Abstract
Objective
Small RNA (sRNA) sequencing has revealed new sRNA classes beyond microRNAs (miRNAs). These sRNAs can regulate genes and act as biomarkers. The aim of this study was to determine if the endogenous plasma sRNA landscape is altered in patients with rheumatoid arthritis (RA) compared with control subjects and to determine its association with disease‐related parameters in RA.
Methods
sRNA sequencing was performed on plasma from 165 RA and 90 control subjects who were frequency‐matched for age, race, and sex. Endogenous sRNAs, such as miRNAs, isomiRs, sRNAs derived from small nuclear RNAs (snDRs), small nucleolar RNAs (snoDRs), Y RNAs (yDRs), transfer‐derived RNAs (tDRs), long noncoding RNAs (lncDRs) as well as miscellaneous sRNAs (miscRNAs), were quantified using Tools for Integrative Genome analysis of Extracellular sRNAs (TIGER). Individual and categories of sRNAs were compared between RA and controls, and significantly altered sRNAs and sRNA categories were correlated with disease activity and general laboratory measures in RA.
Results
Patients with RA had more miRNAs (1.42‐fold, P = 0.01), more tDRs (1.14‐fold, P = 0.04), and fewer yDRs (−1.41‐fold, P = 0.009) compared with control subjects. Disease duration was inversely associated with yDRs. Disease‐related parameters, such as Disease Activity Score‐28 (DAS28), swollen joint count, and inflammatory markers were significantly positively associated with tDRs and miscRNAs, and miR‐22‐3p and related sequences and isomiRs were most significantly associated with DAS28.
Conclusion
Endogenous plasma sRNAs are altered in RA compared with control subjects. Although individual miRNAs have been well studied and many are excellent biomarkers in RA, several non‐miRNA sRNAs were significantly associated with disease‐related parameters as classes and may represent novel biomarkers for RA.
Introduction
Small RNAs (sRNAs) are short noncoding RNAs that can fine tune gene expression and be used as disease biomarkers 1, 2 and therapeutic targets. These sRNAs are detected within cells and extracellularly in plasma and other body fluids 3. Most sRNA investigation in rheumatoid arthritis (RA) has focused on canonical microRNAs (miRNAs); however, sequence variations in miRNAs (such as isomiRs) can modify function. There are also other sRNA classes of similar size to miRNAs that have been studied rarely in autoimmunity. Some of these include transfer RNA–derived fragments and halves (tDRs), fragments derived from Y RNAs (yDRs), ribosomal RNAs (rDRs), small nuclear RNAs (snDRs), small nucleolar RNAs (snoRNAs), and long noncoding RNAs (lncDRs).
A miRNA typically regulates gene expression by binding the 3ʹ untranslated region of messenger RNA through complementary seed region located at approximately nucleotide 2 to 8 at the 5ʹ end of the miRNA 4. Although miRNA structure and function provide a framework for understanding other sRNAs, sRNAs can regulate genes in different ways and may have different significance as circulating biomarkers. For example, some tDRs may act as miRNAs with seed region binding 5, others may loosen hairpin structures of specific mRNAs to increase protein production 6, and others (typically transfer RNA halves) cause global downregulation of translation by displacing translation initiation factors 7. Another sRNA class, yDRs, are released during apoptosis and can stimulate Toll‐like receptor 7 (TLR7) and promote further apoptosis 8, 9. Although functions of many of the other parent RNAs, such as ribosomal, small nuclear, small nucleolar, and long noncoding RNAs, are established, their shorter sRNA fragment forms—rDRs, snDRs, snoDRs, and lncDRs—are poorly understood. This area of research, however, is evolving rapidly.
The altered expression of canonical miRNAs has been extensively studied as biomarkers that also provide insight into disease pathogenesis. In contrast, research on isomiRs and non‐miRNA sRNA classes has only developed recently. Some findings include serum tDR and yDR composition as a biomarker of breast cancer 10 and coronary artery disease 11, and bone marrow tDRs as a biomarker for myelodysplastic syndrome 12. A prior study compared serum sRNAs, including some non‐miRNA sRNA classes such as yDRs and tDRs, in four patients with early RA and four control subjects 13, but to our knowledge there is no prior study describing the plasma non‐miRNA sRNA composition or its clinical associations in RA.
The purpose of this study was to characterize the landscape of human (endogenous) plasma sRNA classes of patients with RA compared with control subjects, present differential expression data of individual sRNAs across this landscape to serve as a resource for future studies on sRNAs in RA, and determine the relationship between the various sRNA classes and disease‐related parameters in RA.
Methods
Study population
The study included 167 patients with RA and 91 control subjects who were frequency‐matched for age, race, and sex from a prior cross‐sectional study for which recruitment and study procedures have been described previously 14. Three subjects who had insufficient depth of sequencing were excluded from analysis, leaving a total of 165 patients with RA and 90 control subjects. All subjects were 18 years of age or older, and patients with RA met classification criteria for RA 15. The study was approved by the Vanderbilt Institutional Review Board, and all subjects gave written informed consent (IRB# 000567).
Disease‐related variables
RA disease activity was assessed by a 28‐joint‐count disease activity score (DAS28) using erythrocyte sedimentation rate (ESR) 16. Functional status was assessed with the modified Health Assessment Questionnaire 17. Complete blood counts, creatinine, albumin, high‐sensitivity C‐reactive protein (CRP) concentrations, and ESR were measured by the Vanderbilt University Medical Center Clinical Laboratory. Interleukin 6 (IL‐6) and tumor necrosis factor alpha (TNF‐α) were measured by multiplex enzyme‐linked immunosorbent assay (ELISA; Millipore). These disease‐related variables were correlated with sRNA categories (as reads per million). The Spearman correlations were presented using heatmap with hierarchical clustering.
