Abstract
Alternative splicing (AS) plays a significant role in regulating gene expression at the transcriptional level in eukaryotes. Flexibility and diversity of transcriptome and proteome can be significantly increased through alternative splicing of genes. In the present study, transcriptome data of peripheral immune organs including spleen and inguinal lymph nodes (ILN) were used to identify AS difference between PRRSV-resistant Tongcheng (TC) pigs and PRRSV-susceptible Large White (LW) pigs artificially infected with porcine reproductive and respiratory syndrome virus (PRRSV) in vivo. The results showed that PRRSV infection induced global alternative splicing events (ASEs) with different modes. Among them, 373 genes and 595 genes in the spleen and ILN of TC pigs, while 458 genes and 560 genes in the spleen and ILN of LW pigs had significantly differential ASEs. Alternative splicing was subject to tissue-specific and lineage-specific regulation in response to PRRSV infection. Enriched GO terms and pathways showed that genes with differential ASEs played important roles in transcriptional regulation, immune response, metabolism, and apoptosis. Furthermore, a splicing factor associated with apoptosis, SRSF4, was significantly upregulated in LW pigs. Functional analysis on apoptosis associated genes was validated by RT-PCR and DNA sequencing. These findings revealed different response to PRRSV between PRRSV-resistant TC pigs and PRRSV-susceptible LW pigs at the level of alternative splicing, suggesting the potential relationship between AS and disease resistance to PRRSV.
1. Introduction
Porcine reproductive and respiratory syndrome (PRRS) has devastated the swine industries in Africa, Asia, Europe, and North America for many years. Since 2006, highly pathogenic PRRS (HP-PRRS) has emerged in China and then spread rapidly to other Asian countries [1, 2]. No doubt, PRRS presents a major health challenge and economic burdens for the swine industry worldwide [3]. The major clinical symptoms of PRRS include severe respiratory diseases in pigs of any age and reproductive disorder in sows [1, 4]. The respiratory symptom is characterized by fever, interstitial pneumonia, and severe lesions in lung and lymph nodes, which usually cause high mortality in nursery pigs. The reproductive disorder is typically shown increased rates of abortions, mummies, and stillbirth in sows [1, 4]. The pathogen of PRRS is called the PRRS virus (PRRSV), which is a small positive strand RNA virus belonging to Arterivirus family. Currently, there are two main PRRSV genotypes based on geographic origins: type I (European) and type II (North American) PRRSV [5]. Type I and type II PRRSV only share approximate 55–70% nucleotide identity and 50–80% amino acid identity of viral genes [6]. The target cells of PRRSV are monocytes and macrophages in different tissues, and the fully differentiated porcine alveolar macrophages (PAMs) are the major target cell [7, 8].
Vaccination is unsuccessful in preventing PRRSV infection due to its rapid evolution and hundreds of new strains emergence with a high mutation rate and recombination rate [9]. As a result, it has been suggested that genetic improvement by enhancing host resistance to PRRSV is a feasible strategy to control PRRS [10]. In order to investigate potential genes or pathways associated with host resistance to PRRSV or pathogenesis of PRRSV, studies have explored whole genome transcriptome analysis of PAMs, lungs or blood samples challenged with different virulent PRRSV strains using microarray, serial analysis of gene expression (SAGE) and RNA-seq [11–14]. Microarray data revealed critical differentially expressed genes in lung tissue between Dapulian (DPL) pigs and Duroc × Landrace × Yorkshire (DLY) pigs after PRRSV infection resulting in different clinical outcomes [14]. Transcriptome analysis of lung dendritic cells (DCs) derived from Pietrain and Duroc revealed Duroc reacted more distinctly and strongly than Pietrain during various periods of PRRSV infection [15]. The transcriptome study of PAMs in Tongcheng (TC) pigs and Large White (LW) pigs showed that two breeds had different immune responses to PRRSV infection [16]. It is mainly manifested that the differentially expressed genes in TC pigs are enriched in activation of leukocyte extravasation and suppression of apoptosis while LW pigs are enriched in the suppression of Gαq and PI3K-AKT signaling [17]. Although those transcriptome studies have provided useful genes and pathways information related to the pathogenesis of PRRSV and host immune response, they have not investigated the complexity of transcriptome arising from RNA processing.
Alternative splicing (AS) is one of the most important posttranscriptional RNA processing, which can significantly change the sequence of RNA transcript [18]. AS plays a critical role in regulating gene expression at the transcriptional level in eukaryotes. Flexibility and diversity of transcriptome and proteome can be significantly increased through alternative splicing of genes [19, 20]. Alternative splicing events (ASEs) mainly include skipped exon (SE), retained intron (RI), an alternative to 5′ splicing site (AS5), an alternative to 3′ splicing site (AS3), and mutually exclusive exon (MXE) [21]. Virus-host interaction studies of a wide range of viruses have identified that AS can change many biological processes in host cells upon viral infection, including activation of host immune response, modulation of synthesis system, and cellular protein quality control [22–24]. Poliovirus protease 2A (2Apro) can impair RNA splicing to modulate host gene expression and avoid antiviral response [25]. Influenza A virus inhibits the cellular gene expression through interaction with the spliceosome complex by NS1 protein [26]. Moreover, the genetic variability of different individuals or breeds has been proven to affect alternative splicing outcomes [27, 28]. Therefore, the global profiling of alternative splicing differences among different breeds in response to PRRSV infection will provide us new insights of host resistance to PRRSV. In the present study, transcriptome data of peripheral immune organs including the spleen and inguinal lymph nodes (ILN) were used to identify AS difference between PRRSV-resistant TC pigs and PRRSV-susceptible LW pigs artificially infected with PRRSV in vivo. The results will enhance the understanding of the molecular mechanism of genetic resistance to PRRSV infection and provide detailed information for future studies.
2. Materials and Methods
2.1. Animals, Experimental Design, and Sample Collection
The PRRSV challenge experiments were conducted by our previous study [16]. Briefly, a total of twenty-four five-week-old pigs including twelve TC pigs and twelve LW pigs were selected to perform the artificial challenge experiment as described previously. All selected pigs are free of PRRSV, porcine circovirus (PCV), and pseudorabies virus (PRV). Six TC pigs and six LW pigs were intramuscularly challenged with PRRSV WuH3 strain at a viral dose of 105 CCID50/mL (3 mL/15 kg), and the rest control pigs were challenged with the same amount of RPMI-1640 (Gibco, Grand Island, NY, USA). Then, all pigs were humanely euthanized for sample collection on day 7 after challenging. In this study, spleen and inguinal lymph nodes were collected for total RNA extraction and RNA sequencing library construction. All animal procedures were supervised and approved by the Ethical Committee for Animal Experiments at Huazhong Agricultural University (permit number: HZAUSW-2013-005).
