Abstract
A few studies have highlighted the importance of the respiratory microbiome in modulating the frequency and outcome of viral respiratory infections. However, there are insufficient data on the use of microbial signatures as prognostic biomarkers to predict respiratory disease outcomes. In this study, we aimed to evaluate whether specific bacterial community compositions in the nasopharynx of children at the time of hospitalization are associated with different influenza clinical outcomes. We utilized retrospective nasopharyngeal (NP) samples (n=36) collected at the time of hospital arrival from children who were infected with influenza virus and had been symptomatic for less than 2 days. Based on their clinical course, children were classified into two groups: patients with mild influenza, and patients with severe respiratory or neurological complications. We implemented custom 16S rRNA gene sequencing, metagenomic sequencing and computational analysis workflows to classify the bacteria present in NP specimens at the species level. We found that increased bacterial diversity in the nasopharynx of children was strongly associated with influenza severity. In addition, patients with severe influenza had decreased relative abundance of Staphylococcus aureus and increased abundance of Prevotella (including P. melaninogenica), Streptobacillus, Porphyromonas, Granulicatella (including G. elegans), Veillonella (including V. dispar), Fusobacterium and Haemophilus in their nasopharynx. This pilot study provides proof-of-concept data for the use of microbial signatures as prognostic biomarkers of influenza outcomes. Further large prospective cohort studies are needed to refine and validate the performance of such microbial signatures in clinical settings.
Keywords: microbiome, influenza, biomarker, children, emergency unit, intensive care
Introduction
Seasonal influenza in children is a major burden on public health. It is estimated that between 20 to 30 % of children are infected with influenza each winter (WHO). While most of the children develop uncomplicated influenza illness, between 1.5 to 4 % of infected children are hospitalized for influenza complications [1] including pneumonia, asthma exacerbations, neurological complications and death [2]. Risk factors for hospital admission in children include age <2 years and having underlying medical conditions (such as neurological disorders, prematurity, immunosuppression, diabetes, or sickle cell disease) [3]. However, about half of paediatric hospitalizations from seasonal influenza occur in children without a high-risk medical condition [4].
From a clinical perspective, failure to identify influenza-infected patients that have a higher risk of developing severe disease delays the delivery of appropriate treatment, and may have profound consequences for the recovery and long-term health of the patient. It has been reported, in an adult population, that a delay of 1 day from onset of symptoms to hospital admission increased the risk of death from influenza-caused disease by 5.5 % [5]. Several attempts have been made to develop prognostic indicators that help to identify patients at risk of severe influenza disease progression. Influenza viral load is not predictive of clinical outcome [6], but increased serum levels for several cytokines in adults infected with 2009 H1N1 influenza virus are correlated with disease progression [7], and increased nasal levels of two proinflammatory cytokines are associated with severe influenza [6]. However, at present, there are no biomarkers readily available to the physician at the time of hospital admission that are able to predict the likelihood of disease progression in all patients.
Improving severe influenza prognosis and treatment requires a better understanding of influenza disease. Influenza severity is determined by a complex interplay between viral, environmental and host factors, which is not yet fully understood and which cannot be comprehended without looking at the system as a whole [8]. One factor that has been poorly studied and needs to be integrated in influenza pathogenesis models is the microbiome, defined as the microbial communities colonizing the human body (microbiota) together with the surrounding environment [9, 10]. Secondary bacterial pneumonia is a well-known cause of pulmonary complications resulting from influenza infection, and was reported as a major cause of mortality during the 1918 pandemic [11]. However, the idea that the microbiota might also be involved in influenza pathogenesis is a recent concept. Leung et al. recently reported that the oropharyngeal microbiota of a pool of patients with H1N1 pneumonia was different from that of patients with non-H1N1 pneumonia [12]. A second metagenomic study observed that patients infected with H1N1 had a higher amount of Proteobacteria in the upper respiratory tract than was observed in previously published normal reference nasopharyngeal (NP) microbial profiles [13]. This study also showed a variation in NP microbiota depending on patient age. However, sequencing was performed on pools of anonymized samples with no associated clinical data and therefore could not link specific changes in microbiota composition with different influenza outcomes.
In addition, several studies using mice have shown that the gut microbiota protects against influenza disease. Antibiotic-treated mice exhibit impaired antiviral immune responses following influenza virus infection, resulting in delayed viral clearance [14], while oral administration of probiotics in mice prior to influenza infection reduces viral titres [15]. Finally, a recent mouse study showed that Staphylococcus aureus colonization in the upper respiratory mucosa attenuated influenza infection compared with germ-free mice [16]. However, germ-free mice may not be appropriate to study the role of the microbiome in influenza, as they have important physiological defects, such as altered immune systems, which can perturb responses to influenza infection (reviewed in [17]). Further, there are significant differences between the microbiomes of mice and humans [17].
Because of the heterogeneity of the respiratory microbiome in humans, and the low proportion of severe cases in seasonal influenza, large prospective studies are needed to link the respiratory microbiome with influenza severity in patients. As a proof-of-concept, we utilized NP specimens from a retrospective collection of influenza-positive samples from children infected with influenza virus with different disease outcomes, mild or severe, and tested whether specific bacterial species in the nasopharynx of patients at the time of hospitalization correlated with influenza disease severity. We determined a differential microbial signature discriminating patients with severe and mild influenza. Such a signature could be used as a prognostic biomarker for early diagnosis of severe influenza.
Results
Selected patients
We profiled the microbial communities of 36 NP samples collected at the time of hospital arrival from patients classified retrospectively into two groups based on their clinical course: patients with mild influenza (n=22) or severe influenza (n=14) outcomes. Patients had been symptomatic for less than 2 days before hospital admission. None of the patients received antibiotics for up to 3 weeks before sample collection. After sampling, severe patients were treated pre-emptively. The demographics and clinical parameters of the 36 patients are summarized in Table 1 (baseline characteristics for Table 1a and follow-up characteristics for Table 1b). Among the patients who developed severe influenza, 4 patients had severe respiratory complications requiring ventilatory support and hospitalization in an intensive care unit (ICU), and 10 patients had neurological complications requiring hospitalization in the ICU (n=6 from 10; 60 %) or in other units. The most common neurological complications were coma (n=4 from 10; 40 %) and epilepsy (n=5 from 10; 50 %). The average time of hospitalization was similar among the two severe groups (7.4 and 9.8 days for the patients with neurological and respiratory complications, respectively). Patients with mild influenza were either discharged or hospitalized in emergency short-stay units. Their median time of hospitalization of 1 day corresponds to a monitoring of more than 6 h in emergency service, which is considered in France to be a hospitalization. Because of the insufficiently large number of patients included, no age- and sex-matched analyses could be performed on this cohort with adequate power. As the respiratory microbiome changes with age and sex [18], our results have been adjusted for these parameters in the rest of the study using multivariate analysis (see the Methods section). Finally, cycle threshold (Ct) values from influenza real-time RT-PCR, as described in the Methods section, were used as a proxy measure of viral load and were not significantly different between the groups, suggesting that there is no relationship between initial viral load and disease outcome, as previously described [6].