The association between individual sRNAs that were significantly altered comparing RA and control subjects and DAS28 were assessed by linear regression and presented using dot plots of P values.
sRNA sequencing
The sRNA sequencing libraries were prepared as previously described 2 in batches containing samples from both RA and control subjects. Briefly, 200 ul of plasma was used to extract RNA with Total RNA purification kits (Norgen), and complementary DNA libraries were prepared using TruSeq Small RNA Library Preparation Kits (Illumina). The Vanderbilt Technologies for Advanced Genomics (VANTAGE) core facility assessed library quality and performed size selection of sRNAs by Pippin Prep (Sage Science). VANTAGE performed sequencing using the Illumina NextSeq500.
sRNA sequence alignments and analysis
Sequence data were analyzed using Tools for Integrative Genome analysis of Extracellular sRNAs (TIGER) 18. Detailed methods for this pipeline have been previously reported 2, 18. Briefly, to detect miRNA with nontemplated nucleotide additions (NTAs), three potential NTA isoforms of each sRNA sequencing read were generated by removing one, two, or three bases from the 3ʹ end. All four isoforms, including three NTA isoforms and the original read, were mapped to a custom database using Bowtie 19 (version 1.1.2), allowing one mismatch. The custom database included the human genome (hg19), and several mature transcript databases (transfer RNAs: http://gtrnadb.ucsc.edu/GtRNAdb2/; ribosomal RNAs: http://archive.broadinstitute.org/cancer/cga/rnaseqc_download). Read alignment to different sRNA classes was filtered based on the following criteria: tDRs, snDRs, snoDRs, yDRs, and miscellaneous (miscRNAs) required 1 or fewer mismatches, 16 or greater nucleotide length, and 90% or greater overlap with parent RNAs; lncDRs and rDRs required perfect match, 20 or longer nucleotide length, and 90% or greater overlap with parent RNAs; miRNAs required 1 or fewer mismatches and 16 or longer nucleotide length. For miRNAs, the 5ʹ end offset of −2, −1, 0, 1, or 2 nucleotides (Supplementary Figure 1) based on the miRNA genome coordinates from miRBase (version 21) was included to analyze isomiRs. An average of approximately 9.7 million reads were obtained from sequencing, with an average of approximately 7.2 million reads available for mapping after trimming. An average of approximately 3 million reads (range 0.05‐31 million) were mapped to the human genome or human sRNA databases as above.
The sRNA read counts were normalized to reads per million total reads (RPM); thus, throughout read counts represent the proportion of reads to total reads. Differential expression of sRNAs were assessed by DESeq2 20, adjusting for age, race, sex, and batch effect. Because of the multiple comparisons, Benjamini‐Hochberg's method was used to control false discovery rate (FDR) at 5%. The differential expression data for each sRNA category is provided in supplementary tables as counts aligning to particular parent RNA (designated in supplementary table tabs as “xx_counts”) and as counts of a specific sequence (designated as “xx_sequence counts”). These supplementary tables include the sRNAs that have at least a median of five read counts in either RA or control subject groups. Within tables and the text, sRNA nucleotide sequences are presented as DNA sequences as convention when referring to the sequencing results. For presenting average, fold difference, and percentage of sRNA categories in RA versus control subjects, sRNA RPM data were log‐transformed and presented as geometric mean (95% confidence interval [CI]) due to skewness.
General statistical methods
Descriptive statistics were calculated as median (interquartile range) for continuous variables and as frequency and proportion for categorical variables. Wilcoxon's rank sum tests were used to compare continuous variables, and Pearson's chi‐square test was used to compare categorical variables. Spearman correlation was used to assess the relationship between sRNA categories and disease‐related variables. In linear regression analyses, variables were log‐transformed as needed to normalize residuals.
Results
Subject characteristics
Subject characteristics are presented in Table 1 . Patients with RA and control subjects were of similar age, race, and sex. Median age was approximately 54 years, and subjects were predominantly Caucasian and female. Among RA patients, median disease activity was moderate (DAS28‐ESR = 3.91 units), and median disease duration was 3 years.
Table 1.
Patient demographicsa
| Demographics | RA (N = 165) | Control (N = 90) | P |
|---|---|---|---|
| Age, years | 54 [45, 63] | 53 [44, 59] | 0.35 |
| Race, # (%) Caucasian | 146 (88) | 76 (84) | 0.49 |
| Sex, # (%) female | 113 (68) | 56 (62) | 0.31 |
| RF, # (%) positive | 113 (72) | … | … |
| Disease duration, years | 3 [2, 18] | … | … |
| DAS28‐ESR, score | 3.91 [2.62, 4.91] | … | … |
| Tender joint count | 2 [0, 7] | … | … |
| Swollen joint count | 3 [0, 8] | … | … |
| ESR, mm/h | 15 [7, 36] | … | … |
| hsCRP, mg/l | 4.00 [1.20, 10.75] | 0.55 [0.20, 1.59] | <0.001 |
| Methotrexate use, # (%) current | 117 (71) | … | … |
| Leflunomide use, # (%) current | 29 (18) | … | … |
| Corticosteroids use, # (%) current | 87 (47) | 2 (2) | <0.001 |
| Anti‐TNFα use, # (%) current | 33 (20) | … | … |
Abbreviation: DAS28, Disease Activity Score‐28; ESR, erythrocyte sedimentation rate; hsCRP, high‐sensitivity C‐reactive protein; RA, rheumatoid arthritis; RF, rheumatoid factor; TNFα, tumor necrosis factor alpha.
Data presented as median [interquartile range] or # number (%).
Differences in the sRNA landscape in RA vs Control subjects
miRNAs. Total miRNA reads (RPM) were 1.42‐fold higher in patients with RA compared to controls (P = 0.01), and 175 miRNAs were significantly differentially expressed in RA after adjustment for age, race, sex, batch, and multiple comparisons (Supplemental Figure 2 and Supplemental Tables 1 and 2 show total miRNA counts and individual sequence counts), with the majority being increased. The top 15 most significantly differentially expressed miRNAs comparing patients with RA to control subjects were previously reported 2 and vary slightly from our previously published data due to DESeq2 program updates. One of the most abundant and significantly altered miRNAs in RA versus control plasma was miR‐22‐3p (2.71‐fold increased, P adj = 2.67E‐11).
isomiRs. The proportion of 5ʹ isomiRs (which would change the miRNA seed region and thus possibly target binding as illustrated in Supplementary Figure 1) to total miRNAs was not significantly different in patients with RA (geometric mean 2.6% [95%CI: 2.5%, 2.7%]) versus control subjects (2.5% [95%CI: 2.4%, 2.7%], P = 0.54) (Supplemental Figure 3). However, some isomiRs were disproportionately altered in RA. For example, the proportion of this isomiR miR‐30e‐5p_+_1 to total miR‐30e‐5p was increased to a greater extent among those with RA (geometric mean 15.4% [95%CI: 14.1%, 16.8%]) compared to control subjects (9.6% [95%CI: 8.5%, 11%]) (1.6‐fold increase in proportion of isomiR to total miRNA counts, P < 0.001; Supplemental Figure 4). NTAs to the 3ʹ end (Supplementary Figure 1) can modify function. For example, uridylated miR‐26b‐5p (designated NTA_T, NTA_TT, or NTA_TTT), which can suppress the miRNA's ability to downregulate IL‐6 21, was not significantly altered in patients with RA versus control subjects (P adj > 0.05) (Supplementary Table 3). The proportion of uridylated to total miR‐26b‐5p, however, was significantly increased in the plasma of patients with RA (1.32‐fold, P = 0.01). Neither uridylated miR‐26b‐5p as RPM nor as a percentage of total miR‐26b‐5p was associated with plasma IL‐6 expression (P > 0.05) in RA.
