Abstract
We present here the differential analysis of metabolite–metabolite association networks constructed from an array of 24 serum metabolites identified and quantified via nuclear magnetic resonance spectroscopy in a cohort of 825 patients of which 123 died within 2 years from acute myocardial infarction (AMI). We investigated differences in metabolite connectivity of patients who survived, at 2 years, the AMI event, and we characterized metabolite–metabolite association networks specific to high and low risks of death according to four different risk parameters, namely, acute coronary syndrome classification, Killip, Global Registry of Acute Coronary Events risk score, and metabolomics NOESY RF risk score. We show significant differences in the connectivity patterns of several low-molecular-weight molecules, implying variations in the regulation of several metabolic pathways regarding branched-chain amino acids, alanine, creatinine, mannose, ketone bodies, and energetic metabolism. Our results demonstrate that the characterization of metabolite–metabolite association networks is a promising and powerful tool to investigate AMI patients according to their outcomes at a molecular level.
Keywords: metabolite−metabolite association networks, nuclear magnetic resonance, metabolomics, network inference, acute myocardial infarction
1. Introduction
Acute myocardial infarction (AMI) is associated with high mortality and (co)morbidity.1,2 A favorable outcome of the patients is directly linked to a rapid and effective management of the condition, while a deferred diagnosis can result in severe clinical conditions. Unfortunately, stratification of AMI patients in different risk categories and their management remain a clinical and a research challenge.3−6 Several studies evaluated the clinical usefulness of prognostic biomarkers for (early) identification of patients at risk of a negative clinical outcome. Several studies have reported prognostic utility of high levels of inflammatory markers like CRP (C-reactive protein) and IL-8 (interleukin-8) in patients with acute coronary syndrome (ACS) who underwent coronary revascularization,7−9 often in combination with metabolite profiling.10−12
Nuclear magnetic resonance (NMR)-based high-throughput metabolomics analysis of hundreds of different metabolites present in a biological specimen13−15 such as blood, urine, and tissue can provide a global picture of the many metabolic and biomolecular processes underlying complex and multifactorial diseases16−20 such as acute coronary syndrome.
Through an in-depth NMR metabolomics analysis of serum samples of AMI patients in the acute phase, we provided recently a very promising discrimination between patients with different clinical outcomes.21 Multivariate and univariate analyses were applied to infer a pattern of metabolic alterations that identifies high risk of death within 2 years after AMI, but mechanistic understanding of the underlying risk factors was not investigated in a systemic fashion,22 which can enhance comprehension and characterization of both the biological process that are perturbed and the key regulatory mechanisms underlying disease pathophysiology.
Integrative systems biology approaches offer a holistic representation of the structural and functional properties of living organisms and can help shed light on the relationships and interaction among molecular entities governing the system behavior. In this framework, network representations and network analysis provide a unique view to understand biological systems, where not only the individual components are considered but also their interconnections and their functions as a whole, and constitute a valuable integration to classical multivariate data analysis tools.23
Network analysis has proven to be a powerful tool to analyze, understand, and interpret the complex patterns observed in metabolomics data.23−27 The rationale underlying the use of network analysis is that metabolite concentrations (like genes and proteins) measured in blood or other biofluids exhibit association patterns that can be considered, to a certain extent, related to the structure of the underlying biological and metabolic networks.
In the metabolomics context, networks are best exploited when compared across different conditions in a so-called differential network analysis approach:23,28 different network characteristics and different patterns of association between metabolites can highlight possibly affected molecular mechanisms. In this work, we attempt with our previous metabolomics investigation21 a more systematic approach using the analysis of metabolite–metabolite association networks to investigate the possible molecular mechanisms underlying different clinical outcomes (i.e., death as a consequence of adverse cardiovascular event) in AMI patients treated by percutaneous coronary intervention (PCI). The patients of this study are a subset of the AMI-Florence II cohort.21,29
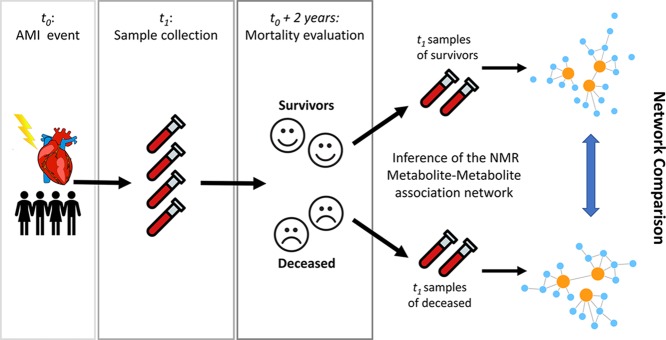
The first objective was to investigate, from a metabolite–metabolite association network point of view, the differences in the baseline characteristic of patients who, after 2 years, had survived the AMI with respect to those who died. To this aim, metabolite–metabolite association networks were built starting from the serum metabolites quantified using NMR spectra of serum samples that have been collected from patients in the 24–48 h after the AMI: samples were then split into two groups (alive at 2 years from AMI and deceased), and metabolite networks were compared as shown in Figure 1.
Figure 1.
Graphical overview of the analysis design to investigate differences between metabolite–metabolite association networks of patients who survived and did not survive 2 years after AMI. The AMI event is recorded at time t0, and blood samples, analyzed in this study and the previous one, were collected at time t1 (24–48 h after percutaneous coronary intervention and overnight fasting). Survival status was evaluated after 2 years (time t1 + 2 years), and samples are retrospectively split into two groups according to the survival status (alive vs deceased). Metabolite–metabolite association networks were inferred from the two groups using the PCLRC algorithm23 and compared to detect metabolites with differential connectivity with respect to the survival status.
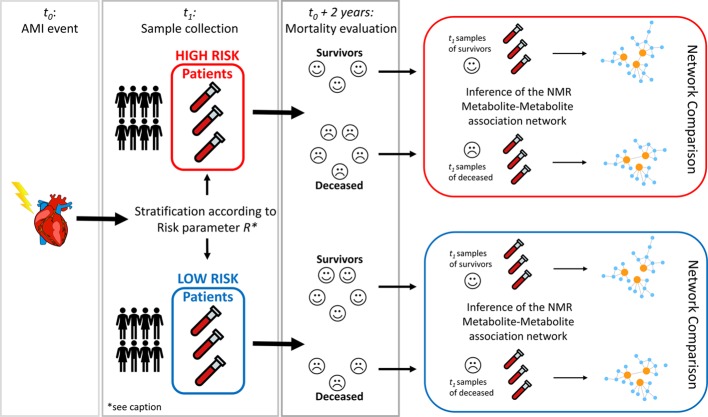
The second objective of the study was to investigate the (possibly different) molecular mechanisms underlying different risk scores used in clinical practice to stratify patients of different categories of risk of mortality after AMI; therefore, we aimed to investigate if patients at low- or high-risk mortality after AMI were characterized by risk-level-specific metabolite–metabolite association networks.
Different risk parameters like the ACS classification,30 the Killip index,31 and the GRACE (Global Registry of Acute Coronary Events) score32 are routinely used to stratify different classes of risks of mortality after the AMI, and since they are based on different input parameters, they reflect different patient characteristics and may assign patients to different risk categories. We investigate the differences in the metabolite–metabolite networks underlying these three risk parameters in addition to the metabolomics NOESY RF risk score, which was developed in our previous study,21 taking into account also whether the patients survived or not 2 years post-AMI. Practically, we compared the networks of the patients who survived the AMI at 2 years within both risk classes (low and high) for every risk parameter considered. The overall analysis design is graphically illustrated in Figure 2.
Figure 2.
Graphical overview of the analysis design to investigate differences between metabolite–metabolite association networks underlying different mortality risk factors. The AMI event is recorded at time t0, and blood samples, analyzed in this study and the previous one, were collected at time t1 (24–48 h after percutaneous coronary intervention and overnight fasting). Given a risk factor R (with R equal to Killip, STEMI, GRACE, and Metabolomics RF), patient samples are divided into two groups, high risk of mortality and low risk of mortality. Patients in the high-risk group are then evaluated for mortality at 2 years after AMI, and samples are split according to the survival status (alive vs deceased.) Metabolite–metabolite association networks were inferred from the two groups (alive vs deceased) using the PCLRC algorithm23 and compared to detect metabolites with differential connectivity with respect to the survival status within patients initially classified to be at high risk of mortality. The same analysis was performed on the sample of patients in the low-risk group. Overall, the analysis resulted in 16 different networks as shown in Figure 3. *The generic risk factor R assumes values: Killip, STEMI, GRACE, and Metabolomics RF (see Methods for more details).
