Abstract
Combination therapies consisting of multiple short therapeutic RNAs, such as small interfering RNA (siRNA) and microRNA (miRNA), have enormous potential in cancer treatment as they can precisely silence specific set of oncogenes and target multiple related disease pathways. However, clinical use of siRNA/miRNA combinations is limited by the availability of safe and efficient systemic delivery systems with sufficient tumor penetrating and endosomal escaping capabilities. This study reports on the development of multifunctional tumor-penetrating mesoporous silica nanoparticles (iMSNs) for simultaneous delivery of siRNA (siPlk1) and miRNA (miR-200c), using encapsulation of a photosensitizer indocyanine green (ICG) to facilitate endosomal escape and surface conjugation of iRGD peptide to enable deep tumor penetration. Increased cell uptake of the nanoparticles was observed in both 3D tumor spheroids in vitro and in orthotopic MDA-MB-231 breast tumors in vivo. Using a galectin-8 recruitment assay, we showed that reactive oxygen species generated by ICG upon light irradiation functioned as an endosomolytic stimulus that caused release of the siRNA/miRNA combination from endosomes. Co-delivery of the therapeutic RNAs displayed combined cell killing activity in cancer cells. Systemic intravenous treatment of metastatic breast cancer with the iMSNs loaded with siPlk1 and miR-200c resulted in a significant suppression of the primary tumor growth and in marked reduction of metastasis upon short light irradiation of the primary tumor. This work demonstrates that siRNA-miRNA combination assisted by photodynamic effect and tumor penetrating delivery system may provide a promising approach for metastatic cancer treatment.
Keywords: siRNA-miRNA combination, photodynamic effect, mesoporous silica nanoparticle, iRGD, tumor penetration, metastasis
Graphical Abstract
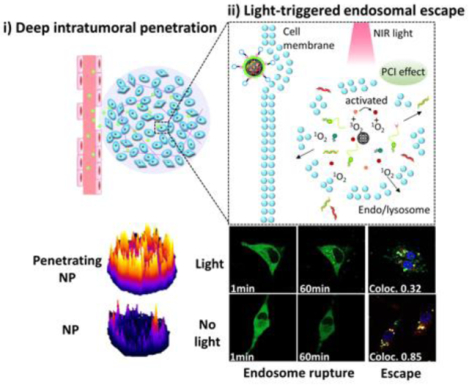
1. INTRODUCTION
Nucleic acids have been investigated as a potential class of therapeutic agents for the treatment of various human diseases, including ongoing clinical trials focused on cancer, liver infections, ocular and retinal disorders, hematologic disorders, and urinary system diseases.1–4 Nucleic acids can selectively silence the expression of cancer-related genes and gene products, alter messenger RNA (mRNA) splicing, express genes, edit the genome, and regulate the pathways involved in the development and progression of cancer. Due to their potential to act against many otherwise un-druggable targets, RNA-based therapeutics including small-interfering RNA (siRNA), microRNA (miRNA), synthetic mRNA, and gene editing technologies (i.e., CRISPR/Cas9) have attracted considerable attention.5–10 Multiple clinical trials of RNA-based agents have already been conducted and achieved significant progress in the last 10 years,11–12 culminating in the 2018 FDA approval of the first siRNA lipid nanoparticle drug Onpattro (Alnylam) to treat polyneuropathy caused by hereditary transthyretin-mediated amyloidosis.13 There are currently hundreds of patients enrolled in clinical trials testing RNA therapeutics to fight against cancer, making this research concept into clinical reality.14
siRNA-based therapeutic approaches introduce a synthetic short double-stranded RNA into the cytoplasm of cells to elicit RNA interference (RNAi), thereby guiding the cleavage of a specific mRNA to inhibit transcription of the target protein.4, 15–16 The specificity of siRNA makes it attractive for personalized cancer treatment and thus more siRNA candidates have entered into clinical trials than any other RNA drugs.17–18 In contrast to siRNA, which are engineered to recognize only one mRNA, miRNAs have naturally evolved to regulate networks of genes by degrading transcripts which share a partially complimentary motif on the 3' untranslated region of the mRNA. While miRNAs can themselves be dysregulated in cancer, miRNAs can also be used to block oncogenic pro-survival gene networks.19–22.
Despite the promise of siRNA and miRNA for cancer therapy, various hurdles need to be confronted before bringing them into clinical use. Due to the similar physicochemical properties, some of these challenges, like poor in vivo stability and off-target effects, are similar for both types of RNA molecules.23 Appropriate delivery systems should protect siRNA and miRNA from premature nuclease degradation and facilitate cellular uptake at target sites.24 Current viral vectors, non-viral vectors (polymers, lipids), and chemically modified RNAs have demonstrated effectiveness to some extent, but they all face different problems such as safety concerns, production costs, and generally poor correlation between in vitro and in vivo efficacy, making their development a significant challenge.25–27
The vast majority of nanosized RNA delivery systems is routed to the endosomal pathway upon cellular uptake from which they have to be liberated to reach the cytoplasm.21 In the past decades, researchers explored various strategies to overcome this barrier by getting across endosomal lipid bilayers, including rationally designing cationic materials to respond to the changes in pH during endosomal processing. These strategies face concerns of in vivo safety, stability and sensitivity of response, to achieve the goal of effective and controllable cytosolic delivery.28–30 Multiple studies showed that photochemical internalization (PCI) can improve cytoplasmic delivery of small RNAs by disrupting endo/lysosomal membranes through light-activated generation of reactive oxygen species (ROS).31–32 Applying the photochemical effect in the design of RNA delivery systems can increase the bioavailability of the therapeutic macromolecules in the tumor microenvironment. Further, potential phototoxicity of the photodynamic effect can be precisely controlled by regulating the exposure conditions and used to achieve a therapeutic advantage in cancer treatment.
Another obstacle for small RNA therapeutics in cancer is restricted vascular access and limited tissue permeability due to altered extracellular matrix, increased collagen content and density, and high interstitial fluid pressure in solid tumors.33 Our previous study demonstrated that tumor-homing and penetrating peptide iRGD, which interacts with αν integrins and neuropilin-1 receptor, can enhance nanoparticle penetration and distribution in orthotopic breast tumors via the recently suggested three-step endocytotic transport pathway.34–35 After iRGD binding to integrins, the peptide exposes RXXK/R motif for neuropilin-1 (NRP-1) receptor, which induces further penetration into tumor area and cells. Integrating the iRGD peptide into an RNA delivery system will likely be beneficial for delivery efficiency and therapeutic performance.
Here, we utilized mesoporous silica nanoparticles (MSNs) stabilized by a surface lipid layer conjugated to the iRGD peptide, as a tumor-penetrating RNA delivery carrier (Scheme 1). A near-infrared (NIR)-responsive photosensitizer, indocyanine green (ICG) was loaded into the MSNs for the local generation of ROS to enhance cytosolic RNA delivery. The large internal surface area and pore volume of MSNs ensure easy adsorption and high loading of various therapeutic agents.36–37 Further, simple cargo-loading and preparation methods are likely to be beneficial for future clinical translation and expanding therapeutic applications. More importantly, MSN are biodegradable and many have been demonstrated safe in animal testing.38–39 As displayed in Scheme 1, we hypothesized that iRGD-modified lipid-coated MSN (iMSN) for co-delivery of siPlk1 and miR-200c mimic would display enhanced tumor penetration and superior anticancer activity in a metastatic breast cancer model. Polo-like kinase 1 (Plk1) is a well-established mitotic regulator of the cell cycle, which is overexpressed in various types of human tumors and suggested to control cancer development and progression.40 Silencing Plk1 expression inhibits cell proliferation and induces apoptosis.41 MiR-200c broadly suppresses cancer development via hindering epithelial-to-mesenchymal transition (EMT), which promotes epithelial cancer cells to acquire the migratory, invasive, and apoptosis-resistant activities of mesenchymal cancer cells. Increased levels of miR-200c can effectively decrease the extent of EMT.42–44 Moreover, recent studies in mouse models revealed the crucial role of the activation of the EMT program for the dissemination of primary tumor cells to the lung and the formation of metastases.45 Combining siPlk1and miR-200c is thus expected to achieve improved therapeutic effect by acting both on the primary tumor and metastatic process by attacking multiple targets and cellular pathways in breast cancer. We hypothesized that the rational combination of siPlk1 and miR-200c is capable of enhancing overall antitumor performance by simultaneously targeting both primary tumor and metastasis.
Scheme 1.
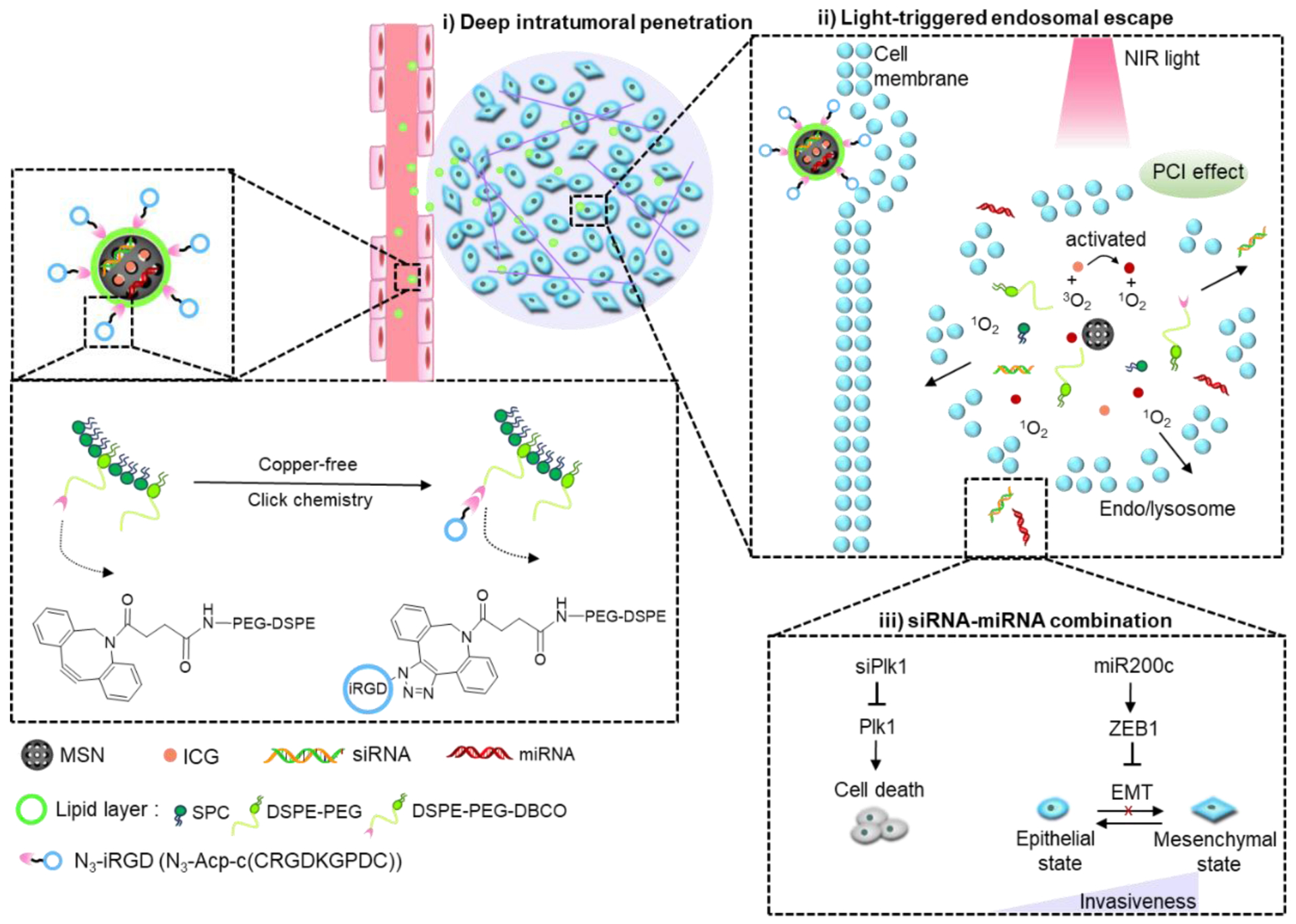
Schematic illustration of light-triggered RNA delivery by tumor-penetrating iMSNs for siPlk1/miR-200c combination therapy.