Y RNA fragments (yDRs). The most abundant endogenous sRNA class in the plasma of both patients with RA and control subjects was yDRs (Figure 1), and these were decreased among patients with RA versus controls (−1.41‐fold, P = 0.009) (Table 2). Humans have four Y RNAs (hY1, hY3, hY4, and hY5) and Y RNA pseudogenes. Most of the yDRs in plasma are derived from the 5ʹ and 3ʹ ends of the Y RNA (Supplementary Figure 5).
Figure 1.
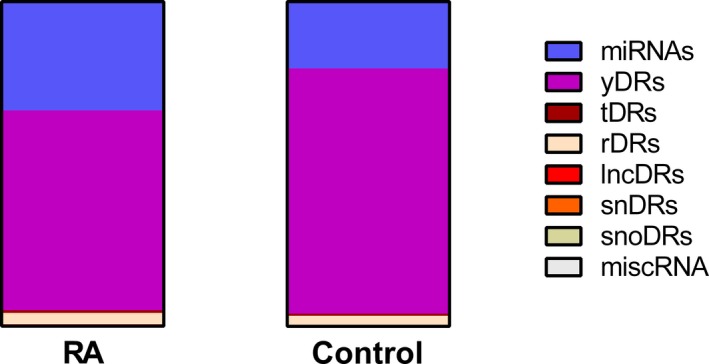
Proportions of endogenous plasma small RNAs (sRNAs) by classes in rheumatoid arthritis (RA) and control subjects. Proportions were calculated using the geometric mean of the sRNA classes due to skewness. Abbreviation: lncDR, long noncoding RNA–derived sRNA; miRNAs, microRNA; miscRNA, miscellaneous RNA; RA, rheumatoid arthritis; rDR, ribosomal RNA–derived sRNA; snDR, small nuclear RNA–derived sRNA; snoDR, small nucleolar RNA–derived sRNA; tDR, transfer RNA–derived sRNA; yDR, Y RNA–derived sRNA.
Table 2.
Plasma small RNA classes in patients with RA and control subjectsa
| sRNA Class | RA (N = 165) | Control (N = 90) | Fold Difference | P |
|---|---|---|---|---|
| miRNA | 100 814 (86 923, 116 925) | 70 982 (57 236, 88 029) | 1.42 | 0.01 |
| yDR | 185 245 (159 332, 215 372) | 260 447 (217 449, 311 947) | −1.41 | 0.009 |
| tDR | 2690 (2399, 3017) | 2370 (1876, 2995) | 1.14 | 0.04 |
| rDR | 10 772 (9319, 12 451) | 9521 (7473, 12 130) | 1.13 | 0.21 |
| lncDR | 708 (615, 815) | 810 (623, 1053) | −1.14 | 0.67 |
| snDR | 311 (267, 363) | 344 (277, 427) | −1.11 | 0.39 |
| snoDR | 56 (47, 66) | 49 (37, 64) | 1.14 | 0.22 |
| miscRNA | 108 (89, 130) | 79 (63, 99) | 1.37 | 0.06 |
Abbreviation: lncRNA, long noncoding RNA–derived small RNA; miRNA, microRNA; miscRNA, miscellaneous RNA; RA, rheumatoid arthritis; rDR, ribosomal RNA–derived small RNA; snDR, small nuclear RNA–derived small RNA; snoDR, small nucleolar RNA–derived small RNA; tDR, transfer RNA–derived small RNA; yDR, Y RNA–derived small RNA.
Data are presented as the geometric mean with 95% confidence intervals of the geometric mean due to skewness of the data.
Differential expression of yDRs in patients with RA versus control subjects is presented in Supplementary Figure 6 and Supplementary Tables 4 and 5. Based on alignment to the Y RNA, RNY3P6 was most significantly increased in patients with RA versus control subjects (2.03‐fold, P adj = 2.85E‐08) but was low in abundance. Among the top differentially expressed read alignments to Y RNAs, yDRs aligning to RNY5 were abundant and significantly decreased in patients with RA compared to control plasma (1.42‐fold decreased, P adj = 0.002) (Supplementary Table 4). Among the top differentially expressed yDR sequences, 5ʹ‐AGTTGGTCCGAGTGTTGTGGGTTATTGTTA‐3ʹ was abundant and significantly reduced (1.59‐fold, P adj = 9.48E‐05) in patients with RA compared to control plasma along with several other similar sequences with 3ʹ end length variations (Supplementary Table 5).
tDRs. Plasma tDRs were increased in patients with RA compared to control subjects (1.14‐fold, P = 0.04) (Table 2), and encompass less than 1% of the endogenous plasma sRNAs (Figure 1). Most of the tDRs arise from the 5ʹ side of the tRNA in both subjects with RA and control subjects and are more commonly 5ʹ halves (Supplementary Figure 7). The tDRs from naturally occurring suppressor tRNAs were increased 1.71‐fold in patients with RA versus control subjects (P adj = 2.73E‐05, Supplementary Table 6), and the main contributing isoacceptor was tRNA‐Sup‐TTA, which was increased 1.69‐fold (P adj = 5.1E‐05, Supplementary Table 7) in patients with RA versus control subjects.
tDRs from tRNAs encoding for asparagine, isoleucine, and aspartic acid were decreased significantly in patients with RA (−1.68‐fold to −1.47‐fold, P adj < 0.001) (Supplementary Figure 8, Supplementary Table 6), with each having tDR alignments to a predominant isoacceptor that was significantly decreased in RA: tRNA‐Asn‐GTT, tRNA‐Ile‐AAT, and tRNA‐Asp‐GTC (−1.67‐fold to −1.46‐fold, P adj < 0.05). Additionally, tDRs from an isoacceptor for threonine, tRNA‐Thr‐TGT, were also significantly decreased in patients with RA compared to controls (−1.48‐fold, P adj = 0.04) (Supplementary Figure 8, Supplementary Tables 6 and 7).
Most of the significantly altered tDR sequences were low in abundance, but a tDR (5ʹ‐GCATTGGTGGTTCAGTGGTAGAATTCTCGCCT‐3ʹ) derived from the 5ʹ half of glycine‐encoding tRNAs was abundant and significantly increased in patients with RA (1.40‐fold, P adj = 0.03) (Supplementary Table 8), although overall tDRs aligning to tRNAs encoding glycine were not significantly altered (Supplementary Table 6).
