Abstract
Cilia and flagella are evolutionarily conserved organelles whose motility relies on the outer and inner dynein arm complexes (ODAs and IDAs). Defects in ODAs and IDAs result in primary ciliary dyskinesia (PCD), a disease characterized by recurrent airway infections and male infertility. PCD mutations in assembly factors have been shown to cause a combined ODA-IDA defect, affecting both cilia and flagella. We identified four loss-of-function mutations in TTC12, which encodes a cytoplasmic protein, in four independent families in which affected individuals displayed a peculiar PCD phenotype characterized by the absence of ODAs and IDAs in sperm flagella, contrasting with the absence of only IDAs in respiratory cilia. Analyses of both primary cells from individuals carrying TTC12 mutations and human differentiated airway cells invalidated for TTC12 by a CRISPR-Cas9 approach revealed an IDA defect restricted to a subset of single-headed IDAs that are different in flagella and cilia, whereas TTC12 depletion in the ciliate Paramecium tetraurelia recapitulated the sperm phenotype. Overall, our study, which identifies TTC12 as a gene involved in PCD, unveils distinct dynein assembly mechanisms in human motile cilia versus flagella.
Keywords: TTC12, cilia, dynein arm assembly, primary ciliary dyskinesia, sperm flagella, CRISPR-Cas9
Introduction
Primary ciliary dyskinesia (PCD [MIM: 244400]) is a group of clinically and genetically heterogeneous disorders wherein dysfunction of motile cilia leads to impaired mucociliary clearance and early recurrent airway infections. About one person in 10,000 individuals is affected,1 and given the key role of motile cilia in the establishment of left-right asymmetry during embryogenesis,2 nearly 50% of those individuals display situs inversus and have Kartagener syndrome. In addition, because the microtubule-based structure of motile cilia, the axoneme, is close to that of sperm flagella, most affected male individuals are also infertile. The axoneme consists of nine peripheral outer microtubule doublets circularly arranged around two central microtubules surrounded by a central sheath (“9+2” pattern). Attached all along the microtubule length, the outer dynein arms (ODAs), and the inner dynein arms (IDAs) are multiprotein complexes that carry an ATPase activity and provide the sliding force for motility.
In humans, ODAs are composed of two axonemal dynein heavy chains (HCs), namely the β and γ chains, which are attached to a large intermediate chain/light chain complex (IC/LC). Two types of ODAs have been described in cilia: the type 1 ODAs are located at the proximal part of the cilium and contain DNAH5 (γ chain) associated with DNAH11 (β chain), and the type 2 ODAs are located at the distal part of the cilium and contain DNAH5 (γ chain) associated with DNAH9 (β-chain). It is worth noting that recent studies performed in humans revealed that the ODA composition of spermatozoa differs from that found in cilia:3 in spermatozoa, the γ and β chains consist of DNAH8 and DNAH17, respectively, which are both specifically expressed in sperm cells. As for IDAs, their exact structure and composition is virtually unknown in humans. Most of the available knowledge was provided by studies in the flagellated alga Chlamydomonas reinhardtii, where seven IDA subspecies have been identified. Six distinct single-headed IDA subspecies (called IDA a, b, c, d, e, and g), each including only one HC, have been characterized, and one dimeric IDA complex (called IDA f or IDA-I1), which contains two HCs, has been identified.4, 5, 6, 7 There is growing interest in identifying the key players involved in the process of ODA and IDA assembly and transport. In this regard, one of the most frequent ciliary ultrastructural defects found in PCD is the absence of both dynein arms (DAs)8 as a result of mutations in genes encoding proteins involved in ODA and IDA assembly, docking, and transport (i.e., such genes include DNAAF2/KTU9 [MIM: 612517], DNAAF1/LRRC5010 [MIM: 613190], DNAAF311 [MIM: 614566], CCDC10312 [MIM: 614677], DNAAF5/HEATR213 [MIM: 614864], LRRC614 [MIM: 614930], DNAAF4/DYX1C115 [MIM: 608709], ZMYND1016 [MIM: 607070], SPAG117 [MIM: 603395], CFAP298/C21orf5918 [MIM: 615494], PIH1D319 [MIM: 300933], and CFAP300/C11orf7020 [MIM: 618058]).
Although several of these genes have been renamed with the acronym DNAAF, which stands for dynein axonemal assembly factor, their precise role in the assembly and/or transport machinery is not known. As shown recently, multiciliated cells contain cytoplasmic liquid-like organelles that concentrate DNAAFs with dynein chains and chaperones.21 To date, very few investigations have been performed on spermatozoa from PCD-affected individuals; in all cases, the sperm ultrastructure defects revealed by transmission electron microscopy (TEM) were always found to be identical to those reported in the cilia. In this regard, the recent observation of differences in the protein composition of ODAs between human cilia and sperm flagella3 raises the question as to whether the assembly and transport mechanisms of ODA and IDA components are conserved between human cilia and flagella.
In the present study, we identified independent families in which affected individuals displayed a peculiar PCD phenotype characterized by the absence of IDAs in respiratory cilia; this phenotype was in contrast with the absence of both ODAs and IDAs in sperm. Such an unusual observation prompted us to search for the molecular basis of the PCD phenotype in those individuals and to decipher the consequences of the identified molecular defects on cilia and flagella assembly by developing an in vivo model in the ciliate Paramecium tetraurelia and a CRISPR-Cas9-mediated genome-editing approach in human primary airway epithelial cells (AECs).
Material and Methods
Affected Individuals
We obtained written informed consent from all affected individuals and/or their parents according to protocols approved by the Comité de Protection des Personnes (CPP) Ile de France III (CPP02748) and the institutional review board of the French Institute of Health and Medical Research (CEEI-IRB: no. 15-259).
Genetic Analyses
Identification of TTC12 (GenBank: NM_017868.4) sequence variations was performed from genomic blood DNA, either by whole-exome sequencing (WES) or by parallel sequencing with a custom targeted-capture panel. More precisely, in individual DCP791, WES was performed with the Agilent SureSelect V5 target enrichment system on a HiSeq sequencing machine (Illumina). In individuals DCP153 and DCP1606, WES was performed with the SeqCap EZ MedExome target-enrichment kit on a NextSeq sequencing machine (Illumina). The DNA of individual 18GM00157 was analyzed on a MiSeq sequencer (Illumina) with a custom targeted-capture panel (SeqCap EZ Choice, Roche Diagnostics) that encompasses the 45 genes involved in PCD and 250 candidate genes for PCD. The libraries were prepared according to the manufacturer’s instructions. Data were analyzed through an in-house double pipeline based on Bowtie2 and BWA tools. Reads were visualized with the Integrative Genomics Viewer (IGV, Broad Institute). Copy-number variation was analyzed via a depth-ratio comparison between subjects sequenced in the same run, and depth ratios obtained for each of the four individuals were represented on a graph built with GraphPad Prism 5 software.
Sanger sequencing was performed on genomic blood DNA with the BigDye Terminator v3.1 system (Thermo Fisher) after PCR amplification with the Go-Taq Green Master Mix (Promega) at an annealing temperature of 60°C and purification with ExoSAP-IT(USB), according to the manufacturer’s instructions. Fragments were analyzed on a 3730XL device (Thermo Fisher) after Sephadex G-50 Superfine purification (GE Healthcare). Amplification and sequencing primers are listed in Table S1. Sequences were analyzed with SeqScape software (Thermo Fisher).
TEM Analysis of Ciliary and Flagella Ultrastructure
AECs obtained from nasal or bronchial biopsies were fixed in 2.5% glutaraldehyde to be processed for TEM as previously described.22 Ciliary ultrastructural results were quantified as a percentage of abnormal cilia among the total number of analyzed cilia. Semen was fixed in 2.0% v/v glutaraldehyde in phosphate buffer, washed for 15 min in fresh buffer containing 4% w/v sucrose, and embedded in 2% agar. Post-fixation lasted 1 h in 1% w/v osmic acid in phosphate buffer. After dehydration in a graded series of ethanol, small pieces of agar containing sperm cells were embedded in Epon resin (Polysciences). Sections were cut on a Reichert OmU2 ultramicrotome (Reichert-Jung AG) with a diamond knife. 70 nm sections were collected on nickel grids and stained with uranyl acetate (2% in 70% ethanol, 20 min) and Reynolds lead citrate (10 min) and examined with a Zeiss transmission electron microscope 902 (Leo). Images were acquired with a Gatan Orius SC1000 CCD camera (Gatan France).
Immunoblotting
AECs obtained by airway brushing from individuals 18GM00157 and DCP1606 and control individuals, as well as spermatozoa from a control donor, were suspended in ice‐cold lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X‐100, and Complete Protease Inhibitor Cocktail [Roche Diagnostics]). Lysates were sonicated and cleared by 10 min of centrifugation at 1,000 × g at 4°C.
Cell lysates were loaded on 10% SDS-PAGE; proteins were then transferred for 2 h to a PVDF membrane, and analyzed via immunoblotting with TTC12 antibody (Santa Cruz sc-390229, 1/200), DNAJB13 antibody (HPA052465, 1/300), or DNAI1 antibody (HPA021649, 1/400), all incubated overnight at 4°C in PBS containing 0.1% Tween 20 and 5% milk. The secondary antibody (rabbit-HRP, SIGMA, A0545) was incubated for 1 h at room temperature at a final dilution of 1:5000 in PBS containig 0.1% Tween 20 and 5% milk. Proteins were detected with Amersham ECL Select Western Blotting Detection Reagent (GE healthcare) according to the manufacturer’s recommendations.
Immunofluorescence Analyses on AECs
All antibodies are listed in Table S2. AECs obtained via nasal brushing from individuals 18GM00157, DCP1606, and control individuals (i.e., individuals in whom the diagnosis of PCD was excluded based on the absence of sinopulmonary symptoms and of normal values of nasal nitric oxide) were suspended in FertiCult IVF medium. Samples were cytocentrifugated onto glass slides, air dried, and directly used for immunostaining. Cells were first fixed with 4% paraformaldehyde for 15 min at room temperature, washed three times with PBS++ (i.e., PBS supplemented with 0.49 mM MgCl2 and 0.9 mM CaCl2), incubated for 10 min with PBS++ 50 mM NH4Cl, and permeabilized with 0.1% Triton X-100 for 10 min. Then, cells were incubated for 2 h at room temperature with primary antibodies before being revealed by secondary antibodies at room temperature for 1 h. All antibodies were incubated in PBS++ 1% BSA. Finally, cells were washed three times with PBS++ and mounted in ProLong Gold Antifade Reagent with DAPI (Cell Signaling #8961) so that nuclei would be stained, and images were taken with an ECLIPSE 80i microscope (Nikon) with a Retiga-2000R camera (QImaging).