2. MATERIALS AND METHODS
2.1. Materials.
Propylamine-functionalized silica particles (NH2-MSN) and soy phosphatidylcholine (SPC) were purchased from Sigma-Aldrich (St. Louis, MO). Indocyanine green (ICG) was obtained from Polysciences (Warrington, PA). MicroRNA-200c mimic (mature microRNA sequence: 5′-UCACAACCUCCUAGAAAGAGUAGA-3′), siRNA targeting Plk1 (siPlk1, sense strand, 5′-CAACCAAAGUCGAAUAUGAUU-3′), siRNA silencing luciferase (siluc, sense strand, 5′-UCGAAGUACUCAGCGUAAGdTdT-3′), negative control siRNA non-specific to any human gene (siNC, sense strand, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) and carboxyfluorescein (FAM) labeled siRNA were obtained from Dharmacon (Lafayette, CO). Soy phosphatidylcholine (SPC) and cholesterol (Chol) were obtained from Sigma-Aldrich. 1,2-distearoyl-sn-glycero-3-phosphoethanol-amine-N-[methyl ether (polyethylene glycol)-2000] (DSPE-mPEG) was purchased from Laysan Bio (Arab, AL). 1,2-distearoyl-sn-glycero-3-phospho-ethanolamine-N-[dibenzocyclooctyl (polyethylene glycol)-2000] (DSPE-PEG-DBCO), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-distearoyl-sn-glycero-3- phosphocholine were obtained from Avanti (Alabaster, AL). N-terminal azido-functionalized iRGD peptide (N3-Ac (CRGDKGPDC)) was synthesized by GL Biochem (China). Eagle's Minimum Essential Medium (EMEM) was obtained from ATCC (Manassas, VA). Dulbecco’s modified eagle medium (DMEM), FluroBrite DMEM media, phosphate buffered saline (PBS), Penicillin-Streptomycin (Pen-Strep) solution and fetal bovine serum (FBS) were from Thermo Scientific (Waltham, MA). XenoLight D-luciferin potassium salt was provided by PerkinElmer (Waltham, MA). All other reagents were obtained from Fisher Scientific and used as provided unless otherwise stated.
2.2. Preparation of nanoparticles.
Amine-functionalized MSNs were purchased from Sigma with average pore size of 4 nm and particle size range from 100–200 nm. To load both siRNA and miRNA in the silica nanoparticles, NH2-MSNs were soaked in siRNA and miRNA-containing solution (NH2-MSN: small RNAs, 30:1, w/w) for 0.5 h. Then, free ICG dissolved in deionized water was added and stirred for 1 h. Excess ICG was removed by centrifugation at 3500 rpm for 10 min and washed twice with DI water. By screening different lipid compositions to improve nanoparticles colloidal stability, we prepared lipid film with SPC/DSPE-mPEG/DSPE-PEG-DBCO at weight ratio of 2:2:1. The stabilization effect of lipids was recorded as the time period for which the nanoparticles remained colloidally stable. These lipid components were dissolved in a round-bottom flask containing 5 mL chloroform. The solvent was then evaporated in a vacuum rotary evaporator, which led to the formation of a thin lipid film. Continuous vacuum drying was performed overnight to remove any traces of organic solvents. To coat lipid films onto drug-loaded NH2-MSNs, particle aqueous suspension was added to the hydrated film (lipids: MSN, 2:1, w/w) and sonicated for 30 min. Lipid coated particles (MSN/siRNA+miRNA+ICG) were obtained by further removal of excess lipids through centrifugation. For iRGD-conjugation to the nanoparticles, azido functionalized iRGD (DSPE-PEG-DBCO: iRGD, molar ratio 1:1) was added to MSN/siRNA+miRNA+ICG. Post 30 min reaction at 37 °C, the iRGD-modified silica particles (iMSN/siRNA+miRNA+ICG) were centrifuged for collection and washed with DI water. FAM-siRNA or siluc loaded particles were prepared with the same method. ICG and small RNAs ratio loaded in the nanoparticle was fixed at 1 μg/mL: 200 nM for all the formulations in this work.
2.3. Characterization of nanoparticles.
The binding of NH2-MSN to small RNAs was tested using agarose gel electrophoresis containing 0.5 μg/mL ethidium bromide (EtBr). NH2-MSN/siRNA+miRNA+ICG was prepared in pH 7.4 10 mM HEPES (siRNA: miRNA, 1:1, w/w). 20 μL of the nanoparticle solutions (0.5 μg of siRNA+miRNA) formed at different weight ratios were loaded for running in 0.5 × Tris/Borate/EDTA buffer (100 V, 15 min). The gels were observed using a KODAK Gel Logic 100 imaging system. The absorbance spectrum and concentration of free ICG, NH2-MSN, NH2-MSN/siRNA+miRNA and NH2-MSN/siRNA+miRNA+ICG were measure by microplate reader at 790 nm (Molecular Devices, CA). Dynamic light scattering was used to determine zeta potential of the nanoparticles (Malvern, U.S.A.). Nanoparticle morphology was observed using TEM (Tecnai G2 Spirit, FEI Company, U.S.A.). To improve clarity, image contrast and sharpness were adjusted. Heparin displacement assay was conducted to study the release of siRNA and miRNA from nanoparticles. Increasing concentrations of heparin were incubated with NH2-MSN/siRNA+miRNA+ICG and iMSN/siRNA+miRNA+ICG for 0.5 h. Agarose gel electrophoresis was used to analyze the samples (20 μL, 0.5 μg of siRNA+miRNA). For the stability assay against serum degradation, siRNA+miRNA solution and iMSN/siRNA+miRNA+ICG solution were treated by FBS (50% v/v) at 37 °C. Sample was collected at each time point (0.5, 1, 2, 4, 6, and 24 h) for electrophoresis analysis. The production of ROS was evaluated by measuring the reduction in absorbance of ROS indicator (DPBF). 20 μL 10 mM DPBF (ethanol) was added to a solution of iMSN/miRNA+siRNA+ICG (ICG, 5 μg/mL) and exposed to a near-infrared 808 nm laser (2 W/cm2, Meig Waei, China). The absorption spectra of the mixture were obtained by the microplate reader (Molecular Devices, CA) after the designed period of time of laser exposure. To assess the potential deleterious effect of ROS on stability of RNA molecules, transfection activity was measured using iMSN/siLuc+ICG with different concentrations in B16F10.Luc cells. iMSN/siLuc+ICG was pre-irradiated with light for 1 h before incubation with cells. After 24 h transfection, luciferase expression was measured. iMSN/siLuc+ICG without light irradiation (-l) or irradiated 4 h after incubation were also included as controls.
2.4. Cellular uptake and intracellular trafficking.
2.4.1. Cell culture.
Human breast carcinoma cell line MDA-MB-231 stably expressing luciferase (MDA-MB-231.Luc) was purchased from PerkinElmer and cultured in EMEM containing 10% FBS. MDA-MB-231 expressing galectin-8-YFP (Gal8) was generated by Dr. Craig Duvall’s lab and cultured in DMEM containing 10% FBS, 1% Pen-Strep and Blasticidin (10 μg/mL). Mouse melanoma cell line B16F10 expressing luciferase (B16F10.Luc) was purchased from PerkinElmer and cultured in RPMI containing 10% FBS.
2.4.2. Intracellular trafficking.
Intracellular localization of iMSN/miRNA+siRNA+ICG was observed by confocal microscope (LSM800, Zeiss). MDA-MB-231 cells were seeded in 20 mm glass-bottom dish (2 × 104 cells/dish). After 24 h, the medium was aspirated and replaced with serum-free medium containing iMSN/FAM-siRNA+ICG (FAM-siRNA 300 nM, ICG 1.5 μg/mL). Incubating for 4 h, cells were washed and irradiated for 30 s 808 nm laser. Then, the cells were stained with LysoTracker® Red DND-99 (Life Technology, USA) and Hoechst 33342 for live cell imaging. Images were acquired by confocal microscope and processed by Zen 2014. Colocalization coefficients were calculated by Coloc 2 in Fiji-ImageJ.
2.4.3. Gal8 recruitment assay.
For Gal8 recruitment assay, MDA-MB-231 expressing galectin-8-YFP (Gal8) were cultured in the glass-bottom dish (1 × 104 cells/dish) and grown in culture medium for one day. Cells were exposed to iMSN/ICG+NC for 4 h then rinsed with PBS and changed to FluroBrite DMEM media supplemented with 10% FBS. Short time laser irradiation (30 s) was performed before live cell imaging. All the images were acquired by laser scanning confocal microscope and processed by Image J (automated counting by ITCN plugin).
2.5. Tumor penetration in 3D tumor spheroids.
To prepare MDA-MB-231 spheroids, cells (5000 cells/well in 5% matrigel) were added to a low-attachment 96-well spheroid microplate (Corning, USA) and cultured for further use. The growth of spheroids was observed using a microscope (AMG) until the diameter of spheroids were around 400 μm. Free FAM-siRNA+ICG, MSN/FAM-siRNA+ICG, iRGD+MSN/FAM-siRNA+ICG and iMSN/FAM-siRNA+ICG (FAM-siRNA 400 nM, ICG 2 μg/mL) were added and incubated with spheroids for 12 h. Then, the spheroids were washed and fixed with 4% paraformaldehyde. The fluorescent signal of ICG and FAM-siRNA in spheroids were observed using XY-stack method of the confocal microscope (10 μm intervals for each sample). The images were then converted to surface-plot images using ImageJ software. For quantitative measurement, the spheroids were trypsinized to obtain single-cell suspension which were subjected to flow cytometry analysis.
2.6. Delivery efficacy in vitro.
2.6.1. Luciferase knockdown.
Transfection study was conducted on MDA-MB-231.Luc cells and spheroids and B16F10.Luc cells. Cells or spheroids were cultured in a 48-well plate 24 h prior to transfection. Nanoparticles loaded with siLuc or siNC were prepared as described before. The cells or spheroids were treated with different nanoparticles (siRNA 400 nM, ICG 2 μg/mL) for 24 hours with or without laser irradiation. To measure luciferase expression, the cells were rinsed and lysed in 0.5× cell culture lysis reagent buffer (Promega, Madison, WI). 0.5 mM luciferin solution was injected into each well with cell lysate (20 μL) and the luminescence was measured with Synergy 2 Microplate Reader (BioTek, VT). Total protein was measured by the BCA protein assay (Pierce, Rockford, IL). Transfection activity of different nanoparticles was expressed as relative luciferase activity (%) which was normalized by the same nanoparticles loaded with siNC. For comparison, branched PEI (Mw=25k, Sigma) and Lipofectamine 3000 (Thermal Fisher) were applied as positive control. Small RNAs were condensed with PEI at weight ratio of 1.5. Lipofectamine 3000 was used according to user instruction.
2.6.2. Quantitative real-time polymerase chain reaction (qRT-PCR).
Trypsinized cells were added to 24-well plates (5 × 104 cells/well) before treatment. After 24 h, the cells were treated with various nanoparticles (siRNA 200 nM, miRNA 200 nM, ICG 2 μg/mL) for 48 hours with or without laser irradiation. Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion, USA). The levels of miR-200c were analyzed by TaqMan qRT-PCR. RNA (10 ng) was converted into cDNA with miR-200c primer (or internal control Z30 primer) and TaqMan microRNA reverse transcription kit (Applied Biosystems). PCR reaction was performed using miR-200c or Z30 primers and TaqMan Universal Master Mix II, No AmpErase UNG (2×) (Applied Biosystems, CA) on a Rotor-Gene Q (QIAGEN). MiRNA were quantified according to the comparative threshold cycle (Ct) method. Levels of the Plk1 and ZEB1 were measured using SYBR Green RT-PCR. Total RNA (0.5 μg) was reverse-transcribed to cDNA with QuantiTect reverse transcription kit (Qiagen). The relative level of mRNA was quantified by RT-PCR with QuantiFast SYBR Green PCR kit (Qiagen) on a Rotor-Gene Q (QIAGEN). The following primers were used: Plk1 (forward 5′-ATTTCCGCAATTACATGAGC; reverse, 5′-TCCTGGAAGAAGTTGATCTG), ZEB1 (forward 5′-AAAGATGATGAATGCGAGTC; reverse, 5′-TCCATTTTCATCATGACCAC), GAPDH (forward 5′-ATCAAGAAGGTGGTGAAGCAGGCA; reverse, 5′-TGGAAGAGTGGGAGTTGCTGTTGA). Relative levels of mRNA were calculated based on the Ct values.
