Abstract
Liver dysfunction and type 2 diabetes (T2D) are consistently associated. However, it is currently unknown whether liver dysfunction contributes to, results from, or is merely correlated with T2D due to confounding. We used Mendelian randomization to investigate the presence and direction of any causal relation between liver function and T2D risk including up to 64,094 T2D case and 607,012 control subjects. Several biomarkers were used as proxies of liver function (i.e., alanine aminotransferase [ALT], aspartate aminotransferase [AST], alkaline phosphatase [ALP], and γ-glutamyl transferase [GGT]). Genetic variants strongly associated with each liver function marker were used to investigate the effect of liver function on T2D risk. In addition, genetic variants strongly associated with T2D risk and with fasting insulin were used to investigate the effect of predisposition to T2D and insulin resistance, respectively, on liver function. Genetically predicted higher circulating ALT and AST were related to increased risk of T2D. There was a modest negative association of genetically predicted ALP with T2D risk and no evidence of association between GGT and T2D risk. Genetic predisposition to higher fasting insulin, but not to T2D, was related to increased circulating ALT. Since circulating ALT and AST are markers of nonalcoholic fatty liver disease (NAFLD), these findings provide some support for insulin resistance resulting in NAFLD, which in turn increases T2D risk.
Introduction
Observational studies have repeatedly reported that liver dysfunction and type 2 diabetes (T2D) are associated (1–3). Since the liver plays a core role in the regulation of glucose homeostasis, it is hypothesized that liver dysfunction might increase T2D risk by exacerbating hepatic insulin resistance, leading to overstimulation of hepatic gluconeogenesis (4). Alternatively, it is suggested that insulin resistance and T2D might disturb liver function, possibly via an effect of chronic inflammation and immunological changes (5,6), as well as by directly upregulating hepatic lipogenesis (4). It is currently unknown whether liver dysfunction contributes to, or results from, T2D development or whether there is a genuine bidirectional relationship, where insulin resistance facilitates fat accumulation in the liver, which in turn leads to hepatic insulin resistance and increased fasting glucose (7–9). As most evidence to date is from observational studies, it is possible that the association between liver dysfunction and T2D reflects underlying common causes, such as obesity or lifestyle characteristics.
Plasma concentrations of liver enzymes (i.e., alanine aminotransferase [ALT], aspartate aminotransferase [AST], alkaline phosphatase [ALP], and γ-glutamyl transferase [GGT]) are routinely measured clinical markers that represent different dimensions of liver dysfunction. ALT, located in the cytosol, and AST, located in the mitochondria, are released from damaged hepatic cells into the blood after hepatocellular injury or death. ALT and AST are potentially useful surrogates for alcohol-induced liver disease and nonalcoholic fatty liver disease (NAFLD), defined as hepatic steatosis in the absence of excessive alcohol consumption. ALP is present in the ducts of the liver, and GGT is located on liver cell membranes. The combined elevation of ALP and GGT can indicate obstructive or cholestatic liver disease, where bile is not properly transported from the liver because of obstruction of the bile duct. GGT is also an indicator of alcohol use (10).
Classical observational studies show that plasma concentrations of these enzymes, even within the normal range, are positively associated with T2D (1–3). Mendelian randomization (MR), where genetic variants that are strongly associated with a risk factor of interest are used to test its causal effect on an outcome, can help to distinguish causal effects from associations due to confounding or reverse causality (11,12). Previous MR studies do not support a link between circulating GGT or ALP on T2D risk or glycemic status in Europeans (13,14) or of ALT on T2D risk or glycemic status in Chinese (15). In contrast, MR studies reported some evidence of a positive association of circulating GGT with T2D risk in South Koreans (16) and insulinemia in Europeans (17) and of circulating ALT with T2D risk in Europeans (14). To the best of our knowledge, no MR study has investigated the effect of predisposition to T2D or to insulin resistance on liver function markers.
We have used the largest available data sets to interrogate the potential effect of liver dysfunction, proxied by multiple biomarkers (ALT, AST, ALP, and GGT), on T2D risk (64,094 T2D case and 607,012 control subjects), as well as on related outcomes (fasting glucose, insulin, and lipids). In addition, we have investigated whether predisposition to T2D and insulin resistance affects circulating liver function markers (ALT, AST, ALP, and GGT).
Research Design and Methods
Study Design
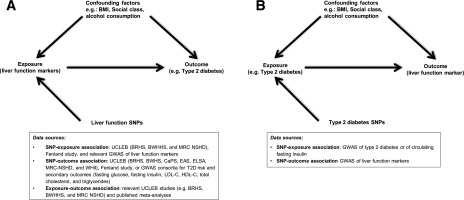
We explored the relationship of four liver function markers (plasma concentration of ALT, AST, ALP, and GGT) with T2D (primary outcome) and with six related metabolic traits (secondary outcomes) reflecting hyperglycemia, assessed by fasting glucose, insulin resistance; assessed by fasting insulin, and dyslipidemia; assessed by LDL cholesterol (LDLc), HDL cholesterol (HDLc), total cholesterol, and triglycerides, using two approaches: multivariable regression and MR. We also used MR to investigate whether predisposition to T2D and to insulin resistance is likely to have an impact on circulating ALT, AST, ALP, and GGT. The hypotheses, study design, and data sources used are detailed in Fig. 1.
Figure 1.
Study design and data sources used to investigate the effect of liver dysfunction (proxied by biomarkers: ALT, AST, ALP, and GGT) on T2D or secondary outcomes (fasting glucose, fasting insulin, LDLc, HDLc, total cholesterol, and triglycerides) (A) and the effect of predisposition to T2D or insulin resistance on circulating liver function biomarkers (B). As shown in A, the multivariable association of liver function markers with T2D risk (or related outcomes) was estimated by meta-analyzing results from each data source using logistic regression models (or linear regression models in the case of secondary outcomes) with participant-level data from relevant studies within the UCLEB consortium (BRHS, BWHHS, MRC NSHD) and summary-level data from the published meta-analyses of Kunutsor et al. (2013) (2) and Fraser et al. (2009) (27). We also estimated the association of liver function markers with T2D risk (or secondary outcomes) using an MR approach. In MR analysis, we used different data sources to estimate the SNP–liver function marker association (UCLEB consortium [BRHS, BWHHS, and MRC NSHD], Fenland study, and GWAS of liver function markers [Chambers et al., 2011] [28]) and SNP–T2D risk association (UCLEB consortium [BRHS, BWHS, CaPS, EAS, ELSA, MRC-NSHD, and WHII] and GWAS consortium) or SNP–secondary outcomes. As shown in B, the summary-level data for the association of SNP–T2D risk and SNP–fasting insulin for the reverse MR was extracted from GWAS consortia, and the association of SNP–liver function marker was extracted from Chambers et al. (2011) (28).
