Abstract
The coordination of progenitor self-renewal, neuronal production and migration is essential to the normal development and evolution of the cerebral cortex. Numerous studies have shown that the Notch, Wnt/beta-catenin and Neurogenin pathways contribute separately to progenitor expansion, neurogenesis, and neuronal migration, but it is unknown how these signals are coordinated. In vitro studies suggested that the mastermind-like 1 (MAML1) gene, homologue of the Drosophila Mastermind, plays a role in coordinating the afore-mentioned signaling pathways, yet its role during cortical development remains largely unknown. Here we show that ectopic expression of dominant-negative MAML (dnMAML) causes exuberant neuronal production in the mouse cortex without disrupting neuronal migration. Comparing the transcriptional consequences of dnMAML and Neurog2 ectopic expression revealed a complex genetic network controlling the balance of progenitor expansion versus neuronal production. Manipulation of MAML and Neurog2 in cultured human cerebral stem cells exposed interactions with the same set of signaling pathways. Thus, our data suggest that evolutionary changes that affect the timing, tempo and density of successive neuronal layers of the small lissencephalic rodent and large convoluted primate cerebral cortex depend on similar molecular mechanisms that act from the earliest developmental stages.
Keywords: cortical evolution, neuronal migration, mastermind-like 1, notch, neurogenin 2, in utero electroporation
Introduction
The mammalian cerebral cortex is built from distinct layers of neurons generated in distant proliferative areas (Angevine and Sidman, 1961; Sidman and Rakic, 1973; Desai and McConnell, 2000; Noctor et al., 2001; Caviness et al., 2009; Johnson et al., 2015). In the past forty years, numerous studies have shown that the stepwise inside-out production, migration, and ultimate settlement of neurons is a tightly woven process [reviewed in (Ayala et al., 2007; Rakic et al., 2007; Gertz and Kriegstein, 2015)]. Evolutionary changes to these processes led to species specific growth in surface, areal pattern, density of neurons, and thickness of the cortical sheet (Geschwind and Rakic, 2013). Our understanding of transcriptional machinery that coordinates the regimented production of neurons is poor despite its significance to evolutionary changes in cortical size and role in numerous cortical pathologies (Gleeson and Walsh, 2000; Galaburda et al., 2006; Geschwind and Levitt, 2007; Meechan et al., 2011; Carrion-Castillo et al., 2013).
The regulation of neurogenesis and migration by Neurogenins (Neurog), the Wnt/beta-catenin and Notch pathways the subject of numerous studies therefore a detailed expose is beyond the limits of this article. These pathways exert strong influence on neural progenitor maintenance, neuronal migration, positioning and differentiation but it is unknown how their signals are coordinated. Neurog is one of earliest molecular markers of neuronal birth and migration but its role the generation of outer radial glia (oRG) in the ferret and in the human neocortex remains controversial (Scardigli et al., 2003; Mattar et al., 2004; Johnson et al., 2015; Pollen et al., 2015; Thomsen et al., 2016). Experimental manipulation of the Wnt/β-catenin pathway in mice causes expansion of progenitor pool (Chenn and Walsh, 2002; Mutch et al., 2009; Munji et al., 2011; Boitard et al., 2015). Similarly, the Notch signaling pathway controls increases in population size and diversity of cortical progenitors (Corbin et al., 2008; Shimojo et al., 2008; Dominguez et al., 2013). The Notch co-factor mastermind-like (MAML) was shown to be necessary for the activation of downstream Notch genes (Nam et al., 2006; Wu et al., 2007){Nam, 2006 #69;Wu, 2007 #70}{Nam, 2006 #69;Wu, 2007 #70}{Nam, 2006 #69;Wu, 2007 #70}. Initial observations showed that dominant-negative MAML (dnMAML) phenocopies the effect of Notch inhibition on cortical progenitors in vivo (Maillard et al., 2004; Yoon et al., 2008; Dominguez et al., 2013). However, it is unknown whether disturbing MAML in vivo affects the assignment of neuronal fate, the migration process, as well as long-term migration and laminar defects.
Despite recent advances, untangling gene networks controlling various processes in the developing brain continue to be a challenge because the cellular composition of transient zones changes over time and space (Ayoub et al., 2011; Aprea et al., 2013; Reilly et al., 2015). To address this knowledge deficit, we first show that we can manipulate neurogenesis without detectable long-term adverse effects on neuronal migration. We then deduced a neurogenesis program by comparing normal and experimental developmental transcriptomes. Next, we confirmed that cultured human fetal glia undergo a comparable program. Our results show that the mastermind-like gene coordinates neurogenic and migration programs under tight post-transcriptional regulatory control in both the mouse developing brain and in human cells.
Material and Methods
Animals Tissue
Timed-pregnant C57BL6j mice were purchased from Charles River Laboratories. Vaginal plug date was considered E0.5. Animals were perfused at E15.5, E16.5, P0 and P21 using ice-cold PBS followed by 4% paraformaldehyde as described earlier (Burns et al. 2007; Ayoub et al. 2005). Tissue collected for next generation sequencing experiments was embedded in O.C.T., then immediately frozen in isopentane on powdered dry ice. The institutional animal care and use committee at Yale University reviewed and approved all procedures.
In utero electroporations
Timed-pregnant mice were anesthetized (Ketamine/Xylazine mix; 0.1ml/10g body weight), then the uterus was gently exteriorized and one cerebral ventricle injected with mixtures of the pCAG-TagBFP_V5_NLS, which contains the TagBFP coding sequence (Evrogen) and with a C-terminal V5 epitope tag (‘GKPIPNPLLGLDST’) followed by a 3x NLS (‘DPKKKRKV’; derived from the SV40 large T-antigen) cloned into the pCAGEN vector, pEGFP-dnMAML and pCAG-Ngn2-IRES-. A post-operative analgesic was supplied in the drinking water for up to two days. Brains (n=5) were dissected in ice-cold RNAse-free buffer from consistent electroporations then processed for subsequent experiments.
Human Tissue
The institutional review committee approved the procurement of postmortem human tissue, which was collected from the Human Developmental Biology Resource at Newcastle University following approved laws and ethical guidelines. Samples (n=3 each) from gestational week 14 were assessed from developmental abnormalities before performing subsequent experiments.