2. Methods
2.1. Study Population
The study population consists of patients admitted to the Coronary Unit of the six hospitals (five community hospitals, namely, Santa Maria Annunziata Hospital, Santa Maria Nuova Hospital, Nuovo San Giovanni di Dio Hospital, Mugello Hospital, and Figline Hospital, and one University hospital, the Careggi Hospital) of the Florence health district (Tuscany, Italy) who participated in the AMI-Florence 2 registry29 between April 2008 and April 2009.
Demographic information, medical history, clinical characteristics, treatments, and outcomes during hospitalization were collected and are reported in Table 1. Inclusion criteria and details about patient treatment are available to the reader in the publication describing the original study.21
Table 1. Demographic and Clinical Characteristicsa.
| parameters | survivor patients (702) | deceased patients (123) | P-value adjusted |
|---|---|---|---|
| demographic characteristics | |||
| age (years), median (IQR) | 73 (63–80) | 82 (78–87) | 1.15 × 10–20 |
| gender, females, n (%) | 253 (36.0%) | 58 (47.1) | 2.08 × 10–01 |
| medical history, n (%) | |||
| chronic heart failure | 29 (4.1%) | 20 (16.3%) | 1.73 × 10–06 |
| diabetes | 169 (24.1%) | 49 (39.8%) | 2.91 × 10–03 |
| hypertension | 457 (65.1%) | 88 (71.5%) | 1.00 × 10+00 |
| dyslipidemia | 240 (34.2%) | 26 (21.1%) | 7.47 × 10–02 |
| cerebrovascular disease | 42 (6.0%) | 24 (19.5%) | 3.83 × 10–06 |
| risk features | |||
| ACS classification, STEMI, n (%) | 257 (36.6%) | 26 (21.1%) | 9.41 × 10–03 |
| Killip II-III, n (%) | 101 (14.4%) | 52 (42.3%) | 2.33 × 10–12 |
| GRACE score ≥ 170, n (%) | 501 (71.4%) | 116 (94.3%) | 7.13 × 10–07 |
| NOESY RF risk score ≥ 0.454, n (%) | 197 (28.1%) | 92 (74.8%) | 1.34 × 10–22 |
IQR, interquartile range; ACS, acute coronary syndrome; STEMI, ST-segment elevation myocardial infarction; GRACE, Global Registry of Acute Coronary Events risk score; NOESY RF, nuclear Overhauser effect spectroscopy random forest risk score. A P value adjusted with the Bonferroni correction <0.05 is deemed significant.
We considered only complete cases from the original study:21 153 subjects were excluded because the data set was missing relevant clinical information necessary for the calculation of the risk scores, leaving 825 patients available for analysis: 311 females (37.7%) and 514 males (62.3%), with a median age of 75 years, of which 123 (14.9%) died within 2 years from the AMI event (labeled “deceased patients” in the remainder of the paper) and 702 (85.1%) patients have lived for at least 2 years after the AMI event (labeled “survivor patients”).
Survival status after 2 years from the AMI event was obtained by consultation of the registry office’s city of residence of the patients.
2.2. Ethical Issues
The study received approval on March 19, 2008 from the Ethical Committee of the University of Florence and the Careggi Hospital (study number 11/2008). Informed, written consent was obtained from all participants. The study adheres to the directives of the Declaration of Helsinki (1964) and its later amendments.
2.3. Overview of Mortality Risk Scores
The Global Registry of Acute Coronary Events (GRACE) risk score and the metabolomics nuclear Overhauser effect spectroscopy random forest risk score were calculated as described in the original publication.21 For the network analyses here proposed, the ACS classification, Killip classification, GRACE score, and NOESY RF score were considered in order to obtain high- and low-risk classes for cardiovascular risk of death after AMI.
AMI patients are subdivided into ST elevation myocardial infarction (STEMI) and Non-ST elevation myocardial infarction (NSTEMI) if they exhibit, or not, significant ST segment elevations on the electrocardiogram. STEMI is characterized by total occlusion in a coronary artery, whereas in NSTEMI, the occlusion is partial; for these reasons, NSTEMI are considered at a lower risk of death with respect to STEMI patients.
The classification or index of heart failure severity proposed by Killip and Kimball31 in AMI patients aims at assessing the risk of in-hospital death and the potential benefit of specialized care in coronary care units. Patients are classified into four classes based on physical examination: patients in class I demonstrate no clinical signs of heart failure (HF); class II patients exhibit findings consistent with mild to moderate HF; class III patients display overt pulmonary edema; and patients in class IV are in cardiogenic shock or arterial hypotension with evidence of peripheral vasoconstriction. We considered Killip class I patients low risk and all the other classes high risk.
The Global Registry of Acute Coronary Events hospital discharge risk score (GRACE score), developed from a multinational registry involving all subsets of ACS, predicts long-term mortality post-ACS.32 Variables measured to calculate the score include age, heart rate, systolic blood pressure, renal function, Killip class, ST-segment deviation, cardiac arrest, and elevated cardiac enzyme levels. AMI patients with a GRACE score >170 are considered high risk.
The present cohort of AMI patients was previously utilized for large-scale NMR-based profiling of serum samples aiming at the metabolomics characterization of acute myocardial infarction patients to define a metabolomics score able to predict 2 year post-AMI mortality.21 A random forest (RF) classifier33 was built to discriminate the NOESY NMR spectra of patients who died or survived the AMI event within two years. A patient-specific score, called “NOESY RF risk score”, was defined in the training set by quantifying the similarity of the blood NMR fingerprint of a given patient to those of patients who died; patients with NOESY RF score ≥ 0.454 are deemed at a high risk of death 2 years after AMI.
2.4. NMR Sample Collection and Preparation
Blood samples were collected in the 24–48 h after the PCI and overnight fasting. Serum samples were obtained by centrifuging blood sample at 2000g for 10 min at 4 °C and then stored in aliquots at −80 °C until analysis.
Samples were prepared for NMR experiments as detailed in Bernini et al.34
2.5. Experimental Methods
2.5.1. NMR Experiments
One-dimensional 1H NMR spectra were acquired on a Bruker 600 MHz spectrometer (Bruker BioSpin) operating at 600.13 MHz and equipped with a 5 mm cryoprobe, an automatic tuning-matching (ATM), and an automatic sample changer.
A water-suppressed Carr–Purcell–Meiboom–Gill (CPMG)35 spin echo pulse was used to obtain spectra with attenuated broad signals from lipids and proteins. More details on instrument configuration and setting and on NMR experiments can be found in the original publication.21
2.5.2. Metabolite Identification and Quantification
Metabolites were identified on the CPMG spectra using AMIX 3.8.4, the BBIOREFCODE (Bruker BioSpin), and the Human Metabolome Database (HMDB).36
The metabolite selection was purely based on an experimental ground and not because they were a priori known or supposed to be associated with AMI or post-AMI death. The quantification (in arbitrary units) of metabolites above the detection limit (1 μM) was carried on using software developed in-house based on standard line-shape analysis methods.
Data have been deposited in the MetaboLights database (www.ebi.ac.uk/metabolights) under the accession number MTBLS395 from where they can be freely retrieved.
2.6. Network Analysis
Differential network analysis was used to investigate the changes in metabolite–metabolite association patterns between patients who survived and those who did not survive the AMI event at 2 years (see Figure 1) and to investigate differences among high- and low-mortality-risk patients (see Figure 2).
The AMI event is recorded at time t0, and blood samples analyzed in this study and in a previous one21 were collected at time t1 (between 24–48 h after percutaneous coronary intervention and overnight fasting, see Figure 1). Survival status was evaluated after 2 years (time t1 + 2 years), and samples were retrospectively split into two groups according to the survival status (alive vs deceased). Network inference, comparison, and validation were performed as detailed in Section 2.6.1 and 2.6.2.
To investigate the differences between metabolite–metabolite association networks underlying different mortality risk factors, the samples collected at t1 were retrospectively stratified to groups with high risk of mortality and low risk of mortality according to a given mortality risk index (Killip, STEMI, GRACE, or NOESY RF scores; see Figure 2 for a graphical overview of the analysis setting and see Section 2.3 for more details on the risk scores). For each of the two different risk groups (high/low), patients were then retrospectively evaluated for mortality at 2 years after AMI, and samples were divided according to the survival status (alive vs deceased); metabolite–metabolite association networks were inferred from the two groups (alive vs deceased) as detailed in Section 2.6.1. This resulted in 16 different networks: 4 mortality risk factors × 2 risk statuses (high/low) × 2 survival at 2 years after AMI statuses (alive/deceased) = 16.
2.6.1. Network Reconstruction
The probabilistic context likelihood of relatedness (PCLRC)23 algorithm was used to build metabolite–metabolite correlation networks. This algorithm is a modification of the context likelihood of relatedness (CLR) algorithm (using correlation instead of mutual information to measure similarity between metabolite profiles) and on iteratively sampling the data set resulting in a weighted adjacency matrix containing an estimate of the association likeliness between any two metabolites. The default values of 25–75% data split and 90% confidence level have been used. An R implementation of this algorithm is available at semantics.systemsbiology.nl.
