Abstract
The family Caliciviridae includes viruses with single-stranded, positive-sense RNA genomes of 7.4–8.3 kb. The most clinically important representatives are human noroviruses, which are a leading cause of acute gastroenteritis in humans. Virions are non-enveloped with icosahedral symmetry. Members of seven genera infect mammals (Lagovirus, Norovirus, Nebovirus, Recovirus, Sapovirus, Valovirus and Vesivirus), members of two genera infect birds (Bavovirus and Nacovirus), and members of two genera infect fish (Minovirus and Salovirus). This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Caliciviridae, which is available at ictv.global/report/caliciviridae.
Keywords: ICTV Report, taxonomy, Caliciviridae, norovirus
Virion
Calicivirus virions are 27–40 nm in diameter, non-enveloped with icosahedral symmetry (Table 1). The capsid is composed of 90 dimers of the major structural protein VP1 arranged on a T=3 icosahedral lattice (Fig. 1) [1]. Caliciviruses are characterised by a capsid architecture with 32 distinct cup-shaped depressions. Generally, caliciviruses are stable in the environment and enteric caliciviruses are acid-stable.
Table 1.
Characteristics of members of the family Caliciviridae
|
Typical member: |
Norwalk virus (M87661), species Norwalk virus, genus Norovirus |
|---|---|
|
Virion |
Non-enveloped with icosahedral symmetry, 27–40 nm in diameter |
|
Genome |
Single-stranded, positive-sense genomic RNA of 7.4–8.3 kb, with a 5′-terminal virus protein, genome-linked (VPg) and 3′-terminal poly(A) |
|
Replication |
Cytoplasmic |
|
Translation |
From genome-sized (non-structural proteins) and 3′-terminal subgenomic (structural proteins) mRNAs |
|
Host range |
Mammals (Lagovirus, Norovirus, Nebovirus, Recovirus, Sapovirus, Valovirus and Vesivirus), birds (Bavovirus, Nacovirus), fish (Minovirus, Salovirus) |
|
Taxonomy |
Realm Riboviria; more than ten genera |
Fig. 1.

The structure of the calicivirus capsid exemplified by a cryo-image reconstruction of recombinant Norwalk virus-like particles (left). X-ray structure of the Norwalk virus capsid (right) with the shell, protruding 1 and protruding 2 domains shown in blue, red and yellow, respectively. (Courtesy of B. V. Prasad.)
Genome
Caliciviruses have a single-stranded, positive-sense genomic RNA of 6.4–8.5 kb organized into either two or three major ORFs, while a further ORF4 of murine norovirus encodes virulence factor 1 (VF1). A protein [virus protein, genome-linked (VPg), 10–15 kDa] is covalently linked to the 5′-terminus of genomic RNAs, which are also polyadenylated at their 3′-termini (Fig. 2). Genus-specific conserved nucleotide motifs are found at the 5′-terminus of ORF1 and at the junction of the coding sequences for the non-structural/structural proteins.
Fig. 2.
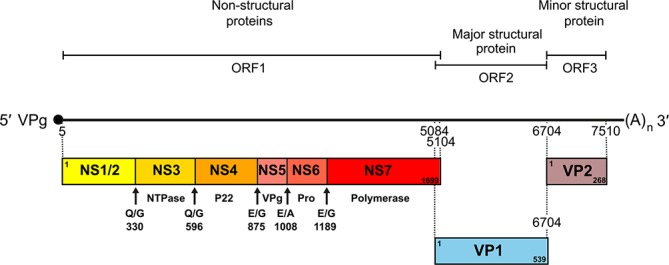
Genome organization of human calicivirus MD-145 (AY032605, 7556 nt, genus Norovirus). Protein VPg is covalently linked to the 5′-end of genomic RNA and is depicted by a black circle. Cleavage sites in the ORF1 polyprotein are indicated by arrows; the flanking residues and amino acid coordinates are indicated, although these vary within the family. Pro: protease.