2.2. RNA Preparation and Sequencing
Total RNA extraction was performed as described previously [17]. RNA degradation and contamination were monitored by electrophoresis. RNA purity, concentration, and integrity were measured by Agilent 2100 Bioanalyzer (Agilent, Santa Clara, California, USA). Total RNA of the spleen and ILN from twelve selected pigs (three pigs selected randomly from each group) were used for RNA sequencing libraries construction by using the NEBNext® UltraTM RNA Library Prep kit for Illumina® (NEB, Ipswich, MA, USA), and all the procedures and standards were performed following the manufacturer's protocols. After quality control, all libraries were sequenced on an Illumina Hiseq 2500 platform and 150 bp paired-end reads were generated. The clean reads were obtained by using FastQC software version 1.3 with the default parameters [29].
2.3. Alternative Splicing Analysis
The replicate multivariate analysis of transcript splicing (rMATS) v4.0.2 [21] was used to screen alternative splicing events across different samples. First, all clean reads were aligned to the Sus scrofa reference genome using TopHat v2.0.13 with default parameters [30]. Second, the aligned reads were run on rMATS v4.0.2 for alternative splicing analysis. The obtained ASEs were classified into five types of ASEs including skipped exon (SE), retained intron (RI), alternative 5′ splice sites (A5SS), alternative 3′ splice sites (A3SS), and mutually exclusive exons (MXE). The differential ASEs between the PRRSV infected group and the control group were identified by change in the percent in (∆PSI), which is a common parameter to describe the degree of alternative splicing. The value of ∆PSI were estimated as in the following formula:
| (1) |
I represents the count of reads specific to the splicing transcript. S represents the count of reads specific to the reference transcript. lI represents the length of splicing transcript, and lS represents the length of reference transcript. |ΔPSI| means the absolute value of the change in the percent-spliced-in PSIInfection represents the mean PSI value of an ASE in the PRRSV infected group. PSIControl represents the mean PSI value of an ASE in the control group.
For pairwise comparison, we performed the –c 0.0001 parameter to compute P value of ASEs in rMATs running and then extracted the ASEs with a P value <0.05 and a |ΔPSI| ≥ 10% for the following functional enrichment analysis.
2.4. Enrichment of GO Category and KEGG Pathway
The Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 (https://david.ncifcrf.gov/) was used and Bonferroni correction was applied to obtained adjusted P values. The genes containing differential ASEs were sorted by the enrichment of Gene ontology (GO) categories and KEGG pathway database. The human homologous genes were used to do functional analysis in this study.
2.5. Alternative Splicing Factor Analysis
All porcine splicing factor genes were collected on ENSEMBL database (http://asia.ensembl.org/index.html) and analyzed their expression by fragments per kilobase of transcript per million mapped reads (FPKM) value. Genes with an adjusted P values <0.05 and an absolute fold change >1.5 were considered as significantly differentially expressed in this study.
2.6. Validation of Alternative Splicing Events
The expression of candidate genes with ASEs was calculated by FPKM value, and cDNA was reverse-transcribed using the PrimeScript RT Reagent Kit (TaKaRa Biotechnology Co. Ltd., Beijing, China) according to the manufacturer's protocol. Primers for semiquantitative PCR were designed using Primer Premier version 5. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for normalization. A list of all primers and product sizes is included in . Semiquantitative PCR reaction mixture contained the following: 1 × PCR buffer (Mg2+), 0.1 mM dNTP mixture, 1 unit of TaKaRa Taq (TaKaRa Biotechnology Co. Ltd., Beijing, China), 100 nM of forward and reverse primers, 1 μL cDNA template, and sterile water to reach 10 μL. The PCR cycling conditions were as follows: 95°C for 5 min, followed by variable cycles (20 to 30) of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final extension of 3 min at 72°C. Then the PCR products were electrophoresed in 2% agarose gel. DNA molecules were recovered from the gel and sequenced in TsingKe biological technology company.
3. Results
3.1. Increasing Global Alternative Splicing Events in Peripheral Immune Organs in response to PRRSV Infection
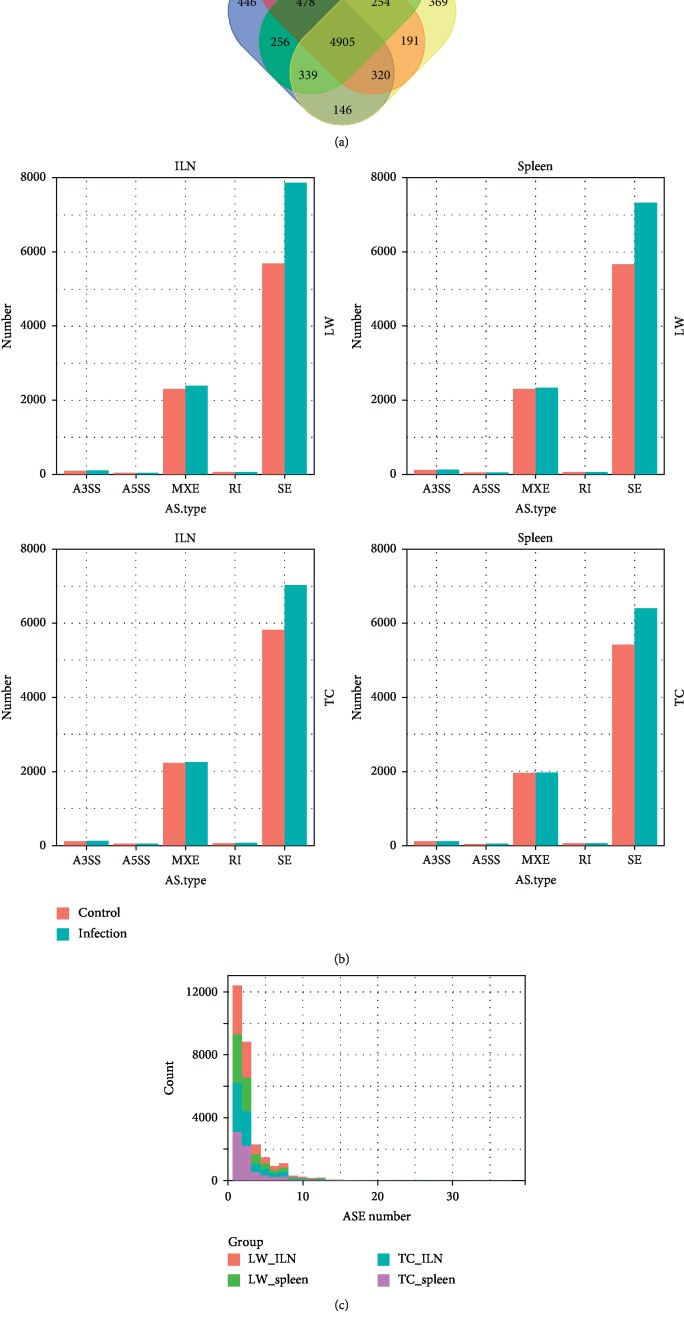
In total, twenty-four RNA-seq libraries were used to analyze alternative splicing events in peripheral immune organs in response to PRRSV infection. The ILN transcriptome dataset contains twelve samples from the control group (LW_ILN_C and TC_ILN_C) and the PRRSV infected group (LW_ILN_I and TC_ILN_I), while the spleen transcriptome dataset also contains twelve samples from the control group (LW_S_C and TC_S_C) and the PRRSV infected group (LW_S_I and TC_S_I), and each group contained three individuals for analysis. Furthermore, we have identified 20781 and 19475 ASEs, belonging to 7175 and 7040 genes in the ILN of LW pigs and TC pigs, respectively. Meanwhile, 19730 and 17888 ASEs were identified in the spleen of LW pigs and TC pigs, which were assigned to 6991 and 6766 genes, respectively. The Venn diagram showed the four datasets had 4905 genes in common, accounting for average of 70%, while only about 5.6% ASE genes were unique in each dataset (Figure 1(a) and Table 1). Generally, ASEs were categorized as five types including A3SS, A5SS, MXE, RI, and SE. It can be noticed that the number of SE accounted for the highest proportion in all four datasets (average 72%), followed by MXE (25%), but there were less than 3% ASEs belonging to A3SS, A5SS, and RI (Figure 1(b) and Table 1). Among the five AS types, SE in PRRSV infected groups was significantly more than in control groups regardless of the tissues and breeds. Besides, the percentage of SE in LW pigs was much higher in the ILN (38.4%) and the spleen (29.3%), compared to only 20.7% and 18.0% in TC pigs. However, the difference of ASE genes between the ILN and the spleen was barely noticeable. The number of ASEs per gene, ranging from 1 to 37, experienced a gradual downward trend in the frequency. It was clear that the single ASE genes accounted for the largest rate, about 46.6%, and almost 98.2% ASE genes had less than ten ASEs. Furthermore, the maximum number of ASEs per gene was found in EIF4G1, which underwent 37 ASEs in total.