Data Sources
Participant-Level Data
The UCL-LSHTM-Edinburgh-Bristol (UCLEB) consortium consists of 12 prospective observational studies comprising >30,000 participants (18). For the current study, data from up to seven UCLEB studies were included in multivariable and MR analyses: the British Regional Heart Study (BRHS) (19), British Women’s Heart and Health Study (BWHHS) (20), Caerphilly Prospective Study (CaPS) (21), Edinburgh Artery Study (EAS) (22), English Longitudinal Study of Ageing (ELSA) (23), Whitehall II study (WHII) (24), and Medical Research Council (MRC) National Survey of Health and Development (NSHD) (25). Full details of the studies included in the UCLEB consortium have previously been published (18). For the multivariable analyses of liver marker–T2D associations, we used data from up to 6,593 individuals (728 T2D case subjects) from up to five UCLEB studies (BRHS, BWHHS, EAS, CaPS, and NSHD). For the MR analyses of the effect of liver function on T2D risk, we used up to 11,790 individuals (1,202 T2D case subjects) from up to seven UCLEB studies (BRHS, BWHHS, CaPS, EAS, ELSA, NSHD, and WHII) where information on genotypes and a liver function marker/s, and/or information on genotypes and outcome measure/s, was available.
The Fenland study is a U.K. population-based study based in the East Cambridgeshire and Fenland areas and has previously been described in detail (26). Data from Fenland study participants were included in both the multivariable (up to 9,968 individuals) and the MR analyses of the liver marker-lipid outcome associations (up to 9,982 individuals) except for AST.
Full details of the exposure, outcome, and confounder variables available in each UCLEB and Fenland study are given in Supplementary Table 1 and participant characteristics in Supplementary Table 2.
Summary-Level Data
For the multivariable analysis, we pooled our individual participant-level results (from UCLEB and Fenland studies) with those from two published meta-analyses of the association of liver function markers and T2D risk: Kunutsor et al. (2) for ALT and AST (up to 60,359 participants including 3,890 incident T2D cases) and Fraser et al. (27) for GGT (up to 63,285 including 2,805 incident T2D cases).
For the MR analysis, we also used publicly available summary-level data from genome-wide association studies (GWAS) for the association of single nucleotide polymorphisms (SNPs) with exposures and outcomes from the relevant GWAS consortia. Summary statistics for the association between SNP and liver function markers were extracted from the study by Chambers et al. (28), including 61,089 individuals with information of ALT, ALP, AST, and GGT plasma concentration. Summary data for the association between SNPs and T2D were extracted from a consortium (29) including 62,892 case and 596,424 control subjects, mostly of European descent. In cases where a liver function SNP could not be found in this latest T2D GWAS, data were extracted from a previous T2D GWAS from the DIAGRAM consortium including 34,840 case and 114,981 control subjects (30). Summary statistics for the association of SNPs with fasting glucose and with fasting insulin were obtained from MAGIC (the Meta-Analyses of Glucose and Insulin-related traits Consortium) (31,32), which included up to 133,010 and 108,557 participants of European ancestry without diabetes, respectively. Summary data for the association of SNPs with LDLc, HDLc, total cholesterol, and triglycerides were extracted from the GLGC (Global Lipids Genetics Consortium) (33), including 188,577 individuals mostly of European ancestry. We excluded Fenland participants who had been included in the meta-analyses of the GLGC to avoid having duplicated information from these participants.
Definition of Diabetes
In UCLEB studies, T2D definition varied by study and included self-report, medical history review, use of glucose-lowering medication, or having a fasting glucose value of ≥7 mmol/L (18). For the MR analysis, we used both prevalent and incident cases of T2D to maximize power, as the prevalent diabetes cases cannot influence genetic variation, which is fixed at conception.
In the GWAS, criteria for defining T2D differed across studies and included previous diagnosis, fasting glucose ≥7 mmol/L, treatment with glucose-lowering medication, or self-reported T2D status (29,30,34).
Genotyping and Quality Control
Genotyping in all studies in the UCLEB consortium was done using the Illumina Cardio-MetaboChip (Illumina, San Diego, CA). Details on the genotyping and imputation quality control criteria used have previously been published (18,35). Genotyping and imputation of missing genotypes in the published GWAS studies used here are described in the original publications (28–37).
Statistical Analyses
All continuous variables in the UCLEB and Fenland studies that were not normally distributed were natural log transformed. All continuously measured traits were standardized within each study to allow comparison between studies. In any published GWAS that did not report effects in SD units, effects were standardized based on the GWAS reported SD or, where this was not available, the median SD across UCLEB studies.
For analyses involving fasting glucose and fasting insulin, we removed individuals with T2D (defined as a clinical diagnosis, fasting glucose values ≥7.0 mmol/L, or taking glucose-lowering drugs). For analyses involving lipid outcomes, we removed individuals on lipid-lowering medications.
Analyses were performed using Stata/SE version 14.0 (StataCorp, Brownsville, TX) and R version 3.4.4 (R Foundation for Statistical Computing, Vienna, Austria).
Multivariable Regression Analysis
We estimated the association between each liver function marker and T2D in each UCLEB study using logistic regression. For continuous outcomes, we used linear regression models. We adjusted the logistic/linear regression models for age and sex (if relevant) and for as many potential confounders available in each study from the following: BMI, waist circumference, alcohol consumption, smoking, and social class. We then used DerSimonian and Laird random effects model meta-analysis to combine these estimates with results from the published meta-analyses of Kunutsor et al. (2) and Fraser et al. (27) for the associations between ALT, AST, and GGT with T2D, excluding any UCLEB studies that had contributed to the published meta-analyses. In the previously published meta-analyses, all (27) or some (2) of the participating studies expressed T2D results as hazard ratios. Given the overall proportion of T2D cases was <10%, hazard ratios and odds ratios (ORs) were assumed to approximate to the same measure of T2D relative risk in our meta-analysis.
MR Analysis
For the MR analysis, we used multiple genetic variants robustly associated with circulating ALT, AST, ALP, or GGT as genetic instruments for each liver function marker to investigate their effect on T2D and other outcomes. We also applied MR analysis to assess the effect of predisposition to T2D and fasting insulin (a marker of insulin resistance) on circulating ALT, AST, ALP, or GGT by using genetic variants robustly associated with T2D risk and fasting insulin. We used a “two-sample” analysis strategy in which the genetic variant–exposure and genetic variant–outcome associations are estimated from different data sources with comparable populations (38).
Selection of Genetic Instruments for Liver Function Markers
Genetic instruments for each liver function marker were defined as independent SNPs (R2 < 0.3) associated with each liver function marker at genome-wide levels of significance (P < 5 × 10−8) from GWAS in the NIHR GWAS catalog (available at www.ebi.ac.uk/gwas/home). At the time the study was conducted, there were limited GWAS data available for AST. Therefore, we conducted a genome-wide association analysis in up to 6,647 individuals from five UCLEB studies (BRHS, BWHHS, CaPS, EAS, and ET2DS) to identify novel variants for AST, as this biomarker had only been assessed in one previous GWAS with <1,000 individuals (37). We selected GWAS studies, which had been primarily conducted in populations of European ancestry. In total, we selected 4, 3 (including the two novel SNPs identified from our own GWAS), 15, and 26 independent SNPs associated with ALT, AST, ALP, and GGT, respectively (Supplementary Table 3). UCLEB data were excluded from the MR analysis of AST (as an exposure) to avoid any bias from winner’s curse (39).
