr1 accounts for 25% of the doxorubicinol-forming activity in mouse liver cytosol but that the majority of the doxorubicinol-forming activity remains unidentified.
SIGNIFICANCE STATEMENT
Earlier studies suggested carbonyl reductase (Cbr) 1 plays a dominant role in converting chemotherapeutic doxorubicin to cardiotoxic doxorubicinol, but a new immunoclearing approach described herein shows that Cbr1 accounts for only 25% of the doxorubicinol-forming activity in mouse liver cytosol, that two other candidate enzymes—Cbr3 and thioredoxin reductase 1—play no role, and that the majority of the activity remains unidentified. Thus, targeting Cbr1 is necessary but not sufficient to eliminate doxorubicinol-associated cardiotoxicity; identification of the additional doxorubicinol-forming activity is an important next challenge.
Introduction
Doxorubicin (Dox), also known as Adriamycin, is used to treat a wide array of cancers, but its effectiveness is limited by dose-dependent cardiotoxicity, which manifests as congestive heart failure (Singal and Iliskovic, 1998; Swain et al., 2003; Kremer and Caron, 2004). The mechanism underlying cardiotoxicity is not fully understood, but doxorubicinol (Doxol), a metabolite generated by reduction of Dox at the C-13 carbonyl position (Fig. 1A) (Joerger et al., 2005) likely plays a significant role, as Doxol is far more potent than Dox in inhibiting several cardiac ion transporters (Boucek et al., 1987; Olson et al., 1988; Dodd et al., 1993; Hanna et al., 2014), and Doxol causes systolic and diastolic dysfunction in isolated rabbit heart muscle (Boucek et al., 1987; Olson et al., 1988).
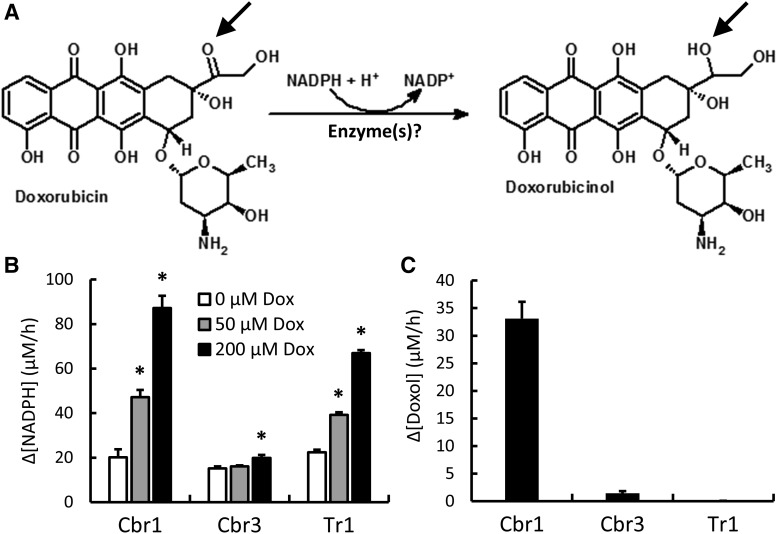
Fig. 1.
Reduction of Dox to its cardiotoxic metabolite Doxol. (A) Diagram showing the formation of Doxol via the two-electron reduction of the C-13 carbonyl group of Dox to an alcohol (arrows). (B) Dox-dependent NADPH oxidation by purified Cbr1, Cbr3, and Tr1. Reaction conditions were 200 µM NADPH, indicated concentrations of Dox, and 1 µM Cbr1, 1.9 µM Cbr3, or 1.9 µM Tr1 (Cbr3 and Tr1 histograms were normalized to 1 µM enzyme). Change in A340 was monitored for 1 hour at 37°C, and the rate of NADPH oxidation was calculated from the linear phase of the time course. (C) NADPH-dependent Doxol formation by purified Cbr1, Cbr3, and Tr1. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 1 µM enzyme; incubations were for 1 hour at 37°C. Doxol levels were measured by LC-MS/MS. Error bars represent 1 S.D.; n = 3 for all reactions; *P < 0.05 by Student’s t test.
Compared with its parent compound, Doxol displays minimal antineoplastic activity (Olson et al., 1988; Chang et al., 1989; Bernardini et al., 1991; Heibein et al., 2012). This is critical, as it suggests that the antitumor properties of Dox are not inherently linked to its cardiotoxicity. Preventing the metabolism of Dox to Doxol could thus increase the safety of the drug while retaining its effectiveness. As such, the goal of this study was to directly assess the contributions of three cytosolic enzymes to Doxol formation in liver, the most dominant location of xenobiotic metabolism. The enzymes carbonyl reductase (Cbr) 1, Cbr3, and thioredoxin reductase 1 (Tr1) were selected for the following reasons.
Cbr1 can catalyze the NADPH-dependent reduction of Dox to Doxol in vitro, and drugs that inhibit purified Cbr1 activity also inhibit Doxol formation by liver cytosol (Kassner et al., 2008; Bains et al., 2010), suggesting Cbr1 is the predominant Dox reductase in liver cytosol. Genetic evidence also suggests Cbr1 plays a major role in Doxol formation. Mice with a single null allele of Cbr1 show decreased sensitivity to Dox-induced cardiotoxicity, whereas mice that overexpress Cbr1 show increased sensitivity (Forrest et al., 2000; Olson et al., 2003). Additionally, cancer cell lines that overexpress carbonyl reductase are resistant to the antiproliferative effects of Dox (Bains et al., 2013). Pharmacological and genetic studies alone, however, do not confirm a major role for Cbr1 in Doxol formation, as drugs are often nonspecific and genetic interventions often have secondary effects on gene expression.
Cbr3 is a syntenic homolog of Cbr1. Although less well studied, it has also been shown to catalyze Doxol formation in vitro (Blanco et al., 2008; Bains et al., 2010). In vivo evidence for Cbr3 playing a role in Doxol formation derives from work done on knockout mice missing the γ-glutamyl-cysteine ligase modulatory subunit (Gclm) gene encoding the glutamate-cysteine ligase modifier subunit. These mice overexpress Cbr3 mRNA, and to a lesser extent Cbr1 mRNA (Haque et al., 2010), and show elevated formation of Doxol by liver cytosol and isolated hepatocytes (Schaupp et al., 2015).
Tr1 is known to reduce many substrates besides thioredoxin, including the small molecules lipoate (Arnér et al., 1996), lipid hydroperoxides (Björnstedt et al., 1995), dehydroascorbate (May et al., 1997), and menadione (Luthman and Holmgren, 1982). Liver-specific deletion of the thioredoxin reductase 1 ( Txnrd1)gene encoding Tr1 also causes massive overexpression of Cbr3 (Bondareva et al., 2007; Suvorova et al., 2009), potentially as a compensatory response to the loss of Tr1. Because of its broad substrate specificity, as well as a potential substrate overlap with Cbr3, we evaluated Tr1 as another candidate for playing a role in Doxol formation.
The aldo-keto reductase (AKR) 1C3 can catalyze Doxol formation in vitro; however, we did not include it in our investigations because the inhibition profile of AKR1C3 does not match that of liver cytosol (Tanaka et al., 2005; Kassner et al., 2008).
Initially, purified recombinant enzymes were tested for their ability to catalyze Dox-dependent NADPH oxidation and NADPH-dependent Doxol formation in vitro. Although all three enzymes catalyzed Dox-dependent NADPH oxidation, only Cbr1 and Cbr3 catalyzed Doxol formation. Instead of Doxol formation, Tr1 catalyzed Dox-dependent generation of H2O2, probably via redox cycling. To directly test for a physiologic role of the three enzymes in Dox metabolism, antibodies to the three proteins were developed and used to immunoclear the respective proteins from liver cytosol. Immunoclearing Cbr3 or Tr1 had no effect on Doxol formation by liver cytosol, suggesting these enzymes played no physiologic role in Doxol formation. In contrast, immunoclearing Cbr1 eliminated 25% of the Doxol-forming activity, verifying that Cbr1 contributes to Doxol formation in vivo. Importantly, however, the results also showed that most of the Doxol-forming activity in liver cytosol was not Cbr1.
Materials and Methods
Mouse Liver Fractionation.