Immunofluorescence Analyses of Human Spermatozoa
Study participants provided semen samples via masturbation after a period of 2–7 days of sexual abstinence, evaluated according to World Health Organization (WHO) guidelines,23 and then frozen until further analysis. 10 μL of frozen semen samples were spread onto a Superfrost Plus slide (Menzel Glasbearbeitungswerk) and fixed by incubation with 4% paraformaldehyde for 10 min. The slides were incubated 20 min at 95°C in citrate buffer (H-3300, VectorLabs), treated with 0.2% Triton in PBS for permeabilization, and then blocked by incubation in 1% BSA for 1 h. The slides were then incubated with primary antibodies overnight at 4°C and then with secondary antibodies for 1 h at room temperature. The slides were mounted in Vectashield medium (Vector Laboratories) supplemented with 0.5 mg/mL DAPI and were subsequently analyzed with a Zeiss Axiophot epifluorescence microscope. Digital images were acquired with a cooled charge-coupled device (CCD) camera (Hamamatsu) under identical instrument settings with MetaMorph software (Molecular Devices).
Paramecium Experiments
All experiments were carried out with entirely homozygous mutant strains: 7S strain for feeding experiments and nd7, which carries a recessive monogenic mutation blocking trichocyst discharge,24 for Paramecium transformation.
Paramecium Strain and Cultivation
Cells were grown at 27°C in a wheat grass powder (BHB from GSE Vertrieb GmbH) infusion medium bacterized with Klebsiella pneumoniae and supplemented with 0.8 μg/mL β-sitosterol according to standard procedures.25
Paramecium Gene-Knockdown Experiments
We used the feeding procedure previously described to achieve RNAi gene silencing. 26 We obtained the plasmids used in RNAi silencing experiments by cloning PCR products, which were amplified from the coding region of each gene (Table S3), in L4440 vector27 via Gibson technology (Gibson DJ 2009). These plasmids were then transformed in an HT115 DE3 Escherichia coli strain to produce T7Pol-driven dsRNA as previously described.26
Microinjection of a Transgene Expressing a GFP-Tagged TTC12a Protein
To create a version of TTC12a that would be resistant to RNAi silencing, we replaced the sequence from nucleotide 471 to 745 (corresponding to dsRNA sequence) of the TTC12a coding region in vitro by a modified synthetic DNA sequence coding for the same amino acid sequence (Figure S1). The GFP coding sequence was fused at the 3′ end of WT or TTC12a RNAi-resistant versions and cloned (via Gibson technology [Gibson DJ 2009)) in the XhoI site of the pPXV-GFP vector under the control of the constitutive regulators of the Paramecium calmodulin gene.28 Twelve nucleotides coding for four glycine amino acids were introduced between the TTC12a-coding region and the GFP-coding sequence. The plasmid was linearized by Sfi1 digestion and microinjected along with ND7-complementing plasmid into the macronucleus of nd7-1 mutant cells unable to discharge their trichocysts.24 Transformants were first screened for their ability to discharge trichocysts, and if they had this ability, they were further analyzed. Microinjection was performed under an inverted Nikon phase-contrast microscope via a Narishige micromanipulation device and an Eppendorf air pressure microinjector.
Paramecium TEM Analysis
For ultrastructural observations, paramecia were permeabilized in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 2 mM MgCL2, adjusted to pH 6.9 with NaOH) with 1% Triton X-100 for 1 min, rinsed in PHEM buffer, and fixed in 1% (v/v) tannic acid and 1% (v/v) glutaraldehyde in PHEM buffer for 30 min. Cells were then rinsed in PHEM and 0.05 M cacodylate buffers before being postfixed in 1% OsO4 (v/v) 0.05 M cacodylate buffer at pH 7.4 for 30 min. After being rinsed, cells were embedded into 2% agarose. Agar blocks were then dehydrated in graded series of ethanol and propylene oxide and embedded in Epon. Ultrathin sections were contrasted with uranyl acetate and lead citrate. The sections were examined with a Jeol transmission electron microscope 1400 (at 120 kV). The number of ODAs and IDAs were counted in about 140 cilia cross-sections obtained from TTC12-knocked-down or control cells.
Paramecium Swimming Speed and Cell Trajectory
We analyzed paramecia swimming behavior of TTC12-depleted and control cells by transferring 8–10 paramecia into a drop of conditioned BHB medium and imaging them for 10 s every 0.3 s to assess the swimming pattern under dark-field microscopy by using MetaVue software. Image analysis and the measurement of swimming velocity were then performed with the ImageJ software.
Paramecium Fluorescence-Microscopy Analysis
Immunofluorescence (IF) was performed as follows: paramecia were fixed in 2% paraformaldehyde in PHEM buffer for 15 min. Cells were then permeabilized for 15 min in 1% Triton X-100 in PHEM and washed three times in PBS supplemented with 3% BSA. Immunostaining was performed with a polyglutamylated tubulin antibody (kindly provided by Carsten Janke, 1/500) as previously described.29 Cells were observed with a confocal Leica SP8 microscope, and image stacks were processed with the ImageJ and Photoshop softwares.
Paramecium Deciliation, Protein Extraction, and Immunoblot
Cilia were purified30 and recovered by centrifugation at 28,000 × g for 30 min, and protein was directly extracted in SDS-Laemmli buffer. Total protein content from paramecia or paramecia cell bodies (deciliated paramecia) was directly boiled in 10% SDS, followed by dilution in 2XSDS Laemmli buffer. Proteins were then separated on SDS-PAGE, blotted on a nylon membrane, and probed with an anti-GFP antibody (1:1,000, Interchim).
Air-Liquid Interface Culture and Infection
AECs obtained either from surgical material from non PCD-affected individuals suffering from nasal polyps requiring surgery or from nasal brushing of PCD-affected individuals were cultured at the air-liquid interfacw (ALI) as previously described.31 In brief, epithelial cells were seeded in 12-well collagen IV-coated transwell inserts (Costar, corning) and cultured in PneumaCult-Exp Plus medium (#05040, Stem Cell) until confluence was reached. The medium was then removed from the apical surface and replaced in the basal chamber by the PneumaCult-ALI medium (#5001, Stem Cell) so that cells would be exposed to an air-liquid interface that promoted ciliogenesis during the following 21 days. Lentiviral transduction of AECs was performed as previously described.32 In brief, viral particles together with a Rho-associated protein kinase inhibitor were added to the medium during the expansion phase before a cell-selection step with the use of puromycine (2 μg/mL). Resistant cells were then cultured at the ALI as described above. Cells infected with an empty vector were used as a control.
Results
Next-Generation Sequencing-Based Approaches Identify Homozygous Loss-of-Function TTC12 Mutations in Four Independent PCD-Affected Individuals with Distinct Ciliary and Flagellar DA Defects
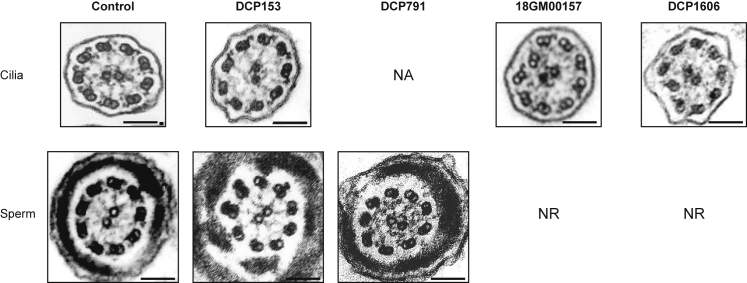
To uncover novel genetic causes responsible for PCD, we used a next-generation sequencing (NGS)-based approach in four independent probands with a PCD phenotype that was not explained by mutations in genes known to be involved in PCD. As summarized in Table 1, individuals DCP791, 18GM00157, and DCP1606 originate from North Africa and were born from a consanguineous union, whereas the fourth individual, DCP153, originates from Europe and has parents with no known consanguinity. All four individuals displayed sinopulmonary symptoms with rhino-sinusitis and had normal situs position. Individuals 18GM00157 and DCP1606 also displayed bronchiectasis, and individual DCP1606 suffered from neonatal respiratory distress. Nasal nitric oxide (NO) concentrations were recorded for those four individuals and showed low levels for two of them. High-speed videomicroscopy (HSVM) performed on nasal brushings of three individuals showed variable proportions of immotile cilia (70%, 50%, and 10% in 18GM00157, DCP1606, and DCP153, respectively); a normal beat frequency and moderate beating defects (for the portion of motile cilia) consisting of a reduced beating angle; and a lower distance swept by the cilium tip (Table S1). Representative videos showing immotile and beating cilia are shown in the supplemental data (Videos S1 and S2; S3 and S4; and S5 and S6 for individuals 18GM00157, DCP1606, and DCP153, respectively, and Video S7 for a control individual). TEM analysis of respiratory cilia was performed for individuals DCP153, 18GM00157, and DCP1606 and showed the absence of IDAs associated with some axonemal disorganization (2%, 22%, and 24%, respectively) but also, importantly, the presence of ODAs in all examined cilia (Table 1) (Figure 1).
Table 1.
Phenotype of PCD-Affected Individuals with Identified TTC12 Mutations
| Individuals (Origin) | Known Consanguinity | Gender (Age at Diagnosis) | SI |
Airway Disease |
Fertility | Nasal NO (nL/min) |
TEM Defect (%) |
Allele 1 | Allele 2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Cilia | Sperm | ||||||||
| DCP791 (Turkey) | yes | male (24 years) | no | minima | sinusitis | infertility | lowa | NA | IDAs+ ODAs (100%) | c.1614+3A>T (p.?) | c.1614+3A > T (p.?) |
| 18GM00157 (Morocco) | yes | female (50 years) | no | asthma bronchiectasis | rhino-sinusitis | not tested | 37.6 | IDAs (100%) disorganization (22%) | NR | c.1678C>T (p.Arg560∗) | c.1678C>T (p.Arg560∗) |
| DCP1606 (Tunisia) | yes | male (11 years) | no | NNRD, bronchiectasis, pneumopathies | rhino-sinusitis otitis | NR | 146 | IDAs (100%) disorganization (24%) | NR | c.607del (p.Ile203∗) | c.607del (p.Ile203∗) |
| DCP153 (Europe) | no | male (33 years) | no | bronchitis | rhino-sinusitis | Infertility | 186 | IDAs (100%) disorganization (2%) | IDAs+ODAs (100%) | c.1700T>G (p.Met567Arg) | c.1700T>G (p.Met567Arg) |
Abbreviations are as follows: NNRD, neonatal respiratory distress; NR, not relevant; NO, nitric oxide; NA, not available; TEM, transmission electron microscopy; IDAs, inner dynein arms, and ODAs, outer dynein arms. PCD nasal NO < 77 nL/min.8
Exact value not available.
