Abstract
High content screening (HCS) technology combining automated microscopy and quantitative image analysis can address biological questions in academia and the pharmaceutical industry. Various HCS experimental applications have been utilized in the research field of in vitro toxicology. In this review, we describe several HCS application approaches used for studying the mechanism of compound toxicity, highlight some challenges faced in the toxicological community, and discuss the future directions of HCS in regards to new models, new reagents, data management, and informatics. Many specialized areas of toxicology including developmental toxicity, genotoxicity, developmental neurotoxicity/neurotoxicity, hepatotoxicity, cardiotoxicity, and nephrotoxicity will be examined. In addition, several newly developed cellular assay models including induced pluripotent stem cells (iPSCs), three-dimensional (3D) cell models, and tissues-on-a-chip will be discussed. New genome editing technologies (e.g., CRISPR/Cas9), data analyzing tools for imaging, and coupling with high-content assays will be reviewed. Finally, the applications of machine learning to image processing will be explored. These new HCS approaches offer a huge step forward in dissecting biological processes, developing drugs, and making toxicology studies easier.
Keywords: high-content screen, toxicology, HCS, Tox21
Introduction
Cellular imaging has become a popular research tool, integrating with cell biology, compound safety, and drug discovery. Previously, cellular imaging was considered a descriptive science, solely amenable to a low-throughput format [1]. High-throughput imaging (HTI) enables visualization and quantification by capturing many cellular features on a large scale, using automated microscopy and image analysis platforms [2]. High-content screening (HCS), or automated microscope-based screening, is one of the HTI approaches, combining multi-parametric microscopy with quantitation of large data sets of morphological features. Several terms similar to HCS, including HCA (high-content analysis), HCI (high-content imaging) and IC (image cytometry), generally refer to low- or medium-throughput automated microscope-based assays [3].
High-content screening was first introduced in the mid-1990s as a promising approach to facilitate drug discovery via study of the complex physiology of a cell or organism [4]. Recently, HCS has been widely used due to the unprecedented development of automatic microscopes with autofocusing, image acquisition and real-time analysis of cellular samples in multi-well microtiter plates, single-cell informatics approaches, and a biology toolbox enriched with antibodies, dyes, and chemical probes [5, 6]. Boutros et al. described the application of HCS in diverse processes, including changes in protein-localization, vulnerabilities in cancer cells, and phenotypes of complex organisms. Considerations were also discussed for the HCS experimental designs, such as fluorescent probes, miniaturization and automation, image acquisition, image analysis, image data, image processing and segmentation, quantification of phenotypic features, and image analysis [7].
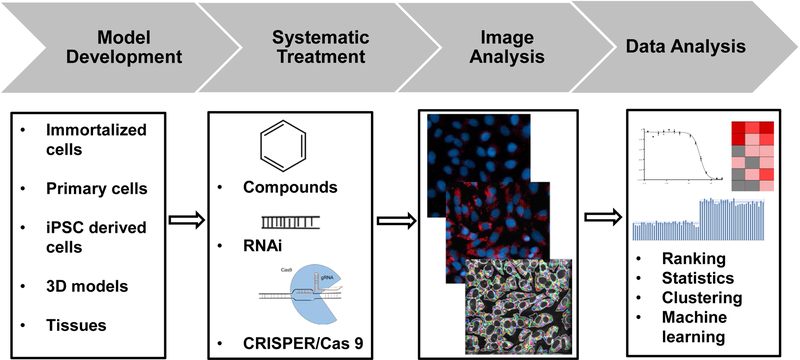
General steps for the development of a high content screening (HCS) experiment are shown in Figure 1. First, cell models (immortalized, stem cells, 3D models) were developed or selected depending on the assays to be performed. Second, treatment including chemicals, RNAi reagents or CRISPER/Cas9 was conducted. Third, the images were acquired based on the use of live cell dyes, fluorescent-labeled antibodies, GFP-labeled protein or immunofluorescence. Images parameters were selected and analyzed for quantification. Finally, the generated data needs to be formatted and organized for ranking, clustering, statistics, and machine learning. Models, treatments, readouts, and data analysis are developed iteratively with HCS technology, depending on the specific questions asked. Several commercial, automated imagers enable HCS and offer the flexibility of different imaging modalities—see Table 1. These high-content readers equipped with confocal imaging provide solutions for complex disease models, such as 3D and micro-tissues. Selection of an imaging platform is often based on the study needs. Much non-commercial online software is available for phenotypic image analysis, for which strengths and weaknesses, recent developments, and future perspective have been examined and discussed in detail [8]. Incremental HCS improvements and knowledge accumulation in the scientific community led to breakthroughs in scientific fields including drug discovery and compound safety.
Figure 1.
Flow chart of high-content screening. The design of a HCS assay includes development or selection of cell models, incubation with test compounds or genetic reagents, and image acquisition and analysis. The final step is to analyze the data and interpret the results.
Table 1.