Table 1.
(a) Baseline demographic and clinical characteristics of all patients included in the study and (b) clinical evolution and therapeutic management of all patients included in the study
| (a) | ||||
|---|---|---|---|---|
| Baseline demographics and clinical characteristics | Mild influenza | Severe influenza | ||
| Neurological | Respiratory | |||
| Baseline demographics | No. of patients | 22 | 10 | 4 |
| Sex : male | 7 (31.8 %) | 4 (40 %) | 3 (75 %) | |
| Median age in months (Q1–Q3) | 3 (2–4) | 16.5 (15–27.8) |
17.5 (4.8–56.8) |
|
| Characteristics at the time of admission | Sample nature : NP aspirate | 17 (77.3 %) | 8 (80 %) | 4 (100 %) |
| Sample nature : NP swab | 5 (22.7 %) | 2 (20 %) | 0 (0 %) | |
| Days since symptom onset : 0 | 6 (27.3 %) | 2 (20 %) | 0 (0 %) | |
| Days since symptom onset : 1 | 12 (54.5 %) | 4 (40 %) | 1 (25 %) | |
| Days since symptom onset : 2 | 4 (18.2 %) | 4 (40 %) | 3 (75 %) | |
| Influenza B virus | 3 (13.6 %) | 2 (20 %) | 1 (25 %) | |
| Influenza A virus (H1N1) | 6 (27.3 %) | 4 (40 %) | 1 (25 %) | |
| Influenza A virus (H3N2) | 12 (54.5 %) | 4 (40 %) | 2 (50 %) | |
| Median influenza RT-PCR Ct (Q1–Q3) | 20 (18.4–23.5) | 21.5 (21–23.5) |
28.5 (23.8–34.6) |
|
| Comorbidities*: neurological disorder | 0 (0 %) | 2 (20 %) | 0 (0 %) | |
| Comorbidities* : other (not known as a risk factor for severe influenza) | 1 (4.5 %) | 0 (0 %) | 3 (75 %) | |
| Comorbidities : none | 21 (95.5 %) | 8 (80 %) | 1 (25 %) | |
| (b) | ||||
| Outcome | Hospitalized in ICU | 0 (0 %) | 6 (60 %) | 4 (100 %) |
| Median days hospitalized (Q1–Q3) | 1 (0–3) | 3 (2 - 3) | 8 (7–10.8) | |
| Median days hospitalized in ICU (Q1–Q3) | 0 (0–0) | 2 (0–3) | 7.5 (4.8–10.8) | |
| Ventilation | 0 (0 %) | 3 (30 %) | 4 (100 %) | |
| Signs of respiratory distress† | 6 (27.3 %) | 0 (0 %) | 4 (100 %) | |
| Bacterial pneumonia complicating influenza | 0 (0 %) | 1 (10 %) | 1 (25 %) | |
| Coma | 0 (0 %) | 4 (40 %) | 0 (0 %) | |
| Encephalitis | 0 (0 %) | 2 (20 %) | 0 (0 %) | |
| Epilepsy | 0 (0 %) | 5 (50 %) | 0 (0 %) | |
| Multiple or prolonged seizure | 0 (0 %) | 5 (50 %) | 0 (0 %) | |
| Death | 0 (0 %) | 0 (0 %) | 0 (0 %) | |
| Treatment | No antiviral | 20 (90.9 %) | 3 (30 %) | 0 (0 %) |
| Antiviral : aciclovir | 0 (0 %) | 3 (30 %) | 0 (0 %) | |
| Antiviral : Tamiflu | 2 (9.1 %) | 4 (40 %) | 4 (100 %) | |
| No antibiotic | 20 (90.9 %) | 7 (70 %) | 1 (25 %) | |
| Antibiotherapy | 2 (9.1 %) | 3 (30 %) | 3 (75 %) | |
*None of these comorbidities were previously associated with changes in NP microbiota composition. Neurological disorder: viral encephalitis sequelae and tuberous sclerosis of Bourneville. Other comorbidities: Down's syndrome for one patient in the mild influenza group and one patient in the respiratory complication group; Prader–Willi syndrome for one patient in the respiratory complication group; myopathy for one patient in the respiratory complication group.
†Signs of respiratory distress included: modification of breathing rate, increased heart rate, colour changes, grunting, nose flaring, retractions, sweating wheezing, stridor, accessory muscle use, or changes in alertness.
NP microbiota composition of children infected with influenza virus
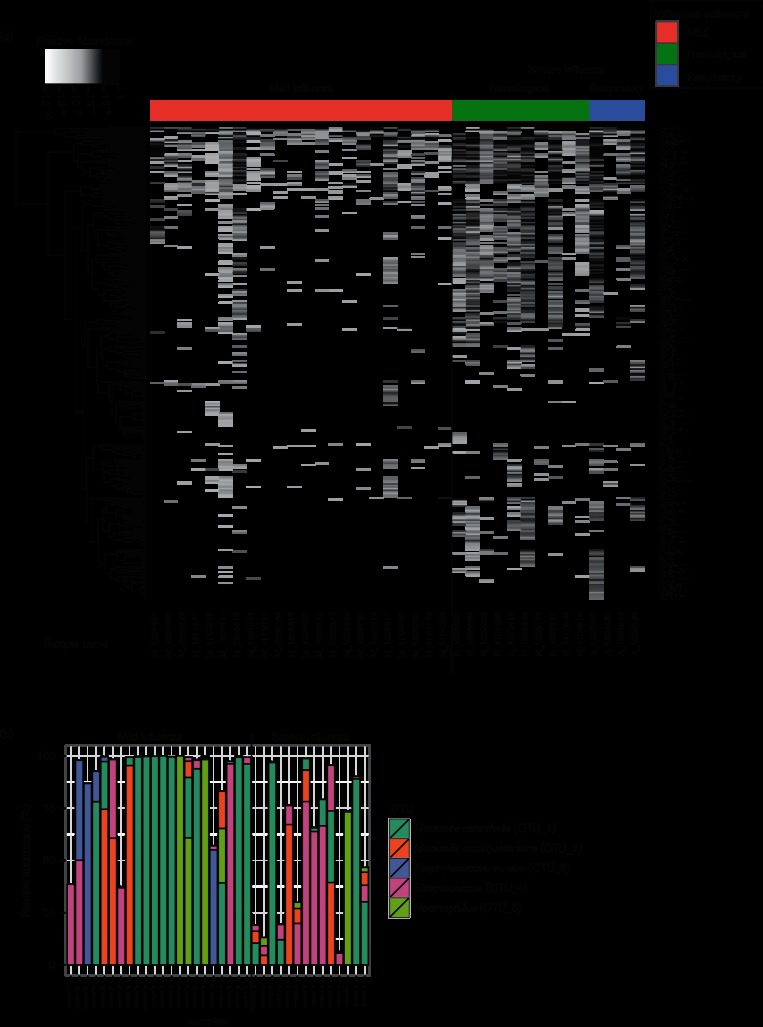
Across all 36 respiratory samples, >5.2 million high-quality 16S RNA sequences were classified into 300 operational taxonomic units (OTUs), with 101 OTUs supported by >1000 reads each (Fig. S1, available in the online Supplementary Material). All OTUs were classified up to the genus level and 49 % were classified up to the species level. The relative abundance is given for all 300 OTUs in each sample (Fig. 1a). Five OTUs dominated the NP bacterial profiles (defined as a relative abundance >50 % in at least one sample). Among these five OTUs, the genera were Moraxella, Staphylococcus, Streptococcus and Haemophilus (Fig. 1b). Moraxella catharrhalis was found as the dominant species in 11 out of 36 samples (30 %). Moraxella nonliquefaciens was also detected and was the dominant species in four samples. Staphylococcus aureus was the dominant species in two samples. OTU_4 was only classified to the genus level – Streptococcus – and was the dominant species in five samples. Finally, OTU_5 (Haemophilus) was the dominant OTU in four samples. In the remaining 10 (28 %) samples, there was more than one dominant OTU: for three samples, there were two major OTUs (with a relative abundance of between 25 and 50 %), and for seven samples the bacterial profiles were more complex. Importantly, the presence of a dominant OTU was not significantly associated with influenza outcomes after adjustment with covariates (Table S1). To validate the bacteria taxonomy classifications inferred from the 16S ribosomal profiling, we performed shotgun metagenomic sequencing on a subset of NP specimens obtained from patients with mild (n=10) and severe (n=10) influenza. Comparable bacterial compositions were found in the 16S targeted sequencing and shotgun metagenomic datasets (Fig. S2).
