Summary
TRIM21 is an interferon‐stimulated E3 ligase that controls the activity of pattern‐recognition signaling via ubiquitination of interferon regulatory factors and DDX41. Previous studies on the role of TRIM21 in innate immune responses have yielded contradictory results, suggesting that the role of TRIM21 is cell specific. Here, we report that bone‐marrow‐derived macrophages (BMDMs) generated from Trim21 −/− mice have reduced expression of mature macrophage markers. Reflecting their reduced differentiation in response to macrophage colony‐stimulating factor (M‐CSF), Trim21 −/− BMDMs had decreased expression of M‐CSF signature genes. Although Trim21 −/− BMDMs responded normally to Toll‐like receptor 9 (TLR9) activation, they produced lower levels of pro‐inflammatory cytokines in response to the TLR2 agonist PAM3CSK4. In line with this, the response to infection with the Bacillus Calmette–Guérin strain of Mycobacterium bovis was also diminished in Trim21 −/− BMDMs. Our results indicate that TRIM21 controls responses to TLR2 agonists.
Keywords: macrophage, Toll‐like receptor 2, TRIM21
Trim21‐deficient bone marrow‐derived macrophages had a reduced response to the Toll‐like receptor 2 agonists PAM3CSK4 and Bacillus Calmette–Guérin. This was reflected by a reduced production of pro‐inflammatory cytokines and iNOS.

Abbreviations
- APC
allophycocyanin
- BCG
Bacillus Calmette–Guérin
- BMDM
bone marrow‐derived macrophage
- ERK1/2
extracellular signal‐regulated kinase 1/2
- FCS
fetal calf serum
- HRP
horseradish peroxidase
- IFN
interferon
- IL‐6
interleukin‐6
- IRF3
interferon regulatory factor 3
- M‐CSF
macrophage colony‐stimulating factor
- PBS
phosphate‐buffered saline
- TLR
Toll‐like receptor
- TRIM
Tripartite‐motif
Introduction
Innate immune responses are under firm control by a large number of negative and positive regulators, enabling strong responses against pathogens while at the same time minimizing tissue damage and promoting timely resolution.1 A group of proteins involved in regulating innate immune responses is the Tripartite‐motif (TRIM) family, comprising > 70 members.2 Many TRIM proteins are E3 ligases that control pattern‐recognition receptor responses via ubiquitination‐dependent degradation or activation of signaling mediators.3, 4 TRIM21 is an interferon (IFN) ‐stimulated E3 ligase that post‐translationally modifies IFN regulatory factor 3 (IRF3), IRF5, IRF7, IRF8 and DDX41 with ubiquitin.5, 6 Trim21 −/− mice develop strong T helper type 17‐mediated contact hypersensitivity responses and have enhanced type I IFN responses against DNA viruses resulting in reduced viral titers.7, 8 TRIM21 has also been shown to be important for IgG‐mediated antiviral responses.9 Intriguingly, several studies on the role of TRIM21 in innate immune responses have yielded contradictory results.7, 8, 9, 10, 11, 12, 13 This may be due to the use of different cell types, different protocols for triggering of innate immune signaling, or to differences between the two commonly used Trim21 −/− strains.11 Trim21 −/− mouse embryonic fibroblasts have strongly increased responses to poly(I:C) and lipopolysaccharide, whereas Trim21 −/− bone‐marrow‐derived macrophages (BMDMs) do not demonstrate enhanced reactivity to the same Toll‐like receptor (TLR) ligands.11 This suggests that the phenotype of Trim21 −/− BMDMs is masked by redundancy or compensatory mechanisms, or that TRIM21 is necessary for proper differentiation of BMDMs.11 To better understand the role of TRIM21 in BMDMs, we have compared gene expression profiles and TLR responses between Trim21 −/− and wild‐type BMDMs.
Material and methods
Cell culture
We isolated bone marrow cells from Trim21 −/− 7 and wild‐type mice on the C57BL/6J background, followed by erythrocyte lysis and differentiation into macrophages. For time–course experiments with PAM3CSK4 and Mycobacterium bovis Bacillus Calmette–Guérin (BCG), we used complete Dulbecco's modified Eagle's medium supplemented with supernatant from L929 cell cultures as previously described.14 In other quantitative RT‐PCR (qRT‐PCR) experiments, and in ELISA and immunoblotting experiments, BMDMs were instead generated using 20 ng/ml recombinant mouse macrophage colony‐stimulating factor (M‐CSF) (R&D Systems, Minneapolis, MN) in complete Dulbecco's modified Eagle's medium. The study was approved by the Ethical Review Committee North, Stockholm County.