High content screen instrumentation
| Instrument names | Applications and biological models | Image analysis software | Website |
|---|---|---|---|
| ImageXpress Micro Confocal (Molecular Devices) |
|
MetaXpress® | https://www.moleculardevices.com |
| Opera Phenix™ HCS System (PerkinElmer) | Harmony | http://www.perkinelmer.com | |
| IN Cell Analyzer 6500HS (GE Health) | IN Cell Investigator & IN Carta | https://www.gelifesciences.com | |
| CellInsight™ CX7 LZR (Thermo Fisher) | HCS studio | https://www.thermofisher.com | |
| CellVoyager® CV8000 HCS (YOKOGAWA) | CellPathfinder | https://www.yokogawa.com |
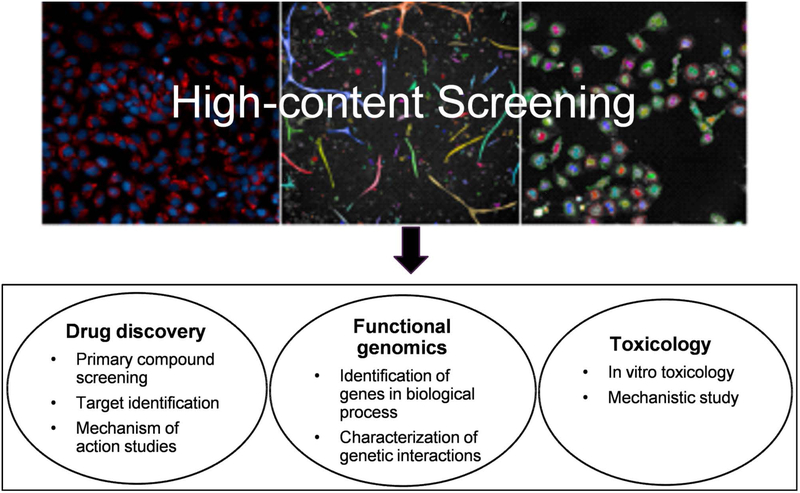
High-content screening applications have been utilized within drug discovery and functional genomics, including lead optimization, compound library enrichment, functional annotation of genes/alleles, identification of small molecule modulators of gene activity, and disease-specific phenotypes [9], as shown in Figure 2. Currently, HCS is considered as an important tool in supporting drug discovery and development, including target identification, primary compound screening, secondary confirmation, mechanism of action (MOA) studies, and in vitro toxicology [1, 5, 9–11]. HCS can be used to identify genes required for specific biological processes from a genetic perturbation screen using a genome-wide RNA interference (RNAi) screen [12–14]. In addition, HCS has been used as an in vitro toxicological tool to test compound toxicity and to elucidate MOAs, greatly reducing the use of animals in toxicological testing. Animal studies are costly, low-throughput and sometimes inconsistent in predicting human toxicity [15]. Target-specific, mechanism-oriented in vitro assays combined with computational models are promising for elucidating compound mechanism of toxicity and prioritizing compounds for further in-depth toxicological testing [16–18]. Compared with high-throughput screening (HTS), which measures compound activities in an entire assay wells, HCS provides detailed information at the cellular level with insights into the spatial distribution and the dynamic of responses within a biological system. Therefore, HCS is a promising tool to address the challenges of the “Toxicity testing in the 21st century” approach [19]. Table 2 lists the HCS assays which have been evaluated and validated in the toxicological field. The details of HCS applications in different toxicological areas are described in the following sections.
Figure 2.
Applications of high content screening. HCS has been applied at all stages of drug discovery and development processes. HCS can also be used in genetic screens for identifying genes required for a specific biological process or proteome-wide changes. Moreover, HCS can be used as an in vitro tool to prioritize compounds for toxicological and mechanistic studies.
Table 2.
High content screens for toxicological applications
| Toxicology areas | Assay models | Assay readouts |
|---|---|---|
| Developmental toxicity |
|
|
| Genotoxicity |
|
|
| Neurotoxicity |
|
|
| Hepatotoxicity |
|
|
| Cardiotoxicity |
|
|
| Nephrotoxicity |
|
|
HCS in Developmental Toxicity
Traditional testing for developmental toxicity has relied on in vivo animal models for data collection [20, 21]. However, animal tests are time consuming and expensive, and sometimes fail to predict human toxicity [16]. Recently, in vitro cellular models have become more popular for the assessment of developmental toxicity; these models include mouse embryonic stem cells, rat embryos, and zebrafish embryos [22, 23]. Of these models, the zebrafish embryo assay is amenable to high-throughput and high-content approaches. High content assays using transgenic zebrafish embryos with morphology and behavioral endpoints have been used for toxicity screening. Sixteen chemicals from the US Environmental Protection Agency’s (EPA’s) ToxCast Phase-I library were evaluated; two compounds, abamectin and emamectin benzoate, were found to abolish movement of the developing embryos without gross malformations [24]. An image-based high content screening (HCS) assay was developed by Lantz-McPeak et al. to identify compounds toxic to zebrafish embryos. Embryo length was used as a statistically quantifiable endpoint of toxicity. The assay was also validated by evaluating the effects of a group of known developmental toxic compounds (e.g., ethanol, nicotine, ketamine, and caffeine) on zebrafish embryo models. These HCS developmental assays promise evaluation of the effects of developmental toxins in a high throughput manner [25].
HCS in Genotoxicity
Genotoxic chemicals can damage DNA, potentially leading to genetic mutations, which in turn increase the risk of tumor formation [26]. Several in vitro assays, such as the Ames test, the mouse lymphoma assay, the micronucleus test, and the comet assay have been used to assess chemically induced DNA damage [27]. Some of the assays, such as the Vitotox™ assay measuring mutagenicity, the RadarScreen assay measuring clastogenicity, and the comet assay evaluating DNA breaks, have been optimized for medium to high-throughput formats [28–31]. Compared with these methods, HCS assays provide more insight into the mechanism of action of genotoxic compounds because of their ability to measure multiple parameters (e.g., γH2AX and micronucleus) [32]. In addition, HCS facilitates the filtering out of false positives. The isogenic chicken DT40 B-lymphocyte cell line, deficient in DNA repair pathways, was utilized for screening genotoxic compounds on a high-throughput screening platform by measuring differential cytotoxicity in the US Tox21 program. Many well-known genotoxins (e.g., adriamycin, melphalan) were confirmed in this study and several potential genotoxic agents, such as 2-oxiranemethanamine, AD-67, and tetraphenylolethane glycidyl ether, were identified as genotoxic agents using an imaging-based, immunocytological γH2AX (serine-139-phosphorylated histone) assay and micronucleus assay [33]. These HCS assays are often used to evaluate genotoxicity. Measurement of γH2AX and micronucleus using high-throughput flow cytometry also received attention for predicting genotoxic potential [34–37]. Garcia-Canton et al. validated the HCS γH2AX assay using a group of known genotoxic and non-genotoxic compounds in a human bronchial epithelial cell line BEAS-2B. This assay showed high accuracy (86%) with 86–92% sensitivity and 80–88% specificity [38]. HCS micronucleus assays were developed with the rodent cell line CHO-K1 and the human hepatoma cell line HepG2 used in regulatory genotoxicity assays. The sensitivity and specificity was 80% and 88% for the CHO-K1 cell line, and 60% and 88% for the HepG2 cell line, respectively [39]. Recently, an HCS micronucleus assay in a 384-well plate format was developed for evaluating genotoxicity in CHO-K1 cells [40].