Selection of Genetic Instrument for Type 2 Diabetes and Fasting Insulin
We used multiple independent SNPs strongly associated (P < 5 × 10−8) with T2D risk (n = 139 SNPs) (29) and with fasting insulin (n = 14 SNPs) (32). T2D SNPs were identified as reported by the original GWAS publication (29). Fasting insulin SNPs were selected from data published by Scott et al. (32) using the R package TwoSampleMR excluding any correlated SNPs (R2 > 0.001) (40).
Main Analysis
In the main MR analyses, we used the conventional inverse variance weighted (IVW) estimator. The IVW method consists of a weighted regression of the SNP-outcome regression coefficients on the SNP-exposure regression coefficients constraining the intercept to be zero. IVW weights are the inverse of the variance of the SNP-outcome regression coefficients. For a dichotomous outcome such as a T2D status, the regression coefficient of the SNP-outcome association is a log OR from a logistic regression model. The resulting regression coefficient from the IVW regression represents an increase/decrease in the outcome per unit increase in the exposure.
Sensitivity Analyses
We performed several sensitivity analyses to test whether the MR IVW estimates are likely to be biased by unbalanced horizontal pleiotropic effects (i.e., due to genetic variants that affect the outcome independently of the exposure of interest). We used the MR-Egger regression method (41) and the weighted median estimator (42), both of which are more robust to pleiotropic genetic variants, to test the extent to which any unbalanced pleiotropy may have biased the IVW result. The MR-Egger method is similar to the IVW except that the model allows the intercept to vary. The intercept of the MR-Egger regression will reflect the average pleiotropic effect across genetic variants and the slope coefficient will provide an estimate of the causal effect provided that the InSIDE (Instrument Strength Independent of Direct Effect) assumption holds, which requires that there is no correlation between SNP-exposure association and any direct (pleiotropic) effects of SNP on outcome (41). In contrast, the weighted median estimator gives consistent estimates even if up to 50% of weight in the analysis comes from invalid genetic instruments (42). For the MR analysis of the effect of insulin resistance, proxied by fasting insulin, on liver markers, we have performed an additional sensitivity analysis, in which we used multivariable MR (43) to adjust results by BMI (32). In all analyses with GWAS data, we have excluded SNPs with C/G or A/T genotypes and minor allele frequency >0.42 due to ambiguity problems they could introduce in harmonizing of SNP-exposure and SNP-outcome data sets (44).
Results
Genetic Instruments for Liver Function Markers, T2D Risk, and Fasting Insulin
Results for the association of genetic instruments with the respective liver function marker are given in Table 1. Most of the genetic instruments previously identified in GWAS consortia replicated (had consistent direction and magnitude) in UCLEB and Fenland, seven were not available in UCLEB but were replicated in Fenland, seven were null or in the opposite direction in UCLEB but consistent in Fenland, and one was null in Fenland (but consistent in UCLEB). Overall, CIs in UCLEB and Fenland included the point estimate from the published GWAS.
Table 1.
The associations of individual SNPs (used as genetic instruments in MR analyses) with the relevant liver function markers in UCLEB studies, Fenland study, and GWAS
| SNP | Liver function marker | Locus* | Effect size in SD units in UCLEB studies (95% CI)† | Effect size in SD units in Fenland study (95% CI)† | Effect size in SD units from published GWAS (95% CI)‡ |
|---|---|---|---|---|---|
| rs6834314 | ALT | HSD17B13, MAPK10 | 0.05 (0.01, 0.09) | 0.05 (0.02, 0.07) | 0.06 (0.04, 0.08) |
| rs11597390 | ALT | CPN1 | 0.09 (0.05, 0.13) | 0.03 (0.01, 0.06) | 0.04 (0.03, 0.06) |
| rs2143571 | ALT | SAMM50 | 0.14 (0.09, 0.19) | 0.07 (0.04, 0.12) | 0.09 (0.07, 0.11) |
| rs2954021 | ALT | TRIB1n | 0.03 (−0.01, 0.07) | 0.02 (−0.00, 0.04) | 0.04 (0.02, 0.05) |
| rs17109512 | AST | GOT1 | −0.04 (−0.18, 0.11) | N/A | 0.03 (−0.05, 0.1) |
| rs738407 | AST | PNPLA3 | 0.13 (0.08, 0.19) | N/A | 0.09 (0.06, 0.12) |
| rs738408 | AST | PNPLA3 | 0.12 (0.08, 0.17) | N/A | 0.19 (0.16, 0.22) |
| rs1780324 | ALP | NBPF3-ALPL | 0.07 (0.02, 0.12) | 0.08 (0.05, 0.10) | 0.09 (0.07, 0.11) |
| rs16856332 | ALP | ABCB11 | −0.02 (−0.20, 0.15) | 0.06 (−0.01, 0.13) | 0.14 (0.09, 0.19) |
| rs9467160 | ALP | GPLD1 | 0.06 (0.01, 0.12) | 0.10 (0.07, 0.13) | 0.1 (0.08, 0.13) |
| rs514708 | ALP | ABO | 0.06 (0.00, 0.12) | 0.08 (0.05, 0.11) | 0.1 (0.08, 0.13) |
| rs2236653 | ALP | ST3GAL4 | 0.01 (−0.05, 0.07) | 0.07 (0.04, 0.10) | 0.06 (0.04, 0.08) |
| rs7186908 | ALP | HPR, PMFBP1 | −0.02 (−0.09, 0.05) | 0.02 (−0.01, 0.05) | 0.07 (0.05, 0.1) |
| rs7267979 | ALP | ABHD12, GINS1, PYGB | 0.03 (−0.02, 0.08) | 0.07 (0.04, 0.10) | 0.05 (0.03, 0.07) |
| rs281377 | ALP | ABO | 0.00 (−0.05, 0.06) | 0.06 (0.04, 0.09) | 0.07 (0.05, 0.09) |