Fig. 1.
NP microbiota composition associated with each respiratory sample. (a) heat map displaying the relative abundance of the 300 OTUs detected in each NP sample. Relative abundance is shown with a black to white gradient scale, with OTUs that were not quantified in white, and OTUs with a relative abundance of 1 in black. The samples were ordered based on patient group and sample number. OTUs were clustered based on their relative abundance values across samples using Euclidian distances and a complete linkage function. (b) Bar plot showing relative abundance of the five most abundant OTUs in each sample, coloured by OTUs. The samples were ordered as in (a).
NP microbiota composition differentiates influenza outcomes
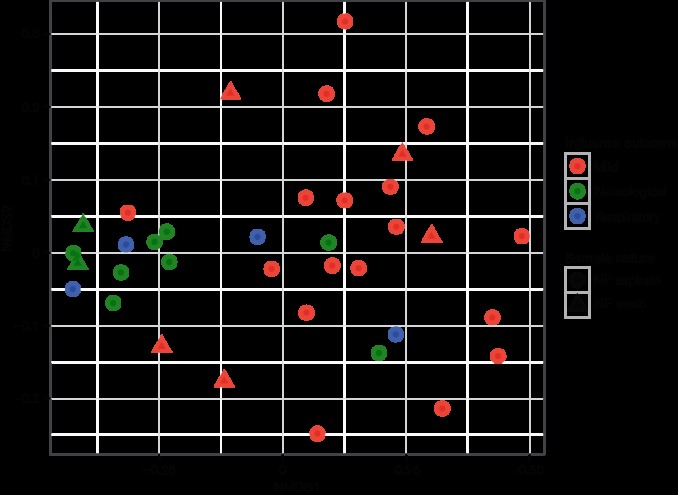
To examine whether variations in the composition of NP bacterial communities were related to the clinical and/or demographic characteristics of the patients, a UniFrac distance matrix was calculated and used in PerMANOVA analyses against five clinical variables (clinical outcome, days between sampling and symptom onset, patient’s age, patient’s sex and nature of sample). This analysis revealed that differences in NP microbiota composition were strongly associated with influenza outcomes (P-value=0.00025) (Fig. 2 and Table S2). Similarly, unsupervised hierarchical clustering of the samples based on whole-microbiota profiles, was able to segregate mild and severe cases, with all but one patient with mild influenza belonging in the first cluster, and all but four patients with severe influenza belonged in the second cluster (Fig. S3). The age of the patients was also associated with NP microbiota composition in PerMANOVA analysis (P-value=0.03450), but less strongly than influenza outcome. There was no evidence of significant interaction between age and clinical outcome in terms of an effect on NP microbiota composition (Table S2). Finally, we tested whether there was a difference in microbiota composition between patients with respiratory or neurological complications. After adjusting for covariates, the type of severe influenza complication was not significantly associated with whole-microbiota profiles (Table S3).
Fig. 2.
The NP microbiota composition differentiates influenza severity. Non-metric multidimensional scaling (NMDS) plot comparing the global NP microbial profiles with distances calculated using Unifrac distance. Each dot represents a sample: samples with similar microbial profiles are close together in the NMDS plot, while increasing distances in the plot between samples suggest divergent microbial profiles. Patients’ groups are depicted with different colours, and the sample natures (NP aspirate or swab) with different shapes. Stress=0.1102
Patients with mild influenza have a less diverse NP microbiota than patients that develop severe influenza
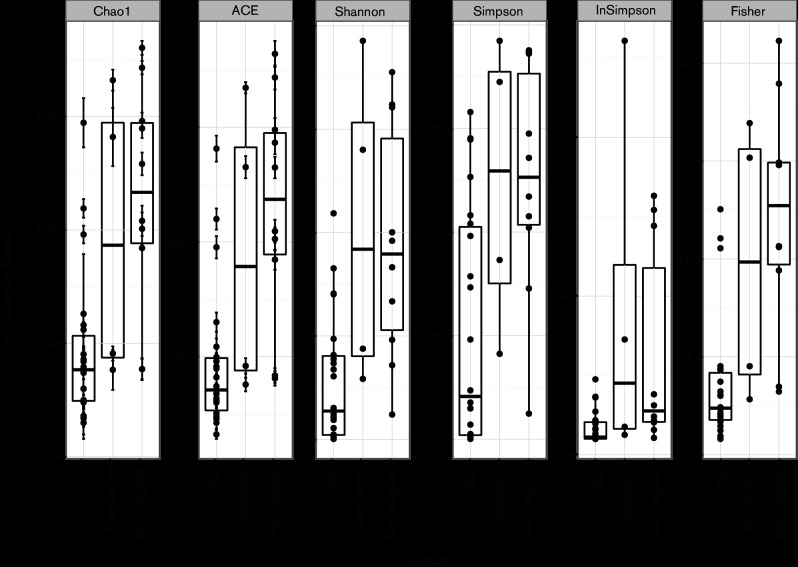
We further tested whether NP microbiota diversity (alpha diversity) was also associated with the patients’ demographics and/or clinical features. Several diversity indices were calculated (Fig. 3) and were further used in statistical analysis to examine whether species diversity in NP samples were related to five variables (clinical outcome, days between sampling and symptom onset, patient’s age, patient’s sex, nature of sample). Using multiway ANOVA, we found that NP microbiota diversity, as estimated by the Shannon diversity index (H), ACE, Chao1, Fisher and Simpson indices, was strongly associated with influenza outcome (Table S4). Simpson’s reciprocal index was the only one that was not significantly associated with influenza outcome (Table S4). This observation could be linked to the size of the cohort, with this index being the most sensitive to a lack of power. Patients with mild influenza had significantly lower bacterial diversity (average H=0.57) than patients with severe influenza (average H=2.03). The NP microbiota diversity was weakly associated with patient age (Table S4). There was no evidence of significant interaction between age and clinical outcome, in terms of an effect on NP microbiota diversity (Table S4). In addition, there was no significant difference in microbiota diversity between patients with neurological and respiratory complications (Table S5). Overall, we found that increased bacterial diversity in the upper respiratory tract was associated with influenza severity.