Other sRNAs. The rDRs, snDRs, snoDRs, other sRNAs (including lncDRs and miscRNAs) as classes were not significantly altered in patients with RA compared to control subjects (Table 2). However, the composition of these sRNAs based on sequencing counts aligning to a specific parent RNA and individual sRNA sequence counts were significantly altered in RA (Supplementary Figures 9‐12, Supplementary Tables 9‐16). For example, rDRs aligning to several 5S ribosomal pseudogenes were altered in the plasma patients with RA (RNA5SP442: 1.58‐fold increased, P adj = 1.43E‐04; RNA5SP336: 1.66‐fold decreased, P adj = 0.007; RNA5SP216: 1.36‐fold increased, P adj = 0.002) compared to control subjects (Supplementary Table 9). The plasma abundance of these sRNA classes was low at less than 5% of endogenous sRNAs (Figure 1).
Relationship between sRNAs and disease‐related parameters
sRNAs by classes and disease‐related parameters. DAS28 was significantly positively associated with miscRNAs (Rho = 0.22, P = 0.005) and tDRs (Rho = 0.17, P = 0.03) (Figure 2). Swollen joint count was similarly positively associated with miscRNAs (Rho = 0.21, P = 0.007), tDRs (Rho = 0.18, P = 0.03) as well as snDRs (Rho = 0.21, P = 0.007) and snoDRs (Rho = 0.19, P = 0.02). Tender joint count was only correlated with miscRNAs (Rho = 0.19, P = 0.01). ESR was significantly associated with tDRs (Rho = 0.21, P = 0.006) and lncDRs (Rho = 0.15, P = 0.049). CRP was significantly associated with miscRNAs (Rho = 0.16, P = 0.04) (Figure 2). Although miRNAs as a class were not significantly associated with DAS28, the ratio of 5ʹ isomiRs to total miRNAs was inversely associated with DAS28 (Rho = −0.23, P = 0.003) and disease duration (Rho = −0.27, P < 0.001) (Figure 2). These associations were independent of each other and age (DAS28, P adj = 0.03; disease duration, P adj = 0.003). Disease duration was inversely associated with yDRs (Rho = −0.34, P = 8.5E‐06) and to a lesser extent with rDRs (Rho = −0.18, P = 0.02; Figure 2). The association remained independent of age with disease duration for yDRs (P = 8.8E‐05) but not rDRs.
Figure 2.
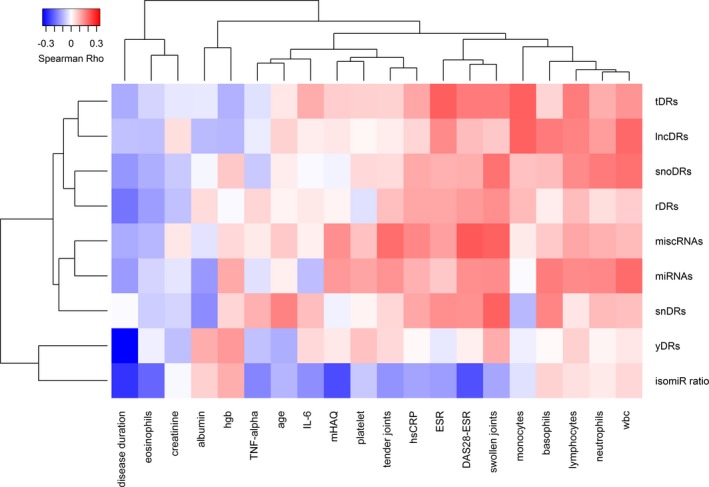
Heatmap with hierarchical clustering demonstrating the relationship between sequencing reads of plasma small RNAs (sRNAs) based on category, disease‐associated variables, and laboratory measures. Abbreviation: DAS28, Disease Activity Score‐28; ESR, erythrocyte sedimentation rate; hgb, hemoglobin; hsCRP, high‐sensitivity C‐reactive protein; IL‐6, interleukin 6; isomiR ratio, ratio of 5ʹ isomiRs to total miRNAs; lncDR, long noncoding RNA–derived sRNA; mHAQ, modified Health Assessment Questionnaire score; miRNAs, microRNAs; miscRNA, miscellaneous RNA; rDR, ribosomal RNA–derived sRNA; snDR, small nuclear RNA–derived sRNA; snoDR, small nucleolar RNA–derived sRNA; tDR, transfer RNA–derived sRNA; TNF, tumor necrosis factor; wbc, white blood cell count; yDRs, Y RNA–derived sRNA.
Individual sRNAs and DAS28. We broadly surveyed the association between RA disease activity by DAS28 and sRNAs, which are differentially expressed in patients with RA versus control subjects (Figure 3). For miRNAs, we included those significantly altered in patients with RA versus control subjects after adjustment for FDR, but for the remaining sRNA classes, we included those significantly altered using an unadjusted P value (P < 0.05) to include a larger number of these less‐studied sRNAs. Despite a lack of association between miRNAs as a sRNA category with disease activity, individual miRNAs were more significantly associated with disease activity than other sRNAs (Figure 3). Sequences related to miR‐22‐3p were most robustly represented. For example, the sequence 5ʹ‐AAGCTGCCAGTTGAAGAACTG‐3ʹ was most significantly associated with disease activity. This sequence aligns to the canonical miR‐22‐3p but is short by one nucleotide at the 3ʹ end. The second most significant sequence, 5ʹ‐AAGCAGCCAGTTGAAGAACTGT‐3ʹ, is miR‐22‐3p with a mismatch at the fifth base (T‐>A), which would change the seed region of the miRNA and potentially mRNA targeting. Although sequences related to miR‐22‐3p are highly correlated, the individual sequences were not uniformly correlated with disease activity (Supplementary Table 17).
Figure 3.