Microarray
BMDMs were harvested in cold phosphate‐buffered saline (PBS) and lyzed in RLT buffer (Qiagen, Hilden, Germany). RNA isolation and quality control were performed at the Bioinformatics and Expression Analysis (BEA) facility at Karolinska Institutet, followed by standard protocol for hybridization to Mouse Gene Chip 1·0 ST (Affymetrix, Santa Clara, CA). CEL files from microarrays were preprocessed and normalized with robust multi‐array average using the R package ‘oligo’.15, 16 Expression data were filtered using the R package ‘genefilter’, significance analysis of microarrays was performed using the R package ‘siggenes’, and volcano plots were generated using the R packages ‘ggplot2’, ‘ggrepel’ and ‘gghighlight’. Heat maps for microarray data were generated using the multi experiment viewer (MeV) software.17
Quantitative RT‐PCR
Total RNA was isolated using TRIzol followed by reversed transcription using the iScript cDNA Synthesis kit (Bio‐Rad, Hercules, CA). The qRT‐PCR was performed using the iQ SYBR Green Supermix (Bio‐Rad) or TaqMan assays (ThermoFisher, Waltham, MA). For SYBR based qRT‐PCR, we used the following primers: Nos2‐F CAGCTGGGCTGTACAAACCTT, Nos2‐R CATTGGAAGTGAAGCGTTTCG, Il12b‐F TCATCAGGGACATCATCAAACC, Il12b‐R GTGCTCCAGGAGTCAGGGTACT, Il6‐F GAGGATACCACTCCCAACAGACC, Il6‐R AAGTGCATC‐ATCGTTGTTCATACA, and Hprt‐F CTCATGGACTGATTATGGACAGGAC, Hprt‐R GCAGGTCAGCAAAGAACTTATAGCC. For quantification of Trim21, we used a taqman assay (Mm00447364_m1) targeting Trim21 exons that are deleted in the Trim21 −/− mice (exons 5 and 6), and normalized the expression to Hprt (Mm01545399_m1) (ThermoFisher Scientific).
TLR stimulation experiments
To determine the expression genes by qRT‐PCR, 2 × 106 BMDMs per well were seeded in triplicates for each time‐point. Cells were either infected with M. bovis BCG at a multiplicity of infection of 5, or stimulated with 0·1 µg/ml PAM3CSK4 (Invivogen, San Diego, CA), 1 µg/ml poly(I:C) (Invivogen) or 1 µg/ml CpG‐ODN M362 (Alexis Biochemicals, San Diego, CA) with 100 U/ml IFN‐γ (R&D Systems). Cells were lyzed in TRIzol after 3, 6, 24 and 48 hr, and kept at −80° until total RNA isolation followed by qRT‐PCR. To detect secreted cytokines, 1 × 105 BMDMs were seeded in 48‐well plates and stimulated with 0·1 µg/ml PAM3CSK4 (Invivogen, San Diego, CA) for 24 hr. Supernatants were collected and assayed for interleukin‐6 (IL‐6) and IL‐12‐p40 using the Mouse IL‐12 p40 NonAllele‐specific Quantikine ELISA or Mouse IL‐6 NonAllele‐specific Quantikine ELISA kits (R&D Systems, Minneapolis, MN).
Gene‐set enrichment analysis
Gene‐set enrichment analysis was performed using the GenePattern module (Broad Institute, Cambridge, MA) and visualized using the replotGSEA script in R.18 Gene sets were downloaded from the Molecular Signature Database v5.2 (Broad Institute, Cambridge, MA). We used the following gene signatures for gene‐set enrichment analysis: GSE5099_UNSTIM_VS_MCSF_TREATED_MONOCYTE_DAY7_UP (M‐CSF signature), GSE17721_CTRL_VS_PAM3CSK4_6H_BMDC_UP (PAM3CSK signature) and GSE22935_UNSTIM_VS_12H_MBOVIS_BCG_STIM_MACROPHAGE_UP (BCG signature).
Flow cytometry
For isolation of splenic dendritic cells and macrophages, mouse spleens were perfused with 400 U/ml of collagenase D (Roche, Basel, Switzerland) in Hanks' balanced salt solution and incubated for 45 min at 37° followed by mechanical dissociation. Splenocytes were first incubated with anti‐CD16/32 (Fc‐block) (Biolegend, San Diego, CA) in PBS [1 mm EDTA, 2% fetal calf serum (FCS)] at 4° for 15 min, and were then stained with anti‐CD11c‐allophycocyanin (APC) (BD Biosciences, San Jose, CA) or anti‐F4/80‐APC (BD Biosciences, San Jose, CA) at 4° in PBS with 1 mm EDTA, 2% FCS. The BMDMs were first incubated with anti‐CD16/32 (Fc‐block) (Biolegend, San Diego, CA) in PBS (1 mm EDTA, 2% FCS) at 4° for 15 min. Cells were then stained with the following panel for 30 min at 4° in PBS (1 mm EDTA, 2% FCS): TLR2‐APC (Biolegend, San Diego, CA), CD206‐phycoerythrin/Cy7 (Biolegend, San Diego, CA), CD38‐BV510 (BD Biosciences, San Jose, CA) and F4/80‐APC/Cy7 (Biolegend, San Diego, CA). After washing twice, the cells were acquired using a Gallios flow cytometer (Beckman Coulter, Brea, CA) followed by data analysis using flowjo v10 (FlowJo, Ashland, OR).