Mice were handled using procedures approved by Oregon State University Institutional Animal Care and Use Committee. All mice were strain C57Bl6. A mixture of male and female mice between 3 and 9 months of age were used. No sex- or age-specific trends in the results obtained were observed. The ∆Gclm knockout line and liver-specific ∆Txnrd1 knockout line were described previously (Bondareva et al., 2007; Haque et al., 2010). Mice were fasted overnight to minimize variability caused by recent feeding activity and euthanized by cervical dislocation. Livers were perfused in situ with PBS (150 mM NaCl, 50 mM NaPO4 pH 7.4) to reduce blood content, removed to a petri dish, minced, and homogenized (Elvehjem, five strokes) in 10 volumes cold MOPS isolation buffer (200 mM sucrose, 10 mM Tris/4-morpholinepropanesulfonic acid pH 7.4, 1 mM EGTA) (Frezza et al., 2007). Homogenates were centrifuged for 10 minutes at 12,000g to remove nuclei and undisrupted cells. Clarified lysates were centrifuged for 30 minutes at 45,000g to remove mitochondria. The 45,000g supernatants were centrifuged for 1 hour at 100,000g to obtain microsomes (100,000g pellet) and cytosols (100,000g supernatant). Microsomes were resuspended in MOPS isolation buffer. Protein concentrations were determined by the Bradford method using bovine serum albumin as standard.
Ammonium sulfate fractionation was initiated by adding solid (NH4)2SO4 to cytosol prepared from 13 pooled mouse livers to achieve a salt solution at 40% of saturation (1.64 M). After stirring 30 minutes at 4°C, the mixture was centrifuged for 30 minutes at 12,000g to obtain a 40% pellet and 40% supernatant. Solid (NH4)2SO4 was added to the 40% supernatant, and the above steps were repeated to achieve first a 50% pellet and supernatant and subsequently a 60% pellet and supernatant. Precipitates were resuspended in 150 mM KCl, 50 mM KPO4 pH 7.4. Fractions were stored at −20°C until assay.
Protein Expression and Purification.
Using cDNA plasmids purchased from Open Biosystems (Huntsville, AL) as templates, polymerase chain reaction fragments containing the complete coding regions of mouse Cbr1, Cbr3, and Tr1 and a synthetic Nde1 site overlapping the start codon were blunt end-cloned into EcoR5-cleaved pBluescript KS and subcloned as Nde1/Hind3 fragments into the expression vector pET28a (Novagen, La Jolla, CA) such that expressed proteins had a histidine tag and thrombin cleavage site at the N terminus and the native amino acid at the C terminus. BL21 bacteria transformed with the expression plasmids were shaken at 37°C in LB medium containing 50 µg/ml kanamycin. When absorbance at 600 nm (A600) reached 0.6, 1 mM isopropyl-β-D-thiogalactoside was added to induce protein expression, and incubation continued for 6 hours at 25°C or overnight at 18°C. Cells were harvested by centrifugation at 3300g, resuspended in 20 ml chilled extraction buffer (150 mM NaCl, 50 mM NaPO4 pH 7.4), and lysed using an M-110P Microfluidizer (Microfluidics, Westwood, MA). Lysates were clarified by centrifugation at 15,000 rpm for 30 minutes in a Sorvall SS-34 rotor. Clarified lysate was mixed with 1 ml TALON metal affinity resin (Clontech, Mountain View, CA), and the mixture incubated for 1 hour with tumbling at 4°C. Resin containing bound protein was collected by centrifugation for 1 minute at 2000g, washed four times with 40 ml chilled extraction buffer, transferred to a gravity column, and washed with 10 ml extraction buffer and 5 ml of extraction buffer with 5 mM imidazole. Bound protein was eluted in 500-µl increments using 5 ml elution buffer (extraction buffer with 50 mM imidazole). Protein concentration was determined by A280 using extinction coefficients of 20,970 M−1cm−1 for Cbr1 and Cbr3 and 58,330 M−1cm−1 for Tr1. Protein purity, assessed by SDS-PAGE and Coomassie staining, was greater than 95%; thus no further purification was required. Gels representative of the purity achieved are shown in Supplemental Material (Supplemental Fig. 1). Proteins were stored at 4°C in elution buffer containing 0.1 mM dithiolthreitol. Freeze/thawing or storage at concentrations above 25 µM led to protein precipitation and was avoided. When precipitation occurred, fresh preparations of enzyme were made.
Coupling of Cbr1 and Cbr3 to Affi-gel 15 and Tr1 to Affi-gel 10 was done using the aqueous protocol described by the vendor (Bio-Rad, Hercules, CA). Prior to coupling, imidazole and dithiolthreitol were removed by gel filtration using 150 mM KCl, 50 mM KPO4 pH 8.0 as exchange buffer.
Immunologic Procedures.
Antisera were generated in New Zealand White rabbits, using full-length mouse Cbr1, Cbr3, and Tr1 protein with N-terminal histidine tags as antigen, expressed and purified as described above. Cbr3 and Tr1 immunizations and bleeds were done at the Oregon State University Laboratory Animal Resources Center. Cbr1 immunizations and bleeds were done by Pacific Immunology (Ramona, CA). Two rabbits were used for each protein. Antisera identification codes were assigned based on the source rabbit (Cbr1, 12081 and 12082; Cbr3, OB2 and OB4; Tr1, OB1 and OB3). Terminal bleeds (100–120 ml) were done by heart puncture. Raw antisera were prepared by allowing blood to coagulate at 4°C overnight, centrifuging the mixture at 5000g for 15 minutes, and storing 10-ml aliquots of supernatant at −70°C. Woring aliquots were stored at 4°C, with 0.1% NaN3 added as a preservative.
Raw Cbr3 antiserum cross-reacted with Cbr1 and raw Cbr1 antiserum cross-reacted with Cbr3 when used to probe immunoblots. To increase specificity, raw Cbr1 antiserum (12081) was cross-adsorbed to Cbr3-conjugated Affi-gel 15 (Bio-Rad) to obtain Cbr1-specific antiserum, and raw Cbr3 antiserum (OB4) was cross-adsorbed to Cbr1-conjugated Affi-gel to obtain Cbr3-specific antiserum. Comparison of immunoblots probed with raw Cbr3 antiserum, Cbr3-specific antiserum, and Cbr1-specific antiserum showed that Cbr1 and Cbr3 migrated as a triplet during SDS-PAGE analysis of mouse liver cytosol, with the slower two bands representing Cbr1 and the faster band representing Cbr3.
To remove bulk immunoglobulins that would compete for Protein A binding during immunoclearing, Cbr1- and Cbr3-specific antisera were affinity-purified. Briefly, 10 ml of specific antisera were incubated with 5 ml of Cbr1-conjugated or Cbr3-conjugated Affi-gel 15 beads overnight at 4°C, the beads were rinsed several times with PBS, and bound antibodies were eluted with 150 mM NaCl, 20 mM HCl into tubes containing 100 µl of neutralizing 1 M KPO4 pH 7.5 per 1.5 ml of eluate.
Immunoclearing of Cbr1 or Cbr3 was done using a noncovalent bridge of affinity-purified Cbr1-specific or Cbr3-specific antibody bound to Protein A magnetic beads (Bio-Rad). Beads were preincubated overnight with antibodies and washed several times with PBS immediately before use. Beads preincubated only with PBS were used for mock-cleared controls. About 50 µl of washed beads was used to clear Cbr1 or Cbr3 from 120 µg of cytosolic liver protein.
Immunoclearing of Tr1 was done using affinity-purified anti-Tr1 antibody that was covalently coupled to Affi-gel 10. Covalent coupling was necessary because the molecular weights of Tr1 and IgG heavy chain are similar (50 kDa) and unavoidable leaching of some IgG using the noncovalent Protein A bridge method would complicate interpretation of immunoblots designed to measure the efficiency of Tr1 removal. Ethanolamine-coupled Affi-gel was used for mock-cleared controls. About 50 µl of anti-Tr1–coupled Affi-gel was used to clear Tr1 from 300 µg of cytosolic liver protein.
After the efficiency of immunoclearing was determined by immunoblotting, 12-µg equivalents of immunocleared or mock-cleared cytosolic protein were assayed for Dox-dependent NADPH oxidation activity and NADPH-dependent Doxol forming activity as described below.
For immunoblot analyses, proteins were separated by SDS-PAGE and electroblotted to nitrocellulose membrane (Bio-Rad). Membranes were blocked overnight at 4°C in TBST (20 mM Tris pH 7.4, 150 mM NaCl, 0.1% Tween) containing 5% nonfat dried milk (NFDM). Primary antibody incubations were for at least 2 hours at 25°C, using a 1:1000 dilution of raw antiserum or 1:300 dilution of affinity-purified antibody in TBST 5% NFDM. After four 5-minute washes with TBST 5% NFDM, blots were incubated with secondary antibody for at least 1 hour at 25°C, using a 1:5000 dilution of goat anti-rabbit horseradish peroxidase–conjugated IgG (Bio-Rad). Following four TBST washes, blots were developed with Western Lightning enhanced chemiluminescent substrate (Santa Cruz Biotechnology, Dallas, TX) and imaged using a ChemiGenius Bio-imaging system (Syngene, Frederick, MD). Protein band intensities were determined using Syngene Genetools software.
Enzyme Assays.