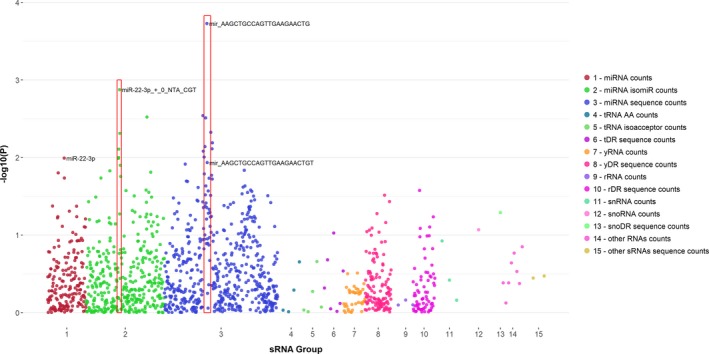
Plot of the significance of association between individual small RNAs (sRNAs) and rheumatoid arthritis (RA) disease activity. The individual sRNAs that were significantly altered in patients with RA compared to control subjects were used in the analysis. The sRNAs by parent RNA and sequences are plotted across the x‐axis. For sequence counts, the sequences aligning to a particular parent RNA are clustered together. Red boxes highlight sRNAs related to miR‐22‐3p. “Other RNAs counts” includes counts aligning to long noncoding RNAs (lncRNAs) and miscellaneous RNAs (miscRNAs). “Other sRNAs sequence counts” refers to lncRNA‐derived sRNA (lncDR) and miscRNA sequence counts. Abbreviation: miRNA, microRNA; rDR, rRNA‐derived sRNA; rRNA, ribosomal RNA; snRNA, small nuclear RNA; snoDR, snoRNA‐derived sRNA; snoRNA, small nucleolar RNA; tDR, tRNA‐derived sRNA; tRNA, transfer RNA; yDR, Y RNA–derived sRNA.
Discussion
We found that the endogenous plasma sRNAome is altered in patients with RA, with differences in the proportions of individual sRNA classes and their composition. Specifically, plasma miRNAs and tDRs were significantly increased among patients with RA, whereas plasma yDRs were significantly decreased among patients with RA compared with control subjects. All sRNA classes examined possessed altered composition, whether by sequencing counts aligning to parent RNA, individual sRNA sequence counts, or both, among patients with RA compared to control subjects. The sRNA classes tDRs and miscRNAs, the ratio of isomiRs to total miRNAs, and individual sRNAs related to miR‐22‐3p were most strongly correlated with RA disease activity.
We 1, 2 and others 22 have previously shown that plasma miRNA composition is altered in patients with RA. Moreover, as we showed in the current study, miRNAs as a class were increased in the plasma of patients with RA 22. We hypothesize that this shift is due to broad changes in miRNA processing. For example, Dicer and Drosha, which cleave miRNAs to maturity, are upregulated in RA peripheral blood mononuclear cells 23. This appears to serve a homeostatic role in RA because this increase in Dicer and Drosha causes a decrease in TNF‐α expression 23, likely due to an increase in multiple miRNAs regulating inflammatory pathways.
In the current study, as in others 24, miR‐22‐3p was both highly abundant and significantly increased in RA plasma. Moreover, canonical miR‐22‐3p and some variants of miR‐22‐3p (isomiRs, sequences with mismatches, etc.) were most significantly associated with DAS28 in patients with RA. Given the large number of comparisons, these associations were not significant after adjustment for FDR; however, the reproducibility of the findings across multiple variations of the canonical miR‐22‐3p suggests a relationship to RA disease activity. We previously showed that a panel including miR‐22‐3p along with miR‐24‐3p, miR‐96‐5p, miR‐134‐5p, miR‐140‐3p, and miR‐627‐5p robustly differentiated patients with RA from control subjects in two cohorts 2. Additionally, we previously showed that miR‐22‐3p was significantly increased in plasma from patients with systemic lupus erythematosus (SLE) compared to control subjects, and inhibition of this miRNA decreased lupus nephritis and Th1 T cell differentiation in a murine model of SLE (D.L. Michell, A. Faust, J.L. Moore, B.D. Appleton, M. Ormseth, M. Ramirez‐Solano, Q. Sheng, J.F. Solus, C.M. Stein, K.C. Vickers, A.S. Major, under review). This demonstrates that not only is miR‐22‐3p significant in mediating autoimmunity but also that plasma sRNAs are biomarkers that can provide mechanistic insight into disease.
To our knowledge, this is the first report on isomiRs in RA. These miRNA variants can alter canonical miRNA function. For example, because of imprecise cutting of the miRNA, the 5ʹ end may be offset, shifting the seed region sequence and changing the genes that the miRNA regulates. IsomiRs formed by nontemplated additions to the 3ʹ end of the miRNA can alter the miRNA's regulatory capacity and increase miRNA processing 5, 6. It is challenging to quantitively detect isomiRs and other sequence modifications by more commonly used methods, such as PCR or array‐based approaches. These technologies typically measure all isomiRs and canonical miRNAs together, as we present in the total miRNA reads. On a broad scale, although isomiRs read counts were increased in patients with RA compared to control subjects, the proportion of isomiRs to total miRNAs was not altered between patients with RA and control subjects. However, we did find that some individual isomiRs were disproportionately increased in RA plasma. In the example discussed, miR‐30e‐5p and its isomiR, miR‐30e‐5p_+_1, the isomiR may cause a change in gene regulation due to shifting of the canonical seed region sequence from GUAAACA to UAAACAU. Further studies will be necessary to determine if the disproportionate increase in some isomiRs leads to quantitative differences in target mRNA gene expression in patients with RA.
We questioned whether the production of 5ʹ isomiRs might be increased in the setting of high RA disease activity related possibly to proliferative pressure and imprecise endonuclease cutting. We found the opposite. Higher DAS28 was associated with a smaller ratio of isomiR to total miRNAs. We speculate that this could be homeostatic, and in the setting of inflammation, more precise processing could yield better downregulation of inflammatory processes. Alternatively, some isomiRs, with their altered seed region binding, may dampen inflammation more effectively.
The tDRs were modestly increased in patients with RA versus control plasma. Broadly, this may serve a homeostatic role to decrease cellular response to stresses. For example, tDRs, particularly tRNA halves, are increased through upregulation of the ribonuclease angiogenin in the setting of cellular stress such as oxidative damage, infection, hypoxia, and amino acid deficiency to repress translation and dampen cellular activities 7, 25, 26. tDRs arising from the 5ʹ end of the parent tRNA are capable of inhibiting translation and a GG sequence motif at positions 17‐18 or 18‐19 are necessary 27. In this study, all tDR sequences that were significantly altered in the plasma of patients with RA compared to control subjects are derived from the 5ʹ end of the parent tRNA and possess a GG motif at this location.
The tDRs arising from naturally occurring suppressor tRNAs were significantly increased in RA. To our knowledge, the effect of suppressor tRNAs and their tDRs on RA and other autoimmune diseases is unknown. As with other tDRs 5, 6, 7, 25, 26, 27, these sequences may play a regulatory role through specific base pairing or displacing translational machinery. Another consideration is whether patients with RA are producing more of these naturally occurring suppressor tRNAs. If so, this could be a novel source of antigens in RA. Because suppressor tRNAs incorporate an amino acid at a stop codon and cause continued readthrough of the mRNA transcript 28, increased suppressor tRNAs could lead to more abnormal, potentially antigenic proteins.
