Abstract
Objective
Numerous genetic variants from meta-analyses of observational studies and GWAS were reported to be associated with migraine susceptibility. However, due to the random errors in meta-analyses, the noteworthiness of the results showing statistically significant remains doubtful. Thus, we performed this field synopsis and re-analysis study to evaluate the noteworthiness using a Bayesian approach in hope of finding true associations.
Methods
Relevant meta-analyses from observational studies and GWAS examining correlation between all genetic variants and migraine risk were included in our study by a PubMed search. Identification of noteworthy associations were analyzed by false-positive rate probability (FPRP) and Bayesian false discovery probability (BFDP). Using noteworthy variants, GO enrichment analysis were conducted through DAVID online tool. Then, the PPI network and hub genes were performed using STRING database and CytoHubba software.
Results
As for 8 significant genetic variants from observational studies, none of which showed noteworthy at prior probability of 0.001. Out of 47 significant genetic variants in GWAS, 36 were noteworthy at prior probability of 0.000001 via FPRP or BFDP. We further found the pathways “positive regulation of cytosolic calcium ion concentration” and “inositol phosphate-mediated signaling” and hub genes including MEF2D, TSPAN2, PHACTR1, TRPM8 and PRDM16 related to migraine susceptibility.
Conclusion
Herein, we have identified several noteworthy variants for migraine susceptibility in this field synopsis. We hope these data would help identify novel genetic biomarkers and potential therapeutic target for migraine.
Keywords: Genetic variant, Migraine, Meta-analysis, FPRP, BFDP, GWAS
Introduction
Migraine is a complex and incapacitating neurologic condition with more than one billion individuals suffering from and imposes a huge socioeconomic burden worldwide [1, 2]. It is characterized by recurrent episodes of unilateral throbbing pain often accompanied with nausea, phonophobia and photophobia, leading to a decline in life quality or even disability [3]. In the Global Burden of Disease (GBD) 2015, migraine ranked the seventh among the leading causes of years lived with disability for all ages and the third for ages 15 to 49 years [4]. Recently, it has been reported that, in the United States, migraine affected almost 15% adults and the annual economic cost was over $ 2600 each person diagnosed with episodic migraine and $ 8000 for those with chronic migraine [5, 6]. Thus, a large amount of studies was performed to explore the risk factors and pathogenesis of migraine over the past decades.
Except for the common risk factors such as obesity, medication overuse, poor sleep, caffeine and stressful life events, the genetic factors for migraine susceptibility was drawing more and more attention [7, 8]. Numerous genetic polymorphisms from meta-analyses of observational studies and GWAS were reported to be associated with migraine susceptibility [9]. However, due to the random errors in meta-analyses leading to false-positive results, the noteworthiness of the results with statistical significance remains doubtful. Therefore, in this field synopsis, we summarized and re-analyzed all significant genetic variants from meta-analyses of observational studies and GWAS, then assessed their noteworthiness using Bayesian procedures including false-positive rate probability (FPRP) and Bayesian false discovery probability (BFDP) and discussed possible molecular mechanisms for migraine occurrence.
Methods
Search strategy and data extraction
A comprehensive literature search was conducted in the PubMed database up to 31 July 2019, by using the following terms: “migraine/ headache” and “meta-analysis” and “polymorphism/genome-wide association study/SNP/GWAS/variant/allele/genotype”. Studies were all selected according to the following criteria: (1) meta-analysis design study; (2) evaluating the association between genetic polymorphisms and migraine risk; (3) raw data available including odds ratios (ORs), 95% confidence intervals (CIs) or other information necessary for FPRP and BFDP calculation; (4) studies published in English. Data including author, published year, genetic variant, genetic model, OR, 95% CI, P-value, ethnicity, type of migraine, number of cases and controls, heterogeneity and publication bias were extracted from the meta-analyses included according to above criteria.
Assessment methods for meta-analysis
In order to figure out the noteworthiness of meta-analysis on the association between genetic polymorphisms and migraine susceptibility, two novel statistic methods, FPRP and BFDP, were applied in our study. FPRP is the probability that no true association exists between genetic variant and disease drawing a statistically significant finding [10]. The magnitude of the FPRP is determined by prior probability, statistical power and observed P-value. Owing to the highly subjective prior probability, we analyzed a wide range of prior probability. We calculated FPRP values using two levels of prior probabilities: at 0.05/0.001 (medium/low prior level) that would be expected for candidate SNPs, and at 0.001/0.000001 (medium/low prior level) for GWAS SNPs. Moreover, the lower the prior probability, the more reliable the result. Besides, we used statistical power to detect ORs of 1.2 and 1.5 for computing FPRP at each prior probability. For the statistically significant SNPs (95% CI that excluded 1 and P-value which was lower than 5 × 10− 8 for meta-analysis of GWAS or 0.05 for observational studies), we calculated FPRP by using the Excel spreadsheet offered by Wacholder (http://jncicancerspectum.oupjournals.org/jnci/content/vol96/issue6) [10]. FPRP values lower than 0.2 were considered to be noteworthy.
BFDP is the fact that if an association is reported as noteworthy, BFDP is the probability of a false discovery [11]. Relevant data and prior probability applied in the calculation of BFDP were same as it of FPRP. The BFDP was estimated by using the excel Calculation Spreadsheet (http://faculty.washington.edu/jonno/cv.html) [11]. And BFDP with the values of less than 0.8 were considered to be a noteworthy significant association. Different from FPRP, BFDP is a new statistical method based on logistic regression model rather than standard normal distribution and doesn’t rely on the statistical power. Thus, BFDP has a sounder methodological basis. Nevertheless, Wakefield admits that there is no significant difference in the overall behavior between these two approaches [11]. Therefore, we presented the both results of FPRP and BFDP which allowed readers to reach a more comprehensive judgment.
Besides, for the meta-analysis of observational studies, summary evidence was also evaluated using Venice criteria, which have been described in detail previously [12, 13]. Briefly, we classified the strength of credibility into A, B and C grades that were separately characterized as strong, moderated and weak in three parameters including amount of evidence, replication of association and the protection from bias. According to this criteria, high credibility was defined as including A grades only, intermediate credibility was composed of A and B grades and low credibility was one or more C grades.
Joint population attributable risk calculation
We evaluated the cumulative effect of all noteworthy SNPs on migraine susceptibility. We used the minor allelic frequency (MAF) to calculated the population attributable risk (PAR) and further estimated the Joint PAR% for the SNPs showing noteworthiness during the computing of BFDP (BFDP < 0.8) or FPRP (FPRP< 0.2) at a prior probability of 10− 6 assumed for GWAS SNPs and 10− 3 for candidate SNPs at a statistical power to detect the OR of 1.5.
GO and enrichment pathway analysis and PPI network construction
Gene ontology (GO) analysis is a useful bioinformatic method for annotating genes. In our study, GO enrichment analyses were carried out using a list of genes with noteworthy SNPs through DAVID online tool (http://david.abcc.ncifcrf.gov/). Then, we applied the STRING 11.0 network database to construct a protein-protein interaction (PPI) network. We set the minimum required interaction score at 0.15, no more than five interactors and four active interaction sources (experiments, text mining, co-expression and database) for PPI construction. Finally, we used the Cytoscape (version 3.4.2) and cytoHubba to detect hub genes meanwhile the cut-off criterion of hub genes was setting as degree ≥7.
Results
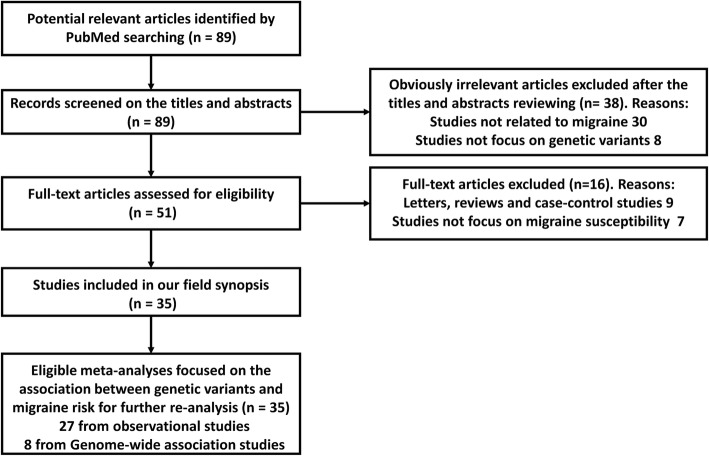
First, a total of 89 articles were identified according to our search strategy. Second, 38 obviously irrelevant articles were excluded by screening the titles and abstracts. After reviewing the full-text articles, 16 were further excluded, among which 9 articles were not meta-analyses and 7 were not correlated with migraine susceptibility. At last, 35 articles were selected into our study [14–48]. The screening process of the articles was performed by two independent researchers and shown in Fig. 1. Overall data retrieved from meta-analyses of observational studies and GWAS on risk of migraine were summarized in Additional file 1: Table S1 and Additional file 2: Table S2. Third, when more than one meta-analysis was available for certain SNP, we only included the results from more recent meta-analysis with larger sample size. Finally, the statistically significant results with P < 0.05 for observational studies and P < 5 × 10− 8 for GWAS were summarized in Tables 1 and 2.
Fig. 1.
The flow chart of the articles screening and selection
Table 1.