Figure 1.
Ciliary and Flagellar Ultrastructural Defects in PCD-Affected Individuals with TTC12 Mutations
Electromicrographs of cross-sections of ciliary axonemes (upper panel) from a control individual and individuals DCP153, 18GM00157, and DCP1606 are compared to those of cross-sections of sperm flagellar axonemes (lower panel) from a control individual and from the infertile PCD-affected individuals DCP153 and DCP791. In each individual, the ciliary section shows an abnormal configuration with missing IDAs and an associated mild axonemal disorganization in individuals 18GM00157 and DCP1606, but ODAs are clearly present. The flagellar sections show a lack of both IDAs and ODAs. Black scale bars represent 0.1 μm. Abbreviations are as follows: NR, not relevant; NA, not available.
Representative Video Showing Immotile Cilia of Individual 18GM00157
Representative Video Showing Beating Cilia of Individual 18GM00157
Representative Video Showing Immotile Cilia of Individual DCP1606
Representative Video Showing Beating Cilia of Individual DCP1606
Representative Video Showing Immotile Cilia of Individual DCP153
Representative Video Showing Beating Cilia of Individual DCP153
Representative Video Showing Beating Cilia of Control Individual
Among the three male individuals, individual DCP1606 was a child (11 years old), and individuals DCP791 and DCP153 were adults known to be infertile as a result of total asthenozoospermia (Table 2). Surprisingly, TEM analysis of sperm cells from the two infertile individuals revealed the total lack of both IDAs and ODAs, an ultrastructural phenotype clearly different from that observed in cilia from respiratory cells where only IDAs were missing.
Table 2.
Semen Characteristics of Infertile Individuals with Identified TTC12 Mutations
| Flagellum Defects | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individuals | pH | Volume (ml) | Sperm Count (106/mL) | Total Motility (%) | Progressive Motility (%) | Vitality (%) | Typical Forms (%) | Absent | Short | Irregular Caliber | Coiled | Multiple |
| DCP791 | ND | ND | 21 | 0b | 0b | 75 | 36 | 7 | 0 | 0 | 10 | 0 |
| DCP153 | 7.9 | 4.5 | 81 | 0b | 0b | 68 | 33 | 2 | 1 | 1 | 6 | 1 |
| Reference valuesa | 7.2 | 1.5 | 15 | 40 | 32 | 58 | 23 | 5 | 1 | 2 | 17 | 1 |
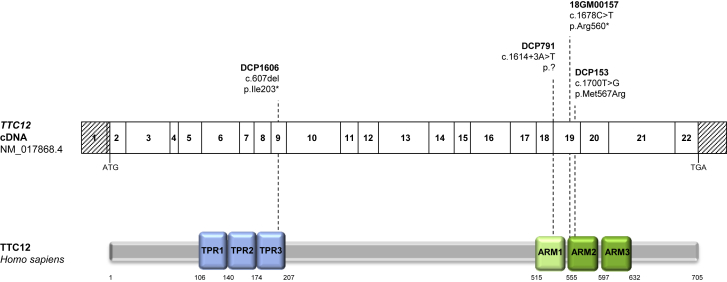
NGS analyses led to the identification of homozygous mutations in TTC12 [MIM: 610732], a gene that has so far not been associated with PCD. TTC12 is located on chromosome 11 and consists of 22 coding exons (GenBank: NM_017868.4), suggesting a protein of 705 residues (UniProt: Q9H892). Individuals DCP1606 and 18GM00157 were shown to carry the nonsense mutations c.607del (p.Ile203∗) and c.1678C>T (p.Arg560∗), respectively. Individual DCP791 carried a splice mutation (c.1614+3A>T, p.?) and individual DCP153 was homozygous for the missense mutation c.1700T>G (p.Met567Arg). Two of these mutations, c.1614+3A>T and c.607del, are not reported in gnomAD, and the c.1678C>T and c.1700T>G mutations are found at very low frequencies: 4 × 10−5 and 6 × 10−5, respectively (Table S4). The presence of all these TTC12 mutations was confirmed by Sanger sequencing, as illustrated in Figures S2 and S3. The expected consequences of the identified variants at the protein level are shown in Figure 2.
Figure 2.
TTC12 Mutations and Their Putative Impact at the Protein Level in Individuals with PCD
The mutations are shown within the exonic organization of the human TTC12 cDNA (top) and a domain-organization model of the corresponding protein (bottom) according to the SMART webtool and CDD NCBI database. Abbreviations are as follows: TPR, tetratricopeptide repeat; and ARM, armadillo repeat. The light green box represents a degenerated ARM domain, and the two dark green boxes correspond to typical ARM domains.
TTC12 Is a Cytoplasmic Protein of Ciliated Cells
TTC12 contains three tetratricopeptide repeat (TPR) domains in its N-terminal part and three armadillo repeat domains (ARM1, ARM2, and ARM3) in its C-terminal part. A TPR domain consists of a motif of ~34 amino acids organized in two antiparallel alpha helices known to mediate protein-protein interactions and the assembly of multiprotein complexes.34 ARM domains, which are approximately 40 amino acids long, have also been implicated in protein-protein interactions through their superhelix structure.35 To date, the biological functions of TTC12 are unknown. To gain insight into its role, we first checked TTC12 tissue expression by RT-PCR in AECs and in testis tissue (Figure S4A) with a primer set (located in exons 2 and 22) bracketing the entire coding region (Table S1). Sequencing of the resulting RT-PCR products allowed the identification of the same TTC12 isoform (GenBank: NM_017868.4) in both samples. As shown by real-time PCR performed with primers located in exons 11 and 12 of TTC12 (Table S1), a similar expression level of this transcript was found in AEC and testis samples (Figure S4B). The corresponding TTC12 75 KDa protein was also easily detectable by immunoblot in both cell types (Figure S4C). We next investigated the subcellular localization of TTC12 in ciliated cells. Given the absence of available anti-TTC12 antibodies suitable for IF studies, we performed immunoblot experiments on protein extracts from ciliary or cytoplasmic fractions of cultured AECs. This analysis showed that TTC12 is virtually exclusively detected in the cytosolic fraction; in contrast, DNAJB13, a component of radial spokes, was mainly detected in the ciliary fraction, as expected (Figure S4D). These results are in line with data from quantitative proteomic analyses, which did not identify TTC12 in purified preparations of axonemes from human airway cells.36 Overall, the cytoplasmic localization of TTC12 indicates that this protein is not a structural component of the axoneme but might play a role in the assembly and/or the transport of axonemal components.
Functional Consequences of the Identified TTC12 Mutations
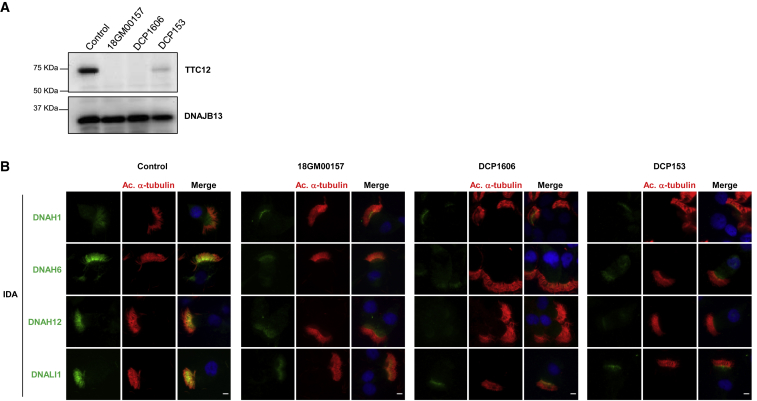
The nonsense mutations carried by individuals 18GM00157 and DCP1606 might generate truncated proteins lacking the ARM superhelix structure of TTC12 (Figure 2). Such premature stop codons might also trigger the nonsense-mediated mRNA decay pathway and therefore might lead to the absence of protein. To test these hypotheses, we cultured AECs from individuals 18GM00157 and DCP1606 and from a control individual at the ALI as previously described.31 Analysis of total protein extracts by immunoblotting with an antibody raised against the N-terminal part of TTC12 led to the detection of TTC12 in the control but not in samples from individuals 18GM00157 and DCP1606 (Figure 3A). These results clearly indicate that the two nonsense mutations identified in individuals 18GM00157 and DCP1606 are associated with TTC12 loss of function.
Figure 3.
TTC12 Loss-of-Function Mutations Cause Defective Single-Headed IDA Assembly in Respiratory Cilia
(A) Protein lysates extracted from AECs of a control individual and individuals 18GM00157, DCP1606, and DCP153 were resolved by SDS-PAGE and analyzed by immunoblotting with an anti-TTC12 antibody. The expression of TTC12 is virtually absent or dramatically reduced in PCD-affected individuals. DNAJB13, a component of radial spokes, was used as a positive control.
(B) IF microscopy analysis of IDA markers in respiratory epithelial cells from a control and from individuals 18GM00157, DCP1606, and DCP153 homozygous for TTC12 mutations. Cells were double-labeled with an anti-acetylated α-tubulin antibody (red), a marker of ciliary axonemes, and anti-DNAH6, anti-DNAH12, or anti-DNALI1 antibodies as markers of IDAs (green). Nuclear staining was performed with DAPI (blue). In affected individuals, DNAH12 is completely absent in the cilia but seems to accumulate at the subapical part of the cytoplasm. By contrast, DNALI1 and DNAH6 appear to be strongly reduced but are partially present at the proximal part of cilia. (DNAH2 and DNAH10, two markers of the double-headed IDA-I1, were shown to be normally expressed in the individuals; see Figure S7). Scale bars represent 5 μm. IDA: inner dynein arm.
As for individual DCP153, he carries a missense variation replacing methionine 567, located in the second ARM domain of TTC12, by an arginine residue. Several lines of evidence support the involvement of the p.Met567Arg variation in the PCD phenotype of this individual. First, as mentioned above, this variation is reported in gnomAD at a very low frequency (6 × 10−5) and only in the heterozygous state (Table S5). Second, it predicts changes in both the polarity and the charge of this amino acid that is highly conserved in mammals. In zebrafish and Xenopus, this methionine residue is replaced by a leucine, an amino acid that is also apolar (Figure S5A). In addition, alignments of the well-conserved ARM2 and ARM3 domains of TTC12 with the ten ARM domains found in ARMC4—also involved in PCD37—unveil (1) that the methionine found in ARM2 at position 567 also corresponds to a methionine in the ARM3 domain (Met610) (Figure S5B) and (2) the high conservation among all those ARM domains of a hydrophobic residue at a position corresponding to Met567 in TTC12 (Figure S5C). Third and most importantly, in immunoblot analyses on protein extracts from AECs and from spermatozoa of individual DCP153, who carries the missense mutation in the homozygous state, the amount of TTC12 was found to be dramatically reduced (Figures 3A and 4B), thereby attesting to the pathogenicity of the p.Met567Arg mutation.