In the current study, yDRs were the most abundant sRNA in the plasma and were decreased significantly in patients with RA versus control subjects. Y RNAs have significance in autoimmune disease. Y RNAs are incorporated into Ro60 and La ribonucleoprotein complexes 29, 30. These complexes are the targets of antibodies commonly present in the circulation of patients with autoimmune diseases such as Sjogren's syndrome, SLE, immune‐mediated myositis, and others. The 5ʹ stem and 3ʹ polyuridine tail of Y RNAs are the binding sites for Ro60 and La, respectively 31, and it is the protected 5ʹ and 3ʹ ends that were observed as yDRs in the plasma in this study.
The yDRs are increased and released during apoptosis, and yDRs complexed to Ro60 can serve as TLR7 ligands and promote inflammatory cytokine production and further apoptosis 9, 32, 33. During apoptosis, blebs form at the cell surface containing Ro60 and La 34, which is where these antigens are accessible to antibodies 35. In the process, Y RNA bound to Ro60 is degraded to the shorter yDR form and remains bound to Ro60 36. Although yDRs as a class were significantly decreased in RA, reads aligning to a RNY3 gene were most significantly increased in RA. Relatively little is known about specific Y RNA functions, but in murine cells, exposure of Ro60 on the membrane of apoptotic cells is dependent on RNY3 37. This exposure permits antibody binding and engulfment of the apoptotic cells. Subsequently, the yDR‐Ro60 complex can be delivered to the endosomal compartment to be a ligand for TLR7, creating a cycle of inflammation and apoptosis 38.
One of the most abundant yDR sequences that was significantly decreased in patients with RA was a 5ʹ fragment of RNY5, 5ʹ‐AGTTGGTCCGAGTGTTGTGGGTTATTGTTA‐3ʹ along with variations at the 3ʹ end. This particular yDR sequence was abundant in extracellular vesicles from a human myelogenous leukemia cell line and caused dose‐dependent increase in cell death of noncancer cells but not cancer cells 9. This further demonstrates the importance of the yDRs to cell death. Further studies will be important to determine if a lower abundance of plasma yDRs could contribute to the defective apoptosis of autoreactive T and B cells observed in RA.
The abundance of other sRNA classes such as rDRs, snDRs, snoDRs, and lncDRs were not significantly altered in patients with RA compared to controls, but their compositions differed. The functions of these sRNA classes as well as the functions of the individual sRNAs that comprise them, are still largely unknown. Some snoDRs are associated with argonaute proteins, and may target specific mRNAs, which suggest that these may act in the same way as miRNAs 39. The snDRs, like yDRs, may have specific relevance to autoimmune diseases: snRNAs complexed with ribonucleoproteins can serve as antigens for some autoimmune diseases. For example, smith is a protein that complexes with snRNAs, and anti‐smith antibodies are found in patients with SLE. Further work is needed to determine the role of these additional sRNA classes.
This study has some limitations. It is a cross‐sectional study that includes patients with predominately established, treated RA, and endogenous plasma sRNAs may change over the course of disease and with treatment. Also, this study was limited to evaluation of sRNAs approximately 35 nucleotides in length and shorter. Some sRNAs, such as piwi‐interacting RNA, were not assessed because of their rarity in plasma 40.
A limitation of sRNA sequencing is that it provides data on the proportion of sRNA classes relative to other sRNAs within an individual sample rather than providing absolute quantification. Additionally, sRNA library preparation kits can have certain biases to enable amplification of particular sRNAs over others 41, 42. Moreover, some kits are designed specifically to enhance sequencing of miRNAs and deplete ribosomal RNAs and Y RNAs from the sample. Thus, the library preparation method should be taken under consideration when comparing different studies. Similarly, we studied plasma rather than serum, which can affect the sRNA composition and should be considered when comparing studies 43. When examining individual sRNAs as potential biomarkers, it is ideal to validate sequencing findings with another method. In the same sequencing data used for the present study, we have previously performed quantitative polymerase chain reaction (qPCR) validation of the top 15 canonical miRNAs that were altered in patients with RA versus control subjects and also validated the qPCR findings in an external cohort in a study that sought to define an miRNA panel that differentiates between patients with RA and control subjects 2. It would not be feasible to separately validate whole classes of sRNAs with qPCR because we have identified thousands of sRNAs; moreover, methods other than sequencing cannot readily distinguish between isomiRs or other sRNAs with a very limited sequence variability.
In conclusion, endogenous plasma sRNAs are altered in patients with RA compared to control subjects. Shifts in the proportion of several sRNA classes as well as the composition of sRNAs within these classes were altered in patients with RA. A microRNA, miR‐22‐3p, and variants of the canonical sequence were most strongly associated with disease activity. However, microRNAs as a class were less closely associated with disease activity as compared with the ratio of isomiRs to total miRNAs and other sRNAs, such as tDRs and miscRNAs. These novel endogenous sRNA classes are a rich area of future research to define the mechanistic and prognostic effects of differential sRNA expression in RA.
Author Contributions
All authors were involved in drafting the manuscript or revision it critically for intellectual content, and all authors approved the final version to be published.
Study conception and design
Ormseth, Stein, Sheng.
Acquisition of data
Ormseth, Solus, Oeser, Stein.
Analysis and interpretation of data
Ormseth, Sheng, Ye, Song, Wu, Guo, Oeser, Allen, Vickers, Stein.
Supporting information
Supplementary Table 1. miRNA counts in RA vs Control subjects
Supplementary Table 2. miRNA sequence counts in RA vs Control subjects
Supplementary Table 3. isomiR counts in RA vs Control subjects
Supplementary Table 4. YRNA counts in RA vs Control subjects
Supplementary Table 5. yDR sequence counts in RA vs Control subjects
Supplementary Table 6. tRNA counts based on coding amino acid in RA vs Control subjects
Supplementary Table 7. tRNA counts based on isoacceptor in RA vs Control subjects
Supplementary Table 8. tDR sequence counts in RA vs Control subjects
Supplementary Table 9. rRNA counts in RA vs Control subjects
Supplementary Table 10. rDR sequence counts in RA vs Control subjects
Supplementary Table 11. snRNA counts in RA vs Control subjects
Supplementary Table 12. snDR sequence counts in RA vs Control subjects
Supplementary Table 13. snoRNA counts in RA vs Control subjects
Supplementary Table 14. snoDR counts in RA vs Control subjects
Supplementary Table 15. Other RNA counts in RA vs control subjects
Supplementary Table 16. Other sRNA sequence counts in RA vs Control subjects
Supplementary Table 17. Association between sRNAs which are altered in RA vs control subjects and DAS28 score
Supplementary Figure 1. IsomiR and non‐templated addition nomenclature. miRNAs designated as miR‐xx_+_0 have the typical 5’ nucleotide coordinate per miRbase.org (the canonical miRNA sequence). When designated as miR‐xx_+_1, the miRNA starts at the second 5’ nucleotide coordinate of the canonical miRNA sequence. Non‐templated additions (NTA) to the 3’ miRNA end are designated after the isomiR nomenclature for the miRNA.