Fig. 3.
Patients developing severe influenza have more diverse NP bacterial communities than patients with mild influenza. Box plots showing different alpha diversity measures for the three groups of patients (mild, neurological complications and respiratory complications).
Differential microbial abundance between patients with mild or severe influenza
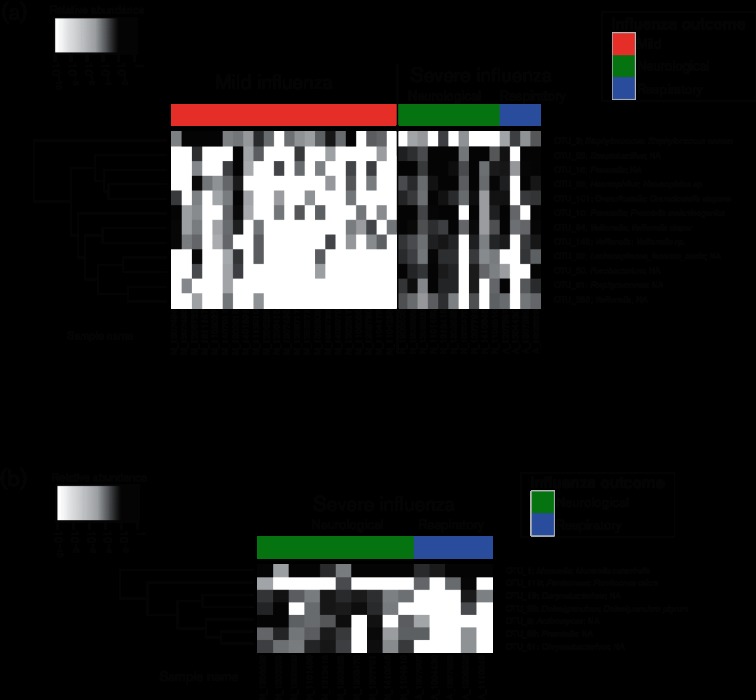
Statistical analysis to characterize the differential microbial species abundance between patients with mild influenza and patients with severe influenza was performed using a generalized linear model, adjusting for age, sex, time since symptom onset and sample nature. Twelve OTUs were found to be differentially abundant between the two groups of patients (Fig. 4a). The average abundance in each group for the 12 differential OTUs is given Table 2. Among the differential OTUs, one was among the five most abundant OTUs detected in the respiratory microbiome profiles: Staphylococcus aureus was more abundant in patients with mild influenza than in patients with severe influenza (Table 2). The remaining 11 differential OTUs had limited abundance (<3 % on average) and were all more abundant in patients with severe influenza. The genera of these 11 OTUs were: Prevotella for two of them, Streptobacillus, Porphyromonas and Veillonella for three of them, and Fusobacterium, Haemophilus, Lachnospiracea incertae sedis and Granulicatella. Some of these OTUs were detected almost exclusively in patients with severe influenza, for example Porphyromonas (OTU_31) was detected in 11 (76 %) patients with severe influenza, but only 2 (9 %) patients with mild influenza.
Fig. 4.
Bacterial species that were differentially abundant between patients with mild and severe influenza (a) and between patients with respiratory and neurological complications. (b) The heat map shows the relative abundance of 12 OTUs that were found to be differentially abundant between patients with mild or severe disease after adjustment for covariates (P-value <0.05). Relative abundance is shown with a black to white gradient scale, with OTUs that were not quantified in white, and OTUs with a relative abundance of 1 in black. The samples were ordered based on patient group and sample number. OTUs were clustered based on their relative abundance values across samples using Euclidian distances and a complete linkage function.
Table 2.
Taxonomy and average relative abundance for the OTUs differentially abundant between mild and severe influenza groups after adjusting for covariates (age, sex, sample nature and time since symptom onset)
| Genus | Species | Average relative abundance in mild influenza (%) | Average relative abundance in severe influenza (%) | P-value | |
|---|---|---|---|---|---|
| OTU_3 | Staphylococcus | Staphylococcus aureus | 9.4164 | 0.0094 | 0.0136 |
| OTU_10 | Prevotella | Prevotella melaninogenica | 0.2967 | 2.3501 | 0.0381 |
| OTU_16 | Prevotella | na | 0.3789 | 2.1213 | 0.0011 |
| OTU_22 | Streptobacillus | na | 0.2686 | 2.0337 | 0.0021 |
| OTU_31 | Porphyromonas | na | 0.0001 | 0.3730 | 0.0220 |
| OTU_64 | Veillonella | Veillonella dispar | 0.0527 | 0.6623 | 0.0234 |
| OTU_80 | Fusobacterium | na | 0.0040 | 0.3361 | 0.0082 |
| OTU_85 | Haemophilus | na | 0.0404 | 0.6245 | 0.0065 |
| OTU_92 | Lachnospiracea_incertae_sedis | na | 0.0358 | 0.0954 | 0.0381 |
| OTU_101 | Granulicatella | Granulicatella elegans | 0.0621 | 0.2280 | 0.0082 |
| OTU_148 | Veillonella | na | 0.0381 | 0.1316 | 0.0409 |
| OTU_355 | Veillonella | na | 0.0003 | 0.0152 | 0.0442 |
In addition, we performed a similar statistical analysis on the severe influenza group only, to determine whether there were differences between patients with neurological and respiratory complications. Seven OTUs (Moraxella catarrhalis, Actinomyces, Corynebacterium, Dolosigranulum pigrum, Chryseobacterium, Prevotella and Parvimonas micra) were found to be differentially abundant (Fig. 4b and Table 3). Except for Moraxella catarrhalis and Parvimonas micra, these OTUs were significantly more abundant in neurological severe forms than in respiratory severe forms. As already described, Moraxella catarrhalis remain abundant in both severe forms (17.39 % vs 29.89 %), unlike all other OTUs, with limited abundance in both groups (<1 % in average).