Immunoblotting
Cell lysates for immunoblotting were prepared using CelLytic M (Sigma Aldrich, St Louis, MO) supplemented with the Halt™ Protease and Phosphatase Inhibitor Cocktail (ThermoFisher Scientific). Proteins were separated using 4%–20% Mini‐PROTEAN TGX Precast Protein Gels (Bio‐Rad). This was followed by the transfer of proteins to Amersham Hybond polyvinylidene fluoride membranes (GE Healthcare, Chalfont St Giles, UK), and blocking of membranes in 5% non‐fat milk in 0·1% Tween–TBS for 1 hr. For immunoblotting, we used the following antibodies: anti‐extracellular signal‐regulated kinase 1/2 (anti‐ERK1/2; #9102; Cell Signaling Technologies, Danvers, MA), anti‐phospho‐ERK1/2 (#9106; Cell Signaling Technologies). The following secondary antibodies were used: anti‐mouse IgG‐horseradish peroxidase (HRP) (#7076; Cell Signaling Technologies), and anti‐rabbit IgG‐HRP (#7074S; Cell Signaling Technologies). The binding of HRP‐conjugated antibodies was visualized using Clarity Western ECL Substrate (Bio‐Rad). All antibodies were used at concentrations recommended by the manufacturers. Quantification of bands was performed using ImageJ (National Institutes of Health, Bethesda, MA).
Results
Expression of Trim21 in macrophages and dendritic cells
To verify that Trim21 is expressed in macrophages and dendritic cells, we used the EGFP reporter inserted into the Trim21 locus. We used heterozygous Trim21 −/+ mice in flow cytometry and could demonstrate that splenic dendritic cells (CD11c+) and splenic macrophages (F4/80+) both express Trim21 (Fig. 1a,b). To verify that BMDMs generated in vitro also express Trim21, we analyzed EGFP fluorescence by flow cytometry and found that Trim21 −/− BMDMs were EGFP positive (Fig. 1c). To validate the expression of Trim21 with an additional method, we used qRT‐PCR to quantify Trim21 expression in BMDMs and in splenic macrophages (Fig. 1d). By analyzing the expression of Trim21 in mononuclear myeloid cells using a public RNA‐seq data set (GSE122108), we found that the highest expression of Trim21 is in yolk sac macrophages (data not shown).
Figure 1.
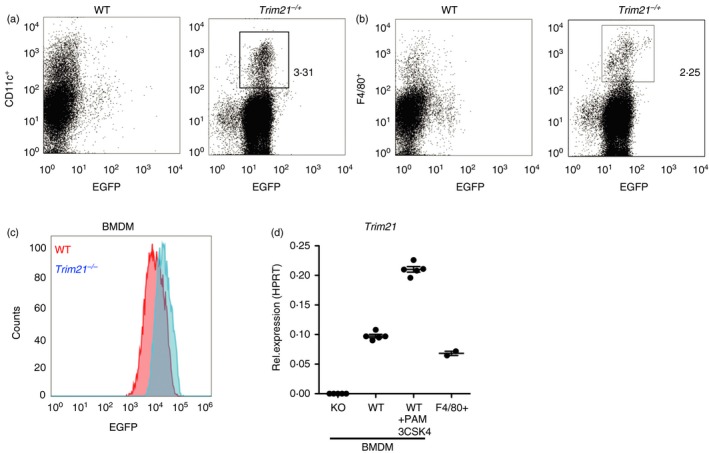
Trim21 expression in macrophages and dendritic cells. Trim21 expression was determined by EGFP fluorescence in splenocytes from wild‐type (WT) and Trim21 −/+ mice stained for CD11c (a) and F4/80 (b). Trim21 expression was determined by EGFP fluorescence in WT and Trim21 −/− bone‐marrow‐derived macrophages (BMDMs) (c). The expression of Trim21 in BMDMs from Trim21 −/− (knockout, KO) and WT mice, and in WT splenic macrophages (F4/80+), was determined by quantitative RT‐PCR (d).