Tissue Culture, Cell sorting, and Transfections
Tissue was homogenized in HBSS containing 1% Pen/Strep (GIBCO) then trypsinized, treated with DNAse and centrifuged. Progenitor cells were magnetically sorted based on binding the CD15 antibody microbeads in MACS columns (Miltenyi Biotec). Subsequently cells were grown in proliferation media containing the B27 supplement, bFGF and EGF. Cell viability was assessed using the LIVE/DEAD viability/cytotoxicity kit (Molecular Probes). Cells were passaged when flasks reached 90% confluence. Growth factors were removed from the culture media to induce differentiation of cells. Transfections with equimolar solutions of dnMAML, Neurog2, or EGFP plasmids were performed in quadruplets using the Amaxa Nucleofector following manufacturer protocols (Lonza Grp Ltd). Culture experiments were repeated in three independent biological replicates. Immunohistochemistry and microscopy were performed after fixation in 4% PFA. Automated cell counting on coded images of randomly acquired 200μm x 200μm fields was performed using CellProfiler 2. Multiple comparisons of count data were performed using a two-way ANOVA analysis followed by post-hoc test in Graphpad Prism 7.
Immunohistochemistry and Confocal Imaging
Frozen tissue sections were stained using a battery of markers specific for stem cells, cortical progenitors, immature neurons, and differentiated neurons as detailed in previous publications (Ayoub et al., 2011; Dominguez et al., 2013). Sections for counting were chosen by systematic random sampling from n=5 replicates for mouse experiments then coded by a colleague to prevent experimenter bias. Data sets were exported as tiffs for cell counting in Stereoinvesitgator (Microbrightfield Inc). In sections acquired from in utero electroporations, the contralateral un-injected hemisphere was used as an internal control for the distribution of fate markers. Statistical analysis of cell to layer distributions and nuclei counts were tested in 2-way ANOVA with Tukey’s multiple comparison tests in Prism 7 (GraphPad Inc) with a significance set at p<0.001.
Laser Microdissection
All tissue for laser microdissection (LMD) was dissected in RNAse-free ice cold 0.1M PBS then immersed in O.C.T compound (TissueTek) before freezing at −60°C in cooled isopentane and dry ice slurry. Small groups of cells were collected from in utero electroporated or normal tissue using LMD6000 microscope by visualizing GFP labeling (Leica Microsystems). LMD dissectate from at least four littermates per condition and age group were combined into each replica. Dissected LMD tissues were stored at −80C until RNA extractions in RNeasey micro kit (Qiagen) according to standard LMD protocol. We then tested the concentration of collected RNA on Nanodrop 1000 (Thermo Scientific). A portion of extracted RNA from each replica was saved for RT-qPCR. Total RNA and miRNA were purified using the miRNeasy Micro Kit (Qiagen) following standard protocol. We analyzed data using the DDCt method followed by statistical analysis in GraphPad Prism 7 as described previously (Ayoub et al. 2011).
Next-Generation Sequencing
RNA quality was assessed on an Agilent BioAnalyzer. At least two technical replicates per group (with RIN>8) were sequenced on an Illumina Hiseq2000 using the standard Illumina protocols for 75bp paired-end sequencing. Library preparations and sequencing were performed at the Yale Center for Genome Analysis. We performed all subsequent computations on the Yale Biomedical HPC clusters as previously described (Ayoub et al., 2011; Reilly et al., 2015). Furthermore, we compared our samples to publicly available data. Raw sequence reads for additional samples from studies GSE38805, GSE66217, and GSE30765 were downloaded from the gene expression omnibus and then processed in a similar fashion. Statistical analysis was performed in R to identify differentially expressed genes. We applied p value Benjamini-Hochberg correction following multiple testing. We used BH p≤10 −3 and Log2 fold change as criteria for choosing differentially expressed genes. BED interval files containing genomic coordinates for cis-regulatory elements were downloaded from GSE63649 (Reilly et al., 2015). We searched for a single nearest gene close to the cis-regulatory elements and calculated their hypergeometric p-value. The list of genes near human-specific enhancers is provided in table S2.
Network analysis
We used Log2 normalized expression values as input for subsequent processing. Analysis with the self-organizing maps algorithm was performed in R using oposSOM package. Fuzzy clustering of genes and groups after self-organizing map analysis was performed in AutoSOME. The gene-set Z statistics (GSZ) score for over-represented and under-represented gene groups are presented in table S1. Additional gene-set enrichment analysis and network mapping were performed in Cytoscape and the GeneMania plugin. Identification of transcription factor binding sites, using position-specific weighed matrices in proximal promoters, and microRNA seed sites was performed by extracting the list of annotated regulators of SOM-derived ranked list of differentially expressed genes from publicly available databases (Ensembl, miRBase, microrna.org, TargetScan, miRTarBase and others). Analysis of overrepresentation is performed using a Fisher’s exact test with a 2 × 2 contingency table. Predictions of transcription factor Geneset enrichment data (Fig. S10), as well as a list transcription factors and microRNAs are provided in table S2.
Results
Disrupting MAML in vivo initiates neuronal differentiation and migration
First, we determined the spatial and temporal expression of MAML in the developing murine cortex. Immune labeling showed diffuse cytoplasmic staining in M-phase cells and punctate staining in the interphase cells in the VZ (Fig. 1A). In situ hybridization showed a decreasing gradient from the VZ, to the subventricular zone (SVZ), intermediate zone (IZ), and the cortical plate (CP at E15 (fig S1–A). Analysis of previously published mRNAseq data (Ayoub et al 2011) showed a similar decreasing gradient, with highest enrichment of MAML1 and MAML2 in the VZ at E14.5 (Fig. S1–C). On the other hand, mRNAseq data showed that MAML3 is expressed at very low levels at this stage of the developing mouse cortex. We further confirmed this expression by performing qRT-PCR on laser microdissected cells from E14.5 mouse cortex (Fig. S1–D). We detected the highest expression for both MAML1 and MAML2 in the VZ but failed to detect MAML3 in the same LMD samples.
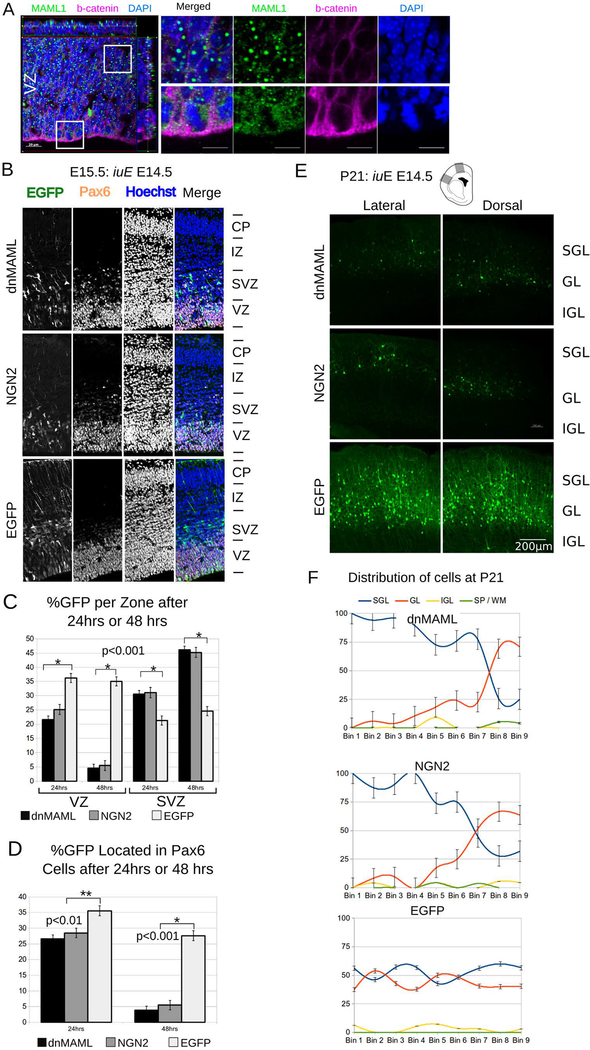
Fig. 1. MAML regulates neurogenesis in cortical progenitors.