| rs314253 | ALP | ASGR1o, DLG4n | 0.02 (−0.04, 0.07) | 0.04 (0.01, 0.07) | 0.08 (0.06, 0.1) |
| rs579459 | ALP | ABO | 0.08 (0.02, 0.14) | 0.25 (0.22, 0.28) | 0.31 (0.28, 0.34) |
| rs6984305 | ALP | PPP1R3B | −0.03 (−0.11, 0.05) | 0.08 (0.04, 0.12) | 0.1 (0.07, 0.13) |
| rs7923609 | ALP | JMJD1Cnce, NRBF2e | 0.05 (0.00, 0.10) | 0.09 (0.06, 0.11) | 0.08 (0.06, 0.1) |
| rs174601 | ALP | C11orf10e, FADS1e, FADS2ne | −0.00 (−0.06, 0.05) | 0.06 (0.03, 0.08) | 0.06 (0.04, 0.09) |
| rs2954021 | ALP | TRIB1 | 0.07 (0.01, 0.12) | 0.04 (0.01, 0.06) | 0.05 (0.03, 0.07) |
| rs10819937 | ALP | ALDOBo, C9orf125n | Not in UCLEB | 0.06 (0.02, 0.09) | 0.09 (0.06, 0.12) |
| rs1497406 | GGT | RSG1, EPHA2 | 0.05 (0.01, 0.09) | 0.07 (0.04, 0.09) | 0.06 (0.04, 0.07) |
| rs12145922 | GGT | CCBL2, PKN2 | −0.02 (−0.05, 0.02) | 0.01 (−0.02, 0.03) | 0.04 (0.03, 0.06) |
| rs1335645 | GGT | CEPT1, DENND2D | Not in UCLEB | 0.03 (−0.01, 0.07) | 0.07 (0.04, 0.09) |
| rs10908458 | GGT | DPM3, EFNA1, PKLR | 0.04 (0.01, 0.07) | 0.05 (0.02, 0.07) | 0.06 (0.04, 0.07) |
| rs13030978 | GGT | MYO1B, STAT4 | 0.07 (0.02, 0.12) | 0.03 (0.01, 0.06) | 0.06 (0.04, 0.08) |
| rs2140773 | GGT | EFHD1, LOC100129166 | −0.01 (−0.04, 0.03) | 0.02 (−0.01, 0.04) | 0.05 (0.03, 0.06) |
| rs4547811 | GGT | ZNF827 | 0.08 (0.03, 0.13) | 0.09 (0.06, 0.12) | 0.1 (0.08, 0.12) |
| rs4074793 | GGT | ITGA1 | 0.14 (0.07, 0.20) | 0.07 (0.02,0.11) | 0.08 (0.05, 0.12) |
| rs9296736 | GGT | MLIP | 0.03 (0.00, 0.07) | 0.04 (0.02, 0.07) | 0.05 (0.03, 0.06) |
| rs754466 | GGT | DLG5 | Not in UCLEB | 0.07 (0.04, 0.10) | 0.05 (0.03, 0.07) |
| rs8038465 | GGT | NPTN -CD276 | 0.04 (−0.00, 0.09) | 0.00 (−0.02, 0.03) | 0.04 (0.02, 0.05) |
| rs4581712 | GGT | DYNLRB2 | 0.09 (−0.00, 0.18) | 0.07 (0.04, 0.10) | 0.05 (0.03, 0.07) |
| rs1076540 | GGT | MICAL3 | 0.05 (0.01, 0.09) | 0.06 (0.03, 0.09) | 0.07 (0.06, 0.09) |
| rs2739330 | GGT | DDT, DDTL, GSTT1, GSTT2BMIF | 0.06 (0.02, 0.10) | 0.03 (0.01, 0.06) | 0.06 (0.04, 0.08) |
| rs4820599 | GGT | GGT1 | 0.13 (0.10, 0.17) | 0.09 (0.06, 0.12) | 0.13 (0.11, 0.15) |
| rs10513686 | GGT | SLC2A2nc | 0.06 (0.01, 0.11) | 0.04 (0.01, 0.08) | 0.08 (0.05, 0.1) |
| rs1260326 | GGT | C2orf16e, GCKRnc | 0.03 (−0.00, 0.06) | 0.05 (0.02, 0.07) | 0.05 (0.03, 0.07) |
| rs17145750 | GGT | MLXIPLnce | 0.08 (0.03, 0.12) | 0.07 (0.03, 0.10) | 0.07 (0.04, 0.1) |
| rs339969 | GGT | RORAn | 0.05 (0.02, 0.09) | 0.05 (0.03, 0.08) | 0.07 (0.05, 0.09) |
| rs516246 | GGT | FUT2 | 0.04 (0.01, 0.07) | 0.04 (0.01, 0.06) | 0.04 (0.02, 0.05) |
| rs7310409 | GGT | HNF1Anc, C12orf27e | 0.11 (0.08, 0.15) | 0.10 (0.08, 0.13) | 0.1 (0.09, 0.12) |
| rs944002 | GGT | C14orf73nc | 0.12 (0.08, 0.15) | 0.09 (0.06, 0.12) | 0.1 (0.08, 0.12) |
| rs6888304 | GGT | CDH6n | Not in UCLEB | 0.05 (0.02, 0.08) | 0.04 (0.02, 0.06) |
| rs9913711 | GGT | FLJ37644e, SOX9n | Not in UCLEB | 0.05 (0.03, 0.08) | 0.04 (0.02, 0.05) |
| rs12968116 | GGT | ATP8B1ncg | Not in UCLEB | 0.05 (0.01, 0.09) | 0.07 (0.04, 0.1) |
| rs4503880 | GGT | NEDD4Ln | Not in UCLEB | 0.07 (0.04, 0.10) | 0.06 (0.03, 0.08) |
N/A, not applicable.
*Mapped gene for each SNP is given as reported in the original GWAS publication.
†All SNP–liver function marker associations in the UCLEB studies and the Fenland study were adjusted for age and sex (if relevant). Individual study estimates in UCLEB were combined using fixed-effect meta-analyses.
‡Effect sizes extracted from Chambers et al., 2011 (28). GWAS effect sizes for ALT, ALP, and GGT were converted to SD units using the median SD from the UCLEB and Fenland studies.
For the known variant for AST (rs17109512), only P values were provided by the original GWAS, and therefore it was not possible to compare effect estimates with those from the previous GWAS. The two novel variants for AST, identified in our GWAS conducted in five UCLEB studies, were replicated in independent data sources (Table 1 and Supplementary Table 4). In meta-analysis of UCLEB and Fenland studies, 4, 13, and 22 variants were replicated from 4, 15, and 26 variants associated with ALT, ALP, and GGT, and the remaining variants were directionally consistent with the previous reports (data only shown for UCLEB and Fenland separately).
Results for the association of the 139 T2D genetic instruments with T2D risk are given in Supplementary Table 5. Results for the association of the 14 fasting insulin genetic instruments with fasting insulin (in SD log pmol/L) are given in Supplementary Table 6.
Association of Genetic Variants Related to Liver Function Markers With Potential Confounders of the Exposure-Outcome Association
Some individual SNPs used as instruments in the MR analysis were associated with potential confounders of the exposure-outcome association (e.g., age, BMI, waist circumference, waist-to-hip ratio, and other liver function markers). However, when these were combined into a single instrument using fixed-effect meta-analysis, there was no strong evidence that genetic instruments were associated with potential confounders, except for the ALT instrument, which was also associated with AST and GGT, and the AST instrument, which was associated with ALT (Supplementary Table 7).
