Abstract
Members of the family Filoviridae produce variously shaped, often filamentous, enveloped virions containing linear non-segmented, negative-sense RNA genomes of 15–19 kb. Several filoviruses (e.g., Ebola virus) are pathogenic for humans and are highly virulent. Several filoviruses infect bats (e.g., Marburg virus), whereas the hosts of most other filoviruses are unknown. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on Filoviridae, which is available at www.ictv.global/report/filoviridae.
Keywords: Filoviridae, filovirus, ICTV Report, ebolavirus, marburgvirus, taxonomy
Virion
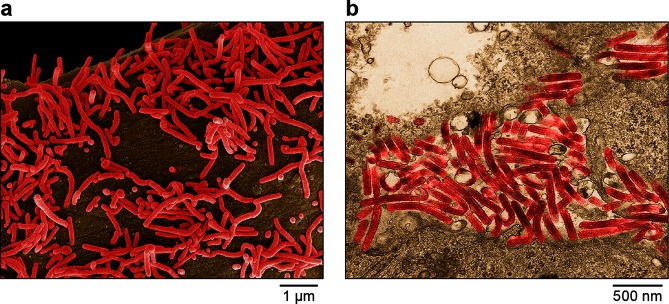
Virions are enveloped and diverse in shape and can appear as branched, toroid, U- or 6-shaped, and long filamentous forms (Table 1, Fig. 1). Virions contain ribonucleoprotein (RNP) complexes composed of genomic RNA and, typically, the structural proteins nucleoprotein (NP), polymerase co-factor (VP35), transcriptional activator (VP30), RNP-associated protein (VP24) and RNA-dependent RNA polymerase (L). The matrix protein (VP40) forms a regular layer beneath the viral envelope. Surface spikes formed by glycoproteins (GP1,2) are approximately 7 nm in diameter and cover the virion surface at approximately 10-nm intervals [1–3]. Some filoviruses do not have discernable glycoproteins and may have different RNP complexes [4].
Table 1. Characteristics of members of the family Filoviridae .
| Typical member: | Marburg virus (DQ217792), species Marburg marburgvirus, genus Marburgvirus |
|---|---|
| Virion | Enveloped, variously shaped but predominantly filamentous, typically with a single nucleocapsid |
| Genome | Approximately 15–19 kb of linear, negative-sense, non-segmented RNA |
| Replication | Antigenomic RNA is a replication intermediate. The genome and antigenome form ribonucleoprotein complexes, which serve as templates |
| Translation | From multiple 5′-capped and 3′-polyadenylated mRNAs |
| Host range | Primates (e.g., ebolaviruses, marburgviruses), bats (e.g., marburgviruses), domestic pigs (e.g., Reston virus) and probably fish (e.g., striaviruses, thamnoviruses) become naturally infected |
| Taxonomy | Realm Riboviria, phylum Negarnaviricota, subphylum Haploviricotina, class Monjiviricetes, order Mononegavirales; family includes multiple genera |
Fig. 1.
Electron microscopic images of Marburg virus particles budding from infected Vero E6 cells, (a) scanning EM, (b) transmission EM. Images are colourized for clarity. Courtesy of John G. Bernbaum and Jiro Wada, IRF-Frederick.
Genome
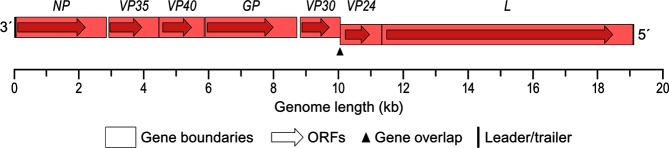
Filovirus genomes are approximately 15–19 kb (Fig. 2) without a 5′-cap or 3′-poly(A). Terminal leader and trailer sequences contain the replication and transcription promoters. Marburgvirus genomes contain seven separate, continuous open reading frames (ORFs) flanked by 3′- and 5′-terminal non-coding regions that contain transcription initiation and termination sites. These ORFs encode the virion structural proteins. Cuevavirus and ebolavirus genomes encode homologues of the marburgvirus structural proteins. However, the marburgvirus GP gene encodes only GP1,2, whereas the primary expression product of cuevavirus and ebolavirus GP gene transcription is a soluble glycoprotein. Co-transcriptional editing is used to express GP1,2 and additional non-structural proteins. Striaviruses and thamnoviruses encode some, but not all, marburgvirus protein homologues and several proteins of unknown function [4, 5].
Fig. 2.
Schematic representation of the genome organization of Marburg virus. Courtesy of Jiro Wada, IRF-Frederick.
Replication
Virus proteins are translated from mRNAs that are synthesized by successive, polar transcription from RNP complexes containing genomic RNA. Replication occurs in the cytoplasm through the synthesis of RNP complexes containing antigenomes that are templates for genomic RNA production. Replication and transcription enzymes include L and VP35. VP30 serves as a transcription enhancer for ebolaviruses and probably cuevaviruses, but its function in marburgvirus infection is less defined. Virion assembly, including acquisition of the GP1,2-containing lipid envelope, occurs by VP40-mediated budding at the plasma membrane [4, 5].
Taxonomy
Filoviruses form a family in the haploviricotine order Mononegavirales. Within this order, filoviruses are most closely related to members of the families Paramyxoviridae, Pneumoviridae and Sunviridae. The family Filoviridae includes multiple genera for viruses that differ in geographic and host range and genomic organization.
Resources
Full ICTV Report on the family Filoviridae: www.ictv.global/report/filoviridae.
Funding information
Production of this summary, the online chapter and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA). This work was supported in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I (J.H.K.). This project was funded in part with federal funds from the National Cancer Institute, National Institutes of Health under Contract No. HHSN261200800001E. This work was also funded in part under Contract No. HSHQDC-15-C-00064 awarded by Department of Homeland Security (DHS S&T) for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a federally funded research and development center (V.W.). The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of the Army, the US Department of Defense, the US Department of Health and Human Services, or of the institutions and companies affiliated with the authors. In no event shall any of these entities have any responsibility or liability for any use, misuse, inability to use or reliance upon the information contained herein. The US departments do not endorse any products or commercial services mentioned in this publication.
Acknowledgements
Members of the ICTV (10th) Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Jens H. Kuhn. We thank Laura Bollinger (IFR-Frederick) for critically editing the manuscript and the associated report chapter.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: L, RNA-dependent RNA polymerase; NP, nucleoprotein; RNP, ribonucleoprotein.
References
- 1.Bharat TAM, Riches JD, Kolesnikova L, Welsch S, Krähling V, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M. Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 Å resolution. Nature. 2018;563:137–140. doi: 10.1038/s41586-018-0630-0. [DOI] [PubMed] [Google Scholar]
- 3.Kirchdoerfer RN, Wasserman H, Amarasinghe GK, Saphire EO. Filovirus structural biology: the molecules in the machine. Curr Top Microbiol Immunol. 2017;411:381–417. doi: 10.1007/82_2017_16. [DOI] [PubMed] [Google Scholar]
- 4.Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 5.Brauburger K, Deflubé LR, Mühlberger E. Filovirus transcription and replication. In: Pattnaik AK, Whitt MA, editors. Biology and Pathogenesis of Rhabdo- and Filoviruses. Singapore: World Scientific Publishing; 2015. pp. 515–555. (editors) [Google Scholar]