The PCLRC outputs a matrix containing a probabilistic measure pij for each correlation rij (which is the ith and jth element of the Spearman correlation matrix R) between any two metabolites i and j. We retain correlations for which pij > 95%, setting to 0 all other correlations:
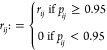 |
For each metabolite i in network a, the connectivity is defined as
For the metabolite i, the differential connectivity Δi calculated among two networks a and b is
2.6.2. Assessing the Significance of Differential Connectivity
The significance of the differential connectivity was assessed implementing a permutation test. First, each column of each data matrix X is permuted independently so that the column values x1, x2, ..., xn are replaced by xp(1), xp(2), ..., xp(n) where p(1), p(2), ..., p(n) are random permutations of 1, 2, ..., n. In this way, the distributional properties of every column of X (mean and variance) are preserved but the relationships among the variables of different columns are destroyed. The permuted version of X is collected in the matrix Xk.
The overall network estimation using the PCLR algorithm is performed on the permuted versions of the data matrix, obtaining the corresponding correlation matrix Rk, which are then used to compute, for each metabolite, the “permuted” connectivity
and the permuted differential connectivity
The permutation procedure is repeated k = 1, 2, ...., K times to build a distribution Di of permuted differential connectivity values for metabolite i, which is used to compute the significance of the differential connectivity of metabolite i among two networks a and b. The significance is expressed as a P-value calculated as
where #(|Di|>|Δia, b|) indicates the number of elements of Di whose absolute value is larger than the absolute value of the differential connectivity calculated from the original data.
2.7. Statistical Methods
2.7.1. Univariate Analysis
Group comparisons were carried out using the Wilcoxon–Mann–Whitney test38 (for metabolite concentrations), t test (demographic and clinical variables), and chi-square test (categorical clinical variables). All P-values were adjusted for multiple test correction using either the Benjamini–Hochberg39 method (metabolites) or Bonferroni correction37 (all other variables).
An adjusted P-value <0.05 was deemed significant.
2.7.2. Simultaneous Analysis of Adjacency Matrices
Covariance simultaneous component analysis (COVSCA) is a recently introduced model to analyze communalities and differences across a set of Sk (k = 1,2,..., K) covariance matrices simultaneously.39 As an adjacency matrix can be considered a particular case of the covariance matrix, this method can be used to model communalities and differences between adjacency matrices. In COVSCA, the matrices are approximated as a combination of a limited number (L ≪ K) of low dimensional prototypes
where ckl ≥ 0 (l = 1, 2, ..., L) are weight coefficients and ZlZlT are the prototypical covariance matrices that consist of loading Z of size J × Rl that hold simultaneously for all Sk. We have chosen to fit our model with two rank-2 prototype matrices as the best compromise between fit (70%) and model complexity. We refer the reader to the original publication for more details on model derivation and implementation.
COVSCA scores were analyzed with the k-nearest neighbor classification algorithm to obtain the discrimination accuracy, and the result was validated using a permutation test using 103 permutations.
3. Results and Discussion
3.1. Differential Network Analysis of Survivor- and Deceased-Specific Metabolite–Metabolite Association Networks
The analysis of some baseline patient characteristics identifies multiple factors that are significantly different between who had or had not died within 2 years since their AMI presentation (see Table 1) and constitute possible confounding factors when comparing the survival status of the groups. For this reason, metabolite profiles were corrected for age and history of chronic heart failure, diabetes, and cerebrovascular disease prior network inference.
The specific serum metabolite association networks for deceased and survivor patients estimated using the PCLRC23 algorithm are shown in Figure 3A (survivors, constructed using n = 702 serum samples) and Figure 3B (deceased, constructed using n = 123 serum samples). Network nodes were arranged and colored according to the metabolite degree of connections, whereas the width of network edges represents metabolite–metabolite correlations and edge colors represent edge betweenness.
Figure 3.
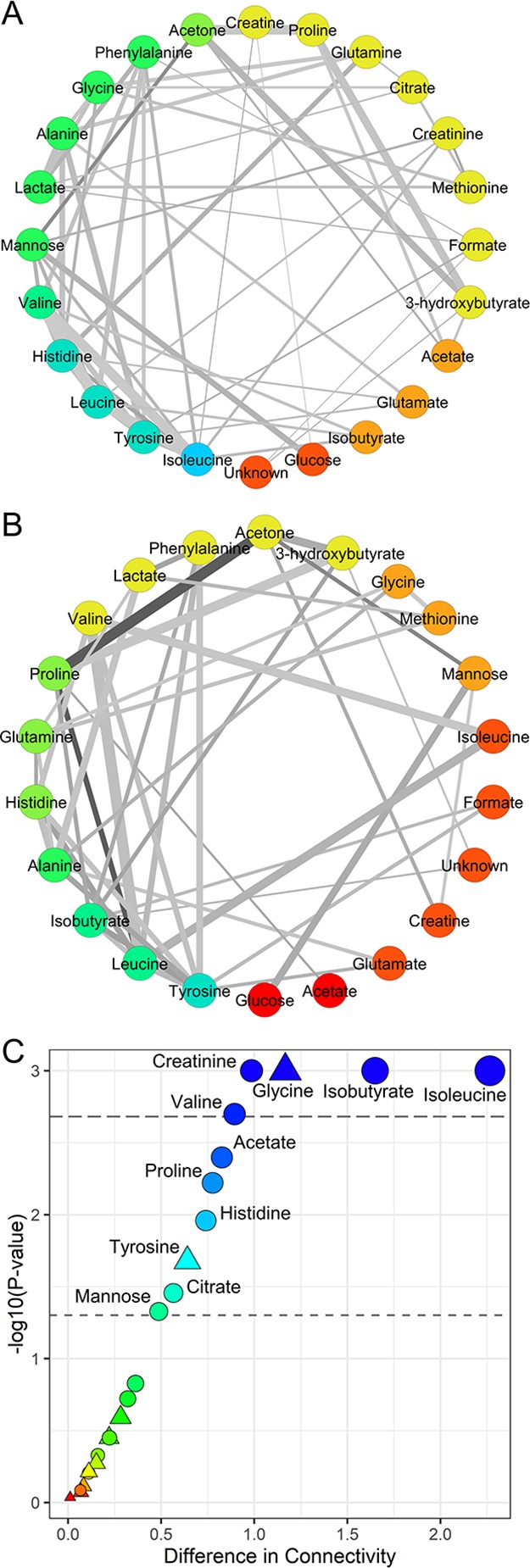
Metabolite–metabolite association networks reconstructed using the PCLRC algorithm from the serum metabolite concentration profiles. (A) Survived and (B) deceased AMI patients within 2 years from the cardiovascular event; vertexes are colored according to the metabolite degree of connectivity. (C) Differences in terms of connectivities in networks of survived and deceased are reported against each metabolite’s P-value. The thresholds for significance at 0.05 and after Bonferroni correction are given. The colors red to blue encode for the increasing difference. Significance in terms of metabolite levels is also reported: circles are metabolites that are statically different between survived and deceased patients, whereas triangles encode for the not significant ones.
We observed that the metabolite network specific to patients who survived the AMI at 2 years tends to be more connected with respect to the ones of deceased patients. This different topology is likely to reflect underlying metabolic differences, but it could be also affected by the differences in the sample size.
We assessed the significance of the observed differences in connectivity using a permutation test: isobutyrate, glycine, creatinine, isoleucine, and valine present statistically different connectivities (P-value <0.05 after Bonferroni correction) in survivor and deceased patient-specific networks. Among these metabolites, only glycine (Figure 3C) did not show also statistically significant differences in terms of concentrations (P-value <0.05 after Benjamini–Hochberg correction).