Reduced maturation of Trim21 −/− BMDMs in response to M‐CSF
Members of the IRF transcription factor family are important for proper differentiation and polarization of macrophages.19, 20, 21, 22 As TRIM21 is an E3 ubiquitin ligase for several IRFs, we hypothesized that the maturation of Trim21 −/− BMDMs might be affected. We performed microarray experiments (GeneChip Mouse Gene 1·0 ST Array) to identify changes in gene expression between wild‐type and Trim21 −/− BMDMs. The top up‐regulated and down‐regulated genes are displayed in heat maps (Fig. 2a), and as a volcano plot (Fig. 2b). Analyzing the gene expression data, we found that Trim21 −/− BMDMs had an altered expression of genes typical of mature and undifferentiated (immature) macrophages (Fig. 2c). This included reduced expression of Cd38 (cyclic ADP ribose hydrolase) and the mannose receptor Mrc1 (MRC1/CD206), and increased expression of immature genes such as Cd34, Nanos1 and Id1. However, we did not observe any differences in morphology and cell numbers when comparing the differentiation of wild‐type and Trim21 −/− BMDMs (data not shown). Since minor changes in mRNA levels do not always translate into changes in protein levels, we tried to verify these observations on the protein level using flow cytometry. We could only verify a reduced surface expression of MRC1 in Trim21 −/− BMDMs (Fig. 2d,e), and not for, for example, CD38 (data not shown). We found a reduced M‐CSF signature in Trim21 −/− BMDMs (Fig. 2f), suggesting that Trim21 −/− bone marrow myeloid progenitors have a reduced response to M‐CSF and therefore do not differentiate efficiently to BMDMs in vitro. To test whether Trim21 −/− BMDMs retained the ability to mature, we treated BMDMs with CpG‐ODN and IFN‐γ for 24 hr. The expression of macrophage maturation markers was similar between wild‐type and Trim21 −/− BMDMs after TLR9 activation, suggesting that Trim21 −/− BMDMs matured normally in response to CpG‐ODN (Fig. 2g). Also, treating Trim21 −/− BMDMs with CpG‐ODN and IFN‐γ led to reduced expression of immature markers (Fig. 2h). In all, these results indicate that Trim21 −/− BMDMs have a slightly reduced response to M‐CSF.
Figure 2.
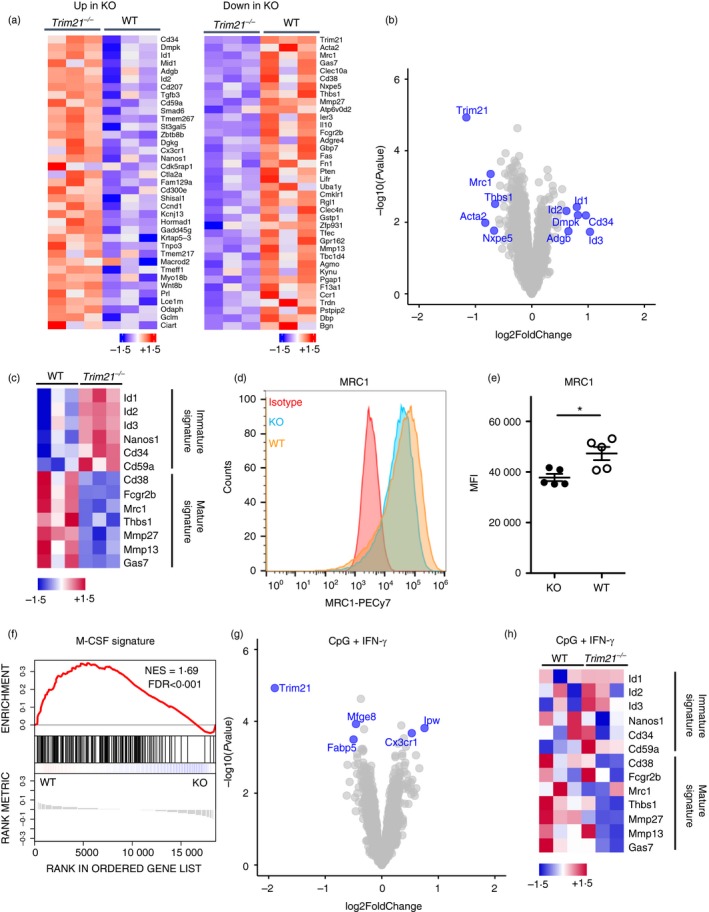
Reduced expression of macrophage maturation genes in Trim21 −/− bone‐marrow‐derived macrophages (BMDMs). Heat map of top up‐regulated and down‐regulated genes in Trim21 −/− (knockout; KO) BMDMs (a). Volcano plot of gene expression in Trim21 −/− BMDMs versus wild‐type (WT) BMDMs (b). Expression of macrophage progenitor and maturation genes in unstimulated WT and Trim21 −/− BMDMs (c). Surface expression of MRC1 (CD206) on BMDMs was determined by flow cytometry (n = 5) (d, e). Gene‐set enrichment analysis of macrophage colony‐stimulating factor (M‐CSF) signature genes in unstimulated WT and Trim21 −/− BMDMs (f). Volcano plot of gene expression in stimulated Trim21 −/− BMDMs versus stimulated WT BMDMs [CpG + interferon‐γ (IFN‐γ), 24 hr] (g). Expression of macrophage progenitor and maturation genes in WT and Trim21 −/− BMDMs stimulated with CpG + IFN‐γ for 24 hr (h). Heat maps display individual expression values as z‐scores. Gene‐set enrichment was calculated as a normalized enrichment score. Statistical testing for flow cytometry data was performed using two‐sided Mann–Whitney test (*P < 0·05). Multiple testing was corrected using false‐discovery rate calculated based on gene‐set permutations.
