Abstract
Multi-tasking is the pivotal feature in the next generation chemo- or bio-analyses. However, simultaneous analyses rarely exceed over three different tasks, which is ascribed to limited space to accommodate analyzing units and compromised signal-to-noise (S/N) level as the number of tasks increases. Here, by leveraging superior S/N of single-molecule techniques, we analyzed five microRNA biomarkers by spatially encoding miRNA recognition units with nanometers resolution in a DNA template while decoding the analyte binding temporally in seconds. The hairpin stem is interspersed by internal loops to encode recognition units for miRNA. By mechanical unfolding of the hairpin, individual internal loops are sequentially interrogated for the binding of each miRNA. Using this so-called topochemical spatiotemporal analysis, we were able to achieve sub-picomolar detection limits of miRNAs. We anticipate this new single-molecule topochemical analysis can massively analyze single-molecule targets.
Graphical Abstract
INTRODUCTION
Rapid development of internet of things (IOT)1 calls for analytical devices with multiplexing capabilities to collect and analyze electrical, chemical, or biological signals from targets2. In line with this growing demand, next generation chemo- and bio-analyses require the capability to process large-scale of targets. For example, in the emerging lab-on-a-molecule field, multiple chemicals or ions are analyzed on a molecular template3–5. However, constrained space in a template sets an upper limit of three analyzing units5. In addition, as the number of analytes grows, ensemble signals become crowded and indistinguishable from each other due to signal-to-noise (S/N) issues. Compared to ensemble approaches, single-molecule methods provide superior S/N to analyze samples with low concentrations.
In single-molecule sensing, the ultimate mass detection limit of individual molecules can be reached. However, multiplex analyses of different analytes are often limited in single-molecule methods using AFM6–9 or optical tweezers10–12 technique, in which molecules are processed one at a time. Parallel processing of many single-molecule templates can be achieved by magnetic tweezers13–15 or fluorescence microscopy16–19. In these methods, encoding of different recognition units and decoding of the corresponding binding events become challenging. To address these coding problems, substrate surfaces in magnetic tweezers or fluorescence microscopies can be patterned so that each region hosts only one particular analyte. However, involved steps are necessary for surface patterning, which also suffers from limited number of different analytes that can be accommodated in a typical pattern with micrometer resolutions (microchips20, 21).
Recently, we exploited an alternative approach to increase the multiplexing capacity in single-molecule analyses22. In this so-called single-molecule mechanochemical sensing method, we expanded the size of a template capable of incorporating multiple sensing units. For example, we used DNA origami nanoassembly as the sensing backbone22. Not only can multiple recognition elements be encoded in the DNA origami, the binding events can also be differentiated (or decoded) by leveraging the 3D configuration of the origami structure. Since such coding strategy utilizes the space of the sensing template, it has limited multiplexing capability as the size of the template becomes a bottleneck when the number of analytes increases.
To achieve large-scale multiplexing tasks, highly efficient coding approaches are therefore critical. As discussed above, current strategies suffer from limited space that can be exploited for both encoding and decoding of different analytes. We rationalize that by performing encoding/decoding in both space and time, unlimited multiplex tasks can be achieved. To demonstrate this goal, we incorporated many recognition units with ~10-nm spacing in a DNA hairpin that is interspersed with internal loops23, 24. During mechanical unzipping of the hairpin, each internal loop induces a transient drop in the unzipping force, partitioning the unzipping process into discrete steps. Due to the sequential unzipping nature of the hairpin that propagates from the base of the stem to the loop, the resultant sawtooth unzipping force pattern allows us to differentiate individual internal loops that encode specific recognition units for analytes. Decoding is achieved by varied size or different stability of the fully-addressed recognition units during their mechanical unfolding, which reveals the binding status of each analyte. Since both encoding and decoding exploit topological arrangement of recognition units, we named our approach as single-molecule topochemical analysis, a term closely related to the topochemistry, which investigates localized chemical reactions under the influence of the topology of the reaction environment. As a proof of concept, we demonstrated that such a spatiotemporal coding scheme was able to specifically detect five different miRNAs with detection limits on the order of 100 fM in buffer and 1 pM in 10 times diluted serum within 25 minutes. Since any number of internal loops can be prepared with a fully addressable fashion based on the location and sequential arrangement of internal loops, such a spatiotemporal coding system can readily characterize massively different analyte molecules.
EXPERIMENTAL SECTION
Materials and Reagents.
All DNA oligomers were purchased from Integrated DNA Technologies (www.idtdna.com) and purified with denaturing polyacrylamide gel electrophoresis (PAGE). HPLC purified miRNA 151 and miRNA let 7a were purchased from Integrated DNA Technologies (www.idtdna.com). The miRNA 369, miRNA 21 and miRNA 222 were synthesized using MEGAscript™ T7 Transcription Kit (www.thermofisher.com). All chemicals including Tris-HCl, KCl, EDTA, and DEPC were purchased from either Fisher Scientific or Sigma with purities > 99.0 %. Unless specified otherwise, all enzymes were obtained from New England Biolabs (www.neb.com). The streptavidin or anti-digoxigenin antibody coated polystyrene beads for laser tweezers experiments were purchased from Spherotech (Lake Forest, Illinois, USA). Human serum and bovine serum albumin (BSA, > 98.0%) were purchased from Germini (West Sacramento, CA) and Amresco (Solon, Ohio, USA), respectively.
Synthesis of the DNA Constructs Used in Single-Molecule Topochemical Experiments.
The DNA construct for single-molecule topochemical sensing of nucleic acid fragments (DNA/miRNA) (Table S1) was prepared by sandwiching a DNA hairpin between 2028 and 2690 bp DNA handles. The DNA hairpin contains multiple internal loops and stems as analyte recognition and signal reporting units for multiplex topochemical sensing of miRNAs (Figure S1). The 2690 bp dsDNA handle was synthesized by SacI (NEB) and EagI (NEB) digestions of a pEGFP plasmid (Clontech, Mountain View, CA). The digested 2690 bp handle was gel purified using a kit (Midsci, St. Louis, MO). The handle was subsequently labeled at the 3′ end by digoxigenin (Dig) using 18 μM Dig-dUTP (Roche, Indianapolis, IN) and terminal transferase (Fermentas, Glen Burnie, MD). The biotin labeled 2028 bp dsDNA handle was prepared by PCR amplification of pBR322 plasmid (New England Biolab, NEB) using a 5’ biotinylated reverse primer (IDT, Coralville, IA). The PCR product was purified with QIAquick PCR purification kit (Qiagen, Germantown, MD) and subsequently digested with XbaI restriction enzyme (NEB). The digested 2028 bp handle was gel purified using a QIAquick gel extraction kit (Qiagen, Germantown, MD).