Peculiar is the behavior of creatinine: in survivor-specific network, creatinine shows four connections with mannose, methionine, leucine and isoleucine, whereas creatinine in deceased patient-specific network is completely disconnected; previous studies have demonstrated the association between compromised myocardial flow and negative prognosis for 1 year survival and a high level of creatinine in patients.40,41
Correlation between creatinine and mannose has been previously observed,42 suggesting an association between insulin resistance and diabetic kidney disease,43 which is a well-established risk factor for CVD as well as for overt diabetic kidney disease. Interestingly, results in our data set confirm this observation only in survivor patients: creatinine correlates significantly with mannose as well as with leucine, isoleucine, and methionine only in survivor patients, whereas it does not have any significant correlation in deceased patients. Disruption of this correlation could indicate a possible impairment or re-wiring of mannose metabolism, which has been considered as a potential biomarker for the prediction of CVD. Possible implications of mannose are further discussed in Section 3.2. As already shown in our previous study,21 creatinine presents statistically higher concentrations in patients who died with respect to survivors (Figure 3C). Both these pieces of evidence suggest a central metabolic role of creatinine, which may condition the prognosis of AMI patients. Moreover, increase of serum creatinine levels has been related to adverse outcomes, including an increased risk of in-hospital mortality, in patients undergoing primary percutaneous coronary intervention for AMI. It has been proposed that adopting interventions in the patient management able to reduce rises in creatinine should improve outcomes of AMI patients.44 Moreover, creatinine has been shown to be inversely associated with incident CVD while mannose was found to be directly associated with incident CVD.45
Glycine does not show any significant change in concentrations between survivor and deceased patients, whereas it shows significantly higher connections in survivors (Table 2). Previous studies demonstrated that high plasma glycine levels were associated with an overall favorable coronary heart disease risk profile.46 We may assume that a higher glycine degree of connectivity probably reflects a major contribution of glycine metabolism in the AMI patient prognosis, and indeed, serum glycine has been correlated with anti-inflammatory effects in animal models.47,48
Table 2. Metabolite Univariate Analysisa.
| molecule | survivors (702) | deceased (123) | P-value | P-value adjusted |
|---|---|---|---|---|
| creatinine | 196.6 ± 52.2 | 261.0 ± 108.5 | 1.05 × 10–10 | 2.53 × 10–09 |
| proline | 115.2 ± 60.0 | 154.8 ± 95.3 | 3.43 × 10–07 | 4.11 × 10–06 |
| formate | 9.4 ± 3.4 | 11.4 ± 3.8 | 3.10 × 10–06 | 2.48 × 10–05 |
| unknown | 10.2 ± 11.6 | 19.8 ± 21.2 | 5.94 × 10–06 | 3.56 × 10–05 |
| valine | 1122.6 ± 237.2 | 1026.2 ± 257.4 | 6.67 × 10–05 | 3.20 × 10–04 |
| 3-hydroxybutyrate | 329.1 ± 320.1 | 487.1 ± 523.7 | 4.83 × 10–04 | 1.93 × 10–03 |
| mannose | 105.4 ± 34.5 | 1229 ± 55.9 | 8.27 × 10–04 | 2.84 × 10–03 |
| histidine | 114.2 ± 21.6 | 106.9 ± 23.7 | 3.84 × 10–03 | 1.15 × 10–02 |
| acetate | 78.9 ± 47.1 | 97.9 ± 67.2 | 7.54 × 10–03 | 1.85 × 10–02 |
| acetone | 735.2 ± 559.6 | 900.44 ± 824.9 | 7.71 × 10–03 | 1.85 × 10–02 |
| isobutyrate | 30.3 ± 12.0 | 35.54 ± 14.6 | 9.18 × 10–03 | 2.00 × 10–02 |
| citrate | 89.7 ± 29.8 | 99.84 ± 39.6 | 1.23 × 10–02 | 2.47 × 10–02 |
| glutamine | 188.3 ± 46.5 | 203.89 ± 71.3 | 1.69 × 10–02 | 2.98 × 10–02 |
| glucose | 2765.7 ± 745.9 | 3037.1 ± 1067.3 | 1.74 × 10–02 | 2.98 × 10–02 |
| isoleucine | 162.0 ± 42.4 | 151.6 ± 41.0 | 2.55 × 10–02 | 4.08 × 10–02 |
| phenylalanine | 227.84 ± 58.7 | 231.1 ± 63.9 | 4.85 × 10–02 | 7.27 × 10–02 |
| alanine | 1488.5 ± 364.0 | 1395.3 ± 323.2 | 7.04 × 10–02 | 9.93 × 10–02 |
| glutamate | 205.5 ± 105.7 | 181.0 ± 114.1 | 8.69 × 10–02 | 1.16 × 10–01 |
| glycine | 522.18 ± 162.0 | 478.2 ± 157.4 | 1.21 × 10–01 | 1.53 × 10–01 |
| methionine | 109.6 ± 34.8 | 115.9 ± 43.2 | 2.54 × 10–01 | 3.04 × 10–01 |
| leucine | 520.9 ± 119.0 | 508.6 ± 146.7 | 3.33 × 10–01 | 3.80 × 10–01 |
| lactate | 1515.5 ± 416.4 | 1578.3 ± 611.9 | 8.55 × 10–01 | 8.98 × 10–01 |
| tyrosine | 169.1 ± 36.9 | 169.5 ± 45.7 | 8.60 × 10–01 | 8.98 × 10–01 |
| creatine | 101.9 ± 60.0 | 100.1 ± 80.7 | 9.61 × 10–01 | 9.61 × 10–01 |
List of metabolites assigned and quantified in the serum NMR spectra, reported as median with median absolute deviation (arbitrary units). A P-value adjusted with the Benjamini–Hochberg correction <0.05 is deemed significant.
For 3-hydroxybutyrate, we found increased serum levels (Table 2) in deceased patients, whereas no difference in connectivity is present. A recent study indicates that ketone body consumption is reduced under conditions of myocardial ischemia, probably suggesting a shift in the energy metabolism to anaerobic pathways.49,50 We can hypothesize that the onset of metabolic ketosis may influence the patient prognosis but whether the increase of ketone bodies is an adaptive response necessary for supporting cell metabolism or it is a trigger for disease progression21 has to be ascertained. In addition, it is interesting to mention that we observed differences in 3-hydroxybutyrate connectivity if we did not adjust the matrix of correlations for age. We already observed in our previous study on a healthy population that 3-hydroxybutirate is more connected in younger people;27 thus, we can hypothesize that the pattern of connections of this metabolite is directly influenced by aging.
In deceased patients, we observed a significant reduction of histidine serum levels and a concomitant reduction of histidine connectivity (from 8 to 5 connections, not significant after Bonferroni correction). This finding is of particular interest if compared with the available literature: previous studies have shown that histidine was significantly reduced during the acute phase of thrombotic myocardial infarction47 and that histidine can reduce platelet aggregation after vascular injury in animal models.51 DeFilippis and coauthors47 hypothesized that the simultaneous increase of steroid hormones and decrease of histidine in the plasma of acute thrombotic MI patients, but not in the plasma of non-thrombotic MI patients, could imply a contribution of these metabolites in a pathological coronary thrombosis setting; this hypothesis is in line with the decrease of histidine that we observed in deceased patients.
3.2. Differential Network Analysis or Risk-Associated Metabolite Networks
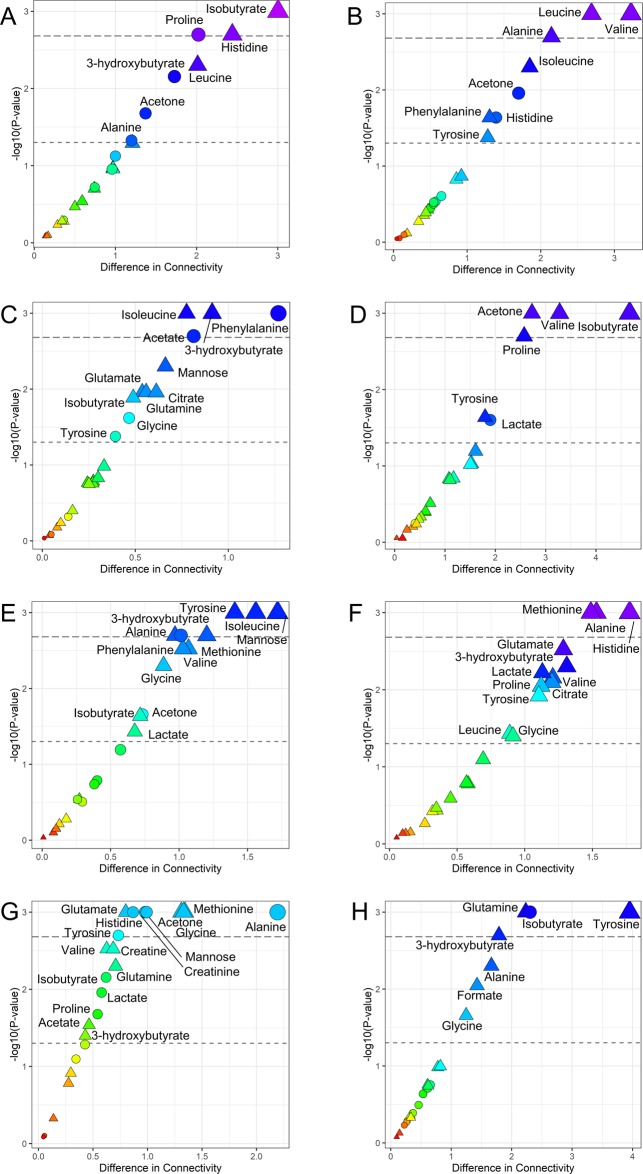
For each risk parameter, namely ACS, Killip, GRACE, and metabolomics NOESY RF classifications, serum metabolite–metabolite association networks distinct for high and low risk of death were estimated using the PCLRC23 algorithm considering the two subgroups of survivor and deceased patients for a total of 16 different networks. Differences in terms of connectivity in each couple of networks (survivors vs deceased patients) were analyzed for each risk parameters according to the low (Figure 4A,C,E,G) or high (Figure 4B,D,F,H) risk of death at 2 years. Metabolites for which connectivity does not change with the phenotype considered (i.e., high/low risk of mortality) can be considered to be unrelated to that specific trait and being involved in “housekeeping” biological reactions.52 On the other hand, it is known that metabolites participating in many metabolic processes tend to have higher levels of correlation and connectivity;53 thus, decreased connectivity indicates reduced activity of certain pathways where metabolites with significantly altered connectivity are involved. Thus, changes in connectivity can be seen as a proxy for changes and/or alteration in metabolically functional modules.