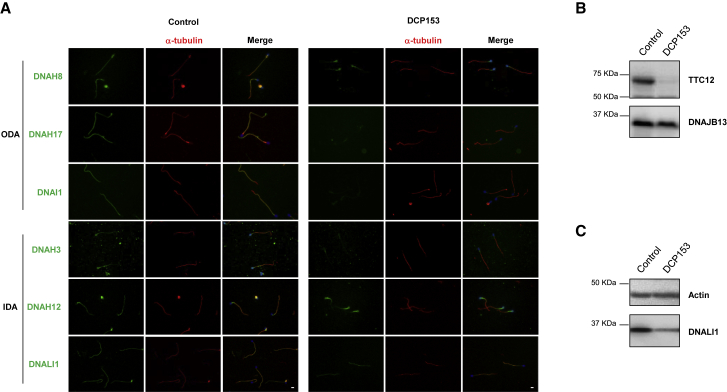
Figure 4.
TTC12 Mutations Impact the Assembly of Both ODAs and IDAs in Spermatozoa
(A) IF microscopy analysis of ODA and IDA markers in spermatozoa from a control and the infertile individual DCP153 carrying bi-allelic TTC12 mutations. Cells were double-labeled with a marker of ciliary axonemes and anti-α-tubulin antibody (red), together with anti-DNAH8, anti-DNAH17, or anti-DNAI1 antibodies as markers of ODAs (green) and anti-DNAH3, anti-DNAH12, or anti-DNALI1 antibodies as markers of IDAs (green). Nuclear staining was performed with DAPI (blue). In individual DCP153, DNAH8, DNAH17, DNAI1, and DNAH3 are absent along the flagellar axoneme. By contrast, DNAH12 remains exclusively present in the proximal part of flagella, and DNALI1 appears to be present but reduced along the axoneme. Scale bars represent 5 μm.
(B) Spermatozoa protein lysates of a control individual and individual DCP153 were resolved by SDS-PAGE and analyzed by immunoblotting with an anti-TTC12 antibody, showing the absence of TTC12 expression in the individual. Very low amounts of TTC12 were detected in spermatozoa from the infertile individual DCP153. DNAJB13, a component of radial spokes, was used as a positive control.
(C) Spermatozoa protein lysates of a control individual and individual DCP153 were resolved by SDS-PAGE and analyzed by immunoblotting with an anti-DNALI1 antibody, showing the reduced amounts of DNALI1 in flagella from the affected individual. Actin was used as a positive loading control. Abbreviations are as follows: ODA, outer dynein arm; and IDA, inner dynein arm.
The last individual, DCP791, carries the c.1614+3A>T transversion involving the splice-donor site of TTC12 intron 18. As predicted by the MaxEntScan webtool, this nucleotide substitution is expected to have a strong impact on the splicing of TTC12 transcripts (score decreasing from 9.49 to 2.03). In addition, a recent study described a deep neural network that precisely models mRNA splicing from a genomic sequence and accurately predicts the effects of all possible genomic SNVs on splicing.38 On the basis of this dataset, the probability that the TTC12 c.1614+3A>T mutation alters the intron 17 canonical splice site is 88%. To further confirm the deleterious effect of this mutation on splicing, we generated minigene constructs containing a genomic DNA region spanning exons 17 to 19 of TTC12, with or without the A>T transversion. Analysis of the corresponding transcripts in HEK293 cells transfected with those plasmid constructs showed that the mutation induces a complete skipping of exon 18 (Figures S6A and S6B). This in-frame deletion of exon 18 spares the exons encoding the ARM2 and ARM3 domains but, as shown with the NCBI CDD webtool, the prediction of those domains is lost (Figure S6C).
TTC12 Mutations Affect a Subset of Single-Headed IDAs in Respiratory Cilia
To further investigate the function of TTC12 in respiratory cilia and in sperm flagella, we first used high-resolution IF to compare the localization of different axonemal components in AECs from three individuals (18GM00157, DCP1606, and DCP153) to their localization in controls. We used DNAH5 and DNAI1 as markers of the ODAs; DNAH2 and DNAH10 for the double-headed IDA-I1 complex; RSPH3 and RSPH4a for radial spokes (RSs); GAS8 for the nexin-dynein regulatory complex (N-DRC); and CCDC40 for the ciliary ruler.39 As proposed by Kollmar,40 the single-headed IDA subspecies (a, b, c, d, e, and g) would result from gene duplications followed by the generation of highly divergent N termini and the formation, in Chlamydomonas but not in mammals, of chimeric single-headed heavy dynein chains. This model, which relies on thorough phylogenetic analyses, readily explains the fact that the single-headed IDAs a, c, and d bind the light-chain DNALI1 (both in Chlamydomonas and in humans), whereas the heavy chains belonging to another subgroup (b, e, and g) bind centrin in Chlamydomonas, and in humans, the b and e heavy chains, which are not chimeric, would bind DNALI1. We therefore performed immunostaining with antibodies directed against DNALI1 (corresponding to IDA subspecies a, c, and d and potentially to IDA subspecies b and e), DNAH1 (corresponding to IDA subspecies d), DNAH6 (corresponding to IDA subspecies g), and DNAH12 and DNAH3, both corresponding to other IDA subspecies (a, b, c, or e). We found that, in the respiratory cilia of the three individuals with TTC12 mutations, the subcellular localization patterns of the markers for ODAs, radial spokes (RSs), the nexin-dynein regulatory complex (N-DRC), and the ciliary ruler were similar to those found in controls (Figure S7). Interestingly with regard to IDAs, although DNAH2 and DNAH10, corresponding to the double-headed IDA-I1, were found to be normal in those same three individuals (Figure S7), all the markers of the single-headed IDA subspecies (i.e., DNALI1, DNAH1, DNAH6, and DNAH12) displayed an abnormal pattern (Figure 3B). DNALI1, DNAH1, and DNAH6 were present at low levels in the very proximal part of the cilia from the three individuals and absent from the rest of the axoneme, whereas DNAH12 was not detectable all along the cilia and was found to accumulate at the subapical part of the cytoplasm of ciliated cells. Overall, in cilia, TTC12 mutations do not affect ODAs or IDA-I1 but compromise a single-headed-IDA subset, which comprises DNALI1, DNAH1, DNAH6, and DNAH12. Of note, DNAH3 was not detected in cilia from control individuals (data not shown).
TTC12 Mutations Affect ODAs and a Distinct Subset of Single-Headed IDAs in Sperm Flagella
In line with the TEM data, IF analyses in spermatozoa from the infertile PCD-affected individual DCP153 showed that DNAI1 as well as DNAH8 and DNAH17, the two dynein HCs specific to ODAs in sperm flagella,3 were undetectable (Figure 4A), thus clearly confirming that TTC12 mutations induce a loss of ODAs in sperm cells but not in airway cilia. Because very little information on the IDA subtypes and their HC composition is available in human sperm cells, we analyzed the pattern of all described IDA HCs in humans (except DNAH7 and DNAH14, for which no antibodies are so far suitable for IF). Interestingly, as observed in cilia, markers of the double-headed IDA-I1 complex (i.e., DNAH2 and DNAH10) were found unchanged in individual DCP153. Regarding markers of single-headed IDAs, although the dynein HC DNAH3 (a, b, c, or e) was not detected in control respiratory cilia, it was found along the axoneme of control spermatozoa. Remarkably, this dynein HC was totally absent in sperm from individual DCP153. Additionally, DNAH12 (also corresponding to the single-headed IDA subspecies a, b, c, or e) was detected along the full length of the flagellar axoneme in a control but displayed an expression pattern restricted to the proximal part of the flagella in the affected individual DCP153 (Figure 4A). By contrast, the dynein HCs of the single-headed IDA subspecies d and g (i.e., DNAH1 and DNAH6) displayed a normal expression pattern (Figure S8). As for DNALI1, the light chain associated with the IDA subspecies a, c, and d, it was found to be normally localized along the flagellar axoneme of spermatozoa from the affected individual DCP153; however, the IF signal intensity appeared to be reduced (Figure 4A), a result confirmed by an immunoblot performed with an anti-DNALI1 antibody on protein extracts from the individual’s spermatozoa (Figure 4C). Taken together, these data demonstrate that in sperm cells from individuals with TTC12 mutations, the assembly and/or the transport of ODAs and an IDA subset comprising DNALI1, DNAH3, and DNAH12 are compromised, whereas IDA-I1 remains intact.
TTC12-Depleted Paramecia Reproduce the Spermatozoa Phenotype Observed in PCD-Affected Individuals
To further investigate the function of TTC12 in cilia motility, we undertook studies in the free-living ciliate Paramecium tetraurelia, which is a powerful model organism with which to characterize potential functions of PCD candidate genes41,42 because gene functional analysis by RNA interference silencing is rapid, efficient, and easy to perform. Gene homology analyses performed on the P. tetraurelia genome28 identified four genes putatively encoding TTC12; these genes are GSPATT00036969001, GSPATT00034215001, GSPATT00035675001, and GSPATT00037155001, hereafter named TTC12a, TTC12b, TTC12-like a, and TTC12-like b, respectively. Interpro software analysis detected both TPR and ARM repeats in TTC12a and TTC12b only. Therefore, we focused our analysis on these two proteins, which are the closest orthologs of human TTC12 (Figure S9). RNA-seq analysis showed that TTC12a is highly expressed in comparison to TTC12b and is the only paralog overexpressed during ciliogenesis (Table S6)43,44. By generating a GFP-tagged TTC12a protein in paramecia, we showed that, as observed in human cells, TTC12a is a cytoplasmic protein (Figure S10). We used the dsRNA feeding method to silence TTC12a, TTC12b, and the combination TTC12a+TTC12b. The knockdown (KD) of TTC12a alone or in association with TTC12b led to a dramatic defect in the swimming velocity (Figures 5A and 5B) and subsequent cell death. By contrast, cells that were knocked down for TTC12b showed no obvious abnormal swimming phenotype, indicating that only TTC12a is essential for cilia motility (Figures 5A and 5B). The swimming velocity and cell death observed in TTC12a-depleted cells were rescued by the expression of an RNAi-resistant version of the gene (Figures 5C 5D, and 5E), confirming that the observed phenotypes are specific to TTC12 knockdown. Cilia-staining IF and TEM indicated that in Paramecium TTC12a+b KD cilia were globally intact (Figure 6A) but contained a reduced number of ODAs and/or IDAs per cilium (Figure 6B). Similar results were obtained after depletion of the TTC12a protein alone (data not shown). Taken together, these data indicate that, similarly to what we found in human spermatozoa, the assembly and/or transport of both ODAs and IDAs was affected by the absence of TTC12a in the Paramecium model, which suggests that the function of this protein is at least partly conserved throughout evolution.