HCS in Developmental Neurotoxicity and Neurotoxicity
Developmental neurotoxicity (DNT) and neurotoxicity (NT) have been linked to neurological diseases including attention deficit hyperactivity disorder (ADHD), autism, Alzheimer’s, and Parkinson’s disease [41–43]. Measuring neuronal morphology and connectivity can assess the toxicity of compounds on DNT and NT [44]. Automated neurite imaging and analysis platforms as well as algorithms specifically for measuring various aspects of neurite outgrowth are commercially available [45–47]. Measuring neurite outgrowth is commonly used for in vitro assessment of NT[44].
Several reports state that HCS neurite outgrowth assays can distinguish neurotoxic compounds from general cytotoxic compounds. One high-content imaging assay allowed the simultaneous evaluation of both neurite outgrowth and cell viability within each well of a plate. This dual-readout assay can distinguish neurite outgrowth inhibitors from compounds detrimental to cell viability [48–53]. Several studies evaluated larger sets of environmental compounds as well as drugs using an HCS neurite outgrowth assay [54–57]. Most of the assays utilized immunostaining or calcein acetoxymethyl staining for measuring neurite outgrowth. Recently, Li et al. developed an HCS neurite outgrowth assay using induced pluripotent stem cell (iPSC) derived neurons labeled with GFP (green fluorescent protein), enabling live and time-lapse imaging of neurite outgrowth [58]. Many protocols detail the preparation of different neuronal types and specific workflows for utilizing them in HCS [59]. Three dimensional (3D) models have been developed and used in neurite outgrowth assays for predicting neurotoxicity, such as the human pluripotent stem cell-derived 3D model [60], the 3D neurospheres [61], and the 3D dopaminergic neuronal model [62]. The 3D neurospheres from human neural progenitor cells mimic basic processes of brain development and are useful for identification of developmental neurotoxicity hazard. Omnisphere software was also developed to assess multiple endpoints in the 3D neurosphere assays with user-assisted parameter optimization procedures [61]. Amenable to HCS, these new models promise to assist in profiling compounds for neurotoxicity and developmental neurotoxicity.
HCS in Hepatotoxicity
Hepatotoxicity is a great concern in drug development and clinical use, since the liver plays a central role in the biotransformation of drugs and toxicants [63]. A group of in vitro assays were applied to assess the hepatotoxicity of a large set of marketed drugs [64]. A variety of cell models have been employed in hepatotoxicity studies, such as the HepG2 cell line, primary hepatocytes, and hepatocyte-like cells derived from pluripotent stem cells [65]. The endpoints for predicting hepatotoxicity include mitochondrial function inhibition, calcium homeostasis disturbance, apoptosis activation, oxidative stress, and inhibition of specific enzymes and transporters [66–68].
However, several mechanisms implicated in hepatotoxicity and the assays using cytotoxicity as a single endpoint may lead to poor sensitivity. A panel of pre-lethal mechanistic cellular assays, such as oxidative stress, cholestasis, steatosis, phospholipidosis, mitochondrial membrane potential, and drug interactions were recommended, since these assays are perceived as more sensitive than general cytotoxicity in detecting an MOA for specific drug toxicities [66]. In the pharmaceutical industry, HCS assays have become a standard tool for identifying the mechanism implicated in toxicity by encompassing multiple endpoints, such as cell viability, mitochondrial integrity, and apoptosis. For example, there have been several multi-parametric HCS assays that were developed to screen and classify hepatotoxic compounds. This multi-parametric strategy using HCS can identify early and late events during the hepatotoxic process. More importantly, the mechanisms implicated in the toxicity suggest a stratification of compounds according to their particular degree of injury [69–74]. In addition, HCS assays using 3D models formed with iPSC-derived hepatocytes and HepG2 were evaluated using 48 known hepatotoxic compounds; significant differences were observed in the hepatotoxicity assessment of compounds between the two cell types [75].
HCS in Cardiotoxicity
Cardiotoxicity is one of the leading causes for drug failure during clinical development, accounting for 22–28% post-marketing drug withdrawal [76, 77]. The new strategies for detecting drug-induced cardiotoxicity were explored by Gintant et al. using in vitro ion channel assays, in silico reconstructions, and human stem cell-derived cardiomyocytes [78]. Inhibition of the human ether-a-go-go-related gene (hERG) is a routinely used to assess cardiotoxicity. Cell-based hERG assays, such as thallium influx [79] and hERG channel current [80] have 78% and 73% accuracies, respectively, in predicting cardiotoxicity. In vitro phenotypic assays such as action potential, field potential, impedance, and Ca2+ dynamics predict cardiotoxicity with 80–100% accuracy [81].
HCS assays for nuclear remodeling, mitochondrial status, apoptosis, and necrosis were also developed for predicting in vitro cardiotoxicity using human pluripotent stem cell-derived cardiomyocytes [82]. With kinetic imaging cytometry, an HCS assay for Ca2+ dynamics was developed for assessment of cardiotoxicity [83]. Alternations in permeability of the ion channel (Na+/K+-pump), intracellular Ca2+ levels, and induction of cell death were also coupled with an HCS system to evaluate the cardiotoxic effects of compounds [84]. Combined phenotypic assays (Ca2+ flux) and HCS assays (cell viability, mitochondrial integrity, and reactive oxygen species) can not only identify potential cardiotoxic compounds, but also explore their MOAs [85, 86]. An HCS assay, utilizing transgenic zebrafish expressing enhanced GFP (eGFP) in vascular endothelial cells, was also developed and optimized to evaluate potential cardiotoxic chemicals. Following treatment, body length, circulation, heart rate, pericardial area, and intersegmental vessel area were quantified using custom image analysis protocols [87]. Moreover, a 3D models including engineered heart tissue and human iPSC-based cardiac micro-physiological system (MPS) were also developed for evaluating cardiotoxic compounds [88, 89]. The engineered heart tissue was generated in a 24-well plate format using differentiated human embryonic stem cells; the tissue showed chronotropic responses to calcium and the β-adrenergic agonist isoprenaline [88]. The cardiac MPS kept stem cell-derived cardiac tissue viable and functional for several weeks. Compared with cellular level studies, the data from cardiac MPS is more consistent with data from whole organs than those done with in vivo models [89].