Figure 1.
The number and distribution of alternative splicing events. (a) Number of ASEs detected in the ILN and spleen of LW pigs and TC pigs; (b) distribution of five ASE types in the ILN and spleen of LW pigs and TC pigs; (c) distribution of ASE among genes in the ILN and spleen of LW pigs and TC pigs. LW_ILN: inguinal lymph nodes of LW pigs, LW_spleen: spleen of LW pigs, TC_ILN: inguinal lymph nodes of TC pigs, TC_spleen: spleen of TC pigs, A3SS: alternative 3′ splice site, A5SS: alternative 5′ splice sites, MXE: mutually exclusive exons, RI: retained intron, SE: skipped exon.
Table 1.
Alternative splicing events in the ILN and spleen of LW pigs and TC pigs.
| Tissue | ASE type | LW-I | LW-C | TC-I | TC-C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L3 | L4 | L5 | L6 | T1 | T2 | T3 | T4 | T5 | T6 | ||
| ILN | A3SS | 109 | 95 | 103 | 90 | 92 | 91 | 120 | 120 | 111 | 115 | 121 | 102 |
| A5SS | 46 | 42 | 40 | 40 | 36 | 38 | 52 | 43 | 45 | 46 | 42 | 40 | |
| MXE | 2375 | 2421 | 2374 | 2325 | 2299 | 2286 | 2251 | 2247 | 2250 | 2235 | 2230 | 2203 | |
| RI | 60 | 50 | 63 | 59 | 59 | 59 | 63 | 62 | 63 | 60 | 62 | 57 | |
| SE | 7600 | 8351 | 7663 | 6136 | 5446 | 5484 | 7406 | 6980 | 6686 | 6979 | 5934 | 4539 | |
|
| |||||||||||||
| Spleen | A3SS | 126 | 127 | 117 | 123 | 113 | 112 | 106 | 115 | 103 | 111 | 113 | 108 |
| A5SS | 50 | 49 | 49 | 46 | 48 | 45 | 43 | 43 | 40 | 38 | 37 | 37 | |
| MXE | 2341 | 2326 | 2331 | 2313 | 2307 | 2306 | 1935 | 1979 | 1968 | 1949 | 1949 | 1966 | |
| RI | 61 | 58 | 64 | 66 | 61 | 60 | 58 | 60 | 56 | 55 | 58 | 57 | |
| SE | 7095 | 7863 | 7051 | 6014 | 5349 | 5658 | 5653 | 7273 | 6263 | 4986 | 5346 | 5933 | |
L1-L6 represents different individuals from LW pigs; T1-T6 represents different individuals from TW pigs.
3.2. Comparison of Differential Alternative Splicing Events between TC Pigs and LW Pigs in response to PRRSV Infection
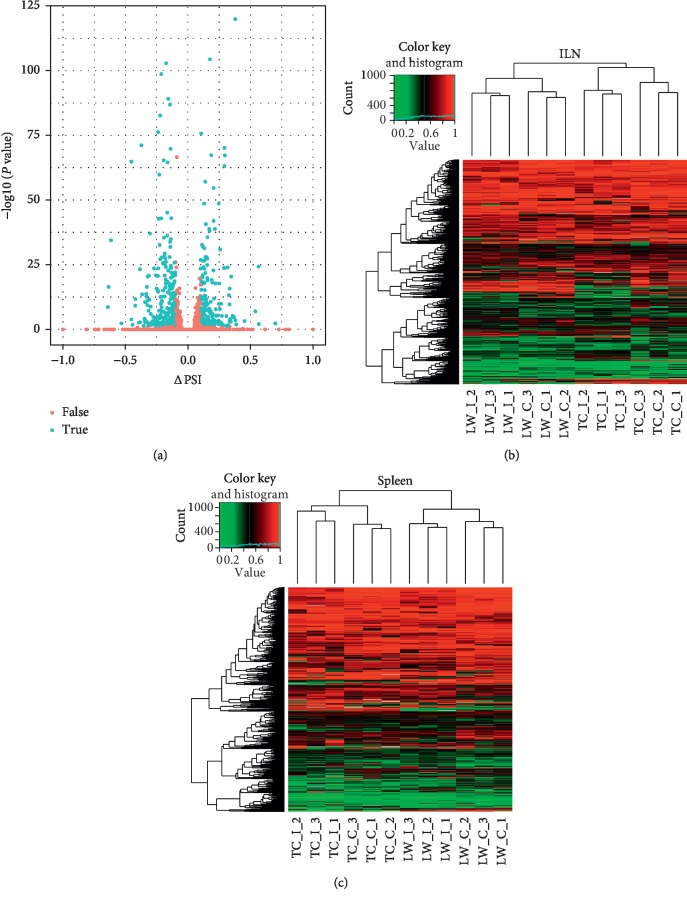
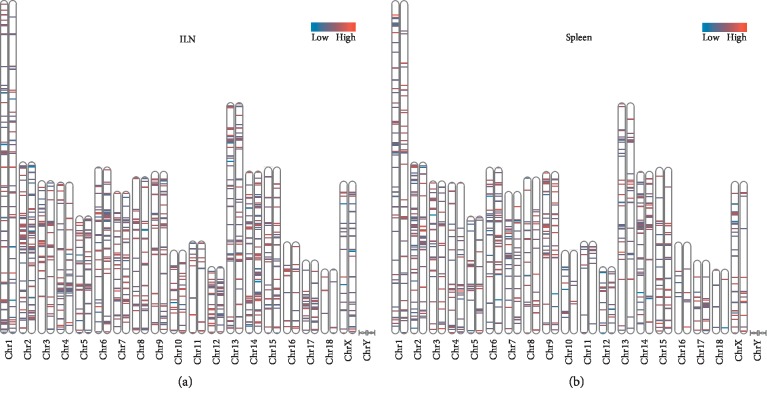
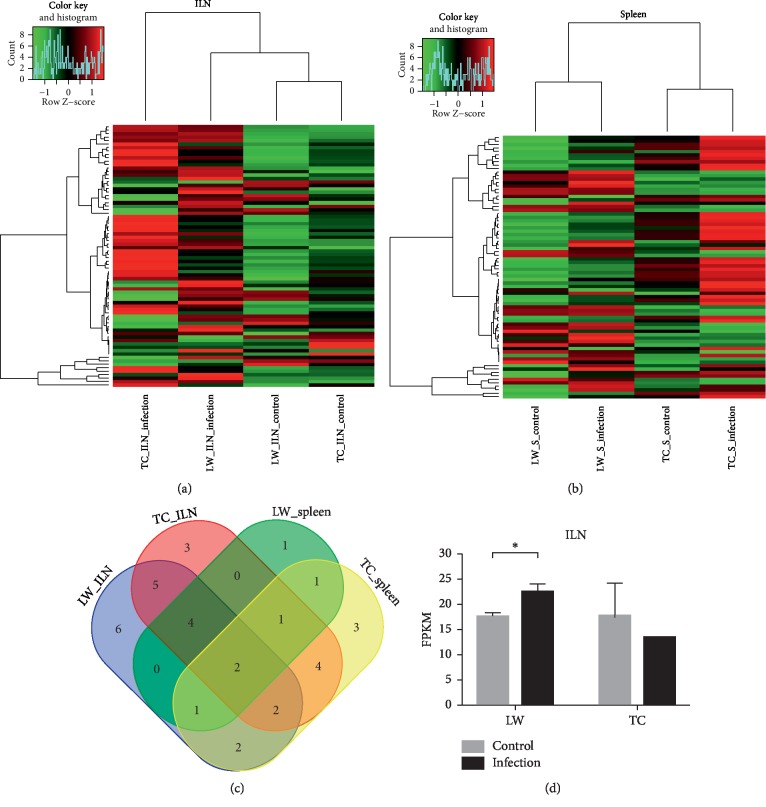
In order to identify the differential ASE pattern between TC pigs and LW pigs in response to PRRSV infection, a stringent cutoff (|ΔPSI| ≥ 10% and P value <0.05) was set to select differential ASEs (Figure 2(a)). Six hundred and fifty and 530 ASEs in the ILN and spleen of LW pigs were selected as differential ASEs, as well as 697 and 434 differential ASEs in the ILN and spleen of TC pigs were selected. Moreover, there were 560 genes and 458 genes containing significant differential ASEs in the ILN and spleen of LW pigs, while TC pigs had 595 differential alternative splicing genes (ASGs) and 373 differential ASGs in the ILN and spleen. One hundred and fifty-seven common differential ASGs were identified in both of the ILN and spleen of LW pigs (Tables and ). However, 403 and 301 differential ASGs were