Table 3.
Taxonomy and average relative abundance for the seven OTUs that were differentially abundant between patients with respiratory and neurological complications after adjustment for covariates (age, sex, nature of sample, time since onset of symptoms)
| Genus | Species | Average relative abundance in patients with neurological complications (%) | Average relative abundance in patients with respiratory complications (%) | P-value | |
|---|---|---|---|---|---|
| OTU_1 | Moraxella | Moraxella catarrhalis | 17.3854 | 29.8941 | 0.0249 |
| OTU_9 | Actinomyces | na | 0.7323 | 0.0001 | 0.0107 |
| OTU_19 | Corynebacterium | na | 0.1472 | 0.0627 | 0.0118 |
| OTU_28 | Dolosigranulum | Dolosigranulum pigrum | 0.3227 | 0.0094 | 0.0425 |
| OTU_61 | Chryseobacterium | na | 0.1308 | 0.0004 | 0.0124 |
| OTU_89 | Prevotella | na | 0.1176 | 0.0040 | 0.0301 |
| OTU_113 | Parvimonas | Parvimonas micra | 0.0037 | 0.2264 | 0.0424 |
Minimal microbial signatures classify mild versus severe cases
To determine whether a smaller subset of OTUs could efficiently classify patients into mild and severe groups, we used an association rule mining algorithm among differentially abundant OTUs. To test the signatures’ predictive performance, we used the trained rules to determine the influenza disease severity of patients excluded from the training set [leave-one-out cross-validation (LOOCV)]. In addition, only rules that were found in all cross-validation instances were considered to be robust. This analysis revealed 5 robust association rules comprising two to three OTUs able to classify mild versus severe patients with cross-validated sensitivity of up to 93 % and specificity of up to 100 % (Table 4). For instance, the quantification of OTU_22 (Streptobacillus) at a relative abundance higher than 8.8e-06 and the quantification of OTU_85 (Haemophilus) at a relative abundance higher than 4.8e-05 predict that the patient will develop a severe influenza with a sensitivity of 92.86 % and a specificity of 90.91 %. The performance of these different rules needs to be validated in prospective cohort studies, but these data indicate that microbial signatures comprising very few bacterial species have the potential to predict influenza outcomes with substantial accuracy.
Table 4.
LOOCV performance characteristics of minimal microbial signatures in discriminating patients with mild versus severe influenza outcomes
Only the rules predicted in all cross-validation instances are displayed in this table. Sensitivity and specificity were calculated using out-of-sample predictions. The rules are sorted based on specificity values, and are placed in descending order.
| Patient classified as severe influenza if the following OTUs are detected (with relative abundance) | Sensitivity | Specificity |
|---|---|---|
| OTU_16 (>7.6e-05)&OTU_22 (>8.8e-06)&OTU_148 (>3.8e-05) | 78.57 | 100 |
| OTU_22 (>8.8e-06)&OTU_101 (>1.3e-05)&OTU_148 (>3.8e-05) | 78.57 | 95.45 |
| OTU_22 (>8.8e-06)&OTU_85 (>4.8e-05) | 92.86 | 90.91 |
| OTU_22 (>8.8e-06)&OTU_101 (>1.3e-05) | 85.71 | 90.91 |
| OTU_22 (>8.8e-06)&OTU_64 (>5.2e-05) | 85.71 | 90.91 |
Altogether, these results demonstrate that differences in NP microbiota composition early after infection are associated with influenza severity and provide proof-of-concept data for the use of microbial signatures as prognostic biomarkers of influenza outcomes in a clinical setting.
Discussion
Influenza virus transmission and pathogenesis in humans is unpredictable at the population and individual level in terms of clinical severity. While rapidly identifying patients at risk of influenza complications is crucial for patient management and survival, there is currently no host biomarker assay available to predict influenza severity at the time of infection. In this study, we used a retrospective collection of early NP samples from children infected with influenza virus to test the feasibility of using a prognostic microbial signature to predict influenza disease outcome in a clinical setting. By comparing the relative abundance and diversity of microbial populations in the upper respiratory tract of children at the time of hospitalization, we identified bacterial profiles that differentiated children that developed severe disease versus non-progressive mild influenza infection using five different diversity indexes. All but one showed statistical differences between these two groups. This difference was not significant for Simpson’s reciprocal index, the most dependent on cohort size (as reviewed in [19]). To our knowledge, this is the first study associating influenza severity with NP bacterial composition in children.
There are a limited number of studies describing the composition of the respiratory tract microbiota in healthy and diseased children. Studies profiling the NP microbiota have reported that healthy children younger than 2 years old have their upper respiratory tract colonized by one predominant commensal bacterial genera consisting of Moraxella, Haemophilus, Streptococcus, Prevotella, Dolosigranulum, Staphylococcus, Corynebacterium, Flavobacteria or Neisseria [18, 20–22]. Similarly to these studies, we found that most of the children infected with influenza had microbial compositions in their nasopharynx that were dominated by one bacterial species, with Moraxella, Streptococcus, Haemophilus and Staphylococcus representing the most abundant bacterial genera. However, the nasopharynx microbiomes of children infected with influenza were not colonized with Gram-positive commensal species (Anoxybacillus, Dolosigranulum, Corynebacterium and Lactococcus). It is possible that influenza virus induced a decrease in the abundance of these species early after infection. Interestingly, these Gram-positive commensal species have been shown to control the abundant colonization of pathobionts such as H. influenza, S. pneumonia, S. aureus, or M. catarrhalis and enhanced risk of acquiring acute otitis media [21, 23], pneumonia and bronchiolitis [24]. In future studies, it will be interesting to follow the respiratory microbiome dynamics longitudinally prior and post-influenza infection to characterize early virus–microbiome interactions.