Trim21 −/− BMDMs are hypo‐responsive to stimulation with the TLR2 ligand PAM3CSK4
To identify altered TLR signaling pathways in Trim21 −/− BMDMs, we compared tonic TLR signaling between Trim21 −/− and wild‐type BMDMs. Tonic TLR signaling occurs as the result of bacterial contaminants in serum, as previously reported.23, 24, 25 By comparing the expression of TLR‐induced genes at baseline, we discovered a reduced TLR2‐activation (PAM3CSK4) gene signature in Trim21 −/− BMDMs (Fig. 3a). This suggested that TRIM21 is necessary for optimal responses to TLR2 agonists. To verify this, we generated Trim21 −/− and wild‐type BMDMs and stimulated them with the synthetic TLR2 agonist PAM3CSK4 for 6 hr. Indeed, the induction of Il6 (IL6) and Nos2 (iNOS) was reduced in Trim21 −/− BMDMs after stimulation with PAM3CSK4 (Fig. 3b). We then generated new Trim21 −/− and wild‐type BMDMs and stimulated them with PAM3CSK4 for between 3 and 48 hr and found a clearly reduced response in Trim21 −/− BMDMs at all time‐points (Fig. 3c). The secretion of IL‐6 and IL‐12‐p40 was also diminished in stimulated Trim21 −/− BMDMs as determined by ELISA (Fig. 3d). Furthermore, the ratio of phosphorylated ERK1/2 (p‐ERK1/2) to total ERK1/2 was reduced in stimulated Trim21 −/− BMDMs (Fig. 3e). The reduced response to TLR2 activation was not to the result of lower expression of TLR2 at the cell surface (Fig. 3f,g). In contrast, the TLR3 agonist poly(I:C) induced similar expression of Nos2 and Il6 in wild‐type and Trim21 −/− BMDMs (Fig. 3h). The decreased response of Trim21 −/− BMDMs in response to PAM3CSK4 was of a similar magnitude to what have been reported after knockdown of other E3 ligases.26, 27 In all, these data indicate that Trim21 −/− BMDMs have a deficient response to TLR2 agonists.
Figure 3.
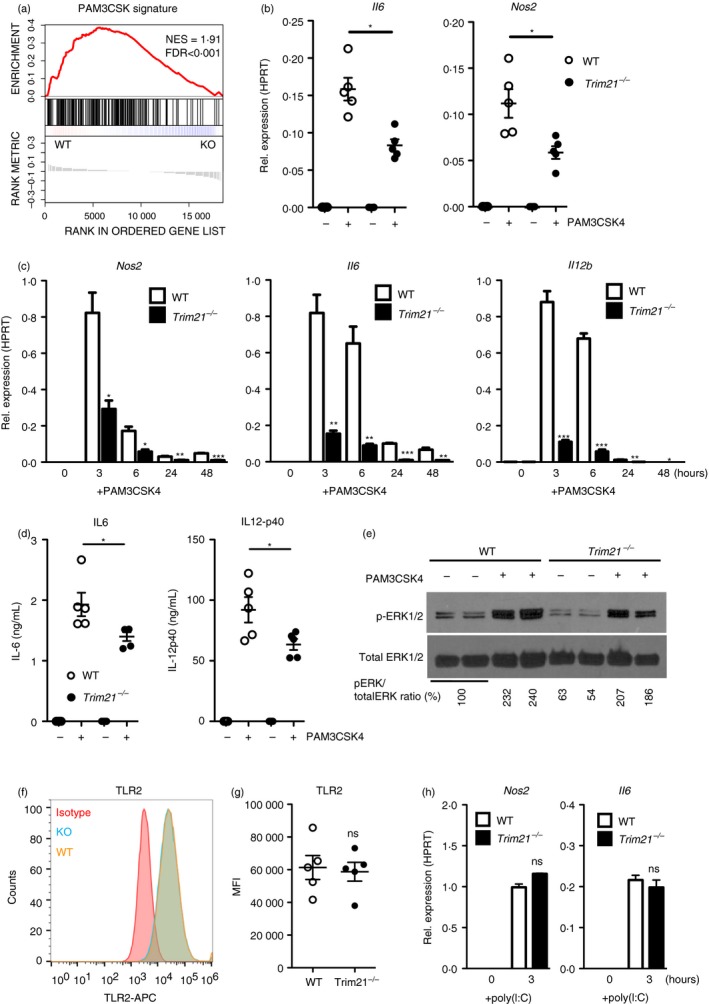
Trim21 −/− bone‐marrow‐derived macrophages (BMDMs) are hyporesponsive to stimulation with PAM3CSK4. Gene‐set enrichment analysis of Toll‐like receptor 2 (TLR2) ‐activation (PAM3CSK4) signature genes in Trim21 −/− (knockout; KO) and wild‐type (WT) BMDMs at baseline (a). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 6 hr followed by quantitative RT‐PCR against Nos2 (iNOS) and Il6 (IL‐6) (n = 5) (b). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 3–48 hr followed by quantitative RT‐PCR against Nos2 (iNOS), Il6 (IL‐6) and Il12b (IL‐12‐p40) (n = 3) (c). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 24 hr, followed by ELISA to measure secreted IL‐6 and IL‐12‐p40 protein (n = 5) (d). Immunoblotting against phosphorylated EKR1/2 (p‐ERK1/2) and total ERK1/2 in BMDMs stimulated with 0·1 µg/ml PAM3CSK4 for 6 hr (e). Trim21 −/− and WT BMDMs were stimulated with 1·0 µg/ml poly(I:C) for 3 hr followed by quantitative RT‐PCR against Nos2 and Il6 (n = 3) (f). Surface expression of TLR2 on BMDMs was determined by flow cytometry (n = 5) (g, h). Statistics for gene expression data were calculated using the Student's t‐test, and statistical testing for flow cytometry data was performed using two‐sided Mann–Whitney test (*P < 0·05, **P < 0·01, ***P < 0·005). The experiment was performed three times with similar results. Gene‐set enrichment was calculated as a normalized enrichment score. Multiple testing was corrected using false‐discovery rate calculated based on gene‐set permutations.
Trim21 −/− BMDMs are hyporesponsive to stimulation with BCG
PAM3CSK4 is a synthetic triacylated lipopeptide that mainly activates TLR2; therefore, we asked whether TRIM21 was also necessary for full responses to more complex stimuli. To this end, we compared the responses of Trim21 −/− and wild‐type BMDMs infected with M. bovis BCG. By comparing the expression of BCG‐induced genes at baseline, we discovered a reduced BCG‐activation gene signature in Trim21 −/− BMDMs (Fig. 4a). Trim21 −/− BMDMs infected with BCG showed reduced expression of Il12b, Il6 and Nos2 transcripts compared with wild‐type BMDMs (Fig. 4b‐d).
Figure 4.
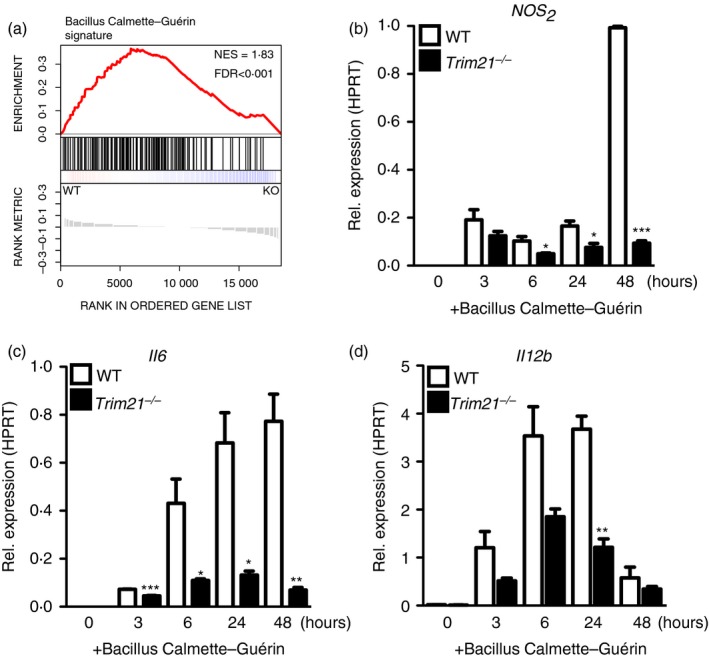
Trim21 −/− bone‐marrow‐derived macrophages (BMDMs) are hyporesponsive to Mycobacterium bovis Bacillus Calmette–Guérin (BCG). Gene‐set enrichment analysis of BCG‐activation signature genes in Trim21 −/− and wild‐type (WT) BMDMs at baseline (a). Trim21 −/− and WT BMDMs were stimulated with M. bovis BCG (multiplicity of infection = 5) for 3–48 hr followed by quantitative RT‐PCR against Nos2 (iNOS), Il6 (IL‐6) and Il12b (IL‐12‐p40) (b–d). Statistics were calculated using the Student's t‐test. The experiment was performed twice with similar results (n = 3) (*P < 0·05, **P < 0·01, ***P < 0·005). Gene‐set enrichment was calculated as a normalized enrichment score. Multiple testing was corrected using false‐discovery rate calculated based on gene‐set permutations.