HCS in Nephrotoxicity
The kidney is particularly vulnerable to chemical- and drug-induced injury because of its role in filtration. Acute renal failure can result from exposure to a variety of drugs, natural products, and industrial and environmental chemicals. Several in vitro cell models have been used for nephrotoxicity evaluations including human embryonic kidney 293 (HEK293), porcine kidney (LLC-PK1), human kidney-2 (HK-2), hTERT immortalized human renal proximal tubular epithelial cell line (RPTEC/TERT1), modified human primary renal proximal tubule epithelial cell (SA7K), and human iPSC-derived renal cells [90–94]. The renal cell types differentiated from pluripotent stem cells, and microfluidic and 3D culture systems are more physiologically relevant, improving the ability to identify and characterize the nephrotoxicants [95]. Several biomarkers, such as Kim-1 and clusterin, indicating renal damage, have been identified and used to predict compound nephrotoxicity [96]. Ma et al. used an HCS assay measuring cell viability, nuclear area, nuclear roundness, mitochondrial mass, and mitochondrial membrane potential in HEK293 cells to evaluate thirteen herbs. The compounds, cantharidin, triptolide, diosbulbin-B, and sophocarpine, present in traditional Chinese medicines, were confirmed to cause nephrotoxicity [97]. By quantifying 129 image-based phenotypic features, chromatin and cytoskeletal characteristics proved to be highly predictable with 82% (primary renal proximal tubular cells) and 89% (immortalized renal proximal tubular cells) accuracies with evaluation of 44 reference compounds [98]. Recently, bioengineered 3D platforms, replicating the complex 3D architecture of a nephron and organ-on-a-chip technologies have shown an increase in sensitivity during the evaluation of toxic agents [99–101]. Future studies should focus on implementation of these complex models in an HCS format, to increase accurate prediction of nephrotoxicity.
Perspectives of HCS technology in Toxicology
Tox21
Toxicology in the 21st Century (Tox21) is a federal collaboration among the Environmental Protection Agency (EPA), the National Center for Advancing Translational Sciences (NCATS), the National Toxicology Program (NTP) at the National Institute of Environmental Health Sciences (NIEHS), and the Food and Drug Administration (FDA) [102]. One of the goals of Tox21 is to develop and validate alternative, non-animal methods to quickly and efficiently test hundreds and thousands of chemicals for potential health effects. HCS promises to address challenges confronted by Toxicity Testing in the 21st Century. To better understand pathways and mechanisms of toxicity in human cell systems, the role of HCS needs to expand [19].
Since 2008, more than 70 quantitative high-throughput assays have been optimized and then used to screen approximately 10,000 chemicals, including approved and investigational drugs, industrial chemicals, and consumer products, such as food additives and chemical mixtures. Many cell-based HTS assays were used to screen compounds with validation done in 384-well or 1536-well plate format. Readouts for these assays include angiogenesis [103, 104], micronucleus [40], phospholipdosis [105], and neurite outgrowth assays [58]. These HCS assays will be incorporated into the Tox21 screening program. The newly published Tox21 strategic plan focuses on assays for molecular events in high priority adverse outcome pathways (AOPs), including previously inaccessible signal types (e.g. ion channel signaling), high-content microscopy, and two-photon imaging of 3D cellular organoid structures [106]. Thus, HCS will play a critical role in bringing Tox21 goals to fruition.
3D Modeling and CRISPR/Cas9 Systems
Advances in cellular models, more closely resembling in vivo cell environments, such as engineered cells, stem cells, 3D cellular models or organisms, have been applied to HCS [5]. The availability of HCS instrumentation to image 3D models and tissues in a 384-well or 1536-well plates, enhances the role of HCS in drug discovery, biological sciences, and toxicological testing. Moreover, methods using 3D cell models and tissues have greatly increased the depth to which tissues can be imaged in situ [107]. Complex 3D systems can be detected and quantified using multiplexed probes [108–110]. Challenges in using 3D models in high-throughput robotic screening system arise because of the tissue’s complexity.
A novel gene-editing approach, CRISPR/Cas9-mediated gene editing can identify new perturbation reagents and build new cell lines [111, 112]. CRISPR/Cas9-mediated gene editing generated cell lines expressing GFP. In addition, CRISPR/Cas9 can be employed to make mutated primary cells, increasing disease relevance. HCS can generate millions of data points with its multi-parametric readouts as well as capturing thousands of images, each associated with metadata. A widely adopted algorithm employed in HCS is t-distributed stochastic neighbor embedding (t-SNE) [113, 114]. HCS employing fluorescence-based readouts can identify a number of false-positives due to compound optical interference [115]. The majority of HCS assays require fluorescent readouts using GFP, potentially inadvertently altering native biological conditions. The first label-free (non-GFP) HCS assay targeting cellular lipid accumulation was developed and validated by combining Bessel beam illumination with sparse sampling acquisition in hyperspectral coherent anti-Stokes Raman scattering (CARS) imaging [116]. This label-free technology has an advantage over fluorescent labeling by not artificially disrupting the biology of the system, improving the biological relevance of HCS.
Machine Learning
Analyzing large image data sets is a challenge, requiring hours of hands-on work filled with subjective pitfalls. With minimal user input, machine learning has the capacity to identify cellular features and phenotypes, greatly reducing the labor required to interpret HCS readouts. Machine learning strategies include supervised and unsupervised approaches. Unsupervised machine learning can derive biologically meaningful information by clustering data points into patterns [117]. Supervised machine learning is typically used to classify problems based on pre-defined classes, and to predict to in which category a new object belongs. Supervised learning can be applied to predicting the MOA with an overall accuracy of 94% [118], predicting compounds-target activities based on data from high-throughput image assays [119]. In addition, supervised learning can also identify small molecules for reverting disease phenotype, such as cerebral cavernous malformation disease caused by loss-of-function of the CCM2 gene [120]. Finally, HCS can also predict gene function using genome-wide RNAi screens, identifying hundreds of genes involved in cellular functions [121].