2.7. Anticancer activity in vitro.
2.7.1. Cell killing activity in monolayer cells.
Cell killing activity of the nanoparticles was assessed by Cell Titer Blue assay. Cells were seeded in 96-well plate (0.5 × 104 cells/well) and cultured for one day. Increasing doses of nanoparticles loaded with ICG, siRNA and miRNA were added to the cells and exposed to laser irradiation (30 s) after 4 h nanoparticle exposure. After 48 h incubation, medium was aspirated and replaced with fresh medium containing Cell Titer Blue reagent (20%, v/v). Fluorescence intensity was measured on the microplate reader. The relative cell viability of different samples was normalized to the viability of untreated cells and shown as the means ± SD (n=3).
2.7.2. Serum-induced migration.
Cells were seeded in 6-well plate (2.5 × 105 cells/well) and cultured for one day. Then, the cells were treated with various nanoparticles (siRNA 150 nM, miRNA 150 nM, ICG 1.5 μg/mL) for 24 hours with or without laser irradiation. Next, cells were treated by trypsin and suspended in serum-free medium. 5×104 cells were added to the transwell inserts and placed in a 24-well plate containing complete culture medium in each well. The cells were cultured at 37 °C incubator and allowed migration through the insert membrane for 10 h. Non-migrated cells (on the upper chamber) were removed by swabs. Migrated cells were fixed and stained with 0.2% Crystal Violet solution and imaged under an EVOS microscope. Three random fields were visualized (20×) per insert. Each group was performed in triplicate inserts. Data were shown as average number of migrated cells/image ± SD (n=3).
2.7.3. Clonogenic assay.
Cells were transfected with different formulations (miRNA 300 nM, siRNA 300 nM, ICG 3 μg/mL) for 4 h. We accordingly increased RNA and ICG concentration in this assay to improve the quantity of nanoparticles delivered into cells during the short period of treatment. With or without 30 s laser irradiation, transfected cells were reseeded in 12-well plates (200 cell/well) in triplicates and allowed to grow for 2 weeks. After that, the cells were fixed and stained by 1% w/v crystal violet. Plates were rinsed with water followed by photography. The colony number in each well was counted.
2.7.4. Cell killing activity in tumor spheroids.
Cell killing activity of particles in tumor spheroids was evaluated via a Live/Dead Kit (Biotium, USA). Briefly, spheroids with a diameter of about 400 μm were exposed to different formulations for 48 h (siRNA 300 nM, miRNA 300 nM, ICG 3 μg/mL,). Light irradiation (30 s) was performed 4 h after particle exposure. The spheroids were rinsed and treated by Live/Dead Kit. All the images were captured by the confocal microscope.
2.8. Biodistribution of nanoparticles.
All animal studies were performed with a protocol approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee. IVIS (Xenogen 200) imaging was applied to investigate the biodistribution of nanoparticles in tumor-bearing mice. MDA-MB-231.Luc cells (2.5 x106 cells in 200 μL of 50:50 matrigel/PBS) were injected into the mammary fat pad of immunocompromised female NCG mice. When the tumor was around 200 mm3 in size, free ICG+FAM-siRNA, MSN/ICG+FAM-siRNA, iRGD+MSN/FAM-siRNA+ICG and iMSN/ICG+FAM-siRNA were administered via tail injection (2 mg/kg FAM-siRNA, ICG 720 μg/kg). All mice were anesthetized and imaged at 4, 24 and 48 h post-injection. After 48 h, mice were sacrificed and tumors together with other major organs (heart, liver, spleen, lung, kidney) were collected for ex vivo imaging. The fluorescent signals from different organs were analyzed by IVIS software. Excised tumor tissues were embedded in O.C.T reagent then cut into frozen sections (10 μm). Blood vessels were stained with CD 31 antibody (Abcam) and imaged by confocal microscopy.
2.9. Therapeutic efficacy.
Spontaneous metastasis breast cancer model was established as described above. Luciferase-expressing MDA-MB-231 cells suspended in matrigel/PBS (2.5 million/injection site) were injected into the mammary fat pad of the NCG mice. When the tumor volume reached 100 mm3 in size, mice were assigned into 7 groups (n=5) and administered i.v. with saline, iMSN/NC+ICG, iMSN/Plk1+NC+ICG, iMSN/NC+200c+ICG, MSN/Plk1+200c+ICG, iMSN/Plk1+200c+ICG twice a week for 3 weeks (1 mg/kg siPlk1, 1 mg/kg miR-200c, ICG 720 μg/kg). 5 groups of mice received 5 min laser irradiation 4 h-post administration except for iMSN/Plk1+200c+ICG (- light) and saline-treated groups. Laser source was set about 1 cm above the mice tumor. The length and width axes of tumors was measured by calipers and the tumor volume was calculated according to the formula: volume (mm3) = length × width2/2. Animal body weight was recorded every 6 days. For bioluminescence imaging, mice were anesthetized and given 3 mg/0.1 mL of D-luciferin by i.p. administration. Fifteen minutes post administration, bioluminescence was evaluated with IVIS system. Bioluminescent signal from relative optical intensity was defined manually, and data were shown as radiance (p/sec/cm2/sr). All the animals were euthanized on day 45 from tumor inoculation, tumors and major organs were obtained, imaged and used for staining and histological analysis. Tumor sections were also immunostained with anti-Plk1 (abcam 35206) and anti-ZEB1 (Cell Signaling Technology 3396) using immunohistochemistry (IHC). Blood was withdrawn in heparin tubes and analyzed using Vetscan VS (Abaxis). Creatinine, blood urea nitrogen (BUN), alanine transaminase (ALT), and aspartate transaminase (AST) were measured by assay kit (Bioassay systems, CA, USA). Tumors were also sectioned for Ki-67 staining proliferation analysis. Lungs were inflated using sucrose and immersed in Bouin’s solution for 18 h then placed in 70% ethanol for storage. Pulmonary lobes were separated. The lung surface tumors was counted using a dissecting microscope. Lungs were sectioned for H&E staining.
2.10. Statistical analysis.
All the results are expressed as mean ± SD. The statistical significance was performed with Prism using ANOVA. P< 0.05 was considered statistically significant.
3. RESULTS AND DISSCUSSION
3.1. Preparation and characterization of nanoparticles.
The nanoparticles developed here are composed of a mixture of siRNA and miRNA adsorbed on the surface of the mesoporous silica core loaded with ICG and further stabilized with a supporting lipid layer modified with azide-terminated iRGD peptide by copper-free click-chemistry (Scheme 1). Detailed nanoparticle preparation steps were outlined in Figure 1A. Gel retardation assay indicated that both siRNA and miRNA were immobilized onto the bare NH2-MSN nanoparticles at MSN/RNA w/w ratio of 30 (Figure 1B). The RNA cargo loading was calculated as approximately 2.57 nmol/mg MSN. Absorbance spectrum of NH2-MSN/siRNA+miRNA+ICG and free ICG confirmed successful loading of ICG, with a high encapsulation efficiency of 91% (Figure 1C). After the efficient loading of the cargos, lipids were coated onto the nanoparticles to improve colloidal stability. The most stable lipid film formulation was selected as SPC/mDSPE-PEG/DSPE-PEG-DBCO (w/w/w, 2:2:1) after various lipid species screening (Table 1). The optimal lipid layers effectively stabilized cargo-loaded MSN in aqueous solution. The change of the nanoparticles zeta potential provided an easy qualitative validation of the loading of cargos and coating with lipids. The zeta potential of bare NH2-MSN was 34.3 mV and loading of the RNA and ICG cargos decreased the value to 21.6 mV (NH2-MSN/siRNA+miRNA+ICG). The fact that the particles retained its overall positive charge suggested that MSN surface was not saturated by the negatively charged molecules owing to its high loading capacity. We observed no replacement of the small RNA by ICG after its loading. After cargo loading, providing the particles with a stabilizing layer composed of phospholipids and PEGylated lipids further decreased the zeta potential to negative −15.7 mV. The zeta potential of the final iRGD-modified nanoparticles (iMSN/siRNA+miRNA+ICG) was −19.0 mV (Figure 1D). TEM was used to validate the deposition of the lipid layer on the surface of MSN. As shown in Figure 1E, the observed thickness of the lipid layer was ~10 nm. Release of siRNA and miRNA from the nanoparticles was analyzed by heparin replacement test (Figure 1F). Without lipid layer shielding, NH2-MSN/siRNA+miRNA+ICG released the RNAs when the heparin concentration exceeded 100 μg/mL. In contrast, the lipid layer stabilized iMSN/siRNA+miRNA+ICG particles and no RNA release was observed even when incubated with 4000 μg/mL heparin. Serum stability of iMSN/siRNA+miRNA+ICG was studied in 50% FBS at 37 °C using gel electrophoresis (Figure 1G). Most of the naked siRNA/miRNA mixture was degraded within 0.5 h of FBS incubation. iMSN improved serum stability as suggested by at least 6 h RNA protection.
Figure 1.
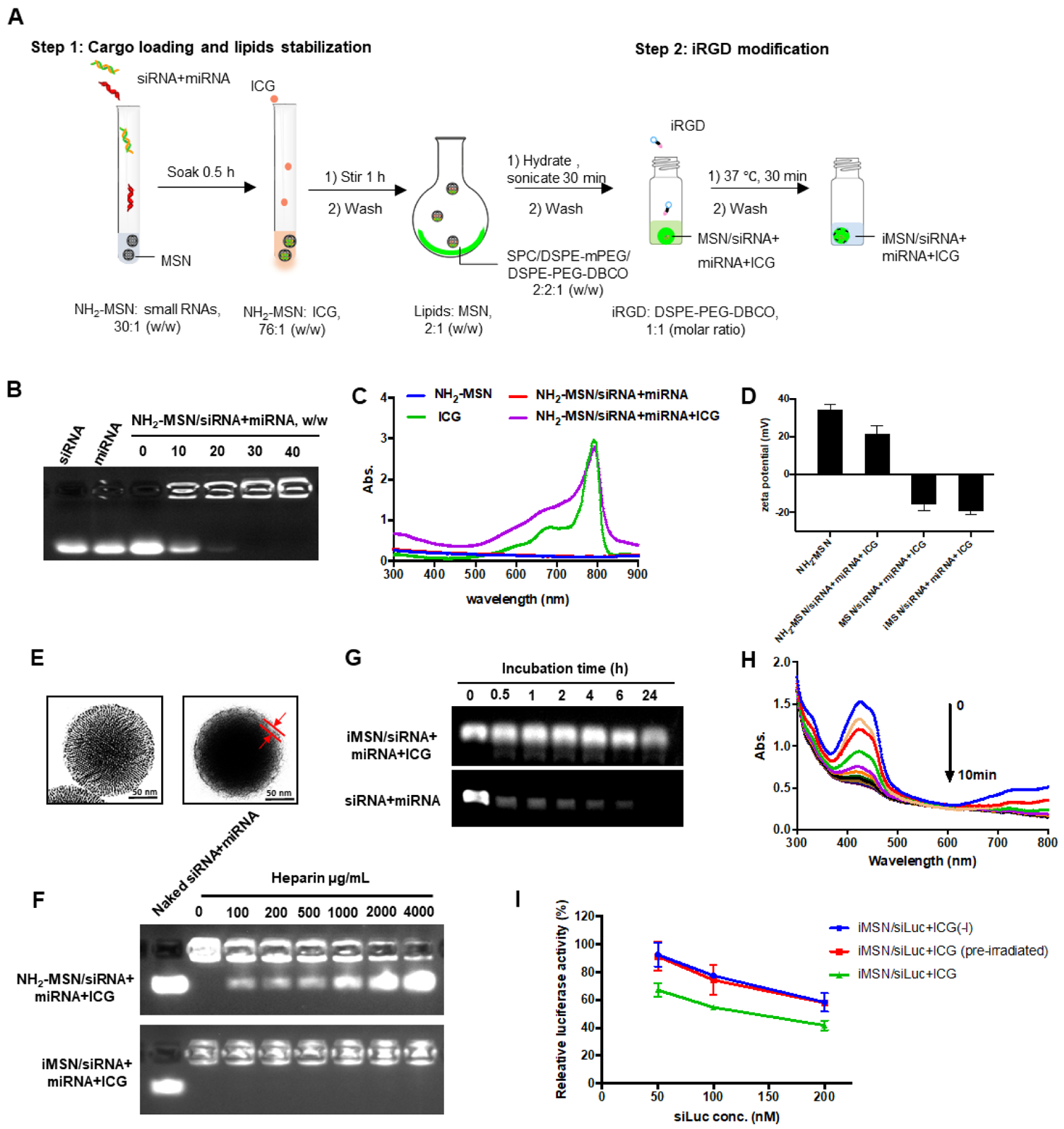
(A) Schematic depiction of the steps used in the preparation of the nanoparticles. (B) Loading of siRNA/miRNA in NH2-MSN assessed by agarose gel retardation assay. (C) Absorbance spectrum of ICG and MSN particles. (D) Zeta potential of NH2-MSN, NH2-MSN loaded with siRNA/miRNA and ICG (NH2-MSN/siRNA+miRNA+ICG), lipid-coated MSN loaded with siRNA/miRNA and ICG (MSN/siRNA+miRNA+ICG), and iRGD-modified lipid-coated MSN loaded with siRNA/miRNA and ICG (iMSN/siRNA+miRNA+ICG). Data are expressed as mean ± SD (n = 3). (E) TEM images of bare NH2-MSN (left) and iMSN/siRNA+miRNA+ICG (right). (F) Heparin-induced RNA release from NH2-MSN/siRNA+miRNA+ICG and iMSN/siRNA+miRNA+ICG with increasing concentration of heparin. (G) Stability of iMSN/siRNA+miRNA and naked siRNA+miRNA mixture against serum. (H) ROS generation by iMSN/siRNA+miRNA+ICG. Normalized absorption spectra of iMSN/siRNA+miRNA+ICG with singlet oxygen probe (DPBF) after near-infrared light exposure (2 W/cm2). (I) Luc silencing efficacy of siLuc loaded in iMSN on B16F10.Luc cells with light irradiation or pre-irradiated by light before treatment (pre-irradiated) or without light (-l). Data are expressed as mean ± SD (n = 3).