Multivariable Analysis Between Liver Function Markers and T2D and Related Continuous Outcomes
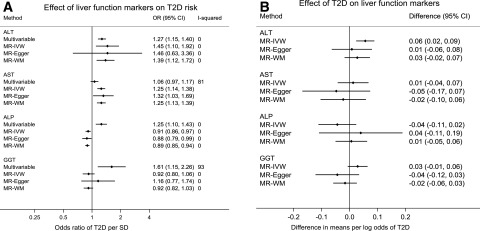
Pooled results from multivariable analyses across the relevant UCLEB and Fenland studies and published meta-analyses are given in Fig. 2A. Most liver function markers were positively associated with T2D risk. The OR for T2D was 1.27 (95% CI 1.15, 1.40) for ALT, 1.06 (95% CI 0.97, 1.17) for AST, 1.25 (95% CI 1.10, 1.43) for ALP, and 1.61 (95% CI 1.15, 2.26) for GGT (per standard unit increase in the liver function marker). Heterogeneity across studies was low for ALT and ALP (I2 = 0%) but high for AST (I2 = 81%) and GGT (I2 = 93%) (Fig. 2A).
Figure 2.
Multivariable and MR analysis of the effect of liver function on T2D (A) and MR analysis of the effect of T2D on liver function markers (B). Results from Fig. 1A correspond to OR of T2D per unit increase in standardized liver function markers (and 95% CI). Results from Fig. 1B correspond to change in standardized liver function markers per unit increase in log odds of T2D (and 95% CI). I-squared indicates between-study heterogeneity and is only presented when estimates for more than one study were available.
In meta-analyses of the continuous outcomes, ALT was positively associated with insulin and triglycerides. AST was positively associated with insulin and HDLc. ALP was negatively related with HDLc. GGT was positively associated with all continuous outcomes (Supplementary Table 8).
Effect of Liver Function Markers on T2D and Related Continuous Outcomes Using MR
Pooled results from the MR across UCLEB, Fenland study, and GWAS are given in Fig. 2A and Supplementary Table 8. In the main MR analysis (IVW), the OR for T2D was 1.45 (95% CI 1.10, 1.92) for ALT, 1.25 (95% CI 1.14, 1.38) for AST, 0.91 (95% CI 0.86, 0.97) for ALP, and 0.92 (95% CI 0.80, 1.06) for GGT (per each standard unit increase in the liver function marker) (Fig. 2A). The other MR methods (MR-Egger and weighted median) used as sensitivity analyses were consistent with IVW estimates for ALT, AST, and ALP. The inverse point estimate for the IVW association between GGT and T2D changed in direction when the MR-Egger method was used. There was no clear evidence of unbalanced horizontal pleiotropy in any liver function marker–T2D associations based on the intercept for MR-Egger method (all P values ≥0.38). Heterogeneity between UCLEB and GWAS estimates was low for all liver markers. For AST, only GWAS data were available for analysis (Fig. 2A).
In meta-analyses of the continuous outcomes, there was some evidence across different MR methods that genetically predicted liver markers were negatively related to blood lipids (Supplementary Table 8). There was some evidence of unbalanced horizontal pleiotropy of ALT instruments in relation to HDLc, LDLc, total cholesterol, and triglycerides (Supplementary Table 9).
Effect of T2D and Insulin Resistance on Liver Function Markers Using MR
Overall, findings from MR analysis did not consistently support a reverse causal effect of T2D predisposition on any of the liver function markers assessed (ALT, AST, ALP, or GGT). In the main MR analysis (IVW), higher predisposition to T2D (each increase in 1 log odds) was related to an increase of 0.06 SD units of ALT (95% CI 0.02, 0.09) but not of AST (0.01 [95% CI −0.04, 0.07]), ALP (−0.04 [95% CI −0.11, 0.02]), or GGT (0.03 [95% CI −0.01, 0.06]). Results for ALT were substantially attenuated with use of MR methods that are more robust to pleiotropic variants: 0.01 (95% CI −0.06, 0.08) for MR-Egger and 0.03 (95% CI −0.02, 0.07) for the weighted median estimator (Fig. 2B).
On the other hand, there was evidence from the MR-IVW findings that greater insulin resistance, proxied by fasting insulin, increases circulating ALT. Results were consistent with the weighted median and BMI-adjusted estimates. MR-Egger point estimates substantially differed, but 95% CIs were very wide. Results for the other liver markers were less consistent across different MR methods, and 95% CIs were wide (Table 2).
Table 2.
MR analysis of the effect of insulin resistance (proxied by circulating fasting insulin) on liver function markers
| Outcome | Method | β | 95% CI | P |
|---|---|---|---|---|
| ALT | IVW | 0.46 | 0.22, 0.69 | 0.0001 |
| MR-Egger | 0.14 | −1.15, 1.42 | 0.84 | |
| Weighted median | 0.43 | 0.10, 0.76 | 0.01 | |
| IVW adjusted by BMI | 0.47 | 0.21, 0.72 | 0.0004 | |
| AST | IVW | 0.31 | −0.1, 0.71 | 0.14 |
| MR-Egger | −1.01 | −3.31, 1.29 | 0.41 | |
| Weighted median | 0.32 | −0.26, 0.89 | 0.28 | |
| IVW adjusted by BMI | 0.25 | −0.22, 0.72 | 0.30 | |
| ALP | IVW | −0.14 | −0.78, 0.49 | 0.66 |
| MR-Egger | 3.39 | 0.37, 6.41 | 0.05 | |
| Weighted median | −0.40 | −0.88, 0.07 | 0.09 | |
| IVW adjusted by BMI | −0.21 | −0.6, 0.18 | 0.29 | |
| GGT | IVW | 0.28 | −0.24, 0.8 | 0.3 |
| MR-Egger | −2.35 | −4.95, 0.25 | 0.10 | |
| Weighted median | 0.51 | 0.18, 0.84 | 0.003 | |
| IVW adjusted by BMI | 0.22 | −0.1, 0.54 | 0.18 |
Results correspond to mean difference (in SD units) of log10 liver function marker (95% CI) per 1 SD increase in fasting insulin (in pmol/L).
Discussion
More than a century ago, a link between liver disease and diabetes was described (45). Since then, multiple observational studies have repeatedly reported that liver dysfunction and T2D are associated (1–3), as broadly replicated by our multivariable analysis using clinical biomarkers (i.e., circulating ALT, AST, ALP, and GGT) as proxies of liver dysfunction. Nevertheless, it is still unclear whether this association reflects causation, and if so, whether liver dysfunction represents a cause or a consequence of T2D.
Our study expands on previous MR analyses investigating the effect of liver dysfunction on T2D risk by including a more comprehensive set of liver function markers routinely used in clinical practice in the largest available data sets and by applying bidirectional MR to investigate whether predisposition to T2D and to insulin resistance might instead lead to liver dysfunction.
