Abstract
Live yeast probiotics and yeast cell wall components (paraprobiotics) may serve as an alternative to the use of antibiotics in prevention and treatment of infections caused by pathogenic bacteria. Probiotics and paraprobiotics can bind directly to pathogens, which limits binding of the pathogens to the intestinal cells and also facilitates removal from the host. However, knowledge of bacterial binding, specificity, and/or capability is limited with regard to probiotics or paraprobiotics. The goal of this study was to characterize the qualitative and quantitative nature of two Saccharomyces cerevisiae probiotics and three S. cerevisiae paraprobiotics to adhere to thirteen different pathogenic bacteria using scanning electron miscroscopy and filtration assays. On average, the yeast probiotics (LYA and LYB) exhibited overall greater (P < 0.05) adhesion to the pathogenic bacteria tested (41% and 34%) in comparison to paraprobiotics (23%, 21%, and 22%), though variations were observed between pathogens tested. The ability of Salmonella and Listeria to utilize components of the yeast as a nutrient source was also tested. Bacteria were cultured in media with limited carbon and supplemented with cell free extracts of the probiotics and paraprobiotics. Salmonella exhibited growth, indicating these pathogens could utilize the yeast lysates as a carbon source. Listeria monocytogenes had limited growth in only one of the lysates tested. Together, these data indicate that the interaction between probiotics and paraprobiotics occurs in a strain dependent mechanism. Administration of probiotics and paraprobiotics as therapeutics therefore needs to be specific against the bacterial pathogen target.
Keywords: adhesion, direct antagonism, paraprobiotics, pathogenic bacteria, yeast probiotics
INTRODUCTION
The use of antibiotics as a means for treating and preventing illness in livestock has impacted animal health and performance (Dunlop et al., 1998). However, current trends in consumer preference and government regulation through pending directives (i.e., the Veterinary Feed Directive) have sparked interest in potential antibiotic alternatives, such as pro- and paraprobiotics. Probiotics, as defined by The Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO), are “live organisms which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO, 2002). Common probiotics recognized by the FAO and WHO include Lactobacillus sp., Bacillus sp., Enterococcus sp., Saccharomyces sp., and Aspergillus sp. (Chaucheyras-Durand and Fonty, 2002; Reid et al., 2003). Of these, the yeast Saccharomyces boulardii and S. cerevisiae have been the most commonly used among livestock (Martins et al., 2005; Duarte et al., 2012). Yeast probiotics that have been dried/fragmented into probiotic components have also been shown to confer a health benefit to a host (Middelbos et al., 2007). These products are referred to as “paraprobiotics” (Taverniti and Guglielmetti, 2011). These products are primarily composed of cell wall fragments and contain β-(1,3)-D–glucans, β-(1,6)-D–glucans, chitin, and mannoproteins. These products have also been shown to exert immunological benefits to the host (Kollar et al., 1997; Taverniti and Guglielmetti, 2011). Yeast based products have been used in dairy and beef cattle (Thrune et al., 2009; Sanchez et al., 2014), swine (Braude et al., 1944; van der Peet-Schwering et al., 2007), lambs (Tripathi and Karim, 2011), and poultry (Dawson, 2001) to enhance growth performance and animal health. Additionally, their usage has increased due to the fact that use of live microorganisms poses a potential risk to an immunocompromised host.
Yeast probiotics have multiple mechanisms of action by which they confer a health benefit to the host, including direct binding to toxins produced by pathogens and also stimulation of the host immune system. Additionally, probiotics have the potential to prevent colonization of bacteria to the mucosal surface of the intestine through either direct antagonism or through competitive inhibition (Shoaf-Sweeney and Hutkins, 2009). This inhibition is hypothesized to be due to the ability of certain pathogenic bacteria with mannose-binding fimbriae to bind mannoproteins within yeast cell walls (Ofek et al., 1977). Direct adhesion to the bacteria could facilitate removal from the digestive tract as well as limit adhesion to the intestinal epithelial cells (Gedek, 1999). Despite previous research on the binding effects of pathogenic bacteria to yeast, it is not known whether probiotics and paraprobiotics bind equally to bacteria (Korhonen et al., 1981; Gedek, 1999; Martins et al., 2010). Therefore, the objective of this study was to characterize the binding relationship of multiple Saccharomyces cerevisiae probiotic and paraprobiotic products with Gram-positive and Gram-negative pathogenic bacteria.
MATERIALS AND METHODS
Microbial Strains and Growth Conditions
The bacterial strains used in this study are listed in Table 1. Escherichia coli, L. monocytogenes, and Salmonella were grown in tryptic soy agar or broth (TSA/TSB) at 37°C. Clostridium were grown in Clostridial Reinforced Medium (CRM; BD 218081) anaerobically at 37°C. Bacteroides fragilis and Peptostreptococcus assacharolyticus were grown on Brucella broth with Vitamin K and hemin (BRU-BROTH; Anaerobe Systems AS-105) anaerobically at 37°C. Fusobacterium necrophorum and Arcanobacterium pyogenes were grown in Chopped Meat Glucose broth (CMG; Anaerobe Systems AS-813) anaerobically at 37°C. The 2 live Saccharomyces cerevisiae yeast samples used in this study were products commercially available, but produced by 2 different facilities. The 3 yeast cell-wall paraprobiotics analyzed in this study were processed from 3 different facilities. All of the yeast products were reconstituted in Yeast Peptone Dextrose (YPD) media at 37°C at a concentration of 0.1 g/mL (∼2 × 108 CFU/mL). The concentrations of paraprobiotics were based on initial populations of the live yeast probiotics and weighed out similarly. Viability of the probiotics was verified by plating aliquots on YPD agar. Where required, anaerobic conditions were achieved by using a Coy anaerobic chamber with a gas mix of 5% H2 and 95% N2 (Type B, Coy Laboratory Products INC.). Anaerotest strips and an oxygen sensor were used to monitor anaerobiosis throughout the study.