Table 1.
Screening of lipid formulations of MSN/siRNA+miRNA+ICG.
| 1) Single lipid species | ||||||
|---|---|---|---|---|---|---|
| Formulation | (-) | SPC | mDSPE-PEG | Chol | DPPC | DSPC |
| Stabilization effect (h) | 0.1–0.2 | 6–10 | 8–12 | 2–4 | 0.5–1 | 0.5–1 |
| 2) Two lipids species (w/w, 1:1) | |||
|---|---|---|---|
| Formulation | SPC + mDSPE-PEG | SPC + Chol | mDSPE-PEG + Chol |
| Stabilization effect (h) | >48 | 4–6 | 24–48 |
| 3) Ratio of SPC and mDSPE-PEG | |||
|---|---|---|---|
| Ratio (w/w) | 2:1 | 1:1 | 1:2 |
| Stabilization effect (h) | 24–48 | >48 | >48 |
| 4) Three lipids species (w/w, 2:2:1)c | ||
|---|---|---|
| Formulation | SPC + mDSPE-PEG + DSPE-PEG-DBCO | SPC + mDSPE-PEG + Chol |
| Stabilization effect (h) | >48 | 24–48 |
Based on prior evidence, we hypothesized that the ROS produced by light activation of ICG loaded in the nanoparticles will play an important role in endosomal escape of the siRNA/miRNA combination. We determined ROS generation upon irradiation with 808 nm light using an ROS indicator dye DPBF.46 A sustained decrease of DPBF absorption was observed in nanoparticle sample showing 36% reduction of the initial absorbance upon light exposure, suggesting the production of singlet oxygen by ICG upon light exposure (Figure 1H). We also observed effective ROS generation in tumor cells after light exposure (Figure S1). There is a possibility that heat generated during the exposure may further contribute to the observed effects, although we attempted to minimize this effect by short exposure and low ICG dose. Despite some reports suggesting that MSN nanoparticles have inherent antioxidant effect, we have observed no ROS scavenging by the iMSN used in this study, possibly owing to different surface chemistry and particle shape from the previous reports.47–49 Of note, the generated ROS had no significant adverse effect on the biological activity of the delivered RNA molecules under the used irradiating conditions as suggested by no significant changes in the extent of gene silencing activity (Figure 1I).
3.2. Intracellular trafficking of light-triggered endosomal escape.
To study the endosomal escape of the nanoparticles in vitro, we used Lysotracker colocalization and galectin-8 (Gal8) recruitment assay. First, lysosomes were stained red using Lysotracker and cell nuclei in blue with Hoechst 33258 (Figure 2A). Considering the similar physicochemical properties of siRNA and miRNA, we used only FAM-labeled siRNA (FAM-siRNA) in the intracellular distribution studies both in vitro and in vivo. After 4 h incubation of MDA-MB-231 cells with iMSN/FAM-siRNA+ICG, the FAM-siRNA (green) was colocalized with the endo/lysosomal staining (red) and exhibited a typical dot-like pattern in the cells, indicating siRNA entrapment in the acidic vesicles and inefficient endosomal escape. Thus, we sought a specific approach to overcome this key challenge in RNA delivery. We performed a short irradiation (808 nm) of the cells incubated with iMSN/FAM-siRNA+ICG to trigger ROS generation by ICG. After light activation, the green signal of FAM-siRNA dispersed through the entire cell, suggesting the ROS produced by ICG damaged the membrane of the endosomal vesicles and released FAM-siRNA into the cytoplasm. Quantitative analysis showed that the Manders’ colocalization coefficient between FAM-siRNAs and Lysotracker in the irradiated cells was markedly decreased than in the control cells not exposed to the light (Figure 2C).
Figure 2.
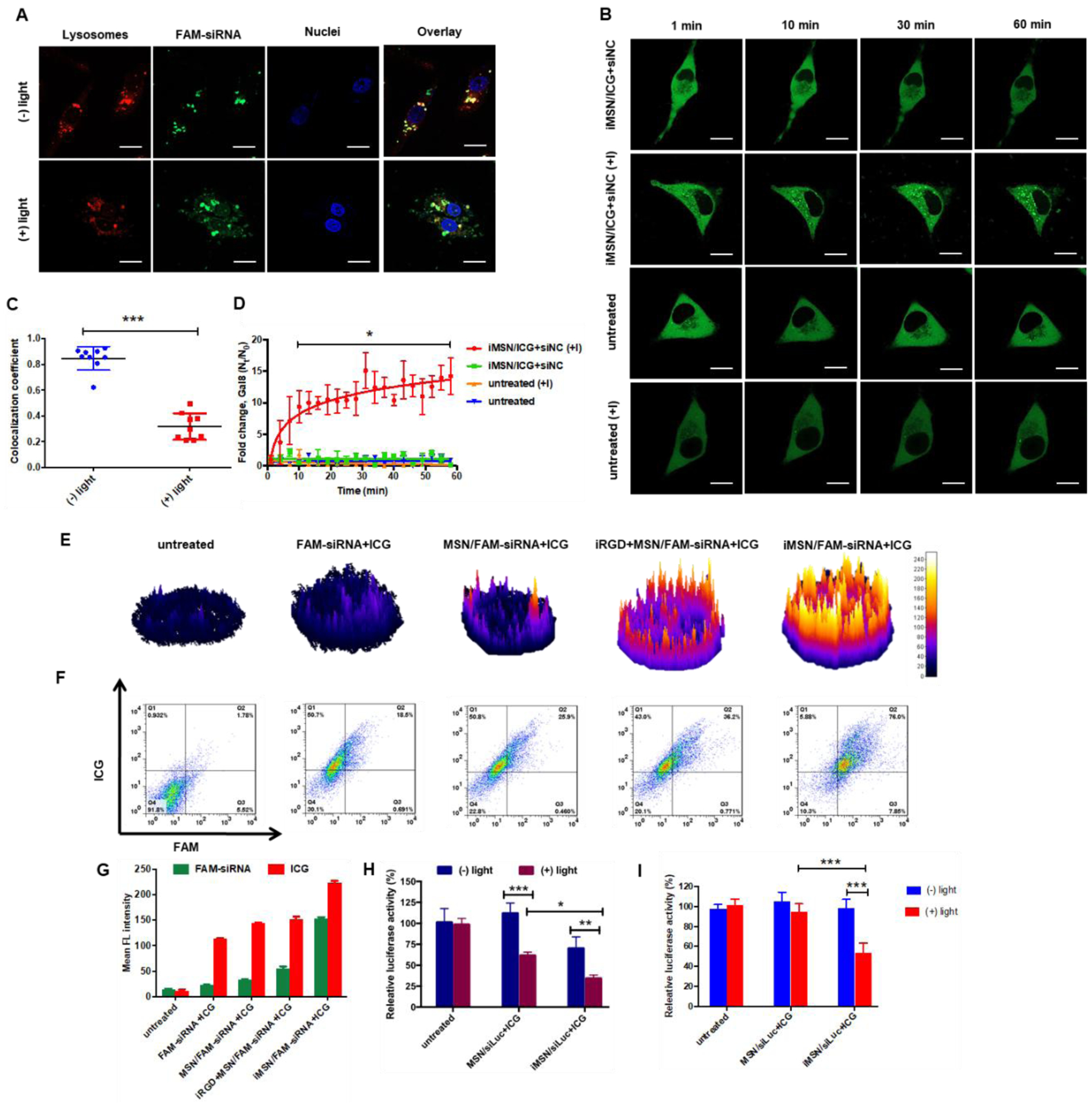
(A) and (C) Intracellular trafficking of iMSN/FAM-siRNA+ICG in MDA-MB-231 cells with (+) or without (-) light irradiation using Lysotracker and Manders’ colocalization coefficient analysis. The value of 0 indicates no colocalization of FAM-siRNA and Lysotracker, value of 1 indicates complete colocalization. (scale bar=20 μm) (B) and (D) Confocal images of untreated MDA-MB-231-Gal8 cells and iMSN/siNC+ICG treated cell (+) or (-) light irradiation at different time points. Endosomal disruption kinetics plotted as fold change in Gal8 punctation. Scale bar=10 μm. (E) Fluorescence (ICG) surface plot images of MDA-MB-231 tumor spheroids treated with free FAM-siRNA+ICG, MSN/FAM-siRNA+ICG, iRGD+MSN/FAM-siRNA+ICG, iMSN/FAM-siRNA+ICG and negative control. (F) and (G) Flow cytometry analysis of single cell suspension from spheroids after different treatments, as indicated with double-positive staining cells and mean fluorescence intensity. (H) Luc silencing efficacy of siLuc loaded in MSN or iMSN (+) or (-) light irradiation in cells. Data expressed as mean ± SD (n = 3). **p<0.01, ***p<0.001. (I) Luciferase silencing efficacy with siLuc loaded in MSNs or iMSNs (+) or (-) light irradiation in MDA-MB-231.Luc spheroids. Data expressed as mean ± SD (n = 3). ***p<0.001.
To enable a real-time monitoring of the endosomal escape process observed above, we utilized the Gal8 visualization method in live MDA-MB-231 cells expressing Gal8-YFP.50 Gal8 binds to glycosylated transmembrane proteins localized on the inner surface of endosomal membranes. When endosomes are disrupted, Gal8 binds and concentrates at the exposed luminal surface of the membrane.51–52 Expression of Gal8-YFP fusion protein can be thus used as a direct and quantitative approach to assess endosomal membrane disruption in living cells.53 Compared to non-irradiated Gal8 cells, cells treated with negative control siRNA (siNC) nanoparticles (iMSN/siNC+ICG) displayed robust Gal8 recruitment to the intracellular vesicles after light irradiation, as indicated by intensive punctate fluorescence (Figure 2B). No Gal8 recruitment was found in the untreated MDA-MB-231-Gal8 cells regardless of the light exposure. The endosomal disruption process triggered by light in the MDA-MB-231-Gal8 cells was quantified by ImageJ software (Figure 2D). Light-irradiated cells displayed significant Gal8 recruitment as early as 10 min after irradiation and reached a 14-fold increase of the recruited Gal8 at 60 min. Non-irradiated and untreated cells regardless of light exposure showed no increase in Gal8 recruitment. These results validate the membrane disruptive role of produced-ROS in the light-triggered endosomal escape of the silica particles. Most likely, the ICG produced ROS under light activation, which subsequently disrupted endosomal membrane by lipid peroxidation,54–55 and allowed the trapped RNA to be liberated and escape from inactivation in lysosomes.