Discussion
The role of TRIM21 in innate immune responses is complex and depends on cell type and the nature of the stimulatory signal. Several studies on the cellular function of TRIM21 have been performed in immortalized cell lines, warranting further studies using primary immune cells. Here we investigated the role of TRIM21 in mouse BMDMs and found that Trim21 −/− BMDMs have a reduced response to M‐CSF. Furthermore, Trim21 −/− BMDMs were hyporesponsive to TLR2 stimulation by both the synthetic ligand PAM3CSK4 and M. bovis BCG, indicating that TRIM21 also plays an important role in response to more complex stimuli. Our results fit well with data from Foltz et al.,28 who identified TRIM21 as a critical factor in the defense against the obligate intracellular parasite Toxoplasma gondii. Since both M. bovis BCG and T. gondii activate TLR2 signaling,29 it is possible that TRIM21 is an important factor for the defense against intracellular parasites that trigger TLR2 signaling. The exact mechanism by which TRIM21 controls TLR2 signaling remains unclear. The E3 ligase activity of TRIM21 might enhance TLR2 signaling by promoting the activity of TLR2 signaling mediators, or by degrading negative regulators of TLR2 signaling. Interestingly, a TLR2‐specific E3 ubiquitin ligase (PPP1R11) has recently been described, demonstrating that TLR2 signaling can directly be controlled by ubiquitination.30 The change in cytokine production after TLR2 activation of Trim21 −/− BMDMs is of a similar moderate magnitude to changes reported after knockdown of other E3 ligases, including TRIM1326 and CHIP.27 This is expected, because E3 ligases are modulators of signaling rather than necessary components of the signaling pathways. Importantly, previous reports have also found that TRIM21 activates TAK1,9, 31 demonstrating that TRIM21 can directly control TLR signaling. There are no added antibodies in our experimental set up, demonstrating that the observed effect of TRIM21 deficiency is antibody independent.
In all, our results indicate that TRIM21 is involved in regulating innate immune responses against TLR2 agonist including lipoproteins, peptidoglycans and lipoteichoic acids. The role of TRIM21 in TLR2 responses in vivo, and the exact molecular mechanism, remain to be investigated.
Author contributions
MS designed experiments and performed cell stimulations and gene expression analysis with Affymetrix microarray and quantitative RT‐PCR; BC designed experiments, and performed cell stimulations and infections with BCG; WN performed quantitative RT‐PCR analysis and immunoblotting on BMDMs. RC performed analysis of gene expression data; MR designed experiments; AE designed experiments and wrote the manuscript.
Disclosure
The authors have no conflicts of interest.
Acknowledgements
This work was supported by the Karolinska Institutet, the Swedish Research Council, Cancerfonden, and Konung Gustaf V:s och Drottning Victorias Frimurarestiftelse. We acknowledge Professor Marie Wahren‐Herlenius for support and critical reading of the manuscript.