Figure 4.
Difference in terms of connectivities in networks of survived and deceased are reported against each metabolite’s P-value for each risk class. The thresholds for significance at 0.05 and after Bonferroni correction are reported. The colors red to blue encode for the increasing difference. Significance in terms of metabolite levels is also reported: circles are metabolites that are statically different between survived and deceased patients, whereas triangles encode for the not significant ones. (A) NSTEMI (low risk); (B) STEMI (high risk); (C) low risk for GRACE score; (D) high risk for GRACE score; (E) Killip class I (low risk); (F) Killip class II-II (high risk); (G) low risk for NOESY RF score; (H) high risk for NOESY RF score.
In many sub-group, acetone and 3-hydroxybutyrate showed statistically significant differences in connectivity in the comparison between networks of survivor and deceased patients. Analysis of the medical history of deceased patients revealed that they were characterized by a higher ratio of chronic heart failure and diabetes (Table 1). It has been shown that ketone body metabolism dysregulation plays a crucial role in both these pathological conditions49,54,55 and acetone has been proposed as a possible early biomarker for metabolic responses to AMI.56
Excluding the ACS classification risk, only low-risk patients show significant differences in the connectivity of mannose: we observed a progressive decrease of the connectivity of mannose with BCAA, alanine, creatinine, and acetone going from survivor low-risk patients to high-risk ones to low-risk deceased and finally to high-risk deceased patients. Only the connection with glucose remains unaffected among all risk classes. Based on the observation that highly connected metabolites participate in many chemical reactions, this decrease in connectivity leads to the hypothesis of decreased metabolic activity for high-risk subjects. This substantiates the previous discussion that mannose may be a pivotal player for the prediction of mortality following AMI and of CVD in general.
It has been suggested that the processing of glycoconjugates composed of glucose-derived mannose and its efflux from the cells account for most of the mannose present in blood and it is responsible for the maintenance of its steady state.57 Mannose plays a central role in glycation processes of lipoproteins that are involved in the initiation and development of atherogenesis. N-Glycans are up-regulated in proinflammatory settings and have been found on the endothelial cell surface in early stages of atherosclerotic plaque development.58
The role of mannose-binding lectin (MBL), an activator of the complement immune system and thereby involved in inflammatory activation, has been investigated in patients with ACS, and it has been shown that ACS patients featured higher MBL and genotypes in comparison with healthy control.59 It has been observed that the lectin complement pathway60 is activated after myocardial ischemia–reperfusion and leads to tissue injury: the blockade of the lectin pathway with inhibitory mAbs protects the heart from ischemia–reperfusion by reducing neutrophil infiltration and attenuating proinflammatory gene expression.61
Investigation of mannose levels in AMI patients provides complementary information to that obtained by network analysis: higher levels of mannose in high-risk patients (see Table 2) suggest that less mannose is bound to MBL, thus reducing the activation of the lecithin pathway, which is one of the three ways in which the complement system is activated.
In addition, differences in metabolite network connectivity between survivor and deceased patients emerged in both high- and low-risk networks, especially regarding branched-chain amino acid, alanine, and aromatic amino acid metabolisms. Alanine is synthetized and secreted by the heart under almost all circumstances, including hypoxia and ischemia:62 it can be transaminated to pyruvate whose oxidation produces three molecules of NADH and one of GTP, and thus it contributes to the maintenance of the energetic metabolism even in hypoxic conditions,63 like those post-AMI. We observed a negative correlation between alanine and acetone levels in survivor patients with low ACS and RF risk scores, while this correlation disappears in high-risk patients, either survived or deceased at 2 years after AMI.
We can explain the observed negative correlation between alanine and acetone, suggesting that in low-risk patients with favorable outcomes (alive at 2 years after AMI), the CORI cycle (also known as the lactic acid cycle) is favored or more active than in patients with negative outcomes, which are also characterized by higher levels of acetone. Indeed, the CORI cycle is more efficient for energy production than the alanine–glucose cycle (Cahill cycle), and this suggests that high-risk patients may switch to alternative ways to produce energy like fatty acid metabolism.
During ischemia, the overall mitochondrial oxidative metabolism is reduced as a consequence of the decrease in oxygen supply to the heart,64 whereas many studies already showed that during myocardial reperfusion, there is an overshoot in the rate of fatty acid oxidation, impaired pyruvate oxidation, and accelerated nonoxidative glycolysis.65 The serum samples analyzed here were collected in the hours immediately after PCI and thus in the condition of increased cardiac fatty acid oxidation rates. It is interesting to note that in high-risk patients and in patients who did not survive the AMI event, the negative correlation between alanine and acetone is lost, indicating that the preferential metabolic activity is lost and substituted by one or more, less efficient alternatives. In fact, in patients with negative outcomes, we observed generally higher levels of ketone bodies and glucose and lower levels of BCAAs, indicating that metabolic pathways alternative to fatty acids oxidation are underutilized. Our results support the evidence that the direct inhibition of mitochondrial fatty acid oxidation64,65 could be a promising target for treatment of ischemic heart diseases.
Branched-chain amino acids (BCAAs) show higher connectivity in deceased patients, in particular isoleucine in low-risk networks and valine in high-risk networks, thus implying a central role of their metabolism in AMI. Previous studies have shown that myocardial infarction leads to impaired BCAA catabolism, directly contributing to post-AMI cardiac dysfunction and remodeling.66,67 This can be reflected in the altered connectivity patterns observed here. Furthermore, in diabetes patients, it was also reported that BCAAs and aromatic amino acids were strongly predictive for the development of cardiovascular diseases.68
3.3. Comprehensive Analysis of AMI Risk-Associated Metabolite–Metabolite Association Networks
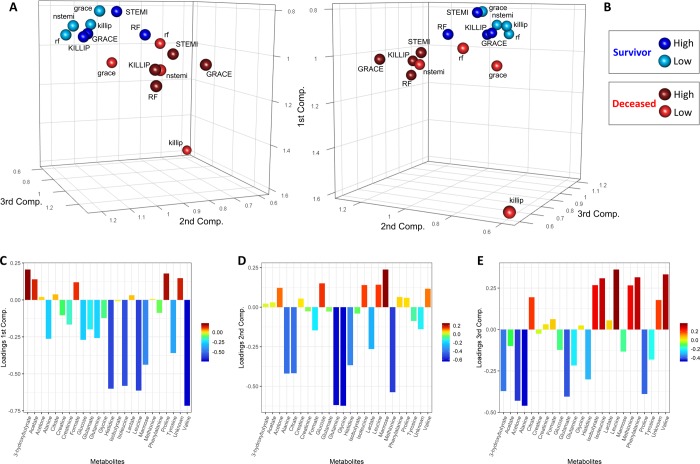
To investigate similarity and dissimilarity across the different networks in a compressive fashion, the 16 networks were analyzed simultaneously using covariance simultaneous component analysis: with this method, each network becomes a point in the COVSCA component space as shown in Figure 5A,B. We observe a clear separation between the networks (discrimination accuracy of 87.5% using k-NN, P-value = 0.0134) pertaining to subjects alive 2 years after AMI (blue dots) and those who died within 2 years (red dots): this indicates that the correlation patterns observed are, overall, different in the two groups. However, within each group, we observe also a separation among networks pertaining to high and low risk, indicating the existence of risk-specific metabolite–metabolite correlation signatures. We notice, in particular, that the network specific to the low risk for the KILLIP parameters for dead subjects within 2 years from AMI is markedly different from the others.
Figure 5.