A. Confocal z-stack for MAML1/2 (green) and beta-catenin (red) in cortical mouse progenitors at E15. DAPI nucleic stain in blue. Insets show MAML1 (green) perinuclear staining in telophase and punctate staining elongated G1/G0 nuclei (Scale bar = 20μm). iuE : in utero electroporation.
B. Representative confocal images show E15.5 mouse dorsal cortices electroporated in utero with dnMAML, NGN2 or control plasmid, then sacrificed after 24 hrs.
C. Graphs shows the percentage of GFP+ cells in the VZ and SVZ region 24hrs or 48hrs after electroporation (n=5). Data were analyzed in a 2way ANOVA with a post hoc test (+/− SEM).
D. Graph shows the percentage of Pax6+ and GFP+ cells in dnMAML, NGN2 and controls 24 or 48 hrs after electroporations (n=5). Data were analyzed in a 2way ANOVA with a post hoc test (+/− SEM).
E. A Panel of representative coronal sections showing lateral and medial aspects from P21 brains. Brains were electroporated in utero with dnMAML, NGN2, or Control EGFP plasmid at E14.5 (scale bar 200 μm). SGL: supragranular layers, GL: granular layer, IGL: infragranular layers.
F. Graphs show the normalized distribution of EGFP cells at P21 after iuE at E14.5. The cortex of dnMAML, NGN2, or Control (n=5 each) at P21 was divided into 9 counting bins. Cells were assigned to one of four categories based on layer position (+/− SEM). SGL: supragranular layers, GL: granular layer, IGL: infragranular layers, SP/WM: subplate and white matter.
To determine the consequences of disturbing MAML, we electroporated dnMAML into the mouse dorsal cortex starting at embryonic day (E) 14.5, with survival from 24hrs up to postnatal day 21 (P21). Because previous studies suggested a disruption of progenitor proliferation or premature exit from the VZ, we decided to contrast the effects of dnMAML to the well-studied effects of overexpressing Neurog2 on neocortical development. Both dnMAML and Neurog2 electroporations at E14.5 caused rapid differentiation and migration out of the VZ as compared to age-matched controls (Fig. 1B). After 24hrs, only 21.5% dnMAML+ and 25.1% of Neurog2+ electroporated cells remained in the VZ versus 36.2% of those that received control EGFP plasmids (fig. 1C; n=5; p<0.0)1, ± SEM). Over the same 24hr period, significantly more cells electroporated with dnMAML (30.5%) and Neurog2 (31.1%) had migrated to the SVZ, compared to only 21.3% of EGFP controls (fig. 1C; n=5; p<0.01, ± SEM). The percentage of cells electroporated with dnMAML and Neurog2 in the VZ dropped sharply after 48hrs of electroporation (Fig. 1C). Approximately 35.1% of cells positive for control EGFP remained in the VZ versus 4.6% of dnMAML and 5.4% of Neurog2+ electroporated cells (Fig. 1C; n=5; p<0.001, ± SEM). This is evident in the sharp reduction of Pax6-positive/GFP positive cells located in the VZ and SVZ after 24hrs (Fig. 1D). In utero electroporation of full-length MAML1 (fl-MAML1) caused a slight increase in cells remaining in the VZ and mild decrease of cells transiting to the SVZ after 24 hrs as compared to control electroporations (Fig. S1C, D). In contrast to dnMAML in utero electroporation of the Notch intracellular domain (NICD) elicited progenitor maintenance at the expense of neurogenesis (Fig. S1A,B). Similar to dnMAML, RBPj-mut caused a significant decrease in the proportion of cells in the VZ after 24hrs (fig. S1C,D). Forty-eight hours after electroporation with dnMAML, neuronal exodus continued at a much faster rate than in controls (Fig. 1D).
In utero electroporation of Neurog2 at E14.5 led to swift neuronal migration out of the VZ as observed after ectopic expression of dnMAML (Fig. 1 and S1). We examined E12.5 electroporations of Neurog2 at 24 or 48hrs post-electroporation. Cells forced to exit the VZ by virtue of Neurog2 overexpression at E12.5 had largely done so by E14.5, or 48hrs post-electroporation. In contrast to controls, only 57% of dnMAML - or 82% Neurog2-expressing cells remained in the VZ 24hrs post-electroporations (fig. S1). These data indicate that exogenous dnMAML only accelerated the neurogenic process and exit from the VZ, thus mimicking a loss of Notch phenotype (Mizutani et al., 2007).
To examine effect on neuronal migration we allowed a cohort of age-matched animals that received dnMAML, Neurog2, or EGFP at E14.5 (n=5) to survive until P21. An analysis of electroporated brains of all conditions using systematic random sampling showed no significant migration defects. Occasional EGFP-positive cells trapped in the cortical white matter were not significantly different between experimental conditions and controls. The vast majority of neurons produced in the proliferative zone on or after E14.5 migrated to the upper-layers of postnatal neocortex (Fig. 1E, F). Control electroporated EGFP+ cells were scattered from layer 4 to layer 2 regardless of their lateral or medial position. On the other hand, Neurog2 or dnMAML electroporation exhibited stepwise laminar positioning of EGFP+ cells, in layer 2 laterally and towards layer 4 medially (Fig. S3). Interestingly, forced acceleration of neuronal exodus due to the overexpression of Neurog2 or dnMAML did not cause any noticeable defects in laminar positioning at P21.