Dox-dependent NADPH oxidation was measured in UV-transparent 96-well plates by tracking A340 using a Biotek Synergy 2 Multi-Mode Microplate Reader (Winooski, VT). Reaction cocktails contained 50 mM KPO4 pH 7, 0–200 µM Dox, 1–2 µM recombinant enzyme or 50–100 µg of liver protein, and 200 µM NADPH (added last) in a total volume of 150 µl. Absorbance was monitored for 1 hour at 37°C, and the molar amount of NADPH oxidized was calculated using an extinction coefficient of 6220 M−1cm−1 and estimated pathlength of 3 mm. The rate of NADPH oxidation in the absence of Dox was subtracted from rates measured in the presence of Dox.
NADPH-dependent Doxol formation was measured by liquid chromatography–mass spectrometry/collision-induced dissociation mass spectrometry (LC-MS/MS). Reaction cocktails were prepared as described above. After a 1-hour incubation at 37°C, reactions were stopped and proteins precipitated by adding chilled sulfosalicylic acid to 0.5%. Precipitated protein was removed by centrifugation for 10 minutes at 12,000g. Supernatants were analyzed using a SCIEX 3200 Q TRAP LC-MS/MS system equipped with Shimadzu LC-20AD pumps, SIL-20A model autosampler, CTO-10ASvp model column oven, and CBM-20A model system controller. Dox was separated from Doxol using an Agilent Poroshell 120 PFP reversed-phase column (Santa Clara, CA), 10 µl sample injection volume, 0.3 ml/min flow rate, and 10%–90% acetonitrile gradient applied over 10 minutes. For Dox, the Q1 parent peak was 544.4 Da, and the Q3 peaks were 397.3, 379.3, and 130.1 Da. For Doxol, the Q1 parent peak was 546.4 Da, and Q3 peaks were 399.3, 381.3, and 130.1 Da. The 399.3 peak was used for quantitation of Doxol, and the other peaks were used as quality controls. Representative mass spectra are shown in Supplemental Material (Supplemental Fig. 2).
Kinetic parameters were calculated using nonlinear least squares regression. For mouse liver cytosol, Vmax was calculated instead of the catalytic constant (kcat). Raw data are provided in Supplemental Material (Supplemental Table 1).
H2O2 formation was measured using an Amplex red-based assay kit (Invitrogen, Carlsbad, CA). Purified Cbr1 and Tr1 (1 µM) were mixed with 50 µM Dox in 50 mM KPO4 pH 7.4. NADPH (200 µM) was added to initiate the reaction and bring the final volume to 100 µl. Samples were combined with 100 µl of Amplex red working solution (100 µM Amplex red, 200 U/ml horseradish peroxidase, 50 mM KPO4 pH 7.4). Using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA), samples were excited at 470 nm, and emission in the far-red range (665–720 nm) was tracked. A standard curve of 0 to 1 mM H2O2 was used to quantify H2O2 production by samples. The same procedure was followed to measure H2O2 production by 30 µg of liver cytosolic or microsomal protein.
Results
Dox-Dependent NADPH Oxidation and NADPH-Dependent Doxol Formation by Purified Cbr1, Cbr3, and Tr1.
As described above, the enzymes Cbr1, Cbr3, and Tr1 were potential candidates for catalyzing reduction of Dox to Doxol (Fig. 1A). As an initial assessment of these candidate enzymes, recombinant mouse Cbr1, Cbr3, and Tr1 were purified and tested for their ability to catalyze Dox-dependent NADPH oxidation. All three enzymes were active (Fig. 1B). Kinetic parameters for Dox-dependent NADPH oxidation by these enzymes were determined and are summarized in Table 1. Cbr1 had a kcat 10-fold greater than Cbr3 and 1.5-fold greater than Tr1. Tr1 had the highest catalytic efficiency (kcat/Km).
TABLE 1.
Kinetic parameters Km, kcat, and kcat/Km for Dox-dependent NADPH oxidation by purified Cbr1, Cbr3, and Tr1
| Enzyme | R2 | Parameter | ||
|---|---|---|---|---|
| Km | kcat | kcat/Km | ||
| µM | s−1 × 10−3 | µM−1 s−1 × 10−6 | ||
| Cbr1 | 0.989 | 130 | 15 | 120 |
| Cbr3 | 0.920 | 41 | 1.4 | 35 |
| Tr1 | 0.969 | 36 | 10 | 290 |
Although spectrophotometric analysis of NADPH oxidation was an expedient way of assessing whether an oxidoreductase used Dox as a substrate, it did not establish that the product of the reaction was Doxol. For example, ring keto groups on Dox could alternatively serve as electron acceptors. We thus used LC-MS/MS to directly measure the production of Doxol by Cbr1, Cbr3, and Tr1. Pilot analyses with standards established that Doxol could be distinguished from its precursor and that the Doxol mass/charge (M/Z) peaks were proportional to the amount of Doxol added to the mixture (Supplemental Fig. 2). Using the LC-MS/MS assay, the amount of Doxol formed by each enzyme during a 1-hour incubation with 200 µM NADPH and Dox was determined (Fig. 1C). Both Cbr1 and Cbr3 produced Doxol. In contrast, despite showing a high rate of Dox-dependent NADPH oxidation, Tr1 produced no Doxol during the reaction. Thus, tracking Dox-dependent NADPH oxidation alone was not sufficient to draw conclusions regarding the formation of Doxol. Kinetic parameters for Doxol formation by Cbr1 and Cbr3 were determined and compared with kinetic parameters for Doxol formation by mouse liver cytosol (Table 2). Cbr1 and Cbr3 had similar Km values (99 and 74 µM, respectively), but the catalytic efficiency (kcat/Km) for Doxol formation by Cbr1 was 280-fold higher than by Cbr3. Importantly, the Km values for Doxol formation exhibited by purified Cbr1 and Cbr3 were similar to those exhibited by liver cytosol (75 µM), consistent with these enzymes playing a role in vivo.
TABLE 2.
Kinetic parameters Km, kcat, and kcat/Km for NADPH-dependent Doxol formation by purified Cbr1 and Cbr3, as well as Km and Vmax for NADPH-dependent Doxol formation by mouse liver cytosol
| Enzyme | R2 | Parameter | |||
|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Vmax | ||
| µM | s−1 × 10−3 | µM−1 s−1 × 10−6 | µM/h | ||
| Cbr1 | 0.997 | 99 | 36 | 360 | — |
| Cbr3 | 0.855 | 74 | 0.1 | 1.3 | — |
| Cytosol (0.55 µg/µl) | 0.976 | 75 | — | — | 8.7 |
Doxorubicin-Dependent Redox Cycling Catalyzed by Tr1.
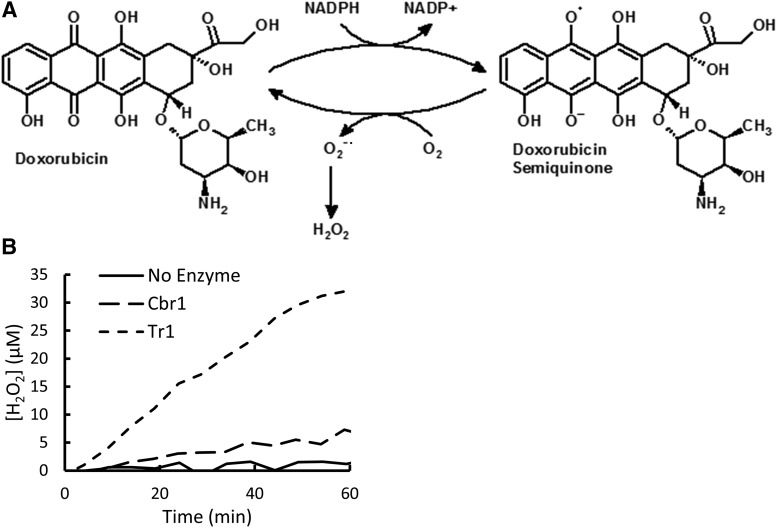
Because Dox-dependent NADPH oxidation by Tr1 was not associated with Doxol formation, we investigated whether Tr1 was catalyzing NADPH oxidation by a redox cycling mechanism (Fig. 2A). In this model, an electron equivalent from NADPH is transferred to Dox to form a semiquinone but is subsequently transferred to another entity, presumably solvent oxygen, which results in regeneration of Dox and the sequential formation of superoxide radical and H2O2. To test this hypothesis, Tr1 and Cbr1 were incubated with Dox and NADPH, and the amount of H2O2 produced over the course of the reaction was measured using Amplex Red reactivity as an indicator. The results (Fig. 2B) showed that in the presence of Dox and NADPH, Tr1 generated significant amounts of H2O2. Cbr1 also produced H2O2, but at a much slower rate. The efficient formation of H2O2, along with the lack of Doxol formation, suggested Tr1 was indeed catalyzing Dox-dependent redox cycling.