References
- 1. Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol 2009; 9:692–703. [DOI] [PubMed] [Google Scholar]
- 2. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al The tripartite motif family identifies cell compartments. EMBO J 2001; 20:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McNab FW, Rajsbaum R, Stoye JP, O'Garra A. Tripartite‐motif proteins and innate immune regulation. Curr Opin Immunol 2011; 23:46–56. [DOI] [PubMed] [Google Scholar]
- 4. Meroni G, Diez‐Roux G. TRIM/RBCC, a novel class of 'single protein RING finger' E3 ubiquitin ligases. BioEssays 2005; 27:1147–57. [DOI] [PubMed] [Google Scholar]
- 5. Oke V, Wahren‐Herlenius M. The immunobiology of Ro52 (TRIM21) in autoimmunity: a critical review. J Autoimmun 2012; 39:77–82. [DOI] [PubMed] [Google Scholar]
- 6. Sjostrand M, Ambrosi A, Brauner S, Sullivan J, Malin S, Kuchroo VK et al Expression of the immune regulator tripartite‐motif 21 is controlled by IFN regulatory factors. J Immunol 2013; 191:3753–63. [DOI] [PubMed] [Google Scholar]
- 7. Espinosa A, Dardalhon V, Brauner S, Ambrosi A, Higgs R, Quintana FJ et al Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL‐23‐Th17 pathway. J Exp Med 2009; 206:1661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double‐stranded DNA. Nat Immunol 2013; 14:172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody‐bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 2013; 14:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vinter H, Langkilde A, Ottosson V, Espinosa A, Wahren‐Herlenius M, Raaby L et al TRIM21 is important in the early phase of inflammation in the imiquimod‐induced psoriasis‐like skin inflammation mouse model. Exp Dermatol 2017; 26:713–20. [DOI] [PubMed] [Google Scholar]
- 11. Ozato K, Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC 3rd. Comment on "Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF‐κB‐dependent cytokine expression in fibroblasts". J Immunol 2009; 183:7619; author reply 720–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S et al TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol 2009; 182:3782–92. [DOI] [PubMed] [Google Scholar]
- 13. Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN‐β production post‐pathogen recognition by polyubiquitin‐mediated degradation of IRF3. J Immunol 2008; 181:1780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carow B, Gao Y, Teran G, Yang XO, Dong C, Yoshimura A et al CISH controls bacterial burden early after infection with Mycobacterium tuberculosis in mice. Tuberculosis 2017; 107:175–80. [DOI] [PubMed] [Google Scholar]
- 15. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010; 26:2363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N et al TM4: a free, open‐source system for microarray data management and analysis. Biotechniques 2003; 34:374–8. [DOI] [PubMed] [Google Scholar]
- 18. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chistiakov DA, Myasoedova VA, Revin VV, Orekhov AN, Bobryshev YV. The impact of interferon‐regulatory factors to macrophage differentiation and polarization into M1 and M2. Immunobiology 2018; 223:101–11. [DOI] [PubMed] [Google Scholar]
- 20. Tsujimura H, Nagamura‐Inoue T, Tamura T, Ozato K. IFN consensus sequence binding protein/IFN regulatory factor‐8 guides bone marrow progenitor cells toward the macrophage lineage. J Immunol 2002; 169:1261–9. [DOI] [PubMed] [Google Scholar]
- 21. Tamura T, Nagamura‐Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 2000; 13:155–65. [DOI] [PubMed] [Google Scholar]
- 22. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N et al IRF5 promotes inflammatory macrophage polarization and TH1‐TH17 responses. Nat Immunol 2011; 12:231–8. [DOI] [PubMed] [Google Scholar]
- 23. Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Molinaro R, Mukherjee T, Flick R, Philpott DJ, Girardin SE. Trace levels of peptidoglycan in serum underlie the NOD‐dependent cytokine response to endoplasmic reticulum stress. J Biol Chem 2019; 294:9007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H et al Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008; 4:28–39. [DOI] [PubMed] [Google Scholar]
- 26. Huang B, Baek SH. Trim13 potentiates toll‐like receptor 2‐mediated nuclear factor κB activation via K29‐linked polyubiquitination of tumor necrosis factor receptor‐associated factor 6. Mol Pharmacol 2017; 91:307–16. [DOI] [PubMed] [Google Scholar]
- 27. Yang M, Wang C, Zhu X, Tang S, Shi L, Cao X et al E3 ubiquitin ligase CHIP facilitates Toll‐like receptor signaling by recruiting and polyubiquitinating Src and atypical PKCζ . J Exp Med 2011; 208:2099–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foltz C, Napolitano A, Khan R, Clough B, Hirst EM, Frickel EM. TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP‐positive parasite vacuoles. Sci Rep 2017; 7:5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mun HS, Aosai F, Norose K, Chen M, Piao LX, Takeuchi O et al TLR2 as an essential molecule for protective immunity against Toxoplasma gondii infection. Int Immunol 2003; 15:1081–7. [DOI] [PubMed] [Google Scholar]
- 30. McKelvey AC, Lear TB, Dunn SR, Evankovich J, Londino JD, Bednash JS et al RING finger E3 ligase PPP1R11 regulates TLR2 signaling and innate immunity. eLife 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sokolova O, Kahne T, Bryan K, Naumann M. Interactome analysis of transforming growth factor‐β‐activated kinase 1 in Helicobacter pylori‐infected cells revealed novel regulators tripartite motif 28 and CDC37. Oncotarget 2018; 9:14366–81. [DOI] [PMC free article] [PubMed] [Google Scholar]