Meta-analyses results of genetic variants with statistically significance (P-value< 0.05) from observational studies
| Author, year | Gene/variant | Comparison | OR (95%CI) | P-Value | Ethnicity | No. of cases/controls | FPRP Values at Prior probability | BFDP 0.05 | BFDP 0.001 | Venice criteria | Venice criteria score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||||
| 0.05 | 0.001 | 0.05 | 0.001 | |||||||||||
| Liu L, 2019 [48] | MTHFR/rs1801133 | T vs. C | 1.19 (1.06–1.33) | 0.004 | Overall 26 (Caucasian 20, Asian 6) | 10,228/28608 | 0.069 | 0.795 | 0.040 | 0.685 | 0.476 | 0.980 | A + C + C | Low |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CT + CC | 1.29 (1.06–1.56) | 0.010 | Overall 26 (Caucasian 20, Asian 6) | 10,228/28608 | 0.419 | 0.974 | 0.149 | 0.902 | 0.726 | 0.993 | A + C + C | Low |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT + CT vs. CC | 1.17 (1.02–1.35) | 0.027 | Overall 26 (Caucasian 20, Asian 6) | 10,228/28608 | 0.485 | 0.980 | 0.375 | 0.969 | 0.880 | 0.997 | A + C + C | Low |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CC | 1.32 (1.07–1.64) | 0.011 | Overall 26 (Caucasian 20, Asian 6) | 10,228/28608 | 0.543 | 0.984 | 0.209 | 0.933 | 0.778 | 0.995 | A + C + C | Low |
| Liu L, 2019 [48] | MTHFR/rs1801131 | CC vs. AC + AA | 1.82 (1.09–3.04) | 0.022 | Overall 5 (Caucasian 4, Asian 1) | 1368/1411 | 0.883 | 0.998 | 0.647 | 0.990 | 0.899 | 0.998 | A + C + C | Low |
| Liu L, 2019 [48] | MTHFR/rs1801131 | CC vs. AA | 1.78 (1.03–3.07) | 0.038 | Overall 5 (Caucasian 4, Asian 1) | 1368/1411 | 0.903 | 0.998 | 0.729 | 0.993 | 0.917 | 0.998 | A + C + C | Low |
| Gao X, 2018 [47] | GRIA1/rs2195450 | CT vs. CC | 1.23 (1.02–1.48) | 0.03 | Overall 4 (Caucasian 3, Asian 1) | 963/1167 | 0.576 | 0.986 | 0.354 | 0.966 | 0.862 | 0.997 | A + B + A | Intermediate |
| Terrazzino S, 2017 [44] | BDNF/rs6265 | A vs. G | 1.17 (1.03–1.34) | 0.014 | Overall 5 (Caucasian 5) | 2884/3760 | 0.408 | 0.973 | 0.307 | 0.959 | 0.856 | 0.997 | A + A + A | High |
| Terrazzino S, 2017 [44] | BDNF/rs6265 | AA + GA vs. GG | 1.22 (1.05–1.41) | 0.011 | Overall 5 (Caucasian 5) | 2884/3760 | 0.247 | 0.945 | 0.119 | 0.877 | 0.694 | 0.992 | A + A + A | High |
| Cai X, 2017 [42] | BDNF/rs2049046 | A vs. T | 0.88 (0.79–0.98) | 0.02 | Overall 4 (Caucasian 4) | 1260/1380 | 0.311 | 0.960 | 0.275 | 0.952 | 0.855 | 0.997 | A + A + C | Low |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TA + TT | 0.80 (0.67–0.96) | 0.02 | Overall 4 (Caucasian 4) | 1260/1380 | 0.486 | 0.980 | 0.243 | 0.944 | 0.809 | 0.996 | A + A + A | High |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TT | 0.78 (0.62–0.97) | 0.02 | Overall 4 (Caucasian 4) | 1260/1380 | 0.637 | 0.989 | 0.345 | 0.965 | 0.853 | 0.997 | A + A + A | High |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TA | 0.81 (0.67–0.99) | 0.03 | Overall 4 (Caucasian 4) | 1260/1380 | 0.658 | 0.990 | 0.436 | 0.976 | 0.887 | 0.998 | A + A + A | High |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CC | 1.51 (1.15–1.99) | 0.003 | Overall 5 (Caucasian 4, Asian 1) | 2027/1919 | 0.559 | 0.985 | 0.119 | 0.877 | 0.644 | 0.990 | A + A + A | High |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CG + CC | 1.52 (1.16–1.98) | 0.002 | Overall 5 (Caucasian 4, Asian 1) | 2027/1919 | 0.477 | 0.980 | 0.073 | 0.805 | 0.542 | 0.984 | A + B + A | Intermediate |
| Li L, 2015 [37] | ESR1/rs2228480 | AG vs. GG | 1.14 (1.01–1.28) | 0.030 | Overall 6 (Caucasian 5, Asian 1) | 2293/2026 | 0.385 | 0.971 | 0.336 | 0.964 | 0.877 | 0.997 | A + A + A | High |
| Li L, 2015 [37] | ESR1/rs2228480 | AA + AG vs. GG | 1.13 (1.00–1.26) | 0.003 | Overall 6 (Caucasian 5, Asian 1) | 2293/2026 | 0.381 | 0.970 | 0.346 | 0.965 | 0.885 | 0.998 | A + B + A | Intermediate |
| Liu H, 2011 [23] | 5-HTT/VNTR | Stin2.12 allele | 1.34 (1.09–1.64) | 0.006 | Overall 4 (Caucasian 3, Asian1) | 495/729 | 0.377 | 0.970 | 0.090 | 0.840 | 0.628 | 0.989 | A + A + NA | NA |
| Liu H, 2011 [23] | 5-HTT/VNTR | 12/12 genotype | 1.55 (1.17–2.05) | 0.002 | Overall 4 (Caucasian 3, Asian1) | 495/729 | 0.526 | 0.983 | 0.090 | 0.838 | 0.557 | 0.986 | A + A + NA | NA |
OR odds radio, CI confidence interval, FPRP false-positive rate probability, BFDP Bayesian false discovery probability, NA not available
Table 2.
Meta-analyses results of genetic variants with statistically significance (P-value< 5*10−8) from GWAS
| Author, year | Gene/variant | Comparison | OR (95%CI) | P-Value | Ethnicity | No. of cases/controls | FPRP Values at Prior probability | BFDP 0.001 | BFDP 0.000001 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | |||||||||
| Chang X, 2018 [45] | NMUR2/rs1946225 | G vs. T | 2.29 (1.73–3.05) | 9.55E-09 | Overall 3 (European 1, African 2) | 1212/13494 | 0.717 | 1.000 | 0.006 | 0.851 | 0.027 | 0.965 |
| Chang X, 2018 [45] | NMUR2/rs72793414 | A vs. G | 2.44 (1.85–3.23) | 3.81E-10 | Overall 3 (European 1, African 2) | 1212/13494 | 0.545 | 0.999 | 0.001 | 0.541 | 0.003 | 0.743 |
| Gormley P, 2016 [40] | LRP1/rs11172113 | C vs. T | 0.90 (0.89–0.91) | 5.6E-49 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 6.004E-72 | 6.011E-69 |
| Gormley P, 2016 [40] | PRDM16/rs10218452 | G vs. A | 1.11 (1.10–1.13) | 5.3E-38 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 8.348E-25 | 8.356E-22 |
| Gormley P, 2016 [40] | FHL5/rs67338227 | T vs. A | 1.09 (1.08–1.11) | 2.0E-27 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 4.430E-15 | 4.435E-12 |
| Gormley P, 2016 [40] | TSPAN2/rs2078371 | C vs. T | 1.11 (1.09–1.13) | 4.1E-24 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 8.348E-25 | 8.356E-22 |
| Gormley P, 2016 [40] | TRPM8/rs10166942 | C vs. T | 0.94 (0.89–0.99) | 1.0E-23 | Overall (Caucasian 22) | 59,674/316078 | 0.951 | 1.000 | 0.951 | 1.000 | 0.998 | 1.000 |
| Gormley P, 2016 [40] | PHACTR1/rs9349379 | G vs. A | 0.93 (0.92–0.95) | 5.8E-22 | Overall (Caucasian 22) | 59,674/316078 | 2.308E-08 | 2.310E-05 | 2.308E-08 | 2.310E-05 | 4.007E-06 | 3.995E-03 |
| Gormley P, 2016 [40] | MEF2D/rs1925950 | A vs. G | 1.07 (1.06–1.09) | 9.1E-22 | Overall (Caucasian 22) | 59,674/316078 | 8.014E-10 | 8.022E-07 | 8.014E-10 | 8.022E-07 | 1.694E-07 | 1.695E-04 |
| Gormley P, 2016 [40] | SLC24A3/rs4814864 | C vs. G | 1.07 (1.06–1.09) | 2.2E-19 | Overall (Caucasian 22) | 59,674/316078 | 8.014E-10 | 8.022E-07 | 8.014E-10 | 8.022E-07 | 1.694E-07 | 1.695E-04 |
| Gormley P, 2016 [40] | FGF6/rs1024905 | A vs. G | 1.06 (1.04–1.08) | 2.1E-17 | Overall (Caucasian 22) | 59,674/316078 | 9.959E-07 | 9.959E-04 | 9.959E-07 | 9.959E-04 | 1.767E-04 | 0.150 |
| Gormley P, 2016 [40] | C7orf10/rs186166891 | T vs. A | 1.09 (1.07–1.12) | 9.7E-16 | Overall (Caucasian 22) | 59,674/316078 | 4.993E-07 | 4.935E-04 | 4.993E-07 | 4.935E-04 | 6.430E-05 | 6.048E-02 |
| Gormley P, 2016 [40] | PLCE1/rs10786156 | G vs. C | 0.95 (0.94–0.96) | 2.0E-14 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 3.842E-16 | 3.846E-13 |
| Gormley P, 2016 [40] | KCNK5/rs10456100 | T vs. C | 1.06 (1.04–1.07) | 6.9E-13 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 3.372E-28 | 3.376E-25 |
| Gormley P, 2016 [40] | ASTN2/rs6478241 | T vs. A | 1.05 (1.04–1.07) | 1.2E-12 | Overall (Caucasian 22) | 59,674/316078 | 4.011E-04 | 0.287 | 4.011E-04 | 0.287 | 0.055 | 0.983 |
| Gormley P, 2016 [40] | MRVI1/rs4910165 | G vs. C | 0.94 (0.91–0.98) | 2.9E-11 | Overall (Caucasian 22) | 59,674/316078 | 0.783 | 1.000 | 0.783 | 1.000 | 0.993 | 1.000 |