3.3. Enhanced cellular uptake and penetration in vitro.
Targeting and tumor penetrating ability of iMSNs were studied in MDA-MB-231 cells and multicellular tumor spheroids. We used iMSN loaded with FAM-siRNA and ICG to quantify the uptake of the two cargos. In conventional 2D cell culture conditions, cell uptake was higher in cells treated with iMSN/FAM-siRNA+ICG than all the used control groups (Figure S2). The tumor penetration ability of the nanoparticles was assessed by observing the fluorescence of ICG and FAM-siRNA in MDA-MB-231 tumor spheroids via confocal microscopy. As shown in the confocal images converted to surface plots (Figure 2E, S3), the highest fluorescent signal was found throughout the spheroids incubated with iMSN/FAM-siRNA+ICG. Quantitative analysis of the penetration in the spheroids was performed by measuring both ICG and FAM-siRNA fluorescence intensity in a single cell suspension collected from the spheroids by FACS (Figure 2F, G). Consistent with the confocal observations, cells treated by iMSN/FAM-siRNA+ICG had the highest uptake of both cargos as indicated by 76% of double-positive cells (FAM+/ICG+). In contrast, the physical mixture of iRGD peptide and nanoparticles (iRGD+MSN/FAM-siRNA+ICG) showed only 36.2 % of FAM+/ICG+ cells, MSN/FAM-siRNA+ICG without the iRGD peptide showed 25.9%. Free FAM-siRNA+ICG mixture had 18.5% of double-positive cells and this diffusion effect possibly results from the size advantage. These data suggested that iRGD modification increases the internalization of iMSNs in the tumor spheroids and thereby improves inward distribution and accumulation of RNA.
3.4. Improved delivery efficacy in vitro.
The light-activated endosomal escape and tumor penetrating ability of iMSNs were expected to boost RNA transfection. We determined the gene silencing efficacy of the nanoparticles carrying siRNA against luciferase (siLuc) in MDA-MB-231.Luc monolayer and 3D spheroids. The results showed that iMSN loaded with siLuc and ICG (iMSN/siLuc+ICG) exhibited better silencing activity than MSN/siLuc+ICG without iRGD in both 2D and 3D settings regardless of light exposure (Figure 2H–I). Light irradiation significantly enhanced the silencing effect of both siLuc-loaded MSN and iMSN in cells. After irradiation, iMSN/siLuc+ICG effectively decreased luciferase expression by 64%, which was much higher than control PEI/siRNA polyplexes (19%) and Lipofectamine 3000 (27%) at the same siLuc dose (Figure S4). In tumor spheroids (Figure 2I), iMSN/siLuc+ICG with light irradiation significantly reduced luciferase expression (47%), while other treatment groups failed to silence luciferase expression at all. These results confirmed that iMSNs with light-triggered endosomal escape and tumor penetrating abilities effectively improved RNA delivery efficacy.
To confirm efficacy of therapeutic siRNA/miRNA delivery, iMSNs carrying ICG, siPlk1, and miR-200c mimic were incubated with MDA-MB-231 cells. The cellular levels of Plk1 mRNA, as well as the levels of miR-200c and one of its main downstream targets the zinc finger E-box-binding homeobox 1 (ZEB1) mRNA were quantified using qRT-PCR. Considering the potential activity of ROS on mRNA level, iMSN/NC+ICG (+light) was included to evaluate the light-induced effect on gene expression. A significant decrease of Plk1 mRNA expression following transfection with iMSN/Plk1+NC+ICG (+light) and iMSN/Plk1+200c+ICG (+light) was achieved when compared to untreated cells. The silencing effect of iMSN/Plk1+200c+ICG against Plk1 was significantly smaller without laser irradiation (Figure 3A). Incubation of cells with iMSN/Plk1+200c+ICG greatly increased the delivery of miR-200c mimic. Interestingly, iMSN/Plk1+NC+ICG plus light also slightly, but not statistically significantly, increased miR-200c levels in the cells (Figure 3B), which was possibly an indirect effect of blocking the multiple growth pathways driven by Plk1.56 iMSN/NC+ICG had no effect on Plk1 and miR-200c expression after light exposure, suggesting limited adverse effects of the ROS generation on the bioactivity of RNA. After endosomal escape and arrival at cytoplasm, miR-200c mimic can downregulate ZEB1, which is an EMT inducer, to reduce cancer cell migration and invasion. ZEB1 expression was significantly inhibited after treatment with iMSN/200c+NC+ICG (+light) and iMSN/Plk1+200c+ICG (+light). iMSN/Plk1+200c+ICG (-light) failed to reduce ZEB1 levels in cells due to endo/lysosomal entrapment (Figure 3C). These results demonstrated effective delivery of therapeutic siRNA/miRNA into cancer cells by iMSNs (+light). Though siPlk1 and miR-200c were not able to significantly regulate mutual targets, a more comprehensive signaling pathway study could be performed to delineate the signaling mechanism of this combination.
Figure 3.
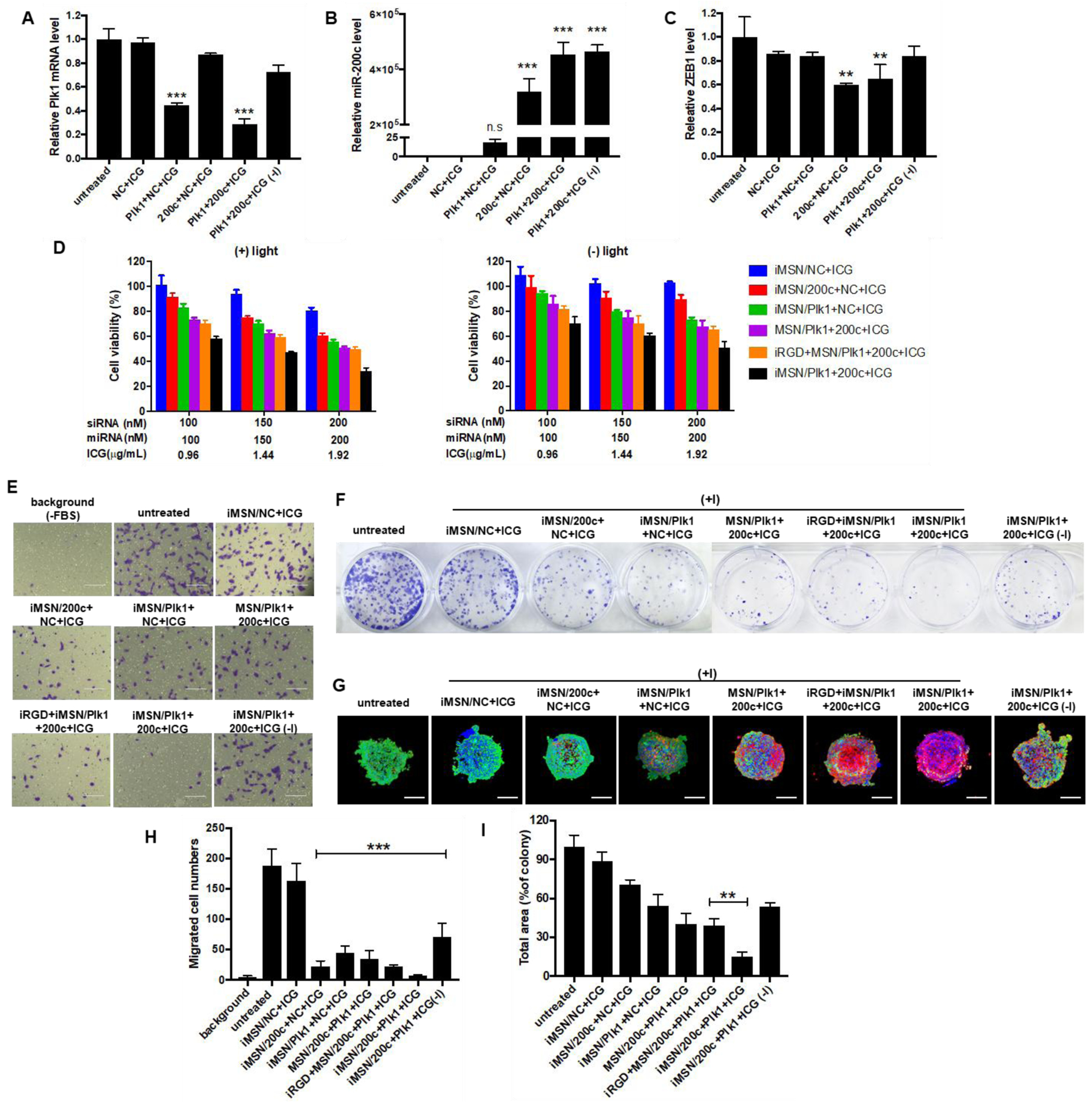
(A) qRT-PCR analysis of Plk1 mRNA level in MDA-MB-231 cells following treatment with iMSN/NC+ICG, iMSN/200c+NC+ICG, iMSN/Plk1+NC+ICG, iMSN/Plk1+200c+ICG plus light irradiation and iMSN/Plk1+200c+ICG (-) light. ***p<0.001 vs untreated. (B) and (C) Relative miR-200c and ZEB1 mRNA levels in cells transfected with different groups were detected by qRT-PCR. ***p<0.001 vs untreated, **p<0.01 vs untreated. (D) Cell viability of cells after different treatments for 48 h (+) or (-) light irradiation. iRGD+MSN/Plk1+200c+ICG represents the co-administration formulation of iRGD peptide and nanoparticle. Data expressed as mean ± SD (n = 3). (E) Inhibition of FBS-induced cancer cell migration following various treatments. “Background” represents randomly migrating cells without FBS and “untreated” represents migrating cells with FBS. (F) Representative images of colonies treated by different nanoparticles from the clonogenic assay at 14 days. (G) Representative images of live/dead viability assays in MDA-MB-231 spheroids following treatments (+) or (-) light irradiation. The dead cells were stained red by EthD-1 and live cells were stained green with calcein AM. Nuclei were stained with Hoechst (blue). Scale bar=200 μm. (H) The number of migrated cells was counted under microscope and results are expressed as average number of migrated cells/20× view ± SD (n =9). “Background” represented migrated cells in FBS-free media without any treatment. ***p<0.001 vs untreated. (I) Quantification of colonies from the clonogenic assay at 14 days. Data expressed as mean ± SD (n = 3). **p<0.01 vs iRGD+MSN/200c+Plk1+ICG.
3.5. Anticancer activity in vitro.
Though intensive studies have evaluated the application of individual siRNAs and miRNAs in cancer therapies, the combination of both siRNA and miRNA has been rarely pursued. Such combination holds great promise as a novel therapeutic strategy to achieve an enhanced therapeutic effect through simultaneously silencing specific oncogenes by siRNA and regulating multiple genes by miRNA.22 Knockdown of Plk1 was reported to suppress the proliferation of cancer cells. miR-200c has been identified as inhibitor of growth and metastasis of breast cancer by reversing the EMT.57–58 Thus, we determined the anticancer activity of the siPlk1/miR-200c combination delivered by iMSNs in vitro. The viability of cells treated by iMSNs loaded with siRNA and miRNA was assessed by Cell Titer Blue assay. As shown in Figure 3D, the RNA combination treatment significantly reduced the viability of cancer cells upon light activation (down to 32% at 200 nM siPlk1, 200 nM miR-200c, 2 μg/mL ICG concentration). The treatment was more effective than NC+ICG (81%), 200c+NC+ICG (61%) and Plk1+NC+ICG (56%) at the same dose. Moreover, we found that iMSNs provided enhanced delivery efficacy with more cell death than MSNs without the iRGD (51%) or when co-incubating MSN with iRGD (50%). All the treated groups showed dose-dependent cytotoxicity after light irradiation. These results indicated light-triggered iMSNs delivery system benefited the therapeutic activity of the siPlk1+miR-200c combination treatment. Cell migration was evaluated by a transwell assay (Figure 3E, 3H). The siPlk1+miR-200c combination delivered by iMSNs showed the most effective inhibition of FBS-induced cell migration upon light irradiation. Colony formation was applied to assess the inhibiting ability of the combination treatment against tumorigenesis. The siPlk1+miR-200c group (+light) significantly reduced the number of MDA-MB-231 colonies (by 85%) when compared to the untreated cells and control groups (Figure 3F, 3I). Live/dead assay was used to assess the anticancer activity of the combination treatment in 3D tumor spheroids (Figure 3G). Dead cells were indicated by red fluorescence and live cells by green. siPlk1+miR-200c (+light) caused large extent of cell death in tumor spheroids and there were less live cells than in other treatment groups. These results validated the combinational anticancer activity of siPlk1-miR-200c in vitro mediated by iMSNs upon light irradiation.