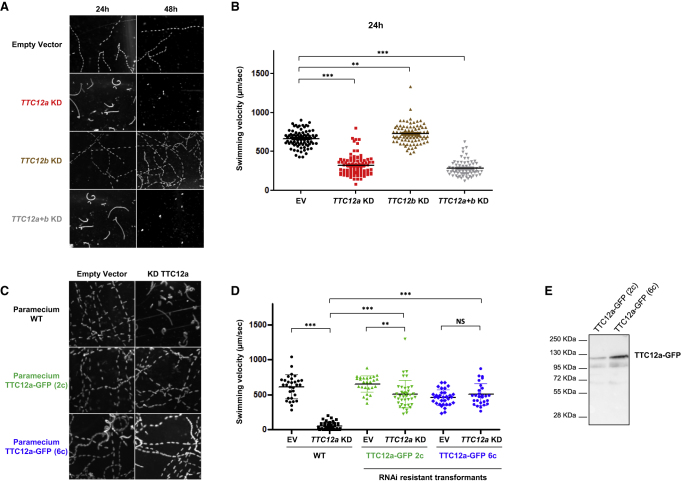
Figure 5.
Knockdown of TTC12a Affects the Swimming Velocity of Paramecia
(A) Z projection of track recording paramecia swimming under a dark-field microscope with a 10× objective lens after 24 h or 48 h of RNAi treatment. At 24 h of silencing, swimming behavior was affected in TTC12a- and TTC12a+b-silenced cells, whereas the silencing of TTC12b did not show any swimming defect. At 48 h, TTC12a- and TTC12a+b-silenced cells did not swim anymore, and they died at 72 h.
(B) Analysis of the swimming velocity of TTC12a, TTC12b, and TTC12a+b KD cells as well as control paramecia (i.e., empty vector silenced paramecia). Note that TTC12b-depleted cells swam slightly faster (725mm/s) than control cells (EV, 665 mm/s). The swimming velocity of TTC12a (317 mm/s) and TTC12a+b (282 mm/s) KD cells was reduced to 50% of that of control cells at 24 h. Data are represented as the mean ± SEM. Statistical analyses were performed with GrahPad Prism v5 software through the use of two-tailed unpaired t tests, ∗∗∗p < 0.001, ∗∗p < 0.01, (n = 69–88 per experiment).
(C) Z projection of track recording paramecia swimming under a dark-field microscope equipped with a 10× objective lens. Clones transformed by an RNAi-resistant version of TTC12a (2c and 6c) as well as non-transformed paramecia were grown for 36 h in an empty-vector silencing medium or in a TTC12a-silencing medium. TTC12a-depleted cells barely swam, as shown previously, whereas cells expressing an RNAi-resistant version of TTC12a again swam similarly to control cells. This indicates that TTC12a-depleted phenotypes are rescued by the TTC12a RNAi-resistant version.
(D) Analysis of the swimming velocity of transformed and non-transformed cells grown in an empty-vector silencing medium or in a TTC12a-silencing medium. As expected, TTC12a silencing in non-transformed cells dramatically reduced the swimming velocity (52 mm/s). However, the expression of the TTC12 RNAi-resistant version rescues this phenotype (88% of complementation). Note that the swimming velocity of transformed 2c cells (653 mm/s) was similar to that of non-transformed paramecia (612 mm/s), while transformed 6c cells swam slower (458 mm/s). Data are represented as the mean ± SEM. Statistical analyses were performed with GrahPad Prism v5 software and two-tailed unpaired t tests. ∗∗∗p < 0.001, ∗∗p < 0.01, NS (non-significant) p > 0.1 (n = 26-48 per experiment).
(E) Immunoblot analysis on total protein extract of transformed 2c and 6c cells showing that both clones express TTC12a-GFP. Abbreviations are as follows: EV, empty vector; and KD, knockdown.
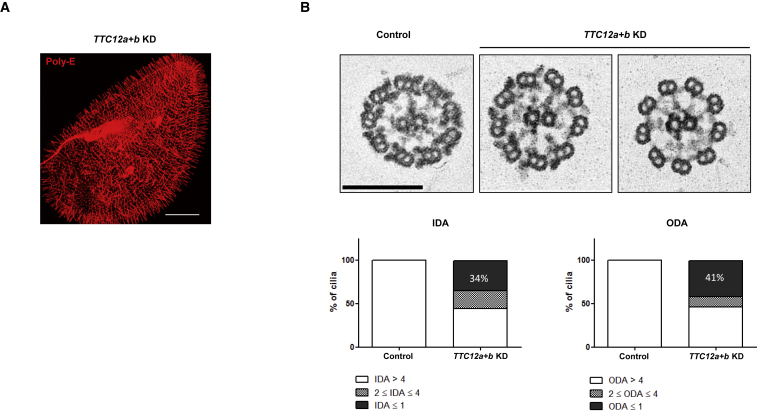
Figure 6.
Knockdown of TTC12a+b in Paramecia Leads to the Loss of Both ODAs and IDAs
(A) Depletion of TTC12a+b in Paramecium does not affect cilia number or length. TTC12a+b-silenced paramecia were fixed and immunostained for cilia (anti-polyglutamylated tubulin antibodies) after 48 h of RNAi. The scale bar represents 20 μm.
(B) Upper panel: Eelectromicrographs of cross-sections of cilia from TTC12a+b KD paramecia. Control cilia show the normal distribution of both ODAs and IDAs. By contrast, TTC12a+b KD cilia show a partial or total lack of ODAs and IDAs. The scale bar represents 200 nm.
Lower panel: quantitative TEM analysis of the number of IDAs and ODAs on 140 cross-sections from a control and TTC12a+b co-depleted cells. ODAs and IDAs were normally distributed in 100% of cilia from control cells and in about 50% of the co-depleted TTC12a+b cells. One must interpret this result while taking into account that, in paramecia, half of cilia are renewed at each cell division. Therefore, after the first division, paramecia display half of normal cilia and half of TTC12a+b-silenced cilia. The former half (generated before silencing) did not display the phenotype, whereas the new one did (generated after silencing).
Abbreviations are as follows: ODA, outer dynein arm; IDA, inner dynein arm; and KD, knockdown.
CRISPR-Cas9-Mediated TTC12 Invalidation in AECs Leads to Single-Headed IDA Subspecies Defects Mimicking the Phenotype of AECs from Individuals with TTC12 Mutations
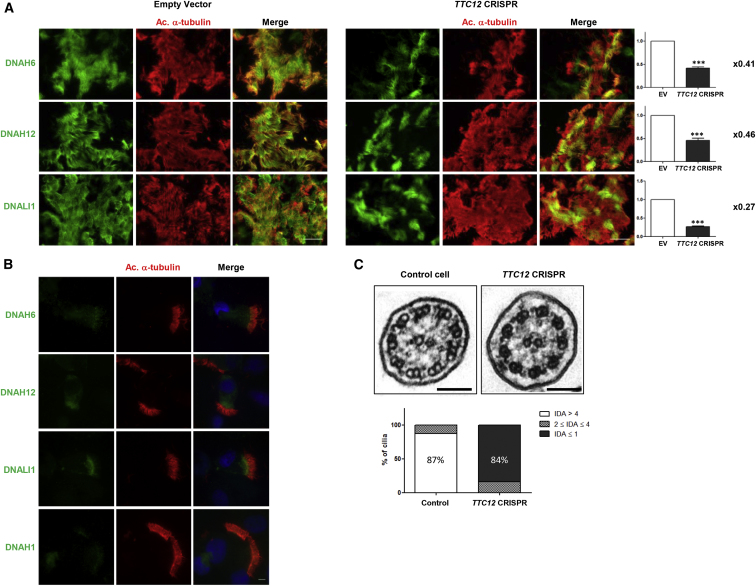
In order to further assess the consequences of TTC12 loss-of-function mutations in airway ciliated cells, we invalidated TTC12 in the model of primary AECs cultured at ALI31 through a CRISPR-Cas9 approach. Control AECs were transduced with lentiviruses expressing plentiCRISPRv2 carrying Cas9 and RNA guides (gRNA) targeting TTC12. Two gRNAs targeting exon 3 were tested, and the most effective one, gRNA 1 (i.e., with the higher level of insertions or deletions as assessed by TIDE webtool), was used for further experiments. Genomic DNA (gDNA) was then extracted from ALI cultures transduced with gRNA 1. We found that 82% of gDNA carried indels, including 28% of single-nucleotide insertions (Figure S11A and S11B). TTC12 invalidation in AECs was confirmed by immunoblot (Figure S11C), thereby allowing us to investigate the functional effects of such invalidation on the main axonemal components. IF staining of TTC12-invalidated cultured epithelial cells revealed a phenotype similar to that found in primary cells from individuals with TTC12 mutations; only axonemal components specific to single-headed IDAs (i.e., DNAH1, DNAH6, DNAH12, and DNALI1) were found to be affected (Figures 7A and 7B). More specifically, the analysis of ciliated cells cultured in ALI by high-resolution IF confirmed the invalidation of TTC12 in clusters with a combination of ciliated cells lacking DNAH6, DNAH12, and DNALI1 along the cilia length and ciliated cells with normal staining (Figure 7A). Moreover, analysis of isolated ciliated cells confirmed the data obtained in AECs from individuals with TTC12 mutations; the abnormal localization of DNAH1, DNAH6, and DNALI1 was found at very low levels at the base of the cilia, and DNAH12 was completely absent all along the cilia (Figure 7B). All the other tested axonemal components displayed a normal staining (i.e., DNAH5, DNAH9, DNAH11, and DNAI1 for ODAs; DNAH2 and DNAH10 for IDA-I1; RSPH1, RSPH3, RSPH4a, RSPH9, and RSPH11 for RSs; GAS8 for the N-DRC; and CCDC39 and CCDC40 for the ciliary ruler) indicating that all other axonemal complexes were normally assembled (Figure S12). To further study the impact of TTC2 invalidation on ciliary axonemal components, we analyzed cross-sections of cilia from TTC12-invalidated epithelial cells by TEM. As shown in Figure 7C, those cilia displayed an abnormal ultrastructure; 84% of cilia lacked IDAs, whereas a non-infected culture harbored only 3% of abnormal cilia. Again, this ultrastructural phenotype correlates with the one found in individuals with TTC12 mutations.