Disrupting MAML in vivo does not affect laminar fate commitment
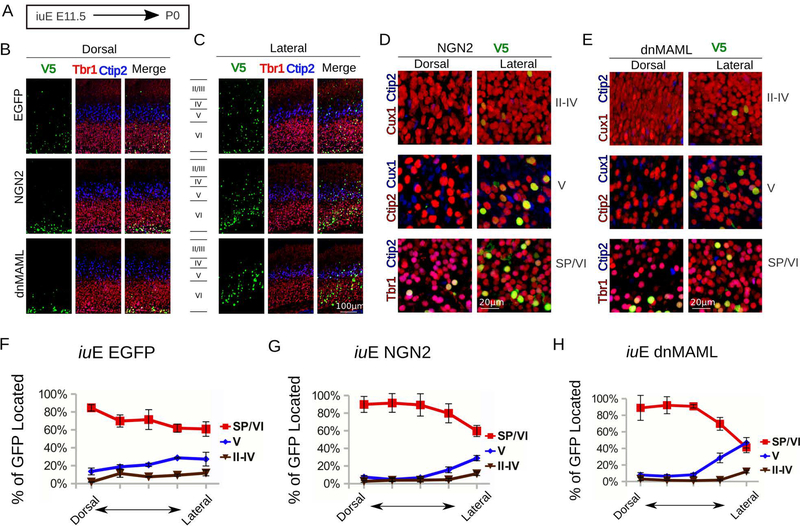
To test whether dnMAML alters the assignment of neuronal fate, we electroporated at E11.5, E12.5, and E14.5 (Fig. 2 and S2). We assessed neuronal fate at P0 by quantifying the co-localization of CUX1, a marker of upper neuronal layers, CTIP2 and Tbr1, markers of deep neuronal layers, with V5-positive electroporated nuclei at P0 (fig. 2 and S2). Cells electroporated with control EGFP dispersed in several laminae at P0, which is possibly due to labeled progenitors re-entering the cell cycle and producing multiple daughter neurons. In contrast to controls, dnMAML electroporations resulted in a narrow band of electroporated cells that migrated to their proper laminar destination according to the dorsal-to-lateral neurogenesis gradient for each specified age. We observed this effect in brains electroporated at E11.5 (Fig 2 A–H), E12.5 and E13.5. For instance, in utero electroporations with dnMAML at E11.5 produced layer IV cells on the dorsomedial aspect of the cortex but layer II/III cells on the lateral aspect of the cortex (Fig. 2E and H). Electroporations of dnMAML at E11.5 overproduced subplate cells dorsally at the expense of layers 5 and 6 cells, as observed in controls (Fig. 2F–G). Lastly, electroporation with dnMAML at E12.5 produced deep layer cells dorsomedially but upper-layer cells on the lateral aspect of the cortex, as shown at P0 (Fig. S2).
Fig. 2. MAML affects Notch and beta-catenin signaling without altering neurogenic gradients.
A. Experimental paradigm for panels H to M. We performed in utero electroporation at E11.5 and sacrificed mice at P0. We mixed pCAG-V5-NLS at 1/4th the concentration of pCAG-EGFP, dnMAML, or NGN2 to all plasmids to co-localize electroporated nuclei layer markers.
B. View of dorsal cortical plate with V5+ nuclei (green), Tbr1 (red) and Ctip2 (blue). Note that, unlike controls, dnMAML and NGN2 electroporations cause a strong bias (>90%) towards SP and Layer VI fate.
C. View of lateral cortical plate with V5+ nuclei (green), Tbr1 (red) and Ctip2 (blue). Note that, unlike in controls and dorsal CP, dnMAML and NGN2 in lateral plate produce a mixture of fates including upper layers, however SP and Layer VI fates predominate.
D. Higher magnification views of NGN2 electroporation showing layers II-IV (V5+ in green), Cux1 (red), and Ctip2 (blue). Higher magnification view of layer V (V5+ in green), Cux1 (blue) and Ctip2 (red). Higher magnification of the SP and layer VI with (V5+ in green), Tbr1 (Red) and Ctip2 (blue). Scale bar = 20 μm.
E. Higher magnification views of dnMAML electroporation showing layers II-IV (V5+ in green), Cux1 (red), and Ctip2 (blue). Higher magnification view of layer V (V5+ in green), Cux1 (blue) and Ctip2 (red). Higher magnification of the SP and layer VI with (V5+ in green), Tbr1 (Red) and Ctip2 (blue). Scale bar = 20 μm.
F, G, H. Graphs show the layer distribution of electroporated cells at P0 after iuE at E11.5. The CP of dnMAML, NGN2, or Control (n=5 each) at P0 was divided into 5 adjacent 200 μm -wide bins. Bin1 starts at the dorsal aspect of the cortex while Bin5 is at the lateral aspect of the cortex. Cells were assigned to one of three categories based on layer position (+/− SEM). SP/VI: subplate and layer VI, V: layer V, II-IV: layers II-IV.
Similarly, we quantified the distribution of CUX1 and CTIP2 at P0 to test whether Neurog2 over-expression affected laminar fate and positioning of electroporated cells (Fig. 2B,C,D–G). We found that the largest proportion of electroporated cells expressed CTIP2 if electroporated at E12.5, but CUX1 if electroporated at E13.5 (Fig. S2). Thus, in the mouse, forced neuronal migration produces discrete laminar assignment and stratification. Furthermore, our data suggests that committed differentiating cells in the mouse may remain in the VZ for more than 24hrs, which is sufficient time for intrinsic specification among adjacent “self-renewing” progenitors, as they shift from deep- to upper-layer fates. These findings also explain why cells pre-destined for upper-layers in this species exist alongside deeper cells that are still differentiating before E14.5.
Transcriptome analysis of dnMAML-expressing migrating neurons
To isolate the complete genetic network coordinated by the mastermind-like gene in vivo, we performed transcriptome analysis at 24hrs and 48hrs on laser-microdissected cells after in utero electroporation with dnMAML, Neurog2, or control vector (Fig. 3A). Because compatible published datasets are scarce, we generated new transcriptome data from LMD tissue at E11.5, E12.5, and E17.5. Groups of cells were dissected from the frontal and occipital aspect of the dorsal cortex including cellular subdivision from the VZ, SVZ-IZ and CP.
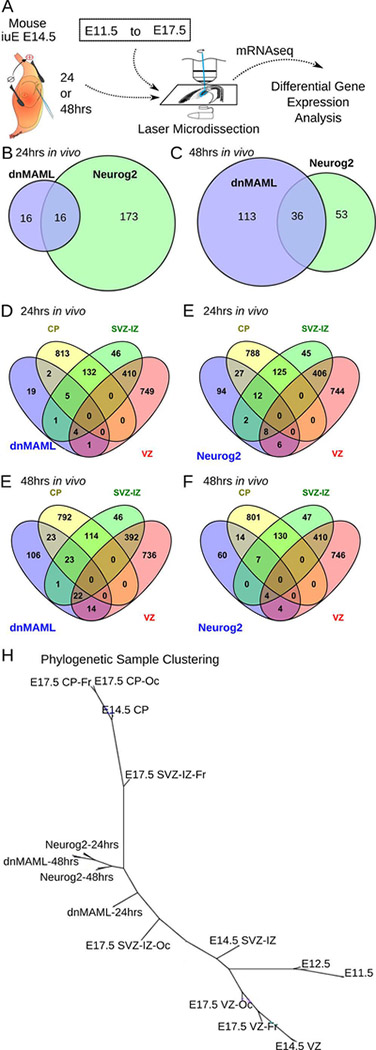
Fig. 3. Differentially expressed genes that drive neurogenesis and migration in vivo.