Different DNA constructs were synthesized by stepwise ligation of different DNA oligomers (Table S2) with two long dsDNA handles (2028 bp and 2690 bp) using T4 DNA ligase (NEB) (see the schemes in Figure S1). In general, to synthesize the DNA construct 1 (Figure S1A), an oligonucleotide 5´ CTAG TG CAT TAG GAA GCA GCC CAG TAG TAG GAA AAA AAA GAT CG AAC TAT ACA AC C TAC TAC CTC A 3´ (Oligo-1) which contained 11-nt toehold to bind with miRNA let 7a (Table S1) and a part of the hairpin stem (11 bp, underlined) was annealed with an oligonucleotide 5´ TTT TTT TTC CTA CTA CTG GGC TGC TTC CTA ATG CA 3´ (Oligo-2) at 97 °C for 5 min and slowly cooled to room temperature for 2 hours. This fragment was ligated with the 2028 bp DNA handle by T4 DNA ligase (NEB) and gel purified using a QIAquick gel extraction kit (Qiagen, Germantown, MD). On the other side of the DNA construct, oligonucleotides 5´ GGAC T GAG GTA GTA G CA ACA TAT CAA GC TAG GCC AGC AAG ACG TAG CCC AGC GCG TC 3´ (Oligo-3) containing another part of the hairpin stem (15 bp, underlined) was annealed with oligonucleotides, 5´ GGCC GAC GCG CTG GGC TAC GTC TTG CTG GC 3´ (Oligo-4). This fragment was ligated with the 2690 bp handle and gel purified. The final DNA construct was synthesized using T4 DNA ligase (NEB) through three-piece ligation of the 2028 and 2690 bp DNA handles and an ssDNA fragment, 5´ GTC CGG ACC CTG TTTT CAG GGT CC 3´(Oligo-5), which contained a tetra thymine loop with underlined regions representing the complementary regions of the hairpin stem. To prepare DNA constructs 2, 3 and 4 (Figure S1B–D), the 2028-bp and 2690-bp handles were synthesized using a similar strategy described above by employing oligonucleotides listed in Table S2. The final DNA constructs 2, 3 and 4 were ligated after self-assembly of appropriate DNA oligomers listed in Table S2 according to the schemes shown in Figure S1B–D. The DNA construct for large-scale target analyses (Figure 6) was synthesized according to the scheme described in Figure S10.
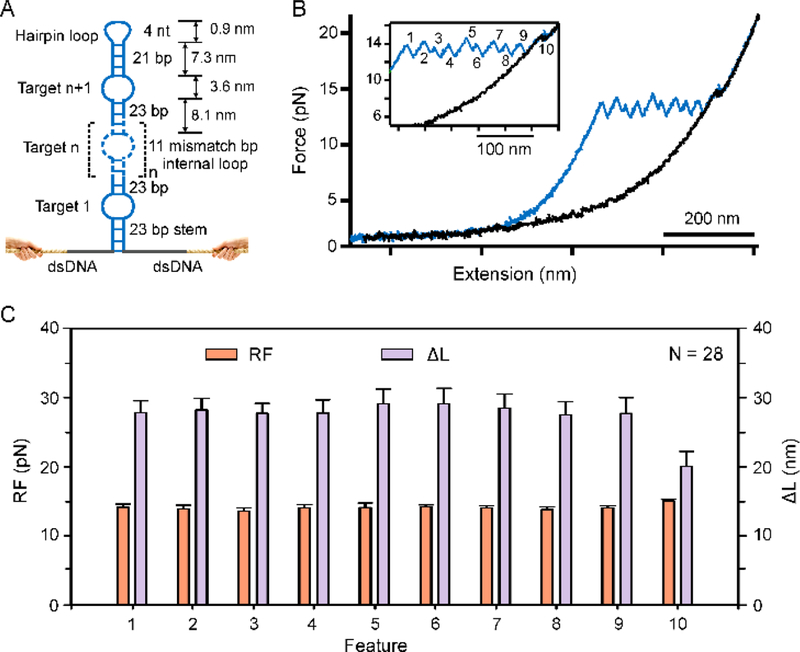
Figure 6.
Single-molecule probe capable of massive multiplex evaluation of targets. (A) Schematic of the single-molecule topochemical probe consisting of n recognition units. (B) Representative F-X curves in 10 mM Tris pH 7.4 supplemented with 100 mM KCl showed sawtooth unfolding features correspond to ten recognition units. (C) The RF and ΔL diagrams [Mean ± SD, N = 28]. The observed ΔL for different unfolding features is close to the theoretical ΔL as calculated by equation S1(see SI Material and Methods).
Preparation of Four-Channel Microfluidic Chambers.
The single-molecule mechanochemical studies were performed in a four-channel microfluidic chamber in laser tweezers instrument. CorelDraw program (Corel Corporation, Ottawa, Canada) was used to prepare microfluidic patterns (Figure S2) and imprinted into the Para-film (BEMIS, Neenah, WI) on a glass cover slip by a laser cutter (VL-200, Universal Laser Systems, Scottsdale, AZ). The imprinted para-film was thermally sealed at 85 °C between two #1 VWR cover glass slips. Two microcapillary tubes (ID 25 μm, OD 100 μm, King Precision Glass, Inc. California) were used to make an internal conduit between middle channels and the top and bottom channels to transport digoxigenin-antibody− and streptavidin− coated beads into the buffer and target channels, respectively. The single-molecule topochemical experiments were performed in the middle of the buffer and the target channels to avoid the variation of flow rates from the edge effects. The distance between the buffer and the target channels through the conduit was about 500 μm (see Figure S2).
Single-Molecule Topochemical Experiments.