Score and loading plots of the COVSCA model for the metabolite correlation networks obtained using the PCLRC algorithm. (A, B) Score plots of the first three components. Each sphere represents a network that corresponds to each mortality risk parameter analyzed. Blue spheres indicate patients that survive within 2 years after AMI, whereas red spheres indicate deceased patients. Light colors denote networks of patients at low risk of mortality, while dark colors are for networks of patients with high risk. (C, D, E) Loading plots of the first three components.
Since the COVSCA model is a component model, the relative metabolite contribution to explain the observed clustering patterns can be obtained by inspecting the loading plots (Figure 5C–E) as in standard PCA. The first and third components account mostly for the separation among survivor and deceased patients and are dominated by amino acids (histidine, leucine, isoleucine, and valine). Leucine, isoleucine, and valine are branched-chain amino acids (BCAAs), which are primarily catabolized in the extrahepatic tissues, notably the cardiac muscle, and are essential for normal growth and function at the cellular and the organ levels.69,70 BCAA catabolism plays a fundamental role in normal cardiac physiology and cellular viability, and defective BCAA catabolism has been reported to promote dilated cardiomyopathy in humans.69,71 Furthermore, our results are in line with the ones of a previous study that identifies in the decrease of plasma amino acid levels an important clinical consequence of thrombotic AMI.50 The second component partially accounts for the subclustering with the two major clusters with larger contributions from glutamine, leucine, methionine, glycine, alanine, which are glucogenic amino acids, and citrate. The observed clustering of the metabolite association network specific to the ACS classification, Killip classification, GRACE score, and NOESY RF score suggests that these scores may describe similar mechanisms, albeit with different levels of resolution.
4. Conclusions
We have presented network reconstruction and analysis of experimentally identified relationships between metabolites and applied a differential network approach to analyze AMI patients who survived more than 2 years after the cardiovascular event with respect to those who did not survive. Our results show significant differences in the connectivity patterns of several low-molecular-weight molecules, implying variations in the regulation of several metabolic pathways. In particular, creatinine and mannose emerged as possible prognostic biomarkers.
The network approach seems to provide more insights than the standard approach, showing a decrease of the connectivity in the network of deceased patients. However, the biological implications of our result need to be further investigated.
The large number of patients studied (n = 825) and the long period of follow-up are major strength points of this study. However, information about biochemical mechanisms underlying the transition to the quiescent phase could not be obtained because samples were collected only during the acute phase of the disease. This aspect is of crucial importance, and thus further efforts in this direction are guaranteed. Moreover, samples were collected not at the time of presentation but after the therapeutic intervention, and at this point, all acute ischemia (the etiology of the event) has resolved. In conclusion, this study provides important information on how the metabolic networks of AMI patients change according to the patient outcomes.
Acknowledgments
The authors acknowledge the support and the use of resources of Instruct-ERIC, a Landmark ESFRI project, and specifically the CERM/CIRMMP Italy Centre. A.V. was supported by an AIRC fellowship for Italy.
Author Contributions
C.L., E.S., A.V., and L.T. designed the study. A.M.G., B.G., R.M., S.V., N.C., D.B., A.B., N.M., and G.F.G. recruited patients, collected samples, and managed biological material and clinical data collection. E.S. and A.V. performed statistical data analysis. C.L., E.S., L.T., and A.V. interpreted data and results and prepared the manuscript. All authors read and approved the final version of the manuscript and were responsible for its final content.
The authors declare no competing financial interest.
References
- Wright R. S.; Anderson J. L.; Adams C. D.; Bridges C. R.; Casey D. E. Jr.; Ettinger S. M.; Fesmire F. M.; Ganiats T. G.; Jneid H.; Lincoff A. M.; et al. 2011 ACCF/AHA Focused Update Incorporated into the ACC/AHA 2007 Guidelines for the Management of Patients with Unstable Angina/Non-ST-Elevation Myocardial Infarction. J Am. Coll. Cardiol. 2011, 57, e215–e367. 10.1016/j.jacc.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D.; Benjamin E. J.; Go A. S.; Arnett D. K.; Blaha M. J.; Cushman M.; de Ferranti S.; Després J.-P.; Fullerton H. J.; Howard V. J.; et al. Heart Disease and Stroke Statistics--2015 Update: A Report from the American Heart Association. Circulation 2015, 131, e29–e322. 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Brogan R. A.; Malkin C. J.; Batin P. D.; Simms A. D.; McLenachan J. M.; Gale C. P. Risk Stratification for ST Segment Elevation Myocardial Infarction in the Era of Primary Percutaneous Coronary Intervention. World J. Cardiol. 2014, 6, 865–873. 10.4330/wjc.v6.i8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccheri S.; D’Arrigo P.; Franchina G.; Capodanno D. Risk Stratification in Patients with Coronary Artery Disease: A Practical Walkthrough in the Landscape of Prognostic Risk Models. Interv. Cardiol. Rev. 2018, 13, 112–120. 10.15420/icr.2018.16.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber Y.; Weston S. A.; Enriquez-Sarano M.; Jaffe A. S.; Manemann S. M.; Jiang R.; Roger V. L. Contemporary Risk Stratification After Myocardial Infarction in the Community: Performance of Scores and Incremental Value of Soluble Suppression of Tumorigenicity-2. J. Am. Heart Assoc. 2017, 6, e005958 10.1161/JAHA.117.005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle D. Risk Stratification in Post-Myocardial Infarction Patients. Eur. Cardiol. Rev. 2010, 6, 22. 10.15420/ecr.2010.6.3.22. [DOI] [Google Scholar]
- Cavusoglu E.; Marmur J. D.; Yanamadala S.; Chopra V.; Hegde S.; Nazli A.; Singh K. P.; Zhang M.; Eng C. Elevated Baseline Plasma IL-8 Levels Are an Independent Predictor of Long-Term All-Cause Mortality in Patients with Acute Coronary Syndrome. Atherosclerosis 2015, 242, 589–594. 10.1016/j.atherosclerosis.2015.08.022. [DOI] [PubMed] [Google Scholar]
- Kaptoge S.; Di Angelantonio E.; Lowe G.; Pepys M. B.; Thompson S. G.; Collins R.; Danesh J. C-Reactive Protein Concentration and Risk of Coronary Heart Disease, Stroke, and Mortality: An Individual Participant Meta-Analysis. Lancet 2010, 375, 132–140. 10.1016/S0140-6736(09)61717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zairis M. N.; Adamopoulou E. N.; Manousakis S. J.; Lyras A. G.; Bibis G. P.; Ampartzidou O. S.; Apostolatos C. S.; Anastassiadis F. A.; Hatzisavvas J. J.; Argyrakis S. K.; et al. The Impact of Hs C-Reactive Protein and Other Inflammatory Biomarkers on Long-Term Cardiovascular Mortality in Patients with Acute Coronary Syndromes. Atherosclerosis 2007, 194, 397–402. 10.1016/j.atherosclerosis.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Trainor P. J.; Yampolskiy R. V.; DeFilippis A. P. Wisdom of Artificial Crowds Feature Selection in Untargeted Metabolomics: An Application to the Development of a Blood-Based Diagnostic Test for Thrombotic Myocardial Infarction. J. Biomed. Inform. 2018, 81, 53–60. 10.1016/j.jbi.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran A.; Aliani M.; Ravandi A. Metabolomic Characterization of Myocardial Ischemia-Reperfusion Injury in ST-Segment Elevation Myocardial Infarction Patients Undergoing Percutaneous Coronary Intervention. Sci. Rep. 2019, 9, 11742. 10.1038/s41598-019-48227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M.; Han Y.; Zhang Y.; Zhang S.; Wei C.; Cong Z.; Du W. Metabolomics Study of the Biochemical Changes in the Plasma of Myocardial Infarction Patients. Front. Physiol. 2018, 9, 1017. 10.3389/fphys.2018.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart A. D.; Beebe K.; Milburn M. Metabolomics as a Key Integrator for “Omic” Advancement of Personalized Medicine and Future Therapies. Clin. Transl. Sci. 2012, 5, 285–288. 10.1111/j.1752-8062.2011.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli A.; Ghini V.; Meoni G.; Licari C.; Takis P. G.; Tenori L.; Turano P.; Luchinat C. High-Throughput Metabolomics by 1D NMR. Angew. Chem., Int. 2019, 58, 968–994. 10.1002/anie.201804736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takis P. G.; Ghini V.; Tenori L.; Turano P.; Luchinat C. Uniqueness of the NMR Approach to Metabolomics. TrAC, Trends Anal. Chem. 2019, 115300. 10.1016/j.trac.2018.10.036. [DOI] [Google Scholar]