The most common genus found in our population was Moraxella, with two main species, M. catarrhalis and M. nonliquefaciens. High abundance of M. nonliquefaciens was previously associated with viral pneumonia caused by adenovirus, rhinovirus or enterovirus, respiratory syncytial virus, or human metapneumovirus infections [20]. In addition, Moraxella versus Alloiococcus colonization is associated with increased respiratory syncytial virus infections and severity [22]. As we could not profile the NP microbiota of healthy children in parallel with influenza-infected children, we could not determine whether Moraxella colonization was also associated with influenza infection, but we found that M. catarrhalis and M. nonliquefaciens relative abundance were not predictive of influenza severity after adjustment for covariates, but were predictive of neurological versus respiratory complications. Further evaluation of the effect of specific virus–microbiome interactions is necessary to better define the respiratory microbiota and its influence on clinical disease outcomes for acute respiratory infections in children.
In our study, severe influenza infection was predicted by increased bacterial diversity in the nasopharynx of children. In addition, a microbial signature of 12 OTUs was able to discriminate patients developing severe versus mild influenza. Eleven of these 12 OTUs had higher abundance in severe patients, and included the following genera: Prevotella, Streptobacillus, Porphyromonas, Granulicatella, Veillonella, Fusobacterium, Lachnospiracea incertae sedis and Haemophilus spp. Except for Haemophilus, these genera all belong to obligate or facultative anaerobic bacteria. The species that were found to be more abundant in patients with severe influenza included Prevotella melaninogenica, and Veillonella dispar – which are anaerobic Gram-negative bacteria that are part of the normal oral [25] and respiratory tract microbiota [26] and can cause anaerobic pulmonary infections [25] or meningitis and endocarditis [27] – and Granulicatella elegans, a member of the nutritionally variant streptococci, which are common components of the oral microbiota that have been associated with endocarditis and septicaemia [28]. Only one OTU was more abundant in patients with mild symptoms: Staphylococcus aureus. This is in line with the study of Wang et al., which reported that S. aureus priming prevents influenza-mediated lung injury in a mouse model [16]. In the proposed mechanism S. aureus induces monocyte polarization into M2 alveolar macrophages via Toll-like receptor 2 signalling, which inhibits influenza-mediated lethal inflammation [16]. Based on our pilot study, the higher abundance of S. aureus in mild non-progressors could explain the lack of severe clinical manifestations observed in our patient cohort, while the presence of a diverse nasopharynx microbiota containing multiple pathobionts significantly increased the risk of acquiring severe influenza-related disease. Furthermore, we compared each of the two severe influenza groups. Seven OTUs were significantly more diverse in one group, the respiratory group for two of them (Moraxella catarrhalis and Parvimonas micra), and the neurological group for the rest of them (Actinomyces, Corynebacterium, Dolosigranulum pigrum, Chryseobacterium and Prevotella). Physiologically, all of these bacteria were commensal of the respiratory tract and oral cavity. They were mainly described in pneumonia and periodontitis infections, but could also be the cause of central nervous system infections, bone infections and endocarditis [29–31]. Interestingly, Dolosigranulum pigrum, which is associated with the development of kerato-conjunctivitis, has already been characterized in the respiratory tract microbiome as belonging to a health-associated community that has been developing recently in children born by caesarean section [32]. Note that this analysis remains to a large extent underpowered because of the small number of patients in each severe group in this study.
It is noteworthy that the use of microbial signatures to predict clinical outcomes for influenza could be reduced to only two–three OTUs. If this proves to be robust in future studies, such signatures could open up the possibility of developing a real-time clinical assay using current technology. Rapid PCR panels that accurately quantify both bacterial and viral species [33] could facilitate a practical implementation of prognostic microbial signature-based assays simultaneously with acute respiratory virus diagnostics in a clinical setting.
Our study has limitations that are inherent in its retrospective nature. First, despite the large number of samples (n=372) from children infected with influenza virus available in our collection, only 36 samples (9.7 %) met our selection criteria. Because of our relatively small cohort, we could not match the patient groups in sex and age. As age and sex were described as being important factors that modify the microbiota in the nasopharynx [18, 22], we had to adjust for these potential confounders in statistical models. We found that the main factor influencing the NP microbiota was age. Sex was not associated with microbiota composition and diversity, as expected for young prepubescent children. The microbiome was also described as changing with seasons [18]. However in our study, samples were collected over different years, but all were collected during the influenza season (January–March), so there was no seasonal effect to account for. Finally, due to the retrospective nature of our pilot study we could not compare the performance of our signature with existing clinical severity scores, such as the pediatric risk of mortality (PRISM III). The differences in number between the neurological and respiratory groups (n=10 vs n=4) could be explained by the administration of antimicrobial therapeutics in patients with respiratory complications before the NP sample was collected. Indeed, children with respiratory distress were most likely to have had pre-emptive antibiotics administered at hospital admission before sampling, and so were excluded from analyses. In contrast, when children developing neurological complication had antibiotics administered, this was mainly during hospitalization after NP sample was collected, and the treatments administered prior to admission were mostly anti-epileptic treatments.
Future studies will involve the enrolment of prospective cohorts of children admitted in the emergency departments with longitudinal sample and clinical data collection to validate our microbial signature performance and analyse host–virus–microbiome interactions. Studies that integrate microbiome analysis with a systems biology approach to influenza pathogenesis will accelerate the development of novel diagnostic tools and personalized therapeutics to combat influenza virus infections in children.
Methods
Ethical statement
Respiratory samples (NP aspirate or swab) were collected for regular disease management during hospital stays and no additional samples were taken. For the purpose of this study, patient confidentiality was strictly protected and informed consent was obtained. This study was approved by the ethical committee of Hospices Civils de Lyon on 14 October 2014. The study was also approved by the University of Washington IRB under expedited category 5 (human subjects application #49811).
Patient selection
We analysed 372 clinical records from children (≤15 years) who were admitted to a paediatric hospital (Hôpital Femme-Mère-Enfant, Hospices Civils de Lyon, France) in 2011–2014. All of the analysed samples tested positive for influenza A or B virus using the respiratory multi-well system MWS r-gene kit (bioMérieux, Marcy-l’étoile, France) during routine testing in the virology department of the University Hospital of Lyon. Ct could be obtained and were considered to be an estimated viral load in the sample.