In summary, to bring toxicological related screening into the twenty-first century, the application of HCS in many areas of toxicology and drug discovery holds much promise. With the development of automated microscopy platforms and image-based informatics, HCS uncovers even subtle phenotypic changes within cell models or tissues in response to perturbations of a chemical or genetic nature. As new methods develop, with better biological insights, HCS stands as a front runner in delivering necessary information for drug discovery in addition to toxicology screening. HCS will significantly contribute to drug discovery, profiling compounds in specific toxicological pathways, and dissecting diverse biological questions.
Acknowledgements
This study was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health. The authors thank Dr. DeeAnn Visk for assistance of editing the manuscript. The views expressed in this review are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the National Center for Advancing Translational Sciences, the NIH. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Lang P, et al. , Cellular imaging in drug discovery. Nat Rev Drug Discov, 2006. 5(4): p. 343–56. [DOI] [PubMed] [Google Scholar]
- 2.Pegoraro G and Misteli T, High-Throughput Imaging for the Discovery of Cellular Mechanisms of Disease. Trends Genet, 2017. 33(9): p. 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchser W, et al. , Assay Development Guidelines for Image-Based High Content Screening, High Content Analysis and High Content Imaging, in Assay Guidance Manual, Sittampalam GS, et al. , Editors. 2004: Bethesda (MD). [PubMed] [Google Scholar]
- 4.Giuliano KA, et al. , High-content screening: A new approach to easing key bottlenecks in the drug discovery process. Journal of Biomolecular Screening, 1997. 2(4): p. 249–259. [Google Scholar]
- 5.Fraietta I and Gasparri F, The development of high-content screening (HCS) technology and its importance to drug discovery. Expert Opin Drug Discov, 2016. 11(5): p. 501–14. [DOI] [PubMed] [Google Scholar]
- 6.Taylor DL, A personal perspective on high-content screening (HCS): from the beginning. J Biomol Screen, 2010. 15(7): p. 720–5. [DOI] [PubMed] [Google Scholar]
- 7.Boutros M, Heigwer F, and Laufer C, Microscopy-Based High-Content Screening. Cell, 2015. 163(6): p. 1314–25. [DOI] [PubMed] [Google Scholar]
- 8.Smith K, et al. , Phenotypic Image Analysis Software Tools for Exploring and Understanding Big Image Data from Cell-Based Assays. Cell Syst, 2018. 6(6): p. 636–653. [DOI] [PubMed] [Google Scholar]
- 9.Caicedo JC, Singh S, and Carpenter AE, Applications in image-based profiling of perturbations. Curr Opin Biotechnol, 2016. 39: p. 134–142. [DOI] [PubMed] [Google Scholar]
- 10.Bickle M, The beautiful cell: high-content screening in drug discovery. Anal Bioanal Chem, 2010. 398(1): p. 219–26. [DOI] [PubMed] [Google Scholar]
- 11.Niu G and Chen X, Has molecular and cellular imaging enhanced drug discovery and drug development? Drugs R D, 2008. 9(6): p. 351–68. [DOI] [PubMed] [Google Scholar]
- 12.Mattiazzi Usaj M, et al. , High-Content Screening for Quantitative Cell Biology. Trends Cell Biol, 2016. 26(8): p. 598–611. [DOI] [PubMed] [Google Scholar]
- 13.Hayer A and Meyer T, High-content imaging. Nature Biotechnology, 2010. 28(5): p. 424–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffat J, et al. , A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell, 2006. 124(6): p. 1283–98. [DOI] [PubMed] [Google Scholar]
- 15.Rangarajan A and Weinberg RA, Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer, 2003. 3(12): p. 952–9. [DOI] [PubMed] [Google Scholar]
- 16.Huang R, et al. , Modelling the Tox21 10 K chemical profiles for in vivo toxicity prediction and mechanism characterization. Nat Commun, 2016. 7: p. 10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MT, et al. , Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data. Environmental Health Perspectives, 2016. 124(5): p. 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo DP, et al. , Nonanimal Models for Acute Toxicity Evaluations: Applying Data-Driven Profiling and Read-Across. Environ Health Perspect, 2019. 127(4): p. 47001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Vliet E, et al. , Current approaches and future role of high content imaging in safety sciences and drug discovery. ALTEX, 2014. 31(4): p. 479–93. [DOI] [PubMed] [Google Scholar]
- 20.Ema M, et al. , Historical control data on prenatal developmental toxicity studies in rabbits. Congenit Anom (Kyoto), 2012. 52(3): p. 155–61. [DOI] [PubMed] [Google Scholar]
- 21.Ema M, et al. , Historical control data on developmental toxicity studies in rodents. Congenit Anom (Kyoto), 2014. 54(3): p. 150–61. [DOI] [PubMed] [Google Scholar]
- 22.de Jong E, et al. , Comparison of the mouse Embryonic Stem cell Test, the rat Whole Embryo Culture and the Zebrafish Embryotoxicity Test as alternative methods for developmental toxicity testing of six 1,2,4-triazoles. Toxicol Appl Pharmacol, 2011. 253(2): p. 103–11. [DOI] [PubMed] [Google Scholar]
- 23.Augustine-Rauch K, Zhang CX, and Panzica-Kelly JM, In vitro developmental toxicology assays: A review of the state of the science of rodent and zebrafish whole embryo culture and embryonic stem cell assays. Birth Defects Res C Embryo Today, 2010. 90(2): p. 87–98. [DOI] [PubMed] [Google Scholar]
- 24.Raftery TD, et al. , High-content screening assay for identification of chemicals impacting spontaneous activity in zebrafish embryos. Environ Sci Technol, 2014. 48(1): p. 804–10. [DOI] [PubMed] [Google Scholar]
- 25.Lantz-McPeak S, et al. , Developmental toxicity assay using high content screening of zebrafish embryos. Journal of Applied Toxicology, 2015. 35(3): p. 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciccia A and Elledge SJ, The DNA damage response: making it safe to play with knives. Mol Cell, 2010. 40(2): p. 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendriks G, et al. , Cellular-signaling pathways unveil the carcinogenic potential of chemicals. J Appl Toxicol, 2013. 33(6): p. 399–409. [DOI] [PubMed] [Google Scholar]
- 28.Knight AW, et al. , Evaluation of high-throughput genotoxicity assays used in profiling the US EPA ToxCast chemicals. Regul Toxicol Pharmacol, 2009. 55(2): p. 188–99. [DOI] [PubMed] [Google Scholar]