| Gormley P, 2016 [40] | HPSE2/rs12260159 | A vs. G | 0.92 (0.89–0.94) | 3.2E-10 | Overall (Caucasian 22) | 59,674/316078 | 2.979E-11 | 2.982E-08 | 2.979E-11 | 2.982E-08 | 5.906E-09 | 5.912E-06 |
| Gormley P, 2016 [40] | CFDP1/rs77505915 | A vs. T | 1.05 (1.03–1.06) | 3.3E-10 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 3.476E-18 | 3.479E-15 |
| Gormley P, 2016 [40] | RNF213/rs17857135 | C vs. T | 1.06 (1.04–1.08) | 5.2E-10 | Overall (Caucasian 22) | 59,674/316078 | 9.959E-07 | 9.959E-04 | 9.959E-07 | 9.959E-04 | 1.767E-04 | 0.150 |
| Gormley P, 2016 [40] | NRP1/rs2506142 | G vs. A | 1.06 (1.04–1.07) | 1.5E-09 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 3.372E-28 | 3.376E-25 |
| Gormley P, 2016 [40] | GPR149/rs13078967 | C vs. A | 0.87 (0.83–0.91) | 1.8E-09 | Overall (Caucasian 22) | 59,674/316078 | 1.300E-06 | 1.299E-03 | 1.260E-06 | 1.260E-03 | 1.117E-04 | 0.101 |
| Gormley P, 2016 [40] | JAG1/rs111404218 | G vs. C | 1.05 (1.03–1.07) | 2.0E-09 | Overall (Caucasian 22) | 59,674/316078 | 4.011E-04 | 0.287 | 4.011E-04 | 0.287 | 0.055 | 0.983 |
| Gormley P, 2016 [40] | SPINK2/rs7684253 | T vs. C | 0.96 (0.94–0.97) | 2.5E-09 | Overall (Caucasian 22) | 59,674/316078 | 1.153E-11 | 1.154E-08 | 1.153E-11 | 1.154E-08 | 4.525E-09 | 4.530E-06 |
| Gormley P, 2016 [40] | ZCCHC14/rs4081947 | G vs. A | 1.03 (1.00–1.06) | 2.5E-09 | Overall (Caucasian 22) | 59,674/316078 | 0.978 | 1.000 | 0.978 | 1.000 | 1.000 | 1.000 |
| Gormley P, 2016 [40] | HEY2/rs1268083 | C vs. T | 0.96 (0.95–0.97) | 5.3E-09 | Overall (Caucasian 22) | 59,674/316078 | 1.153E-11 | 1.154E-08 | 1.153E-11 | 1.154E-08 | 4.525E-09 | 4.530E-06 |
| Gormley P, 2016 [40] | WSCD1/rs75213074 | T vs. C | 0.89 (0.86–0.93) | 7.1E-09 | Overall (Caucasian 22) | 59,674/316078 | 2.044E-04 | 0.170 | 2.040E-04 | 0.170 | 0.015 | 0.937 |
| Gormley P, 2016 [40] | GJA1/rs28455731 | T vs. G | 1.06 (1.04–1.08) | 7.3E-09 | Overall (Caucasian 22) | 59,674/316078 | 9.959E-07 | 9.959E-04 | 9.959E-07 | 9.959E-04 | 1.767E-04 | 0.150 |
| Gormley P, 2016 [40] | TGFBR2/rs6791480 | T vs. C | 1.04 (1.03–1.06) | 7.8E-09 | Overall (Caucasian 22) | 59,674/316078 | 0.052 | 0.982 | 0.052 | 0.982 | 0.863 | 1.000 |
| Gormley P, 2016 [40] | ITPK1/rs11624776 | C vs. A | 0.96 (0.94–0.97) | 7.9E-09 | Overall (Caucasian 22) | 59,674/316078 | 1.153E-11 | 1.154E-08 | 1.153E-11 | 1.154E-08 | 4.525E-09 | 4.530E-06 |
| Gormley P, 2016 [40] | ADAMTSL4/rs6693567 | T vs. C | 1.05 (1.03–1.06) | 1.2E-08 | Overall (Caucasian 22) | 59,674/316078 | – | – | – | – | 3.476E-18 | 3.479E-15 |
| Gormley P, 2016 [40] | CCM2L/rs144017103 | T vs. C | 0.85 (0.76–0.96) | 1.2E-08 | Overall (Caucasian 22) | 59,674/316078 | 0.934 | 1.000 | 0.899 | 1.000 | 0.993 | 1.000 |
| Gormley P, 2016 [40] | YAP1/rs10895275 | A vs. T | 1.04 (1.03–1.06) | 1.6E-08 | Overall (Caucasian 22) | 59,674/316078 | 0.052 | 0.982 | 0.052 | 0.982 | 0.863 | 1.000 |
| Gormley P, 2016 [40] | MED14/rs12845494 | G vs. C | 0.96 (0.95–0.97) | 1.7E-08 | Overall (Caucasian 22) | 59,674/316078 | 1.153E-11 | 1.154E-08 | 1.153E-11 | 1.154E-08 | 4.525E-09 | 4.530E-06 |
| Gormley P, 2016 [40] | DOCK4/rs10155855 | T vs. A | 1.08 (1.05–1.12) | 2.1E-08 | Overall (Caucasian 22) | 59,674/316078 | 0.032 | 0.971 | 0.032 | 0.971 | 0.688 | 1.000 |
| Gormley P, 2016 [40] | LRRIQ3/rs1572668 | G vs. A | 1.04 (1.02–1.05) | 2.1E-08 | Overall (Caucasian 22) | 59,674/316078 | 8.873E-13 | 8.882E-10 | 8.873E-13 | 8.882E-10 | 4.186E-10 | 4.190E-07 |
| Gormley P, 2016 [40] | CARF/rs138556413 | G vs. A | 0.88 (0.84–0.92) | 2.3E-08 | Overall (Caucasian 22) | 59,674/316078 | 1.748E-05 | 0.017 | 1.733E-05 | 0.017 | 1.395E-03 | 0.583 |
| Gormley P, 2016 [40] | ARMS2/rs2223089 | C vs. G | 0.93 (0.91–0.95) | 3.0E-08 | Overall (Caucasian 22) | 59,674/316078 | 2.308E-08 | 2.310E-05 | 2.308E-08 | 2.310E-05 | 4.007E-06 | 3.995E-03 |
| Gormley P, 2016 [40] | IGSF9B/rs561561 | T vs. A | 0.94 (0.92–0.96) | 3.4E-08 | Overall (Caucasian 22) | 59,674/316078 | 8.384E-06 | 0.008 | 8.384E-06 | 0.008 | 1.254E-03 | 0.557 |
| Gormley P, 2016 [40] | MPPED2/rs11031122 | C vs. T | 1.04 (1.03–1.06) | 3.5E-08 | Overall (Caucasian 22) | 59,674/316078 | 0.052 | 0.982 | 0.052 | 0.982 | 0.863 | 1.000 |
| Gormley P, 2016 [40] | NOTCH4/rs140002913 | A vs. G | 0.91 (0.88–0.94) | 3.8E-08 | Overall (Caucasian 22) | 59,674/316078 | 1.204E-05 | 0.012 | 1.204E-05 | 0.012 | 1.229E-03 | 0.552 |
| Anttila V, 2013 [29] | PRDM16/rs2651899 | C vs. T | 1.09 (1.07–1.12) | 3.28E-14 | Overall (Caucasian 19) | 23,285/95425 | 4.933E-07 | 4.935E-04 | 4.933E-07 | 4.935E-04 | 6.430E-05 | 0.0605 |
| Anttila V, 2013 [29] | TSPAN2/rs12134493 | A vs. C | 1.14 (1.10–1.18) | 6.71E-14 | Overall (Caucasian 19) | 23,285/95425 | 9.555E-11 | 9.565E-08 | 9.538E-11 | 9.548E-08 | 1.305E-08 | 1.307E-05 |
| Anttila V, 2013 [29] | MEF2D/rs2274316 | C vs. A | 1.07 (1.04–1.09) | 3.14E-08 | Overall (Caucasian 19) | 23,285/95425 | 8.014E-10 | 8.022E-07 | 8.014E-10 | 8.022E-07 | 1.694E-07 | 1.695E-04 |
| Anttila V, 2013 [29] | TRPM8/rs7577262 | A vs. G | 0.87 (0.84–0.90) | 3.27E-13 | Overall (Caucasian 19) | 23,285/95425 | 8.231E-13 | 8.239E-10 | 8.180E-13 | 8.188E-10 | 1.259E-10 | 1.260E-07 |
| Anttila V, 2013 [29] | FHL5/rs13208321 | A vs. T | 1.18 (1.13–1.24) | 2.15E-12 | Overall (Caucasian 19) | 23,285/95425 | 8.181E-08 | 8.189E-05 | 6.109E-08 | 6.115E-05 | 5.781E-06 | 5.753E-03 |
| Anttila V, 2013 [29] | c7orf10/rs4379368 | T vs. C | 1.11 (1.08–1.15) | 1.46E-09 | Overall (Caucasian 19) | 23,285/95425 | 7.560E-06 | 7.511E-03 | 7.560E-06 | 7.511E-03 | 7.339E-04 | 0.424 |
| Anttila V, 2010 [16] | MTDH/rs1835740 | C vs. T | 1.18 (1.13–1.24) | 1.60E-11 | Overall (Caucasian 7) | 5950/50809 | 1.109E-06 | 1.109E-03 | 8.138E-07 | 8.139E-04 | 6.811E-05 | 0.064 |
GWAS genome-wide association studies, OR odds radio, CI confidence interval, FPRP false-positive rate probability, BFDP Bayesian false discovery probability
As shown in Table 1, 5 genes with 8 genetic variants from observational studies were found to be significant after excluding the overlapping data. At a prior probability of 0.05, we identified the genetic variants, MTHFR/rs1801133, noteworthy via FPRP estimation with a statistical power to detect OR of 1.2. Likewise, 4 genetic variants including MTHFR/rs1801133, BDNF/rs6265, ESR1/rs1801132 and 5-HTT/VNTR showed noteworthy FPRP values to detect OR of 1.5. As for the statistical method BFDP, the same 4 genetic variants were noteworthy. However, when it comes to the re-analysis at a prior probability of 0.001, no noteworthy relationship between genetic variants and migraine risk could be detected via FPRP and BFDP.
Moreover, we performed subgroup analysis of observational studies based on the migraine subtypes (Table 3). In the migraine with aura subgroup, only 1 (TNF-α/rs1800629) and 3 (MTHFR/rs1801133, ESR1/rs1801132 and TNF-α/rs1800629) genetic variants was noteworthy in FPRP at the prior probability of 0.05 with a statistical power to detect an OR of 1.2 and 1.5, respectively. And all the variants with noteworthy FPRP values were also noteworthy in BFDP estimation. Similarly, none was identified to be noteworthy in FPRP and BFDP when the prior probability was 0.001. In migraine without aura, we did not observe any noteworthy SNPs. Subsequent subgroup analyses based on ethnicity were performed and displayed in Table 4. Compared with 4 noteworthy SNPs identified at the prior probability of 0.05 containing MTHFR/rs1801133, BDNF/rs6265, ESR1/rs1801132 and ESR1/rs2228480 for Caucasian population, none of candidate polymorphisms was considered to be noteworthy in non-Caucasian population.
Table 3.