Fig. 2.
Tr1-catalyzed redox cycling of Dox. (A) Diagram depicting reduction of Dox to a semiquinone, which decays back to Dox, generating superoxide, H2O2, and other free radicals. (B) H2O2 formation by Cbr1 and Tr1. Reaction conditions were 200 µM NADPH, 50 µM Dox, and 1 µM enzyme; incubations were for 1 hour at room temperature.
Subcellular Localization of Dox-Metabolizing Activities in Liver.
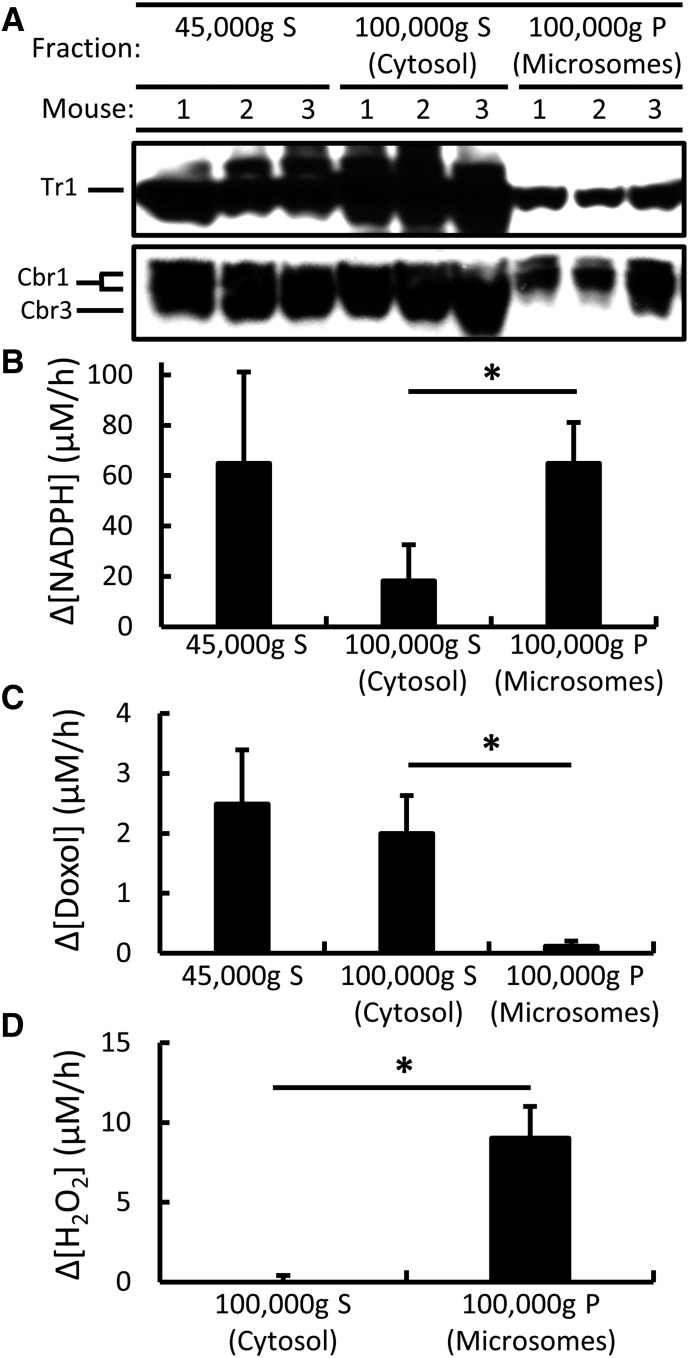
To confirm the cytosolic localization of Cbr1, Cbr3, and Tr1 and to determine whether the enzymes catalyzing Dox-dependent NADPH oxidation and Doxol formation were similarly cytosolic, 45,000g liver supernatants were fractionated into their microsomal and cytosolic components by centrifugation at 100,000g. Equal amounts of protein from each component were analyzed for Cbr1, Cbr3, and Tr1 protein by immunoblotting, Dox-dependent NADPH oxidation by spectrophotometry, and Doxol-forming activity by LC-MS/MS. Immunoblot analyses (Fig. 3A) confirmed that all three proteins were primarily cytosolic. However, when Dox-dependent NADPH oxidation activity was assayed, most of the activity present in the original 45,000g supernatant was recovered in the microsomal fraction, and only 30% of the activity was recovered in the cytosolic fraction (Fig. 3B). In contrast, when Doxol formation was assayed, almost all of the activity present in the 45,000g supernatant was recovered in the cytosolic fraction (Fig. 3C). To test whether the high levels of Dox-dependent NADPH oxidation activity observed in microsomal fractions was due to redox cycling, we measured the production of H2O2 by cytosolic and microsomal fractions. As shown in Fig. 3D, microsomal fractions produced significant amounts of H2O2, whereas cytosolic fractions did not, suggesting that Dox-dependent redox cycling was occurring in the microsomes.
Fig. 3.
Subcellular localization of Cbr1, Cbr3, and Tr1 protein; Dox-dependent NADPH oxidation activity; NADPH-dependent Doxol-forming activity; and Dox-dependent H2O2-forming activity in mouse liver. (A) Immunoblot analysis of Cbr1, Cbr3, and Tr1 protein in indicated supernatants (S) and pellets (P): 45,000g S (a mixture of cytosol and microsomes), 100,000g S (cytosol), and 100,000g P (microsomes). ∆Gclm lysates were used to facilitate Cbr3 localization. (B) Dox-dependent NADPH oxidation by indicated fractions of wild-type liver lysates. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 50 µg fraction protein; incubations were for 1 hour at 37°C. (C) NADPH-dependent Doxol formation by indicated fractions of wild-type liver lysates. Reaction conditions and incubations were as described in (B). Doxol levels were measured by LC-MS/MS. (D) H2O2 formation by indicated fractions of wild-type liver lysates. Reaction conditions were 200 µM NADPH, 50 µM Dox, and 30 µg fraction protein; incubations were for 1 hour at 25°C. Error bars represent 1 S.D.; n = 3 for all reactions; *P < 0.05 by Student’s t test.
Effect of Immunoclearing Cbr1 on Doxol-Forming Activity in Mouse Liver Cytosol.
As demonstrated above, purified Cbr1 and Cbr3 were able to catalyze NADPH-dependent Doxol formation, with Cbr1 catalyzing the reaction much more efficiently than Cbr3. However, these in vitro analyses did not establish the extent to which these enzymes contributed to Doxol formation in the complicated milieu of the cytosol. Posttranslational modifications, accessory proteins, and small molecule effectors could impact the formation of Doxol by these enzymes in vivo. Additionally, other Doxol-forming enzymes might exist in the cytosol, diluting the contributions of Cbr1 and Cbr3 to overall activity. Genetic or drug interventions are often used to study the role of specific proteins within an organism or cell; however, genetic interventions often have unpredictable effects on the transcriptome, and “specific” drugs often target many enzymes in addition to the one being studied. We thus developed an immunoclearing approach to analyze the roles of Cbr1, Cbr3, and Tr1 in Doxol formation. This approach avoids complications that could arise from genetic or drug interventions while still requiring the enzymes to function within the context of the cytosol.
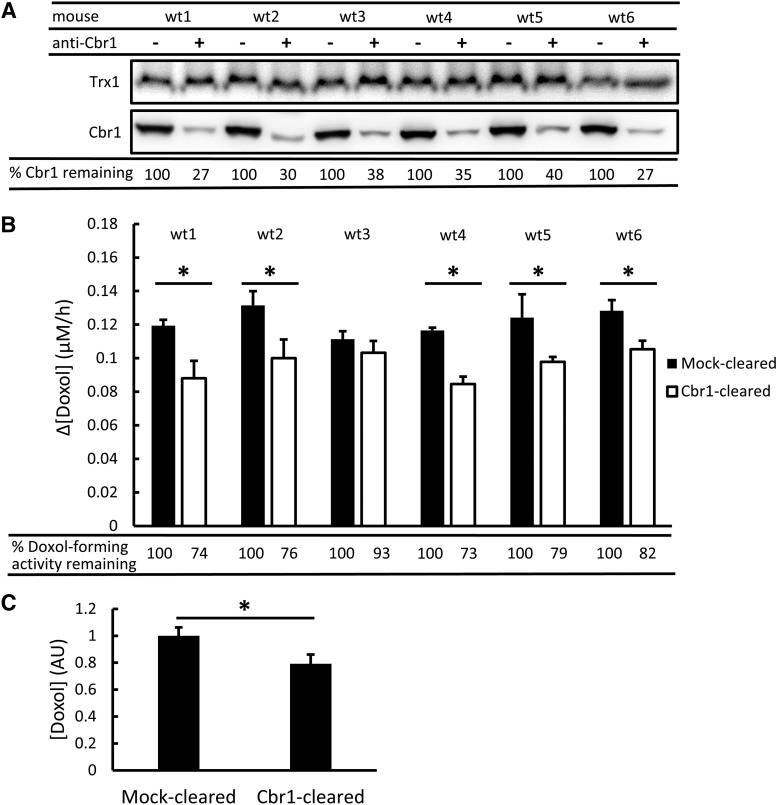
Protein A magnetic beads preincubated with Cbr1-specific antibody were used to immunoclear Cbr1 from six mouse liver cytosols. Beads preincubated only with buffer served as mock-cleared controls. The amount of Cbr1 present in Cbr1-cleared and mock-cleared cytosols was determined by immunoblot analysis (Fig. 4A). Densitometry showed that on average 67% of Cbr1 was cleared. To confirm the specificity of the immunoclearing reaction, immunoblots were reprobed with antibody against the small redox protein thioredoxin (Trx1). As expected, there was no significant difference in the amount of Trx1 present in Cbr1-cleared and mock-cleared cytosols (Fig. 4A).