- 29.Westerink WM, et al. , Evaluation of the Vitotox and RadarScreen assays for the rapid assessment of genotoxicity in the early research phase of drug development. Mutat Res, 2009. 676(1–2): p. 113–30. [DOI] [PubMed] [Google Scholar]
- 30.Azqueta A, et al. , A comparative performance test of standard, medium- and high-throughput comet assays. Toxicol In Vitro, 2013. 27(2): p. 768–73. [DOI] [PubMed] [Google Scholar]
- 31.Watson C, et al. , High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using CometChip technology. ACS Nano, 2014. 8(3): p. 2118–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motoyama S, et al. , Advantages of evaluating gammaH2AX induction in non-clinical drug development. Genes Environ, 2018. 40: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishihara K, et al. , Identification of genotoxic compounds using isogenic DNA repair deficient DT40 cell lines on a quantitative high throughput screening platform. Mutagenesis, 2016. 31(1): p. 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smart DJ, et al. , Genotoxicity screening via the gammaH2AX by flow assay. Mutat Res, 2011. 715(1–2): p. 25–31. [DOI] [PubMed] [Google Scholar]
- 35.Bryce SM, et al. , Interlaboratory evaluation of a flow cytometric, high content in vitro micronucleus assay. Mutat Res, 2008. 650(2): p. 181–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thougaard AV, et al. , Validation of a high throughput flow cytometric in vitro micronucleus assay including assessment of metabolic activation in TK6 cells. Environ Mol Mutagen, 2014. 55(9): p. 704–18. [DOI] [PubMed] [Google Scholar]
- 37.Bryce SM, et al. , Genotoxic mode of action predictions from a multiplexed flow cytometric assay and a machine learning approach. Environ Mol Mutagen, 2016. 57(3): p. 171–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Canton C, Anadon A, and Meredith C, Assessment of the in vitro gamma H2AX assay by High Content Screening as a novel genotoxicity test. Mutation Research-Genetic Toxicology and Environmental Mutagenesis, 2013. 757(2): p. 158–166. [DOI] [PubMed] [Google Scholar]
- 39.Westerink WM, et al. , Development and validation of a high-content screening in vitro micronucleus assay in CHO-k1 and HepG2 cells. Mutat Res, 2011. 724(1–2): p. 7–21. [DOI] [PubMed] [Google Scholar]
- 40.Shahane SA, Nishihara K, and Xia M, High-Throughput and High-Content Micronucleus Assay in CHO-K1 Cells. Methods Mol Biol, 2016. 1473: p. 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grandjean P and Landrigan PJ, Developmental neurotoxicity of industrial chemicals. Lancet, 2006. 368(9553): p. 2167–2178. [DOI] [PubMed] [Google Scholar]
- 42.Grandjean P and Landrigan PJ, Neurobehavioural effects of developmental toxicity. Lancet Neurol, 2014. 13(3): p. 330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chin-Chan M, Navarro-Yepes J, and Quintanilla-Vega B, Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Frontiers in Cellular Neuroscience, 2015. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radio NM and Mundy WR, Developmental neurotoxicity testing in vitro: models for assessing chemical effects on neurite outgrowth. Neurotoxicology, 2008. 29(3): p. 361–76. [DOI] [PubMed] [Google Scholar]
- 45.Weaver CM, et al. , An algorithm for neurite outgrowth reconstruction. J Neurosci Methods, 2003. 124(2): p. 197–205. [DOI] [PubMed] [Google Scholar]
- 46.Wang DD, et al. , HCA-Vision: Automated Neurite Outgrowth Analysis. Journal of Biomolecular Screening, 2010. 15(9): p. 1165–1170. [DOI] [PubMed] [Google Scholar]
- 47.Charoenkwan P, et al. , HCS-Neurons: identifying phenotypic changes in multi-neuron images upon drug treatments of high-content screening. BMC Bioinformatics, 2013. 14 Suppl 16: p. S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiegler NV, et al. , Assessment of chemical-induced impairment of human neurite outgrowth by multiparametric live cell imaging in high-density cultures. Toxicol Sci, 2011. 121(1): p. 73–87. [DOI] [PubMed] [Google Scholar]
- 49.Krug AK, et al. , Evaluation of a human neurite growth assay as specific screen for developmental neurotoxicants. Arch Toxicol, 2013. 87(12): p. 2215–31. [DOI] [PubMed] [Google Scholar]
- 50.Harrill JA, et al. , Use of high content image analyses to detect chemical-mediated effects on neurite sub-populations in primary rat cortical neurons. Neurotoxicology, 2013. 34: p. 61–73. [DOI] [PubMed] [Google Scholar]
- 51.Sirenko O, et al. , High-content high-throughput assays for characterizing the viability and morphology of human iPSC-derived neuronal cultures. Assay Drug Dev Technol, 2014. 12(9–10): p. 536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrill JA, et al. , Quantitative assessment of neurite outgrowth in human embryonic stem cell-derived hN2 cells using automated high-content image analysis. Neurotoxicology, 2010. 31(3): p. 277–90. [DOI] [PubMed] [Google Scholar]
- 53.Radio NM, et al. , Assessment of chemical effects on neurite outgrowth in PC12 cells using high content screening. Toxicol Sci, 2008. 105(1): p. 106–18. [DOI] [PubMed] [Google Scholar]
- 54.Ryan KR, et al. , Neurite outgrowth in human induced pluripotent stem cell-derived neurons as a high-throughput screen for developmental neurotoxicity or neurotoxicity. Neurotoxicology, 2016. 53: p. 271–281. [DOI] [PubMed] [Google Scholar]
- 55.Delp J, et al. , A high-throughput approach to identify specific neurotoxicants/ developmental toxicants in human neuronal cell function assays. ALTEX, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nyffeler J, et al. , Combination of multiple neural crest migration assays to identify environmental toxicants from a proof-of-concept chemical library. Arch Toxicol, 2017. 91(11): p. 3613–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirenko O, et al. , In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicology and Applied Pharmacology, 2017. 322: p. 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li S, Zhang L, Huang R, Xu T, Behl M, Parham F, Xia M, Evaluation of Chemical Compounds that Inhibit Neurite Outgrowth Using GFP-labeled iPSC-derived Human Neurons. NeuroToxicology, 2019. (revision submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Ali H, et al. , High Content Screening with Primary Neurons, in Assay Guidance Manual, Sittampalam GS, et al. , Editors. 2004: Bethesda (MD). [Google Scholar]