Table 1.
Bacterial strains used in study
| Bacteria | Strain/Source1 | Growth condition2 |
|---|---|---|
| Arcanobacterium pyogenes | 19411/ATCC | CMG, anaerobic |
| Bacteroides fragilis | 25285/ATCC | BRU-BROTH, anaerobic |
| Clostridium difficile | NR-32882/ATCC | CRM, anaerobic |
| Clostridium perfringens | 13124/ATCC | CRM, anaerobic |
| Escherichia coli O157:H7 | 43895/ATCC | TSB |
| Fusobacterium necrophorum | 25286/ATCC | CMG, anaerobic |
| Listeria monocytogenes | F2365/MSU | TSB |
| Porphyromonas asacharolyticus | 25260/ATCC | BRU-BROTH, anaerobic |
| Salmonella enterica Dublin | NR-28793/ATCC | TSB |
| Salmonella enterica Enteriditis | 13076/ATCC | TSB |
| Salmonella enterica Heidelberg | 8326/ATCC | TSB |
| Salmonella enterica Typhi | 6539/ATCC | TSB |
| Salmonella enterica Typhimurium | 13311/ATCC | TSB |
ATCC: American Type Culture Collection; MSU: Mississippi State University.
CMG: Chopped meat glucose medium; BRU-Broth: Brucella Broth medium; CRM: Clostridial Reinforced medium; TSB: Tryptic Soy Broth.
Scanning Electron Microscopy Adhesion Assay
Overnight cultures of the pathogenic bacteria and the yeast products were cultured at 37°C with constant agitation. A cover slip (Nunc Thermonox) was placed in each well of a 6-well culture plate. Overnight cultures of the yeast probiotics and paraprobiotics (2 mL, ∼4x108 CFU/ml) were added to cover slips and incubated at 37°C for 16 h; cover slips were then washed three times with 1X phosphate buffered saline (PBS). Overnight bacterial cultures (5 mL) were pelleted for 5 min at 13,000 x g and resuspended in TSB at a concentration of 2 × 1010 CFU/mL, at which point 1 mL of bacteria was added to the yeast cover slips. The co-culture of yeast and bacteria was incubated for 4 h at 37°C, after which each cover slip was washed with 1X PBS 3 times. After these washings, 2 mL of 2.5% glutaraldehyde in PO4 fixative was added to each well. Each cover slip was rinsed with distilled water, post-fixed with 2% osmium tetroxide (OsO4), rinsed with distilled water, and then dehydrated in a graded ethanol series (Merritt and Donaldson, 2009). Each cover slip was critical point dried, mounted on aluminum stubs with double sided carbon tape, and coated with 15nm platinum. The cover slips were then viewed under a JEOL JSM-6500F scanning electron microscope (SEM) at 5 Kv. Per cover slip, 40 yeast probiotic cells were counted, and of that count, the number of yeast cells found with bacteria bound was used in calculating the percent adherence per sample.
Membrane Filtration Adhesion Assay
Overnight bacterial and yeast cultures were prepared similarly as for SEM analysis. Yeast were cultured for 16 h at 37°C in 50 mL conical tubes, after which 0.05 mL (∼1 × 106 CFU/mL) was added to 4.9 mL of YPD, and co-cultured with 0.05 mL of bacteria (∼1x108 CFU/mL). The yeast-bacteria (YB) co-culture was vortexed and incubated for 4 h at 37°C. For controls, 0.05 mL of the bacterial culture was added to 4.95 mL of YPD and incubated for 4 h at 37°C. Following the 4 h incubation, 0.05 mL of YB co-culture or bacteria only control was added to 1.45 mL of PBS. Membrane filters (3.0µm, Millipore) were first washed with 1.5 mL of PBS, then the bacteria or YB mix was vacuum filtered, followed by a wash with 2 mL of PBS. The 3μm membrane filter was verified to contain pores small enough to trap the probiotics and paraprobiotics, but large enough to allow non-adhered bacteria to pass. The resulting filtrate (5 mL) was serially diluted in PBS and plated onto TSA. Viable bacterial colonies were counted on the plates following a 24 h incubation at 37°C. A minimum of 3 independent experiments were conducted.
Yeast Probiotic and Paraprobiotic Supernatant Effect Assay
Bacterial and yeast cultures were prepared similarly to the membrane filtration assay. Yeast products were cultivated in YPD for 16 h at 37°C and vacuum filtered using 3µm membrane filters. Resulting filtrate (0.05 mL) was added to 4.9 mL of YPD and co-cultured with 0.05 mL of bacteria (1 × 108 CFU/mL). The supernatant + bacteria (SB) co-culture was then vortexed and incubated for 4 h at 37°C. For controls, 0.05 mL of the bacterial culture was added to 4.95 mL of YPD and incubated for 4 h at 37°C. Following the 4 h incubation, 0.05 mL of the SB co-culture was serially diluted in PBS and plated onto TSA. Viable bacterial colonies were counted on the plates following a 24 h incubation at 37°C. A minimum of 3 independent experiments were conducted.
Yeast Lysate Growth Analysis
All yeast probiotics and paraprobiotics were reconstituted at 0.2g into 5 mL mineral salts medium (MSM) without glucose. The medium per 1000 mL contained 9.0g Na2HPHO4, 1.5g KH2PO4, 1.0g NH4Cl, 0.2g MgSO47∙H2O, 0.02g CaCl2∙2H20, 1.2mg FeNH4–citrate, and 2 mL Hoagland's Solution, pH 6.9 (Donaldson et al., 2014). Yeast products were lysed on ice using a sonicator (Fisherbrand Sonic Dismembrator Model 100, setting 3) for eight 1-min intervals, with 1 min cooling on ice between intervals. Yeast lysates were collected after centrifugation for 2 min at 12,000 x g and filtered using a 0.2 μm syringe filter.