A. Comparison of differential gene expression after electroporation to normal embryonic zones from E11.5 until E17.5. Embryos were electroporated at E14.5 with EGFP, dnMAML or Neurog2 and were sacrificed after 24 hrs or 48 hrs. RNA was extracted from electroporated cells using laser-microdissection, followed by mRNAseq and differential gene expression analysis.
B, C. Venn diagrams contrast the number of differentially expressed genes after 24hrs or 48hrs in dnMAML and NGN2 electroporations.
D, E, F, and G. Venn diagrams compare dnMAML and Neurog2 electroporations after 24hrs and 48hrs in relation to the transcriptome of normal developmental zones at E14.5. VZ: ventricular zone; SVZ-IZ: Subventricular and Intermediate Zones; CP: Cortical Plate.
H. Phylogenetic clustering of samples using the neighbor-joining tree algorithm based on Eucledian distance between self-organizing map clusters. A list of log normalized differentially expressed genes combined from dnMAML-24hrs, dnMAML-48hrs, Neurog2–24hrs, and Neurog2–48hrs over controls in addition to the high-resolution transcriptome from E11.5 to E17.5 was used as input for this analysis. Of interest is the progressive shift in dnMAML-positive and Neurog2-positive cells compared to cells isolated from the subventricular and intermediate zone (SVZ-IZ). Twenty-four hours after dnMAML and Neurog2 electroporation at E14.5, cells acquire characteristics closer to E17.5 occipital or frontal SVZ-IZ.
Next-generation sequencing data was mapped to the mouse genome after quality control assessments following methods published previously (Ayoub et al. 2011; Benoit et al 2015). We calculated normalized gene expression values and then performed statistical analysis to isolate differentially expressed genes in experimental conditions as compared to controls. Genes whose expression changed over one fold (log2-fold change; p<0.05 Benjamini Hochberg corrected p) in all replicates were considered to be differentially expressed. As compared to controls, 32 genes in dnMAML and 189 genes in Neurog2 were differentially expressed in the 24hrs group (Fig. 3B). Of these genes, 16 genes were differentially expressed in both experimental conditions. Interestingly, we observed a significant down-regulation of neuronal differentiation genes such as Fezf2 and Nrp2 in the 24hrs group electroporated with dnMAML, not Neurog2. The difference between dnMAML and Neurog2 increased to 36 genes in samples microdissected after 48hrs (Fig. 3C). Satb2, Neurod2, Neurod6, and Foxg1 changed significantly in both conditions after 48hrs (table S1). Numerous neuronal migration and fate genes exhibited shifting expression between conditions from 24hrs to 48hrs including Tbr1, Pou3f3, Pou3f2, and Id2. These genes were upregulated by Neurog2, not dnMAML, at 24hrs, then upregulated by the dnMAML electroporation, but not the Neurog2 condition, after 48hrs. Surprisingly, Tbr2 (Eomes), a known downstream target of Neurog2 and typically expressed in intermediate progenitors, was not changed in either conditions versus controls. It is possible that one or both treatments affected Tbr2 expression before the 24hrs sampling window, but we did not test this scenario. Furthermore, we found genes typically expressed in the interneuronal lineage, such as Nkx2–1 and Dlx1, changed after dnMAML but not Neurog2 electroporation, indicating that dnMAML may coordinate multiple pathways. However, its role is not exclusive to dorsal cortical neurogenesis.
To verify these findings, we collected in utero electroporated cells by laser-microdissection from all three conditions after 48 hrs of survival, and then performed real-time quantitative PCR. As evident in Fig. S3, these data supported the results obtained after mRNAseq analysis.
In utero disruption of MAML accelerates neuronal differentiation and migration
MAML disruption directed cells to exit the proliferative zones and migrate towards the cortical plate (Fig. 2D, E). To understand how this experimental manipulation compares to normal developmental processes, we compared the transcriptome of dnMAML-electroporated cells to the normal transcriptome of cortical zones at E14.5, which we generated previously (Ayoub et al. 2011). We found that dnMAML upregulated genes normally expressed in cells transitioning through the IZ and residing in the CP (see Venn diagram; Fig. 3D). Gene ontology analysis revealed an enrichment of differentiation and migration genes (e.g. Id2, Tcf4, Satb2, Nr2f1, Nr2f2) in dnMAML-electroporated cells without causing the appearance of heterotopias or migration defects.
Subsequently, we compared the transcriptional consequences dnMAML-electroporation to Neurog2 electroporation, which was shown to causes excessive neuronal migration, and alters the time course of normal mouse corticogenesis (Fig. 3E, F). Transcriptome data were processed using the same bioinformatics pipeline and statistical analysis described previously. We then contrasted genes differentially expressed in dominant-negative MAML and Neurog2 electroporated cells (compared to electroporation controls) to the developmental mouse transcriptome from E11.5 to E17.5 (Fig. 3A). We used the self-organizing map (SOM) machine-learning algorithm to isolate significantly enriched sets of genes. Phylogenetic sample clustering after SOM analysis showed that electroporated cells occupied an intermediate state between VZ and CP samples (Fig. 3B). Both dnMAML and Neurog2 electroporated cells showed progressive change from 24hrs to 48hrs, while in a transcriptional state between E14.5 SVZ-IZ and occipital E17.5 SVZ-IZ on one side and E14.5 CP and frontal E17.5 SVZ-IZ on the other. Indeed, gene-set enrichment analysis showed that in utero manipulations accelerated fate commitment and neuronal migration in experimental conditions.