Subgroup analysis of genetic variants with statistically significance (P-value< 0.05) from observational studies based on migraine subtype
| Author, year | Gene/variant | Comparison | OR (95%CI) | P-value | Subtypes | No. of cases/controls | FPRP Values at Prior probability | BFDP 0.05 | BFDP 0.001 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||
| 0.05 | 0.001 | 0.05 | 0.001 | |||||||||
| Liu L, 2019 [48] | MTHFR/rs1801133 | T vs. C | 1.28 (1.09–1.51) | 0.003 | MA | 4313/28092 | 0.226 | 0.939 | 0.063 | 0.778 | 0.560 | 0.985 |
| Liu L, 2019 [48] | MTHFR/rs1801133 | CT + TT vs. CC | 1.20 (1.00–1.44) | 0.049 | MA | 4313/28092 | 0.655 | 0.990 | 0.489 | 0.981 | 0.904 | 0.998 |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CT + CC | 1.46 (1.10–1.95) | 0.010 | MA | 4313/28092 | 0.682 | 0.991 | 0.256 | 0.948 | 0.788 | 0.995 |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CC | 1.51 (1.09–2.08) | 0.012 | MA | 4313/28092 | 0.735 | 0.993 | 0.314 | 0.960 | 0.812 | 0.996 |
| Terrazzino S, 2017 [44] | BDNF/rs6265 | GA + AA vs. GG | 1.22 (1.00–1.47) | 0.047 | MA | 717/1593 | 0.617 | 0.988 | 0.413 | 0.974 | 0.882 | 0.997 |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CC | 1.59 (1.17–2.15) | 0.003 | MA | 1427/1919 | 0.593 | 0.987 | 0.123 | 0.880 | 0.632 | 0.989 |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CG + CC | 1.58 (1.18–2.13) | 0.002 | MA | 1427/1919 | 0.590 | 0.987 | 0.122 | 0.880 | 0.633 | 0.989 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | AA+GA vs. GG | 1.17 (1.05–1.30) | 0.004 | MA | 1763/21837 | 0.089 | 0.837 | 0.062 | 0.777 | 0.582 | 0.987 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | A vs. G | 1.13 (1.03–1.24) | 0.010 | MA | 1763/21837 | 0.173 | 0.917 | 0.159 | 0.908 | 0.783 | 0.995 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | GA vs. GG | 1.17 (1.05–1.31) | 0.005 | MA | 1763/21837 | 0.115 | 0.906 | 0.110 | 0.866 | 0.694 | 0.992 |
| Chen M, 2015 [35] | NOS3/rs1799983 | TT vs. GG | 1.61 (1.12–2.31) | 0.010 | MA | 440/881 | 0.770 | 0.994 | 0.345 | 0.965 | 0.816 | 0.996 |
| Chen M, 2015 [35] | NOS3/rs1799983 | TT vs. GT + GG | 1.50 (1.08–2.09) | 0.016 | MA | 440/881 | 0.771 | 0.994 | 0.387 | 0.971 | 0.843 | 0.996 |
| Liu H, 2011 [23] | 5-HTT/VNTR | 12/other | 1.33 (1.01–1.75) | 0.042 | MA | 176/629 | 0.774 | 0.994 | 0.496 | 0.981 | 0.891 | 0.998 |
| Liu H, 2011 [23] | 5-HTT/VNTR | 12/12 vs. 12/other +other | 1.58 (1.07–2.33) | 0.021 | MA | 176/629 | 0.829 | 0.996 | 0.501 | 0.981 | 0.873 | 0.997 |
| Schurks M, 2010 [20] | ACE/rs1799752 | II vs. ID + DD | 0.71 (0.55–0.93) | 0.011 | MA | 1761/22310 | 0.667 | 0.991 | 0.266 | 0.950 | 0.801 | 0.995 |
| Liu L, 2019 [48] | MTHFR/rs1801131 | C vs. A | 1.43 (1.06–1.92) | 0.018 | MO | 159/1477 | 0.730 | 0.993 | 0.345 | 0.965 | 0.835 | 0.996 |
| Liu L, 2019 [48] | MTHFR/rs1801131 | CC vs. AC + AA | 2.74 (1.46–5.14) | 0.002 | MO | 159/1477 | 0.864 | 0.997 | 0.515 | 0.982 | 0.842 | 0.996 |
| Liu L, 2019 [48] | MTHFR/rs1801131 | CC vs. AA | 2.83 (1.30–6.16) | 0.009 | MO | 159/1477 | 0.916 | 0.998 | 0.752 | 0.994 | 0.991 | 0.998 |
| Liu H, 2011 [23] | 5-HTT/VNTR | 12/other | 1.30 (1.02–1.67) | 0.037 | MO | 319/697 | 0.741 | 0.993 | 0.467 | 0.979 | 0.887 | 0.998 |
| Liu H, 2011 [23] | 5-HTT/VNTR | 12/12 vs. 12/other +other | 1.55 (1.11–2.16) | 0.010 | MO | 319/697 | 0.737 | 0.993 | 0.302 | 0.958 | 0.801 | 0.995 |
| Schurks M, 2010 [20] | ACE/rs1799752 | II vs. ID + DD | 0.84 (0.70–0.99) | 0.049 | MO | 2853/22310 | 0.570 | 0.986 | 0.417 | 0.974 | 0.888 | 0.998 |
MA migraine with aura, MO migraine without aura
Table 4.
Subgroup analysis of genetic variants with statistically significance (P-value< 0.05) from observational studies based on ethnicity
| Author, year | Gene/variant | Comparison | OR (95%CI) | P-Value | Ethnicity | No. of cases/controls | FPRP Values at Prior probability | BFDP 0.05 | BFDP 0.001 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||
| 0.05 | 0.001 | 0.05 | 0.001 | |||||||||
| Liu L, 2019 [48] | MTHFR/rs1801133 | T vs. C | 1.18 (1.04–1.34) | 0.012 | Caucasian 20 | 9635/27592 | 0.253 | 0.947 | 0.169 | 0.915 | 0.766 | 0.994 |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CT + CC | 1.25 (1.02–1.53) | 0.035 | Caucasian 20 | 9635/27592 | 0.626 | 0.989 | 0.376 | 0.969 | 0.867 | 0.997 |
| Liu L, 2019 [48] | MTHFR/rs1801133 | TT vs. CC | 1.28 (1.02–1.60) | 0.036 | Caucasian 20 | 9635/27592 | 0.667 | 0.991 | 0.384 | 0.968 | 0.886 | 0.997 |
| Dong H, 2018 [46] | eNOS/rs2070744 | CC vs. TC + TT | 1.62 (1.03–2.56) | 0.04 | Caucasian 4 | 435/344 | 0.881 | 0.997 | 0.665 | 0.991 | 0.908 | 0.998 |
| Terrazzino S, 2017 [44] | BDNF/rs6265 | A vs. G | 1.17 (1.03–1.34) | 0.014 | Caucasian 5 | 2884/3760 | 0.408 | 0.973 | 0.307 | 0.959 | 0.856 | 0.997 |
| Terrazzino S, 2017 [44] | BDNF/rs6265 | AA + GA vs. GG | 1.22 (1.05–1.41) | 0.011 | Caucasian 5 | 2884/3760 | 0.247 | 0.945 | 0.119 | 0.877 | 0.694 | 0.992 |
| Cai X, 2017 [42] | BDNF/rs2049046 | A vs. T | 0.88 (0.79–0.98) | 0.02 | Caucasian 4 | 1260/1380 | 0.311 | 0.960 | 0.275 | 0.952 | 0.855 | 0.997 |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TA + TT | 0.80 (0.67–0.96) | 0.02 | Caucasian 4 | 1260/1380 | 0.486 | 0.980 | 0.243 | 0.944 | 0.809 | 0.996 |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TT | 0.78 (0.62–0.97) | 0.02 | Caucasian 4 | 1260/1380 | 0.637 | 0.989 | 0.345 | 0.965 | 0.853 | 0.997 |
| Cai X, 2017 [42] | BDNF/rs2049046 | AA vs. TA | 0.81 (0.67–0.99) | 0.03 | Caucasian 4 | 1260/1380 | 0.658 | 0.990 | 0.436 | 0.976 | 0.887 | 0.976 |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CC | 1.63 (1.20–2.22) | 0.002 | Caucasian 4 | 1693/1719 | 0.586 | 0.987 | 0.110 | 0.866 | 0.600 | 0.988 |
| Li L, 2015 [37] | ESR1/rs1801132 | GG vs. CG + CC | 1.63 (1.21–2.21) | 0.001 | Caucasian 4 | 1693/1719 | 0.564 | 0.986 | 0.096 | 0.848 | 0.572 | 0.986 |
| Li L, 2015 [37] | ESR1/rs2228480 | AG vs. GG | 1.19 (1.04–1.35) | 0.009 | Caucasian 5 | 1959/1826 | 0.192 | 0.926 | 0.116 | 0.873 | 0.696 | 0.992 |
| Li L, 2015 [37] | ESR1/rs2228480 | AA + AG vs. GG | 1.17 (1.04–1.33) | 0.016 | Caucasian 5 | 1959/1826 | 0.323 | 0.962 | 0.237 | 0.942 | 0.821 | 0.996 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | A vs. G | 1.74 (1.13–2.67) | 0.012 | Non-Caucasian 5 | 985/956 | 0.828 | 0.996 | 0.462 | 0.978 | 0.851 | 0.997 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | AA+GA vs. GG | 1.82 (1.15–2.87) | 0.010 | Non-Caucasian 5 | 985/956 | 0.838 | 0.996 | 0.483 | 0.980 | 0.854 | 0.997 |
| Chen M, 2015 [35] | TNF-α/rs1800629 | GA vs. GG | 1.78 (1.17–2.72) | 0.007 | Non-Caucasian 5 | 985/956 | 0.810 | 0.996 | 0.405 | 0.973 | 0.828 | 0.996 |
| Chen M, 2015 [35] | NOS3/rs1799983 | TT vs. GT + GG | 1.84 (1.02–3.33) | 0.043 | Non-Caucasian 3 | 504/339 | 0.914 | 0.998 | 0.770 | 0.994 | 0.923 | 0.998 |
| Chen M, 2015 [35] | NOS3/rs1799983 | TT vs. GG | 2.10 (1.14–3.88) | 0.018 | Non-Caucasian 3 | 504/339 | 0.902 | 0.998 | 0.706 | 0.992 | 0.907 | 0.998 |
| Liu R, 2014 [32] | TNF-β/rs909253 | GG vs. AG + AA | 1.38 (1.04–1.84) | 0.027 | Non-Caucasian 3 | 746/717 | 0.759 | 0.994 | 0.428 | 0.975 | 0.869 | 0.997 |
In addition to FPRP and BFDP, we also used the Venice criteria to evaluate the credibility of the meta-analyses characterized by low, intermediate and high level (Table 1). We observed a consistency between the noteworthiness measured by FPRP and BFDP and Venice criteria score for candidate SNPs, that is, most of the noteworthy SNPs observed in our re-analysis were with high or intermediate level of evidence, with the exception of MTHFR/rs1801133.