only expressed in the ILN and spleen, respectively. Moreover, TC pigs also displayed the same trend that only 122 differential ASGs were shared by the ILN and spleen, accounting for 20% in the ILN dataset and 33% in the spleen dataset. In the ILN dataset, TC pigs and LW pigs shared 170 differential ASGs, accounting for 28% in TC pigs and 30% in LW pigs. In the spleen dataset, TC pigs and LW pigs shared 109 differential ASGs, accounting for 29% in TC pigs and 24% in LW pigs. Our results clearly showed lineage-specific and tissue-specific alternative splicing regulation in host response to PRRSV infection (Figures 2(b) and 2(c)). Then, we analyzed the ASE types in the above differential ASGs (Tables and ), and the results showed that the SE was still dominated in differential ASGs, but its proportion decreased (53.5% in the ILN of LW pigs, 63.2% in the spleen of LW, 46.6% in the ILN of TC pigs, and 64.5% in the spleen of TC pigs). Besides, the proportion of MXE increased significantly (44.3% in the ILN of LW pigs, 33.6% in the spleen of LW, 50.8% in the ILN of TC pigs, and 33.9% in the spleen of TC pigs), and the frequency of the other three ASE types (A3SS, A5SS, and RI) in each breed and tissue is less than or equal to 10 (). Furthermore, we have investigated the distribution of differential ASEs across pig genome. As shown in Figure 3, the distribution of genes varies from different chromosomes. For example, the largest number of differential ASGs was located on chromosome 2, harboring 71 differential ASGs in the ILN of LW pigs, 69 differential ASGs in the ILN of TC pigs, 62 differential ASGs in the spleen of LW pigs, and 43 differential ASGs in the spleen of TC pigs. Chromosome 18 contains differential ASGs at the lowest proportion for only 4 differential ASGs in the ILN of LW pigs, 6 differential ASGs in the ILN of TC pigs, 4 differential ASGs in the spleen of LW pigs and 6 differential ASGs in the spleen of TC pigs.
Figure 2.
Differential alternative splicing events analysis on splicing level in response to PRRSV infection. (a) Volcano plots analysis of the differential ASEs in response to PRRSV infection. ASEs were detected and quantified using the percent-spliced-in (PSI) metric, and the blue dots indicated differential ASEs; (b) heatmap analysis of the PSI distribution of differential ASEs in the ILN; (c) heatmap analysis of the PSI distribution of differential ASEs in the spleen.
Figure 3.
Distribution of differential alternative splicing genes among chromosomes in response to PRRSV infections. (a) Mapping ΔPSI of differential ASGs on chromosomes in the ILN; (b) mapping ΔPSI of differential ASGs on chromosomes in the spleen; the left chromosome on each paired chromosome represents LW pigs, and the right one represents TC pigs. Lines on chromosomes represent differential ASEs.
