Abstract
The family Artoviridae was created in 2018 for the established monospecific genus Peropuvirus and six new species of invertebrate viruses that had all been discovered by high-throughput sequencing. Artoviruses have negative-sense RNA genomes of about 12 kb and produce enveloped, spherical particles that are 100–130 nm in diameter. Hosts include parasitoid wasps, barnacles, pillworms, woodlice, copepods and odonates. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Artoviridae, which is available at www.ictv.global/report/artoviridae.
Keywords: ICTV Profile, taxonomy, Artoviridae
Virion
Virions of Pteromalus puparum negative-strand RNA virus 1 are enveloped and spherical [1] with a diameter of 100–130 nm (Table 1, Fig. 1). Virion information is not available for other artoviruses, which are only known from genomic sequence data [2, 3].
Table 1.
Characteristics of members of the family Artoviridae
|
Typical member: |
Pteromalus puparum negative-strand RNA virus 1 (KX431032), species Pteromalus puparum peropuvirus, genus Peropuvirus |
|---|---|
|
Virion |
Enveloped, spherical particles, 100–130 nm in diameter |
|
Genome |
Negative-sense, unsegmented RNA of about 12 kb |
|
Replication |
Nuclear: the RNA-directed RNA polymerase engages with the ribonucleoprotein complex at the genome 3′-end |
|
Translation |
Individual, putatively polyadenylated mRNAs are translated in the cytoplasm |
|
Host range |
Barnacles, copepods, odonates, parasitoid wasps, pillworms, woodlice |
|
Taxonomy |
Realm Riboviria, phylum Negarnaviricota, subphylum Haploviricotina, class Monjiviricetes, order Mononegavirales. The genus Peropuvirus includes several species |
Fig. 1.
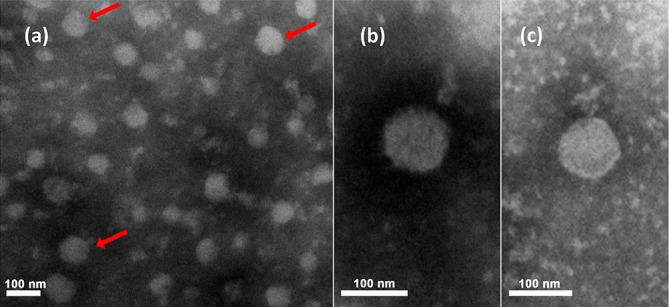
Electron micrographs of purified Pteromalus puparum negative-strand RNA virus 1 particles from follicular cells. (a) Electron micrographs of purified particles. (b) and (c) are magnifications from (a). Red arrows indicated viral particles. Reproduced from [1].
Spherically shaped Pteromalus puparum negative-strand RNA virus 1 particles are present in follicular cells of the ovaries of infected parasitoid wasps (Fig. 1). Similarly, virion-like particles stacked in intracellular vesicles have been observed in cells of the digestive tract. The spherical particles of Pteromalus puparum negative-strand RNA virus 1 are similar to particles produced by nyamiviruses [1, 4].
Genome
Artovirus negative-sense RNA genomes are of up to 12 kb. All known artoviruses have unsegmented genomes (Fig. 2). The Pteromalus puparum negative-strand RNA virus 1 genome contains five large, independently transcribed, non-overlapping open reading frames (ORFs) encoding three hypothetical proteins of unknown function (U1, U2 and U3), a putative glycoprotein (G) and a large protein (L) encoding an RNA-directed RNA polymerase domain. The genome 3′-leader and 5′-trailer regions have terminal nucleotides that do not exhibit obvious complementarity. Typical conserved transcription initiation and termination motifs are identified upstream and downstream, respectively, of each putative ORF.
Fig. 2.
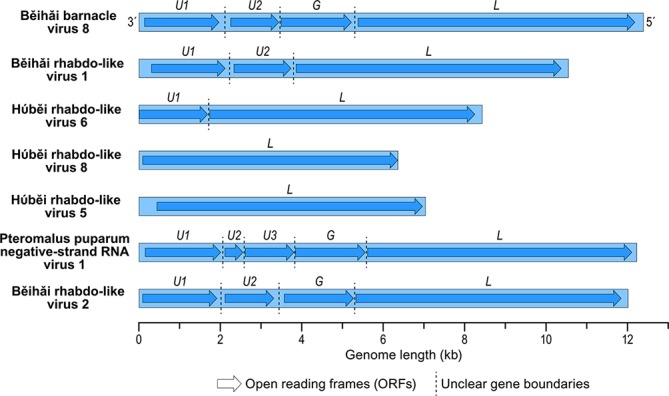
Artovirus genome organization. Some virus genome sequences may be incomplete.
Replication
Knowledge of artovirus replication is limited.
Pathogenicity
Pteromalus puparum negative-strand RNA virus 1 was originally isolated from a laboratory parasitoid strain of a pteromalid wasp [Pteromalus puparum (Linnaeus, 1758)]. The virus is present in various tissues and life stages of the parasitoid and is transmitted vertically through females and males. Virus infection increases adult longevity and impairs several fitness parameters of the wasp, but has no influence on successful parasitism, although it moderates the offspring sex ratio by decreasing female offspring numbers [1].
Taxonomy
Artoviruses form a family in the haploviricotine order Mononegavirales [5]. Within this order, artoviruses are most closely related to members of the families Bornaviridae, Lispiviridae, Mymonaviridae and Nyamiviridae. Like most other mononegaviruses, artoviruses (i) have unsegmented, negative-sense RNA genomes; (ii) encode proteins with high sequence identity; (iii) have five conserved motifs (A–E) in the amino acid sequence of their RNA-directed RNA polymerases; (iv) produce enveloped virions; and (v) do not rely on cap-snatching but instead cap their own mRNAs.
Resources
Full ICTV Report on the family Artoviridae: www.ictv.global/report/artoviridae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors. This work was funded in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under contract no. HHSN272200700016I (J. H. K.).
Acknowledgements
Members of the ICTV (10th) Report Consortium are Elliot J. Lefkowitz, Andrew J. Davison, Stuart G. Siddell, Peter Simmonds, Sead Sabanadzovic, Donald B. Smith, Richard J. Orton and Jens H. Kuhn.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Wang F, Fang Q, Wang B, Yan Z, Hong J, et al. A novel negative-stranded RNA virus mediates sex ratio in its parasitoid host. PLoS Pathog. 2017;13:e1006201. doi: 10.1371/journal.ppat.1006201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li CX, Shi M, Tian JH, Lin XD, Kang YJ, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 4.Dietzgen RG, Ghedin E, Jiāng D, Kuhn JH, Song T, et al. ICTV virus taxonomy profile: Nyamiviridae . J Gen Virol. 2017;98:2914–2915. doi: 10.1099/jgv.0.000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, et al. Taxonomy of the order Mononegavirales: update 2019. Arch Virol. 2019;161 doi: 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