As shown in Table 2, within the data extracted from the meta-analyses of GWAS, 40 genes with 47 genetic variants were statistically significant (P < 5 × 10− 8). According to the results of the re-analyses, 32 and 26 SNPs were found to be noteworthy via FPRP estimation at the statistical power to detect the OR of 1.2 with the prior probability of 0.001 and 0.000001. 34 and 26 SNPs were identified as noteworthy to detect the OR of 1.5 via FPRP. As for BFDP, 40 and 35 SNPs were noteworthy under the prior probability of 0.001 and 0.000001. Almost all noteworthy variants calculated through FPRP also showed noteworthy BFDP values. Only three variants (YAP1/rs10895275, TGFBR2/rs6791480 and MPPED2/ rs11031122) were noteworthy in the calculation of FPRP, but not via BFDP. In addition, we detected 9 noteworthy SNPs (LRP1/rs11172113, PRDM16/rs10218452, FHL5/rs67338227, TSPAN2/rs2078371, PLCE1/ rs10786156, KCNK5/rs10456100, CFDP1/rs77505915, NRP1/rs2506142 and ADAMTSL4/rs6693567) in the computation of BFDP rather than FPRP, which could be explained by the fact that, in some cases, noteworthiness could not be assessed on account of a mathematical error during the calculation of the inverse of cumulative normal distribution in FPRP. Furthermore, in the subgroup analysis, we only performed the re-analysis of migraine without aura but not migraine with aura owing to the lack of raw data. We found that all ten statistically significant variants within 8 genes were noteworthy in migraine without aura subgroup (Table 5). In addition, due to the population included in the GWAS were almost Caucasians, subgroup re-analysis based on ethnicity cannot be performed.
Table 5.
Subgroup analysis results of genetic variants with statistically significance (P-value< 5*10−8) from GWAS studies based on migraine subtype
| Author, year | Gene/variant | Comparison | OR (95%CI) | P-value | Subtypes | No. of cases/controls | FPRP Values at Prior probability | BFDP 0.001 | BFDP 0.000001 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR 1.2 | OR 1.5 | |||||||||||
| 0.001 | 0.000001 | 0.001 | 0.000001 | |||||||||
| Gormley P, 2016 [40] | LRP1/rs11172113 | C vs. T | 0.85 (0.82–0.89) | 4.3E-16 | MO | 8348/139622 | 5.365E-09 | 5.370E-06 | 4.295E-09 | 4.300E-06 | 4.578E-07 | 4.580E-04 |
| Gormley P, 2016 [40] | FHL5/rs7775721 | T vs. A | 1.15 (1.11–1.20) | 1.1E-12 | MO | 8348/139622 | 1.253E-07 | 1.254E-04 | 1.222E-07 | 1.223E-04 | 1.210E-05 | 0.012 |
| Gormley P, 2016 [40] | ASTN2/rs6478241 | G vs. A | 1.14 (1.09–1.18) | 1.2E-10 | MO | 8348/139622 | 9.555E-11 | 9.565E-08 | 9.538E-11 | 9.548E-08 | 1.305E-08 | 1.307E-05 |
| Gormley P, 2016 [40] | TRPM8/rs6724624 | G vs. C | 0.86 (0.82–0.90) | 1.1E-09 | MO | 8348/139622 | 8.654E-08 | 8.662E-05 | 7.899E-08 | 7.906E-05 | 7.681E-06 | 7.630E-03 |
| Gormley P, 2016 [40] | PHACTR1/rs9349379 | G vs. A | 0.88 (0.85–0.92) | 2.1E-09 | MO | 8348/139622 | 1.748E-05 | 0.017 | 1.733E-05 | 0.017 | 1.395E-03 | 0.583 |
| Gormley P, 2016 [40] | FGF6/rs1024905 | A vs. G | 1.12 (1.08–1.16) | 2.5E-09 | MO | 8348/139622 | 2.451E-07 | 2.453E-04 | 2.451E-07 | 2.453E-04 | 2.680E-05 | 0.026 |
| Gormley P, 2016 [40] | TSPAN2/rs2078371 | C vs. T | 1.18 (1.12–1.25) | 7.4E-09 | MO | 8348/139622 | 2.525E-05 | 0.025 | 1.808E-05 | 0.018 | 1.276E-03 | 0.561 |
| Anttila V, 2013 [29] | TRPM8/rs6741751 | A vs. G | 0.80 (0.75–0.86) | 8.64E-11 | MO | 7107/69427 | 1.094E-05 | 0.011 | 1.469E-06 | 1.469E-03 | 1.143E-04 | 0.103 |
| Anttila V, 2013 [29] | LRP1/rs11172113 | C vs. T | 0.87 (0.84–0.91) | 9.96E-11 | MO | 7107/69427 | 1.300E-06 | 1.300E-03 | 1.260E-06 | 1.260E-03 | 1.117E-04 | 0.101 |
| Anttila V, 2013 [29] | PHACTR1/rs9349379 | G vs. A | 0.86 (0.82–0.90) | 2.81E-10 | MO | 7107/69427 | 8.654E-08 | 8.662E-05 | 7.899E-08 | 7.906E-05 | 7.681E-06 | 7.630E-03 |
| Anttila V, 2013 [29] | FHL5/rs11759769 | A vs. G | 1.18 (1.13–1.24) | 1.58E-12 | MO | 7107/69427 | 8.181E-08 | 8.189E-05 | 6.109E-08 | 6.115E-05 | 5.781E-06 | 5.753E-03 |
| Anttila V, 2013 [29] | MMP16/rs10504861 | T vs. C | 0.86 (0.81–0.90) | 1.17E-08 | MO | 7107/69427 | 8.654E-08 | 8.662E-05 | 7.899E-08 | 7.906E-05 | 7.681E-06 | 7.630E-03 |
GWAS genome-wide association studies, MO migraine without aura
In order to provide readers with the predictive potential of the migraine risk, we calculated the joint PAR which was recently argued to be more credible than P-values or ORs by combining 36 noteworthy variants via FPRP or BFDP. And the value of joint PAR in our study was 44.2%, indicating that these involved SNPs together may attribute to an obvious increment in the risk of migraine and this method may be a useful way for screening migraine susceptibility and identify at-risk populations (Table 6).
Table 6.
Combination of genetic polymorphisms to predict risk of migraine. Calculation of joint PAR% using the SNPs showing noteworthiness during the computing of BFDP (BFDP < 0.8) or FPRP (FPRP< 0.2) at a statistical power to detect the OR of 1.5
| Author, year | Gene/variant | Comparison | MAF | OR (95%CI) | P-value | Ethnicity | No. of cases/controls | PAR (%) | Joint PAR (%) |
|---|---|---|---|---|---|---|---|---|---|
| Chang X, 2018 [45] | NMUR2/rs72793414 | A vs. G | 0.1082 | 2.44 (1.85–3.23) | 3.81E-10 | Overall 2 | 1212/13494 | 13.4804396578 | 44.2094776354 |
| Gormley P, 2016 [40] | LRP1/rs11172113 | C vs. T | 0.3894 | 0.90 (0.89–0.91) | 5.6E-49 | Caucasian 22 | 59,674/316078 | 4.0517761638 | |
| Gormley P, 2016 [40] | PRDM 16/rs10218452 | G vs. A | 0.2264 | 1.11 (1.10–1.13) | 5.3E-38 | Caucasian 22 | 59,674/316078 | 2.4298861162 | |
| Gormley P, 2016 [40] | FHL5/rs67338227 | T vs. A | 0.0220 | 1.09 (1.08–1.11) | 2.0E-27 | Caucasian 22 | 59,674/316078 | 0.1976087347 | |
| Gormley P, 2016 [40] | TSPAN2/rs2078371 | C vs. T | 0.1252 | 1.11 (1.09–1.13) | 4.1E-24 | Caucasian 22 | 59,674/316078 | 1.3584908638 | |
| Gormley P, 2016 [40] | PHACTR1/rs9349379 | G vs. A | 0.3774 | 0.93 (0.92–0.95) | 5.8E-22 | Caucasian 22 | 59,674/316078 | 2.7134848426 | |
| Gormley P, 2016 [40] | MEF2D/rs1925950 | A vs. G | 0.4277 | 1.07 (1.06–1.09) | 9.1E-22 | Caucasian 22 | 59,674/316078 | 2.9068711836 | |
| Gormley P, 2016 [40] | SLC24A3/rs4814864 | C vs. G | 0.3021 | 1.07 (1.06–1.09) | 2.2E-19 | Caucasian 22 | 59,674/316078 | 2.0709065394 | |
| Gormley P, 2016 [40] | FGF6/rs1024905 | A vs. G | 0.3165 | 1.06 (1.04–1.08) | 2.1E-17 | Caucasian 22 | 59,674/316078 | 1.8636100452 | |
| Gormley P, 2016 [40] | C7orf10/rs186166891 | T vs. A | 0.1631 | 1.09 (1.07–1.12) | 9.7E-16 | Caucasian 22 | 59,674/316078 | 1.4466644131 | |
| Gormley P, 2016 [40] | PLCE1/rs10786156 | G vs. C | 0.4852 | 0.95 (0.94–0.96) | 2.0E-14 | Caucasian 22 | 59,674/316078 | 2.4863180765 | |
| Gormley P, 2016 [40] | KCNK5/rs10456100 | T vs. C | 0.1813 | 1.06 (1.04–1.07) | 6.9E-13 | Caucasian 22 | 59,674/316078 | 1.0760942468 | |
| Gormley P, 2016 [40] | HPSE2/rs12260159 | A vs. G | 0.1673 | 0.92 (0.89–0.94) | 3.2E-10 | Caucasian 22 | 59,674/316078 | 1.3565561475 | |
| Gormley P, 2016 [40] | CFDP1/rs77505915 | A vs. T | 0.4898 | 1.05 (1.03–1.06) | 3.3E-10 | Caucasian 22 | 59,674/316078 | 2.3904576911 | |
| Gormley P, 2016 [40] | RNF213/rs17857135 | C vs. T | 0.2115 | 1.06 (1.04–1.08) | 5.2E-10 | Caucasian 22 | 59,674/316078 | 1.2530981840 | |
| Gormley P, 2016 [40] | NRP1/rs2506142 | G vs. A | 0.1865 | 1.06 (1.04–1.07) | 1.5E-09 | Caucasian 22 | 59,674/316078 | 1.1066169563 | |
| Gormley P, 2016 [40] | GPR149/rs13078967 | C vs. A | 0.0110 | 0.87 (0.83–0.91) | 1.8E-09 | Caucasian 22 | 59,674/316078 | 0.1432047828 | |
| Gormley P, 2016 [40] | SPINK2/rs7684253 | T vs. C | 0.4499 | 0.96 (0.94–0.97) | 2.5E-09 | Caucasian 22 | 59,674/316078 | 1.8325790934 | |
| Gormley P, 2016 [40] | HEY2/rs1268083 | C vs. T | 0.4419 | 0.96 (0.95–0.97) | 5.3E-09 | Caucasian 22 | 59,674/316078 | 1.7994063059 | |
| Gormley P, 2016 [40] | WSCD1/rs75213074 | T vs. C | 0.0094 | 0.89 (0.86–0.93) | 7.1E-09 | Caucasian 22 | 59,674/316078 | 0.1035070263 | |
| Gormley P, 2016 [40] | GJA1/rs28455731 | T vs. G | 0.1302 | 1.06 (1.04–1.08) | 7.3E-09 | Caucasian 22 | 59,674/316078 | 0.7751445706 | |
| Gormley P, 2016 [40] | ITPK1/rs11624776 | C vs. A | 0.2278 | 0.96 (0.94–0.97) | 7.9E-09 | Caucasian 22 | 59,674/316078 | 0.9195792057 | |