Fig. 4.
Immunoclearing of Cbr1 and its effect on Doxol-forming activity in mouse liver cytosol. (A) Immunoblot analyses of Cbr1 in mock-cleared (−) and Cbr1-cleared (+) cytosols from six wild-type (wt) mice. Cbr1 protein levels were determined by densitometry, and the % Cbr1 protein remaining after immunoclearing, relative to mock-cleared samples, is shown at bottom. Trx1 levels were determined in parallel as a control. (B) NADPH-dependent Doxol-forming activity in mock-cleared and Cbr1-cleared cytosols. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 12-µg equivalents of cytosolic protein; incubations were for 1 hour at 37°C. Doxol levels were measured by LC-MS/MS. The % Doxol-forming activity remaining after immunoclearing, relative to mock-cleared samples, is shown at bottom. Error bars represent 1 S.D.; n = 3 technical replicates for all reactions; *P < 0.05 by Student’s t test. (C) Mock-cleared and Cbr1-cleared Doxol-forming activity for the six biologic replicates. The concentration of Doxol generated is reported in arbitrary units (AU), where the average level in mock-cleared samples is set to 1. rror bars represent 1 S.D.; *P < 0.005 by Student’s t test.
Equivalent amounts of Cbr1-cleared and mock-cleared cytosolic protein were incubated with NADPH and Dox, and the amount of Doxol produced during a 1-hour reaction was measured by LC-MS/MS (Fig. 4B). Five of six cytosols showed a statistically significant reduction in the amount of Doxol produced by Cbr1-cleared cytosol, compared with mock-cleared cytosol. When averaged, Doxol formation by the Cbr1-cleared cytosols was 21% lower than by the mock-cleared group (Fig. 4C). Because the immunoblotting analyses showed that the immunoclearing procedure removed only 67% of the Cbr1 in the cytosol, we calculated that 31% of the Doxol-forming activity in mouse liver cytosol was attributable to Cbr1. When the immunoclearing analyses were repeated using the same cytosols and independently prepared new cytosols (n = 12), slightly different estimates for the amount of activity attributable to Cbr1 were obtained, ranging from an average of 20% to 31%. From all experiments, we calculated that on average 23.4% ± 13.7% of Doxol-forming activity (roughly 25%) was attributable to Cbr1. The immunoclearing results thus provided unequivocal evidence that Cbr1 contributed significantly to Doxol formation in liver. Importantly, however, even with the majority of Cbr1 cleared from the cytosol, most of the Doxol-forming activity remained.
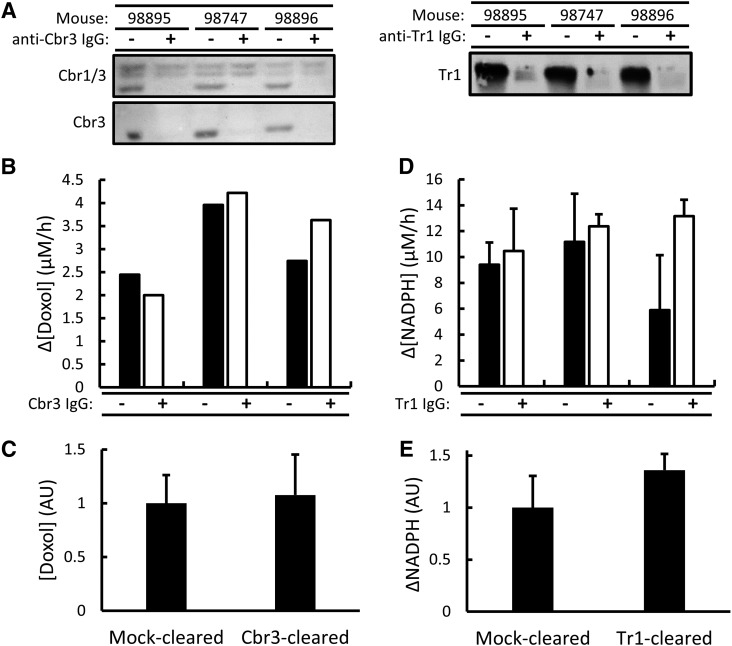
Effect of Immunoclearing Cbr3 and Tr1 on Dox-Dependent NADPH Oxidation and Doxol-Forming Activity in Mouse Liver Cytosol.
The above results showed that Cbr1 accounted for about 25% of the total Doxol-forming activity in liver cytosol. To investigate whether Cbr3 and Tr1 contributed to Dox metabolism, these proteins were also selectively immunocleared from mouse liver cytosol. Immunoblot analyses showed that Cbr3 and Tr1 were efficiently removed by the immunoclearing procedure (Fig. 5A). Despite near total clearing of Cbr3, the levels of Doxol-forming activity were unaffected (Fig. 5, B and C). Thus, Cbr3 did not contribute to the Doxol-forming activity detected in liver cytosol. Dox-dependent NADPH oxidation activity was also measured in Tr1-cleared and mock-cleared samples, and no significant difference was observed (Fig. 5, D and E). Thus, although purified Tr1 efficiently catalyzed Dox-dependent NADPH oxidation in vitro, Tr1 did not contribute to Dox-dependent NADPH oxidation in the context of the cytosol.
Fig. 5.
Immunoclearing Cbr3 and Tr1 from mouse liver cytosols and effect on Dox-dependent NADPH oxidation and NADPH-dependent Doxol formation. (A) Immunoblot analysis of Cbr3 and Tr1 in mock-cleared (−) and immunocleared (+) mouse liver cytosols. The Cbr3 immunoblots were probed with either raw anti-Cbr3 antiserum (top) or Cbr3-specific antiserum (bottom). (B) NADPH-dependent Doxol formation by mock-cleared and Cbr3-cleared cytosols. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 45-µg equivalents of cytosolic protein; incubations were for 1 hour at 37°C. Doxol levels were measured by LC-MS/MS. Triplicate incubations for each cytosol were pooled and analyzed as one sample. (C) Mock-cleared and Cbr3-cleared Doxol-forming activity for the three biologic replicates. The concentration of Doxol generated is reported in arbitrary units (AU), where the average level in mock-cleared samples is set to 1. Error bars represent 1 S.D.; there was no significant difference. (D) Dox-dependent NADPH oxidation by mock-cleared and Tr1-cleared cytosols. Reaction conditions were as described in (B). Error bars represent 1 S.D.; n = 3 technical replicates for all reactions. (E) Mock-cleared and Tr1-cleared Dox-dependent NADPH oxidation for the three biologic replicates. The change in NADPH is reported in arbitrary units (AU), where the average change in mock-cleared samples is set to 1. Error bars represent 1 S.D.; there was no significant difference.
Doxol Formation in ΔTxnrd1 Mouse Liver Cytosol.