- 60.Clarke KE, et al. , A robust and reproducible human pluripotent stem cell derived model of neurite outgrowth in a three-dimensional culture system and its application to study neurite inhibition. Neurochem Int, 2017. 106: p. 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmuck MR, et al. , Omnisphero: a high-content image analysis (HCA) approach for phenotypic developmental neurotoxicity (DNT) screenings of organoid neurosphere cultures in vitro. Arch Toxicol, 2017. 91(4): p. 2017–2028. [DOI] [PubMed] [Google Scholar]
- 62.Harris G, et al. , Toxicity, recovery, and resilience in a 3D dopaminergic neuronal in vitro model exposed to rotenone. Arch Toxicol, 2018. 92(8): p. 2587–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bjornsson ES, Drug-induced liver injury: an overview over the most critical compounds. Arch Toxicol, 2015. 89(3): p. 327–34. [DOI] [PubMed] [Google Scholar]
- 64.Dambach DM, Andrews BA, and Moulin F, New technologies and screening strategies for hepatotoxicity: use of in vitro models. Toxicol Pathol, 2005. 33(1): p. 17–26. [DOI] [PubMed] [Google Scholar]
- 65.Donato MT, Gomez-Lechon MJ, and Tolosa L, Using high-content screening technology for studying drug-induced hepatotoxicity in preclinical studies. Expert Opinion on Drug Discovery, 2017. 12(2): p. 201–211. [DOI] [PubMed] [Google Scholar]
- 66.Xu JJ, Diaz D, and O’Brien PJ, Applications of cytotoxicity assays and pre-lethal mechanistic assays for assessment of human hepatotoxicity potential. Chem Biol Interact, 2004. 150(1): p. 115–28. [DOI] [PubMed] [Google Scholar]
- 67.Sakamuru S, et al. , Application of a homogenous membrane potential assay to assess mitochondrial function. Physiol Genomics, 2012. 44(9): p. 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia MH, et al. , Comprehensive Analyses and Prioritization of Tox21 10K Chemicals Affecting Mitochondrial Function by in-Depth Mechanistic Studies. Environmental Health Perspectives, 2018. 126(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M, et al. , A testing strategy to predict risk for drug-induced liver injury in humans using high-content screen assays and the ‘rule-of-two’ model. Arch Toxicol, 2014. 88(7): p. 1439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tolosa L, et al. , Development of a multiparametric cell-based protocol to screen and classify the hepatotoxicity potential of drugs. Toxicol Sci, 2012. 127(1): p. 187–98. [DOI] [PubMed] [Google Scholar]
- 71.Kim JA, et al. , Real-time concurrent monitoring of apoptosis, cytosolic calcium, and mitochondria permeability transition for hypermulticolor high-content screening of drug-induced mitochondrial dysfunction-mediated hepatotoxicity. Toxicol Lett, 2012. 214(2): p. 175–81. [DOI] [PubMed] [Google Scholar]
- 72.Persson M, et al. , A high content screening assay to predict human drug-induced liver injury during drug discovery. J Pharmacol Toxicol Methods, 2013. 68(3): p. 302–13. [DOI] [PubMed] [Google Scholar]
- 73.Sirenko O, et al. , High-Content Assays for Hepatotoxicity Using Induced Pluripotent Stem Cell-Derived Cells. Assay and Drug Development Technologies, 2014. 12(1): p. 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tolosa L, et al. , High-content screening of drug-induced mitochondrial impairment in hepatic cells: effects of statins. Arch Toxicol, 2015. 89(10): p. 1847–60. [DOI] [PubMed] [Google Scholar]
- 75.Sirenko O, et al. , Phenotypic Characterization of Toxic Compound Effects on Liver Spheroids Derived from iPSC Using Confocal Imaging and Three-Dimensional Image Analysis. Assay Drug Dev Technol, 2016. 14(7): p. 381–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Onakpoya IJ, Heneghan CJ, and Aronson JK, Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: a systematic review of the world literature. Bmc Medicine, 2016. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilke RA, et al. , Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov, 2007. 6(11): p. 904–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gintant G, Sager PT, and Stockbridge N, Evolution of strategies to improve preclinical cardiac safety testing. Nat Rev Drug Discov, 2016. 15(7): p. 457–71. [DOI] [PubMed] [Google Scholar]
- 79.Xia M, et al. , Identification of quaternary ammonium compounds as potent inhibitors of hERG potassium channels. Toxicol Appl Pharmacol, 2011. 252(3): p. 250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beacham DW, et al. , Cell-based potassium ion channel screening using the FluxOR assay. J Biomol Screen, 2010. 15(4): p. 441–6. [DOI] [PubMed] [Google Scholar]
- 81.Li X, et al. , Cardiotoxicity screening: a review of rapid-throughput in vitro approaches. Arch Toxicol, 2016. 90(8): p. 1803–16. [DOI] [PubMed] [Google Scholar]
- 82.Mioulane M, et al. , Development of high content imaging methods for cell death detection in human pluripotent stem cell-derived cardiomyocytes. J Cardiovasc Transl Res, 2012. 5(5): p. 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cerignoli F, et al. , High throughput measurement of Ca(2)(+) dynamics for drug risk assessment in human stem cell-derived cardiomyocytes by kinetic image cytometry. J Pharmacol Toxicol Methods, 2012. 66(3): p. 246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim MJ, et al. , High-content screening of drug-induced cardiotoxicity using quantitative single cell imaging cytometry on microfluidic device. Lab Chip, 2011. 11(1): p. 104–14. [DOI] [PubMed] [Google Scholar]
- 85.Sirenko O, et al. , In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol Appl Pharmacol, 2017. 322: p. 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grimm FA, et al. , High-Content Assay Multiplexing for Toxicity Screening in Induced Pluripotent Stem Cell-Derived Cardiomyocytes and Hepatocytes. Assay Drug Dev Technol, 2015. 13(9): p. 529–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yozzo KL, et al. , High-content screening assay for identification of chemicals impacting cardiovascular function in zebrafish embryos. Environ Sci Technol, 2013. 47(19): p. 11302–10. [DOI] [PubMed] [Google Scholar]