3.6. In vivo biodistribution of nanoparticles.
Following the encouraging in vitro results, we investigated the in vivo delivery efficiency of iMSN. All the nanoparticles (loaded with FAM-siRNA and ICG) were administered via tail vein into mice implanted with orthotopic MDA-MB-231 tumors. IVIS imaging of ICG at designated time-points was performed to gain initial estimate of the nanoparticle distribution (Figure 4A). Consistent with our previous study, free ICG mainly accumulated in the liver and followed by rapid clearance from the body.34 Strong and sustained (at least 48 h) ICG fluorescence signal was observed at tumor sites in iMSNs-injected mice. In contrast, the ICG signal at tumor sites disappeared rapidly in mice injected with MSNs and a mixture of iRGD peptide and MSN. Following animal sacrifice, we further analyzed the fluorescence intensity in ex vivo images of tumors and major organs (Figure S5). Stronger fluorescence intensity was found in the tumors of mice injected with iMSNs than in the other groups. In addition to the abundant tumor accumulation, we also found nanoparticle distributed in other major organs, especially in the liver but not in other eliminating organs like kidney. To evaluate the intratumoral distribution of FAM-siRNA (green) and ICG (red), tumor sections from different groups were obtained and stained with anti-CD31 antibody to label blood vessels (Figure 4B, white). iMSN/FAM-siRNA+ICG-treated group displayed higher ICG and FAM-siRNA fluorescence intensity than free drug combination, non-modified nanoparticle and the mixture of iRGD and nanoparticle, indicating a broader and robust distribution of both cargos to the tumors. Thus, iRGD-modified MSN nanoparticles have been validated as a tumor-penetrating carrier, suitable for effective systemic delivery of small RNAs. Co-administration of free iRGD peptide and MSN nanoparticles was significantly less effective than using the nanoparticles with covalently attached iRGD. This result was consistent with our previous work34 and based on the lack of effective tumor accumulation, we have not pursued the iRGD co-administration any further in the therapeutic in vivo work.
Figure 4.
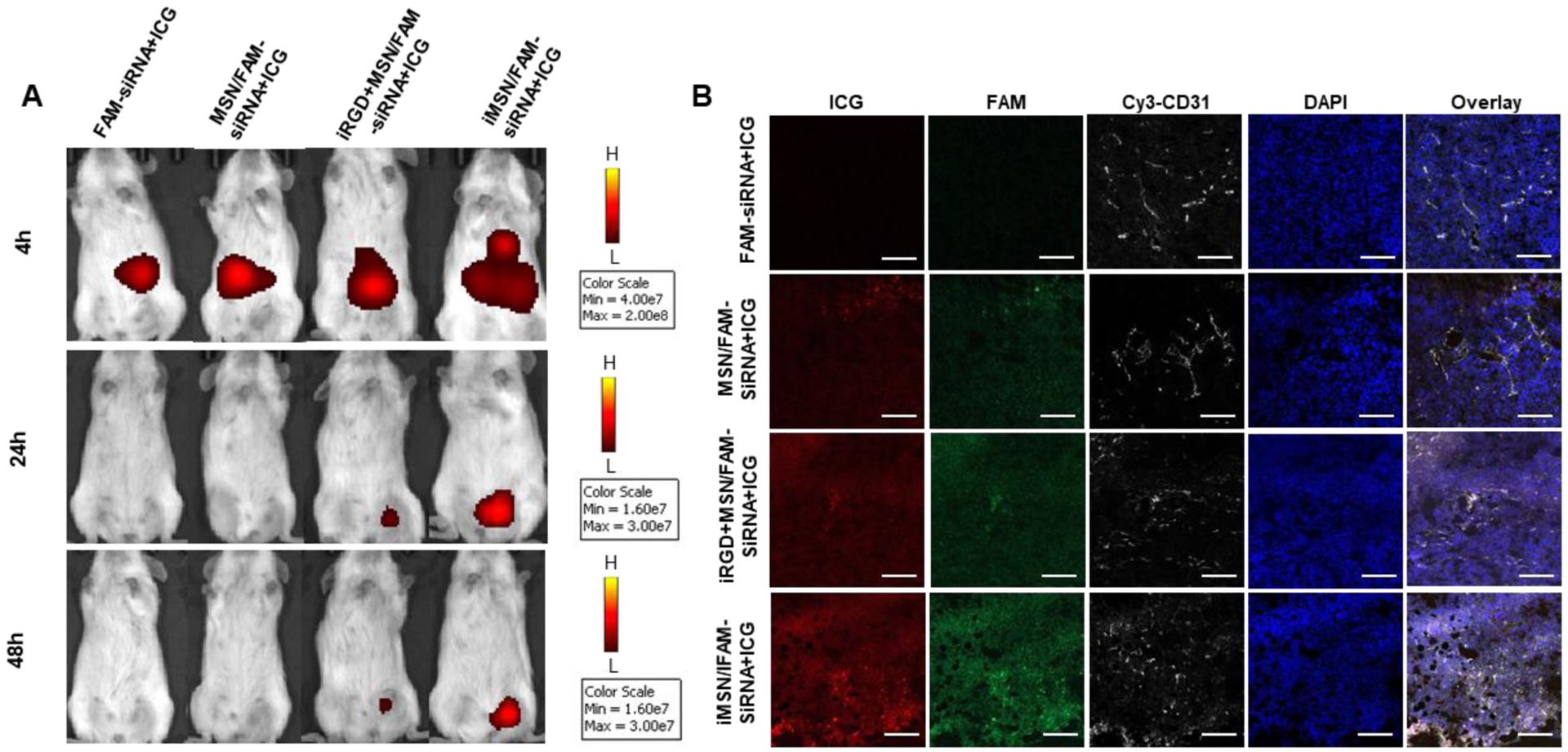
(A) IVIS imaging of mice at 4, 24, and 48 h after i.v. administration of free drug and nanoparticles. Fluorescence signal was from ICG. (B) Confocal microscopic images of tumor cryosections. Tumor vasculature was stained with Cy3-CD31 antibody (white). ICG was shown in red, FAM-siRNA was in green and the nuclei in blue (DAPI). Scale bar=100 μm.
3.7. In vivo therapeutic efficacy.
To demonstrate the therapeutic effect of the RNA-combination (siPlk1+miR-200c) delivered by tumor-penetrating iMSNs in vivo, the antitumor effect was assessed in a highly metastatic orthotopic MDA-MB-231.Luc breast tumor model using a treatment schedule depicted in Figure 5A. Tumor formation and metastatic spread were monitored at predetermined time points by IVIS bioluminescence imaging, and quantitative analysis was performed by determining bioluminescence signal in the region of interest (ROI) using IVIS software. We found the bioluminescence signal at primary tumor sites in untreated tumor-bearing mice increased over time initially but became saturated (1010-1011 radiance) 18 days after tumor implantation (Figure S6A), possibly due to necrosis and blocked oxygen supply associated with the tumor growth. Bioluminescence signal from metastatic lesions increased gradually until the end of the study (107-1011 radiance, Figure S6B). Therefore, bioluminescence imaging signal was used to monitor metastasis progression in this study, while primary tumor growth was assessed by physically measuring tumor dimensions. Tumor-bearing mice were assigned into 7 groups and intravenously injected with different combination treatments (i) iMSN/Plk1+200c+ICG (-light), (ii) iMSN/NC+ICG, (iii) iMSN/Plk1+NC+ICG, (iv) iMSN/200c+NC+ICG, (v) MSN/Plk1+200c+ICG, (vi) iMSN/Plk1+200c+ICG with light irradiation and (vii) an untreated group administered with saline. Tumor growth monitoring revealed that iMSN/Plk1+200c+ICG (+light) exhibited the most effective tumor growth inhibition (Figure 5B). Without light activation, the antitumor activity of iMSN/Plk1+200c+ICG (-light) was significantly decreased and reached an inferior tumor suppressive effect that was comparable to other control treatments (iMSN/Plk1+NC+ICG, iMSN/200c+NC+ICG, MSN/Plk1+200c+ICG). Mice were sacrificed on day 45 post-tumor implantation, and excised tumors were photographed and weighed (Figure 5C). The results confirmed that siPlk1+miR-200c combination assisted by ICG-mediated photodynamic effect, delivered via tumor-penetrating iMSN caused regression of the primary tumor.
Figure 5.
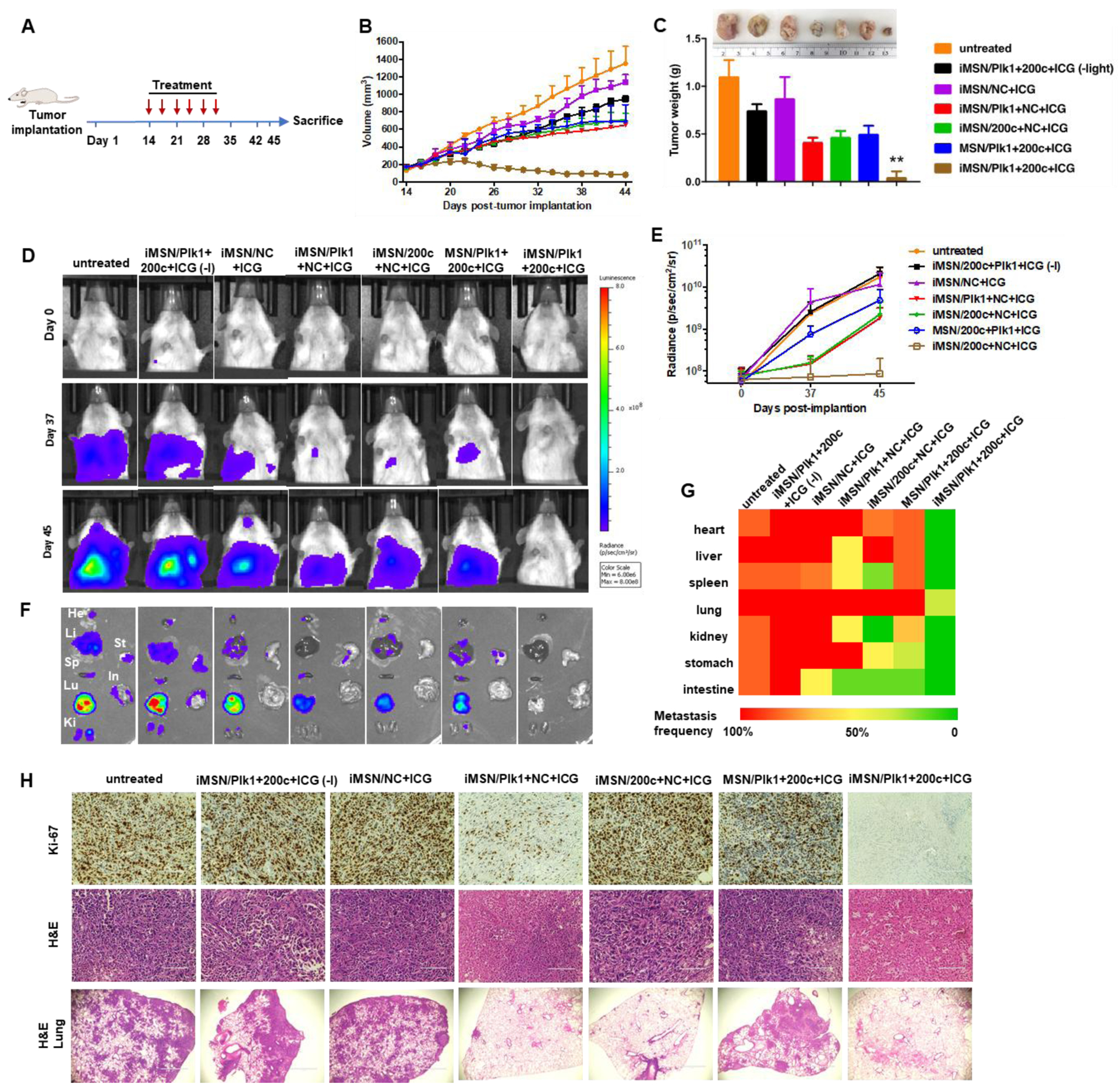
(A) Treatment schedule to assess efficacy of the siPlk1+miR-200c combination treatment in the orthotopic MDA-MB-231 breast cancer model. (B) Tumor growth curves in mice treated with saline, iMSN/NC+ICG, iMSN/Plk1+200c+ICG (-light), iMSN/Plk1+NC+ICG, iMSN/200c+NC+ICG, MSN/Plk1+200c+ICG and iMSN/Plk1+200c+ICG (+) or (-) light irradiation. (C) Representative images and weight of the isolated tumors from different groups. **p<0.01 vs all the groups. (D) Representative bioluminescence images of mice at day 0, 37, and 45 post-tumor implantation. (E) Quantitative analysis of bioluminescence signal in tumor-bearing mice during treatments. (F) Ex vivo images of bioluminescence signal in isolated organs. (G) Heat map showing the summary of the comparative analysis of tumor spread measured by quantitative ex vivo imaging in (F). (H) Ki-67 and H&E staining images of tumor sections. Scale bar=100 μm. Representative images of lung tissue sections stained with H&E. Scale bar=1000 μm. Results are shown as mean ± SD (n = 5).