Regulatory control of neuronal migration
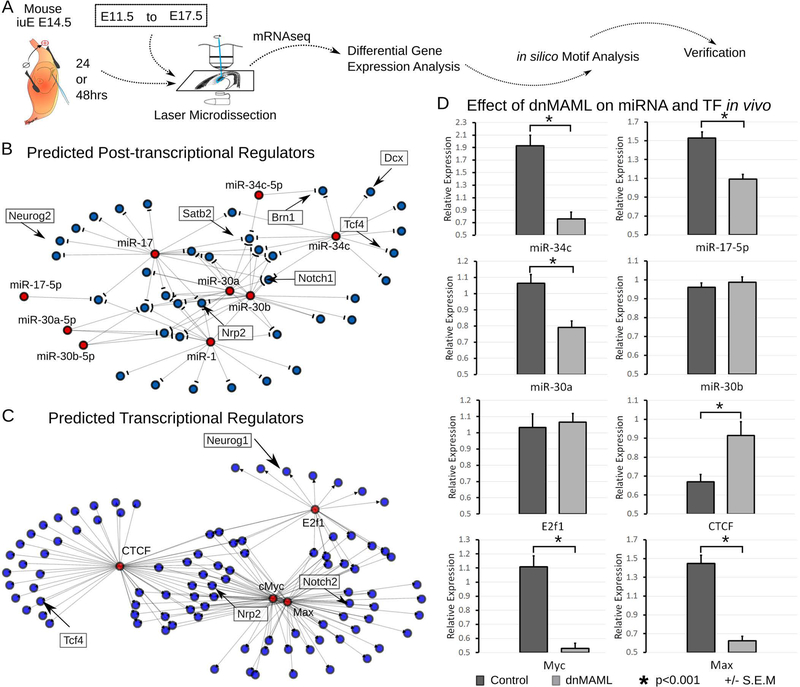
To understand the regulatory architecture governing neuronal migration during mouse development, we analyzed the group of genes differentially expressed in dnMAML and Neurog2 samples for shared transcription factor binding motifs as well as microRNA binding sites (Fig. 4A). To our surprise, we found that four microRNAs (p<0.05), including MIR-30 and MIR-34, are potentially regulating the post-transcriptional activity of differentially expressed genes in both datasets (Fig. 4B). To verify the presence of these microRNAs, we conducted qRT-PCR on microdissected samples, as for the mRNAseq experiments (Fig. 4D). We found that the expression of miR-34c, miR-17, miR-30a, but not miR-30b, were influenced by the dnMAML. Interestingly, miR-30 microRNA has been implicated in regulating self-renewal and apoptosis of cancer cells (Yu et al., 2010). To our knowledge, little is known about the role of miR-30 and miR-34 during cortical development, and they have not demonstrated in vivo.
Fig. 4. Transcriptional networks that drive neurogenesis and migration in vivo.
A. Schematic of the strategy used to manipulate and isolate the transcriptional network responsible for neuronal fate commitment and neuronal migration. To gain insight into the regulatory mechanisms controlling fate commitment and neuronal migration networks, we searched for common transcription factor and micro-RNA binding sites near genes differentially expressed after dnMAML or Neurog2 overexpression.
B. Network representation shows in silico predicted post-transcriptional regulation of key neuronal fate and migration genes, such as Brn1, Satb2, Dcx, and Tcf4, by microRNAs.
C. Network representation shows that as few as 4 transcription factors may regulate the entire gene network. Numerous target genes are important contributors to cortical development, including Nrp2, Tcf4, Neurog1 and Notch2.
D. Verification of expression and effect of dnMAML on predicted microRNAs and transcription factors. Real-time QPCR was performed on RNA isolated from electroporated cortical cells by LMD. dnMAML affected miR-34c, miR-17–5p, and miR-30a but not miR-30b. dnMAML caused a significant reduction in the transcription factors Myc and Max, increased CTCF but did not change E2F1. (ANOVA, *p<0.001, +/− SEM).
We identified motif enrichment for five transcription factors (p<0.05) in the group of differentially expressed genes (Fig. 4C). These transcription factors have restricted species- and developmental expression. For example, Klf4 has higher expression in human apical and outer radial glia, as well as migrating neurons, compared to other human and mouse samples. E2f1 has higher expression in progenitors than in migrating neurons or the cortical plate. We verified the expression of E2f1, CTCF, Myc, and Max in microdissected tissue from experimental and control conditions (Fig. 4D). Electroporation of dnMAML caused an increase in the expression of CTCF, a decrease in Myc and Max expression, but no significant effect on E2f1.
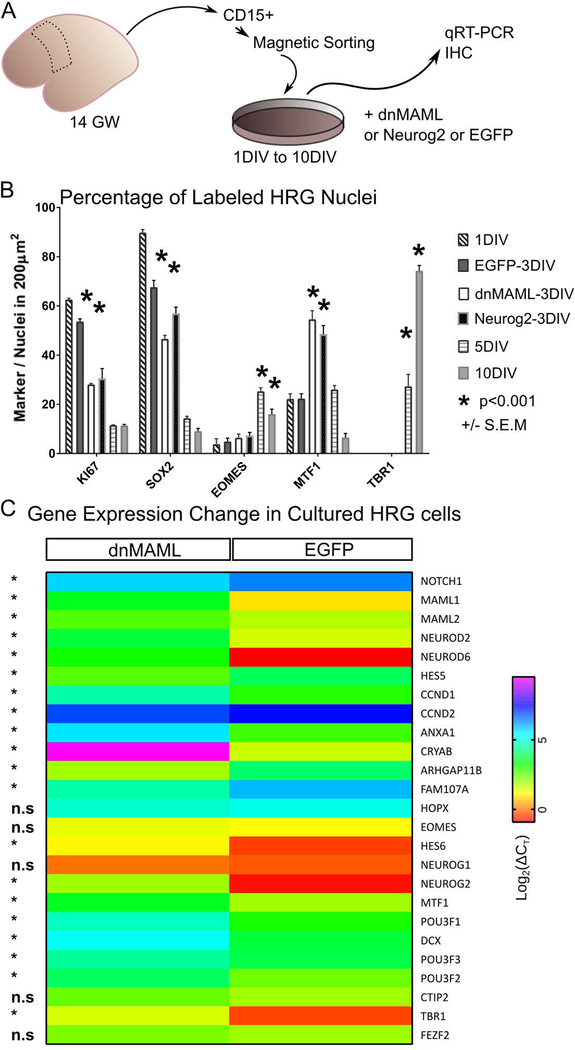
Ectopic expression dnMAML in human progenitors
To investigate the effect of dnMAML and Neurog2 on human progenitors in vitro we isolated human radial glia (HRG) from 14GW dorsal cortex by magnetic sorting using CD15-beads (Fig. 5A). We compared the cellular composition of the dnMAML, Neurog2, and GFP transfected cultures by immunocytochemistry utilizing cell type-specific markers such as Ki67 and SOX2 to identify neural stem cells, TBR2 (EOMES) for intermediate progenitors and TBR1 to label postmitotic neurons (fig. 5B). As expected, the percentage of cells positive for the stem cell markers Ki67 and SOX2 gradually decline in cultures exposed to differentiation media for 5 days in vitro (DIV) and 10DIV (Fig. 5B). HRG cultures transfected with dnMAML and Neurog2 (3DIV) showed a drastic reduction in the percentage of cells double-positive for Ki67 and SOX2 as compared to 1DIV and control EGFP-3DIV conditions. In contrast, the number of TBR2 and TBR1+ cells showed an increase in differentiation media after 5DIV and 10DIV but not in the dnMAML-3DIV or Neurog2–3DIV (Fig. 5B). To assess the transfection efficiency, we also tested MTF1 (mouse gene Myt1), which was induced by in utero electroporations of dnMAML and Neurog2 (Fig. 3). We found that the Neurog2 downstream target MTF1 was significantly increased following transfection with dnMAML or Neurog2 after 3DIV. Additionally, we performed quantitative real-time PCR to detect the expression of a battery of dorsal proliferation and differentiation genes in cultured control and transfected HRG cells (Fig. 5C). These data showed that dnMAML depresses the expression of most genes typically expressed in progenitors, such as NOTCH1, HES5, CRYAB, and ARHGAP11B in cultured HRG cells. On the other hand, dnMAML increased the expression of POU3F3/2, an upper-layer neuron determinant, and the neuronal migration genes POU3F1, DCX, and NEUROD6. Among the genes identifying subtypes of RG progenitors, dnMAML transfection had a significant effect on ANXA1, CRYAB, FAM107A but not HOPX gene.