3.3. Functional Enrichment Analysis of Alternative Splicing Genes between TC Pigs and LW Pigs in response to PRRSV Infection
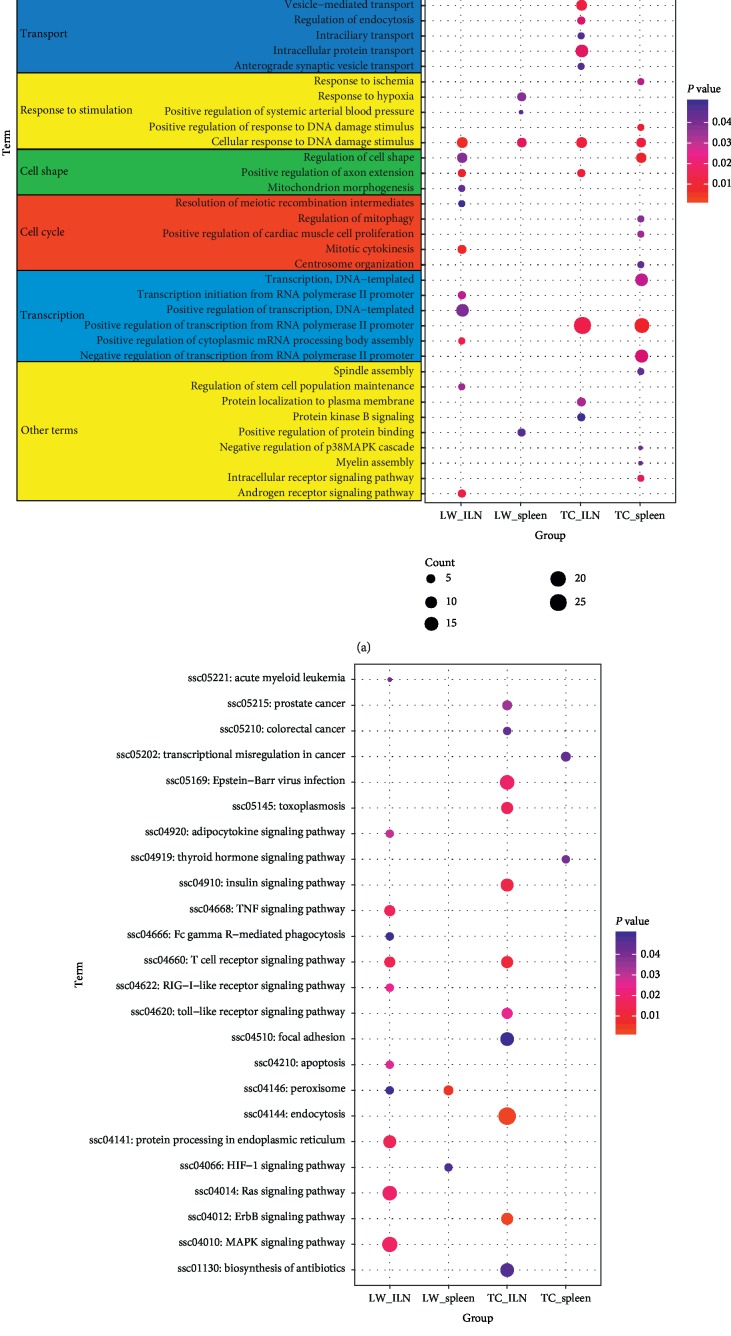
To further investigate the biological functions of genes with differential ASEs, we performed GO analysis and KEGG enrichment analysis by DAVID. The results of GO functional enrichment were shown in Figure 4(a); the differential ASGs of LW pigs in response to PRRSV infection were significantly enriched in important protein posttranslational modifications including protein phosphorylation, autophosphorylation, and ubiquitination. The differential ASGs also were enriched in programmed cell death including response to tumor necrosis factor (MAP2K7, YTHDC2, CHI3L1), extrinsic apoptotic signaling pathway (HSF4, HUNTINGTIN, ZYMND11, and PELI3) and autophagosome assembly. These GO terms play vital roles in host cell apoptosis and protein metabolism in response to virus infection. While the differential ASE genes of TC pigs were assigned to GO terms related to tissue development including brain, embryonic organ, and nervous system development, and biological process involved in material transport such as anterograde synaptic vesicle transport, intracellular protein transport, intraciliary transport, endocytosis, and vesicle-mediated transport, which were associated with the host immune response to PRRSV infection. Among LW and TC pigs, differential ASGs were enriched in GO terms related to changes in cell morphology, especially in axon extension, mitochondrion morphogenesis, cell shape, and response to intracellular environmental stimuli such as hypoxia, ischemia, and DNA damage stimulus. In addition, differential ASGs were also widely involved in cell proliferation and transcription related GO terms in both of LW pigs and TC pigs (). Meanwhile, the KEGG pathway analysis identified that there are great differences among different breeds and tissues. As shown in Figure 4(b), differential ASGs in the ILN were significantly enriched in T cell receptor signaling pathway in both LW pigs (MAP2K7, IKBKG, TEC, PAK1, FYN, AKT2, and CD4) and TC pigs (MAP2K7, IKBKG, NFKBIE, PIK3CA, and DLG1). Differential ASGs in the ILN of LW pigs were uniquely enriched in TNF signaling pathway (RIPK1, AKT2, TRAF2, IKBKG, MAP2K7, FLIP-L, and CASP10), apoptosis (RIPK1, AKT2, TRAF2, IKBKG, FLIP-L, and CASP10), MAPK signaling pathway, and RIG-I-like receptor signaling pathway (RIPK1, TRAF2, SIKE1, IKBKG, and CASP10). Compared with LW pigs, KEGG pathway analysis in the ILN of TC pigs revealed significant enrichment in endocytosis, ErbB signaling pathway, Toll-like receptor signaling pathway (RIPK1, TLR4, CD40, MAPK8, TLR8, IKBKG, MAP2K7, and PIK3CA) and Epstein-Barr virus infection (RIPK1, SLA-6, NFKBIE, PSMC6, CD40, MAPK8, ENTPD1, GSK3B, IKBKG, MAP2K7, POLR2B, and PIK3CA). Besides, differential ASGs in the spleen of LW pigs tended to involve peroxisome and HIF-1 signaling pathway, while the thyroid hormone signaling pathway and transcriptional misregulation in cancer were enriched in TC pigs ().
Figure 4.
GO and KEGG pathway enrichment of differential alternative splicing genes. (a) GO enrichment analysis results; (b) KEGG pathway enrichment analysis results. The X-axis represents four groups, including ILN_LW (ILN of LW pigs), ILN_TC (ILN of TC pigs), spleen_LW (spleen of LW pigs), spleen_TC (spleen of TC pigs). The color represents the P value, and the size represents the number of gene.