Overnight (2 mL) cultures of Salmonella were centrifuged at 13,000 x g for 2 min, washed twice with 1 mL MSM (no glucose), then resuspended in 2 mL of MSM (no glucose). For analysis of Listeria monocytogenes, overnight cultures were centrifuged, washed twice with 1 mL of glucose limited mineral media (GLMM) and resuspended in 2 mL of GLMM without glucose (Schneebeli and Egli, 2013). The yeast lysates were added to a 96-well plate in 20 μL increments to 2 μL of bacterial cells and 180 μL of MSM (no glucose); as a control for growth, bacteria were added to MSM supplemented with 3% glucose. Growth of the bacteria was monitored using a PowerWave plate reader (BioTek, Winooski, VT), with OD600 collected every 1 h for 16 h. Growth was analyzed in a minimum of three replicates.
Statistical Analysis
The data from the SEM and the membrane filtration adherence assays were analyzed using the GLIMMIX Procedure of SAS (SAS Inst. Inc., Cary, NC). Presence/absence of binding was analyzed across replications as categorical data using logistic regression as a proportion of bound samples to total bound samples. Quantified concentrations were log normalized prior to analysis. Fixed effects included probiotic or paraprobiotic, bacterial strain, and their interactions. LSmeans were separated using α = 0.05 using the Tukey-Kramer option.
RESULTS
Qualitative Assessment of Binding Capabilities of Probiotics and Paraprobiotics to Pathogens
To determine if variations existed in the direct interaction between pathogens and different probiotics, a qualitative assessment of the adhesion capability was determined by SEM. Figure 1 represents images from yeast probiotics and paraprobiotics that were directly bound to bacteria. Yeast probiotics (LYA and LYB) exhibited overall greater (P < 0.05) adhesion to bacteria (41% and 34%) in comparison to the overall amount of adhesion by the paraprobiotics (23%, 21%, and 22%; Tables 2 and 3). Escherichia coli O157:H7 and L. monocytogenes exhibited the least binding potential to probiotics or paraprobiotics when compared to other bacteria (P < 0.05; Tables 2 and 3). Differences were observed between the 2 species of Clostridium analyzed in this study. Clostridium perfringens adhered to all products tested, while C. difficile adhered best to probiotics in comparison to paraprobiotics. Porphyromonas assacharolyticus and Arcanobacterium pyogenes only adhered to the live yeast probiotics.
Figure 1.
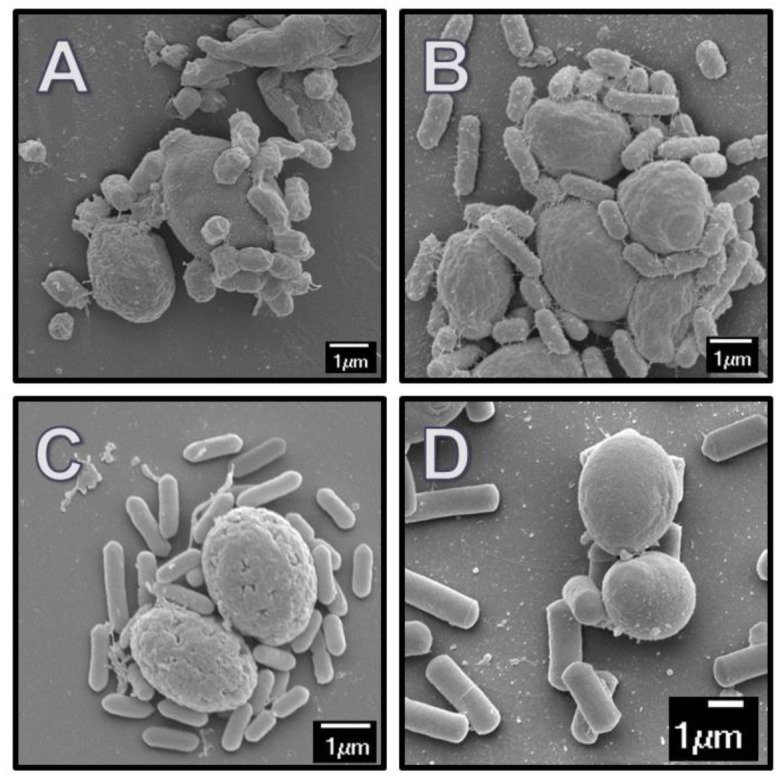
SEM images of pathogenic bacteria adhered to yeast probiotics and paraprobiotics. (A) E. coli O157:H7 bound to yeast paraprobiotic CWC; (B) S. enterica Typhimurium bound to yeast paraprobiotic CWC; (C) L. monocytogenes F2365 bound to yeast paraprobiotic CWA; and (D) C. perfringens bound to yeast probiotic LYB. Images are representative of a minimum of 40 yeast cells observed.
Table 2.