Our findings from the MR analyses show evidence that genetic predisposition to higher circulating ALT and AST is related to higher risk of T2D. No strong evidence of a causal effect of genetically predicted GGT on T2D and evidence of a modest negative effect of genetically predicted ALP on T2D were found. Genetic predisposition to T2D did not appear to influence blood concentration of any of the studied liver function markers (ALT, AST, ALP, and GGT), whereas genetic predisposition to insulin resistance, proxied by fasting insulin, seems to increase ALT (effects on other liver markers are uncertain).
Our results are broadly consistent with two previous MR studies, of largely European origin participants (using a study sample that partially overlaps with ours), which reported strong evidence for a positive effect of ALT on T2D (14) but not for ALP (14) or GGT (13,14). Results for ALP (14) were directionally consistent with our findings, but we were better powered to test for the association between ALP and T2D risk given the substantially larger number of T2D case and control subjects included in our analyses. In non-European populations, MR studies suggest that ALT does not relate to T2D risk in Chinese adults (15) but that higher GGT increases T2D risk in Koreans (16). However, the latter result might be explained by statistical overfitting, since instruments were selected from a GWAS that included the population used in the MR analyses (∼20% of the GWAS discovery sample).
The different liver biomarkers reflect different aspects of liver dysfunction. High circulating ALT and AST are widely used proxies of NAFLD, while high circulating ALP and GGT (in combination) are more related to obstructive or cholestatic liver disease. The positive association between genetically predicted liver function markers and T2D in MR analyses was robust for ALT and AST, but not as apparent for ALP or GGT, which suggests that NAFLD might be the primary type of liver dysfunction driving these associations.
In agreement with that, a genetic variant (rs738409) in perfect linkage disequilibrium (R2 = 1.0 for 1000 Genomes European population [GRCh37]) with one of our instruments for AST (rs738408) has previously been reported to be associated with computed tomography–measured hepatic steatosis (46,47). NAFLD is the most common cause of chronic liver disease in Western countries owing to the rapid increase in obesity prevalence (48,49). NAFLD affects 70% of patients with T2D in contrast to ∼20% of the general population (48,49).
To our knowledge, previous MR studies have not examined the causal effect of predisposition to T2D or insulin resistance on liver dysfunction. We found supportive evidence that insulin resistance increases circulating ALT. Our combined findings that insulin resistance (but not T2D) may cause elevated ALT (marker of NAFLD) and that ALT and AST are in turn related to increased T2D risk (but not insulin resistance) are consistent with the twin-cycle hypothesis (8), which postulates that there is a vicious cycle between hepatic insulin resistance and β-cell dysfunction. According to the twin-cycle hypothesis, elevated insulin (due to insulin resistance) stimulates de novo lipogenesis in the liver, which promotes hepatic insulin resistance leading to overstimulation of hepatic gluconeogenesis and increased fasting glucose. The resulting increased output of triglycerides and glucose by the liver to the circulation would impair β-cell function, eventually leading to T2D (9).
On the other hand, it is worth emphasizing that the relation between ALT/AST and T2D risk might be explained by factors other than NAFLD, since circulating ALT/AST are not specific markers of NAFLD and can also increase in response to liver injury from other causes, such as drug toxicity, infection, and alcohol consumption (50,51). In addition, there is some evidence that hepatic triglyceride accumulation by itself may not necessarily cause metabolic changes increasing the risk of cardiometabolic complications (52) and that there might be multiple T2D subtypes that differ in terms of disease presentation and responsiveness to interventions (53), which we were unable to tease out due to the predominant use of summary-level data. Finally, given the key roles of ALT and AST in the intermediary metabolism of glucose and amino acids, we cannot fully discard that they might be directly implicated in T2D development.
Although MR can substantially improve causal inference in epidemiological studies (54), it is important to note that the reliability of MR findings depends on the three core assumptions of instrumental variable (IV) analysis, which require that genetic instruments are strongly associated with the exposure (IV1), are not related to exposure-outcome confounders (IV2), and only influence the outcome through the exposure (IV3).
To avoid violations of IV1, we selected genetic variants strongly associated with liver function markers (P < 5 × 10−8) that broadly replicated in independent data sets. It should also be noted that our genetic instruments were selected to be strongly associated with each liver function marker in the largest available GWAS (P < 5 × 10−8) and that some variants relevant to specific forms of liver dysfunction might not have met our inclusion criteria, as is the case of variants near TM6SF2, previously reported to be associated with NAFLD in a GWAS (55) and with alcohol-related cirrhosis in an exome-wide association study (56). To minimize the risk of population stratification, which could violate IV2, we mostly restricted our analyses to individuals of European ancestry. IV3 could be violated in the presence of horizontal pleiotropy. We have attempted to address that by examining the relation of the genetic variants with several social, behavioral, and metabolic phenotypes and by using methods that are more robust to violations of this assumption in sensitivity analyses (i.e., MR-Egger and weighted median estimator). Overall, there was no strong suggestion that horizontal pleiotropy could have biased our results. Importantly, our liver function instruments were not associated with general adiposity measures such as BMI, and therefore it is unlikely that these associations are driven by general adiposity. However, it is important to note that we cannot fully discard that our genetic instruments may be associated with exposure-outcome confounders that we have not tested for and that the sensitivity analyses used (i.e., MR-Egger and weighted median) are of limited use for exposures instrumented by few genetic variants (as in the analyses of ALT and AST as exposures).
Two-sample MR makes the additional assumption that data from two independent (but comparable) populations are used. In our study, there was some overlap between participants used to estimate SNP-exposure and SNP-outcome associations. However, this is unlikely to bias the study results, since the overlap is very low in proportion to the overall sample size (<7% of T2D cases in the main analysis) (57).
Finally, effect estimates for the relation of each liver function marker on T2D risk should be interpreted with caution given these biomarkers are unlikely to be the causal factors for T2D risk but proxies of liver function.
In conclusion, MR findings indicate that increased circulating ALT and AST are related to higher T2D risk, while increased circulating ALP is associated with lower T2D risk. In addition, higher fasting insulin (but not predisposition to T2D) is related to higher circulating ALT. Since circulating ALT and AST are markers of NAFLD, these findings provide some support for insulin resistance resulting in NAFLD, which in turn increases T2D risk.
Supplementary Material
Article Information
Acknowledgments. The UCLEB consortium consists of 12 studies: Northwick Park Heart Study II (NPHS II), BRHS, WHII, ELSA, MRC NSHD, 1958 Birth cohort (1958BC), CaPS, BWHHS, EAS, Edinburgh Heart Disease Prevention Study (EHDPS), Edinburgh Type 2 Diabetes Study (ET2DS), and Asymptomatic Atherosclerosis Aspirin Trial (AAAT). The authors thank all participants and staff who have contributed to those studies that contributed to the current study (BRHS, BWHHS, CaPS, EAS, ELSA, ET2DS, NSHD, WHII, Fenland study, and NFBC1966). The authors thank Naveed Sattar (Institute of Cardiovascular and Medical Sciences, University of Glasgow, U.K.) for the insightful comments on the interpretation of findings. The authors acknowledge the BRHS team for data collection.