3.4. Differential Expression of Splicing Factors between TC Pigs and LW Pigs
In order to determine whether alternative splicing factors play important roles in regulating the transcriptome difference between TC pigs and LW pigs, the expression of alternative splicing factors was investigated in this study. We identified 22 and 10 differentially expressed splicing factors in the ILN and spleen of LW pigs after PRRSV infection, respectively. Meanwhile, we also identified 23 and 16 differentially expressed splicing factors in the ILN and spleen of TC pigs after PRRSV infection, respectively. Among them, HNRNPU and DHX38 were upregulated in the ILN and spleen of both breeds, while HNRNPLL is downregulated. Interestingly, most of the differentially expressed splicing factors were upregulated after PRRSV infection, but there were different responses between TC pigs and LW pigs to PRRSV infection. For example, SRSF4 was upregulated in the ILN of LW pigs but had no significant expression differences in the ILN of TC pigs and downregulated in the spleen of TC pig after PRRSV infection (Figure 5(d)). Expression information of splicing factors is presented in .
Figure 5.
Differentially expressed splicing factors analysis in response to PRRSV infection. (a) Cluster analysis of differentially expressed alternative splicing factors in the ILN; (b) cluster analysis of differentially expressed alternative splicing factors in spleen; (c) comparison of differentially expressed alternative splicing factors in the ILN and spleen of LW pigs and TC pigs; (d) expression of SRSF4 in the ILN of TC pigs and LW pigs. Expression of each ASE was converted to Z scores.
3.5. Verification of Differential ASEs in TC Pigs and LW Pigs
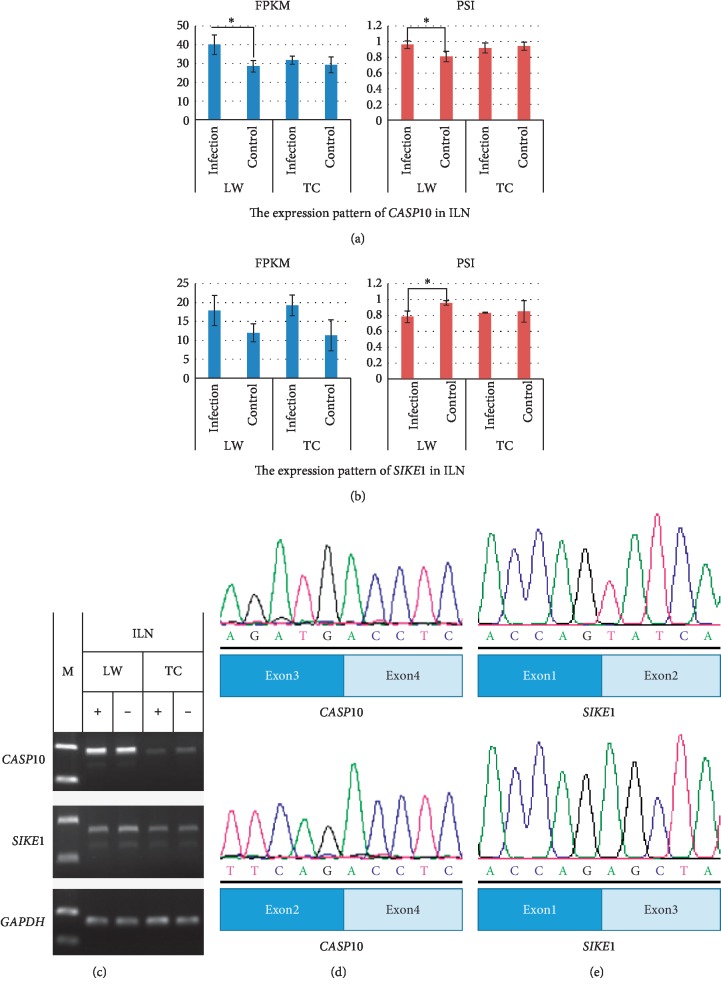
The ASEs reported in this study were validated via PCR and DNA sequencing. Genes involving apoptotic and immune response pathways including CASP10 and SIKE1 were selected for validation. CASP10 encodes Caspase-10 and directly affects the apoptotic process in different cells [31, 32]. Through PCR reaction and DNA sequencing, we found that exon 3 of CASP10 (ENSSSCT00000053464) was deleted in the spliced transcripts, which resulted in frame-shifting mutation and lost its original function (Figure 6(c)). From transcriptome data of LW pigs, both the expression level of functional transcripts and the value of PSI in CASP10 were increased in the infected group but no significant change occurred in TC pigs after PRRSV infection (Figure 6(a)). SIKE1, a critical member in the RIG-I-like receptor signaling pathway, acts as a suppressor of TLR3 and virus-triggered interferon activation pathways by interacting with IKK-epsilon and TBK1 [33]. The spliced transcript of SIKE1 (ENSSSCT00000045016) generated a truncated protein with the loss of exon 2. Moreover, its splicing levels were significantly different between the infection and control group in LW pigs (Figure 6(b)). After PCR validation, we sequenced PCR products and aligned them with transcript reference sequences. The results showed that the transcript sequence generated by splicing was consistent with the predictions (Figures 6(d) and 6(e)). The sequencing results are shown in .
Figure 6.
Verification of alternative splicing events in CASP10 and SIKE1. (a) Semiquantitative PCR of CASP10 and SIKE1 in the ILN; (b) sequencing results of splicing sites in different transcripts of CASP10; (c) DNA sequencing results of splicing sites in different transcripts of SIKE1. (d) The expression of CASP10 in the ILN; (e) the expression of SIKE1 in the ILN.
4. Discussion
Alternative splicing has been reported to play a critical role in producing protein diversity, regulating development and growth, and controlling responses to different stimuli by regulating gene expression and increasing the diversity of transcripts. Many viruses can widely modify splicing patterns of many cell transcripts involved in gene expression and RNA processing [34–38]. Alternative splicing related to viral infection was usually caused by manipulation of the spliceosome or host response to viral infection [39]. In this study, we used the transcriptome data of different peripheral immune organs to reveal that alternative splicing events were significantly increased in response to PRRSV infection, especially the increase in the frequency of SE type. Previous reports have revealed that SE was always the most frequent alternative splicing event in a large number of animal studies [20, 40]. It has been reported that the splicing level of host RNA will be changed when the virus invades the organism, which may be caused by the direct effect of the virus and the role of the immune response of the organism [4]. Therefore, we can speculate that PRRSV will significantly affect the host alternative splicing regulation.