| Gormley P, 2016 [40] | ADAMTSL4/rs6693567 | T vs. C | 0.3037 | 1.05 (1.03–1.06) | 1.2E-08 | Caucasian 22 | 59,674/316078 | 1.4957864823 | |
| Gormley P, 2016 [40] | MED14/rs12845494 | G vs. C | 0.4114 | 0.96 (0.95–0.97) | 1.7E-08 | Caucasian 22 | 59,674/316078 | 1.6731330779 | |
| Gormley P, 2016 [40] | LRRIQ3/rs1572668 | G vs. A | 0.4930 | 1.04 (1.02–1.05) | 2.1E-08 | Caucasian 22 | 59,674/316078 | 1.9338641980 | |
| Gormley P, 2016 [40] | CARF/rs138556413 | G vs. A | 0.0104 | 0.88 (0.84–0.92) | 2.3E-08 | Caucasian 22 | 59,674/316078 | 0.1249559450 | |
| Gormley P, 2016 [40] | ARMS2/rs2223089 | C vs. G | 0.1288 | 0.93 (0.91–0.95) | 3.0E-08 | Caucasian 22 | 59,674/316078 | 0.9098027819 | |
| Gormley P, 2016 [40] | IGSF9B/rs561561 | T vs. A | 0.0827 | 0.94 (0.92–0.96) | 3.4E-08 | Caucasian 22 | 59,674/316078 | 0.4986744225 | |
| Gormley P, 2016 [40] | NOTCH4/rs140002913 | A vs. G | 0.1150 | 0.91 (0.88–0.94) | 3.8E-08 | Caucasian 22 | 59,674/316078 | 1.0458242813 | |
| Anttila V. 2013 [29] | PRDM16/rs2651899 | C vs. T | 0.4708 | 1.09 (1.07–1.12) | 3.28E-14 | Caucasian 19 | 23,285/95425 | 4.0649595346 | |
| Anttila V. 2013 [29] | TSPAN2/rs12134493 | A vs. C | 0.0709 | 1.14 (1.10–1.18) | 6.71E-14 | Caucasian 19 | 23,285/95425 | 0.9828442876 | |
| Anttila V. 2013 [29] | MEF2D/rs2274316 | C vs. A | 0.4289 | 1.07 (1.04–1.09) | 3.14E-08 | Caucasian 19 | 23,285/95425 | 2.9147892814 | |
| Anttila V. 2013 [29] | TRPM8/rs7577262 | A vs. G | 0.2356 | 0.87 (0.84–0.90) | 3.27E-13 | Caucasian 19 | 23,285/95425 | 3.1595713513 | |
| Anttila V. 2013 [29] | FHL5/rs13208321 | A vs. T | 0.2949 | 1.18 (1.13–1.24) | 2.15E-12 | Caucasian 19 | 23,285/95425 | 5.0406331131 | |
| Anttila V. 2013 [29] | c7orf10/rs4379368 | T vs. C | 0.1961 | 1.11 (1.08–1.15) | 1.46E-09 | Caucasian 19 | 23,285/95425 | 2.1115517179 | |
| Anttila V. 2013 [29] | MTDH/rs1835740 | C vs. T | 0.3510 | 1.18 (1.13–1.24) | 1.60E-11 | Caucasian 7 | 5950/50809 | 5.9425497094 |
MAF minor allelic frequency, PAR population attributable risk
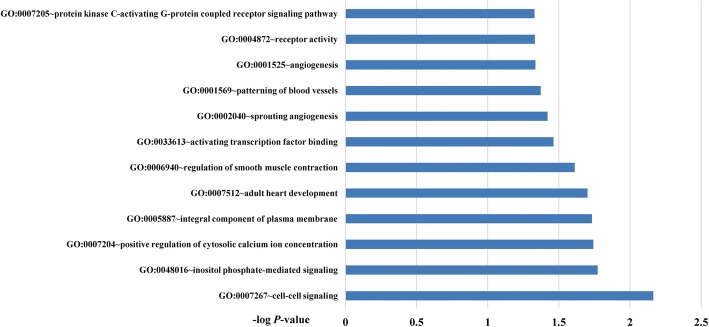
To explore the potential biology mechanism for migraine, GO enrichment analysis was performed. As shown in Fig. 2, twelve GO terms were identified including “cell-cell signaling” (GO: 0007267), “inositol phosphate-mediated signaling” (GO:0048016), “positive regulation of cytosolic calcium ion concentration” (GO:0007204), “integral component of plasma membrane” (GO:0005887), “adult heart development” (GO:0007512), “regulation of smooth muscle contraction” (GO:0006940), “activating transcription factor binding” (GO:0033613), “sprouting angiogenesis” (GO:0002040), “patterning of blood vessels” (GO:000156), “angiogenesis” (GO:0001525), “receptor activity” (GO:0004872) and “protein kinase C-activating G-protein coupled receptor signaling pathway” (GO:0007205). To detect the interactive relationships and potential hub nodes, PPI network of all genes with noteworthy SNPs were constructed and displayed in Additional file 3: Figure S1. Moreover, when degree ≥7 as the cut-off criterion, 5 genes (MEF2D, TSPAN2, PHACTR1, TRPM8 and PRDM16) were selected as hub genes by using CytoHubba software.
Fig. 2.
Gene ontology enrichment analysis of migraine. The cut-off P-value for this analysis was 0.05 (−log P-value = 1.3)
Discussion
Despite the serious impact of migraine on human health and its burden on public, understanding of the pathogenesis, valid prevention and treatment remains limited [49, 50]. Recent years, the genetic component of migraine has received widespread attention. Moreover, a growing number of meta-analyses from observational studies and GWAS were performed and further identified numerous significant genetic variants, which displayed important insights into the mechanisms underlying migraine development. However, due to false-positive possibility in meta-analyses, we performed the first re-analysis of meta-analyses of genetic association studies in migraine in hope of finding noteworthy associations. We used Bayesian methods and Venice criteria to evaluate the credibility of genetic associations. In this work, we synthesized all relevant data from the meta-analyses which detected the association between genetic polymorphisms and migraine risk before 31 July 2019.
As for the candidate genes, we found 4 genetic variants noteworthy at prior probability of 0.05 which were consistent with the results of Venice criteria. However, when we raised the prior probability standard from 0.05 to 0.001, we did not find any gene noteworthy under FPRP or BFDP, which indicated that the results of observational studies should be interpreted with caution. In comparison to the observational studies, according to GWAS, out of the 47 significant genetic variants, 36 were considered to be noteworthy at prior probability of 0.000001 via FPRP or BFDP. Most significant variants identified in GWAS remained noteworthy at the prior probability of 0.000001 demonstrated that the results from GWAS were more reliable.
We also detected the pathways involved in migraine by conducting functional enrichment analysis and further explore the possible molecular mechanisms. We found several significant pathways (cell-cell signaling, inositol phosphate-mediated signaling, positive regulation of cytosolic calcium ion concentration, integral component of plasma membrane and adult heart development, etc.) and 5 hub genes (MEF2D, TSPAN2, PHACTR1, TRPM8 and PRDM16) which were considered to play vital roles in migraine occurrence.
To date, the pathophysiology of migraine is partially understood that the headache of migraine is associated with activation and sensitization of trigeminovascular system [51, 52]. This hypothesis is based on a fact that migraine attack was originated from the activation of nociceptors that are initialed from trigeminal ganglion and innervate arachnoid, pial, blood vessels of dural, large cerebral arteries and sinuses [53]. These activated nociceptors released some inflammatory mediators and vasoactive neuropeptides such as calcitonin gene-related peptide (CGRP), neurokinin A and substance P causing vasodilation of dural and pial vessels and neurogenic inflammation which finally results in central sensitization causing headache of migraine to some extent [53–55].
PLCE1 and NMUR2, whose variants showed noteworthy association with migraine susceptibility, were found to be involved in two pathways called “positive regulation of cytosolic calcium ion concentration” and “inositol phosphate-mediated signaling” detected in our study. PLCE1 is a gene encodes an enzyme called phospholipase C that promotes the generation of inositol triphosphate (IP3) and further direct the calcium mobilization by initiate the release of calcium ion stored in the endoplasmic reticulum through IP3 receptor leading to the increment of cytosolic calcium concentration in neurons [56, 57]. As for NMUR2, an impaired pain response was observed in NMUR2-deficient mice, supporting the hypothesis that nociceptive effects may be partially mediated through NMUR2 [58]. Similar with PLCE1, NMUR2 is abundantly expressed in the central nervous system and considered as a regulator for intracellular calcium mobilization via IP3 as well [59]. Furthermore, the increasing of cytosolic calcium may cause the release of CGRP that finally results in migraine development [60, 61].
Within the hub genes detected in our study, TRPM8 gene was found to play an important role in the pathophysiology of migraine. As a cation channel, TRPM8 is characterized as a cold temperature detector firstly [62, 63]. However, recently, it is also discovered as a parameter for ongoing persistent pain with several evidence. TRPM8 null mice showed a significant decrease in the injury-induced response [64]. On the opposite, the activation of TRPM8 can lead to the depolarization for nerve endings and afferent impulse into central nervous system. Moreover, these activated neurons by noxious cold temperature have properties similar with nociceptors [65]. Mechanistically, a functional linkage was observed between TRPM8 and CGRP, that is, CGRP release was deficient in neurons without TRPM8 triggering and the release of CGRP was closely related to neurogenic inflammation and future migraine occurrence [66].