Fig. 5. Effect of forced neurogenesis in human fetal cortical progenitors.
A. Schematic shows how human radial glial (HRG) progenitors were isolated from gestational week (GW) 14 postmortem tissue. We dissociated and bound cells to the CD15 antibody coated beads then propagated them after magnetic sorting. Cells were seeded in proliferation media (PM) until reaching 90% confluence, which is considered the start of the experiment. We transfected HRG with dnMAML, Neurog2, or EGFP for 3 days in vitro (3DIV). Control coverslips were taken from 1DIV as well as from coverslips on differentiation media (DM) for 5DIV and 10DIV.
B. We quantified the proportion of HRG marker proteins expressed in vitro under different conditions. Immunohistochemistry verification and quantification was performed on coverslips stained for the proliferation marker KI67, the stem cell marker SOX2, intermediate progenitor marker EOMES (also known as Tbr2), the Neurog2 target MTF1, and the neuronal differentiation marker TBR1. The percentage of EOMES and TBR1positive nuclei is significantly higher after 5DIV and 10DIV in differentiation media. On the other hand, the percentage of MTF1 nuclei is significantly higher after dnMAML and Neurog2 transfections.
C. Heat map showing qRT-PCR verification of neurogenesis and fate commitment genes in cultured HRG after dnMAML or EGFP transfection (3DIV). Notice that dnMAML transfection triggers significant change in numerous fate commitment and radial glial genes. Data analyzed and plotted as Log2(ΔCt) (*p<0.01).
Discussion
Stem cell amplification and progeny diversification are mechanisms that are essential to the evolutionary enlargement and cytoarchitectural divergence of mammalian brains. We found that the transcriptional co-activator MAML, initially identified in Drosophila, coordinates several stem cell maintenance pathways, including Notch and Beta-catenin, in mammals as diverse as rodents and primates. It was shown recently that dnMAML accelerates neurogenesis in iPSCs but it is not known whether the observed acceleration adheres faithfully to the neurogenic timetable and gradients observed in vivo. We here show that neurogenesis in native VZ progenitors can be accelerated without altering neuronal fate assignment and laminar destination. Indeed, forced exit from the VZ via ectopic expression of dnMAML or Neurog2 allowed cells to reach their final laminar destination as predicted by developmental neurogenic gradients. These data indicate that neurogenesis and fate assignment, which move in lockstep, are served by distinct, yet closely coordinated, gene networks. Our in silico data show that a regulatory logic including both trans-acting regulatory mediators and microRNAs appears to exert tight control over this developmental mechanism. Further experiments are necessary to explain how the combinatorial action of transcription factors and microRNAs can affect neurogenesis in a way that induces the emergence of additional, novel cell types during evolution.
A long-standing hypothesis in the cortical development field predicts that migrating neurons settle in their areal, layer and columnar position within the cortex based on the time and position of their origin (Rakic,1988). A recent elaboration of this model posits that some neurons transfer horizontally to nearby radial fascicles to provide for proper mixture of the polyclonal functional columns (Torii et la., 2009). Numerous studies support this view, which also explains the expansion of cerebral cortex as a sheet rather than a lump (Noctor at al., 2001; Reviewed in Geschwind and Rakic, 2013). In addition, our data indicate that transient blockade of Notch and b-catenin pathways, via dnMAML, increases the expression of most outer radial glia (oRG) genes in cultured human progenitors. We speculate that this transient increase facilitates the emergence of oRG from apical RG. A sustained blockade of Notch and b-catenin beyond five days in vitro favors IPs and final neuronal differentiation. These results also hint at a possible divergence in the neurogenic and migration pathways downstream of Neurog2 (NEUROG2 > TBR2 or NEUROG2 > MTF1), thus allowing finer control of cell type production in human RGs.
A recent alternative hypothesis suggests that introduction of the outer subventricular zone (oSVZ) in human is the main factor for expansion of cerebral cortex and formation of convolutions during development and evolution (e.g.Stahl et al., 2013; Namba and Huttner, 2017). However, recent data from developing macaques (Old World primates) show that the oSVZ does indeed add some neurons to the superficial layers initially, but that after neurogenesis stops, it mainly produces glial cells, which in the primates outnumber neurons and settle in the cortex as well as in large subcortical white matters (Rash et al., 2019). Thus, our present data bridge the gap between the two hypotheses by showing that neurons migrate and settle in the cortex not only based on their time of origin, but also based on the region of origin at any given time point. In our experiments, neurons settled in the upper layers on the lateral side of cortex and in the deep layers on the medial aspect of the cortex, even when forced to exit the VZ at E12.5, which is the peak of deep layer genesis in the mouse. The proportion of neurons settling in the upper layers increased in the lateral and dorsal aspect of the cortex, as shown at P21, when cells were forced to exit the VZ at E14.5. Therefore, we expect gradual intermixing of fate-restricted progenitors in the VZ as the wave of neurogenesis sweeps across the cortex. A second notable result is that the accelerated tempo of development in the mouse reduces our ability to resolve fast acting genetic mechanisms. This issue is highlighted by the inability to detect TBR2 and MTF1 expression, both downstream targets of Neurog2, after 24hrs or 48hrs of electroporation. However, both genes were detectable in human progenitors after 3-days in vitro. The procession of neuronal migration in different areas of the cortex is more difficult to dissect in the mouse brain than in the larger primate and human brain. By taking these regional differences into consideration, future studies may discover intrinsic differences between progenitors at a given time point but in different areas of the cortex.
Conclusions
During mammalian evolution the cerebral cortex expands greatly in surface area with little change in thickness, but undergoes a marked increase in complexity of neuronal organization and diversity of cytoarchitectonic maps. In spite of these species-specific differences, the cellular behavior follows the same basic principles of the radial unit model. Here we show that molecular mechanisms involved in these complex events, including stem cell proliferation, neuronal determination, and migration to the proper final positions, use the same signaling systems in humans and mice. Moreover, some of these developmental mechanisms are orchestrated by a homologue of a gene identified in Drosophila—an organism that does not have a cortex. Our findings illustrate the unity of the evolutionary process, but how species-specific differences are initiated and timed remains elusive and needs additional studies.