Scanning electron microscopy averages for adherence of pathogenic bacteria to yeast probiotics LYA and LYB and paraprobiotics CWA, CWB and CWC
| Bacteria | LYA | LYB | CWA | CWB | CWC |
|---|---|---|---|---|---|
| % Adhere | % Adhere | % Adhere | % Adhere | % Adhere | |
| A. pyogenes | 39.05 | 17.11 | 0.00 | 0.00 | 0.00 |
| B. fragilis | 55.32 | 13.51 | 37.50 | 15.22 | 4.55 |
| C. difficile | 17.24 | 20.97 | 0.00 | 5.00 | 0.00 |
| C. perfringens | 35.61 | 37.32 | 41.23 | 30.09 | 75.00 |
| E. coli O157:H7 | 10.76 | 9.93 | 15.04 | 1.49 | 12.04 |
| F. necrophorum | 13.51 | 55.32 | 37.50 | 15.22 | 4.55 |
| L. monocytogenes | 8.69 | 6.25 | 1.96 | 9.37 | 5.88 |
| P. assacharolytica | 85.63 | 31.79 | 0.00 | 0.00 | 0.00 |
| S. Dublin | 34.00 | 9.09 | 12.00 | 0.00 | 6.00 |
| S. Enteritidis | 49.09 | 55.22 | 16.67 | 0.00 | 11.90 |
| S. Heidelberg | 29.30 | 30.43 | 24.99 | 46.43 | 22.22 |
| S. Typhi | 65.08 | 58.83 | 21.53 | 59.32 | 50.00 |
| S. Typhimurium | 92.31 | 96.67 | 88.89 | 85.71 | 98.11 |
| Average | 41.20% | 34.03% | 22.87% | 20.60% | 22.33% |
Table 3.
Statistical analysis of binding potentials of pathogenic bacteria between each yeast probiotic (LYA and LYB) and paraprobiotic (CWA, CWB, and CWC) based on SEM observations
| Bacteria | LYA vs. LYB | LYA vs. CWA | LYA vs. CWB | LYA vs. CWC | LYB vs. CWA | LYB vs. CWB | LYB vs. CWC | CWA vs. CWB | CWA vs. CWC | CWB vs CWC |
|---|---|---|---|---|---|---|---|---|---|---|
| A. pyogenes | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.036 | 0.039 | 0.042 | 1.000 | 1.000 | 1.000 |
| B. fragilis | < 0.001 | 0.222 | < 0.001 | < 0.001 | < 0.001 | 0.997 | 0.176 | 0.033 | < 0.001 | 0.106 |
| C. difficile | 0.946 | 0.007 | 0.1114 | 0.0109 | < 0.001 | 0.013 | < 0.001 | 0.865 | 1.000 | 0.876 |
| C. perfringens | 0.995 | 0.860 | 0.8584 | < 0.001 | 0.952 | 0.657 | < 0.001 | 0.409 | < 0.001 | < 0.001 |
| E. coli O157:H7 | 0.998 | 0.794 | 0.0435 | 0.9970 | 0.607 | 0.058 | 0.974 | 0.011 | 0.967 | 0.036 |
| F. necrophorum | < 0.001 | 0.009 | 0.997 | 0.1756 | 0.222 | < 0.001 | < 0.001 | 0.033 | < 0.001 | 0.106 |
| L. monocytogenes | 0.965 | 0.598 | 0.9999 | 0.9647 | 0.802 | 0.944 | 1.000 | 0.567 | 0.854 | 0.945 |
| P. assacharolytica | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 1.000 | 1.000 | 1.000 |
| S. Dublin | 0.001 | 0.004 | < 0.001 | < 0.001 | 0.989 | 0.560 | 0.986 | 0.298 | 0.868 | 0.868 |
| S. Enteritidis | 0.919 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.191 | 0.974 | 0.642 |
| S. Heidelberg | 0.999 | 0.989 | 0.345 | 0.9296 | 0.972 | 0.372 | 0.876 | 0.200 | 0.998 | 0.104 |
| S. Typhi | 0.950 | < 0.001 | 0.967 | 0.4914 | < 0.001 | 1.000 | 0.876 | < 0.001 | 0.016 | 0.866 |
| S. Typhimurium | 0.858 | 0.972 | 0.8199 | 0.6970 | 0.522 | 0.301 | 0.999 | 0.985 | 0.421 | 0.268 |
Variations were also observed in the binding efficiencies between subspecies of Salmonella enterica. While S. Typhimurium and S. Heidelberg did not exhibit any difference in adherence between the products tested (P > 0.05), S. Enteritidis and S. Dublin exhibited greater binding to the live yeast probiotics (Tables 2 and 3). Salmonella Typhi exhibited binding to all yeast products, with the least amount of binding to yeast paraprobiotic CWA (Table 2).
Quantitative Assessment of the Binding Efficiency of Probiotics and Paraprobiotics
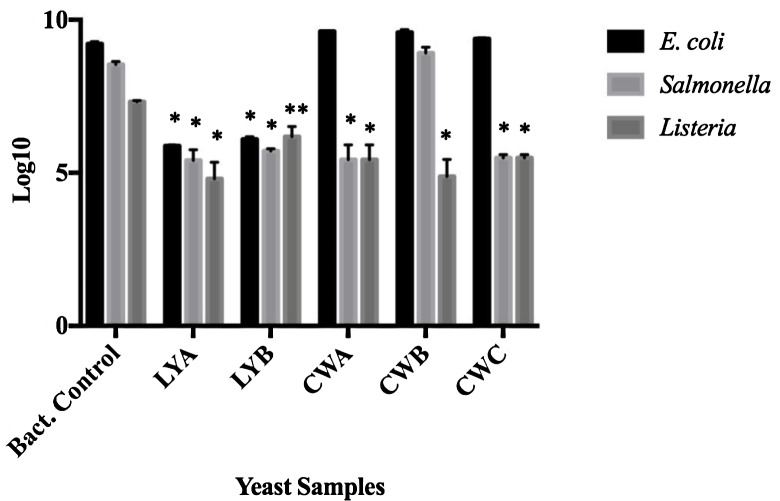
The membrane filtration adhesion assay was used to quantify the binding potential of the yeast probiotics and paraprobiotics to bacteria. To quantitate the pathogenic bacteria that remained unadhered to the yeast probiotics, filtrates from the co-incubations were analyzed by viable plate counts and compared to bacteria that were membrane filtered in the absence of yeast (Fig. 2). A decrease in the amount of E. coli O157:H7 in the filtrate was evident when co-cultured with the live yeast probiotics (LYA and LYB), indicating that this bacterium bound to both probiotics. A decrease of viable L. monocytogenes F2365 was observed in the filtrates following co-incubation with all yeast samples (P < 0.05) when compared to bacterial controls, indicating that L. monocytogenes bound to all products tested. Salmonella Typhimurium adhered to all yeast samples except for the paraprobiotic CWB.