As for PHACTR1, it might be involved in migraine attack by regulating vasomotor tone. Concretely, PHACTR1 could bind with protein phosphatase 1 (PP1) and its gene silencing has been demonstrated to decrease the activity of PP1 [67]. In arteries, PP1 may play an important role in vasomotor tone by mediating calcium cycling and contractility in smooth muscle cells [68]. Also, PHACTR1 could regulate dendritic morphology and synaptic activity by interacting with PP1 in nervous system, and was further thought to be implicated with the pathophysiology of migraine [69, 70].
Different with the hub genes mentioned above, PRDM16, a zinc-finger nuclear protein, works as an important activator of brown adipogenesis, and depletion of PRDM16 may lead to a significant loss in brown adipocyte identity [71]. Recently, brown adipose tissue has been in the focus of metabolism research, which can dissipate energy by the regulation of uncoupling protein-1, increase fatty acid oxidation and heat production and counteract obesity [72, 73]. Moreover, PRDM16 rs2651899 is an intron variant that may affect the splicing of PRDM16 and its downstream regulatory elements, reduce PRDM16 expression and thus increase body mass index [74]. Large cohort studies suggest that obesity is a risk factor for migraine even after adjusting for comorbidities [75]. In obese individuals, the expression of many inflammatory mediators was increased including CGRP and interleukins, which in turn could cause central sensitization in migraine pathophysiology [76, 77]. In addition, Obesity is a state of sympathetic activation, that may contribute to increase in migraine attack [78].
For other hub genes for migraine, TSPAN2 is highly expressed in oligodendrocyte cell lines and may regulate the differentiation process of oligodendrocytes to myelin-forming glia [79]. As a transcription factor in neurons, MEF2D is concerned to be involved in neurogenesis, neuronal survival and differentiation by controlling MEF2D-dependent gene transcription [80]. However, up till now, the potential role of TSPAN2 and MEF2D in migraine development still remains unknown. Further functional studies are required to explore the underlying biological mechanisms.
Although our study included a large number of relevant articles on migraine, some limitations should be acknowledged in our re-analysis. First, most of original studies did not consider the potential confounders such as gene-gene and gene-environment interactions. Second, there exist some inherent methodological flaws in observational studies, such as selection bias, publication bias and small sample size. Although Venice criteria were applied, these potential biases were difficult to measure. Additionally, owing to the inadequate raw data, Venice Score cannot be applied for GWAS. Third, the GWAS involved in our report were mostly performed in Caucasian population, thus, the results of which was restricted to be applied in diverse populations. Also, GWAS included in our study were lack of subgroup analysis on migraine subtype. Thus, we could only study the noteworthiness of GWAS SNPs in migraine without aura subgroup. Lastly, we constructed a PPI network to explore the underlying biological mechanism for migraine. However, the criteria for PPI construction was relatively subjective, so the result of PPI might need to be interpreted with caution.
Conclusion
The current findings identify several noteworthy variants for migraine susceptibility. We hope this field synopsis and systematic re-analysis would help identify novel genetic biomarkers and potential therapeutic target for migraine.
Supplementary information
Additional file 1: Table S1. Overall summary of results from meta-analyses of observation studies on risk of migraine (including duplicates, statistically significant and non-significant results).
Additional file 2: Table S2. Overall summary of results from meta-analyses of GWAS on risk of migraine (including duplicates, statistically significant and non-significant results).
Additional file 3: Figure S1. Protein-protein interaction network of noteworthy genes related with migraine. The active interaction sources included text mining, experiments, databases as well as co-expression. MCL clustering method was used with the inflation parameter setting at 3.
Acknowledgements
Not applicable.
Authors’ contributions
XL conceived and designed the study. XL YZ RZ performed the database search, data extraction, statistical analysis and bioinformatics analysis. XL YZ TX wrote the draft and revised the whole manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81400950, 81501006) and Natural Science Foundation of Liaoning Province (Grant No. 2019-MS-365, 2019-MS-364).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s10194-020-01087-5.
References
- 1.Diener HC, et al. Integrated care for chronic migraine patients: epidemiology, burden, diagnosis and treatment options. Clin Med (Lond) 2015;15(4):344–350. doi: 10.7861/clinmedicine.15-4-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collaborators GBDH. Global, regional, and national burden of migraine and tension-type headache, 1990-2016: a systematic analysis for the global burden of Disease study 2016. Lancet Neurol. 2018;17(11):954–976. doi: 10.1016/S1474-4422(18)30322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younger DS. Epidemiology of migraine. Neurol Clin. 2016;34(4):849–861. doi: 10.1016/j.ncl.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Disease, G.B.D., I. Injury, and C. Prevalence Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the global burden of Disease study 2015. Lancet. 2016;388(10053):1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacGregor EA. Migraine. Ann Intern Med. 2017;166(7):ITC49–ITC64. doi: 10.7326/AITC201704040. [DOI] [PubMed] [Google Scholar]
- 6.Messali A, et al. Direct and indirect costs of chronic and episodic migraine in the United States: a web-based survey. Headache. 2016;56(2):306–322. doi: 10.1111/head.12755. [DOI] [PubMed] [Google Scholar]
- 7.Lipton RB, Bigal ME. Migraine: epidemiology, impact, and risk factors for progression. Headache. 2005;45(Suppl 1):S3–S13. doi: 10.1111/j.1526-4610.2005.4501001.x. [DOI] [PubMed] [Google Scholar]
- 8.Bohm PE, Stancampiano FF, Rozen TD. Migraine headache: updates and future developments. Mayo Clin Proc. 2018;93(11):1648–1653. doi: 10.1016/j.mayocp.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Kondratieva N, et al. Biomarkers of migraine: part 1 - genetic markers. J Neurol Sci. 2016;369:63–76. doi: 10.1016/j.jns.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Wacholder S, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81(2):208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khoury MJ, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170(3):269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langevin SM, et al. Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines. Pharmacogenet Genomics. 2010;20(10):586–597. doi: 10.1097/FPC.0b013e32833c3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oterino A, et al. Genetic association study and meta-analysis of the HTR2C Cys23Ser polymorphism and migraine. J Headache Pain. 2007;8(4):231–235. doi: 10.1007/s10194-007-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubino E, et al. Association of the C677T polymorphism in the MTHFR gene with migraine: a meta-analysis. Cephalalgia. 2009;29(8):818–825. doi: 10.1111/j.1468-2982.2007.01400.x. [DOI] [PubMed] [Google Scholar]
- 16.Anttila V, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet. 2010;42(10):869–873. doi: 10.1038/ng.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schurks M, Rist PM, Kurth T. Sex hormone receptor gene polymorphisms and migraine: a systematic review and meta-analysis. Cephalalgia. 2010;30(11):1306–1328. doi: 10.1177/0333102410364155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schurks M, Rist PM, Kurth T. 5-HTTLPR polymorphism in the serotonin transporter gene and migraine: a systematic review and meta-analysis. Cephalalgia. 2010;30(11):1296–1305. doi: 10.1177/0333102410362929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schurks M, Rist PM, Kurth T. STin2 VNTR polymorphism in the serotonin transporter gene and migraine: pooled and meta-analyses. J Headache Pain. 2010;11(4):317–326. doi: 10.1007/s10194-010-0230-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schurks M, Rist PM, Kurth T. MTHFR 677C>T and ACE D/I polymorphisms in migraine: a systematic review and meta-analysis. Headache. 2010;50(4):588–599. doi: 10.1111/j.1526-4610.2009.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chasman DI, et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet. 2011;43(7):695–698. doi: 10.1038/ng.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligthart L, et al. Meta-analysis of genome-wide association for migraine in six population-based European cohorts. Eur J Hum Genet. 2011;19(8):901–907. doi: 10.1038/ejhg.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, et al. Association of 5-HTT gene polymorphisms with migraine: a systematic review and meta-analysis. J Neurol Sci. 2011;305(1–2):57–66. doi: 10.1016/j.jns.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Samaan Z, et al. Methylenetetrahydrofolate reductase gene variant (MTHFR C677T) and migraine: a case control study and meta-analysis. BMC Neurol. 2011;11:66. doi: 10.1186/1471-2377-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurks M, et al. Tumour necrosis factor gene polymorphisms and migraine: a systematic review and meta-analysis. Cephalalgia. 2011;31(13):1381–1404. doi: 10.1177/0333102411419022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu L, et al. The TNF-alpha-308G/a polymorphism is associated with migraine risk: a meta-analysis. Exp Ther Med. 2012;3(6):1082–1086. doi: 10.3892/etm.2012.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao J, Wang F, Fang Y. Association of 231G>a polymorphism of endothelin type a receptor gene with migraine: a meta-analysis. J Neurol Sci. 2012;323(1–2):232–235. doi: 10.1016/j.jns.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Tammimaki A, Mannisto PT. Catechol-O-methyltransferase gene polymorphism and chronic human pain: a systematic review and meta-analysis. Pharmacogenet Genomics. 2012;22(9):673–691. doi: 10.1097/FPC.0b013e3283560c46. [DOI] [PubMed] [Google Scholar]
- 29.Anttila V, et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet. 2013;45(8):912–917. doi: 10.1038/ng.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esserlind AL, et al. Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur J Neurol. 2013;20(5):765–772. doi: 10.1111/ene.12055. [DOI] [PubMed] [Google Scholar]