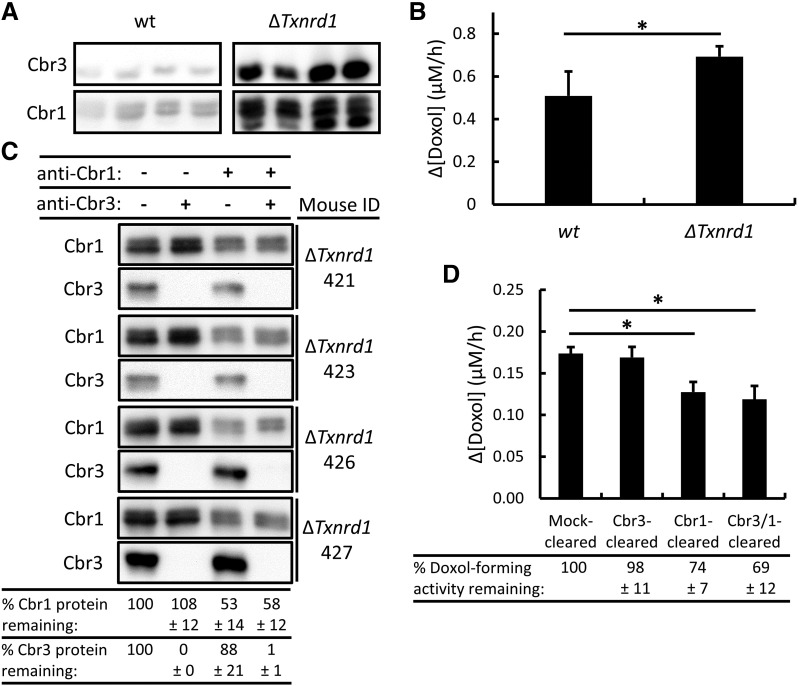
Cbr3 was implicated as a potential Dox reductase when it was observed that ∆Gclm null mice, which have elevated levels of Cbr3 mRNA and protein, have higher rates of Doxol formation (Schaupp et al., 2015). Interestingly, liver-specific deletion of the Txnrd1 gene also resulted in greatly elevated levels of Cbr3 and mildly elevated levels of Cbr1 (Fig. 6A). We investigated whether Cbr3 contributed significantly to Doxol formation in tissues that overexpressed Cbr3. We found that ΔTxnrd1 cytosols produced significantly more Doxol, but the effect was small (35% increase), relative to the large increase in Cbr3 protein (Fig. 6A).
Fig. 6.
Immunoclearing of Cbr1 and/or Cbr3 and its effect on Doxol-forming activity in ΔTxnrd1 liver cytosols that overexpress Cbr3. (A) Immunoblot analysis of Cbr3 and Cbr1 in wild-type (wt) and ΔTxnrd1 mouse liver cytosols; 18 µg of total cytosolic protein loaded per lane. The top panel shows a blot probed with Cbr3-specific antibody. The bottom panel shows the same blot reprobed with Cbr1-specific antibody; because the blot was not stripped, the fast-migrating Cbr3 signal remained visible. (B) NADPH-dependent Doxol formation by wild-type and ΔTxnrd1 cytosols. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 12-µg equivalents of cytosolic protein; incubations were for 1 hour at 37°C. Doxol levels were measured by LC-MS/MS. (C) Immunoblot analysis of immunocleared ΔTxnrd1 cytosols. Cbr1 and Cbr3 protein levels were determined by densitometry, and the % protein remaining after immunoclearing, relative to mock-cleared samples, is shown at bottom. (D) NADPH-dependent Doxol formation by mock-cleared, Cbr3-cleared, Cbr1-cleared, and Cbr3- and Cbr1-cleared ΔTxnrd1 cytosols. Reaction conditions and incubations were as described in (B), except 50 µM Dox was used. Doxol levels were measured by LC-MS/MS. The % Doxol-forming activity remaining after immunoclearing, relative to mock-cleared samples, is shown at bottom. Error bars represent 1 S.D.; n = 4 for all reactions; *P < 0.05 by Student’s t test.
To ascertain whether the increased Doxol-forming activity observed in ∆Txnrd1 null cytosols was attributable to Cbr3, Cbr1, or both enzymes, we immunocleared Cbr3 and Cbr1 individually and simultaneously from four ΔTxnrd1 cytosols (Fig. 6C). Total clearing of Cbr3 was achieved in all cytosols, whereas clearing of Cbr1 was less thorough (on average, about 55%). Clearing Cbr3 had no effect on Doxol formation, whereas clearing Cbr1 again led to a reduction in Doxol formation. Clearing Cbr1 and Cbr3 simultaneously did not lead to any difference in Doxol formation when compared with clearing Cbr1 alone (Fig. 6D). This suggests that even when Cbr3 was highly overexpressed, it was not responsible for any of the observed Doxol-forming activity in liver cytosol. The increased Doxol formation observed in ΔTxnrd1 livers was thus likely due to the small increase in Cbr1 or some as yet unidentified enzyme.
Ammonium Sulfate Fractionation of Doxol-Forming Activity in Liver Cytosol.
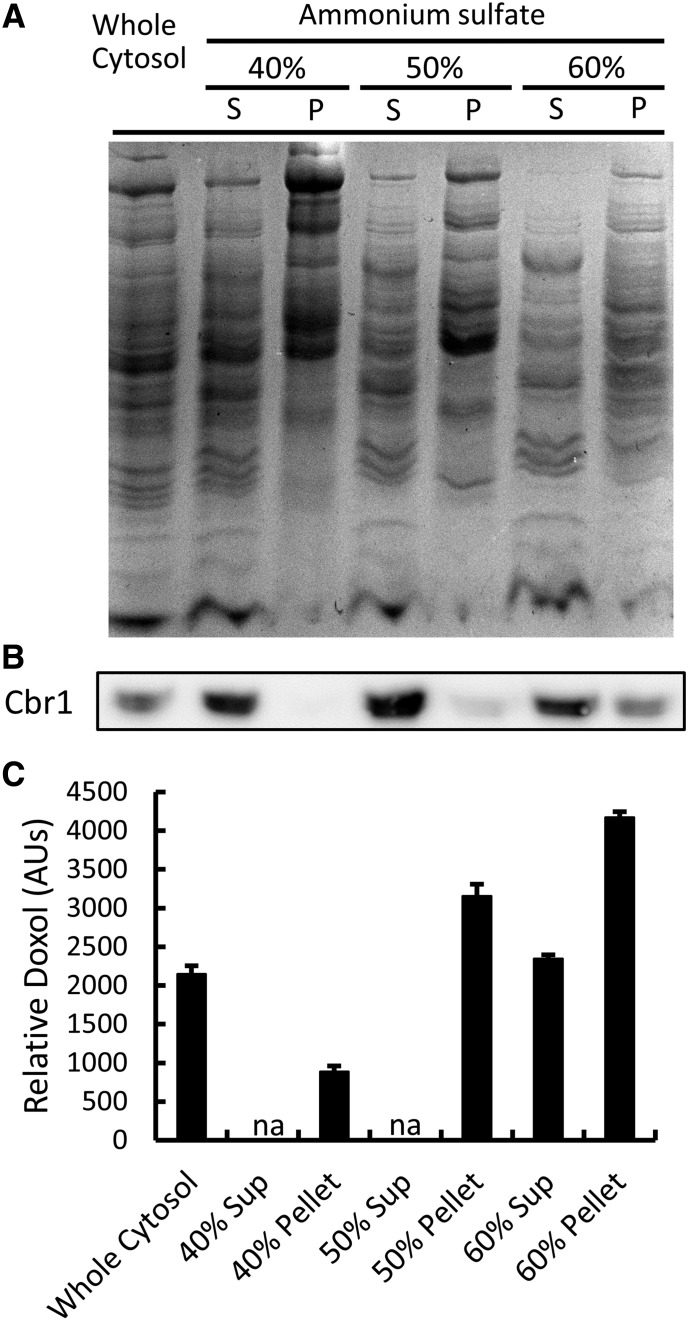
Having shown that only 25% of the Doxol-forming activity in liver cytosol was attributable to Cbr1, most of the total Doxol-forming activity remained unidentified. As an initial means of characterizing this remaining activity, we fractionated liver cytosol by ammonium sulfate precipitation. SDS-PAGE analysis revealed that each fraction had a distinct protein profile (Fig. 7A). Immunoblot analysis revealed that Cbr1 was found primarily in the 60% supernatant (Fig. 7B), whereas Doxol-forming activity was found primarily in the 60% pellet and 50% pellet (Fig. 7C). The finding that peak Doxol-forming activity was found in an ammonium sulfate fraction containing relatively little Cbr1 independently corroborated the conclusion of the immunoclearing experiment that enzymes other than Cbr1 comprised the majority of the Doxol-forming activity in liver cytosol.
Fig. 7.
Cbr1 levels and Doxol-forming activity in ammonium sulfate-fractionated mouse liver cytosol. (A) Coomassie-stained SDS-PAGE analysis of proteins in ammonium sulfate fractions; 10 µg of protein loaded per lane. P, pellet; S, supernatant. (B) Immunoblot analysis of Cbr1 in ammonium sulfate fractions; 10 µg of protein loaded per lane. (C) Relative amount of Doxol produced by each fraction. Reaction conditions were 200 µM NADPH, 200 µM Dox, and 12 µg fraction protein; incubations were for 1 hour at 37°C. Doxol levels were determined by LC-MS/MS. The relative level of Doxol generated is reported in arbitrary units (AU), where the average level in whole cytosol is set to 2200. Error bars represent 1 S.D.; n = 3 for all reactions; na, not assayed; sup, supernatant.