Supplementary Figure 2 A and B. Volcano plots of differential expression of miRNAs (A) and individual miRNA sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual miRNA or sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 3. Proportion of plasma isomiRs to total miRNAs is not significantly altered in patients with RA compared to control subjects. Error bars indicate geometric mean and 95% confidence intervals.
Supplementary Figure 4. Proportion of plasma miR‐30e‐5p_+_1 to total miR‐30e‐5p is increased significantly in patients with RA compared to control subjects. Error bars indicate geometric mean and 95% confidence intervals.
Supplementary Figure 5. Plasma yDR position across select YRNA transcripts.
Supplementary Figure 6 A and B. Volcano plots of differential expression of YRNA derived fragments based on alignment to a YRNA (A) and individual yDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual YRNA or yDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 7. tDR position across tRNA transcript among select isoacceptors.
Supplementary Figure 8 A, B and C. Volcano plots of differential expression of tRNA derived fragments and halves (tDRs) counts aligning to tRNAs based on amino acid (A), tDR counts aligning to tRNAs based on issoacceptor (B), and individual tDR sequence counts (C). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual amino acid encoding tRNA, tRNA isoaccptor, or tDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 9 A and B. Volcano plots of differential expression of sequencing counts aligning to rRNAs (A) and individual rDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual rRNA or rDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 10 A and B. Volcano plots of differential expression of sequencing counts aligning to snRNAs (A) and individual snDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to individual snRNA or snDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 11 A and B. Volcano plots of differential expression of sequencing counts aligning to snoRNAs (A) and individual snoDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual snoRNA or snoDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 12 A and B. Volcano plots of differential expression of sequencing counts aligning to other RNAs (e.g. lincRNAs and miscRNAs, which also include vaultRNAs) (A) and individual sRNA sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual RNA or sRNA sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Funding
Veterans Health Administration CDA IK2CX001269, Arthritis Foundation Delivering on Discovery grant, Alpha Omicron Pi, NIH Grants: P60AR056116, P01HL116263, and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
No potential conflicts of interest relevant to this article were reported.
References
- 1. Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM. Utility of select plasma microRNA for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. J Rheumatol 2015;42:1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ormseth MJ, Solus JF, Sheng Q, Ye F, Wu Q, Guo Y, et al. Development and validation of a microRNA panel to differentiate between patients with rheumatoid arthritis and control subjects. J Rheumatol 2019. 10.3899/jrheum.181029. E‐pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep 2017;7:44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008;455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, et al. tRNA‐derived microRNA modulates proliferation and the DNA damage response and is down‐regulated in B cell lymphoma. Proc Natl Acad Sci USA 2013;110:1404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, et al. A transfer‐RNA‐derived small RNA regulates ribosome biogenesis. Nature 2017;552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin‐induced tRNA fragments inhibit translation initiation. Mol Cell 2011;43:613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kabeerdoss J, Sandhya P, Danda D. Y RNA derived small RNAs in Sjögren's syndrome: candidate biomarkers? [Review]. Int J Rheum Dis 2017;20:1763–6. [DOI] [PubMed] [Google Scholar]
- 9. Chakrabortty SK, Prakash A, Nechooshtan G, Hearn S, Gingeras TR. Extracellular vesicle‐mediated transfer of processed and functional RNY5 RNA. RNA 2015;21:1966–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DI. Deep sequencing of serum small RNAs identifies patterns of 5’ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer 2014;6:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Repetto E, Lichtenstein L, Hizir Z, Tekaya N, Benahmed M, Ruidavets JB, et al. RNY‐derived small RNAs as a signature of coronary artery disease. BMC Med 2015;13:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo Y, Strickland SA, Mohan S, Li S, Bosompem A, Vickers KC, et al. MicroRNAs and tRNA‐derived fragments predict the transformation of myelodysplastic syndromes to acute myeloid leukemia. Leuk Lymphoma 2017;58:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunaeva M, Blom J, Thurlings R, Pruijn GJ. Circulating serum miR‐223‐3p and miR‐16‐5p as possible biomarkers of early rheumatoid arthritis. Clin Exp Immunol 2018;193:376–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chung CP, Oeser A, Raggi P, Gebretsadik T, Shintani AK, Sokka T, et al. Increased coronary‐artery atherosclerosis in rheumatoid arthritis: relationship to disease duration and cardiovascular risk factors. Arthritis Rheum 2005;52:3045–53. [DOI] [PubMed] [Google Scholar]
- 15. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. [DOI] [PubMed] [Google Scholar]
- 16. Prevoo ML, van ‘t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty‐eight‐joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum 1995;38:44–8. [DOI] [PubMed] [Google Scholar]
- 17. Pincus T, Summey JA, Soraci SA Jr, Wallston KA, Hummon NP. Assessment of patient satisfaction in activities of daily living using a modified Stanford Health Assessment Questionnaire. Arthritis Rheum 1983;26:1346–53. [DOI] [PubMed] [Google Scholar]
- 18. Allen RM, Zhao S, Ramirez Solano MA, Zhu W, Michell DL, Wang Y, et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J Extracell Vesicles 2018;7:1506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones MR, Quinton LJ, Blahna MT, Neilson JR, Fu S, Ivanov AR, et al. Zcchc11‐dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol 2009;11:1157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Churov AV, Oleinik EK, Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev 2015;14:1029–37. [DOI] [PubMed] [Google Scholar]
- 23. Wang S, Yuan M, Song L, Zhang X, Geng Q, Zhang H, et al. Expression of Dicer in rheumatoid arthritis is associated with disease activity and balances the production of TNF‐α. Mol Med Rep 2017;16:1590–5. [DOI] [PubMed] [Google Scholar]
- 24. Ouboussad L, Hunt L, Hensor EM, Nam JL, Barnes NA, Emery P, et al. Profiling microRNAs in individuals at risk of progression to rheumatoid arthritis. Arthritis Res Ther 2017;19:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S, Xu Z, Sheng J. tRNA‐derived small RNA: a novel regulatory small non‐coding RNA. Genes (Basel) 2018;9:E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress‐induced translational repression. J Cell Biol 2009;185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sobala A, Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013;10:553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 2001;29:4767–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol 1981;1:1138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruijn GJ, Wingens PA, Peters SL, Thijssen JP, van Venrooij WJ. Ro RNP associated Y RNAS are highly conserved among mammals. Biochim Biophys Acta 1993;1216:395–401. [DOI] [PubMed] [Google Scholar]