The single-molecule topochemical experiments were performed in the four-channel microfluidic chamber (Figure S2) in laser tweezers instrument. The detailed description of the laser tweezers instrument and methods used for the single-molecule experiments have been reported elsewhere25. Briefly, to perform single-molecule experiment, diluted DNA construct (~1 ng) was incubated with 1 μL of 0.1 % solution of digoxigenin(Dig)-antibody coated polystyrene beads (diameter: 2.10 μm) for about 30 minutes at room temperature (25ᵒC), which immobilized the Dig-labelled DNA construct on the bead surface through Dig-anti-Dig complex formation. The incubated sample was further diluted to 1 mL in a 10 mM Tris buffer (pH 7.4, supplemented with 100 mM KCl). Streptavidin-coated polystyrene beads (1 μL, diameter: 1.87μm) were also dispersed into the same buffer (1 mL) and injected into the microfluidic chamber (Figure S2). A 10 mM Tris buffer (pH 7.4) containing 100 mM KCl, 1× BSA, and RNase inhibitors (Murine) (www.neb.com) without and with target miRNA was flowed in the top (buffer) and bottom (target) channels, respectively. The streptavidin-coated bead was trapped in the target channel and moved to the buffer channel where the antibody-coated bead was trapped by another laser beam. The DNA construct initially on the surface of the antibody-coated bead was tethered between the two types of the beads in the buffer channel by escorting one of the beads closer to another using a steerable mirror (Madcity Labs Inc., Madison, WI). In the ramping-force detection mode (F-X mode), the antibody–coated bead was moved away from the streptavidin-coated bead with a loading speed of ~5.5 pN/s by using the steerable mirror. The hairpin structure was unfolded when the tension inside the tether was gradually increased. The unfolding of the hairpin structure was depicted by a sudden change in the end-to-end distance during the process. Single-molecule tether was confirmed by a DNA overstretching plateau at 65 pN or a single-step breakage event in the force-extension (F-X) curves. The F-X curve for each tether was recorded in a Labview 8 program (National Instruments Corp., Austin, TX), and data treatment was performed using Matlab (The MathWorks, Natick, MA) and Igor (WaveMetrics, OR, USA) programs. The unfolding force was measured directly from the F-X curves while the change-in-contour-length (ΔL) due to the unfolding was calculated by the two data points flanking a rupture event using an extensible worm-like chain (WLC) model (Equation 1)26, 27,
| (1) |
where Δx is the change in extension between the data points of the stretching and relaxing curves at the same force (F), kB is the Boltzmann constant, T is absolute temperature, P is the persistent length (50.8 ± 1.5 nm)28, F is the force, and S is the elastic stretch modulus (1243 ± 63 pN)28.
RESULTS AND DISCUSSION
Strategy of Single-Molecule Mechanochemical Sensing for Nucleic Acid Fragment.
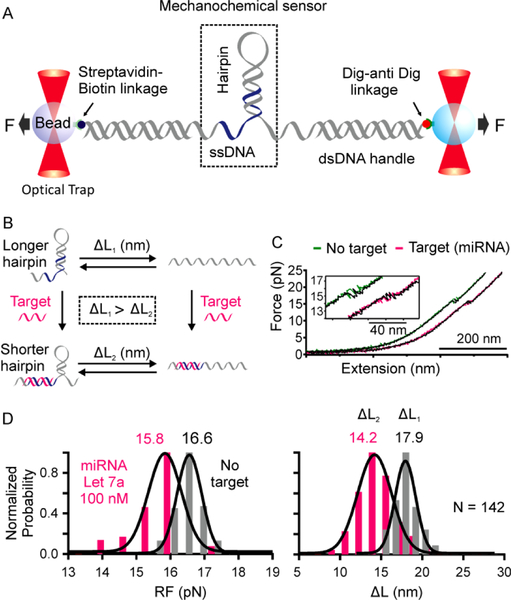
Single-molecule mechanochemical biosensor to detect miRNA in a laser tweezers setup is shown in Figure 1. The sensor consists of a DNA hairpin juxtaposed by an ssDNA segment. The sensing probe is sandwiched between two long dsDNA handles (Figures 1A and S1A), which are tethered to two optically trapped beads via affinity linkages for mechanical unfolding and refolding of the single-molecule sensor in laser tweezers. The ssDNA serves as a toehold29, 30 recognition site whereas the DNA hairpin functions as both the receptor and signal reporter for the binding of complementary miRNA targets (Figure 1A). Target binding to the ssDNA toehold region leads to partial unfolding of the hairpin, which causes a smaller change-in-contour-length (ΔL) or decreased unfolding force compared to the control DNA construct without targets (Figure 1B, see Figure S3 for ΔL comparison).
Figure 1.
One-tiered mechanochemical analysis of single miRNA molecule in laser tweezers. (A) A single-molecule mechanochemical sensing construct consists of a DNA hairpin flanked by a single-stranded DNA, which is sandwiched between two long dsDNA handles. (B) The miRNA target first binds to the ssDNA toehold region and invades to the DNA hairpin stem (blue). After mechanical unfolding, the target bound DNA hairpin shows smaller change-in-contour-length (ΔL) (bottom right) compared to the free DNA probe (top right, see Figure S3 for detailed calculation). (C) Representative F-X curves of the DNA construct in the Tris buffer without (green) and with (pink) 100 nM miRNA Let 7a. (D) The RF and ΔL histograms of the DNA construct in the Tris buffer without (grey) and with (pink) 100 nM miRNA Let7a.
To characterize the single-molecule mechanochemical sensor, we first performed force-ramping experiments on the sensor in a 10 mM Tris buffer (pH 7.4) with 100 mM KCl at room temperature without and with 100 nM miRNA Let 7a31 (Figure 1C, see Table S1 for the sequence) in a four-channel microfluidic chamber in a laser tweezers instrument (Figure S2). The Force-Extension (F-X) curves clearly showed that the target-bound DNA hairpin was unfolded at lower force with a smaller step size compared to the control DNA hairpin (Figure 1C, inset). To determine the nature of these unfolding transitions, we analyzed the change-in-contour-length (ΔL) and the rupture force (RF) of the hairpin mechanophore with and without target miRNA (Figure 1D and Equation 1 in Experimental Section). The observed ΔL (14.2 ± 0.3 nm) and RF (15.8 ± 0.1 pN) (Figure 1D, pink) for target-bound DNA hairpin were lower compared to those for free DNA hairpin (ΔL:17.9 ± 0.2 nm; RF:16.6 ± 0.1 pN; Figure 1D, Grey). The decrease in the ΔL value was expected by theoretical calculations (see Equation S1 in Supporting Methods and Figure S3) whereas the decrease in RF is due to the shorter stem size of the hairpin32 upon binding of the miRNA target.