- 31.Liu R, et al. MTHFR C677T polymorphism and migraine risk: a meta-analysis. J Neurol Sci. 2014;336(1–2):68–73. doi: 10.1016/j.jns.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, et al. Effects of tumor necrosis factor-beta (TNF-beta) 252A>G polymorphism on the development of migraine: a meta-analysis. PLoS One. 2014;9(6):e100189. doi: 10.1371/journal.pone.0100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng JM, et al. Meta-analysis of 5-hydroxytryptamine type 2A receptor polymorphisms and migraine susceptibility. Int J Neurosci. 2014;124(12):882–889. doi: 10.3109/00207454.2014.888425. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, et al. Association between polymorphisms of DRD2, COMT, DBH, and MAO-A genes and migraine susceptibility: a meta-analysis. Medicine (Baltimore) 2015;94(47):e2012. doi: 10.1097/MD.0000000000002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, et al. Tumor necrosis factor (TNF) -308G>a, nitric oxide synthase 3 (NOS3) +894G>T polymorphisms and migraine risk: a meta-analysis. PLoS One. 2015;10(6):e0129372. doi: 10.1371/journal.pone.0129372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobsen KK, et al. Genome wide association study identifies variants in NBEA associated with migraine in bipolar disorder. J Affect Disord. 2015;172:453–461. doi: 10.1016/j.jad.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, et al. Impact of ESR1 gene polymorphisms on migraine susceptibility: a meta-analysis. Medicine (Baltimore) 2015;94(35):e0976. doi: 10.1097/MD.0000000000000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippi G, Mattiuzzi C, Cervellin G. Meta-analysis of factor V Leiden and prothrombin G20210A polymorphism in migraine. Blood Coagul Fibrinolysis. 2015;26(1):7–12. doi: 10.1097/MBC.0000000000000188. [DOI] [PubMed] [Google Scholar]
- 39.Miao J, et al. Association of the Apolipoprotein E polymorphism with migraine: a meta-analysis. BMC Neurol. 2015;15:138. doi: 10.1186/s12883-015-0385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gormley P, et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet. 2016;48(8):856–866. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan D, et al. Association between angiotensin-converting enzyme insertion/deletion polymorphism and migraine: a meta-analysis. Int J Neurosci. 2016;126(5):393–399. doi: 10.3109/00207454.2015.1025395. [DOI] [PubMed] [Google Scholar]
- 42.Cai X, et al. The association between brain-derived neurotrophic factor gene polymorphism and migraine: a meta-analysis. J Headache Pain. 2017;18(1):13. doi: 10.1186/s10194-017-0725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao YJ, Jiang JR, Jin SQ. The association between COMT Val158Met polymorphism and migraine risk: a meta-analysis. Cephalalgia. 2017;37(6):592–598. doi: 10.1177/0333102416649758. [DOI] [PubMed] [Google Scholar]
- 44.Terrazzino S, et al. Brain-derived Neurotrophic factor Val66Met gene polymorphism impacts on migraine susceptibility: a meta-analysis of case-control studies. Front Neurol. 2017;8:159. doi: 10.3389/fneur.2017.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang X, et al. Common variants at 5q33.1 predispose to migraine in African-American children. J Med Genet. 2018;55(12):831–836. doi: 10.1136/jmedgenet-2018-105359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong H, et al. Endothelial nitric oxide synthase (−786T>C) polymorphism and migraine susceptibility: a meta-analysis. Medicine (Baltimore) 2018;97(36):e12241. doi: 10.1097/MD.0000000000012241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Wang J (2018) Quantitative assessment of the association between GRIA1 polymorphisms and migraine risk. Biosci Rep 38(6). 10.1042/BSR20181347 [DOI] [PMC free article] [PubMed]
- 48.Liu L, et al. Effects of MTHFR C677T and A1298C polymorphisms on migraine susceptibility: a meta-analysis of 26 studies. Headache. 2019;59(6):891–905. doi: 10.1111/head.13540. [DOI] [PubMed] [Google Scholar]
- 49.Dodick DW, Gargus JJ. Why migraines strike. Sci Am. 2008;299(2):56–63. doi: 10.1038/scientificamerican0808-56. [DOI] [PubMed] [Google Scholar]
- 50.Wessman M, et al. Migraine: a complex genetic disorder. Lancet Neurol. 2007;6(6):521–532. doi: 10.1016/S1474-4422(07)70126-6. [DOI] [PubMed] [Google Scholar]
- 51.Olesen J, et al. Origin of pain in migraine: evidence for peripheral sensitisation. Lancet Neurol. 2009;8(7):679–690. doi: 10.1016/S1474-4422(09)70090-0. [DOI] [PubMed] [Google Scholar]
- 52.Pietrobon D, Striessnig J. Neurobiology of migraine. Nat Rev Neurosci. 2003;4(5):386–398. doi: 10.1038/nrn1102. [DOI] [PubMed] [Google Scholar]
- 53.Goadsby PJ, et al. Pathophysiology of migraine: a disorder of sensory processing. Physiol Rev. 2017;97(2):553–622. doi: 10.1152/physrev.00034.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noseda R, Burstein R. Migraine pathophysiology: anatomy of the trigeminovascular pathway and associated neurological symptoms, CSD, sensitization and modulation of pain. Pain. 2013;154(Suppl 1):S44–S53. doi: 10.1016/j.pain.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peck KR, Johnson YL, Smitherman TA. Migraine. Handb Clin Neurol. 2016;138:283–293. doi: 10.1016/B978-0-12-802973-2.00016-1. [DOI] [PubMed] [Google Scholar]
- 56.Gargus JJ. Genetic calcium signaling abnormalities in the central nervous system: seizures, migraine, and autism. Ann N Y Acad Sci. 2009;1151:133–156. doi: 10.1111/j.1749-6632.2008.03572.x. [DOI] [PubMed] [Google Scholar]
- 57.Shi Y, et al. Genetic variation in the calcium/calmodulin-dependent protein kinase (CaMK) pathway is associated with antidepressant response in females. J Affect Disord. 2012;136(3):558–566. doi: 10.1016/j.jad.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 58.Torres R, et al. Mice genetically deficient in neuromedin U receptor 2, but not neuromedin U receptor 1, have impaired nociceptive responses. Pain. 2007;130(3):267–278. doi: 10.1016/j.pain.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 59.Liu JJ, et al. Discovery and pharmacological characterization of a small-molecule antagonist at neuromedin U receptor NMUR2. J Pharmacol Exp Ther. 2009;330(1):268–275. doi: 10.1124/jpet.109.152967. [DOI] [PubMed] [Google Scholar]
- 60.Han F, et al. Paeoniflorin protects diabetic mice against myocardial ischemic injury via the transient receptor potential vanilloid 1/calcitonin gene-related peptide pathway. Cell Biosci. 2016;6:37. doi: 10.1186/s13578-016-0085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao Y, Richter JA, Hurley JH. Release of glutamate and CGRP from trigeminal ganglion neurons: role of calcium channels and 5-HT1 receptor signaling. Mol Pain. 2008;4:12. doi: 10.1186/1744-8069-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuzuki K, et al. Menthol-induced Ca2+ release from presynaptic Ca2+ stores potentiates sensory synaptic transmission. J Neurosci. 2004;24(3):762–771. doi: 10.1523/JNEUROSCI.4658-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peier AM, et al. A TRP channel that senses cold stimuli and menthol. Cell. 2002;108(5):705–715. doi: 10.1016/S0092-8674(02)00652-9. [DOI] [PubMed] [Google Scholar]
- 64.Colburn RW, et al. Attenuated cold sensitivity in TRPM8 null mice. Neuron. 2007;54(3):379–386. doi: 10.1016/j.neuron.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 65.Dussor G, Cao YQ. TRPM8 and migraine. Headache. 2016;56(9):1406–1417. doi: 10.1111/head.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Jong PR, et al. TRPM8 on mucosal sensory nerves regulates colitogenic responses by innate immune cells via CGRP. Mucosal Immunol. 2015;8(3):491–504. doi: 10.1038/mi.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allain B, et al. Neuropilin-1 regulates a new VEGF-induced gene, Phactr-1, which controls tubulogenesis and modulates lamellipodial dynamics in human endothelial cells. Cell Signal. 2012;24(1):214–223. doi: 10.1016/j.cellsig.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 68.Patel RS, et al. A genetic risk variant for myocardial infarction on chromosome 6p24 is associated with impaired central hemodynamic indexes. Am J Hypertens. 2012;25(7):797–803. doi: 10.1038/ajh.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allen PB, et al. Phactrs 1-4: a family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci U S A. 2004;101(18):7187–7192. doi: 10.1073/pnas.0401673101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winsvold BS, et al. Shared genetic risk between migraine and coronary artery disease: a genome-wide analysis of common variants. PLoS One. 2017;12(9):e0185663. doi: 10.1371/journal.pone.0185663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kajimura S. Promoting brown and beige adipocyte biogenesis through the PRDM16 pathway. Int J Obes Suppl. 2015;5(Suppl 1):S11–S14. doi: 10.1038/ijosup.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fenzl A, Kiefer FW. Brown adipose tissue and thermogenesis. Horm Mol Biol Clin Invest. 2014;19(1):25–37. doi: 10.1515/hmbci-2014-0022. [DOI] [PubMed] [Google Scholar]
- 73.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.AlAmrani A, AbdelKarim M, AlZoghaibi M. PRDM16 gene polymorphism is associated with obesity and blood lipids profiles in Saudi population. J Clin Med. 2018;7(6):141. doi: 10.3390/jcm7060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bigal ME, Lipton RB. Putative mechanisms of the relationship between obesity and migraine progression. Curr Pain Headache Rep. 2008;12(3):207–212. doi: 10.1007/s11916-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 76.Bigal ME, et al. Obesity, migraine, and chronic migraine: possible mechanisms of interaction. Neurology. 2007;68(21):1851–1861. doi: 10.1212/01.wnl.0000262045.11646.b1. [DOI] [PubMed] [Google Scholar]
- 77.Zelissen PM, et al. Calcitonin gene-related peptide in human obesity. Peptides. 1991;12(4):861–863. doi: 10.1016/0196-9781(91)90147-H. [DOI] [PubMed] [Google Scholar]
- 78.Peroutka SJ. Migraine: a chronic sympathetic nervous system disorder. Headache. 2004;44(1):53–64. doi: 10.1111/j.1526-4610.2004.04011.x. [DOI] [PubMed] [Google Scholar]
- 79.Fang J, et al. Involvement of the Tetraspanin 2 (TSPAN2) gene in migraine: a case-control study in Han Chinese. Front Neurol. 2018;9:714. doi: 10.3389/fneur.2018.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang P, et al. Dual-specificity tyrosine-phosphorylation regulated kinase 1A gene transcription is regulated by Myocyte enhancer factor 2D. Sci Rep. 2017;7(1):7240. doi: 10.1038/s41598-017-07655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Overall summary of results from meta-analyses of observation studies on risk of migraine (including duplicates, statistically significant and non-significant results).
Additional file 2: Table S2. Overall summary of results from meta-analyses of GWAS on risk of migraine (including duplicates, statistically significant and non-significant results).
Additional file 3: Figure S1. Protein-protein interaction network of noteworthy genes related with migraine. The active interaction sources included text mining, experiments, databases as well as co-expression. MCL clustering method was used with the inflation parameter setting at 3.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.