Replication
Replication of caliciviruses occurs in the cytoplasm in complexes on intracellular membranes by a VPg-mediated translation initiation process unique to the virus family and that uses genomic positive-sense RNA as the template to translate a large polyprotein that undergoes post-translational cleavage by a virus-encoded protease to form at least six mature non-structural proteins (NS1/2, NS3, NS4, NS5, NS6 and NS7) [2]. Subgenomic-sized, positive-sense RNA, co-terminal with the 3′- terminus of the genome, is the template for translation of VP1 as well as the 3′-terminal ORF product VP2 [3]. A dsRNA corresponding in size to full-length genomic RNA has been identified in feline calicivirus, murine norovirus and San Miguel sea lion virus-infected cells, indicating that replication occurs via a negative-sense intermediate. All caliciviruses require VPg; some require the function of eIF4E (feline calicivirus, porcine sapovirus), and some do not (murine norovirus).
Pathogenicity
Caliciviruses cause species-specific infections, with most noroviruses, sapoviruses and neboviruses restricted to the gastro-intestinal tract; some lagoviruses, saloviruses and vesiviruses cause severe systemic infections in their natural hosts.
Taxonomy
Viruses of seven genera (Lagovirus, Norovirus, Nebovirus, Recovirus [4], Sapovirus, Valovirus [5] and Vesivirus) infect a wide range of mammals, members of two genera infect birds (Bavovirus [6] and Nacovirus [7]) and members of two genera infect fish (Minovirus [8] and Salovirus [9]), while caliciviruses have also been detected in the greater green snake and frogs [10], highlighting the wide host range of viruses in the family. Caliciviruses are similar to picornaviruses in the presence of VPg and in sequence similarity of their RNA-directed RNA polymerase and protease proteins.
Resources
Current ICTV Report on the family Caliciviridae: ictv.global/report/caliciviridae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Members of the ICTV Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Nick J. Knowles.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: VPg, virus protein, genome-linked.
References
- 1.Prasad BVV, Hardy ME, Dokland T, Bella J, Rossmann MG. X-Ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 2.Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in norovirus biology. Cell Host Microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asanaka M, Atmar RL, Ruvolo V, Crawford SE, Neill FH, et al. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc Natl Acad Sci U S A. 2005;102:10327–10332. doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farkas T, Sestak K, Wei C, Jiang X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae . J Virol. 2008;82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L’Homme Y, Sansregret R, Plante-Fortier Étienne, Lamontagne A-M, Ouardani M, et al. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae . Virus Genes. 2009;39:66–75. doi: 10.1007/s11262-009-0360-3. [DOI] [PubMed] [Google Scholar]
- 6.Wolf S, Reetz J, Otto P. Genetic characterization of a novel calicivirus from a chicken. Arch Virol. 2011;156:1143–1150. doi: 10.1007/s00705-011-0964-5. [DOI] [PubMed] [Google Scholar]
- 7.Day JM, Ballard LL, Duke MV, Scheffler BE, Zsak L. Metagenomic analysis of the turkey gut RNA virus community. Virol J. 2010;7:313. doi: 10.1186/1743-422X-7-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mor SK, Phelps NBD, Ng TFF, Subramaniam K, Primus A, et al. Genomic characterization of a novel calicivirus, FHMCV-2012, from baitfish in the USA. Arch Virol. 2017;162:3619–3627. doi: 10.1007/s00705-017-3519-6. [DOI] [PubMed] [Google Scholar]
- 9.Mikalsen AB, Nilsen P, Frøystad-Saugen M, Lindmo K, Eliassen TM, et al. Characterization of a novel calicivirus causing systemic infection in atlantic salmon (Salmo salar L.): proposal for a new genus of Caliciviridae . PLoS One. 2014;9:e107132. doi: 10.1371/journal.pone.0107132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi M, Lin X-D, Chen X, Tian J-H, Chen L-J, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]