In this study, we have reported 9184 alternative splicing genes, which indicated that ASEs were widespread in organisms and related to most biological activities. EIF4G1 was identified as the gene with most ASEs among all alternative splicing genes. The protein encoded by EIF4G1 is a component of the multisubunit protein complex EIF4F. The complex can inhibit viral replication in many viral infections [41]. Alternative splicing is subject to tissues-specific and lineage-specific regulation in primates [42]. We found that the number of differential ASGs in the spleen was less than that in the ILN in both of TC pigs and LW pigs. Similar results have been observed in previous studies. Human transcriptome reported that 10–30% of alternative splicing genes showed evidence of tissue-specific splice form [43]. Human virus infection studies indicated specific alternative splicing in different tissues in response to viral infection [20]. Tissue-specific alternative splicing is regulated by splicing regulatory elements and tissues-specific RNA-binding factors, which result in generating tissue-specific proteins. SE was considered to be one of the most important ASEs in regulating tissue-specific alternative splicing events between different tissues [20]. Previous studies have demonstrated that exons regulation transcripts mediated by tissue-specific alternative splicing can significantly remodel protein-protein interaction [44, 45]. Moreover, the shared differential ASGs of TC pigs and LW pigs in response to PRRSV infection were about 170 genes, almost 30% of total differential ASGs, which indicated lineage-specific alternative splicing regulation between TC pigs and LW pigs. Researches have shown that alternative splicing was tightly regulated and the expression of different splice forms was frequently changed between species [42]. Porcine myoneurin (MYNN) was identified to express different alternative splicing isoforms in LW pigs and Mashen (MS) pigs [28]. Functional enrichment of differential ASGs in different tissues and different breeds also proved tissue-specific and linage-specific alternative splicing regulation in response to PRRSV infection. Differential ASGs in the ILN of both TC pigs and LW pigs have enriched more functional pathways compared with differential ASGs in the spleen of both TC pigs and LW pigs. The functional pathway was mainly enriched in the TNF signaling pathway, apoptotic pathway, RIG-I receptor signaling pathway, and MAPK signaling pathway in the ILN of LW pigs. However, ASE genes in the ILN of TC pigs were mainly enriched in the Toll-like receptor signaling pathway. Several studies have shown that alternative splicing could affect host immune response mainly through apoptosis-related biological processes [46], DNA damage response [47, 48], and RIG-I pathway [49, 50]. These results were consistent with our previous results that TC pigs and LW pigs had difference immune response to PRRSV infection [16].
Splicing factors are critical for alternative splicing regulation because they can bind to short regulatory motifs on the pre-mRNA and activate alternative splicing events [51]. SRSF4 was significantly upregulated only in the ILN of LW pigs in response to PRRSV infection, but there was no significant change in TC pigs. SRSF4 was reported to act as a splicing regulator of Caspase-8 and mediates Caspase-8 induced apoptosis as a proapoptotic protease [52]. SRSF4 can regulate alternative splicing induced by cisplatin and contribute to apoptosis [53]. SRSF4 was significantly upregulated only in the ILN of LW pigs, which was consistent with the result that TC pigs and LW pigs performed different apoptosis regulation in response to PRRSV infection in our previous result [17].
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31572374 and 31802040), the National Key Technology R&D Program (2015BAD03B02), Frontier Special Project of Applied Foundation in Wuhan City (2018020401011306), and Fundamental Research Funds for the Central Universities (2662018QD004).
Contributor Information
Xiang Zhou, Email: zhouxiang@mail.hzau.edu.cn.
Bang Liu, Email: liubang@mail.hzau.edu.cn.
Data Availability
The raw RNA-seq data for this study have been submitted to SRA database (https://www.ncbi.nlm.nih.gov/sra) under accession no. PRJNA488960.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Xiang Zhou and Bang Liu conceived and supervised the study. Yu Zhang, Liyao Xue, Hang Xu, Wang Liang, and Qingqing Wu performed the experiments and analyzed the data. Xiang Zhou, Yu Zhang, and Qingqing Wu drafted the manuscript. All authors read and approved the manuscript.
Supplementary Materials
Table S1: PCR Primers used in the validation of alternative splicing transcripts. Table S2: differential ASE Statistics upon PRRSV infection in different groups. Table S3: information of differential ASEs upon PRRSV infection. Table S4: detailed information of enriched GO terms belonging to biological process by ASE genes. Table S5: description of KEGG pathways enrichment by ASE genes. Table S6: expression levels of splicing factors in the ILN and spleen of TC pigs and LW pigs upon PRRSV infection. Figure S1: (a) CASP10.SPLICING.fasta; (b) SIKE1.SPLICING.fasta.
References
- 1.Tian K., Yu X., Zhao T., et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2(6):p. e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An T.-Q., Tian Z. J., Leng C. L., et al. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerging Infectious Diseases. 2011;17(9):1782–1784. doi: 10.3201/eid1709.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holtkamp D. J., Kliebenstein J. B., Neumann E. J., et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. Journal of Swine Health and Production. 2013;21(2):72–84. [Google Scholar]
- 4.Albina E., Madec F., Cariolet R., Torrison J. Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Veterinary Record. 1994;134(22):567–573. doi: 10.1136/vr.134.22.567. [DOI] [PubMed] [Google Scholar]
- 5.Nelsen C. J., Murtaugh M. P., Faaberg K. S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. Journal of Virology. 1999;73(1):270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsberg R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Molecular Biology and Evolution. 2005;22(11):2131–2134. doi: 10.1093/molbev/msi208. [DOI] [PubMed] [Google Scholar]
- 7.Duan X., Nauwynck H. J., Pensaert M. B. Virus quantification and identification of cellular targets in the lungs and lymphoid tissues of pigs at different time intervals after inoculation with porcine reproductive and respiratory syndrome virus (PRRSV) Veterinary Microbiology. 1997;56(1-2):9–19. doi: 10.1016/s0378-1135(96)01347-8. [DOI] [PubMed] [Google Scholar]
- 8.Dokland T. The structural biology of PRRSV. Virus Research. 2010;154(1-2):86–97. doi: 10.1016/j.virusres.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi M., Lam T. T. Y., Hon C. C., et al. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Research. 2010;154(1-2):7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Lunney J. K., Chen H. B. Genetic control of host resistance to porcine reproductive and respiratory syndrome virus (PRRSV) infection. Virus Research. 2010;154(1-2):161–169. doi: 10.1016/j.virusres.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P., Zhai S., Zhou X., et al. Molecular characterization of transcriptome-wide interactions between highly pathogenic porcine reproductive and respiratory syndrome virus and porcine alveolar macrophages in vivo. International Journal of Biological Sciences. 2011;7(7):947–959. doi: 10.7150/ijbs.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Z. H., Zhou X., Michal J. J., et al. Reactomes of porcine alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059229.e59229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao S. Q., Jia J. Y., Mo D. L., et al. Understanding PRRSV infection in porcine lung based on genome-wide transcriptome response identified by deep sequencing. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing J. Y., Xing F., Zhang C. H., et al. Genome-wide gene expression profiles in lung tissues of pig breeds differing in resistance to porcine reproductive and respiratory syndrome virus. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0086101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proll M. J., Neuhoff C., Schellander K., et al. Transcriptome profile of lung dendritic cells after in vitro porcine reproductive and respiratory syndrome virus (PRRSV) infection. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0187735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang W., Li Z., Wang P., et al. Differences of immune responses between Tongcheng (Chinese local breed) and large white pigs after artificial infection with highly pathogenic porcine reproductive and respiratory syndrome virus. Virus Research. 