In order to define a prognostic signature, only patients who were symptomatic for less than 2 days and had a respiratory sample collected at the time of hospital arrival were selected. Based on their clinical course after sample collection, patients were classified into two groups: patients with mild influenza or patients with respiratory or neurological complications (the severe influenza group). The criteria used for classifying children in the severe influenza group with respiratory complications were: utilization of invasive or non-invasive ventilation, blood gas alteration (hypoxemia <95 % in arterial sample) and hospitalization in the ICU for respiratory complications. Neurological complications were defined as symptoms that affected the central or peripheral nervous system, including seizures, encephalopathy, encephalitis, or any focal neurological symptom [34]. Patients with neurological complications were hospitalized either in the ICU or in neurology units specializing in the management of encephalitis or epilepsy. We excluded patients with incomplete clinical files, as well as patients who had received antibiotics prior to respiratory sample collection, as this treatment likely perturbs the microbiome. Other exclusion criteria included known risk factors for severe influenza (defined as chronic respiratory diseases, cardiac diseases, metabolic diseases, immunosuppression, etc.) and known chronic disease associated with respiratory microbiota dysbiosis (such as severe asthma, etc.) [3]. Note that two patients had Down’s syndrome, one in the mild influenza group and one in the severe influenza group. In addition, in the respiratory group one patient presented with Prader–Willi syndrome and another presented with an uncategorized myopathy. While individuals with Down's syndrome, Prader–Willi syndrome and myopathy are known to be at risk of respiratory problems, these comorbidities have not been associated with modifications in the NP microbiome, and so we did not exclude these patients. In total, 72 patients were selected, including 42 patients with mild influenza, 11 patients with respiratory distress symptoms and 19 patients with neurological complications. We extracted DNA and RNA using Nuclisens EasyMag. Only 36 patients had samples with sufficient quantity and quality of DNA for sequencing, including 22 patients with mild influenza, 4 patients with respiratory distress symptoms and 10 patients with neurological complications.
16S rRNA sequencing and analysis
Genomic DNA was extracted from de-identified patient respiratory samples (n=36). Custom primers were synthesized to amplify a 510 bp fragment containing variable regions V1–V3 of the 16S ribosomal gene. Briefly, 5 ng of genomic DNA from each sample was used as a starting template to generate the V1–V3 amplicon libraries. All genomic DNA was subjected to 20 cycles of PCR (Failsafe, Epicentre) and the 16S amplicons were cleaned using 0.8X AMPure XP beads (Beckman Couter, Inc.) following the manufacturer’s instructions. Nextera dual index adaptors (Nextera XT adaptors, Illumina, Inc.) were incorporated by performing 10 PCR cycles (Failsafe, Epicentre), cleaned using 1.1X AMPure XP beads (Beckman Couter, Inc,), quantified using a qubit (DNA high sensitivity, Life Technologies) and multiplexed using equal molar ratios of DNA for each sample. The final 16S libraries were loaded on a MiSeq sequencer at 2 pM with 5 % PhiX control and sequenced using custom Illumina read primers to eliminate the V1–V3 16S primer sequences (Table S6). Each sample had an average of 150K read depth (300 bp, paired end) and all 16S ribosomal sequences were classified using the UPARSE metagenomic pipeline [35]. Briefly, this pipeline removed low-quality reads, merged the paired reads to generate ~430 bp fragments, removed sequence artifacts (chimaeric sequences and sequence errors), mapped full-length reads to a highly curated 16S ribosomal database derived from the RDP 16S training set (v14), identified sequence reads to genus/species level with a 97 % cut-off based on OTU classification and generated summary tables for downstream statistical analyses. In total, all samples passed our cut-off of at least 50 000 high-quality 16S DNA sequence reads per sample and all OTUs were classified to the genus level (Fig. S1). Rarefaction curves were performed to verify that sampling had exhausted the diversity at the sequence read depth cut-off of 50 000 reads per sample (Fig. S1c). For classification to the species level, we blasted the OTU representative sequences against the silva Ref NR99 database (release 119), from which uncultured species were removed. We used blastn with the following parameters: ‘-evalue 1e-50 -perc_identity 99. The blast results were further filtered to only keep results that were identical (up to the genus level) to the UPARSE classification.
The raw sequence data were deposited at SRA (SUB1938413).
Metagenome sequencing and analysis
Genomic DNA (5 ng) from 10 mild and 10 severe influenza patients were fragmented and tagged using Nextera transposase following the manufacturer’s instructions (Nextera XT DNA library kit, Illumina). Dual-index adaptors were incorporated by PCR (14 cycles) and the final libraries were multiplexed for sequencing. The final library was loaded on a NextSeq 500 using a 300 cycle high-output kit. All reads were demultiplexed, the adaptor sequences were removed and low-quality reads were discarded. High-quality reads were aligned to a reference mammalian ribosomal RNA database and the human hg19 genome using STAR aligner. Unmapped nonhuman reads were classified using Kraken metagenome analysis software and the MiniKraken DB using the default settings [33]. Genus/species level identification, including total read counts for each bacterial group, was determined.
Statistical analysis
All statistical analyses were performed using R statistical Software. The R ‘phyloseq’ package [36] was used to calculate abundance-based richness estimators (e.g. Chao1 and ACE) and diversity indices (e.g. Shannon, Simpson and Fisher). Multi-way ANOVA (type III) was used to identify the covariates associated with each diversity index. The R phyloseq package was also used to calculate the distance metrics (Bray–Curtis and weighted Unifrac) and visualize the samples in two dimensions using non-metric multidimensional scaling (NMDS). Beta diversity analysis was performed on relative abundance data (i.e. proportions, the counts are divided by total library size). Permutational multivariate analysis of variance (PerMANOVA) was performed using the adonis function in the R ‘vegan’ package [37] to test the ability of variables (patient group, sex and age, nature of sample, days since onset of symptoms) to account for the observed variance in microbial profiles. Finally, statistical analyses for the differential abundance of mapped OTU reads were conducted using the ALDEx2 R package with a generalized linear model (GLM) using the aldex.glm function [38]. We modified the aldex.glm function to enable the effect of several variables in the glm model to be tested, rather than only testing one variable, as in the default aldex.glm function. The covariates that were taken into account were age, sex, time since the onset of symptoms and nature of sample. Differential OTUs were defined using a P-value cut-off of 0.05.
To determine whether small sets of OTUs could classify the patients, we used classification based on association rules using the R ‘arules’ package. For this analysis, the relative abundance matrix of differentially abundant OTUs was transformed into a binary matrix based on the median relative of each OTU, with 1 meaning that the relative abundance of OTU_x in sample (i) is greater than the median value of OTU_x across all samples. The rules were mined with support >0.3 and confidence >0.8. Redundant rules were further pruned. We judged the robustness of the rules using an LOOCV approach. For each subject, association rules were predicted on all other subjects, excluding the one for which we made a prediction. After we had performed this procedure for all subjects, we selected the rules that were found in all instances and calculated their sensitivities and specificities from predicted versus true classifications.
Supplementary Data
Supplementary File 1
Funding information
This work was supported by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health and the Department of Health and Human Services under Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract no. HHSN272201400005C. The funding sources did not play any role in the study or in the preparation of the article or the decision to publish
Acknowledgements
The authors thank Marcus Korth and Bruno Simon for valuable feedback on the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ACE, abundance-based coverage estimator; ANOVA, analysis of variance; CCEIRS, Centers of Excellence for Influenza Research and Surveillance; Ct, cycle threshold; DNA, desoxyribo nucleic acid; glm, generalized linear model; H, Shannon diversity index; ICU, intensive care unit; IRB, institutional review board; LOOCV, leave-one-out cross-validation; NMDS, non-metric multidimensional scaling; NP, nasopharyngeal; OTU, operational taxonomic unit; PRISM, paediatric risk of mortality; RNA, ribonucleic acid; V1/V3, variable region of the 16S DNA.