Figure 2.
Adhesion between bacteria and yeast probiotics and paraprobiotics based on filtration analyses. Probiotics (LYA and LYB) and paraprobiotics (CWA, CWB, CWC) were co-incubated with E. coli, Salmonella, or L. monocytogenes and filtered using a 3µm membrane filter. Resulting filtrates were diluted and plated. Values represent the average filtrates from three independent experiments. Error bars represent standard error. * P < 0.001; ** P < 0.05.
To confirm that the decrease in bacterial concentration observed following the membrane filtration adhesion assay was due to bacterial binding to the yeast samples and not due to extracellular components of the yeast impeding the viability of the bacteria, the supernatants of the yeast products were co-incubated with the bacteria and viability was assessed after 4 h. None of the supernatants exhibited an effect on bacterial growth, indicating that the decrease in viable counts observed from the co-incubation was due to adherence to the yeast (data not shown).
Utilization of Yeast Probiotic Lysate as a Carbon Source for Bacterial Growth
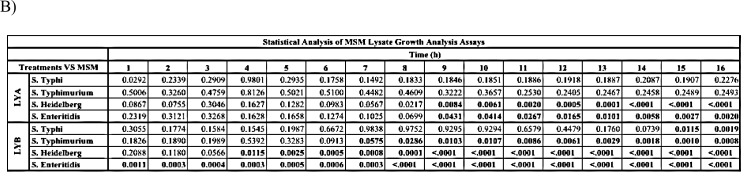
From the filtrate analysis, co-incubation of Salmonella Typhimurium with the paraprobiotic CWB resulted in a slight increase in viable bacteria. This was unexpected considering the bacteria were in a buffer that was devoid of nutrients. Therefore, to determine whether this increase was due to Salmonella utilizing components of the yeast product as a carbon source, nonviable forms of the yeast products were supplemented to media devoid of carbon and growth of Salmonella was assessed over a 16 h growth period (Fig. 3). All 4 subspecies of Salmonella tested were able to grow in the presence of LYB lysates (Fig. 3). Growth in the presence of lysates prepared from the probiotic LYA only occurred with S. Heidelberg and S. Enteriditis (Fig. 3B).
Figure 3.
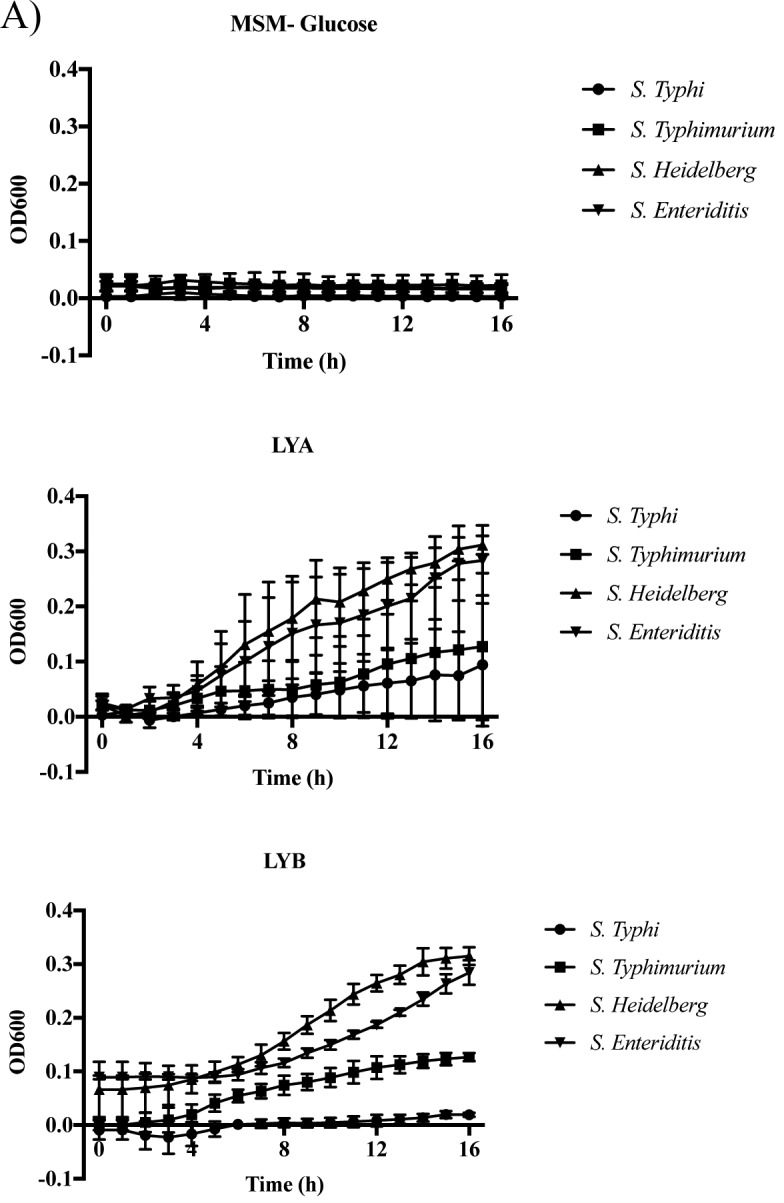
Utilization of cell-free yeast probiotic extracts as a carbon source by Salmonella. Each Salmonella strain was cultured in mineral salts media (MSM) supplemented with a cell-free lysate of each yeast probiotic (LYA and LYB). A control of each Salmonella strain was cultured in MSM (-Glucose). (A) Values represent the average OD600 from three independent replications. Error bars represent the standard error. (B) Statistical analysis of bacterial growth in lysate supplemented media in comparison to growth in MSM. Bold numbers indicate P < 0.05.