It is clear from Figure 1D that in 100 nM miRNA target concentration, the RF and ΔL histograms for target and control have some overlaps. These problems may be solved by optimizing the stem length of the DNA hairpin. For an example, the length of the DNA stem can be made shorter than miRNA sequence so that the latter can effectively invade through the entire DNA stem due to increased thermodynamic stability. This will dissolve the hairpin to generate distinct histogram patterns with and without targets. To determine the limit of detection (LOD) of miRNA target using current sensing template, we compared RF vs ΔL plots of individual F-X traces from the same sensing molecule with and without targets (Figure 2). First, we obtained more than 15 ΔL and RF measurements of a sensor in the buffer channel to calculate average ΔLaverage and RFaverage with standard deviations (σ). The same DNA sensor was then transported to the miRNA channel with specific concentrations to collect 50 F-X traces for about 25 minutes. Binding of the miRNA was depicted by decreased values (3σ) of either ΔL, or RF, or both, with respect to ΔLaverage or RFaverage (Figure 2, yellow region, 99.7% confidence level). Compared to ΔL, the RF was not as sensitive to report the binding of miRNA. After evaluating the ΔL–RF plots at different miRNA concentrations (see 100 and 10 fM in Figure 2 for example), we found Let 7a miRNA had a LOD of 100 fM in 25 minutes (≥ 50% detection probability, n ≥ 4). Further concentration decreases to 10 fM did not yield binding events (Figure 2, bottom panel).
Figure 2.
Rupture force (RF) versus change-in-contour-length (ΔL) plots of the unfolding event in each F-X curve to determine the limit of detection (LOD) of Let 7a miRNA. Green ellipses indicate the data range of 95% confidence level in the buffer channel.
Single-Molecule Mechanochemical Sensing for Multiplexing Tasks.
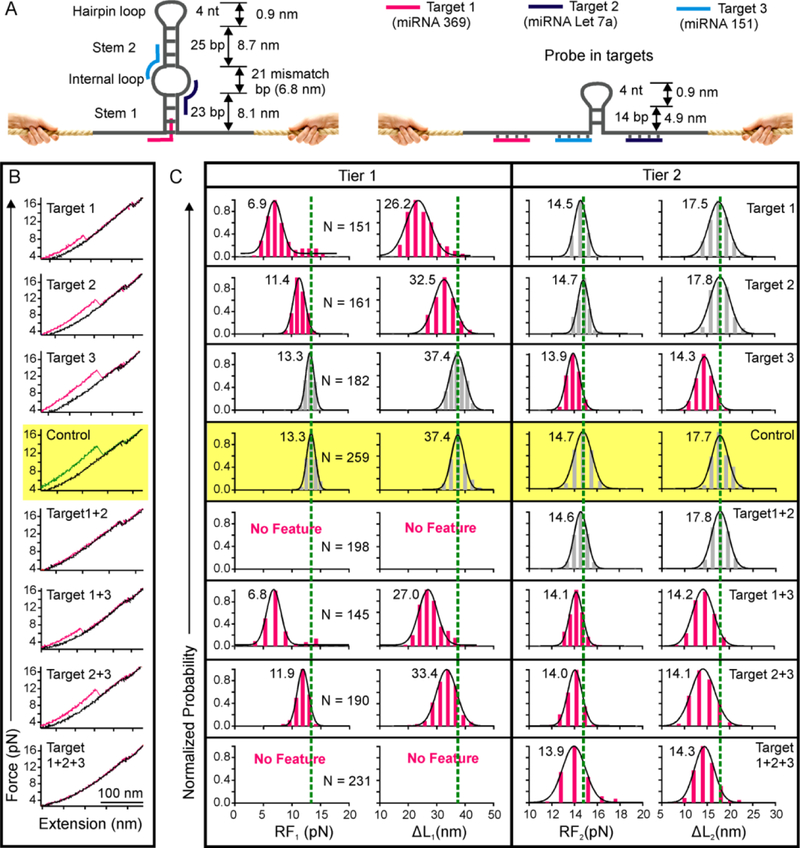
To demonstrate multiplex tasking of the mechanochemical sensor, we incorporated a symmetrical internal loop in the DNA hairpin probe (Figure 3A). The internal loop contains 21 mismatch base pairs, which separate the hairpin into two stems of 23 (stem 1) and 25 (stem 2) bp in length (See Figure S1C for details). The size of the internal loop (21 nt) was determined by the best binding efficiency between the 22-nt ssDNA (DNA Let 7a, see Table S1), a surrogate for miRNA Let 7a (Table S1), and a 11-nt toehold29, 30 region in DNA loops of different size (11, 22, or 33 nt, see Figures S1B and S4).
Figure 3.
Two-tiered topochemical analyses of three miRNAs. (A) The probe consists of a DNA hairpin with a symmetrical internal loop and an adjacent ssDNA fragment serving as toeholds. Target 1 binds with the ssDNA toehold and invades into the stem 1. Targets 2 and 3 bind with the internal loop toeholds and then hybridize to the stems 1 and 2, respectively. Binding of all three targets destabilize the internal loop and both stems, yielding a much smaller hairpin structure (right). (B) Representative F-X curves of the DNA probe with (pink) and without (green) 50 nM miRNA targets in the 10 mM Tris buffer supplemented with 100 mM KCl at pH 7.4. Binding of targets reduces the change-in-contour-length (ΔL) and rupture force (RF) of respective unfolding features. (C) The RF and ΔL histograms with (pink) and without (grey) 50 nM miRNA targets. Dotted green lines are the guides to the eye.
Specific miRNA targets first partially hybridize with the 11-nt toehold either in the flanking ssDNA or the internal loop (Figure 3A).This is followed by full hybridization to the duplex hairpin stem (~11 bp) by displacing one of the complementary strands. As a result, the remaining hairpin becomes weakened, reducing both rupture force (RF) and ΔL (Figure 3A, right). Sensing experiments confirmed these expected patterns. Mechanical unfolding of the free mechanochemical sensor produced two features. While the first feature corresponds to the unfolding of the stem 1 and the internal loop, the second feature is for the unfolding of the stem 2 and the hairpin loop (Figure 3B, middle, yellow background). Binding of either 50 nM target 1 (miRNA 369)33 or target 2 (miRNA Let 7a)31 reduced the RF and ΔL of the first feature only (Figure 3 B&C, Tier 1), whereas binding of 50 nM target 3 (miRNA 151)34 reduced RF and ΔL of the second feature only (Figure 3 B&C, Tier 2).