Funding. This work was supported by funds from the U.K. MRC (grant G0801456) and a British Heart Foundation (BHF) grant (AA/18/7/34219). N.M.G.D.S., M.-R.J., and S.S. received support from the European Union’s Horizon 2020 research and innovation programs DynaHEALTH (grant 633595) and LifeCycle (grant 733206). M.C.B., T.R.G., and D.A.L. work in a unit that receives funding from the U.K. MRC (grants MC_UU_00011/6 and MC_UU_00011/4). M.C.B. is supported by a Skills Development Fellowship from the U.K. MRC (grant MR/P014054/1). A.D.H. is a National Institute for Health Research (NIHR) Senior Investigator. X.Z. is supported by a scholarship from the China Scholarship Council (grant 201406220101). A.W. and D.K. are funded by the U.K. MRC (MC UU 12019/1). D.A.L. is a U.K. National Institute for Health Research (NIHR) Senior Investigator (grant NF-0616-10102). The UCLEB consortium is supported by BHF Programme grant RG/10/12/28456 and the UCL Hospitals NIHR Biomedical Research Centre. BRHS is supported by BHF grants (RG/08/013/25942 and RG/13/16/30528). BWHHS is supported by the BHF (PG/13/66/30442). CaPS was funded by the MRC and undertaken by the former MRC Epidemiology Unit (South Wales). The CaPS DNA bank was established with funding from an MRC project grant. The EAS is supported by the BHF and by the Chief Scientist Office of Scotland. ELSA is supported by the National Institute on Aging (NIA)/National Institutes of Health (grant 5 R01 AG017644-16) and a consortium of the U.K. government departments coordinated by the Economic and Social Research Council (ESRC). WHII is supported by grants from the MRC (K013351); BHF (RG/07/008/23674); the Stroke Association; the National Heart, Lung, and Blood Institute (5RO1 HL036310); the NIA (5RO1AG13196); the Agency for Healthcare Research and Quality (HS06516); and the John D. and Catherine T. MacArthur Foundation Research Networks on Successful Midlife Development and Socio-economic Status and Health. NSHD is funded by the U.K. MRC (MC UU 12019/1). The Fenland study is funded by the Wellcome Trust and the MRC (grants MC_U106179471 and MC_UU_12015/1).
The funders had no role in the design, analyses, interpretation of results, or writing of the manuscript. The views expressed in this article are those of the authors and not necessarily any of the funders. The BHF had no role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Duality of Interest. T.R.G. receives support from GlaxoSmithKline, Biogen, and Sanofi for research unrelated to this study. D.A.L. has received support from Medtronic and Roche Diagnostics for biomarker research unrelated to this study. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. D.A.L. designed the study and initial analysis plan, with further input from N.M.G.D.S., M.C.B., and T.R.G. J.E., T.S., X.Z., J.L., C.L., A.W., D.K., J.C.C., W.Z., T.R.G., and D.A.L. collected and/or managed data from one or more of the contributing studies. N.M.G.D.S. and M.C.B. undertook statistical analyses with supervision from T.R.G. and D.A.L. N.M.G.D.S., M.C.B., and D.A.L. wrote the first draft of the manuscript, with all other co-authors contributing to revisions. N.M.G.D.S. and M.C.B. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Data and Resource Availability. Data on T2D have been contributed by the T2D GWAS published by Xue et al. (2018) (29) and have been downloaded from http://cnsgenomics.com/data.html. Data on glycemic traits have been contributed by MAGIC investigators and have been downloaded from www.magicinvestigators.org. Data on lipid traits have been contributed by the GLGC and have been downloaded from http://csg.sph.umich.edu/abecasis/public/lipids2013/. Data on anthropometric traits have been contributed by the Genetic Investigation of ANthropometric Traits (GIANT) consortium and have been downloaded from http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files. UCLEB data are available upon request. Data on mortality and cancer events were routinely provided from NHS Digital to the BWHHS under data sharing agreement MR104a-Regional Heart Study (Female Cohort). BWHHS data are available to bona fide researchers for research purposes. Please refer to the BWHHS data sharing policy at www.ucl.ac.uk/british-womens-heart-health-study. The CaPS data archive is maintained by the University of Bristol.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1048/-/DC1.
N.M.G.D.S. and M.C.B. contributed equally as joint first authors. T.R.G. and D.A.L. contributed equally as joint senior authors.
References
- 1.Ballestri S, Zona S, Targher G, et al. . Nonalcoholic fatty liver disease is associated with an almost twofold increased risk of incident type 2 diabetes and metabolic syndrome. Evidence from a systematic review and meta-analysis. J Gastroenterol Hepatol 2016;31:936–944 [DOI] [PubMed] [Google Scholar]
- 2.Kunutsor SK, Apekey TA, Walley J. Liver aminotransferases and risk of incident type 2 diabetes: a systematic review and meta-analysis. Am J Epidemiol 2013;178:159–171 [DOI] [PubMed] [Google Scholar]
- 3.Wannamethee SG, Shaper AG, Lennon L, Whincup PH. Hepatic enzymes, the metabolic syndrome, and the risk of type 2 diabetes in older men. Diabetes Care 2005;28:2913–2918 [DOI] [PubMed] [Google Scholar]
- 4.Smith BW, Adams LA. Nonalcoholic fatty liver disease and diabetes mellitus: pathogenesis and treatment. Nat Rev Endocrinol 2011;7:456–465 [DOI] [PubMed] [Google Scholar]
- 5.Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology 1998;114:842–845 [DOI] [PubMed] [Google Scholar]
- 6.Utzschneider KM, Kahn SE. Review: the role of insulin resistance in nonalcoholic fatty liver disease. J Clin Endocrinol Metab 2006;91:4753–4761 [DOI] [PubMed] [Google Scholar]
- 7.Loria P, Lonardo A, Anania F. Liver and diabetes. A vicious circle. Hepatol Res 2013;43:51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor R. Pathogenesis of type 2 diabetes: tracing the reverse route from cure to cause. Diabetologia 2008;51:1781–1789 [DOI] [PubMed] [Google Scholar]
- 9.Taylor R, Al-Mrabeh A, Zhyzhneuskaya S, et al. . Remission of human type 2 diabetes requires decrease in liver and pancreas fat content but is dependent upon capacity for β cell recovery. Cell Metab 2018;28:667. [DOI] [PubMed] [Google Scholar]
- 10.Green RM, Flamm S. AGA technical review on the evaluation of liver chemistry tests. Gastroenterology 2002;123:1367–1384 [DOI] [PubMed] [Google Scholar]
- 11.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163 [DOI] [PubMed] [Google Scholar]
- 12.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noordam R, Smit RA, Postmus I, Trompet S, van Heemst D. Assessment of causality between serum gamma-glutamyltransferase and type 2 diabetes mellitus using publicly available data: a Mendelian randomization study. Int J Epidemiol 2016;45:1953–1960 [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Au Yeung SL, Lin SL, Leung GM, Schooling CM. Liver enzymes and risk of ischemic heart disease and type 2 diabetes mellitus: a Mendelian randomization study. Sci Rep 2016;6:38813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Jiang CQ, Lam TH, et al. . Mendelian randomization estimates of alanine aminotransferase with cardiovascular disease: Guangzhou Biobank Cohort study. Hum Mol Genet 2017;26:430–437 [DOI] [PubMed] [Google Scholar]