To identify the role of RNA combination in tumor metastasis, the progression of metastasis was monitored over time by bioluminescence imaging. Representative IVIS images and quantitative analysis (Figure 5D, E) showed that bioluminescence intensity rapidly increased in the thorax and abdomen of all the mice and wide-spread signal was observed 45 days post implantation, except for iMSN/Plk1+200c+ICG (+light) treated group. Ex vivo imaging of the bioluminescence intensity from isolated major organs showed the treatment impact of different groups on tumor metastasis (Figure 5F). Visible metastatic spread was found in lung, liver, spleen, kidney, heart, stomach and intestine in the saline-treated groups, as well as iMSN/Plk1+200c+ICG (-light) and iMSN/NC+ICG groups. The heat map shows treatment impact on tumor metastasis frequency in different organs (Figure 5G). The rest of siPlk1 and miR-200c treatment groups with light irradiation decreased the metastasis, as suggested by less metastatic foci. Of note, iMSN/Plk1+200c+ICG (+light) greatly decreased the metastasis burden and nearly eliminated metastases. The lung metastasis frequency in mice treated by iMSN/Plk1+200c+ICG (+light) was reduced to 40% while all the other groups still showed 100% lung metastasis. All the mice tolerated the combination treatments well, with no significant weight loss (Figure 6A). Immunohistochemical staining of anti-Plk1 and anti-ZEB1 showed specific down-regulation of these two targets in the tumors after Plk1+200c treatment (Figure 6C). The result confirmed the effective delivery of siPlk1 and miR-200c in vivo. Tumor cell proliferation was evaluated by staining the tumor sections with Ki-67 antigen. Low Ki-67 expression was observed in the iMSN/Plk1+200c+ICG (+light) group, suggesting successful effect on tumor cell proliferation with the siRNA/miRNA treatment (Figure 5H). Similarly, H&E analysis of the tumor sections showed increased necrotic area in the Plk1+200c combination group. From the number of surface lung metastases and H&E staining results, we found significantly reduced number of lung metastatic lesions in the iMSN/Plk1+200c+ICG (+light) treatment group (Figure 5H, 6B, 6D). Other major organs were also analyzed by H&E staining and no metastases were found in the iMSN/Plk1+200c+ICG (+light) group (Figure 6E, S7). The effect of the treatment on blood cell counts and blood biochemistry were also studied, and no toxic effects were observed (Table S1). These results validate that siPlk1-miR-200c combination therapy delivered by the endosomal escaping and tumor penetrating iMSN nanoparticles is a highly effective and safe approach for metastatic cancer therapy. Even though we successfully achieved inhibitory effect on tumor cell growth and metastasis both in vitro and in vivo, a more comprehensive study is needed to fully understand the signaling pathways involved and the overall biological mechanism of action of the siPlk1 and miR-200c combination.
Figure 6.
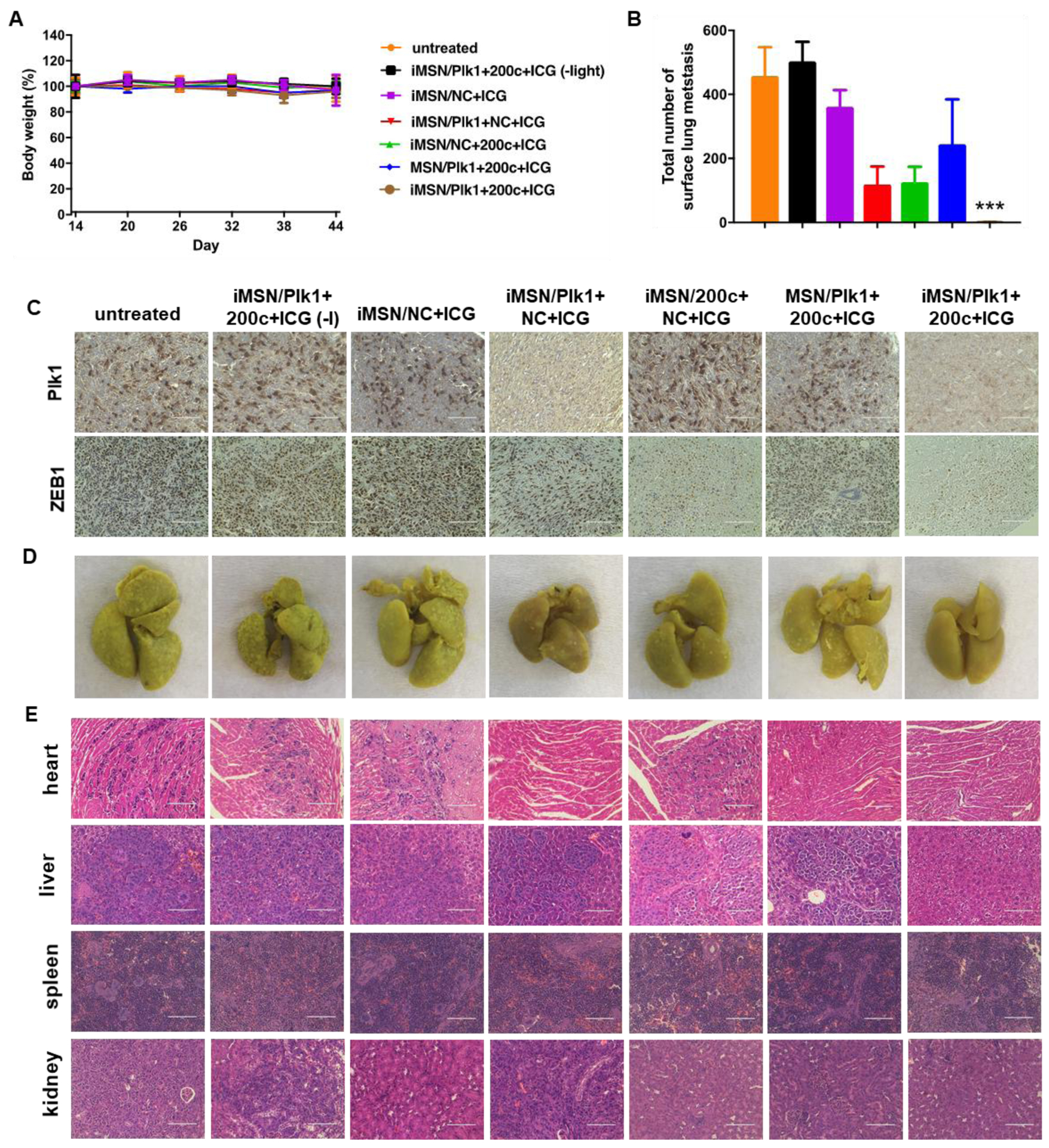
(A) Mice body weight during anticancer treatment. Data are expressed as means ± SD (n = 5). (B) Surface lung metastases from mice treated with different groups. Data represented the number of metastases counted by microscopy (n = 5). ***p<0. 001 vs all the groups. (C) Representative immunostaining images of Plk1 and ZEB1 of tumors obtained from different treatments. Scale bar=100 μm. (D) Photos of the lungs isolated from mice after treatment. (E) Histological observation of tissue sections from major organs of mice was performed after the treatment. Hematoxylin and eosin (H&E) staining was performed. Scale bar=100 μm.
4. CONCLUSIONS
In summary, we have successfully developed iRGD-modified tumor-penetrating mesoporous silica nanoparticles for effective systemic delivery of small RNA combinations. Light triggered photodynamic effect of ICG generated ROS and resulted in the disruption of endo-lysosomal membranes followed by the release of entrapped therapeutic RNAs. The results showed that iRGD-conjugation to the nanoparticle improved the accumulation and penetration of loaded cargos at tumor sites. Furthermore, light activation triggered small RNAs to exert combinational therapeutic effects on tumor cells. Upon light irradiation, iMSN/Plk1+200c+ICG effectively inhibited tumor growth and exhibited superior antimetastatic activity in an orthotopic breast cancer model. The described silica nanoparticles may be promising in the future anticancer treatment.
Supplementary Material
ACKNOWLEDGMENT
Financial support from the National Institutes of Health (R01 EB015216, R01 CA224241) and the University of Nebraska Medical Center are gratefully acknowledged. We thank Janice Taylor and James Talaska for help with confocal microscopy and Jiang Jiang and Lijun Sun for tissue sectioning and staining.
Footnotes
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website. Additional data about ROS generation in tumor cells, microscopic images of cellular uptake, Luc silencing efficacy, biodistribution of nanoparticles in mice tissues, quantitative analysis of bioluminescence signal from untreated tumor-bearing mice, H&E staining of major organs, hematology analysis (PDF).
The authors declare no competing financial interest.