Figure 7.
TTC12 Is Required for the Assembly of Single-Headed IDAs
(A) TTC12 was invalidated in AEC cultures with TTC12 CRISPR-Cas9 lentiviruses and immunostained with an anti-acetylated α-tubulin antibody (red), a marker of ciliary axonemes, and anti-DNAH6, anti-DNAH12, or anti-DNALI1 antibodies as markers of single-headed IDAs (green). IF analysis in clusters of ciliated cells shows the partial invalidation of TTC12 in culture with a loss of DNAH6, DNAH12, and DNALI1 in cells lacking TTC12. Scale bars represent 25 μm. DNAH6-, DNAH12-, and DNALI1-labeled areas were measured with ImageJ software and normalized against tubulin-labeled area. The dynein/tubulin ratios obtained in TTC12-CRISPR-transduced cells were then normalized to ratios obtained in EV-transduced cells. Values of those final ratios are indicated on the right of the histograms and showed the decrease of dynein-chain-labeling signal compared to tubulin. Data are represented as the mean ± SEM. Statistical analyses were performed with GrahPad Prism v5 software and two-tailed unpaired t tests. ∗∗∗p < 0.001 (n = 3).
(B) IF images of isolated ciliated cells lacking TTC12 show the complete loss of DNAH12 along the cilia and its accumulation in the subapical region of the cytoplasm, whereas DNAH6 and DNALI1 expression appears to be strongly decreased but still present at the base of cilia. Nuclear staining was performed with DAPI (blue). The scale bar represents 25 μm.
(C) Electromicrographs of cross-sections of cilia axonemes from a non-infected AEC culture and a TTC12-CRISPR-infected cell culture show an abnormal arrangement lacking IDAs in the TTC12-invalidated culture. Proportions of cilia harboring more than four IDAs, between two and four IDAs, and up to one IDA,in electromicrograph cross-sections, were reported in a histogram comparing control cells to TTC12-invalidated cells. Scale bars represent 5 μm.
Abbreviations are as follows: ODA, outer dynein arm; IDA, inner dynein arm; and EV, empty vector.
Discussion
We report mutations in TTC12, a gene encoding a protein involved in the cytoplasmic assembly and/or transport of axonemal DAs, in PCD-affected individuals. A thorough analysis of the phenotype of sperm flagella and respiratory cilia from TTC12-deficient individuals revealed that the loss of function of TTC12 affects both ODAs and a subset of single-headed IDAs in sperm flagella, but only a different subset of single-headed IDAs in respiratory cilia. These data, which are further supported by two cellular models of the pathology (i.e., the ciliate Paramecium tetraurelia depleted in TTC12 and human AECs differentiated in vitro after a CRISPR-Cas9-mediated invalidation of TTC12), reveal the existence of distinct dynein assembly mechanisms in human sperm flagella and respiratory cilia.
The biological function of TTC12 is still elusive. One study showed an upregulation of TTC12 transcripts during mouse ciliogenesis in vitro and a centrosomal localization of TTC12 fused to GFP.45 In paramecia, TTC12a, the ortholog of human TTC12, was also found to be overexpressed during ciliogenesis (ParameciumDB). Although some TTC proteins have been involved in the so-called intraflagellar transport (IFT)46, 47, 48 the following points support TTC12’s possible role as a co-chaperone: (1) TTC12 contains three TPR domains, three ARM domains, and two structural motifs with alpha helices known to be involved in protein-protein interactions, and TPR-containing proteins are part of several multiprotein complexes that include co-chaperones;34 and (2) notably, in humans, only two other proteins, UNC45A and UNC45B, are known to contain both TPR and ARM domains, and these have been shown to interact with HSP90 and to act as molecular co-chaperones.35,49, 50, 51 These data, together with the cytoplasmic localization of TTC12, are therefore consistent with a role of TTC12 as a co-chaperone involved in dynein assembly.
The cytoplasmic assembly of DAs is a complex process involving a wide and still incompletely resolved chaperone-mediated protein network. DA components are first synthetized and pre-assembled into multi-subunit structures in the cytoplasm before being transported by IFT into the axoneme for microtubule attachment.52,53 Except for HSP90, all the proteins known to be involved in the axonemal dynein pre-assembly network have been implicated in PCD (i.e., such proteins include CFAP300, CFAP298, CCDC103, DNAAF1, DNAAF2, DNAAF3, DNAAF4, DNAAF5, LRRC6, PIH1D3, SPAG1, and ZMYND10). HSP90 is known to interact with TPR domains of various co-chaperones to promote their function.54 Two so-called dynein-assembly factors contain TPR domains: DNAAF4 and SPAG1. In recent studies, DNAAF4 was found to interact with both HSP90 and DNAAF2 through its TPR domains to form an R2TP-like complex.55,56 PIH1D3, whose corresponding gene has recently been found to be involved in PCD, might also interact with HSP90 through a TPR-containing protein, which could be DNAAF4.55,56 Interestingly, DNAAF4, which seems to play a central role in this network, was found to interact with ZMYND10, a partner of LRRC6 and CFAP298,16,57 the latter of which also interacts with DNAAF1.58 In this assembly network, it has been reported that DNAAF2 and PIH1D3 interact with DNAI2,9,56 a dynein intermediate chain known to assemble early in the human ODA assembly process.59 As for DNAAF5, it has been shown to interact with DNAI2 but would not bind HSP90.59 The functions of CFAP300, DNAAF3, SPAG1, and CCDC103 remain unclear, and there are currently no reported interactions of these proteins with HSP90 or other chaperones. In this context, TTC12, with its three TPR domains that suggest an interaction with HSP90 and/or other chaperones, emerges as a player in this complex pre-assembly network.
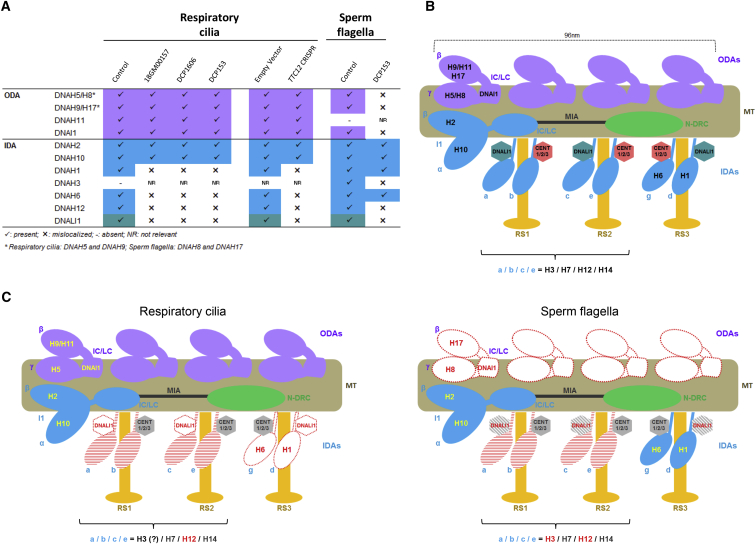
In most studies, the absence of both DAs in PCD-affected individuals has been identified by TEM and high-resolution IF mainly through the use of DNAH5 and DNALI1 antibodies directed, respectively, against the ODA γ-heavy chain and the IDA subspecies a, c, and d. However, these two approaches do not allow determination of which ODA type(s) and which IDA subspecies are affected. For DNAAF4, DNAAF5, and CCDC103 mutations, which are responsible for an absence of both DAs, the proximal staining of DNAH5 and DNAI2 and/or the loss of DNAH9 staining suggests that only type 2 ODAs are affected.9,12,15
In order to accurately decipher the role played by TTC12 in DA assembly, we aimed to identify which single-headed IDA subspecies are affected in the respiratory cilia of individuals with TTC12 mutations. Besides the presence of ODAs, as attested to by TEM and IF analyses, IDA staining revealed that IDA-I1 remained present, whereas according to the Chlamydomonas model, four of the six single-headed IDA subspecies (a, c, d, and g) were affected. No conclusion could be raised on the b and e subspecies because the heavy chain DNAH12 is not yet assigned to a specific subspecies (a, b, c, or e), and no efficient antibodies are available to detect the two remaining IDA heavy chains (i.e., DNAH7 and DNAH14) (Figure 8). Of note, proteomic analyses performed on human airway cilia and spermatozoa showed that DNAH7 is present in both proteomes; in contrast, NAH14 is undetected in both of them.36,60, 61, 62 Like in cilia, spermatozoa from individuals with TTC12 mutations conserve the dimeric IDA-I1. Nonetheless, unlike in cilia, those spermatozoa displayed an absence of ODAs, as revealed by TEM analyses and confirmed by the absence of staining of the ODA HCs (DNAH8 and DNAH17) that are specifically expressed in flagella.3 Interestingly, as for single-headed IDA components, our results showed that the IDA heavy chain DNAH3 (present in the IDA subspecies a, b, c, or e) is detected in flagellar axonemes but is not detectable in cilia; these results are in line with what would be expected from proteomic data.36,60 Additionally, spermatozoa from an individual with bi-allelic TTC12 mutations displayed a loss of DNAH3 as well as a mislocalization of DNAH12 (another monomeric IDA heavy chain that is not specifically assigned to subspecies a, b, c, or e), thereby unveiling the role of TTC12 in the proper expression of DNAH3 and DNAH12 in sperm cells.
Figure 8.
Dynein Chains Affected by the Absence of TTC12 and Diagrammatic Representation of the Axonemal Components Impacted by TTC12 Loss-of-Function Mutations in Respiratory Cilia and Sperm Flagella
(A) Table summarizing the dynein chains studied by IF in AECs of a control and of individuals 18GM00157, DCP1606, DCP153, in AEC cultures invalidated or not for TTC12 by a CRISPR-Cas9 approach, and in spermatozoa from a control and from individual DCP153. ODA labeling was performed with anti-DNAH5 and anti-DNAH9 antibodies in AECs and anti-DNAH8 and anti-DNAH17 antibodies in spermatozoa. Black check marks represent normal localization, minus signs represent an absence of protein, and black cross signs indicate mislocalization. NR: not relevant.