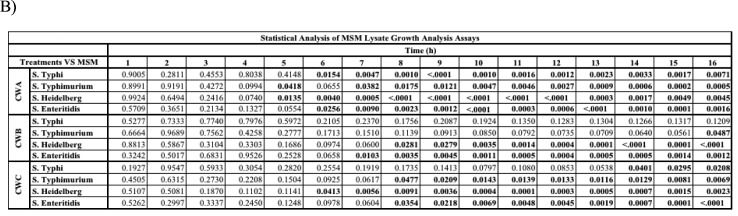
To determine whether variations existed in the use of paraprobiotics as the carbon source, growth of Salmonella Typhi, S. Typhimurium, S. Heidelberg, and S. Enteriditis was analyzed in carbon-free media supplemented with lysates from three different yeast paraprobiotics CWA, CWB, and CWC. All 4 strains of Salmonella exhibited an increase in viability over the 16 h growth period analyzed, though growth rates were different for CWA and CWC supplemented media. Ironically, S. Typhi was not able to utilize lysate CWB, whereas all other strains analyzed had an increase in growth in comparison to S. Typhi (P < 0.05; Fig. 4).
Figure 4.

Utilization of yeast paraprobiotic lysates as a carbon source by Salmonella. Each Salmonella strain was cultured in mineral salts media (MSM) media supplemented with the lysate of each yeast paraprobiotic (CWA, CWB, and CWC). A control of each Salmonella strain was cultured in MSM media (-Glucose). (A) Values represent the average OD600 from three independent replications. Error bars represent the standard error. (B) Statistical analysis of bacterial growth in lysate supplemented media in comparison to growth in MSM. Bold numbers indicate P < 0.05.
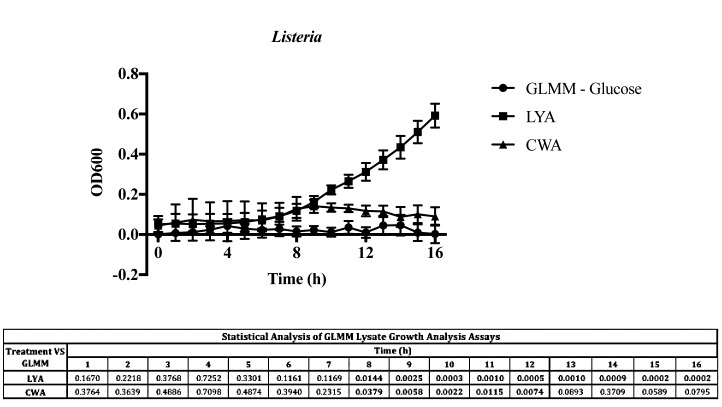
To determine whether the impact that yeast probiotics and paraprobiotics had on growth was limited to Salmonella, growth of L. monocytogenes was also analyzed in carbon-free media supplemented with these products' lysates (Fig. 5). Yeast probiotic LYA and paraprobiotic CWA were selected due to their efficiency in binding in the membrane filtration adhesion assay. Listeria monocytogenes F2365 exhibited an increase in growth with the probiotic LYA lysate (P < 0.05), but was not able to sustain growth in the presence of the paraprobiotic CWA lysate (Fig. 5).
Figure 5.
Utilization of yeast probiotics and paraprobiotics as a carbon source by Listeria monocytogenes. Listeria monocytogenes strain F2365 was cultured in three different glucose limited mineral media (GLMM): GLMM (no glucose), GLMM (with lysate of yeast paraprobiotic CWA), and GLMM (with lysate of live yeast probiotic LYA). Values represent the average OD600 from three independent replications. Error bars represent the standard error. Statistical analysis is presented for growth in media supplemented with LYA or CWA in comparison to GLMM. Bold numbers indicate P < 0.05.
DISCUSSION
To characterize the relationship between pathogenic bacteria and probiotics/paraprobiotics, qualitative and quantitative assays were used to assess the binding potential of various pathogens to probiotics and paraprobiotics. Salmonella Typhimurium and Escherichia coli have both been found to express mannose-specific adhesins that allow for direct interaction with yeast cell walls (Sharon, 1987). Previous studies have suggested a correlation between the administration of probiotics and the decrease in prevalence of E. coli O157:H7 in mature ruminants and in vitro in sheep fecal suspensions (Chaucheyras-Durand et al., 2005). Escherichia coli has also been reported to attach to the surface of S. boulardii via fimbriae interacting with mannose on the yeast cell's surface (Gedek, 1999). Variations have also been reported in the binding potential of S. boulardii and S. cerevisiae to E. coli and Salmonella. In this previous study, variations were observed in the binding potential between Gram-negative bacteria to yeast and that preference was given to S. boulardii due to the greater amount of mannose on this yeast's cell wall (Tiago et al., 2012). These previous studies provided evidence in support of the mechanism that yeast probiotics bind to pathogens, which facilitates clearance from the host and reduces binding to the intestine. However, these studies indicate that there is variation in the binding capability of pathogens to yeast probiotics and paraprobiotics. In vivo binding of these pathogens in the gastrointestinal tract may provide a plethora of benefits with regards to not only animal health, but also growth performance. Additionally, some of these pathogens are foodborne pathogens and the removal of some of these pathogens may lead to reduced carcass contamination from gastrointestinal content.