REFERENCES
- (1).Shen J; Zhang W; Qi R; Mao Z-W; Shen H Engineering Functional Inorganic–Organic Hybrid Systems: Advances in siRNA Therapeutics. Chem. Soc. Rev 2018, 47, 1969–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chen Y; Zhu X; Zhang X; Liu B; Huang L Nanoparticles Modified with Tumor-Targeting scFv Deliver siRNA and miRNA for Cancer Therapy. Mol. Ther 2010, 18, 1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gibori H; Eliyahu S; Krivitsky A; Ben-Shushan D; Epshtein Y; Tiram G; Blau R; Ofek P; Lee JS; Ruppin E; et al. Amphiphilic Nanocarrier-Induced Modulation of PLK1 and miR-34a Leads to Improved Therapeutic Response in Pancreatic Cancer. Nat. Commun 2018, 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bobbin ML; Rossi JJ RNA Interference (RNAi)-Based Therapeutics: Delivering on the Promise? Ann. Rev. Pharm. Toxicol 2016, 56, 103–122. [DOI] [PubMed] [Google Scholar]
- (5).Dowdy SF Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol 2017, 35, 222–229. [DOI] [PubMed] [Google Scholar]
- (6).Kent O; Mendell J A Small Piece in the Cancer Puzzle: MicroRNAs as Tumor Suppressors and Oncogenes. Oncogene 2006, 25, 6188–6196. [DOI] [PubMed] [Google Scholar]
- (7).Shalem O; Sanjana NE; Hartenian E; Shi X; Scott DA; Mikkelson T; Heckl D; Ebert BL; Root DE; Doench JG; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rupaimoole R; Calin GA; Lopez-Berestein G; Sood AK miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discovery 2016, 6, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Xie Y; Qiao H; Su Z; Chen M; Ping Q; Sun M PEGylated Carboxymethyl Chitosan/Calcium Phosphate Hybrid Anionic Nanoparticles Mediated hTERT siRNA Delivery for Anticancer Therapy. Biomaterials 2014, 35, 7978–7991. [DOI] [PubMed] [Google Scholar]
- (10).Xie Y; Wang Y; Li J; Hang Y; Jaramillo L; Wehrkamp CJ; Phillippi MA; Mohr AM; Chen Y; Talmon GA; et al. Cholangiocarcinoma Therapy with Nanoparticles That Combine Downregulation of MicroRNA-210 with Inhibition of Cancer Cell Invasiveness. Theranostics, 2018, 8, 4305–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ginn SL; Amaya AK; Alexander IE; Edelstein M; Abedi MR Gene Therapy Clinical Trials Worldwide to 2017: An Update. J. Gene. Med 2018, 20, e3015. [DOI] [PubMed] [Google Scholar]
- (12).Voutila J; Reebye V; Roberts TC; Protopapa P; Andrikakou P; Blakey DC; Habib R; Huber H; Saetrom P; Rossi JJ; et al. Development and Mechanism of Small Activating RNA Targeting CEBPA, a Novel Therapeutic in Clinical Trials for Liver Cancer. Mol. Ther 2017, 25, 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Garber K Alnylam Launches Era of RNAi Drugs. Nat. Biotechnol 2018, 36, 777–778. [DOI] [PubMed] [Google Scholar]
- (14).Sullenger BA; Nair S From the RNA World to the Clinic. Science 2016, 352, 1417–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hannon GJ RNA Interference. Nature 2002, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- (16).Deng Y; Wang CC; Choy KW; Du Q; Chen J; Wang Q; Li L; Chung TKH; Tang T Therapeutic Potentials of Gene Silencing by RNA Interference: Principles, Challenges, and New Strategies. Gene 2014, 538, 217–227. [DOI] [PubMed] [Google Scholar]
- (17).Barata P; Sood AK; Hong DS RNA-Targeted Therapeutics in Cancer Clinical Trials: Current Status and Future Directions. Cancer Treat. Rev 2016, 50, 35–47. [DOI] [PubMed] [Google Scholar]
- (18).Li Y; Lee RJ; Yu K; Bi Y; Qi Y; Sun Y; Li Y; Xie J; Teng L Delivery of siRNA Using Lipid Nanoparticles Modified with Cell Penetrating Peptide. ACS Appl. Mater. Interfaces 2016, 8, 26613–26621. [DOI] [PubMed] [Google Scholar]
- (19).Devulapally R; Sekar NM; Sekar TV; Foygel K; Massoud TF; Willmann J. r. K.; Paulmurugan R Polymer Nanoparticles Mediated Codelivery of AntimiR-10b and AntimiR-21 for Achieving Triple Negative Breast Cancer Therapy. ACS Nano 2015, 9, 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Takamizawa J; Konishi H; Yanagisawa K; Tomida S; Osada H; Endoh H; Harano T; Yatabe Y; Nagino M; Nimura Y; et al. Reduced Expression of the Let-7 microRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [DOI] [PubMed] [Google Scholar]
- (21).Rupaimoole R; Slack FJ MicroRNA Therapeutics: towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discovery 2017, 16, 203–222. [DOI] [PubMed] [Google Scholar]
- (22).Chen Y; Gao D-Y; Huang L In Vivo Delivery of miRNAs for Cancer Therapy: Challenges and Strategies. Adv. Drug Delivery Rev 2015, 81, 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Conde J; Oliva N; Atilano M; Song HS; Artzi N Self-Assembled RNA-Triple-Helix Hydrogel Scaffold for microRNA Modulation in the Tumour Microenvironment. Nat. Mater 2016, 15, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Xue W; Dahlman JE; Tammela T; Khan OF; Sood S; Dave A; Cai W; Chirino LM; Yang GR; Bronson R; et al. Small RNA Combination Therapy for Lung Cancer. Proc. Natl. Acad.f Sci 2014, 111, E3553–E3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Rubinson DA; Dillon CP; Kwiatkowski AV; Sievers C; Yang L; Kopinja J; Rooney DL; Zhang M; Ihrig MM; McManus MT; et al. A Lentivirus-Based System to Functionally Silence Genes in Primary Mammalian Cells, Stem Cells and Transgenic Mice by RNA Interference. Nat. Genet 2003, 33, 401–406. [DOI] [PubMed] [Google Scholar]
- (26).Schipper H; Alla V; Meier C; Nettelbeck DM; Herchenröder O; Pützer BM Eradication of Metastatic Melanoma through Cooperative Expression of RNA-Based HDAC1 Inhibitor and p73 by Oncolytic Adenovirus. Oncotarget 2014, 5, 5893–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zuris JA; Thompson DB; Shu Y; Guilinger JP; Bessen JL; Hu JH; Maeder ML; Joung JK; Chen Z-Y; Liu DR Cationic Lipid-Mediated Delivery of Proteins Enables Efficient Protein-Based Genome Editing in Vitro and in Vivo. Nat. Biotechnol 2015, 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zhou Z; Liu X; Zhu D; Wang Y; Zhang Z; Zhou X; Qiu N; Chen X; Shen Y Nonviral Cancer Gene Therapy: Delivery Cascade and Vector Nanoproperty Integration. Adv. Drug Delivery Rev 2017, 115, 115–154. [DOI] [PubMed] [Google Scholar]
- (29).Huang J; Lin C; Fang J; Li X; Wang J; Deng S; Zhang S; Su W; Feng X; Chen B; et al. pH-Sensitive Nanocarrier-Mediated Codelivery of Simvastatin and Noggin siRNA for Synergistic Enhancement of Osteogenesis. ACS Appl. Mater. Interfaces 2018, 10, 28471–28482. [DOI] [PubMed] [Google Scholar]
- (30).Yan H; Zhu D; Zhou Z; Liu X; Piao Y; Zhang Z; Liu X; Tang J; Shen Y Facile Synthesis of Semi-Library of Low Charge Density Cationic Polyesters from Poly (Alkylene Maleate) s for Efficient Local Gene Delivery. Biomaterials 2018, 178, 559–569. [DOI] [PubMed] [Google Scholar]
- (31).Mackowiak SA; Schmidt A; Weiss V; Argyo C; von Schirnding C; Bein T; Braäuchle C Targeted Drug Delivery in Cancer Cells with Red-Light Photoactivated Mesoporous Silica Nanoparticles. Nano Lett. 2013, 13, 2576–2583. [DOI] [PubMed] [Google Scholar]
- (32).Han K; Lei Q; Jia HZ; Wang SB; Yin WN; Chen WH; Cheng SX; Zhang XZ A Tumor Targeted Chimeric Peptide for Synergistic Endosomal Escape and Therapy by Dual‐Stage Light Manipulation. Adv. Funct. Mater 2015, 25, 1248–1257. [Google Scholar]
- (33).Ren Y; Cheung HW; von Maltzhan G; Agrawal A; Cowley GS; Weir BA; Boehm JS; Tamayo P; Karst AM; Liu JF; et al. Targeted Tumor-Penetrating siRNA Nanocomplexes for Credentialing the Ovarian Cancer Oncogene ID4. Sci. Trans. Med 2012, 4, 147ra112–147ra112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wang Y; Xie Y; Li J; Peng Z-H; Sheinin Y; Zhou J; Oupickyý D Tumor-Penetrating Nanoparticles for Enhanced Anticancer Activity of Combined Photodynamic and Hypoxia-Activated Therapy. ACS Nano 2017, 11, 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sugahara KN; Teesalu T; Karmali PP; Kotamraju VR; Agemy L; Greenwald DR; Ruoslahti E Coadministration of A Tumor-Penetrating Peptide Enhances the Efficacy of Cancer Drugs. Science 2010, 328, 1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tang F; Li L; Chen D Mesoporous Silica Nanoparticles: Synthesis, Biocompatibility and Drug Delivery. Adv. Mater 2012, 24, 1504–1534. [DOI] [PubMed] [Google Scholar]
- (37).Liu X; Situ A; Kang Y; Villabroza KR; Liao Y; Chang CH; Donahue T; Nel AE; Meng H Irinotecan Delivery by Lipid-Coated Mesoporous Silica Nanoparticles Shows Improved Efficacy and Safety over Liposomes for Pancreatic Cancer. ACS Nano 2016, 10, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tarn D; Ashley CE; Xue M; Carnes EC; Zink JI; Brinker CJ Mesoporous Silica Nanoparticle Nanocarriers: Biofunctionality and Biocompatibility. Acc. Chem. Res 2013, 46, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Durfee PN; Lin Y-S; Dunphy DR; Munñiz AJ; Butler KS; Humphrey KR; Lokke AJ; Agola JO; Chou SS; Chen I-M; et al. Mesoporous Silica Nanoparticle-Supported Lipid Bilayers (Protocells) for Active Targeting and Delivery to Individual Leukemia Cells. ACS Nano 2016, 10, 8325–8345. [DOI] [PubMed] [Google Scholar]
- (40).Cholewa BD; Liu X; Ahmad N The Role of Polo-Like Kinase 1 in Carcinogenesis: Cause or Consequence? Cancer Res. 2013, 73, 6848–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zhao J; Mi Y; Feng S-S Targeted Co-delivery of Docetaxel and siPlk1 by Herceptin-Conjugated Vitamin E TPGS Based Immunomicelles. Biomaterials 2013, 34, 3411–3421. [DOI] [PubMed] [Google Scholar]
- (42).Xie Y; Wehrkamp CJ; Li J; Wang Y; Wang Y; Mott JL; Oupickyý D Delivery of miR-200c Mimic with Poly (Amido Amine) CXCR4 Antagonists for Combined Inhibition of Cholangiocarcinoma Cell Invasiveness. Mol. Pharm 2016, 13, 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Shibue T; Weinberg RA EMT, CSCs, and Drug Resistance: the Mechanistic Link and Clinical Implications. Nat. Rev. Clin. Oncol 2017, 14, 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cortez MA; Valdecanas D; Zhang X; Zhan Y; Bhardwaj V; Calin GA; Komaki R; Giri DK; Quini CC; Wolfe T; et al. Therapeutic Delivery of miR-200c Enhances Radiosensitivity in Lung Cancer. Mol. Ther 2014, 22, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Smith BN; Bhowmick NA Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med 2016, 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Wang Y; Wang C; Ding Y; Li J; Li M; Liang X; Zhou J; Wang W Biomimetic HDL Nanoparticle Mediated Tumor Targeted Delivery of Indocyanine Green for Enhanced Photodynamic Therapy. Colloids. Surf. B 2016, 148, 533–540. [DOI] [PubMed] [Google Scholar]
- (47).Hao N; Yang H; Li L; Li L; Tang F The Shape Effect of Mesoporous Silica Nanoparticles on Intracellular Reactive Oxygen Species in A375 Cells. N. J. Chem 2014, 38, 4258–4266. [Google Scholar]
- (48).Morry J; Ngamcherdtrakul W; Gu S; Goodyear SM; Castro DJ; Reda MM; Sangvanich T; Yantasee W Dermal Delivery of HSP47 siRNA with NOX4-Modulating Mesoporous Silica-Based Nanoparticles for Treating Fibrosis. Biomaterials 2015, 66, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Morry J; Ngamcherdtrakul W; Gu S; Reda M; Castro DJ; Sangvanich T; Gray JW; Yantasee W Targeted Treatment of Metastatic Breast Cancer by PLK1 siRNA Delivered by An Antioxidant Nanoparticle Platform. Mol. Cancer Ther 2017, 16, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kilchrist KV; Dimobi SC; Jackson MA; Evans BC; Werfel TA; Dailing EA; Bedingfield SK; Kelly IB; Duvall CL Gal8 Visualization of Endosome Disruption Predicts Carrier-mediated Biologic Drug Intracellular Bioavailability. ACS Nano 2019, 13, 1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Thurston TL; Wandel MP; von Muhlinen N; Foeglein Á; Randow F Galectin 8 Targets Damaged Vesicles for Autophagy to Defend Cells against Bacterial Invasion. Nature 2012, 482, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wittrup A; Ai A; Liu X; Hamar P; Trifonova R; Charisse K; Manoharan M; Kirchhausen T; Lieberman J Visualizing Lipid-Formulated siRNA Release from Endosomes and Target Gene Knockdown. Nat. Biotechnol 2015, 33, 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kilchrist KV; Evans BC; Brophy CM; Duvall CL Mechanism of Enhanced Cellular Uptake and Cytosolic Retention of MK2 Inhibitory Peptide Nano-Polyplexes. Cell. Mol. Bioeng 2016, 9, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Rwei AY; Lee J-J; Zhan C; Liu Q; Ok MT; Shankarappa SA; Langer R; Kohane DS Repeatable and Adjustable on-Demand Sciatic Nerve Block with Phototriggerable Liposomes. Proc. Nat. Acad. Sci 2015, 112, 15719–15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Ruskowitz ER; DeForest CA Photoresponsive Biomaterials for Targeted Drug Delivery and 4D Cell Culture. Nat. Rev. Mater 2018, 3, 17087. [Google Scholar]
- (56).Dimri M; Cho J-H; Kang M; Dimri GP PLK1 Inhibition Down-Regulates Polycomb Group Protein BMI1 via Modulation of the miR-200c/141 Cluster. J. Biol. Chem 2015, 290, 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Fu Z; Wen D The Emerging Role of Polo-Like Kinase 1 in Epithelial-Mesenchymal Transition and Tumor Metastasis. Cancers 2017, 9, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yu J; Ohuchida K; Mizumoto K; Sato N; Kayashima T; Fujita H; Nakata K; Tanaka M MicroRNA, hsa-miR-200c, Is an Independent Prognostic Factor in Pancreatic Cancer and Its Upregulation Inhibits Pancreatic Cancer Invasion but Increases Cell Proliferation. Mol. Cancer 2010, 9, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