- 16.Lee YS, Cho Y, Burgess S, et al. . Serum gamma-glutamyl transferase and risk of type 2 diabetes in the general Korean population: a Mendelian randomization study. Hum Mol Genet 2016;25:3877–3886 [DOI] [PubMed] [Google Scholar]
- 17.Conen D, Vollenweider P, Rousson V, et al. . Use of a Mendelian randomization approach to assess the causal relation of gamma-glutamyltransferase with blood pressure and serum insulin levels. Am J Epidemiol 2010;172:1431–1441 [DOI] [PubMed] [Google Scholar]
- 18.Shah T, Engmann J, Dale C, et al.; UCLEB Consortium . Population genomics of cardiometabolic traits: design of the University College London-London School of Hygiene and Tropical Medicine-Edinburgh-Bristol (UCLEB) Consortium. PLoS One 2013;8:e71345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaper AG, Pocock SJ, Walker M, Cohen NM, Wale CJ, Thomson AG. British Regional Heart Study: cardiovascular risk factors in middle-aged men in 24 towns. Br Med J (Clin Res Ed) 1981;283:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawlor DA, Bedford C, Taylor M, Ebrahim S. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women’s Heart and Health Study. J Epidemiol Community Health 2003;57:134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bainton D, Miller NE, Bolton CH, et al.; The Caerphilly and Speedwell Collaborative Heart Disease Studies . Plasma triglyceride and high density lipoprotein cholesterol as predictors of ischaemic heart disease in British men. Br Heart J 1992;68:60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fowkes FG, Housley E, Cawood EH, Macintyre CC, Ruckley CV, Prescott RJ. Edinburgh Artery Study: prevalence of asymptomatic and symptomatic peripheral arterial disease in the general population. Int J Epidemiol 1991;20:384–392 [DOI] [PubMed] [Google Scholar]
- 23.Marmot M, Banks J, Blundell R, Lessof C, Nazroo J. Health, Wealth and Lifestyles of the Older Population in England: The 2002 English Longitudinal Study of Ageing. London, Institute for Fiscal Studies, 2003 [Google Scholar]
- 24.Marmot MG, Smith GD, Stansfeld S, et al. . Health inequalities among British civil servants: the Whitehall II study. Lancet 1991;337:1387–1393 [DOI] [PubMed] [Google Scholar]
- 25.Wadsworth M, Kuh D, Richards M, Hardy R. Cohort profile: the 1946 National Birth Cohort (MRC National Survey of Health and Development). Int J Epidemiol 2006;35:49–54 [DOI] [PubMed] [Google Scholar]
- 26.Burgoine T, Forouhi NG, Griffin SJ, Wareham NJ, Monsivais P. Associations between exposure to takeaway food outlets, takeaway food consumption, and body weight in Cambridgeshire, UK: population based, cross sectional study. BMJ 2014;348:g1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser A, Harris R, Sattar N, Ebrahim S, Davey Smith G, Lawlor DA. Alanine aminotransferase, gamma-glutamyltransferase, and incident diabetes: the British Women’s Heart and Health Study and meta-analysis. Diabetes Care 2009;32:741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers JC, Zhang W, Sehmi J, et al.; Alcohol Genome-wide Association (AlcGen) Consortium; Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study; Genetic Investigation of Anthropometric Traits (GIANT) Consortium; Global Lipids Genetics Consortium; Genetics of Liver Disease (GOLD) Consortium; International Consortium for Blood Pressure (ICBP-GWAS); Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) . Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue A, Wu Y, Zhu Z, et al.; eQTLGen Consortium . Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun 2018;9:2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Schmidt EM, Sengupta S, et al.; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voight BF, Scott LJ, Steinthorsdottir V, et al.; MAGIC investigators; GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan X, Waterworth D, Perry JR, et al. . Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 2008;83:520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen H, Damcott C, Shuldiner SR, et al. . Genome-wide association study identifies genetic variants in GOT1 determining serum aspartate aminotransferase levels. J Hum Genet 2011;56:801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawlor DA. Commentary: two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol 2016;45:908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraft P. Curses--winner’s and otherwise--in genetic epidemiology. Epidemiology 2008;19:649–651; discussion 657–658 [DOI] [PubMed] [Google Scholar]
- 40.Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 10 December 2018 [Epub ahead of print]. DOI: 10.1093/ije/dyy262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol 2016;45:1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naunyn B. Glykosurie und Diabetes durch experimentelle Insulte und Krankheiten der Leber. In Der Diabetes Mellitus. Naunyn B, Ed. Vienna, A. Holder, 1898, p. 38–49 [Google Scholar]
- 46.Speliotes EK, Yerges-Armstrong LM, Wu J, et al.; NASH CRN; GIANT Consortium; MAGIC Investigators; GOLD Consortium . Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 2011;7:e1001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauridsen BK, Stender S, Kristensen TS, et al. Liver fat content, non-alcoholic fatty liver disease, and ischaemic heart disease: Mendelian randomization and meta-analysis of 279 013 individuals. Eur Heart J 2018;39:385–393 [DOI] [PubMed] [Google Scholar]
- 48.Chalasani N, Younossi Z, Lavine JE, et al.; American Association for the Study of Liver Diseases; American College of Gastroenterology; American Gastroenterological Association . The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Am J Gastroenterol 2012;107:811–826 [DOI] [PubMed] [Google Scholar]
- 49.Sattar N, Forrest E, Preiss D. Non-alcoholic fatty liver disease. BMJ 2014;349:g4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dufour DR, Lott JA, Nolte FS, Gretch DR, Koff RS, Seeff LB. Diagnosis and monitoring of hepatic injury. II. Recommendations for use of laboratory tests in screening, diagnosis, and monitoring. Clin Chem 2000;46:2050–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dufour DR, Lott JA, Nolte FS, Gretch DR, Koff RS, Seeff LB. Diagnosis and monitoring of hepatic injury. I. Performance characteristics of laboratory tests. Clin Chem 2000;46:2027–2049 [PubMed] [Google Scholar]
- 52.Sliz E, Sebert S, Würtz P, et al. . NAFLD risk alleles in PNPLA3, TM6SF2, GCKR and LYPLAL1 show divergent metabolic effects. Hum Mol Genet 2018;27:2214–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahlqvist E, Storm P, Käräjämäki A, et al. . Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 2018;6:361–369 [DOI] [PubMed] [Google Scholar]
- 54.Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med 2007;4:e352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozlitina J, Smagris E, Stender S, et al. . Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buch S, Stickel F, Trépo E, et al. . A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443–1448 [DOI] [PubMed] [Google Scholar]
- 57.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.