In presence of all three targets 1, 2, and 3, the first feature disappeared due to the binding of targets 1 & 2 while the second feature had smaller RF and ΔL due to the target 3 binding. During binary target binding, targets 1 & 2 led to disappearance of the first feature whereas the second feature was unaffected. In presence of targets 1 & 3 or 2 & 3, both the first and the second features showed reduced RF and ΔL values (Figure 3B&C, see Tier 1 and Tier 2). These experiments clearly demonstrated that any combination of the three miRNA targets can be differentiated by this multi-tasking single-molecule sensor.
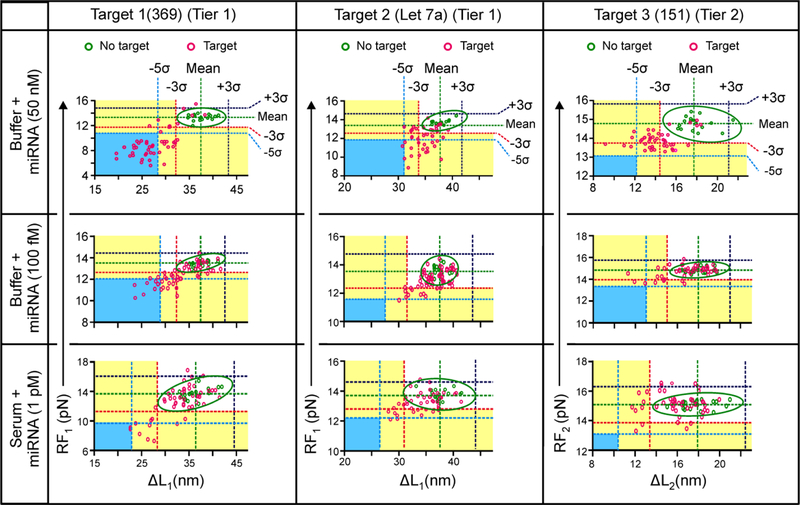
After specific recognition of the three miRNAs (Figure 3) at relatively high concentration (50 nM) using ΔL or RF histograms, next, we proceeded to determine the LOD of each miRNA. At rather low concentrations, most F-X curves did not show binding events, generating a histogram indistinguishable with that of the control without any targets. To address this issue, similar to Figure 2, RF vs ΔL plots of individual F-X curves with higher signal-to-noise ratio were used for LOD determination.
To increase the confidence level, we also evaluated the recognition events at the 5σ levels (99.9% confidence level, see Figure 4). First, we obtained RF vs ΔL plots to identify positive target binding events using 50 nM miRNA targets. For miRNA target 1, most bound data were beyond 5σ of both ΔLaverage and RFaverage from the 1st unfolding feature (Tier 1 detection) whereas they were located beyond 3σ for miRNA target 2 (Figure 4). In contrast, for miRNA target 3, bound features were concentrated beyond 3σ of ΔLaverage for the second feature only (Figure 4, right). We therefore used these criteria as the thresholds in the multi-tiered detection schemes to identify binding events of specific miRNA at low concentration levels (see Figure 4 middle row for 100 fM miRNAs). We found LOD of 100 fM for each of the three miRNAs (miRNA 369, Let 7a, and 151) in the Tris buffer; and 1 pM (Figure 4, third row) in 10 times diluted serum within a time window of 25 minutes (≥ 50% detection probability, n ≥ 4).
Figure 4.
Pattern recognition in the 2-tiered topochemical analyses for miRNAs. Binding of individual targets is recognized in the RF vs ΔL plot of either the 1st (Tier 1) or the 2nd (Tier 2) unfolding feature of individual F-X curves. Green and pink depict without and with miRNA targets, respectively. Ellipses indicate the data range at 95% confidence level without target.
To evaluate the specificity of miRNA target binding, RF vs ΔL was also plotted at the 2nd unfolding feature (Tier 2 detection) for miRNA 369 & Let 7a, as well as at the 1st unfolding feature (Tier 1 detection) for miRNA 151 in buffer and in 10 times diluted serum (Figure S5). We did not observe any target binding event during Tier 2 detection for miRNA 369 & Let 7a or Tier 1 detection for miRNA 151 according to the criteria as discussed above, which indicates that the 2-tiered sensing probe can specifically bind with different miRNAs. The 100 fM LOD was also observed in any target combinations among the three miRNAs (see Figures S6 and S7). It is significant that these LOD’s are already lower than the natural miRNA concentration in human serum35, 36, suggesting our single-molecule sensing can be directly applied for field samples. Based on the methods described in our previous report37, we expect that our topochemical sensor can differentiate single nucleotide polymorphism or variation (SNP or SNV). Indeed, when we evaluated the LOD’s of DNA surrogates of microRNA Let 7a and Let 7c (5′-TGAGGTAGTAGGTTGTATGGTT, underlined sequence depicts the SNV in the Let 7c) 38 (Table S1) using the two tiered sensor shown in Figures 3, we found that the Let7a has an LOD ~ 5 orders of magnitude low than the Let 7c (Figure S8).
To quantify miRNAs using our topochemical approach, we constructed a calibration curve by plotting observation time for the first binding event against known concentrations of miRNA Let 7a. Immediately after the sensing probe was switched from the buffer channel to the target channel within 10s, F-X curves were recorded at 20s intervals. The time for miRNA binding was determined when either RF or ΔL of the hairpin reporter was smaller than (RFaverage-3σ) or (ΔLaverage-3σ) (Figure 4). Using two-tiered topochemical sensing strategy (Figure 3), we obtained a linear range of 100 fM-100 nM for miRNA Let 7a within a time window of ~25 minutes (Figure S9) in 10 mM Tris buffer (pH 7.4) supplemented with 100 mM KCl. From the first binding time of the target, the unknown concentration can be estimated from the calibration curve. Due to the similar binding time at specific concentration for different miRNAs used in this report (Table S3), we anticipate similar quantification ranges exist among different miRNAs.