Discussion
Although several purified enzymes have been shown to catalyze Dox reduction, past efforts to test the biologic relevance of such findings have relied exclusively on pharmacological and genetic approaches. However, these approaches share the fundamental weakness of nonspecificity. Enzyme inhibitors rarely are specific for only one enzyme, and altering the genome for the purpose of over- or underexpressing a specific gene often has unpredictable effects on the expression of other genes. To avoid these issues, we developed a method of studying Doxol formation that takes advantage of the truly specific binding between antigen and antibody. We raised antibodies against Cbr1, the most studied Dox reductase, and against Cbr3 and Tr1, two other enzymes potentially involved in Dox reduction, and used the antibodies to immunoclear the respective proteins from mouse liver cytosol. By selectively removing each of these specific enzymes, we were able to quantify the contribution of each enzyme to Dox metabolism in the quasi in vivo context of liver cytosol.
Our initial characterization of the three enzymes revealed that Cbr3 and Cbr1 were both capable of producing Doxol but that Tr1 was not, despite efficiently catalyzing Dox-dependent NADPH oxidation. Further analyses revealed that purified Tr1 catalyzed Dox-dependent redox cycling, generating significant amounts of H2O2. However, subcellular fractionation analyses showed that Dox-dependent redox cycling activity in liver lysates was primarily microsomal, whereas Tr1 protein was almost exclusively cytosolic. Furthermore, immunoclearing Tr1 from cytosol had no effect on the observed redox cycling activity. Tr1 was thus not a significant contributor to the Dox-dependent redox cycling activity observed in liver lysates. Identifying the Dox-dependent redox cycling activity is important because redox cycling may contribute to the chemotherapeutic and/or cardiotoxic activities of Dox. For example, Dox is known to intercalate into DNA, and if it were to participate in redox cycling reactions from that location, it could create localized high concentrations of DNA-damaging reactive oxygen species and thereby arrest replication.
When Cbr3 was immunocleared from liver cytosol, no reduction in Doxol-forming activity was observed, indicating that this enzyme did not contribute to Doxol formation by liver cytosol. In contrast, when Cbr1 was immunocleared from liver cytosol, 25% of the Doxol-forming activity was removed, indicating that 25% of the Doxol-forming activity in liver was attributable to the Cbr1 polypeptide. Importantly, 75% of the Doxol-forming activity in liver cytosol remained after Cbr1 was removed by immunoclearing, indicating that other enzymes constituted the majority of the Doxol-forming activity. This conclusion was further supported by ammonium sulfate fractionation results that showed that peak levels of Doxol-forming activity were detected in fractions that contained little Cbr1.
Cbr3 mRNA is the most highly induced message in both ∆Txnrd1 null (Bondareva et al., 2007; Suvorova et al., 2009) and ΔGclm null tissues (Haque et al., 2010). We earlier suggested that Cbr3 played a role in Doxol formation based on the observations that cytosols from ΔGclm livers and isolated hepatocytes had elevated levels of Cbr3 protein and elevated levels of Doxol-forming activity, that ΔGclm hepatocytes incubated with Dox produced a substance toxic to myocytes and produced higher levels of Doxol, and that purified Cbr3 was able to reduce Dox to Doxol in vitro (Schaupp et al., 2015). Our current findings show that purified Cbr1 was 280-fold more efficient than Cbr3 in converting Dox to Doxol and that immunoclearing Cbr1, but not Cbr3, from either wild-type or ∆Txnrd1 cytosols removed a significant fraction of the measurable Doxol-forming activity. In view of the current findings, we suspect that the increase in Doxol-forming activity previously reported to occur in ΔGclm livers (Schaupp et al., 2015) was due to induction of Cbr1, rather than induction of Cbr3. Cbr1 is induced in both ΔGclm and ∆Txnrd1 livers, although not to the same extent as Cbr3.
Olson et al. (2003) showed that mice systemically heterozygous for a ∆Cbr1 deletion mutation are more resistant to the cardiotoxic effects of Dox. This result suggested that halving the amount of Cbr1 in the cytosol was cardioprotective. As our results indicated that only 25% of the total Doxol-forming activity in wild-type cytosol was attributable to Cbr1, heterozygosity for the ∆Cbr1 null mutation would be expected to result in only a 12.5% reduction in total Doxol-forming activity. Thus, more experiments are necessary to understand the cardioprotective effect of ∆Cbr1 heterozygosity during Dox treatment.
Others have shown (Jo et al., 2017) that coadministration of the Cbr1-inhibiting drug 3-(7-isopropyl-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidin-5yl)phenol (hydroxy-PP-Me) during Dox treatment of rodents results in greater chemotherapeutic activity against implanted tumors and lower cardiotoxicity, and they concluded that Cbr1 plays a major role in converting Dox to cardiotoxic Doxol. Our current results are consistent with a role for Cbr1 in Doxol formation, but it is difficult to explain how inhibition of Cbr1 alone results in reduced cardiotoxicity when Cbr1 constitutes only 25% of the total Doxol-forming in liver cytosol. It is possible that Cbr1 constitutes a much higher percentage of total Doxol-forming activity in heart or other tissues, but we consider it more likely that the hydroxy-PP-Me is not specific for Cbr1 and instead inhibits a broader spectrum of NADPH-dependent oxidoreductases, including the unidentified enzymes responsible for 75% of the Doxol-forming activity in liver cytosol. Nevertheless, the hydroxy-PP-Me study is extremely important because it suggests that development of drugs that inhibit Doxol-forming enzymes represents a promising strategy for combating cardiotoxicity during Dox chemotherapy.
Identification of the enzyme or enzymes that constitute the remaining unidentified Doxol-forming activity is important. Another enzyme capable of catalyzing Doxol formation in vitro is Akr1C3 (Kassner et al., 2008). However, the Doxol-forming activity of Akr1C3 is 10-fold more sensitive to the inhibitor 4-amino-1-tert-butyl-3-(2-hydroxyphenyl)pyrazolo[3,4-d]pyrimidine than the activity detected in cytosol (Tanaka et al., 2005; Kassner et al., 2008), reducing the likelihood that Akr1C3 plays a physiologic role in Doxol formation. Other aldo-keto reductases potentially play a role in Dox metabolism. For example, pharmacogenetic studies have suggested Akr1B10 or Akr1B1 may mediate the cardiotoxic effects of Dox (Morikawa et al., 2015; Sonowal et al., 2017). However, as the drugs used in these studies likely targeted other oxidoreductases, it is unclear whether the described effects were mediated by Akr1B10 or Akr1B1. The microsomal enzyme 11β-hydroxysteroid dehydrogenase 1 (11β-HSD1) was also recently linked to Doxol formation (Yang et al., 2018). The study showed that Doxol formation in human liver hepatocytes is reduced by treatment with an inhibitor of 11β-HSD1. Because the inhibitor used was not specific to 11β-HSD1, inhibition of other oxidoreductases may have been responsible for the effect of the drug on Doxol-forming activity in vivo. Furthermore, the current study showed Doxol formation to be predominantly cytosolic rather than microsomal. Moreover, although the inhibitor used was not specific for 11β-HSD1, it was shown to not inhibit Cbr1. Thus, the reduction of Doxol formation seen following treatment with the inhibitor is further evidence for the existence of enzymes other than Cbr1 that play an important role in Doxol formation.
The current study showed that although Cbr1 contributed significantly to Doxol formation in liver cytosol, currently unidentified enzyme or enzymes were responsible for the majority of the Doxol formation activity. The immunoclearing approach outlined here provides a method for evaluating the contributions of other potential Dox reductases such as AKR1C3 or 11β-HSD1. However, rather than evaluating specific candidate enzymes using an immunoclearing approach, identification of the enzyme(s) responsible for the unaccounted portion of hepatic Doxol formation likely will require exhaustive fractionation of cytosolic proteins to achieve purity, biochemical identification of specific proteins, and subsequent immunoclearing validation. The ammonium sulfate fractionation of mouse liver cytosol described in this study represents a potential first step in this process and demonstrates that fractionation and tracking Doxol-forming activity is a viable method for identifying unknown Dox reductases.
Acknowledgments
We thank Jeff Morre of the Oregon State University Mass Spectrometry Facility for assistance in developing the Doxol assay, Oregon State University undergraduates Brittany Marshal and Blake Migaki for assistance in constructing expression plasmids and fractionating liver cytosol, and University of Washington undergraduate Claire Chisholm for assistance in assaying protein levels. The primary laboratory was that of G.F.M.
Abbreviations
- 11β-HSD1
11β-hydroxysteroid dehydrogenase 1
- Akr
aldo-keto reductase
- Cbr
carbonyl reductase
- Dox
doxorubicin
- Doxol
doxorubicinol
- Gclm
γ-glutamyl-cysteine ligase modulatory subunit
- hydroxy-PP-Me
3-(7-isopropyl-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidin-5yl)phenol
- LC-MS/MS
liquid chromatography–mass spectrometry/collision-induced dissociation mass spectrometry
- NFDM
nonfat dried milk
- Tr1
thioredoxin reductase 1
- Trx1
thioredoxin1
Authorship Contributions
Participated in research design: Breysse, Schaupp, Kavanagh, Schmidt, G.F. Merrill.