Sequence Accession Number: The Sequence Read Archive (SRA) accession number for the raw sequences is SUB1938413.
Six supplementary tables and three supplementary figures are available with the online Supplementary Material.
References
- 1.Belongia EA, Irving SA, Waring SC, Coleman LA, Meece JK, et al. Clinical characteristics and 30-day outcomes for influenza A 2009 (H1N1), 2008-2009 (H1N1), and 2007-2008 (H3N2) infections. JAMA. 2010;304:1091–1098. doi: 10.1001/jama.2010.1277. [DOI] [PubMed] [Google Scholar]
- 2.Dawood FS, Chaves SS, Pérez A, Reingold A, Meek J, et al. Complications and associated bacterial coinfections among children hospitalized with seasonal or pandemic influenza, United States, 2003-2010. J Infect Dis. 2014;209:686–694. doi: 10.1093/infdis/jit473. [DOI] [PubMed] [Google Scholar]
- 3.Gill PJ, Ashdown HF, Wang K, Heneghan C, Roberts NW, et al. Identification of children at risk of influenza-related complications in primary and ambulatory care: a systematic review and meta-analysis. Lancet Respir Med. 2015;3:139–149. doi: 10.1016/S2213-2600(14)70252-8. [DOI] [PubMed] [Google Scholar]
- 4.Committee on Infectious Diseases, American Academy of Pediatrics Recommendations for prevention and control of influenza in children, 2012-2013. Pediatrics. 2012;130:780–792. doi: 10.1542/peds.2012-2308. [DOI] [PubMed] [Google Scholar]
- 5.Campbell A, Rodin R, Kropp R, Mao Y, Hong Z, et al. Risk of severe outcomes among patients admitted to hospital with pandemic (H1N1) influenza. Can Med Assoc J. 2010;182:349–355. doi: 10.1503/cmaj.091823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshansky CM, Gartland AJ, Wong SS, Jeevan T, Wang D, et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am J Respir Crit Care Med. 2014;189:449–462. doi: 10.1164/rccm.201309-1616OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey RT, Lynfield R, Dwyer DE, Losso MH, Cozzi-Lepri A, et al. The association between serum biomarkers and disease outcome in influenza A(H1N1)pdm09 virus infection: results of two international observational cohort studies. PLoS One. 2013;8:e57121. doi: 10.1371/journal.pone.0057121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josset L, Tisoncik-Go J, Katze MG. Moving H5N1 studies into the era of systems biology. Virus Res. 2013;178:151–167. doi: 10.1016/j.virusres.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung RK, Zhou JW, Guan W, Li SK, Yang ZF, et al. Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin Microbiol Infect. 2013;19:930–935. doi: 10.1111/1469-0691.12054. [DOI] [PubMed] [Google Scholar]
- 13.Chaban B, Albert A, Links MG, Gardy J, Tang P, et al. Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLoS One. 2013;8:e69559. doi: 10.1371/journal.pone.0069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori T, Kiyoshima J, Shida K, Yasui H. Augmentation of cellular immunity and reduction of influenza virus titer in aged mice fed Lactobacillus casei strain Shirota. Clin Diagn Lab Immunol. 2002;9:105–108. doi: 10.1128/CDLI.9.1.105-108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Li F, Sun R, Gao X, Wei H, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. From meta-omics to causality: experimental models for human microbiome research. Microbiome. 2013;1:14. doi: 10.1186/2049-2618-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao A, Chiu C-H, Jost L, Diversity US, Diversity P. Functional diversity, and related similarity and differentiation measures through hill numbers. Annu Rev Ecol Evol Syst. 2014;45:297–324. [Google Scholar]
- 20.Sakwinska O, Bastic Schmid V, Berger B, Bruttin A, Keitel K, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol. 2014;52:1590–1594. doi: 10.1128/JCM.03280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012;205:1048–1055. doi: 10.1093/infdis/jis024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teo SM, Mok D, Pham K, Kusel M, Serralha M, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, et al. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol. 2012;78:6262–6270. doi: 10.1128/AEM.01051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188:1246–1252. doi: 10.1164/rccm.201302-0215OC. [DOI] [PubMed] [Google Scholar]
- 25.Schreiner A. Anaerobic pulmonary infections. Scand J Infect Dis Suppl. 1979;19:77–79. [PubMed] [Google Scholar]
- 26.De Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loughrey AC, Chew EW. Endocarditis caused by Veillonella dispar. J Infect. 1990;21:319–321. doi: 10.1016/0163-4453(90)94197-8. [DOI] [PubMed] [Google Scholar]
- 28.Yacoub AT, Krishnan J, Acevedo IM, Halliday J, Greene JN. Nutritionally variant streptococci bacteremia in cancer patients: a retrospective study, 1999–2014. Mediterr J Hematol Infect Dis. 2015;7:2015030. doi: 10.4084/mjhid.2015.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abou Abdallah R, Cimmino T, Baron S, Cadoret F, Michelle C, et al. Description of Chryseobacterium timonianum sp. nov., isolated from a patient with pneumonia. Antonie van Leeuwenhoek. 2017:1121–1132. doi: 10.1007/s10482-017-0885-8. [DOI] [PubMed] [Google Scholar]
- 30.Cobo F, Rodríguez-Granger J, Sampedro A, Aliaga-Martínez L, Navarro-Marí JM. Pleural effusion due to parvimonas micra. A case report and a literature review of 30 cases. Rev Esp Quimioter. 2017;30:285–292. [PubMed] [Google Scholar]
- 31.Wong VK, Turmezei TD, Weston VC. Actinomycosis. BMJ. 2011;343:d6099. doi: 10.1136/bmj.d6099. [DOI] [PubMed] [Google Scholar]
- 32.Bosch AA, Levin E, van Houten MA, Hasrat R, Kalkman G, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine. 2016;9:336–345. doi: 10.1016/j.ebiom.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khandaker G, Zurynski Y, Buttery J, Marshall H, Richmond PC, et al. Neurologic complications of influenza A(H1N1)pdm09: surveillance in 6 pediatric hospitals. Neurology. 2012;79:1474–1481. doi: 10.1212/WNL.0b013e31826d5ea7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 36.Mcmurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oksanen J, Kindt R, Legendre P, O’Hara RB. Vegan: community ecology package. Bioconductor. 2015 [Google Scholar]
- 38.Fernandes AD, Reid JN, Macklaim JM, Mcmurrough TA, Edgell DR, et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:15. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1