Spatiotemporal Coding Capable of Large-Scale miRNA Analyses.
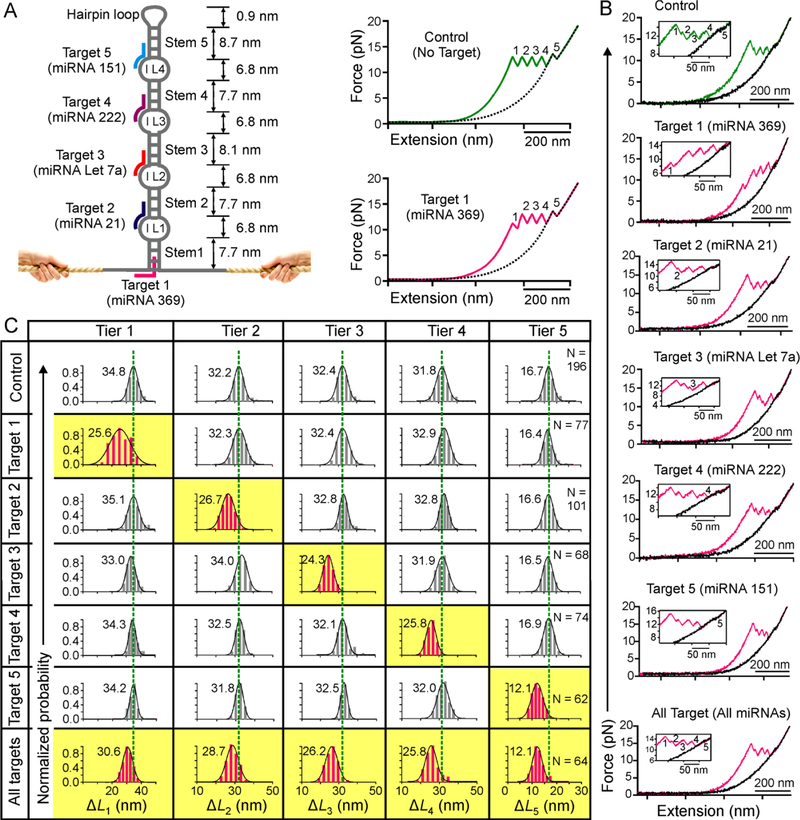
With successful demonstration of multiplex sensing of three miRNA targets, we set out to explore the possibility of large-scale miRNA analyses. We used multiplex sensing of 5 miRNAs as a proof-of-concept. To this end, we expanded the spatiotemporal coding by introducing a total of four internal loops in the hairpin, which separate the hairpin stem into five regions (Figures 5A and S1D). Each internal loop serves as a separate toehold to initiate different miRNA binding. The probe also contains a flanking ssDNA in the hairpin base to serve as an additional toehold (Figure 5A). Without any target, the DNA probe is expected to produce five sawtooth unfolding signals corresponding to the sequential unfolding of five stems that are interrupted by the four internal loops (Figure 5A, top right). When Target 1 (miRNA 369)33 is present, it first binds to the 11-nt ssDNA toehold via complementary base pairing, followed by invading into the stem 1 by 11-bp hybridization. The resultant stem 1 becomes shorter with decreased unfolding force and smaller ΔL (Figure 5A, bottom right). Similarly, for Targets 2–5 (miRNA 21, Let 7a, 222, and 151, respectively)31, 34, 38, 39, bindings start with the 11-nt toeholds in the internal loops 1–4 respectively, which are followed by the 11-bp hybridizations to the corresponding stems 2–5 (Figure 5A, left panel).
Figure 5.
Single-molecule probe capable of large-scale topochemical analyses of miRNAs. (A) Schematic of the hairpin sensor that contains four internal loops (IL1-IL4) and 5 stems. Target 1 (pink) binds with the toehold at the base of the probe and invades into stem 1. Targets 2–4 bind with internal loop toeholds and half-length of the corresponding stems. Without target, the DNA construct produces five well addressed unfolding features (top right). With target 1, both RF and ΔL of the first feature are expected to be smaller (bottom right). Dotted curves depict relaxing F-X traces. Note feature 5 indicates the transition for the stem linked to the hairpin tetraloop. (B) Representative F-X curves of the probe without (green) and with (pink) 50 nM miRNAs in 10 mM Tris buffer (pH 7.4 with 100 mM KCl). (C) Change-in-contour-length (ΔL) histograms of a particular unfolding feature without (grey) and with (pink) 50 nM miRNA targets. Dotted green lines are the guides to the eye.
These predictions were exactly observed when 50 nM of individual miRNA targets were tested in the Tris buffer. While the F-X curve showed five distinctive unfolding features (Figure 5B, green) without targets, binding of target 1 (miRNA 369) reduced both the rupture force (RF) and the size (ΔL) of the stem 1 in the first unfolding feature (Figure 5B&C, pink). Likewise, binding of miRNA 21, miRNA Let 7a, miRNA 222, and miRNA 151 reduced the size (ΔL) of the stems 2, 3, 4, and 5, respectively (Figure 5 B&C). Similar to previous probes, the decrease in the RF was not as significant as ΔL for these miRNA targets. When five miRNAs at 50 nM were tested together, each target was bound with the corresponding internal loop-stem region, producing expected ΔL signal in the 5-tiered analyzing system (Figure 5C, bottom row).We anticipate that using the approach described in Figure 4, we could be able to detect all five miRNAs in fM concentrations.