Supplementary Material
Table 1.
List, sources and concentrations of the of the antibodies used
| Antibody | Source | concentration | Antigen retrieval | cell specificity |
|---|---|---|---|---|
| Rb-Pax6 | Millipore (AB2237) | 1/1000 | yes | dorsal progenitors |
| Gt-Pou3f2 (Brn2) | SCBT (sc-6029) | 1/200 | yes | upper layer neurons |
| MTF1 (Myt1) | SIGMA (HPA006303) | 1/250 | yes | post-mitotic neurons |
| Ms-Ki67 | BD Pharmingen (550609) | 1/50 | yes | S-M phase |
| Ms-Blbp | Abcam (ab27171) | 1/1000 | not necessary | radial glia |
| Gt-Sox2 | SCBT (sc-17320) | 1/250 | yes | stem cells/progenitors |
| Rb-pH3 | Millipore/Upstat e (07–424) | 1/300 | not necessary | M phase |
| Rt-CTIP2 | Abcam (ab18465) | 1/1000 | yes | depends on age; Prenatal Layer 5 |
| Rb-Tbr1 | Abcam (ab31940) | 1/500 | yes | deep layer neurons |
| Rb-CDP (M222; Cux1) | SCBT (sc-13024) | 1/100 | yes | upper layer neurons |
| Chk-Tbr2 | Millipore (AB15894) | 1/500 | yes | Intermediate progenitors |
| Chk-V5 | Abcam (ab9113) | 1/200 | not necessary | v5-tag |
Acknowledgements
The authors thank Mariamma Pappy for technical assistance and members of the Rakic lab for critical comments on the manuscript. This work was supported by National Institutes of Health Grants DA02399 and EY002593, and the Kavli Institute for Neuroscience at Yale University. We thank Christopher Castaldi, David Harrison, and Kaya Bilguvar for assistance with RNAseq at the Yale Center for Genome Analysis. The Yale University Biomedical High Performance Computing Center is supported by NIH Grant RR19895
Footnotes
Disclosure Statement: The authors declare that they have no competing financial interest.
References
- Angevine JB Jr., Sidman RL (1961) Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192:766–768. [DOI] [PubMed] [Google Scholar]
- Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D, Lesche M, Dahl A, Groszer M, Hiller M, Calegari F (2013) Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. The EMBO journal 32:3145–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH (2007) Trekking across the brain: the journey of neuronal migration. Cell 128:29–43. [DOI] [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P (2011) Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci U S A 108:14950–14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitard M, Bocchi R, Egervari K, Petrenko V, Viale B, Gremaud S, Zgraggen E, Salmon P, Kiss JZ (2015) Wnt signaling regulates multipolar-to-bipolar transition of migrating neurons in the cerebral cortex. Cell Rep 10:1349–1361. [DOI] [PubMed] [Google Scholar]
- Carrion-Castillo A, Franke B, Fisher SE (2013) Molecular genetics of dyslexia: an overview. Dyslexia 19:214–240. [DOI] [PubMed] [Google Scholar]
- Caviness VS Jr., Nowakowski RS, Bhide PG (2009) Neocortical neurogenesis: morphogenetic gradients and beyond. Trends Neurosci 32:443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA (2002) Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297:365–369. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E, Haydar TF (2008) Regulation of neural progenitor cell development in the nervous system. J Neurochem 106:2272–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AR, McConnell SK (2000) Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development 127:2863–2872. [DOI] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P (2013) POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex 23:2632–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM, LoTurco J, Ramus F, Fitch RH, Rosen GD (2006) From genes to behavior in developmental dyslexia. Nat Neurosci 9:1213–1217. [DOI] [PubMed] [Google Scholar]
- Gertz CC, Kriegstein AR (2015) Neuronal Migration Dynamics in the Developing Ferret Cortex. J Neurosci 35:14307–14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P (2007) Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol 17:103–111. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Rakic P (2013) Cortical evolution: judge the brain by its cover. Neuron 80:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Walsh CA (2000) Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci 23:352–359. [DOI] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA (2015) Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci 18:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS (2004) Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood 104:1696–1702. [DOI] [PubMed] [Google Scholar]
- Mattar P, Britz O, Johannes C, Nieto M, Ma L, Rebeyka A, Klenin N, Polleux F, Guillemot F, Schuurmans C (2004) A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Developmental biology 273:373–389. [DOI] [PubMed] [Google Scholar]
- Meechan DW, Maynard TM, Tucker ES, LaMantia AS (2011) Three phases of DiGeorge/22q11 deletion syndrome pathogenesis during brain development: patterning, proliferation, and mitochondrial functions of 22q11 genes. Int J Dev Neurosci 29:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N (2007) Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449:351–355. [DOI] [PubMed] [Google Scholar]
- Munji RN, Choe Y, Li G, Siegenthaler JA, Pleasure SJ (2011) Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci 31:1676–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch CA, Funatsu N, Monuki ES, Chenn A (2009) Beta-catenin signaling levels in progenitors influence the laminar cell fates of projection neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 29:13710–13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC (2006) Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973–983. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR (2001) Neurons derived from radial glial cells establish radial units in neocortex. Nature 409:714–720. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, Lim DA, Leyrat AA, West JA, Kriegstein AR (2015) Molecular identity of human outer radial glia during cortical development. Cell 163:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Hashimoto-Torii K, Sarkisian MR (2007) Genetic determinants of neuronal migration in the cerebral cortex. Novartis Found Symp 288:45–53; discussion 53–48, 96–48. [DOI] [PubMed] [Google Scholar]
- Rash BG, Duque A, Morozov YM, Arellano J, Micali N, Rakic P. 2019. Gliogenesis in the outer subventricular zone promotes enlargement and gyrification of the primate cerebrum. Proc Natl Acad Sci (USA) 116: 7089–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP (2015) Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347:1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I (2003) Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130:3269–3281. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R (2008) Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58:52–64. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P (1973) Neuronal migration, with special reference to developing human brain: a review. Brain Res 62:1–35. [DOI] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, Ramanathan S (2016) Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat Methods 13:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Maillard I, Nakamura M, Pear WS, Griffin JD (2007) The transcriptional coactivator Maml1 is required for Notch2-mediated marginal zone B-cell development. Blood 110:3618–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY (2008) Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron 58:519–531. [DOI] [PubMed] [Google Scholar]
- Yu F, Deng H, Yao H, Liu Q, Su F, Song E (2010) Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene 29:4194–4204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.