(B) Schematic structure of a normal 96-nm-long axonemal unit. Each unit, which repeats along the microtubule doublets, consists of four ODAs represented in purple and IDAs shown in blue. Because the exact composition of IDAs is virtually unknown in humans, the IDA representation is based on data from Chlamydomonas, in which the seven IDA subspecies include six single-headed DAs (a, b, c, e, g, and d) and one double-headed DA (IDA-I1).
(C) Phenotype of TTC12-deficient respiratory cilia versus TTC12-deficient sperm flagella. The proteins unaffected by the loss of TTC12 are represented in full color and written in yellow, and the proteins affected by the loss of TTC12 are represented in white with a red dotted line and written in red. The proteins found to be affected by TTC12 mutations, but for which the exact localization is unknown in humans, are represented in a hatched red pattern and are written in red. Proteins with lower but non-null expression are represented in a hatched gray pattern and are written in red. The proteins that could not be tested are represented and written in gray.
Abbreviations are as follows: ODA, outer dynein arm; IDA, inner dynein arm; N-DRC, nexin-dynein regulatory complex; MIA, modifier of inner arms; IC/LC, intermediate chain/light chain, RS, radial spoke; and MT, microtubules.
Surprisingly, although DNAH1 (d) and DNAH6 (g) are present in both normal cilia and flagella, they were not impacted by TTC12 mutations in spermatozoa, in contrast with what was observed in cilia.
Moreover, in those spermatozoa, DNALI1 amounts were found to be reduced but present (Figure 8).
Taken together, these results show that, in spermatozoa, TTC12 defects lead to the complete absence of ODAs and of a set of monomeric IDA subspecies; this set is different from the set of monomeric IDA subspecies impacted in respiratory cilia. Although the absence of ODAs in flagella could be explained by differences in ODA composition between cilia and flagella (DNAH5, DNAH9, and DNAH11 in cilia; DNAH8 and DNAH17 in flagella), the differences affecting single-headed IDAs are not exclusively due to distinct dynein HC composition (e.g., DNAH1 and DNAH6 are both present in cilia and flagella).
In paramecia, TTC12 knock down led both to the absence of DAs and to a drastic impairment of the swimming velocity. This suggests similarities in the ODA and IDA assembly process at play in this protist and in human sperm flagella. In human cilia, the defects of all known co-chaperones, except for TTC12, affect both ODAs and IDAs. The situation is very different in Chlamydomonas, where several of the algal homolog genes are important for assembly of one DA type; DNAAF1 and HEATR2 homologs (i.e., ODA7 and HEATR2, respectively) govern the assembly of ODAs only, whereas the DNAAF4 homolog (i.e., PF23) contributes only to the assembly of some IDAs.63
Because paramecia mimicked the human flagellar phenotype but not the ciliary one, we looked for an additional cellular model to further strengthen our conclusions on the role of TTC12 in cilia. We developed a human differentiated cell-culture model based on CRISPR-Cas9 invalidation of TTC12. Primary AECs cultured under ALI conditions constitute a strong epithelial barrier that is very difficult to efficiently transfect, especially without inhibiting ciliogenesis. In this study, we showed that transduction of AECs during their expansion phase not only is highly efficient but, most importantly, also preserves ciliogenesis and thus provides large amounts of ciliated cells from which DNA, RNA, and protein expression can be studied. After puromycin selection, 100% of cultured cells were infected, but four populations with different genotypes could have been present: no indel in TTC12, indels within one TTC12 allele, distinct indels within each of the two TTC12 alleles, or the same indel within the two TTC12 alleles; these genotypes are equivalent to a wild-type genotype at the TTC12 locus and to a TTC12 loss-of-function mutation present in the heterozygous, compound heterozygous, and homozygous state, respectively. Because AECs cannot be dissociated and isolated for selection, contrary to what is done for cell lines, we had to work with a heterogeneous population of cells that were either invalidated or not invalidated for TTC12. Nevertheless, our results revealed that TTC12 CRISPR AECs perfectly mimic the phenotype observed in cilia from individuals carrying TTC12 mutations, whereas TTC12-KD paramecia displayed a ciliary phenotype closer to the flagellar phenotype observed in human spermatozoa. From a general viewpoint, this demonstrates that CRISPR-Cas9-engineered AECs represent a valuable model for studying the impact of PCD mutations on ciliary axonemes.
As for clinical aspects, the diagnosis of PCD is difficult to establish in individuals with mild respiratory symptoms and no laterality defect. In such situations, low concentrations of nasal NO, an abnormal ciliary beat frequency, and/or the presence of ciliary ultrastructural defects is essential if clinicians are to confirm or deny the suspicion of PCD. In the current study, all the individuals were exempt from laterality defects and displayed motile cilia with a normal beat frequency. In two of the individuals, nasal NO concentrations were normal. Furthermore, the ciliary ultrastructural defect (i.e., the isolated absence of IDAs with an inconstant and mild axonemal disorganization) was also difficult to assert by TEM because of the usual low contrast of IDAs. The diagnosis of PCD was, however, much easier to establish in the two adult male individuals because of their infertility due to a sperm immotility related to the flagellar absence of both DAs, an ultrastructural defect easily identified by TEM. It is therefore tempting to speculate that PCD could be underdiagnosed in female individuals with TTC12 mutations, in young male individuals, or in adult males for whom no TEM analyses of their spermatozoa is performed, thereby underscoring the major benefit of molecular analyses in such individuals.
In conclusion, TTC12 emerges as a cytoplasmic player in DA assembly and/or transport. So far, all the mutations in genes encoding cytoplasmic pre-assembly factors cause a combined ODA and IDA defect. However, TTC12 mutations lead to distinct ultrastructural defects in cilia (i.e., absence of single-headed IDAs) and in sperm flagella (i.e., absence of both DAs), which unveils its role as a possible co-chaperone in charge of monomeric IDAs in both organelles but not of ODAs in cilia. More precisely, TTC12 is in charge of different dynein heavy chains in monomeric IDAs from cilia and from flagella. Furthermore, our findings should be of particular help to establish the diagnosis of PCD in individuals with isolated IDA defects in respiratory cilia. The development of a cellular model based on human AECs differentiated in vitro into ciliated cells after a CRISPR-cas9-mediated invalidation of genes of interest should open new avenues to decipher both the role of the numerous proteins expressed during ciliogenesis and the pathophysiology of diseases of the airway epithelium.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We are grateful to the affected persons and their families, whose cooperation made this study possible. We acknowledge the contributions of the CELPHEDIA Infrastructure, especially the center AniRA in Lyon and Gisèle Froment, Didier Nègre, and Caroline Costa from the lentivectors production facility (UMS3444/US8, SFR Biosciences Gerland-Lyon Sud) and RaDiCo, funded by the French National Research Agency under the specific program “Investments for the Future,” (Cohort grant agreement ANR-10-COHO-0003) and the Chancellerie des Universités de Paris (Legs Poix grant). We thank Lucas Fares-Taie and Araksya Izmiryan at IMAGINE Institute for their advice in setting up the CRISPR-Cas9 technology. We thank Anne Loyens from INSERM U1172 at Lille University for TEM technical assistance and Feng Zhang from the Massachusetts Institute of Technology for the gift of the plentiCRISPRv2 plasmid.
Published: January 23, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.12.010.
Contributor Information
Aminata Touré, Email: aminata.toure@inserm.fr.
Serge Amselem, Email: serge.amselem@inserm.fr.
Web Resources
CELPHEDIA Infrastructure, http://www.celphedia.eu/
Conserved Domain Database (NCBI CDD webtool), https://www.ncbi.nlm.nih.gov/cdd/
Clustal Omega program, https://www.ebi.ac.uk/Tools/msa/clustalo/
CRISPOR TEFOR, http://crispor.tefor.net/
EMBL EBI Ensembl, https://www.ensembl.org/index.html
EMBL EBI Expression Atlas, https://www.ebi.ac.uk/gxa/home
InterPro, https://www.ebi.ac.uk/interpro/
OMIM, https://www.omim.org/
ParameciumDB, https://paramecium.i2bc.paris-saclay.fr
Uniprot, https://www.uniprot.org/
Supplemental Data
References
- 1.Lucas J.S., Burgess A., Mitchison H.M., Moya E., Williamson M., Hogg C., National P.C.D., National PCD Service, UK Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 2014;99:850–856. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dasgupta A., Amack J.D. Cilia in vertebrate left-right patterning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150410. doi: 10.1098/rstb.2015.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield M., Thomas L., Bequignon E., Schmitt A., Stouvenel L., Montantin G., Tissier S., Duquesnoy P., Copin B., Chantot S. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to Asthenozoospermia. Am. J. Hum. Genet. 2019;105:198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui K.H., Yagi T., Yamamoto R., Kamiya R., Ishikawa T. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 2012;198:913–925. doi: 10.1083/jcb.201201120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J., Yin W., Smith M.C., Song K., Leigh M.W., Zariwala M.A., Knowles M.R., Ostrowski L.E., Nicastro D. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat. Commun. 2014;5:5727. doi: 10.1038/ncomms6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viswanadha R., Sale W.S., Porter M.E. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol. 2017;9:a018325. doi: 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hom E.F.Y., Witman G.B., Harris E.H., Dutcher S.K., Kamiya R., Mitchell D.R., Pazour G.J., Porter M.E., Sale W.S., Wirschell M. A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken) 2011;68:555–565. doi: 10.1002/cm.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas J.S., Barbato A., Collins S.A., Goutaki M., Behan L., Caudri D., Dell S., Eber E., Escudier E., Hirst R.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017;49:1601090. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N.T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.-M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchison H.M., Schmidts M., Loges N.T., Freshour J., Dritsoula A., Hirst R.A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012;44:381–389. doi: 10.1038/ng.1106. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panizzi J.R., Becker-Heck A., Castleman V.H., Al-Mutairi D.A., Liu Y., Loges N.T., Pathak N., Austin-Tse C., Sheridan E., Schmidts M. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horani A., Druley T.E., Zariwala M.A., Patel A.C., Levinson B.T., Van Arendonk L.G., Thornton K.C., Giacalone J.C., Albee A.J., Wilson K.S. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kott E., Duquesnoy P., Copin B., Legendre M., Dastot-Le Moal F., Montantin G., Jeanson L., Tamalet A., Papon J.-F., Siffroi J.-P. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarkar A., Loges N.T., Slagle C.E., Francis R., Dougherty G.W., Tamayo J.V., Shook B., Cantino M., Schwartz D., Jahnke C., UK10K DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zariwala M.A., Gee H.Y., Kurkowiak M., Al-Mutairi D.A., Leigh M.W., Hurd T.W., Hjeij R., Dell S.D., Chaki M., Dougherty G.W. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles M.R., Ostrowski L.E., Loges N.T., Hurd T., Leigh M.W., Huang L., Wolf W.E., Carson J.L., Hazucha M.J., Yin W. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am. J. Hum. Genet. 2013;93:711–720. doi: 10.1016/j.ajhg.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin-Tse C., Halbritter J., Zariwala M.A., Gilberti R.M., Gee H.Y., Hellman N., Pathak N., Liu Y., Panizzi J.R., Patel-King R.S. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paff T., Loges N.T., Aprea I., Wu K., Bakey Z., Haarman E.G., Daniels J.M.A., Sistermans E.A., Bogunovic N., Dougherty G.W. Mutations in PIH1D3 cause X-Linked primary ciliary dyskinesia with outer and inner dynein arm defects. Am. J. Hum. Genet. 2017;100:160–168. doi: 10.1016/j.ajhg.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höben I.M., Hjeij R., Olbrich H., Dougherty G.W., Nöthe-Menchen T., Aprea I., Frank D., Pennekamp P., Dworniczak B., Wallmeier J. Mutations in C11orf70 cause primary ciliary dyskinesia with randomization of left/right body asymmetry due to defects of outer and inner dynein arms. Am. J. Hum. Genet. 2018;102:973–984. doi: 10.1016/j.ajhg.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huizar R.L., Lee C., Boulgakov A.A., Horani A., Tu F., Marcotte E.M., Brody S.L., Wallingford J.B. A liquid-like organelle at the root of motile ciliopathy. eLife. 2018;7:e38497. doi: 10.7554/eLife.38497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verra F., Fleury-Feith J., Boucherat M., Pinchon M.-C., Bignon J., Escudier E. Do nasal ciliary changes reflect bronchial changes? An ultrastructural study. Am. Rev. Respir. Dis. 1993;147:908–913. doi: 10.1164/ajrccm/147.4.908. [DOI] [PubMed] [Google Scholar]
- 23.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W.G., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 24.Skouri F., Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonneborn T.M. Chapter 12: Methods in Paramecium ResearchIn Methods in Cell Biology. In: Prescott D.M., editor. Academic Press; 1970. pp. 241–339. [Google Scholar]
- 26.Galvani A., Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 27.Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 28.Hauser K., Haynes W.J., Kung C., Plattner H., Kissmehl R. Expression of the green fluorescent protein in Paramecium tetraurelia. Eur. J. Cell Biol. 2000;79:144–149. doi: 10.1078/S0171-9335(04)70016-3. [DOI] [PubMed] [Google Scholar]
- 29.Rüdiger A.H., Rüdiger M., Wehland J., Weber K. Monoclonal antibody ID5: epitope characterization and minimal requirements for the recognition of polyglutamylated alpha- and beta-tubulin. Eur. J. Cell Biol. 1999;78:15–20. doi: 10.1016/s0171-9335(99)80003-x. [DOI] [PubMed] [Google Scholar]
- 30.Adoutte A., Ramanathan R., Lewis R.M., Dute R.R., Ling K.Y., Kung C., Nelson D.L. Biochemical studies of the excitable membrane of Paramecium tetraurelia. III. Proteins of cilia and ciliary membranes. J. Cell Biol. 1980;84:717–738. doi: 10.1083/jcb.84.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coste A., Brugel L., Maître B., Boussat S., Papon J.F., Wingerstmann L., Peynègre R., Escudier E. Inflammatory cells as well as epithelial cells in nasal polyps express vascular endothelial growth factor. Eur. Respir. J. 2000;15:367–372. doi: 10.1034/j.1399-3003.2000.15b24.x. [DOI] [PubMed] [Google Scholar]
- 32.Horani A., Nath A., Wasserman M.G., Huang T., Brody S.L. Rho-associated protein kinase inhibition enhances airway epithelial Basal-cell proliferation and lentivirus transduction. Am. J. Respir. Cell Mol. Biol. 2013;49:341–347. doi: 10.1165/rcmb.2013-0046TE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
- 34.Blatch G.L., Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 35.Tewari R., Bailes E., Bunting K.A., Coates J.C. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Blackburn K., Bustamante-Marin X., Yin W., Goshe M.B., Ostrowski L.E. Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J. Proteome Res. 2017;16:1579–1592. doi: 10.1021/acs.jproteome.6b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 39.Osinka A., Poprzeczko M., Zielinska M.M., Fabczak H., Joachimiak E., Wloga D. Ciliary proteins: Filling the gaps. recent advances in deciphering the protein composition of motile ciliary complexes. Cells. 2019;8:730. doi: 10.3390/cells8070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollmar M. Fine-tuning motile cilia and flagella: Evolution of the dynein motor proteins from plants to humans at high resolution. Mol. Biol. Evol. 2016;33:3249–3267. doi: 10.1093/molbev/msw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fassad M.R., Shoemark A., Legendre M., Hirst R.A., Koll F., le Borgne P., Louis B., Daudvohra F., Patel M.P., Thomas L. Mutations in outer dynein arm heavy chain DNAH9 cause motile cilia defects and situs inversus. Am. J. Hum. Genet. 2018;103:984–994. doi: 10.1016/j.ajhg.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fassad M.R., Shoemark A., le Borgne P., Koll F., Patel M., Dixon M., Hayward J., Richardson C., Frost E., Jenkins L. C11orf70 mutations disrupting the intraflagellar transport-dependent assembly of multiple axonemal dyneins cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2018;102:956–972. doi: 10.1016/j.ajhg.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaiz O., Malinowska A., Klotz C., Sperling L., Dadlez M., Koll F., Cohen J. Cildb: a knowledgebase for centrosomes and cilia. Database (Oxford) 2009;2009:bap022. doi: 10.1093/database/bap022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnaiz O., Cohen J., Tassin A.M., Koll F. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia. 2014;3:9. doi: 10.1186/2046-2530-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoh R.A., Stowe T.R., Turk E., Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y., Cao J., Huang S., Feng D., Zhang W., Zhu X., Yan X. Characterization of tetratricopeptide repeat-containing proteins critical for cilia formation and function. PLoS ONE. 2015;10:e0124378. doi: 10.1371/journal.pone.0124378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taschner M., Bhogaraju S., Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisa N.H., Jilani Y., Kainth K., Redd P., Lu S., Bougrine O., Abdul Sater H., Patwardhan C.A., Shull A., Shi H. The co-chaperone UNC45A is essential for the expression of mitotic kinase NEK7 and tumorigenesis. J. Biol. Chem. 2019;294:5246–5260. doi: 10.1074/jbc.RA118.006597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mooneyham A., Iizuka Y., Yang Q., Coombes C., McClellan M., Shridhar V., Emmings E., Shetty M., Chen L., Ai T. UNC-45A Is a Novel Microtubule-Associated Protein and Regulator of Paclitaxel Sensitivity in Ovarian Cancer Cells. Mol. Cancer Res. 2019;17:370–383. doi: 10.1158/1541-7786.MCR-18-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholls P., Bujalowski P.J., Epstein H.F., Boehning D.F., Barral J.M., Oberhauser A.F. Chaperone-mediated reversible inhibition of the sarcomeric myosin power stroke. FEBS Lett. 2014;588:3977–3981. doi: 10.1016/j.febslet.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi D., Takeda H. Ciliary motility: the components and cytoplasmic preassembly mechanisms of the axonemal dyneins. Differentiation. 2012;83:S23–S29. doi: 10.1016/j.diff.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 54.Assimon V.A., Southworth D.R., Gestwicki J.E. Specific binding of tetratricopeptide repeat proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry. 2015;54:7120–7131. doi: 10.1021/acs.biochem.5b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pal M., Morgan M., Phelps S.E.L., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., Prodromou C. Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 2014;22:805–818. doi: 10.1016/j.str.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olcese C., Patel M.P., Shoemark A., Kiviluoto S., Legendre M., Williams H.J., Vaughan C.K., Hayward J., Goldenberg A., Emes R.D. X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 2017;8:14279. doi: 10.1038/ncomms14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho K.J., Noh S.H., Han S.M., Choi W.-I., Kim H.-Y., Yu S., Lee J.S., Rim J.H., Lee M.G., Hildebrandt F., Gee H.Y. ZMYND10 stabilizes intermediate chain proteins in the cytoplasmic pre-assembly of dynein arms. PLoS Genet. 2018;14:e1007316. doi: 10.1371/journal.pgen.1007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaffe K.M., Grimes D.T., Schottenfeld-Roames J., Werner M.E., Ku T.S., Kim S.K., Pelliccia J.L., Morante N.F.C., Mitchell B.J., Burdine R.D. c21orf59/kurly controls both cilia motility and polarization. Cell Rep. 2016;14:1841–1849. doi: 10.1016/j.celrep.2016.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diggle C.P., Moore D.J., Mali G., zur Lage P., Ait-Lounis A., Schmidts M., Shoemark A., Garcia Munoz A., Halachev M.R., Gautier P. HEATR2 plays a conserved role in assembly of the ciliary motile apparatus. PLoS Genet. 2014;10:e1004577. doi: 10.1371/journal.pgen.1004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G., Guo Y., Zhou T., Shi X., Yu J., Yang Y., Wu Y., Wang J., Liu M., Chen X. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics. 2013;79:114–122. doi: 10.1016/j.jprot.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Amaral A., Castillo J., Estanyol J.M., Ballescà J.L., Ramalho-Santos J., Oliva R. Human sperm tail proteome suggests new endogenous metabolic pathways. Mol. Cell. Proteomics. 2013;12:330–342. doi: 10.1074/mcp.M112.020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vandenbrouck Y., Lane L., Carapito C., Duek P., Rondel K., Bruley C., Macron C., Gonzalez de Peredo A., Couté Y., Chaoui K. Looking for missing proteins in the proteome of human spermatozoa: An update. J. Proteome Res. 2016;15:3998–4019. doi: 10.1021/acs.jproteome.6b00400. [DOI] [PubMed] [Google Scholar]
- 63.Desai P.B., Dean A.B., Mitchell D.R. Chapter 4: Cytoplasmic preassembly and trafficking of axonemal dyneins. In: King S.M., editor. Dyneins. Second Edition. Academic Press; 2018. pp. 140–161. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative Video Showing Immotile Cilia of Individual 18GM00157
Representative Video Showing Beating Cilia of Individual 18GM00157
Representative Video Showing Immotile Cilia of Individual DCP1606
Representative Video Showing Beating Cilia of Individual DCP1606
Representative Video Showing Immotile Cilia of Individual DCP153
Representative Video Showing Beating Cilia of Individual DCP153
Representative Video Showing Beating Cilia of Control Individual