Conducted experiments: Breysse, Boone, Long, M.E. Merrill, Schaupp, G.F. Merrill.
Contributed new reagents or analytic tools: White, Kavanagh, Schmidt, G.F. Merrill.
Performed data analysis: Breysse.
Wrote or contributed to the writing of the manuscript: Breysse, Boone, M.E. Merrill, G.F. Merrill.
Footnotes
G.F.M. was supported by an Oregon Health Sciences University Medical Research Foundation grant and an Oregon State University General Research Fund small grant. E.E.S. was supported by US National Institutes of Health National Cancer Institute [Grants CA152559-R21 and CA215784-RO1] and National Institute on Aging [Grants AG040020-RO1 and AG055022-R21], the Montana Agricultural Experiment Station [Grant MONB00443], and the Montana State University Department of Microbiology & Immunology.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Arnér ES, Nordberg J, Holmgren A. (1996) Efficient reduction of lipoamide and lipoic acid by mammalian thioredoxin reductase. Biochem Biophys Res Commun 225:268–274. [DOI] [PubMed] [Google Scholar]
- Bains OS, Karkling MJ, Lubieniecka JM, Grigliatti TA, Reid RE, Riggs KW. (2010) Naturally occurring variants of human CBR3 alter anthracycline in vitro metabolism. J Pharmacol Exp Ther 332:755–763. [DOI] [PubMed] [Google Scholar]
- Bains OS, Szeitz A, Lubieniecka JM, Cragg GE, Grigliatti TA, Riggs KW, Reid RE. (2013) A correlation between cytotoxicity and reductase-mediated metabolism in cell lines treated with doxorubicin and daunorubicin. J Pharmacol Exp Ther 347:375–387. [DOI] [PubMed] [Google Scholar]
- Bernardini N, Giannessi F, Bianchi F, Dolfi A, Lupetti M, Zaccaro L, Malvaldi G, Del Tacca M. (1991) Comparative activity of doxorubicin and its major metabolite, doxorubicinol, on V79/AP4 fibroblasts: a morphofunctional study. Exp Mol Pathol 55:238–250. [DOI] [PubMed] [Google Scholar]
- Björnstedt M, Hamberg M, Kumar S, Xue J, Holmgren A. (1995) Human thioredoxin reductase directly reduces lipid hydroperoxides by NADPH and selenocystine strongly stimulates the reaction via catalytically generated selenols. J Biol Chem 270:11761–11764. [DOI] [PubMed] [Google Scholar]
- Blanco JG, Leisenring WM, Gonzalez-Covarrubias VM, Kawashima TI, Davies SM, Relling MV, Robison LL, Sklar CA, Stovall M, Bhatia S. (2008) Genetic polymorphisms in the carbonyl reductase 3 gene CBR3 and the NAD(P)H:quinone oxidoreductase 1 gene NQO1 in patients who developed anthracycline-related congestive heart failure after childhood cancer. Cancer 112:2789–2795. [DOI] [PubMed] [Google Scholar]
- Bondareva AA, Capecchi MR, Iverson SV, Li Y, Lopez NI, Lucas O, Merrill GF, Prigge JR, Siders AM, Wakamiya M, et al. (2007) Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic Biol Med 43:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucek RJ, Jr, Olson RD, Brenner DE, Ogunbunmi EM, Inui M, Fleischer S. (1987) The major metabolite of doxorubicin is a potent inhibitor of membrane-associated ion pumps. A correlative study of cardiac muscle with isolated membrane fractions. J Biol Chem 262:15851–15856. [PubMed] [Google Scholar]
- Chang BK, Brenner DE, Gutman R. (1989) Cellular pharmacology of doxorubicinol alone and combined with verapamil in pancreatic cancer cell lines. Anticancer Res 9:341–345. [PubMed] [Google Scholar]
- Dodd DA, Atkinson JB, Olson RD, Buck S, Cusack BJ, Fleischer S, Boucek RJ., Jr (1993) Doxorubicin cardiomyopathy is associated with a decrease in calcium release channel of the sarcoplasmic reticulum in a chronic rabbit model. J Clin Invest 91:1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest GL, Gonzalez B, Tseng W, Li X, Mann J. (2000) Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res 60:5158–5164. [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2:287–295. [DOI] [PubMed] [Google Scholar]
- Hanna AD, Lam A, Tham S, Dulhunty AF, Beard NA. (2014) Adverse effects of doxorubicin and its metabolic product on cardiac RyR2 and SERCA2A. Mol Pharmacol 86:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque JA, McMahan RS, Campbell JS, Shimizu-Albergine M, Wilson AM, Botta D, Bammler TK, Beyer RP, Montine TJ, Yeh MM, et al. (2010) Attenuated progression of diet-induced steatohepatitis in glutathione-deficient mice. Lab Invest 90:1704–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibein AD, Guo B, Sprowl JA, Maclean DA, Parissenti AM. (2012) Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization. BMC Cancer 12:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo A, Choi TG, Jo YH, Jyothi KR, Nguyen MN, Kim JH, Lim S, Shahid M, Akter S, Lee S, et al. (2017) Inhibition of carbonyl reductase 1 safely improves the efficacy of doxorubicin in breast cancer treatment. Antioxid Redox Signal 26:70–83. [DOI] [PubMed] [Google Scholar]
- Joerger M, Huitema AD, Meenhorst PL, Schellens JH, Beijnen JH. (2005) Pharmacokinetics of low-dose doxorubicin and metabolites in patients with AIDS-related Kaposi sarcoma. Cancer Chemother Pharmacol 55:488–496. [DOI] [PubMed] [Google Scholar]
- Kassner N, Huse K, Martin HJ, Gödtel-Armbrust U, Metzger A, Meineke I, Brockmöller J, Klein K, Zanger UM, Maser E, et al. (2008) Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab Dispos 36:2113–2120. [DOI] [PubMed] [Google Scholar]
- Kremer LC, Caron HN. (2004) Anthracycline cardiotoxicity in children. N Engl J Med 351:120–121. [DOI] [PubMed] [Google Scholar]
- Luthman M, Holmgren A. (1982) Rat liver thioredoxin and thioredoxin reductase: purification and characterization. Biochemistry 21:6628–6633. [DOI] [PubMed] [Google Scholar]
- May JM, Mendiratta S, Hill KE, Burk RF. (1997) Reduction of dehydroascorbate to ascorbate by the selenoenzyme thioredoxin reductase. J Biol Chem 272:22607–22610. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Kezuka C, Endo S, Ikari A, Soda M, Yamamura K, Toyooka N, El-Kabbani O, Hara A, Matsunaga T. (2015) Acquisition of doxorubicin resistance facilitates migrating and invasive potentials of gastric cancer MKN45 cells through up-regulating aldo-keto reductase 1B10. Chem Biol Interact 230:30–39. [DOI] [PubMed] [Google Scholar]
- Olson LE, Bedja D, Alvey SJ, Cardounel AJ, Gabrielson KL, Reeves RH. (2003) Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res 63:6602–6606. [PubMed] [Google Scholar]
- Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, Boucek RJ., Jr (1988) Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci USA 85:3585–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp CM, White CC, Merrill GF, Kavanagh TJ. (2015) Metabolism of doxorubicin to the cardiotoxic metabolite doxorubicinol is increased in a mouse model of chronic glutathione deficiency: a potential role for carbonyl reductase 3. Chem Biol Interact 234:154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal PK, Iliskovic N. (1998) Doxorubicin-induced cardiomyopathy. N Engl J Med 339:900–905. [DOI] [PubMed] [Google Scholar]
- Sonowal H, Pal PB, Wen JJ, Awasthi S, Ramana KV, Srivastava SK. (2017) Aldose reductase inhibitor increases doxorubicin-sensitivity of colon cancer cells and decreases cardiotoxicity. Sci Rep 7:3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Lucas O, Weisend CM, Rollins MF, Merrill GF, Capecchi MR, Schmidt EE. (2009) Cytoprotective Nrf2 pathway is induced in chronically txnrd 1-deficient hepatocytes. PLoS One 4:e6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Whaley FS, Ewer MS. (2003) Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer 97:2869–2879. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, et al. (2005) An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol 3:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hua W, Ryu S, Yates P, Chang C, Zhang H, Di L. (2018) 11β-hydroxysteroid dehydrogenase 1 human tissue distribution, selective inhibitor, and role in doxorubicin metabolism. Drug Metab Dispos 46:1023–1029. [DOI] [PubMed] [Google Scholar]