- 88.Schaaf S, et al. , Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One, 2011. 6(10): p. e26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mathur A, et al. , Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep, 2015. 5: p. 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shaw G, et al. , Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb Journal, 2002. 16(6): p. 869–+. [DOI] [PubMed] [Google Scholar]
- 91.Ryan MJ, et al. , HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int, 1994. 45(1): p. 48–57. [DOI] [PubMed] [Google Scholar]
- 92.Wilmer MJ, et al. , Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening. Trends Biotechnol, 2016. 34(2): p. 156–70. [DOI] [PubMed] [Google Scholar]
- 93.Li Y, et al. , Identification of nephrotoxic compounds with embryonic stem-cell-derived human renal proximal tubular-like cells. Mol Pharm, 2014. 11(7): p. 1982–90. [DOI] [PubMed] [Google Scholar]
- 94.Li S, et al. , Development and Application of Human Renal Proximal Tubule Epithelial Cells for Assessment of Compound Toxicity. Curr Chem Genom Transl Med, 2017. 11: p. 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Soo JY, et al. , Advances in predictive in vitro models of drug-induced nephrotoxicity. Nat Rev Nephrol, 2018. 14(6): p. 378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fuchs TC and Hewitt P, Biomarkers for drug-induced renal damage and nephrotoxicity-an overview for applied toxicology. AAPS J, 2011. 13(4): p. 615–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma Z, et al. , Establishment and Validation of an In Vitro Screening Method for Traditional Chinese Medicine-Induced Nephrotoxicity. Evid Based Complement Alternat Med, 2018. 2018: p. 2461915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su R, et al. , High-throughput imaging-based nephrotoxicity prediction for xenobiotics with diverse chemical structures. Arch Toxicol, 2016. 90(11): p. 2793–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davies J, Engineered renal tissue as a potential platform for pharmacokinetic and nephrotoxicity testing. Drug Discovery Today, 2014. 19(6): p. 725–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peloso A, et al. , Prospect for kidney bioengineering: shortcomings of the status quo. Expert Opin Biol Ther, 2015. 15(4): p. 547–58. [DOI] [PubMed] [Google Scholar]
- 101.Jang KJ, et al. , Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol (Camb), 2013. 5(9): p. 1119–29. [DOI] [PubMed] [Google Scholar]
- 102.Attene-Ramos MS, et al. , The Tox21 robotic platform for the assessment of environmental chemicals--from vision to reality. Drug Discov Today, 2013. 18(15–16): p. 716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li SZ, et al. , Identification of Angiogenesis Inhibitors Using a Co-culture Cell Model in a High-Content and High-Throughput Screening Platform. Slas Technology, 2018. 23(3): p. 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saili Katerine S., F. JA, Baker Nancy C., Ellis-Hutchings Robert G., Settivari Raja S., Carney Edward W., Spencer Richard, Zurlinden Todd J., Kleinstreuer Nicole C., Li Shuaizhang, Xia Menghang, Knudsen Thomas B., Systems Modeling of Developmental Vascular Toxicity. Current Opinion in Toxicology, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shahane SA, et al. , Detection of phospholipidosis induction: a cell-based assay in high-throughput and high-content format. J Biomol Screen, 2014. 19(1): p. 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thomas RS, et al. , The US Federal Tox21 Program: A strategic and operational plan for continued leadership. ALTEX, 2018. 35(2): p. 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Azaripour A, et al. , A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem, 2016. 51(2): p. 9–23. [DOI] [PubMed] [Google Scholar]
- 108.Duchi S, et al. , A new holistic 3D non-invasive analysis of cellular distribution and motility on fibroin-alginate microcarriers using light sheet fluorescent microscopy. PLoS One, 2017. 12(8): p. e0183336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei L, et al. , Super-multiplex vibrational imaging. Nature, 2017. 544(7651): p. 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Senutovitch N, et al. , Fluorescent protein biosensors applied to microphysiological systems. Exp Biol Med (Maywood), 2015. 240(6): p. 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gilbert LA, et al. , Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell, 2014. 159(3): p. 647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lackner DH, et al. , A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun, 2015. 6: p. 10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kraus OZ and Frey BJ, Computer vision for high content screening. Critical Reviews in Biochemistry and Molecular Biology, 2016. 51(2): p. 102–109. [DOI] [PubMed] [Google Scholar]
- 114.van der Maaten L and Hinton G, Visualizing Data using t-SNE. Journal of Machine Learning Research, 2008. 9: p. 2579–2605. [Google Scholar]
- 115.Ibanez G, et al. , Evaluation of Compound Optical Interference in High-Content Screening. SLAS Discov, 2018. 23(4): p. 321–329. [DOI] [PubMed] [Google Scholar]
- 116.Masia F, et al. , Bessel-Beam Hyperspectral CARS Microscopy with Sparse Sampling: Enabling High-Content High-Throughput Label-Free Quantitative Chemical Imaging. Anal Chem, 2018. 90(6): p. 3775–3785. [DOI] [PubMed] [Google Scholar]
- 117.Reisen F, et al. , Linking Phenotypes and Modes of Action Through High-Content Screen Fingerprints. Assay and Drug Development Technologies, 2015. 13(7): p. 415–427. [DOI] [PubMed] [Google Scholar]
- 118.Ljosa V, et al. , Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J Biomol Screen, 2013. 18(10): p. 1321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simm J, et al. , Repurposing High-Throughput Image Assays Enables Biological Activity Prediction for Drug Discovery. Cell Chem Biol, 2018. 25(5): p. 611–618 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gibson CC, et al. , Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation, 2015. 131(3): p. 289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neumann B, et al. , Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature, 2010. 464(7289): p. 721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]