CONCLUSIONS
As each specific transition event in the sawtooth unfolding pattern exactly corresponds to a particular recognition unit, such a single-molecule template contains probing units whose identities are fully-addressed. Since the interaction of an analyzing unit and the corresponding analyte occurs at a particular location whereas their interaction status is probed according to their localized arrangement with respect to other interactions, we call this single-molecule method as topochemical analysis. This strategy avoids any ambiguity presented in conventional designs in which a signal variation may correspond to more than one analyte. For example, bindings of miRNA 369 and Let 7a cannot be differentiated at low concentrations by the design described in Figure 4. In current strategy, by interrogating characteristic ΔL features for the 1st and the 3rd unfolding events (Figure 5A), bindings of these two miRNAs can be readily determined. Another special feature of this spatiotemporal coding is its high density. Current design allows a spacing of ~10 nm between adjacent analyzing units. This spacing can be further reduced if the targets to be analyzed are small molecules, instead of nucleic acids that require relatively bulky toehold or hybridization regions29, 30. In the case that aptamer probes40, 41 are used for small molecule analytes, ~5 nm/unit coding density is expected to be achieved. For a hairpin made of lambda DNA (48.5 kbp)42 for example, this suggests ~3000 analyzing units can be incorporated for target analyses. Further increase in the unit number can be achieved readily with molecular biology approaches to generate longer templates. It is significant that the spatial resolution in this spatiotemporal coding well exceeds that of super-resolution fluorescence microscopy16–19. Such small spacing between analyzing units is already equivalent to the smallest transistors incorporated in the silicon wafer by the most advanced lithography nowadays43. However, to reach the reading and writing efficiency of the microelectronics, the temporal resolution in the decoding needs to be improved, which can be partially achieved by increasing the pulling speed for the hairpin probe.
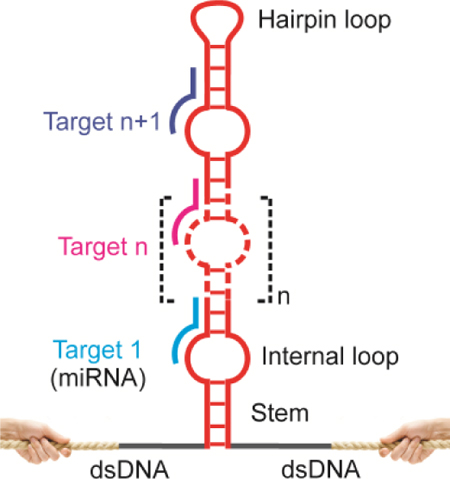
By adopting a coding strategy that exploits both space and time to differentiate analytes, we have successfully demonstrated a single-molecule topochemical platform capable of massive analyses of different single-molecules (single-molecule topochemical analyses). The spatiotemporal coding principle used here is generic. It can be readily extended to analyze massive number of single-molecule targets. For example, by using molecular biology methods, hairpins can be easily prepared with a large set of internal loops each serving as a recognition element (spatial encoding) (see Figure 6 for a DNA construct with 10 recognition elements; see Figure S10 for its sequence and synthesis). The analyte binding is then decoded by mechanical unfolding events that are sequential in time (temporal decoding). When the internal loops are replaced by aptamers, analyses of massive number of small-molecules or macromolecules can be accomplished. The spatiotemporal coding can also be used for massive screening of bioactive chemicals if the internal loops are replaced by peptides or biomolecules other than nucleic acids.
Supplementary Material
ACKNOWLEDGMENT
H.M. thanks financial support from National Science Foundation [CHE-1609514] and National Institutes of Health [NIH 1R01CA236350] (in part).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Synthesis of miRNAs, Calculation of change-in contour length (ΔL) and probe density. Table S1-S3 and Figures S1-S10.
The authors declare no competing financial interest.
REFERENCES
- 1.Atzori L; Iera A; Morabito G, The Internet of Things: A survey. Comput. Netw. 2010, 54 (15), 2787–2805. [Google Scholar]
- 2.Gubbi J; Buyya R; Marusic S; Palaniswami M, Internet of Things (IoT): A vision, architectural elements, and future directions. Future Gener. Comput. Syst. 2013, 29 (7), 1645–1660. [Google Scholar]
- 3.Magri DC; Brown GJ; McClean GD; de Silva AP, Communicating Chemical Congregation: A Molecular AND Logic Gate with Three Chemical Inputs as a “Lab-on-a-Molecule” Prototype. J. Am. Chem.Soc. 2006, 128 (15), 4950–4951. [DOI] [PubMed] [Google Scholar]
- 4.Magri DC; Camilleri Fava M; Mallia CJ, A sodium-enabled ‘Pourbaix sensor’: a three-input AND logic gate as a ‘lab-on-a-molecule’ for monitoring Na+, pH and pE. Chem.Comm. 2014, 50 (8), 1009–1011. [DOI] [PubMed] [Google Scholar]
- 5.Chen K; Shu Q; Schmittel M, Design strategies for lab-on-a-molecule probes and orthogonal sensing. Chem. Soc. Rev. 2015, 44 (1), 136–160. [DOI] [PubMed] [Google Scholar]
- 6.Rief M; Gautel M; Oesterhelt F; Fernandez JM; Gaub HE, Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276 (5315), 1109–12. [DOI] [PubMed] [Google Scholar]
- 7.Oberhauser AF; Marszalek PE; Erickson HP; Fernandez JM, The molecular elasticity of the extracellular matrix protein tenascin. Nature 1998, 393 (6681), 181–5. [DOI] [PubMed] [Google Scholar]
- 8.Florin E; Moy V; Gaub H, Adhesion forces between individual ligand-receptor pairs. Science 1994, 264 (5157), 415–417. [DOI] [PubMed] [Google Scholar]
- 9.Moy V; Florin E; Gaub H, Intermolecular forces and energies between ligands and receptors. Science 1994, 266 (5183), 257–259. [DOI] [PubMed] [Google Scholar]
- 10.Ashkin A; Dziedzic JM; Bjorkholm JE; Chu S, Observation of a single-beam gradient force optical trap for dielectric particles. Opt. Lett. 1986, 11 (5), 288–290. [DOI] [PubMed] [Google Scholar]
- 11.Smith SB; Cui YJ; Bustamante C, Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996, 271 (5250), 795–799. [DOI] [PubMed] [Google Scholar]
- 12.Svoboda K; Schmidt CF; Schnapp BJ; Block SM, Direct observation of kinesin stepping by optical trapping interferometry. Nature 1993, 365, 721. [DOI] [PubMed] [Google Scholar]