Scanning electron microscopy was used to qualitatively observe the adherence of the bacteria to the yeast products. The method utilized in this current study was unique from other studies that have measured adherence between yeast and bacteria in that the binding between the 2 microorganisms was conducted on cover slips. Cover slips were extensively washed to remove non-adhered bacteria and yeast, ensuring that the only interactions observed were due to direct adherence between the yeast and the bacteria and not potential artifacts due to processing. The live yeast probiotics exhibited the greatest amount of binding to all bacteria when compared to the control. Salmonella Typhimurium bound well to all paraprobiotics except for CWB (P < 0.01). Although it was not different when compared to the control, the binding potential of S. Typhimurium with paraprobiotic CWB was different when compared to the binding potentials of S. Typhimurium with the other yeast samples, suggesting strain specificity of S. Typhimurium to yeast probiotics and paraprobiotics. This was unexpected as S. Typhimurium bound well to all yeast samples ( > 86%; Table 2). Therefore, components of the yeast paraprobiotic CWB may have served as a source of nutrients, positively affecting the growth of S. Typhimurium.
To analyze the impact that CWB had on the viability of S. Typhimurium, 4 different strains of Salmonella were cultivated in MSM media with cell free lysates of yeast provided as the only potential carbon source. Salmonella Heidelberg and S. Enteritidis exhibited an increase in growth with lysates from both probiotics (P < 0.05), while S. Typhimurium only exhibited growth with the LYB lysates (P < 0.05) and S. Typhi had no differential growth with either probiotic lysate. The yeast paraprobiotics were also subjected to the same lysis procedure even though some may have already been fractionated due to processing. When cultivated in the MSM supplemented with paraprobiotic lysates, all Salmonella strains displayed a similar increase in growth with most of the yeast paraprobiotics. When Listeria monocytogenes was also used in this assay, the yeast probiotic LYA yielded an increase in growth, while the yeast paraprobiotic CWA could not sustain growth. These results support the hypothesis that the cellular components of these yeasts improve the viability of certain bacteria by providing a source of nutrients, but the amount of growth differs between bacteria. Further research is needed to determine how various strains of yeast affect the growth of bacteria.
Conclusion
Infectious diseases caused by pathogenic bacteria are treated by administration of antibiotics. However, with the increase in prevalence of antibiotic resistant pathogens and pending government regulation, alternative therapeutic strategies are being heavily explored. Yeast probiotics and paraprobiotics offer much promise as therapies and preventative feed additives against infectious agents, though little is known in regards to the interaction between these products and pathogens. This study analyzed the adherence between probiotics and paraprobiotics against a variety of Gram-positive and Gram-negative bacteria that are responsible for animal diseases. Although adhesion was observed with all bacterial strains, the binding potentials were strain-specific and yeast sample type-specific. Further research is warranted to conclude the best strategies for designing probiotic therapies against infectious diseases.
Footnotes
This work was supported Phileo-Lesaffre Feed Additives. Work in the laboratory of JRD and JAT is also supported by the National Institutes of Health, award number P20GM103646.
LITERATURE CITED
- Braude R., Kon S. K., White E. G.. 1944. Yeast as a protein supplement for pigs: Further observations on its rachitogenic effect. J. Comp. Pathol. Ther. 54:88–96. doi: 10.1016/S0368-1742(44)80009-1 [DOI] [Google Scholar]
- Chaucheyras-Durand F., Fonty G.. 2002. Influence of a probiotic yeast (Saccharomyces cerevisiae CNCM I-1077) on microbial colonization and fermentations in the rumen of newborn lambs. Microb. Ecol. Health Dis. 14:30–36. doi: 10.1080/089106002760002739 [DOI] [Google Scholar]
- Chaucheyras-Durand F., Masseglia S., Fonty G.. 2005. Effect of the microbial feed additive Saccharomyces cerevisiae CNCM I-1077 on protein and peptide degrading activities of rumen bacteria grown in vitro. Curr. Microbiol. 50:96–101. doi: 10.1007/s00284-004-4433-1 [DOI] [PubMed] [Google Scholar]
- Dawson K. A. 2001. The application of yeast and yeast derivatives in the poultry industry. Proc. Aust. Poult. Sci. Sym. 13:100–105. [Google Scholar]
- Donaldson J. R., Shields-Menard S., Bernard J. M., Revellame E., Hall J. I., Lawrence A., Wilson J. G., Lipzen A., Martin J., Schackwitz W., Woyke T., Shapiro N., Biddle K. S., Holmes W. E., Hernandez R., French W. T.. 2014. Characterization of the novel Enterobacter cloacae strain JD6301 and a genetically modified variant capable of producing extracellular lipids. Agricul. Food Anal. Bacteriol. 4:212–223. [Google Scholar]
- Duarte K. M. R., Gomes L. H., Sampaio A. C. K., Issakowicz J., Rocha F., Granato T. P., Terra S. R.. 2012. Saccharomyces Cerevisiae used as probiotic: Strains characterization And cell viability. J. Agricul. Vet. Sci. 1:17–19. doi: 10.9790/2380-0121719 [DOI] [Google Scholar]