- McCartney A.; Vignoli A.; Biganzoli L.; Love R.; Tenori L.; Luchinat C.; Di Leo A. Metabolomics in Breast Cancer: A Decade in Review. Cancer Treat. Rev. 2018, 67, 88–96. 10.1016/j.ctrv.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Hart C. D.; Vignoli A.; Tenori L.; Uy G. L.; van To T.; Adebamowo C.; Hossain S. M.; Biganzoli L.; Risi E.; Love R. R.; et al. Serum Metabolomic Profiles Identify ER-Positive Early Breast Cancer Patients at Increased Risk of Disease Recurrence in a Multicenter Population. Clin. Cancer Res. 2017, 23, 1422–1431. 10.1158/1078-0432.CCR-16-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montuschi P.; Santini G.; Mores N.; Vignoli A.; Macagno F.; Shoreh R.; Tenori L.; Zini G.; Fuso L.; Mondino C.; et al. Breathomics for Assessing the Effects of Treatment and Withdrawal With Inhaled Beclomethasone/Formoterol in Patients With COPD. Front. Pharmacol. 2018, 9, 258. 10.3389/fphar.2018.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli A.; Orlandini B.; Tenori L.; Biagini M. R.; Milani S.; Renzi D.; Luchinat C.; Calabrò A. S. Metabolic Signature of Primary Biliary Cholangitis and Its Comparison with Celiac Disease. J. Proteome Res. 2019, 18, 1228–1236. 10.1021/acs.jproteome.8b00849. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Rodio D. M.; Bellizzi A.; Sobolev A. P.; Anzivino E.; Mischitelli M.; Tenori L.; Marini F.; Priori R.; Scrivo R.; et al. NMR-Based Metabolomic Approach to Study Urine Samples of Chronic Inflammatory Rheumatic Disease Patients. Anal. Bioanal. Chem. 2017, 409, 1405–1413. 10.1007/s00216-016-0074-z. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Tenori L.; Giusti B.; Takis P. G.; Valente S.; Carrabba N.; Balzi D.; Barchielli A.; Marchionni N.; Gensini G. F.; et al. NMR-Based Metabolomics Identifies Patients at High Risk of Death within Two Years after Acute Myocardial Infarction in the AMI-Florence II Cohort. BMC Med. 2019, 17, 3. 10.1186/s12916-018-1240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato A.; Tenori L.; Cascante M.; De Atauri Carulla P. R.; Martins dos Santos V. A. P.; Saccenti E. From Correlation to Causation: Analysis of Metabolomics Data Using Systems Biology Approaches. Metabolomics 2018, 14, 37. 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti E.; Suarez-Diez M.; Luchinat C.; Santucci C.; Tenori L. Probabilistic Networks of Blood Metabolites in Healthy Subjects as Indicators of Latent Cardiovascular Risk. J. Proteome Res. 2015, 14, 1101–1111. 10.1021/pr501075r. [DOI] [PubMed] [Google Scholar]
- Krumsiek J.; Suhre K.; Illig T.; Adamski J.; Theis F. J. Gaussian Graphical Modeling Reconstructs Pathway Reactions from High-Throughput Metabolomics Data. BMC Syst. Biol. 2011, 5, 21. 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J.; Mittelstrass K.; Do K. T.; Stückler F.; Ried J.; Adamski J.; Peters A.; Illig T.; Kronenberg F.; Friedrich N.; et al. Gender-Specific Pathway Differences in the Human Serum Metabolome. Metabolomics 2015, 11, 1815–1833. 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti E.; Menichetti G.; Ghini V.; Remondini D.; Tenori L.; Luchinat C. Entropy-Based Network Representation of the Individual Metabolic Phenotype. J. Proteome Res. 2016, 15, 3298–3307. 10.1021/acs.jproteome.6b00454. [DOI] [PubMed] [Google Scholar]
- Vignoli A.; Tenori L.; Luchinat C.; Saccenti E. Age and Sex Effects on Plasma Metabolite Association Networks in Healthy Subjects. J. Proteome Res. 2018, 17, 97–107. 10.1021/acs.jproteome.7b00404. [DOI] [PubMed] [Google Scholar]
- Suarez-Diez M.; Adam J.; Adamski J.; Chasapi S. A.; Luchinat C.; Peters A.; Prehn C.; Santucci C.; Spyridonidis A.; Spyroulias G. A.; et al. Plasma and Serum Metabolite Association Networks: Comparability within and between Studies Using NMR and MS Profiling. J. Proteome Res. 2017, 16, 2547–2559. 10.1021/acs.jproteome.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari F.; Marcucci R.; Gori A.; Caporale R.; Fanelli A.; Casola G.; Balzi D.; Barchielli A.; Valente S.; Giglioli C.; et al. Reticulated Platelets Predict Cardiovascular Death in Acute Coronary Syndrome Patients. Thromb. Haemost 2013, 109, 846–853. 10.1160/TH12-09-0709. [DOI] [PubMed] [Google Scholar]
- Ren L.; Ye H.; Wang P.; Cui Y.; Cao S.; Lv S. Comparison of Long-Term Mortality of Acute ST-Segment Elevation Myocardial Infarction and Non-ST-Segment Elevation Acute Coronary Syndrome Patients after Percutaneous Coronary Intervention. Int. J. Clin. Exp. Med. 2014, 7, 5588–5592. [PMC free article] [PubMed] [Google Scholar]
- Killip T. III; Kimball J. T. Treatment of Myocardial Infarction in a Coronary Care Unit. A Two Year Experience with 250 Patients. Am. J. Cardiol. 1967, 20, 457–464. 10.1016/0002-9149(67)90023-9. [DOI] [PubMed] [Google Scholar]
- Tang E. W.; Wong C.-K.; Herbison P. Global Registry of Acute Coronary Events (GRACE) Hospital Discharge Risk Score Accurately Predicts Long-Term Mortality Post Acute Coronary Syndrome. Am. Heart J. 2007, 153, 29–35. 10.1016/j.ahj.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001, 45, 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Bernini P.; Bertini I.; Luchinat C.; Nincheri P.; Staderini S.; Turano P. Standard Operating Procedures for Pre-Analytical Handling of Blood and Urine for Metabolomic Studies and Biobanks. J. Biomol. NMR 2011, 49, 231–243. 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- Meiboom S.; Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev. Sci. Instrum. 1958, 29, 688–691. 10.1063/1.1716296. [DOI] [Google Scholar]
- Wishart D. S.; Jewison T.; Guo A. C.; Wilson M.; Knox C.; Liu Y.; Djoumbou Y.; Mandal R.; Aziat F.; Dong E.; et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni C. E.Il Calcolo Delle Assicurazioni Su Gruppi Di Teste. In Studi in Onore del Professore Salvatore Ortu Carboni; Bardi: Rome, 1935; pp 13–60. [Google Scholar]
- Neuháuser M.Wilcoxon–Mann–Whitney Test. In International Encyclopedia of Statistical Science; Springer: Berlin, Heidelberg, 2011; pp 1656–1658, 10.1007/978-3-642-04898-2_615. [DOI] [Google Scholar]
- Smilde A. K.; Timmerman M. E.; Saccenti E.; Jansen J. J.; Hoefsloot H. C. J. Covariances Simultaneous Component Analysis: A New Method within a Framework for Modeling Covariances. J. Chemom. 2015, 29, 277–288. 10.1002/cem.2707. [DOI] [Google Scholar]
- Zhao L.; Wang L.; Zhang Y. Elevated Admission Serum Creatinine Predicts Poor Myocardial Blood Flow and One-Year Mortality in ST-Segment Elevation Myocardial Infarction Patients Undergoing Primary Percutaneous Coronary Intervention. J. Invasive Cardiol. 2009, 21, 493–498. [PubMed] [Google Scholar]