- 31. Kowalski MP, Krude T. Functional roles of non‐coding Y RNAs. Int J Biochem Cell Biol 2015;66:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicolas FE, Hall AE, Csorba T, Turnbull C, Dalmay T. Biogenesis of Y RNA‐derived small RNAs is independent of the microRNA pathway. FEBS Lett 2012;586:1226–30. [DOI] [PubMed] [Google Scholar]
- 33. Hizir Z, Bottini S, Grandjean V, Trabucchi M, Repetto E. RNY (YRNA)‐derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis 2017;8:e2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Casciola‐Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med 1994;179:1317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miranda‐Carús ME, Askanase AD, Clancy RM, Di Donato F, Chou TM, Libera MR, et al. Anti‐SSA/Ro and anti‐SSB/La autoantibodies bind the surface of apoptotic fetal cardiocytes and promote secretion of TNF‐α by macrophages. J Immunol 2000;165:5345–51. [DOI] [PubMed] [Google Scholar]
- 36. Rutjes SA, van der Heijden A, Utz PJ, van Venrooij WJ, Pruijn GJ. Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J Biol Chem 1999;274:24799–807. [DOI] [PubMed] [Google Scholar]
- 37. Reed JH, Sim S, Wolin SL, Clancy RM, Buyon JP. Ro60 requires Y3 RNA for cell surface exposure and inflammation associated with cardiac manifestations of neonatal lupus. J Immunol 2013;191:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reed JH, Gordon TP. Autoimmunity: Ro60‐associated RNA takes its toll on disease pathogenesis. Nat Rev Rheumatol 2016;12:136–8. [DOI] [PubMed] [Google Scholar]
- 39. Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA‐like functions. Mol Cell 2008;32:519–28. [DOI] [PubMed] [Google Scholar]
- 40. Godoy PM, Bhakta NR, Barczak AJ, Cakmak H, Fisher S, MacKenzie TC, et al. Large differences in small RNA composition between human biofluids. Cell Rep 2018;25:1346–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dard‐Dascot C, Naquin D, d'Aubenton‐Carafa Y, Alix K, Thermes C, van Dijk E. Systematic comparison of small RNA library preparation protocols for next‐generation sequencing. BMC Genomics 2018;19:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeri A, Courtright A, Danielson K, Hutchins E, Alsop E, Carlson E, et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics 2018;19:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the microRNA spectrum between serum and plasma. PloS One 2012;7:e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. miRNA counts in RA vs Control subjects
Supplementary Table 2. miRNA sequence counts in RA vs Control subjects
Supplementary Table 3. isomiR counts in RA vs Control subjects
Supplementary Table 4. YRNA counts in RA vs Control subjects
Supplementary Table 5. yDR sequence counts in RA vs Control subjects
Supplementary Table 6. tRNA counts based on coding amino acid in RA vs Control subjects
Supplementary Table 7. tRNA counts based on isoacceptor in RA vs Control subjects
Supplementary Table 8. tDR sequence counts in RA vs Control subjects
Supplementary Table 9. rRNA counts in RA vs Control subjects
Supplementary Table 10. rDR sequence counts in RA vs Control subjects
Supplementary Table 11. snRNA counts in RA vs Control subjects
Supplementary Table 12. snDR sequence counts in RA vs Control subjects
Supplementary Table 13. snoRNA counts in RA vs Control subjects
Supplementary Table 14. snoDR counts in RA vs Control subjects
Supplementary Table 15. Other RNA counts in RA vs control subjects
Supplementary Table 16. Other sRNA sequence counts in RA vs Control subjects
Supplementary Table 17. Association between sRNAs which are altered in RA vs control subjects and DAS28 score
Supplementary Figure 1. IsomiR and non‐templated addition nomenclature. miRNAs designated as miR‐xx_+_0 have the typical 5’ nucleotide coordinate per miRbase.org (the canonical miRNA sequence). When designated as miR‐xx_+_1, the miRNA starts at the second 5’ nucleotide coordinate of the canonical miRNA sequence. Non‐templated additions (NTA) to the 3’ miRNA end are designated after the isomiR nomenclature for the miRNA.
Supplementary Figure 2 A and B. Volcano plots of differential expression of miRNAs (A) and individual miRNA sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual miRNA or sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 3. Proportion of plasma isomiRs to total miRNAs is not significantly altered in patients with RA compared to control subjects. Error bars indicate geometric mean and 95% confidence intervals.
Supplementary Figure 4. Proportion of plasma miR‐30e‐5p_+_1 to total miR‐30e‐5p is increased significantly in patients with RA compared to control subjects. Error bars indicate geometric mean and 95% confidence intervals.
Supplementary Figure 5. Plasma yDR position across select YRNA transcripts.
Supplementary Figure 6 A and B. Volcano plots of differential expression of YRNA derived fragments based on alignment to a YRNA (A) and individual yDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual YRNA or yDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 7. tDR position across tRNA transcript among select isoacceptors.
Supplementary Figure 8 A, B and C. Volcano plots of differential expression of tRNA derived fragments and halves (tDRs) counts aligning to tRNAs based on amino acid (A), tDR counts aligning to tRNAs based on issoacceptor (B), and individual tDR sequence counts (C). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual amino acid encoding tRNA, tRNA isoaccptor, or tDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 9 A and B. Volcano plots of differential expression of sequencing counts aligning to rRNAs (A) and individual rDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual rRNA or rDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 10 A and B. Volcano plots of differential expression of sequencing counts aligning to snRNAs (A) and individual snDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to individual snRNA or snDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 11 A and B. Volcano plots of differential expression of sequencing counts aligning to snoRNAs (A) and individual snoDR sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual snoRNA or snoDR sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.
Supplementary Figure 12 A and B. Volcano plots of differential expression of sequencing counts aligning to other RNAs (e.g. lincRNAs and miscRNAs, which also include vaultRNAs) (A) and individual sRNA sequence counts (B). DESeq2 analysis comparing RA and controls was adjusted for age, race, sex, batch, and multiple comparisons. Each dot represents counts aligning to an individual RNA or sRNA sequence. Larger dots represent greater abundance. Red indicates increased in RA and blue indicates increased in control subjects.