- Dunlop R. H., McEwen S. A., Meek A. H., Clarke R. C., Black W. D., Friendship R. M.. 1998. Associations among antimicrobial drug treatments and antimicrobial resistance of fecal Escherichia coli of swine on 34 farrow-to-finish farms in Ontario, Canada. Prev. Vet. Med. 34:283–305. doi: 10.1016/S0167-5877(97)00095-0 [DOI] [PubMed] [Google Scholar]
- FAO/WHO 2002. Guidelines for the evaluation of probiotics in food. Report of a Joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food; Ontario, Canada: (Accessed 15 August 2016.) ISBN: 92-5-105513-0 [Google Scholar]
- Gedek B. R. 1999. Adherence of Escherichia coli serogroup O 157 and the Salmonella Typhimurium mutant DT 104 to the surface of Saccharomyces boulardii. Mycoses 42:261–264. doi: 10.1046/j.1439-0507.1999.00449.x [DOI] [PubMed] [Google Scholar]
- Kollar R., Reinhold B. B., Petráková E., Yeh H. J., Ashwell G., Drgonová J., Kapteyn J. C., Klis F. M., Cabib E.. 1997. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→)3-glucan, and chitin. J. Biol. Chem. 272:17762–17775. doi: 10.1074/jbc.272.28.17762 [DOI] [PubMed] [Google Scholar]
- Korhonen T. K., Leffler H., Svanborg Eden C.. 1981. Binding specificity of piliated strains of Escherichia coli and Salmonella Typhimurium to epithelial cells, Saccharomyces cerevisiae cells, and erythrocytes. Infect. Immun. 32:796–804. doi: 0019-9567/81/050796-09$02.00/0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F. S., Dalmasso G., Arantes R. M. E., Doye A., Lemichez E., Lagadec P., Imbert V., Peyron J. F., Rampal P., Nicoli J. R., Czerucka D.. 2010. Interaction of Saccharomyces boulardii with Salmonella enterica serovar Typhimurium protects mice and modifies T84 cell response to the infection. PLoS One 5:e8925. doi: 10.1371/journal.pone.0008925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F. S., Nardi R. M., Arantes R. M., Rosa C. A., Neves M. J., Nicoli J. R.. 2005. Screening of yeasts as probiotic based on capacities to colonize the gastrointestinal tract and to protect against enteropathogen challenge in mice. J. Gen. Appl. Microbiol. 51:83–92. doi: 10.2323/jgam.51.83 [DOI] [PubMed] [Google Scholar]
- Merritt M. E., Donaldson J. R.. 2009. Effect of bile salts on DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 58:1533–1541. doi: 10.1099/jmm.0.014092-0 [DOI] [PubMed] [Google Scholar]
- Middelbos I. S., Godoy M. R., Fastinger N. D., Fahey G. C. Jr. 2007. A dose–response evaluation of spray-dried yeast cell wall supplementation of diets fed to adult dogs: Effects on nutrient digestibility, immune indices, and fecal microbial populations. J. Anim. Sci. 85:3022–3032. doi: 10.2527/jas.2007-0079 [DOI] [PubMed] [Google Scholar]
- Ofek I., Mirelman D., Sharon N.. 1977. Adherence of Escherichia coli to human mucosal cells mediated by mannose receptors. Nature 265:623–625. doi: 10.1038/265623a0 [DOI] [PubMed] [Google Scholar]
- Reid G., Jass J., Sebulsky M. T., McCormick J. K.. 2003. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 16:658–672. doi: 10.1128/CMR.16.4.658-672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez N. C., Young T. R., Carroll J. A., Corley J. R., Rathmann R. J., Johnson B. J.. 2014. Yeast cell wall supplementation alters the metabolic responses of crossbred heifers to an endotoxin challenge. Innate Immun. 20:104–112. doi: 10.1177/1753425913482152 [DOI] [PubMed] [Google Scholar]
- Schneebeli R., Egli T.. 2013. A defined, glucose-limited mineral medium for the cultivation of Listeria spp. Appl. Environ. Microbiol. 79:2503–2511. doi: 10.1128/AEM.03538-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N. 1987. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Letters. 217:145–157. doi: 10.1016/0014-5793(87)80654-3 [DOI] [PubMed] [Google Scholar]
- Shoaf-Sweeney K. D., Hutkins R. W.. 2009. Adherence, anti-adherence, and oligosaccharides preventing pathogens from sticking to the host. Adv. Food Nutr. Res. 55:101–61. doi: 10.1016/S1043-4526(08)00402-6 [DOI] [PubMed] [Google Scholar]
- Taverniti V., Guglielmetti S.. 2011. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes Nutr. 6:261–274. doi: 10.1007/s12263-011-0218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrune M., Bach A., Ruiz-Moreno M., Stern M. D.. 2009. Effects of Saccharomyces cerevisiae on ruminal pH and microbial fermentation in dairy cows: Yeast supplementation on rumen fermentation. Livest. Sci. 124:261–265. doi: 10.1016/j.livsci.2009.02.007 [DOI] [Google Scholar]
- Tiago F. C., Martins F. S., Souza E. L., Pimenta P. F., Araujo H. R., Castro I. M., Brandäo R. L., Nicoli J. R.. 2012. Adhesion to the yeast cell surface as a mechanism for trapping pathogenic bacteria by Saccharomyces probiotics. J. Med. Microbiol. 61:1194–1207. doi: 10.1099/jmm.0.042283-0 [DOI] [PubMed] [Google Scholar]
- Tripathi M. K., Karim S. A.. 2011. Effect of yeast cultures on supplementation on live weight change, rumen fermentation, ciliate protozoa population, microbial hydrolytic enzymes status and slaughtering performance of growing lamb. Liv. Sci. 135:17–25. doi: 10.1016/j.livsci.2010.06.007 [DOI] [Google Scholar]
- van der Peet-Schwering C. M., Jansman A. J., Smidt H., Yoon I.. 2007. Effects of yeast culture on performance, gut integrity, and blood cell composition of weanling pigs. J. Anim. Sci. 85:3099–3109. doi: 10.2527/jas.2007-0110 [DOI] [PubMed] [Google Scholar]