2016;215:84–93. doi: 10.1016/j.virusres.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Liang W., Ji L. K., Zhang Y., et al. Transcriptome differences in porcine alveolar macrophages from Tongcheng and large white pigs in response to highly pathogenic porcine reproductive and respiratory syndrome virus (PRRSV) infection. International Journal of Molecular Sciences. 2017;18(7) doi: 10.3390/ijms18071475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornblihtt A. R., Schor I. E., Allo M., et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nature Reviews Molecular Cell Biology. 2013;14(5) doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 19.Pan Q., Shai O., Lee L. J., et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2009;41(6):p. 762. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 20.Wang E. T., Sandberg R., Luo S., et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen S., Park J. W., Lu Z.-X., et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proceedings of the National Academy of Sciences. 2014;111(51):E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy P. D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nature Reviews Microbiology. 2012;10(2):137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudreault S., Martenon-Brodeur C., Caron M., et al. Global profiling of the cellular alternative RNA splicing landscape during virus-host interactions. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0161914.e0161914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh D., Mathews M. B., Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harbor Perspectives in Biology. 2013;5(1) doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Álvarez E., Castelló A., Carrasco L., Izquierdo J. M. Alternative splicing, a new target to block cellular gene expression by poliovirus 2A protease. Biochemical and Biophysical Research Communications. 2011;414(1):142–147. doi: 10.1016/j.bbrc.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 26.De Maio F. A., Risso G., Iglesias N. G., et al. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathogens. 2016;12(8) doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao Y.-H. E., Bahn J. H., Lin X., Chan T.-M., Wang R., Xiao X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Research. 2016;26(4):440–450. doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., Li M., Gao P., et al. Novel splice isoforms of pig myoneurin and their diverse mRNA expression patterns. Asian-Australasian Journal of Animal Sciences. 2018;31(10):1581–1590. doi: 10.5713/ajas.17.0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis M. P. A., van Dongen S., Abreu-Goodger C., Bartonicek N., Enright A. J. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63(1):41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Roberts A., Goff L., et al. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nature Protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wachmann K., Pop C., van Raam B. J., et al. Activation and specificity of human caspase-10. Biochemistry. 2010;49(38):8307–8315. doi: 10.1021/bi100968m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh J. E., Kim M. S., Ahn C. H., et al. Mutational analysis of CASP10 gene in colon, breast, lung and hepatocellular carcinomas. Pathology. 2010;42(1):73–76. doi: 10.3109/00313020903434371. [DOI] [PubMed] [Google Scholar]
- 33.Chau T.-L., Gioia R., Gatot J.-S., et al. Are the IKKs and IKK-related kinases TBK1 and IKK-ɛ similarly activated? Trends in Biochemical Sciences. 2008;33(4):171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Sessions O. M., Tan Y., Goh K. C., et al. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Neglected Tropical Diseases. 2013;7(3) doi: 10.1371/journal.pntd.0002107.e2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batra R., Stark T. J., Clark E., et al. RNA-binding protein CPEB1 remodels host and viral RNA landscapes. Nature Structural & Molecular Biology. 2016;23(12):1101–1110. doi: 10.1038/nsmb.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B., Li X., Huo Y., et al. Cellular responses to HSV-1 infection are linked to specific types of alterations in the host transcriptome. Scientific Reports. 2016;6:p. 28075. doi: 10.1038/srep28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu B., Huo Y., Yang L., et al. ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virology Journal. 2017;14(1):p. 217. doi: 10.1186/s12985-017-0882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B., Huo Y., Yang L., et al. Correction to: ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virology Journal. 2019;16(1):p. 17. doi: 10.1186/s12985-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashraf U., Benoit-Pilven C., Lacroix V., Navratil V., Naffakh N. Advances in analyzing virus-induced alterations of host cell splicing. Trends in Microbiology. 2019;27(3):268–281. doi: 10.1016/j.tim.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Montes M., Sanford B. L., Comiskey D. F., Chandler D. S. RNA splicing and disease: animal models to therapies. Trends in Genetics. 2019;35(1):68–87. doi: 10.1016/j.tig.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montero H., García-Román R., Mora S. eIF4E as a control target for viruses. Viruses. 2015;7(2):739–750. doi: 10.3390/v7020739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blekhman R., Marioni J. C., Zumbo P., Stephens M., Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Research. 2010;20(2):180–189. doi: 10.1101/gr.099226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Research. 2002;30(17):3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buljan M., Chalancon G., Eustermann S., et al. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Molecular Cell. 2012;46(6):871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis J. D., Barrios-Rodiles M., Çolak R., et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Molecular Cell. 2012;46(6):884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 46.Paronetto M. P., Passacantilli I., Sette C. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell Death & Differentiation. 2016;23(12):1919–1929. doi: 10.1038/cdd.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson K., Wu C. J., Schwartz S. Role of the DNA damage response in human papillomavirus RNA splicing and polyadenylation. International Journal of Molecular Sciences. 2018;19(6) doi: 10.3390/ijms19061735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shkreta L., Chabot B. The RNA splicing response to DNA damage. Biomolecules. 2015;5(4):2935–2977. doi: 10.3390/biom5042935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P.-H., Fung S.-Y., Gao W.-W., et al. A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Research. 2018;46(8):4054–4071. doi: 10.1093/nar/gky186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gee P., Chua P. K., Gevorkyan J., et al. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. Journal of Biological Chemistry. 2008;283(14):9488–9496. doi: 10.1074/jbc.m706777200. [DOI] [PubMed] [Google Scholar]
- 51.Kosti I., Radivojac P., Mandel-Gutfreund Y. An integrated regulatory network reveals pervasive cross-regulation among transcription and splicing factors. PLoS Computational Biology. 2012;8(7) doi: 10.1371/journal.pcbi.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan W., Wang W., Ma Q. Physiological and pathological function of serine/arginine-rich splicing factor 4 and related diseases. BioMed Research International. 2018;2018:6. doi: 10.1155/2018/3819719.3819719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabriel M., Delforge Y., Deward A., et al. Role of the splicing factor SRSF4 in cisplatin-induced modifications of pre-mRNA splicing and apoptosis. BMC Cancer. 2015;15:p. 227. doi: 10.1186/s12885-015-1259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: PCR Primers used in the validation of alternative splicing transcripts. Table S2: differential ASE Statistics upon PRRSV infection in different groups. Table S3: information of differential ASEs upon PRRSV infection. Table S4: detailed information of enriched GO terms belonging to biological process by ASE genes. Table S5: description of KEGG pathways enrichment by ASE genes. Table S6: expression levels of splicing factors in the ILN and spleen of TC pigs and LW pigs upon PRRSV infection. Figure S1: (a) CASP10.SPLICING.fasta; (b) SIKE1.SPLICING.fasta.
Data Availability Statement
The raw RNA-seq data for this study have been submitted to SRA database (https://www.ncbi.nlm.nih.gov/sra) under accession no. PRJNA488960.