- 13.Smith SB; Finzi L; Bustamante C, Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 1992, 258, 1122–1126. [DOI] [PubMed] [Google Scholar]
- 14.Assi F; Jenks R; Yang J; Love C; Prentiss M, Massively parallel adhesion and reactivity measurements using simple and inexpensive magnetic tweezers. Journal of Applied Physics 2002, 92 (9), 5584–5586. [Google Scholar]
- 15.Ribeck N; Saleh OA, Multiplexed single-molecule measurements with magnetic tweezers. Rev Sci Instrum 2008, 79 (9), 2981687. [DOI] [PubMed] [Google Scholar]
- 16.Dickson RM; Cubitt AB; Tsien RY; Moerner WE, On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 1997, 388, 355. [DOI] [PubMed] [Google Scholar]
- 17.Betzig E; Chichester RJ, Single Molecules Observed by Near-Field Scanning Optical Microscopy. Science 1993, 262 (5138), 1422–1425. [DOI] [PubMed] [Google Scholar]
- 18.Hell SW; Wichmann J, Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19 (11), 780–782. [DOI] [PubMed] [Google Scholar]
- 19.Rust MJ; Bates M; Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 2006, 3, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proudnikov D; Timofeev E; Mirzabekov A, Immobilization of DNA in Polyacrylamide Gel for the Manufacture of DNA and DNA–Oligonucleotide Microchips. Anal. Biochem. 1998, 259 (1), 34–41. [DOI] [PubMed] [Google Scholar]
- 21.Arenkov P; Kukhtin A; Gemmell A; Voloshchuk S; Chupeeva V; Mirzabekov A, Protein Microchips: Use for Immunoassay and Enzymatic Reactions. Anal. Biochem. 2000, 278 (2), 123–131. [DOI] [PubMed] [Google Scholar]
- 22.Koirala D; Shrestha P; Emura T; Hidaka K; Mandal S; Endo M; Sugiyama H; Mao H, Single-molecule mechanochemical sensing using DNA origami nanostructures. Angew. Chem. Int. Ed. 2014, 53, 8137–8141. [DOI] [PubMed] [Google Scholar]
- 23.Kahn JD; Yun E; Crothers DM, Detection of localized DNA flexibility. Nature 1994, 368 (6467), 163–166. [DOI] [PubMed] [Google Scholar]
- 24.Prislan I; Lee H-T; Lee C; Marky LA, The Size of the Internal Loop in DNA Hairpins Influences Their Targeting with Partially Complementary Strands. J. Phys. Chem. B 2015, 119 (1), 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao H; Luchette P, An integrated laser-tweezers instrument for microanalysis of individual protein aggregates. Sens. Actuators B 2008, 129, 764–771. [Google Scholar]
- 26.Baumann CG; Smith SB; Bloomfield VA; Bustamante C, Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA 1997, 94, 6185–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Z; Mao H, Non-B DNA structures show diverse conformations and complex transition kinetics comparable to RNA or proteins ― a perspective from mechanical unfolding and refolding experiments. Chem. Rec. 2013, 13, 102–116. [DOI] [PubMed] [Google Scholar]
- 28.Dhakal S; Cui Y; Koirala D; Ghimire C; Kushwaha S; Yu Z; Yangyuoru PM; Mao H, Structural and mechanical properties of individual human telomeric G-quadruplexes in molecularly crowded solutions. Nucleic Acids Res. 2013, 41, 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yurke B; Turberfield AJ; Mills AP; Simmel FC; Neumann JL, A DNA-fuelled molecular machine made of DNA. Nature 2000, 406 (6796), 605–608. [DOI] [PubMed] [Google Scholar]
- 30.Zhang DY; Seelig G, Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3 (2), 103–113. [DOI] [PubMed] [Google Scholar]
- 31.Takamizawa J; Konishi H; Yanagisawa K; Tomida S; Osada H; Endoh H; Harano T; Yatabe Y; Nagino M; Nimura Y; Mitsudomi T; Takahashi T, Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64 (11), 3753–3756. [DOI] [PubMed] [Google Scholar]
- 32.Woodside MT; Behnke-Parks WM; Larizadeh K; Travers K; Herschlag D; Block SM, Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U S A 2006, 103 (16), 6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan S; Tong Y; Steitz JA, Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318 (5858), 1931–1934. [DOI] [PubMed] [Google Scholar]
- 34.Fu H; Tie Y; Xu C; Zhang Z; Zhu J; Shi Y; Jiang H; Sun Z; Zheng X, Identification of human fetal liver miRNAs by a novel method. FEBS Lett 2005, 579 (17), 3849–54. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell PS; Parkin RK; Kroh EM; Fritz BR; Wyman SK; Pogosova-Agadjanyan EL; Peterson A; Noteboom J; O’Briant KC; Allen A; Lin DW; Urban N; Drescher CW; Knudsen BS; Stirewalt DL; Gentleman R; Vessella RL; Nelson PS; Martin DB; Tewari M, Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U S A 2008, 105 (30), 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R; Zhang C; Hu Z; Li G; Wang C; Yang C; Huang D; Chen X; Zhang H; Zhuang R; Deng T; Liu H; Yin J; Wang S; Zen K; Ba Y; Zhang C-Y, A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur. J. Cancer 2011, 47 (5), 784–791. [DOI] [PubMed] [Google Scholar]
- 37.Koirala D; Yu Z; Dhakal S; Mao H, Detection of single nucleotide polymorphism using tension-dependent stochastic behavior of a single-molecule template. J. Am. Chem. Soc. 2011, 133 (26), 9988–9991. [DOI] [PubMed] [Google Scholar]
- 38.Lagos-Quintana M; Rauhut R; Lendeckel W; Tuschl T, Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294 (5543), 853–858. [DOI] [PubMed] [Google Scholar]
- 39.Lewis BP; Shih I.h. ; Jones-Rhoades MW; Bartel DP; Burge CB, Prediction of Mammalian MicroRNA Targets. Cell 2003, 115 (7), 787–798. [DOI] [PubMed] [Google Scholar]
- 40.Lu Y; Liu J, Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers. Curr. Opin. Biotechnol. 2006, 17 (6), 580–588. [DOI] [PubMed] [Google Scholar]
- 41.Dieckmann T; Fujikawa E; Xhao X; Szostak J; Feigon J, Structural Investigations of RNA and DNA aptamers in Solution. J. Cell. Biochem. 1995, 59, 56. [Google Scholar]
- 42.Daniels DL; Schroeder JL; Szybalski W; Sanger F; Coulson AR; Hong GF; Hill DF; Petersen GB; Blattner FR, APPENDIX II Complete Annotated Lambda Sequence. 1983. [Google Scholar]
- 43.Monica Chen; Hsinchu Shen, J., TSMC ramping up 7nm chip production. DIGITIMES 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.