- Cakar M. A.; Gunduz H.; Vatan M. B.; Kocayigit I.; Akdemir R. The Effect of Admission Creatinine Levels on One-Year Mortality in Acute Myocardial Infarction. Sci. World J. 2012, 2012, e186495 10.1100/2012/186495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A.; Stančáková A.; Lotta L. A.; Kuusisto J.; Boren J.; Blüher M.; Wareham N. J.; Ferrannini E.; Groop P. H.; Laakso M.; et al. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell Metab. 2017, 26, 281–283. 10.1016/j.cmet.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Hsu C.-C.; Chang H.-Y.; Huang M.-C.; Hwang S.-J.; Yang Y.-C.; Tai T.-Y.; Yang H.-J.; Chang C.-T.; Chang C.-J.; Li Y.-S.; et al. Association between Insulin Resistance and Development of Microalbuminuria in Type 2 Diabetes: A Prospective Cohort Study. Diabetes Care 2011, 34, 982–987. 10.2337/dc10-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer J.; Verbrugge F. H.; Lamm G. Editor’s Choice- What Do Small Serum Creatinine Changes Tell Us about Outcomes after Acute Myocardial Infarction?. Eur. Heart J. Acute Cardiovasc. Care 2018, 7, 739–742. 10.1177/2048872617728721. [DOI] [PubMed] [Google Scholar]
- Tzoulaki I.; Castagné R.; Boulangé C. L.; Karaman I.; Chekmeneva E.; Evangelou E.; Ebbels T. M. D.; Kaluarachchi M. R.; Chadeau-Hyam M.; Mosen D.; et al. Serum Metabolic Signatures of Coronary and Carotid Atherosclerosis and Subsequent Cardiovascular Disease. Eur. Heart J. 2019, 40, 2883–2896. 10.1093/eurheartj/ehz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Svingen G. F. T.; Pedersen E. R.; Gregory J. F.; Ueland P. M.; Tell G. S.; Nygård O. K. Plasma Glycine and Risk of Acute Myocardial Infarction in Patients With Suspected Stable Angina Pectoris. J. Am. Heart Assoc. 2016, 5, e002621 10.1161/JAHA.115.002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis A. P.; Trainor P. J.; Hill B. G.; Amraotkar A. R.; Rai S. N.; Hirsch G. A.; Rouchka E. C.; Bhatnagar A. Identification of a Plasma Metabolomic Signature of Thrombotic Myocardial Infarction That Is Distinct from Non-Thrombotic Myocardial Infarction and Stable Coronary Artery Disease. PLoS One 2017, 12, e0175591 10.1371/journal.pone.0175591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty M. F.; Barroso-Aranda J.; Contreras F. The Hyperpolarizing Impact of Glycine on Endothelial Cells May Be Anti-Atherogenic. Med. Hypotheses 2009, 73, 263–264. 10.1016/j.mehy.2008.12.021. [DOI] [PubMed] [Google Scholar]
- Lim G. B. Ketone Body Utilization in Myocardial Ischaemia. Nat. Rev. Cardiol. 2019, 16, 133. 10.1038/s41569-019-0161-x. [DOI] [PubMed] [Google Scholar]
- Trainor P. J.; Hill B. G.; Carlisle S. M.; Rouchka E. C.; Rai S. N.; Bhatnagar A.; DeFilippis A. P. Systems Characterization of Differential Plasma Metabolome Perturbations Following Thrombotic and Non-Thrombotic Myocardial Infarction. J. Proteomics 2017, 160, 38–46. 10.1016/j.jprot.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. Q.; Zhao G.; Li J.; Qian W. Effect of Histidine on Myocardial Mitochondria and Platelet Aggregation during Thrombotic Cerebral Ischemia in Rats. Zhongguo Yao Li Xue Bao 1998, 19, 493–496. [PubMed] [Google Scholar]
- Cui X.; Yu X.; Sun G.; Hu T.; Likhodii S.; Zhang J.; Randell E.; Gao X.; Fan Z.; Zhang W. Differential Metabolomics Networks Analysis of Menopausal Status. PLoS One 2019, 14, e0222353 10.1371/journal.pone.0222353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T.; Soyer O. S.; Bonhoeffer S. The Evolution of Connectivity in Metabolic Networks. PLoS Biol. 2005, 3, e228 10.1371/journal.pbio.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa T.; Sugano Y.; Shimouchi A.; Shibata A.; Nakayama T.; Ohara T.; Jinno N.; Kanzaki H.; Anzai T. A Case of Acute Decompensated Heart Failure Evaluated by Series of Exhaled Acetone Concentrations as Noninvasive Biomarker of Heart Failure Severity. Int. J. Cardiol. 2016, 204, 112–113. 10.1016/j.ijcard.2015.11.173. [DOI] [PubMed] [Google Scholar]
- Klein M. S.; Shearer J. Metabolomics and Type 2 Diabetes: Translating Basic Research into Clinical Application. J. Diabetes Res. 2016, 2016, 1. 10.1155/2016/3898502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S. E.; Farag M. A.; Holvoet P.; Hanafi R. S.; Gad M. Z. A Comparative Metabolomics Approach Reveals Early Biomarkers for Metabolic Response to Acute Myocardial Infarction. Sci. Rep. 2016, 6, 36359. 10.1038/srep36359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.; Freeze H. H. Mannose Efflux from the Cells A POTENTIAL SOURCE OF MANNOSE IN BLOOD. J. Biol. Chem. 2011, 286, 10193–10200. 10.1074/jbc.M110.194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. W.; Chen J.; Chacko B. K.; Traylor J. G. Jr.; Orr A. W.; Patel R. P. Role of Endothelial N-Glycan Mannose Residues in Monocyte Recruitment During Atherogenesis. Arterioscler., Thromb., Vasc. Biol. 2012, 32, e51–e59. 10.1161/ATVBAHA.112.253203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesonen E.; Hallman M.; Sarna S.; Andsberg E.; Haataja R.; Meri S.; Persson K.; Puolakkainen M.; Öhlin H.; Truedsson L. Mannose-Binding Lectin as a Risk Factor for Acute Coronary Syndromes. Ann. Med. 2009, 41, 591–598. 10.1080/07853890903110994. [DOI] [PubMed] [Google Scholar]
- Walport M. J. Complement. N. Engl. J. Med. 2001, 344, 1140–1144. 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- Jordan J. E.; Montalto M. C.; Stahl G. L. Inhibition of Mannose-Binding Lectin Reduces Postischemic Myocardial Reperfusion Injury. Circulation 2001, 104, 1413–1418. 10.1161/hc3601.095578. [DOI] [PubMed] [Google Scholar]
- Drake K. J.; Sidorov V. Y.; McGuinness O. P.; Wasserman D. H.; Wikswo J. P. Amino Acids as Metabolic Substrates during Cardiac Ischemia. Exp. Biol. Med. 2012, 237, 1369–1378. 10.1258/ebm.2012.012025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taegtmeyer H.; Peterson M. B.; Ragavan V. V.; Ferguson A. G.; Lesch M. De Novo Alanine Synthesis in Isolated Oxygen-Deprived Rabbit Myocardium. J. Biol. Chem. 1977, 252, 5010–5018. [PubMed] [Google Scholar]
- Fillmore N.; Mori J.; Lopaschuk G. D. Mitochondrial Fatty Acid Oxidation Alterations in Heart Failure, Ischaemic Heart Disease and Diabetic Cardiomyopathy. Br. J. Pharmacol. 2014, 171, 2080–2090. 10.1111/bph.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley W. C.; Lopaschuk G. D.; Hall J. L.; McCormack J. G. Regulation of Myocardial Carbohydrate Metabolism under Normal and Ischaemic Conditions Potential for Pharmacological Interventions. Cardiovasc. Res. 1997, 33, 243–257. 10.1016/s0008-6363(96)00245-3. [DOI] [PubMed] [Google Scholar]
- Wang W.; Zhang F.; Xia Y.; Zhao S.; Yan W.; Wang H.; Lee Y.; Li C.; Zhang L.; Lian K.; et al. Defective Branched Chain Amino Acid Catabolism Contributes to Cardiac Dysfunction and Remodeling Following Myocardial Infarction. Am. J. Physiol.-Heart Circ. Physiol. 2016, 311, H1160–H1169. 10.1152/ajpheart.00114.2016. [DOI] [PubMed] [Google Scholar]
- Du X.; You H.; Li Y.; Wang Y.; Hui P.; Qiao B.; Lu J.; Zhang W.; Zhou S.; Zheng Y.; et al. Relationships between Circulating Branched Chain Amino Acid Concentrations and Risk of Adverse Cardiovascular Events in Patients with STEMI Treated with PCI. Sci. Rep. 2018, 8, 15809. 10.1038/s41598-018-34245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M.; Lewis G. D.; Ericson U.; Orho-Melander M.; Hedblad B.; Engström G.; Östling G.; Clish C.; Wang T. J.; Gerszten R. E.; et al. A Diabetes-Predictive Amino Acid Score and Future Cardiovascular Disease. Eur. Heart J. 2013, 34, 1982–1989. 10.1093/eurheartj/ehs424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajeda-Iglesias C.; Aviram M. Specific Amino Acids Affect Cardiovascular Diseases and Atherogenesis via Protection against Macrophage Foam Cell Formation: Review Article. Rambam Maimonides Med. J. 2018, 9, e0022 10.5041/RMMJ.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper A. E.; Miller R. H.; Block K. P. Branched-Chain Amino Acid Metabolism. Annu. Rev. Nutr. 1984, 4, 409–454. 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Sun H.; Olson K. C.; Gao C.; Prosdocimo D. A.; Zhou M.; Wang Z.; Jeyaraj D.; Youn J.-Y.; Ren S.; Liu Y.; et al. Catabolic Defect of Branched-Chain Amino Acids Promotes Heart Failure. Circulation 2016, 133, 2038–2049. 10.1161/CIRCULATIONAHA.115.020226. [DOI] [PMC free article] [PubMed] [Google Scholar]